Abstract
Background Integration of electronic health records (EHRs) data across sites and access to that data remain limited.
Objective We developed an EHR-based pediatric inpatient repository using nine U.S. centers from the National Institute of Child Health and Human Development Pediatric Trials Network.
Methods A data model encompassing 147 mandatory and 99 optional elements was developed to provide an EHR data extract of all inpatient encounters from patients <17 years of age discharged between January 6, 2013 and June 30, 2017. Sites received instructions on extractions, transformation, testing, and transmission to the coordinating center.
Results We generated 177 staging reports to process all nine sites' 147 mandatory and 99 optional data elements to the repository. Based on 520 prespecified criteria, all sites achieved 0% errors and <2% warnings. The repository includes 386,159 inpatient encounters from 264,709 children to support study design and conduct of future trials in children.
Conclusion Our EHR-based data repository of pediatric inpatient encounters utilized a customized data model heavily influenced by the PCORnet format, site-based data mapping, a comprehensive set of data testing rules, and an iterative process of data submission. The common data model, site-based extraction, and technical expertise were key to our success. Data from this repository will be used in support of Pediatric Trials Network studies and the labeling of drugs and devices for children.
Keywords: pediatric clinical trials, electronic heath record reuse, NICHD Pediatric Trials Network, inpatient repository, data extraction
Background and Significance
Electronic health record (EHR) systems hold great promise for pediatric clinical research. 1 2 3 4 Primarily designed to improve clinical care, EHRs store vast amounts of routinely generated patient and provider data. 5 Through dissemination and development of sophisticated clinical research informatics tools, it is increasingly feasible to reuse EHR data for research. 6 7 This strategy is particularly attractive for the study of rare diseases including those affecting children, the study of uncommonly used drugs, or the characterization of infrequent adverse drug events in adults and children. 8 9 10 11 12 13
Despite the value of retrospective studies, prospective trials remain essential to drug development. 14 In response to the gap in pediatric drug labeling, the Eunice Kennedy Shriver National Institute for Child Health and Human Development (NICHD) created the Pediatric Trials Network (PTN), tasked under the Best Pharmaceuticals for Children Act with the design and conduct of prospective off-patent drug and device trials in children. 15 16 The PTN has enrolled >7,000 children in 38 phase 1 to phase 4 studies in 18 therapeutic areas.
From its inception, the PTN recognized the value of EHR data to support clinical trials. The network leveraged the Mednax Clinical Data Warehouse, the American Academy of Pediatrics-supported CER 2 (Comparative Effectiveness Research through Collaborative Electronic Reporting) database, and individual site EHRs to identify target drugs and study populations, inform protocol development, and support findings from prospective trials. 17 18 19 20 21 22
Objective
To take full advantage of data generated at sites, and because of our initial positive experience with EHR-generated data, the PTN sought to develop a multicenter pediatric inpatient data repository: The Pediatric Trials Network Data Repository. Data from this repository will be used in support of PTN trials and the labeling of drugs and devices for children. Herein, we describe the design and creation of this EHR-derived repository and share lessons learned in the process.
Methods
Coordinating Center Team
The coordinating center team included a project leader, principal investigator, two clinical research informaticists, and an information technology (IT) team including a senior data modeler, senior extract-transform-load (ETL) developer, a quality assurance tester, and a project manager. At each participating site, the coordinating center team identified a principal investigator and at least one technical lead. Site principal investigators were proposed by the participating sites, and included a combination of pediatric trialists and investigators with experience in data science or outcomes research. Formal IT or informatics training was not required for the principal investigator. The technical leads were required to hold IT- or informatics-based positions at their site and be actively involved in the management of the site EHR. Whenever possible, we encouraged the inclusion of a local informaticist. Sites were compensated for their efforts with payments based on the completion of prespecified study milestones including site activation and data transfers.
Site Selection
The 30 highest enrolling sites across the PTN were screened for participation. These include a mix of academic and private practice sites with inpatient pediatric services across North America ( www.pediatrictrials.org ). Enrollment was defined as the cumulative number of children enrolled across all PTN studies conducted since the inception of the network in 2010. A Web-based feasibility survey was administered for the following capabilities: (1) presence of an inpatient EHR system, (2) availability of qualified site technical personnel to analyze EHR data and map them according to the PTN specifications, (3) accessibility of IT infrastructure to produce extract files, and (4) general interest in the study. Based on feasibility results, 15 individual sites completed a secondary telephone interview with the coordinating center focused on 21 additional technical factors (see Supplementary Table S1 for a list of interview questions, available in the online version). Sites were then ranked based on equally weighted scores assigned by all coordinating center team members, resulting in the selection of 10 sites. One site was not able to complete the contracting process within the time constraints of the study, resulting in nine participating sites.
