Abstract
Petunia (Petunia hybrida) is an important ornamental plant with a wide range of corolla colors. Although pale-yellow-flowered cultivars, with a low amount of carotenoids in their corollas, are now available, no deep-yellow-flowered cultivars exist. To find why petunia cannot accumulate enough carotenoids to have deep-yellow flowers, we compared carotenoid profiles and expression of carotenoid metabolic genes between pale-yellow-flowered petunia and deep-yellow-flowered calibrachoa (Calibrachoa hybrida), a close relative. The carotenoid contents and the ratios of esterified xanthophylls to total xanthophylls in petunia corollas were significantly lower than those in calibrachoa, despite similar carotenoid components. A lower esterification rate of trans-xanthophylls than of cis-xanthophylls in petunia suggests that petunia xanthophyll esterase (XES) has low substrate specificity for trans-xanthophylls, which are more abundant than cis-xanthophylls in petunia corolla. The expression of genes encoding key enzymes of carotenoid biosynthesis was lower and that of a carotenoid catabolic gene was higher in petunia. XES expression was significantly lower in petunia. The results suggest that low biosynthetic activity, high cleavage activity, and low esterification activity cause low carotenoid accumulation in petunia corollas.
Keywords: Calibrachoa hybrida, carotenoid, flower color, gene expression, Petunia hybrida, xanthophyll esterase
Introduction
Carotenoids are important pigments responsible for petal colors ranging from yellow to red. The main role of carotenoids in flowers is the attraction of insect and bird pollinators. In the green tissues of higher plants, carotenoids have important functions in photosynthesis and protection against photooxidative damage (Robert et al. 2004, Ruban et al. 2007). There is a wide variation in carotenoid composition in petals among plant species; most carotenoids are xanthophylls (Kishimoto et al. 2007, Ohmiya 2011). Carotenoids in petals are biosynthesized through multiple enzymatic steps and accumulate in chromoplasts (Cazzonelli and Pogson 2010, Hirschberg 2001). The condensation of two molecules of geranylgeranyl diphosphate in the first committed and rate-limiting step produces phytoene. Through desaturation, isomerization, and cyclization steps, α- and β-carotene are produced via lycopene. These carotenoids from phytoene to α- and β-carotene are “carotenes” defined as carotenoids composed only of hydrogen and carbon atoms. The hydroxylation of α- and β-carotene produces “xanthophylls”, defined as carotenoids containing oxygen atoms, and those are followed by epoxidation. A number of studies showed that carotenoid accumulation in petals is regulated at the transcriptional level of carotenoid biosynthetic genes (reviewed by Ohmiya 2013). Overall carotenoid accumulation is regulated through degradation by carotenoid cleavage dioxygenase (CCD) family enzymes (Ohmiya 2009, Ohmiya et al. 2006).
In green leaves, all xanthophylls occur in free form, while in petals they are mostly esterified (Ariizumi et al. 2014, Breithaupt et al. 2002, Goodwin 1980, Maoka et al. 2011, Yamamizo et al. 2010). In some carotenoid-rich petals and fruits, various types of carotenoid-containing bodies, including fibrillar, globular, and tubular, are formed inside the chromoplasts (Camara et al. 1995, Ljubesić et al. 1991). Deruère et al. (1994) reported that esterified xanthophylls were more efficient for fibril assembly than free xanthophylls in vitro. They presumed that carotenoid sequestration by the formation of carotenoid-containing bodies prevents harmful effects of excess carotenoids on cellular functions. Recently, Ariizumi et al. (2014) identified a gene encoding xanthophyll esterase (XES) in the pale yellow petal 1 (pyp1) mutants of tomato. They showed that disruption of PYP1 caused complete loss of esterified xanthophylls, reduction in total carotenoid content, and abnormal chromoplast development in petals. The results suggest that esterification is important for mass accumulation of carotenoids in petals.
Petunia (Petunia hybrida) is an important ornamental plant and an excellent model for investigating floral pigmentation (Tornielli et al. 2008), which ranges wide from pink to red to purple, due mainly to anthocyanins (Muszyński 1964). Several pale-yellow-flowered petunia cultivars are now available, but no deep-yellow cultivars exist. The main pigments in the corollas of the pale-yellow cultivars are carotenoids, albeit at a lower content than in deep-yellow flowers of other species (Kishimoto et al. 2018, Murakami et al. 2003, Nielsen and Bloor 1997). We previously reported that carotenoid accumulation in pale-yellow petunia corollas was due to higher expression of several biosynthetic genes than in white cultivars and lower catabolic activity due to complete lack of CCD4a expression (Kishimoto et al. 2018).
Here, to find why petunia does not accumulate high amounts of carotenoids in its corollas, we compared carotenoid composition and metabolic gene expression between pale-yellow-flowered petunia and deep-yellow-flowered calibrachoa (Calibrachoa hybrida), a close relative (Jędrzejuk et al. 2017, Olmstead et al. 2008), whose corolla color is derived from carotenoids (Murakami et al. 2004). We show that low expression of carotenoid biosynthetic genes, high expression of a carotenoid catabolic gene, and low esterification activity are the main reasons for the low carotenoid accumulation in petunia corollas.
Materials and Methods
Plant materials
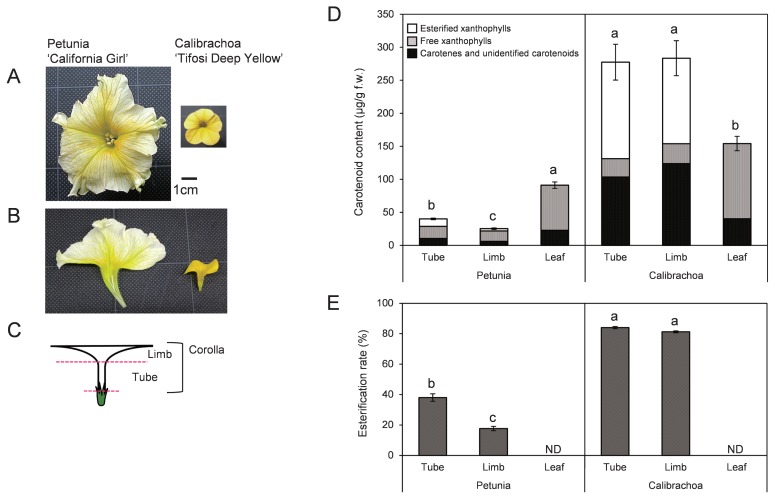
Pale-yellow-flowered petunia ‘California Girl’ and deep-yellow-flowered calibrachoa ‘Tifosi Deep Yellow’ were grown in greenhouses at the NARO Institute of Vegetable and Floriculture Science (Tsukuba, Japan). Tubes and limbs of corollas on flowering day and mature leaves were sampled (Fig. 1A–1C) and stored at −80°C until use.
