 As a well-established human carcinogen, arsenic has increased the risk of lung cancer over the past decades.
As a well-established human carcinogen, arsenic has increased the risk of lung cancer over the past decades.
Abstract
As a well-established human carcinogen, arsenic has increased the risk of lung cancer over the past decades. Wide exposure to arsenic in the environment has attracted the attention of scientists. Its carcinogenicity at early life stages has been observed in certain animal studies already, yet current evidence is insufficient to extrapolate this to humans. Although the mechanisms of lung cancer induced by arsenic remain unclear, most of them are related to the biotransformation of arsenic, which would further provide target sites for precaution and therapy. This review comprehensively summarizes current studies associated to arsenic exposure and lung cancer and the mechanism of its carcinogenesis in lung cancer in three sections, namely, epidemiological studies, experimental studies, and mechanistic studies. In addition, prevention and treatment strategies as well as directions for future studies are discussed.
1. Introduction
Arsenic is a naturally occurring semi-metal and is widely distributed on a global scale. It has been estimated that more than 100 million people are exposed to high concentrations of arsenic (>50 μg L–1) via drinking water, and millions of individuals are significantly exposed to arsenic via food globally.1 Additionally, it is reported that lung cancer is one of the major causes of death in people exposed to high levels of arsenic via drinking water.2 The International Agency for Research on Cancer (IARC) classifies arsenic as a Group I carcinogen, which is capable of inducing human malignant lung tumors. A considerable number of people around the world are under high risk of lung cancer caused by arsenic, especially nonsmokers.3 The most common type of lung cancer caused by arsenic exposure is the squamous cell carcinoma.4,5 This review comprehensively summarizes current studies on the mechanisms of arsenic exposure that cause lung cancer in three stages including epidemiology, animal studies, and molecular mechanism investigations. In addition, prevention and treatment strategies as well as directions for future studies are included.
2. Epidemiological studies
Arsenic in drinking water was determined as a cause for human lung cancer by the IARC in 2004. Currently, most of the studies are performed in areas with higher concentrations of arsenic (up to several hundred micrograms per liter) in drinking water, such as regions in Taiwan, Chile, Japan and Argentina.6 According to the linear extrapolation of cancer risk observed at higher doses, the World Health Organization (WHO) set a threshold value of 10 μg L–1 for arsenic in drinking water.7 However, there is still a controversy among epidemiological studies on whether low to moderate arsenic concentrations possess any potential threats.7–9 In addition, one meta-analysis and two recent meta-regression studies failed to reach consensus on this issue.10–12
Considering the varying results from previous studies, our group recently performed a dose-responsive meta-analysis on data extracted from 6 eligible case-control studies based on our inclusion criteria.13 The study identified an evident lung cancer risk even at the standard limit of 10 μg L–1. There was a linear association between arsenic concentration in drinking water and logarithmically transformed lung cancer risk (p for nonlinearity = 0.47). Previous systematic reviews concluded that people exposed to high levels of arsenic had added risk of lung cancer.14 However, the association between lung cancer risk and low to moderate arsenic concentration (<100 μg L–1) is still inconclusive. The conclusion from our study differs from previous reports, which could be related to different statistical methods and inclusion criterion.11,12 Further epidemiological studies are still needed to confirm our conclusion and update the safe threshold of arsenic concentration in drinking water.
3. Experimental studies
3.1. Animal studies
Arsenic or its metabolite, as human carcinogens, fails to exhibit any tumorigenic effects on the lungs of immunocompetent animals.15 However, positive results were observed in transgenic mice, which were hypersensitive to carcinogens.16,17 In addition, it has been reported that arsenic enhanced the carcinogenic effects of other toxicants.18 Thus, arsenic is regarded as a kind of oncogenic promoter without direct genotoxicity, possibly by inhibiting DNA repair and/or increasing cell proliferation.15
Moreover, arsenic was proposed as a complete transplacental carcinogen by a series of animal studies with utero exposure. Pregnant mice were orally treated with sodium arsenate during a short period of gestation. Dose-dependent tumors were observed in the lung tissues of their offsprings.19 Further extending the arsenic exposure from the embryo stage to the whole life of its offspring could induce even malignant tumors at much lower doses.20 Conversely, another recently published study focused on 9 early life arsenic human exposure cases, showing inconsistent results for transplacental carcinogenesis.21
3.2. Cellular studies
The capability of arsenic caused lung cancer has been extensively studied in cell-based transformation studies as well.