Data Model
After reviewing existing national standardization efforts of multiple EHR data systems, 23 24 25 26 27 the coordinating team decided to pattern the first version of the PTN repository data points using the National Patient-Centered Clinical Research Network (PCORnet) data model (version 2.0). 26 28 This model was chosen because it included several of the inpatient data elements relevant to pediatric drug trials, including demographics and medication administration information, and used standard terminology and coding systems for healthcare. PCORnet was developed in part based on the Observational Medical Outcomes Partnership (OMOP), which has provided data for pharmacoepidemiologic research similar to the type of projects planned by the PTN. 29 30 31 In addition, the Duke Clinical Research Institute (DCRI) supports the PCORnet Coordinating Center (in partnership with Harvard Pilgrim Health Care Institute and the Genetic Alliance), providing access to personnel with experience working with this data model. 32 To ensure utility of the developed repository, pediatric clinical trialists and outcomes researchers from within the PTN were consulted to identify key data domains of interest. Whenever possible, the identified domains were aligned with PCORnet domains. Data perceived to be of particular relevance for early phase pharmacokinetic and pharmacodynamic trials commonly conducted by the PTN, including vital signs, inpatient assessments captured on flow sheets (e.g., pain and sedation scores), daily enteral and parenteral fluid intake and output, and respiratory support information, were added to the data model. After selection of the domains, all data points and their definitions were reviewed by pediatric thought leaders for completeness and applicability to the project. Following this review, a data master list was created and rules were developed for validation of each data point. These validation rules were added to the data master list together with coded value sets, data formats, a clinical description of the data point, and an indicator of whether the field was mandatory or optional. The data master list was then released to sites to perform an initial inventory assessment. As the funding sponsor of the PTN, NICHD has ownership of the data. In its role as the PTN Coordinating Center, DCRI will maintain the data and provide data science and statistical analysis support to PTN investigators, who will propose data queries through the PTN Web site ( www.pediatrictrials.org ). All queries will be reviewed by PTN leadership for approval, and requests for publication will follow the established PTN publication policy. Analysis results will be shared with requesting sites, while only the coordinating center, funding sponsor (NICHD), and regulators may access the original data. Physical access to the database server and associated analytical tools is mandated via the Duke Health Security and Federal Information Security Management Act (FISMA) 33 policies. This data-sharing approach was approved by the institutional review board of Duke University (as the coordinating center), and all participating sites.
Repository Design
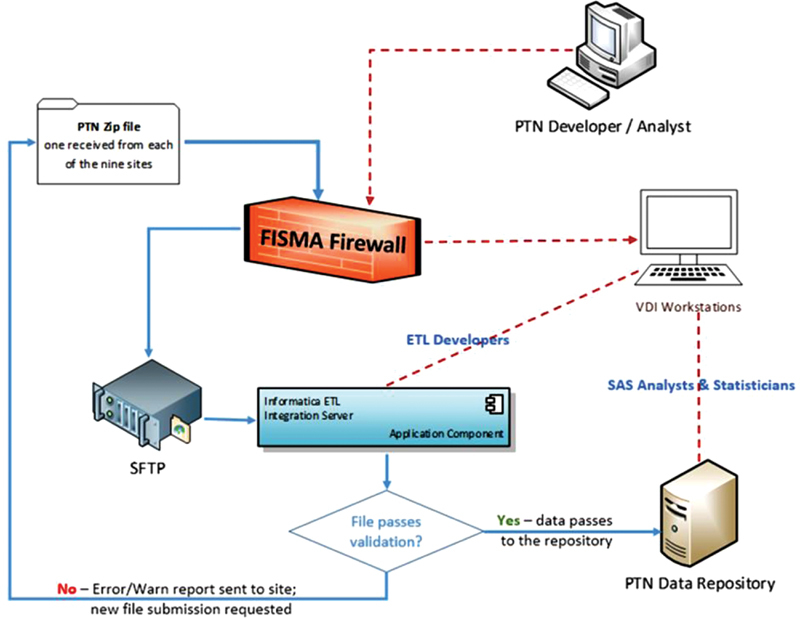
Each participating site was provided with an implementation guide, including instructions on how to perform EHR extractions, transformation, and testing, and how to package the data for submission. The transmissions were performed via secured file transfer protocol (FTP) delivery method. Data were downloaded from the secured FTP drop site and processed using 15 Informatica (Informatica, Redwood City, California, United States) ETL workflow processes, and stored in an Oracle repository (Oracle Corp., Redwood Shores, California, United States). All data were secured within a medium-level FISMA-compliant environment ( Fig. 1 ). The data intake process was designed to be agnostic to the specific type of EHR run at each site. Rather, sites were required to map their data according to the PTN specifications.
Fig. 1.

Pediatric Trials Network (PTN) contextual data model.
