In plants, chaperonin containing T-complex polypeptide-1 is a chaperonin protein complex of eight subunits that participates in the folding of tubulins and the catalytic subunit of protein phosphatase 4.
Keywords: CCT chaperonin, PP4 catalytic subunit, Tap46, TOR signaling pathway, tubulin biogenesis, virus-induced gene silencing
Abstract
Chaperonin containing T-complex polypeptide-1 (CCT) is an evolutionarily conserved chaperonin multi-subunit complex that mediates protein folding in eukaryotes. It is essential for cell growth and survival in yeast and mammals, with diverse substrate proteins. However, only a few studies on plant CCT have been reported to date, due to the essentiality of CCT subunit genes and the large size of the complex. Here, we have investigated the structure and function of the Arabidopsis CCT complex in detail. The plant CCT consisted of eight subunits that assemble to form a high-molecular-mass protein complex, shown by diverse methods. CCT-deficient cells exhibited depletion of cortical microtubules, accompanied by a reduction in cellular α- and β-tubulin levels due to protein degradation. Cycloheximide–chase assays suggested that CCT is involved in the folding of tubulins in plants. Furthermore, CCT interacted with PPX1, the catalytic subunit of protein phosphatase 4, and may participate in the folding of PPX1 as its substrate. CCT also interacted with Tap46, a regulatory subunit of PP2A family phosphatases, but Tap46 appeared to function in PPX1 stabilization, rather than as a CCT substrate. Collectively, our findings reveal the essential functions of CCT chaperonin in plants and its conserved and novel substrates.
Introduction
Chaperonin containing T-complex polypeptide-1 (CCT), also known as T-complex polypeptide-1 ring complex (TRiC), is a eukaryotic cytosolic group II chaperonin and the homolog of archaeal thermosome (Yébenes et al., 2011). Chaperonins mediate protein folding by encapsulating nascent polypeptides inside their central chamber and providing a sequestered environment. As shown by studies in yeast and mammals, CCT chaperonin is a large hetero-oligomeric complex of approximately 1000 kDa, which consists of eight subunits (CCT1–CCT8) located at fixed positions to form double rings stacked back to back (Hartl et al., 2011; Joachimiak et al., 2014). An ATP-driven conformational cycle of CCT mediates the opening and closing of the lid to encapsulate and release the substrate (Meyer et al., 2003; Cong et al., 2012; Reissmann et al., 2012).
The cortical microtubules (MTs) that line the cortex of interphase plant cells are essential in guiding the cellulose synthase complex and for regulating cell morphogenesis (Chen et al., 2016). In addition to dynamic polymerization and depolymerization to form various MT structures, tubulins undergo a complex process of autoregulation through transcription, post-translational modification, and degradation (Lundin et al., 2010). Furthermore, nascent tubulin polypeptides cannot fold spontaneously; thus, tubulin biogenesis requires various chaperone factors that have co-evolved with tubulins in eukaryotes (Szymanski, 2002; Bertrand et al., 2005).
It has been demonstrated that CCT chaperonin is essential for the folding of tubulin and actin in yeast and mammals (Llorca et al., 2000; Muñoz et al., 2011). Tubulin and actin are among the most abundant proteins in eukaryotic cells, and cells exhibit dramatic phenotypes when they are deficient. Therefore, a search for additional substrate proteins of the CCT complex has been hindered by the predominant cytoskeletal defects in CCT-deficient mutants. Nevertheless, an estimated 7% of nascent polypeptides have been reported to bind to the CCT complex after or during synthesis from ribosomes, suggesting that CCT likely has many other substrates (Yam et al., 2008). Indeed, recent studies have shown that CCT is involved in the folding of cell cycle regulators (CDC20 and CDH1), tumor suppressors (VHL and p53), and a telomerase cofactor (TCAB1), suggesting a vital role for the CCT chaperonin in animal cell growth and survival (Feldman et al., 1999; Camasses et al., 2003; Trinidad et al., 2013; Freund et al., 2014; Lopez et al., 2015). In contrast to the extensive characterization of CCT’s structure and functions in other eukaryotes, information on plant CCT and its de novo substrates is scarce, partly due to the difficulty in obtaining loss-of-function mutants since CCT’s functions are essential for cell survival. Earlier studies in maize and oats using anti-CCT antibodies showed co-sedimentation of tubulin and CCT subunits in sucrose fractionation and co-immunoprecipitation of CCTε (CCT5) with β-tubulin (Himmelspach et al., 1997; Moser et al., 2000). More recently, Xu et al. (2011), after analysing weak cct8 mutant alleles, reported that CCT is essential for cell-to-cell trafficking and stem cell function of the KNOTTED1 homeobox family of transcript factors. The difficulty in purifying the CCT complex has hindered identification of new CCT substrates in plants.
Plant Tap46 and its homologs, Tap42 and α4/IGBP1, in yeast and mammals, respectively, are regulatory subunits of PP2A family phosphatases (PP2A, PP4, and PP6), which act by direct association with the phosphatase catalytic subunits to form a heterodimer (Chen et al., 1998; Wang et al., 2003; Ahn et al., 2011). Tap46, Tap42, and α4 are all essential for cell survival. In yeast, Tap42 is phosphorylated by the target of rapamycin (TOR) kinase, and Tap42-regulated protein phosphatase activities constitute a major mechanism for regulation of the downstream effectors of the TOR pathway (Düvel and Broach, 2004). In plants, TOR phosphorylated recombinant Tap46 protein in vitro, and Tap46 depletion reproduced the signature phenotypes of TOR inactivation, supporting a functional link between Tap46 and TOR (Ahn et al., 2011). Tap46 has a particularly strong interaction with the PP4 catalytic subunit (PP4c), among the PP2A family members (Ahn et al., 2011). Kong et al. (2009) reported that α4 plays a critical role in maintaining cellular PP2A activity by stabilizing PP2A family catalytic subunits. It was proposed that α4 acts as a scaffold/chaperone protein and protects the catalytic subunits from degradation until the assembly of the functional phosphatase complex is finished.
Interestingly, tandem affinity purification tagging and mass spectrometry indicated that PP4c and α4 interact with multiple CCT subunits in mammals (Gingras et al., 2005; Herzog et al., 2012). However, a functional role of those interactions has not been elucidated. In this study, we investigated the complex formation and in vivo functions of CCT chaperonin in Arabidopsis. The CCT complex, made of eight subunits, is essential for plant growth. The CCT complex is involved in the folding of tubulins, and silencing of the CCT subunit genes resulted in cortical MT defects among other pleiotropic phenotypes. Furthermore, our results suggest that PPX1 (PP4c) may be a novel substrate of plant CCT. Tap46, which interacts with PPX1 and CCT, may play a role in stabilization of PPX1. Thus, CCT appears to link to the TOR signaling pathway through biogenesis of PP4 catalytic subunits.
Materials and methods
Plant materials and growth conditions
Arabidopsis plants (ecotype Col-0) were grown in a 22°C growth chamber under long-day conditions (16 h light–8 h dark) with light intensity of 100–150 μmol m−2 s−1. Nicotiana benthamiana plants were grown in a 23°C growth chamber under long-day conditions (16 h light–8 h dark) with 80 μmol m−2 s−1 light intensity. GFP–TUB6 OE (CS6550) and GFP–TUA6 OE seeds (CS6551) were obtained from the Arabidopsis Biological Resources Center (ABRC).
Bimolecular fluorescence complementation
Protein coding regions were PCR-amplified and cloned into the pSPYNE vector containing the N-terminal region (amino acid residues 1–155) of yellow fluorescent protein (YFP) or into pSPYCE vector containing the C-terminal region (residues 156–239) of YFP. The pSPYNE and pSPYCE fusion constructs were agroinfiltrated together into the leaves of 3-week-old N. benthamiana plants as described (Walter et al., 2004). After 48 h, protoplasts were generated and the YFP signal was detected using a confocal microsope (Zeiss LSM510).
Transient protein expression in N. benthamiana plants
One-month-old N. benthamiana seedlings were used for transient infiltration. Agrobacterium cells carrying constructs of interest and p19 were grown overnight in selective YEP medium (rifampicin/kanamycin) at 28°C. After centrifugation at 200 g for 15 min, cells were resuspended in the induction medium (10 mM MgSO4, 10 mM MES–KOH, pH 5.7, and 1 mM acetosyringone) and incubated for 1–2 h. Agrobacterium containing the expression constructs was mixed with Agrobacterium containing p19, and the final OD600 value of each strain was set to 1~1.5. The mixed media were then infiltrated into N. benthamiana seedlings with a needle-less syringe. Leaf samples were harvested at 2 d after infiltration (DAI).
Virus-induced gene silencing in Arabidopsis plants
Virus-induced gene silencing (VIGS) was performed in Arabidopsis plants as described previously (Ahn et al., 2015b). TRV2 vectors (Burch-Smith et al., 2006) harboring fragments of CCT2, CCT3, TUA6, ACT2, Tap46, and PPX1/2 cDNA were used for VIGS. Agrobacterium transformed with the cloned TRV2 vectors and Agrobacterium containing TRV1 (pBINTRA) vectors were inoculated in LB medium with 10 mM MES–KOH (pH 5.7) and 20 μM acetosyringone and grown overnight in 28°C. After centrifugation at 200 g for 15 min, cells were harvested and resuspended in the infiltration medium (10 mM MgCl2, 10 mM MES–KOH, pH 5.7, and 200 μM acetosyringone). Then TRV2 and TRV1 infiltration media were mixed at a 1:1 ratio at OD600=1 and incubated for 3–4 h, followed by infiltration into Arabidopsis seedlings at approximately 10 d after germination (DAG) using needle-less syringes. Phenotypes were observed at 15–18 DAI.
Trichome isolation
Trichomes were isolated from TRV2 and TRV2:CCT2 sample leaves using previously published methods (Marks et al., 2008). Five to six rosette leaves were harvested and put into 50 ml tubes. Fifteen milliliters of trichome isolation buffer (1× PBS, 50 mM EGTA, pH 7.5) with 0.01 g of 425–600 μm glass beads (Merck) was added to the tubes. The tubes were vortexed four times at the maximum speed for 30 s, and during the intervals the tubes were kept on ice. After vortexing, the supernatant was transferred to new tubes. The remaining beads and leaves were rinsed with PBS to isolate the trichomes still attached to the beads and the leaves. The supernatant was then passed through a 70 μm cell strainer (SPL Life Sciences), trapping the trichomes inside the cell strainer. Then the cell strainer containing the trichomes was inverted in a new cell culture dish. Ten milliliters of PBS was added to detach the trichomes from the cell strainer. PBS solution containing the trichomes was then transferred to 15 ml tubes and centrifuged at 100 g for 5 min to remove supernatants. The number of branches in each trichome were counted using a light microscope.
Microtubule density measurement
Microtubule density was determined by measuring green fluorescent protein (GFP) signal intensity in rosette leaf cells of GFP–TUB6 overexpression (OE) lines as previously described (Kirik et al., 2012). First, microtubule array images of leaf epidermal cells were collected using a confocal microscope (LSM 700, Carl Zeiss) with 1.5 μm sections. For imaging, a pinhole was set to 77.1, and 9–10 optical sections were taken for each sample, which were then Z-stacked to create a three-dimensional image. The stacked images were analysed using ImageJ. Background noise signals were subtracted using a 15-pixel diameter rolling ball filter, followed by passing through a Fourier bandpass filter limited to 3–4 pixels. Then the Otsu algorithm was applied for blind thresholding. Ten cells were analysed for each sample, and the percentage of the cell area occupied by the GFP signal was measured.
