Abstract
Human cells lacking DNA polymerase η (polη) are sensitive to platinum-based cancer chemotherapeutic agents. Using DNA combing to directly investigate the role of polη in bypass of platinum-induced DNA lesions in vivo, we demonstrate that nascent DNA strands are up to 39% shorter in human cells lacking polη than in cells expressing polη. This provides the first direct evidence that polη modulates replication fork progression in vivo following cisplatin and carboplatin treatment. Severe replication inhibition in individual platinum-treated polη-deficient cells correlates with enhanced phosphorylation of the RPA2 subunit of replication protein A on serines 4 and 8, as determined using EdU labelling and immunofluorescence, consistent with formation of DNA strand breaks at arrested forks in the absence of polη. Polη-mediated bypass of platinum-induced DNA lesions may therefore represent one mechanism by which cancer cells can tolerate platinum-based chemotherapy.
Despite recent advances in the development of targeted therapies, DNA damaging agents such as ionizing radiation and platinum-based chemotherapeutic drugs continue to play a major role in cancer treatment. Cisplatin [cis-diamminedichloroplatinum (II)], the first platinum-based anticancer agent to be introduced, is in wide clinical use, in particular in the treatment of testicular and ovarian cancers, as well as small cell lung cancer1. Cisplatin analogues including carboplatin [cis-diammine(1,1-cyclobutanedicarboxylate)-platinum(II)] have been developed, with reduced side-effects and improved selectivity towards cancer cells1. However, further investigation of the cellular processes that modulate the response of human cells to platinum-induced DNA damage is required, in order to understand the basis of platinum resistance, the development of secondary tumours in surviving cells, and to identify novel targets to potentiate the cytotoxic effects of these agents2.
Cisplatin and carboplatin induce DNA damage in the form of monoadducts, as well as intrastrand and interstrand cross-links (ICLs), primarily at guanine residues3. While the cytotoxic effects of platinum-based drugs are largely attributable to the formation of ICLs, the majority (>90%) of platinum adducts in the DNA are intrastrand adducts rather than ICLs4,5. Platinum-DNA adducts block both transcription and DNA replication, leading to cell cycle arrest and cell death3. The process of translesion synthesis (TLS), a cellular DNA damage tolerance pathway that allows cells to continue DNA replication in the presence of unrepaired damage in the genome, may contribute to tumour cell survival following platinum-induced DNA damage1.
TLS is carried out by specialized, low-fidelity DNA polymerases, including DNA polymerase η (polη), a 78 kDa protein encoded by the human POLH gene6,7,8. Mutations in POLH underlie the skin cancer-prone genetic disease xeroderma pigmentosum variant (XPV)6,7. Polη normally carries out error-free replication at sites of UV-induced di-thymine cyclobutane pyrimidine dimers (CPDs)9. In the absence of polη in XPV patients, mutations accumulate in the genome as a result of error-prone lesion bypass carried out by other DNA polymerases10,11. In addition to the major biological role of polη in bypass of CPDs, it also plays a role in cisplatin tolerance. Consistent with this, purified polη can bypass the most abundant cisplatin-induced intrastrand guanine-guanine adducts in vitro9,12, and human cells lacking polη are more sensitive to platinum-based drugs13,14,15. While there is evidence that polη can also bypass interstrand platinum-DNA adducts in an error-prone fashion in vitro16,17 other studies indicate it is dispensable for the processing of cisplatin-induced ICLs in vivo18.
Activation of the DNA damage response (DDR) plays an important role in determining the outcome of replication arrest induced by DNA damaging agents19,20. Replication arrest and DNA strand break formation activate phosphatidylinositol-3-kinase-related protein kinases (PIKKs) which phosphorylate downstream target proteins to orchestrate the DNA damage response20,21,22. It has been previously shown by our group and others that following treatment with platinum-based drugs, DNA damage response signalling is enhanced in cells lacking polη15,23,24. In the present study, we have used DNA combing to directly investigate the effect of polη-deficiency on replication of genomic DNA containing platinum lesions in vivo. We show that in the absence of polη, the length of nascent DNA strands is reduced following cisplatin- and carboplatin-induced DNA damage. We also show that damage-induced phosphorylation of the RPA2 subunit of replication protein A, a key PIKK target, directly correlates with the extent of replication inhibition in individual cells. Our data provide new insights into the role of polη and downstream signalling pathways in the processing of DNA damage induced by platinum-based anti-cancer drugs.
Results
Enhanced cisplatin- and carboplatin-induced replication inhibition in polη-deficient cells
To directly investigate the role of polη in replication of platinum-damaged DNA in vivo, XP30RO and TR30-2 cells were either mock-treated or treated with cisplatin (1.7 μM) or carboplatin (50 μM)25. Cell cycle distribution was determined using flow cytometry, and replication fork progression was analysed by DNA combing. Cisplatin and carboplatin induced S-phase arrest in both cell lines. Consistent with our previous report25, S-phase accumulation was significantly higher in polη-deficient XP30RO cells compared to TR30-2 cells expressing polη.
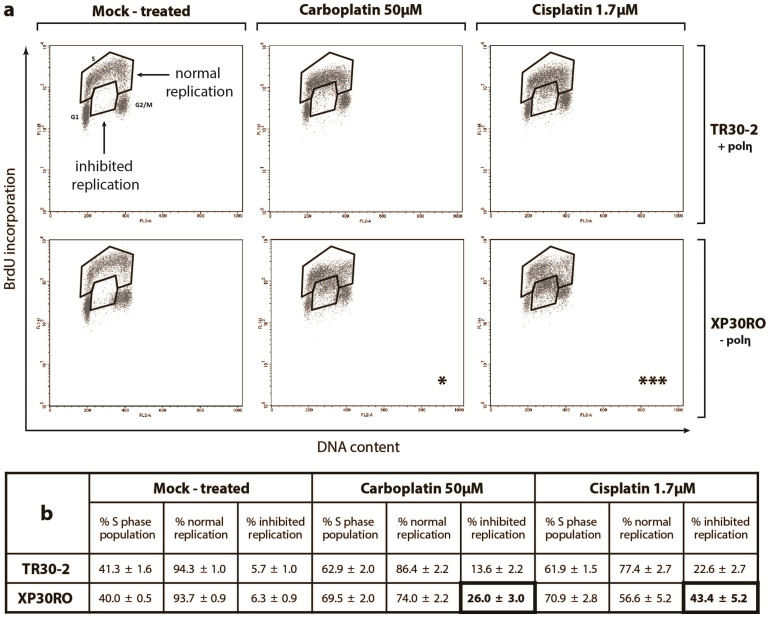
Accumulation of cells in S-phase is a result of inhibition of DNA replication, as determined by analysis of BrdU incorporation (Fig. 1a, b). While the percentage of cells in S-phase increased in both cell lines following treatment with cisplatin or carboplatin, two separate populations of S-phase cells could be identified (Fig. 1a). Thus, in addition to BrdU-positive S-phase cells, a population of cells having S-phase DNA content but with greatly decreased BrdU incorporation could be identified (Fig. 1a). These two cell populations have been termed ‘normal replication’ and ‘inhibited replication’, respectively (Fig. 1a). The percentage of S-phase cells showing inhibited replication was significantly higher in polη-deficient XP30RO cells compared to TR30-2 cells expressing polη, as determined using two-way ANOVA (Fig. 1b). Thus, after treatment of XP30RO cells with 50 μM carboplatin, 26% of S-phase cells showed inhibited replication, while this value was 43.4% in cells treated with 1.7 μM cisplatin (Fig. 1a, b). The increase in the percentage of cells with inhibited replication observed in the absence of polη was DNA damage-dependent, since polη expression did not influence the percentage of cells in S-phase in the absence of platinum treatment (Fig. 1a, b, mock-treated). Thus, following exposure to platinum-based DNA damaging agents, the extent of DNA replication inhibition in S-phase cells is dependent on polη expression.