Via a priori method, we defined a set of 520 rules designed to detect errors or warnings in the submitted data. We created an automated script that was built into the ETL process to check submitted data against all 520 rules. Errors were generated and logged if a data point violated a critical rule (e.g., invalid formats, required value not present, or nonunique primary key). Warnings were generated and logged if a data point violated a noncritical rule (e.g., certain reasonable logical inconsistencies, nonrequired value missing, or dates outside of expected sequences). Each new data submission was tested against all 520 rules, and a warning and error report was generated and shared with sites. For a data submission to be considered acceptable, 0% errors and <2% warnings were required across all records.
The database and ETL logic were first created and tested within development and validation environments. After successful loads in both, the same logic was migrated to the production environment. All three environments were constructed in an identical manner.
Results
We included all children <17 years of age at the time of admission between June 1, 2013, and June 30, 2017, whose admission source records were available in the EHR. The type of EHR used varied by site and represented two different platforms: Epic (Epic Systems Corporation, Verona, Wisconsin, United States) and Cerner (Cerner Corporation, North Kansas City, Missouri, United States). We excluded outpatient encounters and inpatient admissions without an EHR record. Our final data model included 147 mandatory and 99 optional data elements across 14 domains ( Fig. 2 ). Patient and site identity and admission date were used as primary keys to each file related to an admission, while patient and site identity were used as primary keys for all files not related to an admission. In addition to 10 PCORnet domains, we created four custom data domains because of their perceived importance to PTN trial design and execution: vital signs including weight, enteral and parenteral fluids, flowsheet assessments, and respiratory support. We included any flowsheet assessment provided with a tool name, result, and date and time of assessment.
Fig. 2.
Repository design and infrastructure.
All data mapping was conducted by the sites using the implementation guide. The mapping was iterative, with each site first completing a trial submission on a small subset of their records across all 14 domains. Following each data submission and testing against rules, a report of warnings and errors was generated and shared with sites. Individual conference calls between each site and the coordinating center technical team were then scheduled to review results of each submission and discuss strategies for resolution if needed. Over a 14-month period, 177 data submissions were received. Final transfer of a site file to the repository required approval of the entire coordinating center team.
An average of 20 submissions per site (range 13–29) was required before data were moved into the repository. Site participation from kickoff to final data submission ranged from 8 to 14 months. The first data submissions were received within 2 months of sending out the implementation guide, and the time from first to final submission ranged from 6 to 10 months. Reasons for repeat submissions varied by site and submission, and were systematically recorded in site-specific project trackers. Most common reasons for repeat submissions included: failure of site to supply a required data element, failure of site to submit data in correct format, and failure of site to submit a required domain. Time to process the final complete site files ranged from 30 to 36 hours. All initial data submissions contained at least one error. However, all sites achieved 0% errors and <2% warnings.
The complete repository includes 386,159 inpatient encounters from 264,709 children across 9 participating sites. The encounters consist of 563,886,147 data points. Encounters were evenly distributed across the study period, but varied by site as expected based on hospital admission volume. Among the 14 data domains, 2 domains account for the majority of data points: “assessments” (33%) and “vital signs” (31%). Clinical and demographic data are shown in Table 1 .
Table 1. Summary of clinical data from participants in the repository.
| Variables | Counts, median (range), or % |
|---|---|
| Patients | 264,709 |
| Encounters | 386,159 |
| Patient age | 2.3 (0–17) |
| Patient sex | |
| Female | 53% |
| Male | 47% |
| Patient race | |
| American Indian or Alaska Native | <1% |
| Asian | 4% |
| Black or African American | 19% |
| Native Hawaiian or Other Pacific Islander | <1% |
| White | 58% |
| Multiple races | 2% |
| Other | 9% |
| Unknown | 6% |
| Non-Hispanic | 82% |
| Patient ethnicity | |
| Hispanic | 18% |
| Encounters by discharge year | |
| 2013 | 11% |
| 2014 | 25% |
| 2015 | 26% |
| 2016 | 26% |
| 2017 | 12% |
| Encounters by site | |
| 01 | 12% |
| 02 | 21% |
| 03 | 15% |
| 04 | 7% |
| 05 | 6% |
| 06 | 9% |
| 07 | 11% |
| 08 | 7% |
| 09 | 12% |
| Common drug administration records | |
| Albuterol sulfate | 974,551 |
| Acetaminophen | 656,159 |
| Lorazepam | 494,086 |
| Furosemide | 482,635 |
| Morphine | 385,013 |
| Common diagnosis records | |
| Feeding difficulties/intolerance | 40,767 |
| Gastroesophageal reflux disease | 38,178 |
| Fever, unspecified | 25,388 |
| Dehydration | 18,143 |
| Other dyspnea and respiratory anomaly | 18,039 |
Discussion
We created a FISMA-compliant data repository extracted from inpatient pediatric EHR encounters at nine U.S. hospitals participating in the NICHD PTN. Our approach utilized a customized data model heavily influenced by the PCORnet format, site-based data mapping, a comprehensive set of data testing rules, and an iterative process of data submission. Crucial to our success were the close-working relationships between dedicated teams with expertise in IT, informatics, and clinical medicine at both the coordinating center and participating sites.
