Abstract
Background
Polymorphisms of the prion protein gene may influence scrapie susceptibility in small ruminants through modified protein conformation. At least 47 amino acid substitutions and 19 silent polymorphisms have been described in goat PRNP reported from several countries. The objective of this study was to investigate PRNP polymorphisms of native Ethiopian goat breeds and compare the results with other goat breeds.
Results
The analysis of the prion protein gene PRNP in 229 goats belonging to three of the main Ethiopian native goat breeds showed a remarkably high frequency (> 34.6%) of p.(Asn146Ser) in these breeds, a variant involved in scrapie resistance in Cyprus. In addition, two novel amino-acid substitutions p.(Gly127Ala) and p.(Thr193Ile), with frequencies ranging from 1.5 to 7.3% were detected. Both amino acids are well conserved in prion proteins (PrP) of most species and these changes have never been reported before in goats worldwide. Residue 127 is within the N–terminal domain of PrP and is probably involved in the recruitment of neural cell adhesion molecules (NCAM). Residue 193 is within the highly conserved string of 4 threonines that plays a role in determining the efficiency of prion protein conversion towards its pathological form.
Conclusion
Two novel coding polymorphisms and a high frequency of a scrapie protective variant indicate a high level of genetic diversity in PRNP of Ethiopian goats. This finding increases the interest in exploring PRNP polymorphisms of native goat breeds in areas where cross breeding with foreign goats has rarely occurred.
Electronic supplementary material
The online version of this article (10.1186/s12917-019-1870-4) contains supplementary material, which is available to authorized users.
Keywords: Ethiopian native goats; novel polymorphisms, PRNP, PrP, Scrapie
Background
Scrapie is a fatal, neurodegenerative disease that affects small ruminants world-wide. It belongs to the group of prion diseases or transmissible spongiform encephalopathies (TSEs), such as bovine spongiform encephalopathy (BSE) in cattle, chronic wasting disease (CWD) in cervids and Creutzfeldt–Jakob disease (CJD) in humans. Recently, a new prion disease, Camel Prion Disease (CPD) of unknown origin was detected in dromedary camels (Camelus dromedarius) in Algeria enlarging the spectrum of animal species susceptible to prion disease [1].
The conversion of normal prion protein (PrP) into pathogenic PrP conformers is central to prion diseases. Polymorphisms of the prion protein gene (PRNP) sequences may influence scrapie susceptibility in sheep and goats through modified protein conformation [2]. Although small changes in the amino acid sequence do not significantly alter the overall structure of the cellular PrP [3] they can severely alter prion transmissibility [4].
Goat PRNP displays at least 47 amino acid substitutions and 19 silent polymorphisms that were described in several countries [5]. The amino acid substitution p.(Gln222Lys) has been described as a protective allele in experimental and epidemiological studies in several European goat breeds [6–10]. An important factor in the genetic resistance to scrapie is related to the variants p.(Asn146Ser) or (Asn146Asp) which are described particularly on the island of Cyprus [11]. Recently, the impact of these variants on prion infectivity and disease outcome has been confirmed through experimental infections [12].
Ethiopia possesses about 29.1 million sheep and 29.3 million goats [13] raised in various agro-ecological zones. Goat production is economically and socially important and the native goats are widely distributed in all regions of the country in relation to environmental adaptation and survival in different areas. In lowland areas, goats are raised for milk and meat production, while in the highlands they are mainly kept for the meat. In addition, goats are also a source of non-food products such as skin for local leather industries [14]. Based on their phenotypic characteristics and genetic differences at the DNA level, four distinct families and 12 breeds of goats have been identified in Ethiopia [15, 16]. A family is a group of breeds that are genetically more related and physically more similar than breeds outside the group. Central Highland and Western Highland breeds of goat belong to the small East African family while the Long-eared Somali breed belongs to the Somali family [15, 16].
As is the case for most African countries, no surveillance for scrapie is performed and no history of TSE is available in Ethiopia. The objective of this study was to investigate PRNP polymorphisms of native Ethiopian goat breeds and to compare the results with available data reported in other goat breeds.
