Abstract
The classical view of the molecular clock is based on interlocked transcriptional-translational feedback loops. Since a substantial fraction of the mammalian genome is expressed in a circadian manner, chromatin remodelling has been proposed to play a critical role in clock function. Here, we show that K4 tri-methylation of histone H3 is rhythmic and follows the same profile as previously described H3K9/K14 acetylation on circadian promoters. MLL1, a mammalian homologue of Drosophila trithorax, is a H3K4 specific methyltransferase implicated in transcriptional control. We demonstrate that MLL1 is essential for circadian transcription and cyclic H3K4 tri-methylation. MLL1 is in a complex with CLOCK:BMAL1 and contributes to their rhythmic recruitment to circadian promoters and to H3 acetylation. Remarkably, MLL1 fails to interact with CLOCKΔ19, providing an explanation for this mutations dominant negative function. Our results favor a scenario in which H3K4 tri-methylation by the MLL1 histone methyltrasferase are required to establish a chromatin permissive state for circadian transcription.
Circadian rhythms govern a wide array of physiological, behavioral and metabolic functions in organisms ranging from cyanobacteria to humans1. In mammals, circadian rhythms are generated in pacemaker neurons within the suprachiasmatic nuclei (SCN) of the hypothalamus2,3. Peripheral clocks are present in most tissues, and their rhythmicity is synchronized by the SCN using a series of ill-defined systems2,4. Disruption of circadian rhythms can have a profound influence on human health and has been linked to insomnia, depression, coronary heart diseases, various neurodegenerative disorders and cancer5–8. The molecular basis underpinning such extensive influence of the circadian clock is global regulation of gene expression. It is established that up to 10% of all mammalian transcripts follow a circadian rhythm in expression9–11. Recent studies suggest that an even higher proportion of genes may be expressed in a circadian manner, especially in metabolic tissues12 or depending on the stringency of the data analysis13.
The classical view of the molecular clock is based on interlocked transcriptional-translational feedback loops14. Various core circadian clock genes have been identified in mammals: Clock, Bmal1, Casein kinase I epsilon (CKIε), Cryptochromes 1 and 2 (Cry1, Cry2), Period 1, 2 and 3 (Per1, Per2, Per3), and Rev-erb-α2. CLOCK and BMAL1 are basic-helix-loop-helix (bHLH)-PAS transcription activators which heterodimerize and induce the expression of Per and Cry genes via binding to E-box elements present in their promoters. PER and CRY form a complex which inhibits CLOCK:BMAL1-mediated transcription through direct protein-protein interactions15,16. Importantly, the CLOCK:BMAL1 heterodimer regulates the transcription of many clock-controlled genes (CCGs), which in turn influence a wide array of physiological functions, including food intake, hormonal synthesis and release, body temperature and metabolism17–19.
Accumulating evidence places dynamic changes in chromatin transitions as critical events for the proper timing and extent of circadian gene expression20–23. Rhythmic histone modifications have been shown to be associated with the cyclic transcription of several clock-controlled genes, including Dbp, Per1 and Per2 (refs. 22, 24–27) with most of the attention focused on histone acetylation. The finding that CLOCK itself has an intrinsic histone acethyltransferase (HAT) activity necessary for circadian function28, provided a conceptually novel avenue of investigation. Indeed, the subsequent finding that the NAD+-dependent histone deacetylase SIRT1 associates with CLOCK and controls the deacetylation of both H3 (refs. 22, 29–31) and BMAL132,33, extended the link of circadian acetylation to signaling pathways governing cellular metabolism8,19.
In addition to acetylation, the N-terminal tails of histones undergo various other posttranslational modifications (PTMs), including phosphorylation, methylation and ubiquitination34–36. Specific combinations of these PTMs correspond to distinct nuclear functions and physiological responses34,37. Unlike acetylation, which generally correlates with transcriptional activation, methylation of histones occupies a pivotal position and is accociaated with either activation or repression, depending on the sites of modification38. Notably, lysine methylation of histone H3 at K4 has been intimately linked to transcriptional activation18,35. Lysine residues can be mono-, di - or tri-methylated at the ε-amino group, with each state correlating with a distinct functional effect39. H3K4me2 occurs at both inactive and active euchromatic genes, whereas K4me3 is present prominently at actively transcribed genes40, and is widely accepted as an unique epigenetic mark defining an active chromatin state in most eukaryotes41,42. Importantly, H3K4 methylation has been shown to be often associated with specific H3K9/14 and H4K16 acetylation, both ‘marks’ associated with active gene expression18.
Mixed lineage leukemia 1 (MLL1) is a histone methyltransferase (HMT) that specifically promotes tri-methylation of histone H3 at K4 and regulates transcriptional activation43,44. MLL1 is a mammalian homolog of the Drosophila trithorax gene, with which it shares several functional domains, including the conserved C-terminal region. The most C-terminal 250 amino acids of MLL1 include a SET-domain, which displays H3K4 methyltransferase activity44,45. MLL1 was first reported as a transcriptional coactivator involved in the maintenance of selected Hox genes expression in morphogenesis44. MLL1 is an element of a large chromatin remodeling complex that includes other critical regulators, including, among others WDR5, Ash2L, Menin and Rbbp545–47. Some components of the MLL1 complex have been shown to function as subunits of other nuclear complexes, suggestive of coordinated events of chromatin remodeling.
Here we show that H3K4me3 is circadian and that it is elicited by MLL1. Our studies demonstrate that this histone modification directs the circadian acetylation at H3K9/14 and that MLL1 allows the recruiting of CLOCK:BMAL1 to chromatin. Thus, MLL1 is implicated in establishing a chromatin permissive state for circadian transcription.
RESULTS
H3K4me3 is circadian at clock-controlled promoters
To investigate whether methylation of histone H3K4 exhibits rhythmic changes, we performed chromatin immunoprecipitation (ChIP) assays at different circadian times in mouse embryonic fibroblasts (MEFs) entrained either by dexamethasone or serum shock. As previously reported22, H3K9/14 acetylation levels on the Dbp promoter oscillates, being high at circadian time (CT) 18 and low at CT 30 (Fig. 1a). H3K4me1 and me2 levels do not cycle, being equivalent between CT18 and CT30 30 (Fig. 1a and Supplementary Fig. 1a). On the other hand, tri-methylation levels show a robust oscillation, displaying apparent amplitude comparatively higher than K9/K14 acetylation (Fig. 1b and Supplementary Fig. 1b). Moreover, H3K9/K14 acetylation and K4 methylation show a highly similar profile of daily changes on the Dbp promoter (Fig. 1b). These circadian histone modifications were also observed on the Bmal1 promoter with an antiparallel phase with respect to Dbp (Supplementary Fig 1b), corresponding to the profile of Bmal1 transcription. Importantly, no cyclic H3 modifications were observed within the 3’-UTR of either Dbp or Bmal1 at any circadian time point (Supplementary Fig. 1c). In short both K9/K14 acetylation and H3K4 methylation occur sequentially, at specific times on actively transcribed regions. Moreover, our ChIP analyses show that most H3K4me1/2 on the Dbp promoter are not cycling throughout the circadian cycle, whereas me3 levels robustly oscillate (Supplementary Fig. 1a). Thus, the HMT specifically involved in the conversion of di- to tri-methylation at H3K4 could be implicated in the control of circadian transcription.
