Abstract
We derived novel, testable predictions from a mathematical model of the budding yeast cell cycle. A key qualitative prediction of bistability was confirmed in a strain simultaneously lacking cdc14 and G1 cyclins. The model correctly predicted quantitative dependence of cell size on gene dosage of the G1 cyclin CLN3, but it incorrectly predicted strong genetic interactions between G1 cyclins and the anaphase- promoting complex specificity factor Cdh1. To provide constraints on model generation, we determined accurate concentrations for the abundance of all nine cyclins as well as the inhibitor Sic1 and the catalytic subunit Cdc28. For many of these we determined abundance throughout the cell cycle by centrifugal elutriation, in the presence or absence of Cdh1. In addition, perturbations to the Clb-kinase oscillator were introduced, and the effects on cyclin and Sic1 levels were compared between model and experiment. Reasonable agreement was obtained in many of these experiments, but significant experimental discrepancies from the model predictions were also observed. Thus, the model is a strong but incomplete attempt at a realistic representation of cell cycle control. Constraints of the sort developed here will be important in development of a truly predictive model.
INTRODUCTION
The eukaryotic cell cycle is controlled by cyclin-dependent kinase activity, where the activity of the kinases is controlled by abundance of the positive regulatory cyclin subunits and by phosphorylation of the kinase catalytic subunit (Morgan, 1997). Cyclins are regulated transcriptionally and proteolytically; this regulation is interdigitated with control of chromosome replication and segregation (Nasmyth, 1996; Zachariae and Nasmyth, 1999) and spindle morphogenesis (Haase et al., 2001).
Chen et al. (2000) presented a mathematical model of the budding yeast cell cycle that formulates a great deal of genetic and biochemical data, in terms of chemical kinetic rate equations. The model contains a number of simplifications. All cyclins are implicitly assumed to be nuclear (Novak et al., 1998), although this is not always the case (Miller and Cross, 2000). A number of nonessential cyclins are omitted (CLN1, CLB1,3,4,6). The model lacks modeling of control of mitotic exit by the Cdc14 phosphatase and the mitotic exit network that controls it (Jaspersen et al., 1998; Shou et al., 1999; Visintin et al., 1999; Bardin et al., 2000).
An important component in the model is the delayed activation of the anaphase-promoting complex (APC) specificity factor Cdc20 due to checkpoint/surveillance mechanisms dependent on chromosome replication and alignment on the metaphase spindle. Because such surveillance mechanisms are at least individually dispensable for viability (Zhao et al., 1998; Alexandru et al., 1999; Vallen and Cross, 1999; Bardin et al., 2000), it is unlikely that a delay in Cdc20 activation due to damage surveillance is an essential component of the cell cycle oscillator.
Despite these limitations, the model implements an interesting concept of the cell cycle as an alternation of two states: a low-Clb state in which Clb inhibitors and degradation are high and a high-Clb state in which the reverse is true (Nasmyth, 1996). Well-characterized pathways are proposed to make these states self-maintaining. For example, the Sic1 inhibitor of B-type cyclin-dependent kinase activity is proteolyzed after its ubiquitination, and ubiquitination is in turn dependent on cyclin-dependent kinase phosphorylation of Sic1 (Verma et al., 1997a, 1997b). Thus, the inhibitor will be degraded, and the kinase will therefore not be inhibited, if and only if the kinase starts at a high activity level. A similar pattern exists for Cdh1/Hct1 (Schwab et al., 1997; Visintin et al., 1998), which activates ubiquitination and subsequent proteolysis of some B-type cyclins. Phosphorylation of Cdh1 by cyclin-dependent kinases prevents its ability to cause cyclin ubiquitination by the APC (Zachariae et al., 1998; Jaspersen et al., 1999).
In the model, the Cln-dependent kinases drive transition from the low-Clb to the high-Clb state (in part by phosphorylating both Sic1 and Cdh1), and Cdc20 drives the reverse transition by initiating Clb proteolysis. Hysteresis is predicted in the transitions between these states, such that the forces driving the transition must push for a while before the transition occurs, making the transitions irreversible.
The model accounts for an impressive number of mutant situations (Chen et al., 2000), but all of these situations were used as input information to generate the model and so were not independent confirmation. Here we derive and test new predictions from the model. The results of these studies suggest the need for more empirically based parameters for future modeling efforts. In the second part of this article we obtain absolute quantitative information on the abundance of most cell cycle regulators through the cell cycle. Such information is likely to provide constraints that will make future models significantly more realistic and may lead to the development of mathematical models usable as predictive tools for cell cycle control.
MATERIALS AND METHODS
Strain and Plasmid Constructions
In Figures 1–3, strains were BF264-15D background. bck2::ARG4 strains (Epstein and Cross, 1994) were transformed with an integrating TRP1-CLN3 plasmid containing ∼3.5 kb of 5′ and 1.2 kb of 3′ information, targeted to trp1 by BglII digestion or an identical plasmid lacking CLN3. Transformants were mated to a cln3::URA3 strain and meiotic segregants identified with and without BCK2, endogenous CLN3, and the CLN3 transgene. Transgene copy number was established by digestion of DNA (Holm et al., 1986) from Trp+ segregants with BglII, yielding a 7.1-kb endogenous CLN3 band and an 11.6-kb transgene band, quantitated by Southern hybridization and Phosphorimager. Ratios of transgene to endogenous signal (duplicate meiotic segregants for each initial transformant) indicate transgene copy number. A correction was required for apparently lower recovery or transfer of the 11.6-kb transgene fragment. Strains with the minimum ratio detected gave a ratio of ∼0.5 rather than 1. We assume these to be single copy. This ratio was the most commonly detected (3/7), and these are stable integrants in which effectively one copy of CLN3 is functioning, based on essentially equal cell volume of cln3::URA3 cells containing the transgene to CLN3 cells not containing the transgene. Therefore, to obtain CLN3 copy number in the transgene array, we multiplied the (11.6 kb/7.1 kb) signal ratio by 2 and rounded to the nearest integer. The total CLN3 copy number in a strain is this number, plus one for strains containing endogenous CLN3.
Figure 1.
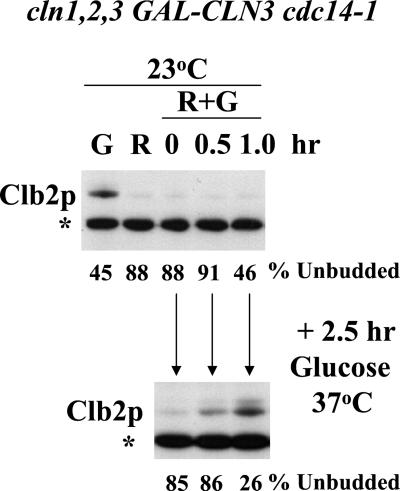
Hysteresis in the cell cycle. A strain of genotype cln1 cln2 cln3 GAL-CLN3 cdc14-1 was grown to log phase in YEPGal (galactose medium, GAL-CLN3 on) at 23°C and blocked due to CLN deficiency by incubation in YEPRaff (raffinose medium, GAL- CLN3 off) at 23°C for 6 h. Galactose was then added to 3% final concentration to induce GAL- CLN3. At the indicated times after galactose addition, aliquots of the culture were removed. Some of the aliquot was immediately extracted for immunoblotting with anti-Clb2 antibody. To the rest of the aliquot glucose was added to 2% final concentration to repress GAL-CLN3, and the aliquot was incubated at 37°C for 2.5 h. The aliquot was then extracted for immunoblotting. The percentages of unbudded cells at the time of aliquot removal and after the 2.5-h incubation in glucose at 37°C were determined microscopically. Top: Clb2 immunoblot for cells in continuous galactose (G), after the raffinose block (R), and after the indicated amounts of time after galactose addition (R+G; all at 23°C). Bottom: Clb2 immunoblot for cells incubated in R+G for the indicated amount of time and then shifted to glucose at 37°C for 2.5 h. *An unidentified background band used for standardizing protein loading. The percentage of unbudded cells at the time of protein harvest is indicated below the gel lanes. The top and bottom immunoblots were from the same membrane and film exposure.
Figure 3.
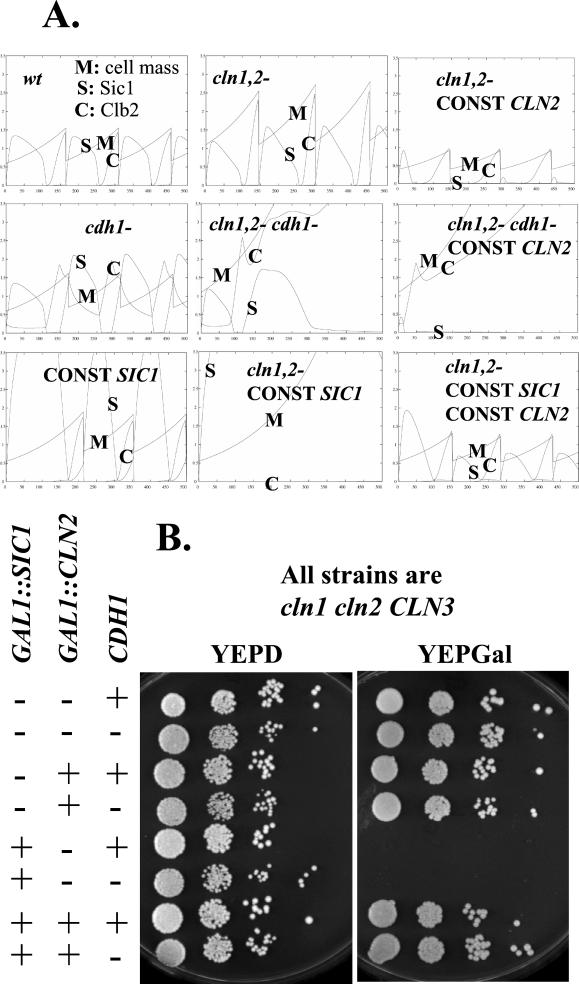
Interaction between G1 cyclins and mitotic regulators Sic1 and Cdh1. (A) Model predictions for various genotypes. Parameters: M, cell mass; C, Clb2 levels; S, Sic1 levels. Standard parameters from (Chen et al., 2000) were used throughout, except for the following changes to model the mutants: cln1,2-: ksn2" = 0; CONST CLN2: ksn2′ = 0.05, ksn2" = 0; cdh1-: kdb2" = 0.01; CONST SIC1: ksc1′ = 0.1, ksc1" = 0.1. The model predicts lethality (cell cycle arrest and failure of cell division) in the cln1,2 cdh1, the CONST CLN2 cdh1 (with or without CONST SIC1), and the cln1,2−CONST SIC1 (with or without CDH1) situations. The CONST CLN2 cdh1 CONST SIC1 simulation (not shown) is similar to the CONST CLN2 cdh1 simulation. The cln1,2−CONST SIC1 cdh1 simulation differs from the cln1,2−CONST SIC1 simulation, because Clb2 levels ultimately rise because of cdh1 deletion, but in both cases inviability is predicted. (B) Genetic test of the predictions in A. Strains (BF264-15D background) of the indicated genotypes were constructed by tetrad analysis, using GAL- SIC1/URA3 and GAL-CLN2/TRP1 cassettes and using a cdh1::LEU2 deletion (Schwab et al., 1997) backcrossed five times into the BF264- 15D background. All strains were cln1 cln2 CLN3. cln1 cln2 CLN3 corresponds to absence of Cln2 in the model, and cdh1:: LEU2 corresponds to absence of Cdh1 (called Hct1 in Chen et al., 2000) in the model. The presence of the GAL-SIC1 and GAL-CLN2 cassettes corresponds to CONST SIC1 and CONST CLN2, respectively. Tenfold serial dilutions of cultures of strains of each genotype, grown to stationary phase in YEPD, were spotted on YEPD or YEPGal plates and grown at 30°C for 2 and 3 d, respectively. Another set of strains of these genotypes behaved identically. CLN1,2 controls (wild type, cdh1, and GAL-SIC1) are not shown here but all are viable under these conditions (our unpublished data). The model predicts lethality due to cln1 cln2 cdh1 (YEPD plates, all cdh1- strains, 2nd, 4th, 6th, 8th rows), but this is not observed. It also predicts lethality due to cln1 cln2 cdh1 GAL-CLN2 (YEPGal plates, 4th row), but this is not observed. The model correctly predicts lethality due to constitutive Sic1 expression in the absence of cln1,2 and its rescue by constitutive CLN2, in the presence or absence of cdh1 (YEPGal plates, 3rd, 4th, 7th, and 8th rows).
The CLN3myc integration vector pMM162 was constructed by moving the SalI-SacII cassette of pMM99 (Miller and Cross, 2000) containing CLN3 promoter driven CLN3myc into the SalI-SacII site of the integration vector pRS404. This vector was targeted for integration at CLN3 by digestion with EcoRI, resulting in introduction of a C-terminal Cln3-myc epitope followed by TRP1 and untagged CLN3.
Protein A (PrA) tagging (W303 background) was performed by the PCR-based method (Aitchison et al., 1995) using pBXAHIS5 (Wach et al., 1997). Integration was verified by PCR using flanking oligonucleotides. Myc-epitope tagging was described for Cln2, Cln3, and Clb5 (Jacobson et al., 2000; Miller and Cross, 2000).
