Abstract
Four cobalt(III) complexes of the general formula [Co(Schiff base)(L)2]+, where L is ammonia (NH3) or 3-fluorobenzylamine (3F-BnNH2), were synthesized. The complexes were characterized by NMR spectroscopy, mass spectrometry, and X-ray crystallography. Their electrochemical properties, ligand substitution mechanisms, and ligand exchange rates in aqueous buffer were investigated. These physical properties were correlated to the cellular uptake and anticancer activities of the complexes. The complexes undergo sequential, dissociative ligand exchange, with the exchange rates depending heavily on the axial ligands. Eyring analyses revealed that the relative ligand exchange rates were largely impacted by differences in the entropy, rather than enthalpy, of activation for the complexes. Performing the substitution reactions in the presence of ascorbate led to a change in the reaction profile and kinetics, but no change in the final product. The cytotoxic activity of the complexes correlates with both the ligand exchange rate and reduction potential, with the more easily reduced and rapidly substituted complexes showing higher toxicity. These relationships may be valuable for the rational design of Co(III) complexes as anticancer or antiviral prodrugs.
Introduction
Cobalt is an essential element for human health predominately in its role as the central metal ion in the cofactor vitamin B12.1,2 The in vivo compatibility of cobalt has motivated the investigation of coordination complexes of this metal as less toxic alternatives to conventional precious metal-based therapeutic agents, such as cisplatin.3–5 The medicinal applications of cobalt complexes have been explored in the context of imaging agents,6–9 drug-delivery scaffolds,6,10–17 anticancer agents,14,18–22 enzyme inhibitors,23–32 and antiviral drugs.33–36 The unique chemical properties of cobalt, in comparison to other first row transition metal ions, render it particularly useful for medicinal applications. In the +3 oxidation state, for example, this ion is kinetically inert, undergoing ligand substitution reactions on a much slower timescale than its neighbors on the periodic table.37–39 In contrast, the +2 oxidation state of cobalt is labile. The dichotomy between the inertness of these two oxidation states has enabled the development of Co(III) prodrugs that undergo reduction in biological systems to form labile Co(II) complexes, which subsequently release their ligands as a cytotoxic payload.40–43 This reductive release strategy has been investigated for several different classes of Co(III) complexes with the ultimate objective of selectively targeting hypoxic environments.6,10,12,13,17,44–48
In addition to exploiting the Co(III) redox properties for developing redox-activated prodrugs, the slow ligand exchange kinetics of Co(III) have been used to design enzyme inhibitors.24,26 Within this class of Co(III) complexes, the most studied are those of the general structural formula [Co(acacen)(L)2]+ where acacen = N,N′-bis(acetylacetone)ethylenediamine and L is usually a neutral N-donor ligand. These octahedral [Co(acacen)(L)2]+ complexes undergo ligand substitution reactions with histidine residues in the active sites of proteins to irreversibly inhibit them.25,29,49–51 Because of their efficacy against serine proteases, these complexes have been explored for the treatment of viruses, such as HIV and herpes, which rely heavily on these enzymes.33–35 For example, the compound [Co(acacen)(2-methylimidazole)2]+ is currently in clinical trials for the treatment of herpes.3,4 Functionalization of the ligand scaffold of these complexes allows for fine tuning of the ligand substitution rate, which can dramatically affect their enzyme-inhibitory activities.52–54 Furthermore, the facile synthesis of these complexes has allowed for the design of light-activated31,32 and targeted versions of these inhibitors to improve their selectivity.24,25,27
In this study, we aimed to investigate the physical properties, ligand exchange kinetics, and biological activity of a small subset of [Co(Schiff base)(L)2]+ complexes with the objective of understanding ligand substitution processes in this class of compounds. The complexes shown in Chart 1, bearing either 3-fluorosalicylaldehyde ethylenediamine- (3F-salen) or trifluoroacetylacetone ethylenediamine-(tfacen) based equatorial ligands and NH3 or 3-fluorobenzylamine (3F-BnNH2) axial ligands, were chosen to investigate the role of both the equatorial and axial ligands on their activation mechanisms. The use of these fluorinated ligands was motivated by several reasons. First, the presence of fluorine in these complexes provides a useful NMR handle for studying their speciation in solution via 19F NMR spectroscopy.9,55–58 Second, equatorial ligands containing electron-withdrawing groups, such as fluorine, give rise to Co(III) complexes with slow ligand substitution kinetics,50 which may be more therapeutically useful. The kinetic inertness of fluorinated complexes in comparison to their non-fluorinated analogues makes them much easier to isolate, and these slow kinetics are also much more amenable to measurement. Finally, novel imaging opportunities are available for fluorine-containing compounds via 19F magnetic resonance imaging or 18F positron emission tomography.9,57–61 The ease of detection and altered pharmacokinetics of fluorinated compounds make this element ideal for use in drugs, as reflected by the fact that over 20% of currently administered drugs contain fluorine.62
Chart 1.
Structures of the [Co(Schiff base)(L)2]+ Complexes Investigated in This Work
After synthesizing the complexes and confirming their basic structure and stability, the complexes were analyzed via cyclic voltammetry to ascertain the favorability and reversibility of reduction, and their ligand exchange rates, activation parameters, and reaction mechanisms were evaluated. These properties were then correlated with the relative cytotoxicity and uptake of the complexes in cancer cells. The combined results provide valuable insight into the both the thermal and electrochemical reactivity of this class of complexes and may help future researchers to design Co(III) complexes as enzyme inhibitors or cytotoxic agents.
Experimental
Materials and Methods
CoCl2·6H2O, IRA410(Cl) anion exchange resin, and N-methylimidazole (MeIm) were obtained from Alfa Aesar (Tewksbury, MA) and used as received. The compound 3-fluorobenzylamine was obtained from Oakwood Chemical (Estill, SC) and used as received. The Schiff base ligands 3F-salen63 and tfacen50 were synthesized according to previously published procedures. The Co(III) complexes, [Co(3F-salen)(MeIm)2]Cl and [Co(tfacen)(MeIm)2]Cl, were synthesized following a previously described procedure.23 These complexes were used only to confirm the identities of the final products obtained in our ligand substitution kinetic studies. Solvents were of ACS grade or higher. CHAPS (3-[(3-chloamidopropyl)dimethylammonio]-1-propanesulfonate) lysis buffer was prepared using 1% CHAPS buffer by mass, 5 mM ethylenediaminetetraacetic acid (EDTA), 50 mM tris(hydroxymethyl)aminomethane (Tris), and 110 mM NaCl; the pH was adjusted to 7.4 using dilute HCl or NaOH as necessary. Ultrapure 3-(N-morpholino)propanesulfonic acid (MOPS) buffer was obtained from VWR Life Sciences (Radnor, PA). All reactions were performed under ambient atmospheric conditions without any attempts to exclude oxygen or water.
Physical Measurements
NMR spectra were acquired on a 500 MHz Bruker AV 3HD-spectrometer equipped with a broadband Prodigy cryoprobe. 1H and 13C{1H} NMR spectra in MeOD-d4 were referenced to the residual solvent peaks at 3.31 and 49.00 ppm, respectively, and 1H and 13C{1H} NMR spectra in D2O were referenced to a p-dioxane internal standard at 3.75 and 67.19 ppm, respectively.64 19F NMR spectra were referenced internally to sodium trifluoroacetate (NaTFA) in H2O at –75.51 ppm,65,66 and 59Co NMR spectra were referenced to an external standard of K3[Co(CN)6] in D2O at 0 ppm.67 Samples for IR spectroscopy were prepared as KBr pellets and were analyzed on a Nicolet Avatar 370 DTG FTIR spectrometer. Graphite furnace atomic absorption spectroscopy (GFAAS) measurements were performed with a PerkinElmer PinAAcle 900z instrument. Electrochemical measurements were carried out using a Pine WaveNow potentiostat with a three-electrode setup consisting of a glassy carbon working electrode, a platinum counter electrode, and an Ag wire quasi-reference electrode. Complexes were dissolved in anhydrous DMF with 0.10 M [Bu4N][PF6] (TBAP) as the supporting electrolyte. Potentials were referenced using an internal standard of the ferrocene/ferricenium couple at 0.45 V vs the saturated calomel electrode (SCE).68,69 The sample cell was deoxygenated by bubbling nitrogen gas through the solution prior to analysis and maintained under a blanket of nitrogen during the experiment. Analytical high-performance liquid chromatography (HPLC) was performed using a Shimadzu LC20-AT HPLC with an Ultra Aqueous C18 column, 100 Å, 5 µm, 250 mm x 4.6 mm (Restek, Bellefonte, PA) and an SPD-20AV UV/vis detector monitoring at 220 and 260 nm. The flow rate for all HPLC analyses was 1 mL/min. Gradient elution for purity analysis was performed as follows: 10% methanol in water containing 0.1% TFA to 100% methanol containing 0.1% TFA over 20 min. Gradient elution for kinetics experiments was performed using 10% methanol in water containing 0.1% TFA to 100% methanol containing 0.1% TFA over 15 min, then continuing at 100% methanol for 10 minutes. Isocratic elution was used to determine retention factors. The isocratic mobile phase for all complexes was 65% methanol in water containing 0.1% TFA. The dead time of the instrument was determined using thiourea as a solute. Retention factors (RF) were calculated using the equation RF = (tr – t0)/t0, where tr is the retention time of the complex and t0 is the retention time of thiourea.70,71 High-resolution mass spectra (HRMS) were recorded on an Exactive Orbitrap mass spectrometer in positive electrospray ionization (ESI) mode (ThermoFisher Scientific, Waltham, MA). Elemental analyses (CHN) were performed by Atlantic Microlab Inc., Norcross, GA, USA.
[Co(3F-salen)(3F-BnNH2)2]Cl
Solid CoCl2·6H2O (0.203 g, 0.854 mmol) was added to a suspension of 3F-salen (0.26 g, 0.854 mmol) in isopropanol (10 mL). The yellow suspension of 3F-salen immediately turned dark green, and the mixture was heated at 70 °C with stirring for 1 h. The axial ligand 3F-BnNH2 (0.63 mL, 5.5 mmol) was added, and the mixture was heated with stirring for 2 h, during which time it slowly turned red. The resulting red-brown suspension was cooled to room temperature, and the precipitate was collected by filtration. The solid product was washed with water, isopropanol, and diethyl ether and dried under vacuum to yield a dark red solid. Yield 0.15 g (27%). 1H NMR (500 MHz, MeOD-d4) δ 8.25 (s, 2H), 7.16–7.28 (m, 6H), 6.92 (td, 2H, J = 8.6, 2.5 Hz), 6.86 (d, 2H, J = 8 Hz), 6.82 (d, 2H, J = 10 Hz), 6.61–6.67 (m, 2H), 4.05 (s, 4H), 3.27 (s, 2H). 19F NMR (470 MHz, H2O) δ –113.26 (m, 2F), –135.64 (d, 2F, JH-F = 12 Hz). 13C{1H} NMR (126 MHz, MeOD-d4) δ 170.9 (d, JF-C = 3 Hz), 164.2 (d, JF-C = 248 Hz), 157.7 (d, JF-C = 243 Hz), 155.0 (d, JF-C = 13 Hz), 141.60 (d, JF-C = 7 Hz), 141.57 (d, JF-C = 7 Hz), 131.5 (d, JF-C = 8 Hz), 131.2 (d, JF-C = 4 Hz), 125.5 (d, JF-C = 3 Hz), 122.0 (d, JF-C = 5 Hz), 121.0 (d, JF-C = 19 Hz), 116.4 (d, JF-C = 21 Hz), 115.8 (d, JF-C = 7 Hz), 115.6 (d, JF-C = 21 Hz), 59.6, 45.8. IR (KBr, cm−1): 3338 w, 3271 w, 3100 m, 2944 w, 2907 m, 2857 w, 1636 s, 1614 m, 1551 w, 1453 s, 1409 w, 1395 w, 1337 w, 1351 w, 1307 s, 1298 s, 1240 s, 1099 m, 1057 m, 865 m, 778 m, 734 s. ESI-MS (positive ion mode): m/z [M]+ 611.147, calcd 611.148. Anal. Calcd for [Co(3F-salen)(3F-BnNH2)2]Cl (C30H28N4O2F4CoCl): C, 55.70; H, 4.36; N, 8.66. Found: C, 55.44; H, 4.63; N, 8.83.