First, short-term arsenic exposure exhibited genotoxic properties in primary human lung epithelial cells, of which the chromosome damage and DNA double strand breaks would induce tumor initiation.22 The increased levels of ROS with arsenic exposure caused angiogenesis, which further contributed to tumor growth and migration.23 Activation of AKT/p38 signals and phosphorylation of ERK1/2, as well as the increased expression of hypoxia-inducible factor 1 (HIF-1) and vascular endothelial growth factor (VEGF), were also reported to contribute to tumor initiation.23 The transformation from human lung bronchial epithelial cells (HBE) to mesnimenchymal cells (EMT) is the most common consequence of long-term arsenic exposure.24 Low dose arsenic-induced sub-lethal stress was reported to be the additional reason for the transformation of cells from HBE to cancer stem cells (CSS).25
Second, miR-21 mediated by the ROS-dependent signal transducer and activator of transcription 3 (STAT3) could suppress the tumorigenesis inhibitor programmed cell death 4 (PCD4) in arsenic-transformed HBE.26 The polarization of macrophages towards tumorigenesis-related activation status, HIF-2α-mediated lung cells inflammation, E-cadherin decrease and chromatin structure change have also been reported to facilitate arsenic-induced HBE malignant transformations.24,27–29 Moreover, a newly identified lung cancer-linked oncogene, MDIG (mineral dust-induced gene), was overexpressed in arsenic-transformed HBE by mediating the JNK and STAT3 signaling pathways.30 Besides that, the oncogenic properties of transformed cells are also correlated with the overexpression of antioxidant enzymes and antiapoptotic proteins, mediated by nuclear factor erythroid 2-related factor (Nrf2) and P62.31
Third, a cellular energy metabolism disorder induced by arsenic exposure also plays a critical role in promoting carcinogenesis through the change in its characteristic transformation from mitochondrial respiration to glycolysis. This metabolic shift is related to the elevated Polo-like kinase 1 through the activation of the PI3K/AKT/mTOR pathway. The activation of the AKT/STAT3 signaling pathway was also reported to enhance arsenic-transformed cell migration.32–34 Moreover, long-term arsenic exposure increased the process of glycolysis by stimulating the HIF-1 alpha translocation in the hypoxia status. Thus, arsenic-induced metabolic transformations observed in HBE cells are cellular anchorage-independent growth.35
4. Molecular mechanisms
The field of arsenic-caused lung cancer has attracted significant interest, but the published results have been too complicated to be consistent with each other. The detailed mechanism is still needed to understand the relationship. The purpose of this review is to sort out the underlying mechanisms based on the previous studies, and provide conclusions that will be helpful to future theoretical researches and clinical therapies.
4.1. Biotransformation of arsenic and its associated consequences
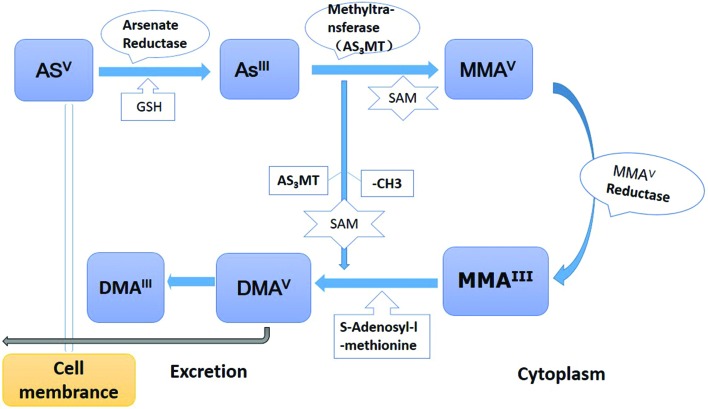
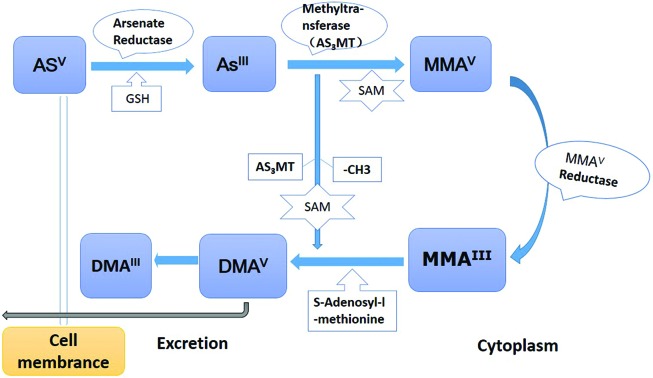
The metabolism of ingested inorganic arsenic is shown in Fig. 1: 1. Arsenate (Asv) is reduced to arsenite (AsIII) by arsenate reductase and glutathione (GSH); 2. AsIII is methylated by arsenic methyltransferase (AS3MT) to form monomethylarsonic acid (MMAV) using S-adenosylmethionine (SAM) as the methyl group donor; 3. MMAV reductase reduces MMAV to monomethylarsonous acid (MMAIII), then MMA3+ is further methylated to dimethylarsine acid (DMAV),36 which is methylated and reduced to form dimethlyarsinic acid (DMAIII).37 The whole biotransformation process is dependent on an individual's methylation capacity. Epidemiological studies demonstrated that inter-individual differences in arsenic metabolism accounted for the susceptibility of arsenic-related cancer.38,39
Fig. 1. Biotransformation process of arsenic in the human body. 1. Inorganic arsenic compounds are actively transported into cells by a transporter. The transported arsenate (AsV) would also be reduced to arsenite (AsIII) by arsenate reductase and glutathione (GSH). 2. AsIII is methylated by arsenic methyltransferase (AS3MT) to form monomethylarsonic acid (MMAV) with the methyl group donor (S-adenosylmethionine, SAM). 3. MMAV reductase reduces MMAV to monomethylarsonous acid (MMAIII). Following the MMAIII methylated with AS3MT, it was reduced to dimethlyarsinic acid (DMAIII).