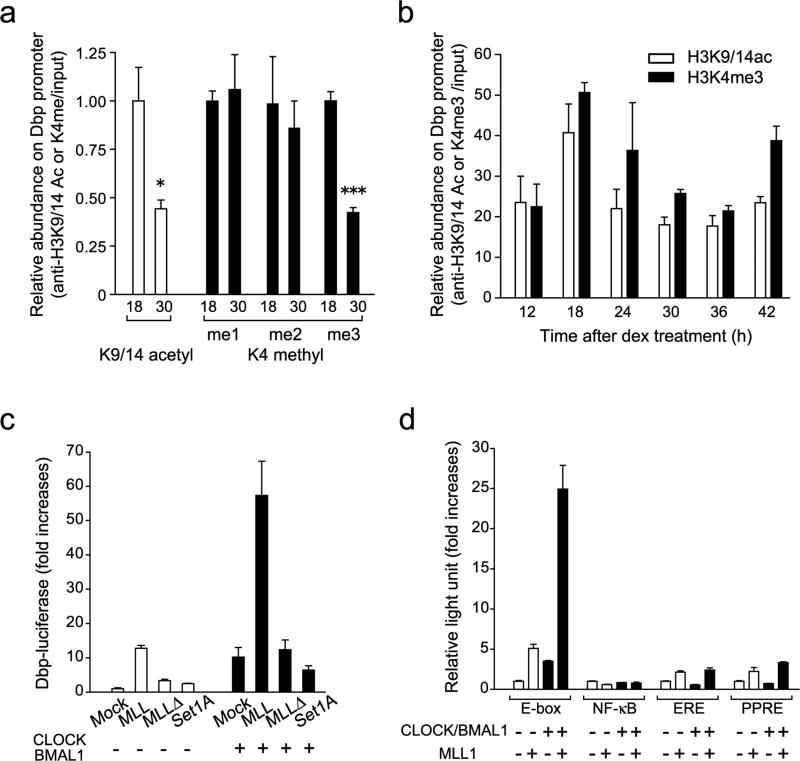
Figure 1. Histone H3K4 methyltransferase MLL1 synergistically activate CLOCK:BMAL1-mediated gene transcription.
(a) Me3 but not me1/2 level of H3K4 changes between two time points on the Dbp promoter E-box region. ChIP analyses were performed in MEFs at 18 or 30 hr after dexamethasone synchronization using antibodies against acetylated H3K9 and K14 (white) and me1, me2 or me3 H3K4 (black). (Means ±SEM of three independent samples, *p<0.05, and ***p<0.0001). The amount of highest modification time point (18 hr) was set to 1. (b) H3K4me3 follows same circadian rhythmicity as acetylation of H3K9/K14 on the Dbp promoter E-box region. ChIP analyses were performed in MEFs after dexamethasone synchronization. (Means ±SEM of three independent samples from a representative experiment. Analogous results were observed in three independent experiments). (c) Histone H3K4 specific-methyltransferase MLL1 and SET1A were transiently transfected with or without CLOCK:BMAL1 in 293 cells. MLL1 but not SET1A showed substantial increases of the dbp-luc transcription. On the other hand, methyltransferase-dead mutant of MLL1 (MLL1Δ)) abolished transcriptional activity. (Means ±SEM of eight samples in two independent experiments) (d) Synergistic transcriptional activation on E-box motif by MLL1 together with CLOCK:BMAL1. E-box motif-containing sequences (E-box), NF-κB-responsive element (NF-κB), estrogen responsive element (ERE) or PPARresponsive element (PPRE) luciferase reporter genes were transfected in presence or in absence of CLOCK:BMAL1 in 293 cells. (Means ±SEM of eight samples from two independent experiments).
MLL1 elevates CLOCK:BMAL1-mediated transcriptional activation
To test if HMTs have a role in circadian gene transcription, we analyzed CLOCK:BMAL1-mediated activation in 293 cells on a Dbp promoter-based luciferase reporter vector. Together with CLOCK:BMAL1 we co-expressed either Set1A or MLL1, two different H3K4 specific HMT, which belong to same MLL family48, or other unrelated HMTs, such as Suv39 and PRMT1, enzymes that specifically modify residues other than H3K4.
While Set1A (Fig. 1c), Suv39 and PRMT1 (Supplementary Fig. 2a), have little or no effect on Dbp promoter activity, co-expression of MLL1 elicited a robust increase of transcriptional activity, which was abolished when the SET methyltransferase domain was deleted (mutant MLL1Δ), demonstrating the critical role of the chromatin modification activity. When MLL1 was co-expressed with CLOCK and BMAL1, its activation was further enhanced about 7–10 fold, leading to an average of about 60-fold activation (Fig. 1c). Similar results were obtained using Per1 and Per2 reporters (not shown). Again, use of the MLL1Δ mutant demonstrated the absolute requirement of the SET domain for full activity and cooperation with CLOCK:BMAL1. These results show that MLL1 potentiates CLOCK:BMAL1-dependent gene activation in a synergistic, rather than additional, manner (Fig. 1c). Finally, specificity of activation was validated by the use of luciferase reporters driven by different promoter elements. While MLL1 elicits robust transactivation of an E-box motif sequence, NF-κB-responsive element (NFκB), estrogen responsive element (ERE), or PPAR-responsive element (PPRE), are poorly activated (Fig. 1d). Synergistic activation of CLOCK:BMAL1 is observed uniquely on E-box based promoters. Also, specificity was confirmed since MLL1-mediated synergistic activation was not observed in combination with PPARγ (peroxisome proliferator-activated receptor γ) and the respective ligand on a PPRE-reporter (Supplementary Fig. 2b).
MLL1 is required for transcriptional activation by CLOCK:BMAL1
MLL1 appears to have the unique property of powerfully enhancing transcriptional activation by CLOCK:BMAL1. Next, we sought to establish whether MLL1 would be required by CLOCK:BMAL1 to elicit activation. To do so, we compared the capacity of CLOCK:BMAL1 to activate transcription directed from Dbp and Per2 promoters in MEFs derived from wild-type (WT) versus MLL1-null mice44. In striking contrast to WT MEFs, activation by CLOCK:BMAL1 is drastically reduced in MLL1-deficient MEFs (Fig. 2a). Importantly, ectopic expression of MLL1 in MLL1-null MEFs rescued activation by CLOCK:BMAL1 (Fig. 2a), while the MLL1Δ mutant is not able to do so (not shown). Hence, MLL1 appears to be required to establish a permissive state for CLOCK:BMAL1 mediated transcriptional activation.