[Co(3F-salen)(NH3)2]Cl
CoCl2·6H2O (0.52 g, 2.17 mmol) was added to 3F-salen (0.66 g, 2.17 mmol) in methanol (35 mL). The yellow suspension of 3F-salen immediately turned dark green, and the mixture was heated at 60 °C with stirring for 1 h. Concentrated NH4OH (1 mL, 18 mmol) was added, and the mixture was stirred at room temperature for 2 h. The resulting red solution was filtered, and excess aqueous NaPF6 was added to the filtrate, which resulted in the precipitation of a red-brown solid. The solid was washed sequentially with water and diethyl ether, dissolved in methanol (20 mL), and stirred with approximately 5 g of anion exchange resin at room temperature for 2 h. The dark red solution was filtered to remove resin. The filtrate was evaporated to dryness and further dried under high vacuum to yield a dark red solid. Yield 0.31 g (33%). 1H NMR (500 MHz, D2O) δ 8.23 (s, 2H), 7.21 (d, 2H JH-H = 7.9 Hz), 6.97 (m, 2H), 6.58 (m, 2H), 4.07 (s, 4H). 19F NMR (470 MHz, H2O) δ –135.97 (d, 2F, JH-F = 9.7 Hz). IR (KBr, cm−1): 3300 w, 3017 w, 1652 m, 1629 s, 1587 w, 1453 s, 1324 m, 1312 m, 1241 m, 1227 m, 871 w, 792 w, 738 m. ESI-MS (positive ion mode): m/z [M]+ 395.072, calcd 395.073. Anal. Calcd for [Co(3F-salen)(NH3)2]Cl·0.2H2O (C16H18.4N4O2.2F2CoCl): C, 44.25; H, 4.27; N, 12.90. Found: C, 44.02; H, 4.25; N, 12.99.
[Co(tfacen)(3F-BnNH2)2]Cl
CoCl2·6H2O (0.265 g, 1.11 mmol) was added to tfacen (0.37 g, 1.11 mmol) in methanol (10 mL). The white suspension of tfacen immediately turned dark green, and the mixture was heated at 60 °C with stirring for 1 h. The axial ligand 3F-BnNH2 (0.6 mL, 5.2 mmol) was added, and the mixture was stirred at room temperature overnight. The resulting red solution was filtered, and excess aqueous NaPF6 was added to the filtrate, which resulted in the precipitation of a red-brown solid. The solid was washed with water and diethyl ether, then dissolved in methanol (20 mL) and stirred with approximately 5 g anion exchange resin at room temperature for 2 h. The dark red solution was filtered to remove resin, evaporated to dryness, and further dried under high vacuum to yield a light brown solid. Yield 0.35 g (47%). 1H NMR (500 MHz, MeOD-d4) δ 7.34 (m, 2H), 7.03 (m, 6H), 5.78 (s, 2H), 3.68 (s, 4H). 19F NMR (470 MHz, H2O) δ –72.33 (s, 6F), –113.13 (m, 2F). 13C{1H} NMR (126 MHz, MeOD-d4) δ 175.41, 164.30 (d, JF-C = 245.3 Hz), 161.05 (q, JF-C = 32.5 Hz), 141.63 (d, JF-C = 7.2 Hz), 131.65 (d, JF-C = 8.3 Hz), 125.65 (d, JF-C = 2.8 Hz), 118.83 (d, JF-C = 280.6 Hz), 116.54 (d, JF-C = 21.8 Hz), 115.75 (d, JF-C = 21.3 Hz), 96.54, 54.77, 45.49, 23.96. IR (KBr, cm−1): 3440 br m, 3052 br m, 1617 s, 1584 m, 1543 m, 1470 m, 1361 w, 1287 s, 1248 s, 1187 s, 1200 s, 900 m, 780 m, 800 m. ESI-MS (positive ion mode): m/z [M]+ 639.141, calcd 639.142. Anal. Calcd for [Co(tfacen)(3F-BnNH2)2]Cl (C26H28N4O2F8CoCl): C, 46.27; H, 4.18; N, 8.30. Found: C, 46.00; H, 4.29; N, 8.14.
[Co(tfacen)(NH3)2]Cl
CoCl2·6H2O (0.269 g, 1.13 mmol) was added to tfacen (0.375 g, 1.13 mmol) in methanol (15 mL). The white suspension of tfacen immediately turned dark green, and the mixture was heated at 60 °C with stirring for 1 h. Concentrated NH4OH (0.5 mL, 9 mmol) was added, and the mixture was stirred at room temperature for 2 h. The resulting solution was filtered, and excess aqueous NaPF6 was added to the filtrate, which resulted in the precipitation of a red-brown solid. The solid was washed with water and diethyl ether, then dissolved in methanol (10 mL) and stirred with approximately 5 g anion exchange resin at room temperature for 2 h. The dark red solution was filtered to remove resin, evaporated to dryness, and further dried under high vacuum to yield a light brown solid. Yield 0.20 g (39%). 1H NMR (500 MHz, D2O) δ 8.23 (s, 2H), 7.21 (d, J = 8.0 Hz, 2H), 7.03 – 6.96 (m, 2H), 6.60 (td, J = 7.9, 4.5 Hz, 4H), 4.07 (s, 4H), 3.76 (s). 19F NMR (470 MHz, H2O) δ –72.37 (s, 6F). 13C{1H} NMR (126 MHz, D2O): δ 187.59, 172.35 (q, JF-C = 32.6 Hz), 131.09 (q, JF-C = 280.0 Hz), 109.52 (q, JF-C = 3.1 Hz), 80.33, 36.46. IR (KBr, cm−1): 3682 w, 3513 w, 3304 w, 3104 w, 1635 m, 1543 m, 1461 m, 1304 s, 1187 s, 1117 s, 900 s, 787 s. ESI-MS (positive ion mode): m/z [M]+ 423.066, calcd 423.067. Anal. Calcd for [Co(tfacen)(NH3)2]Cl·1.3H2O (C12H20.6N4O3.3F6CoCl): C, 29.90; H, 4.31; N, 11.62. Found: C, 30.40; H, 4.16; N, 11.14.
X-ray Crystallography
Single crystals of the tfacen complexes were obtained by vapor diffusion of diethyl ether into methanol solutions of these compounds. Diffusion of diethyl ether into a dimethylformamide (DMF) solution of [Co(3F-salen)(3F-BnNH2)2]Cl afforded suitable crystals for single-crystal X-ray diffraction studies. Low-temperature X-ray diffraction data for [Co(3F-salen)(3F-BnNH2)2]Cl, [Co(tfacen)( 3F-BnNH2)2]Cl, and [Co(tfacen)(NH3)2]Cl were collected on a Rigaku XtaLAB Synergy diffractometer coupled to a Rigaku Hypix detector with Mo Kα radiation (λ = 0.71073 Å) from a PhotonJet micro-focus X-ray source at 100 K. The structures were solved through intrinsic phasing using SHELXT72 and refined against F2 on all data by full-matrix least squares with SHELXL73 following established refinement strategies.74 All non-hydrogen atoms were refined anisotropically. All hydrogen atoms bound to carbon were included in the model at geometrically calculated positions and refined using a riding model. Hydrogen atoms bound to nitrogen were located in the difference Fourier synthesis map and subsequently refined semi-freely with the help of distance restraints. The isotropic displacement parameters of all hydrogen atoms were fixed to 1.2 times the Ueq value of the atoms they are linked to (1.5 times for CH3 or NH3 groups). The structure of [Co(3F-salen)(3F-BnNH2)2]Cl contained a disordered DMF molecule; the two disordered components were refined with appropriate similarity restraints, allowing the occupancy of each component to refine freely with net occupancy of both components summing to one. Likewise, in the structure of [Co(3F-salen)(3F-BnNH2)2]Cl, one of the axial 3F-BnNH2 ligands is rotationally disordered about the Co–N axis. The two disordered components were refined as described above. The complex [Co(tfacen)(3F-BnNH2)2]+ resides on a crystallographic 2-fold axis. Additionally, this compound crystallized in a chiral space group. Refinement of the absolute structure revealed a Flack parameter of –0.006(8), indicating that this crystal was enantiomerically pure, rather than a racemic twin. Details of the data quality and a summary of the residual values of the refinements are listed in Table 1.
Table 1.
X-ray Crystallographic Data Collection and Refinement Parameters
| [Co(3F-salen)(3F-BnNH2)2]Cl·DMF | [Co(tfacen)(3F-BnNH2))2]Cl·Et2O | [Co(tfacen)(NH3)2]Cl·MeOH | |
|---|---|---|---|
| formula | C33H35CoN5O3F4Cl | C30H38CoN4O3F8Cl | C13H32CoN4O3F6Cl |
| Fw | 720.04 | 749.02 | 490.72 |
| space group | P21/n | P 43212 | |
| a, Å | 10.9482(2) | 15.4493(3) | 7.9661(2) |
| b, Å | 22.6458(5) | 15.4493(3) | 11.0108(2) |
| c, Å | 13.1929(3) | 14.4740(5) | 11.9024(2) |
| α, deg | 90 | 90 | 65.768(2) |
| β, deg | 100.046(2) | 90 | 89.306(2) |
| γ, deg | 90 | 90 | 82.092(2) |
| V, Å3 | 3220.78(12) | 3454.67(18) | 941.82(4) |
| Z | 4 | 4 | 2 |
| ρcalcd g·cm−3 | 1.485 | 1.440 | 1.730 |
| T, K | 100.00(10) | 100.00(10) | 100.00(10) |
| µ(Mo Kα), mm−1 | 0.682 | 0.654 | 1.133 |
| θ range, deg | 2.092 to 26.373 | 2.336 to 27.103 | 1.879 to 29.128 |
| completeness to θ, % | 100.0 | 99.9 | 100.0 |
| total no. of data | 32486 | 16169 | 23740 |
| no. of unique data | 6588 | 3800 | 5022 |
| no. of param | 496 | 217 | 277 |
| no. of restraints | 196 | 483 | 7 |
| R1a, % | 2.95 | 5.92 | 3.31 |
| wR2b, % | 7.06 | 15.47 | 6.75 |
| GOFc | 1.053 | 1.048 | 1.058 |
| max, min peaks e·Å−1 | 0.577, −0.271 | 0.790, –0.520 | 0.499, –0.406 |
R1 = Σ||Fo| − |Fc||/Σ|Fo| for all data.
wR2 = {Σ[w(Fo2 – Fc2)2]/Σ[w(Fo2)2]}1/2 for all data.
