Abstract
Chemokines are a family of small cytokines, which are signalling proteins secreted by cells. The principal role of chemokines is to serve as chemoattractants to guide the migration of their target cells. Chemokine C-X3-C motif ligand 1 (CX3CL1) is a protein-coding gene of fractalkine, which serves as a ligand for chemokine C-X3-C motif receptor 1 (CX3CR1) and integrins. However, the roles of CX3CL1 in different pathological types of lung cancer remain poorly understood. The present study aimed to investigate the potential clinical and biological function of CX3CL1 mRNA expression in patients with lung cancer. In the present study, lung cancer data obtained from the Gene Expression Omnibus database and The Cancer Genome Atlas were downloaded and analysed, and the results demonstrated that an increased CX3CL1 mRNA expression in tumour tissues from lung adenocarcinoma (LUAD) was associated with improved overall survival. However, no significant association was identified between CX3CL1 expression and the prognosis of lung squamous cell carcinoma (LUSC). Furthermore, the genes whose expression levels were correlated with CX3CL1 expression were subjected to enrichment analysis, and the results for the LUAD data demonstrated that the most significant biological processes included ‘positive regulation of cell adhesion’, ‘leukocyte cell-cell adhesion’, ‘leukocyte migration’ and ‘T cell activation’, whereas, the important highly ranked pathways included ‘cell adhesion molecules (CAMs)’, ‘leukocyte transendothelial migration’ and ‘natural killer cell-mediated cytotoxicity’. However, in the patients with LUSC, the genes that were highly correlated with CX3CL1 were not enriched for any biological processes or signalling pathways. Based on the data of the present study, it was hypothesised that CX3CL1 may serve as a prognostic marker for LUAD.
Keywords: lung cancer, chemokines, chemokine C-X3-C motif ligand 1, prognosis, pooled analysis
Introduction
Lung cancer is a common type of malignant tumour and a leading cause of cancer-associated mortality worldwide. Every year, lung cancer occurs in ~1.8 million individuals and results in 1.6 million mortalities. Despite advances in early diagnosis and multimodal treatment, over one-half of the patients who are diagnosed with lung cancer do not survive beyond a year, and the 5-year survival rate is only ~18% (1). The invasive and metastatic mechanisms of lung cancer remain unclear. Chemokines and their receptors have emerged as pivotal regulators of tumour growth, progression and metastasis (2). Therefore, elucidating the mechanistic roles of chemokines in the recurrence and metastasis of lung cancer may improve the diagnosis and treatment of lung cancer.
Chemokine C-X3-C motif ligand 1 (CX3CL1), a protein-coding gene of fractalkine, serves as a ligand for chemokine C-X3-C motif receptor 1 (CX3CR1) and integrins. When CX3CL1 binds to CX3CR1 and integrins, CX3CR1-dependent and CX3CR1-independent signal pathways may be activated. In contrast to other chemokines, CX3CL1 has two different forms, a soluble form and a membrane-bound form, and each form mediates distinct biological actions. The soluble form is a strong activator of chemotaxis and causes migration of natural killer cells, cytotoxic T lymphocytes and macrophages (3). The membrane-bound form promotes leukocyte-endothelial cell adhesion, and may serve a role in the processes of leukocyte adhesion and migration at the endothelium (4,5). CX3CL1 and its receptor are involved in a number of inflammatory processes, including allergic asthma, rheumatoid arthritis, Crohn's disease and atherosclerosis (3,6,7). These previous studies demonstrated that CX3CL1 may be expressed in different tissues and may contribute to a number of inflammatory diseases by promoting the accumulation of CX3CR1-positive immune cells at inflammation sites (8,9). However, under pathogenic conditions with abnormal local and systemic immune responses, CX3CL1 may additionally induce potent antitumour and tissue-protective effects (10,11).
It was hypothesised that CX3CL1/CX3CR1 are involved in cancer pathogenesis and therapeutic approaches targeting this ligand-receptor pair have additionally demonstrated promising results in experimental settings. In epithelial ovarian cancer cells, CX3CL1 promoted cancer cell proliferation by binding to CX3CR1 and subsequently activating protein kinase B (12). Furthermore, ovarian carcinoma cells migrated towards CX3CL1, and silencing of CX3CR1 reduced their migration (13). Li et al (14), demonstrated that CX3CL1 silencing in the HepG2 cell line inhibited angiogenesis in vitro and in vivo. However, CX3CL1 has a range of effects in breast cancer. It is involved in breast cancer metastasis (15,16); however, increased CX3CL1 expression is positively correlated with prognosis and tumour-infiltrating lymphocytes (TIL) levels (16).
The association between CX3CL1 and clinicopathological parameters in lung cancer remains unclear, and correlations between its expression levels and its prognostic value in different lung cancer subtypes require further study. In the present study, datasets from various public databases were analysed using statistical models. The prognostic effects identified in the included studies were pooled to determine the significance of CX3CL1 in different lung cancer subtypes.
Materials and methods
Search strategy and data extraction
The Cancer Genome Atlas (TCGA, cancergenome.nih.gov) and the Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/gds) were searched using the key words ‘Lung cancer’ and ‘Homo sapiens’. Subsequently, a preliminary screening based on the title content was conducted. A total of two independent researchers were asked to read the contents of each dataset. Datasets with a small sample size (<50), incomplete clinicopathological parameters or no clinical data were excluded. In addition, the staging information of included datasets was based on the 7th Tumor-Node-Metastasis (TNM) staging system (17). For the screening results on which the researchers disagreed, a third researcher was responsible for the final decisions. In total, six datasets downloaded from the GEO database [GSE30219 (18), GSE37745 (19), GSE42127 (20), GSE50081 (21), GSE68465 (22) and GSE14814 (23)] and two TCGA datasets: lung adenocarcinoma [LUAD, https://xenabrowser.net/datapages/?cohort=GDC%20TCGA%20 Lung%20Adenocarcinoma%20(LUAD)&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443] and lung squamous cell carcinoma [LUSC, http://xenabrowser.net/datapages/?cohort=GDC%20TCGA%20Lung%20Squamous%20Cell%20Carcinoma%20(LUSC)&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443] were included in the present study. The sub-datasets, including CX3CL1 mRNA expression and associated clinical data, were extracted for further statistical analysis.
Statistical analysis
All statistical analyses were performed using R v3.5.0 (R Foundation for Statistical Computing, Vienna, Austria), RStudio 1.1.456 (RStudio, Boston, MA, USA) and GraphPad Prism 5.0 software package (GraphPad Software, Inc., La Jolla, CA, USA). The CX3CL1 expression levels were divided into two groups; high expression and low expression. A web-based function called Cutoff Finder (http://molpath.charite.de/cutoff) was used to determine a cutoff point (24). Independent samples t-test was used to compare means between two groups, whereas one-way analysis of variance and the Tukey honest significant difference post hoc test were used for multiple comparisons. Kaplan-Meier analysis and a log-rank test were used for comparing survival curves. Cox models were used to calculate the hazard ratios (HRs) and the 95% confidence intervals (CIs) based on the CX3CL1 expression levels and their corresponding clinical parameters. HRs and their 95% CIs from all datasets were pooled, and the heterogeneity of this pooled analysis based on CX3CL1 mRNA expression level was appraised by the Cochran Q test and the I2 test. A random-effects model (the DerSimonian-Laird method) was applied when significance was P<0.1 or I2>50%; otherwise, a fixed-effects model (the Mantel-Haenszel method) was used. P<0.05 was considered to indicate a statistically significant difference.
