Abstract
Plants produce strigolactones (SLs) in roots in response to nitrogen or phosphate deficiency. To evaluate SL levels under other mineral deficiencies in rice, we cultivated rice seedlings in hydroponic media without nitrogen, phosphorus, potassium, sulfur, calcium, magnesium, and iron. Tiller bud outgrowth was stimulated under calcium deficiency because of low SL levels. SL levels increased under sulfur deficiency, in addition to phosphate, and nitrogen deficiencies. To explore which genes are key regulators of SL production under sulfur deficiency, we analyzed the expression of SL‐related genes in sulfur‐sufficient and sulfur‐deficient conditions. An SL biosynthesis gene, DWARF27 (D27), was strongly expressed under sulfur deficiency, and its expression was decreased by sulfur supply. The levels of D10, D17, and OsMAX1 transcripts did not differ between sulfur‐sufficient and sulfur‐deficient conditions. These results suggest that the increased SL levels under sulfur deficiency are due to a high expression of D27. A combination of nitrogen, phosphorus, and sulfur deficiencies had no additive synergistic effect on SL production. Under combined phosphorus and sulfur deficiency, the expression levels of most SL biosynthesis genes were elevated. The number of tiller buds in the d27 mutant was higher than in the wild type, but lower than in other d mutants. Under sulfur deficiency, the chlorophyll content of d27 was lower than those of other d mutants. These results indicate that D27 plays an important role in adaptation to sulfur deficiency in rice.
Keywords: D27, leaf senescence, macronutrient deficiency, Oryza sativa, shoot branching, strigolactones
1. INTRODUCTION
Strigolactones (SLs) are a class of terpenoid lactones that were originally discovered as stimulants of seed germination in root parasitic plants such as Striga spp., Orobanche spp., and Phelipanche spp. (for reviews, see Xie, Yoneyama, & Yoneyama, 2010; Yoneyama, Awad, Xie, Yoneyama, & Takeuchi, 2010). Later, SLs were found to induce hyphal branching of arbuscular mycorrhizal (AM) fungi, which support the acquisition of P and N in soil by the host plants (Akiyama, Matsuzaki, & Hayashi, 2005). Thus, SLs are thought to act as communication signals to recognize host plants for parasitism and symbiosis in the rhizosphere. In addition to the roles in the rhizosphere, SLs inhibit shoot branching (Gomez‐Roldan et al., 2008; Umehara et al., 2008). Because SL‐related mutants show pleiotropic phenotypes including enhanced shoot branching, SLs have recently been recognized as plant hormones that control plant growth at various developmental stages. In vascular plants, SLs stimulate stem thickening, leaf senescence, root hair elongation, and primary root growth and suppress adventitious root formation (Agusti et al., 2011; Kapulnik et al., 2011; Rasmussen et al., 2012; Ruyter‐Spira et al., 2011; Ueda & Kusaba, 2015; Yamada et al., 2014). In the moss Physcomitrella patens, SLs suppress chloronema branching and colony expansion (Proust et al., 2011).
Carlactone (CL), a biosynthetic precursor of SLs, is synthesized from β‐carotene through consecutive reactions catalyzed by the β‐carotene isomerase DWARF27 (D27) and carotenoid cleavage dioxygenases 7 and 8 (CCD7 and CCD8) (Alder et al., 2012; Seto et al., 2014). In Arabidopsis, CL is converted to carlactonoic acid via oxidation by a cytochrome P450 encoded by MORE AXILLARY GROWTH1 (MAX1) (Abe et al., 2014). In rice, D17/HTD1 encodes CCD7, and D10 encodes CCD8 (Arite et al., 2007; Zou et al., 2006), and there are five MAX1 homologs: Os01g0700900 (Os900), Os01g0701400 (Os1400), Os01g0701500 (Os1500), Os02g0221900 (Os1900), and Os06g0565100 (Os5100) (Nelson, Schuler, Paquette, Werck‐Reichhart, & Bak, 2004) (Figure 1). Os900 catalyzes the oxidation of CL into the parent SL 4‐deoxyorobanchol (4DO), and Os1400 catalyzes the hydroxylation of 4DO to orobanchol (Zhang et al., 2014). Canonical SLs have tricyclic lactone and methylbutenolide moieties connected by an enol ether bond. This bond and the methylbutenolide structure are essential for shoot branching inhibition (Boyer et al., 2012; Umehara et al., 2015). Configuration of the methylbutenolide moiety affects the biological activity of SLs in rice and Arabidopsis but not in pea (Boyer et al., 2012; Umehara et al., 2015). In SL signaling, D14 is a probable SL receptor and belongs to the α/β‐fold hydrolase superfamily, which also includes the gibberellin receptor GID1 (Arite et al., 2009) (Figure 1). D14 is transported through the phloem to tiller buds in rice (Kameoka et al., 2016). D3 is a leucine‐rich‐repeat F‐box protein that acts as a recognition subunit in a SKP1–CUL1–F‐box‐protein complex and binds target proteins for proteasomal degradation (Ishikawa et al., 2005) (Figure 1).
