Abstract
Fertilization is an intricate cascade of events that irreversibly alter the participating male and female gamete and ultimately lead to the union of paternal and maternal genomes in the zygote. Fertilization starts with sperm capacitation within the oviductal sperm reservoir, followed by gamete recognition, sperm–zona pellucida interactions and sperm–oolemma adhesion and fusion, followed by sperm incorporation, oocyte activation, pronuclear development and embryo cleavage. At fertilization, bull spermatozoon loses its acrosome and plasma membrane components and contributes chromosomes, centriole, perinuclear theca proteins and regulatory RNAs to the zygote. While also incorporated in oocyte cytoplasm, structures of the sperm tail, including mitochondrial sheath, axoneme, fibrous sheath and outer dense fibers are degraded and recycled. The ability of some of these sperm contributed components to give rise to functional zygotic structures and properly induce embryonic development may vary between bulls, bearing on their reproductive performance, and on the fitness, health, fertility and production traits of their offspring. Proper functioning, recycling and remodeling of gamete structures at fertilization is aided by the ubiquitin–proteasome system (UPS), the universal substrate-specific protein recycling pathway present in bovine and other mammalian oocytes and spermatozoa. This review is focused on the aspects of UPS relevant to bovine fertilization and bull fertility.
Keywords: fertilization, artificial insemination, sperm capacitation, acrosome, zygote
Implications
Fertilization is an intricate cascade of events that irreversibly alter the participating male and female gamete and ultimately lead to the union of paternal and maternal genomes in the zygote. Proper functioning, recycling and remodeling of gamete structures at fertilization is aided by the ubiquitin–proteasome system (UPS), the universal substrate-specific protein recycling pathway present in bovine and other mammalian oocytes and spermatozoa. Research focused on the UPS-regulated aspects of the fertilization process has a potential to impact the management of bull fertility, the optimization of artificial insemination, genomic selection for production and fertility-related traits, and the improvement of sperm viability after sexing and cryopreservation in cattle.
Introduction: prelude to sperm–oocyte interactions
However, for the semen to exert its effect on the eggs, it must enter their little bodies, as it seems unlikely that it could animate them by the mere contact with their skin (Spallanzani, 1780).
Quoted above, Italian pioneer of reproductive research and father of fertilization biology Lazzaro Spallanzani (1729–99), known for conducting the first recorded experiments with artificial insemination in frogs and canines, captured the essence of the fertilization process long before his successors were able to observe the actual union of a spermatozoon and an oocyte and decipher the molecular machineries that guide it. To paraphrase the words of another luminary of fertilization research, C.R. Moore, fertilization is a precisely orchestrated cascade of irreversible events that starts long before the male and female gametes meet. This intricate wedding dance starts with the mature gametes reaching the site of fertilization within the female oviduct. Spermatozoa have to bypass multiple hurdles crossing from the site of deposition though the female cervix (in vaginal depositors), uterus and uterotubal junction, to finally reach the site of oviductal sperm reservoir and bind, transiently, to its apical epithelia surfaces (Suarez, 2015). In bulls, sperm transport is believed to take between 4 and 16 h. The sperm–oviductal epithelium binding is mediated by acrosomal surface ligands and the fucose-containing glycans of the plasma membrane annexins, expressed by the oviductal epithelial cells. The sperm release upon capacitation requires proteolytic cleavage and shedding of the acrosomal binder of sperm proteins, particularly binder of sperm protein 1 (BSP1) (Suarez, 2015).
Upon ovulation, signals from the ovary, oocyte–cumulus complex and its accompanying follicular fluid may be responsible for the capacitation and hyperactivation of spermatozoa bound to the oviductal sperm reservoir. Capacitation endows the reservoir-bound spermatozoa with fertilizing ability and helps them detach from the reservoir epithelium. As in other species, bull sperm capacitation encompasses cholesterol efflux, calcium influx, pH increase, remodeling of plasma membrane, and increase in tyrosine phosphorylation of sperm head and sperm tail proteins (Parrish, 2013). Such structural and molecular remodeling brings about hyperactivation, manifested by an increased amplitude and frequency of flagellar movement. Chemoattractants issued from the oocyte and its surrounding cumulus cells are likely responsible for gamete recognition in mammals. Although the exact identity of the attractants involved in bovine gamete recognition is not known, progesterone from cumulus cells has been shown to fulfill such a role in other species (Oren-Benaroya et al., 2008).
Ubiquitin–proteasome system and fertilization
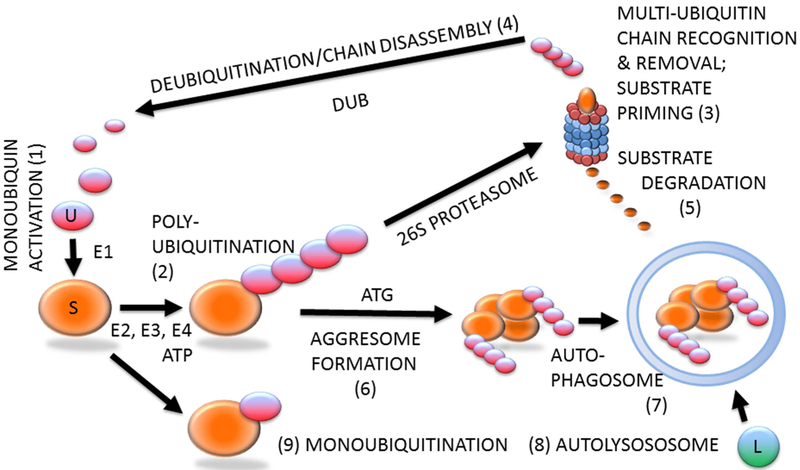
Ubiquitin–proteasome system (Figure 1) is likely involved in all steps of the reproductive process, including gameto-genesis, fertilization, zygotic and preimplantation embryonic development, implantation, and beyond (Sutovsky, 2003). Protein ubiquitination is a stable, covalent post-translational modification by which the substrate proteins can be targeted for degradation by 26S proteasome, lysosome or autophagosome (Cohen-Kaplan et al., 2016). Alternatively, protein ubiquitination, which is reversible by deubiquitinases, can serve a regulatory function, for example, through receptor endocytosis and histone code modification. Ubiquitination is a substrate-specific process that requires an energy donor (ATP) and a set of specific activator, carrier and ligase enzymes; it starts with the activation of single ubiquitin molecules by the ubiquitin-activating enzyme E1 (HUGO acronym UBA1). The activated ubiquitin is passed onto one of several carrier enzymes (E2 or UBE2) and ligated to an internal lysine residue of a substrate protein by substrate-specific ubiquitin ligases (E3 enzymes or UBE3), of which there is a great variety. Additional ubiquitin molecules are linked to the substrate-bound monoubiquitin by E3 enzymes, sometimes assisted by chain elongation factors (E4 enzymes). Substrate proteins linked to multi-ubiquitin chains of four or more ubiquitin molecules are degraded by the 26S proteasome, a multisubunit holoenzyme composed of a 20S proteolytic core capped with a 19S proteasomal regulatory complex responsible for the recognition and removal of the multi-ubiquitin chain, and for the priming of the substrate protein before its transport to the 20S core.
Figure 1.
(Colour online) Protein ubiquitination and degradation. Unconjugated monoubiquitin (U; 1) binds covalently to its substrate (S) protein, catalyzed by ubiquitin-activating and -conjugating enzymes (E1, E2, E3) and fueled by ATP, to form a multi-ubiquitin chain (2), which is recognized by the 26S proteasome (3). The multi-ubiquitin chain is removed by the 19S proteasomal regulatory complex to be disassembled by deubiquitinating enzymes (DUB) and re-enter the pool of monoubiquitin available for conjugation (4). The substrate is degraded in the 20S proteasomal core (5). Proteasomal degradation has been implicated in sperm–zona pellucida binding, acrosomal exocytosis and sperm–zona penetration. Proteasomal proteolysis also regulates cell cycle progression, and pronuclear development and apposition in the zygote. During autophagy, which has been implicated in the regulation of early embryo development and post-fertilization sperm mitophagy, polybiquitinated substrate molecules are aggregated by autophagy-associated proteins (ATG) to form an aggresome (6). Aggresomes are recognized and engulfed by the autophagophore that encloses them to form an autophagosome (7), which upon fusion with a lysososome (L) becomes an autolysosome (8) capable of degrading the entire aggresome, or an ubiquitinated organelle such as sperm mitochondrion. The ubiquitin–proteasome system-regulated post-fertilization sperm mitophagy mediates clonal, maternal inheritance of mammalian mitochondrial genes. Monoubiquitination (9) is reversible and serves regulatory purposes relevant to gametogenesis, fertilization and early development, such as histone modification to establish the epigenetic histone code, or plasma membrane receptor internalization to change cell responsiveness to specific external stimuli/ligands.
While the proteasomes have been studied in spermatozoa of a number of mammalian species, as well as in other vertebrate and invertebrate animals (Sutovsky, 2011), little is known about the role of UPS in the early events of ungulate fertilization. Involvement of the sperm-borne proteasomes in sperm capacitation has been described in human and porcine spermatozoa (reviewed by Kerns et al., 2016). A recent study details the role of proteasomal degradation of the A-kinase anchoring fibrous sheath protein AKAP3 (A-kinase anchoring protein 3) in bull sperm capacitation, which was correlated with capacitation and acrosomal exocytosis ability that varied among individual bulls (Hillman et al., 2013). More is known about the involvement of UPS in later events of fertilization, as will be discussed next.
