Abstract
Inactivation is a fundamental characteristic of Na+ channels, and small changes cause skeletal muscle paralysis and myotonia, epilepsy, and cardiac arrhythmia. Brain Nav1.2a channels have faster inactivation than cardiac Nav1.5 channels, but minor differences in inactivation gate structure are not responsible. We constructed chimeras in which the C termini beyond the fourth homologous domains of Nav1.2a and Nav1.5 were exchanged. Replacing the C-terminal domain (CT) of Nav1.2a with that of Nav1.5 (Nav1.2/1.5CT) slowed inactivation at +40 mV ≈2-fold, making it similar to Nav1.5. Conversely, replacing the CT of Nav1.5 with that of Nav1.2a (Nav1.5/1.2CT) accelerated inactivation, making it similar to Nav1.2a. Activation properties were unaffected. The voltage dependence of steady-state inactivation of Nav1.5 is 16 mV more negative than that of Nav1.2a. The steady-state inactivation curve of Nav1.2a was shifted +12 mV in Nav1.2/1.5CT, consistent with destabilization of the inactivated state. Conversely, Nav1.5/1.2CT was shifted −14 mV relative to Nav1.5, consistent with stabilization of the inactivated state. Although these effects of exchanging C termini were consistent with their effects on inactivation kinetics, they magnified the differences in the voltage dependence of inactivation between brain and cardiac channels rather than transferring them. Thus, other parts of these channels determine the basal difference in steady-state inactivation. Deletion of the distal half of either the Nav1.2 or Nav1.5 CTs accelerated open-state inactivation and negatively shifted steady-state inactivation. Thus, the C terminus has a strong influence on kinetics and voltage dependence of inactivation in brain Nav1.2 and cardiac Nav1.5 channels and is primarily responsible for their differing rates of channel inactivation.
Voltage-gated Na+ channels play an essential role in most excitable cells where they transiently increase the Na+ conductance in response to membrane depolarization (1). Brain Na+ channels generate the action potential that is the basis for neuronal electrical excitability. Cardiac Na+ channels cause the initial depolarization of the cardiac action potential that is responsible for the rapid spread of excitation through the atria and ventricles, generating synchronous cardiac contraction.
The principal component of the voltage-gated Na+ channel is a 260-kDa α subunit, which forms the pore and is responsible for the voltage-sensitive properties of the channel (2–4). The primary sequence of the α subunit contains four homologous domains (I–IV) each containing six transmembrane α-helices (S1–S6) as well as a segment connecting S5 and S6 of each domain that forms the narrow outer end of the ion-conducting pore.
Na+ channels respond to membrane depolarization by activating, opening, allowing Na+ current to flow, and then rapidly inactivating. Fast inactivation is a fundamental property of Na+ channels, and minor modifications cause inherited skeletal muscle myotonia and paralysis, cardiac arrhythmia, and epilepsy (5, 6). Nav1.2a, a principal Na+ channel isoform in adult brain (7, 8), has faster inactivation kinetics and more positive voltage dependence of inactivation than the cardiac Nav1.5 channel (9).
The fast inactivation gate is formed by the intracellular loop between domains III and IV (10–12), which blocks the ion conducting pore a few milliseconds after the channel opens (13). However, replacement of the Nav1.5 inactivation gate with that of Nav1.2a does not transfer the inactivation properties (14). Therefore, the molecular determinants governing these inactivation properties must lie elsewhere in the channel.
One possible location is the C-terminal domain (CT) of the α subunit beyond segment IVS6. Multiple mutations that cause human diseases related to inactivation have been identified in the CT of the cardiac sodium channel (15–20). In these experiments, we have studied chimeric channels exchanging Nav1.2a and Nav1.5 C termini. Substituting only the CT transferred inactivation kinetics from one channel to the other and had additional major effects on the voltage dependence of steady-state inactivation. Further analysis using truncated brain and heart channels indicated that the distal half of the CT of both channels inhibited inactivation. Overall, our findings indicate that the CT plays a critical role in inactivation and is responsible for the differences in inactivation rate between cardiac and brain Na+ channels.
Experimental Procedures
Construction of Chimeras and Mutagenesis.
Plasmid, pCDM8-rH1, encoding the cardiac Nav1.5 α subunit, has been described (21). Plasmid pCDM8-rIIA encoding the Nav1.2a α subunit (22) contains a unique, silent XhoI restriction site at the junction between IVS6 and the C-terminal cytoplasmic region (amino acid positions L1776–E1777). An equivalent restriction site is present in pCDM8-rH1. Chimeric Na+ channels were constructed by exchanging a restriction fragment between this XhoI site and a vector ClaI site including the entire CT of Nav1.2a and Nav1.5 Na+ channels. The resulting chimeric channels Nav1.2-E1777-Nav1.5-E1775CT and Nav1.5E1775-Nav1.2E1777CT were designated Nav1.2/1.5CT and Nav1.5/1.2CT, respectively. The truncation mutations deleting segments of the CT of the Nav1.2a channel α subunit were constructed by standard PCR mutagenesis by insertion of a stop codon. Mutagenic oligonucleotides were designed in reverse orientation with an in-frame TAA stop codon followed by ClaI restriction site; the forward oligonucleotide was designed from sequences in the IVS6 transmembrane segment, upstream of the XhoI site. The PCR fragments were each digested with XhoI and ClaI and subcloned into the identical sites of pCDM8-rIIA. This procedure yielded the truncated Nav1.2a channel constructs Nav1.2ΔE1777, Nav1.2ΔK1863, Nav1.2ΔI1877, Nav1.2ΔK1890, Nav1.2ΔA1909, Nav1.2ΔS1929, Nav1.2ΔT1951, Nav1.2ΔS1977, and Nav1.2ΔK1998.