Figure 1.
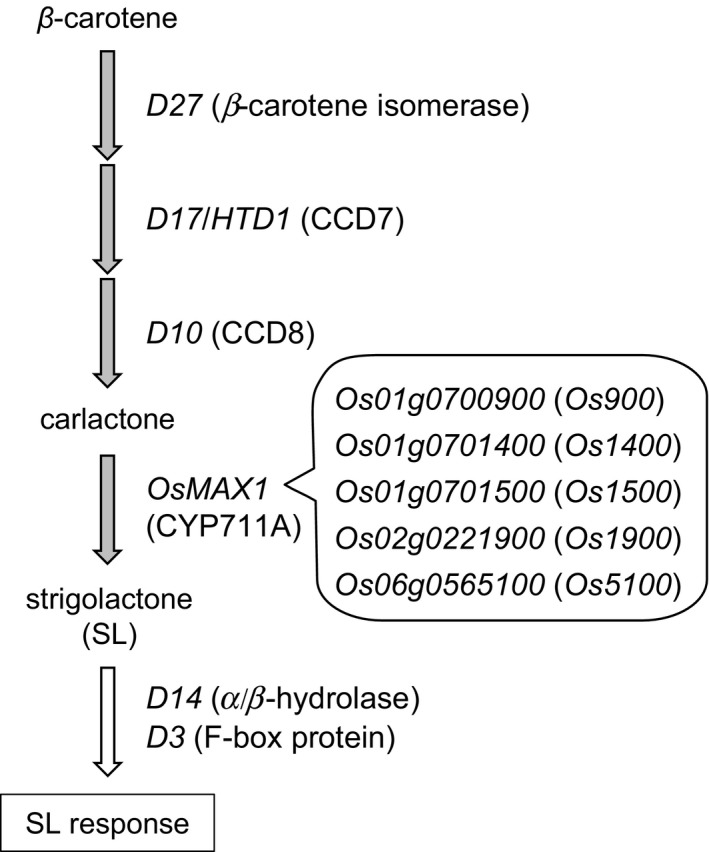
SL pathway in rice. Gray and white arrows indicate the SL biosynthesis and signaling, respectively
Several types of SLs have been identified; these are mainly produced in roots in response to P deficiency in various plant species including red clover, tomato, sorghum, Lotus japonicus, alfalfa, and rice (Lopez‐Raez et al., 2008; Sugimoto & Ueyama, 2008; Umehara et al., 2008; Yoneyama, Xie, et al., 2007; Yoneyama, Yoneyama, Takeuchi, & Sekimoto, 2007; Yoneyama et al., 2012). In rice, the levels of D27, D17, D17, and Os900 mRNAs are highly elevated in roots under P deficiency and contribute to the high levels of SLs, indicating that most SL biosynthesis genes are upregulated in response to P deficiency (Umehara, Hanada, Magome, Takeda‐Kamiya, & Yamaguchi, 2010). Tiller bud outgrowth is not inhibited under P‐sufficient conditions because SL levels are low in roots. Under P deficiency, SL levels in roots are highly elevated and SLs are probably transported to tiller buds to inhibit the outgrowth. However, shoot branching is not suppressed under P deficiency in SL mutants of rice and Arabidopsis (Kohlen et al., 2011; Umehara et al., 2010). In some plant species such as lettuce, marigold, sorghum, and rice, SL levels in roots increase under both N deficiency and P deficiency (Sun et al., 2014; Yoneyama, Xie, et al., 2007; Yoneyama, Yoneyama, et al., 2007), and N and P fertilization suppresses SL production and exudation in roots (Umehara et al., 2010; Yoneyama, Xie, Kisugi, Nomura, & Yoneyama, 2013; Yoneyama et al., 2012). Thus, SLs are thought to be key regulators of efficient nutrient allocation and adaption to nutrient‐deficient environments (Umehara, 2011).