Most of the current arsenic-carcinogenic studies are based on this metabolic model.40 First, the higher toxicity metabolites (MMAIII and DMAIII) were accumulated with their closer affinity to sulfhydryl groups in enzymes and receptors. Second, depletion of the essential antioxidants and free radical scavengers, such as GSH, could not maintain the low levels of reactive oxygen and nitrogen products (ROS/RNS), which caused the oxidative stress in lung tissues.41 Meanwhile, the increased arsenic methylated metabolites and mitochondrial impairment facilitated the production of excess ROS/RNS.42 Third, followed by the depletion of the methyl group donor (SAM), the original epigenetics of DNA and protein changed.43
4.2. Genetic changes
4.2.1. Direct genetic impairment
DNA damage is the most representative of direct genetic impairment. Long-term arsenic exposure could induce DNA double-strands breakage, which leads to DNA damage and chromosomal instability. The continuing discussions regarding the levels of DNA damage and the doses of arsenic provide some guidance in preventing the lung cancer induced by arsenic.44–46
In addition to directly inducing DNA damage (DNA mutation, DNA deletion, DNA double stranded break, DNA–DNA or DNA–protein crosslink), the induction of ROS levels by arsenic exposure could disrupt the chromosome structure and stability, resulting in end-to-end fusion, abnormal sister chromatid separation and aneuploidy.47–50 Based on a previous study, arsenic caused chromosome abnormalities were derived from not only oxidative damages but also the interaction between arsenic methylated metabolites and the sulfhydryl group of tubulin.50 In addition, studies also reported that the reactive nitrogen products (such as RNS) could induce DNA alkylation and deamination.51,52
4.2.2. DNA repair dysfunction
Multiple studies have demonstrated that arsenic impairs the DNA repair process by binding to DNA repair proteins. For example, through binding to the zinc finger motifs of xeroderma pigmentosum complementation group A (XPA) and poly (ADP-ribose) polymerase 1 (PARP1), arsenic suppressed the nucleotide excision repair and base excision repair, respectively.53–55 Similarly, arsenic hindered the DSB repair process by binding to the RING finger domains of RNF20/RNF40 histone E3 ubiquitin ligase, which leads to the damaged sites of compacted chromatin that are less accessible to repair enzymes.56 Moreover, a recent study found that DNA double strand crosslink repair could also be disrupted by the coordination between arsenic and the RING finger domain of FANCL E3 ubiquitin ligase.57
Consequently, the genomic instability and absence of DNA repair could activate the proto-oncogenes and/or suppress tumor suppression genes, which conferred a high risk of carcinogenesis on the cells.
4.2.3. Epigenetic changes
Because SAM donates methyl groups to many crucial cellular biomolecules, its depletion could cause gene methylation dysregulation and chromatin remodeling, resulting in tumor-related gene expressions and cellular malignant transformations. For example, hypo-methylation near the transcriptional start site of oncogenes HYAL1 and S100P was observed during the process of inorganic arsenic-induced cell malignant transformation.58 Studies have also found that the hyper-methylation of CpG islands in some promoters of onco-related genes (such as P53, CDKN2A and RASSF1A) contribute to the carcinogenesis induced by arsenic.59,60
4.2.4. MicroRNAs expression alterations
As a major family of non-coding RNAs, microRNAs translationally regulate the gene expression by complementarily binding with the specific sequence on the messenger RNA (mRNA).61 A series of studies have reported that there were numbers of microRNAs with aberrant expressions in cells and tissues with long-term arsenic exposure.62 Overexpressed miR-222, miR-21 and miR-155 played a role in facilitating arsenic-induced HBE malignant transformation.26,63–65 The elevated miR-21 caused the activation of MEK/ERK/NF-κB pathways to promote cell proliferation, while the increased miR-155 may enhance Nrf2-mediated antioxidative stress.63,64
4.2.5. Histone structure changes
Histones are important for DNA packing and compacting in nucleosome core particles, which stabilize the chromosome structure.66 Other non-chromatin related histones could bind to DNA by regulating DNA transcription, DNA repair and chromosome segregation.67 Studies demonstrated that arsenic exposure caused histone modifications (acetylation, methylation and phosphorylation), leading to structural alteration of chromosomes and onco-associated gene expressions.44 Arsenic significantly increased H3K9 dimethylation (H3K9me2) in human lung carcinoma A549 cells and decreased stem-loop binding proteins (SLBP; essential for canonical histone mRNA synthesis) in HBE by inducing histone deacetylation, which caused the aberrant polyadenylation on canonical histone H3.1 mRNA and cell anchorage-independent growth.68,69
4.3. Non-genotoxic arsenic-induced carcinogenesis
4.3.1. Signal transduction pathways alternation
