Abstract
Rearrangement of actin filaments by polymerization, depolymerization, and severing, is important for cell locomotion, membrane trafficking, and many other cellular functions. Cofilin and actin-interacting protein 1 (AIP1, also known as WDR1) are evolutionally conserved proteins that cooperatively sever actin filaments. However, little is known about the biophysical basis of the actin-filament severing by these proteins. Here, we performed single molecule kinetics analyses of fluorescently labeled AIP1 during the severing process of cofilin-decorated actin filaments. Results demonstrated that binding of a single AIP molecule was sufficient to enhance filament severing. After AIP1 binding to a filament, severing occurred with a delay of 0.7 s. Kinetics of binding and dissociation of a single AIP1 molecule to/from actin filaments followed a second-order and a first-order kinetics scheme respectively. AIP1 binding and severing were detected preferentially at the boundary between the cofilin-decorated and bare regions on actin filaments. Based on the kinetic parameters explored in this study, we propose a possible mechanism behind the enhanced severing by AIP1.
Keywords: AIP1, Actin, Cofilin, single molecule imaging, severing process
Dynamic reorganization of the actin cytoskeleton is crucial for a number of cellular processes including cytokinesis, cell motility, and morphogenesis [1]. However, filamentous actin alone is relatively stable, and acceleration of filament disassembly is often required to promote actin cytoskeletal dynamics [2–4]. Actin-depolymerizing factor (ADF)/cofilin is one of the major factors to accelerate disassembly of actin filaments by enhancing filament fragmentation and monomer dissociation from the pointed end. However, in vitro studies indicate that ADF/cofilin alone severs actin filaments only weakly and optimally when a cluster of ADF/cofilin molecules bind to an actin filament at low densities [5, 6]. When ADF/cofilin binds to the side of actin filaments at high densities, fragmentation of the filaments is rather infrequent [7]. These biochemical properties suggest that ADF/cofilin alone may not be sufficient to account for rapid turnover of actin filaments observed in live cells. It should be noted the marked diversity in the severing action of cofilin exists in the field of biology. Some cofilin at concentrations lower than the Kd for binding severs actin filaments optimally [8]. The severing is affected by restricting the flexibility of the filament [9] or by the ATP hydrolysis of actin filaments [10]. Under a certain condition severing is observed in the filaments saturated with cofilin [11].
Actin-interacting protein 1 (AIP1) is a conserved protein that promotes disassembly of actin filaments in the presence of ADF/cofilin [12, 13]. In vitro, AIP1 preferentially interacts with ADF/cofilin-decorated actin filaments and induces rapid disassembly of the filaments when ADF/cofilin is present at high densities [14, 15]. Thus, the combination of ADF/cofilin and AIP1 is an efficient mechanism to disassemble actin filaments. Genetic studies have shown that AIP1 is essential for viability in multicellular organisms including mice [16], nematodes [17], insects [18], and plants [19]. At cellular levels, AIP1 plays important roles in cytokinesis, cell migration [20–22], epithelial/epidermal morphogenesis [23–25].
The molecular process of the enhancement of actin filament disassembly is, however, largely unknown; especially, the kinetics of AIP1 binding, severing, and dissociation to/from the filament has not been directly analyzed. This could be due to that discrimination between severing and depolymerization of actin filaments is very hard for analyses depending on the ensemble average of number of molecules (such as conventional biochemistry), analyses with snapshot type image acquisition [26, 27] , or relatively slow time lapse imaging [28]. By contrast, high-speed single molecule time lapse imaging of molecules (e.g., actin, AIP1 and cofilin) can distinguish severing and depolymerization in a single AIP1 molecule level under adequate conditions [5, 29], and enable the kinetics analysis of AIP1, which may have multiple actions, e.g., binding, severing and end-binding [28, 30, 31].
We made a video-rate time lapse imaging of single Alexa-labeled-AIP1 molecules that bind to an actin filament, which showed that severing of actin filaments was greatly enhanced when a single AIP1 bound to an actin filament. The kinetics of AIP1 binding, severing, and transient end-binding (capping) was analyzed at a single molecule level, which showed that AIP1 bindings could be well described by the binding and dissociation and the end-binding, which could be described by Michaelis-Menten type state transitions.
Results
Binding of a single GST2-AIP12 substantially enhances the severing of actin filaments
Disassembly of cofilin-decorated actin filaments by GST2-AIP12 (GST-Alexa-488-AIP1, Alexa-488 labeled nematode AIP1, UNC-78), which is presumably dimeric, was imaged. The enhancement of severing of nematode cofilin (UNC-60B) decorated single actin (from rabbit skeletal muscle) filaments were analyzed. Cofilin (UNC-60B) modestly severs rabbit muscle actin filaments at a relatively high concentration (2 μM), which is significantly enhanced by AIP1 (UNC-78) [26, 27]. AIP1 (UNC-78) disassembles C. elegans actin in the presence of cofilin (UNC-60B) more efficiently than rabbit muscle actin, but the differences were subtle [32]. This could be due to actin being evolutionally conserved across species. We used rabbit skeletal muscle actin, since it is well documented in literature.
Rhodamine-labeled actin filaments were anchored to glass surface via anti-biotin antibodies with 1–2 μm intervals of tethering points, which allowed for fluctuations of the filament between tethering points as described previously [5]; the amplitude of fluctuations was estimated at a few tens nm in z-axis. A mixture of cofilin (2 μM) and GST2-AIP12 (10 nM) was applied to these fluorescently labeled actin filaments by gentle pressure ejection through a micro pipette system (see details in Experimental Procedures). Under this condition, cofilin and GST2-AIP12 efficiently disassembled actin filaments as demonstrated in the previous study [12], and binding of single GST2-AIP1 2 molecules to the actin filament and severing were imaged and analyzed, as shown in Fig. 1, and Movie S1. Monomeric AIP1 labeled with Alexa-488 showed similar binding and severing of actin filaments as that of GST2-AIP12 (see below), the mean duration of binding of monomeric AIP1 (Ts, see definition in figure 1) was 0.8 ± 0.3 s (n=29), and severing was detected in 27% of side-bindings (n=26), supporting the idea that GST2-AIP12 behaves like native monomeric AIP1.
Figure 1.
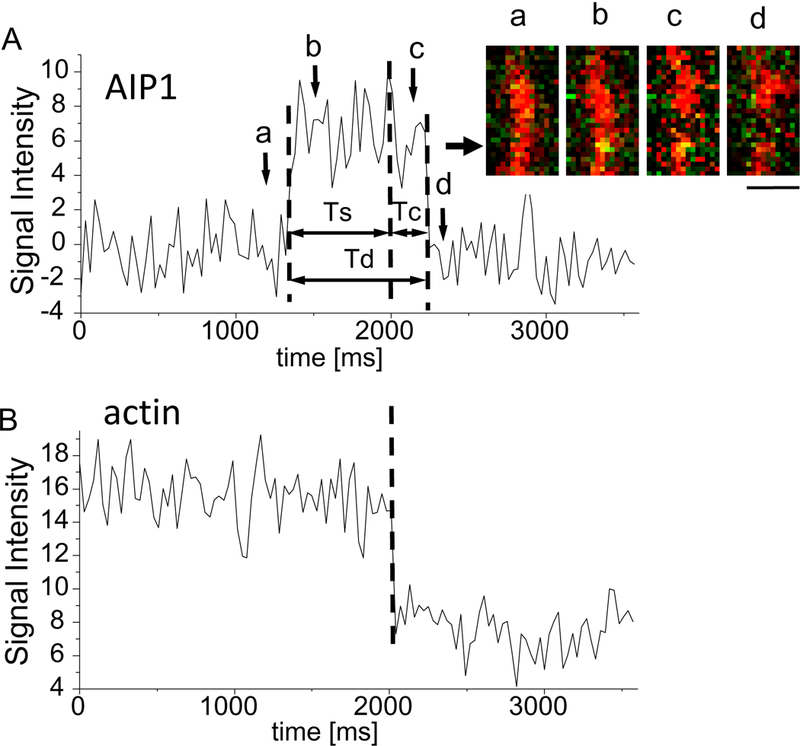
Sequential observation of single GST2-AIP12 binding and severing of an actin filament in the presence of cofilin. A, signal intensity of AIP1 fluorescence was plotted against time (ms). The signal intensities of AIP1 (A) and actin (B) are measured at the 600 nm diameter circle area on the actin filament; the center of the area is shown by the arrow besides the time lapse image in the inset. The time from AIP1 binding to severing (Ts), and the duration of AIP1 binding (Td) are shown by double headed arrows. The inset shows the time lapse fluorescence image of the actin filament (red) and AIP1 (green, which turned yellow on the filament, images b and c in panel A). AIP1 binds to the side of the filament (a, b), and severing and transient end-binding (c) of the filament are detected, and the AIP1 is dissociated (d) from the filament. Time of acquisition of these images (a–d) are shown by arrows and letters in A. Bar 1 μm. B, the signal intensity of fluorescence from the region of the actin filament was plotted against time (ms), which decreased suddenly when the filament was severed and began to fluctuate; the abrupt stepwise decrease in actin fluorescence intensity was detected due to the 600 nm diameter image circle contained a part of the daughter filament that fluctuated after severing. The duration of AIP1 on the filament following the severing, which denotes the duration of transient end-binding, is shown by the double headed arrow (Tc). A yellow dot on an actin filament denotes fluorescent AIP1, and small green dots flanking the actin filaments were mostly the noise of the imaging system. See also a streaming video of AIP1 and actin filaments (supplemental video 1). Approximately 10 nM GST2-AIP1 2 and 2 μM cofilin in the solution (see details in Experimental Procedures).
