Abstract
Numerous studies have shown that mitochondrial dysfunction contributes to consequential phenotypes of Huntington’s disease (HD), a fatal and inherited neurodegenerative disease caused by the expanded CAG repeats in the N-terminus of the huntingtin (Htt) gene. To maintain proper function, mitochondria develop a dedicated protein quality control mechanism by activating a stress response termed the mitochondrial unfolded protein response (UPRmt). Defects in the UPRmt have been linked to aging and are also associated with neurodegenerative diseases. However, little is known about the role of the UPRmt in HD. In this study, we find that ABCB10, a mitochondrial transporter involved in the UPRmt pathway, is downregulated in HD mouse striatal cells, HD patient fibroblasts, and HD R6/2 mice. Deletion of ABCB10 causes increased mitochondrial reactive oxygen species (ROS) production and cell death, whereas overexpression of ABCB10 reduces these aberrant events. Moreover, the mitochondrial chaperone HSP60 and mitochondrial protease Clpp, two well-established markers of the UPRmt, are reduced in the in vitro ABCB10-deficienct HD models. CHOP, a key transcription factor of HSP60 and Clpp, is regulated by ABCB10 in HD mouse striatal cells. Furthermore, we find that mutant huntingtin (mtHtt) inhibits the mtUPR by impairing ABCB10 mRNA stability. These findings demonstrate a suppression of the UPRmt by mtHtt, suggesting that disturbance of mitochondrial protein quality control may contribute to the pathogenesis of HD.
Keywords: Huntington’s disease, ABCB10, mitochondrial unfolded protein response, mitochondrial dysfunction
1. Introduction
Huntington’s disease (HD) is a fatal and inherited neurodegenerative disorder that progresses for 15–20 years after initial onset [1]. The main cause of HD is the expanded CAG repeats encoding polyglutamine (polyQ) in the N-terminus of the huntingtin (Htt) protein [2,3]. The genetic cause of HD (mutant Htt, mtHtt) has been identified for more than 20 years. However, the underlying mechanisms occurring in HD remain elusive.
Accumulating evidence suggests that mitochondrial dysfunction plays an important role in the pathogenesis of HD [2]. For instance, mtHtt associates with the outer mitochondrial membrane in different HD models, resulting in mitochondrial permeability transition pore opening, calcium disturbance, reduced ATP production, mitochondrial membrane potential loss, increased ROS production, and release of cytochrome c [4]. PGC-1α is a transcription co-activator that regulates the genes involved in mitochondrial biogenesis [5]. mtHtt interacts with PGC-1α and suppresses its activity in HD mouse striatal cells as well as in HD transgenic mouse models [6]. Mitochondria are highly dynamic organelles, and the balance between fission and fusion is important for maintaining normal mitochondrial function. mtHtt interacts with Drp1, the master regulator of mitochondrial fission, leading to Drp1 hyperactivation and mitochondrial fragmentation [7]. We previously reported that treatment with peptide P110 (a specific peptide inhibitor of Drp1) decreased mtHtt-induced mitochondrial fragmentation, corrected defects in mitochondrial function, and reduced neuronal cell death in both in vivo and in vitro HD models [8]. Moreover, in vivo and in vitro HD models exhibit accelerated mitochondrial outer membrane protein degradation and excessive mitophagy [9,10]. Collectively, these findings highlight the mitochondria as a promising therapeutic target for the treatment of HD.
When damaged protein accumulates in the mitochondrial matrix and exceeds the maximal capacity of the protein folding apparatus, the defense mechanism called the mitochondrial unfolded protein response (UPRmt) is activated to process the cellular stress occurring in the mitochondrial matrix [11,12]. Upon UPRmt activation, mitochondrial chaperones are induced and imported into mitochondria to refold the damaged proteins [11,12]. On the other hand, the mitochondrial matrix protease Clpp (ATP-dependent Clp protease proteolytic subunit) cleaves unfolded or misfolded proteins inside the mitochondria into polypeptides [13,14]. In worms, activating transcription factor associated with stress-1 (ATFS-1), a leucine zipper transcription factor, is imported into the mitochondrial matrix for degradation under normal physiological conditions [15]. Damaged proteins are then cleaved into short peptides, which are exported to the cytosol via the inner membrane transporter HAF-1, leading to ATFS-1 nuclear translocation [13, 15]. Consequently, ATFS1 facilitates transcriptional activation of UPRmt target genes [15]. It has recently been reported that the UPRmt is activated in diseases such as Friedreich’s ataxia [16], spastic paraplegia [17], cancer [18,19], and aging [20]. However, little is known about the role of the UPRmt in the pathogenesis of HD.
ABCB10 is one of the components of the UPRmt pathway in mammalian cells [21]. In this study, we found that mtHtt suppressed the expression of ABCB10 in various HD models by impairing its mRNA stability. Deletion of ABCB10 induced ROS production and cell death in HD mouse striatal cells. Moreover, ABCB10 was required for CHOP activation under mitochondrial stress. We also showed that HSP60 and Clpp, two downstream genes of CHOP [22], were decreased in HD cells. These data suggest a dysregulation of UPRmt in HD, revealing a novel mechanism of mitochondrial dysfunction in the pathogenesis of this devastating disease.