Sperm–zona pellucida interactions
Bull and other mammals’ spermatozoa bind to oocyte zona pellucida (ZP) by the apical ridge of their acrosome, composed of the acrosomal matrix sandwiched between the inner and outer acrosomal membranes (IAM and OAM, respectively), with the IAM laid atop subacrosomal perinuclear theca (PT) (Oko and Sutovsky, 2009). Bovine ZP is composed of three heavily glycosylated proteins, ZPA (homolog of human ZP2), ZPB (homolog of human ZP4/ZP3β) and ZPC (homolog of human ZP3/ZP3α), with the ZPC/ZP3 and ZPB/ZP4 being the predicted primary receptors for sperm binding and induction of acrosomal exocytosis (Yonezawa, 2014). In this scenario, ZPA/ZP2 is proposed to be responsible for secondary/sustained sperm–zona binding (Gadella, 2010).
During fertilization, the spermatozoa primed by acrosomal membrane remodeling during capacitation bind to ZP surface, undergo acrosomal exocytosis enacting the loss of OAM and acrosomal matrix in all parts of the acrosome except the equatorial segment, and penetrate the ZP with only the IAM left intact after exocytosis (Gerton, 2002). The OAM and acrosomal matrix do not merely disperse from zona surface as previously thought; they remain associated with it in the form of acrosomal shroud. In addition to enabling sperm–zona penetration, acrosomal exocytosis prepares spermatozoa for fusion with the oolemma, by exposing oolemma-binding proteins on the sperm head equatorial segment (Cuasnicu et al., 2016). Based on imaging of live mouse spermatozoa with green fluorescent acrosomes, it was proposed that acrosomal exocytosis is initiated before sperm contact with the oocyte, that is, during sperm migration along the female oviduct (La Spina et al., 2016). It is not clear whether such concept applies to bovine fertilization. Furthermore, one has to be cautious about interpreting such observations as the true acrosomal exocytosis, as opposed to the remodeling of acrosomal membranes induced by sperm capacitation.
The signaling cascade of acrosomal exocytosis, induced by the binding of sperm head ligands to ZPC, activates signaling through phospholipases, and both the tyrosine and serine/threonine protein kinases which leads to the influx of external calcium, depolymerization of the actin microfilaments scaffolding the acrosome, and the activation of membrane fusion pathways leading to the fusion and hybridizing vesiculation of plasma membrane and OAM, resulting in the formation of acrosomal shroud (Belmonte et al., 2016; Romarowski et al., 2016).
Bull spermatozoa display all three major proteasomal core enzymatic activities (Pizarro et al., 2004). Proteasomes are localized in all three layers of the mammalian sperm acrosome, including OAM, matrix and IAM (Sutovsky et al., 2004; Zimmerman et al., 2011). Acrosomal proteasomes may serve a dual purpose during sperm–ZP interactions, first as the mediators of acrosomal exocytosis and subsequently as proteases contributing to localized degradation of the zona matrix that results in the formation of the fertilization slit (Zimmerman et al., 2011; Miles et al., 2013). Bull spermatozoa capacitated in the presence of proteasomal inhibitor epoxomicin had reduced ability to undergo acrosomal exocytosis induced in vitro by progesterone and to fertilize oocytes in vitro (Sanchez et al., 2011). Addition of proteasomal inhibitor MG132 into in vitro fertilization (IVF) medium significantly reduced the rate of bovine fertilization (Rawe et al., 2008). Solubilized ZP protein degradation by bull sperm proteasomes has not been described, to our knowledge, but the degradation of solubilized ZP proteins by sperm proteasomes has been shown in humans (Saldivar-Hernandez et al., 2015) and Japanese quail (Sasanami et al., 2012). In agreement with their proposed role in sperm–zona interactions, a subpopulation of proteasomes are exposed on the bull sperm surface and prominently represented in the sperm surface proteome (Byrne et al., 2012), as has also been described in other mammals. It is thus possible that a third function could be assigned to them: that of secondary or alternative receptor for ZP. In agreement the proteasomes of human and porcine spermatozoa are part of a large multimeric zona-binding complex that also contains chaperonins, zona-binding receptors, acrosin and its binding protein, and seminal plasma proteins (Redgrove et al., 2011; Kongmanas et al., 2014). In summary, UPS is likely involved in multiple events during sperm interactions with oocyte ZP.
Gamete adhesion and fusion, and sperm incorporation
Commonly referred to as gamete fusion, the process by which the fertilizing spermatozoon contacts and passes through the oocyte plasma membrane, the oolema, is likely more complex than originally thought. It is thus prudent to distinguish between its adhesion and fusion phase (reviewed in Sutovsky, 2009). Moreover, important is to note that gamete adhesion and fusion is enabled by sperm capacitation and acrosomal exocytosis during which proteins involved in the sperm–oolemma interactions become exposed on the sperm head equatorial segment (e.g. equatorin/MN-9 (Manandhar and Toshimori, 2001), CD9 (Ito et al., 2010) and IZUMO1 (Satouh et al., 2012)).
Early studies implicated sperm disintegrins and oolemma integrins in sperm–oolemma adhesion, though none such receptors have fusogenic properties. Gene knock-out studies demonstrated that these molecules are either non-essential or compensable in mouse fertilization (Primakoff and Myles, 2007). Alternatively proposed sperm receptors on the oollema, tetraspanins CD9 and CD81 were knocked out, resulting in reduced fertility of single gene knock-out and complete infertility of double knock-out (Benammar et al 2016). The most convincing evidence for gamete adhesion receptors came with the discovery of the sperm-specific immunoglobulin family cell adhesion protein IZUMO (Inoue et al., 2005) and its oocyte-binding partner, the folate-binding receptor JUNO (Bianchi et al., 2014). Mice null in either of these proteins are infertile. Both IZUMO (Fukuda et al., 2016) and JUNO (Zhao et al., 2017) are present in bovine gametes, but there is not yet a direct proof of their involvement in bovine gamete adhesion. A plausible model for bovine/mammalian sperm–oolemma interaction is thus the tetraspanin web on the oolemma, in which said tetraspanins are the organizers of various receptors for sperm equatorial segment ligands, including integrins, JUNO and the downstream signaling proteins that relay the adhesion signals to oocyte cortical cytoskeleton (Sutovsky, 2009; Wright and Bianchi, 2015). Accordingly, microfilaments in the bovine oocyte microvilli (Sutovsky et al., 1996) and cortex are required for physical incorporation of the sperm head and tail in the ooplasm and probably for signaling in response to the initial adhesion between sperm and oocyte plasma membranes, preceding their fusion, as exemplified by gamete adhesion induced signaling via oocyte proline-rich tyrosine kinase (PYK2) in zebra fish (Sharma and Kinsey, 2012). Consequently, microfilament disrupting drug cytochalasin B prevents sperm incorporation without interfering with sperm–oolema fusion or with the release of sperm PT proteins that trigger oocyte activation (Sutovsky et al., 1996; Sutovsky et al., 1997; Sutovsky et al., 2003).
Cortical actin cytoskeleton appears to be regulated in part by UPS. Interference with ubiquitin C-terminal hydrolase (UCHL1), enriched in the mammalian oocyte cortex can thus alter polyspermy rate in vitro (Yi et al., 2007; Mtango et al., 2011b). The expression of Uchl1 gene in bovine zygotes was altered by heat stress (Sakatani et al., 2015), wherein the UCHL1 inhibition impaired migration and exocytosis of cortical granules, the major line of defense against polyspermic fertilization (Susor et al., 2010). A related deubiquitinase, UCHL3, is associated with oocyte meiotic spindle and may regulate oocyte maturation (Mtango et al., 2014 and 2011a). Altogether, it is likely that the UPS mainly regulates the later events of sperm–oocyte adhesion and fusion; in particular, sperm incorporation and activation of the anti-polyspermy defense.
Oocyte activation
While adhesion and fusion between oocyte and sperm plasma membranes initially occurs within the sperm head equatorial segment, it quickly extends distally to post-acrosomal sheath (Sutovsky et al., 2003). The current paradigm of oocyte activation assumes that a soluble sperm factor(s) released from post-acrosomal PT triggers calcium release from oocyte endoplasmic reticulum that triggers a signaling cascade guiding the completion of oocyte meiosis, mobilization of anti-polyspermy defense, zygotic/pronuclear development and ultimately embryo cleavage (Oko et al., 2017). Initial studies of this process in mammals were conducted in rodent and bovine IVF systems. It was demonstrated that freeze-thaw sperm extracts activate hamster oocytes (Swann, 1990). The removal and solubilization of sperm PT was shown to occur during bovine IVF even when sperm incorporation was prevented by treatment with actin depolymerizing drug cytochalasin B (Sutovsky et al., 1997).
While the dissolution of PT can be hindered by protease inhibitors (Perry et al., 2000), it is not clear whether oocyte proteasomes are directly involved. However, UPS clearly regulates zygotic cell cycle by mediating the oocyte spindle exit from metaphase II through degradation of EMI1 (synonym F-boax protein 5/FBOX5) protein that maintains the activity of oocyte cytostatic factor and by degrading the cyclin component of the M-phase promoting factor to assure successful metaphase–anaphase transition (Glotzer et al., 1991; Tung et al., 2005; Karabinova et al., 2011).
