The Phosphorylated Pathway of l-Ser Biosynthesis in Arabidopsis thaliana provides Ser for thiol assimilation in heterotrophic tissues such as roots.
Abstract
Although the plant Phosphorylated Pathway of l-Ser Biosynthesis (PPSB) is essential for embryo and pollen development, and for root growth, its metabolic implications have not been fully investigated. A transcriptomics analysis of Arabidopsis (Arabidopsis thaliana) PPSB-deficient mutants at night, when PPSB activity is thought to be more important, suggested interaction with the sulfate assimilation process. Because sulfate assimilation occurs mainly in the light, we also investigated it in PPSB-deficient lines in the day. Key genes in the sulfate starvation response, such as the adenosine 5′phosphosulfate reductase genes, along with sulfate transporters, especially those involved in sulfate translocation in the plant, were induced in the PPSB-deficient lines. However, sulfate content was not reduced in these lines as compared with wild-type plants; besides the glutathione (GSH) steady-state levels in roots of PPSB-deficient lines were even higher than in wild type. This suggested that PPSB deficiency perturbs the sulfate assimilation process between tissues/organs. Alteration of thiol distribution in leaves from different developmental stages, and between aerial parts and roots in plants with reduced PPSB activity, provided evidence supporting this idea. Diminished PPSB activity caused an enhanced flux of 35S into thiol biosynthesis, especially in roots. GSH turnover also accelerated in the PPSB-deficient lines, supporting the notion that not only biosynthesis, but also transport and allocation, of thiols were perturbed in the PPSB mutants. Our results suggest that PPSB is required for sulfide assimilation in specific heterotrophic tissues and that a lack of PPSB activity perturbs sulfur homeostasis between photosynthetic and nonphotosynthetic tissues.
The amino acid l-Ser plays an important function in metabolism and has been suggested to be a signal for transcriptional and/or metabolic regulation in both mammals and plants (Ye et al., 2012; Timm et al., 2013). In mammals, the Phosphorylated Pathway of Ser Biosynthesis (PPSB) is the primary route for the biosynthesis of this amino acid (Snell, 1984; Tabatabaie et al., 2010; Locasale et al., 2011). In plants the glycolate pathway, associated with photorespiration, is considered the most important source of Ser, at least in photosynthetic cells (Tolbert, 1980, 1985). However, the existence of the so-called nonphotorespiratory pathways (PPSB and glycerate pathways) raised the question of whether the glycolate pathway is sufficient to supply Ser to all plant cell types (heterotrophic/autotrophic), and under different environmental (i.e. day/night) and developmental conditions (Ros et al., 2013, 2014).
The plant PPSB, which is plastid-localized, was first proposed by Handford and Davies (1958). The PPSB defines a branch point for the 3-phosphoglycerate (3-PGA) produced in plastidial glycolysis, and comprises three sequential reactions catalyzed by 3-PGA dehydrogenase (PGDH; EC 1.1.195), 3-phosphoSer aminotransferase (PSAT; EC 2.6.1.52), and 3-phosphoSer phosphatase (PSP; EC 3.1.3.3; Fig. 1). Biochemical evidence to support the activity of PPSB enzymes in plants emerged in the 1960s to 1970s (Slaughter and Davies, 1968; Larsson and Albertsson, 1979). In the late 1990s, several genes coding for PGDH, PSAT, and PSP isoforms were cloned, and the encoded proteins were demonstrated to possess catalytic activity in vitro (Ho et al., 1998, 1999a, 1999b; Ho and Saito, 2001). Three genes encode for PGDHs (At4g34200, PGDH1; At1g17745, PGDH2; At3g19480, PGDH3), two for PSAT (At4g35630, PSAT1; At2g17630; PSAT2), and one for PSP (At1g18640, PSP1; Ros et al., 2014). The recent functional characterization of PPSB genes demonstrated that this pathway is essential for male gametophyte and embryo development, and for root growth (Benstein et al., 2013; Cascales-Miñana et al., 2013; Toujani et al., 2013; Wulfert and Krueger, 2018). Knock-out mutants of PGDH1 and PSP1 are embryo-lethal, and microspore development in conditional mutants arrests at the polarized stage (Benstein et al., 2013; Cascales-Miñana et al., 2013; Toujani et al., 2013). It was concluded that PPSB is the only Ser source for specific cell types poorly connected to the vasculature (Cascales-Miñana et al., 2013; Toujani et al., 2013; Ros et al., 2014). Yet PPSB genes are not only expressed in very specific heterotrophic cell types, and some such as, for example, PGDH3 are even preferentially, if not exclusively, expressed in green organs (Benstein et al., 2013; Cascales-Miñana et al., 2013). The fact that Ser levels in PPSB-deficient mutants showed dramatic changes in neither the aerial parts (AP) nor in roots (Benstein et al., 2013; Cascales-Miñana et al., 2013; Toujani et al., 2013; Wulfert and Krueger, 2018) led us to postulate that the PPSB could also be important for metabolic reactions other than Ser biosynthesis.
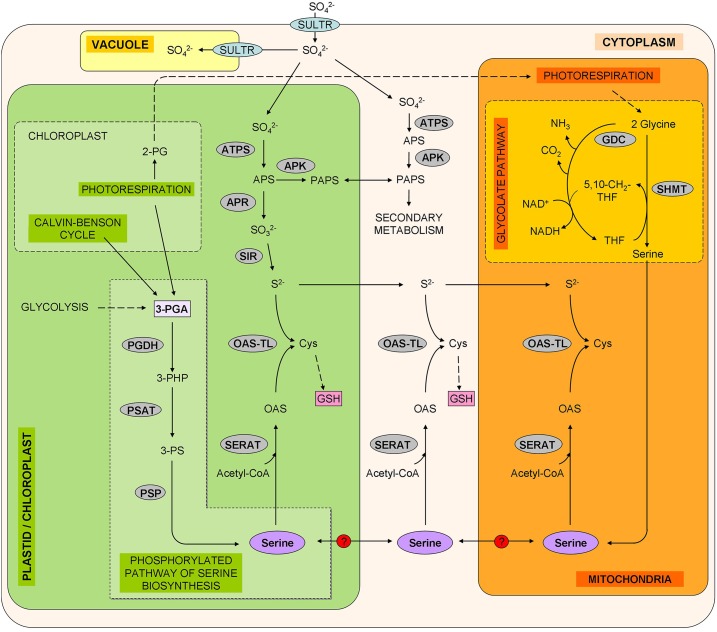
Figure 1.
Schematic representation of the possible contribution of the glycolate and Phosphorylated Pathways of Ser Biosynthesis (PPSB) to the sulfur metabolism. Enzymes and transporters are highlighted in gray and blue, respectively. Glycolate pathway: GDC, Gly decarboxylase; SHMT, Ser hydroxymethyl transferase. Sulfate assimilation: APK, APS kinase; ATPS, ATP sulfurylase; SIR, sulfite reductase; SULTR, sulfate transporter. Metabolites: 2-PG, 2-phosphoglycolate; 3-PHP, 3-phosphohydroxypyruvate; 3-PS, 3-phospho-Ser; 5,10-CH2-THF, 5.10-methylene-tetrahydrofolate; APS, adenosine 5′-phosphosulfate; Cys, Cysteine; PAPS, 3′-phosphoadenosine 5′-phosphosulfate; THF, tetrahydrofolate.
In the biosynthesis of Cys, the flux of carbon/nitrogen metabolism converges, via Ser, with the flux of reduced sulfur. Thus, regulation and coordination of this crossroad is of vital importance for plant metabolism and growth (Dong et al., 2017). It has recently been reported that sulfide availability is regulated by the rapamycin pathway, whereas the carbon/nitrogen precursors required for sulfide incorporation into Cys are sensed by the General Control Nondepressible 2 kinase (Dong et al., 2017). Sulfate is reduced to sulfide by the three-step serial reactions catalyzed by ATP sulfurylase, adenosine 5′phosphosulfate reductase (APR), and sulfite reductase (Fig. 1). Then, sulfide is incorporated into O-acetylSer (OAS) by OAS(thiol)lyase (OAS-TL) to form Cys. Ser is the carbon source of Cys biosynthesis since OAS is formed from Ser and acetyl-CoA in a reaction catalyzed by Ser Acetyltransferase (SERAT). Some enzymes that participate in sulfur assimilation, such as APRs, are exclusively located in the chloroplast (Fig. 1), whereas others, like SERAT and OAS-TL, occur in all compartments capable of protein synthesis (i.e. cytosol, plastid, and mitochondria; Heeg et al., 2008; Watanabe et al., 2008; Krueger et al., 2009). It has been postulated that the different subcellular compartments involved in sulfur assimilation are interconnected to allow the flux of sulfide, OAS, Cys, or their derivatives between them, and compensate for the loss of any of these metabolites in each compartment (Heeg et al., 2008; Watanabe et al., 2008; Krueger et al., 2009). In this sense, genes from SERAT and OAS-TL are in part functionally redundant in vivo (Heeg et al., 2008; Watanabe et al., 2008; Krueger et al., 2009). Analysis of Cys synthesis in Arabidopsis (Arabidopsis thaliana) mutants indicated that under regular growth conditions, chloroplasts provide the substrate sulfide, the mitochondria synthesize most of the substrate OAS, and the cytosol synthesizes most of the Cys, although all compartments are, in principle, able to produce this amino acid (Haas et al., 2008; Heeg et al., 2008; Watanabe et al., 2008; Krueger et al., 2009).
To date, sulfur homeostasis at the tissue and organ level has not been studied as deeply as at the cellular level. Specially, the interactions between autotrophic and heterotrophic tissues are poorly understood. The AP is presumed to be the main site of sulfur assimilation in plants (Wilson, 1962; Takahashi, 2010). In this sense, Ser synthesized by photorespiration in the mitochondria of photosynthetic cells has been linked to sulfur assimilation via the synthesis of the thiols Cys and glutathione (GSH; Noctor et al., 1998, 1999). The substantial amounts of GSH and S-methyl-Met in the phloem lead to the conclusion that reduced sulfur is translocated from mesophyll to heterotrophic sink organs to meet their demands (Bourgis et al., 1999; Lappartient et al., 1999; Durenkamp and De Kok, 2004). However, all the enzymes required for sulfate reduction and thiol biosynthesis are also present in heterotrophic organs such as roots (Pate, 1965; Yonekura-Sakakibara et al., 1998; Heiss et al., 1999; Lappartient et al., 1999; Saito, 2000), where the glycolate pathway is absent. Besides, the activity of key enzymes in Cys biosynthesis, such as SERAT or OAS-TL, is of the same order of magnitude in roots and leaves (Krueger et al., 2009). Yet, the role of local thiol biosynthesis in heterotrophic tissue and specifically that of PPSB as source of Ser for these processes remains largely unknown. In this work, we examined plants with reduced PPSB activity with respect to plant sulfur metabolism. We show that PPSB interacts with the sulfate assimilation pathway and affects sulfur metabolism in both AP and roots. We conclude that PPSB plays an important role in sulfide assimilation in heterotrophic tissues and that lack of PPSB activity perturbs sulfur homeostasis between photosynthetic and nonphotosynthetic cells.
RESULTS
Phenotypic Characterization of PPSB-Deficient Lines
To study the function of PPSB we targeted the two known essential genes, PGDH1 and PSP1, which code for the first and last enzyme of the pathway, respectively (Fig. 1). For this purpose, we used deficient lines of both genes. The PSP1 conditional mutant (c-psp1) is a psp1 knock-out mutant (psp1.1psp1.1) in which a complementary DNA (cDNA) of PSP1 is expressed under the control of a heat shock induced promoter (Cascales-Miñana et al., 2013). This mutant has low background levels of PSP1 expression under noninductive conditions (Cascales-Miñana et al., 2013), which makes it viable and thus suitable for studying the PSP1 function during plant vegetative development. As PGDH1 deficient lines (ts-pgdh1), two previously described PGDH1-silenced lines (ts-pgdh1.1 and ts-pgdh1.2) were used (Benstein et al., 2013; Krueger et al., 2017). Either c-psp1 or ts-pgdh1 or both genetic backgrounds were selected depending on the experiment being performed.
