Abstract
Doppel (Dpl) is a paralog of the mammalian prion protein (PrP); it is abundant in testes but expressed at low levels in the adult central nervous system. In two Prnp-deficient (Prnp0/0) mouse lines (Ngsk and Rcm0), Dpl overexpression correlated with ataxia and death of cerebellar neurons. To determine whether Dpl overexpression, rather than the dysregulation of genes neighboring the Prn gene complex, was responsible for the ataxic syndrome, we placed the mouse Dpl coding sequence under the control of the Prnp promoter and produced transgenic (Tg) mice on the Prnp0/0-ZrchI background (hereafter referred to as ZrchI). ZrchI mice exhibit neither Dpl overexpression nor cerebellar degeneration. In contrast, Tg(Dpl)ZrchI mice showed cerebellar granule and Purkinje cell loss; the age of onset of ataxia was inversely proportional to the levels of Dpl protein. Crosses of Tg mice overexpressing wild-type PrP with two lines of Tg(Dpl)ZrchI mice resulted in a phenotypic rescue of the ataxic syndrome, while Dpl overexpression was unchanged. Restoration of PrP expression also rendered the Tg(Dpl) mice susceptible to prion infection, with incubation times indistinguishable from non-Tg controls. Whereas the rescue of Dpl-induced neurotoxicity by coexpression of PrP argues for an interaction between the PrP and Dpl proteins in vivo, the unaltered incubation times in Tg mice overexpressing Dpl in the central nervous system suggest that Dpl is unlikely to be involved in prion formation.
Prion diseases are transmissible neurodegenerative diseases that include scrapie, bovine spongiform encephalopathy (BSE), and Creutzfeldt-Jakob disease (CJD). During prion infection, the normal, cellular form of the prion protein (PrPC), a conserved host-encoded glycoprotein, undergoes a posttranslational conversion to a disease-specific, protease-resistant and detergent-insoluble isoform, PrPSc, which accumulates within the central nervous system (CNS; refs. 1 and 2). The function of PrPC remains unknown, and initial studies with PrP-deficient (Prnp0/0) mice failed to reveal any abnormalities (3, 4). Subsequently, late-onset ataxia associated with loss of cerebellar Purkinje cells was described in two independently generated Prnp0/0 lines, Ngsk (5) and Rcm0 (6, 7).
These seemingly disparate observations were interpreted as evidence that PrP might have a role in the long-term survival of Purkinje neurons (8) but were later resolved with the discovery of Prnd, a PrP-like gene situated 16 Kb downstream of the Prnp gene (7) that codes for doppel (Dpl). The levels of Dpl mRNA are very low in the CNS of normal adult mice and ZrchI mice, in contrast to elevated levels in the Ngsk and Rcm0 lines. These findings suggested that an increase in Dpl mRNA levels was an unexpected consequence of the targeted inactivation of the Prnp locus, and that the loss of Purkinje neurons and ataxia in Ngsk and Rcm0 mice were caused by Dpl overexpression.
Because we were unable to exclude dysregulation of other neighboring genes contributing to the ataxic phenotype in the Ngsk and Rcm0 Prnp0/0 lines (9, 10), we introduced a Dpl transgene into the ZrchI mice. The transgenic (Tg)(Dpl)ZrchI mice overexpressed Dpl at levels of one to eight times those found in wild-type (wt) mice and developed severe neurodegeneration and ataxia with an age of onset inversely proportional to the level of Dpl expression, arguing that the Dpl protein is highly neurotoxic. Because the phenotype of Ngsk mice could be rescued by coexpression of wt PrP (11), we introduced a wt PrP transgene into the Tg(Dpl)ZrchI animals. Coexpression of PrP resulted in the phenotypic rescue of mice expressing high levels of Dpl in the CNS. These observations argue that interactions between the PrPC and Dpl proteins can occur under certain circumstances.
Materials and Methods
Generation of Tg Mice Overexpressing Dpl.
The murine Dpl ORF was excised from pcDNA3-Dpl (7) by digestion with XbaI and EcoRI and blunted before ligation into the EcoRI site of pEGFP-CI (CLONTECH) to provide XhoI and SalI sites. The Dpl ORF was excised with XhoI and SalI and ligated into SalI-digested CosSHa.tet hamster Prnp expression vector (12–14). This CosSHa.Dpl construct was purified from vector sequences and microinjected into FVB/N ZrchI zygotes. Tg mice were detected by hybridization of tail DNA samples with the hamster PrP 3′ untranslated region probe, as described (14). Tg lines were established by breeding viable Tg(Dpl) founders to FVB/N.Prnp0/0N10F1 mice.
Clinical Assessment.
All mice were assessed three times a week for neurological deficits such as abnormal gait, circling behavior, hyperactivity, and general signs of illness (15).
Neuropathology.
