A comprehensive, high-resolution overview is presented of the molecular, ultrastructural, and physiological events involved in chloroplast differentiation and thylakoid biogenesis.
Abstract
Upon exposure to light, plant cells quickly acquire photosynthetic competence by converting pale etioplasts into green chloroplasts. This developmental transition involves the de novo biogenesis of the thylakoid system and requires reprogramming of metabolism and gene expression. Etioplast-to-chloroplast differentiation involves massive changes in plastid ultrastructure, but how these changes are connected to specific changes in physiology, metabolism, and expression of the plastid and nuclear genomes is poorly understood. Here, we describe a new experimental system in the dicotyledonous model plant tobacco (Nicotiana tabacum) that allows us to study the leaf deetiolation process at the systems level. We have determined the accumulation kinetics of photosynthetic complexes, pigments, lipids, and soluble metabolites and recorded the dynamic changes in plastid ultrastructure and in the nuclear and plastid transcriptomes. Our data describe the greening process at high temporal resolution, resolve distinct genetic and metabolic phases during deetiolation, and reveal numerous candidate genes that may be involved in light-induced chloroplast development and thylakoid biogenesis.
The thylakoid membranes of mature chloroplasts contain the protein complexes required for the light reactions of photosynthesis and, as such, house the reactions that fuel nearly all life on Earth. Mature chloroplasts develop from proplastids, which, remarkably, contain little or no membrane structures and are found, in dicotyledons, exclusively within a few tens of cells in the shoot apical meristem (Charuvi et al., 2012). Alternatively, chloroplasts may develop from etioplasts, a type of plastid that is formed in the absence of light. Etioplasts form under laboratory conditions but have also been identified in natural contexts, including emerging seedlings, inner leaves of white cabbage (Brassica oleracea ‘Capitata’) and lettuce (Lactuca sativa; Solymosi et al., 2004), and the leaf buds of various tree species (Solymosi and Böddi, 2006; Solymosi et al., 2006). The precise processes underlying the transition from etioplast to chloroplast, while of vital importance to the productivity of the plant, remain poorly understood.
Unlike proplastids, etioplast contain a complex membrane structure, consisting primarily of a semicrystalline network of tetrahedrally branched tubules, referred to as the prolamellar body (PLB). Like thylakoid membranes, PLBs are formed from the lipids monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), sulfoquinovosyldiacylglycerol (SQDG), and phosphatidylglycerol (PG; Bahl et al., 1976; Galey et al., 1980; Murata et al., 1990) and additionally contain carotenoids (Park et al., 2002). Etioplast proteins include many found in mature chloroplasts, such as those involved in protein synthesis and carbohydrate and amino acid metabolism, as well as chaperones, redox regulators/protectants, and proteases. Furthermore, many photosynthetic proteins, including the Rubisco complex (and its chaperones and activators), assembled ATP synthase and cytochrome b6f (cyt b6f) complexes, and various non-chlorophyll-binding PSII subunits, are present in etioplasts, although generally in lower amounts than seen in chloroplasts (Komatsu et al., 1999; von Zychlinski et al., 2005; Blomqvist et al., 2006, 2008; Kanervo et al., 2008; Reisinger et al., 2008; Plöscher et al., 2009, 2011). The predominant protein in the etioplast is the light-dependent NADPH:protochlorophyllide oxidoreductase (POR; Sperling et al., 1998; Franck et al., 2000), the accumulation of which is associated with both the formation and size of PLBs. Associated with POR is its cofactor NADPH and the chlorophyll a precursor, protochlorophyllide. The cosequestration of POR and protochlorophyllide allows the nearly instantaneous synthesis of chlorophyll upon lighting (Reinbothe et al., 1999; Domanskii et al., 2003), marking the first step of deetiolation and the transition from etioplasts to mature chloroplasts.
Deetiolation involves the concerted and synchronized activity of a highly complex biogenesis program. Thylakoid membranes must develop from disassembling PLBs and prothylakoids and from newly synthesized lipids. Large protein complexes containing dozens of protein subunits and hundreds of pigments and cofactors must be inserted into the budding membrane in strictly defined stoichiometric ratios. The protein complexes consist of polypeptides originating from two evolutionarily distinct compartments, the nucleus and the plastid, which must be expressed, processed, targeted, and inserted into the membrane in a highly coordinated manner. These processes are dependent upon and controlled by a wide range of assembly chaperones and other biogenesis factors, which are not or only poorly understood (Adam et al., 2011). In spite of the complexity of thylakoid biogenesis, the etioplast-to-chloroplast transition can occur astoundingly rapidly. Upon illumination, PSI activity may appear in as little as 15 min, and PSII and ATP synthase activity appear after 2 to 3 h (Baker and Butler, 1976; Wellburn and Hampp, 1979). Although it is clear that the genetic, biochemical, and physiological processes involved in this transition must occur in a highly timed manner, little is known about the kinetics of the various processes and activities mediating the development of functional thylakoids and the regulators and assembly factors involved.
The complexity of the processes required for this developmental transition and their precise timing underscore the need for highly time-resolved, systems biology analyses of the deetiolation process. Although many studies have investigated specific aspects of deetiolation, the use of diverse experimental conditions, systems, and plant species has prevented comparison and integration of data sets. Many studies have focused on monocotyledons (Lonosky et al., 2004; Blomqvist et al., 2008; Cahoon et al., 2008; Li et al., 2010; Plöscher et al., 2011), in which the transition from proplastids to chloroplasts occurs along the longitudinal axis of the developing leaf. By contrast, dicotyledons, despite being the largest angiosperm group, are much less studied (Rudowska et al., 2012; Kowalewska et al., 2016; Skupień et al., 2017). Finally, most studies have primarily focused on a single technique, a small subset of processes, and/or only a few time points.
Here, we have developed a system to study both the deetiolation process and the process of photosynthetic maturation in leaves of tobacco (Nicotiana tabacum) at high temporal resolution. Targeted and nontargeted approaches were undertaken to define the dynamic changes in the transcriptomes of the nucleus and the plastid. In addition, the accumulation kinetics of pigments, lipids, soluble metabolites, and photosynthetic proteins and their activities were determined and correlated with the physical changes in membrane ultrastructure. Our work provides a comprehensive systems-level description of thylakoid development and the etioplast-to-chloroplast differentiation process and also reveals candidate genes involved in chloroplast biogenesis and the acquisition of photosynthetic competence.
RESULTS
A System to Observe Both Deetiolation and Development of Photosynthetic Competence in Tobacco Leaves
The few deetiolation studies in dicots that are currently available have investigated only certain aspects of the greening process and have predominantly observed greening in cotyledons grown on (often sucrose-supplemented) media under aseptic conditions (Sperling et al., 1998; Philippar et al., 2007). However, elements of the developmental program of cotyledons may differ from or even oppose those of true leaves (Trémolières and Lepage, 1971), while application of exogenous sugars influences plastid number and morphology as well as photosynthetic activity and the expression of nuclear and plastid genes (Eckstein et al., 2012; Häusler et al., 2014; Van Dingenen et al., 2016). We therefore developed and optimized a system that allows deetiolating leaf tissue to be observed in soil-grown dicotyledonous plants.
Mature tobacco plants grown under long-day conditions accumulate large amounts of starch, which is not fully depleted during a single night and can be used to fuel new growth. We optimized the initial growth conditions and age of the plant at treatment and the length of the extended dark period to maximize the growth of new tissue during darkness while minimizing the onset of starvation-induced senescence responses. In our system, nearly mature long-day (16-h photoperiod, 350 μmol photons m2 s−1)-grown tobacco plants were subjected to an extended night of 56 h (one night followed by two dark day/night cycles). Placement of the entire plant in darkness was selected to prevent the newly forming leaves from (1) receiving primary or secondary light signals during the dark period, (2) developing under an altered microenvironment compared with the rest of the plant, and (3) undergoing dark-induced senescence, which is accelerated when individual leaves are darkened (Weaver et al., 1998). During the extended night, cell division and expansion of the smallest, developing leaves resulted in the growth of new, pale tissue.
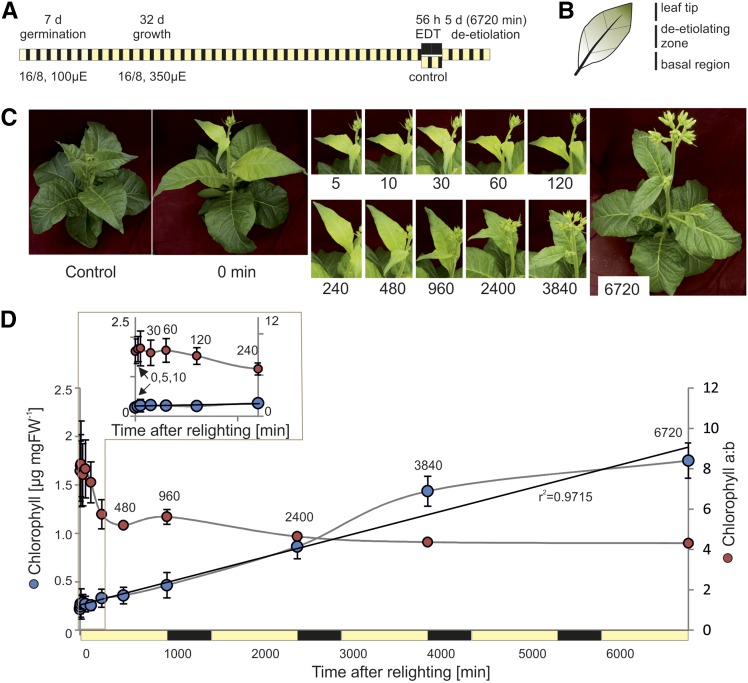
To analyze both rapid changes associated with de-etiolation and the more gradual development of photosynthetic maturity, samples were taken densely throughout the first photoperiod following the extended night (0 min, 5 min, 10 min, 30 min, 1 h, 2 h, 4 h, 8 h, and 16 h, where the final sample corresponded with the end of the photoperiod) and at the end of the photoperiod on days 2, 3, and 5 after lighting (Fig. 1A). These time points are referred to throughout as minutes following relighting: 0, 5, 10, 30, 60, 120, 240, 480, 960, 2,400, 3,840, and 6,720 min. Tissue was also collected directly, prior to relighting, from plants that had not been subjected to extended darkness (control). The wedge-shaped leaf tip containing green tissue that developed prior to the extended night and the basal region containing newly dividing cells were excised and excluded from analysis (Fig. 1B). Each biological repeat consisted of homogenized pooled tissue taken from each of the two youngest leaves of each of three replicate plants. These pools were used for genome copy number, transcript, pigment, protein, and metabolite measurements, with in vivo photosynthetic measurements and samples for microscopy taken separately.
Figure 1.
A system to observe deetiolation in leaves of tobacco. A, Experimental design. Following extended dark treatment (EDT) of nearly mature tobacco plants, the dark-grown leaf sectors of young leaves were sampled across 5 d (6,720 min) of deetiolation, with particular focus on early time points. B, The measured leaf sector (deetiolating zone) was separated from the leaf tip (grown in part prior to EDT) and the basal region (newly growing tissue) following rapid freezing in liquid nitrogen. Only the deetiolating zone was used for subsequent analyses. C, Visual phenotypes of young leaves immediately following the dark treatment (0 min) and throughout the greening time course. The control plant was kept under standard growth conditions and harvested at t = 0. Numbers under the images represent time after lighting in minutes. D, Total chlorophyll content and chlorophyll a/b ratio in etiolated and deetiolating samples. Error bars indicate sd (n = 3). The black line indicates a linear trend, fitted to changing chlorophyll content, with the r2 value of the regression shown. The first 240 min are shown as an inset. Time points 960, 2,400, 3,840, and 6,720 min correspond with the end of the photoperiod on days 1, 2, 3, and 5 after lighting, respectively. FW, Fresh weight.
Relative to the developing leaves of control plants, those of dark-treated plants were pale at 0 min (t = 0) and both visibly greened and grew in size during the time course (Fig. 1C). To assess the greening kinetics, chlorophyll content and chlorophyll a/b ratio were measured spectroscopically (Porra et al., 1989). Chlorophyll content per fresh weight rose in a nearly linear fashion during the first photoperiod, consistent with previous reports (Boasson and Laetsch, 1969; Baker and Butler, 1976), and continued to increase in a similar manner throughout the subsequent day-night periods (r20-6720 min = 0.97; Fig. 1D). Chlorophyll b accumulated less rapidly than chlorophyll a during greening, resulting in a high chlorophyll a/b ratio shortly after lighting, which then rapidly decreased within the first hours of greening, as previously shown (Thorne and Boardman, 1971; Porra et al., 1994). This decrease occurred primarily between 120 and 240 min of greening (Fig. 1D) and then continued more gradually, to reach a final ratio of 4.3, consistent with that seen in fully expanded tobacco leaves grown under standard conditions (Schöttler et al., 2017).
Unlike completely dark-grown cotyledons, leaf tissue subject to our conditions contained some chlorophyll at t = 0 (Fig. 1D). This likely arose due to the formation of dark-grown tissue via cellular division and expansion from the leaf basal region (Nelissen et al., 2016), which, although largely shielded from light in the early stages of leaf formation, may receive some light signals prior to the extended dark treatment. Nonetheless, the ∼8-fold increase in chlorophyll content (and presence of PLBs in the plastids after dark treatment; Fig. 2) supports the presence of a dominant greening process in the sampled tissue.
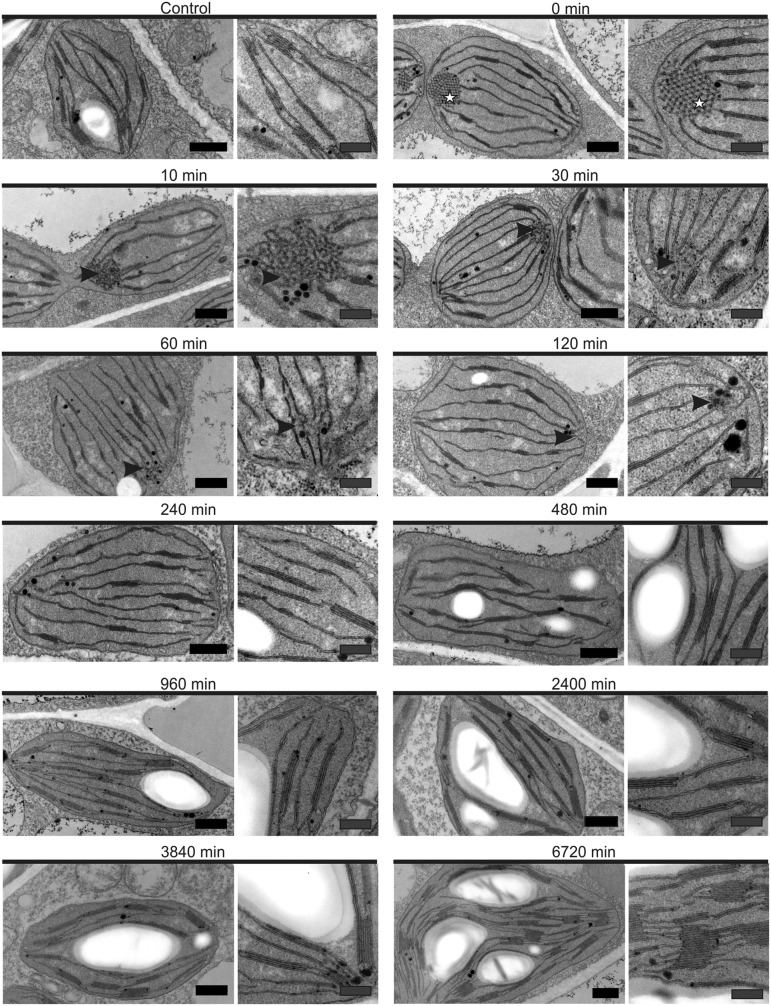
Figure 2.
Transmission electron micrographs of plastids present in chemically fixed deetiolating tobacco leaves. Etiolated leaf tissue contained plastids with structured, paracrystalline PLBs, in contrast with control plastids, harvested at the same time but not subjected to extended dark treatment. After just 10 min, the PLBs (stars) took on a less uniform, more irregular shape (arrowheads) and simultaneously began to disintegrate: residual PLBs were absent after 120 min of light. Subsequent time points were marked by increased abundance and size of grana membranes and the gradual accumulation of starch. Images at right reveal membrane structures at higher magnification. See Figure 3 and Supplemental Table S1 for quantification. For technical reasons (sample processing time), the 5 min time point was excluded from microscopy analysis. See Supplemental Figure S1 for comparison between chemical fixation and high-pressure freezing and Supplemental Figure S2 for further images of the etiolated plastids. Black bars = 500 nm and gray bars = 250 nm.
The Etioplast-to-Chloroplast Transition as Revealed by Transmission Electron Microscopy
To confirm our system as suitable for observing the etioplast-to-chloroplast transition, and to follow the changes in chloroplast ultrastructure over the time course of the deetiolation, transmission electron microscopy (TEM) was undertaken. Due to technical limitations of sample handling time, the 5 min time point was excluded from microscopy analysis. To control for fixation artifacts, particularly at early time points (at which infiltration of fixatives may be limiting relative to subsequent time points), samples were simultaneously prepared by both chemical fixation and the instantaneous high-pressure freezing method (Dahl and Staehelin, 1989). As a high degree of similarity was seen between samples collected by the two methods (Supplemental Fig. S1), the chemically fixed samples, which showed greater contrast of the membrane structures, were used for further analysis.
Plastids from control plants that were not subjected to extended dark treatment contained well-developed thylakoid membrane structures with stroma lamellae and stacked membranes and clearly visible starch granules (Fig. 2). By contrast, plastids from t = 0 contained a PLB with prothylakoids and no visible starch granules. Quantification from single-plane ultra-thin sections revealed that 63% of plastids at t = 0 had one or more visible PLBs in etioplast cross sections (Supplemental Table S1). Of these, 83% contained a single PLB visible in plastid cross sections, 16% contained two PLBs, and 0.6% contained three PLBs (Supplemental Table S1). This indicated that zero to at least three PLBs might be present in a single etioplast. The TEM random cross-section method (2D representation of a 3D structure) likely misestimates both PLB presence and PLB number within the complete volume of the plastid (Mostowska, 1985): plastids from dark-germinated and -grown seedlings similarly do not all show visible PLBs in a single 2D plane (Supplemental Fig. S2). Therefore, we undertook statistical analysis based on the TEM data at t = 0 and numerical simulations of cross sections through the plastid (Supplemental Information S1). From this analysis, we were able to establish that the majority of observed etioplasts contained at least one PLB.
The PLBs in t = 0 samples were highly organized and repetitive structures forming paracrystalline tubular networks (Fig. 2), consistent with those seen in etioplasts of dark-germinated and -grown dicot seedlings (Kowalewska et al., 2016). Therefore, despite the presence of some stacked membrane lamellae, likely to have assembled prior to the extended night period, we conclude that the system is suitable for investigation of the deetiolation processes. For simplicity, the initial time point is referred to as etiolated hereafter.
