Abstract
Plant genetic transformation heavily relies on the bacterial pathogen Agrobacterium tumefaciens as a powerful tool to deliver genes of interest into a host plant. Inside the plant nucleus, the transferred DNA is capable of integrating into the plant genome for inheritance to the next generation (i.e. stable transformation). Alternatively, the foreign DNA can transiently remain in the nucleus without integrating into the genome but still be transcribed to produce desirable gene products (i.e. transient transformation). From the discovery of A. tumefaciens to its wide application in plant biotechnology, numerous aspects of the interaction between A. tumefaciens and plants have been elucidated. This article aims to provide a comprehensive review of the biology and the applications of Agrobacterium-mediated plant transformation, which may be useful for both microbiologists and plant biologists who desire a better understanding of plant transformation, protein expression in plants, and plant-microbe interaction.
INTRODUCTION
Agrobacterium tumefaciens is a soil phytopathogen that naturally infects plant wound sites and causes crown gall disease via delivery of transferred (T)-DNA from bacterial cells into host plant cells through a bacterial type IV secretion system (T4SS). Through the advancement and innovation of molecular biology technology during the past few decades, various important bacterial and plant genes involved in tumorigenesis were identified. With the help of more comprehensive knowledge of how A. tumefaciens interacts with host cells, A. tumefaciens has become the most popular plant transformation tool to date. Any gene of interest can now easily be used to replace the oncogenes in the T-DNA region of various types of binary vectors to perform plant genetic transformation with A. tumefaciens. Arabidopsis, the most-studied model plant with powerful genetic and genomic resources, is readily transformable by A. tumefaciens for stable and transient transformation in several ecotypes tested, although variable transformation efficiencies in different accessions were observed.
The Agrobacterium-plant interaction is a complex process that can be divided into several functional steps, which have been reviewed extensively (Bhattacharya et al., 2010; Gelvin, 2010b, 2012; Lacroix and Citovsky, 2013; Pitzschke, 2013; Christie et al., 2014). In this article, we summarize the major achievements in the field in two main areas: first, the key genes and molecular events of Agrobacterium-plant interactions; and second, current status and methods of Agrobacterium-mediated stable and transient transformation methods. The aim of this article is to provide an overview for scientists with microbiology or plant background to have a better understanding for the other side, which could help them pursue their research interests in a more comprehensive way.
THE BIOLOGY OF AGROBACTERIUM-PLANT INTERACTIONS AND T-DNA TRANSFORMATION
The initial research interest in the genus Agrobacterium resulted from the search for the causative agent of plant diseases including crown gall, cane gall, and hairy root. Agrobacterium tumefaciens harboring the tumor-inducing (Ti) plasmid induces galls on roots and crowns of numerous dicot angiosperm species and some gymnosperms; whereas Agrobacterium rhizogenes harboring the root-inducing (Ri) plasmid induces abnormal root production on host plants. The neoplastic tumor-like cell growth is induced by expression of the oncogenes residing in the transferred-DNA (TDNA) transported from these bacteria into the plant nucleus and integrated into the plant genome.
The A. tumefaciens-mediated plant genetic transformation process requires the presence of two genetic components located on the bacterial Ti-plasmid. The first essential component is the T-DNA, defined by conserved 25-base pair imperfect repeats at the ends of the T-region called border sequences. The second is the virulence (vir) region, which is composed of at least seven major loci (virA, virB, virC, virD, virE, virF, and virG) encoding components of the bacterial protein machinery mediating T-DNA processing and transfer. The VirA and VirG proteins are two-component regulators that activate the expression of other vir genes on the Ti-plasmid. The VirB, VirC, VirD, VirE and perhaps VirF are involved in the processing, transfer, and integration of the TDNA from A. tumefaciens into a plant cell. Figure 1 shows the major steps of the Agrobacterium-mediated plant transformation process. The current knowledge and recent findings regarding the key events of Agrobacterium-mediated plant transformation process are reviewed in the following sections. Table 1 summarizes the major transformation steps and known plant factors participating in these steps.
Figure 1.
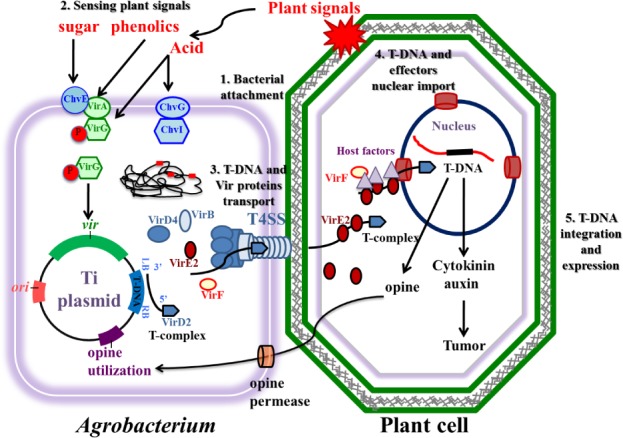
Major steps of the Agrobacterium tumefaciens-mediated plant transformation process.
(1) Attachment of A. tumefaciens to the plant cells. (2) Sensing plant signals by A. tumefaciens and regulation of virulence genes in bacteria following transduction of the sensed signals. (3) Generation and transport of T-DNA and virulence proteins from the bacterial cells into plant cells. (4) Nuclear import of T-DNA and effector proteins in the plant cells. (5) T-DNA integration and expression in the plant genome.
Table 1.
Plant factors involved in Agrobacterium-mediated plant transformation process
| Known or possible functions | Plant factors | AGI locus identifier1 | References |
|---|---|---|---|
| Attachment of A. tumefaciens to plant cells | Vitronectin | N/A | Wagner and Matthysse, 1992 |
| PVN1 (plant vitronectin-like protein 1) | N/A | Zhu et al., 1993; Zhu et al., 1994 | |
| AGP17 (RAT1, arabinogalactan protein) | At2g23120 | Zhu et al., 2003b; Gaspar et al., 2004 | |
| CSLA (RAT4, cellulose synthase-like A9) | At5g03760 | Zhu et al., 2003a, b | |
| MTF1 (Myb family transcription factor) | At2g40970 | Sardesai et al., 2013 | |
| AT14A (similar to integrins) | At3g28290 | Sardesai et al., 2013 | |
| Plant hormone compounds (salicylic acid, SA) | N/A | Anand et al., 2008; Yuan et al., 2007a; Yuan et al., 2007b | |
| Plant signals sensed by A. tumefaciens and/or regulating vir gene expression | Phenolic compounds | N/A | Stachel et al., 1985 |
| Sugar compounds | N/A | Stachel et al., 1985; Ankenbauer and Nester, 1990; Shimoda et al., 1993; Doty et al., 1996; Hu et al., 2012 | |
| HDMBOA (2-hydroxy-4,7-dimethoxybenzoxazin-3-one, maize seedling root exudate) | N/A | Zhang et al., 2000 | |
| Plant hormone compounds (auxin, cytokinin, salicylic acid, and ethylene) | N/A | Liu and Nester, 2006; Anand et al., 2007; Yuan et al., 2007a; Yuan et al., 2007b; Nonaka et al., 2008; Hwang et al., 2010; Hwang et al., 2013b | |
| Acidity | N/A | Li et al., 2002; Liu et al., 2005; Yuan et al., 2007a; Hu et al., 2012; Wu et al., 2012; Heckel et al., 2014 | |
| T-DNA/virulenceprotein transport or initial contact of A. tumefaciens to plants | RTNLB1 (reticulon-like protein B-1) | At4g23630 | Hwang and Gelvin, 2004 |
| RTNLB2 (reticulon-like protein B-2) | At4g11220 | Hwang and Gelvin, 2004 | |
| RTNLB4 (reticulon-like protein B-4) | At5g41600 | Hwang and Gelvin, 2004 | |
| RAB8B (small GTPase) | At3g53610 | Hwang and Gelvin, 2004 | |
| Cytoplasmic trafficking and nuclear import of T-DNA and effector proteins | ROC1 (cyclophilin proteins, peptidyl-prolyl cistrans isomerase CYP1) | At4g38740 | Deng et al., 1998 |
| PP2C (type 2C protein phosphatase, ABI1) | At4g26080 | Tao et al., 2004 | |
| KAPα/IMPA-1 (Karyopherin protein, importin alpha isoform 1 involved in nuclear import) | At3g06720 | Ballas and Citovsky, 1997; Bhattacharjee et al., 2008 | |
| IMPA-2 (importin alpha isoform 2) | At4g16143 | Bhattacharjee et al., 2008; Lee et al., 2008 | |
| IMPA-3 (importin alpha isoform 3) | At4g02150 | Bhattacharjee et al., 2008; Lee et al., 2008 | |
| IMPA-4 (importin alpha isoform 4) | At1g09270 | Bhattacharjee et al., 2008; Lee et al., 2008 | |
| importin β-3 | N/A | Zhu et al., 2003a | |
| VIP1 (VirE2-interacting plant protein 1) | At1g43700 | Tzfira, 2001; Tzfira et al., 2002 | |
| VIP2 (VirE2-interacting plant protein 2) | At5g59710 | Tzfira, 2001; Tzfira et al., 2002; Anand et al., 2007 | |
| MPK3 (mitogen-activated protein kinase 3) | At3g45640 | Djamei et al., 2007 | |
| Microtubules and kinesin | N/A | Zhu et al., 2003a; Salman et al., 2005 | |
| Microfilament and ACT2 (actin gene) | At3g18780 | Zhu et al., 2003a; Yang et al., 2017 | |
| ACT7 (actin gene) | At5g09810 | Zhu et al., 2003a | |
| CAK2M kinase (cyclin-dependent kinase-activating kinases) | N/A | Bako et al., 2003 |
Table 1.
(continued)
| Known or possible functions | Plant factors | AGI locus identifier1 | References |
|---|---|---|---|
| Integration and/or expression of T-DNA | AtLIG4 (DNA ligase IV) | At5g57160 | Ziemienowicz et al., 2000; Friesner and Britt, 2003; van Attikum et al., 2003; Zhu et al., 2003a; Nishizawa-Yokoi et al., 2012; Park et al., 2015 |
| KU80 | At1g48050 | van Attikum et al., 2001; Friesner and Britt, 2003; Gallego et al., 2003; Li et al., 2005b; Nishizawa-Yokoi et al., 2012; Jia et al., 2012; Mestiri et al., 2014; Park et al., 2015 | |
| KU70 | At1g16970 | van Attikum et al., 2001; Li et al., 2005b; Nishizawa-Yokoi et al., 2012; Jia et al., 2012; Mestiri et al., 2014; Park et al., 2015 | |
| MRE11 (meiotic recombination 11) | At5g54260 | van Attikum et al., 2001; Jia et al., 2012; | |
| XRCC1 (homolog of X-ray repair cross complementing 1) | At1g80420 | Mestiri et al., 2014; Park et al., 2015 | |
| XRCC2 (homolog of X-ray repair cross complementing 2) | At5g64520 | Mestiri et al., 2014; Park et al., 2015 | |
| XRCC4 (homolog of X-ray repair cross complementing 4) | At3g23100 | Vaghchhipawala et al., 2012; Park et al., 2015 | |
| XPF/RAD1/UVH1 (ultraviolet hypersensitive 1) | At5g41150 | Nam et al., 1998; Mestiri et al., 2014; Park et al., 2015 | |
| PARP1 (poly(ADP-ribose) polymerases 1) | At2g31320 | Jia et al., 2012; Park et al., 2015 | |
| HTA1 (histone H2A) | At5g54640 | Nam et al., 1999; Mysore et al., 2000a, b; Yi et al., 2002, 2006; Zhu et al., 2003a; Tenea et al., 2009 | |
| HTR11 (histone H3-11) | At5g65350 | Zhu et al., 2003a; Tenea et al., 2009 | |
| VIP1 (VirE2-interacting plant protein 1) | At1g43700 | Tzfira, 2001; Tzfira et al., 2002; Li et al., 2005a; Loyter et al., 2005; Lacroix et al., 2008 | |
| VIP2 (VirE2-interacting plant protein 2) | At5g59710 | Tzfira, 2001; Tzfira et al., 2002; Anand et al., 2007 | |
| HDA1/HDA19 (histone deacetylase 1/histone deacetylase 19) | At4g38130 | Zhu et al., 2003a; Crane and Gelvin, 2007; Gelvin and Kim, 2007 | |
| HDA2/HDT1 (histone deacetylase 2A) | At3g44750 | Zhu et al., 2003a; Crane and Gelvin, 2007; Gelvin and Kim, 2007 | |
| HD2B/HDT2 (histone deacetylase 2B) | At5g22650 | Zhu et al., 2003a; Crane and Gelvin, 2007; Gelvin and Kim, 2007 | |
| SGA1/ASF1B (anti-silencing function 1B) | At5g38110 | Crane and Gelvin, 2007 | |
| FAS1 (fasciata 1/nucleosome/chromatin assembly factor group B/chromatin assembly factor-1 (CAF-1) subunit) | At1g65470 | Endo et al., 2006 | |
| FAS2 (fasciata 2/nucleosome/chromatin assembly factor group B/chromatin assembly factor-1 (CAF-1) subunit) | At5g64630 | Endo et al., 2006 | |
| ASK1/SKP1 (SKP1 homologue 1) | At1g75950 | Schrammeijer et al., 2001; Zaltsman et al., 2010a, b, 2003; Anand et al., 2012; Tzfira et al., 2002, 2004 | |
| ASK2 (SKP-like 2) | At5g42190 | Schrammeijer et al., 2001; Zaltsman et al., 2010a, b, 2003; Anand et al., 2012; Tzfira et al., 2002, 2004 | |
| ASK10 (SKP-like 10) | At3g21860 | Schrammeijer et al., 2001; Zaltsman et al., 2010a, b, 2003; Anand et al., 2012; Tzfira et al., 2002, 2004 | |
| SGT1a (suppressor of the G2 allele of SKP1) | At4g23570 | Anand et al., 2012 | |
| SGT1b/EDM1 (Enhanced downy mildew 1) | At4g11260 | Anand et al., 2012 | |
| VBF (VIP1-binding F-box protein) | At1g56250 | Ditt et al., 2006; Zaltsman et al., 2010a, b, 2013; Wang et al., 2014; Niu et al., 2015 | |
| TEBICHI/POLQ (DNA polymerase theta, Pol θ) | At4g32700 | van Kregten et al., 2016 |
1N/A: not available
Attachment of A. tumefaciens to plant cells
Bacterial attachment to host cells is a crucial early step in disease development for many plant and animal pathogens. The isolation of A. tumefaciens mutants unable to bind to plant cells led to the identification of several chromosomal virulence (chv) genes, chvA, chvB, and pscA, required for the attachment processes. chvA, chvB, and pscA are involved in the synthesis, processing, and export of cyclic β-1,2-glucan and other sugars (Thomashow et al., 1987; Zorreguieta et al., 1988; Cangelosi et al., 1989; O'Connell and Handelsman, 1989). These mutants are either avirulent or highly attenuated in virulence under many inoculation conditions (Douglas et al., 1982; Douglas et al., 1985; Matthysse, 1987). During attachment, the A. tumefaciens cells synthesize cellulose fibrils that entrap large numbers of bacteria at the wounded sites (Matthysse et al., 1981; Matthysse, 1983). The cellulose deficient A. tumefaciens mutant is still able to attach to the carrot cell surface but cannot form aggregates (Matthysse, 1983). These mutants are virulent but are more susceptible to being washed off the plant cells (Matthysse et al., 1981; Matthysse, 1983). Thus, the major role of cellulose fibrils may be in aiding attachment of A. tumefaciens to the plant cell.
Aside from the cellulose and cyclic β-1,2-glucan, A. tumefaciens produces an additional exopolysaccharide, unipolar polysaccharide (UPP), that participates in bacterial attachment (Tomlinson and Fuqua, 2009; Li et al., 2011; Xu et al., 2012; Heindl et al., 2014). The unipolar polysaccharide helps A. tumefaciens cells attach to the plant surfaces in a polar fashion because the UPP is mainly produced at the cell pole. The UPP is rarely observed in A. tumefaciens planktonic cells or in colonies on solid media, suggesting that the production of UPP is a signature of A. tumefaciens permanent attachment (Li et al., 2011; Xu et al., 2012; Xu et al., 2013). The UPP consists of at least two types of sugars, N-acetylglucosamine and N-acetylgalactosamine (Tomlinson and Fuqua, 2009; Xu et al., 2012). The A. tumefaciens UPP biosynthetic mutant uppE is defective in both bacterial attachment and biofilm formation, supporting the idea that the UPP plays an important role in bacterial attachment (Xu et al., 2012; Heindl et al., 2014). Interestingly, while A. tumefaciens attachment to plant tissues often leads to biofilm formation and is required for virulence, A. tumefaciens strains lacking UPP or impaired in biofilm formation remain efficient in T-DNA transfer. Thus, biofilm formation and UPP-mediated bacterial attachment may not play a role during Agrobacterium T-DNA translocation process, or their importance may not be discernible using the current infection as-says under laboratory conditions.
Plant factors are also required for bacterial attachment to the plant cell surface. One such surface factor may belong to a vitronectin protein family. In animal cells, vitronectin, a component of the extracellular matrix, is utilized as a specific receptor by several pathogenic bacterial strains (Paulsson, and Wadstrom, 1990). Human vitronectin and anti-vitronectin antibodies both inhibited the binding of A. tumefaciens to cultured plant cells (Wagner and Matthysse, 1992). Further, A. tumefaciens chvB, pscA, and att mutants that are unable to bind to plant cells exhibit reduced binding to vitronectin (Wagner and Matthysse, 1992). Plant proteins immunologically related to the animal substrate adhesion molecule vitronectin, named PVN1 (plant vitronectin-like protein 1), have been identified in tobacco (Zhu et al., 1993; Zhu et al., 1994). PVNl is localized in the cell wall and may mediate cell adhesion (Zhu et al., 1994). However, whether PVN1 is a receptor for Agrobacterium binding remains to be investigated.
Nam et al. (1997) identified several Arabidopsis ecotypes that are deficient in binding A. tumefaciens and are resistant to A. tumefaciens root mediated transformation. Two of these eco-types, Bl-1 and Petergof, showed lower transient transformation efficiency than a more susceptible ecotype, suggesting that the transformation process was blocked at step(s) prior to integration. Further, microscopic analysis revealed that fewer bacteria bound to the root surface of B1-1 and Petergof than to a highly transformable ecotype such as Aa-0. In addition, Nam et al. (1999) identified three Arabidopsis T-DNA insertion mutants in the ecotype Ws with resistance to Agrobacterium transformation (rat mutants). The root explants of rat1, rat3, and rat4 mutants are deficient in binding A. tumefaciens. RAT1 encodes an arabinogalactan protein (AtAGP17, At2g23120) that is found in the extracellular matrix associated with the plant cell wall (Gaspar et al., 2004). Interestingly, although the Arabidopsis rat1 and rat3 mutants and the Bl-1 and Petergof ecotypes are recalcitrant to A. tumefaciens root-mediated transformation, they are transformed as well as are the corresponding wild-type plants and highly transformable ecotypes using a flower vacuum infiltration method (Mysore et al., 2000a). These results suggested that plant proteins needed for transformation of somatic tissues may not be required for the transformation of the female gametophyte. In addition, a genetic screen to identify Arabidopsis hyper-susceptible to A. tumefaciens transformation (hat) mutants uncovered the heterozygous mutant mtf1-1, in which the mRNA transcript of the MTF1 (myb family transcription factor, At2g40970) gene exhibited more than a 12-fold reduction as compared to wild-type plants (Sardesai et al., 2013). The increased A. tumefaciens attachment to roots and improved stable and transient transformation efficiencies in mtf1-1 suggested that the MTF1 played a negative role in mediating the A. tumefaciens transformation process. Interestingly, the presence of A. tumefaciens-secreted cytokinin caused decreased MTF1 gene expression via the cytokinin response regulator ARR3-mediated signaling pathway. Because gene expression of AT14A (At3g28290), encoding a putative transmembrane receptor for a cell adhesion molecule, was increased in the mtf1 mutant and the at14a mutant showed decreased transformation rates and A. tumefaciens attachment to plant roots, AT14A might be an anchor point for A. tumefaciens binding (Sardesai et al., 2013).
Sensing and regulation of virulence genes of A. tumefaciens in response to plant signals
In nature, wounded plant tissues secrete a wide range of chemical compounds that can function as chemotactic agents to attract A. tumefaciens to the wounded sites for infection in plants. Wounded plants secrete sap with a characteristic acidic pH (5.0 to 5.8) and high content of different phenolic compounds, including lignin and flavonoid precursors. While these compounds can be used to ward off microbial attachment, these conditions stimulate A. tumefaciens virulence gene expression. The best studied and most effective vir gene inducers are monocyclic phenolics such as 3,5-dimethoxy acetophenone (acetosyringone, AS), as shown by induction of vir::lacZ fusions (Stachel et al., 1985). Several other environmental conditions are also important for optimal vir gene induction, including acidity, low phosphate, temperature, and sugars. However, phenolics are the only signal that is absolutely required (Brencic et al., 2003; Gao and Lynn, 2005; Mc-Cullen and Binns, 2006; Lin et al., 2008; Subramoni et al., 2014).
A. tumefaciens uses the VirA/VirG two-component regulatory system to regulate vir gene induction (Stachel et al., 1986; Jin et al., 1990a, 1990b, 1990c). To initiate the signaling pathway, plant phenolics interact either directly or indirectly with the trans-membrane sensory protein VirA. Although two chromosomally encoded proteins, p10 and p21, but not VirA, are able to bind the phenolic signal in vitro (Lee et al., 1992), genetic evidence strongly suggests that VirA directly senses the phenolic compounds required for vir gene activation (Lee et al., 1995; Lee et al., 1996). ChvE, a chromosomally encoded glucose/galactose binding protein, interacts with VirA and enhances vir gene activation by binding to sugars (Shimoda et al., 1993; Doty et al., 1996). A. tumefaciens also uses the ChvE protein to recognize and bind various plant sugars that may help the bacterium to establish its host range (Hu et al., 2012). In addition, the affinity of the ChvE protein for sugar acids is increased at low pH, suggesting a link between the sugar and acidity response in A. tumefaciens (Hu et al., 2012). A. tumefaciens utilizes the ChvG/ChvI two-component system to activate the transcription of virG and induces expression of several other bacterial genes (Mantis and Winans, 1992; Chang and Winans, 1996), including genes involved in chemo-taxis and motility, several chromosome-encoded virulence genes, succinoglycan biosynthesis genes, and the gene cluster encoding the type VI secretion system (T6SS) (Yuan et al., 2007a; Wu et al., 2008; Wu et al., 2012). Recent studies further identified ExoR as a periplasmic negative regulator acting upstream of the ChvG sensor kinase to repress acid-inducible gene expression (Wu et al., 2012; Heckel et al., 2014) and uncovered a role for the T6SS in interbacterial competition activity during the plant colonization process (Ma et al., 2014). These data strongly suggested that these acid-inducible genes may play a role in A. tumefaciens survival and competitive fitness during Agrobacterium-plant interactions.
Because phenolic molecules are bacteriostatic at high concentration, A. tumefaciens also harbors cytochrome P450-like enzymes such as VirH2 to detoxify a phenolic vir gene inducer, ferulic acid (Kalogeraki et al., 1999; Brencic et al., 2003), suggesting that virH may be involved in the detoxification of harmful phenolics secreted by the wounded plants (Kanemoto et al., 1989). On the other hand, plant factors can also act as inhibitors of the A. tumefaciens sensory machinery. A major organic exudate of maize seedling roots, 2-hydroxy-4,7-dimethoxybenzoxazin-3-one (HDMBOA), has been demonstrated to provide innate immunity and hinder successful A. tumefaciens infection of maize plants (Zhang et al., 2000); an o-imidoquinone decomposition intermediate, (3Z)-2,2-dihydroxy-N-(4-methoxy-6-oxocyclohexa-2,4-dienylidene) acetamide, is a specific inhibitor of signal perception in A. tumefaciens (Maresh et al., 2006; Lin et al., 2008). These inhibitors may account for the recalcitrance of some plants to Agrobacterium-mediated transformation.
Several studies revealed that two plant hormones, auxin and cytokinin, not only contribute to tumor growth, but also affect agrobacterial physiology and gene expression (Liu and Nester, 2006; Hwang et al., 2010; Hwang et al., 2013b). It has been shown that IAA, the plant hormone auxin overproduced upon T-DNA integration, affects bacterial growth and inhibits vir gene induction by competing with phenolic inducers for interaction with the VirA protein. It has been proposed that relatively low auxin levels may promote transformation during the initial stages of A. tumefaciens infection, whereas the higher level of auxin produced in the crown gall tissues may prevent further transformation by inhibiting bacterial growth and virulence. In addition to auxin, A. tumefaciens-secreted cytokinin also affects bacterial virulence by regulating vir promoter activity and bacterial growth (Hwang et al., 2010, 2013b). Other studies also indicate that the plant defense molecule salicylic acid (SA) influences the A. tumefaciens infection process by inhibiting the expression of vir genes, bacterial growth, bacterial attachment to plant cells, and virulence (Anand et al., 2008; Yuan et al., 2007a, 2007b). The plant defense roles of SA against bacterial infections are further supported by genetic studies showing that mutant plants with SA overproduction are resistant to A. tumefaciens infection, whereas mutant plants with lower SA accumulation have a higher percentage of tumor formation (Anand et al., 2008; Yuan et al. 2007b; Lee et al., 2009). Another plant hormone, ethylene, can repress vir gene expressions in A. tumefaciens but shows no significant inhibitory effects on bacterial growth and population size (Nonaka et al., 2008). In an Arabidopsis ethylene-insensitive mutant, the A. tumefaciens-mediated transformation efficiency increased (Nonaka et al., 2008). In summary, intricate temporal and spatial regulation and a delicate balance of various plant hormones likely play important roles in modulating A. tumefaciens infection efficiency (Gohlke and Deeken, 2014; Subramoni et al., 2014).
