Abstract
Genetic or epigenetic inactivation of one of the DNA mismatch repair (MMR) genes in tumor precursor cells causes a profound mutator phenotype, known as the microsatellite mutator phenotype (MMP). This mutator phenotype induces mutations not only in cancer genes that drive tumorigenesis but also in other DNA repair genes. The functional significance of these successive DNA repair gene mutations, however, has not been substantiated. Here we show that the concomitant inactivation of two DNA MMR genes (hMLH1 and hMSH6) increases the mutator phenotype. We isolated cell clones of the SW48 MMP-positive cell line with either active or inactive hMSH6. All of these clones lacked expression of hMLH1 because of promoter hypermethylation. Compared with inactivation of hMLH1 alone, the additional inactivation of hMSH6 produced a higher mutation rate and a different spectrum of mutations in the endogenous hprt gene. These results confirm our model that the mutator phenotype can increase during tumorigenesis by the consecutive inactivation of different members of the DNA MMR system. Thus, a stronger mutator phenotype accelerates the accumulation of mutations in target cancer genes, which, in turn, speeds up tumor progression. The results of this study also have significant impact on our understanding of the mechanism of DNA MMR.
Inactivation of the DNA mismatch repair (MMR) machinery in tumor precursor cells causes a profound mutator phenotype, which leads to the accumulation of mutations in cancer genes and to the development of cancer (1, 2). Germ-line mutations in the hMSH2, hMLH1, and less frequently in the hMSH6 and hPMS2 DNA MMR genes confer susceptibility to hereditary nonpolyposis colorectal cancer (3–8). In addition to these genes, somatic mutations in the hMSH3 DNA MMR gene have been found in sporadic tumors of different tissue origin (9, 10).
This mutator phenotype was discovered through the presence of deletion/insertion mutations in microsatellite repeat sequences (microsatellite instability, MSI) (11–13), which are particularly prone to errors caused by slippage by strand misalignment during DNA replication (14). In this respect, microsatellites could be regarded as mutational targets for the mutator phenotype, which, for this reason, we often call the microsatellite mutator phenotype (MMP).
The prevalence of the deletions/insertions in repetitive sequences in MMP-positive cells may be responsible for the involvement of different sets of cancer genes in the development of MMP-positive and -negative tumors. Some genes involved in cell growth and survival (such as TGF-βRII, and BAX) commonly have mutations in mononucleotide repeat sequences located in their coding regions in MMP-positive tumors (15–17). Thus, a distinctive tumorigenic pathway, characteristic of MMP-positive cancer, leads to the development of tumors with different features in genotype and phenotype compared with tumors without microsatellite instability (11, 12, 18–22).
Along with cell growth/survival genes, some DNA MMR genes as well as other genes involved in the maintenance of genome integrity contain nucleotide repeat sequences in their coding regions. Frameshift mutations in these sequences have been found in the hMSH3, hMSH6, RAD50, MBD4, DNA-PKcs, BLM, BRCA1, BRCA2, ATR, and DNA helicase genes in MMP-positive tumors (17, 23–26). However, the functional significance of these mutations has not been shown.
Based on the discovery of frameshift mutations in the mononucleotide repeat sequences of the hMSH3 and hMSH6 genes in MMP-positive tumors, we proposed a model of the “mutator that mutates another mutator,” which describes a cascade increase of the mutator phenotype during tumorigenesis of MMP cancer (22, 23, 27). We hypothesized that genetic or epigenetic inactivation of a primary DNA MMR gene (such as hMLH1 and hMSH2) leads to inactivating mutations in secondary DNA repair genes, which could be involved in MMR or another type of DNA repair. Such consecutive mutational inactivation of DNA repair genes would unfold genetic instability during tumor progression. The goal of this study was to test this hypothesis.
We report here that, compared with inactivation of hMLH1 alone, the additional inactivation of hMSH6 results in a higher mutation rate and a different spectrum of mutations in the endogenous hprt gene. While the majority of mutations in the cells with inactivated hMLH1 were deletions/insertions, cells with concomitant inactivation of hMLH1 and hMSH6 produced more nucleotide substitutions characteristic of hMSH6 inactivation.
Materials and Methods
Cell Lines and Media.
The SW48, LS180, and DLD-1 human colon carcinoma cell lines were obtained from the American Type Culture Collection. Cells were grown on 80-mm Nunc tissue culture plates in DMEM supplemented with 15% fetal bovine serum (FBS; Tissue Culture Biologicals, Tulare, CA). To select against hprt mutants, the medium was supplemented with HAT (300 μM hypoxanthine/1 μM aminopterin/50 μM thymidine; Sigma). To select for hprt mutants, the medium was supplemented with 15 μg/ml 6-thioguanine (6-TG; Sigma). The demethylating agent 5-aza-2′-deoxycytidine (Sigma) in concentrations of 2 μM and 10 μM was used to restore hMLH1 gene expression.