Arsenic exposure caused the dysregulation of several signals, which were related to the process of carcinogenesis. In this review, we concluded the major pathways in lung tumorigenesis as follows:
The EGFR signaling pathway plays an important role in regulating cell survival, cell cycle progression and tumor invasion. Several studies have reported that arsenic exposure activated this pathway: (a) ROS induced the mutation of the tyrosine kinase domain in EGFR; (b) the dysfunction of its negative regulators; (c) increased levels of cellular tyrosine phosphorylation and the activation of non-receptor tyrosine kinase c-Src.70 Then, its downstream gene (Shc) was reported to activate another onco-associated pathway (MAPK signals).71 Moreover, arsenic-induced ROS also could upregulate a series of EGFR-related genes’ expression, such as RAS, RAF, MEK and ERK, which facilitated the initiation and development of lung tumors.72
The hedgehog (HH) signaling pathway was also reported to be associated with cancer stem cell survival and proliferation in lung cancer cells. With arsenic long-term exposure, it can be activated by decreasing the stability of the transcription factor GLI3.73,74
As a kind of transcription factor inhibited by Keap1 in the cytoplasm, Nrf2 is responsible for the generation of cytoprotective proteins that abolish the toxic species in the cells. However, the Nrf2-dependent self-protective function can be hijacked by cancer cells, and the constitutive activation of Nrf2 has been observed in arsenic-induced lung cancers. It has been reported that arsenic enhances the Nrf2 protein level via inhibiting autophagy, which caused the accumulation of P62 in the cytosome. The P62 protein contained the ‘pSTGE’ domain, which could competitively bind with Keap1 and activate the Nrf2 signals accordingly.75
4.3.2. Autophagy dysfunction
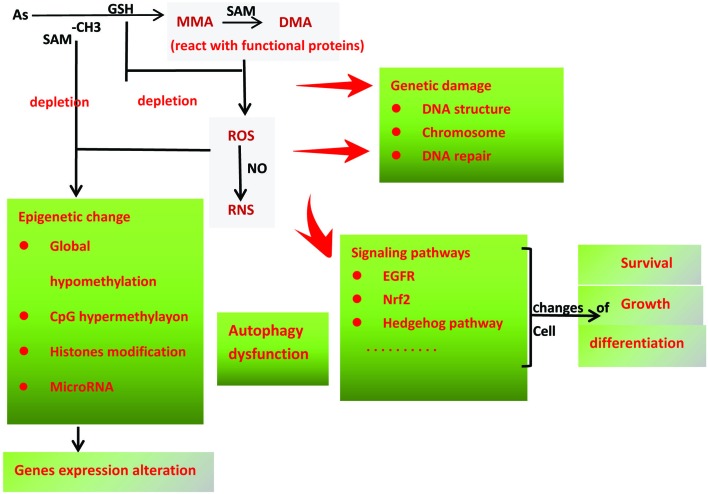
Autophagy is a self-protective activity that isolates the intracellular harmful components within double-membrane vesicles, and its contents degrade by fusing with lysosomes.76 Autophagy can also promote tumor progression through prevention against intracellular ROS and its environmental stress.77 In human bronchial epithelial cells, the overexpression of pro-inflammatory interleukin6 (IL-6) was found to inhibit autophagy degradation.78 Moreover, Lau et al. found that arsenic could enhance the cancer-promoting effect and block autophagic influx by activating the Nrf2-Keap1 pathway. Furthermore, arsenic-mediated autophagy dysfunction can be alleviated by sulforaphane (SF) and tert-butylhydroquinone (tBHQ).79 The molecular mechanisms discussed above were integrated in Fig. 2.
Fig. 2. Molecular mechanisms of arsenic-induced lung cancer. 1. The arsenic biotransformation consumes GSH, which indirectly increases the levels of ROS and RNS in the cells; 2. During the transformation from MMA to DMA, the reaction of the arsenic species with the functional proteins also could directly cause the up-regulation of ROS and RNS; 3. The depletion of the methyl group donor SAM causes the endogenous gene and protein's epigenetic changes, such as global hypo-methylation, CpG islands hyper-methylation, and histone modification. 4. Numbers of microRNA have been reported to deregulate in arsenic-transformed cells as well. 5. Arsenic inhibits autophagy degradation, which caused the extra accumulation of dysfunctional proteins and complexes in the cells; 6. Arsenic exposure disrupts the chromosome structure and affects its stability, causes the DNA double-strands breakage directly or impairs DNA repair; 7. Long-term arsenic exposure dysregulates a series of signaling pathways, such as the EGFR pathway, NRF2 and Hedgehog Pathway, which lead to changes in the cell's survival, proliferation and differentiation.
5. Prevention, diagnosis and treatment
Because the risk factors are obvious, we can decrease arsenic exposure levels to alleviate lung cancer induced by arsenic. Moreover, its specific pathogenesis can also provide some target sites for diagnosis and treatment.
With regard to prevention, some traditional methods and newly-developed economic techniques should be used primarily to decrease the levels of arsenic and its species in drinking water, such as nanoparticle and electrodynamic technologies.80 Along with setting more strict standards of arsenic in the drinking water, the arsenic concentrations in rice should be controlled.81 Studies revealed that there is a positive association between the risk of arsenic-induced lung cancer and body mass index (BMI),82 which hinted that weight-control should be incorporated in the health education programs conducted in high arsenic exposure areas.
The detection of specific arsenic methylated metabolites in urine is an effective diagnostic method for arsenic-related diseases at preclinical stages. Recent studies reported that the risk of arsenic-induced cancers is related to the levels of MMA and DMA in the bodies.83 People with low levels of MMA and DMA are genetically more susceptible to the toxicity of arsenic due to their innate poor methylation capacity.