GoF = {Σ[w(Fo2 − Fc2)2]/(n − p)}1/2, where n is the number of data and p is the number of refined parameters.
Ligand Exchange Experiments
To probe the axial ligand substitution of these complexes, solutions were prepared to contain 0.5 mM Co complex, 0.5 mM NaTFA, 100 mM MOPS buffer (pH 7.4), and 12.5 mM MeIm as an entering ligand. The pH of the resulting solutions was measured to verify that the MOPS buffer fixed it at 7.4. Immediately after preparation, samples were analyzed via 19F NMR spectroscopy using the NaTFA signal as an internal standard or by HPLC using the relative integration of each peak. 19F NMR spectra or HPLC chromatograms were acquired periodically to monitor the ligand substitution reaction with MeIm. For reactions carried out at temperatures greater than 37 °C, aliquots were removed and flash-frozen prior to analysis by HPLC. Pseudo first-order rate constants for the decay of the starting complex were calculated by plotting the peak integration vs time and fitting these data to the appropriate first-order integrated rate law using the Magic Plot Pro software. Rate constants for the reaction of the intermediate species were calculated using the appropriate integrated rate laws for this process.75,76 The reported values at 18 and 37 °C are the average of at least three independent replicates, whereas data at higher temperatures were collected for a single kinetics run.
The ligand exchange reactions of all complexes with MeIm were evaluated in the presence of ascorbate at 37 °C. Solutions were prepared containing 0.5 mM Co complex, 0.5 mM NaTFA, 20 mM ascorbate, 100 mM MOPS buffer (pH 7.4), and 20 mM MeIm as an entering ligand. The exchange reactions were monitored via HPLC. The first order rate constants and half-life for the decay of the starting material were calculated using the Magic Plot Pro software by fitting the date to a standard exponential decay curve.
Cell Culture Conditions
A549 (human lung cancer) cells were obtained from American Type Culture Collection (ATCC). Cells were cultured as adherent monolayers in an incubator at 37 °C with a humidified atmosphere of 5% CO2. The cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Cells were checked for mycoplasma contamination monthly using the PlasmoTest™ mycoplasma detection kit from InvivoGen.
Cytotoxicity Assay
The colorimetric MTT assay was used to evaluate cytotoxicity.77 Trypsinized cells were plated at 2000 cells/well with 100 µL/well in a 96-well plate and incubated for 24 h. The media was then removed and replaced with 200 μL of growth media containing varying concentrations of the complexes. After 24 h, the culture medium containing the complex was removed, 200 μL of fresh media was added, and plates were incubated an additional 48 h. After this time, the media was removed again, and a solution of thiazolyl blue tetrazolium bromide (MTT) in DMEM (200 µL, 1 mg/mL) was added to each well. Upon incubation for 4 h, the DMEM/MTT solutions were aspirated, and the purple formazan crystals were dissolved in 200 μL of an 8:1 mixture of DMSO: pH 10 glycine buffer. The absorbance of each well at 570 nm was measured using a BioTek Synergy HT platereader. Absorbance values were normalized to the untreated wells and plotted as concentration of cobalt complex vs % viability. The resulting dose-response curves were analyzed using a logistic sigmoid function.78 Reported IC50 values represent the average of three independent experiments, each carried out with six replicates per concentration level. Stock solutions of the cobalt complexes were prepared fresh in 18.2 MΩ·cm H2O prior to serial dilution in the cell culture medium. Stock solutions of free ligands were prepared in DMSO and diluted to less than 1% DMSO by volume with cell culture medium.
Uptake Experiments
Cellular uptake of cobalt was determined by implementing slight modifications to previously reported protocols.79,80 Trypsinized cells (2 × 106) were seeded in six 75 cm2 culture dishes and incubated for 24 h. The media was removed, and the cells were treated with either 0 or 100 µM cobalt complex for 24 h. Dishes containing no cells were also incubated with 100 µM of the cobalt complex to correct for non-specific adsorption of cobalt to the plastic. Media was removed from all dishes, and the cells were rinsed with 3 mL PBS and detached with trypsin (3 mL). The cells were centrifuged at 1000 rpm for 10 min. The supernatant was discarded, and the pellet was resuspended in 1 mL PBS. Samples were centrifuged and resuspended twice more using the same conditions to remove extracellular cobalt and centrifuged a final time to pellet cells. The pellet was resuspended in ice-cold, ultrapure 1× CHAPS lysis buffer, and samples were gently agitated for 30 min. The protein concentration in each sample was then determined using the Thermo Fisher Bicinchoninic Acid Protein Assay Kit according to the manufacturer’s instructions. The cobalt concentration in each sample was determined using GFAAS. Results were reported as the mass ratio of cobalt to protein (pg/µg) in each sample.
Results
Synthesis of [Co(Schiff base)(L)2]Cl Complexes
The four complexes, shown in Chart 1, were targeted to study the effects of both the equatorial and axial ligands on the nature of the biological activity of this class of [Co(Schiff base)(L)2]+ complexes. The syntheses were straightforward, following closely with procedures found in the literature for related complexes.23 Briefly, a mixture of CoCl2·6H2O and 1 equiv of the Schiff base ligand was heated to reflux in methanol for 1 h to yield a dark green solution. This dark green solution, which contains an uncharacterized intermediate, was treated with an excess of the axial ligand to form the desired red-brown Co(III) complexes. As observed during the synthesis of related Co(III) complexes, the oxidation of the initial Co(II) starting material is most likely mediated by atmospheric oxygen. The low-spin diamagnetic nature of these complexes was confirmed by 1H NMR spectroscopy, which showed sharp resonances within the range of 0 to 10 ppm. Further characterization of these complexes was provided by mass spectrometry, showing the expected ion peaks for the intact cationic complexes. 13C{1H} NMR spectroscopy further confirmed the structure and symmetry of the complexes. However, no suitable 13C{1H} NMR spectrum could be obtained for [Co(3F-salen)(NH3)2]+ due to its poor solubility. In addition to these spectroscopic methods, elemental analysis and HPLC further verified these compounds to be greater than 95% pure. (Fig. S1–S2I†).
X-Ray Crystallography
The complexes [Co(3F-salen)(3F-BnNH2)2]+, [Co(tfacen)(3F-BnNH2)2]+, and [Co(tfacen)(NH3)2]+ were further characterized by single-crystal X-ray diffraction. The crystal structures are shown in Fig. 1, and selected interatomic distances and angles are collected in Table 2. The Co(III) center attains the expected pseudo-octahedral geometry comprising the four donor atoms of the Schiff base ligand in the equatorial plane and two of the monodentate nitrogen donors in a trans arrangement. In these three structures, the equatorial ligand does not reside in a perfectly planar configuration, a feature that is most notable for [Co(3F-salen)(3F-BnNH2)2]+. The equatorial Co–N and Co–O distances vary slightly among the three complexes. These distances are longest for [Co(tfacen)(NH3)2]+, which range from 1.904 to 1.910 Å, and shortest for [Co(3F-salen)(3F-BnNH2)]+, with values between 1.894 to 1.899 Å. The longer distances observed for [Co(tfacen)(NH3)2]+ may be a result of the strong electron donating character of the axial NH3 ligand, which decreases the Lewis acidity of the metal center.81 In comparing the axial ligand distances, this trend is the opposite. Namely, the axial Co–N distances of [Co(tfacen)(3F-BnNH2)2]+ (1.972 Å) are longer than those of [Co(tfacen)(NH3)2]+ (1.967 and 1.961 Å). The smaller steric profile of the NH3 may partly account for the shorter Co–Naxial distances in [Co(tfacen)(NH3)2]+ compared to [Co(tfacen)(3F-BnNH2)2]+.
Fig. 1.
X-ray crystal structures of [Co(3F-salen)(3F-BnNH2)2]+, [Co(tfacen)(3F-BnNH2)2]+, and [Co(tfacen)(NH3)2]+. Ellipsoids are drawn at the 50% probability level. Non-acidic hydrogen atoms, counterions, and solvent molecules are omitted for clarity.
Table 2.
Selected Interatomic Distances (Å) and Angles (°) of [Co(3F-salen)(3F-BnNH2)2]+, [Co(tfacen)(3F-BnNH2)2]+, and [Co(tfacen)(NH3)2]+a
| Interatomic Distance | [Co(3F-salen)(3F-BnNH2)2]+ | [Co(tfacen)(3F-BnNH2)2]+b | [Co(tfacen)(NH3)2]+ |
|---|---|---|---|
| Co–O1 | 1.8966(9) | 1.898(3) | 1.9106(9) |
| Co–O2 | 1.8938(9) | 1.9069(10) | |
| Co–N1 | 1.8942(11) | 1.902(4) | 1.9044(12) |
| Co–N2 | 1.8990(11) | 1.9056(11) | |
| Co–N3 | 1.9762(12) | 1.972(4) | 1.9670(12) |
| Co–N4 | 1.9803(12) | 1.9614(12) | |
| Angle | [Co(3F-salen)(3F-BnNH2)2]+ | [Co(tfacen)(3F-BnNH2)2]+ | [Co(tfacen)(NH3)2]+ |
| O1–Co–O2 | 86.98(4) | 84.17(19) | 84.08(4) |
| O1–Co–N1 | 93.25(4) | 178.40(15) | 95.35(4) |
| O1–Co–N2 | 178.48(5) | 94.74(15) | 178.66(5) |
| O1–Co–N3 | 89.76(5) | 90.47(16) | 88.81(5) |
| O1–Co–N4 | 89.68(4) | 87.95(15) | 88.72(5) |
| O2–Co–N1 | 179.13(5) | 179.07(5) | |
| O2–Co–N2 | 94.50(5) | 94.61(5) | |
| O2–Co–N3 | 90.24(5) | 90.75(5) | |
| O2–Co–N4 | 88.52(5) | 89.11(5) | |
| N1–Co–N2 | 85.27(5) | 86.3(3) | 85.97(5) |
| N1–Co–N3 | 90.60(5) | 90.88(16) | 89.96(5) |
| N1–Co–N4 | 90.63(5) | 90.67(16) | 90.15(5) |
| N2–Co–N3 | 89.90(5) | 90.94(5) | |
| N2–Co–N4 | 90.69(5) | 91.54(5) | |
| N3–Co–N4 | 178.67(5) | 177.9(2) | 177.52(5) |
Atoms are labeled as indicated in Fig. 1.
This molecule resides on a crystallographic 2-fold rotation axis. Redundant distances and angles are omitted from the table.
59Co NMR Spectroscopy
All four complexes were characterized by 59Co NMR spectroscopy. The quadrupolar 59Co nucleus is characterized by a ground state nuclear spin of I = 7/2 and is 100% naturally abundant. By virtue of its large quadrupole moment, this nucleus gives rise to broad NMR signals with linewidths ranging from a few hundred to over 20,000 Hz.67 Both the chemical shift and linewidth of 59Co NMR spectra convey valuable information about properties of the complex being measured, such as ligand field splitting and reduction potential.67,82–87 In particular, upfield chemical shifts arise from complexes with larger ligand field splitting, indicating that the chemical shift is a useful parameter for quantifying the donor strength of a given set of ligands. The signal linewidth is dictated by the quadrupolar relaxation rate of the 59Co nucleus, which is a function of the solution tumbling time and electric field gradient imposed by the ligand environment.88 The 59Co NMR chemical shifts and linewidths of all complexes are reported in Table 3, and representative 59Co NMR spectra are shown in Fig. 2. Other spectra are deposited in the Supporting Information (Fig. S22–S25†).
