Abstract
Many metabolic and physiological processes display circadian oscillations. We have shown that the core circadian regulator, CLOCK, is a histone acetyltransferase whose activity is counterbalanced by the NAD+-dependent histone deacetylase SIRT1. Here we show that intracellular NAD+ levels cycle with a 24 h rhythm, an oscillation driven by the circadian clock. CLOCK:BMAL1 regulate the circadian expression of NAMPT (nicotinamide phosphoribosyltransferase), a rate limiting step enzyme in the NAD+ salvage pathway. SIRT1 is recruited to the Nampt promoter and contributes to the circadian synthesis of its own coenzyme. Using the specific inhibitor FK866, we demonstrate that NAMPT is required to modulate circadian gene expression. Our findings reveal an interlocked transcriptional-enzymatic feedback loop that governs the molecular interplay between cellular metabolism and circadian rhythms.
Accumulating evidence reveal intriguing links between the circadian clock and cellular metabolism (1–3), which at least in part rely on epigenetic control and chromatin remodeling (4). The circadian regulator CLOCK has an intrinsic acetyltransferase activity, which enables circadian chromatin remodeling by acetylation of histones (5) and non-histone proteins, including its own partner BMAL1 (6). The histone deacetylase (HDAC) that counterbalances the HAT function of CLOCK is SIRT1 (7, 8), an enzyme whose activity is dependent on intracellular NAD+ levels (9). While NAD+ is SIRT1’s natural cosubstrate, the reduced form NADH and the by-product of NAD+ consumption, nicotinamide (NAM), repress the activity of SIRT1 (9), generating an enzymatic feedback loop on the HDAC function of this enzyme. Two main systems determine NAD+ levels in the cell, the de novo biosynthesis from tryptophan and the NAD+ salvage pathway (10). A critical step of this latter pathway is controlled by the enzyme nicotinamide phosphoribosyltransferase (NAMPT), also known as visfatin or PBEF (11), which catalyzes the first step in the biosynthesis of NAD+ from NAM. Evidence shows that NAMPT is implicated in cellular metabolism, senescence and survival in response to genotoxic stress (12–14).
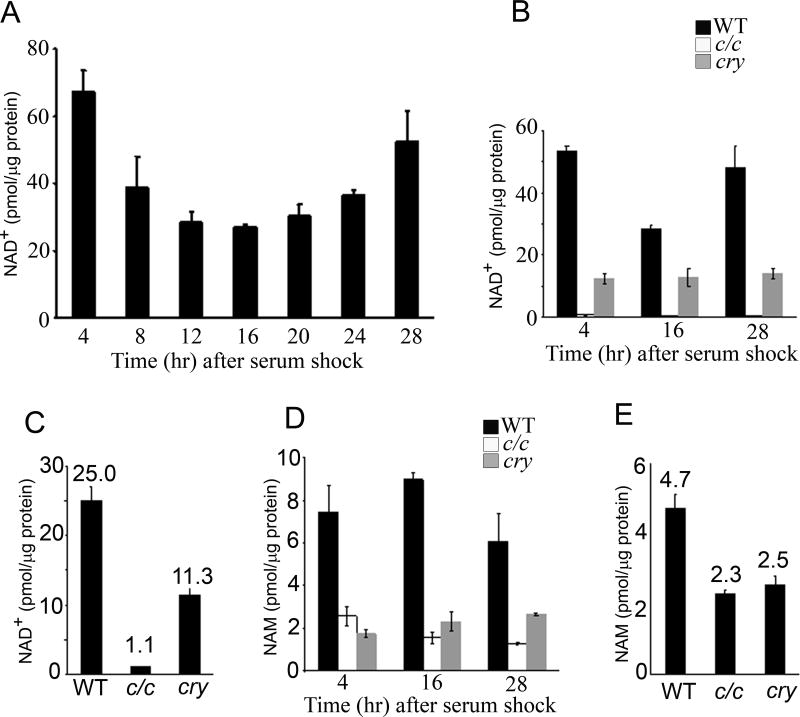
Because of the role of SIRT1 in modulating clock function (7) , we reasoned that circadian tuning may be achieved by NAD+ oscillating levels. Wild type mouse embryo fibroblasts (MEFs) were serum-entrained and cellular NAD+ levels measured by liquid chromatography coupled to tandem mass spectrometry (LC/MSn) from total extracts prepared at various time intervals after serum shock. Strikingly, cellular NAD+ showed a reproducible circadian oscillation of about 2.5 fold (Fig. 1A). The average NAD+ concentration of 25 pmol/µg protein (approximately 60 µM) found in MEFs is in keeping with recent reports for rat axons using LC coupled to ultraviolet detector (9 pmol/µg protein) (15), mouse erythrocyte using LC/MS/MS (368 µM) (16) and HEK293 cells using LC/MALDI/MS (365 µM) (14). Interestingly, cellular NAD+ oscillation is in phase with SIRT1 deacetylase activity and is in opposite phase to acetylation of histone H3 and BMAL1 (7).
