How self-DNA avoids unchecked activation of innate DNA sensors is poorly understood. Gratia et al. show that BLM protein, mutated in Bloom syndrome (BS), restrains interferon-stimulated gene induction and micronuclei recognition by cGAS. Consistently, BS patients exhibit an interferon signature.
Abstract
Cellular innate immune sensors of DNA are essential for host defense against invading pathogens. However, the presence of self-DNA inside cells poses a risk of triggering unchecked immune responses. The mechanisms limiting induction of inflammation by self-DNA are poorly understood. BLM RecQ–like helicase is essential for genome integrity and is deficient in Bloom syndrome (BS), a rare genetic disease characterized by genome instability, accumulation of micronuclei, susceptibility to cancer, and immunodeficiency. Here, we show that BLM-deficient fibroblasts show constitutive up-regulation of inflammatory interferon-stimulated gene (ISG) expression, which is mediated by the cGAS–STING–IRF3 cytosolic DNA–sensing pathway. Increased DNA damage or down-regulation of the cytoplasmic exonuclease TREX1 enhances ISG expression in BLM-deficient fibroblasts. cGAS-containing cytoplasmic micronuclei are increased in BS cells. Finally, BS patients demonstrate elevated ISG expression in peripheral blood. These results reveal that BLM limits ISG induction, thus connecting DNA damage to cellular innate immune response, which may contribute to human pathogenesis.
Introduction
The innate immune system plays a critical role in maintaining the health of the host. This is mediated, in part, by innate immune sensors of DNA that induce inflammatory signals in response to non–self-DNA from pathogens or damaged self-DNA (Härtlova et al., 2015). These responses are under tight control, and imbalances in these processes can cause a wide range of diseases, including immunodeficiency, autoinflammation, and autoimmunity.
Cyclic GMP-AMP synthase (cGAS) is an innate immune sensor of DNA that recognizes cytosolic DNA, resulting in the activation of the signaling adaptor stimulator of interferon genes (STING), the transcription factor interferon regulatory factor 3 (IRF3) and the expression of type I/III IFN and IFN-stimulated genes (ISGs; Chen et al., 2016). cGAS recognizes cytosolic DNA derived from invading microbes (Gao et al., 2013; Lahaye et al., 2013; Sun et al., 2013) and also self-DNA from engulfed tumor cells (Woo et al., 2014), damaged mitochondria (Rongvaux et al., 2014; White et al., 2014; West et al., 2015), and nuclear DNA damage (Dou et al., 2017; Glück et al., 2017; Harding et al., 2017; Mackenzie et al., 2017; Yang et al., 2017). After damage, nuclear DNA can accumulate in the cytosol in the form of micronuclei and chromatin fragments. Activation of cGAS by damaged nuclear DNA contributes to auto-tumor immunity, cellular senescence, and autoinflammatory disorders (Li and Chen, 2018). While multiple studies have demonstrated that induction of inflammatory cytokines upon exogenous DNA damage is mediated through the cGAS–STING pathway, the factors that normally keep endogenous DNA damage–mediated inflammation in check remain poorly understood.
Monogenic human diseases that collectively belong to a group referred to as type I interferonopathies have led to the identification of several factors that normally limit pathogenic inflammation (Crow and Manel, 2015). These interferonopathies are characterized by a stable ISG signature in blood samples and clinical manifestations of inflammation, with certain features restricted to mutant genotypes (Meyts and Casanova, 2016). These observations raise the possibility that other monogenic diseases may involve aspects of pathology that are, albeit currently unrecognized, related to enhanced type I IFN signaling, thereby potentially revealing the normal function of such genes in keeping inflammation in check.
Several studies have documented the link between deficiencies in genome integrity and induction of an IFN signature. Patients with monogenic mutations in Artemis show an IFN signature in blood, and this was attributed to cell extrinsic effects through NETosis (Gul et al., 2018). Fibroblasts from Werner syndrome, mutated in the RecQ helicase WRN, demonstrate an increased transcriptional level of IFNB (Yu et al., 2015). Rats with a missense mutation in Atm show an IFN signature in the brain (Quek et al., 2017), and mice knockout for Atm show a Sting-dependent IFN response (Härtlova et al., 2015). DNase II deficiency can cause an interferonopathy in human patients (Rodero et al., 2017b) and Dnase2-deficient murine cells accumulate extranuclear DNA that activates a Sting-dependent inflammatory response (Lan et al., 2014). Whether monogenic mutations in genes required for genome integrity can increase micronuclei frequency resulting in an inflammatory response is not known.
Bloom syndrome (BS) is a rare human autosomal recessive disorder characterized by marked genetic instability associated with an increased predisposition to all types of cancers affecting the general population (reviewed in Amor-Guéret, 2006). BS patients also present a pre- and postnatal growth retardation with normal proportioning, frequently associated with sun-sensitive facial erythema and immunodeficiency manifest by recurrent respiratory tract and gastrointestinal infections (Cunniff et al., 2017). BS is caused by mutations in both copies of the BLM gene, encoding the BLM RecQ–like helicase (Ellis et al., 1995). BLM protein displays ATP and Mg2+-dependent 3′-5′-DNA helicase activity and plays an essential role in maintaining genome integrity at the crossroads of the response pathways induced by DNA damage and stalled replication forks (Karow et al., 2000; Wu and Hickson, 2003; Amor-Guéret, 2006; Ralf et al., 2006; Bugreev et al., 2007; Chan et al., 2007; Rao et al., 2007). The BS cellular phenotype includes chromosome breaks, a high rate of sister chromatid exchange (SCE), a slowing of the replication fork speed associated with endogenous activation of the ATM–Chk2–γH2AX pathway, and an increase in the frequency of blocked replication forks and anaphase bridges (Amor-Guéret, 2006; Rao et al., 2007; Chan and Hickson, 2011). BS cells also accumulate micronuclei (Yankiwski et al., 2000).
Here we identify an inflammatory ISG signature in BLM-deficient cells and determine how BLM connects DNA damage to innate immune-sensing pathways.
Results
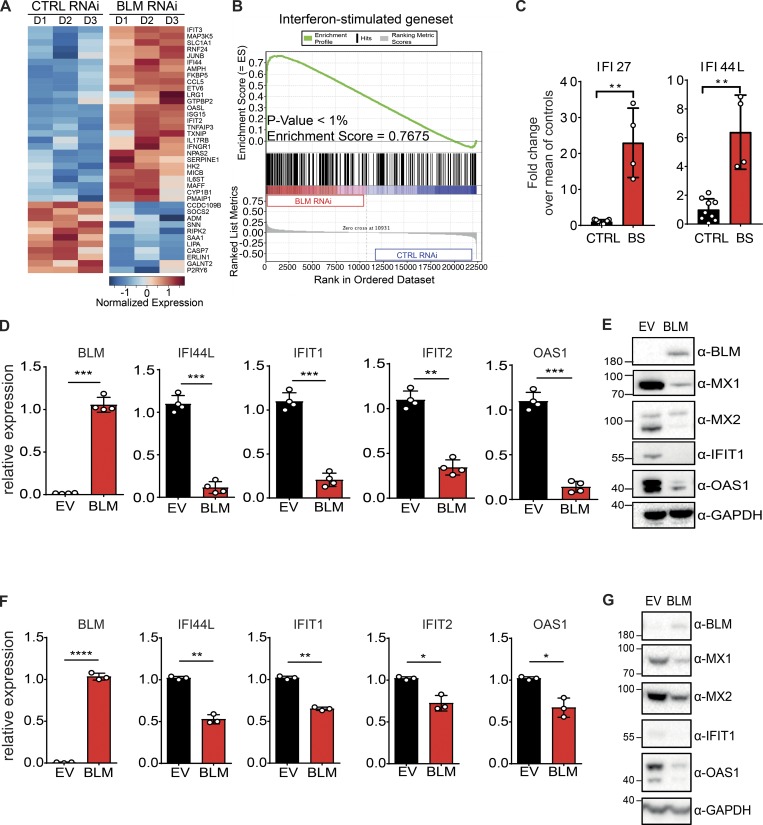
We examined the transcriptional profile of cells following a previously reported down-regulation of BLM (Chabosseau et al., 2011). Expression of ISGs was induced upon down-regulation of BLM (Fig. 1 A and Fig. S1; Schoggins et al., 2011). ISG expression was significantly enriched in BLM-depleted cells, as shown by a gene set enrichment analysis (Fig. 1 B).
Figure 1.
BS cells exhibit an ISG signature. (A) Heatmap representing the Z-score for normalized expression values of 38 ISGs between control (CTRL RNAi) and BLM-depleted (BLM RNAi) HeLa cells. ISGs were selected from another study (Schoggins et al., 2011). (B) Gene set enrichment analysis of ISGs in control (CTRL RNAi) and BLM-depleted (BLM RNAi) HeLa cells (see Materials and methods). (C) mRNA level expression of IFI44L and IFI27 determined by RT-qPCR in primary CTRL and BS fibroblasts. Data are the combination of three independent experiments (nCTRL = 2, 3, and 4, and nBS = 1, 2, and 1, respectively, for each experiment). Values were normalized to the average of controls within each experiment (mean with SD; Mann-Whitney, **P < 0.01). (D) mRNA level expression of BLM, IFI44L, IFIT1, IFIT2, and OAS1 determined by RT-qPCR in SV40-immortalized BS cells transduced with BLM or EV (mean with SD of n = 4 independent experiments; paired t test, **P < 0.01, ***P < 0.001). (E) Protein levels of BLM, MX1, MX2, IFIT1, OAS1, and GAPDH in cells, as in D (representative of n = 4 independent experiments). (F) Expression of BLM, IFI44L, IFIT1, IFIT2, and OAS1 in primary BS cells transduced with BLM or EV (mean with SD of n = 3 independent experiments; paired t test, *P < 0.05, **P < 0.01, ****P < 0.0001). (G) Protein levels of BLM, MX2, MX1, IFIT1, OAS1, and GAPDH in cells, as in F (representative of n = 3 independent experiments).
These data suggested the possibility that ISGs could also be induced in BLM-deficient cells from BS patients. We examined ISG expression in primary fibroblasts from a BS patient carrying a homozygous mutation that generates a premature termination codon at position 784 (AGS721; Table S1 and Fig. S2, A–C). As expected, mutant cells demonstrated an increased frequency of SCE (Fig. S2, D and E). BLM protein could not be detected in these slow-dividing cells due to its S-phase restricted expression (Dutertre et al., 2000). Expression of IFI44L and IFI27 was increased in BLM-deficient cells as compared with control cells (Fig. 1 C).