Transport of T-DNA and virulence proteins via type IV secretion system (T4SS)
Virulence gene expression leads to the production of a single-stranded T-DNA, termed the T-strand, which is then transported into the host cell. VirD1 and VirD2 proteins function together as a site- and strand-specific endonuclease which binds to the supercoiled Ti plasmid at the T-DNA borders, relaxes it and nicks the bottom T-DNA strand between the third and fourth bases of the T-DNA borders (Stachel et al., 1986; Yanofsky and Nester, 1986; Albright et al., 1987; Jayaswal et al., 1987; Stachel et al., 1987; Wang et al., 1987; Filichkin and Gelvin, 1993). This process results in the generation of single-strand T-DNA molecules (T-strand) covalently attached to VirD2 at the 5′ end of the T-strand at the right border nick (Herrera-Estrella et al., 1988; Ward and Barnes, 1988; Young and Nester, 1988; Howard et al., 1989). Recently, three VirD2-binding proteins (VBP) from A. tumefaciens were identified by pull-down assays using AS-treated A. tumefaciens crude extract (Guo et al., 2007a). These proteins are functionally redundant, but removal of all three vbp genes in A. tumefaciens affects bacterial virulence on Kalanchoe plants (Guo et al., 2007a). The VBP1 protein not only interacts with the VirD2-bound T-strand, but also interacts with three components of the Type IV secretion system (T4SS), VirD4, VirB4, and VirB11, via two independently binding domains (Guo et al., 2007b). The VBP may form a dimer in the A. tumefaciens cytoplasm and recruit the VirD2-bound T-strand to the T4SS apparatus through interactions with the VirD4 proteins (Guo et al., 2007b; Padavannil et al., 2014).
T-DNA export requires the vir-encoded T4SS for delivery across the bacterial envelope and the plasma membrane of the host plant cell (Zechner et al., 2012; Bhatty et al., 2013; Chandran, 2013; Christie et al., 2014). This A. tumefaciens T4SS consists of 12 proteins (VirB1-B11 and VirD4), which form two functional components: a filamentous T-pilus and a membrane associated transporter complex that is responsible for translocating substrates across both bacterial cell membranes (Alvarez-Martinez and Christie, 2009; Fronzes et al., 2009a; Waksman and Fronzes, 2010; Wallden et al., 2010). This transport complex can be further subdivided into four functional subassemblies: the VirD4 coupling protein as substrate receptor, an inner-membrane translocase (VirB3-B4-B6-B8-B11), an outer-membrane core complex (VirB7-B9-B10), and an extracellular T-pilus (VirB2-B5) (Christie et al., 2014). While it is clear that each of the VirB2-11 and VirD4 proteins is essential for T-DNA and effector transport, it remains unknown whether the extracellular T-pilus is part of the translocation channel.
The VirD4 protein is an integral inner membrane protein with ATPase activity functioning as a coupling protein responsible for presentation of DNA and protein substrates to the VirB transporter. Studies using a transfer DNA immunoprecipitation (TrIP) assay revealed that the transferring T-DNA substrates make close contacts with the VirD4 protein, further supporting the function of VirD4 as the substrate receptor (Cascales and Christie, 2004a). Genetic studies of the virD4 mutants also revealed that DNA substrate binding of VirD4 protein and delivery to VirB11 both activate the ATP hydrolysis activities of VirD4 and VirB11. These signals may cause a conformational change of VirB10 which plays an important role in regulating DNA substrate transfer through the T4SS apparatus opening and assembly (Atmakuri et al., 2004; Cascales and Christie, 2004b; Cascales et al., 2013).
VirB4 is an ATPase and contains a Walker A nucleotide tri-phosphate binding motif essential for virulence (Christie et al., 1989; Fullner et al., 1994). The VirB4 ATPase also directly participates in T-pilus biogenesis. VirB4 can interact with the VirB2 T-pilin and induce changes in the VirB2 membrane topological state via an ATP-dependent mechanism, suggesting that the VirB4 ATPase may function as a dislocase to mediate membrane extraction of pilin monomers during pili biogenesis (Kerr and Christie, 2010; Wallden et al., 2010). In addition, VirB11 may function with VirB4 to coordinate a structural change in VirB2 pilin protein and mediate VirB2 binding to other T4SS components, which in turn affects pilus polymerization (Kerr and Christie, 2010). Accumulating genetic studies and recent structural studies support a model in which VirB4 dimers or homomultimers contribute both structural information for the assembly of a trans-envelope channel competent for DNA transfer and an ATP-dependent activity for configuring this channel as a dedicated export machine (Fronzes et al., 2009a; Durand et al., 2010, 2011; Kerr and Christie, 2010; Li et al., 2012; Pena et al., 2012; Wallden et al., 2012). In addition, the TrIP assay results showed that translocating DNA substrates first interact with the VirD4 receptor, followed by VirB11 ATPase, suggesting VirB11 may function together with VirB4 and deliver the T-DNA substrate to the transfer channel of the T4SS (Cascales and Christie, 2004a; Bhatty et al., 2013; Chandran, 2013; Christie et al., 2014).
Recent electron microscopy imaging has revealed the structure of purified VirB3-10 complex encoded by R388 T4SS, in which VirB3, VirB4, and likely VirB5, VirB6, and VirB8 are part of the inner membrane complex (Low et al., 2014). This result is consistent with a predicted inner-membrane translocase complex consisting the VirB3, VirB4, VirB6, VirB8, and VirB11 based on subcellular localization, protein-protein interaction, and biochemical studies (Christie et al., 2014). This study also revealed the outer membrane complex consisting of VirB7-B9-B10, which is in agreement with previous cryoelectron microscopy and crystallography of purified VirB7, VirB9, and VirB10 proteins encoded from pKM101 (Chandran et al., 2009; Fronzes et al., 2009b). This outer-membrane core complex contains 14 copies each of VirB7, VirB9, and VirB10; and is inserted in both the inner and outer membranes (Fronzes et al., 2009a, 2009b). The structure of the core complex is cylindrical and contains two layers, the inner (I) and outer (O) layers (Fronzes et al., 2009b; Wallden et al., 2012; Bhatty et al., 2013). Upon sensing ATP energy utilization by the VirD4 and VirB11 ATPases, the VirB10 protein conformation changes and may help regulate the O layer hole opening to allow substrate transfer through the T4SS channel (Cascales and Christie, 2004b). While the majority of T4SS components can be assembled into subcomplexes when expressed in a heterologous E. coli system, an alpha-crystallin-type small heat shock protein, HspL was shown to be important for stability of several VirB proteins and efficient T-DNA transfer and tumorigenesis (Tsai et al., 2009). HspL expression is induced by AS in response to VirB protein expression and functions as a VirB8 chaperone (Tsai et al., 2009, 2010). Moreover, overexpression of HspL in A. tumefaciens enhanced transient transformation efficiencies of both susceptible and recalcitrant Arabidopsis ecotypes. A. tumefaciens virulence during heat shock was also considerably improved by over-expression of the HspL, revealing the importance of the VirB8 chaperone HspL during A. tumefaciens infections (Hwang et al., 2015).
The extracellular T-pilus consists of major subunit VirB2 and minor subunit VirB5, which is located at the pilus tip (Lai and Kado, 1998; Schmidt-Eisenlohr et al., 1999; Aly and Baron, 2007). Genetic and environmental studies show that each virB gene is required for T-pilus biogenesis; however, only the virB2 gene is required for VirB2 pro-pilin processing and peptide cyclization (Eisenbrandt et al., 1999; Lai et al., 2000, 2002). Non-polar virB mutants abolish T-pilin transport and T-pilus biogenesis, and support a model in which the VirB-specific transporter is not only used for translocating the T-complex, but also for exporting cyclic T-pilin subunits to the bacterial cell surface, perhaps as a scaffold to facilitate efficient assembly of the T-pilus (Lai and Kado, 2000; Christie et al., 2014). However, several amino acid substitutions in the T4SS components VirB6, VirB9, VirB10, and VirB11 have been demonstrated to block the polymerization of VirB2 pilin to form the extracellular T-pilus but do not significantly affect substrate transfer. These “uncoupling” mutations still require intracellular VirB2 for substrate transfer (Zhou and Christie, 1997; Sagulenko et al., 2001; Jakubowski et al., 2003, 2005, 2009; Banta et al., 2011; Garza and Christie, 2013). Wu et al. (Wu et al., 2014a) further generated several uncoupling virB2 mutants with single amino acid substitutions in the VirB2 protein that retain wild-type levels of tumor formation on tomato stems but show reductions in transient transformation efficiencies of Arabidopsis seedlings. Taken together, these results suggest the auxiliary role of the filamentous T-pilus in optimizing the A. tumefaciens transformation efficiency of certain plant species or infection conditions.
Biochemical studies show that VirB5 co-fractionates as a minor component in pilus preparations and contributes to T-pilus assembly (Schmidt-Eisenlohr et al., 1999). Additionally, immunogold staining demonstrated that VirB5 is located at the tips of the cell-bound T-pili and the ends of detached T-pili, suggesting the VirB5 might be involved in host-bacteria contact (Aly and Baron, 2007). This notion is supported by two lines of evidence. One is that the VirB5 homolog CagL is localized on the T4SS pilus tip of Helicobacter pylori and functions in binding host human cells, likely by interacting with the integrin receptor (Kwok et al., 2007; Backert et al., 2008). Evidence that the CagL protein, the VirB5 homologue in the H. pylori, functions as a specialized adhesin binding to the human integrin ß1 and may activate the downstream host cell response, suggested a putative host cell receptor for the bacterial T4SS protein (Kwok et al., 2007; Backert et al., 2008). However, whether A. tumefaciens VirB5 also serves as an adhesin remains to be determined. Secondly, addition of exogenous VirB5 or expression of secreted VirB5 in the tobacco transgenic plant was able to enhance T-DNA expression (Lacroix and Citovsky, 2011). Interestingly, the VirB5 protein interacts with the cytokinin biosynthetic enzyme Tzs (trans-zeatin synthesizing) protein in yeast and in vitro (Aly et al., 2008). The tzs gene in the nopaline strains of A. tumefaciens shares significant homology with the ipt gene, is located outside the T-DNA region and is not incorporated into the host plant genome during infection (Akiyoshi et al., 1985, 1987; Beaty et al., 1986; Powell et al., 1988). The loss of Tzs protein expression and trans-zeatin secretion by the tzs mutants correlates with reduced stable and transient transformation efficiencies on Arabidopsis roots, suggesting the Tzs, by synthesizing trans-zeatin at early stage(s) of the infection process, modulates plant transformation efficiency by A. tumefaciens (Hwang et al., 2010; Hwang et al., 2013b). Because Tzs was detected in the A. tumefaciens membrane fraction (Lai et al., 2006; Aly et al., 2008) and a functional T4SS is required for Tzs localization on the cell surface (Aly et al., 2008), Tzs may function extracellularly, likely at the interface of the Agrobacterium-plant interaction. Since AT14A has partial sequence similarity to the animal integrin receptor, has been shown to mediate the cell wall-plasma membrane-cytoskeleton continuum in A. thaliana cells (Lu et al., 2012), and is critical for A. tumefaciens attachment to plant roots (Sardesai et al., 2013), it is plausible to hypothesize that T-pilus tip protein VirB5 may interact with AT14A for A. tumefaciens binding to the plant cell surface.
VirB1 may have two functions in the VirB transporter. The amino-terminal region of VirB1 contains a signal peptide and a motif homologous to that of lytic transglycosylases and lysozymes (Mushegian et al., 1996; Baron et al., 1997), suggesting that VirB1 may function to prepare sites in the bacterial envelope for transporter assembly by localized lysis of the peptidoglycan layer. The virB1 non-polar mutant is highly attenuated in virulence and cannot assemble T-pili (Berger and Christie, 1993; Lai and Kado, 2000), suggesting that pilus assembly across an intact peptidoglycan layer is inefficient or unstable. VirB1 is processed to a 73-residue, carboxy-terminal fragment termed VirB1*. The secreted VirB1* may play a direct role in T-pilus assembly, either by stabilizing VirB5 or by mediating VirB2-VirB5 interaction (Llosa et al., 2000; Zupan et al., 2007). VirB5 may directly interact with VirB2 or confer its effect by means of other proteins, e.g., VirB7. Energy for T-pilus assembly may be provided by VirB11, and it may be transduced to the pilus assembly complex in the outer membrane via VirB6 and VirB7 (Krall et al., 2002; Christie et al., 2005). This current view of the T4SS structure is in accordance with the T-DNA transfer route in the T4SS that was proposed based on the results of TrIP assays (Cascales and Christie, 2004a). The T-DNA substrate contacts six of the 12 components of the T4SS machine, including the VirD4 receptor, followed by the VirB11 ATPase, VirB6/VirB8, and VirB2 pilin/VirB9 proteins (Cascales and Christie, 2004a). However, whether the VirB2 protein in contact with T-DNA is part of T4SS translocation channel or filamentous pilus has not been determined.
A. tumefaciens cells attach to plant cells at the bacterial poles and several VirB proteins, as well as VirD4, were observed at cell poles, suggesting that the VirB/D4 transfer apparatus localizes at the poles (Matthysse, 1987; Kumar and Das, 2001; Kumar et al., 2002; Jakubowski et al., 2004; Judd et al., 2004, 2005). However, recent studies using very high resolution deconvolution microscopy suggested that the A. tumefaciens VirB/D4 transfer apparatus forms helically arranged foci around the bacteria cells to help make intimate lateral contact with plant cells, which might maximize substrate transfer through the T4SS (Aguilar et al., 2010, 2011; Cameron et al., 2012). Several possible functions could be assigned to the T-pilus. First, the T-pilus could serve as a conduit for several components needed for virulence, including the T-pilin subunit, VirE2 proteins and single-stranded T-DNA that is piloted by the covalently linked VirD2 protein (Cascales and Christie, 2004a; Cascales et al., 2013). Second, the T-pilus could serve as a bridge to bring the bacterium and the host cell into close proximity while T-DNA is transferred into the host cell through T4SS translocation channel. Kelly and Kado (2002) described a presumably coiled thread-like interconnection with an average width of approximately 30 nm between A. tumefaciens and Streptomyces lividans cells during A. tumefaciens transformation. This interconnecting structure is dependent on virB genes and appears only under the same conditions as those required for T-pilus formation. One possible model derived from this observation is that the T-pilus may retract and subsequently draw the bacterial cell into sufficiently close contact with the target host cells for T-DNA transfer to occur.
So far, limited knowledge exists about what kind of plant proteins might be involved in the initial contact of A. tumefaciens T4SS components with host cell surfaces. Two kinds of plant proteins that interact with VirB2 protein were identified. The first class is composed of three related proteins with unknown function, RTNLB1 (reticulon-like protein B-1, At4g23630), RTNLB2 (At4g11220), and RTNLB4 (At5g41600) proteins; the second class is an AtRAB8B GTPase (At3g53610). The level of RTNLB1 protein is transiently increased immediately after A. tumefaciens infection. Overexpression of RTNLB1 in transgenic Arabidopsis results in plants that are hypersusceptible to Agrobacterium-mediated transformation while transgenic RTNLB and AtRAB8 antisense and RNA interference Arabidopsis plants are less susceptible to transformation by A. tumefaciens than are wild-type plants. These data suggested that the three RTNLB proteins and AtRAB8 are involved in the initial interaction of A. tumefaciens with plant cells (Hwang and Gelvin, 2004).
Cytoplasmic trafficking and nuclear import of T-DNA and effector proteins in plant cells
The VirD2 protein attaches to the 5′ end of the T-strand and serves as a pilot protein to guide the T-DNA from the bacteria into the plant cell. In addition to T-strands, several virulence effector proteins can be exported from the bacterial cells independently of each other and of the T-DNA. In addition to VirD2, VirE2, VirD5, VirE3, VirF, MobA, and Atu6154 in A. tumefaciens, and the GALLS protein in A. rhizogenes, have been shown to be transferred from bacteria into plant cells via the A. tumefaciens transfer apparatus, using the Cre recombinase reporter assay for translocation (CRAfT) (Vergunst et al., 2000, 2003, 2005). C-terminal clusters of positively-charged amino acids (specifically an Arg-x-Arg motif) of T4SS effectors function as the secretion signal for the A. tumefaciens transfer apparatus. In addition to genetic evidence, split-GFP studies directly visualized VirE2 protein translocation in plant and yeast cells (Li et al., 2014b; Sakalis et al., 2014). However, active movement of linear and filament VirE2 and transgene expression only occur in tobacco cells but not in yeast cells, suggesting that active VirE2 trafficking is important for efficient T-DNA expression. Because translocated VirE2 forms the filamentous structure in the absence of T-DNA, the associations and assistance of host plant cell proteins may be required for VirE2 movement (Li et al., 2014b; Sakalis et al., 2014). Indeed, using split-GFP technology to visualize VirE2 movement in agroinfiltrated N. benthamiana leaf epidermal cells, recent studies showed that VirE2 is transported into plant cells via clathrin-mediated endocytosis (Li and Pan, 2017) and trafficked via an ER/actin network that is powered by myosin XI-K (Yang et al., 2017).
Both VirD2 and VirE2 proteins contain plant-active nuclear localization signal (NLS) sequences that are believed to be involved in T-DNA import into the nucleus. By the yeast interaction trap (two-hybrid) approach, several plant proteins that interact with either VirD2 or VirE2 proteins were identified (Ballas and Citovsky, 1997; Deng et al., 1998; Bako et al., 2003; Tao et al., 2004; Bhattacharjee et al., 2008). Two VirD2-interacting proteins, ROC1 (peptidyl-prolyl cis-trans isomerase CYP1, At4g38740) and the CyP A proteins of Arabidopsis, belong to a large cyclophilin family of peptidyl-prolyl cis-trans isomerases, which are highly conserved in plants, animals, and prokaryotes (Deng et al., 1998). The VirD2 domain interacting with the cyclophilins is in the central region of VirD2 protein, which is distinct from the nuclear localization signal domains (Deng et al., 1998). Although the biological role of cyclophilins in A. tumefaciens infection is unclear, they were proposed to maintain the proper conformation of VirD2 within the host cell cytoplasm and/or nucleus during T-DNA nuclear import and/or integration (Deng et al., 1998; Bako et al., 2003). Another cellular factor encoded by a tomato protein phosphatase 2C (PP2C) interacted with the VirD2 NLS region (Tao et al., 2004). An Arabidopsis abi1 (At4g26080) mutant showed higher sensitivity to A. tumefaciens transformation than did the wild-type plant, whereas over-expression of PP2C in tobacco protoplasts inhibited nuclear import of a ß-glucuronidase-VirD2 NLS fusion protein (Tao et al., 2004). These data suggested that phosphorylation of the VirD2 protein near the NLS region may influence the import of the T-complex.
The VirD2 protein was also found to interact with an Arabidopsis protein, AtKAPα/IMPA-1 (At3g06720), which belongs to a large karyopherin family known to mediate nuclear import of NLS-containing proteins (Ballas and Citovsky, 1997). AtKAPα-VirD2 interactions, which occur both in the yeast two-hybrid system and in vitro, require the presence of the VirD2 carboxyl-terminal NLS (Ballas and Citovsky, 1997). There are nine importin α members in Arabidopsis. The IMPA-2 (At4g16143), IMPA-3 (At4g02150), and IMPA-4 (At1g09270) show interactions with VirD2 and VirE2 with different strengths in yeast, in vitro, and in plants (Bhattacharjee et al., 2008; Lee et al., 2008). In addition, IMPA-1 interacts with the VirE2 protein in the plant cytoplasm and IMPA-4 interacts with VirE2 predominantly in the plant nucleus (Lee et al., 2008). Stable and transient transformation assay results showed that an IMPA1, IMPA2, or IMPA3 T-DNA insertion mutant remains susceptible to Agrobacterium-mediated transformation, whereas the IMPA4 T-DNA insertion mutant is resistant to Agrobacterium-mediated root transformation (Bhattacharjee et al., 2008). The resistant phenotype of the impa4 mutant could be complemented by the IMPA4 gene but could not be complemented by other importin α genes under the control of their native promoters, which might be due to the different promoter expression profiles of importin α genes. Additionally, an Arabidopsis T-DNA insertion mutant of the importin β-3 exhibits lower Agrobacterium-mediated transformation efficiencies, suggesting that importin β-like transportins might be also involved in Agrobacterium-mediated transformation (Zhu et al., 2003b). Collectively, these data indicate that after the TDNA and Vir proteins enter the plant cells, the host cell nuclear import machinery may be hijacked to mediate the nuclear targeting of T-DNA (Gelvin, 2010b, 2012; Lacroix and Citovsky, 2013).
Yeast two-hybrid assays identified two VirE2-interacting plant proteins, VIP1 (At1g43700) and VIP2 (At5g59710) (Tzfira et al., 2001, 2002; Anand et al., 2007). The VIP1 protein shows homology to a class of basic leucine zipper (bZIP) proteins, and is known to localize to the cell nucleus (Tzfira et al., 2001). VIP1 anti-sense transgenic tobacco plants exhibit significantly reduced nuclear import of GUS-VirE2 but not of GUS-VirD2, suggesting a role for VIP1 in VirE2 nuclear import (Tzfira et al., 2001). The early stage of T-DNA expression is blocked in VIP1 anti-sense tobacco transgenic plants, whereas the over-expression of VIP1 in transgenic tobacco plants enhances the early steps of the A. tumefaciens infection process (Tzfira et al., 2001, 2002). Further, the evidence of VIP1 interacting with VirE2–ssDNA complexes in vitro (Tzfira et al., 2001) with VirE2 and importin α to form a ternary complex (Citovsky et al., 2004) suggested that VIP1 might function as an adaptor between VirE2 and the conventional nuclear import machinery of plant cells. Nuclear targeting of the VIP1-VirE2 complex could be triggered by the phosphorylation of VIP1 on serine-79 by the mitogen-activated protein kinase 3 (MPK3, At3g45640) (Djamei et al., 2007). Substitution of the serine-79 to alanine, mimicking the non-phosphorylated form of VIP1, resulted in the cytoplasmic localization of VIP1 (Djamei et al., 2007). These data led to the proposed ‘Trojan horse’ model, which suggested that A. tumefaciens infection activates MPK3-mediated phosphorylation of VIP1 in plant cells, and thereby promotes VIP1-VirE2-ssDNA complex trafficking to the plant nucleus (Djamei et al., 2007). However, a recent study by Shi et al. (2014) indicated that transformation efficiencies of the Arabidopsis vip1 mutant and the VIP1 overexpression transgenic plants were not altered when infected by numerous tested A. tumefaciens strains. Furthermore, overexpression of several forms of VIP1 did not alter the subcellular localization of VirE2 (Shi et al., 2014). The conflicting results from these VIP1 studies might be due to different plant systems, experiment setups, and/or analysis methods (Tzfira et al., 2001; Li et al., 2005a; Shi et al., 2014). Thus, a modified model of VirE2-VIP1 interactions in A. tumefaciens infection process has been proposed, in which the nuclear import of T-DNA is largely affected by the VirD2, and VirE2 may play an accessory structural role to facilitate T-DNA targeting to the plant nucleus. VirE2 proteins may interact with VIP1 in the cytoplasm regions and prevent some endogenous VIP1 from activating plant defense genes, thus promoting A. tumefaciens infection (Pitzschke, 2013; Shi et al., 2014).
As described above, A. tumefaciens may utilize the host cell nuclear import system to help T-DNA and effector proteins to enter plant nucleus. The plant cytoskeleton may also play a role in T-complex cytoplasmic trafficking. Salman et al. (2005) utilized single-particle-tracking methods to follow the movement of fluorescently labeled VirE2–ssDNA complexes in a cell-free system. Chemical disruptions of microtubule networks and dynein motors blocked VirE2 complex movement, suggesting the possible involvement of microtubules in A. tumefaciens T-complex nuclear targeting (Salman et al., 2005; Tzfira, 2006). However, cytoskeleton inhibitor treatment together with real time VirE2 trafficking imaging experiments in N. benthamiana demonstrated that cytoplasmic movement of VirE2 does not require microtubules but does rely on actin powered by myosin (Yang et al., 2017). The involvement of actin in VirE2 trafficking is also supported by the findings that Arabidopsis mutant plants impaired in actin genes (ACT2, At3g18780 and ACT7, At5g09810) expressed in the roots were resistant to stable and transient transformation, suggesting the involvement of the actin cytoskeleton in Agrobacterium-mediated transformation (Zhu et al., 2003b). In addition, both VirD2 and VirE2 proteins interact with filamentous actin in an in vitro cosedimentation assay. Inhibitors of the actin cytoskeleton lowered the A. tumefaciens transformation efficiency of tobacco BY-2 cells (Gelvin, 2012).
Integration into the plant genome and expression of the T-DNA
T-DNA integration into the plant cell genome is the final step of the Agrobacterium-mediated transformation process. Unlike other mobile DNA elements such as transposons and retroviruses, the T-DNA does not encode enzymatic activities required for integration. Thus, T-DNA insertion into the plant DNA must be mediated by proteins transported from A. tumefaciens itself, namely VirD2 and VirE2, and/or host cell factors.