Sucrose density gradient sedimentation
Sucrose density gradient sedimentation was performed as described in Cho et al. (2013) with modifications. Frozen seedlings (0.2 g) of Flag–CCT2 OE, Flag–PPX1 OE and HA–Tap46 OE lines were ground into fine powder in liquid nitrogen and thawed without vortexing in 1 ml fractionation buffer (FB) (200 mM Tris-Cl, pH 8.4, 50 mM KCl, 25 mM MgCl2, and half concentration of protease inhibitor cocktail). Cell debris was removed by centrifugation at 10 000 g for 8 min. Supernatants were filtered through a layer of nylon mesh, and were then loaded onto 11.6 ml of 10–30% sucrose gradient in FB. Then the samples were centrifuged at 150 000 g for 3.5 h in 4°C. Fractions (400 μl) were collected, followed by protein precipitation using methanol–chloroform (Wessel et al 1984). Then the proteins in each fraction were analysed by immunoblotting.
Size exclusion chromatography
GFP–CCT2 was transiently expressed in N. benthamiana leaves by agroinfiltration. At 2 DAI, samples were harvested, and proteins were extracted using 2 ml of gel chromatography buffer as described (Kim et al., 2017). After centrifugation at 21 000 g for 15 min twice, samples were passed through a layer of nylon mesh, and subsequently filtered through a 0.22 μm filter (MILLEX-HP; Merck). A Sephacryl s300 gel filtration column (Hiprep 16/60; GE Healthcare) was equilibrated with filter-passed equilibration/washing buffer (50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 1 mM MgCl2, 2 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride). After equilibration, 1 ml of samples was loaded onto the gel filtration column. Fractions of 1 ml were collected, and the proteins in the fractions collected between 41 ml and 51 ml elution volumes were precipitated by the methanol–chloroform method. After elution of the precipitated proteins in 2× SDS sample buffer, samples were loaded onto SDS-PAGE gels for immunoblot analyses.
MG132 treatment
GFP–TUB6 OE or Flag–PPX1 OE plants were infiltrated with Agrobacterium cells containing TRV2, TRV2:CCT2 or TRV2:TUA6, and their rosette leaves were collected at DAI 15. The leaves were submerged in 20 μM MG132 with 0.01% Tween-20 for 4 h. Proteins were extracted from these samples and subjected to immunoblotting.
Immunoblotting
Plant samples were ground in liquid nitrogen and proteins were extracted. After native- or SDS-PAGE, separated proteins were transferred to polyvinylidene difluoride (PVDF) membranes. After blocking with 5% skim milk for 30 min, immunoblotting was performed. The following antibodies were used for immunoblotting analyses: anti-Myc (1:5000, cat. G019, abm; 1:10000, cat. A5598, Merck), anti-HA (1:5000, cat. G036, abm), anti-GFP (1:500 for immunoprecipitation, cat. G095, abm; 1:5000 for IB, cat. 632381, Clontech), anti-Flag (1:5000, cat. G191, abm), anti-RPN6 (1:5000, cat. BML-PW8370-0025, Enzo Life Sciences), anti-α-tubulin (1:5000, T9026, Merck), anti-β-tubulin (1:5000, cat. ab15568, Abcam), anti-chloroplast HSP70 (1:10000, cat. AS08 348, Agrisera), anti-Flag conjugated resin (cat. F2426, Merck), anti-Myc conjugated resin (cat. E6654, Merck), anti-PP2Ac (1:5000, cat. 2038S, Cell Signaling Technology), and anti-PP4c (1:2000, cat. A300-835A, Bethyl Laboratories).
Native PAGE
Native-PAGE was performed as described (Frydman et al., 1992) with minor modification. Proteins were extracted using the native-PAGE buffer (50 mM HEPES–KOH, pH 7.4, 50 mM KCl, and 2 mM DTT). After centrifugation, samples were mixed in 2× native-PAGE loading buffer (Freund et al., 2014) and run at 120 V for 120 min in 4°C in native-PAGE gels. Proteins were then transferred to PVDF membranes by the wet transfer method (65 V, 50 min) at 4°C, followed by immunoblotting. For two-dimensional PAGE (2D-PAGE), native-PAGE (1D) gel strips were cut and incubated for 1 h in 1× SDS buffer containing 1% β-mercaptoethanol. The strips were rinsed several times with 1× SDS buffer and mounted on SDS-PAGE running gels. 2D-PAGE gels were transferred onto PVDF membranes via the semi-dry transfer method (75 V, 45 min) for immunoblot analysis.
Cycloheximide–chase assay
Seedlings were grown in MS medium for 5 d after germination. The seedlings were harvested and treated with 2 mg ml−1 cycloheximide (CHX) for 4 h at 25°C. Proteins were extracted from the samples with native-PAGE buffer and subjected to native-PAGE. SDS-PAGE was also performed with the identical extracts.
Results
Identification of eight CCT subunits in Arabidopsis
CCT is a conserved chaperonin complex in eukaryotes, consisting of eight paralogous subunits (CCT1 to CCT8) with mutual sequence identity of ~30% (Archibald et al., 2001). We analysed sequence similarity in the CCT subunit genes from Arabidopsis, soybean (Glycine max), maize (Zea mays), rice (Oryza sativa), tomato (Solanum lycopersicum), Nicotiana benthamiana, budding yeast (Saccharomyces cerevisiae), and human (Homo sapiens) (see Supplementary Table S1 at JXB online). The phylogenetic tree was generated by the maximum likelihood method using the CCT subunit sequences (Supplementary Fig. S1). The paralogous CCT subunits of different species were clustered together. The strong paralogy of CCT subunits indicates that CCT subunits diverged very early, prior to eukaryotic speciation. However, isoforms of each CCT subunit were clustered together within species, suggesting diversification of each isoform after the speciation event. Collectively, these results suggest that CCT subunits are highly conserved in plants, but species-specific modifications have occurred during plant evolution. The conservation of the CCT subunits in eukaryotic organisms suggests conserved functions of the CCT chaperonin.
Detection of the high-molecular-mass CCT complex in plant cells
We observed that all CCT subunits are mainly localized to the cytosol, but were also detected in the nucleoplasm, but not in the nucleolus (see Supplementary Fig. S2). To detect the whole high-molecular-mass CCT complex in plant cells, diverse experimental techniques were employed. First, we performed size exclusion chromatography to determine the approximate size of the plant CCT complex (Fig. 1A). GFP–CCT2 was expressed in N. benthamiana leaves via agroinfiltration, and the leaf protein extract was separated by gel chromatography, followed by immunoblotting with anti-GFP antibody to detect GFP–CCT2. GFP–CCT2 was mainly detected in fractions 5 and 6 between the two size markers of 669 and 2000 kDa, consistent with the predicted size (943 kDa) of the plant CCT complex.
Fig. 1.
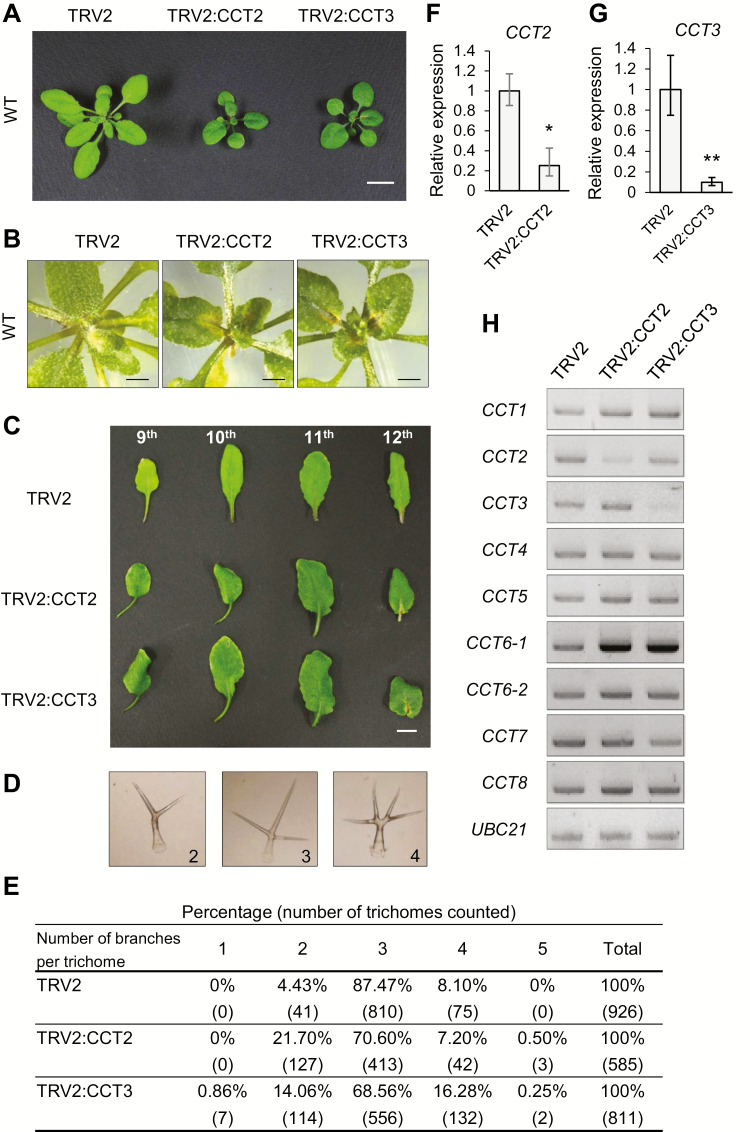
CCT subunits function as a complex. (A) Size exclusion chromatography using N. benthamiana leaf extracts that expressed GFP–CCT2. Eleven fractions were collected between the elution volumes of 41 and 51 ml, and subjected to immunoblotting using anti-GFP antibody. GFP–CCT2 is approximately 83 kDa in size (arrow). (B) Sucrose density gradient sedimentation using Arabidopsis Flag–CCT2 overexpression (OE) plants. Extracts of the seedlings at 7 d post-germination (DAG) were mounted on a 10–30% sucrose gradient for ultracentrifugation, followed by immunoblotting of the fractions with anti-Flag and anti-RPN6 antibodies. RPN6 is a subunit of 26S proteasome. Flag–CCT2 is approximately 67 kDa in size (arrow). (C) Native-PAGE and SDS-PAGE. Flag–CCT subunits were expressed transiently in N. benthamiana leaves. The CCT complex (asterisk) and monomeric CCT subunits (bracket) are indicated. (D) Two-dimensional PAGE using N. benthamiana extracts that co-expressed Flag–CCT2 and Flag–CCT4. After native-PAGE (1D) (3.5% gel) and SDS-PAGE (2D), immunoblotting was performed with anti-Flag antibody. The CCT complex (arrow), Flag–CCT2 (white arrowhead) and Flag–CCT4 (black arrowhead) are indicated. (E) Plant phenotypes after virus-induced gene silencing (VIGS) of CCT3 (TRV2:CCT3) in Flag–CCT2 OE plants at 15 DAI. TRV2-infiltrated plants served as the control. Scale bars: 1 cm. (F) RT-qPCR of CCT3 mRNA in the VIGS plants shown in (E). Transcript levels are normalized to those of UBC10 mRNA, and expressed relative to those of TRV2 samples. Values represent mean ±standard error (SE) derived from three biological replicates. Asterisks denote statistical significance of the differences between TRV2 control and TRV2:CCT3 plants calculated using Student’s t-test (**P≤0.01). (G) Native-PAGE and SDS-PAGE using leaf extracts of the VIGS plants shown in (E). Immunoblotting was performed with anti-Flag antibody. Ponceau-stained rbcL was used as a loading control for native-PAGE. The asterisk indicates the CCT complex. (This figure is available in color at JXB online.)
