Abstract
Recent studies have highlighted the role of gene networks in disease biology. To formally assess this, we constructed a broad set of pathway, network, and pathway+network annotations and applied stratified LD score regression to 42 diseases and complex traits (average N = 323K) to identify enriched annotations. First, we analyzed 18,119 biological pathways. We identified 156 pathway-trait pairs whose disease enrichment was statistically significant (FDR < 5%) after conditioning on all genes and 75 known functional annotations (from the baseline-LD model), a stringent step that greatly reduced the number of pathways detected; most significant pathway-trait pairs were previously unreported. Next, for each of four published gene networks, we constructed probabilistic annotations based on network connectivity. For each gene network, the network connectivity annotation was strongly significantly enriched. Surprisingly, the enrichments were fully explained by excess overlap between network annotations and regulatory annotations from the baseline-LD model, validating the informativeness of the baseline-LD model and emphasizing the importance of accounting for regulatory annotations in gene network analyses. Finally, for each of the 156 enriched pathway-trait pairs, for each of the four gene networks, we constructed pathway+network annotations by annotating genes with high network connectivity to the input pathway. For each gene network, these pathway+network annotations were strongly significantly enriched for the corresponding traits. Once again, the enrichments were largely explained by the baseline-LD model. In conclusion, gene network connectivity is highly informative for disease architectures, but the information in gene networks may be subsumed by regulatory annotations, emphasizing the importance of accounting for known annotations.
Keywords: pathway analysis, network analysis, gene network, network connectivity, hub genes, pathway, heritability enrichment, functional annotations, genetic architecture, baseline LD
Introduction
Human diseases and complex traits are heritable and highly polygenic, potentially involving a large number of disease-associated genes connected by dense cellular networks.1, 2, 3, 4, 5 Recent work has employed several approaches to infer gene interaction networks, including protein-protein interaction networks,6, 7, 8 tissue-specific co-expression networks,9, 10 and tissue-specific regulatory networks.11, 12, 13 An appealing extension of traditional genome-wide association studies (GWASs) is to identify genes and gene pathways associated with disease by leveraging gene networks and network connectivity between disease-associated genes.9, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22 However, despite considerable progress on inferring gene interaction networks and applying newly developed methods for network connectivity-informed GWASs to identify specific genes and gene pathways associated to disease, an overall assessment and interpretation of the contribution of gene networks to the genetic architecture of disease has remained elusive. In particular, the extent to which this contribution can be explained by disease enrichments of known functional annotations23, 24, 25, 26, 27, 28 is unknown.
Here, we sought to answer three questions. First, what is the contribution of disease-associated gene pathways2, 14, 29, 30, 31, 32, 33, 34, 35, 36, 37 to disease heritability, irrespective of network connectivity? Second, what is the contribution of genes with high network connectivity in known gene networks8, 9, 10, 12 to disease heritability? Third, what is the contribution of genes with high network connectivity to disease-associated gene pathways to disease heritability?
To answer these questions, we constructed a broad set of pathway, network, and pathway+network annotations. The pathway annotations were constructed from known gene pathways by including 100 kb windows around each gene; the network annotations were constructed by quantifying network connectivity using closeness centrality, a measure of how close a gene is to other genes in the network;38, 39 and the pathway+network annotations were constructed by annotating genes with high network connectivity to the input pathway, again quantified using closeness centrality. We applied stratified LD score regression23, 24 to quantify the contribution of the pathway, network, and pathway+network annotations to disease heritability. We conditioned our analyses on all genes and on the baseline-LD model, which includes a broad set of coding, conserved, regulatory, and LD-related annotations.23 In each case, we compared results before and after conditioning on the baseline-LD model, to assess the extent to which the disease enrichments that we identified could be explained by known functional annotations.
Material and Methods
Genomic Annotations and the Baseline-LD Model
We define a genomic annotation as an assignment of a numeric value to each SNP in a predefined reference panel (e.g., 1000 Genomes;40 see Web Resources). Continuous-valued annotations can have any real value. Probabilistic annotations can have any real value between 0 and 1. Binary annotations can have value 0 or 1 only. A binary annotation can be viewed as a subset of SNPs (the set of SNPs with annotation value 1). Annotations that correspond to known or predicted function are referred to as functional annotations.
The baseline-LD model23 (v.1.1) contains 75 functional annotations (see Web Resources). These annotations include binary coding, conserved, and regulatory annotations (e.g., promoter, enhancer, histone marks, transcription factor [TF] binding sites) and continuous-valued linkage disequilibrium (LD)-related annotations.
Excess Overlap between Binary and/or Probabilistic Annotations
To study which annotation(s) from the baseline-LD model captures information of focal annotation, excess (fold) overlap of SNPs in annotations can be computed for any pair of binary and/or probablistic annotations. We define excess (fold) overlap between a pair of annotations as the fraction of overlap between the annotations divided by the amount of overlap expected by chance, computed as the following:
| (Equation 1) |
where M is the total number of SNPs (5,961,159). When there is excess overlap, the excess fold overlap is >1; when there is depletion, the excess fold overlap is <1. We did not compute excess overlap for continuous-valued annotations that are not probabilistic (e.g., those that can have negative values). A separate definition of excess overlap between gene sets is provided below.
Effect Size and Enrichment Metrics Estimated by S-LDSC
We used stratified LD score regression (S-LDSC23, 24) to estimate the enrichment and the standardized effect size of an annotation. Let acj represent the annotation value of the SNP j for the annotation c. S-LDSC assumes the variance of per normalized genotype effect sizes is a linear additive contribution to the annotation c:
| (Equation 2) |
where is the per-SNP contribution of the annotation c. S-LDSC estimates using the following equation:
| (Equation 3) |
where N is the sample size of the GWAS and is the LD score of the SNP j to the annotation c. The LD score is computed as follow: where rjk is the correlation between the SNPs j and k. The standardized effect size , the proportionate change in per-SNP heritability associated with a one standard deviation increase in the value of the annotation (conditional on all the other annotations in the model), is defined as the following by a previous study:23
| (Equation 4) |
where sd(C) is the standard deviation of the annotation c, is the estimated SNP-heritability, and M is the number of variants used to compute (in our experiment, M is equal to 5,961,159). Unlike enrichment, quantifies effects that are unique to the focal annotation.
Enrichment of the annotation is the fraction of heritability explained by SNPs in the annotation divided by the proportion of SNPs in the annotation. The definition of enrichment could be extended to continuous annotations23, 41 as the following:
| (Equation 5) |
where is the heritability captured by the cth annotation. When the annotation is enriched for trait heritability, the enrichment is >1; the overlap is greater than one would expect given the trait heritability and the size of the annotation.
The significance for enrichment for each annotation is computed using the block jackknife as mentioned in previous studies.24, 41, 42 The significance for the effect size for each annotation, as mentioned in previous studies,23, 41 is computed as , assuming that follows a normal distribution with zero mean and unit variance.
In all our analyses, we used the European samples in 1000G40 (see Web Resources) as reference SNPs. Regression SNPs were obtained from HapMap 343 (see Web Resources). SNPs with marginal association statistics >80 and SNPs in the major histocompatibility complex (MHC) region were excluded. Unless stated otherwise, we included the baseline-LD model23 in all primary analyses using S-LDSC, both to minimize the risk of bias in enrichment estimates due to model mis-specification23, 24 and to estimate effect sizes conditional on known functional annotations.
Pathway Annotations
We define a pathway as a set of genes (gene set). We considered 18,119 pathways from five sources: 2,118 biological pathways from the BioSystem (BS) database44 (which includes pathways from BioCyc,45 Kyoto Encyclopedia of Genes and Genomes [KEGG],46 Pathway Interaction Database [PID],47 REACTOME,48 WikiPathways49), 1,927 biological pathways from the Pathway Commons (PC) database50 (which includes pathways from HumanCyc,51 Integrating Network Objects with Hierarchies [INOH],52 KEGG,46 PANTHER,53 PID,47 REACTOME,48 SMPDB,54 NetPath55), 7,209 protein-protein interaction gene sets from the InWeb database,8 3,903 mouse phenotype gene sets from the Mouse Genome Informatics (MGI; sets of genes whose orthologs are associated to mouse phenotypes) database56 (i.e., sets of genes whose orthologs are associated to mouse phenotypes), and 2,961 gene ontology gene sets from the Genome Ontology (GO) database.57 This set of pathways, which contain at least 10 genes and at most 500 genes (consistent with previous studies31, 37), significantly overlap with mSigDB58 and pathways analyzed in a previous study.31 The complete list of pathways is provided in Table S1, and a histogram of the number of genes in pathways from each of the five sources is provided in Figure S1.
For each of 18,119 pathways, we constructed a binary pathway annotation by annotating a value of 1 for variants around the protein-coding genes in a given pathway (±100 kb as in previous work37, 42) and 0 for all other variants. To evaluate the contribution of each pathway to disease/trait heritability, we applied S-LDSC23, 24 to 18,119 pathway annotations across 42 independent diseases and complex traits (average N = 323K; including 30 UK Biobank traits; see Table S2). We conditioned on the 75 functional annotations from the baseline-LD model23 (which includes a broad set of coding, conserved, regulatory, and LD-related annotations), as well as an “all-genes” annotation representing the set of all 19,031 protein-coding genes (±100 kb). We removed 129 pathway-trait pairs whose annotated SNPs are less than 0.02% of the reference genome (European samples from the 1000 Genomes Project;40 see Web Resources) as S-LDSC is not well equipped for annotations that span very small proportion of the genome.
For each of 760,869 pathway-trait pairs (roughly 18,119 pathways × 42 traits), we assessed the statistical significance of the pathway annotations based on global FDR < 5% on the pathway annotation’s standardized effect size p value, defined as the proportionate change in per-SNP heritability associated to a one standard deviation increase in the value of the annotation, conditioned on other annotations included in the model. We note that controlling FDR for each trait and for all traits did not make a major difference in the number of identified enriched pathway-trait pairs. We further note that our choice of two-tailed test on the significance is conservative, partially attributing to the reduced number of significantly associated pathway-trait pairs. Among significantly enriched pathway-trait pairs, we calculated a pairwise correlation for every pair of annotations and retained the more significant pathway for correlated pathways with r ≥ 0.5 (as in a previous study38). If correlated pathways were enriched for different traits, we retained both pathways.
For each of 156 significantly enriched pathway-trait pairs, we also constructed pathway annotations excluding genes implicated by GWAS. First, we downloaded all GWAS associations from the GWAS Catalog (see Web Resources); we restricted to significant associations (p value ≤ 5e−8). For each of the 34 traits, we defined genes implicated by GWASs by including genes mapped to the lead SNP; if the SNP was intergenic, we included the nearest upstream and downstream genes. (We note that the nearest gene might not be the correct target gene.59) Then, for each of the 156 enriched pathway-trait pairs, we removed trait-specific GWAS significant genes (5% of the genes, on average across 156 pathway-trait pairs) and rebuilt our pathway annotations with a 100 kb window. For pathways significant for multiple traits, we built unique pathway annotations excluding genes implicated by GWAS for the corresponding traits.