Cell Culture and Transfection.
The tsA-201 cell subclone of HEK293 cells was maintained as described (23). The plasmid containing cDNA for the Na+ channel was cotransfected with pIRES-EYFP (CLONTECH) encoding the yellow variant of green fluorescent protein by CaPO4 precipitation as described (24). Twelve hours after transfection, the cells were replated at low density for electrophysiological recordings. Transfected cells were identified by fluorescence.
Electrophysiology and Data Analysis.
Whole-cell patch-clamp recordings were performed at room temperature by using an Axopatch 200B amplifier (Axon Instruments, Union City, CA) with PCLAMP 6 software (Axon Instruments). Capacitative currents were minimized by using the amplifier circuitry. Seventy percent prediction and 90–95% series resistance compensation were routinely used. Remaining linear capacity and leakage currents were eliminated by P/4 subtraction. The intracellular solution contained 120 mM CsAspartate, 5 mM NaCl, 2 mM MgCl2, 10 mM EGTA, 10 mM Hepes, pH 7.3 with CsOH. The extracellular solution contained 140 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 10 mM Hepes, pH 7.4 with NaOH. Current signals were filtered at 5 or 10 kHz before sampling.
Conductance-voltage (g-V) relationships (activation curves) were calculated from the current-voltage (I–V) relationships according to g = INa/(V − ENa), where INa was the peak Na+ current measured at potential, V, and ENa, the calculated equilibrium potential.
Normalized activation and inactivation curves were fit to Boltzmann relationships of the form: y = 1/{1 + exp [(V − V1/2)/k]} + A, where y is normalized gNa or INa, A, the baseline, V, the membrane potential, V1/2, the voltage of half-maximal activation, Va, or inactivation, Vh, and k is a slope factor. In fitting the activation curves, A was fixed at 0. Inactivation kinetics were evaluated by fitting the decay of the current from the peak to a point 60 ms from the beginning of the voltage stimulus, with the sum of two exponentials and a baseline. Statistical results are given as mean ± SEM. Statistical analyses were performed by using PRISM 3 (GraphPad, San Diego) software. The threshold P value for statistical significance was 0.05.
Nav1.2a and its mutants generate a variable amount of persistent Na+ current in different transfected cells, which was not observed for Nav1.5 and its mutants (25).† Only cells with less than 8% persistent current were selected for analysis to permit comparison with Nav1.5.
Results
Inactivation of Na+ Channels from Closed and Open States.
After strong depolarization, Na+ channels are
thought to move through a series of closed states (Cn; Scheme
1), reach the open state (O), and rapidly inactivate
(I).
![]()
These state changes are driven by voltage-dependent movement of gating charges in the channel structure. At positive membrane potentials, Na+ channels rapidly open and the rate of decay of the Na+ current reflects the inactivation of open channels (O → I). For weaker depolarizations, Na+ channels transit rightward through the series of close states (Cn) but usually inactivate by transition into one of the CnI states before opening (Scheme 1), causing steady-state inactivation. The molecular transitions governing rapid inactivation from the open state and slower, steady-state inactivation from closed states are distinct (Scheme 1), and we show below that the CT affects these aspects of inactivation differentially.
Substitution of the CTs Does Not Alter the Voltage Dependence of Activation.
The activation properties of Nav1.2a (rIIA; refs. 26 and 27) and Nav1.5 (rH1; refs. 28–30) channels transiently expressed in tsA-201 cells are compared in Fig. 1. Nav1.2a currents activate positive to −30 mV and peak near +10 mV (Fig. 1 A and E). Nav1.5 currents activate at more negative potentials. They are first detectable at −50 mV and peak near −10 mV (Fig. 1 B and F).
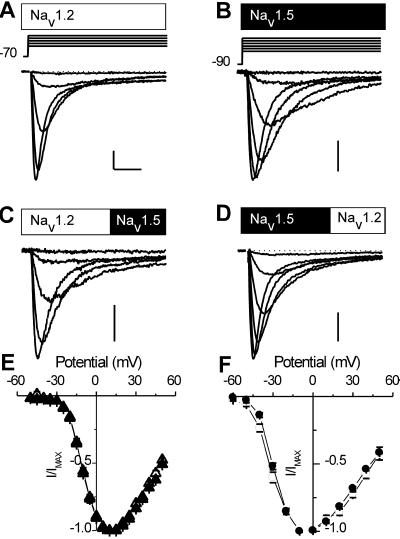
Figure 1.
Na+ currents in tsA-201 cells transfected with wild-type Nav1.2a, Nav1.5, or chimeric Na+ channels. (A–E) Na+ current traces recorded in response to depolarizations to the indicated voltages from representative cells transfected with either brain Nav1.2a α subunits (A), cardiac Nav1.5 α subunits (B), Nav1.2/1.5CT chimeric α subunits (C), or Nav1.5/1.2CT chimeric α subunits (D). Traces were obtained in response to stimuli at 10-mV intervals as indicated by the voltage protocols above each column. Calibration bars: 1 nA, 3 ms. (E) Mean normalized current-voltage (I–V) curves for Nav1.2a (▴; n = 9) and Nav1.2/1.5CT (▵, n = 11). (F) Mean normalized I–V curves for Nav1.5 (●, n = 7) and Nav1.5/1.2CT (○, n = 6). Chimeric channels generated smaller Na+ currents than the parental channels. Mean current densities were: Nav1.2a, 550 ± 50 pA/pF (n = 46); Nav1.2/1.5CT, 310 ± 50 pA/pF (n = 15); Nav1.5, 300 ± 100pA/pF (n = 7); Nav1.5/1.2CT, 140 ± 40 pA/pF (n = 7).