Figure 1. Cell cycle progression and DNA replication analysis in carboplatin- and cisplatin-treated cells lacking or expressing DNA polymerase η.
Cells expressing (TR30-2) or lacking (XP30RO) polη were treated for 24 h with the indicated doses of cisplatin and carboplatin. Cells were incubated with 10 μM BrdU before fixing and staining with propidium iodide (PI) and anti-BrdU-FITC antibody as described in Materials and Methods. (a) Representative flow cytometry (PI- and BrdU-stained) dot plots are presented. Populations of cells with S-phase DNA content were divided in two groups termed normal replication and inhibited replication, based on BrdU staining. Asterisks indicate statistically significant differences in the percentage of cells exhibiting replication inhibition, determined using the two-way ANOVA test. (b) The mean percentage of cells in the indicated groups +/− SEM values, derived from four independent experiments, is presented in Table format.
Effect of polη expression on nascent DNA strand length following DNA damage
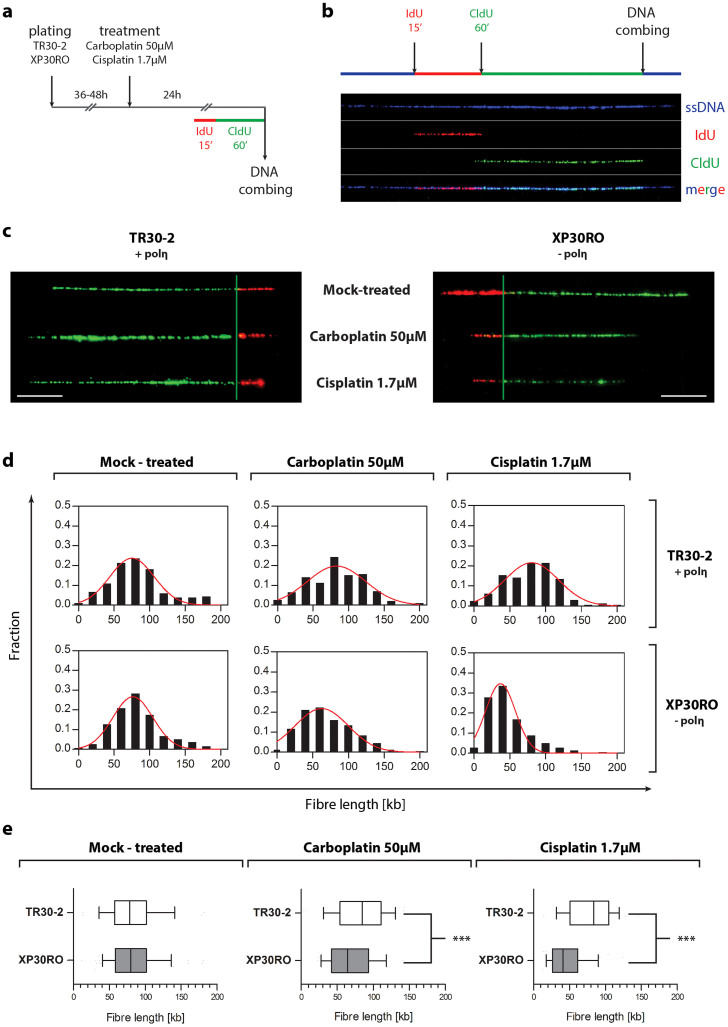
To directly determine whether the decrease in BrdU incorporation in cisplatin- and carboplatin-treated XP30RO cells (Fig. 1) was due to a deficiency in polη during replication of DNA containing platinum-induced lesions in vivo, the length of individual nascent DNA strands was analysed using DNA combing in polη-expressing and polη-deficient fibroblasts exposed to cisplatin or carboplatin (Fig. 2). Before harvesting, cells were pulse-labelled for 15 min with IdU, followed by 60 min labelling with CldU (Fig. 2a). Differential labelling of nascent DNA using two modified nucleosides allowed strands derived from newly-fired origins and from stalled replication forks to be distinguished (Fig. 2a, b). For each experimental condition, individual DNA fibres were uniformly stretched on glass coverslips, and visualized by fluorescence microscopy using specific antibodies to detect IdU, CldU and total DNA (Fig. 2b). Only fibres that were stained for both IdU (green) and CldU (red) were included in the subsequent analysis (Fig. 2c). Merged replicons and origins that fired during the second pulse (characterised by fibres stained for CldU but not for IdU) were therefore excluded from the analysis. For each fibre, only the length of the CldU track was measured (Fig. 2c, green tracks).
Figure 2. Analysis of DNA replication by DNA combing.
(a) A schematic outline of the DNA combing protocol is shown. (b) DNA fibres were visualized using specific primary antibodies and fluorescently-labelled secondary antibodies. A representative fibre is shown. Single-stranded DNA is shown in blue, IdU-labelled DNA in red, and CldU-labelled DNA in green. (c) Representative, individual DNA fibres for each experimental condition are presented. (d) The frequency distribution of fibres of particular lengths is presented as histograms plots. Data represent the population of fibres obtained by measuring at least 150 individual DNA fibres for selected experimental conditions, in two independent experiments. The red line indicates a Gaussian curve fitted to the data. (e) Data are presented as inbox-and-whiskers graphs. Each box represents the 25th and 75th percentile (the lower and upper quartiles, respectively). The median (50th percentile) is marked as the line near the middle of the box. Data not included between the whiskers are plotted as outliers (dots). Data was analyzed by the use of the Mann-Whitney test. Asterisks indicate statistically significant differences.