To determine if ISG expression was the result of BLM deficiency, we rescued expression of WT BLM in immortalized BS cells (Fig. 1 D). Of note, the induction of ISGs in BS cells was subject to a clonal effect, and we used polyclonal conditions hereafter (data not shown). Transcriptional levels of IFI44L, IFIT1, IFIT2, and OAS1 were significantly reduced upon rescue with WT BLM (Fig. 1 D). At the protein level, we found that MX1, MX2, IFIT1, and OAS1 are expressed in immortalized BS cells (Fig. 1 E). Rescue of WT BLM expression reduced the levels of these proteins, confirming the ISG transcriptional signature of BLM-deficient cells (Fig. 1 E). Similar to immortalized cells, rescuing WT BLM expression in primary BS cells significantly reduced expression of IFI44L, IFIT1, IFIT2, and OAS1 at the RNA level (Fig. 1 F). Protein levels of MX1, MX2, IFIT1, and OAS1 were also reduced by WT BLM expression in BS cells (Fig. 1 G). We conclude that BS cells are characterized by an increased expression of ISGs that can be attenuated by reintroducing WT BLM.
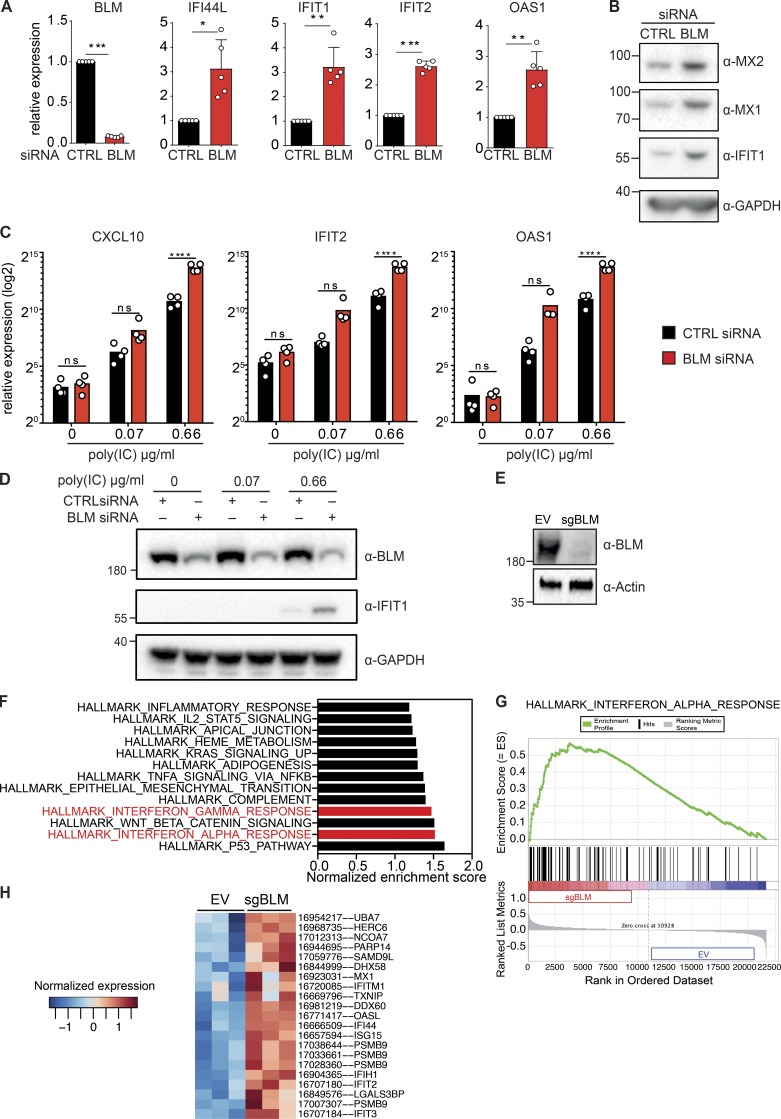
We next sought to determine if BLM was required in WT cells to suppress ISG expression. We inhibited BLM expression in primary fibroblasts using siRNA (Fig. 2 A). As a result, RNA levels of IFI44L, IFIT1, IFIT2, and OAS1 were significantly increased (Fig. 2 A). Consistently, protein levels of MX1, MX2, and IFIT1 were also increased upon transient down-regulation of BLM (Fig. 2 B). Of note, a baseline level of MX1, IFIT1, and OAS1 proteins was also detected in control cells, which could result from the siRNA transfection. These results were reproduced in an independent source of primary human fibroblasts (Fig. S3). ISGs comprise multiple innate immune sensors and signaling components (Schoggins et al., 2011), and cells with a higher level of ISG expression are expected to show an increased sensitivity to stimulation with double strand RNA (Kaiser et al., 2004; Tissari et al., 2005; Tohyama et al., 2005). To test if ISG induction upon BLM down-regulation results in such functional sensitization, we reduced BLM protein expression with siRNA and treated cells with increasing concentrations of poly(I:C) (polyinosinic–polycytidylic acid). As expected, CXCL10, IFIT2, and OAS1 transcripts were induced upon poly(I:C) treatment in a dose-dependent manner in control cells. This induction was significantly increased with concomitant down-regulation of BLM (Fig. 2 C). Accordingly, IFIT1 protein was induced by poly(I:C) treatment and further increased with down-regulation of BLM (Fig. 2 D). We conclude that BLM is required to limit functional sensitization to an innate immune stimulus in cells, consistent with the up-regulation of ISGs in BLM-deficient cells. To determine if the ISG signature was present at the transcriptome level in BLM-deficient cells, we knocked out BLM using clustered regularly interspaced short palindromic repeat (CRISPR) lentivectors in SV40-immortalized fibroblasts to perform a microarray gene expression analysis (Fig. 2 E). Two significant hallmark gene sets corresponding to ISGs were enriched in the transcriptome of BLM knockout cells (Fig. 2, F and G). Among genes that were significantly deregulated in BLM-deficient cells over control cells, all gene set ISGs were up-regulated (Fig. 2 H). We conclude that the ISG signature was significantly enriched in BLM-deficient cells at the transcriptome level.
Figure 2.
Down-regulation of BLM induces ISGs. (A) mRNA level expression of BLM, IFI44L, IFIT1, IFIT2, and OAS1 determined by RT-qPCR in primary human fibroblasts treated with control (CTRL) or BLM siRNA for 72 h (mean with SD of n = 5 independent experiments; one-sample t test, *P < 0.05, **P < 0.01, ***P < 0.001). (B) Protein levels of MX2, MX1, IFIT1, and GAPDH in primary human fibroblasts treated with control or BLM siRNA for 72 h (representative of n = 5 independent experiments). (C) mRNA level expression of CXCL10, IFIT2, and OAS1 determined by RT-qPCR in primary human fibroblasts treated with control or BLM siRNA for 48 h followed by 24 h of treatment with poly(I:C) (mean with SD of n = 4 independent experiments; one-way ANOVA with Tukey post-test, ****P < 0.0001). (D) Protein levels of BLM, IFIT1, and GAPDH in primary human fibroblasts treated with control or BLM siRNA for 48 h followed by 24 h of treatment with poly(I:C) (representative of n = 4 independent experiments). (E) Protein levels of BLM and Actin in SV40-immortalized cells after transduction with CRISPR-Cas9 lentivector targeting BLM (sgBLM) and the corresponding control lentivectors (EV; representative of three independent replicates). (F) Normalized enrichment score of the hallmark gene sets that were significantly enriched in BLM knockout cells over control cells, as in E. Microarray-based analysis of three independent replicates. (G) Enrichment of the hallmark IFNα response gene set in BLM knockout cells. (H) Heatmap representing the Z-score for normalized expression values of the hallmark IFNα response genes that were significantly deregulated at the gene level between BLM knockout cells and control cells (P < 0.05).
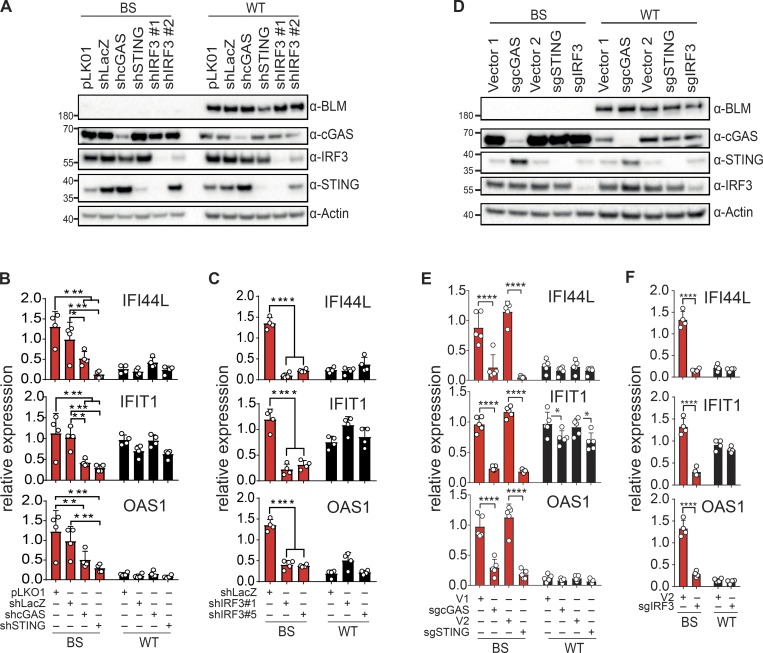
We next aimed to identify the signaling pathway involved in ISG expression upon BLM deficiency. Since BLM functions in DNA repair, we hypothesized that the cGAS–STING–IRF3 pathway could be activated as a result of BLM deficiency. We inhibited the expression of cGAS, STING, and IRF3 with shRNA vectors in SV40-immortalized BS and control cells (Fig. 3 A and Fig. S4, A and B). As previously observed, expression of IFI44L, IFIT1, and OAS1 was increased in BS cells. Inhibition of cGAS, STING, or IRF3 expression significantly inhibited expression of these genes (Fig. 3, B and C). Of note, one of the shRNA targeting IRF3 also reduced the expression of STING in this cell type. To confirm the implication of the cGAS–STING–IRF3 pathway, we knocked out these genes using CRISPR lentivectors in SV40-immortalized BS and control cells (Fig. 3 D and Fig. S4, C and D). Similar to shRNA-mediated down-regulation, expression of IFI44L, IFIT1, and OAS1 was significantly reduced in knockout cells (Fig. 3, E and F). We conclude that BLM is required to limit ISG expression resulting from engagement of the cGAS–STING–IRF3 pathway.