Publication bias was assessed by Begg's rank correlation method. The significance levels of statistical tests were determined according to two-tailed P-values and all pooled analyses were performed using the STATA software package (v12.0; StataCorp LP, College Station, TX, USA).
Functional enrichment analysis
For each dataset, the Pearson correlation coefficients between the expression level of CX3CL1 and those of other genes were calculated, and the coefficients were merged by gene name across all datasets. The genes that had highly ranked positive or negative correlation coefficients with CX3CL1 were selected, and functional enrichment analysis was performed using the R package ‘clusterProfiler’ (www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html). Dot plots of biological processes and Kyoto Encyclopedia of Genes and Genomes (KEGG, www.genome.jp/kegg/pathway.html) pathways were drawn using the R functions ‘enrichGO’ and ‘enrichKEGG’ in ‘clusterProfiler’. Enrichment maps for the enrichment results of over-representation tests or gene set enrichment analysis were generated using the R function ‘emapplot’.
Results
Study characteristics
In the present study, six GEO datasets and two TCGA datasets were included. In the primary screening, 1,092 relevant datasets were selected using the key word ‘Lung cancer’. The search filters were set as follows: The sample type was set as ‘tissue’, selecting 326 datasets, and the sample size was set as >50, selecting 126 datasets. Subsequent to reading the titles, abstracts and clinical outcomes of these datasets, a total of six datasets from GEO met the inclusion criteria and two from TCGA, for a total of eight datasets in the present study.
The baseline characteristics of all the included studies is presented in Table I. Datasets covering 2,443 patients from France, Sweden, Canada and the USA were included in the present analysis. All of those datasets included overall survival (OS), and the majority of them contained sex, age, pathological type, clinical stage and treatment information; the mRNA expression levels of CX3CL1 and associated genes were integrated into the information mentioned above.
Table I.
Baseline characteristics of the eight included datasets for pooled-analysis.
| Clinicopathological parameters | GSE30219 | GSE37745 | GSE42127 | GSE50081 | GSE68465 | GSE14814 | TCGA lung adenocarcinoma | TCGA lung squamous cell carcinoma |
|---|---|---|---|---|---|---|---|---|
| Public date | 24-May-13 | 12-Oct-12 | 30-Jan-13 | 22-Dec-13 | 2-May-15 | 9-Sep-10 | 24-Feb-15 | 24-Feb-15 |
| Country | France | Sweden | USA | Canada | USA | Canada | USA | USA |
| Case number | 307 | 196 | 173 | 181 | 443 | 132 | 509 | 502 |
| Age (Range), years | 62.50 | 65.00 | 66.30 | 69.77 | 65.00 | 61.80 | 66.00 | 68.00 |
| (15.00–84.00) | (39.00–84.00) | (42.30–86.20) | (40.16–87.93) | (33.00–87.00) | (35.40–81.30) | (38.00–88.00) | (39.00–90.00) | |
| Sex | ||||||||
| Male | 250 | 107 | 91 | 98 | 223 | 90 | 235 | 364 |
| Female | 43 | 89 | 82 | 83 | 220 | 42 | 274 | 127 |
| Pathology | ||||||||
| Adenocarcinoma | 85 | 106 | 131 | 127 | 443 | 70 | 509 | NR |
| Squamous cell carcinoma | 61 | 66 | 42 | 42 | NR | 52 | NR | 502 |
| Large cell carcinoma | 3 | 24 | NR | 7 | NR | NR | NR | NR |
| Small cell carcinoma | 21 | NR | NR | NR | NR | NR | NR | NR |
| Large cell neuroendocrine | 56 | NR | NR | NR | NR | NR | NR | NR |
| Basal squamous cell carcinoma | 39 | NR | NR | NR | NR | NR | NR | NR |
| Other | 28 | NR | NR | 5 | NR | 10 | NR | NR |
| Tumour stage | ||||||||
| T1 | 166 | NR | NR | 57 | NR | NR | 167 | 113 |
| T2 | 69 | NR | NR | 122 | NR | NR | 274 | 287 |
| T3 | 31 | NR | NR | 2 | NR | NR | 47 | 069 |
| T4 | 21 | NR | NR | NR | NR | NR | 19 | 022 |
| TX | 6 | NR | NR | NR | NR | NR | 2 | NR |
| Node stage | ||||||||
| N0 | 198 | NR | NR | 129 | NR | NR | 326 | 313 |
| N1 | 53 | NR | NR | 52 | NR | NR | 96 | 128 |
| N2 | 50 | NR | NR | NR | NR | NR | 74 | 039 |
| N3 | 10 | NR | NR | NR | NR | NR | 2 | 005 |
| NX | 2 | NR | NR | NR | NR | NR | 10 | 006 |
| Metastasis stage | ||||||||
| M0 | 282 | NR | NR | 181 | NR | NR | 343 | 403 |
| M1 | 8 | NR | NR | 0 | NR | NR | 25 | 7 |
| MX | 3 | NR | NR | NR | NR | NR | 137 | 75 |
| Tumour Node Metastasis stage | ||||||||
| I | NR | 130 | 109 | 127 | 276 | 72 | 276 | 241 |
| II | NR | 35 | 32 | 54 | 95 | 60 | 122 | 154 |
| III | NR | 27 | 30 | NR | 69 | NR | 84 | 086 |
| IV | NR | 4 | 1 | NR | NR | NR | 26 | 007 |
| Treatment | ||||||||
| Yes | NR | 29 | 49 | NR | 89 | 132 | 126 | 115 |
| No | NR | 71 | 124 | NR | 340 | NR | 258 | 264 |
| Unknown | NR | 96 | NR | NR | 13 | NR | 127 | 123 |
| Differentiation | ||||||||
| Well | NR | NR | NR | NR | 60 | NR | NR | NR |
| Moderate | NR | NR | NR | NR | 209 | NR | NR | NR |
| Poorly | NR | NR | NR | NR | 166 | NR | NR | NR |
| Unknown | NR | NR | NR | NR | 7 | NR | NR | NR |
NR, not reported; TCGA, The Cancer Genome Atlas.