2. Results
2.1. ABCB10 is reduced in HD models
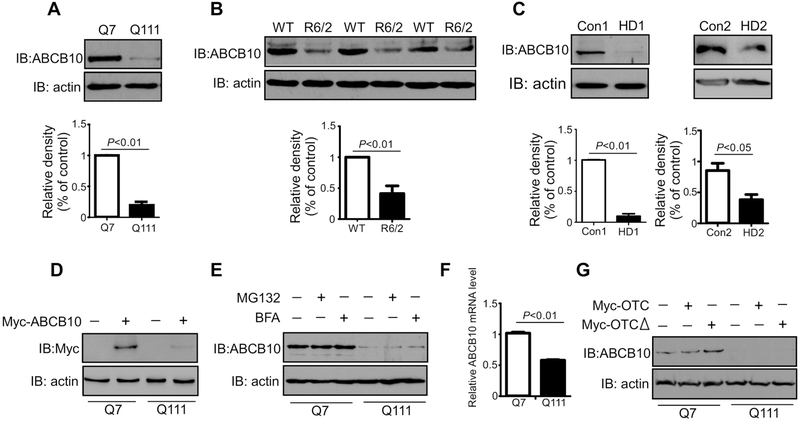
To determine if the UPRmt is perturbed in HD, we first examined ABCB10 protein level in the HdhQ111 and HdhQ7 mouse striatal cells. Western blot analysis showed that the protein level of ABCB10 was dramatically decreased in HdhQ111 mutant mouse striatal cells, when compared to that in HdhQ7 cells (Fig. 1A). Moreover, the protein level of ABCB10 was much lower in the striatum of HD R6/2 mice than that in wild-type littermates (Fig. 1B). Consistently, downregulation of ABCB10 was observed in the dermal fibroblasts of two HD patients (GM04693, GM21756) (Fig. 1C). We next expressed Myc-ABCB10 in HdhQ7 and HdhQ111 cells, and we found that the expression of Myc-ABCB10 in HdhQ111 was less than that in HdhQ7 cells (Fig. 1D). Reduction of ABCB10 in HdhQ111 cells was not blocked by treatment with either the proteasome inhibitor MG132 or the lysosome inhibitor bafilomycin A (BFA), indicating that a decrease in ABCB10 protein level is not the result of the ubiquitin-proteasome system (UPS)-mediated degradation nor autophagy (Fig. 1E). Furthermore, we performed real-time PCR to determine the gene expression of ABCB10. The mRNA expression of ABCB10 was significantly decreased in HdhQ111 cells when compared to that in HdhQ7 wildtype cells (Fig. 1F). OTCΔ is a mutant of ornithine transcarbamylase lacking amino acids 30–114. Overexpression of OTCΔ has been shown to induce the UPRmt [22–24]. We found that expression of OTCΔ in HdhQ7 wildtype cells induced ABCB10 expression, but the same induction was not seen in HdhQ111 mutant cells (Fig. 1G). Taken together, these data demonstrate that ABCB10 was suppressed in multiple models of HD ranging from mouse to patient.
Fig.1: ABCB10 is downregulated in models of Huntington’s disease.
(A)Whole cell extracts of HdhQ7 and HdhQ111 mouse striatal cells were harvested. Western blot (WB) analysis was performed to determine the protein levels of ABCB10 using the indicated antibodies. Actin was used as a loading control. The data shown in the histogram are mean ± SE from three independent experiments (n=3). (B) Total lysates were harvested from the striatum of HD R6/2 and wildtype mice. ABCB10 protein levels were determined by WB analysis. The shown blots are from three independent experiments (n=6). (C) HD patient fibroblasts (GM04693 and GM21756) and normal fibroblasts (HDFa and HDFn) were harvested. ABCB10 was detected by WB analysis. The data are mean ± SE of three independent experiments (n=3). (D) HdhQ7 or HdhQ111 cells were transfected with Myc-ABCB10 plasmids for 24h followed by immunoblotting with anti-Myc antibody (n=3). (E) HdhQ7 or HdhQ111 cells were treated for 12 hours with proteasome inhibitor MG132 or 4 hours with lysosomal inhibitor BFA. Protein extracts were immunoblotted with ABCB10 antibody. The shown blots are representative of three independent experiments (n=3). (F) Total RNA was extracted from HdhQ7 or HdhQ111 cells. ABCB10 and GAPDH mRNA levels were determined by quantitative real-time PCR. The data are mean ± SE from three independent experiments (n=3). (G) Total cell lysates were immunoblotted by indicated antibodies after transfection of control vector, myc-OTC, or myc-OTCΔ for 24h. The blots shown are from three independent experiments (n=3).
2.2. Deletion of ABCB10 causes increased mitochondrial ROS production and cell death
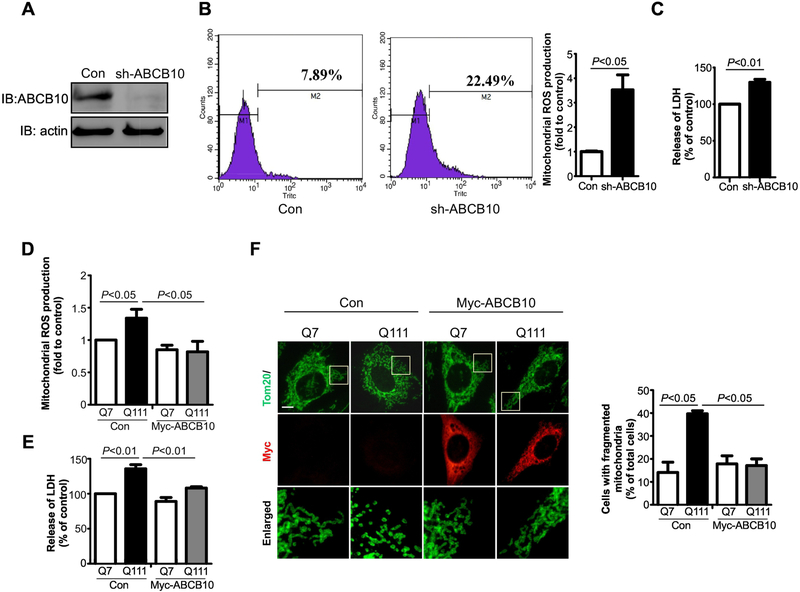
Deletion of ABCB10 increases reactive oxygen species (ROS) production in an ischemia-reperfusion model [25] and activates ROS suppressing genes in HepG2 cells [21]. However, the physio-pathological function of ABCB10 in neurodegenerative diseases like HD is unknown. To elucidate the functional role of ABCB10 in HD, we lentivirally expressed pLKO-sh-ABCB10 in wildtype HdhQ7 cells to downregulate the level of ABCB10, in order to mimic the decrease in ABCB10 observed in our HD models. The expression of pLKO-sh-ABCB10 efficiently reduced the level of ABCB10 (Fig. 2A). We observed a great increase in the production of mitochondrial ROS in the ABCB10-depleting cells, relative to that in cells with control vector (Fig. 2B). Moreover, increased LDH release, an indicator of cell death, was seen in the ABCB10 knockdown HdhQ7 cells (Fig. 2C). In contrast, overexpression of Myc-ABCB10 reduced mitochondrial ROS and blocked LDH release in HdhQ111 cells (Figs. 2D and 2E). Next, we assessed mitochondrial morphology in both Myc-ABCB10 and control vector expressing HD striatal cells by staining cells with anti-Tom20 antibody. As shown in Fig. 2F, expression of Myc-ABCB10 reduced the number of cells exhibiting fragmented mitochondria when compared to those cells expressing control vector (Fig. 2F). These data suggest that a decrease in ABCB10 level in HdhQ111 mouse striatal cells causes mitochondrial dysfunction and subsequent neuronal damage.
