Abstract
Cryptococcus neoformans (Cn) is a soil fungus that causes life-threatening meningitis in immunocompromised patients and is a facultative intracellular pathogen capable of replication inside macrophages. The mechanism by which environmental fungi acquire and maintain virulence for mammalian hosts is unknown. We hypothesized that the survival strategies for Cn after ingestion by macrophages and amoebae were similar. Microscopy, fungal and amoebae killing assays, and phagocytosis assays revealed that Cn is phagocytosed by and replicates in Acanthamoeba castellanii, which leads to death of amoebae. An acapsular strain of Cn did not survive when incubated with amoebae, but melanization protected these cells against killing by amoebae. A phospholipase mutant had a decreased replication rate in amoebae compared with isogenic strains. These observations suggest that cryptococcal characteristics that contribute to mammalian virulence also promote fungal survival in amoebae. Intracellular replication was accompanied by the accumulation of polysaccharide containing vesicles similar to those described in Cn-infected macrophages. The results suggest that the virulence of Cn for mammalian cells is a consequence of adaptations that have evolved for protection against environmental predators such as amoebae and provide an explanation for the broad host range of this pathogenic fungus.
Keywords: Acanthamoeba castellanii‖phagocytosis‖melanin‖capsule
The encapsulated yeast Cryptococcus neoformans (Cn) causes life-threatening meningitis in immunocompromised patients (1). Cn is found predominately in soil contaminated by pigeon excreta, and infections are presumably acquired from the environment by means of the inhalation of infectious particles (2). Cn rarely causes clinically apparent infections in normal hosts, but once established it has the capacity for latency and persistence, inside macrophages (3–5). Cn is a facultative intracellular pathogen in vivo and in vitro (6, 7). The Cn polysaccharide capsule, composed primarily of glucuronoxylomannan (GXM), plays both a defensive and offensive role in pathogenesis by protecting against phagocytosis and promoting intracellular pathogenesis through cytotoxic release of polysaccharide into macrophage vacuoles, respectively (6). Because Cn does not require an animal host for replication, the question arises as to why this soil organism possesses an unusual and sophisticated strategy for intracellular parasitism. Apart from Cn, other free-living fungal pathogens such as Histoplasma capsulatum and Blastomyces dermatitidis manifest intricate virulence strategies in mammalian hosts, although an animal vector is not required for replication or dissemination (8). The mechanisms responsible for the emergence and maintenance of virulence among endemic human pathogenic fungi are not understood.
Several macrophage-tropic intracellular bacterial pathogens are capable of growing inside free-living soil amoebae, such as Acanthamoeba castellanii (Ac) (9). Legionella pneumophila was the first human pathogen demonstrated to multiply and persist in both macrophages and amoebae (10). Amoebae and macrophages share common properties; both phagocytose particles into vacuoles and secrete lysosomal enzymes that digest the particles (11). The intracellular events after L. pneumophilia infection of amoebae and macrophages are similar (11). Hence, we hypothesized that Cn was capable of intracellular growth in amoebae and that the mechanism used for subversion of macrophage function was selected for surviving interactions with environmental phagocytic cells.
Amoebae of the genera Acanthamoeba feed on both bacteria and fungi (12). Ac is a fresh water and soil protozoa (13) that was initially isolated as a cryptococcal culture contaminate (14). Ac has been used to study bacteria–amoeba interactions. Bunting et al. (15) showed that Acanthamoeba phagocytosed and killed 78–99% of three Cn clinical isolates. In this study, we investigated the interaction of Cn with Ac. Cn was demonstrated to replicate in Ac and to escape killing by Ac in a mechanism similar to that used to survive in macrophages. These results are consistent with the view that certain aspects of cryptococcal pathogenesis derive from mechanisms used by the fungus to survive in the presence of environmental amoebae.
Materials and Methods
Organisms and Culture Conditions.
Ac strain 30324 was obtained from the American Type Culture Collection (ATCC) and cultured in peptone-yeast extract-glucose broth, PYG (ATCC medium 354), as described (16, 17) at 28°C.
The Cn strains used are listed in Table 1. Candida albicans SC5314 was a gift from M. Ghannoun (Case Western Reserve, Cleveland) and Saccharomyces cerevisiae LM23–3az was obtained from L. Marsh (Albert Einstein College of Medicine, Bronx, NY). Yeast cultures were maintained and grown in Sabauraud dextrose (SAB) broth as described (6). For melanization studies, the cells were grown in minimal media with or without L-dopa (100 mg/liter) for 7 days (18).