Little is known about how SL levels change in roots under deficiencies of macronutrients other than P and N. To address this question, here we examined the effects of deficiency in N, P, K, S, Ca, Mg, or Fe on SL production in rice. Here, we show that tiller buds of rice seedlings outgrow under Ca deficiency because SL levels decrease and that SL levels increase under S deficiency as well as N or P deficiency. We also show that D27 is strongly expressed under S deficiency, and its expression is reduced by S supply, whereas the expression levels of D10, D17, and OsMAX1 are not affected by S supply. These results suggest that SL production under S deficiency is due to a marked increase in the expression of D27. Under S deficiency, tiller bud outgrowth in a d27 mutant was strongly suppressed compared with that in other d mutants, and chlorophyll content was lower than that in other d mutants, indicating that D27 expression plays an important role in survival and adaptation to S deficiency.
2. MATERIALS AND METHODS
2.1. Plant materials and growth conditions
We used the rice (Oryza sativa L.) cultivar Shiokari as the WT and tillering dwarf mutants, d10, d14, and d17 in the Shiokari background (Ishikawa et al., 2005). Seeds were provided by Dr. Junko Kyozuka (Tohoku University) and were propagated in a glass room at the Research Center for Life and Environmental Sciences, Toyo University. Seedlings were grown hydroponically as described previously (Umehara et al., 2008). Surface‐sterilized seeds were incubated in sterile water at 27°C in the dark for 1 day. Germinated seeds were put in hydroponic culture media solidified with 0.6% agar and cultured under fluorescent white light (130–180 μmol m−2 s−1; 16 hr light at 25°C, 8 hr dark at 23°C) for 7 days. The seedlings were then transferred to glass vials containing hydroponic culture medium (Kamachi, Yamaya, Mae, & Ojima, 1991). To evaluate the effect of nutrient deficiency on SL production, we prepared hydroponic culture media as shown in Table S1. All media contained 1 mM 2‐(N‐morpholino) ethanesulfonic acid (MES) buffer adjusted to pH 5.7. Only tillers growing over 2 mm were counted (Umehara et al., 2008). Relative chlorophyll contents were measured as SPAD values using a leaf chlorophyll meter, SPAD‐502Plus (Konica Minolta Inc., Japan) (Ata‐Ul‐Karim et al., 2016).
2.2. SL analysis
To measure the amounts of 4DO released from roots, hydroponic media were extracted with ethyl acetate twice after adding d 1‐labeled 4DO as an internal standard. The ethyl acetate phase was evaporated to dryness under nitrogen gas. The extracts were dissolved in ethyl acetate: n‐hexane (15:85) and loaded onto 1‐ml Sep‐Pak Silica cartridges (Waters, MA, USA), washed with ethyl acetate: n‐hexane (15:85) and then eluted with ethyl acetate: n‐hexane (35:65).
To measure 4DO levels in roots, roots (ca. 650 mg) were homogenized in 10 ml acetone containing d 1‐labeled 4DO; the homogenates were filtered with Bond Elute reservoirs (Agilent, CA, USA) and evaporated to dryness under nitrogen gas. The extracts were dissolved in water adjusted to pH 2–3 with 1 N HCl and extracted with ethyl acetate twice. The ethyl acetate phase was evaporated to dryness under nitrogen gas. The extracts were dissolved in 10% acetone, loaded onto Oasis HLB 3‐ml cartridges (Waters), washed with 10% acetone, and eluted with 60% acetone. The eluates were dissolved in ethyl acetate: n‐hexane (15:85), loaded onto Sep‐Pak Silica 1‐ml cartridges (Waters), washed with ethyl acetate: n‐hexane (15:85), and eluted with ethyl acetate: n‐hexane (35:65).