VirD2 might play a dual role in the T-DNA integration process, ensuring both its fidelity and efficiency. The integration of the 5′ end of the T-strand into plant DNA is generally precise. This may result from the linkage of VirD2 to the 5′ end of the T-strand, which provides protection from exonucleases inside the plant cells (Durrenberger et al., 1989; Tinland et al., 1995; Rossi et al., 1996). A region in the C-terminal portion of VirD2 called the ω domain is implicated in the integration process (Shurvinton et al., 1992; Tinland et al., 1995; Mysore et al., 1998). However, it is not clear how exactly the ω domain of VirD2 is involved in T-DNA integration because the pattern of integration of the T-DNA is not affected by deletion of the ω sequence (Shurvinton et al., 1992; Tinland et al., 1995; Bravo-Angel et al., 1998; Mysore et al., 1998). Involvement of VirD2 in protecting the integrity of the right border was also demonstrated in a fungal transformation system (Michielse et al., 2004); this study further showed that VirE2 similarly protects the left end of the T-DNA, while VirC2 helps ensure correct processing. Importantly, VirC2 is also required for single-copy integration in this system (Michielse et al. 2004).
A report by Bako et al. (2003) demonstrated that VirD2 interacts with, and is phosphorylated both in vitro and in alfalfa cells by, the nuclear CAK2Ms kinase, a conserved plant ortholog of cyclin-dependent kinase-activating kinases. VirD2 is also found in tight association with the TATA box-binding protein (TBP) in vitro and in Agrobacterium-transformed Arabidopsis cells (Bako et al., 2003). The TBP protein is implicated in the control of transcription-coupled repair (TCR), which ensures preferential and effective removal of DNA lesions from transcribed genes (Bako et al., 2003). CAK2Ms isolated by immunoprecipitation phosphorylate cyclin-dependent kinase (CDK2) and the C-terminal domain (CTD) of the RNA polymerase II largest subunit, which may serve as a TBP-binding platform (Bako et al., 2003). The functional conservation of TBP and CDK-activating protein kinase (CAK2) orthologs in eukaryotes indicates that these nuclear VirD2-binding factors may provide a link between T-DNA integration and transcription-coupled repair.
Ziemienowicz et al (2000) demonstrated that VirD2 protein does not possess general ligase activity using an in vitro ligation assay. Enzyme(s) present in plant extracts, likely DNA ligase, and E. coli T4 DNA ligase were able to ligate the 5′ end of T-DNA from an artificial T-DNA-VirD2 complex to a partly double-stranded oligonucleotide (Ziemienowicz et al., 2000). An Arabidopsis orthologue of the yeast and mammalian DNA ligase IV gene termed AtLIG4 (At5g57160) is required for the repair of DNA damage because seedlings of the Atlig4 mutant are hyper-sensitive to the DNA-damaging agents methyl methanesulfonate and X-rays (Friesner and Britt, 2003; van Attikum et al., 2003). However, using tumorigenesis and germline transformation assays, the Arabidopsis Atlig4 mutant is not impaired in T-DNA integration. Thus, DNA ligase IV may not be required for T-DNA integration in certain plant tissues. Similarly, the ku80 Arabidopsis mutant plant is hypersensitive to the DNA-damaging agent methyl methane sulphonate and has a reduced capacity to carry out nonhomologous end joining recombination (Gallego et al., 2003). ku80 (At1g48050) mutant plants show no significant defect in the efficiency of floral dip transformation of plants with A. tumefaciens (Friesner and Britt, 2003; Gallego et al., 2003). However, in other studies, contradictory data indicated that Arabidopsis ku80 or ku70 (At1g16970) mutant plants and rice plant lines down-regulated in ku70, ku80, or lig4 showed different transformation responses (van Attikum et al., 2001; Friesner and Britt, 2003; Gallego et al., 2003; Li et al., 2005b; Jia et al., 2012; Nishizawa-Yokoi et al., 2012; Vaghchhipawala et al., 2012; Mestiri et al., 2014; Park et al., 2015). These discrepancies might be the result of different techniques and different plant tissues used to examine transformation efficiency, or they may reveal more complex and redundant pathways for T-DNA integration mechanisms during A. tumefaciens infections (Tzfira et al., 2004a; Citovsky et al., 2007; Gelvin, 2010a, 2010b; Magori and Citovsky, 2012; Lacroix and Citovsky, 2013).
The contradictory conclusions for the involvement of NHEJ for T-DNA integration implied that an alternative DNA repair pathway might be responsible for T-DNA integration. A recent breakthrough showed that T-DNA integration in A. thaliana requires the polymerase theta (Pol θ, At4g32700) (van Kregten et al., 2016). This discovery was inspired by the similar signature of double-strand breaks (DSBs) of T-DNA integration with other systems. A kind of genome scar ‘filler DNA’ is usually found in the site of T-DNA integration as well as the Pol θ-mediated error-prone DSB repair in mammalian cells (Mateos-Gomez et al., 2015). Pol θ mutant plants tebichi/polq showed resistant to T-DNA integration but were susceptible to Agrobacterium-mediated transient transformation demonstrated via both floral dip and root transformation. Based on the knowledge of molecular action of Pol θ, it has been proposed that T-DNA could target the random DSBs in the genome and the microhomology between the ends of T-DNA and the DSBs is required for the integration of T-DNA. Enhanced understanding of the mechanism of T-DNA integration may allow researchers to improve transgene targeting by introducing artificial breaks in the plant genome, which ideally will lead to specific integration of DNA fragments in a designated location (van Kregten et al., 2016).
Identification of additional cellular factors involved in the TDNA interaction comes from genetic approaches. Nam et al. (1999) identified several Arabidopsis rat mutants deficient in TDNA integration. One mutant, rat5, which harbors a mutation in the H2A histone gene (HTA1, At5g54640), is deficient in stable but not transient T-DNA expression, suggesting the involvement of HTA1 in T-DNA integration (Mysore et al., 2000b). The rat5 mutant plant is highly recalcitrant to stable transformation by root inoculation. However, the mutant plant is efficiently transformed by flower vacuum infiltration (Mysore et al., 2000a). The highest levels of HTA1 gene expression are detected in the elongation zone of the root, a region that is most susceptible to transformation (Yi et al., 2002). The HTA1 promoter activity is induced by wounding or by A. tumefaciens infections (Yi et al., 2006). The HTA1 cDNA not only complements the rat5 mutant, but was also shown to increase transient and stable transformation rates of wild-type plants, indicating that histone H2A-1 may play an role in transformation (Mysore et al., 2000b; Yi et al., 2006; Tenea et al., 2009). Tenea et al. (2009) demonstrated that overexpression of Arabidopsis cDNAs encoding seven tested histone H2A (HTA), histone H4 (HFO), and histone H3-11 (HTR11, At5g65350) genes increased both Agrobacterium-mediated transformation and transgene expression; whereas none of the seven tested histone H2B (HTB) cDNAs, nor other tested histone H3 (HTR) cDNAs, increased transformation. Overexpression of HTA, HFO, or HTR11 cDNA increased expression of incoming single-stranded or double-stranded forms of a gusA gene in plant cells. These results suggested that several histone proteins may enhance transgene expressions within the plant cells and therefore may increase A. tumefaciens infection efficiency (Tenea et al., 2009; Gelvin, 2010b).
The VirE2-interacting protein, VIP1, may also participate the T-DNA integration step, possibly by creating a link to the host cell chromatin component histone protein. VIP1 shows interaction with plant histone H2A-1 in vivo, and with purified Xenopus core histones H2A, H2B, H3 and H4 in vitro (Li et al., 2005a; Loyter et al., 2005). In addition, the VIP1 protein associates with purified plant nucleosomes in vitro (Lacroix et al., 2008). VIP1 is also able to provide links between plant nucleosomes with VirE2 and with a synthetic minimal T-complex in vitro by forming quaternary nucleosome-VIP1-VirE2-ssDNA complexes (Lacroix et al., 2008). Another VirE2-interacting protein, VIP2, is a basic domain/leucine zipper motif-containing protein. Virus-induced gene silencing of VIP2 in Nicotiana benthamiana and the Arabidopsis vip2 mutant are both defective in T-DNA integration but not in transient T-DNA expression. The transcriptome analyses of Arabidopsis vip2 mutant revealed that expression of several histone genes was decreased, suggesting VIP2 may function as a transcriptional regulator to affect histone gene expression, thereby participating in the T-DNA integration step indirectly (Anand et al., 2007). When the T-DNA is inserted into the host chromosomal DNA wrapped in chromatin, the chromatin structure may need to be remodeled or removed to accommodate external T-DNA insertions. Several genetic studies indicated that T-DNA integration may be influenced by several chromatin proteins, including histone deacetylase (HDA1/HDT19, At4g38130; HDA2/HDT1, At3g44750; HD2B/HDT2, At5g22650), and two subunits of the chromatin assembly factor 1 (FAS1, At1g65470; FAS2, At5g64630) (Zhu et al., 2003b; Endo et al., 2006; Crane and Gelvin, 2007; Gelvin and Kim, 2007).
After the T-complex enters the plant nucleus, the associated bacterial virulence proteins and host proteins may need to be removed from the T-DNA to allow efficient T-DNA integration. The disassembly of the T-complex is likely mediated by targeted proteolysis, which is accomplished by the VirF protein and the plant ubiquitin proteasome complex (Tzfira et al., 2004b; Zaltsman et al., 2010a, 2010b). virF mutant bacterial strains show weak virulence on some plant species, including Nicotiana glauca and tomato, but can efficiently transform other plant species, suggesting the VirF protein may function as a host-range factor (Melchers et al., 1990; Regensburg-Tuïnk and Hooykaas, 1993). The virF mutant can be complemented by expressing VirF in transgenic plants, showing that the VirF protein functions in plant cells (Regensburg-Tuïnk and Hooykaas, 1993). VirF is an F-box protein and interacts with three host Skp-1 related proteins, ASK1 (At1g75950), ASK2 (At5g42190), and ASK10 (At3g21860) proteins (Schrammeijer et al., 2001). Both F-box proteins and Skp1 proteins are core components of the SCF (Skp1-Cullin-F-box protein) family of E3 ubiquitin ligases and serve to mediate targeted protein destabilization by the 26S proteasome. The VirF protein can interact with the VIP1 protein and mediate VirE2 protein degradation in the presence of VIP1 in yeast and plant cells, suggesting the VirF-mediating protein degradation is important for efficient transformation (Tzfira et al., 2004b). The involvement of members of the host SCF-E3 ligase complex were further deciphered by gene silencing and gene expression studies (Anand et al., 2012). The SCF-E3 ligase is composed of four different proteins: an F-box protein that recruits substrates; SKP1 that functions as an adaptor protein; Cullin (CUL1); and a RING-finger protein (RBX1) that interacts with both Cullin and E2 conjugating enzymes (Anand et al., 2012; Magori and Citovsky, 2012). Microarray analysis results showed that only ASK1, ASK2, and ASK20, among 21 Arabidopsis SKP1-LIKE (ASK) genes, are induced after A. tumefaciens infections. The Arabidopsis ask1 and ask2 mutants showed lower A. tumefaciens infection efficiencies, suggesting the SKP1 proteins may be positively involved in A. tumefaciens infection (Anand et al., 2012; Magori and Citovsky, 2012). However, the transformation rates of both RBX1 and CUL1c gene silenced N. benthamiana were not significantly affected (Anand et al., 2012). The Arabidopsis SGT1a (suppressor of the G2 allele of SKP1, At4g23570) and SGT1b (At4g11260) genes that encode accessory proteins interacting with the SCF-complex were induced following A. tumefaciens infection (Anand et al., 2012). Only the sgt1b mutant showed lower tumor formation efficiencies on roots when infected with A. tumefaciens, suggesting the potential role of SGT1 in the A. tumefaciens-plant interaction (Anand et al., 2012). Another plant SCF-E3 ligase complex member, the VIP1-binding F-box protein (VBF, At1g56250), is able to substitute for VirF function in plants and its expression is also induced by A. tumefaciens infection (Ditt et al., 2006; Zaltsman et al., 2010b). The VBF protein interacts with VIP1 and targets VIP1 and VIP1-VirE2 complexes for proteasomal destabilization via the SCFVBF complex (Zaltsman et al., 2010b). Additionally, the involvement of the VBF protein in uncoating a synthetic single-stranded DNA packaged by VirE2 was demonstrated in a cell-free proteasomal degradation system (Zaltsman et al., 2013). This proteasomal uncoating of synthetic minimal T-complexes is dependent on the presence of functional VBF and VIP1 proteins; and is inhibited by a known selective inhibitor of proteasomal activity, MG132 (Lacroix and Citovsky, 2013; Zaltsman et al., 2013).
The VirF protein may have multiple functions during A. tumefaciens infection, in addition to mediating the host cell ubiquitinproteasome system (UPS) to unmask the T-complex and help T-DNA become integrated into the host genome (Gelvin, 2010a; Magori and Citovsky, 2012; Lacroix and Citovsky, 2013; Pitzschke, 2013). As noted above, upon A. tumefaciens infection, the VIP1 protein is phosphorylated by the mitogen-activated protein kinase 3 (MPK3) and functions as a transcription factor that induces expression of several stress-responsive genes (Djamei et al., 2007; Pitzschke, 2013). A. tumefaciens may utilize VirF to repress the VIP1-mediated host defense responses via VIP1 protein degradation. At the same time, premature or excessive degradation of VirE2-VIP1 protein complexes by VirF might also hinder T-DNA nuclear import and integration. In order to work against host cell UPS-mediated VirF degradation, A. tumefaciens exports another effector protein, VirD5, into plant cells; this protein interacts with VirF and protects it from rapid degradation by the defensive action of the host UPS (Magori and Citovsky, 2011). Similarly, VirD5 interacts with the Arabidopsis VIP1 protein and is able to form a ternary complex of VirD5-VIP1-VirE2 (Wang et al., 2014). VirD5 can compete with VBF for VIP1 binding and help stabilize VIP1 and VirE2 proteins, supporting the idea that the VirD5 protein may target T-complex to prevent its degradation in the plant cell nucleus (Wang et al., 2014). Finally, the VirE3 protein can be transferred into plant cells (Vergunst et al., 2000, 2003, 2005; Schrammeijer et al., 2003). Interestingly, VirE3 may interact with VirE2 and expression of VirE3 in VIP1 antisense tobacco plants can complement both nuclear import of VirE2 and transformation susceptibility (Lacroix et al., 2005). Thus, the A. tumefaciens may use the VirE3 effector protein as a substitute for VIP1 in plants to facilitate VirE2 nuclear targeting during infection. Collectively, these examples illustrate how the A. tumefaciens-mediated plant transformation process represents an impressive arms race, in which the plant cells try to fight off pathogen infections while the pathogen utilizes bacterial effectors to evade or outweigh plant defense responses.
APPLICATIONS OF AGROBACTERIUM-MEDIATED TRANSFORMATION
Stable transformation
A. tumefaciens can genetically transform various host cells including numerous dicot and some monocot angiosperm species and gymnosperms (De Cleene and De Ley, 1976). In addition, A. tumefaciens can transform non-plant organisms, including fungi, algae, sea urchin embryos, human cells, and the gram positive bacterium Streptomyces lividans (Bundock et al., 1995, 1999; Piers et al., 1996; de Groot et al., 1998; Kunik et al., 2001; Kelly and Kado, 2002; Hooykaas, 2004; Kumar et al., 2004; Pelczar et al., 2004; Michielse et al., 2005; Bulgakov et al., 2006; Lacroix et al., 2006). In order to perform effective and large-scale functional genomic studies, highly efficient and simple genetic transformation methods for the studied organisms are crucial. The A. tumefaciens-mediated transformation method has several advantages over other transformation methods; it is easy to use, relatively inexpensive, and generally results in low copy number and well-defined DNA insertions into the host cell chromosome (Koncz et al., 1994; Pawlowski and Somers, 1996; Hansen et al., 1997; Enríquez-Obregón et al., 1998; Shou et al., 2004; Travella et al., 2005; Zhang et al., 2005; Gao et al., 2008). The traditional transformation methods such as protoplast transformation by electroporation, microinjection, or polyethylene glycol fusion are not as well-suited for production of transgenic plants, because the regeneration of plants from protoplasts is time-consuming and low efficiency (Fromm et al., 1985, 1986; Uchimiya et al., 1986; de la Peña et al., 1987; Newell, 2000; van den Eede et al., 2004; Banta and Montenegro, 2008). Microprojectile bombardment, also called biolistics, is the most important alternative to Ti plasmid DNA delivery systems for plants (Sanford, 2000). Spherical gold or tungsten particles are coated with DNA, accelerated to high speed with a particle gun, such that they penetrate into plant tissues (Kikkert et al., 2004). Microprojectile bombardment can be used to introduce foreign DNA into various plant tissues of a wide range of plant species. The gene of interest needs not to be cloned into a specialized transformation vector in order to be transformed into plant cells (Herrera-Estrella et al., 2005). However, the bombardment process tends to cause multiple-copy DNA insertions and the loss of molecular integrity of the DNA, and there are limitations on the size of the DNA. The complex DNA integration patterns usually result in genetic instability and/or epigenetic silencing of the transgene in the transgenic plants (Newell, 2000; Kikkert et al., 2004; Lorence and Verpoorte, 2004; van den Eede et al., 2004; Altpeter et al., 2005; Herrera-Estrella et al., 2005). Due to these limitations and drawbacks of physical and chemical transformation methods, the A. tumefaciens-mediated gene transfer method is still the most frequently used and most popular method for generation of transgenic plants and genetically modified crops (Table 2).
Table 2.
Summary of different plant transformation methods
| Method | Advantages | Disadvantages |
|---|---|---|
| Agrobacterium tumefaciens-mediated transformation | Minimal equipment and facility required and less expensive; easy to set up and use; generally low transgene copy number; minimal DNA rearrangements; fewer undesired mutations-so-maclonal variation; relatively higher stability of transferred gene | Several elite monocot crops, legumes, and woody plants are recalcitrant to A. tumefaciens-mediated transformation |
| Microprojectile bombardment | A wide range of plants and tissues can be used; an excellent and efficient system for transient expression; simultaneous delivery of large numbers of different genetic elements; effective system for organelle transformation | Multiple copies of DNA insertions; fragmentation and loss of molecular integrity of DNA insertions; possible emergence of chimeric plants |
| Microinjection | Only few successful reports in rye, barley, and other plants | Limited to protoplasts or few plant cell types; requirement for highly skilled personnel; difficult to regenerate viable plants |
| Electroporation | An alternative system to obtain transient expression in plants; convenient, simple, and fast | Limited to protoplasts or few plant cell types; difficult to regenerate viable plants |
| Polyethyleneglycol (PEG)-mediated gene transfer | Direct gene transfer to cells circumvents the host range limitation; easy to perform with relatively low cost | Limited to protoplasts or few plant cell types; difficulties and slow process in regeneration of viable plants |
| Pollen tube pathway | Elimination of tissue culture systems and is not limited by the ability to regenerate from transformed plant tissues, cells or protoplasts; only few successful reports in rice, maize, cotton, soybean, wheat, and other plants | Difficulty in establishing standard protocols and low success rates; only works with plants that flower and readily produce seeds; can only be performed during the flowering period |
The transformation methods can be categorized into two types: transient transformation and stable transformation. For transient transformation, T-DNA integration into the host genome is not required, and T-DNA expression usually lasts for a few days (Wroblewski et al., 2005; Marion et al., 2008; Jones et al., 2009; Kim et al., 2009; Li et al., 2009; Tsuda et al., 2012; Wu et al., 2014b). It presents useful advantages, in that it allows results of experimental treatments to be observed in a short period of time, as discussed below. On the other hand, stable transformation is a long process that often requires established tissue culture techniques to promote whole plant growth from the transformed cells or tissues. For stable transformation, the T-DNA must integrate in the host cell genome, so that it is subsequently passed on to the next generation (Bent, 2006; Gelvin, 2006; Tague and Mantis, 2006; Rivero et al., 2014).
In order to utilize pathogenic A. tumefaciens to generate transgenic plants, several obstacles need to be overcome. First, the oncogenes from the wild-type T-DNA are removed to eliminate the pathogenicity of the bacterium, and the strain becomes nonpathogenic (disarmed) without affecting its ability to transfer T-DNA. Second, the gene of interest and selection markers for transgenic plants need to be introduced into the T-DNA, and molecular biology tools are needed for DNA cloning in vitro. The Ti plasmid size is large and usually low-copy number, which makes isolation and cloning of the Ti plasmid quite challenging. In order to overcome these difficulties, most research teams utilize a binary vector system, in which the T-DNA region is carried on a broad-host range replicon and the vir genes required for T-DNA transfer are located on the disarmed Ti-plasmid (Barton et al., 1983; de Framond et al., 1983; Hoekema et al., 1983; Zambryski et al., 1983; Bevan, 1984). This binary vector system has provided the plant research community enormous versatility and flexibility, and has enabled an upsurge in the production of transgenic plants.
With advances in recombinant DNA technology, T-DNA binary vectors have evolved over the past years to be highly developed and more specialized for different purposes (Hellens et al., 2000; Chung et al., 2005; Komari et al., 2006; Lee and Gelvin, 2008; Mehrotra and Goyal, 2012; Anami et al., 2013). T-DNA-based binary vectors generally consist of several features, including (1) the T-DNA right and left border repeat sequences that define the T-DNA region; (2) selectable marker genes to function in bacteria (E. coli and A. tumefaciens) and in plants; (3) restriction endonuclease sites within the T-DNA to allow insertion of one or more gene(s) of interest; (4) origin(s) of replication to allow plasmids to replicate in bacteria. In order to identify the transformed cells or plants, it is essential to be able to detect the foreign DNA that has been integrated into plant chromosome. Various antibiotic or herbicide resistance genes have been inserted into the T-DNA region of the binary vectors and used for transgenic plant selection. The neomycin phosphotransferase (NPTII) gene for kanamycin resistance and hygromycin phosphotransferase (HPT), conferring resistance to hygromycin, are the most widely used selectable marker gene systems in plants. Several other frequently used selection systems utilize other antibiotics, including chloramphenicol, gentamicin, streptomycin, bleomycin, blasticidin; herbicides, including phosphinothricin, gluphosinate, glyphosate, bromoxynil; or metabolic markers, including acetolactate synthase, threonine dehydratase, phosphomannose isomerase (Gheysen et al., 1998; Newell, 2000; Banta and Montenegro, 2008; Lee et al., 2008; Lee and Gelvin, 2008; Anami et al., 2013). Furthermore, in plant gene functional studies and other quantification studies, the use of reporter genes allows transformed cells to be quantified. The frequently used reporter gene products, such as ß-D-glucuronidase (GUS), both firefly and bacterial luciferases (LUC), green fluorescent protein (GFP) and other fluorescent proteins, can be detected in transformed plant tissues. The GUS activity in the transformed plant tissues can be observed by the blue staining after hydrolysis of the 5-bromo-4-chloro-3-indolyl ß-D-glucuronic acid; or can be quantitatively assayed by a fluorometric analysis that requires hydrolysis of the 4-methylumbelliferyl ß-D-glucuronide. The GFP protein is a useful in vivo marker to examine gene expression or recombinant protein localization, because it can be excited with either ultraviolet or blue light without addition of any substrates or cofactors.
Based on the agrobacterial chromosomal background and the resident Ti plasmids, the commonly used disarmed A. tumefaciens strains originated from two representative wild isolates: C58 (Lin and Kado, 1977) and Ach5 (Kovacs and Pueppke, 1994). The A. tumefaciens strains AGL1, EHA101, EHA105, and GV3101(pMP90), have the nopaline type C58 chromosomal background and the modified Ti plasmids from either the pTiBo542 or the pTiC58 plasmids (Hood et al., 1986; Koncz and Schell, 1986; Lazo et al., 1991). The A. tumefaciens strain LBA4404 has the octopine type Ach5 chromosomal background and the octopine-type Ti plasmid pAL4404 derived from the pTiAch5 plasmid (Ooms et al., 1982). Genetic background and modified Ti plasmid combinations of the disarmed A. tumefaciens strains noticeably affect the bacterial host range and the transformation efficiencies of various plant species (Hwang et al., 2010; Hwang et al., 2013a) (Table 3).
Table 3.
Frequently used disarmed Agrobacterium strains
| Strain Name | Chromosome background | Ti-plasmid types | Antibiotic Resistance1 | Reference |
|---|---|---|---|---|
| LBA4404 | Ach5 | a disarmed octopine-type Ti plasmid pAL4404 | RmR | Hoekema et al., 1983 |
| EHA101 | C58 | a disarmed agropine-type Ti plasmid pEHA101 (pTiBo542ΔT-DNA) | RmR, KmR | Hood et al., 1986 |
| EHA105 | C58 | a disarmed agropine-type Ti plasmid pEHA105 (pTiBo542ΔT-DNA) | RmR | Hood et al., 1993 |
| C58C1 | C58 | Cured of Ti plasmid | RmR | Deblaere et al., 1985 |
| C58C1(pTiB6S3ΔT, pCH32) | C58 | a disarmed octopine-type Ti plasmid pTiB6S3ΔT-DNA and a helper plasmid pCH32 | RmR, CbR, TcR | McBride and Summerfelt, 1990 |
| GV3101 | C58 | Cured of Ti plasmid | RmR | Holsters et al., 1980 |
| GV3101(pMP90) | C58 | a disarmed nopaline-type pTiC58ΔT-DNA | RmR, GmR | Koncz and Schell, 1986 |
| A136 | C58 | Cured of Ti plasmid | RmR | Watson et al., 1975 |
| AGL-1 | C58 | a disarmed pTiBo542ΔT-DNA | RmR, CbR | Lazo et al., 1991 |
| C58-Z707 | C58 | a disarmed nopaline-type pTiC58-Z707 | KmR | Hepburn et al., 1985 |
| NTL4(pKPSF2) | C58 | a disarmed chrysopine-type pTiChry5ΔT-DNA | EmR | Palanichelvam et al., 2000 |
1Cb, carbenicillin; Em, erythromycin; Gm, gentamicin; Km, kanamycin; Rm, rifampicin, Tc: tetracycline.
