Abstract
Antimicrobial peptides (AMPs) carry great potential as new antibiotics against “superbugs.” Dermcidin (DCD), a broad-spectrum AMP in human sweat, has been recently crystallized in its oligomeric state and showed channel-like properties. In this work, we performed multiscale molecular dynamics simulations to study how the membrane composition influences the behavior of a transmembrane pore formed by the DCD oligomer in the hope of revealing the origin of the membrane selectivity of this AMP toward bacteria. Our results indicate that bilayers composed of various lipids (DMPC, DPPC, and DSPC) with different thicknesses result in different orientations of the DCD oligomer when embedded in lipid bilayers. The thicker the bilayer, the less tilted the channel. Cholesterol makes the bilayers more rigid and thicker, which also affects the orientation of the channel. Furthermore, we observed that the predicted conductance of the channel from computational electrophysiology simulations is related to its orientation in the lipid bilayer: the larger the tilt, the larger the conductance. Our results indicate that the membrane composition has a significant influence on the activity of the DCD channel, with thicker, cholesterol-rich membranes showing lower conductance than that of thinner membranes.
Introduction
Because of the overuse and misuse of existing antibiotics, as well as the slow development of new antibiotics, many pathogenic species of bacteria are developing resistance to existing antibiotics, which has become one of the world’s most pressing public health concerns (1). Antimicrobial peptides (AMPs), which widely exist in various forms of lives from plant to mammals, exhibit great potential as a new generation of antibiotics. Many kinds of AMPs kill bacteria by interacting with their membranes rather than with specific proteins and thereby make it harder for bacteria to develop resistance (2, 3). Therefore, it is of great interest to understand how AMPs interact with membranes, with a long-term goal of understanding the detailed mechanisms of the action of AMPs enabling development of potential new antibiotics.
The various types of AMPs differ in their sequence, length, structure, net charge, and other properties (4). It has been proposed that some AMPs function as metabolic inhibitors, whereas others can increase the permeability of membranes by various mechanisms (3). Three permeation models have dominated the discussion of AMPs: the barrel-stave, toroidal, and carpet models (3). It is likely that the detailed pore-forming mechanism varies between different AMPs. A number of computational studies have attempted to address possible mechanisms of pore formation and/or interaction with membranes (5, 6, 7, 8, 9, 10, 11, 12, 13). Such studies can reveal aspects of the underlying mode(s) of action of AMPs and thereby provide a guide for design of novel antibiotics (2, 14, 15, 16, 17).
Among recently discovered AMPs, dermcidin (DCD) was initially discovered in human sweat in 2001 and found to have broad-spectrum antimicrobial and antifungal activities under a wide range of conditions (18). Significantly, deficiency of DCD in the sweat of patients with atopic dermatitis correlates with an impaired resistance to infection of human skin (19). There have been several studies performed to investigate its functional mechanism (19, 20, 21, 22, 23, 24, 25). DCD is a 47-residue peptide, the monomeric structure of which was determined by NMR and shown to be a kinked α-helix (26). The DCD monomer is negatively charged, which is unusual for an AMP, many of which are cationic (3, 27) to facilitate peptide/lipid interactions. Based on structure-activity studies of DCD-1L, it was proposed to form oligomeric complexes, stabilized by Zn2+ ions, which form ion-permeable pores (i.e., channels in the bacterial membrane) (18, 28).
A crystal structure of a channel-like helix-bundle oligomer of DCD in combination with molecular dynamics (MD) simulations and ion channel current measurements was used to propose a detailed mechanism for the action of this unusual AMP (28). It was suggested that pore formation by DCD involves the divalent ion Zn2+ such that the oligomeric complex is neutral. This oligomeric structure can form a stable pore when embedded in a lipid bilayer in accord with the barrel-stave model, and the pore shows high water and ion permeability. This pore-forming mechanism may therefore explain how DCD kills bacteria (28).
To both understand and potentially engineer selectivity of AMPs, it is important to understand how membrane lipid composition (which differs profoundly between bacterial and mammalian cells) influences the behavior of DCD. In this study, we performed multiscale MD simulations to study the effect of the membrane lipid composition on the orientation and predicted conduction properties of DCD when embedded in various lipid bilayers. The orientation of DCD relative to the bilayer is shown to be correlated with the lipid bilayer thickness: the thicker the bilayer, the less the tilt of the channel assembly relative to the bilayer plane. The conductance of the channel exhibits a correlation with the tilt of DCD such that more tilted channels exhibit a larger conductance. This suggests that the biological activity of the DCD pore will be strongly influenced by the nature of the membrane within which the DCD oligomer is embedded.
Methods
Selection of the simulation systems
We took the crystal structure of the hexameric DCD pore (Protein Data Bank [PDB]: 2YMK) as the initial structure for our MD simulations. To study the effect of membrane thickness and composition on the DCD activity, we selected three kinds of lipids with different lengths, namely DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine), DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine), and DSPC (1,2-distearoyl-sn-glycero-3-phosphocholine), which have 14, 16, and 18 carbon atoms in the fatty acid chains, respectively. Moreover, we added cholesterol into some of the simulation systems to study its influence for a given lipid type. Therefore, we studied the following systems: DCD in DMPC, DCD in DPPC, and DCD in DSPC bilayers, with 0, 20, and 40% cholesterol in the each of the respective lipid bilayers.
CG simulations
To investigate the orientation of the DCD oligomer on various lipid bilayers, we performed coarse-grained (CG) MD simulations (29) to study the self-assembly processes of the DCD oligomer into lipid bilayers. Initially, the DCD oligomer was positioned along the z axis or perpendicular to the z axis and was surrounded by randomly positioned lipids and water molecules (and cholesterol as well for some cases) in a simulation box of 120 × 120 × 80 Å3, which was sandwiched into two extrawater layers with the thickness of 20 Å along the z axis. As a consequence, the initial simulation system was a box of 120 × 120 × 120 Å 3 as shown in Fig. 1, A and B. In all the CG simulations, we included the six Zn2+ ions within the DCD hexamer because these have been shown to be important for the stability of the oligomeric structure (28). There are not well-established Zn2+ parameters in the Martini force field, so in the CG, we used generic divalent cation parameters represented by Ca2+. We consider this strategy to be reasonable because an elastic network was applied to the DCD-Zn2+ complex as a whole, thus ensuring its conformational stability.
