Abstract
Purpose
Male infertility is a multifactorial syndrome encompassing a wide variety of disorders. A previous Chinese genome-wide single-nucleotide polymorphism (SNP) association studies have identified four SNPs (rs12097821 in PRMT6 gene, rs2477686 in PEX10 gene, rs6080550 in SIRPA-SIRPG, and rs10842262 in SOX5 gene) as being significantly associated with risk factors for nonobstructive azoospermia (NOA). However, the results were not fully repeated in later studies, which calls for further investigations.
Methods
We here performed a case-control study in a central Chinese population to explore the association between the four SNPs and male infertility, which included 631 infertile men (NOA and oligozoospermia) and 720 healthy fertile men. The genotyping was performed using the polymerase chain reaction–restriction fragment length polymorphism and confirmed by sequencing.
Results
The results showed that rs12097821 and rs10842262 were strongly associated with the risk of NOA but not total male infertility or oligozoospermia, while rs2477686 and rs6080550 were not associated with the risk of total male infertility, NOA, or oligozoospermia. To improve the statistical strength, a meta-analysis was conducted. The results suggested that rs2477686, rs6080550, and rs10842262 were significantly associated with male infertility, especially with NOA, while rs12097821 was only found to be associated with total male infertility.
Conclusions
Collectively, the rs2477686, rs6080550, and rs10842262 may indeed be the genetic risk factors for NOA, which requires further investigation using larger independent sets of samples in different ethnic populations.
Keywords: Single-nucleotide polymorphism (SNP), Male infertility, Han Chinese, Meta-analysis
Introduction
Male infertility is a major problem worldwide that affects approximately 10–15% of couples and roughly half of these cases are due to male-factor etiology. The main cause of male infertility is spermatogenic failure such as nonobstructive azoospermia (NOA) and oligozoospermia [1]. Interestingly, genetic factors have been regarded as contributing to male infertility [2], and few genetic variations, such as chromosome number defects, Y chromosome microdeletions, and autosomal chromosome variations, have been found to be associated with male infertility [3, 4]. However, the genetic basis of male infertility remains largely unknown.
Recently, Hu et al. performed a GWAS (genome-wide association study) of NOA in Han Chinese men by genotyping 906,703 SNPs in 1000 individuals with NOA (cases) and 1703 male controls using Affymetrix Genome-Wide Human SNP Array 6.0 chips [5]. They identified that the four SNPs (rs1207821 in PRMT6 gene, rs2477686 in PEX10 gene, rs6080550 in SIRPA-SIRPG, and rs10842262 in SOX5 gene) were significantly associated with NOA. However, the follow-up studies in Japanese and Chinese men failed to replicate these results [6–9], except for that Zou et al. repeatedly found that rs10842262 in SOX5 gene was significantly associated with NOA risk in Chinese men [10]. In this view, the association between the four SNPs and NOA still needs further investigation. On the other side, genetic associations in male infertility, such as NOA and/or oligozoospermia, are still not very clear. The same disease symptoms may share the same mechanisms or pathways in different ethnic groups. Therefore, the previous GWAS-linked SNPs with NOA (rs12097821, rs2477686, rs10842262, and rs6080550) may also contribute to the genetic susceptibility to oligozoospermia, which is subject to verification. To this end, a hospital-based case-control study was carried out to evaluate the association between the four SNPs and susceptibility to NOA and oligozoospermia in a Chinese population of Hubei province.
Nowadays, meta-analysis has been a popular method for resolving discrepancies in genetic association studies. Specifically, meta-analysis combines results from different studies on the same topic and thus increases statistical strength and precision in estimating effects [11]. Hence, a meta-analysis was performed, combining the results from previously published literatures and present case-control study, to provide a more precise estimation of the association between the four SNPs and male infertility.
Material and methods
Participants
This study was approved by the Ethical Committees of Huazhong University of Science and Technology, and written informed consent for the genetics analysis was obtained from all participants or their guardians.
In this study, we recruited a total of 630 infertile males (301 NOA cases and 330 oligozoospermia cases) and 720 normal controls. The infertile males recruited in this study were strictly screened by clinicians from Wuhan Tongji Reproductive Medicine Hospital. The control group consisted of 720 fertile males, who had fathered at least one child. All of the fertile controls had normal semen parameters. Men who exhibited normal semen parameters but with unknown fertilization status were not included in this study.
To diagnose the NOA and oligozoospermia, a standard clinical examination procedure was performed, including medical history, physical examination, semen analysis, serum hormone analysis, ultrasound evaluation, and genetic examination (e.g., karyotype testing and Y chromosome microdeletion detection). Then, patients were excluded if they had testicular dysplasia, hormonal abnormalities, infections, history of diseases affecting fertility (e.g., varicocele), genital tract pathologies, karyotyping anomalies, or Y-chromosome deletions. Semen samples were collected by masturbation and examined after liquefaction for 30 min at 37 °C. Semen analysis was performed using the computer-assisted semen analysis system (WLJY-9000; Weili New Century Science and Tech Development, Beijing, China) with semen parameters determined according to the guidelines of World Health Organization 2010 [12].
The genotyping of the four SNPs
The peripheral blood samples (5 ml per participant) were collected into blood vacuum tubes containing EDTA and stored at 4 °C. Then, genomic DNA was extracted from blood samples using the TIANamp Blood DNA Kit (DP348; TianGen Biotech, Beijing, China) according to the manufacturer’s instructions, and stored at − 20 °C before use. Next, the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technique was conducted to genotype the four candidate polymorphisms. The primers and restriction enzymes used in this study were previously described in the study of Sato et al. [6]. For quality control, the PCR-RFLP assay was repeated twice for all subjects, and the results were 100% concordant. Moreover, 20% randomly selected PCR-amplified DNA samples were examined by DNA sequencing, the results were also 100% concordant.
Statistical analysis
All the statistical analysis was performed using Statistical Program for Social Sciences (SPSS, version 15.0, Chicago, IL, USA). All numerical data (age, abstinence time, and semen parameters) were presented as means ± SD (standard deviation of the mean). Differences in these numerical data were assessed by a one-way ANOVA (analysis of variance). Genotypic frequencies of the four SNPs (rs12097821, rs2477686, rs10842262, and rs6080550) in normal controls were tested for departure from HWE (Hardy-Weinberg equilibrium). Logistic regression analysis was used to estimate the association between the four SNPs and male infertility risk. P values less than 0.05 were considered significant for all statistical analyses. To adjust for multiple comparisons, we applied the Bonferroni method [13], which is made by dividing the P value threshold by the number of comparisons made (0.05/n).