Multiple subspecialties have successfully leveraged clinical data from the EHR for research applications, including cardiovascular medicine, emergency care, epidemiology, and pediatrics. 8 9 22 34 35 36 37 38 39 PEDSnet is a PCORnet clinical data research network developed to provide the digital infrastructure for a national pediatric learning health system. 40 41 This objective requires PEDSnet to have extensive capabilities including dual methods for data submission and sharing (i2b2 and OMOP), terminology harmonization across sites and existing networks, a link to NICHD terminology efforts, the ability to integrate patient-reported outcomes and biospecimen data, and partnerships with national data partners including pharmacy benefits − managing companies and multipayer administrative databases. 42 Even though the scope of the PTN repository is much narrower, we were able to adapt features of the PEDSnet model including the use of a centralized data coordinating center, a predefined set of ETL conventions, and an iterative building process with prespecified data quality checks. 43 Similar to the PEDSnet approach, we initially focused on completeness of data and conformity with predefined conventions. As the repository evolves, we will gradually shift our focus toward assessing plausibility. Despite the considerable breadth and reach of PCORnet data, we chose to develop our own, modified data model. This approach was motivated by small but important differences in the anticipated use of the data, including the desire to capture clinical data commonly recorded on inpatient flow sheets, such as pain and sedation scores and vital signs, and data on daily fluid intake and output. These data are of particular importance for early phase pharmacokinetic and pharmacodynamic trials commonly conducted by the PTN. Further, not all PTN sites are members of the PCORnet distributed research network, and while we acknowledge that joint participation of all pediatric research sites in one data-sharing network would be preferable, this goal is beyond the current scope of the PTN.
The applications of EHR data to clinical research are diverse, and include protocol design, participant selection, translational research efforts, safety analyses, and trial execution. 44 45 46 47 In its effort to secure pediatric labeling of off-patent drugs and devices, the PTN has successfully reused EHR data collected by collaborating networks and individual participating sites. Analyzing EHR data, we described drug utilization practices and target molecules for study, reported on real-world drug safety, and performed comparative effectiveness studies. 18 48 49 50 51 The PTN has also leveraged EHR data as an innovative tool in pharmacometric analyses, a critically important step in pediatric drug development in concert with the Food and Drug Administration. 17 52 53 54 55 56
Despite these successes, more widespread application of EHR data within the PTN is contingent upon access to detailed clinical information including drug administration, laboratory results, vital signs, diagnoses and procedures, and clinical assessments. Data sources from previous PTN collaborating networks including the Pediatrix Medical Group Clinical Data Warehouse do not routinely capture this level of detail, collaborations with CER 2 were focused on outpatient data, and site-based collection of data from the EHR for a specific study is resource-consuming and inefficient. This multicenter repository provides access to detailed data, including new domains not routinely collected in registries such as vital signs, fluid intake and outputs, flowsheet assessments, and respiratory support variables.
This repository creates an opportunity to advance the PTN mission to design and conduct off-patent drug and device trials in children. Specific applications made possible by the data include clinical trial simulation, cohort enrichment strategies, synthetic control groups of patients whose data are captured in the EHR and used for comparison against prospective trial populations, retrospective safety assessments, and comparative effectiveness. To meet these goals, a team of clinicians, IT and informatics experts, and biostatisticians has been assembled to design and implement appropriate analytics, quality control, and data curation tools. The team will also leverage the repository to explore solutions to critical real-world data aresues including record linkage, missing data elements, and quality. 57 58 59 To maximize the pediatric public health benefit of the repository while maintaining data privacy and confidentiality, PTN investigators will be offered the opportunity to propose studies and research questions through a Web-based interface ( www.pediatrictrials.org ). Further, we will explore opening the repository to participation by other PTN sites.
We anticipate that through ongoing use of the repository, errors, discrepancies, and limitations of the data will become apparent. Whenever possible, we will remediate these with the assistance of our sites.
At this stage, several limitations and important lessons have been identified. By design, we did not include textual data, which is challenging to collect, deidentify, and analyze. As new natural language process methods become available, however, textual data reuse for clinical research will grow. 60 The repository also focused on inpatient data only. Collection of ambulatory data may require additional considerations of its formats, linking to inpatient encounters, and availability of qualified personnel to perform data collection. A successful example of outpatient EHR data use for research is the American Academy of Pediatrics CER 2 network. 22 To ensure success, CER 2 also relies on a common data model (derived from the OMOP model), and was built by allying existing outpatient practice networks, rather than recruiting individual clinics and sites. Because our data model relied heavily on the PCORnet model, which is itself derived from OMOP, our model can be viewed as yet another extension of OMOP focused on the inpatient setting. A similar effort within the PTN was beyond the scope of this work but could be implemented in future iterations of this repository. Site-based mapping, iterative submission and error reporting, and intense bidirectional communication was ultimately successful in creating a repository, but may be challenging to scale up to a larger number of sites. At this time, prospective new sites would have to undergo the same data submission process. While lessons learned to date may help identify errors and warnings prior to submission, we would anticipate a high workload for site research teams to process multiple rounds of submission prior to the elimination of all errors. In our experience, the majority of site-based efforts were concentrated on the mapping of data elements, a process that requires considerable informatics expertise. This is in turn associated with significant costs at the site level, which would need to be carefully considered and potentially shared between sites and coordinating institutions to ensure success. Finally, once a data repository is created, comprehensive high-quality analyses of these data will be performed by trained experts not traditionally involved in clinical research networks including biostatistics, IT, informatics, machine learning and advanced analytics, and systems medicine. Integration of this workforce into clinical trial teams will help maximize the value of EHR data.
