Abstract
Early-life exposures are believed to influence the incidence of pediatric acute lymphoblastic leukemia (ALL). Archived neonatal blood spots (NBS), collected within the first days of life, offer a means to investigate small molecules that reflect early-life exposures. Using untargeted metabolomics, we compared abundances of small-molecule features in extracts of NBS punches from 332 children that later developed ALL and 324 healthy controls. Subjects were stratified by early (1–5 y) and late (6–14 y) diagnosis. Mutually-exclusive sets of metabolic features - representing putative lipids and fatty acids -were associated with ALL, including 9 and 19 metabolites in the early- and late-diagnosis groups, respectively. In the late-diagnosis group, a prominent cluster of features with apparent 18:2 fatty-acid chains suggested that newborn exposure to the essential nutrient, linoleic acid, increased ALL risk. Interestingly, abundances of these putative 18:2 lipids were greater in infants who were fed formula rather than breast milk (colostrum) and increased with the mother’s pre-pregnancy body mass index. These results suggest possible etiologic roles of newborn nutrition in late-diagnosis ALL.
Keywords: lipids, breastfeeding, maternal BMI, pre-B ALL, t(12;21) translocation
1. Introduction
Acute lymphoblastic leukemia (ALL) is the most common form of childhood cancer in the U.S. and other developed countries[1,2] with national incidence rates between <20 and 60 cases per million children per year[3]. In a recent review of the etiology of childhood ALL, Greaves[4] summarized compelling evidence that the most common subtype of this disease, B cell precursor ALL (BCP-ALL), is caused by two distinct events. First, a pre-leukemic clone is initiated in utero by fusion-gene formation in approximately 1% of newborns. Then, about 1% of those children with pre-leukemic clones progress to overt leukemia, with a peak of incidence occurring at two to five years of age. Greaves concluded that the latency period (extending to ~15 years of age) for transition to BCP-ALL pointed to secondary genetic changes, notably those caused by early-life exposures to commensal microbes, infections and diet. Therefore, objective measures of early-life exposures and their biological imprints could point the way to discovering causes of ALL, at least in children diagnosed after one year of age[4,5].
Because microbiota, infections, diet and other potential environmental risk factors generate molecules that circulate in blood, a promising avenue for discovering causes of ALL involves comparisons of metabolomes between ALL cases and controls in prediagnostic blood[6]. Archived neonatal blood spots (NBS, also known as Guthrie cards), which are collected from virtually all live births in the U.S. to test for metabolic defects, offer avenues for detecting exposures that occur near birth. Since 1982 the State of California has archived unused NBS at -20°C for epidemiological investigations[7].
We recently developed an untargeted metabolomics method with NBS via liquid chromatography high-resolution mass spectrometry (LC-HRMS)[8]. Here we report results from analyses of 656 archived NBS from ALL cases, diagnosed after one year of age, and matched controls who participated in the California Childhood Leukemia Study (CCLS)[9]. Because the age at diagnosis has been shown to affect the strength of associations with several ALL risk factors[10–12], we stratified case-control pairs by early diagnosis (1–5 years) coinciding with peak incidence, and late diagnosis (6–14 years)[4,13]. Data were filtered and normalized, focusing on 869 abundant features that were detected in most NBS.
2. Materials and Methods
2.1. Neonatal blood spots.
The CCLS is a case–control study conducted between 1995 and 2015 to identify risk factors for childhood leukemia. Incident cases of childhood leukemia, diagnosed up to 14 years of age, were ascertained across California, generally within 72 hours of diagnosis[9]. Archived NBS for CCLS participants were obtained from the California biobank program (Sacramento, CA). We used 4.7-mm punches (equivalent to ~8 μL of whole blood) from 656 NBS collected between 1985 and 2005, with ALL cases and controls matched by date of birth, sex, ethnicity (one or both parents being Hispanic) and maternal race. Information on socio-demographic characteristics was obtained from parent interviews. Summary statistics are available in Supplementary Table 1, available as Supplementary Materials. An additional set of blank punches was obtained from adjacent portions of the same Guthrie cards.
2.2. Metabolomic analysis.
A total of 656 NBS punches were analyzed (Supplementary Table 1). Briefly, samples were extracted with water and assayed for potassium[8] (batch 1) or hemoglobin[14] (batches 2–4) to adjust for blood volume (see Supplementary Figure 1). Then, acetonitrile was added to precipitate proteins and extracts were analyzed by LC-HRMS[8]. Data processing was performed in the R statistical programming environment using methods described elsewhere[15]. Detection of sample outliers, beyond a proportional expansion value of 1.2 for Hotelling’s ellipse (PC1 and PC2), resulted in removal of two cases and 10 controls. Of the 61,945 features detected in NBS and blank punches, filtering features by blank samples left 25,261, excluding features with more than 20% missing values left 1,606, and excluding features with intraclass correlation coefficients less than 0.2 left 869 features to be examined for their associations with ALL. Missing values were imputed abundances based on k-nearest neighbor imputation using k=5. Features were annotated by comparing masses, isotope patterns, and MSMS fragmentation spectra[16,17], and confirmed (when possible) with authentic standards. Annotation confidence was evaluated with criteria reported by Schymanski et al. [18] (Table 1).
Table 1.
Annotation of metabolites associated with early diagnosis and late diagnosis of ALL.