Meta-analysis
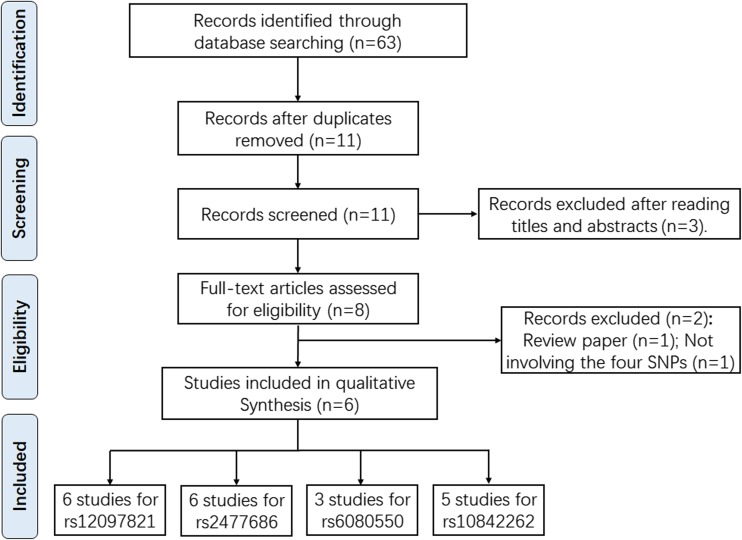
We performed a comprehensive literature search updated to September of 2018 in PubMed, EMBASE, ISI Web of Science, and CNKI and Wanfang databases without language restriction. The search terms used were as follows: “PRMT6, rs12097821, and cancer/tumor/carcinoma”, “PEX10, rs2477686, and cancer/tumor/carcinoma”, “SIRPA-SIRPG, rs6080550, and cancer/tumor/carcinoma”, and “SOX5, rs10842262, and cancer/tumor/carcinoma”. References listed in retrieved articles were also checked for missing information. Enrolled studies should meet the following criteria: (1) studies on humans; (2) investigation of the PRMT6 rs12097821, PEX10 rs2477686, SIRPA-SIRPG rs6080550 or SOX5 rs10842262, and male infertility risk; (3) case-control study design; (4) sufficient data (allele and genotype frequencies) were accessible to estimate the OR and its 95%CI; and (5) HWE equilibrium should be established in controls. Figure 1 showed us the flowchart of the search strategy and article selection for this meta-analysis. Different ethnicity descents were categorized as Asian and Caucasian. STATA 14.0 (Stata Corp, College Station, TX) was employed to calculate all the statistical analyses. The Cochran’s Q test and I2 were used to assess the heterogeneity of included studies [14]. If Pheterogeneity ≥ 0.1, the fixed-effect model was applied to calculate the combined OR [15], otherwise, random-effects model was conducted [16]. The significance of combined OR was determined by the Z test. A P value < 0.05 was considered significantly, and the Bonferroni correction for multiple testing was applied.
Fig. 1.
Flow diagram of the literature review process for PRMT6 rs1207821 polymorphism, PEX10 rs2477686 polymorphism, SIRPA-SIRPG rs6080550 polymorphism, and SOX5 rs10842262 polymorphism and male infertility susceptibility
Results
Participants
As shown in Table 1, there were no statistical differences between case group and control group in individual characteristics, including age, duration of marriage, and abstinence time. However, semen concentration and total semen count per ejaculate were significantly lower in oligozoospermia group.
Table 1.
Comparison of individual characteristics and semen parameters in study groups
| Characteristics | Patients (n = 631) | Control (n = 720) | ||
|---|---|---|---|---|
| Total cases (n = 631) | NOAa (n = 301) | Oligozoospermia (n = 330) | ||
| Age, year | 33.16 ± 4.85 | 32.86 ± 5.47 | 34.06 ± 6.85 | 33.87 ± 5.76 |
| Duration of marriage, year | 7.16 ± 5.37 | 7.85 ± 3.87 | 6.26 ± 6.97 | 6.97 ± 5.63 |
| Abstinence, day | 5.85 ± 2.37 | 6.23 ± 2.87 | 5.16 ± 3.24 | 5.35 ± 2.63 |
| Semen parameters | ||||
| Ejaculate volume, ml | 2.73 ± 1.37 | 2.25 ± 1.87 | 2.89 ± 2.23 | 2.64 ± 1.18 |
| Total sperm/ejaculate × 106 | – | 0 ± 0 | 27.67 ± 13.852 | 207.43 ± 131.61 |
| Sperm concentration, × 106/ml | – | 0 ± 0 | 12.84 ± 4.67b | 85.36 ± 47.65 |
All data were presented as means ± SD (standard deviation of the mean)
aNOA, nonobstructive azoospermia
bP < 0.001 compared with the control group by one-way ANOVA (analysis of variance)
The association between the four SNPs and male infertility susceptibility
In this study, we performed comparisons of allele and genotype frequencies of the four SNPs and applied six genetic model (allele model, carrier model, homozygote model, heterozygote model, recessive model, dominant model) analysis between infertile males (total cases, NOA cases or oligozoospermia cases) and normal controls (Table 2). There were no deviations from HWE observed within the control group for all studied SNPs. After Bonferroni correction (P < 0.0084, 0.05/6), we found that PRMT6 rs12097821 polymorphism and SOX5 rs10842262 polymorphism were strongly associated with risk of NOA, but not with risk of total male infertility or oligozoospermia. For SNP 12097821, allele G was a significant predisposition allele of NOA (G vs. T, P = 0.002, OR = 0.71, 95% CI = 0.58–0.88), and individuals with genotype GG had a higher risk for NOA compared with GT + TT (GG vs. GT + TT, P = 0.005, OR = 0.66, 95% CI = 0.50–0.87). For SNP rs10842262, G allele had a higher risk for NOA than those carrying the C allele (G vs. C, P = 0.004, OR = 1.35, 95% CI = 1.10–1.64), and GG genotype conferred higher risk for NOA relative to CC genotype (GG vs. CC, P = 0.008, OR = 1.80, 95% CI = 1.15–2.80). Moreover, we observed that PEX10 rs2477686 polymorphism and SIRPA-SIRPG rs6080550 polymorphism were not associated with the susceptibility to total male infertility, NOA, or oligozoospermia under none of the six genetic models.
Table 2.