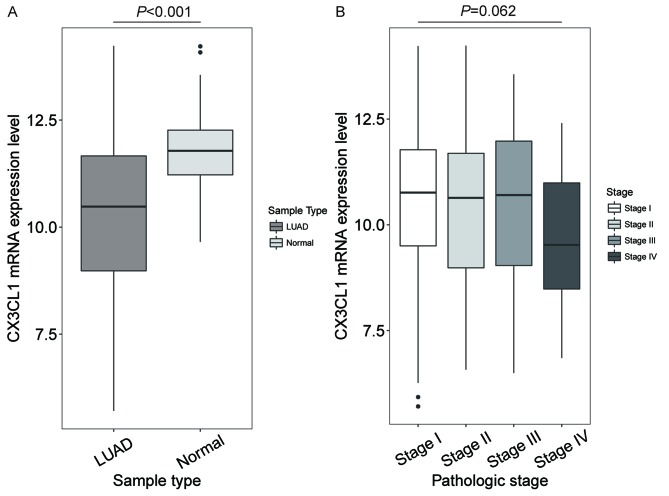
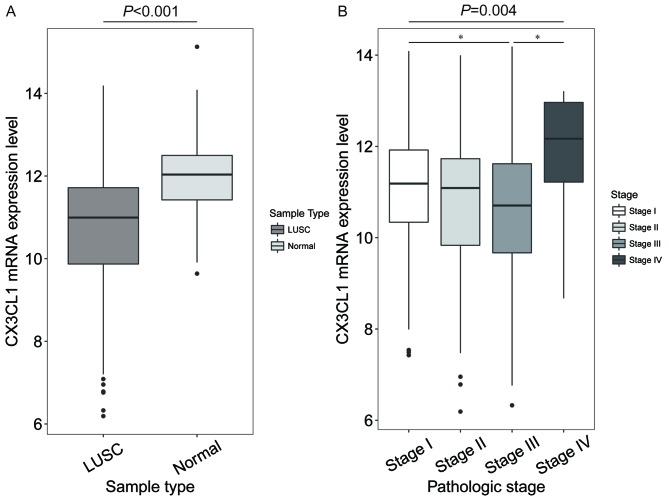
CX3CL1 mRNA expression levels in different tissue and TNM stages
In patients with LUAD, the CX3CL1 mRNA expression levels in tumour tissue were significantly decreased compared with normal tissue (t=10.259, P<0.001; Fig. 1A), but no significant difference was shown in different clinical stages (F=3.512, P=0.062; Fig. 1B). In patients with LUSC, the CX3CL1 mRNA expression levels were significantly decreased compared with normal tissue (t=7.762, P<0.001; Fig. 2A). In addition, the CX3CL1 mRNA expression levels in stage III samples were significantly different from those in stage I and IV samples (F=4.432, P=0.004; stage III vs. stage I, P=0.021; stage III vs. stage IV, P=0.036; Fig. 2B).
Figure 1.
CX3CL1 mRNA expression levels in LUAD and at Tumour Node Metastasis stages. (A) Patients with LUAD had significantly decreased mRNA expression levels of CX3CL1 in tumour tissues compared with normal tissue (t=10.259, P<0.001). (B) No significant difference was shown in different clinical stages (F=3.512, P=0.062). CX3CL1, chemokine C-X3-C motif ligand 1; LUAD, lung adenocarcinoma.
Figure 2.
CX3CL1 mRNA expression levels in LUSC and at Tumour Node Metastasis stages. (A) In patients with LUSC, the CX3CL1 mRNA expression levels in normal tissue were significantly increased compared with tumour tissue (t=7.762, P<0.001). (B) CX3CL1 mRNA expression levels in stage III were significantly different compared with in stages I and IV (F=4.432, P=0.004; stage III vs. stage I P=0.021, stage III vs. stage IV P=0.036). *P<0.05. CX3CL1, chemokine C-X3-C motif ligand 1; LUSC, lung squamous cell carcinoma.
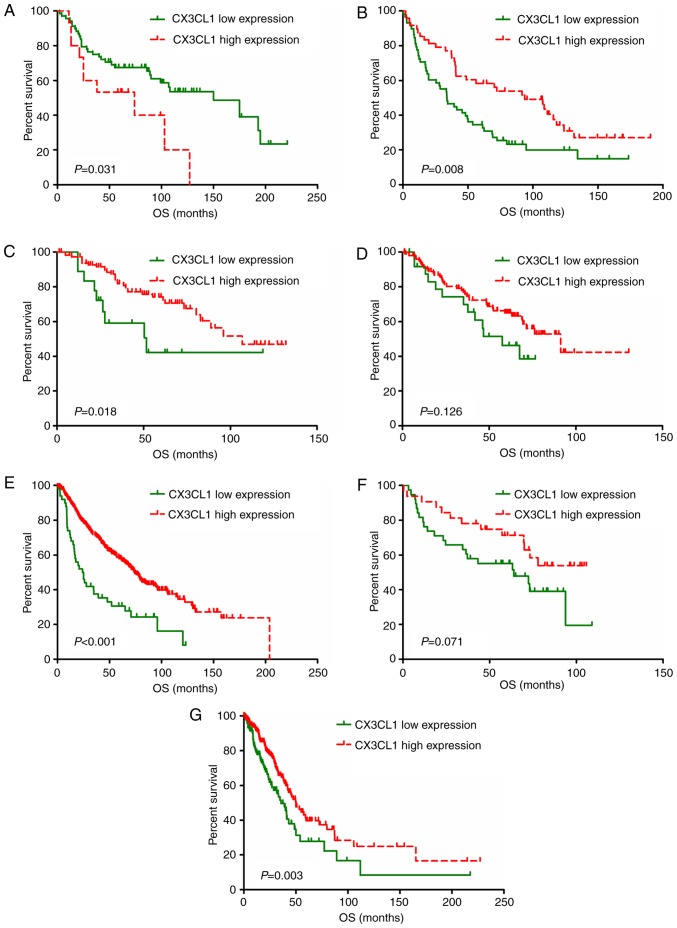
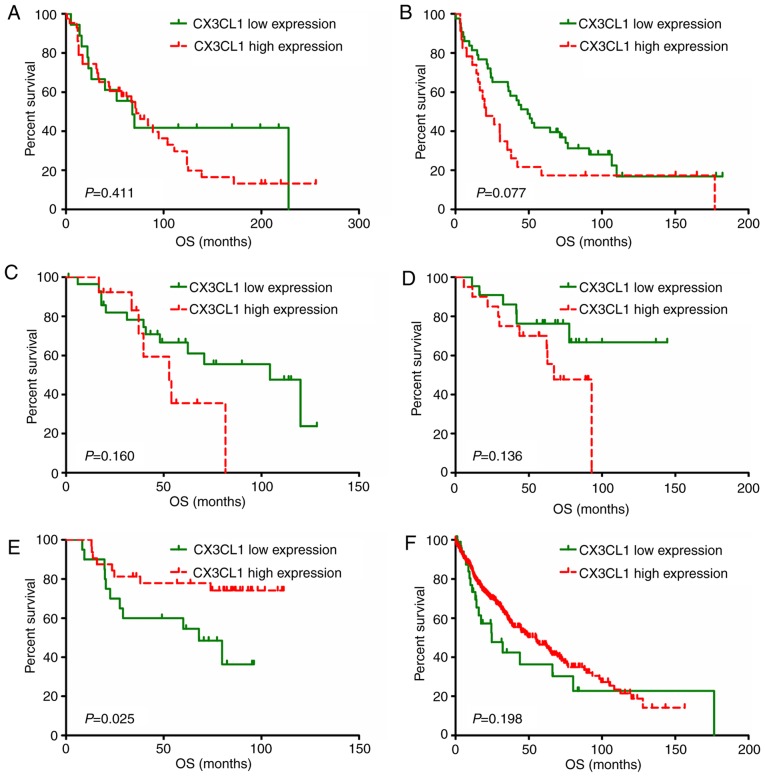
Survival analysis based on CX3CL1 expression level
A total of eight studies, including 2,443 patients, were used to establish univariate and multivariate Cox models. P-values, HRs and 95% CIs for each study based on LUAD and LUSC are presented in Tables II and III. In patients with LUAD, univariate and multivariate Cox model analyses of the GSE37745, GSE42127, GSE68465 and TCGA datasets revealed that higher CX3CL1 mRNA expression was significantly associated with improved survival (HR<1, P<0.05); however, results of the analyses in the GSE30219 dataset revealed an opposite trend (HR>1, P<0.05). In patients with LUSC, univariate Cox model analysis of the GSE14814 dataset revealed that CX3CL1 mRNA expression exhibited a significant protective effect on prognosis (HR<1, P<0.05). Conversely, multivariate Cox model analysis of the GSE37745 and GSE42127 datasets indicated that higher CX3CL1 mRNA expression was a risk factor for LUSC prognosis (HR>1, P<0.05). Survival plots of each dataset are presented in Figs. 3 and 4. In six out of seven datasets, high expression of CX3CL1 was associated with a decreased risk of mortality in patients with LUAD (Fig. 3). Amongst patients with LUSC, two datasets demonstrated that CX3CL1 was associated with a decreased risk of mortality, whereas, in four datasets, CX3CL1 was associated with an increased risk of mortality (Fig. 4). These findings indicated that CX3CL1 may be a candidate prognostic indicator for patients with LUAD but not for patients with LUSC.