The fluorescence of a single GST2-AIP12 molecule was detected along the tethered actin filaments. The intensity profile of the AIP1 fluorescence signal (Fig. S2) was fitted with two Gaussian distributions (basal noise and single GST2-AIP12 bindings, respectively). This denotes that the binding of single GST2-AIP12 molecules was detected (Fig. 1). A typical sequence of AIP1 binding and actin-filament severing is shown in Fig. 1; a single GST2-AIP12 bound to an actin filament (Fig. 1A, inset), and a severing of the filament (Fig. 1B, dashed line). In this case, AIP1 binding was detected 690 ms before severing (Fig. 1A, Ts) and AIP1 stayed bound on the severed filament for 210 ms following the severing (Fig. 1A, Tc). Severing of filaments occurred in 11.7 % of total AIP1 binding events (the time lapse images with the lowest background noise were used for analysis, and 102 AIP1 binding events were detected and analyzed). The duration of AIP1 binding that did not sever the filament was fitted well by a single exponential function with decay constant of 289 ms as shown in Fig. 5A. The value 289 ms was nearly three times smaller than that of the binding associated with severing (see more in the later section). As discussed in the later section 93% of the severing events are enhanced by AIP1 binding. It is highly likely that AIP1 binding in Fig. 1 enhanced severing.
Figure 5.
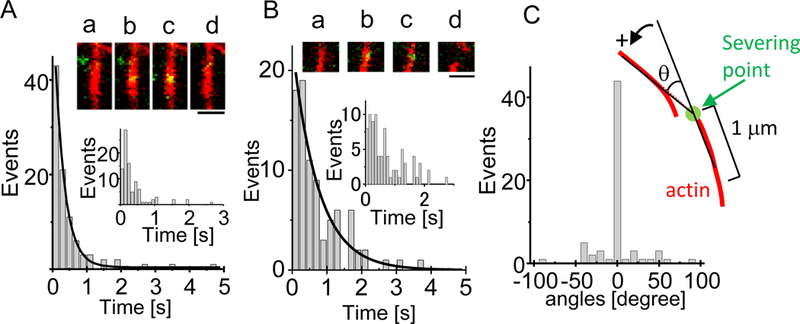
Distribution of the duration of GST2-AIP12 binding to actin filaments in the presence of cofilin. Panel A shows the distribution of AIP1 binding that did not sever the actin filament. The inset shows the time lapse fluorescence images of the actin filament and AIP1. AIP1 binds (b, c) to a filament (a), and dissociates from the filament (d). Bar, 1 μm. The line denotes the single exponential function fitting for the distribution of binding; time constant of the decay is 289 ± 9 ms. Panel B shows the distribution of the duration of AIP1 binding (Ts) that was associated with severing. The inset shows the time lapse fluorescence image of the actin filament and AIP1. AIP1 binds to a filament (b, c), and severing of the filament is detected (d). Transient end-binding by AIP1 was not detected in this case. The distribution is magnified with smaller bins (inset). Notation of the inset is same as Fig. 1A. Cofilin is present in the assays for AIP1 severing. Bar, 1 μm. Panel C shows distribution of the angle of bending at severing. Notation of the angle measurement is shown in the inset; the angle is measured in the anti-clockwise direction. Severing point is shown by an arrow and the filaments are approximated by 1μm straight lines. The experimental conditions were the same as in Fig. 1.
The above ejection method may have introduced slight uncertainty in the concentration of cofilin and AIP1. Therefore, we also performed experiments with a precise concentration of GST2-AIP12 (10 nM) and cofilin (1 μM) to estimate the rate of severing and on rate of GST2-AIP12 binding (see following sections). In this solution the rate of severing was (4.0 ± 2.1, mean ± SD) ×10−2 severing events per μm of filament per s (n=3 image fields, see the materials and methods section), which is 14.0 times higher than the rate by 1 μM cofilin alone (2.8± 4. 4)×10−3 severing events per μm of filament per s (n=7 imaging fields), suggesting the presence of the “spontaneous” severing of cofilin-occupied actin filaments in the absence of AIP1; i.e., GST2-AIP12 (10 nM) substantially (14 times) enhances the severing of cofilin-occupied actin filaments as previously suggested [28][8]. Statistically, 93% of the severing events are strongly enhanced by AIP1 binding and 7% of the severing events might be “spontaneous”. On the other hand, GST2-AIP12 (10 nM) alone never induced severing of actin filaments.
Kinetics of GST2-AIP12 binding, severing and transient end-binding
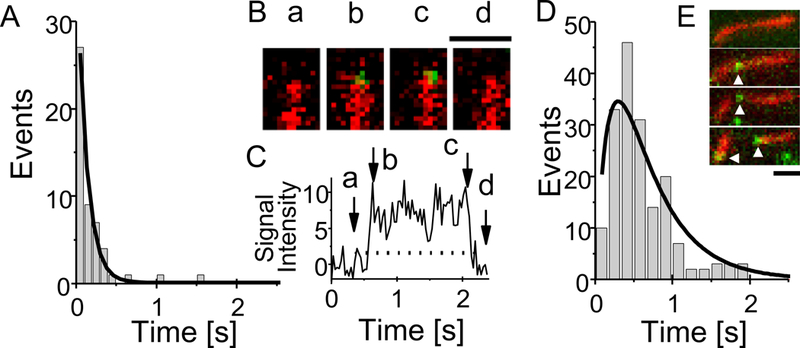
The duration of typical GST2-AIP12 binding events associated with severing are shown in Fig. 2A; analysis of the time-dependent distribution of AIP1 binding associated with severing shows that AIP1 binding was detected most frequently immediately before the severing events, and the chance of finding AIP1 on the filament decayed slowly with a time constant of 711 ± 18 ms (90 severing events) (Fig. 2); more detailed analysis shows that AIP1 was found on the filament at 30 ms prior to the time of severing in 79 % of cases (Fig. S4). AIP1 remained bound at the end of the daughter filaments in 50 % of the severing events, and the frequency of detecting AIP1 there was decreased exponentially with a time constant of 164 ± 5 ms (n=90) (Fig. 2) (see more in Discussion).
Figure 2.
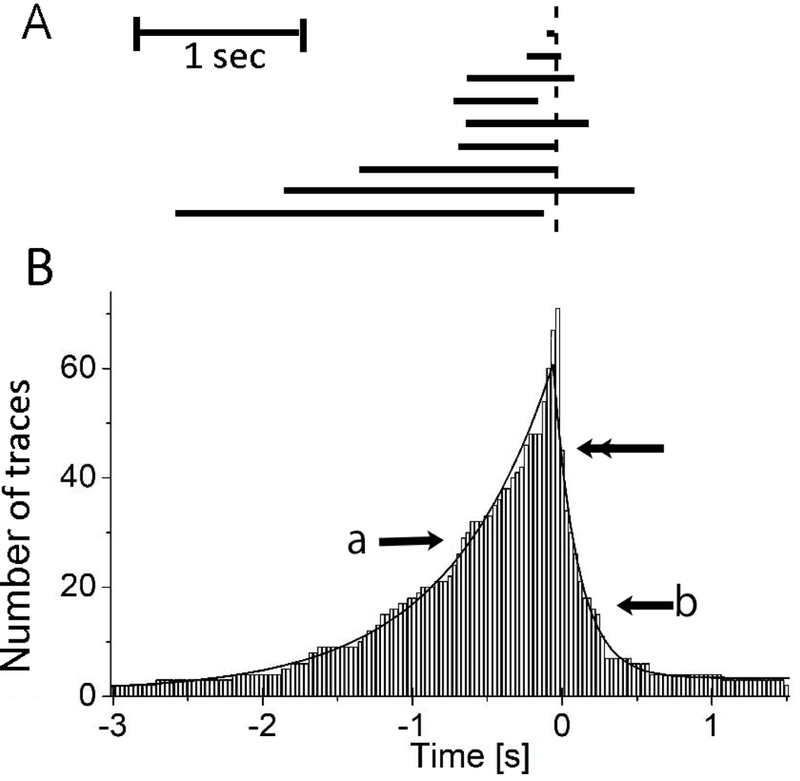
Time dependent distribution of GST2-AIP12 stay bound to the actin filament which associated with severing in the presence of cofilin. Panel A shows typical examples of duration time lines where AIP1 was found on the actin filament; horizontal bars denote the duration of AIP1 binding (Td). Vertical dotted line denotes the time of serving. A scale bar denotes 1 s. Note that AIP1 dissociated from the filament prior to the severing in 4th and 9th traces. For making Panel B, we found a severing event and collected all AIP1 binding events before and after the severing and repeated this procedure. Panel B shows the number of time traces (vertical axis) where AIP1 stayed bound at given time (horizontal axis). Ninety time-traces of AIP1 binding were analyzed. Time zero corresponds to the time of severing filaments. Seventy-one traces were counted at zero; i.e., AIP1 stayed on the filament at severing in 71 traces out of 90, denoting AIP1 were dissociated briefly before the severing in 19 traces (see supplemental Fig. 4S for detail). In 2 cases AIP1 stayed bound for more than 3 s before severing. Forty-five traces were counted (shown by the double headed arrow) at 30 ms; i.e., AIP1 stays bound at the end of daughter filaments in a half of cases. The count declines with time. The lines pointed by (a) and (b) denote the single exponential function fitting for the binding before and after severing respectively. The time constant shown by the arrow (a) and (b) are 711 ± 18 ms (n=90) and 164 ± 5 ms (n=45), respectively. The error on the time constants is the error of the fit. “Simple binding events which were not associated with severing” as well as “AIP1 binding events which resulted in severing (e.g., the data in figure 1)” (see Discussion) are used to construct the histogram. The “simple binding events” will produce a very small plateau at −3 s and 1.5 s, since the “simple binding events” distribute uniformly if it follows the Michaelis-Menten (M-M) type kinetics of binding. The plateau did not affect the estimation of the time constants. The experimental conditions were the same as in Fig. 1.