Fig. 1.
(A) Flowers of petunia ‘California Girl’ and calibrachoa ‘Tifosi Deep Yellow’. (B) Vertical sections of corollas. (C) Sampling positions of tube and limb. (D) Carotenoid content in tubes, limbs, and leaves of petunia and calibrachoa. (E) Ratio of esterified xanthophylls to total xanthophylls. Analyses were performed in triplicate; means ± SE are shown. The same letter indicates no significant difference by Tukey’s test (P < 0.05).
Cloning of carotenoid metabolic genes
We isolated partial-length cDNAs of carotenoid metabolic genes and actin of calibrachoa, and full-length cDNAs encoding putative xanthophyll esterase (XES), an ortholog of tomato PYP1 (SlPYP1), of both species, as described by Kishimoto et al. (2018). Primer sequences are shown in Supplemental Tables 1 (RT-PCR) and 2 (RACE of XES).
cDNA sequences were deposited in DDBJ/EMBL/GenBank under the following accession numbers: 1-deoxy-D-xylulose 5-phosphate synthase (ChDXS), LC335780; isopentenyl diphosphate isomerase (ChIPI), LC335781; geranylgeranyl pyrophosphate synthase (ChGGPS1), LC335782; phytoene synthase 1 (ChPSY1), LC335783; ChPSY2, LC335784; phytoene desaturase (ChPDS), LC335785; 15-cis-ζ-carotene isomerase (ChZ-ISO), LC369596; ζ-carotene desaturase (ChZDS), LC335786; carotenoid isomerase (ChCRTISO), LC369595; lycopene β-ring cyclase (ChLCYB), LC335787; lycopene ɛ-ring cyclase (ChLCYE), LC335788; β-ring hydroxylase 1 (ChCHYB1), LC335789; ChCHYB2, LC335790; cytochrome P450-type β-ring hydroxylase (ChCHYB/CYP97A), LC361460; cytochrome P450-type ɛ-ring hydroxylase (ChCHYE/CYP97C), LC361461; zeaxanthin epoxygenase (ChZEP), LC335791; ChXES, LC335779; carotenoid cleavage dioxygenase 1 (ChCCD1), LC335792; carotenoid cleavage dioxygenase 4 (ChCCD4a), LC335793; 9-cis-epoxycarotenoid dioxygenase (ChNCED), LC335794; ChActin, LC335795, PhXES, LC335778. Other cDNA sequences of petunia were previously reported (Kishimoto et al. 2018). Similarity of cDNA sequences encoding each gene between petunia and calibrachoa and putative amino acid sequences among PhXES, ChXES, and SlPYP1 were analyzed by clustalW (Larkin et al. 2007). Prediction of the presence of chloroplast/chromoplast transit peptides (cTP) in the protein sequences and the location of potential cTP cleavage sites were carried out using the ChloroP 1.1 server (Emanuelsson et al. 1999).
Quantitative real-time PCR analysis
Total RNA was isolated with Trizol reagent in a PureLink RNA mini column (both Thermo Fisher Scientific). Reverse-transcription quantitative real-time PCR (RT-qPCR) was performed as described by Kishimoto et al. (2018). Primers for carotenoid metabolic genes were designed from sequences of partial-length cDNAs conserved between petunia and calibrachoa in Oligo software (Supplemental Table 3). RT-qPCR analyses were performed in technical triplicate. Statistically significant differences were determined by Tukey’s test at the 5% level.
HPLC analysis of carotenoids
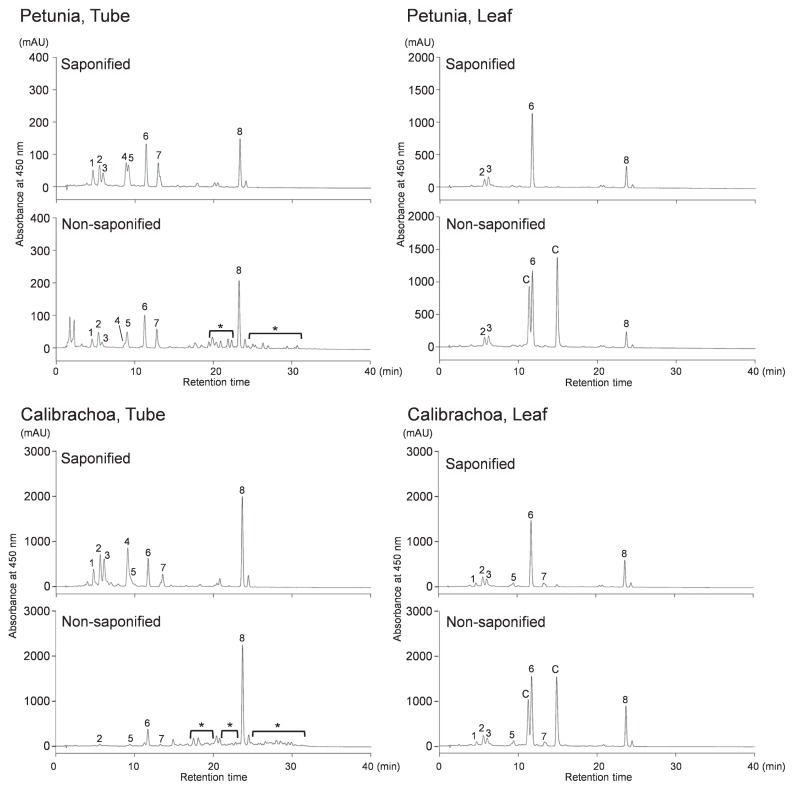
An acetone extract of frozen sample (0.1 g) was partitioned between diethyl ether and aqueous NaCl. The organic layer was washed with 5 mM Tris-HCl (pH 8.0) and was divided into two portions. One portion was saponified with an equivalent volume of 5% KOH/MeOH (w/v) for 1 h at room temperature; the other was dried and dissolved in 10% methyl tert-butyl ether (MTBE)/MeOH (v/v) (non-saponified sample). The KOH-treated sample was extracted with diethyl ether and washed with water. The organic layer was dried and dissolved in 10% MTBE/MeOH (saponified sample). Samples were analyzed by HPLC as described by Liu et al. (2013). Peaks were identified by comparing the retention times and absorbance spectra with those of carotenoids previously identified in the petals of chrysanthemum (Kishimoto et al. 2004) and tomato (Ariizumi et al. 2014). The contents of individual and total carotenoids were estimated from the peak areas of the HPLC chromatograms. The contents of esterified xanthophylls in tubes and limbs were calculated from the difference in peak areas between saponified and non-saponified samples. Measurements were performed in biological triplicate. Statistically significant differences were determined by Tukey’s test at the 5% level.