Animals were killed, their brains were removed and either frozen at −80°C or immersion-fixed in 10% (vol/vol) buffered formalin. Formalin-treated samples were paraffin-embedded, sectioned at 10 μm, and processed for hematoxylin/eosin (H&E) staining, glial fibrillary acidic protein (GFAP) immunostaining and in situ end labeling (ISEL). GFAP immunostaining was performed by using anti-bovine-GFAP rabbit antiserum (DAKO), as described (16). Histoblots were performed as described (17), and Dpl expression was detected with the anti-Dpl rabbit polyclonal serum E6977. For ISEL, brains were formalin-fixed for a period that did not exceed 1 week (18). ISEL was performed as described (19). The DNA-strand breaks were identified by using the terminal deoxynucleotidyl transferase with fast red as the substrate.
Dpl Polyclonal Antisera Production.
A bacterial construct for the expression of MoDpl 26–155 in the expression vector pET11d (Novagen) was generated by standard procedures. Subsequent to transformation into E. coli BL21(DE3) cells, fermented cultures were processed in a microfluidizer, recombinant (rec) Dpl was purified on a Vydac C4-HPLC column, and peak fractions were lyophilized. Rabbits (Western Oregon Rabbit Company, Philomath, OR) were immunized and boosted with 200 μg of SDS-denatured murine recDpl RIBI suspension in PBS. Bleeds were assessed by SDS/PAGE and compared with testes and brains from wt FVB/N animals, as well as to brains from Rcm0 animals.
Detection of Dpl and PrP.
Detection of Dpl and PrP in 50-μg crude tissue homogenates (unless otherwise noted) was performed according to standard procedures. We used anti-Dpl(26–155) polyclonal antiserum E6977 (1:1,000) and horseradish peroxidase (HRP)-conjugated donkey anti-rabbit IgG (Amersham Pharmacia; 1:5,000) for Dpl detection, the 3C5 monoclonal antibody (Chemicon; 1:5,000) with a secondary anti-mouse IgG (1:5,000) for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) detection. We used mAb D13 (1 μg/ml) and HRP-conjugated goat anti-human IgG F(ab′)2 fragment (ICN; 1:5,000) for PrP detection.
Results
Dpl Expression in wt and PrP-Deficient Mice.
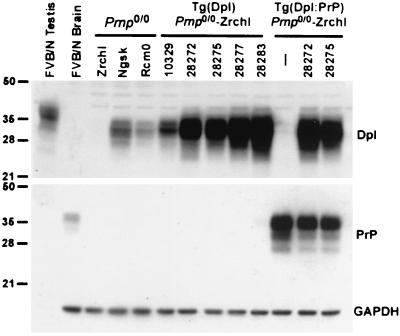
By using an anti-mouse Dpl polyclonal antiserum raised against recDpl 26–155, strong staining was seen in the testis (Fig. 1), but expression was not detectable in other tissues of wt mice (data not shown). We showed previously that Dpl mRNA levels were increased in the brains of the Ngsk and Rcm0 Prnp0/0 mice, which developed ataxia and Purkinje cell loss (7). These brains stained intensely with the Dpl antiserum, whereas Dpl was undetectable in the brains of wt and Prnp0/0-ZrchI mice (Fig. 1).
Figure 1.
Dpl and PrP expression in mouse brain and testis. (Top) Dpl expression in wt and Tg mice. (Middle) A PrP reprobe of the membrane shown in Top. (Bottom) GAPDH reprobe showing similar loadings in all lanes. All samples are 50-μg protein loadings from crude tissue homogenates. Lane 1 is testis and all other lanes are brain. Lane 1, wt FVB/N; lane 2, wt FVB/N; lane 3, ZrchI; lane 4, Ngsk; lane 5, Rcm0. Lanes 6 to 10 are from five independent Tg(Dpl)ZrchI lines: 10329, 28272, 28275, 28277, 28283, respectively. Lanes 11 to 13 are from mice expressing both Dpl and PrP. Lane 11, Tg(SHaPrP)7 hemizygote; lane 12, mice double-hemizygous for Tg(Dpl)28272/ZrchI and Tg(SHaPrP)7 transgenes; lane 13, mice double-hemizygous for the Tg(Dpl)28275/ZrchI and Tg(SHaPrP)7 transgenes.
Ataxia in Tg Mice Expressing Dpl from the PrP Promoter.
A Dpl transgene was generated by introducing the murine Dpl ORF into the Syrian hamster (SHa) PrP cosmid transgene, CosSHa.tet (20), resulting in Dpl expression under the control of the PrP promoter. The transgene was microinjected into Prnp0/0-ZrchI zygotes (3), which do not overexpress Dpl in the CNS. An extensive microinjection series yielded seven founders (Table 1), two of which developed ataxia and died at 21 and 40 days. Five Tg(Dpl) founders survived and were bred with Prnp0/0-ZrchI mice to establish Tg lines, designated Tg(Dpl)ZrchI. F1 mice from four founders developed ataxia between 21 to 84 days of age. F1 mice from founder Tg(Dpl)10329/ZrchI developed ataxia at ≈375 days of age (Table 1). All F1 pups from the five Tg(Dpl)ZrchI lines developed ataxia, indicating a highly penetrant phenotype. None of the nontransgenic ZrchI littermates developed ataxia.
Table 1.
