Abstract
The inclusion of exons during the splicing process depends on the binding of splicing factors to short low-complexity regulatory sequences. The relationship between exonic splicing regulatory sequences and coding sequences is still poorly understood. We demonstrate that exons that are coregulated by any given splicing factor share a similar nucleotide composition bias and preferentially code for amino acids with similar physicochemical properties because of the nonrandomness of the genetic code. Indeed, amino acids sharing similar physicochemical properties correspond to codons that have the same nucleotide composition bias. In particular, we uncover that the TRA2A and TRA2B splicing factors that bind to adenine-rich motifs promote the inclusion of adenine-rich exons coding preferentially for hydrophilic amino acids that correspond to adenine-rich codons. SRSF2 that binds guanine/cytosine-rich motifs promotes the inclusion of GC-rich exons coding preferentially for small amino acids, whereas SRSF3 that binds cytosine-rich motifs promotes the inclusion of exons coding preferentially for uncharged amino acids, like serine and threonine that can be phosphorylated. Finally, coregulated exons encoding amino acids with similar physicochemical properties correspond to specific protein features. In conclusion, the regulation of an exon by a splicing factor that relies on the affinity of this factor for specific nucleotide(s) is tightly interconnected with the exon-encoded physicochemical properties. We therefore uncover an unanticipated bidirectional interplay between the splicing regulatory process and its biological functional outcome.
Alternative splicing is a cellular process involved in the regulated inclusion or exclusion of exons during the processing of mRNA precursors. Alternative splicing is the rule rather than the exception in human because 95% of human genes produce several splicing variants (Pan et al. 2008; Wang et al. 2008). The exon selection process relies on RNA binding proteins or splicing factors that enhance or repress exon inclusion following two main principles. First, splicing factors bind to short intronic or exonic motifs (or splicing regulatory sequences) that are often low-complexity sequences composed of the repetition of the same nucleotide or dinucleotide (Fu and Ares 2014). In addition, the interaction of splicing factors with their cognate binding motifs often depends on the sequence context and on the presence of clusters of related binding motifs (Zhang et al. 2013; Cereda et al. 2014; Fu and Ares 2014; Dominguez et al. 2018; Jobbins et al. 2018). The second principle states that the splicing outcome (i.e., exon inclusion or skipping) depends on where splicing factors bind on pre-mRNAs with respect to the regulated exons. For example, HNRNP-like splicing factors often repress the inclusion of exons they bind, but they enhance exon inclusion when they do not bind exons but instead bind to introns (Erkelenz et al. 2013; Fu and Ares 2014; Geuens et al. 2016). Meanwhile, the exonic binding of SR-like splicing factors (SRSFs) usually enhances exon inclusion (Erkelenz et al. 2013; Fu and Ares 2014).
Because some splicing regulatory sequences lie within protein-coding sequences, a major challenge is to understand how coding sequences accommodate this overlapping layer of information (Itzkovitz et al. 2010; Lin et al. 2011; Savisaar and Hurst 2017a,b). To date, a general assumption is that protein-coding sequences can accommodate overlapping information or “codes” (including the “splicing code”) as a direct consequence of the redundancy of the genetic code that allows the same amino acid to be encoded by several codons differing only on their third “wobble” nucleotide (Goren et al. 2006; Itzkovitz and Alon 2007; Itzkovitz et al. 2010; Lin et al. 2011; Shabalina et al. 2013; Savisaar and Hurst 2017a,b). Therefore, coding and exonic splicing regulatory sequences could evolve independently because of the variation of the third nucleotides of codons. In this setting, it has recently been shown that splicing regulatory sequence selection severely constrains coding sequence evolution (Savisaar and Hurst 2018). In addition, it has been reported that some amino acids are preferentially encoded near exon–intron junctions because of the presence of general splicing consensus sequences near splicing sites (Parmley et al. 2007; Warnecke et al. 2008; Smithers et al. 2015). Finally, recent evidence has suggested that exons that are coregulated in specific pathophysiological conditions may code for protein regions engaged in similar cellular processes (Irimia et al. 2014; Tranchevent et al. 2017). These observations raised the possibility that exons regulated by the same splicing regulatory process code for similar biological information. So far, the lack of large sets of coregulated exons has limited the studies addressing the interplay between the splicing regulatory process and peptide sequences encoded by splicing-regulated exons. By focusing on exons coregulated by different splicing factors, we uncover a bidirectional interplay between the physicochemical protein features encoded by exons and their regulation by splicing factors.
Results
Nucleotide composition bias of coregulated exons
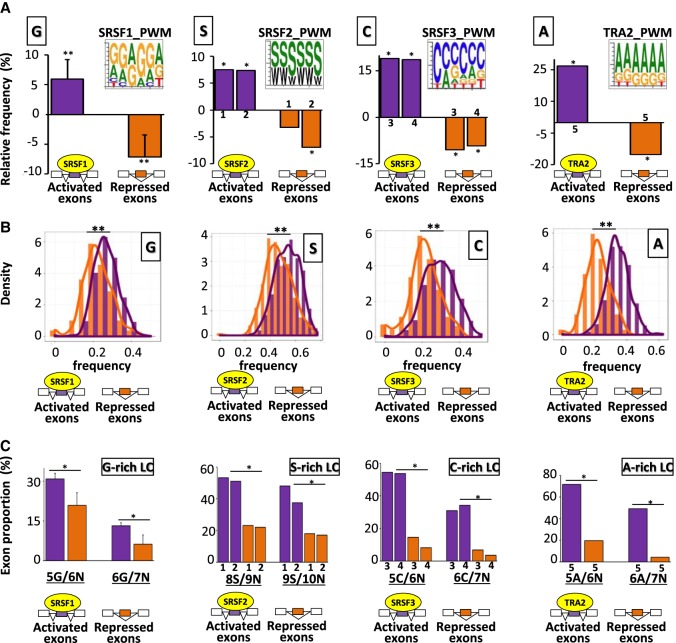
To investigate the relationship between exonic splicing regulatory sequences and coding sequences, we analyzed publicly available RNA-seq data sets generated from different cell lines transfected with siRNAs or shRNAs targeting specific splicing factors (e.g., SRSF1, SRSF3, TRA2A, TRA2B) or transfected with an SRSF2-expression vector (Supplemental Table S1). Because TRA2A and TRA2B are paralogous and have been codepleted in the analyzed data sets, we will refer both of these factors as TRA2. SRSF1, SRSF2, SRSF3, and TRA2 belong to the family of Arg/Ser (RS) domain-containing splicing factors (SRSFs). Each splicing factor regulated a common set of exons in several cell lines, but many exons were regulated on a cell line–specific mode (Supplemental Fig. S1). SRSF1, SRSF2, SRSF3, and TRA2 bind to GGA-rich motifs, SSNG motifs (where S = G or C), C-rich and G-poor motifs, and AGAA-like motifs, respectively (Grellscheid et al. 2011; Tsuda et al. 2011; Änkö et al. 2012; Pandit et al. 2013; Ray et al. 2013; Best et al. 2014; Fu and Ares 2014; Anczuków et al. 2015; Hauer et al. 2015; Giudice et al. 2016; Luo et al. 2017). As expected, hexanucleotides enriched in SRSF1-, SRSF2-, SRSF3-, or TRA2-activated exons across different cell lines were enriched in purine-rich, S-rich, C-rich, or A-rich hexanucleotides, respectively, when compared to control exons and in contrast to exons repressed by the same factor (position weight matrices [PWM]) (Fig. 1A; Supplemental Fig. S2A; Supplemental Table S2).
Figure 1.
Nucleotide composition bias of coregulated exons. (A) Position weight matrices (PWM) using the 10 most enriched hexanucleotides in SRSF1-, SRSF2-, SRFS3-, or TRA2-activated exons. The histograms represent the relative frequency (%) when compared to sets of control exons of G, S, C, and A nucleotides in SRSF1-, SRSF2-, SRSF3-, and TRA2-regulated exons, respectively, identified in different cell lines. The average values obtained from four data sets are represented in the case of SRSF1: (**) Mann–Whitney U test P-value < 0.03. The sets of the other SRSF-regulated exons originated from K562 (1), Huh7 (2), HepG2 (3), GM19238 (4), and MDA-MB-231 (5) cell lines: (*) randomization test FDR < 0.03. (B) Density chart of G, S, C, and A nucleotides in SRSF1-, SRSF2-, SRSF3-, and TRA2-regulated exons, respectively: (**) Kolmogorov–Smirnov (K–S) test < 1 × 10−13. (C) Proportion of exons containing at least one low-complexity (LC) sequence of 6, 7, 9, or 10 nt. In a sliding window of N nucleotides, the number of the same nucleotide (G, S, C, or A) must be equal to or greater than (N-1). The x-axis is labeled to indicate the number of single nucleotides identified in a given window. For example, “5G/6N” means that a sequence of 6 consecutive nucleotides (6N) is composed of at least 5 Gs (5G). The average values obtained from four data sets are represented in the case of SRSF1. The sets of the other SRSF-regulated exons originated from K562 (1), Huh7 (2), HepG2 (3), GM19238 (4), and MDA-MB-231 (5) cell lines. A logistic regression analysis was performed to test if the presence of low-complexity sequences was different between activated and repressed exons by a given splicing factor while accounting for cell line variations: (*) P-value < 3 × 10−7.