Results
Comparison of carotenoid content and composition between pale-yellow-flowered petunia and deep-yellow-flowered calibrachoa
The total average carotenoid content of calibrachoa was 6.9× that of petunia in tubes, 11.0× in limbs, and 1.7× in leaves (Fig. 1D). Carotenoid components in saponified samples of limbs and tubes were similar between petunia and calibrachoa; we detected mainly (all-E)-neoxanthin, (all-E)-violaxanthin, (9′Z)-neoxanthin, (9Z)-violaxanthin, (all-E)-lutein, (all-E)-zeaxanthin, (all-E)-antheraxanthin, and (all-E)-β-carotene (Fig. 2). However, the composition differed between petunia and calibrachoa: the ratios of (9Z)-violaxanthin and (all-E)-β-carotene were higher in calibrachoa, and (all-E)-lutein and (all-E)-antheraxanthin + (all-E)-zeaxanthin were higher in petunia (Table 1).
Fig. 2.
HPLC chromatograms of carotenoid extracts from tubes and leaves of petunia and calibrachoa. Saponified and non-saponified carotenoid extracts from 0.025 g f.w. of tubes and leaves were analyzed. Peaks: 1, (all-E)-neoxanthin; 2, (all-E)-violaxanthin; 3, (9′Z)-neoxanthin; 4, (9Z)-violaxanthin; 5, unknown xanthophyll; 6, (all-E)-lutein; 7, (all-E)-antheraxanthin + (all-E)-zeaxanthin; 8, (all-E)-β-carotene; C, chlorophylls. *Esterified xanthophylls.
Table 1.
Carotenoid contents and compositions in tubes, limbs, and leaves of petunia and calibrachoa
| Species | Tissue | Carotenoid component | Total carotenoid content (μg/g f.w.) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| (all-E)-Neoxanthin | (all-E)-Violaxanthin | (9′Z)-Neoxanthin | (9Z)-Violaxanthin | Unknown xanthophyll | (all-E)-Lutein | (all-E)-Zeaxanthin + (all-E)-antheraxanthin | (all-E)-β-Carotene | Unidentified | |||
| Petunia “California Girl” | Tube (μg/g f.w.) | 3.59 ± 0.49b | 4.29 ± 0.51 | 2.95 ± 0.07 | 4.11 ± 0.21 | 2.93 ± 0.38 | 6.82 ± 0.51 | 3.36 ± 0.58 | 5.78 ± 0.20 | 6.18 ± 0.50 | 40.01 ± 1.20 cd |
| (%)a | 8.96 ± 1.24 ac | 10.73 ± 1.27 a | 7.38 ± 0.19 ab | 10.27 ± 0.53 b | 7.33 ± 0.96 a | 17.05 ± 1.26 bc | 8.41 ± 1.45 ab | 14.4 ± 0.50 c | 15.44 ± 1.24 abc | ||
|
| |||||||||||
| Limb (μg/g f.w.) | 1.23 ± 0.24 | 2.11 ± 0.51 | 1.70 ± 0.05 | 1.49 ± 0.15 | 2.37 ± 0.04 | 6.29 ± 0.27 | 3.94 ± 0.88 | 3.98 ± 0.40 | 2.54 ± 0.13 | 25.65 ± 0.90 d | |
| (%) | 4.83 ± 0.95 b | 8.22 ± 2.01 a | 6.62 ± 0.20 b | 5.81 ± 0.57 c | 9.23 ± 0.15 a | 27.54 ± 1.04 b | 15.35 ± 3.42 a | 15.5 ± 1.57 abc | 9.90 ± 0.52 c | ||
|
| |||||||||||
| Leaf (μg/g f.w.) | 1.64 ± 0.45 | 9.31 ± 2.13 | 9.26 ± 0.62 | 1.96 ± 0.31 | 0.31 ± 0.31 | 51.33 ± 1.26 | 1.23 ± 0.27 | 12.9 ± 0.15 | 12.60 ± 0.76 | 100.23 ± 5.40 bc | |
| (%) | 1.63 ± 0.45 bc | 9.29 ± 2.13 a | 9.24 ± 0.62 ab | 1.96 ± 0.31 cd | 0.31 ± 0.31 c | 51.21 ± 1.25 a | 1.23 ± 0.27 b | 12.9 ± 0.15 c | 12.57 ± 0.75 bc | ||
|
| |||||||||||
| Calibrachoa “Tifosi Deep Yellow” | Tube (μg/g f.w.) | 17.64 ± 1.29 | 31.14 ± 1.51 | 30.40 ± 4.15 | 41.78 ± 2.75 | 12.75 ± 0.30 | 23.62 ± 1.89 | 13.46 ± 1.55 | 61.18 ± 10.01 | 45.30 ± 4.64 | 277.29 ± 27.13 a |
| (%) | 6.36 ± 0.47 ab | 11.23 ± 0.55 a | 10.96 ± 1.50 a | 15.07 ± 0.99 a | 4.60 ± 0.11 b | 8.52 ± 0.68 c | 4.86 ± 0.56 b | 22.07 ± 3.61 ab | 16.34 ± 1.67 abc | ||
|
| |||||||||||
| Limb (μg/g f.w.) | 13.23 ± 1.42 | 41.87 ± 4.72 | 20.50 ± 2.53 | 29.38 ± 4.31 | 12.80 ± 1.32 | 29.89 ± 3.40 | 10.40 ± 1.22 | 67.86 ± 5.28 | 57.37 ± 4.50 | 283.31 ± 26.50 a | |
| (%) | 4.67 ± 0.50 b | 14.78 ± 1.67 a | 7.24 ± 0.89 ab | 10.37 ± 1.52 b | 4.52 ± 0.47 b | 10.55 ± 1.20 c | 3.67 ± 0.43 b | 23.95 ± 1.86 a | 20.25 ± 1.59 a | ||
|
| |||||||||||
| Leaf (μg/g f.w.) | 5.47 ± 0.11 | 13.66 ± 1.18 | 13.84 ± 1.68 | 2.78 ± 0.27 | 4.82 ± 0.32 | 79.00 ± 8.09 | 6.19 ± 0.54 | 25.08 ± 1.00 | 24.43 ± 2.74 | 170.45 ± 15.20 b | |
| (%) | 3.21 ± 0.06 b | 8.01 ± 0.69 a | 8.12 ± 0.98 ab | 1.63 ± 0.16 d | 2.83 ± 0.19 b | 46.35 ± 4.74 a | 3.63 ± 0.32 b | 14.71 ± 0.59 bc | 14.33 ± 1.60 ab | ||
Ratio of carotenoid component to total carotenoids (%).