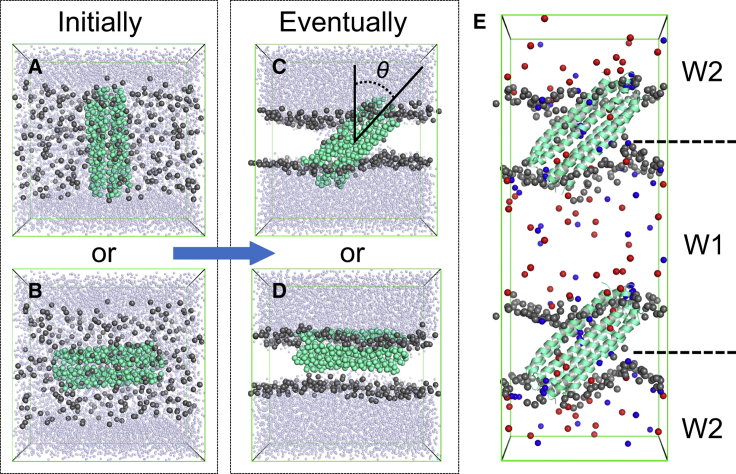
Figure 1.
Coarse-grained (CG) self-assembly simulations of DCD/lipid bilayer systems and the atomistic computational electrophysiology (CE) simulation system. (A) and (B) show two initial systems for the CG simulations, with the helix bundle (green-cyan) perpendicular or parallel, respectively, to the plane of the initial slab containing lipid molecules (dark gray). (C) and (D) show two possible resultant orientations of the DCD oligomer relative to the self-assembled bilayer (lipid phosphate particles only are shown in dark gray) at the end of the 0.5-μs CG simulations. The tilt angle is indicated by θ in (C). The small transparent light-blue spheres are water particles. (E) The atomistic CE simulation system is shown in which the green-cyan cartoons represent the DCD oligomer, the gray spheres are the phosphorus atoms of the lipid (DMPC) headgroups, and the red and blue spheres are Cl− and Na+ ions, respectively. W1 and W2 represent the two aqueous compartments between which a voltage difference of ∼250–450 mV was maintained in the CE simulations. To see this figure in color, go online.
The system was then heated up to 345 K, with all the molecules in the simulations box being allowed to move freely. Within 200 ns, the system was able to reach a well-equilibrated condition in which the DCD oligomer self-inserted into a well-shaped lipid bilayer that was sandwiched by extra- and intrawater layers representing the extra- and intracellular environment (Fig. 1 C), or in rare cases, the oligomer was found to be partly embedded into the lipid bilayer and partly exposed to water, with the pore axis parallel to the bilayer surface as shown in Fig. 1 D. The temperature of 345 K was chosen to make sure all the lipids would stay in the fluid phase.
For the CG simulations, we used GROMACS 4.6 (30) to perform the MD simulations with the Martini force field (29, 31). A time step of 20 fs was adopted. We used the Berendsen barostat (32) and the v-rescale thermostat (33), with a time constant of 1.0 ps for both. The short-range van der Waals and electrostatic interactions were calculated with a cutoff of 11 Å. The electrostatic interactions were calculated with the “reaction field” method. The dielectric constant was set to be 15. More details about the self-assembly CG simulations can also be found in a recently established automated method (34).
Atomistic CE simulations
Atomistic computational electrophysiology (CE) simulation is a recently developed method, which is highly suitable for the study of ion conduction properties of channels or pores (35, 36). In such a simulation, we set up a double-layer system in which the simulation box is divided into three segments by two lipid bilayers. Effectively, there are only two water compartments (named W1 and W2) in the simulation system because the top and lower water sections are actually treated as continuous with one another in the simulations because of the existence of periodic boundary conditions, as shown in Fig. 1 E. Different numbers of ions can be put into the two water compartments, which can generate a transmembrane ion gradient and transmembrane potential because of the imbalance of the ion distribution. By adjusting the ion number difference in the two water compartments, we can apply a desired transmembrane potential to the simulation system.
The DCD hexamer and the six Zn2+ ions within the channel were extracted from PDB:2YMK to build the all-atom simulation system. The atomistic MD simulations were performed with GROMACS 4.6 (30) and the CHARMM36 force field (37). Using g_membed (38), we first embedded the DCD oligomer into a lipid bilayer built with CHARMM-GUI (39) and further solvated the system by water molecules and a certain number of ions to have an ion (NaCl) concentration of 0.15 M. The resulting system is overall neutral. We adopted a time step of 2 fs. The semi-isotropic NPT barostat was applied with the Parrinello-Rahman method at 1 bar with a coupling time constant of 1.0 ps (40, 41), and the thermostat was applied with the Nose-Hoover method at 345 K with a coupling time constant of 0.5 ps (42, 43). The short-range electrostatic interaction cutoff was set to be 14 Å, and the particle mesh Ewald method was utilized for calculating long-range coulomb interactions (44, 45). The van der Waals interactions were cut off at 12 Å and turned off smoothly from 8 to 12 Å with the “shift” method.
