Abstract
The occurrence of parallel speciation strongly implies the action of natural selection. However, it is unclear how general a phenomena parallel speciation is since it was only shown in a small number of animal species. In particular, the adaptive process and mechanisms underlying the process of parallel speciation remain elusive. Here, we used an integrative approach incorporating population genomics, common garden, and crossing experiments to investigate parallel speciation of the wild rice species Oryza nivara from O. rufipogon. We demonstrated that O. nivara originated multiple times from different O. rufipogon populations and revealed that different O. nivara populations have evolved similar phenotypes under divergent selection, a reflection of recurrent local adaptation of ancient O. rufipogon populations to dry habitats. Almost completed premating isolation was detected between O. nivara and O. rufipogon in the absence of any postmating barriers between and within these species. These results suggest that flowering time is a “magic” trait that contributes to both local adaptation and reproductive isolation in the origin of wild rice species. Our study thus demonstrates a convincing case of parallel ecological speciation as a consequence of adaptation to new environments.
Keywords: parallel speciation, reproductive isolation, adaptation, divergent selection, wild rice
Introduction
The occurrence of parallel evolution, or the repetition of evolutionary changes in similar environments strongly implicates the action of natural selection in biological diversification because random genetic drift is unlikely to generate such a pattern (Rundle et al. 2000; Schluter 2009; Nosil 2012; Ostevik et al. 2012). As a special form of parallel evolution, parallel speciation involves independent formations of reproductive isolation in separate but closely related lineages/populations as by-products of adaptation to similar divergent environments, providing a convincing case for ecological speciation and for natural selection in generating reproductive isolation (Schluter and Nagel 1995; Johannesson 2001; Nosil 2012).
Nevertheless, demonstrating parallel speciation has been a challenging task because it requires in-depth analyses involving phylogenetics, phenotypes, ecology, and particularly the repeated formation of reproductive isolation between species (Schluter and Nagel 1995; Johannesson 2001; Rieseberg and Willis 2007; Nosil 2012). As noted in previous studies, a convincing example of parallel speciation would meet the following criteria (Schluter and Nagel 1995; Abbott and Comes 2007; Nosil 2012; Ostevik et al. 2012): 1) the populations in similar environments must be phylogenetically distinct and must arise from multiple origins rather than from gene flow caused by secondary contact of allopatric populations; 2) reproductive isolation must have established between descendent and ancestral populations; 3) reproductive isolation must not have been established between descendent populations; and 4) the shared characteristics in descendent populations must be proved to have evolved by natural selection. Despite well-characterized cases of parallel speciation in animals such as sticklebacks (Rundle et al. 2000; Colosimo et al. 2005), stick insects (Nosil et al. 2002; Soria-Carrasco et al. 2014), finches (Ryan et al. 2007), marine snails (Butlin et al. 2014; Ravinet et al. 2016), and cichlid fishes (Elmer et al. 2014; Meier et al. 2017), only a few cases have been documented in plants (Abbott and Comes 2007; Roda et al. 2013; Richards et al. 2016; Comes et al. 2017; Trucchi et al. 2017). Moreover, the adaptive process and mechanisms underlying parallel speciation remain largely unknown (Rundle et al. 2000; Schluter 2009; Nosil 2012).
The wild rice Oryza rufipogon and O. nivara offer a unique opportunity for studying adaptive divergence and ecological speciation (Sang and Ge 2007; Zheng and Ge 2010). They are the most closely related species in the rice genus Oryza and are collectively regarded as the wild progenitors of cultivated rice (Oryza sativa L.) (Morishima et al. 1992; Sang and Ge 2007; Vaughan et al. 2008). As a perennial, O. rufipogon is widely distributed throughout southern China, South and Southeast Asia, Papua New Guinea, and northern Australia, and grows in areas with year-round water, such as swamps and lakes; whereas O. nivara is an annual found within a more restricted distribution in South and Southeast Asia and usually inhabits ponds and swamps that dry up completely during the dry season (Sharma and Shastry 1965; Morishima et al. 1992; Vaughan et al. 2008). Within the same geographical area, the two species can be found to occur in close physical proximity, ranging from several kilometers to <100 m (Morishima et al. 1992; Kuroda et al. 2007; Vaughan et al. 2008). Therefore, the geographic distribution of the two species is of typical sympatry or parapatry, where the populations of the species pairs are found in different habitats of the same localities. Biologically, O. rufipogon is characterized by its tall stature, a mixed mating system, photoperiod sensitivity (late flowering), and profligate vegetative reproduction. In contrast, O. nivara is characterized by its short stature, predominant self-fertilization, and photoperiod insensitivity (early flowering) (Morishima et al. 1992; Sang and Ge 2007; Banaticla-Hilario et al. 2013). Despite marked differences in gross morphology and ecology, the two species are cross-compatible (Lu et al. 2000; Banaticla-Hilario et al. 2013) and exhibit little interspecific genetic differentiation (Barbier 1989; Zhu et al. 2007; Zheng and Ge 2010); thus their speciation process is incomplete.
Many lines of evidence suggest that the annual O. nivara originated from the perennial O. rufipogon as an adaptation to dry habitats (Barbier 1989; Morishima et al. 1992; Zheng and Ge 2010). In this process, many changes such as flowering time and mating system promoted assortative mating and acted as premating barriers to genetic exchanges between species (Morishima et al. 1992; Zheng and Ge 2010; Liu et al. 2015). A recent population genetic study (Liu et al. 2015) suggested that O. nivara might have originated multiple times, implicative of parallel speciation. Here, we studied all four criteria that must be fulfilled for parallel speciation using multiple approaches and provide a convincing case of parallel speciation in wild rice. In addition, our genomic and phenotypic data, in conjunction with demographic modeling demonstrate that multiple origins of O. nivara populations are associated with habit shifts during the last glaciation and that a change of flowering time in the descendant species is a key step during this process.
Results
Whole-Genome Resequencing and SNP Validation
We performed whole-genome resequencing of pooled samples (Pool-Seq) for 15 O. rufipogon and 9 O. nivara populations (fig. 1; supplementary table S1, Supplementary Material online; see Materials and Methods). We chose over 20 individuals per population for DNA pooling, except for seven populations for which 14–18 individuals were available (supplementary table S1, Supplementary Material online). To acquire sufficient high-quality mapping data, we used the latest version IRGSP 1.0 of the Oryza sativa genome (Kawahara et al. 2013) as the reference and retained reads that were uniquely mapped to this reference with a mapping quality score >30. We produced 268-Gb high-quality data after trimming with an average sequencing depth of 30× per pooled sample and finally retained 6,168,151 polymorphic sites for subsequent analyses (supplementary table S2, Supplementary Material online; see Materials and Methods).
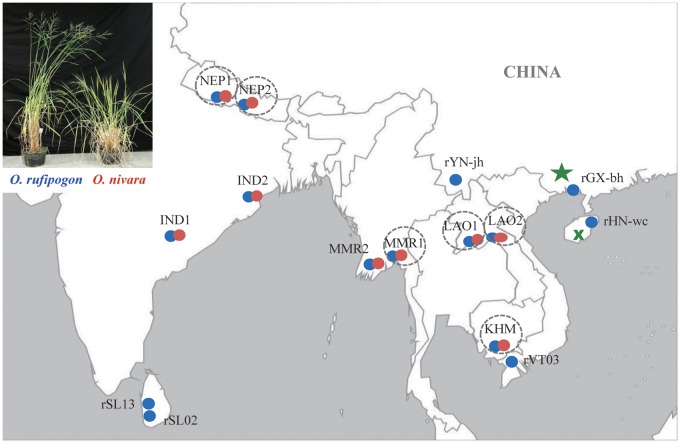
Fig. 1.
Sampling locations of Oryza rufipogon (blue) and O. nivara (red) populations used in this study. Inset photo of perennial O. rufipogon (left) and annual O. nivara (right) plants originally published in Journal of Systematics and Evolution (2013, 51: Cover) and adapted with permission. Circles denote the six population pairs of O. rufipogon and O. nivara used in the common garden experiment, of which three pairs (i.e., NEP1, LAO2, and KHM) were used for artificial crossing. The green star and cross show the locations where the common garden experiment and the artificial crossing study were conducted, respectively. Detailed information on all the populations is listed in supplementary table S1, Supplementary Material online.
To assess the potential mapping bias in our data, we compared the mapping rate, genome coverage (with sites where the mapping depth ≥10), mismatch, and indel rate between mapped reads and the reference (supplementary table S2, Supplementary Material online) and did not find significant differences between species in these features (t-test, P = 0.075 for mapping rate, P = 0.203 for genome coverage, P = 0.317 for mismatch, and P = 0.172 for indels). Previous studies also showed that the two species had comparable genetic distances to the reference O. sativa without interspecies mapping bias (Huang et al. 2012; Guo et al. 2016). These observations indicated that mapping bias did not influence SNP calling.
To further validate the reliability of our SNP calling and, hence, our allele frequency estimation, we genotyped 271 individuals sampled from four population pairs (i.e., NEP1, NEP2, KHM, and LAO1) and one Chinese O. rufipogon population (rGX-bh) using Illumina Infinium BeadChip (Illumina Inc.). After excluding the sites in which calling rate <90% or Hardy–Weinberg equilibrium was rejected (Silva-Junior and Grattapaglia 2015), we successfully genotyped a total of 1,096 sites that were also detected by the Pool-Seq approach. For this subset of SNPs, we compared allele frequency estimates by Pool-Seq with estimates based on genotyping, and found high consistency between the two estimates (Pearson’s correlation coefficient r = 0.90, ranging from 0.86 to 0.91 for individual populations; determination coefficient in linear regression R2 = 0.82, ranging from 0.74 to 0.98), which validated the accuracy of allele frequencies determined by the Pool-Seq approach (Rellstab et al. 2013). On the other hand, because of the inherent limitation of distinguishing rare alleles from sequencing errors in Pool-Seq (Futschik and Schlötterer 2010; Lynch et al. 2014), we also applied different cut-off levels (i.e., 0.03, 0.05, 0.07) to rare alleles of the Pool-Seq data and obtained the same results in terms of the tree topology.