Each set of splicing factor–regulated exons had a specific nucleotide composition bias. Indeed, SRSF1-activated exons (but not SRSF1-repressed exons) were enriched in G when compared to control exons (Mann–Whitney U test P-value 0.029) (Fig. 1A; see also Supplemental Fig. S2B; Supplemental Table S2). SRSF2-activated exons (but not SRSF2-repressed exons) were enriched in S (G or C; randomization test FDR < 1 × 10−3) (Fig. 1A). SRSF3-activated exons were enriched in C (randomization test FDR < 1 × 10−4) (Fig. 1A) and impoverished in G (Supplemental Fig. S2C). Finally, TRA2-activated exons were enriched in A (randomization test FDR < 1 × 10−4), unlike TRA2-repressed exons (randomization test FDR < 1 × 10−4), when compared to control exons (Fig. 1A). Accordingly, a high density of G, S, C, or A nucleotides was more frequent in SRSF1-, SRSF2-, SRSF3-, or TRA2-activated exons, respectively, when compared to the corresponding repressed exons (Kolmogorov–Smirnov [K–S] test P-value < 1 × 10−14) (Fig. 1B).
Although the enriched nucleotides within splicing factor–regulated exons could be randomly distributed across exons, we observed an increased frequency of specific dinucleotides and low-complexity sequences. For example, GG, SS, CC, or AA dinucleotides were more frequent in SRSF1-, SRSF2-, SRSF3-, or TRA2-activated exons, respectively, than in control exons or in the corresponding repressed exons (Supplemental Fig. S2D; Supplemental Table S2). We next performed a logistic regression analysis to test differences in low-complexity sequence content between activated and repressed exons for a given splicing factor while accounting for cell line variations. As shown in Figure 1C, a larger proportion of SRSF1-, SRSF2-, SRSF3-, or TRA2-activated exons contained G-, S-, C-, or A-rich low-complexity sequences, respectively, when compared to the corresponding repressed exons (logistic regression analysis P-value < 3 × 10−7).
Amino acid composition bias of SRSF-coregulated exons
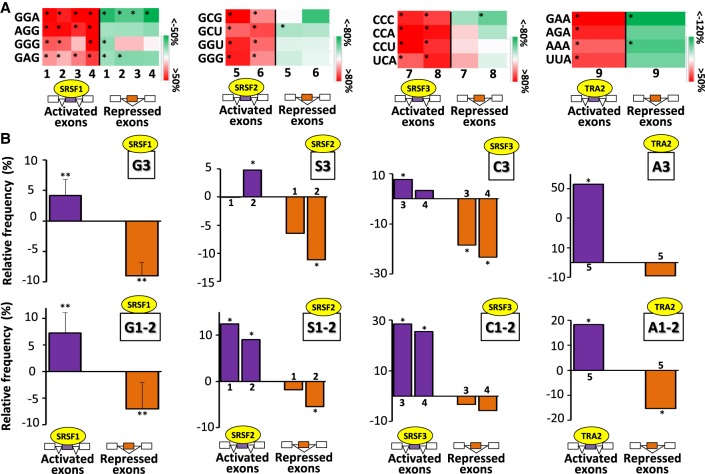
We next analyzed the codon content of exons regulated by SRSF-related splicing factors. In agreement with the nucleotide composition bias described previously, SRSF1-, SRSF2-, SRSF3-, or TRA2-activated exons were enriched in G-, S-, C-, or A-rich codons, respectively, compared to both sets of control exons or the corresponding repressed exons (randomization test FDR < 0.05) (Fig. 2A; see also Supplemental Fig. S3; Supplemental Table S2). In this setting, it has been proposed that coding sequences can accommodate exonic splicing regulatory sequences through variation of the third codon position (“Introduction”). Accordingly, the nucleotide composition bias observed at the whole exon level (Fig. 1) was also observed on the third codon position across some data sets and for a subset of codons (Fig. 2B, upper panels; Supplemental Table S2; Supplemental Fig. S4). However, the identity of the nucleotide at the first or second codon positions was systematically biased (Fig. 2B, lower panels; Supplemental Table S2). This raises the possibility that different sets of SRSF-regulated exons may preferentially code for different amino acids.
Figure 2.
Nucleotide composition bias of codons of coregulated exons. (A) Color code of the relative frequency (%) compared with sets of control exons of some codons in SRSF-activated and SRSF-repressed exons across different cell lines: K562 (1), HepG2 (2), GM19238 (3), HeLa (4), K562 (5), Huh7 (6), HepG2 (7), GM19238 (8), and MDA-MB-231 (9). The frequency of each codon was calculated in SRSF-activated and SRSF-repressed exons and expressed as the percentage of the average frequency calculated in sets of control exons. Red and green colors indicate when the codon frequency is higher and lower, respectively, in the sets of regulated exons when compared with sets of control exons. Only some enriched codons identified in SRSF-activated exons are represented: (*) randomization test FDR < 0.05. (B) The upper panels represent the relative frequency (%) compared with sets of control exons of G (G3), S (S3), C (C3), or A (A3) nucleotides at the third codon positions in SRSF-activated and SRSF-repressed exons. The lower panels represent the relative frequency (%) compared with sets of control exons of G (G1-2), S (S1-2), C (C1-2), or A (A1-2) nucleotides at the first and second codon positions in SRSF-activated and SRSF-repressed exons. The average values obtained from four data sets are represented in the case of SRSF1: (**) Mann–Whitney U test P-value < 0.03. The sets of the other SRSF-regulated exons originated from K562 (1), Huh7 (2), HepG2 (3), GM19238 (4), and MDA-MB-231 (5) cell lines: (*) randomization test FDR < 0.03.
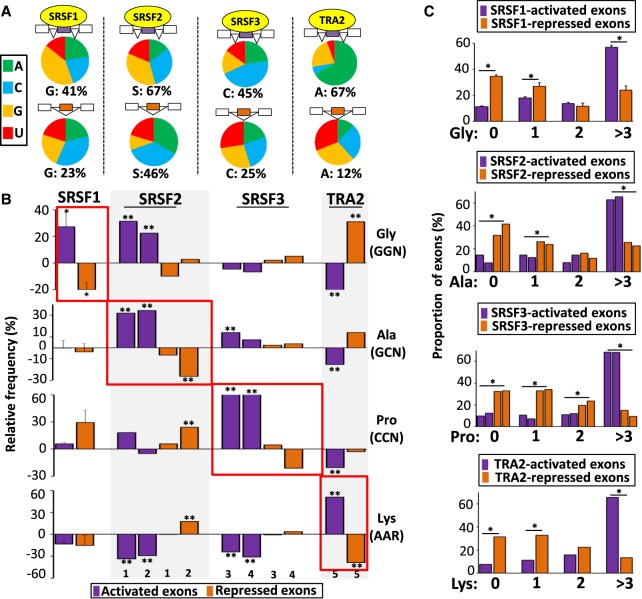
As shown in Figure 3A (upper panels), amino acids more frequently encoded by SRSF1-, SRSF2-, SRSF3-, and TRA2-activated exons corresponded to G-, S-, C-, and A-rich codons, respectively (see also Supplemental Fig. S5; Supplemental Table S2). This was in sharp contrast to the corresponding repressed exons (Fig. 3A, lower panels). For example, glycine (GGN codons) was more frequently encoded by SRSF1-activated exons (Mann–Whitney U test P-value 0.029) than by control exons and SRSF1-repressed exons (Fig. 3B). A counting of glycines encoded within SRSF1-activated versus SRSF1-repressed exons showed a mirrored distribution: A large proportion of activated exons (∼60%) coded for more than three glycines, whereas nearly 70% of repressed exons coded for a maximum of one glycine (Fig. 3C). Similarly, alanine (GCN codons), proline (CCN codons), and lysine (AAR codons) were more frequently encoded by SRSF2-, SRSF3-, and TRA2-activated exons, respectively (Fig. 3B,C).
Figure 3.