In the absence of cisplatin or carboplatin, the mean fibre length in both cell lines was approximately 82 kb (Fig. 2e; Suppl. Fig. 1a). Compared to mock-treated cells, there was no statistically significant difference in the mean length of nascent DNA strands, or in the replication fork rate, when TR30-2 cells expressing polη were treated with 1.7 μM cisplatin or 50 μM carboplatin (Fig. 2a, b; Suppl. Fig. 1a). However, following treatment of polη-deficient XP30RO cells for 24 h with 50 μM carboplatin, the frequency of shorter fibres was increased, visible as a shift towards shorter fibre lengths on the frequency distribution graphs (Fig. 2d). The mean fibre length decreased to 69.1 kb, compared to 82.3 kb and 82.0 kb in mock-treated XP30RO and TR30-2 cells, respectively. The difference in mean fibre length was greater in XP30RO cells treated with 1.7 μM cisplatin (Fig. 2e; Suppl. Fig. 1a). In this case, the mean DNA strand length was 48.2 kb in XP30RO cells, compared to 78.9 kb in TR30-2 cells, representing a 39% decrease in fibre length in the absence of polη. The replication fork rate was also decreased in XP30RO cells treated with cisplatin or carboplatin (Suppl. Fig. 1a). This data provides the first direct evidence in vivo that polη-deficiency results in a statistically significant reduction (Fig. 2e; Mann-Whitney test, p < 0.0001) in the mean length of nascent DNA strands in XP30RO cells treated with cisplatin or carboplatin.
Cisplatin- and carboplatin-induced DNA damage responses
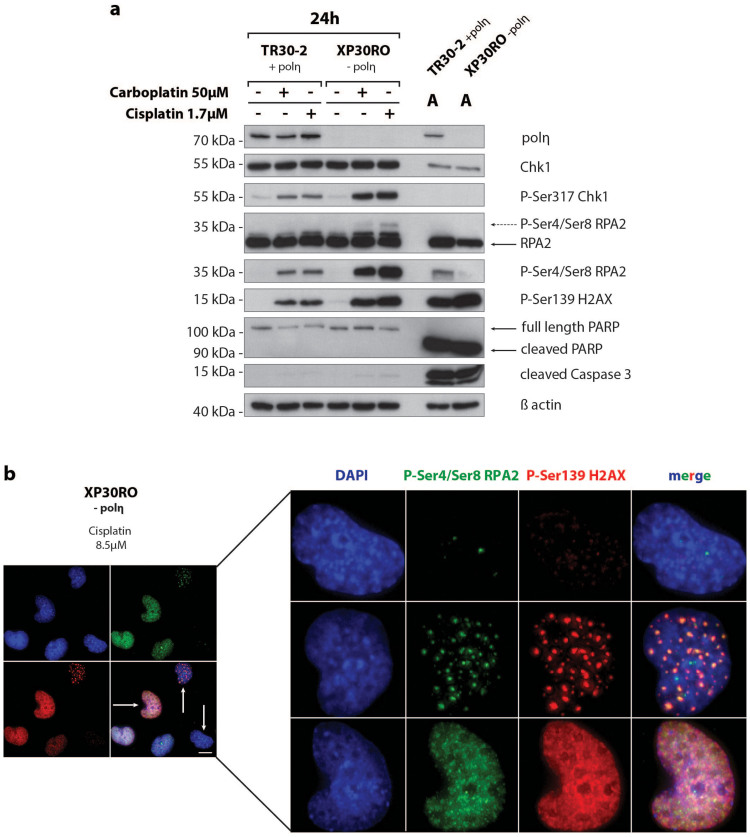
Since polη deficiency leads to replication arrest and a reduction in the length of nascent strands in cells treated with cisplatin or carboplatin, we investigated the relationship between inhibition of DNA synthesis and activation of DNA damage responses in XP30RO and in TR30-2 cells. Protein samples from mock-treated cells and cells exposed to cisplatin and carboplatin were analysed by western blotting for polη expression and for key DNA damage responses, including phosphorylation of Chk1, RPA2 and H2AX. Consistent with POLH being mutated in XP30RO cells6,7, polη protein was undetectable in XP30RO cell extracts but was expressed in TR30-2 cells (Fig. 3a).
Figure 3. Carboplatin- and cisplatin-induced DNA damage responses and apoptosis induction in cells lacking and expressing DNA polymerase η.
(a) Polη-deficient XP30RO cells and polη-expressing TR30-2 cells were treated with carboplatin or cisplatin and harvested 24 h post-treatment. Polη, DNA damage response proteins and apoptosis-related proteins were analysed by western blotting using specific antibodies, as described in Materials and Methods. For direct comparison, samples from both cell lines were run on the same gel. As a control for apoptosis induction, proteins, obtained from floating cells following treatment of cells with 5 μg/ml cisplatin for 24 h, were loaded in the lanes marked A. (b) Polη-deficient XP30RO cells were treated for 24 h with 8.5 μM cisplatin. Immunofluorescence microscopy was carried out using phospho-specific primary antibodies against γH2AX and phospho-Ser4/Ser8 RPA2, and Alexa 594- and Alexa 488-labeled secondary antibodies. DNA was counterstained with DAPI. Scale bar corresponds to 25μm.
The PIKK ATR is activated in an ATRIP-dependent manner in response to single-stranded DNA generated at arrested replication forks26,27. ATR-mediated phosphorylation of Chk1 on serine 317 directly modulates the DNA damage response by regulating S-phase arrest and mitotic entry28. Compared to mock-treated cells, Chk1 phosphorylation on Ser317 was elevated in the polη-deficient XP20RO cell line (Fig. 3a). Chk1 phosphorylation was detectable by 6 h after exposure to both cisplatin and carboplatin, and peaked at 24 h post-treatment in both cell lines (Suppl. Fig. 2), consistent with sustained replication arrest in these cells (Fig. 1). During this time period, the level of total Chk1 did not change (Suppl. Fig. 2). At 36 h and 48 h post-treatment, the level of phosphorylated Chk1 and total Chk1 decreased (Suppl. Fig. 2). Based on analysis of the apoptotic markers, cleaved caspase 3 and PARP-1, there was little evidence of apoptosis 24 h after treatment of cells with 1.7 μM cisplatin or 50 μM carboplatin (Fig. 3a). The observed decrease in Chk1 levels at 36 h and 48 h post-treatment may reflect proteolytic degradation of Chk1 in cells that are undergoing apoptosis as a result of prolonged drug exposure29.
Following replication arrest, PIKK-dependent phosphorylation of histone H2AX on Ser139 (generating γH2AX) was found to stabilize stalled forks and prevent their collapse20 as well as to function in DNA strand break signalling30. In the present study the levels of γH2AX were found to be elevated in polη-deficient cell lines compared to cells expressing polη (Fig. 3a). Slight phosphorylation of H2AX was detectable at 6 h post-treatment with cisplatin and carboplatin, and the level of γH2AX increased with time up to 48 h post-exposure (Suppl. Fig. 2). Induction of H2AX phosphorylation occurred earlier and to a higher level in response to 1.7 μM cisplatin compared to 50 μM carboplatin (Suppl. Fig. 2). In contrast to Chk1 phosphorylation, γH2AX formation is a later event, and is significantly enhanced from 24 h post-drug exposure, especially in XP30RO cells (Suppl. Fig. 2). This could be due to prolonged replication fork arrest, and fork collapse when polη-deficient cells attempt to progress through S-phase in the presence of damaged DNA19.