Clinical Relevance Statement
We created a multicenter, pediatric inpatient, EHR-derived data repository to support PTN trials and the labeling of drugs and devices for children. In our multicenter model, clinical, informatics-trained, and information technology personnel performed site-based data mapping with central research support and intense bidirectional communication for the successful creation of our data repository.
Multiple Choice Questions
-
The data model of the newly developed PTN repository was based on which of the following national data standardization efforts:
Health Level Seven International (HL7).
Fast Healthcare Interoperability Resources (FHIR).
Biomedical Research Integrated Domain Group (BRIDG).
Patient-Centered Clinical Research Network (PCORnet).
Correct Answer: The correct answer is option d, patient-Centered Clinical Research Network (PCORnet). We chose to pattern the first version of the PTN repository data points using the PCORnet data model. This choice was made after careful review of all existing data models, and alignment of their content with the requirement of our repository to help support drug and device trials in children. Despite PCORnet's strengths and significant advantages, the data model did not encompass all domains felt to be of relevance to pediatric drug and device development. Additional fields were therefore added, including vital signs, fluids, flowsheet assessments, and respiratory support.
-
Which of the following strategies were implemented in support of site-based teams responsible for data mapping?
Financial incentives.
Threat of study termination.
An implementation guide.
Written instructions only.
Correct Answer: The correct answer is option c, an implementation guide was one of the strategies employed to support site-based research teams. Other strategies included individual conference calls, and customized error and warning reports. An early decision during the development process was to employ site-based rather than centralized data mapping. This decision was made based on several perceived advantages given the overall constraints of the project, but immediately raised the question of how best to support sites during the mapping efforts. The PTN team ultimately decided on a combination of written instructions in the form of an implementation guide, combined with individual conference calls with technical support teams, customized error and warning reports, and iterative submission processes designed to incrementally submit data and correct errors. Overall, the individual communication with sites was essential to the success of this project, although it raises some concerns regarding scalability of these efforts.
Acknowledgments
The Best Pharmaceuticals for Children Act—Pediatric Trials Network Publication Committee: Gary Furda, Duke Clinical Research Institute, Durham, NC; Danny Benjamin, Duke Clinical Research Institute, Durham, NC; Edmund Capparelli, University of California San Diego, San Diego, CA; Gregory L. Kearns, Arkansas Children's Hospital Research Institute, Little Rock, AR; Ian M. Paul, Penn State College of Medicine, Hershey, PA; Christoph Hornik, Duke Clinical Research Institute, Durham, NC; Kelly Wade, Children's Hospital of Philadelphia, Philadelphia, PA.
The Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD): Perdita Taylor-Zapata.
The Pediatric Trials Network Repository Study Teams:
Children's Hospital of Philadelphia: Kevin J. Downes, Mark Ramos, Shawn O'Connor, and Robert W. Grundmeier.
Medical University of South Carolina: Andrew Atz, Leslie Lenert, John Clark, and Kalyan Chundru.
University of Minnesota: Brian A. Harvey and Sonya Grillo.
University of Michigan: Debbie S. Gipson, Samara Attala, Richard Eickstadt, Erin Kaleba, Don Liamini, Jamie Estill, Jeremy Jared, and Peter Bow.
The University of North Carolina at Chapel Hill: Jennifer Talbert, MS, BSN, RN, RDH, CCRP, and Cindy Clark, RN.
University of Louisville (Kosair Charities Pediatric Clinical Research Unit): Janice E. Sullivan, MD; and Norton Healthcare: Steve Heilman; K.P. Singh; Satish Vuyyuri; Jeff Schwitters; and Don Stone.
Loma Linda School of Medicine: Francis Chen and Stephanie Fan.
Funding Statement
Funding This work was funded under National Institute for Child Health and Human Development (NICHD) contract HHSN27520100–003I for the Pediatric Trials Network (PI: Danny Benjamin). C.P.H. receives support for research from NICHD grant K23HD090239. M.L. receives funding from FDA grant RO1 5R01FD005101–03.
Conflict of Interest K.D. has received research support from Merck, Inc. and Pfizer, Inc. M.L. has received research support from United Therapeutics. The remaining authors have nothing to disclose.