Second, we performed sucrose density gradient sedimentation using transgenic Arabidopsis plants that expressed Flag-fused CCT2 under the control of the 35S promoter (Flag–CCT2 OE) (Fig 1B; Fig. S3A, B). The transgenic plants exhibited no visible phenotypes under normal growth conditions. RT-PCR showed that CCT2 transcript levels increased in the plants, but expression of other CCT subunit genes remained unchanged. The leaf extract of the Flag–CCT2 OE plants was fractionated on a 10–30% sucrose density gradient, and fractions were subjected to immunoblot analyses with anti-Flag antibody (Fig. 1B). As the control for fractionation, immunoblotting was also performed with antibody against RPN6, a component of the 26S proteasome. Flag–CCT2 was most abundant in fractions 2 and 3, whereas RPN6 was most strongly detected in fraction 4. Thus, the Arabidopsis CCT2 complex was distributed in fractions lighter than those containing 26S proteasomes, consistent with the observation that the yeast CCT complex migrates at 20S through the sucrose gradient (Camasses et al., 2003).
Third, we performed native-PAGE analysis. Flag-tagged CCT subunits were expressed in N. benthamiana leaves, and the leaf protein extracts were subjected to native-PAGE on a 4–10% gradient gel, followed by immunoblotting with anti-Flag antibody. All CCT subunits were detected as a slow-migrating band in the upper position of the native gel (asterisk), and also as smeared bands (bracket) at the lower position of the gel (Fig. 1C, upper). The upper single band may represent the CCT complex, while the lower smeared bands may represent the monomeric form of each subunit. SDS-PAGE and immunoblotting detected each CCT subunit in the molecular mass range of 60–70 kDa (Fig. 1C, lower).
To determine that the slow-migrating band in native-PAGE consists of the CCT subunits, two-dimensional PAGE was performed (Fig. 1D). Flag–CCT2 and Flag–CCT4 were expressed in N. benthamiana leaves, and the proteins were first resolved by native-PAGE (1D-PAGE). The 1D gel strip was horizontally loaded onto a SDS-PAGE gel (2D-PAGE) to separate the individual proteins. The gels were then subjected to immunoblotting with anti-Flag antibody. In 1D-PAGE, three protein bands were detected: the CCT complex form (Fig. 1D, upper, arrow), and monomeric Flag–CCT2 (Fig. 1D, upper, white arrowhead) and Flag–CCT4 (Fig. 1D, upper, black arrowhead). The migration rate of monomeric Flag–CCT4 was lower than that of monomeric Flag–CCT2 due to its high isoelectric point (pI 7.8), although Flag–CCT4 was slightly smaller than Flag–CCT2 in molecular mass. The monomeric forms of Flag–CCT4 and Flag–CCT2 in 1D-PAGE were separated in 2D-PAGE according to their sizes, with Flag–CCT4 (Fig. 1D, lower, black arrowhead) migrating faster than Flag–CCT2 (Fig. 1D, lower, white arrowhead). In 2D-PAGE, two protein bands of different sizes were detected at the position of the CCT complex (Fig. 1D, lower, black arrow), and the two bands resolved from the CCT complex matched in size with those of the monomeric Flag–CCT4 and Flag–CCT2 (Fig. 1D).
If the CCT complex requires all the CCT subunits, then the depletion of one of the subunits may result in loss of the whole complex. To test this hypothesis, we performed virus-induced gene silencing (VIGS) of CCT3 in Flag–CCT2 OE Arabidopsis plants (Fig. 1E–G; Supplementary Fig. S3A). We cloned a 450-bp cDNA fragment of Arabidopsis CCT3 into the tobacco rattle virus 2 (TRV2) vector, and infiltrated the leaves of the Flag–CCT2 OE plants with Agrobacterium containing this plasmid. VIGS of CCT3 caused growth arrest in the plants at 15 d after infiltration (DAI), accompanied by a reduction in CCT3 mRNA levels to approximately 8% of the TRV2 levels, as determined by RT-qPCR (Fig. 1E, F; Supplementary Table S2). The leaf protein extracts prepared from the VIGS plants were divided into two halves for native-PAGE and SDS-PAGE, followed by immunoblotting with anti-Flag antibody to detect Flag–CCT2 (Fig 1G). Ponceau-stained Rubisco large subunit (rbcL) was used as the control. CCT3 silencing did not change Flag–CCT2 protein levels, as detected by SDS-PAGE, but significantly decreased the level of CCT complex (Fig 1G, asterisk) in native-PAGE, suggesting that the CCT3 subunit is essential for CCT complex formation. Collectively, these results suggest that plant CCT subunits form a high-molecular-mass complex, and the size of the whole complex is similar to that of the complex in other eukaryotic organisms (approximately 1000 kDa).
VIGS of CCT subunit genes results in pleiotropic phenotypes in Arabidopsis
Null mutations of CCT subunits are lethal in Arabidopsis (Xu et al., 2011), as in yeast and mammals (Horwich et al., 2007). To circumvent the absence of available null mutants, we used VIGS for the silencing of CCT2 (TRV2:CCT2) and CCT3 (TRV2:CCT3) in Arabidopsis WT plants. VIGS of CCT2 and CCT3 resulted in a similar phenotype of retarded plant growth and the formation of necrotic lesions at 15 DAI (Fig. 2A). These lesions were formed in new leaves near the leaf base at 15 DAI, and the inflorescence failed to elongate (Fig. 2B). Interestingly, the petioles of TRV2:CCT2 and TRV2:CCT3 leaves showed a right-handed twist at 21 DAI (Fig. 2C). The handedness of petioles and cell files in hypocotyls and roots are indicative measures of microtubule defects (Ishida et al., 2007).
Fig. 2.
Depletion of CCT subunits leads to pleiotropic defects in Arabidopsis. (A) Phenotypes after VIGS of CCT2 or CCT3 at 15 DAI. Scale bar: 1 cm. (B) Closer views of the necrotic lesions around the shoot apex of VIGS plants at 15 DAI. Scale bar: 5 mm. (C) Right-handed twist in leaf petioles of the VIGS plants at 21 DAI. The 9th to 12th leaves from cotyledons are shown. Scale bar: 1 cm. (D) Representative light microscopic images of trichomes with different branch numbers. (E) Quantification of trichome branch numbers. Trichomes were harvested from leaves and counted under a light microscope. The percentages of each branch number and numbers of trichomes counted (in parentheses) are shown. (F, G) RT-qPCR of CCT2 (F) and CCT3 (G) mRNAs in VIGS plants. Transcript levels are normalized to those of UBC10 mRNA, and expressed relative to those of TRV2 samples. Values represent mean ±SE derived from three biological replicates. Asterisks denote statistical significance of the differences between TRV2 control and other VIGS plants calculated using Student’s t-test (*P≤0.05; **P≤0.01). (H) Semi-quantitative RT-PCR for CCT subunit genes in the VIGS plants. UBC21 mRNA served as the loading control. (This figure is available in color at JXB online.)
VIGS of CCT2 and CCT3 also caused trichome defects. Trichomes were isolated from the leaves and the number of their branches was counted under a light microscope (Fig. 2D). In TRV2 control leaves, the vast majority of trichomes (87.47%) contained three branches, and 4.43% and 8.10% of trichomes had two and four branches, respectively (Fig. 2E). However, TRV2:CCT2 and TRV:CCT3 trichomes showed larger variations in branch number; in particular, higher amounts of two-branched trichomes and occasional five-branched trichomes were notable. It is well known that cortical microtubules play a role in trichome cell morphogenesis (Mathur and Chua, 2000; Tian et al., 2015). Indeed, trichome branch number is affected in various mutants with microtubule defects, such as mutants of tubulin-folding cofactors (Kirik et al., 2002a, b). The right-handed twist of the petioles and the abnormal trichome branch number suggest microtubule defects in CCT VIGS plants.
RT-qPCR analyses demonstrated that CCT2 and CCT3 mRNA levels were reduced to 25% and 7% of the TRV2 control level in TRV2:CCT2 and TRV2:CCT3 plants, respectively (Fig. 2F, G). However, VIGS of CCT2 and CCT3 genes did not decrease mRNA levels of other CCT subunit genes based on semi-quantitative RT-PCR, suggesting specificity of the gene silencing (Fig. 2H). The only significant anomaly was CCT6-1 mRNA levels that were elevated in TRV2:CCT2 and TRV2:CCT3 plants, although the reason for this was unclear. The similar silencing phenotypes of CCT2 and CCT3 suggest that their encoded proteins function together as subunits of the CCT complex.
CCT silencing causes defects in cortical microtubule organization and tubulin accumulation in Arabidopsis
To investigate how the deficiency of CCT function affects cortical MT organization, silencing of CCT2 was performed in transgenic Arabidopsis plants that expressed β-tubulin isomer 6 in GFP fusion (GFP–TUB6 OE). In these transgenic plants, the GFP-tagged β-tubulin isoform was incorporated into cortical MTs, making them visible under confocal microscopy (Fujita et al., 2013). Control VIGS was performed with TUA6 encoding Arabidopsis α-tubulin isoform 6 and TRV2 vector in the GFP–TUB6 OE plants. Multiple α-tubulin genes were expected to be simultaneously silenced by VIGS of TUA6 due to high sequence homology between α-tubulin isoforms. Silencing of TUA6 caused severe growth arrest with tiny leaves being developed around the shoot apex at 15 DAI (Fig. 3A). Inflorescences did not form and flower development was aborted in these plants. Unlike TRV2:CCT2 plants, TRV2:TUA6 plants did not develop necrotic lesions in the leaves, suggesting that MT deficiency is not the cause of necrotic lesion formation. RT-qPCR indicated that TUA6 mRNA levels were reduced to 8% of TRV2 control levels in TUA6 VIGS plants (Fig. 3B).
Fig. 3.