| Accurate mass (m/z)a | Nameb | CLc | Chemical class | Fold changed | p-valuee | Clusterf |
|---|---|---|---|---|---|---|
| Early diagnosis (1–5 years) | ||||||
| 363.3263 | Tetracosadienoic acid | 4 | Unsaturated fatty acid | 0.94 | 0.0027 | 1 |
| 431.3151 | Unknown | 5 | 1.12 | 0.0063 | 2 | |
| 570.3401 | Unknown | 5 | 1.09 | 0.0072 | 2 | |
| 604.3610 | LysoPC(20:3) | 4 | Glycerophospholipid | 1.07 | 0.0489 | 2 |
| 638.2760 | Unknown | 5 | 1.08 | 0.0034 | 2 | |
| 696.3573 | Unknown | 5 | 1.07 | 0.0177 | 2 | |
| 766.5589 | PS(16:0/16:0) | 4 | Glycerophospholipid | 1.08 | 0.0037 | 2 |
| 775.2985 | Unknown | 5 | 1.09 | 0.0164 | 2 | |
| 884.6004 | PC(18:0/20:4 (OH)) | 3 | Oxidized glycerophospholipid | 1.09 | 0.0210 | 2 |
| Late diagnosis (6–14 years) | ||||||
| 251.2011 | Hexadecadienoic acid | 4 | Unsaturated fatty acid | 0.89 | 0.0097 | 5 |
| 277.2170 | Linolenic acidg | 1 | Unsaturated fatty acid | 1.22 | 0.0016 | 3 |
| 279.2329 | Linoleic acid | 1 | Unsaturated fatty acid | 1.14 | 0.0092 | 3 |
| 377.1419 | Unknown | 5 | 1.20 | 0.0102 | 3 | |
| 387.3264 | C26H44O2 | 4 | 1.07 | 0.0548 | 4 | |
| 449.3629 | Unknown | 5 | 0.91 | 0.0530 | 5 | |
| 476.2778 | LysoPE(18:2) | 2 | Glycerophospholipid | 1.18 | 0.0032 | 3 |
| 500.2782 | LysoPE(20:4) | 2 | Glycerophospholipid | 1.07 | 0.0222 | 4 |
| 530.3234 | Unknown PE | 5 | Glycerophospholipid | 1.14 | 0.0161 | 4 |
| 552.2956 | Unknown | 5 | 0.91 | 0.0441 | 5 | |
| 564.5344 | C36H71NO3 | 4 | Sphingolipid | 0.90 | 0.0390 | 5 |
| 578.3458 | LysoPC (18:2) | 2 | Glycerophospholipid | 1.22 | 0.0064 | 3 |
| 592.3238 | Unknown | 5 | 0.91 | 0.0391 | 5 | |
| 760.5126 | PS(16:0/18:1) | 3 | Glycerophospholipid | 1.08 | 0.0387 | 4 |
| 789.6123 | SM(d16:1/20:0) | 3 | Sphingolipid | 0.95 | 0.0497 | 5 |
| 824.5793 | PC(P16:0/20:4) | 4 | Glycerophospholipid | 1.07 | 0.0519 | 4 |
| 844.6069 | PC(18:0/18:2) | 3 | Glycerophospholipid | 1.10 | 0.0154 | 3 |
| 897.6298 | Unknown | 5 | 0.93 | 0.0381 | 5 | |
| 965.7626 | C64H102O6 | 4 | 1.16 | 0.0464 | 3 | |
Feature identifier given by accurate mass (m/z).
Common names as used in the Human Metabolome Database (HMDB): lysophosphatidylcholine, LysoPC; phosphaditylcholine, PC; lysophosphatidylethanolamine, LysoPE; phosphatidylserine, PS; sphingomyelin, SM.
Confidence level of annotation consistent with the scheme of Schymanski et al [18] (1=highest, 5=lowest confidence).
Case/control fold change of feature abundances.
Nominal p-value for the case-control coefficient (β1) from Model 1.
Correlation cluster (Figure 1)
It was not possible to distinguish between alpha- and gamma- isomers.
2.3. Feature selection.
Feature selection was performed separately for early- and late-diagnosis of ALL. Peak areas were log transformed and normalized with the Bioconductor R package ‘scone’ [15,19,20], which implemented and evaluated different scaling and regression-based-normalization methods for removing unwanted variation while preserving differences in case status. The normalization scheme selected by ‘scone’ used DESeq scaling and accounted for the following unwanted sources of variation: NBS age, blood volume, run order, and batch.
To capture different types of associations between metabolites and ALL, features were selected using a combination of methods based on multivariate linear regression, regularized logistic regression and random forest[21]. First, the following linear regression model was used for a given logged and scaled feature Y in the ith subject:
| [1] |
where: caco denotes the binary case-control status, NBS age, sex, and ethnicity are matching variables, blood volume represents the volume of blood in the NBS punch, and run order and batch adjust for technical variation. Features were ranked by their nominal unadjusted p-values for the case-control coefficient (β1), and the case/control fold change was estimated as exp(β1). Second, a regularized logistic regression (lasso)[22,23] model was fit to normalized feature abundances over 500 bootstrapped datasets, with caco as the outcome variable and the following independent variables: normalized logged intensities for all 869 features and matching variables (NBS age, sex and ethnicity). The percentage of time that each feature was selected by lasso across the bootstrap iterations was used to rank the association with ALL. Features that were ranked in the top 5% for both linear regression p-value and lasso were joined with those of high importance from random forest (separated by more than a 20% increase in importance) to include possible non-linear associations with ALL[24–26]. After removing selected features with poor peak morphology, ion suppression, as well as minor isotopes and electrospray adducts, the ensemble of variable selection methods was repeated iteratively until a final set of 28 features was selected, 9 for early diagnosis and 19 for late diagnosis. Correlations between normalized abundances of the selected features were displayed with agglomerative hierarchical clustering using complete linkage and Spearman correlation (‘hclust’ function in R). Apparent clusters from hierarchical clustering were validated with the partitioning around medoids (PAM) method (k=2, …, 6 ‘pam’ function in R) also using Spearman correlations. Associations between selected features and covariates were visualized using scatter plots with loess smoothing.
2.4. Evaluation of covariates.
To investigate factors that could potentially confound relationships between predictive features and ALL status, the continuous covariates breastfeeding duration (weeks), birthweight (g) and mother’s BMI (kg/m2; prenatal) were evaluated because breastfeeding, maternal diet, gestational diabetes, and birthweight have been suggested as risk factors for ALL[27,28]. Confounding by household income (income, binary, with a cut point of $60,000) was also considered because the number of cases with low income (n=185) was higher than that of controls (n=126) (Supplementary Table 1). To investigate possible confounders and relative strengths of associations with ALL, a random forest model was used to rank all selected features and covariates by their importance in predicting ALL[29,30].
