Abstract
Deep neural networks are increasingly being used in both supervised learning for classification tasks and unsupervised learning to derive complex patterns from the input data. However, the successful implementation of deep neural networks using neuroimaging datasets requires adequate sample size for training and well-defined signal intensity based structural differentiation. There is a lack of effective automated diagnostic tools for the reliable detection of brain dysmaturation in the neonatal period, related to small sample size and complex undifferentiated brain structures, despite both translational research and clinical importance. Volumetric information alone is insufficient for diagnosis. In this study, we developed a computational framework for the automated classification of brain dysmaturation from neonatal MRI, by combining a specific deep neural network implementation with neonatal structural brain segmentation as a method for both clinical pattern recognition and data-driven inference into the underlying structural morphology. We implemented three-dimensional convolution neural networks (3D-CNNs) to specifically classify dysplastic cerebelli, a subset of surface-based subcortical brain dysmaturation, in term infants born with congenital heart disease. We obtained a 0.985 +/− 0. 0241-classification accuracy of subtle cerebellar dysplasia in CHD using 10-fold cross-validation. Furthermore, the hidden layer activations and class activation maps depicted regional vulnerability of the superior surface of the cerebellum, (composed of mostly the posterior lobe and the midline vermis), in regards to differentiating the dysplastic process from normal tissue. The posterior lobe and the midline vermis provide regional differentiation that is relevant to not only to the clinical diagnosis of cerebellar dysplasia, but also genetic mechanisms and neurodevelopmental outcome correlates. These findings not only contribute to the detection and classification of a subset of neonatal brain dysmaturation, but also provide insight to the pathogenesis of cerebellar dysplasia in CHD. In addition, this is one of the first examples of the application of deep learning to a neuroimaging dataset, in which the hidden layer activation revealed diagnostically and biologically relevant features about the clinical pathogenesis. The code developed for this project is open source, published under the BSD License, and designed to be generalizable to applications both within and beyond neonatal brain imaging.
Keywords: Deep Learning, Congenital Heart Disease, Neonatal Imaging, Structural MR
1. Introduction
Deep neural networks, or deep learning, are a set of machine learning algorithms that use nested layers of linear combinations of the original input allowing for the approximation of highly complex non-linear functions.1 This property can be used in both supervised learning for classification tasks and unsupervised learning to derive complex patterns from the input data.
Deep neural networks have recently gained traction across a variety of domains, but none more than in imaging and computer vision. Naturally, the application of deep neural networks to clinical inference, biomarker discovery, and automated diagnosis in neuroimaging presents innumerable opportunities. Variants of deep learning have already been used extensively in neuroimaging applications, primarily in intensity-based classification tasks. Kleesiek et al. successfully implemented a 3-D Convolutional Neural Network (CNN) that outperforms the state-of-the-art algorithms in skull stripping, generalizing well to multi-modal inputs including contrast-enhanced images.2 Brosch et al. used 3-D CNNs to segment white matter lesions in brain MRI from patients with multiple sclerosis.3 Gupta et al. used sparse auto-encoders and CNNs to detect lesions in structural MRIs using a large public dataset of patients with Alzheimer’s disease (ADNI).4,5 Payan et. al using a similar technique, but with 3-dimensional CNNs, was able to marginally improve the classification accuracy.6 Housseini-Asl et al. implemented 3-D convolutional auto-enconders to achieve excellent classification results using multi-modal imaging in this same dataset.7 Rajpurkar et al.8 achieved comparable results to expert radiologists on diagnostic chest x-ray images, and were able to localize the regions of the image that most contributed to the disease classification. For a comprehensive review of deep learning applications in a variety of multi-modal imaging on datasets including patients with Alzheimer’s disease, schizophrenia, and various mild cognitive impairments, please see the review by Vieira et al.9 Of note, there is little data supporting the use of deep learning techniques in the neonatal neuroimaging domain, mainly due to small testable sample sizes and relatively increased complexity of brain structure.
As deep learning matures in this domain, CNNs have become an increasingly dominant approach to achieving great classification accuracies in the setting of small samples size, given appropriate constraints,10 and relatively increased complexity of brain structures as seen in neonatal neuroimaging.3 CNNs are a specialized variant of neural networks that leverages the inherent structural information encoded in images. Instead of indiscriminately using the entire image as the input to a given neuron, CNNs look for localized patterns across the image, recording the spatial location of an individual feature encoded by each neuron. This has two major advantages over traditional neural networks. First, CNNs decrease computational complexity, resulting in less encoding of overall parameters. Second, CNNs incorporate the spatial location of shared features within the input data. Taken together, these features open the possibility for the application of this technique to more limited, sparse datasets as seen in the setting of neonatal neuroimaging. In addition, 3D-CNNs have characteristic properties that make them exceptionally suitable for the morphological characterization of subtle structural differences, as seen in neonatal neuroimaging, particularly brain dysmaturation.
Neonatal brain dysmaturation refers to aberrant brain development in infants at risk for perinatal brain injury including those born prematurely or with congenital heart disease.11 Neonatal brain dysmaturation can be classified into two broad types based on neuroimaging structural acquisition: reduced volume and/or shape distrubances.12 Currently, most patterns of neonatal brain dysmaturation have been characterized by reduced volume, which can be the result of hypoplasia (lag in development) and/or a destructive mechanism multi-factorial in etiology (i.e. genetic, environmental or acquired injury). In contrast, subtle shape abnormalities can be relatively more difficult to visually assess and quantify, requiring either a trained expert in neonatal neuroimaging and/or sophisticated post-processing methods that are difficult to implement clinically. Volumetric information alone is not sufficient for accurate characterization. We have recently described a subset of term neonates with congenital heart disease that demonstrates brain dysplasia in subcortical structures including the olfactory bulb, cerebellum and hippocampus, as an example of brain dsymaturation related to visual shape disturbances.13 The application of an automated machine learning technique, like CNNs, to detecting these types of neonatal brain dysplasia while also differentiating from volumetric abnormalities, would not only help facilitate basic discovery research related to etiological underpinnings, but also improve clinical detection for individualized patient care.
Here, we pair CNNs with previously developed neonatal structural brain segmentation methods to overcome some of the technical and biological limitations described above. We attempt to overcome limitations related to small sample size by first extracting each brain substructure of interest individually and transforming them into a standard space. This greatly reduces the search space required to learn the subtle abnormalities associated with a given pathology, making it feasible to implement a 3-D CNN as the classification algorithm. Additionally, enforcing spatial localization in the input data by pre-registering the structures into a standard space results in the feature maps retaining their spatial relation to the original structural morphology. The benefit of this approach is two-fold. First, once the model is derived from proper tuning of its parameters based on the training data, its implementation as a classification tool is straightforward, providing a feasible application of automated diagnostic systems in medical imaging. Second, by registering the input substructures into a standard space, the algorithm generates human-interpretable class activation maps of the hidden layers learned by the network, thereby retaining the original 3-D structural relationships. This refined algorithm gives us a data-driven model of the features within the dataset that contribute to the final classification, providing further insight into the structural morphology associated with the classification criteria and underlying pathology. We chose the cerebellum as the test substructure to test our algorithm because: (1) its relatively simple structure to segment compared to other regions of the brain, including the cerebral cortex and other subcortical structures including the hippocampus; (2) there are known neuroradiological criteria for cerebellar dysplasia in the developing human infant;14–16 and (3) anatomic related-sub regions of the developing cerebellum are important mediators of both genetics factors,17,18 and downstream pathways associated with neurodevelopmental impairment.19,20 We tested our algorithm on a dataset of term neonates born with CHD at high risk for brain dysmaturation, including recently described cerebellar abnormalities, including both hypoplasia and dysplasia.13
Materials and Methods
Subjects
We prospectively recruited 90 term-born neonates with congenital heart disease and 40 term-born healthy controls at Children’s Hospital of Pittsburgh of UPMC with parent consent, as part of an ongoing Institutional Review Board approved study. Infants were scanned at close to term equivalent age, or when deemed clinically stable. 54 infants in the CHD cohort were scanned prior to their first surgical intervention, with the remaining 36 scanned post-surgical intervention. Infants were scanned on a 3T Siemens Skyra MRI (Siemens, Erlangen, Germany) without sedation using a 32-channel head coil. 18 infants in the control group were scanned on a 3T GE HDXT. Only infants who completed the volumetric imaging portion of the protocol were included in this study, and a cut-off of 52 weeks post-menstrual age was used to control for age-related morphological changes. Imaging parameters were as follows: (1) volumetric T1 Magnetization-Prepared Rapid Gradient-Echo at echo time (TE)/repetition time (TR): 418/3100 ms, 1.0 × 1.0 × 1.0 mm3, and matrix size 320 × 320; (2) volumetric T2 Sampling Perfection with Application optimized Contrasts using different flip angle Evolution sequence at TE/TR: 2.56/2400 ms, 1.0 × 1.0 × 1.0 mm3, and matrix size 256 × 196; (3) axial T2 Weighted Fast Spin Echo (FSE) at TE/TR: 3/2400 ms, slice thickness 2.0 mm with 0 skip, and in-plane matrix resolution 200 × 256.