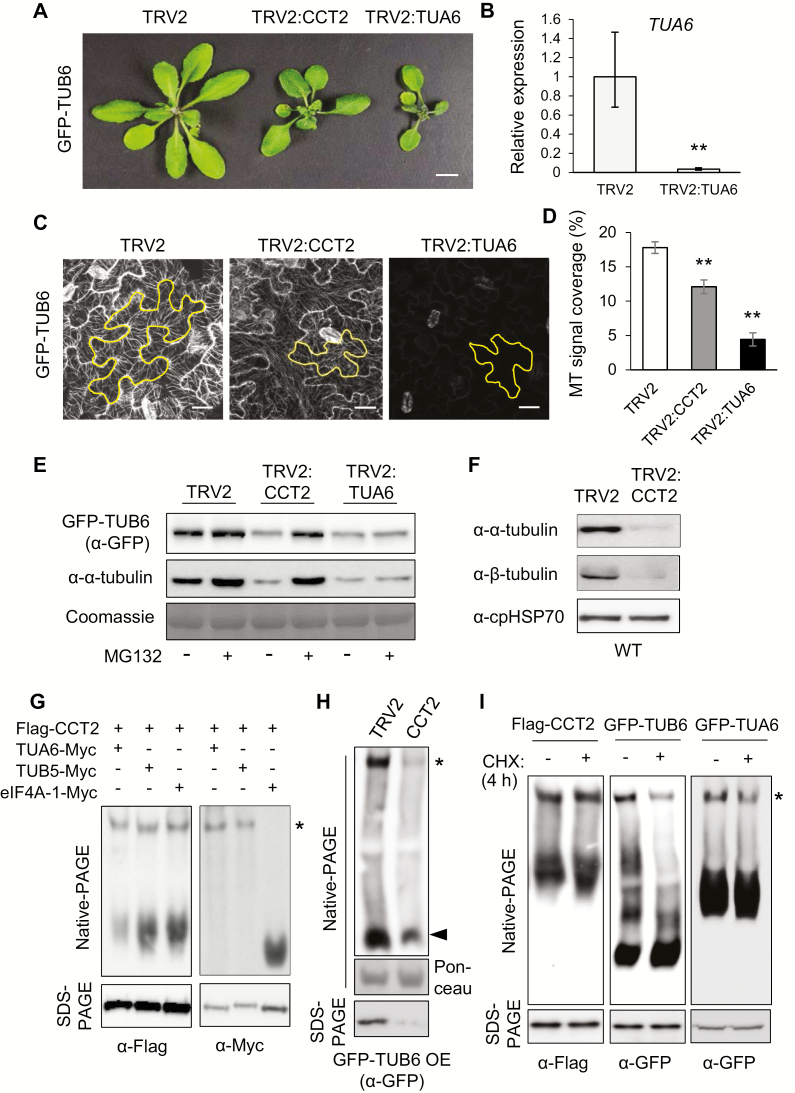
Tubulins are substrates of the CCT complex in plants. (A) VIGS phenotype of CCT2 and TUA6 in GFP–TUB6 background at 15 DAI. TUA6 encodes α-tubulin isoform 6. Scale bar: 1 cm. (B) RT-qPCR of TUA6 mRNA in TRV2 and TRV2:TUA6 plants shown in (A) at 15 DAI. Transcript levels are normalized to those of UBC10 mRNA, and expressed relative to those of TRV2 samples. Values represent mean ±SE derived from three biological replicates (**P≤0.01). (C) Confocal microscopy of cortical MT arrays in leaf epidermis of the VIGS plants shown in (A) at 18 DAI. The images are projections of a z-stack. One cell in each image is outlined. Scale bar: 20 μm. (D) Cortical MT array density of the VIGS plants. MT density was quantified using ImageJ with 10 cells per sample (**P≤0.01). (E) Immunoblotting with anti-GFP and anti-α-tubulin antibodies in the VIGS plants shown in (A) with or without 4 h treatment with 20 μM MG132. The Rubisco large subunit (rbcL) was stained with Coomassie as the loading control. (F) Immunoblotting with anti-α-tubulin and anti-β-tubulin antibodies in VIGS plants in WT background. Chloroplast HSP70 (cpHSP70) served as the loading control. (G) Native-PAGE and SDS-PAGE of N. benthamiana extracts co-expressing Flag–CCT2 with TUA6–Myc, TUB5–Myc, or eIF4A-1–Myc. eIF4A-1 is the cytosol-localized isoform of eukaryotic translation initiation factor 4A. Immunoblotting was performed with anti-Myc and anti-Flag antibodies. Monomeric TUA6–Myc and TUB5–Myc ran out of the gel due to the long running time. (H) Native-PAGE and SDS-PAGE analyses of VIGS plants in GFP–TUB6 OE background, followed by immunoblotting with anti-GFP antibody. Ponceau-stained rbcL served as the loading control for native-PAGE. (I) Cycloheximide (CHX)–chase assays. Five-day-old Flag–CCT2 OE, GFP–TUB6 OE, and GFP–TUA6 OE seedlings were treated with water (−CHX) or 2 mg ml−1 CHX for 4 h, and subjected to native-PAGE and SDS-PAGE, followed by immunoblotting with anti-Flag and anti-GFP antibodies. (This figure is available in color at JXB online.)
Confocal microscopy revealed that TRV2 control plants contained ordered MTs in puzzle-shaped pavement cells in the leaf epidermis (Fig. 3C). TRV2:TUA6 plants with α-tubulin deficiency lacked visible cortical MTs, because αβ-tubulin dimers are required to form MT protofilaments. It was noted that the pavement cells of TRV2:TUA6 plants were small and had a simpler structure. TRV2:CCT2 pavement cells also showed reduced MT arrays. The MT density was quantified by measuring GFP signal intensity in the form of strands within a cell. MT signal coverage was reduced to 68% and 22% of TRV2 control in TRV2:CCT2 and TRV2:TUA6, respectively (Fig. 3D). These results suggest that CCT chaperonin is required for normal cortical MT array organization.
To examine cellular tubulin levels in the VIGS plants, immunoblotting was performed with anti-GFP and anti-α-tubulin antibodies using Coomassie-stained rbcL as the control (Fig. 3E). The results showed that both GFP–TUB6 and α-tubulin levels were reduced in TRV2:CCT2 and TRV2:TUA6 plants. We next tested whether the reduced accumulation of tubulins is caused by protein degradation. Leaves were treated with the proteasome inhibitor MG132 (20 μM) or control DMSO for 4 h. MG132 treatment restored the reduced cellular levels of β-tubulin (GFP–TUB6) and α-tubulin in TRV2:CCT2 plants back to TRV2 control levels (Fig. 3E). These results suggest that CCT depletion in a cell results in the accumulation of misfolded nascent tubulin polypeptides, which are subsequently eliminated by 26S proteasome-mediated protein degradation. However, GFP–TUB6 and α-tubulin levels were constant following MG132 treatment in TRV2:TUA6 plants. It is noteworthy that GFP–TUB6 levels in TRV2:TUA6 plants, in which multiple α-tubulin genes are silenced, were lower than that of the TRV2 control in the absence of MG132 treatment. The reason for this is unclear, but we speculate that the reduction of cellular α-tubulins may have affected β-tubulin expression, since αβ-tubulins are obligate heterodimers for MT assembly (Lundin et al., 2010).
To confirm the results obtained with GFP–TUB6 OE lines, we performed VIGS of CCT2 in WT Arabidopsis plants, followed by immunoblotting with anti-α- and anti-β-tubulin antibodies using chloroplast heat shock protein 70 (cpHSP70) as the loading control (Fig. 3F). The endogenous levels of α- and β-tubulin proteins were significantly reduced in TRV2:CCT2 leaves as compared with TRV2 control leaves, consistent with the results in GFP–TUB6 OE plants. Collectively, these results suggest that CCT chaperonin may be required for protein folding of newly synthesized α- and β-tubulins in plants.
Tubulins are conserved substrates of plant CCT complex
To determine whether tubulins are substrates of CCT chaperonin in plants, we first examined the association of tubulins with the CCT complex in vivo using native-PAGE. Flag–CCT2 was co-expressed with Myc-fused TUA6, TUB5, or cytosolic eukaryotic translation initiation factor eIF4A-1 in N. benthamiana leaves by agroinfiltration. The leaf protein extracts were then subjected to native-PAGE analysis using a 4% gel; the extracts were divided into two halves and each was loaded on the same gel for immunoblotting with anti-Flag and anti-Myc antibodies. Anti-Flag antibody detected the CCT complex as an upper slow-migrating band (asterisk) and CCT2 monomers as a lower smeared band in each lane (Fig. 3G, left). TUA6–Myc and TUB5–Myc were detected by anti-Myc antibody as single upper bands that exactly coincided with the CCT complex (Fig. 3G, right). Low-molecular-mass monomeric tubulins ran out of the gel due to the long running time. Conversely, control eIF4A-1–Myc was detected only as a fast-migrating band in the lower part of the gel. These results suggest the association of α- and β-tubulins with the CCT complex.
Native-PAGE and immunoblotting in GFP–TUB6 OE plants showed that CCT2 silencing caused a significant decrease in both the CCT-associated (asterisk) and monomeric forms (black arrowhead) of GFP–TUB6 proteins, suggesting the possibility that CCT chaperonin is required for folding of GFP–TUB6 (Fig. 3H). We thus performed a cycloheximide (CHX)–chase assay (Fig. 3I). CHX is a protein translation inhibitor. A brief CHX treatment would decrease the production of nascent polypeptides for a short period of time, and if a nascent polypeptide is a substrate of the CCT complex, the treatment may reduce the CCT-associated form of the substrate. GFP–TUB6 OE and Flag–CCT2 OE seedlings were treated with CHX (2 mg ml−1) or water control for 4 h, and then subjected to native- and SDS-PAGE analyses. Immunoblotting with anti-Flag antibody revealed that the 4 h CHX treatment did not change total CCT2 or CCT complex-associated CCT2 levels (Fig. 3I, left). CHX treatment also did not change the total TUB6 levels in SDS-PAGE, but visibly decreased CCT complex-associated TUB6 levels (Fig. 3I, middle). A CHX–chase assay in GFP–TUA6 OE plants expressing α-tubulin isomer 6 in GFP fusion also showed similar results (Fig. 3I, right). Combined with the results described above, these results suggest that tubulins are folding substrates of CCT chaperonin in plants, similar to the findings in yeast and mammals (Dekker et al., 2008; Muñoz et al., 2011).
CCT complex interacts with PP4 catalytic subunit and Tap46, which are components of the TOR signaling pathway
Multiple CCT subunits have been pulled down with PP4c or α4 (the mammalian homolog of Tap46) as bait by tandem affinity purification (Gingras et al., 2005; Herzog et al., 2012). Furthermore, CCT interactions with PP4c and Tap42 (the yeast homolog of Tap46) have been suggested by the yeast interactome study (Breitkreutz et al., 2010). However, it is unknown whether such an interaction network is conserved in plants, and furthermore, the nature of the interactions between the CCT complex and these protein phosphatase subunits has not been revealed yet in any eukaryotic system. Tap46 is a component of the TOR signaling pathway in plants and interacts with PP4 catalytic subunits with high affinity (Ahn et al., 2011). RT-qPCR analyses suggested that CCT gene expression under normal conditions and in response to sugar feeding is modulated by TOR (see Supplementary Fig. S4A, B).
We first performed sucrose density gradient sedimentation (10–20%) to determine whether PPX1 and Tap46 are co-fractionated with the CCT complex (Fig. 4A). Arabidopsis PP4c is encoded by PPX1 and PPX2 (Pujol et al., 2000). We previously generated transgenic Arabidopsis lines that express HA-fused Tap46 and Flag-fused PPX1 under the control of the CaMV35S promoter. HA–Tap46 OE lines exhibited enhanced early plant growth (Ahn et al., 2015a), but Flag–PPX1 OE lines showed no visible phenotypes under normal growth conditions (see Supplementary Fig. S3C, D). Whereas Flag–CCT2 was most abundant in fractions 2–4, Flag–PPX1 and HA–Tap46 were most abundant in fractions 1 and 2, but also detected in fractions 3 and 4, thus overlapping with the distribution of Flag–CCT2.
Fig. 4.