The cdh1::LEU2 (hct1::LEU2) allele was from W. Seufert (Schwab et al., 1997) in the W303 background, and for the experiment in Figure 3 was backcrossed six times into BF264-15D. The GALL-HA-HCT1- m11 mutant expressing unphosphorylatable Cdh1/Hct1 (Zachariae et al., 1998; W303 background) was provided by M. Shirayama. (Note: the standard name for this locus according to the Stanford Saccharomyces Genome database is CDH1, with HCT1 listed as a nonstandard alias; see http://genome-www4.stanford.edu/cgi-bin/SGD/locus.pl?locus=cdh1. We will use the standard name throughout this article, although the Hct1 name was used in Chen et al. [2000] as well as in many other publications. We hope this will cause neither confusion nor offense).
Protein Methods
9XMyc or PrA-containing DNA was cloned into NotI-cut pET42a (Novagen, Madison, WI), encoding GST-HIS. The PrA NotI fragment was obtained from pMM53, constructed by replacing the 3× HA epitope of pKL001 (Levine et al., 1996) with a PCR-amplified NotI fragment containing PrA.
BL21-DE3 with these plasmids was grown to OD600 0.5, induced with 1 mM IPTG for 5 h and lysed by sonication in 100 mM NaH2PO4, 10 mM Tris, pH 8.0, 8 M urea, 0.5 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml leupeptin, and 0.2% aprotinin. Nickel resin (QIAGEN, Santa Clarita, CA) was added to cleared lysate, rocked at room temperature for 30 min, and washed using the same buffer at pH 6.3. Fusions were eluted using the same buffer with imidazole (0.2 M), pH 4.5.
Escherichia coli expressing MBP-Clb2 (from P. Kaldis) was grown at 37°C in LB + 100 μg/ml ampicillin, 0.2% glucose. After 30 min at 23°C cells were induced with 0.3 mM IPTG for 6 h, suspended in ice-cold column buffer (20 mM Tris 7.5, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, 200 μg/ml PMSF), frozen at −20°C overnight, thawed on ice, and sonicated with a Misonix XL2020 sonicator microtip (setting 5, 16 × 15-s bursts, 1-min rests on ice; Farmingdale, NY), centrifuged for 10 min at 9000 rpm at 4°C, added to 1 ml amylose resin (New England Biolabs, Beverly, MA), agitated 4°C for 8 h, washed three times with ice-cold column buffer, and eluted three times with 1 ml ice-cold column buffer containing 10 mM maltose.
Fusion proteins (GST-PrA, GST-Myc, and MBP- Clb2) were quantified as follows. SDS-PAGE gels of the fusions along with BSA standards were stained with Coomassie Brilliant Blue R-250 (ICN Biomedicals, Costa Mesa, CA). The predominant band in each aliquot corresponded to the purified recombinant protein. The mass corresponding to this band was estimated by comparing intensity with the BSA standards. A mass ratio for the full-length fusion protein over total protein in the lane was estimated at between 25 and 50%. Protein concentrations were assayed by a Bradford (Pierce, Rockford, IL) and a Lowry (DC Protein Assay; Bio-Rad, Hercules, CA) assay, and the protein concentrations were corrected for impurities using the ratio estimated from the gel. These values were averaged to determine the concentration of the fusion proteins.
Yeast proteins were extracted with glass bead/SDS extraction (Levine et al., 1996) or NaOH/TCA extraction as follows. Pelleted cells were resuspended in 500 μl of 1.85 N NaOH, 7.4% BME, incubated on ice for 1 h, and then precipitated with 500 μl 50% TCA at 0°C for 1 h. Precipitates were pelleted at 14,000 rpm at 4°C for 1 h, washed with acetone at −20°C, and resuspended in 100 μl 0.5 M Tris, 5% SDS by sonicating. One hundred microliters of 75% glycerol, 250 mM DTT, and 0.05% bromphenol blue were added, and samples were incubated at 95°C for 15 min and centrifuged to pellet debris. For data in Tables 1 and 2, both methods were used as indicated. For other experiments, the glass bead method was used. These methods were compared in parallel and found to be approximately equally efficient at cell breakage and protein yields (our unpublished data).
Table 1.
Quantitation of cyclins, Sic1, and Cdc28
| Protein A-tagged protein | Copies per diploid cell (asynchronous) |
|---|---|
| Cln1 | 995 ± 231 (4) |
| Cln2 | 2011 ± 504 (5) |
| Cln3 | 216 ± 44 (6) |
| Clb1 | 497 ± 146 (4) |
| Clb2 | 1128 ± 231 (4) |
| Clb3 | 861 ± 231 (4) |
| Clb4 | 445 ± 135 (4) |
| Clb5 | 784 ± 193 (5) |
| Clb6 | 92 ± 17 (4) |
| Sic1 | 214 ± 42 (5) |
| Cdc28 | 12,263 ± 3,130 (5) |
Extracts from log-phase diploid cultures (W303 background) using the NaOH/TCA method were analyzed for number of copies per cell of the indicated protein fused to protein A, as described in MATERIALS AND METHODS. Note that these are unsynchronized cultures, so the number of copies per cell measured is less than the peak number of copies per cell. Values are mean ± SEM, with the number of determinations in parentheses. Figure 4 presents a sample experiment in this series.
Table 2.
Cross-check of quantitations using different antibodies and tags
| Protein | Antibody | Standard | Copies per diploid cell (asynchronous) |
|---|---|---|---|
| Clb2 | Anti-Clb2 | MBP- Clb2 | 2700 ± 306 (3) |
| Clb2-protein A | Rabbit IgG | GST- protein A | 1128 ± 231 (4) |
| Clb5-myc | Anti-myc | GST- myc | 1040 ± 85 (2) |
| Clb5-protein A | Rabbit IgG | GST-protein A | 784 ± 193 (5) |
| Cln2- myc | Anti-myc | GST-myc | 3250 ± 636 (2) |
| Cln2- protein A | Rabbit IgG | GST-protein A | 2011 ± 504 (5) |
| Cln3-myc | Anti-myc | GST-myc | 185 ± 78 (2) |
| Cln3-protein A | Rabbit IgG | GST-protein A | 216 ± 44 (6) |
All data are derived from diploid strains. The protein A data are reprinted from Table 1 to allow comparison. Experiments determining the level of endogenous Clb2 using anti-Clb2 and MBP-Clb2 as standard, and determining myc-tagged Cln2, Clb5, and Cln3 with GST-myc using anti-myc and GST-myc as standard are presented. The myc-tagged genes were expressed in heterozygous diploids in the BF264-15D background. For the determinations of myc-tagged protein and endogenous Clb2 levels, one culture (two for Clb2) was extracted with the glass bead/SDS method and one with the NaOH/TCA method. These two methods gave similarly efficient extraction and similar results. Values are mean ± SEM, with number of determinations in parentheses.
For quantitation, diploid strains expressing PrA fusions were grown in YEPD to 1–2 × 107 cells/ml. Triplicate hemocytometer cell counts were used to determine number of cell equivalents of protein analyzed. Serial dilutions of cell extracts and of the recombinant GST-PrA were made in carrier cell extracts obtained from control untagged cultures. Samples were run on 5–20% acrylamide gels and analyzed by Western blot on the same piece of membrane. PrA detection was with rabbit IgG (ICN) followed by donkey anti- rabbit, HRP-coupled antibody (Amersham, Arlington Heights, IL), with chemiluminescent detection. Films with exposures in the linear range were analyzed for signal intensities after background subtraction, using a digital camera and pixel quantifying software (Alpha Innotech, San Leandro, CA). From the dilution of standard and its concentration, a conversion for signal intensity to number of molecules was determined, yielding an estimate for the number of copies per cell (see Figure 4 legend). For myc-tagged proteins, GST-myc standard and polyclonal anti-Myc antibody were used (Santa Cruz Biotechnology, Santa Cruz, CA). For untagged Clb2, the standard was MBP-Clb2, and the dilutions were made in extract from a clb2 deletion strain. Blots were probed with anti-Clb2 antibody (Santa Cruz).
Figure 4.
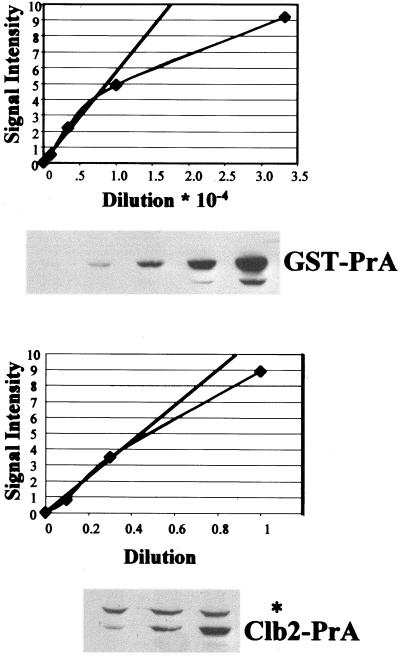
Method of quantitation: sample data for Clb2-PrA. To estimate the number of copies of a PrA fusion per yeast cell, threefold serial dilutions of the GST-PrA standard and of the cell extract from the tagged strain were made in wild-type (untagged) carrier cell extract as described in MATERIALS AND METHODS. SDS-PAGE followed by Western blot analysis was performed with these samples. In addition to the PrA fusion protein band, all blots generated a band at ∼115 kDa (asterisk), derived from the carrier wild-type cell extract. Signal intensities were plotted against dilution factors and points were chosen in the linear parts of the curve (straight line in the figure) for the calculations described in MATERIALS AND METHODS. The points in the graph correspond to the densitometric quantitation of the bands in the autoradiograph. In this example, the middle lane on the GST- PrA gel was determined to contain 1.4 × 109 molecules of GST-PrA based on quantitation of the standard and the dilution factor, and it gave a signal intensity of 2.2, in the linear range of detection. The middle lane of the Clb2-PrA gel derived from 4 × 106 yeast cells and gave a signal intensity of 3.5, also in the linear range of detection. The calculated Clb2-PrA/cell from this example is then (3.5/2.2) × 1.4 × 109 molecules/4 × 106 cells = 557 molecules/cell. Because the strain analyzed was a heterozygous diploid, this number is doubled to give 1114 molecules/cell. Numerous independent determinations of this type (each representing a different protein extraction and quantitation) were averaged to yield the final numbers in Table 1.
Densitometry was used similarly for quantifying results from the elutriation experiments. Because of the large number of samples, the serial dilution strategy was not used, but the exposures were in the linear range of detection. In these experiments, signal from the PrA fusion was standardized by determination of Pgk1 protein levels in the fractions, using anti-Pgk1 antibody (Molecular Probes, Eugene, OR) in parallel immunoblots.
Competition Growth Assay
Fresh stationary phase plate stocks of variously marked gene disruptions were mixed in approximately equal proportions in water. The suspension was streaked out on nonselective YEPD solid medium, and the suspension was also inoculated at ∼200 cells/ml in YEPD, and flasks were incubated with shaking for 2 d at 30°C, to stationary phase. The frequencies of prototrophs for the disruption markers before and after culture growth were determined by plating. The selective disadvantage in this one-step growth regimen was determined as follows: Tm/Tw = ln(I ∗ fWend/fWbeg)/ln( I ∗ fMend/fMbeg), where I is the fold increase in cell number through the experiment (500,000), fWend is the frequency of wild types at end, fWbeg is the frequency of wild types at beginning; and fMend and fMbeg are frequencies for the mutants. This parameter will reflect differential growth rates in exponential growth, if time of exit and entry into stationary phase and differential survival in the stationary phase are ignored. We have not evaluated the latter possibilities.
As controls for the disruptions, we tested W303 strains that were HIS3, LEU2, TRP1, or URA3 in competition with normal W303. The final calculated selective disadvantage due to the cyclin disruption is the disadvantage of the disrupted strain compared with wild type, divided by the selective advantage of the appropriate control strain compared with the reference wild type.
Elutriation
Elutriation was carried out in a Beckman J6 M elutriating centrifuge (40-ml chamber) at 4°C and 2700 rpm. One-liter cultures in YEPD medium (OD660, 1.0) were collected by filtration, resuspended in 100 ml of 0°C water, sonicated three times for 1 min at maximum microtip power in a Misonix XL2020 sonicator, and loaded on the elutriating rotor. Four hundred-milliliter fractions of increasing cell volume were harvested by sequential 10% increments in pump speed, with 0°C water in the pump reservoir. Cell volume was determined using a Coulter Channelyzer calibrated with 68 fl latex beads (Coulter, Hialeah, FL).
Computer Modeling
The WinPP program (ftp://ftp.math.pitt.edu/pub/bardware/winpp.zip, by Bard Ermentrout; see also http://www.math.pitt.edu/∼bard/xpp/xpp.html) was run with a file provided by Kathy Chen that implemented the equation set in Chen et al. (2000). CLN3 gene dosage was varied using the Dn3 parameter. bck2 deletion was simulated by setting BCK2 to zero. CDH1 deletion was simulated by setting kdb2" to 0.01. To simulate GAL promoter driven expression, it was assumed that constitutive expression was equal to peak regulated expression of the endogenous gene (the results of the simulations were not very sensitive to this parameter). To simulate GAL-CLB2db, it was assumed that neither Cdc20-dependent nor Cdh1-dependent degradation could operate (kdb2‴ = 0.01, kdb2p = 0). To model GAL-HCT1-m11 (unregulated Cdh1), GAL promoter expression was neglected, because CDH1 expression is not considered in the model. Instead the nonphosphorylatable status of the mutant Cdh1 encoded by this construct was reflected by setting kit1" to zero, eliminating the effect of Cdk phosphorylation.