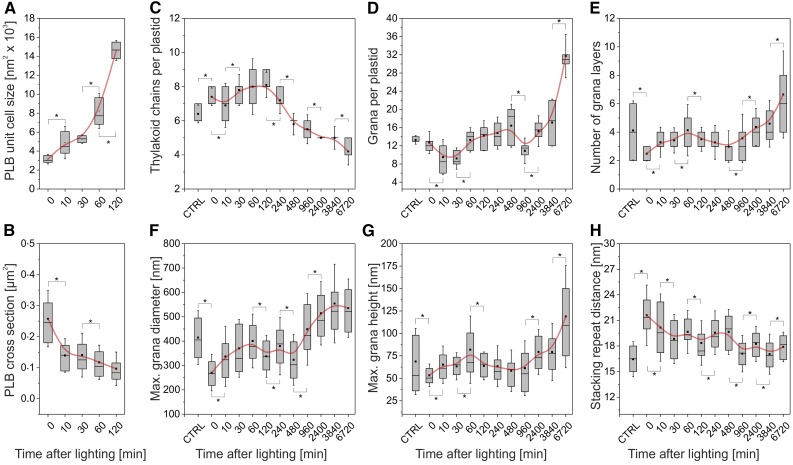
After just 10 min of lighting, the PLBs were more amorphous and irregular, relative to those at t = 0 (Fig. 2). This change was associated with a significantly (P < 0.05) reduced PLB surface area (from, on average, 0.258 to 0.14 µm2) and compactness, visible as an increase in PLB cross-sectional unit cell size (from, on average, 3,161.3 to 4,889.4 nm; Fig. 3A), and loss of regular connections between neighboring unit cells (Figs. 2 and 3). Together, these data imply a nearly immediate outflowing of the PLBs and their direct transformation to prothylakoids. Supporting this interpretation, the loss of PLB compactness, which was particularly pronounced in the 0- to 10-min period, showed strong negative correlation with the changes in the maximum diameter, height, and average number of layers of the grana (r20-60 min = −0.986, −0.947, and −0.997, respectively). Furthermore, the number of thylakoid chains per plastid increased only until 120 min after lighting, coinciding with the visual presence of the last PLB remnants (Fig. 3; Supplemental Table S1), with the subsequent decrease indicating increased connectivity of the grana.
Figure 3.
Quantitative changes in key structural parameters of the plastid membrane system during greening. The x axis indicates time after lighting in minutes. Note the nonlinearity of the time scale. CTRL indicates the control sample, taken at t = 0 but not subjected to extended dark treatment. Box plots show median and interquartile range, with averages represented by the dots. Asterisks indicate significant changes (P < 0.05) between subsequent time points, as calculated by Student’s t test. A, Increasing PLB cross-sectional unit cell size during the first 120 min of deetiolation. After this time point, PLBs were not visible in greening plastids (Fig. 2). n = 20 PLBs per time point. B, Decreasing size of the PLB. Size was estimated from 50 fields of view under 8,000× magnification (∼500 organelles) for each time point. C and D, Number of thylakoid chains (C) and grana (D) per plastid. The decrease in number of thylakoid chains after 120 min likely arises due to increased connectivity between chains. n = 20 plastids at each time point. E, Number of thylakoid layers per granum. n = 30 to 50 grana for each time point. F and G, Maximum grana diameter (F) and height (G). n = 30 to 50 grana for each time point. H, Stacking repeat distance. n = 30 to 50 grana for each time point.
Following the loss of the PLB, both the size and number of grana stacks per plastid generally increased throughout the greening time course (Fig. 3D; Supplemental Table S1). An exception to this is a decrease in the number of grana per plastid between 480 and 960 min, which may be due to increased plastid division during this time period or the merging of multiple grana into a single, larger whole. The latter is supported by the trend of increased maximum grana height (Fig. 3G) and significant increase in the number of grana layers (Fig. 3E).
Grana diameter increased predominantly within the first 60 min post lighting and showed more gradual changes after the first day of light (960–6,720 min; Fig. 3F). By contrast, increases in the maximum height (Fig. 3G) and number of grana layers were particularly large in the last 2 d of greening (3,840–6,720 min). This is in line with previous findings, which demonstrate that thylakoids reach horizontal (diameter) maturity prior to further growth in the vertical direction (stacking; Kowalewska et al., 2016). The few grana stacks present at early time points had a large stacking repeat distance of 21.6 nm (Fig. 3H), consistent with levels seen in the earliest forming grana stacks in deetiolating first true leaves of runner bean (Phaseolus coccineus; 21 nm; Kowalewska et al., 2016), suggesting photosynthetic immaturity (Fig. 3; Supplemental Table S1). The stacking repeat distance decreased to reach a steady value of ∼18 nm by the end of the first day.
Qualitative increases in the size of the plastid were also apparent throughout the greening time course (Fig. 2). However, it is difficult to distinguish the cause of this growth, which may be attributable to thylakoid development but also to massive accumulation of starch granules (Fig. 2; Supplemental Table S1). Comparison between physical attributes of the plastid and membrane structures between control samples (not subjected to extended darkness) and late-stage deetiolating samples (960 min +) indicates that the young control leaves are photosynthetically immature and supports the notion that our time course covers both a deetiolation and photosynthetic maturation (discussed in the section “Photosynthetic Complex Abundance Starts to Increase after 240 min of Lighting”). Together, these data show that our time course faithfully captures the physical transition from the etioplast-like state to nearly mature chloroplasts.
Photosynthetic Complex Abundance Starts to Increase after 240 min of Lighting
To understand the acquisition of photosynthetic competence in our system, we measured the accumulation of photosynthetic proteins and complexes and their activities. Many standard procedures for spectroscopic quantification of complexes and activities require isolation of thylakoid membranes (Schöttler and Tóth, 2014), which could not be undertaken here due to the limited amount of material available and the lack of precise knowledge regarding the selective isolation properties of the PLB and emerging thylakoids. Therefore, we first conducted in planta measurements and complemented these methods with various in vitro measurements using homogenized tissue pools. Due to technical constraints of the spectroscopic methods, observations were not made at time points 5, 10, or 30 min, and the magnitude of changes represented throughout the time course is likely underestimated (detailed in Supplemental Table S2).
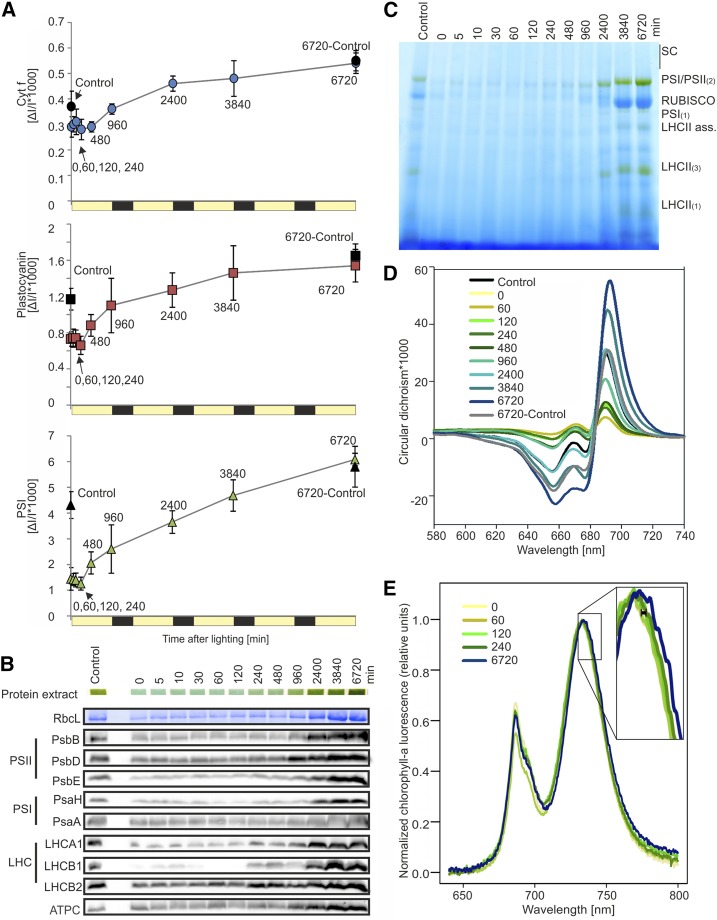
On a leaf area basis, the content of redox-active PSI, cyt b6f, and plastocyanin (PCy) remained unchanged for the first 120 min of lighting, suggesting the presence of a steady state in darkened tissue that was maintained in early deetiolating leaves. Both cyt f, as a proxy for redox-active cyt b6f, and PCy were already accumulated in etiolated tissue to high levels relative to those at 6,720 min: 54% and 47%, respectively (Fig. 4A). This finding is consistent with previous reports of the accumulation of one or both of these components in etioplasts of wheat (Triticum aestivum; Blomqvist et al., 2006, 2008), rice (Oryza sativa; von Zychlinski et al., 2005), pea (Pisum sativum; Kanervo et al., 2008), and barley (Hordeum vulgare; Takabe et al., 1986; Reisinger et al., 2008). By contrast, PSI content in etiolated tissue was less than 25% of that at the final time point (Fig. 4A). PSI, cyt f, and PCy all showed the largest change in abundance within the first day, particularly between 240 and 960 min. Accumulation of PSI then continued linearly (r2960-6720 = 0.988) throughout the remainder of the time course. By contrast, rates of accumulation of both cyt b6f and PC decreased through subsequent time points (Fig. 4A).
Figure 4.
Accumulation of photosynthetic proteins and complexes. A, Accumulation of cyt b6f, PCy, and PSI as measured in vivo by difference absorbance spectroscopy. PSI is quantified from the difference transmission signal of P700, and cyt f is used as a proxy for redox-active cyt b6f. Time points 5, 10, and 30 min were excluded for technical reasons (duration of the measurements). Control plants, which did not undergo extended dark treatment, were measured at both t = 0 (Control) and at the end of the time course (6720-Control). For all three components, minimal changes were seen in the first 240 min, with a large change in rate of accumulation noted before the end of the first day (240–960 min). Subsequently, PSI abundance increased quasilinearly (r2960-6720 = 0.988), while PCy and cyt f demonstrated a generally decreasing rate of accumulation with time. B, Immunoblots with antibodies against selected proteins associated with the light reactions of photosynthesis. Samples were taken from omics tissue pools and loaded on an equal fresh weight basis. RbcL shows the selected band on a Coomassie Blue-stained membrane. Protein extract shows the solubilized protein (in loading dye) within gel wells prior to running. The complete data set, including replicates and quantification, is included as Supplemental Figure S2. C, BN-PAGE. Samples were taken from omics pools and loaded on an equal fresh weight basis. SC represents supercomplexes, and numbers in parentheses indicate monomers (1), dimers (2), or trimers (3); LHCII ass. refers to LHCII assembly complexes, which have been proposed to be a solubilization-induced dissociation product of the PSII-LHCII supercomplexes (Järvi et al., 2011). PSI/PSII dimers, Rubisco, and the LHCs increase in abundance after approximately 480 to 960 min of lighting and continue to increase throughout the time course. D, CD spectroscopy, measured by equal leaf surface area. The control plant, which did not undergo extended dark treatment, was also measured at the end of the time course (6720-Control). The amplitude of the (+)690-nm band, representative of LHCII-PSII supercomplexes, and of the (−)655-nm band, linked to accumulation of LHCII timers, first increased between 60 and 120 min after lighting and continued to increase throughout the time course. Numbers in the legend represent minutes after lighting. E, Changes in the wavelength of the PSI chlorophyll a fluorescence emission peak (∼733 nm) under 77K conditions. An initial shift, indicating attachment of the LHCs to the PS core, occurred between 60 and 120 min after lighting (black bar in enlargement), with further shifts occurring throughout the time course. The complete data set is included as Supplemental Figure S3.
To assess the acquisition of photosynthetic maturity versus deetiolation in our systems, control plants (nondark treated) were measured at both the start and end of the time course. Interestingly, although control plants began with higher PSI, PCy, and cyt f than etiolated plants and also accumulated these components during the time course, deetiolated and control leaves were effectively equal in these parameters at 6,720 min (Fig. 4A). Furthermore, at 6,720 min, chlorophyll content per leaf area, leaf absorptance, chlorophyll a/b ratio (Supplemental Table S2), and maximum photochemical efficiency of PSII (Fv/Fm; Supplemental Fig. S3) were similar between control and deetiolated leaves and were also in good agreement with values obtained previously for mature leaves of tobacco plants grown under similar growth conditions (Schöttler et al., 2017). We, therefore, conclude that control and deetiolated plants reached a photosynthetically mature state at the end of the time course and that our time course effectively captures both the process of deetiolation and the development of photosynthetic maturity.
To support the spectroscopic data and to assess the accumulation kinetics of the ATP synthase and PSII, which could not be quantified in planta, we probed total protein extracted from omics tissue pools with antibodies against diagnostic subunits of photosynthetic protein complexes (Fig. 4B; Supplemental Fig. S4). The reduced dynamic range of this technique allows less precise quantitation than spectroscopic measurements but, nonetheless, reveals the general trends of protein accumulation over the time course. Both PSII proteins, represented by the chlorophyll-binding D2 (PsbD) and CP47 (PsbB) as well as the cytochrome b559 subunit PsbE, and PSI proteins, represented by a small subunit PsaH and the chlorophyll-binding PsaA, were largely evident at later time points (after 480 min). A similar pattern was seen for the ATP synthase, represented by ATPC. By contrast, antenna proteins LHCA1 and LHCB1 were barely detectable at early time points and showed an initial increase between 120 and 240 min (Supplemental Fig. S4). Overall, these data support the quantitative increase in photosynthesis-related complexes measured by spectroscopic data seen throughout the greening time course.
To efficiently capture light, photosystems associate with light-harvesting complexes and form multimeric complexes, which can be visualized after mild solubilization and separation by blue-native (BN)-PAGE. We utilized a total-protein BN method after Järvi et al. (2011) for visualization of both thylakoid membrane complexes and the abundant stromal protein complex Rubisco. Small amounts of PSI and/or PSII dimers are present in the etiolated tissue, consistent with the presence of some thylakoids, as seen by TEM (Fig. 4C). Increases in most membrane protein complexes, as well as the LHC monomers and trimers, were visibly apparent only after 960 min and were most highly accumulated at the last time point. Similarly, Rubisco accumulated predominantly in the last 2 d of greening.
Circular dichroism (CD) spectroscopy can be used to study PSII supramolecular organization in intact leaves. The (+)690-nm polymer and salt-induced (psi)-type band, which arises from long-range interactions between domains within PSII-LHCII supercomplexes (Garab and van Amerongen, 2009), has been shown to arise during greening (Demeter et al., 1976). Here, the initial increase in amplitude of this band was seen between 60 and 120 min, coinciding with the earliest decrease in the chlorophyll a/b ratio (Fig. 1D). Subsequently, a much larger increase in the (+)690-nm psi-type band occurred between 480 and 960 min after lighting, putatively reflecting assembly and accumulation of the PSII-LHCII supercomplexes (Fig. 4D). These changes occurred concurrent with increases in the absolute amplitude of a (−)655-nm band, linked to accumulation of the LHCII trimers (Tóth et al., 2016; Fig. 4D). At 77K, chlorophyll a fluorescence is largely independent of electron transport and the number of occupied vibrational modes decreases, facilitating separation of PSI emission fluorescence (at 733 nm) from that of PSII (686 nm) and uncoupled LHCII and LHCI (682 and 710 nm, respectively; Krause and Weis, 1984, 1991). We observed a shift of the PSI 77K emission maximum from ∼732 nm at early time points to ∼734 nm, indicating attachment of LHCs to the PS core. This shift occurred initially between 60 and 120 min after lighting, with further progressive movement occurring throughout the greening time course (Fig. 4E; Supplemental Fig. S5).
Nearly Immediate Activation of Photosynthetic Activity following Lighting
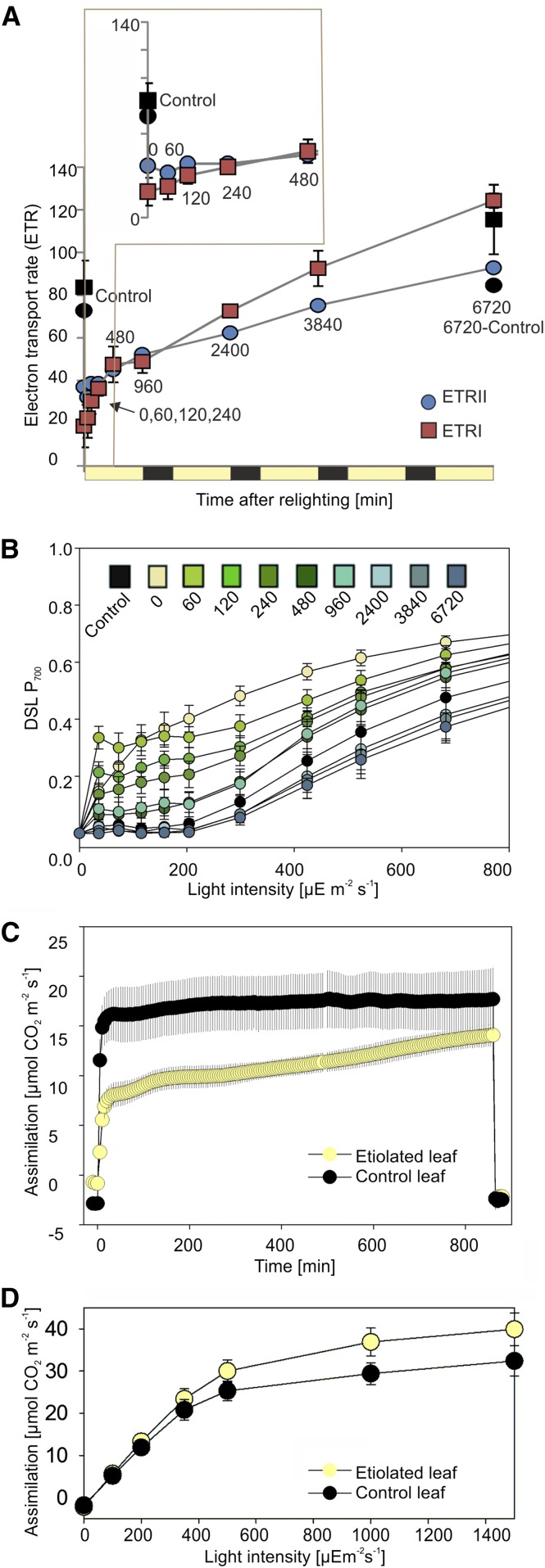
While the accumulation of photosynthetic complexes in the thylakoid membrane may have direct impact on the physical formation of the membrane systems (Daum et al., 2010), protein complex levels cannot act as a proxy for photosynthetic capacity or activity. The photosystems, for example, are in large stoichiometric excess relative to cyt b6f in green tobacco leaves (Schöttler et al., 2017). Hence, we measured the electron transport rate through PSII (ETRII) and PSI (ETRI) and the carbon assimilation capacity of the deetiolating leaf sectors. As with in vivo measurements of photosynthetic complexes, several of the earliest time points (5, 10, and 30 min) could not be measured due to technical limitations. Both ETRII and ETRI, measured after Genty et al. (1989), increased throughout the greening time course, with ETRI exceeding ETRII after the first day of lighting (Fig. 5A). ETR values were corrected for leaf absorptance, measured individually for each sample (Supplemental Table S2). These increases can be attributed to (1) the newly forming membrane complexes and/or (2) increased efficiency or activity of the complexes. We note that the higher ETRII, compared with ETRI calculated for t0-240min (Fig. 5A, inset), is unlikely to be biologically upheld but rather suggests that the assumption of equal light distribution between the two photosystems is not correct in early-stage deetiolating tissue. If we assert that ETRII must not exceed ETRI, we calculate that PSII receives just half the light of PSI at the earliest time points, suggesting greater abundance of PSI or more efficient PSI light capture (e.g. due to associations with antenna proteins). Consistent with a more rapid increase in PSI (content and/or activity) relative to PSII, we observed more rapid P700 (PSI) donor side limitation under increasing light intensity for deetiolating tissue (60 and 120 min) relative to etiolated plants (t = 0; Fig. 5B) and decreased Fv/Fm at early time points (Supplemental Fig. S3), which is largely recovered after 120 min of light. Together, our data support a more rapid increase in PSI activity (occurring within the first 60 min) compared with PSII activity (occurring only after 240 min), consistent with previous reports (Baker and Butler, 1976; Wellburn and Hampp, 1979).