Means ± SE (n = 3).
The same letter within a component indicates no significant difference by Tukey’s test (P < 0.05).
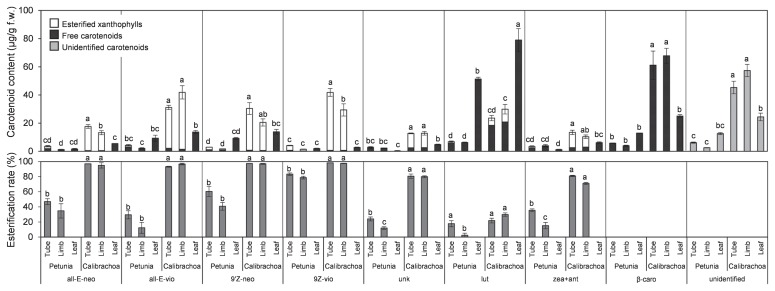
HPLC analysis detected esterified xanthophylls in the non-saponified samples at a retention time of approximately 20 to 30 min in all limbs and tubes tested but not in leaves (Fig. 2). Ratios of esterified to total xanthophylls were >80% in calibrachoa, but <40% in petunia (Fig. 1E). The ratio of esterification of each xanthophyll also differed between species (Fig. 3): in calibrachoa, those of all xanthophylls except for lutein were >70%, but in petunia, those of most xanthophylls except for (9′Z)-neoxanthin in tubes and (9Z)-violaxanthin in tubes and limbs were <50%.
Fig. 3.
Contents and ratios of esterified xanthophylls in tubes, limbs, and leaves of petunia and calibrachoa. all-E-neo, (all-E)-neoxanthin; all-E-vio, (all-E)-violaxanthin; 9′Z-neo, (9′Z)-neoxanthin; 9Z-vio, (9Z)-violaxanthin; unk, unknown xanthophyll; lut, (all-E)-lutein; zea + ant, (all-E)-zeaxanthin and (all-E)-antheraxanthin; β-caro, (all-E)-β-carotene; unidentified, unidentified carotenoids. Analyses were performed in triplicate; means ± SE are shown. The same letter within a component indicates no significant difference by Tukey’s test (P < 0.05).
Comparison of carotenoid metabolic gene expression between pale-yellow-flowered petunia and deep-yellow-flowered calibrachoa
We compared the expressions of Actin between petunia and calibrachoa (Supplemental Fig. 1). Although the expression levels were different among tissues in both petunia and calibrachoa, they showed similar levels when we compared the same tissue in both plants. Similarities in cDNA sequences encoding each gene were very high (>90%) for both the plants (Supplemental Table 4). Therefore, we compared the expressions of each gene between petunia and calibrachoa with the same primers for RT-qPCR and conducted subsequent analyses.
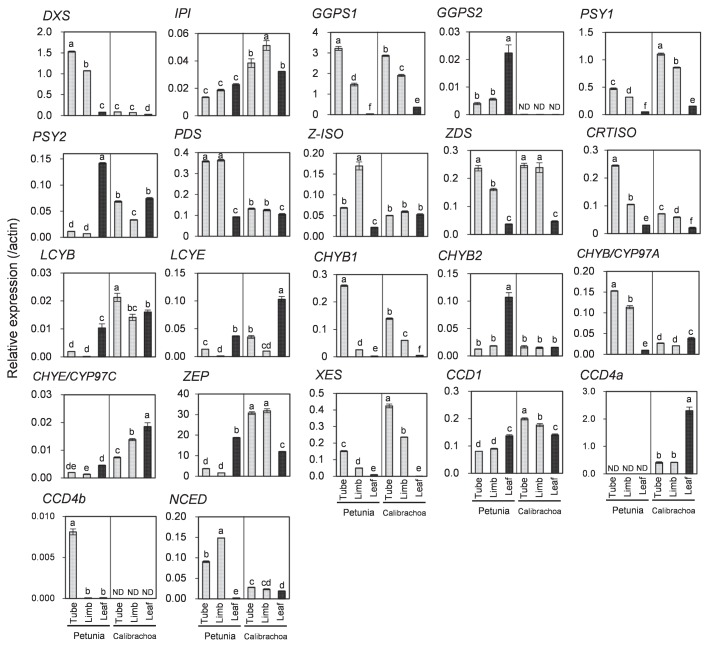
Among the 18 carotenoid biosynthetic genes tested, expression levels of DXS, PDS, CRTISO, and CHYB/CYP97A in tubes and limbs were significantly higher in petunia than in calibrachoa (Fig. 4). In contrast, those of IPI, PSY1, PSY2, LCYB, CHYE/CYP97C, ZEP, and XES in tubes and limbs were significantly lower in petunia than in calibrachoa. Especially, LCYB and ZEP expression in petunia was extremely lower than that in calibrachoa. There was no clear correlation between carotenoid content and expression levels of GGPS1, Z-ISO, ZDS, LCYE, CHYB1, and CHYB2 (Fig. 4). We analyzed GGPS2 expression in calibrachoa using primers for petunia GGPS2 because we could not isolate GGPS2 cDNA from calibrachoa, but we could not detect GGPS2 expression in calibrachoa; this result suggests either that there is no ortholog of PhGGPS2 in calibrachoa or that the RT-qPCR primers did not match the calibrachoa GGPS2.
Fig. 4.
Expression analysis of carotenoid metabolic genes in tubes, limbs, and leaves of petunia and calibrachoa. RT-qPCR analyses were performed in triplicate; means ± SE are shown. The same letter within a gene indicates no significant difference by Tukey’s test (P < 0.05).
Among the 4 carotenoid cleavage genes tested, the expression of NCED in tubes and limbs was significantly higher and that of CCD1 in tubes and limbs was significantly lower in petunia than in calibrachoa, and CCD4a in petunia and CCD4b in calibrachoa were not detected (Fig. 4). We previously reported that CCD4a was expressed in white-flowered petunia cultivars but not in pale-yellow-flowered cultivars because of genomic insertions in the CCD4a promoter and coding regions (Kishimoto et al. 2018). We could not isolate CCD4b cDNA from calibrachoa and instead used the primers for PhCCD4b. It is possible that there is no ortholog of PhCCD4b in calibrachoa or that the RT-qPCR primers did not match the calibrachoa CCD4b.
