Abstract
Daily rhythms are generated by the circadian timekeeping system, which is orchestrated by the master circadian clock in the suprachiasmatic nucleus (SCN) of mammals. Circadian timekeeping is endogenous and does not require exposure to external cues during development. Nevertheless, the circadian system is not fully formed at birth in many mammalian species and it is important to understand how SCN development can affect the function of the circadian system in adulthood. The purpose of the current review is to discuss the ontogeny of cellular and circuit function in the SCN, with a focus on work performed in model rodent species (i.e., mouse, rat, hamster). Particular emphasis is placed on the spatial and temporal patterns of SCN development that may contribute to the function of the master clock during adulthood. Additional work aimed at decoding the mechanisms that guide circadian development is expected to provide a solid foundation upon which to better understand the sources and factors contributing to aberrant maturation of clock function.
Keywords: suprachiasmatic nucleus, circadian clock, postnatal development, clock genes
Graphical Abstract
Daily rhythms are generated by the circadian timekeeping system, which is orchestrated by the master circadian clock in the suprachiasmatic nucleus (SCN) of mammals. The SCN is a neural network of cellular clocks that interact with one another to determine the emergent properties of the system. Like other important neural circuits, the development of the SCN network is a gradual process that spans both embryonic and postnatal ages. This review discusses SCN development at the cellular and circuit levels, with a focus on work performed in model rodent species (i.e., mouse, rat, hamster). Particular emphasis is placed on the spatial and temporal patterns of SCN development that may contribute to clock function in adulthood.
I. Introduction
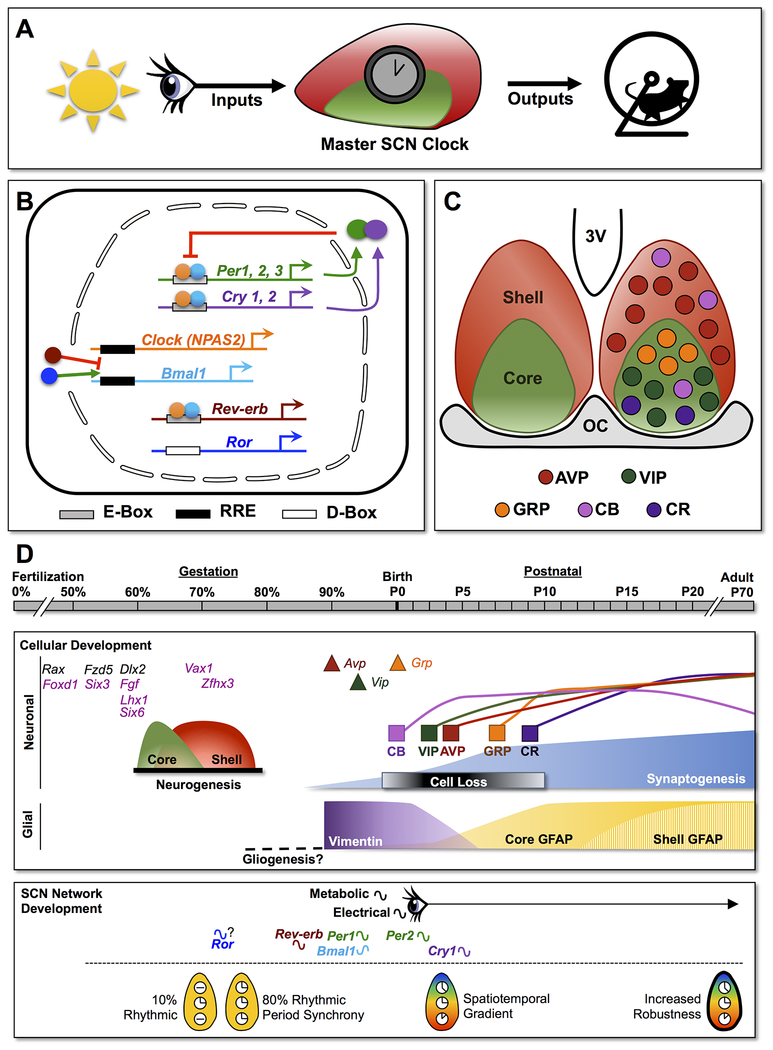
The circadian system generates daily rhythms that anticipate changes in the environment caused by the Earth’s rotation (Pittendrigh, 1960; Hut & Beersma, 2011). The master circadian clock in mammals is located in the suprachiasmatic nucleus (SCN) of the anterior hypothalamus (Weaver, 1998), which receives photic cues and provides daily signals to downstream tissues to coordinate the timing of overt rhythms (Figure 1A, (Mohawk et al., 2012)). Daily rhythms are endogenously generated and not “learned” through exposure to external cues. Nevertheless, evidence from a variety of mammalian species suggests that the circadian system undergoes postnatal maturation much like the rest of the brain. Human newborns start to display daily rhythms gradually over the first few months of life, with body temperature rhythms developing in the first week after birth, sleep:wake rhythms emerging in the first 1-2 months, and hormone rhythms appearing around 3 months of age (Kleitman & Engelmann, 1953; Price et al., 1983; Kennaway et al., 1996; McGraw et al., 1999; Rivkees, 2007). Further, circadian development can be affected by perinatal environmental conditions, which can influence health outcomes. For instance, 24/7 lighting in Neonatal Intensive Care Units decreases weight gain in babies born prematurely, increases latency to discharge from the hospital, and delays the timing of sleep:wake rhythm consolidation (Mann et al., 1986; Brandon et al., 2002; Vasquez-Ruiz et al., 2014). In addition, season of birth is associated with individual differences in chronotype (Mongrain et al., 2006; Natale & Di Milia, 2011), which can increase the risk of developing metabolic and mood disorders in adulthood (Erren et al., 2012; Tonetti et al., 2012; Merikanto et al., 2013; Yu et al., 2015). These studies indicate that the circadian system is sensitive during the perinatal period, and underscore the importance of understanding how development of the circadian system affects its operation in adulthood. In this review, we describe SCN development, with a focus on work performed in model rodent species (i.e., mouse, rat, hamster). Particular emphasis is placed on the spatial and temporal patterns of SCN development at the cellular and network level that may contribute to the function of the master clock during adulthood.
Figure1.
Development of the circadian timekeeping system. A. In adulthood, the suprachiasmatic nucleus (SCN) receives light input via the retinohypothalamic tract and provides daily outputs signals to downstream tissues to coordinate the timing of overt rhythms. B. Simplified model of the circadian molecular clock in mammals, which is composed of self-sustained transcriptional-translational feedback loops that regulate daily expression of clock genes and their protein products. In SCN neurons, the transcription factors CLOCK and BMAL1 dimerize and activate Period (Per) and Cryptochrome (Cry) expression during the day. After translation, PER and CRY dimers repress their own transcription at night. This core feedback loop is interlocked with other transcriptional loops that stabilize and augment circadian function. For example, REV-ERB and ROR regulate the daily expression of Bmal1. C. The shell-core model of the adult SCN network illustrating the spatial location of five neuronal subclasses in mice. AVP: Arginine Vasopressin, VIP: Vasoactive Intestinal Polypeptide, GRP: Gastrin-Releasing Peptide, CB: Calbindin; CR: Calretinin. Note: all five peptides are also expressed in rats and hamsters although the expression of CB and CR differs among rodent species. D. Timeline of SCN development. Important milestones are illustrated for SCN development in rodents. Most information illustrated on the timeline derives from studies using mice, but detailed information on the timing of fetal metabolic/electrical rhythms, Rev-erb rhythms, synaptogenesis, and glial maturation are only available for rats. Early genetic markers of SCN differentiation are labeled purple to represent those for which effects on SCN development have been reported. Color version of this figure can be viewed online.
In adulthood, the SCN is a network of cellular clocks that display daily rhythms in cellular physiology, including metabolism, electrical activity, gene/protein expression, and peptide release (Hastings et al., 2018). Circadian rhythms are generated at the cellular level by self-sustained transcriptional-translational feedback loops that regulate daily expression of clock genes and their protein products (Figure 1B, Takahashi, 2017). Briefly, the transcription factors CLOCK and BMAL1 dimerize to activate expression of Period (Per1, Per2) and Cryptochrome (Cry1, Cry2) genes, which are translated into protein products that repress their own transcription. This core feedback loop is interlocked with other transcriptional loops that stabilize and augment circadian function (Figure 1B, Takahashi, 2017). This molecular clock operates in a heterogeneous population of SCN cells that communicate with one another to function as a unit. A majority of SCN neurons express the transmitter GABA, but they can be classified into subclasses based on neuropeptide expression (Moore & Speh, 1993; Abrahamson & Moore, 2001). Classic models organize the SCN into two complementary compartments, known as the shell and the core, which are often distinguished using arginine vasopressin (AVP) in the SCN shell and vasoactive intestinal polypeptide (VIP) in the SCN core (Figure 1C, Moore et al., 2002; Antle et al., 2003). In general, this organizational scheme is consistent across many mammalian species, although it is a simplified model (Morin, 2007). In addition to neurochemical differences, SCN neurons in the shell and core regions differ in clock function and photic responses (Antle et al., 2003; Hastings et al., 2018), which may be established during network development under the guidance of cell-intrinsic and/or cell-extrinsic factors. Development of the circadian system has been covered in several excellent reviews published recently (Sumova et al., 2012; Landgraf et al., 2014; Bedont & Blackshaw, 2015), and the current review seeks to provide an update on recent progress made in this area (Figure 1D, see also Honma review, same issue).
II. SCN Cellular Development
The SCN is located within the ventral subdivision of the anterior hypothalamus. Similar to other parts of the brain, cellular development in the hypothalamus proceeds along several stages including proliferation and differentiation. Initial stages of cellular development in the rodent hypothalamus are largely completed during gestation, but latter stages can continue after birth. Embryonic age in rodents is defined typically by the presence of a vaginal plug on the morning after fertilization, which is labeled embryonic day 0.5 (E0.5) or embryonic day 1 (E1) depending on the conventions adopted by the laboratory. Because rodent models differ in the duration of gestation (i.e., mouse: 20 days, rat: 21-23 days, and hamster: 16 days), species differences in the timing and patterning of SCN development are considered below.
A. Hypothalamic Development
The progenitor cells that give rise to hypothalamus are located in the matrix layer surrounding the third ventricle (Bedont & Blackshaw, 2015). Hypothalamic development generally follows an outside-in pattern where neurons born early are displaced outward by neurons born later (Ifft, 1972; Shimada & Nakamura, 1973; Altman & Bayer, 1978), although this model may not apply to all hypothalamic regions (Markakis & Swanson, 1997; Padilla et al., 2010). In general, the anatomy and development of the hypothalamus is fairly well conserved across vertebrate species (Ware et al., 2014; Xie & Dorsky, 2017). Further, the timing of hypothalamic neurogenesis is fairly consistent across rodent species when embryonic age is normalized to the duration of the gestational period (GP). For instance, hypothalamic neurogenesis occurs in mice between E10-E16 (50-80% GP) and in rats between E11-E19 (50-85% GP).
During early stages of development, signals secreted by the ventral floor plate pattern the hypothalamus into discrete regions (Bedont & Blackshaw, 2015). Sonic hedgehog (encoded by Shh) is a lipid-linked polypeptide signal released from the ventral floor plate that is necessary for induction of the hypothalamus and drives Shh expression in the anterior hypothalamus. In mice, Shh is expressed by the anterior hypothalamus between E8.5-E10.5 (Alvarez-Bolado et al., 2012). Deletion of Shh from the basal hypothalamus eliminates expression of early genetic markers of the ventral anterior hypothalamus (Shimogori et al., 2010), including members of the LIM homeobox family (Lhx). In addition to SHH, the ventral floor plate releases other secreted factors that contribute to hypothalamic patterning, such as modulators of Wingless/Integrated (WNT) signaling (Rowitch & Kriegstein, 2010; Bedont & Blackshaw, 2015). These morphogenic cues can modulate several important processes during hypothalamic induction, including neurogenesis, differentiation and axon guidance.
During neurogenesis, the ventral anterior hypothalamus expresses a variety of genetic markers, including Lhx2, Retinal and anterior neural fold homeobox (Rax), Forkhead domain 1 (Foxd1), and NK2 homeobox 2 (Nkx2.2). The expression of these early markers is transient, starting before the onset of neurogenesis and ending with its conclusion (Shimogori et al., 2010; Bedont & Blackshaw, 2015; Newman et al., 2018). For example, Rax is expressed in the ventral hypothalamus of the mouse between E10.5-E14.5 (Pak et al., 2014). Deletion of Rax prior to E8.5 disrupts patterning of the mediobasal hypothalamus (Orquera et al., 2016), but later deletion causes aberrant rewiring in a specific progenitor pool (Lu et al., 2013). These observations suggest that early genetic markers are critical for the patterning of the hypothalamus and play more specific roles during later stages by affecting differentiation. Key events regulated by early genetic programs include the activation of additional transcription factors, such as Ventral anterior homeobox 1 (Vax1), Six homeobox 3 (Six3), and Six6 (Hallonet et al., 1999; Jean et al., 1999; Li et al., 2002; Lagutin et al., 2003), which are often retained after differentiation to maintain cell-type specificity. The SCN is similar to other regions of the hypothalamus in its early expression of many of the genes described above, and the influence of these genetic programs on SCN development is discussed in the following sections.
B. SCN Neurogenesis
The bulk of SCN neurogenesis occurs over a 3-4 day interval corresponding to 60-80% GP in mice, rats, and hamsters (Altman & Bayer, 1978; Davis et al., 1990; Kabrita & Davis, 2008). In the mouse, SCN neurogenesis occurs during E11.5-15.5 (58-78% GP), with a peak at E13.5 (Shimada & Nakamura, 1973; Kabrita & Davis, 2008). Similarly, SCN neurogenesis occurs between E13.5-E17 (58-75% GP) in rats (Ifft, 1972; Altman & Bayer, 1978), and between E9.5-E13.5 (60-84% GP) in hamsters (Crossland & Uchwat, 1982; Davis et al., 1990). Following the interval of neurogenesis, the SCN is clearly defined as a cluster of densely packed neurons with small soma (Moore & Bernstein, 1989).
Prior to SCN neurogenesis, Lhx2, Rax, Foxd1, and Nkx2.2 are expressed in the mouse hypothalamus, but these markers are absent after SCN neurogenesis is complete at E16 (Shimogori et al., 2010; Vandunk et al., 2011; Ferran et al., 2015). As in other hypothalamic regions, morphogen signaling is thought to influence SCN neurogenesis and development. Consistent with this idea, the WNT receptor Frizzled 5 (Fzd5) is detected at E10.5-13.5 specifically in cells that lack postmitotic markers (Vandunk et al., 2011). To our knowledge, the contribution of Fzd5 to SCN development has not been examined directly, but represents an interesting area for future work given its role in other circuits of the brain (Liu et al., 2008; Sarin et al., 2018). In addition, members of the Fibroblast growth factor (Fgf) family are expressed in the ventral anterior hypothalamus of the mouse at E11.5 (Ferran et al., 2015). Loss of Fgf signaling has been shown to reduce VIP expression and SCN volume at P0 (Miller et al., 2016), but consequences for circadian behavior after birth have not been explored.
The spatial patterning of neurogenesis across the ventrodorsal axis is consistent across rodent species, with SCN neurons in the ventral core of the network having an earlier birthdate than their counterparts in the dorsal shell (Altman & Bayer, 1978; Davis et al., 1990; Kabrita & Davis, 2008). In the mouse, SCN core neurons are born first (peak at E12, 60%GP), and SCN shell neurons are born at a later age (peak at E13.5, 68% GP) corresponding to the peak of proliferation in AVP neurons at E14 (Okamura et al., 1983; Kabrita & Davis, 2008). Similar ventral-to-dorsal patterning occurs in rats and hamsters (Altman & Bayer, 1978; Davis et al., 1990). The neurogenesis of specific types of SCN neurons has been examined most extensively in the hamster (Antle et al., 2005b). At E10.5-E11.5 (66-72% GP), neurogenesis occurs in SCN core neurons that later express VIP, Gastrin-Releasing Peptide (GRP), Substance P, or Calbindin. In contrast, genesis of AVP neurons occurs over a wider timeframe that extends into later embryonic ages (peak E10.5-E12.5, 66-78% GP). It has been suggested that SCN core and shell neurons derive from distinct progenitor pools in the neuroepithelium (Altman & Bayer, 1986), but the precise mechanisms that regulate cell-type differences in the timing of SCN neurogenesis are not well understood. On one hand, it is possible that cell fate is determined by intrinsic programs operating in neuronal precursors derived from distinct pools of progenitor cells. Another possibility is that extrinsic cues present in the microenvironment determine the maturation of SCN neurons proliferating at different times from the same progenitor pool. Which of these two models is most accurate for SCN development remains an open question, but both intrinsic and extrinsic factors have been shown to influence differentiation of GABA neurons in other brain regions (Quattrocolo et al., 2017).
There is also a spatial gradient of neurogenesis across the anteroposterior SCN, where the duration of neurogenesis is longer and ends later in the anterior SCN (Altman & Bayer, 1986; Davis et al., 1990; Kabrita & Davis, 2008). These studies indicate that SCN shell neurons are produced in at least two neurogenic waves, with those in the middle-posterior regions generated before those in the anterior pole. A similar posterior-to-anterior patterning of the SCN shell has been detected in mouse, hamster, and rat SCN (Altman & Bayer, 1986; Davis et al., 1990; Antle et al., 2005b; Kabrita & Davis, 2008), although the opposite gradient was reported initially in the rat (Altman & Bayer, 1978). In addition, work suggests that there may be sex- and/or hormonally driven differences in the timing of neurogenesis across this axis (Abizaid et al., 2004). Anteroposterior patterning of SCN neurogenesis is of potential interest because photoperiodic encoding occurs along this axis in adulthood (Hazlerigg et al., 2005; Inagaki et al., 2007). Thus, developmental patterning along the anteroposterior axis may influence photoperiodic modulation of behavior and physiology later in life.
The large majority of SCN neurogenesis is complete during gestation, but the number of SCN neurons increases by 50% over the first 2-5 days after birth (Muller & Torrealba, 1998; Ahern et al., 2013). Consistent with these findings, a low rate of cytogenesis in the rat SCN has been observed in the first week after birth (Seress, 1985). Further, neurogenesis has been noted to occur in the anterior SCN during puberty and adulthood in female rats using 5-bromo-2’-deoxyuridine (BrdU) to label proliferating neurons (Mohr et al., 2017), suggesting that the SCN may be an additional neurogenic niche in hypothalamus of adult mammals (Yoo & Blackshaw, 2018). Although BrdU can produce false positives (Duque & Rakic, 2011), inhibiting cell proliferation decreases adult SCN neurogenesis and delays the timing of the luteinizing surge (Mohr et al., 2017). Because reproductive rhythms are regulated by the SCN (Williams & Kriegsfeld, 2012), this raises the possibility that SCN neurons proliferating during adulthood may integrate into clock circuits to influence circadian function. The potential for adult SCN neurogenesis is especially interesting given the unique transcriptional profile displayed by adult SCN neurons, including low levels of NeuN-immunoreactivity (Morin et al., 2011) and continued expression of genes more commonly detected in pluripotent progenitor cells (Sato et al., 2011; Vandunk et al., 2011; Saaltink et al., 2012; Hoefflin & Carter, 2014; Brown et al., 2017; Beligala et al., 2018). Given this pattern of results, it would be interesting to test the potential role of these factors in the adult SCN and whether their expression is modulated by environmental factors that influence clock function (e.g., light).