Phylogenetic and Population Analyses Revealed Multiple Origins of O. nivara
We performed phylogenetic analyses to distinguish between single and multiple origin hypotheses of O. nivara. Under the single origin (SO) hypothesis, populations of the two species should indicate reciprocal monophyly; in contrast, for the multiple origin (MO) hypothesis, populations from the same region should form a distinct clade (Schluter and Nagel 1995; Quesada et al. 2007). First, we conducted phylogenetic analyses based on Pool-Seq data and revealed that the O. nivara populations did not form a monophyletic clade but rather grouped together in accordance to the geographic region where they were collected (fig. 2a). This result did not change when different levels of missing data (supplementary fig. S1, Supplementary Material online) and various distance parameters (supplementary fig. S2, Supplementary Material online; see Materials and Methods) were used, suggesting that O. nivara might have originated multiple times in different regions.
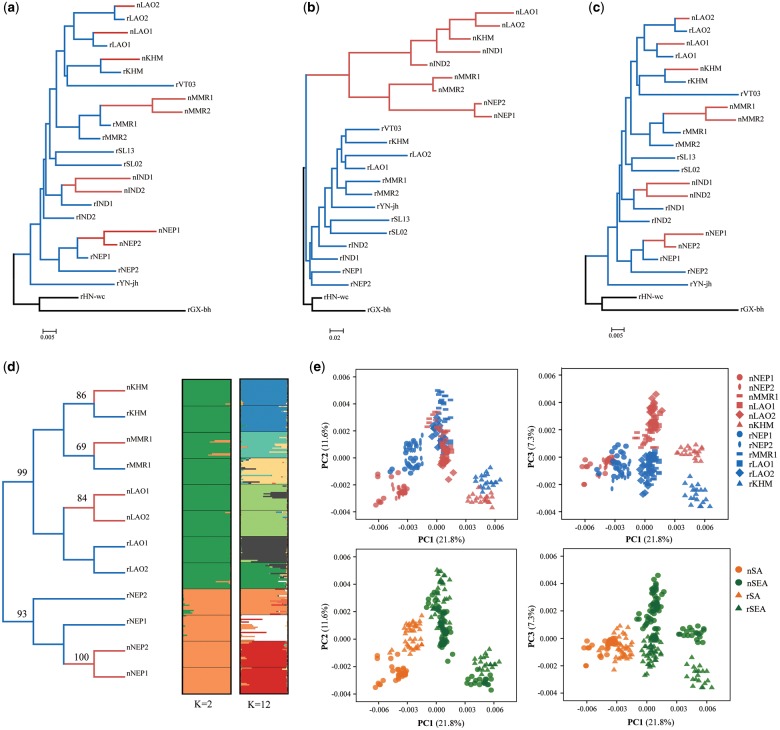
Fig. 2.
Phylogeny of 15 Oryza rufipogon (blue) and 9 O. nivara (red) populations based on whole-genome and population sequencing data. (a–c) Neighbor-joining trees constructed with Wright’s FST distance based on neutral (a), outlier (b), and linked (c) SNPs of whole-genome resequencing data. (d) Neighbor-joining tree inferred based on concatenated sequences of 16 nuclear neutral genes (bootstrap values are shown for nodes with >60% support), and the model-based population assignment at K = 2 set to the highest ΔK value and K = 12 set to the number of populations, where each horizontal line represents an individual, with its assignment probability to genetic clusters represented by different colors. (e) Principal coordinate analysis (PCoA) of all individuals of six population pairs based on sequences of 16 neutral genes indicate that 12 populations form two groups corresponding to geography rather than to species. SA and SEA represent South Asia and Southeast Asia, respectively. Population names correspond to the population IDs in figure 1 and supplementary table S1, Supplementary Material online.
Given the potential limitation of the Pool-Seq approach in detecting low-frequency alleles (Lynch et al. 2014; Fracassetti et al. 2015), we then used the combined sequences of 16 neutral genes to reconstruct the phylogenetic relationship of six population pairs of the two species (see Materials and Methods). The resulting trees showed that the O. nivara populations did not form a monophyletic clade; instead, they were more closely related to geographically neighboring O. rufipogon populations (fig. 2d and supplementary fig. S3, Supplementary Material online). Separate analyses on individual genes clearly indicated that none of genes supports the reciprocal monophyly of the two species (supplementary fig. 4a, Supplementary Material online) although exact relationships among populations were unclear due to low resolutions of genes, which arose from recent species divergence and ancient polymorphisms (Zheng and Ge 2010). In addition, all O. nivara populations clustered with the neighbored O. rufipogon populations in the species tree that inferred from gene trees (supplementary fig. 4b and c, Supplementary Material online). STRUCTURE analysis showed that the best supported clustering (K = 2) suggested the deepest divergence between two regions (South Asia vs. Southeast Asia) rather than between the two species (fig. 2d and supplementary fig. S5a, Supplementary Material online), again supporting the MO hypothesis of the O. nivara origin. Principal coordinate analysis (PCoA) based on these sequences also indicated that all the samples were organized into two distinct groups that corresponded to their geographic origins rather than their species identity (fig. 2e), suggesting that the O. nivara populations have evolved in parallel in different regions. Together, these results indicated that at least two independent origins of O. nivara occurred, with one in South Asia and the other in Southeast Asia.
Test for Alternative Hypotheses for Nonmonophyly
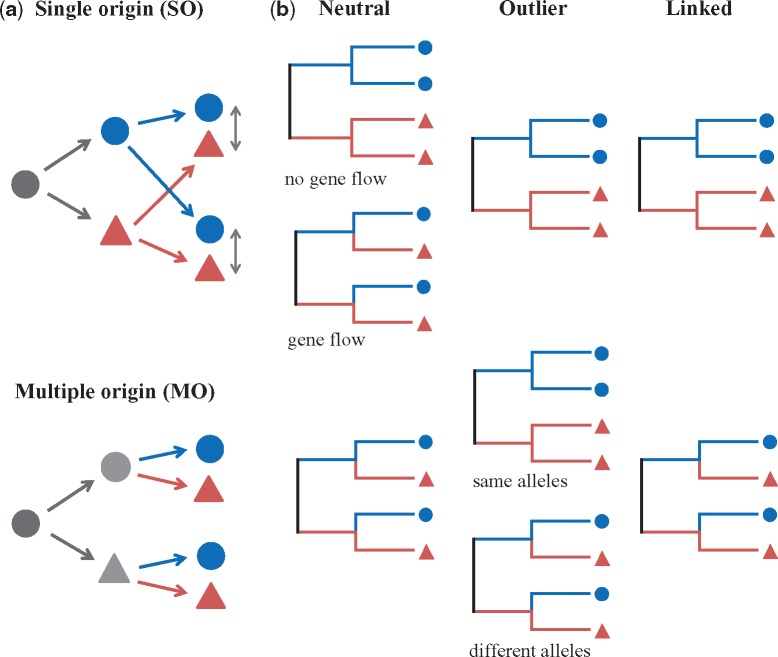
Although nonmonophyly of populations, that is, clustering of individuals from different populations by region rather than by species, is generally interpreted as multiple (parallel) evolution, a single origin with subsequent gene flow between species cannot be excluded entirely (Johannesson 2001; Schluter 2009; Nosil 2012). As shown in figure 3a, under the SO model, an ancestral population diverged into different species before they colonized in multiple regions, with or without an initial period of allopatry; in contrast, under the MO model, an ancestral population colonized multiple regions and then diverged into different species within regions.
Fig. 3.
Schematic diagram and test for single origin (SO) and multiple origin (MO) models. (a) Species divergence may arise in face of gene flow after secondary contact between two species that were already divergent in allopatry (the SO model) or may occur multiple times in sympatry (the MO model). (b) Test for alternative models of species origin. Both neutral and outlier SNPs generate similar phylogenies because of gene flow and different alleles at a locus; whereas linked SNPs give rise to different phylogenetic trees under different models of origin. Blue dots and red triangles represent Oryza rufipogon and O. nivara populations, respectively. Single-headed arrows stand for colonization or migration, and double-headed arrows indicate the gene flow between populations.
To distinguish between the alternative models, we followed the methods by Roda et al. (2013) and Ravinet et al. (2016) to investigate the phylogenetic patterns at neutral, outlier, and linked loci which were assumed to reflect different origin models (fig. 3b). The outlier loci (candidates of selection) were defined as the sites where allele frequency was significantly different between species and the linked loci were those tightly linked to outlier loci (see Materials and Methods). The results indicate that the two rice species show reciprocal monophyly in the tree constructed from the outlier SNPs (fig. 2b); whereas the tree that was constructed based on linked SNPs (fig. 2c) generated a similar phylogeny to that based on neutral SNPs (fig. 2a), that is, populations from the same region clustered together, which is a pattern consistent with the MO model (fig. 3b). These results remained the same when different genetic distances, different criteria of grouping SNPs, and various missing levels were used (supplementary figs. S1, S2, and S6, Supplementary Material online; see Materials and Methods).