Figure 5.
Changing activity of the photosystems during greening. A, ETRII and ETRI. ETRs were measured on an equal leaf surface area basis and calculated using measured absorbance (averages shown in Supplemental Table S2). The inset shows early time points (0–40 min). A nondarkened control sample was measured at t = 0, in agreement with sampling for omics pools, as well as at the end of the time course, shown in black (6720-Control). Error bars, where visible, show sd for nonabsorbance-corrected measurements. n = 5. B, Light response curve for the donor side limitation (DSL) of the PSI reaction center chlorophyll P700. A DSL of 0 means that all P700 are in their reduced state, while a DSL of 1 means that P700 is fully oxidized (P700+) and photochemically inactive. C, Carbon assimilation during the first 800 min (14 h) of lighting in a leaf exposed to extended dark treatment (Etiolated leaf) and a leaf grown under standard conditions (Control leaf). Measurements were made under growth light (350 µE m−2 s−1) and saturating CO2 (2,000 µL L−1). The first and last data points show leaf respiration in darkness. D, Light response curve for carbon assimilation following 100 h of lighting (4 h into the photoperiod of the fifth day).
Carbon assimilation was measured during the first 800 min (∼14 h) of lighting in both control and etiolated plants (Fig. 5C) under growth light (350 µE m−2 s−1) and saturating CO2 (2,000 µL L−1). Etiolated leaves assimilated carbon at approximately half the rate of control leaves at early time points but reached nearly 80% of the control leaf values within 800 min of lighting. Notably, however, carbon assimilation in control leaves also increased throughout the time course (0–800 min [Fig. 5C] versus 0–6,000 min [Fig. 5D]; 350 μE m−2 s−1), suggesting that control leaves were photosynthetically immature at the beginning of the time course.
Interestingly, after 6,000 min (4 h into the photoperiod of day 5), leaf assimilation capacity in saturating light was higher in deetiolating leaves than in control plant leaves (Fig. 5D). This may indicate either compensatory accumulation of more photosynthetic productivity in the former or the onset of postphotosynthetic maturity decline (and/or feedback repression of photosynthesis) in the latter.
Nucleus-Encoded Transcripts of the Photosynthetic Apparatus Show Two-Phased Accumulation
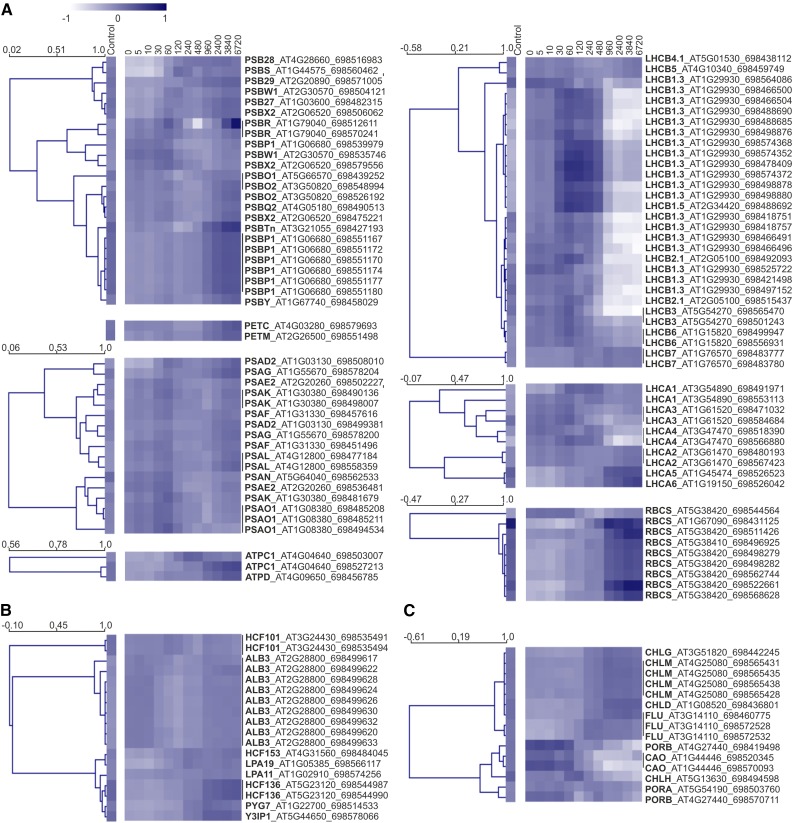
RNA sequencing (RNAseq) was undertaken for all three biological replicates of each of the 13 deetiolation time points. To simplify identification, and because we aimed to identify evolutionarily conserved features, transcripts were mapped to the transcriptome of the maternal progenitor of tobacco, Nicotiana sylvestris, which is more completely retained than that of the paternal progenitor, Nicotiana tomentosiformis, and is also the donor of the plastid and mitochondrial genomes (Renny-Byfield et al., 2011; Sierro et al., 2013). As a diploid species for which plastid and nuclear transformation technologies are available, N. sylvestris is also an ideal model for photosynthetic manipulations (Ort et al., 2015). To better understand the establishment of the photosynthetic apparatus, we observed the abundance patterns of transcripts for the nucleus-encoded subunits of the photosynthetic complexes (Fig. 6A) and selected factors involved in complex biogenesis (Fig. 6B) or chlorophyll synthesis (Fig. 6C). Within the first group (Fig. 6A), nearly all transcripts displayed a two-phase pattern of accumulation (relative to the median): an initial increase at 30 to 60 min and subsequent and progressive increase after 480 min. Excepted from this trend were the majority of LHCII and LHCI transcripts, which mostly decreased following a single peak around 60 to 120 min after lighting. These kinetics are in line with our observed delay in accumulation of antenna proteins relative to photosystem core proteins and changes in chlorophyll a/b ratios (primarily 120–240 min). Transcripts for ATP synthase subunits and the small subunit of Rubisco, both of which have products that accumulate in etiolated tissue but nonetheless increase with greening (Blomqvist et al., 2006), also showed a strong increase after 480 min of lighting and continued to accumulate throughout the time course.
Figure 6.
Accumulation of nucleus-encoded transcripts related to the accumulation of the photosynthetic complexes. Average normalized expression profiles of transcripts are shown for the nucleus-encoded subunits of photosynthetic protein complexes (A), selected complex biogenesis and assembly factors (B), and selected enzymes and accessory proteins of the chlorophyll biosynthesis pathway (C). Transcripts were quantified by RNAseq, with mapping to the tobacco maternal progenitor, N. sylvestris. Transcripts are identified by their common gene name, Arabidopsis Gene Initiative identifier, and N. sylvestris gene identifier, with Arabidopsis Gene Initiative identifiers assigned based on BLAST homology to N. sylvestris transcripts (for details, see “Materials and Methods”). Heat map colors indicate deviation from the log10 transcript median value (0) across the time course, shown here averaged for the three biological repeats. Hierarchical clustering (Pearson, average linkage) was undertaken in Multiple Experiment Viewer: cluster distances are indicated (scale bars show node height). Samples (except the control) are named for time after lighting in minutes.
Hierarchical clustering of selected factors involved in complex biogenesis revealed separation of ALB3, a general membrane insertase (van der Laan et al., 2005), and HCF101, a PSI-specific assembly factor required for insertion of 4Fe-4S clusters (Lezhneva et al., 2004; Stöckel and Oelmüller, 2004), from other PSII- and PSI-related auxiliary factors (Fig. 6B). These two factors show only limited change in abundance throughout the time course. The remaining assembly factors, such as the PSII assembly factor HCF136 (Plücken et al., 2002) and the PSI assembly factors PYG7 (Stöckel et al., 2006) and Y3IPI (Albus et al., 2010), generally increased predominantly in the later phases of the time course, thus displaying ATP synthase/Rubisco transcript-like patterns. Transcripts with products involved in the synthesis of chlorophyll similarly increased throughout the time course (Fig. 6C). The exceptions to this were the various POR isoforms, required for the light-dependent conversion of protochlorophyllide, and chlorophyll a oxygenase (CAO), which acts downstream of POR (Espineda et al., 1999). These transcripts accumulated massively in etiolated tissue and predominantly decreased after 30 min of light.
Plastid Transcript Abundance and Relative Plastid Genome Content, But Not Transcript Abundance per Plastome, Increase with Greening
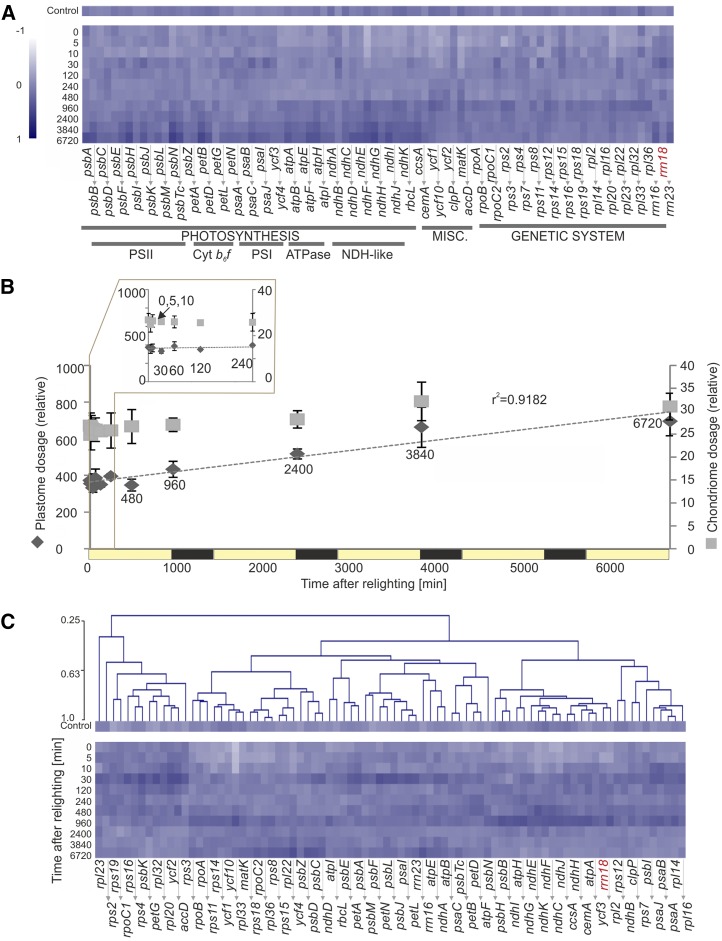
The abundances of 81 plastid genome-encoded transcripts, including mRNAs and the 16S and 23S rRNAs but excluding tRNAs, were quantified by reverse transcription quantitative PCR (RT-qPCR) as values relative to the content of highly stable nuclear reference transcripts. Consistent with previous findings (Krupinska and Apel, 1989), plastid transcript accumulation generally increased with greening, but on average only approximately 2.6-fold (Fig. 7A). Differentially accumulated transcripts were defined throughout as those with more than 2.5-fold change between at least two time points in the greening time course and showing significant change (P < 0.05, Benjamini-Hochberg corrected), as calculated by ANOVA or EDGE (for details, see “Materials and Methods”). Differentially expressed transcripts (25.6% of all transcripts by ANOVA, 45.1% by EDGE) included most ndh transcripts, the psbB-T-H-petB-D operon, the essential conserved reading frames ycf1 and ycf2, and several other transcript species (Supplemental Data Set S1).
Figure 7.
Accumulation of plastid-encoded transcripts with greening. Transcript accumulation was quantified by RT-PCR with normalization to highly stable nuclear transcripts, as defined by GeNorm, with heat maps and clustering (Pearson, average linkage) created with Multiple Experiment Viewer from log10-transformed median-normalized averages of three biological repeats. Samples (except the control) are named for time after lighting in minutes. Heat map color indicates abundance relative to the median (value of 0). A, Transcript accumulation shown by functional grouping. Relative abundance of plastid transcripts changes very little during the time course (less than 2.5-fold in either direction from the median). The nucleus-encoded 18S rRNA (rrn18) is shown in red. B, Quantification of plastome and chondriome copy numbers, relative to the allotetraploid tobacco nuclear genome. While chondriome copies per genome increased just 24% during the time course, plastome copies nearly doubled (from 350 to 695 chondriome copies per genome). C, Further normalization of transcript abundances to plastome copy numbers reveals the extremely limited change in plastid transcript accumulation during greening. This suggests that, generally, the overall increase in plastid transcript abundance arises due to plastome duplication, as opposed to increased transcriptional activity. Nonetheless, a trend of transiently increased transcript abundance occurs for nearly all genes 30 min after lighting.
Since it has been shown that (1) nuclear genome and plastid genome (plastome) replication are unlinked (Heinhorst et al., 1985), (2) plastome copy number is lower in nongreen plastids (Zhang et al., 2012) and increases with deetiolation (Green and Hollingsworth, 1992), and (3) relative plastome copy number can correlate with plastid transcript levels (Thompson et al., 1983; Aguettaz et al., 1987; Hosler et al., 1989; Udy et al., 2012), we quantified relative plastome, as well as chondriome contents during greening, by qPCR (Fig. 7B). We subsequently normalized plastid transcript abundances to plastome copies per tetraploid genome, which nearly doubled across the time course (from 350 to 695). Interestingly, plastome copies per tetraploid genome-normalized data (Fig. 7C) revealed transient increases in abundances of nearly all transcripts after 30 min of lighting, similar to patterns seen in nucleus-encoded transcript data (Fig. 6). Nonetheless, just 23% of the plastid transcripts were differentially expressed on a per-plastome basis by EDGE analysis, with none found to be significantly changing by ANOVA (Supplemental Data Set S1). Together, these data suggest that transcript abundance changes during greening largely occur due to changes in plastome dosage (Fig. 7C).
Lipid Changes Occur after 120 min of Lighting and Are Dominated by Increases in the Plastid Lipids MGDG and DGDG and Triacylglycerol Storage Lipids
Fatty acid biosynthesis and the accumulation of the various lipid species are tightly connected with both the accumulation of the photosynthetic apparatus and the energetic output of photosynthesis itself. The lipids (primarily MGDG and DGDG but also SQDG and PG) form the physical thylakoid membrane structures and are integral parts of the photosynthetic complexes (Doyle and Yu, 1985; Gounaris and Barber, 1985; Pick et al., 1985; Tremolieres et al., 1994; Sato et al., 1995, 2003; Kruse et al., 2000; Fromme et al., 2001; Gombos et al., 2002). However, resources [ATP and NAD(P)H] required for fatty acid biosynthesis are most commonly sourced from photosynthesis (Ohlrogge and Jaworski, 1997), and the de novo synthesis of fatty acids is almost completely inhibited in the dark via redox-controlled repression of acetyl-CoA carboxylase activity (Browse et al., 1981; Sasaki et al., 1997).
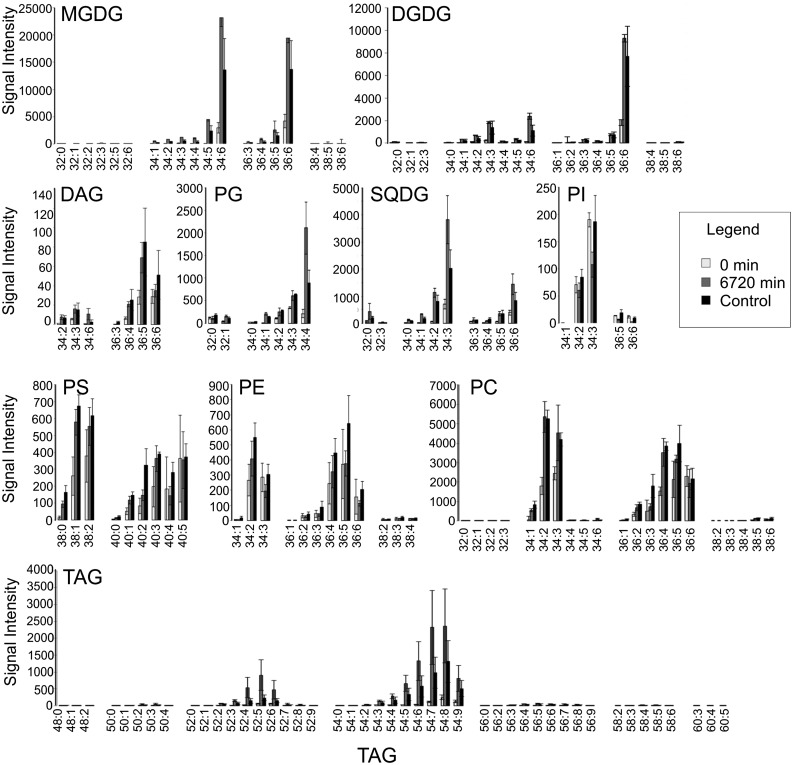
An untargeted ultra-performance liquid chromatography (UPLC)-mass spectrometry (MS) approach applied to our time series identified 177 apparent lipid species. These species belonged to the major plant lipid classes: the galactolipids MGDG and DGDG; the phospholipids phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidylinositol (PI), and PG; and SQDG, diacylglycerol (DAG), and triacylglycerol (TAG). By this method, the variable ionization efficiency of different lipids, defined primarily by the lipid head group, prevents comparison of lipid abundances across classes; however, quantitative comparison within lipid classes and comparison of the kinetics of accumulation are possible.
Throughout the time course, MGDG and DGDG were present predominantly as 34:6 or 36:6 carbon species (i.e. 16-C/18-C or 18-C/18-C with a total of six double bonds), while PG and SQDG preferentially contained 34:4 and 34:3 species (Fig. 8), respectively. This is consistent with known accumulation patterns in Arabidopsis (Arabidopsis thaliana; Burgos et al., 2011). As expected, pools of these lipids were more abundant in green, as opposed to etiolated, tissue, while pools of nonplastidial membrane phospholipids (PI, PS, PE, and PC) displayed fewer changes (Fig. 8).