A case-case analysis of features predictive of t(12;21) was investigated in the early diagnosis group where 44 of the 214 cases (21%) had this cytogenetic abnormality (Supplementary Table 1). Additional stratification of the early or late diagnosis group was performed to investigate potential confounding by the pre-B cell phenotype [cases with t(12;21) translocations, hyperdiploidy and T-cell phenotypes].
3. Results
3.1. Features associated with ALL.
Table 1 lists all features, identified by accurate mass (m/z value), that were selected for associations with either early diagnosis (n=9) or late diagnosis (n=19) of ALL. Effect sizes were modest, with case/control fold changes ranging from 0.94 to 1.11 for early diagnosis and from 0.89 to 1.22 for late diagnosis. Based on annotations (Table 1 and Supplementary Table 2), features that discriminated ALL cases from controls were lipids and unsaturated fatty acids.
3.2. Correlations of features.
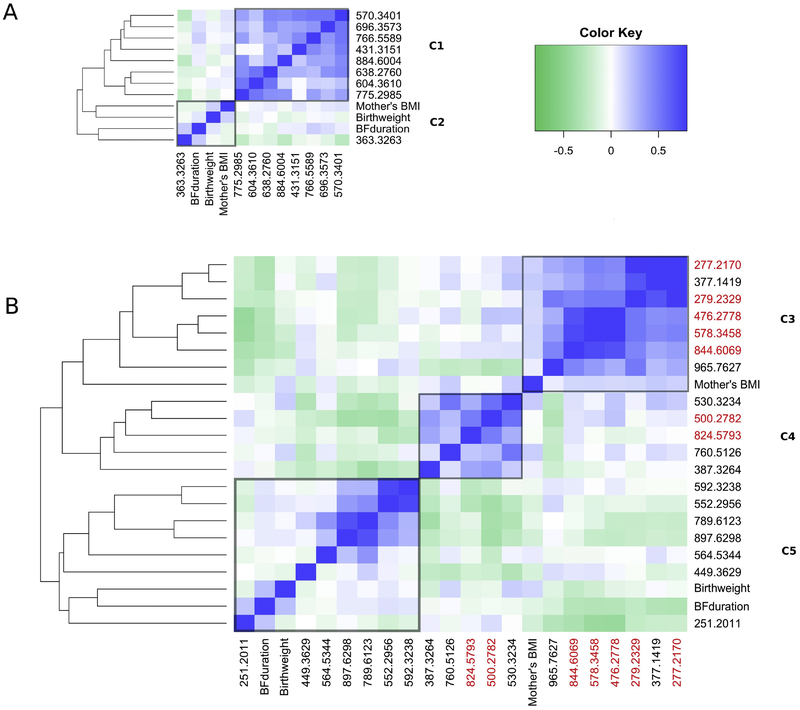
Heat maps were used to visualize clusters of features associated with ALL in the early- and late-diagnosis groups (Figures 1A and 1B, respectively). Results for PAM clustering were consistent (Supplementary Figure 2). For the early-diagnosis group two clusters were observed (C1 and C2 in Figure 1 and Table 1). Cluster C1 contained 8 features, all of which were more abundant in cases than controls (Table 1); three of these features were putatively annotated as glycerophospholipids (604.3610, 766.5589, and 884.6004). Only feature 363.3263 (putative tetracosadienoic acid) in cluster C2 was present at lower levels in cases.
Figure 1.
Agglomerative hierarchical clustering using complete linkage and Spearman correlation (‘hclust’ function in R). Clusters of features predictive of (A) early diagnosis and (B) late diagnosis of ALL, with distinct clusters labeled C1-C6. Metabolites containing 18:2, 18:3 or 20:4 fatty acid chains are highlighted in red.
There were three clusters in the late-diagnosis group (C3 – C5 in Figure 1B and Table 1). All features in clusters C3 and C4 were more abundant in cases while all of those in C5 were less abundant in cases. Annotations in clusters C3 and C4 included linolenic acid (277.2170), linoleic acid (279.7627) and several putative glycerophospholipids (476.2778, 500.2782, 530.3234, 578.3458, 760.5126, 824.5793 and 844.6069). Many putative metabolites in cluster C3 contained 18:2 fatty acid chains, including unequivocally identified linoleic acid. Thus, we speculate that unknown metabolites 377.1419 and 965.7627 may also contain 18:2 moieties. Cluster C4 contained two putative metabolites with arachidonic acid side chains (20:4). The putative annotations in cluster C5 included sphingolipids (789.6123 and 564.5344), and a metabolite of conjugated linoleic acid (hexadecadienoic acid, 251.2011).
