The plant’s response to heavy metals requires glutamate and mitochondrial pyruvate carriers.
Abstract
Cadmium (Cd) is a major heavy metal pollutant, and Cd toxicity is a serious cause of abiotic stress in the environment. Plants protect themselves against Cd stress through a variety of pathways. In a recent study, we found that mitochondrial pyruvate carriers (MPCs) are involved in Cd tolerance in Arabidopsis (Arabidopsis thaliana). Following the identification of MPCs in yeast (Saccharomyces cerevisiae) in 2012, most studies have focused on the function of MPCs in animals, as a possible approach to reduce the risk of cancer developing. The results of this study show that AtMPC protein complexes are required for Cd tolerance and prevention of Cd accumulation in Arabidopsis. AtMPC complexes are composed of two elements, AtMPC1 and AtMPC2 (AtNRGA1 or AtMPC3). When the formation of AtMPCs was interrupted by the loss of AtMPC1, glutamate could supplement the synthesis of acetyl-coenzyme A and sustain the TCA cycle. With the up-regulation of glutathione synthesis following exposure to Cd stress, the supplementary pathway could not efficiently drive the tricarboxylic acid cycle without AtMPC. The ATP content decreased concomitantly with the deletion of tricarboxylic acid activity, which led to Cd accumulation in Arabidopsis. More importantly, ScMPCs were also required for Cd tolerance in yeast. Our results suggest that the mechanism of Cd tolerance may be similar in other species.
Heavy metal stress has become increasingly severe in the world in the past several decades. Cadmium (Cd) pollution is the representative heavy metal contamination, as Cd2+ is readily absorbed by leafy vegetables, fruits, and grain crops (Wagner, 1993; Prasad, 1995; Clemens, 2006; DalCorso et al., 2008). Many Cd2+-chelating materials and transporters have been discovered, and the tolerance mechanism has been applied to plant engineering. Glutathione (GSH) is a major redox buffer in eukaryotic cells (Quastel et al., 1923; Stewart and Tunnicliffe, 1925; Griffith et al., 1978), and its oligomers known as phytochelatins can conjugate Cd2+ and alleviate Cd toxicity (Rauser, 1990; Steffens, 1990; Marrs, 1996; Cobbett et al., 1998; Sun et al., 2005a). Many transporters have also been found to function in Cd resistance; for example, ATP-binding cassette (ABC) transporters can pump Cd into vacuoles or out of plasma membranes in yeast and Arabidopsis (Arabidopsis thaliana; Li et al., 1997; Song et al., 2003; Kim et al., 2007). The P1B-ATPase subfamily, such as heavy metal-associated (HMA) proteins, also plays an important role in Cd allocation or detoxification by isolating metal ion into vacuole (Williams and Mills, 2005; Morel et al., 2009; Liu et al., 2017). Besides the ATP-consuming transporters, natural resistance-associated macrophage proteins, acting as proton-coupled metal ion transporters, can also transport heavy metal ions (Thomine et al., 2000; Cailliatte et al., 2009). Although a variety of pathways are involved in Cd tolerance, the main pathways have yet to be discovered.
Mitochondrial pyruvate carrier (MPC) is a major checkpoint between glycolysis and the tricarboxylic acid cycle. MPC can transport pyruvate into mitochondrial matrix to generate acetyl-CoA for the tricarboxylic acid cycle. The function of MPC has been investigated using genetic and biochemical approaches since its discovery in 2012 (Bricker et al., 2012; Herzig et al., 2012). Many groups have focused on MPCs in animal cells or yeasts to understand their functions in metabolism-related human diseases such as cancer and type 2 diabetes (Bender and Martinou, 2016). In 2014, a homologous gene (MPC2-like gene) of the MPC family, namely, NRGA1, was shown to be negatively involved in abscisic acid (ABA)-regulated guard cell signaling and drought stress response in Arabidopsis (Li et al., 2014). Recently, the MPC1 homologous gene was also shown to mediate the ABA-regulated stomatal closure and drought response by regulating the pyruvate content in Arabidopsis (Shen et al., 2017). Although these two MPC proteins were reported in plants, neither of them has been shown to be involved in the pyruvate metabolism and the tricarboxylic acid cycle.
Here, we screened the functions of MPCs in plant Cd responses. Our results showed that the MPC complexes, composed of MPC1 and NRGA1 (MPC2 or MPC3), could prevent Cd stress by sustaining the tricarboxylic acid cycle and ATP production and alleviating the consumption of Glu to generate GSH in Arabidopsis. More importantly, a similar Cd-sensitive phenotype was found in the yeast MPC mutants. Considering the similar regulation manners of MPC in the pyruvate metabolism, tricarboxylic acid cycle, and Glu oxidation pathway in microorganisms and animals, the Cd tolerance pathway controlled by MPC could be evolutionarily conserved from yeasts to plants.
RESULTS
MPCs Function in a Conservative Process
The initial BLAST analysis using yeast MPC1 protein (YGL080W) predicated three homologous proteins in the yeast genome, four in the Arabidopsis genome, and three in the human genome (Supplemental Fig. S1, A and B). For the MPC homologous proteins in Arabidopsis, since At4g14695 is referred to as MPC2 and the MPC2-like protein (MPC2L [At4g05590]; Herzig et al., 2012) is named as Negative Regulator of Guard Cell ABA Signaling1 (NRGA1; Liu et al., 2017), we named At4g22310 as MPC3 and At5g20090 as MPC1.
Alignment analysis showed that MPCs shared conserved domains (Supplemental Fig. S1A). Interestingly, during the evolutionary process of all MPC members, MPC1 conducted an independent evolutionary pathway that is different from other MPC members (Supplemental Fig. S1B). These results suggested that MPC1 may have specific functions different from other MPCs.
AtMPC1 Is Required for Cd Tolerance in Arabidopsis
To identify AtMPCs that participate in Cd tolerance, we ordered a number of Arabidopsis T-DNA mutants, mpc1-1, mpc1-2, nrga1, and mpc3-1, and generated two MPC2 mutants, mpc2-1 and mpc2-2, using the CRISPR/Cas9 technology (Yan et al., 2015). Mutant lines were confirmed by reverse transcription (RT)-PCR, RT-quantitative PCR, and sequencing (Supplemental Figs. S2, A and B, and S3A; Li et al., 2014; Shen et al., 2017) and used for Cd treatment bioassays. Without the Cd stress, the phenotype of the mutants was indistinguishable from that of the wild type. When cultured with 50 µm CdCl2, the root length of mpc1-1 and mpc1-2 was substantially shorter than that of the wild-type plants and other mutants (Fig. 1, A and B). To further confirm whether this Cd-sensitive phenotype was caused by the loss of AtMPC1, we constructed pAtMPC1::AtMPC1 to generate mpc1-1(AtMPC1) complementation transgenic lines. With the Cd stress, the root length-sensitive phenotype of mpc1-1 was recovered by AtMPC1 (Fig. 1, C and D). We also tested the germination rate and biomass, both of which were affected by the loss of AtMPC1 (Supplemental Fig. S4, A–C). These results suggested that AtMPC1 is required for Cd tolerance in Arabidopsis.
Figure 1.
Cd tolerance test. A, Arabidopsis plants grown on 0.5× MS plates vertically without or with 50 µm CdCl2 for 10 d. Representative results from three reproducible experiments are shown. B, Average root length of seedlings cultured under the same growth condition as in A. The root length of five seedlings of each class was measured as the mean value (removing the top and lowest values). Error bars indicate sd from three independent experiments. C, Arabidopsis plants germinated on 0.5× MS plates vertically for 3 d were transferred to plates without or with 50 µm CdCl2 for another 7 d. Representative results from three reproducible experiments are shown. C-1 and C-2 are two independent complementation lines. D, Average root length of seedlings cultured under the same growth condition as that in C. The root length of five seedlings of each class was measured as the mean value (removing the top and lowest values). Error bars indicate sd from three independent experiments. P values from Student’s t test were determined for mutants or transgenic plants compared with wild-type (WT) plants: ***, P < 0.001.
Loss of Function of AtMPC1 Promotes Cd2+ Influx and Accumulation in Arabidopsis
In order to explore the loss of Cd tolerance in mpc1-1, we measured the Cd content of 10-d-old seedlings grown on 0.5× Murashige and Skoog (MS) medium with 50 µm CdCl2. A higher Cd2+ accumulation was found in mpc1-1 compared with that in the wild type and the complementation lines (Fig. 2A). Since the Cd content in the seeds of crops and in shoots of vegetables is very imperative, we also measured the Cd content of shoots and seeds in Arabidopsis. The Cd content in shoots and seeds also showed a significant increase in mpc1-1 compared with that in the wild type and the complementation lines (Fig. 2B). These results indicated that the Cd-sensitive phenotype of mpc1-1 is likely to be caused by the Cd accumulation.