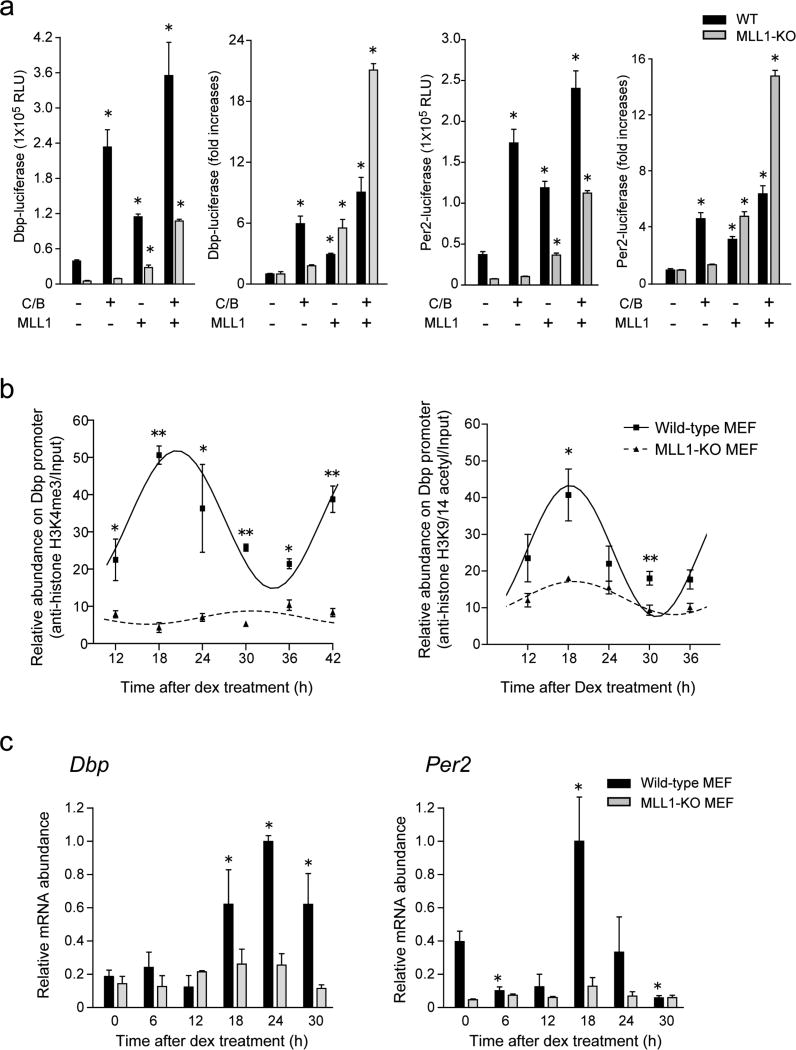
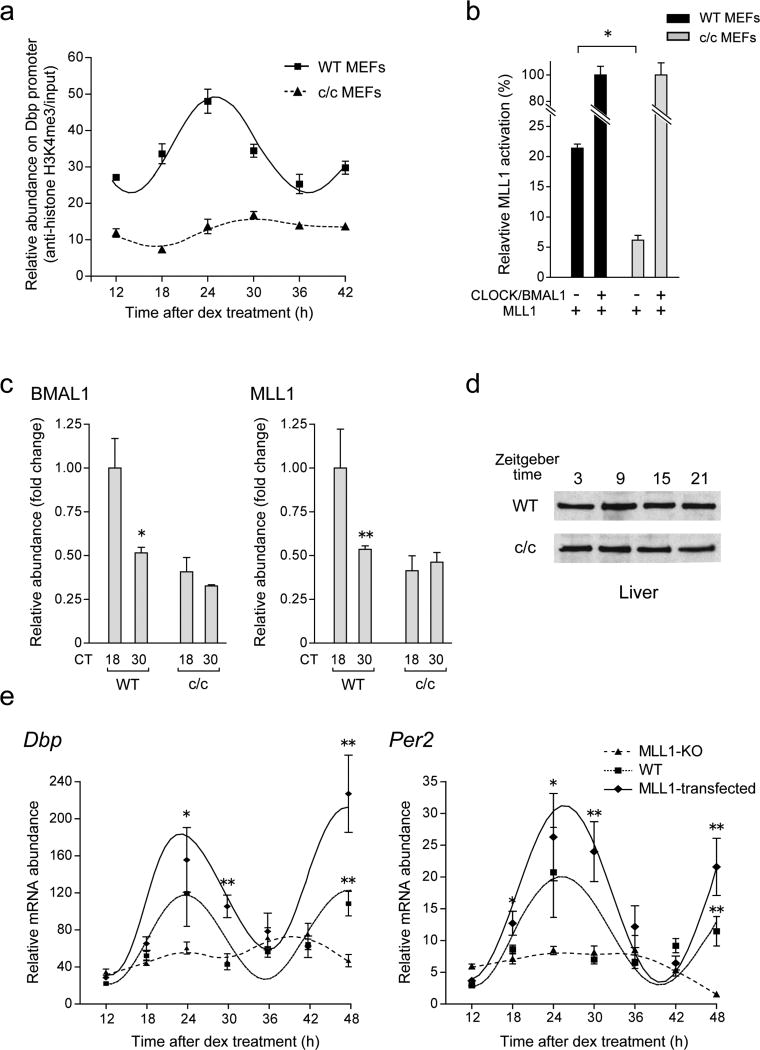
Figure 2. MLL1 governs circadian oscillation of histone modifications and gene expression.
(a) MLL1 deficient MEFs demonstrate impaired CLOCK:BMAL1 mediated transcriptional activities. MLL1 was expressed with or without CLOCK:BMAL1 in MEFs derived from wild type (black) or MLL1-KO (gray) mice. In MLL1-KO MEFs, CLOCK:BMAL1-mediated Dbp and Per2 promoter transactivation was basically abolished, however ectopic expression of MLL1 rescued the expression. (Means ±SEM of seven samples from two independent experiments, *p<0.001). (b) Circadian H3K4me3 is completely impaired in MLL1-KO MEFs. Dexamethasone treated wild type (square) and MLL1-KO (triangle) MEFs were analyzed by ChIP using anti-H3K4me3 (left) or anti-acetylated H3K9/14 antibodies (right). (Means ±SEM of three independent samples, ANOVA p<0.001, and *p<0.05, **p<0.001 at t-test) (c) Circadian rhythmic expression of Dbp and Per2 were dramatically decreased in MLL1-KO MEFs. mRNA was extracted from wild type (black) or MLL1-KO (gray) MEFs after dexamethasone shock and analyzed by quantitative real-time PCR. The amount of highest expression time point was set to 1. (Means ±SEM of three samples, ANOVA p<0.001, and *p<0.05 at t-test)
MLL1 is required for both circadian K4 methylation and K9/K14 acetylation
The above results prompted us to analyze whether MLL1 is required for circadian methylation at H3K4. We performed chromatin immunoprecipitation (ChIP) analyses and monitored H3K4me3 levels on the Dbp promoter at different times of the circadian cycle, comparing WT and MLL1-deficient MEFs. While methylation robustly oscillates in WT MEFs, lack of MLL1 elicited an overall decrease of H3K4me3 levels and a complete abolishment of its oscillation (Fig. 2b, left panel).
H3K4 methylation is often associated with other histone PTMs, such as H3K9/K14 acetylation, both modifications linked to active gene expression18,35. Since we have previously shown that CLOCK directs circadian acetylation at H3K9/K14 (refs. 22, 28), we explored whether MLL1 may contribute to these acetylation events. Interestingly, though MLL1 does not possess HAT activity, oscillation in H3K9/K14 acetylation was virtually abolished in MLL1-null MEFs (Fig. 2b right). Notably, deletion of MLL1 does not have detectable consequence on the global levels of both H3K4me3 and K9/K14 acetylation (Supplementary Fig. 3).
MLL1 is essential for circadian gene expression
The critical role of MLL1 in controlling both H3K4 methylation and K9/K14 acetylation prompted us to analyze its function in circadian gene expression. First, we sought to establish if the oscillatory expression of known circadian genes requires MLL1. We entrained WT or MLL1-deficient MEFs and monitored Dbp and Per2 endogenous expression at various circadian times. Circadian oscillation in mRNAs levels for both genes, readily detectable in WT MEFs, was totally abolished in MLL1-null MEFs (Fig. 2c).