C. Early Stages of SCN Differentiation
Early differentiation requires transcriptional changes that serve to restrict pluripotency and specify cell fate (Achim et al., 2014). One of the most well studied pathways contributing to this process involves Notch signaling (Ware et al., 2014). Notch signaling maintains progenitor cell pools and activates self-renewal genes, such as Hairy enhancer of split (Hes), which is a class E basic helix-loop-helix (bHLH) transcription factor that inhibits differentiation by suppressing the expression of pro-neuronal genes. Inactivation of Notch signaling antagonizes factors related to progenitor cell identity (e.g., Soxb1), causes exit from the cell cycle, and leads to up-regulation of pro-neuronal bHLH transcription factors in neuronal precursors. One such pro-neuronal factor is Achaete-Scute Family bHLH Transcription Factor 1 (Ascl1, also known as Mash1), which is expressed in precursors of hypothalamic GABA neurons between E11.5-13.5 (Allen Brain Atlas). Among its many gene targets, Ascl1 activates expression of Distal-less homeobox 1 (Dlx) and Dlx2 (Poitras et al., 2007; Castro et al., 2011), which are selector genes important for GABA neuron development (Pla et al., 2017). Dlx2 is expressed in the developing SCN of the mouse between E11.5-E14.5 (Vandunk et al., 2011), which is consistent with GABA synthesis in adult SCN neurons (Moore & Speh, 1993; Abrahamson & Moore, 2001).
During early stages of differentiation, the SCN expresses several genetic markers also found in other hypothalamic nuclei. As mentioned earlier, Lhx2, Rax, Foxd1, and Nkx2.2 are absent after the completion of SCN neurogenesis at E16 (Shimogori et al., 2010; Vandunk et al., 2011; Ferran et al., 2015). Despite being temporally restricted in expression, these genes activate genetic programs important for later stages of SCN development. For instance, Foxd1 is broadly expressed in the developing hypothalamus at E11.5, but loss of Foxd1 function causes developmental defects in the anterior hypothalamus that appear after Foxd1 levels have declined (Newman et al., 2018). SCN development is severely disrupted in Foxd1 deficient mice, with reduced expression of many genetic markers, loss of nuclear organization, and agenesis by P0.5 (Newman et al., 2018). The exact mechanisms by which the loss of Foxd1 leads to delayed defects in SCN development are unknown, and consequences for clock function are difficult to assess because Foxd1 deficient mice die within 24 h after birth (Hatini et al., 1996; Newman et al., 2018). But Foxd1 deficient mice display reduced expression of Vax1 and Six3 at E12.5 (Newman et al., 2018), indicating that early genetic programming is disrupted. Vax1, Six3, and Six6 are expressed diffusely across the ventral anterior hypothalamus in mice from E11.5-14.5, but the expression of these genes becomes more restricted to the SCN region by E15.5-16.5 (Shimogori et al., 2010; Vandunk et al., 2011; Ferran et al., 2015; Newman et al., 2018). Consistent with developmental defects evident in Foxd1 deficient mice, loss of Vax1, Six3 or Six6 disrupts SCN differentiation and circadian-control of behavior later in life (Vandunk et al., 2011; Clark et al., 2013; Hoffmann et al., 2016). Collectively, this work establishes that early hypothalamic markers are necessary for normal SCN development, likely by controlling the activation of genetic programs important for its later specification and maintenance.
The first selective SCN marker in the mouse is Lhx1, which is expressed throughout the SCN at E11.5 and becomes restricted to the central SCN by P0 (Vandunk et al., 2011). Earlier transcriptional activators are necessary for Lhx1 expression because loss of Lhx2, Foxd1, or Six3 will disrupt its normal patterning during SCN development (Vandunk et al., 2011; Roy et al., 2013; Newman et al., 2018). Lhx1 directly activates transcription of Vip (Hatori et al., 2014), and Lhx1 binding occurs at many other genes with enriched SCN expression (Bedont et al., 2014). In line with this, loss of Lhx1 in the ventral anterior hypothalamus reduces SCN levels of several neuropeptides known to be important regulators of master clock function, such as Vip, Avp, Prokineticin 2 (Prok2), and Grp (Bedont et al., 2014; Hatori et al., 2014). In addition, Lhx1 deficient mice display a 40% loss of SCN neurons by P4 due to increased cell death over the first few days after birth (Bedont et al., 2014). Consistent with reduced neuronal complement and neuropeptide expression, loss of Lhx1 disrupts SCN molecular and electrical rhythms by attenuating phase coherence among SCN neurons (Bedont et al., 2014; Hatori et al., 2014). Lastly, Lhx1 deficient mice display pronounced changes in circadian behavior, with loss of daily rhythms under free-running conditions, altered responses to photic stimuli, and changes in sleep (Bedont et al., 2014; Hatori et al., 2014; Bedont et al., 2017). Collectively, these studies reveal that the early activation of Lhx1 is critical for the proper development of the SCN network, and that its loss results in a failure to maintain terminal differentiation and prevent death of SCN neurons after birth.
A number of late genetic markers are expressed at E13.5 or later, including Zinc finger homeobox 3 (Zfhx3), POU Class 2 homeobox (Pou2f2, also known as Oct2), and the clock genes RAR-related orphan receptor a (Rora) and Rorb (Allen Brain Atlas, Rivkees et al., 1992; Vandunk et al., 2011). Recent work has revealed that Zfhx3 regulates the expression of Vip and other SCN transcripts, the period of free-running rhythms, and the function of the sleep homeostat (Parsons et al., 2015; Balzani et al., 2016). However, effects of the Zfhx3 mutation do not require its expression during development (Wilcox et al., 2017), indicating that Zfhx3 acts to regulate SCN function in adulthood by activating transcription at AT motifs (Parsons et al., 2015). Lastly, the clock genes Rora and Rorb are late stage markers of SCN development. Rora is expressed in the ventral SCN starting at E14.5, throughout the SCN at E17.5, and in a pattern more restricted to the SCN shell by P21 (Vandunk et al., 2011; Newman et al., 2018). Appropriate expression of Rora/b depends on earlier transcriptional programs, including Foxd1 and Six3 (Vandunk et al., 2011; Newman et al., 2018). But loss of Rora itself does not disrupt terminal differentiation of VIP or AVP neurons in the SCN (Vandunk et al., 2011), consistent with the later timing of Rora expression relative to other genetic markers. In contrast, a loss of function mutation in the Clock gene reduces the postnatal expression of VIP and AVP (Herzog et al., 2000). This is quite interesting given that CLOCK and BMAL are members of the bHLH class C transcription factor family (Takahashi, 2017), and clock gene expression has been implicated in the control of neurogenesis and differentiation in neurogenic zones of the adult brain (Borgs et al., 2009; Bouchard-Cannon et al., 2013). For instance, adult Per2 and Bmal1 knockout mice display an increased number of newly born and undifferentiated cells in neurogenic zones (Borgs et al., 2009; Bouchard-Cannon et al., 2013). In addition, neurospheres and stem cells cultured in vitro develop molecular rhythms after neuronal differentiation (Yagita et al., 2010; Malik et al., 2015), which is abrogated upon cellular reprogramming back into pluripotent cells (Yagita et al., 2010). Further, loss of Clock function adversely affects maturation of neocortical circuits (Kobayashi et al., 2015). Collectively, this work suggests that the molecular clock can regulate cellular differentiation and circuit formation, but it is also possible that clock genes contribute to this process through non-circadian related mechanisms. It remains unknown whether the onset of molecular clock function gates SCN development and/or maturation beyond its role in cellular timekeeping.
At E16, SCN progenitors and post-mitotic neuronal precursors display distinct transcriptional profiles (Shimogori et al., 2010). Many genes important for SCN differentiation are maintained after birth (Vandunk et al., 2011; Brown et al., 2017), and expression of Six3, Six6, Lhx1, and Rora fluctuates in a rhythmic manner under either entrained or constant conditions (Vandunk et al., 2011; Clark et al., 2013; Hatori et al., 2014). One of the final stages of terminal differentiation is the adoption of a specific subclass fate, typically defined by neuropeptide expression in SCN neurons (Moore & Speh, 1993; Abrahamson & Moore, 2001; Southey et al., 2014). The timing of GABA and neuropeptide expression will be discussed in a later section because intercellular signaling among SCN neurons is essential for cohesive clock function at the tissue level.
D. Process Elongation and Synaptogenesis
Newly differentiated neurons undergo elongation of axonal and dendritic processes, ultimately culminating in the genesis of synapses (Tretter & Moss, 2008; Robichaux & Cowan, 2014). These two interrelated stages are orchestrated by a variety of mechanisms (e.g., signaling gradients, contact receptors, cell adhesion molecules), which prompt reorganization of the cytoskeletal proteins actin and tubulin in developing neurons. It has been estimated that the adult rat SCN contains at least 10 × 107 synapses (Guldner, 1976; Moore & Bernstein, 1989), and that many of these connections arise from neurons located within the nucleus itself (van den Pol, 1980; van den Pol & Tsujimoto, 1985). To our knowledge, this developmental stage has not been examined in the mouse, but there are a handful of anatomical studies investigating synaptogenesis in the rat or hamster SCN (Lenn et al., 1977; Moore & Bernstein, 1989; Laemle et al., 1991; Speh & Moore, 1993). These studies suggest that there is a progressive increase in the number and complexity of synapses over the early postnatal period, with two intervals of rapid growth corresponding to a large increase in synapse diversity from P0-P6 and a rapid increase in synapse number from P6-P10 (Lenn et al., 1977; Moore & Bernstein, 1989).
At E19, SCN neurons in the rat are surrounded by sparse neuropil containing dendritic growth cones and occasional bundles of unmyelinated axons (Moore & Bernstein, 1989). The neuropil and primary dendrites have matured by P6 (Moore & Bernstein, 1989), although further increases in complexity have been noted after P14 (Lenn et al., 1977) possibly due to glial maturation (Munekawa et al., 2000). From P2-P6, the diversity of synapses increases, with the appearance of symmetrical, asymmetrical, and intermediate synapses containing clear and/or dense core vesicles abutting dendritic processes that are variable in size (Lenn et al., 1977). However, at P6, less than 50% of axon terminals and less than 30% of synapses are present compared to the adult rat SCN (Moore & Bernstein, 1989). During this early postnatal interval, 80% of dissociated SCN neurons collected from 1-3 day old rats will develop spontaneous inhibitory currents within 4 days in vitro (Welsh et al., 1995). These electrical responses may be supported either by a low percentage of synapses or could be driven by non-synaptic release of GABA (Demarque et al., 2002). By P10, the complement of axon terminals reaches adult levels and the number of synapses has risen to 70% (Moore & Bernstein, 1989). The remaining synapses may develop in a third wave between P21-P35 (Lenn et al., 1977). Similarly, synaptogenesis in the hamster SCN occurs from E16-P4, but has not been investigated as extensively as in the rat (Speh & Moore, 1993).
It is not clear what mechanisms drive process elongation and synaptogenesis in the developing SCN (Tretter & Moss, 2008; Robichaux & Cowan, 2014). Cell adhesion molecules have been implicated in the process of establishing synaptic connections, but whether these systems direct formation and/or maintenance of SCN circuits remains unclear. In the rat SCN, expression of the cell adhesion molecules NCAM and L1 increases between P7-P14 (Yamada et al., 2003). Interestingly, disrupting the function of polysialylated NCAM or L1 in adulthood can impair behavioral rhythms of rats and mice free-running under constant conditions (Yamada et al., 1999; Shen et al., 2001). Further, loss of NCAM-180 increases the number of VIP neurons in the mouse SCN by 2-3 fold, thus raising the possibility that polysialylated NCAM regulates signaling mechanisms important for SCN function. These studies suggest that cell adhesion molecules regulate SCN development and influence cellular interactions in adulthood either directly or indirectly by changing circuit architecture. In addition, gap junctions have been implicated in the development of chemical synapses in other neural systems (Hormuzdi et al., 2004), and SCN cells can be linked by gap junctions (Welsh & Reppert, 1996; Colwell, 2000; Long et al., 2005). Connexin36 is a subunit of gap junctions specifically expressed in neurons, and it has been shown that inhibiting the function of gap junctions eliminates electrical coupling among SCN neurons and deleting connexin36 decreases the amplitude of daily rhythms in behavior (Long et al., 2005; Wang et al., 2014). However, recent work reveals that loss of connexin-36 in adult mice does not impair circadian function at the behavioral or molecular levels (Diemer et al., 2017). Collectively, these results suggest that signaling through connexin36-expression gap junctions modulates short-term electrical coupling among SCN neurons, but this mechanism is not necessary for long-term synchronization among SCN neurons at the level of the molecular clock. Overall, much remains unknown about SCN synaptogenesis, but recent advances in imaging and genetic approaches may provide deeper insight into how these important circuits are constructed and stabilized.
E. Neuronal Loss
It has been estimated that approximately 50% of neurons produced in the vertebrate brain are lost during development due to a variety of mechanisms (Fricker et al., 2018). The activation of caspase and pro-apoptotic members of the Bcl-2 family is a common cause of cell loss during the post-mitotic phase of development, which overlaps with synaptogenesis. Both cell-intrinsic programs and cellular interactions can activate apoptotic pathways during this time. A large number of SCN neurons is lost during the perinatal period, but the mechanisms that activate apoptotic programs in SCN neurons are unclear. In hamsters, there is a 40% decline in neuronal number over P3-P6 that reduces SCN volume to that typical of adults (Muller & Torrealba, 1998). A similar pattern of developmental cell loss is observed in the mouse SCN (peak P3-P5) using caspase-3 activation (Ahern et al., 2013) or DNA fragmentation as a marker of cell death (Bedont et al., 2014). The timing and patterning of SCN cell death is comparable to other hypothalamic nuclei (e.g., paraventricular nucleus, ventromedial nucleus), suggesting that this is a general process in this region (Ahern et al., 2013). Although the most precipitous cell death occurs in the mouse SCN by P7, an additional 20% cells are lost by adulthood (Bedont et al., 2014). Early genetic programs influence perinatal cell loss because Lhx1 deletion causes an additional 40% reduction in the number of SCN neurons between P0-4 (Bedont et al., 2014). Cell loss in the Lhx1 mutant is preceded by reduced peptide expression, suggesting that cell survival is dependent on terminal differentiation. However, it is unknown if specific types of SCN neurons are lost during normal cell death and whether the function of the network changes due to their failure to integrate into the circuit.
F. Glial Development
Throughout the central nervous system, glia are generated after the period of neurogenesis (Rowitch & Kriegstein, 2010). This temporal sequence is thought to reflect that radial glia produce different classes of progenitor pools at early versus late stages of development, with early pools of progenitors producing neurons and later pools producing glia (Tabata, 2015). Earlier-born neural cells influence gliogenesis by releasing extrinsic signals, such as cytokines and Notch ligands (Rowitch & Kriegstein, 2010). Lastly, macroglia can be divided into three distinct types: protoplasmic astrocytes (Aldhl1+, GFAP+ or GFAP-), fibrous astrocytes (AldhL1+, GFAP++), and oligodendrocytes (MBP+, PLP+, APC+), which may be generated from common or independent progenitor pools (Rowitch & Kriegstein, 2010).
SCN astrocytes have been detected in adult mice, rats, and hamsters using either GFAP or Aldh1L1 expression as markers (Guldner, 1983; Morin et al., 1989; Brancaccio et al., 2017; Tso et al., 2017). Morphological analyses indicate that the rat SCN contains both protoplasmic and fibrous astrocytes that can make appositions onto VIP+ neurons and AVP+ neurons (Tamada et al., 1998). Much like neurons, astrocytes can display daily rhythms in cellular physiology, including those in structural protein expression (Gerics et al., 2006; Lindley et al., 2008), morphological arrangements (Becquet et al., 2008), metabolic function (Womac et al., 2009; Burkeen et al., 2011) and clock gene/protein levels (Prolo et al., 2005; Cheng et al., 2009; Duhart et al., 2013; Tso et al., 2017). Recent work indicates that SCN astrocytes can influence GABA and glutamate signaling in the SCN (Barca-Mayo et al., 2017; Brancaccio et al., 2017), and that SCN astrocytes affect SCN function in vitro (Brancaccio et al., 2017; Tso et al., 2017). Further supporting a glial role in clock function, modulating the molecular clock specifically in astrocytes will alter the period of circadian rhythms under constant conditions (Barca-Mayo et al., 2017; Tso et al., 2017). Lastly, anatomical and functional evidence suggests that SCN astrocytes may influence photic resetting (Lavaille & Serviere, 1995; Tamada et al., 1998; Moriya et al., 2000; Lavialle et al., 2001; Girardet et al., 2010), but deficits in photic responses have not been reported in recent studies genetically manipulating the astrocyte clock.
Although there is a growing appreciation that astrocytes contribute to clock function, the development of SCN glia is not well understood. Most studies have used structural markers to track the development of glia in the SCN. For example, vimentin expression (a marker of radial glia) is high in the SCN prior to birth, but decreases markedly from P3-P6 in hamsters and rats (Botchkina & Morin, 1995; Munekawa et al., 2000). This likely reflects loss of radial glial progenitors, and its timing is consistent with the postnatal period of cell death in the hamster (Muller & Torrealba, 1998). In both species, there is a postnatal increase in GFAP+ processes that complements the decreasing expression of vimentin. In the hamster SCN, GFAP is first expressed at E15, but large numbers of GFAP+ processes are not detected until P0 (Botchkina & Morin, 1995). From P3-P10, the number of GFAP+ astrocytes increases further, with expansion continuing into adulthood. Similar development of GFAP expression occurs in the rat SCN, with GFAP expression first evident at E20 and postnatal expansion of GFAP+ processes (Munekawa et al., 2000). Interestingly, this latter study found that GFAP expression appeared in the core and shell SCN at different ages. Specifically, GFAP+ processes first appeared in the ventral SCN and increased from P3-4, but GFAP+ processes did not appear in the dorsal SCN until P12. From P20-P25, GFAP+ increased in both ventral and dorsal SCN, but this process can be affected by changes in light input (Munekawa et al., 2000; Yamazaki et al., 2002; Ikeda et al., 2003; Cambras et al., 2005). Collectively, these studies suggest that different pools of SCN astrocytes developing at discrete postnatal stages are sensitive to changes in the perinatal environment. Going forward, deeper insight into SCN macroglia can be gained from fate mapping studies that track astrocyte lineage and transcriptional programming (Rowitch & Kriegstein, 2010; Tabata, 2015). A greater understanding of SCN gliogenesis may allow for more precise ways to manipulate SCN macroglia and their progenitors at critical stages of development.