Dysplasia Classification
We have previously described a pattern of dysmaturation in a subset of this population13. Each neonate’s MRI was reviewed by an expert neuroradiologist blinded to their clinical history and classified as “normal” or “dysplastic” following existing qualitative imaging criteria developed by Barkovich, et. al and others.14–16,21 The cerebellar dysplasia abnormalities are characterized by diffuse shape disturbance relative to brainstem/supratentorial structures and by abnormal orientation of the cerebellar fissures. Although the cerebellar vermis and cerebellar lobes were scored independently, we binarized the presence of dysplasia if it involved either cerebellar hemisphere or vermis. Infants identified to have hypoplastic substructures, but no evidence of structural dysplasia were not classified as abnormal in this study. For a subset of patients, we had two senior neuroradiologist score the brain dysplasia as previously described with a kappa score ranging between .86–.91 for subcortical structures.13
Experimental Design
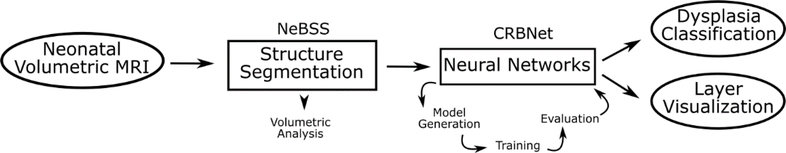
The full framework is summarized in Figure 1. The pipeline takes as input the neonatal volumetric MRI, and segments 50 individual brain substructures. These substructures can be independently used for volumetric analysis. The substructure chosen for the classification task is then linearly registered onto a standard space to remove any size confounder, while retaining the shape information. This is the input to the neural network (Figure 2). The initial architecture of the model to be evaluated is heuristically determined, followed by training and measurement of its performance on a small, randomly selected subset (30%) of the full dataset to prevent overfitting. This is iteratively performed, with incremental changes to the hyperparameters of the model, until satisfactory results are achieved for training on the full dataset with a fixed set of hyperparameters. This step is necessarily separate from validation of the architecture’s final performance to prevent overfitting.22 The final performance evaluation of the architecture is measured by cross-validation. While we did not perform nested cross-validation due to the limited data size and disproportionate amount of normal and dysplastic subjects, we minimized hyper-parameter tuning by starting with a heuristically determined architecture and restricting the epoch size of each parameter testing iteration. No hyperparameters were modified during cross-validation, and the accuracies reported are using the previously fixed architecture, measured on newly sampled partitions of the dataset. The output of the framework is a classifier able to detect structural dysplasia, and the hidden layers of the chosen network can be used to infer the morphological properties that contribute to the final classifier.
Figure 1.
Experimental Design and overview of the framework. The pipeline takes as input the neonatal volumetric MRI, and segments 50 individual brain substructures. These substructures can be independently used for volumetric analysis. These structures are then registered onto a standard space to remove any size confounder but retain the shape information. This is the input to the neural network. The architecture of a given network is heuristically determined, followed but training and evaluation of its performance. This is iteratively performed until satisfactory results are achieved. The output of the framework is a classifier able to detect structural dysplasia, and the hidden layers of the chosen network can be used to infer the morphological properties that contribute to the final classifier.
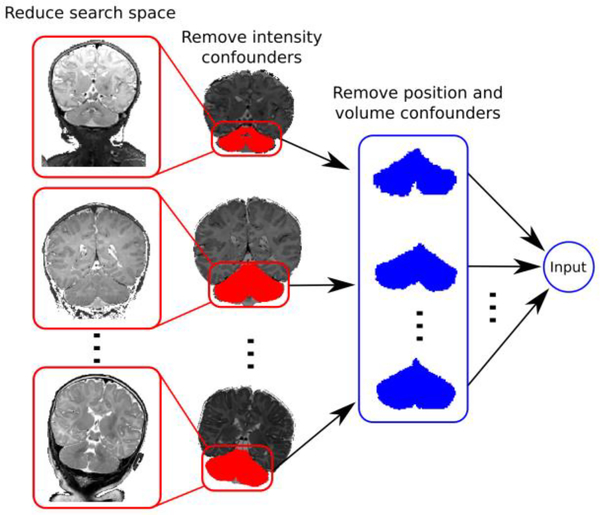
Figure 2.
Dimensionality reduction of neonatal structural MRI decreases the search space by removing intensity, positional, and size confounders.
Substructure Segmentation
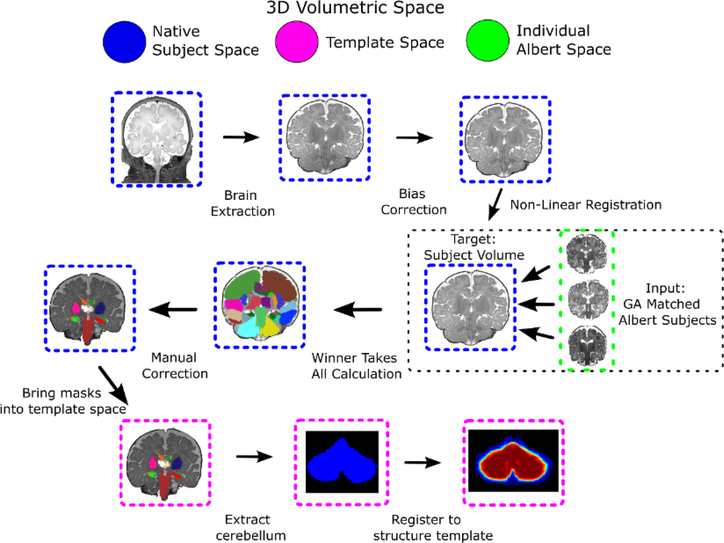
Each infant’s cerebellum is extracted using a semi-automated processing pipeline developed in-house. This pipeline is written in Python using the Nipype framework,23 and the code is freely available at www.github.com/PIRCImagingTools/NeBSS. Figure 3 shows an overview of the pipeline. The preferred input is the neonate’s volumetric T2 image. However, this acquisition tends to be susceptible to motion artifacts, rendering much of the data inadequate for segmentation. As an alternative, we can register the T2 FSE images to the volumetric T1 3D images, utilizing the better T2 tissue contrast while still retaining accurate volumetric information. We first run a brain extraction with FSL’s Brain Extraction Tool (BET),24 followed by FSL’s bias correction algorithm25,26 to remove any intensity gradient artifact due to field inhomogeneity. We then use the ALBERT neonatal parcellation dataset27,28 as the source template to propagate onto the subject space 50 discrete brain substructures modeled by the atlas using the Advanced Normalization Tools (ANTS) algorithm.29,30 ANTS calculates a symmetric, non-linear transformation between the target image and the source image. We choose as source four ALBERT subjects closest in gestation age to our subject to increase our registration accuracy and yield segmentations that more closely match the developmental stage of the subject. We then perform a voxel-wise winner-takes-all calculation across the four transformed ALBERT subject labels to determine the structure classification at each voxel of our subject. If there is a tie, the classification is randomly selected. We are left with 50 non-overlapping discrete regions mapped onto our subject space. This is followed by manual correction step to ensure anatomical accuracy of our label propagation. Finally, the cerebellum is linearly registered into a common template space, effectively removing any confounding volumetric information, but retaining the shape features of the structure. This serves the purpose of narrowing the search space and better enhancing the subtle morphological variations across the subjects.
Figure 3.
Neonatal cerebellum structure extraction pipeline. We use an atlas based image registration pipeline to delineate the desired brain structures, which become the input to the classification task.
As the output of our pipeline is highly dependent on the accuracy of the substructure extraction, we tested our reproducibility using the Dice Similarity Coefficient (DSC)31 between two images:
We assessed the inter-rater reliability by having two independent users perform manual correction on the same set of six subjects (3 Controls and 3 CHD). For intra-rater reliability, one user performed a repeated round of corrections on the same set of subjects.
Volumetric Analysis
To first investigate whether the volumetric information extracted from our pipeline is sufficient for accurate prediction of structural dysplasia, we tested three logistic regression models, and evaluated their accuracy using Leave-One-Out cross-validation (LOOCV). LOOCV iterates through the dataset, removing one sample from the set, and using the remaining samples to train a classifier. The classifier is then tested on the left out single sample. The averaged classification accuracy for each model is then calculated. This method of validation is designed to reduce overfitting of a classifier primarily by reducing its dependence on outliers, while still retaining a large training set for learning. The first model we tested was as follows:
Where PMA is the post-menstrual age at time of scan, cerebellar volume is the manually corrected volume extracted from the pipeline, and CHD is the subject’s CHD/control classification. This model included the entire dataset. Additionally, we tested this model without the CHD classification term to better represent a naïve classifier agnostic to the patient’s clinical diagnosis, as well as training only on the subjects with CHD to minimize noise from healthy controls, as only patients in the CHD cohort were identified to have cerebellar dysplasia.
Neural Networks
A neural network is comprised of units (neurons) that apply an arbitrary, non-linear activation function σ to a linear combination of the inputs and a set of learned weights and bias:
where w is a vector of weights, x is the input and b is the bias term. In a deep neural network, the output of each layer is fed into the next layer. Any given layer in a deep neural network can have an arbitrary number of neurons, each learning their own weights and bias, allowing for a high-dimensional approximation of any non-linear function, given enough neurons and layers. Learning the weights and biases of a neural network is done by an optimization algorithm known as Stochastic Gradient Descent (SGD). The first requirement in gradient descent is the selection of a cost function. This cost function is simply a measure of the accuracy of our classification task. Common cost functions used in neural networks include mean squared error (MSE), negative log-likelihood, and cross-entropy. The negative log-likelihood cost function is particularly powerful in conjunction with a final softmax activation layer:
where z is the weighted sum of the outputs of the previous layer, and K is the total number of neurons in the final layer (the classifiers). Softmax normalizes the output of each neuron of the final layer in the network over all possible output neurons, which are the desired classifiers. The output of the softmax for each neuron (j) in the final layer is then the likelihood of the given input being classified as that particular label. Therefore, minimizing negative log-likelihood function is equivalent to increasing the prediction accuracy of our network. Gradient descent works by calculating the gradient of the cost function given the current parameters of the network, and then changing these parameters in the opposite direction of the gradient, scaled by a small factor - called the learning rate.
SGD uses a simple update rule to calculate the new parameters V’:
where V are the parameters of the network, η is the learning rate, and ∇C is the gradient of the function given the current parameters. The gradient of the cost function is analogous to its slope, generalized to more than two dimensions, and indicates the direction in which the function is increasing. Thus, by making small incremental changes in the weights and bias opposite of their gradient, we iteratively minimize the cost function. Given a small enough learning rate and enough iteration, SGD is guaranteed to converge on a (local) minimum.
Convolutional Neural Network
Traditional neural networks consist of fully connected layers, taking as input a flattened vector of the data. While this architecture has proven powerful across numerous domains, from genomics to text mining, they do not take advantage of the intrinsic spatial information contained in images. Such a network only learns patterns of activation as they appear in a fixed order of the training data. A proposed solution to this limitation is the use of Convolutional Neural Networks (CNNs). Instead of feeding the entire image into each neuron, a CNN convolves a filter of reduced size, the Local Receptive Field (LRF), with the input image. The result of this convolution is a set of neurons, which take as input only the regions of the input image within their respective LRF, but encode the spatial location of the feature in the original image space. This is able to identify position invariant, reusable features by having one full set of neurons in a hidden layer (called a feature map), share the same parameters. This has two advantages: (1) we greatly reduce the number of parameters needed to compute at each layer, and (2) the feature map is now a spatial representation of the features present in our data. Convolution layers are often followed by max-pooling layers, which take the maximum value of each parameter within the selected pooling filter. This effectively de-noises the data while also reducing the dimensionality of the subsequent layer.