Amino acid composition bias encoded by coregulated exons. (A) Nucleotide composition of codons corresponding to amino acids more frequently encoded, when compared to sets of control exons, by SRSF1-, SRSF2-, SRSF3-, or TRA2-activated (upper) and -repressed exons (lower). (B) Relative frequency (%) compared with sets of control exons of glycine (Gly corresponding to GGN codons), alanine (Ala corresponding to GCN codons), proline (Pro corresponding to CCN codons), and lysine (Lys corresponding to AAR codons) encoded by SRSF1-, SRSF2-, SRSF3-, or TRA2-activated and -repressed exons. The average values obtained from four data sets are represented in the case of SRSF1: (*) Mann–Whitney U test P-value < 0.03. The sets of the other SRSF-regulated exons originated from K562 (1), Huh7 (2), HepG2 (3), GM19238 (4), and MDA-MB-231 (5) cell lines: (**) randomization test FDR < 0.03. (C) Proportion (%) of exons from SRSF1-, SRSF2-, SRSF3, and TRA2-regulated exons encoding for 0, 1, 2, and more than 3 Gly, Ala, Pro, or Lys amino acids, respectively. The average values obtained from four data sets are represented in the case of SRSF1. A logistic regression analysis was performed to test if the presence or absence of an amino acid at a given level was different between activated and repressed exons for a given splicing factor while accounting for cell line variations: (*) P-value < 0.05.
SRSF-coregulated exons code for amino acids with similar physicochemical properties
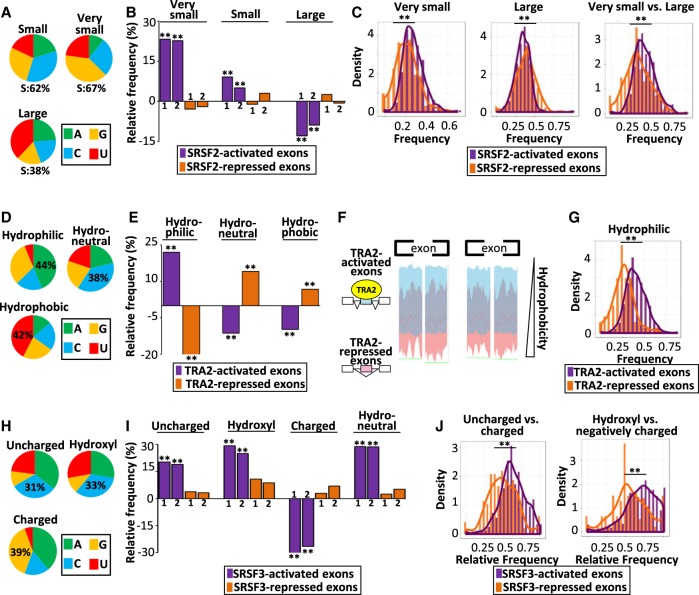
The preceding observations revealed a nucleotide composition bias of splicing factor–regulated exons and a bias regarding the nature of the amino acids that are encoded by these exons. In this setting, it is well established that amino acids sharing similar physicochemical properties (e.g., size, hydropathy, charge) are encoded by similar codons (i.e., codons composed of the same nucleotides) (Woese 1965; Wolfenden et al. 1979; Taylor and Coates 1989; Biro et al. 2003; Prilusky and Bibi 2009). For example, small amino acids (Ala, Asn, Asp, Cys, Gly, Pro, Ser, Thr)—in particular, very small amino acids (Ala, Gly, Ser, Cys)—are encoded by S-rich codons, whereas large amino acids (Arg, Ile, Leu, Lys, Met, Phe, Trp, Tyr) are encoded by S-poor codons (Fig. 4A). SRSF2 binds to SSNG motifs and SRSF2-activated exons are S-rich (Fig. 1). The two sets of analyzed SRSF2-activated exons encoded more frequently for very small and small amino acids when compared to control exons, in contrast to SRSF2-repressed exons (randomization test FDR < 0.0003) (Fig. 4B; Supplemental Table S2). Conversely, large amino acids were less frequently encoded by SRSF2-activated exons (randomization test FDR < 1 × 10−4) (Fig. 4B). Accordingly, a high density of very small amino acids was more frequent in SRSF2-activated when compared to SRSF2-repressed exons, whereas a high density of large amino acids was more frequent in SRSF2-repressed when compared to SRSF2-activated exons (K–S test P-value < 5 × 10−6) (Fig. 4C).
Figure 4.
SRSF-coregulated exons code for amino acids with similar physicochemical properties. (A) Nucleotide composition of codons encoding small, very small, and large amino acids: S = G or C. (B) Relative frequency (%), when compared to sets of control exons, of very small, small, and large amino acids encoded by two sets of SRSF2-activated and SRSF2-repressed exons identified in the K562 (1) and Huh7 (2) cell lines: (**) randomization test FDR < 0.0003. (C) Density chart of SRSF2-activated and SRSF2-repressed exons identified in K562 cells coding for very small and large amino acids: (**) K–S test < 5 × 10−6. (D) Nucleotide composition of codons encoding hydrophobic, neutral, and hydrophilic amino acids. (E) Relative frequency (%), when compared to sets of control exons, of hydrophilic, neutral, and hydrophobic amino acids encoded by TRA2-activated and TRA2-repressed exons: (**) randomization test FDR < 0.005. (F) Hydrophobic scales of TRA2-activated and TRA2-repressed exons. The green line (bottom) indicates the Mann–Whitney U test P-value < 0.05 at each amino acid position. (G) Density chart of TRA2-activated or TRA2-repressed exons coding for hydrophilic amino acids: (**) K–S test < 1 × 10−13. (H) Nucleotide composition of codons encoding polar uncharged, hydroxyl-containing, and charged amino acids. (I) Relative frequency (%), when compared to sets of control exons, of polar uncharged, hydroxyl-containing, charged, or neutral (in terms of hydropathy) amino acids encoded by two sets of SRSF3-activated and SRSF3-repressed exons identified from HepG2 (1) and GM19238 (2) cell lines: (**) randomization test FDR < 1 × 10−4. (J, left) Density chart of SRSF3-activated and SRSF3-repressed exons describing the frequencies of polar uncharged amino acids compared to all polar amino acids. (Right) Density chart of SRSF3-activated and SRSF3-repressed exons describing the frequencies of hydroxyl amino acids compared to negatively charged amino acids. Note that hydroxyl amino acids can be negatively charged after phosphorylation: (**) K–S test < 3 × 10−12.
Amino acids can be classified in three families in regard to their hydropathy, and each family is encoded by codons having different features. Hydrophilic amino acids (Arg, Asn, Asp, Gln, Glu, Lys) are encoded by A-rich codons, hydrophobic amino acids (Ala, Cys, Ile, Leu, Met, Phe, Val) are encoded by U-rich and A-poor codons, and ambivalent or neutral amino acids (Gly, His, Pro, Ser, Thr, Tyr) are encoded by C-rich codons (Fig. 4D; Kyte and Doolittle 1982; Engelman et al. 1986; Chiusano et al. 2000; Biro et al. 2003; Pommié et al. 2004; Prilusky and Bibi 2009; Zhang and Yu 2011). TRA2 that binds to AGAA-like motifs activates the inclusion of A-rich exons (Fig. 1). TRA2-activated exons encoded hydrophilic amino acids more frequently (randomization test FDR < 1 × 10−4) and neutral or hydrophobic amino acids less frequently than control exons (randomization test FDR < 1 × 10−4 ) (Fig. 4E; Supplemental Table S2). Similar results were obtained by using different hydrophobicity propensity scales (Fig. 4F). Accordingly, a high density of hydrophilic amino acids was more frequently encoded by TRA2-activated when compared to TRA2-repressed exons (K–S test P-value < 1 × 10−13) (Fig. 4G).
Polar uncharged amino acids (Asn, Gln, Ser, Thr, Tyr), including hydroxyl-containing amino acids (e.g., Ser and Thr) correspond to C-rich and G-poor codons, whereas polar charged amino acids (Asp, Glu, Lys, Arg) correspond to G-rich and C-poor codons (Fig. 4H; Biro et al. 2003; Zhang and Yu 2011). SRSF3 binds to C-rich motifs and activates C-rich and G-poor exons (Fig. 1). Two sets of SRSF3-activated exons encoded uncharged (including hydroxyl-containing) amino acids more frequently than control exons (randomization test FDR < 1 × 10−4) or SRSF3-repressed exons (Fig. 4I; Supplemental Table S2). They also encoded more frequently hydropathically neutral amino acids (randomization test FDR < 1 × 10−4 for both cell lines) (Fig. 4I) that correspond to C-rich codons (Fig. 4D). As shown in Figure 4J, a high relative density of uncharged versus charged and hydroxyl versus negatively charged amino acids was more frequent in SRSF3-activated when compared to SRSF3-repressed exons (K–S test P-value < 3 × 10−12). These observations suggest a link between splicing-related nucleotide composition bias of splicing-regulated exons and physicochemical properties of the exon-encoded amino acids.