Table 1.
Properties of Cn strains used to study Ac:Cn interactions
| Strain | Serotype | Phenotype | Source or reference |
|---|---|---|---|
| 24067 | D | WT | ATCC |
| 3501 | D | WT | ATCC |
| Cap67 | D | Acapsular mutant of 3501 | ref. 35 |
| H99 | A | WT | J. Perfect, DUMC |
| Plb− | A | H99 phospholipase mutant | ref. 33 |
| Rec1 | A | WT (reconstituted phospholipase mutant) | ref. 33 |
| 2E-Tu | A | Laccase-deficient | ref. 30 |
| 2E-Tuc | A | WT (reconstituted Laccase mutant) | ref. 30 |
WT, wild type; ATCC, American Type Culture Collection; DUMC, Duke University Medical Center.
Phagocytosis Assay.
Ac cells in PYG were added to 96-well tissue culture plates at 105 cells/well and allowed to adhere for 2 h at 28°C before the addition of Cn at a 2:1 effector-to-target ratio. The plates were incubated for 2 h at 28°C. The media were aspirated, and the cells were fixed with ice-cold methanol for 30 min at 4°C and washed three times with PBS, stained with Giemsa diluted 1:10 in PBS for 2 h. The plates were viewed with a microscope at ×200 magnification, and eight wells per experimental condition were used to determine the percentage of phagocytic cells. The phagocytic index is the number of Ac with internalized yeast per 100 Ac cells (19).
Transmission Electron Microscopy (TEM).
Ac monolayers were infected with Cn 24067 or 3501 at an effector-to-target ratio of 1:1 and incubated at 28°C for 1.5 and 24 h, respectively. The monolayers were washed five times with PYG and were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate at room temperature. The samples were prepared as described and viewed with a model 102 electron microscope (Siemens, Iselin, NJ) (20).
Immunogold TEM was performed by using the mAb 2H1 specific for Cn GXM (21) as described (22). Primary Ab staining was done overnight at 4°C with 25 μg/ml mAb 2H1 or murine IgG (Sigma) as a control.
Scanning Electron Microscopy (SEM).
Cn 24067 cells from a 24-h Sabauraud dextrose (SAB) broth culture were incubated with Ac at a 1:10 effector-to-target ratio for 30, 60, or 90 min at room temperature with intermittent rocking. The SEM protocol was adapted from Cleare and Casadevall (23).
Fluorescence Microscopy.
Cn cells were labeled directly with FITC (24). The fungal cells were incubated with 5 μg/ml FITC (Molecular Probes) for 2 h at 37°C and washed with PBS. Amoebae and FITC-labeled Cn cells were incubated at a 1:1 ratio in PYG media for 2 h. The media were aspirated, and the cells were fixed with 2.5% gluteraldehyde for 1 h and washed with PBS. A coverslip was mounted by using a solution of 50% glycerol, 0.1 M n-propyl gallate (Sigma) in PBS. The slides were viewed with a FITC filter-equipped Zeiss axiophote microscope.
For Ab detection of GXM, Ac and Cn cells were incubated in eight-chamber culture slides in PYG media for 2 h. The media were aspirated, and the slides were fixed with ice-cold methanol for 30 min at 4°C. The slides were washed three times with PBS. The slides were incubated with 2H1 (10 μg/ml), a mAb specific to Cn GXM, for 2 h (21). The slides were washed with PBS and incubated with 10 μg/ml FITC-labeled goat anti-mouse IgG1 (Southern Biotechnology Associates) for 1 h at room temperature. The slides were washed, fixed, and viewed as described above.
Budding Index.
Ac and Cn cells were incubated at an effector-to-target ratio of 4:1. After 2 h, the media were replaced with fresh media. At 2, 6, and 24 h, cells were fixed with ice-cold methanol, washed with PBS, and stained with a 1:10 dilution of Giemsa in PBS for 2 h. The budding index is the number of internalized Cn cells with buds (defined as daughter cells with cell walls continuous with that of the parental cell) divided by the total number of amoebae with internal fungal cells (150 amoeba cells were counted per well, and two wells were counted per experimental condition) (25).
Cn Killing Assays.