The single-layer system was firstly equilibrated for 500 ns until the channel found its most stable orientation in the lipid bilayer. The resulting system has a size of around 80 × 80 × 110 Å3. The single-layer system was then duplicated along the z direction. We set the number of Cl− to be 45 and 39 and the number of Na+ to be 39 and 45 in W1 and W2, respectively, which leads to a transmembrane potential of ∼300–450 mV. Then, we ran multiple 200-ns CE simulations for each system. From these production CE simulations, we were able to observe continuous ion permeation along the DCD oligomer. In the end, we calculated the average transmembrane potential and the current during the simulation time (divided into 200-ns segments) and thereby obtained the conductance of the channel. We performed 200-ns atomistic CE simulations for DCD in DMPC, DPPC, DSPC, and DMPC + cholesterol bilayers. All the simulations are summarized in Table S4.
Calculation of the channel conductance
After finishing the CE simulations, we analyzed the transmembrane potential, which was generated from the charge imbalance Δq between the two water compartments. Only the charge distribution perpendicular to the membrane is relevant for calculating the potential drop ΔU, so we integrated twice over the charge distribution with intervals along the z direction (the bilayer normal direction) to obtain the transmembrane potential by using the implementation of the Poisson equation in the GROMACS g potential tool. We then counted the number of ions that permeated through the channel in the 200-ns MD trajectories with which we calculated the current with the equation: , where was the charge that passed through the channel and t was the simulation time. With ΔU and I known, the conductance was simply calculated with the equation: C = I/ΔU. The above calculation was performed for each 200-ns CE simulation.
Results
The orientation of the DCD oligomer is membrane dependent
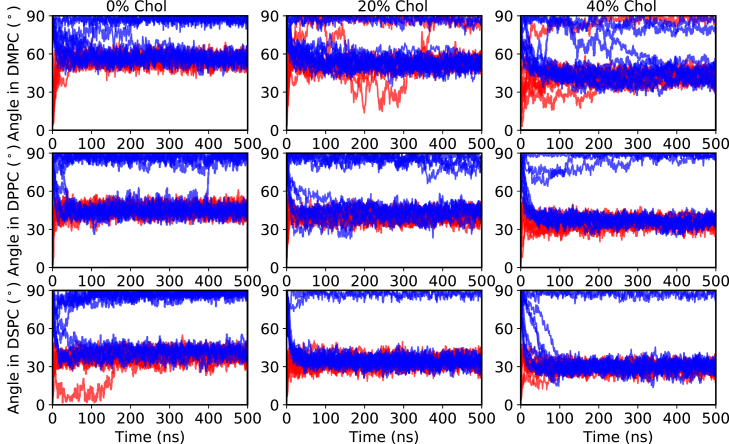
In the CG MD simulations, the lipids (and cholesterol) self-assembled into a bilayer around the DCD oligomer. Most of the self-assembly processes were completed within 200 ns, as indicated by a well-defined lipid bilayer forming around the DCD oligomer (Fig. 1, C and D) and the tilt angle reaching a stable value (Fig. 2). If the DCD oligomer was initially parallel to the z axis (red plot, initial tilt angle 0°), its final orientation was mostly embedded and tilted in the self-assembled lipid bilayer. The exceptions were 2 out of 10 simulations with 80%DMPC:20%Chol (cholesterol), and 1 out of 10 simulations with 60%DMPC:40%Chol, where the DCD was only partially embedded into the bilayer and its axis was parallel to the bilayer surface. If the DCD oligomer was initially normal to the z axis (blue plot, initial tilt angle 90°), the final orientation can be either embedded and tilted in the lipid bilayer or partially embedded into the bilayer and parallel to the bilayer surface.
Figure 2.
DCD helix bundle tilt angle evolution CG simulations in the presence of DMPC (upper row), DPPC (middle row), or DSPC (lower row) lipids, with 0, 20, and 40% cholesterol. The red lines correspond to simulations with the DCD oligomer axis initially parallel to the z axis (see Fig. 1A), whereas the blue lines correspond to simulations with the DCD oligomer initially perpendicular to the z axis (see Fig. 1B). For each case, 10 independent simulations of 0.5-μs duration were performed. To see this figure in color, go online.
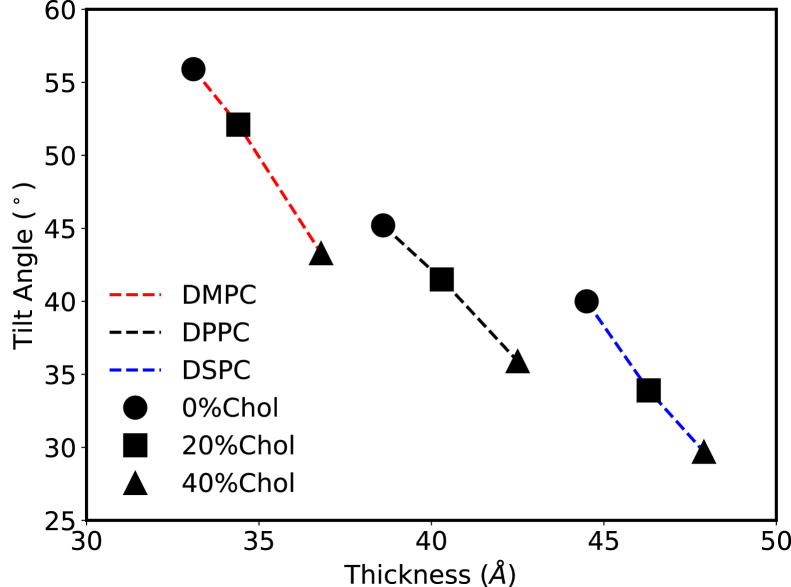
In all of the above simulations, the embedded and tilted orientation was preferred, and the tilt angle was system dependent. We analyzed the tilt angle distribution for all the CG simulations, disregarding the first 200-ns assembly trajectories. We observe two possible orientations, as shown in Figs. 2 and S1. The preferred orientation is the DCD oligomer embedded and tilted in the bilayer, and the tilt angle depends on the composition of the bilayer. We found that the thinner the bilayer, the larger the tilt (Fig. 3). For different types of lipids, the lipids with longer tails (length: DSPC > DPPC > DMPC) form a thicker bilayer, which lead to a less tilted orientation of the DCD oligomer in the bilayer (tilt angle: DSPC < DPPC < DMPC). For the same type of lipids, adding cholesterol results in thicker bilayers, which subsequently leads to a less tilted orientation of the DCD oligomer in the bilayer. The other orientation was the DCD channel axis parallel to the bilayer surface and only partially embedded into the lipid bilayer (Fig. 1 D). Because this orientation did not eventually lead to a pore formation in the bilayers, we think it is not the functional orientation of the DCD oligomer. Therefore, we focused on the study of the tilted orientation.