Trimeric replication protein A (RPA), the main single-stranded DNA binding protein in human cells, plays a role in all aspects of DNA metabolism including replication, recombination and repair. Phosphorylation of the 34 kDa RPA2 subunit occurs in a cell cycle-dependent manner, and in response to DNA damage15,31,32. In cells undergoing replication stress, RPA-coated single-stranded DNA activates ATRIP-dependent ATR signalling31. Following replication arrest, ATR phosphorylates RPA2 on Ser3327 and the protein is also phosphorylated on Thr21 by other PIKKs31. We have previously shown that RPA2 is phosphorylated on Ser33 by ATR15 in response to platinum-based chemotherapeutic agents. In response to UVR-, IR- and platinum-induced DNA damage, RPA2 is further phosphorylated on Ser4/Ser8 in a DNA-PK-dependent manner, generating a distinct hyperphosphorylated form of the protein with reduced mobility upon SDS-polyacrylamide gel electrophoresis15,23,33,34. Hyperphosphorylated RPA2 can be detected using anti-RPA2 antibody as a band corresponding to a slower mobility form (Fig. 3, dashed arrow). Here, the induction of RPA2 phosphorylation on Ser4/Ser8 was confirmed by the use of an anti-phospho-Ser4/Ser8 RPA2 antibody. Consistent with a role for RPA2 phosphorylation in the cellular response to replication arrest25, the level of RPA2 phosphorylated on Ser4/Ser8 was significantly higher in polη-deficient XP30RO cells compared to TR30-2 cells that express polη (Fig. 3a). The timing of RPA2 phosphorylation was similar to that of H2AX phosphorylation rather than Chk1 phosphorylation, being detected at later times post-treatment. Thus, phosphorylation was first detected 12 h post-treatment with cisplatin, and 24 h post-treatment with carboplatin, with the peak signals being detected at 24 h and 36 h post-exposure, respectively (Suppl. Fig. 2). The timing of RPA2 phosphorylation on Ser4/Ser8 was also confirmed by quantitative immunofluorescence (data not shown). Phosphorylated RPA2 and γH2AX were found to co-localise in nuclear foci in cisplatin-treated XP30RO cells (Fig. 3b), consistent with a role for phosphorylated RPA2 at sites of DNA strand breaks possibly arising from replication arrest and fork collapse, as we and others have previously proposed15,23,24.
Overall, following exposure of polη-deficient XP30RO cells to cisplatin and carboplatin, inhibition of DNA synthesis leads to enhanced intra-S checkpoint activation and phosphorylation of DNA damage response proteins including H2AX and RPA2 (Fig. 3).
Quantitative analysis of the association between replication inhibition and RPA2 phosphorylation on Ser4/Ser8
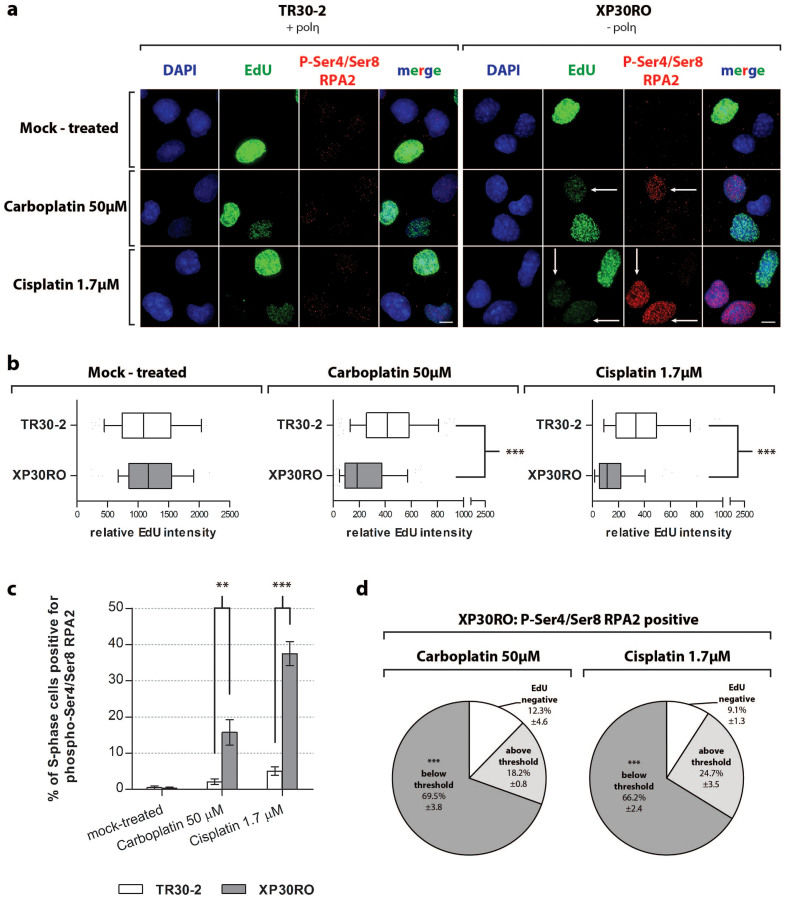
Given that platinum-induced phosphorylation of RPA2 on Ser4/Ser8 was enhanced in cells lacking polη (Fig. 3) in which replication was also strongly inhibited (Fig. 1, 2), we used quantitative immunofluorescence to investigate the relationship between replication inhibition and RPA2 phosphorylation on Ser4 and Ser8 in individual cells. This was studied directly using ‘click chemistry’ to monitor incorporation of the modified nucleoside EdU during DNA replication, and immunofluorescence to determine the extent of RPA2 phosphorylation on Ser4/Ser8 (Fig. 4a).
Figure 4. Analysis of DNA replication by quantitative immunofluorescence.
Cells expressing (TR30-2) or lacking (XP30RO) polη were grown on glass coverslips and treated with the indicated doses of cisplatin or carboplatin. 75 minutes before fixing, cells were incubated with 10 μM EdU. Click chemistry was used to visualise EdU incorporation, and immunofluorescence staining was carried out using anti-phospho-Ser4/Ser8 RPA2 primary antibody and Alexa 594-coupled secondary antibody. DNA was counterstained with DAPI. (a) Representative images from three independent experiments are presented. The scale bar corresponds to 25 μm. Arrows show representative cells which exhibit decreased EdU incorporation and increased phospho-Ser4/Ser8 RPA2 staining. (b) The EdU fluorescence signal was quantified as described in Materials and Methods. Relative EdU fluorescence values were analysed using the Mann-Whitney test; significant values are marked with asterisks. In the inbox-and-whiskers graphs, the top and bottom of each box represents the 25th and 75th percentile (the lower and upper quartiles, respectively). The median (50th percentile) is marked by the line near the middle of the box. (c) Cells positive for phospho-Ser4/Ser8 RPA2 were counted in the EdU-positive cell population. Data represent the mean from three independent experiments +/− SEM. Statistically significant differences are marked with asterisks, as determined using a two-way ANOVA test. (d) EdU staining in phospho-Ser4/Ser8 RPA2-positive cells was analysed under conditions where RPA2 phosphorylation on Ser4/Ser8 is enriched. The intensity of the EdU signal in phospho-Ser4/Ser8 RPA2-positive cells was further analysed, and categorised into values above and below the threshold value. For each condition, the threshold value was defined as the mean relative EdU intensity value from the entire EdU-positive cell population, minus twice the SEM value. Data represent the mean percentage of cells in defined categories obtained in three independent experiments +/− SEM. Statistically significant differences for both the mean EdU intensity in each category as well as mean percentage of each category are marked with asterisks.