Gene Networks and Data Processing
A gene network is defined by an edge weight (which we normalized to lie between 0 and 1) for each pair of genes, representing their connectivity in the network. We considered four gene networks: two co-expression networks (Saha10 and Greene9), one protein-protein interaction network (InWeb8), and one regulatory network (Sonawane12). We note that the Sonawane regulatory network contains nonzero edge weights only for pairs of genes in which at least one of the genes is a known TF.
We processed gene networks into a uniform format. Each gene network is represented by an N × 3 matrix, where N is the number of edges in the network. Three columns represent gene 1, gene 2, and the edge weight between gene 1 and gene 2. We used Entrez IDs as gene identifiers. We used the Ensembl biomart tool (GRCh37 assembly; see Web Resources) for gene identifier conversion. We note that the Saha and Sonawane networks can have negative edge weights. These were transformed to positive edge weights (see below) to avoid calculating connectivity metrics on negative edge weights. In all analyses, we considered protein-coding genes only.
Saha Network
We downloaded Saha transcriptome-wide networks (TWN) for each of 16 tissues (see Web Resources). (We did not consider the Saha tissue-specific networks [TSN] in our primary analyses, as they contained very few edges and small numbers of genes for computing connectivity metrics.) We collapsed isoforms by converting Ensembl transcript to Entrez gene ID; no conflicting edges (multiple non-zero edges between a pair of mapped genes) were found. We treated all edges equally regardless edge types (TE-TE [total expression], TE-IR [isoform ratio], or IR-IR). We transformed negative edge weights to positive edge weights by taking the absolute value.
Greene Network
We downloaded top edges gene networks for each of 144 tissues (see Web Resources). These networks contain edges with evidence supporting a tissue-specific functional interactions. We used Greene networks as downloaded without any further modification.
InWeb Network
We downloaded the InWeb protein-protein interaction network (v.20160912; see Web Resources). We re-formated the network to follow the consistent format as other gene networks with three columns (Entrez ID 1, Entrez ID 2, edge weight) and used the confidence score between a pair of genes as the edge weight.
Sonawane Network
We downloaded Sonawane networks for each of 38 tissues (see Web Resources). We transformed edge weights using the formula to avoid negative edge weights as stated in Sonawane et al.,12 downweighting large negative edge weights. We intersected “net” and “nets” to obtain edge weights of tissue-specific edges and concatenated with a set of edges to construct three-column matrix (TF, gene, weight). Then, we converted genes to Entrez gene ID.
Network Annotations
For each network, we constructed seven different probabilistic annotations based on the following network connectivities (centralities, i.e., how connected the gene is to other genes in the network), which we computed using Graph-tool (see Web Resources).
Let G be a weighted, undirected graph with a set of vertices (V) and a set of edges (E).
Closeness centrality (v) means how close gene v is to all other genes in the network. It is defined as where dvw is the weighted distance from v to w. If there is no path between two genes, the distance of zero is used.
Degree (v) is the number of vertices connected to v; i.e., the number of neighboring genes for each gene in the graph.
Maximum edge weight (v) is the maximum of the weights of all edges connected to v.
Sum edge weight (v) is the sum of the weights of all edges connected to v.
Betweenneses centrality (v) is the number of shortest paths that pass through gene v. It is defined as where is the number of shortest paths from node u to w that pass through v, and is the total number of shortest paths from u to w.
Eigenvector centrality (v) is as follows: intuitively, the eigencentrality of v is proportional to the sum of the centralities of its neighbors.60 It is defined as the solution of of , where x is the eigenvector of the weighted adjacency matrix A with the largest eigenvalue λ.
Pagerank (v) is similar to eigenvector centrality, except it contains a damping factor, the probability that the person who randomly visit genes will continue, under the assumption that more important genes are more likely to receive more visits. It is defined as where d is a damping factor, N(v) are the in-neighbors of v, and is the out-degree of u. Because G is an undirected graph, pagerank treats it as a directed graph by making edges bidirectional.
After computing network connectivities for all genes that exist in a network, we linearly transformed scores to lie between 0 to 1 and annotated variants around genes (±100 kb). When a variant is spanned by multiple genes with the 100 kb window, we assigned the maximum connectivity score. For each of 168 network-trait pairs (4 networks × 42 traits), we applied S-LDSC and assessed the statistical significance of the network annotation’s conditioned on other annotations. We also computed the enrichment, which naturally extends to probabilistic annotations.41 As a secondary analysis, we constructed network annotations with different window sizes (±10 kb and 1 Mb).
Comparison of Closeness Centrality to 18 Gene Sets
To compare closeness centrality to other metrics that quantify the biological importance of each gene, we considered 18 gene sets that reflect a broad range of gene essentiality metrics61 (see Web Resources). The 18 gene sets are provided in Table S3 and briefly described below; the number of genes corresponds to protein-coding genes with an Entrez ID.
All genes: 19,031 genes with protein product according to HGNC62 (HUGO Gene Nomenclature Committee) that have an Entrez ID.
MGI essential genes:63, 64, 65 2,371 genes for which homozygous knockout in mice results in pre-, peri-, or post-natal lethality.
Autosomal-dominant genes:66, 67 698 genes among OMIM disease genes that are deemed to follow autosomal-dominant inheritance.
Haploinsufficient genes:68 174 genes of severe, moderate, and mild haploinsufficiency, where having only a single functioning copy of a gene is not enough for normal function.
High pLI genes:68 3,104 loss-of-function (LoF) genes with pLI > 0.9, i.e., strongly depleted for protein-truncating variants.
High shet genes:69 2,853 constrained genes with shet > 0.1, i.e., strong selection against protein-truncating variants.
High Phi genes:70 588 LoF-constrained genes with probability of haploinsufficiency (Phi) > 0.95.
High missense Z genes:71 1,440 constrained genes strongly depleted for missense mutations, with exp_syn ≥ 5, syn_z_sign < 3.09, and mis_z_sign > 3.09, as retrieved in Lek et al.68
ClinVar genes:72 5,428 genes with a pathogenic or likely pathogenic variant with no conflict among studies.
OMIM disease genes:73 2,266 genes deposited in the Online Mendelian Inheritance in Man (OMIM), as retrieved in Petrovski et al.74
GWAS nearest genes:75 6,271 genes nearest to peak GWAS significant loci (p value ≤ 5e−8) in the GWAS Catalog.
Transcription factors:76 1,610 human transcription factors.
DrugBank genes:77 373 genes whose protein products are human targets of FDA-approved drugs with known mechanisms of action.
High EDS genes:78 2,664 genes among top 3,000 genes highly scored in enhancer domain score (EDS).
Olfactory receptors:79 369 olfactory receptor genes.
eQTL-deficient genes:80 604 genes with no significant variant-gene association in all 48 tissues in GTEx v.7 single-tissue cis-eQTL data.
Genes with more independent SNPs: 1,884 genes, defined as the top 10% of genes ranked based on the number of independent (r2 < 0.1) SNPs near the gene (±100 kb) relative to the length of the gene. We created PLINK files from 1000 Genomes40 European Phase 3 reference genome individuals (see Web Resources) and SNPs with minor allele frequency (MAF) ≥ 5%. We filtered SNPs by applying LD-pruning to retain SNPs with r2 < 0.1 using PLINK (see Web Resources) with the window size of 50 kp and the step size of 10 kp. We excluded SNPs in the major histocompatibility complex (MHC) region, in our other analyses. We computed the number of pruned independent SNPs near each protein-coding gene (±100 kb) and divided by the length of the gene. We obtained highly correlated gene sets with different r2 thresholds (< 0.3 and 0.5) and using a 10 kb window.
Genes with more SNPs: 1,884 genes, defined as the top 10% of genes ranked based on the total number of SNPs near the gene (±100 kb) relative to the length of the gene. We obtained highly correlated gene sets using a 10 kb window.
We extend the definition of excess overlap of annotations provided earlier defined earlier to a definition of excess overlap between gene sets. Let “gene set 1” denote one of the 18 gene sets defined above and “gene set 2” denote a given decile bin of closeness centrality for a given network. We define:
| (Equation 6) |
where = and = The standard error for the excess overlap is similarly scaled:
| (Equation 7) |
When there is excess overlap, the excess fold overlap is >1; when there is depletion, the excess fold overlap is <1. More generally, excess overlap can be computed for any pair of gene sets.
Assessing whether Genes with High Closeness Centrality Are Heavily Regulating Other Genes or Are Heavily Regulated by Other Genes
To assess whether genes with high closeness centrality are heavily regulating other genes or are heavily regulated by other genes, we performed three analyses. First, we assessed the excess overlap of 1,610 known human TFs76 in the top decile of closeness centrality for each network.
Second, we used the ENCODE ChIP-Seq Significance Tool81 (see Web Resources) to assess the excess overlap of TF binding sites in the promoters of high closeness centrality genes (top 10% of genes for each gene network analyzed). We considered 220 TFs from the union of all 91 available ENCODE28 cell lines. We used all protein-coding genes with an Entrez ID as background regions and defined promoters based on ±1 kb from the TSS. We report TFs that have significiant excess overlap with the promoters of high closeness centrality genes (Benjamini-Hochberg adjusted p value < 0.05) and calculated the significance of the excess overlap using the hypergeometric test. In addition, for each network, we computed excess overlap of the ENCODE TF binding sites annotations28 (from the baseline-LD model) in 10 deciles of closeness centrality (±100 kb).
Third, we used DAVID82 (see Web Resources) to assess GO enrichments of high closeness centrality genes (top decile) for each of four gene networks. We used all protein-coding genes with an Entrez ID as a background set. We considered three GO categories: biological process, cellular component, and molecular function. We reported significant GO terms (Benjamini-Hochberg adjusted p value < 0.05).
Assessing the Impact of Noise in Gene Networks
We performed three analyses to assess the impact of noise in gene networks on our results. First, we performed a network perturbation analysis by randomly removing a subset of edges (from 10% to 90% with 10% increment) using an established protocol.83 We performed five separate perturbation analyses for each value of the proportion of edges removed. For each network with edges removed, we computed network connectivity metrics and applied S-LDSC to estimate disease heritability enrichment and .
Second, we applied the diffusion state distance algorithm (DSD;84 see Web Resources) to de-noise networks. (We note that the DSD algorithm has been shown to be effective in de-noising networks; see Wang et al.85 for performance in application to networks from Greene et al. networks.9) We ran DSD using default parameters. Because smaller values in the DSD output correspond to stronger interactions, we used the inverse of DSD output as new edge weights. We computed network connectivity metrics on the transformed gene networks and applied S-LDSC to estimate disease heritability enrichment and .
Third, we constructed two consensus networks by intersecting edges in either (1) the Greene and InWeb networks or (2) the Greene, InWeb, and Sonawane networks. We note that the intersection of all four networks did not contain any edges, as the Saha network is very sparse. We computed consensus networks both including and excluding the Sonawane network, which contains nonzero edge weights only for pairs of genes in which at least one of the genes is a known TF. For tissue-specific networks, we used the same tissue as in our primary analysis (Greene thyroid and Sonawane testis). For edges weights, we used the mean edge weight. As each consensus network contained disjoint connected components, we computed closeness centrality using the connected component with the largest number of edges. We applied S-LDSC to estimate disease heritability enrichment and .