There is no closed form method of designing an optimal neural network. Collectively, the parameters that make up the architecture: learning rate, number of layers, feature maps per layer, regularization coefficient, LRF, and max-pooling size, are known as the hyper parameters of the model. The nature of deep neural networks makes it so that any given network is never optimal, as there exist an infinitely large number of network architectures that may perform equally or better. Instead, we strive to design an effective network, and iteratively try to improve on it. We can use heuristic approaches to expedite the search for ideal parameters and establish a reasonable starting point before committing to fully training a specific network architecture, which can take weeks or months depending on the size and complexity of the dataset. We can instead perform quicker cycles of learning on a subset of the data without reaching convergence or saturation. Here, we applied a combination of random and grid search32 to cycle over viable hyper parameters, giving us an estimation of what hyper parameters are likely to work.
Overtraining is always a concern with highly complex machine learning algorithms. Overtraining occurs when the model implicitly learns features specific to the training data, effectively “memorizing it”, but does not generalize to external datasets. To reduce these effects, we can impose restrictions on our cost function and hidden layers. We used two complementary approaches to overtraining prevention: L2 regularization and layer dropout. L2 regularization adds an additional term to the cost function:
where λ is an additional hyperparameter, n is the number of subjects in the training batch and w are the weights. This method adds a penalty for higher weight values, attenuating runaway effects that can lead to over fitting the data. Similarly, layer dropout is a method of ensuring more generalizability in the classification test. Layer dropout randomly removes a pre-determined number of neurons from the final fully connected layers at each training iteration. This prevents the learning algorithm from relying too heavily on any one neuron, enforcing more generalization distributed across the entire network space instead of localized neurons.
Validation
To evaluate our model, we performed a 10-fold cross validation. This is done by partitioning the dataset into 10 independent sets, and at each iteration using 9 sets as the training set and the remaining data for validation of the classification accuracy. We trained each validation run for a total of 100 epochs. The best performing parameters from the validation runs were then used in a final learning run of 700 epochs to generate the activation maps described in the next section.
Hidden Layer Visualization
Interpreting the features learned by a CNN could potentially provide insight into the biological and structural features that contribute to a given substructure’s malformation. However, the parameters learned in the hidden layers of a neural network are traditionally treated as a black box. We have no direct control on the features learned at each layer, and it has been shown that each individual unit does not often hold any meaningful semantic information, but rather the combination of features within the entire space hold this higher level of abstraction.33 Several methods have been proposed in attempts to visualize the information contained in the hidden layers. Most notably, Zeiler and Fergus used a Deconvolutional Network (deconvnet)34,35 to project the hidden layer activations back into pixel space as a form of pre-training and quality control. This method generates a projection in pixel space that reflects the features at each layer that mostly contribute to the final classifier, independent of spatial location. This approach has shown to be beneficial, particularly when analyzing images with positionally varied features across samples.
In our work, we approximate this method by directly visualizing the mean activation maps at each hidden layer for each cohort. By implementing a 3-D CNN, we retain the volumetric structure of the input data, as each hidden layer’s feature map will encode the local features within the input that contribute to the final classification. As each subject’s cerebellum is linearly registered into a common space, we directly impose structural information into the input data. This allows us to project the mean layer activations for each group into a 3-D space and view them as a proxy for the anatomical features that lead to the final diagnosis. One drawback of this approach, however, is that this loses spatial information at each max pooling step, which can be particularly undesirable when using binarized, sparse data as input.
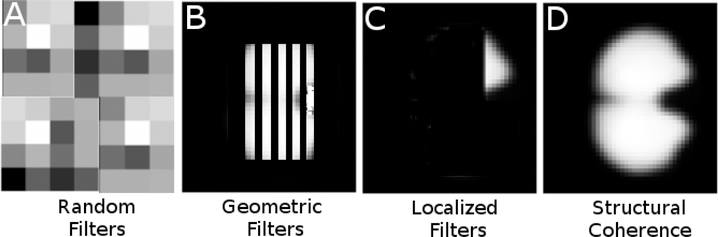
To further illustrate the benefits of spatially constrained filters, Figure 4 shows synthetic examples of possible filters that can be learned by a CNN. Randomly distributed filters (Figure 4–A) may still result in good classification accuracy, but provide no anatomically interpretable benefits. Random filters may also be symptomatic of an overfitting algorithm. Geometric filters (Figure 4–B) are often seen in image classification algorithms with no structural coherence of the input data. These filters are typically found in earlier layers of deep networks and are thought to be used in further layer abstractions to combine to form more complex features. While mechanistically interpretable, they also would not provide useful anatomical insight in this use case. Localized filters (Figure 4–C), where only a subsection of the input image activates the filter, can be useful for identifying specific regions of the input image that highly contribute to the final classification. While potentially desirable due to their specificity, localized filters are not likely to be observed when classifying complex structural morphology but would theoretically be useful in lesion classification or other general discrete feature detection. Structurally coherent activation maps, as imposed as a constraint in this work, (Figure 4–D) retain the structure of the input data, and selectively activate regions of the input dataset that contribute to the final classifier. In this work, we attempt to leverage this property to identify complex patterns within the input structure that ultimately differentiate normal and dysplastic subtypes.
Figure 4.
Non-exhaustive, synthetic examples of possible learned filter outcomes when training CNNs. Randomly distributed filters (A) may still result in good classification accuracy, but provide no anatomically interpretable benefits. Geometric filters (B) are often seen in image classification algorithms with no structural coherence of the input data. These filters are typically found in earlier layers of deep networks, and are thought to be used in further layer abstractions to combine to form more complex features. Localized filters (C) can be useful for identifying specific regions of the input image that highly contribute to the final classification. Structurally coherent activation maps, as are generated by our methods due to structural constraints, (D) retain the structure of the input data, and selectively activate regions of the input dataset that contribute to the final classifier.
Building upon this concept, our final visualization approach aims to implement a similar method proposed by Zhou et al.36 by generating Class Activation Maps (CAMs) to spatially locate the regions of the input image that most contribute to the final classifier at the each convolutional layer. Zhou et al. used a global average pooling (GAP) step prior to the final classifier. This has the advantage of assigning one weight to a spatial map prior to the classifier, providing a measure of importance of the specific feature encoded by each feature map, at the cost of losing spatial coherence. However, by propagating this weight back onto the final 3D convolutional layer, they were able to create a weighted heat map of the spatial location of the most important features to the classifier for each training sample. For each training sample, the CAM is then defined as:
Where (x,y) are the downsampled pixel coordinates at the last convolutional layer of samples of class c, and wkc is the final weight given to the kth feature map fk after the GAP step. This approach has proven powerful for spatially variant, multi-labeled classification problems. Here, since we have fully connected layers prior to the final classifier rather than a GAP layer directly connected to a classifier, we lose the direct connection of an individual feature map’s contribution to the final classifier. However, we can still extract salient anatomical information by generating layer specific CAMs at each convolutional layer, for both normal and dysplastic structural variants, here denoted as within-class average activation maps (wCAMs).
Implementation
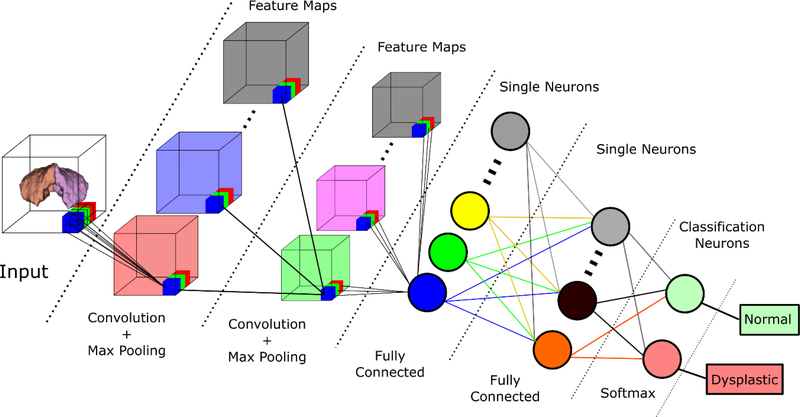
Figure 5 shows a simplified overview of the general architecture used as the basis for the 3D CNN. The computational framework was developed using Python and the Theano37,38 library, with customized routines for the 3D convolution and neuroimaging processing. The pre-processing of the input to the network uses the Neuroimaging in Python (Nipy) library.23 The full code can be viewed at: www.github.com/PIRCImagingTools/CRBNet.
Figure 5.
3-D Convolutional Neural Network Overview. Simplified architecture of the generated CNN. The algorithm takes as input the extracted, binarized cerebellum and outputs a classification of dysplastic or normal structure.
Results
Subjects
Mean gestational age was 38.0 weeks (+/−2.9) in the group of infants with CHD, and 41.2 weeks (+/− 3.8) in the healthy control group. Mean post-menstrual age (PMA) at time of scan was 42.4 (+/−6.9) and 43.5 (+/− 5.5) weeks for infants with CHD and healthy controls, respectively. Infants within the control group showed higher PMA-adjusted cerebellar volumes when compared to neonates with CHD, with a mean cerebellar volume 5,201.5 (+/− 7,930.6) mm3 larger than the dataset mean volume, with neonates with CHD showing on average 1,035.4 (+/− 5,591.7) mm3 smaller than the dataset mean volume (p < 0.000). We saw no statistically significant difference in PMA-adjusted cerebellar volume when comparing infants who had pre-op imaging and infants with post-op imaging, with pre-op MRI’s having on average 941.51 (+/− 3,214.5) mm3 larger cerebelli than the entire CHD cohort mean, and post-op MRI’s, showing 1,439.7 (+/− 7,720.9) mm3 smaller cerebelli than the CHD cohort mean (p < 0.053). The cerebellar data included in this analysis showed no evidence of perinatal injury as we excluded cases with cerebellar infarcts and cerebellar hemorrhage.