Figure 3.
Mean tilt angles of the DCD helix bundle relative to the bilayer normal in lipid bilayers with different thicknesses. The red, black, and blue dashed lines represent the simulation results with DMPC, DPPC, and DSPC lipid bilayers, respectively. The circle, square, and triangle symbols represent the simulations with 0, 20, and 40% cholesterol, respectively. The standard errors of the means of the tilt angles and membrane thicknesses for each case are smaller than or comparable to the size of the symbols, so they are not shown here for clarity. Please refer to the Supporting Materials and Methods for more detailed data. To see this figure in color, go online.
The orientation of the DCD oligomer from atomistic MD simulations
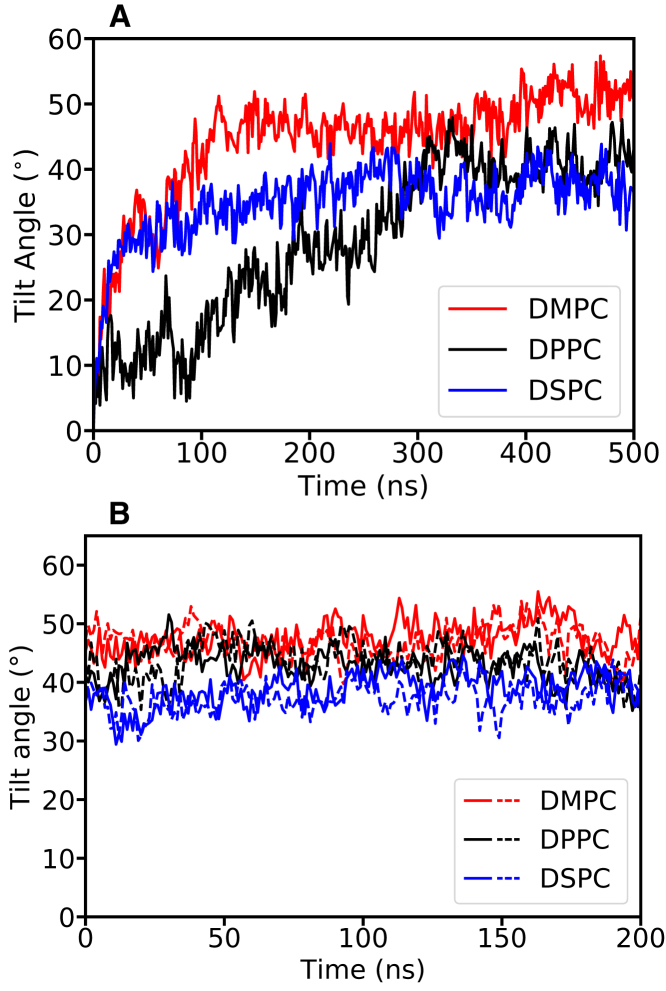
Our atomistic simulations show a similar trend to the CG MD simulations. We examined the tilt angle evolution of the DCD oligomer in three different types of lipid bilayers: DMPC, DPPC, and DSPC. Initially, the DCD oligomer was embedded into the bilayers with the g_membed method (38), and the channel axis was parallel to the z axis (tilt angle 0°). During the equilibration period, the channel spontaneously and gradually tilted in all of the three lipid bilayers (Fig. 4 A). We also examined the tilt angle evolution in the double-layer CE simulations. Because the initial configurations of the double-layer systems were taken from the equilibrated single-layer systems, we found the tilt angles were stable throughout the 200-ns simulations (Fig. 4 B), indicating that the single-layer systems were indeed well equilibrated. All these atomistic MD results show that the tilt angle is dependent on the lipid type: the thicker the bilayer (thickness: DSPC > DPPC > DMPC), the smaller the tilt (tilt angle: DSPC < DPPC < DMPC), as shown in Fig. 4. The tilt angles converged in less than 350 ns for all of the three simulations. The final stable tilt angles are around 51, 41, and 38° for DMPC, DPPC, and DSPC, respectively, which are close to the results obtained in CG MD simulations. Please refer to Tables S1 and S3 and Fig. S1 for a more detailed analysis.
Figure 4.
Tilt angle evolution of the DCD oligomer in DMPC (red), DPPC (black), and DSPC (blue) bilayers as observed in the atomistic simulations. (A) The tilt angle evolution in the single-layer simulations is shown, with the initial channel axis normal to the bilayer surface and (B) the tilt angle evolution in the double-layer simulations in which the initial configurations were taken from the single-layer simulations at 200 ns for DMPC and DSPC and at 400 ns for DPPC, as shown in (A). The solid and dashed lines in (B) represent the two channels in the two bilayers of the double-patch CE systems, respectively. To see this figure in color, go online.