The outcome of the interaction between Ac and the yeast strains was evaluated by using a modification of a L. pneumophilia assay (17). Ac cells were counted with a hemocytometer and suspended at 105 cells/ml. Ac viability was determined by trypan blue staining, and initial viability was always greater than 98% (data not shown). Fungal cells were added to cultures of Ac in 96-well tissue culture plates to yield a 1:1 effector-to-target ratio and were incubated at 28°C. Quantification of viable yeast cells was determined at 0, 6, 24, and 48 h by measuring colony-forming units (cfu). At each time interval, the amoebae and yeast cells were agitated from the bottom of the selected culture wells, and Ac cells were lysed by forcibly pulling the culture through a 27-gauge needle 5–7 times (16). The shear stresses lysed the amoebae and had no effect on Cn viability as determined by the comparison of initial hemocytometer counts and cfu counts. At each time interval, five tissue culture wells per strain were used to ascertain cfu. For each well, serial dilutions were plated in duplicate onto SAB agar plates, which were incubated at 30°C for 48 h.
Acapsular mutants of Cn can bind exogenous GXM (26). GXM was bound to Cap67 cells as described (27). The Cap67 cells with bound GXM were used in the Ac killing assay as described above.
Amoebae Killing.
Quantification of viable Ac cells was done at 0, 6, 24, and 48 h by using a trypan blue exclusion assay. At each time interval, 96-well plates containing Ac and Cn cells at a 1:1 ratio were incubated with a 1:10 dilution of trypan blue in PBS. The plates were then centrifuged at 220 × g for 5 min. The 96-well plates were viewed at ×200, and percent of dead amoebae was determined by counting the number of amoebae cells unable to exclude dye per total amoebae counted.
Statistical Analysis.
The graphs were compiled in MICROSOFT EXCEL 2000. Statistical analysis by using the Student's t test was performed in MICROSOFT EXCEL 2000.
Results
Ac Ingests Cn.
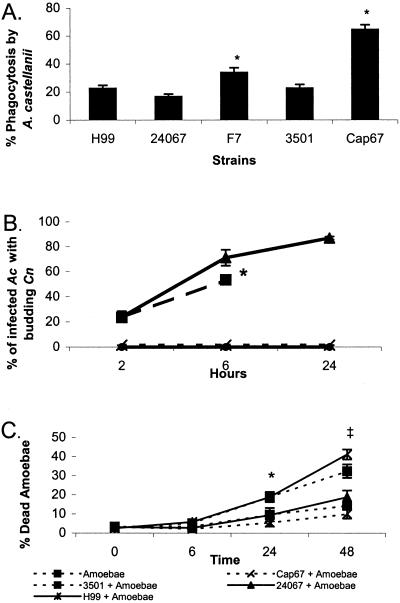
Addition of Cn to monolayers of Ac resulted in phagocytosis of yeast cells by amoebae. The efficacy of phagocytosis was measured by determining the number of amoebae that engulfed at least one fungal cell. The results are depicted in Fig. 1A. For the three encapsulated strains H99, 24067, and 3501, the phagocytic indices ranged from 17% to 23%. F7, the pseudohyphal variant of 24067, and Cap67, the acapsular mutant derived from strain 3501, were phagocytosed more efficiently than the parental strains, with phagocytosis indexes of 34% and 65%, respectively. Therefore, acapsular Cn cells were phagocytosed at a significantly higher rate than other strains tested.
Figure 1.
Properties of the infection of Ac by Cn. (A) Phagocytic index for five Cn strains incubated with Ac in PYG media. Error bars denote 1 SD. For each experiment, 100 Ac cells were counted in eight separate wells, and the number of measurements was eight. Similar results were obtained from an additional independent experiment. * indicates P < 0.001 for both Cap67 and F7 when compared with H99, 24067, or 3501. (B) Percent of infected amoebae with budding Cn cells. Error bars denote 1 SD. * denotes the absence of intact amoebae after this time. (C) Percent of Ac cells that do not exclude trypan blue after incubation with different strains of Cn. Error bars denote 1 SD. Similar results were obtained from an additional independent experiment. * indicates P = 0.001 when 3501 and H99 were compared with amoeba alone at 24 h. ‡ indicates P < 0.001 at 48 h when 3501 and H99 were compared with amoebae alone.
Cn Cells Replicate Inside Ac Cells.
Fig. 1B depicts the results of the budding assay. The number of buds increased over time for strains 3501 and H99. H99 budding is only counted through 6 h because, at 24 h, there were few intact amoebae cells remaining. After 24 h, the amoebae cells incubated with strain 3501 were also lysed. The trend for both strains showed an increase in buds over time. In contrast, strain 24067 and Cap67 showed no significant budding.
Ac Response to Cn.