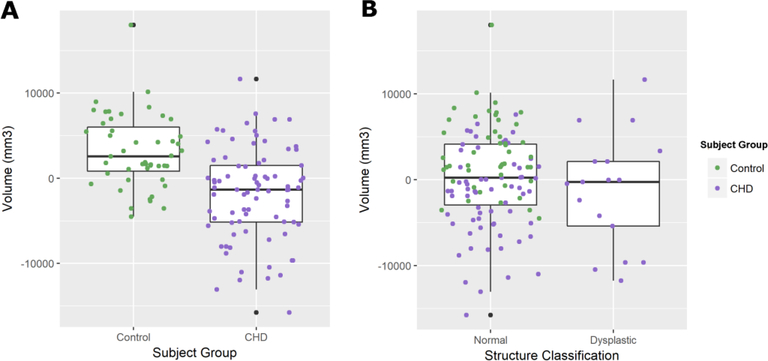
The logistic regression approach was not able to discriminate structural dysplasia using age and volumetric information. While the patient’s CHD classification yielded a very high odds ratio of 1.0×108 (as only patients with CHD had dysplastic cerebelli), it was not statistically significant (p < 0.991) and did not yield any discriminatory power. Cerebellar volume yielded odds ratios near 1.00 for all three models tested and was not statistically significant. Similarly, we saw no statistically significant difference in cerebellar volumes between subjects classified as structurally normal and subjects with dysplastic cerebelli (p < 0.321), shown in figure 6.
Figure 6.
Boxplots showing post-menstrual age corrected cerebellar for between A) control subjects and patients with CHD and B) Subjects classified as having structurally normal cerebelli and subjects diagnosed with dysplastic cerebelli. Only subjects with CHD were diagnosed with dysplastic cerebelli. There was a statistically significant difference in volumes between neonates with CHD and controls (p < 0.000) but no difference between dysplastic structures and normal structures (p < 0.321), suggesting that hypoplasia is independent from structural dysplasia.
Among the infants with CHD, 17 (18.9%) were diagnosed with a dysplastic cerebellum. The total incidence rate in the entire dataset was 13.1%. To attenuate the effects of the low incidence of abnormal cases in the training set, we bootstrapped the data by randomly sampling from the set of abnormal cases and artificially inflating the dataset to contain a more proportional ratio of controls to abnormal cases, with a final training dataset containing 47% dysplastic structures. To prevent overtraining of the algorithm by oversampling the same small subset of cases, we introduce a small amount of random translation (3–5 voxels) in 3 directions to each case in the dataset. This helps prevent the introduction of fixed-position based artifacts and aids in generalization of the parameters, but still retains the spatial integrity of the input data.
Substructure Segmentation
Table 1 shows the Dice coefficients for inter- and intra-rater reliability of the manual corrections performed on a subset of infants. Users were blinded to the infant’s cohort and dysplasia classification prior to correction. Measures were highly consistent across both inter- and intra-raters, with the lowest mean Dice coefficient of 0.941 (+/− 0.03) between one rater measuring the right cerebellum and highest mean Dice coefficient of 0.951 (+/− 0.02) between multiple raters measuring the same substructure.
Table 1.
Intra- and Inter-rater reliability of manual corrections in structure extraction.
| Structure |
||
|---|---|---|
| L Cerebellum | R Cerebellum | |
| Measure | mean (sd) | mean (sd) |
| Inter-Rater | 0.943 (0.01) | 0.951 (0.02) |
| Intra-Rater | 0.952 (0.03) | 0.941 (0.03) |
CNN parameters
Table 2 shows the final parameters for our chosen architecture. It consists of a total of 7 hidden layers, with 4 initial convolutional layers, followed by 2 fully connected layers and a final softmax classification layer. Each convolutional layer is followed by a max-pooling procedure. The initial learning rate (η) was set to 0.005 with a scheduled rate decay of 0.5* η every 40 epochs. We used the negative log-likelihood cost function, with an added L2 regularization hyperparameter (λ) set to 0.01. We used a layer dropout parameter of 0.3. Finally, we implemented a momentum update method with an initial μ value of 0.5, increased to 0.9 after a stabilization period of 15 epochs.
Table 2.
3-D CNN Architecture.
| Layer | Layer Type | Input Dimension | # Neurons/Feature Maps | LRF | Pooling |
|---|---|---|---|---|---|
| 1 | Convolutional/Max Pool | 100 × 90 × 70 | 10 | 4 × 4 × 4 | 2 × 2 ×2 |
| 2 | Convolutional/Max Pool | 48 × 43 × 33 × 10 | 15 | 2 × 2 × 2 | 2 × 2 ×2 |
| 3 | Convolutional/Max Pool | 23 × 21 × 16 × 15 | 25 | 2 × 2 × 2 | 2 × 2 ×2 |
| 4 | Convolutional/Max Pool | 11 × 10 × 7 × 25 | 50 | 2 × 2 × 2 | 2 × 2 ×2 |
| 5 | Fully Connected | 5 × 4 × 3 × 50 = 3,000 | 300 | - | - |
| 6 | Fully Connected | 300 | 100 | - | - |
| 7 | Softmax | 100 | 2 | - | - |
Modern computer vision CNNs have achieved excellent results and improved learning speed using the Rectified Linear Units function (ReLU), and more recently Exponential Linear Units (ELU) as the activation function.39 However, we achieved rather mediocre results with ReLU in our application, with runs never achieving convergence. This is likely a result of the sparse, binarized nature of the inputs, which can lead to exploding and/or vanishing gradients during training. Instead, our final architecture uses the tanh function:
which has traditionally performed well in many classification tasks, at the cost of slower learning at the saturation extreme compared to newer activation functions such as ReLU and ELU.
Cross-Validation
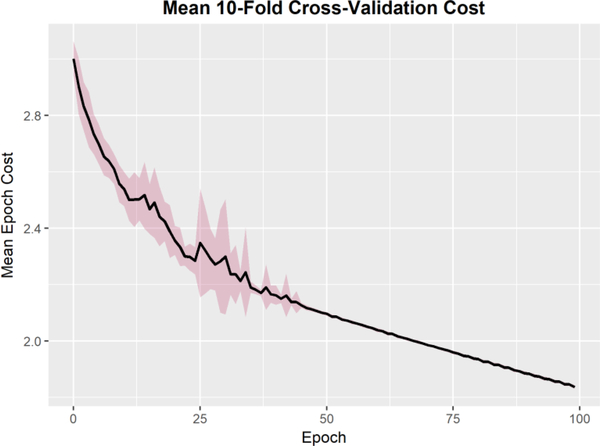
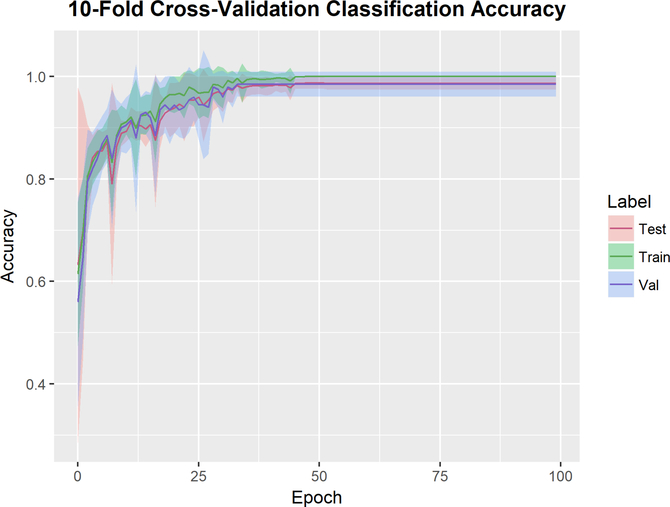
Figure 7 shows the mean cost at each epoch the 10 cross-validation runs. To ensure that the training and validation sets remain independent, the resampled subjects from the bootstrapping pre-processing remained within their own validation sets without crossover into the remaining dataset. While some variance is expected due to the random initialization of weights, all runs converge within 50 epochs. Figure 8 shows the classification accuracy for each run. The training set is a bootstrapped dataset with an inflated incidence of abnormal cases. The data is partitioned into 10 independent sets, where at each run 9 sets are used to train the network and the final set is used as the validation set. The test set is the original dataset (without the bootstrapped cases added in). All runs achieved 100% classification accuracy in the training set within 50 epochs. The average classification accuracy on the validation set was 0.985 +/− 0.0241, with several folds reaching 100% classification accuracy. To inspect the cross-validation results for any subject specific artifacts that may erroneously contributed to the interpretability of the hidden layers, we generated class activation maps for each convolutional layer of each cross-validation run. The wCAMs for layers 3 and 4 are shown in supplemental figures 1 and 2. While some variance is observed in activation values outside of the cerebellar structure, particularly in layer 3, the contribution of the anterior and inferior regions of the cerebellum, as well as the vermis, are consistently present. In layer 4, some variance is observed in features constructed in the center of the structure across folds, but the features differentiating dysplastic and normal structures in the region of the vermis and cerebellar hemispheres is consistent across folds.
Figure 7.
10-Fold Cross validation mean and standard deviation cost across all runs. Cost function was the negative log-likelihood function with an L2 regularization parameter of 0.01. While some variance is expected due to the random initialization of weights, all runs converge within 50 epochs.
Figure 8.
10-Fold Cross validation mean and standard deviation classification error for each dataset. The training set is a bootstrapped dataset with an inflated incidence of abnormal cases. The data is partitioned into 10 independent sets, where at each run 9 are used to train the network and the final set is used as the validation set. All runs achieved 100% classification accuracy in the test set within 50 epochs.