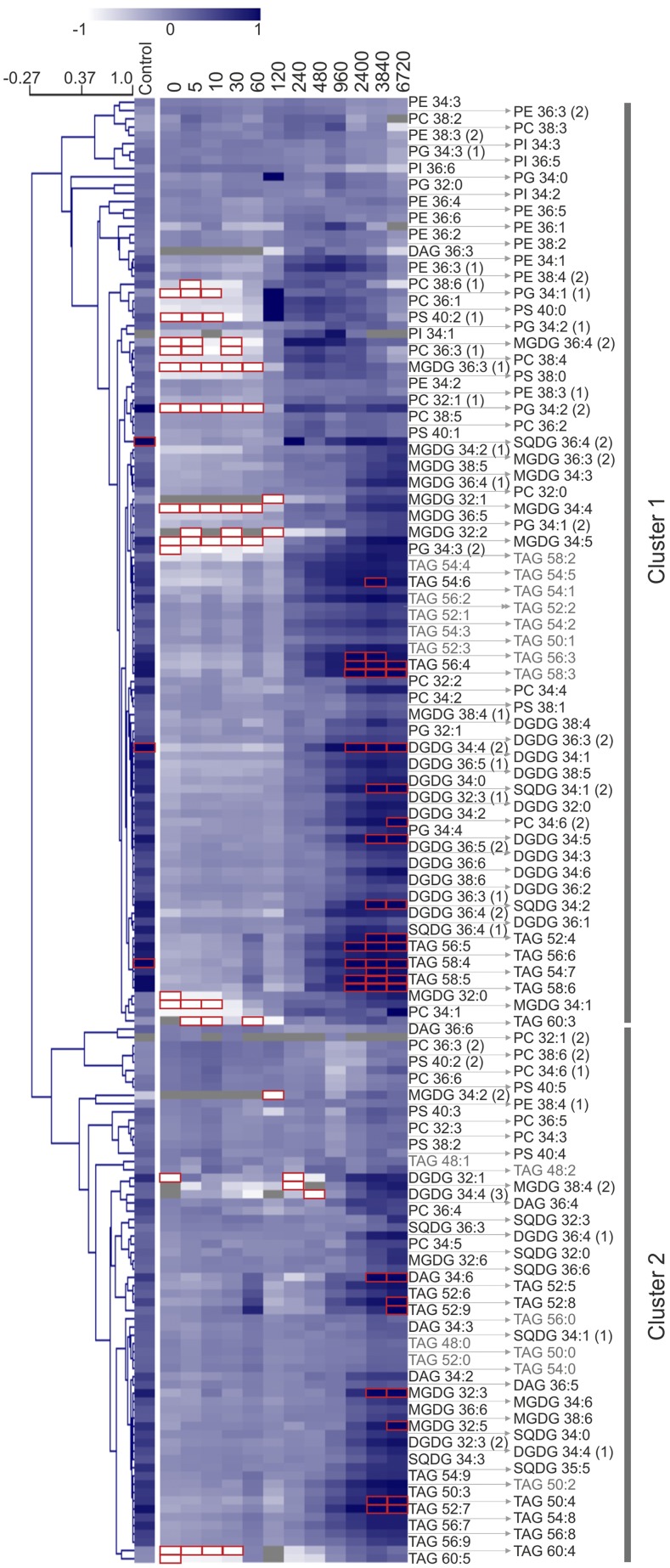
Figure 8.
Normalized signal of the identified apparent lipid species in etiolated (0 min), green (6720 min), and control tissue. The y axis indicates signal intensity, normalized to sample fresh weight and the internal control, PC 34:0. The x axis indicates apparent species. Values are averages of three biological repeats with associated sd values. Note that MGDGs, DGDGs, SQDGs, and TAGs showed massive signal increase between etiolated and green tissue.
TAGs, predominantly used as storage compounds, were absent or very lowly abundant in etiolated tissues, particularly the longest chain species (60-C). Saturated or only weakly desaturated (one, two, and three double bond-containing) MGDG, DGDG, and TAG species were also underrepresented in etiolated, relative to green, tissue. Given that de novo fatty acid biosynthesis involves the formation of saturated species that must then be progressively desaturated by fatty acid desaturases (Siebertz and Heinz, 1977; Miquel and Browse, 1992), this trend likely reflects a limitation of de novo synthesis in etiolated tissue and subsequent activation throughout the greening time course.
More than two-thirds (82.5% by ANOVA, 70.6% by EDGE, with 85% agreement between the methods) of all lipid species differentially accumulated (P < 0.05, Benjamini-Hochberg corrected, greater than 2.5-fold change) during greening, with 50.8% of species exhibiting greater than 10-fold changes in abundance (Supplemental Data Set S2). Unsurprisingly, nonresponsive species primarily included the largely extraplastidial phospholipids, while nearly all species of the thylakoid membrane-building MGDGs and DGDGs changed significantly and abundantly. Hierarchical clustering of all species revealed two large groups of lipids with differential behavior (Figs. 9 and 10). Both clusters contained diverse lipids of different classes, carbon chain length, and degree of saturation. Nonetheless, within the clusters, separation into functional classes, particularly separation between phospholipids (including PG) and other lipid classes, was widely observed. In general, most phospholipid species peaked prior to the end of the time course (2,400 or 3,840 min; Fig. 9), while thylakoid lipids, including PG, as well as TAGs, continued to increase throughout the time course. Globally, a general increase in total lipid pools was observed.
Figure 9.
Accumulation of lipids with greening. Lipid abundances were quantified by liquid chromatography-tandem mass spectrometry with normalization to fresh weight and the internal standard (PC 34:0) and are represented as averages of three biological repeats. The average accumulation profiles of the identified clusters (1 and 2) are shown in Figure 10. Species found also in blank samples are shown in gray (see “Materials and Methods”), and values higher than 1 or lower than −1 (outside of the color range limits) are boxed in red. See Figure 6 for heat map and clustering details.
Figure 10.
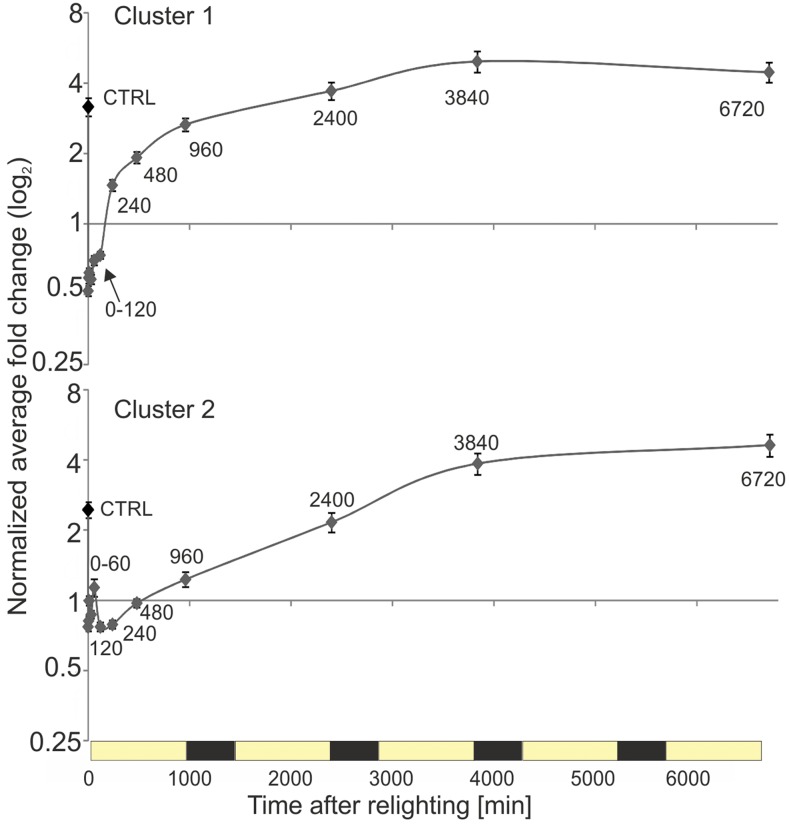
Average accumulation kinetics for lipids belonging to clusters 1 and 2. Normalized average fold changes for all apparent species belonging to clusters 1 and 2 are shown. Values are shown with associated se. Cluster assignment is based on hierarchical clustering (Pearson, average linkage; Fig. 9). On average, members of both clusters showed limited variation within the first 120 min of lighting. Cluster 1 species increased primarily between 120 and 240 min and then generally continued to increase, although at a progressively decreasing rate, until the end of day 3 (3,840 min). By contrast, cluster 2 species increased rapidly between 120 and 3,840 min. Both clusters contained a diverse mix of lipids representing different classes, carbon chain lengths, and degrees of saturation (compare with Fig. 9).
Cluster 1 lipids, which included approximately two-thirds of all measured species, displayed notable increases only after 120 min of lighting, nearly doubling (on average) between 120 and 240 min, before accumulating further at a progressively decreasing rate until the end of day 3 (3,840 min; Fig. 10). Intriguingly, this timing coincides with the disappearance of the PLB (Fig. 2), suggesting that chloroplast membrane biogenesis during the early phase of deetiolation draws upon the lipid reservoir provided by PLBs. By contrast, cluster 2 lipids showed slightly increased transient accumulation 60 min after lighting (which could be largely attributed to the accumulation of several TAG species; Fig. 9). However, as with cluster 1 lipids, the dominant trend was again accumulation only following 120 min of lighting, in this case as a rapid increase that continued until 3,840 min (the end of day 3).
Deetiolation Differentially Reprograms Carbohydrate and Amino Acid Metabolism
Like lipids, the soluble metabolites of the cell represent both the building blocks of the photosynthetic apparatus (e.g. proteinogenic amino acids) and the output of photosynthetic activity (e.g. soluble sugars). To follow the kinetics of metabolic reprogramming throughout deetiolation, we profiled a soluble metabolite fraction that is enriched for primary metabolites and small, specialized (secondary) compounds. We monitored 99 identified metabolites that were included in further statistical analyses (Fig. 11). Of these, over half showed differential accumulation (P < 0.05, Benjamini-Hochberg corrected, greater than 2.5-fold change) throughout the greening time course: 57.6% by ANOVA and 56.6% by EDGE, with high agreement between the two methods (99% similarity). Responsive metabolite species primarily included a wide variety of sugars and amino acids as well as nicotine, polyamines, and some phenylpropanoids (Supplemental Data Set S3).
Figure 11.
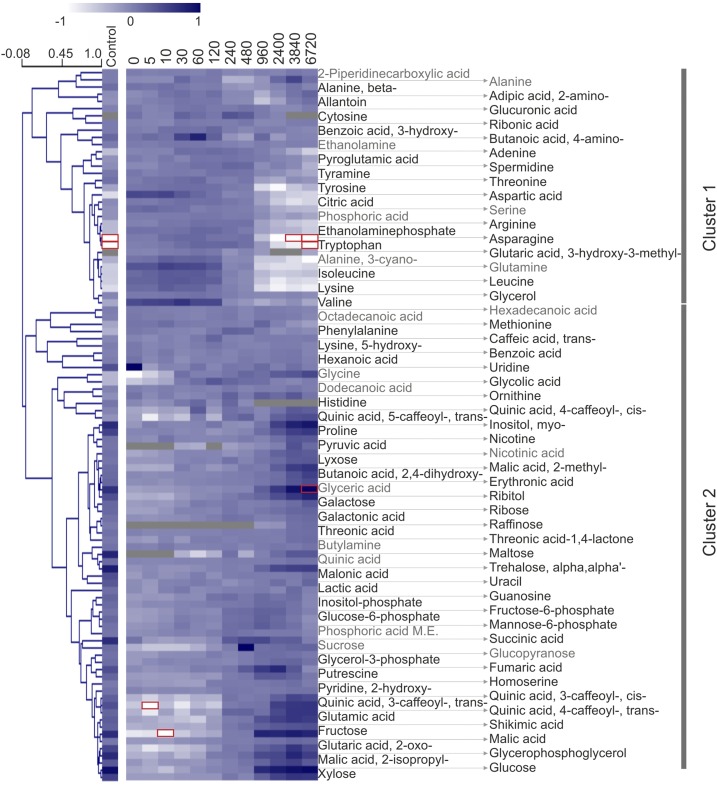
Accumulation of soluble metabolites with greening. Soluble metabolites were quantified by gas chromatography-tandem mass spectrometry (GC-MS) with normalization to fresh weight and the internal standard, [13C6]sorbitol, and are represented as a median-normalized averages of three biological repeats. Phosphoric acid M.E. indicates phosphoric acid monomethyl ester. Hierarchical clustering revealed separation of amino acids from products and intermediates of central metabolism. The former group largely decreased (cluster 1) while the latter group largely increased (cluster 2) with greening. The average accumulation profiles of the identified clusters (1 and 2) are shown in Figure 12. Species found also in blank samples are shown in gray (see “Materials and Methods”), and values higher than 1 or lower than −1 (outside of the color range limits) are boxed in red. See Figure 6 for heat map and clustering details.
Hierarchical clustering (Pearson, average linkage) identified two main patterns of response to greening (distance threshold of 1.04). Metabolite levels of cluster 1 decreased in the course of deetiolation. By contrast, a slight majority of metabolites fall into cluster 2 and increased (Fig. 12). Interestingly, metabolites of both clusters display a discontinuity at the 240- to 480-min mark (Fig. 12). This apparent metabolic pause may arise due to the biological processes that are associated with initiated dry weight deposit, which starts at 240 min (Supplemental Fig. S6), coinciding with the first major increases in lipid accumulation (Fig. 9). Etiolated samples (0 min) have only one-third of the dry mass of fully deetiolated, green plants (6.6% versus 18.5%). For this study, we normalized metabolic profiles to sample fresh weight, since it is difficult to separate the accumulation of dry weight from the accumulation of the measured metabolites themselves.
Figure 12.
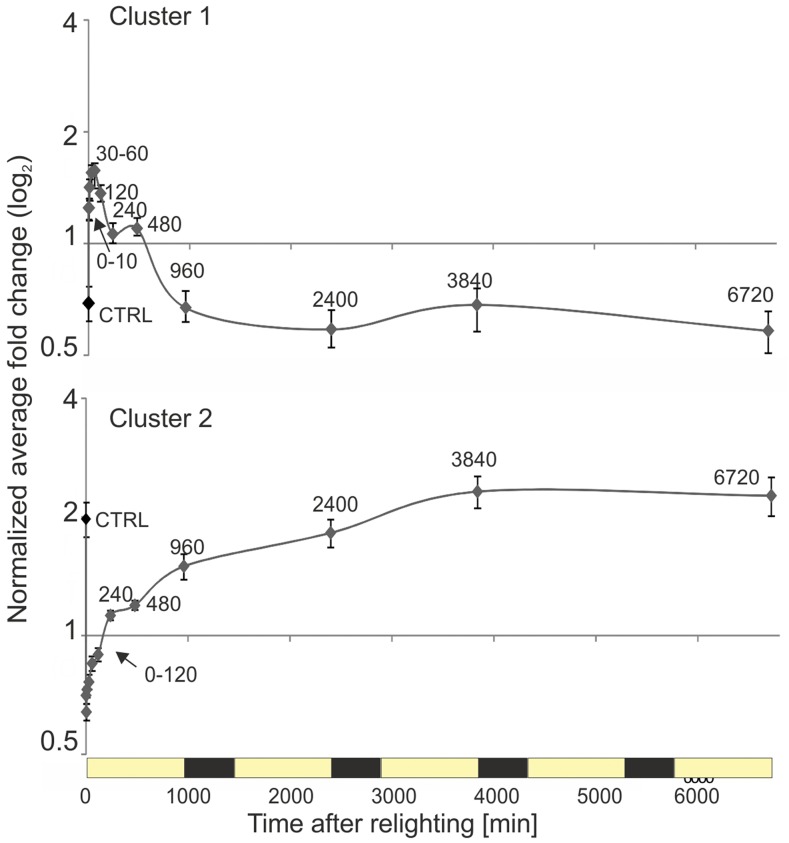
Average accumulation kinetics for soluble metabolites belonging to clusters 1 and 2. Normalized average fold change for all apparent species belonging to cluster 1 and cluster 2. Values are shown with associated se. Cluster assignment is based on hierarchical clustering (Pearson, average linkage; Fig. 11). Cluster 1 predominately contains amino acids. Cluster 2 contains various products and intermediates of primary metabolism including many sugars. The apparent metabolic pause between 240 and 480 min may arise due to changes in the fresh to dry weight ration during this period: soluble metabolites were normalized to fresh weight, and the first major increase in the dry weight to fresh weight ratio occurred between 240 and 480 min (see Supplementary Fig. S4)
In agreement with the initiation of starch accumulation (Fig. 2; Supplemental Table S1) and increasing assimilation (Fig. 5C), cluster 2 contained mostly soluble carbohydrates, which increased primarily during the first photoperiod at 960 min. These include maltose (the first starch breakdown product), Suc (the main soluble carbohydrate store and transport sugar), as well as Glc, Glc-6-P, Fru, Fru-6-P, and trehalose (Figs. 11 and 12). The onset of photorespiratory metabolism was indicated by glycolic acid accumulation that peaked at 120 min and was followed by Gly and glyceric acid accumulation, which both peaked later than 960 min. Specialized metabolites, such as caffeoylquinic acids (e.g. chlorogenic acid or 3-transcaffeoylquinic acid), but not their precursors (i.e. quinic acid and transcaffeic acid), increased strongly after 960 min. Nicotine increased only slightly at the last two monitored time points, consistent with the site of nicotine production being (nonphotosynthetic) root tissue, from where this alkaloid is transported upward into the aerial tissues of the plant.
By contrast, cluster 1 members had high relative abundances in etiolated tissue and decreased mostly after 120 or 480 min of lighting (Figs. 11 and 12). Again, the dominant changes occurred within the first photoperiod. Most of the proteinogenic amino acids were part of cluster 1. The progressive depletion of free amino acids at 240 and 960 min is consistent with stimulation of translation that is required for the synthesis of Rubisco, the photosystems, and their light-harvesting complexes (Fig. 4). By contrast, the nonproteinogenic amino acid homoserine (Aubert et al., 1998) accumulated with greening (Figs. 11 and 12, cluster 2).
A notable exception from the general amino acid depletion was Pro, which accumulated up to 25-fold in green (6,720 min) relative to etiolated (0 min) tissue. Pro has been shown to accumulate independently of other proteinogenic amino acids, not only in response to a variety of stresses but also during leaf development (Verbruggen and Hermans, 2008). Pro has been proposed to act as a reactive oxygen species scavenger and provide stress protection and is also an important store of carbon and nitrogen. Consistent with the latter function of Pro as a nitrogen buffer, Glu and 2-oxoglutaric acid were also found in cluster 2. 2-Oxoglutaric acid provides the carbon backbone of nitrogen assimilation from the tricarboxylic acid cycle and is also the substrate for Glu synthesis via the GS/GOGAT cycle. This activity may also support the accumulation of putrescine, which, in contrast to spermidine of cluster 1, accumulated toward 960 min.
Taken together, and consistent with a high demand for proteinogenic amino acids, nitrogen assimilation was likely increased toward the end of and following the first light cycle. In summary, the dominant changes during greening reflect the expected activation and remodeling of central metabolism and are consistent with the observed increased assimilation and the requirement for increased protein biosynthesis.
The Major Switch from Etiolated to Green State Occurs on Different Time Scales for Transcripts, Lipids, and Soluble Metabolites
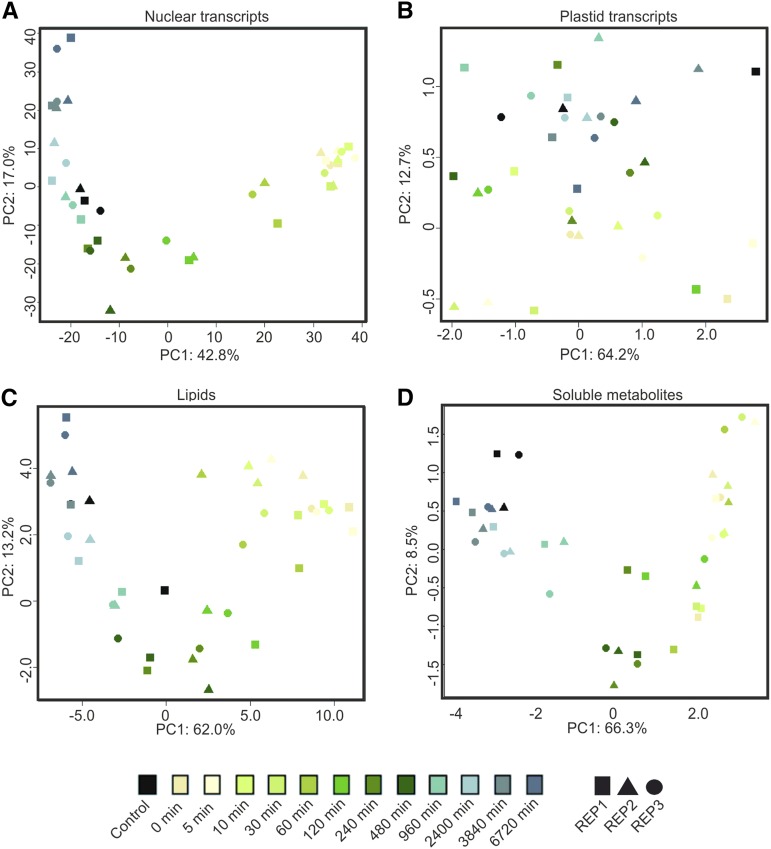
Plastid transcript, nuclear transcript, lipid, and soluble metabolite data sets were subjected to principal component analysis (PCA; Fig. 13), which allows visualization of the main variation across and between data points. Consistent with the lack of significant changes in plastid transcripts during greening, plastid-encoded transcripts did not clearly separate in a time course-dependent manner across principal component 1 (PC1; 64.2% of variation; Fig. 13B). However, they did loosely distinguish as early and late samples of the deetiolation time course by PC2 (12.7% of variation). No discernible pattern of function or genomic location (e.g. operon association) could be seen within the transcript species most strongly contributing to this separation (Supplemental Table S3). However, the splicing factor matK and several intron-containing genes (rpl2, ndhB, ycf3, petD, clpP, and rps12) were represented in this group, which may reflect the detection by RT-qPCR of only spliced transcripts (see “Materials and Methods”).