To further verify that the nonmonophyly of the O. nivara populations represents multiple origins rather than local gene flow due to secondary contact, we used the sequence data of 16 neutral genes to perform approximate Bayesian computation (ABC) for distinguishing the alternative models of origin for two groups of populations: one group consists of two population pairs in Nepal (NEP1 and NEP2) (hereafter, South Asia) and the other group consists of three population pairs in Cambodia and Laos (KHM, LAO1, and LAO2) (hereafter, Southeast Asia). The two groups of populations, geographically separated by >2,000 km (fig. 1), are independent lineages in the trees and thus represent two independent origins of O. nivara (fig. 2d). For either MO or SO model, we considered different scenarios involving population split events only (Standard scenarios), fluctuating population sizes through time (Demography scenarios), and gene flow between paired populations (Migration scenarios) (supplementary fig. S7, Supplementary Material online; see Materials and Methods). We applied these models to different combinations of population pairs in the two regions (i.e., South Asia and Southeast Asia) from which two independent origins of O. nivara were assumed. In all six combinations of population pairs between regions, the posterior probability (PP) was much higher for the MO models than for the SO models (supplementary table S3, Supplementary Material online; see Materials and Methods). To further evaluate which MO model was more likely, we compared the PP values among three MO models and found that the Standard scenario (mean PP of 0.47, ranging from 0.31 to 0.64) and Demography scenario (mean PP of 0.51, ranging from 0.33 to 0.67) were equally supported with much greater possibility than that of the Migration scenario (mean PP was only 0.02), suggesting a negligible level of gene flow between the paired populations.
Adaptive Divergence of Phenotypic Traits and the Role of Natural Selection
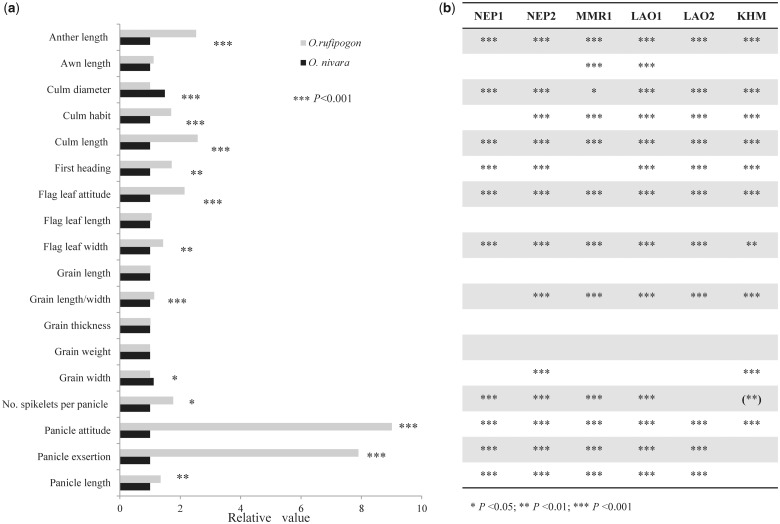
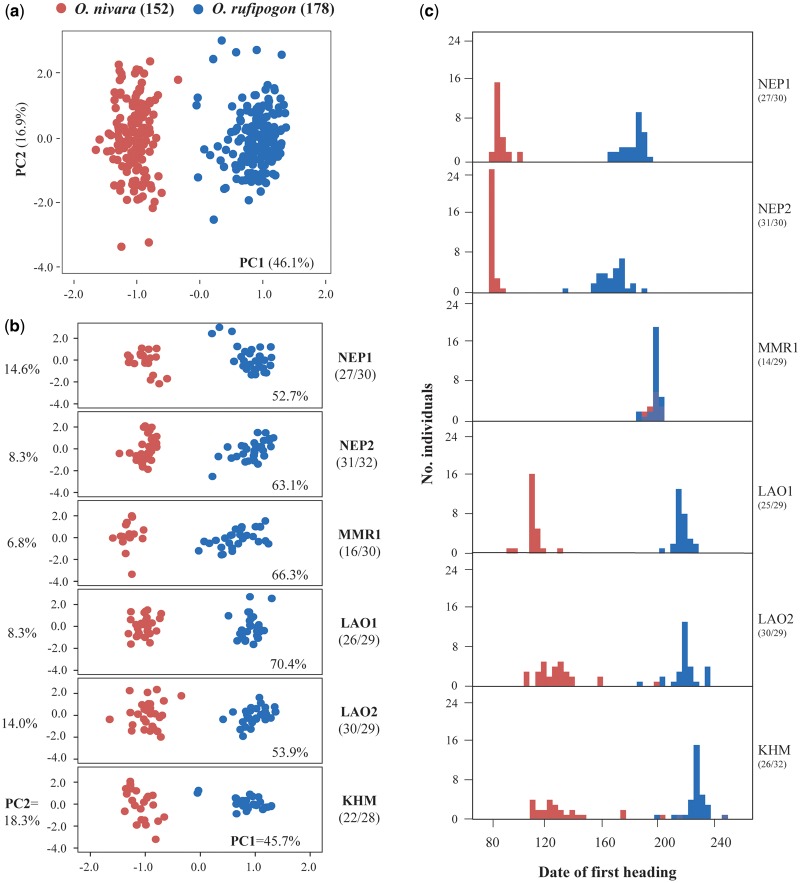
To examine which phenotypic traits are significantly differentiated between species and if the divergence is an adaptive response to divergent habitats, we conducted a common garden experiment for six pairs of populations sampled from different localities (fig. 1). Linear mixed-effect models detected that 14 of 19 traits showed significant differences, and only three grain traits (weight, length, and thickness of grains), awn length, and the flag leaf length were not significantly different between species (fig. 4a andsupplementary table S4, Supplementary Material online). Similarly, PCA showed that the combined samples are organized into two distinct groups (or species), with PC1 and PC2 accounting for 46.1% and 16.9% of the variance, respectively (fig. 5a). A three-way analysis of variance (ANOVA) (Butlin et al. 2014) on first two PCs and all 18 traits further indicate that all traits except for PC2 showed significant differences between species and the differences remain significant after correction for multiple testing, except for three traits (panicle exsertion, number of spikelets per panicle, and attitude of flag leaf). By contrast, no significant differentiation was found between regions within species and between localities within regions for all traits (table 1). In general, relative to the perennial O. rufipogon, the annual O. nivara is shorter and thicker, flowers earlier, has shorter and less exserted panicles with more compactness, and has more spikelets per panicle with shorter anthers and awns (fig. 4a), consistent with previous reports (Grillo et al. 2009; Banaticla-Hilario et al. 2013; Guo et al. 2016).
Fig. 4.
Phenotypic divergence between Oryza rufipogon and O. nivara based on 18 quantitative traits recorded in the common garden experiment. (a) Divergence in mean phenotypic values of the combined samples. The species differences were standardized to the smaller value of the two species and thus the scale represents the relative difference between species for individual traits. (b) Significance (after correction for multiple testing) and direction of phenotypic divergence in six population pairs of O. rufipogon and O. nivara. Parentheses indicate that the direction of phenotypic divergence was opposite to that found in the combined samples.
Fig. 5.
Phenotypic variation and premating reproductive isolation between species. (a and b) Principal component analysis (PCA) of six pairs of Oryza rufipogon (blue) and O. nivara (red) populations based on 19 morphological traits recorded in the common garden experiment. All combined samples (a) and separate analyses on each of six paired populations (b) show the clear parallel phenotypic differentiation between species. Number of individuals (O. nivara/O. rufipogon) observed for the paired populations are in parentheses. (c) Frequency distribution of first heading of six population pairs in the common garden experiment. Bars indicate the number of individuals initiating flowering for O. rufipogon (blue) and O. nivara (red). Number of individuals (O. nivara/O. rufipogon) observed for the paired populations are in parentheses.
Table 1.
Three-Way ANOVA for PC1, PC2, and 18 Phenotypic Traits.
| Traits | Mean ± SE |
Source of Variation |
|||||
|---|---|---|---|---|---|---|---|
| Oryza rufipogon | O. nivara | Species/Habitat | Region | Locality | Species × Region | Species × Locality | |
| PC1 | 0.67±0.34 | −1.31±0.36 | 2.08E-04***(*) | 0.7 | 0.97 | 0.39 | 4.76E-11***(***) |
| PC2 | 0±0.82 | 0.01±1.29 | 0.99 | 0.56 | 0.95 | 0.31 | 1.38E-16***(***) |
| First heading | 203.69±22.62 | 118.61±35.51 | 9.58E-05***(**) | 0.25 | 0.33 | 0.05 | 1.68E-03**(-) |
| Attitude of panicle branches | 40.82±12.81 | 4.53±12.29 | 3.40E-04***(*) | 0.26 | 0.32 | 0.2 | 0.18 |
| Panicle exsertion | 10.43±5.58 | 1.32±3.46 | 5.81E-03**(-) | 0.87 | 0.67 | 0.17 | 8.21E-04***(-) |
| Anther length | 0.48±0.07 | 0.19±0.04 | 2.56E-07***(***) | 0.55 | 0.96 | 0.07 | 0.21 |
| Panicle length | 27.56±4.38 | 20.41±4.51 | 3.20E-04***(*) | 0.88 | 0.81 | 0.38 | 3.33E-12***(***) |
| Awn length | 5.62±1.4 | 5.05±1.3 | 4.90E-05***(**) | 0.41 | 0.95 | 0.12 | 9.04E-03**(-) |
| Culm length | 179.78±32.13 | 69.77±22.09 | 5.37E-05***(**) | 0.72 | 0.56 | 0.3 | 1.44E-12***(***) |
| No. spikelets per panicle | 76.77±34.96 | 43.44±15.59 | 2.37E-03**(-) | 0.76 | 0.62 | 0.48 | 3.52E-16***(***) |
| Grain weight | 0.13±0.02 | 0.13±0.04 | 1.67E-04***(*) | 0.4 | 0.51 | 0.37 | 6.82E-04***(-) |
| Grain length | 8.03±0.5 | 7.94±0.69 | 7.32E-05***(**) | 0.98 | 0.33 | 0.18 | 3.80E-06***(***) |
| Grain width | 2.08±0.17 | 2.33±0.22 | 6.61E-06***(***) | 0.8 | 0.95 | 0.43 | 1.38E-10***(***) |
| Grain length/width | 3.91±0.37 | 3.44±0.19 | 0.0***(***) | 0.89 | 0.39 | 0.96 | 1.16E-06***(***) |
| Grain thickness | 1.49±0.1 | 1.47±0.15 | 1.30E-06*** | 0.65 | 0.52 | 0.59 | 1.31E-07***(***) |
| Culm habit | 79.33±13.65 | 46.67±15.15 | 1.89E-05***(**) | 0.44 | 0.63 | 0.44 | 1.29E-03**(-) |
| Culm diameter at basal internode | 0.51±0.11 | 0.76±0.17 | 6.34E-06***(***) | 0.72 | 0.4 | 0.22 | 0.03*(-) |
| Attitude of flag leaf | 73.71±22.97 | 34.44±19.52 | 8.43E-04***(-) | 0.82 | 0.55 | 0.76 | 3.87E-05***(**) |
| Flag leaf length | 25.04±5 | 23.85±8.79 | 1.42E-04***(*) | 0.7 | 0.74 | 0.23 | 8.44E-07***(***) |
| Flag leaf width | 1.23±0.22 | 0.86±0.16 | 1.07E-05***(***) | 0.38 | 0.67 | 0.07 | 0.04*(-) |
Significance after correction for multiple testing is in the parentheses.