Figure 1. Circadian oscillation of NAD+ is controlled by the clock.
(A and B) Cellular NAD+ was extracted from serum-entrained MEFs derived from wild type (wt) (A), clock/clock mutant mice (c/c) and Cry1/Cry2 double KO mice (cry) (B) at indicated time points and analyzed by LC/MSn. Three independent experiments were performed and representative results are shown. All data are means ± SEM of three independent samples. (C) Average NAD+ levels in wt, c/c and cry MEFs. Data are shown as means ± SEM of >50 independent samples. (D) Cellular nicotinamide (NAM) was extracted from serum-entrained MEFs derived from wt, c/c and cry mice at indicated time points and analyzed by LC/MSn. (E) Average NAM levels in wt, c/c and cry MEFs. Data are shown as means ± SEM of >50 independent samples.
Importantly, NAD+ levels do not oscillate in entrained MEFs originated from the clock/clock mutant mice (c/c) and from the arrhythmic Cry1/Cry2 double KO mice (cry) (Figs. 1B and S1), demonstrating that NAD+ oscillation is driven by the circadian clock. Total cellular NAM levels measured by LC/MSn also showed oscillation, albeit more moderate, with a phase opposite to the one of NAD+. Also NAM oscillation was abolished in the MEFs from c/c and cry mice (Fig. 1D). Thus, the circadian clock controls the levels of these metabolites. This analysis provided also another important clue: NAD+ and NAM levels are significantly lower in mutant MEFs as compared to wild type (wt) cells (for NAD+ only 1.1 pmol/µg protein in c/c MEFs, about 5% of that in wt MEFs; Figs. 1C and E). This notion points to an involvement of the NAD+ salvage pathway and, because of its dynamic regulation, specifically to NAMPT whose prominent function in NAD+ production has been demonstrated (17–19). Furthermore, increased flux through the NAD+ salvage pathway is responsible for sirtuin-dependent responses even under conditions of unaltered steady-state NAD+ levels (12).
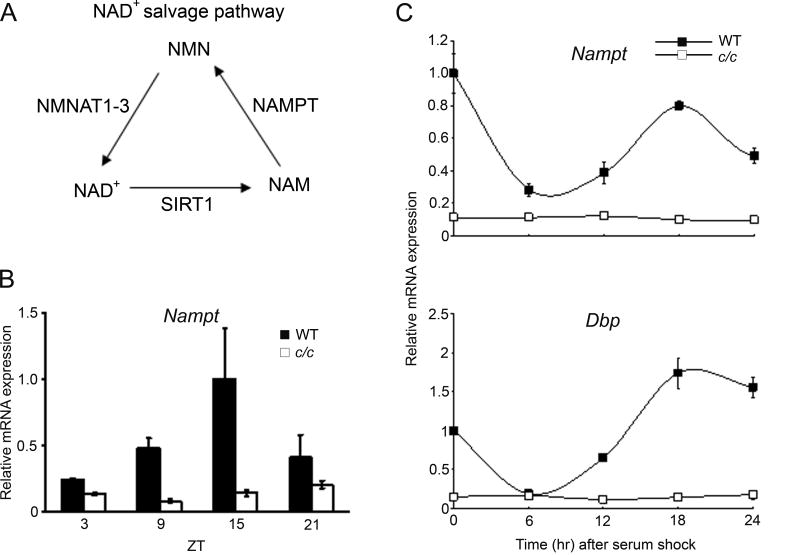
Next we analyzed the expression of NAD+ salvage pathway metabolic enzymes. The nicotinamide mononucleotide adenylyltransferase, NMNAT1, NMNAT2 and NMNAT3, are central in NAD biosynthesis, catalyzing the adenylation of nicotinamide mononucleotide (NMN) or nicotinic acid mononucleotide (NaMN) using the AMP moiety of ATP to form NAD+ or NaAD, respectively (Fig. 2A). The expression of these three enzymes is mildly oscillatory (Fig. S2). Thus, our attention focused on NAMPT, which operates as rate limiting enzyme in NAD+ production within the NAD+ salvage pathway (10, 14, 20). Expression of Nampt is robustly circadian in livers from wt mice, while oscillation is virtually absent in c/c mice (Fig. 2B). Rhythmic expression is observed also in serum-entrained wt MEFs, paralleling Dbp oscillation. Again, Nampt oscillation is abolished in c/c MEFs (Fig. 2C).