To examine the role of the CT, we constructed chimeras between Nav1.2a and Nav1.5 in which the CTs of the channels were exchanged. In Nav1.2/1.5CT, the Nav1.5 CT, beginning with E1775, replaced the CT of Nav1.2a, which had been truncated before E1777. In Nav1.5/1.2CT, the Nav1.2a tail from E1777 replaced the CT of the Nav1.5 channel, beginning with the E1775. The voltage dependence of activation was unaffected in either chimeric channel, indicating that the CT is not responsible for the markedly different voltage dependence of activation of the Nav1.2a and Nav1.5 channels.
To examine the differences in Na+ current time course more closely, mean Na+ currents from the wild-type and chimeric channels were compared (Fig. 2A). The Nav1.5 channel activates more slowly than the Nav1.2a channel, even at the strongly positive potential of +40 mV. These differing activation kinetics were unaffected when the CT tail was replaced by that of the other channel. This finding was confirmed at a range of potentials by measuring the time to half activation (Fig. 2B). At all positive potentials, the time to half activation was determined by the identity of the parental channel rather than by the identity of the CT.
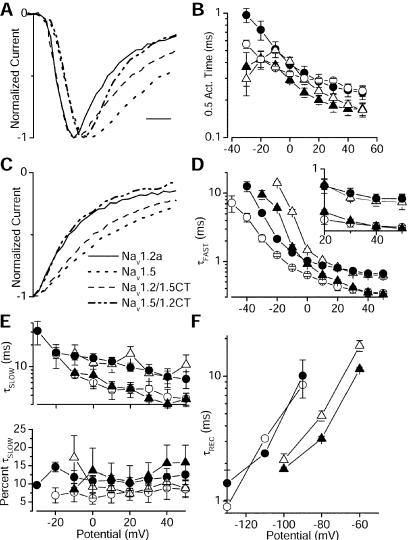
Figure 2.
Kinetics of the Na+ current. (A) Averages of current traces at +40mV for each channel type. (B) Semilogarithmic plot of times of half activation at the indicated potentials for Nav1.2a (▴; n = 8), Nav1.2/1.5CT (▵; n = 9), Nav1.5 (●; n = 7), and Nav1.5/1.2CT (○; n = 5). (C) Traces from A shifted in time so that their peaks are aligned to allow a direct comparison of inactivation kinetics. (D and E) Parameters derived from fits of the sum of two exponential functions to the decay of the current traces during depolarizations to the indicated potentials. Semilogarithmic plot of the voltage dependence of τFast (D), τSlow (E Upper), and fraction of the decay represented by τSlow (E Lower). (D Inset) Replot of the time constants between +20 and +50 mV on a linear scale. (F) Time constants for recovery from inactivation at the indicated potentials. The points for Nav1.5/1.2CT were obtained from a single cell because of difficulties in expressing that construct.
The CT Determines the Kinetics of Inactivation.
Nav1.2a currents decay faster than Nav1.5 currents (compare Fig. 1 A and B). In contrast to the lack of effect on activation, substitution of the Nav1.5 tail in the Nav1.2a channel slowed the decay of the current (compare Fig. 1 A and C). The converse effect is observed in Nav1.5/1.2CT, which has faster inactivation than the parental Nav1.5 channel (compare Fig. 1 B and D).
Whereas Nav1.5 and the Nav1.5/1.2CT activate with identical kinetics (Fig. 2 A and B), the two traces diverge at the peak with the Nav1.5/1.2CT current decaying significantly more rapidly than Nav1.5. Similarly, Nav1.2/1.5CT inactivates more slowly than Nav1.2a. These effects are seen clearly when the peaks of the current traces are aligned so that inactivation kinetics can be compared directly (Fig. 2C). Nav1.5/1.2CT inactivates with the kinetics of the Nav1.2a channel. Nav1.2/1.5CT inactivates with the kinetics of the Nav1.5 channel. To quantify its time course, current decay was fit with the sum of two exponential components (Fig. 2 D and E). Although the second, slower exponential was required, >80% of the decay was described by the faster exponential component (Fig. 2E). The Nav1.2/1.5CT channel inactivates more slowly than the parental Nav1.2a channel at all potentials, whereas the Nav1.5/1.2CT channel inactivates more rapidly than the parental Nav1.5 channel at all potentials (Fig. 2D). In addition, at potentials positive to +20 mV the time constant of Na+ current decay became identical to that of the channel from which the CT was derived (Fig. 2D Inset). At these positive potentials current decay reflects inactivation of open channels (Scheme I) and the differences in inactivation kinetics are determined entirely by the CT. The slow time constants were also determined by the identity of the CT (Fig. 2E). Thus, the isoform-specific rates of inactivation of open channels are determined primarily by the identity of the CT for Nav1.2a and Nav1.5.
Recovery from Inactivation Is Unaffected by the Identity of the CT.
We also determined the rate of recovery from inactivation. Interestingly, recovery from inactivation was unaffected by exchange of the CTs (Fig. 2F).
The CT Modifies the Voltage Dependence of Steady-State Inactivation.