Table II.
Univariate and multivariate Cox model analysis of the prognostic effect of CX3CL1 based on the lung adenocarcinoma datasets.
| Univariate analysis | Multivariate analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene expression Omnibus datasets | Cases | Cutoff value | HR | LCI | UCI | P-value | HR | LCI | UCI | P-value |
| GSE30219 | 83 | 7.091 | 2.192 | 1.056 | 4.55 | 0.035a | 2.296 | 1.098 | 4.802 | 0.027a |
| GSE37745 | 106 | 7.241 | 0.538 | 0.339 | 0.854 | 0.009b | 0.468 | 0.285 | 0.769 | 0.003b |
| GSE42127 | 131 | 6.680 | 0.42 | 0.199 | 0.884 | 0.022a | 0.463 | 0.217 | 0.991 | 0.047a |
| GSE50081 | 127 | 5.686 | 0.612 | 0.325 | 1.154 | 0.129 | 0.659 | 0.346 | 1.256 | 0.205 |
| GSE68465 | 442 | 68.560 | 0.429 | 0.303 | 0.608 | <0.001c | 0.424 | 0.294 | 0.612 | <0.001c |
| GSE14814 | 70 | 6.825 | 0.534 | 0.267 | 1.066 | 0.075 | 0.561 | 0.273 | 1.154 | 0.116 |
| The cancer genome Atlas | 483 | 7.530 | 0.363 | 0.2 | 0.66 | <0.001c | 0.322 | 0.176 | 0.59 | <0.001c |
P<0.05
P<0.01
P<0.001. HR, hazard ratio; LCI, lower confidence interval; UCI, upper confidence interval.
Table III.
Univariate and multivariate Cox model analysis of the prognostic effect of CX3CL1 based on the lung squamous cell carcinoma datasets.
| Univariate analysis | Multivariate analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene expression Omnibus datasets | Cases | Cutoff value | HR | LCI | UCI | P-value | HR | LCI | UCI | P-value |
| GSE30219 | 61 | 6.172 | 1.342 | 0.674 | 2.67 | 0.403 | 1.613 | 0.756 | 3.442 | 0.216 |
| GSE37745 | 66 | 7.529 | 1.657 | 0.941 | 2.916 | 0.080 | 1.989 | 1.078 | 3.67 | 0.028a |
| GSE42127 | 42 | 9.755 | 1.977 | 0.75 | 5.211 | 0.168 | 2.916 | 1.029 | 8.263 | 0.044a |
| GSE50081 | 42 | 7.033 | 2.138 | 0.769 | 5.941 | 0.145 | 2.044 | 0.732 | 5.706 | 0.172 |
| GSE14814 | 52 | 7.055 | 0.365 | 0.145 | 0.916 | 0.032a | 0.401 | 0.158 | 1.016 | 0.054 |
| The cancer genome Atlas | 474 | 8.728 | 0.727 | 0.447 | 1.184 | 0.200 | 0.705 | 0.429 | 1.159 | 0.168 |
P<0.05. HR, hazard ratio; LCI, lower confidence interval; UCI, upper confidence interval.
Figure 3.
Survival analysis of patients with lung adenocarcinoma by CX3CL1 mRNA expression levels. Data was split into two groups based on CX3CL1 expression levels using the web application Cutoff Finder (http://molpath.charite.de/cutoff). Kaplan-Meier analysis and survival curve generation were performed based on the datasets of (A) GSE30219, (B) GSE37745, (C) GSE42127, (D) GSE50081, (E) GSE68465 and (F) GSE14814, in addition to a dataset from (G) The Cancer Genome Atlas. CX3CL1, chemokine C-X3-C motif ligand 1; OS, overall survival.
Figure 4.
Survival analysis of different CX3CL1 mRNA expression levels in patients with lung squamous cell carcinoma. The expression level of CX3CL1 was divided into two groups using the web application Cutoff Finder (http://molpath.charite.de/cutoff). Kaplan-Meier analysis and survival curve generation were performed based on the datasets of (A) GSE30219, (B) GSE37745, (C) GSE42127, (D) GSE50081 and (E) GSE14814, in addition to a dataset from (F) The Cancer Genome Atlas. CX3CL1, chemokine C-X3-C motif ligand 1; OS, overall survival.
Pooled analysis based on the results of survival analysis
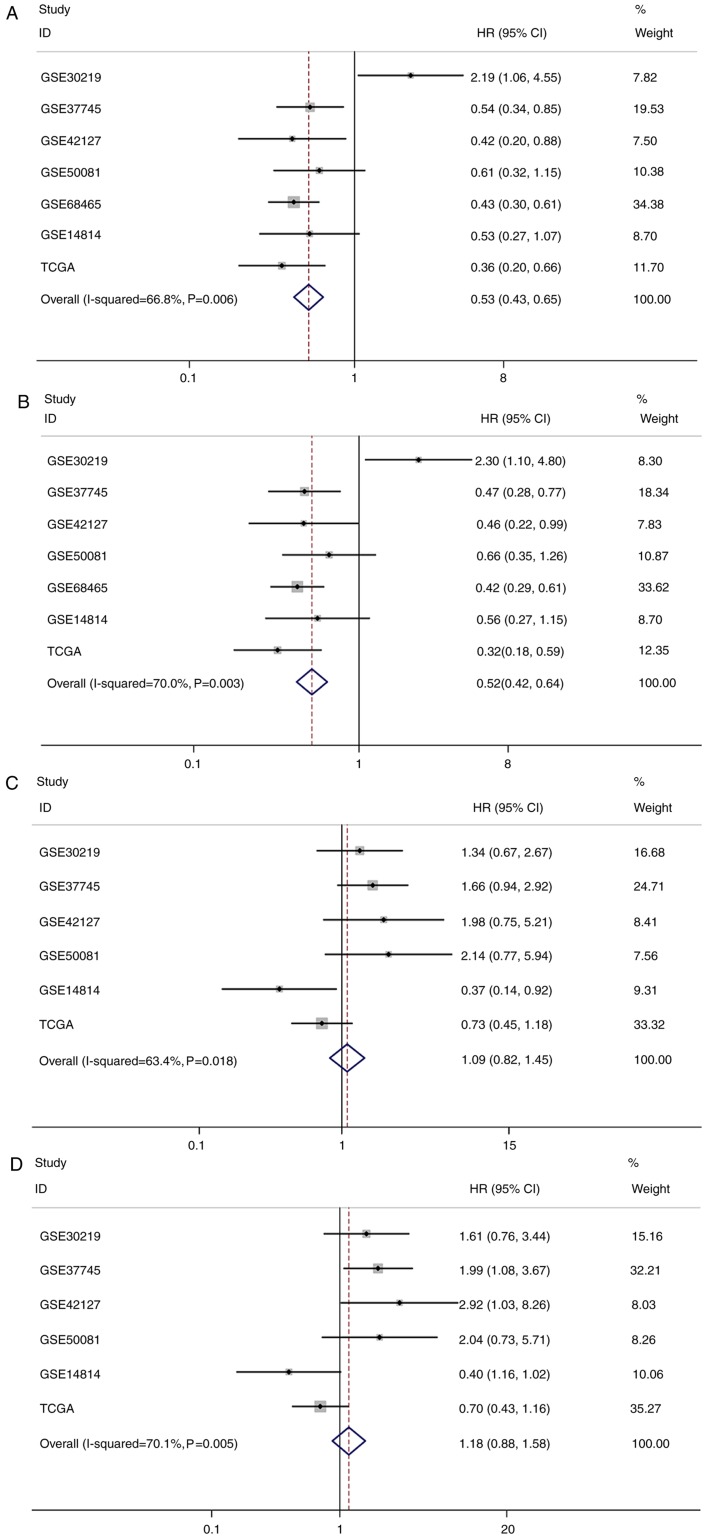
Pooled analysis results for LUAD and LUSC are presented in Tables IV and V. Given the statistical heterogeneity in univariate and multivariate analysis of LUAD and LUSC data (I2 value >50% and P<0.1), random-effect models were used to pool the HRs and 95% CIs. The results of this pooled analysis demonstrated that increased expression of CX3CL1 mRNA was significantly associated with an improved OS in patients with LUAD (univariate Cox model: Pooled HR=0.53; 95% CI=0.43–0.65; P<0.001; multivariate Cox model: Pooled HR=0.52; 95% CI=0.42–0.64; P<0.001; Table IV); however, there was no significant association between the CX3CL1 mRNA expression level and OS in patients with LUSC (univariate Cox model: Pooled HR=1.09; 95% CI=0.82–1.45; P=0.536; multivariate Cox model: Pooled HR=1.18; 95% CI=0.88–1.58; P=0.282; Table V). The forest plots of this pooled analysis are presented in Fig. 5.