3.3. Correlations with covariates.
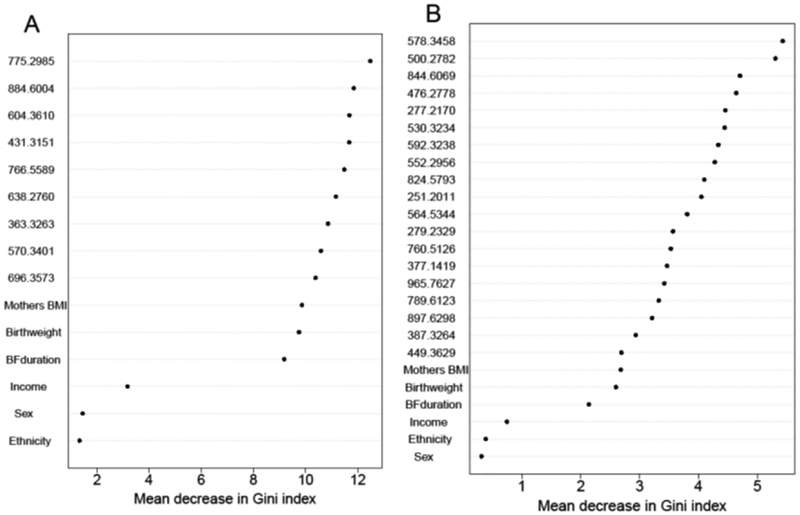
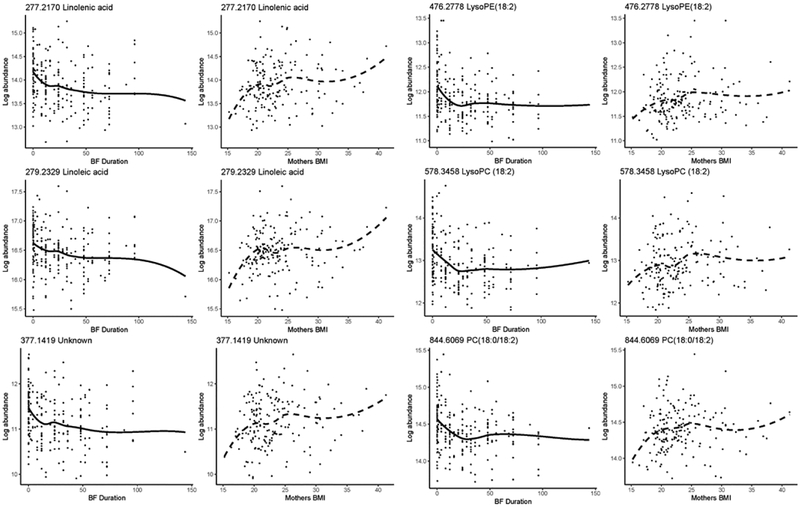
Random forest variable-importance plots, shown in Figure 2, indicate that mother’s BMI, birthweight and breastfeeding duration ranked more highly for classifying ALL cases than income for both early- and late-diagnosis, but were ranked lower than all metabolomic features. This suggests that predictive metabolites were more discriminating for case status than any of the tested covariates and that selection of the predictive metabolites was not unduly influenced by these covariates. Nonetheless, the heat maps in Figure 1 show that breastfeeding duration and mother’s BMI were consistently correlated with six features in cluster C3, including those with known or putative 18:2 fatty acid moieties. Interestingly, the directions of these correlations were reversed for all six features, i.e., negative correlations with breastfeeding duration and positive correlations with mother’s BMI. Scatter plots with loess smoothing further elucidated these relationships, as shown in Figure 3.
Figure 2.
Random Forest variable importance plots for (A) early diagnosis and (B) late diagnosis of ALL. BF duration, breastfeeding duration.
Figure 3.
Scatter plots with loess smoothing of feature abundances from cluster C3 that were correlated with breastfeeding (BF) duration (weeks) and mother’s pre-pregnancy body mass index (BMI, kg/m2).
3.4. Stratified analyses.
ALL cases at early-diagnosis had a higher percentage of t(12;21) translocations (46/205 = 21%) than those at late-diagnosis (6/106 = 5%) (Supplementary Table 1). To assess the associations between t(12;21) and the 9 features predictive of early diagnosis, the variable selection method was repeated by comparing cases with and without t(12;21). This resulted in 13 features predictive of t(12;21) (Supplementary Table 3). Only feature 604.3620 [LysoPC(20:3)] overlapped with those in Table 1 for the early-diagnosis group.
Removing cases with T-cell leukemia (19 in total, 8 from the early group and 11 from the late group) from the analysis did not change the results.
Removing cases with ‘early pre-B’ cell phenotypes [those with t(12;21) translocations, hyperdiploidy, and T-cell phenotypes] from the late-diagnosis group resulted in 63 cases with ‘other B-cell’ phenotypes (Supplementary Table 1). Comparing these to controls (n=117) slightly improved effect sizes as determined by Model 1 for 17 out of 19 metabolites predictive of late diagnosis (0.1–6.3%). This increase was not observed when evaluated in the early-diagnosis group.
4. Discussion
Because ALL risks had previously been shown to be affected by age at diagnosis[10–13], we stratified cases by early (1–5 years) and late diagnosis (6–14 years) and, indeed, discovered mutually-exclusive sets of predictive metabolomic features (Table 1). These 28 metabolites were mainly putative lipids (Table 1), some of which have been found to be perturbed in diagnostic blood for a number of malignancies [31,32] including childhood and adult acute leukemias [33,34]. Our study is unique in that the findings are based on pre-diagnostic blood collected at birth.
Almost all of the putative glycerophospholipids predictive of ALL, including PCs and LysoPCs, were more abundant in cases than controls (Table 1). This finding is consistent with reports that PCs and LysoPCs are overexpressed in some cancers [35,36] and can influence both cell proliferation and apoptosis[37,38]. Also, choline modulation has been shown to be a source of PCs and LysoPCs in diagnostic blood from AML cases compared to controls and other tumors[39]. At the cellular level, LysoPCs have been reported to be more abundant in CCRF-CEM leukemia cells after drug treatment due to increased oxidative stress[40]. Interestingly, a putative oxidized PC [PC(18:0/20:4(OH), 884.6004] that was more abundant in early ALL cases (Table 1) represents a class of biomarkers of oxidative stress[41,42].
Nine metabolites were predictors of early-ALL diagnosis and all but of one were more abundant in cases than controls (Table 1). The eight positively-associated features were correlated (cluster C1 of Figure 1A), and those with putative annotations were products of glycerophospholipid metabolism. The other feature, 363.3263 (putative tetracosadienoic acid, (cluster C2 of Figure 1A), was less abundant in cases than controls and was positively correlated with breastfeeding duration (Table S3).
In the late-diagnosis group, 19 metabolomic features discriminated ALL cases from controls, including a putative ceramide (564.5344) and sphingomyelin [SM(d16:1/20:0), 789.6123)], which were less abundant in cases (Table 1) and were correlated with each other (C5, Figure 1B). This could reflect the effects of acid ceramidase, which catalyzes ceramide breakdown and has been shown to be overexpressed in AML[43], and of altered sphingolipid metabolism that has been implicated with cancer progression[44].