Visualization results
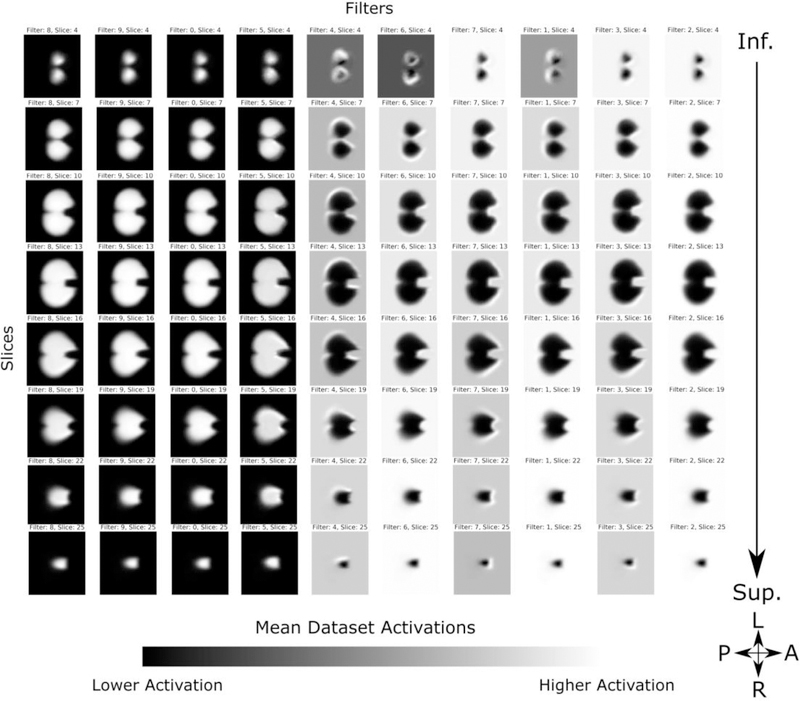
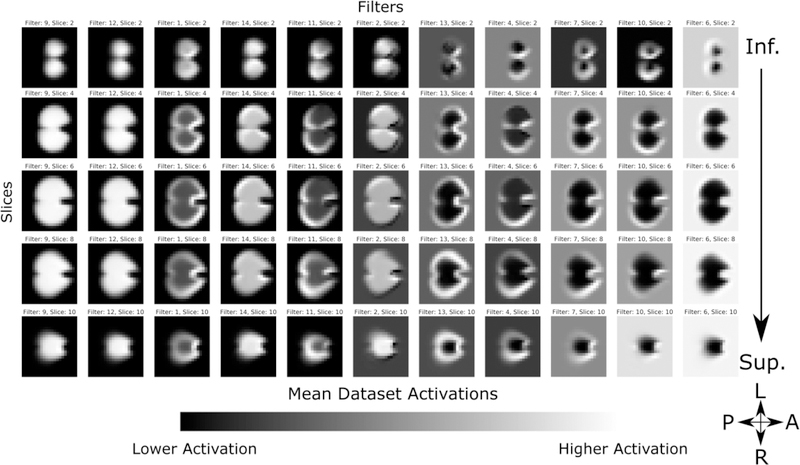
The first layer’s mean activations for the entire dataset is shown in Figure 9. As the algorithm parameters are randomly initiated, there is no intrinsic information to the order of the learned filters; therefore the filters are selectively sorted for visual clarity. Intensities are scaled to show the contrast in activation range in each filter. We see that, as expected due to the structural coherence imposed by linear registration and binarized input, each filter distinctly delineates the cerebellum, with some filters showing higher activations limited to the perimeters of the structure. This corresponds to the cerebellar cortex of the bilateral cerebellar hemispheres, and serves as a potential edge detectors. Comparatively, the activations in layer 2 (Figure 10) show more dramatic delineations of the superior surface of the cerebellum, co-localizing to the posterior subdivision of the bilateral cerebellar cortical hemispheres and also to the midline vermis. Note that filters with visually similar activations are not shown for clarity. Subsequent layers show a similar increase in the range of activations within each filter, however, as they are down-sampled at each hidden layer, the features become less interpretable in an anatomical context.
Figure 9.
First convolutional layer mean activations. Filters are selectively sorted for visual clarity and intensities are scaled to show the contrast in activation range in each filter. Each filter distinctly delineates the cerebellum, with some filters showing higher activations limited to the perimeters of the structure, co-localizing to the cerebellar cortex of the bilateral cerebellar hemispheres, serving as potential edge detectors.
Figure 10.
Second convolutional layer mean activation. Filters are selectively sorted for visual clarity and intensities are scaled to show the contrast in activation range in each filter. Compared to the first layer, activations show more dramatic delineations of the superior surface of the cerebellum, co-localizing to the posterior subdivision of the bilateral cerebellar hemispheres and midline vermis.
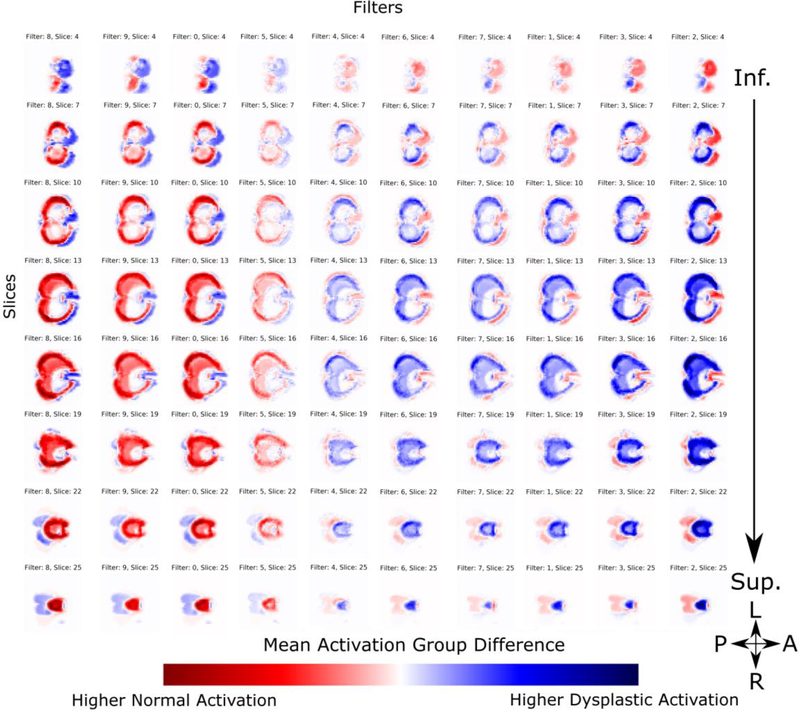
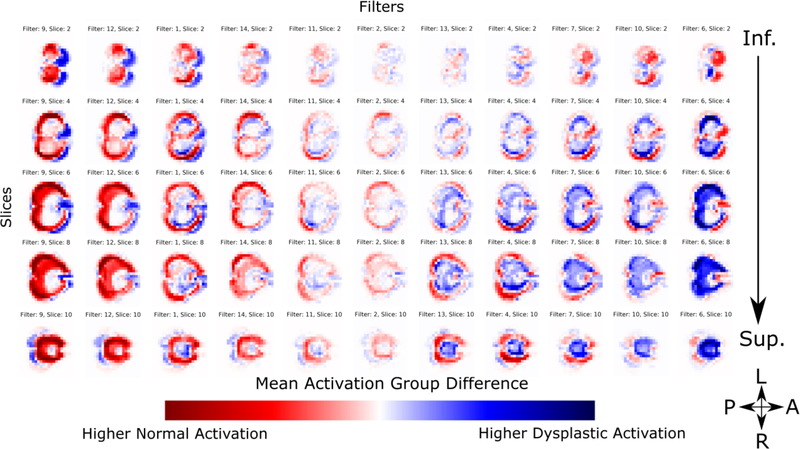
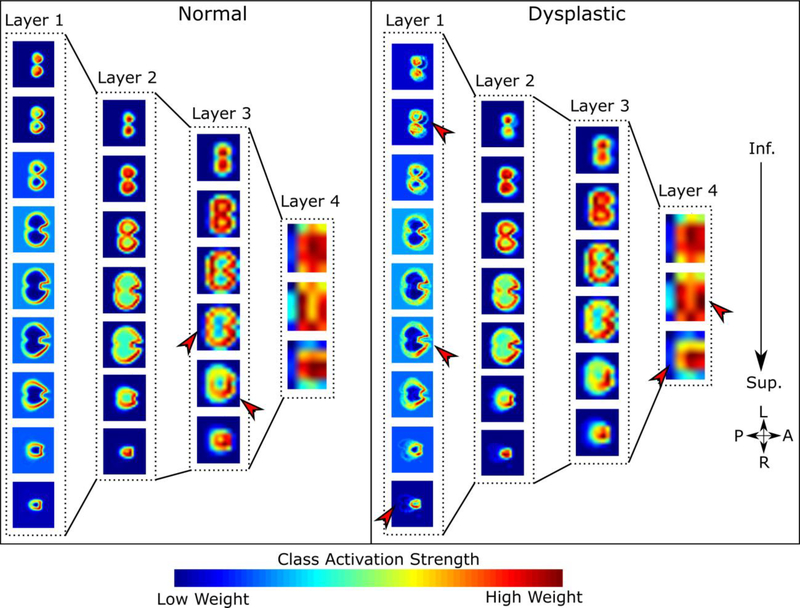
We can then look at the difference in activations between the dysplastic and normal cohorts. The difference map of the first layer is shown in Figure 11. Blue regions are areas in which we see higher activation in the dysplastic cerebelli, and red is increased in the normal substructures. White regions showed low activations equally in both cohorts, and are thus uninformative to the classification task. As with the previous figures, filters are sorted for visual clarity to separate the filters that show higher activations in normal cerebelli contrasted with dysplastic inputs. Each filter shows a predilection for primarily classifying either normal or dysplastic substructures, however, some overlap is observed. We see a clear pattern where the normal substructures show more defined delineation of the inferior and lateral surface of the cerebellar lobes while the dysplastic substructures show more defined superior surface of the cerebellum that anatomically correspond to the posterior lobe of the cerebellar hemispheres and the midline vermis. Similar to the mean activation maps, the first layer primarily serves the purpose of global delineation of the cerebellum. In the second layer’s difference maps (Figure 12), we see more regional delineation of key cerebellar regions that discriminate between dysplastic and normal cerebellar tissue. Specifically, the superior surface of the cerebellum (corresponding anatomically to the posterior lobe subdivision and the midline cerebellar vermis) shows significant discriminatory power, seen as strongly differentiated regions of the activation maps, between dysplastic and normal tissue structure. No lateralization difference (between right and left) was appreciated in the cerebellar hemispheres or midline vermis. In contrast, deep cerebellar regions (including the flocculonodular lobe subdivision) were not informative to the final classifier as shown as white regions in the activation group differences. As with the mean activation maps, subsequent layers lose anatomic interpretability due to down sampling. This is further supported by the Class Activation Maps shown in Figure 13. Notable regional differences between normal and dysplastic variants are shown with red arrows. The normal structural cohort shows a predilection for more defined posterior cerebellar hemispheres, while stronger discriminator power in the dysplastic cohort comes from less developed anterior cerebellar structures nearing the brainstem.