Isolation of SW48 Subclones with Mutations in the hMSH6 Gene.
To isolate subclones carrying mutations in the hMSH6 gene, SW48 cells were inoculated into 96-well plates at an average density of one cell per well. When the colonies reached an average size of about 1 mm the cells were trypsinized and split into two plates. The cells in one of the duplicate plates were lysed by adding 100 μl of TE buffer (10 mM Tris/1 mM EDTA, pH 7.4) supplemented with 150 μg/ml proteinase K and heating at 65°C for 1 h. The lysates were used for PCR amplification of the hMSH6 gene region containing a poly(C) sequence as described (23). Clones with frameshift mutations in the poly(C) sequence were subjected to a second round of subcloning to ensure their single-cell origin.
Selection of hprt Gene Mutants and Calculation of the Mutation Rate.
Twenty independent cultures for each of the subclone cell lines with MSH6(+/+) or MSH6(−/−) genotype were grown from an inoculum of 2 × 104 cells. The cells were grown in HAT medium to eliminate any preexisting hprt− cells. When the cell count reached ≈105 per plate, the HAT medium was replaced with fresh medium without HAT. The cells were grown for approximately two more cell duplications, at which point 6-TG was added to the medium to select for hprt− mutants. The 6-TG-resistant colonies were counted in 2 to 3 weeks. The rate of mutation was calculated according to the formula R = N20/[20 × (N − N0)], where N20 is the total number of 6-TG-resistant colonies on the 20 plates, N0 is the number of cells per plate at the time the HAT supplement was removed, and N is the number of cells per plate at the time 6-TG was added.
Identification of hprt Gene Mutations.
Detection of mutations in the hprt gene was performed as described in ref. 28. Briefly, mutants with splicing aberrations were detected by measuring the length of the entire hprt gene coding region and analyzed for the splice donor and acceptor sites by single strand conformation polymorphism (SSCP) (29). Mutations in the coding region were detected by the SSCP analysis of the reverse transcriptase–PCR (RT-PCR) fragments amplified from four overlapping regions of the gene. The DNA fragments exhibiting mobility shifts on SSCP gels were extracted from the gel, reamplified with appropriate primers, and additionally purified by agarose gel electrophoresis. The DNA fragments were purified from agarose by QIAquick Gel Extraction Kit (Qiagen) and used as templates for sequencing by Dye Terminator Cycle Sequencing Kit (Applied Biosystems).
Analysis of hMLH1 Gene Expression.
Total RNA was isolated from SW48 cells or their derivatives by RNA STAT-60 kit (Tel-Test, Friendswood, TX) followed by cDNA synthesis using Moloney murine leukemia virus reverse transcriptase and random hexamer primers. The cellular content of the hMLH1 mRNA was measured by multiplexed RT-PCR amplification of the gene's region spanning exons 13 through 16 and the hprt gene region spanning exons 3 through 7. The following primers were used for PCR amplification of the 338-bp hMLH1 fragment: forward, 5′-GAAGATTCTGATGTGGAAATGG-3′; reverse, 5′-ATGGCAAGGTCAAAGAGCGGT-3′. The 240-bp hprt gene fragment was amplified with the following primers: forward, 5′-TTATCAGACTGAAGAGCTATTG-3′; reverse, 5′-CTTATATCCAACACTTCGTGG-3′.
Results
Isolation of Cell Lines with hMLH1−hMSH6+ and hMLH1−hMSH6− Genotypes.
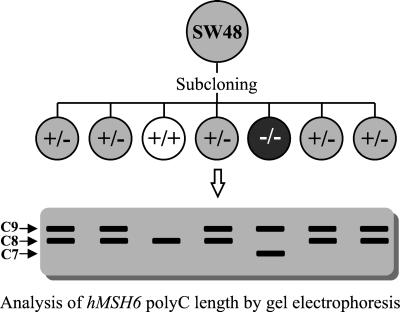
The SW48 human colon adenocarcinoma cell line exhibits the MMP. SW48 cells lack expression of the hMLH1 gene because of hypermethylation of the gene promoter (30) and carry a heterozygous 1-bp insertion frameshift mutation in the C8 sequence (23) encompassing codons 1084–1086 of the hMSH6 protein (31). We have previously shown that this mononucleotide repeat is a hot spot for frameshift mutations in MMP-positive tumors (23). We hypothesized that some SW48 cells should contain insertion/deletion mutations in this sequence. By analyzing about 1,000 subclones of the SW48 cell line (the strategy is illustrated in Fig. 1), we found 5 clones with a 1-nucleotide deletion mutation in the C9 sequence of the mutant allele of the hMSH6 gene reversing it to the wild type [final repeat length in both alleles is C8, designated as MSH6(+/+), homozygous wild type] and 3 clones with a 1-nucleotide deletion mutation in the wild-type allele [repeat length is C7 in one allele and C9 in the other allele, designated as MSH6(−/−), homozygous mutant]. Other mutations increasing the C9 to C10 and C11 were also detected in the screening. They were not analyzed because they retained their heterozygosity (C8/C10 and C8/C11). No hMSH6 protein was detected in the cell lines with homozygous hMSH6 gene mutations by Western blot analysis (32).