Figure 2.
Cd content measurement and Cd2+ flux assay. A, Cd content in 10-d-old seedlings. Seedlings were germinated on 0.5× MS for 3 d and transferred to 0.5× MS with 50 µm CdCl2 for 7 d. B, Cd content in shoot and seeds in mature soil-grown plants. Plants were grown in normal soil until they were 4 weeks old before 50 µm CdCl2 was applied. C, Cd2+ fluxes in the roots of 10-d-old seedlings, which germinated in 0.5× MS medium for 3 d and were then transferred to 0.5× MS with 50 µm CdCl2 application, were recorded every 6 s for 3 min after the seedlings were exposed to measuring solution with 50 μm CdCl2. The number of plants measured was as follows: wild type (WT), five; mpc1-1, five; C-1, three; and C-2, three. D, The mean Cd2+ fluxes in roots of 10-d-old seedlings, which germinated in 0.5× MS medium and were then transferred to 0.5× MS with 50 µm CdCl2, were measured at different time points for 3 min after the seedlings were exposed to measuring solution with 50 μm CdCl2. Error bars indicate sd from three independent experiments. P values from Student’s t test were determined for mutants or transgenic plants compared with wild-type plants: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. DW, Dry weight.
To test whether the Cd accumulation is due to direct absorbance of Cd2+ from plant roots, Cd2+ flux was measured near the root epidermal zone (3 μm) at 400 µm from the root tip (Supplemental Fig. S5) using the Noninvasive Microtest Technology. The ionic fluxes of Cd were calculated based on Fick’s law of diffusion, J = −D0 ⋅ (dc/dx), where J is the ionic flux, dc is the concentration gradient, dx is the distance between the two points, and D0 is the diffusion constant. Under the treatment of 50 µm CdCl2, the Cd2+ influx in roots of mpc1-1 was much higher than that of the wild type and the complementation lines at different time points and the mean values at 3 min (Fig. 2, C and D). These results suggested that AtMPC1 can prevent the accumulation of Cd2+ in the plants, and the Cd2+ accumulation of mpc1-1 is probably due to the enhancement of Cd2+ absorbance or the decrease of Cd2+ exclusion.
AtMPC Complexes Are Required for Cd Tolerance in Arabidopsis
Recently, studies found that MPC1 can form a protein complex with MPC2 in mammals and with MPC2 or MPC3 in yeast (Bricker et al., 2012; Herzig et al., 2012). In Arabidopsis, there are four MPC members, and AtMPC1 was found to interact with AtNRGA1 (Shen et al., 2017). However, there was no evidence showing how the MPC members interact and function with each other in plants. From the phylogenetic analysis, we found that MPC1 evolved in a more similar pathway that was independent of other MPCs in the three species (Supplemental Fig. S1B). It is likely that MPC1s share conserved functions or interaction patterns. Thus, a bimolecular fluorescence complementation (BiFC) system was developed to test the interaction between AtMPC1 and AtMPC2 or AtMPC3. MLO1 and CaM1, which were identified to interact with each other, were used as the positive control (Kim et al., 2002; Gookin and Assmann, 2014). As shown in Figure 3A, the fluorescence signals were detected in the positive control and both of the tested protein groups, while the candidates interacting with empty vectors did not show any signaling. As the BiFC system cannot avoid self-activation fluorescence, we further performed in vivo coimmunoprecipitation (Co-IP) assay using Nicotiana benthamiana leaves simultaneously harboring p35S::FLAG:AtMPC1 and p35S::C-MYC:AtMPC2 or p35S::C-MYC:AtMPC3. Total proteins extracted from the plant leaves were used for precipitation and western blot. Similarly, the interactions were detected between AtMPC1 and AtMPC2 and between AtMPC1 and AtMPC3 (Fig. 3B). In yeast, the ScMPC complexes are composed of ScMPC1 and either ScMPC2 or ScMPC3; ScMPC2 and ScMPC3 have not been found to coexist in the same protein complex (Bricker et al., 2012; Herzig et al., 2012). Considering that MPC1s could share interaction patterns dependent on their conserved evolutionary pathway in yeast and Arabidopsis (Supplemental Fig. S1B), AtMPC1 could interact with one other AtMPC (AtNRGA1, AtMPC2, or AtMPC3) to form three AtMPC complexes in Arabidopsis, similar to the interactions of MPCs in yeast and animals.
Figure 3.
MPC complex is required for Cd tolerance in Arabidopsis. A, BiFC assay of interactions between AtMPC1 and AtMPC2 (left) and AtMPC3 (right). The fluorescence indicates interaction between the indicated partner proteins. The images were obtained from the GFP channel, differential interference contrast (DIC) channel, or a merged image of the two channels. The positive control was CaM1:pDOE01N-MLO1:pDOE01N, and the negative control was AtMPC1:pDOE01N-pDOE01C and pDOE01N-AtMPC2 (AtMPC3):pDOE01C. EV, Empty vector. Bars = 100 µm. B, Co-IP assay for interaction between AtMPC1 with AtMPC2 (AtMPC3) in vivo. Total protein extracted from leaves of N. benthamiana was coinfiltrated with GV3101 harboring 35S::FLAG:AtMPC1 and 35S::C-MYC:AtMPC2 (35S::C-MYC:AtMPC3) after about 2 d. The anti-FLAG antibody was used to detect FLAG::AtMPC1, and the anti-C-MYC antibody was used to detect C-MYC::AtMPC2 and C-MYC::AtMPC3. IB, Immunoblotting; IP, immunoprecipitation. C and D, Arabidopsis plants germinated on 0.5× MS plates for 3 d were transferred to plates without or with 50 µm CdCl2 for culturing for another 7 d. Representative results from three reproducible experiments are shown. WT, Wild type.
According to the interaction pattern of AtMPCs, it is possible that the functional MPC complexes are required for Cd tolerance in Arabidopsis. To explore the function of AtMPCs for Cd tolerance in plants, we generated mpc1-1/nrga1 and mpc1-1/mpc3-1 with a normal hybridization method and mpc1-1/mpc2-3, mpc1-1/mpc2-4, nrga1/mpc2-5/mpc3-2, nrga1/mpc2-6/mpc3-3, mpc1-1/nrga1/mpc2-7/mpc3-4, and mpc1-1/nrga1/mpc2-8/mpc3-5 using the CRISPR/Cas9 system (Yan et al., 2015; Supplemental Fig. S3, A–C). Under the control condition, none of the mutants showed an aberrant phenotype different from that of the wild type (Fig. 3, C and D). In the presence of Cd, the double and quadruple mutants, in which AtMPCs including AtMPC1 were knocked out, grew shorter and weaker compared with the wild-type plants. However, multiple mutants did not enhance the Cd sensitivity compared with mpc1-1 (Fig. 3D). The triple mutants, in which AtMPCs except AtMPC1 were knocked out, were also sensitive to the Cd treatment, showing a similar phenotype to mpc1-1 (Fig. 3C). These results indicated that other AtMPCs other than AtMPC1 are also necessary in Cd tolerance. Because of the redundancy of other AtMPCs except AtMPC1, the single mutants did not show the Cd-sensitive phenotype (Fig. 1A). In conclusion, these results showed that, in Arabidopsis, the AtMPC complex is composed of AtMPC1 and other AtMPCs (AtNRGA1, AtMPC2, and AtMPC3) and exists in three forms. All three AtMPC complexes play a main role in Cd tolerance in Arabidopsis. Moreover, both AtMPC1 and other AtMPCs (AtNRGA1, AtMPC2, and AtMPC3) are essential subunits of the AtMPC complexes and required for Cd tolerance in Arabidopsis.
ScMPC Mutants Showed Similar Cd Sensitivity in Yeast
Yeast (Saccharomyces cerevisiae) is a common eukaryotic model organism for fundamental studies of cell biology. As such, yeast was employed to understand the functions of MPCs in this study. As shown in Figure 4A, the growth of yeast cells of mpc1Δ was similar to that of the wild-type strain JRY472 in the absence of Cd. However, in the presence of Cd, the cell population of mpc1Δ was significantly less than that of the wild-type strain (Fig. 4A). To confirm the function of MPC1 in Cd tolerance in yeast, the yeast expressing recombination vector pScMPC1::ScMPC1 was constructed. A serial dilution assay was conducted to test Cd tolerance. As shown in Figure 4B, ScMPC1 could recover the Cd sensitivity of mpc1Δ. We also tested all the yeast mutants of ScMPCs under Cd treatment. The mpc1Δ, mpc2Δmpc3Δ, and mpc1Δmpc2Δmpc3Δ mutants grew weaker than the wild-type JRY472 and other mutants (Fig. 4C; Supplemental Fig. S6, A–F). These results indicated that both ScMPC1 and other ScMPCs (ScMPC2 and ScMPC3) are necessary elements in Cd tolerance in yeast (Fig. 4C; Supplemental Fig. S6E), which is similar to the Cd-tolerant phenotype in Arabidopsis (Figs. 1A, 3, C and D, and 4, A and C; Supplemental Fig. S6, A–F). Considering that the MPC mutants showed similar Cd tolerance in Arabidopsis and yeast, the Cd tolerance mechanism regulated by MPC could be similar.