In the treatment stages, the proper nucleotide synthesis and DNA methylation of folate acid are both important, which was proved to reduce the level of arsenic in blood in a randomized controlled clinical trial (RCT).84 A large number of natural and synthetic antioxidants, such as vitamin E, vitamin C, emblica officinalis and taurine, have exhibited their abilities of reversing arsenic-induced oxidative stress in numerous animal studies.6,85–87 Moreover, dietary selenium can also neutralize chronic arsenic toxicity in animal models; the further RCT study is being carried out in Bangladesh currently.29,88 Additionally, some arsenic-altered signal pathways offer feasible targets for therapeutic treatment. For example, sulforaphane (SF) can be used to prevent arsenic-caused lung damage by activating the Nrf2-defense response. HH signaling pathway inhibitors have also been proposed as a therapy for arsenic-caused lung cancer.89,90 Interestingly, as a kind of clinical cancer chemotherapeutic drug for acute promyelocytic leukemia, arsenic trioxide also has cytotoxic effects on non-small cell lung cancer, and its anti-tumor effects can be amplified by the combination with some other anti-cancer drugs, such as sulindac and cisplatin.22,91,92 Additionally, a recent review listed a series of reagents that had the potential to provide protection against arsenic-caused damages, such as metal chelators, cysteine, zinc, glutathione, acetyl-l-carnitine, and alpha-lipoic acid.93
6. Future study
In the future, more studies are needed to investigate the potential carcinogenesis of low to moderate concentrations of arsenic exposure. Moreover, considering the inconsistent results about the transplacental carcinogenic effect of inorganic arsenic, additional animal studies with different species and human-relevant arsenic doses are required to resolve this controversy.21 Besides, these studies are also expected to make a further effort to elucidate the underlying mechanism of arsenic-induced lung cancer, which will be implicated in designing effective prevention and treatment strategies. Finally, evaluating potential anti-arsenic therapeutic reagents may extend their studies from experiments to humans, and improve the specific safe ranges and effective values of these therapies.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
The study was supported by the grants as follows: Chinese National Nature Science Foundation (Grant ref: 81703205); Youth Foundation of Jiangsu Province (Grant ref: BK20160333); Natural Science Research in Colleges and Universities of Jiangsu Province (Grant ref: 16KJB330008); China Postdoctoral Science Foundation (Grant ref: 2016M600440); Postdoctoral Science Foundation of Jiangsu Province (Grant ref: 1601079C); China Postdoctoral Science Foundation (Grant ref: 2017T100402).
References
- Oberoi S., Barchowsky A., Wu F. Cancer Epidemiol., Biomarkers Prev. 2014;23:1187–1194. doi: 10.1158/1055-9965.EPI-13-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M. M., Chowdhury U. K., Mukherjee S. C., Mondal B. K., Paul K., Lodh D., Biswas B. K., Chanda C. R., Basu G. K., Saha K. C., Roy S., Das R., Palit S. K., Quamruzzaman Q., Chakraborti D. J. Toxicol., Clin. Toxicol. 2001;39:683–700. doi: 10.1081/clt-100108509. [DOI] [PubMed] [Google Scholar]
- Couraud S., Zalcman G., Milleron B., Morin F., Souquet P. J. Eur. J. Cancer. 2012;48:1299–1311. doi: 10.1016/j.ejca.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Guo H. R., Wang N. S., Hu H., Monson R. R. Cancer Epidemiol., Biomarkers Prev. 2004;13:638–643. [PubMed] [Google Scholar]
- Steinmaus C. M., Ferreccio C., Romo J. A., Yuan Y., Cortes S., Marshall G., Moore L. E., Balmes J. R., Liaw J., Golden T., Smith A. H. Cancer Epidemiol., Biomarkers Prev. 2013;22:623–630. doi: 10.1158/1055-9965.EPI-12-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal M., Flora S. J. Drug Chem. Toxicol. 2007;30:263–281. doi: 10.1080/01480540701380075. [DOI] [PubMed] [Google Scholar]
- Dauphine D. C., Smith A. H., Yuan Y., Balmes J. R., Bates M. N., Steinmaus C. Int. J. Environ. Res. Public Health. 2013;10:3310–3324. doi: 10.3390/ijerph10083310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ippoliti D., Santelli E., De Sario M., Scortichini M., Davoli M., Michelozzi P. PLoS One. 2015;10:e0138182. doi: 10.1371/journal.pone.0138182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdosi H., Dissen E. K. J. Environ. Public Health. 2016;2016:1602929. doi: 10.1155/2016/1602929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum M., Horowitz J., Hossain M. I. Asia Pac. J. Public Health. 2015;27:Np20–Np35. doi: 10.1177/1010539512466568. [DOI] [PubMed] [Google Scholar]