Stepwise extraction of PT from mouse sperm heads progressively eliminated their ability to trigger oocyte activation after intracytoplasmic sperm injection, and the activity was shown to be associated with specific fractions of PT that had differential extraction resistance (Kimura et al., 1998; Perry et al., 2000). Eventually, the currently favored sperm-borne oocyte factor (SOAF) candidate, phospoholipase C zeta 1 (PLCZ1) was identified in silico and shown to trigger oocyte calcium oscillations when a recombinant protein or its messenger RNA (mRNA) were injected in metaphase II oocytes (Saunders et al., 2002). Persistent concerns about localization of PLCZ outside of post-acrosomal sheath (PAS) and its promiscuity with regard to oocyte-activating sperm cytosol fractions prompted a search for alternative or synergistic sperm SOAF components, chief among them male germline-specific WW-domain-binding WBP2NL (ww-domain binding protein 2 N-terminal like) (alias post-acrosomal WW-domain binding protein (PAWP)) and its somatic cousin WBP2 (Wu et al., 2007b; Oko et al., 2017). As in other mammals, PAWP is prominent in bull sperm PT where it is transported from the cytoplasmic lobe along the caudal manchette in the elongating spermatid phase of spermiogenesis, coinciding with the spermatids’ acquisition of oocyte-activating ability (Wu et al., 2007a). In the mouse, targeting neither the Plcz1 (Hachem et al., 2017) nor the Wbp2nl (Satouh et al., 2015) gene appears to eliminate male fertility. Intriguingly, increased expression of Wbp2nl gene is observed in the testis of Plcz1 null mice (Hachem et al., 2017), suggesting compensation in the knock-out mouse and possible synergy between the two gene products in wild type animals.
With regard to bull fertility, individual sperm levels of WBP2NL vary between sires and within individual ejaculates. Excessive amounts of WBP2NL were found in macrocephalic spermatozoa with abnormally large sperm head and PAS, while certain morphologically deviant spermatozoa, particularly the ones with abnormally shaped sperm heads and those positive for ubiquitin, in this case applied as a sperm quality biomarker (Sutovsky et al., 2015), lacked WBP2NL altogether (Kennedy et al., 2014). High abundance of Plcz1 mRNA in spermatozoa was also associated with fertility of Holstein bulls (Kasimanickam et al., 2012), a breed in which Plcz1 gene polymorphism also appears to be a contributing factor (Pan et al., 2013). In conclusion, while the identity of the sperm-borne oocyte-activating factor is still being researched, there appears to be a consensus that it is a partially soluble protein (or several proteins) in the sperm head PT.
Zygotic centrosome assembly and pronuclear apposition
Somatic cell centrosome is a cellular microtubule organizing center composed of two perpendicularly oriented, barrel-shaped (pinwheel-like on cross-section) centrioles and pericentriolar material (PCM) with the ability to nucleate tubulins to polymerize into microtubules. In bovine gametes, the centrosome is reduced in a complementary fashion, with oocytes retaining PCM but devoid of centrioles, and spermatozoa carrying a single, duplication-competent centriole (or no centrioles, as in rodents) and little, if any PCM (Sathananthan et al., 1997; Manandhar et al., 2005). As in other non-rodent Eutherian mammals, bull sperm centrosome is reduced to a single centriole with little or no PCM during spermiogenesis/spermatid elongation in the testis. This centriole is housed within the complex structure of the sperm tail connecting piece and thus termed the proximal centriole (Sutovsky et al., 1996; Simerly et al., 1999). The distal centriole is degraded during spermiogenesis, and only some remnants of centrosomal material may be found within the distal centriolar vault (Manandhar et al., 2005). Failure of proper centrosome remodeling during spermiogenesis has been associated with the tail stump defect in infertile bulls (Blom and Birch-Andersen, 1980).
At fertilization, soon after sperm head and sperm tail midpiece incorporation, the striated columns caging the centriole break up, a disassembly process required for the formation of zygotic centrosome and leading up to the organization of microtubule-made sperm aster responsible for the apposition of paternal and maternal pronuclei (Sutovsky et al., 1996; Sutovsky and Schatten, 1997; Sutovsky, 2010) (Figure 2). The gamma-tubulin (TUBG) containing, microtubule-nucleating PCM is recruited from bovine oocyte cytoplasm by the liberated sperm centriole (Shin and Kim, 2003). The ability of bull spermatozoa to form sperm aster after IVF has been correlated with the developmental potential of bovine IVF embryos in artificial insemination (AI) bulls (Navara et al., 1996; Hara et al., 2015; Hochi, 2016). Inspired by those findings, bovine oocytes have been injected with human spermatozoa as a way of testing for human male infertility due to sperm centrosomal insufficiency (Nakamura et al., 2001). It was noted that in both human and bovine zygote, proteasomal activity is required for the formation of sperm aster, possibly because proteasomes are present in the sperm tail connecting piece and necessary for its disassembly that liberates the sperm centriole and makes it competent to recruit centrosomal proteins from ooplasm (Rawe et al., 2008). Altogether, it is well documented that bull spermatozoa contribute crucial components of the zygotic centrosome at fertilization, and such a mode of centrosomal inheritance is similar to primates and other ungulates, but different from rodents.
Figure 2.
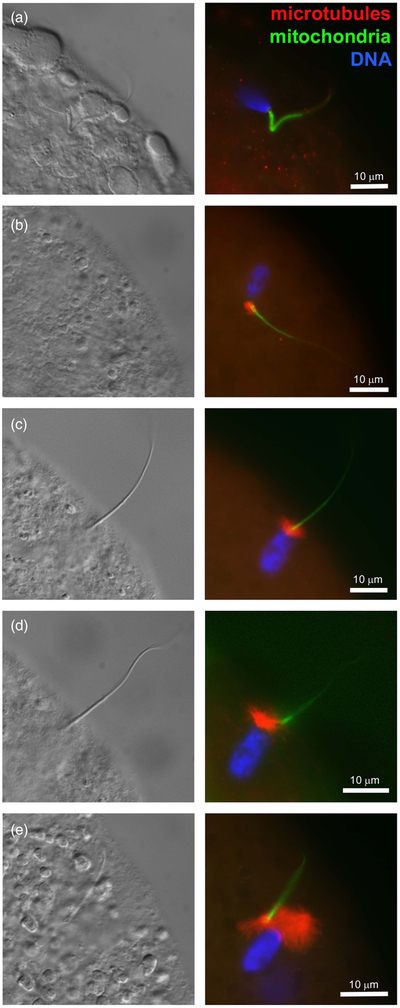
(Colour online) Progression of sperm incorporation, sperm nucleus decondensation and sperm aster formation during bovine fertilization in vitro. In all panels, sperm aster microtubules were labeled with monoclonal antitubulin antibody E7 (red), sperm mitochondria were pre-labeled with vital stain MitoTracker Green FM (green) before IVF and DNA in the sperm nucleus (blue) was counterstained with 2-(4-amidinophenyl)-1H-indole-6-carboxamidine. Left columns show parfocal bright-field images acquired with differential interference contrast optics (DIC). The samples were examined and photographed under a Nikon Eclipse 800 epifluorescence microscope (Nikon Instruments Inc., Melville, NY, USA) with Cool Snap camera (Roper Scientific, Tucson, AZ, USA) and MetaMorph software (Universal Imaging Corp., Downingtown, PA, USA). Images were edited and contrast balanced by Adobe Photoshop CS5 (Adobe Systems Inc., San Jose, CA, USA).
Pronuclear development
Before sperm–oolemma fusion, bull sperm head found within oocyte periviteline space is composed of a nucleus covered with a reduced nuclear envelope and PT all around, and IAM in the acrosomal region and plasma membrane over the equatorial segment and post-acrosomal region. With the exception of a small contingent of somatic and sperm-specific histones in the nucleus and PT (Aul and Oko, 2002; Tovich and Oko, 2003) sperm DNA is packaged with protamines, mainly protamine 1 (PRM1) (Balhorn, 2007). Proper sperm protamination correlates with bull fertility in AI service (Dogan et al., 2015).
The sperm nucleus is demembranated in a stepwise fashion at sperm–oolemma fusion and entry in the ooplasm, when the sperm plasma membrane comingles with oolemma, PT, first in the post-acrosomal and then in the subacrosomal region spreads across the ooplasm, and the sperm nuclear envelope, devoid of nuclear pore complexes and nuclear lamina, dissolves (Sutovsky et al., 1997; Sutovsky et al., 1998). Next, protamines, inserted during sperm nucleus compaction at spermiogenesis, are removed and replaced with oocyte-specific histones, which themselves will later be replaced with somatic cell type histones (Gao et al., 2004). At the time of protamine–histone exchange reversal, the nuclear envelope is reconstituted by membrane vesicle fusion and nuclear lamina formation on both maternal and paternal chromatin, and nuclear pores are reinserted, possibly from a premade stock of ooplasmic annulate lamellae (Sutovsky et al., 1998).
As the maternal and paternal pronuclei grow, maternally stored nucleolar proteins re-enter the nuclear compartment to form nucleolus precursor bodies that will be remodeled into active ribosomal RNA producing nucleoli at the time of major zygotic genome activation (four to eight cells in bovine) (Maddox-Hyttel et al., 2007; Ogushi et al., 2008). The zygote-driven remodeling of bovine sperm nucleus is facilitated by multiple ooplasmic factors, including the disulfide bond reducing peptide glutathione, histones, lamins, nucleoporins and nucleoplasmin (Sutovsky and Schatten, 1997; Burns et al., 2003). Such an extensive restructuring is aided by oocyte proteasomes that accumulate in both pronuclei from the onset of pronuclear development (Figure 3). In addition to pronuclear accumulation, proteasomes are abundant in mammalian zygote cytoplasm, as shown in the zygotes of a transgenic pig expressing proteasomes tagged with green fluorescent protein (Miles et al., 2013).
Figure 3.