Genotype and allele distributions of SNPs and the association with the risk of male infertility
| Genotype | 1. Total cases | 2. NOAa | 3. Oligozoospermia | 4. Normal controls | HWE b | Logistic Regression [P, OR(95% CI)]c | |||
|---|---|---|---|---|---|---|---|---|---|
| Genetic model | 1 vs. 4 | 2 vs. 4 | 3 vs. 4 | ||||||
| PRMT6 rs12097821 polymorphism | |||||||||
| G | 925 (73.3%) | 425 (70.6%) | 500 (75.8%) | 1110 (77.1%) | G vs. T | 0.023, 0.82 (0.69–0.97) | 0.002, 0.71 (0.58–0.88) | 0.505, 0.93 (0.75–1.15) | |
| T | 337 (26.7%) | 177 (29.4%) | 160 (24.2%) | 330 (22.9%) | GG vs. TT | 0.085, 0.68 (0.42–1.06) | 0.014, 0.53 (0.31–0.90) | 0.686, 0.90 (0.49–1.56) | |
| GG | 340 (53.9%) | 150 (49.8%) | 190 (57.6%) | 428 (59.4%) | 0.968 | GG vs. GT | 0.085, 0.80 (0.63–1.05) | 0.019, 0.73 (0.50–0.97) | 0.616, 0.91 (0.70–1.21) |
| GT | 246 (40.0%) | 125 (41.5%) | 121 (36.7%) | 254 (35.3%) | GT vs. TT | 0.398, 0.80 (0.53–1.31) | 0.234, 0.71 (0.40–1.25) | 0.873, 0.97 (0.51–1.70) | |
| TT | 45 (7.1%) | 26 (8.7%) | 19 (5.8%) | 38 (5.3%) | GG vs. GT + TT | 0.040, 0.78 (0.65–0.97) | 0.005, 0.66 (0.50–0.87) | 0.568, 0.91 (0.70–1.24) | |
| GG + GT vs. TT | 0.158, 0.75 (0.49–1.15) | 0.045, 0.61 (0.37–0.98) | 0.750, 0.93 (0.50–1.58) | ||||||
| PEX10 rs2477686 polymorphism | |||||||||
| G | 146 (10%) | 85 (14.1%) | 61 (9.2%) | 171 (11.9%) | G vs. C | 0.103, 0.82 (0.65–1.04) | 0.163, 1.22 (0.92–1.61) | 0.075, 0.76 (0.56–1.03) | |
| C | 1316 (90%) | 517 (85.9%) | 599 (90.8%) | 1269 (88.1%) | GG vs. CC | 0.969, 1.04 (0.40–2.55) | 0.430, 1.50 (0.53–4.20) | 0.465,0.61 (0.16–2.25) | |
| GG | 9 (1.4%) | 6 (2%) | 3 (1%) | 10 (1.4%) | 0.957 | CG vs. CC | 0.758, 0.97 (0.72–1.24) | 0.228, 1.23 (0.90–1.70) | 0.096, 0.73 (0.50–1.07) |
| GC | 128 (20.3%) | 73 (24.2%) | 55 (16.7%) | 151 (21.0%) | GG vs. CG | 0.900, 1.07 (0.40–2.70) | 0.687, 1.25 (0.45–3.56) | 0.774, 0.80 (0.21–3.13) | |
| CC | 494 (78.3%) | 222 (73.8%) | 272 (82.3%) | 559 (77.6%) | GG vs. GC + CC | 0.954, 1.01 (0.40–2.55) | 0.481, 1.45 (0.50–4.03) | 0.517, 0.67 (0.20–2.35) | |
| GG + CG vs. CC | 0.774, 0.98 (0.75–1.27) | 0.182, 1.22 (0.90–1.71) | 0.073, 0.73 (0.51–1.05) | ||||||
| SIRPA-SIRPG rs6080550 polymorphism | |||||||||
| A | 368 (29.2%) | 172 (28.6%) | 196 (54.4%) | 363 (25.0%) | A vs. G | 0.021, 1.22 (1.03–1.45) | 0.115, 1.19 (0.96–1.47) | 0.031, 1.25 (1.02–1.54) | |
| G | 894 (70.8%) | 430 (71.4%) | 464 (45.6%) | 1077 (75.0%) | AA vs. GG | 0.076, 1.49 (0.99–2.25) | 0.247, 1.36 (0.80–2.34) | 0.081, 1.58 (0.97–2.60) | |
| AA | 53 (8.4%) | 24 (8.0%) | 29 (8.8%) | 46 (6.4%) | 0.961 | AA vs. AG | 0.414, 1.22 (0.80–1.85) | 0.611, 1.17 (0.69–1.95) | 0.410, 1.27 (0.74–2.08) |
| AG | 261 (41.4%) | 123 (40.9%) | 138 (41.8%) | 271 (37.6%) | AG vs. GG | 0.078, 1.20 (0.99–1.55) | 0.233, 1.21 (0.93–1.60) | 0.100, 1.25 (0.97–1.68) | |
| GG | 317 (50.2%) | 154 (51.1%) | 163 (49.4%) | 403 (56.0%) | AA vs. AG + GG | 0.182, 1.31 (0.86–2.01) | 0.362, 1.25 (0.79–2.15) | 0.163, 1.44 (0.89–2.31) | |
| AA + AG vs. GG | 0.035, 1.25 (1.00–1.58) | 0.160, 1.20 (0.95–1.61) | 0.047, 1.33 (1.01–1.67) | ||||||
| SOX5 rs10842262 polymorphism | |||||||||
| G | 437 (34.6%) | 222 (36.9%) | 215 (32.5%) | 436 (30.3%) | G vs. C | 0.017, 1.22 (1.04–1.43) | 0.004, 1.35 (1.10–1.64) | 0.301, 1.11 (0.91–1.35) | |
| C | 826 (65.4%) | 380 (63.1%) | 446 (67.5%) | 1004 (69.7%) | GG vs. CC | 0.032, 1.50 (1.02–2.17) | 0.008, 1.80 (1.15–2.80) | 0.356, 1.21 (0.75–1.99) | |
| GG | 76 (12.0%) | 41 (13.6%) | 35 (10.6%) | 66 (9.2%) | 0.991 | GG vs. GC | 0.272, 1.20 (0.81–1.75) | 0.181, 1.37 (0.90–2.05) | 0.648, 1.15 (0.74–1.79) |
| GC | 285 (45.2%) | 140 (46.5%) | 145 (44.0%) | 304 (42.2%) | GC vs. CC | 0.092, 1.20 (0.95–1.50) | 0.045, 1.31 (1.00–1.75) | 0.446, 1.09 (0.81–1.45) | |
| CC | 270 (42.8%) | 120 (39.9%) | 150 (45.4%) | 350 (48.6%) | GG vs. GC + CC | 0.086, 1.33 (0.92–1.90) | 0.035, 1.59 (1.01–2.34) | 0.463, 1.15 (0.79–1.80) | |
| GG + GC vs. CC | 0.032, 1.25 (1.05–1.59) | 0.011, 1.42 (1.10–1.85) | 0.342, 1.11 (0.85–1.40) | ||||||
aNOA, nonobstructive azoospermia
bGenotypic frequency of SNPs in normal controls was tested for departure from Hardy-Weinberg equilibrium (HWE) using the χ2 test
cThe P value was calculated using two-sided χ2 test
OR (95% CI) was estimated by logistic regression analysis, and adjusted for age, marriage, abstinence time and semen parameters
Meta-analysis of the four SNPs and male infertility susceptibility
The characteristics of included studies for this meta-analysis were presented in Table 3. In the control groups of included studies, the genotypic frequencies of rs12097821, rs2477686, rs6080550, and rs10842262 were all concordant with the HWE. For the four SNPs, their association with male infertility susceptibility was evaluated in total group of infertile men, as well as in subtypes of infertile men (NOA and oligozoospermia).
