Abstract
Summary
Coevolutionary sequence analysis has become a commonly used technique for de novo prediction of the structure and function of proteins, RNA, and protein complexes. We present the EVcouplings framework, a fully integrated open-source application and Python package for coevolutionary analysis. The framework enables generation of sequence alignments, calculation and evaluation of evolutionary couplings (ECs), and de novo prediction of structure and mutation effects. The combination of an easy to use, flexible command line interface and an underlying modular Python package makes the full power of coevolutionary analyses available to entry-level and advanced users.
Availability and implementation
1 Introduction
Coevolutionary sequence analysis presents a promising new approach to the long-standing problem of de novo prediction of the 3D structure of proteins and RNAs. In this approach, pairwise graphical models are used to identify evolutionary couplings (ECs) between sites, which frequently correspond to physical contacts in the molecule’s 3D structure. ECs have been used to successfully predict the residue contacts (Balakrishnan et al., 2011; Ekeberg et al., 2013; Marks et al., 2011; Morcos et al., 2011) and full 3D structure of proteins (Hopf et al., 2012; Marks et al., 2011; Ovchinnikov et al., 2015), RNAs (Weinreb et al., 2016), complexes (Hopf et al., 2014; Ovchinnikov et al., 2014; Weigt et al., 2009), as well as effects of mutations (Figliuzzi et al., 2015; Hopf et al., 2017). However, these applications require integrating multiple tools, data sources and extensive data processing. Available software in this field provides high-performance reimplementations of EC inference tools (Kaján et al., 2014; Seemayer et al., 2014; Weinreb et al., 2016), integration of multiple signals to improve prediction accuracy (Jones et al., 2015; Skwark et al., 2014), and a library targeted at format conversion between the different approaches (Simkovic et al., 2017). To make these methods accessible to a general biological audience, we present a flexible, open source application and Python package for end-to-end evolutionary coupling analysis. EVcouplings, making use of external tools, covers all necessary functionality, including alignment generation, EC calculation, de novo structure and mutation effect prediction, visualization of results, and comparison of predictions to experimental structures.
2 EVcouplings framework
The EVcouplings framework integrates the functionality of the previously published methods EVfold (Hopf et al., 2012; Marks et al., 2011), EVcomplex (Hopf et al., 2014) and EVmutation (Hopf et al., 2017). It provides (i) an easy-to-use command-line application and (ii) a modular Python package containing all functions, data structures and pipelines that comprise the application.
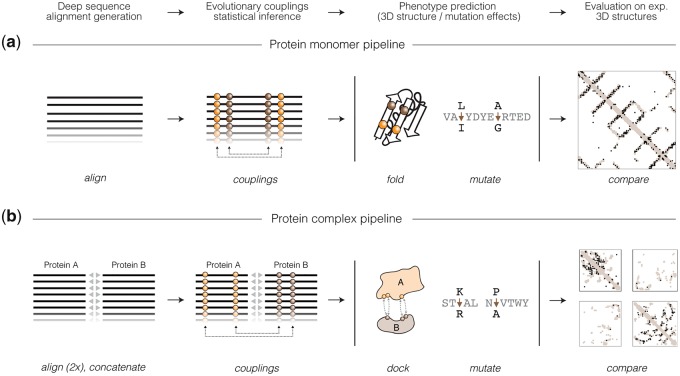
Command-line application: The command-line application allows users to obtain predictions for their proteins and complexes of interest by running the respective EVcouplings pipelines (Fig. 1). Each pipeline is comprised of a series of modular stages that can be configured using a YAML file, which aids reproducibility by documenting all parameters. The pipelines are parallelized and support local multi-process execution as well as commonly used cluster systems, and automatically handles job submission and monitoring. The steps of the prediction pipelines are: align, which generates and processes sequence alignments,concatenate, which pairs putatively interacting sequences for the protein complex pipeline, couplings, which calculates ECs, compare, which compares ECs to experimental structures, mutate, which predicts the effects of mutations, and fold, which generates de novo 3D models.