Figure 4.
Cd tolerance test in yeast. A, Yeast wild-type (WT) strain and mpc1Δ cells were grown at 30°C in adenine-supplemented yeast extract peptone dextrose (YPAD) liquid medium and exposed to 50 µm CdCl2 at the concentration of 0.3 OD600. Cell density was monitored with the A600 at 12, 15, 18, 21, 24, and 27 h after treatment. Error bars indicate sd from three independent experiments. B, Yeast dilution bioassay with the wild-type strain, mpc1Δ transformed with pRS416, and pRS416 expressing ScMPC1 in synthetic complete medium. C, Yeast dilution bioassay with MPC mutants and the wild-type strain in YPAD medium. Triangles represent serial 10-fold dilutions (starting concentration of 0.3 OD600). Representative results from three reproducible experiments are shown. EV, Empty vector.
Loss of Function of AtMPC1 Undermines the Tricarboxylic Acid Activity during Cd Stress
As AtMPC1 is the essential component of these three AtMPC complexes and the loss of function of AtMPC1 showed comparable Cd sensitivity to those mutants with the loss of all AtMPCs in Arabidopsis (Fig. 3D), we used mpc1-1 instead of the AtMPC quadruple mutants for further investigation of the molecular mechanism of the AtMPC complexes in response to Cd stress. Previously, AtNRGA1 has been demonstrated to be localized in mitochondria (Li et al., 2014). Since AtNRGA1 interacts with AtMPC1 and AtMPC1 interacts with other AtMPCs, all three AtMPC complexes might localize and work in mitochondria in Arabidopsis. In yeast and some animals, deletion of MPC can affect the transport of pyruvate from the cytoplasm into the mitochondrial matrix to generate acetyl-CoA and drive the tricarboxylic acid cycle. In plants, there is no evidence showing that MPC is involved in these processes. To verify this, the wild-type seedlings and mpc1-1 without or with the Cd treatment were collected for the acetyl-CoA measurement. As shown in Figure 5A, the wild-type and mutant plants had a comparable acetyl-CoA content under the control condition. However, in the presence of Cd, the acetyl-CoA content significantly decreased mpc1-1 compared with the wild-type plants. Metabolic analyses revealed that the concentration of pyruvate was elevated in mpc1-1 without or with Cd stress, whereas the levels of tricarboxylic acid cycle intermediates were significantly reduced in mpc1-1 with Cd stress (Fig. 5, B–G).
Figure 5.
Analysis of tricarboxylic acid-related metabolites in Arabidopsis and isolated mitochondria. A to G, Acetyl-CoA (A), pyruvate (B), oxaloacetate (C), citrate (D), succinate (E), fumarate (F), and malate (G) content in Arabidopsis seedlings germinated on 0.5× MS plates for 3 d and transferred to plates without or with 50 µm CdCl2 for culturing for another 7 d. H and I, Acetyl-CoA (H) and pyruvate (I) content in mitochondria isolated from Arabidopsis leaves treated without or with 50 µm CdCl2. Error bars indicate sd of three independent experiments. P values from Student’s t test were determined for mpc1-1 lines compared with the wild-type (WT) plants without or with 50 µm CdCl2: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
To further identify the difference at the organelle level, the measurement of the acetyl-CoA and pyruvate content was conducted in the isolated Arabidopsis mitochondria. As shown in Figure 5, H and I, both acetyl-CoA and pyruvate concentrations were significantly decreased in mpc1-1 with Cd stress. These results indicated that the AtMPC1 mutants could not generate acetyl-CoA efficiently under Cd stress. Since acetyl-CoA is the predominant substrate of the tricarboxylic acid cycle, the lack of acetyl-CoA will interrupt the tricarboxylic acid metabolic activity. In the absence of Cd, the acetyl-CoA concentration in mpc1-1 was normal, which is possibly due to the existence of a compensatory bypass pathway for acetyl-CoA generation. However, the compensatory pathway could be interrupted by Cd stress. Recently, several alternative pathways have been found to sustain acetyl-CoA generation and tricarboxylic acid activity in the absence of MPC function in animal cells. Ala can be transported across the mitochondrial inner membrane by unidentified Ala carriers and converted to pyruvate by Ala aminotransferase in the mitochondrial matrix (McCommis et al., 2015). Glu can be oxidized by Glu dehydrogenase to supplement α-ketoglutarate (Du et al., 2013; Bender and Martinou, 2016). The mitochondrial malic enzyme can convert malate to pyruvate and support pyruvate, which generates acetyl-CoA inside mitochondria (Sauer et al., 1980; Pongratz et al., 2007). All of them could provide pyruvate and drive the tricarboxylic acid cycle without MPC, which transports pyruvate from cytosols directly.
MPCs Balance Glc and Glu Consumption to Maintain Tricarboxylic Acid Activity in Arabidopsis
GSH is a well-known chelator of Cd2+ and thus alleviates Cd stress in the eukaryotic cells (Marrs, 1996; Cobbett et al., 1998). GSH also functions as a major redox buffer for oxidative stress tolerance caused by Cd in plants (Yadav, 2010). As shown in Figure 6A, in the absence of Cd, the GSH content decreased significantly in mpc1-1, indicating that the synthesis of GSH was interrupted by the loss of function of the AtMPC complexes. In the presence of Cd, the GSH level in mpc1-1 was much higher than that in the wild-type plants (Fig. 6B). The GSH elevation was probably stimulated by Cd accumulation (Fig. 2A). Glu is the major source of GSH (Quastel et al., 1923; Stewart and Tunnicliffe, 1925; Griffith et al., 1978). In mammal cells, Glu oxidation is the necessary bypass for maintaining the tricarboxylic acid cycle during the impaired mitochondrial pyruvate transport (Yang et al., 2014). In this pathway, a portion of Glu can be converted by glutaminase and then deaminated by Glu dehydrogenase to drive the tricarboxylic acid cycle (Du et al., 2013; Bender and Martinou, 2016). However, in plants, there is no evidence of the Glu supplement pathway.
Figure 6.
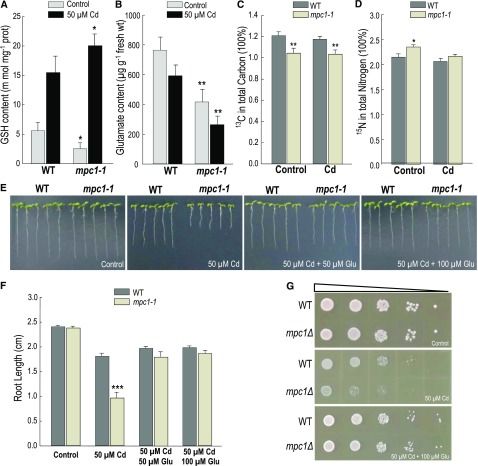
The provision of Glu to MPC1 mutants restored the wild-type (WT) level of Cd tolerance. A and B, GSH (A) and Glu (B) content in Arabidopsis seedlings that were germinated on 0.5× MS plates horizontally for 3 d and transferred to plates without or with 50 µm CdCl2 for culture for another 7 d. Error bars indicate sd of three independent experiments. C and D, Isotopic tracing analysis of stable isotopes 13C and 15N in 10-d-old seedlings. Arabidopsis seedlings were hydroponically germinated and grown on 0.5× MS solution containing 50 µm [13C]Glc and [15N]Glu without or with 50 µm CdCl2 for 10 d. E, Arabidopsis plants germinated on 0.5× MS plates for 3 d were transferred to plates without or with 50 µm CdCl2 or 50 µm CdCl2 plus 50 µm Glu for another 7 d. Representative results from three reproducible experiments are shown. F, Average root length of seedlings cultured in the same growth condition as that in E. The root length of five seedlings of each class was measured as the mean value (removing the top and lowest values). Error bars indicate sd from three independent experiments. P values from Student’s t test were determined for mpc1-1 lines compared with the wild-type plants without or with 50 µm CdCl2: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. G, Yeast dilution bioassay with ScMPC1 mutant and the wild-type strain in YPAD medium. Triangles represent serial 10-fold dilutions (starting concentration of 0.3 OD600).