Consistent with data above (Fig. 1), the mean relative EdU intensity was reduced in cells treated with cisplatin or carboplatin, compared to mock-treated cells (Fig. 4b and Suppl. Tab. 1a). EdU incorporation was inhibited to a greater extent in polη-deficient XP30RO cells compared to TR30-2 cells. Thus, in XP30RO cells treated with 50 μM carboplatin, the mean relative EdU intensity decreased by 5.7-fold compared to mock-treated cells, while the same treatment resulted in a 2.7-fold decrease in EdU intensity in TR30-2 cells (Fig. 4b, and Suppl. Table 1). Following cisplatin treatment, the difference between polη-deficient and polη-expressing cells was more pronounced. When compared to mock-treated samples, the mean relative EdU intensity decreased by 7.3-fold in XR30RO cells, and by 3.2-fold in TR30-2 cells (Suppl. Table 1), further supporting a role of polη in replication of platinum-damaged DNA in vivo. We have shown previously that in polη-deficient XP30RO cells, carboplatin-induced RPA2 phosphorylation on Ser4/Ser8 occurred primarily in cells in S-phase, and to a lesser extent in G2/M-phase cells25. Treatment with cisplatin or carboplatin for 24 h leads to S-phase arrest (Fig. 1) and to RPA2 phosphorylation on Ser4/Ser8 (Fig. 3). In order to directly investigate the correlation between DNA replication inhibition and phosphorylation of RPA2 on Ser4/Ser8, we determined the fraction of EdU-positive S-phase cells that were also positive for phospho-Ser4/Ser8 RPA2. In mock-treated cells, these values were below 0.5% for each cell line, reflecting the fact that phosphorylation of RPA2 on Ser4/Ser8 is DNA damage-dependent (Fig. 4c). Consistent with western blotting data (Fig. 3), cisplatin- and carboplatin-induced RPA2 phosphorylation was strongly associated with polη deficiency (Fig. 4c). 15% of S-phase XP30RO cells were positive for phospho-Ser4/Ser8 RPA2 following carboplatin treatment, while up to 37% of cells were positive following treatment with cisplatin (Fig. 4c). In contrast, in TR30-2 cells expressing polη, 24 h following DNA damage only 2-5% of S-phase cells were positive for phosphorylated RPA2 (Fig. 4c).
To directly correlate DNA replication status with phosphorylation of RPA2 on Ser4/Ser8, we carried out quantitative analysis of the level of EdU incorporation in carboplatin- or cisplatin-treated XP30RO cells that were positive for phospho-Ser4/Ser8 RPA2. XP30RO cells were treated for 24 h with the indicated doses of cisplatin or carboplatin, conditions which generate elevated levels of phospho-Ser4/Ser8 RPA2-positive nuclei. Around 90% of phospho-Ser4/Ser8 RPA2-positive cells were also positive for EdU incorporation (Fig. 4d), consistent with the our previous report that DNA damage-induced RPA phosphorylation on Ser4/Ser8 occurs primarily in S-phase cells25. However, analysis of the intensity of EdU staining in individual cells revealed that phosphorylation of RPA2 on Ser4/Ser8 occurred mainly in cells with the lowest level of EdU incorporation, that is, in which DNA replication was strongly inhibited. As shown in Fig. 4d, using the mean relative EdU staining intensity for the entire cell population as a threshold value, it was found that following carboplatin treatment, 69.5% of phospho-Ser4/Ser8 RPA2-positive cells had a level of EdU incorporation below the threshold value (Fig. 4d, Suppl. Tab. 1c). Similarly, following cisplatin treatment, 66.2% of phospho-Ser4/Ser8 RPA2-positive XP30RO cells were also in the low EdU staining category (Fig. 4d). This data provides the first direct evidence at the level of individual cells that RPA2 phosphorylation on Ser4/Ser8 occurs in cells where DNA replication is severely inhibited (Fig. 4d).
Discussion
Given that cells lacking polη are more sensitive to platinum-based chemotherapeutic drugs13 and that the level of endogenous polη can modulate the sensitivity of non-small lung cancer cells to platinum-based drugs35 a better understanding of the role of polη in replication of DNA containing platinum-induced damage is important for cancer chemotherapy. Elucidation of the structure of polη, crystallized in the presence of an intrastrand cisplatin adduct formed between two adjacent guanines, has provided a biochemical basis for these observations36,37. Although the major biological role of polη is in bypass of UV-induced thymine-thymine CPDs in DNA, the open active site cleft of polη can also accommodate a platinum adduct without major rearrangement of the structure of the protein37.
Consistent with our previous reports15,25, following treatment with cisplatin and carboplatin, polη-deficient XP30RO cells accumulate in S-phase, while BrdU incorporation is strongly inhibited as a consequence of replication arrest in the presence of platinum-induced DNA damage. In order to elucidate the role of polη expression in determining the extent of replication inhibition, we used DNA combing to analyse DNA replication at the level of individual nascent DNA strands. We found that after treatment with cisplatin or carboplatin for 24 h nascent DNA fibres were up to 39% shorter in XP30RO cells lacking polη when compared to TR30-2 cells expressing polη. The observation that the fork rate is also reduced in the absence of polη in platinum-treated cells (Suppl. Fig. 1a) supports a role for polη during bypass of platinum-induced lesions at the arrested fork, rather a model involving repriming and lesion bypass by polη during a subsequent gap-filling step38. There was no difference between the mean lengths of DNA fibres in mock-treated polη-expressing cells and polη-deficient cells, consistent with previous reports that, while polη plays a role in DNA replication at fragile sites, it not required for DNA synthesis on most non-damaged templates24,39,40. The present data provides the first direct evidence that polη expression modulates DNA synthesis on platinum-damaged DNA in human cells in vivo. Targeting polη-dependent TLS could therefore represent one potential approach to improving the efficacy of platinum-based drugs in cancer therapy13,14,37,41. Given that polη has also been reported to play a role in the processing of platinum-induced ICLs16, further investigation is required to establish the relative contribution of each class of lesion to the replication arrest and increased phosphorylation of DNA damage response proteins observed in platinum-treated polη-deficient cells.