AIP1 enhanced severing, and generated two daughter filaments as shown in Fig. 1. One of the daughter filaments often remained near the glass surface (92 %, 83 out of 90), while the other became undetectable in the remaining imaging sequence by the large fluctuations of the daughter filament (Fig. 1d, Fig. 4 and Supplemental Movie S1); the typical length of fluctuating part of the daughter filaments was 1–2 μm. In rare cases, both daughter filaments fluctuated (6 %), or both disappeared from the TIRF illumination range (2 %). The analysis of kinetics of transient end-binding of AIP1 was done on filaments that did not show large fluctuations in the later section.
Figure 4.
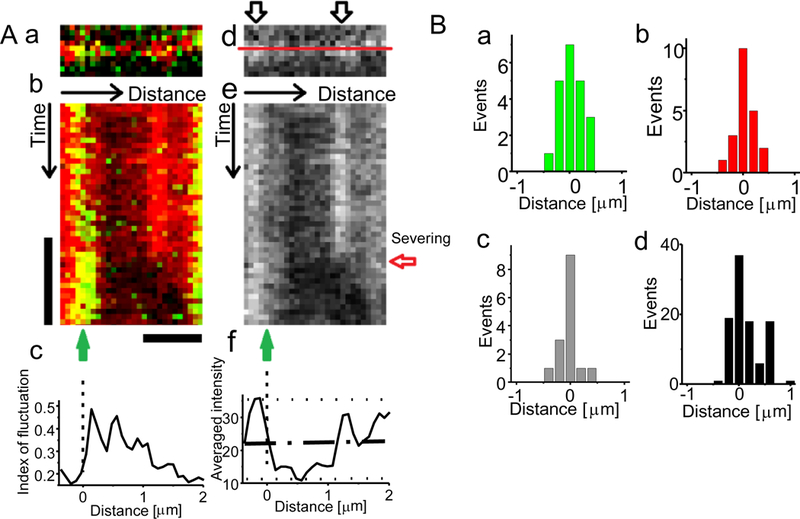
Sites of GST 2-AIP12 binding and severing along the cofilin decorated actin filament. A, (a) A TIRF image of an actin filament (red) and AIP1 (green). (b) A kymograph of the actin filament and AIP1 along the line superimposed on an actin filament in panel d is shown, which is a graphical representation of TIRF fluorescence profile of the actin filament along the “Distance arrow” over time shown by the “Time arrow”. The position of AIP1 binding and severing are shown by a green arrow (binding) and a horizontal red arrow (severing) in panel e. Several brief bindings of AIP1 were detected before the stable binding, and severing was observed at 0.1 s from the start of the stable binding, where 1 μm of the actin fluorescence in the right side of the AIP1 binding disappeared by the severing and fluctuations of the filament. (c) The magnitude of the fluctuations at each pixel, evaluated as “index of fluctuation” (see materials and methods) varied along the filament. (d) The fluorescence image of the actin filament. The intensely illuminated regions of the actin filament are shown by outlined arrows. (e) A kymograph of the same actin filament. (f) The mean intensity of the fluorescence of the filament (from top to the severing). The zero position is chosen to be the middle (50%) of the intensity of actin fluorescence that declines from the peak (the basal and the peak levels are shown by dotted lines and the middle is shown by an interrupted line). Bars denote 0.5 s, and 1 μm. We set the boundary as zero in the contour axis (see panel 4Af). B, The distribution of the position of AIP1 bindings accompanied by severing (a), the position of severing (b), the position of AIP1 bindings without severing (c), and the position of cofilin bindings (d). The experimental conditions were the same as in Fig. 1.
The analysis of the intensity profile of the AIP1 binding and severing events showed that a single GST2-AIP12 bound to a filament and the filament was severed in a sequential manner in 43 % (15 out of 35) of severing events (Fig. 3Aa). Simultaneous binding of two GST2-AIP12 molecules in a vicinity (< 600 nm diameter) was sometimes detected before severing, and one dissociated and the other remained until severing in 31 % of cases (Fig. 3Ab). These denote that in 74 % of cases one GST2-AIP12 molecule bound to a filament at the time of severing (Fig. 3B, a plus b), showing that binding of a single GST2-AIP12 molecule is sufficient to facilitate the severing as has been suggested [22]. This type of AIP1 binding and severing were analyzed mainly in this study as shown in Fig 2. In the remaining 26 % of cases, two or more AIP1 were detected in the vicinity of severing points (Fig. 3B c plus d). The duration of binding, Ts (see Fig.1), of one, two, and three AIP1 bindings were 0.73 ± 0.76 s (n=22), 2.62 ± 2.9 s (n=20), and 1.40± 1.4 s, (n=6) respectively; multiple binding in the vicinity did not shorten the Ts (one-way ANOVA test).
Figure 3.
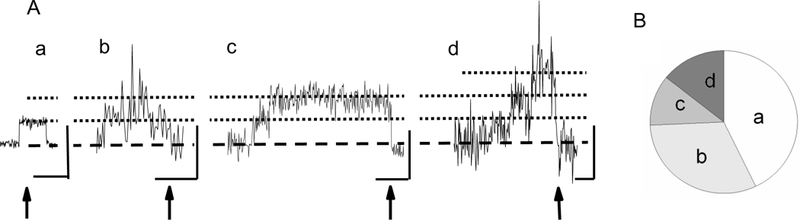
The number of GST2-AIP12 molecules bound to actin filaments at the time of actin filament severing in the presence of cofilin. Panel A shows changes in the signal intensity of AIP1 measured at the 600 nm diameter area. Typical examples of AIP1 fluorescence signal intensity changes are shown; (a) a single binding, (b) a single binding at the severing with additional short binding in the middle of the trace, (c) two bindings detected at the severing, and (d) more than three bindings detected at the severing (three bindings case is shown in this panel). The time of severing is shown by arrows. Scales denote 1 s and 5 counts of photon (photon was detected in the 30 ms time interval). Panel B shows the distribution of (a)–(b)–(c)–(d) types of AIP1 binding (n=35 severing events, d denotes more than three bindings). AIP1 dimer was labeled by zero to five Alexa-488 molecules (see Fig. 3S). Single molecule experiments detected different intensities, but we chose records which showed stepwise changes with a similar amplitude. Note that the amplitude of the initial stepwise increase in panel d is slightly smaller than others presumably due to that the AIP1 was labeled with a smaller number of Alexa-488 molecules. The chance to detect fluorescence quenching was quite low during the imaging because the specimen was weakly illuminated. Therefore, the steps in the panels in A (horizontal dotted lines) correspond to the number of GST2-AIP12. The experimental conditions were the same as in Fig. 1.
Sites of AIP1 binding and severing of actin filaments
Binding of AIP1 to an actin filament is dependent on the preceding cofilin binding to the filament [26, 28], and cofilin tends to bind to a fluctuating part of actin filaments [5]. Presumably, AIP1 binds preferentially to a fluctuating region of actin filaments in the presence of cofilin, and enhances severing.
The site of AIP1 binding and filament severing were tracked by using kymographs drawn along the contours of individual actin filaments (Fig. 4). The brightly-fluorescing regions are tethered to the surface at filament attachment points, whereas the dimly fluorescing regions are located further (~30 nm) away from the surface in the z-direction in TIRF imaging (Fig. 4Ad and e), since actin filaments were nearly uniformly labeled with rhodamine as shown in Fig. 1A (inset a) and Fig. 6E, and the fluorescence intensity under TIRF illumination exponentially decreases with the relative separation distance from the glass surface. Thus, changes in the fluorescence intensity reflect the fluctuations of the filament as shown in Figure 4Ac. The frequency of AIP1 binding was not uniformly distributed along actin filaments. Contrary to what we expected, AIP1 binding was most frequently detected at the boundary between intensely and less intensely illuminated regions (a typical case is shown in Fig. 4A), and severing occurred within 500 nm from the boundary (Fig. 4Ba and b). These results suggest that the filament is severed at the boundary between fluctuating and non-fluctuating regions where AIP1 binds. Similar distribution of AIP1 binding was found when the AIP1 binding was not associated with severing (Fig. 4Bc). The extent of filament decoration is roughly estimated ca.10% according to the previous report [5](see materials and methods).
Figure 6.
Distribution of the duration of GST2-AIP12 transient end-binding in the presence of cofilin. Panel A shows the distribution of the duration of AIP1 transient end-binding (Tc) after severing actin filaments. The line denotes the single exponential function fitting for the distribution of binding; time constant of the decay is 120 ± 6 ms. Panel B shows the time lapse fluorescence images of the actin filament and bound AIP1 at the bare end of the filament. AIP1 binds (b, c) to the end of a bare filament (a), and dissociated from the end (d). Panel C shows the signal intensity of AIP1 measured in the 600 nm diameter circle area at the end of the actin filament. The time of image acquisition is shown by arrows (a-d). Bar, 1 μm. Panel D shows the distribution of the duration of AIP1 transient end-binding to a bare end of filaments (n=174 binding events). The line indicates the change described by the equation (3) in the materials and methods. Panel E shows two monomeric-AIP1 bindings at the same side of a severed filament, which are dedicated by white triangles. Cofilin is present in the assays. The experimental conditions were the same as in Fig. 1.