Figure 2. The circadian clock controls Nampt gene expression.
(A) Scheme of the NAD+ salvage pathway in mammals. NAM, nicotinamide; NMN, nicotinamide mononucleotide; NAMPT, nicotinamide phosphoribosyltransferase; NMNAT1-3, nicotinamide mononucleotide adenyltransferase. (B) Nampt gene expression in livers from light-entrained wt and c/c mutant mice was quantified by q-PCR. Nampt gene expression at ZT 15 in liver from wt mice was set to 1. (C) Nampt and Dbp gene expressions in serum-entrained MEFs from wt and c/c mutant mice were quantified by q-PCR. Expression at time 0 in wt MEFs was set to 1. All data are means ± SEM of three independent samples.
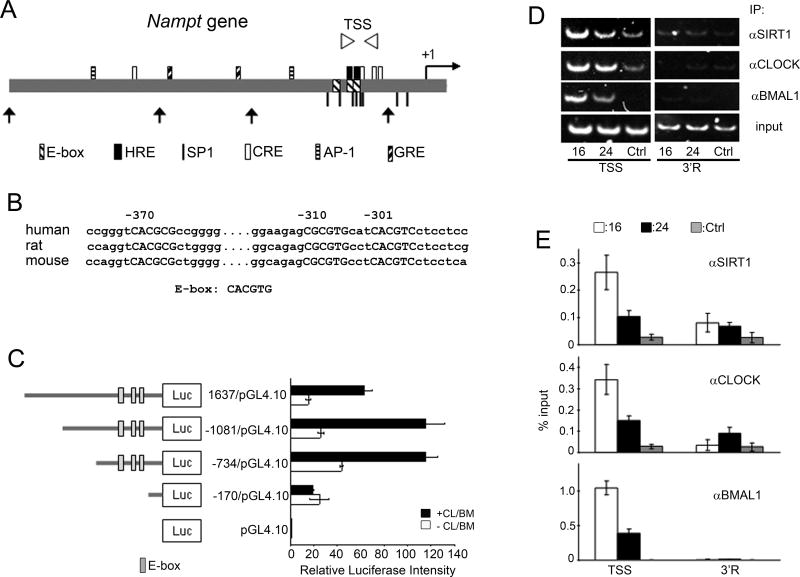
The Nampt promoter contains three putative, highly conserved, E-boxes (Fig. 3A, B). Using promoter mutagenesis and transient expression in cultured cells, we demonstrate that the Nampt promoter is readily activated by CLOCK:BMAL1 through the E-boxes (Fig. 3C). Next, using chromatin immunoprecipitation (ChIP) we showed that CLOCK:BMAL1 physically and specifically associate to the E-boxes on the Nampt promoter in a time-dependent manner (Figs. 3D and E), consistent with Nampt circadian expression. We have recently demonstrated that SIRT1 is in a complex with CLOCK:BMAL1 (7). Dual cross-linking ChIP assays showed that SIRT1 binds to the E-boxes in a time-dependent manner, following the circadian timing of CLOCK:BMAL1 recruitment (Figs. 3D and E). Thus, as previously shown for Dbp and Per2 (7) , CLOCK and SIRT1 contribute to circadian chromatin remodeling at the Nampt promoter. As NAD+ intracellular levels directly influence the HDAC activity of SIRT1 (21–23), an enzymatic/transcription feedback loop seems to operate, in which NAD+ levels determine the oscillatory synthesis of NAMPT, the key enzyme in the NAD+ salvage pathway.
Figure 3. Regulation of the Nampt promoter by CLOCK:BMAL1 and SIRT1.