Figure 11.
First convolutional layer activation difference between control and dysplastic cerebelli. Blue regions are areas in which we see higher activation in the dysplastic cerebelli, and red is increased in the normal substructures. White regions showed low activations equally in both cohorts, and are thus uninformative to the classification task. Filters are selectively sorted for visual clarity to highlight the differences in filters that show higher activations in normal cerebelli contrasted with dysplastic inputs. Each filter shows a predilection for primarily classifying either normal or dysplastic cerebelli, however, some overlap is observed.
Figure 12.
Second convolutional layer activation difference between control and dysplastic cerebelli. Blue regions are areas in which we see higher activation in the dysplastic cerebelli, and red is increased in the normal substructures. White regions showed low activations equally in both cohorts, and are thus uninformative to the classification task. Filters are selectively sorted for visual clarity to highlight the differences in filters that show higher activations in normal cerebelli contrasted with dysplastic inputs. Compared to the first layer, we see more local delineation of regional cerebellar subdivisions. The superior surface of the cerebellum shows significant discriminatory power, seen as strongly differentiated regions of activation in the outer perimeter of the activation maps, which includes the posterior subdivision of the cerebellar lobes and the midline vermis.
Figure 13.
Within-Class Average Activation Maps for normal and dysplastic cohorts. Within-Class Average Activation Maps (wCAMs) are constructed at each convolutional layer by calculating a weighted sum of the activations learned for each feature map at that layer. This generates a weighted spatial map across all feature maps that most contribute to the classifier at each layer. Since we have fully connected layers prior to the final classifier, we lose the direct connection of each individual feature map’s contribution to the final classifier. However, it allows us to extract salient anatomical information at each convolutional layer. Notable regional differences between normal and dysplastic variants are shown with red arrows.
Discussion
We have introduced a computational framework for the application of 3D Convolutional Neural Networks on a dataset with a small sample size, showing excellent performance using cross-validation for assessment of subcortical neonatal brain dysmaturation. Despite gain in popularity, neural networks can still be prohibitively difficult to implement in a large subset of classification tasks. While computationally accessible, large datasets and deep architectures still require powerful hardware and long computation times to learn predictive models or classifiers. More importantly, the accuracy of the classifier is highly dependent on the variance encoded within the training dataset. Intensity-based segmentation tasks have the advantage of dense training datasets, with relatively low complexity modeled by the feature sets. In contrast, structural morphology-based classification tasks, as presented in this study, suffer from sparse data and the need for features of higher complexity to model the structural intricacies of key brain structures, including the cerebral cortex and subcortical structures such as the hippocampus or cerebellum. As our logistic regression results show, the available simple volumetric and conventional metrics were insufficient for detecting these complex morphological patterns using a linear model. This does not prove that linear models are inherently insufficient, as a higher dimensional vector using feature construction, or additional clinical or structural information may prove to be an adequate classifier using simpler linear models. However, more sophisticated non-linear approaches can also be explored using the structural information directly extracted from volumetric imaging.
For clinical populations, training sets are likely to be small, limiting the ability to apply 3D-CNN to more rare disorders. This is particularly difficult when implementing CNNs in medical imaging, as there can often be an insufficiently low incidence of abnormal cases in the training dataset, especially in neonatal and pediatric populations. These limitations can significantly slow down training, or impede it altogether, as the algorithm sees a disproportionate amount of normal cases through each iteration. Here we introduce a novel algorithm for neonatal brain dysmaturation that overcomes these challenges by first extracting the substructures of interest and registering them into a common space. This greatly reduces the computational search space and increases the observed effect size, leading to a relatively trivial application of deep neural networks as the final step. Most of the application of machine learning methods in neonatal neuroimaging to date have either been applied to actual segmentation of neonatal brain structures40–42 or functional resting BOLD imaging.43
A secondary challenge encountered in deep learning is the inherently obfuscated nature of the features learned by the classifier. Classically, deep learning is considered a “black box”, where the set of features used by the algorithm are not known or interpretable by the user.44 However, in medical imaging it is often desirable to generate mechanistic models of the phenotypical observations, rather than simply using a learned pattern to classify a disease. This necessitates the interpretability of the features learned within each hidden layer. Additionally, the coherence of the generated activation maps with known biological significance are added support for the specificity of structural morphology and subsequent clinical interpretation.
Although we have implemented a supervised learning method of classification, we can interpret the activation maps generated by the algorithm as data-driven learned features that provide insight into the underlying morphology of cerebellar dysplasia. The mean activation maps are effectively a lower dimensionality representation of the input data, with strength of activation in each group indicative of which regions in the image contribute most to the final classification task. Therefore, as we move further down each convolutional layer, the mean activations delineate further abstractions of the input images corresponding to the anatomical sub-regions of the cerebellum that are used for the final dysplasia classification. This technique is complementary to existing morphology based inference and classification methods, such as simple logistic regression, or deformation based statistical models,45 including tensor based morphometry.46,47 Deformation based methods look at voxel-wise or point-mesh differences between groups based on deformation onto a common space, with statistical analysis performed on the parametrized deformation mapping variances across the set of images. These models have shown to be very powerful in the analysis of morphological variations in clinical populations, and generally require lower computational and dataset requirements. Such models may result in comparable classification accuracies, and may even be a more efficient model in comparison. Future work aimed at delineating the possible synergy between our proposed method and surface deformation methods is warranted. However, these methods are dependent on both the mesh and deformation algorithms used to compute their parameters, and introduce further model generation complexity. Therefore, for this present study, we chose to naively input the full structure into our network to prevent any bias from being introduced in a normalization or mesh-generating step. As part of our pre-processing, we only linearly register the input into a common space, but retain the native structure input. It was our expectation that as the algorithm learned the necessary features for classification, the weights associated with the interior structure would be reduced to zero, as can be seen in the activation maps. While this may be computationally more expensive during the learning phase, the end result is effectively comparable, and helps validate the anatomical relevance of the learned parameters. As such, not only can this technique be used for rigorous clinical translational mechanistic research studies, but can also be applied to clinical recognition of dysplastic abnormalities in individual neonatal patients.
Most deep learning and CNN applied to neuroimaging data up to date have not revealed insight into the neuropathologic underpinnings of the disease process studies.9 In contrast, our algorithm revealed important regional vulnerability of cerebellar dysplasia in CHD infants that has both clinical and biological relevance. Concordant with the criteria that the clinical pediatric neuroradiologists used to classify the cerebellar dysplasia based on outer structural morphology, deep cerebellar regions (including the flocculonodular lobe) were not informative to the final classifier, shown as white regions in the activation group differences and cold regions in the class activation maps, compared to the superior surface of the cerebellum which anatomically corresponded to the posterior cerebellar cortical hemispheres and midline vermis. This supports the potential use of our automated technique in identifying cerebellar dysplasia in infants with CHD in the clinical setting and suggests that shape-related morphological disturbances in patients with CHD are present. To date, most morphological studies in CHD have focused on volumetric measurements, showing decreased brain volumes both globally and regionally present at birth and in the first year of life, 48,49 with volumetric abnormalities in specific brain structures being predictive of neurobehavior.50 Our regional cerebellar dysplasia is concordant with our recent work in fetal MRI which observed shape abnormalities in patients diagnosed with CHD,51 specifically in cerebellar vermis.
Our technique revealed important regional vulnerability of the cerebellum in CHD infants, with the superior surface (which includes the posterior subdivision and the midline vermis) providing greater discriminatory differences between dysplastic and normal appearing tissue. This finding is clinically relevant as clinical pediatric neuroradiology experts typically use the midline vermis and posterior aspect of the cerebellum to help discriminate between normal and dysplastic tissue, and could provide the basis by which our technique worked in the setting of a relatively small clinical sample size. This finding is also concordant with other genetic-based neuroimaging studies that have documented cerebellar dysplasia localized to the posterior cerebellar hemispheres and the midline vermis in other non-CHD patients, including autistic subjects.17 In addition, the vermis and posterior cerebellar hemispheres have been shown to be phenotypically abnormal in certain genetic animal models of cerebellar dysplasia including hypomorphic Foxc1 mutant mice that have both granule and Purkinje cell abnormalities.18 Interestingly, we have recently described cerebellar dysplasia in CHD infants correlated with ciliary motion assessed by nasal scrapes for ciliary motion video microscopy.13 Brain tissue from human fetuses with Joubert syndrome, a well-known ciliopathy associated with cerebellar dysplasia, showed impaired Shh-dependent cerebellar development, particularly in the vermis and posterior subdivisions, also concordant with our findings.52 Similarly, regional cerebellar dysplasia, also involving the vermis and posterior subdivision is frequently seen in our mouse CHD mutants harboring cilia related mutations.53,54 Together these findings suggest cilia defects may play an important role in the pathogenesis of cerebellar dysplasia observed in CHD patients. In future work, we will aim to combine our deep learning techniques with our recently described novel computer vision approach for assessing the elemental ciliary motion parameters with the computation of optical flow in the digital videos,55 to provide a more sensitive and robust phenotypic characterization of cerebellar dysplasia secondary to ciliary dysfunction.
The regional vulnerability of the CHD cerebellar dysplasia detects with our technique is likely to have downstream neurological and neurobehavioral consequences, which have been documented in CHD patients.56,57 The primary function of the cerebellum has classically been associated with motor learning, muscle tone, coordination and language. However, more recent development indicates cerebellar modulation of higher cognitive functions, including working memory, processing speed, and executive functioning.58 Eloquent cerebellar structure-function studies have demonstrated a topographic organization for motor regulation vs. cognitive and affective processing;59–62 for example, Bolduc et al showed decreased volume in the lateral cerebellar hemisphere was associated with impaired cognitive, expressive language, and motor control. While a reduction in vermal volume was related to impaired global development as well as behavior problems and a higher positive autism spectrum screening test.63 Furthermore, aberrant cerebellar development has been proposed to influence social and affect regulation, a term labeled cerebellar cognitive affective syndrome (CCAS).64 These deficits have long term consequences, and early intervention is imperative.65 Developing more sensitive methods for early detection of cerebellar impairment is an important step in the treatment and supportive neurodevelopmental care of infants with congenital heart disease. It is becoming more evident that localized injury to cerebellar subdivisions have varying effects in downstream neurodevelopment and behavior.66 Current guidelines rely on qualitative observations that lack the sensitivity for more subtle morphological malformations that may have clinical impact in long-term development. By more clearly defining key regions of the cerebellum, which are vulnerable to dysmaturation, we can more aptly create objective diagnostic guidelines. Future works will examine the role of CNN detection of regional vulnerability of cerebellar dysplasia and neurobehavioral outcomes in CHD patients.