Results
Two novel amino acid variations were detected in all Ethiopian goat breeds in two important and well-conserved regions of PrP (Fig. 1): p.(Gly127Ala) and p.(Thr193Ile). All detected genotypes are reported in Table 1. The allelic frequencies are reported in Table 2 and no statistically significant difference was found regarding their distribution between the analysed breeds (P > 0.05). The variant p.(Gly127Ala) showed a frequency of 4.1% in Western Highland goats and lower frequencies in Central Highland and Long-eared Somali goats respectively. The variant p.(Thr193Ile) showed higher frequencies in Western and Central Highland breeds and a lower frequency in the Long-eared Somali goat (Table 2). Additional four amino acid substitutions: p.(His143Arg), p.(Asn146Ser), p.(Arg154His), and p.(Ser240Pro) were detected. SNPs at codon 42 (ccg > cca) and codon 138 (agc > agt) resulted in silent protein changes p.(Pro42=) and p.(Ser138=). At codon 240 a higher frequency of proline compared to serine was observed in all Ethiopian breeds (Table 2) as in other goats worldwide [17]. The resistant allele p.(Asn146Ser), was observed in all breeds in both homozygous and heterozygous combinations. However p. (Ser146Ser) was found only in combination with p.(Pro240Pro) (Additional file 2). The other resistant variant p.(Asn146Asp) was not detected. The variant p.(Arg154His) was found in one Western Highland goat and four Long-eared Somali goats and in this latter breed, a single goat with p. (His143Arg) was also detected (Table 1).
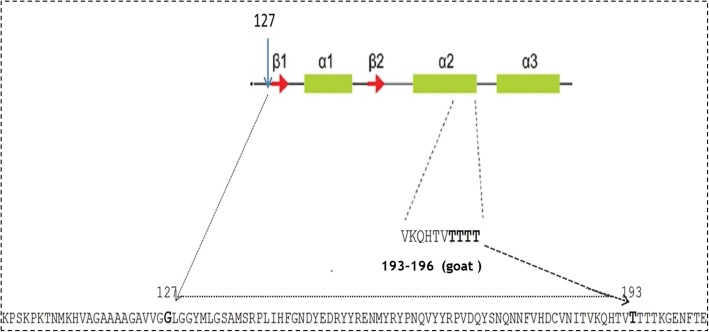
Fig. 1.
Schematic of PrP with the two novel PrP amino acid substitutions in bold. The green areas represent the 3 α-helixes; ß1 and ß2 (in red) are the ß-sheets of the PrP. The amino acid sequence of PrP corresponding to residues 104–206 are from the PrP of Capra hircus sequence (accession number P52113). The sequence is expressed with one letter code of each amino acid (see abbreviations list)
Table 1.
Frequency of all genotypes detected in Ethiopian goats
| Genotypes | Western Highland (N) % | Central Highland (N) % | Long Eared Somali (N) % |
|---|---|---|---|
| p.(Gly127Gly) | (101) 91.82 | (50) 96.15 | (65) 97.01 |
| p.(Gly127Ala) | (9) 8.18 | (2) 3.85 | (2) 2.99 |
| p.(Ala127Ala) | – | – | – |
| p.(His143His) | (110) 100.00 | (52) 100.00 | (66) 98.51 |
| p.(His143Arg) | – | – | (1) 1.49 |
| p.(Arg143Arg) | – | – | – |
| p.(Asn146Asn) | (38) 34.55 | (23) 44.23 | (22) 32.84 |
| p.(Asn146Ser) | (55) 50.00 | (22) 42.31 | (37) 55.22 |
| p.(Ser146Ser) | (17) 15.45 | (7) 13.46 | (8) 11.94 |
| p.(Arg154Arg) | (109) 99.09 | (52) 100.00 | (63) 94.03 |
| p.(Arg154His) | (1) 0.91 | – | (4) 5.97 |
| p.(His154His) | – | – | – |
| p.(Thr193Thr) | (94) 85.46 | (45) 86.54 | (62) 92.54 |
| p.(Thr193Ile) | (16) 14.54 | (7) 13.46 | (5) 7.46 |
| p.(Ile193Ile) | – | – | – |
| p.(Ser240Ser) | (4) 3.64 | (1) 1.92 | (2) 2.99 |
| p.(Ser240Pro) | (20) 18.18 | (22) 42.31 | (16) 23.88 |
| p.(Pro240Pro) | (86) 78.18 | (29) 55.77 | (49) 73.13 |
Table 2.