RESULTS
Hysteresis: Is the Cell Cycle Characterized by Bistability?
A central aspect of the model derives from the bistability concept (Nasmyth, 1996), in which the cell cycle is considered as an alternation between two stable self-maintaining states, one in which Clb kinase is low (G1) and one in which Clb kinase is high (S/M). The components in the model causing switching between the two states are the Cln kinases for low-to-high and Cdc20 for high-to-low. The Cln kinases switch from the low to the high state by phosphorylating Sic1 and Cdh1, allowing accumulation of Clb kinases. Clb kinases can subsequently maintain the high state by continuing Sic1 and Cdh1 phosphorylation. Cln kinases can reverse the low state because they are immune to Sic1 and Cdh1 regulation, but once the Clb kinases are high, Cln kinases are dispensable (and indeed are predicted to be deleterious; see below). Conversely, Cdc20 activates the high-to-low transition by inducing degradation of Clb5 and initial degradation of Clb2; once Clb kinases have been pushed below a threshold level, Sic1 and Cdh1 phosphorylation become inefficient, and they then take over from Cdc20 to push Clb kinase activity to a very low level. At this point Cdc20 is no longer needed to maintain the low-Clb kinase state.
The role of Cdc20 in the model is more limited than current information indicates. Cdc20 not only leads to degradation of Clb2, but also to degradation of Pds1, and Pds1 is thought to inhibit release of the Cdc14 phosphatase from the nucleolus (see INTRODUCTION). Cdc14 is thought to dephosphorylate and hence activate Cdh1 and Sic1. For purposes of this discussion, because the available model does not include Cdc14 and its associated regulatory machinery, we can consider Cdc20 as a component that somehow encompasses both Cdc20 and Cdc14 activities, and these jointly drive Clb kinase from the high to the low state.
The model thus contains initiator activities (Cln kinases, primarily Cln2 in the model) and terminator activities (Cdc20, Cdc14, et al.; Cdc20 in the model). These initiator and terminator activities antagonize each other with respect to activation or inactivation of Cdh1 and Sic1, which are the main final enforcers of the low-Clb state. Intriguingly, simultaneous absence of initiator and terminator activities does not result in a unique predicted final outcome; rather, hysteresis is predicted. “Hysteresis occurs in systems with multiple steady states and refers to the fact that the observed state of the system depends not only on its parameter values but also on its history (how the system is prepared)” (Novak et al., 1998). Thus, if the neutral situation lacking initiator and terminator is encountered from a prior history of a low-Clb state, this state will be maintained indefinitely; conversely, encountering neutral coming from the high-Clb state means that the high-Clb state will be maintained. This formulation makes the prediction that a third steady state, with intermediate values of Clb-dependent kinases, is mathematically possible but unstable.
To try to experimentally realize the neutral state lacking both initiator and terminator, we constructed a strain of the genotype cln1 cln2 cln3 GAL-CLN3 cdc14-1. The strain is viable on galactose medium at 23°C, because galactose provides CLN function by keeping GAL-CLN3 on, and the cdc14-1 temperature-sensitive allele functions at 23°C. The strain is inviable at 37°C on galactose and is inviable without galactose at any temperature. Glucose medium at 37°C is the experimental approximation of the neutral state lacking initiator and terminator simultaneously. What is the phenotype of this strain in this neutral condition, and does this phenotype indeed depend on the prior history of the culture?
We blocked the strain in G1 by CLN deprivation, by turning off GAL-CLN3 by incubation in raffinose medium at 23°C for 6 h. Under these conditions ∼90% of the cells were unbudded (a morphological marker of the pre-Start state; Cross, 1995), and Clb2 protein in the culture was very low (Figure 1). We then induced GAL-CLN3 transcription with galactose. At intervals we removed aliquots of the culture, added glucose to block further GAL-CLN3 transcription, and shifted to 37°C for 2.5 h to inactivate cdc14-1 (CLN3 RNA and functional Cln3 protein disappear within minutes of GAL-CLN3 shutoff; Cross, 1990; Tyers et al., 1992; Cross and Blake, 1993). The aliquots were then analyzed for percentage of unbudded cells and Clb2 levels. The results were consistent with the bistability prediction. Shifting the culture to 37°C + glucose without prior galactose addition (our unpublished data) or immediately after galactose addition (time zero) resulted in stable retention of the low-Clb state, and cells did not bud in the 2.5-h incubation in 37°C+glucose. In contrast, incubation in galactose at 23°C for 1 h before shift to 37°C+glucose resulted in acquisition of a significant level of Clb2 at the end of the 2.5-h 37°C+glucose incubation, with most cells arrested in the characteristic large-budded morphology observed with cdc14-1 arrest (Figure 1). This was so even though before the shift, Clb2 protein levels were low. These results indicate that the cln1,2,3 arrest does not require CDC14 function for its maintenance, and the cdc14-1 arrest does not require CLN function for its maintenance. The phenotype resulting from simultaneous absence of CDC14 and CLN function depends on the prior history of the system, and a relatively short exposure to CLN function is sufficient to commit the system to later entrance into the high-Clb state. In the absence of initiator or terminator, the system can reside in either of two states (high- or low- Clb), and which state the system adopts depends on its prior history.
Thus, Cdc14 activity is not required for maintenance of G1 arrest with low-Clb2 levels. In contrast, Cdh1 and Sic1, which are activated by Cdc14-dependent dephosphorylation, are required for maintenance of low-Clb2 G1 blocks due to cln deprivation (Tyers, 1996) or α-factor treatment (Schwab et al., 1997). Similarly, we have observed that cln-deficient cdh1 mutants are inviable but arrest in glucose medium with high Clb2 levels (our unpublished data). APC activity (presumably Cdh1- dependent) is also required for maintenance of an α- factor G1 block (Irniger and Nasmyth, 1997). This distinction between activities (such as Cdc14) required to enter a new state and activities (such as Cdh1 and Sic1) required to maintain the state is expected, based on the bistability hypothesis (Nasmyth, 1996; Chen et al., 2000).
Control of Cell Cycle Start by CLN3
In the model of Chen et al. (2000), cell cycle initiation or “Start” is coupled to cell size by the following mechanism. The Cln3 G1 cyclin is assumed to accumulate in total cellular abundance in parallel to total cell mass. It is assumed to concentrate in the nucleus (or in principle any cell compartment of constant volume) so that as its cellular abundance increases, its nuclear concentration increases. Past a certain threshold level it triggers G1/S transcription by activating SBF/MBF (Koch and Nasmyth, 1994), turning on the more downstream-acting G1 cyclins Cln1 and Cln2 along with other genes. Consistent with this model, we found recently that Cln3 does indeed accumulate in the nucleus. Also, moving Cln3 from the nucleus to the cytoplasm significantly reduces its function (Miller and Cross, 2000; Miller and Cross, submitted).
In the simplest version of the idea that cells read their size based on Cln3 nuclear abundance, one might expect that doubling Cln3 levels should result in cells reading their size as twice the actual size, thus halving the cell volume at which Start occurs. In fact, the cell volume response to doubling CLN3 gene dosage is much more modest (Nash et al., 1988; Cross, 1989; Figure 2). The model primarily accounts for this using the properties of Bck2, which acts genetically as a parallel system to Cln3 activating SBF/MBF- regulated genes (Epstein and Cross, 1994; Di Como et al., 1995). In the absence of Bck2, Cln3 becomes essential, and in the absence of both Cln3 and Bck2, SBF/MBF-regulated genes are expressed at very low levels (Epstein and Cross, 1994; Di Como et al., 1995). The presence of the BCK2 gene provides backup and blunts the response to CLN3 gene dosage. Therefore, according to the model, deleting BCK2 should result in highly elevated responsiveness of cell size to CLN3 gene dosage.
Figure 2.
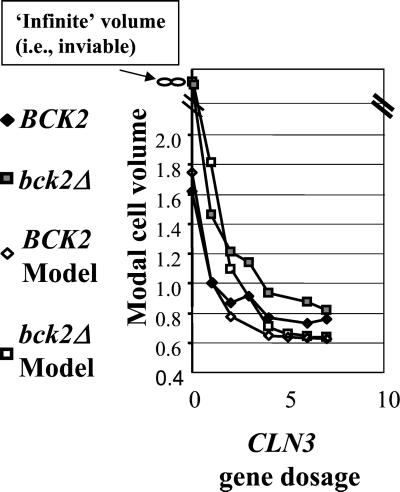
The effect of CLN3 gene dosage on cell size control. For the model predictions, standard parameters from Chen et al. (2000) were used. Changes in CLN3 gene dosage were simulated by changing the DN3 parameter. bck2 deletion was simulated by reducing the BCK parameter to 0. The modal cell volume from the model was estimated as the midpoint between the size at birth and at cell division, relative to a wild-type value of 1. For the experimental observations, cultures of the appropriate genotype (see MATERIALS AND METHODS) were grown to log phase in YEPD, and the peak modal cell size was determined relative to a wild-type control, set to a value of 1. The total CLN3 gene dosage was taken to be the calculated number of CLN3 genes in the transgene array for strains that were cln3:: URA3 at the endogenous CLN3 locus. CLN3 gene dosage was incremented by one for strains with wild-type CLN3 at this position.
To test this idea, we constructed a series of strains with or without BCK2, in which the endogenous CLN3 gene was either present or absent and additionally containing ectopic copies of the CLN3 gene inserted at the TRP1 locus. A 6.2-kb chromosomal segment containing CLN3 was stably integrated at trp1 in one or multiple copies (quantitated by Southern hybridization). The size of the segment makes it likely that expression levels will be little affected by the site or copy number of integration. Indeed, we observed a similar cell volume in cln3::URA3 strains containing a single- copy CLN3 transgene to the cell volume of wild- type cells (our unpublished data), whereas CLN3+ strains containing a single-copy transgene or cln3::URA3 strains containing multiple- copy transgenes were smaller than wild type (Figure 2).
Predictions from the model for approximate modal cell volume were taken as the midpoint between predicted birth size and division size, relative to wild type. Modal cell volumes for the constructed strain set were determined by electronic cell volume measurements, relative to wild type. A reasonable correspondence between model and experiment was observed in the BCK2 and bck2 backgrounds (Figure 2). A minor difference may be that the model predicts a more extreme response to CLN3 dosage than was actually observed; thus the size control system (with or without BCK2) may be more robust with respect to these genetic perturbations than predicted. (It is also possible that the effects of CLN3 gene dosage saturate at higher levels due to limitation of some other factor). It is important to note that although limited information on the relationship between CLN3 gene dosage and cell size was used as input information in formulating the model, these data were all in a BCK2 background. Therefore, the bck2 results presented here are independent confirmation of the model.
An interesting feature of CLN3 expression is that it is under moderate cell cycle regulation, with RNA expression peaking in late M/early G1 (McInerny et al., 1997). This feature is not implemented in the model, and it is unclear how its implementation would affect these size control predictions.
Predicted Interactions between G1 Cyclin Function and Mitotic Regulators Sic1 and Cdh1
The model makes critical use of Cln2 as an initiator activity to drive cells from a low-Clb to a high-Clb state, because Cln2-dependent phosphorylation is assumed to be able to reverse two independent controls that reinforce the low-Clb state, Sic1 stability, and Cdh1 function. Cdh1 (Hct1 in the model; see MATERIALS AND METHODS for a note on nomenclature) is thought to control Clb2 degradation in mitotic exit, and most specifically in the G1, low-Clb state (Schwab et al., 1997; Visintin et al., 1998). Cdh1 is dispensable for viability, presumably because Sic1 is sufficient to control Clb kinase levels, as evidenced by specific lethality of cdh1 sic1 double mutants (Schwab et al., 1997; Visintin et al., 1998).
SIC1 expression is also transcriptionally controlled (Knapp et al., 1996; Toyn et al., 1996), and this is implemented in the model. This control is helpful but not essential in the model: making SIC1 transcription constitutive at the level of peak regulated expression is not lethal (Figure 3A, CONST SIC1), because first Cln2 and later Clb5 and Clb2 can keep Sic1 protein levels low. Deleting CLN2 in the model results in lethality of constitutive SIC1 expression (cln1,2−CONST SIC1), as has been observed experimentally (cln1 cln2 GAL-SIC1 strains are inviable; Tyers, 1996).
CLN2 is turned off transcriptionally by Clb2 (Amon et al., 1993; Koch et al., 1996). This regulation is implemented in the model, where it is helpful but not essential: constitutive CLN2 expression (at the level of peak regulated Cln2 expression) is not predicted to be lethal (CONST CLN2). Constitutive CLN2 expression in the cln1,2 background is predicted to rescue inviability due to constitutive SIC1 expression (CONST CLN2, CONST SIC1).