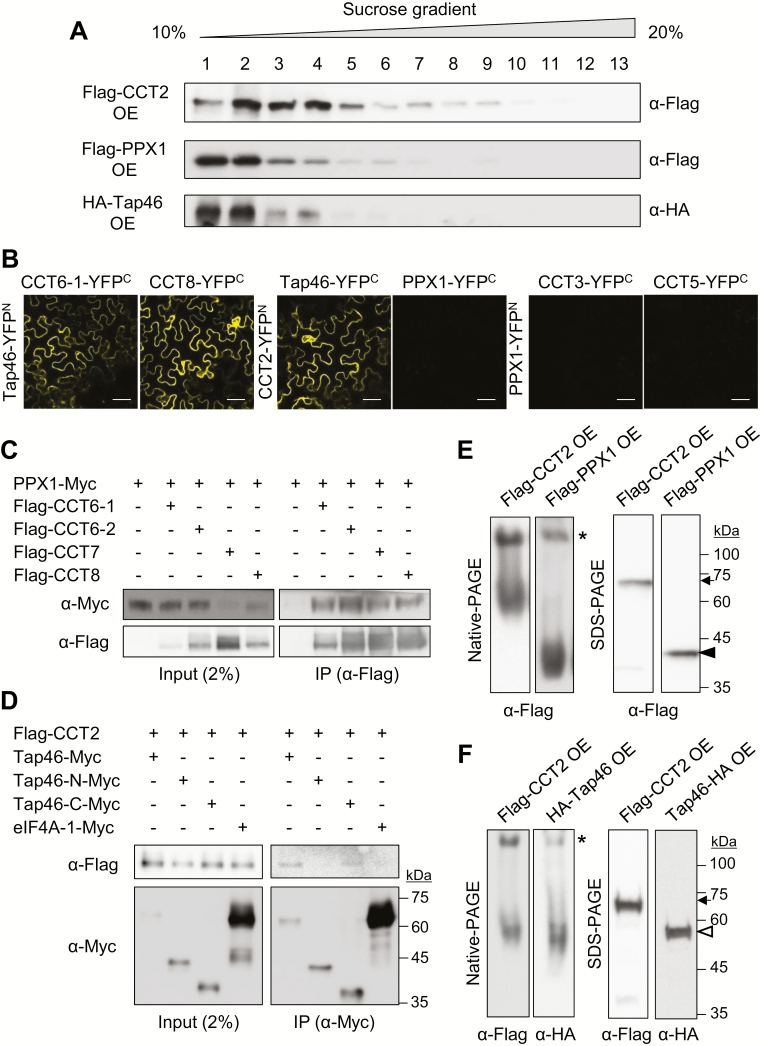
PPX1 and Tap46 interact with CCT complex. (A) Sucrose density gradient sedimentation using Flag–CCT2 OE, Flag–PPX1 OE, and HA–Tap46 OE plants. Seven-day-old seedling extracts were mounted on a 10–20% sucrose gradient for ultracentrifugation. Thirteen fractions were collected for immunoblotting with anti-Flag or anti-HA antibody. (B) BiFC. YFP signals were observed from N. benthamiana leaf epidermal cells at 2 DAI. Scale bar: 50 μm. Negative controls and protein expression are shown in Supplementary Fig. S5. (C) Co-immunoprecipitation. PPX1–Myc was co-expressed with Flag–CCT6-1, Flag–CCT6-2, Flag–CCT7, or Flag–CCT8 in N. benthamiana leaves. Immunoprecipitation was performed with anti-Flag antibody-conjugated resin, followed by immunoblotting with anti-Myc and anti-Flag antibodies. (D) Co-immunoprecipitation. Flag–CCT2 was co-expressed with Myc-tagged full-length Tap46 (Tap46–Myc), the N-terminal region (Tap46-N–Myc; 1–223 residues), the C-terminal region (Tap46-C–Myc; 224–405 residues), or eIF4A-1 in N. benthamiana leaves. Immunoprecipitation was performed with anti-Myc antibody-conjugated resin, followed by immunoblotting with anti-Myc and anti-Flag antibodies. (E) Native-PAGE and SDS-PAGE using 7-day-old Flag–CCT2 OE and Flag–PPX1 OE seedlings, followed by immunoblotting with anti-Flag antibody. The asterisk, arrow, and black arrowhead indicate the CCT complex, Flag–CCT2, and Flag–PPX1, respectively. Predicted molecular masses of Flag–CCT2 and Flag–PPX1 are ~67 and ~43 kDa, respectively. (F) Native-PAGE and SDS-PAGE using 7-day-old Flag–CCT2 OE and HA–Tap46 OE seedlings, followed by immunoblotting with anti-Flag and anti-HA antibodies. The asterisk, arrow, and white arrowhead indicate CCT complex, Flag–CCT2, and HA–Tap46, respectively. Predicted molecular mass of HA–Tap46 is ~55 kDa. (This figure is available in color at JXB online.)
To test CCT interaction with PP4c or Tap46, we performed bimolecular fluorescence complementation (BiFC) using the pSPYNE and pSPYCE vectors that contain YFPN (1–155 aa) and YFPC (156–239 aa), respectively (Walter et al., 2004). Various YFPN- and YFPC-fused proteins were expressed in N. benthamiana leaves via agroinfiltration, and leaf epidermal cells were observed for yellow fluorescence under a confocal microscope (Fig. 4B). BiFC suggested that Tap46 interacts with CCT subunits, including CCT2, CCT3, CCT5, CCT6-1, and CCT8, in the cytosol; however, PPX1 (PP4c isoform) does not interact with any of those CCT subunits, despite the high protein expression in leaves (Fig. 4B; Supplementary Figs S5, S6).
To confirm the results, we next performed co-immunoprecipitation (co-IP) (Fig. 4C, D). Myc-tagged PPX1 and Flag-tagged CCT6-1, CCT6-2, CCT7, and CCT8 were co-expressed in N. benthamiana leaves using agroinfiltration. Flag-tagged CCT subunits were immunoprecipitated from leaf extracts using anti-Flag antibody-conjugated resin, followed by immunoblotting with anti-Myc antibody to detect co-immunoprecipitated PPX1–Myc proteins. PPX1–Myc was clearly detected in all immunoprecipitates, suggesting the interaction of PPX1 with CCT subunits (Fig. 4C), despite the negative BiFC data. To confirm the data, we performed another set of co-IP experiments (see Supplementary Fig. S7). PPX1–Myc and eIF4A-1–Myc were co-expressed with Flag–CCT2 and Flag–CCT7 in N. benthamiana leaves. Immunoprecipitation was performed with anti-Myc antibody-conjugated resin, followed by immunoblotting with anti-Myc and anti-Flag antibodies to detect immunoprecipitated and co-immunoprecipitated proteins, respectively. CCT2 and CCT7 were immunoprecipitated with PPX1–Myc, but not with eIF4A-1–Myc (asterisks), despite its abundant expression and successful immunoprecipitation (Supplementary Fig. S7). These results suggest specific interactions between PPX1 and CCT subunits.
We performed co-IP with full-length Tap46, its N-terminal region (1–223 residues, Tap46-N), and its C-terminal region (224–405 residues, Tap46-C) (Fig. 4D). Myc-tagged eIF4A-1 was used as a negative control. Myc-tagged proteins were immunoprecipitated from leaf extracts using anti-Myc antibody-conjugated resin, followed by immunoblotting with anti-Flag antibody. Flag–CCT2 was immunoprecipitated with full-length Tap46 and Tap46-C proteins, but not with Tap46-N or eIF4A-1 (Fig. 4D). It has been reported that the N-terminal region of α4 interacts with PP2A catalytic subunits in mammals (LeNoue-Newton et al., 2011). These results suggest that Tap46 interacts with CCT2 via the C-terminal region.
To elucidate the association of PPX1 and Tap46 with the CCT complex, native-PAGE and immunoblotting were performed. Both Flag–PPX1 and HA–Tap46 were detected in two forms in the native gel: as a slow-migrating band coinciding with the CCT complex (asterisk) detected in Flag–CCT2 OE samples, and as a lower fast-migrating band corresponding to their mature forms (Fig. 4E, F). SDS-PAGE and immunoblotting detected the expression of CCT2, PPX1, and Tap46 proteins in the corresponding OE lines. Taken together, the data suggest that PPX1 and Tap46 interact with the CCT complex in vivo. Since proteins must be fully folded to emit fluorescence in BiFC, some substrates of CCT may not be detected by BiFC, but could be still associated with CCT complex. Thus, we next explored the possibility of PPX1 and Tap46 as substrates of CCT chaperonin.
PP4c may be a novel substrate of plant CCT complex
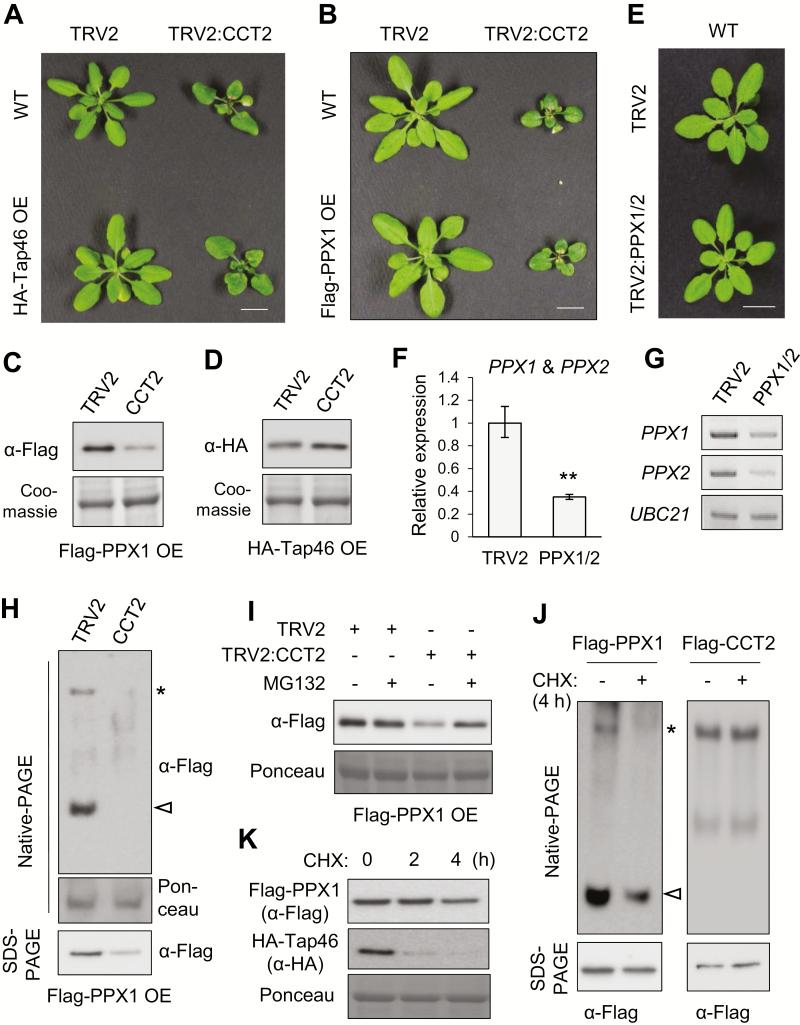
We performed VIGS with TRV2 vector and TRV2:CCT2 constructs in WT, Flag–PPX1 OE, and HA–Tap46 OE plants. There were no significant differences in CCT2 VIGS phenotypes in different plant backgrounds (Fig. 5A, B). Immunoblotting using Coomassie-stained rbcL as the control showed that CCT2 silencing significantly decreased PPX1 protein levels (Fig. 5C). However, HA–Tap46 protein levels were slightly increased or remained constant after CCT2 silencing (Fig. 5D). To examine PP4c-deficient phenotypes, we performed VIGS in Arabidopsis (Fig. 5E–G). To silence both PPX1 and PPX2 genes encoding PP4c, partial cDNA fragments of PPX1 (250 bp) and PPX2 (265 bp) were fused and inserted into the TRV2 vector. Silencing using this fusion construct reduced the PPX1/2 transcript levels to approximately 38% of the TRV2 control levels, based on RT-qPCR using primers recognizing both genes (Fig. 5F). Semi-quantitative RT-PCR with gene-specific primers detected reductions in PPX1 and PPX2 mRNA levels in PPX1/2 VIGS plants (Fig. 5G), although the plants showed no visible phenotypes under normal conditions (Fig. 5E).
Fig. 5.