Allelic frequency calculated over the total genotypes of Ethiopian goats
| Western Highland (N) | Central Highland (N) | Long Eared Somali (N) | All (N) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| p.(Gly127Gly) | p.(His143His) | p.(Asn146Asn) | p.(Arg154Arg) | p.(Thr193Thr) | p.(Ser240Ser) | 0.104 (23) | 0.135 (14) | 0.104 (14) | 0.111 (51) |
| p.(Gly127Ala) | – | – | – | – | p.(Ser240Pro) | 0.041 (9) | 0.019 (2) | 0.015 (2) | 0.028 (13) |
| – | p.(His143Arg) | – | – | – | p.(Ser240Pro) | 0 | 0 | 0.007 (1) | 0.002 (1) |
| – | – | p.(Asn146Ser) | – | – | p.(Pro240Pro) | 0.405 (89) | 0.346 (36) | 0.396 (53) | 0.389 (178) |
| – | – | – | p.(Arg154His) | – | p.(Ser240Pro) | 0.005 (1) | 0 | 0.030 (4) | 0.011 (5) |
| – | – | – | – | p.(Thr193Ile) | p.(Pro240Pro) | 0.073 (16) | 0.067 (7) | 0.037 (5) | 0.061 (28) |
| – | – | – | – | – | p.(Pro240Pro) | 0.372 (82) | 0.433 (45) | 0.410 (55) | 0.397 (182) |
| 1.00 (220) | 1.00 (104) | 1.00 (134) | 1.00 (458) | ||||||
Discussion
In this paper two novel coding polymorphisms and a relatively high frequency of p.(Asn146Ser) are reported for Ethiopian goat breeds. The novel variant p. (Gly127Ala) is located within the N terminal domain of PrP involved in the interaction with the neuronal cell adhesion molecule (NCAM) [18]. A variant with serine at the same position p.(Gly127Ser) was already described in Europe, America and Asia [5], and related to a delay in the clinical onset of the disease through epidemiological and experimental studies [19–21]. The other variant p.(Thr193Ile), is found within the highly conserved string of 4 threonines located at the C-terminus of the α2-helix of PrP conferring a high instability to the protein, with great propensity to form β-sheets [22]. The first threonine (T) of the string (Thr193 in goat), has been proposed to initiate the pathologic PrP conversion through an increased formation of fibrils [23]. A substitution in the same amino acid string with proline at codon 194 was already described in mixed Southern Italian goats [24] but its role in scrapie resistance or susceptibility remains unknown. A study on mouse PrP, reported a replacement with valine (V) at codon 189 (equivalent to codon 193 in goat) that prevents prion disease-related pathologic conversion by conferring more stability to the prion protein [25]. The two novel variants p.(Gly127Ala) and p.(Thr193Ile) detected in this study might have similar effects to the above-mentioned p.(Gly127Ser) in other goats and p.(Thr189Val) in mouse, since all are radical replacements. A high prevalence of proline at codon 240 was also detected as in other goat breeds worldwide [17] but amino acid variations at codon 240 have no impact on scrapie disease [26]. However the majority of variations in other codons were associated to p.(Ser240Pro) in this study (Additional file 2). The variant p.(Asn146Ser) was detected at a frequency of 40.5% in Western Highland goats, 34.6% in Central Highland goats and 39.6% in Long-eared Somali goats (Table 2). This scrapie protective variant has been described already in other native breeds in Tanzania at a frequency of 16.3% [27], and 28.1% in Halep goats in Turkey [28]. Frequencies similar to Ethiopian goats have been reported in imported Boer goats in Great Britain (24.5–35.5%) [29], in the USA (35.2%) [30], and in the Netherlands (31%) [31], suggesting that this resistance-associated polymorphism has been maintained in the Damascus-related breeds independently from the presence of scrapie [28]. However, serine at codon 146 is present at frequency of 16.5% in Damascus goat of Cyprus [32] but absent in Derivata di Siria, another Damascus related goat breed in Sicily, that showed conversely a 15% frequency of the resistant variant p.(Gln222Lys) [33]. Both islands are considered as endemic areas for scrapie, so it could be possible that different alleles in native breeds were positively selected in relation to the local circulating scrapie strains in specific geographic areas [34].