In contrast, the model predicts that constitutive CLN2 expression should be lethal in the absence of Cdh1, presumably because then the cell becomes highly sensitive to the ability of even low-level Cln2 to destabilize Sic1 by phosphorylation (CONST CLN2 cdh1-). Interestingly, at the other extreme, the model predicts that complete absence of Cln2 should be lethal in the absence of Cdh1 (cln1,2−cdh1-). This lethality is predicted because in the absence of Cln2, cell cycle Start occurs at abnormally large size. Therefore, when Clb2 cyclin accumulates, it is driven by the large cell mass to levels that require the presumed catalytic activity of Cdh1 for effective Clb2 disposal. Thus, the model makes two predictions: that Cln2 constitutive expression should be lethal in the absence of Cdh1 and that simultaneous removal of Cln2 and Cdh1 should also be lethal. In these simulations, constitutive Cln2 expression from the GAL promoter is set to be equal to peak expression of the endogenous gene, but the results are not very sensitive to this level (our unpublished data).
To test these predicted interactions, we constructed a diploid with the genotype cdh1::LEU2/CDH1 GAL1::CLN2::TRP1/trp1 GAL1::SIC1::URA3/ura3 cln1/cln1 cln2/cln2. Segregants of this diploid were tested for viability on glucose or galactose medium (where the GAL-controlled cassettes were off or on). As expected, GAL-SIC1 expression resulted in inviability in the cln1,2 background (Tyers, 1996), and this inviability was rescued by GAL-CLN2 expression (Figure 3B, +GAL-SIC1, vs. +GAL-SIC1 +GAL- CLN2). These observations meet the expectation of the model (Figure 3A, cln1,2−CONST SIC1 vs. cln1,2−CONST SIC1 CONST CLN2). In contrast to the expectations of the model, this CLN2 overexpression cassette, which effectively rescued inviability due to GAL-SIC1, did not cause lethality in the absence of CDH1 (Figure 3A, cln1,2−cdh1- CONST CLN2; Figure 3B, +GAL-CLN2, +GAL-CDH1). Also, there was no reduction of viability of the cln1 cln2 segregants (without GAL1::CLN2 or GAL1::SIC1 expression) due to cdh1 deletion, although the model predicts absolute inviability of cdh1 cln1 cln2 strains (Figure 3A, cln1,2 cdh1-; Figure 3B, all cdh1- strains on glucose medium).
One possible explanation for the viability of GAL- CLN2 cdh1 strains, that Cln2 expressed late in the cell cycle is unable to form an active complex with Cdc28 kinase, is contradicted by previous experimental evidence (Amon et al., 1993).
Fiddling with the parameter set for the model can remedy some of these incorrect predictions. For example, increasing the rate of Sic1 expression threefold rescues lethality due to constitutive Cln2 expression in the absence of Cdh1, but does not rescue inviability due to lack of Cln2 and Cdh1. Lowering Clb2 synthesis rates twofold rescues inviability due to lack of Cln2 and Cdh1, but does not rescue lethality due to constitutive Cln2 in the absence of Cdh1. These two changes in the parameter set work essentially by increasing the Sic1/Clb2 ratio and thus help the model to inactivate Clb2 kinase even in the absence of Cdh1-mediated Clb2 degradation. Alternatively, increasing the ability of Cdc20 to degrade Clb2 (by increasing kdb2p from 0.05 to 0.5) rescues inviability due to lack of Cdh1 combined with either Cln2 overexpression or Cln2 absence. This change works by reducing the importance of Cdh1 in controlling Clb2 abundance. A related solution (K. Chen and J. Tyson, personal communication) is to increase Sic1 expression twofold and also to increase Cdc20-dependent Clb2 degradation fourfold. Possible empirical justification for increasing Cdc20-dependent Clb2 degradation are discussed below (see DISCUSSION), but the case is still unclear.
These findings emphasize a significant problem with the modeling approach: in the absence of empirical constraints on parameters, one is free to propose any parameters that fit the available data. Therefore, it appears likely that before this or any future model can be forcefully tested, more of the parameters need to be based on empirical data. An initial step toward accumulating a suitable data set is the subject of the remainder of this article.
Quantitative Analysis of Abundance of Cell Cycle Regulators
There is a large amount of information available on regulation of abundance of cyclins through the yeast cell cycle (summarized in Chen et al., 2000). A nearly universal deficit in this data set is that one can almost never compare quantities of one cyclin to another, and absolute abundance of these proteins have never been determined. The work of Tyers on the G1 cyclins (Tyers et al., 1993) is an exception to the first point. Tyers et al. tagged the three CLN cyclins identically with the HA epitope tag, such that after immunoprecipitation and Western analysis, the abundance of the three tagged cyclins could be compared with each other. A problem with this analysis was that detection of Cln3 (clearly the least abundant) was so low that the exact reduction in its abundance could not be determined. A related problem was that for some of Tyers' experiments, immunoprecipitation was required before Western analysis, with unknown losses in this step.
PrA Tagging
We constructed strains in which endogenous cyclin genes were C-terminally tagged with protein A, with expression from the endogenous promoter and chromosomal location. It is important to confirm that any epitope tag addition does not significantly affect function of the tagged protein. To address this, we performed a range of tests on most of the PrA-tagged genes.
Cdc28-PrA–expressing haploids were viable; because Cdc28 is essential, the PrA tag cannot have inactivated function. All the PrA-tagged haploid strains had essentially normal FACS profiles (our unpublished data). In contrast, clb5, clb2, or sic1 deleted strains have increased proportions of cells between 1 and 2C DNA content (clb5; Epstein and Cross, 1992) or increased proportions of 2C DNA content cells (sic1, clb2; Surana et al., 1991; Schwob et al., 1994). This indicates approximately normal function of the tagged Cdc28, Clb5, Clb2, and Sic1. sic1::HIS3/+ diploids and sic1::HIS3/SIC1-PrA diploids had FACS profiles indistinguishable from wild type, in contrast to the defective profile of sic1::HIS3 homozygous diploids (with few or no 1C DNA content cells). This indicates full function of the PrA-tagged Sic1 even under conditions potentially limiting for Sic1 (our unpublished data).
clb3 and clb1 inactivation do not have an identified phenotype. Therefore, we tested the PrA- tagged versions by crossing them to a clb2::LEU2 strain, because clb1 clb2 and clb2 clb3 double mutants are inviable (Fitch et al., 1992). In parallel we crossed a clb1::URA3 and a clb3::TRP1 strain to the clb2::LEU2 strain. In tetrad analysis from these diploids, we confirmed inviability of clb1 clb2 and clb2 clb3 double mutants. In contrast, CLB1-PrA clb2 and CLB3-PrA clb2 double mutants were recovered at the expected frequency and did not have a significant slow-growth phenotype compared with clb2 single mutants, although this was not evaluated quantitatively (our unpublished data).
clb2 deletion results in a significant delay in the cell cycle after DNA replication (Surana et al., 1991), and CLB1 and CLB3 are both partially redundant with CLB2 (Fitch et al., 1992). Therefore, if PrA-tagged Clb1 or Clb3 were reduced in function, then clb2 CLB1-PrA or clb2 CLB3-PrA strains might be expected to have an exacerbated postreplicative delay relative to that in clb2 CLB1 CLB3 strains. We compared the phenotypes of clb2 CLB1-PrA and clb2 CLB3- PrA strains to clb2 CLB1 CLB3 strains by FACS analysis and observed little difference, although the clb2 CLB3-PrA strains may have had a moderate decrease in the proportion of 1C cells compared with clb2 CLB1 CLB3 strains (our unpublished data). Overall, these data indicate that the tagged Clb1 and Clb3 have a significant degree of biological function.
clb5 CLB6 strains exhibit a lengthened period of DNA replication and a compensating decrease in the population of cells with 1C DNA content. Deletion of clb6 in the clb5 background results in a long delay before replication and a large increase in the population of cells with 1C DNA content (Epstein and Cross, 1992; Schwob and Nasmyth, 1993). This is due to activation of early but not late origins of replication by Clb6 in the absence of Clb5; when both Clb5 and Clb6 are deleted, neither class of origins is activated until Clb1,2,3,4 are activated later in the cell cycle (Donaldson et al., 1998). We therefore tested CLB6- PrA in a clb5 background by FACS analysis, to test the ability of CLB6-PrA to promote early origin activation. We found that clb5 CLB6-PrA strains had FACS profiles similar to clb5 CLB6 strains, lacking the strong accumulation of 1C DNA content cells seen in clb5 clb6 strains, suggesting significant ability of Clb6-PrA to activate early origins of replication (our unpublished data). The population of cells with 1C DNA content was slightly increased in clb5 CLB6-PrA strains compared with clb5 CLB6 strains, suggesting a moderate reduction of Clb6-PrA function compared with Clb6.
cln3 disruption results in a cell volume increase of at least 50% (Cross, 1988; Nash et al., 1988), while CLN3-PrA strains exhibited at most a 10% increase in cell volume (our unpublished data). CLN3-PrA also rescued cln1 cln2 cln3 inviability about as well as did wild-type CLN3 (the latter assay was performed using low-copy-number plasmids, expressing CLN3 or CLN3-PrA from the CLN3 promoter; our unpublished data). Thus, Cln3-PrA was functional.
As a further functional test, we tested Cln2-PrA, Clb5- PrA, and Clb2-PrA for binding to Cdc28 by constructing strains expressing both the PrA-tagged cyclin and HA-tagged Cdc28 and purifying the PrA-tagged cyclin on IgG-agarose. Although the result was not quantitated, all three cyclins bound Cdc28-HA roughly in accordance with the abundance of the cyclin (our unpublished data). For all nine cyclins, we also were able to recover IgG-agarose–purified histone H1 kinase activity, indicating that the tagged cyclins were able to activate enzymatic activity of bound Cdc28.
Thus, the PrA fusions generally exhibit significant biological and biochemical function and in most cases function similarly to the untagged wild-type genes. Moderate reductions in function cannot be ruled out in most cases, and this leads to a caveat in the use of the tagged proteins for quantitation. An additional subtle caveat could be that if the PrA addition simultaneously weakens biological function but increases protein stability, the net effect could be to hide the loss of activity, while confounding the quantitative measurements of protein abundance.
Average Copies per Cell in Asynchronous Culture
To determine copies per cell of the PrA-tagged proteins, we used the following procedure. We produced recombinant His-GST-PrA fusions in E. coli, purified the fusion on nickel beads, and quantitated the yield. We then performed serial dilutions of the recombinant protein and compared the signal obtained to that from serial dilutions of yeast protein extracts from known numbers of yeast cells. We used dilutions yielding signal in a linear range of detection using digital camera detection from exposed film (Figure 4). The results of this quantitation are presented in Table 1.
Validation of the Quantitation
As an independent test of our data set, we constructed a recombinant GST-myc standard and quantitated myc-tagged Cln2, Cln3, and Clb5 (Table 2). As a second independent test of our data set, we compared the abundance of endogenous Clb2 to recombinant standard MBP-Clb2, using anti-Clb2 antibody (Table 2). These independent comparisons agree with the PrA data set, within a factor of two or three. Given the number of experimental manipulations and calculations involved, we consider this agreement reasonable.
We have been able to find only one literature value to compare with our data: for Cdc28, 10 ng/107 haploid cells (Funakoshi et al., 1997), translating to 16,000 copies per haploid cell. We calculate 12,000 copies per diploid cell (Table 1). Diploids have two copies of the Cdc28 gene and are about twice as big as haploids. One might therefore expect to find twice as much Cdc28 in diploid cells (although a systematic examination of the consequences of ploidy changes on individual protein levels has not been carried out to our knowledge). Thus, our estimate is in a similar range to the published one, although probably a few-fold lower.
Cells simultaneously expressing Clb2 and Clb5 C-terminally tagged with an HA epitope, from the endogenous promoters, show a moderate (although unquantitated) excess of Clb2 over Clb5 (Schwab et al., 1997), consistent with our results (Tables 1 and 2).
The approximately twofold difference between Cln2 and Cln1 levels that we detect is slightly greater than might be expected, based on the nearly identical levels of Cln1- and Cln2-associated kinase activity reported previously using HA-tagged cyclins (Tyers et al., 1993). Tyers et al. (1993) reported a 200-fold difference between Cln2-associated and Cln3- associated kinase activity, compared with a 15-fold difference in protein abundance detected in our experiments. Cln3-associated kinase activity is relatively low under the extraction conditions used by Tyers, and different conditions improve Cln3-associated kinase compared with Cln2 (Jeoung et al., 1998; Miller and Cross, 2000). Tyers et al. (1993) did not quantitate their Western signal for Cln3 compared with Cln1 and Cln2, but a value of 7% does not seem unreasonable from inspection of their data. Thus, overall we consider our G1 cyclin quantitation to be in reasonable agreement with published data.