The host species A. thaliana is characterized by several traits, including small plant size, short generation time, large quantity of seeds, relatively small genome size, and ease of transformation by A. tumefaciens, which has led it to become the model system for plant biologists to address questions of basic and applied technology. Several Agrobacterium-mediated transfer methods for A. thaliana were developed using various A. thaliana ecotypes, plant tissue types, bacterial strains, binary vectors, and selection marker (Lloyd et al., 1986; Feldmann and Marks, 1987; Sheikholeslam and Weeks, 1987; Valvekens et al., 1988; Akama et al., 1992; Clarke et al., 1992; Huang and Mā, 1992; Sangwan et al., 1992; Chang et al., 1994). Here, two major stable transformation methods of A. thaliana are described as follows.
Stable transformation using root explants
Root explant is one of the plant tissues that is frequently used to genetically transform A. thaliana (van den Elzen et al., 1985; Valvekens et al., 1988; Akama et al., 1992; Clarke et al., 1992; Kemper et al., 1992; Czako et al., 1993; DeNeve et al., 1997; De Buck et al., 1998, 2000, 2009). There are several advantages of root explants for genetic transformation by A. tumefaciens. A. thaliana root cells are competent for regeneration, can be easily acquired in large quantities, and can be maintained in sustained cultures (Valvekens et al., 1988; Sangwan et al., 1992; Czako et al., 1993). Furthermore, a relatively low percentage of transformants show polyploidy when root explants are used for A. tumefaciens transformation, in comparison to leaf transformation or direct gene transfer methods, such as microprojectile bombardment (Altmann et al., 1994). Additionally, root transformations of Arabidopsis tend to result in higher percentage of single T-DNA insertions when compared with leaf-disc transformations (Grevelding et al., 1993). The root transformation assay also provides a reproducible and quantitative assay system for plant biologists to examine transformation efficiencies of somatic cells (Gelvin, 2006). Several studies have utilized quantitative root transformation assays to determine virulence of different A. tumefaciens strains and relative transformation efficiencies of different A. thaliana mutants or ecotypes. Based on the results obtained from these root transformation assays, bacterial and plant proteins that are involved in Agrobacterium-mediated plant transformation process can be identified and characterized; several of these were described above (Nam et al., 1997, 1998, 1999 Bundock et al., 1999; Mysore et al., 2000a; Zhu et al., 2003a, 2003b; Tsai et al., 2009; Gelvin, 2010a, 2010b; Hwang et al., 2010; Tsai et al., 2010; Gelvin, 2012; Hwang et al., 2013b, 2015).
It is known that different factors affect transformation efficiency, including plant growth temperature, plant-bacteria co-incubation time and temperature, light intensity and periods, addition of AS, pretreatment with phytohormones, different A. thaliana eco-types, different A. tumefaciens helper strains, and other factors (Vergunst et al., 1998; Zambre et al., 2003; Gelvin, 2006). The optimum growth temperature for Arabidopsis is 25°C. It is recommended that night temperature can be 2 to 4°C lower than the day temperature. During co-cultivation of plant root explants with A. tumefaciens, the transformation efficiencies and plant regeneration frequencies were relatively higher at 25°C in comparison to 21°C (Vergunst et al., 1998). On the other hand, although A. tumefaciens grows well at 28°C, accumulation of some of the VirB proteins constituting the T4SS T-DNA delivery machinery, as well as virulence on the host species Kalanchoe, is strongly compromised at this temperature (Fullner and Nester 1996; Banta, et al. 1998; Baron et al., 2001); these data indicate that pre-induction with AS (see below) should be performed at 19–21°C. Furthermore, A. tumefaciens can lose virulence when grown at 37°C due to loss of the Ti plasmid in the bacteria (Hamilton and Fall, 1971; Kerr, 1971). Light conditions during Agrobacterium-mediated plant transformation also affect transformation efficiencies significantly. The transformation efficiency is higher under continuous light than under a 16 hour light/8 hour dark photoperiod (Vergunst et al., 1998; Zambre et al., 2003). Another important factor for obtaining high transformation frequency is addition of 50–200 μM AS, a natural wound response molecule, to the A. tumefaciens culture prior to or during the co-incubation period in order to fully induce virulence gene expression in bacteria (Sheikholeslam and Weeks, 1987; Vergunst et al., 1998). It has also been suggested that coincubation periods should not exceed two days in order to avoid overgrowth of bacteria (Vergunst et al., 1998; Gelvin, 2006). After bacteria and root explant coincubation, different antibiotics, such as carbenicillin, cefotaxime, vancomycin HCl, or timentin, may be added into the selection media to eliminate A. tumefaciens. Timentin is an antibacterial combination consisting of the semisynthetic antibiotic ticarcillin disodium and the β-lactamase inhibitor clavulanate potassium and has frequently been used in A. thaliana root transformation methods (Nam et al., 1997, 1998, 1999; Vergunst et al., 1998; Bundock et al., 1999; Mysore et al., 2000a; Zhu et al., 2003a).
Different opine-type A. tumefaciens strains show different degrees of virulence on different plant species, partly because the tzs and virF genes exist in only one type of Ti-plasmid. It has been suggested that nopaline-type A. tumefaciens strains are preferable for transformation of A. thaliana and other plants from the Brassicaceae family (Nam et al., 1997; Gelvin, 2006; Hwang et al., 2013a). Similarly, different wild-type A. thaliana ecotypes show different competence for A. tumefaciens transformation (Nam et al., 1997; Chateau, 2000; Mysore et al., 2000a; Hwang et al., 2013a); ecotypes Aua/Rhon (Aa-O), Bensheim/Bergstrasse (Be-O), Wassilewskija (Ws), Columbia (Col-O), and C-24 are more susceptible to A. tumefaciens root transformation (Nam et al., 1997). Furthermore, competence of several A. thaliana eco-types can be enhanced when the explants are pre-cultured on media containing the phytohormones auxin and cytokinin (Valvekens et al., 1988; Sangwan et al., 1991, 1992; Hwang et al., 2013b; Sardesai et al., 2013).
Stable transformation by floral dipping
The transformation and regeneration of plants is a vital tool in both basic and applied plant biology research fields. The typical plant transformation method includes delivery of target gene into plant cells by A. tumefaciens transformation, followed by selection and regeneration of transgenic plants from the transformed cells through tissue culture techniques. Established plant tissue culture systems and sterile conditions are essential to regenerate plants. Suitable plant growth regulators are added to help regeneration of transgenic plants. However, several difficulties exist in the typical molecular transformation method. First, some A. thaliana ecotypes, such as Antwerpen (An-1), Angleur (Ang-0), Bologna (Bl-1), Blanes/Gerona (Bla-2), Calver (Cal-0), and Dijon-G, and mutant lines, are resistant to transformation and/or regeneration (Nam et al., 1997; Chateau, 2000; Mysore et al., 2000a; Hwang et al., 2013a). In addition, regeneration of whole plants from transformed somatic cells sometimes leads to somatic mutations. The presence of phytohormones in the tissue culture during regeneration also increases the chance of somatic mutations (Bent, 2000; Bent, 2006; Tague and Mantis, 2006). In order to resolve these difficulties, several plant transformation methods have been established and improved (Feldmann and Marks, 1987; Bechtold et al., 1993; Chang et al., 1994; Katavic et al., 1994; Clough and Bent, 1998; Richardson et al., 1998; Martinez-Trujillo et al., 2004; Zhang et al., 2006). Feldmann and Marks (1987) first reported that imbibition of A. thaliana germinating seeds with an appropriate A. tumefaciens strain can generate transformed progeny in the next generation. This method was applied to provide the first insertion mutagenized lines of A. thaliana. Another whole plant transformation method was presented by Chang et al. (1994) and by Katavic et al. (1994) in which reproductive inflorescences were cut off and the wounded sites were inoculated with A. tumefaciens. The treated plants were grown to maturity, harvested, and the progeny screened for transformants under selection. These methods avoid tissue culture steps and the resulting somaclonal variation, and a relatively short time is needed to acquire transformed progeny. However, the frequency of obtaining transformants in the next generation is highly variable and sometimes inconsistent for routine use.
An improved in planta transformation method was later proposed by Bechtold et al. (1993) in which the uprooted A. thaliana plants were vacuum infiltrated with the appropriate A. tumefaciens culture, infiltrated plants were then immediately replanted, and harvested seeds were finally screened for successful transformants. This method was developed to preclude altogether the requirement for plant tissue culture or regeneration steps (Bechtold et al., 1993; Bechtold and Pelletier, 1998). However, the removal and replanting of plants constrain the usefulness of this method. It was later discovered that submergence of inflorescences in the early flowering stages in a A. tumefaciens suspension is sufficient to obtain successful transformants (Clough and Bent, 1998). Furthermore, the vacuum infiltration process can be eliminated; simple dipping of developing floral tissues into the bacterial suspension is enough to acquire transformants in the next generation (Clough and Bent, 1998; Richardson et al., 1998; Bent, 2000). A. tumefaciens strains are first cultured in rich media with appropriate antibiotic selection until stationary phase (OD600= 0.8–1.0). The A. tumefaciens cultures are then collected by centrifugation and the pellet is resuspended in a solution containing 5% sucrose and the surfactant Silwet L-77 (Clough and Bent, 1998; Bent, 2000). Both sucrose and surfactant 0.05% Silwet L-77 are important for the success of the floral dip method (Clough and Bent, 1998; Richardson et al., 1998; Bent, 2000; Bent, 2006). However, Silwet L-77 can be toxic to plants at high concentrations, and 0.02% L-77 or as low as 0.005% L-77 can be used in the floral dip method. Repeated applications of A. tumefaciens can enhance the transformation efficiency (Clough and Bent, 1998; Martinez-Trujillo et al., 2004). After bacterial inoculation, covering plants for 1 day to maintain humidity also increase the transformation rates (Bent, 2000; Bent, 2006). Spraying of the A. tumefaciens is a useful alternative to transform Arabidopsis, and this method may provide the possibility of in planta transformation of plant species that are too large for dipping or vacuum infiltration (Chung et al., 2000). With these floral dip or spray methods, 0.1–3.0% of the seeds is typically found to be transgenic. This floral dip method is simple and can be easily performed by nonspecialists, which avoids extensive labor and the issues and equipment associated with regeneration. This method has been successfully used on a large number of A. thaliana ecotypes and mutants that might be resistant to transformation by other methods targeting somatic cells or tissues (Mysore et al., 2000a). The most striking contribution of this method is the high-throughput generation of A. thaliana T-DNA insertion mutant lines (Alonso et al., 2003; Alonso and Stepanova, 2003). Plant researchers can now easily obtain various T-DNA insertion mutants for most Arabidopsis genes from various stock centers (Table 4) (Scholl et al., 2000; Pan, 2003; Li et al., 2014a).
Table 4.
Selected Arabidopsis stock and database resources.
| Database/Center | Websites | Description | References | |
|---|---|---|---|---|
| 1 | Arabidopsis Biological Resource Center (ABRC)/The Arabidopsis Information Resource (TAIR) | https://www.arabidopsis.org/portals/mutants/stockcenters.jsp https://abrc.osu.edu/ | Various seed and clone stocks can be searched and ordered from the ABRC stock center. Useful tools and resources for Arabidopsis researchers are provided through the website. | Huala et al., 2001; Garcia-Hernandez et al., 2002; Rhee et al., 2003; Swarbreck et al., 2008; Lamesch et al., 2011 |
| 2 | The European Arabidopsis Stock Centre/The Nottingham Arabidopsis Stock Center (NASC) | http://arabidopsis.info/ | Various seed and clone stocks can be searched and ordered from the NASC. | Scholl et al., 2000 |
| 3 | AGRIKOLA | http://www.agrikola.org/index.php?o=/agrikola/main | Various RNAi knock-down lines for Arabidopsis can be searched by the website. Various seed and clone stocks can be ordered through the NASC | Hilson et al., 2004 |
| 4 | The French National Institute for Agricultural research (INRA) Arabidopsis Resource Center for Genomics/The Versailles Arabidopsis Stock Center | http://publiclines.versailles.inra.fr/ | Various seed and clone stocks can be searched and ordered from the stock center. | |
| 6 | The Lehle Seeds Company | http://www.arabidopsis.com/ | The private company sells Arabidopsis seeds and consumables. | |
| 7 | The RIKEN Biological Resource Center Experimental Plant Division (Japan) | http://epd.brc.riken.jp/en | Various seed and clone stocks can be searched and ordered from the stock center. | |
| 8 | Bielefeld University | http://www.gabi-kat.de/ | Various seeds of Arabidopsis T-DNA insertion lines (GABI-Kat lines) and clone stocks can be searched and ordered through the websites. | Li et al., 2007; Kleinboelting et al., 2012 |
| 9 | SIGnAL (Salk Institute Genomic Analysis Laboratory): T-DNA Express | http://signal.salk.edu/cgi-bin/tdnaexpress | Various T-DNA insertion lines of Arabidopsis can be searched through the website. | Alonso et al., 2003 |
| 10 | GARNet | http://www.garnetcommunity.org.uk/resources/mutant-collections | Useful tools and resources for Arabidopsis researchers are provided through the website. | Beale et al., 2002 |
In the A. thaliana vacuum infiltration method, most of the primary transformants are hemizygous at any T-DNA insertion site, suggesting that A. tumefaciens transformation may occur late in the floral development, possibly after the divergence of male and female gametophyte cell lineages (Bechtold et al., 1993; Bent, 2000; Bent, 2006). It was later discovered that the majority of transgenic plants result from transformation of the female gametophyte; this conclusion is based on the following observations (Ye et al., 1999; Bechtold et al., 2000; Desfeux et al., 2000; Bechtold et al., 2003): First, no transformed progeny were obtained after inoculation of A. tumefaciens on pollen donor plants, while 0.48% of transformed progeny were produced after inoculation of A. tumefaciens on pollen recipient plants (Desfeux et al., 2000). Second, various promoters fused to a GUS marker gene were used to observe sites of delivery of T-DNA (Ye et al., 1999; Bechtold et al., 2000; Desfeux et al., 2000). GUS staining was mainly observed in the ovules of the flowers and only rarely in pollen, suggesting that ovules are the primary target for A. tumefaciens transformation (Ye et al., 1999; Desfeux et al., 2000). In A. thaliana flowers, the gynoecium is an open and vase-like structure, which can fuse to form closed locules almost three days prior to anthesis. Based on this observation, the timing of A. tumefaciens infection is important to allow A. tumefaciens to enter the interior of the gynoecium prior to locule closure and ensure successful transformation (Desfeux et al., 2000). Third, most of the transformants contain T-DNA that is derived from the maternal chromosome set, based on a genetic linkage study (Bechtold et al., 2000). The floral dip or infiltration transformation methods have been successfully applied to wheat, radish, Medicago truncatula, and some other members from the Brassicaceae, such as pakchoi (Brassica campestris), Shepherd's purse (Capsella bursa-pastoris), Lepidium campestre, and Arabidopsis lasiocarpa (Liu et al., 1998; Trieu et al., 2000; Curtis and Nam, 2001; Tague, 2001; Wang et al., 2003; Inan, 2004; Agarwal et al., 2008; Bartholmes et al., 2008; Zale et al., 2009; Lenser and Theißen, 2014). Other bacteria, including Ensifer adhaerens, Rhizobium sp. NGR234, Sinorhizobium meliloti, and Mesorhizobium loti, contained the foreign vir genes and T-DNA regions on separate plasmids and have been used for floral dip transformation of A. thaliana (Broothaerts et al., 2005; Wendt et al., 2012). Recently, the Rhizobium etli CFN42 genome analysis results showed that it has functional and high homologs of vir genes in its p42a plasmid. The R. etli can both stably and transiently transform tobacco plants when the T-DNA region is introduced into the R. etli on a binary vector (Lacroix and Citovsky, 2016). Although these bacteria are not naturally able to transform plants because they lack native T-DNA and/or vir genes, most of them belong to Rhizobiaceae family, specifically the Alpha-Proteobacteria class.
The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) technology is a recent advance and powerful tool for genome editing in eukaryotic cells. The technique utilizes Streptococcus pyogenes Cas9 endonuclease and single chimeric guide RNA (sgRNA) to generate DNA double-stranded breaks in targeted genome sequences. It has been successfully used to edit genomes in animal systems (Cong et al., 2013; Hwang et al., 2013c; Wang et al., 2013) and was recently applied in plants as well. CRISPR can create genome-edited transgenic plants if a stable transformation approach (such as floral dip) is used (Zhang et al., 2015). In summary, the floral dip or infiltration transformation method has now become the most important and frequently used Agrobacterium-mediated transformation method in Arabidopsis research.
Transient transformation
In general, months are required to generate a transgenic line, and even longer if a homozygous genotype is desirable. Agrobacterium-mediated transient transformation is a rapid alternative to assay gene function, promoter behaviour and protein function when generation of a stable transgenic line is unnecessary. A binary vector carrying a reporter system (i.e. promoter-of-interest fused with a GUS/luciferase/fluorescence protein gene) or a fusion protein driven by a constitutive promoter (e.g. for subcellular localization, protein activity or protein-protein interaction study) can be naturally processed to generate single stranded T-DNA and delivered into the nucleus of the host. In the host nucleus, the single stranded T-DNA can be synthesized into double-stranded T-DNA, which is readily transcribed and translated into the reporter protein/fusion protein for analysis without genome integration. Transient transformation can be also used for gene silencing by expressing small interfering RNAs (siRNAs) and microRNAs (miRNAs) in plant tissues (McHale et al., 2013). As integration is not the main purpose in the transient system, a plant selectable marker is usually not required in the construct to be delivered. The drawback however, is the highly variable transformation efficiency for individual Agrobacterium strains, target plant species (including different A. thaliana ecotypes) and specific tissues. Although A. thaliana is not the easiest species to transiently transform, due to its importance in plant biology, extensive optimizations have been carried out. Below, we have summarized the reported methods (an overview can be seen in Table 5) that facilitate the transient transformation in A. thaliana.
Table 5.
Summary of Agrobacterium-mediated transient transformation assays in plants
| Method Type | Tissue/Reporter (genes)/Applications | Key findings | References |
|---|---|---|---|
| Agroinfiltration of adult leaves | Leaves of adult plants in vegetative stage/Expression of GUS reporter gene | No pre-induction or co-cultivation with acetosyringone is required, C58C1(pTiB6S3ΔT, pCH32) was the strain with the best efficiency | Wroblewski et al., 2005 |
| Expression of nahG increases the efficiency of Agrobacterium-mediated transient transformation | Rosas-Díaz et al., 2017 | ||
| Leaves of 4-week-old adult plants/GUS/Quantitative analysis of Agrobacterium-mediated transient transformation efficiency | Use of efr mutant for enhanced transient expression | Zipfel et al., 2006 | |
| Leaves of 6-week-old adult plants/GUS | Application of acetosyringone and 0.01% Triton X-100 with LBA4044 showed better transformation efficiency than C58C1(pTiB6S3ΔT, pCH32) | Kim et al., 2009 | |
| Leaves of 4- to 5-week-old adult plants/GUS, CFP/subcellular localization, protein–protein interaction | AvrPto suppresses immune responses triggered by multiple receptors, more susceptible than efr mutant | Tsuda et al., 2012 | |
| Leaves of 4-week-old adult plants/GFP | Pre-induction of vir genes by acetosyringone in AB-MES followed by infiltration solution containing acetosyringone | Mangano et al., 2014 | |
| Seedling transformation | Seedlings (4-day-old)/GFP/subcellular localization | (FAST)Vacuum infiltration of Agrobacterium directly into young Arabidopsis seedlings | Marion et al 2008 |
| Seedlings within 1-week-old/fluorescent proteins /subcellular localization, protein-protein interaction | Use of surfactant Silwet L-77 | Li et al., 2009 | |
| Seedlings (4-day-old)/GUS/gene function analysis | (AGROBEST) Pre-induction of vir genes by acetosyringone, optimization with AB salts in plant culture medium buffered to pH 5.5 during Agrobacterium infection | Wu et al., 2014b | |
| Seedlings/GFP, SYP121/membrane transport analysis | Use of Ubiquitin-10 promoter based bicistronic vector to co-express gene of interest and GFP | Chen et al., 2011. | |
| Root transformation | Root segments/GUS/distinguish transient expression from T-DNA integration | Co-cultivation with Agrobacterium for 2 days followed by incubation in callus induction medium for 4 days/Quantitative analysis of Agrobacterium-mediated transient transformation efficiency |
Nam et al. 1997 Zhu et al., 2003a Hwang and Gelvin, 2004 |
| Seedlings/tagged fluorescence proteins/study of root–rhizosphere interactions | Use of Agrobacterium rhizogenes strain MSV440 to effectively transform roots | Campanoni et al., 2007 | |
| RNA silencing via agroinfiltration | Seedlings at two- to three- leaf stage/AtPDS/knocking down genes involved in metabolism and defence | Use of tobacco rattle virus(TRV)-based VIGS vector for gene silencing | Burch-Smith et al., 2006 |
| Seedlings/number of transformed seeds/increased stable transformation rate | Transient down-regulation of the AGO2 and NRPD1a increased stable transformation efficiency | Bilichak et al.,2014 |
Transient transformation in roots
Root explants of various A. thaliana ecotypes respond differently to Agrobacterium-mediated transient transformation. Some eco-types such as Bl-1, Petergof, Cal-0, and Dijon-G are poorly transformed (Nam et al., 1997). Attachment assays revealed that the wild type A. tumefaciens virulent strain C58 binds poorly to roots of the Bl-1 and Petergof ecotypes, in contrast to highly efficient binding to roots of susceptible ecotypes Aa-0 and WS. This indicates that the binding ability of agrobacteria to the root tissue could be directly related to transformation efficiency. The Arabidopsis rat4 mutant, with a deletion of the cellulose synthase-like gene CSLA9 (At5g03760), showed a reduction in transient transformation efficiency (Zhu et al., 2003a). It has been shown that cellulose is not essential but important for efficient binding of A. tumefaciens cells to host cells for subsequent infection (Zhu et al., 2003a; Matthysse et al., 2005). The CSLA9 promoter is specifically active in the elongation zone of roots, the root tissue that is most susceptible to Agrobacterium-mediated transient transformation (Zhu et al., 2003a). RTNLB1 (reticulon-like protein B-1, At4g23630), which was found to interact with the A. tumefaciens T-pilus protein VirB2, is important for stable and transient transformation efficiency of root explants (Hwang and Gelvin, 2004).
Agrobacterium rhizogenes is a relative of A. tumefaciens which cause adventitious root development at the site of infection (also known as “hairy root disease”) and can be manipulated to co-transfer the T-DNA borne on a binary vector into root cells (Limpens et al., 2004). A. rhizogenes-mediated transformation appears to be a fast and effective tool to study the function of genes involved in root biology. It has been employed in RNA interference (RNAi) studies in roots (Limpens et al., 2004) and the disarmed strain can also be used for transient expression at the root epidermis without affecting the root morphology (Campanoni et al., 2007).
Transient transformation in adult leaves via agrofiltration
Due to the anatomical specificity, root tissues may not be suitable for experiments related to defense, chloroplasts, or for protein purification. Vegetative tissues like leaves are more abundant and sacrificing the plant is not necessary. For species Nicotiana benthamiana and lettuce, which can reach ~80% mesophyll cells, it is possible to achieve nearly 100% transient transformation efficiency in some regions of the leaf (Wroblewski et al., 2005). In contrast, successful transient transformation in A. thaliana leaves by the same method can be challenging and depends on ecotype and A. tumefaciens strain. As mentioned above, most laboratory A. tumefaciens strains are derivatives of two wild type virulent strains, C58 and Ach5. For instance, GV3101(pMP90) from C58 and LBA4404 from Ach5 are routinely used for transient transformation of various plant species (Kim et al., 2009). Different A. tumefaciens strains produce variable opines: nopaline from pTiC58, octopine from pTiAch5, agropine from A281 and succinamopine from EHA105 (Dessaux et al., 1992; Frandsen, 2011). Although different opine-producing strains seem to work better in certain plant species, it remains unclear whether the specificity of opine utilization contributes to different transformation efficiencies in specific plant species. Strain C58 cured of pTiC58, also known as C58C1, harbouring the octopine-type pTiB6S3DT-DNA and a pCH32 helper plasmid is regarded as the best; in a study of more than 10 A. tumefaciens wild type and disarmed strains, it achieved the highest transient transformation efficiency of A. thaliana, lettuce and tomato leaves (Wroblewski et al., 2005). Phenolics have been reported to influence transient transformation efficiency. As is true for stable transformation, it is now routine to add AS during transformation. Pre-treating A. tumefaciens with AS before agroinfiltration of in A. thaliana leaves can also increase the transient transformation efficiency (Mangano et al., 2014). Detergents/surfactants are also used to improve cuticular penetration of fluid on leaf surfaces. Silwet L-77 is one of the widely used surfactants in transient transformation while Triton X-100 was also shown to significantly enhance the efficiency (Kim et al., 2009).