Similarity of amino acid sequences among PhXES, ChXES, and SlPYP1
We isolated full-length cDNAs encoding XES from corollas of petunia (PhXES) and calibrachoa (ChXES). The deduced amino acid sequences of PhXES and ChXES consisted of 710 amino acids (Supplemental Fig. 2), showing 86% similarity. Similarity between PhXES and SlXES was 81% and that between ChXES and SlXES was 77%. BLAST searches against protein databases indicated that the polypeptide of PhXES and ChXES include α/β hydrolase fold domain (amino acids 132–383 in PhXES) and lysophospholipid acyltransferase (LPAT)-like domain (amino acids 407–679 in PhXES). Both XES contained chloroplast transit peptides (cTPs) at the N-terminus, indicating that they might be transported to the chromoplast. The predicted lengths of the cTPs were 29 amino acids in both PhXES and ChXES.
Discussion
Although a wide range of flower colors has been developed in petunia cultivars, there are no deep-yellow-flowered cultivars. In corollas of both petunia and calibrachoa, eight kinds of xanthophylls including two kinds of 9-cis isomers and β-carotene were mainly detected. These carotenoids had similar absorption maxima (Britten 1995). Therefore, the corolla color tone of petunia and calibrachoa might be determined not by the ratio of carotenoid components but by the total accumulation level. To find out why petunia corollas cannot accumulate enough carotenoids to express deep-yellow color, we compared carotenogenic gene expressions between pale-yellow-flowered petunia and deep-yellow-flowered calibrachoa. We found several genes whose expression levels differed significantly between them (Figs. 4, 5).
Fig. 5.
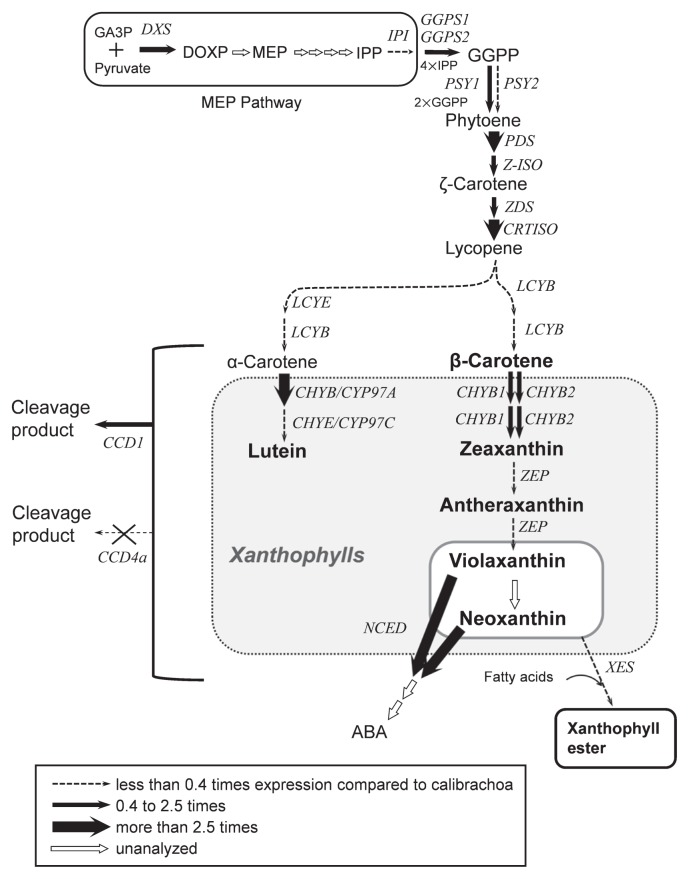
Putative carotenoid biosynthetic pathway in corollas of petunia. GA3P, glyceraldehyde 3-phosphate; DXS, 1-deoxy-D-xylulose 5-phosphate synthase; DOXP, 1-deoxy-D-xylulose 5-phosphate; MEP, 2-C-methyl-D-erythritol-2,4-cyclodisphosphate; IPP, isopentenyl diphosphate; IPI, IPP isomerase; GGPS, GGPP synthase; GGPP, geranylgeranyl diphosphate; PSY, phytoene synthase; PDS, phytoene desaturase; Z-ISO, 15-cis-ζ-carotene isomerase; ZDS, ζ-carotene desaturase; CRTISO, carotenoid isomerase; LCYB, lycopene β-ring cyclase; LCYE, lycopene ɛ-ring cyclase; CHYB, β-ring hydroxylase; CHYE, ɛ-ring hydroxylase; CHYB/CYP97A, cytochrome P450-type β-ring hydroxylase; CHYE/CYP97C, cytochrome P450-type ɛ-ring hydroxylase; ZEP, zeaxanthin epoxidase; NCED, 9-cis-epoxy carotenoid dioxygenase; ABA, abscisic acid; CCD1, carotenoid cleavage dioxygenase 1; CCD4a, carotenoid cleavage dioxygenase 4a; XES, xanthophyll esterase. Major carotenoids accumulated in corollas of petunia and calibrachoa are indicated in bold letters. Thickness of arrows indicates the level of gene expression in tubes relative to calibrachoa.
Among biosynthetic genes tested, PSY1, PSY2, LCYB, and LCYE had significantly lower expression in petunia than in calibrachoa. PSY is a key enzyme of carotenoid biosynthesis functioning upstream in the pathway (Fig. 5; Cazzonelli and Pogson 2010, Ohmiya 2013). A low carotenoid content in petals is attributed to low PSY expression in carnation (Ohmiya et al. 2013), eustoma (Liu et al. 2013), Japanese morning glory (Yamamizo et al. 2010), and marigold (Moehs et al. 2001). We reported previously that PSY1 expression in petunia differed significantly between white-flowered and pale-yellow-flowered cultivars, but PSY2 expression did not (Kishimoto et al. 2018). Here, we showed that expression of both PSY1 and PSY2 was significantly lower in petunia than in calibrachoa. Therefore, low PSY expression is likely to be one of the causes of low carotenoid content in corollas of petunia.
LCYB and LCYE catalyze formation of β- and ɛ-rings, respectively (Fig. 5; Cunningham et al. 1996, Hugueney et al. 1995). The balance between LCYB and LCYE activities determines the ratio of β,β-carotenoids (β-carotene and its derivatives) to β,ɛ-carotenoids (α-carotene and its derivatives) (Cunningham et al. 1996, Ohmiya 2013). In petals of Oncidium (Chiou et al. 2010) and tubers of potato (Diretto et al. 2006), higher expression of LCYB than of LCYE causes the higher accumulation of β,β-carotenoids. In contrast, in petals of marigold (Moehs et al. 2001) and chrysanthemum (Kishimoto and Ohmiya 2006), higher expression of LCYE than of LCYB causes the higher accumulation of β,ɛ-carotenoids. In tomato, the expression of both LCYB and LCYE is decreased during fruit development, resulting in the low accumulation of cyclic carotenoids and the high accumulation of lycopene (Ronen et al. 1999). In corollas of petunia, the expression of both LCYB and LCYE was significantly lower than in calibrachoa, but lycopene was undetectable (Figs. 2, 4). Plant species accumulating lycopene in their petals are very rare (Ohmiya 2011). It is likely that most species, including petunia, are unable to accumulate lycopene in petals. Therefore, it is possible that lack of ability to accumulate lycopene and the low level of lycopene cyclization activity in petunia cause not only low accumulation of cyclic carotenoids, but also low levels of total carotenoids.