Figure 3.
ISG signature induction in BS cells requires the cGAS–STING–IRF3 pathway. (A) Protein levels of BLM, cGAS, STING, IRF3, and Actin in SV40-immortalized control (WT) or BS cells transduced with control shRNA lentivector (pLKO1, shLacZ) or shRNA lentivectors against cGAS, STING, or IRF3 (representative of n = 4 independent experiments). (B) mRNA level expression of IFI44L, IFIT1, and OAS1 determined by RT-qPCR in cells transduced with control shRNAs or shRNA against cGAS or STING, as in A (mean with SD of n = 4 independent experiments; two-way ANOVA with Tukey post-test, *P < 0.05, **P < 0.01, ***P < 0.001). (C) mRNA level expression of IFI44L, IFIT1, and OAS1 determined by RT-qPCR in cells transduced with control shRNAs or shRNAs against IRF3, as in A (mean with SD of n = 4 independent experiments; two-way ANOVA with Tukey post-test, ****P < 0.0001). (D) Protein levels of BLM, cGAS, STING, IRF3, and Actin in SV40-immortalized control (WT) or BS cells transduced with CRISPR-Cas9 lentivectors directed against cGAS, STING, or IRF3 or control lentivectors (V1 or V2; representative of n = 5 independent experiments). (E) mRNA level expression of IFI44L, IFIT1, and OAS1 determined by RT-qPCR in SV40-immortalized control (WT) or BS cells transduced with CRISPR-Cas9 lentivectors directed against cGAS, STING, or control lentivectors (V1 or V2), as in D (mean with SD of n = 5 independent experiments; two-way ANOVA with Tukey post-test, *P < 0.05, ****P < 0.0001). (F) mRNA level expression of IFI44L, IFIT1, and OAS1 determined by RT-qPCR in SV40-immortalized control (WT) or BS cells transduced with CRISPR-Cas9 lentivector directed against IRF3 or control lentivector (V2), as in D (mean with SD of n = 4 independent experiments; two-way ANOVA with Sidak post-test, ****P < 0.0001).
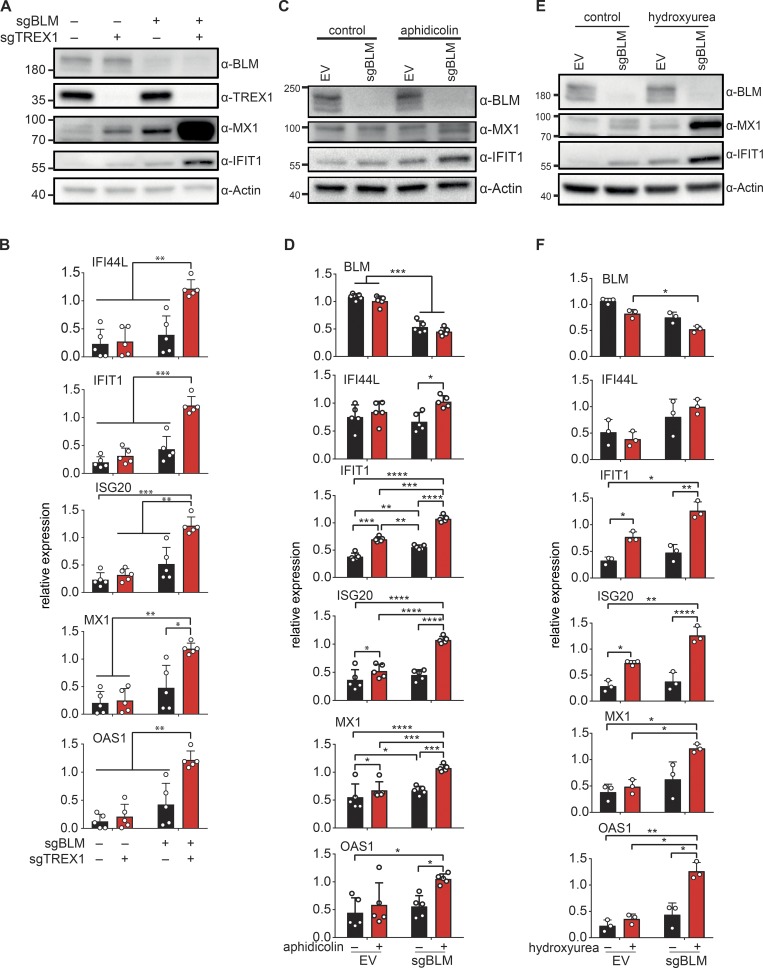
The cGAS–STING–IRF3 pathway is triggered by the accumulation of cytosolic DNA that activates cGAS. Accumulation of cytosolic DNA is counterbalanced by the activity of the DNaseIII TREX1, and TREX1 deficiency leads to increased ISG expression in a cGAS-dependent manner (Ablasser et al., 2014). To determine if cytosolic DNA might be implicated in cGAS activation in BLM-deficient cells, we knocked out TREX1 in control and in BLM knockout cells (Fig. 4 A). Inhibiting TREX1 expression in BLM-deficient cells increased expression of IFI44L, IFIT1, ISG20, MX1, and OAS1 (Fig. 4 B). At the protein level, MX1 and IFIT1 were strongly increased upon TREX1 disruption in BLM knockout cells (Fig. 4 A). These results suggested that TREX1-sensitive DNA was responsible for the cGAS-dependent ISG expression in BLM-deficient cells.
Figure 4.
DNA damage and down-regulation of TREX1 enhance ISG expression in BLM-deficient cells. (A) Protein levels of BLM, TREX1, MX1, IFIT1, and Actin in SV40-immortalized cells after transduction with CRISPR-Cas9 lentivector targeting BLM (sgBLM) and/or TREX1 (sgTREX1) or corresponding control lentivectors (–; representative of n = 5 experiments). (B) mRNA level expression of IFI44L, IFIT1, ISG20, MX1, and OAS1 determined by RT-qPCR in cells, as in A (mean with SD of n = 5 independent experiments; two-way ANOVA with Tukey post-test, *P < 0.05, **P < 0.01, ***P < 0.001). (C) Protein levels of BLM, MX1, IFIT1, and Actin in SV40-immortalized control cells (EV) and BLM-deficient cells (sgBLM) treated with 0.4 µM of aphidicolin for 24 h (representative of n = 5 independent experiments). (D) mRNA level expression of BLM, IFI44L, IFIT1, ISG20, MX1, and OAS1 determined by RT-qPCR in cells, as in C (mean with SD of n = 5 independent experiments; two-way ANOVA with Tukey post-test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (E) Protein levels of BLM, MX1, IFIT1, and Actin in SV40-immortalized control cells (EV) and BLM-deficient cells (sgBLM) treated with 0.5 µM of hydroxyurea for 24 h (representative of n = 3 independent experiments). (F) mRNA level expression of BLM, IFI44L, IFIT1, ISG20, MX1, and OAS1 determined by RT-qPCR in cells, as in E (mean with SD of n = 3 independent experiments; one-way ANOVA with Tukey post-test, *P < 0.05, **P < 0.01, ****P < 0.0001).
We next sought to determine if activation of a cGAS-dependent ISG expression in BLM-deficient cells could be the result of their characteristic DNA replication stress (Rao et al., 2007). We used aphidicolin and hydroxyurea to induce replication-dependent DNA damage in control and BS cells. We first treated BLM knockout cells and control cells with aphidicolin (Fig. 4 C). Aphidicolin treatment alone slightly increased ISG expression, which was significant for IFIT1, ISG20, and MX1 but not for IFI44L and OAS1 in control cells (Fig. 4 D). In contrast, aphidicolin treatment in BLM knockout cells significantly increased IFIT1, ISG20, MX1, IFI44L, and OAS1 expression (Fig. 4 D). IFIT1 and MX1 protein levels were also increased by aphidicolin treatment in BLM knockout cells (Fig. 4 C). We next treated BLM knockout cells with hydroxyurea (Fig. 4 E). Hydroxyurea significantly increased the expression of ISG20 and IFIT1, but not MX1 or OAS1 in control cells, indicative of a slight induction of ISG expression (Fig. 4 F). In BLM knockout cells, hydroxyurea treatment further increased expression of IFIT1, ISG20, MX1, and OAS1 as compared with untreated BLM knockout cells (Fig. 4 F). At the protein level, induction of MX1 and IFIT1 was also enhanced by hydroxyurea in BLM knockout cells (Fig. 4 E). Altogether, these results show that BLM is required to limit ISG expression induced by DNA damage generated during replication.