The ion conduction through the DCD oligomer is correlated with its orientation
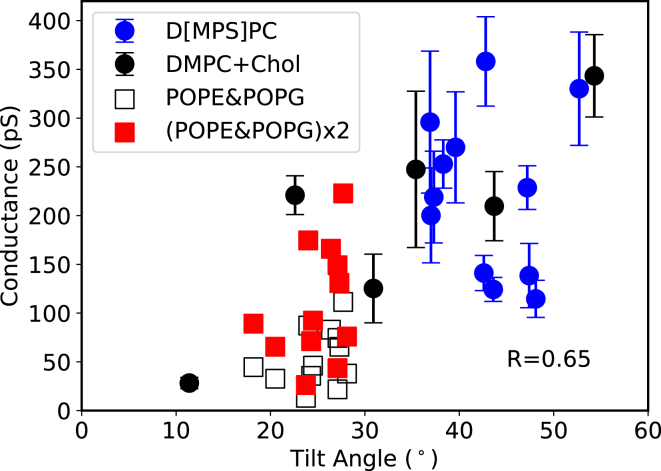
In our atomistic CE simulations, we counted how many ions permeated through the DCD channel within the simulation time. Together with the average transmembrane potential across the membrane, this allowed us to calculate the predicted conductance of the channel. Also, from the simulation trajectory, we obtained the stable tilt angle of the DCD oligomer in the lipid bilayer. When the conductance values and the tilt angles are plotted in Fig. 5, although there are local variations resulting in a relatively weak correlation coefficient of 0.65, we can see a clear overall trend between the conductance and the tilt angle of the DCD oligomer: the larger the tilt angle, the larger the conductance.
Figure 5.
Correlation between the average single channel conductance as estimated by CE simulations and the corresponding tilt angle of the DCD oligomer. Each scatter was obtained from a 200-ns atomistic molecular dynamics (MD) simulation under a transmembrane potential of ∼250–450 mV as described in the method section. The blue circles were derived from the CE simulations with DMPC, DPPC, or DSPC and denoted as D[MPS]C. The black circles were obtained from the CE simulations with DMPC and cholesterols (DMPC/Chol = 8:2). The red plots were derived from our previous simulations of DCD in a bilayer composed of POPE/POPG (3:1) at 310 K (28). Because previous studies showed that the conductance of ion channels at 345 K is less than twice of that at 310 K (47, 48), we simply doubled the conductance of DCD in POPE and POPG at 310 K (empty squares) to roughly estimate the conductance at 345 K (red squares). The Pearson correlation coefficient was 0.65 based on the calculations for the results at 345 K. The error bars represent the standard errors of means. For the detailed data, including the error estimation, please refer to Table S5. To see this figure in color, go online.
It is also notable that the addition of cholesterol has a significant effect on the DCD tilt angle (black scatters in Fig. 3) as well as on the conductance (Fig. 5). From our simulations, the addition of cholesterol makes the lipid bilayer stiffer and thicker, which in turn leads to a smaller tilt angle and a smaller conductance.
Ion permeation path and selectivity
The majority of the permeating ions are anions. From our analysis, we found that ∼90% of the permeating ions were Cl−, indicating the DCD oligomer is anion selective. This is consistent with our previous simulation results in POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine)/POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol) bilayers in which ∼88% permeated ions were anions. Although the DCD peptide is overall negatively charged, there are six Zn2+ ions forming a complex with the peptides upon oligomerization, which makes the DCD-Zn2+ complex overall neutral. Moreover, the Zn2+ ions are located near both the end and side openings of the channel, which may facilitate anion permeation. As shown in Fig. S2, the density of Cl− is significantly larger than that of Na+ at the side entrance of the channel, as well as throughout the interior of the channel. The major permeation pathway was from side openings rather than channel end openings, resulting a zigzag permeation path as we illustrated before (28).
The influence of the DCD oligomer on the surrounding lipid bilayer
The insertion and tilt of the DCD oligomer strongly influences the lipid bilayer properties around it. Unlike most of the membrane proteins, which have a symmetric configuration around the z axis, the tilted DCD oligomer introduces a unique distortion on the lipid bilayer.
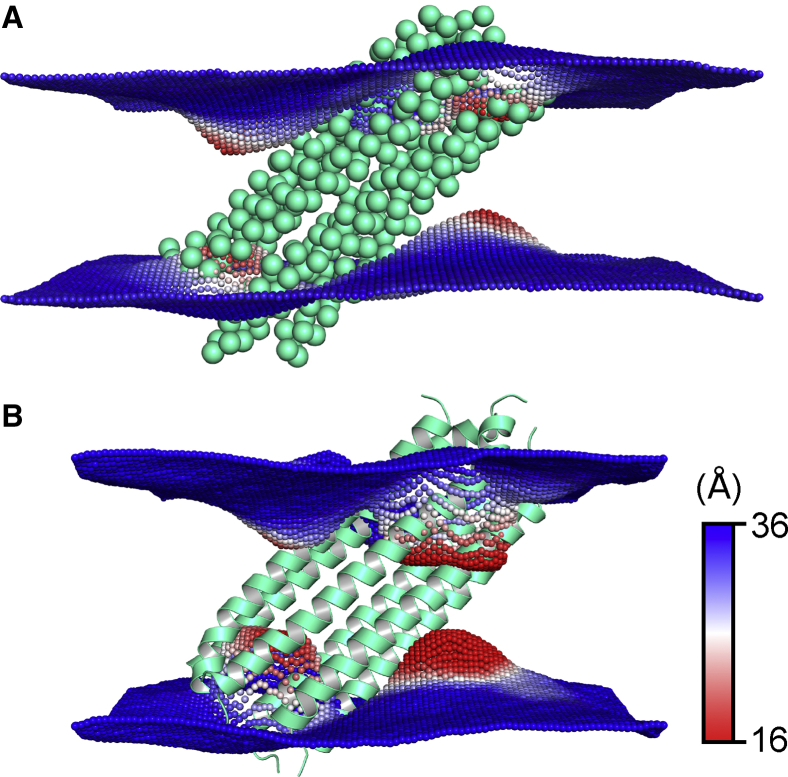
As shown in Fig. 6, the tilted DCD oligomer introduces local bending and thinning of the lipid bilayer around it, both in atomistic and CG MD simulations. In the simulation trajectories, we noticed that some lipid headgroups around the DCD oligomer were able to move near the center of the lipid bilayer along the hydrophilic gap on the DCD oligomer outer surface, which effectively thinned the bilayer nearby. It is also notable that the bilayer distortion is not completely identical in atomistic and CG simulations. In fact, the distortion is more pronounced in the atomistic simulations than in CG simulations, which is understandable because the CG models have a smoother surface than that of atomistic models.