As shown in Figure 6B, Glu decreased in mpc1-1 compared with the wild-type plants, and the decrease was more significant in mpc1-1 with the presence of Cd. Considering that pyruvate, which is mainly generated by Glc, needs MPC to directly participate in the tricarboxylic acid cycle and Glu is a putative necessary nutrient element, isotopic tracing analysis of Glc and Glu metabolites was conducted by feeding stable isotope [13C]Glc and [15N]Glu in 10-d-old Arabidopsis seedlings on 0.5× MS hydroponic medium without or with 50 μm Cd. As shown in Figure 6, C and D, without Cd stress, the abundance of 13C decreased in mpc1-1 compared with that of the wild-type plants. However, the abundance of 15N increased in mpc1-1 compared with that in the wild-type lines. These results indicated that the consumption of Glu was elevated to replenish the loss of Glc, which is the main source of pyruvate, caused by the impaired AtMPCs in Arabidopsis. Upon Cd stress, the abundance of 13C also decreased in mpc1-1 compared with that in the wild-type lines. Nonetheless, the abundance of 15N in mpc1-1 was similar to that in the wild-type lines (Fig. 6, C and D). It is probable because the Cd stress reduced the efficiency of Glu utilization. Taken together, in the presence of Cd, without AtMPC, the bypass provided Glu to produce more GSHs to cope with Cd stress and could not replenish the tricarboxylic acid cycle efficiently. The growth rate of mpc1-1 was affected by the decrease of the tricarboxylic acid activity, which is at the center of cellular energy metabolism and chemical synthesis. To further confirm this, we added a certain concentration (50 or 100 μm) of Glu to 0.5× MS and YPAD solid medium with 50 μm Cd. Under the same tolerance test condition, both MPC1 mutants in Arabidopsis and yeast significantly recovered their Cd tolerance (Fig. 6, E–G). In conclusion, the AtMPC complexes were the key element in maintaining the acetyl-CoA production and driving the tricarboxylic acid cycle and thus released the Glu supplementary pathway to generate GSH to alleviate Cd stress in Arabidopsis.
AtMPC Is Required for Cd2+ Exclusion through ATP Support
Cd2+ transporters play a major role in plant Cd2+ accumulation or exclusion. Various types of Cd2+ transporters have been identified in plants, such as ABC transporters and HMAs (Mills et al., 2005; Kim et al., 2007; Morel et al., 2009). These transporters can be divided into two groups according to their subcellular localization: cell membrane- and tonoplast-Cd2+ transporters; the former group includes AtHMA4 and AtPDR8, which can transport Cd2+ out of cells (Mills et al., 2005; Kim et al., 2007), and the latter includes AtHMA3, which can drive Cd2+ into vacuoles (Morel et al., 2009). All these Cd2+ transporters consume ATP to remove Cd2+ from cytosols to protect basic metabolism from Cd2+ interruption. ATP is primarily produced through the tricarboxylic acid cycle, and acetyl-CoA is the main source. As shown in Figure 5A, the acetyl-CoA content decreased in mpc1-1 under Cd stress and consequently reduced the ATP production. Considering that root is the predominant tissue to absorb or exclude Cd2+, we measured the ATP content of root protoplasts of mpc1-1 and the wild-type plants at the same growth condition. Consistent with the change of the acetyl-CoA content, the ATP level also decreased in mpc1-1 under the Cd treatment (Fig. 7A). With the deletion of ATP, Cd2+ transporters in the cellular membrane system could not drive Cd2+ sufficiently. Some Cd2+ transporters localized in root epidermal cell membranes, such as AtPDR8, can transport Cd2+ out of plantlets from root directly (Kim et al., 2007). Previously, exogenous ATP has been identified to be associated with plant physiological reactions in plant roots (Kim et al., 2006). Thus, to verify whether ATP affects Cd2+ absorption or exclusion in epidermal cells of roots, Cd2+ flux was measured in the same place of wild-type plants and mpc1-1 without or with 1 mm ATP (Supplemental Fig. S7). As shown in Figure 7, B and C, the Cd2+ influxes were significantly influenced and changed into effluxes by adding 1 mm ATP in the roots of both the wild-type lines and mpc1-1. Moreover, the Cd fluxes in the wild-type lines changed more significantly than those in mpc1-1. The Cd flux changed from about −10 to about 70 pmol cm−2 s−1 compared with that from about −20 to about 20 pmol cm−2 s−1 (Fig. 7, B and C). It seems that ATP helps to export Cd2+ more efficiently in the presence of MPC. In conclusion, our results explain why the Cd2+ influx was elevated in the root of mpc1-1 (Fig. 2, C and D), as the activity of the Cd2+ transporters localized in the epidermal cells of roots were interrupted by the decrease of the ATP level caused by the loss of function of the AtMPC complexes under the treatment of Cd.
Figure 7.
ATP changes Cd2+ flux in Arabidopsis roots. A, ATP content in root cells of Arabidopsis seedlings that were germinated on 0.5× MS plates for 3 d and transferred to plates without or with 50 µm CdCl2 for culture for another 7 d. Error bars indicate sd of three independent experiments. P values from Student’s t test were determined for mpc1-1 lines compared with the wild-type (WT) plants without or with 50 µm CdCl2: *, P < 0.05. B, The mean Cd2+ fluxes in the roots of 10-d-old seedlings, which were germinated in 0.5× MS medium and then transferred to 0.5× MS with 50 µm CdCl2, were measured at different time points for 3 min after exposing the seedlings to measuring solution with 50 μm CdCl2 or 50 μm CdCl2 plus 1 mm ATP. Error bars indicate sd from three independent experiments. P values from Student’s t test were determined for mpc1-1 compared with the wild-type plants. C, Cd2+ fluxes in the roots of 10-d-old seedlings, which were germinated in 0.5× MS medium and then transferred to 0.5× MS with 50 µm CdCl2, were recorded every 6 s for 3 min after the seedlings were exposed to measuring solution with 50 μm CdCl2 or 50 μm CdCl2 plus 1 mm ATP. The number of plants measured was as follows: wild type treated with Cd, five; mpc1-1 treated with Cd, five; wild type treated with Cd and ATP, five; and mpc1-1 treated with Cd and ATP, five.
DISCUSSION
Loss of function of all MPCs in Arabidopsis and yeast is viable in 0.5× MS and rich medium (Figs. 3D and 4C; Supplemental Fig. S6F). These results were consistent with previous studies in yeast (Bricker et al., 2012; Herzig et al., 2012; Timón-Gómez et al., 2013), which is due to the bypasses involving Glu utilization (Du et al., 2013; Vacanti et al., 2014; Yang et al., 2014). In this study, without Cd stress, even the AtMPC pathway is interrupted. It appears that the Glu bypass could complement the loss of acetyl-CoA and ATP through the tricarboxylic acid cycle (Fig. 8A). In the presence of Cd, AtMPC maintains the tricarboxylic acid cycle and ATP production. Moreover, AtMPC alleviates the consumption of Glu for tricarboxylic acid activity, which generates more GSH to enhance Cd tolerance in plants (Fig. 8B). Without AtMPC, the main pathway for the generation of acetyl-CoA is interrupted, and Glu is shared to generate GSH to regulate Cd stress. With the consumption of Glu, there is insufficient Glu to supplement the tricarboxylic acid cycle, leading to a decreased tricarboxylic acid activity and ATP production (Fig. 8B). Additionally, since most Cd transporters in plants, such as ABC transporters, HMA family members, vacuolar pumps, and a set of tonoplast transporters, need ATP to accomplish their functions (Finkemeier et al., 2003; Sharma et al., 2004, 2016; Kim et al., 2007; Morel et al., 2009), the deletion of ATP will interrupt the absorbance or exclusion of Cd2+. In the long-time evolutionary process, plants have evolved plenty of transporters, which can be used to prevent the invasion of heavy metals or metal ions by segregating them mainly into vacuoles and removing them from cells or even the plant.
Figure 8.
Proposed model of MPC function in Cd tolerance. Loss of MPC in the control situation (A) and in the presence of MPC or loss of MPC under Cd stress (B) is shown. Thick black full lines indicate enhanced pathways; black full lines indicate normal pathways; black segment lines indicate reduced pathways; black dotted lines indicate substantially reduced pathways; red full lines indicate increased content; and yellow full lines indicate reduced content or activity.