When grown on one-half-strength Murashige and Skoog medium (1/2 MS) plates, c-psp1 displayed a significant reduction in AP fresh weight (around 40% to 50%) compared with wild-type plants (Fig. 2A; Supplemental Fig. S1). However, the severest growth phenotype of c-psp1 was the reduction of primary root growth rate (Fig. 2, A and B). Both AP fresh weight and primary root growth rate were recovered by supplementing the culture medium with physiological concentrations of Ser (Fig. 2, A and B; Supplemental Fig. S1). When the c-psp1 lines were grown in 1/5 MS, the same drastic root phenotype was found (Fig. 2, A and B). Notwithstanding, the root growth rate in this medium was significantly higher than in 1/2 MS. Given the extreme short root phenotype of c-psp1 lines (Fig. 2C), in order to increase the root biomass to enable experimental manipulation, the 1/5 MS medium was selected to conduct further studies with this mutant.
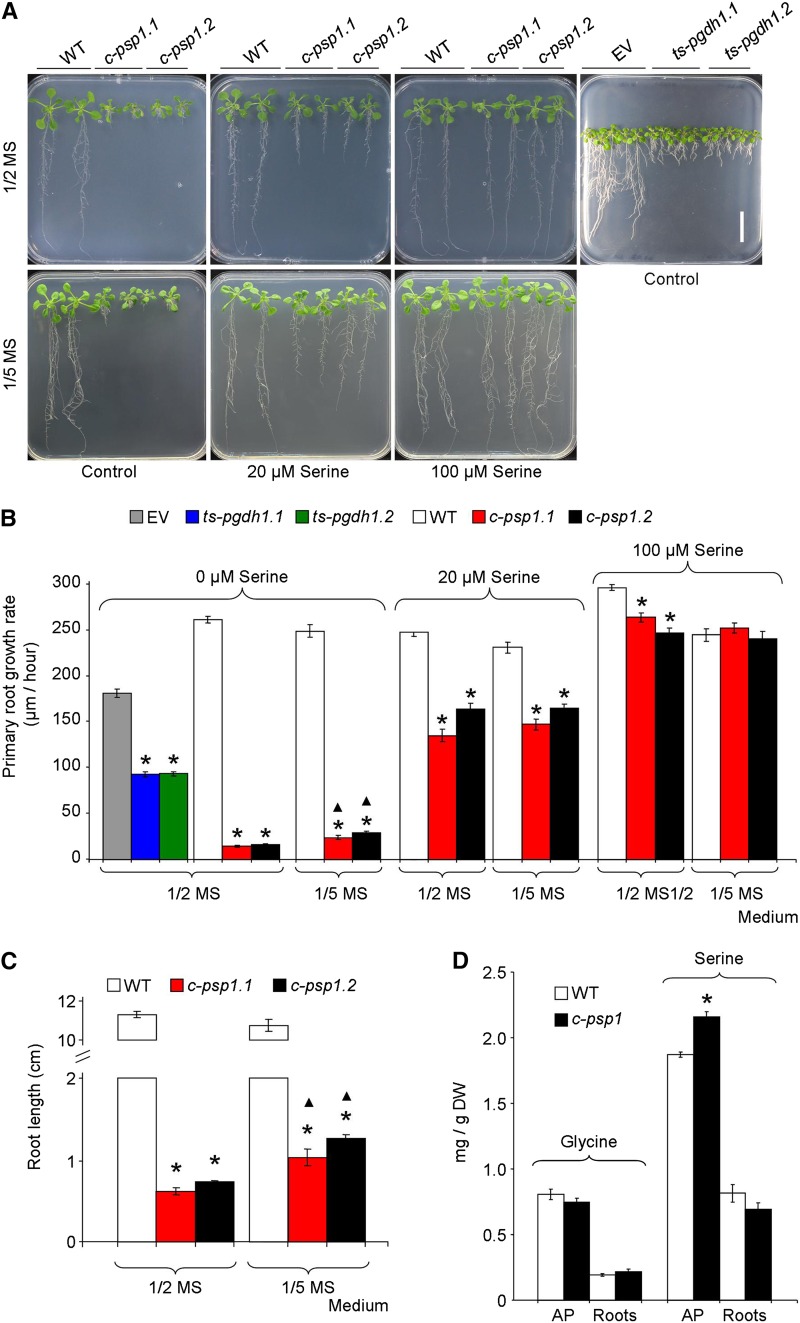
Figure 2.
Phenotype of PPSB-deficient lines in different growth media. A, Picture of c-psp1 (c-psp1.1 and c-psp1.2), ts-pgdh1 (ts-pgdh1.1 and ts-pgdh1.2), and control representative individuals grown in 1/2 MS or 1/5 MS plates supplemented with 0, 20 and 100 µM Ser. B, Primary root growth rate from seedlings grown on 1/2 MS or 1/5 MS plates supplemented with l-Ser as indicated in (A). C, Root length of 18-d-old c-psp1 grown on 1/2 MS or 1/5 MS plates. D, Gly and Ser contents in c-psp1 AP and roots. As control lines, wild-type plants (WT) or wild type transformed with the EV were used. Values represent the means ± se. In (B) and (C), n > 30 plants; in (D), n > 3 pools of 30 plants. In (D), data presented are the mean from pools of two different transgenic events. *Significantly different to control lines; ▴Significant differences between growth media (P < 0.05, Student’s t test). Scale bar = 2 cm. DW, dry weight.
Despite being less drastic than the c-psp1 lines, the ts-pgdh1 lines also displayed impaired primary root growth in Ser-free 1/2 MS medium (Fig. 2, A and B) and recovered their root growth in a medium supplemented with Ser (Benstein et al., 2013). The impaired root growth phenotype was not observed in the mutants of the other two genes of the PGDH family, PGDH2 and PGDH3 (Toujani et al., 2013), whose expression is much weaker than PGDH1 (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) and have a different expression pattern (Toujani et al., 2013).
Although Ser supplementation was able to rescue root growth, levels of Ser and its direct derivative Gly did not lower in c-psp1, and those of Ser were even higher in the AP (Fig. 2D) similarly to what occurred with ts-pgdh1 lines (Benstein et al., 2013). These results indicate that reduced PPSB activity might affect the flux through Ser or local Ser levels, but does not lower the global amino acid steady-state levels, suggesting that other metabolites and metabolic processes derived from Ser could be affected by lack of PPSB activity.
Search for the Molecular Targets Affected by PPSB Deficiency
We followed a transcriptomics analysis of c-psp1 lines as a first approach to target important genes affected by PPSB deficiency. Because the PPSB has been assumed to be more important at night, and because some of the PPSB genes are induced by darkness (Ros et al., 2013, 2014; Toujani et al., 2013), we performed this experiment with material collected at the end of the night period. Quantitative reverse transcription-PCR (qRT-PCR) analysis confirmed that more than 90% of the tested genes followed the same expression pattern in the qRT-PCR and RNA-sequencing (RNA-seq) experiments (Supplemental Fig. S2A). The principal component analysis of wild type and c-psp1 root and AP gene expression profiles showed that the widest variation in their profiles was due to organ type (root versus AP, 58% of variation), whereas the second largest component was genotype (psp1 versus wild type, 12% of variation; Supplemental Fig. S2B). Lack of PSP1 activity affected the expression profile of both organs, although the effect was stronger in roots than in AP. The expression analysis of the transcriptomic data resulted in 280 differentially expressed genes in the c-psp1 AP and 2,936 differentially expressed genes in the c-psp1 roots compared with the wild type (q < 0.01; edgeR classic mode with FDR; Supplemental Table S1). Both c-psp1 organs shared 50 transcripts that were more abundant, and 36 that were less abundant as compared with the wild type (Fig. 3A; Supplemental Table S1). Functional enrichment analysis of four conditions (significantly up- and down-regulated in AP and roots; q < 0.01; Fisher’s exact test, 1992, with Benjamini Yekutieli correction to account for dependence) revealed that many of the most significantly enriched categories associated with down-regulated genes in the AP and roots were related to processes requiring assimilated sulfur (S-glycoside, glucosinolate, Cys, and sulfur biosynthetic processes; or sulfur compound metabolic processes; Fig. 3B; Supplemental Table S1). These data indicated that sulfide-derived metabolism is likely affected, pointing to a sulfur limitation in psp1 under these conditions.
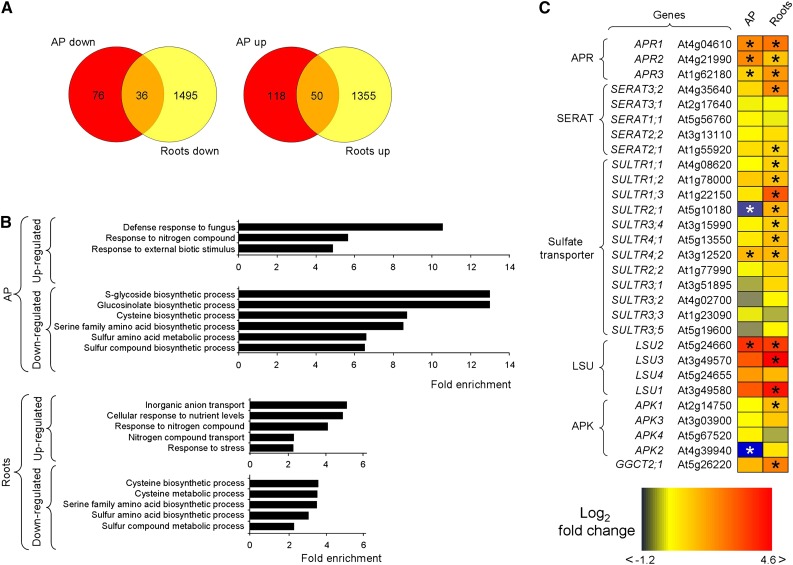
Figure 3.
Transcriptomics profile of c-psp1 at the end of the night period. A, Venn diagrams. B, Most significantly enriched categories associated with up- and down-regulated genes in the AP (AP down and AP up) and roots (Roots down and Roots up) of c-psp1. C, Most relevant changes in the expression of c-psp1 genes related to sulfur transport and metabolism. Three biological replicates (pools of about 100 plants per replicate) were used for the analysis. Transgenic plants proceed from pools of two different transgenic events. *Significantly different to wild type (q < 0.01). The q value was calculated with edgeR in classic mode (Robinson et al., 2010) followed by Benjamini Hochberg correction (Benjamini and Hochberg, 1995). AP, aerial parts; APK, APS kinase; GGCT, γ-glutamyl cyclotransferase; LSU, genes belonging to the response to Low SUlfate family.
A detailed analysis of transcriptomics data obtained by RNA-seq showed that many of the transcripts of genes involved in sulfate assimilation and transport were more abundant in both the c-psp1 roots and AP than in the wild type (Fig. 3C; Supplemental Table S1). These include six genes considered to be read-outs of sulfate deficiency (At4g04610, At1g62180, At4g21990, At5g24660, At5g10180, and At3g12520; Kopriva, 2006). At4g04610, At1g62180, and At4g21990 code for the three Arabidopsis isoforms of APR (APR1, APR2, and APR3), a key regulatory enzyme in the plastidic sulfate assimilation pathway (Fig. 1). Genes belonging to the Response to Low SUlfate (LSU) and sulfate transporter families (At5g24660, LSU2; At5g10180, SULTR2;1 and At3g12520, SULTR4;2) were also deregulated in both organs (Fig. 3C). Overall, the transcriptional response of sulfur-related genes was stronger in roots than in AP. Of the 12 sulfate transporters described in Arabidopsis, seven were up-regulated in roots. This includes the three high affinity sulfate transporters belonging to group 1 (SULTR1;1, SULTR1;2, SULTR1;3), the low affinity sulfate transporter SULTR2;1, the vacuolar efflux sulfate transporters SULTR4;1 and SULTR4;2, and group 3 transporter SULTR3;4 (Bohrer and Takahashi, 2016). Three out of the four LSU genes found in Arabidopsis are induced by sulfur deficiency (Sirko et al., 2015). All three LSU genes (LSU1, LSU2, LSU3) were highly induced in c-psp1 roots, whereas LSU2 was also induced in the AP. Finally, another read-out of sulfate deficiency, a gene coding for a γ-glutamyl cyclotransferase (At5g26220, GGCT2;1), was also highly induced in roots.
