Genes associated with stone cell formation, fruit size, and sugar content underwent directional selection during pear domestication and improvement, which contributed to drastic changes of cultivated pears.
Abstract
Knowledge of the genetic changes that occurred during the domestication and improvement of perennial trees at the RNA level is limited. Here, we used RNA sequencing analysis to compare representative sets of wild, landrace, and improved accessions of pear (Pyrus pyrifolia) to gain insight into the genetic changes associated with domestication and improvement. A close population relationship and similar nucleotide diversity was observed between the wild and landrace groups, whereas the improved group had substantially reduced nucleotide diversity. A total of 11.13 Mb of genome sequence was identified as bearing the signature of selective sweeps that occurred during pear domestication, whereas a distinct and smaller set of genomic regions (4.04 Mb) was identified as being associated with subsequent improvement efforts. The expression diversity of selected genes exhibited a 20.89% reduction from the wild group to the landrace group, but a 23.13% recovery was observed from the landrace to the improved group, showing a distinctly different pattern with variation of sequence diversity. Module-trait association analysis identified 16 distinct coexpression modules, six of which were highly associated with important fruit traits. The candidate trait-linked differentially expressed genes associated with stone cell formation, fruit size, and sugar content were identified in the selected regions, and many of these could also be mapped to the previously reported quantitative trait loci. Thus, our study reveals the specific pattern of domestication and improvement of perennial trees at the transcriptome level, and provides valuable genetic sources of fruit traits that could contribute to pear breeding and improvement.
Human selection has modified many plant traits that distinguish cultivated accessions from their wild forms, including organ size, shape, and the quantities of seeds and fruit that are useful to humans. Among these traits, the fruit size of many species has been impressively increased from wild to cultivated plants. Therefore, a comparative analysis of wild and cultivated plants can provide insights into the evolutionary process underlying the typical traits that have been subjected to intense human selection. Attempting to study plant domestication based on a single gene and very few markers has limitations that have made this technique unpopular. With the development of next-generation sequencing techniques, increasing reports of sequencing and resequencing, as well as transcriptome analyses, have provided vast amounts of information from the whole genome, deepening our understanding of plant domestication.
To date, numerous resequencing studies of plant domestication have been carried out in annual crops (Xu et al., 2011; Hufford et al., 2012; Zhou et al., 2015). In rice (Oryza sativa sp. japonica) and soybean (Glycine max), 49% and 52% reductions in nucleotide diversity, respectively, have been proposed as a result of domestication (Lam et al., 2010; Xu et al., 2011). Domesticated common bean (Phaseolus vulgaris) showed a drastic reduction in nucleotide diversity for coding sequence (CDS) regions (∼60%) and for gene expression (18%) compared with wild progenitors. It has also been suggested that 9% of the genes associated with abiotic stress responses and flowering time were actively selected during domestication (Bellucci et al., 2014; Schmutz et al., 2014). In tomato (Solanum lycopersicum), 50 genes under positive selection were detected based on sequence differences between cultivated tomato and five wild progenitor species (Koenig et al., 2013). In maize (Zea mays sp. mays), the presence of 11% less nucleotide diversity than the ancestral public US lines has been proposed to result from domestication (Jiao et al., 2012), and 2% to 4% of the genes and 7.6% of the maize genome were shown to have experienced artificial selection during domestication (Wright et al., 2005; Hufford et al., 2012).
Among perennial woody species, the domestication of peach (Prunus persica) has been studied through resequencing at the DNA level (Cao et al., 2014). Recently, whole genome resequencing of 113 pear (Pyrus sp.) accessions was reported, and selected regions and genes associated with important traits have been detected (Wu et al., 2018). RNA sequencing (RNA-seq) differs from resequencing, as only a fraction of the genome is transcribed, and this technique can narrow the target range for genes of interest to those in the expressed genomic regions, thereby rendering the effects of variations of gene expression easier to observe. At present, few studies have focused on plant domestication and improvement in perennial trees via transcriptome analysis, but such analyses could be expected to yield insights about the nature of different domestication routes and traits of interest, some of which would be expected to differ markedly from those experienced by annual crops. Thus, to better establish the genome-wide consequences of domestication and improvement, it is necessary to extend domestication studies to additional plant species at both the DNA and RNA levels.
In pear, large phenotypic differences (e.g. fruit size) have appeared between cultivated varieties and wild accessions, and these differences can be used as a good medium to explore domestication and improvement. Worldwide, at least 22 pear species are recognized; however, only five of these species are major cultivated species, while the others are wild species. It was found that only wild ancestors of Pyrus pyrifolia and Pyrus ussuriensis have been indisputably recognized (Kikuchi, 1946; Bell et al., 1996). P. pyrifolia, also called sand pear in China, is mainly distributed in the south of China, the original location of the ancestral pear species, which holds a comprehensive collection of germplasm, including wild genotypes, landraces, and improved cultivars, showing high genetic diversity. P. ussuriensis is distributed in the cold northeastern region of China, and comparatively few genotypes were conserved or selected because of the requirement for tolerance of severe cold.
Here, 41 pear accessions of P. pyrifolia, including wild progenitors, landraces, and improved genotypes, were selected for a comparative analysis, with one wild P. ussuriensis accession as the outgroup. We conducted RNA-seq analysis to investigate the expression differences and sequence diversity responsible for the phenotypic variations from domestication and improvement among these accessions. These analyses revealed that human selection has caused dramatic differences in the transcriptomes among wild, landrace, and improved pears. The genes in the candidate selective sweeps identified during pear domestication and improvement provide a basis for exploring the functions of key candidate genes that apparently control important fruit quality traits, and our extensive transcriptome data for this perennial tree represent a valuable reference for plant breeding and improvement efforts.
RESULTS
Qualitative Evaluation of RNA-Seq from Wild, Landrace, and Improved Pears
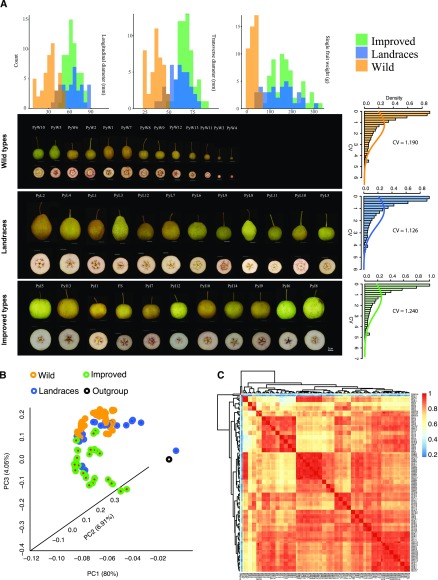
To investigate the transcriptomic changes that occur during the domestication and improvement of pear, a total of 41 P. pyrifolia species, including 14 wild accessions, 12 landraces, and 15 improved accessions that showed dramatic phenotypic variations (Fig. 1A), with one wild accession 'Shanlisanhao' ('Outgroup') of P. ussuriensis as an outgroup, were analyzed with RNA-seq. In our analysis, we deemed the 14 wild accessions as PyW1–PyW14, the 12 landraces as PyL1–PyL12, and the 15 improved accessions as PyI1–PyI15 (Supplemental Table S1). As shown in Figure 1A, these three groups (wild, landrace, and improved) displayed abundant phenotypic differences in fruit size and fruit quality (e.g. stone cell content), suggesting that the panel we selected represents a substantial amount of the phenotypic diversity present among the wild, landrace, and improved pear accessions of P. pyrifolia.
Figure 1.
The phenotype and expression pattern of all sequencing accessions. A, The phenotypic features of fruit and images of its transverse section with dyed stone cell from wild, landraces, and cultivated pear accessions. The fruits were digitally extracted for comparison. At top, the phenotypic distribution of longitudinal diameter, transverse diameter, and single fruit weight from three groups are presented. At right, the distribution of CV within each group is presented. B, The three-dimensional plot of PCA from all gene RPKMs of the 41 accessions. Orange points represent wild accessions; blue points represent landraces; green points represent improved accessions; black point represents the outgroup. C, The heatmap of PCCs from all 41 accessions. The color scale at the right represents PCC values. Red color represents the high correlation; blue represents the low correlation. The text on the right and bottom represent the sample names. Wild, wild type; PC, principal component.
A total of 124 RNA-seq libraries were constructed and sequenced (Supplemental Table S1). After the adaptor sequences and low-quality reads were removed, the average amount of clean bases per library was 8.11 Gb, with an average of 16× coverage of the pear genome. Using the ‘Dangshansuli’ genome as the reference genome, we mapped an average of 76.91%, 77.80%, and 78.20% of clean reads for the wild, landrace, and improved libraries, respectively (Supplemental Table S1), suggesting a fair basis for comparison among these three groups. A total of 37,490 genes were expressed (reads per kilobase per million mapped reads [RPKM] > 0) in at least one accession among the 41 accessions. There were 36,125, 34,740, and 34,889 genes expressed in at least one accession of the wild, landrace, or improved groups, respectively. In addition, there were 1,168, 428, and 482 unique genes that were only expressed in the wild, landrace, or improved groups, respectively. Gene annotation suggested that the wild group had enrichment for genes involved in stress resistance among its unique expressed genes, whereas the landraces and improved groups exhibited enrichment for genes related to sugar metabolism (Supplemental Table S2).