For Cd transporters localized in tonoplast or other membrane systems in the cytosol, the deletion of ATP will affect their ability to transport Cd2+ into vacuoles or membrane systems. For Cd2+ transporters localized within the cell membrane, the deletion of ATP will affect their ability to remove Cd2+ from cells. Since some Cd2+ transporters localized in root epidermal cells, such as AtPDR8, exclude Cd2+ from the root directly (Kim et al., 2007), the deletion of ATP will interrupt the Cd2+ efflux from the root. This explains why the Cd2+ influx increased near root epidermal cells (Figs. 2, C and D, and 7, B and C). The GSH level was elevated in mpc1-1, suggesting that more damage inside cells was caused by the higher Cd content (Sun et al., 2005b; Tang et al., 2005) compared with the wild-type plants, and the cells need to produce more GSH to remove the excessive Cd2+ (Figs. 2A and 6A). Supposedly, as with the deletion of ATP, both of the types of Cd2+ transporters cannot transport Cd2+ efficiently and cause more damage to the cytosolic metabolism. Overall, the Cd toxicity and loss of pyruvate, which import from cytosol, result in the decreases of acetyl-CoA and ATP content in the AtMPC1 mutants. More importantly, MPC is also required for Cd tolerance in yeast. However, the Cd2+ accumulation did not change in the types of yeasts tested with 50 μm Cd (Supplemental Fig. S8). Compared with the plant, yeast is the primary monad and has its Cd transporters, such as YOR1P or YCF1 (Li et al., 1997; Nagy et al., 2006). As the ATP level decreases, both Cd transporters will be affected, and the Cd content of the whole cell will not be changed. For Arabidopsis, the transporters have evolved more advanced functions and been distributed in different tissues, such as AtPDR8, which specifically outflows Cd2+ in root epidermal cells (Kim et al., 2007). Since the root epidermis is the main tissue for plants to absorb Cd2+ and the first line of defense for plants to exclude Cd2+, the root-specific Cd transporter will change the Cd content of the whole plant. Although the Cd transporters are different in plants and yeasts, the basic Cd tolerance mechanism should be similar.
Previous studies on heavy metal tolerance have been conducted in animals, plants, and microorganisms. The pathways and elements that regulate Cd stress are significantly different in each species. Chelating materials, transporters, and systems maintaining the cell environment are involved in heavy metal detoxification. Although some Cd tolerance pathways have been well established in plants and microorganisms, they are not consistent with those in animals. For example, some transporters and Glc can transport Cd2+ into vacuoles to alleviate Cd stress (Li et al., 1997; Song et al., 2003; Shi et al., 2015), and some Cd resistance genes are specifically expressed in roots (Kim et al., 2007; He et al., 2016). Since MPC functions that refer to tricarboxylic acid activities and Glu supplementary pathways should be conserved in plants and animals (Jacoby et al., 2012; Zhang and Fernie, 2018), the tolerance mechanism is possibly appropriate for animals as well. Diseases caused by dietary intake of Cd are a significant human health concern (McLaughlin et al., 1999; Waalkes, 2003). Thus, our results also provide a basis for developing strategies to prevent Cd toxicity in animal bodies.
The MPC proteins were firstly identified in 2012 (Bricker et al., 2012; Herzig et al., 2012). The major biological energy source is Glc, which can be converted to ATP and pyruvate in glycolysis. During the past several years, studies mainly focused on the understanding of the MPC function in human cancers and related diseases in animals and microorganisms. Our group found that AtNRGA1 (AtMPC2L) and AtMPC1, two members of the AtMPC family, interact with each other and are involved in the ABA-related stoma regulation (Li et al., 2014; Shen et al., 2017). These AtMPC proteins may regulate drought tolerance in Arabidopsis. However, there are no studies on the function of pyruvate metabolism and the tricarboxylic acid cycle in plants. Technically, it is difficult to test the difference of growth phenotypes and tricarboxylic acid products in plants by using nutritional selection for the selection of the metabolism pathways (Bricker et al., 2012; Herzig et al., 2012). In this study, the results indicated that AtMPC was required for Cd tolerance in plants. Both of them are imperative in plant growth and development, including seed germination and root elongation. In the presence of Cd, with the deletion of acetyl-CoA, nearly all metabolites produced in the tricarboxylic acid cycle would be deleted. Taken together, this tolerance regulation mechanism involves multiple important elements. Considering the conserved regulation manner of MPC in plants, microorganisms, and animals, MPC is supposedly the conserved regulator of Cd resistance. Manipulation of the expression of MPC can be used as a strategy to regulate Cd stress in plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) wild-type Columbia-0 (SALK_6000) and T-DNA insertion mutant nrga1 (SALK_050950) have been described previously (Li et al., 2014). Single mutant mpc1-1 (SALK_008945) and AtMPC1 complement lines (C-1 and C-2) driven by its native promoter were kindly provided by Jianlin Shen as described previously (Shen et al., 2017). Mutants mpc1-2 (SALK_007363C) and mpc3-1 (SALK_11024) were obtained from the Arabidopsis Biological Resource Center and confirmed for the T-DNA insertion through PCR analysis. Mutants mpc2-1 and mpc2-2 were created using the CRISPR/Cas9 system with the Columbia background. Double mutants mpc1-1nrga1 and mpc1-1mpc3 were generated by a normal hybridization method. Double mutants mpc1-1mpc2-3 and mpc1-1mpc2-4 were created using the CRISPR/Cas9 system with the mpc1-1 background. Triple and quadruple mutants nrga1mpc2-5mpc3-2, nrga1mpc2-6mpc3-3, mpc1-1nrga1mpc2-7mpc3-4, and mpc1-1nrga1mpc2-7mpc3-5 were also generated using the CRISPR/Cas9 system, including double target adopter cassettes with the nrga1 and mpc1-1nrga1 background. Arabidopsis plants were grown in a controlled environment at 22°C/20°C in a 16-h-light/8-h-dark photoperiod. Seeds used for phenotypic assays were harvested at the same time. The tolerance test was performed on plates with 0.5× MS solid medium without or with 50 µm CdCl2 or first cultured seeds were germinated on plates without stress for 3 d and then transferred to plates with 50 µm CdCl2 for another 7 d.
For the Cd2+ concentration measurements, Arabidopsis plants were grown in soil for 4 weeks and then bottom flooded once with 0.4 L of 50 µm CdCl2 solution per pot (pot size of 0.4 L) followed by normal watering without Cd until maturity. Shoots and seeds were collected for sampling. For 10-d-old seedlings, the Cd2+ concentration was measured as previously described (Gong et al., 2003) with minor modification. The Cd2+ contents were measured with an atomic absorption spectrometer (Shimadzu; AA-7000).
Nicotiana benthamiana plants were grown in soil in a controlled environment at 28°C with a 14-h-light/10-h-dark photoperiod. Infiltration was done on 3- to 4-week-old plants. Single clones of GV3101 carrying different vectors were inoculated in the yeast extract peptone agar medium containing 10 μg mL−1 rifampicin and 50 μg mL−1 kanamycin and then grown for 1 to 2 d at 28°C. A total of 1 mL of near-saturation Agrobacterium tumefaciens was inoculated in 20 mL of fresh yeast extract peptone agar medium containing 10 mg mL−1 rifampicin and 50 mg mL−1 kanamycin and then grown for about 6 h (until the absorbance [OD600] reached 0.8). Cells were collected by centrifugation (4,000 rpm, 5 min), resuspended in 10 mL of 0.5× MS medium with 50 µm acetosyringone, and continued to incubate for another 2 h. An equal volume of infiltration buffer (10 mm MgSO4, 200 µm acetosyringone, and 10 mm MES) was mixed with the samples before the infiltration was performed using a syringe. For the Co-IP assay, two different samples with different proteins were mixed equally and then mixed with infiltration buffer. After infiltration, plants were kept in a dark chamber with high humidity for one night and then placed in a normal growth chamber for about 2 d.
Molecular Construct
Binary vectors p35S::FLAG:AtMPC1 and p35S::C-MYC:AtNRGA1 for Co-IP were provided by Jianlin Shen as previously described (Shen et al., 2017). For the constructs AtMPC2 and AtMPC3, the open reading frame (ORF) without the ATG codon was PCR amplified from cDNA with the primer sets AtMPC2-1F/AtMPC2-1R and AtMPC3-1F/AtMPC3-1R, respectively. The fragments were inserted into pCAMBIA1307 with C-MYC using the XbaI and XhoI sites to yield p35S::C-MYC:AtMPC2 and p35S::C-MYC:AtMPC3.
For the BiFC vectors, the AtMPC1 ORF without the stop codon was amplified with the primer set AtMPC1-F/AtMPC1-R and then inserted into the BamHI site on pDOE01 (Gookin and Assmann, 2014) using the In-Fusion HD cloning kit (catalog no. 011614; Clontech) to yield the vector AtMPC1:NYFP-CYFP. The AtMPC2 and AtMPC3 ORFs without the stop codon were amplified with the primer sets AtMPC2-2F/AtMPC2-2R and AtMPC3-2F/AtMPC3-2R and then inserted into SnaBI on pDOE01 harboring AtMPC1:NYFP or pDOE01 separately to yield the constructs AtMPC1:NYFP-AtMPC2:CYFP, AtMPC1:NYFP-AtMPC3:CYFP, NYFP-AtMPC2:CYFP, and NYFP-AtMPC3:CYFP.
For the yeast (Saccharomyces cerevisiae) expression vectors, the ScMPC1 ORF was PCR amplified from cDNA of S. cerevisiae strain JRY472 with the primer set ScMPC1-F/ScMPC1-R and then inserted into the BamHI site on pRS416 using the In-Fusion HD cloning kit (catalog no. 011614; Clontech).