We find it interesting that two essential fatty acids, linoleic acid (18:2n6, 279.2329) and linolenic acid (277.2169), were more abundant in late-diagnosis cases (Table 1), suggesting that maternal and/or newborn nutrition were involved in the early-life etiology of ALL. This conjecture is supported by correlations of putative features in cluster C3, including several with 18:2 fatty acid chains, with mother’s BMI and with breastfeeding duration. As shown by the scatter plots in Figure 3, abundances of the features in cluster C3 increased with mother’s BMI, reinforcing a previous finding that newborns’ levels of linoleic acid were positively correlated with the mothers’ BMI[45]. The negative correlations between breastfeeding duration and the same putative 18:2 features in cluster C3 (Figure 1), are informative because NBS were typically obtained between 24 and 48 hours post-delivery after infants had received multiple feedings of either breastmilk (colostrum) or formula. Since levels of linoleic and linolenic acid have been shown to be lower in colostrum than formula,[46–48] post-delivery breast feeding arguably led to reduced abundances of these 18:2 and 18:3 fatty acids in NBS and also served as sentinels of breastfeeding duration, which has been shown to reduce risks of ALL in the CCLS and other studies[12,28,49,50]. Conversely, breastfeeding duration was positively correlated with putative hexadecadienoic acid (16:2n6, 251.2011), a metabolite of conjugated linoleic acid that has been shown to be anti-carcinogenic [51,52] and is more abundant in colostrum than formula[46,53]. The gut microbiome, including Bifidobacterium and Lactobacillus, is involved in the conversion of linoleic acid to conjugated linoleic acid[54].
It is worth mentioning that Shu et al. [55] observed a lower odds ratio for the association of breastfeeding with ‘pre-B ALL’ than ‘early pre-B ALL’ phenotypes. Since ‘early pre-B ALL’ includes B cell cases with t(12;21) translocations and hyperdiploidy, we may be observing a similar effect because removal of t(12;21), hyperdiploidy, and T-cell ALL cases from the late diagnosis group led to modest increases in the effect sizes of the same metabolites. Scatter plots (Figure 3) also show that the relationships between feature abundances and breastfeeding duration occurred predominately in the first 26 weeks (< 6 months). Thus, the correlations between metabolites selected for ALL in the late-diagnosis group and breastfeeding duration may be driven by a specific phenotype consistent with the findings of Shu et al.
Because most of the predictive metabolites of late-diagnosis ALL were positively correlated (Figure 1B), they may represent a single underlying pathway or network. The predominant pathways associated with late diagnosis had case/control fold changes greater than one (Table 1) and were related to parent (linoleic and linolenic) fatty acids, while the corresponding conjugated linoleic acid metabolite (putative hexadecadienoic acid) had a fold change less than one. Higher fold changes were also observed for structural lipids, including putative PC, LysoPC and LysoPE species that contained linoleic (18:2n6) and arachidonic acid (20:4n6) side chains. Fatty acids, including linoleic acid (n-6) are converted by desaturases to long-chain PUFAs including arachidonic acid[56]. Increased arachidonic acid levels can result in increased eicosanoid production (e.g., prostaglandins), which can influence cancer progression, possibly through an immune response[57,58]. These findings suggest that early nutritional intake in the form of fatty acid consumption is associated with ALL and may involve the downstream biosynthetic machinery including desaturase/elongase enzymes and/or eicosanoid synthesis.
We recognize that this discovery study is limited to a single sample of ALL cases and matched controls and will require validation in independent cohorts. Annotations of the lipid features predictive of ALL in our study were limited by the LC-HRMS platform used for untargeted metabolomics, and by online databases that are particularly lacking in negative mode data, especially with acetic acid as an additive. Because of this, only linoleic acid and linolenic acid were unambiguously identified based on comparisons with reference standards. We encourage future studies to employ methods that can discriminate fatty acid isomers (e.g., gas chromatography–mass spectrometry).
In summary, fetal metabolomics of NBS revealed putative lipid modifications associated with childhood leukemia that differed between early and late diagnosis of ALL, notably lipids containing 18:2 moieties derived from dietary linoleic acid that were more abundant in late-diagnosis cases than controls. Interestingly, these same putative lipids were negatively correlated with breastfeeding duration, thus supporting epidemiological findings that breastfeeding is protective for ALL. This work should encourage efforts to elucidate systems biology that links lipidomic pathways with early-life nutrition and the associated ALL risks.
Supplementary Material
Highlights.
Metabolites from archived neonatal blood spots revealed distinct ALL phenotypes.
Putative phosphatidylcholines and sphingolipids were associated with pediatric ALL.
Associations of ALL with linoleic and linolenic acids suggest roles for early nutrition.
Acknowledgements.
We appreciate the generous loan of LC-HRMS equipment by Agilent Technologies (Santa Clara, CA). We thank the families for their participation. We also thank the clinical investigators at the following collaborating hospitals for help in recruiting patients: University of California Davis Medical Center (Dr. Jonathan Ducore), University of California San Francisco (Drs. Mignon Loh and Katherine Matthay), Children’s Hospital of Central California (Dr. Vonda Crouse), Lucile Packard Children’s Hospital (Dr. Gary Dahl), Children’s Hospital Oakland (Dr. James Feusner), Kaiser Permanente Roseville (former Sacramento) (Drs. Kent Jolly and Vincent Kiley), Kaiser Permanente Santa Clara (Drs. Carolyn Russo, Alan Wong and Denah Taggar), Kaiser Permanente San Francisco (Dr. Kenneth Leung) and Kaiser Permanente Oakland (Drs. Daniel Kronish and Stacy Month). We also thank the entire California Childhood Leukemia Study staff.
Funding. This work was supported by the National Institute for Environmental Health Sciences at the National Institutes of Health [P01 ES018172, P50 ES018172, P01 ES018172, U2C ES026561]; the United States Environmental Protection Agency [RD83615901, RD83451101]; Children with Cancer U.K. [partial support for cytogenetic characterization], as well as a post-doctoral fellowship by the Environment and Health Fund, Israel. C.E.W. was supported by the Swedish Heart-Lung Foundation [20170736, 20170603]. The biospecimens used in this study were obtained from the California Biobank Program, California Department of Public Health (SIS request number 26), in accordance with Section 6555(b), 17 CCR. The NIEHS, USEPA, and California Department of Public Health are not responsible for the results or conclusions drawn by the authors of this publication.
Ethics approval: The study was approved by the University of California Committee for the Protection of Human Subjects, the California Health and Human Services Agency Committee for the Protection of Human Subjects, and the institutional review boards of all participating hospitals.