Plant defense responses also play a key role in limiting transient transformation. Arabidopsis senses A. tumefaciens elongation factor EF-Tu through a transmembrane Leucine-rich-repeat (LRR) receptor kinase, EFR (At5g20480), to trigger downstream immune responses. A. thaliana efr mutants fail to elicit defence responses effectively against A. tumefaciens, and as a result the mutants are more susceptible to transient transformation in agroinfiltrated Arabidopsis leaves, as assessed using GUS as a reporter (Zipfel et al., 2006). Effector AvrPto from the plant bacterial pathogen Pseudomonas syringae weakens the immunity of Arabidopsis by targeting multiple pattern recognition receptors (PRRs) (Hauck et al., 2003; Xiang et al., 2008). The effector was engineered into A. thaliana, such that expression was driven by a dexamethasone (DEX)-inducible promoter. In the transgenic line, AvrPto was expressed only when DEX was applied. Pre-treating the plant with DEX before A. tumefaciens infiltration suppressed the plant immunity and hence the transient expression efficiency was significantly enhanced (Tsuda et al., 2012). Salicylic acid (SA) plays an important role in defense against bacteria, a recent report shows that use of nahG (a bacterial SA hydroxylase) expressing Arabidopsis plants can also enhance the transient transformation efficiency in Arabidopsis leaves. (Rosas-Díaz et al., 2017)
Leaf transient transformation has been employed for virus-induced gene silencing (VIGS). VIGS uses viral vectors to generate double-stranded RNA of a gene of interest to be silenced. While performing VIGS in Solanaceous plant species, including N. benthamiana, tomato, pepper, potato and petunia is straightforward, it is relatively difficult in A. thaliana due to the inconsistent transformation efficiency. Optimization of Tobacco rattle virus (TRV)-based VIGS in A. thaliana involved minimal modification of the N. benthamiana protocol, but transformation has to be done in seedlings at the two- to three-leaf stage (Burch-Smith et al., 2006). Light intensity and temperature were shown to affect systemic movement of the silencing signal in transient agroinfiltration in N. benthamiana. High light intensities (≥ 450 μE/m2/s) and temperatures (≥ 30°C) localized the silencing signal in the leaf tissue that was infiltrated. In contrast, lower light intensities with a constant temperature of 25°C supported strong systemic movement of the silencing signal (Patil and Fauquet, 2015). On the other hand, the transient VIGS technique can be applied to improve overall stable transformation efficiency. A. thaliana AGO2 (At1g31280) and NRPD1A (At1g63020) are inhibitors of Agrobacterium-mediated transformation. Transient transformation with AGO2 and NRPD1a silencing VIGS vectors by agroinfiltration of leaves prior to transformation with a gene of interest by floral dip was shown to increase the number of transgenic seeds by 6.0- and 3.5-fold, respectively (Bilichak et al., 2014).
Transient transformation in seedlings
Transient transformation of seedlings has the advantage of allowing analysis in the whole-plant cellular context, in contrast to localized agroinfiltration in adult leaves. Most seedling optimization has aimed to provide fast and convenient protocols for a preliminary analysis of uncharacterised genes and constructs. Vacuum infiltration was introduced to facilitate whole A. thaliana seedling adsorption of agrobacterial cells (Marion et al., 2008), while the Fast Agrobacterium-mediated seedling transformation (FAST) protocol optimized the cell density (OD600=0.5) and concentration of Silwet L-77 (0.005%) without the need for vacuuming (Li et al., 2009). FAST was demonstrated to work for Catharanthus roseus seedling transformation. However, it is worth noting that the FAST method strongly induces host de-fence genes Zct1 and Orca3; this should be taken into account in experimental design (Weaver et al., 2014). Optimization of A. tumefaciens culture conditions can also improve seedling transformation efficiency. The AGROBEST (Agrobacterium-mediated enhanced seedling transformation) method achieves high transient transformation efficiency in whole seedlings, allowing gain-of-function studies, by using a pH-buffered medium supplemented with AB salts and glucose. It has been shown that transient expression of GUS or LUC of Col-0 or efr-1 seedlings was significantly enhanced when AGROBEST was used during A. tumefaciens inoculation (Wu et al., 2014b). Similar to what was observed for agroinfiltration in adult leaves (Wroblewski et al., 2005), C58C1(pTiB6S3ΔT-DNA, pCH32) achieves higher transient transformation efficiency than the wild type C58 virulent strain or the C58-derived disarmed strain GV3101(pMP90) in A. thaliana seedlings (Wu et al., 2014b). This study also showed an inverse association of root growth inhibition and transient expression efficiency, suggesting that C58C1(pTiB6S3ΔT-DNA, pCH32) may circumvent a plant defense barrier to enable high transient expression levels in A. thaliana seedlings. However, the genetic factors and mechanisms underlying the ability of this chimeric disarmed A. tumefaciens strain to achieve the highest transient transformation efficiency await future investigations. By expressing a gene of interest separately with a GFP marker in one bicistronic vector, fluorescent GFP signals are used to identify and locate the cells that are successfully transformed by the vector; in the same cells, the signals also indicates expression of the gene of interest. It has been demonstrated that the bicistronic vector transiently expressing SYP121 (At3g11820) was able to rescue the syp121 mutant inward K+ channel current phenotype (Chen et al., 2011).
CONCLUSIONS AND FUTURE DIRECTIONS
From the discovery of Agrobacterium tumefaciens to the marketing of genetically modified (GM) crops, tremendous knowledge in plant gene function, cell biology, plant-microbe interaction and bio-technology has been unveiled. In particular, the optimized protocols for A. thaliana transformation have provided an irreplaceable tool for studying plant genetics by generation of random T-DNA insertion mutants. Seed stocks with various genomic disruptions by T-DNA insertion are available to order and have been used by plant scientists around the world. This enormous resource has greatly transformed plant research by streamlining studies of genes of interest. The convenience of Agrobacterium-mediated transformation in A. thaliana is also an ideal pilot scheme to test the effects of gene constructs before applying them in other plant species such as crops. At the same time, due to its role in plant applications, the biology of A. tumefaciens has been extensively studied. The most notable area is the study of the mechanism of T4SS, which mediates gene transfer from A. tumefaciens to plants. By comprehensively reviewing the functions of key genes involved in Agrobacterium-plant interactions and the transformation process including the recent breakthrough discovery of Pol θ in T-DNA integration, we hope to draw the attention of plant community to the importance of A. tumefaciens research and to the many unresolved questions about T-DNA translocation and integration process that await future studies.
Although A. tumefaciens is an excellent tool for dicot transformation, the efficiency in monocots, including in some important crop plants, is lagging far behind. Among the reasons for the poor efficiency may be the unique defense mechanisms in monocots and the absence of essential (efficient) components in monocots for T-DNA transfer and integration. One future direction could be investigating the defence signalling pathways in monocots against A. tumefaciens and introducing essential components into monocots. Combining the efficient transformation by A. tumefaciens with the CRISPR technique is also a growing trend in genome editing; this enables the removal of unwanted gene fragments such as antibiotic markers and non-essential foreign genes in the genome. The unmarked genome should help reduce public concerns about the potential toxicity of the current GM crops because of whole T-DNA integration. Apart from T4SS, A. tumefaciens possesses the T6SS which carries antibacterial activity for interbacterial competition; its secretion activity was shown to be suppressed when virulence proteins including T4SS were massively expressed (Wu et al., 2012). Some bacterial T6SS effectors are able to manipulate host immunity in animal systems, but there is no solid evidence to suggest that A. tumefaciens or other plant pathogens can use their T6SS effectors to manipulate host immunity in plants. Future efforts to identifying bacterial T6SS effectors targeting plants could be beneficial to enhance transformation efficiency. Given the increasing demands for plant engineering in medicine, environmental protection and food security, it is expected that A. tumefaciens will continue to play an important role in plant sciences, and understanding the biology of agrobacteria as well as its interactions with plants will remain essential, contributing to scientific developments throughout the plant domain.
Acknowledgments
We thank the anonymous reviewers for careful reading and valuable suggestions of this review. We also thank the Hwang and Lai laboratory members for suggestions and Miss Shin-Fei Chi for help with the figure. This work is made possible by grants from Academia Sinica and Ministry of Science and Technology (MOST 104-2311-B-001-025-MY3), Taiwan, to E.-M.L. and from Ministry of Education and Ministry of Science and Technology (MOST 105-2313-B-005-008), Taiwan, to H.-H.H. M.Y has been awarded a postdoctoral fellowship from Academia Sinica.
REFERENCES
- Agarwal S, Loar S, Steber C, Zale J. Floral transformation of wheat. Methods Mol. Biol. 2008:105–113. doi: 10.1007/978-1-59745-379-0_6. [DOI] [PubMed] [Google Scholar]
- Aguilar J, Cameron T.A, Zupan J, Zambryski P. Membrane and core periplasmic Agrobacterium tumefaciens virulence type IV secretion system components localize to multiple sites around the bacterial perimeter during lateral attachment to plant cells. 2011:e00218–00211. doi: 10.1128/mBio.00218-11. mBio. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar J, Zupan J, Cameron T.A, Zambryski P.C. Agrobacterium type IV secretion system and its substrates form helical arrays around the circumference of virulence-induced cells. Proc. Natl. Acad. Sci. USA. 2010:3758–3763. doi: 10.1073/pnas.0914940107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akama K, Shiraishi H, Ohta S, Nakamura K, Okada K, Shimura Y. Efficient transformation of Arabidopsis thaliana comparison of the efficiencies with various organs, plant ecotypes and Agrobacterium strains. Plant Cell Reps. 1992:7–11. doi: 10.1007/BF00232413. [DOI] [PubMed] [Google Scholar]
- Akiyoshi D.E, Regier D.A, Gordon M.P. Cytokinin production by Agrobacterium and Pseudomonas spp. J. Bacteriol. 1987:4242–4248. doi: 10.1128/jb.169.9.4242-4248.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi D.E, Regier D.A, Jen G, Gordon M.P. Cloning and nucleotide sequence of thetzsgene from Agrobacterium tumefaciens strain T37. Nucleic Acids Res. 1985:2773–2788. doi: 10.1093/nar/13.8.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albright L.M, Yanofsky M.F, Leroux B, Ma D.Q, Nester E.W. Processing of the T-DNA of Agrobacterium tumefaciens generates border nicks and linear, single-stranded T-DNA. J. Bacteriol. 1987:1046–1055. doi: 10.1128/jb.169.3.1046-1055.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M, Stepanova A.N. T-DNA mutagenesis in Arabidopsis. Methods Mol. Biol. 2003:177–188. doi: 10.1385/1-59259-413-1:177. [DOI] [PubMed] [Google Scholar]
- Alonso J.M, Stepanova A.N, Leisse T.J, Kim C.J, Chen H, Shinn P, Stevenson D.K, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers C.C, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter D.E, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby W.L, Berry C.C, Ecker J.R. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Altmann T, Damm B, Frommer W, Martin T, Morris P, Schweizer D, Willmitzer L, Schmidt R. Easy determination of ploidy level in Arabidopsis thaliana plants by means of pollen size measurement. Plant Cell Reports. 1994:652–656. doi: 10.1007/BF00232939. [DOI] [PubMed] [Google Scholar]
- Altpeter F, Baisakh N, Beachy R, Bock R, Capell T, Christou P, Daniell H, Datta K, Datta S, Dix P.J, Fauquet C, Huang N, Kohli A, Mooibroek H, Nicholson L, Nguyen T.T, Nugent G, Raemakers K, Romano A, Somers D.A, Stoger E, Taylor N, Visser R. Particle bombardment and the genetic enhancement of crops: myths and realities. Mol. Breed. 2005:305–327. [Google Scholar]
- Alvarez-Martinez C.E, Christie P.J. Biological diversity of prokaryotic type IVsecretion systems. Microbiol. Mol. Biol. Rev. 2009:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly K.A, Baron C. The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens. Microbiology. 2007:3766–3775. doi: 10.1099/mic.0.2007/010462-0. [DOI] [PubMed] [Google Scholar]
- Aly K.A, Krall L, Lottspeich F, Baron C. The type IV secretion system component VirB5 binds to the trans-zeatin biosynthetic enzyme Tzs and enables its translocation to the cell surface of Agrobacterium tumefaciens. J. Bacteriol. 2008:1595–1604. doi: 10.1128/JB.01718-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anami S, Njuguna E, Coussens G, Aesaert S, Van Lijsebettens M. Higher plant transformation: principles and molecular tools. Int. J. Dev. Biol. 2013:483–494. doi: 10.1387/ijdb.130232mv. [DOI] [PubMed] [Google Scholar]
- Anand A, Krichevsky A, Schornack S, Lahaye T, Tzfira T, Tang Y, Citovsky V, Mysore K.S. Arabidopsis VIRE2 INTERACTING PROTEIN2 is required for Agrobacterium T-DNA integration in plants. Plant Cell. 2007:1695–1708. doi: 10.1105/tpc.106.042903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A, Uppalapati S.R, Ryu C.M, Allen S.N, Kang L, Tang Y, Mysore K.S. Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens. Plant Physiol. 2008:703–715. doi: 10.1104/pp.107.111302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A, Rojas C.M, Tang Y, Mysore K.S. Several components of SKP1/Cullin/F-box E3 ubiquitin ligase complex and associated factors play a role in Agrobacteriummediated plant transformation. New Phytol. 2012:203–216. doi: 10.1111/j.1469-8137.2012.04133.x. [DOI] [PubMed] [Google Scholar]
- Ankenbauer R.G, Nester E.W. Sugar-mediated induction of Agrobacterium tumefaciens virulence genes: structural specificity and activities of monosaccharides. J. Bacteriol. 1990:6442–6446. doi: 10.1128/jb.172.11.6442-6446.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Christie P.J. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 2004:1199–1211. doi: 10.1111/j.1365-2958.2004.04345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Fronzes R, Waksman G. VirB2 and VirB5 proteins: specialized adhesins in bacterial type-IV secretion systems? Trends Microbiol. 2008:409–413. doi: 10.1016/j.tim.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Bako L, Umeda M, Tiburcio A.F, Schell J, Koncz C. The VirD2 pilot protein of Agrobacteriumtransferred DNA interacts with the TATA box-binding protein and a nuclear protein kinase in plants. Proc. Natl. Acad. Sci. USA. 2003:10108–10113. doi: 10.1073/pnas.1733208100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Citovsky V. Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc. Natl. Acad. Sci. USA. 1997:10723–10728. doi: 10.1073/pnas.94.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta L.M, Montenegro M. Agrobacterium and plant biotechnology. In: Tzfira T, Citovsky V, editors. Agrobacterium From Biology to Biotechnology. Springer Science+Business Media; Berlin: 2008. pp. 73–147. [Google Scholar]
- Banta L.M, Bohne J, Lovejoy S.D, Dostal K. Stability of the Agrobacterium tumefaciens VirB10 protein is modulated by growth temperature and periplasmic osmoadaption. J. Bacteriol. 1998:6597–6606. doi: 10.1128/jb.180.24.6597-6606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta L.M, Kerr J.E, Cascales E, Giuliano M.E, Bailey M.E, McKay C, Chandran V, Waksman G, Christie P.J. An Agrobacterium VirB10 mutation conferring a Type IV secretion system gating defect. J. Bacteriol. 2011:2566–2574. doi: 10.1128/JB.00038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron C, Domke N, Beinhofer M, Hapfelmeier S. Elevated temperature differentially affects virulence, VirB protein accumulation, and T-pilus formation in different Agrobacterium tumefaciens and Agrobacterium vitis strains. J. Bacteriol. 2001:6852–6861. doi: 10.1128/JB.183.23.6852-6861.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron C, Llosa M, Zhou S, Zambryski P.C. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens is processed to a C-terminal secreted product, VirB1. J. Bacteriol. 1997:1203–1210. doi: 10.1128/jb.179.4.1203-1210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholmes C, Nutt P, Theiβen G. Germline transformation of Shepherd's purse (Capsella bursa-pastoris) by the ‘floral dip’ method as a tool for evolutionary and developmental biology. Gene. 2008:11–19. doi: 10.1016/j.gene.2007.10.033. [DOI] [PubMed] [Google Scholar]
- Barton K.A, Binns A.N, Matzke A.J.M, Chilton M.-D. Regeneration of intact tobacco plants containing full length copies of genetically engineered T-DNA, and transmission of T-DNA to R1 progeny. Cell. 1983:1033–1043. doi: 10.1016/0092-8674(83)90288-x. [DOI] [PubMed] [Google Scholar]
- Beale M, Dupree P, Lilley K, Beynon J, Trick M, Clarke J, Bevan M, Bancroft I, Jones J, May S, van de Sande K, Leyser O. GARNet, the genomic Arabidopsis resource network. Trends Plant Sci. 2002:145–147. doi: 10.1016/s1360-1385(01)02224-5. [DOI] [PubMed] [Google Scholar]
- Beaty J.S, Powell G.K, Lica L, Regier D.A, MacDonald E.M.S, Hommes N.G, Morris R.O. Tzs a nopaline Ti plasmid gene from Agrobacterium tumefaciens associated with transzeatin bio-synthesis. Mol. Gen. Genet. 1986:274–280. [Google Scholar]
- Bechtold N, Pelletier G. In planta Agrobacterium mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In-planta Agrobacteriummediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris, Life Sci. 1993:1194–1199. [Google Scholar]
- Bechtold N, Jaudeau B, Jolivet S, Maba B, Vezon D, Voisin R, Pelletier G. The maternal chromosome set is the target of the T-DNA in the in planta transformation of Arabidopsis thaliana. Genetics. 2000:1875–1887. doi: 10.1093/genetics/155.4.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Jolivet S, Voisin R, Pelletier G. The endo-sperm and the embryo of Arabidopsis thaliana are independently transformed through infiltration by Agrobacterium tumefaciens. Transgenic Res. 2003:509–517. doi: 10.1023/a:1024272023966. [DOI] [PubMed] [Google Scholar]
- Bent A.F. Arabidopsis in planta transformation. Uses, mechanisms, and prospects for transformation of other species. Plant Physiol. 2000:1540–1547. doi: 10.1104/pp.124.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A.F. Arabidopsis thaliana floral dip transformation method. Methods Mol. Biol. 2006:87–104. doi: 10.1385/1-59745-130-4:87. [DOI] [PubMed] [Google Scholar]
- Berger B.R, Christie P.J. The Agrobacterium tumefaciens VirB4 gene-product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J. Bacteriol. 1993:1723–1734. doi: 10.1128/jb.175.6.1723-1734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Lee L.Y, Oltmanns H, Cao H, Veena, Cuperus J, Gelvin S.B. AtImpa-4, an Arabidopsis importin α isoform, is preferentially involved in Agrobacteriummediated plant transformation. Plant Cell. 2008:2661–2680. doi: 10.1105/tpc.108.060467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Sood P, Citovsky V. The roles of plant phenolics in defence and communication during Agrobacterium and Rhizobium infection. Mol. Plant Pathol. 2010:705–719. doi: 10.1111/j.1364-3703.2010.00625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatty M, Laverde Gomez J.A, Christie P.J. The expanding bacterial type IV secretion lexicon. Res. Microbiol. 2013:620–639. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilichak A, Yao Y.L, Kovalchuk I. Transient down-regulation of the RNA silencing machinery increases efficiency of Agrobacteriummediated transformation of Arabidopsis. Plant Biotechnol. J. 2014:590–600. doi: 10.1111/pbi.12165. [DOI] [PubMed] [Google Scholar]
- Bravo-Angel A.M, Hohn B, Tinland B. The omega sequence of VirD2 is important but not essential for efficient transfer of T-DNA by Agrobacterium tumefaciens. Mol. Plant Microbe Interact. 1998:57–63. doi: 10.1094/MPMI.1998.11.1.57. [DOI] [PubMed] [Google Scholar]
- Brencic A, Eberhard A, Winans S.C. Signal quenching, detoxification and mineralization of vir gene-inducing phenolics by the VirH2 protein of Agrobacterium tumefaciens. Mol. Microbiol. 2003:1103–1115. doi: 10.1046/j.1365-2958.2003.03887.x. [DOI] [PubMed] [Google Scholar]
- Broothaerts W, Mitchell H.J, Weir B, Kaines S, Smith L.M.A, Yang W, Mayer J.E, Roa-Rodríguez C, Jefferson R.A. Gene transfer to plants by diverse species of bacteria. Nature. 2005:629–633. doi: 10.1038/nature03309. [DOI] [PubMed] [Google Scholar]
- Bulgakov V.P, Kiselev K.V, Yakovlev K.V, Zhuravlev Y.N, Gontcharov A.A, Odintsova N.A. Agrobacteriummediated transformation of sea urchin embryos. Biotechnol. J. 2006:454–461. doi: 10.1002/biot.200500045. [DOI] [PubMed] [Google Scholar]
- Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas P.J.J. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995:3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P, Mroczek K, Winkler A.A, Steensma H.Y, Hooykaas P.J.J. T-DNA from Agrobacterium tumefaciens as an efficient tool for gene targeting in Kluyveromyces lactis. Mol. Gen. Genet. 1999:115–121. doi: 10.1007/s004380050948. [DOI] [PubMed] [Google Scholar]
- Burch-Smith T.M, Schiff M, Liu Y, Dinesh-Kumar S.P. Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol. 2006:21–27. doi: 10.1104/pp.106.084624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron T.A, Roper M, Zambryski P.C. Quantitative image analysis and modeling indicate the Agrobacterium tumefaciens type IV secretion system is organized in a periodic pattern of foci. PLoS ONE. 2012 doi: 10.1371/journal.pone.0042219. e42219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanoni P, Sutter J.U, Davis C.S, Littlejohn G.R, Blatt M.R. A generalized method for transfecting root epidermis uncovers endosomal dynamics in Arabidopsis root hairs. Plant J. 2007:322–330. doi: 10.1111/j.1365-313X.2007.03139.x. [DOI] [PubMed] [Google Scholar]
- Cangelosi G.A, Martinetti G, Leigh J.A, Lee C.C, Theines C, Nester E.W. Role for Agrobacterium tumefaciens ChvA protein in export of beta-1,2-glucan. J. Bacteriol. 1989:1609–1615. doi: 10.1128/jb.171.3.1609-1615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie P.J. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004a:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie P.J. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc. Natl. Acad. Sci. USA. 2004b:17228–17233. doi: 10.1073/pnas.0405843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Atmakuri K, Sarkar M.K, Christie P.J. DNA substrate-induced activation of the Agrobacterium VirB/VirD4 type IV secretion system. J. Bacteriol. 2013:2691–2704. doi: 10.1128/JB.00114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran V. Type IV secretion machinery: molecular architecture and function. Biochem. Soc. Trans. 2013:17–28. doi: 10.1042/BST20120332. [DOI] [PubMed] [Google Scholar]
- Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009:1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.H, Winans S.C. Resection and mutagenesis of the acid pH-inducible P2 promoter of the Agrobacterium tumefaciens virG gene. J. Bacteriol. 1996:4717–4720. doi: 10.1128/jb.178.15.4717-4720.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.S, Park S.K, Kim B.C, Kang B.J, Kim D.U, Nam H.G. Stable genetic transformation of Arabidopsis thaliana by Agrobacterium inoculation in planta. Plant J. 1994:551–558. [Google Scholar]
- Chateau S. Competence of Arabidopsis thaliana genotypes and mutants for Agrobacterium tumefaciensmediated gene transfer: role of phytohormones. J. Exp. Bot. 2000:1961–1968. doi: 10.1093/jexbot/51.353.1961. [DOI] [PubMed] [Google Scholar]
- Chen Z.H, Grefen C, Donald N, Hills A, Blatt M.R. A bicistronic, Ubiquitin-10 promoter-based vector cassette for transient transformation and functional analysis of membrane transport demonstrates the utility of quantitative voltage clamp studies on intact Arabidopsis root epidermis. Plant Cell Environ. 2011:554–564. doi: 10.1111/j.1365-3040.2010.02262.x. [DOI] [PubMed] [Google Scholar]