Trypan blue exclusion by amoebae cells was used to measure amoebae death after incubation with Cn. The results in Fig. 1C show no difference in amoeba viability when incubated with and without Cn strain 24067 or Cap67. However, incubation of Ac with Cn strains H99 and 3501 resulted in death of amoebae reaching 41% and 34%, respectively, at 48 h.
Cn Interacts with Ac.
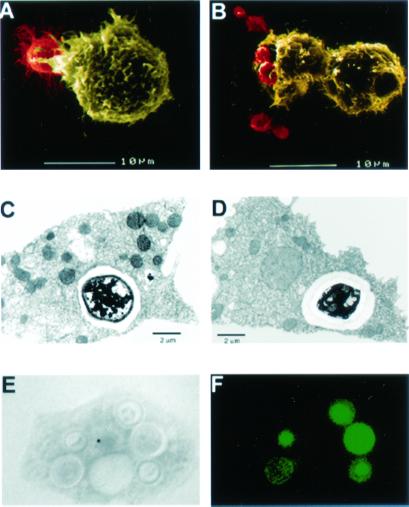
SEM analysis was used to visualize the interaction of Cn with amoebae. Fig. 2 A and B shows Cn cells directly interacting with Ac. TEM was used to demonstrate that amoebae internalized Cn cells. Fig. 2 C and D shows cells of Cn strains 3501 and 24067, respectively, enclosed in membrane-bound vesicles inside Ac. Fig. 2 E and F depicts a light microscopy image and the corresponding IF staining of Cn strain 3501 inside an Ac cell.
Figure 2.
Microscopy of Ac incubated with Cn in PYG media. Each field is representative of amoeba/Cn interactions at a given time. For both SEM figures, Cn cells are depicted in red and Ac cells are depicted in yellow: (A) SEM 30-min postincubation with Cn 24067 and (B) SEM 90-min postincubation with Cn 24067. (C) TEM 24-h postincubation with Cn 3501. (D) TEM 1.5-h postincubation with Cn 24067. (E and F) Corresponding light microscopy (E) and immunofluorescent (F) pictures 2 h postincubation of Ac with FITC-labeled Cn 3501. (Magnifications: ×1,000.)
Growth of the Cn Incubated with Ac.
When incubated in PBS alone, there were generally 1–2 replication cycles of all Cn strains tested (Fig. 3). Of the three virulent strains tested, 3501, 24067, and H99, only 24067 showed little growth difference when incubated in PBS alone compared with incubation with amoebae, with PBS-grown cells having a slightly increased cfu. However, when grown with amoebae, strains H99 and 3501 displayed significant replication over cells incubated in PBS at 48 h (Fig. 3 A and C). For strain F7, a pseudohyphal variant of 24067, cfu decreased during the 48-h time course when grown with Ac (data not shown). Also, to determine the effect of amoebae on nonpathogenic and nonenvironmental strains of fungi, S. cerevisiae and C. albicans were incubated with Ac. C. albicans and S. cerevisiae cells were both all killed when incubated with amoebae by 48 h (data not shown).
Figure 3.
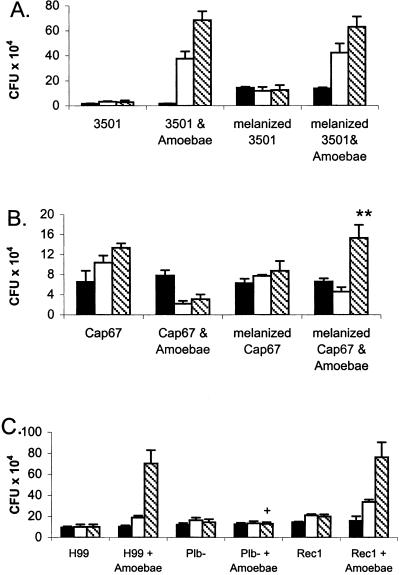
cfu counts of wild-type and mutant Cn strains after incubation with or without Ac in PBS. Bars represent average cfu at different times. Solid bars denote 0 h, white bars denote 24 h, and cross-hatched bars denote 48 h. (A and B) cfu counts of melanized and nonmelanized Cn cells incubated with or without Ac in PBS. (A) Strain 3501 cells. (B) Strain Cap67 cells; ** denotes melanized Cap67 with amoebae has greater growth than Cap67 with amoebae (P < 0.001). (C) Comparison of the cfu counts of Cn between the parental strain, H99, the phospholipase-deficient strain, plb−, and the reconstituted mutant, rec1, strains after incubation with or without Ac in PBS. + indicates a P < 0.001 for plb− with amoebae versus H99 with amoebae. There is no significant difference between H99 and amoebae versus rec1 with amoebae. Error bars denote 1 SD. For each experiment, the number of repetitions was five, and similar results were obtained from two additional independent experiments.