A large difference between the voltage dependence of inactivation of the Nav1.2a-derived channels (Nav1.2a and Nav1.2/1.5CT) and the Nav1.5-derived channels (Nav1.5 and Nav1.5/1.2CT) is expected because of the difference in voltage dependence of activation (Figs. 1 E and F and 3). The voltage dependence of steady-state inactivation is set by multiple factors, including the rate and stability of the inactivation process. Thus, most mutations that slow inactivation from the open state also cause a positive shift in steady-state inactivation (e.g., refs. 31 and 32). Consistent with this finding, when the CT of Nav1.2a is substituted for that of Nav1.5 in Nav1.5/1.2CT, the faster rate of inactivation is accompanied by a shift of steady-state inactivation toward more negative potentials, from −60.5 ± 0.2 mV to −75.7 ± 0.1 mV (Fig. 3). Conversely, steady-state inactivation of the Nav1.2a channel shifts to more positive potentials, from −44.1 ± 0.2 mV to −32.4 ± 0.3 mV, when the Nav1.5 CT is substituted in Nav1.2/1.5CT. Thus, steady-state inactivation of Nav1.2a from closed states is weakened by substitution of the CT of Nav1.5, and steady-state inactivation of Nav1.5 is strengthened by substitution of the Nav1.2a CT. The identity of the CT also determines the slope of the inactivation curve, which is shallower for Nav1.5 than for Nav1.2a channels (Fig. 3). These effects parallel the changes in rates of inactivation of these chimeras from the open state (Figs. 1 and 2). Despite this parallel, the large initial differences in the voltage dependence of steady-state inactivation between the channel isoforms are only increased by exchanging the C-terminal tails, becoming more negative in Nav1.5/1.2CT and more positive in Nav1.2/1.5CT. This finding indicates that molecular features of these two channel types other than the CT and the inactivation gate itself are dominant determinants of the voltage dependence of steady-state inactivation.
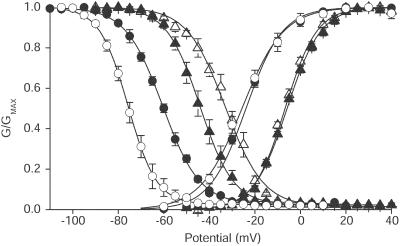
Figure 3.
Voltage dependence of activation and inactivation. Mean voltage dependence of activation and inactivation for Nav1.2a (▴), Nav1.2/1.5CT (▵), Nav1.5 (●), and Nav1.5/1.2CT (○). For Nav1.2a and Nav1.2/1.5CT the inactivation protocol used a test pulse to 0 mV, preceded by 100-ms long prepulses to the indicated potentials. The holding potential was −100 mV. For Nav1.5 and Nav1.5/1.2CT the test potential was −20 mV and the holding potential was −120 mV. The continuous lines are mean Boltzmann fits to the individual experiments with the following mean parameters: Activation—Nav1.2a (n = 9), Va = −5.9 ± 0.5, k = 7.2 ± 0.3; Nav1.2/1.5CT (n = 11), Va = −6.7 ± 0.6, k = 6.8 ± 0.3; Nav1.5 (n = 11), Va = −25.9 ± 1.0, k = 7.6 ± 0.3; Nav1.5/1.2CT (n = 8), Va = −23.7 ± 1.9, k = 9.0 ± 0.5; Inactivation—Nav1.2a (n = 7), Vh = −43.9 ± 1.7, k = 6.8 ± 0.3; Nav1.2/1.5CT (n = 10), Vh = −32.6 ± 1.5, k = 8.1 ± 0.3; Nav1.5 (n = 7), Vh = −60.4 ± 0.8, k = 8.4 ± 0.3; Nav1.5/1.2CT (n = 6), Vh = −75.1 ± 1.6, k = 6.3 ± 0.5. ANOVA followed by Tukey's post test showed that the activation V1/2 and k values for the Nav1.2a-derived channels differed from those of the Nav1.5-derived channels. The k value for Nav1.5/1.2CT differed significantly from the k values for the other constructs. For inactivation curves, the V1/2 values for all constructs differed significantly. The inactivation curve k values of Nav1.2a and Nav1.5/1.2CT were similar to each other and significantly different from Nav1.5 or Nav1.2/1.5CT.
The Distal CTs of Nav1.2a and Nav1.5 Inhibit Inactivation.
The studies with chimeric channels define the effects of the CTs of the Nav1.2a and Nav1.5 channels relative to each other but they do not reveal the nature of the effect of the Na+ channel CT per se. To obtain this type of information, the simplest experiment would be to delete the CT entirely, thereby removing its influence. However, truncation of Nav1.2a at E1777 produced a channel that did not give rise to detectable currents. Smaller truncations at K1863 and I1877 gave rise to Na+ currents too small to distinguish from endogenous currents that are sometimes observed in tsA-201 cells (33, 34).
The channel with the largest C-terminal deletion that produced adequate Na+ currents for analysis was Nav1.2ΔK1890. Although the activation curve of Nav1.2ΔK1890 did not differ from Nav1.2, the inactivation curve was shifted −7.3 mV (Fig. 4A). Inactivation was more likely at each potential. Inactivation during depolarizations also was faster. The fast time constant for inactivation of Nav1.2ΔK1890 became significantly faster than Nav1.2a at potentials more positive than +10 mV (e.g., 1.4-fold at +40 mV; Fig. 4B Inset). The equivalent deletion of the CT of Nav1.5 (Nav1.5ΔK1888; Fig. 4D) also had an unchanged activation curve (Fig. 4C), but the steady-state inactivation curve was shifted −9.7 mV relative to Nav1.5. Inactivation of Nav1.5ΔK1888 was also accelerated (1.3-fold at +40 mV; Fig. 4D). Because truncating either Nav1.2a at K1890 or Nav1.5 at the equivalent K1888 enhances inactivation, the C-terminal tail distal to this point normally inhibits inactivation.