The coefficient of expression variation (CV) within a group for all of the genes showed no significant difference among the three groups, with an average of 1.190 in wild, 1.126 in the landrace, and 1.240 in the improved groups (Fig. 1A; Supplemental Table S3). Principal component analysis (PCA) was performed using the RPKM values of genes from the 41 accessions, which showed that the wild and improved groups were well separated, whereas the landrace group was mixed among the wild and improved groups, but was closer to the wild group (Fig. 1B). In addition, it was notable that the PyL7-1 sample grouped closer to the outgroup, suggesting that perhaps a sequencing or sampling error had occurred with PyL7-1. The heatmaps of Pearson correlation coefficient (PCC) values showed that all samples have similar expression patterns and high correlations (Fig. 1C; Supplemental Table S4); especially, the three biological repeats of each accession showed an extremely high PCC value, up to 0.95 (Supplemental Fig. S1), except for PyL7-1 (Average PCC = 0.26; Supplemental Fig. S1), which grouped together with the outgroup. This is consistent with the result of the PCA. Therefore, sample PyL7-1 was discarded from all subsequent analyses.
Divergence of Gene Expression during Domestication and Improvement of Pear
When exploring divergence of gene expression during pear domestication and improvement, we compared the differentially expressed genes (DEGs) in the comparisons of wild versus landraces and landraces versus improved groups. A total of 2,118 DEGs (fold change ≥ 2 and false discovery rate [FDR] ≤ 0.001), occupying 5.65% of total expressed genes, were identified in the wild versus landraces group, whereas more genes (3,517 DEGs, 9.38%) were identified in the landraces versus improved group (Supplemental Fig. S2, a and b; Supplemental Table S5). In addition, DEGs were also identified in the comparison of wild versus improved group. We found that 3,695 DEGs (9.86%) were identified in the wild versus improved group comparison, similar to the landraces versus improved group comparison. Gene Ontology–based enrichment analysis was performed for all three comparisons, which indicated that genes with annotations relating to photosynthesis and light harvesting were significantly enriched (Supplemental Fig. S3) among the DEGs from the wild versus improved and the wild versus landraces group comparisons, but that no pathways were significantly enriched for the landraces versus improved group comparison.
To explore whether the effects of human selection have contributed to a biased genomic distribution of DEGs, we calculated a p-value based on a matrix of DEGs/non-DEGs on one subjected chromosome versus DEGs/non-DEGs on the remaining other chromosomes using a χ2 test. The results revealed that genes on chromosome 15 were more likely (p-value < 0.01) to be differentially expressed between wild and landraces pears (Supplemental Table S6), whereas chromosome 1 was significantly differentially targeted between landraces and improved pears. Further, a window-based χ2 test was performed to detect the significant enrichment distribution of DEGs based on each 500-kb slide window across the whole pear genome. A total of 777 windows were subjected to the χ2 test. For DEGs between wild and landraces pears, 16 (2.06%) windows were significantly enriched (p-value < 0.01) for 142 DEGs (Supplemental Table S6), and the highest number (4 windows, including 35 DEGs) of significantly enriched windows was observed on chromosome 15. This is consistent with the observation from the chromosome-based χ2 test. Although DEGs between landrace and improved pears were significantly enriched in 15 windows, the highest number of significantly enriched windows was observed on chromosome 11, with windows thus identified from chromosome 1.
Population Relationships, Diversity, and Linkage Disequilibrium in the Wild, Landrace, and Improved Groups
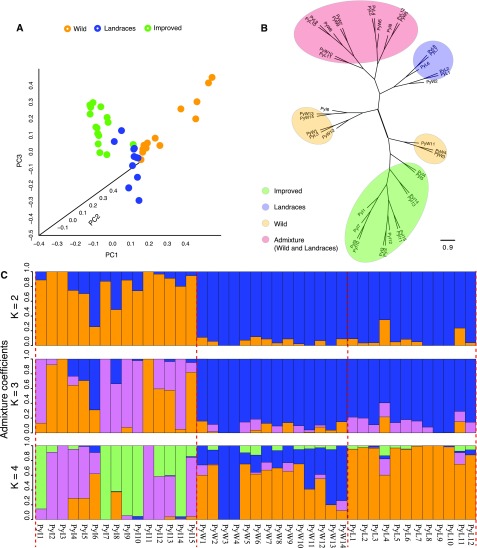
A total of 875,319 high-quality single nucleotide polymorphisms (SNPs) were identified in 41 pear accessions. Using the SNP data of all samples, we performed PCA, which indicated that the wild and improved groups were clearly separated, whereas the landrace accessions clustered closer to the wild group (Fig. 2A). This is consistent with the population relationship displayed in the PCA based on the expression data (Fig. 1B). These results suggested that the wild and landrace accessions have apparently similar genetic backgrounds as compared with the improved accessions. In addition, as shown in Figure 2A, it also revealed that the wild and landrace groups were characterized by a higher diversity (a wider distribution in the PCA scores plot) compared with the improved group. A maximum-likelihood phylogenetic tree showed that the accessions of the improved group clustered together as a branch, whereas the wild group accessions were split into three branches, including a branch consisting of the wild and the landraces accession (Fig. 2B). Population structure (K = 2–4) showed that the improved pear accessions exhibited a higher extent of admixture than the wild and landrace groups, likely due to recent hybridizations during pear improvement (Fig. 2C).
Figure 2.
The population relationship, clustering, and structure of 41 pear accessions from three groups inferred from SNPs. A, The three-dimensional plot of PCA using SNP data from all 41 pear accessions. Orange points represent wild accessions; blue points represent landraces; green points represent improved accessions. B, The phylogenetic tree based on SNP data. The light-green circle indicates the improved group; light blue indicates the landraces group; light yellow indicates the wild group; pink indicates the admixture of wild and landrace accessions. C, The population structure (K = 2–4) of all 41 pear accessions. Each bar represents an accession. PyW, PyL, and PyI indicate wild, landrace, and improved accessions, respectively. Wild, wild type; PC, principal component.
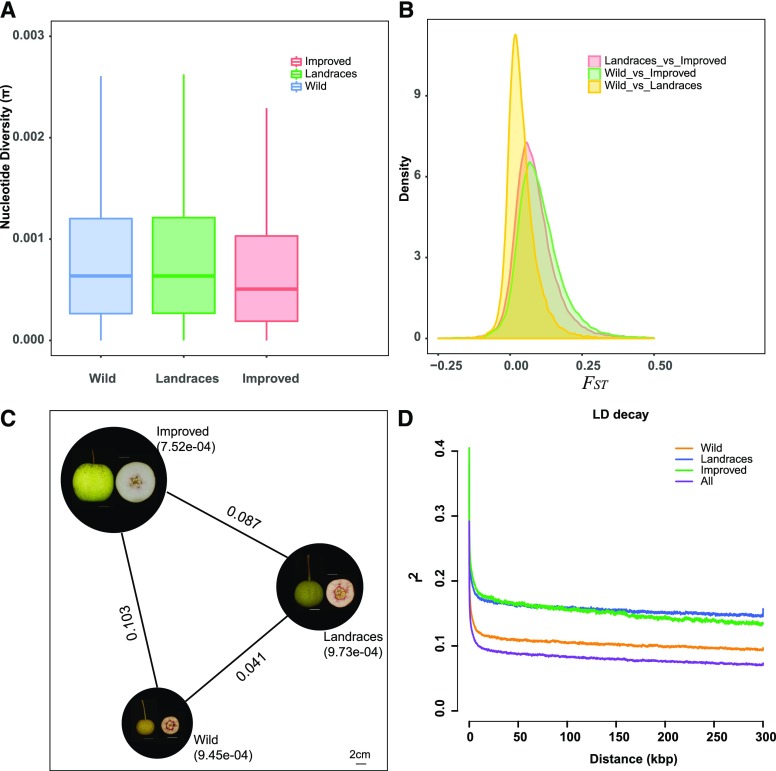
Further, we calculated the nucleotide diversity (π) within the three pear groups using the 875,319 high-quality SNPs and evaluated the divergence level by calculating the fixation index (FST) value between the three groups. We found that the wild and landrace groups had similar extents of genetic diversity, with average π values of 9.45e-04 and 9.73e-04, respectively; a similar result, albeit for genomic sequence diversity, was reported in a previous pear resequencing study (Wu et al., 2018). In the improved group, a lower average π value (7.52e-04) was observed, with 22.71% less nucleotide diversity than the landrace group (Fig. 3, A and C; Supplemental Table S7). We also found that the average FST value (0.041) between the wild and the landrace groups was smaller than that between the landrace and the improved groups (0.087) and smaller than that between the wild and improved groups (0.103; Fig. 3, B and C; Supplemental Table S8). These findings indicate that humans have apparently contributed weakly to the genetic diversity in landrace pear accessions during pear domestication, and a slightly stronger impact in improved accessions during pear improvement.
Figure 3.
Nucleotide diversity, divergence, and LD within and between three groups. A, The boxplots of nucleotide diversity (π) within three groups. Blue indicates the wild group; green indicates the landraces group; red indicates the improved group. B, The distribution of divergence level measured using the FST value between the three groups. Yellow displays the distribution of FST between the wild and landrace group; red displays the distribution of FST between the landraces and improved group; green displays the distribution of FST between the wild and improved group. C, The average value of π and FST among the three pear groups. The fruits were digitally extracted for comparison. The values beside the line represent the average FST; the values in brackets represent the average π; three representative fruit phenotypes from wild, landraces, and improved groups are displayed in black circles. D, LD decay determined by correlation of allele frequencies (r2) against distance (kb) among polymorphic SNP sites in different pear groups, including wild (orange), landraces (blue), improved (green), and all 41 accessions (purple). Wild, wild type.