Table 3.
Characteristics of previous studies and the present study
| References (author, year) | Country (ethnicity) | Male infertility | Genotyping assay | Case, control (n) | HWE | ||
|---|---|---|---|---|---|---|---|
| Total | Allele | Genotype | |||||
| PRMT6 rs12097821 polymorphism | G/T | GG/GT/TT | |||||
| Sato et al. 2013[6] | Japan (Asian) | NOAa | PCR-RFLP | 490, 1167 | 795/185, 1917/417 | 322/151/17, 788/341/38 | 0.881 |
| Hu et al., 2012[12] | China (Asian) | NOA | SNP Array Chip | 2920, 5727 | 4106/1734, 8562/2892 | 1456/1194/270, 3205/2152/370 | 0.731 |
| Zou et al., 2014[10] | China (Asian) | NOA | Sequencing | 525, 512 | 735/315, 731/293 | 265/205/55, 255/221/36 | 0.200 |
| Tu et al., 2015[7] | China (Asian) | NOA | TaqMan assay | 545, 632 | 786/304, 905/359 | 280/226/39, 317/271/44 | 0.172 |
| Liu-1 et al., 2017[8] | China (Asian) | Male infertility | Snapshot PCR | 184, 51 | 266/102, 78/24 | 99/68/17, 29/20/2 | 0.522 |
| NOA | Snapshot PCR | 97, 51 | 136/58, 78/24 | 48/40/9, 29/20/2 | 0.522 | ||
| Oligozoospermia | Snapshot PCR | 87, 51 | 130/44, 78/24 | 51/28/8, 29/20/2 | 0.522 | ||
| Liu-2 et al., 2017[9] | China (Asian) | Male infertility | MassARRAY | 134, 454 | 182/86, 657/251 | 61/60/13, 232/193/29 | 0.182 |
| Asthenozoospermia | MassARRAY | 95, 454 | 130/60, 657/251 | 45/40/10, 232/193/29 | 0.182 | ||
| Oligoasthenozoospermia | MassARRAY | 21, 454 | 25/17, 657/251 | 6/13/2, 232/193/29 | 0.182 | ||
| Oligozoospermia | MassARRAY | 18, 454 | 27/9, 657/251 | 10/7/1, 232/193/29 | 0.182 | ||
| This study, 2018 | China (Asian) | Male infertility | RFLP-PCR | 631, 720 | 925/337, 1110/330 | 340/246/45, 428/254/38 | 0.968 |
| NOA | RFLP-PCR | 301, 720 | 425/177, 1110/330 | 150/125/26, 428/254/38 | 0.968 | ||
| Oligozoospermia | RFLP-PCR | 330, 720 | 500/160, 1110/330 | 190/121/19, 428/254/38 | 0.968 | ||
| PEX10 rs2477686 polymorphism | G/C | GG/GC/CC | |||||
| Sato et al., 2013[6] | Japan (Asian) | NOA | PCR-RFLP | 485, 1161 | 131/839, 313/2009 | 9/113/363, 18/277/866 | 0.436 |
| Hu et al., 2012[12] | China (Asian) | NOA | SNP Array Chip | 2882, 5729 | 837/4927, 1225/10233 | 88/661/2133, 76/1073/4580 | 0.146 |
| Zou et al., 2014[10] | China (Asian) | NOA | Sequencing | 524, 518 | 138/910,117/919 | 13/112/399,11/95/412 | 0.054 |
| Tu et al., 2015[7] | China (Asian) | NOA | TaqMan assay | 545, 632 | 154/936, 160/1104 | 14/126/405, 11/138/483 | 0.753 |
| Liu-1 et al., 2017[8] | China (Asian) | Male infertility | Snapshot PCR | 183, 51 | 36/330, 17/85 | 1/34/148, 0/17/34 | 0.153 |
| NOA | Snapshot PCR | 96, 51 | 22/170, 17/85 | 1/20/75, 0/17/34 | 0.153 | ||
| Oligozoospermia | Snapshot PCR | 87, 51 | 14/160, 17/85 | 0/14/73, 0/17/34 | 0.153 | ||
| Liu-2 et al., 2017[9] | China (Asian) | Male infertility | MassARRAY | 135, 456 | 38/232, 114/798 | 7/24/104, 5/104/347 | 0.363 |
| Asthenozoospermia | MassARRAY | 95, 456 | 22/168, 114/798 | 3/16/76, 5/104/347 | 0.363 | ||
| Oligoasthenozoospermia | MassARRAY | 22, 456 | 13/31, 114/798 | 4/5/13, 5/104/347 | 0.363 | ||
| Oligozoospermia | MassARRAY | 18, 456 | 3/33, 114/798 | 0/3/15, 5/104/347 | 0.363 | ||
| This study, 2018 | China (Asian) | Male infertility | PCR-RFLP | 631, 720 | 146/1316, 171/1269 | 9/128/494, 10/151/559 | 0.957 |
| NOA | PCR-RFLP | 301, 720 | 85/517, 171/1269 | 6/73/222, 10/151/559 | 0.957 | ||
| Oligozoospermia | PCR-RFLP | 330, 720 | 61/599, 171/1269 | 3/55/272, 10/151/559 | 0.957 | ||
| SIRPA-SIRPG rs6080550 polymorphism | A/G | AA/AG/GG | |||||
| Sato et al., 2013[6] | Japan (Asian) | NOA | RFLP-PCR | 481, 1158 | 134/828, 333/1983 | 8/118/355, 30/273/855 | 0.148 |
| Hu et al.,2012[12] | China (Asian) | NOA | SNP Array Chip | 2923, 5731 | 1340/4506, 2177/9285 | 154/1032/1737, 215/1747/3769 | 0.478 |
| Liu-2 et al., 2017[9] | China (Asian) | Male infertility | MassARRAY | 135, 454 | 113/157, 402/506 | 27/59/49, 84/234/136 | 0.343 |
| Asthenozoospermia | MassARRAY | 95, 454 | 85/105, 402/506 | 21/43/31, 84/234/136 | 0.343 | ||
| Oligoasthenozoospermia | MassARRAY | 22, 454 | 17/27, 402/506 | 2/13/7, 84/234/136 | 0.343 | ||
| Oligozoospermia | MassARRAY | 18, 454 | 11/25, 402/506 | 4/3/11, 84/234/136 | 0.343 | ||
| This study, 2018 | China (Asian) | Male infertility | RFLP-PCR | 631, 720 | 368/894, 363/1077 | 53/261/317, 46/271/403 | 0.961 |