Figure 13.
PCA of nucleus-encoded transcripts (A), plastid-encoded transcripts (B), lipids (C), and soluble metabolites (D). Control indicates samples not treated with extended dark treatment, taken at t = 0. Rep1, Rep2, and Rep3 indicate samples taken from the independently repeated experiments. PC1, which defines 40% to 60% of the variation in the nuclear transcript, lipid, and soluble metabolite data sets, separates data points in a manner that reflects the deetiolation time course. By contrast, the primary variance in plastid-encoded transcripts (PC1, 64.2%) does not reflect the greening process. The shift from the etiolated to the green state does not proceed in an even manner; instead, rapid shifts are seen, such as the change between 60 and 120 min for nuclear transcripts and between 480 and 960 min for metabolites. Importantly, control samples, which were harvested at the same time as samples 0, group rather with later time points, which have a similar green status. Probabilistic PCAs were undertaken in R studios using the pcaMethods package (Stacklies et al., 2007).
In contrast to plastid transcripts, the first principal component of RNAseq (Fig. 13A), lipid (Fig. 13C), and soluble metabolite (Fig. 13D) data, representing 42.8%, 62%, and 66.3%, respectively, of the variation in these data sets, clearly reflected the greening status of the samples. Consistent with this observation, control samples, which were harvested at the same time of day as sample 0, grouped with later time points (960–6,720 min) rather than early time points. Furthermore, separation across PC1 was associated with a major increase in the thylakoid galactolipid species MGDG and DGDG and the storage lipid TAG in the lipidomic data sets (Supplemental Data Set S2), while separation by PC1 of soluble metabolite data were determined by the observed increases in sugars and decreases in amino acids as well as an increase in a group of phenylpropanoids, consistent with our preceding analyses (Figs. 11 and 12).
The greening transition observable across PC1 in the nuclear transcripts, lipids, and soluble metabolites did not proceed evenly based on time in light and showed different dynamics for the different data sets. The shift in lipid species becomes apparent after 120 min of lighting, while soluble metabolites show the greatest shift between 480 and 960 min. In both cases, the majority of the rearrangement occurs within the first day after relighting (up to 960 min). For nuclear transcripts, a major shift is seen after 30 min of lighting and is largely completed by 240 min (Fig. 13).
The Etioplast-to-Chloroplast Transition Is Defined by Changes in Nuclear Transcript Abundances between 30 and 240 min after Lighting
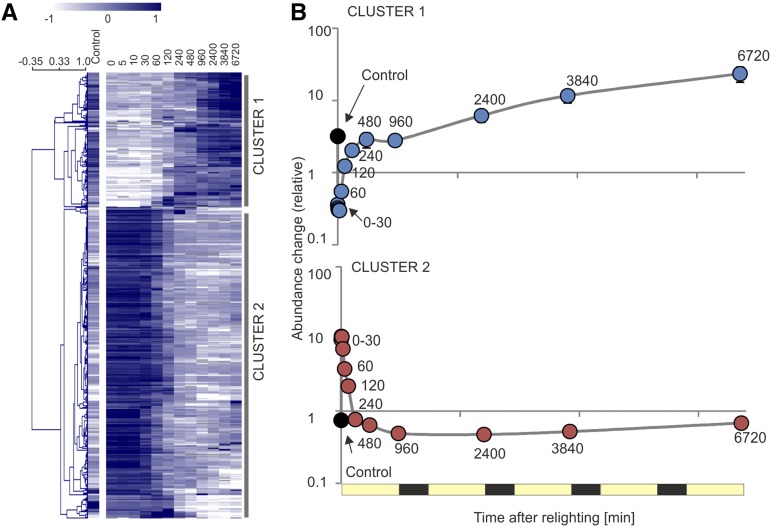
To further understand the accumulation of nuclear transcripts contributing to separation across PC1, and therefore, putatively linked to the greening process, the 500 transcripts with the highest absolute PC1 loadings were extracted (Supplemental Data Set S4). Hierarchical clustering of these transcripts identified two clear clusters (Fig. 14A), with transcripts in both clusters showing, on average, little variation within the first 30 min and between 240 and 960 min (Fig. 14B). Instead, the dominant changes in abundances occur between 30 and 240 min in both clusters.
Figure 14.
Accumulation of nucleus-encoded transcripts contributing to separation across PC1. A, Heat map (Pearson, average linkage) showing clustering of the top 500 nuclear transcripts contributing to separation across PC1 (Fig. 13). Note that 10.2% of elements are above (3.1%) or below (7.1%) the color scale limits. B, Average accumulation of transcripts in clusters defined by A. Points represent averages of all transcripts belonging to the clusters, with associated se.
Cluster 1 transcripts increased on average 5-fold between the two static phases (30–240 min) and then increased following the second stasis (240–960 min), to reach an average of 60-fold the abundance in etiolated tissue by the end of the greening time course (Fig. 14B). This cluster of early responders included transcripts encoding proteins associated with central metabolism and some highly abundant photosynthesis-related proteins (Supplemental Data Set S4). By contrast, cluster 2 transcripts decreased within the 30- to 240-min period by approximately 15-fold, after which transcript abundances did not change. This cluster contained transcripts with homology to Arabidopsis etiolated state transcripts, with products linked to light sensing and deetiolation, such as a phytochrome-interacting factor (PIF3; AT2G20180), an unknown protein whose overexpression leads to pseudoetiolation in the light (AT3G55240), a light quality-responsive homeodomain Leu zipper protein (AT4G16780), a phototrophic response signal transducer (AT2G30520), and the CAO enzyme catalyzing chlorophyll b biosynthesis (AT1G44446). We propose that transcripts found in this top 500 list have an increased likelihood to play an active role in the etioplast-to-chloroplast transition; however, due to the nature of PCA, these top 500 are also biased toward transcripts showing high variance across the entire time course.
Interestingly, the 240-min time point, which corresponded with the end of the transition, also corresponded with the point at which transcripts, on average, reached an accumulation state comparable to that seen in the nonetiolated control sample. Given these results, and in line with the ultrastructural changes seen by electron microscopy (Fig. 2) and the changes in photosynthetic complex accumulation (Fig. 4), we define the time period between 0 and 240 min as the deetiolation phase, during which the dominant transition from etioplasts to chloroplasts takes place, while the time period from 960 to 6,720 min is defined as a building phase, in which photosynthetic capacity is further increased.
Identification of Putative Candidates Involved in the Etioplast-to-Chloroplast Transition
Global analysis of RNAseq data identified many transcripts (48.1% by ANOVA, 33.6% by EDGE) that were differentially expressed (P < 0.05, Benjamini-Hochberg corrected, greater than 2.5-fold change) throughout the time course (Supplemental Data Set S5). This high proportion of responsive transcripts is, perhaps, unsurprising given the dominant role of the chloroplast in defining the physiological status of the plant and the subsequent large changes in global development occurring across our relatively long time course. A previous report has found large differences in gene expression between light-grown and dark-grown Arabidopsis seedlings at a single age time point (32% of transcripts differentially expressed with more than 2-fold change in 6-d-old seedlings; Ma et al., 2001). Interestingly, however, of the differentially expressed transcripts found in our study, just 4,380 (13.75% by ANOVA) or 2,314 (7.3% by EDGE) changed more than 5-fold during the time course (Supplemental Data Set S5).
In the context of our experimental design, it cannot be excluded that a subset of these transcripts show differential accumulation due to diurnal or light signals unrelated to deetiolation, circadian signals, or cell growth and leaf development. In an attempt to identify transcripts more likely associated with deetiolation, we focused on changes occurring within the first 240 min (the deetiolation phase). In total, 7,996 (25.1%) and 6,723 (21.1%) transcripts were differentially expressed, as analyzed by ANOVA and EDGE, respectively, with strong agreement between the two methods. Less than 2,000 transcripts (1,869 by ANOVA, 1,232 by EDGE) displayed more than 5-fold changes during the deetiolation phase, with 99.4% of the transcripts shown to be significant by EDGE also identified by ANOVA. These 1,224 common transcripts could be associated with 740 unique Arabidopsis gene putative orthologs, of which 117 (15.8%) had products with plastid localization (Supplemental Data Set S6), as defined by SUBA4 consensus (Hooper et al., 2017).
Among these genes were many encoding nucleic acid-binding proteins (primarily pentatricopeptide repeat proteins but also RNA recognition motif- and zinc finger domain-containing proteins), transcriptional and translational regulators, chaperones, and some protein kinases and phosphatases. Several transcripts known to be associated with the etiolated state or with the development of photosynthetic competency could also be identified, such as those encoding an early light-inducible chlorophyll a/b-binding family protein (AT3G22840), PSBS (AT1G44575) and PGR5 (AT2G04030), both involved in photoprotection, a member of the LHC family (AT1G34000), Rubisco activase (AT2G39730), and a Rubisco folding chaperone (AT2G28000). Finally, the 740 Arabidopsis orthologs were further filtered based on conservation in green lineages as defined by the GreenCut list (GreenCut includes 710 associated Arabidopsis loci; Grossman et al., 2010; Karpowicz et al., 2011; Heinnickel and Grossman, 2013). Of those with products predicted to be targeted to the plastid, 23 transcripts were members of GreenCut, with a further eight GreenCut members found as part of the 740 orthologs but not having a predicted plastid localization. These genes, which may be considered as putative chloroplast biogenesis factors, are to be characterized further in future functional studies using reverse genetic approaches.
DISCUSSION
Thylakoid membrane biogenesis represents one of the most intricately complex and important biological processes on earth. The complexity and interconnection of the many cellular processes involved in the etioplast-to-chloroplast transition mean that many features, and the timing of the transition, remain poorly defined and suggest it as a perfect subject for systems biology analysis. Systems biology is a field still in its infancy and, to date, has largely been undertaken in single-celled organisms (Escherichia coli, Saccharomyces cerevisiae, and Chlamydomonas reinhardtii) or included postexperimental gathering of data collected from diverse experimental contexts (Bassel et al., 2012). Here, transcriptomic, metabolomic, physiological, and ultrastructural analyses were performed within a single system: greening leaves of the dicot model plant tobacco in the process of acquiring photosynthetic competence. In addition to providing detailed understanding of various aspects of the transition, the linked nature of the data sets obtained facilitates the direct comparison and integration of data sets. Furthermore, the dense sampling in our study has allowed us to resolve the timing of the genetic, developmental, and metabolic changes underlying the deetiolation process (Fig. 15). A major finding from the integration of our data sets is that the developmental switch from etioplasts to chloroplasts (1) occurs on different time scales for transcripts, lipids, and soluble metabolites and (2) involves different levels of genetic regulation in the nucleus versus the chloroplast.
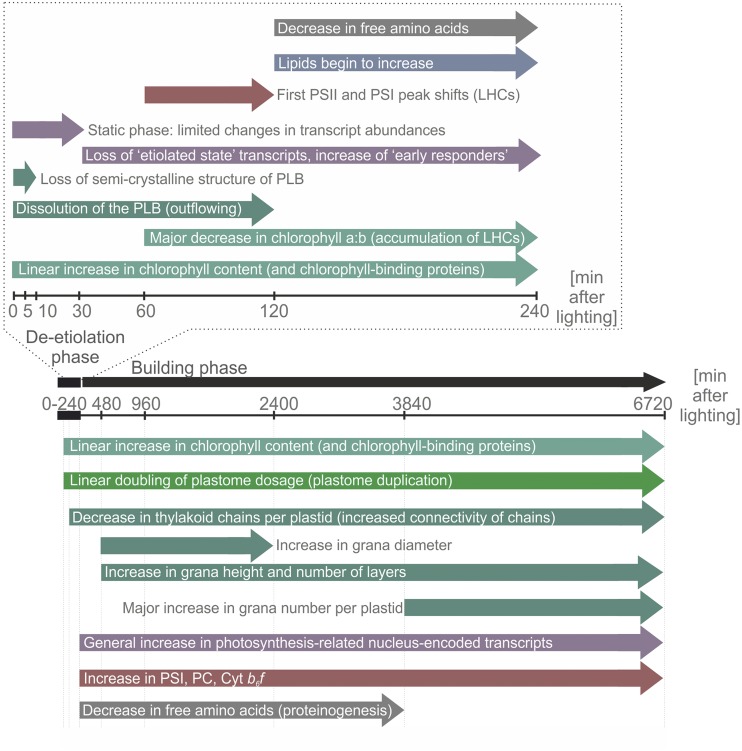
Figure 15.
Summary of the timing of changes associated with the greening process. The model presents a summary of the major changes observed during 6720 min (5 d) of greening following extended dark treatment of tobacco. The time course can be divided into two phases: a deetiolation phase, in which the plastid PLB is disassembled, and a building phase, which largely involves the accumulation of thylakoid membranes and associated photosynthetic proteins and capacity.
The Deetiolation Phase (0–240 min)
On being placed in the light, changes in the physical structure of the etioplast PLB occurred almost immediately. After just 10 min of lighting, the regularity of the PLB was already reduced. This loss of semicrystallinity was associated with outward flowing of the PLB membranes, apparent in both a decrease in the compactness of the PLB and a decrease in its size. Although the dissolution of the PLB proceeded throughout the first 120 min of greening, the differences in structural features were most pronounced within the first 10 min, underscoring the nearly immediate nature of this change (Figs. 2 and 3).
On a global scale, the accumulation of (nuclear) transcripts began to shift only after 30 min of lighting (Fig. 13). This is unlikely to be an issue arising solely from the biological requirements involved in RNA synthesis: mRNAs have been shown to be able to be both up- and down-regulated in a time scale of just 20 to 60 s (Suzuki et al., 2015). Transcripts whose products are required for the maintenance of the etiolated state, such as POR, PIFs, and phytochromes, also begin to decrease only after 30 min or more of lighting (Fig. 14, cluster 2), despite the occurrence of major ultrastructural changes within these first 30 min (Fig. 2). As shown previously in Arabidopsis, the accumulation of POR proteins is directly related to not only the formation, but also the size, of the PLB (Sperling et al., 1998; Franck et al., 2000) and of certain PIFs (Stephenson et al., 2009). We, therefore, propose that (1) the early ultrastructural changes are nongenic in nature, largely do not require induction of a specific gene expression program, and are likely explained by rapid light-induced enzyme activation and (2) the transcript changes after 30 min occur initially in response to the cellular changes, as opposed to being responsible for them (Casano et al., 2001). It should be noted that increases in abundances of the components of the photosynthetic electron transport chain (PCy, PSI, and cyt b6f; Fig. 4A) occurred only after 240 min of lighting. However, increases in the activity of at least some components started more rapidly, with the activity increase of PSI occurring already within the first 60 min and preceding that of PSII (Fig. 5). The latter likely represents a sensible safety precaution, given that the accumulation of PSII in the absence of PSI (as acceptor for the electrons generated by PSII) would be particularly dangerous.
After 120 min of lighting, we also noted a decrease in the abundance of nearly all proteinogenic amino acids (Figs. 11 and 12). Given that there is at least one report linking decreased abundance of enzymes involved in amino acid biosynthesis to greening (Kleffmann et al., 2007), a greening-related reduction in amino acid biosynthesis cannot be fully excluded here. However, a decrease in amino acid synthesis is certainly counterintuitive in the context of an increasing demand for de novo protein synthesis (and of activated assimilation and central metabolism), seen throughout our greening time course. Therefore, we consider it more likely that the loss of free amino acids arises from a stimulation of translation beginning after 120 min of lighting. This timing corresponds in our greening time course with a trend of transiently increased nucleus- and plastid-encoded transcripts, which may be linked to increased RNA synthesis and/or increased transcript stability (Deng and Gruissem, 1987; Green and Hollingsworth, 1992; Kim et al., 1993). Although the influence of ribosome loading on plastid transcript stability may vary (Scharff et al., 2017), ribosomal loading is known to increase mRNA stability in both bacteria and eukaryotes (Barkan, 1993; Sullivan and Green, 1993; Mayfield et al., 1995; Barahimipour et al., 2015), and light is known to stimulate plastid (Klein et al., 1988; Klein, 1991; Edhofer et al., 1998; Kanervo et al., 2008) and cytosolic (Liu et al., 2012; Paik et al., 2012) translation. However, it is clear that translation may also proceed (albeit at a slower rate) in etiolated tissue. It has been shown that the etioplast already contains many of the proteins required for the acquisition of photosynthetic competency, with the exception of most chlorophyll-binding proteins that are simply unstable in the absence of their cofactors (Kim et al., 1994; Komatsu et al., 1999; von Zychlinski et al., 2005; Blomqvist et al., 2006, 2008; Kanervo et al., 2008).
As such, the etioplast is a poised and primed prechloroplast, which is limited primarily by the (light-dependent) accumulation of chlorophyll. A reliance on increased transcription, as a barrier to the etioplast-to-chloroplast transition, may imply an additional temporal bottleneck arising from transcript processing, translation, protein import, protein processing, and protein complex assembly. It has been reported that chlorophyll can be created from protochlorophyllide nearly instantaneously (Reinbothe et al., 1999; Domanskii et al., 2003), continuing to rise linearly throughout the first hours of lighting, with PSI accumulation and activity measurable after just 15 min of lighting and PSII activity following after 2 to 3 h (Baker and Butler, 1976; Wellburn and Hampp, 1979). While we observed a linear increase in chlorophyll content throughout the first photoperiod (Fig. 1), we did not observe increases in PSI content, or contents of cyt b6f or PCy, until 240 min after the initiation of greening. This may be a particular feature of our system, which already contained some photosystems at t = 0 (Fig. 4A), but could also represent a difference in the greening of true leaves compared with that of cotyledons. Nonetheless, it is interesting that our data suggest an increase in the accumulation of the LHCs, and their subsequent attachment to (preexisting) photosystem reaction centers, that occurs earlier (60–120 min) than further accumulation of more reaction centers (240 min). This phenomenon suggests a multiphase accumulation of photosynthetic competency in which, following the initial accumulation of a few reaction centers, attachment of LHCs to these reaction centers is prioritized over the further accumulation and assembly of photosystem core complexes and, ultimately, supercomplexes (Figs. 4 and 15). The lower cost of LHC biosynthesis, relative to that of reaction centers, coupled to the increased potential for the capture of more light of varied wavelengths granted by LHCs, would support the selection of such a mechanism in nature. This, in turn, would necessitate prioritization of the production of chlorophyll b at early time points, as seen in our system (dominant shift in the chlorophyll a/b ratio between 60 and 240 min; Fig. 1D).