(A) Schematic diagram of regulatory elements in human Nampt promoter. TSS (Transcription Start Site) primer region for q-PCR is shown as arrowheads. Truncated forms of Nampt promoter used in (C) are shown (arrows). Putative transcription factors binding sites are indicated: HRE, hypoxia-inducible factor-responsible element; SP1, specificity protein 1; CRE, cAMP-response element; AP-1, activator protein 1; GRE, glucocorticoid receptor response element. (B) Conserved E-boxes (capitalized) are shown. Numbers indicate positions from human transcription start site. (C) Schematic diagram of Nampt promoter constructs. Effect of CLOCK:BMAL1 (+CL/BM; black bars) on luciferase activity is shown. Luciferase activity of CLOCK:BMAL1 on the pGL4.10 was set as 1. All data are means ± SEM of three independent samples. (D) Representative results of ChIP assays analyzed by semiquantitative PCR. Dual crosslinked nuclear extracts isolated from MEFs after 16 or 24 hr serum shock and subjected to ChIP assay with anti-SIRT1, anti-CLOCK, anti-BMAL1, or no antibody (ctrl). No antibody and 3’R primers were used as controls for immunoprecipitation and PCR, respectively. (E) Quantification of ChIP by q-PCR. q-PCR was performed on the same samples as described in (D). All data are means ± SEM of three independent samples.
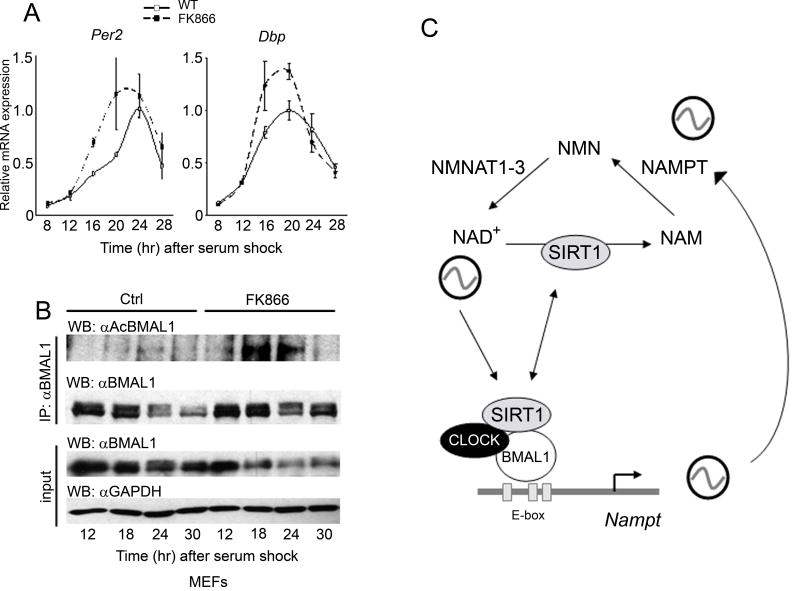
To address whether NAMPT is critical in modulating clock function, we took advantage of FK866, a low molecular weight specific NAMPT inhibitor. FK866 lowers cellular NAD+ and NAM levels over prolonged length of time (17) (Figs. S3 and S4). Pharmacological inhibition of NAMPT significantly modifies the circadian expression of Per2 and Dbp in serum-stimulated MEFs (Fig. 4A), causing an earlier onset of the circadian peak for both genes by 3–4 hours and increasing the amplitude of oscillation by 30–40%. Importantly, this effect of FK866 is highly similar to the one caused by inhibition of SIRT1 (7). Thus, we predicted that blocking NAMPT would modify BMAL1 acetylation, a regulatory event critical to clock function (6). Using the anti-acetyl-BMAL1 antibody recently developed in our laboratory (7), we show that inhibiton of NAMPT induces a considerable increase and a broader peak of K537 acetylation (Fig. 4B). Significantly, this BMAL1 acetylation profile is basically equivalent to the one observed in MEFs and livers from Sirt1-/- mice (7).
Figure 4. NAMPT modulates the circadian clock.
(A) Per2 and Dbp gene expression levels in serum-entrained MEFs treated with 10 nM FK866 or EtOH as control (solvent: ctrl) were analyzed by q-PCR. Highest value for each gene in EtOH treated MEFs was set to 1. All data are means ± SEM of three independent samples. (B) BMAL1 Lys537 acetylation profile in serum-entrained MEFs either treated with 10 nM FK866 or EtOH. Extracts prepared from indicated time points and BMAL1 acetylation was monitored (7). Input samples were probed with anti-BMAL1 and anti-GAPDH antibodies. (C) Scheme of the transcription-enzymatic interplay by which the circadian machinery governs the intracellular levels of NAD+. The NAD+-dependent deacetylase SIRT1 is thereby controlling the oscillatory synthesis of its own coenzyme.