Further, linkage disequilibrium (LD) analysis showed that the transcriptome sequences encoded by pear genomes have short LD distances (869 bp of average distance when LD decayed to ∼50% of its maximum value) and rapid LD decay (Fig. 3D). In the wild group, the average r2 value was 0.104, with 1,104 bp of average LD decay distances. Higher r2 values (0.157 and 0.153) and longer LD decay distances (3,682 bp and 4,141 bp) were observed in the landrace and the improved groups (Supplemental Table S9). When compared with the wild group, in the landrace group we detected slightly longer LD decay distances and more rapid LD decay, further supporting a relatively weak impact of selection in the transcriptomic level during pear domestication.
Evidence for Selective Sweeps during Domestication and Improvement
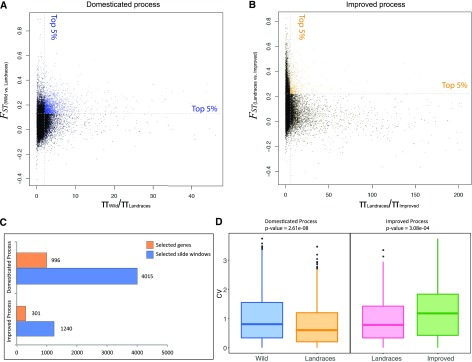
Generally, when compared with wild plants, the nucleotide diversity of cultivated plants is dramatically reduced. The π statistic (Watterson, 1975), which is used to evaluate nucleotide diversity, can be used to identify loci under artificial selection. The FST statistic can also be used to identify loci with a high diversity; loci with high FST values have often undergone selection during human intervention (Weir and Cockerham, 1984; Lam et al., 2010; Fumagalli et al., 2013). Therefore, we used the top 5% of the π ratio (πgroup1/πgroup2) and the top 5% of the FST value as two thresholds to identify candidate selective sweeps that have occurred during pear domestication (wild versus landraces) and improvement (landraces versus improved). In the comparison of the wild versus landrace groups, there were 4,015 slide windows identified as candidate selection regions (πWild/πLandraces ≥ 2.12; FST ≥ 0.13), for a total of 11.13 Mb of the genome sequence identified, and 996 genes were included in these selection sweep regions (Fig. 4, A and C; Supplemental Table S10). A total of 555 out of the 996 candidate genes were assigned annotations by blasting against genes with known function in other plants. Among these, there were genes annotated to be involved in plant cell division (ftsH), auxin synthesis (small auxin up-regulated RNA [SAUR]) and efflux (PIN6), lignin synthesis (cinnamoyl CoA reductase and peroxidase [POD]), photosynthesis (CONSTANS), and stress resistance (leucine-rich repeat [LRR]; Supplemental Table S10; Yano et al., 2000; Sarid-Krebs et al., 2015).
Figure 4.
The identification of selective sweeps during pear domestication and improvement. A, The identification of selective sweeps during pear domestication. Each point represents a 10-kb slide window. Blue points represent the candidate selective sweeps. The x axis indicates the πWild/πLandraces ratio; y axis indicates the FST value. B, the identification of selective sweeps during pear improvement. Each point represents a 10-kb slide window. Orange points represent the candidate selective sweeps. The x axis indicates the πLandraces/πImproved ratio; y axis indicates the FST value. C, The number of selected slide window and genes during domestication and improvement. Blue bar represents the selected slide window; orange bar represents selected genes. D, the CV for selected genes in the domesticated and improved process. Wild, wild type.
In the comparison of the landrace versus improved groups, fewer candidate-selected slide windows were identified (4.04 Mb of the genome sequence; πLandraces/πImproved ≥ 5.35; FST ≥ 0.22; 1,240 windows), and fewer genes were included in these windows (301 genes; Fig. 4, B and C; Supplemental Table S10), including 151 genes with annotated functions. Similarly, these genes also had annotations relating to cell division, auxin, and lignin synthesis. There were no genes with annotations relating to stress resistance, suggesting that breeders have not strongly emphasized resistance in recent pear improvement breeding efforts. There was one gene (Pbr020127.1) associated with sugar transport in this comparison, emphasizing that traits relating to edible and sensory characteristics have been a focus of pear improvement breeding. In addition, there were only seven candidate genes that were common for the selective sweeps in both pear domestication and improvement, clearly suggesting that different genome regions have been selected during pear domestication versus improvement. One of these seven genes, Pbr037862.1 (ftsH), is annotated to be involved in cell division, and the remaining six genes did not have annotations.
We also found that the distributions of the candidate-selected genes for both domestication and improvement were randomly distributed across the whole pear genome (Supplemental Fig. S4). A χ2 test also showed that the number of candidate domesticated genes was significantly correlated with gene density on most of the chromosomes (Supplemental Table S11), whereas there was no significant correlation between the number of candidate selected genes in the improvement process and gene density on chromosomes. These results emphasized that human selection has apparently been a complex process that involves multiple genes and multiple biological pathways from across the entire genome.
Moreover, we calculated the CV for the candidate selected genes from the domestication and improvement processes and found that the candidate domesticated genes presented a 20.89% loss of expression diversity from the wild group (average CV = 1.077) to the landrace group (average CV = 0.852), whereas there was a 23.13% increase in expression diversity from the landrace group (average CV = 1.047) to the improved group (average CV = 1.362; Fig. 4D; Supplemental Table S12). This could be interpreted to suggest that the long domestication process prompted a moderate decrease of gene expression diversity in the landrace group that was followed by a recovery of expression diversity during pear improvement. In addition, candidate selected genes associated with important pear traits, such as stone cells, fruit size, and sugar content, were also characterized by this feature: larger expression variation was observed in the wild pear group for candidate selected genes during domestication, and smaller values were observed in the landraces group for candidate selected genes in the improvement process (Table 1). We also checked if candidate selected genes were differentially expressed in the different pear groups, and the results showed that only 72 and 22 candidate genes identified from the selected regions during the domestication and improvement process, respectively, were differentially expressed in the wild versus the landrace groups and in the landrace versus improved groups (Supplemental Fig. S5).
Table 1. The CV of selected genes associated with important traits in the domestication and improvement process.
| Gene ID | CV | Function | |
|---|---|---|---|
| Wild | Landraces | ||
| Pbr003257.1 | 0.271684173 | 0.20350751 | Cell division |
| Pbr019276.1 | 0.271095022 | 0.178158405 | Cell division |
| Pbr026630.1 | 0.310784855 | 0.214153063 | Cell division |
| Pbr037862.1 | 0.538962117 | 0.470558378 | Cell division |
| Pbr041499.1 | 0.626003563 | 0.315680301 | Cell division |
| Pbr022044.2 | 1.353331585 | 0.51841435 | Light harvest |
| Pbr013295.1 | 1.718277373 | 0.602279064 | Flowering |
| Pbr015108.2 | 2.32545572 | 1.271463218 | Lignin synthesis |
| Pbr015718.1 | 0.956482065 | 0.824749225 | Stress resistance |
| Pbr035421.1 | 2.440465826 | 1.061326853 | Stress resistance |
| Pbr042386.1 | 2.546427328 | 0 | Stress resistance |
| Pbr029126.1 | 0.356787067 | 0.237205414 | Stress resistance |
| Pbr010582.1 | 0.361393451 | 0.361393451 | Auxin synthesis |
| Landraces | Improved | ||
|---|---|---|---|
| Pbr012887.1 | 0.334129791 | 0.442072319 | Cell division |
| Pbr037862.1 | 0.470558378 | 0.891959634 | Cell division |
| Pbr041690.1 | 2.798545745 | 3.872983346 | Cell division |
| Pbr018073.1 | 2.440370626 | 3.872983346 | Flowering |
| Pbr020127.1 | 1.190027884 | 1.374033643 | Sugar transport |
| Pbr000146.1 | 0.938625424 | 1.852144008 | Lignin synthesis |
Dynamic Transcriptome and Coexpression Network Analyses
We next conducted an expression analysis with 6 pear genotypes selected from 41 pear accessions, including three wild accessions, PyW12, PyW13, PyW14, and three landraces accessions, PyL1, PyL2, PyL3, at three key fruit developmental stages (small [a], enlarged [b], and mature [c] stages; Supplemental Fig. S6a). Multidimensional scaling analysis indicated that samples were more likely to be separated by developmental stage. As shown in Supplemental Figure S6b, most of the small and enlarged stage samples clustered into a group, whereas the mature stages clustered to form another group. PCCs showed that most of the coefficients reached 85%, and some were as high as 98% (Supplemental Fig. S7). DEGs were identified between wild and landrace pears with three developmental stages as the biological repeats (Supplemental Fig. S6b; Supplemental Table S13). With increasing fruit maturity, the number of DEGs gradually reduced, and the number of down-regulated genes was far higher than that of up-regulated genes (Supplemental Fig. S6c). A total of 2,905 (6.86%) genes were expressed in at least one developmental stage, which was used for the subsequent analysis.