Figure 1.
Strategy for isolation of SW48 subclones with wild-type or inactivated hMSH6 gene. +/+, wild-type hMSH6; +/−, heterozygous for hMSH6 frameshift mutations; −/−, homozygous for hMSH6 frameshift mutations.
Rate and Spectrum of hprt Gene Mutations in MSH6(+/+) and MSH6(−/−) Cells.
We used the endogenous hprt gene as a reporter to measure the mutation rate and the spectrum of mutations in the MSH6(+/+) and MSH6(−/−) cells. Three independent subclones for each of the hMSH6 genotypes were taken for these experiments to ensure the absence of clonal variability in the mutation rate and spectrum. The mutation rate did not differ considerably among the three subclones with the same genotype and was (0.8 ± 0.2) × 10−5, (0.8 ± 0.3) × 10−5, and (1.0 ± 0.2) × 10−5 per allele per replication for the MSH6(−/−) subclones and (2.0 ± 0.5) × 10−5, (2.5 ± 0.4) × 10−5, and (2.4 ± 0.4) × 10−5 per allele per replication for the MSH6(+/+) subclones. The average rate of the hprt gene mutations was (0.9 ± 0.2) × 10−5 and (2.3 ± 0.4) × 10−5 per allele per replication for MSH6(+/+) and MSH6(−/−) cells, respectively.
To ensure independence of the mutants, only one 6-TG-resistant colony was taken from each of the independent subcloned cultures for identification of hprt gene mutations. Sixteen or 17 mutants were isolated for mutational analysis for each of the six subclones (50 for each genotype). The list of all identified mutations is presented in Table 1. There was no significant difference in the mutation spectra among the three subclones corresponding to the same genotype. The mutation spectrum for the MSH6(+/+) and MSH6(−/−) cells is shown in Fig. 2.
Table 1.
The hprt gene mutations detected in MSH6(+/+) and MSH6(−/−) cell lines
| Position* | Mutation† | Mutation type | Effect | Incidence
|
|
|---|---|---|---|---|---|
| MSH6(+/+) | MSH6(−/−) | ||||
| 28−2 | agAT-ggAT | Transition | Splicing error | 1 | — |
| 65 | TTT-TCT | Transition | Missense (Phe → Ser) | — | 1 |
| 98 | A3-A2 | Deletion | Frameshift | 1 | — |
| 133 | AGG-GGG | Transition | Missense (Arg → Gly) | — | 1 |
| 146 | T2-T3 | Insertion | Frameshift | — | 1 |
| 151 | CGA-TGA | Transition | Nonsense (Arg → stop) | — | 4 |
| 197 | TGT-TAT | Transition | Missense (Cys → Tyr) | 1 | — |
| 202 | CTC-TTC | Transition | Missense (Leu → Phe) | 1 | — |
| 207 | G6–G7 | Insertion | Frameshift | 19 | 7 |
| 207 | G6–G5 | Deletion | Frameshift | 5 | 1 |
| 325 | CCAGT-CGT | Deletion | Frameshift | — | 1 |
| 385−2 | agAA-ggAA | Transition | Splicing error | 1 | 1 |
| 395 | ATT-ACT | Transition | Missense (Ile → Thr) | 1 | — |
| 404 | GAT-GGT | Transition | Missense (Asp → Gly) | 1 | — |
| 419 | GGC-GTC | Transversion | Missense (Gly → Val) | — | 1 |
| 421 | A4–A5 | Insertion | Frameshift | 1 | — |
| 454 | CAG-TAG | Transition | Nonsense (Gln → stop) | 1 | 4 |
| 483 | A2–A1 | Deletion | Frameshift | 1 | — |
| 496 | A4–A3 | Deletion | Frameshift | 1 | — |
| 508 | CGA-TGA | Transition | Nonsense (Arg → stop) | 6 | 17 |
| 532+1 | CTgt-CTat | Transition | Splicing error | — | 1 |
| 533−2 | agTT-ggTT | Transition | Splicing error | 2 | — |
| 533−1 | agTT-atTT | Transversion | Splicing error | 1 | 1 |
| 568 | GGA-TGA | Transversion | Nonsense (Gly → stop) | — | 1 |
| 577 | CTT-ATT | Transversion | Missense (Leu → Ile) | — | 1 |
| 580 | GAC-AAC | Transition | Missense (Asp → Asn) | 1 | — |
| 582 | GAC-GAA | Transversion | Missense (Asp → Glu) | 1 | 1 |
| 610−2 | agCA-ggCA | Transition | Splicing error | 2 | — |
| 610−1 | agCA-aaCA | Transition | Splicing error | 1 | — |
| 610 | CAT-TAT | Transition | Missense (His → Tyr) | 1 | 6 |
| Total | 50 | 50 | |||
Mutated nucleotides in the hprt coding region are indicated relative to the ATG initiation codon. Intron mutations are indicated as the cDNA position minus (−) or plus (+) the appropriate number of nucleotides into the introns.