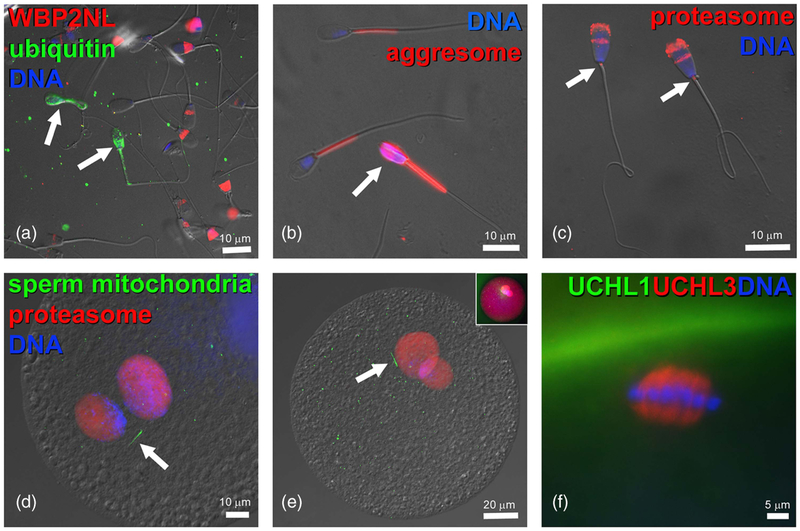
(Colour online) Components of ubiquitin–proteasome system in bovine gametes and embryos. (a) Bull spermatozoa (arrows) with abnormal phenotypes are ubiquitinated on their surface (green) and lack the WBP2NL/PAWP protein (red) present at varied levels in the post-acrosomal sheaths of surrounding morphologically normal spermatozoa. (b) Aggresome-binding ProteoStat probe (red) binds exclusively to mitochondrial sheaths of normal spermatozoa but detects aggresomes in the deformed sperm head of an abnormal spermatozoon (arrow). (c) Proteasomes (red) detected in bull sperm head acrosome and sperm tail connecting piece (arrows) by polyclonal antibody recognizing the 20S proteasomal core subunit PSMB10 (PW8150, Enzo Lifesciences, Ann Arbor, MI, USA). (d,e) Proteasomes (red) in the apposing (d), and apposed (e) maternal and paternal pronuclei of bovine zygotes, detected by a polyclonal antibody recognizing multiple 20S proteasomal core subunits (PW8155, Enzo Lifesciences). Inset in (e) shows the same zygote from which the differential interference contrast layer has been subtracted and red fluorescence brightened to reveal cytoplasmic proteasome labeling. Bovine oocytes were fertilized by spermatozoa pre-labeled with MitoTracker Green FM (green) to detect sperm tail mitochondrial sheaths (arrows). (f) Detection of deubiquitinases ubiquitin C-terminal hydrolase L1 (UCHL1) (green) and UCHL3 (red) in the bovine oocyte cortex and meiotic spindle, respectively. DNA in all panels was counterstained with 2-(4-amidinophenyl)-1H-indole-6-carboxamidine. The samples were examined and photographed under a Nikon Eclipse 800 epifluorescence microscope with Cool Snap camera and MetaMorph software. Images were edited and contrast balanced by Adobe Photoshop CS5.
Proteasomal inhibitors alter male pronuclear chromatin (Huo et al., 2004;Rawe et al., 2008) possibly interfering with protamine removal and/or oocyte–somatic histone swap. The ensemble of epigenetic histone marks, the zygotic histone code, which is reset at the onset of embryo development, is also influenced by UPS (Nevoral and Sutovsky, 2017), possibly through UPS regulation of sirtuin 1 (SIRT1), a histone deacetylase (Adamkova et al., 2017). The involvement of UPS extends beyond the zygote. Genes encoding UPS components such as proteasomal subunits, ubiquitin-conjugating enzymes and ubiquitin ligases likely regulate later events of bovine preimplantation development, including the major zygotic genome activation and blastocyst differentiation (Massicotte et al., 2006; Adjaye et al., 2007). It can be concluded that the UPS regulates multiple facets of early zygotic development in bovines and other mammals.
Mitochondrial inheritance and the fate of sperm tail accessory structures
At fertilization, bull spermatozoa carry into ooplasm several dozen mitochondria organized into a helical structure of the mitochondrial sheath in the sperm tail midpiece. Early studies demonstrated that bull sperm mitochondria are sought out and targeted for degradation by the fertilized oocyte, typically lasting only up to 4-cell stage (Sutovsky et al., 1996). Such targeted mitophagy, not observed in interspecies zygotes of domestic cattle oocytes and gaur (Bos gaurus) spermatozoa (Sutovsky et al., 1996) may be developmentally advantageous due to the high potential for sperm mitochondrial DNA (mtDNA) damage, mutation and deletion during spermatogenesis and fertilization (reviewed in Song et al., 2014). Failure of this mechanism in cattle, causing heteroplasmy by leakage of paternal mtDNA, could affect offspring health and production traits influenced by mitochondrial fitness, such as milk protein content and meat quality (Gibson et al., 1997).
Bovine post-fertilization mitophagy depends on the progression of the zygotic cell cycle, and it is not a coincidence that both processes are co-regulated by UPS. Consequently, sperm mitochondria become ubiquitinated inside the oocyte (Sutovsky et al., 1999) and targeted for degradation by proteasomal proteolysis and lysosomal/autophagic machinery (Sutovsky et al., 2000). Recent studies in domestic pig zygotes indicate that the central role in sperm mitophagy is played by the ubiquitin-binding autophagy receptor SQSTM1 and the proteasome-serving ubiquitinated protein dislocase valosin containing protein (VCP) (Song et al., 2016). The participation of VCP explains why proteasomal inhibitors block sperm mitophagy, but proteasomes seldom associate directly with the sperm mitochondrial sheath inside the zygote. Some mitochondrial membrane and matrix proteins, such as prohibitin (Thompson et al., 2003), are already tagged with ubiquitin in sperm mitochondria in bull spermatozoa, and the properties of the heavily disulfide bond cross-linked, ubiquitinated sperm mitochondrial sheath are similar to those of an aggresome, a ubiquitinated protein aggregate subject to autophagy in somatic cells (Thompson et al., 2003; Kennedy et al., 2014).
Attention has also been given to sperm tail accessory structures including the fibrous sheath covering the principal piece and the axonemal outer dense fibers running the whole length of the sperm tail except the connecting piece, where they are transformed into striated columns. While the fibrous sheath appears to dissolve very quickly within the first few hours of bull sperm incorporation, the outer dense fibers and microtubules may persist past one cell stage (Sutovsky et al., 1996). Given the minute size of bovine sperm tail, its resident proteins are not likely to be the source of nutrition for the embryo, as has been proposed in Drosophila, a species endowed with an enormous sperm tail. However, the densely amalgamated tail accessory structures could harbor paternal RNAs that affect embryo development, as will be discussed below.
Research on sperm ubiquitin has been extended beyond sperm mitochondrion ubiquitination and led to the discovery of sperm surface ubiquitination in defective bull spermatozoa (Sutovsky et al., 2001; Baska et al., 2008), and later on to identification of aggresomes outside bull sperm mitochondrial sheath (Kennedy et al., 2014). Sperm surface ubiquitination has been validated as a sperm quality biomarker in bulls and other species (Sutovsky et al., 2015) and correlated with sperm acrosome (Odhiambo et al., 2011) and DNA integrity (Sutovsky et al., 2002), expression of fertility-related sperm proteins (platelet activating factor receptor) (Sutovsky et al., 2007) and PAWP/WBP2NL, which also correlated with sperm aggresome content in AI bulls (Kennedy et al., 2014). Identification of sperm surface ligands unique to defective spermatozoa enabled the development of nanopurification for the improvement of bull AI semen (Odhiambo et al.,, 2014). At present, there is already enough evidence to conclude that the clonal maternal inheritance of bovine mitochondrial genes is regulated by UPS through proteasomal and autophagosomal protein/organelle degradation.
Regulatory molecules in bull spermatozoa that affect early embryo development
Mammalian spermatozoa carry their own transcriptome (Ostermeier et al., 2004), which is likely reflective of genome activity before germ cell meiosis and particularly early after meiosis, when round spermatids remain transcriptionally active until the process of histone–protamine exchange is initiated. Approximately 3500 species of mRNA alone are found in human spermatozoa (Ostermeier et al., 2002), among which the genes within UPS appear to be dysregulated in teratospermic infertile men (Platts et al., 2007). Recent sequencing effort revealed transcripts of over 13 000 genes in bull spermatozoa (Selvaraju et al., 2017). Beside mRNA, non-coding RNAs such as micro-RNA (miR), long Piwi-interacting RNAs and small nucleolar RNAs could be carried by bull spermatozoa and influence embryo development and inheritance (Hossain et al., 2012). The miR-196b, targeting homeobox genes, appears to be the most abundant miR in bull spermatozoa (Selvaraju et al., 2017). The miR content may vary between a high- and low-motility spermatozoa (Capra et al., 2017), as well as between bulls of varied fertility (Govindaraju et al., 2012; Fagerlind et al., 2015).
Protamine Prm1 is among the most abundant bull sperm mRNAs (Card et al., 2013), probably reflecting its abundance in spermatids, wherein its product is required for the proper packaging of sperm DNA. Sperm levels of several other transcripts, including ubiquitin-conjugating enzyme gene Ube2e3 correlate with bull AI fertility (Parthipan et al., 2017). This and other UPS-related sperm-borne mRNAs could be translated by the zygote, for example, to assist the postfertilization clean-up of maternally stored proteins.
Some studies suggest the lone presence of spermatozoa and seminal plasma can stimulate specific transcriptional response in the female reproductive system that may be favorable for the establishment of pregnancy. Seminal plasma may influence gene expression in uterine epithelia directly and indirectly regulate embryo development through uterine secretion of embryotrophic growth factors (Bromfield et al., 2014). Such findings warrant further inquiry into the transcriptome of bovine seminal plasma, from which some RNA containing protein complexes could be adsorbed on the bull sperm surface and carried past cervix during mating, or be introduced directly in uterus with extended semen during AI.