The presence of p.(Asn146Ser) in Halep (Damascus), Ethiopian and Tanzanian native goats suggests that the allele was naturally selected in the Middle East first, and spread through the central Sahara and Ethiopia later [35, 36] and subsequently into southern parts of the African continent through a different migration route compared to the main Mediterranean and North African routes. As in the Sicilian Damascus breed, p.(Asn146Ser) has not been detected so far in Algerian, Tunisian and Moroccan goats [37, 38]
The frequency of the protective variant p.(Asn146Ser) in three of the main Ethiopian goat breeds and the presence of two novel amino acid substitutions in highly conserved regions of prion protein (PrP), show a major diversity in PRNP of Ethiopian goats compared to other breeds. However, a low genetic variation among indigenous Ethiopian goats has been detected through genetic studies on microsatellites probably due to the ancient and widespread practice of nomadism [39, 40]. This traditional practice might also explain the presence of different goat families that overlap breeds and the similarity of PRNP substitution frequencies among the breeds in this study. In any case extensive cross breeding with goats coming from other countries has never been performed in Ethiopia, except for some pilot studies with Saanen goats to increase milk production [14], so these results reflect the original genetic background of Ethiopian goats.
Conclusion
The two novel amino acid substitutions and the high frequency of the scrapie protective variant p.(Asn146Ser) might indicate a lower risk of scrapie in Ethiopian goats. It could be very important to perform PRNP characterization also in sheep populations of the same areas, especially because mixed holdings are very common. A surveillance plan for the disease in both goats and sheep populations would be also important to understand the true epidemiology of scrapie in the country. The occurrence of a new prion disease in dromedaries, CPD in Algeria, could be an impetus for a surveillance plan for prion diseases in Africa since dromedaries are often bred with sheep and goats simultaneously; sharing common pastures in many African countries such as in Ethiopia. A new prion disease in a farmed animal species requires a thorough risk assessment for implementing policies to control the disease in animals and minimize human exposure [1]. This study can fill gaps on the knowledge of PRNP polymorphisms of goats in economically marginal areas and increase the interest in exploring the genetic variation of the PRNP gene in native goat breeds and sheep as well, in countries where cross-breeding with imported animals has never been performed.
Methods
Goat breeds
Three of the main Ethiopian native goat breeds were analysed for the PRNP gene: Western and Central Highlands and Long-eared Somali. Somatic aspects of the different goats have been already described [15, 16, 41]. Professional veterinarians collected the blood samples aseptically from jugular veins using ethylene diamine tetraacetic acid (EDTA) coated tubes. A total of 229 goats, belonging to 80 herds were sampled. Central Highland breeds (n = 52) were sampled in the Seka, Tiroafata and Omonada districts of Jimma; Western Highland (n = 110) from the Ambo and Bako districts; and Long eared Somali (n = 67) from the Gursum and Jijiga districts of Eastern Hararghe. The samples consisted of 75% males and 25% females with no genetic relation.
Genetic analysis
Polymerase chain reaction (PCR) amplification of the open reading frame of the caprine PRNP (NM_001314247.1 for the cDNA position and NP_001301176.1 for the protein variants) was performed and direct PCR fragment sequencing was carried out as previously described [42].
Raw data of Ethiopian goats genotyping are reported in Additional file 1
Amino acid variants are described following recommendation of Den Dunnen et al. [43]
Allelic frequencies were calculated using the Hardy-Weinberg equation. Statistical differences between the breeds were calculated with a chi-square test.
Additional files
Prion protein gene polymorphism in native Ethiopian goats. Excel file that include all information related to each single goat in the three Ethiopian regions. For each goat, information regarding sex, age in months and the related prion polymorphisms are reported. (XLS 60 kb)
Table of the genotypes combinations of Prion protein gene in native Ethiopian goats.. (PDF 423 kb)
Acknowledgements
The authors wish to thank Dr. Maria La Giglia for the technical assistance.