Table 3.
59Co NMR Chemical Shifts and Linewidthsa
| Complex | δ (ppm) | ν1/2 (Hz) |
|---|---|---|
| [Co(3F-salen)(NH3)2]+b | 8340 | 4900 |
| [Co(3F-salen)(3F-BnNH2)2]+c | 8600 | 10000 |
| [Co(tfacen)(NH3)2]+b | 8540 | 3800 |
| [Co(tfacen)(3F-BnNH2)2]+c | 8740 | 5800 |
Chemical shifts are referenced to K3[Co(CN)6] in D2O at 0 ppm.
Obtained in D2O.
Obtained in MeOD-d4.
Fig. 2.
59Co (120 MHz) NMR of spectra of [Co(tfacen)(NH3)2]+ (red) and [Co(tfacen)(3F-BnNH2)2]+ (blue), obtained in D2O and MeOD-d4, respectively at 295 K. Chemical shifts are reported relative to K3[Co(CN)6] in D2O at 0 ppm.
In comparison to complexes bearing axial 3F-BnNH2 ligands, those with axial NH3 ligands give rise to signals that have narrower linewidths and are shifted approximately 200 ppm upfield. This upfield shift reflects the greater ligand donor strength of NH3 compared to 3F-BnNH2. The equatorial ligands have an equally important effect upon the chemical shift. The 59Co resonance of complexes containing the 3F-salen equatorial ligand are shifted approximately 200 ppm upfield and have broader linewidths compared with their tfacen counterparts.
Cyclic Voltammetry
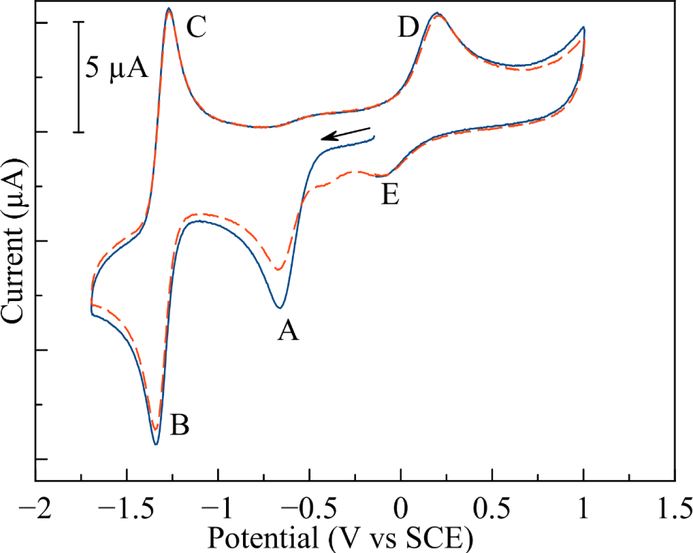
Because of the potential importance of redox activation for this class of compounds, the electrochemical properties of the complexes were analyzed by cyclic voltammetry. The cyclic voltammograms of all complexes were obtained in DMF solution containing 0.1 M TBAP as the supporting electrolyte. A representative voltammogram of the complex [Co(tfacen)(3F-BnNH2)2]+ with redox features labeled is shown in Fig. 3, and relevant reduction potentials for the complexes are reported in Table 4. Cyclic voltammograms of all complexes are shown in Fig. S26–S29†.
Fig. 3.
Cyclic voltammogram of [Co(tfacen)(3F-BnNH2)2]+ in DMF with 0.1 M TBAP supporting electrolyte. Potential is referenced to the SCE based on ferrocene (E1/2 0.45 V vs SCE) as internal standard. The first scan is shown as a solid blue line, while the second scan is shown as a dashed, red line.
Table 4.
Electrochemical Parameters of [Co(Schiff base)(L)2]+ Complexesa
| Complex | Epc Co(III)/Co(II) | E1/2 Co(II)/Co(I) |
|---|---|---|
| [Co(3F-salen)(NH3)2]+ | –0.73 | –1.07 |
| [Co(3F-salen)(3F-BnNH2)2]+ | –0.58 | –1.10 |
| [Co(tfacen)(NH3)2]+ | –0.81 | –1.31 |
| [Co(tfacen)(3F-BnNH2)2]+ | –0.67 | –1.31 |
Potentials are referenced to the SCE. Data were obtained in anhydrous DMF with 0.1 M TBAP using a glassy carbon working electrode. The scan rate was 0.1 V/s, and all data were obtained at 25 °C.
The electrochemical properties of these complexes are comparable to previously studied Co(III) Schiff base complexes.16,23 Upon scanning cathodically, an initial, irreversible reduction (A) is observed. The irreversibility of this feature, which corresponds to the Co(III)/Co(II) couple, arises from the concomitant loss of axial ligands upon the reduction of Co(III). At more negative potentials, a quasi-reversible redox event, corresponding to the Co(II)/Co(I) couple (B/C) is detected. Because at this point the Co(II) complex has lost its axial ligands, the potential of couple B/C is only dependent on the equatorial Schiff base ligand. Scanning anodically, oxidation back to Co(III) (D) results in the formation of a new species, presumed to be the solvent adduct [Co(Schiff base)(DMF)2]+. Subsequent reduction of this solvent-bound species on following scans gives rise to a new reduction event (E), which occurs at a more positive potential compared to the intact complex.
The peak potential of the Co(III)/Co(II) couple is approximately 100 mV more negative for the tfacen complexes compared to those with the 3F-salen ligand. The axial ligands play a more pronounced role in modifying this couple; NH3 complexes are 150 mV more negative than the corresponding 3F-BnNH2 complexes. As mentioned above, the Co(II)/Co(I) (B/C) feature is independent of the axial ligands. This couple is approximately 200 mV more negative for the tfacen complexes than the 3F-salen complexes.
Stability and Ligand Exchange
To assess the kinetic lability of the complexes, they were challenged with 25 equiv of MeIm, a model for a protein histidine side chain, and the ensuing axial ligand substitution reactions were monitored by 19F NMR spectroscopy and RP-HPLC. Under these conditions, all of the complexes exhibit the same general behavior, undergoing two sequential stepwise axial ligand substitution reactions with MeIm (Fig. 4). These reactions are conveniently monitored by 19F NMR spectroscopy, capitalizing on the sensitivity of the 19F resonance of the equatorial Schiff base ligand with respect to the nature of the axial ligands. In monitoring the 19F NMR spectrum over time, an initial new species, the monosubstituted complex, is detected, followed by the much slower formation of the disubstituted product. Representative plots of the concentration of all species over time are shown the ESI (Fig. S30–S33†). To confirm that the final products of these reactions were the disubstituted MeIm complexes [Co(3F-salen)(MeIm)2]Cl and [Co(tfacen)(MeIm)2]Cl, we synthesized them independently and verified that their NMR spectroscopic signatures match those observed in our kinetics experiments (Fig. S34 and S35†). Complexes bearing axial 3F-BnNH2 ligands, which contain fluorine, could also be monitored by the shift in the 19F NMR signal of the 3F-BnNH2. The rate of formation of the 19F resonance of free 3F-BnNH2 is consistent with the rate of decay of the corresponding resonance for the equatorial ligand of the starting complex. HPLC chromatograms of these reactions at different time points reveal the presence of three species, which are assigned to starting material, monosubstituted complex, and disubstituted complex. The relative peak integrations determined by NMR spectroscopy and HPLC are consistent, indicating that both methods are viable for determining the reaction rates.
Fig. 4.
Panel I: Proposed ligand exchange pathway of [Co(Schiff base)(L)2]+ complexes by MeIm. Panel II: 19F NMR spectra of [Co(3F-salen)( 3F-BnNH2)2]+ over time. Chemical shifts are referenced to 0.5 mM NaTFA internal standard at –75.51 ppm.
Changing the concentration of MeIm while maintaining pseudo first-order conditions does not affect the rate of the reaction, indicating that the rate law for this reaction is zero-order with respect to this incoming ligand. For example, incubating with 200 equiv of MeIm rather than 25 equiv yields essentially the same rate constant (Fig. S36†). The observed pseudo first-order rate constants for the decay of the starting material are within an order of magnitude of those reported for Co(III) pentammine halide complexes.37 The independence of the reaction rate on the concentration of MeIm is consistent with a dissociative ligand substitution pathway, a mechanism that has been observed for related [Co(acacen)(L)2]+ complexes.50,52,53 Co(III) ammine complexes are also known to undergo ligand substitution via the conjugate-base mechanism, which requires deprotonation of an ammine ligand to labilize the trans leaving group.38 For this mechanism, the reaction rate is accelerated at higher pH values. When the substitution reaction was carried out at pH 8.3 instead of pH 7.4, no increase in the reaction rate was observed, indicating that the conjugate-base mechanism is not operative within this pH range (Fig. S37†).
The rate constants of these ligand substitution processes were determined by plotting the relative 19F NMR or HPLC peak areas of the starting complex and products versus time and fitting this data to a series of first-order integrated rate laws for a sequential A → B → C reaction pathway. (Equation 1, Fig. S30–S33†).75,76 The resulting pseudo first-order rate constants are given in Table 5, and representative data demonstrating the concentrations of all species over time are shown in Fig. 5. The rate constant for the second ligand substitution step was determined by fitting the concentration vs time data for the monosubstituted complex, [Co(Schiff base)(L)(MeIm)]+, to Equation 2. After solving for the rate constants k1 and k2 using Equations 1 and 2, Equation 3 was used to calculate the predicted product concentration over time. The comparison between the product concentration calculated using Equation 3 and the actual product concentration was used to verify the validity of the calculated rate constants, as shown in Fig. 5. In these equations, A, B, and C refer to the species shown in Fig. 4.
Table 5.
Axial Ligand Substitution Rate Constants of [Co(Schiff base)(L)2]+ Complexes at 37 °C and Activation Parameters Determined by Eyring Analysis
| Complex | k1 × 107 (s−1) (37 °C) | k2 × 107 (s−1) (37 °C) | k1 ∆H‡ (kJ/mol)a | k1 ∆S‡ (J·mol−1 ·K−1) a | k2 ∆H‡ (kJ/mol) a | k2 ∆S‡ (J·mol−1 ·K−1)) a |
|---|---|---|---|---|---|---|
| [Co(3F-salen)(NH3)2]+ | 64.5 ± 0.6 | 33.5 ± 0.2 | 115.3 ± 1.8 | 27.6 ± 0.9 | 130.1 ± 2.9 | 69.9 ± 2.3 |
| [Co(3F-salen)(3F-BnNH2)2]+ | 734 ± 40 | 164 ± 9 | 115.7 ± 3.7 | 50.2 ± 2.4 | 119.3 ± 1.8 | 49.3 ± 1.8 |
| [Co(tfacen)(NH3)2]+ | 39.6 ± 0.2 | 7.98 ± 0.7 | 115.6 ± 0.7 | 24.0 ± 0.2 | 137.4 ± 3.4 | 78.9 ± 2.9 |
| [Co(tfacen)(3F-BnNH2)2]+ | 172 ± 1.0 | 22.5 ± 0.7 | 115.0 ± 3.0 | 35.8 ± 1.3 | 129.4 ± 4.6 | 66.0 ± 3.7 |
Values obtained from Eyring-style plots of reaction rate constant versus temperature. The reported errors are the standard errors based on the least square regression analysis of the data.