Correlation between Closeness and Gene Expression
To assess the correlation between closeness centrality and gene expression, we used gene expression (TPM) across 53 tissues from GTEx RNA sequencing data80 (see Web Resources), restricted to protein-coding genes that are present in a given network. We computed the Pearson correlation between a vectors of gene expression and closeness centrality, for each of 53 GTEx tissues.
Pathway+Network Annotations
In our pathway+network analyses, we considered 156 significant pathway-trait pairs (from our pathway analyses) and four gene networks for a total of 590 (pathway-trait, network) pairs (122 pathway-trait pairs for Saha + 156 pathway-trait pairs × 3 other networks). We constructed pathway+network annotations, specific to an input pathway and a gene network. For each of 141 enriched pathways, we first constructed an adjacency matrix by mapping genes in a pathway to a gene network and identifying the set of neighboring genes outside the pathway, using Graph-tool (see Web Resources). In the adjacency matrix representing a pathway-specific subnetwork, a vector represents an Entrez ID of a gene in a pathway, an Entrez ID of a neighboring gene, and the interaction score (e.g., posterior probability of these two genes). We computed closeness centrality, using the inverse of the interaction score as the cost. Using the set of genes with the linearly transformed closeness scores that lie from 0 to 1, we annotated variants around genes (±100 kb). When a variant is spanned by multiple genes within the 100 kb window, we assigned the maximum closeness score. For each of 590 (pathway-trait, network) pairs, we applied S-LDSC and assessed the statistical significance of the pathway+network annotation’s conditioned on other annotations.
Network Connectivity of a Pathway
Given a pathway and a network, we quantified how tightly connected the pathway is in the network, compared to a null pathway. Let P be a set of genes in the pathway that exist in the network. Let Q be a set of neighboring genes; that is, genes in P are connected with genes in Q with at least one edge, in the network. We calculated three metrics: (1) size of Q (number of neighboring genes), (2) sum of edges among genes in P, and (3) sum of edges between genes in P and genes in Q. We did not consider genes in the pathway that are not coding or do not appear in the network.
For each of 141 enriched pathways, we constructed a corresponding null pathway as follows: (1) randomly choose a pathway from the full set of 18,119 pathways (as shown in Table S1), (2) randomly choose a gene from the sampled pathway, and (3) repeat (1) and (2) N times, where N is the number of genes in the pathway. For each of four gene networks analyzed, we repeated this procedure 10,000 times and reported three connectivity metrics of enriched and null pathways (for null, the mean of 10,000 permutations is reported).
Set of 42 Independent Traits
Analogous to a previous study,41 we considered 89 GWAS summary association statistics, including 34 traits from publicly available sources and 55 traits from the UK Biobank (up to N = 459K); summary association statistics were computed using BOLT-LMM v2.3.86, 87 Among 47 summary statistics with z-scores of total SNP heritability of at least 6 (computed using S-LDSC with the baseline-LD model), we further removed 5 summary statistics that have genetic correlation of at least 0.9 (computed using cross-trait LDSC88). Whenever applicable, meta-analysis across 42 independent traits (Table S2), whose GWAS summary statistics are publicly available (see Web Resources), was performed using a random-effect meta-analysis using the R package rmeta.
Set of 10 Blood-Related and 8 Brain-Related Traits
We analyzed ten independent blood- and autoimmune-related traits: Crohn disease,89 rheumatoid arthritis,90 and ulcerative colitis89 from publicly available datasets and eczema, autoimmune diseases. eosinophil count, platelet count, red blood cell count, white blood cell count, and red blood cell distribution width from the UK Biobank.
We analyzed eight independent brain-related traits: autism spectrum,91 depressive symptoms,92 and schizophrenia93 from publicly available datasets, and age at menarche, body mass index, neuroticism, smoking status, and years of education from the UK Biobank. We selected these traits from 42 independent traits we analyzed, as inferred from heritability enrichment of tissue-specific gene expression and chromatin annotations24, 42 and eQTL annotations.41 We additionally considered autism spectrum based on previous studies94, 95 that the brains of subjects with autism have altered expression.
Results
Enrichment of Disease Heritability in Pathway Annotations
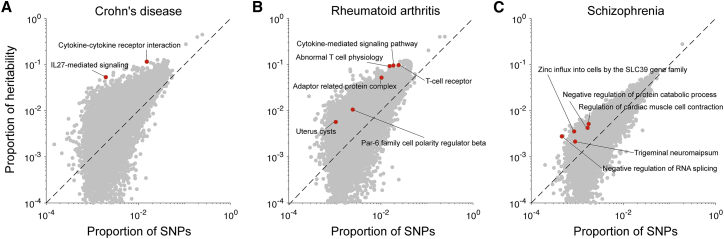
We sought to identify pathways that are enriched for disease heritability. We applied S-LDSC to 760,869 pathway-trait pairs, spanning 18,119 pathways from five sources (Table S1 and Figure S1) and 42 independent diseases and complex traits, including 30 UK Biobank traits (average N = 323K; Table S2). We identified 156 pathway-trait pairs that were significantly enriched after conditioning on the baseline-LD model and the all-genes annotation (FDR < 5% for positive ). The 156 pathway-trait pairs spanned 141 pathways and 34 traits, implying that most pathway enrichments are trait specific. Complete results for three representative traits2—Crohn disease (IBD [MIM: 266600]), rheumatoid arthritis (RA [MIM: 180300]), and schizophrenia (SCZ [MIM: 181500])—are reported in Figure 1, and complete results for all traits are reported in Table S4. The top pathway (most significant ) for each of the 34 traits is reported in Table 1, and the complete set of 156 significant pathway-trait pairs is reported in Table S5. Genes in the 141 enriched pathways had a larger gene size (92 kb on average) compared to all protein-coding genes (58 kb) and genes in all pathways (76 kb) (see Table S6). We meta-analyzed the 156 pathway-trait pairs using random-effect meta-analysis (analogous to previous work23, 24, 41). Both the enrichment (4.13, SE = 0.12; p = 4.74e−158) and (0.15, SE = 0.0061; p = 3.41e−131) were large and highly statistically significant (Table S7); we caution that these p values are slightly inflated because we meta-analyzed across significant pathway-trait pairs only. When we repeated our analysis excluding from each associated pathway all genes harboring genome-wide significant associations for the corresponding trait (average of 2 genes removed per pathway-trait pair; see Material and Methods), only 53 pathway-trait pairs remained significant (Table S8). For each of 156 pathway-trait pairs, genes after excluding GWAS significant genes are provided in Table S5.
Figure 1.
Enriched Pathways for Three Representative Traits
For (A) Crohn disease, (B) rheumatoid arthritis, and (C) schizophrenia, we report the proportion of heritability explained and proportion of SNPs for each of 18,119 pathways analyzed. Red points indicate significantly enriched pathways (FDR < 5%) and gray points indicate non-significant pathways. Numerical results for all 42 diseases and complex traits are reported in Table S4.
Table 1.
Top Enriched Pathway for Each Trait
| Trait | Top Enriched Pathway | Database | # Genes | Enr. (SE) | (SE) |
|---|---|---|---|---|---|
| Age at menarche | SIX homeobox 6 | InWeb | 11 | 3.06 (0.45) | 0.06 (0.02) |
| Auto immune traits | mismatch repair directed by MSH2:MSH6 | BS | 14 | 8.53 (2.05) | 0.28 (0.07) |
| BMI | positive regulation of synapse maturation | GO | 11 | 3.34 (0.25) | 0.07 (0.01) |
| Dermatologic diseases | aldo-keto reductase family 1 member E2 | InWeb | 12 | 7.51 (1.80) | 0.19 (0.05) |
| Eczema | Th17 cell differentiation | BS | 102 | 9.01 (1.56) | 0.64 (0.15) |
| Eosinophil count | Jak-STAT signaling pathway | BS | 152 | 7.67 (1.11) | 0.46 (0.11) |
| Forced vital capacity | absent acrosome | MGI | 11 | 2.21 (0.31) | 0.04 (0.01) |
| Heel T Score | skeletal system development | GO | 142 | 4.43 (0.60) | 0.31 (0.07) |
| Height | abnormal trabecular bone morphology | MGI | 172 | 2.93 (0.39) | 0.20 (0.05) |
| Hypothyroidism | cytokine-mediated signaling pathway | GO | 235 | 4.88 (0.78) | 0.42 (0.11) |
| Morning person | N-acetylglucosamine metabolic process | GO | 13 | 2.07 (0.26) | 0.03 (0.01) |
| Neuroticism | HSF1 activation | BS | 11 | 2.54 (0.27) | 0.04 (0.01) |
| Platelet count | platelet activation | GO | 208 | 4.32 (0.62) | 0.30 (0.09) |
| Red blood cell count | exogenous drug catabolic process | GO | 10 | 3.62 (0.63) | 0.08 (0.02) |
| Red blood cell distribution width | decreased erythrocyte cell number | MGI | 206 | 4.61 (0.74) | 0.39 (0.11) |
| Respiratory/ear-nose-throat diseases | abnormal T cell activation | MGI | 108 | 4.28 (0.75) | 0.26 (0.07) |
| Skin color | melanin biosynthetic process | GO | 11 | 192.14 (55.37) | 5.90 (1.73) |
| Smoking status | fibromodulin | InWeb | 17 | 2.53 (0.48) | 0.06 (0.02) |
| Systolic blood pressure | cGMP-PKG signaling pathway | BS | 164 | 3.41 (0.49) | 0.25 (0.07) |
| Type 2 diabetes | glucuronidation | BS | 30 | 2.74 (0.32) | 0.06 (0.01) |
| Waist-hip ratio | negative regulation of transcription | GO | 496 | 2.60 (0.22) | 0.19 (0.05) |
| White blood cell count | ERK cascade | PC | 15 | 4.65 (0.69) | 0.11 (0.03) |
| Years of education | absent corpus callosum | MGI | 45 | 2.50 (0.33) | 0.08 (0.02) |
| Age first birth | receptor signaling protein tyrosine kinase | GO | 10 | 3.62 (0.58) | 0.10 (0.02) |
| Anorexia | activation of the AP-1 family of TF | BS | 10 | 5.64 (1.04) | 0.15 (0.03) |
| Autism spectrum | GABA-A receptor activity | GO | 15 | 4.01 (0.73) | 0.12 (0.02) |
| Coronary artery disease | sodium-independent organic anion transport | GO | 12 | 7.05 (1.49) | 0.22 (0.04) |
| Crohn disease | cytokine-cytokine receptor interaction | BS | 266 | 7.59 (1.50) | 0.65 (0.18) |
| Depressive symptoms | glycoprotein metabolic process | GO | 12 | 5.22 (1.10) | 0.15 (0.04) |
| LDL | increased erythroblast number | MGI | 21 | 5.63 (1.15) | 0.18 (0.05) |
| Number children even born | lipoxygenase pathway | GO | 12 | 9.81 (1.59) | 0.27 (0.04) |
| Rheumatoid arthritis | Par-6 family cell polarity regulator beta | InWeb | 16 | 4.31 (1.01) | 0.17 (0.04) |
| Schizophrenia | trigeminal neuroma | MGI | 10 | 2.39 (0.31) | 0.03 (0.01) |
| Ulcerative colitis | FGFR2b ligand binding and activation | BS | 10 | 8.00 (1.97) | 0.18 (0.05) |
We report the top enriched pathway (most significant ) for each of 34 traits with at least one significantly enriched pathway. The first 23 traits (above the line) are UK Biobank traits. Enrichment of the “melanin biosynthetic process” pathway for skin color is consistent with previous studies,96, 97 and enrichment of the “absent corpus callosum” pathway for years of education was reported in a previous genetic study.92 The complete set of 156 significant pathway-trait pairs is reported in Table S5.