Funding
This work is supported by EMIDA ERA-NET project: "GOAT-TSE-FREE and Ricerca finalizzata Code: RF-2010-2318525, and IZSSi08/16 RC to MV from Italian Ministry of Health. The funding source had no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Availability of data and materials
Excel file with raw data and a table with combination of some alleles are presented as Additional files 1 and 2.
Abbreviations
- BSE
Bovine spongiform encephalopathy
- CWD
Chronic wasting disease
- PRNP
Prion protein gene
- PrP
Prion protein
- TSE
Transmissible spongiform encephalopathy
Amino acid code
- Ala (A)
Alanine
- Arg (R)
Arginine
- Asn (N)
Asparagine
- Asp (D)
Aspartic acid
- Gln (Q)
Glutamine
- Gly (G)
Glycine
- His (H)
Histidine
- Ile (I)
Isoleucine
- Lys (K)
Lysine
- Pro (P)
Proline
- Ser (S)
Serine
- Thr (T)
Threonine
- Val (V)
Valine
Authors’ contributions
MV, VDMLP, EZG conceived and designed the proposal, participated in the coordination and management of the study. EZG, BT and MA organized the study area and sample collection in Ethiopia. SM and IS made the laboratory testing, participated in data analysis and in writing the manuscript. MT performed data analysis and contributed in writing the manuscript and drawing the Fig. 1. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All animals were handled strictly in accordance with the good animal practice in order to minimize animal sufferings during blood sampling. An informed verbal consent was obtained from the animal owners, due to their illiteracy. This procedure was approved by the ethical committee of the Ambo University since no damage to the animals was involved and no diffusion of private information was implicated. For the purpose of this study, we used the blood samples from animals collected as the routine activity of a research project on surveillance of animal diseases of zoonotic importance that was approved by the ethical committee for animal experimentation of the Ambo University, Ethiopia (Ref. No. RD/AREC/004/2016).
Consent for publication
Not applicable
Competing interests
All authors declare that no conflict of interest is present. The work is an original paper and is not under consideration in other journals.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Maria Vitale, Phone: 39-3665858273, Email: marvitus@yahoo.com.
Sergio Migliore, Email: migliore.sergio@gmail.com.
Berhanu Tilahun, Email: btilahun11@gmail.com.
Mukarim Abdurahaman, Email: mukevet@yahoo.com.
Marco Tolone, Email: marco.tolone@unipa.it.
Ignazio Sammarco, Email: netsam@libero.it.
Vincenzo Di Marco Lo Presti, Email: dimarco.vince@gmail.com.
Endrias Zewdu Gebremedhin, Email: endrias.zewdu@gmail.com.
References
- 1.Babelhadj B, Di Bari M, Pirisinu L, Chiappini B, Gaouar S, Riccardi G, et al. Prion disease in dromedary camels, Algeria. Emerg Infect Dis. 2018;24(6):1029–1036. doi: 10.3201/eid2406.172007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JH, Bae SE, Jung S, Ahn I, Son HS. Discriminant analysis of prion sequences for prediction of susceptibility. Exp Mol Med. 2013;45(10):e48. doi: 10.1038/emm.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lysek DA, Schorn C, Nivon LG, Esteve-Moya V, Christen B, Calzolai L, von Schroetter C, Fiorito F, Herrmann T, Guntert P, Wuthrich K. Prion protein NMR structures of cats, dogs, pigs, and sheep. Proc Natl Acad Sci U S A. 2005;102(3):640–645. doi: 10.1073/pnas.0408937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 5.EFSA Biohaz Panel Genetic resistance to transmissible spongiform encephalopathies (TSE) in goats. EFSA J. 2017;15(8):4962. doi: 10.2903/j.efsa.2017.4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaccari G, Di Bari MA, Morelli L, Nonno R, Chiappini B, Antonucci G, Marcon S, Fazzi P, Palazzini N, Troiano P, Petrella A, Di Guardo G, Agrimi U. Identification of an allelic variant of the goat PrP gene associated with resistance to scrapie. J Gen Virol. 2006;87:1395–1402. doi: 10.1099/vir.0.81485-0. [DOI] [PubMed] [Google Scholar]