Reduced PPSB Activity Induces Sulfate Assimilation Genes in the Light
The up-regulation of genes induced by sulfate starvation indicated an altered sulfur metabolism in c-psp1 at night (Fig. 3, B and C). However, sulfate assimilation is performed fundamentally in the light, and this assimilation is minimal by the end of the dark period (Huseby et al., 2013). Thus, we focused on the interaction of the PPSB and sulfur metabolism in the light. We initially checked the expression of genes involved in sulfate assimilation in both c-psp1 and ts-pgdh1. All three APR genes along with sulfate transporters SULTR2;1 and SULTR4;2 were induced in the c-psp1 roots as compared with controls. APR genes and SULTR4,2 were also induced in the c-psp1 AP (Fig. 4A). Besides, a cytosolic isoform of SERAT (SERAT3;2, At4g35640) was also confirmed to be significantly induced in not only roots, as found at night, but also in the AP. The most important change in expression of all the assayed genes was the expression of SULTR1;3, which was induced up to 9- and 44-fold in the AP and roots of c-psp1, respectively. SULTR1;3 is a high-affinity sulfate transporter described as being required for the uptake of sulfate and maintenance of sulfur metabolism in the sieve element-companion cell complex (Yoshimoto et al., 2003). The presence of Ser in the growing medium attenuated or completely abolished differences in sulfur-related gene expression between c-psp1 and wild type. SULTR1;3 and SULTR2;1 were still significantly induced in c-psp1 roots compared with wild type, but to a much lesser extent than in the medium without Ser (e.g. SULTR1;3 relative expression was reduced by more than 20 times in comparison with a medium without Ser).
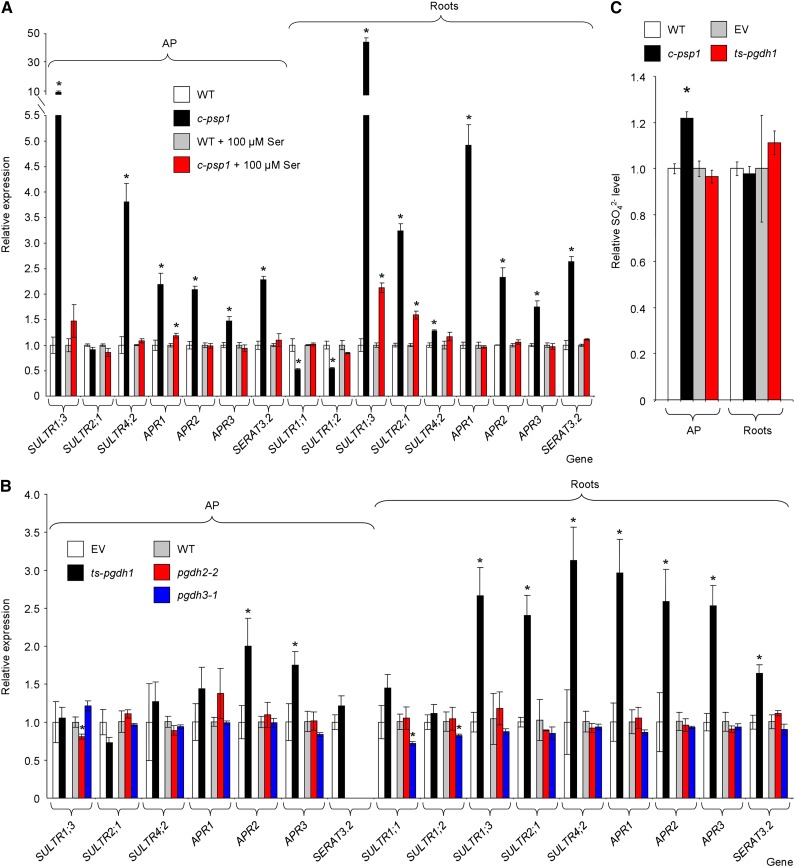
Figure 4.
Expression of sulfur metabolism-related genes in PPSB-deficient lines during the daytime. A, qRT-PCR expression analysis in c-psp1 in the presence or absence of added l-Ser. B, qRT-PCR expression analysis in PGDH family deficient lines (ts-pgdh1, pgdh2-2, and pgdh3-1). C, Sulfate content in AP and roots of c-psp1 and ts-pgdh1. Data for c-psp1, pgdh2-2, and pgdh3-1 are relative values normalized to the mean calculated for wild-type plants (WT). Data for ts-pgdh1 lines were normalized to the values of the wild type transformed with the EV. Samples were collected in the middle of the light period. Values represent the means ± se; n > 3 pools of 30 plants. Both c-psp1 and ts-pgdh1 material proceeded from pools of two different transgenic events. *Significantly different to control lines (P < 0.05, Student’s t test).
The ts-pgdh1 lines also appear to be more sensitive to sulfate limitation than the wild type expressing the empty plasmid (empty vector [EV]), given that anthocyanin accumulation was more severe in these lines (Supplemental Fig. S3). In order to support the notion that sulfate assimilation and transport genes are altered in PPSB-deficient lines, we also investigated the impact of diminished PGDH1 activity on the expression of these genes (Fig. 4B). The expression pattern of sulfur-related genes in ts-pgdh1 was similar to that of c-psp1, especially in roots (Fig. 4B). Thus, the expression of sulfate transporters SULTR1;3, SULTR2;1, SULTR4;2, and that of SERAT3;2 were significantly higher in roots of ts-pgdh1 plants than in the EV (Fig. 4B), as observed in c-psp1. Similarly, the APR genes were also significantly induced in AP (APR2, APR3) and roots (APR1, APR2, APR3) of ts-pgdh1 lines, which further supports the idea that sulfur metabolism is altered in PPSB-deficient lines. The lesser induction pattern of sulfur-related genes in the ts-pgdh1 compared with c-psp1 suggested that other PGDH family genes could partially mask the ts-pgdh1 response. However, mutants from other PGDH family genes did not show induction of sulfur-related genes (Fig. 4B). This finding indicates that PGDH1 is the main gene of the family affecting sulfate assimilation and that ts-pgdh1 lines are a good model to study PPSB function in sulfate metabolism.
The induction in c-psp1 and ts-pgdh1 of key enzymes in sulfate assimilation and transport could indicate a sulfate deficiency. The sulfate levels in the c-psp1 roots were similar to those in wild type, whereas those in the AP were even higher (Fig. 4C). No reduction in the sulfate content in ts-pgdh1 lines was found either, discarding the possibility of a sulfate deficiency in lines with PPSB reduced activity.
Overall, these results indicated that both PGDH1- and PSP1-deficient lines share similar alterations in the sulfur gene expression, which reinforces the idea that PPSB perturbs sulfur metabolism in Arabidopsis in the light. For this reason, further experiments were conducted during the light period.
Reduced PPSB Activity Increases OAS Levels in Both Roots and the AP and GSH Content in Roots
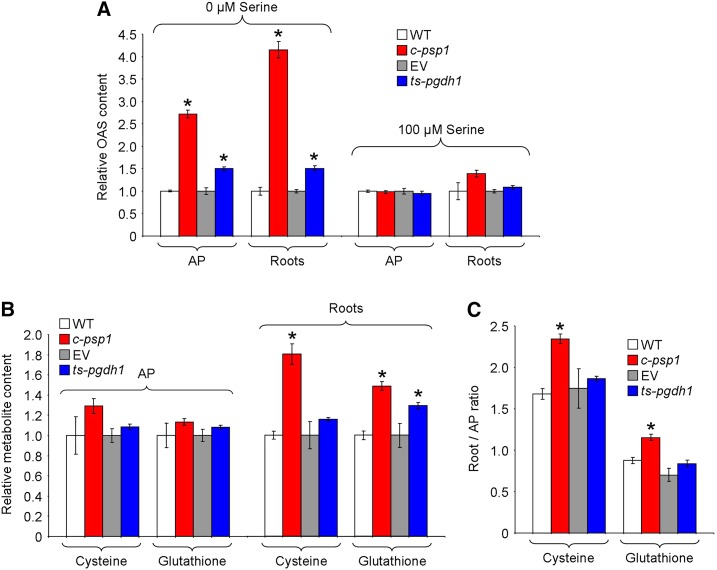
Cys, the first product of sulfide assimilation, is synthesized by the condensation of sulfide with OAS, the activated form of Ser. The induction of sulfate-related genes in PPSB-deficient lines could be associated with an OAS deficiency. However, the OAS level increased in both AP and roots of c-psp1 and ts-pgdh1 compared with controls (up to 2.7- and 4.2-fold the wild-type level, in the AP and roots of c-psp1; Fig. 5A). Interestingly, in the presence of Ser in the growing medium the OAS levels reverted to normal values, as occurred with the expression of genes involved in sulfate assimilation (Fig. 5A).
Figure 5.
Altering expression of PPSB genes affects sulfur metabolism in the light. A, OAS relative contents in c-psp1 and ts-pgdh1 AP and roots as compared with control wild-type plants (WT) or wild type transformed with the EV. B, Relative Cysteine (Cys) and glutathione steady-state contents. C, Cys and glutathione distribution between roots and AP. Samples were collected in the middle of the light period. Values represent the means ± se; n > 5 pools of 30 plants. Transgenic material proceeded from pools of two different transgenic events. *Significantly different to control line (P < 0.05, Student’s t test).
Cys is incorporated into GSH, an important component of the cellular redox buffer system (Meyer et al., 2007). Measurements of the Cys and GSH steady-state levels showed that c-psp1 and ts-pgdh1 had similar levels of these metabolites in the AP compared with control lines (Fig. 5B). However, they contained higher levels of Cys (c-psp1) and GSH (c-psp1 and ts-pgdh1) in roots (Fig. 5B). Accordingly, the Cys and GSH distribution between both organs was altered in PPSB-deficient lines as compared with controls (Fig. 5C).