Uppercase, exon sequences; lowercase, intron sequences; underlined, mutated nucleotides.
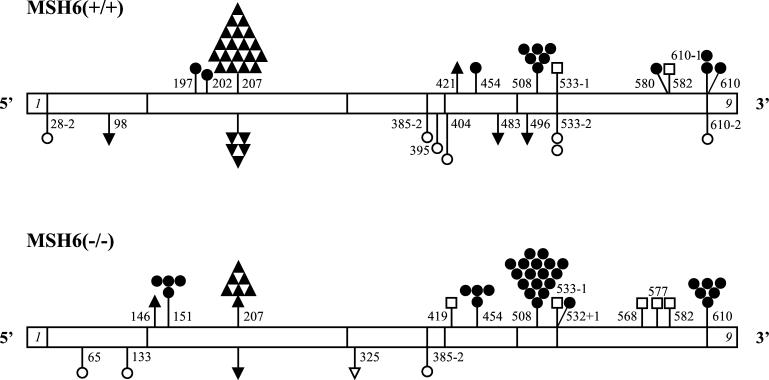
Figure 2.
Spectrum of hprt gene mutations in MSH6(+/+) and MSH6(−/−) cells. cDNA structure of the hprt gene is shown (only exons 1 and 9 are indicated). Exon 1 begins at base pair 1, and exon 9 ends at base pair 657. Mutations are represented by the following symbols: ▴, insertion of one nucleotide; ▾, deletion of one nucleotide; ▿, deletion of two nucleotides; ●, transition at C⋅G base pairs; ○, transition at A⋅T base pairs; □, transversion. Numbers indicate position of the mutated nucleotides. Position of the mutated nucleotides in the intron sequences is indicated as the cDNA position − or + the appropriate number of nucleotides into the introns.
There were no large structural deletions in the hprt sequence, and all alterations detected were single point mutations (Table 1). The spectrum of mutations included frameshifts, as well as nucleotide substitutions in the hprt coding and splicing site sequences. All but one frameshift mutations were a deletion or insertion of a single nucleotide in mononucleotide repeat sequences two or more nucleotides in length. The only two-nucleotide deletion mutation in a nonrepetitive CA nucleotide sequence was found in the MSH6(−/−) cells. All 100 mutations were distributed among 30 sites. There were five sites with more than two identical independent mutations: frameshift mutations at the G6 nucleotide repeat (position 207–212), C-to-T transitions at two CpG sites (positions 151 and 508), and C-to-T transitions at positions 454 and 610 (Fig. 2).
The specificity of these mutational hot spots for cells with different hMSH6 genotypes was, however, different. Transition mutations at position 151 were exclusive for the MSH6(−/−) cells. Transition mutations at positions 454, 508, and 610 were more frequent in the MSH6(−/−) cells than in the MSH6(+/+) cells, whereas frameshift mutations at position 207–212 were more specific for the MSH6(+/+) cells and accounted for about 50% of all mutations. Although MSH6(+/+) and MSH6(−/−) cells shared 8 mutational sites, mutations at 22 sites were present in only one of the cell types. One-nucleotide frameshift mutations in the MSH6(+/+) cells made up 65% of all mutations and were distributed among 5 different sites, whereas in the MSH6(−/−) cells this type of mutations occurred only at 2 sites and made only 20% of all mutational events. The difference between the spectra of the hprt gene mutations in the MSH6(+/+) and MSH6(−/−) cells was statistically significant for transitions and frameshifts at the G6 sequence (Table 2).