There are many other known clinical risk factors thought to contribute to cerebellar dysmaturation, which have been studied more extensively in premature infants, who also demonstrate different forms of brain dysmaturation, including cerebellar hypoplasia. Exposure to glucocorticoids, commonly used to accelerate lung maturation in-utero and to treat hypotension, is known to inhibit sonic hedgehog pathways (SHH) critical to cerebellar development, was not extensive used in our CHD patients.12 Additionally, direct exposure to blood products and hemosiderin from cerebellar hemorrhage can have a direct effect on cerebellar growth.48 Finally, cerebral injury such as intra-ventricular hemorrhage (IVH) and more severe white matter injury (WMI) can lead to downstream disruption in cerebellar development.67 We excluded cases of cerebellar hemorrhage, IVH and WMI from our analysis in the CHD infants, so these are not likely contributing to our results. Future studies will be performed to examine the relationship of important perioperative clinical risk factors and CNN features of cerebellar dysplasia in CHD infants.
This study has several limitations. First, our sample size is small. The incidence of cerebellar dysplasia in this population is low, as there is a clear selection bias against more injured infants who may not have been deemed healthy enough to enroll in the study or have additional injuries that would exclude them, including gross ventriculomegaly and intra-ventricular hemorrhage. We did correct for pre-operative and post-operative status, and we did have few cases that had imaging in both periods. Another current limitation is the inclusion of a manual correction step to ensure accurate segmentation of the cerebellum prior to training the algorithm. This step may introduce user bias into the input, despite the users being blind to clinical diagnosis. Additionally, we validate our results using cross-validation. While cross-validation, along with the inclusion of drop-out layers during training, help prevent overtraining of the algorithm, it does not completely eliminate biases implicit to the dataset, such as population-specific variance and data acquisition parameters. Unfortunately, the use of a hold-out set given the available dataset significantly impacts the availability of abnormal structures used for training, hindering the learning process. As such, our results may still be overfitted. However, our framework’s ability to discriminate subtle dysplastic cerebelli is encouraging, and the inferential results into the biological underpinnings of cerebellar dysmaturation are significant. In the future, we aim to test our method on an independently acquired dataset for external validation. We chose to train a 3D CNN, rather than use transfer learning of existing high-performing models such as AlexNet, to preserve the 3D structure of the input substructures. While using an existing feature rich model trained on a much larger dataset could result in more powerful discriminatory performance, it would require pre-processing of the input data that would diminish the interpretability of the structural morphology associated with the learned classifiers. Therefore, we believe that generating interpretable features is a worthwhile tradeoff. Our future work is geared towards fully automating the structure extraction step of the pipeline to ensure a completely user-independent throughput and broadening the scope to target additional brain structures and populations.
Conclusion
We have introduced a computational framework for the extraction and specific classification of brain dysmaturation of subcortical structures in neonatal MRI, using a 3D convolutional neural network. We achieved a mean classification accuracy of 98.5% using 10-fold cross-validation. Furthermore, the hidden layer activations and class activation maps provide insight into the regional morphological characteristics of these dysplastic substructures in this at-risk population, which has both clinical diagnostic and biological implications. Our finding of the posterior lobe and vermis having the greatest discriminatory power for distinguishing dysplastic tissue from normal tissue in CHD infants, which is likely driven by genetic mechanisms. These findings also likely have important prognosticating implications with regard to neuromotor and neurobehavioral impairments in CHD patients. The code developed for this project is open source and published under the BSD License.
Supplementary Material
Acknowledgments
Funding
This work was supported by the Department of Defense (W81XWH-16–1-0613), the National Heart, Lung and Blood Institute (R01 HL128818–03), the National Institute of Neurological Disorders and Stroke (K23–063371), the National Library of Medicine (5T15LM007059–27), the Pennsylvania Department of Health, the Mario Lemieux Foundation, and the Twenty Five Club Fund of Magee Women’s Hospital.
References
- 1.LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86(11):2278–2323. doi: 10.1109/5.726791. [DOI] [Google Scholar]
- 2.Kleesiek J, Urban G, Hubert A, et al. Deep MRI brain extraction: A 3D convolutional neural network for skull stripping. Neuroimage. 2016;129:460–469. doi: 10.1016/j.neuroimage.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 3.Brosch T, Tang LYW, Yoo Y, Li DKB, Traboulsee A, Tam R. Deep 3D Convolutional Encoder Networks With Shortcuts for Multiscale Feature Integration Applied to Multiple Sclerosis Lesion Segmentation. IEEE Trans Med Imaging. 2016;35(5):1229–1239. doi: 10.1109/TMI.2016.2528821. [DOI] [PubMed] [Google Scholar]
- 4.Mueller SG, Weiner MW, Thal LJ, et al. Ways toward an early diagnosis in Alzheimer’s disease: The Alzheimer’s Disease Neuroimaging Initiative (ADNI). Alzheimer’s Dement. 2005;1(1):55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta A, Ayhan MS, Maida AS. Natural Image Bases to Represent Neuroimaging Data. http://proceedings.mlr.press/v28/gupta13b.pdf. Accessed October 4, 2017.
- 6.Payan A, Montana G. Predicting Alzheimer’s disease: a neuroimaging study with 3D convolutional neural networks. 2015:1–9. doi: 10.1613/jair.301. [DOI] [Google Scholar]
- 7.Hosseini-Asl E, Gimel’farb G, El-Baz A, Gimel’farb G, El-Baz A. Alzheimer’s Disease Diagnostics by a Deeply Supervised Adaptable 3D Convolutional Network. 2016;(502). doi: 10.1109/ICIP.2016.7532332. [DOI] [PubMed] [Google Scholar]
- 8.Rajpurkar P, Irvin J, Zhu K, et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. 2017:3–9. doi:1711.05225. [Google Scholar]
- 9.Vieira S, Pinaya WHL, Mechelli A. Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: Methods and applications. Neurosci Biobehav Rev. 2017;74:58–75. doi: 10.1016/j.neubiorev.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Wagner R, Thom M, Schweiger R, Palm G, Rothermel A. Learning Convolutional Neural Networks From Few Samples. http://geza.kzoo.edu/~erdi/IJCNN2013/HTMLFiles/PDFs/P274-1108.pdf. Accessed January 8, 2018.
- 11.Volpe JJ. Encephalopathy of Congenital Heart Disease- Destructive and Developmental Effects Intertwined. 2014. doi: 10.1016/j.jpeds.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Back SA, Miller SP. Brain injury in premature neonates: A primary cerebral dysmaturation disorder? Ann Neurol. 2014;75(4):469–486. doi: 10.1002/ana.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panigrahy A, Lee V, Ceschin R, et al. Brain Dysplasia Associated with Ciliary Dysfunction in Infants with Congenital Heart Disease. J Pediatr. 2016;178:141–148.e1. doi: 10.1016/j.jpeds.2016.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poretti A, Boltshauser E, Huisman TAGM. Pre- and Postnatal Neuroimaging of Congenital Cerebellar Abnormalities. Cerebellum. 2016;15(1):5–9. doi: 10.1007/s12311-015-0699-z. [DOI] [PubMed] [Google Scholar]
- 15.Doherty D, Millen KJ, Barkovich AJ. Midbrain-Hindbrain Malformations: Advances in Clinical Diagnosis, Imaging, and Genetics. Lancet Neurol. 2013;12(4):381–393. doi: 10.1016/S1474-4422(13)70024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oegema R, Cushion TD, Phelps IG, et al. Recognizable cerebellar dysplasia associated with mutations in multiple tubulin genes. Hum Mol Genet. 2015;24(18):5313–5325. doi: 10.1093/hmg/ddv250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wegiel JJ, Kuchna I, Nowicki K, et al. The neuropathology of autism: Defects of neurogenesis and neuronal migration, and dysplastic changes. Acta Neuropathol. 2010;119(6):755–770. doi: 10.1007/s00401-010-0655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haldipur P, Dang D, Aldinger KA, et al. Phenotypic outcomes in Mouse and Human Foxc1 dependent Dandy-Walker cerebellar malformation suggest shared mechanisms. Elife. 2017;6:1–15. doi: 10.7554/eLife.20898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosemani T, Poretti A. Cerebellar disruptions and neurodevelopmental disabilities. Semin Fetal Neonatal Med. 2016:1–10. doi: 10.1016/j.siny.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Stoodley CJ, Limperopoulos C. Structure–function relationships in the developing cerebellum: Evidence from early-life cerebellar injury and neurodevelopmental disorders. Semin Fetal Neonatal Med. 2016;21(5):1–9. doi: 10.1016/j.siny.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barkovich AJ, Raybaud C. Pediatric Neuroimaging. LWW; 2012. [Google Scholar]
- 22.Varoquaux G, Raamana PR, Engemann DA, Hoyos-Idrobo A, Schwartz Y, Thirion B. Assessing and tuning brain decoders: Cross-validation, caveats, and guidelines. Neuroimage. 2017;145:166–179. doi: 10.1016/J.NEUROIMAGE.2016.10.038. [DOI] [PubMed] [Google Scholar]
- 23.Gorgolewski K, Burns CD, Madison C, et al. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python. Front Neuroinform. 2011;5. doi: 10.3389/fninf.2011.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62(2):782–790. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 26.Vovk U, Pernuš F, Likar B. A review of methods for correction of intensity inhomogeneity in MRI. IEEE Trans Med Imaging. 2007;26(3):405–421. doi: 10.1109/TMI.2006.891486. [DOI] [PubMed] [Google Scholar]
- 27.Gousias IS, Edwards AD, Rutherford MA, et al. Magnetic resonance imaging of the newborn brain: Manual segmentation of labelled atlases in term-born and preterm infants. Neuroimage. 2012;62(3):1499–1509. doi: 10.1016/j.neuroimage.2012.05.083. [DOI] [PubMed] [Google Scholar]
- 28.Gousias IS, Hammers A, Counsell SJ, et al. Magnetic Resonance Imaging of the Newborn Brain: Automatic Segmentation of Brain Images into 50 Anatomical Regions. Rodriguez-Fornells A, ed. PLoS One. 2013;8(4):e59990. doi: 10.1371/journal.pone.0059990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avants B, Tustison N, Song G. Advanced Normalization Tools (ANTS). Insight J. 2009:1–35. ftp://ftp3.ie.freebsd.org/pub/sourceforge/a/project/ad/advants/Documentation/ants.pdf. [Google Scholar]
- 30.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54(3):2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou KH, Warfield SK, Bharatha A, et al. Statistical Validation of Image Segmentation Quality Based on a Spatial Overlap Index. Acad Radiol. 2004;11(2):178–189. doi: 10.1016/S1076-6332(03)00671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao H, Lu Z, Poupart P. Self-adaptive hierarchical sentence model. IJCAI Int Jt Conf Artif Intell. 2015;2015-Janua:4069–4076. doi: 10.1162/153244303322533223. [DOI] [Google Scholar]
- 33.Szegedy C, Zaremba W, Sutskever I, et al. Intriguing properties of neural networks. December 2013. doi: 10.1021/ct2009208. [DOI] [Google Scholar]
- 34.Zeiler MD, Krishnan D, Taylor GW, Fergus R. Deconvolutional networks. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition; 2010:2528–2535. [Google Scholar]
- 35.Zeiler MD, Fergus R. Visualizing and Understanding Convolutional Networks. Comput Vision–ECCV 2014. 2014;8689:818–833. doi: 10.1007/978-3-319-10590-1_53. [DOI] [Google Scholar]
- 36.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning Deep Features for Discriminative Localization. December 2015. doi: 10.1109/CVPR.2016.319. [DOI]
- 37.Theano Development Team. Theano: A Python framework for fast computation of mathematical expressions. arXiv e-prints. 2016:19 http://arxiv.org/abs/1605.02688.