Fig. 2: Deletion of ABCB10 causes increased mitochondrial ROS and cell death.
HdhQ7 cells were cultured and infected with pLKO (Control) or pLKO –sh-ABCB10 lentivirus for ABCB10-knockdown (KD). (A) Total protein was extracted from cells and immunoblotted by using ABCB10 antibody. Actin was used as loading control (n=3). (B) Control cells and ABCB10 knockeddown cells were stained with MitoSOX (red). The mitochondrial ROS was determined by Flow cytometry. The data are mean ± SE from three independent experiments (n=3). (C) Cell death was determined by the release of lactate dehydrogenase (LDH). The data are mean ± SE of three independent experiments (n=3). (D and E) HdhQ7 or HdhQ111 cells were transfected with control vector or Myc-ABCB10 for 48 hours. The cells were stained with MitoSOX (red). The mitochondrial ROS was determined by Flow cytometry. The data are mean ± SE from three independent experiments (n=3). Cell death was determined by LDH release. The data are mean ± SE of three independent experiments (n=3). (F) Control vector or Myc-ABCB10 expressed cells were stained with anti-myc and anti-Tom 20 antibodies. Enlarged mitochondrial morphology were showed in the lower panels. At least 100 cells per group were counted. The data are mean ± SE of three independent experiments (n=3).
2.3. Transcription factor CHOP is regulated by ABCB10
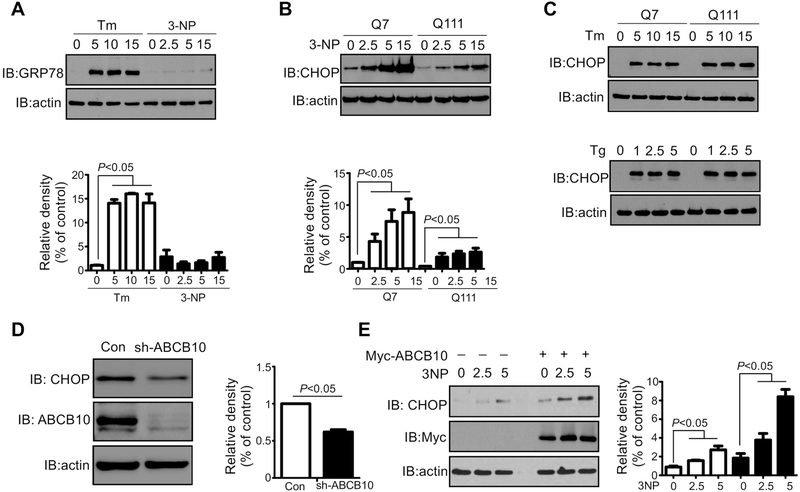
CHOP is a key transcription factor that binds to the MSR element to activate HSP60 and Clpp expression in mammalian cells [22,23]. Overexpression of either OTCΔ or endonuclease G (another protein that can accumulate in the mitochondria), elevates CHOP protein level [22,26,27]. 3-nitropropionic acid (3-NP), an inhibitor of the mitochondrial citric acid cycle, is a well-characterized inducer of ROS and may cause proteotoxic stress [28]. We hypothesize that treatment with 3-NP may activate the mtUPR by increasing the rate of ROS production and promoting protein aggregation in the mitochondrial matrix. Treatment with the ER stress inducer tunicamycin (Tm) but not 3-NP elevated the ER stress marker GRP78 (Fig. 3A), excluding the possibility that 3-NP induces the ER UPR. As shown in Fig. 3B, the protein levels of CHOP are higher in the HdhQ7 relative to that in HdhQ111 cells (Fig. 3B, first lane and fourth lane, respectively). Moreover, treatment of 3-NP caused a greater rate of CHOP induction in HdhQ7 cells (Fig. 3B). These results indicate that mtHtt inhibits 3-NP-induced mtUPR activation. We observed no differences of CHOP protein levels between HdhQ7 and HdhQ111 cells in the presence of two ER stress inducers tunicamycin and thapsigargin (Fig. 3C), suggesting that mtHtt selectively suppresses activation of the mtUPR. Deletion of ABCB10 in HdhQ7 cells resulted in a significant decrease in CHOP protein level (Fig. 3D), whereas overexpression of Myc-ABCB10 increased CHOP expression in HdhQ111 cells (Fig. 3E, first lane and fourth lane, respectively). Furthermore, greater induction of CHOP by 3-NP was seen in Myc-ABCB10-expressing HdhQ111 cells relative to that in control vector-expressing cells (Fig. 3E). These results suggest that ABCB10 is required for mitochondrial stress-induced CHOP activation.
Fig. 3: Transcription factor CHOP is regulated by ABCB10.
(A) HdhQ7 cells were treated with Tm or 3-NP at the indicated concentrations. Whole cell extracts were prepared, and the expression of CHOP was analyzed by WB. The data are mean ± SE of three independent experiments (n=3). (B) HdhQ7 or HdhQ111cells were treated with 3-NP for 16 hours at the indicated concentrations. The levels of CHOP and actin were examined by WB analysis. The data are mean ± SE of three independent experiments (n=3). (C) HdhQ7 or HdhQ111 cells were treated with ER stress inducers Tg or Tm. Total cell lysates were harvested. CHOP and actin expression was determined by WB analysis. The shown blots are representative of three independent experiments (n=3). (D) HdhQ7 cells were infected with pLKO-sh-ABCB10 and pLKO-sh-Con lentivirus. ABCB10 knock-down stable cells were collected after puromycin selection (3 μg/ml, 1 week). ABCB10, CHOP, and actin were analyzed by WB. The data are mean ± SE of three independent experiments (n=3). (E) HdhQ111 cells were transfected with control vector or myc-ABCB10 for 24 hours followed by treatment with 3-NP at 2.5 or 5 mM for 16 hours. Total cell lysates were harvested and analyzed by WB. The data are mean ± SE of three independent experiments (n=3).