HNRNP-corepressed exons code for amino acids with similar physicochemical properties
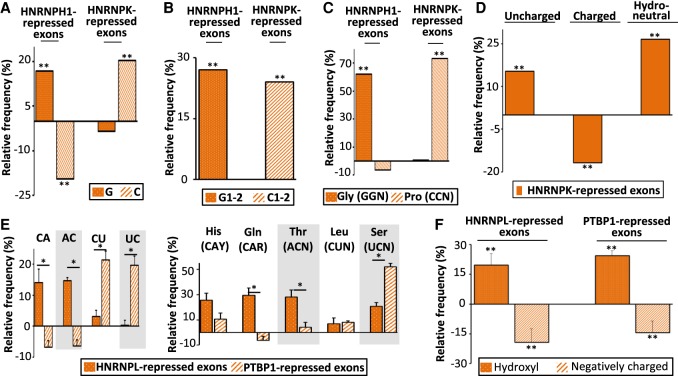
SRSF-like splicing factors activate exons they bind, in contrast to HNRNP-like splicing factors that repress exons they bind. We analyzed publicly available RNA-seq data sets generated from different cell lines transfected with siRNAs or shRNAs targeting HNRNPH1, HNRNPK, HNRNPL, or PTBP1 (Supplemental Table S1; Supplemental Fig. S1). HNRNPH1, HNRNPK, HNRNPL, and PTBP1 bind to G-rich, C-rich, CA-rich, and CU-rich motifs, respectively (Klimek-Tomczak et al. 2004; Katz et al. 2010; Llorian et al. 2010; Ray et al. 2013; Rossbach et al. 2014; Hauer et al. 2015; Giudice et al. 2016). As shown in Figure 5A, HNRNPH1-repressed exons were enriched in Gs (randomization test FDR < 0.005), whereas HNRNPK-repressed exons were enriched in Cs when compared to control exons (randomization test FDR < 1 × 10−4) (Supplemental Fig. S2F). This nucleotide composition bias was observed at the first and second codon positions (randomization test FDR < 0.003) (Fig. 5B; Supplemental Fig. S2G). Accordingly, glycine (GGN codons) and proline (CCN codons) were more frequently encoded by HNRNPH1-repressed and by HNRNPK-repressed exons, respectively (randomization test FDR < 0.05) (Fig. 5C; Supplemental Fig. S2H). As in the case of C-rich SRSF3-activated exons (Fig. 4I), C-rich HNRNPK-repressed exons encoded more frequently uncharged and hydropathically neutral amino acids than control exons (randomization test FDR < 1 × 10−4) (Fig. 5D; Supplemental Fig. S2I).
Figure 5.
HNRNP-corepressed exons code for amino acids with similar physicochemical properties. (A) Relative frequency (%), when compared to sets of control exons, of G and C nucleotides in HNRNPH1- and HNRNPK-repressed exons identified in 293T and GM19238 cells, respectively: (**) randomization test FDR < 0.006. (B) Relative frequency (%), when compared to sets of control exons, of G (G1–2), or C (C1–2) nucleotides at the first and second codon position from HNRNPH1- or HNRNPK-repressed exons identified in 293T and GM19238 cells, respectively: (**) randomization test FDR < 0.003. (C) Relative frequency (%), when compared to sets of control exons, of glycine (Gly corresponding to GGN codons) and proline (Pro corresponding to CCN codons) encoded by HNRNPH1- and HNRNPK-repressed exons identified in 293T and GM19238 cells, respectively: (**) randomization test FDR < 0.05. (D) Relative frequency (%), when compared to sets of control exons, of polar uncharged, charged, or neutral (in terms of hydropathy) amino acids encoded by HNRNPK-repressed exons identified in GM19238 cells: (**) randomization test FDR < 1 × 10−4. (E) The left panel represents the average of the relative frequency (%), when compared to sets of control exons of CA, CT, AC, and TC dinucleotides calculated from four sets from different cell lines of HNRNPL- or PTBP1-repressed exons. The right panel represents the average of the relative frequency (%), when compared to sets of control exons of histidine (His corresponding to CAY codons), glutamine (Gln corresponding to CAR codons), leucine (Leu corresponding to CTN codons), threonine (Thr corresponding to ACN codons), and serine (Ser corresponding to TCN codons) encoded by four sets of HNRNPL- and PTBP1-repressed exons. A Mann–Whitney U test was used to compare whether the real frequencies of those amino acids and dinucleotides between HNRNPL- and PTBP1-repressed exons: (*) P-value < 0.03. (F) Relative frequency (%), when compared to sets of control exons, of hydroxyl-containing and negatively charged amino acids encoded by HNRNPL- or PTBP1-repressed exons. The average values of four data sets are represented for HNRNPL and PTBP1: (**) Mann–Whitney U test P-value < 0.03.
As mentioned above, HNRNPL represses the inclusion of exons containing CA-rich motifs, whereas PTBP1 represses the inclusion of exons containing CU-rich motifs. As shown in Figure 5E (left), HNRNPL repressed the inclusion of exons enriched in CA and AC dinucleotides when compared to PTBP1-repressed exons (Mann–Whitney U test P-value = 0.029), whereas PTBP1 repressed exons enriched in CU and UC dinucleotides unlike HNRNPL (Mann–Whitney U test P-value = 0.029). Glutamine (Gln, CAR codons) and threonine (Thr, ACN codons) were more frequently encoded by HNRNPL-repressed exons than by PTBP1-repressed exons, whereas serine (Ser, UCN codons) was more frequently encoded by PTBP1-repressed exons than by HNRNPL-repressed exons (Mann–Whitney U test P-value = 0.03) (Fig. 5E, right). Both HNRNPL- and PTBP1-repressed exons, respectively, encoded more frequently hydroxyl-containing amino acids and less frequently negatively charged amino acids, compared to control exons (Mann–Whitney U test P-value = 0.03) (Fig. 5F). This is consistent with the fact that hydroxyl-containing and charged amino acids are C-rich and C-poor, respectively (Fig. 4H).
In conclusion, each tested set of SRSF- or HNRNP-coregulated exons has a specific nucleotide composition bias and codes for amino acids with similar physicochemical properties.
Bidirectional interplay between the splicing regulatory process and its functional outcome
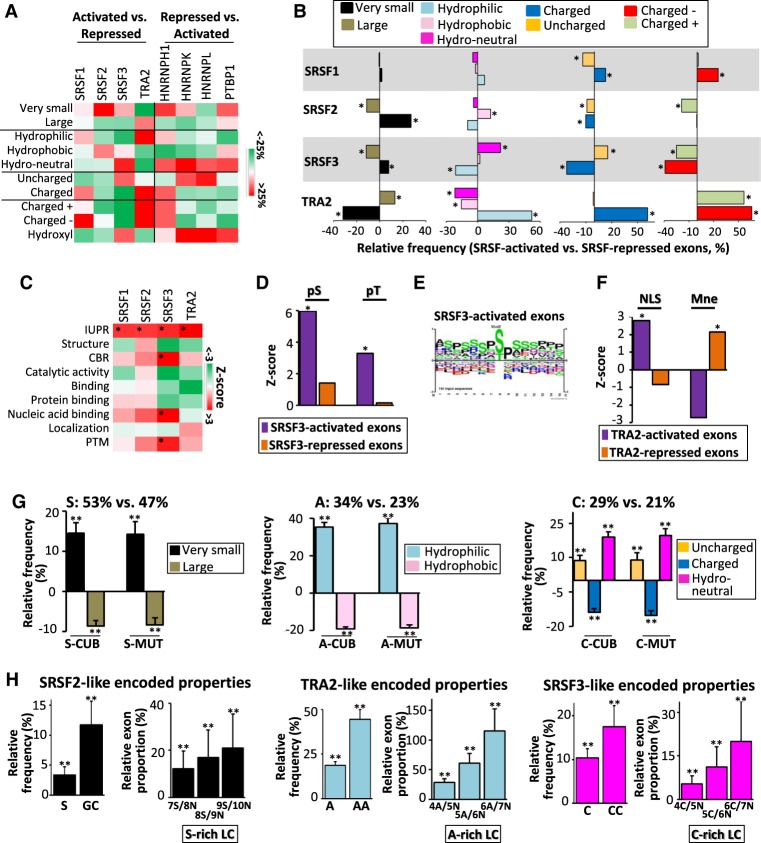
The physicochemical properties of amino acids are often related. For example, hydrophilic amino acids are often charged amino acids. In addition, we observed that exons regulated by a given splicing factor often coded for amino acids that have different physicochemical properties depending on whether the exons are activated or repressed by this factor (Fig. 4). Consequently, each splicing factor induced a shift toward different combinations of protein physicochemical properties encoded by their regulated exons (Mann–Whitney U test FDR < 0.05) (Fig. 6A,B).
Figure 6.