Purified 4DO‐containing fractions were dissolved in 50% acetonitrile and subjected to liquid chromatography–tandem mass spectrometry (LC‐MS/MS) analysis using a system consisting of a quadrupole tandem mass spectrometer (3200 QTRAP; Sciex, MA, USA) and a high‐performance liquid chromatograph (Prominence, Shimadzu, Kyoto, Japan) equipped with a reverse‐phase column (Acquity UPLC BEH‐C18, 2.1 × 50 mm, 1.7 μm, Waters). Data were analyzed in Analyst 1.5.1 and Multi Quant 2.0.2 (Sciex, MA, USA).
2.3. Gene expression analysis
Total RNA was extracted from roots using an RNeasy Maxi kit (Qiagen, Hilden, Germany) and concentrated using an RNeasy Mini kit (Qiagen). A 3‐μg aliquot of total RNA was used for cDNA synthesis with a ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo, Osaka, Japan). qRT‐PCR was performed on a StepOnePlus RT‐PCR system (Thermo Fisher Scientific, MA, USA) using a Thunderbird Probe qPCR Mix (Toyobo, Osaka, Japan), specific primers, and Taq‐Man probes listed in Table S2. Ubiquitin expression was used as internal control. Total RNA extraction, cDNA synthesis, and qRT‐PCR were conducted according to manufacturers' instructions.
2.4. Statistical analysis
Statistical analysis was performed in SPSS 23 (IBM SPSS Inc., Armonk, NY, USA) using Student's t test for pairwise comparisons, ANOVA and Tukey's honestly significant difference test (HSD) for multiple comparisons.
3. RESULTS
3.1. Effect of macronutrient deficiency in hydroponic culture medium on tiller bud outgrowth and SL production in rice
To evaluate the effect of macronutrient deficiency on tiller bud outgrowth in rice, we cultivated WT seedlings in hydroponic media without N, P, K, S, Ca, Mg, or Fe for 1 week and counted the number of outgrowing tiller buds (>2 mm). Interestingly, the second leaf tillers grew in the absence of Ca but not of other nutrients (Figure S1A). Tiller bud outgrowth in the absence of Ca was inhibited by 1 μM GR24 that is a SL synthetic analog (Figure S1B). LC‐MS/MS analysis demonstrated that the 4DO level was approximately 0.05 pg/ml in root exudates and 6.6 pg/g fresh weight (FW) in roots of nutrient‐sufficient control (Figure 2). Under Ca deficiency (–Ca), 4DO was not detected in root exudates and its content in roots (3.7 pg/g FW) was lower than that of control (Figure 2). The tiller bud outgrowth in –Ca plants was probably due to the reduction of 4DO levels in roots. Reduced SL levels under –Ca were reported in root exudates of red clover by liquid chromatography–tandem mass spectrometry (LC‐MS/MS) analysis and seed germination assay of root parasitic plants (Yoneyama, Xie, et al., 2007; Yoneyama, Yoneyama, et al., 2007).
Figure 2.
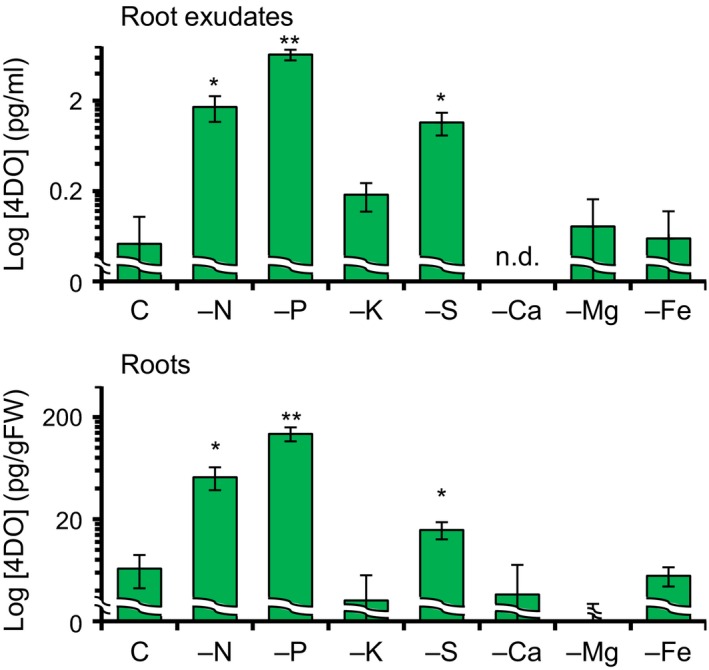
4DO levels in root exudates and roots under deficiency of N, P, K, S, Ca, Mg, or Fe. C, control. Samples were collected on day 7 after transfer to hydroponic media, and 4DO levels were analyzed using LC‐MS/MS. n.d., not detected. Data are means ± SE (n = 4). *p < .05, **p < .01 vs. control (Student's t test)
Under –P conditions, the 4DO levels were highest among all experimental conditions tested (~6.5 pg/ml in root exudates and 137 pg/g FW in roots; Figure 2). Under –N conditions, the 4DO levels were also higher than in control (~1.7 pg/ml in root exudates and 51.5 pg/g FW in roots). Under –S conditions, the 4DO levels increased to ~1.1 pg/ml in root exudates and 15.7 pg/g FW in roots (Figure 2). To further investigate the effects of S deficiency on SL production, we grew rice seedlings for 1 week in hydroponic media with different S concentrations (potassium sulfate: 0, 10, 110, 210, and 310 μM as the S source). 4DO levels were higher at 0 and 10 μM than at 110, 210, and 310 μM potassium sulfate (Figure S2).