| NOA | RFLP-PCR | 301, 720 | 172/430, 363/1077 | 24/123/154, 46/271/403 | 0.961 | ||
| Oligozoospermia | RFLP-PCR | 330, 720 | 196/464, 363/1077 | 29/138/163, 46/271/403 | 0.961 | ||
| SOX5 rs10842262 polymorphism | G/C | GG/GC/CC | |||||
| Sato et al. 2013[6] | Japan (Asian) | NOA | PCR-RFLP | 487, 1155 | 373/601, 827/1483 | 81/211/195, 146/535/474 | 0.794 |
| Hu et al., 2012[12] | China (Asian) | NOA | SNP Array Chip | 2887, 5711 | 2121/3653, 3656/7766 | 338/1445/1104, 581/2494/2636 | 0.802 |
| Zou et al., 2014[10] | China (Asian) | NOA | sequencing | 522, 529 | 351/693, 291/767 | 65/221/236, 41/209/279 | 0.831 |
| Tu et al., 2015[7] | China (Asian) | NOA | TaqMan assay | 545, 632 | 385/705, 401/863 | 58/269/218, 65/271/296 | 0.798 |
| Liu-1 et al., 2017[8] | China (Asian) | Male infertility | Snapshot PCR | 177, 51 | 89/265, 28/74 | 15/59/103, 6/16/29 | 0.129 |
| NOA | Snapshot PCR | 96, 51 | 55/137, 28/74 | 9/37/50, 6/16/29 | 0.129 | ||
| Oligozoospermia | Snapshot PCR | 81, 51 | 34/128, 28/74 | 6/22/53, 6/16/29 | 0.129 | ||
| This study, 2018 | China (Asian) | Male infertility | PCR-RFLP | 631, 720 | 437/826, 436/1004 | 76/285/270, 66/304/350 | 0.998 |
| NOA | PCR-RFLP | 301, 720 | 222/380, 436/1004 | 41/140/120, 66/304/350 | 0.998 | ||
| Oligozoospermia | PCR-RFLP | 330, 720 | 215/446, 436/1004 | 35/145/150, 66/304/350 | 0.998 | ||
aNOA, nonobstructive azoospermia
Meta-analysis of PRMT6 rs12097821 polymorphism
A sum of seven publications for rs12097821 that met the inclusion criteria were finally retrieved. As shown in Table 4, we found that rs12097821 significantly decreased male infertility risk under two models: G vs. T, OR = 0.84, 95% CI = 0.80–0.89, P < 0.001; GG vs. GT, OR = 0.87, 95% CI = 0.81–0.94, P < 0.001. However, our data indicated that rs12097821 was not associated with NOA or oligozoospermia under none of the six genetic models (G vs. T, GG vs. TT, GG vs. GT, GT vs. TT, GG vs. GT + TT or GG + GT vs. TT).
Table 4.
Meta-analysis of associations between PRMT6 rs12097821 polymorphism and male infertility susceptibility
| Genetic model | Heterogeneity test | Summary OR (95% CI) | Hypothesis test | Studies (n) | |||
|---|---|---|---|---|---|---|---|
| Q | P | I 2 | Z | P | |||
| rs12097821 and male infertility | |||||||
| G vs. T | 9.22 | 0.162 | 34.9% | 0.84 (0.80–0.89) | 6.20 | < 0.001 | 7 |
| GG vs. TT | 167.09 | < 0.001 | 96.4% | 1.05 (0.92–1.19) | 0.05 | 0.957 | 7 |
| GG vs. GT | 8.66 | 0.194 | 30.7% | 0.87 (0.81–0.94) | 3.71 | < 0.001 | 7 |
| GT vs. TT | 120.01 | < 0.001 | 95.0% | 1.13 (1.00–1.30) | 0.16 | 0.873 | 7 |
| GG vs. GT + TT | 80.75 | < 0.001 | 92.6% | 0.91 (0.85–0.97) | 0.05 | 0.963 | 7 |
| GG + GT vs. TT | 168.80 | < 0.001 | 96.4% | 0.91 (0.89–0.94) | 0.02 | 0.988 | 7 |
| rs12097821 and nonobstructive azoospermia (NOA) | |||||||
| G vs. T | 11.49 | 0.043 | 56.5% | 0.86 (0.77–0.97) | 2.57 | 0.010 | 6 |
| GG vs. TT | 168.50 | < 0.001 | 97.0% | 1.00 (0.38–2.66) | 0.01 | 0.995 | 6 |
| GG vs. GT | 10.11 | 0.072 | 50.5% | 0.90 (0.79–1.03) | 1.49 | 0.136 | 6 |
| GT vs. TT | 118.93 | < 0.001 | 95.8% | 1.13 (0.48–2.61) | 0.27 | 0.784 | 6 |
| GG vs. GT + TT | 83.72 | < 0.001 | 94.0% | 1.00 (0.70–1.43) | 0.02 | 0.987 | 6 |
| GG + GT vs. TT | 169.03 | < 0.001 | 97.0% | 1.05 (0.41–2.68) | 0.09 | 0.925 | 6 |
| rs12097821 and oligozoospermia | |||||||
| G vs. T | 0.28 | 0.869 | 0% | 0.94 (0.77–1.14) | 0.62 | 0.533 | 3 |
| GG vs. TT | 0.79 | 0.673 | 0% | 0.83 (0.50–1.40) | 0.69 | 0.488 | 3 |
| GG vs. GT | 0.72 | 0.699 | 0% | 0.98 (0.76–1.26) | 0.16 | 0.875 | 3 |
| GT vs. TT | 1.29 | 0.524 | 0% | 0.85 (0.50–1.43) | 0.62 | 0.533 | 3 |
| GG vs. GT + TT | 0.38 | 0.828 | 0% | 0.96 (0.75–1.22) | 0.35 | 0.724 | 3 |
| GG + GT vs. TT | 1.00 | 0.607 | 0% | 0.84 (0.50–1.39) | 0.69 | 0.487 | 3 |
Meta-analysis of PEX10 rs2477686 polymorphism
A sum of seven publications for rs2477686 that met the inclusion criteria were finally retrieved. In Table 5, our results showed that GG genotype significantly increases the susceptibility to male infertility and NOA when compared with GC and/or CC genotypes (GG vs. CC, GG vs. GC, and GG vs. GC + CC). However, the significant association did not remain in oligozoospermia.
Table 5.