The binding of fluorescently labeled nematode cofilin (Alexa-488-labeled, 10 nM) to actin filaments was also examined; cofilin bound to fluctuating regions of filaments, but not to less-fluctuating tethered regions as observed before with mouse cofilin [5]; cofilin binding was detected most frequently at the boundary between the fluctuating and less-fluctuating regions as shown in Fig. 4Bd. These observations suggest that AIP1 binds to actin filaments and enhances the severing at the boundary between the cofilin-decorated and bare actin filaments.
Analysis of on-, off-rates, and duration of AIP1 binding before severing
The most remarkable feature of the AIP1 binding to actin filaments is its statistical nature as shown in Fig. 2A. The duration of the AIP1 binding until severing (Ts) corresponds to the “waiting time (or on-duration)” according to the scheme of the preceding study on single-molecule enzymatic dynamics [33]. The distribution of the “waiting time” before severing was examined in a similar manner that has been conducted in single ion channel studies [34] and enzymes [33]. The advantage of this study is that the time sequence of AIP1 binding, severing (or not-severing), and following transient end-binding was detected non-interruptedly at the single-molecule level; this type of direct single-molecule observation has not been reported.
The binding duration distribution derived from binding events of AIP1 that did not sever the filament is shown in Fig. 5A, which could correspond to a simple binding and dissociation [33]. The duration was fitted well by a single exponential function with decay constant of 289 ± 9 ms (n=94) (Fig. 5A) according to the Poisson statistics [34]; i.e., off-rate of AIP1 binding was estimated 3.46 s−1, and the apparent “on-rate” was 0.21 ± 0.09 μM−1s−1 (n = 9 filaments) with precise 10 nM AIP1, which gives the dissociation constant of AIP1, Kd = 16.6 μM, a value slightly higher than previous estimates for mutant forms of AIP1 [35].
The binding duration (Ts) distribution derived from those of AIP1 binding associated with severing was also fitted by a single exponential with a longer decay constant of 740 ± 81 ms (n=90) (Fig. 5B) than that in the case not associated with severing.
Similar binding and severing of actin filaments were observed by using AIP1-cys-Alexa-488, a monomeric AIP1 labeled with Alexa-488 (Supplemental Movie S2, and Fig. 6E) as mentioned above. These results support again the hypothesis that single AIP1 binding enhances severing of a cofilin decorated actin filament. Detailed kinetic study was not conducted, because the S/N of the images was lower than that of GST2-AIP12.
The angle of filament bending at the AIP1 binding site, and the length of daughter filaments
The bending angle of cofilin decorated actin filaments at the severing point [36] is 31 degrees; note that filaments suspended in the solution were used for analysis. In this study, nearly straight cofilin decorated filaments were severed at the site of AIP1 binding; the bending angle was −0.8 ± 24.0 degree (n = 67) at the severing-point (Fig. 5C); note the large S.D., indicating filaments with various bending were included. We attempted to estimate the bending angle of the filaments at severing point with cofilin alone. Terminal-segments (1–2 μm) that fluctuated were sometimes severed; the chance to observe the severing with 1μM cofilin alone was very low in the tethered filament. The bending angle at the severing point was not determined due to the fluctuations of the terminal-segment.
The length of the shorter daughter filament produced by severing was measured. Daughter filaments with 0.4–3.0 μm length were produced from 3 to 7 μm filaments; the length was nearly uniformly distributed, indicating that there was no apparent preferable length for severing. These daughter filaments were much longer than those (75 nm) observed by EM after 3 hours of AIP1 treatment [14]. The discrepancy could be due to differences in the duration of AIP1 treatment (1–10 s in this study and 3 hours in the EM study) and the methods of observations.
Kinetics of transient end-binding of AIP1 at the severed actin filaments
AIP1 transiently bound to the end of filaments (Fig. 1) as suggested [28, 30, 31] within the limit of optical resolution. In 50 % of severing events (45/90), the AIP1 fluorescence was uninterruptedly detected at the end of daughter filaments that did not apparently fluctuate. In the other 50 % of the cases, the fluorescence of AIP1 was not detected after severing presumably due to the large fluctuations of the daughter filament. The duration (Tc) of the fluorescence signal of AIP1 found at the end following the severing was exponentially decayed with time constant of 120 ± 6 ms (n = 50) (Fig. 6A).
Transient end-binding of AIP1 to pre-existing bare ends was also often observed as shown in Fig. 6B. The duration of the AIP1 transient end-binding showed a non-single-exponential distribution, which was described with the equation [3] in materials and methods with k1=5.3 s−1 and k3=2 s−1 (Fig. 6D) [33, 37, 38]. The difference in kinetic parameters of binding of AIP1 to pre-existing filament ends as compared to severed filament ends could possibly arise from having cofilin-actin on the severed filaments versus actin alone at the ends of bare filaments.
Transient end-binding was often detected only at one of the daughter actin filaments, which is consistent with that AIP1 binds only to the barbed ends of actin filaments [28, 30] (Fig. 6E). The time constant of the dissociation of the AIP1 transient end-binding is ca. 103 times higher than that of capping proteins [39]. The apparent “on-rate” (kf) 112 ± 88 μM−1s−1 (n = 5 image fields), which is in the same range as those for other actin binding proteins, e.g., myosin [40], but is two orders of magnitudes higher than those of capping proteins [41]. These mean that AIP1 has different end-binding properties compared to capping proteins.
Discussion
Single molecule kinetics analyses of the side-binding, severing and end-binding of fluorescently labeled AIP1 (dimeric and monomeric) to/of actin filaments were performed for the first time, which strongly suggests that severing of cofilin-decorated actin filaments is enhanced by a single AIP1 molecule bound to the side of filaments. AIP1 stays at one end of either daughter filaments for a short period. Transient binding to bare ends was also observed. Kinetics of AIP1 binding that was associated with/without severing of the actin filament can be described by a single-exponential decay equation with different time constants. AIP1 binding that was associated with severing was detected most often at the boundary between the tightly tethered and less-tightly tethered (fluctuating) region of actin filaments. In the following sections, we discuss a possible mechanism of the AIP binding and the enhancement of severing by the bound AIP1, and a role of AIP1 in actin filament turnover based on the kinetic parameters obtained in this study.
Kinetics of AIP1 binding and severing of cofilin-decorated actin filaments
Kinetics of enzymes or state transition of proteins have been estimated by single-molecule imaging techniques in a very limited number of cases [42]; e.g., duration of FAD activation [33], β-galactosidase [43], and the dwell time of tension generation by myosin [40], which could be due to technical difficulties to observe directly the enzymatic activation or state transition of proteins.
The rate of severing was (4.0 ± 2.1) ×10−2 severing events per μm of filament per s in AIP1 (10 nM) and cofilin (1 μM), which is 14.0 times higher than the rate by 1 μM cofilin alone. These results denote that AIP1 strongly enhanced the severing of cofilin decorated actin filaments, and suggest that 93% of the severing events is enhanced by AIP1 binding. In other words, 7% of the severing events might be “spontaneous” (i.e., cofilin mediated the severing not by AIP1). We observed one to five AIP1 binding events in a 2–5 μm filament in 15 s of time lapse imaging, and most of the severing events were associated with AIP1 binding, and this observation agrees with the above estimation.
The Michaelis-Menten (M-M) type kinetics of enzymatic binding suggest reversible binding and dissociation are followed by the enzymatic reaction (see equation 3 in the Kinetics analysis), which predicts the “simple binding” without “enzymatic” reactions. Our data showed 88% of the AIP1 binding belongs to this “simple binding”, and 12% of the Aip1 binding events resulted in a severing event. This fits the M-M model and showed that majority of bindings (88%) were classified as “simple binding”.
Here, let us consider the AIP1 bindings and severing of a 5 μm filament in AIP1 (10 nM) and cofilin (1 μM) to evaluate the effect of “simple binding” on the characterization of the enhancement of severing by AIP1. During the 15 s of imaging, there would be 3 severing events associated with AIP1 binding based on the serving rate of 4.0×10−2 severing events per μm of filament per s; e.g., Fig. 6E. There would be 22 AIP1 binding events without severing, since there must be 7.3 times larger number of the “simple binding” behind 3 binding with severing. Total duration of the “simple binding” is 6.38 s, since the mean duration of “simple” bindings is 0.29 s. This means AIP1 is found on the site of the actin filament in 42% of imaging period 15 s; i.e., the chance to find “simple AIP1 binding” at the time of severing is 42%. This value roughly agrees with the chance (26%) to find additional “simple AIP1 binding(s)” during the AIP1 binding associated with severing (Fig. 3B). The probability to find “simple AIP1 binding” at the time of “spontaneous” severing is 3% of all severing events, showing that chance to see the coincidence is low. We assume, in the above calculations, the sites of “simple AIP1 binding” and AIP1 binding with severing are same as shown in Fig. 4B for simplicity and we ignore the simultaneous multi AIP1 bindings, since the chance was low to observe them. In 74 % of cases one AIP1 molecule bound to a filament at the time of severing and these were analyzed. The analysis supports the strong correlation between binding of a single AIP1 molecule to a filament and severing as shown in Fig. 2.
The binding duration of AIP1 that did not sever the filament was fitted well by a single exponential function with decay constant of 289 ms, while the binding duration (Ts) of AIP1 binding associated with severing was also fitted by a single exponential with longer decay constant of 740 ± 81 ms (n=90) (Fig. 5B). The longer decay constant suggests the kinetics of binding without severing and of binding with severing are different; the longer delay may reflect that states during Ts include not only the state of binding but also the state that facilitates the severing.
The facilitating effect of AIP1 binding to the severing may continue after the dissociation of AIP1 from the filament, because substantial fraction of AIP1 (21%) was dissociated prior (several tens ms) to the severing (see Fig. 2A and Supplemental Fig. 4S).