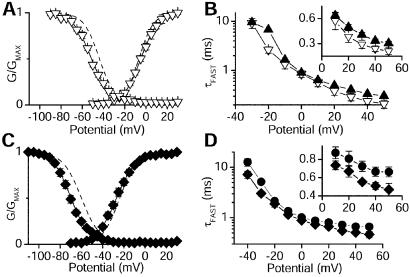
Figure 4.
Properties of the deletion mutants Nav1.2ΔK1890 and Nav1.5ΔK1888. (A) Mean voltage dependence of activation and inactivation for Nav1.2ΔK1890 (▿) and Nav1.2a (dashed lines; from Fig. 3). The mean parameters for Boltzmann fits to individual Nav1.2ΔK1890 experiments were: Va = −4.6 ± 0.2 mV, k = 8.2 ± 0.1 mV (n = 7); Vh = −51.4 ± 0.3 mV, k = 8.6 ± 0.2 mV (n = 6). (B) Semilogarithmic plot of the fast time constants derived from fits of two exponentials to the decay of the current. (Inset) The data for positive potentials on a linear scale. Nav1.2a, ▴; Nav1.2-ΔK1890 (▿). (C) Mean voltage dependence of activation and inactivation for Nav1.5ΔK1888 (⧫) and Nav1.5 (dashed lines; from Fig. 3). Fits to Nav1.5ΔK1888 individual experiments yielded the following mean parameters: Va = −26.9 ± 0.2, k = 9.2 ± 0.2 (n = 9); Vh = −70.2 ± 0.2, k = 8.1 ± 0.2 (n = 8). (D) The fast time constants of two exponential fits to test pulses to the indicated potentials are plotted versus test pulse voltage for Nav1.5 (●) and Nav1.5ΔK1888 (⧫) as in B.
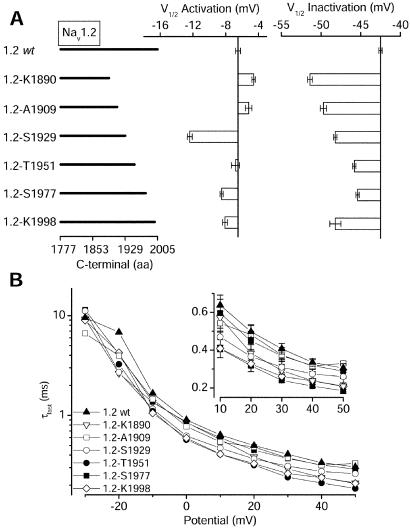
Effects of Progressive Deletions of the Distal CT of Nav1.2a on Inactivation.
To localize the molecular determinants of inactivation in the distal CT, we made a series of channels that were truncated C terminal to K1890 (Fig. 5A). All generated Na+ currents similar in magnitude to Nav1.2a. As expected from the results with the larger truncations, effects on the voltage dependence of activation were small for all constructs except Nav1.2ΔS1929, which produced a substantial negative shift of activation (Fig. 5A Center).
Figure 5.
Voltage-dependent properties of Nav1.2a deletion mutants. (A) Bar graphs showing the mean values of the midpoint of the activation (Center) and inactivation (Right) curves for Nav1.2a (Va = −5.9 ± 0.5 mV, n = 9; Vh = −43.9 ± 1.7 mV, n = 7), Nav1.2 ΔK1890 (Va = −4.6 ± 0.2, n = 7; Vh = −51.4 ± 0.3, n = 6), Nav1.2-ΔA1909 (Va = −5.2 ± 0.4, n = 10; Vh = −49.7 ± 0.4, n = 6), Nav1.2ΔS1929 (Va = −12.4 ± 0.3, n = 6; Vh = −48.2 ± 0.2, n = 6); Nav1.2-ΔT1951 (Va = −6.8 ± 0.5, n = 9; Vh = −48.5 ± 0.2, n = 8); Nav1.2-ΔS1977 (Va = −8.5 ± 0.2, n = 13; Vh = −45.4 ± 0.2, n = 10), and Nav1.2ΔS1998 (Va = −8.1 ± 0.3, n = 9; Vh = −48.2 ± 0.7, n = 7). (B) Semilogarithmic plot of the fast time constants for each mutant as described in Fig. 2. (Inset) The data at positive voltages plotted on a linear scale.
In contrast to activation, each deletion mutation produced a significant negative shift of steady-state inactivation in comparison to wild-type Nav1.2a. Surprisingly, the shifts in inactivation did not decrease monotonically with smaller deletions (Fig. 5A Right). Instead, a negative shift of inactivation was observed when only the last 8 aa were deleted in Nav1.2ΔK1998. This pattern suggests at least two loci in the distal CT that affect inactivation—one between residues 1890 and 1951 and one at the extreme CT between residues 1998 and 2005. Because both activation and inactivation are negatively shifted for Nav1.2Δ1929, the additional amino acid residues having specific effects on inactivation are likely to be located between residues 1890 and 1929. The effects of deletion of the last eight C-terminal residues are apparently increased by additional deletion of residues 1890–1929.
Na+ currents through the mutant channels also decayed more rapidly than wild type at all potentials (Fig. 5B). This is particularly evident at positive potentials, where the effects of activation rate on inactivation are minimal, as illustrated by the time constants plotted on a linear scale in Fig. 5B Inset. The rank orders of the negative shifts in inactivation curves (Fig. 5A) and the speeding of inactivation (Fig. 5B) are approximately correlated, except for Nav1.2Δ1909 and Nav1.2Δ1951. Overall, these results show that the CT of Nav1.2a can selectively inhibit the inactivation, without consistent effects on activation. The effects on inactivation from open states during strong depolarizations and the effects on inactivation from closed states during weaker depolarizations may be affected differently by molecular determinants in the C-terminal tail, as considered in Discussion.