Meta-analysis of associations between PEX10 rs2477686 polymorphism and male infertility susceptibility
| Genetic model | Heterogeneity test | Summary OR (95% CI) | Hypothesis test | Studies (n) | |||
|---|---|---|---|---|---|---|---|
| Q | P | I 2 | Z | P | |||
| rs2477686 and male infertility | |||||||
| G vs. C | 30.65 | < 0.001 | 80.4% | 1.058 (0.86–1.30) | 0.55 | 0.585 | 7 |
| GG vs. CC | 9.88 | 0.130 | 39.3% | 1.973 (1.55–2.51) | 5.51 | < 0.001 | 7 |
| CG vs.CC | 19.14 | 0.004 | 68.6% | 1.026 (0.85–1.24) | 0.27 | 0.788 | 7 |
| GG vs. CG | 7.77 | 0.256 | 22.7% | 1.662 (1.29–2.13) | 3.99 | < 0.001 | 7 |
| GG + CG vs.CC | 21.49 | 0.002 | 72.1% | 1.06 (0.88–1.29) | 0.64 | 0.524 | 7 |
| GG vs. GC + CC | 9.29 | 0.158 | 35.4% | 1.904 (1.50–2.42) | 5.24 | < 0.001 | 7 |
| rs2477686 and nonobstructive azoospermia (NOA) | |||||||
| G vs. C | 14.55 | 0.012 | 65.6% | 1.167 (0.99–1.37) | 1.86 | 0.063 | 6 |
| GG vs. CC | 5.66 | 0.341 | 11.6% | 1.982 (1.55–2.54) | 5.38 | < 0.001 | 6 |
| CG vs.CC | 10.38 | 0.065 | 51.8% | 1.138 (0.97–1.33) | 1.61 | 0.106 | 6 |
| GG vs.CG | 2.93 | 0.710 | 0% | 1.612 (1.25–2.09) | 3.64 | < 0.001 | 6 |
| GG + CG vs.CC | 12.83 | 0.025 | 61.0% | 1.169 (1.08–1.27) | 1.72 | 0.085 | 6 |
| GG vs. GC + CC | 5.00 | 0.416 | 0% | 1.898 (1.48–2.43) | 5.06 | < 0.001 | 6 |
| rs2477686 and oligozoospermia | |||||||
| G vs. C | 1.75 | 0.417 | 0% | 0.698 (0.53–0.92) | 2.53 | 0.011 | 3 |
| GG vs. CC | 0.54 | 0.463 | 0% | 0.712 (0.22–2.35) | 0.56 | 0.577 | 2 |
| CG vs.CC | 2.20 | 0.333 | 9.0% | 0.680 (0.50–0.92) | 2.47 | 0.014 | 3 |
| GG vs.CG | 0.49 | 0.483 | 0% | 0.943 (0.28–3.20) | 0.09 | 0.925 | 2 |
| GG + CG vs.CC | 2.12 | 0.346 | 5.7% | 0.690 (0.59–0.81) | 2.59 | 0.010 | 3 |
| GG vs. GC + CC | 0.57 | 0.450 | 0% | 0.752 (0.23–2.48) | 0.47 | 0.640 | 2 |
Meta-analysis of SIRPA-SIRPG rs6080550 polymorphism
A sum of four publications for rs6080550 that met the inclusion criteria were finally retrieved. As shown in Table 6, rs6080550 was suggested to significantly associate with male infertility and NOA under different genetic models. Specifically, rs6080550 conferred an increased risk to male infertility under AA vs. AG + GG model (OR = 1.31, 95% CI = 1.10–1.55, P = 0.002), while to NOA under AG vs. GG model (OR = 1.24, 95% CI = 1.14–1.35, P < 0.001) and AA vs. AG + GG model (OR = 1.33, 95% CI = 1.10–1.61, P = 0.003). However, we observed that there was no association between rs6080550 and oligozoospermia.
Table 6.
Meta-analysis of associations between SIRPA-SIRPG rs6080550 polymorphism and male infertility susceptibility
| Genetic model | Heterogeneity test | Summary OR (95% CI) | Hypothesis test | Studies (n) | |||
|---|---|---|---|---|---|---|---|
| Q | P | I 2 | Z | P | |||
| rs6080550 and male infertility | |||||||
| A vs. G | 9.96 | 0.019 | 69.9% | 1.12 (0.96–1.30) | 1.39 | 0.164 | 4 |
| AA vs. GG | 7.35 | 0.062 | 59.2% | 1.21 (0.85–1.71) | 1.07 | 0.287 | 4 |
| AA vs. AG | 2.62 | 0.455 | 0.0% | 1.17 (0.98–1.40) | 1.74 | 0.081 | 4 |
| AG vs. GG | 8.89 | 0.031 | 66.3% | 1.11 (0.92–1.35) | 1.11 | 0.267 | 4 |
| AA vs. AG + GG | 4.38 | 0.224 | 31.4% | 1.31 (1.10–1.55) | 3.08 | 0.002 | 4 |
| AA + AG vs. GG | 10.38 | 0.016 | 71.1% | 1.17 (1.09–1.26) | 1.14 | 0.255 | 4 |
| rs6080550 and non-obstructive azoospermia (NOA) | |||||||
| A vs. G | 5.60 | 0.061 | 64.3% | 1.16 (0.99–1.36) | 1.80 | 0.071 | 3 |
| AA vs. GG | 4.55 | 0.103 | 56.0% | 1.27 (0.84–1.92) | 1.12 | 0.263 | 3 |
| AA vs. AG | 2.50 | 0.287 | 19.9% | 1.15 (0.94–1.40) | 1.39 | 0.165 | 3 |
| AG vs. GG | 2.45 | 0.294 | 18.2% | 1.24 (1.14–1.35) | 4.99 | <0.001 | 3 |
| AA vs. AG + GG | 3.83 | 0.147 | 47.8% | 1.33 (1.10–1.61) | 2.95 | 0.003 | 3 |
| AA + AG vs. GG | 4.27 | 0.118 | 53.2% | 1.20 (1.02–1.41) | 2.20 | 0.028 | 3 |
| rs6080550 and oligozoospermia | |||||||
| A vs. G | 4.56 | 0.033 | 78.1% | 0.90 (0.41–1.97) | 0.27 | 0.791 | 2 |
| AA vs. GG | 2.24 | 0.135 | 55.3% | 1.32 (0.83–2.07) | 1.18 | 0.239 | 2 |
| AA vs. AG | 1.81 | 0.178 | 44.8% | 1.38 (0.85–2.22) | 1.31 | 0.191 | 2 |
| AG vs. GG | 9.48 | 0.002 | 89.5% | 0.49 (0.07–3.75) | 0.68 | 0.495 | 2 |
| AA vs. AG + GG | 0.03 | 0.855 | 0% | 1.39 (0.89–2.17) | 1.44 | 0.149 | 2 |
| AA + AG vs. GG | 9.36 | 0.002 | 89.3% | 0.64 (0.14–2.95) | 0.57 | 0.567 | 2 |
Meta-analysis of SOX5 rs10842262 polymorphism
A sum of six publications for rs10842262 that met the inclusion criteria were finally retrieved. In Table 7, we found that rs10842262 was significantly associated with male infertility and NOA under the same genetic models (G vs. C, GG vs. CC, GC vs. CC, GG vs. GC + CC and GG + GC vs. CC). Whereas, no significant association was found for rs10842262 and oligozoospermia.