- Lamm S. H., Ferdosi H., Dissen E. K., Li J., Ahn J. Int. J. Environ. Res. Public Health. 2015;12:15498–15515. doi: 10.3390/ijerph121214990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch H. N., Zu K., Kennedy E. M., Lam T., Liu X., Pizzurro D. M., Loftus C. T., Rhomberg L. R. Environ. Int. 2017;109:195–196. doi: 10.1016/j.envint.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Yuan T., Zhang H., Chen B., Zhang H., Tao S. Toxicol. Res. 2018;7(6):1257–1266. doi: 10.1039/c8tx00177d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamm S. H., Boroje I. J., Ferdosi H. Int. J. Environ. Res. Public Health. 2018;15(6):pii: E1200. doi: 10.3390/ijerph15061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman T. G., Uddin A. N., Burns F. J., Bosland M. C. Environ. Health Perspect. 2002;110(Suppl 5):749–752. doi: 10.1289/ehp.02110s5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Wakai T., Shirai Y., Hatakeyama K., Hirano S. Toxicol. Sci. 2006;91:372–381. doi: 10.1093/toxsci/kfj159. [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Wanibuchi H., Morimura K., Wei M., Nakae D., Arai T., Minowa O., Noda T., Nishimura S., Fukushima S. Cancer Sci. 2007;98:803–814. doi: 10.1111/j.1349-7006.2007.00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C. D., LaDow K., Schumann B. L., Savage Jr. R. E., Caruso J., Vonderheide A., Succop P., Talaska G. Carcinogenesis. 2004;25:493–497. doi: 10.1093/carcin/bgg199. [DOI] [PubMed] [Google Scholar]
- Waalkes M. P., Liu J., Diwan B. A. Toxicol. Appl. Pharmacol. 2007;222:271–280. doi: 10.1016/j.taap.2006.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalkes M. P., Qu W., Tokar E. J., Kissling G. E., Dixon D. Arch. Toxicol. 2014;88:2063–2065. doi: 10.1007/s00204-014-1369-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garry M. R., Santamaria A. B., Williams A. L., DeSesso J. M. Regul. Toxicol. Pharmacol. 2015;73:378–390. doi: 10.1016/j.yrtph.2015.07.023. [DOI] [PubMed] [Google Scholar]
- Xie S. L., Yang M. H., Chen K., Huang H., Zhao X. W., Zang Y. S., Li B. Cell Biochem. Biophys. 2015;71:1325–1333. doi: 10.1007/s12013-014-0352-3. [DOI] [PubMed] [Google Scholar]
- Liu L. Z., Jiang Y., Carpenter R. L., Jing Y., Peiper S. C., Jiang B. H. PLoS One. 2011;6:e20858. doi: 10.1371/journal.pone.0020858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Wang F., Luo F., Chen X., Liang X., Jiang W., Huang Z., Lei J., Shan F., Xu X. Am. J. Transl. Res. 2017;9:416–428. [PMC free article] [PubMed] [Google Scholar]
- Chang Q., Chen B., Thakur C., Lu Y., Chen F. Oncotarget. 2014;5:1290–1303. doi: 10.18632/oncotarget.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratheeshkumar P., Son Y. O., Divya S. P., Wang L., Zhang Z., Shi X. Sci. Rep. 2016;6:37227. doi: 10.1038/srep37227. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cui J., Xu W., Chen J., Li H., Dai L., Frank J. A., Peng S., Wang S., Chen G. Oncotarget. 2017;8:21398–21409. doi: 10.18632/oncotarget.15232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedmann C., Ma Y., Melikishvili M., Godfrey S. G., Zhang Z., Chen K. C., Rouchka E. C., Fondufe-Mittendorf Y. N. BMC Genomics. 2015;16:212. doi: 10.1186/s12864-015-1295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Zhao Y., Xu W., Luo F., Wang B., Li Y., Pang Y., Liu Q. Toxicol. Appl. Pharmacol. 2013;272:542–550. doi: 10.1016/j.taap.2013.06.017. [DOI] [PubMed] [Google Scholar]
- Sun J., Yu M., Lu Y., Thakur C., Chen B., Qiu P., Zhao H., Chen F. Cancer Lett. 2014;346:257–263. doi: 10.1016/j.canlet.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son Y. O., Pratheeshkumar P., Roy R. V., Hitron J. A., Wang L., Divya S. P., Xu M., Luo J., Chen G., Zhang Z., Shi X. J. Biol. Chem. 2015;290:27090–27100. doi: 10.1074/jbc.M115.675371. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu J., Chen B., Lu Y., Guan Y., Chen F. Toxicol. Sci. 2012;129:363–371. doi: 10.1093/toxsci/kfs199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yang J., Fisher T., Xiao H., Jiang Y., Yang C. Environ. Health Perspect. 2012;120:92–97. doi: 10.1289/ehp.1104061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Lu Y., Ahmad N., Strebhardt K., Liu X. Cell Cycle. 2015;14:3030–3039. doi: 10.1080/15384101.2015.1080397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Malm S. W., Hinchman A. N., Li H., Beeks C. G., Klimecki W. T. PLoS One. 2014;9:e114549. doi: 10.1371/journal.pone.0114549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. Y., Lin Y. C., Shiue H. S., Chen W. J., Su C. T., Pu Y. S., Ao P. L., Hsueh Y. M. Toxicol. Lett. 2018;295:64–73. doi: 10.1016/j.toxlet.2018.05.035. [DOI] [PubMed] [Google Scholar]