Fig. 5.
Representative concentration vs time data for the axial ligand substitution of [Co(3F-salen)(3F-BnNH2)2]+ with MeIm at 37 °C. Concentrations of species (A) (solid blue line, circular marker), (B) (broad dashed orange line, diamond marker), and (C) (short dashed green line, square marker) were fit to equations 1, 2, and 3, respectively. Here, (A), (B), and (C) refer to ligand substitution reactants and products shown in Fig. 4.
| Equation 1: |
| Equation 2: |
| Equation 3: |
The resulting rate constants are strongly dependent on both the axial and equatorial ligands. The complexes with 3F-salen equatorial ligands undergo substitution reactions faster than the analogous tfacen complexes. Likewise, complexes with axial 3F-BnNH2 ligands are more labile than the analogous complexes with NH3 axial ligands. The relative inertness of the NH3 complexes is in contrast to similar acacen complexes,52 which were found to undergo fairly rapid substitution. Related bis(thiosemicarbazone) Co(III) complexes, however, are similar to the Schiff base complexes in this study because they are also more stable with axial NH3 ligands compared to benzylamine ligands.16 The axial ligands have a more pronounced effect on the substitution rate constants compared to the equatorial ligands. This result is expected for the proposed dissociative mechanism because the rate of substitution for this mechanistic pathway primarily depends on the strength of the M–L bond of the dissociating ligand. For all complexes, the substitution of the second axial ligand with MeIm is substantially slower than that of the first axial ligand.
The activation parameters for these reactions were determined via Eyring analysis. These values are given in Table 5, and plots of ln(k/T) vs 1/T are shown in Fig. S38 and S39†. The magnitudes of the enthalpies of activation and the positive values of the entropies of activation observed for all compounds are consistent with a dissociative ligand exchange mechanism.89 In comparing the complexes, the enthalpies of activation for the first step (k1) are all similar, sitting near a value of 115 kJ/mol. The entropies of activation vary more widely in a manner that is dependent on both the equatorial and axial ligands. Complexes with 3F-BnNH2 axial ligand exhibit higher entropies of activation than the analogous NH3 complexes. Likewise complexes with equatorial 3F-salen ligands have a larger entropy of activation than tfacen complexes. In all cases, the enthalpy of activation for the second step (k2) is greater than that for the first step.
The ligand exchange reactions were also evaluated in the presence of ascorbate to determine the effects of a reducing environment on the ligand exchange pathway. A comparison of the ligand exchange reaction of [Co(3F-salen)(3F-BnNH2)2]+ in both the presence and absence of ascorbate is shown in Fig. 6. The reactions of other complexes under these reducing conditions are shown in Figs. S40–S43†, and the relative rates of starting material and product formation are shown in Table 6. Because of the increased complexity of this process in the presence of ascorbate, the data in Table 6 as the t1/2 of the starting material and the amount of species C present when t = t1/2. These qualitative values provide a concise way to compare relative rates of these reactions without assuming a specific kinetics model.
Fig. 6.
Representative concentration vs time data for the axial ligand substitution of [Co(3F-salen)(3F-BnNH2)2]+ with MeIm at 37 °C in the absence (left) and presence (right) of ascorbate. Concentrations of species (A) (circular markers), (B) (diamond markers), and (C) (square markers) refer to ligand substitution reactants and products shown in Fig. 4.
Table 6.
Comparison of Ligand Exchange Reactions in the Presence and Absence of Ascorbate
| Compound | t1/2 (h) No Ascorbate | t1/2 (h) With Ascorbate | % C at t1/2 No Ascorbate | % C at t1/2 With Ascorbate |
|---|---|---|---|---|
| [Co(3F-salen)(NH3)2]+ | 29.8 ± 0.3 | 16.8 ± 0.1 | 4 | 30 |
| [Co(3F-salen)(3F-BnNH2)2]+ | 2.62 ± 0.14 | 2.6 ± 0.1 | 9 | 28 |
| [Co(tfacen)(NH3)2]+ | 48.6 ± 0.2 | 35.8 ± 2.6 | 4 | 34 |
| [Co(tfacen)(3F-BnNH2)2]+ | 11.2 ± 0.1 | 11.7 ± 0.6 | 2 | 26 |
In all cases, the addition of ascorbate to the reaction mixture only marginally accelerated the initial decay of the starting material (species A). However, ascorbate had a much more pronounced effect on the rate of formation of the final product (species C). Species C, the disubstituted product, accumulates rapidly, and the buildup of species B, the monosubstituted intermediate, is greatly diminished. Thus, ascorbate dramatically increase the rate of formation of C from B, but has little effect on the decay of A. The similar rate of decay of A suggests that the complex reduction is gated by the loss of one axial ligand. Whereas the reactions of [Co(3F-salen)(3F-BnNH2)2]+, [Co(tfacen)(3F-BnNH2)2]+, and [Co(tfacen)(NH3)2]+ yielded the same products as the thermal reactions, the ligand exchange of [Co(3F-salen)(NH3)2]+ in the presence of ascorbate also resulted in the formation of several side products in smaller amounts.
Cytotoxicity
The cytotoxicity of all complexes and the free ligands were evaluated in A549 lung cancer cells using the MTT assay. The 50% growth inhibitory concentration (IC50) values for all complexes, free ligands, and relevant control compounds are listed in Table 7. Representative dose-response curves for [Co(3F-salen)(3F-BnNH2)2]+ and [Co(3F-salen)(NH3)2]+ are shown in Fig. 7. Additional dose-response curves are deposited in the Supporting Information (Fig. S44–S47†).
Table 7.
Cytotoxicity and Cellular Uptake of [Co(Schiff base)(L)2]+ Complexes in A549 Lung Cancer Cells
| Compound | IC50 (µM) | Cobalt/Protein (pg/µg) | HPLC Retention Factorb |
|---|---|---|---|
| [Co(3F-salen)(NH3)2]+ | >500 | 43.0 ± 8.4 | 0.21 |
| [Co(3F-salen)(3F-BnNH2)2]+ | 50 ± 16 | 175 ± 19 | 2.95 |
| [Co(tfacen)(NH3)2]+ | >500 | 26.7 ± 2.2 | 0.27 |
| [Co(tfacen)(3F-BnNH2)2]+ | 60 ± 17 | 697 ± 13 | 13.8 |
| Co(NO3)2a | >500 | 37 ± 2 | NA |
| Tfacen | >200 | NA | NA |
| 3F-salen | 97 ± 9.2 | NA | NA |
| cisplatina | 5.5 ± 3.2 | NA | NA |
Reported in reference 16.
Retention Factor = (tr – t0)/t0, where tr is the retention time of the complex and t0 is the column dead time.
Fig. 7.
Cell viability in the presence of varying concentrations of [Co(3F-salen)(3F-BnNH2)2]+ (solid red line, square marker) and [Co(3F-salen)(NH3)2]+ (dashed blue line, circular marker) as measured by the MTT assay.
The complexes [Co(3F-salen)(NH3)2]+ and [Co(tfacen)(NH3)2]+ are effectively inactive against this cancer cell line. In contrast, complexes bearing axial 3F-BnNH2 ligands [Co(3F-salen)(3F-BnNH2)2]+ and [Co(tfacen)(3F-BnNH2)2]+ exhibit moderate activity, as characterized by IC50 values of 50 and 60 μM, respectively. This result indicates that the axial ligand plays the major role in determining the activities of this class of complexes, implicating ligand exchange mechanisms as an important component to their activities. The tfacen ligand is non-toxic, and 3F-salen is substantially less potent than the complex [Co(3F-salen)(3F-BnNH2)2]+, indicating that the toxic effects are not arising from dissociation of the equatorial ligands.
Cobalt Uptake
To evaluate the relationship between complex structure and cellular uptake, the cobalt uptake of all complexes by A549 cells was measured. Cells were treated with 100 µM cobalt complexes for 24 h, and the uptake was quantified by the ratio of cobalt to protein in the cell samples (pg/µg), which were determined by GFAAS and the Bicinchonic Acid Assay, respectively. Cobalt uptake for Co(NO3)2, measured previously in our lab, was used as a control to probe the effects of supporting ligands.16 Results of the uptake experiments are shown in Table 7. In general, complexes with the axial ligand 3F-BnNH2 are taken up by cells much more effectively than those with axial NH3 ligands. However, no clear dependence on the equatorial ligands is evidenced. For example, [Co(3F-salen)(NH3)2]+ is taken up more effectively than [Co(tfacen)(NH3)2]+, but [Co(3F-salen)(3F-BnNH2)2]+ is taken up less effectively than [Co(tfacen)(3F-BnNH2)2]+. For related [Co(bis(thiosemicarbazone))(L)2]+ complexes a similar marked dependence on the axial ligand on cell uptake was noted.16 The uptake of all complexes except [Co(tfacen)(NH3)2]+ was greater than that of free Co2+, administered in the form of Co(NO3)2, which enters cells through both ion transporters and by transferrin-mediated pathways.90,91 To determine whether cobalt uptake correlated with the lipophilicity or cytotoxicity of the complexes, the HPLC retention factors of all complexes were measured using an isocratic mobile phase. Large retention factors correlate with high lipophilicity. The retention factors of all complexes are reported in Table 7. In general, the 3F-salen complexes have lower retention factors than the tfacen complexes, and NH3 complexes have much lower retention factors than the analogous 3F-BnNH2 compounds. The complex [Co(tfacen)(3F-BnNH2)2]+ has a much higher retention factor than all other compounds, which may explain its much greater cell uptake.
Discussion
Co(III) Schiff base complexes have been employed for a wide range of biological applications, utilizing their enzyme inhibitory,24,27,49 antiviral,34–36 and anticancer12,13,16,92–94 properties. These complexes have also been studied as reduction-activated prodrugs.81,95 Despite these previous investigations, few studies have systematically evaluated the general activity of the [Co(Schiff base)(L)2]+ scaffold. In this study, a small set of [Co(Schiff base)(L)2]+ complexes was prepared and thoroughly investigated.. Two families of Co(III) Schiff base complexes, bearing either ammonia (NH3) or 3-fluorobenzylamine (3F-BnNH2) axial ligands, were studied. These efforts are specifically focused on how ligand substitution and reduction pathways are affected by both the equatorial and axial ligands, and how these properties manifest in biological activity. The data obtained in this study provides useful insight on the mechanisms of activation of Co(III) drug candidates.