Table 2.
Types of the hprt gene mutations detected in MSH6(+/+) and MSH6(−/−) cell lines
| Mutation type | Incidence
|
P value | |
|---|---|---|---|
| MSH6(+/+) | MSH6(−/−) | ||
| Base substitutions | 22 (44%) | 40 (80%) | <0.0002 |
| Transitions | 20 (40%) | 35 (70%) | <0.0026 |
| G⋅C → A⋅T | 13 (26%) | 32 (64%) | <0.0001 |
| A⋅T → G⋅C | 7 (14%) | 3 (6%) | |
| Transversions | 2 (4%) | 5 (10%) | |
| G⋅C → T⋅A | 2 (4%) | 5 (10%) | |
| Frameshifts | 28 (56%) | 10 (20%) | <0.0002 |
| +/−G | 24 (48%) | 8 (16%) | <0.0006 |
| +/−A | 4 (8%) | 0 (0%) | |
| +T | 0 (0%) | 1 (2%) | |
| −CA | 0 (0%) | 1 (2%) | |
hMLH1 Gene Expression in Subclones of the SW48 Cell Line.
One possible explanation for the differences in the spectrum and the rate of mutations between the MSH6(+/+) and MSH6(−/−) cell lines could be artifactual reactivation of the hMLH1 gene expression in these cells before or during selection for 6-TG resistant clones. To test this hypothesis, we performed RT-PCR analysis of the cellular content of hMLH1 transcript in all types of SW48 subclones.
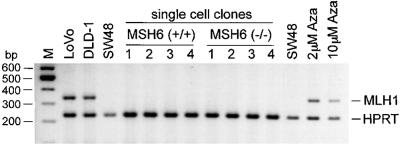
While we detected hMLH1 expression in control cell lines, no expression was found by either regular (data not shown) or multiplexed PCR in the parental SW48, MSH6(+/+), and MSH6(−/−) cell lines, as well as in various hprt-negative daughter clones regardless of the type of the hprt gene mutations (base substitutions or frameshifts) (representative results are illustrated in Fig. 3). Treatment of the SW48 cells with the demethylating agent 5-aza-2′-deoxycytidine, which can partially restore hMLH1 expression (33), led to the detection of the hMLH1 transcript. Therefore, a similar hMLH1 expression reactivation in the SW48 cell derivatives should have been similarly detected by our experimental system.
Figure 3.
Analysis of cellular hMLH1 mRNA content. Multiplexed RT-PCR was performed with primers specific for the hMLH1 and hprt genes. LoVo and DLD-1 cell lines, as well as SW48 cells treated for 48 h with various doses of 5-aza-2′-deoxycytidine (2 μM and 10 μM Aza), were used as positive controls for hMLH1 expression. Four hprt− single cell clones obtained from MSH6 (+/+) or (−/−) cell lines were analyzed for hMLH1 expression. Single-cell clones 1 and 2 are mutants with frameshift hprt mutations and single-cell clones 3 and 4 are mutants with base-substitution hprt mutations.
Discussion
The MMP is generated by a complex mechanism that involves inactivation of different components of the DNA MMR system. The discovery of concomitant mutations of different members of the DNA MMR family in the same tumor cells raised the question of the possible functional role of such mutations in the development of the mutator phenotype (23). We previously reported that MMP-positive tumor cell lines with mutations in different DNA MMR genes exhibited different spectra of mutations (28). This finding suggested that the mutator phenotype could differ, depending on which DNA MMR gene was inactivated. Consistent with this hypothesis, we proposed that the concomitant inactivation of different DNA MMR genes could not only enhance the overall mutation rate in cells but also change the spectrum of mutations by combining mutations that are specific for each of the DNA MMR genes.
Initiation of human DNA MMR occurs through the recognition of mismatches by the MutSα (heterodimer of MSH2 and MSH6 proteins) and MutSβ (heterodimer of MSH2 and MSH3 proteins) complexes. The MutSα complex preferentially binds to mispaired bases and to small loops that result from slippage by misalignment of DNA strands, whereas MutSβ binds to larger loops and exhibits little, if any, affinity to mispaired bases (31, 34–36).
A human analogue of the bacterial MutL activity, called MutLα, was isolated by its ability to restore strand-specific DNA MMR in some MMP-positive cell lines (37). The MutLα complex was shown to be a heterodimer of human homologues of bacterial MutL: hMLH1 and hPMS2 (reviewed in ref. 38). This complex supposedly interacts with MutSα and MutSβ complexes and thus mediates DNA repair after the step of mismatch recognition. It was proposed that defects in the MutLα activity block mismatch correction before or at the excision step in repair (39, 40).