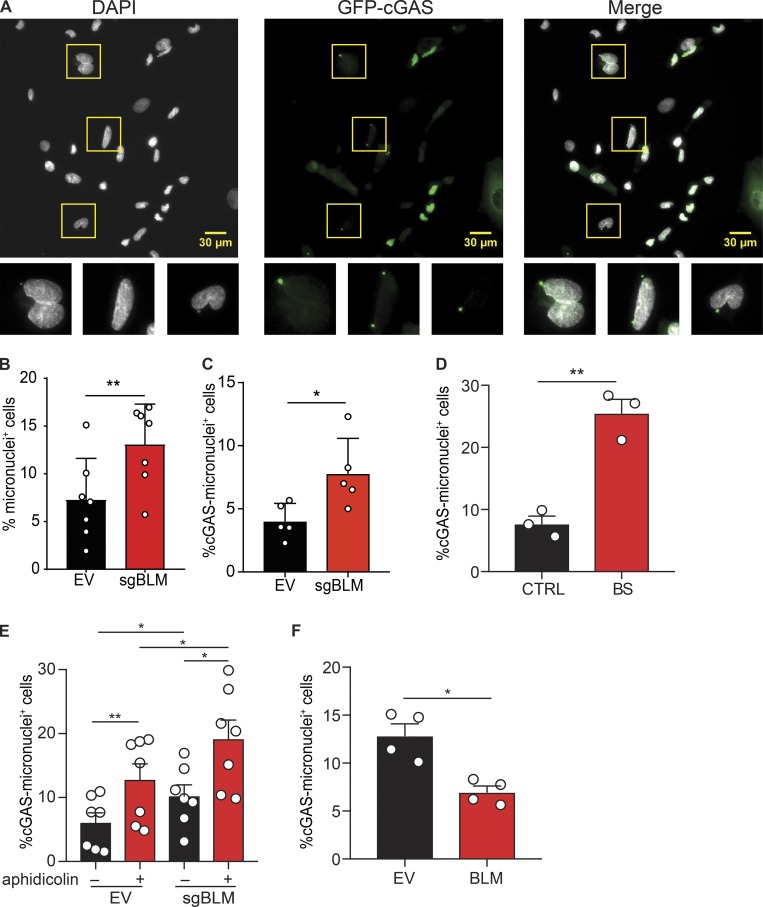
DNA-damaging agents lead to the accumulation of misplaced DNA fragments that activate the cGAS pathway (reviewed in Li and Chen, 2018). Our data suggested that cGAS-dependent ISG expression in BLM-deficient cells could be the result of unresolved DNA damage. BLM-deficient cells are characterized by the accumulation of micronuclei in the cytosol (Yankiwski et al., 2000), and micronuclei resulting from DNA damage activate the cGAS pathway (Harding et al., 2017; Mackenzie et al., 2017). We quantified the frequency of micronuclei and their association with cGAS in BLM-deficient cells using a GFP-cGAS reporter as previously described (Fig. 5 A; Raab et al., 2016; Mackenzie et al., 2017). As expected, the frequency of micronuclei was significantly increased in engineered BLM-deficient cells (Fig. 5 B). This increased level was similar to that in BS cells (Fig. S5 A). We found that cells containing cGAS-positive micronuclei were also increased in BLM-deficient cells (Fig. 5 C). Within micronuclei, the frequency of GFP-cGAS micronuclei was not different in control and BLM-deficient cells (Fig. S5 B), suggesting that BLM determines the rate of micronuclei production but not their susceptibility to cGAS association. Similarly, in primary fibroblasts, the frequency of micronuclei, and of cells with cGAS-positive micronuclei, was higher in BS cells (Fig. 5 D and Fig. S5 C). Treatment with aphidicolin, which increased ISG expression in BS cells (Fig. 4, C and D), increased the frequency of micronuclei, and of cells with cGAS-positive micronuclei, in both control and BLM-deficient cells (Fig. 5 E and Fig. S5 D). Interestingly, the frequency of cells with cGAS-positive micronuclei was increased in BLM-deficient cells over control following aphidicolin treatment (Fig. 5 E). Next, we rescued BLM expression in BS cells. In agreement with the inhibition experiment, rescue of BLM expression significantly reduced the frequency of micronuclei and of cells with cGAS-positive micronuclei (Fig. 5 F and Fig. S5 E). We conclude that cGAS recognizes micronuclei in BLM-deficient cells.
Figure 5.
BLM deficiency increases the frequency of micronuclei positive for cGAS. (A) Identification of total and cGAS-positive micronuclei in SV40-immortalized BLM-knockout cells, as in Fig. 4 A. Cells were transduced with GFP-cGAS E225A-D227A lentivector to visualize cGAS-positive micronuclei. Scale bars, 30 μm. (B) Frequency of cells positive for at least one micronucleus in SV40-immortalized control (EV) or BLM-deficient cells (sgBLM), as in Fig. 4 A (mean with SD of n = 7 combined independent experiments; paired t test, **P < 0.01). (C) Frequency of cells positive for at least for one GFP-cGAS–positive micronucleus in SV40-immortalized control (EV) or BLM-knockout cells (sgBLM), as in Fig. 4 A (mean with SD of n = 5 combined independent experiments; paired t test, *P < 0.05). (D) Frequency of cells positive for at least one GFP-cGAS–positive micronucleus in primary control (CTRL) or BLM-deficient fibroblasts (BS; mean with SEM of n = 3 combined independent experiments; paired t test, **P < 0.01). (E) Frequency of cells positive for at least one GFP-cGAS–positive micronucleus in SV40-immortalized control (EV) or BLM-knockout cells (sgBLM) and treated by aphidicolin (mean with SEM of n = 7 combined independent experiment; treatment with 12 or 30 µM in four and three experiments, respectively; repeated measures one-way ANOVA with a Tukey post-test, *P < 0.05, **P < 0.01). (F) Frequency of cells positive for at least one GFP-cGAS–positive micronucleus in SV40-immortalized control BS cells or BLM-transduced BS cells, as in Fig. 1 D (mean with SEM of n = 4 combined independent experiments; paired t test, *P < 0.05).
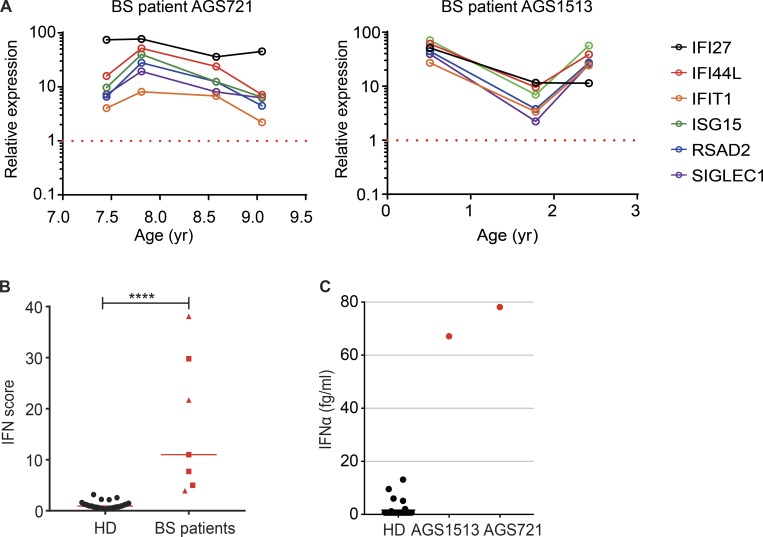
We finally sought to determine whether the expression of ISG in BS cells was physiologically relevant in BS patients. In monogenic interferonopathies, leukocytes are characterized by an increased expression of ISGs that is sustained over time and that can be used to derive a discriminative IFN score (Rice et al., 2013). In addition, elevated IFNα protein can be detected in the serum using a Simoa digital ELISA (Rodero et al., 2017a). We examined these parameters in available samples from two BS patients, AGS721 and AGS1513 (Table S1). AGS1513 carried the homozygous BLM mutation p.C1055S that we previously described (Foucault et al., 1997; Fig. S2 F). Increased ISG expression was consistently detected in three and four independent samples over a period of 1.5 and 2 yr, respectively (Fig. 6 A). The combined IFN score of all samples was significantly higher than that of control samples (Fig. 6 B). We next measured the concentration of IFNα in the plasma from the AGS1513 patient and in the cerebrospinal fluid (CSF) from the AGS721 patient. The plasma IFNα concentration of AGS1513 was increased 97× as compared with the average of all healthy donors (Fig. 6 C). The CSF IFNα concentration of AGS721 was also increased compared with a previously reported set of CSF samples from patients with retinal vasculopathy and cerebral leukodystrophy that is not associated with increased type I IFN signaling (Fig. 6 C; Rodero et al., 2017a). We conclude that BS patients are characterized in vivo by a sustained up-regulation of IFN signaling.
Figure 6.
Increased ISG expression in BS patients. (A) IFI27, IFI44L, IFIT1, ISG15, RSAD2, and SIGLEC1 expression was measured in the whole blood of two BS patients (AGS721 and AGS1513). Measurements were made at different time points and were normalized to 29 control samples (set to 1; red dotted line). (B) IFN score of the two BS patients (red squares: AGS721; red triangles: AGS1513) and control individuals (bar = median; Mann-Whitney test, ****P < 0.0001). (C) IFNα protein concentration in 36 control sera, in serum from the AGS1513 BS patient, and in CSF from the AGS712 BS patient.
Discussion
Here, we show that BLM-deficient cells present an increased expression of inflammatory genes through the cGAS–STING–IRF3 pathway as a result of DNA damage and micronuclei generation and provide evidence that BS patients exhibit an IFN-gene signature. Altogether, these results establish that BLM is essential to prevent the induction of unchecked inflammatory gene responses.
We observed induction of ISGs at the transcriptional and protein levels in peripheral blood cells and plasma from BS patients, in primary BS cells, in SV40-immortalized BS cells and upon CRISPR- and RNA interference (RNAi)–mediated inhibition of BLM expression in wild-type cells. Consistent with this, a previous report noted the up-regulation of an immune gene signature in a panel of BS cells (Nguyen et al., 2014). BS includes an immune deficiency that often leads to life-threatening infections. Patients usually present with low concentrations of one or more plasma immunoglobulins and fail to develop delayed-type hypersensitivity (Hütteroth et al., 1975; Van Kerckhove et al., 1988; Kondo et al., 1992). BS patients are also at risk of developing diabetes mellitus. In mice, the full deficiency of Blm is embryonic lethal (Chester et al., 1998; Luo et al., 2000; Goss et al., 2002). Tissue-specific deletion of Blm has revealed that BLM plays an essential role during B and T lymphocyte development (Babbe et al., 2007, 2009). In agreement with our results, it was previously noted that the levels of Ifnb transcripts are increased in Werner-Bloom-Terc triple-deficient mice (Yu et al., 2015). Chronic IFN production was also detected in ataxia telangiectasia and ATM deficiency (Quek et al., 2017; Gul et al., 2018). However, the cellular basis of the immune deficiency in BS is poorly understood, and the function of the BLM protein in the immune system is not defined. Our work enables investigation of how innate immune sensing of damaged DNA in BS could contribute to immune-related disorders in patients.