Bidirectional interplay between splicing regulatory process and its functional outcome. (A) Color code corresponding to the relative frequency (%) of amino acid physicochemical properties as indicated, when comparing all the SRSF1-, SRSF2-, SRSF3-, and TRA2-activated exons to all the SRSF1-, SRSF2-, SRSF3-, and TRA2-repressed exons, respectively, or when comparing all the HNRNPH1-, HNRNPK-, HNRNPL-, and PTBP1-repressed exons to all the HNRNPH1-, HNRNPK-, HNRNPL-, and PTBP1-activated exons, respectively. The sets of exons used correspond to exons regulated by a given splicing factor in at least one cell line and being regulated in the same manner when regulated in multiple cell lines. (B) Relative frequency (%) of very small, large, hydrophilic, neutral, hydrophobic, charged, uncharged, negatively charged (charged −), and positively charged (charged +) amino acids when comparing all the SRSF1-, SRSF2-, SRSF3-, or TRA2-activated exons to all the SRSF1-, SRSF2-, SRSF3-, or TRA2-repressed exons, respectively. The sets of exons used correspond to exons regulated by a given splicing factor in at least one cell line and being regulated in the same manner when regulated in multiple cell lines: (*) Mann–Whitney U test FDR < 0.05. (C) Color code corresponding to the Z-score of annotated protein features encoded SRSF1-, SRSF2-, SRSF3-, and TRA2-activated exons compared to all human coding exons: (IUPR) intrinsically unstructured regions; (CBR) compositionally biased protein region; (PTM) post-translational modifications; (*) Mann–Whitney U test FDR < 0.05. (D) Z-score of experimentally validated phosphorylated serine (pS) and threonine (pT) encoded by SRSF3-activated and -repressed exons compared to all human coding exons: (*) Mann–Whitney U test FDR < 0.05. (E) Sequence logo generated from PhosphoSitePlus (Hornbeck et al. 2015) using sequences surrounding experimentally validated phosphorylated residues coded by SRSF3-activated exons. (F) Z-score of nuclear localization signal (NLS) and intramembrane peptides (Mne) terms encoded by TRA2-activated and -repressed exons compared to all human coding exons: (*) Mann–Whitney U test FDR < 0.05. (G, left) The relative frequency (%) of very small and large amino acids encoded by 100 sets of 300 generated-exonic sequences containing a high frequency (53%) of the S nucleotide (S-CUB and S-MUT) compared to 100 sets of 300 generated-exonic sequences containing a low S-nucleotide frequency (47%); (middle) the relative frequency (%) of hydrophilic and hydrophobic amino acids encoded by exonic sequences containing a high frequency (34%) of A nucleotide (A-CUB and A-MUT) compared to exonic sequences containing a low A-nucleotide frequency (23%); (right) the relative frequency (%) of polar uncharged, charged, and neutral (in terms of hydropathy) amino acids encoded by exonic sequences containing a high frequency (29%) of Cs (C-CUB and C-MUT) compared to exonic sequences containing a low C-nucleotide frequency (21%); (**) t-test P-value <1 × 10−14. (H, left) The relative frequency (%) of the S nucleotide and GC dinucleotide, as well as the relative proportion (%) of exons with S-rich low-complexity (LC) sequences of 100 sets of 300 mutated exons encoding for the same physicochemical properties as SRSF2-activated exons compared to 100 sets of 300 mutated exons encoding for the same physicochemical properties as SRSF2-repressed exons; (middle) the relative frequency (%) of the A nucleotide and AA dinucleotide, as well as the relative proportion (%) of exons with A-rich low-complexity sequences of mutated exons encoding for the same physicochemical properties as TRA2-activated exons compared to mutated exons encoding for the same physicochemical properties as TRA2-repressed exons: (right) the relative frequency (%) of the C nucleotide and CC dinucleotide, as well as the relative proportion (%) of exons with C-rich low-complexity (LC) sequences of mutated exons encoding for the same physicochemical properties as SRSF3-activated exons compared to mutated exons encoding for the same physicochemical properties as SRSF3-repressed exons; (**) t-test P-value <1 × 10−14.
Because protein features depend on amino acid physicochemical properties, we measured an enrichment Z-score of annotated protein features to determine whether each factor-specific set of exons encodes specific protein features using our recently developed Exon Ontology bioinformatics suite (Tranchevent et al. 2017). As shown in Figure 6C, all sets of SRSF-activated exons preferentially encoded intrinsically unstructured protein regions (IUPR; FDR < 0.05), in agreement with previous reports indicating that alternatively spliced exons often code for intrinsically disordered regions (Tranchevent et al. 2017). However, each set of SRSF-coregulated exons encoded specific sets of annotated protein features. For example, C-rich SRSF3-activated exons encoded peptides that are enriched for experimentally validated phospho-serine and -threonine (“PTM”; FDR < 0.05) (Fig. 6C,D). These phosphorylation sites arise in serine- and proline-rich regions (Fig. 6E). This observation is consistent with the fact that hydroxyl-containing amino acids and proline correspond to C-rich codons (Fig. 4H). Serine- and proline-rich regions have been shown to play a role in RNA-protein interactions that can be regulated by phosphorylation (Wang et al. 2006; Thapar 2015). In this setting, SRSF3-activated exons encoded annotated “Nucleic Acid Binding” activity (FDR < 0.05) (Fig. 6C).
Along the same line, A-rich TRA2-activated exons often encoded for nuclear localization signal (NLS) (Fig. 6F). This is consistent with the fact that they encode hydrophilic amino acids, in particular lysine, that correspond to A-rich codons (Figs. 3B,C, 4D–G), a major amino acid of classical NLS (Marfori et al. 2011). In contrast, A-poor TRA2-repressed exons code for intramembrane protein parts (Mne; FDR < 0.05) (Fig. 6F) that are intrinsically rich in hydrophobic amino acids. This is consistent with the fact that A-poor TRA2-repressed exons encode more frequently hydrophobic amino acids corresponding to A-poor codons (Fig. 4D–G). Collectively, these observations support a model in which a splicing factor–related nucleotide composition bias of exons (Figs. 1–3) affects the physicochemical properties of their encoded amino acids because of the nonrandomness of the genetic code (Fig. 4) with direct consequences on protein features encoded by splicing factor–regulated exons (Fig. 6A–F).
On one hand, splicing factors bind to sequences that have a biased nucleotide composition and on the other hand, amino acids with similar physicochemical properties are encoded by codons having the same nucleotide composition bias. Therefore, we hypothesized that increasing the exonic density of specific nucleotides as measured in splicing factor–regulated exons would increase the density of encoded amino acids sharing the same physicochemical properties, as observed in those exons. To challenge this possibility, we generated random exonic coding sequences enriched in specific nucleotide(s) by following the human codon usage bias (labeled CUB sequences) or by randomly mutating human coding exons (labeled MUT sequences). For example, we generated 100 sets of 300 coding exons containing either 53% or 47% of S nucleotides, as measured in SRSF2-activated and SRSF2-repressed exons, respectively. Increasing by ∼13% the density of S nucleotides in coding exons (S-CUB or S-MUT) increased (by ∼15%) the frequency of encoded very small amino acids, whereas it decreased (by ∼10%) the frequency of encoded large amino acids (“S = 53% vs. 47%”; t-test P-value < 1 × 10−14) (Fig. 6G), as observed when comparing SRSF2-activated and SRSF2-repressed exons (Fig. 4B). Increasing the density of A nucleotides in coding exons (A-CUB and A-MUT) from 23% to 34%, as measured in TRA2-repressed and TRA2-activated exons, respectively, increased (by ∼40%) the frequency of encoded hydrophilic amino acids and it decreased (by ∼20%) the frequency of encoded hydrophobic amino acids (“A = 34% vs. 23%”; t-test P-value < 1 × 10−14) (Fig. 6G), as observed when comparing TRA2-activated and TRA2-repressed exons (Fig. 4E). Finally, increasing the density of C nucleotides in coding exons (C-CUB and C-MUT) from 21% to 29%, as measured in SRSF3-repressed and SRSF3-activated exons, respectively, increased the frequency of encoded uncharged amino acids and neutral amino acids whereas it decreased the frequency of encoded charged amino acids (“C = 29% vs. 21%”; t-test P-value < 1 × 10−14) (Fig. 6G), as observed when comparing SRSF3-activated and SRSF3-repressed exons (Fig. 4I).