How intimate is the link between cellular metabolism and the circadian clock? Specifically, are metabolic pathways regulating the circadian machinery (3, 24, 25), or are molecular elements of the clock controlling metabolism (3, 26, 27)? The findings reported here demonstrate that both pathways exist in the cell, utilizing a discrete molecular mechanism of control in which the enzyme NAMPT occupies a pivotal position. Our results reveal the interlocking of two auto-regulatory systems, in which a classical transcription circadian loop is coupled to an enzymatic feedback loop (Fig. 4C). These findings point to the oscillation of NAD+ as a key regulatory step in the modulation of rhythms in metabolic tissues and peripheral clocks. Interestingly, FK866 may be used for treatment of diseases implicating deregulated apoptosis or as a sensitizer for genotoxic agents (17). Thus, our findings could have multiple implications and may open novel avenues for pharmacological intervention.
Supplementary Material
References
- 1.Wijnen H, Young MW. Annu Rev Genet. 2006;40:409. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 2.Green CB, Takahashi JS, Bass J. Cell. 2008;134:728. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckel-Mahan K, Sassone-Corsi P. Nat. Str. Mol Biol. doi: 10.1038/nsmb.1595. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belden WJ, Dunlap JC. Cell. 2008;134:212. doi: 10.1016/j.cell.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doi M, Hirayama J, Sassone-Corsi P. Cell. 2006;125:497. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 6.Hirayama J, et al. Nature. 2007;450:1086. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 7.Nakahata Y, et al. Cell. 2008;134:329. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asher G, et al. Cell. 2008;134:317. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 9.Bordone L, Guarente L. Nat Rev Mol Cell Biol. 2005;6:298. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 10.Rongvaux A, Andris F, Van Gool F, Leo O. Bioessays. 2003;25:683. doi: 10.1002/bies.10297. [DOI] [PubMed] [Google Scholar]
- 11.Revollo JR, Grimm AA, Imai S. J Biol Chem. 2004;279:50754. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 12.Anderson RM, et al. J Biol Chem. 2002;277:18881. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 13.van der Veer E, et al. J Biol Chem. 2007;282:10841. doi: 10.1074/jbc.C700018200. [DOI] [PubMed] [Google Scholar]
- 14.Yang H, et al. Cell. 2007;130:1095. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, et al. J Cell Biol. 2005;170:349. doi: 10.1083/jcb.200504028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada K, Hara N, Shibata T, Osago H, Tsuchiya M. Anal Biochem. 2006;352:282. doi: 10.1016/j.ab.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 17.Hasmann M, Schemainda I. Cancer Res. 2003;63:7436. [PubMed] [Google Scholar]
- 18.Olesen UH, et al. Biochem Biophys Res Commun. 2008;367:799. doi: 10.1016/j.bbrc.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 19.Rongvaux A, et al. J Immunol. 2008;181:4685. doi: 10.4049/jimmunol.181.7.4685. [DOI] [PubMed] [Google Scholar]
- 20.Revollo JR, et al. Cell Metab. 2007;6:363. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Nature. 2000;403:795. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 22.Landry J, et al. Proc Natl Acad Sci U S A. 2000;97:5807. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith JS, et al. Proc Natl Acad Sci U S A. 2000;97:6658. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damiola F, et al. Genes Dev. 2000;14:2950. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaasik K, Lee CC. Nature. 2004;430:467. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 26.Lamia KA, Storch KF, Weitz CJ. Proc Natl Acad Sci U S A. 2008;105:15172. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turek FW, et al. Science. 2005;308:1043. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thanks to D. Piomelli for insightful discussions and help. Thanks also to L. Guarente, K. Yamagata and members of the Sassone-Corsi's laboratory for help, reagents and support. Y.N. was supported by JSPS Postdoctoral Fellowships for Research Abroad. S.S. was supported by a postdoctoral fellowship from American Heart Association, Western States Affiliate. Work supported by the Cancer Research Coordinating Committee of the University of California and NIH (R01-GM081634-01).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.