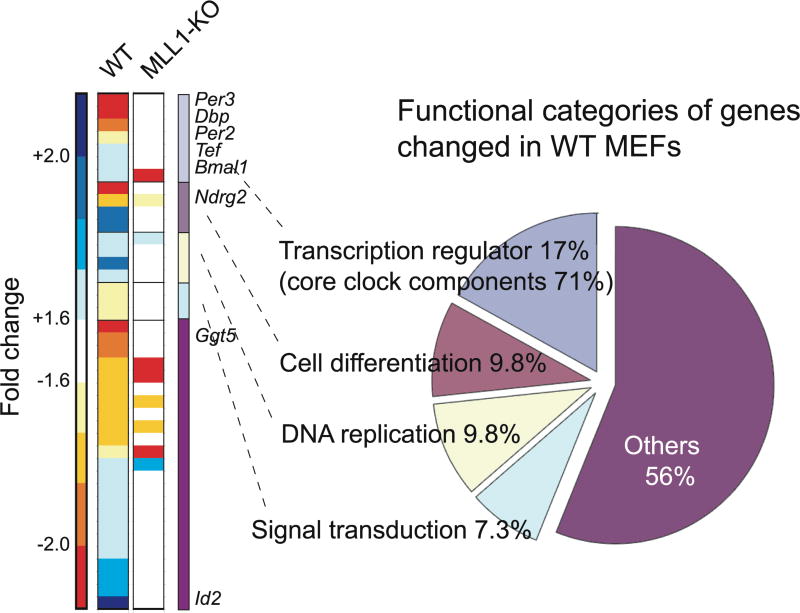
Next, we sought to determine the extent of MLL1 function on circadian gene regulation by performing a comparative microarray analysis of global gene expression levels in WT and MLL1-deficient MEFs, at two different circadian time points (CT18 and CT30). Forty-one genes out of 35557 probes of mouse Gene 1.0 ST array (Affymetrix Inc.) were identified as genes whose expression levels was remarkably changed (>1.6 fold) between CT18 and CT30 in WT MEFs (Fig. 3). These data are in agreement with previous studies that analyzed the proportion of genes oscillating in cultured cells49. Typical clock-regulated genes such as Per2, Per3, Bmal1, or Dbp were found in our analysis (Fig. 3), confirming the accuracy of our approach. The expression of these genes was also confirmed by real time PCR to ensure the accuracy of our array data (Supplementary Fig. 4). Of the eight clock-controlled genes expressed in a circadian manner in WT MEFs, none oscillate in MLL1-null MEFs (for detail see Table 1 and Supplementary Table 1). Several sets of genes were found to be expressed in an oscillatory manner both in WT and MLL1-null MEFs, however, these genes may change their expression responding to serum stimulation, since they are not reported to be regulated by CLOCK:BMAL1. Interestingly, the set of genes that is under MLL1 control highly overlaps with CLOCK-controlled genes. This is demonstrated by the expression profiles in c/c MEFs, in which the CLOCKΔ19 protein, that is unable to interact with MLL1 (see below and Fig. 4a), is expressed. Thus, MLL1 and CLOCK seem to operate in parallel to control the stringency of circadian gene expression. It is noteworthy that MLL1 appears to function quite specifically, since our microarray analysis shows that the expression profile of 93% of the genes does not change more than 2-fold in the MLL1-null cells as compared to WT MEFs (Supplementary Fig. 5). Thus, MLL1 plays a critical role in the control of circadian gene expression.
Figure 3. Global analysis of gene expression profile in MLL1-KO MEFs.
mRNA was extracted from wild type or MLL1-KO MEFs post 18hr or 30hr after serum shock, and microarray analyses were performed using GeneChip Mouse Gene 1.0 ST Array. Forty-one genes out of 35557 probes were identified as genes whose expression levels were significantly changed (>1.6 fold) between two-time point in WT MEFs. Heat map represent the fold changes of 41 genes in WT and MLL1-KO MEFs. Pie chart represent functional categories of genes changing in wild type MEFs. Eight out of 41 gene were listed as typical circadian regulated genes.
Table1.
Eight typical circadian regulated genes listed in microarry analysis
| Gene Description | Mean Intensity ± SEM | Fold change |
Change in MLL1-KO |
|
|---|---|---|---|---|
| CT18 | CT30 | |||
| γ-glutamyltransferase 5 (Ggt5) | 1414 ± 183 | 3150 ± 351 | − 2.23 | 1.04 |
| Period homolog 3 (mPer3) | 524 ± 59 | 1122 ± 160 | − 2.14 | − 1.13 |
| N-myc downstream regulated gene 2 (Ndrg2) | 211 ± 40 | 439 ± 70 | − 2.08 | − 1.40 |
| D site albumin promoter binding protein (Dbp) | 515 ± 24 | 1049 ± 87 | − 2.04 | − 1.22 |
| Drosophila period homolog 2 (mPer2) | 454 ± 26 | 875 ± 26 | − 1.93 | − 1.03 |
| Thyrotroph embryonic factor (Tef) | 1220 ± 53 | 2056 ± 64 | − 1.69 | 1.02 |
| Aryl hydrocarbon receptor nuclear translocator (BMAL1) | 1276 ± 55 | 762 ± 74 | 1.67 | − 1.48 |
| Inhibitor of DNA binding 2 (Id2) | 1478 ± 78 | 858 ± 42 | 1.72 | − 1.15 |
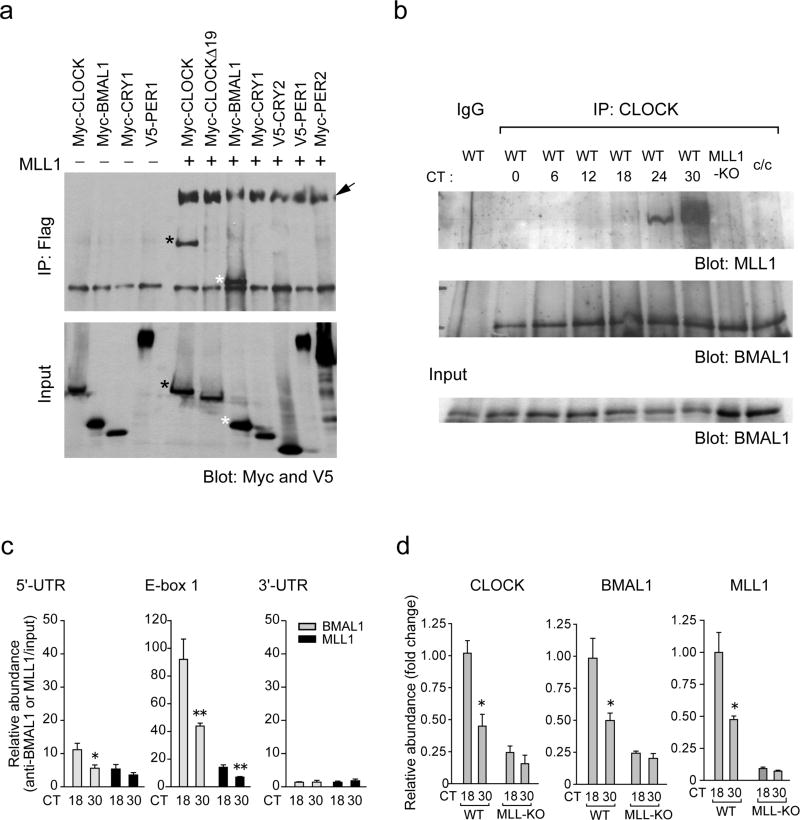
Figure 4. MLL1 interacts with CLOCK and BMAL1 and recruited to chromatin in circadian fashion.