Large phenotypic differences existed between the wild and landraces pear groups, such as fruit size and lignin proportion (Supplemental Table S14). The landrace pears presented larger fruit size, higher sugar content, and lower lignin and acid contents than those of wild pears. Correlation analysis of 15 fruit traits revealed positive correlations between single fruit weight and longitudinal and transverse diameter, as well as between stone cell content and several acids (Supplemental Fig. S8, a and b). In addition, significantly negative correlations between acids and sugars were observed, whereas citric acid presented a highly negative correlation with stone cell and other acids, especially in wild pears (Supplemental Fig. S8a). Interestingly, we also found that sorbitol, a major photosynthetic product in rosaceous fruits, presented a highly positive correlation with stone cell and several acids in landraces pears (Supplemental Fig. S8b). A Student’s t-test was used to evaluate the significance of differences of 15 pear fruit traits between the wild and landrace groups. The results showed that most of the 15 traits were significantly different (P < 0.05; Supplemental Fig. S8c).
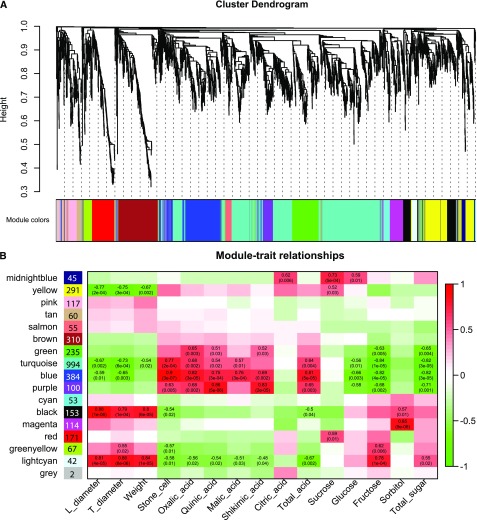
Weighted gene coexpression network analysis (WGCNA; Langfelder and Horvath, 2008) was further performed to identify the candidate trait-linked genes based on 2,905 DEGs from wild versus landraces pears. A total of 16 distinct modules were identified, containing 2,903 DEGs, and the remaining 2 genes were considered outliers and were excluded from the list (Fig. 5A; Supplemental Table S15). Furthermore, we identified modules that were significantly associated with the measured phenotypic traits by quantifying module-trait associations. As shown in Figure 5B, 6 of the 16 coexpression modules comprise genes highly associated with one or two traits (|r| ≥ 0.80, P < 1e-3), e.g. fruit size–related modules (black and lightcyan), a stone cell and acid related module (blue), and a sorbitol-related module (magenta). The fruit size related module (black) including 105 genes was most significantly associated with fruit size (a linear correlation between longitudinal diameter, transverse diameter, and weight). The stone cell and acid related module, which contained 373 genes, was significantly associated with the content of stone cells and acid (Fig. 5B). Gene significance and module membership appeared to be highly correlated in the fruit size–related (cor = 0.79, P < 6.8e-34) and stone cell and acid-related (cor = 0.83, P < 6.8e-99; Supplemental Fig. S9, a and , b) modules.
Figure 5.
WGCNA analyses. A, Clustering dendrogram of genes with dissimilarity based on topological overlap, with assigned module colors. Each tree branch constitutes a module, and each leaf in the branch is one gene. B, Module-trait associations. Each row corresponds to a module. The number of genes in each module is indicated on the left. Each column corresponds to a phenotypic trait, labeled below. Each cell at the row-column intersection contains the correlation coefficient (|r|<0.5 not shown) and the p-value (in brackets) between that module and that trait. A highly positive correlation between a specific module and a trait is indicated by dark red, and green indicates negative correlation. The gray module is reserved for genes outside of all modules.
We also performed a hierarchical cluster analysis with logarithm-transformed RPKM values to explore the gene expression patterns in the trait-related modules. The results showed that diametrically different expression patterns were present in the wild and landraces accessions. Decreased stone cell and acid content, as well as dramatic increases in fruit size, distinguished landraces pear from its wild accessions (Supplemental Table S14). Each of these phenotypes can be well correlated with gene expression profiles. As shown in Supplemental Figure S9c, most of the genes in the stone cell and acid-related module were highly expressed in wild pears. In particular, these genes had higher expression levels at the small fruit stage, with a significant expression difference (p-value = 6.24e-05) from the enlarged (p-value = 0.27) and mature fruit stages (p-value = 0.91). This result is consistent with the physiological indicators of stone cells and acid, which undergo large-scale synthesis during the early developmental stages of pear fruit. Moreover, these results also revealed that most of the genes in the stone cell and acid-related module were involved in lignin and acid synthesis through positive regulation. As expected, in the fruit size–related module, most genes showed higher expression levels during all developmental stages in cultivated pears than that in the wild accessions (Supplemental Fig. S9d).
Functional Analysis of the Fruit Quality–Related Module
Stone cells
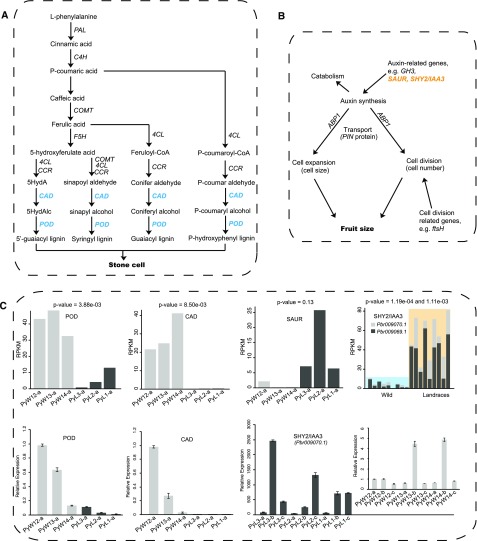
Stone cells represent an important and unique feature of fresh fruit quality in pears and directly affect the taste of pear fruit. As shown in Figure 6A, four important final products are synthesized as stone cell compounds in pear fruit. In this pathway, lignin synthesis is closely related to acid synthesis, which shares regulation with the precursor of lignin synthesis. Therefore, when lignin is synthesized constantly, acid is also produced constantly, precisely explaining the result that lignin and acidity were positively correlated in wild pears. To determine which genes in the stone cell–related module were more likely to regulate stone cell formation, we performed a functional annotation analysis and aligned these protein sequences with those of known genes reported as key regulators of lignin synthesis in previous studies. We found that the genes Pbr000691.1 and Pbr041997.1 were annotated as POD and cinnamyl-alcohol dehydrogenase (CAD), respectively, which were reported in our previous study (Wu et al., 2013). The POD and CAD genes are involved in the lignin synthesis pathway and control the formation of the end-product compounds (Fig. 6A). Furthermore, the genes Pbr000691.1 and Pbr041997.1 were differentially expressed at the small fruit stage, with extremely high significance levels (p-values = 3.88e-03 and 8.50e-03, respectively; Fig. 6C). A higher expression level was present in wild pears than that in the landrace accessions, which is consistent with the wild pear characteristic of strong lignin synthesis at the early developmental stage of pear fruit.
Figure 6.
The biosynthetic pathways of stone cell (A) and fruit size (B) and the expression patterns of genes related to these pathways (C) are shown. Blue text in (A) indicates genes down-regulated in cultivated pears relative to wild accessions at the small fruit stage. PAL, Phe ammonia lyase; C4H, transcinnamate 4-monooxygenase; COMT, caffeic acid 3-O-methyltransferase; F5H, ferulate-5-hydroxylase; 4CL, 4-coumarate-CoA ligase. Yellow text in (B) indicates genes up-regulated in cultivated pears at the small fruit stage or all developmental stages. ABP1, auxin binding protein 1; GH3, gretchen hagen 3; SAUR, small auxin up-regulated RNA. Top in (C) presents the RPKM values of the genes Pbr000691.1 (POD), Pbr041997.1 (CAD), Pbr023270.1 (SAUR), and Pbr009069.1 (SHY2/IAA3; black bar) and Pbr009070.1 (SHY2/IAA3; gray bar); bottom in (C) presents the relative expression levels of Pbr000691.1, Pbr041997.1, and Pbr009070.1. Data shown are the means ± sd from three replications. Wild, wild type.
Fruit size
Larger plant organs, including fruit and leaf size, are major characteristics distinguishing landrace pears from its wild relatives (Supplemental Fig. S6a). Increased fruit size is attributed to cell division and expansion, as shown in Figure 6B, and auxin is an important phytohormone modulating cell expansion. In the fruit size–related module, the gene Pbr023270.1 is a homologous gene of SAUR, a gene involved in the auxin synthesis pathway. Expression analysis showed that Pbr023270.1 was differentially expressed between wild and landrace pears at the small fruit stage, with a higher expression level in landrace pears (Fig. 6C). In addition, the genes Pbr009069.1 and Pbr009070.1 are both annotated to regulate auxin synthesis as an auxin-responsive protein, SHY2/IAA3 (Carraro et al., 2012). These two genes presented higher expression levels in landrace pears during all developmental stages of pear fruit (Fig. 6C), with extremely high significance levels (p-values = 1.19e-04 and 1.11e-03, respectively). Reverse transcription-quantitative PCR (RT-qPCR) analysis showed that Pbr009070.1 presented a higher relative expression level in landrace pears (Fig. 6C), whereas no expression was detected for Pbr023270.1 and Pbr009069.1.