Figure 6.
The local DMPC bilayer thickness around the DCD helix bundle from (A) CG to (B) atomistic MD simulations. The green-cyan spheres/cartoons represent the DCD oligomer. The blue, white, and red spheres represent the average local bilayer thickness averaged across a trajectory. The bilayer thickness was calculated by the distance between the phosphate particles (or phosphorus atoms) of the two leaflets with our in-house analysis tool. To see this figure in color, go online.
Discussion
It is important to question what causes the selective activity of AMPs against bacteria rather than against host cells. Answering this question can not only reveal the detailed function mechanism of AMPs but also help to develop novel selective antibiotics. Because many AMPs execute their function by interacting with membranes, it is a reasonable hypothesis that the composition of membranes may influence the activity of AMPs because it has been shown that the membranes of bacteria and human cells have distinct compositions (4, 46). As a first step toward understanding this question, we studied how different types of lipids affect the orientation and conductance of the DCD oligomer when the DCD oligomer is embedded in a lipid bilayer.
Our results confirm that the DCD oligomer indeed prefers to be embedded into the lipid bilayer, which is expected because about two thirds of the outer surface of the DCD oligomer is hydrophobic. When there is a large exposure of the hydrophobic surface to water (i.e., hydrophobic mismatch), the relative free energy of the system will be higher. By being fully embedded in the lipid bilayer, the hydrophobic mismatch is minimized and the system reaches a more stable state. In such a scenario, changing the lipid tail length, which in turn changes the bilayer thickness, changes the optimal orientation of the DCD oligomer while embedded in the bilayer. As shown in Fig. 3, the stable tilt angle shows a strong correlation with the bilayer thickness in our CG MD simulations: the thicker the bilayer, the less the tilt because the DCD oligomer can tilt to a smaller extent to minimize the hydrophobic mismatch in a thicker bilayer. We found the same trend in the atomistic MD simulations (Fig. 4; Table S3); although, perhaps not surprisingly, the tilt angle values do not exactly match those in CG simulations, given the approximations intrinsic in the CG representation. Therefore, the overall trend is the same in both atomistic and CG models: thicker bilayers lead to a less tilted orientation of the DCD oligomer. Also, it was shown that the addition of cholesterol leads to a more rigid and thicker bilayer and therefore a less tilted orientation of the DCD oligomer in our CG MD simulations. This may lead to a natural hypothesis that the cholesterol may play a role in the selectivity of the AMPs activity because we already know that cholesterol does not exist in bacteria membranes but exists in human cells.
It was interesting that an alternative stable orientation of the DCD oligomer was observed in the CG MD simulations, which was not seen in our atomistic simulations. This does not mean there is a discrepancy between the two methods. But rather, it reflects the fact that the CG simulations are able to provide more complex sampling of protein/membrane interactions. CG self-assembly simulations have been proven to be very reliable at studying the orientation of membrane proteins in lipid bilayer because of its high efficiency of sampling, and a web server has been built recently to show what the most likely orientations of integral membrane proteins in lipid bilayers are (http://memprotmd.bioch.ox.ac.uk/) (34). Because of this sampling efficiency, CG MD was able to find the alternative orientation of the DCD oligomer: it lies on the bilayer surface, is partially embedded into the bilayer, and exposes the hydrophilic gap on the outer surface to the water solution. This way, the oligomer can also reduce the hydrophobic mismatch and find a stable position. We transformed the CG model with this alternative orientation into an atomistic system and ran atomistic MD simulations. The results showed that this alternative orientation is indeed also stable in the atomistic system (Fig. S4). However, this orientation does not fully penetrate the bilayer; therefore, we think it is not a functional pore. We note that the two orientations can interchange within the simulation timescale in our CG MD simulations (Fig. 2), meaning the free-energy barrier between the two orientations can be overcome within a reasonable time. The exchange of the two orientations is rare in our CG simulations however, which is the reason why we did not see the other orientation in our atomistic simulation in which the DCD oligomer was initially embedded in the preformed bilayer.
We have previously observed that the DCD oligomer can act as an ion-permeable pore when embedded in a lipid bilayer, which is likely its mechanism of action (28). Herein, we examined the conductivity of the DCD oligomer by CE simulations in various lipid bilayers in which the peptide adopts different tilt angles. In these simulations, we observed an overall correlation between the predicted conductance and the tilt angle of the DCD channel in various bilayers from a total of ∼3-μs all-atom CE simulations. We found that the larger the tilt angle, the larger the conductance, as shown in Fig. 5. From our atomistic simulation results, one thing notable is that the addition of cholesterol results in a very broad and scattered distribution of tilt angles and conductance values. The wide distribution of the tilt angle is a bit different from what we see in the CG MD simulations in Fig. S1. We believe this is caused by the fact that the free-energy landscape is rougher in atomistic simulations; therefore, there are many more metastable orientations when cholesterol is added. On the one hand, this requires a much longer time to find the most stable and optimal orientation; on the other hand, this gives us an opportunity to study the correlation between the conductance and the tilt angle for a given membrane composition because we have enough time to measure the conductance for a metastable orientation before it spontaneously evolves to the next state and no external restraint is required. Interestingly, the six simulations of the DCD oligomer in the DMPC and cholesterol mixture bilayer showed the same trend as well: the larger the tilt angle, the larger the conductance (Fig. 5, black circles). Taken together, we found an overall correlation between the conductance and the tilt of the channel with a correlation coefficient of 0.65.
We also note that we do not expect this trend to hold at very large tilt angles. In our simulations, we have only seen tilt angles from around 20 to 55°. When embedded in a lipid bilayer, the DCD oligomer will probably take a tilt angle within this range, and we expect the above trend to hold. If the tilt angle further increases, for example, in the transition to the membrane surface-parallel orientation, the trend may be changed.