Our results include eight pathway-trait pairs reported in previous genetic studies (see Tables 1 and S5). These include “pathways in cancer” for height;109 “neuropeptide hormone activity” for BMI;110 “immune response” for both Crohn disease and ulcerative colitis;89 “T cell receptor,” “cytokine-mediated signaling pathway,” and “abnormal T cell physiology” for rheumatoid arthritis;90 and “absent corpus callosum” for years of education.92 In addition, “melanin biosynthetic process” was overwhelmingly enriched for skin color (Table 1), consistent with the fact that genetic variants in constituent genes are strongly associated with skin pigmentation (e.g., MC1R) and other pigmentation traits (e.g., TYR [MIM: 606933], OCA2 [MIM: 611409], SLC45A2 [MIM: 606202]).96, 97
Surprisingly, most pathway-trait pairs reported in recent studies2, 33, 34, 35, 36 using genome-wide polygenic methods24, 30, 37 were not significant in our analysis. Specifically, we considered 95 significant pathway-trait pairs for the six traits (schizophrenia, Crohn disease, rheumatoid arthritis, neuroticism, intelligence, depressive symptoms) analyzed in five previous studies,2, 33, 34, 35, 36 restricting to at most the top 20 pathways per trait per study. We assessed the significance of these 95 pathway-trait pairs in our analysis, based on global FDR < 5% across 18,119 pathways tested ( < 0.000989). Only 15/95 pathway-trait pairs were significant in our primary analysis, after conditioning on the baseline-LD model and all-genes annotation (Table S9A). However, 67/95 were significant when we repeated the S-LDSC analysis conditioning on just the all-genes annotation and not the baseline-LD model (Table S9B). We obtained similar results for a pathway-trait pair reported in a very recent study37 (see Web Resources; Tables S9A and S9B). Enriched pathways that were fully explained by the baseline-LD model could potentially be due to factors that do not play a direct role in trait biology,98 although we caution that our analyses do not resolve which factors are causal (see Discussion).
Our results also highlight pathway-trait pairs that have not previously been reported but are consistent with known biology. These include “GABA-A receptor activity” with autism (Table 1), consistent with the finding that the brains of subjects with autism have altered expression of GABA receptors;94, 95 “Oncostatin M” (OSM [MIM: 165095]) for ulcerative colitis (Table S5), consistent with the finding that inflamed intestinal tissues from patients with inflammatory bowel diseases contained higher expression of OSM and that OSM-deficient mice displayed significantly attenuated colitis;99 and “zinc influx into cells by the SLC39 gene family” with schizophrenia (Table S5), consistent with the finding that SLC39A12A expression in dorsolateral prefrontal cortex is associated with schizophrenia.100, 101
We also analyzed three additional gene sets (distinct from the 18,119 pathways) reflecting genes under strong selection: ExAC68 (high pLI genes; genes strongly depleted for protein-truncating variants), Cassa69 (high shet genes; genes with strong selection against protein-truncating variants), and Samocha71 (high missense Z scores genes; genes strongly depleted for missense mutations), which were previously shown to be enriched for heritability in a meta-analysis across traits.41 We identified 13 significantly enriched gene set-trait pairs (7 for ExAC, 2 for Cassa, and 4 for Samocha) spanning 9 traits, after conditioning on the baseline-LD model and the all-genes annotation (FDR < 5% for positive ; Table S10). In a meta-analysis of these 13 gene set-trait pairs, both the enrichment (1.57, SE = 0.053; p = 7.52e−33) and (0.13, SE = 0.0090; p = 3.30e−48) were highly statistically significant (Table S7).
Enrichment of Disease Heritability in Network Annotations
We sought to assess the hypothesis that genes with high network connectivity are enriched for disease heritability. We constructed probabilistic annotations based on closeness centrality for each of the Saha, Greene, InWeb, and Sonawane networks (see Material and Methods). For three networks that include tissue-specific networks, we selected the Saha-skin (sun-exposed lower leg), Greene-thyroid, and Sonawane-testis networks for our primary analyses, as these tissue-specific networks maximized the correlation of the resulting network annotations with H3K27ac (Table S11); we also considered other criteria for selecting tissue-specific networks (see below). We determined that closeness centrality was independent of gene size (r =0.015 to 0.019) and exon proportion (r =0.18 to 0.008; Figure S2). We note that different tissue-specific networks from the same source were only weakly correlated (r = 0.027 to 0.076 for Saha, 0.17–0.21 for Greene, 0.062–0.24 for Sonawane; see Table S11). The number of genes, number of edges, and distribution of edge weights for each network are reported in Table S12. The Greene network is very dense (25,825 genes with mean degree 6,484), the Saha network is very sparse (2,381 genes with mean degree 7.9), and the InWeb and Sonawane networks have intermediate density.
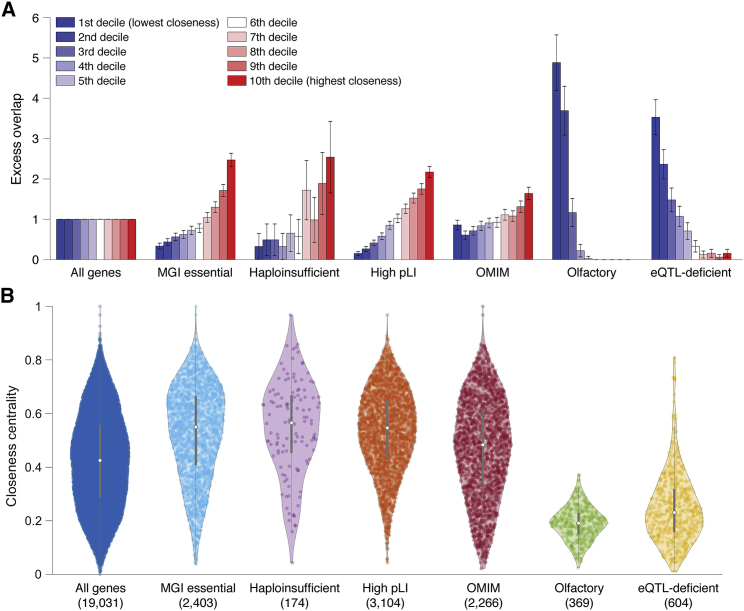
To compare closeness centrality to other metrics that quantify the biological importance of each gene, we computed the excess overlap between genes in each decile bin of closeness centrality (for each network) and 18 other gene sets (Table S3; see Material and Methods). We first describe results for the Greene network. We determined that high closeness centrality genes (top decile of closeness centrality) had high excess overlap with gene sets defined by constraint metrics (e.g., high pLI (ExAC) genes,68 high shet (Cassa) genes69), and essentiality metrics (e.g., MGI essential genes63, 64, 65) as compared to other decile bins (Figure 2A and Table S13), consistent with larger values of closeness centrality in these gene sets (Figure 2B and Table S14; r = 0.22 to 0.27 between closeness centrality and gene set, Table S15). On the other hand, high closeness centrality genes were highly depleted for olfactory receptor genes and eQTL-deficient genes (Figure 2A and Table S13), consistent with smaller values of closeness centrality in these gene sets (Figure 2B and Table S14; r = 0.19 to 0.21, Table S15). High closeness centrality genes were also depleted for genes with more independent SNPs and more total SNPs relative to the length of the gene (which are less functionally important as they are less likely to be under negative selection102) (Figure S3 and Table S13), consistent with smaller values of closeness centrality in these gene sets (Figure S4 and Table S14; r = 0.208 to 0.211, Table S15); however, this was not the case for the Saha, InWeb, and Sonawane networks (Figures S3 and S4, Tables S13 and S14; r = 0.047 to 0.037, Table S15). For other gene sets, results for the InWeb network, but not Saha or Sonawane networks, were generally similar to those for the Greene network (Figures S3 and S4, Tables S13, S14, and S15).
Figure 2.
Comparison of Closeness Centrality to Other Metrics that Quantify the Biological Importance of Each Gene
(A) For each of 7 gene sets, we report the excess overlap of genes in each decile bin of closeness centrality for the Greene thyroid network. Error bars represent 95% confidence intervals.
(B) For each of 7 gene sets, we report the distribution of closeness centrality for the Greene thyroid network. Colored dots denote genes, white dots denote medians, and gray lines denote boxplots. Numbers in parentheses below each gene set denote the number of genes.
Results for all four networks and all 18 gene sets analyzed are reported in Figure S3 and Table S13 (for A) and Figure S4 and Table S14 (for B). Lists of genes for each of the 18 gene sets are provided in Table S3.
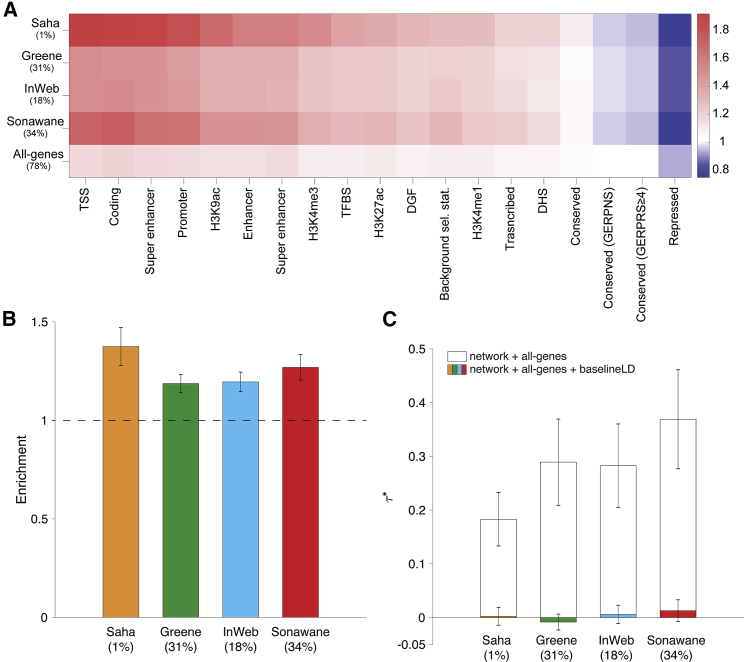
We computed the excess overlap between each of the four network annotations (probabilistic annotations based on closeness centrality) and representative annotations from the baseline-LD model and determined that each of the network annotations had substantial excess overlap with regulatory annotations (e.g., 1.23–1.36 with H3K27ac, 1.35–1.66 with H3K9ac; Figure 3A and Table S16), significantly stronger than the excess overlap between the all-genes annotation and regulatory annotations (e.g., 1.11 with H3K27ac, 1.14 with H3K9ac; Figure 3A); this implies that, for each network, genes with high connectivity to other genes in the network are enriched for the presence of nearby regulatory marks, perhaps because they are regulated by many other genes (see below). Correlations between these annotations produced similar conclusions (Table S17; larger correlation for H3K27ac than H3K9ac), although we consider excess overlap to be a more robust metric because the size of the network annotations varies from 1% to 34% of SNPs. We also report excess overlap and correlations between baseline-LD model annotations in Table S18.