- Christie P.J, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 2005:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P.J, Ward J.E, Gordon M.P, Nester E.W. A gene required for transfer of T-DNA to plants encodes an ATPase with autophosphorylating activity. Proc. Natl. Acad. Sci. USA. 1989:9677–9681. doi: 10.1073/pnas.86.24.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P.J, Whitaker N, Gonzalez-Rivera C. Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta. 2014:1578–1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M.H, Chen M.K, Pan S.M. Floral spray transformation can efficiently generate Arabidopsis transgenic plants. Transgenic Res. 2000:471–476. doi: 10.1023/a:1026522104478. [DOI] [PubMed] [Google Scholar]
- Chung S.-M, Frankman E.L, Tzfira T. A versatile vector system for multiple gene expression in plants. Trends Plant Sci. 2005:357–361. doi: 10.1016/j.tplants.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Citovsky V, Kapelnikov A, Oliel S, Zakai N, Rojas M.R, Gilbertson R.L, Tzfira T, Loyter A. Protein interactions involved in nuclear import of the Agrobacterium VirE2 protein in vivo and in vitro. J. Biol. Chem. 2004:29528–29533. doi: 10.1074/jbc.M403159200. [DOI] [PubMed] [Google Scholar]
- Citovsky V, Kozlovsky S.V, Lacroix B, Zaltsman A, Dafny-Yelin M, Vyas S, Tovkach A, Tzfira T. Biological systems of the host cell involved in Agrobacterium infection. Cell Microbiol. 2007:9–20. doi: 10.1111/j.1462-5822.2006.00830.x. [DOI] [PubMed] [Google Scholar]
- Clarke M.C, Wei W, Lindsey K. High-frequency transformation of Arabidopsis thaliana by Agrobacterium tumefaciens. Plant Mol. Biol. Reporter. 1992:178–189. [Google Scholar]
- Clough S.J, Bent A.F. Floral dip: a simplified method for Agrobacteriummediated transformation of Arabidopsis thaliana. Plant J. 1998:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran F.A, Cox D, Lin S, Barretto R, Habib N, Hsu P.D, Wu X, Jiang W, Marraffini L.A, Zhang F. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane Y.M, Gelvin S.B. RNAi-mediated gene silencing reveals involvement of Arabidopsis chromatin-related genes in Agrobacteriummediated root transformation. Proc. Natl. Acad. Sci. USA. 2007:15156–15161. doi: 10.1073/pnas.0706986104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis I.S, Nam H.G. Transgenic radish (Raphanus sativus L. longipinnatus Bailey) by floral-dip method - plant development and surfactant are important in optimizing transformation efficiency. Transgenic Res. 2001:363–371. doi: 10.1023/a:1016600517293. [DOI] [PubMed] [Google Scholar]
- Czako M.l, Wilson J, Yu X, Morton L.s. Sustained root culture for generation and vegetative propagation of transgenic Arabidopsis thaliana. Plant Cell Rep. 1993:603–606. doi: 10.1007/BF00232807. [DOI] [PubMed] [Google Scholar]
- De Buck S, De Wilde C, Van Montagu M, Depicker A. Determination of the T-DNA transfer and the T-DNA integration frequencies upon cocultivation of Arabidopsis thaliana root explants. Mol. Plant Microbe Interact. 2000:658–665. doi: 10.1094/MPMI.2000.13.6.658. [DOI] [PubMed] [Google Scholar]
- De Buck S, Jacobs A, Van Montagu M, Depicker A. Agrobacterium tumefaciens transformation and cotransformation frequencies of Arabidopsis thaliana root explants and tobacco protoplasts. Mol. Plant Microbe Interact. 1998:449–457. doi: 10.1094/MPMI.1998.11.6.449. [DOI] [PubMed] [Google Scholar]
- De Buck S, Podevin N, Nolf J, Jacobs A, Depicker A. The T-DNA integration pattern in Arabidopsis transformants is highly determined by the transformed target cell. Plant J. 2009:134–145. doi: 10.1111/j.1365-313X.2009.03942.x. [DOI] [PubMed] [Google Scholar]
- De Cleene M, De Ley J. The host range of crown gall. Bot. Rev. 1976:389–466. [Google Scholar]
- de Framond A.J, Barton K.A, Chilton M.-D. Mini–Ti: A new vector strategy for plant genetic engineering. Nat. Biotechnol. 1983:262–269. [Google Scholar]
- de Groot M.J.A, Bundock P, Hooykaas P.J.J, Beijersbergen A.G.M. Agrobacterium tumefaciensmediated transformation of filamentous fungi. Nat. Biotechnol. 1998:839–842. doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- de la Peña A, Lörz H, Schell J. Transgenic rye plants obtained by injecting DNA into young floral tillers. Nature. 1987:274–276. [Google Scholar]
- Deblaere R, Bytebier B, De Greve H, Deboeck F, Schell J, Van Montagu M, Leemans J. Efficient octopine Ti plasmid-derived vectors for Agrobacteriummediated gene transfer to plants. Nucleic Acids Res. 1985:4777–4788. doi: 10.1093/nar/13.13.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNeve M, DeBuck S, Jacobs A, VanMontagu M, Depicker A. T-DNA integration patterns in co-transformed plant cells suggest that T-DNA repeats originate from co-integration of separate TDNAs. Plant J. 1997:15–29. doi: 10.1046/j.1365-313x.1997.11010015.x. [DOI] [PubMed] [Google Scholar]
- Deng W, Chen L, Wood D.W, Metcalfe T, Liang X, Gordon M.P, Comai L, Nester E.W. Agrobacterium VirD2 protein interacts with plant host cyclophilins. Proc. Natl. Acad. Sci. USA. 1998:7040–7045. doi: 10.1073/pnas.95.12.7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desfeux C, Clough S.J, Bent A.F. Female reproductive tissues are the primary target of Agrobacteriummediated transformation by the Arabidopsis floral-dip method. Plant Physiol. 2000:895–904. doi: 10.1104/pp.123.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaux Y, Petit A, Tempe J. Molecular Signals in Plant-Microbe Communications. Boca Raton: CRC Press; 1992. Opines in Agrobacterium biology; pp. 109–136. [Google Scholar]
- Ditt R.F, Kerr K.F, de Figueiredo P, Delrow J, Comai L, Nester E.W. The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol. Plant Microbe Interact. 2006:665–681. doi: 10.1094/MPMI-19-0665. [DOI] [PubMed] [Google Scholar]
- Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H. Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science. 2007:453–456. doi: 10.1126/science.1148110. [DOI] [PubMed] [Google Scholar]
- Doty S.L, Yu M.C, Lundin J.I, Heath J.D, Nester E.W. Mutational analysis of the input domain of the VirA protein of Agrobacterium tumefaciens. J. Bacteriol. 1996:961–970. doi: 10.1128/jb.178.4.961-970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas C, Halperin W, Gordon M, Nester E. Specific attachment of Agrobacterium tumefaciens to bamboo cells in suspension cultures. J. Bacteriol. 1985:764–766. doi: 10.1128/jb.161.2.764-766.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas C.J, Halperin W, Nester E.W. Agrobacterium tumefaciens mutants affected in attachment to plant cells. J. Bacteriol. 1982:1265–1275. doi: 10.1128/jb.152.3.1265-1275.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E, Oomen C, Waksman G. Biochemical dissection of the ATPase TraB, the VirB4 homologue of the Escherichia coli pKM101 conjugation machinery. J. Bacteriol. 2010:2315–2323. doi: 10.1128/JB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E, Waksman G, Receveur-Brechot V. Structural insights into the membrane-extracted dimeric form of the ATPase TraB from the Escherichia coli pKM101 conjugation system. BMC Struct. Biol. 2011:4. doi: 10.1186/1472-6807-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrenberger F, Crameri A, Hohn B, Koukolikova-Nicola Z. Covalently bound VirD2 protein of Agrobacterium tumefaciens protects the T-DNA from exonucleolytic degradation. Proc. Natl. Acad. Sci. USA. 1989:9154–9158. doi: 10.1073/pnas.86.23.9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbrandt R, Kalkum M, Lai E.M, Lurz R, Kado C.I, Lanka E. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 1999:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- Endo M, Ishikawa Y, Osakabe K, Nakayama S, Kaya H, Araki T, Shibahara K.-i, Abe K, Ichikawa H, Valentine L, Hohn B, Toki S. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 2006:5579–5590. doi: 10.1038/sj.emboj.7601434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enríquez-Obregón G.A, Vázquez-Padrón R.I, Prieto-Samsonov D.L, De la Riva G.A, Selman-Housein G. Herbicide-resistant sugarcane (Saccharum officinarum L.) plants by Agrobacteriummediated transformation. Planta. 1998:20–27. [Google Scholar]
- Feldmann K.A, Marks M.D. Agrobacteriummediated transformation of germinating seeds of Arabidopsis thaliana A non-tissue culture approach. Mol. Gen. Genet. 1987:1–9. [Google Scholar]
- Filichkin S.A, Gelvin S.B. Formation of a putative relaxation intermediate during T-DNA processing directed by the Agrobacterium tumefaciens VirD1, D2 endonuclease. Mol. Microbiol. 1993:915–926. doi: 10.1111/j.1365-2958.1993.tb01637.x. [DOI] [PubMed] [Google Scholar]
- Frandsen R.J. A guide to binary vectors and strategies for targeted genome modification in fungi using Agrobacterium tumefaciensmediated transformation. J. Microbiol. Methods. 2011:247–262. doi: 10.1016/j.mimet.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Friesner J, Britt A.B. Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J. 2003:427–440. doi: 10.1046/j.1365-313x.2003.01738.x. [DOI] [PubMed] [Google Scholar]
- Fromm M, Taylor L.P, Walbot V. Expression of genes transferred into monocot and dicot plant cells by electroporation. Proc. Natl. Acad. Sci. USA. 1985:5824–5828. doi: 10.1073/pnas.82.17.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm M, Taylor L.P, Walbot V. Stable transformation of maize after gene transfer by electroporation. Nature. 1986:791–793. doi: 10.1038/319791a0. [DOI] [PubMed] [Google Scholar]
- Fronzes R, Christie P.J, Waksman G. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 2009a:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronzes R, Schafer E, Wang L, Saibil H.R, Orlova E.V, Waksman G. Structure of a type IV secretion system core complex. Science. 2009b:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullner K.J, Nester E.W. Temperature affects the T-DNA transfer machinery of Agrobacterium tumefaciens. J. Bacteriol. 1996:1498–1504. doi: 10.1128/jb.178.6.1498-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullner K.J, Stephens K.M, Nester E.W. An essential virulence protein of Agrobacterium tumefaciens VirB4, requires an intact mononucleotide binding domain to function in transfer of T-DNA. Mol. Gen. Genet. 1994:704–715. doi: 10.1007/BF00297277. [DOI] [PubMed] [Google Scholar]
- Gallego M.E, Bleuyard J.Y, Daoudal-Cotterell S, Jallut N, White C.I. Ku80 plays a role in non-homologous recombination but is not required for T-DNA integration in Arabidopsis. Plant J. 2003:557–565. doi: 10.1046/j.1365-313x.2003.01827.x. [DOI] [PubMed] [Google Scholar]
- Gao C, Long D, Lenk I, Nielsen K.K. Comparative analysis of transgenic tall fescue (Festuca arundinacea Schreb.) plants obtained by Agrobacteriummediated transformation and particle bombardment. Plant Cell Rep. 2008:1601–1609. doi: 10.1007/s00299-008-0578-x. [DOI] [PubMed] [Google Scholar]
- Gao R, Lynn D.G. Environmental pH sensing: resolving the VirA/VirG two-component system inputs for Agrobacterium pathogenesis. J. Bacteriol. 2005:2182–2189. doi: 10.1128/JB.187.6.2182-2189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Hernandez M, Berardini T.Z, Chen G, Crist D, Doyle A, Huala E, Knee E, Lambrecht M, Miller N, Mueller L.A, Mundodi S, Reiser L, Rhee S.Y, Scholl R, Tacklind J, Weems D.C, Wu Y, Xu I, Yoo D, Yoon J, Zhang P. TAIR: a resource for integrated Arabidopsis data. Funct. Integr. Genomics. 2002:239–253. doi: 10.1007/s10142-002-0077-z. [DOI] [PubMed] [Google Scholar]
- Garza I, Christie P.J. A putative transmembrane leucine zipper of Agrobacterium VirB10 is essential for T-Pilus biogenesis but not type IV secretion. J. Bacteriol. 2013:3022–3034. doi: 10.1128/JB.00287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar Y.M, Nam J, Schultz C.J, Lee L.Y, Gilson P.R, Gelvin S.B. Characterization of the Arabidopsis lysine-rich arabinogalactan-protein AtAGP17 mutant (rat1) that results in a decreased efficiency of Agrobacterium Transformation. Plant Physiol. 2004:2162–2171. doi: 10.1104/pp.104.045542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin S.B. Agrobacterium transformation of Arabidopsis thaliana roots. Methods Mol. Biol. 2006:105–114. doi: 10.1385/1-59745-130-4:105. [DOI] [PubMed] [Google Scholar]
- Gelvin S.B. Finding a way to the nucleus. Curr. Opin. Microbiol. 2010a:53–58. doi: 10.1016/j.mib.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Gelvin S.B. Plant proteins involved in Agrobacteriummediated genetic transformation. Annu. Rev. Phytopathol. 2010b:45–68. doi: 10.1146/annurev-phyto-080508-081852. [DOI] [PubMed] [Google Scholar]
- Gelvin S.B. Traversing the Cell: Agrobacterium T-DNA's journey to the host genome. Front. Plant Sci. 2012:52. doi: 10.3389/fpls.2012.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin S.B, Kim S.I. Effect of chromatin upon Agrobacterium T-DNA integration and transgene expression. Biochim. Biophys. Acta. 2007:410–421. doi: 10.1016/j.bbaexp.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Gheysen G, Angenon G, Van Montagu M. Agrobacteriummediated plant transformation: a scientifically intriguing story with significant applications. In: Lindsey K, editor. Transgenic Plant Research. Harwood Academic Publishers; 1998. pp. 1–33. [Google Scholar]
- Gohlke J, Deeken R. Plant responses to Agrobacterium tumefaciens and crown gall development. Front. Plant Sci. 2014:155. doi: 10.3389/fpls.2014.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevelding C, Fantes V, Kemper E, Schell J, Masterson R. Single-copy T-DNA insertions in Arabidopsis are the predominant form of integration in root-derived transgenics, whereas multiple insertions are found in leaf discs. Plant Mol. Biol. 1993:847–860. doi: 10.1007/BF00021539. [DOI] [PubMed] [Google Scholar]
- Guo M, Hou Q, Hew C.L, Pan S.Q. Agrobacterium VirD2-binding protein is involved in tumorigenesis and redundantly encoded in conjugative transfer gene clusters. Mol. Plant Microbe Interact. 2007a:1201–1212. doi: 10.1094/MPMI-20-10-1201. [DOI] [PubMed] [Google Scholar]
- Guo M, Jin S, Sun D, Hew C.L, Pan S.Q. Recruitment of conjugative DNA transfer substrate to Agrobacterium type IV secretion apparatus. Proc. Natl. Acad. Sci. USA. 2007b:20019–20024. doi: 10.1073/pnas.0701738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton R.H, Fall M.Z. The loss of tumor-initiating ability in Agrobacterium tumefaciens by incubation at high temperature. Experientia. 1971:229–230. doi: 10.1007/BF02145913. [DOI] [PubMed] [Google Scholar]
- Hansen G, Shillito R.D, Chilton M.D. T-strand integration in maize protoplasts after codelivery of a T-DNA substrate and virulence genes. Proc. Natl. Acad. Sci. USA. 1997:11726–11730. doi: 10.1073/pnas.94.21.11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck P, Thilmony R, He S.Y. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA. 2003:8577–8582. doi: 10.1073/pnas.1431173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel B.C, Tomlinson A.D, Morton E.R, Choi J.H, Fuqua C. Agrobacterium tumefaciens ExoR controls acid response genes and impacts exopolysaccharide synthesis, horizontal gene transfer, and virulence gene expression. J. Bacteriol. 2014:3221–3233. doi: 10.1128/JB.01751-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindl J.E, Wang Y, Heckel B.C, Mohari B, Feirer N, Fuqua C. Mechanisms and regulation of surface interactions and biofilm formation in Agrobacterium. Front. Plant Sci. 2014:176. doi: 10.3389/fpls.2014.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R, Mullineaux P, Klee H. Technical focus: A guide to Agrobacterium binary Ti vectors. Trends Plant Sci. 2000:446–451. doi: 10.1016/s1360-1385(00)01740-4. [DOI] [PubMed] [Google Scholar]
- Hepburn A.G, White J, Pearson L, Maunders M.J, Clarke L.E, Prescott A.G, Blundy K.S. The use of pNJ5000 as an intermediate vector for the genetic manipulation of Agrobacterium Tiplasmids. J. Gen. Microbiol. 1985:2961–2969. doi: 10.1099/00221287-131-11-2961. [DOI] [PubMed] [Google Scholar]
- Herrera-Estrella A, Chen Z.M, Van Montagu M, Wang K. VirD proteins of Agrobacterium tumefaciens are required for the formation of a covalent DNA--protein complex at the 5′ terminus of T-strand molecules. EMBO J. 1988:4055–4062. doi: 10.1002/j.1460-2075.1988.tb03299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Estrella L, Simpson J, Martínez-Trujillo M. Transgenic plants: an historical perspective. Methods Mol. Biol. 2005:3–32. doi: 10.1385/1-59259-827-7:003. [DOI] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao R.P, Bitton F, Caboche M, Cannoot B, Chardakov V, Cognet-Holliger C, Colot V, Crowe M, Darimont C, Durinck S, Eickhoff H, de Longevialle A.F, Farmer E.E, Grant M, Kuiper M.T, Lehrach H, Leon C, Leyva A, Lundeberg J, Lurin C, Moreau Y, Nietfeld W, Paz-Ares J, Reymond P, Rouze P, Sandberg G, Segura M.D, Serizet C, Tabrett A, Taconnat L, Thareau V, Van Hummelen P, Vercruysse S, Vuylsteke M, Weingartner M, Weisbeek P.J, Wirta V, Wittink F.R, Zabeau M, Small I. Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res. 2004:2176–2189. doi: 10.1101/gr.2544504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekema A, Hirsch P.R, Hooykaas P.J.J, Schilperoort R.A. A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature. 1983:179–180. [Google Scholar]
- Holsters M, Silva B, Van Vliet F, Genetello C, De Block M, Dhaese P, Depicker A, Inzé D, Engler G, Villarroel R, Van Montagu M, Schell J. The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid. 1980:212–230. doi: 10.1016/0147-619x(80)90110-9. [DOI] [PubMed] [Google Scholar]
- Hood E.E, Gelvin S.B, Melchers L.S, Hoekema A. New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 1993:208–218. [Google Scholar]
- Hood E.E, Helmer G.L, Fraley R.T, Chilton M.D. The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J. Bacteriol. 1986:1291–1301. doi: 10.1128/jb.168.3.1291-1301.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooykaas P.J.J. Transformation mediated by Agrobacterium tumefaciens. In: Tkacz J.S, Lange L, editors. Advances in Fungal Biotechnology for Industry, Agriculture, and Medicine. Boston, MA: Springer US; 2004. pp. 41–65. [Google Scholar]
- Howard E.A, Winsor B.A, De Vos G, Zambryski P. Activation of the T-DNA transfer process in Agrobacterium results in the generation of a T-strand-protein complex: Tight association of VirD2 with the 5′ ends of T-strands. Proc. Natl. Acad. Sci. USA. 1989:4017–4021. doi: 10.1073/pnas.86.11.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Zhao J, DeGrado W.F, Binns A.N. Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals. Proc. Natl. Acad. Sci. USA. 2012:678–683. doi: 10.1073/pnas.1215033110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Dickerman A.W, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, Mueller L.A, Bhattacharyya D, Bhaya D, Sobral B.W, Beavis W, Meinke D.W, Town C.D, Somerville C, Rhee S.Y. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001:102–105. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Mā H. An improved procedure for transforming Arabidopsis thaliana (Landsbergerecta) root explant. Plant Mol. Biol. Reporter. 1992:372–383. [Google Scholar]
- Hwang H.H, Gelvin S.B. Plant proteins that interact with VirB2, the Agrobacterium tumefaciens pilin protein, mediate plant transformation. Plant Cell. 2004:3148–3167. doi: 10.1105/tpc.104.026476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H.H, Liu Y.T, Huang S.C, Tung C.Y, Huang F.C, Tsai Y.L, Cheng T.F, Lai E.M. Overexpression of the HspL promotes Agrobacterium tumefaciens virulence in Arabidopsis under heat shock conditions. Phytopathology. 2015:160–168. doi: 10.1094/PHYTO-05-14-0133-R. [DOI] [PubMed] [Google Scholar]
- Hwang H.H, Wang M.H, Lee Y.L, Tsai Y.L, Li Y.H, Yang F.J, Liao Y.C, Lin S.K, Lai E.M. Agrobacteriumproduced and exogenous cytokinin-modulated Agrobacteriummediated plant transformation. Mol. Plant Pathol. 2010:677–690. doi: 10.1111/j.1364-3703.2010.00637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H.H, Wu E.T, Liu S.Y, Chang S.C, Tzeng K.C, Kado C.I. Characterization and host range of five tumorigenic Agrobacterium tumefaciens strains and possible application in plant transient transformation assays. Plant Pathol. 2013a:1384–1397. [Google Scholar]
- Hwang H.H, Yang F.J, Cheng T.F, Chen Y.C, Lee Y.L, Tsai Y.L, Lai E.M. The Tzs protein and exogenous cytokinin affect virulence gene expression and bacterial growth of Agrobacterium tumefaciens. Phytopathology. 2013b:888–899. doi: 10.1094/PHYTO-01-13-0020-R. [DOI] [PubMed] [Google Scholar]
- Hwang W.Y, Fu Y, Reyon D, Maeder M.L, Tsai S.Q, Sander J.D, Peterson R.T, Yeh J.R.J, Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotech. 2013c:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inan G, Zhang Q, Li P, Wang Z, Cao Z, Zhang H, Zhang C, Quist T.M, Goodwin S.M, Zhu J, Shi H, Damsz B, Charbaji T, Gong Q, Ma S, Fredricksen M, Galbraith D.W, Jenks M.A, Rhodes D, Hasegawa P.M, Bohnert H.J, Joly R.J, Bressan R.A, Zhu J.K. Salt Cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol. 2004:1718–1737. doi: 10.1104/pp.104.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski S.J, Cascales E, Krishnamoorthy V, Christie P.J. Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-Pilus biogenesis. J. Bacteriol. 2005:3486–3495. doi: 10.1128/JB.187.10.3486-3495.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski S.J, Kerr J.E, Garza I, Krishnamoorthy V, Bayliss R, Waksman G, Christie P.J. Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol. Microbiol. 2009:779–794. doi: 10.1111/j.1365-2958.2008.06565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski S.J, Krishnamoorthy V, Christie P.J. Agrobacterium tumefaciens VirB6 protein articipates in formation of VirB7 and VirB9 complexes required for type IV secretion. J. Bacteriol. 2003:2867–2878. doi: 10.1128/JB.185.9.2867-2878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski S.J, Krishnamoorthy V, Cascales E, Christie P.J. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J. Mol. Biol. 2004:961–977. doi: 10.1016/j.jmb.2004.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaswal R.K, Veluthambi K, Gelvin S.B, Slightom J.L. Double-stranded cleavage of T-DNA and generation of single-stranded T-DNA molecules in Escherichia coli by a virDencoded border-specific endonuclease from Agrobacterium tumefaciens. J. Bacteriol. 1987:5035–5045. doi: 10.1128/jb.169.11.5035-5045.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q, Bundock P, Hooykaas P.J.J, de Pater S. Agrobacterium tumefaciens T-DNA integration and gene targeting in Arabidopsis thaliana non-homologous end-joining mutants. J. Bot. 2012:1–13. [Google Scholar]
- Jin S, Roitsch T, Ankenbauer R.G, Gordon M.P, Nester E.W. The VirA protein of Agrobacterium tumefaciens is autophosphorylated and is essential for vir gene regulation. J. Bacteriol. 1990a:525–530. doi: 10.1128/jb.172.2.525-530.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S.G, Prusti R.K, Roitsch T, Ankenbauer R.G, Nester E.W. Phosphorylation of the VirG protein of Agrobacterium tumefaciens by the autophosphorylated VirA protein: essential role in biological activity of VirG. J. Bacteriol. 1990b:4945–4950. doi: 10.1128/jb.172.9.4945-4950.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S.G, Roitsch T, Christie P.J, Nester E.W. The regulatory VirG protein specifically binds to a cisacting regulatory sequence involved in transcriptional activation of Agrobacterium tumefaciens virulence genes. J. Bacteriol. 1990c:531–537. doi: 10.1128/jb.172.2.531-537.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H.D, Doherty A, Sparks C.A. Transient transformation of plants. Methods Mol. Biol. 2009:131–152. doi: 10.1007/978-1-59745-427-8_8. [DOI] [PubMed] [Google Scholar]
- Judd P.K, Kumar R.B, Das A. The type IV secretion apparatus protein VirB6 of Agrobacterium tumefaciens localizes to a cell pole. Mol. Microbiol. 2004:115–124. doi: 10.1111/j.1365-2958.2004.04378.x. [DOI] [PubMed] [Google Scholar]
- Judd P.K, Kumar R.B, Das A. Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc. Natl. Acad. Sci. USA. 2005:11498–11503. doi: 10.1073/pnas.0505290102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalogeraki V.S, Zhu J, Eberhard A, Madsen E.L, Winans S.C. The phenolic vir gene inducer ferulic acid is O-demethylated by the VirH2 protein of an Agrobacterium tumefaciens Ti plasmid. Mol. Microbiol. 1999:512–522. doi: 10.1046/j.1365-2958.1999.01617.x. [DOI] [PubMed] [Google Scholar]
- Kanemoto R.H, Powell A.T, Akiyoshi D.E, Regier D.A, Kerstetter R.A, Nester E.W, Hawes M.C, Gordon M.P. Nucleotide sequence and analysis of the plant-inducible locus pinF from Agrobacterium tumefaciens. J. Bacteriol. 1989:2506–2512. doi: 10.1128/jb.171.5.2506-2512.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katavic V, Haughn G.W, Reed D, Martin M, Kunst L. In planta transformation of Arabidopsis thaliana. Mol. Gen. Genet. 1994:363–370. doi: 10.1007/BF00290117. [DOI] [PubMed] [Google Scholar]