Carotenoid degradation is another factor that affects carotenoid accumulation in flowers. We examined the expression of carotenoid catabolic genes to see whether carotenoid degradation is involved in the regulation of carotenoid accumulation in petunia corollas. The expression of NCED was significantly higher in petunia than in calibrachoa (Fig. 4). NCED cleaves (9′Z)-neoxanthin and (9Z)-violaxanthin to xanthoxin, an intermediate in the biosynthesis of abscisic acid (Fig. 5; Schwartz et al. 1997). Kato et al. (2004, 2006) found an inverse relationship between CitNCED2 expression and (9Z)-violaxanthin level in citrus fruits. In corollas of petunia, the low levels of (9′Z)-neoxanthin and (9Z)-violaxanthin might be due to the high NCED expression.
There was no inverse relationship of CCD1 or CCD4a expression with carotenoid content in calibrachoa or petunia. CCD1 cleaves carotenoids at 9,10 (9′,10′) double bonds and contributes to the emission of β- and α-ionones, important fragrance components, in flowers of petunia and Osmanthus fragrans (Baldermann et al. 2010, Simkin et al. 2004b). However, lack of correlation between CCD1 expression and carotenoid content has been demonstrated in petals of Japanese morning glory (Yamamizo et al. 2010), rice endosperm (Ilg et al. 2010), tomato fruit (Simkin et al. 2004a), and citrus fruit (Kato et al. 2006), possibly because CCD1 is located in the cytoplasm and has limited access to its substrates in chromoplasts (Bouvier et al. 2003, McCarty and Klee 2006). We previously reported that CCD1 is constitutively expressed in corollas of both white-flowered and pale-yellow-flowered cultivars (Kishimoto et al. 2018). Therefore, we consider that the expression of CCD1 did not affect the carotenoid content in corollas of petunia and calibrachoa. Ohmiya et al. (2006) revealed that CCD4 plays a key role in cleavage of carotenoids in petals of chrysanthemum. There is increasing evidence to show that CCD4 is involved in the regulation of carotenoid accumulation in chromoplasts of flowers and fruits (Falchi et al. 2013, Gonzalez-Jorge et al. 2013, Hai et al. 2012, Zhang et al. 2015). We have previously shown that an insertion in the putative promoter region and a palindromic sequence in the coding region of the CCD4a genomic sequence of pale-yellow petunia prevents CCD4a expression (Kishimoto et al. 2018). In contrast, white petunia corollas have high CCD4a expression. Therefore, CCD4a activity is predicted to be a major cause of extremely low levels of carotenoids in white-flowered petunia cultivars. Although calibrachoa had substantial CCD4a expression both in tubes and limbs, we expect that biosynthesis greatly exceeds degradation. Therefore, CCD4a is not a key determinant of carotenoid accumulation in calibrachoa corolla. CCD4b expression was detected only in petunia. However, we speculate that CCD4b is not involved in the regulation of carotenoid accumulation in corollas of petunia, because the expression pattern of CCD4b in petunia was not associated with carotenoid content (Kishimoto et al. 2018).
Esterification of xanthophylls is important for mass accumulation of carotenoids in chromoplasts. Ariizumi et al. (2014) showed that disruption of PYP1 (encoding XES) causes not only loss of esterified xanthophylls, but also a drastic decrease in total carotenoid levels in tomato petals. We showed that the ratio of esterified to total xanthophylls was significantly lower in petunia than in calibrachoa (Fig. 1E). We assume that esterification activity is lower in petunia than in calibrachoa because of lower XES expression (Fig. 4). Both cis- and trans-forms of neoxanthin and violaxanthin were almost completely esterified in calibrachoa, but were incompletely esterified in petunia; in particular, ratios of esterification of trans-forms of neoxanthin and violaxanthin were <50% (Fig. 3). These results suggest that petunia XES has higher substrate specificity for cis-xanthophylls than for trans-xanthophylls. It is possible that the substrate specificity in PhXES is due to the amino acids divergence in the conserved domain important for esterase/lipase activity (Supplemental Fig. 2). However, the contents of cis-xanthophylls, such as (9′Z)-neoxanthin and (9Z)-violaxanthin, were low in all pale-yellow-flowered petunia cultivars tested (Kishimoto et al. 2018). The expression of ZEP, which catalyzes epoxidation of zeaxanthin to produce violaxanthin (Fig. 5; Marin et al. 1996), was significantly lower in petunia corollas than in calibrachoa corollas (Fig. 4); therefore, one reason for the low level of cis-forms of neoxanthin and violaxanthin in petunia (Table 1) would be low epoxidation activity. In addition, cleavage activity of NCED to cis-forms of neoxanthin and violaxanthin is likely to be higher in petunia than in calibrachoa, as mentioned above. Recently, Ma et al. (2017) reported that citrus CCD1 and CCD4 can cleave free but not esterified β-cryptoxanthin in vitro. The higher ratio of free forms of xanthophylls in petunia leads us to speculate that carotenoids in petunia corollas are more susceptible to cleavage by CCD family enzymes.
We conclude that low carotenoid accumulation in corollas of petunia is due to low biosynthesis of violaxanthin, high cleavage activity, especially cleavage of violaxanthin and neoxanthin, and low ratios of esterified xanthophylls. We assume that low expression of XES and low substrate specificity of XES for trans-xanthophylls in petunia result in a low ratio of esterification of xanthophylls. The mechanism by which esterification of xanthophylls promotes carotenoid accumulation remains unclear; therefore, further studies will be needed to clarify how.
Supplementary Information
Acknowledgments
This work was supported by JSPS KAKENHI grant number JP25450051.