- Khairul I., Wang Q. Q., Jiang Y. H., Wang C., Naranmandura H. Oncotarget. 2017;8:23905–23926. doi: 10.18632/oncotarget.14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melak D., Ferreccio C., Kalman D., Parra R., Acevedo J., Perez L., Cortes S., Smith A. H., Yuan Y., Liaw J., Steinmaus C. Toxicol. Appl. Pharmacol. 2014;274:225–231. doi: 10.1016/j.taap.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmaus C., Yuan Y., Kalman D., Rey O. A., Skibola C. F., Dauphine D., Basu A., Porter K. E., Hubbard A., Bates M. N., Smith M. T., Smith A. H. Toxicol. Appl. Pharmacol. 2010;247:138–145. doi: 10.1016/j.taap.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubaux R., Becker-Santos D. D., Enfield K. S., Rowbotham D., Lam S., Lam W. L., Martinez V. D. Mol. Cancer. 2013;12:20. doi: 10.1186/1476-4598-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stýblo M., Arsenicals, Glutathione Reductase and Cellular Redox Status, 2015. [Google Scholar]
- Naranmandura H., Xu S., Sawata T., Hao W. H., Liu H., Bu N., Ogra Y., Lou Y. J., Suzuki N. Chem. Res. Toxicol. 2011;24:1094–1103. doi: 10.1021/tx200156k. [DOI] [PubMed] [Google Scholar]
- Rossman T. G. Mutat. Res. 2003;533:37–65. doi: 10.1016/j.mrfmmm.2003.07.009. [DOI] [PubMed] [Google Scholar]
- Audia J. E., Campbell R. M. Cold Spring Harbor Perspect. Biol. 2016;8:a019521. doi: 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm K., Barnard S., Moquet J., Ellender M., Rana Z., Burdak-Rothkamm S. Environ. Mol. Mutagen. 2015;56:491–504. doi: 10.1002/em.21944. [DOI] [PubMed] [Google Scholar]
- Aardema M. J. Environ. Mol. Mutagen. 2013;54:617–620. doi: 10.1002/em.21813. [DOI] [PubMed] [Google Scholar]
- Bau D. T., Wang T. S., Chung C. H., Wang A. S., Wang A. S., Jan K. Y. Environ. Health Perspect. 2002;110(Suppl 5):753–756. doi: 10.1289/ehp.02110s5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchin K. T., Wallace K. Toxicol. Appl. Pharmacol. 2008;232:252–257. doi: 10.1016/j.taap.2008.06.021. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee P., Banerjee M., Giri A. K. Environ. Int. 2013;53:29–40. doi: 10.1016/j.envint.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Toselli P., Li W. Int. J. Environ. Res. Public Health. 2012;9:474–495. doi: 10.3390/ijerph9020474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman J. S., Beckman T. W., Chen J., Marshall P. A., Freeman B. A. Proc. Natl. Acad. Sci. U. S. A. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radi R., Beckman J. S., Bush K. M., Freeman B. A. J. Biol. Chem. 1991;266:4244–4250. [PubMed] [Google Scholar]
- Asmuss M., Mullenders L. H., Eker A., Hartwig A. Carcinogenesis. 2000;21:2097–2104. doi: 10.1093/carcin/21.11.2097. [DOI] [PubMed] [Google Scholar]
- Ding W., Liu W., Cooper K. L., Qin X. J., de Souza Bergo P. L., Hudson L. G., Liu K. J. J. Biol. Chem. 2009;284:6809–6817. doi: 10.1074/jbc.M805566200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerdtle T., Walter I., Hartwig A. DNA Repair. 2003;2:1449–1463. doi: 10.1016/j.dnarep.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Zhang F., Paramasivam M., Cai Q., Dai X., Wang P., Lin K., Song J., Seidman M. M., Wang Y. J. Am. Chem. Soc. 2014;136:12884–12887. doi: 10.1021/ja507863d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Bellani M., Li L., Wang P., Seidman M. M., Wang Y. ACS Chem. Biol. 2017;12:1858–1866. doi: 10.1021/acschembio.6b01135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelch K. E., Tokar E. J., Merrick B. A., Waalkes M. P. Toxicol. Appl. Pharmacol. 2015;286:159–167. doi: 10.1016/j.taap.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro M., Caradonna F., Klein C. B. Environ. Mol. Mutagen. 2016;57:137–150. doi: 10.1002/em.21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarabi M. M., Naghibalhossaini F. Cell Biochem. Funct. 2015;33:427–433. doi: 10.1002/cbf.3126. [DOI] [PubMed] [Google Scholar]
- Bartel D. P. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage A. P., Minatel B. C., Ng K. W., Stewart G. L., Dummer T. J. B., Lam W.W. L.L., Martinez V. D. Oncotarget. 2017;8:25736–25755. doi: 10.18632/oncotarget.15106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Jiang X., Gu S., Zhang Z. Toxicol. Lett. 2017;278:38–47. doi: 10.1016/j.toxlet.2017.07.215. [DOI] [PubMed] [Google Scholar]
- Luo F., Ji J., Liu Y., Xu Y., Zheng G., Jing J., Wang B., Xu W., Shi L., Lu X., Liu Q. Toxicol. Lett. 2015;232:301–309. doi: 10.1016/j.toxlet.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Wang M., Ge X., Zheng J., Li D., Liu X., Wang L., Jiang C., Shi Z., Qin L., Liu J., Yang H., Liu L. Z., He J., Zhen L., Jiang B. H. Oncotarget. 2016;7:17805–17814. doi: 10.18632/oncotarget.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Smith M. M. Cold Spring Harbor Perspect. Biol. 2015;7:a019364. doi: 10.1101/cshperspect.a019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D., Burkhart S. L., Singh R. K., Kabbaj M. H., Gunjan A. Nucleic Acids Res. 2012;40:9604–9620. doi: 10.1093/nar/gks722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Li Q., Arita A., Sun H., Costa M. Toxicol. Appl. Pharmacol. 