Table 7.
Meta-analysis of associations between SOX5 rs10842262 polymorphism and male infertility susceptibility
| Genetic model | Heterogeneity test | Summary OR (95% CI) | Hypothesis test | Studies (n) | |||
|---|---|---|---|---|---|---|---|
| Q | P | I 2 | Z | P | |||
| rs10842262 and male infertility | |||||||
| G vs. C | 4.09 | 0.536 | 0.0% | 1.22 (1.15–1.28) | 7.40 | < 0.001 | 6 |
| GG vs. CC | 4.19 | 0.523 | 0.0% | 1.40 (1.24–1.57) | 5.64 | < 0.001 | 6 |
| GG vs. CG | 7.81 | 0.167 | 36.0% | 1.09 (0.97–1.22) | 1.42 | 0.156 | 6 |
| CG vs. CC | 9.24 | 0.100 | 45.9% | 1.30 (1.20–1.40) | 6.84 | < 0.001 | 6 |
| GG vs. CG + CC | 5.69 | 0.338 | 12.1% | 1.23 (1.10–1.37) | 3.68 | < 0.001 | 6 |
| GG + CG vs. CC | 6.90 | 0.228 | 27.5% | 1.32 (1.23–1.41) | 7.63 | < 0.001 | 6 |
| rs10842262 and nonobstructive azoospermia (NOA) | |||||||
| G vs. C | 3.68 | 0.597 | 0.0% | 1.23 (1.16–1.30) | 7.59 | < 0.001 | 6 |
| GG vs. CC | 4.31 | 0.505 | 0.0% | 1.42 (1.26–1.60) | 5.78 | < 0.001 | 6 |
| GG vs. CG | 8.33 | 0.139 | 40.0% | 1.09 (0.97–1.23) | 1.44 | 0.149 | 6 |
| CG vs. CC | 8.50 | 0.131 | 41.2% | 1.31 (1.22–1.42) | 7.02 | < 0.001 | 6 |
| GG vs. CG + CC | 6.08 | 0.298 | 17.8% | 1.24 (1.11–1.38) | 3.77 | < 0.001 | 6 |
| GG + CG vs. CC | 5.98 | 0.308 | 16.4% | 1.33 (1.24–1.43) | 7.83 | < 0.001 | 6 |
| rs10842262 and oligozoospermia | |||||||
| G vs. C | 2.17 | 0.140 | 54.0% | 1.06 (0.88–1.28) | 0.59 | 0.556 | 2 |
| GG vs. CC | 1.51 | 0.219 | 33.9% | 1.12 (0.73–1.71) | 0.52 | 0.602 | 2 |
| GG vs. CG | 0.36 | 0.546 | 0.0% | 1.06 (0.69–1.63) | 0.27 | 0.787 | 2 |
| CG vs. CC | 0.85 | 0.357 | 0.0% | 1.07 (0.82–1.38) | 0.49 | 0.626 | 2 |
| GG vs. CG + CC | 1.08 | 0.298 | 7.7% | 1.09 (0.72–1.63) | 0.40 | 0.691 | 2 |
| GG + CG vs. CC | 1.57 | 0.211 | 36.2% | 1.07 (0.84–1.37) | 0.56 | 0.577 | 2 |
Discussion
Up to now, several replication studies have been conducted to validate the four GWAS-linked SNPs (rs12097821, rs2477686, rs10842262, and rs6080550) with NOA, while the results remain conflicting rather than conclusive. Sato et al. were the first group to repeat an association study between the four SNPs and NOA in a Japanese population, but showed no significant difference for these four SNPs, which is almost inconsistent with findings of the previous GWAS in the Han Chinese population by Hu et al. [5, 6]. Furthermore, 2 replication studies in the Han Chinese population showed that rs12097821 and rs2477686 were not associated with NOA [7, 10]. However, Zou et al. [10], but not Tu et al. [7], reported that there was a significant association between rs10842262 and NOA. Consistent with the finding of Tu et al., Liu et al. found that the rs12097821, rs2477686, and rs10842262 were not associated with NOA, and also were not associated with oliogozoospermia [8]. Similarly, the significant association of the four SNPs with NOA was partially repeated in the present case-control study, and only rs12097821 and rs10842262 were shown to be associated with NOA in a central Chinese population. Moreover, the association of the four SNPs with idiopathic male infertility (asthenozoospermia, oligozoospermia, and oligoasthenozoospermia) was also explored, and Liu et al. found that rs2477686, but not rs12097821, rs10842262, or rs6080550, was significantly associated with idiopathic male infertility, especially oligoasthenozoospermia [9].
The possible reasons for these discrepancies might be as follows. First, there may be a risk of false positive of the risk loci reported by the original GWAS study in Chinese population, which leads to subsequent validation studies failing to replicate the original result. Second, the interaction of these four SNPs and different genetic backgrounds among different ethnic populations may produce the variation of individual susceptibility to male infertility. Third, the insufficient sample size may cause inadequate statistical strength. In the present study, rs2477686 and rs6080550 were not significantly associated with NOA susceptibility in Chinese population. However, rs2477686 and rs6080550 displayed associations in the same direction (per-genetic comparison ORs > 1) as reported in the previous GWAS (Table 2). Therefore, the four SNPs need to be investigated using more samples in other ethnic populations or in Chinese population from different geographic regions for further confirmation.
To solve the discrepancies and improve the statistical strength, a meta-analysis followed combining results from previously published literature and the present case-control study was conducted. Interestingly, three of the four SNPs (rs2477686, rs10842262, and rs6080550) displayed significant associations with NOA in the same direction (per-allele ORs > 1) in combined cohorts as previously reported in the GWAS, while none of the four SNPs was associated with oligozoospermia. When combing the NOA and oligozoospermia as total male infertility, all the four SNPs were shown to associate with male infertility. However, one limitation of this study should be acknowledged. Since the publication bias can be evaluated for meta-analysis with sufficient numbers of included studies (n > 10), the assessment of publication bias was not performed. Therefore, we could not eliminate the possibility of publication bias in the present meta-analysis.
Previous knowledge suggested that PRMT6 (rs12097821) may regulate POLB and thereby function in synapsis and recombination during meiosis [17–20], PEX10 (rs2477686) has roles in male fertility (e.g., regulating spermatocyte cytokinesis) [21], SIRPA (rs6080550) may modulate engraftment of human hematopoietic stem cells [22], and SOX5 (rs10842262) regulates gene expression during spermatogenesis [23, 24]. These findings collectively suggested that these four SNPs may serve as potential biomarkers for male infertility predisposition in Asian population. However, it cannot rule out the possibility that the four SNPs may not be the causal loci but rather be in linkage disequilibrium with the causal loci. Moreover, the effect of four SNPs on functions of PRMT6, PEX10, SIRPA-SIRPG, and SOX5 was not assessed in spermatogenesis from individuals with different genotypes (in vivo), which should be analyzed in further functional study.