Trait-Related Quantitative Trait Loci Validated Trait-Linked Genes in Selective Sweep Regions
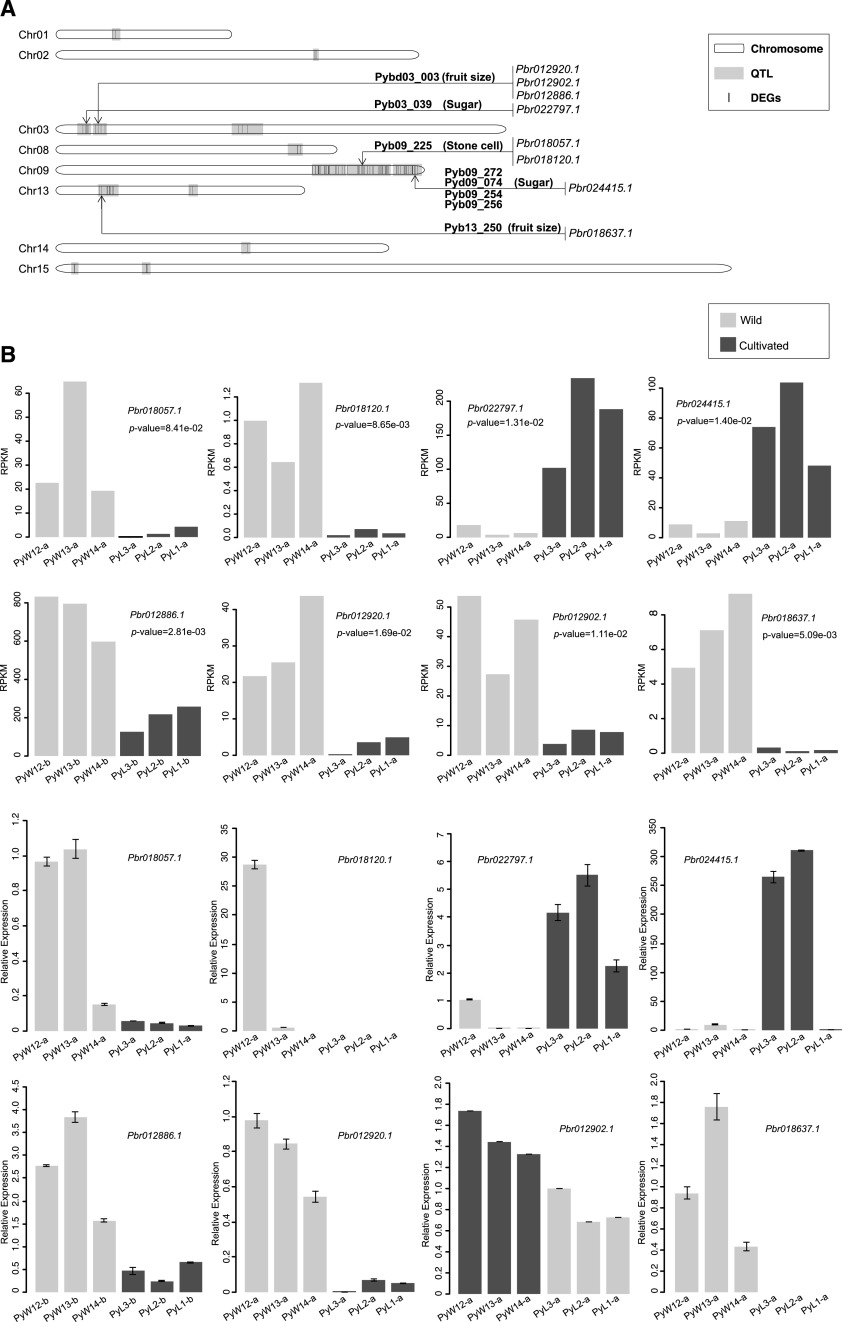
To validate the functions of candidate trait-linked genes during pear domestication, we collected previously reported quantitative trait loci (QTLs) associated with important fruit traits, e.g. sugar, acidity, stone cell, fruit size, and fruit shape, in pear (Yamamoto et al., 2013; Wu et al., 2014; Kumar et al., 2017; Supplemental Table S16). Then, we mapped the 2,903 DEGs against the genomic positions of the QTLs; a total of 98 DEGs were mapped to all 28 QTLs (Supplemental Table S16). Among them, most genes were mapped to QTLs associated with stone cells and sugar (13 genes and 52 genes, respectively). Interestingly, two unannotated genes, Pbr018057.1 and Pbr018120.1, in the stone cell–related module were mapped to the QTL Pyb09_225 associated with stone cell formation on chromosome 9 (Fig. 7A). Meanwhile, expression analysis showed that Pbr018057.1 and Pbr018120.1 were differentially expressed only at the small fruit stage, with a higher expression level in wild pears (Fig. 7B). Therefore, it is reasonable to hypothesize that these two genes are involved in the regulatory pathway of lignin synthesis. As we know from the WGCNA analysis, the genes in the sorbitol-related module were highly associated (r = 0.85; p-value = 9.00e-06) with the level of sorbitol, which is a unique sugar component in pear and Rosaceae fruit. Interestingly, two unannotated genes, Pbr022797.1 and Pbr024415.1, in the sorbitol-related module were mapped to QTLs associated with sugar synthesis, and Pbr024415.1 was mapped to four QTLs associated with sugar (Fig. 7A). Expression analysis also showed that these two genes had higher expression levels in landrace pears at the small fruit stage, with a significant difference between wild and landrace pears (p-value = 1.31e-02 and p-value = 1.40e-02, respectively; Fig. 7B). In addition, four genes, Pbr012886.1, Pbr012920.1, Pbr012902.1, and Pbr018637.1, included in the turquoise module (negatively related to stone cells and acid, as shown in Fig. 5B), were mapped to two QTLs associated with fruit size (Supplemental Table S16). Among them, Pbr012886.1 presented a higher expression level in wild pears at the enlarged fruit stage, whereas the other three genes were more highly expressed at the small fruit stage, suggesting their involvement in the inhibition of fruit enlargement. Further, we performed RT-qPCR analysis to verify the relative expression levels of these eight candidate genes in all samples. The results showed that most gene expression was consistent with that found by RNA-seq, showing similar trends of differential expression (Fig. 7B).
Figure 7.
Overlap mapping between DEGs and QTLs related to fruit traits in pear. A, The mapping of QTLs against DEGs. The light blue blocks indicate QTLs, and the inside black lines indicate DEGs. All chromosomes with QTLs are displayed. The arrows indicate the genes related to important traits; the text beside the horizontal line indicates the QTL name, and the text in the brackets indicates the trait controlled by that QTL. B, The expression levels of eight genes mapped to QTLs controlling stone cell content, sugar content, and fruit size between wild and cultivated pears at the small or enlarged fruit stage. The gray bar represents the wild pear accession, and the black bar represents the cultivated accession. Data shown are the means ± sd from three replications. Wild, wild type.
Further, we found that two candidate domesticated pear–related genes and five improved pear genes were included in these 98 trait-linked DEGs (Table 2). As candidate domesticated pear genes, Pbr035421.1 and Pbr022248.1 were clustered into the turquoise module, in which genes presented a negative correlation with several kinds of sugar, as well as fruit size and single fruit weight, especially for Fru content (r = −0.84, p-value = 1e-05) and transverse diameter (r = −0.73, p-value = 6e-04) of pear fruit (Fig. 5B). These two genes were also validated by two sugar-related QTLs in chromosome 9, Pyb09_202 and Pyb09_228, respectively. Meanwhile, for improved pear genes, three out of five also clustered into the turquoise module. Among them, two genes, Pbr018637.1 and Pbr012886.1, were located in the regions of two fruit size–related QTLs, Pyb13_250 and Pybd03_003, respectively. Pbr041550.1 was located in the QTL Pyd09_074, which was identified in chromosome 9 and associated with the sugar content of pear fruit. Interestingly, Pbr022797.1, hypothesized to be involved in sorbitol synthesis and validated by RNA-seq and RT-qPCR in the wild versus landrace comparison (Fig. 7B), was also identified as one candidate selected gene during pear improvement. In addition, Pbr018100.1, associated with Suc in the red module, was mapped to the stone cell–related QTL Pyb09_225. Five of the seven selected genes were not annotated in pear and other important model or nonmodel plants.