Abbreviations:
- ALL
acute lymphoblastic leukemia
- BCP-ALL
B-cell precursor ALL
- Hb
hemoglobin
- FDR
false discovery rate
- LC-HRMS
liquid chromatography high-resolution mass spectrometry
- NBS
neonatal dried blood spots
- CCLS
California Childhood Leukemia Study
- lasso
least absolute shrinkage and selection operator
- LysoPC
lysophosphatidylcholine
- LysoPE
lysophosphatidylethanolamine
- MS2
tandem MS/MS fragmentation
- PC
phosphaditylcholine
- PS
phosphatidylserine
- PUFA
polyunsaturated fatty acid
- SM
sphingomyelin
- t(12;21) translocation
also known as ETV6–RUNX1 and TEL-AML1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: the authors declare no conflict of interest
Informed consent: Written informed consent was obtained from the parents of all participating subjects.
Appendix A. Supplementary material
Supplementary data are available online.
References
- [1].Cancer Facts & Figures 2014: Special Addition, (n.d.). http://www.cancer.org/acs/groups/content/@research/documents/webcontent/acspc-041787.pdf (accessed March 4, 2016).
- [2].Inaba H, Greaves M, Mullighan CG, Acute lymphoblastic leukaemia, Lancet. 381 (2013) 1943–1955. doi: 10.1016/S0140-6736(12)62187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].W.H.O. Europe, Incidence of Childhood Leukaemia, Eur. Environ. Heal. Inf. Syst 2000 (2009) 5. doi:Code: RPG4_Rad_E1. [Google Scholar]
- [4].Greaves M, A causal mechanism for childhood acute lymphoblastic leukaemia, Nat. Rev. Cancer (2018) 1–14. doi: 10.1038/s41568-018-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Brown P, Treatment of infant leukemias: challenge and promise., Hematology Am. Soc. Hematol. Educ. Program. 2013 (2013) 596–600. doi: 10.1182/asheducation-2013.1.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rappaport S, Redefining environmental exposure for disease etiology, NPJ Syst. Biol. Appl 4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].California Department of Public Health, Background and History of the California Biobank Program (CBP), (2016). https://www.cdph.ca.gov/programs/GDSP/Pages/MoreAboutTheCBP.aspx.
- [8].Petrick L, Edmands W, Schiffman C, Grigoryan H, Perttula K, Yano Y, Dudoit S, Whitehead T, Metayer C, Rappaport S, An untargeted metabolomics method for archived newborn dried blood spots in epidemiologic studies, Metabolomics. 13 (2017) 1–11. doi: 10.1007/s11306-016-1153-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Metayer C, Zhang L, Wiemels JL, Bartley K, Schiffman J, Ma X, Aldrich MC, Chang JS, Selvin S, Fu CH, Ducore J, Smith MT, Buffler P.a, Tobacco smoke exposure and the risk of childhood acute lymphoblastic and myeloid leukemias by cytogenetic subtype, Cancer Epidemiol Biomarkers Prev 22 (2013) 1600–1611. doi: 10.1158/1055-9965.EPI-13-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Paltiel O, Tikellis G, Linet M, Golding J, Lemeshow S, Phillips G, Lamb K, Stoltenberg C, Håberg SE, Strøm M, Granstrøm C, Northstone K, Klebanoff M, Ponsonby AL, Milne E, Pedersen M, Kogevinas M, Ha E, Dwyer T, Birthweight and childhood cancer: Preliminary findings from the international childhood cancer cohort consortium (I4C), Paediatr. Perinat. Epidemiol 29 (2015) 335–345. doi: 10.1111/ppe.12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wallace A, Francis W, Ma X, McKean-Cowdin R, Selvin S, Whitehead T, Barcellos L, Kang A, Morimoto L, Moore T, Wiemels J, Metayer C, Allergies and childhood acute lymphoblastic leukemia: A case-control study and meta-analysis, CEBP. (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rudant J, Lightfoot T, Urayama KY, Petridou E, Dockerty JD, Magnani C, Milne E, Spector LG, Ashton LJ, Dessypris N, Kang AY, Miller M, Rondelli R, Simpson J, Stiakaki E, Orsi L, Roman E, Metayer C, Infante-Rivard C, Clavel J, Childhood acute lymphoblastic leukemia and indicators of early immune stimulation: A childhood leukemia international consortium study, Am. J. Epidemiol 181 (2015) 549–562. doi: 10.1093/aje/kwu298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Westergaard T, Andersen PK, Pedersen JB, Olsen JH, Frisch M, Sorensen HT, Wohlfahrt J, Melbye M, Birth characteristics, sibling patterns, and acute leukemia risk in childhood: a population-based cohort study, J Natl Cancer Inst. 89 (1997) 939–947. doi: 10.1093/jnci/89.13.939. [DOI] [PubMed] [Google Scholar]
- [14].Yano Y, Grigoryan H, Schiffman C, Edmands WM, Petrick L, Hall K, Whitehead T, Metayer C, Rappaport S, Untargeted adductomics of Cys34 modifications to human serum albumin in newborn dried blood spots, Anal. Bioanal. Chem (n.d.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schiffman C, Petrick L, Perttula K, Yano Y, Carlsson H, Whitehead T, Metayer C, Hayes J, Edmands WMB, Rappaport S, Dudoit S, Data-adaptive pipeline for filtering and normalizing metabolomics data., BioRxiv. (2018) 387365. doi: 10.1101/387365. [DOI] [Google Scholar]