We find that cGAS-positive micronuclei are increased in BLM-deficient cells. For recombinant BLM, preferred substrates are structures mimicking replication forks or homologous recombination intermediates, such as D-loops and X-junctions, and G-quadruplex DNA, which could all contribute to micronuclei generation (Amor-Guéret, 2006). Consistent with a role of DNA damage in micronuclei generation in BLM-deficient cells, we find that DNA-damaging agents aphidicolin or hydroxyurea treatment potentiated ISG induction. The increased frequency of cGAS-positive micronuclei in BLM-deficient cells over control, following aphidicolin treatment, suggests the interesting possibility that aphidicolin promotes association of cGAS with micronuclei when BLM is not present. In addition to micronuclei, DNA damage can also lead to release of immunostimulatory single-stranded DNA (ssDNA) in the cytosol (Li and Chen, 2018). Release of ssDNA in the cytosol was recently reported in cells simultaneously depleted for SAMHD1 and treated with hydroxyurea (Coquel et al., 2018). Co-inhibition of BLM expression did not modify the quantity of cytosolic ssDNA in this study, contrasting with its effect on micronuclei. We also find that down-regulation of TREX1, a protein that degrades cytosolic DNA (Stetson et al., 2008), enhances ISG induction in BS cells. Altogether, this suggests that BLM normally contributes to resolving nucleic acid structures that would otherwise lead to the release of TREX1-sensitive, immunostimulatory micronuclei. We found that ISG expression in BS cells required cGAS, STING, and IRF3. A low level of ISG expression was also detectable in wild-type fibroblasts, but that did not require cGAS, STING, or IRF3, and was not enhanced by TREX1 down-regulation. This is consistent with ISG expression resulting from tonic IFN signaling (Gough et al., 2012), which is unlikely to result from accumulation of cytosolic DNA in wild-type fibroblasts in this case.
The IFNα concentrations that we measured in two BS patients are in the range of what has been reported in polygenic diseases such as systemic lupus erythematosus or juvenile-onset dermatomyositis and are lower than in monogenic interferonopathies (Rodero et al., 2017a). How do these data inform our clinical understanding? In monogenic interferonopathies, systemic lupus erythematosus, and juvenile-onset dermatomyositis, there is no strict correlation between the IFN signature and disease (Rodero et al., 2017a), and there is clear distinction in the phenotype of BS. Previously, two constant features (growth delay and susceptibility to all kinds of cancer) and nine nonconstant features have been described in BS (German, 1995). The contribution of the IFN signature as a driver of the BS phenotype requires further interrogation, and understanding the clinical relevance of the DNA damage response genes in controlling innate inflammation and pathological processes will possibly require identification of the anatomical and cellular sites where IFN is produced at the highest levels.
ISGs can be induced by radiotherapy and chemotherapy, and the expression of a subset of ISGs was reported to correlate with resistance to treatment and cancer cell survival in another study (Erdal et al., 2017). In this context, contrasting with our results on BLM expression in primary and immortalized fibroblasts, in an MCF7 cancer cell model, BLM was instead required for the accumulation of cytoplasmic DNA foci, phosphorylation of STAT1, and induction of IFI44, IFI6, ISG15, and MX1 in response to irradiation (Erdal et al., 2017). This could suggest that in specific cancer cell types, where ISG expression might contribute to resistance to treatment, BLM is “repurposed” to enhance the production of inflammatory cytoplasmic DNA species. In contrast, we find that in primary fibroblasts, BLM is required to limit these processes. The mechanisms that enable such putative repurposing of BLM in cancer cells are not known. Taken together, our results highlight the multifaceted role of BLM in health and cancer, in the context of cytoplasmic DNA sensing and inflammation.
Altogether, our results indicate that BLM keeps inflammatory gene induction resulting from DNA damage in check. This extends our basic understanding of how DNA damage is prevented from triggering damaging innate immune activation in sterile conditions and provides new insights into the pathogenesis of rare monogenic human diseases.
Materials and methods
Patients
The study was approved by the Comité de Protection des Personnes (ID-RCB/EUDRACT: 2014-A01017-40). Patients are described in Table S1.
Plasmid and reagents
Plasmids used in the study are listed in Table S2. pTRIP-CMV-puro-2A and pTRIP-CMV-EGFP-FLAG-cGAS E225A-D227A were described elsewhere (Gentili et al., 2015). BLM open reading frame was amplified from pGFP-BLM (Eladad et al., 2005) and inserted in pTRIP-CMV-puro-2A. pCMV-VSVG, psPAX2, pLKO.1-puro, and pLCRISPR-CMV were described elsewhere (Manel et al., 2010; Gentili et al., 2015), and pLentiCRISPRv2 was described in Sanjana et al. (2014). pLentiCRISPRv2 Neo was create by replacing puromycin-resistant gene by neomycin in pLentiCRISPRv2. shRNAs against LacZ and cGAS were previously described (Lahaye et al., 2013). shRNA against STING was purchased from Sigma (TRCN0000163296). shRNAs against IRF3 were transferred from pLKO1gfp to pLKO.1-puro (Manel et al., 2010). The lentivector with a guide RNA (gRNA) against cGAS was previously described (Gentili et al., 2015). gRNA against TREX1 was inserted in pLentiCRISPRv2 Neo. gRNA against STING was previously described (Cerboni et al., 2017). gRNA against BLM and IRF3 was inserted in pLentiCRISPRv2. All plasmids’ DNA was purified using Nucleobond Xtra Midi Plus (Macherey-Nagel) and were sequenced over the full BLM open reading frame or on the shRNA/single guide RNA inserted before use.
Poly(I:C) was purchased from Invivogen (ref tlrl-pic). Aphidicolin (#A0781) and hydroxyurea (#H8627) were purchased from Sigma-Aldrich (#A0781).
Cell culture, transfection, and lentiviral transduction
Primary human BS fibroblasts were skin fibroblasts from patient AGS721. SV40-immortalized BS fibroblasts (GM08505B) were obtained from CORIELL Institute. Primary human control fibroblasts were described elsewhere (Jenkinson et al., 2016). SV40-transformed and human telomerase reverse transcriptase (hTERT) immortalized human control fibroblasts were described elsewhere (Nicolas et al., 1998; Poinsignon et al., 2004). All cells were cultured in DMEM with 10% serum (Euromedex; lot S67113-0217), glutamine, and penicillin/streptomycin. Puromycine (1 µg/ml) or neomycine (500 µg/ml) was added according to lentiviral transduction.
For lentivirus production, packaging vectors pCMV-VSVG (1 µg) and psPAX2 (0.4 µg) were cotransfected in 6-well plates with lentiviral vector (1.6 µg) in HEK293T cells using Jetprime transfection reagent (Ozyme) according to the manufacturer’s protocol. HEK293T cells were washed 24 h after transfection, and the viruses were collected 48 h after transfection. Viruses were filtered (0.45 µm) and directly used for transductions. 8 µg/ml of protamine was added in each virus solution prior to each transduction. Cells were transduced for 24 h and then subjected to antibiotics selection (puromycin or neomycin).
For BLM complementation, human SV40-immortalized BS fibroblasts (GM08505B) and human primary BS fibroblasts (AGS721) were transduced with pTRIP-CMV-puro-2A or pTRIP-CMV-puro-2A-BLM. For shRNAs or CRISPR-Cas9 depletion, human SV40-immortalized BS fibroblasts (GM08505B) and human SV40-immortalized control fibroblasts were transduced with pLKO1-puro, pLCRISPR-CMV, or pLentiCRISPRv2 vectors. Cells were selected for at least 1 wk and harvested for RNA and protein extraction. To visualize cGAS in micronuclei, human SV40-immortalized BS fibroblasts (GM08505B) and human SV40-immortalized control fibroblasts were transduced with pTRIP-CMV-EGFP-FLAG-cGAS E225A-D227A.
For siRNA transfection, human primary control fibroblasts (in-house or from the American Type Culture Collection) or HEK293T cells were reverse transfected by siCtrl or siBLM (ON-targetplus SMART-pool; Dharmacon) at a final concentration of 50 nM by using Dharmafect 4 (Dharmacon). siRNAs are listed in Table S3. Cells were harvested at 72 h after transfection for RNA and protein extraction.
For poly(I:C) treatment, HEK293T cells depleted for BLM by siRNA were transfected with 0.66 or 0.07 µg/ml of poly(I:C) 48 h after siRNA transfection. Cells were harvested 72 h after siRNA transfection for RNA and protein extractions.
For aphidicolin or hydroxyurea treatments, human SV40-immortalized control fibroblasts depleted for BLM by CRISPR-Cas9 were treated by 0.4 µM of aphidicolin or 0.5 µM of hydroxyurea. Cells were washed at 24 h and harvested at 48 h after treatment for RNA and protein extraction.
Affymetrix data
For RNAi-treated HeLa cells, the microarray data were described previously (Chabosseau et al., 2011). For SV40-immortalized fibroblasts, 48 h after seeding of single guide RNA against BLM (sgBLM) and empty vector (EV) cells, total RNA was extracted with a Nucleospin kit and eluted in 40 µl of RNase/DNase free water. Quality of total RNA was assessed using RNA 6000 NanoChips with the Agilent 2100 Bioanalyzer (Agilent). Using 100 ng of total RNA, double-stranded cDNA was synthesized with random hexamers tagged with a T7 promoter sequence. The double-stranded cDNA was subsequently used as a template and amplified by T7 RNA polymerase producing many copies of antisense complementary RNA. In the second cycle of cDNA synthesis, random hexamers are used to prime reverse transcription of the complementary RNA from the first cycle to produce ssDNA in the sense orientation. cDNA was synthesized using the GeneChip WT (Whole Transcript) Sense Target Labeling and Control Reagents kit as described by the manufacturer (Affymetrix). The sense cDNA was then fragmented by uracil DNA glycosylase and apurinic/apyrimidic endonuclease-1 and biotin labeled with terminal deoxynucleotidyl transferase using the GeneChip WT Terminal labeling kit (Affymetrix). Hybridization was performed using 5 µg of biotinylated target, which was incubated with the GeneChip Human gene 2.0 array (Affymetrix) at 45°C for 16–20 h. After hybridization, non–specifically bound material was removed by washing and specifically bound target was detected using the GeneChip Hybridization, Wash and Stain kit, and the GeneChip Fluidics Station 450 (Affymetrix). The arrays were scanned using the GeneChip Scanner 3000 7G (Affymetrix), and raw data were extracted from the scanned images and analyzed with the Affymetrix Power Tools software package. Microarray data analyses were performed with R (version 3.0.0) using packages from Bioconductor. The quality control was performed using the ArrayQualityMetrics package (Kauffmann et al., 2009) without detecting any outlier among the experiment. Data were normalized using the Robust Multi-array Average algorithm from the Oligo package (Carvalho and Irizarry, 2010). Different expression analysis was performed with the Limma package (Ritchie et al., 2015). For RNAi-treated cells, the threshold was an adjusted P value <5% and a −1 > log2 fold-change > 1 (n = 560 probes). ISGs were selected based on a previous study (Schoggins et al., 2011; n = 330 genes). For SV40-immortalized fibroblasts, the threshold was a P value <5%. Gene-set enrichment analyses were performed with javaGSEA software (Subramanian et al., 2005). For SV40-immortalized fibroblasts, hallmark gene sets from MSigDB (n = 50) were tested for enrichment. The data have been deposited into the Gene Expression Omnibus database (accession no. GSE123447).