Protection of Human and Animal Subjects
This study was approved by institutional review boards of Duke University (coordinating center) and each participating site.
Supplementary Material
References
- 1.Park H A. President's Statement. Yearb Med Inform. 2017;26(01):1–2. doi: 10.15265/IY-2016-028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Issa N T, Byers S W, Dakshanamurthy S. Big data: the next frontier for innovation in therapeutics and healthcare. Expert Rev Clin Pharmacol. 2014;7(03):293–298. doi: 10.1586/17512433.2014.905201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Margolis R, Derr L, Dunn M et al. The National Institutes of Health's Big Data to Knowledge (BD2K) initiative: capitalizing on biomedical big data. J Am Med Inform Assoc. 2014;21(06):957–958. doi: 10.1136/amiajnl-2014-002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross M K, Wei W, Ohno-Machado L. “Big data” and the electronic health record. Yearb Med Inform. 2014;9:97–104. doi: 10.15265/IY-2014-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans R S. Electronic health records: then, now, and in the future. Yearb Med Inform. 2016 01:S48–S61. doi: 10.15265/IYS-2016-s006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weng C, Kahn M G. Clinical research informatics for big data and precision medicine. Yearb Med Inform. 2016;25(01):211–218. doi: 10.15265/IY-2016-019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott J H, Grimshaw J, Altman Ret al. Informatics: make sense of health data Nature 2015527(7576):31–32. [DOI] [PubMed] [Google Scholar]
- 8.Sutherland S M, Kaelber D C, Downing N L, Goel V V, Longhurst C A. Electronic health record-enabled research in children using the electronic health record for clinical discovery. Pediatr Clin North Am. 2016;63(02):251–268. doi: 10.1016/j.pcl.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Sward K A, Rubin S, Jenkins T L, Newth C J, Dean J M; Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Collaborative Pediatric Critical Care Research Network (CPCCRN).Case study: semantic annotation of a pediatric critical care research study Comput Inform Nurs 20163403101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, Henriksson A, Asker L, Boström H. Predictive modeling of structured electronic health records for adverse drug event detection. BMC Med Inform Decis Mak. 2015;15 04:S1. doi: 10.1186/1472-6947-15-S4-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silverstein J M, Roe E D, Munir K M et al. Use of electronic health records to characterize a rare disease in the U.S.: treatment, comorbidities, and follow-up trends among patients with a confirmed diagnosis of acromegaly. Endocr Pract. 2018;24(06):517–526. doi: 10.4158/EP-2017-0243. [DOI] [PubMed] [Google Scholar]
- 12.Liu F, Jagannatha A, Yu H. Towards drug safety surveillance and pharmacovigilance: current progress in detecting medication and adverse drug events from electronic health records. Drug Saf. 2019;42(01):95–97. doi: 10.1007/s40264-018-0766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imatoh T, Sai K, Takeyama M et al. Identification of risk factors and development of detection algorithm for denosumab-induced hypocalcaemia. J Clin Pharm Ther. 2019;44(01):62–68. doi: 10.1111/jcpt.12753. [DOI] [PubMed] [Google Scholar]
- 14.Laughon M M, Benjamin D K., Jr Mechanisms to provide safe and effective drugs for children. Pediatrics. 2014;134(02):e562–e563. doi: 10.1542/peds.2014-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.England A, Wade K, Smith P B, Berezny K, Laughon M; Best Pharmaceuticals for Children Act — Pediatric Trials Network Administrative Core Committee.Optimizing operational efficiencies in early phase trials: the Pediatric Trials Network experience Contemp Clin Trials 201647376–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laughon M M, Benjamin D K, Jr, Capparelli E V et al. Innovative clinical trial design for pediatric therapeutics. Expert Rev Clin Pharmacol. 2011;4(05):643–652. doi: 10.1586/ecp.11.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornik C P, Benjamin D K, Jr, Smith P B et al. Electronic health records and pharmacokinetic modeling to assess the relationship between ampicillin exposure and seizure risk in neonates. J Pediatr. 2016;178:125–1290. doi: 10.1016/j.jpeds.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornik C P, Chu P Y, Li J S, Clark R H, Smith P B, Hill K D. Comparative effectiveness of digoxin and propranolol for supraventricular tachycardia in infants. Pediatr Crit Care Med. 2014;15(09):839–845. doi: 10.1097/PCC.0000000000000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Testoni D, Hornik C P, Neely M L et al. Safety of octreotide in hospitalized infants. Early Hum Dev. 2015;91(07):387–392. doi: 10.1016/j.earlhumdev.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmerman K O, Hornik C P, Ku L et al. Sedatives and analgesics given to infants in neonatal intensive care units at the end of life. J Pediatr. 2015;167(02):299–304. doi: 10.1016/j.jpeds.2015.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Autmizguine J, Hornik C P, Benjamin D K, Jr et al. Anaerobic antimicrobial therapy after necrotizing enterocolitis in VLBW infants. Pediatrics. 2015;135(01):e117–e125. doi: 10.1542/peds.2014-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiks A G, Grundmeier R W, Steffes J et al. Comparative effectiveness research through a collaborative electronic reporting consortium. Pediatrics. 2015;136(01):e215–e224. doi: 10.1542/peds.2015-0673. [DOI] [PubMed] [Google Scholar]