2.4. HSP60 and Clpp expression are decreased in HD cell cultures.
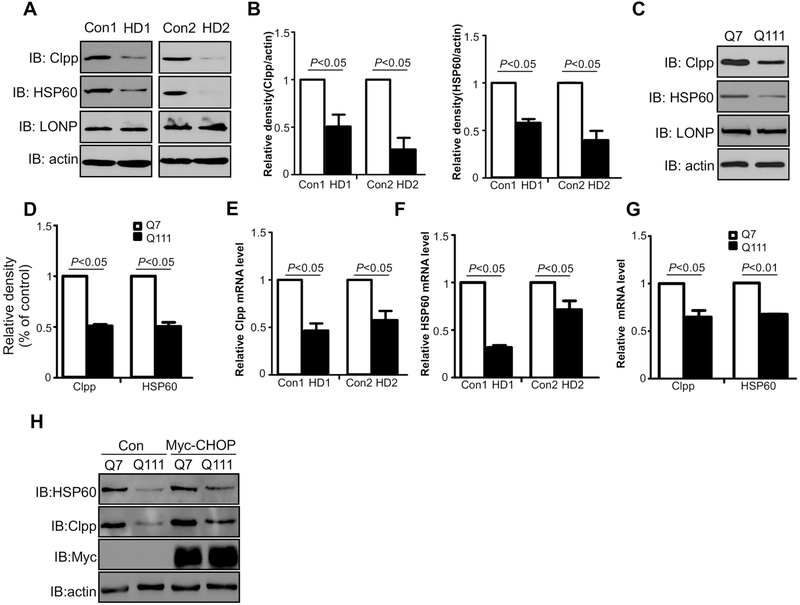
It is well established that CHOP associates with the promoters of HSP60 and Clpp to promote transcriptional activation [22,23]. Because we found CHOP protein level was down-regulated in HD mouse striatal cells, we examined the protein levels of HSP60 and Clpp in two lines of HD patient-derived fibroblasts. The protein levels of HSP60 and Clpp were significantly reduced in HD patient fibroblasts (GM04693 and GM21756) relative to those in control fibroblasts (HDFa and HDFn). However, no change in the protein level of LONP was observed (Figs. 4A and 4B). Consistently, lower protein levels of HSP60 and Clpp were found in HdhQ111 cells than in HdhQ7 cells (Figs. 4C and 4D). The mRNA levels of HSP60 and Clpp were also drastically reduced in HD patient-derived fibroblasts (Figs. 4E and 4F) along with HdhQ111 mutant striatal cells (Fig. 4G). In addition, overexpression of Myc-CHOP enhanced the expression of HSP60 and Clpp (Fig. 4H). Collectively, these results suggest that the mtUPR is inhibited in HD cell cultures
Fig. 4: Mitochondrial UPR components HSP60 and Clpp are decreased in cell culture models of HD.
(A and B) Total cell lysates of HD patient fibroblasts (GM04693 and GM21756) and normal fibroblasts (HDFn and HDFa) were harvested. Mitochondrial chaperone HSP60, proteases Clpp and LONP were examined by WB analysis. The data are mean ± SE of three independent experiments (n=3). (C and D) Protein extracted from HdhQ7 or HdhQ111 cells were used for analysis of HSP60, Clpp, and LONP with indicated antibodies. The data are mean ± SE of three independent experiments (n=3). (E and F) Total RNA was extracted from patient fibroblasts (GM04693 and GM21756) and normal fibroblasts (HDFn and HDFa) cells. HSP60 and Clpp mRNA levels were determined by quantitative real-time PCR. The data are mean ± SE from three independent experiments (n=3). (G)The mRNA levels of ABCB10 in HdhQ7 or HdhQ111 cells were determined by real-time PCR. The data are mean ± SE from three independent experiments (n=3). (H) HdhQ7 or HdhQ111 cells were transfected with Myc or Myc-CHOP plasmids for 48 hours. Total protein levels of HSP60 and Clpp were determined by western blot analysis. Shown blots are from three independent experiments (n=3).
2.5. mtHtt suppresses mtUPR by regulating ABCB10 mRNA stability
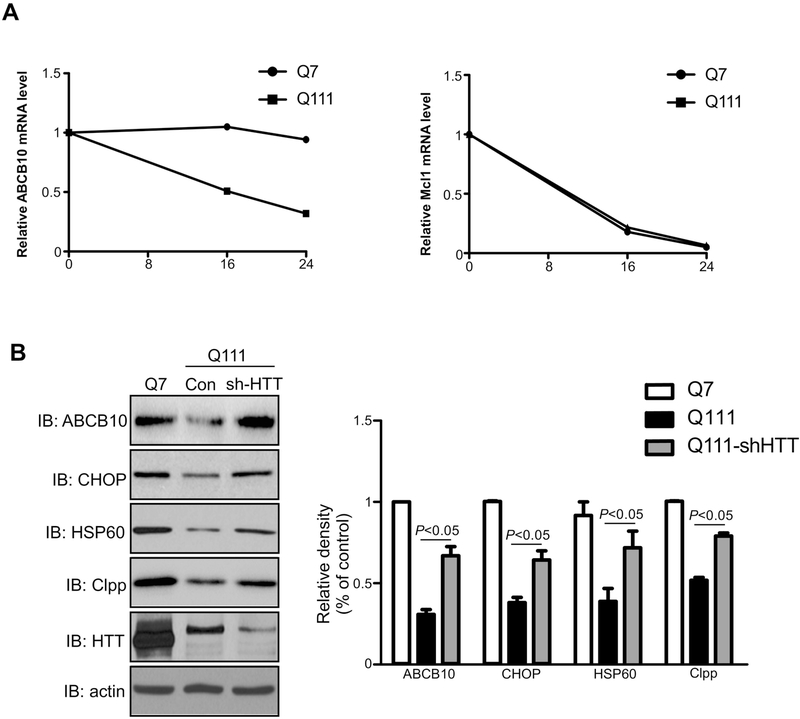
The nuclear accumulated mtHtt interacts with several transcription factors, such as PGC-1α [6], CBP [29–31], TBP [32], NCoR [33], and REST/NRSF [34], leading to transcriptional alteration. We performed reporter gene assays to measure the promoter activity of ABCB10 in HD cell cultures. Surprisingly, there was no apparent difference between wild-type striatal cells and mutant striatal cells (data not shown). To determine whether mtHtt affects ABCB10 by reducing its mRNA stability, we measured steady-state levels of ABCB10 mRNA in the HdhQ7 and HdhQ111 cells by treatment of Actinomycin D, an inhibitor of RNA polymerase [35]. After transcription inhibition, mtHtt accelerated ABCB10 mRNA degradation in HdhQ111 striatal cells (Fig. 5A left panel). By contrast, the mRNA stability of the other mitochondrial membrane protein induced myeloid leukemia cell differentiation protein Mcl1 was not affected by mtHtt (Fig. 5A, right panel). Finally, we examined whether deletion of mtHtt could abolish the defects of the mtUPR in HD cell cultures. Upon knockdown of Htt in HD mutant mouse striatal cells, we found that the decreased mtUPR components were all restored (Fig. 5B). These data support the above findings and suggest that mtHtt inhibits the mtUPR at least in part by impairing ABCB10 mRNA stability.