To ascertain the affect of the capsule on the interactions between Cn and Ac, the killing assay was performed by using an acapsular strain, Cap67, and its parental strain, 3501. At 48 h, incubation of strain 3501 with amoebae resulted in cfu that were >70-fold greater than when the strain was incubated in PBS alone (Fig. 3A). The cfu from strain 3501 grown with Ac more than doubled between 6 and 24 h and continued to increase between 24 and 48 h. However, for the acapsular mutant, Cap67, cfu decreased in the presence of Ac (Fig. 3B). A significant reduction was apparent by 6 h, and this reduction was maintained at 48 h as measured by cfu. Incubation of acapsular strain cells with Cn polysaccharide leads to adherence of the polysaccharide, thereby forming a capsule (28). Cap67 cells incubated with soluble GXM results in a change in cell charge that is consistent with acquiring a capsule (27). Cap67 cells with bound exogenous GXM were protected from killing, although they did not replicate (data not shown). Hence, cfu for encapsulated yeast strains either increased or remained stable in the presence of amoebae, whereas cfu for pseudohyphal and acapsular cells decreased.
Effect of Cn Melanin and Phospholipase on the Interaction with Amoebae.
When comparing melanized and nonmelanized 3501 Cn cells incubated with amoebae, no difference in cfu was detected (Fig. 3A). Similarly, when the laccase-deficient 2eTu strain was compared with the complemented mutant strain, 2eTuc, no difference in cfu was found after incubation with amoebae (data not shown). It was postulated that the absence of an effect might be because of the presence of the capsule, which was sufficient to protect against amoebae. Nonmelanized acapsular Cap67 cells incubated with amoebae were rapidly killed; in contrast, melanized Cap67 cells survived even though they did not replicate (Fig. 3B). In the presence of the amoebae, the cfu counts of melanized Cap67 with amoebae were comparable with Cap67 in PBS alone.
The Cn phospholipase mutant, plb1, and its reconstituted counterpart, rec1, were tested in the amoebae assay. As shown in Fig. 3C, the total cfu from the phospholipase mutant when incubated with Ac did not change during the time course. In contrast, the reconstituted mutant, rec1, and wild-type strain H99 both show almost a 7-fold cfu increase when incubated with Ac.
Cn Polysaccharide Localization in Infected Amoebae.
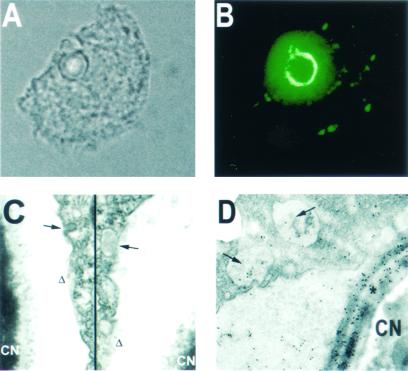
The mAb 2H1 was used in both immunofluorescence microscopy and immunogold TEM. Fig. 4 A and B depicts a single, budding Cn cell inside an amoeba by bright-field and immunofluorescent staining for GXM, respectively. Throughout the amoeba, distant from the Cn cell, there are many discrete fluorescent areas as well as bright fluorescence surrounding the fungal cell reminiscent of polysaccharide in vesicles as has been described in murine macrophages (6). Fig. 4C shows a discontinuous membrane in the amoeba vacuole surrounding Cn with vacuole formation. Finally, Fig. 4D shows an immunogold-labeled TEM of Cn in Ac. The small arrows show immunogold labeling of GXM in vacuoles separated from the Cn cell. Also, note the heavy gold labeling of the GXM capsule, denoted by an asterisk, surrounding the fungal cell.
Figure 4.
Microscopy of Ac cells infected by Cn. Results are indicative of the cellular disruption induced by Cn infection. (A and B) Corresponding bright-field (A) and immunofluorescent (B) microscopy photos. GXM-specific antibody, 2H1, was used for immunofluorescent staining. (C) TEM photograph showing the disruption of the amoebae phagocytic vacuole membrane (delta signs) and of small vacuole formation (arrows). (D) Immunogold-labeled TEM with 2H1 mAb. Small arrows show internal amoebae vacuoles containing Cn polysaccharide; * depicts the fungal capsule. (Magnifications: A and B, ×1,000; C and D, ×22,500.)