III. SCN Network Development
SCN neurons coordinate their cellular rhythms at the population level (Hastings et al., 2018), and thus the appearance of tissue-level rhythms can be used to infer when SCN circuits have matured. The molecular clock generates circadian rhythms at the cellular level, and intercellular interactions among SCN neurons synchronize, amplify, and stabilize these cellular rhythms (Herzog et al., 2004). In addition, interactions among SCN neurons generate spatiotemporal gradients in electrical and molecular activity at the network level, which can be adjusted to encode environmental conditions (Meijer et al., 2010; Evans & Gorman, 2016). These emergent properties reflect intercellular communication, and numerous chemical signals are produced in the SCN that may modulate network function via synaptic and/or non-synaptic signaling (Castel et al., 1996; Yamaguchi et al., 2003; Deery et al., 2009; Maywood et al., 2011). Specification of neurochemical identity is an important step in terminal differentiation, but postnatal changes in chemical signals are not uncommon. Thus, the role of these chemical signals may vary over development in ways that have important consequences for network maturation (see also Honma review, same issue). In this section, we review the ontogeny of SCN rhythms and the development of the specific chemical signals that are best understood at this time.
A. Development of the SCN Clock
A long-standing question in the field is when the SCN first starts to function as clock. Daily fluctuations in metabolic and electrical activity have been detected during late fetal development in rats using a variety of in vivo and acute ex vivo methods. Metabolic rhythms in the rat SCN manifest at E19, with high daytime levels of glucose utilization as in adulthood (Reppert & Schwartz, 1983; 1984; Shibata et al., 1987). Further, SCN slices from fetal rats display higher daytime firing as early as E22 when measured with an ex vivo between-group sampling method (Shibata & Moore, 1987). In this study, daytime firing was low and irregular at E22, but from P1-P14 gradually adopted more adult-like properties (i.e., increase in regular firing, mean firing rate, and rhythm amplitude). Collectively, these studies reveal that daily rhythms in cellular physiology are expressed prior to birth, which is an age when few synapses are present in the rat SCN (Lenn et al., 1977; Moore & Bernstein, 1989). Further, changes in SCN electrical rhythms after birth indicate that additional maturation of the circuit occurs during the postnatal period of synaptic elaboration and diversification.
Daily rhythms of in vivo clock gene expression also appear during late gestation, although clock genes can become rhythmic at different ages. As mentioned earlier, Rora is first expressed in the mouse at E13.5 and is found throughout the SCN by E17.5 in this species (Vandunk et al., 2011), which is consistent with high constitutive expression of its target Bmal1 at E18 (Ansari et al., 2009). By E18, Per1 and PER1 are rhythmically expressed in mouse SCN (Shimomura et al., 2001; Ohta et al., 2003; Ansari et al., 2009), suggesting that the core molecular loop is functional prior to birth. Studies disagree on whether Per2 and PER2 start to cycle before or after birth (Shimomura et al., 2001; Ohta et al., 2003; Ansari et al., 2009; Huang et al., 2010), which may reflect differences in methods or strain/sex of mice used. The last of the core clock components to oscillate appears to be Cry and CRY, with clear daily rhythms only emerging after P2 (Ansari et al., 2009; Huang et al., 2010). This dovetails with work indicating that Cry genes are not required for SCN rhythms until 1-2 weeks after birth (Ono et al., 2013; 2016; see also Honma review, same issue). Similar staging of molecular rhythms is observed in rats and hamsters, with high early expression of Bmal1 and maturation of molecular rhythms spanning into postnatal life (Sladek et al., 2004; Li & Davis, 2005; Kovacikova et al., 2006; Houdek & Sumova, 2014). In studies where both positive and negative clock genes are included, it appears that Per1/2 and Bmal1 oscillate out of phase in vivo when they are both rhythmically expressed (Sladek et al., 2004; Li & Davis, 2005; Kovacikova et al., 2006; Houdek & Sumova, 2014). Interestingly, a recent study found that Rev-erbα is one of the earliest clock genes to oscillate at E19 in the rat SCN (Houdek & Sumova, 2014). Given that Rev-erbα (also known as Nr1d1) negatively regulates Bmal1 expression by competing with Rora (Takahashi, 2017), this suggests that the rhythmic regulation of Bmal1 may be one of the earliest events in the development of the molecular clock. Another consistent pattern across studies and species is that age-related changes in the amplitude and/or phasing of clock gene rhythms manifest in a gene-specific manner, which may reflect further maturation of the molecular clock and/or the influence of external factors (e.g., parturition, light).
Another important question is when the SCN is able to sustain rhythms without maternal influence. This issue has been addressed by examining SCN rhythms over time in culture, which also allows one to study the emergent properties of the network at the cellular and population levels. Pioneering studies employing this approach have demonstrated that SCN neurons collected from P2-P20 mice can sustain electrical and molecular rhythms in vitro (Yamaguchi et al., 2003; Maywood et al., 2006; Ohta et al., 2006; Enoki et al., 2012; Maywood et al., 2013). Recent developmental studies using this ex vivo technique have revealed that SCN molecular rhythms are expressed by E15.5 in mice (Wreschnig et al., 2014; Landgraf et al., 2015; Carmona-Alcocer et al., 2018) and by E20 in rats (Nishide et al., 2014). Similar to the development of SCN electrical rhythms, the strength and precision of molecular rhythms continues to increase with age. Real-time imaging of molecular rhythms revealed that only 10% of SCN neurons oscillate at E14.5, but 70% are able to oscillate with high precision by E15.5 (Carmona-Alcocer et al., 2018). Competence of SCN rhythmicity at this age is consistent with work demonstrating that rhythms in SCN-lesioned adults can be restored 90% of time by SCN transplants collected from E14-15 fetuses (Kaufman & Menaker, 1993; Romero et al., 1993). Further, SCN neurons appear capable of forming a cohesive network early in development because period synchrony is evident at E15.5 and the spatiotemporal gradient emerges by P2 (Carmona-Alcocer et al., 2018). Interestingly, phase relationships among different clock genes may also mature late in development because dual real-time reporters indicate the relative phasing of Bmal1 and Per1 rhythms is less stable at P7 compared to adulthood (Ono et al., 2017). Collectively, this work indicates that the maturation of the SCN clock occurs in stages, with early development of oscillator capacity but later development of network properties. Delayed network development is consistent with findings that the SCN is sensitive to perturbation early in development (Nishide et al., 2008; Wreschnig et al., 2014; Landgraf et al., 2015), which may reflect lack of robustness (Herzog & Huckfeldt, 2003; Buhr et al., 2010). Further, period synchronization at E15.5 does not require GABA or VIP signaling (Wreschnig et al., 2014; Carmona-Alcocer et al., 2018), indicating that novel signaling mechanisms operate early in development (see also Honma review, same issue).
B. Development of Specific Chemical Signals
1. GABA (γ-Aminobutyric Acid)
GABA is the main neurotransmitter produced by SCN neurons (Moore & Speh, 1993; Abrahamson & Moore, 2001), and it is expressed in different peptidergic classes (Francois-Bellan et al., 1990; Castel & Morris, 2000). Synthesis of GABA is controlled by glutamic acid decarboxylase (GAD), and the adult SCN expresses both the GAD65 and GAD67 isoforms (O’Hara et al., 1995; Huhman et al., 1996; Castel & Morris, 2000). Synaptic release of GABA requires packaging by the vesicular GABA transporter VGAT, (encoded by Slc32a1), which is a general marker for GABA neurons (Chen et al., 2017). Lastly, cellular responses to GABA are mediated by GABAA and GABAB receptors. The GABAA receptor is a heteropentameric ligand-gated ion channel permeable to chloride and bicarbonate, with synaptic or extra-synaptic GABAA receptors that differ in subunit composition and functional properties (Albers et al., 2017). In the SCN, many GABA responses are driven by GABAA receptor signaling (Jiang et al., 1997; Strecker et al., 1997; Liu & Reppert, 2000), but the Gαi/o coupled GABAB receptor is also expressed (Gribkoff et al., 2003; Belenky et al., 2008).
GABA signaling modulates properties of the SCN network and photic responses in adulthood (reviewed in Albers et al., 2017), but surprisingly little is known about its development in the SCN. During brain development, GABA signaling regulates the formation of neural circuits by influencing cellular proliferation, neuronal differentiation, dendritic outgrowth, synapse formation, and circuit refinement (Akerman & Cline, 2007). In many developing networks, GABA is the first transmitter to become functional, but this signaling mechanism can operate differently in the perinatal versus adult brain. For instance, GABA can be released as a paracrine signal prior to synaptogenesis, which is facilitated by low transporter activity in perinatal rats and mice (Demarque et al., 2002). In the rat hypothalamus, GABA is expressed in neuronal soma, dendritic growth cones, and growing axons as early as E15 (van den Pol, 1997). Consistent with this embryonic window of GABA expression, Gad1 has been detected in the ventral anterior hypothalamus as early as E16 (Shimogori et al., 2010), and it is likely that the SCN displays postnatal increases in GAD expression similar to that reported for other hypothalamic regions (Puymirat et al., 1982; Popp et al., 2009). Further, Vgat can be detected at low levels in the SCN at E18.5 (Allen Brain Atlas), which is consistent with the timing of Vgat expression in the mediobasal hypothalamus (Kobayashi et al., 2017). After birth, there is a large increase in SCN Vgat expression over the postnatal period of synaptogenesis (Allen Brain Atlas). In general, very little is known about the developmental patterning of GABA receptors in the SCN, however, the postnatal presence of GABAA receptors can be inferred from functional responses to their pharmacological manipulation (Kawahara et al., 1993; Welsh et al., 1995; Ikeda et al., 2003a). Further, other hypothalamic regions display postnatal swapping of GABAA receptor subunits (Fritschy et al., 1994; Davis et al., 2000), but GABAA subunits have been examined in the adult SCN only (Gao et al., 1995; O’Hara et al., 1995; Walton et al., 2017).
One interesting feature of GABA responses during development is that there is a postnatal switch from depolarization to hyperpolarization (Ben-Ari et al., 2012). In the rat hypothalamus, the developmental switch in GABA responses has been estimated to occur from P4-P10 (Obrietan & van den Pol, 1995), but studies tracking developmental changes in SCN responses to GABA are scarce. In one study, application of GABA was found to increase [Ca+2]i in 46% of dissociated SCN neurons collected from E18 rats and cultured for 4 days (Obrietan & van den Pol, 1995). Similarly, application of the GABAA receptor antagonist bicuculline after 6 days in culture depressed [Ca+2]i in 50% of these SCN neurons, suggesting that basal [Ca+2]i is elevated by GABA released from perinatal SCN neurons. By 18 days in culture, however, GABA increased [Ca+2]i in only 13% of SCN neurons. Although dispersed cell cultures may not fully recapitulate the in vivo state, another study has examined [Ca+2]i responses in SCN slices collected from mice at discrete postnatal ages (Ikeda et al., 2003a). Overall, this study demonstrates that GABAA receptor activation increases [Ca+2]i in 60-65% of P6-7 SCN neurons, but this is reduced to 26-40% by P9-P10. Further, the P9-P10 SCN displays a daily rhythm in the [Ca+2]i response to GABAA receptor activation that is not present at P6-P7 (Ikeda et al., 2003a). These results suggest that at least a subset of SCN neurons develop phase-dependent responses to GABAA activation, but it is unknown if this process is influenced by maturation of the molecular clock, cell-type specific programming, the stabilization of intra-network connections, and/or the arrival of afferent inputs. The factors that regulate this developmental process in the SCN are especially intriguing given that excitatory GABA responses are reported in the adult SCN (reviewed in (Albers et al., 2017)) and GABA signaling remains plastic in the adult SCN (Farajnia et al., 2014; Myung et al., 2015). Thus, it is interesting to consider how this developmental process may sculpt GABA responses of adult SCN subclasses and their capacity for plasticity later in life.
2. VIP (Vasoactive Intestinal Polypeptide)
VIP is a member of the secretin family and binds to VPAC1/2 receptors with high affinity (Couvineau & Laburthe, 2011). In the SCN, VIP acts through the VPAC2 receptor, which is expressed by a majority of SCN neurons in the mouse (King et al., 2003; Kallo et al., 2004). When applied to the SCN, VIP and VPAC2 agonists modulate membrane potential, firing rate, [Ca+2]i, and GABA signaling (Reed et al., 2002; Itri & Colwell, 2003; Irwin & Allen, 2010; Kudo et al., 2013). Further, VIP signaling can alter electrical and molecular rhythms in the SCN, which requires activation of PKA, MAPK and PLC signaling pathways (Nielsen et al., 2002; Meyer-Spasche & Piggins, 2004; An et al., 2011). Loss of VIP signaling will hyperpolarize SCN neurons, decrease the number capable of sustaining circadian rhythms, and compromise synchronization among those that remain oscillating (Aton et al., 2005; Brown et al., 2005; Maywood et al., 2006; Brown et al., 2007). The importance of VIP is further illustrated by the wide range of circadian phenotypes displayed by mice deficient in VIP signaling, including low-amplitude, short period under free-running conditions, increased incidence of arrhythmia under constant darkness, accelerated recovery from simulated jetlag, deficits in photoperiodic encoding, and altered responses to constant light (Harmar et al., 2002; Colwell et al., 2003; Aton et al., 2005; Bechtold et al., 2008; Lucassen et al., 2012; An et al., 2013; Loh et al., 2014; Hughes et al., 2015). Collectively, these studies indicate that VIP signaling is required to sustain normal SCN function in adulthood and suggest that insight into its functional role may be gained by examining developmental changes in VIP expression.
The onset of Vip expression occurs early in SCN development for many rodent species. In mice, Vip expression occurs by E18.5, which corresponds to 93% GP (Vandunk et al., 2011), which is 3.5 days after the end of neurogenesis in the SCN core in this species (Kabrita & Davis, 2008). This is followed by expression of VPAC2 at P0 and VIP at P2 (Carmona-Alcocer et al., 2018), with further increases in VIP expression between P6-P30 (Herzog et al., 2000). Rats display similar developmental patterns, with the onset of Vip and VIP expression by E18 (79% GP), increased expression at P0, and two postnatal waves of increased expression between P5-P10 and P10-P20 (Laemle, 1988; Ban et al., 1997; Houdek & Sumova, 2014). In addition, hamsters display VIP by E13-E14 (80-88% GP) with postnatal increases in expression (Romero & Silver, 1990; Botchkina & Morin, 1995). In all three rodent species, there is postnatal expansion of VIP processes, with a dense plexus formed by P30 in mice (Herzog et al., 2000), by P20 in rats (Ban et al., 1997), and by P10 in hamsters (Botchkina & Morin, 1995). Interestingly, VIP signaling does not appear to be required for SCN synchrony at E15.5 (Wreschnig et al., 2014; Carmona-Alcocer et al., 2018), suggesting that other signaling mechanisms support network function at this age. The loss of VIP signaling reduces the expression of other SCN neuropeptides in adulthood, such as Avp and Prok2 (Harmar, 2003; Bedont et al., 2014), which could reflect loss of SCN synchrony during adulthood or a developmental deficit in network maturation. With this in mind, it may be interesting to examine whether levels of these other neuropeptides are rescued by treatments that restore intercellular synchrony in VIP deficient SCN (Aton et al., 2005; Maywood et al., 2006; Brown et al., 2007; Hughes et al., 2015).
The spatial patterning of postnatal changes in VIP expression has been investigated in both mice and rats, with development in both species characterized by VIP expression expanding into the middle and posterior SCN. In the rat, there are also medial-lateral differences in the timing of Vip expression, with two spatially distinct clusters of VIP neurons that differ in daily VIP expression and molecular responses to light (Ban et al., 1997; Kawamoto et al., 2003). This suggests that there may be two subsets of VIP neurons that differ in the timing of maturation and function within the network. Interestingly, recent work has revealed that VIP neurons in mice can be divided into two subsets based on electrical firing (i.e., tonic versus irregular), and that rapid entrainment can be achieved in vitro or in vivo by driving high frequency firing from VIP neurons with optogenetic stimulation (Mazuski et al., 2018). Collectively, these data suggest the VIP neurons can be divided into at least two subclasses, which may differ in transcriptional programming, cellular physiology, and functional roles.
In addition to the developmental increase in VIP levels, there is an age-related change in the daily rhythm of VIP expression. A daily rhythm in Vip expression is observed in rats as soon as this transcript is detected at E19, but the phasing and amplitude of the rhythm is altered by E21 (Houdek & Sumova, 2014). Changes also occur at postnatal ages, with an inversion of the Vip rhythm under free-running conditions between P10 and P20 (Ban et al., 1997) and a reduction in the amplitude of the VIP rhythm under entrained conditions between P4 and P20 (Isobe & Muramatsu, 1995). During this early postnatal window, VIP can be released rhythmically from organotypic SCN slices collected from P5-P6 rats (Shinohara et al., 1995; Nakamura et al., 2001). But in adult rats, the endogenous rhythm of Vip and VIP expression under constant darkness is lost (Shinohara et al., 1993; Ban et al., 1997), suggesting that mechanisms engaged later in life alter the circadian regulation of this peptide. The neurobiological bases and functional implications of these developmental changes are unknown.
3. AVP (Arginine Vasopressin)
Recent work indicates that signaling by AVP neurons regulates SCN function in ways beyond its role as an output to downstream tissues (Kalsbeek et al., 2010). The SCN expresses transcripts for the AVP receptors, V1a and V1b (Bedont et al., 2018), and inhibition of AVP signaling modulates SCN rhythms in vitro by affecting the period and phase relationships of SCN neurons (Edwards et al., 2016; Bedont et al., 2018). In vivo, deletion of V1a/b receptors accelerates behavioral and molecular recovery from simulated jetlag, which is mimicked by injections of V1A and V1B antagonists into the SCN (Yamaguchi et al., 2013). Further, manipulation of the molecular clock specifically in AVP neurons can drive changes in behavioral and SCN rhythms (Mieda et al., 2015; Mieda et al., 2016). Evidence indicates that the role of AVP signaling varies over SCN development (Ono et al., 2016), suggesting that age-related changes in AVP expression influence maturation of the master clock network.