- [16].Brugger B, Erbent G, Sandhoff R, Wieland FT, W.. Lehmann, Quantitative analysis of biological membrane lipids at the low picome level by nano-electrospray ionization tandem mass spectrometry, Proc. Natl. Acad. Sci 94 (1997) 2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Edmands WMB, Petrick L, Barupal DK, Scalbert A, Wilson MJ, Wickli K, Rappaport SM, compMS2Miner: An Automatable Metabolite Identification, Visualization, and Data-Sharing R Package for High-Resolution LC − MS Data Sets, Anal. Chem 89 (2017) 3919–3928. doi: 10.1021/acs.analchem.6b02394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Schymanski EL, Jeon J, Gulde R, Fenner K, Ru M, Singer HP, Hollender J, Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence, Environ. Sci. Technol 48 (2014) 2097–2098. doi: 10.1021/es5002105 |. [DOI] [PubMed] [Google Scholar]
- [19].Cole MB, Risso D, Wagner A, DeTomaso D, Ngai J, Purdom E, Dudoit S, Yosef N, Performance Assessment and Selection of Normalization Procedures for Single-Cell RNA-Seq, BioRxiv. (2017) 235382. doi: 10.1101/235382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Risso D, Ngai J, Speed TP, Dudoit S, Normalization of RNA-seq data using factor analysis of control genes or samples, Nat. Biotechnol 32 (2014) 896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Perttula K, Schiffman C, Edmands WM, Petrick L, Grigoryan H, Cai X, Gunter MJ, Naccarati A, Polidoro S, Dudoit S, Vineis P, Rappaport S, Untargeted lipidomic features associated with colorectal cancer in a prospective cohort, BMC Cancer. (In press) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bach F, Bolasso: model consistent Lasso estimation through the bootstrap, ArXive. (2008). doi: 10.1145/1390156.1390161. [DOI] [Google Scholar]
- [23].Friedman J, Hastie T, Tibshirani R, Journal of Statistical Software, 33 (2010) 1–22. https://www.jstatsoft.org/index. [PMC free article] [PubMed] [Google Scholar]
- [24].Wang H, Yang F, Luo Z, An experimental study of the intrinsic stability of random forest variable importance measures, BMC Bioinformatics. 17 (2016) 1–18. doi: 10.1186/s12859-016-0900-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Calle ML, Urrea V, Letter to the editor: Stability of Random Forest importance measures, Brief. Bioinform 12 (2011) 86–89. doi: 10.1093/bib/bbq011. [DOI] [PubMed] [Google Scholar]
- [26].Breiman L, Random Forests, Mach. Learn 45 (2001) 5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- [27].Dessypris N, Karalexi MA, Ntouvelis E, Diamantaras AA, Papadakis V, Baka M, Hatzipantelis E, Kourti M, Moschovi M, Polychronopoulou S, Sidi V, Stiakaki E, Petridou ET, Association of maternal and index child’s diet with subsequent leukemia risk: A systematic review and meta analysis, Cancer Epidemiol 47 (2017) 64–75. doi: 10.1016/j.canep.2017.01.003. [DOI] [PubMed] [Google Scholar]
- [28].Amitay EL, Dubnov Raz G, Keinan-Boker L, Breastfeeding, Other Early Life Exposures and Childhood Leukemia and Lymphoma, Nutr. Cancer. 68 (2016) 968–977. doi: 10.1080/01635581.2016.1190020. [DOI] [PubMed] [Google Scholar]
- [29].Schneeweiss S, Eddings W, Glynn RJ, Patorno E, Rassen J, Franklin JM, Variable Selection for Confounding Adjustment in High-dimensional Covariate Spaces When Analyzing Healthcare Databases., Epidemiology. 28 (2017) 237–248. doi: 10.1097/EDE.0000000000000581. [DOI] [PubMed] [Google Scholar]
- [30].Lu M, Sadiq S, Feaster D, Ishwaran H, Estimating Individual Treatment Effect in Observational Data Using Random Forest Methods, J. Comput. Graph. Stat 17 (2018) 209–219. doi: 10.1016/j.chemosphere.2012.12.037.Reactivity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Armitage EG, Southam AD, Monitoring cancer prognosis, diagnosis and treatment efficacy using metabolomics and lipidomics, Metabolomics. 12 (2016) 1–15. doi: 10.1007/s11306-016-1093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bandu R, Mok J, Kim K, Phospholips as cancer biomarkers: mass spectrometry-based analysis, Mass Spectrom. Rev 37 (2018) 107–138. doi: 10.1002/mas. [DOI] [PubMed] [Google Scholar]
- [33].Bai Y, Zhang H, Sun X, Sun C, Ren L, Biomarker identification and pathway analysis by serum metabolomics of childhood acute lymphoblastic leukemia., Clin. Chim. Acta 436C (2014) 207–216. doi: 10.1016/j.cca.2014.05.022. [DOI] [PubMed] [Google Scholar]
- [34].Musharraf SG, Siddiqui AJ, Shamsi T, Naz A, Serum metabolomics of acute lymphoblastic leukaemia and acute myeloid leukaemia for probing biomarker molecules, Hematol. Oncol 35 (2017) 769–777. doi: 10.1002/hon.2313. [DOI] [PubMed] [Google Scholar]
- [35].Kim IC, Lee JH, Bang G, Choi SH, Kim YH, Kim KP, Kim HK, Ro J, Lipid profiles for HER2-positive breast cancer, Anticancer Res 33 (2013) 2467–2472. [PubMed] [Google Scholar]
- [36].Ishikawa S, Tateya I, Hayasaka T, Masaki N, Takizawa Y, Ohno S, Kojima T, Kitani Y, Kitamura M, Hirano S, Setou M, Ito J, Increased expression of phosphatidylcholine (16:0/18:1) and (16:0/18:2) in thyroid papillary cancer, PLoS One. 7 (2012). doi: 10.1371/journal.pone.0048873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ridgway ND, The role of phosphatidylcholine and choline metabolites to cell proliferation and survival, Crit. Rev. Biochem. Mol. Biol 48 (2013) 20–38. doi: 10.3109/10409238.2012.735643. [DOI] [PubMed] [Google Scholar]
- [38].Okamoto Y, Aoki A, Ueda K, Jinno H, Metabolomic analysis uncovered an association of serum phospholipid levels with estrogen-induced mammary tumors in female ACI/Seg rats, Toxicol. Lett 288 (2018) 65–70. doi: 10.1016/j.toxlet.2018.02.017. [DOI] [PubMed] [Google Scholar]