Thus, according to the current model (38) total inactivation of the hMLH1 gene should lead to a complete shutdown of the DNA MMR. Consequently, MSH6(+/+) cells, which lack expression of hMLH1, should produce all types of mutations caused by the inability of the DNA MMR to correct replication errors. Our results, however, show that MSH6(−/−) cells, which have inactivated hMSH6 in addition to unexpressed hMLH1, produce a different spectrum of mutations, with the mutation rate about 2.5 times higher than MSH6(+/+) cells. This finding implies that hMLH1 may not be required for all MutL activity in human cells and that an alternative pathway for the MutL function has about equal capacity.
There are, however, a few alternative explanations for our results. One possible explanation could be incomplete repression of hMLH1 expression in the MSH6(+/+) and MSH6(−/−) cells or hprt− mutants. However, we could not detect any traces of hMLH1 mRNA by RT-PCR in the SW48 cells or their derivatives, including the hprt mutant clones. Still there is a chance of a transient expression of the hMLH1 gene during selection for 6-TG resistance. However, the possibility of the occurrence of such phenomenon seems to be very small and not amenable to experimental test. The resemblance of the hprt gene mutation spectrum in the MSH6(+/+) cells to that of the hMLH1-defective cell lines obtained in other studies (approximately equal number of frameshifts and base substitutions) (28, 41, 42) is an additional factor in favor of the absence of hMLH1 expression at any step of our experiments.
Another spurious reason for the differences between the mutator phenotype of the MSH6(+/+) and MSH6(−/−) cells could be mutations accumulated in DNA replication or repair genes other than hMSH6. This would imply that all three MSH6(+/+) subclone cell lines but neither one of the three MSH6(−/−) subclones carry de novo mutations in additional “mutator” genes. The possibility that we isolated for our experiments three subclones with a single mutator gene (hMSH6) and three subclones with two mutator genes (one of which is hMSH6) is also very low: the frequency of single hMSH6 allele mutations was about 10−3, thus, roughly, the frequency of double mutations in the hMSH6 and some other mutator gene (even if we assume that a single allele mutation of this gene is sufficient for triggering a strong mutator phenotype) would be 10−6.
In addition, the mutation spectrum of the MSH6(−/−) cells resembles a combined spectrum of mutations caused by the two individual mutator genes, hMLH1 and hMSH6. We and others showed previously that hMLH1-defective cells were characterized by the prevalence of insertion/deletion types of mutations, whereas hMSH6-defective cells were characterized by the occurrence of base substitutions and almost no frameshift mutations at the hprt gene (28, 41, 42). It is reasonable to assume that involvement of mutator genes other than hMLH1 or hMSH6 would yield a distinguishable spectrum of mutation.
Insertions/deletions of only one nucleotide were the most common type of mutational events in the hMLH1-negative cells MSH6(+/+) (56%). This observation suggests that hMLH1 protein is essential for the repair of single nucleotide loops but less important for the repair of nucleotide mismatches. The latter function of hMLH1 could be complemented by other members of the DNA MMR system. Considering the approximately twice higher mutation rate in MSH6(−/−) cells, additional inactivation of the hMSH6 gene in these cells did not change the number of frameshift mutations and transitions at A⋅T base pairs per cell replication compared with the MSH6(+/+) cells but considerably increased the number of transitions and transversions at G⋅C base pairs (Table 2). This is consistent with the key feature of the MutSα complex, and hence of hMSH6, to correct base mismatches. At the same time, this suggests that hMSH6 protein is more specific for recognition of mismatches at G⋅C base pairs rather than at A⋅T base pairs, which could be more specifically recognized by other, perhaps still unidentified, DNA MMR proteins.
It is possible that not only a complete inactivation of one or several components of the DNA MMR, but their partial inactivation could contribute to the manifestation of the mutator phenotype in tumor cells. The probability of accumulation of monoallelic mutations in different DNA MMR genes might not be lower than a biallelic inactivation of a single DNA MMR gene, especially if a prior mutator phenotype is present, even if it is weak. Hence, one can expect that the steps in the development of the MMP in a tumor cell (or a tumor precursor cell) would include monoallelic mutations in different DNA MMR genes preceding the homozygous inactivation of a single DNA MMR gene.