SCN expression of Avp occurs at a later embryonic age relative to other hypothalamic nuclei. In the mouse, Avp is first detected in the SON at E14, in the PVN at E16, and in the SCN at E18 (Hyodo et al., 1992; Jing et al., 1998; Vandunk et al., 2011). The staging of Avp expression across these three structures corresponds with regional differences in the timing of neurogenesis for AVP neurons (Okamura et al., 1983). In addition, the onset of Avp expression in the SCN may be gated by maturation of the molecular clock since this transcript is a first order clock-controlled gene (Jin et al., 1999). In the mouse SCN, AVP expression is evident by P3-P6, and there is a large postnatal increase in AVP production where adult levels are achieved by P20 (Hyodo et al., 1992; Herzog et al., 2000). Similar developmental patterns of Avp and AVP expression have been reported in both rats and hamsters (de Vries et al., 1981; Reppert & Uhl, 1987; Delville et al., 1994; Houdek & Sumova, 2014). In rats, Avp is expressed rhythmically as soon as the transcript is detected (Reppert & Uhl, 1987; Houdek & Sumova, 2014), but there are subsequent changes in the time of peak transcription from E19-E21 (Houdek & Sumova, 2014) and peak protein expression from P0-P20 (Isobe & Muramatsu, 1995).
Comparison of AVP and VIP specification may speak to whether different peptidergic classes of SCN neurons are similar in the staging of critical developmental events. Transcripts of both peptides are expressed by E19 in the mouse and rat SCN (Vandunk et al., 2011; Houdek & Sumova, 2014). In the mouse SCN, the Avp transcript appears approximately 2 days after the end of neurogenesis in AVP neurons (Okamura et al., 1983), whereas Vip expression occurs within 3-4 days after the end of neurogenesis in the SCN core (Kabrita & Davis, 2008; Vandunk et al., 2011). This suggests that there may be a longer interval to VIP production, but the precise time of neurogenesis in VIP neurons has only been identified in the hamster (Antle et al., 2005b). Similarly, the two transcripts differ in how their rhythms change with age. Rhythmic expression of both transcripts in the rat SCN occurs within 1-2 days after detection, but the phasing and amplitude for both is altered over subsequent days (Houdek & Sumova, 2014). Interestingly, both the Avp and Vip rhythms shift by approximately 6 h over E19-E21, but Avp amplitude increases and Vip amplitude decreases. Both peptides appear early in postnatal development, with detectable levels of VIP at P2 (Carmona-Alcocer et al., 2018) and AVP by P3-P6 (Herzog et al., 2000). Over the subsequent 1-3 weeks, both proteins display a marked increase in expression together with changes in the phasing and/or amplitude of daily expression patterns. These perinatal changes in transcript/protein could occur at the single-cell level or could be influenced by new sets of neurons that have expression later in life. Detailed investigations comparing the timing of specification and maturation of different neuronal subclasses in the same species may provide insight into potential factors regulating developmental staging in different types of SCN neurons.
4. Other Signaling Peptides
Two additional neuropeptides expressed in the SCN core are gastrin-releasing peptide (GRP) and Substance P, both of which are involved in photic responses. GRP is produced in mice, rats, and hamsters (LeSauter et al., 2002; Karatsoreos et al., 2006; Guillaumond et al., 2007), but Substance P expression is more variable across rodent species (Shibata et al., 1992; Silver et al., 1999; LeSauter et al., 2002). Further, the spatial location of receptors for each neuropeptide appears different, with the GRP receptor (the Gq-coupled Bombesin 2 receptor, BB2) most densely expressed in the SCN shell (Aida et al., 2002), and the Substance P receptor (NK-1) largely confined to the SCN core (Piggins et al., 2001). Likewise, there is functional evidence that these two neuropeptides have different roles in photic processing. Exogenous application of either GRP or Substance P can mimic the resetting actions of light, but only GRP elicits light-like responses during both early and late subjective night (Shibata et al., 1992; Albers et al., 1995; McArthur et al., 2000; Aida et al., 2002; LeSauter et al., 2002; Antle et al., 2005a; Piggins et al., 2005; Gamble et al., 2007; Sterniczuk et al., 2010; Kallingal & Mintz, 2014). In addition, GRP is rhythmically expressed in a species- and/or age-dependent manner (Shinohara et al., 1993; Okamura & Ibata, 1994; Isobe & Muramatsu, 1995; McArthur et al., 2000; Karatsoreos et al., 2006; Francl et al., 2010), but levels of Substance P do not appear to fluctuate (Otori et al., 1993).
VIP, GRP, and Substance P are all SCN core peptides that contribute to photic processing, but there are differences in their developmental patterning. In the hamster, VIP, GRP, and Substance P neurons are generated at a similar embryonic age (Antle et al., 2005b), but the timing of neuropeptide expression differs (Romero & Silver, 1990; Botchkina & Morin, 1995; Antle et al., 2005b). Expression of GRP and Substance P occurs between P8-P10 in the hamster SCN, whereas VIP is produced much earlier at E13-E14 (as discussed in above section). Likewise, a developmental delay between VIP and GRP expression occurs in the mouse SCN (VIP: P2, GRP: P7; Drouyer et al., 2010; Carmona-Alcocer et al., 2018), and the rat SCN (VIP: E18-E19, GRP & Substance P: by P4-P5; Takatsuji et al., 1991; Isobe & Muramatsu, 1995; Ban et al., 1997). The stark difference in the timing of VIP and GRP is surprising given their parallels in the adult SCN. Like VIP neurons, GRP neurons in the mature SCN are retino-recipient (Tanaka et al., 1997; Karatsoreos et al., 2004; Drouyer et al., 2010; Lokshin et al., 2015; Fernandez et al., 2016) and display light-induced increases in firing rate, immediate early gene expression, and neuropeptide release (Earnest et al., 1993; Romijn et al., 1996; Guillaumond et al., 2007; Francl et al., 2010; Gamble et al., 2011; Lesauter et al., 2011). In addition, potential interactions between GRP and VIP are illustrated by their co-expression in a subset of SCN neurons (Okamura et al., 1986; Romijn et al., 1998) and synergistic responses to GRP and VIP co-administration (Albers et al., 1991; Albers et al., 1995; Chan et al., 2016). Further, BB2 signaling can synchronize SCN neurons in the absence of VIP signaling (Brown et al., 2005; Maywood et al., 2011). But unlike VIP, there is no evidence that GRP is required for SCN timekeeping because BB2-deficient mice exhibit deficits in photic responses but otherwise normal rhythms in behavior and SCN function (Aida et al., 2002). Thus, it is tempting to speculate that the later emergence of GRP expression during development is related to its more specific role in adult SCN function.
5. Other Cellular Markers
A subset of SCN neurons express the calcium binding proteins, Calbindin and Calretinin, which are intracellular calcium buffers that play a role in calcium sensing and/or transport (Schmidt, 2012; Schwaller, 2014). Calbindin can be used as a cell-type marker, although this buffer is often co-expressed with other core peptides such as VIP, GRP, and Substance P (LeSauter et al., 2002; Drouyer et al., 2010). In adulthood, expression of each calcium buffer can fluctuate over the 24 h day, with higher Calbindin expression during the night (Arvanitogiannis et al., 2000; LeSauter et al., 2009) and higher Calretinin expression during the day (Arvanitogiannis et al., 2000; Moore, 2016). In the adult SCN, the expression of Calbindin and Calretinin varies among rodent species. The hamster displays both Calbindin and Calretinin in the SCN core, with dense expression of Calbindin in a compact subnucleus (Silver et al., 1996; Marshall et al., 2000; Antle et al., 2005). In the adult rat, Calbindin is expressed in the SCN core as well (Arvanitogiannis et al., 2000), but Calretinin is expressed in the SCN shell (Marshall et al., 2000). Lastly, Calbindin is diffusely expressed in the adult mouse SCN, and Calretinin is more concentrated in the SCN core (Silver et al., 1999). Despite species differences in Calbindin expression in adulthood, work conducted to date suggests an important role for this calcium buffer in daily timekeeping in both hamsters and mice. In the hamster, Calbindin-expressing SCN neurons influence daily timekeeping and photic responses (LeSauter & Silver, 1999; Antle et al., 2003; Hamada et al., 2003), and micro-lesions targeting this specific SCN subpopulation has been shown to eliminate overt rhythms (Kriegsfeld et al., 2004). Similarly, Calbindin−/− mice exhibit deficits in photic entrainment together with low amplitude rhythms or arrhythmia under constant darkness (Kriegsfeld et al., 2008), which suggests a role for this calcium buffer in regulating the function of the developing and/or mature SCN.
To our knowledge, development of calcium buffers has been examined only in postnatal hamsters and mice. In the hamster SCN, Calbindin-expressing neurons are generated during a similar embryonic window as other SCN core neurons (Antle et al., 2005b), and Calbindin is present at birth with postnatal increases in expression until adult levels are achieved at P15 (Antle et al., 2005b). The developing mouse SCN also displays postnatal changes in Calbindin expression, but levels decrease with age in this species and the spatial pattern becomes more diffuse (Ikeda & Allen, 2003; Kriegsfeld et al., 2008). From P3-P16, high levels of Calbindin are localized to a central subregion of the mouse SCN core, but this region-specific pattern is lost by adulthood (Silver et al., 1999; Ikeda & Allen, 2003; Kriegsfeld et al., 2008). Further, it has been reported that Calretinin levels increase from P9-P20 (Ikeda & Allen, 2003), suggesting a developmental switch in calcium buffers in the SCN that is opposite to that observed in newborn granule cells of the hippocampus (Todkar et al., 2012). It has been proposed that Calbindin and Calretinin protect neurons against excitotoxicity and apoptotic cell death due to their capacity for “fast” calcium buffering properties (D’Orlando et al., 2001; D’Orlando et al., 2002; Choi et al., 2008). In this light, it is interesting to note that the developmental period of high Calbindin expression in the mouse SCN overlaps with the interval of depolarizing responses to GABA (Obrietan & van den Pol, 1995; Ikeda et al., 2003a), maturation of retinal inputs (Sekaran et al., 2005; McNeill et al., 2011), and cell loss in this species (Ahern et al., 2013; Bedont et al., 2014). Of note, Calbindin−/− mice display a normal complement of VIP neurons, but a decrease in the number of AVP-immunoreactive neurons (Kriegsfeld et al., 2008). Circadian deficits in Calbindin−/− mice relate to the loss of this calcium buffer during a critical period of SCN development (Kriegsfeld et al., 2008), but the precise mechanisms remain unclear.
IV. Conclusions
The development of the SCN network is a gradual process that spans both embryonic and postnatal ages (Figure 1D). Recent work using real-time reporters of molecular rhythms dovetails with classic studies to support the idea that SCN neurons display the capacity for circadian timekeeping soon after the close of neurogenesis. Nevertheless, postnatal maturation of SCN circuits occurs at both the molecular and network levels. At the molecular level, this is best exemplified by evidence indicating that clock genes are expressed and become rhythmic at different developmental stages. At the network level, postnatal maturation of the SCN includes the progressive introduction, elaboration, and rewiring of signaling mechanisms that support master clock function in adulthood. In many respects, the precise mechanisms guiding the development of SCN cells and circuits are not well understood, but the current gaps in understanding provide promising avenues for future research.
Genetic programming and intercellular signaling guide SCN development, but there is still much to learn about the relative contribution and interplay of these two factors. For instance, to what extent do intrinsic genetic programs and intercellular signaling interact to guide induction and refinement of the SCN network? Do SCN cells communicate with one another to influence their mutual development? For example, SCN neurons that mature early in development might produce signals (e.g., GABA, VIP) that could set the stage for the later specification of other cells in a homo- or heterotypic manner. In other regions of the hypothalamus, neuropeptides can influence proliferation, differentiation, and synaptic wiring (Bakos et al., 2016). Thus, it is possible that SCN neuropeptides play a similar role during the development of master clock circuits. In addition, it would be interesting to examine what cues and factors regulate neurite elongation, synaptogenesis, cell survival, and neuronal loss in the developing SCN. This may provide new insights into whether SCN circuits are rearranged and modified after they are initially established. Recent advances in genetic labeling of specific pools of post-mitotic neurons may aid in pursuing the answers to some of these questions.
Another issue that warrants further examination is whether the timing of key developmental checkpoints is generic to all SCN neurons or whether there are cell-type specific programs at play. Some support for the latter model derives from evidence that the onset of neuropeptide expression can differ markedly among classes of SCN neurons that appear to be generated during a similar embryonic window. For example, neurons that will produce VIP, GRP, Substance P, and Calbindin are born at a similar embryonic age in the hamster SCN, but the onset of neuropeptide expression appears to vary substantially among these subclasses. Although it is difficult to precisely pinpoint the exact age at which peptides are produced due to developmental changes in levels or rhythms of expression, it is tempting to speculate based on this evidence that the timing of differentiation and specification varies among distinct classes of SCN neurons. Might there be other phenotypic checkpoints that differ among SCN subclasses, such as the maturation of molecular clock function, neurite elongation, or receptor expression? If subclasses of SCN neurons do mature in a cell-type specific manner, it would be of interest to determine what extent these programs are guided by cell-intrinsic and/or cell-extrinsic factors.
Given evidence that perinatal photic conditions can influence circadian function in both humans and animal models, a key area for future research is to investigate the basis of this effect. Although this review has focused on developmental events within the SCN itself, there is mounting evidence that maturation of the SCN network is influenced by signals from the eye. Light is transmitted to the SCN by the retinohypothalamic tract (RHT), which is formed by axons from intrinsically photosensitive retinal ganglion cells (ipRGCs) that express the photopigment melanopsin and relay signals from the rod and cone photoreceptors (Van Gelder & Buhr, 2016). Neurogenesis of ipRGCs occurs during E11-E19 in the mouse (McNeill et al., 2011), with the expression of melanopsin by E11-E15 in the mouse retina (Tarttelin et al., 2003; McNeill et al., 2011) and by E18 in the rat retina (Fahrenkrug et al., 2004). The first RHT terminals innervate the rat and mouse SCN by P0 (Speh & Moore, 1993; McNeill et al., 2011), and by this age ipRGCs in the mouse are capable of mounting light-evoked electrical responses that can increase immediate early gene expression in the SCN (Hannibal & Fahrenkrug, 2004; Sekaran et al., 2005). Despite early RHT innervation by a small number of terminals, there is a prolonged period of postnatal expansion and elaboration over the first two weeks after birth that corresponds with greater SCN responses to light (Speh & Moore, 1993; Kaufman & Menaker, 1994; Munoz Llamosas et al., 2000; Lupi et al., 2006; Mateju et al., 2009; McNeill et al., 2011). This postnatal window coincides with several critical events in the SCN, including cell loss, synaptogenesis, glial maturation, the switch in GABA responses, and changes in neuropeptide expression. Evidence that the RHT influences SCN development is provided by work demonstrating that enucleation and/or dark-rearing will decrease VIP, increase AVP, decrease Substance P, and modulate GFAP expression (Takatsuji et al., 1991; Isobe & Muramatsu, 1995; Ban et al., 1997; Munekawa et al., 2000; Ikeda et al., 2003b; Cambras et al., 2005; Smith & Canal, 2009). This is matched by work indicating that the perinatal lighting environment can influence SCN function and behavioral rhythms later in life (Cambras et al., 1997; Ciarleglio et al., 2011; Chew et al., 2017), including modulation of key properties such as free-running period length. The neurobiological basis of these effects remain unclear, but may relate directly to changes in neuropeptide expression since these signals serve as both important modulators of the SCN network and as daily outputs to downstream clock tissues. The precise mechanisms by which the retina influences the composition and function of the SCN network are important areas for future research.
Several principles of circadian development gleaned from rodent studies appear to generalize to humans and non-human primates. The emergence of fetal rhythms during embryonic development is common in many mammalian species (reviewed in Mirmiran & Ariagno, 2000; Seron-Ferre et al., 2007). In humans and non-human primates, daily rhythms in fetal heart rate, respiration, hormone production, and body movement can be detected during late gestation. Daily rhythms in body temperature can be detected in 50% of babies born prematurely (Mirmiran & Ariagno, 2000), but body temperature rhythms are low amplitude in most newborns (Glotzbach et al., 1995). Over the ensuing months, daily rhythms re-emerge in the neonate with distinct rhythms emerging at different ages (Kleitman & Engelmann, 1953; Price et al., 1983; Glotzbach et al., 1995; Kennaway et al., 1996; McGraw et al., 1999; Rivkees, 2007). The prenatal emergence of overt rhythms in primates appears to coincide with the onset of molecular clock function in the SCN, although there are very few studies examining this issue. In the capuchin monkey, Bmal1, Per2, Cry2, and Clock are expressed by an age corresponding to 90% gestation (Torres-Farfan et al., 2006). Daily fluctuations in Bmal1 and Per2 are evident at this age, although the phasing and amplitude of these rhythms is not identical to adults. Lastly, sequential expression of VIP and AVP occurs in humans prior to birth, with postnatal increases in the number of SCN neurons until 1-3 years of age (Swaab et al., 1996; Xu et al., 2003). Collectively, these data suggest that SCN development in both primates and rodents is characterized by an extended interval of postnatal maturation at the molecular and network levels. Against this backdrop, many investigators have warned about the potential harm caused by circadian disruption during perinatal development (Mirmiran & Ariagno, 2000; Amaral et al., 2014). There is an increasing appreciation of the health costs of circadian disruption in adulthood (Evans & Davidson, 2013), and circadian disruption during gestation may have detrimental consequences for both mother and child (Amaral et al., 2014). Additional work aimed at decoding the mechanisms that guide SCN development is expected to provide a solid foundation upon which to better understand the sources and effects of circadian disruption early in life. To achieve this goal, it will be important to study the precise mechanisms that guide SCN development in both rodent and non-human primates, as well as the effects of circadian disruption on short- and long-term health outcomes.
Acknowledgements
We acknowledge support from the NIH (R01NS091234).