(a) CLOCK and BMAL1, but not CLOCKΔ19, were co-immunoprecipitated with MLL1. 293 cells were transfected with indicated expression vectors together with or without Flag-MLL1-Myc. Flag-tagged MLL1 N-terminal fragments (320 kDa) were immunoprecipitated by Flag-Agar, and coimmunoprecipitated proteins were determined by western analysis using anti-Myc and anti-V5 antibodies. Together with Myc-tagged 180 kDa MLL1 C-terminal fragments (arrow), Myc-BMAL1 (white asterisk) and Myc-CLOCK (black asterisk) proteins were coimmunoprecipitated with MLL1 N-terminal fragments. On the other hand, CLOCKΔ19, CRY1, CRY2, PER1 and PER2 were not co-immunoprecipitated with MLL1. Lower panel shows the results of total cell lysates as an input. (b) Endogenous MLL1 interacts with CLOCK in a circadian manner. MEFs were entrained by serum shock and harvested at various circadian times, then cellular extracts were immunoprecipitated using anti-CLOCK antibody. Co-immunoprecipitated proteins were determined using anti-BMAL1 and anti-MLL1 antibodies. Lower panel shows the results of total cell lysates as an input. (c) MLL1 recruited to E-box proximal region of the Dbp promoter in a circadian manner. ChIP analyses were performed on dual crosslinked MEFs at 18 or 30 hr after dexamethasone synchronization using antibodies against BMAL1 (gray) and MLL1 (black), and quantitative PCR was performed using 5’-UTR, E-box, or 3’-UTR primers. (Means ±SEM of six independent samples. *p<0.05, and **p<0.01) (d) ChIP analyses on the Dbp promoter E-box region in wild type or MLL1-KO MEFs. Analyses were performed on dual crosslinked MEFs at 18 or 30 hr after dexamethasone treatment using antibodies against BMAL1, CLOCK, and MLL1. The amount of highest binding time point (18 hr) in wild type MEFs was set to 1. (Means ±SEM of six independent samples. *p<0.01)
MLL1 interacts with CLOCK and BMAL1, but not with CLOCK-Δ19
The transcriptional enhancing property of MLL1 on CLOCK:BMAL1 (Fig. 1c, d) suggested that these proteins may interact. To test this possibility, we used 293 cells to co-express tagged proteins, N-terminal Flag- and C-terminal Myc-tagged MLL1 with Myc-tagged CLOCK or BMAL1. Immunoprecipitation assays using Flag antibody-coupled agarose beads revealed that CLOCK and BMAL1 physically interact with the N-terminal region of MLL1 (Fig. 4a, black and white asterisk, respectively). Interaction of CLOCK:BMAL1 and MLL1-ΔSET was also observed, indicating that other domains of MLL1 are implicated in the interaction (Supplementary Fig. 6). However, association of these proteins is specific as shown by the lack of interaction with CRY1, CRY2, PER1, or PER2 (Fig. 4a).
To investigate whether native, endogenous MLL1 protein interacts with CLOCK, we entrained MEFs with either serum or dexamethasone and prepared cellular extracts at various circadian times. Immunoprecipitation assays revealed that, while CLOCK and BMAL1 interact at all circadian times, cellular CLOCK interacts with endogenous MLL1 in a specific, circadian manner. Interaction peaks at CT18 when cells are entrained with dexamethasone (not shown) or slightly later (CT24–CT30) when entrained with serum (since serum entrainment has been observed to have a 6–12 hour delay in a circadian phase compared to dexamethasone treatment) (Fig. 4b). These profiles remarkably parallel the increased transcription of clock-controlled genes (see Per2 and Dbp after dexamethasone shock in Fig. 2b; and Per2, Per3, and Dbp after serum shock in Fig. 3) and the peaks of H3K4me3 and H3K9/K14 acetylation (Fig. 2b).
The clock/clock (c/c) mutant mice are arrhythmic due to a 51 amino acids deletion constituting the exon 19 of the CLOCK protein50. Intriguingly, while DNA binding51, BMAL1 dimerization52, and HAT functions28 of the CLOCK-Δ19 protein are normal, its transcriptional activation potential is drastically impaired51. Notably, the functional and molecular reasons for this defect have remained obscure and are yet not understood. We thereby explored whether the CLOCK-Δ19 mutant protein could associate with N-terminal MLL1. Myc-tagged CLOCK-Δ19 fails to coimmunoprecipitate with MLL1, while the same levels of MLL1 precipitated ensure equivalent immunoprecipitation efficacy between samples (Fig. 4a, arrow). This result parallels the lack of H3K9/K14 acetylation oscillation in c/c MEFs22,28, and provides a plausible explanation for the lack of rhythmicity caused by the deletion of exon 19 (ref. 50). One interpretation of these findings is that interaction with MLL1 may be critical to recruit CLOCK to chromatin and to consequent transcription activation.
Recruitment to chromatin, but not expression, of MLL1 is circadian
Oscillation in H3K4me3 could be brought about by a circadian expression of MLL1. To investigate this question, we prepared RNA and protein extracts from mouse liver at various Zeitgeber times (ZT). Both transcript and protein levels for MLL1 do not oscillate throughout the circadian cycle and remain remarkably constant (Supplementary Fig. 7a, b). The levels of Taspase1, an endopeptidase that specifically cleaves MLL1 (ref. 53), Wdr5, a component of the MLL1 complex which preferentially associates with H3K4me2 (ref. 46), as well as other components of the MLL1 complex (not shown), remain constant and display no circadian expression (Supplementary Fig. 7a).
The circadian H3K4me3 (Fig. 1b, 2b), and the cyclic association of MLL1 with CLOCK (Fig. 4b), begged the question of whether MLL1 would be recruited to target promoters in a circadian manner. To test this possibility, we performed dual cross-linking ChIP analyses using combinations of anti-CLOCK, anti-BMAL1, and anti-MLL1 antibodies. As previously reported, both CLOCK and BMAL1 are recruited to the E-box regions of Dbp and Per2 in a time-dependent manner (Fig. 4c). It is notable that MLL1 is recruited to the same E-box regions as CLOCK and BMAL1, following the same circadian timing, being high at CT18 and low at CT30 (Fig. 4c). This result is in keeping with the circadian interaction of CLOCK and MLL1 native proteins.
These investigations lead to an important question. The dependence on MLL1 for transcriptional activation by CLOCK:BMAL1 (Fig. 2a) and for H3K9/K14 acetylation (Fig. 2b), prompted us to analyze whether efficient and circadian recruiting to chromatin of CLOCK:BMAL1 would depend on MLL1. We have found that the efficacy of recruitment of CLOCK and BMAL1 on E-box sequences is significantly inhibited in the absence of MLL1. Importantly, rhythmicity of recruitment is also affected in MLL1-null MEFs (Fig. 4d). These results indicate that MLL1 directly influences CLOCK:BMAL1 recruiting to chromatin and circadian function.
MLL1 function and chromatin recruitment are clock-controlled
The role of MLL1 in CLOCK:BMAL1 function and recruitment (Fig. 2a, and 4d), and the physical interaction between CLOCK and MLL1 (Fig. 4a), raised the possibility that MLL1 function and recruitment to chromatin could be controlled by the circadian clock. This would parallel the circadian association of the two native CLOCK and MLL1 proteins (Fig. 4b), whose expression is non-oscillatory27,54 (Supplementary Fig. 7a, b). To address this question we first analyzed the levels of H3K4me3 at the level of the Dbp promoter in MEFs from c/c mice, which have a disrupted circadian clock55. In contrast to WT MEFs, rhythmic methylation is abolished in c/c MEFs (Fig. 5a). Furthermore, ectopic expression assays demonstrated that the transcriptional activating function of MLL1 (Fig. 1c, 2a) is drastically decreased in c/c MEFs, indicating that MLL1 alone is not sufficient for activation of clock-controlled promoters (Fig. 5b). Co-expression of CLOCK:BMAL1 restored MLL1-mediated stimulation at equivalent levels in WT and c/c MEFs (Fig. 5b). Finally, by ChIP analysis on the Dbp promoter, we show that efficient and cyclic MLL1 recruitment to chromatin is abolished in c/c MEFs (Fig. 5c), despite the normal levels of MLL1 protein (Fig. 5d) and of the CLOCK-Δ19 protein in these cells. Thus, MLL1 function and recruitment to chromatin requires a functional CLOCK protein.
Figure 5. Impaired H3K4me3 and less recruitment of BMAL1 and MLL1 proteins in c/c mutant MEFs.