Discussion
Our results demonstrate that incubation of Cn and amoebae results in phagocytosis of yeast cells, killing of amoebae, and replication of fungal cells. Despite minimal fungal growth in PBS alone, strains 3501 and H99 grew rapidly when incubated with Ac cells in PBS. Fungal growth in PBS typically continued for 1–2 cycles of replication, thereby completing the growth cycle occurring in culture before incubation in PBS. Throughout the killing assay, 85% of amoebae remain viable in the absence of Cn cells in PBS. Fungal growth was associated with killing of amoebae, which presumably releases nutrients that can sustain fungal replication.
Phagocytosis assays revealed that all Cn strains tested were ingested by amoebae. Pseudohyphal and acapsular strains were more avidly ingested than encapsulated strains. These results indicate that the polysaccharide capsule has antiphagocytic properties for amoebae and presumably functions in a protective role to prevent ingestion of yeast cells by amoebae. Once internalized, Cn resides and replicates in a phagocytic vacuole. Immunofluorescence studies revealed the presence of polysaccharide-containing vacuoles in amoebae that are similar to those found in Cn-infected macrophages (6). The immunogold TEM confirmed that the vacuoles in amoeba with ingested Cn contained polysaccharide. Polysaccharide-containing vesicles are deleterious to macrophage cells (6). Hence, vesicles containing polysaccharide accumulate in both macrophages and amoebae after ingestion of Cn.
We evaluated the impact of various fungal phenotypes associated with mammalian virulence on the fungal–amoebal interaction. The acapsular strain Cap67 was rapidly phagocytosed and killed by amoebae, whereas the parent strain 3501 proliferated in the same conditions. This result is similar to in vivo murine studies that showed most Cap67 cells were found intracellularly in macrophages and neutrophils, and they were killed and eliminated by day 14 (6). The encapsulated strains H99 and 3501 had high budding indices consistent with intracellular growth. In contrast, there was no intracellular budding for Cap67 cells indicative of growth arrest and killing. The inability of Cap67 cells to survive their interactions with Ac strongly suggests a role for the cryptococcal capsule in protection against this environmental predator. Consequently, the addition of exogenous GXM to form an artificial capsule significantly enhanced the survival of Cap67, although the cells did not replicate. Similarly, Ac killed the hypovirulent variant F7 derived from strain 24067. These results closely correlate with the results of previous macrophage cell-line studies (29).
Melanization of Cn has been shown to be protective in macrophage studies and in vivo murine studies (20, 30). In this study, we evaluated the role of melanin in Cn to protect against amoebae. We did not observe any difference in Cn growth in amoebal cultures when comparing melanized and nonmelanized cells of strain 3501, or melanin-deficient strains (2eTu) to its wild-type counterparts (H99 and 2eTuc). Because each of these strains are encapsulated, we hypothesized that the capsule was a dominant virulence factor that would obscure any contribution from melanin to survival of the fungus in amoebae (27). To evaluate this, we compared the survival of melanized and nonmelanized cells from the acapsular strain Cap67 and found the melanized cells were significantly more resistant to killing by amoebae as evidenced by their stable cfu counts. Hence, melanin probably contributes to resistance against amoebae, but in our assay this effect was manifested only in a strain devoid of polysaccharide capsule. This effect may have environmental significance because most environmental isolates of Cn have a small capsule (31) and can be melanized (32).
Phospholipase activity is an important virulence factor for Cn (33). A dramatic decrease in virulence of the Cn phospholipase mutant, plb1, was demonstrated in vivo and in macrophage studies (33). The killing assay results showing that the phospholipase mutant, plb, is unable to grow in Ac indicates that phospholipase is an important virulence factor for infection of Ac as well.
One of the most intriguing observations in this study is that the process of Cn infection of Ac closely resembles Cn infection of macrophages at the cellular level. The interaction of Cn and macrophages was recently described in detail (7). First, Cn is phagocytosed and internalized by both macrophages and amoebae. Second, the internalized fungal cell is then enclosed in a membrane-bound vacuole in which the Cn cells replicate. Third, the formation and accumulation of polysaccharide-filled vacuoles in both macrophages and amoebae accompany fungal cell internalization. Blebbing of cell membranes in contact with polysaccharide occurs in both amoebae and macrophages after ingestion of Cn. These cellular alterations are specifically induced by Cn infection and have been associated with macrophage toxicity. Here, we demonstrate that many virulent Cn strains cause the lysis of amoebae. The deleterious effect of Cn in the murine model and in cell culture depends on several virulence factors including laccase, phospholipase, and capsule production. Our results demonstrate that these virulence factors also play critical roles in Cn infection of amoebae and suggest that the Cn intracellular survival strategy observed in macrophages (6) may have evolved for the survival of this fungus against amoebae. The finding that Cn interacts with Ac in a manner similar to its interaction with macrophages suggests that this model could be useful for studying fungal responses to ingestion at environmental temperatures and conditions.