Mouse studies provide additional insight into potential roles of sperm RNAs in zygotic development and inheritance. Some of the sperm mRNA contributed to zygote could be translated to regulate early development and even to promote the RNA-mediated non-Mendelian inheritance, as suggested for the mouse sperm transmitted mutated Kit allele dominating the mRNA produced from wild type allele (Rassoulzadegan et al., 2006). Another example, miR-34c is present in spermatozoa but absent from oocytes and appears to be required for embryo cleavage (Liu et al., 2011). Pronuclear injection of miR-24 caused paramutation in the mouse zygote coinciding with heritable histone modification associated with promoter region of transcription factor Sox9 gene (Grandjean et al., 2009). Pertinent to the regulation of zygotic development by UPS, injection of eight different sperm derived miR in mouse zygotes caused degradation of maternal, oocyte stored mRNA that, among other effects on gene expression, affected the genes encoding aforementioned, UPS-regulated SIRT1 protein and ubiquitin ligase UBE3A, both involved in histone code modification (Rodgers et al., 2016). Similarly, sperm-borne transfer RNA fragments, acquired during epididymal passage seem to repress expression of specific genes in the murine embryo and may mediate epigenetic influences between father and offspring (Sharma et al., 2017). Altogether, new evidence indicates that RNAs with both the protein coding and regulatory functions are present in bull spermatozoa and contributed to the zygote at fertilization.
Effects of paternal genome on conception rate and offspring fertility
Gene polymorphisms and epigenetic marks transmitted to offspring by the father can directly influence offspring fertility and have an effect on the development of their own offspring. Efforts are under way to identify genes in bulls with extreme high v. low fertility that bear on sperm phenotype and sperm production traits (Taylor et al., 2018). There are already some examples of gene polymorphisms that affect bull sperm phenotype. Genes encoding for sperm head proteins, integrin β 5 (ITGB5) and collagen type I α 2 (COL1A1), have been associated with bull fertility (Feugang et al., 2009). Other polymorphisms associated with bull sperm phenotypes include genes encoding for sperm surface CD9, which may influence sperm motility (Kumar et al., 2015) and TEX11 (testis expressed 11) associated with scrotal circumference (de Camargo et al., 2015). Holstein bull fertility may be influenced by single-nucleotide polymorphisms in the miR target sites of gene encoding spermatid transitional protein 1 (TNP1) (Zhang et al., 2015) and by polymorphism-driven alternative splicing of the sperm flagella 2 (SPEF2) gene (Guo et al., 2013). The copy number in spermatogenesis associated multi-copy genes such as preferentially expressed antigen in melanoma may also result in varied bull fertility (Yue et al., 2013). More attention should be directed to epigenetic marks on sperm-borne genome and chromatin that are generally thought to be erased in the zygote and re-established in the early embryo. Both sperm histone H3 modifications (Kutchy et al., 2017) and sperm DNA methylation (Kropp et al., 2017) have been associated with bull fertility. Finally, genes that are transcribed from paternal allele during early development and may support developmental competence and pregnancy establishment, or guard against pregnancy loss will be examined for potentially detrimental polymorphisms (Cochran et al., 2013). Ongoing genomic studies will likely identify a number of gene mutations that may contribute to fertility or convey subfertility to bulls used in AI service.
Summary
Gamete interactions during bovine fertilization represent an intricate mesh of remodeling, recycling and signaling that results in transformation of gamete-specific structures into zygotic components. Thus, the sperm acrosome is remodeled during capacitation causing the exposure of zona-binding ligands on the OAM, and is partially lost to acrosomal exocytosis induced by the oocyte ZP. The sperm head PT dissolves in ooplasm to release oocyte-activating factors, and the denuded sperm nucleus is turned into a zygotic paternal pronucleus. The reduced sperm centrosome contributes the formation of zygotic centrosomes and initiates nucleation of microtubule sperm aster necessary for maternal and paternal pronucleus apposition. Various sperm-borne RNAs may regulate early embryo development and even impose epigenetic marks. Sperm contributed paternal genome affects developmental competence of embryos and ultimately imposes heritable traits on offspring.
While some sperm accessory structures are simply recycled after fertilization as they become obsolete, the fate of sperm mitochondria is of particular interest as these have to be eliminated to promote clonal, maternal inheritance of mitochondrial genes and avoid heteroplasmy. Similarly, the regulatory influence of sperm-borne RNAs on zygotic and embryonic development warrants further investigation. Altogether, the ability of some sperm contributed structures and molecules to develop into the functional zygotic components and properly induce embryonic development may vary between bulls, bearing on their reproductive performance, and on the fitness, health, fertility and production traits of their offspring.
Acknowledgments
The author wishes to thank Kathryn Craighead for editorial and clerical assistance. Work in the author’s laboratory pertinent to this review article is currently funded by Agriculture and Food Research Initiative Competitive Grant No. 2015-67015-23231 from the USDA National Institute of Food and Agriculture, Grant No. 5 R01 HD084353-02 from NIH National Institute of Child and Human Development, and by seed funding from the Food for The 21st Century Program of the University of Missouri.
Footnotes
Declaration of interest
There is no conflicts of interest to declare.
Ethics statement
This is a review article; consequently, there are no applicable ethics/compliance committee approvals to be listed.
Software and data repository resources
There are no deposited data or models associated with this article.
References
- Adamkova K, Yi YJ, Petr J, Zalmanova T, Hoskova K, Jelinkova P, Moravec J, Kralickova M, Sutovsky M, Sutovsky P and Nevoral J 2017. SIRT1-dependent modulation of methylation and acetylation of histone H3 on lysine 9 (H3K9) in the zygotic pronuclei improves porcine embryo development. Journal of Animal Science and Biotechnology 8, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adjaye J, Herwig R, Brink TC, Herrmann D, Greber B, Sudheer S, Groth D, Carnwath JW, Lehrach H and Niemann H 2007. Conserved molecular portraits of bovine and human blastocysts as a consequence of the transition from maternal to embryonic control of gene expression. Physiological Genomics 31, 315–327. [DOI] [PubMed] [Google Scholar]
- Aul RB and Oko RJ 2002. The major subacrosomal occupant of bull spermatozoa is a novel histone H2B variant associated with the forming acrosome during spermiogenesis. Developmental Biology 242, 376–387. [PubMed] [Google Scholar]
- Balhorn R 2007. The protamine family of sperm nuclear proteins. Genome Biology 8, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baska KM, Manandhar G, Feng D, Agca Y, Tengowski MW, Sutovsky M, Yi YJ and Sutovsky P 2008. Mechanism of extracellular ubiquitination in the mammalian epididymis. Journal of Cellular Physiology 215, 684–696. [DOI] [PubMed] [Google Scholar]
- Belmonte SA, Mayorga LS and Tomes CN 2016. The molecules of sperm exocytosis. Advances in Anatomy, Embryology, and Cell Biology 220, 71–92. [DOI] [PubMed] [Google Scholar]
- Benammar A, Ziyyat A, Lefevre B and Wolf JP 2016. Tetraspanins and mouse oocyte microvilli related to fertilizing ability. Reproductive Science 24, 1062–1069. [DOI] [PubMed] [Google Scholar]
- Bianchi E, Doe B, Goulding D and Wright GJ 2014. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508, 483–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom E and Birch-Andersen A 1980. Ultrastructure of the tail stump sperm defect in the bull. Acta Pathologica Et Microbiologica Scandinavica A 88, 397–405 [DOI] [PubMed] [Google Scholar]
- Bromfield JJ, Schjenken JE, Chin PY, Care AS, Jasper MJ and Robertson SA 2014. Maternal tract factors contribute to paternal seminal fluid impact on metabolic phenotype in offspring. Proceedings of the National Academy of Sciences of the United States of America 111, 2200–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH, Viveiros MM, Ren Y, Wang P, DeMayo FJ, Frail DE, Eppig JJ and Matzuk MM 2003. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science 300, 633–636. [DOI] [PubMed] [Google Scholar]
- Byrne K, Leahy T, McCulloch R, Colgrave ML and Holland MK 2012. Comprehensive mapping of the bull sperm surface proteome. Proteomics 12, 3559–3579. [DOI] [PubMed] [Google Scholar]
- Capra E, Turri F, Lazzari B, Cremonesi P, Gliozzi TM, Fojadelli I, Stella A and Pizzi F 2017. Small RNA sequencing of cryopreserved semen from single bull revealed altered miRNAs and piRNAs expression between high- and low-motile sperm populations. BMC Genomics 18, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card CJ, Anderson EJ, Zamberlan S, Krieger KE, Kaproth M and Sartini BL 2013. Cryopreserved bovine spermatozoal transcript profile as revealed by high-throughput ribonucleic acid sequencing. Biology of Reproduction 88, 49. [DOI] [PubMed] [Google Scholar]