Figure 3.
Heritability Enrichment of Network Annotations
We report (A) excess (fold) overlap between network annotations and baseline-LD functional categories; (B) heritability enrichment of network annotations, meta-analyzed across 42 independent traits; and (C) values of network annotations, conditioned on either just the all-genes annotation, or the all-genes annotation and the baseline-LD model, meta-analyzed across 42 independent traits. The percentage under each bar indicates the proportion of SNPs in each annotation (defined for probabilistic annotations as the average value of the annotation), and error bars represent 95% confidence intervals. Numerical results for (A) are reported in Table S16, and numerical results for (B) and (C) are reported in Table S22. The S-LDSC results for the complete set of 168 network-trait pairs are reported in Table S22.
To investigate whether genes with high closeness centrality are heavily regulated by other genes, or are heavily regulating other genes, we performed three analyses; we did not reach a consistent conclusion. First, we assessed the excess overlap of 1,610 known human TFs76 in the top decile of closeness centrality for each network (see Material and Methods). We determined that TFs (which regulate other genes) were depleted in high closeness centrality genes for the Greene and Sonawane networks but enriched in high closeness centrality genes for the Saha and InWeb networks (Figure S3). Thus, this analysis did not reach a consistent conclusion. Second, we assessed the excess overlap of TF binding sites in the promoters (±1 kb from TSS) of high closeness centrality genes, by applying the ENCODE ChIP-seq significance tool81 to ENCODE ChIP-seq data28 spanning 220 TFs and 91 cell lines (see Material and Methods). We determined that, for each gene network, binding sites for the majority of TFs (132 to 206 out of 220 TFs) had significant excess overlap (FDR < 0.05) in the promoters of high closeness centrality genes (Table S19); we also observed significant excess overlap throughout high closeness centrality genes (±100 kb) (Table S20). Thus, this analysis supports the hypothesis that genes with high closeness centrality are heavily regulated by other genes. Third, we assessed the GO enrichment of high closeness centrality genes using DAVID82 (see Material and Methods). We used all protein-coding genes as a background set and evaluated gene sets corresponding to three GO categories: biological process, cellular component, and molecular function. We determined that high closeness centrality genes from all four networks were often significantly enriched (FDR < 0.05) in protein/TF/DNA/regulatory region binding (e.g., “transcription regulatory region DNA binding [GO:0044212],” “transcription factor binding [GO:0008134]”) and transcriptional regulation (e.g., “positive regulation of transcription [GO:0045944]”) gene sets (Table S21). Thus, this analysis supports the hypothesis that genes with high closeness centrality are heavily regulating other genes. Overall, our analyses did not reach a consistent conclusion on the regulatory role of high closeness centrality genes.
For each of the four network annotations, we applied S-LDSC to the 42 independent diseases and complex traits, conditioning on the baseline-LD model and the all-genes annotation, and meta-analyzed the results across traits using random-effects meta-analysis. We identified strongly significant enrichments for each network annotation: 1.19 (SE = 0.024; p = 2.5e−30) to 1.37 (SE = 0.049; p = 1.6e−17) (Figure 3B and Table S22). However, estimates of , quantifying effects unique to the network annotations, were not significant (p = 0.21 to 0.77) (Figure 3C and Table S22). This implies that the enrichment signal in the network annotations (Figure 3B) is entirely explained by the excess overlap between the network and baseline-LD model annotations (Figure 3A); accordingly, when we repeated the S-LDSC analysis conditioning only on the all-genes annotation and not on the baseline-LD model, estimates were large and highly significant (Figure 3C). We repeated the S-LDSC analysis conditional on one annotation from the baseline-LD model at a time and confirmed that regulatory annotations (primarily histone marks and transcription factor binding sites) reduced estimates of by 74%–100% (Table S23). We note that other annotations did not have this effect (e.g., 16%–20% for conservation annotations). We also partitioned all genes in a given network into 10 deciles of closeness centrality and applied S-LDSC to estimate the heritability enrichments for each decile. For the Greene, InWeb, and Sonewane networks, we determined that the top decile of closeness centrality had the highest enrichment (1.68 [SE 0.062]–1.76 [SE 0.070]) and the bottom decile of closeness centrality had the lowest enrichment (0.92 [SE 0.031]–1.24 [0.038]; see Table S24). For the Saha network, the top decile had the highest enrichment (1.98 [SE 0.099]) but the bottom decile had the fourth lowest enrichment (1.464 [SE 0.071]). We also observed that the top decile had significantly larger correlations with regulatory annotations than the bottom decile (Table S20). On the one hand, these findings represent a negative result for efforts to improve upon the baseline-LD model. On the other hand, these findings provide a strong validation of the baseline-LD model, in that the information about diseases/traits in annotations from other sources that broadly reflect the action of gene regulation are fully captured by the baseline-LD model.
We performed three analyses to assess the impact of noise in gene networks on our results. First, we performed a network perturbation analysis by repeating the S-LDSC analysis on networks with 10%–90% of edges randomly removed, following an established protocol83 (see Material and Methods). We obtained similar results, including significant enrichments and non-significant (Table S25). Second, we de-noised each network by applying the diffusion state distance (DSD) algorithm,84 computed closeness centrality on the transformed networks (see Material and Methods), and repeated the S-LDSC analysis. We determined that the DSD-transformed network annotations were 1.9%–25.4% more enriched, but remained non-significant (Table S26). Third, we constructed two “consensus” networks by intersecting the edges in either (1) the Greene and InWeb networks or (2) the Greene, InWeb, and Sonawane networks (the intersection of all four networks did not contain any edges, as the Saha network is very sparse), and repeated the S-LDSC analysis. We obtained similar results, including significant enrichments and non-significant (Table S27). These analyses support the robustness of our results.
We performed several secondary analyses. First, we examined the correlation between closeness centrality (for each network) and gene expression, using GTEx RNA-seq data80 (see Material and Methods). We determined that closeness centrality in the Saha and Sonawane networks was independent of gene expression (r = 0.045 to 0.030 for each of 53 GTEx tissues), whereas closeness centrality in the Greene and InWeb networks was moderately correlated with gene expression, particularly in brain tissues (e.g., r = 0.17 to 0.27 for cerebellum) (Table S28). Second, we performed heritability analyses on gene sets defined by membership in gene networks (analogous to pathway annotations). We focused our analyses on the Saha networks, which are much sparser than the other networks. We defined gene sets using the 16 Saha transcriptome-wide networks (TWN) used in our primary analyses, as well as the 36 Saha tissue-specific networks (TSN), and applied S-LDSC to these gene sets. All 16 gene sets derived from TWNs and 4 of 36 gene sets derived from TSNs were significantly enriched for disease/trait heritability, but none had a significant conditional on the baseline-LD model (Table S29). Third, we repeated the S-LDSC analysis for the three tissue-specific networks (Saha, Greene, Sonawane) using the tissue that maximized the excess overlap of the High pLI (ExAC) gene set with the top decile of closeness centrality of the tissue-specific network (Table S30; see Material and Methods). We obtained similar results (as compared to our primary analysis in which tissue-specific networks were selected so as to maximize correlation with H3K27ac), including significant enrichments for all three tissue-specific networks but non-significant conditional on the baseline-LD model (Tables S22 and S24). Fourth, for ten blood-related traits and eight brain-related traits, we repeated the S-LDSC analysis for the three tissue-specific networks using the most biologically relevant tissue (as inferred from heritability enrichment of tissue-specific specifically expressed gene and chromatin annotations;42 see Table S2 for list of traits and tissues). For the ten blood-related traits, the resulting tissue-specific networks (blood) produced slightly larger enrichments (significant enrichment for all three networks; significant difference for Sonawane only), but conditional on the baseline-LD model remained non-significant (Table S31; consistent with Table S22). For the eight brain-related traits, both the resulting tissue-specific networks (brain) and the default tissue-specific networks produced non-significant enrichments (Table S31; consistent with Table S22). Finally, we also obtained similar results when we repeated the S-LDSC analysis using six connectivity metrics other than closeness centrality (see Material and Methods; Table S32), using window sizes other than 100 kb (10 kb and 1 Mb; Table S33).
Enrichment of Disease Heritability in Integrated Pathway+Network Annotations
We sought to assess the hypothesis that genes with high network connectivity to genes in enriched pathways are also enriched for disease heritability; these enriched pathways serve as relevant, disease-specific signals that network analysis often lacks. For each of 4 networks and 141 enriched pathways, we constructed probabilistic pathway+network annotations based on closeness centrality within the subnetwork consisting of input genes and their one-degree neighbors. As most of the pathway enrichments that we identified are disease specific, the pathway+network annotations are expected to harbor disease-specific signals. As before, for three networks that include tissue-specific networks, we selected the Saha-skin (sun-exposed lower leg), Greene-thyroid, and Sonawane-testis networks for our primary analyses.