Kinetic properties of transient end-binding following severing
The kinetics of capping protein binding to barbed ends predicts its half-life is of ca. 30 min [41], which is four orders longer than that of AIP1 half-life, suggesting that the dissociation process of AIP1 is quite different from that of capping protein. This idea agrees with the in vivo estimation of AIP1 binding [44].
The on-rate of AIP1 transient end-binding is in the order of 112 ± 88 μM−1 s−1, which may reflect that the binding process of AIP1 is a diffusion-limited process as proposed for other actin-binding proteins, e.g., myosin subfragment-1 [45] , gelsolin [46], filamin [47], and α-actinin [48], and could be very different from capping protein [41].
The Kd of AIP1 end-binding is estimated to be 23.4 nM, which is three orders lower than the Kd of AIP1 binding to the side of actin filaments (16.6 μM), suggesting that the underlying mechanism is quite different from that of the AIP1 binding to the side of actin filaments. It should be noted that optical resolution of our experimental setup (ca. 300 nm) is not enough to distinguish AIP1 binding to the filament end or to the side near actin filament end (<300 nm). However, the Kd value of the end binding is different from that of side binding (>300 nm away from the end), supporting that most of the “apparently” end binding could be classified as the end binding.
AIP1 capping at the barbed ends of cofilin-decorated actin filaments was not detected in preceding studies [28, 49–51] based on filament elongation [30]. This could be due to that the transient end-binding by AIP1 may not result in capping ends or may not be detected with conventional biochemical techniques or with snapshot type image acquisition or with the combination of proteins used. Similar observation of AIP1 that remains bound to the new filament ends was reported with cofilin and Cor1B [28].
The chance to find AIP1 that binds to the end of daughter filaments is 50%, which fits the idea that AIP1 stayed bound to an end of either daughter filaments in all cases, but only half of them were detected because only half of the daughter filaments could be imaged in our experimental conditions.
A burst type disassembly from the end of filaments was reported in a solution containing coronin, cofilin, and AIP1 all together [52], and severing and monomer dissociation from ends were reported within minutes of observation[50]. In this study, facilitation of depolymerization from the end of filaments was detected only in very limited cases. AIP1 binding near the end of filaments enhanced severing in a majority of cases, which agrees with a recent study [28]. These differences could be due to that experiments were performed under different experimental conditions and that the monomer dissociation from ends is relatively slow and is difficult to be detected within a short period (1–10 s) of high-speed imaging.
Possible molecular mechanism behind the enhancement of cofilin-dependent severing by AIP1
Filaments fully decorated with cofilin are stable and severed less readily than partially decorated ones [8, 53, 54]. These studies favor a model that the filament mechanics and conformational dynamics at boundaries of bare and decorated regions increase the chance for severing [53–56]. Similar mechanism may also work in the case of AIP1, which binds to the boundaries of bare (not-fluctuating) and cofilin decorated (fluctuating) regions of actin filaments, and facilitates the severing activity of cofilin.
The force required to sever bare actin filaments is relatively large, estimated as 300 pN [57]. This magnitude of force may be required to provide activation energy for the severing. The required force declines when the filament is twisted, suggesting that the conformational changes in the actin filament affect the activation energy for severing as discussed previously [58]. Gelsolin might sever actin filaments in a similar mechanism [59].
The severing angle of cofilin-decorated actin filaments suspended in solution was 31 degrees [36]; they proposed a hypothesis that the energy accumulation by the bending provides the activation energy for severing. The higher bending angle is presumably required for severing in bare actin filaments [60], which will be the reason why bare actin filaments are not severed spontaneously by intrinsic fluctuations. In other words, the activation energy required for the severing of bare actin filaments is very high without cofilin and/or AIP1.
The severing by cofilin of actin filaments in vitro starts with a considerable delay (tens of seconds) after an application of cofilin [6, 29], which may reflect the time required for spontaneous bending (e.g., 31 degrees) or accumulation of energy at the boundary between the bare and cofilin- decorated regions. The time delay from AIP1 binding to severing will be shorter than that of cofilin alone according to the above idea, which is consistent with our observations; mean Ts of AIP1 is 0.7 s, which is more than 104 shorter than that of human cofilin [8] and mouse cofilin [5].
Severing of cofilin decorated filaments (without AIP1) was only detected at the boundary between the fluctuating terminal-segment and tethered point in this study, suggesting the severing of actin filaments by cofilin alone may require the spontaneous bending due to intrinsic fluctuations of the terminal segment as in the case of suspended filaments.
A recent study by Aggeli and others [61] mentioned a possibility that AIP1 induces cofilin-dependent severing through affecting the actin-cofilin secondary binding site, resulting in the loss of cofilin-mediated stabilization of tilted actin conformers.
Based on the above observations and models, cofilin-dependent AIP1-enhanced severing of actin filaments presumably involves at least two steps; [1] cofilin decoration of a fluctuating region of actin filaments, and [2] side-binding of AIP1 to cofilin decorated actin at or near the boundary between bare and cofilin-decorated regions, which may enhance the dissociation of an actin-actin bond by destabilizing the actin-cofilin secondary binding site [61]. There is another possibility that AIP1 competes with cofilin for binding at/near the boundary, which reduced the occupancy of the filament by cofilin to the range favorable for severing [19]. These models explain that most of the actin filaments were severed in a nearly straight configuration with AIP1 binding. In addition, what makes AIP1 function more efficient may be that after severing, AIP1 binds to the filament barbed end, which may prevent filament re-annealing as proposed for gelsolin [62].
The possible mechanism mentioned above is also favored by the observation that AIP1 dissociates from the severing site before the severing event in 19 of the 90 events in Fig. 2. These transiently bound AIP1 molecules may compete with cofilin for actin filament binding and could be clearing cofilin from the severing site [63]. Once the AIP1 dissociates the ternary complex no longer exists, which may alter the bare-decorated segment boundary density, and facilitate the filament severing activity of cofilin. This could also explain the AIP1-induced severing when the actin filaments are fully decorated by cofilin [50]. AIP1 may compete with cofilin and facilitate severing as described above.
Biological significance of AIP1-dependent enhancement of severing of actin filaments
AIP1 and cofilin are thought to be the best candidates for the cellular actin disassembly mechanism [44], since these proteins collaborate to facilitate filament disassembly fast enough to account for the actin dynamics in the lamellipodia [64]. The value for the severing rate estimated in this study (1.4 s −1) is more than ten times higher than the value independently estimated in the preceding study 0.1–0.02 s−1 [44], This difference may due to the difference in the environment of these molecules or due to the possible involvement of coronin in living cells [28].
EXPERIMENTAL PROCEDURES
Preparation of proteins
Bacterially expressed recombinant UNC-60B (nematode cofilin) and glutathione S-transferase (GST)-tagged UNC-78 (a Caenorhabditis elegans AIP1) were purified as described [35, 65]. Although a thrombin recognition site was present between GST and UNC-78, thrombin did not cleave the site appreciably in our conditions, and a dimer of GST-tagged UNC-78 was presumably formed because GST fusion proteins are generally dimers. GST-tagged UNC-78 was labeled with Alexa-488. Approximately 90% of the AIP1 molecules were labeled with one to five Alexa-488 molecules which enables high-speed and high-S/N imaging (Fig. 1S).
Recombinant monomeric UNC-78 for fluorescence labeling was also prepared; the original expression vector for GST-UNC-78 (pGEX-UNC-78) [35] was engineered to elongate the linker sequence to contain two tandem thrombin recognition sites followed by three cysteine residues for fluorophore labeling; monomeric UNC-78 was labeled with 1.7 Alexa-488 molecules in mean. Rabbit skeletal muscle actin was prepared essentially as described previously [29].
The GST-tagged 3xCys-UNC-78 was bacterially expressed, purified by glutathione-affinity chromatography in the same manner as described for original GST-UNC-78, and dialyzed against phosphate-buffered saline containing 0.2 mM dithiothreitol. The protein was digested by 10 units of bovine thrombin for 8 hours at room temperature. Cleavage of the GST portion was confirmed by SDS-PAGE, and the digested protein was applied to Glutathione-Uniflow column equilibrated with PBS to adsorb free GST and uncleaved GST-fusion protein. The flow-through fraction was dialyzed against buffer containing 0.1 M KCl, 2 mM MgCl2, 20 mM HEPES-KOH, pH 7.5, 0.2 mM DTT, 0.2 mM phenylmethanesulfonylfluoride (PMSF), and 50 % glycerol and stored at −20 °C, and labeled with Alexa-488 before use. Protein concentrations were determined by a BCA Protein Assay Kit (Pierce Biotechnology) using bovine serum albumin as a standard.
Preparation of GST2-AIP12 and rhodamine labeled-actin filaments and AIP1 application
G-actin (1 μM, in the ratio 79% non-labeled: 20% rhodamine labeled actin: 0.1–1% biotin labeled actin) was polymerized in F-buffer (100 mM KCl, 2 mM MgCl2, 0.2 mM ATP, 0.1 mM DTT, 10 mM HEPES, pH 7.5) overnight at 4°C. F-buffer (pH 7.0) contained 100 nM F-actin, 10 mM ascorbic acid. Long actin filaments are attached to the coverslip surface using the anti-biotin antibody cross-linked on the coverslip surface; the average distance between tethering points was in the 1 μm range. 17 μm square area was examined by TIRF microscopy (Fig. 1S).