Table 2. The DEGs associated with trait-related QTLs in selective sweeps.
| Gene ID | π Ratio | FST | Module | QTLs | Traits | Annotation |
|---|---|---|---|---|---|---|
| Domesticated | ||||||
| Pbr035421.1 | 2.41 | 0.18 | Turquoise | Pyb09_202 | Sugar | FLS2 |
| Pbr022248.1 | 3.05 | 0.24 | Turquoise | Pyb09_228 | Sugar | No annotation |
| Improved | ||||||
| Pbr018637.1 | 5.81 | 0.23 | Turquoise | Pyb13_250 | Fruit size | 4-nitrophenyl phosphatase |
| Pbr012886.1 | 7.32 | 0.38 | Turquoise | Pybd03_003 | Fruit size | No annotation |
| Pbr041550.1 | 8.17 | 0.28 | Turquoise | Pyd09_074 | Sugar | No annotation |
| Pbr022797.1 | 5.45 | 5.45 | Magenta | Pyb03_039 | Sugar | No annotation |
| Pbr018100.1 | 6.22 | 0.28 | Red | Pyb09_225 | Stone cell | No annotation |
DISCUSSION
Similar to other annual crops, pear also experienced a long domestication process and a recent improvement process (Hufford et al., 2012; Zhou et al., 2015). Domestication and improvement has imposed sequence diversity and changes in gene expression between wild and landrace types, as well as landraces and improved pears. When compared with the evaluation of genome-wide SNPs in a previous resequencing study (Wu et al., 2018), approximately 1/16th of these genome-wide SNPs, 695,167 SNPs in the wild group, 690,669 in the landrace group, and 569,445 in the improved group of pear, was identified in our transcriptomic evaluation. However, it is consistent that a higher number of SNPs was observed in wild pears compared with the other two groups, and more SNPs were identified in the landrace group compared with the improved group. It is reasonable that domestication and improvement has led to a smaller decrease of variants from wild to landrace types (0.65%) and a greater decrease from landraces to improved pears (17.55%), respectively. This strongly supports a weak domestication selection occurred in pear, which is consistent with the previous observation revealed by whole genome resequencing of pear (Wu et al., 2018). Around 21.57% less nucleotide diversity was observed in the improved group than in the other two groups, which might be due to breeders preferring to new varieties possessing more superior phenotypes for edible quality, such as lower stone cell content and higher sugar content etc., leading to a narrow biodiversity in the improved group. When compared with the previous study (Wu et al., 2018), lower values of nucleotide diversity were observed in our current analysis, which might be due to the fact that only a single pear species (P. pyrifolia) was analyzed. The self-incompatible reproductive system and long generation cycle might contribute to a smaller loss of nucleotide diversity during domestication in pear. For common bean, as an autogamous species, continued self-crossing will gradually produce homozygous genotypes, which will contribute to the more obvious decline of nucleotide diversity (60% loss). Self-pollination also enhances the effects of genetic drift and increases the extent of linkage disequilibrium, leading to large genomic windows affected by genetic sweeps (Glémin and Bataillon, 2009; Bitocchi et al., 2013). This was also confirmed by the resequencing results in other autogamous species, such as soybean and rice (Lam et al., 2010; Xu et al., 2011).
Here, we demonstrated a similar change from domestication over the entire set of genes compared with annual crops and autogamous species, that is, a 20.89% loss in gene expression diversity was associated with domestication in pear. However, we also observed a 23.13% increase of expression diversity in the improved group. This suggested that diversifying selection might play a more active role in modern improvement of pear, and likely that introgression has occurred from wild or landrace relatives. Pears have more than 3,000 years of cultivation history (Lombard and Westwood, 1987). Therefore, it has had a long recovery period for polymorphism. In addition, the self-incompatibility might contribute to the recovery of expression diversity, and favored traits from different pear varieties would help with adapting to new environments and human actions in modern breeding. When compared with pear, in maize a lower degree of recovery of expression diversity was observed (Hufford et al., 2012; Swanson-Wagner et al., 2012). Even though maize is also a widely openly pollinated species, maize has a more stringent selection breeding system, which contributes to slower recovery.
Further, 5.65% and 9.38% of the reference genes were differentially expressed in wild versus landrace pears and landrace versus improved pears, respectively. More genes were down-regulated in landrace pears for the comparison of wild versus landrace pears, supporting that loss-of-function mutations are relatively frequent compared with gain-of-function types as an easily available source of variation that supports selection during rapid environmental change (Olson, 1999). As first stated by Darwin (1868), as plants evolve from wild to cultivated agronomic traits during domestication selection, cultivated traits show recessive inheritance in domesticated plants (Lester, 1989). Moreover, the module-trait association analysis showed that most genes in the module highly associated with acid and stone cells were down-regulated in landrace pears. In contrast, in the fruit size–related module, most genes were up-regulated in landrace pears (Supplemental Fig. S9). These results suggest that acid and stone cell content could be considered as plant traits caused by loss-of-function mutations during pear domestication, whereas the larger fruit size of cultivated pears may be due to gain-of-function mutations selected by humans as traits that improved the usefulness of the fruit.
Meanwhile, high-confidence–selected genes were identified through tight thresholds of π ratio and FST evaluation. A total of 2.35% (996) of genes was identified as the candidate selected genes during pear domestication. When compared with the reported 857 selected genes from pear resequencing (Wu et al., 2018), around 13.65% (136) overlapped genes were observed. These genes will be considered as the important domesticated genes in pear, whereas no overlapped gene was found between the candidate selected genes identified in our current study and the selected genes reported by Kumar et al. (2017). However, only 7.23% of domesticated and 7.31% of improved genes are differentially expressed in the comparison of wild versus landraces pools and landraces versus improved pools, respectively. Additionally, 53 (6.18%) out of 857 selected genes were differentially expressed between the wild and landrace pear group in our study. These results suggested that diversifying selection might have occurred in the landraces groups, with domestication increasing the level of functional diversity.
In pear, the most intuitively apparent domestication-associated trait is the dramatic increase in fruit size (Fig. 1A). Previous studies have shown that fruit size is the most typical of the domestication-associated traits and is controlled by a relatively small number of loci (Grandillo and Tanksley, 1996; Koenig et al., 2013). In addition to fruit size, the phenotypic diversity between cultivated pear and its wild genotypes also specifically included stone cell content and the proportions of sugar and acid. These traits provided an excellent system for comparing gene expression differences between cultivated and wild pears to detect genes associated with domestication and improvement. Wild plants are known to have higher stress resistance than cultivated genotypes. Therefore, during pear domestication, resistance is likely the main target of functional loss. In the stone cell– and acid-related module, 240 of the 373 genes were annotated, and an abundance of genes (35.42%) were involved in the biotic-abiotic response and photosynthetic pathways (Supplemental Table S17), which further supported the hypothesis that plant resistance and light-harvesting ability have changed dramatically between wild and landrace pears. In addition, more sugar-acid related and lignin-related genes were identified; for example, the genes Pbr019305.1, Pbr021220.1, and Pbr040238.1 were predicted to participate in the lignin synthesis pathway as the homologous genes of caffeoyl-CoA 3-O-methyltransferase, alcohol dehydrogenase, and CAD (Wu et al., 2013). Malic acid and sorbitol are the important components of the sugar/acid ratio, which deeply affects pear fruit flavor. In our analysis, interestingly, a malic acid synthesis-related gene, Pbr024269.1, a homologous gene of PMDH1 (Pracharoenwattana et al., 2007), and a sorbitol synthesis-related gene, Pbr013916.1, were identified, and these genes might regulate the synthesis and metabolism of sugar and acid in pear fruit. We also identified two genes, Pbr013295.1 and Pbr028831.1, involved in the regulation of flowering time and the circadian clock as homologs of the gene CONSTANS (Yano et al., 2000; Suárez-López et al., 2001; Sarid-Krebs et al., 2015). Flowering time is very different between wild and cultivated pears, in order to adapt to changing growth environments. Indeed, most genes in the fruit size–related module had unknown functions, and a large number of genes were annotated to participate in biotic or/and abiotic stress responses (Supplemental Table S17). Meanwhile, the conserved domains of each gene were identified as a further predictor of gene function (Supplemental Table S18) using the software HMMER3 (http://hmmer.org/) with p-value ≤ 1e-05.
MATERIALS AND METHODS
Sample Collection
We selected 41 pear (Pyrus pyrifolia) accessions from 14 wild (PyW1-PyW14), 12 landraces (PyL1-PyL12), and 15 improved genotypes (PyI1-PyI15) at the key stage (enlarged) of fruit development for RNA-seq. All genotypes were collected at different dates for the enlarged fruit stage because of different phenological periods. Three independent biological repeats (named as PyW1-1 to PyW1-3, etc.) were collected from three trees of each accession of 41 pears. Meanwhile, one wild pear (Outgroup) of P. ussuriensis, collected at the mature stage, was selected as an outgroup. To assess the transcriptomic and phenotypic data, three representative wild (PyW12, PyW13, PyW14) and three representative landrace (PyL1, PyL2, PyL3) genotypes at three key stages (small, enlarged, and mature) of fruit development, named as PyL1-a to represent the small fruit stage, PyL1-b to represent the enlarged fruit stage, and PyL1-c to represent the mature stage, were collected for associated analyses. To minimize expression differences due to environmental conditions, all samples were grown in the natural environment without artificial regulation and collected in the year 2014 and 2018, and the pulp of all pear samples was frozen for subsequent experiments.
RNA Extraction and Library Construction
According to the manufacturer’s protocol, 100 mg of pear flesh from the mixed pulp sample was used for RNA isolation using the Plant Total RNA Isolation Kit Plus (FOREGENE Co.). The RNA was then treated with RNase-Free DNase I. Qualitative and quantitative control was performed with an Applied Biosystems StepOnePlus Real-Time PCR System and an Agilent 2100 Bioanalyzer. Only RNA samples with an RNA integrity number > 8.0 were used. Three biological replicates of RNA extraction were performed separately and then mixed for later sequencing. RNA from three replicates was mixed in equal amounts, and then 10 μg of combined RNA was used for the construction of anondirectional Illumina RNA-seq library, using the TruSeq RNA sample preparation kits v2 (Illumina), following the manufacturer’s instructions. Libraries were quantified using an Applied Biosystems StepOnePlus Real-Time PCR System, and quality control was performed with the Agilent 2100 Bioanalyzer. RNA-seq was performed with an Illumina HiSeq 2500 Sequencer using the TruSeq SBS v3-HS kits (200 cycles) and TruSeq PE Cluster v3-cBot-HS kits (Illumina) to generate 125-bp paired-end reads.