For the CRISPR/Cas9 vectors, target sequences and primers of the target adopters were designed according to the descriptions on the Web site http://crispr.mit.edu and the published protocol (Yan et al., 2015). For the target adopters of AtMPC2 and AtMPC3, which were generated by annealing, the primers were ligated with the small guide RNA (sgRNA) vector digested with BsaI to yield intermediate vectors separately. Cassettes of the target adopter in fusion with the sgRNA sequence were cut from the intermediate vector with SpeI. For single target modification, the cassette containing the target adopter of AtMPC2 or AtMPC3 was inserted into the SpeI site on pCAMBIA1300-pYAO:Cas9 (Yan et al., 2015) separately. For double target editing, the sgRNA vector with the AtMPC3 target adopter was digested with SpeI and NheI to yield the cassette containing AtMPC3 target adopter with sgRNA, and then the cassette was ligated with the sgRNA vector containing the AtMPC2 target adopter digested with SpeI to form an intermediate construct harboring two types of cassettes. The two ligated cassette fragment was cut with SpeI from the sgRNA vector and inserted into pCAMBIA1300-pYAO:Cas9 to form a double target modification construct. All primers are listed in Supplemental Table S1.
RT-Quantitative PCR
RNA extraction was conducted using TRIzol reagent (Sigma). Reverse transcription was performed using a PrimeScript RT reagent kit with gDNA Eraser (catalog no. RR047A; TaKaRa). RT-quantitative PCR was conducted with SYBR Premix Ex Taq (catalog no. 4913914001; Roche) with Actin2 (At3g18780) as the internal control. All primers are listed in Supplemental Table S1.
Measurements of Net Cd2+ Fluxes
The net Cd2+ fluxes were measured using the Noninvasive Microtest Technology (NMT100; YoungerUSA) and iFluxes/imFluxes 1.0 (YoungerUSA) software at Xuyue (Beijing) Science and Technology. The Cd2+ microsensors were prepared as described previously (Ma et al., 2015) with minor modification. Prepulled and silanized microsensors (Φ1.5 ± 0.5 μm; XY-CGQ-02; YoungerUSA) were first filled with a backfilling solution (50 µm CdCl2 + 0.1 mm KCl) to approximately 1 cm from the tip. The micropipettes were front filled with 40- to 50-μm columns of selective liquid ion-exchange cocktails (Cd2+ LIX, XY-SJ-Cd; YoungerUSA). An Ag/AgCl wire electrode holder (XY-DJGD; YoungerUSA) was inserted into the back of the electrode to make electrical contact with the electrolyte solution. A YG003-Y05 (YoungerUSA) was used as the reference electrode. Before the Cd2+ flux measurement, the microelectrodes were calibrated in 50 µm CdCl2. Only the electrodes with a Nernstian slope greater than 22 mV per decade were used.
Seedlings were germinated on 0.5× MS solid medium for 3 d and then transferred to 0.5× MS with 50 µm CdCl2 for 7 d. Roots were rinsed with double-distilled water and incubated in the measuring solution to equilibrate for 5 min. After that, roots were transferred to a measuring chamber containing 15 mL of a fresh measuring solution. Cd2+ was monitored in the following measuring solutions (0.1 mm KCl, 0.05 mm CdCl2, and 0.3 mm MES, pH 5.8) without or with 1 mm ATP. The measuring chamber was mounted on the micromanipulator, and the electrodes were positioned close to the roots of seedlings at 400 µm from the tip (Fig. 3A). Cd2+ fluxes were measured by moving the Cd2+-selective microelectrode between two positions in a preset excursion of 20 µm in a perpendicular direction to the cell surface, completing a 6-s-per-point cycle for more than 3 min. All experiments were repeated using independent lines at least three times, and their mean values ± sd are presented.
In Vivo Co-IP Assay
N. benthamiana leaves coinfiltrated with two types of constructs were harvested and ground with 2 mL of immunoprecipitation buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 1% [v/v] Triton X-100) with freshly prepared DTT (1 mm) and 1× protease inhibitor cocktail (catalog no. 04693132001; Roche) on ice. Protein extracts were centrifuged at 14,000g for 30 min at 4°C. A total of 100 µL of supernatant was stored at −80°C for input immunoblot, and the remaining 0.8 mL of supernatant was incubated with 20 µL of magnetic beads (catalog no. 88804; Thermo) with or without anti-FLAG antibody (1:400; catalog no. CW0287M; CWBio) overnight at 4°C. The beads were washed four times with the immunoprecipitation buffer and then boiled for 5 min with 30 µL of loading buffer for western blot using anti-C-MYC antibody (1:5,000; catalog no. CW0259M; CWBio).
Western Blot
Input and immunoprecipitated protein samples (20 µL) were loaded onto 15% (w/v) SDS-PAGE gels and transferred to Amersham Hybond-P (catalog no. RPN303B; GE Healthcare) using a Mini-PROTEAN Tetra system (Bio-Rad). The membranes were washed with TBST (20 mm Tris-Cl, 150 mm NaCl, and 0.05% [v/v] Tween 20), blocked with TBST containing 5% (w/v) nonfat milk (TBSTM) for 2 h, and incubated with anti-FLAG antibody (1:5,000; catalog no. CW0287M; CWBio) or anti-C-MYC antibody (1:5,000; catalog no. CW0259M; CWBio) in TBSTM for 5 h at room temperature. The membranes were washed four times (5 min each) and then incubated with goat anti-mouse IgG (catalog no. CW0110s; CWBio) in fresh TBSTM for about 2 h. After the membranes were washed six times with TBST, the bound antibodies were monitored with the enhanced chemiluminescence substrate (catalog no. 32132; Thermo).
BiFC Assays
BiFC constructs including MOL1 and CaM1 (positive control; Gookin and Assmann, 2014) were transiently expressed in tobacco (Nicotiana benthamiana) leaves by A. tumefaciens-mediated infiltration. The YFP fluorescence of tobacco leaves was imaged 2 d after infiltration using a Zeiss LSM780 confocal laser scanning microscope.
Mitochondria Isolation
Arabidopsis plants were grown in soil for 3 weeks and then bottom flooded once using 0.2 L of water without or with 50 μm CdCl2 per pot (pot size of 0.2 L), followed by normal watering without Cd. Mitochondria were isolated from Arabidopsis leaves 1 week after the 50 μm CdCl2 treatment as described (Whelan and Murcha, 2015).
Acetyl-CoA and ATP Quantification
Acetyl-CoA was extracted from the seedlings and isolated mitochondria, and its concentration was estimated as previously described (Xu et al., 2016) with minor revisions. Seedlings were germinated on 0.5× MS in a controlled environment at 22°C/20°C in a 16-h-light/8-h-dark photoperiod for 3 d and transferred to plates with or without 50 μm CdCl2 for 7 d. The 10-d-old seedlings were ground, and the tissue powder (about 0.1 g) was recorded and used for protein quantification (Bradford, 1976). For the measurement of the acetyl-CoA concentration in the isolated mitochondria, 100 μL of mitochondria solution was used for each sample (Bradford, 1976). The acetyl-CoA content in both the whole seedlings and isolated mitochondria was measured according to the manufacturer’s protocols (Sigma; MAK039).
The ATP content was measured using the luciferin-luciferase method (Manfredi et al., 2002) in the isolated root protoplasts. Roots (2 cm from the tip) of the 10-d-old Arabidopsis seedlings were cut into small fragments and incubated in the enzyme solution (1.5% [w/v] cellulase, 0.1% [w/v] Macerozyme, 0.8 m mannitol, 0.2 m KCl, and 0.1 m MES, pH 5.7) at room temperature for 3 h. The enzyme solutions with the root fragments were filtered, and the cells were collected at 100 g for 5 min. After washing with the W5 solution (3 m NaCl, 1 m CaCl2, 0.2 m KCl, 0.1 Glc, and 0.1 m MES, pH 5.7) twice, the cells were incubated with or without 50 μm CdCl2 for 2 h. To avoid metabolic degradation of ATP, cells were suspended in 2.5% (v/v) TCA and then neutralized by 0.75 m Tris-acetate buffer (pH 7.75). Cellular ATP was measured by ENLITEN rLuciferase/Luciferin reagent (Promega) according to the manufacturer’s instructions. Luminescence was measured using a multimode microplate reader (centro XS3 LB 960; Berthold Technologies). For the measurements, a 3-s delay time after rLuciferase/Luciferin reagent loading was used. Another 25 μL of cell suspension was disrupted by ultrasound and used for protein quantification.