Table IV.
Pooled hazard ratio of chemokine C-X3-C motif ligand 1 high expression in patients based on lung adenocarcinoma datasets.
| Random effect model | Heterogeneity | |||
|---|---|---|---|---|
| Analysis | Hazard ratio (95% confidence interval) | P-value | I2, % | P-value |
| Univariate | 0.53 (0.43–0.65) | <0.001 | 66.8 | 0.006 |
| Multivariate | 0.52 (0.42–0.64) | <0.001 | 70.0 | 0.003 |
Table V.
Pooled hazard ratio of chemokine C-X3-C motif ligand 1 high expression in patients based on lung squamous cell carcinoma datasets.
| Random effect model | Heterogeneity | |||
|---|---|---|---|---|
| Analysis | Hazard ratio (95% confidence interval) | P-value | I2, % | P-value |
| Univariate | 1.09 (0.82–1.45) | 0.536 | 63.4 | 0.018 |
| Multivariate | 1.18 (0.88–1.58) | 0.282 | 70.1 | 0.005 |
Figure 5.
Forest plots based on univariate and multivariate survival analysis in patients with LUAD or LUSC. Pooled results of (A) univariate and (B) multivariate Cox model analysis in patients with LUAD. Pooled results of (C) univariate and (D) multivariate Cox model analysis in patients with LUSC. LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; HR, hazard ratio; CI, confidence interval; TCGA, The Cancer Genome Atlas.
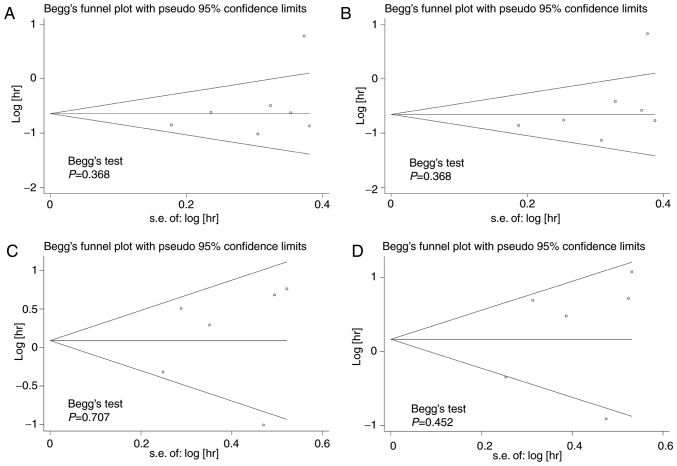
Begg's test was used to test for publication bias among the included studies, and funnel plots were used to demonstrate any publication bias graphically. The results indicated that there was no significant publication bias amongst the included studies; Begg's test demonstrated all P>0.1 for the studies (Fig. 6).
Figure 6.
Funnel plots based on univariate and multivariate survival analysis in patients with LUAD and LUSC. Publication bias tests based on (A) univariate and (B) multivariate survival analysis in patients with LUAD. Publication bias tests based on (C) univariate and (D) multivariate survival analysis in patients with LUSC. LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; hr, hazard ratio; s.e., standard error.
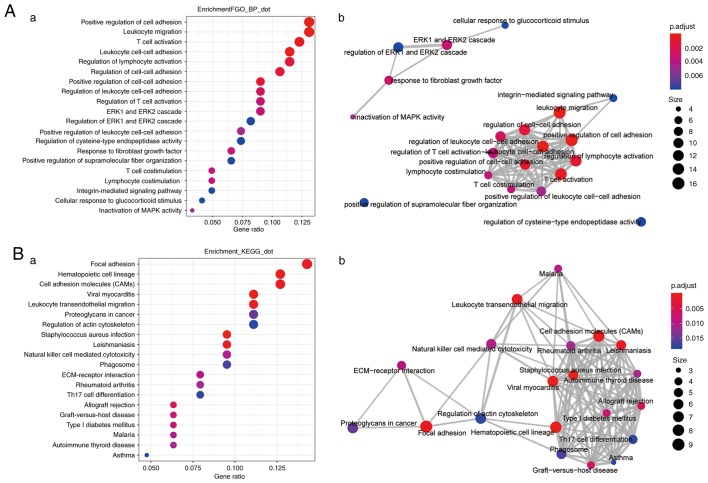
Enrichment analysis of genes whose expression is highly correlated with CX3CL1 expression
In patients with LUAD, genes whose expression correlation coefficients (r)>0.4 with CX3CL1 expression (Table SI) were included in the enrichment analysis. The top 20 enriched biological processes and pathways are presented in Fig. 7. The most significant biological processes included ‘positive regulation of cell adhesion’ (GO:0045785), ‘leukocyte cell-cell adhesion’ (GO:0007159), ‘leukocyte migration’ (GO:0050900) and ‘T-cell activation’ (GO:0042110), and the top 20 important pathways associated with tumour immunity included ‘cell adhesion molecules (CAMS)’ (KEGG: hsa04514), ‘leukocyte transendothelial migration’ (KEGG: hsa04670) and ‘natural killer cell mediated cytotoxicity’ (KEGG: hsa04650). However, in the patients with LUSC, the number of genes that were highly correlated with CX3CL1 (r>0.4) was too small and these genes were not enriched in any biological processes or signalling pathways (Table SII).
Figure 7.