It was of interest to characterise the DNA damage responses that were activated following treatment with cisplatin and carboplatin. While drug exposure did not reduce the length of DNA strands synthesized in TR30-2 cells expressing polη, as determined by DNA combing, incorporation of halogenated nucleosides was significantly inhibited as determined using FACS analysis (Fig. 1a, b) and immunofluorescence (Fig. 4a, b). This could reflect activation of the Chk1-mediated S-phase checkpoint (Fig. 3) in TR30-2 cells, consistent with the report that dissociation of DNA damage-activated Chk1 is crucial for efficient recruitment of polη to sites of damage42. Chk1-mediated activation of the S-phase checkpoint could also allow more time for repair of platinum-DNA lesions present on the template strand21,43. Active checkpoint signalling also inhibits S-phase progression by preventing new origins from firing24,44,45. We found that cisplatin and carboplatin treatment reduced the frequency of origin firing, independent of polη expression, with an up to 37% reduction following cisplatin treatment (Suppl. Fig. 1b). In polη-deficient XP30RO cells, incorporation of halogenated nucleosides is strongly inhibited upon treatment with cisplatin or carboplatin. This may be a cumulative effect of both reduced lesion bypass and checkpoint-dependent inhibition of origin firing24. However, the DNA damage-dependent increase in phosphorylation of RPA2 on Ser4/Ser8 in XP30RO cells depends on inhibition of DNA replication, since the level of damage-induced RPA2 phosphorylation is significantly lower in TR30-2 cells expressing polη. We have previously reported that platinum-induced phosphorylation of RPA2 on Ser4/Ser8 occurs predominantly in S-phase cells25. Here, we directly link RPA2 phosphorylation on Ser4/Ser8 with severe replication inhibition. While the present analysis of EdU fluorescence intensity does not distinguish between early, mid- or late S-phase cells, since the frequency distribution graphs and BrdU incorporation profiles (Fig. 1a, b) show that arrested cells are distributed throughout S-phase, it is unlikely that all XP30RO cells with relative EdU fluorescence intensities below the threshold value represent early S-phase cells.
Based on our observations, we propose that in polη-deficient cells, DNA synthesis on the leading strand is impaired by platinum-induced intrastrand DNA lesions, which can lead to uncoupling of the DNA polymerase and helicase activities at the replication fork. This results in accumulation of ssDNA, enhanced RPA binding and PIKK-mediated checkpoint signalling. In support of this conclusion, cells treated with UV radiation, which, like platinum-based drugs, does not directly induce DSBs, show a similar pattern of accumulation of RPA-coated ssDNA24 and γH2AX formation24. Uncoupling of helicase and polymerase activities during replication of cisplatin-damaged DNA has been reported previously in Xenopus egg extracts46. UV radiation- and cisplatin-induced inhibition of DNA synthesis in human cells lacking polη also leads to accumulation of ssDNA and formation of double strand ends due to collapse of stalled replication forks15,23,24, consistent with co-localisation of phosphorylated RPA2 and γH2AX in nuclear foci in XP30RO cells (Fig. 3B).
Overall, the present data provide novel insights into the cellular processes that modulate the response of human cells to treatment with cisplatin and related platinum-based drugs, and suggest that targeting polη-mediated lesion bypass may represent one approach to potentiate the inhibitory effects of platinum-based drugs on DNA replication in cancer cells.
Methods
Cell culture
The SV40-transformed human skin fibroblast cell lines XP30RO (GM03617A) and TR30-2, were grown in Minimal Essential Medium (MEM), Eagle-Earle BSS supplemented with 10% non-heat inactivated fetal bovine serum (FBS), 2× essential and non-essential amino acids, vitamins and 2 mM L-glutamine. XP30RO cells lack functional polη as result of 13-base pair deletion in POLH6,7. The TR30-2 cell line constitutively expresses wild-type polη, following stable transfection of XP30RO cells with a POLH transgene, as previously described23.
Cell treatment
Cells were treated with cisplatin (1 mg/ml stock solution, Hospira UK Limited) or carboplatin (Sigma; 20 mM stock solution prepared in distilled water), 48 h after plating, at approximately 80% confluence. Drugs were added directly to the cell culture medium. Control cell cultures were treated with the equivalent volume of water only. Cells were treated with cisplatin (1.7 μM; 0.5 μg/ml) and carboplatin (50 μM; 18.6 μg/ml). Apoptotic cells were obtained by collecting floating cells following treatment with a high dose of cisplatin (17 μM) for 24 h.
Western immunoblotting
Whole cell lysates were prepared as described previously23. Protein concentration was determined using the DC assay (BioRad). Proteins were separated by SDS-PAGE, transferred to PVDF membrane, and analyzed by western immunoblotting. Where indicated, membranes were probed overnight at 4°C with one of the following antibodies: anti-DNA polymerase η (1/1000, Abcam), anti-RPA2 (1/4000, Oncogene Research Products), anti-phosphoSer4/Ser8 RPA2 (1/4000, Bethyl Laboratories), anti-Chk1 (1/1000, Sigma), anti-phosphoSer317 Chk1 (1/1000, Cell Signalling), anti-phosphoSer139-H2AX (1/1000, Upstate Technologies), anti-actin (1/5000, Sigma), anti-cleaved caspase-3 (1/1000, Cell Signalling) and anti-cleaved-PARP (1/2000, Cell Signalling). Blots were incubated with horseradish peroxidase-linked secondary antibody (Jackson Immunochemicals) and visualized using the ECL+ chemiluminescence method (Amersham).
Flow cytometry
Cell cycle progression was analysed by flow cytometry. Cells were pulse-labelled with 10 μM bromodeoxyuridine (BrdU) for 75 min before harvesting. Samples were analysed using a FACSCalibur (BD Biosciences), and Cell Quest™ software as described previously23.
DNA combing
Cells were plated in 100 mm dishes, 36–48 h before treatment with cisplatin or carboplatin for 24 h. Before harvesting, cells were pulse-labelled with 20 μM iodo-deoxyuridine (IdU) for 15 min, followed by 1 h incubation with 200 μM chloro-deoxyuridine (CldU) in 7 ml of pre-warmed medium. Cells were then harvested, counted, resuspended in PBS and diluted 1 in 2 in 1% (w/v) low melting point agarose (Molecular Probes) to a final concentration of 8 × 105 cells/ml. Agarose plugs were formed using 100 μl of cells (4 × 104 cells) suspended in agarose. Solidified plugs were incubated in 10 mM Tris-HCl buffer (pH 7.5) containing 50 mM EDTA, 1% (w/v) Sarkosyl (Sigma) and 2 mg/ml proteinase K (Roche) for 48 h, at 50°C in the dark. Further steps were performed as previously described47,48,49. Briefly, DNA fibres extracted from cells were stretched on silanized coverslips and counterstained using a mouse monoclonal antibody against ssDNA (MAB3034; Chemicon; 1/500) and anti-mouse IgG2a coupled to Alexa 647 (A21241, Molecular Probes, 1/50). IdU was detected using B44 mouse anti-BrdU antibody (Becton Dickinson, 1/20) and Alexa-Fluor 546-conjugated anti-mouse IgG1 secondary antibody (A21123, Molecular Probes, 1/50). CldU was detected using BU1/75 anti-BrdU antibody (AbCys, 1/20) and Alexa-Fluor 488-conjugated anti-rat secondary antibody (A11006, Molecular Probes, 1:50). DNA fibres were analysed on a Leica DM6000B microscope (40× objective), equipped with a CoolSNAP HQ CCD camera (Roper Scientific). Data acquisition and fibre length analysis was performed using MetaMorph (Roper Scientific). Only fibres stained for both IdU and CldU were included in the statistical analysis, and only the length of the CldU-labelled region was measured49.