Fig. 5: Mutant Huntingtin regulates ABCB10 mRNA stability.
(A) HdhQ7 cells were treated with 10ug/ml AntinomycinD for 16 or 24 hours. Total RNA was extracted, and the mRNA levels of ABCB10 and Mcl1 were determined by quantitative real-time PCR (n=3). (B) HdhQ111 cells were infected with pLKO (Control) or pLKO-sh-HTT lentivirus for mtHtt-knockdown. Total cell lysates were extracted and immunoblotted with anti-huntingtin, anti-ABCB10, anti-HSP60, anti-Clpp, anti-CHOP, and anti-actin antibodies. The data are mean ± SE from three independent experiments (n=3).
3. Materials and method
3.1. Reagents and antibodies
Anti-ABCB10 (sc-25750, 1:500), anti-HSP60 (sc-13115, 1:1000), and anti-c-Myc (sc-40, 1:1000) antibodies were purchased from Santa Cruz Biotechnology. Anti-Clpp (ab124822, 1:1000), anti-Tom20(ab78547, 1:2000) antibodies were purchased from Abcam. Anti-LONP (15440–1-AP, 1:1000), anti-CHOP (15204–1-AP, 1:1000) antibodies were from Proteintech. Pan-actin (A1978, 1:5000) antibody was from Sigma-Aldrich. Full-length Htt (MAB2166, 1:1000) antibody was from Millipore. Anti-GRP78 (ADI-SPA-826, 1:1,000) was from Enzo Life Sciences. Peroxidase-linked, species-specific antibodies (31430, 31460) were from Thermo Scientific. Proteasome inhibitor MG132 was from Tocris Bioscience. 3-NP(3-nitropropionic acid), Bafilomycin A1 (BFA), Antinomycin D and cocktail of protease inhibitor was purchased from Sigma-Aldrich. Lipofectamine®2000 Transfection Reagent was purchased from Invitrogen.
3.2. Constructs and transfection
OTC and OTCΔ plasmids were gifts from Dr. Hoogenraad (La Trobe University, Melbourne). Lentivirus packaging plasmids were obtained from Addgene. Myc-ABCB10 and Myc-CHOP were generated by inserting PCR-amplified fragments into the pCMV-Myc vector. Control plasmid pLKO, sh-RNA against ABCB10 (TRCN0000113448) and sh-RNA against Huntingtin(TRCN0000100349) were from Sigma-Aldrich. Cells were transfected with Lipofectamine®2000 transfection reagent following the manufacturer’s protocol.
3.3. Cell culture.
Immortalized HdhQ111 mutant striatal cell lines and HdhQ7 wt cells were obtained from the CHDI Foundation. Cells were cultured in DMEM supplemented with 10% FBS, 100 mg/ml penicillin, 100 mg/ml streptomycin, and 400 mg/ml G418. Cells were grown at 33°C in a 5% CO2 incubator. Cells within 14 passages were used in all experiments.
HD patient fibroblasts (HD1: GM04693, HD2: GM21756, purchased from Coriell Institute, USA) and normal fibroblasts (Con 1, fibroblasts from adult, HDFa; Con 2, fibroblasts from juvenile, HDFn; purchased from Sigma-Aldrich) were maintained in MEM supplemented with 15% (vol/vol) FBS and 1% (vol/vol) penicillin/streptomycin. All of the above cells were maintained at 37°C in 5% CO2–95% air.
3.4. Lysate preparation and Western blot analysis
Cells were lysed in a total cell lysate buffer (50mM Tris–HCl, pH 7.5,150 mM NaCl, 1% Triton X-100, and protease inhibitor). Total protein concentration was determined by Bradford assay. The protein was re-suspended in Laemmli buffer, loaded on SDS-PAGE, and transferred onto nitrocellulose membranes. Membrane was probed with the indicated antibodies, followed by visualization with ECL and detection by chemiluminescence imaging system (Tanon 5200).
3.5. Mitochondrial ROS production
To measure mitochondrial ROS generation, cells were incubated with 5μM mitochondrial ROS indicator (MitoSOX™, Invitrogen Life Science) for 10 min at 37 °C and washed for three times with PBS. The cells were collected by centrifugation (400 rcf, 3min) and resuspended in 1 ml of PBS. Fluorescence intensities in cells were analyzed by flow cytometric analysis (BD FACSCalibur™).
3.6. Real-time PCR
Cells were washed twice with ice-cold PBS. Total RNA was isolated from the cells using TRI Reagent®(T9424,Sigma-Aldrich). cDNA was synthesized by reverse transcription using 0.5μg of total RNA with ReverTra Ace® qPCR RT Master Mix (FSQ-201 −201,TOYOBO). For real-time analysis, cDNA was 10-fold diluted. Quantitative real-time PCR reaction was performed using THUNDERBIRD™ SYBR® qPCR mix (QPS-201, TOYOBO) on a LightCycler®96 PCR system (Roche). Relative quantitative plots were constructed for quantity of RNA input and for each gene of interest. The following primers were used for real-time PCR:
| Target gene | Forward primer | Reverse primer |
|---|---|---|
| mouseABCB10 | 5'-GCCGTCTCCTCTCGGATATG-3' | 5'-CTGGCGAACAGGGGATGTG-3' |
| mouse hsp60 | 5'-CACAGTCCTTCGCCAGATGAG-3' | 5'-CTACACCTTGAAGCATTAAGGCT-3' |
| mouse Clpp | 5'-GCCTTGCCGTGCATTTCTC-3' | 5'-CTCCACCACTATGGGGATGA-3' |
| mouse MCL1 | 5'-AAAGGCGGCTGCATAAGTC-3' | 5'-TGGCGGTATAGGTCGTCCTC-3' |
| mouse GAPDH | 5'-TGGCCTTCCGTGTTCCTAC-3' | 5'-GAGTTGCTGTTGAAGTCGCA-3' |
| human Clpp | 5'-TTGCCAGCCTTGTTATCGCA-3' | 5'-GGTTGAGGATGTACTGCATCG-3' |
| human hsp60 | 5'-ATGCTTCGGTTACCCACAGTC-3' | 5'-AGCCCGAGTGAGATGAGGAG-3' |
| human GAPDH | 5'-GGAGCGAGATCCCTCCAAAAT-3' | 5'-GGCTGTTGTCATACTTCTCATGG-3' |
3.7. Cell death assay
Medium from the cultured cells was harvested. Cell death was determined by measuring LDH release into the culture medium, using LDH-Cytotoxicity Assay Kit II (Roche, USA) by following the manufacturer’s instruction.