- 23.FHIR R4. A Standard for Health Care Data Exchange. Available at:http://www.hl7.org/fhir/index.html. Accessed February 1, 2019
- 24.HL7 Standards-Section 3: Clinical and Administrative Domains. Available at:http://www.hl7.org/implement/standards/product_section.cfm?section=3. Accessed February 1, 2019
- 25.CDISC. Biomedical Research Integrated Domain Group (BRIDG) Model. Available at:https://www.cdisc.org/standards/domain-information-module/bridg. Accessed February 1, 2019
- 26.PCORnet Common Data Model (CDM) Specification.Version 2.0. Available at:http://pcornet.org/wp-content/uploads/2015/03/2015-02-27-PCORnet-Common-Data-Model-v2-0-RELEASE.pdf. Accessed February 1, 2019
- 27.U.S. National Library of Medicine. ClinicalTrials.gov. Available at:https://clinicaltrials.gov/ct2/resources/download. Accessed February 1, 2019
- 28.Selby J V, Lipstein S H. PCORI at 3 years--progress, lessons, and plans. N Engl J Med. 2014;370(07):592–595. doi: 10.1056/NEJMp1313061. [DOI] [PubMed] [Google Scholar]
- 29.Zhou X, Murugesan S, Bhullar H et al. An evaluation of the THIN database in the OMOP Common Data Model for active drug safety surveillance. Drug Saf. 2013;36(02):119–134. doi: 10.1007/s40264-012-0009-3. [DOI] [PubMed] [Google Scholar]
- 30.Xu Y, Zhou X, Suehs B T et al. A comparative assessment of Observational Medical Outcomes Partnership and Mini-Sentinel common data models and analytics: implications for active drug safety surveillance. Drug Saf. 2015;38(08):749–765. doi: 10.1007/s40264-015-0297-5. [DOI] [PubMed] [Google Scholar]
- 31.Stang P E, Ryan P B, Racoosin J A et al. Advancing the science for active surveillance: rationale and design for the Observational Medical Outcomes Partnership. Ann Intern Med. 2010;153(09):600–606. doi: 10.7326/0003-4819-153-9-201011020-00010. [DOI] [PubMed] [Google Scholar]
- 32.PCORnet. PCORnet Coordinating Center. Available at:https://pcornet.org/about-pcornet/pcornet-coordinating-center/. Accessed February 6, 2019
- 33.National Institute of Standards and Technology. Federal Information Security Management Act (FISMA) Implementation Project. Available at:https://www.nist.gov/programs-projects/federal-information-security-management-act-fisma-implementation-project. Accessed February 6, 2019
- 34.Antman E M, Benjamin E J, Harrington R A et al. Acquisition, analysis, and sharing of data in 2015 and beyond: a survey of the landscape: a conference report from the American Heart Association Data Summit 2015. J Am Heart Assoc. 2015;4(11):e002810. doi: 10.1161/JAHA.115.002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janke A T, Overbeek D L, Kocher K E, Levy P D. Exploring the potential of predictive analytics and big data in emergency care. Ann Emerg Med. 2016;67(02):227–236. doi: 10.1016/j.annemergmed.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 36.Khoury M J. Planning for the future of epidemiology in the era of big data and precision medicine. Am J Epidemiol. 2015;182(12):977–979. doi: 10.1093/aje/kwv228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J. Big data, health informatics, and the future of cardiovascular medicine. J Am Coll Cardiol. 2017;69(07):899–902. doi: 10.1016/j.jacc.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Sutherland S M, Goldstein S L, Bagshaw S M. Acute kidney injury and big data. Contrib Nephrol. 2018;193:55–67. doi: 10.1159/000484963. [DOI] [PubMed] [Google Scholar]
- 39.Deakyne Davies S J, Grundmeier R W, Campos D A et al. The Pediatric Emergency Care Applied Research Network Registry: a multicenter electronic health record registry of pediatric emergency care. Appl Clin Inform. 2018;9(02):366–376. doi: 10.1055/s-0038-1651496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deans K J, Sabihi S, Forrest C B. Learning health systems. Semin Pediatr Surg. 2018;27(06):375–378. doi: 10.1053/j.sempedsurg.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Forrest C B, Margolis P, Seid M, Colletti R B. PEDSnet: how a prototype pediatric learning health system is being expanded into a national network. Health Aff (Millwood) 2014;33(07):1171–1177. doi: 10.1377/hlthaff.2014.0127. [DOI] [PubMed] [Google Scholar]