To generate images, representative DNA fibres were selected from different fields and processed with Adobe Photoshop, as described previously48,49. Graphs were plotted using Prism v5.0 (GraphPad Software). Frequency distribution graphs represent the fraction of fibres of particular lengths. A Gaussian non-linear regression curve was fitted to the data. Statistical analysis was performed in Prism v5.0 (GraphPad Software) using the non-parametric Mann-Whitney rank sum test. Origin firing analysis was performed by capturing random, distant fields on the ssDNA fluorescence channel and measuring at least 80 Mbp of DNA fibres per experimental condition. Replication origins were scored based on IdU and CldU incorporation, taking incorporation of either IdU, CldU or both labels to be the result of an ongoing fork. The percentage of the measured DNA that is derived from S-phase cells was obtained by multiplying the total length of the analysed DNA by the fraction of cells in S-phase, as determined from BrdU incorporation experiments. The origin density was calculated by dividing the length of the S-phase DNA by the number of origins. Results are expressed as Mb of S-phase DNA per ori [Mb/ori].
Analysis of DNA replication by fluorescence microscopy
DNA replication was analysed using the azide-alkyne Huisgen cycloaddition reaction (‘click chemistry’) and fluorescence microscopy to determine the extent of incorporation of ethynyl-deoxyuridine (EdU) into genomic DNA50. Cells were grown on glass coverslips, treated with cisplatin or carboplatin as described and pulse-labelled with 10 μM EdU for 75 minutes. Cells were pre-permeabilised51 and fixed in 4% (v/v) PFA for 15 minutes at room temperature. Cells were washed three times with PBS, permeabilised with 0.1% (v/v) Triton X-100 in PBS (PBS-TX) for 20 min, and blocked using PBS containing 20% (v/v) NGS (Sigma), 0.5% (w/v) BSA and 0.1% Triton X-100 for 30 min at room temperature. Cells were then stained using rabbit anti-phosphoSer4/Ser8-RPA2 (1/200, Bethyl Laboratories). Coverslips were washed five times with PBS-TX for 5 minutes before the click chemistry reaction was initiated50. Cells were incubated for 30 min at room temperature in PBS containing 10 mM (+)-sodium-L-ascorbate, 0.1 mM 6-carboxyfluorescein-TEG azide and 2 mM copper (II) sulphate. Coverslips were washed and stained using goat anti-rabbit Alexa-Fluor 488-labelled antibody (A11008, Molecular Probes, 1/50). Nuclear DNA was subsequently stained with DAPI, and the cells were mounted on slides in SlowFade (Invitrogen). Images were captured using a DeltaVision Core system (Applied Precision) controlling an interline charge-coupled device camera (Coolsnap HQ2; Roper) mounted on an inverted microscope (IX-71; Olympus). For each sample, images were collected at 2× binning using a 60× oil objective at 0.2 μm z sections, and processed as described previously52. Briefly, images were deconvolved and the maximum intensity was projected using SoftWoRx (Applied Precision). For quantification, unscaled DeltaVision images were used. EdU signal intensity was determined using ImagePro 6.3. Using the DAPI signal to define the nucleus, a mask was generated that was applied to the images in the FITC channel, and the mean fluorescence intensity was measured. For each individual sample, a mean background intensity value, obtained from five different areas outside the ‘nucleus’ mask, was subtracted from the values obtained from nuclear staining. The mean intensity obtained from EdU-negative nuclei (defined based on threshold values and visible EdU staining) was then determined and subtracted from the mean signal in EdU-positive cells.
Phospho-Ser4/Ser8 RPA2 and phospho-Ser139 H2AX foci were analysed using double-staining indirect immunofluorescence. Cells were treated, pre-permeabilised and fixed as described above. Anti-phospho-Ser139-H2AX antibody (Upstate Technologies) was used at 1/8000 dilution, and detected using goat anti-rabbit Alexa-Fluor 594-labelled secondary antibody (Jackson Immunochemicals). The samples were processed and analysed by fluorescence microscopy as described above.
Supplementary Material
Acknowledgments
We would like to thank Dr. Seamus Coyne for development of the TR30-2 cell line, and Dr. Julien Bianco for training in DNA combing. We are grateful to the DNA combing facility of Montpellier for silanized coverslips. We wish to thank Dr. Anna E. Kliszczak and Prof. Corrado Santocanale for reagents and for assistance with EdU labelling experiments. This project was funded by the Irish Research Council (AMS) and the Health Research Board (SCH; MPC). DNA combing experiments were supported by the NUI Galway Beckman Scholarship Fund, and the Thomas Crawford Hayes Scholarship (AMS). Research in the laboratory of Dr. Pasero is supported by the Ligue contre le Cancer (Equipe labellisée).
Footnotes
The authors declare no competing financial interests.
Author Contributions A.M.S., S.C.H., P.P. and M.P.C. conceptualized and designed the study. A.M.S. performed all experiments. A.M.S. and P.P. analyzed DNA combing data. A.M.S. and M.P.C. wrote the manuscript. All authors reviewed and revised the manuscript, and approved the final manuscript.