Grandin and Reed (1993) concluded that Clb3 accounted for about two thirds of the total Cdc28 histone H1 kinase activity in asynchronous cells, based on recovery of Cdc28- associated kinase from a clb3 deletion mutant. This result is not consistent with our finding that Clb3- PrA is present at less than one third the level of the other Clbs added together and at an even lower level when Cln1 and Cln2 are included (Table 1). This discrepancy might suggest that Clb3-PrA levels are under-reporting true Clb3 levels. Alternatively, the effects reported for the clb3 deletion mutant (Grandin and Reed, 1993) could be indirect effects of clb3 deletion on levels of other cyclins, or the Clb3-associated kinase could be unusually active relative to other cyclin- associated Cdc28 kinase because of posttranslational effects. The last explanation is unlikely, although, since using IgG-agarose purification, we recover similar levels of histone H1 kinase activity and similar amounts of PrA- tagged cyclin from cells expressing Clb2-PrA and Clb3-PrA (our unpublished results).
Overall, it appears likely that the data obtained by PrA tagging (Table 1) are reasonably accurate. For purposes of discussion we will take the PrA quantitation literally, although the caveats discussed above (both functional and quantitative) should be kept in mind.
Correlation between Abundance and Functional Importance in B-type Cyclins
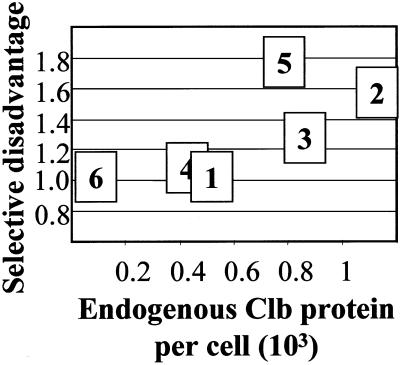
The six B-type cyclins derive by gene duplication from a single ancestor and more recent relationships can be observed. The B-type cyclins can be classed by sequence homology and time of expression in the cell cycle into the CLB5,6, CLB3,4, and CLB1,2 pairs (Fitch et al., 1992; Grandin and Reed, 1993; Schwob and Nasmyth, 1993). Recent work (Lynch and Conery, 2000) suggests that some gene duplications may be found in modern genomes simply as a consequence of their recent generation. To evaluate the functional significance of the six CLB genes, we performed competition growth experiments between various clb gene deletions and wild-type strains. We found that deletion of the three CLB genes with the least abundant products, CLB1, CLB4, and CLB6, resulted in no significant selective disadvantage in competition with wild type, whereas deletion of the three CLB genes with more abundant products, CLB2, CLB3, and CLB5, yielded clear selective disadvantages (Figure 5). (Note that these selective disadvantages are unlikely to be entirely due to differences in exponential growth rate, based on previous data, but we have not attempted to determine the sources of the disadvantages.) This result suggests that although the three sequence classes are functionally distinct and all maintained by natural selection, one member of each class (satisfyingly, in each case the one expressed at a lower level) may not be under strong selection, at least in vegetative culture in rich medium. It is important to note, although, that CLB1 and CLB4 have significant roles in meiosis (Grandin and Reed, 1993; Dahmann and Futcher, 1995), which imposes a distinct selective pressure for their maintenance.
Figure 5.
Selective disadvantage due to clb gene disruption. The selective disadvantage due to the indicated clb gene disruption was determined in a single cycle competitive growth experiment from stationary phase, through a dilution of ∼106-fold, back to stationary phase. The experiment was in liquid YEPD medium, over a 30-h period at 30°C, and the culture initially contained an approximately equal mixture of wild-type and mutant cells. The selective disadvantage parameter will reflect differences in doubling time on the assumption that entry into and exit from stationary phase are identical for all strains, but this has not been tested. In all cases, the clb gene disruption was compared with a control strain with identical auxotrophic markers (see MATERIALS AND METHODS).
Of the mitotic cyclins CLB1,2,3,4, clb2 deletion alone results in a significant cell cycle delay before mitosis, with consequent cell enlargement and reduction of length of G1; in contrast, single deletions of other mitotic CLB genes (CLB1,3,4) have only minor phenotypes. If these cyclins are fully overlapping in all functional aspects and differ only quantitatively, then the data in Table 1 allow the conclusion that clb2-deleted cells should result in a reduction of ∼40% in total mitotic Clb level. This rather moderate reduction can be easily modeled using the Chen et al. (2000) parameters by lowering Clb2 synthesis parameters ksb2′ and ksb2" by 40%, yielding about a 12% increase in predicted cell volume at cell division. This increase is significantly less than is observed with clb2 deletion (Surana et al., 1991). These quantitative considerations may suggest only partial functional overlap among the mitotic cyclins. For example, suppose Clb1 completely overlaps in function with Clb2 (consistent with the high sequence conservation between Clb1 and Clb2), whereas other cyclins are not considered at all. Then the clb2 deletion will result in about a 70% decrease in Clb1/2 functional protein, which is predicted by the model to yield a nearly twofold increase in cell volume at cell division. An increase of this magnitude is more consistent with observation (Surana et al., 1991). Simple quantitative considerations of this sort may therefore have implications for cyclin functional specificity (see DISCUSSION).
Abundance through the Cell Cycle and the Role of Cdh1
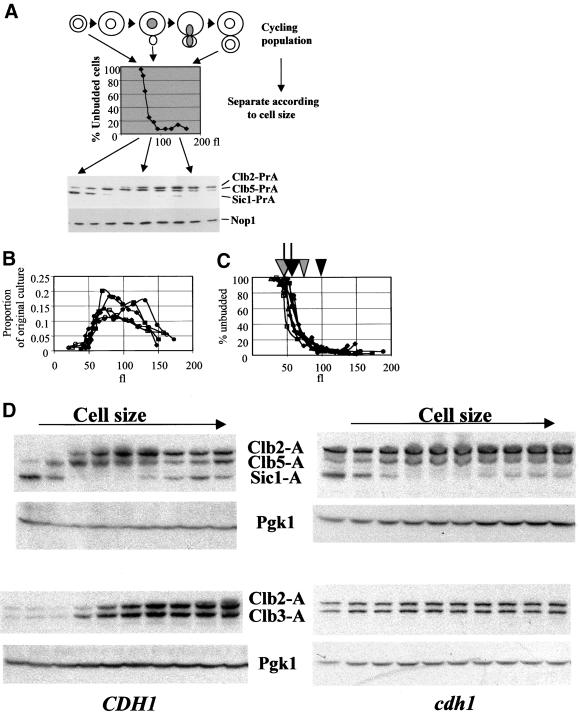
To analyze fluctuations of the PrA-tagged cyclins through the cell cycle, we separated cells on the basis of cell size. In this elutriation method, cultures growing rapidly in rich medium are quickly chilled and then directly fractionated, such that no further physiological response of the culture is required after chilling (Levine et al., 1996; Oehlen et al., 1996). The entire culture is recovered and analyzed in this way. The strategy and sample data are shown in Figure 6A.
Figure 6.
Characterization of the elutriation protocol. Log phase cultures of diploid strains heterozygous for various PrA-tagged genes were elutriated as described in MATERIALS AND METHODS. The entire culture was collected. (A) Schematic of the procedure, and sample immunoblotting data. Top: cartoon of an asynchronous population. Middle: the effect of separation on the elutriator, indicated by percentage of unbudded cells. Bottom: sample blot for detection of Clb2-PrA, Clb5-PrA, and Sic1-PrA (all expressed from heterozygous PrA-tagged genes in the diploid strain analyzed). Nop1: loading control detected with anti- Nop1 antibody. (B) The proportion of the total mass of the culture (indicated by OD660) as a function of the modal cell volume of the fractions determined by Coulter Channelyzer calibrated with standard 68 fl latex beads. Closed symbols: wild-type diploid strains; open symbols: cdh1 diploid strains. Each symbol represents a different elutriation. The cdh1 strains reproducibly yielded significant mass at slightly smaller cell volume than the wild type. In the first few fractions derived from the cdh1 mutant cultures, two peaks were reproducibly observed in the Coulter Channelyzer electronic cell volume analysis. We do not know the meaning of this observation. For the data reported in Figures 6, B and C, and 7 A and C, we use the larger peak for the x-axis values for these samples. (C) The percentage of unbudded cells in the fractions. Closed symbols: wild-type diploid strains; open symbols: cdh1 diploid strains. Each symbol represents a different elutriation. Arrows above: approximate positions of 50% completion of DNA replication in wild- type (black) and cdh1 (gray) strains, based on FACS analysis of the fractions. Arrowheads above: beginning of nuclear division, indicated by initial accumulation of a significant population of binucleate cells, determined by microscopic examination of propidium iodide-stained cells; in earlier fractions, essentially no binucleate cells are observed. (D) PrA detection from diploids dou-bly heterozygous for CLB2- PrA, CLB5-PrA, and SIC1- PrA (top) or for CLB2-PrA and CLB3-PrA (bottom). Both CDH1 (left) and cdh1 (right) diploids were analyzed. Pgk1 was detected using anti-Pgk1 antibody for a loading control. U, extract from untagged culture. Note that densities of exposure are not comparable between experiments.
This elutriation method loses resolution in the larger size cell fractions. This is in part due to loss of accuracy of size resolution in the fractions containing larger cells, which is evident from a somewhat variable increase in the peak width in electronic cell volume measurements (normalized to peak position) for later fractions (our unpublished data). It is also possible that cell size does not correlate as tightly with later cell cycle events. The largest fractions contain cells that have started the next cell cycle before cell separation is complete, as evidenced microscopically by occasional rebudding of already budded cells (our unpublished data). Similarly, FACS analysis shows that the fractions of modal cell volume > 125 fl contain a significant 1C DNA content population upon resonication.
Thus, this method gives an accurate separation of cells in early periods of the cell cycle, from birth until after DNA replication. Fractions with the largest cell sizes (>150 fl) are closer to being asynchronous averages due to loss of resolution of the elutriation.
Multiple diploid strains were analyzed with similar volume distributions and dependence of budding on cell volume (Figure 6, B and C). cdh1 mutant diploid strains reproducibly budded and initiated DNA replication at ∼10 fl smaller volume than the wild type (arrows in Figure 6C) and initiated nuclear division at ∼25 fl smaller than wild type (arrowheads in Figure 6C). An increase in the population of anaphase cdh1 mutants was noted previously (Visintin et al., 1998).
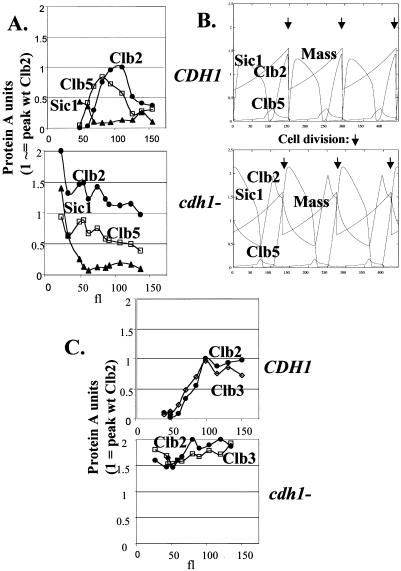
We elutriated a triply heterozygous diploid, in which coding sequences for Clb2, Clb5 and Sic1 were tagged with PrA on one of the two alleles of each. This allowed a direct comparison of the abundance of the three proteins with the same tag, within the same experiment. We express the units in this experiment relative to peak Clb2 concentration. If peak Clb2 expression corresponds to two times the average asynchronous level (Table 1), one unit on these graphs should correspond to ∼35 nM (2400 copies/120 fl cell).
As a cross-check, it is possible to predict asynchronous levels of the tagged proteins by integrating across the elutriation profile, multiplying the observed amount of the protein in the size fractions (Figure 7, A and C) by the proportion of the mass of the culture recovered in these fractions (Figure 6A). This can then be compared with the levels directly obtained in asynchronous cells (Table 1). This calculation from the two elutriations quantitated in Figure 7 yields predicted asynchronous Clb2:Clb3:Clb5:Sic1 ratios of 1:0.91:0.91:0.27, compared with ratios from Table 1 of 1:0.76:0.70:0.19. This agreement suggests that the quantitation of the elutriation is reasonably accurate, with most of the proteins recovered and assigned to the different cell size classes.
Figure 7.
Abundance of Clb2, Clb3, Clb5, and Sic1 through the cell cycle, with and without Cdh1: model and experiment. (A) Quantitation of data from Figure 6D, top; diploids heterozygous for CLB2-PrA, CLB5-PrA, and SIC1-PrA. Top: wild type; bottom: cdh1. Levels of the PrA fusions were determined densitometrically, and corrected for loading by quantitation of anti-Pgk1 signal. The scale was normalized to 1 for peak Clb2 levels in wild-type cells; this is probably ∼35 nM (see text). This scale was transferred to the cdh1 case by comparing a sample with peak Clb2-PrA from a wild-type diploid to a similar sample from a cdh1 diploid and standardizing the PrA signal to the Pgk1 signal, resulting in the determination that the peak in the cdh1 mutant was ∼2 times that in the wild type. This correction is somewhat approximate, compared with the scale within an elutriation, which is internally controlled because of coexpression of single chromosomal identically tagged genes. (B) Model predictions for the levels of the same proteins. Wild type: standard parameters (Chen et al., 2000). cdh1: the parameter kdb2" was reduced from 2 to 0.01. Mass is total cell mass in the simulation. Arrows indicate cell division. (C) Quantitation of data from Figure 6D, bottom; diploids doubly heterozygous for CLB2-PrA and CLB3-PrA. Top: wild type; bottom: cdh1 mutant. Analysis as in A.