Dpl expression provokes an early-onset ataxia in PrP-deficient mice
| Mouse line | Dpl expression level* | Age of onset, days (± SEM) | n/n0† | Phenotype | Reference |
|---|---|---|---|---|---|
| Rcm0 | 0.5× | 611 ± 12 | 34/34 | Ataxia, kyphosis, tremor, weight loss | This paper |
| Ngsk | 1× | ∼500 | N/D | Ataxia, kyphosis | 5 |
| ZrchI | 0× | >600 | |||
| Tg(Dpl)10326/ZrchI‡ | ND | 21 | 1/1 | Ataxia, runted | This paper |
| Tg(Dpl)10328/ZrchI‡ | ND | 40 | 1/1 | Ataxia, runted | This paper |
| Tg(Dpl)10329/ZrchI | 1× | 375 ± 8 | 7/9 | Ataxia | This paper |
| Tg(Dpl)28272/ZrchI | 4× | 32 ± 1 | 4/4 | Ataxia, runted | This paper |
| Tg(Dpl)28275/ZrchI | 3× | 44 ± 4 | 7/7 | Ataxia, runted | This paper |
| Tg(Dpl)28277/ZrchI | ∼4× | 32 ± 32 | 4/4 | Ataxia | This paper |
| Tg(Dpl)28283/ZrchI | ∼8× | 27 ± 0 | 2/2 | Ataxia, runted | This paper |
Relative to levels in FVB/N testis; ND, not determined.
n, number of sick mice; n0, number of inoculated mice.
Founder.
Tg(Dpl)ZrchI lines with high Dpl-expression levels in the CNS had an early onset of ataxia (Fig. 1). Tg(Dpl)28283/ZrchI had the highest expression level (about eight times that of wt testis) and showed disease at 27 days of age (n = 2), whereas Tg(Dpl)28272/ZrchI (n = 4) and Tg(Dpl)28275/ZrchI (n = 7) mice had an approximately three to four times Dpl expression and exhibited ataxia between 32 to 57 days of age (Table 1). Tg(Dpl)10329/ZrchI mice (one times that of testis) developed ataxia at 375 days of age.
Neuroanatomy of Dpl Expression in Tg Mice.
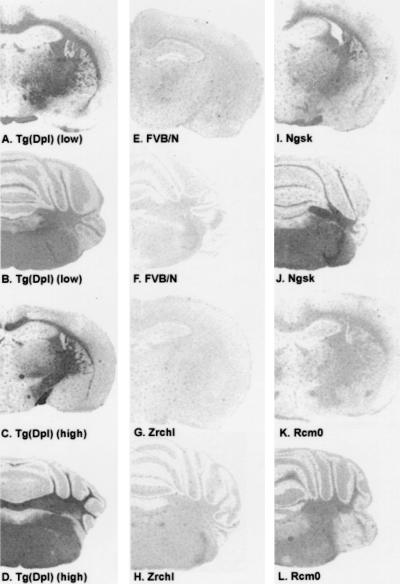
Dpl immunostaining was localized almost exclusively in white matter tracts including the corpus callosum, internal capsule, optic chiasm, optic tract, and cerebellar white matter of Tg(Dpl)ZrchI mice. In the brainstem (Fig. 2 B and D), Dpl was localized to white matter tracts. Relative expression levels were similar to those observed by Western blot analysis (Fig. 1). The low levels of staining in the histoblots of FVB/N mice (Fig. 2 E and F) may represent nonspecific background, given the virtual absence of Dpl in the Western analysis of wt FVB/N mice (Fig. 1; ref. 7), although low levels of basal expression cannot be completely excluded. Elevated Dpl expression correlated with Purkinje cell loss in both Tg(Dpl)ZrchI and various Prnp0/0 mice (Table 1; Fig. 2).
Figure 2.
Neuroanatomical distribution of Dpl expression. Histoblots of paired coronal brain sections stained for Dpl. Each pair includes a section at the level of the dorsal hippocampus/thalamus (A, C, E, G, I, K) and a section at the level of the cerebellum/pons (B, D, F, H, J, L) of Tg(Dpl)ZrchI (A–D), wt FVB/N (E, F), and PrP-deficient mice (G–L). (A and B) Low-expressor Tg(Dpl)10329/ZrchI. (C and D) High-expressor Tg(Dpl)28277/ZrchI. (E and F) Wt FVB/N. (G and H) ZrchI. (I and J) Ngsk. (K and L) Rcm0.
Neuropathology in Tg(Dpl)ZrchI and PrP-Deficient Mice.