(a) Rhythmic H3K4me3 was completely abolished in c/c mutant MEFs. ChIP analyses were performed on the Dbp promoter E-box region in wild-type (square) or c/c mutant (triangle) MEFs at each time point post dexamethasone shock. (b) Luciferase reporter gene assay using the Dbp promoter in c/c mutant MEFs. MLL1 was transiently transfected with or without CLOCK:BMAL1 in wild type (black) or c/c mutant (gray) MEFs, and luciferase activity was measured post 48 hr. Relative light units obtained from each CLOCK:BMAL1 and MLL1 transfected MEFs were set to 100 (%). (Means ±SEM of four independent samples. *p<0.001) (c) Circadian recruitment of BMAL1 and MLL1 on the Dbp promoter E-box regions were abolished in c/c mutant MEFs. Dexamethasone shocked and dual crosslinked wild-type and c/c mutant MEFs were analyzed by ChIP using anti-BMAL1 (left) or anti-MLL1 (right) antibodies. (Means ±SEM of three independent samples. *p<0.05, **p=0.0512) (d) MLL1 protein levels of MLL1 do not change in the liver from c/c mice. Proteins were extracted from each time point of liver from WT or c/c mutant mice and MLL1 expression levels were detected using anti-MLL antibodies. (e) Rescue of circadian mRNA expression of clock genes by stably expressing MLL1 in MLL1-KO MEFs. mRNA was extracted from wild type (square), Flag-MLL1 stably expressed MLL1-KO (diamond), or mock transfected MLL1-KO (triangle) MEFs after dexamethasone shock and Dbp and Per2 expression levels were analyzed by quantitative real-time PCR. (Means ±SEM of four independent samples. *p<0.05, **p<0.001)
Rescue of MLL1 function restores circadian gene expression
To establish whether MLL1 is necessary for circadian gene transcription, we monitored and compared the endogenous expression of Per2 and Dbp along the circadian cycle in MEFs derived from WT and MLL1-null mice littermates. As previously described (Fig. 2c), circadian gene expression is totally abolished in MLL1-deficient MEFs. Next we wanted to determine whether this drastic effect on gene expression is due to the lack of MLL1 or to other ill-defined genetic or epigenetic defects of the MLL1-null MEFs. Thus, we generated cell lines expressing either GFP (Mock) or MLL1, following a strategy previously validated22. Rescue of MLL1 fully restored cyclic gene expression of both Dbp and Per2 genes (Fig. 5e).
DISCUSSION
Enzymes involved in chromatin remodeling exert their coordinated activity to govern nuclear events in a timely fashion. A number of cases of combined histone modifications, which could represent examples of an ‘indexing code’37,56, appear to correspond to distinct activating or silencing states of chromatin18,35. The control exerted by the circadian machinery on a significant portion of the epigenome constitutes an exquisite paradigm of wide regulation by cyclic, time-controlled activities of chromatin remodeling.
Previous work has established that CLOCK, a critical regulator of circadian transcription51, is a HAT28 and is present on circadian promoters in a nuclear complex with its dimerization partner BMAL1 and the NAD+-dependent deacetylase SIRT1 (ref. 22). Here we have demonstrated that the histone methyltransferase MLL1 is recruited to circadian promoters in a cyclic manner (Fig. 4b). MLL1 and CLOCK physically interact at specific circadian times, paralleling the cyclic peaks of transcription. Our results suggest a scenario in which MLL1 favor the recruitment of CLOCK:BMAL1 to chromatin, an event that parallels the cyclic and interdependent acetylation/methylation of the H3 tail (Fig. 2b). MLL1 appears to be implicated in generating a chromatin state permissive for circadian transcription. Based on the dynamic, cyclic CLOCK-MLL1 interaction (Fig. 4b), it is conceivable that additional and diverse nuclear components may be recruited at various circadian times and in different cell types. It will be important to decipher whether these protein-protein interactions may be significantly different in SCN neurons versus peripheral, metabolic tissues.
A compelling aspect of this study is the clock-regulated interdependence of histone modifications on the H3 tail. We provide a first view of how the circadian machinery operates in order to promote rhythmic recruitment of specialized enzymes to chromatin. In this respect, MLL1 plays a critical role as it directs the acetylation at K9/K14 and the efficacy of CLOCK recruitment (Fig. 5a, 5d). In exerting this activity, MLL1 requires a functional CLOCK (Fig. 5b, d), a finding possibly related to the recently reported increase in MLL1 enzymatic activity in vitro on a peptide previously acetylated on H3K9 (ref. 57).
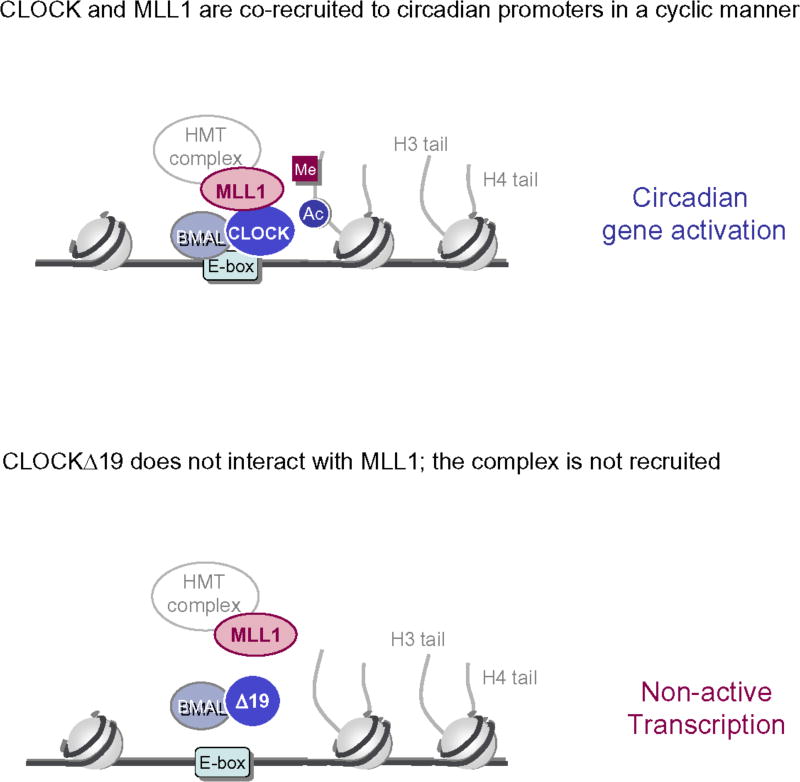
The interaction of MLL1 with CLOCK is of interest for various reasons. First, it is dynamic, following the circadian cycle (Fig. 3b) and being controlled by the clock (Fig. 5b, d). Second, it reveals a helper function of MLL1 on favouring coordinated acetylation (Fig. 2b). Finally, it provides an attractive interpretation of the mechanisms of CLOCK recruitment to circadian gene promoters (Fig. 4c). Related to this last notion, the lack of interaction between MLL1 and the CLOCKΔ19 mutant protein appears of special interest (Fig. 3a). The mutant c/c mice have been used extensively since they display a severe circadian phenotype50, although the molecular reasons responsible for this phenotype have been elusive. The mutant protein CLOCKΔ19 is expressed at normal levels in these animals50, and was reported to have normal dimerization potential, DNA binding and HAT activity28,51,52. We have demonstrated that MLL1 is not able to associate with CLOCKΔ19, and thereby lack of transcriptional activation appears to be caused by an impaired recruitment of the mutant protein (see Figs. 5d, 6).
Figure 6.