Although Sic1 was found at quite low levels in asynchronous culture (Table 1), it was abundant in the smallest cells in the culture, where it was in molar excess over the levels of Clb5 and Clb2 coexpressed in the same cells (Figures 6, A and D, and 7A). These cells contributed a very small proportion of the total mass of the culture (Figure 6B), and this could account for the low relative yield of Sic1 in asynchronous total culture (Table 1). If Sic1 inhibits Clb- Cdc28 complexes in a 1:1 stoichiometric ratio, this suggests that under these growth conditions Sic1 is only present at levels sufficient for Clb inhibition for a brief period early in the cell cycle. The onset of DNA replication in cells of ∼60 fl (black arrow, Figure 6C) correlates with the increase of Clb5 above the Sic1 threshold (Figure 7A). We find that limiting the growth rate of the culture by changing the carbon source from glucose to glycerol increases the duration of the period when Sic1-PrA is high. Thus, growth limitation may expand the high Sic1 pre-Start period of the cell cycle (M.K. and F.C., unpublished data). This expansion is expected based on the known mechanisms for coordinating growth and division in yeast (Hartwell and Unger, 1977; Cross et al., 1989; Cross, 1995).
By comparison with coexpressed cyclins, it appears that the highest concentration of Sic1 found in the smallest cells analyzed is ∼0.4 times the peak concentration of Clb2 (Figure 7A). Clb2 peaks in the vicinity of 2400 copies per cell (estimating that peak concentration is two times the average concentration reported in Table 1), in cells of ∼120 fl (Figure 7A), yielding a concentration of 35 nM. Thus, peak Sic1 concentrations should be ∼15 nM. The Ki determined for purified Sic1 on Clb-Cdc28 kinase activity was 1.6 nM (Mendenhall, 1993). This will allow Sic1 to be effective at inhibiting Clb kinase, provided there is even a moderate excess of Sic1 over Clbs (assuming 1:1 stoichiometry for inhibition). This effect becomes much stronger if Sic1 is concentrated in the nucleus, but we are unaware of data on this point.
The Chen et al. (2000) model predicts qualitatively patterns of accumulation of these different proteins similar to what we observe, with several potentially significant differences (Figure 7B). First, the model predicts a long period of time when Sic1 accumulates stably. We observe, in contrast, very little Sic1 accumulating, for only a short time (translating cell volume increments into time, based on the fact that yeast cells probably increase approximately exponentially in cell mass throughout the cell cycle; Elliott and McLaughlin, 1979). Second, the model calls for a high level of Clb2 compared with Clb5, although we observe nearly comparable levels of these cyclins. Third, Clb2 accumulation appears significantly “peakier” in the model than in the experiment, but this could be a consequence of the poor synchrony in the larger-cell fractions noted above. Overall, the model clearly does a very good job of qualitatively predicting times of accumulation, but the lack of common- scale quantitative information prevented relative levels of different components from being appropriately specified. The more accurate numbers provided here will have consequences for the predicted efficiency with which different cyclins carry out different tasks (see DISCUSSION).
Cdh1 is known to be important for restricting Clb2 protein accumulation, especially in postmitotic cells (Schwab et al., 1997; Visintin et al., 1998; Zachariae et al., 1998). We elutriated a cdh1 strain triply heterozygous for PrA-tagged CLB2, CLB5, and SIC1 genes and found strong deregulation of Clb2 accumulation, with only minor effects on Clb5 and Sic1 accumulation. (The minor fluctuations in Clb2 levels that we observe are not very reproducible; cf. Figure 7A with 7C). It was notable in this background that even in the smallest cells that we could isolate, Sic1 was most likely not in stoichiometric excess over Clb cyclins (because it was not even in clear excess of Clb2 considered alone, without including Clb5, Clb3, and other cyclins). Thus, in the absence of Cdh1, Sic1 regulation of Clb kinase in postmitotic cells may be inefficient. This may account for the entry into DNA replication of these strains at a smaller cell size (gray arrow, Figure 6C).
We approximately standardized the scale of the cdh1 experiments to the CDH1 experiments by determining that the signal from peak Clb2 levels in cdh1 strains was about two times the level in a CDH1 strain. Thus, all the graphs in Figure 7, A and C, are similarly scaled to a value of 1 for peak Clb2 expression in a CDH1 strain. (Note that this is an approximation to allow rough quantitative comparison between the experiments and is intrinsically less accurate than the within-experiment comparisons, which are standardized by coexpression of PrA-tagged genes within the same cell.)
The effects of cdh1 deletion are generally similar to those predicted by the model (Figure 7B), except that a more significant residual regulation of Clb2 is predicted than we observe. These oscillations are predicted because the model assumes very strong transcriptional positive feedback for CLB2 and cyclical degradation of Clb2 by Cdc20. Both of these ideas are supported by experimental data (Amon et al., 1993; Baumer et al., 2000; Yeong et al., 2000), but the strength of one or both of the effects may be overstated in the model. Alternatively, if Cdc20-dependent Clb2 degradation becomes ineffective at low Clb2 levels (Yeong et al., 2000), detection of cdh1-independent degradation could be quite difficult at normal Clb2 expression levels. It will be interesting to implement the proposed biphasic Clb2 degradation (Cdc20-dependent degradation to an intermediate level, followed by Cdh1-dependent degradation; Yeong et al., 2000) in a computational model (see DISCUSSION).
The model predicts oscillations of Clb5 and Sic1 in the cdh1 strain that are similar to wild type, essentially as we observe. The Clb5 oscillations are predicted to be of lower amplitude and those of Sic1 of higher amplitude. These are less than twofold effects, and we are not sure if our data confirm these small changes, especially because of the extra correction involved in putting wild- type and cdh1 data on a common scale (see above). The Sic1 prediction seems better confirmed than the Clb5 prediction (Figure 7B).
The model does not include the Clb3 cyclin. Clb3 overlaps functionally with both the Clb5/6 S cyclins and the Clb1/2 M cyclins (Fitch et al., 1992; Schwob and Nasmyth, 1993) and is also present in asynchronous culture at levels similar to Clb5 and Clb2 (Table 1). We determined the pattern of Clb3 and Clb2 accumulation by elutriating doubly tagged diploid strains and observed similar timing and levels of these two cyclins (Figure 7C). Clb3-PrA reproducibly accumulated in slightly smaller cells than Clb2-PrA, possibly because of earlier transcriptional activation of CLB3 than CLB2 (Fitch et al., 1992; Richardson et al., 1992). Grandin and Reed (1993) reported somewhat earlier accumulation of Clb3 than of Clb2. In the experiment in Figure 7C, we did not observe a significant fall-off of Clb2 or Clb3 levels in the largest cells, unlike the results seen in Figure 7A for Clb2; this difference was not reproducible. In experiments (with the same doubly tagged Clb2-PrA, Clb3-PrA strain) where a fall-off of Clb2 was observed in larger cells, a parallel fall-off of Clb3 was also observed (our unpublished data). We attribute the variability to the loss of resolution of the elutriation method in larger cells (see above).
We determined the pattern of accumulation of Clb1-PrA in a diploid doubly heterozygous for tagged CLB1 and CLB3. Clb1 accumulated with periodicity similar to Clb3, but accumulated to only ∼60% the peak level of Clb3 (our unpublished results), as expected from the asynchronous values in Table 1.
We observed similar deregulation of both Clb2-PrA and Clb3- PrA by cdh1 deletion (Figure 7C). This observation suggests that Cdh1 controls both Clb2 and Clb3 accumulation similarly, in disagreement with another report using induced synchrony and overexpressed Clb3 (Baumer et al., 2000). Consistent with our findings, Zachariae et al. (1998) showed that ectopic Cdh1 can induce Clb3 degradation, and Alexandru et al. (1999) proposed that Clb3 might be under control of both Cdc20 and Cdh1.
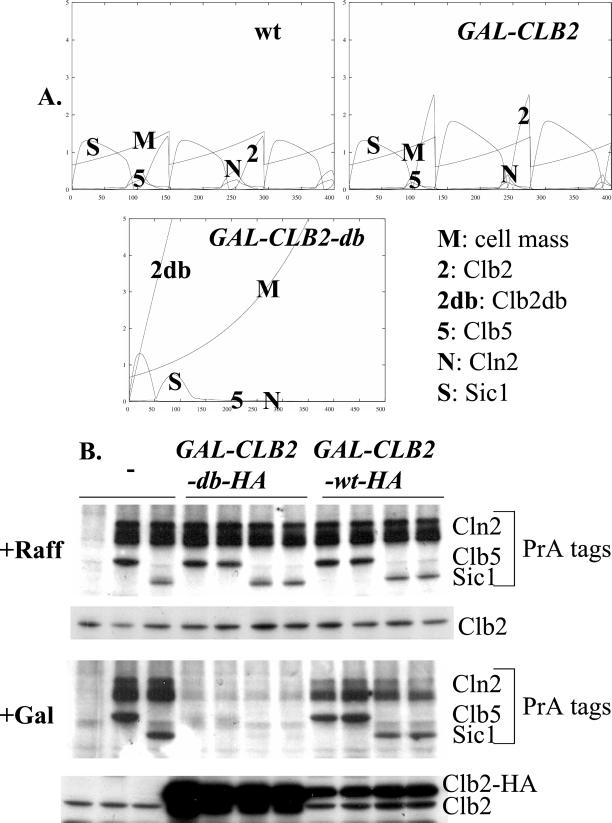
Effects of Constitutive Undegradable Clb2 on Accumulation of Other Cell Cycle Regulators
The model allows explicit predictions to be made about the consequences of interfering with the cell cycle oscillator, not only for cell cycle events but also for accumulation of cell cycle regulators. The availability of a comprehensive set of tagged regulators allows these predictions to be tested. Overexpression of Clb2 lacking its destruction box causes cell cycle arrest late in mitosis (Surana et al., 1993). This genetic manipulation can be simulated in the model (Figure 8A). The model predicts that Clb2-db overexpression should eliminate Cln2, Clb5, and Sic1 proteins. We introduced a GAL-CLB2-db cassette or a control GAL-CLB2 cassette into strains expressing various PrA fusions and tested the effect of a 3.5-h incubation in galactose medium (enough to give efficient cell cycle arrest with GAL-CLB2- db). The predicted disappearance of Cln2, Clb5, and Sic1 (Figure 8A) was observed in this experiment (Figure 8B), whereas expression of GAL-CLB2 containing the destruction box was without significant effect, also as the model predicts.
Figure 8.
Effects of constitutive undegradable Clb2. (A) Model predictions. Loss of transcriptional control of GAL-CLB2 or GAL-CLB2db compared with wild-type CLB2 was modeled by setting ksb2′ = 0.05, ksb2" = 0. Loss of proteolytic control of GAL- CLB2db was modeled by setting kdb2" to 0.01 and kdb2p to 0, eliminating degradation dependent on Cdh1 and Cdc20, respectively. (B) Haploid strains expressing the indicated PrA fusions were transformed with integrating plasmids containing GAL-CLB2-HA or GAL-CLB2db-HA (Jacobson et al., 2000). The untransformed parent and two transformants with each DNA were grown to log phase in YEP- raffinose (noninducing), and then GAL-CLB2- HA or GAL-CLB2db-HA was induced by addition of galactose to 3%. After 3.5 h proteins were extracted for PrA detection using rabbit IgG and for Clb2 and Clb2-HA detection using anti-Clb2 antibody.
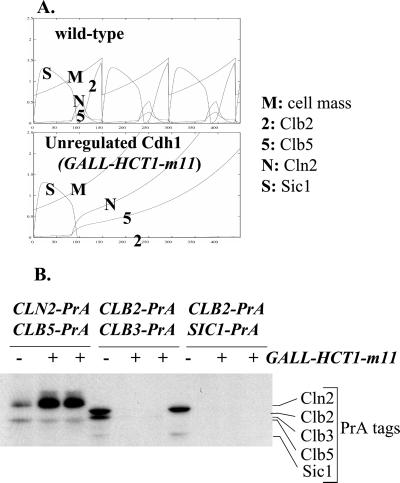
Effects of Unregulated Cdh1
To examine the effect of making Clb2 degradation constitutive, we constructed strains containing various PrA fusions and also expressing an unregulatable Cdh1 mutant under GAL control (GALL-HA3- HCT1-m11::TRP1; Zachariae et al., 1998). The galactose-induced mutant Cdh1 protein expressed from this construct is mutated in its Cdk phosphorylation sites, and so negative control by cyclin-Cdc28 is lost. Thus, this construct results in constitutive Clb2 and Clb3 degradation and blocks the cell cycle (Zachariae et al., 1998). We modeled expression of this construct by simulating Cdh1 not subject to negative regulation by phosphorylation and determined the predicted effects on abundance of Cln2, Clb2, Clb5, and Sic1 (Figure 9A). The model predicts cell cycle arrest with very low levels of Sic1 and Clb2 and very high levels of Cln2 and Clb5. Experimentally, we observed a three- to fourfold increase in Cln2-PrA and disappearance of Clb2-PrA and Sic1-PrA, all in accordance with the model's predictions (Figure 9B). We observed no significant increase in Clb5-PrA, in disagreement with the model, but in agreement with previously published data (Schwab et al., 1997; Zachariae et al., 1998).