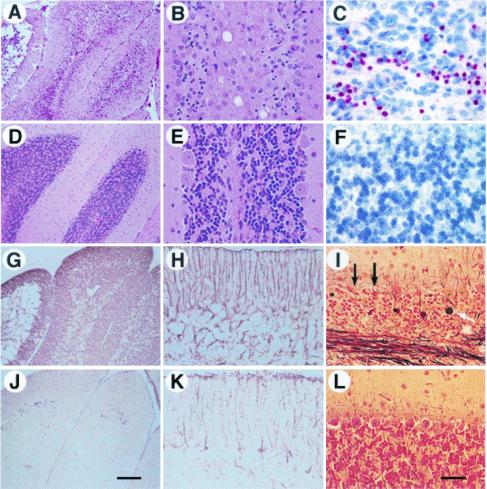
Neurohistological examination of two Tg(Dpl)ZrchI founders and nine ataxic F1 pups from four other founders revealed severe neurodegeneration restricted to the cerebellar cortex and hippocampus (Fig. 3 A–C and G–I). Loss of granule cells in both the lateral cerebellar hemispheres and the vermis (Fig. 3 A and B) correlated with Purkinje cell loss. These observations raise the possibility that Purkinje cell loss is determined in part by the degeneration of granule cells. Also, there was moderate vacuolation of the white-matter tracts and of the deep cerebellar nuclei with focal macrophage infiltration (Fig. 3B). The nuclei of many granule cells stained for fragmented DNA by terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL), characteristic of apoptosis (Fig. 3C). In the hippocampus of Tg(Dpl)ZrchI high-expressor lines, most pyramidal cells of the CA1 and CA2 regions showed degenerative changes, including bright pink cytoplasms indicative of coagulation necrosis. In all Tg(Dpl)ZrchI lines, GFAP immunohistochemistry revealed that reactive astrocytic gliosis was severe in the cerebellar granule cell layer and was associated with intense Bergmann radial gliosis in the molecular layer (Fig. 3 G and H). Astrocytic gliosis also was widespread in other regions of the brain, such as the neocortex, entorhinal cortex, and hippocampus, but to a lesser degree (data not shown).
Figure 3.
Neuropathology in PrP-deficient mice overexpressing Dpl in the CNS. Neurodegenerative changes were most intense in the cerebellum of Tg(Dpl)ZrchI mice overexpressing high levels of Dpl and were not found in ZrchI mice. (A–C and G–I) Ataxic Tg(Dpl)28272/ZrchI mouse. (D–F and J–L) Age-matched ZrchI littermate control. All panels show coronal sections from the lateral cerebellar cortex of 32-day-old mice. H&E staining shows severe granule cell loss in a Tg(Dpl)ZrchI mouse (A and B, low- and high-power views, respectively), compared with similar regions from age-matched controls (D and E). (B) Extensive Purkinje and granule cell loss and moderate vacuolation are apparent in the white matter, compared with E, which shows a normal number of granule cells and large Purkinje cells. (C) A high-power view of the TUNEL-stained vermis of a Tg(Dpl)ZrchI mouse shows a cluster of granule cells undergoing apoptosis in red (hemalun of Mayer counterstain), which contrasts with F, a control section with no evidence of TUNEL-positive granule cells. GFAP staining reveals intense gliosis in several cerebellar lobules (G, low-power view) and intense Bergmann radial gliosis in the molecular layer and gliosis within the granule cell layer (H, high-power view), compared with a normal control (J and K, low- and high-power views, respectively). (I) Bielschowsky silver stain of the vermis from the mouse depicted in A–C shows cerebellar torpedoes (black spheroidal masses distributed horizontally across the center of the panel, white arrow). The black arrows point to sites where Purkinje cells once resided (empty baskets). (L) The cerebellar vermis in a ZrchI mouse shows no cerebellar torpedoes, Purkinje or granule cell loss. (A, D, G, and J) Bar = 100 μm. (B, C, E, F, H, I, K, and L) Bar = 50 μm.
Phenotypic Rescue of Dpl-Induced Ataxia by Coexpression of PrP.
We crossed two Tg(Dpl)ZrchI founders (28272 and 28275) expressing high Dpl levels with Tg(SHaPrP+/+)7/Prnp0/0-ZrchI mice, which are homozygous for the SHaPrP transgene (21). Six mice harboring both transgenes on the Prnp0/0 background were recovered from each Tg(Dpl) line and designated Tg(Dpl:PrP)ZrchI. In all cases, expression of PrP prevented disease in Tg(Dpl:PrP)ZrchI mice (Table 2). Mice from the two high-expressor lines survived in excess of 375 days, which is considerably beyond the age of onset of ataxia in their respective parental Tg(Dpl) lines.
Table 2.
Expression of Syrian hamster PrP rescues Dpl-induced ataxia in Tg mice
| Mouse line* | Dpl expression level† | Age of onset, days | n/n0‡ | Phenotype |
|---|---|---|---|---|
| Tg(Dpl:PrP)28272 | 4× | >400§ | 0/6§ | Normal |
| Tg(Dpl:PrP)28275 | 3× | >375¶ | 0/6¶ | Normal |
| Tg(PrP)‖ | 0× | >400 | 0/9 | Normal |
All lines are ZrchI homozygotes and Tg(SHaPrP)7 hemizygotes.
Relative to levels in FVB/N testis.
n, number of sick mice; n0, number of inoculated mice.
Four mice were killed for analysis at 180 days of age, two others are alive and healthy at 411 and 413 days of age.