Literature Cited
- Ariizumi, T., Kishimoto, S., Kakami, R., Maoka, T., Hirakawa, H., Suzuki, Y., Ozeki, Y., Shirasawa, K., Bernillon, S., Okabe, Y.et al. (2014) Identification of the carotenoid modifying gene PALE YELLOW PETAL 1 as an essential factor in xanthophyll esterification and yellow flower pigmentation in tomato (Solanum lycopersicum). Plant J. 79: 453–465. [DOI] [PubMed] [Google Scholar]
- Baldermann, S., Kato, M., Kurosawa, M., Kurobayashi, Y., Fujita, A., Fleischmann, P. and Watanabe, N. (2010) Functional characterization of a carotenoid cleavage dioxygenase 1 and its relation to the carotenoid accumulation and volatile emission during the floral development of Osmanthus fragrans Lour. J. Exp. Bot. 61: 2967–2977. [DOI] [PubMed] [Google Scholar]
- Bouvier, F., Suire, C., Mutterer, J. and Camara, B. (2003) Oxidative remodeling of chromoplast carotenoids: identification of the carotenoid dioxygenase CsCCD and CsZCD genes involved in crocus secondary metabolite biogenesis. Plant Cell 15: 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breithaupt, D.E., Wirt, U. and Bamedi, A. (2002) Differentiation between lutein monoester regioisomers and detection of lutein diesters from marigold flowers (Tagetes erecta L.) and several fruits by liquid chromatography−mass spectrometry. J. Agric. Food Chem. 50: 66–70. [DOI] [PubMed] [Google Scholar]
- Britten, G. (1995) UV/Visible Spectroscopy. In: Britton, G., Liaaen-Jensen S. and Pfander H. (eds.) Carotenoids Volume 1B: Spectroscopy, Birkhäuser, Basel, Boston, Berlin, pp. 13–62. [Google Scholar]
- Camara, B., Hugueney, P., Bouvier, F., Kuntz, M. and Monéger, R. (1995) Biochemistry and molecular biology of chromoplast development. Int. Rev. Cytol. 163: 175–247. [DOI] [PubMed] [Google Scholar]
- Cazzonelli, C.I. and Pogson, B.J. (2010) Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant. Sci. 15: 266–274. [DOI] [PubMed] [Google Scholar]
- Chiou, C.-Y., Pan, H.-A., Chuang, Y.-N. and Yeh, K.-W. (2010) Differential expression of carotenoid-related genes determines diversified carotenoid coloration in floral tissues of Oncidium cultivars. Planta 232: 937–948. [DOI] [PubMed] [Google Scholar]
- Cunningham, F.X., Jr., Pogson, B., Sun, Z., McDonald, K.A., DellaPenna, D. and Gantt, E. (1996) Functional analysis of the β and ɛ lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 8: 1613–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deruère, J., Römer, S., d’Harlingue, A., Backhaus, R.A., Kuntz, M. and Camara, B. (1994) Fibril assembly and carotenoid overaccumulation in chromoplasts: a model for supramolecular lipoprotein structures. Plant Cell 6: 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diretto, G., Tavazza, R., Welsch, R., Pizzichini, D., Mourgues, F., Papacchioli, V., Beyer, P. and Giuliano, G. (2006) Metabolic engineering of potato tuber carotenoids through tuber-specific silencing of lycopene epsilon cyclase. BMC Plant Biol. 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H. and von Heijne, G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8: 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falchi, R., Vendramin, E., Zanon, L., Scalabrin, S., Cipriani, G., Verde, I., Vizzotto, G. and Morgante, M. (2013) Three distinct mutational mechanisms acting on a single gene underpin the origin of yellow flesh in peach. Plant J. 76: 175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Jorge, S., Ha, S.H., Magallanes-Lundback, M., Gilliland, L.U., Zhou, A., Lipka, A.E., Nguyen, Y.N., Angelovici, R., Lin, H., Cepela, J.et al. (2013) CAROTENOID CLEAVAGE DIOXYGENASE4 is a negative regulator of β-carotene content in Arabidopsis seeds. Plant Cell 25: 4812–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, T.W. (1980) Carotenoids in higher plants. In: The Biochemistry of the Carotenoids Vol. 1 Plants. Chapman and Hall, London and New York, pp. 143–188. [Google Scholar]
- Hai, M.T.L., Masuda, J., Miyajima, I., Thien, N.Q., Mojtahedi, N., Hiramatsu, M., Kim, J.-H. and Okubo, H. (2012) Involvement of carotenoid cleavage dioxygenase 4 gene in tepal color change in Lilium brownii var. colchesteri. J. Japan. Soc. Hort. Sci. 81: 366–373. [Google Scholar]
- Hirschberg, J. (2001) Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 4: 210–218. [DOI] [PubMed] [Google Scholar]
- Hugueney, P., Badillo, A., Chen, H.C., Klein, A., Hirschberg, J., Camara, B. and Kuntz, M. (1995) Metabolism of cyclic carotenoids: a model for the alteration of this biosynthetic pathway in Capsicum annuum chromoplasts. Plant J. 8: 417–424. [DOI] [PubMed] [Google Scholar]
- Ilg, A., Yu, Q., Schaub, P., Beyer, P. and Al-Babili, S. (2010) Overexpression of the rice carotenoid cleavage dioxygenase 1 gene in Golden Rice endosperm suggests apocarotenoids as substrates in planta. Planta 232: 691–699. [DOI] [PubMed] [Google Scholar]
- Jędrzejuk, A., Meyer, L. and Serek, M. (2017) Characterization of interspecific hybrids of Petunia and Calibrachoa by multiplex PCR, DNA content, and chromosome number. J. Hortic. Sci. Biotechnol. 92: 493–501. [Google Scholar]
- Kato, M., Ikoma, Y., Matsumoto, H., Sugiura, M., Hyodo, H. and Yano, M. (2004) Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol. 134: 824–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M., Matsumoto, H., Ikoma, Y., Okuda, H. and Yano, M. (2006) The role of carotenoid cleavage dioxygenases in the regulation of carotenoid profiles during maturation of citrus fruit. J. Exp. Bot. 57: 2153–2164. [DOI] [PubMed] [Google Scholar]
- Kishimoto, S., Maoka, T., Nakayama, M. and Ohmiya, A. (2004) Carotenoid composition in petals of chrysanthemum (Dendranthema grandiflorum (Ramat.) Kitamura). Phytochemistry 65: 2781–2787. [DOI] [PubMed] [Google Scholar]
- Kishimoto, S. and Ohmiya, A. (2006) Regulation of carotenoid biosynthesis in petals and leaves of chrysanthemum (Chrysanthemum morifolium). Physiol. Plant. 128: 437–447. [Google Scholar]