- 42.Forrest C B, Margolis P A, Bailey L C et al. PEDSnet: a national pediatric learning health system. J Am Med Inform Assoc. 2014;21(04):602–606. doi: 10.1136/amiajnl-2014-002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khare R, Utidjian L, Ruth B J et al. A longitudinal analysis of data quality in a large pediatric data research network. J Am Med Inform Assoc. 2017;24(06):1072–1079. doi: 10.1093/jamia/ocx033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hemingway H, Asselbergs F W, Danesh J et al. Big data from electronic health records for early and late translational cardiovascular research: challenges and potential. Eur Heart J. 2018;39(16):1481–1495. doi: 10.1093/eurheartj/ehx487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weng C. Optimizing clinical research participant selection with informatics. Trends Pharmacol Sci. 2015;36(11):706–709. doi: 10.1016/j.tips.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Moor G, Sundgren M, Kalra D et al. Using electronic health records for clinical research: the case of the EHR4CR project. J Biomed Inform. 2015;53:162–173. doi: 10.1016/j.jbi.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Hernandez A F, Fleurence R L, Rothman R L. The ADAPTABLE Trial and PCORnet: shining light on a new research paradigm. Ann Intern Med. 2015;163(08):635–636. doi: 10.7326/M15-1460. [DOI] [PubMed] [Google Scholar]
- 48.Hsieh E M, Hornik C P, Clark R H, Laughon M M, Benjamin D K, Jr, Smith P B; Best Pharmaceuticals for Children Act—Pediatric Trials Network.Medication use in the neonatal intensive care unit Am J Perinatol 20143109811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trembath A, Hornik C P, Clark R, Smith P B, Daniels J, Laughon M; Best Pharmaceuticals for Children Act—Pediatric Trials Network.Comparative effectiveness of surfactant preparations in premature infants J Pediatr 201316304955–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ericson J E, Gostelow M, Autmizguine J et al. Safety of high-dose acyclovir in infants with suspected and confirmed neonatal herpes simplex virus infections. Pediatr Infect Dis J. 2017;36(04):369–373. doi: 10.1097/INF.0000000000001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hornik C P, Herring A H, Benjamin D K, Jr et al. Adverse events associated with meropenem versus imipenem/cilastatin therapy in a large retrospective cohort of hospitalized infants. Pediatr Infect Dis J. 2013;32(07):748–753. doi: 10.1097/INF.0b013e31828be70b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Autmizguine J, Benjamin D K, Jr, Smith P B et al. Pharmacokinetic studies in infants using minimal-risk study designs. Curr Clin Pharmacol. 2014;9(04):350–358. doi: 10.2174/1574884709666140520153308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salerno S, Hornik C P, Cohen-Wolkowiez M et al. Use of population pharmacokinetics and electronic health records to assess piperacillin-tazobactam safety in infants. Pediatr Infect Dis J. 2017;36(09):855–859. doi: 10.1097/INF.0000000000001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hornik C P, Onufrak N J, Smith P B et al. Association between oral sildenafil dosing, predicted exposure, and systemic hypotension in hospitalised infants. Cardiol Young. 2018;28(01):85–92. doi: 10.1017/S1047951117001639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ku L C, Wu H, Greenberg R G et al. Use of therapeutic drug monitoring, electronic health record data, and pharmacokinetic modeling to determine the therapeutic index of phenytoin and lamotrigine. Ther Drug Monit. 2016;38(06):728–737. doi: 10.1097/FTD.0000000000000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ge S, Beechinor R J, Hornik C P et al. External evaluation of a gentamicin infant population pharmacokinetic model using data from a national electronic health record database. Antimicrob Agents Chemother. 2018;62(09):e00669–e18. doi: 10.1128/AAC.00669-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gliklich R, Dreyer N, Leavy M, Velentgas P, Khurana L.Standards in the conduct of registry studies for patient-centered outcomes research. Patient-Centered Outcomes Research Institute;March 15, 2012. Available at:https://www.pcori.org/assets/Standards-in-the-Conduct-of-Registry-Studies-for-Patient-Centered-Outcomes-Research.pdf. Accessed June 6, 2018
- 58.Leavy M B.Multinational registries: challenges and opportunities. Addendum to Registries for Evaluating Patient Outcomes: A User's Guide.3rd ed. Rockville, MD: Agency for Healthcare Research and Quality;2018 [PubMed] [Google Scholar]
- 59.Mack C, Su Z, Westreich D.Managing missing data in patient registries. Addendum to Registries for Evaluating Patient Outcomes: A User's Guide.3rd ed. Rockville, MD: Agency for Healthcare Research and Quality;2018 [PubMed] [Google Scholar]
- 60.Velupillai S, Mowery D, South B R, Kvist M, Dalianis H. Recent advances in clinical natural language processing in support of semantic analysis. Yearb Med Inform. 2015;10(01):183–193. doi: 10.15265/IY-2015-009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