Four mice were killed for analysis at 180 days of age, two others are alive and healthy at 377 days of age.
Non-Tg(Dpl), age-matched control littermates from Tg(Dpl) × Tg(SHaPrP)7 matings.
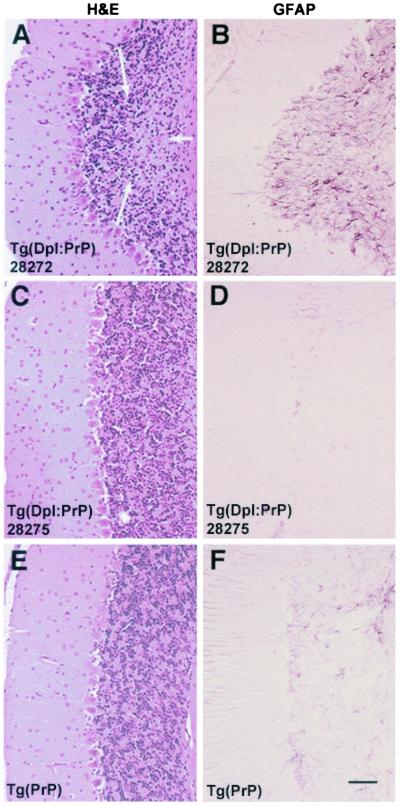
No significant pathological abnormalities were apparent in brains from Tg(Dpl:PrP)28275/ZrchI mice killed at 150 days of age (Fig. 4 C and D); however, limited neurodegeneration was seen in the cerebellar cortex of Tg(Dpl:PrP)28272/ZrchI mice (Fig. 4 A and B). Granule cell loss was associated with reactive astrocytic gliosis of the granule cell layer but not with Bergmann radial gliosis (Fig. 4B). The differences in the ability of PrP to prevent histopathological changes in these two Tg(Dpl:PrP) lines might be related to Dpl-expression levels, as Tg(Dpl)28272 mice express Dpl at slightly higher levels than Tg(Dpl)28275 mice.
Figure 4.
Rescue of Dpl ataxia by PrP coexpression. PrP provides a partial rescue of the cerebellar degeneration in Tg(Dpl)ZrchI high-expressor mice and total rescue in moderate expressors. All panels show coronal cerebellar sections from 170-day-old mice. (A and B) Nonataxic ZrchI mouse hemizygous for both Tg(Dpl)28272 (high expressor) and the Tg(SHaPrP)7 transgenes. (C and D) Nonataxic ZrchI mouse hemizygous for both Tg(Dpl)28275 (moderate expressor) and Tg(SHaPrP)7 transgenes. (E and F) Age-matched, nonataxic ZrchI mouse hemizygous for the Tg(SHaPrP)7 transgene only. (A) H&E staining shows intact Purkinje cell layer with a small area of focal granule cell loss (arrows). The focal granule cell loss is associated with (B) reactive astrocytic gliosis in the granule cell layer (GFAP stain). H&E stain of moderate expressors rescued by the PrP transgene shows an absence of neurodegeneration (C) and reactive gliosis (D). No neuropathological changes were apparent in a control brain except for a mild increase in GFAP immunoreactivity in astrocytes (E) and Bergmann radial glia (F), which may be age-related. The bar in F represents 100 μm and applies to all panels.
Dpl- and PrP-expression levels in the CNS were examined by immunoblotting with Dpl and PrP antisera (Fig. 1). No effects were observed on the expression levels of either protein when both transgenes were coexpressed. When histoblots were performed on the brains of Tg(Dpl:PrP)ZrchI mice to probe the neuroanatomical distribution of Dpl and PrP expression, no alteration in distribution was observed in mice from either line (data not shown).
Prion Challenge to Mice Expressing both Dpl and PrP.
To test the effect of Dpl overexpression on prion replication, Tg(Dpl)10329/Prnp0/0 were crossed to wt FVB/Prnpa/a mice to reintroduce two copies of the endogenous Prnp gene. This procedure generated Tg(Dpl)10329/Prnpa/a mice, which express wt levels of mouse PrP. Groups of Tg(Dpl)10329/Prnpa/a and FVB/Prnpa/a control mice were challenged intracerebrally with Rocky Mountain Laboratory (RML) prions. Tg(Dpl)10329/Prnpa/a mice had mean incubation times of 129 days (±1.2 SEM; n = 20), which were not significantly different from the 131-day incubation times (±4.0 SEM; n = 13) in wt FVB/Prnpa/a controls. No differences in the distribution of neuropathological lesions were seen when comparing FVB and Tg(Dpl)10329/Prnpa/a mice, suggesting that Dpl expression did not influence the accumulation of infectious RML prions.
Discussion
Genetic Modulation of Dpl-Induced Ataxia.