Discussion
The CT Has Minimal Effect On Activation.
Our results show that the overall CT of the Na+ channel has no detectable effect on activation. Neither exchange of the CTs between Nav1.2a and Nav1.5 nor deletion of the distal CT of Nav1.2a had systematic effects on the kinetics or voltage dependence of channel activation. Because activation is initiated by outward movement of the S4 voltage sensors, it is not surprising that the intracellular CT has little effect.
The CT Determines the Difference in Rate of Inactivation Between Brain and Cardiac Na+ Channels.
Substituting the cardiac CT in the brain Na+ channel transfers cardiac inactivation kinetics to the brain channel as observed for chimera Nav1.2/1.5CT. Similarly, substituting the brain Nav1.2a CT in the cardiac Nav1.5 channel transfers brain inactivation kinetics to the cardiac channel, as observed for chimera Nav1.5/1.2CT. Thus, the CT is responsible for the faster inactivation of Nav1.2a brain channels relative to cardiac Na+ channels.
The CT Has an Important Influence on the Voltage Dependence of Inactivation.
The CTs of Nav1.2a and Nav1.5 have a parallel influence on rate of inactivation and on the steady-state inactivation curves. Evidently, inactivation is enhanced in two ways by substitution of the Nav1.2a CT: increased rate of inactivation from the open state and increased steady-state inactivation from the closed states. The CT of the cardiac channel has opposite effects. However, these effects of the CTs magnify the already large difference in steady-state inactivation between brain and cardiac sodium channels rather than exchanging the properties of the two channels. Therefore, the CTs greatly influence steady-state inactivation, but other molecular features of these two channel types are responsible for their different voltage dependences of steady-state inactivation.
Although the CT of Nav1.2a causes stronger inactivation than its cardiac counterpart, the separation between the midpoint voltages of the activation and inactivation is virtually identical, indicating similar efficiency of coupling of activation to inactivation (Fig. 3). This comparison also supports the conclusion that other parts of these two channels determine the strength of coupling of activation to inactivation, thereby conserving the voltage separation between activation and inactivation curves.
The CT Favors Inactivation.
When the distal CT is truncated, inactivation from the open state during strong depolarizations is accelerated, and the steady-state inactivation curve is shifted toward more negative membrane potentials. Thus, the distal CT normally inhibits inactivation. Detailed deletion analysis of the distal C terminal of the brain Nav1.2a channel indicated that this net effect resulted from at least two distinct determinants that inhibited inactivation. Deleting only the final eight amino acid residues in the CT caused a pronounced acceleration of inactivation, which was almost as strong as that caused by truncation of the entire tail. These final eight amino acid residues must normally slow inactivation. Further deletion between residues 1951 and 1891 gave even stronger inactivation, indicating that amino acid residues in this region have an additional inhibitory effect on inactivation. This net inhibitory effect of the distal half of the CT results from the sum of interactions opposing inactivation.
Role of the Membrane Proximal Half of the CT in Inactivation.
The effects of deleting the distal half of the CT of either the brain Nav1.2a or the cardiac Nav1.5 channel are smaller than the effects of exchanging the entire lengths of the tails. Replacing the entire cardiac CT with that of the brain channel caused a 15-mV negative shift in steady-state inactivation. The effects of truncating the distal halves of the C termini of the brain and cardiac channels differed by only 3 mV. Assuming that these effects are additive, −12 mV of the −15-mV shift caused by exchanging CTs is caused by transfer of the membrane-proximal half of the CT. The converse experiment, replacing the CT of the brain channel with that of the cardiac channel, resulted in a +12 mV shift in the steady-state inactivation curve. Of this, 3 mV might be attributed to differential effects of the distal halves of the Nav1.2a and Nav1.5 CTs. The remaining 9 mV must be attributed to differences in the membrane-proximal portion. Thus, 9- to 12-mV of the shifts of steady-state inactivation of the brain versus cardiac Na+ channels can likely be attributed to the membrane proximal halves of the CTs.
For both the brain and cardiac Na+ channels, truncating the CT caused a 1.3- to 1.4-fold increase in the rate of inactivation. In contrast, substituting the Nav1.2a CT in the cardiac channel caused a 1.9-fold increase in the rate of inactivation, while substituting the cardiac CT in the brain channel reduced the rate of inactivation of the brain channel 2-fold. Thus, the membrane-proximal halves of the CTs are likely to have larger effects on both the kinetics and voltage dependence of inactivation.
Molecular Mechanism of Action of the CT in Inactivation.
There are multiple steps in the pathway leading to inactivation of Na+ channels. Channel activation occurs first. It is thought to involve molecular rearrangements in the transmembrane segments of all four homologous domains. These rearrangements link to conformational changes near the extracellular side of homologous domains III and IV (35). Blocking these movements of extracellular loops with toxins (36, 37) or mutations (38) slows channel inactivation. Furthermore, mutations in these extracellular regions appear to prevent coupling of activation to inactivation (38). These movements at the extracellular surface of the Na+ channel are molecularly linked to additional conformational changes on the intracellular face of the channel. The structure responsible for the last step in fast inactivation is the intracellular loop between domain III and IV (LIII-IV) (10, 12, 39), which moves during the inactivation process (13, 40) and blocks conduction through the pore of the channel (41). LIII-IV is thought to act by binding to a receptor region on the intracellular surface of the channel. Candidates for forming the receptor site for LIII-IV include the intracellular loops connecting transmembrane segments S4 and S5 in domains III and IV (42–44). The intracellular end of transmembrane segment IVS6 also appears to play an important role (45).