P < 0.05, **P < 0.01, ***P < 0.001.
Separate analyses on individual pairs of populations based on one-way ANOVA and χ2 test obtained more striking differences between species (fig. 5b and supplementary table S4, Supplementary Material online), suggesting that phenotypic difference between the paired populations is associated with habitat differentiation. Importantly, almost all the traits that diverged between species showed remarkable concordance in the direction of phenotypic differentiation for all six population pairs (fig. 4b). It is noted that the six O. nivara populations consist of two groups originating independently from different areas as indicated earlier. These results imply the role of natural selection during phenotypic divergence between O. rufipogon and O. nivara.
To test the hypothesis that phenotypic differentiation between species is an adaptive response to divergent habitats, we performed QST–FST analysis, a method that has been widely used to examine whether natural selection or genetic drift is the main cause of population differentiation in phenotypic traits (McKay and Latta 2002; Leinonen et al. 2013). Based on the sequences of 16 neutral genes, we obtained an average FST value of 0.300 between species across six paired populations, with individual pair values ranging from 0.195 to 0.393 (supplementary fig. S8, Supplementary Material online). For the combined test, all 18 traits showed significantly higher QST than FST (Z-test, P < 0.0001; supplementary table S5, Supplementary Material online). Separate analyses on individual population pairs revealed that 11 traits showed significantly higher values for QST than for FST across all six pairs (ranging from 14 to all 18 traits) (supplementary fig. S9 and table S5, Supplementary Material online). The QST–FST comparison therefore indicated that the parallel phenotypic divergence between species was an adaptive response to divergent habitats and driven by natural selection.
Premating Isolation Contributes to the Barrier of Gene Flow between Species
To determine whether reproductive isolation is established between the ancestral O. rufipogon and descendent O. nivara populations and between populations within species, we investigated both premating and postmating isolations for various comparisons of populations based on common garden and crossing experiments (see Materials and Methods). We found marked between-species differences in first heading and flowering peak for five out of the six population pairs (fig. 5c). One exception is the population pair in Myanmar (MMR1), where the flowering time of the two species entirely overlapped (fig. 5c and supplementary fig. S10 and table S4, Supplementary Material online). A plausible reason is that the O. nivara populations in Myanmar represent a third origin that is different from the two origins we discussed here (i.e., one in South Asia and the other in Southeast Asia). This explanation is in agreement with the phylogenetic trees based on the Pool-Seq data (fig. 2a), in which two O. nivara populations in Myanmar (nMMR1 and nMMR2) formed a distinct lineage with two O. rufipogon populations in Myanmar (rMMR1 and rMMR2). An in-depth study of the hypothesis on the origin of the Myanmar O. nivara is currently underway and should provide further insights into the origin of this species.
On an average, the O. nivara populations flowered much earlier than the O. rufipogon populations, with the differences ranging from 84.3 (NEP2) to 105.6 (LAO1) days (supplementary table S4, Supplementary Material online). This difference in flowering time between species was consistent in two consecutive years under standard common garden conditions (supplementary fig. S10, Supplementary Material online). Notably, within each species, flowering time showed a latitudinal cline and overlapped, with first heading date depending on the latitude of the populations (fig. 5c), suggesting potential gene flow between populations/localities within species. Further analysis based on a three-way ANOVA revealed significant differences in flowering time for populations between species but not for populations within species whether they were from same or different regions (table 1). The Overlap Index (OI) (Pianka 1973) also shows no significant difference in flowering time between populations within species (supplementary table S6, Supplementary Material online).
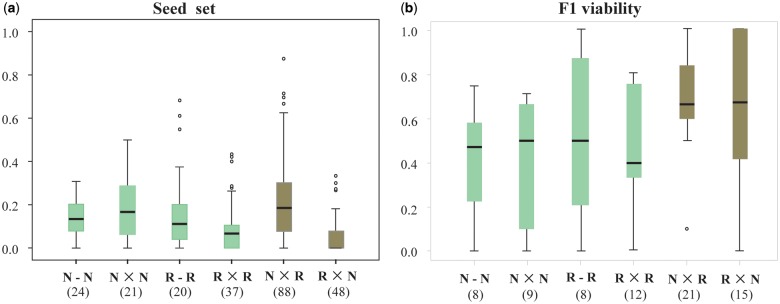
We further chose three population pairs to conduct a crossing experiment to detect potential postmating isolation (see Materials and Methods). Based on a total of 238 artificial crosses involving those within populations, between populations within species, and between species, we found that the average seed set and F1 viability of the crosses between species were comparable to those of the crosses within species, although significant differences in seed set were found between the crosses with O. rufipogon as the maternal parent and those with O. nivara as the maternal parent (fig. 6 and supplementary table S7, Supplementary Material online). In addition, neither premating nor postmating reproductive barriers were observed between populations within both O. rufipogon and O. nivara (figs. 5c and6), in agreement with previous reports (Lu et al. 2000; Banaticla-Hilario et al. 2013). Similarly, statistical analyses did not detect significant differences in average seed set and F1 viability between populations within species (supplementary table S7, Supplementary Material online). Together, these results indicate that the difference between species in flowering time caused almost completed premating reproductive isolation between species and contributed to the divergence of two wild species.
Fig. 6.
Postmating reproductive isolation between species. Boxplots showing average seed set (a) and F1 viability (b) of crosses for various combinations of the three pairs of populations, respectively. Boxes and horizontal bars represent the central 50% and the median of the data, respectively. Dots represent outliers beyond 1.5 times the interquartile range. “N-N” and “R-R” represent crosses between individuals from the same population within Oryza nivara (N) and O. rufipogon (R), respectively. “N × N” and “R × R” represent crosses between individuals from different populations within the same species. “R × N” and “N × R” (dark columns) represent crosses between individuals from different species (with O. rufipogon as the maternal and paternal parents, respectively). Number of crosses for a specific combination is in parentheses.
Discussion
Parallel speciation has been reported in animals (Johannesson 2001; Nosil 2012) and strongly suggests the action of natural selection in the process of speciation. However, few cases of parallel speciation have been demonstrated in plants, which might arise from the possibility that plants are less prone to parallel speciation than animals or reflect a lack of empirical studies involving rigorous testing (Abbott and Comes 2007; Ostevik et al. 2012). This study demonstrates an unequivocal case of parallel speciation in plants, in which all four criteria for parallel speciation are satisfied. One particular challenge to demonstrate parallel speciation is to distinguish between the multiple origin and the single origin following gene flow between species (Quesada et al. 2007; Nosil 2012; Roda et al. 2013; Butlin et al. 2014; Faria et al. 2014), which was overlooked in many previous studies. Using whole-genome resequencing and Sanger sequencing of population samples, we performed both phylogenetic analyses and ABC modeling to obtain solid evidence for supporting the multiple origin of the derived species (i.e., O. nivara). Our study of wild rice, together with an accumulating number of other cases in plants (Roda et al. 2013, 2017; Richards et al. 2016; Ru et al. 2016; Comes et al. 2017; Trucchi et al. 2017) imply that parallel speciation might not be uncommon in plant species.
The evolution of reproductive isolation is the most important step in the formation of new species (Rieseberg and Willis 2007; Nosil 2012). In addition to our common garden study that identified almost complete premating reproductive isolation between species, a literature survey on the phenology of wild populations further indicates marked differences in flowering time between the species across their entire distributional regions, with O. nivara flowering significantly earlier than O. rufipogon (Sharma and Shastry 1965; Barbier 1989; Lu 1998; Kuroda et al. 2006). Such a premating isolation mechanism in plant speciation has been reported in many previous investigations (Hall and Willis 2006; Lowry et al. 2008; Moyers and Rieseberg 2016; Ferris et al. 2017), supporting the notion that prezygotic isolation was either a more important or earlier-evolving barrier to gene flow than was postzygotic isolation in plant speciation (Rieseberg and Willis 2007; Nosil 2012). It is evident that the phenotypic divergence and reproductive isolation between O. rufipogon and O. nivara are associated with habitat differences, as expected in ecological speciation (Schluter 2009; Nosil 2012; Moyers and Rieseberg 2016). Based on a quantitative trait locus (QTL) analysis of eight traits involving life history, mating system, and flowering time, Grillo et al. (2009) found that a total of 30 QTLs, with effect sizes ranging from 2.9% to 36.5%, contributed to the major phenotypic differentiation between O. rufipogon and O. nivara, with >80% QTL alleles of O. nivara acting in the same direction of phenotypic evolution. Our recent genome-wide expression investigation of O. rufipogon and O. nivara (Guo et al. 2016) revealed that of the 21,415 expressed genes across three tissues, ∼8% (1,717 genes) differed significantly in expression levels between species and that 62% of the differentially expressed genes exhibited a signature of directional selection. These results demonstrate a complex genetic basis of the phenotypic divergence between the two species and suggest that these genetic changes were fixed under directional selection.