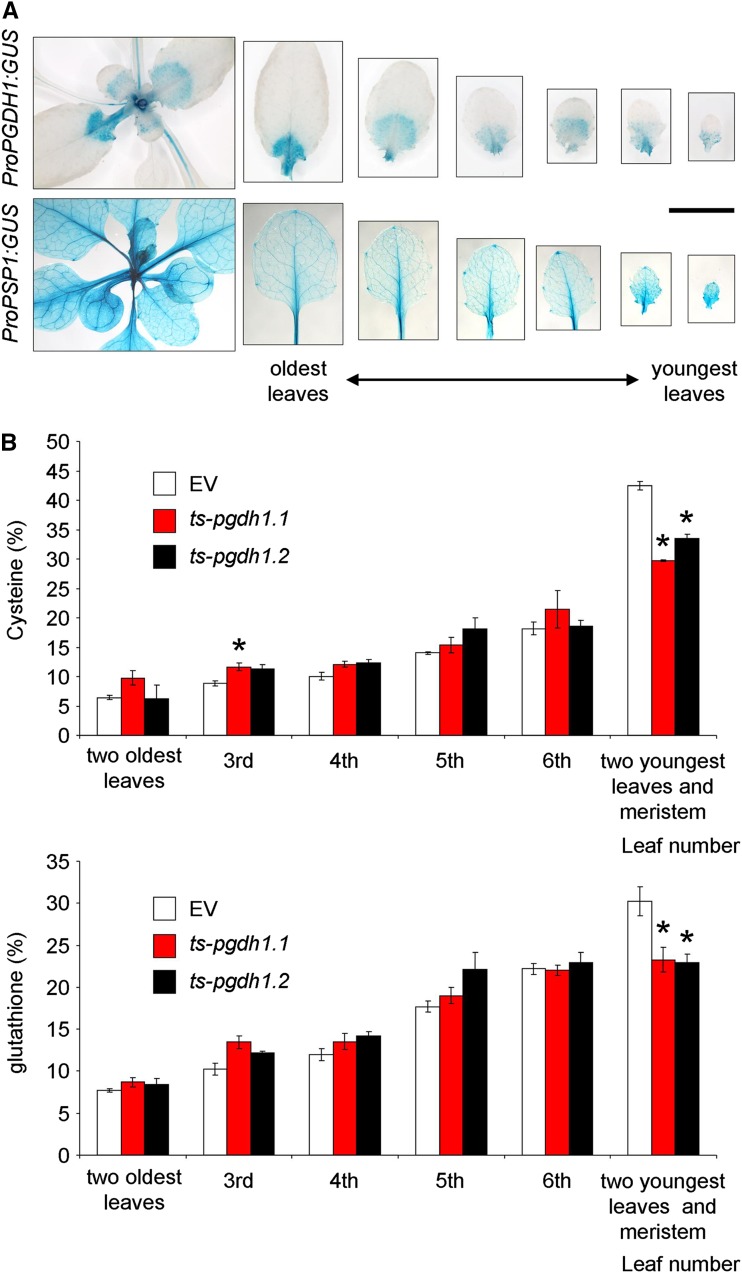
In order to investigate whether thiol distribution in the AP was also affected, we determined Cys and GSH levels in the leaves of both control EV and ts-pgdh1 lines at different developmental stages. Both PGDH1 and PSP1 are highly expressed in heterotrophic tissues, not only in roots (Benstein et al., 2013; Cascales-Miñana et al., 2013), but also in meristematic cells and the leaf vasculature (Fig. 6A; http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). The thiol level in leaves at different developmental stages showed that the distribution of Cys and GSH in the youngest sink leaves and the apical meristem of the AP is higher than in older leaves. The thiol distribution in these youngest leaves and meristems of the ts-pgdh1 lines were significantly lower compared with that of the EV (Fig. 6B). Overall, these data indicate that the PPSB deficiency alter thiol homeostasis in Arabidopsis.
Figure 6.
PGDH1 and PSP1 expression, and thiol distribution in leaves in different developmental stages. A, GUS activity in young rosette leaves expressing either the ProPGDH1:GUS or ProPSP1:GUS. B, Thiol distribution in leaves of different developmental stages. Data presented are means ± se (n = 5). *Significantly different to control EV (P < 0.05, Student’s t test). Scale bar = 1 cm.
Reduced PGDH1 Activity Accelerates Sulfide Assimilation and Alters GSH Redox Potential
To additionally support the role of PPSB in sulfide assimilation, we tested the effect of PPSB deficiency on Cys and subsequently GSH biosynthesis. The incorporation of 35S into Cys and GSH was monitored in AP and roots of plants fed with 35SO42--traced nutrient solution (Fig. 7). Because we demonstrated that both c-psp1 and ts-pgdh1 lines are good models to study the effect of PPSB in sulfur metabolism, we selected ts-pgdh1 to perform these experiments. Our choice was based on using those lines with more root biomass, which were more properly manipulated for transplanting and radioactivity studies. The plant material was harvested after 4 h incubation and the concentration of newly taken up SO42− and synthesized Cys and GSH were measured. The flux experiments revealed a trend to a higher incorporation of 35SO42- in the AP of ts-pgdh1.1, which was significant in ts-pgdh1.2 (Fig. 7A). This result would agree with a higher translocation of sulfate from roots to AP in PPSB-deficient lines. The incorporation of 35S in Cys and GSH in ts-pgdh1 lines was also increased, especially in roots. The amount of newly synthesized Cys was 1.4 (ts-pgdh1.1)- and 1.9 (ts-pgdh1.2)-fold higher in AP and 2.2 (ts-pgdh1.1)- and 2.1 (ts-pgdh1.2)-fold higher in roots of the silenced lines compared with the controlEV (Fig. 7B). Similarly, the amount of newly synthesized GSH was 1.4 (ts-pgdh1.1 and ts-pgdh1.2)-fold higher in AP and 2.0 (ts-pgdh1.1)- and 1.9 (ts-pgdh1.2)-fold higher in roots of ts-pgdh1.1 lines than in the EV (Fig. 7C).
Figure 7.
Thiol fluxes are altered in ts-pgdh1 lines. SO42− (A), Cysteine (Cys; B), and glutathione incorporation (C) in AP and roots of two ts-pgdh1 lines (ts-pgdh1.1 and ts-pgdh1.2) as compared with control wild type transformed with the EV. The total contents were determined by flux analysis using 35S as a tracer. Plants were incubated for 4 h in medium supplemented with 35SO42−, and the distribution of 35S was analyzed. Data presented are means ± se (n = 5). *Significantly different to control EV (P < 0.05, Student’s t test).
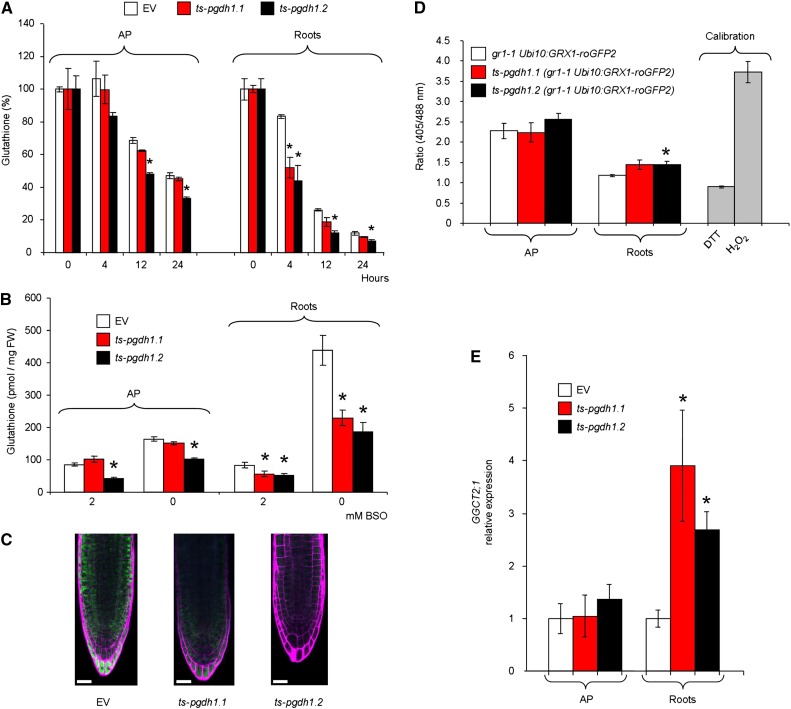
Previous studies have shown that GSH synthesis is similar to its turnover, assuming that the metabolism of GSH is generally high in plants (Noctor et al., 2012). The increased GSH synthesis in ts-pgdh1 lines prompted us to measure GSH turnover. For that purpose GSH biosynthesis was inhibited by treating plants with buthionine sulfoximine (BSO), a potent inhibitor of the Glu-Cys ligase, the first committed enzyme in GSH biosynthesis (Drew and Miners, 1984). The decrease in GSH in the plants was monitored 4, 12, and 24 h after transfer to BSO-containing medium (Fig. 8A). The GSH content in the roots of both silenced lines exhibited a significantly lower GSH level (52%, ts-pgdh1.1 and 44%, ts-pgdh1.2) after 4 h of BSO treatment compared with that in the control plants (83%).
Figure 8.
Glutathione turnover is affected in ts-pgdh1 lines. A, Time course analysis of glutathione content in AP and roots of two ts-pgdh1 lines (ts-pgdh1.1 and ts-pgdh1.2) grown in the presence of 2 mm BSO. B, Glutathione content of plants grown in medium containing 2 mm BSO and retransferred for 24 h to BSO-free or BSO-containing medium. C, In vivo staining with monochlorobimane of glutathione in roots of plants grown in BSO (2 mM) and retransferred for 24 h to BSO-free medium. Scale bar = 20 µM. D, Ratiometric fluorescence measurements of GRX1-roGFP2 redox state in a glutathione reductase 1 background mutant (gr1.1 Ubi10:GRX1-roGFP2) in which PGDH1 was silenced (ts-pgdh1.1 Ubi10:GRX1-roGFP2, ts-pgdh1.2 gr1.1 Ubi10:GRX1-roGFP2). E, Expression of GGCT2;1 in AP and roots of ts-pgdh1 lines. Data presented are means ± se; n = 5. *Significantly different to control EV (P < 0.05, Student’s t test).
To test whether the GSH pool could be completely restored following chemically induced depletion, plants were first transferred for 4 d to medium containing 2 mm BSO and then retransferred for 24 h to BSO-free or BSO-containing medium. AP and roots were then harvested for GSH content determination. After 4 d on medium supplemented with 2 mm BSO, plant growth was arrested and the GSH content was reduced to about 85 pmol mg−1 fresh weight (FW) in AP and roots of control EV (Fig. 8B). Under these conditions, the GSH content was significantly lower in roots of both ts-pgdh1 lines as compared with EV (Fig. 8B). Within 24 h after transfer of the plants to BSO-free medium, 164 pmol mg−1 FW−1 GSH was measured in AP and 438 pmol mg−1 FW−1 in roots of control plants. The root GSH content of both silenced lines was lower than that of EV, as well as that of the AP in ts-pgdh1.2. These results were confirmed by the in situ staining of GSH with monochlorobimane (MCB). Root tips of control plants exhibited substantially stronger fluorescence signal than those of ts-pgdh1 lines after transfer for 24 h from BSO-containing (2 mM) to BSO-free medium (Fig. 8C).
GSH is a well-characterized component of the cellular antioxidant system (Halliwell and Foyer, 1978; Noctor et al., 2012). The perturbed GSH metabolism in ts-pgdh1 plants prompted us to test whether reduced PGDH1 activity influences the cellular GSH redox potential (EGSH). Changes in EGSH can be visualized in vivo with redox-sensitive GFP (roGFP; Meyer et al., 2007). Mutants deficient in cytosolic GSH reductase 1 (gr1) contain increased amounts of GSH disulfide and hence display a less reducing cytosolic redox potential (EGSH) than wild type (Marty et al., 2009). Because roGFP2 is more sensitive in the gr1 background than in wild type, we transformed gr1-1 UBI10:GRX1-roGFP2 plants that express the glutaredoxine 1 (GRX1) fused to roGFP (GRX1-roGFP2 biosensor) in the cytosol with the ts-pgdh1 construct (Benstein et al., 2013) and analyzed the roGFP2 fluorescence following excitation at 405 and 488 nm (Fig. 8D), as described by Gutscher et al. (2008). Treatment of plants with 10 mm dithiothreitol (DTT) results in full reduction of GRX1-roGFP2, whereas treatment with 25 mm H2O2 results in full oxidation of GRX1-roGFP2 in gr1-1 Ubi10:GRX1-roGFP2 lines. Ratiometric analysis of roGFP2 fluorescence in AP of control and ts-pgdh1 plants revealed no significant difference in the fluorescence ratio (Fig. 8D). In contrast, the roGFP2 fluorescence ratio was higher in roots of ts-pgdh1.2 in comparison with the control (Fig. 8D). Thus, roGFP2 is more oxidized in the root tips of plants with a reduced PGDH1 activity.
Altogether, PGDH1 deficiency resulted in an accelerated flux through the sulfur assimilation pathway, but also enhanced GSH turnover and altered the GSH redox potential in roots. It was previously reported that the γ-glutamyl cycle substantially contributes to GSH turnover in plants to recycle its amino acid constituents (Paulose et al., 2013). The Arabidopsis γ-glutamyl cyclotransferase GGCT2;1, which catalyses the second step in the γ-glutamyl cycle, is strongly induced by GSH limitation caused by arsenic treatment or following sulfate limitation (Paulose et al., 2013; Bielecka et al., 2015) and has recently been found to be crucial for GSH turnover (Gerlich et al., 2018; Joshi et al., 2019). The expression of GGCT2;1 was greater in the roots of both c-psp1 and ts-pgdh1 plants (Figs. 3C and 8E) supporting that the GSH turnover is altered in PPSB-deficient lines.