Quantitative PCR of gene expression (RT-qPCR)
Total RNA was extracted from cells using Nucleospin RNA (Macherey-Nagel) according to the manufacturer’s instructions. cDNA was synthetized from 1 µg of total RNA using iScript Advanced cDNA Synthesis Kit (Bio-Rad). Quantitative PCR (qPCR) was done using iTaq Universal SYBR Green (Bio-Rad) on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). ACTB and HMBS were used as internal control genes, except for experiments with aphidicolin or hydroxyurea treatments, where B2M and HPRT were used as internal control genes. A list of the primers used in the study is provided Table S4. Gene expression was analyzed through CFX Manager software (Bio-Rad) according to the 2ΔΔct method.
For comparison of gene expression between BS patient cells and control cultured primary fibroblasts, IFI27 and IFI44L were quantified by RT-qPCR using TaqMan probes (IFI27: Hs01086370_m1; and IFI44L: Hs00199115_m1), the relative abundance of each target transcript normalized to the expression level of HPRT1 (Hs03929096_g1), and assessed with the Applied Biosystems StepOne Software v2.1. Values were normalized to the average of controls within each experiment.
Western blot
Dry cell pellets were resuspended in 10 mM Tris HCl pH 7.4, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 100 mM NaF, 4 mM Na4P2O4, 2 mM Na3VO4, 1% Triton X100, 0.5% NP-40 IGEPAL, and protease inhibitors. Cells lysis was performed for 5 min on ice and the lysate was cleared by centrifugation. An equivalent amount of proteins was loaded in polyacrylamide gel either according to the number of harvested cells or by dosing protein concentration with Qubit Protein Assay Kit (Thermo Fisher Scientific). Lithium dodecyl sulfate (LDS) sample buffer was added to lysates (Nupage LDS Sample Buffer 4X; NP0007; Thermo Fisher Scientific) and protein migration was performed in 25 mM Tris pH 8.3, 192 mM glycine, and 0.1% SDS with MINI-PROTEAN SFX, 4–20% gels (Bio-Rad) at 50 V for 3 h in a Mini-PROTEAN Tetra Vertical Electrophoresis Cell. Protein transfer was performed at 35 V overnight on a polyvinylidene fluoride membrane in 25 mM Tris pH 8.3 and 192 mM glycine. Transfers on membrane were verified by using a Ponçeau staining. Membranes were incubated in Tris-buffered saline (TBS) 0.1% Tween-20 and 5% milk for 1 h, and then with the primary antibodies. Membranes were washed 3 times in TBS 0.1% Tween-20 after primary and secondary antibodies. All the antibodies were diluted in TBS 0.1% Tween and 5% milk at the concentration indicated in Table S5. Protein revelation was performed using Chemidoc XRS+ (Bio-Rad) after incubation with Clarity Western ECL (Bio-Rad).
IFN score (IS)
Blood was collected into PAXgene tubes (PreAnalytix) and, after being kept at room temperature for between 1 and 72 h, frozen at −20°C until extraction. Total RNA was extracted from whole blood using a PAXgene (PreAnalytix) RNA isolation kit. RNA concentration was assessed using a spectrophotometer (FLUOstar Omega; Labtech). RT-qPCR analysis was performed using the TaqMan Universal PCR Master Mix (Applied Biosystems) and cDNA derived from 40 ng total RNA. Using TaqMan probes for IFI27 (Hs01086370_m1), IFI44L (Hs00199115_m1), IFIT1 (Hs00356631_g1), ISG15 (Hs00192713_m1), RSAD2 (Hs01057264_m1), and SIGLEC1 (Hs00988063_m1), the relative abundance of each target transcript was normalized to the expression level of HPRT1 (Hs03929096_g1) and 18S (Hs999999001_s1) and assessed with the Applied Biosystems StepOne Software v2.1 and DataAssist Software v.3.01. For each of the six probes, individual (patient and control) data were expressed relative to a single calibrator (control C25). The median fold-change of the six ISGs, when compared with the median of previously collected 29 healthy controls, was used to create an IS for each individual. Relative quantification is equal to 2−ΔΔCt (i.e., the normalized fold-change relative to the control data). In this way, we define an abnormal IS as being greater than +2 SDs above the mean of this control group (i.e., 2.466).
Single molecule array (Simoa)
The Simoa IFNα assay was developed using a Quanterix Homebrew Simoa assay according to the manufacturer’s instructions and using two autoantibodies specific for IFNα isolated and cloned from two APS1/APECED (autoimmune polyendocrinopathy candidiasis ectodermal dystrophy) patients (Rodero et al., 2017a). The 12H5 antibody clone was used as a capture antibody after coating on paramagnetic beads (0.3 mg/ml), and the 8H1 was biotinylated (biotin-to-antibody ratio = 30:1) and used as the detector.
Fluorescent microscopy and micronuclei counting
Cells were seeded on coverslips and allowed to grow. Cells were then fixed with 4% paraformaldehyde for 20 min at room temperature, then washed three times with PBS and mounted on slides with Prolong Gold DAPI (Thermo Fisher Scientific, Molecular Probes) or Fluoromount-G with DAPI (Invitrogen). For cGAS-positive micronuclei visualization, cells were first transduced 24 h with GFP-cGAS lentivector before seeding on coverslips and were allowed to grow for 4 d.
For Fig. 5, B and C, and Fig. S5, A and B, cells were visualized with an upright widefield Leica microscope using a 63× oil objective mounted with a sCMOS Hamamatsu Orca Flash 4.0 camera (pixel size 6.5 µm). Stacks of images were acquired with Metamorph for DAPI or GFP fluorescence. Nuclei and micronuclei per nuclei were manually counted for each image. To identify cGAS-positive micronuclei, and exclude micronuclei located in a cGAS-positive area of the cytoplasm, we defined cGAS-positive micronuclei as cGASimax/cGASimin >2, where cGASimax is maximal cGAS intensity, cGASimin is minimal cGAS intensity, and measures are performed by automated quantification in a mask comprising each micronucleus and its surrounding cytoplasmic corona.
In Fig. 5, D–F, and Fig. S5, C–E, cells were visualized with a Leica SP8 scanning head on a DMi8 inverted confocal microscope. Images were acquired using a 20× dry objective (numerical aperture = 0.75) with excitation at 405 (for DAPI) and 488 (for GFP) lasers, and fluorescence signals were collected on hybrid detectors. Stacks of images were acquired with Leica LAS X software. Nuclei, micronuclei, and cGAS-positive micronuclei were detected and quantified for each image with an automated in-house macro in Fiji. In details, micronuclei detection was manually corrected to remove false-positive micronuclei. The frequency of cells with at least one micronucleus was calculated for each experiment. GFP-cGAS–positive micronuclei were identified using a global threshold of intensity above cytoplasmic levels of GFP-cGAS. To account for the different rate of GFP-cGAS transduction between samples within an experiment, the frequency of cells with GFP-cGAS micronuclei was determined either by gating in GFP-cGAS–positive cells (Fig. 5, D and F) or by normalizing to the percentage of transduced cells (Fig. 5 E), based on a threshold of GFP-cGAS intensity in the nucleus.
The number of nuclei analyzed for each experiment is reported in Table S6. All images were analyzed in Fiji.
Whole-exome sequencing
DNA was extracted from whole-blood samples using an in-house salting-out method. Agilent SureSelect libraries were prepared from 3 µg of genomic DNA extracted from leukocytes and sheared with a Covaris S2 Ultrasonicator as recommended by the manufacturer. We used a 51-Mb SureSelect Human All Exon kit V5 (Agilent Technologies) for exome capture, with multiplexed molecular barcodes for sample traceability. Pooled barcoded exome libraries were then sequenced on a HiSeq2500 (Illumina) machine, generating 76- + 76-bp paired-end reads, followed by processing using the Genome Analysis Toolkit (GATK), SAMtools, and Picard. Calls were made with the Polyweb application (https://www.polyweb.fr).
Sanger sequencing
Sanger sequencing was performed to confirm the Bloom variants found by exome sequencing.
For P1, Sanger confirmation of the mutation was performed on RNA extracted from whole blood. Whole blood was collected into PAXgene tubes. We used a PreAnalytix RNA isolation kit and RNA concentration, assessed with a spectrophotometer (FLUOstar Omega; Labtech), to extract total RNA. cDNA was generated using Qiagen OneStep RT-PCR kit. The region of the gene was amplified by PCR using specific primers (eurofinsgenomics) designed with Primer3plus (Blm cDNA 2F, 5′-CTCCTGGGGTCACTGTTGTC-3′; Blm cDNA 2R, 5′-AGCCATCACCGGAACAGAAG-3′).
SCEs
The assay was performed according to the method described (Gemble et al., 2015). SCE frequency in HeLa cells was previously described (Lahkim Bennani-Belhaj et al., 2010).
Statistics
Unless indicated, data are represented by the mean with SD bars. Statistical tests are indicated in the legends of each figure.
Online supplemental material
Fig. S1 shows the ISG signature in HeLa cells treated with BLM siRNA. Fig. S2 shows the characterization of the BLM mutations of patients AGS721 and AGS1513. Fig. S3 shows that depletion of BLM induces ISGs in an independent source of primary human fibroblasts. Fig. S4 shows the expression of additional genes related to Fig. 3. Fig. S5 shows additional quantifications related to Fig. 5. Table S1 shows the mutations and clinical features of patients in the study. Tables S2, S3, S4, and S5 describe the plasmids, siRNAs, primers, and antibodies used in the study, respectively. Table S6 shows the number of primary nuclei analyzed in imaging experiments.