- Cochran SD, Cole JB, Null DJ and Hansen PJ 2013. Single nucleotide polymorphisms in candidate genes associated with fertilizing ability of sperm and subsequent embryonic development in cattle. Biology of Reproduction 89, 69. [DOI] [PubMed] [Google Scholar]
- Cohen-Kaplan V, Livneh I, Avni N, Cohen-Rosenzweig C and Ciechanover A 2016. The ubiquitin-proteasome system and autophagy: coordinated and independent activities. International Journal of Biochemistry & Cell Biology 79, 403–418. [DOI] [PubMed] [Google Scholar]
- Cuasnicu PS, Da Ros VG, Weigel Munoz M and Cohen DJ 2016. Acrosome reaction as a preparation for gamete fusion. Advances in Anatomy, Embryology, and Cell Biology 220, 159–172. [DOI] [PubMed] [Google Scholar]
- de Camargo GM, Porto-Neto LR, Kelly MJ, Bunch RJ, McWilliam SM, Tonhati H, Lehnert SA, Fortes MR and Moore SS 2015. Non-synonymous mutations mapped to chromosome X associated with andrological and growth traits in beef cattle. BMC Genomics 16, 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan S, Vargovic P, Oliveira R, Belser LE, Kaya A, Moura A, Sutovsky P, Parrish J, Topper E and Memili E 2015. Sperm protamine-status correlates to the fertility of breeding bulls. Biology of Reproduction 92, 92. [DOI] [PubMed] [Google Scholar]
- Fagerlind M, Stalhammar H, Olsson B and Klinga-Levan K 2015. Expression of miRNAs in bull spermatozoa correlates with fertility rates. Reproduction in Domestic Animals 50, 587–594. [DOI] [PubMed] [Google Scholar]
- Feugang JM, Kaya A, Page GP, Chen L, Mehta T, Hirani K, Nazareth L, Topper E, Gibbs R and Memili E 2009. Two-stage genome-wide association study identifies integrin beta 5 as having potential role in bull fertility. BMC Genomics 10, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Sakase M, Fukushima M and Harayama H 2016. Changes of IZUMO1 in bull spermatozoa during the maturation, acrosome reaction, and cryopreservation. Theriogenology 86, 2179–2188 e2173. [DOI] [PubMed] [Google Scholar]
- Gadella BM 2010. Interaction of sperm with the zona pellucida during fertilization. Society for Reproduction and Fertility 67, 267–287. [DOI] [PubMed] [Google Scholar]
- Gao S, Chung YG, Parseghian MH, King GJ, Adashi EY and Latham KE 2004. Rapid H1 linker histone transitions following fertilization or somatic cell nuclear transfer: evidence for a uniform developmental program in mice. Developmental Biology 266, 62–75. [DOI] [PubMed] [Google Scholar]
- Gerton GL 2002. Function of the sperm acrosome In Fertilization (ed. Hardy DM), pp. 265–302. Academic Press, San Diego, CA, USA. [Google Scholar]
- Gibson JP, Freeman AE and Boettcher PJ 1997. Cytoplasmic and mitochondrial inheritance of economic traits in cattle. Livestock Production Science 47, 115–124. [Google Scholar]
- Glotzer M, Murray AW and Kirschner MW 1991. Cyclin is degraded by the ubiquitin pathway. Nature 349, 132–138. [DOI] [PubMed] [Google Scholar]
- Govindaraju A, Uzun A, Robertson L, Atli MO, Kaya A, Topper E, Crate EA, Padbury J, Perkins A and Memili E 2012. Dynamics of microRNAs in bull spermatozoa. Reproductive Biology and Endocrinology 10, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean V, Gounon P, Wagner N, Martin L, Wagner KD, Bernex F, Cuzin F and Rassoulzadegan M 2009. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development 136, 3647–3655. [DOI] [PubMed] [Google Scholar]
- Guo F, Yang B, Ju ZH, Wang XG, Qi C, Zhang Y, Wang CF, Liu HD, Feng MY, Chen Y, Xu YX, Zhong JF and Huang JM 2013. Alternative splicing, promoter methylation, and functional SNPs of sperm flagella 2 gene in testis and mature spermatozoa of Holstein bulls. Reproduction 147, 241–252. [DOI] [PubMed] [Google Scholar]
- Hachem A, Godwin J, Ruas M, Lee HC, Ferrer Buitrago M, Ardestani G, Bassett A, Fox S, Navarrete F, de Sutter P, Heindryckx B, Fissore R and Parrington J 2017. PLCzeta is the physiological trigger of the Ca2 + oscillations that induce embryogenesis in mammals but conception can occur in its absence. Development 144, 2914–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Tagiri M, Hirabayashi M and Hochi S 2015. Multiple aster formation is frequently observed in bovine oocytes retrieved from 1-day stored ovaries. Zygote 24, 115–120. [DOI] [PubMed] [Google Scholar]
- Hillman P, Ickowicz D, Vizel R and Breitbart H 2013. Dissociation between AKAP3 and PKARII promotes AKAP3 degradation in sperm capacitation. PLoS One 8, e68873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochi S 2016. Microtubule assembly crucial to bovine embryonic development in assisted reproductive technologies. Animal Science Journal 87, 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MM, Sohel MM, Schellander K and Tesfaye D 2012. Characterization and importance of microRNAs in mammalian gonadal functions. Cell and Tissue Research 349, 679–690. [DOI] [PubMed] [Google Scholar]
- Huo LJ, Fan HY, Liang CG, Yu LZ, Zhong ZS, Chen DY and Sun QY 2004. Regulation of ubiquitin-proteasome pathway on pig oocyte meiotic maturation and fertilization. Biology of Reproduction 71, 853–862. [DOI] [PubMed] [Google Scholar]
- Inoue N, Ikawa M, Isotani A and Okabe M 2005. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 434, 234–238. [DOI] [PubMed] [Google Scholar]
- Ito C, Yamatoya K, Yoshida K, Maekawa M, Miyado K and Toshimori K 2010. Tetraspanin family protein CD9 in the mouse sperm: unique localization, appearance, behavior and fate during fertilization. Cell and Tissue Research 340, 583–594. [DOI] [PubMed] [Google Scholar]
- Karabinova P, Kubelka M and Susor A 2011. Proteasomal degradation of ubiquitinated proteins in oocyte meiosis and fertilization in mammals. Cell and Tissue Research 346, 1–9. [DOI] [PubMed] [Google Scholar]
- Kasimanickam V, Kasimanickam R, Arangasamy A, Saberivand A, Stevenson JS and Kastelic JP 2012. Association between mRNA abundance of functional sperm function proteins and fertility of Holstein bulls. Theriogenology 78, 2007–2019e2002. [DOI] [PubMed] [Google Scholar]
- Kennedy CE, Krieger KB, Sutovsky M, Xu W, Vargovic P, Didion BA, Ellersieck MR, Hennessy ME, Verstegen J, Oko R and Sutovsky P 2014. Protein expression pattern of PAWP in bull spermatozoa is associated with sperm quality and fertility following artificial insemination. Molecular Reproduction and Development 81, 436–449. [DOI] [PubMed] [Google Scholar]
- Kerns K, Morales P and Sutovsky P 2016. Regulation of sperm capacitation by the 26S proteasome: an emerging new paradigm in spermatology. Biology of Reproduction 94, 117. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Yanagimachi R, Kuretake S, Bortkiewicz H, Perry AC and Yanagimachi H 1998. Analysis of mouse oocyte activation suggests the involvement of sperm perinuclear material. Biology of Reproduction 58, 1407–1415. [DOI] [PubMed] [Google Scholar]
- Kongmanas K, Kruevaisayawan H, Saewu A, Sugeng C, Fernandes J, Souda P, Angel JB, Faull KF, Aitken RJ, Whitelegge J, Hardy D, Berger T, Baker MA and Tanphaichitr N 2014. Proteomic characterization of pig sperm anterior head plasma membrane reveals roles of acrosomal proteins in ZP3 binding. Journal of Cellular Physiology 230, 449–463. [DOI] [PubMed] [Google Scholar]
- Kropp J, Carrillo JA, Namous H, Daniels A, Salih SM, Song J and Khatib H 2017. Male fertility status is associated with DNA methylation signatures in sperm and transcriptomic profiles of bovine preimplantation embryos. BMC Genomics 18, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Singh U, Deb R, Tyagi S, Mandal DK, Kumar M, Sengar G, Sharma S and Singh R 2015. A SNP (g.358A > T) at intronic region of CD9 molecule of crossbred bulls may associate with spermatozoal motility. Meta Gene 5, 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutchy NA, Menezes ESB, Chiappetta A, Tan W, Wills RW, Kaya A, Topper E, Moura AA, Perkins AD and Memili E 2017. Acetylation and methylation of sperm histone 3 lysine 27 (H3K27ac and H3K27me3) are associated with bull fertility. Andrologia e12915, 10.1111/and.12915. [DOI] [PubMed] [Google Scholar]
- La Spina FA, Puga Molina LC, Romarowski A, Vitale AM, Falzone TL, Krapf D, Hirohashi N and Buffone MG 2016. Mouse sperm begin to undergo acrosomal exocytosis in the upper isthmus of the oviduct. Developmental Biology 411, 172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WM, Pang RT, Chiu PC, Wong BP, Lao K, Lee KF and Yeung WS 2011. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proceedings of the National Academy of Sciences of the United States of America 109, 490–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox-Hyttel P, Svarcova O and Laurincik J 2007. Ribosomal RNA and nucleolar proteins from the oocyte are to some degree used for embryonic nucleolar formation in cattle and pig. Theriogenology 68 (suppl. 1), S63–S70. [DOI] [PubMed] [Google Scholar]
- Manandhar G, Schatten H and Sutovsky P 2005. Centrosome reduction during gametogenesis and its significance. Biology of Reproduction 72, 2–13. [DOI] [PubMed] [Google Scholar]
- Manandhar G and Toshimori K 2001. Exposure of sperm head equatorin after acrosome reaction and its fate after fertilization in mice. Biology of Reproduction 65, 1425–1436. [DOI] [PubMed] [Google Scholar]