3.2. Time course analysis of SL levels and expression of SL‐related genes
To investigate which genes regulate SL production under S deficiency, we analyzed the expression levels of SL‐related genes using quantitative real‐time polymerase chain reaction (qRT‐PCR). We grew WT seedlings in hydroponic medium without S for 1 week, transferred them to fresh +S or −S medium, and analyzed 4DO levels and gene expression levels at 0, 1, and 2 days after transfer (Figures S3A). 4DO levels and expression of SL biosynthetic genes at 1 day were picked up in Figure 3. Although 4DO levels remained high under −S conditions, they were significantly reduced by S supply on days 1 and 2 after transfer (Figures 3b and S3B). D27 was strongly expressed under −S conditions, and its expression was significantly reduced by S supply (Figures 3c and S3C). The expression pattern of D27 resembled the changes in 4DO levels (Figure S3B,C). mRNA levels of D10, D17, and the five MAX1 homologs did not differ significantly between +S and −S conditions (Figures 3c and S3C). On the other hand, the expression levels of the SL signaling genes D3 and D14 strongly increased under +S conditions in comparison with −S (Figure S3C).
Figure 3.
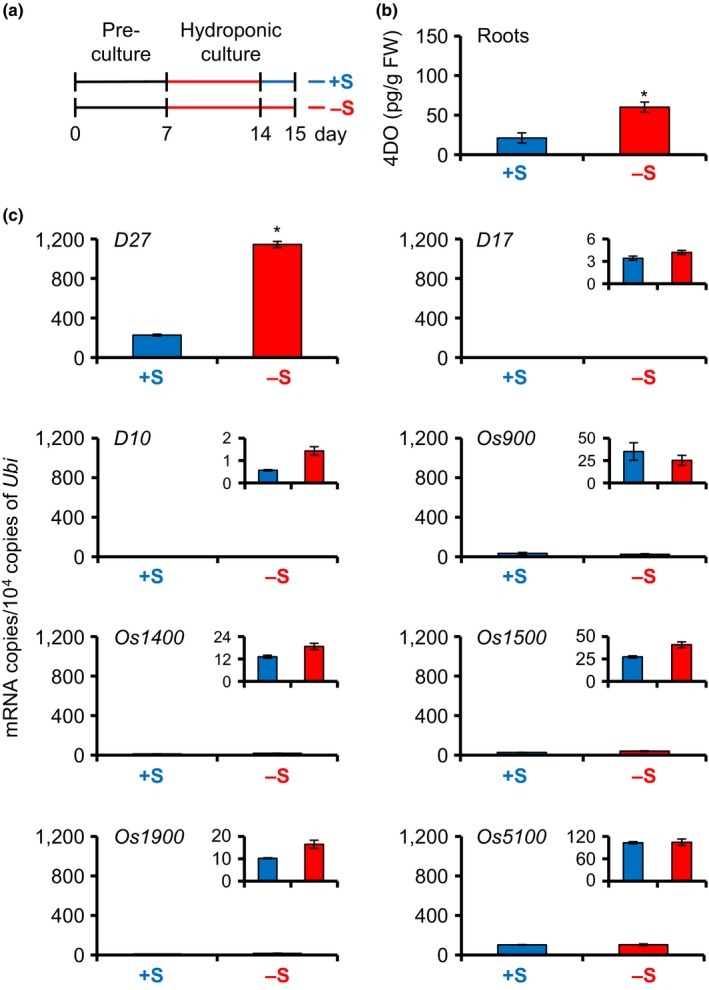
SL levels and expression of SL biosynthesis genes in WT rice seedlings under +S or −S conditions. (a) Schematic diagram showing the experimental conditions. Black and gray bars indicate +S and −S conditions, respectively. Seedlings were transferred to fresh +S or −S medium on day 14 of culture. 4DO levels and gene expression were analyzed on day 15. (b) 4DO levels in roots. Data are means ± SE (n = 4). (c) Transcript levels of SL biosynthesis genes in roots. Data are means ± SE (n = 3). *p < .05 (Student's t test)