The parallel speciation in wild rice raises an interesting question of what drives the formation of new species. Previous studies hypothesized that O. nivara originated from O. rufipogon in association with an ecological shift from a persistently wet to a seasonally dry habitat during recent glaciations (Barbier 1989; Morishima et al. 1992; Zheng and Ge 2010; Banaticla-Hilario et al. 2013; Huang et al. 2013). This hypothesis is consistent with the estimated time of O. nivara origin (Zheng and Ge 2010) and species distribution modeling which suggested that precipitation and temperature were the main climate variables contributing to the distribution of O. nivara (Liu et al. 2015), as well as the fact that many annual grasses evolved in response to the dry climate in monsoonal Asia in recent glaciations (Morishima et al. 1992; Liu et al. 2015). Of the three major mechanisms (escape, avoidance, and tolerance) through which plants adapt to drought, drought escape is to develop rapidly and reproduce before the onset of drought and thus is optimal for annual plants in environments with short growing seasons (Juenger 2013; Kooyers 2015). As such, flowering time is usually investigated as a measure of drought escape because earlier flowering results in greater fitness during the shortened growing season (Kooyers 2015). Flowering time changes can have widespread ecological consequences and adaptive responses to drought through flowering time shifts have been well-characterized in plant species (Hall and Willis 2006; Franks 2015; Kooyers 2015; Moyers and Rieseberg 2016; Ferris et al. 2017). For example, strong selection for earlier flowering was reported in two Brassica rapa populations following drought (Kooyers 2015). In a study of two ecotypes of Mimulus gutttatus, Hall and Willis (2006) found that early flowering was favored in an area where annual plants grew and was characterized by dry soils, demonstrating that divergent selection on flowering time contributed to local adaptation. Therefore, early flowering is a classic adaptation of plants to dry habitats and allows plants to complete reproduction before seasonal drought (Franks 2015; Kooyers 2015; Ferris et al. 2017). Similarly, flowering time in our case is a typical “magic trait” (Servedio et al. 2011; Nosil 2012; Moyers and Rieseberg 2016) that contributes both to adaption and to reproductive isolation. The nearly complete isolation in flowering time together with a difference in mating system (Sang and Ge 2007; Vaughan et al. 2008) provide strong premating barriers to gene flow between species and play critical roles in the O. nivara origin. Previous studies also found that an QTL with relatively large effect contributed to the loss of photoperiod sensitivity in O. nivara (Grillo et al. 2009) and that genetic changes of seed storage protein genes in O. nivara provided an advantage for seedling establishment and survival in dry soil conditions (Huang et al. 2013). To summarize, this system provides an outstanding scenario for testing the underlying mechanisms of ecological speciation as well as what genes and gene networks and to what extent selection acts repeatedly during speciation.
Materials and Methods
Study System and Population Sampling
We sampled 15 O. rufipogon and 9 O. nivara populations that covered the major areas where the two species are sympatric (fig. 1). Of them, 18 populations were sampled by the authors during field collections from 2008 to 2011, in which both the leaf and seed samples were collected individually, with the population sizes ranging from 14 to 32. The remaining six populations were obtained from the International Rice Research Institute (IRRI) in Los Banos, Philippines, with sample sizes >14 individuals per population (supplementary table S1, Supplementary Material online). These IRRI samples were collected from localities with the same latitudes and longitudes and thus were treated as “populations” similar to previous investigations (Zheng and Ge 2010; Banaticla-Hilario et al. 2013; Liu et al. 2015). We conducted whole-genome sequencing of pooled DNA samples (Pool-Seq) for all 24 populations. Of them, six and three pairs of sympatric populations of the two species were used in a common garden experiment in Guangxi Province and an artificial crossing experiment in Hainan Province, respectively (fig. 1 and supplementary table S1, Supplementary Material online).
Common Garden Experiment and Analyses of Morphological Traits
To investigate phenotypic variation in populations within and between species and its correlation with habitats, we conducted a common garden experiment using six sympatric pairs of O. rufipogon and O. nivara populations at the Guangxi Academy of Agricultural Sciences (GAAS) in Nanning City (N22°50.72′, E108°14.85′), Guangxi Province, China, for three consecutive years (2011–2013) (fig. 1). These natural populations were sampled across four localities (Nepal, Myanmar, Laos, and Cambodia) by the authors, with the distance between two populations of each pair ranging from 1.3 to 44.9 km (supplementary table S1, Supplementary Material online).
Over 30 individuals, each descended from separately collected maternal plants, were used for all populations (supplementary table S1, Supplementary Material online). Seeds were germinated at 30°C in an incubator, and the young seedlings were then transplanted to pots at three-leaf stage and finally planted randomly in the experiment garden of GAAS. We measured 19 morphological traits (supplementary table S8, Supplementary Material online) for each individual based on the methods described in Bioversity International et al. (2007). To obtain detailed information on phenotypic variation of all populations, we conducted censuses every day throughout the entire growth season during the three years. All measurements were taken on three tillers/culms and averaged for each trait, with exceptions of first heading, seed traits and stolon trait (supplementary table S8, Supplementary Material online).
We used the linear mixed-effects models (nlme package in R) to analyze divergence of morphological traits at the species level. The species served as fixed effect and population as random effect in linear mixed model. For each population pair, phenotypic divergence was analyzed using ANOVA and χ2 test (Guo et al. 2016). Phenotypic differentiation between species was also assessed by a standard principal component analysis (PCA) based on the combined and individual data sets. To examine how the phenotypic variation is partitioned between species/habitat, among localities within species and among individuals within populations, we further performed a three-way analysis of variance (ANOVA) (Butlin et al. 2014) on first two PCs and all 18 phenotypic traits, with habitat/species and region (Nepal, Myanmar, Laos, and Cambodia) as fixed factors, and locality (six sites where pairs of species populations were sampled) as a random factor within the interaction between fixed factors. To quantitatively assess the extent of flowering time-based reproductive isolation between populations, we calculated an Overlap Index (OI) (Pianka 1973) as detailed in Lobo et al. (2003).
Whole-Genome Resequencing, Variant Calling and SNP Validation
Genomic DNA was extracted from fresh or silica-gel dried leaves using the standard CTAB method. DNA pools were created for each of the 24 populations by mixing equal amount of individual DNA that was quantified using a Qubit 2.0 fluorometer. For sequencing, we sheared the pooled DNA and constructed ∼500-bp paired-end libraries using a Paired-End Sample Preparation Kit (Illumina, San Diego, CA) following the manufacturer’s instructions. After removing the adaptor and primer sequences (Bolger et al. 2014), the reads were trimmed with a minimum base quality score 30 (error rate 0.001) and a minimum length 50 bp (Kofler, Orozco-terWengel, et al. 2011). The reads that lost their mates after trimming were also dropped. As a result, a total of 268-Gb high-quality data with a depth of 1.35× for each individual was generated (supplementary table S2, Supplementary Material online); this amount of data ensures the accurate estimation of allele frequencies according to empirical and theoretical studies (Futschik and Schlötterer 2010; Kofler, Orozco-terWengel, et al. 2011). The summary statistics of the raw data were calculated using FastQC v0.10.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
The mapping and variant calling processes were conducted following the protocol proposed by Schlötterer (Schlötterer et al. 2014). The read mapping tool bwa v0.6.2-r126 (Li and Durbin 2009) and samtools v0.1.19 (Li et al. 2009) were used to map the reads to the reference genome (IRGSP 1.0 of Oryza sativa genome; Kawahara et al. 2013), allowing a maximum of three mismatches and one gap for each mapping read. Only the reads that were uniquely mapped to the reference and had a mapping quality score >30 were retained. Two different pipelines, PoPoolation2 (Kofler, Pandey, et al. 2011) and VarScan 2 (Koboldt et al. 2012) were adopted for variant calling. We removed duplicates showing identical 5′ coordinate (Fracassetti et al. 2015) using Picard (http://broadinstitute.github.io/picard/) and indels together with ten flanking nucleotides to further reduce false positives in variant calling. To validate the reliability of these SNPs, an Illumina Infinium BeadChip (Illumina Inc.) was customized using the probes designed from rice whole-genome resequencing data (Huang et al. 2012). All the manipulations were carried out following the manual. Genotyping calling was analyzed using software Genome Studio (Illumina Inc.). The markers whose call rate <90% or violating Hardy–Weinberg equilibrium were excluded (Silva-Junior and Grattapaglia 2015). The consistency of allele frequencies between Pool-Seq and genotyping data was evaluated by Pearson’s correlation coefficient and linear regression.
Sequencing of 20 Neutral Genes and Population Genetic Analysis
We sequenced fragments of 20 single-copy nuclear genes for 20 individuals from each of the 12 populations that were used for the common garden experiments (supplementary table S1, Supplementary Material online). These nuclear genes are unlinked loci randomly distributed on all 12 chromosomes of rice, and most of them were demonstrated to evolve neutrally in previous studies (Zhu et al. 2007; Zheng and Ge 2010; Liu et al. 2015); thus, they were suitable for a population genetics study. Additional information on the sequenced regions of all loci and the primers used for amplification is provided in supplementary table S9, Supplementary Material online.
Genomic DNA extraction, polymerase chain reaction (PCR) amplification and sequencing of the genes followed our previous studies (Zheng and Ge 2010; Liu et al. 2015). Sequencing was performed on an ABI3730XL automatic sequencer (Applied Biosystems Corp.). Purified PCR products were sequenced either directly or after cloning into pGEM T-easy vectors (Promega Corp.) if dual peaks were found due to some heterozygous individuals (Zhu et al. 2007; Zheng and Ge 2010). Singletons produced by direct sequencing were confirmed through repeated PCR amplification and sequencing (Zheng and Ge 2010; Liu et al. 2015). The sequenced fragments were assembled by ContigExpress (Informax Inc., North Bethesda, MD) and edited manually. All sequences have been deposited in GenBank (http://www.ncbi.nlm.nih.gov/genbank/), with the accession numbers KX275412–KX276089.