Measurement of GSH, Pyruvate, Glu, and Tricarboxylic Acid Intermediates
Arabidopsis seedlings were germinated on 0.5× MS in a controlled environment at 22°C/20°C in a 16-h-light/8-h-dark photoperiod for 3 d and transferred to plates with or without 50 μm CdCl2 for 7 d. Tissues were homogenized in liquid nitrogen and stored at −80°C before the measurement. During the analysis of the contents of GSH, pyruvate (including pyruvate in isolated mitochondria), and tricarboxylic acid intermediates, the protein contents from the same sample loading solution were measured according to a classical method (Bradford, 1976). For the Glu analysis, the same growth condition and treatment were conducted as mentioned above. The fresh weight of seedlings was measured, and the seedlings were frozen quickly in tubes with liquid nitrogen. The measurement of GSH, pyruvate, and Glu in tissues was carried out by Suzhou Comin Biotechnology using reverse-phase HPLC according to published methods (Priscila del Campo et al., 2009; Paulose et al., 2013).
Stable Isotopic Tracing Assay
Stable isotopic tracing analysis was conducted by an IsoPrime100 mass spectrometer (Isoprime) as described with minor revision (Agnihotri et al., 2014). Briefly, Arabidopsis seedlings were hydroponically grown in 0.5× MS liquid solution with 50 μm [13C]Glc and 50 μm [15N]Glu (IsoReag) with or without 50 μm CdCl2. Ten-day-old seedlings were washed five times by clean water and then collected and dried at 60°C for 48 h. The dried samples (50 mg each) were packed in tin cups and then burned in the combustion tube (heated to about 1,000°C). Air generated by burning solid samples was passed through the reduction tube (heated to about 600°C) filled with activated metallic copper and reduced to N2 and CO2. N2 and CO2 were separated by passing through the chromatographic column (70°C for N2, 100°C for CO2) and introduced to the mass spectrometer to measure the abundance of 13C and 15N according to the abundances of standard samples. At the same time, total C and N were measured in a separate procedure. The values were calculated as the percentage of 13C and 15N compared with the total C and N.
Yeast Cd Stress Assay
S. cerevisiae JRY472 wild type and mutants were grown in the YPAD liquid solution, and the transformed yeast cells were grown on SC selection liquid medium. Overnight cultures in the YPAD or SC solutions with essential supplements were diluted to OD600 of about 0.1 and grown to OD600 of 0.3. After 10-fold serial dilution, 3 µL of each sample was spotted onto YPAD solid plates with or without 50 µm Cd and incubated at 30°C for 2 d. For growth curve tests, overnight cultures in the YPAD solutions were diluted to OD600 of about 0.1 and grown to OD600 of about 0.5. The cultures with an OD600 of about 0.5 were diluted to OD600 of 0.3 with or without 50 µm Cd and incubated with shaking. After 12 h, the OD600 of the cultures was measured every 3 h until 27 h.
Statistical Analysis
Student’s t test was used to test the significance of root length, fresh weight, Cd content, Cd2+ flux, metabolite, 13C in total C element (percentage), 15N in total N element (percentage), ATP concentration, and transcript abundance between wild-type lines and AtMPC1 mutants or transgenic lines.
Accession Numbers
Sequence data can be found in The Arabidopsis Information Resource database under accession numbers At5g20090 (AtMPC1), At4g05590 (AtMPC2L/AtNRGA1), At4g22310 (AtMPC3), and At4g14695 (AtMPC2) and in The Saccharomyces Genome database under accession numbers ygl080w (ScMPC1), yhr162w (ScMPC2), and ygr243w (ScMPC3).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. MPC homolog proteins in the indicated species.
Supplemental Figure S2. Transcript abundance identification.
Supplemental Figure S3. Sequences of mutant alleles of MPC2 or MPC3 edited by the CRISPR/Cas9 system.
Supplemental Figure S4. Phenotyping analysis of mpc1-1, wild-type, and AtMPC1 complemental plants without or with Cd.
Supplemental Figure S5. Localization of microelectrodes in roots.
Supplemental Figure S6. The growth curve of wild-type JRY472 and mpc mutants.
Supplemental Figure S7. Localization of microelectrodes in roots.
Supplemental Figure S8. Cd concentration in yeast cells.
Supplemental Table S1. Primer sequences.
Acknowledgments
We thank Dr. Jared Rutter (University of Utah) for yeast mpcΔ strains and the pRS416 vector, Dr. Qi Xie (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for the YAO promoter-driven CRISPR/Cas9 System, and Sarah M. Assmann (Pennsylvania State University) for the BiFC system.
Footnotes
This work was supported by the Foundation for Distinguished Young Scholars of Shandong University (61200088963137) to W.Z., the Special Foundation for Postdoctoral Innovation Projects (201603036) to L.H., the National Key Research and Development Program of China (2017YFD0101801) to J.G., and the Modern Agricultural Industrial Technology System Funding of Shandong Province (SDAIT-05-04) to J.G.
Articles can be viewed without a subscription.
References
- Agnihotri R, Kumar R, Prasad MVSN, Sharma C, Bhatia SK, Arya BC (2014) Experimental setup and standardization of a continuous flow stable isotope mass spectrometer for measuring stable isotopes of carbon, nitrogen and sulfur in environmental samples. Mapan-Journal of Metrology Society of India 29: 195–205 [Google Scholar]
- Bender T, Martinou JC (2016) The mitochondrial pyruvate carrier in health and disease: To carry or not to carry? Biochim Biophys Acta 1863: 2436–2442 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, Chen YC, Cox JE, Cardon CM, Van Vranken JG, Dephoure N, et al. (2012) A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 337: 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cailliatte R, Lapeyre B, Briat JF, Mari S, Curie C (2009) The NRAMP6 metal transporter contributes to cadmium toxicity. Biochem J 422: 217–228 [DOI] [PubMed] [Google Scholar]
- Clemens S. (2006) Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 88: 1707–1719 [DOI] [PubMed] [Google Scholar]
- Cobbett CS, May MJ, Howden R, Rolls B (1998) The glutathione-deficient, cadmium-sensitive mutant, cad2-1, of Arabidopsis thaliana is deficient in gamma-glutamylcysteine synthetase. Plant J 16: 73–78 [DOI] [PubMed] [Google Scholar]
- DalCorso G, Farinati S, Maistri S, Furini A (2008) How plants cope with cadmium: Staking all on metabolism and gene expression. J Integr Plant Biol 50: 1268–1280 [DOI] [PubMed] [Google Scholar]
- Du J, Cleghorn WM, Contreras L, Lindsay K, Rountree AM, Chertov AO, Turner SJ, Sahaboglu A, Linton J, Sadilek M, et al. (2013) Inhibition of mitochondrial pyruvate transport by zaprinast causes massive accumulation of aspartate at the expense of glutamate in the retina. J Biol Chem 288: 36129–36140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I, Kluge C, Metwally A, Georgi M, Grotjohann N, Dietz KJ (2003) Alterations in Cd-induced gene expression under nitrogen deficiency in Hordeum vulgare. Plant Cell Environ 26: 821–833 [DOI] [PubMed] [Google Scholar]
- Gong JM, Lee DA, Schroeder JI (2003) Long-distance root-to-shoot transport of phytochelatins and cadmium in Arabidopsis. Proc Natl Acad Sci USA 100: 10118–10123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gookin TE, Assmann SM (2014) Significant reduction of BiFC non-specific assembly facilitates in planta assessment of heterotrimeric G-protein interactors. Plant J 80: 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith OW, Bridges RJ, Meister A (1978) Evidence that the gamma-glutamyl cycle functions in vivo using intracellular glutathione: Effects of amino acids and selective inhibition of enzymes. Proc Natl Acad Sci USA 75: 5405–5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Ma X, Li Z, Jiao Z, Li Y, Ow DW (2016) Maize OXIDATIVE STRESS2 homologs enhance cadmium tolerance in Arabidopsis through activation of a putative SAM-dependent methyltransferase gene. Plant Physiol 171: 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Raemy E, Montessuit S, Veuthey JL, Zamboni N, Westermann B, Kunji ER, Martinou JC (2012) Identification and functional expression of the mitochondrial pyruvate carrier. Science 337: 93–96 [DOI] [PubMed] [Google Scholar]
- Jacoby RP, Li L, Huang S, Pong Lee C, Millar AH, Taylor NL (2012) Mitochondrial composition, function and stress response in plants. J Integr Plant Biol 54: 887–906 [DOI] [PubMed] [Google Scholar]