UNC-60B (nematode cofilin, 2 μM) and the GST2-AIP12 (10 nM) in F-buffer was applied to actin filaments by gentle ejection through a fine glass capillary (inside diameter 5 μm, Narishige, Japan). AIP1-Alexa and these concentrations were chosen because cofilin almost fully decorates the fluctuating regions of actin filaments, and imaging a fluorescently labeled single GST2-AIP12 bound to actin filaments requires extremely low background fluorescence noise [5]. Ejection of AIP1 and cofilin containing solution (100 nM AIP1 and 20 μM cofilin) was made by a small ejector (Narishige, Japan); nearly no-pressure was applied and the solution diffused to the imaging small chamber. The actin filaments were exposed to cofilin (ca. 2 μM resulting from dilution) for ten seconds, which allows cofilin to decorate the filament, the AIP1 binding to the cofilin decorated filament was imaged and analyzed at nearly equilibrium condition in this study (see more in the following section for imaging).
In some experiments, AIP1 and cofilin containing solution was dropped on the avidin conjugated coverslip (24 × 60 mm) and covered with another coverslip (22 × 22 mm), which served to flatten the solution and reduce the background fluorescence noise. The on-rate of AIP1 binding (side and end of filaments) was estimated in this condition (10 nM AIP1 and 1 μM cofilin). Fluorescence images were recorded for 1–10 s at a rate of 10, 30 or 100 ms per frame. The duration of the typical bindings (<1 s) was shorter than the time for the bleaching of Alexa-488 dye in our recording condition, ensuring the reliable estimation of the period of bindings.
Preparation of anti-biotin-conjugated coverslips
Coverslips were conjugated with anti-biotin antibody to tether the actin and biotin-actin copolymer. Coverslips (24 × 60 mm, Matsunami, Japan) were treated with 2N NaOH for more than a week, washed using milli-Q water, and stored after air-drying. Plasma oxidation was performed on the coverslips, and were treated with 1% (v/v) 3-(triethoxysilyl)propyl isocyanate (Wako, Japan) in 10 mM CH3COOH, 50% ethanol (v/v) for 30 sec, and washed using milli-Q water. Coverslips were dried at 60 °C for 10 min and treated with 10 μg/ml of anti-biotin sheep-IgG (BETHYL LABORATORIES INC. TX, USA Lot: A150–110A-1) in PBS (pH 7.0) for 20 min. Coverslips were blocked by 10 mg/ml bovine serum albumin (BSA)-fraction V (SIGMA-ALDRICH) for 10 min and washed by PBS and replaced by F-buffer containing 10 mM ascorbic acid. The average length of the fluctuating filament segments is 1–2 μm. The length reduced as the density of biotin-conjugated-actin in the actin filaments was increased.
Labeling of UNC-78 (nematode AIP1)
AIP1-GST fusion protein (100 μM) was treated with 1 mM Alexa-488 C5 maleimide (Life Technologies, USA) in 50 mM HEPES-KOH buffer (pH 7.5) for 2 hr on ice. Reaction was attenuated by adding 100 mM of DTT at the final concentration, and the specimen was dialyzed against F-buffer. The enhancement of severing of cofilin decorated actin filaments by nematode Alexa-AIP1 is in the substantially the same range as that by nematode non-labeled AIP1 [27].
The number of Alexa-488 in a presumably dimerized AIP1-GST fusion protein was estimated by a separate experiment (Fig. 3S).
Imaging of GST2-AIP12 and rhodamine labeled-actin filaments
The fluorescence of GST2-AIP12 was observed using total internal reflection microscopy made by modifying an epifluorescence microscope (TE2000-U, Nikon, Japan) equipped with a high-numerical-aperture lens (Plan Apo TIRF, 60x, NA = 1.45, Nikon, Japan) and an ORCA Flash D4 CMOS camera (Hamamatsu Photonics, Japan). The TIRF illumination light was passed through a quarter wave plate to avoid linear polarized laser illumination. Experiments were performed at room temperature (23–25°C) (Fig. S1). The fluorescent level of each actin filament was almost the same among the filaments, and single step severing was observed with AIP1 bindings. These suggest single actin filaments were imaged in this study.
Fluorescence images were recorded for 1–10 s at a rate of 10, 30 or 100 ms per frame. The time constant of the photo-bleaching of GST2-AIP12 (t2) was 5 ± 0.7 s (n = 3; intensity of illumination, 2.5% or 1.25 mW) under our recording conditions, which is long enough to check the number of AIP1 bindings as shown in Fig. 3.
It should be noticed that the intensity of the laser illumination in Fig. S3 is ten times higher (12.5 mW) than the ordinal experiment, which enhanced the photo bleaching of the fluorescence dye as well as increased the photon emission from the dye molecule. These made it easier to estimate the number of fluorophore in an Alexa-488-tagged GST-AIP1.
The dilution of the pipette solution was fitted in the following way. First, we imaged 10 nM fluorescence AIP1 and actin filaments tethered to the glass surface. Second, AIP1 and cofilin was gently ejected from the pipette which contained 100 nM AIP1 and 20 µM cofilin to the actin filaments. We adjusted the distance between the ejection pipette and the imaging region until the density of AIP1 was nearly the same as that of 10 nM AIP1; the concentration of AIP1 was ca.10 nM in most of the cases, but it rarely increased to 15 nM at most, since the number of AIP1 fluorescence spots in the time-lapse images was stochastic. We used this condition in the majority of the experiments. When the AIP1 concentration was increased to 20 nM by displacing the pipette, we observed similar severing events by AIP1, but we did not attempt to analyze them since a very large number of observations will be required to resolve the difference in kinetic parameters between experiments at 10 nM and 20 nM AIP1. When the AIP1 concentration was increased to 100 nM, we observed enhanced severing, but the background fluorescence became higher and disturbed the precise measurements. On the other hand, the chance to find AIP1 binding was very low in 1 nM AIP1 for quantitative analysis; actin filaments were often photo bleached before accumulating the number of measurements. We have not attempted to analyze kinetics under these (1 nM and 100 nM) conditions.
The on-rate of cofilin binding is 0.14 s−1µM−1, and the dissociation constant of cofilin ca. 4 µM. The expected concentration of cofilin at the glass surface was 2 µM in this experiment; the extent of filament decoration at equilibrium is roughly estimated 10–20% accordingly.
The on-rate of cofilin will be 0.28 s−1 for 2 µM cofilin, which means one binding site of cofilin will be occupied 0.28 times within one second, in other words, the binding site will be mostly occupied within 10 s. This suggests the cofilin binding reached nearly equilibrium when measurement started several seconds after the gentle ejection of the pipette solution.
The on-rate was estimated by counting the number of the transient fluorescence increase events along filaments for 1–10 s. The number of events was divided by the number of AIP1 binding sites (366 sites per μm) and by the duration of the measurement to evaluate the on-rate. One to five AIP1 binding events in a 2–5 μm filament in 15 s of time lapse imaging were analyzed. The number of AIP1 molecules binding to each actin filament ranged from several to approximately 10. In typical cases 10–50 filaments were in the image field, and 10–100 fields of time lapse images were analyzed. Analysis of a sequence of time-lapse images of one image field gives one severing rate, and three different sequence of time-lapse images were analyzed to estimate the rate of severing. The image field contained filaments 50–250 μm in total length.
The rate of the association of AIP1 with the glass surface without actin filaments was approximately 100 times lower than the rate of binding to the actin filaments, suggesting that non-specific binding of AIP1 to the glass surface was negligible. No apparent binding of Alexa-488-BSA (1 μg/ml) or Alexa-488-GST (1 μg/ml) to the actin filaments was detected. The duration of non-specific binding was <5 ms in our set up, implying that individual non-specific AIP1 binding events were not detected using our current imaging procedure (30 ms per frame). Background punctate fluorescence noise (ca. 0.1 μm−1s−1) was subtracted from data in the AIP1 on-rate estimation.
The amplitude of the filament fluctuations was ~30 nm in z-axis, while the length of the filament was several μm. Therefore, it is possible to estimate the bending angle in the x-y plane as illustrated in figure 5C.
Fragmentation examined in this study was not a result of photodamage, since filaments could be photographed multiple times without any fragmentation in the absence of AIP1. Photon damage was greatly reduced by 10 mM ascorbic acid in the bath.
The mean duration of AIP1 binding was ca. 700 ms. These results denote that nearly all AIP1 bindings associated with severing were detected in this study. It is, thus, not likely that two (or more) AIP1 binds to the different sites of the filament and a segment of the filament between the AIP1 binding sites was moved away from the original filament (e.g., Fig. 5B and 6E), since we did not detect multi AIP1 bindings at both ends of the severed segment.
Analyses of AIP1 binding to an actin filament
The AIP1 binding number and the on-rate of AIP1 binding to actin filaments were evaluated in every pixel (85 nm) of the CMOS image sensor along the actin filament. Fluctuations in the fluorescence intensity of actin filaments were also examined along the same filament. The magnitude of the fluctuations was evaluated in the same way [28]. The fluorescence intensity of each pixel during the 1–10 s of recording was averaged for each pixel along the filament. The fluorescence intensity of each pixel of the filament was then divided by the average to normalize the fluorescence intensity. The standard deviation of the fluorescence intensity for each pixel was calculated, multiplied by 200, and named “index of fluctuation” (Fig. 4).
Each experiment was independently performed more than three times, and similar results were obtained. Data from transient fluorescence increases were accumulated for typically 100–300 events, and statistical analyses were performed using Origin software (OriginLab Corp., Northampton, MA). Statistical significance was determined using Student’s t-test; differences resulting in P values <0.05 were considered statistically significant.
Binding of AIP1 to actin filaments was observed as shown in Fig. S2. Binding and dissociation of GST2-AIP1 2 (0.3– 3s) to and from an actin filament were observed without large fluorescence fluctuations of the actin filament, indicating that the duration of the binding was measured reliably (as shown in Fig. 1). Temporal fluctuations of actin filaments were much slower than the off-rate of the binding in most of the observations, which also denotes that the duration was measured reliably in most cases.