Abbreviations
- ALDHL1:
Aldehyde Dehydrogenase 1 Family Member L1
- APC:
Adenomatous Polyposis Coli
- ASCL:
Achaete-Scute Family Transcription Factor
- AVP:
Arginine Vasopressin
- BB2:
Bombesin 2 receptor
- BCL-2:
B-Cell Lymphoma 2 Protein
- bHLH:
Basic Helix-Loop-Helix
- Bmal1/BMAL1:
Brain and Muscle Arnt-Like Protein 1 gene/protein
- BrdU:
5-Bromo-2’-Deoxyuridine
- CB:
Calbindin
- Clock/CLOCK:
Circadian Locomotor Output Cycles Kaput gene/protein
- CR:
Calretinin
- Cry/CRY:
Cryptochrome gene/protein
- DLX:
Distal-Less Homeobox
- E#:
Embryonic Day
- FGF:
Fibroblast Growth Factor
- FOXD:
Forkhead Box Domain
- FZD:
The Wingless Receptor, Frizzled
- GABA:
γ-Aminobutyric Acid
- GAD:
Glutamic Acid Decarboxylase
- GFAP:
Glial Fibrillary Acidic Protein
- GP:
Gestational Period
- GRP:
Gastrin-Releasing Peptide
- HES:
Hairy Enhancer of Split
- ipRGCs:
Intrinsically Sensitive Retinal Ganglion Cells
- L1:
L1 protein
- LHX:
LIM Homeobox
- MAPK:
Mitogen-Activated Protein Kinase
- MBP:
Myelin Basic Protein
- NCAM:
Neural Cell Adhesion Molecule
- NeuN:
Neuronal Nuclei Protein
- NK-1:
Substance P receptor
- NKX:
Nkx Homeobox
- NMS:
Neuromedin S
- P#:
Postnatal Day
- Per/PER:
Period gene/protein
- PKA:
Protein Kinase A
- PLC:
Phospholipase C
- PLP:
Proteolipid Protein
- POU2F:
POU Class 2 Homeobox
- PROK:
Prokineticin
- PVN:
Paraventricular Nucleus
- RAX:
Retinal and Anterior Neural Fold Homeobox
- RHT:
Retinohypothalamic Tract
- Ror/ROR:
RAR-Related Orphan Receptor gene/protein
- SCN:
Suprachiasmatic Nucleus
- SHH:
Sonic Hedgehog
- SIX:
Six Homeobox
- SON:
Supraoptic Nucleus
- V1:
Vasopressin Receptor
- VAX:
Ventral Anterior Homeobox
- VGAT:
Vesicular GABA Transporter
- VIP:
Vasoactive Intestinal Polypeptide
- VPAC2:
Vasoactive Intestinal Peptide Receptor 2
- WNT:
Wingless Signaling Pathway
- ZFHX:
Zinc Finger Homeobox
Footnotes
Conflict of Interest
The author declares that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Abizaid A, Mezei G, Sotonyi P & Horvath TL (2004) Sex differences in adult suprachiasmatic nucleus neurons emerging late prenatally in rats. Eur J Neurosci, 19, 2488–2496. [DOI] [PubMed] [Google Scholar]
- Abrahamson EE & Moore RY (2001) Suprachiasmatic nucleus in the mouse: Retinal innervation, intrinsic organization and efferent projections. Brain Res, 916, 172–191. [DOI] [PubMed] [Google Scholar]
- Achim K, Salminen M & Partanen J (2014) Mechanisms regulating GABAergic neuron development. Cell Mol Life Sci, 71, 1395–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahern TH, Krug S, Carr AV, Murray EK, Fitzpatrick E, Bengston L, McCutcheon J, De Vries GJ & Forger NG (2013) Cell death atlas of the postnatal mouse ventral forebrain and hypothalamus: Effects of age and sex. J Comp Neurol, 521, 2551–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida R, Moriya T, Araki M, Akiyama M, Wada K, Wada E & Shibata S (2002) Gastrin-releasing peptide mediates photic entrainable signals to dorsal subsets of suprachiasmatic nucleus via induction of Period gene in mice. Mol Pharmacol, 61, 26–34. [DOI] [PubMed] [Google Scholar]
- Akerman CJ & Cline HT (2007) Refining the roles of GABAergic signaling during neural circuit formation. Trends Neurosci, 30, 382–389. [DOI] [PubMed] [Google Scholar]
- Albers HE, Gillespie CF, Babagbemi TO & Huhman KL (1995) Analysis of the phase shifting effects of gastrin releasing peptide when microinjected into the suprachiasmatic region. Neurosci Lett, 191, 63–66. [DOI] [PubMed] [Google Scholar]
- Albers HE, Liou SY, Stopa EG & Zoeller RT (1991) Interaction of colocalized neuropeptides: Functional significance in the circadian timing system. J Neurosci, 11, 846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers HE, Walton JC, Gamble KL, McNeill J.K.t. & Hummer DL (2017) The dynamics of GABA signaling: Revelations from the circadian pacemaker in the suprachiasmatic nucleus. Front Neuroendocrinol, 44, 35–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman J & Bayer SA (1978) Development of the diencephalon in the rat. I. Autoradiographic study of the time of origin and settling patterns of neurons of the hypothalamus. J Comp Neurol, 182, 945–971. [DOI] [PubMed] [Google Scholar]
- Altman J & Bayer SA (1986) The development of the rat hypothalamus. Adv Anat Embryol Cell Biol, 100, 1–178. [PubMed] [Google Scholar]
- Alvarez-Bolado G, Paul FA & Blaess S (2012) Sonic hedgehog lineage in the mouse hypothalamus: From progenitor domains to hypothalamic regions. Neural Dev, 7, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral FG, Castrucci AM, Cipolla-Neto J, Poletini MO, Mendez N, Richter HG & Sellix MT (2014) Environmental control of biological rhythms: Effects on development, fertility and metabolism. J Neuroendocrinol, 26, 603–612. [DOI] [PubMed] [Google Scholar]
- An S, Harang R, Meeker K, Granados-Fuentes D, Tsai CA, Mazuski C, Kim J, Doyle FJ 3rd, Petzold LR & Herzog ED (2013) A neuropeptide speeds circadian entrainment by reducing intercellular synchrony. Proc Natl Acad Sci U S A, 110, E4355–4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Irwin RP, Allen CN, Tsai CA & Herzog ED (2011) Vasoactive intestinal polypeptide requires parallel changes in adenylate cyclase and phospholipase C to entrain circadian rhythms to a predictable phase. J Neurophysiol, 105, 2289–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari N, Agathagelidis M, Lee C, Korf HW & von Gall C (2009) Differential maturation of circadian rhythms in clock gene proteins in the suprachiasmatic nucleus and the pars tuberalis during mouse ontogeny. Eur J Neurosci, 29, 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antle MC, Foley DK, Foley NC & Silver R (2003) Gates and oscillators: A network model of the brain clock. J Biol Rhythms, 18, 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antle MC, Kriegsfeld LJ & Silver R (2005a) Signaling within the master clock of the brain: Localized activation of mitogen-activated protein kinase by gastrin-releasing peptide. J Neurosci, 25, 2447–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antle MC, LeSauter J & Silver R (2005b) Neurogenesis and ontogeny of specific cell phenotypes within the hamster suprachiasmatic nucleus. Brain Res Dev Brain Res, 157, 8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitogiannis A, Robinson B, Beaule C & Amir S (2000) Calbindin-D28k immunoreactivity in the suprachiasmatic nucleus and the circadian response to constant light in the rat. Neuroscience, 99, 397–401. [DOI] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J & Herzog ED (2005) Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci, 8, 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakos J, Zatkova M, Bacova Z & Ostatnikova D (2016) The role of hypothalamic neuropeptides in neurogenesis and neuritogenesis. Neural Plast, 2016, 3276383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzani E, Lassi G, Maggi S, Sethi S, Parsons MJ, Simon M, Nolan PM & Tucci V (2016) The Zfhx3-mediated axis regulates sleep and interval timing in mice. Cell Rep, 16, 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban Y, Shigeyoshi Y & Okamura H (1997) Development of vasoactive intestinal peptide mRNA rhythm in the rat suprachiasmatic nucleus. J Neurosci, 17, 3920–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barca-Mayo O, Pons-Espinal M, Follert P, Armirotti A, Berdondini L & Tonelli DDP (2017) Astrocyte deletion of Bmal1 alters daily locomotor activity and cognitive functions via GABA signalling. Nat Commun, 8, 14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold DA, Brown TM, Luckman SM & Piggins HD (2008) Metabolic rhythm abnormalities in mice lacking VIP-VPAC2 signaling. Am J Physiol Regul Integr Comp Physiol, 294, R344–351. [DOI] [PubMed] [Google Scholar]
- Becquet D, Girardet C, Guillaumond F, Francois-Bellan AM & Bosler O (2008) Ultrastructural plasticity in the rat suprachiasmatic nucleus. Possible involvement in clock entrainment. Glia, 56, 294–305. [DOI] [PubMed] [Google Scholar]
- Bedont JL & Blackshaw S (2015) Constructing the suprachiasmatic nucleus: A watchmaker’s perspective on the central clockworks. Front Syst Neurosci, 9, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedont JL, LeGates TA, Buhr E, Bathini A, Ling JP, Bell B, Wu MN, Wong PC, Van Gelder RN, Mongrain V, Hattar S & Blackshaw S (2017) An LHX1-regulated transcriptional network controls sleep/wake coupling and thermal resistance of the central circadian clockworks. Curr Biol, 27, 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedont JL, LeGates TA, Slat EA, Byerly MS, Wang H, Hu J, Rupp AC, Qian J, Wong GW, Herzog ED, Hattar S & Blackshaw S (2014) Lhx1 controls terminal differentiation and circadian function of the suprachiasmatic nucleus. Cell Rep, 7, 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedont JL, Rohr KE, Bathini A, Hattar S, Blackshaw S, Sehgal A & Evans JA (2018) Asymmetric vasopressin signaling spatially organizes the master circadian clock. J Comp Neurol. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenky MA, Yarom Y & Pickard GE (2008) Heterogeneous expression of gamma-aminobutyric acid and gamma-aminobutyric acid-associated receptors and transporters in the rat suprachiasmatic nucleus. J Comp Neurol, 506, 708–732. [DOI] [PubMed] [Google Scholar]
- Beligala DH, De A & Geusz ME (2018) A meta-analysis characterizing stem-like gene expression in the suprachiasmatic nucleus and its circadian clock. Biomed Res Int, 2018, 3610603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Y, Woodin MA, Sernagor E, Cancedda L, Vinay L, Rivera C, Legendre P, Luhmann HJ, Bordey A, Wenner P, Fukuda A, van den Pol AN, Gaiarsa JL & Cherubini E (2012) Refuting the challenges of the developmental shift of polarity of GABA actions: GABA more exciting than ever! Front Cell Neurosci, 6, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgs L, Beukelaers P, Vandenbosch R, Nguyen L, Moonen G, Maquet P, Albrecht U, Belachew S & Malgrange B (2009) Period2 regulates neural stem/progenitor cell proliferation in the adult hippocampus. BMC Neurosci, 10, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkina GI & Morin LP (1995) Ontogeny of radial glia, astrocytes and vasoactive intestinal peptide immunoreactive neurons in hamster suprachiasmatic nucleus. Brain Res Dev Brain Res, 86, 48–56. [DOI] [PubMed] [Google Scholar]
- Bouchard-Cannon P, Mendoza-Viveros L, Yuen A, Kaern M & Cheng HY (2013) The circadian molecular clock regulates adult hippocampal neurogenesis by controlling the timing of cell-cycle entry and exit. Cell Rep, 5, 961–973. [DOI] [PubMed] [Google Scholar]
- Brancaccio M, Patton AP, Chesham JE, Maywood ES & Hastings MH (2017) Astrocytes control circadian timekeeping in the suprachiasmatic nucleus via glutamatergic signaling. Neuron, 93, 1420–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon DH, Holditch-Davis D & Belyea M (2002) Preterm infants born at less than 31 weeks’ gestation have improved growth in cycled light compared with continuous near darkness. J Pediatr, 140, 192–199. [DOI] [PubMed] [Google Scholar]
- Brown LA, Williams J, Taylor L, Thomson RJ, Nolan PM, Foster RG & Peirson SN (2017) Meta-analysis of transcriptomic datasets identifies genes enriched in the mammalian circadian pacemaker. Nucleic Acids Res, 45, 9860–9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TM, Colwell CS, Waschek JA & Piggins HD (2007) Disrupted neuronal activity rhythms in the suprachiasmatic nuclei of vasoactive intestinal polypeptide-deficient mice. J Neurophysiol, 97, 2553–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TM, Hughes AT & Piggins HD (2005) Gastrin-releasing peptide promotes suprachiasmatic nuclei cellular rhythmicity in the absence of vasoactive intestinal polypeptide-VPAC2 receptor signaling. J Neurosci, 25, 11155–11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr ED, Yoo SH & Takahashi JS (2010) Temperature as a universal resetting cue for mammalian circadian oscillators. Science, 330, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkeen JF, Womac AD, Earnest DJ & Zoran MJ (2011) Mitochondrial calcium signaling mediates rhythmic extracellular ATP accumulation in suprachiasmatic nucleus astrocytes. J Neurosci, 31, 8432–8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambras T, Canal MM, Torres A, Vilaplana J & Diez-Noguera A (1997) Manifestation of circadian rhythm under constant light depends on lighting conditions during lactation. Am J Physiol, 272, R1039–1046. [DOI] [PubMed] [Google Scholar]
- Cambras T, Lopez L, Arias JL & Diez-Noguera A (2005) Quantitative changes in neuronal and glial cells in the suprachiasmatic nucleus as a function of the lighting conditions during weaning. Brain Res Dev Brain Res, 157, 27–33. [DOI] [PubMed] [Google Scholar]
- Carmona-Alcocer V, Abel JH, Sun TC, Petzold LR, Doyle FJ, Simms CL & Herzog ED (2018) Ontogeny of circadian rhythms and synchrony in the suprachiasmatic nucleus. J Neurosci, 38, 1326–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone VM, Speh JC, Card JP & Moore RY (1988) Comparative anatomy of the mammalian hypothalamic suprachiasmatic nucleus. J Biol Rhythms, 3, 71–91. [DOI] [PubMed] [Google Scholar]
- Castel M, Morris J & Belenky M (1996) Non-synaptic and dendritic exocytosis from dense-cored vesicles in the suprachiasmatic nucleus. Neuroreport, 7, 543–547. [DOI] [PubMed] [Google Scholar]
- Castel M & Morris JF (2000) Morphological heterogeneity of the GABAergic network in the suprachiasmatic nucleus, the brain’s circadian pacemaker. J Anat, 196, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro DS, Martynoga B, Parras C, Ramesh V, Pacary E, Johnston C, Drechsel D, Lebel-Potter M, Garcia LG, Hunt C, Dolle D, Bithell A, Ettwiller L, Buckley N & Guillemot F (2011) A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev, 25, 930–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RK, Sterniczuk R, Enkhbold Y, Jeffers RT, Basu P, Duong B, Chow SL, Smith VM & Antle MC (2016) Phase shifts to light are altered by antagonists to neuropeptide receptors. Neuroscience, 327, 115–124. [DOI] [PubMed] [Google Scholar]
- Chen R, Wu X, Jiang L & Zhang Y (2017) Single-cell RNA-seq reveals hypothalamic cell diversity. Cell Rep, 18, 3227–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Alvarez-Saavedra M, Dziema H, Choi YS, Li A & Obrietan K (2009) Segregation of expression of mPeriod gene homologs in neurons and glia: Possible divergent roles of mPeriod1 and mPeriod2 in the brain. Hum Mol Genet, 18, 3110–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew KS, Renna JM, McNeill DS, Fernandez DC, Keenan WT, Thomsen MB, Ecker JL, Loevinsohn GS, VanDunk C, Vicarel DC, Tufford A, Weng SJ, Gray PA, Cayouette M, Herzog ED, Zhao HQ, Berson DM & Hattar S (2017) A subset of ipRGCs regulates both maturation of the circadian clock and segregation of retinogeniculate projections in mice. eLife, 6, e22861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WS, Lee E, Lim J & Oh YJ (2008) Calbindin-D28K prevents drug-induced dopaminergic neuronal death by inhibiting caspase and calpain activity. Biochem Biophys Res Commun, 371, 127–131. [DOI] [PubMed] [Google Scholar]
- Ciarleglio CM, Axley JC, Strauss BR, Gamble KL & McMahon DG (2011) Perinatal photoperiod imprints the circadian clock. Nat Neurosci, 14, 25–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DD, Gorman MR, Hatori M, Meadows JD, Panda S & Mellon PL (2013) Aberrant development of the suprachiasmatic nucleus and circadian rhythms in mice lacking the homeodomain protein Six6. J Biol Rhythms, 28, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS (2000) Rhythmic coupling among cells in the suprachiasmatic nucleus. J Neurobiol, 43, 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X & Waschek JA (2003) Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am J Physiol Regul Integr Comp Physiol, 285, R939–949. [DOI] [PubMed] [Google Scholar]
- Couvineau A & Laburthe M (2011) VPAC receptors: Structure, molecular pharmacology and interaction with accessory proteins. Br J Pharmacol, 166, 42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossland WJ & Uchwat CJ (1982) Neurogenesis in the central visual pathways of the golden hamster. Brain Res, 281, 99–103. [DOI] [PubMed] [Google Scholar]
- D’Orlando C, Celio MR & Schwaller B (2002) Calretinin and calbindin D-28k, but not parvalbumin protect against glutamate-induced delayed excitotoxicity in transfected N18-RE 105 neuroblastoma-retina hybrid cells. Brain Res, 945, 181–190. [DOI] [PubMed] [Google Scholar]
- D’Orlando C, Fellay B, Schwaller B, Salicio V, Bloc A, Gotzos V & Celio MR (2001) Calretinin and calbindin D-28k delay the onset of cell death after excitotoxic stimulation in transfected P19 cells. Brain Res, 909, 145–158. [DOI] [PubMed] [Google Scholar]
- Davis AM, Penschuck S, Fritschy JM & McCarthy MM (2000) Developmental switch in the expression of GABA(A) receptor subunits alpha(1) and alpha(2) in the hypothalamus and limbic system of the rat. Brain Res Dev Brain Res, 119, 127–138. [DOI] [PubMed] [Google Scholar]
- Davis FC, Boada R & LeDeaux J (1990) Neurogenesis of the hamster suprachiasmatic nucleus. Brain Res, 519, 192–199. [DOI] [PubMed] [Google Scholar]
- de Vries GJ, Buijs RM & Swaab DF (1981) Ontogeny of the vasopressinergic neurons of the suprachiasmatic nucleus and their extrahypothalamic projections in the rat brain--presence of a sex difference in the lateral septum. Brain Res, 218, 67–78. [DOI] [PubMed] [Google Scholar]
- Deery MJ, Maywood ES, Chesham JE, Sladek M, Karp NA, Green EW, Charles PD, Reddy AB, Kyriacou CP, Lilley KS & Hastings MH (2009) Proteomic analysis reveals the role of synaptic vesicle cycling in sustaining the suprachiasmatic circadian clock. Curr Biol, 19, 2031–2036. [DOI] [PubMed] [Google Scholar]
- Delville Y, Mansour KM, Quan EW, Yules BM & Ferris CF (1994) Postnatal development of the vasopressinergic system in golden hamsters. Brain Res Dev Brain Res, 81, 230–239. [DOI] [PubMed] [Google Scholar]