- 38.Nielsen MA. Neural Networks and Deep Learning. Determination Press; 2015. http://neuralnetworksanddeeplearning.com/. [Google Scholar]
- 39.Clevert D-A, Unterthiner T, Hochreiter S. Fast and Accurate Deep Network Learning by Exponential Linear Units (ELUs). November 2015. doi: 10.3233/978-1-61499-672-9-1760. [DOI] [Google Scholar]
- 40.Srhoj-Egekher V, Benders M, Kersbergen KJ, Viergever MA, Isgum I. Automatic segmentation of neonatal brain MRI using atlas based segmentation and machine learning approach. MICCAI Gd Chall Neonatal Brain Segmentation. 2012;2012. [Google Scholar]
- 41.Song Z, Awate SP, Licht D, Gee JC. Clinical Neonatal Brain MRI Segmentation and Intensity-based Markov Priors. Computer (Long Beach Calif). 2007:1–8. doi: 10.1007/978-3-540-75757-3_107. [DOI] [PubMed] [Google Scholar]
- 42.Jaware TH, Khanchandani KB, Zurani A. Multi-kernel support vector machine and Levenberg-Marquardt classification approach for neonatal brain MR images. 2016 IEEE 1st Int Conf Power Electron Intell Control Energy Syst. 2016;8(0):1–4. doi: 10.1109/ICPEICES.2016.7853639. [DOI] [Google Scholar]
- 43.Ball G, Aljabar P, Arichi T, et al. Machine-learning to characterise neonatal functional connectivity in the preterm brain. Neuroimage. 2016;124(Pt A):267–275. doi: 10.1016/j.neuroimage.2015.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erhan D, Bengio Y, Courville A, Vincent P. Visualizing higher-layer features of a deep network. Bernoulli. 2009;(1341):1–13. https://www.researchgate.net/profile/Aaron_Courville/publication/265022827_Visualizing_Higher-Layer_Features_of_a_Deep_Network/links/53ff82b00cf24c81027da530.pdf.
- 45.Cootes TF, Twining CJ, Taylor CJ. Diffeomorphic Statistical Shape Models. http://personalpages.manchester.ac.uk/staff/timothy.f.cootes/Papers/cootes_bmvc04.pdf. Accessed January 8, 2018.
- 46.Wang Y, Panigrahy A, Shi J, et al. Surface multivariate tensor-based morphometry on premature neonates: a pilot study. 2nd MICCAI 2013 Work Clin Image-Based Proced Transl Res Med Imaging. September 2011. [Google Scholar]
- 47.Hua X, Leow AD, Parikshak N, et al. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer’s disease: an MRI study of 676 AD, MCI, and normal subjects. Neuroimage. 2008;43(3):458–469. doi: 10.1016/j.neuroimage.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Rhein M, Buchmann A, Hagmann C, et al. Severe Congenital Heart Defects Are Associated with Global Reduction of Neonatal Brain Volumes. J Pediatr. 2015;167(6):1259–1263.e1. doi: 10.1016/j.jpeds.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 49.Ortinau C, Inder T, Lambeth J, Wallendorf M, Finucane K, Beca J. Congenital heart disease affects cerebral size but not brain growth. Pediatr Cardiol. 2012;33(7):1138–1146. doi: 10.1007/s00246-012-0269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Owen M, Shevell M, Donofrio M, et al. Brain volume and neurobehavior in newborns with complex congenital heart defects. J Pediatr. 2014;164(5):1121–1127.e1. doi: 10.1016/j.jpeds.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong A, Chavez T, O’neil S, et al. Synchronous Aberrant Cerebellar and Opercular Development in Fetuses and Neonates with Congenital Heart Disease: Correlation with Early Communicative Neurodevelopmental Outcomes, Initial Experience. Am J Perinatol Rep. 2017;7(1):17–27. doi: 10.1055/s-0036-1597934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aguilar A, Meunier A, Strehl L, et al. Analysis of human samples reveals impaired SHH-dependent cerebellar development in Joubert syndrome/Meckel syndrome. Proc Natl Acad Sci. 2012;109(42):16951–16956. doi: 10.1073/pnas.1201408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu X, Yagi H, Saeed S, et al. The complex genetics of hypoplastic left heart syndrome. Nat Genet. 2017;49(7):1152–1159. doi: 10.1038/ng.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Klena NT, Gabriel GC, et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 2015;521(7553):520–524. doi: 10.1038/nature14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quinn SP, Zahid MJ, Durkin JR, Francis RJ, Lo CW, Chennubhotla SC. Automated identification of abnormal respiratory ciliary motion in nasal biopsies. Sci Transl Med. 2015;7(299):299ra124–299ra124. doi: 10.1126/scitranslmed.aaa1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bellinger DC, Wypij D, Rivkin MJ, et al. Adolescents with d-transposition of the great arteries corrected with the arterial switch procedure: Neuropsychological assessment and structural brain imaging. Circulation. 2011;124(12):1361–1369. doi: 10.1161/CIRCULATIONAHA.111.026963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bellinger DC, Newburger JW. Neuropsychological, psychosocial, and quality-of-life outcomes in children and adolescents with congenital heart disease. Prog Pediatr Cardiol. 2010;29(2):87–92. doi: 10.1016/j.ppedcard.2010.06.007. [DOI] [Google Scholar]
- 58.D’Angelo E, Casali S. Seeking a unified framework for cerebellar function and dysfunction: from circuit operations to cognition. Front Neural Circuits. 2012;6(January):116. doi: 10.3389/fncir.2012.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Limperopoulos C, Chilingaryan G, Sullivan N, Guizard N, Robertson RL, Du Plessis AJAJAJ. Injury to the premature cerebellum: Outcome is related to remote cortical development. Cereb Cortex. 2014;24(3):728–736. doi: 10.1093/cercor/bhs354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brossard-Racine M, du Plessis AJ, Limperopoulos C. Developmental Cerebellar Cognitive Affective Syndrome in Ex-preterm Survivors Following Cerebellar Injury. Cerebellum. 2015;14(2):151–164. doi: 10.1007/s12311-014-0597-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allin MPG, Salaria S, Nosarti C, Wyatt J, Rifkin L, Murray RM. Vermis and lateral lobes of the cerebellum in adolescents born very preterm. Neuroreport. 2005;16(16):1821–1824. doi: 10.1097/01.wnr.0000185014.36939.84. [DOI] [PubMed] [Google Scholar]
- 62.Limperopoulos C, Chilingaryan G, Guizard N, et al. Cerebellar injury in the premature infant is associated with impaired growth of specific cerebral regions. Pediatr Res. 2010;68(2):145–150. doi: 10.1203/00006450-201011001-00282. [DOI] [PubMed] [Google Scholar]
- 63.Bolduc M-E, du Plessis AJ, Sullivan N, et al. Regional cerebellar volumes predict functional outcome in children with cerebellar malformations. Cerebellum. 2012;11(2):531–542. doi: 10.1007/s12311-011-0312-z. [DOI] [PubMed] [Google Scholar]
- 64.Schmahmann JD, Sherman JC. The cerebellar cognitive affective syndrome. Brain. 1998;121(4):561–579. doi: 10.1093/brain/121.4.561. [DOI] [PubMed] [Google Scholar]
- 65.Calderon J, Bellinger DC. Executive function deficits in congenital heart disease: why is intervention important? Cardiol Young. 2015;(JANUARY):1–9. doi: 10.1017/S1047951115001134. [DOI] [PubMed] [Google Scholar]
- 66.Tiemeier H, Lenroot RK, Greenstein DK, Tran L, Pierson R, Giedd JN. Cerebellum development during childhood and adolescence: A longitudinal morphometric MRI study. Neuroimage. 2010;49(1):63–70. doi: 10.1016/j.neuroimage.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tam EWY. Potential mechanisms of cerebellar hypoplasia in prematurity. Neuroradiology. 2013;55(SUPPL. 2):41–46. doi: 10.1007/s00234-013-1230-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.