The embedding of the DCD oligomer in lipid bilayers strongly influences the bilayers. Unlike most of the membrane proteins that are sitting in the membranes with the structural axis perpendicular to the membrane surface and has a symmetry effect on the membrane around this axis, the tilted orientation does not introduce a symmetric effect around its axis to the lipid bilayers. As shown in Fig. 6, the influence on the lipid bilayer is not symmetric around the z axis. Instead, the distorted bilayer has an inversion center. Both CG and atomistic simulations showed the same feature. The distortion mainly involves a bending and thinning of the membrane around the interface between the DCD oligomer and the bilayers. This is caused by the inward moving of the lipid headgroups toward the bilayer center along the hydrophilic gap on the outer surface of the DCD oligomer. This inward motion of the lipid headgroups also exposes the side opening of the DCD oligomer to the water solution, which can facilitate the ion permeation. Therefore, this unique distortion of the lipid bilayer clears the major ion permeation pathway and makes the ion permeation more efficient.
From our results, we conclude that the composition of the membrane has a significant impact on the orientation and activity of the DCD oligomer. Basically, the thicker the bilayer, the smaller the tilt; and the smaller the tilt, the smaller the conductance. The addition of cholesterol makes the membranes thicker and leads to a lower conductance. However, we should keep in mind that cell membranes are much more complex than the simplified models we used for our study, and we have not studied the oligomerization and insertion of the DCD in membranes, so we cannot conclude the selectivity mechanism of the DCD against bacteria from these data alone. Rather, our work revealed the relation of the membrane thickness and the orientation and conductance of the DCD oligomer after embedded into the membrane. Because the conductance of the DCD oligomer is highly related to its activity, we think the activity of DCD is closely related to the membrane composition after forming pores in the membrane. To reveal the full selectivity mechanism, more realistic membrane models and the pore formation processes should be studied in the future.
Author Contributions
C.S. designed and performed the research. B.L.d.G. and M.S.P.S. supervised the research. C.S., B.L.d.G., and M.S.P.S. wrote the manuscript.
Acknowledgments
We thank Dr. Phillip J. Stansfeld for advice with CG simulations and Dr. Matthieu Chavent for advice on visualization. Part of the molecular dynamics simulation was performed on the Computing Platform of the Center for Life Sciences at Peking University.
The research was supported by a Marie Curie Intra European Fellowship within the 7th European Community Framework Programme (C.S. and M.S.P.S.). Research in M.S.P.S.’s group is supported by the Biotechnology and Biological Sciences Research Council, Engineering and Physical Sciences Research Council, Leverhulme Trust, and Wellcome. C.S. is supported by grants from the Ministry of Science and Technology of China (National Key Research & Development Program of China, 2016YFA0500401), National Natural Science Foundation of China (grant no. 21873006), and the Young Thousand Talents Program of China. B.L.d.G. acknowledges funding from the German Research Foundation Deutsche Forschungsgemeinschaft via SFB803 (project A03).
Editor: Alan Grossfield.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.03.033.
Supporting Material
References
- 1.Ventola C.L. The antibiotic resistance crisis: part 1: causes and threats. P&T. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 2.Hancock R.E., Sahl H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 3.Brogden K.A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 4.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 5.Bennett W.F., Hong C.K., Tieleman D.P. Antimicrobial peptide simulations and the influence of force field on the free energy for pore formation in lipid bilayers. J. Chem. Theory Comput. 2016;12:4524–4533. doi: 10.1021/acs.jctc.6b00265. [DOI] [PubMed] [Google Scholar]
- 6.Mihajlovic M., Lazaridis T. Antimicrobial peptides in toroidal and cylindrical pores. Biochim. Biophys. Acta. 2010;1798:1485–1493. doi: 10.1016/j.bbamem.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thøgersen L., Schiøtt B., Tajkhorshid E. Peptide aggregation and pore formation in a lipid bilayer: a combined coarse-grained and all atom molecular dynamics study. Biophys. J. 2008;95:4337–4347. doi: 10.1529/biophysj.108.133330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Chen C.H., Ulmschneider J.P. Spontaneous formation of structurally diverse membrane channel architectures from a single antimicrobial peptide. Nat. Commun. 2016;7:13535. doi: 10.1038/ncomms13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Schlamadinger D.E., McCammon J.A. Comparative molecular dynamics simulations of the antimicrobial peptide CM15 in model lipid bilayers. Biochim. Biophys. Acta. 2012;1818:1402–1409. doi: 10.1016/j.bbamem.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parton D.L., Akhmatskaya E.V., Sansom M.S. Multiscale simulations of the antimicrobial peptide maculatin 1.1: water permeation through disordered aggregates. J. Phys. Chem. B. 2012;116:8485–8493. doi: 10.1021/jp212358y. [DOI] [PubMed] [Google Scholar]
- 11.Pino-Angeles A., Leveritt J.M., III, Lazaridis T. Pore structure and synergy in antimicrobial peptides of the magainin family. PLOS Comput. Biol. 2016;12:e1004570. doi: 10.1371/journal.pcbi.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cirac A.D., Moiset G., Sengupta D. The molecular basis for antimicrobial activity of pore-forming cyclic peptides. Biophys. J. 2011;100:2422–2431. doi: 10.1016/j.bpj.2011.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leontiadou H., Mark A.E., Marrink S.J. Antimicrobial peptides in action. J. Am. Chem. Soc. 2006;128:12156–12161. doi: 10.1021/ja062927q. [DOI] [PubMed] [Google Scholar]