Supplementary Material
Acknowledgments
We thank Philippe Benaroch for critical reading of the manuscript; Vincent Bondet for the Simoa analysis; Santy Marques-Ladeira for bioinformatics analysis; Xavier Lahaye for molecular biology; Géraldine Buhagiar-Labarchède and Rosine Onclercq-Delic for technical assistance; Aude Vieillefon, Audrey Rapinat, and David Gentien from the Genomics Platform of the translational research department of Institut Curie; and Prof. Dominique Stoppa-Lyonnet, Head of the Department of Genetics at Institut Curie and Professor of Medical Genetics at University Paris Descartes, for stimulating discussions. Y.J. Crow and D. Duffy thank ImmunoQure AG for sharing of antibodies used to assess interferon alpha protein levels in the Simoa assay.
This work was supported by Laboratoire d'Excellence Vaccine Research Institute (ANR-10-LABX-77), Laboratoire d'Excellence Biologie des Cellules Dendritiques (ANR-10-IDEX-0001-02 PSL* and ANR-11-LABX-0043), ACTERIA Foundation, Fondation Schlumberger pour l'Education et la Recherche, Agence Nationale de Recherche sur le Sida et les hépatites virales (ECTZ25472 and ECTZ36691), Sidaction (VIH2016126002), Bristol-Myers Squibb Foundation, European Research Council (grant 309848 HIVINNATE and INSERM) to N. Manel, National Research Agency (ANR-14-CE14-0004) and Institut Curie to N. Manel and M. Amor-Gueret, Institut National du Cancer (2016-1-PLBIO-03-ICR-1), Ligue Nationale Contre le Cancer (Comité de l’Essonne), and Centre National de la Recherche Scientifique to M. Amor-Gueret. Y.J. Crow acknowledges the European Research Council (GA 309449 and 786142 - E-T1IFNs), ERA-NET Neuron (MR/M501803/1), and a state subsidy managed by the National Research Agency (France) under the “Investments for the Future” program bearing the reference ANR-10-IAHU-01. Y.J. Crow and D. Duffy acknowledge the National Research Agency (grant CE17001002).
The authors declare no competing financial interests.
Author contributions: M. Gratia performed most experiments. C. Conrad and E. Bou Samra performed initial experiments. Y.J. Crow, M. Amor-Gueret, and N. Manel designed and piloted the study. M.P. Rodero contributed to experiments with BS cells. M. Maurin designed image analysis methods and wrote scripts. P. Revy characterized the BLM mutation from one patient. D. Duffy performed Simoa IFN measurements. G.I. Rice performed measured ISG expression in patient samples. R.C. Dale and F. Petit provided patient samples.
References
- Ablasser A., Hemmerling I., Schmid-Burgk J.L., Behrendt R., Roers A., and Hornung V.. 2014. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J. Immunol. 192:5993–5997. 10.4049/jimmunol.1400737 [DOI] [PubMed] [Google Scholar]
- Amor-Guéret M. 2006. Bloom syndrome, genomic instability and cancer: the SOS-like hypothesis. Cancer Lett. 236:1–12. 10.1016/j.canlet.2005.04.023 [DOI] [PubMed] [Google Scholar]
- Babbe H., Chester N., Leder P., and Reizis B.. 2007. The Bloom’s syndrome helicase is critical for development and function of the alphabeta T-cell lineage. Mol. Cell. Biol. 27:1947–1959. 10.1128/MCB.01402-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbe H., McMenamin J., Hobeika E., Wang J., Rodig S.J., Reth M., and Leder P.. 2009. Genomic instability resulting from Blm deficiency compromises development, maintenance, and function of the B cell lineage. J. Immunol. 182:347–360. 10.4049/jimmunol.182.1.347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev D.V., Yu X., Egelman E.H., and Mazin A.V.. 2007. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 21:3085–3094. 10.1101/gad.1609007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho B.S., and Irizarry R.A.. 2010. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 26:2363–2367. 10.1093/bioinformatics/btq431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerboni S., Jeremiah N., Gentili M., Gehrmann U., Conrad C., Stolzenberg M.C., Picard C., Neven B., Fischer A., Amigorena S., et al. 2017. Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes. J. Exp. Med. 214:1769–1785. 10.1084/jem.20161674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabosseau P., Buhagiar-Labarchède G., Onclercq-Delic R., Lambert S., Debatisse M., Brison O., and Amor-Guéret M.. 2011. Pyrimidine pool imbalance induced by BLM helicase deficiency contributes to genetic instability in Bloom syndrome. Nat. Commun. 2:368 10.1038/ncomms1363 [DOI] [PubMed] [Google Scholar]
- Chan K.L., and Hickson I.D.. 2011. New insights into the formation and resolution of ultra-fine anaphase bridges. Semin. Cell Dev. Biol. 22:906–912. 10.1016/j.semcdb.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Chan K.L., North P.S., and Hickson I.D.. 2007. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 26:3397–3409. 10.1038/sj.emboj.7601777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Sun L., and Chen Z.J.. 2016. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 17:1142–1149. 10.1038/ni.3558 [DOI] [PubMed] [Google Scholar]
- Chester N., Kuo F., Kozak C., O’Hara C.D., and Leder P.. 1998. Stage-specific apoptosis, developmental delay, and embryonic lethality in mice homozygous for a targeted disruption in the murine Bloom’s syndrome gene. Genes Dev. 12:3382–3393. 10.1101/gad.12.21.3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquel F., Silva M.J., Técher H., Zadorozhny K., Sharma S., Nieminuszczy J., Mettling C., Dardillac E., Barthe A., Schmitz A.L., et al. 2018. SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature. 557:57–61. 10.1038/s41586-018-0050-1 [DOI] [PubMed] [Google Scholar]
- Crow Y.J., and Manel N.. 2015. Aicardi-Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 15:429–440. 10.1038/nri3850 [DOI] [PubMed] [Google Scholar]
- Cunniff C., Bassetti J.A., and Ellis N.A.. 2017. Bloom’s syndrome: clinical spectrum, molecular pathogenesis, and cancer predisposition. Mol. Syndromol. 8:4–23. 10.1159/000452082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z., Ghosh K., Vizioli M.G., Zhu J., Sen P., Wangensteen K.J., Simithy J., Lan Y., Lin Y., Zhou Z., et al. 2017. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature. 550:402–406. 10.1038/nature24050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S., Ababou M., Onclercq R., Delic J., Chatton B., Jaulin C., and Amor-Guéret M.. 2000. Cell cycle regulation of the endogenous wild type Bloom’s syndrome DNA helicase. Oncogene. 19:2731–2738. 10.1038/sj.onc.1203595 [DOI] [PubMed] [Google Scholar]
- Eladad S., Ye T.Z., Hu P., Leversha M., Beresten S., Matunis M.J., and Ellis N.A.. 2005. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum. Mol. Genet. 14:1351–1365. 10.1093/hmg/ddi145 [DOI] [PubMed] [Google Scholar]
- Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., and German J.. 1995. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 83:655–666. 10.1016/0092-8674(95)90105-1 [DOI] [PubMed] [Google Scholar]
- Erdal E., Haider S., Rehwinkel J., Harris A.L., and McHugh P.J.. 2017. A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev. 31:353–369. 10.1101/gad.289769.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucault F., Vaury C., Barakat A., Thibout D., Planchon P., Jaulin C., Praz F., and Amor-Guéret M.. 1997. Characterization of a new BLM mutation associated with a topoisomerase II alpha defect in a patient with Bloom’s syndrome. Hum. Mol. Genet. 6:1427–1434. 10.1093/hmg/6.9.1427 [DOI] [PubMed] [Google Scholar]
- Gao D., Wu J., Wu Y.T., Du F., Aroh C., Yan N., Sun L., and Chen Z.J.. 2013. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 341:903–906. 10.1126/science.1240933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemble S., Ahuja A., Buhagiar-Labarchède G., Onclercq-Delic R., Dairou J., Biard D.S., Lambert S., Lopes M., and Amor-Guéret M.. 2015. Pyrimidine pool disequilibrium induced by a cytidine deaminase deficiency inhibits PARP-1 activity, leading to the under replication of DNA. PLoS Genet. 11:e1005384 10.1371/journal.pgen.1005384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentili M., Kowal J., Tkach M., Satoh T., Lahaye X., Conrad C., Boyron M., Lombard B., Durand S., Kroemer G., et al. 2015. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science. 349:1232–1236. 10.1126/science.aab3628 [DOI] [PubMed] [Google Scholar]
- German J. 1995. Bloom’s syndrome. Dermatol. Clin. 13:7–18. 10.1016/S0733-8635(18)30101-3 [DOI] [PubMed] [Google Scholar]
- Glück S., Guey B., Gulen M.F., Wolter K., Kang T.W., Schmacke N.A., Bridgeman A., Rehwinkel J., Zender L., and Ablasser A.. 2017. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 19:1061–1070. 10.1038/ncb3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss K.H., Risinger M.A., Kordich J.J., Sanz M.M., Straughen J.E., Slovek L.E., Capobianco A.J., German J., Boivin G.P., and Groden J.. 2002. Enhanced tumor formation in mice heterozygous for Blm mutation. Science. 297:2051–2053. 10.1126/science.1074340 [DOI] [PubMed] [Google Scholar]
- Gough D.J., Messina N.L., Clarke C.J., Johnstone R.W., and Levy D.E.. 2012. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 36:166–174. 10.1016/j.immuni.2012.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gul E., Sayar E.H., Gungor B., Eroglu F.K., Surucu N., Keles S., Guner S.N., Findik S., Alpdundar E., Ayanoglu I.C., et al. 2018. Type I IFN-related NETosis in ataxia telangiectasia and Artemis deficiency. J. Allergy Clin. Immunol. 142:246–257 [DOI] [PubMed] [Google Scholar]
- Harding S.M., Benci J.L., Irianto J., Discher D.E., Minn A.J., and Greenberg R.A.. 2017. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 548:466–470. 10.1038/nature23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtlova A., Erttmann S.F., Raffi F.A., Schmalz A.M., Resch U., Anugula S., Lienenklaus S., Nilsson L.M., Kröger A., Nilsson J.A., et al. 2015. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 42:332–343. 10.1016/j.immuni.2015.01.012 [DOI] [PubMed] [Google Scholar]