We found that Dpl-induced ataxia and Purkinje cell loss were modulated by three factors: (i) Dpl expression, (ii) PrPC expression, and (iii) the mouse strain background. The level of Dpl expression in the CNS was inversely proportional to the age of onset of ataxia in Tg(Dpl)ZrchI mice (Table 1). The distribution of Dpl in Tg(Dpl)ZrchI mice observed by histoblotting of brains was similar to that in Ngsk and Rcm0 mice (Fig. 2 I–L). The Dpl-expression level in Ngsk mice was higher than in the Rcm0 line (Fig. 1; ref. 7), which may reflect differences in the structure of the two knockout alleles. The Ngsk line retains a phosphoglycerate kinase (PGK) promoter in the same orientation as Dpl (as part of the PGK/Neo selectable marker within the targeted PrP exon 3) and may lead to higher levels of readthrough transcription into Prnd, whereas the Rcm0 line contains a PGK promoter/hypoxanthine phosphoribosyltransferase minigene in reverse orientation to the Dpl promoter (22). It is possible that the Prn gene locus harbors complex regulatory signals and that the interposition of heterologous genetic elements readily interferes with this transcriptional regulation. Interestingly, replacement of a PrP ORF with an ORF encoding a tet-dependent transactivator was found to up-regulate chimeric mRNAs encoding Dpl (23).
The most potent modulator of Dpl-induced ataxia was PrPC expression (Table 2). Twelve double-hemizygous mice were generated expressing both Dpl and PrP in the CNS; in all cases, phenotypic rescue of the Dpl-induced ataxia was found. Because levels of Dpl mRNA and protein were unaltered by PrP transgene expression (ref. 7 and this study), our data argue for an interaction between the biological activities of PrPC and Dpl. This conclusion is consistent with observations on Prnp0/0-ZrchII mice and mice expressing PrP-derived cosmid vector encoding a tet-transactivator that also favors Dpl overexpression (23).
The genetic background of the mice comprises a third modulator of Dpl-induced ataxia. A large spread in the age of onset of ataxia (370–680 days) was noted for outbred BALB/c × 129/Ola Rcm0 mice (Table 1), segregating both BALB/c and 129/Ola alleles, whereas in an inbred 129/Ola background, Dpl-related ataxia was apparent with a relatively synchronous onset age of 442 days (±5 SEM; n = 22; ref. 6). A recent quantitative trait loci study revealed that prion incubation time is influenced by loci on chromosomes 9 and 11 (24); whether or not similar analyses will identify genes that modify the response to ectopic Dpl expression remains to be established.
Dpl-Induced Neurodegeneration Differs from Prion Disease.
The transgenetic studies reported here distinguish between the diseases caused by two protein products of the Prn gene complex. Our results are of particular interest in view of the structural similarities of two proteins, which exhibit only 25% sequence homology (25). Dpl, like PrP, is a three-helix bundle protein that is anchored to the cell surface by a glycosylphosphatidylinositol (GPI) moiety (26–28). Additionally, NMR structures of recombinant Dpl and PrP are strikingly similar in the globular, C-terminal region of the protein, which is composed of two short β-strands and three α-helices (25). The second and third helices of PrP and Dpl are covalently linked by one and two disulfide bonds, respectively.
Dpl overexpression was neurotoxic in five lines of Tg(Dpl)ZrchI mice expressing different levels of Dpl. Histoblotting for Dpl provided a remarkably consistent neuroanatomical pattern of Dpl expression within the CNS among multiple lines of Tg(Dpl)ZrchI mice (Fig. 2). The pattern of neurotoxicity in all Tg(Dpl)ZrchI lines was almost exclusively cerebellar, with the most striking change being the loss of neurons from the Purkinje and granule cell layers. Extensive areas of TUNEL-positive granule cells were apparent in the cerebellum of Tg(Dpl)ZrchI mice, suggesting that many cells were undergoing apoptosis (Fig. 3C). It is unlikely that this selective pathology is a result of higher Dpl-expression levels in these regions, as histoblotting revealed high Dpl levels in regions devoid of neuronal cell loss (thalamus, corpus callosum, internal capsule, and brainstem). Conversely, the hippocampus, which expresses lower levels of Dpl (Fig. 2 A and C), exhibited some cell death. Although these observations argue for a selective vulnerability of cerebellar and hippocampal neurons, the widespread reactive gliosis in Tg(Dpl)ZrchI mice indicates that Dpl expression also may be toxic elsewhere within the CNS.
Analyses performed on wt mouse tissues with Dpl polyclonal antiserum E6977 revealed an expression pattern markedly different from that of PrP. Western blot analysis indicated substantial Dpl expression in testis but not in adult brain or many other tissues. In contrast, PrP is expressed in many peripheral tissues and is present in the adult CNS at high levels. We conclude that in instances in which PrP and Dpl gene-expression overlap, e.g., in the testis, perhaps during sperm maturation (29), interactions between these two proteins may be particularly important. Other studies suggest that PrPC and Dpl are expressed on the surface of different CNS cells even though PrPC can rescue the Dpl-induced cerebellar degeneration (23). Dpl expressed in the CNS has a more pronounced localization in white-matter tracts than does PrP. Whether the propensity of GPI-anchored proteins to translocate from one cell to another (30) is operative in some rescue studies reported for Dpl and PrP remains to be determined. Although overexpression of wt PrPC causes degeneration of muscle and peripheral nerves (31), this disease is different from naturally occurring prion diseases that are caused by the accumulation of PrPSc. Our findings clearly separate the disease paradigms based upon Dpl overexpression, PrPC overexpression, and experimental prion disease.