RNA-Seq Expression Analysis and SNP Calling
For each sequence library, read quality was evaluated using FastQC software (Andrews, 2010). The Trimmomatic (Bolger et al., 2014) software package was used to remove the adapter sequences and low quality sequences. Clean reads of each library were mapped to the reference genome of ‘Dangshansuli’ pear using HISAT2 version 2.1.0 (Kim et al., 2015) with the following parameters:–min-intronlen, 20;–max-intronlen, 4000; -I, 0; -X, 500. For expression quantification, we used featureCounts (Liao et al., 2014) to count the mapped read counts of each sample, and then calculated the RPKM value using an in-house python script based on the formula: RPKM = (total exon reads)/(mapped reads [millions]*exon length [kb]). The coefficient of variation was calculated using the formula: Cν = σ/μ, where σ represents sd and μ represents mean. Pearson correlation coefficients of expression levels were calculated between each pair of genotypes using R package. A relatively high correlation was expected because the same tissue was harvested across the genotypes; thus, genotypes with R2 values < 0.5 across samples were removed. DEGs were identified using EdgeR package (Robinson et al., 2010) in R software, considering samples within wild, landraces, and improved groups as multiple biological replicates. Significantly DEGs were further filtered using thresholds set as follows: |log2FoldChange|>1 and FDR ≤ 0.001 (Audic and Claverie, 1997; Mortazavi et al., 2008). The chromosome-based χ2 test was performed using the chisq.test() function in R package, and window-based χ2 test was performed using our in-house python scripts.
To call SNPs, we further removed the PCR duplicated reads and multiple mapped reads using rmdup and view functions in SAMtools software (Li et al., 2009), respectively. Alignments for reads that mapped uniquely were processed using the sort, index, and pileup programs within SAMtools version 1.4.1, and called SNPs used BCFtools version 1.4.1 (http://samtools.github.io/bcftools/). A locus was considered polymorphic if at least two alleles had > 5% allele frequency and with a minimum mapping quality threshold of q = 20 were retained. Finally, loci were removed that had > 50% missing data, and 875,319 high-quality SNPs were maintained for subsequent analyses.
Population Structure and Diversity Analyses
We performed PCA to explore the genetic relationships among individuals. Based on the expression data, we used the graphics package to calculate in the R statistical environment. For the SNP dataset, Variant Call Format (VCF) tools (Danecek et al., 2011) and Plink (Purcell et al., 2007) software was used to process the data, and genome-wide complex trait analysis (Yang et al., 2011) software was used for the calculations. The final figures were visualized using ggplot2 package in R. Further, software SNPhylo (Lee et al., 2014) was used to construct the phylogenetic tree based on the SNP data with the parameters sets as follow: -m, 0.05; -M, 0.5; -l, 0.2; -b, 1000. The tree was visualized in FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). The VCF file with SNP data were converted to genotype using our in-house python script, and then the LEA package (http://membres-timc.imag.fr/Olivier.Francois/tutoRstructure.pdf) in R software was used to calculate the admixture coefficients with default parameters: maximum number of iterations, 200; regularization parameter, 100; and tolerance error, 1e-5. The number of clusters (K) was set from 2 to 4 and displayed using the barplot() function in R package. The population diversity statistics including π and FST were computed using software VCFtools v0.1.13. A slide window of 10 kb, along with a step length of 1 kb, was used to estimate the π value. Pairwise FST value was estimated in the same windows and steps to measure the population differentiation between groups. Correlation coefficients (r2) of alleles were calculated and visualized using PopLDdecay (Zhang et al., 2018) to measure LD values in each of the three groups (wild, landraces, and improved group).
Detection of Selective Sweeps during Domestication and Improvement
To identify regions with selective signals, the π ratio (πgroup1/πgroup2) and FST values were calculated in 10-kb slide windows across the entire pear genome. The 10-kb slide windows with significant selective signals were identified using the following criteria: top 5% of FST and 5% of the π ratio. Genes were identified from these regions with selective signals as the candidate selected genes. Further, the coefficient of expression variation of these selected genes was also calculated using the formula mentioned above.
Measurement of Phenotypic Data
Phenotypic data including pear fruit size, sugar and acid content, and stone cell content were determined for each sample at each time point. A Vernier caliper was used to measure the longitudinal diameter and transverse diameter of pear fruit, and an electronic balance was used to measure the fruit weight. Stone cell content was measured through the combined method of HCl separation and freezing processing. First, a 100-g sample of peeled pear flesh was weighed and homogenized with distilled water in a stirrer for 10 min (min). Then, we diluted the homogenate with distilled water and placed the suspension at room temperature for 30 min. Finally, the aqueous phase was decanted, and the sediment was suspended in 0.5 n HCl for 30 min, decanted, and washed with distilled water. This operation was repeated several times until the stone cells were completely separated (Lee et al., 2006; Tao et al., 2009). We used HPLC (Waters 1525 HPLC system) to measure the soluble sugars and organic acids. A Breeze chromatography data system was used to integrate the peak areas according to external standard solution calibrations (standard sugars were purchased from Sigma Chemical Co.). Finally, we used mg/g fresh weight to describe the sugar and acid concentrations (Liu et al., 2016). The correlation analysis was performed using the cor() function in R package, and the significant difference was evaluated using T-test in R. The final figures were plotted using ggplot2 package in R.
Network analysis of Gene Coexpression
Coexpression networks were constructed using the WGCNA (v1.51; Langfelder and Horvath, 2008) package in R. A total of 2,905 DEGs from wild versus landrace accessions were used in the K-means clustering analysis; genes with RPKM were used for the WGCNA unsigned coexpression network analysis. The modules were obtained using the automatic network construction function blockwiseModules with default settings, except that the power was 9, TOMType was signed, minModuleSize was 30, and mergeCutHeight was 0.20. Modules are defined as clusters of highly interconnected genes, and genes within the same cluster have high pairwise correlation coefficients. Using gene significance and module membership measurements, we identified genes with high significance for interesting traits, as well as high module membership in interesting modules. The eigengene value was calculated for each module and used to test the association with each trait type. The kME (for modular membership, also known as eigengene-based connectivity) and kME-P value were calculated for the 2,903 genes, which were clustered into 16 trait-specific modules. The remaining two genes were outliers (gray module) and are not shown in Supplemental Table S5.
RNA Extraction and RT-qPCR Analysis
Total RNA was extracted using the Plant Total RNA Isolation Kit Plus. First-strand cDNA was synthesized using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech Co. Ltd.) according to the manufacturer’s instructions. We designed primers to amplify genes using Primer Premier 5.0 software (Premier Biosoft International). RT-qPCR analysis was carried out using the LightCycler 480 SYBR GREEN I Master (Roche) according to the manufacturer’s protocol. We performed each reaction using a 20 µl mixture containing 10 µl of LightCycler 480 SYBR GREEN I Master Mix, 100 ng of template cDNA, and 0.5 μm of each primer. All reactions were run in 96-well plates, and each cDNA was analyzed in quadruplicate. The RT-qPCR conditions were set as follows: preincubation at 95°C for 5 min; 55 cycles of 95°C for 3 s, 60°C for 10 s, 72°C for 30 s; then extension at 72°C for 3 min; finally, fluorescence data collection was carried out at 60°C. We calculated the average threshold cycle (Ct) of each sample. Pyrus Actin (accession no. AF386514) and Pyrus glyceraldehyde-3-phosphate dehydrogenase were used as the internal control genes, and the relative expression levels were calculated with the 2−ΔΔCt method described by Livak and Schmittgen (2001).
Accession Numbers
The accession number PRJNA157875 is available in National Center for Biotechnology Information and genes/proteins mentioned in our study are also available for download from Pear Genome Project (http://peargenome.njau.edu.cn).
SUPPLEMENTAL DATA
The following supplemental materials are available.
Supplemental Figure S1. The heatmap of Pearson correlation coefficients (PCCs) among three biological repeats of each accession.
Supplemental Figure S2. The summary of differentially expressed genes (DEGs) from three pair comparisons of three groups.
Supplemental Figure S3. The GO enrichment analysis of differentially expressed genes (DEGs).
Supplemental Figure S4. The distribution of 996 (blue bar) and 301 (orange bar) candidate domesticated and improved genes on 17 pear chromosomes.
Supplemental Figure S5. The Venn plot shows the common genes between selected genes and DEGs.
Supplemental Figure S6. The summary of sample phenotypes, number of genes differentially expressed and multidimensional scaling (MDS) analysis.
Supplemental Figure S7. The heatmap of Pearson correlation coefficients (PCC) using expressed genes (RPKM > 5).
Supplemental Figure S8. Correlation and statistical significances of 15 phenological traits in pear fruit.
Supplemental Figure S9. A scatterplot and heatmap of MM vs. GS.
Supplemental Table S1. Summary of the 41 pear accessions, sequencing and mapping based on the reference genome of 'Dangshansuli' (P. bretschneideri).
Supplemental Table S2. The list of gene specifically expressed in wild, landraces and improved group.
Supplemental Table S3. The coefficient of expression variation (CV) within group.
Supplemental Table S4. The Pearson correlation coefficient (PCC) among all samples.
Supplemental Table S5. Differentially expressed genes in the three comparisons.
Supplemental Table S6. Chromosomal distribution of differentially expressed genes on the P. bretschneideri genome.