In conclusion, the rs12097821 and rs10842262 were strongly associated with NOA in Chinese men of Hubei province. In addition, the meta-analysis that followed showed that in Asian population, rs2477686, rs6080550, and rs10842262 were significantly associated with male infertility especially with NOA, while rs12097821 was only associated with total male infertility. Our current findings suggest that rs2477686, rs6080550, and rs10842262 may indeed be the genetic risk factors for NOA. However, cohort expansion and further mechanistic studies on the role of these genetic factors that influence spermatogenesis and sperm progressive motility are necessary for the future.
Acknowledgments
The authors thank all the participants and investigators enrolled in this study.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiuli Gu, Phone: +86 027-82742277, Email: glucose2016@sina.com.
Chengliang Xiong, Phone: +86 027-82742277, Email: clxiong951@sina.com.
References
- 1.Mclachlan RI, de Kretser DM. Male infertility: the case for continued research. Med J Aust. 2001;174(3):116–117. doi: 10.5694/j.1326-5377.2001.tb143180.x. [DOI] [PubMed] [Google Scholar]
- 2.Krausz C, Riera-Escamilla A. Genetics of male infertility. Nat Rev Urol. 2018;15(6):369–384. doi: 10.1038/s41585-018-0003-3. [DOI] [PubMed] [Google Scholar]
- 3.DS D, AM B. The genetics of male infertility. J Urol. 2009;27(02):124–136. doi: 10.1055/s-0029-1202301. [DOI] [PubMed] [Google Scholar]
- 4.Ferlin A, Foresta C. New genetic markers for male infertility. Curr Opin Obstet Gynecol. 2014;26(3):193–198. doi: 10.1097/GCO.0000000000000061. [DOI] [PubMed] [Google Scholar]
- 5.Hu Z, Xia Y, Guo X, Dai J, Li H, Hu H, Jiang Y, Lu F, Wu Y, Yang X, Li H, Yao B, Lu C, Xiong C, Li Z, Gui Y, Liu J, Zhou Z, Shen H, Wang X, Sha J. A genome-wide association study in Chinese men identifies three risk loci for non-obstructive azoospermia. Nat Genet. 2012;44(2):183–186. doi: 10.1038/ng.1040. [DOI] [PubMed] [Google Scholar]
- 6.Sato Y, Jinam T, Iwamoto T, Yamauchi A, Imoto I, Inoue I, Tajima A. Replication study and meta-analysis of human nonobstructive azoospermia in Japanese populations. Biol Reprod. 2013;88(4):87. doi: 10.1095/biolreprod.112.106377. [DOI] [PubMed] [Google Scholar]
- 7.Tu W, Liu Y, Shen Y, Yan Y, Wang X, Yang D, Li L, Ma Y, Tao D, Zhang S, Yang Y. Genome-wide loci linked to non-obstructive azoospermia susceptibility may be independent of reduced sperm production in males with normozoospermia. Biol Reprod. 2015;92(2):41. doi: 10.1095/biolreprod.114.125237. [DOI] [PubMed] [Google Scholar]
- 8.Liu F, Liu Y, Bao J, Jin R, Bai G, Tang D, et al. Relationship between PRMT6, PEX10, SOX5 and CYP19 gene polymorphism and dyszoospermia. Ningxia Med J. 2017;39(2):97–100. [Google Scholar]
- 9.Liu SY, Zhang CJ, Peng HY, Sun H, Lin KQ, Huang XQ, et al. Strong association of SLC1A1 and DPF3 gene variants with idiopathic male infertility in Han Chinese. Asian J Androl. 2017;19(4):486–492. doi: 10.4103/1008-682X.178850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou S, Li Z, Wang Y, Chen T, Song P, Chen J, He X, Xu P, Liang M, Luo K, Zhu X, Tian E, du Q, Wen Z, Li Z, Wang M, Sha Y, Cao Y, Shi Y, Hu H. Association study between polymorphisms of PRMT6, PEX10, SOX5, and nonobstructive azoospermia in the Han Chinese population. Biol Reprod. 2014;90(5):96. doi: 10.1095/biolreprod.113.116541. [DOI] [PubMed] [Google Scholar]
- 11.Munafò MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20(9):439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Esteves SC, Zini A, Aziz N, Alvarez JG, Sabanegh ES Jr, Agarwal A. Critical appraisal of World Health Organization's new reference values for human semen characteristics and effect on diagnosis and treatment of subfertile men. Urology 2012;79(1):16–22. [DOI] [PubMed]
- 13.Sedgwick P. Multiple significance tests: the Bonferroni correction. BMJ 2012;344.
- 14.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127(9):820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 15.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 16.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 17.El-Andaloussi N, Valovka T, Toueille M, Steinacher R, Focke F, Gehrig P, et al. Arginine methylation regulates DNA polymerase beta. Mol Cell. 2006;22(1):51–62. doi: 10.1016/j.molcel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, et al. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379(6561):183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 19.Olsen AK, Bjørtuft H, Wiger R, Holme J, Seeberg E, Bjørås M, Brunborg G. Highly efficient base excision repair (BER) in human and rat male germ cells. Nucleic Acids Res. 2001;29(8):1781–1790. doi: 10.1093/nar/29.8.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plug AW, Clairmont CA, Sapi E, Ashley T, Sweasy JB. Evidence for a role for DNA polymerase beta in mammalian meiosis. Proc Natl Acad Sci U S A. 1997;94(4):1327–1331. doi: 10.1073/pnas.94.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, Liu Z, Huang X. Drosophila models of peroxisomal biogenesis disorder: peroxins are required for spermatogenesis and very-long-chain fatty acid metabolism. Hum Mol Genet. 2010;19(3):494–505. doi: 10.1093/hmg/ddp518. [DOI] [PubMed] [Google Scholar]
- 22.Takenaka K, Prasolava TK, Wang JC, Mortin-Toth SM, Khalouei S, Gan OI, et al. Polymorphism in Sirpa modulates engraftment of human hematopoietic stem cells. Nat Immunol. 2007;8(12):1313–1323. doi: 10.1038/ni1527. [DOI] [PubMed] [Google Scholar]
- 23.Denny P, Swift S, Connor F, Ashworth A. An SRY-related gene expressed during spermatogenesis in the mouse encodes a sequence-specific DNA-binding protein. EMBO J. 1992;11(10):3705–3712. doi: 10.1002/j.1460-2075.1992.tb05455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Budde LM, Wu C, Tilman C, Douglas I, Ghosh S. Regulation of IkappaBbeta expression in testis. Mol Biol Cell. 2002;13(12):4179–4194. doi: 10.1091/mbc.01-07-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]