- Massicotte L, Coenen K, Mourot M and Sirard MA 2006. Maternal housekeeping proteins translated during bovine oocyte maturation and early embryo development. Proteomics 6, 3811–3820. [DOI] [PubMed] [Google Scholar]
- Miles EL, O’Gorman C, Zhao J, Samuel M, Walters E, Yi YJ, Sutovsky M, Prather RS, Wells KD and Sutovsky P 2013. Transgenic pig carrying green fluorescent proteasomes. Proceedings of the National Academy of Sciences of the United States of America 110, 6334–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mtango NR, Latham KE and Sutovsky P 2014. Deubiquitinating enzymes in oocyte maturation, fertilization and preimplantation embryo development. Advances in Experimental Medicine and Biology 759, 89–110. [DOI] [PubMed] [Google Scholar]
- Mtango NR, Sutovsky M, Susor A, Zhong Z, Latham KE and Sutovsky P 2011b. Essential role of maternal UCHL1 and UCHL3 in fertilization and preimplantation embryo development. Journal of Cellular Physiology 227, 1592–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mtango NR, Sutovsky M, Vandevoort CA, Latham KE and Sutovsky P 2011a. Essential role of ubiquitin C-terminal hydrolases UCHL1 and UCHL3 in mammalian oocyte maturation. Journal of Cellular Physiology 227, 2022–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Terada Y, Horiuchi T, Emuta C, Murakami T, Yaegashi N and Okamura K 2001. Human sperm aster formation and pronuclear decondensation in bovine eggs following intracytoplasmic sperm injection using a Piezo-driven pipette: a novel assay for human sperm centrosomal function. Biology of Reproduction 65, 1359–1363. [DOI] [PubMed] [Google Scholar]
- Navara CS, First NL and Schatten G 1996. Phenotypic variations among paternal centrosomes expressed within the zygote as disparate microtubule lengths and sperm aster organization: correlations between centrosome activity and developmental success. Proceedings of the National Academy of Sciences of the United States of America 93, 5384–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevoral J and Sutovsky P 2017. Epigenome modification and ubiquitin-dependent proteolysis during pronuclear development of the mammalian zygote: animal models to study pronuclear development In Animal models and human reproduction (ed. Schatten H and Constantinescu GM), pp. 435–466. Wiley-Blackwell, John Wiley & Sons, Inc., Hoboken, NY, USA. [Google Scholar]
- Odhiambo JF, DeJarnette JM, Geary TW, Kennedy CE, Suarez SS, Sutovsky M and Sutovsky P 2014. Increased conception rates in beef cattle inseminated with nanopurified bull semen. Biology of Reproduction 91, 97. [DOI] [PubMed] [Google Scholar]
- Odhiambo JF, Sutovsky M, DeJarnette JM, Marshall C and Sutovsky P 2011. Adaptation of ubiquitin-PNA based sperm quality assay for semen evaluation by a conventional flow cytometer and a dedicated platform for flow cytometric semen analysis. Theriogenology 76, 1168–1176. [DOI] [PubMed] [Google Scholar]
- Ogushi S, Palmieri C, Fulka H, Saitou M, Miyano T and Fulka J Jr. 2008. The maternal nucleolus is essential for early embryonic development in mammals. Science 319, 613–616. [DOI] [PubMed] [Google Scholar]
- Oko R, Aarabi M, Mao J, Balakier H and Sutovsky P 2017. Sperm specific WW-domain binding proteins In The sperm cell: production, maturation, fertilization, regeneration (ed. DeJonge C and Barratt C), pp. 157–176. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Oko R and Sutovsky P 2009. Biogenesis of sperm perinuclear theca and its role in sperm functional competence and fertilization. Journal of Reproductive Immunology 83, 2–7. [DOI] [PubMed] [Google Scholar]
- Oren-Benaroya R, Orvieto R, Gakamsky A, Pinchasov M and Eisenbach M 2008. The sperm chemoattractant secreted from human cumulus cells is progesterone. Human Reproduction 23, 2339–2345. [DOI] [PubMed] [Google Scholar]
- Ostermeier GC, Dix DJ, Miller D, Khatri P and Krawetz SA 2002. Spermatozoal RNA profiles of normal fertile men. Lancet 360, 772–777. [DOI] [PubMed] [Google Scholar]
- Ostermeier GC, Miller D, Huntriss JD, Diamond MP and Krawetz SA 2004. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature 429, 154. [DOI] [PubMed] [Google Scholar]
- Pan Q, Ju Z, Huang J, Zhang Y, Qi C, Gao Q, Zhou L, Li Q, Wang L, Zhong J, Liu M and Wang C 2013. PLCz functional haplotypes modulating promoter transcriptional activity are associated with semen quality traits in Chinese Holstein bulls. PLoS One 8, e58795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JJ 2013. Bovine in vitro fertilization: in vitro oocyte maturation and sperm capacitation with heparin. Theriogenology 81, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthipan S, Selvaraju S, Somashekar L, Arangasamy A, Sivaram M and Ravindra JP 2017. Spermatozoal transcripts expression levels are predictive of semen quality and conception rate in bulls (Bos taurus). Theriogenology 98, 41–49. [DOI] [PubMed] [Google Scholar]
- Perry AC, Wakayama T, Cooke IM and Yanagimachi R 2000. Mammalian oocyte activation by the synergistic action of discrete sperm head components: induction of calcium transients and involvement of proteolysis. Developmental Biology 217, 386–393. [DOI] [PubMed] [Google Scholar]
- Pizarro E, Pasten C, Kong M and Morales P 2004. Proteasomal activity in mammalian spermatozoa. Molecular Reproduction and Development 69, 87–93. [DOI] [PubMed] [Google Scholar]
- Platts AE, Dix DJ, Chemes HE, Thompson KE, Goodrich R, Rockett JC, Rawe VY, Quintana S, Diamond MP, Strader LF and Krawetz SA 2007. Success and failure in human spermatogenesis as revealed by teratozoospermic RNAs. Human Molecular Genetics 16, 763–773. [DOI] [PubMed] [Google Scholar]
- Primakoff P and Myles DG 2007. Cell-cell membrane fusion during mammalian fertilization. FEBS Letters 581, 2174–2180. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I and Cuzin F 2006. RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature 441, 469–474. [DOI] [PubMed] [Google Scholar]
- Rawe VY, Diaz ES, Abdelmassih R, Wojcik C, Morales P, Sutovsky P and Chemes HE 2008. The role of sperm proteasomes during sperm aster formation and early zygote development: implications for fertilization failure in humans. Human Reproduction 23, 573–580. [DOI] [PubMed] [Google Scholar]
- Redgrove KA, Anderson AL, Dun MD, McLaughlin EA, O’Bryan MK, Aitken RJ and Nixon B 2011. Involvement of multimeric protein complexes in mediating the capacitation-dependent binding of human spermatozoa to homologous zonae pellucidae. Developmental Biology 356, 460–474. [DOI] [PubMed] [Google Scholar]
- Rodgers AB, Morgan CP, Leu NA and Bale TL 2016. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proceedings of the National Academy of Sciences of the United States of America 112, 13699–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romarowski A, Luque GM, La Spina FA, Krapf D and Buffone MG 2016. Role of actin cytoskeleton during mammalian sperm acrosomal exocytosis. Advances in Anatomy, Embryology, and Cell Biology 220, 129–144. [DOI] [PubMed] [Google Scholar]
- Sakatani M, Yamanaka K, Balboula AZ, Takenouchi N and Takahashi M 2015. Heat stress during in vitro fertilization decreases fertilization success by disrupting anti-polyspermy systems of the oocytes. Molecular Reproduction and Development 82, 36–47. [DOI] [PubMed] [Google Scholar]
- Saldivar-Hernandez A, Gonzalez-Gonzalez ME, Sanchez-Tusie A, Maldonado-Rosas I, Lopez P, Trevino CL, Larrea F and Chirinos M 2015. Human sperm degradation of zona pellucida proteins contributes to fertilization. Reproductive Biology and Endocrinology 13, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Deppe M, Schulz M, Bravo P, Villegas J, Morales P and Risopatron J 2011. Participation of the sperm proteasome during in vitro fertilisation and the acrosome reaction in cattle. Andrologia 43, 114–120. [DOI] [PubMed] [Google Scholar]
- Sasanami T, Sugiura K, Tokumoto T, Yoshizaki N, Dohra H, Nishio S, Mizushima S, Hiyama G and Matsuda T 2012. Sperm proteasome degrades egg envelope glycoprotein ZP1 during fertilization of Japanese quail (Coturnix japonica). Reproduction 144, 423–431. [DOI] [PubMed] [Google Scholar]
- Sathananthan AH, Tatham B, Dharmawardena V, Grills B, Lewis I and Trounson A 1997. inheritance of sperm centrioles and centrosomes in bovine embryos. Archives of Andrology 38, 37–48. [DOI] [PubMed] [Google Scholar]
- Satouh Y, Nozawa K and Ikawa M 2015. Sperm postacrosomal WW domain-binding protein is not required for mouse egg activation. Biology of Reproduction 93, 1–7. [DOI] [PubMed] [Google Scholar]
- Satouh Y, Inoue N, Ikawa M and Okabe M 2012. Visualization of the moment of mouse sperm-egg fusion and dynamic localization of IZUMO1. Journal of Cell Science 125, 4985–4990. [DOI] [PubMed] [Google Scholar]
- Saunders CM, Larman MG, Parrington J, Cox LJ, Royse J, Blayney LM, Swann K and Lai FA 2002. PLC zeta: a sperm-specific trigger of Ca(2+) oscillations in eggs and embryo development. Development 129, 3533–3544. [DOI] [PubMed] [Google Scholar]
- Selvaraju S, Parthipan S, Somashekar L, Kolte AP, Krishnan Binsila B, Arangasamy A and Ravindra JP 2017. Occurrence and functional significance of the transcriptome in bovine (Bos taurus) spermatozoa. Scientific Reports 7, 42392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D and Kinsey WH 2012. PYK2: a calcium-sensitive protein tyrosine kinase activated in response to fertilization of the zebrafish oocyte. Developmental Biology 373, 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, Song L, Carone BR, Ricci EP, Li XZ, Fauquier L, Moore MJ, Sullivan R, Mello CC, Garber M and Rando OJ 2017. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin MR and Kim NH 2003. Maternal gamma (gamma)-tubulin is involved in microtubule reorganization during bovine fertilization and parthenogenesis. Molecular Reproduction and Development 64, 438–445. [DOI] [PubMed] [Google Scholar]