3.8. Immunocytochemistry
Cells cultured on coverslips were washed with ice-cold PBS, fixed in 4% (w/v) formaldehyde and permeabilized with 0.1% Triton X-100. After incubation with 1% (v/v) BSA (to block non-specific staining), fixed cells were incubated overnight at 4 °C with antibodies against Myc (1:200 dilution, Santa Cruz) and Tom20 (1:2000 dilution, Abcam). Cells were washed with PBS and incubated with Alexa Fluor® 488-conjugated anti-rabbit and Alexa Fluor® 568-conjugated anti-mouse secondary antibodies (1:500 dilution, Invitrogen) for 60 min. Coverslips were mounted on glass slides and imaged by NIKON Ci-S microscopy.
3.9. Statistical analysis
Data were analyzed by Student’s t-test (for two group comparisons) or a Kruskal–Wallis one-way ANOVA (for more than two group comparisons). In animal studies, we used n=6 mice/group for biochemical analysis. In cell culture studies, we performed each study with at least three independent replications. Data are expressed as mean ± SEM. Statistical significance was considered achieved at value p <0.05.
4. Discussion
Mitochondrial dysfunction is one of the main pathological mechanisms of HD. Perturbation in mitochondrial function causes mitochondrial stress and activates a series of mitochondrial unfolded protein response (mtUPR) signaling events, including CHOP up-regulation and HSP60 and Clpp transactivation, which in turn reduces the excessive mitochondrial proteotoxic stress [22,23]. Therefore, control of the mtUPR may be important for mitochondrial homeostasis. Upon mtUPR activation, CHOP is induced and recruited to the promoter of HSP60 and Clpp for their induction [22,26,27]. Furthermore, the misfolded and/or unfolded mitochondrial protein will be re-folded or degraded by mitochondrial chaperones and proteases [11–14]. A recent study reported that ABCB10 is required for maintaining mitochondrial chaperone and protease expression, suggesting its critical role in controlling the mtUPR. Although the physiological function of the mtUPR has been reported in several diseases [36,37], little is known about the relationship between the mtUPR and HD. In this study, we demonstrated that the protein levels of the mitochondrial inner-membrane ABC transporter ABCB10 were reduced in HD cell cultures and HD R6/2 mice, which in turn led to downregulation of CHOP, HSP60, and Clpp. Notably, overexpression of ABCB10 rescued the levels of the mtUPR related genes, improved mitochondrial function, and attenuated cell death in HdhQ111 mutant striatal cells. Moreover, we found that mtHtt inhibited the mtUPR pathway by promoting ABCB10 mRNA degradation. Thus, inhibition of the mtUPR by mtHtt may be added to the list of proposed mechanisms of mitochondrial dysfunction in HD.
Multiple studies report the disturbance of mtUPR machinery in neurodegenerative diseases. Polyglutamine diseases, also known as PolyQ diseases, are autosomal dominantly inherited diseases, including spinocerebellar ataxia (SCA) [38], dentatorubral pallidoluysian atrophy (DRPLA) [39], Machado– Joseph disease (MJD) [40], and HD [41, 42]. Expansion of the PolyQ is the main cause of these diseases [43]. In the neurons of the model organism Caenorhabditis elegans, PolyQ40 expression is sufficient to activate the mtUPR, but the activated mtUPR can be inhibited by deletion of ATFS-1 [44, 45]. However, we observed that the mtHtt carrying 111 CAG repeats suppressed the mtUPR machinery and induced striatal cell death. It is possible that the length of PolyQ expressed in the neuronal cells disrupts the mtUPR with different mechanisms. In Alzheimer’s disease, Beck et al. revealed that specific markers of the mtUPR are upregulated in both sporadic and familial AD [46]. Expression of ATFS-1ΔMTS exhibits dopaminergic neurodegenerative phenotypes in C. elegans. Hyperactivation of the mtUPR was found in α-synuclein over-expressing animals [47]. In addition, we also found that Clpp was selectively elevated in the LRRK2 mutant expressing HEK293 cells (data not shown). These findings establish a link between the regulation of the mtUPR pathway and Parkinson’s disease. Interestingly, α-synuclein and its mutant induced the mtUPR in a life span dependent manner, implying that the activity of the mtUPR associates with the development of disease and progression of age. Thus, it will of interest to investigate the changes of the UPRmt in a chronic animal model of HD, such as YAC128 mice, which exhibit progressive mitochondrial deficits.
The role of ABCB10 in controlling the mtUPR in HD pathogenesis raises many questions. For instance, silencing of ABCB10 by RNAi reduces expression of mitochondrial chaperones (HSPD1, DNAJA3) as well as mitochondrial protease (LONP), which leads to increased ROS production [21]. Consistently, we found that higher ROS level was detected in both ABCB10 deficient HD mouse striatal cells and in ABCB10 knocked-down wild-type striatal cells. However, mitochondrial protease Clpp, but not LONP, was downregulated significantly in ABCB10 deficient HD mouse striatal cells. It is possible that specific mechanisms might occur in different cell models. Transcription factors such as GATA-1 and those belonging to the E2F family have been reported to regulate the expression of ABCB10 in G1E-ER2 and HEK293 cells [48, 49]. mtHtt suppresses gene expression by blocking the association of transcription factor to response element [6]. In the present study, we examined ABCB10 promoter activity and found no significant changes between HdhQ7 and HdhQ111 cells (data not shown). Interestingly, the half-life of ABCB10 mRNA was greatly reduced in the HD mutant striatal cells, compared with wild-type striatal cells. Thus, a further investigation on how mtHtt promotes ABCB10 mRNA reduction is worthy. In addition, HAF-1 serves as a short peptide transfer channel under mtUPR activation in C.elegans [13], but ABCB10, the mammalian ortholog of HAF-1, has no effect on peptide export from the mitochondria matrix to the cytosol in mammalian cells [21]. How ABCB10 regulates the mtUPR should be more carefully investigated in future studies.