- [39].Wang Y, Zhang L, Chen W-L, Wang J-H, Li N, Li J-M, Mi J-Q, Zhang W-N, Li Y, Wu S-F, Jin J, Wang Y-G, Huang H, Chen Z, Chen S-J, Tang H, Rapid Diagnosis and Prognosis of de novo Acute Myeloid Leukemia by Serum Metabonomic Analysis, J. Proteome Res 12 (2013) 4393–4401. doi: 10.1021/pr400403p. [DOI] [PubMed] [Google Scholar]
- [40].Wang Y, Gao D, Chen Z, Li S, Gao C, Cao D, Liu F, Liu H, Jiang Y, Acridone Derivative 8a Induces Oxidative Stress-Mediated Apoptosis in CCRF-CEM Leukemia Cells: Application of Metabolomics in Mechanistic Studies of Antitumor Agents, PLoS One. 8 (2013). doi: 10.1371/journal.pone.0063572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Fruhwirth GO, Loidl A, Hermetter A, Oxidized phospholipids: From molecular properties to disease, Biochim. Biophys. Acta - Mol. Basis Dis 1772 (2007) 718–736. doi: 10.1016/j.bbadis.2007.04.009. [DOI] [PubMed] [Google Scholar]
- [42].Nakamura T, Hall L, Murphy RC, Oxidation of Arachidonate Containing Glycerophospholipids in Intact Red Blood Cells and Red Blood Cell Membranes with Tert-Butylhydroperoxide, in: Honn KV, Marnett LJ, Nigam S, Dennis EA (Eds.), Eicosanoids Other Bioact. Lipids Cancer, Inflammation, Radiat. Inj 4, Springer; US, Boston, MA, 1999: pp. 539–545. doi: 10.1007/978-1-4615-4793-8_79. [DOI] [PubMed] [Google Scholar]
- [43].Tan S, Liu X, Fox TE, Barth BM, Sharma A, Stephen D, Awwad A, Dewey A, Doi K, Spitzer B, Liao J, Yun J, Kester M, Claxton DF, Wang H, Acid ceramidase is upregulated in AML and represents a novel therapeutic target, Oncotarget. 7 (2016) 83208–83222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ryland LK, Fox TE, Liu X, Loughran TP, Kester M, Dysregulation of sphingolipid metabolism in cancer, Cancer Biol. Ther 11 (2011) 138–149. doi: 10.4161/cbt.11.2.14624. [DOI] [PubMed] [Google Scholar]
- [45].Cinelli G, Fabrizi M, Ravà L, Atti MCD, Vernocchi P, Vallone C, Pietrantoni E, Lanciotti R, Signore F, Manco M, Influence of maternal obesity and gestational weight gain on maternal and foetal lipid profile, Nutrients. 8 (2016) 1–13. doi: 10.3390/nu8060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sinanoglou VJ, Cavouras D, Boutsikou T, Briana DD, Lantzouraki DZ, Paliatsiou S, Volaki P, Bratakos S, Malamitsi-Puchner A, Zoumpoulakis P, Factors affecting human colostrum fatty acid profile: A case study, PLoS One. 12 (2017) 1–14. doi: 10.1371/journal.pone.0175817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yu K, Duchen K, Bjorksten B, Fatty acid composition in colostrum and mature breast milk during the first 6 months of lactation, from allergic and non-allergic mothers, Acta Paediatr. 87 (1998) 720–736. [DOI] [PubMed] [Google Scholar]
- [48].Mendonca MA, Araujo WMC, Borgo LA, Alencar EDR, Lipid profile of different infant formulas for infants, PLoS One. 12 (2017) 1–14. doi: 10.1371/journal.pone.0177812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kwan ML, Buffler PA, Abrams B, Kiley VA, Breastfeeding and the risk of childhood leukemia: A meta-analysis, Public Health Rep 119 (2004) 521–535. doi: 10.1016/j.phr.2004.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Greenop KR, Bailey HD, Miller M, Scott RJ, Attia J, Ashton LJ, Downie P, Armstrong BK, Milne E, Breastfeeding and nutrition to 2 years of age and risk of childhood acute lymphoblastic leukemia and brain tumors, Nutr. Cancer 67 (2015) 431–441. doi: 10.1080/01635581.2015.998839. [DOI] [PubMed] [Google Scholar]
- [51].Arab A, Akbarian SA, Ghiyasvand R, Miraghajani M, The effects of conjugated linoleic acids on breast cancer: A systematic review, Adv. Biomed. Res 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lee KW, Lee HJ, Cho HY, Kim YJ, Role of the conjugated linoleic acid in the prevention of cancer, Crit. Rev. Food Sci. Nutr 45 (2005) 135–144. doi: 10.1080/10408690490911800. [DOI] [PubMed] [Google Scholar]
- [53].McGuire MK, Park Y, Behre R, Harrison LY, Shultz TD, McGuire M.a, Conjugated linoleic acid concentrations of human milk and infant formula, Nutr. Res 17 (1997) 1277–1283. doi: 10.1017/CBO9781107415324.004. [DOI] [Google Scholar]
- [54].Hennessy AA, Ross PR, Fitzgerald GF, Stanton C, Sources and Bioactive Properties of Conjugated Dietary Fatty Acids, Lipids. 51 (2016) 377–397. doi: 10.1007/s11745-016-4135-z. [DOI] [PubMed] [Google Scholar]
- [55].Shu XO, Linet MS, Steinbuch M, Wen WQ, Buckley JD, Joseph P, Potter JD, Gregory H, Robison LL, Breast-Feeding and Risk of Childhood Acute Leukemia, J. Natl. Cancer Inst 91 (1999) 1765–1772. [DOI] [PubMed] [Google Scholar]
- [56].Gibson RA, Muhlhausler B, Makrides M, Conversion of linoleic acid and alpha-linolenic acid to long-chain polyunsaturated fatty acids (LCPUFAs), with a focus on pregnancy, lactation and the first 2 years of life, Matern. Child Nutr 7 (2011) 17–26. doi: 10.1111/j.1740-8709.2011.00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Li F, He B, Ma X, Yu S, Bhave RR, Lentz SR, Tan K, Guzman ML, Zhao C, Xue HH, Prostaglandin E1 and Its Analog Misoprostol Inhibit Human CML Stem Cell Self-Renewal via EP4 Receptor Activation and Repression of AP-1, Cell Stem Cell. 21 (2017) 359–373.e5. doi: 10.1016/j.stem.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Mao Y, Poschke I, Kiessling R, Tumour-induced immune suppression: Role of inflammatory mediators released by myelomonocytic cells, J. Intern. Med 276 (2014) 154–170. doi: 10.1111/joim.12229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.