We have previously shown the presence of monoallelic mutations in the DNA MMR genes in MMP-positive tumors with various frequencies (16, 23, 32, 43). Accumulation of such mutations can lead to haploinsufficiency of MMR function causing a weak mutator phenotype, which would increase with additional mono- or biallelic mutations of the DNA MMR genes until a “maximum” mutator phenotype is reached, after which no further selection would occur. This scenario is particularly applicable to the cases involving the incomplete or gradual inactivation of DNA repair by, for example, epigenetic silencing of DNA MMR genes, which probably occurs in a gradual manner during several mitotic cycles, or by splicing mutations that decrease the amount of correctly spliced transcripts.
This model leads to a somewhat paradoxical situation, because when a “maximum” mutator phenotype is reached, the probability of occurrence of nonfunctional mutations is also increased. Consequently, it is difficult to determine in a primary tumor which of the DNA MMR or other DNA repair gene mutations are functional and which are neutral. This difficulty is magnified because the high mutation frequency in MMP-positive tumors depreciates the presence of a gene mutation as a criterion for its functionality (44) and because mutations in MMR mutator genes do not immediately lead to a growth or territorial expansion advantage over sister cells (45, 46). The reversibility of these mutations, together with the potential deleterious impact of the mutator phenotype on cell viability (47), adds further complication to this complex process.
Nevertheless, we have provided functional evidence that the accumulation of a biallelic mutation in one of the MMR genes in a tumor cell already harboring another inactivated MMR gene is functional because it increases the mutation rate and alters the mutation spectrum of the mutator phenotype. This magnified mutator phenotype would accelerate the accumulation of mutations in the target cancer genes, which, in turn, would speed up tumor progression.
Acknowledgments
We thank Dr. K. Pyssarchuk for excellent technical support. This research was supported by the Cancer Research Fund, under Interagency Agreement 97-12013 (University of California, Contract 98-00924V) with the Department of Health Services, Cancer Research Program, and by National Institutes of Health Grants CA63585 and CA38579.
Abbreviations
- MMR
mismatch repair
- MMP
microsatellite mutator phenotype
- 6-TG
6-thioguanine
- RT-PCR
reverse transcriptase–PCR
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kolodner R. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 2.Shibata D, Aaltonen L A. Adv Cancer Res. 2001;80:83–114. doi: 10.1016/s0065-230x(01)80013-0. [DOI] [PubMed] [Google Scholar]
- 3.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen L A, Nystrom-Lahti M. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 4.Fishel R, Lescoe M K, Rao M R, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 5.Bronner C E, Baker S M, Morrison P T, Warren G, Smith L G, Lescoe M K, Kane M, Earabino C, Lipford J, Lindblom A, et al. Nature (London) 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 6.Papadopoulos N, Nicolaides N C, Wei Y F, Ruben S M, Carter K C, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, et al. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 7.Nicolaides N C, Papadopoulos N, Liu B, Wei Y F, Carter K C, Ruben S M, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, et al. Nature (London) 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 8.Kolodner R D, Tytell J D, Schmeits J L, Kane M F, Gupta R D, Weger J, Wahlberg S, Fox E A, Peel D, Ziogas A, et al. Cancer Res. 1999;59:5068–5074. [PubMed] [Google Scholar]
- 9.Risinger J I, Umar A, Boyd J, Berchuck A, Kunkel T A, Barrett J C. Nat Genet. 1996;14:102–105. doi: 10.1038/ng0996-102. [DOI] [PubMed] [Google Scholar]
- 10.Fishel R, Wilson T. Curr Opin Genet Dev. 1997;7:105–113. doi: 10.1016/s0959-437x(97)80117-7. [DOI] [PubMed] [Google Scholar]
- 11.Ionov Y, Peinado M A, Malkhosyan S, Shibata D, Perucho M. Nature (London) 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 12.Thibodeau S N, Bren G, Schaid D. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 13.Aaltonen L A, Peltomaki P, Leach F S, Sistonen P, Pylkkanen L, Mecklin J P, Jarvinen H, Powell S M, Jen J, Hamilton S R, et al. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 14.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, et al. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 16.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed J C, Perucho M. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 17.Duval A, Rolland S, Compoint A, Tubacher E, Iacopetta B, Thomas G, Hamelin R. Hum Mol Genet. 2001;10:513–518. doi: 10.1093/hmg/10.5.513. [DOI] [PubMed] [Google Scholar]