We next generated exons coding for different proportions of amino acids sharing the same physicochemical features by mutating randomly selected human coding exons. For example, we generated 100 sets of 300 mutated exons encoding different proportions of very small and large amino acids, using their respective frequency measured in SRSF2-activated or SRSF2-repressed exons (Methods). Mutated exons coding more frequently very small rather than large amino acids had a higher frequency of S nucleotides and GC dinucleotides and contained more frequently S-rich low-complexity sequences (“SRSF2-like encoded properties”; t-test P-value < 1 × 10−14) (Fig. 6H), as observed when comparing SRSF2-activated to SRSF2-repressed exons (Fig. 1). Mutated exons encoding more frequently hydrophilic amino acids had a higher frequency of A nucleotides and AA dinucleotides and contained more frequently A-rich low-complexity sequences (“TRA2-like encoded properties”; t-test P-value < 1 × 10−14) (Fig. 6H), as observed when comparing TRA2-activated to TRA2-repressed exons (Fig. 1). Finally, mutated exons encoding more frequently hydropathically neutral amino acids had a higher frequency of C nucleotides and CC dinucleotides and contained more frequently C-rich low-complexity sequences (“SRSF3-like encoded properties”; t-test P-value < 1 × 10−14) (Fig. 6H), as observed when comparing SRSF3-activated to SRSF3-repressed exons (Fig. 1).
Discussion
In this work, we uncover a direct link between the splicing regulatory process and its biological outcome relying on two straightforward principles: (1) Splicing factors bind to exonic sequences that have a nucleotide composition bias with consequences on the nucleotide composition of codons from coregulated exons; and (2) codons having the same nucleotide composition bias encode amino acids with similar physicochemical properties with consequences on protein features encoded by the coregulated exons.
Exons coregulated by a given splicing factor are enriched for specific low-complexity sequences—often composed of a repeated (di)nucleotide—that correspond to the RNA binding sites of the cognate factor (Figs. 1A–C, 5A,E). We showed that each set of exons whose inclusion (or exclusion) is enhanced by a given splicing factor is enriched for specific nucleotide(s) when compared to control exons or to exons repressed (or activated, respectively) by the same factor (Fig. 1A–C). To the best of our knowledge, this splicing-related exonic nucleotide composition bias has not been reported yet. However, it is in agreement with recent observations indicating that the interaction of a splicing factor with a binding motif depends on the sequence context and on the presence of clusters of related binding motifs (Zhang et al. 2013; Cereda et al. 2014; Fu and Ares 2014; Dominguez et al. 2018; Jobbins et al. 2018). For example, increasing the exonic frequency of GGA-like motifs increases the probability of an exon to be regulated by the SRSF1 splicing factor that binds to GGAGGA-like motifs although only one binding site is used (Jobbins et al. 2018). In this setting, we observed similar nucleotide and amino acid composition biases when analyzing exons regulated by a splicing factor or exons containing CLIP-related signals for the same splicing factor (Supplemental Fig. S6).
Coding sequences overlap several kinds of regulatory sequences, including exonic splicing regulatory sequences. To date, it has been assumed that the redundancy of the genetic code permits protein-coding regions to carry this extra information (Goren et al. 2006; Itzkovitz and Alon 2007; Itzkovitz et al. 2010; Lin et al. 2011; Shabalina et al. 2013; Savisaar and Hurst 2017a,b). This means that the sequence constraints imposed by splicing factor binding motifs would accommodate with coding sequences by impacting only the third codon position. In this setting, we observed that nucleotide composition bias of splicing factor–regulated exons impacts not only the third codon position but also the first and second positions (Figs. 2B, 5B). Because amino acids having the same physicochemical properties correspond to codons with similar nucleotide composition bias, a direct consequence of the exonic nucleotide composition bias associated with the splicing regulatory process is that each set of splicing factor–regulated exons preferentially encodes amino acids having similar physicochemical properties (Figs. 4, 5). In addition, because specific local protein features depend on amino acid physicochemical properties, splicing factor–coregulated exons encode specific sets of protein features (Fig. 6A–F).
Therefore, we propose that the interplay between coding and exonic splicing regulatory sequences that we report is based on straightforward principles related to both the nonrandomness of the genetic code and the preferential binding of splicing factors to low-complexity sequences. Owing to these properties, the high exonic density of a specific nucleotide related to splicing factor binding features increases the probability that an exon encodes amino acids with similar physicochemical properties (Fig. 6G). Conversely, the high density of amino acids corresponding to specific physicochemical properties increases the probability of generating exonic nucleotide composition bias and nucleotide low-complexity sequences (Fig. 6H).
A deeper understanding of the interplay between splicing regulatory sequences and coding information will require improving (1) the characterization of splicing factor binding sites, (2) the analysis of exon-encoded protein features, and (3) the identification of exons dependent on several splicing factors. Indeed, splicing factor binding sites are unlikely to be defined by their sole nucleotide composition bias. For example, it has been recently shown that RNA binding sites are within specific context and that some RNA binding sites correspond to spaced “bipartite” short linear motifs (Zhang et al. 2013; Cereda et al. 2014; Fu and Ares 2014; Dominguez et al. 2018; Jobbins et al. 2018). By dissecting each RNA binding site, their flanking nucleotide preferences, their clustering, and the space between them, it would be possible to better characterize the way they can affect codon and amino acid usage. In this setting, although we focused our investigation on general properties of amino acids, it will be interesting to look for a complex pattern of protein-related features. For example, the alternation of amino acids having specific features (e.g., alternation of hydrophilic and hydrophobic amino acids) may allow uncovering specific protein-related properties (e.g., alpha-helix made of periodic alternation of hydrophilic and hydrophobic amino acids). Finally, it will be important to identify exons that are simultaneously regulated by two different splicing factors and to characterize how the combination of different regulatory binding motifs impacts coding sequences. An interesting possibility is that the combinatory regulation of an exon by two splicing factors is associated with specific exonic encoded protein features.
Another challenge will be to link our observations with the known tissue-specific regulation of alternative splicing. Based on this work and previously published observations (Irimia et al. 2014; Tranchevent et al. 2017), it can be anticipated that tissue-specific coregulated exons encode similar protein-related features. In this setting, the function of splicing factors would be not only to regulate the production of individual specialized protein isoforms, but also to more globally control the intracellular content of specific protein regions having specific physicochemical properties. Each splicing factor (or combination of factors) would control a specific combination of exon-encoded protein physicochemical properties accordingly to its (or their) affinity for specific nucleotides. Our work unravels how a complex phenomenon (e.g., the splicing regulatory process and its biological consequences) can rely on straightforward principles.
Methods
RNA-seq data set analyses
Publicly available RNA-seq data sets generated from different human cell lines transfected with siRNAs or shRNAs targeting specific splicing factors or transfected with splicing factor expression vectors were recovered from NCBI Gene Expression Omnibus (GEO) (Supplemental Table S1). These RNA-seq data sets were analyzed using FARLINE, a computational program dedicated to analyze and quantify alternative splicing variations as previously reported (Benoit-Pilven et al. 2018). This pipeline is freely available (http://kissplice.prabi.fr/pipeline_ks_ farline). To determine a set of exons that is regulated by a given splicing factor, we measured the percent-spliced-in (PSI) that corresponds to the exon inclusion rate. Each exon with a PSI variation (deltaPSI) greater than 10% or lower than −10% and a P-value <0.05, when comparing each sample to its corresponding control, is considered to be regulated (Benoit-Pilven et al. 2018). FARLINE analyzes exons annotated from FASTERDB (http://fasterdb.ens-lyon.fr/faster/home.pl) that is based on hg19 annotation. A liftOver from hg19 to GRCh38 recovers the same sequence for 99.94% of the analyzed exons.
Frequency of hexanucleotides, dinucleotides, nucleotides, codons, amino acids, and amino acid physicochemical features in exon sets
Equation (1) was used to compute the frequencies of words (Dn) of size n within a set of exons SN = {y1, …, yN} such that yi is an exon i having a number Li of nucleotides as follows:
| (1) |
in which xi is the number of occurrences of Dn in exon i; and n is set to 6, 2, and 1 for hexanucleotides, dinucleotides, and nucleotides, respectively. For codons, amino acids, and amino acid physicochemical properties, n is set to 3. P = 51 is a penalty size used to decrease the border effects seen in small exons, and N is the number of exons in the set SN. For hexanucleotides and dinucleotides, the occurrences xi of Dn are overlapping, whereas they are contiguous for the others. In the particular case of amino acids and amino physiochemical properties, Dn represents a group of codons encoding the same amino acid or the same physiochemical properties, respectively. To compute the frequencies of a nucleotide D1 at a specific codon position for a set of exon SN , Equation (1) with n ∈{1, 2} is used with small variations. In that case, xi corresponds only to the number of occurrences of D1 at this given codon position and Li corresponds to the number of nucleotides at this codon position for the exon i. When coding phase is mandatory, incomplete codons at exon borders were deleted.
Position weight matrices of the 10 most enriched hexanucleotides (Supplemental Table S2) in SRSF1-, SRSF2-, SRFS3-, or TRA2-activated exons were created using the MEME-Suite (Bailey et al. 2015) website (http://meme-suite.org/index.html).