We tested the neutrality of all 20 loci in our samples using Tajima’ D (Tajima 1989) and Fu and Li’s D* and F* (Fu and Li 1993). Although all these fragments did not show significant derivation from neutrality at the population level, we found significant values for either Tajima’s D or Fu and Li’s D* and F* at one or a few of the specific populations for four loci (GELP80, P14, PK, T4) (supplementary table S10, Supplementary Material online). Despite the fact that significant values for the parameters might arise from different factors other than nonneutrality, we conducted our population genetic analyses by excluding these four loci. Based on the sequences of the 16 neutrally evolved loci, we calculated the number of segregating sites (S), number of haplotypes (h), nucleotide diversity π, θw, and FST by DNASP v5.1 (Librado and Rozas 2009).
To examine the genetic subdivision of six pairs of populations, we conducted Bayesian clustering analysis using STRUCTURE 2.23 (Pritchard et al. 2000) and performed principal coordinate analysis (PCoA) implemented in DARWIN (Perrier and Jacquemoud-Collet 2006). For STRUCTURE analysis, we modeled K values from 1 to 15, with 50,000 burn-in iterations followed by 300,000 Markov chain Monte Carlo (MCMC) iterations for accurate parameter estimates. To verify the consistency of the results, we performed 20 independent runs for each K value using an admixture model with correlated allele frequencies. The uppermost hierarchical level of the genetic structure was estimated using the statistic ΔK following the procedure described by Evanno et al. (2005).
Phylogenetic Analysis
We performed phylogenetic analyses based on the Pool-Seq and population sequencing data sets to reveal the genetic affinities among populations and to distinguish between the SO and MO hypotheses of O. nivara. Under the SO hypothesis, populations of the two species should indicate reciprocal monophyly; in contrast, for the MO hypothesis, populations from the same region should form a distinct clade.
For the Pool-Seq data, we generated the Neighbor-Joining (NJ) tree, as implemented in phylip v3.69 (Felsenstein 1989), based on the neutral SNPs (see below); this method was used because the neutral loci were desirable for inferring the shared ancestry and were free of the distortion role of selection on the affinity among populations (Beaumont and Balding 2004). For generating the population trees, we calculated the widely used genetic distances based on the allele frequency of SNPs, including the classic Wright’s FST (Hudson et al. 1992), Nei’s GST (Nei and Chesser 1983), Hedrick’s standardized GST (generally called G’st) (Hedrick 2005) and Jost’s D (Jost 2008). As these relative measures relied on the diversity within population, the absolute measure of divergence dXY was also calculated by dividing the number of pairwise nucleotide differences by the total length of the variable and invariable sites between populations (Nei and Li 1979). We also generated the trees under different missing levels (from 10% to 60%) that were defined as the proportion of missing data in 276 pairwise comparisons of the 24 populations.
For combined sequences of the 16 neutral genes, population trees were constructed using NJ and UPGMA methods by Mega 5 (Tamura et al. 2011). A maximum likelihood (ML) tree of single genes was also constructed using the RAxML program (Stamatakis 2014), which were used to generate estimated species-tree by using ASTRAL (Mirarab et al. 2014) at the individual and population levels (fig. S4).
Test for Alternative Hypotheses for Nonmonophyly
We used two approaches to distinguish between the SO and MO models. First, we test for the alternative models by generating phylogenies using different types of SNPs across the genome (Roda et al. 2013; Ravinet et al. 2016). Specifically, we divided the genome-wide SNPs into three categories: outlier SNPs (candidates of selection), linked SNPs (tightly linked to outlier SNPs), and neutral SNPs. The outlier SNPs were defined as the sites where allele frequency was significantly different between species for one/three/five pairs of O. rufipogon/O. nivara populations (Fisher’s exact test, FDR-corrected P value ≤0.05). Here, three criteria (one/three/five pairs) were used to define outliers because some SNPs met the criteria by chance. When the criterion was set to one pair, more SNPs would be defined as outlier even they were neutral in most other pairs; however, when five pairs used as the criterion, more real outliers will be omitted. For the linked SNPs, we empirically defined them as the SNPs that were <100/300/500 bp away from the nearest outlier SNPs. The neutral SNPs were then defined as the remaining SNPs. Phylogenetic trees were then constructed using the data sets from different combinations of the SNP grouping criteria. In the MO model, outlier sites would group populations either by species or by geography; whereas the neutral and linked sites would produce a phylogenetic pattern in which populations are clustered by geography rather than by species (Roda et al. 2013) (fig. 3b). We found that data sets with different combinations obtained the same conclusion that the O. nivara populations originated multiple times (supplementary fig. S6, Supplementary Material online). Thus, we retained in the main text only the result from the combination of the criterion “three pairs” and “300 bp”, in which the number of three types of SNPs is 4,292,150 (neutral), 561,218 (outlier), and 1,314,783 (linked), respectively (fig. 2). Furthermore, phylogenetic analyses under different distances and various levels of missing data (from 10% to 60% of 276 pairwise comparisons of 24 populations) did not change our conclusion (supplementary figs. S1 and S2, Supplementary Material online).
In the second method, as described by (Butlin et al. 2014), we performed approximate Bayesian computation (ABC) that is likelihood-free and widely used in the parameter estimation and model selection of complicated evolutionary scenarios (Beaumont 2010; Csilléry et al. 2012) to compare different models for demographic history of the populations of two species based on the sequences of 16 neutral genes. Because two major models of O. nivara origin (fig. 3a) involve different scenarios of demographic history, the extent and direction of gene flow, and the different split times between paired populations, three kinds of scenarios were considered for each major model with a total of 12 models (supplementary fig. S7, Supplementary Material online). In the first scenario, only the split event between populations was modeled (i.e., the standard scenario) (supplementary fig. S7, top panel, Supplementary Material online) and in two additional more complex scenarios, the history of population size (i.e., the demography scenario) and gene flow (i.e., the migration scenario) were modeled separately (supplementary fig. S7, middle and bottom panels, Supplementary Material online). The prior distribution of the model parameter was defined based on previous studies (Zheng and Ge 2010; Ai et al. 2012; Liu et al. 2015) and is listed in supplementary table S11, Supplementary Material online. Shared parameters between models had the same prior, and all the parameters were log uniform, with the exception of the split time and mutation rate, which were uniform. Generation time was set to one year. For each model, we performed 1.0e7 coalescent simulations using fastsimcoal2 (Excoffier et al. 2013) by randomly drawing the parameter sets from the parameter space.
The summary statistics, calculated by arlsumstat in Arlequin v3.11 (Excoffier et al. 2005), were used to compare the simulated and observed data. These statistics included the number of segregating sites (S), nucleotide diversity (π), Tajima’s D, and pairwise FST among populations. As a result, a total of 18 summary statistics were used. To capture the main information in the summary statistics and to avoid the noise introduced by large number of statistics, we tried to transform the summary statistics using partial least squares (PLS) method (Wegmann et al. 2010). However, we found that the information on variation from the summary statistics could not be recovered with subset of the PLS components, probably because of the lack of multicollinearity of our statistics. Thus, the raw summary statistics were used in our ABC analysis.
Selection between the MO and SO models was performed under each scenario by comparing their posterior probabilities (Wegmann et al. 2010; Csilléry et al. 2012; Butlin et al. 2014), which were estimated by the multinomial logistic regression method (“mnlogistic”) with a tolerance rate of 0.001. The posterior probabilities among three MO models were further compared with test which MO model was more likely. As the posterior probability was an approximation in ABC method, we performed additional analysis to test the goodness of fit between our models and the data. We first evaluated whether ABC could distinguish between the MO and SO models by performing cross-validation under each scenario. The confusion matrices of cross-validation indicated that most models were assigned to their own categories, except for the migration scenario, in which the three SO models were slightly confused (supplementary fig. S11, Supplementary Material online). These plots demonstrated that the models were distinguishable by the ABC method in our analysis. We then adopted three methods to test the goodness of fit for the model: 1) to compute a P value according to the null distribution of the mean distance between observed and accepted samples (supplementary table S12, Supplementary Material online); 2) to make the posterior predictive checks by sampling 1,000 sets of parameters from their posterior distribution and simulating the summary statistics a posteriori using fastsimcoal2. The resulting distribution of each summary statistic should reproduce the observed value if the preferred model fits our data well (as judged by the P value) (supplementary table S13, Supplementary Material online); and 3) we plotted the first two PCs based on PCA of the accepted summary statistics of the preferred models (supplementary fig. S12, Supplementary Material online), and the result should match the observed summary statistics if the model fits our data. R package abc (Csilléry et al. 2012) was used to implement the ABC method.
Test for Natural Selection for Phenotypic Divergence between Species
We performed the QST–FST comparison to test whether natural selection or genetic drift was the main cause of population differentiation in the phenotypic traits (McKay and Latta 2002; Leinonen et al. 2013). We first used the sequences of the neutral genes to measure genetic differentiation between species or between paired populations by FST, implemented in DNASP v5.1. Then, we estimated QST, an analogue of FST, to measure the amount of variance among populations relative to the total variance in the quantitative traits (Leinonen et al. 2013). Specifically, we followed the method proposed by Spitze (1993) to calculate QST according to QST=VB/(VB+2VW) for each trait, where VB and VWare the components of variance between and within species, respectively. The QST values were obtained by 1,000 bootstrap iterations with each involving resampling across individuals within populations or species using nested ANOVA. The distribution of QST was used to evaluate the significance of difference between QST and FST. If the QST value significantly exceeded FST, the hypothesis of natural selection driving species divergence was supported; in contrast, the neutral genetic differentiation between species was assumed if the QST values did not differ significantly from FST (McKay and Latta 2002; Whitlock and Guillaume 2009; Leinonen et al. 2013).