2009;236:78–84. doi: 10.1016/j.taap.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocato J., Chen D., Liu J., Fang L., Jin C., Costa M. Biol. Trace Elem. Res. 2015;166:72–81. doi: 10.1007/s12011-015-0296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora S. J. Free Radical Biol. Med. 2011;51:257–281. doi: 10.1016/j.freeradbiomed.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Chen W., Martindale J. L., Holbrook N. J., Liu Y. Mol. Cell. Biol. 1998;18:5178–5188. doi: 10.1128/mcb.18.9.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew A. S., Mason R. A., Memoli V., Duell E. J. Toxicol. Sci. 2009;109:350–357. doi: 10.1093/toxsci/kfp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei D. L., Li H., Kozul C. D., Black K. E., Singh S., Gosse J. A., DiRenzo J., Martin K. A., Wang B., Hamilton J. W., Karagas M. R., Robbins D. J. Cancer Res. 2010;70:1981–1988. doi: 10.1158/0008-5472.CAN-09-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savani M., Guo Y., Carbone D. P., Csiki I. Ther. Adv. Med. Oncol. 2012;4:225–233. doi: 10.1177/1758834012450362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. J., Sun Z., Chen W., Li Y., Villeneuve N. F., Zhang D. D. Toxicol. Appl. Pharmacol. 2008;230:383–389. doi: 10.1016/j.taap.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Klionsky D. J. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Pietrocola F., Bravo-San Pedro J. M., Amaravadi R. K., Baehrecke E. H., Cecconi F., Codogno P., Debnath J., Gewirtz D. A., Karantza V., Kimmelman A., Kumar S., Levine B., Maiuri M. C., Martin S. J., Penninger J., Piacentini M., Rubinsztein D. C., Simon H. U., Simonsen A., Thorburn A. M., Velasco G., Ryan K. M., Kroemer G. EMBO J. 2015;34:856–880. doi: 10.15252/embj.201490784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Zhang M., Li H., Frank J. A., Dai L., Liu H., Zhang Z., Wang C., Chen G. Cancer Res. 2014;74:3740–3752. doi: 10.1158/0008-5472.CAN-13-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A., Zheng Y., Tao S., Wang H., Whitman S. A., White E., Zhang D. D. Mol. Cell. Biol. 2013;33:2436–2446. doi: 10.1128/MCB.01748-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicomel N. R., Leus K., Folens K., Van Der Voort P., Du Laing G. Int. J. Environ. Res. Public Health. 2015;13(1) doi: 10.3390/ijerph13010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve S. BMC Public Health. 2014;14:465. doi: 10.1186/1471-2458-14-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmaus C., Castriota F., Ferreccio C., Smith A. H., Yuan Y., Liaw J., Acevedo J., Perez L., Meza R., Calcagno S., Uauy R., Smith M. T. Environ. Res. 2015;142:594–601. doi: 10.1016/j.envres.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. C., Moon K. A., Wang S. L., Silbergeld E., Navas-Acien A. Environ. Health Perspect. 2017;125:087001. doi: 10.1289/EHP577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B. A., Hall M. N., Liu X., Parvez F., Sanchez T. R., van Geen A., Mey J. L., Siddique A. B., Shahriar H., Uddin M. N., Islam T., Balac O., Ilievski V., Factor-Litvak P., Graziano J. H., Gamble M. V. Environ. Health Perspect. 2015;123:1294–1301. doi: 10.1289/ehp.1409396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh J., Das J., Manna P., Sil P. C. Toxicol. Appl. Pharmacol. 2009;240:73–87. doi: 10.1016/j.taap.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Rana T., Bera A. K., Das S., Pan D., Bandyopadhyay S., Bhattacharya D., De S., Sikdar S., Das S. K. Food Chem. Toxicol. 2010;48:1072–1077. doi: 10.1016/j.fct.2010.01.027. [DOI] [PubMed] [Google Scholar]
- Singh M. K., Yadav S. S., Gupta V., Khattri S. BMC Complementary Altern. Med. 2013;13:193. doi: 10.1186/1472-6882-13-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohn R. M., Raqib R., Akhtar E., Vandenberg A., Smits J. E. Trials. 2016;17:218. doi: 10.1186/s13063-016-1344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe Y., Tanaka N. BioMed Res. Int. 2016;2016:7969286. doi: 10.1155/2016/7969286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Tao S., Lian F., Chau B. T., Chen J., Sun G., Fang D., Lantz R. C., Zhang D. D. Toxicol. Appl. Pharmacol. 2012;265:292–299. doi: 10.1016/j.taap.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. R., Kim E. J., Yang S. H., Jeong E. T., Park C., Kim S. J., Youn M. J., So H. S., Park R. Int. J. Oncol. 2006;28:1401–1408. [PubMed] [Google Scholar]
- Walker A. M., Stevens J. J., Ndebele K., Tchounwou P. B. J. Cancer Sci. Ther. 2016;8:1–9. doi: 10.4172/1948-5956.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao C. V., Pal S., Mohammed A., Farooqui M., Doescher M. P., Asch A. S., Yamada H. Y. Oncotarget. 2017;8:57605–57621. doi: 10.18632/oncotarget.17745. [DOI] [PMC free article] [PubMed] [Google Scholar]