- 7.Barillet F, Mariat D, Amigues Y, Faugeras R, Caillat H, Moazami-Goudarzi K, Rupp R, Babilliot JM, Lacroux C, Lugan S, Schelcher F, Chartier C, Corbière F, Andréoletti O, Perrin-Chauvineau C. Identification of seven haplotypes of the caprine PrP gene at codons 127, 142, 154,211, 222 and 240 in French alpine and Saanen breeds and their association with classical scrapie. J. Gen. Virol. 2009;90:769–776. doi: 10.1099/vir.0.006114-0. [DOI] [PubMed] [Google Scholar]
- 8.Acutis PL, Martucci F, D'Angelo A, Peletto S, Colussi S, Maurella C, Porcario C, Iulini B, Mazza M, Dell’Atti L, Zuccon F, Corona C, Martinelli N, Casalone C, Caramelli M, Lombardi G. Resistance to classical scrapie in experimentally challenged goats carrying mutation K222of the prion protein gene. Vet Res. 2012;43:8–17. doi: 10.1186/1297-9716-43-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguilar-Calvo P, Espinosa JC, Pintado B, Gutiérrez-Adán A, Alamillo E, Miranda A, Prieto I, Bossers A, Andreoletti O, Torres JM. Role of the goat K222-PrP(C) polymorphic variant in prion infection resistance. J.Virol. 2014;88(5):2670–2676. doi: 10.1128/JVI.02074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lacroux C, Perrin-Chauvineau C, Corbière F, Aron N, Aguilar-Calvo P, Torres JM, Costes P, Bremaud I, Lugan S, Schelcher F, Barillet F, Andreoletti O. Genetic resistance to scrapie infection in experimentally challenged goats. J Virol. 2014;88(5):2406–2413. doi: 10.1128/JVI.02872-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papasavva-Stylianou P, Kleanthous M, Toumazos P, Mavrikiou P, Loucaides P. Novel polymorphisms at codons 146 and 151 in the prion protein gene of Cyprus goats, and their association with natural scrapie. Vet J. 2007;173:459–462. doi: 10.1016/j.tvjl.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 12.Papasavva-Stylianou P, Mathieson Simmons M, Ortiz-Pelaez A, Windl O, Spiropoulos J, Soteria G. Effect of polymorphisms at codon 146 of the goat PRNP gene on susceptibility to challenge with classical scrapie by different routes. J Virol. 2017;91:22. doi: 10.1128/JVI.01142-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.FAO . FAOStat official live animals data, food and agriculture organization of the United Nations (FAO) 2017. [Google Scholar]
- 14.Tsegahun A, Lemma S, Sebsbie A, Mekoya A, Sileshi Z. The opportunities and challenges of enhancing goat production in East Africa. Proceedings of a conference held at Debub University, Awassa, Ethiopia from November. 2000. National goat research strategy in Ethiopia; pp. 1–5. [Google Scholar]
- 15.Farm-Africa . Goat types of Ethiopia and Eritrea. Physical description and management systems. 1996. p. 76. [Google Scholar]
- 16.Tucho TA. PhD thesis. Karnal (Haryana) India: National Dairy Research Institute (Deemed University; 2004. Genetic characterization of indigenous goat populations of Ethiopia using microsatellite DNA markers. [Google Scholar]
- 17.Srithayakumar V, Mitchell GB, White BN. Identification of amino acid variation in the prion protein associated with classical scrapie in Canadian dairy goats. BMC Vet Res. 2016;12:59. doi: 10.1186/s12917-016-0684-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slapšak U, Salzano G, Ladan A, Abskharon RNN, Ilc G, Zupančič B, Biljan I, Plavec J, Giachin G, Legname G. The N Terminus of the Prion Protein Mediates Functional Interactions with the Neuronal Cell Adhesion Molecule (NCAM) Fibronectin Domain. J Biol Chem. 2016;291(42):21857–21868. doi: 10.1074/jbc.M116.743435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldmann W, Ryan K, Stewart P, Parnham