The syntheses of the four complexes followed a method that was previously reported.23 This procedure calls for the reaction of the Schiff base ligand and Co(NO3)2, followed by the addition of an excess of axial ligand. Atmospheric oxygen is presumed to be the oxidant for this reaction, affording the diamagnetic Co(III) complexes. Although [Co(3F-salen)(3F-BnNH2)2]Cl precipitated from the reaction mixture as the analytically pure product, the other three complexes required additional purification via precipitation from solution with NaPF6. The PF6− counterion was then replaced with Cl− using anion exchange resin. The identity and purity of the complexes were verified by 1H, 19F, and 13C{1H} NMR spectroscopy, HPLC, elemental analysis, and X-ray crystallography. The X-ray crystal structures of [Co(3F-salen)(3F-BnNH2)2]+, [Co(tfacen)(3F-BnNH2)2]+, and [Co(tfacen)(NH3)2]+ (Fig. 1) reveal the expected octahedral coordination geometries. A notable feature of these structures was the shorter Co–Naxial distances of [Co(tfacen)(NH3)2]+ compared to those for [Co(3F-salen)(3F-BnNH2)2]+ and [Co(tfacen)(3F-BnNH2)2]+. All complexes are mono-cationic, water soluble at millimolar concentrations, and stable with respect to ambient air and light for at least several months.
The complexes were further characterized by cyclic voltammetry and 59Co NMR spectroscopy. Cyclic voltammetry is a useful method that has been employed to investigate the reduction of biologically active Co(III) complexes. Both the relative potential and reversibility of redox processes in Co(III) complexes are key parameters that dictate the suitability of a compound for hypoxia-targeting.10,11 These properties are also important for Co(III) complexes that are intended for activation by ligand substitution; if the redox potential is too positive, undesired reduction to Co(II) may occur, drastically reducing the stability of the complex and possibly generating reactive oxygen species via redox cycling.96,97 The cyclic voltammograms of the four complexes all reveal an initial irreversible reduction from Co(III) to Co(II). The irreversible nature of this redox process is a consequence of dissociation of the axial ligands, which occurs upon population of the σ* dz2 orbital upon reduction. This electrochemical process has been observed for related Co(III) complexes.16,23,95 Consistent with the dz2 LUMO, the potential of the Co(III)/Co(II) couple is most strongly dependent on the nature of the axial ligand, a phenomenon that has been documented for similar complexes.98,99 As expected, the more electron-donating NH3 ligands give rise to more negative reduction potentials compared to the analogous 3F-BnNH2 complexes.100
NMR spectroscopy of NMR-active transition metal nuclei relays important information on the chemical environment of the metal center. Under some circumstances, chemical shifts can be correlated to ligand field splitting,83,86,101 thermodynamic stability,102 and ligand exchange rates.67,84,103–105 For 59Co, a 100% naturally abundant nucleus with I = 7/2, the chemical shift of the 59Co NMR signal is related to the energy of the lowest excited state, which is a major contributor to the paramagnetic shielding term. The linewidth of 59Co NMR signals is a consequence of quadrupolar relaxation and is therefore a function of the electric field gradient at the nucleus and the rotational correlation time of the complex.88,106 For tetragonal complexes, the 59Co chemical shift exhibits an inverse, linear correlation with the ligand field splitting of the complex after adjusting for the nephelauxetic effect of the ligands.82,83,85,86 Thus, 59Co NMR spectroscopy provides a method to probe both the electron donating capacity and the nephelauxetic properties of the ligand scaffolds. Furthermore, these properties that dictate the 59Co NMR chemical shift may also relate to chemical reactivity patterns, enabling correlations between this chemical shift and catalytic activity, for example.107,108 The four complexes studied in this work give rise to clean 59Co NMR spectra. The chemical shifts of these complexes range from 8340 to 8740 ppm, a relatively narrow span considering that a 16,000 ppm window exists for octahedral Co(III) complexes.84 Complexes with axial 3F-BnNH2 ligands resonate approximately 200 ppm downfield from their NH3 counterparts. Per correlations found in the literature,83 this downfield shift reflects weaker ligand field strength of the 3F-BnNH2 complexes compared to the NH3 complexes,109,110 a feature that is most likely a consequence of the greater steric hindrance of the larger 3F-BnNH2 ligand. Because the axial ligands are exclusively sigma-donors, this shift also indicates that the metal-ligand bond is stronger in the NH3 complexes than in the 3F-BnNH2 complexes. The equatorial Schiff base ligands also contribute to the observed chemical shifts. The tfacen complexes resonate approximately 200 ppm downfield from the 3F-salen complexes. The upfield shift of the 3F-salen complexes relative to the tfacen complexes may be due to increased delocalization of the HOMO on the aromatic rings of the 3F-salen ligand. The delocalization reflects the larger nephelauxetic effect of the π-aromatic 3F-salen ligands compared to tfacen, which expands the apparent valence d-electron radius. This property gives rise to greater shielding of the 59Co nucleus.82,84,101
Many bioactive Co(III) Schiff base complexes are proposed to be activated by ligand exchange rather than reduction.12,50,52 To assess the role of ligand substitution in the activation of the four Schiff base complexes in this study, the relative rates of this process and activation parameters were determined. The complexes were challenged with an excess of MeIm to drive formation of [Co(Schiff Base)(MeIm)2]+, and this process was followed by 19F NMR spectroscopy or HPLC. Varying the concentration of MeIm had no effect on the rate of the reaction. The zero-order dependence of this incoming ligand is consistent with a dissociative mechanism.111 A conjugate base mechanism, whereby the acidic NH protons of the axial NH3 or 3F-BnNH2 ligand are first deprotonated, was ruled out by virtue of the fact that there was no acceleration of the reaction rate at a higher pH.38 As expected for a dissociative mechanism, the rate of ligand substitution is highly dependent on the departing axial ligand.37 The more strongly donating NH3 complexes exhibit much slower exchange rates than the 3F-BnNH2 complexes. These results may also be due to steric effects, as dissociation of the bulkier 3F-BnNH2 ligands will be more favorable.112 Although the effect of the equatorial Schiff base is less pronounced, the 3F-salen complexes were consistently more reactive than their tfacen counterparts. In all cases, the rate constant for the first exchange step (k1) was at least three times greater than that for the second (k2). This behavior has been observed previously for the acacen family of complexes, and it may indicate greater stabilization of the monosubstituted intermediate relative to the starting complex.52,53
Eyring analysis of the ligand exchange reactions was performed by measuring the rate constants for each reaction at different temperatures. In all cases, the entropies of activation are positive, a feature that is consistent with a dissociative ligand substitution mechanism 89,113,114 The magnitude of the enthalpies of activation are also consistent with a dissociative mechanism, where the Co–N bond is weakened in the transition state.115 These values for ΔH‡ and ΔS‡ are similar to those reported for structurally related Co(III) haloammine complexes and Co(III)-trans dioximes,116–118 which are known to undergo dissociative ligand substitution.
Trends are apparent in comparing the relative enthalpies and entropies of activation of the complexes. Complexes bearing the bulkier 3F-BnNH2 axial ligands have higher ∆S‡ values than those with NH3 axial ligands. The larger entropy of activation of the 3F-BnNH2 complexes may be a consequence of conformational restriction of these large ligands when bound to the complexes that is relieved as they dissociate from the inner coordination sphere. Likewise, the 3F-salen complexes have higher ∆S‡ values than the corresponding tfacen complexes. As observed in the crystal structures, the 3F-salen ligands can deviate significantly from planarity when bound to Co(III), whereas the tfacen ligand remain strictly planar. As such, we hypothesize that the larger ∆S‡ of these complexes compared to tfacen complexes arises from the ability of these ligands to access more conformations, manifested as deviations from planarity, as the axial ligand departs. The enthalpies of activation for the first ligand substitution step are very similar for all four complexes. In considering the similar pKa values of fluorobenzylammonium and ammonium (9.1 and 9.3, respectively)119,120, their donor strengths as ligands are most likely comparable. Hence the enthalpies of activation, which are expected to be related to the donor strengths of the departing ligand, are also quite similar, and the relative reaction rates are predominantly dictated by the entropy of activation of ligand dissociation. For the substitution of the second axial ligand with MeIm, there is more variation in the ΔH‡ values. In this second step, 3F-BnNH2 complexes exhibit lower ∆H‡ values than the corresponding NH3 complexes, and 3F-salen complexes exhibit lower ∆H‡ values than their tfacen analogues. In all cases, the ∆H‡ values for k2 are greater than those for k1, suggesting that the slower rates for the second ligand substitution step arise from enthalpic, rather than entropic, effects. The larger ∆H‡ values for the monosubstituted complex may arise from the weaker trans-effect induced by MeIm compared to the NH3 and 3F-BnNH2 ligands.
Due to their tendency to undergo irreversible reduction to Co(II), many Co(III) complexes are believed to be activated by reductive mechanism in biological systems. Because the Co-Schiff base complexes studied in this work have moderately high reduction potentials, we considered that reduction might also be a potential mechanism of activation for these complexes. In order to probe the effects of a reducing environment on the ligand exchange kinetics, we performed the ligand exchange assays in the presence of an excess of ascorbate, a mild, biologically relevant reductant. The addition of ascorbate to the reaction mixture had little effect on the rate of decay of the starting material, but it led to much more rapid formation of the final, disubstituted product. We take these data to indicate that reduction by ascorbate is cannot occur until an axial ligand dissociates. This hypothesis is supported by the more positive reduction potential of the mono-aquo complex that results from loss of one axial ligand. The replacement of the axial N-donor ligand by the more poorly donating aquo-ligand decreases the electron density at the Co(III) center, increasing the reduction potential and facilitating the electron transfer reaction with ascorbate. The reduced Co(II) complex is much more labile than its Co(III) counterpart, and may rapidly react with MeIm to form [Co(Schiff base)(MeIm)2], which may then be oxidized by oxygen in solution to yield the final product, [Co(Schiff base)(MeIm)2]+. A similar mechanism for accelerated ligand exchange after reduction was proposed for an analogous Co(III)-Shiff base enzyme inhibitor.32
The structural, electrochemical, and kinetic data described above reveal useful correlations between these properties. The Co–Naxial distances measured in the crystal structures scale directly with the axial ligand exchange rates. These long Co–Naxial distances, in turn, may be related to weaker coordinative bonds. Because the rate-determining step of dissociative ligand substitution is cleavage of the M–L bond, the rates of these reactions will be related to the M–L coordinative bond energy. Another correlation between the Co(III)/Co(II) reduction potential and the ligand substitution rate is observed. Complexes with more negative reduction potentials undergo slower ligand exchange rates compared to complexes with more positive reduction potentials. Although correlations have been reported between reduction potential and electron transfer rate for Co(III) complexes,98 no related examples correlating ligand substitution kinetics and reduction potentials have been reported. Although the origins of this correlation warrant further investigation, it may arise as a consequence of the similarity between the transition state of dissociative ligand substitution pathway and the one-electron reduced compound. Both of these species should attain geometries with substantially elongated axial ligand bond distances. Although interesting, this correlation needs to be explored further in related Co(III) complexes to assess its generality. An implication of this correlation is that easily reduced Co(III) complexes are also more labile. Hence, efforts to tune the complexes for activation by reduction versus ligand exchange may be complicated by this interdependence on one another.