New Directions in Prion Protein Biology.
Demonstration of an interaction between PrP and Dpl is significant because there is a paucity of evidence that PrP-binding proteins identified in vitro interact with PrP in vivo. Conversely, the genes of some PrP-interacting proteins identified on genetic grounds have yet to be identified, such as protein X (32–34). Dpl is noteworthy, as its gene has been cloned and the product interacts with PrP in vivo either directly or indirectly through another protein or a complex of proteins. Identifying the protein or complex of proteins to which Dpl binds and produces neurodegeneration will be important. Whether this protein or a complex is the same as that postulated to mediate the neurotoxicity of N-terminally truncated PrP in Tg mice (35), which, like Dpl toxicity, can be rescued by wt PrP expression, remains to be established.
The structural similarities of Dpl and PrP, with their divergent sequences, offer an interesting opportunity to create chimeric Dpl:PrP proteins. Determining which regions of Dpl are responsible for neurotoxicity by substituting structurally similar segments from PrP might provide a workable transgenetic approach. However, Tg mouse studies comprise a cumbersome approach to the task of identifying those residues required for Dpl-mediated neurotoxicity. Devising a neuronal cell-culture system to study the mechanism of Dpl-induced neurodegeneration might prove more useful and provide important insights into the molecular pathogenesis of prion disease.
Acknowledgments
R.M. was supported by a Long Term Fellowship from the Human Frontier Science program. P.M. is a Peterborough-Burgess fellow of the Alzheimer Society of Ontario. This work was supported by grants from the National Institutes of Health, Canadian Institutes of Health Research, and the Alzheimer Society of Ontario as well as by a gift from the G. Harold and Leila Y. Mathers Foundation.
Abbreviations
- CNS
central nervous system
- wt
wild type
- Tg
transgenic
- GFAP
glial fibrillary acidic protein
- SHa
Syrian hamster
- Dpl
doppel
- PrP
prion protein
- Prnp
prion protein gene
- Prnd
doppel gene
- Prn
gene locus composed of Prnp and Prnd
References
- 1.Bendheim P E, Barry R A, DeArmond S J, Stites D P, Prusiner S B. Nature (London) 1984;310:418–421. doi: 10.1038/310418a0. [DOI] [PubMed] [Google Scholar]
- 2.DeArmond S J, McKinley M P, Barry R A, Braunfeld M B, McColloch J R, Prusiner S B. Cell. 1985;41:221–235. doi: 10.1016/0092-8674(85)90076-5. [DOI] [PubMed] [Google Scholar]
- 3.Büeler H, Fisher M, Lang Y, Bluethmann H, Lipp H-P, DeArmond S J, Prusiner S B, Aguet M, Weissmann C. Nature (London) 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 4.Manson J C, Clarke A R, Hooper M L, Aitchison L, McConnell I, Hope J. Mol Neurobiol. 1994;8:121–127. doi: 10.1007/BF02780662. [DOI] [PubMed] [Google Scholar]
- 5.Sakaguchi S, Katamine S, Nishida N, Moriuchi R, Shigematsu K, Sugimoto T, Nakatani A, Kataoka Y, Houtani T, Shirabe S, et al. Nature (London) 1996;380:528–531. doi: 10.1038/380528a0. [DOI] [PubMed] [Google Scholar]
- 6.Moore R. Ph.D. Thesis. Edinburgh, Scotland: Univ. of Edinburgh; 1997. [Google Scholar]
- 7.Moore R C, Lee I Y, Silverman G L, Harrison P M, Strome R, Heinrich C, Karunaratne A, Pasternak S H, Chishti M A, Liang Y, et al. J Mol Biol. 1999;292:797–817. doi: 10.1006/jmbi.1999.3108. [DOI] [PubMed] [Google Scholar]
- 8.Nishida N, Tremblay P, Sugimoto T, Shigematsu K, Shirabe S, Petromilli C, Erpel S P, Nakaoke R, Atarashi R, Houtani T, et al. Lab Invest. 1999;79:689–697. [PubMed] [Google Scholar]
- 9.Pham C T, MacIvor D M, Hug B A, Heusel J W, Ley T J. Proc Natl Acad Sci USA. 1996;93:13090–13095. doi: 10.1073/pnas.93.23.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olson E N, Arnold H H, Rigby P W, Wold B J. Cell. 1996;85:1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- 11.Katamine S. Rinsho Byori. 2000;48:437–441. [PubMed] [Google Scholar]