Crossing Experiment and Analysis of Postmating Isolation
To examine the strength of postmating isolation, we conducted artificial crossing, including crosses within populations, between populations within species, and between species in 2016 and 2017 in the Lingshui Station (N18°30.6′, E110°2.4′) in Hainan Province, China (fig. 1). For this purpose, we choose to grow the seeds from the three pairs of O. rufipogon and O. nivara populations (i.e., NEP1, LAO2, and KHM) that were used in the common garden experiment, with each population consisting of five individuals (for a total of 30 plants). A total of 238 crosses were performed, including 44 intrapopulation, 58 interpopulation within species, and 136 interspecies crosses. Selfing of the panicles with (28 crosses) and without (40) castration were also conducted (supplementary fig. S13, Supplementary Material online).
Seeds were exposed to 50°C for five days to break dormancy and then soaked in culture dishes after the coats were peeled. Seedlings were transplanted into soil cultures until the trefoil stage and then were planted in buckets (32 cm in diameter and 30 cm in height) filled with field soil. Since O. nivara started flowering >30 days earlier than O. rufipogon in Hainan Province, we grew three lots of O. nivara seeds to ensure the flowering times of the two species matched. We emasculated the mother panicles between ∼7:30 and 10:00 AM, enclosed them in paper bags, and then pollinated emasculated panicles when the spikelets of paternal donors were producing pollens.
We examined the extent of postmating isolation by determining the seed set of crosses and the viability of F1 hybrids following the methods of Sobel and Streisfeld (2015). Seed set was calculated by counting the number of seeds per spikelet, and F1 viability was measured by the survival rate of the F1 hybrids of 73 combinations (399 seeds) representing various crossing types. Seeds that germinated were transplanted into soil culture material in the same way mentioned earlier. Seedlings were transplanted into the garden at the tillering stage. We defined the survival rate of F1 hybrids as the percentage of seeds that germinated and grew to the flowering stage with more than half of tillers heading. We calculated average seed set and F1 viability at the population and species levels and performed the t-test for significance by R language v3.1.2 (https://www.R-project.org/).
Supplementary Material
Acknowledgments
We thank Tao Sang, Bao-Rong Lu, Yong-Qing Zhu, and Disna Ratnasekera for their help in field collections and phenotyping. We also thank the International Rice Research Institute (Los Banos, Philippines) for providing seed samples. This work was supported by the National Natural Science Foundation of China (91731301; 91231201; 31800186), the grants from the Chinese Academy of Sciences (XDB31000000; XDA08020103; CAS/SAFEA International Partnership Program for Creative Research Teams), and China Postdoctoral Science Foundation (2017M620950).
Author Contributions
S.G. designed the study. Z.C., L.Z., N.-N.R, and X.X. performed the research. Z.C. and S.G. analyzed the Pool-Seq data and conducted various simulations. L.Z., N.-N.R., R.L., Y.-S.D., M.-X.W., C.-B.C., H.-Z.Z., and Y.-T.L. performed the common garden experiments in Guangxi. N.-N.R., L.Z., L.H., C.-Y.J., J.G., and F.-M.Z. conducted the analyses of phenotypic and sequencing data. X.X., Q.-L.M., and M.-F.G. conducted the artificial crossing in Hainan. X.-M.Z., H.-F.Z., W.-L.C., R.L., L.Z., X.-H.Z., Y.-L.G., and X.-H.W. joined the field survey, sample collections, and lab assistances. S.G., and Z.C. wrote the manuscript.
References
- Abbott RJ, Comes HP.. 2007. Blowin’ in the wind – the transition from ecotype to species. New Phytol. 175(2):197–200. [DOI] [PubMed] [Google Scholar]
- Ai B, Wang ZS, Ge S.. 2012. Genome size is not correlated with effective population size in the Oryza species. Evolution 66(10):3302–3310. [DOI] [PubMed] [Google Scholar]
- Banaticla-Hilario MCN, Sosef MSM, McNally KL, Hamilton NRS, van den Berg RG.. 2013. Ecogeographic variation in the morphology of two Asian wild rice species, Oryza nivara and Oryza rufipogon. Intern J Plant Sci. 174(6):896–909. [Google Scholar]
- Barbier P. 1989. Genetic variation and ecotypic differentiation in the wild rice species Oryza rufipogon. I. Population differentiation in life-history traits and isozymic loci. Jpn J Genet. 64(4):259–271. [Google Scholar]
- Beaumont MA. 2010. Approximate Bayesian computation in evolution and ecology. Annu Rev Ecol Evol Syst. 41(1):379–406. [Google Scholar]
- Beaumont MA, Balding DJ.. 2004. Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol. 13(4):969–980. [DOI] [PubMed] [Google Scholar]
- Bioversity International, IRRI, WARDA. 2007. Descriptors for wild and cultivated rice (Oryza spp.): Bioversity International, Rome, Italy; International Rice Research Institute, Los Baños, Philippines; WARDA, Africa Rice Center, Cotonou, Benin.
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butlin RK, Saura M, Charrier G, Jackson B, André C, Caballero A, Coyne JA, Galindo J, Grahame JW, Hollander J, et al. 2014. Parallel evolution of local adaptation and reproductive isolation in the face of gene flow. Evolution 68(4):935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D, Kingsley DM.. 2005. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307(5717):1928–1933. [DOI] [PubMed] [Google Scholar]
- Comes HP, Coleman M, Abbott RJ.. 2017. Recurrent origin of peripheral, coastal (sub)species in Mediterranean Senecio (Asteraceae). Plant Ecol Divers. 10(4):253–271. [Google Scholar]
- Csilléry K, François O, Blum MGB.. 2012. abc: an R package for approximate Bayesian computation (ABC). Methods Ecol Evol. 3(3):475–479. [Google Scholar]
- Elmer KR, Fan S, Kusche H, Luise Spreitzer M, Kautt AF, Franchini P, Meyer A.. 2014. Parallel evolution of Nicaraguan crater lake cichlid fishes via non-parallel routes. Nat Commun. 5:5168.. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J.. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 14(8):2611–2620. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M.. 2013. Robust demographic inference from genomic and SNP data. PLoS Genet. 9(10):e1003905.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S.. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinformatics Online. 1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Faria R, Renaut S, Galindo J, Pinho C, Melo-Ferreira J, Melo M, Jones F, Salzburger W, Schluter D, Butlin R.. 2014. Advances in ecological speciation: an integrative approach. Mol Ecol. 23(3):513–521. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1989. PHYLIP – Phylogeny Inference Package (Version 3.2). Cladistics 5:164–166. [Google Scholar]
- Ferris KG, Barnett LL, Blackman BK, Willis JH.. 2017. The genetic architecture of local adaptation and reproductive isolation in sympatry within the Mimulus guttatus species complex. Mol Ecol. 26(1):208–224. [DOI] [PubMed] [Google Scholar]
- Fracassetti M, Griffin PC, Willi Y.. 2015. Validation of pooled whole-genome re-sequencing in Arabidopsis lyrata. PLoS One 10(10):e0140462.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks SJ. 2015. The unique and multifaceted importance of the timing of flowering. Am J Bot. 102(9):1401–1402. [DOI] [PubMed] [Google Scholar]
- Fu YX, Li WH.. 1993. Statistical tests of neutrality of mutations. Genetics 133(3):693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futschik A, Schlötterer C.. 2010. The Next generation of molecular markers from massively parallel sequencing of pooled DNA samples. Genetics 186(1):207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo MA, Li C, Fowlkes AM, Briggeman TM, Zhou A, Schemske DW, Sang T.. 2009. Genetic architecture for the adaptive origin of annual wild rice, Oryza nivara. Evolution 63(4):870–883. [DOI] [PubMed] [Google Scholar]
- Guo J, Liu R, Huang L, Zheng X-M, Liu P-L, Du Y-S, Cai Z, Zhou L, Wei X-H, Zhang F-M, Ge S.. 2016. Widespread and adaptive alterations in genome-wide gene expression associated with ecological divergence of two Oryza species. Mol Biol Evol. 33(1):62–78. [DOI] [PubMed] [Google Scholar]
- Hall MC, Willis JH.. 2006. Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution 60(12):2466–2477. [PubMed] [Google Scholar]
- Hedrick PW. 2005. A standardized genetic differentiation measure. Evolution 59(8):1633–1638. [PubMed] [Google Scholar]
- Huang L, Du YS, Zheng XM, Liu R, Zhou HF, Ge S.. 2013. Nucleotide diversity of 11S seed storage protein gene and its implications for ecological adaptation of Oryza nivara. J Syst Evol. 51(6):641–651. [Google Scholar]
- Huang X, Kurata N, Wei X, Wang Z, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, et al. 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature 490(7421):497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP.. 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 132(2):583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesson K. 2001. Parallel speciation: a key to sympatric divergence. Trends Ecol Evol. 16(3):148–153. [DOI] [PubMed] [Google Scholar]
- Jost L. 2008. GST and its relatives do not measure differentiation. Mol Ecol. 17(18):4015–4026. [DOI] [PubMed] [Google Scholar]
- Juenger TE. 2013. Natural variation and genetic constraints on drought tolerance. Curr Opin Plant Biol. 16(3):274–281. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK.. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Gen Res. 22(3):568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Orozco-terWengel P, De Maio N, Pandey RV, Nolte V, Futschik A, Kosiol C, Schlötterer C.. 2011. PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One 6(1):e15925.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Pandey RV, Schlötterer C.. 2011. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27(24):3435–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooyers NJ. 2015. The evolution of drought escape and avoidance in natural herbaceous populations. Plant Sci. 234:155–162. [DOI] [PubMed] [Google Scholar]
- Kuroda Y, Rao SA, Bounphanousay C, Kanyavong K, Iwata A, Tanaka K, Sata Y.. 2006. Diversity of wild and weedy rice in Laos. In: Schiller JM, editor. Rice in Laos, Los Baños, Philippines, IRRI. p. 215–234. [Google Scholar]
- Kuroda Y, Sato Y-I, Bounphanousay C, Kono Y, Tanaka K.. 2007. Genetic structure of three Oryza AA genome species (O. rufipogon, O. nivara and O. sativa) as assessed by SSR analysis on the Vientiane Plain of Laos. Conserv Genet. 8(1):149–158. [Google Scholar]
- Leinonen T, McCairns RJS, O’Hara RB, Merila J.. 2013. QST-FST comparisons: evolutionary and ecological insights from genomic heterogeneity. Nat Rev Genet. 14(3):179–190. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup GPDP.. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J.. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- Liu R, Zheng XM, Zhou L, Zhou HF, Ge S.. 2015. Population genetic structure of Oryza rufipogon and Oryza nivara: implications for the origin of O. nivara. Mol Ecol. 24(20):5211–5228. [DOI] [PubMed] [Google Scholar]
- Lobo JA, Quesada M, Stoner KE, Fuchs EJ, Herrerias-Diego Y, Rojas J, Saborio G.. 2003. Factors affecting phenological patterns of bombacaceous trees in seasonal forests in Costa Rica and Mexico. Am J Bot. 90(7):1054–1063. [DOI] [PubMed] [Google Scholar]
- Lowry DB, Modliszewski JL, Wright KM, Wu CA, Willis JH.. 2008. The strength and genetic basis of reproductive isolating barriers in flowering plants. Philos Trans R Soc B. 363(1506):3009–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu BR. 1998. A report on NARC-IRRI cooperative collection of wild Oryza species in Nepal. Los Banos, Laguna: Germplasm Specialist Genetic Resources Center, IRRI.