The number of Alexa-488 in single dimeric AIP1 molecules (GST2-AIP12) estimated by single molecule fluorescence changes
Dimerization of GST has been reported, and at least four cysteine residues are found in a single GST [66–68]. There might be a possibility that dimerization occludes or exposes binding surfaces for cofilin or actin, but significant differences in the binding and dissociation to/from actin filaments between dimer AIP1 and monomer AIP1 were not detected, suggesting above possibility is not likely. The number of Alexa-488 in single AIP1 molecules was estimated by the following procedure.
Imaging of fluoresce signal from single GST 2-AIP12 molecules with strong illumination revealed stepwise decreases in the intensity of fluorescence as shown in Fig. 3S. The number of fluorophores in a single GST2-AIP12 was estimated by counting the number of stepwise decreases to the background level during bleaching processes. The number of fluorophores in the single GST2-AIP12 was estimated using the binomial functions as shown in Fig. S3. The chance to detect GST2-AIP12 with 2 fluorophores is highest (40 %) and the chance decreases in other cases, which agrees the bionomical function with five binding sites. The photon count that corresponds to the single fluorophore labeling was ca. 5 in our imaging system. The count will decline 2 or 1 in our experiments when AIP1 bound to actin filaments because the filament fluctuate above the coverslips and actin filaments were tethered to the glass surface via the large (ca. 10 nm) anti-biotin antibodies, or AIP1 was imaged in the less illuminated region of the TIRF imaging (e.g., off-center of the image field).
These results suggest that two GST-AIP1 molecules undergo dimerization, and each of two GST is labeled by zero to five Alexa-488 molecules; in other words, a single GST has 4 cysteine residues, and 1.2 out of 4 cysteine residues (2.4 Alexa molecules per dimer) are labeled with Alexa-488 in mean. This matches that approximately 2 cysteine residues are located in a region for reduced-glutathione binding [61].
The distribution of the number of Alexa-488 bindings with the binomial function (binding sites=5 and p=0.3) suggests the percentage of non-labeled AIP1 is less than 10 % of total AIP1. This agrees our observation that we almost always found AIP1 fluorescence signals in the vicinity of the severing. Therefore we did not take into account the non-labeled AIP1 in our analysis.
We detected the 3–4 alexa-488 labeled AIP1 in this study for high speed imaging (10–30 ms per frame) as a bright fluorescence spot, and used them for kinetic analysis. This bight nature is crucial for high speed and high signal to noise ratio (S/N) imaging of single molecules. We also examined 1–2 Alexa-488 labeled AIP1. By averaging 3 or more series of time lapse images we detected the location of the binding, but these are not included in the kinetic analysis in this study. The number of measurements denotes the number of AIP1 binding events, and SD follows after ±, otherwise mentioned in the text. Statistical tests were made with Student`s t-test and ANOVA (one-way, Origin Lab). Images are rotated to the angle where the left side daughter filament fluctuates after severing, while the other stays in the field of view, which facilitate our analysis.
Kinetic analysis
The kinetics scheme of fluorescence labeled Agonist (A) and Receptor (R) binding can be described by the following equation (“On” and “Off” denote the presence and absence of the fluorescence signal, respectively) [37, 38],
| [1] |
The distribution of on-time duration should be an exponential function with a time constant 1/kb according to the Poisson statistics [31]; off-rate (kb), and the apparent “on-rate” (kf) are shown.
The duration distribution of agonist binding with a state transition of receptor (R*) generally follows the following equation based on the Michaelis-Menten type kinetics:
| [2] |
where k1 and k2 denote first-order rates, and k3 denotes the rate of dissociation; i.e., the level of fluorescence emission is designated as “On” and “Off” in the same manner. The previous study of single enzymology assumes “dissociation (k2)” equals zero (or very small) to simplify the analysis; the probability distribution of “on-times” (Pon) is expressed by the following equation [37, 38]:
| [3] |
This equation is used to fit the data in Fig. 6D. Under a certain condition (e.g., k1 >100 s−1, k2 =0, and k3 <10 s−1) and observation was made under the limited time resolution (e.g., 30 ms), the probability distribution of “on-times” (Pon), i.e., the duration of AIP1 binding associated with severing (Ts, Fig. 1), is apparently expressed by a single exponential equation as shown in Fig. 5. In the above condition the state of the receptor R changes from left to right, and k3 is rate-limiting. The ensemble of data acquired at 30 ms interval is not good enough to distinguish single exponential or multi-exponential distributions, but is apparently expressed by a single exponential equation. We use kinetics scheme of Agonist (A) and Receptor (R) binding instead of Enzyme (E) binding to Substrate (S) as mentioned above, because the enzymatic reactions often involve cleavage of covalent bonds, and on the other hand the severing of actin filaments by cofilin would not involve cleavage of covalent bonds.
Supplementary Material
Acknowledgements
* This work was supported by Grant-in-Aid 15K07025 from the Ministry of Education, Culture, Sports, Science, and Technology Japan, JP18gm5810021h0003 from Advanced Research & Development Programs for Medical Innovation, and Nakatani Foundation (to H. T.), and National Institutes of Health Grant R01AR048615 (to S. O.), and JSPS KAKENHI Grant Number 24657130 (to K. H.), and the grant for collaborative research between Nagoya University and R-Pharm (2614Dj-02b to MS) and JSPS KAKENHI Grant Number JP15H05936 (to MS). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Dr. H. Ogawa and Dr. B. S. Miskowiec (INTERNATIONAL COLLEGE OF TECHNOLOGY, KANAZAWA) for their critical reading the early version of this manuscript.
Footnotes
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- [1].Pollard TD, Cooper JA. Actin, a Central Player in Cell Shape and Movement. Science 2009;326:1208–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brieher W Mechanisms of actin disassembly. Molecular biology of the cell 2013;24:2299–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bugyi B, Carlier MF. Control of actin filament treadmilling in cell motility. Annu Rev Biophys 2010;39:449–70. [DOI] [PubMed] [Google Scholar]
- [4].Ono S Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. IntRevCytol 2007;258:1–82. [DOI] [PubMed] [Google Scholar]
- [5].Hayakawa K, Sakakibara S, Sokabe M, Tatsumi H. Single-molecule imaging and kinetic analysis of cooperative cofilin-actin filament interactions. Proceedings of the National Academy of Sciences of the United States of America 2014;111:9810–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Suarez C, Roland J, Boujemaa-Paterski R, Kang H, McCullough BR, Reymann AC, et al. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Current biology : CB 2011;21:862–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].De La Cruz EM, Martiel JL, Blanchoin L. Mechanical heterogeneity favors fragmentation of strained actin filaments. Biophysical journal 2015;108:2270–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. MolCell 2006;24:13–23. [DOI] [PubMed] [Google Scholar]
- [9].Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin filament severing by cofilin. JMolBiol 2007;365:1350–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Michelot A, Berro J, Guerin C, Boujemaa-Paterski R, Staiger CJ, Martiel JL, et al. Actin-filament stochastic dynamics mediated by ADF/cofilin. CurrBiol 2007;17:825–33. [DOI] [PubMed] [Google Scholar]
- [11].Chan C, Beltzner CC, Pollard TD. Cofilin dissociates Arp2/3 complex and branches from actin filaments. CurrBiol 2009;19:537–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ono S Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1: new blades for twisted filaments. Biochemistry 2003;42:13363–70. [DOI] [PubMed] [Google Scholar]
- [13].Ono S Functions of actin-interacting protein 1 (AIP1)/WD repeat protein 1 (WDR1) in actin filament dynamics and cytoskeletal regulation. Biochem Biophys Res Commun 2017. [DOI] [PMC free article] [PubMed]
- [14].Okada K, Obinata T, Abe H. XAIP1: a Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF/cofilin family proteins. Journal of cell science 1999;112 (Pt 10):1553–65. [DOI] [PubMed] [Google Scholar]
- [15].Rodal AA, Tetreault JW, Lappalainen P, Drubin DG, Amberg DC. Aip1p interacts with cofilin to disassemble actin filaments. The Journal of cell biology 1999;145:1251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kile BT, Panopoulos AD, Stirzaker RA, Hacking DF, Tahtamouni LH, Willson TA, et al. Mutations in the cofilin partner Aip1/Wdr1 cause autoinflammatory disease and macrothrombocytopenia. Blood 2007;110:2371–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ono S, Nomura K, Hitosugi S, Tu DK, Lee JA, Baillie DL, et al. The two actin-interacting protein 1 genes have overlapping and essential function for embryonic development in Caenorhabditis elegans. Mol Biol Cell 2011;22:2258–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ren N, Charlton J, Adler PN. The flare gene, which encodes the AIP1 protein of Drosophila, functions to regulate F-actin disassembly in pupal epidermal cells. Genetics 2007;176:2223–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ketelaar T, Allwood EG, Anthony R, Voigt B, Menzel D, Hussey PJ. The actin-interacting protein AIP1 is essential for actin organization and plant development. Current biology : CB 2004;14:145–9. [DOI] [PubMed] [Google Scholar]
- [20].Chen Q, Courtemanche N, Pollard TD. Aip1 promotes actin filament severing by cofilin and regulates constriction of the cytokinetic contractile ring. Journal of Biological Chemistry 2015;290:2289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kato A, Kurita S, Hayashi A, Kaji N, Ohashi K, Mizuno K. Critical roles of actin-interacting protein 1 in cytokinesis and chemotactic migration of mammalian cells. Biochem J 2008;414:261–70. [DOI] [PubMed] [Google Scholar]