References
- Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer 7, 573–584 (2007). [DOI] [PubMed] [Google Scholar]
- Wernyj R. P. & Morin P. J. Molecular mechanisms of platinum resistance: still searching for the Achilles' heel. Drug Resist Update 7, 227–232 (2004). [DOI] [PubMed] [Google Scholar]
- Wang D. & Lippard S. J. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov 4, 307–320 (2005). [DOI] [PubMed] [Google Scholar]
- Chaney S. G., Campbell S. L., Bassett E. & Wu Y. Recognition and processing of cisplatin- and oxaliplatin-DNA adducts. Crit Rev Oncol/Hematol 53, 3–11 (2005). [DOI] [PubMed] [Google Scholar]
- Kartalou M. & Essigmann J. M. Recognition of cisplatin adducts by cellular proteins. Mutation Res/Fund Mol Mech Mutagen 478, 1–21 (2001). [DOI] [PubMed] [Google Scholar]
- Masutani C. et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature 399, 700–704 (1999). [DOI] [PubMed] [Google Scholar]
- Johnson R. E., Kondratick C. M., Prakash S. & Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285, 263–265 (1999). [DOI] [PubMed] [Google Scholar]
- Burgers P. M. et al. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J Biol Chem 276, 43487–43490 (2001). [DOI] [PubMed] [Google Scholar]
- Masutani C., Kusumoto R., Iwai S. & Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J 19, 3100–3109 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher V. M., Ouellette L. M., Curren R. D. & McCormick J. J. Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature 261, 593–595 (1976). [DOI] [PubMed] [Google Scholar]
- Gueranger Q. et al. Role of DNA polymerases eta, iota and zeta in UV resistance and UV-induced mutagenesis in a human cell line. DNA Rep 7, 1551–1562 (2008). [DOI] [PubMed] [Google Scholar]
- Vaisman A., Masutani C., Hanaoka F. & Chaney S. G. Efficient Translesion Replication Past Oxaliplatin and Cisplatin GpG Adducts by Human DNA Polymerase η. Biochemistry 39, 4575–4580 (2000). [DOI] [PubMed] [Google Scholar]
- Albertella M. R., Green C. M., Lehmann A. R. & O'Connor M. J. A role for polymerase eta in the cellular tolerance to cisplatin-induced damage. Cancer Res 65, 9799–9806 (2005). [DOI] [PubMed] [Google Scholar]
- Chen Y.-W., Cleaver J. E., Hanaoka F., Chang C.-F. & Chou K.-M. A Novel Role of DNA Polymerase eta in Modulating Cellular Sensitivity to Chemotherapeutic Agents. Mol Cancer Res 4, 257–265 (2006). [DOI] [PubMed] [Google Scholar]
- Cruet-Hennequart S., Glynn M. T., Murillo L. S., Coyne S. & Carty M. P. Enhanced DNA-PK-mediated RPA2 hyperphosphorylation in DNA polymerase eta-deficient human cells treated with cisplatin and oxaliplatin. DNA Rep 7, 582–596 (2008). [DOI] [PubMed] [Google Scholar]
- Ho T. V., Guainazzi A., Derkunt S. B., Enoiu M. & Scharer O. D. Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases. Nucleic Acids Res 39, 7455–7464 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A. R., Harbut M. B., Lloyd R. S. & Minko I. G. Replication bypass of N2-deoxyguanosine interstrand cross-links by human DNA polymerases eta and iota. Chem Res Tox 25, 755–762 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J. K. et al. Differential roles for DNA polymerases eta, zeta, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol Cell Biol 30, 1217–1230 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D. & Foiani M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Rep 6, 994–1003 (2007). [DOI] [PubMed] [Google Scholar]
- Chanoux R. A. et al. ATR and H2AX Cooperate in Maintaining Genome Stability under Replication Stress. J Biol Chem 284, 5994–6003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B. B. & Elledge S. J. The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439 (2000). [DOI] [PubMed] [Google Scholar]
- Jackson S. P. & Bartek J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruet-Hennequart S., Coyne S., Glynn M. T., Oakley G. G. & Carty M. P. UV-induced RPA phosphorylation is increased in the absence of DNA polymerase eta and requires DNA-PK. DNA Rep 5, 491–504 (2006). [DOI] [PubMed] [Google Scholar]
- Despras E., Daboussi F., Hyrien O., Marheineke K. & Kannouche P. L. ATR/Chk1 pathway is essential for resumption of DNA synthesis and cell survival in UV-irradiated XP variant cells. Hum Mol Genet 19, 1690–1701 (2010). [DOI] [PubMed] [Google Scholar]
- Cruet-Hennequart S. et al. Characterization of the effects of cisplatin and carboplatin on cell cycle progression and DNA damage response activation in DNA polymerase eta-deficient human cells. Cell Cycle 8, 3039–3050 (2009). [PubMed] [Google Scholar]
- Zou L. & Elledge S. J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 (2003). [DOI] [PubMed] [Google Scholar]
- Olson E., Nievera C. J., Klimovich V., Fanning E. & Wu X. RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. J Biol Chem 281, 39517–39533 (2006). [DOI] [PubMed] [Google Scholar]
- Smith J. et al. in Adv Cancer Res Vol. Volume 108 (ed 2010) 73–112 (Academic Press, 2010). [Google Scholar]
- Zhang Y. W. et al. Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol Cell 19, 607–618 (2005). [DOI] [PubMed] [Google Scholar]
- Revet I. et al. Functional relevance of the histone H2Ax in the response to DNA damaging agents. Proc Natl Acad Sci U S A 108, 8663–8667 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz S. K., Sheehan A. M. & Wold M. S. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Rep 3, 1015–1024 (2004). [DOI] [PubMed] [Google Scholar]
- Stephan H., Concannon C., Kremmer E., Carty M. P. & Nasheuer H. P. Ionizing radiation-dependent and independent phosphorylation of the 32-kDa subunit of replication protein A during mitosis. Nucleic Acids Res 37, 6028–6041 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw H., Lee D. & Myung K. DNA-PK-Dependent RPA2 Hyperphosphorylation Facilitates DNA Repair and Suppresses Sister Chromatid Exchange. PLoS ONE 6, e21424 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley G. G. et al. UV-induced hyperphosphorylation of replication protein a depends on DNA replication and expression of ATM protein. Mol Biol Cell 12, 1199–1213 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceppi P. et al. Polymerase eta mRNA expression predicts survival of non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin Cancer Res 15, 1039–1045 (2009). [DOI] [PubMed] [Google Scholar]
- Zhao Y. et al. Structural basis of human DNA polymerase eta-mediated chemoresistance to cisplatin. Proc Natl Acad Sci U S A 109, 7269–7274 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ummat A. et al. Structural basis for cisplatin DNA damage tolerance by human polymerase eta during cancer chemotherapy. Nat Struct Mol Biol 19, 628–632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles J. T. P., Poli J., Marians K. J. & Pasero P. Rescuing Stalled or Damaged Replication Forks. Cold Spring Harb Perspect Biol 5, a012815 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A. R. et al. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc Natl Acad Sci U S A 72, 219–223 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey L. et al. Human DNA Polymerase η Is Required for Common Fragile Site Stability during Unperturbed DNA Replication. Mol Cell Biol 29, 3344–3354 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt A. et al. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science 318, 967–970 (2007). [DOI] [PubMed] [Google Scholar]
- Speroni J., Federico M. B., Mansilla S. F., Soria G. & Gottifredi V. Kinase-independent function of checkpoint kinase 1 (Chk1) in the replication of damaged DNA. Proc Natl Acad Sci U S A 109, 7344–7349 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J. & Lukas J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol 13, 738–747 (2001). [DOI] [PubMed] [Google Scholar]
- Maya-Mendoza A., Petermann E., Gillespie D. A., Caldecott K. W. & Jackson D. A. Chk1 regulates the density of active replication origins during the vertebrate S phase. EMBO J 26, 2719–2731 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E., Woodcock M. & Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci U S A 107, 16090–16095 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun T. S., Pacek M., Yee M. C., Walter J. C. & Cimprich K. A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 19, 1040–1052 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X. et al. Dynamic Molecular Combing: Stretching the Whole Human Genome for High-Resolution Studies. Science 277, 1518–1523 (1997). [DOI] [PubMed] [Google Scholar]
- Pasero P., Bensimon A. & Schwob E. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev 16, 2479–2484 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco J. N. et al. Analysis of DNA replication profiles in budding yeast and mammalian cells using DNA combing. Methods 57, 149–157 (2012). [DOI] [PubMed] [Google Scholar]
- Kliszczak A. E., Rainey M. D., Harhen B., Boisvert F. M. & Santocanale C. DNA mediated chromatin pull-down for the study of chromatin replication. Sci Rep 1, 95 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard D. J. et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature 453, 677–681 (2008). [DOI] [PubMed] [Google Scholar]
- Prendergast L. et al. Premitotic assembly of human CENPs -T and -W switches centromeric chromatin to a mitotic state. PLoS Biol 9, e1001082 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.