- 14.Giuliani A., Pirri G., Nicoletto S.F. Antimicrobial peptides: an overview of a promising class of therapeutics. Cent. Eur. J. Biol. 2007;2:1–33. [Google Scholar]
- 15.Aoki W., Ueda M. Characterization of antimicrobial peptides toward the development of novel antibiotics. Pharmaceuticals (Basel) 2013;6:1055–1081. doi: 10.3390/ph6081055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Midura-Nowaczek K., Markowska A. Antimicrobial peptides and their analogs: searching for new potential therapeutics. Perspect. Medicin. Chem. 2014;6:73–80. doi: 10.4137/PMC.S13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oyston P.C., Fox M.A., Clark G.C. Novel peptide therapeutics for treatment of infections. J. Med. Microbiol. 2009;58:977–987. doi: 10.1099/jmm.0.011122-0. [DOI] [PubMed] [Google Scholar]
- 18.Schittek B., Hipfel R., Garbe C. Dermcidin: a novel human antibiotic peptide secreted by sweat glands. Nat. Immunol. 2001;2:1133–1137. doi: 10.1038/ni732. [DOI] [PubMed] [Google Scholar]
- 19.Rieg S., Steffen H., Schittek B. Deficiency of dermcidin-derived antimicrobial peptides in sweat of patients with atopic dermatitis correlates with an impaired innate defense of human skin in vivo. J. Immunol. 2005;174:8003–8010. doi: 10.4049/jimmunol.174.12.8003. [DOI] [PubMed] [Google Scholar]
- 20.Rieg S., Garbe C., Schittek B. Dermcidin is constitutively produced by eccrine sweat glands and is not induced in epidermal cells under inflammatory skin conditions. Br. J. Dermatol. 2004;151:534–539. doi: 10.1111/j.1365-2133.2004.06081.x. [DOI] [PubMed] [Google Scholar]
- 21.Lai Y.P., Peng Y.F., Wu Z.R. Functional and structural characterization of recombinant dermcidin-1L, a human antimicrobial peptide. Biochem. Biophys. Res. Commun. 2005;328:243–250. doi: 10.1016/j.bbrc.2004.12.143. [DOI] [PubMed] [Google Scholar]
- 22.Rieg S., Seeber S., Schittek B. Generation of multiple stable dermcidin-derived antimicrobial peptides in sweat of different body sites. J. Invest. Dermatol. 2006;126:354–365. doi: 10.1038/sj.jid.5700041. [DOI] [PubMed] [Google Scholar]
- 23.Senyürek I., Paulmann M., Schittek B. Dermcidin-derived peptides show a different mode of action than the cathelicidin LL-37 against Staphylococcus aureus. Antimicrob. Agents Chemother. 2009;53:2499–2509. doi: 10.1128/AAC.01679-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Senyürek I., Döring G., Schittek B. Resistance to dermcidin-derived peptides is independent of bacterial protease activity. Int. J. Antimicrob. Agents. 2009;34:86–90. doi: 10.1016/j.ijantimicag.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Niyonsaba F., Suzuki A., Okumura K. The human antimicrobial peptide dermcidin activates normal human keratinocytes. Br. J. Dermatol. 2009;160:243–249. doi: 10.1111/j.1365-2133.2008.08925.x. [DOI] [PubMed] [Google Scholar]
- 26.Jung H.H., Yang S.T., Kim J.I. Analysis of the solution structure of the human antibiotic peptide dermcidin and its interaction with phospholipid vesicles. BMB Rep. 2010;43:362–368. doi: 10.5483/bmbrep.2010.43.5.362. [DOI] [PubMed] [Google Scholar]
- 27.Melo M.N., Ferre R., Castanho M.A. Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 2009;7:245–250. doi: 10.1038/nrmicro2095. [DOI] [PubMed] [Google Scholar]
- 28.Song C., Weichbrodt C., Zeth K. Crystal structure and functional mechanism of a human antimicrobial membrane channel. Proc. Natl. Acad. Sci. USA. 2013;110:4586–4591. doi: 10.1073/pnas.1214739110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marrink S.J., Tieleman D.P. Perspective on the Martini model. Chem. Soc. Rev. 2013;42:6801–6822. doi: 10.1039/c3cs60093a. [DOI] [PubMed] [Google Scholar]
- 30.Hess B., Kutzner C., Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 31.Marrink S.J., Risselada H.J., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 32.Berendsen C., Postma P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 33.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 34.Stansfeld P.J., Goose J.E., Sansom M.S.P. MemProtMD: automated insertion of membrane protein structures into explicit lipid membranes. Structure. 2015;23:1350–1361. doi: 10.1016/j.str.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kutzner C., Grubmüller H., Zachariae U. Computational electrophysiology: the molecular dynamics of ion channel permeation and selectivity in atomistic detail. Biophys. J. 2011;101:809–817. doi: 10.1016/j.bpj.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kutzner C., Köpfer D.A., Zachariae U. Insights into the function of ion channels by computational electrophysiology simulations. Biochim. Biophys. Acta. 2016;1858:1741–1752. doi: 10.1016/j.bbamem.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolf M.G., Hoefling M., Groenhof G. g_membed: efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. J. Comput. Chem. 2010;31:2169–2174. doi: 10.1002/jcc.21507. [DOI] [PubMed] [Google Scholar]
- 39.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 40.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 41.Nosé S., Klein M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983;50:1055–1076. [Google Scholar]
- 42.Nosé S.S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984;52:255–268. [Google Scholar]
- 43.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 44.Darden T., York D., Pedersen L. Particle mesh Ewald: an Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 45.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 46.Matsuzaki K. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta. 1999;1462:1–10. doi: 10.1016/s0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- 47.Chimerel C., Movileanu L., Kleinekathöfer U. Transport at the nanoscale: temperature dependence of ion conductance. Eur. Biophys. J. 2008;38:121–125. doi: 10.1007/s00249-008-0366-0. [DOI] [PubMed] [Google Scholar]
- 48.Kuyucak S., Chung S.H. Temperature dependence of conductivity in electrolyte solutions and ionic channels of biological membranes. Biophys. Chem. 1994;52:15–24. doi: 10.1016/0301-4622(94)00034-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.