DISCUSSION
Sulfate Metabolism as Target of Altered PPSB Activity
In this work we used PPSB-deficient lines targeting the first and last enzyme of the pathway, PGDH1 and PSP1. c-psp1 lines displayed a more drastic phenotype than ts-pgdh1 lines, probably because PSP1 is a unique gene, whereas PGDH is coded by two more genes. However, both deficient lines displayed similar alterations in sulfur-related processes. This fact reinforces the idea that PPSB activity affects sulfur metabolism in Arabidopsis.
Transcriptional Analysis Suggests that Sulfur Metabolism Is Perturbed in PPSB-Deficient Plants
Ser biosynthesis is particularly important in plants because it provides the carbon skeleton OAS for the first sulfur-containing organic molecule Cys, and thus, connects nitrogen with sulfur assimilation (Kopriva and Rennenberg, 2004; Wirtz and Hell, 2006). Photorespiration is known to contribute to Cys and GSH biosynthesis by providing the carbon precursors Ser and Gly in autotrophic tissue (Noctor et al., 1998, 1999). However, the role of the PPSB in Cys biosynthesis has barely been studied. A reduction in glucosinolate content, a product of sulfur secondary metabolism, has already been demonstrated in PGDH1 silenced lines (Benstein et al., 2013). It has also been shown that PGDH1 activity can be regulated by sulfur-containing amino acids such as Met and homo-Cys (Okamura and Hirai, 2017; Akashi et al., 2018), which suggests an interconnection between sulfur metabolism and the PPSB.
In this work, untargeted transcriptome analysis of PPSB-deficient plants at the end of night period revealed a strong induction of genes involved in sulfate transport and assimilation, whereas genes related to processes requiring assimilated sulfide were downregulated. Expression analysis of sulfate-related genes indicated that APRs and sulfate transporters were also up-regulated in PPSB-deficient lines during the day, a period where the peak of sulfate assimilation takes place (Huseby et al., 2013), suggesting that PPSB could also be involved in sulfur homeostasis in plants during the light period.
It is known that both APRs and sulfate transporters play predominant roles in controlling the sulfate assimilation flux directed for Cys and GSH biosynthesis and that their coding genes are induced in response to sulfate starvation (Bohrer and Takahashi, 2016). Thus, the PPSB-deficient lines showed a typical response of plants to sulfate limitation. However, the sulfate content of PPSB-deficient plants was not lower, and was even higher in the c-psp1 AP than in the wild type. This result indicated that, at least in c-psp1, sulfate is redistributed between the plant (shoot/root relocation), a result which would agree with the strong induction of sulfate transporters involved in translocation in both PPSB-deficient lines. Both c-psp1 and ts-pgdh1 displayed induction of SULTR2;1 and SULTR4;2 and a very strong induction of SULTR1;3 in roots. SULTR2;1 and SULTR4;2 are involved in sulfate distribution between plant organs and tissues (Takahashi, 2010). SULTR2;1 is expressed mainly in pericycle and xylem parenchyma cells in roots, and also in xylem and phloem parenchyma cells in shoots (Takahashi et al., 1997, 2000). It has been suggested that SULTR2;1 controls loading of sulfate to xylem in roots and to phloem in shoots and that it can facilitate distribution of sulfate to leaf tissues under sulfur-limited conditions (Takahashi et al., 2000; Takahashi, 2010). The vacuolar efflux transporter SULTR4;2 is predominantly expressed in shoot and root vasculature (Kataoka et al., 2004). In sultr4;2 mutant, 35S-sulfate incorporated into roots accumulated in root tissues and its distribution to shoots was significantly restricted (Kataoka et al., 2004), which suggests that SULTR4;2 is required to release the vacuolar sulfate pool in roots when sulfur demands increase in the aerial organs (Kataoka et al., 2004). SULTR1;3 is a high-affinity sulfate transporter localized in the phloem of the AP and roots. Deletion of SULTR1;3 restricted transfer of 35S from cotyledons to shoot meristems and roots in Arabidopsis, which lead to the conclusion that it mediates redistribution of sulfate from source to sink organs in Arabidopsis (Yoshimoto et al., 2003). According to the microarray databases (http://bar.utoronto.ca/eplant/) the maximum level of expression of SULTR1;3 is found in the procambium of the root elongation zone, an area poorly connected to the vasculature, which could well have a local deficiency of reduced sulfur. In summary, induction in c-psp1 and ts-pgdh1 of key genes involved in sulfate translocation suggests that distribution of sulfate between organs rather than uptake from roots is affected in PPSB-deficient lines. This would be additionally supported by (1) the lack of induction of the uptake sulfate transporters SULTR1;1 and SULTR1;2 in roots of both c-psp1 ts-pgdh1 under light conditions; and (2) the higher incorporation of 35SO42- in the AP of ts-pgdh1.2 line, but not in roots.
Despite being similar, the altered expression pattern of sulfur-related genes in ts-pgdh1 was not so strong as in c-psp1, at least in the AP. This could be due to the activity of other PGDH isoforms, which could partly compensate for the PGDH1 deficiency, especially in the AP where PGDH3 is specifically expressed. However, PGDH2 and PGDH3 have a much weaker expression than PGDH1 and mutants of these genes do not present a growth phenotype. Besides, the transcriptional response of PGDH2 and PGDH3 mutants clearly indicated that the sulfur-related genes are mainly, if not only, responding to PGDH1 deficiency.
PPSB Is Important to Balance Sulfate Assimilation between Heterotrophic and Autotrophic Tissues
The induction of APR genes in PPSB-deficient lines, especially in roots but also in the AP, could suggest that PPSB deficiency limits sulfur primary metabolism. However, the steady-state levels of the main thiols Cys and GSH did not lower in these lines, but increased in roots. The accumulation of thiols in PPSB-deficient roots could be explained by their reduced growth. However, the analysis of Cys and GSH synthesis by 35S tracer experiments revealed a significantly higher synthesis rate of both thiols in the AP and roots of PPSB-deficient lines. Besides, the most consistent change observed in both organs was the drastic increase in the Ser derivative OAS, which was more acute in roots than in the AP. Thus, the Ser synthesized by the PPSB does not seem to restrict the overall OAS synthesis. It is known that provision of OAS by SERAT limits Cys synthesis in plants (Watanabe et al., 2008; Krueger et al., 2009). In fact OAS, rather than Cys itself, is sensed by plants to distinguish between carbon/nitrogen limitations and sulfur limitations (Dong et al., 2017). OAS levels increase in plants under sulfate-limiting conditions, because the decreased availability of reduced sulfur for Cys biosynthesis leads to the accumulation of its carbon precursor (Hirai et al., 2003; Nikiforova et al., 2003; Maruyama-Nakashita et al., 2005; Bielecka et al., 2015). Reduced PPSB activity does not produce a deficit in sulfate, yet OAS was higher in PPSB-deficient lines than in the controls. Thus the OAS that increased in PPSB-deficient lines is not related to sulfate deficiency. The increased thiol biosynthetic rate observed in PPSB-deficient lines also differed from what is observed in plants impaired in OAS or Cys biosynthesis, where the flux through sulfur assimilation and Cys biosynthesis pathways is significantly diminished (Haas et al., 2008; Heeg et al., 2008; Watanabe et al., 2018). This difference could be explained by the presence of more than one Ser biosynthesis pathway in plants, and also by their different localization in autotrophic or heterotrophic tissues. It can be assumed that the high OAS content in PPSB-deficient plants results from Ser synthesized via the photorespiratory pathway, which is thought to be the most important Ser source in plants, at least in photosynthetic organs. A recent study found that about 38% of the carbon from the photorespiratory pathway is exported as Ser, and ∼30% of it is used in other pathways, rather than being recycled into the pathway by Ser glyoxylate amino transferase (Busch et al., 2018). Therefore, it is quite likely that lack of Ser for OAS biosynthesis produced by the PPSB in some heterotrophic tissues is compensated by an enhanced removal of Ser from the photorespiratory pathway in autotrophic tissues. Ser synthesized through photorespiration in photosynthetic cells could be supplied to nonphotosynthetic organs by phloem-mediated transport (Riens et al., 1991; Hunt et al., 2010). Thus, transported Ser would be converted into OAS by the SERAT enzymes present in heterotrophic cells (Watanabe et al., 2008). The induction of the SERAT isoforms in roots of PPSB-deficient lines would support this idea and would explain why OAS also accumulates in roots of PPSB-deficient plants. Besides, the general high level of OAS in PPSB-deficient lines might be caused by the elevated flux through the sulfur assimilation pathway, which is supported by our 35S tracer experiments.
The sulfate assimilation impairment observed in PPSB-deficient lines could also be attributed to a Ser deficiency in the plastid. However, the transport of OAS into and out of the mitochondria and plastids have been demonstrated in Arabidopsis using mutant combinations of SERAT isoforms located in different compartments (Heeg et al., 2008; Watanabe et al., 2008; Krueger et al., 2009; Hell and Wirtz, 2011). Therefore, if OAS move freely between these compartments, at least in Arabidopsis, specific Ser requirements for OAS biosynthesis in a specific subcellular compartment are very unlikely. The fact that externally supplied Ser can complement most of sulfur-related alterations in PPSB-deficient lines would also reinforce this idea, and rules out a specific plastidial (compartment) deficiency in Ser.
The AP is assumed to be the main sulfate assimilation site. However, all the enzymes needed for sulfate reduction are present in heterotrophic tissues, and the ability to reduce sulfate has been demonstrated for roots (Pate, 1965; Yonekura-Sakakibara et al., 1998; Heiss et al., 1999; Lappartient et al., 1999; Saito, 2000). Besides, a recent study in the C4 species Flaveria has proposed that sulfate reduction is controlled by roots and connected to the PPSB (Gerlich et al., 2018). Accordingly, we hypothesized that in Arabidopsis, Ser deficiency in those cell/tissue types that strongly depend on PPSB activity might reduce local Cys and GSH pools in the PPSB-deficient lines. This would lead to an enhanced synthesis of both Ser and thiols in cells with no restrictions in Ser supply by photorespiration, which would subsequently be transported as Ser and GSH out of these cells into the deficient tissues. In these heterotrophic tissues, GSH turnover would increase to supply substrates for sulfur-related metabolic processes. This hypothesis would be supported by the strong induction of GGCT2;1 in roots, whose encoding enzyme has already been shown to be a crucial factor in the GSH recycling in plants (Gerlich et al., 2018; Joshi et al., 2019). The following observations in the PPSB-deficient plants would also support our hypothesis: (1) the high Ser content in the AP; (2) the high OAS content not only in the AP but also in roots; (3) the induction of genes involved in sulfate translocation between tissues; (4) the increase in the steady-state content of thiols in roots; (5) the changes observed in GSH and Cys distribution between AP and roots, and between different leaves; and (6) the high GSH turnover.
Although plants possess a wide range of amino acid transporters (Tegeder and Masclaux-Daubresse, 2018), Ser translocation from autotrophic to heterotrophic tissues does not guarantee the supply of this amino acid to each plant cell. This idea would be supported by the existence of different Ser biosynthetic pathways in plants, hence the possibility of a local Ser deficiency in PPSB-deficient mutants should not be ruled out. The local Ser deficiency could include some cells in roots, which could depend on PPSB activity for Ser supply and, thus, for local sulfur assimilation. The fact that OAS levels in PPSB-deficient roots recovered the wild-type control levels when plants were grown in the presence of Ser would support this idea. The local Ser or OAS deficiency could be partly responsible for the induction of the sulfur starvation response observed in PPSB-deficient lines which, in turn, would be responsible for the overall increase in OAS and for the high thiol biosynthesis and turnover rates. This hypothesis is strongly supported by the fact that the cell-type specific expression pattern of the sulfate assimilation pathway genes correlates better with the PPSB pathway genes rather than with those of the photorespiratory Ser biosynthetic pathway (Aubry et al., 2014). Overall, our results suggest that the up-regulation of genes involved in sulfate translocation and assimilation in PPSB-deficient lines is the consequence of an impaired metabolism of sulfur-containing organic molecules rather than of reduced sulfate availability in these plants.