Figure 9.
Effects of unregulatable Cdh1. (A) Model predictions. Loss of negative regulation of Cdh1 was simulated by setting kit1 to zero; this simulates prevention of Cdh1 phosphorylation by all cyclin-Cdc28 complexes. (B) Diploid strains were constructed that were heterozygous for the indicated PrA fusions. For each set of fusions, two diploids (+) were also heterozygous for a double integration of the GALL-HA-HCT1-m11 construct (Zachariae et al., 1998), and one diploid lacked the GALL-HA- HCT1-m11 construct (−), as indicated. GALL-HCT1- m11 encodes a mutant Cdh1 (also called Hct1), lacking all Cdk phosphorylation sites and therefore immune to negative regulation (Zachariae et al., 1998). Strains were grown to log phase in YEP-raffinose (noninducing), and then GALL-HA-HCT1-m11 was induced by addition of galactose to 3%. After 2.5 h proteins were extracted for PrA detection using rabbit IgG. The band in the approximate position of Sic1-PrA in the strain expressing Clb2-PrA and Clb3-PrA is a Clb3-PrA breakdown product. Quantitation of the Cln2-PrA signal (quantitation of an anonymous cross-reacting band from elsewhere in the gel provided a loading control) indicated a three- to fourfold increase in Cln2-PrA and about a 40% decrease in Clb5-PrA, due to induction of GALL-HA-HCT1-m11.
In this experiment, the effects of unregulated mutant Cdh1 (GALL-HA3-HCT1-m11) on Clb2 are likely to be primarily due to direct promotion of Clb2 ubiquitination (leading to its degradation) by Cdh1 (Zachariae et al., 1998). The effects of the mutant Cdh1 on Cln2 and Sic1, in contrast, are likely to be indirect. In the model, unregulated Cdh1 leads to Cln2 accumulation through (1) loss of Clb2, with consequent loss of repression of CLN2 transcription, leading to (2) hyperaccumulation of Cln2. The hyperaccumulated Cln2 contributes to very efficient Sic1 phosphorylation and SCF- dependent ubiquitination, leading to Sic1 proteolysis.
The incorrectly predicted strong increase in Clb5-PrA by unregulated Cdh1 is also an indirect effect in the model and is predicted for two reasons. First, the model implements Clb2-dependent stimulation of Cdc20 synthesis (Prinz et al., 1998), and Cdc20 is required for efficient Clb5 proteolysis in the model. Second, the model assumes that Clb2 is required to turn off CLB5 transcription in the same way as it is required to turn off CLN2 transcription. These two hypothetical mechanisms will make removal of Clb2 by unregulated Cdh1 lead to increased Clb5 levels. There is some information supporting the first assumption (Prinz et al., 1998), but the second assumption is probably incorrect (Amon et al., 1993), as indeed was noted by Chen et al. (2000).
We observed a strong reduction in Clb3-PrA levels upon expression of unregulated Cdh1, consistent with the results of Zachariae et al. (1998), although the reduction was not as effective as the reduction in Clb2-PrA or Sic1-PrA (with longer exposures, in several experiments; our unpublished data).
Thus, for Cln2, Clb2, and Clb3, we observe opposing effects of cdh1 deletion (lowering Cln2 and increasing Clb2 and Clb3) and expression of unregulated Cdh1 (increasing Cln2, and strongly reducing Clb2 and Clb3).
DISCUSSION
Mathematical Modeling of the Cell Cycle
We report empirical tests of the Chen et al. (2000) model for the yeast cell cycle, using genetic and biochemical data not included in the initial model generation. Prediction of dependence of cell size on CLN3 gene dosage and prediction of cyclin and Sic1 abundance in various situations were reasonably accurate. However, the model gave entirely inaccurate predictions concerning interactions between Cdh1 and G1 cyclins. Possible resolutions are discussed in the next section.
The comprehensive set of tagged regulators that we have constructed here will allow tests of quantitative predictions from the model about these levels of these regulators in cycling cultures and at various oscillator blocks (as in Figures 7–9), providing stringent constraints for evaluation of future models.
Of the cyclins not included in the model, CLB3 is a good candidate for inclusion in future. Its functional significance is shown by studies of multiple clb deletion phenotypes (Fitch et al., 1992; Schwob and Nasmyth, 1993), and single clb3 deletion results in a significant selective disadvantage (Figure 5). A difficulty in modeling it is that although its transcription is cell cycle regulated (Fitch et al., 1992; Richardson et al., 1992), nothing is known about the mechanism of this regulation.
Cdh1, Cdc20, and the Control of Clb2 Proteolysis
One way to modify the model to remedy the incorrect predictions on cdh1 (Figure 3) is to propose increased Cdc20-dependent Clb2 degradation. The current model may have an excessive reliance on regulated Cdh1 activity to drive the switch from the high-Clb S/M to the low-Clb G1 state. Regulated Cdh1 may function primarily in maintenance rather than initiation of the low-Clb state, and Cdc20 may be important in removal of Clb's at the end of the cell cycle (Baumer et al., 2000; Yeong et al., 2000). Although we observe little regulation of Clb2 accumulation in cdh1 mutants (Figure 7C), Cdc20 might effectively mediate Clb2 degradation only at higher levels of Clb2 (Yeong et al., 2000). Cdh1 may be required for Clb2 degradation at lower levels. Cdc20-dependent degradation of higher levels of Clb2 might be rapid enough to justify a significant increase in the Chen model's rate constant for Cdc20-dependent Clb2 degradation (kdb2p), but the proposed loss of this rapid Cdc20-dependent degradation as Clb2 levels decline to lower levels (Yeong et al., 2000) would make Cdh1-independent Clb2 degradation hard to detect using the elutriation approach in Figures 6 and 7.
As noted in the INTRODUCTION, checkpoint/surveillance mechanisms are used in the Chen model to introduce oscillations in Cdc20 activity, in an unperturbed cell cycle. We have found, however, that all DNA- and spindle- dependent surveillance mechanisms can be disabled simultaneously (by constructing mad1 bub2 mec1 sml1 quadruple mutants) with no significant effects on viability or growth rate (our unpublished data). The checkpoint mechanisms in the model are essentially a way to provide a delay mechanism between initial Clb2 activation and ultimate resulting Cdc20 activation. This is because lifting the checkpoint is dependent on the SPN variable (simulating spindle morphogenesis) reaching a critical value; SPN is under positive control of Clb2, but if the checkpoints do not regulate Cdc20 in unperturbed cell cycles, a revised model might require another way to provide a delay between Clb2 activation and Cdc20 activation.
Clb2 may activate the Cdc20-dependent form of the APC by phosphorylation of APC subunits (Rudner and Murray, 2000). If this activation proceeded with ultrasensitive switch kinetics (Goldbeter and Koshland, 1982), this would provide a checkpoint-independent delay between Clb2 activation and Cdc20 activation. Such kinetics might also account for the proposed biphasic kinetics of Clb2 proteolysis (Yeong et al., 2000). As Clb2 levels fell because of Cdc20-dependent degradation, at some point Clb2 would pass below the levels required to flip the switch, and this could end the Cdc20-dependent phase of Clb2 degradation, if APC phosphorylation were rapidly reversed. Other ideas could also account for the biphasic kinetics (e.g., a fraction of Clb2 might be protected by an unidentified binding partner; Irniger et al., 1995). More genetic and biochemical data is required to accurately implement biphasic Clb2 degradation in a realistic cell cycle control model.
Cyclin Abundance, Functional Specificity, and ε Factors
Deletions of the different cyclin genes have distinct consequences. The simplest way this could happen is if one cyclin is the most abundant; its deletion might then be expected to have the most severe consequences. For the mitotic B-type cyclins CLB1,2,3,4, the order of severity of phenotypes from deletion analysis (Surana et al., 1991; Fitch et al., 1992) indicates that Clb2 is the most important. clb2 deletion is the only single-CLB deletion in this set with a recognizable phenotype, and all multiple deletion combinations that are lethal or semilethal include clb2. Following CLB2 in importance are CLB1 and CLB3, because clb1 clb2 and clb2 clb3 double mutants are semilethal. CLB4 appears to be of minor importance. The order of importance of Clb1,2,3,4 from these genetic studies: 2 > 1,3 > 4, follows the order of their calculated abundance (Table 1). Determination of selective disadvantage due to clb deficiencies (Figure 5) confirms that only the more abundant ones have a phenotype detectable by this assay. Thus, some apparent cyclin specificity may be due simply to relative abundance. As discussed above, this factor seems unlikely to explain all the data even among these mitotic cyclins, and it is clear that relative abundance cannot universally explain cyclin specificity (reviewed in Miller and Cross, 2001). Instead, some cyclins must be intrinsically specialized, independent of abundance or timing of expression.
To reflect this idea of intrinsic cyclin specialization, Chen et al. (2000) use ε factors to state the efficiency of a given cyclin for a given cell cycle task. With respect to phosphorylation of Sic1 by cyclin-Cdc28, both Cln2- and Clb2- complexes can phosphorylate Sic1 in vitro with broadly similar efficiency (Verma et al., 1997b), although Clb5-Cdc28 may be less effective than Cln2-Cdc28 (Elsasser et al., 1999). The ε factors in the model do not reflect these findings (ε factors for Sic1 inactivation of 1, 1, 0.067 for Cln2, Clb5, and Clb2, respectively). No large differences in Cdh1 phosphorylation in vitro by various Clb complexes were noted (Zachariae et al., 1998), and genetic data suggests that Clb5 may be better than Clb2 at inducing Cdh1 phosphorylation (Shirayama et al., 1999). In contrast, the model parameters suggest twofold more efficient phosphorylation of Cdh1 by Clb2 complexes than by Clb5 complexes.
Genetic experiments where expression and timing of different cyclins have been equalized indicate strong differences in efficiency for inducing DNA replication or for inhibiting mitotic exit for Clb5 versus Clb2 (Cross et al., 1999; Jacobson et al., 2000), whereas both Clb2 and Clb5 appear able to inhibit reloading of replication origins (Nasmyth, 1996, Jacobson et al., 2000). Similar experiments indicate sharp differences in ability to promote various Start-related events for Cln3 versus Cln2 (Levine et al., 1996; e.g., Cln3 is better able to activate SBF-mediated transcription; Cln2 is better at driving bud emergence). These differences in efficiency between Clb2 and Clb5 and between Cln2 and Cln3 are reflected in the model parameters.
Interpreting the Quantitations: How Much is a Lot?
To fully interpret our quantitations (Table 1), subcellular localization must be determined. For example, the model (Chen et al., 2000) proposes a 75-fold greater efficiency of Cln3 compared with Cln1 or Cln2 for activating SBF-regulated transcription. In a compartmentalized model, this extreme efficiency difference may be an unnecessary factor, despite the lower overall cellular concentration of Cln3 compared with Cln1 and Cln2 (Table 1). If Cln2 is uniformly distributed through the cell or excluded from the nucleus, and Cln3 is nuclear- restricted (Miller and Cross, 2000), Cln3 might be at least 2.7-fold more concentrated in the nucleus than Cln2. (216 copies Cln3/4 fl nucleus/2011 copies Cln2/100 fl cell = 2.7, assuming a 4-fl diploid nucleus in a 100-fl cell, because a haploid nucleus is ∼2 fl; Winey et al., 1997).
One of the main roles of Clb5 is to activate DNA replication (Epstein and Cross, 1992; Schwob and Nasmyth, 1993; Donaldson et al., 1998). Clb5 is concentrated in the nucleus (Shirayama et al., 1999; Jacobson et al., 2000). An asynchronous average of 800 copies per diploid cell (Table 1) yields a peak nuclear concentration around 1 μM or 2400 per nucleus, assuming a threefold enrichment at peak over average. Eight percent of the yeast haploid genome contains about 48 origins (Poloumienko et al., 2001), yielding an estimated 1100 origins per diploid genome; similarly, Orc2 (which binds to replication origins as part of the essential ORC complex) was quantitated at ∼600 copies per haploid cell (Donovan et al., 1997). Thus, Clb5-origin affinity may not need to be very high, because Clb5 nuclear concentration is high and Clb5 may be in stoichiometric excess over origins (2400/1110 = 2 Clb5/origin).
In addition to activating replication origin firing, Clb kinases also prevent reloading of replication origins, and this is proposed to contribute to once-per-cell-cycle control of DNA replication (Nasmyth, 1996). Targets for Cdc28-dependent phosphorylation for negative control of replication include Cdc6, the Mcm2–7 complex, and the Orc complex (Nguyen et al., 2001). The abundance of the Orc complex is likely to be similar to that of replication origins (see above), so Clb kinases (considered jointly) are probably significantly more abundant than the Orc complex targets at all times after DNA replication is completed. The Mcm complex may be as much as 50-fold more abundant than the Orc complex (Donovan et al., 1997), so efficient phosphorylation of the Mcm complex may require a more efficient interaction with Clb kinases. Alternatively, since phosphorylation leads to nuclear export of the Mcm complex (Nguyen et al., 2000), even relatively inefficient phosphorylation of the Mcm complex could eventually drain the complex from the nucleus, assuming the Clb kinases remain nuclear.