Very small (Ala, Gly, Ser, Cys), small (Ala, Asn, Asp, Cys, Gly, Pro, Ser, Thr), large (Arg, Ile, Leu, Lys, Met, Phe, Trp, Tyr), polar uncharged (Asn, Gln, Ser, Thr, Tyr), charged (Asp, Glu, Lys, Arg), hydroxyl-containing (Ser, Thr, Tyr), hydrophilic (Arg, Asn, Asp, Gln, Glu, Lys), hydro-neutral (Gly, His, Pro, Ser, Thr, Tyr), and hydrophobic (Ala, Cys, Ile, Leu, Met, Phe, Val) amino acids were classified as previously reported (Kyte and Doolittle 1982; Engelman et al. 1986; Pommié et al. 2004).
The hydrophobicity scale was calculated as defined by Kyte and Doolittle (1982) and Engelman et al. (1986). TRA2-activated or -repressed exons larger than or equal to 30 amino acids were selected to calculate the average of hydrophobicity using a sliding window of five amino acids with a step of one amino acid for the first 30 and last 30 amino acids. Mean and standard deviation of the hydrophobicity values corresponding to each exon set were then calculated for each window position.
Generation of sets of control exons and statistical analyses
To test whether a feature was enriched in a set SN of N exons, a randomization test was made by sampling, from FASTERDB (Mallinjoud et al. 2014), 10,000 sets of control exons, C = {C1, …, C10,000}, with Cl = { yl,1, …, yl,i} such that yl,i is the exon i having a number of Ll,i nucleotides following the constraints:
in which 50 and 300 nt correspond approximately to the 4th and 96th percentile of exon length distribution.
The relative frequency of a feature Dn in SN compared to the sets of control exons C was calculated by the following formula:
in which Freqobs(Dn) is the frequency, as in Equation (1), of a word Dn of size n in SN and
is the average frequency, as in Equation (1), of Dn in C.
To calculate an empirical P-value, the number of control frequencies upper or lower than the frequency in the set of interest is determined. Then, the smaller number between those two is kept and divided by the number of control sets (i.e., 10,000).
All P-values obtained for each set of features have been corrected using the Benjamini–Hochberg procedure (Benjamini and Hochberg 1995). The nucleotide composition of enriched codons or codons corresponding to enriched amino acids was calculated after recovering codons or amino acids whose frequency was 10% higher in the set of exons of interest than their average frequency in sets of control exons.
Low-complexity and random sequences
Low-complexity sequences were defined as sequences of n (n = 5–10) nucleotides containing at least n−1 occurrences of the same nucleotide.
Random exonic sequences (from 50- to 300-nt long) with specific nucleotide composition bias were generated using two strategies. First, random codon sequences respecting the human codon usage bias (CUB exons) were generated. These sequences were then mutated randomly, one nucleotide at a time, to increase or decrease the frequency of a specific nucleotide. Only mutations increasing or decreasing the frequency toward Freqtarget (D1) were kept. Freqtarget (D1) is computed using the following formula:
in which Freqobs(D1) is the nucleotide frequency observed in a specific set of activated or repressed exons by a given splicing factor; Freqcub(D1) is the nucleotide frequency observed in CUB exons before the mutation procedure; y is a random value sampled from , and Lcub corresponds to the number of nucleotides in the CUB exon. The mutation procedure was stopped when
n which Freqmcub(D1) is the nucleotide frequency in CUB exons after the mutation procedure. Second, exonic sequences (MUT exons), selected by sampling human coding exons, were mutated using the same principle used for CUB sequences. In each case, 100 sets of 300 exonic sequences with specific features were generated. A t-test was performed to compare the average frequency of amino acid physicochemical properties between the generated sets.
Exonic sequences encoding for specific amino acid physicochemical properties were generated from sampled human coding exons (MUT exons). The mutation procedure used was similar to the one we applied at the nucleotide level, except that Lcub corresponds in this case to the codon length of the sampled sequences. These sequences were modified by codon substitution to increase the frequency of amino acids encoding for a given physicochemical property P1 and to decrease the frequency of another given physicochemical property P2. Codons that encode P2 were substituted toward codons encoding P1 following the human codon usage bias. SRSF2-like encoded properties were generated using the frequency of very small (0.27) and large (0.34) amino acids measured in SRSF2-activated exons or the frequency of very small (0.21) and large (0.38) amino acids measured in SRSF2-repressed exons. TRA2-like encoded properties were generated using the frequency of hydrophilic (0.4) and hydrophobic (0.33) amino acids measured in TRA2-activated exons or the frequency of hydrophilic (0.26) and hydrophobic (0.39) amino acids measured in TRA2-repressed exons. SRSF3-like encoded properties were generated using the frequency of hydro-neutral (0.38) and charged (0.17) amino acids measured in SRSF3-activated exons or the frequency of hydro-neutral (0.31) and charged (0.22) amino acids measured in SRSF3-repressed exons. The same procedure was used to compare the frequencies of nucleotides or dinucleotides with a t-test.
Density charts
For each exon, the frequency of each nucleotide or each amino acid physicochemical property was calculated and the exonic sequences were parsed using a sliding window (of size 1 and step 1). Truncated codons (at 3′ or 5′ exon extremities) or codons downstream from stop codons were ignored. Frequency histograms were then computed with the R software (R Core Team 2018). Density charts were made using sets of exons regulated by a given splicing factor in at least one cell line and being regulated in the same manner when regulated in multiple cell lines.
Statistical analysis
The Kolmogorov–Smirnov nonparametric test was performed using the R software (command ks.test) (R Core Team 2018) to compare the distributions of nucleotide or amino acid frequency into two data sets (e.g., activated vs. repressed exons). A logistic regression analysis was performed to test if activated or repressed exons by a given splicing factor have a different content in terms of low-complexity sequences (Fig. 1) or in codons encoding particular amino acids (Fig. 3). We modeled the presence or the absence of a given amino acid or the presence or absence of low-complexity sequences according to the cell line and the regulation of the exon (i.e., activated or repressed) using the glm function, with family = binomial (“logit”) in R software. To test the effect of the regulation status of the exon, we used a likelihood ratio test of this model against the null model without this effect (R software, function anova with test = “Chisq”).
Supplementary Material
Acknowledgments
We gratefully acknowledge the support from the Pôle Scientifique de Modélisation Numérique of the ENS de Lyon for the computing resources. We thank the members of the LBMC biocomputing center for their involvement in this project. This work was funded by Fondation ARC (PGA120140200853), Institut National Du Cancer (2014-154), Agence Nationale de la Recherche (CHROTOPAS), Association Française contre les Myopathies–Téléthon, and the Ligue nationale contre le cancer (LNCC). J.-B.C. was supported by Fondation de France.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.241315.118.