- Demarque M, Represa A, Becq H, Khalilov I, Ben-Ari Y & Aniksztejn L (2002) Paracrine intercellular communication by a Ca2+- and SNARE-independent release of GABA and glutamate prior to synapse formation. Neuron, 36, 1051–1061. [DOI] [PubMed] [Google Scholar]
- Diemer T, Landgraf D, Noguchi T, Pan HY, Moreno JL & Welsh DK (2017) Cellular circadian oscillators in the suprachiasmatic nucleus remain coupled in the absence of connexin-36. Neuroscience, 357, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouyer E, LeSauter J, Hernandez AL & Silver R (2010) Specializations of gastrin-releasing peptide cells of the mouse suprachiasmatic nucleus. J Comp Neurol, 518, 1249–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhart JM, Leone MJ, Paladino N, Evans JA, Castanon-Cervantes O, Davidson AJ & Golombek DA (2013) Suprachiasmatic astrocytes modulate the circadian clock in response to TNF-α. J Immunol, 191, 4656–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque A & Rakic P (2011) Different effects of bromodeoxyuridine and 3H-Thymidine incorporation into DNA on cell proliferation, position, and fate. J Neurosci, 31, 15205–15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest DJ, DiGiorgio S & Olschowka JA (1993) Light induces expression of fos-related proteins within gastrin-releasing peptide neurons in the rat suprachiasmatic nucleus. Brain Res, 627, 205–209. [DOI] [PubMed] [Google Scholar]
- Edwards MD, Brancaccio M, Chesham JE, Maywood ES & Hastings MH (2016) Rhythmic expression of cryptochrome induces the circadian clock of arrhythmic suprachiasmatic nuclei through arginine vasopressin signaling. Proc Natl Acad Sci U S A, 113, 2732–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoki R, Kuroda S, Ono D, Hasan MT, Ueda T, Honma S & Honma K (2012) Topological specificity and hierarchical network of the circadian calcium rhythm in the suprachiasmatic nucleus. Proc Natl Acad Sci U S A, 109, 21498–21503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erren TC, Koch MS, Gross JV, Reiter RJ & Meyer-Rochow VB (2012) A possible role of perinatal light in mood disorders and internal cancers: Reconciliation of instability and latitude concepts. Neuro Endocrinol Lett, 33, 314–317. [PubMed] [Google Scholar]
- Evans JA & Davidson AJ (2013) Health consequences of circadian disruption in humans and animal models. Prog Mol Biol Transl Sci, 119, 283–323. [DOI] [PubMed] [Google Scholar]
- Evans JA & Gorman MR (2016) In synch but not in step: Circadian clock circuits regulating plasticity in daily rhythms. Neuroscience, 320, 259–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrug J, Nielsen HS & Hannibal J (2004) Expression of melanopsin during development of the rat retina. Neuroreport, 15, 781–784. [DOI] [PubMed] [Google Scholar]
- Farajnia S, van Westering TL, Meijer JH & Michel S (2014) Seasonal induction of GABAergic excitation in the central mammalian clock. Proc Natl Acad Sci U S A, 111, 9627–9632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez DC, Chang YT, Hattar S & Chen SK (2016) Architecture of retinal projections to the central circadian pacemaker. Proc Natl Acad Sci U S A, 113, 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferran JL, Puelles L & Rubenstein JL (2015) Molecular codes defining rostrocaudal domains in the embryonic mouse hypothalamus. Front Neuroanat, 9, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francl JM, Kaur G & Glass JD (2010) Roles of light and serotonin in the regulation of gastrin-releasing peptide and arginine vasopressin output in the hamster SCN circadian clock. Eur J Neurosci, 32, 1170–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois-Bellan AM, Kachidian P, Dusticier G, Tonon MC, Vaudry H & Bosler O (1990) GABA neurons in the rat suprachiasmatic nucleus: Involvement in chemospecific synaptic circuitry and evidence for GAD-peptide colocalization. J Neurocytol, 19, 937–947. [DOI] [PubMed] [Google Scholar]
- Fricker M, Tolkovsky AM, Borutaite V, Coleman M & Brown GC (2018) Neuronal cell death. Physiol Rev, 98, 813–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritschy JM, Paysan J, Enna A & Mohler H (1994) Switch in the expression of rat GABAA-receptor subtypes during postnatal development: An immunohistochemical study. J Neurosci, 14, 5302–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble KL, Allen GC, Zhou T & McMahon DG (2007) Gastrin-releasing peptide mediates light-like resetting of the suprachiasmatic nucleus circadian pacemaker through cAMP response element-binding protein and Per1 activation. J Neurosci, 27, 12078–12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble KL, Kudo T, Colwell CS & McMahon DG (2011) Gastrin-releasing peptide modulates fast delayed rectifier potassium current in Per1-expressing SCN neurons. J Biol Rhythms, 26, 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Fritschy JM & Moore RY (1995) GABAA-receptor subunit composition in the circadian timing system. Brain Res, 700, 142–156. [DOI] [PubMed] [Google Scholar]
- Gerics B, Szalay F & Hajos F (2006) Glial fibrillary acidic protein immunoreactivity in the rat suprachiasmatic nucleus: Circadian changes and their seasonal dependence. J Anat, 209, 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardet C, Becquet D, Blanchard MP, Francois-Bellan AM & Bosler O (2010) Neuroglial and synaptic rearrangements associated with photic entrainment of the circadian clock in the suprachiasmatic nucleus. Eur J Neurosci, 32, 2133–2142. [DOI] [PubMed] [Google Scholar]
- Glotzbach SF, Edgar DM & Ariagno RL (1995) Biological rhythmicity in preterm infants prior to discharge from neonatal intensive care. Pediatrics, 95, 231–237. [PubMed] [Google Scholar]
- Gribkoff VK, Pieschl RL & Dudek FE (2003) GABA receptor-mediated inhibition of neuronal activity in rat SCN in vitro: Pharmacology and influence of circadian phase. J Neurophysiol, 90, 1438–1448. [DOI] [PubMed] [Google Scholar]
- Guillaumond F, Becquet D, Blanchard MP, Attia J, Moreno M, Bosler O & Francois-Bellan AM (2007) Nocturnal expression of phosphorylated-ERK1/2 in gastrin-releasing peptide neurons of the rat suprachiasmatic nucleus. J Neurochem, 101, 1224–1235. [DOI] [PubMed] [Google Scholar]
- Guldner FH (1976) Synaptology of the rat suprachiasmatic nucleus. Cell Tissue Res, 165, 509–544. [DOI] [PubMed] [Google Scholar]
- Guldner FH (1983) Numbers of neurons and astroglial cells in the suprachiasmatic nucleus of male and female rats. Exp Brain Res, 50, 373–376. [DOI] [PubMed] [Google Scholar]
- Hallonet M, Hollemann T, Pieler T & Gruss P (1999) Vax1, a novel homeobox-containing gene, directs development of the basal forebrain and visual system. Genes Dev, 13, 3106–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, LeSauter J, Lokshin M, Romero MT, Yan L, Venuti JM & Silver R (2003) Calbindin influences response to photic input in suprachiasmatic nucleus. J Neurosci, 23, 8820–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannibal J & Fahrenkrug J (2004) Melanopsin containing retinal ganglion cells are light responsive from birth. Neuroreport, 15, 2317–2320. [DOI] [PubMed] [Google Scholar]
- Harmar AJ (2003) An essential role for peptidergic signalling in the control of circadian rhythms in the suprachiasmatic nuclei. J Neuroendocrinol, 15, 335–338. [DOI] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, Kelly JS, Maywood ES & Hastings MH (2002) The VPAC2 receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell, 109, 497–508. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Maywood ES & Brancaccio M (2018) Generation of circadian rhythms in the suprachiasmatic nucleus. Nat Rev Neurosci, 19, 453–469. [DOI] [PubMed] [Google Scholar]
- Hatini V, Huh SO, Herzlinger D, Soares VC & Lai E (1996) Essential role of stromal mesenchyme in kidney morphogenesis revealed by targeted disruption of Winged Helix transcription factor BF-2. Genes Dev, 10, 1467–1478. [DOI] [PubMed] [Google Scholar]
- Hatori M, Gill S, Mure LS, Goulding M, O’Leary DD & Panda S (2014) Lhx1 maintains synchrony among circadian oscillator neurons of the SCN. eLife, 3, e03357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlerigg DG, Ebling FJ & Johnston JD (2005) Photoperiod differentially regulates gene expression rhythms in the rostral and caudal SCN. Curr Biol, 15, R449–450. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Aton SJ, Numano R, Sakaki Y & Tei H (2004) Temporal precision in the mammalian circadian system: A reliable clock from less reliable neurons. J Biol Rhythms, 19, 35–46. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Grace MS, Harrer C, Williamson J, Shinohara K & Block GD (2000) The role of Clock in the developmental expression of neuropeptides in the suprachiasmatic nucleus. J Comp Neurol, 424, 86–98. [DOI] [PubMed] [Google Scholar]
- Herzog ED & Huckfeldt RM (2003) Circadian entrainment to temperature, but not light, in the isolated suprachiasmatic nucleus. J Neurophysiol, 90, 763–770. [DOI] [PubMed] [Google Scholar]
- Hoefflin S & Carter DA (2014) Neuronal expression of SOX2 is enriched in specific hypothalamic cell groups. J Chem Neuroanat, 61-62, 153–160. [DOI] [PubMed] [Google Scholar]
- Hoffmann HM, Meadows JD, Trang C, Hereford B, Bharti K, Gorman MR & Mellon PL (2016) Deletion of SIX3 or VAX1 in the SCN impairs circadian rhythms and fertility. Endocrine Society’s 98th Annual Meeting and Expo. [Google Scholar]
- Hormuzdi SG, Filippov MA, Mitropoulou G, Monyer H & Bruzzone R (2004) Electrical synapses: A dynamic signaling system that shapes the activity of neuronal networks. Biochim Biophys Acta, 1662, 113–137. [DOI] [PubMed] [Google Scholar]
- Houdek P & Sumova A (2014) In vivo initiation of clock gene expression rhythmicity in fetal rat suprachiasmatic nuclei. PLoS One, 9, e107360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Lu C, Chen S, Hua L & Qian R (2010) Postnatal ontogenesis of clock genes in mouse suprachiasmatic nucleus and heart. Lipids Health Dis, 9, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AT, Croft CL, Samuels RE, Myung J, Takumi T & Piggins HD (2015) Constant light enhances synchrony among circadian clock cells and promotes behavioral rhythms in VPAC2-signaling deficient mice. Sci Rep, 5, 14044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhman KL, Hennessey AC & Albers HE (1996) Rhythms of glutamic acid decarboxylase mRNA in the suprachiasmatic nucleus. J Biol Rhythms, 11, 311–316. [DOI] [PubMed] [Google Scholar]
- Hut RA & Beersma DG (2011) Evolution of time-keeping mechanisms: Early emergence and adaptation to photoperiod. Philos Trans R Soc Lond B Biol Sci, 366, 2141–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyodo S, Yamada C, Takezawa T & Urano A (1992) Expression of provasopressin gene during ontogeny in the hypothalamus of developing mice. Neuroscience, 46, 241–250. [DOI] [PubMed] [Google Scholar]
- Ifft JD (1972) An autoradiographic study of the time of final division of neurons in rat hypothalamic nuclei. J Comp Neurol, 144, 193–204. [DOI] [PubMed] [Google Scholar]
- Ikeda M & Allen CN (2003) Developmental changes in calbindin-D28k and calretinin expression in the mouse suprachiasmatic nucleus. Eur J Neurosci, 17, 1111–1118. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Yoshioka T & Allen CN (2003a) Developmental and circadian changes in Ca2+ mobilization mediated by GABAA and NMDA receptors in the suprachiasmatic nucleus. Eur J Neurosci, 17, 58–70. [DOI] [PubMed] [Google Scholar]
- Ikeda T, Iijima N, Munekawa K, Ishihara A, Ibata Y & Tanaka M (2003b) Functional retinal input stimulates expression of astroglial elements in the suprachiasmatic nucleus of postnatal developing rat. Neurosci Res, 47, 39–45. [DOI] [PubMed] [Google Scholar]
- Inagaki N, Honma S, Ono D, Tanahashi Y & Honma K (2007) Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc Natl Acad Sci U S A, 104, 7664–7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin RP & Allen CN (2010) Neuropeptide-mediated calcium signaling in the suprachiasmatic nucleus network. Eur J Neurosci, 32, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isobe Y & Muramatsu K (1995) Day-night differences in the contents of vasoactive intestinal peptide, gastrin-releasing peptide and Arg-vasopressin in the suprachiasmatic nucleus of rat pups during postnatal development. Neurosci Lett, 188, 45–48. [DOI] [PubMed] [Google Scholar]
- Itri J & Colwell CS (2003) Regulation of inhibitory synaptic transmission by vasoactive intestinal peptide (VIP) in the mouse suprachiasmatic nucleus. J Neurophysiol, 90, 1589–1597. [DOI] [PubMed] [Google Scholar]
- Jean D, Bernier G & Gruss P (1999) Six6 (Optx2) is a novel murine Six3-related homeobox gene that demarcates the presumptive pituitary/hypothalamic axis and the ventral optic stalk. Mech Dev, 84, 31–40. [DOI] [PubMed] [Google Scholar]
- Jiang ZG, Yang Y, Liu ZP & Allen CN (1997) Membrane properties and synaptic inputs of suprachiasmatic nucleus neurons in rat brain slices. J Physiol, 499 141–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ & Reppert SM (1999) A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell, 96, 57–68. [DOI] [PubMed] [Google Scholar]
- Jing X, Ratty AK & Murphy D (1998) Ontogeny of the vasopressin and oxytocin RNAs in the mouse hypothalamus. Neurosci Res, 30, 343–349. [DOI] [PubMed] [Google Scholar]
- Kabrita CS & Davis FC (2008) Development of the mouse suprachiasmatic nucleus: determination of time of cell origin and spatial arrangements within the nucleus. Brain Res, 1195, 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallingal GJ & Mintz EM (2014) Site-specific effects of gastrin-releasing peptide in the suprachiasmatic nucleus. Eur J Neurosci, 39, 630–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallo I, Kalamatianos T, Wiltshire N, Shen S, Sheward WJ, Harmar AJ & Coen CW (2004) Transgenic approach reveals expression of the VPAC2 receptor in phenotypically defined neurons in the mouse suprachiasmatic nucleus and in its efferent target sites. Eur J Neurosci, 19, 2201–2211. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, Fliers E, Hofman MA, Swaab DF & Buijs RM (2010) Vasopressin and the output of the hypothalamic biological clock. J Neuroendocrinol, 22, 362–372. [DOI] [PubMed] [Google Scholar]
- Karatsoreos IN, Romeo RD, McEwen BS & Silver R (2006) Diurnal regulation of the gastrin-releasing peptide receptor in the mouse circadian clock. Eur J Neurosci, 23, 1047–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatsoreos IN, Yan L, LeSauter J & Silver R (2004) Phenotype matters: Identification of light-responsive cells in the mouse suprachiasmatic nucleus. J Neurosci, 24, 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman CM & Menaker M (1993) Effect of transplanting suprachiasmatic nuclei from donors of different ages into completely SCN lesioned hamsters. J Neural Transplant Plast, 4, 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman CM & Menaker M (1994) Ontogeny of light-induced Fos-like immunoreactivity in the hamster suprachiasmatic nucleus. Brain Res, 633, 162–166. [DOI] [PubMed] [Google Scholar]
- Kawahara F, Saito H & Katsuki H (1993) Pharmacological characteristics of GABAA responses in postnatal suprachiasmatic neurons in culture. Neurosci Lett, 160, 45–48. [DOI] [PubMed] [Google Scholar]
- Kawamoto K, Nagano M, Kanda F, Chihara K, Shigeyoshi Y & Okamura H (2003) Two types of VIP neuronal components in rat suprachiasmatic nucleus. J Neurosci Res, 74, 852–857. [DOI] [PubMed] [Google Scholar]
- Kennaway DJ, Goble FC & Stamp GE (1996) Factors influencing the development of melatonin rhythmicity in humans. J Clin Endocrinol Metab, 81, 1525–1532. [DOI] [PubMed] [Google Scholar]
- King VM, Chahad-Ehlers S, Shen S, Harmar AJ, Maywood ES & Hastings MH (2003) A hVIPR transgene as a novel tool for the analysis of circadian function in the mouse suprachiasmatic nucleus. Eur J Neurosci, 17, 822–832. [PubMed] [Google Scholar]
- Kleitman N & Engelmann TG (1953) Sleep characteristics of infants. J Appl Physiol, 6, 269–282. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Shimizu-Okabe C, Kim J, Kobayashi S, Matsushita M, Masuzaki H & Takayama C (2017) Embryonic development of GABAergic terminals in the mouse hypothalamic nuclei involved in feeding behavior. Neurosci Res, 134, 39–48. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Ye Z & Hensch TK (2015) Clock genes control cortical critical period timing. Neuron, 86, 264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacikova Z, Sladek M, Bendova Z, Illnerova H & Sumova A (2006) Expression of clock and clock-driven genes in the rat suprachiasmatic nucleus during late fetal and early postnatal development. J Biol Rhythms, 21, 140–148. [DOI] [PubMed] [Google Scholar]
- Kriegsfeld LJ, LeSauter J & Silver R (2004) Targeted microlesions reveal novel organization of the hamster suprachiasmatic nucleus. J Neurosci, 24, 2449–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegsfeld LJ, Mei DF, Yan L, Witkovsky P, Lesauter J, Hamada T & Silver R (2008) Targeted mutation of the calbindin D28K gene disrupts circadian rhythmicity and entrainment. Eur J Neurosci, 27, 2907–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T, Tahara Y, Gamble KL, McMahon DG, Block GD & Colwell CS (2013) Vasoactive intestinal peptide produces long-lasting changes in neural activity in the suprachiasmatic nucleus. J Neurophysiol, 110, 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemle LK (1988) Vasoactive intestinal polypeptide (VIP)-like immunoreactivity in the suprachiasmatic nucleus of the perinatal rat. Brain Res, 469, 308–312. [DOI] [PubMed] [Google Scholar]
- Laemle LK, Repke KB, Hawkes R & Rice FL (1991) Synaptogenesis in the rat suprachiasmatic nucleus: A light microscopic immunocytochemical survey. Brain Res, 544, 108–117. [DOI] [PubMed] [Google Scholar]