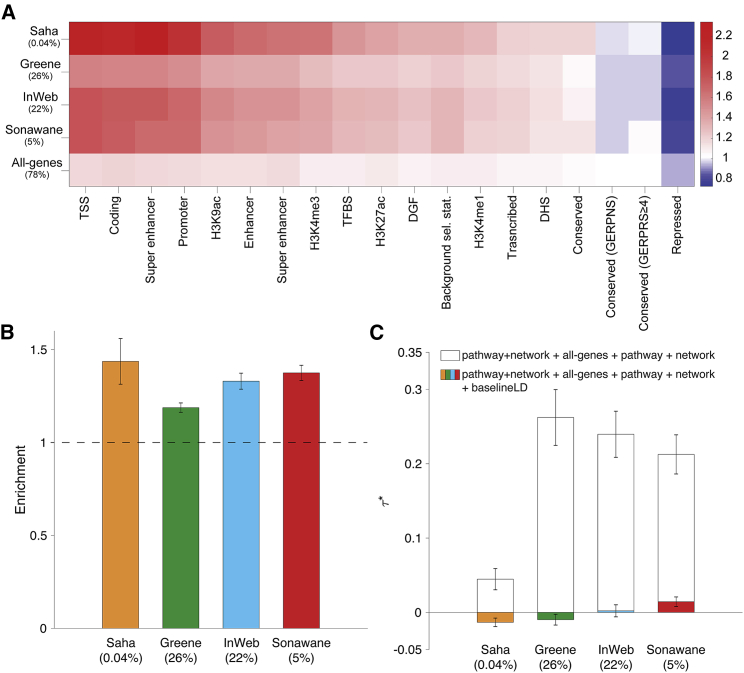
For each network, we computed the excess overlap between the 141 pathway+network annotations and representative annotations from the baseline-LD model and averaged results across the 141 pathways. We observed higher excess overlap with regulatory annotations (e.g., 1.27–1.35 with H3K27ac, 1.42–1.70 with H3K9ac; Figure 4A and Table S16) than the analogous excess overlaps for network annotations (Figure 3A); this implies that, for each network, genes with high network connectivity to genes in input pathways are enriched for the presence of nearby regulatory marks, perhaps because they are regulated by many other genes. Correlations between network, pathway+network, and baseline-LD model annotations are reported in Table S17. We determined that pathway+network annotations were often correlated with network annotations, particularly for the corresponding network (from r = 0.12 for Saha to r = 0.93 for Greene). We further determined that pathway+network annotations constructed using the Greene, InWeb, and Sonawane networks were moderately correlated (r = 0.27 to 0.42), whereas pathway+network annotations constructed using the Saha network were more distinct (r = 0.05 to 0.06), primarily due to its small annotation size (Table S17).
Figure 4.
Heritability Enrichment of Pathway+Network Annotations
We report (A) excess (fold) overlap between pathway+network annotations (averaged across up to 156 pathway-trait pairs); (B) heritability enrichment of pathway+network annotations, meta-analyzed across up to 156 pathway-trait pairs; and (C) values of pathway+network annotations, conditioned on either just the all-genes annotation and the corresponding pathway and network annotations, or the baseline-LD model as well, meta-analyzed across up to 156 pathway-trait pairs. The percentage under each bar indicates the proportion of SNPs in each annotation (defined for probabilistic annotations as the average value of the annotation), and error bars represent 95% confidence intervals. Numerical results for (A) are reported in Table S16, and numerical results for (B) and (C) are reported in Table S34. The S-LDSC results for the complete set of 590 pathway-trait pairs are reported in Table S34.
For each of 590 (pathway-trait,network) pairs (122 pathway-trait pairs for Saha + 156 pathway-trait pairs × 3 other networks; see Material and Methods), we applied S-LDSC to the resulting pathway+network annotation and the corresponding trait, conditioning on the baseline-LD model, the all-genes annotation, and the corresponding pathway and network annotations, and meta-analyzed the results for each network using random-effects meta-analysis. We identified strongly significant enrichments for all of our pathway+network annotations: 1.19 (SE = 0.01; p = 1.5e−49) to 1.44 (SE = 0.06; p = 3.8e−12) (Figure 4B and Table S34). On average, the pathway+network annotations most enriched for trait heritability are those derived from the Saha network (Figure 4B). However, estimates of , quantifying effects unique to the network annotations, were not significant or only weakly significant (p = 0.62 to 4.5e−6) (Figure 4C and Table S34). Once again, this implies that the enrichment signal in the pathway+network annotations is entirely explained by the excess overlap between the pathway+network and baseline-LD model annotations; accordingly, when we repeated the S-LDSC analysis conditioning only on the all-genes annotation and the corresponding pathway and network annotations, and not on the baseline-LD model, estimates were large and highly significant, except for the Saha network (Figure 4C). We repeated the S-LDSC analysis conditional on one annotation from the baseline-LD model at a time and determined that inclusion of regulatory annotations (primarily histone marks and transcription factor binding sites) reduced estimates of by 17%–89% (Table S35).
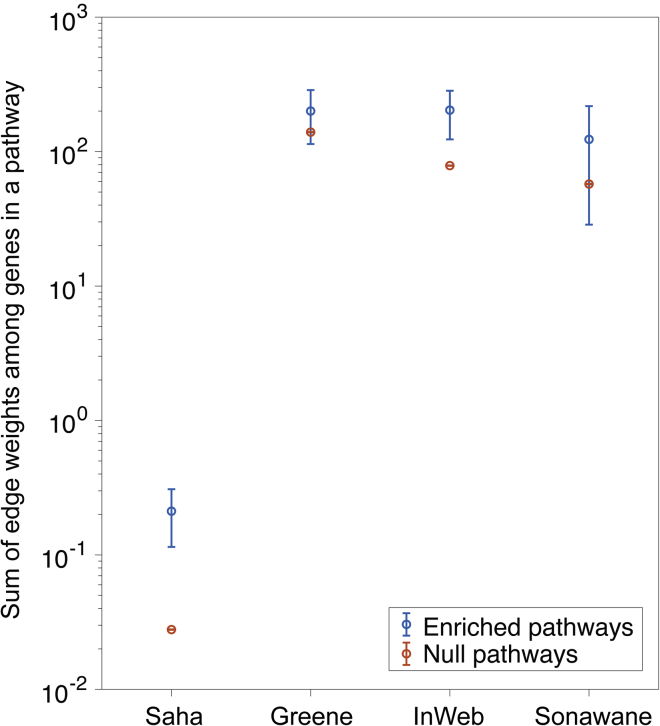
We assessed whether genes in enriched pathways have higher network connectivity than other genes. For each of the four gene networks, for each of the 141 enriched pathways, we assessed both the network connectivity within the pathway and the network connectivity between the pathway and interacting genes (one-degree neighbors) outside the pathway, as compared to 10,000 null pathways with the same number of genes, each randomly sampled from a randomly chosen pathway from the full set of 18,119 pathways (see Material and Methods). We assessed network connectivity using the sum of edge weights between genes. For each network, we averaged results across pathways. We determined that genes in enriched pathways have higher network connectivity within the pathway (1.43×–7.60× more edges; Figure 5 and Table S36), but do not necessarily have higher network connectivity with interacting genes outside the pathway (0.69×–1.56×; Table S36); we note that there is no significant difference in the number of interacting genes between the 141 enriched pathways and the 10,000 null pathways (Table S36).
Figure 5.
Genes in Enriched Pathways Have High Network Connectivity
For each of four networks, we report the sum of edge weights in the network between genes in the pathway, averaged across 141 enriched pathways. For comparison purposes, we report the same quantity averaged across 10,000 null pathways with the same number of genes. Error bars represent 95% confidence intervals (smaller than data points for null pathways). Numerical results and analogous results for network connectivity between a pathway and interacting genes outside the pathway are reported in Table S36.
We repeated the S-LDSC analysis (Figures 4B and 4C) using new pathway+network annotations constructed using a random-forest classifier, Quack,38 that identifies new candidate genes that have similar topological patterns and network centrality metrics as genes in the input pathway. Because genes in enriched input pathways have high network connectivity (Figure 5), this is closely related to our primary strategy of defining pathway+network annotations based on genes with high network connectivity to genes in enriched input pathways. Indeed, Quack annotations were highly correlated with our main pathway+network annotations (r = 0.50–0.67; Table S17) and produced S-LDSC results similar to our main analysis (Figures 4B and 4C), including significant enrichments for all four networks but non-significant conditional on the baseline-LD model (Table S37).
Discussion
We analyzed 42 diseases and complex traits (average N = 323K) to show that genes with high network connectivity are enriched for disease heritability but that it is critical for gene network and pathway analyses to account for known functional annotations, such as those from our baseline-LD model.23 First, in analyses of pathway annotations, we identified 156 pathway-trait pairs with significant heritability enrichment after conditioning on the baseline-LD model, a stringent step that caused a majority of pathway-trait pairs reported in recent studies to become non-significant in our analyses. Second, we determined that network annotations based on closeness centrality, a measure of network connectivity, are strongly enriched for disease heritability, but that these enrichments were fully explained by annotations from the baseline-LD model. Third, for each of the 156 significant pathway-trait pairs, we determined that pathway+network annotations constructed from genes with network connectivity to the input pathway were strongly enriched for the corresponding traits, but that once again these enrichments were largely explained by annotations from the baseline-LD model.
Our findings have important ramifications for studies connecting gene networks and pathways to disease.2, 9, 11, 14, 15, 16, 17, 19, 20, 21, 22, 29, 30, 31, 32, 33, 34, 35, 36, 37 Specifically, it is important to account for known functional annotations when seeking to elucidate biological mechanisms. For some methods, such as S-LDSC,2, 24, 33 it is straightforward to incorporate known functional annotations such as those from the baseline-LD model,23 and we emphasize the importance of doing so. For other methods, it is of high interest to investigate how functional annotations could be incorporated. More generally, it is of broad interest to re-assess previously reported results while accounting for known functional annotations; for example, this could be achieved by running S-LDSC both with and without incorporating functional annotations from the baseline-LD model.
We note several limitations of our work. First, S-LDSC is not well suited to analysis of annotations spanning a very small proportion of the genome24 and does not model sparsity in trait effect sizes, potentially explaining why we did not identify enriched pathways for eight traits that are less polygenic103 (e.g., age at menopause, balding, hair color, sunburn). Nonetheless, our main results attained high statistical significance. Second, we did not explicitly compare S-LDSC to other methods. However, previous work suggests that S-LDSC compares favorably to other gene set enrichment methods, both in simulations and in analyses of real traits.42 Third, interpretation of pathway-trait enrichments is complicated by the possibility that enrichment signals may be driven by a small number of highly significant genes.37 However, we verified that repeating our main pathway+network analyses using the remaining significant pathway-trait pairs (after excluding genes implicated by GWAS) produced similar conclusions (Table S38). Fourth, gene networks may include false-positive interactions, even after correcting for technical confounding.10, 39, 104 However, our network perturbation analysis, DSD-transformed network analysis, and consensus network analysis all support the robustness of our results. Fifth, inferences about components of heritability can potentially be biased by failure to account for LD-dependent architectures.23, 105, 106, 107 All of our main analyses used the baseline-LD model, which includes six LD-related annotations.23 The baseline-LD model is supported by formal model comparisons using likelihood and polygenic prediction methods, as well as analyses using a combined model incorporating alternative approaches;108 however, there can be no guarantee that the baseline-LD model perfectly captures LD-dependent architectures. Sixth, although we showed that many pathway-trait pairs reported in recent studies were fully explained by the baseline-LD model and thus could potentially be due to factors that do not play a direct role in trait biology,98 our analyses do not resolve which factors are causal. Nonetheless, because it is plausible that the regulatory annotations of the baseline-LD model may be the causal factors, accounting for known functional annotations is an appropriate conservative measure to avoid incorrect conclusions. Finally, although we identified many significantly enriched pathways conditional on the baseline-LD model, our results for network and network+pathway annotations represent a negative result for efforts to improve upon the baseline-LD model,23 further emphasizing the importance of accounting for known functional annotations in network and pathway analyses.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We are grateful to Manolis Kellis, Yakir Reshef, Evan Boyle, April Kim, Taibo Li, Marieke Kuijjer, and Xinchen Wang for helpful discussions. This research was funded by NIH grants U01 HG009379, R01 MH109978, R01 MH101244, and R01 MH107649. This research was conducted using the UK Biobank Resource under Application 16549.