Although the inactivation gate formed by LIII-IV ultimately is responsible for inactivation, it is unable to transfer the inactivation properties to the cardiac sodium channel (14, 46). Thus, another region of the channel must modulate its movement, resulting in altered inactivation. Our results suggest that the CT performs this function. Single-channel comparison of inactivation in brain and cardiac channels reveals bursts of openings that are prolonged in cardiac channels, indicating slowed entry into the inactivated state (9, 47). Our data show that these effects require the CT of the channel. The CT could modulate inactivation by interacting directly with the structures involved in the inactivation process. For example, the CT could interact with LIII-IV, in this way modulating the kinetics of its closure. Similarly, the CT could interact with the IIIS4–S5 loop, the IVS4–S5 loop, or the intracellular end of the IVS6 segment and slow their formation of an effective inactivation gate receptor. Alternatively, the CT could exert its effects through allosteric coupling, caused by short or long-range interactions. Further analysis of mutations in the CT that alter inactivation may allow distinction among these possible mechanisms.
Acknowledgments
We thank Elizabeth M. Sharp for making the mutant Nav1.5-K1888. This study was supported by Human Frontier Science Program Fellowship LT-613/98 (to M.M.), a Canadian Institutes of Health Research Fellowship (to F.H.Y.), and National Institutes of Health Grant NS34801 (to T.S.).
Abbreviation
- CT
C-terminal domain
Footnotes
Mantegazza, M. A., Yu, F., Catterall, W. A. & Scheuer, T. (2000) Biophys. J. 78, 88A (abstr.).
References
- 1.Hodgkin A L, Huxley A F. J Physiol (London) 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fozzard H A, Hanck D A. Physiol Rev. 1996;76:887–926. doi: 10.1152/physrev.1996.76.3.887. [DOI] [PubMed] [Google Scholar]
- 3.Marban E, Yamagishi T, Tomaselli G. J Physiol (London) 1998;508:647–657. doi: 10.1111/j.1469-7793.1998.647bp.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catterall W A. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 5.Lehmann-Horn F, Jurkat-Rott K. Physiol Rev. 1999;79:1317–1372. doi: 10.1152/physrev.1999.79.4.1317. [DOI] [PubMed] [Google Scholar]
- 6.Goldin A L. Annu Rev Physiol. 2001;63:871–894. doi: 10.1146/annurev.physiol.63.1.871. [DOI] [PubMed] [Google Scholar]
- 7.Noda M, Ikeda T, Suzuki H, Takeshima H, Takahashi T, Kuno M, Numa S. Nature (London) 1986;322:826–828. doi: 10.1038/322826a0. [DOI] [PubMed] [Google Scholar]
- 8.Gordon D, Merrick D, Auld V, Dunn R, Goldin A L, Davidson N, Catterall W A. Proc Natl Acad Sci USA. 1987;84:8682–8686. doi: 10.1073/pnas.84.23.8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirsch G E, Brown A M. J Gen Physiol. 1989;93:85–99. doi: 10.1085/jgp.93.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vassilev P M, Scheuer T, Catterall W A. Science. 1988;241:1658–1661. doi: 10.1126/science.241.4873.1658. [DOI] [PubMed] [Google Scholar]
- 11.Vassilev P, Scheuer T, Catterall W A. Proc Natl Acad Sci USA. 1989;86:8147–8151. doi: 10.1073/pnas.86.20.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stühmer W, Conti F, Suzuki H, Wang X D, Noda M, Yahagi N, Kubo H, Numa S. Nature (London) 1989;339:597–603. doi: 10.1038/339597a0. [DOI] [PubMed] [Google Scholar]
- 13.Kellenberger S, Scheuer T, Catterall W A. J Biol Chem. 1996;271:30971–30979. doi: 10.1074/jbc.271.48.30971. [DOI] [PubMed] [Google Scholar]
- 14.Hartmann H A, Tiedeman A A, Chen S F, Brown A M, Kirsch G E. Circ Res. 1994;75:114–122. doi: 10.1161/01.res.75.1.114. [DOI] [PubMed] [Google Scholar]
- 15.Wehrens X H, Abriel H, Cabo C, Benhorin J, Kass R S. Circulation. 2000;102:584–590. doi: 10.1161/01.cir.102.5.584. [DOI] [PubMed] [Google Scholar]
- 16.Veldkamp M W, Viswanathan P C, Bezzina C, Baartscheer A, Wilde A A, Balser J R. Circ Res. 2000;86:E91–E97. doi: 10.1161/01.res.86.9.e91. [DOI] [PubMed] [Google Scholar]
- 17.Lupoglazoff J M, Cheav T, Baroudi G, Berthet M, Denjoy I, Cauchemez B, Extramiana F, Chahine M, Guicheney P. Circ Res. 2001;89:E16–E21. doi: 10.1161/hh1401.095087. [DOI] [PubMed] [Google Scholar]
- 18.Baroudi G, Chahine M. FEBS Lett. 2000;487:224–228. doi: 10.1016/s0014-5793(00)02360-7. [DOI] [PubMed] [Google Scholar]
- 19.Baroudi G, Carbonneau E, Pouliot V, Chahine M. FEBS Lett. 2000;467:12–16. doi: 10.1016/s0014-5793(00)01099-1. [DOI] [PubMed] [Google Scholar]