Supplemental Table S7. The value of nucleotide diversity (π) in each 10 kb slide window across the whole pear genome from wild, landraces and improved groups.
Supplemental Table S8. The value of Fst in each 10 kb slide window across the whole pear genome between wild, landraces and improved groups.
Supplemental Table S9. The value of linkage disequilibrium (LD) in wild, landraces, and improved groups and all accessions.
Supplemental Table S10. The list and their annotations of candidate selected genes in the pear domestication and improvement process.
Supplemental Table S11. Chromosomal distribution of selected genes on the P. bretschneideri genome.
Supplemental Table S12. The coefficient of expression variation (CV) for selected genes in the domestication and improvement process.
Supplemental Table S13. The RPKM values and annotation information of DEGs between wild and landrace pear accessions at three developmental stages.
Supplemental Table S14. The phenotypic data from 18 samples.
Supplemental Table S15. The 2,903 DEGs grouped into to 16 important trait-related modules.
Supplemental Table S16. The previously reported QTLs and the QTL mapping against DEGs.
Supplemental Table S17. The annotation information of genes in the stone cell and acid related module and the fruit size related module.
Supplemental Table S18. The analysis of conserved domains of genes in the stone cell related module and the fruit size related module.
Acknowledgments
We thank Dr. Scott Jackson (University of Georgia) and Chunming Xu (University of Georgia) for providing the valuable suggestions for our works. We also thank the group members in the Centre of Pear Engineering Technology Research, State Key Laboratory of Crop Genetics and Germplasm Enhancement at the Nanjing Agricultural University.
Footnotes
This work was supported by the National Key Research and Development Program (2018YFD1000200), the National Science Fund of China (31672111), the Earmarked Fund for the China Agriculture Research System (CARS-28), and the Earmarked Fund for Jiangsu Agricultural Industry Technology System (JATS 2018-277).
Articles can be viewed without a subscription.
References
- Andrews S. (2010) FastQC: A Quality Control Tool For High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (April 10, 2018)
- Audic S, Claverie J-M (1997) The significance of digital gene expression profiles. Genome Res 7: 986–995 [DOI] [PubMed] [Google Scholar]
- Bell RL, Quamme HA, Layne REC, Skirvin RM (1996) Pears. In Janick J and Moore JN, eds, Fruit breeding. John Wiley and Sons, New York, pp 441–514 [Google Scholar]
- Bellucci E, Bitocchi E, Ferrarini A, Benazzo A, Biagetti E, Klie S, Minio A, Rau D, Rodriguez M, Panziera A, et al. (2014) Decreased nucleotide and expression diversity and modified coexpression patterns characterize domestication in the common bean. Plant Cell 26: 1901–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitocchi E, Bellucci E, Giardini A, Rau D, Rodriguez M, Biagetti E, Santilocchi R, Spagnoletti Zeuli P, Gioia T, Logozzo G, et al. (2013) Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol 197: 300–313 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K, Zheng Z, Wang L, Liu X, Zhu G, Fang W, Cheng S, Zeng P, Chen C, Wang X, et al. (2014) Comparative population genomics reveals the domestication history of the peach, Prunus persica, and human influences on perennial fruit crops. Genome Biol 15: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro N, Tisdale-Orr TE, Clouse RM, Knöller AS, Spicer R (2012) Diversification and expression of the PIN, AUX/LAX, and ABCB families of putative auxin transporters in Populus. Front Plant Sci 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. ; 1000 Genomes Project Analysis Group (2011) The variant call format and VCFtools. Bioinformatics 27: 2156–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. (1868) The Variation of Animals and Plants Under Domestication, Vol 2 John Murray, London, United Kingdom [Google Scholar]
- Fumagalli M, Vieira FG, Korneliussen TS, Linderoth T, Huerta-Sánchez E, Albrechtsen A, Nielsen R (2013) Quantifying population genetic differentiation from next-generation sequencing data. Genetics 195: 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S, Bataillon T (2009) A comparative view of the evolution of grasses under domestication. New Phytol 183: 273–290 [DOI] [PubMed] [Google Scholar]
- Grandillo S, Tanksley SD (1996) QTL analysis of horticultural traits differentiating the cultivated tomato from the closely related species Lycopersicon pimpinellifolium. Theor Appl Genet 92: 935–951 [DOI] [PubMed] [Google Scholar]
- Hufford MB, Xu X, van Heerwaarden J, Pyhäjärvi T, Chia JM, Cartwright RA, Elshire RJ, Glaubitz JC, Guill KE, Kaeppler SM, et al. (2012) Comparative population genomics of maize domestication and improvement. Nat Genet 44: 808–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Zhao H, Ren L, Song W, Zeng B, Guo J, Wang B, Liu Z, Chen J, Li W, et al. (2012) Genome-wide genetic changes during modern breeding of maize. Nat Genet 44: 812–815 [DOI] [PubMed] [Google Scholar]
- Kikuchi A. (1946) Speciation and taxonomy of Chinese pears. Collected Records Horticultural Research. 3: 1–8 [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: A fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig D, Jiménez-Gómez JM, Kimura S, Fulop D, Chitwood DH, Headland LR, Kumar R, Covington MF, Devisetty UK, Tat AV, et al. 2013. Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc Natl Acad Sci U S A 110: 2655–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Kirk C, Deng C, Wiedow C, Knaebel M, Brewer L (2017) Genotyping-by-sequencing of pear (Pyrus spp.) accessions unravels novel patterns of genetic diversity and selection footprints. Hortic Res 4: 17015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Xu X, Liu X, Chen W, Yang G, Wong FL, Li MW, He W, Qin N, Wang B, et al. (2010) Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet 42: 1053–1059 [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008) WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Choi JH, Kim WS, Han TH, Park YS, Gemma H (2006) Effect of soil water stress on the development of stone cells in pear (Pyrus pyrifolia cv. ‘Niitaka’) flesh. Sci Hortic (Amsterdam) 110: 247–253 [Google Scholar]
- Lee TH, Guo H, Wang X, Kim C, Paterson AH (2014) SNPhylo: A pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics 15: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester RN. (1989) Evolution under domestication involving disturbance of genic balance. Euphytica 44: 125–132 [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Liu L, Chen CX, Zhu YF, Xue L, Liu QW, Qi KJ, Zhang SL, Wu J (2016) Maternal inheritance has impact on organic acid content in progeny of pear (Pyrus spp.) fruit. Euphytica 209: 305–321 [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lombard P, Westwood M (1987) Pear rootstocks. Rom RC, Carlson RF, eds., Rootstocks for fruit crops. Wiley, New York, pp 145–183. [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Olson MV. (1999) When less is more: Gene loss as an engine of evolutionary change. Am J Hum Genet 64: 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM (2007) Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. Plant J 50: 381–390 [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarid-Krebs L, Panigrahi KC, Fornara F, Takahashi Y, Hayama R, Jang S, Tilmes V, Valverde F, Coupland G (2015) Phosphorylation of CONSTANS and its COP1-dependent degradation during photoperiodic flowering of Arabidopsis. Plant J 84: 451–463 [DOI] [PubMed] [Google Scholar]
- Schmutz J, McClean PE, Mamidi S, Wu GA, Cannon SB, Grimwood J, Jenkins J, Shu S, Song Q, Chavarro C, et al. (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Swanson-Wagner R, Briskine R, Schaefer R, Hufford MB, Ross-Ibarra J, Myers CL, Tiffin P, Springer NM. 2012. Reshaping of the maize transcriptome by domestication. Proc Natl Acad Sci USA 109: 11878-11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao S, Khanizadeh S, Hua Z, Zhang S (2009) Anatomy, ultrastructure and lignin distribution of stone cells in two Pyrus species. Plant Sci 176: 413–419 [Google Scholar]
- Watterson GA. (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7: 256–276 [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370 [DOI] [PubMed] [Google Scholar]
- Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS (2005) The effects of artificial selection on the maize genome. Science 308: 1310–1314 [DOI] [PubMed] [Google Scholar]
- Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S, Khan MA, Tao S, Korban SS, Wang H, et al. (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23: 396–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li L-T, Li M, Khan MA, Li X-G, Chen H, Yin H, Zhang SL (2014) High-density genetic linkage map construction and identification of fruit-related QTLs in pear using SNP and SSR markers. J Exp Bot 65: 5771–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang Y, Xu J, Korban SS, Fei Z, Tao S, Ming R, Tai S, Khan AM, Postman JD, et al. (2018) Diversification and independent domestication of Asian and European pears. Genome Biol 19: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Liu X, Ge S, Jensen JD, Hu F, Li X, Dong Y, Gutenkunst RN, Fang L, Huang L, et al. (2011) Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol 30: 105–111 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Terakami S, Moriya S, Hosaka F, Kurita K, Kanamori H, Katayose Y, Saito T, Nishitani C (2013) DNA markers developed from genome sequencing analysis in Japanese pear (Pyrus pyrifolia). Acta Hortic 976: 477–483 [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet 88: 76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al. (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Dong SS, Xu JY, He W M, Yang T L (2018) PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics bty875. 10.1093/bioinformatics/bty875 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Jiang Y, Wang Z, Gou Z, Lyu J, Li W, Yu Y, Shu L, Zhao Y, Ma Y, et al. (2015) Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol 33: 408–414 [DOI] [PubMed] [Google Scholar]