Published: April 18, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.03.020.
Contributor Information
Samuel S. Kim, Email: sungil@mit.edu.
Alkes L. Price, Email: aprice@hsph.harvard.edu.
Web Resources
1000 Genomes Project Phase 3 data, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502
Baseline-LD annotations, https://data.broadinstitute.org/alkesgroup/LDSCORE/
BOLT-LMM software, https://data.broadinstitute.org/alkesgroup/BOLT-LMM
BOLT-LMM summary statistics for UK Biobank traits, https://data.broadinstitute.org/alkesgroup/UKBB
DAVID (Database for Annotation, Visualization and Integrated Discovery, v.6.8), https://david.ncifcrf.gov
DSD (Diffusion State Distance) algorithm, http://dsd.cs.tufts.edu/capdsd/
ENCODE ChIP-Seq Significance Tool, http://encodeqt.simple-encode.org/
Ensembl biomart, https://grch37.ensembl.org/index.html
GTEx (Release v7), https://www.gtexportal.org/home/datasets
GWAS Catalog (Release v1.0), http://www.ebi.ac.uk/gwas
Graph-tool, https://graph-tool.skewed.de
HumanBase (Greene tissue-specific co-expression networks), https://hb.flatironinstitute.org/
inBio Map (InWeb protein-protein interaction network), https://www.intomics.com/inbio/map
Network Connectivity source codes, gene scores, 18 gene sets, https://github.com/samskim/networkconnectivity
Online Mendelian Inheritance in Man, http://www.omim.org
Pathway, network, and pathway+network annotations: https://data.broadinstitute.org/alkesgroup/LDSCORE/Kim_pathwaynetwork
PLINK software, https://www.cog-genomics.org/plink2
S-LDSC software, https://github.com/bulik/ldsc
Saha transcriptome-wide networks, https://storage.googleapis.com/gtex_analysis_v6p/coexpression_networks/coexpression_networks_v6p.zip
Subset of 18 gene sets, https://github.com/macarthur-lab/gene_lists
UK Biobank, https://www.ukbiobank.ac.uk/
UK Biobank Genotyping and QC Documentation, http://www.ukbiobank.ac.uk/wp-content/uploads/2014/04/UKBiobank_genotyping_QC_documentation-web.pdf
Zenodo (Sonawane gene regulatory networks), https://zenodo.org/record/838734
Zenodo (Zhu & Stephens pathway studies), https://zenodo.org/record/838734#.W89JDxNKiAw
Supplemental Data
We applied S-LDSC to 760,869 pathway-trait pairs, conditioning on all-genes annotation and the baseline-LD model. For each pathway-trait pair, we report a proportion of SNPs, enrichment, and τ.
References
- 1.Visscher P.M., Wray N.R., Zhang Q., Sklar P., McCarthy M.I., Brown M.A., Yang J. 10 years of gwas discovery: biology, function, and translation. Am. J. Hum. Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyle E.A., Li Y.I., Pritchard J.K. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Menche J., Sharma A., Kitsak M., Ghiassian S.D., Vidal M., Loscalzo J., Barabási A.-L. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347:1257601. doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barabási A.-L., Gulbahce N., Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y., Zhu J., Lum P.Y., Yang X., Pinto S., MacNeil D.J., Zhang C., Lamb J., Edwards S., Sieberts S.K. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatr-Aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., Kolas N.K., O’Donnell L., Oster S., Theesfeld C., Sellam A. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45(D1):D369–D379. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li T., Wernersson R., Hansen R.B., Horn H., Mercer J., Slodkowicz G., Workman C.T., Rigina O., Rapacki K., Stærfeldt H.H. A scored human protein-protein interaction network to catalyze genomic interpretation. Nat. Methods. 2017;14:61–64. doi: 10.1038/nmeth.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greene C.S., Krishnan A., Wong A.K., Ricciotti E., Zelaya R.A., Himmelstein D.S., Zhang R., Hartmann B.M., Zaslavsky E., Sealfon S.C. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 2015;47:569–576. doi: 10.1038/ng.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saha A., Kim Y., Gewirtz A.D.H., Jo B., Gao C., McDowell I.C., Engelhardt B.E., Battle A., GTEx Consortium Co-expression networks reveal the tissue-specific regulation of transcription and splicing. Genome Res. 2017;27:1843–1858. doi: 10.1101/gr.216721.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marbach D., Lamparter D., Quon G., Kellis M., Kutalik Z., Bergmann S. Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. Nat. Methods. 2016;13:366–370. doi: 10.1038/nmeth.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonawane A.R., Platig J., Fagny M., Chen C.-Y., Paulson J.N., Lopes-Ramos C.M., DeMeo D.L., Quackenbush J., Glass K., Kuijjer M.L. Understanding tissue-specific gene regulation. Cell Rep. 2017;21:1077–1088. doi: 10.1016/j.celrep.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Wijst M.G.P., de Vries D.H., Brugge H., Westra H.-J., Franke L. An integrative approach for building personalized gene regulatory networks for precision medicine. Genome Med. 2018;10:96. doi: 10.1186/s13073-018-0608-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon S., Nguyen H.C.T., Yoo Y.J., Kim J., Baik B., Kim S., Kim J., Kim S., Nam D. Efficient pathway enrichment and network analysis of GWAS summary data using GSA-SNP2. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gky175. e60–e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cowen L., Ideker T., Raphael B.J., Sharan R. Network propagation: a universal amplifier of genetic associations. Nat. Rev. Genet. 2017;18:551–562. doi: 10.1038/nrg.2017.38. [DOI] [PubMed] [Google Scholar]
- 16.Peters L.A., Perrigoue J., Mortha A., Iuga A., Song W.M., Neiman E.M., Llewellyn S.R., Di Narzo A., Kidd B.A., Telesco S.E. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat. Genet. 2017;49:1437–1449. doi: 10.1038/ng.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taşan M., Musso G., Hao T., Vidal M., MacRae C.A., Roth F.P. Selecting causal genes from genome-wide association studies via functionally coherent subnetworks. Nat. Methods. 2015;12:154–159. doi: 10.1038/nmeth.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerstein M.B., Kundaje A., Hariharan M., Landt S.G., Yan K.-K., Cheng C., Mu X.J., Khurana E., Rozowsky J., Alexander R. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Califano A., Butte A.J., Friend S., Ideker T., Schadt E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat. Genet. 2012;44:841–847. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee I., Blom U.M., Wang P.I., Shim J.E., Marcotte E.M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011;21:1109–1121. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanunu O., Magger O., Ruppin E., Shlomi T., Sharan R. Associating genes and protein complexes with disease via network propagation. PLoS Comput. Biol. 2010;6:e1000641. doi: 10.1371/journal.pcbi.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Köhler S., Bauer S., Horn D., Robinson P.N. Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gazal S., Finucane H.K., Furlotte N.A., Loh P.-R., Palamara P.F., Liu X., Schoech A., Bulik-Sullivan B., Neale B.M., Gusev A., Price A.L. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat. Genet. 2017;49:1421–1427. doi: 10.1038/ng.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.-R., Anttila V., Xu H., Zang C., Farh K., ReproGen Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. RACI Consortium Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trynka G., Sandor C., Han B., Xu H., Stranger B.E., Liu X.S., Raychaudhuri S. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segrè A.V., Groop L., Mootha V.K., Daly M.J., Altshuler D., DIAGRAM Consortium. MAGIC investigators Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6:e1001058. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pers T.H., Karjalainen J.M., Chan Y., Westra H.-J., Wood A.R., Yang J., Lui J.C., Vedantam S., Gustafsson S., Esko T., Genetic Investigation of ANthropometric Traits (GIANT) Consortium Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Leeuw C.A., Neale B.M., Heskes T., Posthuma D. The statistical properties of gene-set analysis. Nat. Rev. Genet. 2016;17:353–364. doi: 10.1038/nrg.2016.29. [DOI] [PubMed] [Google Scholar]
- 33.Pardiñas A.F., Holmans P., Pocklington A.J., Escott-Price V., Ripke S., Carrera N., Legge S.E., Bishop S., Cameron D., Hamshere M.L., GERAD1 Consortium. CRESTAR Consortium Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 2018;50:381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagel M., Jansen P.R., Stringer S., Watanabe K., de Leeuw C.A., Bryois J., Savage J.E., Hammerschlag A.R., Skene N.G., Muñoz-Manchado A.B., 23andMe Research Team Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat. Genet. 2018;50:920–927. doi: 10.1038/s41588-018-0151-7. [DOI] [PubMed] [Google Scholar]
- 35.Savage J.E., Jansen P.R., Stringer S., Watanabe K., Bryois J., de Leeuw C.A., Nagel M., Awasthi S., Barr P.B., Coleman J.R.I. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet. 2018;50:912–919. doi: 10.1038/s41588-018-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wray N.R., Ripke S., Mattheisen M., Trzaskowski M., Byrne E.M., Abdellaoui A., Adams M.J., Agerbo E., Air T.M., Andlauer T.M.F., eQTLGen. 23andMe. Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu X., Stephens M. Large-scale genome-wide enrichment analyses identify new trait-associated genes and pathways across 31 human phenotypes. Nat. Commun. 2018;9:4361. doi: 10.1038/s41467-018-06805-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li T., Kim A., Mercer J., Rosenbluh J., Horn H., Greenfeld L., An D., Zimmer A., Liberzon A., Bistline J. GeNets: A unified web platform for network-based analyses of genomic data. bioRxiv. 2018 doi: 10.1038/s41592-018-0039-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyle E.A., Pritchard J.K., Greenleaf W.J. High-resolution mapping of cancer cell networks using co-functional interactions. Mol. Syst. Biol. 2018;14:e8594. doi: 10.15252/msb.20188594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hormozdiari F., Gazal S., van de Geijn B., Finucane H.K., Ju C.J.-T., Loh P.-R., Schoech A., Reshef Y., Liu X., O’Connor L. Leveraging molecular quantitative trait loci to understand the genetic architecture of diseases and complex traits. Nat. Genet. 2018;50:1041–1047. doi: 10.1038/s41588-018-0148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finucane H.K., Reshef Y.A., Anttila V., Slowikowski K., Gusev A., Byrnes A., Gazal S., Loh P.-R., Lareau C., Shoresh N., Brainstorm Consortium Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 2018;50:621–629. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geer L.Y., Marchler-Bauer A., Geer R.C., Han L., He J., He S., Liu C., Shi W., Bryant S.H. The NCBI BioSystems database. Nucleic Acids Res. 2010;38:D492–D496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caspi R., Billington R., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Midford P.E., Ong Q., Ong W.K. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018;46(D1):D633–D639. doi: 10.1093/nar/gkx935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaefer C.F., Anthony K., Krupa S., Buchoff J., Day M., Hannay T., Buetow K.H. Pid: the pathway interaction database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46(D1):D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pico A.R., Kelder T., van Iersel M.P., Hanspers K., Conklin B.R., Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cerami E.G., Gross B.E., Demir E., Rodchenkov I., Babur O., Anwar N., Schultz N., Bader G.D., Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romero P., Wagg J., Green M.L., Kaiser D., Krummenacker M., Karp P.D. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto S., Sakai N., Nakamura H., Fukagawa H., Fukuda K., Takagi T. INOH: ontology-based highly structured database of signal transduction pathways. Database (Oxford) 2011;2011:bar052. doi: 10.1093/database/bar052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mi H., Thomas P. Humana Press; Totowa, NJ: 2009. PANTHER Pathway: An Ontology-Based Pathway Database Coupled with Data Analysis Tools PANTHER Pathway: An Ontology-Based Pathway Database Coupled with Data Analysis Tools. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jewison T., Su Y., Disfany F.M., Liang Y., Knox C., Maciejewski A., Poelzer J., Huynh J., Zhou Y., Arndt D. Smpdb 2.0: Big improvements to the small molecule pathway database. Nucleic Acids Res. 2014;42:D478–D484. doi: 10.1093/nar/gkt1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kandasamy K., Mohan S.S., Raju R., Keerthikumar S., Kumar G.S.S., Venugopal A.K., Telikicherla D., Navarro J.D., Mathivanan S., Pecquet C. NetPath: a public resource of curated signal transduction pathways. Genome Biol. 2010;11:R3. doi: 10.1186/gb-2010-11-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eppig J.T., Blake J.A., Bult C.J., Kadin J.A., Richardson J.E., Mouse Genome Database Group The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015;43:D726–D736. doi: 10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W., Jansen R., de Geus E.J., Boomsma D.I., Wright F.A. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ozgür A., Vu T., Erkan G., Radev D.R. Identifying gene-disease associations using centrality on a literature mined gene-interaction network. Bioinformatics. 2008;24:i277–i285. doi: 10.1093/bioinformatics/btn182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bartha I., di Iulio J., Venter J.C., Telenti A. Human gene essentiality. Nat. Rev. Genet. 2018;19:51–62. doi: 10.1038/nrg.2017.75. [DOI] [PubMed] [Google Scholar]
- 62.Povey S., Lovering R., Bruford E., Wright M., Lush M., Wain H. The hugo gene nomenclature committee (hgnc) Hum. Genet. 2001;109:678–680. doi: 10.1007/s00439-001-0615-0. [DOI] [PubMed] [Google Scholar]
- 63.Blake J.A., Bult C.J., Kadin J.A., Richardson J.E., Eppig J.T., Mouse Genome Database Group The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 2011;39:D842–D848. doi: 10.1093/nar/gkq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Georgi B., Voight B.F., Bućan M. From mouse to human: evolutionary genomics analysis of human orthologs of essential genes. PLoS Genet. 2013;9:e1003484. doi: 10.1371/journal.pgen.1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu X., Jian X., Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blekhman R., Man O., Herrmann L., Boyko A.R., Indap A., Kosiol C., Bustamante C.D., Teshima K.M., Przeworski M. Natural selection on genes that underlie human disease susceptibility. Curr. Biol. 2008;18:883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berg J.S., Adams M., Nassar N., Bizon C., Lee K., Schmitt C.P., Wilhelmsen K.C., Evans J.P. An informatics approach to analyzing the incidentalome. Genet. Med. 2013;15:36–44. doi: 10.1038/gim.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cassa C.A., Weghorn D., Balick D.J., Jordan D.M., Nusinow D., Samocha K.E., O’Donnell-Luria A., MacArthur D.G., Daly M.J., Beier D.R., Sunyaev S.R. Estimating the selective effects of heterozygous protein-truncating variants from human exome data. Nat. Genet. 2017;49:806–810. doi: 10.1038/ng.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bartha I., Rausell A., McLaren P.J., Mohammadi P., Tardaguila M., Chaturvedi N., Fellay J., Telenti A. The characteristics of heterozygous protein truncating variants in the human genome. PLoS Comput. Biol. 2015;11:e1004647. doi: 10.1371/journal.pcbi.1004647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45(D1):D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T. The human transcription factors. Cell. 2018;172:650–665. doi: 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 77.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang X., Goldstein D.B. Enhancer redundancy predicts gene pathogenicity and informs complex disease gene discovery. bioRxiv. 2018 [Google Scholar]
- 79.Mainland J.D., Li Y.R., Zhou T., Liu W.L.L., Matsunami H. Human olfactory receptor responses to odorants. Sci. Data. 2015;2:150002. doi: 10.1038/sdata.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts. Laboratory, Data Analysis &Coordinating Center (LDACC) NIH program management. Biospecimen collection. Pathology. eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 81.Auerbach R.K., Chen B., Butte A.J. Relating genes to function: identifying enriched transcription factors using the ENCODE ChIP-Seq significance tool. Bioinformatics. 2013;29:1922–1924. doi: 10.1093/bioinformatics/btt316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 83.Yang J., McAuley J., Leskovec J. IEEE 13th international conference on Data Mining (ICDM) IEEE; 2013. Community detection in networks with node attributes; pp. 1151–1156. [Google Scholar]
- 84.Cao M., Zhang H., Park J., Daniels N.M., Crovella M.E., Cowen L.J., Hescott B. Going the distance for protein function prediction: a new distance metric for protein interaction networks. PLoS ONE. 2013;8:e76339. doi: 10.1371/journal.pone.0076339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang B., Pourshafeie A., Zitnik M., Zhu J., Bustamante C.D., Batzoglou S., Leskovec J. Network enhancement as a general method to denoise weighted biological networks. Nat. Commun. 2018;9:3108. doi: 10.1038/s41467-018-05469-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Loh P.-R., Bhatia G., Gusev A., Finucane H.K., Bulik-Sullivan B.K., Pollack S.J., de Candia T.R., Lee S.H., Wray N.R., Kendler K.S., Schizophrenia Working Group of Psychiatric Genomics Consortium Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet. 2015;47:1385–1392. doi: 10.1038/ng.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Loh P.-R., Kichaev G., Gazal S., Schoech A.P., Price A.L. Mixed-model association for biobank-scale datasets. Nat. Genet. 2018;50:906–908. doi: 10.1038/s41588-018-0144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.-R., Duncan L., Perry J.R., Patterson N., Robinson E.B., ReproGen Consortium. Psychiatric Genomics Consortium. Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3 An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., RACI consortium. GARNET consortium Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Okbay A., Beauchamp J.P., Fontana M.A., Lee J.J., Pers T.H., Rietveld C.A., Turley P., Chen G.-B., Emilsson V., Meddens S.F.W., LifeLines Cohort Study Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ripke S., Neale B.M., Corvin A., Walters J.T., Farh K.-H., Holmans P.A., Lee P., Bulik-Sullivan B., Collier D.A., Huang H., Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fatemi S.H., Reutiman T.J., Folsom T.D., Thuras P.D. GABA(A) receptor downregulation in brains of subjects with autism. J. Autism Dev. Disord. 2009;39:223–230. doi: 10.1007/s10803-008-0646-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cellot G., Cherubini E. GABAergic signaling as therapeutic target for autism spectrum disorders. Front Pediatr. 2014;2:70. doi: 10.3389/fped.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Low D., Chen K.-S. UBE3A regulates MC1R expression: a link to hypopigmentation in Angelman syndrome. Pigment Cell Melanoma Res. 2011;24:944–952. doi: 10.1111/j.1755-148X.2011.00884.x. [DOI] [PubMed] [Google Scholar]
- 97.Simeonov D.R., Wang X., Wang C., Sergeev Y., Dolinska M., Bower M., Fischer R., Winer D., Dubrovsky G., Balog J.Z. DNA variations in oculocutaneous albinism: an updated mutation list and current outstanding issues in molecular diagnostics. Hum. Mutat. 2013;34:827–835. doi: 10.1002/humu.22315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.de Leeuw C.A., Stringer S., Dekkers I.A., Heskes T., Posthuma D. Conditional and interaction gene-set analysis reveals novel functional pathways for blood pressure. Nat. Commun. 2018;9:3768. doi: 10.1038/s41467-018-06022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.West N.R., Hegazy A.N., Owens B.M.J., Bullers S.J., Linggi B., Buonocore S., Coccia M., Görtz D., This S., Stockenhuber K., Oxford IBD Cohort Investigators Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat. Med. 2017;23:579–589. doi: 10.1038/nm.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Scarr E., Udawela M., Greenough M.A., Neo J., Suk Seo M., Money T.T., Upadhyay A., Bush A.I., Everall I.P., Thomas E.A., Dean B. Increased cortical expression of the zinc transporter SLC39A12 suggests a breakdown in zinc cellular homeostasis as part of the pathophysiology of schizophrenia. NPJ Schizophr. 2016;2:16002. doi: 10.1038/npjschz.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Takagishi T., Hara T., Fukada T. Recent advances in the role of slc39a/zip zinc transporters in vivo. Int. J. Mol. Sci. 2017;18:2708. doi: 10.3390/ijms18122708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Charlesworth B., Morgan M.T., Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.O’Connor L.J., Schoech A.P., Hormozdiari F., Gazal S., Patterson N., Price A.L. Polygenicity of complex traits is explained by negative selection. bioRxiv. 2018 doi: 10.1016/j.ajhg.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Parsana P., Ruberman C., Jaffe A.E., Schatz M.C., Battle A., Leek J.T. Addressing confounding artifacts in reconstruction of gene co-expression networks. bioRxiv. 2017 doi: 10.1186/s13059-019-1700-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Speed D., Hemani G., Johnson M.R., Balding D.J. Improved heritability estimation from genome-wide SNPs. Am. J. Hum. Genet. 2012;91:1011–1021. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang J., Bakshi A., Zhu Z., Hemani G., Vinkhuyzen A.A., Lee S.H., Robinson M.R., Perry J.R., Nolte I.M., van Vliet-Ostaptchouk J.V., LifeLines Cohort Study Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet. 2015;47:1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Speed D., Cai N., Johnson M.R., Nejentsev S., Balding D.J., UCLEB Consortium Reevaluation of SNP heritability in complex human traits. Nat. Genet. 2017;49:986–992. doi: 10.1038/ng.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gazal S., Marquez-Luna C., Finucane H.K., Price A.L. Reconciling s-ldsc and ldak models and functional enrichment estimates. bioRxiv. 2018 doi: 10.1038/s41588-019-0464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z., Electronic Medical Records and Genomics (eMEMERGEGE) Consortium. MIGen Consortium. PAGEGE Consortium. LifeLines Cohort Study Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., LifeLines Cohort Study. ADIPOGen Consortium. AGEN-BMI Working Group. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GLGC. ICBP. MAGIC Investigators. MuTHER Consortium. MIGen Consortium. PAGE Consortium. ReproGen Consortium. GENIE Consortium. International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
We applied S-LDSC to 760,869 pathway-trait pairs, conditioning on all-genes annotation and the baseline-LD model. For each pathway-trait pair, we report a proportion of SNPs, enrichment, and τ.