- Kim DY, Bovet L, Maeshima M, Martinoia E, Lee Y (2007) The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J 50: 207–218 [DOI] [PubMed] [Google Scholar]
- Kim MC, Panstruga R, Elliott C, Müller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze-Lefert P (2002) Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416: 447–451 [DOI] [PubMed] [Google Scholar]
- Kim SY, Sivaguru M, Stacey G (2006) Extracellular ATP in plants: Visualization, localization, and analysis of physiological significance in growth and signaling. Plant Physiol 142: 984–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CL, Wang M, Ma XY, Zhang W (2014) NRGA1, a putative mitochondrial pyruvate carrier, mediates ABA regulation of guard cell ion channels and drought stress responses in Arabidopsis. Mol Plant 7: 1508–1521 [DOI] [PubMed] [Google Scholar]
- Li ZS, Lu YP, Zhen RG, Szczypka M, Thiele DJ, Rea PA (1997) A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc Natl Acad Sci USA 94: 42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhao H, Wu L, Liu A, Zhao FJ, Xu W (2017) Heavy metal ATPase 3 (HMA3) confers cadmium hypertolerance on the cadmium/zinc hyperaccumulator Sedum plumbizincicola. New Phytol 215: 687–698 [DOI] [PubMed] [Google Scholar]
- Ma J, Cai H, He C, Zhang W, Wang L (2015) A hemicellulose-bound form of silicon inhibits cadmium ion uptake in rice (Oryza sativa) cells. New Phytol 206: 1063–1074 [DOI] [PubMed] [Google Scholar]
- Manfredi G, Yang L, Gajewski CD, Mattiazzi M (2002) Measurements of ATP in mammalian cells. Methods 26: 317–326 [DOI] [PubMed] [Google Scholar]
- Marrs KA. (1996) The functions and regulation of glutathione S-transferases in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 127–158 [DOI] [PubMed] [Google Scholar]
- McCommis KS, Chen Z, Fu X, McDonald WG, Colca JR, Kletzien RF, Burgess SC, Finck BN (2015) Loss of mitochondrial pyruvate carrier 2 in the liver leads to defects in gluconeogenesis and compensation via pyruvate-alanine cycling. Cell Metab 22: 682–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin MJ, Parker DR, Clarke JM (1999) Metals and micronutrients: Food safety issues. Field Crops Res 60: 143–163 [Google Scholar]
- Mills RF, Francini A, Ferreira da Rocha PS, Baccarini PJ, Aylett M, Krijger GC, Williams LE (2005) The plant P1B-type ATPase AtHMA4 transports Zn and Cd and plays a role in detoxification of transition metals supplied at elevated levels. FEBS Lett 579: 783–791 [DOI] [PubMed] [Google Scholar]
- Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, Vavasseur A, Richaud P (2009) AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol 149: 894–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Montigny C, Leverrier P, Yeh S, Goffeau A, Garrigos M, Falson P (2006) Role of the yeast ABC transporter Yor1p in cadmium detoxification. Biochimie 88: 1665–1671 [DOI] [PubMed] [Google Scholar]
- Paulose B, Chhikara S, Coomey J, Jung HI, Vatamaniuk O, Dhankher OP (2013) A γ-glutamyl cyclotransferase protects Arabidopsis plants from heavy metal toxicity by recycling glutamate to maintain glutathione homeostasis. Plant Cell 25: 4580–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongratz RL, Kibbey RG, Shulman GI, Cline GW (2007) Cytosolic and mitochondrial malic enzyme isoforms differentially control insulin secretion. J Biol Chem 282: 200–207 [DOI] [PubMed] [Google Scholar]
- Prasad MNV. (1995) Cadmium toxicity and tolerance in vascular plants. Environ Exp Bot 35: 525–545 [Google Scholar]
- Priscila del Campo C, Garde-Cerdán T, Sánchez AM, Maggi L, Carmona M, Alonso GL (2009) Determination of free amino acids and ammonium ion in saffron (Crocus sativus L.) from different geographical origins. Food Chem 114: 1542–1548 [Google Scholar]
- Quastel JH, Stewart CP, Tunnicliffe HE (1923) On glutathione. IV. Constitution. Biochem J 17: 586–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauser WE. (1990) Phytochelatins. Annu Rev Biochem 59: 61–86 [DOI] [PubMed] [Google Scholar]
- Sauer LA, Dauchy RT, Nagel WO, Morris HP (1980) Mitochondrial malic enzymes: Mitochondrial NAD(P)+-dependent malic enzyme activity and malate-dependent pyruvate formation are progression-linked in Morris hepatomas. J Biol Chem 255: 3844–3848 [PubMed] [Google Scholar]
- Sharma SS, Kaul S, Metwally A, Goyal KC, Finkemeier I, Dietz KJ (2004) Cadmium toxicity to barley (Hordeum vulgare) as affected by varying Fe nutritional status. Zhiwu Kexue Xuebao 166: 1287–1295 [Google Scholar]
- Sharma SS, Dietz KJ, Mimura T (2016) Vacuolar compartmentalization as indispensable component of heavy metal detoxification in plants. Plant Cell Environ 39: 1112–1126 [DOI] [PubMed] [Google Scholar]
- Shen JL, Li CL, Wang M, He LL, Lin MY, Chen DH, Zhang W (2017) Mitochondrial pyruvate carrier 1 mediates abscisic acid-regulated stomatal closure and the drought response by affecting cellular pyruvate content in Arabidopsis thaliana. BMC Plant Biol 17: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YZ, Zhu XF, Wan JX, Li GX, Zheng SJ (2015) Glucose alleviates cadmium toxicity by increasing cadmium fixation in root cell wall and sequestration into vacuole in Arabidopsis. J Integr Plant Biol 57: 830–837 [DOI] [PubMed] [Google Scholar]
- Song WY, Sohn EJ, Martinoia E, Lee YJ, Yang YY, Jasinski M, Forestier C, Hwang I, Lee Y (2003) Engineering tolerance and accumulation of lead and cadmium in transgenic plants. Nat Biotechnol 21: 914–919 [DOI] [PubMed] [Google Scholar]
- Steffens JC. (1990) The heavy-metal binding peptides of plants. Annu Rev Plant Physiol Plant Mol Biol 41: 553–575 [Google Scholar]
- Stewart CP, Tunnicliffe HE (1925) Glutathione: Synthesis. Biochem J 19: 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Wang XR, Ding SM, Yuan XF (2005a) Effects of interaction between cadmium and plumbum on phytochelatins and glutathione production in wheat (Triticum aestivum L.). J Integr Plant Biol 47: 435–442 [DOI] [PubMed] [Google Scholar]
- Sun X, Sun XM, Yang ZM, Li SQ, Wang J, Wang SH (2005b) Expression of Brassica napus L. γ-glutamylcysteine synthetase and low- and high-affinity sulfate transporters in response to excess cadmium. J Integr Plant Biol 47: 243–250 [Google Scholar]
- Tang CF, Liu YG, Zeng GM, Li X, Xu WH, Li CF, Yuan XZ (2005) Effects of exogenous spermidine on antioxidant system responses of Typha latifolia L. under Cd2+ stress. J Integr Plant Biol 47: 428–434 [Google Scholar]
- Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI (2000) Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci USA 97: 4991–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timón-Gómez A, Proft M, Pascual-Ahuir A (2013) Differential regulation of mitochondrial pyruvate carrier genes modulates respiratory capacity and stress tolerance in yeast. PLoS ONE 8: e79405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacanti NM, Divakaruni AS, Green CR, Parker SJ, Henry RR, Ciaraldi TP, Murphy AN, Metallo CM (2014) Regulation of substrate utilization by the mitochondrial pyruvate carrier. Mol Cell 56: 425–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalkes MP. (2003) Cadmium carcinogenesis. Mutat Res 533: 107–120 [DOI] [PubMed] [Google Scholar]
- Wagner GJ. (1993) Accumulation of cadmium in crop plants and its consequences to human health. Adv Agron 51: 173–212 [Google Scholar]
- Whelan J, Murcha MW (2015) Plant Mitochondria: Methods and Protocols. Humana Press, Totowa, NJ [Google Scholar]
- Williams LE, Mills RF (2005) P(1B)-ATPases: An ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci 10: 491–502 [DOI] [PubMed] [Google Scholar]
- Xu YS, Liang JJ, Wang Y, Zhao XJ, Xu L, Xu YY, Zou QC, Zhang JM, Tu CE, Cui YG, et al. (2016) STAT3 undergoes acetylation-dependent mitochondrial translocation to regulate pyruvate metabolism. Sci Rep 6: 39517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav SK. (2010) Heavy metals toxicity in plants: An overview on the role of glutathione and phytochelatins in heavy metal stress tolerance of plants. S Afr J Bot 76: 167–179 [Google Scholar]
- Yan L, Wei S, Wu Y, Hu R, Li H, Yang W, Xie Q (2015) High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant 8: 1820–1823 [DOI] [PubMed] [Google Scholar]
- Yang C, Ko B, Hensley CT, Jiang L, Wasti AT, Kim J, Sudderth J, Calvaruso MA, Lumata L, Mitsche M, et al. (2014) Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol Cell 56: 414–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fernie AR (2018) On the role of the tricarboxylic acid cycle in plant productivity. J Integr Plant Biol 60: 1199–1216 [DOI] [PubMed] [Google Scholar]