Enrichment analysis of genes highly correlated with CX3CL1. The functional enrichment analysis of CX3CL1 and its correlated genes was performed using the R package, clusterProfiler. (A-a) Top 20 enriched biological processes for CX3CL1 and its correlated genes. (A-b) Enrichment map of the inter-relation of the top 20 enriched biological processes. (B-a) Top 20 enriched pathways for CX3CL1 and its correlated genes. (B-b) Enrichment map of the inter-relation of the top 20 enriched pathways. CX3CL1, chemokine C-X3-C motif ligand 1; MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase; ECM, extracellular matrix.
Discussion
Chemokines are a superfamily of proteins that regulate the transmission and involvement of leukocytes in vivo and are involved in inflammatory responses, including the migration processes of lymphocytes, dendritic cells, macrophages and stem cells (25,26). CX3CL1 (additionally termed fractalkine) is a large cytokine protein of 373 amino acids; it contains multiple domains and is the only known ligand of the CX3C chemokine family. It is synthesised as a membrane-bound form that may be released by proteolytic cleavage (5,27). Membrane-bound CX3CL1 serves as a molecule that promotes adhesion of leukocytes to endothelial cells, whereas, soluble CX3CL1 serves as a potent chemoattractant of T-cells and monocytes, causing them to move towards sites of inflammation (28). In contrast to other chemokines, CX3CL1 has a cell-adhesion function in addition to its chemotactic function. CX3CL1 is able to attract immune effector cells to the tumour location site and exert an antitumour immune effect (29). CX3CR1, the receptor of CX3CL1, is expressed on human natural killer (NK) cells, monocytes, T-lymphocytes and mast cells (5). CX3CL1 may additionally promote the adhesion of CX3CR1-positive tumour cells to target organs, causing the migration of tumour cells, thus promoting tumourigenesis (13). This may, in theory, help to elucidate how CX3CL1 serves seemingly opposing roles in a number of tumour types.
A previous study demonstrated that CX3CR1 expression was upregulated in solid tumours, including breast cancer and prostate cancer (15); its overexpression may promote the migration of tumour cells to the brain and bones due to the high expression of soluble CX3CL1 in these tissues. CX3CL1 expression in human bone marrow endothelial cells and osteoblasts is involved in the process of prostate cancer metastasis to bone marrow (30). Blocking CX3CL1 with a specific antibody significantly reduced the migration of prostate cancer cells to the bone marrow epithelium (31). A previous study on pancreatic adenocarcinoma models additionally demonstrated that CX3CR1 was involved in the process of metastatic spread of tumour cells to specific tissues that had increased expression of CX3CL1 (21). Therefore, high expression of CX3CR1 in pancreatic ductal adenocarcinoma (PDAC) may be one of the mechanisms that promote the spread of pancreatic cancer cells along the peripheral nerves, which results in a risk of early recurrence in patients with PDAC (32). Gaudin et al (12) demonstrated that epithelial cells from the surface and fallopian tubes of healthy ovaries, and from benign, borderline and malignant tumours, all stained positive for CX3CL1. Additionally, an increased expression of CX3CL1 was closely associated with rapid tumour growth (12). Zhou et al (33) demonstrated that the expression of cytokine SCM-1 β and CX3CL1 was significantly increased in lung cancer compared with adjacent matched normal tissues, and was correlated with pathological stage. Interactions between CX3CL1 and CX3CR1 mediated the process of cell migration and adhesion between EOC cells and peritoneal mesothelial cells, thereby promoting EOC cell proliferation (13). Similarly, previous studies on the mechanism of CX3CL1 in glioma and neuroblastoma demonstrated its negative regulatory function in these tumours (34–36).
However, CX3CL1 additionally has tumour-suppressive activity. Vitale et al (37) investigated the antitumour effect of fractalkine in its three molecular forms on animal models, and compared the extent of tumour development between C26 colon cancer cells with no fractalkine expression, and those expressing either native, soluble or membrane-bound fractalkine; the results demonstrated that native fractalkine exhibited the strongest antitumour effect, reducing tumours size by 93 and 99% in skin and orthotopic models, respectively. The effects depend on a critical balance between the soluble and membrane-bound forms. The soluble form exerted a marked effect in reducing liver and lung metastases, whereas, the membrane-bound form had very little impact in the liver and promoted tumour growth in the lungs (37). Xin et al (38) transduced CX3CL1 into mouse mesenchymal stem cells (MSCs) ex vivo using an adenoviral vector with the Arg-Gly-Asp-4C peptide in the fibre knob. Systemic administration of CX3CL1-expressing MSCs to the mice bearing lung metastases of C26 and B16F10 cells significantly inhibited the development of lung metastases and thus, improved the survival of these tumour-bearing mice. Transduction of CX3CL1 and CX3CR1 in colorectal cancer (CRC) cell lines induced cell aggregation that markedly inhibited in vitro migration in chemotaxis assays, and overexpression of CX3CL1-CX3CR1 inhibited the spread of cancer to the liver in a mouse model of spleen-liver metastasis (39). Ohta et al (11) demonstrated a significant correlation between CX3CL1 expression with TIL recruitment and prognosis in CRC cases. Hyakudomi et al (10) suggested that patients with gastric adenocarcinoma with increased levels of CX3CL1 expression had elevated levels of cluster of differentiation 8 T-cells and NK cells, which may induce innate and adaptive immunity, and are correlated with a more favourable prognosis. In hepatocellular carcinoma, CX3CL1 overexpression was additionally associated with a favourable prognosis. Intra- and extrahepatic recurrence rates were significantly reduced in tumours with high expression of CX3CL1 and CX3CR1 (40).
Lung cancer, with its rapidly growing morbidity and mortality rates in recent years, is one of the principal threats to human health and life amongst all the malignant tumours (1). Therefore, the identification of a biomarker for the treatment and prognosis of lung cancer may be of considerable value. At present, the roles of CX3CL1 in lung cancer and the mechanisms of those roles remain unclear. In the present study, the results demonstrated that the CX3CL1 and CX3CR1 mRNA expression levels in tumour tissue were significantly decreased compared with levels in normal tissue in patients with LUAD or LUSC. The pooled analysis results demonstrated that higher CX3CL1 mRNA expression was significantly associated with improved OS in patients with LUAD; however, no significant association was identified between CX3CL1 mRNA expression and OS in patients with LUSC. In all the LUAD datasets, the genes that were highly correlated (r>0.4) with CX3CL1 were included for the enrichment analysis. The most significantly altered biological processes included ‘positive regulation of cell adhesion’ (GO:0045785), ‘leukocyte cell-cell adhesion’ (GO:0007159), ‘leukocyte migration’ (GO:0050900) and ‘T cell activation’ (GO:0042110), and the top 20 important pathways associated with tumour immunity included ‘cell adhesion molecules (CAMS)’ (KEGG: hsa04514), ‘leukocyte transendothelial migration’ (KEGG: hsa04670) and ‘natural killer cell mediated cytotoxicity’ (KEGG: hsa04650). However, in the patients with LUSC, the genes that were highly correlated with CX3CL1 were not enriched for any biological processes or signalling pathways. These results demonstrated that CX3CL1 may be a positive prognostic indicator in LUAD. CX3CL1 may promote the adhesion of CX3CR1-positive tumour cells to target organs, causing migration of tumour cells (31,41). However, in LUAD, CX3CL1 overexpression may lead to increased chemotactic efficiency and increased immune effector cell infiltration, which results in an improved prognosis.