The role of the physical properties of these four Schiff base complexes on their biological activities was investigated. The in vitro anticancer activity of these complexes in A549 lung cancer cells was evaluated, revealing a disparate range of IC50 values from 50 µM to higher than 500 µM. The NH3 complexes are all inactive, whereas the 3F-BnNH2 complexes are moderately toxic. With respect to cellular uptake, the complexes bearing NH3 ligands were taken up by cells to a much smaller extent than the 3F-BnNH2. Presumably, this lower cell uptake is a consequence of the lower lipophilicity of the NH3 ligand compared to 3F-BnNH2, which is consistent with the lower HPLC retention factors of the NH3 complexes. The lower toxicity of the NH3 complexes, therefore, may be a factor of both diminished cell uptake and slower ligand exchange kinetics. The increased toxicity of the 3F-BnNH2 complexes may be due to intracellular reduction; related Co(II) salen complexes oxidatively cleave DNA in vitro.121 The overall cytotoxicity of these four complexes is moderate, and it should be emphasized that the 3F-BnNH2 complexes are more active than related Co Schiff base complexes122 and unmodified Co(II) salen complexes, which are inactive.20
Conclusion
Four [Co(Schiff base)(L)2]+ complexes were synthesized and characterized. The complexes undergo ligand exchange in aqueous solution by a dissociative mechanism. Eyring analysis indicates that the relative reaction rates are primarily dictated by differences in the entropy of activation for the complexes. The rate of ligand exchange correlates with the Co(III)/Co(II) reduction potential of the complexes. The ligand exchange reaction mechanism is altered in the presence of ascorbate, leading to more rapid formation of the final disubstituted product. The anticancer activity of the complexes depends heavily on the axial ligand, with the more labile, easily reduced 3F-BnNH2 complexes showing over an order of magnitude higher activity than the inert NH3 complexes.
Supplementary Material
Acknowledgements
This research was supported by Cornell University. A. Paden King thanks the National Institute of Health, National Institute of General Medical Sciences, for a Chemical Biology Interface (CBI) Training Grant (grant number T32GM008500). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. Hendryck A. Gellineau thanks the Howard Hughes Medical Institute grant: Mentored Learning for Groups Underrepresented in Biomedical Research (Project ID 52008135), and the Frank L. and Lynnet Douglas Fellowship Award for summer research support. Dr. Ivan R. Keresztes is thanked for assistance with 59Co and 19F NMR spectroscopy. This work made use of the NMR facility and the Cornell Center for Material Research (CCMR) at Cornell University, which are supported by the NSF under award numbers CHE-1531632 and DMR-1120296, respectively.
Footnotes
Electronic Supplementary Information (ESI) Available: physical characterization data for all complexes, representative ligand exchange plots, Eyring plots, and cytotoxicity data. CCDC 1864183–1864185. For ESI and crystallographic data in CIF or other electronic format see DOI: XXXX
Conflicts of interest
The authors declare no competing financial interests.
Notes and References
- 1.Banerjee R, Chemistry and biochemistry of B12, Wiley, New York, NY, 1999. [Google Scholar]
- 2.Brown KL, Chem. Rev, 2005, 105, 2075–2150. [DOI] [PubMed] [Google Scholar]
- 3.Heffern MC, Yamamoto N, Holbrook RJ, Eckermann AL and Meade TJ, Curr. Opin. Chem. Biol, 2013, 17, 189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munteanu CR and Suntharalingam K, Dalton Trans, 2015, 44, 13796–13808. [DOI] [PubMed] [Google Scholar]
- 5.Renfrew AK, O’Neill ES, Hambley TW and New EJ, Coord. Chem. Rev, 2018, 375, 221–233. [Google Scholar]
- 6.Kim BJ, Hambley TW and Bryce NS, Chem. Sci, 2011, 2, 2135–2142. [Google Scholar]
- 7.Mastren T, V Marquez B, Sultan DE, Bollinger E, Eisenbeis P, Voller T and Lapi SE, Mol. Imaging, 2015, 14, 526–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsitovich PB, Cox JM, Benedict JB and Morrow JR, Inorg. Chem, 2016, 55, 700–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu M, Xie D, Phan KP, Enriquez JS, Luci JJ and Que EL, Chem. Commun, 2016, 52, 13885–13888. [DOI] [PubMed] [Google Scholar]
- 10.Ware DC, Palmer BD, Wilson WR and Denny WA, J. Med. Chem, 1993, 36, 1839–1846. [DOI] [PubMed] [Google Scholar]
- 11.Ware DC, Palmer HR, Pruijn FB, Anderson RF, Brothers PJ, Denny WA and Wilson WR, Anticancer Drug Des, 1998, 13, 81–103. [PubMed] [Google Scholar]
- 12.Yamamoto N, Renfrew AK, Kim BJ, Bryce NS and Hambley TW, J. Med. Chem, 2012, 55, 11013–11021. [DOI] [PubMed] [Google Scholar]
- 13.Karnthaler-Benbakka C, Groza D, Kryeziu K, Pichler V, Roller A, Berger W, Heffeter P and Kowol CR, Angew. Chem. Int. Ed, 2014, 53, 12930–12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renfrew AK, Bryce NS and Hambley T, Chem. - A Eur. J, 2015, 21, 15224–15234. [DOI] [PubMed] [Google Scholar]
- 15.de Souza ICA, Faro LV, Pinheiro CB, Gonzaga DTG, da Silva F. de C., Ferreira VF, Miranda F. da S., Scarpellini M and Lanznaster M, Dalton Trans, 2016, 45, 13671–13674. [DOI] [PubMed] [Google Scholar]
- 16.King AP, Gellineau HA, Ahn J-E, MacMillan SN and Wilson JJ, Inorg. Chem, 2017, 56, 6609–6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Neill ES, Kaur A, Bishop DP, Shishmarev D, Kuchel PW, Grieve SM, Figtree GA, Renfrew AK, Bonnitcha PD and New EJ, Inorg. Chem, 2017, 56, 9860–9868. [DOI] [PubMed] [Google Scholar]
- 18.Kathrin R. Schmidt; Manfred Jung; Roland Keilitz; Beate Scnurr; Gust, Inorg. Chim. Acta, 2000, 306, 6–16. [Google Scholar]
- 19.Ott I, Schmidt K, Kircher B, Schumacher P, Wiglenda T and Gust R, J. Med. Chem, 2005, 48, 622–629. [DOI] [PubMed] [Google Scholar]
- 20.Gust R, Ott I, Posselt D and Sommer K, J. Med. Chem, 2004, 47, 5837–5846. [DOI] [PubMed] [Google Scholar]
- 21.Saghatforoush L, Moeini K, Hosseini-Yazdi SA, Mardani Z, Hajabbas-Farshchi A, Jameson HT, Telfer SG and Woollins JD, RSC Adv, 2018, 8, 35625–35639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smolko L, Černák J, Kuchár J, Sabolová D and Boča R, Chem. Pap, 2018, 72, 877–882. [Google Scholar]
- 23.Böttcher A, Takeuchi T, Hardcastle KI, Meade TJ, Gray HB, Cwikel D, Kapon M and Dori Z, Inorg. Chem, 1997, 36, 2498–2504. [Google Scholar]
- 24.Takeuchi T, Böttcher A, Quezada CM, Simon MI, Meade TJ and Gray HB, J. Am. Chem. Soc, 1998, 120, 8555–8556. [Google Scholar]
- 25.Harney AS, Lee J, Manus LM, Wang P, Ballweg DM, LaBonne C and Meade TJ, Proc. Natl. Acad. Sci. U. S. A, 2009, 106, 13667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomco D, Schmitt S, Ksebati B, Heeg MJ, Dou QP and Verani CN, J. Inorg. Biochem, 2011, 105, 1759–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurtado RR, Harney AS, Heffern MC, Holbrook RJ, Holmgren RA and Meade TJ, Mol. Pharm, 2012, 9, 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verani CN, J. Inorg. Biochem, 2012, 106, 59–67. [DOI] [PubMed] [Google Scholar]
- 29.Heffern MC, Kurutz JW and Meade TJ, Chem. - A Eur. J, 2013, 19, 17043–17053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierroz V, Joshi T, Leonidova A, Mari C, Schur J, Ott I, Spiccia L, Ferrari S and Gasser G, J. Am. Chem. Soc, 2012, 134, 20376–20387. [DOI] [PubMed] [Google Scholar]
- 31.Peterson MD, Holbrook RJ, Meade TJ and Weiss EA, J. Am. Chem. Soc, 2013, 135, 13162–13167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holbrook RJ, Weinberg DJ, Peterson MD, Weiss EA and Meade TJ, J. Am. Chem. Soc, 2015, 137, 3379–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asbell RM, P. A.; Epstein Seth P.; Wallace James A.; Epstein Donald; Stewart Claudia C.; Burger, Cornea, 1998, 17, 550–557. [DOI] [PubMed] [Google Scholar]
- 34.Burger OS, Richard M; Stewart CC; Winicov I; Bourne N; Stanberry LR; Bonnez W; Weislow, AIDS, 2001, 15, 1.11192849 [Google Scholar]
- 35.Schwartz JA, Lium EK and Silverstein SJ, J. Virol, 2001, 75, 4117–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Epstein SP, Pashinsky YY, Gershon D, Winicov I, Srivilasa C, Kristic KJ and Asbell PA, BMC Ophthalmol, 2006, 6, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langford HB, Cooper H; Gray, Ligand Substitution Processes, W. A. Benjamin, Inc., New York, NY, 1966. [Google Scholar]
- 38.Richens DT, Chem. Rev, 2005, 105, 1961–2002. [DOI] [PubMed] [Google Scholar]
- 39.V Wegner S and Spatz JP, Angew. Chem. Int. Ed, 2013, 52, 7593–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall MD, Failes TW, Yamamoto N and Hambley TW, Dalton Trans, 2007, 3983–3990. [DOI] [PubMed]
- 41.Reisner E, Arion VB, Keppler BK and Pombeiro AJL, Inorg. Chim. Acta, 2008, 361, 1569–1583. [Google Scholar]
- 42.Jungwirth U, Kowol CR, Keppler BK, Hartinger CG, Berger W and Heffeter P, Antioxid. Redox Signal, 2011, 15, 1085–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graf N and Lippard SJ, Adv. Drug Deliv. Rev, 2012, 64, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denny WA and Wilson WR, Cancer Metastasis Rev, 1993, 12, 135–151. [DOI] [PubMed] [Google Scholar]
- 45.Ware DC, Brothers PJ, Clark GR, Denny WA, Palmer BD and Wilson WR, J. Chem. Soc. Dalt. Trans, 2000, 925–932.