The Building Phase (240–6,720 min)
The accumulation of the thylakoid lipid species, primarily MGDG and DGDG, began for the most part between 120 and 240 min of lighting. Given that the PLB was completely disassembled following approximately 120 min of lighting, it can be supposed that the majority of the lipids for early membrane biogenesis come directly from the PLB. This assumption is in agreement with the spatial analysis of PLB transformation that revealed direct rearrangement of PLB tubules into flat thylakoid lamellae, without signs of intermediate dispersion into vesicles or degradation of membrane networks. Once the PLB is disassembled, new lipids must be synthesized. At this point, we observed that the previously increasing number of thylakoid chains per plastid began to decrease, indicative of increasing connectivity between the chains. There is some evidence that the signal for de novo synthesis of lipids occurs before this 240-min time point. For example, cluster 2 lipids, including MGDG, DGDG, and TAGs, showed an intermediate increase at 60 min (Fig. 9). Perhaps even more relevant, we noticed an increase in saturated and lowly desaturated lipid species (mostly phospholipids; cluster 2), which showed a first increase between 60 and 120 min. The specific increase in these lipids likely represents an increase in the activity of de novo lipid synthesis at this time point.
Approximately 120 min after lighting also represents a major shift in metabolic productivity. Between 120 and 240 min after lighting, we observed a global trend of decreased free amino acid abundance (indicative of stimulated de novo protein synthesis) and increased output from central metabolism (Figs. 11 and 12). Subsequent to this point, between 240 and 480 min, we see the first major increase in the photosynthetic cores (PSI but also cyt b6f and PCy; Fig. 4). Between 240 and 480 min, the leaves begin to accumulate biomass (Supplemental Fig. S6), likely not only in the form of lipids, soluble metabolites (Figs. 9 and 11), and proteins but also in nonsoluble polymers such as starch (Figs. 2 and 3) and cellulose. The time between 240 and 480 min, therefore, likely represents the switch from sink to source status.
Following the first day of greening, the photosynthetic membrane lipids (Fig. 9) and the protein complexes residing in the thylakoid membranes continue to accumulate (Fig. 4, A–C). The increase in photosynthetic competency occurs in a largely linear manner across the remaining 4 d of greening, as evidenced by the steadily rising ETR (Fig. 5A). Nonetheless, a particularly pronounced surge in the rate of chlorophyll accumulation was seen in the third day of lighting (between 2,400 and 3,840 min; Fig. 1D). Given that a corresponding rise in the rate of PSI accumulation was not seen at this time point, it can be assumed that this increase in chlorophyll is largely associated with an elevated abundance of LHCs. In support of this theory, a large shift in the peak height of the (+)690-nm and (−)655-nm psi-type bands (representing PSII-LHCII and LHCIIs, respectively) was seen between these time points (Fig. 4D). This occurs despite the expression of most LHC transcripts largely peaking between 240 and 480 min (with the exception of the minor forms LHCA5 and LHCA6, which have more specialized functions in processes such as cyclic electron flux; Fig. 6A), indicating a disconnect between transcript and protein (or protein complex) accumulations. Subsequent to the increase in LHCs, between 3,840 and 6,720 min, we observed a gain in grana height (Fig. 3G), which is likely a direct result of proliferating layers of grana (Fig. 3E) as well as an increase in the number of grana per plastid (Fig. 3D). Grana formation has been proposed to arise due to electrostatic interactions between PSII-LHCII complexes (Daum et al., 2010; Johnson et al., 2011), and it, therefore, can be argued that the ultrastructural changes seen arise largely due to the assembly of the chlorophyll-binding LHCs into supercomplexes and megacomplexes (Fig. 4C). This interpretation is also in agreement with the decrease in stacking repeat distance (Fig. 3H), as interactions of the positively charged N terminus of LHCII on one side of the partition gap with the flat LHCII trimer surface on the other side of the stromal gap stabilize the dense stacking of grana membranes (Standfuss et al., 2005). Interestingly, the observed surge in LHC and chlorophyll accumulation between 2,400 and 3,840 min does not lead to a proportional increase in ETR capacity. The likely reason is that, at that time, the rate-limiting cyt b6f (and PCy) have already almost reached their maximum accumulation levels. It, therefore, can be suggested that the additional LHCs and reaction centers (and the resulting PSI and PSII complexes) are largely in surplus, developed to optimize light absorption and excitation energy distribution at light-limited conditions.
Although the most rapid changes in the accumulation of photosystems occur within the first day of lighting, there is nonetheless a continued increase in both the accumulation of thylakoid membrane systems (Fig. 3) and photosynthetic activity (Fig. 5) throughout the time course. Overall, by 6,720 min of greening, the photosynthetic state of the plants, as measured by chlorophyll content (Supplemental Table S2), CO2 assimilation (Fig. 5D), and ETRI and ETRII (Fig. 5A), largely reached that of fully expanded tobacco leaves not subjected to etiolation (Rott et al., 2011; Schöttler et al., 2017). This indicates that our time course covers not only the transition from etioplasts to chloroplasts but also largely captures the complete maturation of the photosynthetic apparatus.
A Comprehensive Description of the Etioplast-to-Chloroplast Transition and Identification of Candidate Genes for Further Analysis
Our systems biology approach allowed us to identify specific phases of deetiolation and thylakoid development that are summarized in the model presented in Figure 15. From these phases, we could deduce a likely time of differential expression of genes involved specifically in the etioplast-to-chloroplast transition.
In addition to providing a comprehensive description of the deetiolation process and revealing candidate genes implicated in chloroplast development and thylakoid biogenesis, our study also sheds fresh light on the genetic regulation of the deetiolation process. Our data suggest that the increase in chloroplast transcript abundance is, largely, not due to light-stimulated transcription of plastome-encoded genes but rather to a (light-induced) increase in genome dosage. By contrast, nuclear gene expression is subject to extensive transcriptional regulation, with large groups of genes being strongly up-regulated or down-regulated (Fig. 14).
Although our short list of candidate genes (Supplemental Data Set S6) for new chloroplast biogenesis-related functions contains some factors known to be involved in the accumulation of photosynthetic competency, the majority of genes had minimal or no annotation. Among these are both genes whose protein products are targeted to the plastid (which could putatively be directly involved in PLB disassembly and/or thylakoid development) and genes whose products are targeted to the nucleus, which might more likely play a regulatory role (e.g. in the reprogramming of nuclear gene expression). This short list of genes will act as a basis for future functional studies. It will also be worthwhile to investigate the role of the various metabolites associated with the deetiolation process, especially the phenylpropanoids (e.g. of the chlorogenic acid family). These were shown to increase during greening and may provide protection from light/UV stress (Clé et al., 2008) during the sensitive phase when the photosynthetic apparatus needs to be assembled from parts and intermediates that are highly prone to photooxidative destruction.
In summary, our work described here (1) provides a comprehensive description of the molecular events involved in chloroplast differentiation and thylakoid biogenesis in a dicot model, (2) reveals the existence of distinct physiological phases in the greening process and their molecular basis, (3) provides fresh insight into the limiting factors and regulatory levels involved in the establishment of photosynthetic competence, and (4) provides large, highly time-resolved omics data sets and a short list of genes for new biogenesis-related functions that will serve as a rich platform for future functional studies.
MATERIALS AND METHODS
Plant Growth and Collection of Material
Tobacco (Nicotiana tabacum ‘Petit Havana’) was raised under nursery (100 μE m−2 s−1, 16/8 h of light/dark, 24°C/20°C, 70%/70% humidity) conditions for 1 week and then transferred to standard conditions (350 μE m−2 s−1, 16/8 h of light/dark, 22°/18°C, 75%/70% humidity) for further growth (Fig. 1A). Extended dark treatment conditions followed standard conditions without illumination. Plants older than 3 weeks were supplemented by watering as required with 1 g L−1 Hakaphos fertilizer. Leaves were immediately snap frozen under liquid nitrogen, and the newly expanded tissue was excised from the leaf tip (grown prior to dark treatment) and the extreme basal region (grown after dark treatment for later time points; Fig. 1B). To minimize biological variation arising from differences in leaf size and age, tissue sectors sampled from the smallest and second smallest leaves of three replicate plants were homogenized and pooled as a single biological repeat, with these pools then used for all subsequent measurements (omics pools), excluding in vivo measurements of photosynthetic complex content and activity, 77K fluorescence emission, CD spectroscopy, and microscopy.
Isolation of Nucleic Acids
Total plant DNA (gDNA) was isolated following the cetyltrimethylammonium bromide-based isolation (Doyle and Doyle, 1990). RNA was isolated using the Nucleospin RNA Plant kit, according to the manufacturer’s instructions (Macherey-Nagel), including the second, optional DNase digest and gDNA depletion confirmed by PCR. Nucleic acid concentrations and purity were analyzed by Nanodrop 1000 (PeqLab), and RNA quality was further assessed by 2010 Bioanalyser (Agilent Technologies): sufficient quality RNA was defined by an RNA integrity number of 6.5 or greater and a 25S:18S greater than 1.
RNAseq
RNA samples were sequenced at the Max Planck Genome Centre with an Illumina HiSeq2500 (Illumina). Single-end, 100-bp reads to a minimum of 30 million (±10%) reads per sample were undertaken using a TruSeq RNA library prepared by the Max Planck Genome Centre. The quality of obtained Illumina single-end sequences was tested with FastQC v0.10.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). As no overrepresented sequences were detected, an additional 3′ trimming step was skipped. To reduce complexity, and because the project aimed to define and identify evolutionarily conserved features, we mapped to the tobacco maternal (Renny-Byfield et al., 2011) progenitor Nicotiana sylvestris (ftp://ftp.ncbi.nlm.nih.gov/genomes/Nicotiana_sylvestris/RNA/rna.fa.gz; file date October 24, 2014), as opposed to tobacco (Sierro et al., 2014). Mapping was undertaken using BWA v0.70 in aln mode with default parameters (Li and Durbin, 2009). Samtools v1.0 was used to create, sort, and index the alignment data in BAM format and calculate transcript counts (Li et al., 2009). Transcripts were normalized to reads per kilobase of transcript per million mapped reads, and species with values greater than 2 for all samples were removed from further analysis. Homologous Arabidopsis (Arabidopsis thaliana) genes were identified from translated N. sylvestris sequences using BLASTX (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The top hit was defined as orthologous (the term is used throughout for simplicity) if the match had a maximum e-value of 10−5.
qPCR
Synthesis of complementary DNA was undertaken using the SuperScript III Reverse Transcriptase kit (Thermo Fisher Scientific) with mixed oligo(dT18) and random hexamers. All qPCR measurements were made in technical triplicate using the LightCycler 480 Real-Time PCR System and LightCycler SYBR Green reaction mixtures (Roche Applied Science), and data were analyzed using the associated software. Raw run fluorescence history was processed with LinRegPCR (Ramakers et al., 2003; Ruijter et al., 2009) to define and allow correction by PCR efficiency. Samples were normalized to the geometric mean of three stable nucleus-encoded transcripts, the number and identity of which were defined by GeNorm software (Vandesompele et al., 2002). Housekeeping genes were chosen from those previously defined for tobacco (Schmidt and Delaney, 2010) and Arabidopsis (Czechowski et al., 2005). The plastid transcript-specific primers used in RT-qPCR cover exon-exon junctions, where present, and as such recognize only spliced transcripts. The design of the platform meant that the 60-min time point was excluded from analysis. All primers, including those used to quantify relative genome copy numbers, are provided in Supplemental Table S4.
Protein Methods
Proteins for BN- and SDS-PAGE were extracted from a mixture containing equal tissue mass from each of the three repeats. Protein extraction for BN-PAGE was performed after Järvi et al. (2011) to permit the complete isolation of membrane and soluble protein complexes from small masses of homogenized frozen leaf tissue. Briefly, tissue was thoroughly resuspended in 25BTHG (25 mm BisTris with 20% [v/v] glycerol) prior to addition of 5% (w/v) n-dodecyl β-d-maltopyranoside in 25BTHG, in a tissue:solution:detergent ratio of 1:3.2:0.8 (w/v/v). For SDS-PAGE, total tissue was solubilized for 30 min at 40°C by addition of 1% (v/w) loading buffer in 25BTHG to a final concentration of 1× 50 mm Tris, pH 6.8, 1.2% (v/v) glycerol, 0.5% (w/v) SDS, 0.15% (w/v) DTT, and 0.005% Coomassie Blue G250 (w/v). Samples were separated on a 10% Tris-Tricine gel (Schägger and von Jagow, 1987) and transferred to activated polyvinylidene difluoride membranes prior to hybridization with antibodies obtained from Agrisera. Luminescence was detected using the ECL Prime Western Blotting Detection Reagents (GE Healthcare) following the manufacturer’s instructions and the G:Box Chemi XT4 (Syngene). Signal intensity was quantified using ImageJ software (Schneider et al., 2012).
Concurrent Extraction for Soluble Metabolite and Lipid Measurements
Extraction from 50 mg (±10%) frozen, homogenized tobacco tissue was undertaken using a methanol:methyl-tertiary-butyl-ether:water extraction method as previously described (Giavalisco et al., 2011; Salem et al., 2016). Briefly, metabolites were extracted from tissue with 500 μL of precooled methyl-tertiary-butyl-ether:methanol extraction solution by mixing and sonication. After addition of 250 μL of 1:3 methanol:water, phases were separated by centrifugation (2 min, room temperature, 16,000g). The nonpolar, upper phase (350 μL) and the polar, lower phase (75 μL) were extracted for lipid (liquid chromatography-tandem mass spectrometry) and primary metabolite (GC-MS) analysis, respectively.
Lipidomics: Sample Preparation, Execution, and Peak Identification
UPLC separation was performed using a Waters Acquity UPLC system with an HSS T3 C18 reverse-phase column at 60°C, with injection volume of 2 μL as previously described (Pant et al., 2015) with MS recorded by a Thermo Exactive device (Thermo Fisher Scientific). By this method, due to the similar mass and chromatographic behavior of certain fatty acid combinations (e.g. 18:2/16:2 PC and 18:3/16:1 PC), lipids are identified as apparent species named for the summed carbon chains and double bonds (i.e. 34:4 PC, containing a total 34 carbons with four double bonds). Chromatogram raw file preprocessing (baseline correction, chemical noise subtraction, alignment, and peak detection) was undertaken with GeneData software (https://www.genedata.com/), as previously described (Giavalisco et al., 2011), gathering detected peak retention time and mass-to-charge ratio pair and a matrix with respective intensities for each sample. Lipids were identified using an R (https://www.r-project.org/) script developed in house by Dr. Alvaro Cuadros Inostroza, which corrects retention times based on identified markers (Giavalisco et al., 2011) and searches for compounds by comparing specific mass-to-charge ratio within user-given ranges (here, mass tolerance of 10 µL L−1 and retention time deviation of 0.2 min). Further confirmation was achieved by manual inspection of chromatograms. Presence was assumed only if intensity was above 5 × 104 units. Data were normalized to sample fresh weight and to the internal standard, PC 34:0, which did not appear to accumulate in a biased way based on time point. Isomers, defined as compounds with the same mass but slightly different retention times, are noted with numbers in brackets. Species found consistently (more than three or six) and to high levels (greater than 50% of the experimental minimum) within concurrently analyzed blank samples, generally representing lowly abundant species, are indicated in figures but were not removed from further analyses.
Metabolomics of Soluble Metabolites: Sample Preparation, Execution, and Peak Identification
A soluble metabolite fraction that was enriched for primary metabolites and small specialized (secondary) metabolites was profiled by GC-MS using an Agilent 6890N24 gas chromatograph coupled to a Pegasus III time-of-flight mass spectrometer, with FactorFour VF column, as previously described (Wagner et al., 2003; Erban et al., 2007). Metabolites were methoxyaminated and trimethylsilylated manually prior to GC-electron impact/time of flight-MS. An mixture of C10, C12, C15, C18, C19, C22, C28, C32, and C36 was added to each sample to calibrate retention indices as described (Strehmel et al., 2008). Following visual control and baseline correction, chromatograms were exported in NetCDF format using ChromaTOF. The Golm Metabolome Database (http://gmd.mpimpgolm.mpg.de/; Kopka et al., 2005; Schauer et al., 2005) and the NIST08 database (https://www.nist.gov/) were used to identify compounds with TagFinder software (Luedemann et al., 2008), with reliable analytes defined by low retention index deviation (less than 1%) and the presence of at least three mass fragments per compound. Representative analytes were hand picked in instances where multiple analytes represented a single metabolite. Data were normalized to sample fresh weight and to the internal standard, [13C6]sorbitol. Analytes found consistently (three or more of six blank samples) and to a high level (50% or greater of the minimum abundance found across all experimental samples) were marked in figures (pale gray) but not removed from further analyses.
Quantification, Statistical Testing, Data Analysis, and Visualization
Statistical analysis for photosynthetic data was conducted by ANOVA using a pairwise multiple comparison procedure (Holm-Sidak method) in SigmaPlot (https://systatsoftware.com/products/sigmaplot/). For lipidomics, transcriptomics, and soluble metabolite analyses, values below the detection level were treated as missing values, which were substituted with an artificially low number (10−5) for significance testing but not for PCA. Probabilistic PCA was undertaken using the pcaMethods package (http://bioconductor.org/biocLite.R; Stacklies et al., 2007), and transcript correlation was defined by the cor function, both conducted using version 3.2.3 of the open-access R software (http://www.r-project.org; R Foundation for Statistical Computing). PCAs were conducted on median-normalized log10-transformed data. Statistical significance was defined by one-way ANOVA (P < 0.05) with adjusted Benjamini-Hochberg false correction using log10-transformed, normalized data in R studio. As ANOVA is a suboptimal choice of analysis for time-course data, due to the necessary assumption of independence between variables (time points), we also performed statistical analysis by EDGE (adjusted P < 0.05), which has been developed as a method specific for the identification of differential expression in the context of time-course data (Storey et al., 2005). Control samples were excluded from differential gene expression analysis. Heat maps and hierarchical clustering (Pearson, average linkage) were made with version 4.9.0 of the Multiple Experiment Viewer (Saeed et al., 2003, 2006).
Chlorophyll Quantification
Frozen tissues of defined fresh weight or leaf discs of defined surface area were extracted by the addition of 80% (v/v) acetone and measured according to Porra et al. (1989).
Light-Induced Difference Absorbance Spectroscopy
Light-induced difference transmittance signals (ΔI/I) of cyt f and P700, the chlorophyll a dimer in the PSI reaction center, were used as measures for changes in the contents per leaf area of redox-active cyt b6f and PSI, respectively. Additionally, light-induced difference transmittance changes of PCy were determined. Difference transmittance signals of cyt f were measured between 505 and 570 nm wavelength using a KLAS-100 spectrophotometer (Heinz Walz) and deconvoluted from the electrochromic shift signal peaking at 520 nm wavelength, the scattering signal peaking at 535 nm wavelength, and signals arising from other cytochromes as described previously (Klughammer et al., 1990; Rott et al., 2011). Leaves were preilluminated for 5 min with saturating light to fully activate the Calvin-Benson cycle, avoid an acceptor-side limitation of PSI, and ensure that the high-potential chain including cyt f could be fully oxidized. Depending on the chlorophyll content during the deetiolation kinetic, the saturating actinic light intensity was adjusted to ensure that illumination was sufficient to fully oxidize cyt f. This steady-state illumination with saturating light was interrupted by short intervals of darkness, to allow for complete reduction of cyt f. After a maximum of 500 ms in darkness, the fully reduced state of cyt f was reached. Ten cycles of light-dark redox kinetics were measured and averaged, and the amplitude of the difference transmission signal between the fully oxidized state at the end of actinic illumination and the fully reduced state in darkness was used as a measure of redox-active cyt f per leaf area.