CLOCK and MLL1 physically interact at the level of clock-controlled promoters (with E-boxes). Together induce PTMs on histone N-terminal tails associate to circadian transcriptional activation. The mutant protein CLOCKΔ19 fails to associate with MLL1 (lower figure), providing a molecular explanation of its impaired activation potential. As MLL1 and CLOCKΔ19 do not interact, there is also lack of PTMs on the H3 tail, leading to lack of circadian transcription.
Previous studies have shown that acute induction of Per1 and Per2 genes by a light pulse is normal in the SCN of CLOCKΔ19 mice58, this despite circadian expression of core clock genes is drastically impaired in constant darkness51. Likewise, MEFs from c/c mice show no cyclic expression of clock genes, but display the acute response of gene expression after serum shock55. We report a dramatic reduction of circadian regulated transcripts in MLL1-deficient MEFs (Figs. 2c, 3), although dexamethasone-induced early response seems to be normal. Indeed, we have observed acute induction of Per2 expression 2hr post dexamethasone stimulation (Supplementary Fig. 8). The first wave of transcriptional activation in response to a mitogenic or hormonal stimulus is thought to be mediated by CREB or other stimulus-responsive transcription factors59,60, rather than CLOCK:BMAL1. Thus, it would appear that MLL1 contributes in an essential manner to establishing and maintaining oscillation in cooperation with CLOCK:BMAL1.
MLL1 is present in chromatin complexes with a number of important regulators, including Taspase1, Wdr5 and Rbbp5 (refs. 46, 47). We report that the expression of these proteins is not cyclic (Supplementary Fig. 7), and future studies will explore if and how these elements may contribute to the circadian function of MLL1 and CLOCK. It would be of interest to unravel the timing of recruitment of all these elements to circadian promoters. These analyses could provide important information on their potential role in circadian control.
The role of MLL1 in a number of critical gene expression programs, including development, neurogenesis and cancer44,61–63, also begs the question of how these processes may interplay with the regulation exerted by the circadian machinery. Some relevant examples in this sense have already been reported62,64, providing fascinating routes of future exploration. We hypothesize that through its specific interaction with selected regulators, MLL1 may operate within unique transcriptional systems. Here we have uncovered one of them: MLL1 and CLOCK interact and coordinately control CCGs. Our findings underscore the critical function that this highly specific, clock-controlled interaction between two chromatin regulators has in circadian control. Indeed, the role of MLL1 and H3K4 methylation is central as it establishes a chromatin permissive state required for circadian gene expression.
ONLINE METHODS
Plasmids
N-terminal Flag-tagged MLL1/pCXN2 and MLL1 mutant with SET domain deletion (Flag-MLL1-ΔSET/pCXN2) are kind gifts from Dr. J Hess. N-terminal Flag and C-terminal Myc -tagged MLL1 (Flag-MLL1-Myc/ pCXN2) was kind gifts from Dr J. Hsieh. The N-terminal Myc-tagged plasmids (mCLOCK, mCLOCKΔ19, mBMAL1, mCRY1, and mPER2) and V5-tagged plasmids (mCRY2, and mPER1) have been described previously1. mDbp promoter fused to luciferase (mDbp/pGL3) was kind gift from Dr. Yagita. mPer2 promoter-luciferase (mPer2/pGL3) has been described previously2.
Antibodies
Antibodies against H3K4me2 and me3 (ab7766 and ab8580) are from Abcam, Myc and H3K4me1 (07–436) are from Millipore, CLOCK was from Santa Cruz Biotechnology, Flag (M2) was from Sigma, and acetyl-histone H3K9/K14 was from Cell Signaling Technology. Antibodies against mBMAL1 was described earlier3. Antibodies against N-terminal 320 kDa fragment of MLL1 was from Bethyl Laboratories and C-terminal 180 kDa was kind gift from Dr. R. Roeder.
Cell Culture
HEK293 cells were grown in DMEM (4.5 g/l glucose) supplemented with 10% (vol/vol) newborn calf serum (NCS) and antibiotics. MEFs from c/c mutant and MLL1 knockout mice were cultured in DMEM supplemented with 10% FBS or with 2.5% NCS and 7.5% FBS respectively.
Luciferase Reporter Gene Assay
293 cells or MEFs were seeded as monolayer in 24-well plates. Semi-confluent cells (70–80%) were transfected with 25 ng of luciferase reporter and β-galactosidase reporter genes respectively, together with or without 50 ng of CLOCK and BMAL1 or 500 ng of MLL1 by using BioT (Bioland) (1.5 µl per 1 µg DNA). The total amount of applied DNA per well was adjusted by adding pcDNA3 vector. Amount of each transfected cDNAs was determined by normalizing with molecular sizes and promoter strength of each vector. Luciferase activities were measured 48 hr post transfection as described earlier3.
RNA Extraction and GeneChip Analyses
MEFs from wild type and MLL1 knockout mice cultured in DMEM supplemented with 2.5% NCS and 7.5% FBS were plated on 14.5 cm dish and grown in same media for 6 days. In this process, each MEFs were leave at confluent for at least 4 days. For serum synchronization, cells were treated with DMEM supplemented with 50% horse serum (this time refer to t=0) for 2hr and then kept in normal medium for 16hr (t=18) or 28hr (t=30). For each time point, we prepared three dishes to perform in triplicate, and isolated total RNA samples were processed as recommended by Affymetrix, Inc. (Transcript Sense Target Labeling Assay Manual, available at www.affymetrix.com.). Briefly, total RNA was extracted using TRIzol (Invitrogen), and passed through a PureLink RNA mini kit column (Invitrogen) for further clean up. Probe synthesis and chip hybridization were performed at the UCI DNA Microarray Core Facility. Hundred nanograms of total RNA per sample were used as a template to obtained cDNA using the GeneChip® WT cDNA synthesis Kit (Affymetrix, Inc.). Mouse Gene 1.0 ST array (Affymetrix, Inc.) was used to characterize the molecular features of circadian dependent gene expression in WT and MLL1-KO MEFs. Cyber-T statistics program (http://cybert.microarray.ics.uci.edu/) was used to identify the genes of our interest. Genes were considered as increased or decreased, if the change between two time points is above 1.6 fold (with P < 0.0001). This value was used because in MEFs, observed amplitude changes in microarray analyses were generally low4, 5. For further analyses, GeneCodis 2.0 sowtware (http://genecodis.dacya.ucm.es/) was used to monitor co-occurring annotations such as in KEGG pathways.
Chromatin Immunoprecipitation (ChIP) Assays
Conventional ChIP assay was used for histones from MEFs. For non-histone proteins, dual crosslinking ChIP assay was used as described previously with some minor modifications. Briefly, after 15 minutes of 100nM Dexamethasone treatment, cells were washed two times with room temperature PBS, and incubated with normal media for indicated hours. To collect samples, cells were washed two times with PBS and then 1 mM MgCl2/PBS was added. Disuccinimidyl Glutalate (DSG [Pierce]) was added to a final concentration of 2 mM for crosslinking. After 45 min incubation at room temperature, formaldehyde was added to a final concentration of 1%(v/v) and cells were incubated for 10 min. After dual crosslinking, glycine was added to a final concentration of 0.1 M and incubated for 10 min to quench formaldehyde out. After harvesting, cells were lysed in 1.5 ml of icecold cell lysis buffer (50 mM Tris/HCl [pH 8.0], 2 mM EDTA, 0.1% NP-40, 10% Glycerol and Protease inhibitor cocktail [Roche]) for 15 min on ice. Nuclei were precipitated by centrifugation (800 g for 5 min), and resuspended in 0.5 ml of ice-cold SDS lysis buffer (50 mM Tris/HCl [pH 8.0], 10 mM EDTA [pH 8.0], 1% SDS, and Protease inhibitor cocktail [Roche]), and incubated on ice for 15 min. Sonication was done to obtain DNA fragments 200–500 bp in length.