Fig. 1.
The EVcouplings Python framework. (a) The protein monomer EVcouplings pipeline entails multiple sequence alignment generation (align stage), EC inference (couplings stage), de novo folding (fold stage), mutation effect prediction (mutate stage) and comparison to experimental structure (compare stage). (b) The protein complex pipeline extends the monomer pipeline to protein interactions by pairing putatively interacting homologs (concatenate stage) and providing restraints for molecular docking (dock stage)
EVcouplings Python package: The command-line application is built on the underlying evcouplings Python package, whose modular architecture and comprehensive documentation facilitate the development of new stages and pipelines. Additionally, the package serves as a toolbox for handling and analyzing EC-related data. Examples for interactive usage are provided in Jupyter notebooks (Kluyver et al., 2016) distributed with the package, and extensive documentation is available on the web (http://evcouplings.readthedocs.io).
3 Conclusion
EVcouplings is an open source, integrated pipeline for evolutionary couplings analyses. The underlying API serves as a modular basis for data analysis and will allow developers to rapidly create new workflows.
Acknowledgements
We thank the members of Marks and Sander labs for helpful comments and testing, and HMS Research Computing for computational resources and support.
Funding
This work has been supported by NSF GRFP DGE1144152 (AGG), DOE CSGF fellowship DE-FG02-97ER25308 (AJR), Pathway Commons U41 HG006623, NRNB P41 GM103504 and R01 GM106303.
Conflict of Interest: none declared.
References
- Balakrishnan S. et al. (2011) Learning generative models for protein fold families. Proteins, 79, 1061–1078. [DOI] [PubMed] [Google Scholar]
- Ekeberg M. et al. (2013) Improved contact prediction in proteins: using pseudolikelihoods to infer Potts models. Phys. Rev. E Stat. Nonlin. Soft Matter. Phys., 87, 012707. [DOI] [PubMed] [Google Scholar]
- Figliuzzi M. et al. (2015) Coevolutionary landscape inference and the context-dependence of mutations in beta-lactamase TEM-1. Mol. Biol. Evol., 33, 268–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf T.A. et al. (2012) Three-dimensional structures of membrane proteins from genomic sequencing. Cell, 149, 1607–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf T.A. et al. (2017) Mutation effects predicted from sequence co-variation. Nat. Biotechnol., 35, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf T.A. et al. (2014) Sequence co-evolution gives 3D contacts and structures of protein complexes. Elife, 3, e03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T. et al. (2015) MetaPSICOV: combining coevolution methods for accurate prediction of contacts and long range hydrogen bonding in proteins. Bioinformatics, 31, 999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaján L. et al. (2014) FreeContact: fast and free software for protein contact prediction from residue co-evolution. BMC Bioinformatics, 15, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluyver T. et al. (2016) Jupyter Notebooks-a publishing format for reproducible computational workflows In: ELPUB, pp. 87–90. [Google Scholar]
- Marks D.S. et al. (2011) Protein 3D structure computed from evolutionary sequence variation. PloS One, 6, e28766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcos F. et al. (2011) Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. USA, 108, E1293–E1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov S. et al. (2014) Robust and accurate prediction of residue–residue interactions across protein interfaces using evolutionary information. Elife, 3, e02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov S. et al. (2015) Large-scale determination of previously unsolved protein structures using evolutionary information. Elife, 4, e09248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemayer S. et al. (2014) CCMpred–fast and precise prediction of protein residue-residue contacts from correlated mutations. Bioinformatics, 30, 3128–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkovic F. et al. (2017) ConKit: a python interface to contact predictions. Bioinformatics, 33, 2209–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skwark M.J. et al. (2014) Improved contact predictions using the recognition of protein like contact patterns. PLoS Comput. Biol., 10, e1003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigt M. et al. (2009) Identification of direct residue contacts in protein–protein interaction by message passing. Proc. Natl. Acad. Sci. USA, 106, 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb C. et al. (2016) 3D RNA and functional interactions from evolutionary couplings. Cell, 165, 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]