Difference absorptance signals originating from P700 and PCy were measured between 830 and 950 nm wavelength using the PCy-P700-version of the Dual-PAM instrument (Heinz Walz) as described previously (Schöttler et al., 2007). To avoid problems arising from an acceptor-side limitation of PSI by the Calvin-Benson cycle, leaves were preilluminated for 5 min in saturating light followed by a short interval of darkness. Then, PCy and P700 were fully oxidized by the application of far-red light preferentially exciting PSI (730 nm wavelength, 8 s length) followed by a short saturating pulse of red light (660 nm wavelength, 250 ms length) to activate linear electron flux and rapidly rereduce the high-potential chain during the following short interval of darkness. The amplitude of the difference transmittance signal between the fully oxidized state at the beginning of the saturating light pulse and the fully reduced state of the high-potential chain (reached after 150 to 300 ms in darkness) was used as a measure for the content of PCy and PSI. The intensity of the saturating light pulses was adjusted between 2,000 and 5,000 µE m−2 s−1, dependent on the increasing chlorophyll content of the leaf during the deetiolation kinetic, to ensure that the pulse was saturating but at the same time avoid artifacts due to detector oversaturation in weakly absorbing leaves during the early time points of the deetiolation kinetics. After the maximum light-induced amplitude of the difference transmittance signals had been established, light response curves were measured in 20 steps between 0 and 2,000 µE m−2 s−1 actinic light intensity. The donor-side limitation of PSI by electron transport from PSII via cyt b6f and PCy was calculated according to Schreiber and Klughammer (2016).
Chlorophyll a Fluorescence Measurements and Leaf Absorptance
In vivo measurements of chlorophyll a fluorescence parameters at 22°C were performed using a Dual-PAM instrument (Heinz Walz). Minimum fluorescence, maximum chlorophyll a fluorescence, and light response curves of linear electron flux were measured after 20 min of dark adaptation. The light intensity was stepwise increased from 0 to 2,500 µE m−2 s−1, with a measuring time of 150 s for each light intensity under light-limited conditions and 60 s under light-saturated conditions. While an equal distribution of absorbed light energy between both photosystems was initially assumed to occur, linear electron transport was corrected for leaf absorptance, which was calculated from leaf transmittance and reflectance spectra as 100% minus transmittance (%) minus reflectance (%). Spectra were measured between 400 and 700 nm wavelength using an integrating sphere attached to a photometer (V650; Jasco). The spectral bandwidth was set to 1 nm, and the scanning speed was 200 nm min−1.
CD Spectroscopy
CD spectra of intact leaves were recorded between 800 and 400 nm wavelength using a J-815 spectrophotometer (Jasco) equipped with a special red-sensitive head-on photomultiplier to position the leaf directly in front of the photomultiplier window, thus reducing loss of signal due to light scattering in the leaf. The scanning speed was 100 nm min−1, and a monochromator bandwidth setting of 5 nm was used. The signal in the wavelength range between 775 and 800 nm, where no CD signal arises from leaves, was normalized to zero. The psi-type CD signal occurring between 690 and 676 nm wavelength [called the (+)690 band] was used as a proxy for the accumulation of PSII-LHCII supercomplexes, as was the negative band peaking at 655 nm (Kovács et al., 2006; Garab and van Amerongen, 2009). Prior to all measurements, leaves were dark adapted for 20 min to avoid possible perturbations arising from reversible light-induced structural changes in the thylakoid membrane associated with rapidly reversible nonphotochemical quenching.
Gas Exchange
Gas-exchange measurements were performed with a GFS-3000 open gas-exchange system equipped with the light-emitting diode array unit 3056-FL as actinic light source (Heinz Walz). On the first day of deetiolation, changes in assimilation rates in deetiolated plants and control plants, which had stayed in the normal day-night rhythm prior to the measurements, were followed at the growth light intensity of 350 µE m−2 s−1 at 22°C cuvette temperature with 17,500 µL L−1 humidity and a saturating CO2 concentration of 2,000 µL L−1, to fully repress photorespiration. Prior to the beginning of illumination and at the end of the day, respiration was measured in darkness. On day 5 of deetiolation, light response curves of CO2 assimilation were measured with deetiolated plants and control plants kept in the normal light-dark cycle. After leaf respiration had been determined in darkness, the actinic light intensity was stepwise increased to 2,000 µE m−2 s−1. At each light intensity, gas exchange was recorded until the steady state of evaporation and leaf assimilation was reached. Cuvette temperature was 22°C, with 17,500 µL L−1 humidity and a saturating CO2 concentration of 2,000 µL L−1. For all measurements, a leaf area of 8 cm2 was used.
77K Chlorophyll a Fluorescence Emission Spectra Measurements
Leaf samples were ground in liquid nitrogen and diluted with ice to a concentration of less than 5 μg chlorophyll mL−1 as described by Weis (1985). Samples were excited using a blue (405 nm) laser diode, and emission spectra were recorded using a spectrophotometer (ASEQ Instruments; LR1) with a bifurcated fiber optic. Spectra were normalized to the PSI emission maximum.
TEM of Chloroplasts
For technical reasons, the 5-min time point was excluded from microscopy analysis, and control and 0 min were sampled, under green light, within the 1-h period prior to dawn. Samples for high-pressure freezing and chemical fixation were taken simultaneously, from the same leaf, to allow comparison of the techniques. For high-pressure freezing, samples were frozen by Leica EM HPM100 in aluminum sample holders (300–400 nm) following hexadecane infiltration, then transferred to freeze substitution solution containing 1% osmium tetroxide and 0.1% uranyl acetate in acetone. Samples were simultaneously fixed, dehydrated, and brought to room temperature in the Leica EM AFS2 (−120°C, 30 min; −120°C to −90°C, 2 h; −90°C, 36 h; −90°C to −80°C, 5 h; −80°C, 24 h; −80°C to −30°C, 24 h; −30°C to 0°C, 2 h; and 0°C, 20 h), then washed five times with anhydrous acetone and infiltrated with Agar 100 resin (24:9:15 [w/w/w] Agar 100:dodecenylsuccinic anhydride:methyl nadic anhydride ) in acetone in the following series: one drop resin mL−1 acetone, 1 h; two drops mL−1, 1 h; three drops mL−1, 1 h; four drops mL−1, 1 h; 10% (v/v), overnight; 20% (v/v), 3 h; 40% (v/v), 3 h; 60% (v/v), overnight; 80% (v/v), 3 h; 100% (v/v), 3 h; and fresh 100% (v/v), overnight. For chemical fixation, samples were vacuum infiltrated for ∼1 h in 2.5% (v/v) glutaraldehyde and 12.5 mm CaCl2 in 0.05 m cacodylate buffer, pH 7.4, the solution was refreshed, and samples were incubated (overnight, 4°C). Following washing (three times in cacodylate buffer), samples were postfixed (1.5 h) with 1% (w/v) OsO4 and 0.8% (w/v) K3Fe(CN)6 in cacodylate buffer and washed again (three times in deionized water). Subsequently, samples were poststained (1.5 h) with 2% uranyl acetate in cacodylate buffer, and the samples were slowly dehydrated with the following ethanol sequence: 10 min, 30% (v/v); 15 min, 50% (v/v); 30 min, 70% (v/v); 40 min, 80% (v/v); 40 min, 90% (v/v); and 3 × 40 min, 100%. Dehydrated samples were stored overnight at 4°C or for longer periods at −20°C, and the ethanol was subsequently substituted with propylene oxide by incubation for 30 min in ethanol:propylene oxide (1:1) and two incubations (30 min) with pure propylene oxide. Samples were then infiltrated with resin in propylene oxide in the following series: 4 h, 3:1 propylene oxide:resin; 4 h, 1:1 propylene oxide:resin; 20 h, 1:3 propylene oxide:resin; and 27 h, pure resin.
Blocks were cut with a Leica UCT ultramicrotome into 90-nm sections and stained with uranyl acetate. Specimens on nickel formvar-coated grids were examined with High Performance Biology Transmission Electron Microscope JEM 1400 (JEOL) equipped with 11 Megapixel TEM Camera MORADA G2 (EMSIS) in the Nencki Institute of Experimental Biology of the Polish Academy of Sciences in Warsaw. Number (and size) of visible PLBs in plastid sections was estimated in 50 fields of view in 8,000× magnification (approximately 500 organelles) in each of the first five developmental stages harvested for TEM (0, 10, 30, 60, and 120 min). Counting of other visible plastid features (number of grana and thylakoid chains) was performed at higher magnifications (30,000×–60,000× according to the size of the plastid) for 20 plastids in each developmental stage. Estimation of the grana sizes (50 for each developmental stage) and starch granule sizes (20 for each developmental stage where starch grains are visible) was performed using ImageJ software (Abramoff et al., 2004). Additionally, we measured the density of the PLB grid (20 in each developmental stage) using ImageJ by measuring the number of unit cells per 500 nm2. For estimation of the PLB distribution in etioplasts, see Supplemental Information S1. Significance was calculated by Student’s t test.
Accession Numbers
The RNAseq data discussed here have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE128049.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparison of tissue-processing techniques for TEM.
Supplemental Figure S2. Uniformity of etiolated samples (t = 0).
Supplemental Figure S3. Fv/Fm.
Supplemental Figure S4. Immunoblot replicates.
Supplemental Figure S5. 77K chlorophyll a fluorescence emission spectra at all time points.
Supplemental Figure S6. Changes in dry weight-to-fresh weight ratio during greening.
Supplemental Information S1. Statistical analysis to verify the probability distribution of PLBs.
Supplemental Data Set S1. Accumulation of plastid-encoded transcripts during greening.
Supplemental Data Set S2. Accumulation of lipid transcripts during greening.
Supplemental Data Set S3. Accumulation of soluble metabolites during greening.
Supplemental Data Set S4. Top 500 transcript species with the highest loading for PC1.
Supplemental Data Set S5. Accumulation of nucleus-encoded transcripts during greening.
Supplemental Data Set S6. Arabidopsis gene associated with tobacco transcripts showing significant changes.
Acknowledgments
We thank the Max Planck Institute of Molecular Plant Physiology (MPI-MP) GreenTeam for help with plant cultivation and Dr. Michael Tillich (MPI-MP) for helpful discussions regarding RNAseq data. We thank Ines Fehrle and Änne Michaelis (MPI-MP) for excellent technical assistance.
Footnotes
This work was supported by the Human Frontier Science Program to R.B. and Z.R. (RGP0005/2013).
Articles can be viewed without a subscription.
References
- Abramoff MD, Magalhães PJ, Ram SJ (2004) Image processing with ImageJ. Biophoton Int 11: 36–42 [Google Scholar]
- Adam Z, Charuvi D, Tsabari O, Knopf RR, Reich Z (2011) Biogenesis of thylakoid networks in angiosperms: Knowns and unknowns. Plant Mol Biol 76: 221–234 [DOI] [PubMed] [Google Scholar]
- Aguettaz P, Seyer P, Pesey H, Lescure AM (1987) Relations between the plastid gene dosage and the levels of 16S rRNA and rbcL gene transcripts during amyloplast to chloroplast change in mixotrophic spinach cell suspensions. Plant Mol Biol 8: 169–177 [DOI] [PubMed] [Google Scholar]
- Albus CA, Ruf S, Schöttler MA, Lein W, Kehr J, Bock R (2010) Y3IP1, a nucleus-encoded thylakoid protein, cooperates with the plastid-encoded Ycf3 protein in photosystem I assembly of tobacco and Arabidopsis. Plant Cell 22: 2838–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert S, Curien G, Bligny R, Gout E, Douce R (1998) Transport, compartmentation, and metabolism of homoserine in higher plant cells: Carbon-13- and phosphorus-31-nuclear magnetic resonance studies. Plant Physiol 116: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl J, Francke B, Monéger R (1976) Lipid composition of envelopes, prolamellar bodies and other plastid membranes in etiolated, green and greening wheat leaves. Planta 129: 193–201 [DOI] [PubMed] [Google Scholar]
- Baker NR, Butler WL (1976) Development of the primary photochemical apparatus of photosynthesis during greening of etiolated bean leaves. Plant Physiol 58: 526–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barahimipour R, Strenkert D, Neupert J, Schroda M, Merchant SS, Bock R (2015) Dissecting the contributions of GC content and codon usage to gene expression in the model alga Chlamydomonas reinhardtii. Plant J 84: 704–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1993) Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Gaudinier A, Brady SM, Hennig L, Rhee SY, De Smet I (2012) Systems analysis of plant functional, transcriptional, physical interaction, and metabolic networks. Plant Cell 24: 3859–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomqvist LA, Ryberg M, Sundqvist C (2006) Proteomic analysis of the etioplast inner membranes of wheat (Triticum aestivum) by two-dimensional electrophoresis and mass spectrometry. Physiol Plant 128: 368–381 [Google Scholar]
- Blomqvist LA, Ryberg M, Sundqvist C (2008) Proteomic analysis of highly purified prolamellar bodies reveals their significance in chloroplast development. Photosynth Res 96: 37–50 [DOI] [PubMed] [Google Scholar]
- Boasson R, Laetsch WM (1969) Chloroplast replication and growth in tobacco. Science 166: 749–751 [DOI] [PubMed] [Google Scholar]
- Browse J, Roughan PG, Slack CR (1981) Light control of fatty acid synthesis and diurnal fluctuations of fatty acid composition in leaves. Biochem J 196: 347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos A, Szymanski J, Seiwert B, Degenkolbe T, Hannah MA, Giavalisco P, Willmitzer L (2011) Analysis of short-term changes in the Arabidopsis thaliana glycerolipidome in response to temperature and light. Plant J 66: 656–668 [DOI] [PubMed] [Google Scholar]
- Cahoon AB, Takacs EM, Sharpe RM, Stern DB (2008) Nuclear, chloroplast, and mitochondrial transcript abundance along a maize leaf developmental gradient. Plant Mol Biol 66: 33–46 [DOI] [PubMed] [Google Scholar]
- Casano LM, Martín M, Sabater B (2001) Hydrogen peroxide mediates the induction of chloroplastic Ndh complex under photooxidative stress in barley. Plant Physiol 125: 1450–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charuvi D, Kiss V, Nevo R, Shimoni E, Adam Z, Reich Z (2012) Gain and loss of photosynthetic membranes during plastid differentiation in the shoot apex of Arabidopsis. Plant Cell 24: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clé C, Hill LM, Niggeweg R, Martin CR, Guisez Y, Prinsen E, Jansen MAK (2008) Modulation of chlorogenic acid biosynthesis in Solanum lycopersicum: Consequences for phenolic accumulation and UV-tolerance. Phytochemistry 69: 2149–2156 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl R, Staehelin LA (1989) High-pressure freezing for the preservation of biological structure: Theory and practice. J Electron Microsc Tech 13: 165–174 [DOI] [PubMed] [Google Scholar]
- Daum B, Nicastro D, Austin J II, McIntosh JR, Kühlbrandt W (2010) Arrangement of photosystem II and ATP synthase in chloroplast membranes of spinach and pea. Plant Cell 22: 1299–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter S, Mustardy L, Machowicz E (1976) The development of the intense circular-dichroic signal during granum formation in greening etiolated maize. Biochem J 156: 469–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Gruissem W (1987) Control of plastid gene expression during development: The limited role of transcriptional regulation. Cell 49: 379–387 [DOI] [PubMed] [Google Scholar]
- Domanskii V, Rassadina V, Gus-Mayer S, Wanner G, Schoch S, Rüdiger W (2003) Characterization of two phases of chlorophyll formation during greening of etiolated barley leaves. Planta 216: 475–483 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL (1990) A rapid total DNA preparation procedure for fresh plant tissue. Focus 12: 13–15 [Google Scholar]
- Doyle MF, Yu CA (1985) Preparation and reconstitution of a phospholipid deficient cytochrome b6f complex from spinach chloroplasts. Biochem Biophys Res Commun 131: 700–706 [DOI] [PubMed] [Google Scholar]
- Eckstein A, Zieba P, Gabrys H (2012) Sugar and light effects on the condition of the photosynthetic apparatus of Arabidopsis thaliana cultured in vitro. J Plant Growth Regul 31: 90–101 [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edhofer I, Mühlbauer SK, Eichacker LA (1998) Light regulates the rate of translation elongation of chloroplast reaction center protein D1. Eur J Biochem 257: 78–84 [DOI] [PubMed] [Google Scholar]
- Erban A, Schauer N, Fernie AR, Kopka J (2007) Nonsupervised construction and application of mass spectral and retention time index libraries from time-of-flight gas chromatography-mass spectrometry metabolite profiles. Methods Mol Biol 358: 19–38 [DOI] [PubMed] [Google Scholar]
- Espineda CE, Linford AS, Devine D, Brusslan JA (1999) The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 10507–10511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck F, Sperling U, Frick G, Pochert B, van Cleve B, Apel K, Armstrong GA (2000) Regulation of etioplast pigment-protein complexes, inner membrane architecture, and protochlorophyllide a chemical heterogeneity by light-dependent NADPH:protochlorophyllide oxidoreductases A and B. Plant Physiol 124: 1678–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme P, Jordan P, Krauss N (2001) Structure of photosystem I. Biochim Biophys Acta 1507: 5–31 [DOI] [PubMed] [Google Scholar]
- Galey J, Francke B, Bahl J (1980) Ultrastructure and lipid composition of etioplasts in developing dark-grown wheat leaves. Planta 149: 433–439 [DOI] [PubMed] [Google Scholar]
- Garab G, van Amerongen H (2009) Linear dichroism and circular dichroism in photosynthesis research. Photosynth Res 101: 135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty B, Briantais JM, Baker NR (1989) The relationship between the quantum yield of photosynthetic electron-transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta 990: 87–92 [Google Scholar]
- Giavalisco P, Li Y, Matthes A, Eckhardt A, Hubberten HM, Hesse H, Segu S, Hummel J, Köhl K, Willmitzer L (2011) Elemental formula annotation of polar and lipophilic metabolites using 13C, 15N and 34S isotope labelling, in combination with high-resolution mass spectrometry. Plant J 68: 364–376 [DOI] [PubMed] [Google Scholar]
- Gombos Z, Várkonyi Z, Hagio M, Iwaki M, Kovács L, Masamoto K, Itoh S, Wada H (2002) Phosphatidylglycerol requirement for the function of electron acceptor plastoquinone QB in the photosystem II reaction center. Biochemistry 41: 3796–3802 [DOI] [PubMed] [Google Scholar]
- Gounaris K, Barber J (1985) Isolation and characterization of a photosystem-II reaction center lipoprotein complex. FEBS Lett 188: 68–72 [Google Scholar]
- Green CD, Hollingsworth MJ (1992) Expression of the large ATP synthase gene cluster in spinach plastids during light-induced development. Plant Physiol 100: 1164–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AR, Karpowicz SJ, Heinnickel M, Dewez D, Hamel B, Dent R, Niyogi KK, Johnson X, Alric J, Wollman FA, et al. (2010) Phylogenomic analysis of the Chlamydomonas genome unmasks proteins potentially involved in photosynthetic function and regulation. Photosynth Res 106: 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häusler RE, Heinrichs L, Schmitz J, Flügge UI (2014) How sugars might coordinate chloroplast and nuclear gene expression during acclimation to high light intensities. Mol Plant 7: 1121–1137 [DOI] [PubMed] [Google Scholar]