Quantitative Real-Time RT-PCR
Each quantitative real-time RT-PCR was performed using the Chromo4 real time detection system (BIO-RAD). The PCR primers for mPer2 and mDbp mRNA were describe previously6. PCR primers for MLL1, Wdr5, and Taspase were designed as follow. MLL1-Sense ATCCTCTCAGACCCATCTGTGT, MLL1-Antisense GTAGGAGGTCTTCCTCTCTTC, Wdr5-Sense ATCAGCCACACAGAGCAAGCC, Wdr5-Antisense CCACGCTACATCAGATATTCCCAG, Taspase-Sense CACAGCTGTGAGTACCTCAGGATG, and Taspase-Antisense CCACAAGCAGTGTCTGTTTATCTTGG. For a 20 µl PCR, 50 ng of cDNA template was mixed with the primers to final concentrations of 200 nM and 10 µl of iQ SYBR Green Supermix (BIO-RAD). The reaction was first incubated at 95 °C for 3 min, followed by 40 cycles at 95°C for 30 sec and 60°C for 1 min.
Supplementary Material
Acknowledgments
We thank Dr. James Hsieh (Washington University) for the MEFs derived from the MLL1-null mice and the Flag-MLL1-Myc cDNA, Dr. Robert Roeder (Rockefeller University) for MLL1 (C-terminal) antibody and MLL1 fragment sequences, Dr. Kazuhiro Yagita (Osaka University) for the mDbp promoter-Luciferase, Dr. David G. Skalnik (Indiana University) for Flag-hSet1A cDNA, and Dr. Jay Hess (University of Pennsylvania) for providing the Flag-MLL1 and Flag-MLL1DSET cDNA. We thank all members of the Sassone-Corsi laboratory for help, reagents and discussions. This work was supported in part by grants from the Japan Society for the Promotion of Science (JSPS), Postdoctoral Fellowships for Research Abroad to S. K., and from the National Institute of Health (R01-GM081634 and R21-AG033888), Inserm (France) and Sirtris Pharmaceutical Inc. to P. S.-C.
Footnotes
Author Contributions
S. K. conceived the project, designed and conducted the experiments, and wrote the manuscript. P.S.-C. conceived the project, provided conceptual support and contributed to writing the manuscript.
References
- 1.Dunlap JC, Loros JJ, Liu Y, Crosthwaite SK. Eukaryotic circadian systems: cycles in common. Genes Cells. 1999;4:1–10. doi: 10.1046/j.1365-2443.1999.00239.x. [DOI] [PubMed] [Google Scholar]
- 2.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 3.Bell-Pedersen D, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–56. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–22. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 5.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–61. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 6.Willis GL. Parkinson's disease as a neuroendocrine disorder of circadian function: dopamine-melatonin imbalance and the visual system in the genesis and progression of the degenerative process. Rev Neurosci. 2008;19:245–316. doi: 10.1515/revneuro.2008.19.4-5.245. [DOI] [PubMed] [Google Scholar]
- 7.Shaw E, Tofler GH. Circadian rhythm and cardiovascular disease. Curr Atheroscler Rep. 2009;11:289–95. doi: 10.1007/s11883-009-0044-4. [DOI] [PubMed] [Google Scholar]
- 8.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–96. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 9.Akhtar RA, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–50. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 10.Duffield GE, et al. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–7. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 11.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 12.Loboda A, et al. Diurnal variation of the human adipose transcriptome and the link to metabolic disease. BMC Med Genomics. 2009;2:7. doi: 10.1186/1755-8794-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ptitsyn AA, Zvonic S, Gimble JM. Digital signal processing reveals circadian baseline oscillation in majority of mammalian genes. PLoS Comput Biol. 2007;3:e120. doi: 10.1371/journal.pcbi.0030120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–90. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 15.King DP, Takahashi JS. Molecular genetics of circadian rhythms in mammals. Annu Rev Neurosci. 2000;23:713–42. doi: 10.1146/annurev.neuro.23.1.713. [DOI] [PubMed] [Google Scholar]
- 16.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–15. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 17.Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–48. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 18.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–94. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckel-Mahan K, Sassone-Corsi P. Metabolism control by the circadian clock and vice versa. Nat Struct Mol Biol. 2009;16:462–7. doi: 10.1038/nsmb.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masri S, Sassone-Corsi P. Plasticity and specificity of the circadian epigenome. Nat Neurosci. 2010 doi: 10.1038/nn.2668. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eissenberg JC, Elgin SC. Marking time. Nat Genet. 2006;38:276–7. doi: 10.1038/ng0306-276. [DOI] [PubMed] [Google Scholar]
- 22.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belden WJ, Loros JJ, Dunlap JC. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell. 2007;25:587–600. doi: 10.1016/j.molcel.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci. 2000;3:1241–7. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- 25.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–82. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 26.Curtis AM, et al. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J Biol Chem. 2004;279:7091–7. doi: 10.1074/jbc.M311973200. [DOI] [PubMed] [Google Scholar]
- 27.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–74. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 28.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 29.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 30.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–7. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–4. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirayama J, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–90. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 33.Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34:483–90. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–71. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 35.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 36.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 37.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–74. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–49. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 39.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–18. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 40.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–11. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 41.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 42.Schneider R, et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–7. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 43.Dou Y, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–85. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 44.Milne TA, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–17. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 45.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–9. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 46.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–72. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 47.Ansari KI, Mishra BP, Mandal SS. MLL histone methylases in gene expression, hormone signaling and cell cycle. Front Biosci. 2009;14:3483–95. doi: 10.2741/3466. [DOI] [PubMed] [Google Scholar]
- 48.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 49.Delaunay F, Laudet V. Circadian clock and microarrays: mammalian genome gets rhythm. Trends Genet. 2002;18:595–7. doi: 10.1016/s0168-9525(02)02794-4. [DOI] [PubMed] [Google Scholar]
- 50.King DP, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–53. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gekakis N, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–9. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 52.Zhao WN, et al. CIPC is a mammalian circadian clock protein without invertebrate homologues. Nat Cell Biol. 2007;9:268–75. doi: 10.1038/ncb1539. [DOI] [PubMed] [Google Scholar]
- 53.Hsieh JJ, Cheng EH, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115:293–303. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 54.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–67. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 55.Pando MP, Morse D, Cermakian N, Sassone-Corsi P. Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell. 2002;110:107–17. doi: 10.1016/s0092-8674(02)00803-6. [DOI] [PubMed] [Google Scholar]
- 56.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 57.Southall SM, Wong PS, Odho Z, Roe SM, Wilson JR. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell. 2009;33:181–91. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 58.Vitaterna MH, et al. The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc Natl Acad Sci U S A. 2006;103:9327–32. doi: 10.1073/pnas.0603601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ginty DD, et al. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science. 1993;260:238–41. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 60.Shiromani PJ, Schwartz WJ. Towards a molecular biology of the circadian clock and sleep of mammals. Adv Neuroimmunol. 1995;5:217–30. doi: 10.1016/0960-5428(95)00011-p. [DOI] [PubMed] [Google Scholar]
- 61.Canaani E, et al. ALL-1/MLL1, a homologue of Drosophila TRITHORAX, modifies chromatin and is directly involved in infant acute leukaemia. Br J Cancer. 2004;90:756–60. doi: 10.1038/sj.bjc.6601639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–7. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 63.Lim DA, et al. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–33. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu H, Takeda S, Cheng EH, Hsieh JJ. Biphasic MLL takes helm at cell cycle control: implications in human mixed lineage leukemia. Cell Cycle. 2008;7:428–35. doi: 10.4161/cc.7.4.5426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.