D, Xicohtencatl R, et al. Caprine prion gene polymorphisms are associated with decreased incidence of classical scrapie in goat herds in the United Kingdom. Vet Res. 2011;42:110–117. doi: 10.1186/1297-9716-42-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acín C, Martín-Burriel I, Monleón E, Lyahyai J, Pitarch JL, Serrano C, Monzón M, Zaragoza P, Badiola JJ. Prion protein gene variability in Spanish goats. Inference through susceptibility to classical scrapie strains and pathogenic distribution of peripheral PrP(sc.) PLoS One. 2013;8(4):e61118. doi: 10.1371/journal.pone.0061118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dassanayake RP, White SN, Madsen-Bouterse SA, Schneider DA, O'Rourke KI. Role of the PRNP S127 allele in experimental infection of goats with classical caprine scrapie. Anim Genet. 2015;46(3):341. doi: 10.1111/age.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salamat K, Moudjou M, Chapuis J, Herzog L, Jaumain E, Béringue V, Rezaei H, Pastore A, Laude H, Dron M. Integrity of helix 2-helix 3 domain of the PrP protein is not mandatory for prion replication. J Biol Chem. 2012;287:18953–18964. doi: 10.1074/jbc.M112.341677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji HF, Zhang HY. β-Sheet constitution of prion proteins. Trends Biochem Sci. 2010;35:129. doi: 10.1016/j.tibs.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Acutis PL, Colussi S, Santagada G, Laurenza C, Maniaci MG, Riina MV, Peletto S, Goldmann W, Bossers A, Caramelli M, Cristoferi I, Maione S, Sacchi P, Rasero R. Genetic variability of the PRNP gene in goat breeds from northern and southern Italy. J Appl Microbiol. 2008;104(6):1782–1789. doi: 10.1111/j.1365-2672.2007.03703.x. [DOI] [PubMed] [Google Scholar]
- 25.Barron RM, Baybutt H, Tuzi NL, McCormack J, King D, Moore RC, Melton DW, Manson JC. Polymorphisms at codons 108 and 189 in murine PrP play distinct roles in the control of scrapie incubation time. J Gen Virol. 2005;86:859–868.27. doi: 10.1099/vir.0.80525-0. [DOI] [PubMed] [Google Scholar]
- 26.Corbière F, Perrin-Chauvineau C, Lacroux C, Costes P, Thomas M, Brémaud I, Martin S, Lugan S, Chartier C, Schelcher F, Barillet F, Andreoletti O. PrP-associated resistance to scrapie in five highly infected goat herds. J Gen Virol. 2013;94:241–245. doi: 10.1099/vir.0.047225-0. [DOI] [PubMed] [Google Scholar]
- 27.Kipanyula MJ. Prion protein gene haplotypes distribution among indigenous goat ecotypes in Tanzania. Livestock Research for Rural Development. 2016;28. Retrieved April 24, 2019, from http://www.lrrd.org/lrrd28/2/kipa28015.html.
- 28.Meydan H, Pehlivan E, Özkan MM, Yildiz MA, Goldmann W. Prion protein gene polymorphisms in Turkish native goat breeds. J Genet. 2017;96(2):299–305. doi: 10.1007/s12041-017-0763-1. [DOI] [PubMed] [Google Scholar]
- 29.Goldmann W, Marier E, Stewart P, Konold T, Street S, Langeveld J, Windl O, Ortiz-Pelaez A. Prion protein genotype survey confirms low frequency of scrapie- resistant K222 allele in British goat herds. Vet Rec. 2016;178:168. doi: 10.1136/vr.103521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White S, Herrmann-Hoesing L, O’Rourke K, Waldron D, Rowe J, Alverson J. Prion gene (PRNP) haplotype variation in United States goat breeds (open access publication) Genet Sel Evol. 2008;40(5):553–561. doi: 10.1186/1297-9686-40-5-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Windig JJ, Hoving RA, Priem J, Bossers A, van Keulen LJ, Langeveld JP. Variation in the prion protein sequence in Dutch goat breeds. J Anim Breed Genet. 2016;133(5):366–374. doi: 10.1111/jbg.12211. [DOI] [PubMed] [Google Scholar]
- 32.European Food Safety Authority Scientific and technical assistance on the provisional results of the study on genetic resistance to classical scrapie in goats in Cyprus. EFSA J. 2012;10(11):2972. doi: 10.2903/j.efsa.2012.2972. [DOI] [Google Scholar]