PPX1 may be a substrate of plant CCT complex. (A, B) Phenotypes of CCT2 VIGS in HA–Tap46 OE (A) and Flag–PPX1 OE plants (B) at 18 DAI. Scale bar=1 cm. (C, D) Immunoblotting to detect Flag–PPX1 (C) and HA–Tap46 (D) protein levels after silencing of CCT2 (15 DAI), using anti-HA and anti-Flag antibodies, respectively. Coomassie-stained rbcL served as the loading control. (E) Phenotypes of PPX1/2 VIGS in WT plants at 18 DAI. The fusion construct of PPX1 and PPX2 cDNAs was used for VIGS. Scale bar: 1 cm. (F) RT-qPCR of PPX1 and PPX2 mRNAs in the VIGS plants shown in (E). Transcript levels are normalized to those of UBC10 mRNA, and expressed relative to those of TRV2 samples. The PCR primers recognized both PPX1 and PPX2 mRNAs. Values represent mean ±SE derived from three biological replicates (**P≤0.01). (G) Semi-quantitative RT-PCR using gene-specific primers for PPX1 and PPX2. UBC21 mRNA served as the loading control. (H) Native-PAGE and SDS-PAGE of VIGS plants in Flag–PPX1 OE background at 15 DAI. Immunoblotting was performed with anti-Flag antibody. Ponceau-stained rbcL served as the loading control. (I) Immunoblotting with anti-Flag antibody in VIGS plants with or without 4 h treatment with 20 μM MG132. (J) CHX–chase assays. Five-day-old Flag–PPX1 OE and Flag–CCT2 OE seedlings were treated with water (−CHX) or 2 mg ml−1 CHX for 4 h, and subjected to native-PAGE and SDS-PAGE, followed by immunoblotting with anti-Flag antibody. The asterisk and white arrowhead indicate the CCT complex-associated form and mature form of Flag–PPX1, respectively. (K) Effects of CHX treatments on PPX1 and Tap46 levels. Five-day-old Flag–PPX1 OE and HA–Tap46 OE seedlings were treated with water or 2 mg ml−1 CHX for 0, 2, and 4 h, and subjected to SDS-PAGE and immunoblotting with anti-Flag and anti-HA antibodies. (This figure is available in color at JXB online.)
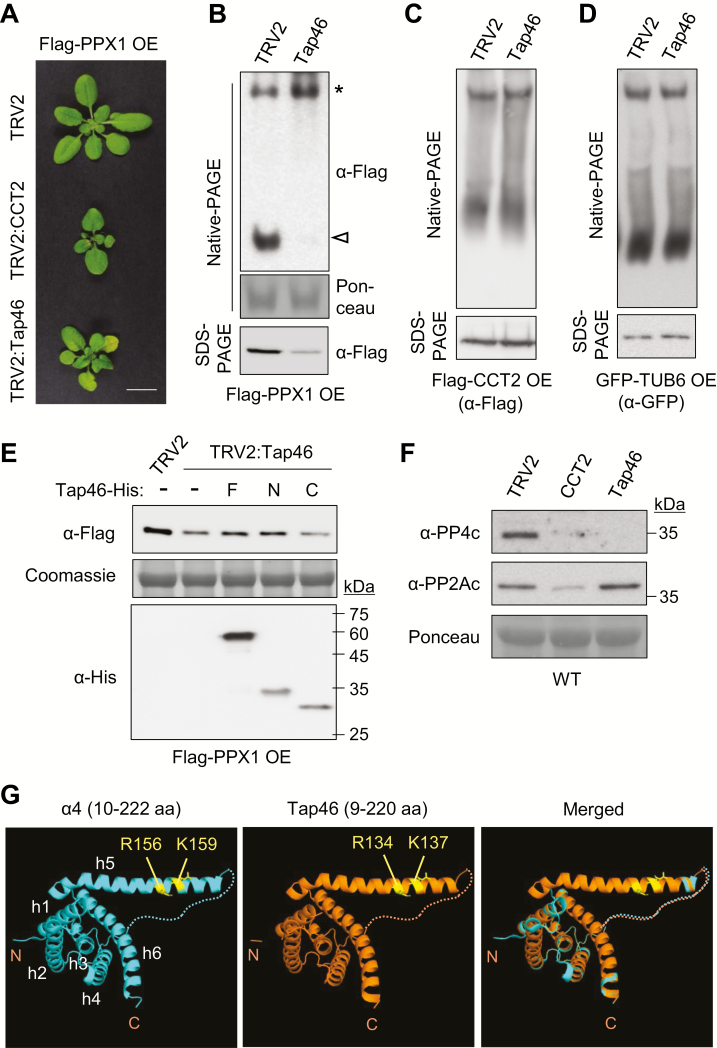
Native-PAGE and immunoblotting showed that CCT2 silencing caused the disappearance of both CCT-associated (asterisk) and mature Flag–PPX1 protein bands (white arrowhead), compared with TRV control (Fig. 5H; Supplementary Fig. S8). MG132 (20 μM) treatment for 4 h did not increase the mature Flag–PPX1 level in TRV:CCT2 plants in native-PAGE, while it produced smeary bands in the upper region of the gel (see Supplementary Fig. S8). SDS-PAGE showed that the MG132 treatment did not change the Flag–PPX1 protein level in TRV control, but increased the level, at least partially, in TRV2:CCT2 samples (Fig. 5I; Supplementary Fig. S8). These results collectively suggest that PPX1 proteins undergo degradation upon CCT deficiency. Finally, in order to demonstrate that PPX1 is a substrate of the CCT complex, we performed a CHX–chase assay using Flag–PPX1 OE seedlings. CHX treatment for 4 h decreased the levels of both CCT-associated Flag–PPX1 and its mature form in native-PAGE (Fig. 5J, left), whereas the total Flag–PPX1 protein level was relatively constant with only a slight decrease after a 4 h treatment in SDS-PAGE (Fig. 5J, K). In contrast, the amount of CCT complex remained constant following CHX treatment (Fig. 5J, right). Furthermore, the presence of PPX1 in the CCT upper band was further supported by 2D-PAGE (Supplementary Fig. S9). Flag–CCT2 was expressed in N. benthamiana leaves along with Flag–PPX1, and the proteins were resolved by native-PAGE (1D-PAGE), followed by SDS-PAGE (2D-PAGE). The gels were then subjected to immunoblotting with anti-Flag antibody. In 2D-PAGE, Flag–CCT2 (black arrowhead) and Flag–PPX1 (white arrowhead) were detected at the position of the CCT complex (Supplementary Fig. S9). Taken together, these results suggest that CCT chaperonin may mediate the folding of nascent PPX1 polypeptides. Furthermore, Tap46 appeared to associate with the CCT complex, but not as a folding substrate.
Tap46 may participate in stabilization of PP4c
Previously, yeast two-hybrid assays have suggested a strong interaction between Tap46 and PPX1 (Ahn et al., 2011). α4, the mammalian homolog of Tap46, is required for the stabilization of nascent catalytic subunits of PP2A family phosphatases (Kong et al., 2009; Jiang et al., 2013; LeNoue-Newton et al., 2016). To understand the functions of Tap46 in relation to PPX1 and CCT2, we performed VIGS of Tap46 and CCT2 in Flag–PPX1 OE plants (Fig. 6A). VIGS of Tap46 caused retarded plant growth and premature senescence. Based on native-PAGE and immunoblotting, Tap46 silencing significantly reduced mature Flag–PPX1 proteins, but consistently increased its CCT-associated form (Fig. 6B). In contrast, CCT2 silencing decreased both forms of Flag–PPX1 (Fig. 5H). SDS-PAGE revealed a decrease in total Flag–PPX1 protein levels in the TRV2:Tap46 sample, compared with those in TRV2 control (Fig. 6B). VIGS of Tap46 in Flag–CCT2 OE and GFP–TUB6 OE plants suggested that Tap46 deficiency does not inhibit CCT complex formation or β-tubulin biogenesis (Fig. 6C, D). Collectively, these results suggest that Tap46 is required for the stability of PPX1. A high turnover rate of Tap46 proteins shown by the CHX treatment (Fig. 5K) might have contributed to the reduction in the mature form of Flag–PPX1 upon CHX treatment (Fig. 5J, left).
Fig. 6.
Tap46 stabilizes PPX1 proteins. (A) VIGS phenotypes of CCT2 and Tap46 in Flag–PPX1 OE plants (15 DAI). Scale bar: 1 cm. (B) Native-PAGE and SDS-PAGE were performed using Flag–PPX1 OE plants after silencing of Tap46 (15 DAI), followed by immunoblotting with anti-Flag antibody. Ponceau-stained rbcL served as the loading control. (C, D) Native-PAGE and SDS-PAGE were performed with Flag–CCT2 OE (C) and GFP–TUB6 OE plants (D) after silencing of Tap46 (15 DAI), followed by immunoblotting with anti-Flag and anti-GFP antibodies, respectively. (E) Leaf extracts of TRV2 and Tap46 VIGS plants were incubated for 2 h with recombinant Tap46–His (F), Tap46-N–His (N), or Tap46-C–His (C) proteins. Immunoblotting was performed with anti-Flag and anti-His antibodies. Coomassie-stained rbcL served as the loading control. (F) Immunoblotting with anti-PP4c and anti-PP2Ac antibodies in CCT2 and Tap46 VIGS plants in WT background. Ponceau-stained rbcL was used as the control. (G) Computational modeling of Tap46 N-terminal region. The tertiary structure of Tap46 N-terminal region (9–220 residues) was predicted using the automated homology modeling server PHYRE2 (Protein Homology/analogY Recognition Engine V 2.0) with the N-terminal domain of α4 (10–222 residues) as template. The PDB file name for α4 N-terminal domain is 3QC1. The predicted molecular model was edited using the PyMOL molecular graphics system (version 1.1). The N- and C-terminus of each sequence input are indicated in the figure. The PP2Ac-binding residues in the extended α-helix 5 (h5) are marked. The disordered loop between h4 and h5 is shown as dotted lines. (This figure is available in color at JXB online.)
We tested if the in vitro addition of recombinant Tap46 proteins can stabilize PPX1 in leaf cell extracts (Fig. 6E). Recombinant proteins of the full-length Tap46 (Tap46-F), Tap46-N, and Tap46-C in His-tag fusion were purified from E. coli. The recombinant proteins (5 μM) were added to the leaf cell extracts of TRV2 and TRV2:Tap46 plants in the Flag–PPX1 OE background. After a 2 h incubation in native-PAGE buffer at 30°C, the whole cell extracts were subjected to immunoblotting with anti-Flag and anti-His antibodies. The TRV2:Tap46 sample contained lower amounts of Flag–PPX1, which was partially restored by the addition of Tap46-F–His and Tap46-N–His, but not by addition of Tap46-C–His. Collectively, these results suggest that Tap46 may stabilize PPX1 via protein interaction, preventing its degradation in the leaf cell extracts.
To detect changes in endogenous PP4c levels, we performed VIGS of CCT2 and Tap46 in WT Arabidopsis plants, followed by immunoblotting with anti-PP4c antibody and as a control anti-PP2Ac antibody (Fig. 6F). The endogenous PP4c protein level was significantly reduced in both CCT2 and Tap46 VIGS plants compared with TRV2 control, consistent with the results in Flag–PPX1 OE plants. This result suggests that both CCT2 and Tap46 are important for PP4c biogenesis in plants. Interestingly, cellular PP2Ac levels also decreased upon CCT2 silencing, albeit less significantly, suggesting PP2Ac as a CCT substrate candidate (Fig. 6F). However, Tap46 silencing did not change PP2Ac levels. Thus, Tap46 may not be critical for PP2Ac stability in plants, which is consistent with the results obtained from dexamethasone-inducible Tap46 RNAi in Arabidopsis (Ahn et al., 2011).