- 12.Scott M, Foster D, Mirenda C, Serban D, Coufal F, Wälchli M, Torchia M, Groth D, Carlson G, DeArmond S J, et al. Cell. 1989;59:847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 13.Scott M R D, Butler D A, Bredesen D E, Wälchli M, Hsiao K K, Prusiner S B. Protein Eng. 1988;2:69–76. doi: 10.1093/protein/2.1.69. [DOI] [PubMed] [Google Scholar]
- 14.Prusiner S B, Tremblay P, Safar J, Torchia M, DeArmond S J. In: Prion Biology and Diseases. Prusiner S B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 113–145. [Google Scholar]
- 15.Carlson G A, Ebeling C, Yang S-L, Telling G, Torchia M, Groth D, Westaway D, DeArmond S J, Prusiner S B. Proc Natl Acad Sci USA. 1994;91:5690–5694. doi: 10.1073/pnas.91.12.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muramoto T, DeArmond S J, Scott M, Telling G C, Cohen F E, Prusiner S B. Nat Med. 1997;3:750–755. doi: 10.1038/nm0797-750. [DOI] [PubMed] [Google Scholar]
- 17.Taraboulos A, Jendroska K, Serban D, Yang S-L, DeArmond S J, Prusiner S B. Proc Natl Acad Sci USA. 1992;89:7620–7624. doi: 10.1073/pnas.89.16.7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davison F D, Groves M, Scaravilli F. Histochem J. 1995;27:983–988. [PubMed] [Google Scholar]
- 19.Adle Biassette H, Levy Y, Colombel M, Poron F, Natchev S, Keohane C, Gray F. Neuropathol Appl Neurobiol. 1995;21:218–227. doi: 10.1111/j.1365-2990.1995.tb01053.x. [DOI] [PubMed] [Google Scholar]
- 20.Scott M R, Köhler R, Foster D, Prusiner S B. Protein Sci. 1992;1:986–997. doi: 10.1002/pro.5560010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prusiner S B, Scott M, Foster D, Pan K-M, Groth D, Mirenda C, Torchia M, Yang S-L, Serban D, Carlson G A, et al. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 22.Moore R C, Redhead N J, Selfridge J, Hope J, Manson J C, Melton D W. Biotechnology. 1995;13:999–1004. doi: 10.1038/nbt0995-999. [DOI] [PubMed] [Google Scholar]
- 23.Rossi D, Cozzio A, Flechsig E, Klein M A, Rulicke T, Aguzzi A, Weissmann C. EMBO J. 2001;20:694–702. doi: 10.1093/emboj/20.4.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephenson D A, Chiotti K, Ebeling C, Groth D, DeArmond S J, Prusiner S B, Carlson G A. Genomics. 2000;69:47–53. doi: 10.1006/geno.2000.6320. [DOI] [PubMed] [Google Scholar]
- 25.Mo H, Moore R C, Cohen F E, Westaway D, Prusiner S B, Wright P E, Dyson H J. Proc Natl Acad Sci USA. 2001;98:2352–2357. doi: 10.1073/pnas.051627998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stahl N, Borchelt D R, Hsiao K, Prusiner S B. Cell. 1987;51:229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 27.Silverman G L, Qin K, Moore R C, Yang Y, Mastrangelo P, Tremblay P, Prusiner S B, Cohen F E, Westaway D. J Biol Chem. 2000;275:26834–26841. doi: 10.1074/jbc.M003888200. [DOI] [PubMed] [Google Scholar]
- 28.Lu K, Wang W, Xie Z, Wong B, Li R, Petersen R B, Sy M, Chen S G. Biochemistry. 2000;39:13575–13583. doi: 10.1021/bi001523m. [DOI] [PubMed] [Google Scholar]
- 29.Shaked Y, Rosenmann H, Talmor G, Gabizon R. J Biol Chem. 1999;274:32153–32158. doi: 10.1074/jbc.274.45.32153. [DOI] [PubMed] [Google Scholar]
- 30.Kooyman D L, Byrne G W, McClellan S, Nielsen D, Tone M, Waldmann H, Coffman T M, McCurry K R, Platt J L, Logan J S. Science. 1995;269:89–92. doi: 10.1126/science.7541557. [DOI] [PubMed] [Google Scholar]
- 31.Westaway D, DeArmond S J, Cayetano-Canlas J, Groth D, Foster D, Yang S-L, Torchia M, Carlson G A, Prusiner S B. Cell. 1994;76:117–129. doi: 10.1016/0092-8674(94)90177-5. [DOI] [PubMed] [Google Scholar]
- 32.Kaneko K, Vey M, Scott M, Pilkuhn S, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1997;94:2333–2338. doi: 10.1073/pnas.94.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perrier V, Wallace A C, Kaneko K, Safar J, Prusiner S B, Cohen F E. Proc Natl Acad Sci USA. 2000;97:6073–6078. doi: 10.1073/pnas.97.11.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zulianello L, Kaneko K, Scott M, Erpel S, Han D, Cohen F E, Prusiner S B. J Virol. 2000;74:4351–4360. doi: 10.1128/jvi.74.9.4351-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shmerling D, Hegyi I, Fischer M, Blattler T, Brandner S, Gotz J, Rulicke T, Flechsig E, Cozzio A, von Mering C, et al. Cell. 1998;93:203–214. doi: 10.1016/s0092-8674(00)81572-x. [DOI] [PubMed] [Google Scholar]