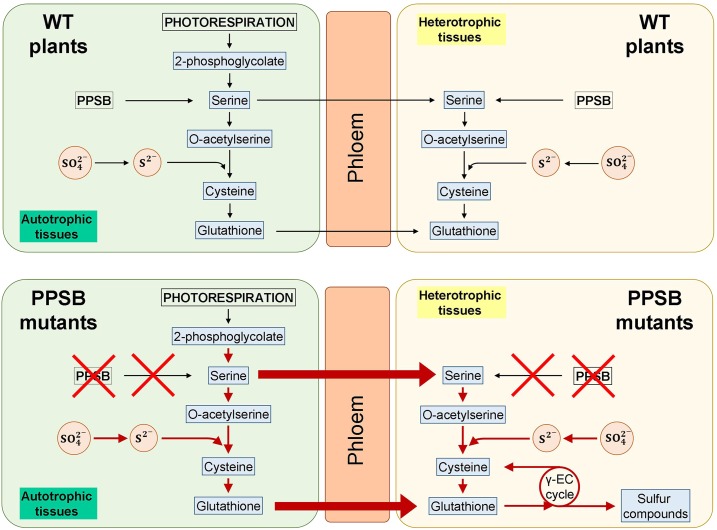
According to the discussed results, we postulate a model for sulfur assimilation in wild type and PPSB-deficient mutants (Fig. 9). In control (wild-type) plants, the photosynthetic cells in the AP represent the main site for Ser biosynthesis and sulfate assimilation, but Ser produced by the PPSB in heterotrophic tissue (or in photosynthetic tissues at night) also contributes to local GSH biosynthesis. In PPSB-deficient plants, local GSH deficiency in heterotrophic tissues stimulates sulfur assimilation in photosynthetic cells, as well as the transport of both Ser and GSH to heterotrophic tissues. In these heterotrophic cells, GSH is degraded by the γ-glutamyl cycle to fulfill the requirements of organic bound reduced sulfur. Our results suggest a clear interaction between the photorespiratory glycolate pathway and the PPSB to maintain plant Ser homeostasis. Besides, interactions of PPSB with other Ser metabolic branches should not be discounted. Future research at the transcriptional, translational, and posttranslational level on mutants of different pathways would be required to unravel these interactions.
Figure 9.
Model for the metabolic response to deficiency in the PPSB. In control (wild-type [WT]) plants, the photosynthetic cells in the AP represent the main site of Ser biosynthesis and sulfate assimilation, although Ser produced by the PPSB in heterotrophic tissue (or in photosynthetic tissues at night) would also contribute to local GSH biosynthesis. In the PPSB-deficient plants, local GSH deficiency in heterotrophic tissues stimulates sulfur assimilation in photosynthetic cells, as well as transport of both Ser and GSH to heterotrophic tissues. In these heterotrophic cells GSH is degraded by the γ-glutamyl (γ-EC) cycle to fulfill the requirements of organic bound reduced sulfur.
Concluding Remarks
All the alterations of sulfur metabolism in the PPSB-deficient plants indicate that Ser supply through this pathway participates in sulfur assimilation in plants. Our results indicate that lack of PPSB affects vegetative growth in roots and the AP, and results in a transcriptional response that mimics sulfate starvation. Although the plant responds by displaying slower growth and, thus, its requirements for reduced sulfur (S−2) are lower, major imbalances in sulfur metabolism persist. We found that reducing Ser biosynthesis through the PPSB results in an overall elevated OAS content and a higher flux through the sulfur assimilation pathway, which could mask a deficiency in specific cell types and tissues. These results clearly point out that studying how cell compartmentation and transport between organs regulate metabolism are outstanding questions that will contribute to our understanding of the primary metabolism in plants. Our results further show that the PPSB is a key pathway in the metabolic networks that connect carbon, nitrogen, and sulfur metabolism and as such could represent a promising target for crop improvement.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Original Arabidopsis (Arabidopsis thaliana) seed stocks (ecotype Columbia-0) were supplied by the European Arabidopsis Stock Center (Scholl et al., 2000). The conditional PSP1 mutant lines c-psp1.1 and c-psp1.2 were previously obtained by transforming a psp1 mutant (SALK_062391) with a construct carrying the PSP-GFP cDNA under the control of a heat shock inducible promoter of gene HSP18.2 (At5g59720; Cascales-Miñana et al., 2013). PGDH2 and PGDH3 mutants (pgdh2-2, SALK_149747; pgdh3-1, SM_337584;) were described by Toujani et al. (2013). The ts-pgdh1.1 and ts-pgdh1.2 lines were described previously (Benstein et al., 2013; Krueger et al., 2017). Wild-type plants expressing the EV were used as control for these silenced lines (Benstein et al., 2013). The expression pattern of PGDH1 and PSP1 was studied using the lines expressing the promoter regions of genes PGDH1 and PSP1 fused to the GUS coding sequence (ProPGDH1:GUS or ProPSP1:GUS) previously described (Benstein et al., 2013; Cascales-Miñana et al., 2013). To measure the EGSH, the gr1-1 UBI10:GRX1-roGFP2 plant line background (Marty et al., 2009) was used to silence PGDH1 as previously described (Benstein et al., 2013).
Unless otherwise stated, seeds were sterilized and sown on 0.8% (w/v) agar plates containing either one-fifth-strength MS (1/5 MS) medium with Gamborg vitamins (c-psp1 lines) or half-strength MS basic salt medium (1/2 MS), buffered with 0.9 g/l MES (adjusted to pH 5.7 with Tris). After 2–4 d of stratification at 4°C, plates were vertically placed in a growth chamber at 20–22°C under a 16 h day/8 h night photoperiod, 100 μmol m−2 s−1.
To determine GSH recovery, plants were germinated and grown on 1/2 MS before transfer to plates supplemented with 2 mm BSO (l-buthionine-[S,R] sulfoximine; Sigma Aldrich) for 4 d. Plant material was harvested from plants retransferred to medium with or without BSO for 24 h. For GSH turnover analysis, plants were grown as described for the analysis of GSH recovery. Plants were transferred to plates supplemented with 2 mm BSO, and plant material was harvested 4, 12, and 24 h following transfer.
The experimental material proceeded from seedlings grown on plates for 2 to 3 weeks. In all experiments presented, individuals from two different transgenic events were used. In some experiments values presented are the averages of both lines.
RNA-seq data and qRT-PCR
For the gene expression analysis by RNA-seq, AP and roots of 21-d-old wild-type and c-psp1 plants vertically grown on 1/5 MS plates were used. Three independent biological replicates of each sample were harvested at the end of the night period for the analysis. Total RNA was extracted using NucleoSpin RNA II kit (Macherey-Nagel). With use of 3–15 µg of RNA as starting material, a mRNA enrichment was performed with the MicroPoly(A) Purist kit (AMBION). To prepare the RNA-Seq library, the SOLID Total RNA-seq kit (Life Technologies) was used. After the library was obtained, an equimolar mixture of it was used to perform an emulsion PCR using the automatic system of EZ Beads (Life Technologies). Then, the bead enrichment was performed, followed by its deposition in the sequencing wells. The sequencing step was performed using a SOLID 5500XL equipment of 75 nucleotides using the Exact Call Chemistry. Single end reads in fastq format were mapped against the TAIR10 Arabidopsis transcriptome (www.arabidopsis.org) using kallisto (v0.43.0 with default parameters except l = 200 and s = 30; Bray et al., 2016). Data were combined in R (v3.3.2) and analyzed. Principal component analysis was carried out with prcomp (scale = T). Differential expression was assessed with edgeR in classic mode (Robinson et al., 2010) followed by Benjamini Hochberg correction (Benjamini and Hochberg, 1995). Enrichment analysis were conducted using topGO (R package version 2.22.0) with Fisher’s Exact Test (Fisher, 1922) followed by Benjamini Yekutieli correction (Benjamini and Yekutieli, 2001). The level of significance was fixed at 0.01 (1%) after correction.
qRT-PCR was performed as previously described (Benstein et al., 2013; Cascales-Miñana et al., 2013). Each reaction was performed in triplicate with 1 µL of the first-strand cDNA in a total volume of 25 µL. Data are the mean of at least three biological replicates samples. The specificity of the PCR amplification was confirmed with a heat dissociation curve (from 60°C to 95°C). Efficiency of the PCR reaction was calculated and different internal standards were selected (Czechowski et al., 2005) depending on the efficiency of the primers. Primers used are listed in Supplemental Table S2.
Metabolite Determination and Sulfate Analysis
The AP and roots of 2- to 3-week-old plants grown on plates were used to determine metabolite contents. Gly and Ser contents were determined by HPLC after derivatization with diethyl ethoxymethylenemalonate (Alaiz et al., 1992). OAS was extracted from freeze-dried plant material (20–50 mg fresh weight) in three sequential steps. The homogenized material was suspended in 200 μL 80% (v/v) ethanol containing 0.5 mm HEPES (pH 6.8) and extracted for 20 min at 25°C. After centrifugation (10,000 g for 10 min at 4°C), the remaining pellet was resuspended in 200 μL 50% (v/v) ethanol containing 1.25 mm HEPES pH 6.8 and re-extracted for 20 min at 25°C. After centrifugation, the pellet was again extracted for 20 min at 25°C with 100 μL 80% (v/v) ethanol and 0.5 mm HEPES (pH 6.8), and the supernatants from all three extractions were combined and diluted 1:10 with water. OAS was automatically precolumn derivatized within the autosampler by mixing 25 µL extract with 25 µL 1 × OPA solution (Grace Davison Discovery Sciences) and 5.5 µL 1 m borate buffer (pH 10.7). After 90-s incubation, 20 µL of derivatization mixture was injected into the HPLC system and separated onto a HyperClone 3 µm ODS(C18)120 150 × 4.6 mm HPLC column (Phenomenex) by a discontinuous gradient 0-2 min, 0% B; 2-21 min, 13% B; 21-28.25 min, 15% B; 28.25-37.3 min, 50% B; 37.3-48.2 min, 60% B; 48.3-54.3 min, 100% B; 54.3-56.3 min, 100% B; 56.3-63.3 min, 0% B; 63.3-65 min 0% B consisting of solvent A (8.8 mm Na3PO4 [pH 6.8] and 0.6% [v/v] tetrahydrofuran) and solvent B (18.7 mm Na3PO4 [pH 6.8], 32.7% [v/v] MeOH, and 20.6% [v/v] acetonitrile). OAS derivatives were detected at 480 nm after excitation by 380 nm using a RF 2000 fluorescence detector (Dionex) in the “mid” sensitivity mode. Alternatively, OAS content was determined in derivatized methanol extracts by gas chromatography-mass spectrometry using the protocol defined in Lisec et al. (2006). Here, OAS was identified in comparison with database entries of authentic standards (Kopka et al., 2005). Chromatograms and mass spectra were evaluated with the Chroma TOF 1.0 (LECO) and TagFinder 4.0 software (Luedemann et al., 2008).
Thiol content (Cys and GSH) in plant tissues was analyzed according to Krueger et al. (2009). Thiols were extracted from freeze-dried homogenized plant material with 1 mL 0.1 m HCl for 40 min at 25°C. After centrifugation for 5 min at 14,000 g and 4°C, thiols in the supernatant were reduced by mixing 120 μL of the supernatant with 200 μL 2-(cyclohexylamino)ethanesulfonic acid (0.25 M, pH 9.4) and 70 μL DTT (10 mM). The mixture was incubated at 25°C for 40 min. Thiols were derivatized by adding 10 μL (25 mm) monobromobimane (Merck). Derivatization was stopped by adding 220 μL methane sulfonic acid (100 mm). Twenty microliters of the derivatization mix were further used for HPLC analysis using the Dionex Ultimate 3000 HPLC System. Derivatized thiols were separated in a Eurosphere 100-3 C18, 150×4 mm column (Knauer), and were detected by fluorescence detection (excitation 380 nm/emission 480 nm).