- Kelly B.A, Kado C.I. Agrobacteriummediated T-DNA transfer and integration into the chromosome of Streptomyces lividans. Mol. Plant Pathol. 2002:125–134. doi: 10.1046/j.1364-3703.2002.00104.x. [DOI] [PubMed] [Google Scholar]
- Kemper E, Grevelding C, Schell J, Masterson R. Improved method for the transformation of Arabidopsis thaliana with chimeric dihydrofolate reductase constructs which confer methotrexate resistance. Plant Cell Rep. 1992:118–121. doi: 10.1007/BF00232162. [DOI] [PubMed] [Google Scholar]
- Kerr A. Acquisition of virulence by non-pathogenic isolates of Agrobacterium radiobacter. Physiol. Plant Pathol. 1971:241–246. [Google Scholar]
- Kerr J.E, Christie P.J. Evidence for VirB4-mediated dislocation of membrane-integrated VirB2 Pilin during biogenesis of the Agrobacterium VirB/VirD4 type IV secretion system. J. Bacteriol. 2010:4923–4934. doi: 10.1128/JB.00557-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkert J.R, Vidal J.R, Reisch B.I. Stable transformation of plant cells by particle bombardment/biolistics. Methods Mol. Biol. 2004:61–78. doi: 10.1385/1-59259-827-7:061. [DOI] [PubMed] [Google Scholar]
- Kim M.J, Baek K, Park C.M. Optimization of conditions for transient Agrobacteriummediated gene expression assays in Arabidopsis. Plant Cell Rep. 2009:1159–1167. doi: 10.1007/s00299-009-0717-z. [DOI] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B. GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 2012:D1211–D1215. doi: 10.1093/nar/gkr1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komari T, Takakura Y, Ueki J, Kato N, Ishida Y, Hiei Y. Binary vectors and super-binary vectors. Methods Mol. Biol. 2006:15–41. doi: 10.1385/1-59745-130-4:15. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 1986:383–396. [Google Scholar]
- Koncz C, Németh K, Rédei G.P, Schell J. Homologous Recombination and Gene Silencing in Plants. Springer Nature; 1994. Homology recognition during T-DNA integration into the plant genome; pp. 167–189. [Google Scholar]
- Kovacs L.s.G, Pueppke S.G. Mapping and genetic organization of pTiChry5, a novel Ti plasmid from a highly virulent Agrobacterium tumefaciens strain. Mol. Gen. Genet. 1994:327–336. doi: 10.1007/BF00280423. [DOI] [PubMed] [Google Scholar]
- Krall L, Wiedemann U, Unsin G, Weiss S, Domke N, Baron C. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA. 2002:11405–11410. doi: 10.1073/pnas.172390699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R.B, Das A. Functional analysis of the Agrobacterium tumefaciens T-DNA transport pore protein VirB8. J. Bacteriol. 2001:3636–3641. doi: 10.1128/JB.183.12.3636-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R.B, Xie Y.-H, Das A. Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol. Microbiol. 2002:608–617. doi: 10.1046/j.1365-2958.2000.01876.x. [DOI] [PubMed] [Google Scholar]
- Kumar S.V, Misquitta R.W, Reddy V.S, Rao B.J, Rajam M.V. Genetic transformation of the green alga—Chlamydomonas reinhardtii by Agrobacterium tumefaciens. Plant Sci. 2004:731–738. [Google Scholar]
- Kunik T, Tzfira T, Kapulnik Y, Gafni Y, Dingwall C, Citovsky V. Genetic transformation of HeLa cells by Agrobacterium. Proc. Natl. Acad. Sci. USA. 2001:1871–1876. doi: 10.1073/pnas.041327598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, König W, Backert S. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007:862–866. doi: 10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- Lacroix B, Citovsky V. Extracellular VirB5 enhances T-DNA transfer from Agrobacterium to the host plant. PLoS ONE. 2011 doi: 10.1371/journal.pone.0025578. e25578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, Citovsky V. The roles of bacterial and host plant factors in Agrobacteriummediated genetic transformation. Int. J. Dev. Biol. 2013:467–481. doi: 10.1387/ijdb.130199bl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, Citovsky V. A functional bacterium-to-plant DNA transfer machinery of Rhizobium etli. PLoS Pathog. 2016 doi: 10.1371/journal.ppat.1005502. e1005502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, Loyter A, Citovsky V. Association of the Agrobacterium T-DNA-protein complex with plant nucleosomes. Proc. Natl. Acad. Sci. USA. 2008:15429–15434. doi: 10.1073/pnas.0805641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, Tzfira T, Vainstein A, Citovsky V. A case of promiscuity: Agrobacterium's endless hunt for new partners. Trends Genet. 2006:29–37. doi: 10.1016/j.tig.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Lacroix B.t, Vaidya M, Tzfira T, Citovsky V. The VirE3 protein of Agrobacterium mimics a host cell function required for plant genetic transformation. EMBO J. 2005:428–437. doi: 10.1038/sj.emboj.7600524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E.M, Kado C.I. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 1998:2711–2717. doi: 10.1128/jb.180.10.2711-2717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E.M, Kado C.I. The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 2000:361–369. doi: 10.1016/s0966-842x(00)01802-3. [DOI] [PubMed] [Google Scholar]
- Lai E.M, Chesnokova O, Banta L.M, Kado C.I. Genetic and environmental factors affecting T-Pilin export and T-Pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J. Bacteriol. 2000:3705–3716. doi: 10.1128/jb.182.13.3705-3716.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E.M, Eisenbrandt R, Kalkum M, Lanka E, Kado C.I. Biogenesis of T pili in Agrobacterium tumefaciens requires precise VirB2 propilin cleavage and cyclization. J. Bacteriol. 2002:327–330. doi: 10.1128/JB.184.1.327-330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E.M, Shih H.-W, Wen S.R, Cheng M.W, Hwang H.H, Chiu S.H. Proteomic analysis of Agrobacterium tumefaciens response to the vir gene inducer acetosyringone. Proteomics. 2006:4130–4136. doi: 10.1002/pmic.200600254. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Berardini T.Z, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander D.L, Garcia-Hernandez M, Karthikeyan A.S, Lee C.H, Nelson W.D, Ploetz L, Singh S, Wensel A, Huala E. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo G.R, Stein P.A, Ludwig R.A. A DNA transformation–competent Arabidopsis genomic library in Agrobacterium. Nat. Biotechnol. 1991:963–967. doi: 10.1038/nbt1091-963. [DOI] [PubMed] [Google Scholar]
- Lee C.W, Efetova M, Engelmann J.C, Kramell R, Wasternack C, Ludwig-Muller J, Hedrich R, Deeken R. Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana. Plant Cell. 2009:2948–2962. doi: 10.1105/tpc.108.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Dudley M.W, Hess K.M, Lynn D.G, Joerger R.D, Binns A.N. Mechanism of activation of Agrobacterium virulence genes: identification of phenol-binding proteins. Proc. Natl. Acad. Sci. USA. 1992:8666–8670. doi: 10.1073/pnas.89.18.8666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L.Y, Gelvin S.B. T-DNA binary vectors and systems. Plant Physiol. 2008:325–332. doi: 10.1104/pp.107.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L.Y, Fang M.J, Kuang L.Y, Gelvin S.B. Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods. 2008:24. doi: 10.1186/1746-4811-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.W, Jin S, Sim W.S, Nester E.W. Genetic evidence for direct sensing of phenolic compounds by the VirA protein of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA. 1995:12245–12249. doi: 10.1073/pnas.92.26.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.W, Jin S, Sim W.S, Nester E.W. The sensing of plant signal molecules by Agrobacterium genetic evidence for direct recognition of phenolic inducers by the VirA protein. Gene. 1996:83–88. doi: 10.1016/s0378-1119(96)00328-9. [DOI] [PubMed] [Google Scholar]
- Lenser T, Theißen G. Floral dip transformation in Lepidium campestre. Bio-protocol. 2014 e1201. [Google Scholar]
- Li D, Dreher K, Knee E, Brkljacic J, Grotewold E, Berardini T.Z, Lamesch P, Garcia-Hernandez M, Reiser L, Huala E. Arabidopsis database and stock resources. Methods Mol. Biol. 2014a:65–96. doi: 10.1007/978-1-62703-580-4_4. [DOI] [PubMed] [Google Scholar]
- Li F, Alvarez-Martinez C, Chen Y, Choi K.J, Yeo H.J, Christie P.J. Enterococcus faecalis PrgJ, a VirB4-like ATPase, mediates pCF10 conjugative transfer through substrate binding. J. Bacteriol. 2012:4041–4051. doi: 10.1128/JB.00648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Brown P.J.B, Tang J.X, Xu J, Quardokus E.M, Fuqua C, Brun Y.V. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol. Microbiol. 2011:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Krichevsky A, Vaidya M, Tzfira T, Citovsky V. Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc. Natl. Acad. Sci. USA. 2005a:5733–5738. doi: 10.1073/pnas.0404118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Vaidya M, White C, Vainstein A, Citovsky V, Tzfira T. Involvement of KU80 in T-DNA integration in plant cells. Proc. Natl. Acad. Sci. USA. 2005b:19231–19236. doi: 10.1073/pnas.0506437103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.F, Park E, von Arnim A.G, Nebenfuhr A. The FAST technique: a simplified Agrobacteriumbased transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods. 2009:6. doi: 10.1186/1746-4811-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Jia Y, Hou Q, Charles T.C, Nester E.W, Pan S.Q. A global pH sensor: Agrobacterium sensor protein ChvG regulates acid-inducible genes on its two chromosomes and Ti plasmid. Proc. Natl. Acad. Sci. USA. 2002:12369–12374. doi: 10.1073/pnas.192439499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang Q, Tu H, Lim Z, Pan S.Q. Direct visualization of Agrobacterium -delivered VirE2 in recipient cells. Plant J. 2014b:487–495. doi: 10.1111/tpj.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Pan S.Q. Agrobacterium delivers VirE2 protein into host cells via clathrin-mediated endocytosis. Sci. Adv. 2017 doi: 10.1126/sciadv.1601528. e1601528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Rosso M.G, Viehoever P, Weisshaar B. GABI-Kat Simple Search: an Arabidopsis thaliana T-DNA mutant database with detailed information for confirmed insertions. Nucleic Acids Res. 2007:D874–D878. doi: 10.1093/nar/gkl753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R. RNA interference in Agrobacterium rhizogenestransformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 2004:983–992. doi: 10.1093/jxb/erh122. [DOI] [PubMed] [Google Scholar]
- Lin B.C, Kado C.I. Studies on Agrobacterium tumefaciens VII. avirulence induced by temperature and ethidium bromide. Can. J. Microbiol. 1977:1554–1561. doi: 10.1139/m77-229. [DOI] [PubMed] [Google Scholar]
- Lin Y.-H, Gao R, Binns A.N, Lynn D.G. Capturing the VirA/VirG TCS of Agrobacterium tumefaciens. Adv. Exp. Med. Biol. 2008:161–177. doi: 10.1007/978-0-387-78885-2_11. [DOI] [PubMed] [Google Scholar]
- Liu F, Cao M.Q, Yao L, Li Y, Robaglia C, Tourneur C. In planta transformation of pakchoi (Brassica campestris L. ssp. Chinensis) by infiltration of adult plants with Agrobacterium. Acta Hort. 1998:187–192. [Google Scholar]
- Liu P, Nester E.W. Indoleacetic acid, a product of transferred DNA, inhibits vir gene expression and growth of Agrobacterium tumefaciens C58. Proc. Natl. Acad. Sci. U.S.A. 2006:4658–4662. doi: 10.1073/pnas.0600366103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Wood D, Nester E.W. Phosphoenolpyruvate carboxykinase is an acid-induced, chromosomally encoded virulence factor in Agrobacterium tumefaciens. J. Bacteriol. 2005:6039–6045. doi: 10.1128/JB.187.17.6039-6045.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llosa M, Zupan J, Baron C, Zambryski P. The N- and C-terminal portions of the Agrobacterium VirB1 protein independently enhance tumorigenesis. J. Bacteriol. 2000:3437–3445. doi: 10.1128/jb.182.12.3437-3445.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A.M, Barnason A.R, Rogers S.G, Byrne M.C, Fraley R.T, Horsch R.B. Transformation of Arabidopsis thaliana with Agrobacterium tumefaciens. Science. 1986:464–466. doi: 10.1126/science.234.4775.464. [DOI] [PubMed] [Google Scholar]
- Lorence A, Verpoorte R. Gene transfer and expression in plants. Methods Mol. Biol. 2004:329–350. doi: 10.1385/1-59259-774-2:329. [DOI] [PubMed] [Google Scholar]
- Low H.H, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, Lu F, Redzej A, Fronzes R, Orlova E.V, Waksman G. Structure of a type IV secretion system. Nature. 2014:550–553. doi: 10.1038/nature13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyter A, Rosenbluh J, Zakai N, Li J, Kozlovsky S.V, Tzfira T, Citovsky V. The plant VirE2 Interacting Protein 1. A molecular link between the Agrobacterium T-Complex and the host cell chromatin? Plant Physiol. 2005:1318–1321. doi: 10.1104/pp.105.062547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Wang J, Zhang Y, Wang H.C, Liang J.S, Zhang J.H. AT14A mediates the cell wall-plasma membrane-cytoskeleton continuum in Arabidopsis thaliana cells. J. Exp. Bot. 2012:4061–4069. doi: 10.1093/jxb/ers063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L.S, Hachani A, Lin J.S, Filloux A, Lai E.M. Agrobacterium tumefaciens deploys a superfamily of type VI secretion DN-ase effectors as weapons for interbacterial competition in planta. Cell Host Microbe. 2014:94–104. doi: 10.1016/j.chom.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magori S, Citovsky V. Agrobacterium counteracts host-induced degradation of Its effector F-Box protein. Sci. Signal. 2011 doi: 10.1126/scisignal.2002124. ra69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magori S, Citovsky V. The role of the ubiquitin-proteasome system in Agrobacterium tumefaciensmediated genetic transformation of plants. Plant Physiol. 2012:65–71. doi: 10.1104/pp.112.200949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangano S, Gonzalez C.D, Petruccelli S. Agrobacterium tumefaciensmediated transient transformation of Arabidopsis thaliana leaves. Methods Mol. Biol. 2014:165–173. doi: 10.1007/978-1-62703-580-4_8. [DOI] [PubMed] [Google Scholar]
- Mantis N.J, Winans S.C. The Agrobacterium tumefaciens vir gene transcriptional activator virG is transcriptionally induced by acid pH and other stress stimuli. J. Bacteriol. 1992:1189–1196. doi: 10.1128/jb.174.4.1189-1196.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresh J, Zhang J, Lynn D.G. The innate immunity of maize and the dynamic chemical strategies regulating two-component signal transduction in Agrobacterium tumefaciens. ACS Chem. Biol. 2006:165–175. doi: 10.1021/cb600051w. [DOI] [PubMed] [Google Scholar]
- Marion J, Bach L, Bellec Y, Meyer C, Gissot L, Faure J.D. Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J. 2008:169–179. doi: 10.1111/j.1365-313X.2008.03596.x. [DOI] [PubMed] [Google Scholar]
- Martinez-Trujillo M, Limones-Briones V, Cabrera-Ponce J.L, Herrera-Estrella L. Improving transformation efficiency of Arabidopsis thaliana by modifying the floral dip method. Plant Mol. Biol. Rep. 2004:63–70. [Google Scholar]
- Mateos-Gomez P.A, Gong F, Nair N, Miller K.M, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature. 2015:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse A.G. Role of bacterial cellulose fibrils in Agrobacterium tumefaciens infection. J. Bacteriol. 1983:906–915. doi: 10.1128/jb.154.2.906-915.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse A.G. Characterization of nonattaching mutants of Agrobacterium tumefaciens. J.Bacteriol. 1987:313–323. doi: 10.1128/jb.169.1.313-323.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse A.G, Holmes K.V, Gurlitz R.H. Elaboration of cellulose fibrils by Agrobacterium tumefaciens during attachment to carrot cells. J. Bacteriol. 1981:583–595. doi: 10.1128/jb.145.1.583-595.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse A.G, Marry M, Krall L, Kaye M, Ramey B.E, Fuqua C, White A.R. The effect of cellulose overproduction on binding and biofilm formation on roots by Agrobacterium tumefaciens. Mol. Plant Microbe. Interact. 2005:1002–1010. doi: 10.1094/MPMI-18-1002. [DOI] [PubMed] [Google Scholar]
- McBride K.E, Summerfelt K.R. Improved binary vectors for Agrobacteriummediated plant transformation. Plant Mol. Biol. 1990:269–276. doi: 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- McCullen C.A, Binns A.N. Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu. Rev. Cell Dev. Biol. 2006:101–127. doi: 10.1146/annurev.cellbio.22.011105.102022. [DOI] [PubMed] [Google Scholar]
- McHale M, Eamens A.L, Finnegan E.J, Waterhouse P.M. A 22-nt artificial microRNA mediates widespread RNA silencing in Arabidopsis. Plant J. 2013:519–529. doi: 10.1111/tpj.12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S, Goyal V. Agrobacteriummediated gene transfer in plants and biosafety considerations. Appl. Biochem. Biotechnol. 2012:1953–1975. doi: 10.1007/s12010-012-9910-6. [DOI] [PubMed] [Google Scholar]
- Melchers L.S, Maroney M.J, den Dulk-Ras A, Thompson D.V, van Vuuren H.A.J, Schilperoort R.A, Hooykaas P.J.J. Octopine and nopaline strains of Agrobacterium tumefaciens differ in virulence; molecular characterization of the virF locus. Plant Mol Biol. 1990:249–259. doi: 10.1007/BF00018565. [DOI] [PubMed] [Google Scholar]
- Mestiri I, Norre F, Gallego M.E, White C.I. Multiple host-cell recombination pathways act in Agrobacteriummediated transformation of plant cells. Plant J. 2014:511–520. doi: 10.1111/tpj.12398. [DOI] [PubMed] [Google Scholar]
- Michielse C.B, Hooykaas P.J.J, van den Hondel C.A, Ram A.F. Agrobacteriummediated transformation as a tool for functional genomics in fungi. Curr. Genet. 2005:1–17. doi: 10.1007/s00294-005-0578-0. [DOI] [PubMed] [Google Scholar]
- Michielse C.B, Ram A.F, Hooykaas P.J.J, van den Hondel C.A. Agrobacteriummediated transformation of Aspergillus awamori in the absence of full-length VirD2, VirC2, or VirE2 leads to insertion of aberrant T-DNA structures. J. Bacteriol. 2004:2038–2045. doi: 10.1128/JB.186.7.2038-2045.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian A.R, Fullner K.J, Koonin E.V, Nester E.W. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc. Natl. Acad. Sci. USA. 1996:7321–7326. doi: 10.1073/pnas.93.14.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore K.S, Bassuner B, Deng X.B, Darbinian N.S, Motchoulski A, Ream W, Gelvin S.B. Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol. Plant Microbe Interact. 1998:668–683. doi: 10.1094/MPMI.1998.11.7.668. [DOI] [PubMed] [Google Scholar]
- Mysore K.S, Kumar C.T, Gelvin S.B. Arabidopsis eco-types and mutants that are recalcitrant to Agrobacterium root transformation are susceptible to germ-line transformation. Plant J. 2000a:9–16. doi: 10.1046/j.1365-313x.2000.00646.x. [DOI] [PubMed] [Google Scholar]
- Mysore K.S, Nam J, Gelvin S.B. An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. Proc. Natl. Acad. Sci. USA. 2000b:948–953. doi: 10.1073/pnas.97.2.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J, Matthysse A.G, Gelvin S.B. Differences in susceptibility of Arabidopsis ecotypes to crown gall disease may result from a deficiency in T-DNA integration. Plant Cell. 1997:317–333. doi: 10.1105/tpc.9.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J, Mysore K.S, Gelvin S.B. Agrobacterium tumefaciens transformation of the radiation hypersensitive Arabidopsis thaliana mutants uvh1 and rad5. Mol. Plant Microbe Interact. 1998:1136–1141. doi: 10.1094/MPMI.1998.11.11.1136. [DOI] [PubMed] [Google Scholar]
- Nam J, Mysore K.S, Zheng C, Knue M.K, Matthysse A.G, Gelvin S.B. Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol. Gen. Genet. 1999:429–438. doi: 10.1007/s004380050985. [DOI] [PubMed] [Google Scholar]
- Newell C.A. Plant transformation technology: Developments and applications. Mol. Biotechnol. 2000:53–66. doi: 10.1385/MB:16:1:53. [DOI] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A, Nonaka S, Saika H, Kwon Y.-I, Osakabe K, Toki S. Suppression of Ku70/80 or Lig4 leads to decreased stable transformation and enhanced homologous recombination in rice. New Phytol. 2012:1048–1059. doi: 10.1111/j.1469-8137.2012.04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X, Zhou M, Henkel C.V, van Heusden G.P, Hooykaas P.J.J. The Agrobacterium tumefaciens virulence protein VirE3 is a transcriptional activator of the F-box gene VBF. Plant J. 2015:914–924. doi: 10.1111/tpj.13048. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Yuhashi K.-I, Takada K, Sugaware M, Minamisawa K, Ezura H. Ethylene production in plants during transformation suppresses vir gene expression in Agrobacterium tumefaciens. New Phytol. 2008:647–656. doi: 10.1111/j.1469-8137.2008.02400.x. [DOI] [PubMed] [Google Scholar]
- O'Connell K.P, Handelsman J. chvA locus may be involved in export of neutral cyclic β-1,2-linked D-Glucan from Agrobacterium tumefaciens. Mol. Plant Microbe Interact. 1989:11–16. [PubMed] [Google Scholar]
- Ooms G, Hooykaas P.J.J, Van Veen R.J.M, Van Beelen P, Regensburg-Tuïnk T.J.G, Schilperoort R.A. Octopine Tiplasmid deletion mutants of Agrobacterium tumefaciens with emphasis on the right side of the T-region. Plasmid. 1982:15–29. doi: 10.1016/0147-619x(82)90023-3. [DOI] [PubMed] [Google Scholar]
- Padavannil A, Jobichen C, Qinghua Y, Seetharaman J, Velazquez-Campoy A, Yang L, Pan S.Q, Sivaraman J. Dimerization of VirD2 binding protein is essential for Agrobacterium induced tumor formation in plants. PLoS Pathog. 2014 doi: 10.1371/journal.ppat.1003948. e1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanichelvam K, Oger P, Clough S.J, Cha C, Bent A.F, Farrand S.K. A second T-Region of the soybean-supervirulent chrysopine-type Ti plasmid pTiChry5, and construction of a fully disarmed vir helper plasmid. Mol. Plant Microbe Interact. 2000:1081–1091. doi: 10.1094/MPMI.2000.13.10.1081. [DOI] [PubMed] [Google Scholar]
- Pan X, Liu H, Clarke J, Jones J, Bevan M, Stein L. ATIDB: Arabidopsis thaliana insertion database. Nucleic Acids Res. 2003:1245–1251. doi: 10.1093/nar/gkg222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y, Vaghchhipawala Z, Vasudevan B, Lee L.-Y, Shen Y, Singer K, Waterworth W.M, Zhang Z.J, West C.E, Mysore K.S, Gelvin S.B. Agrobacterium T-DNA integration into the plant genome can occur without the activity of key non-homologous end-joining proteins. Plant J. 2015:934–946. doi: 10.1111/tpj.12779. [DOI] [PubMed] [Google Scholar]
- Patil B.L, Fauquet C.M. Light intensity and temperature affect systemic spread of silencing signal in transient agroinfiltration studies. Mol. Plant Pathol. 2015:484–494. doi: 10.1111/mpp.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson M, Wadstrom T. Vitronectin and type-I collagen binding by Staphylococcus aureus and coagulase-negative staphylococci. FEMS Microbiol. Immunol. 1990:55–62. doi: 10.1111/j.1574-6968.1990.tb03479.x. [DOI] [PubMed] [Google Scholar]
- Pawlowski W.P, Somers D.A. Transgene inheritance in plants genetically engineered by microprojectile bombardment. Mol. Biotechnol. 1996:17–30. doi: 10.1007/BF02762320. [DOI] [PubMed] [Google Scholar]
- Pelczar P, Kalck V, Gomez D, Hohn B. Agrobacterium proteins VirD2 and VirE2 mediate precise integration of synthetic TDNA complexes in mammalian cells. EMBO Rep. 2004:632–637. doi: 10.1038/sj.embor.7400165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena A, Matilla I, Martin-Benito J, Valpuesta J.M, Carrascosa J.L, de la Cruz F, Cabezon E, Arechaga I. The hexameric structure of a conjugative VirB4 protein ATPase provides new insights for a functional and phylogenetic relationship with DNA translocases. J. Biol. Chem. 2012:39925–39932. doi: 10.1074/jbc.M112.413849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piers K.L, Heath J.D, Liang X, Stephens K.M, Nester E.W. Agrobacterium tumefaciensmediated transformation of yeast. Proc. Natl. Acad. Sci. USA. 1996:1613–1618. doi: 10.1073/pnas.93.4.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A. Agrobacterium infection and plant defense-transformation success hangs by a thread. Front. Plant Sci. 2013:519. doi: 10.3389/fpls.2013.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell G.K, Hommes N.G, Kuo J, Castle L.A, Morris R.O. Inducible expression of cytokinin biosynthesis in Agrobacterium tumefaciens by plant phenolics. Mol. Plant Microbe Interact. 1988:235–242. doi: 10.1094/mpmi-1-235. [DOI] [PubMed] [Google Scholar]
- Regensburg-Tuïnk A.J.G, Hooykaas P.J.J. Transgenic N. glauca plants expressing bacterial virulence gene virF are converted into hosts for nopaline strains of A. tumefaciens. Nature. 1993:69–71. doi: 10.1038/363069a0. [DOI] [PubMed] [Google Scholar]