Cln2 accumulates to a level greater than Sic1 (Table 1). Sic1 is a proposed enzymatic target for Cln2-Cdc28 complexes (Schwob et al., 1994; Verma et al., 1997a, 1997b), and these calculations suggest that a high affinity of Cln2-Cdc28 for Sic1 may not be required for effective phosphorylation of Sic1.
It is interesting that none of the calculations above for relative levels of cyclins and phosphorylation targets imply the need for high-affinity interactions between the cyclin and the target. This could have implications for the tightness of interaction required in general between potential substrate-targeting regions in cyclins (e.g., Cross and Jacobson, 2000) and the phosphorylation targets.
The kinase catalytic subunit Cdc28 was detected at higher levels than any cyclin, about 12,000 copies per cell. Cdk's are generally thought to be in excess of cyclins; the level of Cdc28 calculated here, although in excess, is not tremendously so. This could have implications for experiments in which cyclins are overexpressed, since at high cyclin expression levels competition for Cdc28 could become a significant factor.
The apparently low level of Sic1 in measurements from asynchronous culture (Table 1) nevertheless allows for effective inhibition of Clb kinases in small newborn cells, as predicted (Schwob et al., 1994; see RESULTS).
CONCLUSION
An ideal chemical kinetic model would replace dimensionless concentration terms with molarities in the appropriate subcellular compartment and would replace ε factors with association, dissociation, and enzymatic rate constants. Although this ideal will not be achieved for cell cycle control in the immediate future, the attempt to meet it will generate testable quantitative hypotheses that might be hard to obtain by intuition. Empirical studies targeted at model testing should allow efficient development of more realistic quantitative models, perhaps ultimately leading to true predictive tools.
ACKNOWLEDGMENTS
The authors thank Caihong Li for expert technical assistance, especially in the challenging experiment in Figure 1; John Tyson and Kathy Chen for help in understanding and using the model; Mike Rout and Brian Chait for useful discussions; Peter Schwartz and Jung-Im Lee for preliminary work in the system presented in Figure 2; Phillip Kaldis and Mike Rout for plasmids; Angelika Amon, Bruce Futcher, and Masaki Shirayama for strains; and Mike Rout for the anti-Nop1 antibody. This work was supported by Public Health Service grant GM47238.
Footnotes
DOI:10.1091/mbc.01–05-0265.
REFERENCES
- Aitchison J, Rout M, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins NUP170p and NUP157p: complementation with the vertebrate homologue NUP155p and functional interactions with the yeast nuclear pore-membrane protein POM152p. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandru G, Zachariae W, Schleiffer A, Nasmyth K. Sister chromatid separation and chromosome re- duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 1999;18:2707–2721. doi: 10.1093/emboj/18.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- Bardin A, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Baumer M, Braus G, Irniger S. Two different modes of cyclin clb2 proteolysis during mitosis in Saccharomyces cerevisiae. FEBS Lett. 2000;468:142–148. doi: 10.1016/s0014-5793(00)01208-4. [DOI] [PubMed] [Google Scholar]
- Chen K, Csikasz-Nagy A, Gyorffy B, Val J, Novak B, Tyson J. Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol Biol Cell. 2000;11:369–391. doi: 10.1091/mbc.11.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F, Jacobson M. Conservation and function of a potential substrate-binding domain in the yeast Clb5 B-type cyclin. Mol Cell Biol. 2000;20:4782–4790. doi: 10.1128/mcb.20.13.4782-4790.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F, Roberts J, Weintraub H. Simple and complex cell cycles. Annu Rev Cell Biol. 1989;5:341–395. doi: 10.1146/annurev.cb.05.110189.002013. [DOI] [PubMed] [Google Scholar]
- Cross FR. Cell cycle arrest caused by CLN gene deficiency in Saccharomyces cerevisiaeresembles START-I arrest and is independent of the mating-pheromone signaling pathway. Mol Cell Biol. 1990;10:6482–6490. doi: 10.1128/mcb.10.12.6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR. Further characterization of a size control gene in Saccharomyces cerevisiae. J Cell Sci. 1989;94(Suppl 12):117–127. doi: 10.1242/jcs.1989.supplement_12.10. [DOI] [PubMed] [Google Scholar]
- Cross FR. Starting the cell cycle: what's the point? Curr Opin Cell Biol. 1995;7:790–797. doi: 10.1016/0955-0674(95)80062-x. [DOI] [PubMed] [Google Scholar]
- Cross FR, Blake CM. The yeast Cln3 protein is an unstable activator of Cdc28. Mol Cell Biol. 1993;13:3266–3271. doi: 10.1128/mcb.13.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR, Yuste-Rojas M, Gray S, Jacobson M. Specialization and targeting of B-type cyclins. Mol Cell. 1999;4:11–19. doi: 10.1016/s1097-2765(00)80183-5. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Futcher B. Specialization of B- type cyclins for mitosis or meiosis in S. cerevisiae. Genetics. 1995;140:957–963. doi: 10.1093/genetics/140.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como CJ, Chang H, Arndt KT. Activation of CLN1 and CLN2G1 cyclin gene expression by BCK2. Mol Cell Biol. 1995;15:1835–1846. doi: 10.1128/mcb.15.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Raghuraman MK, Friedman KL, Cross FR, Brewer BJ, Fangman WL. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell. 1998;2(2):173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto prereplicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;27:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott S, McLaughlin C. Synthesis and modification of proteins during the cell cycle of the yeast Saccharomyces cerevisiae. J Bacteriol. 1979;137:1185–1190. doi: 10.1128/jb.137.3.1185-1190.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S, Chi Y, Yang P, Campbell J. Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Mol Biol Cell. 1999;10:3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CB, Cross FR. CLB5: A novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- Epstein CB, Cross FR. Genes that can bypass the CLN requirement for Saccharomyces cerevisiaecell cycle START. Mol Cell Biol. 1994;14:2041–2047. doi: 10.1128/mcb.14.3.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch I, Dahmann C, Surana U, Amon A, Nasmyth K, Goetsch L, Byers B, Futcher B. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol Biol Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi M, Sikder H, Ebihara H, Irie K, Sugimoto K, Matsumoto K, Hunt TN, T, Kobayashi H. Xenopuscyclin A1 can associate with Cdc28 in budding yeast, causing cell-cycle arrest with an abnormal distribution of nuclear DNA. Genes Cells. 1997;2:329–43. doi: 10.1046/j.1365-2443.1997.1230320.x. [DOI] [PubMed] [Google Scholar]
- Goldbeter A, Koshland DE., Jr An amplified sensitivity arising from covalent modification in biological systems. Proc Natl Acad Sci USA. 1981;78:6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Reed SI. Differential function and expression of Saccharomyces cerevisiaeB-type cyclins in mitosis and meiosis. Mol Cell Biol. 1993;13:2113–2125. doi: 10.1128/mcb.13.4.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase S, Winey M, Reed S. Multi-step control of spindle pole body duplication by cyclin-dependent kinase. Nat Cell Biol. 2001;3:38–42. doi: 10.1038/35050543. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Unger MW. Unequal division in Saccharomyces cerevisiaeand its implications for the control of cell division. J Cell Biol. 1977;75:422–435. doi: 10.1083/jcb.75.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C, Meeks-Wagner DW, Fangman WL, Botstein D. A rapid, efficient method for isolating DNA from yeast. Gene. 1986;42:169–173. doi: 10.1016/0378-1119(86)90293-3. [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Irniger S, Nasmyth K. The anaphase promoting complex is required in G1 arrested cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S phase. J Cell Sci. 1997;110:1523–1531. doi: 10.1242/jcs.110.13.1523. [DOI] [PubMed] [Google Scholar]
- Jacobson MD, Gray S, Yuste-Rojas M, Cross FR. Testing cyclin specificity in the exit from mitosis. Mol Cell Biol. 2000;20:4483–93. doi: 10.1128/mcb.20.13.4483-4493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S, Charles J, Morgan D. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- Jaspersen S, Charles J, Tinker-Kulberg R, Morgan D. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeoung D, Oehlen LJWM, Cross FR. Cln3- associated kinase activity in Saccharomyces cerevisiaeis regulated by the mating factor pathway. Mol Cell Biol. 1998;18:433–441. doi: 10.1128/mcb.18.1.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp D, Bhoite L, Stillman DJ, Nasmyth K. The transcription factor Swi5 regulates expression of the cyclin kinase inhibitor p40SIC1. Mol Cell Biol. 1996;16:5701–5707. doi: 10.1128/mcb.16.10.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Nasmyth K. Cell cycle regulated transcription in yeast. Curr Opin Cell Biol. 1994;6:451–459. doi: 10.1016/0955-0674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Koch C, Schleiffer A, Ammerer G, Nasmyth K. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at Start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 1996;10:129–141. doi: 10.1101/gad.10.2.129. [DOI] [PubMed] [Google Scholar]
- Levine K, Huang K, Cross FR. Saccharomyces cerevisiaeG1 cyclins differ in their intrinsic functional specificities. Mol Cell Biol. 1996;16:6794–6803. doi: 10.1128/mcb.16.12.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and Consequences of Duplicate Genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- McInerny CJ, Partridge JF, Mikesell GE, Creemer DP, Breeden LL. A novel Mcm1- dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 1997;11:1277–1288. doi: 10.1101/gad.11.10.1277. [DOI] [PubMed] [Google Scholar]
- Mendenhall MD. An inhibitor of p34CDC28 protein kinase activity from Saccharomyces cerevisiae. Science. 1993;259:216–219. doi: 10.1126/science.8421781. [DOI] [PubMed] [Google Scholar]
- Miller M, Cross FR. Cyclin specificity: how many wheels do you need for a unicycle? J Cell Sci. 2001;114:1811–1820. doi: 10.1242/jcs.114.10.1811. [DOI] [PubMed] [Google Scholar]
- Miller ME, Cross FR. Distinct subcellular localization patterns contribute to functional specificity of the Cln2 and Cln3 cyclins of Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:542–555. doi: 10.1128/mcb.20.2.542-555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Nash R, Tokiwa G, Anand S, Erickson K, Futcher AB. The WHI1+ gene of Saccharomyces cerevisiaetethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. At the heart of the budding yeast cell cycle. Trends Genet. 1996;12:405. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Irie K, Li JJ. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2–7. Curr Biol. 2000;10:195–205. doi: 10.1016/s0960-9822(00)00337-7. [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ. Cyclin- dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- Novak B, Csikasz-Nagy A, Gyorffy B, Nasmyth K, Tyson J. Model scenarios for evolution of the eukaryotic cell cycle. Philos Trans R Soc Lond B Biol Sci. 1998;353:2063–2076. doi: 10.1098/rstb.1998.0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlen LJWM, McKinney JD, Cross FR. Ste12 and Mcm1 regulate cell cycle dependent transcription of FAR1. Mol Cell Biol. 1996;16:2830–2837. doi: 10.1128/mcb.16.6.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poloumienko A, Dershowitz A, De J, Newlon CS. Completion of replication map of Saccharomyces cerevisiae chromosome III. Mol Biol Cell. 2001;12:3317–3327. doi: 10.1091/mbc.12.11.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S, Hwang E, Visintin R, Amon A. The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr Biol. 1998;8:750–760. doi: 10.1016/s0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- Richardson H, Lew DJ, Henze M, Sugimoto K, Reed SI. Cyclin- B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev. 1992;6:2021–2034. doi: 10.1101/gad.6.11.2021. [DOI] [PubMed] [Google Scholar]
- Rudner A, Murray A. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase- promoting complex. J Cell Biol. 2000;149:1377–1390. doi: 10.1083/jcb.149.7.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Schwob E, Boehm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Toth A, Galova M, Nasmyth K. APCCDC20 promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- Shou W, Seol J, Shevchenko A, Baskerville C, Moazed D, Chen Z, Jang J, Shevchenko A, Charbonneau H, Deshaies R. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher AB, Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- Toyn JH, Johnson AL, Donovan JD, Toone WM, Johnston LH. The Swi5 transcription factor of Saccharomyces cerevisiaehas a role in exit from mitosis through induction of the cdk-inhibitor Sic1 in telophase. Genetics. 1996;145:85–96. doi: 10.1093/genetics/145.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for CLNG1 cyclin function at Start. Proc Natl Acad Sci USA. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiaeis regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen E, Cross F. Interaction between the MEC1-dependent DNA synthesis checkpoint and G1 cyclin function in Saccharomyces cerevisiae. Genetics. 1999;151:459–471. doi: 10.1093/genetics/151.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1Cdk required for its degradation and entry into S phase. Science. 1997a;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- Verma R, Feldman RMR, Deshaies RJ. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol Biol Cell. 1997b;8:1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang E, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Visintin R, Hwang E, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: A family of substrate-specific activators of APC-dependent proteolysis. Science. 1998;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Winey M, Yarar D, Giddings TJ, Mastronarde D. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiaecell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol Biol Cell. 1997;8:2119–2132. doi: 10.1091/mbc.8.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeong F, Lim H, Padmashree C, Surana U. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol Cell. 2000;5:501–511. doi: 10.1016/s1097-2765(00)80444-x. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;15:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- Zhao X, Muller E, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;1998 2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]