Sulfate was measured using the sulfate cell test kit (1.14548.0001 Merck) following the manufacturer´s instructions.
GUS Activity Assays
GUS activity was assayed as previously described in Cascales-Miñana et al. (2013).
Isolation and Quantification of [35S]-Labeled Sulfated Compounds
Uptake and incorporation of 35S was investigated in 14-d-old plants grown under long-day condition as previously described in Koprivova et al. (2000). After germination, seedlings were grown on vertical plates containing 1/2 MS medium for 6 d. To avoid local nutrient limitation, plants were transferred to vertical plates containing freshly prepared 1/2 MS medium for an additional 8 d. To analyze 35S uptake and incorporation, plants were transferred to hydroponic culture containing 1/2 MS liquid medium containing 0.2 mm 32SO42-. To avoid alterations in plant metabolism due to the transfer of the plants from solid to liquid medium, plants were first acclimatized for 2 d on liquid medium before experiments were started. After acclimatization, the medium was replaced with fresh 1/2 MS medium supplemented with ∼ 10,000 counts per minute 35SO42- µL−1. Plants were incubated for 4 h before samples were taken. The harvested plants were washed with demineralized H2O, surface-dried with tissue and separated into AP and root samples. Ten individual plants were combined for each replicate (30–100 mg). The samples were immediately frozen in liquid nitrogen and stored overnight at −80°C.
The distribution of 35S was analyzed as described by Koprivova et al. (2000) with minor modifications. Plant tissue was homogenized using a micropistil and extracted with 10 µL 0.1 m HCl mg−1 fresh weight. Extracts were centrifuged at 20,000 g for 5 min, and the supernatant was used for further analysis. Extract (10 µL) was diluted in 2-mL scintillation solution (Rotiszint eco plus; Roth), and radioactivity was measured in a scintillation counter (Beckman) to determine the total 35S uptake. To investigate the incorporation of 35S into thiols, 50 µL extract was mixed with 50 µL of 0.1 m DTT. After 15 min incubation, 5 µL of 0.1 m monobromobimane (MBB) was added for the derivatization and incubated for 15 min in the dark. The derivatization reaction was stopped by adding 11 µL 50% (v/v) acetic acid. For HPLC analysis, the derivatized samples were diluted 1:1 with 0.25 m Tris (pH 8) before injection. The derivatized sample (100 µL) was injected into the HPLC (Dionex Ultimate 3000 HPLC System) equipped with a 250 × 4.6 mm ID; 5 µm particle size; Spheriscorb ODS2 C18 column (Waters). Radioactively labeled compounds were detected in parallel with fluorescent-labeled thiols using a radiation detector (FlowStar LB 513; Berthold Technology) coupled with a fluorescence detector (Dionex). Solvent A (10% [v/v] MeOH; 0.25% [v/v] acetic acid; pH 4.3 [NaOH]) and solvent B (90% [v/v] MeOH; 0.25% [v/v] acetic acid; pH 4.3 [NaOH]) was used to generate the binary gradient.
Live Cell Imaging of GSH and the GSH Redox Status
Plants were grown for 4 d on 1/2 MS agar plates containing 1 mm BSO before being transferred for 24 h to 1/2 MS agar plates without BSO to initiate GSH recovery. For comparison of the absolute GSH levels in intact roots of EV controls and ts-pgdh1 lines, seedlings were stained with 100 µM MCB for 30 min and subsequently with 50 µM propidium iodide (PI) for 5 min. After washout of adherent-free dye in water, seedlings were transferred to a glass slide, covered with a cover slip and placed on the stage of an inverted microscope with an attached confocal scan head (Axio Observer.Z1 with LSM780, Carl Zeiss Microscopy). Images were collected with a 25× lens (Plan-Apochromat 25×/0.8 NA water immersion) with 405 nm excitation for MCB and excitation at 543 nm for PI. Fluorescence was measured at 500–550 nm for MCB and 610–700 nm for PI. All measurements were performed with an averaging of four repetitive scans per line for improved signal-to-noise ratios.
For direct visualization of the EGSH, plants harboring the Grx1-roGFP2 construct were transferred to a glass slide for ratiometric CLSM imaging. roGFP2 was excited in multitracking mode and line switching between 488 nm and 405 nm excitation for roGFP2 measurements. Emission was restricted to the 505–530 nm emission band. Images were taken with a 25× lens (Plan-Apochromat 25x/0.8 NA water immersion) with an averaging of four scans per line. To subtract autofluorescence bleeding into the fluorescence channels, autofluorescence excited at 405 nm was collected from the 430–480 nm band and recorded separately.
Ratiometric analysis for roGFP2 was performed using a custom-written MatLab (www.mathworks.de) program (Fricker, 2016). Analysis was carried out on a pixel-by-pixel basis as I405/I488 following spatial averaging in (x,y) using a 3 × 3 kernel. First, background signals and correction for I405 autofluorescence bleeding into the 405 nm channel were subtracted from the roGFP2 fluorescence recorded after excitation at 405 nm. The mean background value was taken from outside the plant root. Calibration of roGFP2 was carried out by incubating seedlings in 10 mm DTT or 20 mm H2O2 to fully reduce or oxidize the sensor, respectively. False-color images were created, using a spectral color scale ranging from blue (reduced) to red (oxidized) with its limits set by the calibration.
Bioinformatics and Statistics
Experimental values represent the mean values and se; n represents the number of independent samples. Significant differences as compared with wild type were analyzed by Student’s t tests algorithms (two-tailed) using Microsoft Excel. Unless otherwise stated, the level of significance was fixed at 5% (0.05).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers of Arabidopsis genes used in this article are as follows: At1g18640 (PSP1), At4g34200 (PGDH1), At1g7745 (PGDH2), At3g19480 (PGDH3). RNA-seq raw data files were submitted to the database Gene Expression Omnibus at National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/geo/info/submission.html) with the accession number GSE125675.
SUPPLEMENTAL DATA
The following supplemental materials are available:
Supplemental Figure S1. Fresh weight of the aerial parts of c-psp1 and wild type seedlings grown on 1/2 MS plates supplemented with 0, 20 and 100 µM Ser.
Supplemental Figure S2. A, Validation of RNA-Seq by qRT-PCR of selected genes in aerial parts and roots of c-psp1. B, Principal component analysis of root and shoot expression profiles in WT and c-psp1.
Supplemental Figure S3. Anthocyanin content measured in ts-pgdh1 and wild type controls transformed with the empty vector.
Supplemental Table S1. Transcriptomic analysis of c-psp1 aerial parts and roots.
Supplemental Table S2. List of primers used in this work.
Acknowledgments
The authors acknowledge Rainer Schwacke (Research Centre, Jülich) and John Chandler (University of Cologne) for critical comments and discussion. The authors thank Klara Kernig for excellent assistance and the Servei Central de Suport a la Investigació Experimental of the Universitat de València for technical assistance.
Footnotes
This work was supported by the Cluster of Excellence on Plant Sciences (CEPLAS) (to S. Kopriva and S. Krueger); Deutsche Forschungsgemeinschaft (DFG) (grant Kr4245/1-1; Kr4245/2-1 to S. Krueger); the DFG within the Research Training Group GRK 2064 and grants ME1567/9-1/2 within the Priority Program SPP1710; European Union and Ministry of Economy and Competitiveness (FEDER/ BFU2015-64204R); Generalitat Valenciana (Regional Government of Valencia) PROMETEO II/2014/052 (to R.R.); and the University of Valencia (V segles fellowship to M.F.-T.).
References
- Akashi H, Okamura E, Nishihama R, Kohchi T, Hirai MY (2018) Identification and biochemical characterization of the serine biosynthetic enzyme 3-phosphoglycerate dehydrogenase in Marchantia polymorpha. Front Plant Sci 9: 956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaiz M, Navarro JL, Girón J, Vioque E (1992) Amino acid analysis by high-performance liquid chromatography after derivatization with diethyl ethoxymethylenemalonate. J Chromatogr A 591: 181–186 [DOI] [PubMed] [Google Scholar]
- Aubry S, Smith-Unna RD, Boursnell CM, Kopriva S, Hibberd JM (2014) Transcript residency on ribosomes reveals a key role for the Arabidopsis thaliana bundle sheath in sulfur and glucosinolate metabolism. Plant J 78: 659–673 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300 [Google Scholar]
- Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in mutiple testing under dependency. Ann Stat 29: 1165–1188 [Google Scholar]
- Benstein RM, Ludewig K, Wulfert S, Wittek S, Gigolashvili T, Frerigmann H, Gierth M, Flügge UI, Krueger S (2013) Arabidopsis phosphoglycerate dehydrogenase1 of the phosphoserine pathway is essential for development and required for ammonium assimilation and tryptophan biosynthesis. Plant Cell 25: 5011–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielecka M, Watanabe M, Morcuende R, Scheible WR, Hawkesford MJ, Hesse H, Hoefgen R (2015) Transcriptome and metabolome analysis of plant sulfate starvation and resupply provides novel information on transcriptional regulation of metabolism associated with sulfur, nitrogen and phosphorus nutritional responses in Arabidopsis. Front Plant Sci 5: 805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohrer AS, Takahashi H (2016) Compartmentalization and regulation of sulfate assimilation pathways in plants. Int Rev Cell Mol Biol 326: 1–31 [DOI] [PubMed] [Google Scholar]
- Bourgis F, Roje S, Nuccio ML, Fisher DB, Tarczynski MC, Li C, Herschbach C, Rennenberg H, Pimenta MJ, Shen TL, et al. (1999) S-methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell 11: 1485–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527 [DOI] [PubMed] [Google Scholar]
- Busch FA, Sage RF, Farquhar GD (2018) Plants increase CO2 uptake by assimilating nitrogen via the photorespiratory pathway. Nat Plants 4: 46–54 [DOI] [PubMed] [Google Scholar]
- Cascales-Miñana B, Muñoz-Bertomeu J, Flores-Tornero M, Anoman AD, Pertusa J, Alaiz M, Osorio S, Fernie AR, Segura J, Ros R (2013) The phosphorylated pathway of serine biosynthesis is essential both for male gametophyte and embryo development and for root growth in Arabidopsis. Plant Cell 25: 2084–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Silbermann M, Speiser A, Forieri I, Linster E, Poschet G, Allboje Samami A, Wanatabe M, Sticht C, Teleman AA, et al. (2017) Sulfur availability regulates plant growth via glucose-TOR signaling. Nat Commun 8: 1174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Drew R, Miners JO (1984) The effects of buthionine sulphoximine (BSO) on glutathione depletion and xenobiotic biotransformation. Biochem Pharmacol 33: 2989–2994 [DOI] [PubMed] [Google Scholar]
- Durenkamp M, De Kok LJ (2004) Impact of pedospheric and atmospheric sulphur nutrition on sulphur metabolism of Allium cepa L., a species with a potential sink capacity for secondary sulphur compounds. J Exp Bot 55: 1821–1830 [DOI] [PubMed] [Google Scholar]
- Fisher RA. (1922) On the interpretation of χ2 from contingency tables, and the calculation of P. J R Stat Soc 85: 87–94 [Google Scholar]
- Fricker MD. (2016) Quantitative redox imaging software. Antioxid Redox Signal 24: 752–762 [DOI] [PubMed] [Google Scholar]
- Gerlich SC, Walker BJ, Krueger S, Kopriva S (2018) Sulfate metabolism in C4 Flaveria species is controlled by the root and connected to serine biosynthesis. Plant Physiol 178: 565–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutscher M, Pauleau AL, Marty L, Brach T, Wabnitz GH, Samstag Y, Meyer AJ, Dick TP (2008) Real-time imaging of the intracellular glutathione redox potential. Nat Methods 5: 553–559 [DOI] [PubMed] [Google Scholar]