- Hütteroth T.H., Litwin S.D., and German J.. 1975. Abnormal immune responses of Bloom’s syndrome lymphocytes in vitro. J. Clin. Invest. 56:1–7. 10.1172/JCI108058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson E.M., Rodero M.P., Kasher P.R., Uggenti C., Oojageer A., Goosey L.C., Rose Y., Kershaw C.J., Urquhart J.E., Williams S.G., et al. 2016. Mutations in SNORD118 cause the cerebral microangiopathy leukoencephalopathy with calcifications and cysts. Nat. Genet. 48:1185–1192. 10.1038/ng.3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser W.J., Kaufman J.L., and Offermann M.K.. 2004. IFN-alpha sensitizes human umbilical vein endothelial cells to apoptosis induced by double-stranded RNA. J. Immunol. 172:1699–1710. 10.4049/jimmunol.172.3.1699 [DOI] [PubMed] [Google Scholar]
- Karow J.K., Constantinou A., Li J.L., West S.C., and Hickson I.D.. 2000. The Bloom’s syndrome gene product promotes branch migration of holliday junctions. Proc. Natl. Acad. Sci. USA. 97:6504–6508. 10.1073/pnas.100448097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann A., Gentleman R., and Huber W.. 2009. arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics. 25:415–416. 10.1093/bioinformatics/btn647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N., Motoyoshi F., Mori S., Kuwabara N., Orii T., and German J.. 1992. Long-term study of the immunodeficiency of Bloom’s syndrome. Acta Paediatr. 81:86–90. 10.1111/j.1651-2227.1992.tb12088.x [DOI] [PubMed] [Google Scholar]
- Lahaye X., Satoh T., Gentili M., Cerboni S., Conrad C., Hurbain I., El Marjou A., Lacabaratz C., Lelièvre J.D., and Manel N.. 2013. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity. 39:1132–1142. 10.1016/j.immuni.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Lahkim Bennani-Belhaj K., Rouzeau S., Buhagiar-Labarchède G., Chabosseau P., Onclercq-Delic R., Bayart E., Cordelières F., Couturier J., and Amor-Guéret M.. 2010. The Bloom syndrome protein limits the lethality associated with RAD51 deficiency. Mol. Cancer Res. 8:385–394. 10.1158/1541-7786.MCR-09-0534 [DOI] [PubMed] [Google Scholar]
- Lan Y.Y., Londoño D., Bouley R., Rooney M.S., and Hacohen N.. 2014. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Reports. 9:180–192. 10.1016/j.celrep.2014.08.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., and Chen Z.J.. 2018. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 215:1287–1299. 10.1084/jem.20180139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Santoro I.M., McDaniel L.D., Nishijima I., Mills M., Youssoufian H., Vogel H., Schultz R.A., and Bradley A.. 2000. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 26:424–429. 10.1038/82548 [DOI] [PubMed] [Google Scholar]
- Mackenzie K.J., Carroll P., Martin C.A., Murina O., Fluteau A., Simpson D.J., Olova N., Sutcliffe H., Rainger J.K., Leitch A., et al. 2017. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 548:461–465. 10.1038/nature23449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel N., Hogstad B., Wang Y., Levy D.E., Unutmaz D., and Littman D.R.. 2010. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature. 467:214–217. 10.1038/nature09337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyts I., and Casanova J.L.. 2016. A human inborn error connects the α’s. Nat. Immunol. 17:472–474. 10.1038/ni.3420 [DOI] [PubMed] [Google Scholar]
- Nguyen G.H., Tang W., Robles A.I., Beyer R.P., Gray L.T., Welsh J.A., Schetter A.J., Kumamoto K., Wang X.W., Hickson I.D., et al. 2014. Regulation of gene expression by the BLM helicase correlates with the presence of G-quadruplex DNA motifs. Proc. Natl. Acad. Sci. USA. 111:9905–9910. 10.1073/pnas.1404807111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas N., Moshous D., Cavazzana-Calvo M., Papadopoulo D., de Chasseval R., Le Deist F., Fischer A., and de Villartay J.P.. 1998. A human severe combined immunodeficiency (SCID) condition with increased sensitivity to ionizing radiations and impaired V(D)J rearrangements defines a new DNA recombination/repair deficiency. J. Exp. Med. 188:627–634. 10.1084/jem.188.4.627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinsignon C., Moshous D., Callebaut I., de Chasseval R., Villey I., and de Villartay J.P.. 2004. The metallo-beta-lactamase/beta-CASP domain of Artemis constitutes the catalytic core for V(D)J recombination. J. Exp. Med. 199:315–321. 10.1084/jem.20031142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek H., Luff J., Cheung K., Kozlov S., Gatei M., Lee C.S., Bellingham M.C., Noakes P.G., Lim Y.C., Barnett N.L., et al. 2017. Rats with a missense mutation in Atm display neuroinflammation and neurodegeneration subsequent to accumulation of cytosolic DNA following unrepaired DNA damage. J. Leukoc. Biol. 101:927–947. 10.1189/jlb.4VMA0716-316R [DOI] [PubMed] [Google Scholar]
- Raab M., Gentili M., de Belly H., Thiam H.R., Vargas P., Jimenez A.J., Lautenschlaeger F., Voituriez R., Lennon-Duménil A.M., Manel N., and Piel M.. 2016. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 352:359–362. 10.1126/science.aad7611 [DOI] [PubMed] [Google Scholar]
- Ralf C., Hickson I.D., and Wu L.. 2006. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 281:22839–22846. 10.1074/jbc.M604268200 [DOI] [PubMed] [Google Scholar]
- Rao V.A., Conti C., Guirouilh-Barbat J., Nakamura A., Miao Z.H., Davies S.L., Saccá B., Hickson I.D., Bensimon A., and Pommier Y.. 2007. Endogenous gamma-H2AX-ATM-Chk2 checkpoint activation in Bloom’s syndrome helicase deficient cells is related to DNA replication arrested forks. Mol. Cancer Res. 5:713–724. 10.1158/1541-7786.MCR-07-0028 [DOI] [PubMed] [Google Scholar]
- Rice G.I., Forte G.M., Szynkiewicz M., Chase D.S., Aeby A., Abdel-Hamid M.S., Ackroyd S., Allcock R., Bailey K.M., Balottin U., et al. 2013. Assessment of interferon-related biomarkers in Aicardi-Goutières syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. Lancet Neurol. 12:1159–1169. 10.1016/S1474-4422(13)70258-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodero M.P., Decalf J., Bondet V., Hunt D., Rice G.I., Werneke S., McGlasson S.L., Alyanakian M.A., Bader-Meunier B., Barnerias C., et al. 2017a Detection of interferon alpha protein reveals differential levels and cellular sources in disease. J. Exp. Med. 214:1547–1555. 10.1084/jem.20161451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodero M.P., Tesser A., Bartok E., Rice G.I., Della Mina E., Depp M., Beitz B., Bondet V., Cagnard N., Duffy D., et al. 2017b Type I interferon-mediated autoinflammation due to DNase II deficiency. Nat. Commun. 8:2176 10.1038/s41467-017-01932-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A., Jackson R., Harman C.C., Li T., West A.P., de Zoete M.R., Wu Y., Yordy B., Lakhani S.A., Kuan C.Y., et al. 2014. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 159:1563–1577. 10.1016/j.cell.2014.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., and Rice C.M.. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 472:481–485. 10.1038/nature09907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson D.B., Ko J.S., Heidmann T., and Medzhitov R.. 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 134:587–598. 10.1016/j.cell.2008.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wu J., Du F., Chen X., and Chen Z.J.. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 339:786–791. 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissari J., Sirén J., Meri S., Julkunen I., and Matikainen S.. 2005. IFN-alpha enhances TLR3-mediated antiviral cytokine expression in human endothelial and epithelial cells by up-regulating TLR3 expression. J. Immunol. 174:4289–4294. 10.4049/jimmunol.174.7.4289 [DOI] [PubMed] [Google Scholar]
- Tohyama M., Dai X., Sayama K., Yamasaki K., Shirakata Y., Hanakawa Y., Tokumaru S., Yahata Y., Yang L., Nagai H., et al. 2005. dsRNA-mediated innate immunity of epidermal keratinocytes. Biochem. Biophys. Res. Commun. 335:505–511. 10.1016/j.bbrc.2005.07.105 [DOI] [PubMed] [Google Scholar]
- Van Kerckhove C.W., Ceuppens J.L., Vanderschueren-Lodeweyckx M., Eggermont E., Vertessen S., and Stevens E.A.. 1988. Bloom’s syndrome: clinical features and immunologic abnormalities of four patients. Am. J. Dis. Child. 142:1089–1093. 10.1001/archpedi.1988.02150100083032 [DOI] [PubMed] [Google Scholar]
- West A.P., Khoury-Hanold W., Staron M., Tal M.C., Pineda C.M., Lang S.M., Bestwick M., Duguay B.A., Raimundo N., MacDuff D.A., et al. 2015. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 520:553–557. 10.1038/nature14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.J., McArthur K., Metcalf D., Lane R.M., Cambier J.C., Herold M.J., van Delft M.F., Bedoui S., Lessene G., Ritchie M.E., et al. 2014. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 159:1549–1562. 10.1016/j.cell.2014.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S.R., Fuertes M.B., Corrales L., Spranger S., Furdyna M.J., Leung M.Y., Duggan R., Wang Y., Barber G.N., Fitzgerald K.A., et al. 2014. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 41:830–842. 10.1016/j.immuni.2014.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., and Hickson I.D.. 2003. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 426:870–874. 10.1038/nature02253 [DOI] [PubMed] [Google Scholar]
- Yang H., Wang H., Ren J., Chen Q., and Chen Z.J.. 2017. cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. USA. 114:E4612–E4620. 10.1073/pnas.1705499114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankiwski V., Marciniak R.A., Guarente L., and Neff N.F.. 2000. Nuclear structure in normal and Bloom syndrome cells. Proc. Natl. Acad. Sci. USA. 97:5214–5219. 10.1073/pnas.090525897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Katlinskaya Y.V., Carbone C.J., Zhao B., Katlinski K.V., Zheng H., Guha M., Li N., Chen Q., Yang T., et al. 2015. DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function. Cell Reports. 11:785–797. 10.1016/j.celrep.2015.03.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.