- Lu BR, Naredo EB, Amita BJ, Jackson MT.. 2000. Preliminary studies on taxonomy and biosystematics of the AA genome Oryza species (Poaceae). In: Jacobs SWL, Everett J, editors. Grasses, systematics and evolution. Collingwood: CSIRO Publishing. [Google Scholar]
- Lynch M, Bost D, Wilson S, Maruki T, Harrison S.. 2014. Population-genetic inference from pooled-sequencing data. Gen Biol Evol. 6(5):1210–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay JK, Latta RG.. 2002. Adaptive population divergence: markers, QTL and traits. Trends Ecol Evol. 17(6):285–291. [Google Scholar]
- Meier JI, Sousa VC, Marques DA, Selz OM, Wagner CE, Excoffier L, Seehausen O.. 2017. Demographic modelling with whole-genome data reveals parallel origin of similar Pundamilia cichlid species after hybridization. Mol Ecol. 26(1):123–141. [DOI] [PubMed] [Google Scholar]
- Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T.. 2014. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30(17):i541–i548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima H, Sano Y, Oka HI.. 1992. Evolutionary studies in cultivated rice and its wild relatives. Oxford Sur Evol Biol. 8:135–184. [Google Scholar]
- Moyers BT, Rieseberg LH.. 2016. Remarkable life history polymorphism may be evolving under divergent selection in the silverleaf sunflower. Mol Ecol. 25(16):3817–3830. [DOI] [PubMed] [Google Scholar]
- Nei M, Chesser RK.. 1983. Estimation of fixation indices and gene diversities. Ann Hum Genet. 47(3):253–259. [DOI] [PubMed] [Google Scholar]
- Nei M, Li WH.. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 76(10):5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P. 2012. Ecological speciation. Oxford: OUP. [Google Scholar]
- Nosil P, Crespi BJ, Sandoval CP.. 2002. Host-plant adaptation drives the parallel evolution of reproductive isolation. Nature 417(6887):440–443. [DOI] [PubMed] [Google Scholar]
- Ostevik KL, Moyers BT, Owens GL, Rieseberg LH.. 2012. Parallel ecological speciation in plants? Intern J Ecol. 2012:1. [Google Scholar]
- Perrier X, Jacquemoud-Collet JP.. 2006. DARwin software. http://darwin.cirad.fr
- Pianka ER. 1973. Niche overlap and diffuse competition. Proc Natl Acad Sci U S A. 71:2141–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P.. 2000. Inference of population structure using multilocus genotype data. Genetics 155(2):945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada H, Posada D, Caballero A, Morán P, Rolán-Alvarez E.. 2007. Phylogenetic evidence for multiple sympatric ecological diversification in a marine snail. Evolution 61(7):1600–1612. [DOI] [PubMed] [Google Scholar]
- Ravinet M, Westram A, Johannesson K, Butlin R, André C, Panova M.. 2016. Shared and nonshared genomic divergence in parallel ecotypes of Littorina saxatilis at a local scale. Mol Ecol. 25(1):287–305. [DOI] [PubMed] [Google Scholar]
- Rellstab C, Zoller S, Tedder A, Gugerli F, Fischer MC.. 2013. Validation of SNP allele frequencies determined by pooled next-generation sequencing in natural populations of a non-model plant species. PLoS One 8(11):e80422.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TJ, Walter GM, McGuigan K, Ortiz-Barrientos D.. 2016. Divergent natural selection drives the evolution of reproductive isolation in an Australian wildflower. Evolution 70(9):1993–2003. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Willis JH.. 2007. Plant speciation. Science 317(5840):910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roda F, Ambrose L, Walter GM, Liu HL, Schaul A, Lowe A, Pelser PB, Prentis P, Rieseberg LH, Ortiz-Barrientos D.. 2013. Genomic evidence for the parallel evolution of coastal forms in the Senecio lautus complex. Mol Ecol. 22(11):2941–2952. [DOI] [PubMed] [Google Scholar]
- Roda F, Walter GM, Nipper R, Ortiz‐Barrientos D.. 2017. Genomic clustering of adaptive loci during parallel evolution of an Australian wildflower. Mol Ecol. 26(14):3687–3699. [DOI] [PubMed] [Google Scholar]
- Ru D, Mao K, Zhang L, Wang X, Lu Z, Sun Y.. 2016. Genomic evidence for polyphyletic origins and interlineage gene flow within complex taxa: a case study of Picea brachytyla in the Qinghai‐Tibet Plateau. Mol Ecol. 25(11):2373–2386. [DOI] [PubMed] [Google Scholar]
- Rundle HD, Nagel L, Boughman JW, Schluter D.. 2000. Natural selection and parallel speciation in sympatric sticklebacks. Science 287(5451):306–308. [DOI] [PubMed] [Google Scholar]
- Ryan PG, Bloomer P, Moloney CL, Grant TJ, Delport W.. 2007. Ecological speciation in South Atlantic island finches. Science 315(5817):1420–1423. [DOI] [PubMed] [Google Scholar]
- Sang T, Ge S.. 2007. Genetics and phylogenetics of rice domestication. Curr Opin Genet Dev. 17(6):533–538. [DOI] [PubMed] [Google Scholar]
- Schlötterer C, Tobler R, Kofler R, Nolte V.. 2014. Sequencing pools of individuals – mining genome-wide polymorphism data without big funding. Nat Rev Genet. 15(11):749–763. [DOI] [PubMed] [Google Scholar]
- Schluter D. 2009. Evidence for ecological speciation and its alternative. Science 323(5915):737–741. [DOI] [PubMed] [Google Scholar]
- Schluter D, Nagel LM.. 1995. Parallel speciation by natural selection. Am Nat. 146(2):292–301. [Google Scholar]
- Servedio MR, Doorn GSV, Kopp M, Frame AM, Nosil P. 2011. Magic traits in speciation: ‘magic’ but not rare? Trends Ecol Evol. 26:389–397. [DOI] [PubMed] [Google Scholar]
- Sharma S, Shastry S.. 1965. Taxonomic studies in genus Oryza. III. O. rufipogon Griff. sensu stricto and O. nivara Sharma et Shastry nom nov. Indian J Genet Plant Breed. 25:157–167. [Google Scholar]
- Silva-Junior OB, Grattapaglia D.. 2015. Genome-wide patterns of recombination, linkage disequilibrium and nucleotide diversity from pooled resequencing and single nucleotide polymorphism genotyping unlock the evolutionary history of Eucalyptus grandis. New Phytol. 208(3):830–845. [DOI] [PubMed] [Google Scholar]
- Sobel JM, Streisfeld MA.. 2015. Strong premating reproductive isolation drives incipient speciation in Mimulus aurantiacus. Evolution 69(2):447–461. [DOI] [PubMed] [Google Scholar]
- Soria-Carrasco V, Gompert Z, Comeault AA, Farkas TE, Parchman TL, Johnston JS, Buerkle CA, Feder JL, Bast J, Schwander T, et al. 2014. Stick insect genomes reveal natural selection’s role in parallel speciation. Science 344(6185):738–742. [DOI] [PubMed] [Google Scholar]
- Spitze K. 1993. Population structure in Daphnia obtusa: quantitative genetic and allozymic variation. Genetics 135(2):367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3):585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S.. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucchi E, Frajman B, Haverkamp THA, Schönswetter P, Paun O.. 2017. Genomic analyses suggest parallel ecological divergence in Heliosperma pusillum (Caryophyllaceae). New Phytol. 216(1):267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan DA, Lu B-R, Tomooka N.. 2008. The evolving story of rice evolution. Plant Sci. 174(4):394–408. [Google Scholar]
- Wegmann D, Leuenberger C, Neuenschwander S, Excoffier L.. 2010. ABCtoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinformatics 11:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC, Guillaume F.. 2009. Testing for Spatially Divergent selection: comparing QST to FST. Genetics 183(3):1055–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XM, Ge S.. 2010. Ecological divergence in the presence of gene flow in two closely related Oryza species (Oryza rufipogon and O. nivara). Mol Ecol. 19(12):2439–2454. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Zheng X, Luo J, Gaut BS, Ge S.. 2007. Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: severe bottleneck during domestication of rice. Mol Biol Evol. 24(3):875–888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.