- 46.Chang JY-C, Stevenson RJ, Lu G-L, Brothers PJ, Clark GR, Denny WA and Ware DC, Dalton Trans, 2010, 39, 11535–50. [DOI] [PubMed] [Google Scholar]
- 47.Chang JY-C, Lu G-L, Stevenson RJ, Brothers PJ, Clark GR, Botting KJ, Ferry DM, Tercel M, Wilson WR, Denny WA and Ware DC, Inorg. Chem, 2013, 52, 7688–7698. [DOI] [PubMed] [Google Scholar]
- 48.Renfrew AK, Bryce NS and Hambley TW, Chem. Sci, 2013, 4, 3731–3739. [Google Scholar]
- 49.Louie AY and Meade TJ, Proc. Natl. Acad. Sci. U. S. A, 1998, 95, 6663–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeuchi T, Böttcher A, Quezada CM, Meade TJ and Gray HB, Bioorg. Med. Chem, 1999, 7, 815–819. [DOI] [PubMed] [Google Scholar]
- 51.Heffern MC, Velasco PT, Matosziuk LM, Coomes JL, Karras C, Ratner MA, Klein WL, Eckermann AL and Meade TJ, ChemBioChem, 2014, 15, 1584–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manus LM, Holbrook RJ, Atesin TA, Heffern MC, Harney AS, Eckermann AL and Meade TJ, Inorg. Chem, 2013, 52, 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matosziuk LM, Holbrook RJ, Manus LM, Heffern MC, Ratner MA and Meade TJ, Dalton Trans, 2013, 42, 4002–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heffern MC, Reichova V, Coomes JL, Harney AS, Bajema EA and Meade TJ, Inorg. Chem, 2015, 54, 9066–9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tengel T, Fex T, Emtenäs H, Almqvist F, Sethson I and Kihlberg J, Org. Biomol. Chem, 2004, 2, 725–731. [DOI] [PubMed] [Google Scholar]
- 56.Jordan JB, Poppe L, Xia X, Cheng AC, Sun Y, Michelsen K, Eastwood H, Schnier PD, Nixey T and Zhong W, J. Med. Chem, 2012, 55, 678–687. [DOI] [PubMed] [Google Scholar]
- 57.Xie D, King TL, Banerjee A, Kohli V and Que EL, J. Am. Chem. Soc, 2016, 138, 2937–2940. [DOI] [PubMed] [Google Scholar]
- 58.Xie D, Kim S, Kohli V, Banerjee A, Yu M, Enriquez JS, Luci JJ and Que EL, Inorg. Chem, 2017, 56, 6429–6437. [DOI] [PubMed] [Google Scholar]
- 59.Adam MJ and Wilbur DS, Chem. Soc. Rev, 2005, 34, 153. [DOI] [PubMed] [Google Scholar]
- 60.Ametamey SM, Honer M and Schubiger PA, Chem. Rev, 2008, 108, 1501–1516. [DOI] [PubMed] [Google Scholar]
- 61.Venkatachalam TK, Pierens GK, Bernhardt PV, Stimson DHR, Bhalla R, Lambert L and Reutens DC, Aust. J. Chem, 2016, 69, 1033. [Google Scholar]
- 62.Zhou Y, Wang J, Gu Z, Wang S, Zhu W, Aceña JL, Soloshonok VA, Izawa K and Liu H, Chem. Rev, 2016, 116, 422–518. [DOI] [PubMed] [Google Scholar]
- 63.Doctrow SR, Huffman K, Marcus CB, Tocco G, Malfroy E, Adinolfi CA, Kruk H, Baker Keith, Lazarowych N, Mascarenhas Jand Malfroy B, J. Med. Chem, 2002, 45, 4549–4558. [DOI] [PubMed] [Google Scholar]
- 64.Fulmer GR, Miller AJM, Sherden NH, Gottlieb HE, Nudelman A, Stoltz BM, Bercaw JE and Goldberg KI, Organometallics, 2010, 29, 2176–2179. [Google Scholar]
- 65.Yu J-X, Hallac RR, Chiguru S and Mason RP, Prog. Nucl. Magn. Reson. Spectrosc, 2013, 70, 25–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosenau CP, Jelier BJ, Gossert AD and Togni A, Angew. Chem. Int. Ed, 2018, 57, 9528–9533. [DOI] [PubMed] [Google Scholar]
- 67.Yamasaki A, J. Coord. Chem, 1991, 24, 211–260. [Google Scholar]
- 68.Connelly NG and Geiger WE, Chem. Rev, 1996, 96, 877–910. [DOI] [PubMed] [Google Scholar]
- 69.V Pavlishchuk V and Addison AW, Inorg. Chim. Acta, 2000, 298, 97–102. [Google Scholar]
- 70.Valkó K, Snyder LR and Glajch JL, J. Chromatogr. A, 1993, 656, 501–520. [Google Scholar]
- 71.Ayouni L, Cazorla G, Chaillou D, Herbreteau B, Rudaz S, Lantéri P and Carrupt P-A, Chromatographia, 2005, 62, 251–255. [Google Scholar]
- 72.Sheldrick GM, Acta Cryst, 2008, A64, 112–122. [Google Scholar]
- 73.Sheldrick GM, Acta Cryst, 2015, A71, 3–8. [Google Scholar]
- 74.Müller P, Crystallogr. Rev, 2009, 15, 57–83. [Google Scholar]
- 75.Pilling PW, Michael J; Seakins, Reaction Kinetics, Oxford University Press, New York, NY, 1995. [Google Scholar]
- 76.Gellene GI, J. Chem. Educ, 1995, 72, 196. [Google Scholar]
- 77.Freshney RI, Culture of Animal Cells, John Wiley & Sons, Inc., Hoboken, NJ, USA, 5th edn., 2005. [Google Scholar]
- 78.Xia X, Owen MS, Lee REC and Gaudet S, Cell Death Dis, 2014, 5, e1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kirin SI, Ott I, Gust R, Mier W, Weyhermüller T and Metzler-Nolte N, Angew. Chem. Int. Ed, 2008, 47, 955–959. [DOI] [PubMed] [Google Scholar]
- 80.Egger AE, Rappel C, Jakupec MA, Hartinger CG, Heffeter P and Keppler BK, J. Anal. At. Spectrom, 2009, 24, 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garcia CV, Parrilha GL, Rodrigues BL, Barbeira PJS, Clarke RM, Storr T and Beraldo H, Polyhedron, 2017, 124, 86–95. [Google Scholar]
- 82.Juranic N, Inorg. Chem, 1980, 19, 1093–1095. [Google Scholar]
- 83.Bramley R, Brorson M, Sargeson AM and Schaeffer CE, J. Am. Chem. Soc, 1985, 107, 2780–2787. [Google Scholar]
- 84.Mason J, Chem. Rev, 1987, 87, 1299–1312. [Google Scholar]
- 85.Juranic N, J. Am. Chem. Soc, 1988, 110, 8341–8343. [Google Scholar]
- 86.Juranić N, Coord. Chem. Rev, 1989, 96, 253–290. [Google Scholar]
- 87.Medek A, Frydman V and Frydman L, Proc. Natl. Acad. Sci. U. S. A, 1997, 94, 14237–14242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Au-Yeung CFS and Eaton DR, J. Magn. Reson, 1983, 52, 366–373. [Google Scholar]
- 89.Ablov AV and Samus NM, Coord. Chem. Rev, 1975, 17, 253–279. [Google Scholar]
- 90.Chikh Z, Hémadi M, Miquel G, Ha-Duong N-T and El Hage Chahine J-M, J. Mol. Biol, 2008, 380, 900–916. [DOI] [PubMed] [Google Scholar]
- 91.Simonsen LO, Harbak H and Bennekou P, Blood Cells, Mol. Dis, 2011, 47, 214–225. [DOI] [PubMed] [Google Scholar]
- 92.Failes TW, Cullinane C, Diakos CI, Yamamoto N, Lyons JG and Hambley TW, Chem. - A Eur. J, 2007, 13, 2974–2982. [DOI] [PubMed] [Google Scholar]
- 93.Jagadeesan S, Balasubramanian V, Baumann P, Neuburger M, Häussinger D and Palivan CG, Inorg. Chem, 2013, 52, 12535–12544. [DOI] [PubMed] [Google Scholar]
- 94.Cressey PB, Eskandari A, Bruno PM, Lu C, Hemann MT and Suntharalingam K, ChemBioChem, 2016, 17, 1713–1718. [DOI] [PubMed] [Google Scholar]
- 95.Zhang C, Sutherland M, Herasymchuk K, Clarke RM, Thompson JR, Chiang L, Walsby CJ and Storr T, Can. J. Chem, 2018, 96, 110–118. [Google Scholar]
- 96.Chandel PT, N.S.; Maltepe E; Goldwasser E; Mathieu CE; Simon MC; Schumacker, Proc. Natl. Acad. Sci. U. S. A, 1998, 95, 11715–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chandel PT, Navdeep S; McClintock David S.; Feliciano Carlos E.; Wood Teresa M.; Melendez JA; Schumaker, J. Biol. Chem, 2000, 275, 25130–25138. [DOI] [PubMed] [Google Scholar]
- 98.Rillema RC, Paul D; Endicott John F.; Patel, J. Am. Chem. Soc, 1972, 94, 394–401. [Google Scholar]
- 99.Rillema JR, Paul D; Endicott John F.; Barber, J. Am. Chem. Soc, 1973, 95, 6987–6992. [Google Scholar]
- 100.Lever ABP, Inorg. Chem, 1990, 29, 1271–1285. [Google Scholar]
- 101.Juranic N, Inorg. Chem, 1983, 22, 521–525. [Google Scholar]
- 102.Öhrström L, Comments Inorg. Chem, 1996, 18, 305–323. [Google Scholar]
- 103.Koller M and von Philipsborn W, Organometallics, 1992, 11, 467–469. [Google Scholar]
- 104.von Philipsborn W, Chem. Soc. Rev, 1999, 28, 95–105. [Google Scholar]
- 105.Kramer J and Koch KR, Inorg. Chem, 2007, 46, 7466–7476. [DOI] [PubMed] [Google Scholar]
- 106.Ho KKW, Choy WY, Yu-Xin C and Au-Yeung SCF, J. Magn. Reson. Ser. A, 1994, 108, 196–200. [Google Scholar]
- 107.Bönnemann H, Brijoux W, Brinkmann R, Meurers W, Mynott R, von Philipsborn W and Egolf T, J. Organomet. Chem, 1984, 272, 231–249. [Google Scholar]
- 108.Bönnemann H, Angew. Chem. Int. Ed, 1985, 24, 248–262. [Google Scholar]
- 109.Gănescu I, Teodorescu M and Lepădatu CI, Zeitschrift für Phys. Chemie, 1971, 74, 11–16. [Google Scholar]
- 110.Brown DR and Pavlis RR, J. Chem. Educ, 1985, 62, 807–808. [Google Scholar]
- 111.Basolo F and Pearson RG, Adv. Inorg. Chem. Radiochem, 1961, 3, 1–89. [Google Scholar]
- 112.Darensbourg DJ, Adv. Organomet. Chem, 1982, 21, 113–150. [Google Scholar]
- 113.Wendt OF, Robert A, Deeth J and Elding LI, Inorg. Chem, 2000, 39, 5271–5276. [DOI] [PubMed] [Google Scholar]
- 114.Hill D, Delaney C, Clark M, Eaton M, Hassan B, Hendricks O, Dang DK and Kirss RU, RSC Adv, 2017, 7, 34425–34434. [Google Scholar]
- 115.Helm L and Merbach AE, Chem. Rev, 2005, 105, 1923–1960. [DOI] [PubMed] [Google Scholar]
- 116.Poon CK, Coord. Chem. Rev, 1973, 10, 1–35. [Google Scholar]
- 117.Ablov AV and Samus NM, Coord. Chem. Rev, 1975, 17, 253–279. [Google Scholar]
- 118.Benzo F, Bernhardt PV, González G, Martinez M and Sienra B, J. Chem. Soc. Dalt. Trans, 1999, 3973–3979.
- 119.Blackwell LF, Fischer A, Miller IJ, Topsom RD and Vaughan J, J. Chem. Soc, 1964, 3588.
- 120.Maeda M and Kato K, J. Chem. Eng. Data, 1995, 40, 253–256. [Google Scholar]
- 121.Bhattacharya S and Mandal SS, J. Chem. Soc., Chem. Commun, 1995, 2489–2490.
- 122.Kirubavathy SJ, Velmurugan R, Karvembu R, Bhuvanesh NSP, Parameswari K and Chitra S, Russ. J. Coord. Chem, 2015, 41, 345–352. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.