- Simerly C, Zoran SS, Payne C, Dominko T, Sutovsky P, Navara CS, Salisbury JL and Schatten G 1999. Biparental inheritance of gamma-tubulin during human fertilization: molecular reconstitution of functional zygotic centrosomes in inseminated human oocytes and in cell-free extracts nucleated by human sperm. Molecular Biology of the Cell 10, 2955–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WH, Ballard JW, Yi YJ and Sutovsky P 2014. Regulation of mitochondrial genome inheritance by autophagy and ubiquitin-proteasome system: implications for health, fitness, and fertility. Biomed Research International 2014, 981867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WH, Yi YJ, Sutovsky M, Meyers S and Sutovsky P 2016. Autophagy and ubiquitin-proteasome system contribute to sperm mitophagy after mammalian fertilization. Proceedings of the National Academy of Sciences of the United States of America 113, E5261–E5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallanzani L 1780. Dissertazioni Di Fisica Animale E Vegetabile, volume II Modena. [Google Scholar]
- Suarez SS 2015. Mammalian sperm interactions with the female reproductive tract. Cell and Tissue Research 363, 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susor A, Liskova L, Toralova T, Pavlok A, Pivonkova K, Karabinova P, Lopatarova M, Sutovsky P and Kubelka M 2010. Role of ubiquitin C-terminal hydrolase-L1 in antipolyspermy defense of mammalian oocytes. Biology of Reproduction 82, 1151–1161. [DOI] [PubMed] [Google Scholar]
- Sutovsky P 2003. Ubiquitin-dependent proteolysis in mammalian spermatogenesis, fertilization, and sperm quality control: killing three birds with one stone. Microscopy Research and Technique 61, 88–102. [DOI] [PubMed] [Google Scholar]
- Sutovsky P 2009. Sperm-egg adhesion and fusion in mammals. Expert Reviews in Molecular Medicine 11, e11. [DOI] [PubMed] [Google Scholar]
- Sutovsky P 2010. Sperm capacitation, the acrosome reaction, and fertilization In Reproductive endocrinology and infertility: integrating modern clinical and laboratory practice (ed. Douglas Carrell MP), pp. 389–421. Springer Science + Business Media, LLC, New York, NY, USA. [Google Scholar]
- Sutovsky P 2011. Sperm proteasome and fertilization. Reproduction 142, 1–14. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Aarabi M, Miranda-Vizuete A and Oko R 2015. Negative biomarker based male fertility evaluation: sperm phenotypes associated with molecular-level anomalies. Asian Journal of Andrology 17, 554–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutovsky P, Navara CS and Schatten G 1996. Fate of the sperm mitochondria, and the incorporation, conversion, and disassembly of the sperm tail structures during bovine fertilization. Biology of Reproduction 55, 1195–1205. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Neuber E and Schatten G 2002. Ubiquitin-dependent sperm quality control mechanism recognizes spermatozoa with DNA defects as revealed by dual ubiquitin-TUNEL assay. Molecular Reproduction and Development 61, 406–413. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Oko R, Hewitson L and Schatten G 1997. The removal of the sperm perinuclear theca and its association with the bovine oocyte surface during fertilization. Developmental Biology 188, 75–84. [DOI] [PubMed] [Google Scholar]
- Sutovsky P and Schatten G 1997. Depletion of glutathione during bovine oocyte maturation reversibly blocks the decondensation of the male pronucleus and pronuclear apposition during fertilization. Biology of Reproduction 56, 1503–1512. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Simerly C, Hewitson L and Schatten G 1998. Assembly of nuclear pore complexes and annulate lamellae promotes normal pronuclear development in fertilized mammalian oocytes. Journal of Cell Science 111 (Pt 19), 2841–2854. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Manandhar G, Wu A and Oko R 2003. Interactions of sperm perinuclear theca with the oocyte: implications for oocyte activation, antipolyspermy defense, and assisted reproduction. Microscopy Research and Technique 61, 362–378. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Moreno R, Ramalho-Santos J, Dominko T, Thompson WE and Schatten G 2001. A putative, ubiquitin-dependent mechanism for the recognition and elimination of defective spermatozoa in the mammalian epididymis. Journal of Cell Science 114, 1665–1675. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C and Schatten G 1999. Ubiquitin tag for sperm mitochondria. Nature 402, 371–372. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C and Schatten G 2000. Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biology of Reproduction 63, 582–590. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Plummer W, Baska K, Peterman K, Diehl JR and Sutovsky M 2007. Relative levels of semen platelet activating factor-receptor (PAFr) and ubiquitin in yearling bulls with high content of semen white blood cells: implications for breeding soundness evaluation. Journal of Andrology 28, 92–108. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Manandhar G, McCauley TC, Caamano JN, Sutovsky M, Thompson WE and Day BN 2004. Proteasomal interference prevents zona pellucida penetration and fertilization in mammals. Biology of Reproduction 71, 1625–1637. [DOI] [PubMed] [Google Scholar]
- Swann K 1990. A cytosolic sperm factor stimulates repetitive calcium increases and mimics fertilization in hamster eggs. Development 110, 1295–1302. [DOI] [PubMed] [Google Scholar]
- Taylor J, Schnabel RD and Sutovsky P 2018. Genomics of bull fertility. Animal 12(S1), (forthcoming). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WE, Ramalho-Santos J and Sutovsky P 2003. Ubiquitination of prohibitin in mammalian sperm mitochondria: possible roles in the regulation of mitochondrial inheritance and sperm quality control. Biology of Reproduction 69, 254–260. [DOI] [PubMed] [Google Scholar]
- Tovich PR and Oko RJ 2003. Somatic histones are components of the perinuclear theca in bovine spermatozoa. Journal of Biological Chemistry 278, 32431–32438. [DOI] [PubMed] [Google Scholar]
- Tung JJ, Hansen DV, Ban KH, Loktev AV, Summers MK, Adler JR 3rd and Jackson PK 2005. A role for the anaphase-promoting complex inhibitor Emi2/XErp1, a homolog of early mitotic inhibitor 1, in cytostatic factor arrest of Xenopus eggs. Proceedings of the National Academy of Sciences of the United States of America 102, 4318–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright GJ and Bianchi E 2015. The challenges involved in elucidating the molecular basis of sperm-egg recognition in mammals and approaches to overcome them. Cell and Tissue Research 363, 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AT, Sutovsky P, Manandhar G, Xu W, Katayama M, Day BN, Park KW, Yi YJ, Xi YW, Prather RS and Oko R 2007b. PAWP, a sperm-specific WW domain-binding protein, promotes meiotic resumption and pronuclear development during fertilization. Journal of Biological Chemistry 282, 12164–12175. [DOI] [PubMed] [Google Scholar]
- Wu AT, Sutovsky P, Xu W, van der Spoel AC, Platt FM and Oko R 2007a. The postacrosomal assembly of sperm head protein, PAWP, is independent of acrosome formation and dependent on microtubular manchette transport. Developmental Biology 312, 471–483. [DOI] [PubMed] [Google Scholar]
- Yi YJ, Manandhar G, Sutovsky M, Li R, Jonakova V, Oko R, Park CS, Prather RS and Sutovsky P 2007. Ubiquitin C-terminal hydrolase-activity is involved in sperm acrosomal function and anti-polyspermy defense during porcine fertilization. Biology of Reproduction 77, 780–793. [DOI] [PubMed] [Google Scholar]
- Yonezawa N 2014. Posttranslational modifications of zona pellucida proteins. Advances in Experimental Medicine and Biology 759, 111–140. [DOI] [PubMed] [Google Scholar]
- Yue XP, Chang TC, DeJarnette JM, Marshall CE, Lei CZ and Liu WS 2013. Copy number variation of PRAMEY across breeds and its association with male fertility in Holstein sires. Journal of Dairy Science 96, 8024–8034. [DOI] [PubMed] [Google Scholar]
- Zhang S, Zhang Y, Yang C, Zhang W, Ju Z, Wang X, Jiang Q, Sun Y, Huang J, Zhong J and Wang C 2015. TNP1 functional SNPs in bta-miR-532 and bta-miR-204 target sites are associated with semen quality traits in Chinese Holstein bulls. Biology of Reproduction 92, 139. [DOI] [PubMed] [Google Scholar]
- Zhao XM, Wang N, Hao HS, Li CY, Zhao YH, Yan CL, Wang HY, Du WH, Wang D, Liu Y, Pang YW and Zhu HB 2017. Melatonin improves the fertilization capacity and developmental ability of bovine oocytes by regulating cytoplasmic maturation events. Journal of Pineal Research, doi: 10.1111/jpi.12445, Published online 11 October 2017. [DOI] [PubMed] [Google Scholar]
- Zimmerman SW, Manandhar G, Yi YJ, Gupta SK, Sutovsky M, Odhiambo JF, Powell MD, Miller DJ and Sutovsky P 2011. Sperm proteasomes degrade sperm receptor on the egg zona pellucida during mammalian fertilization. PLoS One 6, e17256. [DOI] [PMC free article] [PubMed] [Google Scholar]