- Rhee S.Y, Beavis W, Berardini T.Z, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, Miller N, Mueller L.A, Mundodi S, Reiser L, Tacklind J, Weems D.C, Wu Y, Xu I, Yoo D, Yoon J, Zhang P. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson K, Fowler S, Pullen C, Skelton C, Morris B, Putterill J. T-DNA tagging of a flowering-time gene and improved gene transfer by in planta transformation of Arabidopsis. Aust. J. Plant Physiol. 1998:125–130. [Google Scholar]
- Rivero L, Scholl R, Holomuzki N, Crist D, Grotewold E, Brkljacic J. Handling Arabidopsis plants: growth, preservation of seeds, transformation, and genetic crosses. Methods Mol. Biol. 2014:3–25. doi: 10.1007/978-1-62703-580-4_1. [DOI] [PubMed] [Google Scholar]
- Rosas-Díaz T, Cana-Quijada P, Amorim-Silva V, Botella M.A, Lozano-Durán R, Bejarano E.R. Arabidopsis NahG plants as a suitable and efficient system for transient expression during Agrobacterium tumefaciens. Mol. Plant. 2017:353–356. doi: 10.1016/j.molp.2016.11.005. [DOI] [PubMed] [Google Scholar]
- Rossi L, Hohn B, Tinland B. Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 1996:126–130. doi: 10.1073/pnas.93.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko E, Sagulenko V, Chen J, Christie P.J. Role of Agrobacterium VirB11 ATPase in T-Pilus assembly and substrate selection. J. Bacteriol. 2001:5813–5825. doi: 10.1128/JB.183.20.5813-5825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakalis P.A, van Heusden G.P.H, Hooykaas P.J.J. Visualization of VirE2 protein translocation by the Agrobacterium type IV secretion system into host cells. Microbiology. 2014:104–117. doi: 10.1002/mbo3.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman H, Abu-Arish A, Oliel S, Loyter A, Klafter J, Granek R, Elbaum M. Nuclear localization signal peptides induce molecular delivery along microtubules. Biophys. J. 2005:2134–2145. doi: 10.1529/biophysj.105.060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J.C. The development of the biolistic process. In Vitro Cell. Dev. Biol.-Plant. 2000:303–308. [Google Scholar]
- Sangwan R, Bourgeois Y, Brown S, Vasseur G.r, Sangwan-Norreel B. Characterization of competent cells and early events of Agrobacteriummediated genetic transformation in Arabidopsis thaliana. Planta. 1992:439–456. doi: 10.1007/BF00192812. [DOI] [PubMed] [Google Scholar]
- Sangwan R.S, Bourgeois Y, Sangwan-Norreel B.S. Genetic transformation of Arabidopsis thaliana zygotic embryos and identification of critical parameters influencing transformation efficiency. Mol. Gen. Genet. 1991:475–485. doi: 10.1007/BF00280305. [DOI] [PubMed] [Google Scholar]
- Sardesai N, Lee L.Y, Chen H, Yi H, Olbricht G.R, Stirnberg A, Jeffries J, Xiong K, Doerge R.W, Gelvin S.B. Cytokinins secreted by Agrobacterium promote transformation by repressing a plant Myb transcription factor. Sci. Signal. 2013 doi: 10.1126/scisignal.2004518. ra100. [DOI] [PubMed] [Google Scholar]
- Schmidt-Eisenlohr H, Domke N, Angerer C, Wanner G, Zambryski P.C, Baron C. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 1999:7485–7492. doi: 10.1128/jb.181.24.7485-7492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl R.L, May S.T, Ware D.H. Seed and molecular resources for Arabidopsis. Plant Physiol. 2000:1477–1480. doi: 10.1104/pp.124.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrammeijer B, den Dulk-Ras A, Vergunst A.C, Jurado Jácome E, Hooykaas P.J.J. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 2003:860–868. doi: 10.1093/nar/gkg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrammeijer B, Risseeuw E, Pansegrau W, Regensburg-Tuïnk T.J.G, Crosby W.L, Hooykaas P.J.J. Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr. Biol. 2001:258–262. doi: 10.1016/s0960-9822(01)00069-0. [DOI] [PubMed] [Google Scholar]
- Sheikholeslam S.N, Weeks D.P. Acetosyringone promotes high efficiency transformation of Arabidopsis thaliana explants by Agrobacterium tumefaciens. Plant Mol. Biol. 1987:291–298. doi: 10.1007/BF00021308. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lee L.Y, Gelvin S.B. Is VIP1 important for Agrobacterium -mediated transformation? Plant J. 2014:848–860. doi: 10.1111/tpj.12596. [DOI] [PubMed] [Google Scholar]
- Shimoda N, Toyoda-Yamamoto A, Aoki S, Machida Y. Genetic evidence for an interaction between the VirA sensor protein and the ChvE sugar-binding protein of Agrobacterium. J. Biol. Chem. 1993:26552–26558. [PubMed] [Google Scholar]
- Shou H, Frame B.R, Whitham S.A, Wang K. Assessment of transgenic maize events produced by particle bombardment or Agrobacteriummediated transformation. Mol. Breed. 2004:201–208. [Google Scholar]
- Shurvinton C.E, Hodges L, Ream W. A nuclear localization signal and the C-terminal omega sequence in the Agrobacterium tumefaciens VirD2 endonuclease are important for tumor formation. Proc. Natl. Acad. Sci. USA. 1992:11837–11841. doi: 10.1073/pnas.89.24.11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S.E, Messens E, Van Montagu M, Zambryski P. Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature. 1985:624–629. [Google Scholar]
- Stachel S.E, Timmerman B, Zambryski P. Generation of single-stranded T-DNA molecules during the initial stages of T-DNA transfer from Agrobacterium tumefaciens to plant cells. Nature. 1986:706–712. [Google Scholar]
- Stachel S.E, Timmerman B, Zambryski P. Activation of Agrobacterium tumefaciens vir gene expression generates multiple single-stranded T-strand molecules from the pTiA6 T-region: requirement for 5′ virD gene products. EMBO J. 1987:857–863. doi: 10.1002/j.1460-2075.1987.tb04831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramoni S, Nathoo N, Klimov E, Yuan Z.-C. Agrobacterium tumefaciens responses to plant-derived signaling molecules. Front. Plant Sci. 2014:322. doi: 10.3389/fpls.2014.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini T.Z, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, Radenbaugh A, Singh S, Swing V, Tissier C, Zhang P, Huala E. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tague B.W. Germ-line transformation of Arabidopsis lasiocarpa. Transgenic Res. 2001:259–267. doi: 10.1023/a:1016633617908. [DOI] [PubMed] [Google Scholar]
- Tague B.W, Mantis J. In planta Agrobacterium mediated transformation by vacuum infiltration. Methods Mol. Biol. 2006:215–223. doi: 10.1385/1-59745-003-0:215. [DOI] [PubMed] [Google Scholar]
- Tao Y, Rao P.K, Bhattacharjee S, Gelvin S.B. Expression of plant protein phosphatase 2C interferes with nuclear import of the Agrobacterium T-complex protein VirD2. Proc. Natl. Acad. Sci. USA. 2004:5164–5169. doi: 10.1073/pnas.0300084101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenea G.N, Spantzel J, Lee L.Y, Zhu Y, Lin K, Johnson S.J, Gelvin S.B. Overexpression of several Arabidopsis histone genes increases Agrobacteriummediated transformation and transgene expression in plants. Plant Cell. 2009:3350–3367. doi: 10.1105/tpc.109.070607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M.F, Karlinsey J.E, Marks J.R, Hurlbert R.E. Identification of a new virulence locus in Agrobacterium tumefaciens that affects polysaccharide composition and plant cell attachment. J. Bacteriol. 1987:3209–3216. doi: 10.1128/jb.169.7.3209-3216.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinland B, Schoumacher F, Gloeckler V, Bravoangel A.M, Hohn B. The Agrobacterium tumefaciens virulence D2 protein is responsible for precise integration of T-DNA into the plant genome. EMBO J. 1995:3585–3595. doi: 10.1002/j.1460-2075.1995.tb07364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A.D, Fuqua C. Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens. Curr. Opin. Microbiol. 2009:708–714. doi: 10.1016/j.mib.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travella S, Ross S.M, Harden J, Everett C, Snape J.W, Harwood W.A. A comparison of transgenic barley lines produced by particle bombardment and Agrobacteriummediated techniques. Plant Cell Rep. 2005:780–789. doi: 10.1007/s00299-004-0892-x. [DOI] [PubMed] [Google Scholar]
- Trieu A.T, Burleigh S.H, Kardailsky I.V, Maldonado-Mendoza I.E, Versaw W.K, Blaylock L.A, Shin H, Chiou T.-J, Katagi H, Dewbre G.R, Weigel D, Harrison M.J. Transformation of Medicago truncatula via infiltration of seedlings or flowering plants with Agrobacterium. Plant J. 2000:531–541. doi: 10.1046/j.1365-313x.2000.00757.x. [DOI] [PubMed] [Google Scholar]
- Tsai Y.L, Chiang Y.R, Narberhaus F, Baron C, Lai E.M. The small heat-shock protein HspL is a VirB8 chaperone promoting type IV secretion-mediated DNA transfer. J. Biol. Chem. 2010:19757–19766. doi: 10.1074/jbc.M110.110296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.L, Wang M.H, Gao C, Klusener S, Baron C, Narberhaus F, Lai E.M. Small heat-shock protein HspL is induced by VirB protein(s) and promotes VirB/D4-mediated DNA transfer in Agrobacterium tumefaciens. Microbiology. 2009:3270–3280. doi: 10.1099/mic.0.030676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Qi Y, Nguyen le V, Bethke G, Tsuda Y, Glazebrook J, Katagiri F. An efficient Agrobacteriummediated transient transformation of Arabidopsis. Plant J. 2012:713–719. doi: 10.1111/j.1365-313X.2011.04819.x. [DOI] [PubMed] [Google Scholar]
- Tzfira T. On tracks and locomotives: the long route of DNA to the nucleus. Trends Microbiol. 2006:61–63. doi: 10.1016/j.tim.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Tzfira T, Li J, Lacroix B, Citovsky V. Agrobacterium T-DNA integration: molecules and models. Trends Genet. 2004a:375–383. doi: 10.1016/j.tig.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001:3596–3607. doi: 10.1093/emboj/20.13.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V. Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis nuclear protein VIP1. Proc. Natl. Acad. Sci. USA. 2002:10435–10440. doi: 10.1073/pnas.162304099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004b:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- Uchimiya H, Fushimi T, Hashimoto H, Harada H, Syono K, Sugawara Y. Expression of a foreign gene in callus derived from DNA-treated protoplasts of rice (Oryza sativa L.) Mol. Gen. Genet. 1986:204–207. [Google Scholar]
- Vaghchhipawala Z.E, Vasudevan B, Lee S, Morsy M.R, Mysore K.S. Agrobacterium may delay plant nonhomologous end-joining DNA repair via XRCC4 to favor T-DNA integration. Plant Cell. 2012:4110–4123. doi: 10.1105/tpc.112.100495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Montagu M.V, Van Lijsebettens M. Agrobacterium tumefaciensmediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA. 1988:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Bundock P, Hooykaas P.J.J. Non-homologous end-joining proteins are required for Agrobacterium T-DNA integration. EMBO J. 2001:6550–6558. doi: 10.1093/emboj/20.22.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Bundock P, Overmeer R.M, Lee L.Y, Gelvin S.B, Hooykaas P.J.J. The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Res. 2003:4247–4255. doi: 10.1093/nar/gkg458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Eede G, Aarts H.J, Buhk H.J, Corthier G, Flint H.J, Hammes W, Jacobsen B, Midtvedt T, van der Vossen J, von Wright A, Wackernagel W, Wilcks A. The relevance of gene transfer to the safety of food and feed derived from genetically modified (GM) plants. Food Chem. Toxicol. 2004:1127–1156. doi: 10.1016/j.fct.2004.02.001. [DOI] [PubMed] [Google Scholar]
- van den Elzen P.J.M, Townsend J, Lee K.Y, Bedbrook J.R. A chimaeric hygromycin resistance gene as a selectable marker in plant cells. Plant Mol. Biol. 1985:299–302. doi: 10.1007/BF00020627. [DOI] [PubMed] [Google Scholar]
- van Kregten M, de Pater S, Romeijn R, van Schendel R, Hooykaas P.J.J, Tijsterman M. T-DNA integration in plants results from polymerase-θ-mediated DNA repair. Nat. Plants. 2016 doi: 10.1038/nplants.2016.164. 16164. [DOI] [PubMed] [Google Scholar]
- Vergunst A.C, Schrammeijer B, den Dulk-Ras A, de Vlaam C.M, Regensburg-Tuink T.J, Hooykaas P.J.J. VirB/D4-Dependent protein translocation from Agrobacterium into plant cells. Science. 2000:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- Vergunst A.C, van Lier C.M, den Dulk Ras A, Hooykaas P.J.J. Recognition of the Agrobacterium tumefaciens VirE2 translocation signal by the VirB/D4 transport system does not require VirE1. Plant Physiol. 2003:978–988. doi: 10.1104/pp.103.029223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergunst A.C, van Lier M.C, den Dulk-Ras A, Grosse Stuve T.A, Ouwehand A, Hooykaas P.J.J. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. USA. 2005:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergunst A.C, Waal E.C, Hooykaas P.J.J. Root transformation by Agrobacterium tumefaciens. Methods Mol. Biol. 1998:227–244. doi: 10.1385/0-89603-391-0:227. [DOI] [PubMed] [Google Scholar]
- Wagner V.T, Matthysse A.G. Involvement of a vitronectin-like protein in attachment of Agrobacterium tumefaciens to carrot suspension culture cells. J. Bacteriol. 1992:5999–6003. doi: 10.1128/jb.174.18.5999-6003.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman G, Fronzes R. Molecular architecture of bacterial type IV secretion systems. Trends Biochem. Sci. 2010:691–698. doi: 10.1016/j.tibs.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Wallden K, Rivera-Calzada A, Waksman G. Microreview: Type IV secretion systems: versatility and diversity in function. Cell. Microbiol. 2010:1203–1212. doi: 10.1111/j.1462-5822.2010.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallden K, Williams R, Yan J, Lian P.W, Wang L, Thalassinos K, Orlova E.V, Waksman G. Structure of the VirB4 ATPase, alone and bound to the core complex of a type IV secretion system. Proc. Natl. Acad. Sci. U.S.A. 2012:11348–11353. doi: 10.1073/pnas.1201428109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila Chikdu S, Dawlaty, Meelad M, Cheng, Albert W, Zhang F, Jaenisch R. One-Step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Stachel S.E, Timmerman B, Van Montagu M, Zambryski P.C. Site-specific nick in the T-DNA border sequence as a result of Agrobacterium vir gene expression. Science. 1987:587–591. doi: 10.1126/science.235.4788.587. [DOI] [PubMed] [Google Scholar]
- Wang W.C, Menon G, Hansen G. Development of a novel Agrobacteriummediated transformation method to recover transgenic Brassica napus plants. Plant Cell Rep. 2003:274–281. doi: 10.1007/s00299-003-0691-9. [DOI] [PubMed] [Google Scholar]
- Wang Y, Peng W, Zhou X, Huang F, Shao L, Luo M. The putative Agrobacterium transcriptional activator-like virulence protein VirD5 may target T-complex to prevent the degradation of coat proteins in the plant cell nucleus. New Phytol. 2014:1266–1281. doi: 10.1111/nph.12866. [DOI] [PubMed] [Google Scholar]
- Ward E.R, Barnes W.M. VirD2 protein of Agrobacterium tumefaciens very tightly linked to the 5′ end of T-strand DNA. Science. 1988:927–930. [Google Scholar]
- Watson B, Currier T.C, Gordon M.P, Chilton M.D, Nester E.W. Plasmid required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 1975:255–264. doi: 10.1128/jb.123.1.255-264.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver J, Goklany S, Rizvi N, Cram E.J, Lee-Parsons C.W. Optimizing the transient fast Agro-mediated seedling transformation (FAST) method in Catharanthus roseus seedlings. Plant Cell Rep. 2014:89–97. doi: 10.1007/s00299-013-1514-2. [DOI] [PubMed] [Google Scholar]
- Wendt T, Doohan F, Mullins E. Production of Phytophthora infestansresistant potato (Solanum tuberosum) utilising Ensifer adhaerens OV14. Transgenic Res. 2012:567–578. doi: 10.1007/s11248-011-9553-3. [DOI] [PubMed] [Google Scholar]
- Wroblewski T, Tomczak A, Michelmore R. Optimization of Agrobacteriummediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 2005:259–273. doi: 10.1111/j.1467-7652.2005.00123.x. [DOI] [PubMed] [Google Scholar]
- Wu C.F, Lin J.S, Shaw G.C, Lai E.M. Acid-induced type VI secretion system is regulated by ExoR-ChvG/ChvI signaling cascade in Agrobacterium tumefaciens. PLoS Pathog. 2012 doi: 10.1371/journal.ppat.1002938. e1002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.Y, Chen C.Y, Lai E.M. Expression and functional characterization of the Agrobacterium VirB2 amino Acid substitution variants in T-pilus biogenesis, virulence, and transient transformation efficiency. PLoS ONE. 2014a doi: 10.1371/journal.pone.0101142. e101142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.Y, Chung P.C, Shih H.W, Wen S.R, Lai E.M. Secretome analysis uncovers an Hcp-family protein secreted via a yype VI secretion system in Agrobacterium tumefaciens. J. Bacteriol. 2008:2841–2850. doi: 10.1128/JB.01775-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.Y, Liu K.H, Wang Y.C, Wu J.F, Chiu W.L, Chen C.Y, Wu S.H, Sheen J, Lai E.M. AGROBEST: an efficient Agrobacteriummediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods. 2014b:19. doi: 10.1186/1746-4811-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, Zhou J.M. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 2008:74–80. doi: 10.1016/j.cub.2007.12.020. [DOI] [PubMed] [Google Scholar]
- Xu J, Kim J, Danhorn T, Merritt P.M, Fuqua C. Phosphorus limitation increases attachment in Agrobacterium tumefaciens and reveals a conditional functional redundancy in adhesin biosynthesis. Res. Microbiol. 2012:674–684. doi: 10.1016/j.resmic.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Kim J, Koestler B.J, Choi J.-H, Waters C.M, Fuqua C. Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motileto-sessile switch. Mol. Microbiol. 2013:929–948. doi: 10.1111/mmi.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.H, Li X.Y, Tu H, Pan S.Q. Agrobacteriumdelivered virulence protein VirE2 is trafficked inside host cells via a myosin XI-K-powered ER/actin network. Proc. Natl. Acad. Sci. USA. 2017:2982–2987. doi: 10.1073/pnas.1612098114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky M.F, Nester E.W. Molecular characterization of a host-range-determining locus from Agrobacterium tumefaciens. J. Bacteriol. 1986:244–250. doi: 10.1128/jb.168.1.244-250.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye G.N, Stone D, Pang S.Z, Creely W, Gonzalez K, Hinchee M. Arabidopsis ovule is the target for Agrobacterium in planta vacuum infiltration transformation. Plant J. 1999:249–257. doi: 10.1046/j.1365-313x.1999.00520.x. [DOI] [PubMed] [Google Scholar]
- Yi H, Mysore K.S, Gelvin S.B. Expression of the Arabidopsis histone H2A-1 gene correlates with susceptibility to Agrobacterium transformation. Plant J. 2002:285–298. doi: 10.1046/j.1365-313x.2002.01425.x. [DOI] [PubMed] [Google Scholar]
- Yi H, Sardesai N, Fujinuma T, Chan C.W, Veena, Gelvin S.B. Constitutive expression exposes functional redundancy between the Arabidopsis histone H2A gene HTA1 and other H2A gene family members. Plant Cell. 2006:1575–1589. doi: 10.1105/tpc.105.039719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C, Nester E.W. Association of the virD2 protein with the 5′ end of T strands in Agrobacterium tumefaciens. J. Bacteriol. 1988:3367–3374. doi: 10.1128/jb.170.8.3367-3374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z.C, Edlind M.P, Liu P, Saenkham P, Banta L.M, Wise A.A, Ronzone E, Binns A.N, Kerr K, Nester E.W. The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone-quenching genes in Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 2007b:11790–11795. doi: 10.1073/pnas.0704866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z.C, Liu P, Saenkham P, Kerr K, Nester E.W. Transcriptome profiling and functional analysis of Agrobacterium tumefaciens reveals a general conserved response to acidic conditions (pH 5.5) and a complex acid-mediated signaling involved in AgrobacteriumPlant interactions. J. Bacteriol. 2007a:494–507. doi: 10.1128/JB.01387-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zale J.M, Agarwal S, Loar S, Steber C.M. Evidence for stable transformation of wheat by floral dip in Agrobacterium tumefaciens. Plant Cell Rep. 2009:903–913. doi: 10.1007/s00299-009-0696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A, Krichevsky A, Kozlovsky S.V, Yasmin F, Citovsky V. Plant defense pathways subverted by Agrobacterium for genetic transformation. Plant Signal Behav. 2010a:1245–1248. doi: 10.4161/psb.5.10.12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A, Krichevsky A, Loyter A, Citovsky V. Agrobacterium induces expression of a host F-Box protein required for tumorigenicity. Cell Host Microbe. 2010b:197–209. doi: 10.1016/j.chom.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A, Lacroix B, Gafni Y, Citovsky V. Disassembly of synthetic Agrobacterium T-DNA-protein complexes via the host SCFVBF ubiquitin-ligase complex pathway. Proc. Natl. Acad. Sci. U.S.A. 2013:169–174. doi: 10.1073/pnas.1210921110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambre M, Terryn N, De Clercq J, De Buck S, Dillen W, Van Montagu M, Van Der Straeten D, Angenon G. Light strongly promotes gene transfer from Agrobacterium tumefaciens to plant cells. Planta. 2003:580–586. doi: 10.1007/s00425-002-0914-2. [DOI] [PubMed] [Google Scholar]
- Zambryski P, Joos H, Genetello C, Leemans J, Vanmontagu M, Schell J. Ti-plasmid vector for the Introduction of DNA into plant-cells without alteration of their normal regeneration capacity. EMBO J. 1983:2143–2150. doi: 10.1002/j.1460-2075.1983.tb01715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner E.L, Lang S, Schildbach J.F. Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R Soc. Lond. B: Biol. Sci. 2012:1073–1087. doi: 10.1098/rstb.2011.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Boone L, Kocz R, Zhang C, Binns A.N, Lynn D.G. At the maize/Agrobacterium interface: natural factors limiting host transformation. Chem. Biol. 2000:611–621. doi: 10.1016/s1074-5521(00)00007-7. [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin S.S, Niu Q.W, Chua N.H. Agrobacteriummediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yin X, Yang A, Li G, Zhang J. Stability of inheritance of transgenes in maize (Zea mays L.) lines produced using different transformation methods. Euphytica. 2005:11–22. [Google Scholar]
- Zhang Z, Mao Y, Ha S, Liu W, Botella J.R, Zhu J.K. A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. 2015:1519–1533. doi: 10.1007/s00299-015-1900-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.R, Christie P.J. Suppression of mutant phenotypes of the Agrobacterium tumefaciens VirB11 ATPase by overproduction of VirB proteins. J. Bacteriol. 1997:5835–5842. doi: 10.1128/jb.179.18.5835-5842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K, Damsz B, Kononowicz A.K, Bressan R.A, Hasegawa P.M. A higher plant extracellular vitronectin-like adhesion protein is related to the translational elongation factor-1 alpha. Plant Cell. 1994:393–404. doi: 10.1105/tpc.6.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K, Shi J, Singh U, Wyatt S.E, Bressan R.A, Hasegawa P.M, Carpita N.C. Enrichment of vitronectin- and fibronectin-like proteins in NaCI-adapted plant cells and evidence for their involvement in plasma membrane-cell wall adhesion. Plant J. 1993:637–646. [PubMed] [Google Scholar]
- Zhu Y, Nam J, Carpita N.C, Matthysse A.G, Gelvin S.B. Agrobacteriummediated root transformation is inhibited by mutation of an Arabidopsis cellulose synthase-like gene. Plant Physiol. 2003a:1000–1010. doi: 10.1104/pp.103.030726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Nam J, Humara J.M, Mysore K.S, Lee L.Y, Cao H, Valentine L, Li J, Kaiser A.D, Kopecky A.L, Hwang H.H, Bhattacharjee S, Rao P.K, Tzfira T, Rajagopal J, Yi H, Veena, Yadav B.S, Crane Y.M, Lin K, Larcher Y, Gelvin M.J, Knue M, Ramos C, Zhao X, Davis S.J, Kim S.I, Ranjith-Kumar C.T, Choi Y.J, Hallan V.K, Chattopadhyay S, Sui X, Ziemienowicz A, Matthysse A.G, Citovsky V, Hohn B, Gelvin S.B. Identification of Arabidopsis rat Mutants. Plant Physiol. 2003b:494–505. doi: 10.1104/pp.103.020420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemienowicz A, Tinland B, Bryant J, Gloeckler V, Hohn B. Plant enzymes but not Agrobacterium VirD2 mediate T-DNA ligation in vitro. Mol. Cell Biol. 2000:6317–6322. doi: 10.1128/mcb.20.17.6317-6322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones J.D, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacteriummediated transformation. Cell. 2006:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Zorreguieta A, Geremia R.A, Cavaignac S, Cangelosi G.A, Nester E.W, Ugalde R.A. Identification of the product of an Agrobacterium tumefaciens chromosomal virulence gene. Mol. Plant Microbe Interact. 1988:121–127. doi: 10.1094/mpmi-1-121. [DOI] [PubMed] [Google Scholar]
- Zupan J, Hackworth C.A, Aguilar J, Ward D, Zambryski P. VirB1* promotes T-pilus formation in the virType IV secretion system of Agrobacterium tumefaciens. J. Bacteriol. 2007:6551–6563. doi: 10.1128/JB.00480-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