Recently, there has been much published work concerning bacterial species infecting amoebae cells and its influence on virulence; however, no data have been shown demonstrating interactions between fungal and amoeba cells. Our results support the hypothesis that virulent Cn strains can use Ac cells as a replication niche. Our results suggest an explanation for the curious observation that Cn can efficiently subvert macrophage killing despite the fact that this environmental organism does not depend on an animal parasitic cycle for survival. In contrast to the findings with Cn, both C. albicans and S. cerevisiae were killed by Ac as indicated by a dramatic reduction in fungal cfu after incubation of amoebae and fungal cells. This experiment provides an important control for our studies because it indicates that the amoebae used are fungicidal to other fungal species and supports our proposal that the toxicity of Cn for amoebae is a specific interaction that arose from environmental selection. C. albicans is a human commensal and does not have an environmental reservoir; the S. cerevisiae strain used is a lab-adapted strain.
Although the theme of our findings is similar to the observations made with L. pneumophilia, there seem to be several important differences. Unlike L. pneumophilia that use amoebae as a host for replication, such that hundreds of bacteria can parasitize a single amoeba, the lytic effect of Cn on amoebae seems to be primarily the result of a defense mechanism that allows survival after ingestion. The observation that the intracellular survival strategy of Cn in amoebae and macrophages are very similar strongly suggests that selective pressures placed by amoebae on this fungal species is a critical factor in the maintenance of its virulence characteristics for animal hosts. These results suggest that similar interactions may underlie the phenomenon of virulence among other pathogenic fungi that reside in soils, such as H. capsulatum. Furthermore, the ability of Cn to parasitize amoebae suggests an explanation for the remarkable host range of this fungus that has been reported to cause disease in many species of mammals and birds (34). On the basis of our observations, we propose that Cn is a pathogen for each of these species because it can subvert macrophage fungicidal activity by using strategies that were selected for surviving interactions with environmental predators such as amoebae. Comparative studies of traits permissive for amoebae and macrophage survival may reveal additional important insights into the pathogenesis of Cn infection.
Acknowledgments
We are grateful to Dr. J. D. Nosanchuk for assistance with the SEM and for his valuable and constructive criticism. We thank Yvonne Kress for assistance with electron microscopy. J.N.S. is supported by National Institutes of Health Training Grant T32GM 07491. A.C. is supported by National Institutes of Health Awards AI33774, AI3342, and HL-59842-01. H.S. is supported by National Institutes of Health Award AI23549.
Abbreviations
- Ac
A. castellanii
- Cn
C. neoformans
- GXM
glucuronoxylomannan
- TEM
transmission electron microscopy
- SEM
scanning electron microscopy
- cfu
colony-forming units
- PYG
peptone-yeast extract-glucose
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 14760.