- Lagutin OV, Zhu CC, Kobayashi D, Topczewski J, Shimamura K, Puelles L, Russell HR, McKinnon PJ, Solnica-Krezel L & Oliver G (2003) Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev, 17, 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf D, Achten C, Dallmann F & Oster H (2015) Embryonic development and maternal regulation of murine circadian clock function. Chronobiol Int, 32, 416–427. [DOI] [PubMed] [Google Scholar]
- Landgraf D, Koch CE & Oster H (2014) Embryonic development of circadian clocks in the mammalian suprachiasmatic nuclei. Front Neuroanat, 8, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavaille M & Serviere J (1995) Developmental study in the circadian clock of the golden hamster: A putative role of astrocytes. Brain Res Dev Brain Res, 86, 275–282. [DOI] [PubMed] [Google Scholar]
- Lavialle M, Begue A, Papillon C & Vilaplana J (2001) Modifications of retinal afferent activity induce changes in astroglial plasticity in the hamster circadian clock. Glia, 34, 88–100. [DOI] [PubMed] [Google Scholar]
- Lenn NJ, Beebe B & Moore RY (1977) Postnatal development of the suprachiasmatic hypothalamic nucleus of the rat. Cell Tissue Res, 178, 463–475. [DOI] [PubMed] [Google Scholar]
- LeSauter J, Bhuiyan T, Shimazoe T & Silver R (2009) Circadian trafficking of calbindin-ir in fibers of SCN neurons. J Biol Rhythms, 24, 488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeSauter J, Kriegsfeld LJ, Hon J & Silver R (2002) Calbindin-D(28K) cells selectively contact intra-SCN neurons. Neuroscience, 111, 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeSauter J & Silver R (1999) Localization of a suprachiasmatic nucleus subregion regulating locomotor rhythmicity. J Neurosci, 19, 5574–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesauter J, Silver R, Cloues R & Witkovsky P (2011) Light exposure induces short- and long-term changes in the excitability of retinorecipient neurons in suprachiasmatic nucleus. J Neurophysiol, 106, 576–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X & Davis FC (2005) Developmental expression of clock genes in the Syrian hamster. Brain Res Dev Brain Res, 158, 31–40. [DOI] [PubMed] [Google Scholar]
- Li X, Perissi V, Liu F, Rose DW & Rosenfeld MG (2002) Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science, 297, 1180–1183. [DOI] [PubMed] [Google Scholar]
- Lindley J, Deurveilher S, Rusak B & Semba K (2008) Transforming growth factor-alpha and glial fibrillary acidic protein in the hamster circadian system: Daily profile and cellular localization. Brain Res, 1197, 94–105. [DOI] [PubMed] [Google Scholar]
- Liu C & Reppert SM (2000) GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron, 25, 123–128. [DOI] [PubMed] [Google Scholar]
- Liu C, Wang Y, Smallwood PM & Nathans J (2008) An essential role for Frizzled5 in neuronal survival in the parafascicular nucleus of the thalamus. J Neurosci, 28, 5641–5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh DH, Kuljis DA, Azuma L, Wu Y, Truong D, Wang HB & Colwell CS (2014) Disrupted reproduction, estrous cycle, and circadian rhythms in female mice deficient in vasoactive intestinal peptide. J Biol Rhythms, 29, 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokshin M, LeSauter J & Silver R (2015) Selective distribution of retinal input to mouse SCN revealed in analysis of sagittal sections. J Biol Rhythms, 30, 251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long MA, Jutras MJ, Connors BW & Burwell RD (2005) Electrical synapses coordinate activity in the suprachiasmatic nucleus. Nat Neurosci, 8, 61–66. [DOI] [PubMed] [Google Scholar]
- Lu F, Kar D, Gruenig N, Zhang ZW, Cousins N, Rodgers HM, Swindell EC, Jamrich M, Schuurmans C, Mathers PH & Kurrasch DM (2013) Rax is a selector gene for mediobasal hypothalamic cell types. J Neurosci, 33, 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucassen EA, van Diepen HC, Houben T, Michel S, Colwell CS & Meijer JH (2012) Role of vasoactive intestinal peptide in seasonal encoding by the suprachiasmatic nucleus clock. Eur J Neurosci, 35, 1466–1474. [DOI] [PubMed] [Google Scholar]
- Lupi D, Sekaran S, Jones SL, Hankins MW & Foster RG (2006) Light-evoked FOS induction within the suprachiasmatic nuclei (SCN) of melanopsin knockout (Opn4−/−) mice: A developmental study. Chronobiol Int, 23, 167–179. [DOI] [PubMed] [Google Scholar]
- Malik A, Jamasbi RJ, Kondratov RV & Geusz ME (2015) Development of circadian oscillators in neurosphere cultures during adult neurogenesis. PLoS One, 10, e0122937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann NP, Haddow R, Stokes L, Goodley S & Rutter N (1986) Effect of night and day on preterm infants in a newborn nursery: Randomised trial. Br Med J (Clin Res Ed), 293, 1265–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markakis EA & Swanson LW (1997) Spatiotemporal patterns of secretomotor neuron generation in the parvicellular neuroendocrine system. Brain Res Rev, 24, 255–291. [DOI] [PubMed] [Google Scholar]
- Marshall ST, Fa’anunu AI & Bult A (2000) Calretinin is not a marker for subdivisions within the suprachiasmatic nucleus. Brain Res, 854, 216–219. [DOI] [PubMed] [Google Scholar]
- Mateju K, Bendova Z, El-Hennamy R, Sladek M, Sosniyenko S & Sumova A (2009) Development of the light sensitivity of the clock genes Period1 and Period2, and immediate-early gene c-fos within the rat suprachiasmatic nucleus. Eur J Neurosci, 29, 490–501. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Chesham JE, O’Brien JA & Hastings MH (2011) A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc Natl Acad Sci U S A, 108, 14306–14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Drynan L, Chesham JE, Edwards MD, Dardente H, Fustin JM, Hazlerigg DG, O’Neill JS, Codner GF, Smyllie NJ, Brancaccio M & Hastings MH (2013) Analysis of core circadian feedback loop in suprachiasmatic nucleus of mCry1-luc transgenic reporter mouse. Proc Natl Acad Sci U S A, 110, 9547–9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O’Neill JS, O’Brien JA, McMahon DG, Harmar AJ, Okamura H & Hastings MH (2006) Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol, 16, 599–605. [DOI] [PubMed] [Google Scholar]
- Mazuski C, Abel JH, Chen SP, Hermanstyne TO, Jones JR, Simon T, Doyle FJ 3rd & Herzog ED (2018) Entrainment of circadian rhythms depends on firing rates and neuropeptide release of VIP SCN neurons. Neuron, 99, 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur AJ, Coogan AN, Ajpru S, Sugden D, Biello SM & Piggins HD (2000) Gastrin-releasing peptide phase-shifts suprachiasmatic nuclei neuronal rhythms in vitro. J Neurosci, 20, 5496–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw K, Hoffmann R, Harker C & Herman JH (1999) The development of circadian rhythms in a human infant. Sleep, 22, 303–310. [DOI] [PubMed] [Google Scholar]
- McNeill DS, Sheely CJ, Ecker JL, Badea TC, Morhardt D, Guido W & Hattar S (2011) Development of melanopsin-based irradiance detecting circuitry. Neural Dev, 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer JH, Michel S, Vanderleest HT & Rohling JH (2010) Daily and seasonal adaptation of the circadian clock requires plasticity of the SCN neuronal network. Eur J Neurosci, 32, 2143–2151. [DOI] [PubMed] [Google Scholar]
- Merikanto I, Lahti T, Puolijoki H, Vanhala M, Peltonen M, Laatikainen T, Vartiainen E, Salomaa V, Kronholm E & Partonen T (2013) Associations of chronotype and sleep with cardiovascular diseases and type 2 diabetes. Chronobiol Int, 30, 470–477. [DOI] [PubMed] [Google Scholar]
- Meyer-Spasche A & Piggins HD (2004) Vasoactive intestinal polypeptide phase-advances the rat suprachiasmatic nuclei circadian pacemaker in vitro via protein kinase A and mitogen-activated protein kinase. Neurosci Lett, 358, 91–94. [DOI] [PubMed] [Google Scholar]
- Mieda M, Okamoto H & Sakurai T (2016) Manipulating the cellular circadian period of arginine vasopressin neurons alters the behavioral circadian period. Curr Biol, 26, 2535–2542. [DOI] [PubMed] [Google Scholar]
- Mieda M, Ono D, Hasegawa E, Okamoto H, Honma K, Honma S & Sakurai T (2015) Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron, 85, 1103–1116. [DOI] [PubMed] [Google Scholar]
- Miller AV, Kavanaugh SI & Tsai PS (2016) Disruption of the suprachiasmatic nucleus in fibroblast growth factor signaling-deficient mice. Front Endocrinol, 7, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirmiran M & Ariagno RL (2000) Influence of light in the NICU on the development of circadian rhythms in preterm infants. Semin Perinatol, 24, 247–257. [DOI] [PubMed] [Google Scholar]
- Mohawk JA, Green CB & Takahashi JS (2012) Central and peripheral circadian clocks in mammals. Annu Rev Neurosci, 35, 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr MA, DonCarlos LL & Sisk CL (2017) Inhibiting production of new brain cells during puberty or adulthood blunts the hormonally induced surge of luteinizing hormone in female rats. eNeuro, 4, 0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrain V, Paquet J & Dumont M (2006) Contribution of the photoperiod at birth to the association between season of birth and diurnal preference. Neurosci Lett, 406, 113–116. [DOI] [PubMed] [Google Scholar]
- Moore RY (2016) Calretinin neurons in the rat suprachiasmatic nucleus. J Biol Rhythms, 31, 406–410. [DOI] [PubMed] [Google Scholar]
- Moore RY & Bernstein ME (1989) Synaptogenesis in the rat suprachiasmatic nucleus demonstrated by electron microscopy and synapsin I immunoreactivity. J Neurosci, 9, 2151–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RY & Speh JC (1993) GABA is the principal neurotransmitter of the circadian system. Neurosci Lett, 150, 112–116. [DOI] [PubMed] [Google Scholar]
- Moore RY, Speh JC & Leak RK (2002) Suprachiasmatic nucleus organization. Cell Tissue Res, 309, 89–98. [DOI] [PubMed] [Google Scholar]
- Morin LP (2007) SCN organization reconsidered. J Biol Rhythms, 22, 3–13. [DOI] [PubMed] [Google Scholar]
- Morin LP, Hefton S & Studholme KM (2011) Neurons identified by NeuN/Fox-3 immunoreactivity have a novel distribution in the hamster and mouse suprachiasmatic nucleus. Brain Res, 1421, 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin LP, Johnson RF & Moore RY (1989) Two brain nuclei controlling circadian rhythms are identified by GFAP immunoreactivity in hamsters and rats. Neurosci Lett, 99, 55–60. [DOI] [PubMed] [Google Scholar]
- Moriya T, Yoshinobu Y, Kouzu Y, Katoh A, Gomi H, Ikeda M, Yoshioka T, Itohara S & Shibata S (2000) Involvement of glial fibrillary acidic protein (GFAP) expressed in astroglial cells in circadian rhythm under constant lighting conditions in mice. J Neurosci Res, 60, 212–218. [DOI] [PubMed] [Google Scholar]
- Muller C & Torrealba F (1998) Postnatal development of neuron number and connections in the suprachiasmatic nucleus of the hamster. Brain Res Dev Brain Res, 110, 203–213. [DOI] [PubMed] [Google Scholar]
- Munekawa K, Tamada Y, Iijima N, Hayashi S, Ishihara A, Inoue K, Tanaka M & Ibata Y (2000) Development of astroglial elements in the suprachiasmatic nucleus of the rat: With special reference to the involvement of the optic nerve. Exp Neurol, 166, 44–51. [DOI] [PubMed] [Google Scholar]
- Munoz Llamosas M, Huerta JJ, Cernuda-Cernuda R & Garcia-Fernandez JM (2000) Ontogeny of a photic response in the retina and suprachiasmatic nucleus in the mouse. Brain Res Dev Brain Res, 120, 1–6. [DOI] [PubMed] [Google Scholar]
- Myung J, Hong S, DeWoskin D, De Schutter E, Forger DB & Takumi T (2015) GABA-mediated repulsive coupling between circadian clock neurons in the SCN encodes seasonal time. Proc Natl Acad Sci U S A, 112, E3920–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura W, Honma S, Shirakawa T & Honma K (2001) Regional pacemakers composed of multiple oscillator neurons in the rat suprachiasmatic nucleus. Eur J Neurosci, 14, 666–674. [DOI] [PubMed] [Google Scholar]
- Natale V & Di Milia L (2011) Season of birth and morningness: Comparison between the northern and southern hemispheres. Chronobiol Int, 28, 727–730. [DOI] [PubMed] [Google Scholar]
- Newman EA, Kim DW, Wan J, Wang J, Qian J & Blackshaw S (2018) Foxd1 is required for terminal differentiation of anterior hypothalamic neuronal subtypes. Dev Biol, 439, 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HS, Hannibal J & Fahrenkrug J (2002) Vasoactive intestinal polypeptide induces Per1 and Per2 gene expression in the rat suprachiasmatic nucleus late at night. Eur J Neurosci, 15, 570–574. [DOI] [PubMed] [Google Scholar]
- Nishide SY, Hashimoto K, Nishio T, Honma K & Honma S (2014) Organ-specific development characterizes circadian clock gene Per2 expression in rats. Am J Physiol Regul Integr Comp Physiol, 306, R67–74. [DOI] [PubMed] [Google Scholar]
- Nishide SY, Honma S & Honma K (2008) The circadian pacemaker in the cultured suprachiasmatic nucleus from pup mice is highly sensitive to external perturbation. Eur J Neurosci, 27, 2686–2690. [DOI] [PubMed] [Google Scholar]
- O’Hara BF, Andretic R, Heller HC, Carter DB & Kilduff TS (1995) GABAA, GABAC, and NMDA receptor subunit expression in the suprachiasmatic nucleus and other brain regions. Brain Res Mol Brain Res, 28, 239–250. [DOI] [PubMed] [Google Scholar]
- Obrietan K & van den Pol AN (1995) GABA neurotransmission in the hypothalamus: Developmental reversal from Ca2+ elevating to depressing. J Neurosci, 15, 5065–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H, Honma S, Abe H & Honma K (2003) Periodic absence of nursing mothers phase-shifts circadian rhythms of clock genes in the suprachiasmatic nucleus of rat pups. Eur J Neurosci, 17, 1628–1634. [DOI] [PubMed] [Google Scholar]
- Ohta H, Mitchell AC & McMahon DG (2006) Constant light disrupts the developing mouse biological clock. Pediatr Res, 60, 304–308. [DOI] [PubMed] [Google Scholar]
- Okamura H, Fukui K, Koyama E, Tsutou HL, Tsutou T, Terubayashi H, Fujisawa H & Ibata Y (1983) Time of vasopressin neuron origin in the mouse hypothalamus: Examination by combined technique of immunocytochemistry and [3H]thymidine autoradiography. Brain Res, 285, 223–226. [DOI] [PubMed] [Google Scholar]
- Okamura H & Ibata Y (1994) GRP immunoreactivity shows a day-night difference in the suprachiasmatic nuclear soma and efferent fibers: Comparison to VIP immunoreactivity. Neurosci Lett, 181, 165–168. [DOI] [PubMed] [Google Scholar]
- Okamura H, Murakami S, Uda K, Sungano T, Takahashi Y, Yanaihara C, Yanaihara N & Ibata Y (1986) Coexistence of vasoactive intestinal polypeptide (VIP)-, peptide histidine isoleucine amide (PHI)-, and gastring releasing peptide (GRP)-like immunoreactivity in neurons of the rat suprachiasmatic nucleus. Biomed Res, 7, 295–299. [Google Scholar]
- Ono D, Honma S & Honma K (2013) Cryptochromes are critical for the development of coherent circadian rhythms in the mouse suprachiasmatic nucleus. Nat Commun, 4, 1666. [DOI] [PubMed] [Google Scholar]
- Ono D, Honma S & Honma K (2016) Differential roles of AVP and VIP signaling in the postnatal changes of neural networks for coherent circadian rhythms in the SCN. Sci Adv, 2, e1600960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono D, Honma S, Nakajima Y, Kuroda S, Enoki R & Honma K (2017) Dissociation of Per1 and Bmal1 circadian rhythms in the suprachiasmatic nucleus in parallel with behavioral outputs. Proc Natl Acad Sci U S A, 114, E3699–E3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orquera DP, Nasif S, Low MJ, Rubinstein M & de Souza FSJ (2016) Essential function of the transcription factor Rax in the early patterning of the mammalian hypothalamus. Dev Biol, 416, 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otori Y, Tominaga K, Fukuhara C, Yang J, Yamazaki S, Cagampang FR, Okamura H & Inouye ST (1993) Substance P-like immunoreactivity in the suprachiasmatic nucleus of the rat. Brain Res, 619, 271–277. [DOI] [PubMed] [Google Scholar]
- Padilla SL, Carmody JS & Zeltser LM (2010) Pomc-expressing progenitors give rise to antagonistic neuronal populations in hypothalamic feeding circuits. Nat Med, 16, 403–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak T, Yoo S, Miranda-Angulo AL, Wang H & Blackshaw S (2014) Rax-CreERT2 knock-in mice: A tool for selective and conditional gene deletion in progenitor cells and radial glia of the retina and hypothalamus. PLoS One, 9, e90381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MJ, Brancaccio M, Sethi S, Maywood ES, Satija R, Edwards JK, Jagannath A, Couch Y, Finelli MJ, Smyllie NJ, Esapa C, Butler R, Barnard AR, Chesham JE, Saito S, Joynson G, Wells S, Foster RG, Oliver PL, Simon MM, Mallon AM, Hastings MH & Nolan PM (2015) The regulatory factor ZFHX3 modifies circadian function in SCN via an AT motif-driven axis. Cell, 162, 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggins HD, Goguen D & Rusak B (2005) Gastrin-releasing peptide induces c-Fos in the hamster suprachiasmatic nucleus. Neurosci Lett, 384, 205–210. [DOI] [PubMed] [Google Scholar]
- Piggins HD, Samuels RE, Coogan AN & Cutler DJ (2001) Distribution of substance P and neurokinin-1 receptor immunoreactivity in the suprachiasmatic nuclei and intergeniculate leaflet of hamster, mouse, and rat. J Comp Neurol, 438, 50–65. [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS (1960) Circadian rhythms and the circadian organization of living systems. Cold Spring Harb Symp Quant Biol, 25, 159–184. [DOI] [PubMed] [Google Scholar]
- Pla R, Stanco A, Howard MA, Rubin AN, Vogt D, Mortimer N, Cobos I, Potter GB, Lindtner S, Price JD, Nord AS, Visel A, Schreiner CE, Baraban SC, Rowitch DH & Rubenstein JLR (2017) Dlx1 and Dlx2 promote interneuron GABA synthesis, synaptogenesis, and dendritogenesis. Cereb Cortex, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poitras L, Ghanem N, Hatch G & Ekker M (2007) The proneural determinant MASH1 regulates forebrain Dlx1/2 expression through the I12b intergenic enhancer. Development, 134, 1755–1765. [DOI] [PubMed] [Google Scholar]