- 33.Vitale M, Migliore S, La Giglia M, Alberti P, Di Marco Lo Presti V, Langeveld JP. BMC scrapie incidence and PRNP polymorphisms: rare small ruminant breeds of Sicily with TSE protecting genetic reservoirs. BMC Vet Res. 2016;12(1):141. doi: 10.1186/s12917-016-0766-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Migliore S, Agnello S, D’Avola S, Goldmann W, Di Marco Lo Presti V, Vitale M. Cross-sectional study of PRNP gene in two native Sicilian goat populations in Italy; a relation between prion gene polymorphisms and scrapie incidence. J Genet. 2017;96(2):319–325. doi: 10.1007/s12041-017-0776-9. [DOI] [PubMed] [Google Scholar]
- 35.Clutton-Brock J. Cattle, sheep, and goats south of the Sahara: an archaezoological perspective. In: Blench RM, MacDonald KC, editors. The origins and development of African livestock: archaeology, genetics, linguistics and ethnography. London: UCL Press; 2000. pp. 30–37. [Google Scholar]
- 36.Smith AB. Origins and spread of pastoralism in Africa. Annu Rev Anthropol. 1992;21:125–141. doi: 10.1146/annurev.an.21.100192.001013. [DOI] [Google Scholar]
- 37.Fantazi K, Migliore S, Kdidi S, Racinaro L, Tefiel H, Boukhari R, Federico G, Di Marco Lo Presti V, Gaouar SBS, Vitale M. Analysis of differences in prion protein gene (PRNP) polymorphisms between Algerian and southern Italy’s goats. Ital J Ani Sci. 2018;17(3):578–585. doi: 10.1080/1828051X.2017.1420430. [DOI] [Google Scholar]
- 38.Serrano C, Hammouchi M, Benomar A, Lyahyai J, Ranera B, Acìn C, el Hamidi M, Monzòn M, Badiola JJ, Tligui N, Zaragoza P, Martín-Burriel I. PRNP haplotype distribution in Moroccan goats. Anim Gen. 2009;40:565–568. doi: 10.1111/j.1365-2052.2009.01873.x. [DOI] [PubMed] [Google Scholar]
- 39.Hassen H, Lababidi S, Rischkowsky B, Lababidi S, Rischkowsky B, Baum M, Tibbo M. Molecular characterization of Ethiopian indigenous goat populations. Trop Anim Health Prod. 2012;44:1239. doi: 10.1007/s11250-011-0064-2. [DOI] [PubMed] [Google Scholar]
- 40.Mekuriaw G. Molecular characterization of Ethiopian indigenous goat populations: Genetic diversity and structure, demographic dynamics and assessment of the kisspeptin gene polymorphism, PhD thesis in Applied Genetics. Addis Ababa: Addis Ababa University; 2016. http://hdl.handle.net/10568/76315
- 41.Awgichew K, Abegaz S. Breed of sheep and goats. In: Yami A, Merkel RC, editors. Sheep and goat production handbook for Ethiopia: Ethiopia Sheep and Goat Productivity Improvement Program (ESGPIP); 2008. p. 5–26. http://www.igadhost.com/igaddata/docs/Sheep%20and%20Goat%20Production%20Hand%20Book%20for%20ETHIOPIA.pdf.
- 42.Migliore Sergio, Agnello Stefano, Chiappini Barbara, Vaccari Gabriele, Mignacca Sebastian A., Di Marco Lo Presti Vincenzo, Di Domenico Francesco, Vitale Maria. Biodiversity and selection for scrapie resistance in goats: Genetic polymorphism in “Girgentana” breed in Sicily, Italy. Small Ruminant Research. 2015;125:137–141. doi: 10.1016/j.smallrumres.2015.01.029. [DOI] [Google Scholar]
- 43.Den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, Roux A, Smith T, Antonarakis SE, Taschner PEM. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;25(37):564–569 HGVS. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prion protein gene polymorphism in native Ethiopian goats. Excel file that include all information related to each single goat in the three Ethiopian regions. For each goat, information regarding sex, age in months and the related prion polymorphisms are reported. (XLS 60 kb)
Table of the genotypes combinations of Prion protein gene in native Ethiopian goats.. (PDF 423 kb)
Data Availability Statement
Excel file with raw data and a table with combination of some alleles are presented as Additional files 1 and 2.