References
- 1.Mitchell T G, Perfect J R. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellis D H, Pfeiffer T J. J Clin Microbiol. 1990;28:1642–1644. doi: 10.1128/jcm.28.7.1642-1644.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldman D L, Lee S C, Mednick A J, Montella L, Casadevall A. Infect Immun. 2000;68:832–838. doi: 10.1128/iai.68.2.832-838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Littman M L. Am J Med. 1959;27:976–988. doi: 10.1016/0002-9343(59)90181-0. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Hermoso D, Janbon G, Dromer F. J Clin Microbiol. 1999;37:3204–3209. doi: 10.1128/jcm.37.10.3204-3209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feldmesser M, Kress Y, Novikoff P, Casadevall A. Infect Immun. 2000;68:4225–4237. doi: 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldmesser M, Tucker S, Casadevall A. Trends Microbiol. 2001;9:273–278. doi: 10.1016/s0966-842x(01)02035-2. [DOI] [PubMed] [Google Scholar]
- 8.Hogan L H, Klein B S, Levitz S M. Clin Microbiol Rev. 1996;9:469–488. doi: 10.1128/cmr.9.4.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barker J, Brown M R. Microbiology. 1994;140:1253–1259. doi: 10.1099/00221287-140-6-1253. [DOI] [PubMed] [Google Scholar]
- 10.Rowbotham T J. J Clin Pathol. 1980;33:1179–1183. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swanson M S, Hammer B K. Annu Rev Microbiol. 2000;54:567–613. doi: 10.1146/annurev.micro.54.1.567. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez-Zaragoza S. Crit Rev Microbiol. 1994;20:225–241. doi: 10.3109/10408419409114556. [DOI] [PubMed] [Google Scholar]
- 13.Visvesvara G S, Balamuth W. J Protozool. 1975;22:245–256. doi: 10.1111/j.1550-7408.1975.tb05860.x. [DOI] [PubMed] [Google Scholar]
- 14.Castellani A. J Trop Med Hyg. 1930;33:160. [Google Scholar]
- 15.Bunting L A, Neilson J B, Bulmer G S. Sabouraudia. 1979;17:225–232. doi: 10.1080/00362177985380341. [DOI] [PubMed] [Google Scholar]
- 16.Moffat J F, Tompkins L S. Infect Immun. 1992;60:296–301. doi: 10.1128/iai.60.1.296-301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bozue J A, Johnson W. Infect Immun. 1996;64:668–673. doi: 10.1128/iai.64.2.668-673.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williamson P R, Wakamatsu K, Ito S. J Bacteriol. 1998;180:1570–1572. doi: 10.1128/jb.180.6.1570-1572.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brummer E, Stevens D A. Cell Immunol. 1994;157:1–10. doi: 10.1006/cimm.1994.1200. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Aisen P, Casadevall A. Infect Immun. 1995;63:3131–3136. doi: 10.1128/iai.63.8.3131-3136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casadevall A, Mukherjee J, Devi S J, Schneerson R, Robbins J B, Scharff M D. J Infect Dis. 1992;165:1086–1093. doi: 10.1093/infdis/165.6.1086. [DOI] [PubMed] [Google Scholar]
- 22.Casadevall A, Cleare W, Feldmesser M, Glatman-Freedman A, Goldman D L, Kozel T R, Lendvai N, Mukherjee J, Pirofski L A, Rivera J, et al. Antimicrob Agents Chemother. 1998;42:1437–1446. doi: 10.1128/aac.42.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cleare W, Casadevall A. Clin Diagn Lab Immunol. 1998;5:125–129. doi: 10.1128/cdli.5.2.125-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levitz S M, Nong S H, Seetoo K F, Harrison T S, Speizer R A, Simons E R. Infect Immun. 1999;67:885–890. doi: 10.1128/iai.67.2.885-890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz D A. Ann Clin Lab Sci. 1988;18:388–397. [PubMed] [Google Scholar]
- 26.Small J M, Mitchell T G. Infect Immun. 1986;54:742–750. doi: 10.1128/iai.54.3.742-750.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nosanchuk J D, Casadevall A. Infect Immun. 1997;65:1836–1841. doi: 10.1128/iai.65.5.1836-1841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozel T R, Gotschlich E C. J Immunol. 1982;129:1675–1680. [PubMed] [Google Scholar]
- 29.Neilson J B, Fromtling R A, Bulmer G S. Mycopathologia. 1981;73:57–59. doi: 10.1007/BF00443015. [DOI] [PubMed] [Google Scholar]
- 30.Liu L, Tewari R P, Williamson P R. Infect Immun. 1999;67:6034–6039. doi: 10.1128/iai.67.11.6034-6039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emmons C W. Lab Invest. 1962;11:1026–1032. [Google Scholar]
- 32.Nosanchuk J D, Rudolph J, Rosas A L, Casadevall A. Infect Immun. 1999;67:5477–5479. doi: 10.1128/iai.67.10.5477-5479.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox G M, McDade H C, Chen S C, Tucker S C, Gottfredsson M, Wright L C, Sorrell T C, Leidich S D, Casadevall A, Ghannoum M A, Perfect J R. Mol Microbiol. 2001;39:166–175. doi: 10.1046/j.1365-2958.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- 34.Casadevall A, Perfect J R. Cryptococcus neoformans. Washington, DC: Am. Soc. Microbiol.; 1998. [Google Scholar]
- 35.Fromtling R A, Shadomy H J, Jacobson E S. Mycopathologia. 1982;79:23–29. doi: 10.1007/BF00636177. [DOI] [PubMed] [Google Scholar]