In conclusion, we present new evidence that mtHtt inhibits ABCB10 expression by reducing its mRNA stability. Deficiency of ABCB10 causes mtUPR dysregulation and subsequent mitochondrial damage and cell death. Our findings thus provide a new insight into the mechanism by which mitochondria become dysfunctional in HD.
Highlights.
ABCB10 is decreased in HD cell cultures and HD R6/2 mice, which in turn leads to inhibition of the mtUPR.
Overexpression of ABCB10 rescues the levels of the mtUPR related genes, improves mitochondrial function, and attenuates cell death in HD cell cultures.
Mutant Htt inhibits the mtUPR by impairing ABCB10 mRNA stability.
Acknowledgments
The work is supported by the Chinese National Natural Science Foundation Projects (31471326) (to X.G.), Jiangsu Specially Appointed Professor program (to X.G.) and National Institutes of Health grant (NIH R01 NS088192) (to X.Q.). We thank Dr. Hoogenraad (La Trobe University, Melbourne) for providing the OTC and OTCΔ plasmids.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Landles C, Bates GP, Huntingtin and the molecular pathogenesis of Huntington’s disease. Fourth in molecular medicine review series, EMBO reports. 5 (2004) 958–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Costa V, Scorrano L, Shaping the role of mitochondria in the pathogenesis of Huntington’s disease, The EMBO journal. 31 (2012) 1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].The Huntington’s Disease Collaborative Research Group, A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes., Cell. 72 (1993) 971–983. [DOI] [PubMed] [Google Scholar]
- [4].Bossy-Wetzel E, Petrilli A, Knott AB, Mutant huntingtin and mitochondrial dysfunction, Trends in neurosciences. 31 (2008) 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Spiegelman BM, Transcriptional control of mitochondrial energy metabolism through the PGC1 coactivators, Novartis Foundation symposium. 287 (2007) 60–63; [PubMed] [Google Scholar]
- [6].Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D, Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration, Cell. 127 (2006) 59–69. [DOI] [PubMed] [Google Scholar]
- [7].Song W, Chen J, Petrilli A, Liot G, Klinglmayr E, Zhou Y, Poquiz P, Tjong J, Pouladi MA, Hayden MR, Masliah E, Ellisman M, Rouiller I, Schwarzenbacher R, Bossy B, Perkins G, Bossy-Wetzel E, Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity, Nature medicine. 17 (2011) 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Guo X, Disatnik MH, Monbureau M, Shamloo M, Mochly-Rosen D, Qi X, Inhibition of mitochondrial fragmentation diminishes Huntington’s disease-associated neurodegeneration, The Journal of clinical investigation. 123 (2013) 5371–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Guo X, Qi X, VCP cooperates with UBXD1 to degrade mitochondrial outer membrane protein MCL1 in model of Huntington’s disease, Biochimica et biophysica acta. Molecular basis of disease. 1863 (2017) 552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Guo X, Sun X, Hu D, Wang YJ, Fujioka H, Vyas R, Chakrapani S, Joshi AU, Luo Y, Mochly-Rosen D, Qi X, VCP recruitment to mitochondria causes mitophagy impairment and neurodegeneration in models of Huntington’s disease, Nature communications. 7 (2016) 12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jovaisaite V, Auwerx J, The mitochondrial unfolded protein response-synchronizing genomes, Current opinion in cell biology. 33 (2015) 74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jovaisaite V, Mouchiroud L, Auwerx J, The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease, The Journal of experimental biology. 217 (2014) 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D, The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans, Molecular cell. 37 (2010) 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D, ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans, Developmental cell. 13 (2007) 467–480. [DOI] [PubMed] [Google Scholar]
- [15].Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM, Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation, Science. 337 (2012) 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Di Bella D, Lazzaro F, Brusco A, Plumari M, Battaglia G, Pastore A, Finardi A, Cagnoli C, Tempia F, Frontali M, Veneziano L, Sacco T, Boda E, Brussino A, Bonn F, Castellotti B, Baratta S, Mariotti C, Gellera C, Fracasso V, Magri S, Langer T, Plevani P, Di Donato S, Muzi-Falconi M, Taroni F, Mutations in the mitochondrial protease gene AFG3L2 cause dominant hereditary ataxia SCA28, Nature genetics. 42 (2010) 313–321. [DOI] [PubMed] [Google Scholar]
- [17].Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, Filla A, Cocozza S, Marconi R, Durr A, Fontaine B, Ballabio A, Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease, Cell. 93 (1998) 973–983. [DOI] [PubMed] [Google Scholar]
- [18].Wang S, Kaufman RJ, The impact of the unfolded protein response on human disease, The Journal of cell biology. 197 (2012) 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hipp MS, Park SH, Hartl FU, Proteostasis impairment in protein-misfolding and -aggregation diseases, Trends in cell biology. 24 (2014) 506–514. [DOI] [PubMed] [Google Scholar]
- [20].Guarente L, Sirtuins, aging, and metabolism, Cold Spring Harbor symposia on quantitative biology. 76 (2011) 81–90. [DOI] [PubMed] [Google Scholar]
- [21].Yano M, ABCB10 depletion reduces unfolded protein response in mitochondria, Biochemical and biophysical research communications. 486 (2017) 465–469. [DOI] [PubMed] [Google Scholar]