The present study has certain limitations; specific datasets from the GEO database did not provide complete clinical information, meaning that further hierarchical analysis was difficult to achieve. The distribution of clinical features amongst the patients was uneven. Certain datasets included in this analysis were not based on the same detection platform; thus, a certain degree of heterogeneity existed between different datasets. Despite a number of limitations, the present study may provide insight for other researchers; the prognostic value of CX3CL1 using seven similar datasets in patients with LUAD was demonstrated.
In conclusion, the results of the present study demonstrated that increased CX3CL1 mRNA expression levels in patients with LUAD may be associated with improved prognosis. However, this was not observed in patients with LUSC. The favourable results of increased expression levels of CX3CL1 in patients with LUAD may occur through biological processes and signalling pathwats, including ‘T cell activation’ (GO:0042110), ‘leukocyte cell-cell adhesion’ (GO:0007159), ‘leukocyte migration’ (GO:0050900), ‘cell adhesion molecules (CAMS)’ (KEGG: hsa04514), ‘leukocyte transendothelial migration’ (KEGG: hsa04670) and ‘natural killer cell mediated cytotoxicity’ (KEGG: hsa04650). Further prospective studies are required to confirm the prognostic value of CX3CL1 in patients with LUAD. In addition, the detailed mechanisms of the action of CX3CL1 in LUAD require confirmation by further experimental studies.
Supplementary Material
Acknowledgements
The authors would like to thank Mr. Bin Xu (Department of Tumor Biological Treatment, The Third Affiliated Hospital of Soochow University, Changzhou, Jiangsu, China) for his expertise and useful suggestions in the statistical and bioinformatics analysis.
Funding
The present study was supported by a grant from The International Science and Technology Cooperation Project of the Changzhou Science and Technology Bureau (grant no. CZ20140016; China).
Availability of data and materials
The datasets used and/or analysed during the present study are available from The Cancer Genome Atlas and Gene Expression Omnibus.
Authors' contributions
JL and CL conceived and designed the study. YL, JL and XQZ contributed to data cleansing, the statistical and bioinformatics analysis, and manuscript writing. QL, JX, XHL, SH and WDP contributed to the literature search, data collection, figure and table generation, and manuscript writing. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zappa C, Mousa SA. Non-small cell lung cancer: Current treatment and future advances. Transl Lung Cancer Res. 2016;5:288–300. doi: 10.21037/tlcr.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borsig L, Wolf MJ, Roblek M, Lorentzen A, Heikenwalder M. Inflammatory chemokines and metastasis-Tracing the accessory. Oncogene. 2014;33:3217–3224. doi: 10.1038/onc.2013.272. [DOI] [PubMed] [Google Scholar]
- 3.Liu W, Jiang L, Bian C, Liang Y, Xing R, Yishakea M, Dong J. Role of CX3CL1 in diseases. Arch Immunol Ther Exp. 2016;64:371–383. doi: 10.1007/s00005-016-0395-9. [DOI] [PubMed] [Google Scholar]
- 4.Pan Y, Lloyd C, Zhou H, Dolich S, Deeds J, Gonzalo JA, Vath J, Gosselin M, Ma J, Dussault B, et al. Neurotactin, a membrane-anchored chemokine upregulated in brain inflammation. Nature. 1997;387:611–617. doi: 10.1038/42491. [DOI] [PubMed] [Google Scholar]
- 5.Bazan JF, Bacon KB, Hardiman G, Wang W, Soo K, Rossi D, Greaves DR, Zlotnik A, Schall TJ. A new class of membrane-bound chemokine with a CX3C motif. Nature. 1997;385:640–644. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- 6.Julia V, Staumont-Salle D, Dombrowicz D. Role of fractalkine/CX3CL1 and its receptor CX3CR1 in allergic diseases. Med Sci (Paris) 2016;32:260–266. doi: 10.1051/medsci/20163203010. (Article in French) [DOI] [PubMed] [Google Scholar]
- 7.Thomas S, Baumgart DC. Targeting leukocyte migration and adhesion in Crohn's disease and ulcerative colitis. Inflammopharmacology. 2012;20:1–18. doi: 10.1007/s10787-011-0104-6. [DOI] [PubMed] [Google Scholar]
- 8.Yoneda O, Imai T, Goda S, Inoue H, Yamauchi A, Okazaki T, Imai H, Yoshie O, Bloom ET, Domae N, Umehara H. Fractalkine-mediated endothelial cell injury by NK cells. J Immunol. 2000;164:4055–4062. doi: 10.4049/jimmunol.164.8.4055. [DOI] [PubMed] [Google Scholar]
- 9.Gu X, Tang XD, Gu SY, Yang SQ, Zhou PJ, Tan JM. Expression of fractalkine and its receptor in acute cardiac allografts rejection. Zhonghua wai ke za zhi. 2003;41:139–142. (In Chinese) [PubMed] [Google Scholar]
- 10.Hyakudomi M, Matsubara T, Hyakudomi R, Yamamoto T, Kinugasa S, Yamanoi A, Maruyama R, Tanaka T. Increased expression of fractalkine is correlated with a better prognosis and an increased number of both CD8+ T cells and natural killer cells in gastric adenocarcinoma. Ann Surg Oncol. 2008;15:1775–1782. doi: 10.1245/s10434-008-9876-3. [DOI] [PubMed] [Google Scholar]
- 11.Ohta M, Tanaka F, Yamaguchi H, Sadanaga N, Inoue H, Mori M. The high expression of Fractalkine results in a better prognosis for colorectal cancer patients. Int J Oncol. 2005;26:41–47. [PubMed] [Google Scholar]
- 12.Gaudin F, Nasreddine S, Donnadieu AC, Emilie D, Combadière C, Prévot S, Machelon V, Balabanian K. Identification of the chemokine CX3CL1 as a new regulator of malignant cell proliferation in epithelial ovarian cancer. PLoS One. 2011;6:e21546. doi: 10.1371/journal.pone.0021546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim M, Rooper L, Xie J, Kajdacsy-Balla AA, Barbolina MV. Fractalkine receptor CX(3)CR1 is expressed in epithelial ovarian carcinoma cells and required for motility and adhesion to peritoneal mesothelial cells. Mol Cancer Res. 2012;10:11–24. doi: 10.1158/1541-7786.MCR-11-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li F, Wang Z, Liu Y, Li J. Down-regulation of fractalkine inhibits the in vitro and in vivo angiogenesis of the hepatocellular carcinoma HepG2 cells. Oncol Rep. 2010;24:669–675. [PubMed] [Google Scholar]
- 15.Andre F, Cabioglu N, Assi H, Sabourin JC, Delaloge S, Sahin A, Broglio K, Spano JP, Combadiere C, Bucana C, et al. Expression of chemokine receptors predicts the site of metastatic relapse in patients with axillary node positive primary breast cancer. Ann Oncol. 2006;17:945–951. doi: 10.1093/annonc/mdl053. [DOI] [PubMed] [Google Scholar]