- [22].Konzok A, Weber I, Simmeth E, Hacker U, Maniak M, Muller-Taubenberger A. DAip1, a Dictyostelium homologue of the yeast actin-interacting protein 1, is involved in endocytosis, cytokinesis, and motility. J Cell Biol 1999;146:453–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chu D, Pan H, Wan P, Wu J, Luo J, Zhu H, et al. AIP1 acts with cofilin to control actin dynamics during epithelial morphogenesis. Development 2012;139:3561–71. [DOI] [PubMed] [Google Scholar]
- [24].Lechuga S, Baranwal S, Ivanov AI. Actin-interacting protein 1 controls assembly and permeability of intestinal epithelial apical junctions. American journal of physiology Gastrointestinal and liver physiology 2015;308:G745–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Luxenburg C, Heller E, Pasolli HA, Chai S, Nikolova M, Stokes N, et al. Wdr1-mediated cell shape dynamics and cortical tension are essential for epidermal planar cell polarity. Nature cell biology 2015;17:592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ono S, Mohri K, Ono K. Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/Cofilin-bound actin filaments. The Journal of biological chemistry 2004;279:14207–12. [DOI] [PubMed] [Google Scholar]
- [27].Nomura K, Hayakawa K, Tatsumi H, Ono S. Actin-interacting Protein 1 Promotes Disassembly of Actin-depolymerizing Factor/Cofilin-bound Actin Filaments in a pH-dependent Manner. The Journal of biological chemistry 2016;291:5146–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jansen S, Collins A, Chin SM, Ydenberg CA, Gelles J, Goode BL. Single-molecule imaging of a three-component ordered actin disassembly mechanism. Nature communications 2015;6:7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hayakawa K, Tatsumi H, Sokabe M. Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. The Journal of cell biology 2011;195:721–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Okada K, Blanchoin L, Abe H, Chen H, Pollard TD, Bamburg JR. Xenopus actin-interacting protein 1 (XAip1) enhances cofilin fragmentation of filaments by capping filament ends. The Journal of biological chemistry 2002;277:43011–6. [DOI] [PubMed] [Google Scholar]
- [31].Balcer HI, Goodman AL, Rodal AA, Smith E, Kugler J, Heuser JE, et al. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Current biology : CB 2003;13:2159–69. [DOI] [PubMed] [Google Scholar]
- [32].Mohri K, Ono S. Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. Journal of cell science 2003;116:4107–18. [DOI] [PubMed] [Google Scholar]
- [33].Lu HP, Xun L, Xie XS. Single-molecule enzymatic dynamics. Science 1998;282:1877–82. [DOI] [PubMed] [Google Scholar]
- [34].Sakmann B, and Neher E. Single-channel recording: Plenum Press; 1995. [Google Scholar]
- [35].Mohri K, Vorobiev S, Fedorov AA, Almo SC, Ono S. Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. The Journal of biological chemistry 2004;279:31697–707. [DOI] [PubMed] [Google Scholar]
- [36].McCullough BR, Grintsevich EE, Chen CK, Kang H, Hutchison AL, Henn A, et al. Cofilin-linked changes in actin filament flexibility promote severing. Biophysical journal 2011;101:151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Xie XS. Single-molecule approach to dispersed kinetics and dynamic disorder: Probing conformational fluctuation and enzymatic dynamics. Journal of Chemical Physics 2002;117:11024–32. [Google Scholar]
- [38].Xie SN. Single-molecule approach to enzymology. Single Molecules 2001;2:229–36. [Google Scholar]
- [39].Schafer DA, Jennings PB, Cooper JA. Dynamics of capping protein and actin assembly in vitro: Uncapping barbed ends by polyphosphoinositides. Journal of Cell Biology 1996;135:169–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chuan P, Spudich JA, Dunn AR. Robust mechanosensing and tension generation by myosin VI. Journal of molecular biology 2011;405:105–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Schafer DA, Jennings PB, Cooper JA. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J Cell Biol 1996;135:169–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bavishi K, Hatzakis NS. Shedding light on protein folding, structural and functional dynamics by single molecule studies. Molecules 2014;19:19407–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rotman B Measurement of activity of single molecules of beta-D-galactosidase. Proceedings of the National Academy of Sciences of the United States of America 1961;47:1981–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tsuji T, Miyoshi T, Higashida C, Narumiya S, Watanabe N. An order of magnitude faster AIP1-associated actin disruption than nucleation by the Arp2/3 complex in lamellipodia. PloS one 2009;4:e4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Blanchoin L, Didry D, Carlier MF, Pantaloni D. Kinetics of association of myosin subfragment-1 to unlabeled and pyrenyl-labeled actin. J Biol Chem 1996;271:12380–6. [DOI] [PubMed] [Google Scholar]
- [46].Allen PG, Janmey PA. Gelsolin displaces phalloidin from actin filaments. A new fluorescence method shows that both Ca2+ and Mg2+ affect the rate at which gelsolin severs F-actin. J Biol Chem 1994;269:32916–23. [PubMed] [Google Scholar]
- [47].Goldmann WH, Isenberg G. Analysis of filamin and alpha-actinin binding to actin by the stopped flow method. FEBS Lett 1993;336:408–10. [DOI] [PubMed] [Google Scholar]
- [48].Kuhlman PA, Ellis J, Critchley DR, Bagshaw CR. The kinetics of the interaction between the actin-binding domain of alpha-actinin and F-actin. FEBS Lett 1994;339:297–301. [DOI] [PubMed] [Google Scholar]
- [49].Gressin L, Guillotin A, Guerin C, Blanchoin L, Michelot A. Architecture dependence of actin filament network disassembly. Curr Biol 2015;25:1437–47. [DOI] [PubMed] [Google Scholar]
- [50].Nadkarni AV, Brieher WM. Aip1 destabilizes cofilin-saturated actin filaments by severing and accelerating monomer dissociation from ends. Current biology : CB 2014;24:2749–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chen Q, Courtemanche N, Pollard TD. Aip1 promotes actin filament severing by cofilin and regulates constriction of the cytokinetic contractile ring. The Journal of biological chemistry 2015;290:2289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kueh HY, Charras GT, Mitchison TJ, Brieher WM. Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. The Journal of cell biology 2008;182:341–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].De La Cruz EM. How cofilin severs an actin filament. BiophysRev 2009;1:51–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].De La Cruz EM. Cofilin binding to muscle and non-muscle actin filaments: isoform-dependent cooperative interactions. JMolBiol 2005;346:557–64. [DOI] [PubMed] [Google Scholar]
- [55].McCullough BR, Blanchoin L, Martiel JL, De La Cruz EM. Cofilin increases the bending flexibility of actin filaments: implications for severing and cell mechanics. JMolBiol 2008;381:550–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Prochniewicz E, Janson N, Thomas DD, De La Cruz EM. Cofilin increases the torsional flexibility and dynamics of actin filaments. JMolBiol 2005;353:990–1000. [DOI] [PubMed] [Google Scholar]
- [57].Tsuda Y, Yasutake H, Ishijima A, Yanagida T. Torsional rigidity of single actin filaments and actin-actin bond breaking force under torsion measured directly by in vitro micromanipulation. Proc Natl Acad Sci U S A 1996;93:12937–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gumpp H, Puchner EM, Zimmermann JL, Gerland U, Gaub HE, Blank K. Triggering enzymatic activity with force. Nano Lett 2009;9:3290–5. [DOI] [PubMed] [Google Scholar]
- [59].Bearer EL. Direct observation of actin filament severing by gelsolin and binding by gCap39 and CapZ. The Journal of cell biology 1991;115:1629–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Arai Y, Yasuda R, Akashi K, Harada Y, Miyata H, Kinosita K Jr., et al. Tying a molecular knot with optical tweezers. Nature 1999;399:446–8. [DOI] [PubMed] [Google Scholar]
- [61].Aggeli D, Kish-Trier E, Lin MC, Haarer B, Cingolani G, Cooper JA, et al. Coordination of the filament stabilizing versus destabilizing activities of cofilin through its secondary binding site on actin. Cytoskeleton (Hoboken) 2014;71:361–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Andrianantoandro E, Blanchoin L, Sept D, McCammon JA, Pollard TD. Kinetic mechanism of end-to-end annealing of actin filaments. J Mol Biol 2001;312:721–30. [DOI] [PubMed] [Google Scholar]
- [63].Elam WA, Kang H, De La Cruz EM. Competitive displacement of cofilin can promote actin filament severing. Biochemical and biophysical research communications 2013;438:728–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Miyoshi T, Tsuji T, Higashida C, Hertzog M, Fujita A, Narumiya S, et al. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J Cell Biol 2006;175:947–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ono S, Benian GM. Two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins, encoded by the unc-60 gene, differentially regulate actin filament dynamics. The Journal of biological chemistry 1998;273:3778–83. [DOI] [PubMed] [Google Scholar]
- [66].McTigue MA, Williams DR, Tainer JA. Crystal structures of a schistosomal drug and vaccine target: glutathione S-transferase from Schistosoma japonica and its complex with the leading antischistosomal drug praziquantel. J Mol Biol 1995;246:21–7. [DOI] [PubMed] [Google Scholar]
- [67].Suguoka Y, Kano T, Okuda A, Sakai M, Kitagawa T, Muramatsu M. Cloning and the nucleotide sequence of rat glutathione S-transferase P cDNA. Nucleic Acids Res 1985;13:6049–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Shen H, Tsuchida S, Tamai K, Sato K. Identification of cysteine residues involved in disulfide formation in the inactivation of glutathione transferase P-form by hydrogen peroxide. Arch Biochem Biophys 1993;300:137–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