- Heinhorst S, Cannon G, Weissbach A (1985) Plastid and nuclear DNA synthesis are not coupled in suspension cells of Nicotiana tabacum. Plant Mol Biol 4: 3–12 [DOI] [PubMed] [Google Scholar]
- Heinnickel ML, Grossman AR (2013) The GreenCut: Re-evaluation of physiological role of previously studied proteins and potential novel protein functions. Photosynth Res 116: 427–436 [DOI] [PubMed] [Google Scholar]
- Hooper CM, Castleden IR, Tanz SK, Aryamanesh N, Millar AH (2017) SUBA4: The interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res 45: D1064–D1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosler JP, Wurtz EA, Harris EH, Gillham NW, Boynton JE (1989) Relationship between gene dosage and gene expression in the chloroplast of Chlamydomonas reinhardtii. Plant Physiol 91: 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järvi S, Suorsa M, Paakkarinen V, Aro EM (2011) Optimized native gel systems for separation of thylakoid protein complexes: Novel super- and mega-complexes. Biochem J 439: 207–214 [DOI] [PubMed] [Google Scholar]
- Johnson MP, Brain AP, Ruban AV (2011) Changes in thylakoid membrane thickness associated with the reorganization of photosystem II light harvesting complexes during photoprotective energy dissipation. Plant Signal Behav 6: 1386–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanervo E, Singh M, Suorsa M, Paakkarinen V, Aro E, Battchikova N, Aro EM (2008) Expression of protein complexes and individual proteins upon transition of etioplasts to chloroplasts in pea (Pisum sativum). Plant Cell Physiol 49: 396–410 [DOI] [PubMed] [Google Scholar]
- Karpowicz SJ, Prochnik SE, Grossman AR, Merchant SS (2011) The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J Biol Chem 286: 21427–21439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Eichacker LA, Rudiger W, Mullet JE (1994) Chlorophyll regulates accumulation of the plastid-encoded chlorophyll proteins P700 and D1 by increasing apoprotein stability. Plant Physiol 104: 907–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Christopher DA, Mullet JE (1993) Direct evidence for selective modulation of psbA, rpoA, rbcL and 16S RNA stability during barley chloroplast development. Plant Mol Biol 22: 447–463 [DOI] [PubMed] [Google Scholar]
- Kleffmann T, von Zychlinski A, Russenberger D, Hirsch-Hoffmann M, Gehrig P, Gruissem W, Baginsky S (2007) Proteome dynamics during plastid differentiation in rice. Plant Physiol 143: 912–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RR. (1991) Regulation of light-induced chloroplast transcription and translation in eight-day-old dark-grown barley seedlings. Plant Physiol 97: 335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RR, Mason HS, Mullet JE (1988) Light-regulated translation of chloroplast proteins. I. Transcripts of psaA-psaB, psbA, and rbcL are associated with polysomes in dark-grown and illuminated barley seedlings. J Cell Biol 106: 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klughammer C, Kolbowski J, Schreiber U (1990) LED array spectrophotometer for measurement of time resolved difference spectra in the 530-600 nm wavelength region. Photosynth Res 25: 317–327 [DOI] [PubMed] [Google Scholar]
- Komatsu S, Muhammad A, Rakwal R (1999) Separation and characterization of proteins from green and etiolated shoots of rice (Oryza sativa L.): Towards a rice proteome. Electrophoresis 20: 630–636 [DOI] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kovács L, Damkjaer J, Kereïche S, Ilioaia C, Ruban AV, Boekema EJ, Jansson S, Horton P (2006) Lack of the light-harvesting complex CP24 affects the structure and function of the grana membranes of higher plant chloroplasts. Plant Cell 18: 3106–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalewska Ł, Mazur R, Suski S, Garstka M, Mostowska A (2016) Three-dimensional visualization of the tubular-lamellar transformation of the internal plastid membrane network during runner bean chloroplast biogenesis. Plant Cell 28: 875–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause GH, Weis E (1984) Chlorophyll fluorescence as a tool in plant physiology. II. Interpretation of fluorescence signals. Photosynth Res 5: 139–157 [DOI] [PubMed] [Google Scholar]
- Krause GH, Weis E (1991) Chlorophyll fluorescence and photosynthesis: The basics. Annu Rev Plant Physiol 42: 313–349 [Google Scholar]
- Krupinska K, Apel K (1989) Light-induced transformation of etioplasts to chloroplasts of barley without transcriptional control of plastid gene-expression. Mol Gen Genet 219: 467–473 [Google Scholar]
- Kruse O, Hankamer B, Konczak C, Gerle C, Morris E, Radunz A, Schmid GH, Barber J (2000) Phosphatidylglycerol is involved in the dimerization of photosystem II. J Biol Chem 275: 6509–6514 [DOI] [PubMed] [Google Scholar]
- Lezhneva L, Amann K, Meurer J (2004) The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J 37: 174–185 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Liu MJ, Wu SH, Chen HM, Wu SH (2012) Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol Syst Biol 8: 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonosky PM, Zhang X, Honavar VG, Dobbs DL, Fu A, Rodermel SR (2004) A proteomic analysis of maize chloroplast biogenesis. Plant Physiol 134: 560–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedemann A, Strassburg K, Erban A, Kopka J (2008) TagFinder for the quantitative analysis of gas chromatography-mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 24: 732–737 [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield SP, Yohn CB, Cohen A, Danon A (1995) Regulation of chloroplast gene-expression. Annu Rev Plant Physiol 46: 147–166 [Google Scholar]
- Miquel M, Browse J (1992) Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis: Biochemical and genetic characterization of a plant oleoyl-phosphatidylcholine desaturase. J Biol Chem 267: 1502–1509 [PubMed] [Google Scholar]
- Mostowska A. (1985) Stereometrical analysis of number and size of prolamellar bodies during pea chloroplast development. Acta Soc Bot Pol 54: 53–63 [Google Scholar]
- Murata N, Higashi S, Fujimura Y (1990) Glycerolipids in various preparations of photosystem II from spinach-chloroplasts. Biochim Biophys Acta 1019: 261–268 [Google Scholar]
- Nelissen H, Gonzalez N, Inzé D (2016) Leaf growth in dicots and monocots: So different yet so alike. Curr Opin Plant Biol 33: 72–76 [DOI] [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG (1997) Regulation of fatty acid synthesis. Annu Rev Plant Physiol Plant Mol Biol 48: 109–136 [DOI] [PubMed] [Google Scholar]
- Ort DR, Merchant SS, Alric J, Barkan A, Blankenship RE, Bock R, Croce R, Hanson MR, Hibberd JM, Long SP, et al. (2015) Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc Natl Acad Sci USA 112: 8529–8536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, Yang S, Choi G (2012) Phytochrome regulates translation of mRNA in the cytosol. Proc Natl Acad Sci USA 109: 1335–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Burgos A, Pant P, Cuadros-Inostroza A, Willmitzer L, Scheible WR (2015) The transcription factor PHR1 regulates lipid remodeling and triacylglycerol accumulation in Arabidopsis thaliana during phosphorus starvation. J Exp Bot 66: 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ (2002) Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14: 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar K, Geis T, Ilkavets I, Oster U, Schwenkert S, Meurer J, Soll J (2007) Chloroplast biogenesis: The use of mutants to study the etioplast-chloroplast transition. Proc Natl Acad Sci USA 104: 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick U, Gounaris K, Weiss M, Barber J (1985) Tightly bound sulfolipids in chloroplast CF0-CF1. Biochim Biophys Acta 808: 415–420 [Google Scholar]
- Plöscher M, Granvogl B, Zoryan M, Reisinger V, Eichacker LA (2009) Mass spectrometric characterization of membrane integral low molecular weight proteins from photosystem II in barley etioplasts. Proteomics 9: 625–635 [DOI] [PubMed] [Google Scholar]
- Plöscher M, Reisinger V, Eichacker LA (2011) Proteomic comparison of etioplast and chloroplast protein complexes. J Proteomics 74: 1256–1265 [DOI] [PubMed] [Google Scholar]
- Plücken H, Müller B, Grohmann D, Westhoff P, Eichacker LA (2002) The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett 532: 85–90 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous-equations for assaying chlorophyll a and chlorophyll b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic-absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Porra RJ, Schäfer W, Cmiel E, Katheder I, Scheer H (1994) The derivation of the formyl-group oxygen of chlorophyll b in higher plants from molecular oxygen: Achievement of high enrichment of the 7-formyl-group oxygen from 18O2 in greening maize leaves. Eur J Biochem 219: 671–679 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Reinbothe C, Lebedev N, Reinbothe S (1999) A protochlorophyllide light-harvesting complex involved in de-etiolation of higher plants. Nature 397: 80–84 [Google Scholar]
- Reisinger V, Hertle AP, Plöscher M, Eichacker LA (2008) Cytochrome b6f is a dimeric protochlorophyll a binding complex in etioplasts. FEBS J 275: 1018–1024 [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S, Chester M, Kovařík A, Le Comber SC, Grandbastien MA, Deloger M, Nichols RA, Macas J, Novák P, Chase MW, et al. (2011) Next generation sequencing reveals genome downsizing in allotetraploid Nicotiana tabacum, predominantly through the elimination of paternally derived repetitive DNAs. Mol Biol Evol 28: 2843–2854 [DOI] [PubMed] [Google Scholar]
- Rott M, Martins NF, Thiele W, Lein W, Bock R, Kramer DM, Schöttler MA (2011) ATP synthase repression in tobacco restricts photosynthetic electron transport, CO2 assimilation, and plant growth by overacidification of the thylakoid lumen. Plant Cell 23: 304–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudowska L, Gieczewska K, Mazur R, Garstka M, Mostowska A (2012) Chloroplast biogenesis: Correlation between structure and function. Biochim Biophys Acta 1817: 1380–1387 [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM (2009) Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J (2006) TM4 microarray software suite. Methods Enzymol 411: 134–193 [DOI] [PubMed] [Google Scholar]
- Salem MA, Jüppner J, Bajdzienko K, Giavalisco P (2016) Protocol: A fast, comprehensive and reproducible one-step extraction method for the rapid preparation of polar and semi-polar metabolites, lipids, proteins, starch and cell wall polymers from a single sample. Plant Methods 12: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Kozaki A, Hatano M (1997) Link between light and fatty acid synthesis: Thioredoxin-linked reductive activation of plastidic acetyl-CoA carboxylase. Proc Natl Acad Sci USA 94: 11096–11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Sonoike K, Tsuzuki M, Kawaguchi A (1995) Impaired photosystem II in a mutant of Chlamydomonas reinhardtii defective in sulfoquinovosyl diacylglycerol. Eur J Biochem 234: 16–23 [DOI] [PubMed] [Google Scholar]
- Sato N, Aoki M, Maru Y, Sonoike K, Minoda A, Tsuzuki M (2003) Involvement of sulfoquinovosyl diacylglycerol in the structural integrity and heat-tolerance of photosystem II. Planta 217: 245–251 [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Scharff LB, Ehrnthaler M, Janowski M, Childs LH, Hasse C, Gremmels J, Ruf S, Zoschke R, Bock R (2017) Shine-Dalgarno sequences play an essential role in the translation of plastid mRNAs in tobacco. Plant Cell 29: 3085–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al. (2005) GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Schmidt GW, Delaney SK (2010) Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol Genet Genomics 283: 233–241 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöttler MA, Tóth SZ (2014) Photosynthetic complex stoichiometry dynamics in higher plants: Environmental acclimation and photosynthetic flux control. Front Plant Sci 5: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöttler MA, Flügel C, Thiele W, Stegemann S, Bock R (2007) The plastome-encoded PsaJ subunit is required for efficient photosystem I excitation, but not for plastocyanin oxidation in tobacco. Biochem J 403: 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöttler MA, Thiele W, Belkius K, Bergner SV, Flügel C, Wittenberg G, Agrawal S, Stegemann S, Ruf S, Bock R (2017) The plastid-encoded PsaI subunit stabilizes photosystem I during leaf senescence in tobacco. J Exp Bot 68: 1137–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber U, Klughammer C (2016) Analysis of photosystem I donor and acceptor sides with a new type of online-deconvoluting kinetic LED-array spectrophotometer. Plant Cell Physiol 57: 1454–1467 [DOI] [PubMed] [Google Scholar]
- Siebertz HP, Heinz E (1977) Labelling experiments on the origin of hexa- and octadecatrienoic acids in galactolipids from leaves. Z Naturforsch 32c: 193–205 [Google Scholar]
- Sierro N, Battey JND, Ouadi S, Bovet L, Goepfert S, Bakaher N, Peitsch MC, Ivanov NV (2013) Reference genomes and transcriptomes of Nicotiana sylvestris and Nicotiana tomentosiformis. Genome Biol 14: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierro N, Battey JND, Ouadi S, Bakaher N, Bovet L, Willig A, Goepfert S, Peitsch MC, Ivanov NV (2014) The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun 5: 3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skupień J, Wójtowicz J, Kowalewska Ł, Mazur R, Garstka M, Gieczewska K, Mostowska A (2017) Dark-chilling induces substantial structural changes and modifies galactolipid and carotenoid composition during chloroplast biogenesis in cucumber (Cucumis sativus L.) cotyledons. Plant Physiol Biochem 111: 107–118 [DOI] [PubMed] [Google Scholar]
- Solymosi K, Böddi B (2006) Optical properties of bud scales and protochlorophyll(ide) forms in leaf primordia of closed and opened buds. Tree Physiol 26: 1075–1085 [DOI] [PubMed] [Google Scholar]
- Solymosi K, Martinez K, Kristof Z, Sundqvist C, Boddi B (2004) Plastid differentiation and chlorophyll biosynthesis in different leaf layers of white cabbage (Brassica oleracea cv. capitata). Physiol Plant 121: 520–529 [Google Scholar]
- Solymosi K, Bóka K, Böddi B (2006) Transient etiolation: Protochlorophyll(ide) and chlorophyll forms in differentiating plastids of closed and breaking leaf buds of horse chestnut (Aesculus hippocastanum). Tree Physiol 26: 1087–1096 [DOI] [PubMed] [Google Scholar]
- Sperling U, Franck F, van Cleve B, Frick G, Apel K, Armstrong GA (1998) Etioplast differentiation in Arabidopsis: Both PORA and PORB restore the prolamellar body and photoactive protochlorophyllide-F655 to the cop1 photomorphogenic mutant. Plant Cell 10: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacklies W, Redestig H, Scholz M, Walther D, Selbig J (2007) pcaMethods--a bioconductor package providing PCA methods for incomplete data. Bioinformatics 23: 1164–1167 [DOI] [PubMed] [Google Scholar]
- Standfuss J, Terwisscha van Scheltinga AC, Lamborghini M, Kühlbrandt W (2005) Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 A resolution. EMBO J 24: 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson PG, Fankhauser C, Terry MJ (2009) PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci USA 106: 7654–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöckel J, Oelmüller R (2004) A novel protein for photosystem I biogenesis. J Biol Chem 279: 10243–10251 [DOI] [PubMed] [Google Scholar]
- Stöckel J, Bennewitz S, Hein P, Oelmüller R (2006) The evolutionarily conserved tetratrico peptide repeat protein pale yellow green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiol 141: 870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW (2005) Significance analysis of time course microarray experiments. Proc Natl Acad Sci USA 102: 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehmel N, Hummel J, Erban A, Strassburg K, Kopka J (2008) Retention index thresholds for compound matching in GC-MS metabolite profiling. J Chromatogr B Analyt Technol Biomed Life Sci 871: 182–190 [DOI] [PubMed] [Google Scholar]
- Sullivan ML, Green PJ (1993) Post-transcriptional regulation of nuclear-encoded genes in higher plants: The roles of mRNA stability and translation. Plant Mol Biol 23: 1091–1104 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Devireddy AR, Inupakutika MA, Baxter A, Miller G, Song L, Shulaev E, Azad RK, Shulaev V, Mittler R (2015) Ultra-fast alterations in mRNA levels uncover multiple players in light stress acclimation in plants. Plant J 84: 760–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabe T, Takabe T, Akazawa T (1986) Biosynthesis of P700-chlorophyll a protein complex, plastocyanin, and cytochrome b6/f complex. Plant Physiol 81: 60–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WF, Everett M, Polans NO, Jorgensen RA, Palmer JD (1983) Phytochrome control of RNA levels in developing pea and mung-bean leaves. Planta 158: 487–500 [DOI] [PubMed] [Google Scholar]
- Thorne SW, Boardman NK (1971) Formation of chlorophyll B, and the fluorescence properties and photochemical activities of isolated plastids from greening pea seedlings. Plant Physiol 47: 252–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth TN, Rai N, Solymosi K, Zsiros O, Schröder WP, Garab G, van Amerongen H, Horton P, Kovács L (2016) Fingerprinting the macro-organisation of pigment-protein complexes in plant thylakoid membranes in vivo by circular-dichroism spectroscopy. Biochim Biophys Acta 1857: 1479–1489 [DOI] [PubMed] [Google Scholar]
- Tremolieres A, Dainese P, Bassi R (1994) Heterogenous lipid distribution among chlorophyll-binding proteins of photosystem II in maize mesophyll chloroplasts. Eur J Biochem 221: 721–730 [DOI] [PubMed] [Google Scholar]
- Trémolières A, Lepage M (1971) Changes in lipid composition during greening of etiolated pea seedlings. Plant Physiol 47: 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udy DB, Belcher S, Williams-Carrier R, Gualberto JM, Barkan A (2012) Effects of reduced chloroplast gene copy number on chloroplast gene expression in maize. Plant Physiol 160: 1420–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M, Nouwen NP, Driessen AJM (2005) YidC: An evolutionary conserved device for the assembly of energy-transducing membrane protein complexes. Curr Opin Microbiol 8: 182–187 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dingenen J, De Milde L, Vermeersch M, Maleux K, De Rycke R, De Bruyne M, Storme V, Gonzalez N, Dhondt S, Inzé D (2016) Chloroplasts are central players in sugar-induced leaf growth. Plant Physiol 171: 590–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C (2008) Proline accumulation in plants: A review. Amino Acids 35: 753–759 [DOI] [PubMed] [Google Scholar]
- von Zychlinski A, Kleffmann T, Krishnamurthy N, Sjölander K, Baginsky S, Gruissem W (2005) Proteome analysis of the rice etioplast: Metabolic and regulatory networks and novel protein functions. Mol Cell Proteomics 4: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Wagner C, Sefkow M, Kopka J (2003) Construction and application of a mass spectral and retention time index database generated from plant GC/EI-TOF-MS metabolite profiles. Phytochemistry 62: 887–900 [DOI] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM (1998) A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37: 455–469 [DOI] [PubMed] [Google Scholar]
- Weis E. (1985) Chlorophyll fluorescence at 77 K in intact leaves: Characterization of a technique to eliminate artifacts related to self-absorption. Photosynth Res 6: 73–86 [DOI] [PubMed] [Google Scholar]
- Wellburn AR, Hampp R (1979) Appearance of photochemical function in prothylakoids during plastid development. Biochim Biophys Acta 547: 380–397 [DOI] [PubMed] [Google Scholar]
- Zhang J, Ruf S, Hasse C, Childs L, Scharff LB, Bock R (2012) Identification of cis-elements conferring high levels of gene expression in non-green plastids. Plant J 72: 115–128 [DOI] [PubMed] [Google Scholar]