- 16.Jamieson-Gladney WL, Zhang Y, Fong AM, Meucci O, Fatatis A. The chemokine receptor CX(3)CR1 is directly involved in the arrest of breast cancer cells to the skeleton. Breast Cancer Res. 2011;13:R91. doi: 10.1186/bcr3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edge SB, Compton CC. The American joint committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 18.Rousseaux S, Debernardi A, Jacquiau B, Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY, Lantuejoul S, Hainaut P, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med. 2013;5:186ra166. doi: 10.1126/scitranslmed.3005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Botling J, Edlund K, Lohr M, Hellwig B, Holmberg L, Lambe M, Berglund A, Ekman S, Bergqvist M, Pontén F, et al. Biomarker discovery in non-small cell lung cancer: Integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res. 2013;19:194–204. doi: 10.1158/1078-0432.CCR-12-1139. [DOI] [PubMed] [Google Scholar]
- 20.Tang H, Xiao G, Behrens C, Schiller J, Allen J, Chow CW, Suraokar M, Corvalan A, Mao J, White MA, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res. 2013;19:1577–1586. doi: 10.1158/1078-0432.CCR-12-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Der SD, Sykes J, Pintilie M, Zhu CQ, Strumpf D, Liu N, Jurisica I, Shepherd FA, Tsao MS. Validation of a histology-independent prognostic gene signature for early-stage, non-small-cell lung cancer including stage IA patients. J Thorac Oncol. 2014;9:59–64. doi: 10.1097/JTO.0000000000000042. [DOI] [PubMed] [Google Scholar]
- 22.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, et al. Gene expression-based survival prediction in lung adenocarcinoma: A multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, Thomas RK, Naoki K, Ladd-Acosta C, Liu N, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28:4417–4424. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Budczies J, Klauschen F, Sinn BV, Győrffy B, Schmitt WD, Darb-Esfahani S, Denkert C. Cutoff Finder: A comprehensive and straightforward web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7:e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao X, Qi L, Chen X, Du J, Zhang Z, Liu S. Expression of CX3CR1 associates with cellular migration, metastasis, and prognosis in human clear cell renal cell carcinoma. Urol Oncol. 2014;32:162–170. doi: 10.1016/j.urolonc.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 26.Johnson LA, Jackson DG. The chemokine CX3CL1 promotes trafficking of dendritic cells through inflamed lymphatics. J Cell Sci. 2013;126:5259–5270. doi: 10.1242/jcs.135343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umehara H, Bloom ET, Okazaki T, Nagano Y, Yoshie O, Imai T. Fractalkine in vascular biology: From basic research to clinical disease. Arterioscler Thromb Vasc Biol. 2004;24:34–40. doi: 10.1161/01.ATV.0000095360.62479.1F. [DOI] [PubMed] [Google Scholar]
- 28.Fong AM, Robinson LA, Steeber DA, Tedder TF, Yoshie O, Imai T, Patel DD. Fractalkine and CX3CR1 mediate a novel mechanism of leukocyte capture, firm adhesion, and activation under physiologic flow. J Exp Med. 1998;188:1413–1419. doi: 10.1084/jem.188.8.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park MH, Lee JS, Yoon JH. High expression of CX3CL1 by tumor cells correlates with a good prognosis and increased tumor-infiltrating CD8+ T cells, natural killer cells, and dendritic cells in breast carcinoma. J Surg Oncol. 2012;106:386–392. doi: 10.1002/jso.23095. [DOI] [PubMed] [Google Scholar]
- 30.Jamieson WL, Shimizu S, D'Ambrosio JA, Meucci O, Fatatis A. CX3CR1 is expressed by prostate epithelial cells and androgens regulate the levels of CX3CL1/fractalkine in the bone marrow: Potential role in prostate cancer bone tropism. Cancer Res. 2008;68:1715–1722. doi: 10.1158/0008-5472.CAN-07-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shulby SA, Dolloff NG, Stearns ME, Meucci O, Fatatis A. CX3CR1-fractalkine expression regulates cellular mechanisms involved in adhesion, migration, and survival of human prostate cancer cells. Cancer Res. 2004;64:4693–4698. doi: 10.1158/0008-5472.CAN-03-3437. [DOI] [PubMed] [Google Scholar]
- 32.Marchesi F, Piemonti L, Fedele G, Destro A, Roncalli M, Albarello L, Doglioni C, Anselmo A, Doni A, Bianchi P, et al. The chemokine receptor CX3CR1 is involved in the neural tropism and malignant behavior of pancreatic ductal adenocarcinoma. Cancer Res. 2008;68:9060–9069. doi: 10.1158/0008-5472.CAN-08-1810. [DOI] [PubMed] [Google Scholar]
- 33.Zhou B, Xu H, Ni K, Ni X, Shen J. Expression of chemokine XCL2 and CX3CL1 in lung cancer. Med Sci Monit. 2016;22:1560–1565. doi: 10.12659/MSM.895985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Locatelli M, Boiocchi L, Ferrero S, Martinelli Boneschi F, Zavanone M, Pesce S, Allavena P, Maria Gaini S, Bello L, Mantovani A. Human glioma tumors express high levels of the chemokine receptor CX3CR1. Eur Cytokine Netw. 2010;21:27–33. doi: 10.1684/ecn.2009.0184. [DOI] [PubMed] [Google Scholar]
- 35.Erreni M, Solinas G, Brescia P, Osti D, Zunino F, Colombo P, Destro A, Roncalli M, Mantovani A, Draghi R, et al. Human glioblastoma tumours and neural cancer stem cells express the chemokine CX3CL1 and its receptor CX3CR1. Eur J Cancer. 2010;46:3383–3392. doi: 10.1016/j.ejca.2010.07.022. [DOI] [PubMed] [Google Scholar]
- 36.Sciume G, Soriani A, Piccoli M, Frati L, Santoni A, Bernardini G. CX3CR1/CX3CL1 axis negatively controls glioma cell invasion and is modulated by transforming growth factor-β1. Neuro Oncol. 2010;12:701–710. doi: 10.1093/neuonc/nop076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vitale S, Cambien B, Karimdjee BF, Barthel R, Staccini P, Luci C, Breittmayer V, Anjuère F, Schmid-Alliana A, Schmid-Antomarchi H. Tissue-specific differential antitumour effect of molecular forms of fractalkine in a mouse model of metastatic colon cancer. Gut. 2007;56:365–372. doi: 10.1136/gut.2005.088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xin H, Kanehira M, Mizuguchi H, Hayakawa T, Kikuchi T, Nukiwa T, Saijo Y. Targeted delivery of CX3CL1 to multiple lung tumors by mesenchymal stem cells. Stem Cells. 2007;25:1618–1626. doi: 10.1634/stemcells.2006-0461. [DOI] [PubMed] [Google Scholar]
- 39.Erreni M, Siddiqui I, Marelli G, Grizzi F, Bianchi P, Morone D, Marchesi F, Celesti G, Pesce S, Doni A, et al. The fractalkine-receptor axis improves human colorectal cancer prognosis by limiting tumor metastatic dissemination. J Immunol. 2016;196:902–914. doi: 10.4049/jimmunol.1501335. [DOI] [PubMed] [Google Scholar]
- 40.Matsubara T, Ono T, Yamanoi A, Tachibana M, Nagasue N. Fractalkine-CX3CR1 axis regulates tumor cell cycle and deteriorates prognosis after radical resection for hepatocellular carcinoma. J Surg Oncol. 2007;95:241–249. doi: 10.1002/jso.20642. [DOI] [PubMed] [Google Scholar]
- 41.Marchesi F, Locatelli M, Solinas G, Erreni M, Allavena P, Mantovani A. Role of CX3CR1/CX3CL1 axis in primary and secondary involvement of the nervous system by cancer. J Neuroimmunol. 2010;224:39–44. doi: 10.1016/j.jneuroim.2010.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the present study are available from The Cancer Genome Atlas and Gene Expression Omnibus.