- 20.Rivolta I, Abriel H, Tateyama M, Liu H, Memmi M, Vardas P, Napolitano C, Priori S G, Kass R S. J Biol Chem. 2001;276:30623–30630. doi: 10.1074/jbc.M104471200. [DOI] [PubMed] [Google Scholar]
- 21.Qu Y, Rogers J, Tanada T, Scheuer T, Catterall W A. Proc Natl Acad Sci USA. 1995;92:11839–11843. doi: 10.1073/pnas.92.25.11839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linford N J, Cantrell A R, Qu Y, Scheuer T, Catterall W A. Proc Natl Acad Sci USA. 1998;95:13947–13952. doi: 10.1073/pnas.95.23.13947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herlitze S, Garcia D E, Mackie K, Hille B, Scheuer T, Catterall W A. Nature (London) 1996;380:258–262. doi: 10.1038/380258a0. [DOI] [PubMed] [Google Scholar]
- 24.Margolskee R F, McHendry-Rinde B, Horn R. BioTechniques. 1993;15:906–911. [PubMed] [Google Scholar]
- 25.Ma J Y, Catterall W A, Scheuer T. Neuron. 1997;19:443–452. doi: 10.1016/s0896-6273(00)80952-6. [DOI] [PubMed] [Google Scholar]
- 26.Auld V J, Goldin A L, Krafte D S, Marshall J, Dunn J M, Catterall W A, Lester H A, Davidson N, Dunn R J. Neuron. 1988;1:449–461. doi: 10.1016/0896-6273(88)90176-6. [DOI] [PubMed] [Google Scholar]
- 27.Auld V J, Goldin A L, Krafte D S, Catterall W A, Lester H A, Davidson N, Dunn R J. Proc Natl Acad Sci USA. 1990;87:323–327. doi: 10.1073/pnas.87.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogart R B, Cribbs L L, Muglia L K, Kephart D A, Kaiser M W. Proc Natl Acad Sci USA. 1989;86:8170–8174. doi: 10.1073/pnas.86.20.8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White M M, Chen L Q, Kleinfield R, Kallen R G, Barchi R L. Mol Pharmacol. 1991;39:604–608. [PubMed] [Google Scholar]
- 30.Qu Y, Rogers J, Tanada T, Scheuer T, Catterall W A. Proc Natl Acad Sci USA. 1994;91:3289–3293. doi: 10.1073/pnas.91.8.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kellenberger S, West J W, Scheuer T, Catterall W A. J Gen Physiol. 1997;109:589–605. doi: 10.1085/jgp.109.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kellenberger S, West J W, Catterall W A, Scheuer T. J Gen Physiol. 1997;109:607–617. doi: 10.1085/jgp.109.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cummins T R, Zhou J, Sigworth F J, Ukomadu C, Stephan M, Ptacek L J, Agnew W S. Neuron. 1993;10:667–678. doi: 10.1016/0896-6273(93)90168-q. [DOI] [PubMed] [Google Scholar]
- 34.Ukomadu C, Zhou J, Sigworth F J, Agnew W S. Neuron. 1992;8:663–676. doi: 10.1016/0896-6273(92)90088-u. [DOI] [PubMed] [Google Scholar]
- 35.Cha A, Ruben P C, George A L, Jr, Fujimoto E, Bezanilla F. Neuron. 1999;22:73–87. doi: 10.1016/s0896-6273(00)80680-7. [DOI] [PubMed] [Google Scholar]
- 36.Rogers J C, Qu Y, Tanada T N, Scheuer T, Catterall W A. J Biol Chem. 1996;271:15950–15962. doi: 10.1074/jbc.271.27.15950. [DOI] [PubMed] [Google Scholar]
- 37.Benzinger G R, Kyle J W, Blumenthal K M, Hanck D A. J Biol Chem. 1998;273:80–84. doi: 10.1074/jbc.273.1.80. [DOI] [PubMed] [Google Scholar]
- 38.Chahine M, George A L, Jr, Zhou M, Ji S, Sun W, Barchi R L, Horn R. Neuron. 1994;12:281–294. doi: 10.1016/0896-6273(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 39.West J W, Patton D E, Scheuer T, Wang Y, Goldin A L, Catterall W A. Proc Natl Acad Sci USA. 1992;89:10910–10914. doi: 10.1073/pnas.89.22.10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vedantham V, Cannon S C. J Gen Physiol. 1998;111:83–93. doi: 10.1085/jgp.111.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eaholtz G, Scheuer T, Catterall W A. Neuron. 1994;12:1041–1048. doi: 10.1016/0896-6273(94)90312-3. [DOI] [PubMed] [Google Scholar]
- 42.Lerche H, Peter W, Fleischhauer R, PikaHartlaub U, Malina T, Mitrovic N, Lehmann-Horn F. J Physiol (London) 1997;505:345–352. doi: 10.1111/j.1469-7793.1997.345bb.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith M R, Goldin A L. Biophys J. 1997;73:1885–1895. doi: 10.1016/S0006-3495(97)78219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McPhee J C, Ragsdale D S, Scheuer T, Catterall W A. J Biol Chem. 1998;273:1121–1129. doi: 10.1074/jbc.273.2.1121. [DOI] [PubMed] [Google Scholar]
- 45.McPhee J C, Ragsdale D S, Scheuer T, Catterall W A. J Biol Chem. 1995;270:12025–12034. doi: 10.1074/jbc.270.20.12025. [DOI] [PubMed] [Google Scholar]
- 46.Makita N, Bennett P B, Jr, George A L., Jr Circ Res. 1996;78:244–252. doi: 10.1161/01.res.78.2.244. [DOI] [PubMed] [Google Scholar]
- 47.Berman M F, Camardo J S, Robinson R B, Siegelbaum S A. J Physiol (London) 1989;415:503–531. doi: 10.1113/jphysiol.1989.sp017734. [DOI] [PMC free article] [PubMed] [Google Scholar]