- 18.Lothe R A, Peltomaki P, Meling G I, Aaltonen L A, Nystrom-Lahti M, Pylkkanen L, Heimdal K, Andersen T I, Moller P, Rognum T O, et al. Cancer Res. 1993;53:5849–5852. [PubMed] [Google Scholar]
- 19.Peltomaki P, Lothe R A, Aaltonen L A, Pylkkanen L, Nystrom-Lahti M, Seruca R, David L, Holm R, Ryberg D, Haugen A, et al. Cancer Res. 1993;53:5853–5855. [PubMed] [Google Scholar]
- 20.Kim H, Jen J, Vogelstein B, Hamilton S R. Am J Pathol. 1994;145:148–156. [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B, Nicolaides N C, Markowitz S, Willson J K, Parsons R E, Jen J, Papadopolous N, Peltomaki P, de la Chapelle A, Hamilton S R, et al. Nat Genet. 1995;9:48–55. doi: 10.1038/ng0195-48. [DOI] [PubMed] [Google Scholar]
- 22.Perucho M. Biol Chem. 1996;377:675–684. [PubMed] [Google Scholar]
- 23.Malkhosyan S, Rampino N, Yamamoto H, Perucho M. Nature (London) 1996;382:499–500. doi: 10.1038/382499a0. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto H, Sawai H, Perucho M. Cancer Res. 1997;57:4420–4426. [PubMed] [Google Scholar]
- 25.Riccio A, Aaltonen L A, Godwin A K, Loukola A, Percesepe A, Salovaara R, Masciullo V, Genuardi M, Paravatou-Petsotas M, Bassi D E, et al. Nat Genet. 1999;23:266–268. doi: 10.1038/15443. [DOI] [PubMed] [Google Scholar]
- 26.Loukola A, Vilkki S, Singh J, Launonen V, Aaltonen L A. Am J Pathol. 2000;157:347–352. doi: 10.1016/S0002-9440(10)64546-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perucho M. Nat Med. 1996;2:630–631. doi: 10.1038/nm0696-630. [DOI] [PubMed] [Google Scholar]
- 28.Malkhosyan S, McCarty A, Sawai H, Perucho M. Mutat Res. 1996;316:249–259. doi: 10.1016/s0921-8734(96)90007-7. [DOI] [PubMed] [Google Scholar]
- 29.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Proc Natl Acad Sci USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kane M F, Loda M, Gaida G M, Lipman J, Mishra R, Goldman H, Jessup J M, Kolodner R. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 31.Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D'Arrigo A, Truong O, Hsuan J J, Jiricny J. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 32.Ohmiya N, Matsumoto S, Yamamoto H, Baranovskaya S, Malkhosyan S R, Perucho M. Gene. 2001;272:301–313. doi: 10.1016/s0378-1119(01)00517-0. [DOI] [PubMed] [Google Scholar]
- 33.Lengauer C, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1997;94:2545–2550. doi: 10.1073/pnas.94.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Acharya S, Wilson T, Gradia S, Kane M F, Guerrette S, Marsischky G T, Kolodner R, Fishel R. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drummond J T, Li G M, Longley M J, Modrich P. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 36.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 37.Li G M, Modrich P. Proc Natl Acad Sci USA. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiricny J. Nat Genet. 2000;24:6–8. doi: 10.1038/71698. [DOI] [PubMed] [Google Scholar]
- 39.Parsons R, Li G M, Longley M J, Fang W H, Papadopoulos N, Jen J, de la Chapelle A, Kinzler K W, Vogelstein B, Modrich P. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 40.Kat A, Thilly W G, Fang W H, Longley M J, Li G M, Modrich P. Proc Natl Acad Sci USA. 1993;90:6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhattacharyya N P, Ganesh A, Phear G, Richards B, Skandalis A, Meuth M. Hum Mol Genet. 1995;4:2057–2064. doi: 10.1093/hmg/4.11.2057. [DOI] [PubMed] [Google Scholar]
- 42.Ohzeki S, Tachibana A, Tatsumi K, Kato T. Carcinogenesis. 1997;18:1127–1133. doi: 10.1093/carcin/18.6.1127. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto H, Sawai H, Weber T K, Rodriguez-Bigas M A, Perucho M. Cancer Res. 1998;58:997–1003. [PubMed] [Google Scholar]
- 44.Zhang L, Yu J, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Cancer Res. 2001;61:3801–3805. [PubMed] [Google Scholar]
- 45.Perucho M, Peinado M A, Ionov Y, Casares S, Malkhosyan S, Stanbridge E. Cold Spring Harbor Symp Quant Biol. 1994;59:339–348. doi: 10.1101/sqb.1994.059.01.038. [DOI] [PubMed] [Google Scholar]
- 46.Cahill D P, Kinzler K W, Vogelstein B, Lengauer C. Trends Cell Biol. 1999;9:57–60. [PubMed] [Google Scholar]
- 47.Shibata D, Peinado M A, Ionov Y, Malkhosyan S, Perucho M. Nat Genet. 1994;6:273–281. doi: 10.1038/ng0394-273. [DOI] [PubMed] [Google Scholar]