- Popp A, Urbach A, Witte OW & Frahm C (2009) Adult and embryonic GAD transcripts are spatiotemporally regulated during postnatal development in the rat brain. PLoS One, 4, e4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DA, Close GC & Fielding BA (1983) Age of appearance of circadian rhythm in salivary cortisol values in infancy. Arch Dis Child, 58, 454–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolo LM, Takahashi JS & Herzog ED (2005) Circadian rhythm generation and entrainment in astrocytes. J Neurosci, 25, 404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puymirat J, Faivre-Bauman A, Bizzini B & Tixier-Vidal A (1982) Prenatal and postnatal ontogenesis of neurotransmitter-synthetizing enzymes and [125I]tetanus toxin binding capacity in the mouse hypothalamus. Brain Res, 255, 199–206. [DOI] [PubMed] [Google Scholar]
- Quattrocolo G, Fishell G & Petros TJ (2017) Heterotopic transplantations reveal environmental influences on interneuron diversity and maturation. Cell Rep, 21, 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed HE, Cutler DJ, Brown TM, Brown J, Coen CW & Piggins HD (2002) Effects of vasoactive intestinal polypeptide on neurones of the rat suprachiasmatic nuclei in vitro. J Neuroendocrinol, 14, 639–646. [DOI] [PubMed] [Google Scholar]
- Reppert SM & Schwartz WJ (1983) Maternal coordination of the fetal biological clock in utero. Science, 220, 969–971. [DOI] [PubMed] [Google Scholar]
- Reppert SM & Schwartz WJ (1984) The suprachiasmatic nuclei of the fetal rat: Characterization of a functional circadian clock using 14C-labeled deoxyglucose. J Neurosci, 4, 1677–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM & Uhl GR (1987) Vasopressin messenger ribonucleic acid in supraoptic and suprachiasmatic nuclei: appearance and circadian regulation during development. Endocrinology, 120, 2483–2487. [DOI] [PubMed] [Google Scholar]
- Rivkees SA (2007) The development of circadian rhythms: From animals to humans. Sleep Med Clin, 2, 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivkees SA, Weaver DR & Reppert SM (1992) Circadian and developmental regulation of Oct-2 gene expression in the suprachiasmatic nuclei. Brain Res, 598, 332–336. [DOI] [PubMed] [Google Scholar]
- Robichaux MA & Cowan CW (2014) Signaling mechanisms of axon guidance and early synaptogenesis. Curr Top Behav Neurosci, 16, 19–48. [DOI] [PubMed] [Google Scholar]
- Romero MT, Lehman MN & Silver R (1993) Age of donor influences ability of suprachiasmatic nucleus grafts to restore circadian rhythmicity. Brain Res Dev Brain Res, 71, 45–52. [DOI] [PubMed] [Google Scholar]
- Romero MT & Silver R (1990) Time course of peptidergic expression in fetal suprachiasmatic nucleus transplanted into adult hamster. Brain Res Dev Brain Res, 57, 1–6. [DOI] [PubMed] [Google Scholar]
- Romijn HJ, Sluiter AA, Pool CW, Wortel J & Buijs RM (1996) Differences in colocalization between Fos and PHI, GRP, VIP and VP in neurons of the rat suprachiasmatic nucleus after a light stimulus during the phase delay versus the phase advance period of the night. J Comp Neurol, 372, 1–8. [DOI] [PubMed] [Google Scholar]
- Romijn HJ, Sluiter AA, Wortel J, Van Uum JF & Buijs RM (1998) Immunocytochemical evidence for a diurnal rhythm of neurons showing colocalization of VIP with GRP in the rat suprachiasmatic nucleus. J Comp Neurol, 391, 397–405. [PubMed] [Google Scholar]
- Rowitch DH & Kriegstein AR (2010) Developmental genetics of vertebrate glial-cell specification. Nature, 468, 214–222. [DOI] [PubMed] [Google Scholar]
- Roy A, de Melo J, Chaturvedi D, Thein T, Cabrera-Socorro A, Houart C, Meyer G, Blackshaw S & Tole S (2013) LHX2 is necessary for the maintenance of optic identity and for the progression of optic morphogenesis. J Neurosci, 33, 6877–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saaltink DJ, Havik B, Verissimo CS, Lucassen PJ & Vreugdenhil E (2012) Doublecortin and doublecortin-like are expressed in overlapping and non-overlapping neuronal cell population: implications for neurogenesis. J Comp Neurol, 520, 2805–2823. [DOI] [PubMed] [Google Scholar]
- Sarin S, Zuniga-Sanchez E, Kurmangaliyev YZ, Cousins H, Patel M, Hernandez J, Zhang KX, Samuel MA, Morey M, Sanes JR & Zipursky SL (2018) Role for Wnt signaling in retinal neuropil development: Analysis via RNA-Seq and in vivo somatic CRISPR mutagenesis. Neuron, 98, 109–126 e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Mizoro Y, Atobe Y, Fujimoto Y, Yamaguchi Y, Fustin JM, Doi M & Okamura H (2011) Transportin 1 in the mouse brain: Appearance in regions of neurogenesis, cerebrospinal fluid production/sensing, and circadian clock. J Comp Neurol, 519, 1770–1780. [DOI] [PubMed] [Google Scholar]
- Schmidt H (2012) Three functional facets of calbindin D-28k. Front Mol Neurosci, 5, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaller B (2014) Calretinin: from a “simple” Ca2+ buffer to a multifunctional protein implicated in many biological processes. Front Neuroanat, 8, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekaran S, Lupi D, Jones SL, Sheely CJ, Hattar S, Yau KW, Lucas RJ, Foster RG & Hankins MW (2005) Melanopsin-dependent photoreception provides earliest light detection in the mammalian retina. Curr Biol, 15, 1099–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seress L (1985) Postnatal neurogenesis in the rat hypothalamus. Brain Res, 354, 156–160. [DOI] [PubMed] [Google Scholar]
- Seron-Ferre M, Valenzuela GJ & Torres-Farfan C (2007) Circadian clocks during embryonic and fetal development. Birth Defects Res C Embryo Today, 81, 204–214. [DOI] [PubMed] [Google Scholar]
- Shen H, Watanabe M, Tomasiewicz H & Glass JD (2001) Genetic deletions of NCAM and PSA impair circadian function in the mouse. Physiol Behav, 73, 185–193. [DOI] [PubMed] [Google Scholar]
- Shibata S & Moore RY (1987) Development of neuronal activity in the rat suprachiasmatic nucleus. Brain Res, 431, 311–315. [DOI] [PubMed] [Google Scholar]
- Shibata S, Newman GC & Moore RY (1987) Effects of calcium ions on glucose utilization in the rat suprachiasmatic nucleus in vitro. Brain Res, 426, 332–338. [DOI] [PubMed] [Google Scholar]
- Shibata S, Tsuneyoshi A, Hamada T, Tominaga K & Watanabe S (1992) Effect of Substance P on circadian rhythms of firing activity and the 2-deoxyglucose uptake in the rat suprachiasmatic nucleus in vitro. Brain Res, 597, 257–263. [DOI] [PubMed] [Google Scholar]
- Silver R, Romero MT, Besmer HR, Leak R, Nunez JM & LeSauter J (1996) Calbindin-D28K cells in the hamster SCN express light-induced Fos. Neuroreport, 7, 1224–1228. [DOI] [PubMed] [Google Scholar]
- Shimada M & Nakamura T (1973) Time of neuron origin in mouse hypothalamic nuclei. Exp Neurol, 41, 163–173. [DOI] [PubMed] [Google Scholar]
- Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, Qi L, Qian J & Blackshaw S (2010) A genomic atlas of mouse hypothalamic development. Nat Neurosci, 13, 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura H, Moriya T, Sudo M, Wakamatsu H, Akiyama M, Miyake Y & Shibata S (2001) Differential daily expression of Per1 and Per2 mRNA in the suprachiasmatic nucleus of fetal and early postnatal mice. Eur J Neurosci, 13, 687–693. [DOI] [PubMed] [Google Scholar]
- Shinohara K, Honma S, Katsuno Y, Abe H & Honma K (1995) Two distinct oscillators in the rat suprachiasmatic nucleus in vitro. Proc Natl Acad Sci U S A, 92, 7396–7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara K, Tominaga K, Isobe Y & Inouye ST (1993) Photic regulation of peptides located in the ventrolateral subdivision of the suprachiasmatic nucleus of the rat: Daily variations of vasoactive intestinal polypeptide, gastrin-releasing peptide, and neuropeptide Y. J Neurosci, 13, 793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver R, Sookhoo A, LeSauter J, Stevens P, Jansen H & Lehman M (1999) Multiple regulatory elements result in regional specificity in circadian rhythms of neuropeptide expression in mouse SCN. Neuroreport, 10, 3165–3174. [DOI] [PubMed] [Google Scholar]
- Sladek M, Sumova A, Kovacikova Z, Bendova Z, Laurinova K & Illnerova H (2004) Insight into molecular core clock mechanism of embryonic and early postnatal rat suprachiasmatic nucleus. Proc Natl Acad Sci U S A, 101, 6231–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L & Canal MM (2009) Expression of circadian neuropeptides in the hypothalamus of adult mice is affected by postnatal light experience. J Neuroendocrinol, 21, 946–953. [DOI] [PubMed] [Google Scholar]
- Southey BR, Lee JE, Zamdborg L, Atkins N Jr., Mitchell JW, Li M, Gillette MU, Kelleher NL & Sweedler JV (2014) Comparing label-free quantitative peptidomics approaches to characterize diurnal variation of peptides in the rat suprachiasmatic nucleus. Anal Chem, 86, 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speh JC & Moore RY (1993) Retinohypothalamic tract development in the hamster and rat. Brain Res Dev Brain Res, 76, 171–181. [DOI] [PubMed] [Google Scholar]
- Sterniczuk R, Colijn MA, Nunez M & Antle MC (2010) Investigating the role of Substance P in photic responses of the circadian system: Individual and combined actions with gastrin-releasing peptide. Neuropharmacology, 58, 277–285. [DOI] [PubMed] [Google Scholar]
- Stopa EG, King JC, Lydic R & Schoene WC (1984) Human brain contains vasopressin and vasoactive intestinal polypeptide neuronal subpopulations in the suprachiasmatic region. Brain Res, 297, 159–163. [DOI] [PubMed] [Google Scholar]
- Strecker GJ, Wuarin JP & Dudek FE (1997) GABAA-mediated local synaptic pathways connect neurons in the rat suprachiasmatic nucleus. J Neurophysiol, 78, 2217–2220. [DOI] [PubMed] [Google Scholar]
- Sumova A, Sladek M, Polidarova L, Novakova M & Houdek P (2012) Circadian system from conception till adulthood. Prog Brain Res, 199, 83–103. [DOI] [PubMed] [Google Scholar]
- Swaab DF, Van Someren EJ, Zhou JN & Hofman MA (1996) Biological rhythms in the human life cycle and their relationship to functional changes in the suprachiasmatic nucleus. Prog Brain Res, 111, 349–368. [DOI] [PubMed] [Google Scholar]
- Tabata H (2015) Diverse subtypes of astrocytes and their development during corticogenesis. Front Neurosci, 9, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS (2017) Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet, 18, 164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji K, Miguel-Hidalgo JJ & Tohyama M (1991) Substance P-immunoreactive innervation from the retina to the suprachiasmatic nucleus in the rat. Brain Res, 568, 223–229. [DOI] [PubMed] [Google Scholar]
- Tamada Y, Tanaka M, Munekawa K, Hayashi S, Okamura H, Kubo T, Hisa Y & Ibata Y (1998) Neuron-glia interaction in the suprachiasmatic nucleus: A double labeling light and electron microscopic immunocytochemical study in the rat. Brain Res Bull, 45, 281–287. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Hayashi S, Tamada Y, Ikeda T, Hisa Y, Takamatsu T & Ibata Y (1997) Direct retinal projections to GRP neurons in the suprachiasmatic nucleus of the rat. Neuroreport, 8, 2187–2191. [DOI] [PubMed] [Google Scholar]
- Tarttelin EE, Bellingham J, Bibb LC, Foster RG, Hankins MW, Gregory-Evans K, Gregory-Evans CY, Wells DJ & Lucas RJ (2003) Expression of opsin genes early in ocular development of humans and mice. Exp Eye Res, 76, 393–396. [DOI] [PubMed] [Google Scholar]
- Todkar K, Scotti AL & Schwaller B (2012) Absence of the calcium-binding protein calretinin, not of calbindin D-28k, causes a permanent impairment of murine adult hippocampal neurogenesis. Front Mol Neurosci, 5, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonetti L, Fabbri M, Martoni M & Natale V (2012) Season of birth and mood seasonality in late childhood and adolescence. Psychiatry Res, 195, 66–68. [DOI] [PubMed] [Google Scholar]
- Torres-Farfan C, Rocco V, Monso C, Valenzuela FJ, Campino C, Germain A, Torrealba F, Valenzuela GJ & Seron-Ferre M (2006) Maternal melatonin effects on clock gene expression in a nonhuman primate fetus. Endocrinology, 147, 4618–4626. [DOI] [PubMed] [Google Scholar]
- Tretter V & Moss SJ (2008) GABAA receptor dynamics and constructing GABAergic synapses. Front Mol Neurosci, 1, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso CF, Simon T, Greenlaw AC, Puri T, Mieda M & Herzog ED (2017) Astrocytes regulate daily rhythms in the suprachiasmatic nucleus and behavior. Curr Biol, 27, 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol AN (1980) The hypothalamic suprachiasmatic nucleus of rat: Intrinsic anatomy. J Comp Neurol, 191, 661–702. [DOI] [PubMed] [Google Scholar]
- van den Pol AN (1997) GABA immunoreactivity in hypothalamic neurons and growth cones in early development in vitro before synapse formation. J Comp Neurol, 383, 178–188. [DOI] [PubMed] [Google Scholar]
- van den Pol AN & Tsujimoto KL (1985) Neurotransmitters of the hypothalamic suprachiasmatic nucleus: Immunocytochemical analysis of 25 neuronal antigens. Neuroscience, 15, 1049–1086. [DOI] [PubMed] [Google Scholar]
- Van Gelder RN & Buhr ED (2016) Ocular photoreception for circadian rhythm entrainment in mammals. Annu Rev Vis Sci, 2, 153–169. [DOI] [PubMed] [Google Scholar]
- Vandunk C, Hunter LA & Gray PA (2011) Development, maturation, and necessity of transcription factors in the mouse suprachiasmatic nucleus. J Neurosci, 31, 6457–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez-Ruiz S, Maya-Barrios JA, Torres-Narvaez P, Vega-Martinez BR, Rojas-Granados A, Escobar C & Angeles-Castellanos M (2014) A light/dark cycle in the NICU accelerates body weight gain and shortens time to discharge in preterm infants. Early Hum Dev, 90, 535–540. [DOI] [PubMed] [Google Scholar]
- Walton JC, McNeill J.K.t., Oliver KA & Albers HE (2017) Temporal regulation of GABAA receptor subunit expression: Role in synaptic and extrasynaptic communication in the suprachiasmatic nucleus. eNeuro, 4, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MH, Chen N & Wang JH (2014) The coupling features of electrical synapses modulate neuronal synchrony in hypothalamic superachiasmatic nucleus. Brain Res, 1550, 9–17. [DOI] [PubMed] [Google Scholar]
- Ware M, Hamdi-Roze H & Dupe V (2014) Notch signaling and proneural genes work together to control the neural building blocks for the initial scaffold in the hypothalamus. Front Neuroanat, 8, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver DR (1998) The suprachiasmatic nucleus: A 25-year retrospective. J Biol Rhythms, 13, 100–112. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M & Reppert SM (1995) Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron, 14, 697–706. [DOI] [PubMed] [Google Scholar]
- Welsh DK & Reppert SM (1996) Gap junctions couple astrocytes but not neurons in dissociated cultures of rat suprachiasmatic nucleus. Brain Res, 706, 30–36. [DOI] [PubMed] [Google Scholar]
- Wilcox AG, Vizor L, Parsons MJ, Banks G & Nolan PM (2017) Inducible knockout of mouse Zfhx3 emphasizes its key role in setting the pace and amplitude of the adult circadian clock. J Biol Rhythms, 32, 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams WP 3rd & Kriegsfeld LJ (2012) Circadian control of neuroendocrine circuits regulating female reproductive function. Front Endocrinol (Lausanne), 3, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womac AD, Burkeen JF, Neuendorff N, Earnest DJ & Zoran MJ (2009) Circadian rhythms of extracellular ATP accumulation in suprachiasmatic nucleus cells and cultured astrocytes. Eur J Neurosci, 30, 869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreschnig D, Dolatshad H & Davis FC (2014) Embryonic development of circadian oscillations in the mouse hypothalamus. J Biol Rhythms, 29, 299–310. [DOI] [PubMed] [Google Scholar]
- Xie Y & Dorsky RI (2017) Development of the hypothalamus: conservation, modification and innovation. Development, 144, 1588–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Hu XY, Wu L & Zhou JN (2003) Neurotensin expressing neurons developed earlier than vasoactive intestinal polypeptide and vasopressin expressing neurons in the human suprachiasmatic nucleus. Neurosci Lett, 335, 175–178. [DOI] [PubMed] [Google Scholar]
- Yagita K, Horie K, Koinuma S, Nakamura W, Yamanaka I, Urasaki A, Shigeyoshi Y, Kawakami K, Shimada S, Takeda J & Uchiyama Y (2010) Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc Natl Acad Sci U S A, 107, 3846–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Takayama Y, Okada M & Nagai K (1999) Effect of continuous infusion of anti-L1 antibody into the third cerebral ventricle above the suprachiasmatic nucleus on the circadian rhythm of locomotor activity in rats. Biol Rhythm Res, 30, 573–582. [Google Scholar]
- Yamada S, Takayama Y, Seki T, Okada M & Nagai K (2003) Changes in L1 and NCAM expression in the rat suprachiasmatic nucleus during growth and after orbital enucleation. Brain Res Dev Brain Res, 143, 189–198. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M & Okamura H (2003) Synchronization of cellular clocks in the suprachiasmatic nucleus. Science, 302, 1408–1412. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Suzuki T, Mizoro Y, Kori H, Okada K, Chen Y, Fustin JM, Yamazaki F, Mizuguchi N, Zhang J, Dong X, Tsujimoto G, Okuno Y, Doi M & Okamura H (2013) Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science, 342, 85–90. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Alones V & Menaker M (2002) Interaction of the retina with suprachiasmatic pacemakers in the control of circadian behavior. J Biol Rhythms, 17, 315–329. [DOI] [PubMed] [Google Scholar]
- Yoo S & Blackshaw S (2018) Regulation and function of neurogenesis in the adult mammalian hypothalamus. Prog Neurobiol, S0301-0082, 30200–30209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, Yun CH, Ahn JH, Suh S, Cho HJ, Lee SK, Yoo HJ, Seo JA, Kim SG, Choi KM, Baik SH, Choi DS, Shin C & Kim NH (2015) Evening chronotype is associated with metabolic disorders and body composition in middle-aged adults. J Clin Endocrinol Metab, 100, 1494–1502. [DOI] [PubMed] [Google Scholar]