References
- Anczuków O, Akerman M, Cléry A, Wu J, Shen C, Shirole NH, Raimer A, Sun S, Jensen MA, Hua Y, et al. 2015. SRSF1-regulated alternative splicing in breast cancer. Mol Cell 60: 105–117. 10.1016/j.molcel.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Änkö ML, Müller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. 2012. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol 13: R17 10.1186/gb-2012-13-3-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Johnson J, Grant CE, Noble WS. 2015. The MEME Suite. Nucleic Acids Res 43: W39–W49. 10.1093/nar/gkv416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300. 10.2307/2346101 [DOI] [Google Scholar]
- Benoit-Pilven C, Marchet C, Chautard E, Lima L, Lambert MP, Sacomoto G, Rey A, Cologne A, Terrone S, Dulaurier L, et al. 2018. Complementarity of assembly-first and mapping-first approaches for alternative splicing annotation and differential analysis from RNAseq data. Sci Rep 8: 4307 10.1038/s41598-018-21770-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best A, Dalgliesh C, Kheirollahi-Kouhestani M, Danilenko M, Ehrmann I, Tyson-Capper A, Elliott DJ. 2014. Tra2 protein biology and mechanisms of splicing control. Biochem Soc Trans 42: 1152–1158. 10.1042/BST20140075 [DOI] [PubMed] [Google Scholar]
- Biro JC, Benyó B, Sansom C, Szlávecz A, Fördös G, Micsik T, Benyó Z. 2003. A common periodic table of codons and amino acids. Biochem Biophys Res Commun 306: 408–415. 10.1016/S0006-291X(03)00974-4 [DOI] [PubMed] [Google Scholar]
- Cereda M, Pozzoli U, Rot G, Juvan P, Schweitzer A, Clark T, Ule J. 2014. RNAmotifs: prediction of multivalent RNA motifs that control alternative splicing. Genome Biol 15: R20 10.1186/gb-2014-15-1-r20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiusano ML, Alvarez-Valin F, Di Giulio M, D'Onofrio G, Ammirato G, Colonna G, Bernardi G. 2000. Second codon positions of genes and the secondary structures of proteins. Relationships and implications for the origin of the genetic code. Gene 261: 63–69. 10.1016/S0378-1119(00)00521-7 [DOI] [PubMed] [Google Scholar]
- Dominguez D, Freese P, Alexis MS, Su A, Hochman M, Palden T, Bazile C, Lambert NJ, Van Nostrand EL, Pratt GA, et al. 2018. Sequence, structure, and context preferences of human RNA binding proteins. Mol Cell 70: 854–867.e9. 10.1016/j.molcel.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman DM, Steitz TA, Goldman A. 1986. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem 15: 321–353. 10.1146/annurev.bb.15.060186.001541 [DOI] [PubMed] [Google Scholar]
- Erkelenz S, Mueller WF, Evans MS, Busch A, Schoneweis K, Hertel KJ, Schaal H. 2013. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA 19: 96–102. 10.1261/rna.037044.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Ares M Jr. 2014. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 15: 689–701. 10.1038/nrg3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuens T, Bouhy D, Timmerman V. 2016. The hnRNP family: insights into their role in health and disease. Hum Genet 135: 851–867. 10.1007/s00439-016-1683-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice G, Sánchez-Cabo F, Torroja C, Lara-Pezzi E. 2016. ATtRACT—a database of RNA-binding proteins and associated motifs. Database (Oxford) 2016: baw035 10.1093/database/baw035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. 2006. Comparative analysis identifies exonic splicing regulatory sequences—the complex definition of enhancers and silencers. Mol Cell 22: 769–781. 10.1016/j.molcel.2006.05.008 [DOI] [PubMed] [Google Scholar]
- Grellscheid SN, Dalgliesh C, Rozanska A, Grellscheid D, Bourgeois CF, Stévenin J, Elliott DJ. 2011. Molecular design of a splicing switch responsive to the RNA binding protein Tra2β. Nucleic Acids Res 39: 8092–8104. 10.1093/nar/gkr495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauer C, Curk T, Anders S, Schwarzl T, Alleaume AM, Sieber J, Hollerer I, Bhuvanagiri M, Huber W, Hentze MW, et al. 2015. Improved binding site assignment by high-resolution mapping of RNA–protein interactions using iCLIP. Nat Commun 6: 7921 10.1038/ncomms8921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. 2015. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 43: D512–D520. 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M, Quesnel-Vallières M, Tapial J, Raj B, O'Hanlon D, et al. 2014. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 159: 1511–1523. 10.1016/j.cell.2014.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkovitz S, Alon U. 2007. The genetic code is nearly optimal for allowing additional information within protein-coding sequences. Genome Res 17: 405–412. 10.1101/gr.5987307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkovitz S, Hodis E, Segal E. 2010. Overlapping codes within protein-coding sequences. Genome Res 20: 1582–1589. 10.1101/gr.105072.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobbins AM, Reichenbach LF, Lucas CM, Hudson AJ, Burley GA, Eperon IC. 2018. The mechanisms of a mammalian splicing enhancer. Nucleic Acids Res 46: 2145–2158. 10.1093/nar/gky056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7: 1009–1015. 10.1038/nmeth.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimek-Tomczak K, Wyrwicz LS, Jain S, Bomsztyk K, Ostrowski J. 2004. Characterization of hnRNP K protein–RNA interactions. J Mol Biol 342: 1131–1141. 10.1016/j.jmb.2004.07.099 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132. 10.1016/0022-2836(82)90515-0 [DOI] [PubMed] [Google Scholar]
- Lin MF, Kheradpour P, Washietl S, Parker BJ, Pedersen JS, Kellis M. 2011. Locating protein-coding sequences under selection for additional, overlapping functions in 29 mammalian genomes. Genome Res 21: 1916–1928. 10.1101/gr.108753.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, et al. 2010. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol 17: 1114–1123. 10.1038/nsmb.1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Cheng Y, Liu Y, Chen L, Liu L, Wei N, Xie Z, Wu W, Feng Y. 2017. SRSF2 regulates alternative splicing to drive hepatocellular carcinoma development. Cancer Res 77: 1168–1178. 10.1158/0008-5472.CAN-16-1919 [DOI] [PubMed] [Google Scholar]
- Mallinjoud P, Villemin JP, Mortada H, Polay Espinoza M, Desmet FO, Samaan S, Chautard E, Tranchevent LC, Auboeuf D. 2014. Endothelial, epithelial, and fibroblast cells exhibit specific splicing programs independently of their tissue of origin. Genome Res 24: 511–521. 10.1101/gr.162933.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfori M, Mynott A, Ellis JJ, Mehdi AM, Saunders NF, Curmi PM, Forwood JK, Bodén M, Kobe B. 2011. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim Biophys Acta 1813: 1562–1577. 10.1016/j.bbamcr.2010.10.013 [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415. 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, Huang J, Yeo GW, Ares M Jr, Fu XD. 2013. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell 50: 223–235. 10.1016/j.molcel.2013.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmley JL, Urrutia AO, Potrzebowski L, Kaessmann H, Hurst LD. 2007. Splicing and the evolution of proteins in mammals. PLoS Biol 5: e14 10.1371/journal.pbio.0050014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommié C, Levadoux S, Sabatier R, Lefranc G, Lefranc MP. 2004. IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. J Mol Recognit 17: 17–32. 10.1002/jmr.647 [DOI] [PubMed] [Google Scholar]
- Prilusky J, Bibi E. 2009. Studying membrane proteins through the eyes of the genetic code revealed a strong uracil bias in their coding mRNAs. Proc Natl Acad Sci 106: 6662–6666. 10.1073/pnas.0902029106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. 10.1038/nature12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossbach O, Hung LH, Khrameeva E, Schreiner S, König J, Curk T, Zupan B, Ule J, Gelfand MS, Bindereif A. 2014. Crosslinking-immunoprecipitation (iCLIP) analysis reveals global regulatory roles of hnRNP L. RNA Biol 11: 146–155. 10.4161/rna.27991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savisaar R, Hurst LD. 2017a. Both maintenance and avoidance of RNA-binding protein interactions constrain coding sequence evolution. Mol Biol Evol 34: 1110–1126. 10.1093/molbev/msx061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savisaar R, Hurst LD. 2017b. Estimating the prevalence of functional exonic splice regulatory information. Hum Genet 136: 1059–1078. 10.1007/s00439-017-1798-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savisaar R, Hurst LD. 2018. Exonic splice regulation imposes strong selection at synonymous sites. Genome Res 28: 1442–1454. 10.1101/gr.233999.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Spiridonov NA, Kashina A. 2013. Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res 41: 2073–2094. 10.1093/nar/gks1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithers B, Oates ME, Gough J. 2015. Splice junctions are constrained by protein disorder. Nucleic Acids Res 43: 4814–4822. 10.1093/nar/gkv407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor FJ, Coates D. 1989. The code within the codons. Biosystems 22: 177–187. 10.1016/0303-2647(89)90059-2 [DOI] [PubMed] [Google Scholar]
- Thapar R. 2015. Structural basis for regulation of RNA-binding proteins by phosphorylation. ACS Chem Biol 10: 652–666. 10.1021/cb500860x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranchevent LC, Aubé F, Dulaurier L, Benoit-Pilven C, Rey A, Poret A, Chautard E, Mortada H, Desmet FO, Chakrama FZ, et al. 2017. Identification of protein features encoded by alternative exons using Exon Ontology. Genome Res 27: 1087–1097. 10.1101/gr.212696.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Someya T, Kuwasako K, Takahashi M, He F, Unzai S, Inoue M, Harada T, Watanabe S, Terada T, et al. 2011. Structural basis for the dual RNA-recognition modes of human Tra2-β RRM. Nucleic Acids Res 39: 1538–1553. 10.1093/nar/gkq854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XS, Wang DL, Zhao J, Qu MH, Zhou XH, He HJ, He RQ. 2006. The proline-rich domain and the microtubule binding domain of protein τ acting as RNA binding domains. Protein Pept Lett 13: 679–685. 10.2174/092986606777790566 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke T, Parmley JL, Hurst LD. 2008. Finding exonic islands in a sea of non-coding sequence: splicing related constraints on protein composition and evolution are common in intron-rich genomes. Genome Biol 9: R29 10.1186/gb-2008-9-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR. 1965. Order in the genetic code. Proc Natl Acad Sci 54: 71–75. 10.1073/pnas.54.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden RV, Cullis PM, Southgate CC. 1979. Water, protein folding, and the genetic code. Science 206: 575–577. 10.1126/science.493962 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yu J. 2011. On the organizational dynamics of the genetic code. Genomics Proteomics Bioinformatics 9: 21–29. 10.1016/S1672-0229(11)60004-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Lee KY, Swanson MS, Darnell RB. 2013. Prediction of clustered RNA-binding protein motif sites in the mammalian genome. Nucleic Acids Res 41: 6793–6807. 10.1093/nar/gkt421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.