Finally, computational modeling of the Tap46 N-terminal region (9–220 residues) was performed using the automated protein homology modeling server PHYRE2 (Protein Homology/analogY Recognition Engine V 2.0; http://www.sbg.bio.ic.ac.uk/phyre2/html) with α4 N-terminal domain (10–222 residues) as template (Fig. 6G). The N-terminal domain of α4/Tap42 binds to PP2Ac with a stable secondary structure, while its C-terminal domain is intrinsically disordered and sensitive to protease degradation (Yang et al., 2007; LeNoue-Newton et al., 2011). Despite low sequence identity (~27%), the N-terminal region of Tap46, mostly composed of α-helices, was very similar in structure to that of α4 (Fig. 6G). The PP2Ac-binding residues (R156 and K159) in the extended α-helix 5 of α4 (Yang et al., 2007) are also conserved in Tap46 (R134 and K137). This structural conservation suggests that the Tap46 N-terminal region is involved in binding to catalytic subunits of PP2A family phosphatases in plants. Interestingly, Yang et al. (2007) reported that the α4/Tap42 N-terminal domain was very similar in structure to 14-3-3 and tetratricopeptide repeat proteins, which function as scaffold or adaptor proteins.
Discussion
CCT chaperonin has a stacked double-ring structure, composed of eight different subunits in each ring (Kalisman et al., 2012; Leitner et al., 2012). Phylogenetic analysis of nucleotide sequences from diverse plant species, as well as yeast and human revealed deep paralogy of the CCT subunits, suggesting that CCT evolved very early in eukaryotic evolution, likely in the common ancestor (see Supplementary Fig. S1). Among plant sequences, it was noticed that diploid plant species with larger genome size, such as rice and maize, have increased numbers of CCT subunit genes, in comparison with the nine genes in Arabidopsis (Supplementary Table S1). Archibald et al. (2001) suggested that a large expansion of subunit genes in eukaryotic CCT is driven by gene duplication, and that the duplicate CCT subunits might have coevolved with their target substrates. Recently, Joachimiak et al. (2014) reported on the structural basis of substrate recognition by CCT. CCT contacts full-length substrates combinatorially in a subunit-specific manner, ensuring the specificity and plasticity for diverse substrates. Thus, the binding site of each CCT subunit has a distinct, conserved pattern of residues specifying recognition of different substrate motifs. Collectively, these results suggest that subunit diversification positively correlates with the size of the proteome. In allotetraploid N. benthamiana and allopolyploid soybean, genome duplication and subsequent proteome expansion may have increased CCT subunit gene numbers by more than two-fold (Supplementary Table S1).
Silencing of CCT2 and CCT3 by VIGS resulted in growth arrest in Arabidopsis, with small and round leaves (Fig. 2A). Abnormal trichome branch numbers and right-handed twist of leaf petioles in the VIGS plants are characteristic phenotypes of MT defects (Fig. 2C–E), and indeed cortical MT density was significantly reduced upon CCT2 silencing, as observed upon TUA6 silencing (Fig. 3D). Furthermore, diverse biochemical experiments suggested that the plant CCT complex participates in the folding of tubulins (Fig. 3E–I). Thus, tubulins may be evolutionarily conserved substrates of CCT chaperonin in eukaryotes. Interestingly, the weak allele of cct8 with a site-specific mutation did not cause MT-related defects (Xu et al., 2011), suggesting that the mutation in the CCT8 subunit did not interfere with tubulin binding/folding by the CCT complex. Another prominent phenotype of CCT2 and CCT3 silencing was necrosis around the shoot apex (Fig. 2A, B). Since this phenotype was never observed in TRV2:TUA6 plants despite the severity of their symptoms, it may be caused by the defective folding of other CCT substrates.
Tandem affinity purification using PP4c or α4 as bait co-purified multiple CCT subunits in mammals (Gingras et al., 2005). Recently, Herzog et al. (2012) probed the PP2A network in human cells by analysing endogenous protein complexes. TAP tagging followed by chemical cross-linking and mass spectrometry supported strong interactions of CCT subunits with PP4c and α4. This analysis also identified the PP2A regulatory subunit, 2ABG, as a CCT interactor. Subsequent cryoelectron microscopy revealed the topology of CCT chaperonin interacting with its substrate 2ABG, localized to the inner cavity of the CCT ring (Herzog et al., 2012). The global protein kinase and phosphatase interaction network in budding yeast, based on mass spectrometric analysis of protein complexes, also supported CCT interaction with PP4c (Breitkreutz et al., 2010). Despite these repeated findings on the interaction of CCT with PP4c and α4, the functional significance of these interactions has not been revealed.
In this study, we demonstrated that Arabidopsis PP4c (PPX1) and Tap46 interact with individual CCT subunits and co-localized with the high-molecular-mass CCT complex in native-PAGE and sucrose density gradient sedimentation (Fig. 4). Thus, interactions of PP4c and Tap46 with the CCT complex also occur in plant cells. Furthermore, CCT deficiency significantly reduced cellular PPX1 protein levels but not Tap46 levels, and a brief CHX treatment reduced the CCT-associated forms of PPX1 (Fig. 5). These results suggest that PPX1, but not Tap46, may be a novel substrate of plant CCT. The possibility that PPX1 binds to the CCT complex for dephosphorylation is inconsistent with either the disappearance of mature PPX1 proteins upon CCT2 silencing (Fig. 5C, H) or the disappearance of CCT complex-associated PPX1 upon CHX treatment (Fig. 5J). Future structural studies may reveal unique features of PP4c that require folding by CCT chaperonin.
Tap46 interacts with PP2A family catalytic subunits, with a particularly strong interaction with PPX1 (Ahn et al., 2011). Tap46 silencing significantly decreased cellular PPX1 levels (Fig. 6B, F), but not the protein levels of PP2Ac (Fig. 6F; Ahn et al., 2011), suggesting a more stringent requirement for Tap46 by PPX1. In mammals, depletion of α4 led to the loss of all PP2A family phosphatases (Kong et al., 2009), although knockdown of α4 with a different method preferentially decreased protein levels of PP4c and PP6c, rather than PP2Ac (LeNoue-Newton et al., 2016). Based on the structure of α4 bound to the N-terminal fragment of PP2Ac, it was proposed that α4 is a scavenger chaperone that binds to and stabilizes partially folded PP2Ac for stable latency (Jiang et al., 2013). The PP2Ac–α4 interface sterically hinders a ubiquitination site, preventing proteasome-mediated degradation of PP2Ac (McConnell et al., 2010). In this study, Tap46 was critical for the maintenance of cellular PPX1 levels (Fig. 6B, F), and the negative effect on PPX1 by Tap46 deficiency was partially rescued by the exogenous addition of recombinant Tap46 proteins (Fig. 6E). These results suggest that Tap46 may stabilize PPX1 and prevent its degradation via physical interaction until the assembly of the functional PP4 complex takes place. The finding that a decrease in Tap46 proteins by CHX treatment led to a significant reduction in mature PPX1 proteins (Fig. 5J, K) may suggest the importance of Tap46 for PPX1 stability. Furthermore, it was consistently observed that the CCT complex-associated form of PPX1 increases under Tap46-depleted conditions (Fig. 6B). The reason for this is unknown, but it is tempting to speculate that Tap46 may bind to PPX1 as it exits from the CCT complex, facilitating the process as a post-chaperonin factor. Future studies are needed to address the molecular mechanisms of Tap46 function with respect to PPX1 and CCT.
We did not observe any visible plant phenotypes after the co-silencing of PPX1 and PPX2, despite repeated attempts. The unavailability of knockout mutants of PPX1 and PPX2 and the failure to generate their knockdown transgenic lines have been recently described (Kataya et al., 2017). PP4 often functions as a heterotrimeric complex, consisting of PP4c and two types of regulatory subunits (Gingras et al., 2005). Recently, two reports have suggested functions of PP4 phosphatase in plants, through studying its regulatory subunits. Su et al. (2017) reported that SMEK1/PSY2L (the Arabidopsis homolog of PP4 regulatory subunit PP4R3) interacted with PPX1 and PPX2 in the nucleus, and together dephosphorylated HYL1, a major regulator of miRNA processing. The smek1 mutants displayed dwarfism and abnormal floral organs, resembling the phenotypes of miRNA processing mutants. In another study, smek1 mutants showed growth defects and sensitivity to the DNA-damaging agent cisplatin (Kataya et al., 2017). Recent studies have revealed that PP4 is involved in various cellular processes, including DNA damage repair, histone acetylation, and cell cycle progression in mammals (Cohen et al., 2005; Zhang et al., 2005; Chowdhury et al., 2008; Lee et al., 2012; Shaltiel et al., 2014). We speculate that moderate silencing of PPX1 and PPX2 may be the reason for the lack of observed phenotypes. In future studies, it will be important to elucidate the functions of the PP4 complex associated with different regulatory subunits.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Phylogenetic tree analysis of CCT subunits.
Fig. S2. Subcellular localization of Arabidopsis CCT subunits.
Fig. S3. Characterization of Arabidopsis Flag–CCT2 overexpression (OE) and Flag–PPX1 OE plants under the control of CaMV35S promoter.
Fig. S4. Expression of Arabidopsis CCT subunit genes in estradiol-inducible TOR RNAi (es-tor1) plants.
Fig. S5. Negative controls and immunoblotting analyses for BiFC experiments shown in Fig. 4B.
Fig. S6. BiFC analyses of CCT interactions with Tap46 and PPX1.
Fig. S7. Control experiments for co-IP between PPX1 and CCT subunits.
Fig. S8. Native-PAGE and SDS-PAGE of Flag–PPX1 OE plants after CCT2 VIGS with or without MG132 treatment.
Fig. S9. Two-dimensional PAGE using N. benthamiana extracts that co-expressed Flag–CCT2 and Flag–PPX1.
Table S1. Accession and loci numbers of CCT subunit genes (A) and other genes (B) used in this study.
Table S2. List of primers used in this study.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
The authors wish to thank Dr Jen Sheen (Harvard Medical School, USA) for providing seeds of the estradiol-inducible TOR RNAi lines. This research was supported by the Cooperative Research Program for Agriculture Science & Technology Development [Project numbers PJ013212 (PMBC) and PJ013227 (SSAC)] from the Rural Development Administration, and Mid-Career Researcher Program (NRF-2016R1A2B4013180) from the National Research Foundation (NRF) of the Republic of Korea. H-KA was funded by the Global PhD fellowship (2011-0031010).