- Kishimoto, S., Sumitomo, K., Yagi, M., Nakayama, M. and Ohmiya, A. (2007) Three routes to orange petal colour via carotenoid component in 9 Compositae species. J. Japan. Soc. Hort. Sci. 76: 250–257. [Google Scholar]
- Kishimoto, S., Oda-Yamamizo, C. and Ohmiya, A. (2018) Regulation of carotenoid pigmentation in corollas of petunia. Plant Mol. Biol. Rep. 36: 632–642. [Google Scholar]
- Larkin, M.A., Blackshields, G., Brown, N.P., Chenna, R., McGettigan, P.A., McWilliam, H., Valentin, F., Wallace, I.M., Wilm, A., Lopez, R.et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Liu, H., Kishimoto, S., Yamamizo, C., Fukuta, N. and Ohmiya, A. (2013) Carotenoid accumulations and carotenogenic gene expressions in the petals of Eustoma grandiflorum. Plant Breed. 132: 417–422. [Google Scholar]
- Ljubesić, N., Wrischer, M. and Devidé, Z. (1991) Chromoplasts–the last stages in plastid development. Int. J. Dev. Biol. 35: 251–258. [PubMed] [Google Scholar]
- Ma, G., Zhang, L., Iida, K., Madono, Y., Yungyuen, W., Yahata, M., Yamawaki, K. and Kato, M. (2017) Identification and quantitative analysis of β-cryptoxanthin and β-citraurin esters in Satsuma mandarin fruit during the ripening process. Food Chem. 234: 356–364. [DOI] [PubMed] [Google Scholar]
- Maoka, T., Etoh, T., Kishimoto, S. and Sakata, S. (2011) Carotenoids and their fatty acid esters in the petals of Adonis aestivalis. J. Oleo. Sci. 60: 47–52. [DOI] [PubMed] [Google Scholar]
- Marin, E., Nussaume, L., Quesada, A., Gonneau, M., Sotta, B., Hugueney, P., Frey, A. and Marion-Poll, A. (1996) Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 15: 2331–2342. [PMC free article] [PubMed] [Google Scholar]
- McCarty, D.R. and Klee, H.J. (2006) Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 45: 982–993. [DOI] [PubMed] [Google Scholar]
- Moehs, C.P., Tian, L., Osteryoung, K.W. and DellaPenna, D. (2001) Analysis of carotenoid biosynthetic gene expression during marigold petal development. Plant Mol. Biol. 45: 281–293. [DOI] [PubMed] [Google Scholar]
- Murakami, Y., Fukui, Y., Watanabe, H., Kokubun, H., Toya, Y. and Ando, T. (2003) Distribution of carotenoids in the flower of non-yellow commercial petunia. J. Hortic. Sci. Biotechnol. 78: 127–130. [Google Scholar]
- Murakami, Y., Fukui, Y., Watanabe, H., Kokubun, H., Toya, Y. and Ando, T. (2004) Floral coloration and pigmentation in Calibrachoa cultivars. J. Hortic. Sci. Biotechnol. 79: 47–53. [Google Scholar]
- Muszyński, S. (1964) A survey of anthocyanidins in Petunia. Physiol. Plant. 17: 975–979. [Google Scholar]
- Nielsen, K.M. and Bloor, S.J. (1997) Analysis and developmental profile of carotenoid pigments in petals of three yellow petunia cultivars. Sci. Hortic. 71: 257–266. [Google Scholar]
- Ohmiya, A., Kishimoto, S., Aida, R., Yoshioka, S. and Sumitomo, K. (2006) Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 142: 1193–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya, A. (2009) Carotenoid cleavage dioxygenases and their apocarotenoid products in plants. Plant Biotechnol. 26: 351–358. [Google Scholar]
- Ohmiya, A. (2011) Diversity of carotenoid composition in flower petals. Jpn. Agric. Res. Q. 45: 163–171. [Google Scholar]
- Ohmiya, A. (2013) Qualitative and quantitative control of carotenoid accumulation in flower petals. Sci. Hortic. 163: 10–19. [Google Scholar]
- Ohmiya, A., Tanase, K., Hirashima, M., Yamamizo, C. and Yagi, M. (2013) Analysis of carotenogenic gene expression in petals and leaves of carnation (Dianthus caryophyllus L.). Plant Breed. 132: 423–429. [Google Scholar]
- Olmstead, R.G., Bohs, L., Migid, H.A., Santiago-Valentin, E., Garcia, V.F. and Collier, S.M. (2008) A molecular phylogeny of the Solanaceae. Taxon 57: 1159–1181. [Google Scholar]
- Robert, B., Horton, P., Pascal, A.A. and Ruban, A.V. (2004) Insights into the molecular dynamics of plant light-harvesting proteins in vivo. Trends Plant Sci. 9: 385–390. [DOI] [PubMed] [Google Scholar]
- Ronen, G., Cohen, M., Zamir, D. and Hirschberg, J. (1999) Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J. 17: 341–351. [DOI] [PubMed] [Google Scholar]
- Ruban, A.V., Berera, R., Ilioaia, C., van Stokkum, I.H.M., Kennis, J.T.M., Pascal, A.A., van Amerongen, H., Robert, B., Horton, P. and van Grondelle, R. (2007) Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 450: 575–578. [DOI] [PubMed] [Google Scholar]
- Schwartz, S.H., Tan, B.C., Gage, D.A., Zeevaart, J.A.D. and McCarty, D.R. (1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276: 1872–1874. [DOI] [PubMed] [Google Scholar]
- Simkin, A.J., Schwartz, S.H., Auldridge, M., Taylor, M.G. and Klee, H.J. (2004a) The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles β-ionone, pseudoionone, and geranylacetone. Plant J. 40: 882–892. [DOI] [PubMed] [Google Scholar]
- Simkin, A.J., Underwood, B.A., Auldridge, M., Loucas, H.M., Shibuya, K., Schmelz, E., Clark, D.G. and Klee, H.J. (2004b) Circadian regulation of the PhCCD1 carotenoid cleavage dioxygenase controls emission of β-ionone, a fragrance volatile of petunia flowers. Plant Physiol. 136: 3504–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornielli, G., Koes, R. and Quattrocchio, F. (2008) The genetics of flower color. In: Gerats, T. and Strommer J. (eds.) Petunia: Evolutionary, Developmental and Physiological Genetics. Springer, New York, pp. 325–342. [Google Scholar]
- Yamamizo, C., Kishimoto, S. and Ohmiya, A. (2010) Carotenoid composition and carotenogenic gene expression during Ipomoea petal development. J. Exp. Bot. 61: 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., Liu, C., Wang, Y., Yao, X., Wang, F., Wu, J., King, G.J. and Liu, K. (2015) Disruption of a CAROTENOID CLEAVAGE DIOXYGENASE 4 gene converts flower colour from white to yellow in Brassica species. New Phytol. 206: 1513–1526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.