- [22].Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ, A mitochondrial specific stress response in mammalian cells, The EMBO journal. 21 (2002) 4411–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Horibe T, Hoogenraad NJ, The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response, PloS one. 2 (2007) e835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lamech LT, Haynes CM, The unpredictability of prolonged activation of stress response pathways, The Journal of cell biology. 209 (2015) 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liesa M, Luptak I, Qin F, Hyde BB, Sahin E, Siwik DA, Zhu Z, Pimentel DR, Xu XJ, Ruderman NB, Huffman KD, Doctrow SR, Richey L, Colucci WS, Shirihai OS, Mitochondrial transporter ATP binding cassette mitochondrial erythroid is a novel gene required for cardiac recovery after ischemia/reperfusion, Circulation. 124 (2011) 806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rath E, Berger E, Messlik A, Nunes T, Liu B, Kim SC, Hoogenraad N, Sans M, Sartor RB, Haller D, Induction of dsRNA-activated protein kinase links mitochondrial unfolded protein response to the pathogenesis of intestinal inflammation, Gut. 61 (2012) 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Papa L, Germain D, SirT3 regulates the mitochondrial unfolded protein response, Molecular and cellular biology. 34 (2014) 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liot G, Bossy B, Lubitz S, Kushnareva Y, Sejbuk N, Bossy-Wetzel E, Complex II inhibition by 3-NP causes mitochondrial fragmentation and neuronal cell death via an NMDA- and ROS-dependent pathway, Cell death and differentiation. 16 (2009) 899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nucifora FC Jr., Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson VL, Dawson TM, Ross CA, Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity, Science. 291 (2001) 2423–2428. [DOI] [PubMed] [Google Scholar]
- [30].Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H, Wanker EE, Bates GP, Housman DE, Thompson LM, The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription, Proceedings of the National Academy of Sciences of the United States of America. 97 (2000) 6763–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].McCampbell A, Taylor JP, Taye AA, Robitschek J, Li M, Walcott J, Merry D, Chai Y, Paulson H, Sobue G, Fischbeck KH, CREB-binding protein sequestration by expanded polyglutamine, Human molecular genetics. 9 (2000) 2197–2202. [DOI] [PubMed] [Google Scholar]
- [32].Huang CC, Faber PW, Persichetti F, Mittal V, Vonsattel JP, MacDonald ME, Gusella JF, Amyloid formation by mutant huntingtin: threshold, progressivity and recruitment of normal polyglutamine proteins, Somatic cell and molecular genetics. 24 (1998) 217–233. [DOI] [PubMed] [Google Scholar]
- [33].Boutell JM, Thomas P, Neal JW, Weston VJ, Duce J, Harper PS, Jones AL, Aberrant interactions of transcriptional repressor proteins with the Huntington’s disease gene product, huntingtin, Human molecular genetics. 8 (1999) 1647–1655. [DOI] [PubMed] [Google Scholar]
- [34].Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, Rigamonti D, Cattaneo E, Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes, Nature genetics. 35 (2003) 76–83. [DOI] [PubMed] [Google Scholar]
- [35].Stein GS, Burtner DL, Gene activation in human diploid cells. Age-dependent modifications in the stability of messenger RNAs for nonhistone chromosomal proteins, Biochimica et biophysica acta. 390 (1975) 56–68. [PubMed] [Google Scholar]
- [36].De I, Dogra N, Singh S, The Mitochondrial Unfolded Protein Response: Role in Cellular Homeostasis and Disease, Current molecular medicine. 17 (2017) 587–597. [DOI] [PubMed] [Google Scholar]
- [37].Shpilka T, Haynes CM, The mitochondrial UPR: mechanisms, physiological functions and implications in ageing, Nature reviews. Molecular cell biology. 19 (2018) 109–120. [DOI] [PubMed] [Google Scholar]
- [38].Klein C, Westenberger A, Genetics of Parkinson’s disease, Cold Spring Harbor perspectives in medicine. 2 (2012) a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shao J, Diamond MI, Polyglutamine diseases: emerging concepts in pathogenesis and therapy, Human molecular genetics. 16 Spec No. 2 (2007) R115–123. [DOI] [PubMed] [Google Scholar]
- [40].Li F, Macfarlan T, Pittman RN, Chakravarti D, Ataxin-3 is a histone-binding protein with two independent transcriptional corepressor activities, The Journal of biological chemistry. 277 (2002) 45004–45012. [DOI] [PubMed] [Google Scholar]
- [41].Gasset-Rosa F, Chillon-Marinas C, Goginashvili A, Atwal RS, Artates JW, Tabet R, Wheeler VC, Bang AG, Cleveland DW, Lagier-Tourenne C, Polyglutamine-Expanded Huntingtin Exacerbates Age-Related Disruption of Nuclear Integrity and Nucleocytoplasmic Transport, Neuron. 94 (2017) 48–57 e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fu Y, Wu P, Pan Y, Sun X, Yang H, Difiglia M, Lu B, A toxic mutant huntingtin species is resistant to selective autophagy, Nature chemical biology. 13 (2017) 1152–1154. [DOI] [PubMed] [Google Scholar]
- [43].Chung CG, Lee H, Lee SB, Mechanisms of protein toxicity in neurodegenerative diseases, Cellular and molecular life sciences : CMLS. 75 (2018) 3159–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Berendzen KM, Durieux J, Shao LW, Tian Y, Kim HE, Wolff S, Liu Y, Dillin A, Neuroendocrine Coordination of Mitochondrial Stress Signaling and Proteostasis, Cell. 166 (2016) 1553–1563 e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Brignull HR, Moore FE, Tang SJ, Morimoto RI, Polyglutamine proteins at the pathogenic threshold display neuron-specific aggregation in a pan-neuronal Caenorhabditis elegans model, The Journal of neuroscience : the official journal of the Society for Neuroscience. 26 (2006) 7597–7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Beck JS, Mufson EJ, Counts SE, Evidence for Mitochondrial UPR Gene Activation in Familial and Sporadic Alzheimer’s Disease, Current Alzheimer research. 13 (2016) 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Martinez BA, Petersen DA, Gaeta AL, Stanley SP, Caldwell GA, Caldwell KA, Dysregulation of the Mitochondrial Unfolded Protein Response Induces Non-Apoptotic Dopaminergic Neurodegeneration in C. elegans Models of Parkinson’s Disease, The Journal of neuroscience : the official journal of the Society for Neuroscience. 37 (2017) 11085–11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Karwaciak I, Pulaski L, Ratajewski M, Regulation of the human ABCB10 gene by E2F transcription factors, Genomics. 104 (2014) 520–529. [DOI] [PubMed] [Google Scholar]
- [49].Shirihai OS, Gregory T, Yu C, Orkin SH, Weiss MJ, ABC-me: a novel mitochondrial transporter induced by GATA-1 during erythroid differentiation, The EMBO journal. 19 (2000) 2492–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]