References
- Ahn CS, Ahn HK, Pai HS. 2015a Overexpression of the PP2A regulatory subunit Tap46 leads to enhanced plant growth through stimulation of the TOR signalling pathway. Journal of Experimental Botany 66, 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn CS, Han JA, Lee HS, Lee S, Pai HS. 2011. The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. The Plant Cell 23, 185–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn HK, Kang YW, Lim HM, Hwang I, Pai HS. 2015b Physiological functions of the COPI complex in higher plants. Molecules and Cells 38, 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM, Blouin C, Doolittle WF. 2001. Gene duplication and the evolution of group II chaperonins: implications for structure and function. Journal of Structural Biology 135, 157–169. [DOI] [PubMed] [Google Scholar]
- Bertrand S, Barthelemy I, Oliva MA, Carrascosa JL, Andreu JM, Valpuesta JM. 2005. Folding, stability and polymerization properties of FtsZ chimeras with inserted tubulin loops involved in the interaction with the cytosolic chaperonin CCT and in microtubule formation. Journal of Molecular Biology 346, 319–330. [DOI] [PubMed] [Google Scholar]
- Breitkreutz A, Choi H, Sharom JR, et al. 2010. A global protein kinase and phosphatase interaction network in yeast. Science 328, 1043–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Liu Y, Dinesh-Kumar SP. 2006. Efficient virus-induced gene silencing in Arabidopsis. Plant Physiology 142, 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camasses A, Bogdanova A, Shevchenko A, Zachariae W. 2003. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Molecular Cell 12, 87–100. [DOI] [PubMed] [Google Scholar]
- Chen J, Peterson RT, Schreiber SL. 1998. α4 associates with protein phosphatases 2A, 4, and 6. Biochemical and Biophysical Research Communications 247, 827–832. [DOI] [PubMed] [Google Scholar]
- Chen X, Wu S, Liu Z, Friml J. 2016. Environmental and endogenous control of cortical microtubule orientation. Trends in Cell Biology 26, 409–419. [DOI] [PubMed] [Google Scholar]
- Cho HK, Ahn CS, Lee HS, Kim JK, Pai HS. 2013. Pescadillo plays an essential role in plant cell growth and survival by modulating ribosome biogenesis. The Plant Journal 76, 393–405. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Xu X, Zhong X, Ahmed F, Zhong J, Liao J, Dykxhoorn DM, Weinstock DM, Pfeifer GP, Lieberman J. 2008. A PP4-phosphatase complex dephosphorylates γ-H2AX generated during DNA replication. Molecular Cell 31, 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen PT, Philp A, Vázquez-Martin C. 2005. Protein phosphatase 4 – from obscurity to vital functions. FEBS Letters 579, 3278–3286. [DOI] [PubMed] [Google Scholar]
- Cong Y, Schröder GF, Meyer AS, et al. 2012. Symmetry-free cryo-EM structures of the chaperonin TRiC along its ATPase-driven conformational cycle. The EMBO Journal 31, 720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C, Stirling PC, McCormack EA, Filmore H, Paul A, Brost RL, Costanzo M, Boone C, Leroux MR, Willison KR. 2008. The interaction network of the chaperonin CCT. The EMBO Journal 27, 1827–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düvel K, Broach JR. 2004. The role of phosphatases in TOR signaling in yeast. Current Topics in Microbiology and Immunology 279, 19–38. [DOI] [PubMed] [Google Scholar]
- Feldman DE, Thulasiraman V, Ferreyra RG, Frydman J. 1999. Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Molecular Cell 4, 1051–1061. [DOI] [PubMed] [Google Scholar]
- Freund A, Zhong FL, Venteicher AS, Meng Z, Veenstra TD, Frydman J, Artandi SE. 2014. Proteostatic control of telomerase function through TRiC-mediated folding of TCAB1. Cell 159, 1389–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU. 1992. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. The EMBO Journal 11, 4767–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S, Pytela J, Hotta T, et al. 2013. An atypical tubulin kinase mediates stress-induced microtubule depolymerization in Arabidopsis. Current Biology 23, 1969–1978. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Caballero M, Zarske M, Sanchez A, Hazbun TR, Fields S, Sonenberg N, Hafen E, Raught B, Aebersold R. 2005. A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Molecular & Cellular Proteomics 4, 1725–1740. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. 2011. Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332. [DOI] [PubMed] [Google Scholar]
- Herzog F, Kahraman A, Boehringer D, et al. 2012. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352. [DOI] [PubMed] [Google Scholar]
- Himmelspach R, Nick P, Schäfer E, Ehmann B. 1997. Developmental and light-dependent changes of the cytosolic chaperonin containing TCP-1 (CCT) subunits in maize seedlings, and the localization in coleoptiles. The Plant Journal 12, 1299–1310. [DOI] [PubMed] [Google Scholar]
- Horwich AL, Fenton WA, Chapman E, Farr GW. 2007. Two families of chaperonin: physiology and mechanism. Annual Review of Cell and Developmental Biology 23, 115–145. [DOI] [PubMed] [Google Scholar]
- Ishida T, Kaneko Y, Iwano M, Hashimoto T. 2007. Helical microtubule arrays in a collection of twisting tubulin mutants of Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 104, 8544–8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Stanevich V, Satyshur KA, Kong M, Watkins GR, Wadzinski BE, Sengupta R, Xing Y. 2013. Structural basis of protein phosphatase 2A stable latency. Nature Communications 4, 1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachimiak LA, Walzthoeni T, Liu CW, Aebersold R, Frydman J. 2014. The structural basis of substrate recognition by the eukaryotic chaperonin TRiC/CCT. Cell 159, 1042–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisman N, Adams CM, Levitt M. 2012. Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. Proceedings of the National Academy of Sciences, USA 109, 2884–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataya ARA, Creighton MT, Napitupulu TP, Sætre C, Heidari B, Ruoff P, Lillo C. 2017. PLATINUM SENSITIVE 2 LIKE impacts growth, root morphology, seed set, and stress responses. PLoS One 12, e0180478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Cho SK, Oh TR, Ryu MY, Yang SW, Kim WT. 2017. MPSR1 is a cytoplasmic PQC E3 ligase for eliminating emergent misfolded proteins in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 114, E10009–E10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik A, Ehrhardt DW, Kirik V. 2012. TONNEAU2/FASS regulates the geometry of microtubule nucleation and cortical array organization in interphase Arabidopsis cells. The Plant Cell 24, 1158–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Grini PE, Mathur J, Klinkhammer I, Adler K, Bechtold N, Herzog M, Bonneville JM, Hülskamp M. 2002a The Arabidopsis TUBULIN-FOLDING COFACTOR A gene is involved in the control of the α/β-tubulin monomer balance. The Plant Cell 14, 2265–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Mathur J, Grini PE, Klinkhammer I, Adler K, Bechtold N, Herzog M, Bonneville JM, Hülskamp M. 2002b Functional analysis of the tubulin-folding cofactor C in Arabidopsis thaliana. Current Biology 12, 1519–1523. [DOI] [PubMed] [Google Scholar]
- Kong M, Ditsworth D, Lindsten T, Thompson CB. 2009. α4 is an essential regulator of PP2A phosphatase activity. Molecular Cell 36, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Goodarzi AA, Adelmant GO, Pan Y, Jeggo PA, Marto JA, Chowdhury D. 2012. Phosphoproteomic analysis reveals that PP4 dephosphorylates KAP-1 impacting the DNA damage response. The EMBO Journal 31, 2403–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner A, Joachimiak LA, Bracher A, et al. 2012. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 20, 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeNoue-Newton ML, Wadzinski BE, Spiller BW. 2016. The three Type 2A protein phosphatases, PP2Ac, PP4c and PP6c, are differentially regulated by Alpha4. Biochemical and Biophysical Research Communications 475, 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeNoue-Newton M, Watkins GR, Zou P, Germane KL, McCorvey LR, Wadzinski BE, Spiller BW. 2011. The E3 ubiquitin ligase- and protein phosphatase 2A (PP2A)-binding domains of the Alpha4 protein are both required for Alpha4 to inhibit PP2A degradation. The Journal of Biological Chemistry 286, 17665–17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, Martín-Benito J, Ritco-Vonsovici M, Grantham J, Hynes GM, Willison KR, Carrascosa JL, Valpuesta JM. 2000. Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. The EMBO Journal 19, 5971–5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez T, Dalton K, Frydman J. 2015. The mechanism and function of group II chaperonins. Journal of Molecular Biology 427, 2919–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin VF, Leroux MR, Stirling PC. 2010. Quality control of cytoskeletal proteins and human disease. Trends in Biochemical Sciences 35, 288–297. [DOI] [PubMed] [Google Scholar]
- Marks MD, Betancur L, Gilding E, Chen F, Bauer S, Wenger JP, Dixon RA, Haigler CH. 2008. A new method for isolating large quantities of Arabidopsis trichomes for transcriptome, cell wall and other types of analyses. The Plant Journal 56, 483–492. [DOI] [PubMed] [Google Scholar]
- Mathur J, Chua NH. 2000. Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. The Plant Cell 12, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JL, Watkins GR, Soss SE, Franz HS, McCorvey LR, Spiller BW, Chazin WJ, Wadzinski BE. 2010. Alpha4 is a ubiquitin-binding protein that regulates protein serine/threonine phosphatase 2A ubiquitination. Biochemistry 49, 1713–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AS, Gillespie JR, Walther D, Millet IS, Doniach S, Frydman J. 2003. Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell 113, 369–381. [DOI] [PubMed] [Google Scholar]
- Moser M, Schäfer E, Ehmann B. 2000. Characterization of protein and transcript levels of the chaperonin containing tailless complex protein-1 and tubulin during light-regulated growth of oat seedlings. Plant Physiology 124, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz IG, Yébenes H, Zhou M, et al. 2011. Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nature Structural & Molecular Biology 18, 14–19. [DOI] [PubMed] [Google Scholar]
- Pujol G, Baskin TI, Casamayor A, Cortadellas N, Ferrer A, Ariño J. 2000. The Arabidopsis thaliana PPX/PP4 phosphatases: molecular cloning and structural organization of the genes and immunolocalization of the proteins to plastids. Plant Molecular Biology 44, 499–511. [DOI] [PubMed] [Google Scholar]
- Reissmann S, Joachimiak LA, Chen B, Meyer AS, Nguyen A, Frydman J. 2012. A gradient of ATP affinities generates an asymmetric power stroke driving the chaperonin TRIC/CCT folding cycle. Cell Reports 2, 866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaltiel IA, Aprelia M, Saurin AT, Chowdhury D, Kops GJPL, Voest EE, Medema RH. 2014. Distinct phosphatases antagonize the p53 response in different phases of the cell cycle. Proceedings of the National Academy of Sciences, USA 111, 7313–7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Li Z, Cheng J, Li L, Zhong S, Liu L, Zheng Y, Zheng B. 2017. The protein phosphatase 4 and SMEK1 complex dephosphorylates HYL1 to promote miRNA biogenesis by antagonizing the MAPK cascade in Arabidopsis. Developmental Cell 41, 527–539.e5. [DOI] [PubMed] [Google Scholar]
- Szymanski D. 2002. Tubulin folding cofactors: half a dozen for a dimer. Current Biology 12, R767–R769. [DOI] [PubMed] [Google Scholar]
- Tian J, Han L, Feng Z, Wang G, Liu W, Ma Y, Yu Y, Kong Z. 2015. Orchestration of microtubules and the actin cytoskeleton in trichome cell shape determination by a plant-unique kinesin. eLife 4, e09351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinidad AG, Muller PA, Cuellar J, Klejnot M, Nobis M, Valpuesta JM, Vousden KH. 2013. Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Molecular Cell 50, 805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, et al. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang X, Jiang Y. 2003. Interaction with Tap42 is required for the essential function of Sit4 and type 2A phosphatases. Molecular Biology of the Cell 14, 4342–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D, Flügge UI. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Analytical Biochemistry 138, 141–143. [DOI] [PubMed] [Google Scholar]
- Xu XM, Wang J, Xuan Z, Goldshmidt A, Borrill PG, Hariharan N, Kim JY, Jackson D. 2011. Chaperonins facilitate KNOTTED1 cell-to-cell trafficking and stem cell function. Science 333, 1141–1144. [DOI] [PubMed] [Google Scholar]
- Yam AY, Xia Y, Lin HT, Burlingame A, Gerstein M, Frydman J. 2008. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nature Structural & Molecular Biology 15, 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Roe SM, Prickett TD, Brautigan DL, Barford D. 2007. The structure of Tap42/α4 reveals a tetratricopeptide repeat-like fold and provides insights into PP2A regulation. Biochemistry 46, 8807–8815. [DOI] [PubMed] [Google Scholar]
- Yébenes H, Mesa P, Muñoz IG, Montoya G, Valpuesta JM. 2011. Chaperonins: two rings for folding. Trends in Biochemical Sciences 36, 424–432. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ozawa Y, Lee H, Wen YD, Tan TH, Wadzinski BE, Seto E. 2005. Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes & Development 19, 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.