- Haas FH, Heeg C, Queiroz R, Bauer A, Wirtz M, Hell R (2008) Mitochondrial serine acetyltransferase functions as a pacemaker of cysteine synthesis in plant cells. Plant Physiol 148: 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Foyer CH (1978) Properties and physiological function of a glutathione reductase purified from spinach leaves by affinity chromatography. Planta 139: 9–17 [DOI] [PubMed] [Google Scholar]
- Handford J, Davies DD (1958) Formation of phosphoserine from 3-phophoglycerate in higher plants. Nature 182: 532–533 [Google Scholar]
- Heeg C, Kruse C, Jost R, Gutensohn M, Ruppert T, Wirtz M, Hell R (2008) Analysis of the Arabidopsis O-acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. Plant Cell 20: 168–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss S, Schäfer HJ, Haag-Kerwer A, Rausch T (1999) Cloning sulfur assimilation genes of Brassica juncea L.: Cadmium differentially affects the expression of a putative low-affinity sulfate transporter and isoforms of ATP sulfurylase and APS reductase. Plant Mol Biol 39: 847–857 [DOI] [PubMed] [Google Scholar]
- Hell R, Wirtz M (2011) Molecular biology, biochemistry and cellular physiology of cysteine metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K (2003) Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J 33: 651–663 [DOI] [PubMed] [Google Scholar]
- Ho CL, Saito K (2001) Molecular biology of the plastidic phosphorylated serine biosynthetic pathway in Arabidopsis thaliana. Amino Acids 20: 243–259 [DOI] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito M, Yamazaki M, Saito K (1998) Molecular characterization of plastidic phosphoserine aminotransferase in serine biosynthesis from Arabidopsis. Plant J 16: 443–452 [DOI] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito K (1999a) Plastidic pathway of serine biosynthesis. Molecular cloning and expression of 3-phosphoserine phosphatase from Arabidopsis thaliana. J Biol Chem 274: 11007–11012 [DOI] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito M, Saito K (1999b) Regulation of serine biosynthesis in Arabidopsis. Crucial role of plastidic 3-phosphoglycerate dehydrogenase in non-photosynthetic tissues. J Biol Chem 274: 397–402 [DOI] [PubMed] [Google Scholar]
- Hunt E, Gattolin S, Newbury HJ, Bale JS, Tseng HM, Barrett DA, Pritchard J (2010) A mutation in amino acid permease AAP6 reduces the amino acid content of the Arabidopsis sieve elements but leaves aphid herbivores unaffected. J Exp Bot 61: 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huseby S, Koprivova A, Lee BR, Saha S, Mithen R, Wold AB, Bengtsson GB, Kopriva S (2013) Diurnal and light regulation of sulphur assimilation and glucosinolate biosynthesis in Arabidopsis. J Exp Bot 64: 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NC, Meyer AJ, Bangash SAK, Zheng ZL, Leustek T (2019) Arabidopsis gamma-glutamylcyclotransferase affects glutathione content and root system architecture during sulfur starvation. New Phytol 221:1387–1397 [DOI] [PubMed] [Google Scholar]
- Kataoka T, Hayashi N, Yamaya T, Takahashi H (2004) Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol 136: 4198–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kopriva S. (2006) Regulation of sulfate assimilation in Arabidopsis and beyond. Ann Bot 97: 479–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Rennenberg H (2004) Control of sulphate assimilation and glutathione synthesis: Interaction with N and C metabolism. J Exp Bot 55: 1831–1842 [DOI] [PubMed] [Google Scholar]
- Koprivova A, Suter M, den Camp RO, Brunold C, Kopriva S (2000) Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiol 122: 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger S, Niehl A, Lopez Martin MC, Steinhauser D, Donath A, Hildebrandt T, Romero LC, Hoefgen R, Gotor C, Hesse H (2009) Analysis of cytosolic and plastidic serine acetyltransferase mutants and subcellular metabolite distributions suggests interplay of the cellular compartments for cysteine biosynthesis in Arabidopsis. Plant Cell Environ 32: 349–367 [DOI] [PubMed] [Google Scholar]
- Krueger S, Benstein RM, Wulfert S, Anoman AD, Flores-Tornero M, Ros R (2017) Studying the function of the phosphorylated pathway of serine biosynthesis in Arabidopsis thaliana. Methods Mol Biol 1653: 227–242 [DOI] [PubMed] [Google Scholar]
- Lappartient AG, Vidmar JJ, Leustek T, Glass AD, Touraine B (1999) Inter-organ signaling in plants: Regulation of ATP sulfurylase and sulfate transporter genes expression in roots mediated by phloem-translocated compound. Plant J 18: 89–95 [DOI] [PubMed] [Google Scholar]
- Larsson C, Albertsson E (1979) Enzymes related to serine synthesis in spinach chloroplasts. Physiol Plant 45: 7–10 [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. (2011) Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet 43: 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedemann A, Strassburg K, Erban A, Kopka J (2008) TagFinder for the quantitative analysis of gas chromatography--mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 24: 732–737 [DOI] [PubMed] [Google Scholar]
- Marty L, Siala W, Schwarzländer M, Fricker MD, Wirtz M, Sweetlove LJ, Meyer Y, Meyer AJ, Reichheld JP, Hell R (2009) The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc Natl Acad Sci USA 106: 9109–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Inoue E, Yamaya T, Takahashi H (2005) Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J 42: 305–314 [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, Hell R (2007) Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 52: 973–986 [DOI] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R (2003) Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: Interlacing of biosynthetic pathways provides response specificity. Plant J 33: 633–650 [DOI] [PubMed] [Google Scholar]
- Noctor G, Arisi A, Jouanin L, Kunert K, Rennenberg H, Foyer C (1998) Glutathione: Biosynthesis, metabolism and relationship to stress tolerance explored in transformed plants. J Exp Bot 49: 623–647 [Google Scholar]
- Noctor G, Arisi ACM, Jouanin L, Foyer CH (1999) Photorespiratory glycine enhances glutathione accumulation in both the chloroplastic and cytosolic compartments. J Exp Bot 50: 1157–1167 [Google Scholar]
- Noctor G, Mhamdi A, Chaouch S, Han Y, Neukermans J, Marquez-Garcia B, Queval G, Foyer CH (2012) Glutathione in plants: An integrated overview. Plant Cell Environ 35: 454–484 [DOI] [PubMed] [Google Scholar]
- Okamura E, Hirai MY (2017) Novel regulatory mechanism of serine biosynthesis associated with 3-phosphoglycerate dehydrogenase in Arabidopsis thaliana. Sci Rep 7: 3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pate JS. (1965) Roots as organs of assimilation of sulfate. Science 149: 547–548 [DOI] [PubMed] [Google Scholar]
- Paulose B, Chhikara S, Coomey J, Jung HI, Vatamaniuk O, Dhankher OP (2013) A γ-glutamyl cyclotransferase protects Arabidopsis plants from heavy metal toxicity by recycling glutamate to maintain glutathione homeostasis. Plant Cell 25: 4580–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riens B, Lohaus G, Heineke D, Heldt HW (1991) Amino Acid and sucrose content determined in the cytosolic, chloroplastic, and vacuolar compartments and in the Phloem sap of spinach leaves. Plant Physiol 97: 227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros R, Cascales-Miñana B, Segura J, Anoman AD, Toujani W, Flores-Tornero M, Rosa-Téllez S, Muñoz-Bertomeu J (2013) Serine biosynthesis by photorespiratory and non-photorespiratory pathways: An interesting interplay with unknown regulatory networks. Plant Biol (Stuttg) 15: 707–712 [DOI] [PubMed] [Google Scholar]
- Ros R, Muñoz-Bertomeu J, Krueger S (2014) Serine in plants: Biosynthesis, metabolism, and functions. Trends Plant Sci 19: 564–569 [DOI] [PubMed] [Google Scholar]
- Saito K. (2000) Regulation of sulfate transport and synthesis of sulfur-containing amino acids. Curr Opin Plant Biol 3: 188–195 [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirko A, Wawrzyńska A, Rodríguez MC, Sęktas P (2015) The family of LSU-like proteins. Front Plant Sci 5: 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter JC, Davies DD (1968) The isolation and characterization of 3-phosphoglycerate dehydrogenase from peas. Biochem J 109: 743–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell K. (1984) Enzymes of serine metabolism in normal, developing and neoplastic rat tissues. Adv Enzyme Regul 22: 325–400 [DOI] [PubMed] [Google Scholar]
- Tabatabaie L, Klomp LW, Berger R, de Koning TJ (2010) L-serine synthesis in the central nervous system: A review on serine deficiency disorders. Mol Genet Metab 99: 256–262 [DOI] [PubMed] [Google Scholar]
- Takahashi H. (2010) Regulation of sulfate transport and assimilation in plants. Int Rev Cell Mol Biol 281: 129–159 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Yamazaki M, Sasakura N, Watanabe A, Leustek T, Engler JA, Engler G, Van Montagu M, Saito K (1997) Regulation of sulfur assimilation in higher plants: A sulfate transporter induced in sulfate-starved roots plays a central role in Arabidopsis thaliana. Proc Natl Acad Sci USA 94: 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K (2000) The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J 23: 171–182 [DOI] [PubMed] [Google Scholar]
- Tegeder M, Masclaux-Daubresse C (2018) Source and sink mechanisms of nitrogen transport and use. New Phytol 217: 35–53 [DOI] [PubMed] [Google Scholar]
- Timm S, Florian A, Wittmiß M, Jahnke K, Hagemann M, Fernie AR, Bauwe H (2013) Serine acts as a metabolic signal for the transcriptional control of photorespiration-related genes in Arabidopsis. Plant Physiol 162: 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert NE. (1980) Photorespiration. In Davies DD, ed, The biochemistry of plants. Academic Press, New York, pp 488–525 [Google Scholar]
- Tolbert NE. (1985) The oxidative photosynthetic carbon cycle and peroxisomal glycolate metabolism. In Ludden PW and Burris JE, eds, Nitrogen fixation and CO2 metabolism. Elsevier, New York, pp 333–342 [Google Scholar]
- Toujani W, Muñoz-Bertomeu J, Flores-Tornero M, Rosa-Téllez S, Anoman AD, Alseekh S, Fernie AR, Ros R (2013) Functional characterization of the plastidial 3-phosphoglycerate dehydrogenase family in Arabidopsis. Plant Physiol 163: 1164–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Mochida K, Kato T, Tabata S, Yoshimoto N, Noji M, Saito K (2008) Comparative genomics and reverse genetics analysis reveal indispensable functions of the serine acetyltransferase gene family in Arabidopsis. Plant Cell 20: 2484–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Tohge T, Fernie AR, Hoefgen R (2018) The effect of single and multiple SERAT mutants on serine and sulfur metabolism. Front Plant Sci 9: 702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L-G. (1962) Metabolism of sulfate: Sulfate reduction. Annu Rev Plant Physiol 13: 201–224 [Google Scholar]
- Wirtz M, Hell R (2006) Functional analysis of the cysteine synthase protein complex from plants: Structural, biochemical and regulatory properties. J Plant Physiol 163: 273–286 [DOI] [PubMed] [Google Scholar]
- Wulfert S, Krueger S (2018) Phosphoserine Aminotransferase1 is part of the phosphorylated pathways for serine biosynthesis and essential for light and sugar-dependent growth promotion. Front Plant Sci 9: 1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Mancuso A, Tong X, Ward PS, Fan J, Rabinowitz JD, Thompson CB (2012) Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Natl Acad Sci USA 109: 6904–6909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Ashikari T, Tanaka Y, Kusumi Ta, Hase T (1998) Molecular characterization of tobacco sulfite reductase: Enzyme purification, gene cloning, and gene expression analysis. J Biochem 124: 615–621 [DOI] [PubMed] [Google Scholar]
- Yoshimoto N, Inoue E, Saito K, Yamaya T, Takahashi H (2003) Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol 131: 1511–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]