3.3. SL production under N, P, and S deficiencies
To investigate the effects of combinations of N, P, and S deficiencies on 4DO levels, we grew WT seedlings under nutrient‐sufficient conditions (control), single macronutrient deficiency, or combined deficiencies. 4DO levels increased significantly in all the treatments in comparison with control (Figure 4). The highest 4DO levels were observed in both root exudates and roots under P deficiency, but this increase was reduced in combinations of P deficiency with deficiency in N and/or S (Figure 4).
Figure 4.
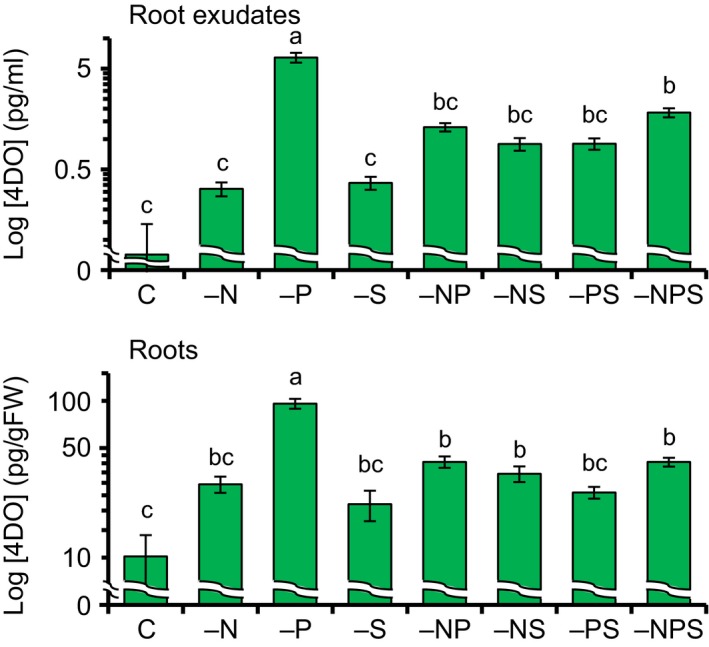
Effects of combined N, P, and S deficiencies on 4DO levels. The samples were collected on the day 7 after transfer to hydroponic culture media, and 4DO levels were analyzed using LC‐MS/MS. Data are means ± SE (n = 4). *p < .05, **p < .01 (Student's t test)
To evaluate how the expression of SL biosynthesis genes changes in response to a combination of nutrient deficiencies, we analyzed the 4DO levels and expression of SL biosynthesis genes under combined P and S deficiency (−PS). We grew WT seedlings in −PS medium for 1 week and then transferred them to fresh +PS or −PS medium (Figure 5a). 4DO levels and gene expression were analyzed 1 day after the transfer. 4DO levels were slightly higher in the roots of seedlings grown in −PS medium than in those of seedlings grown in −S medium and were significantly decreased by PS supply (Figures 3b, 4, and 5b). D27 expression was lower under −PS than under −S conditions, whereas the expression of other SL biosynthesis genes such as D17, D10, and OsMAX1s was higher (Figures 3c and 5c). On the other hand, the expression levels of the SL signaling genes D3 and D14 strongly increased under +PS conditions in comparison with −PS (Figure S4).
Figure 5.
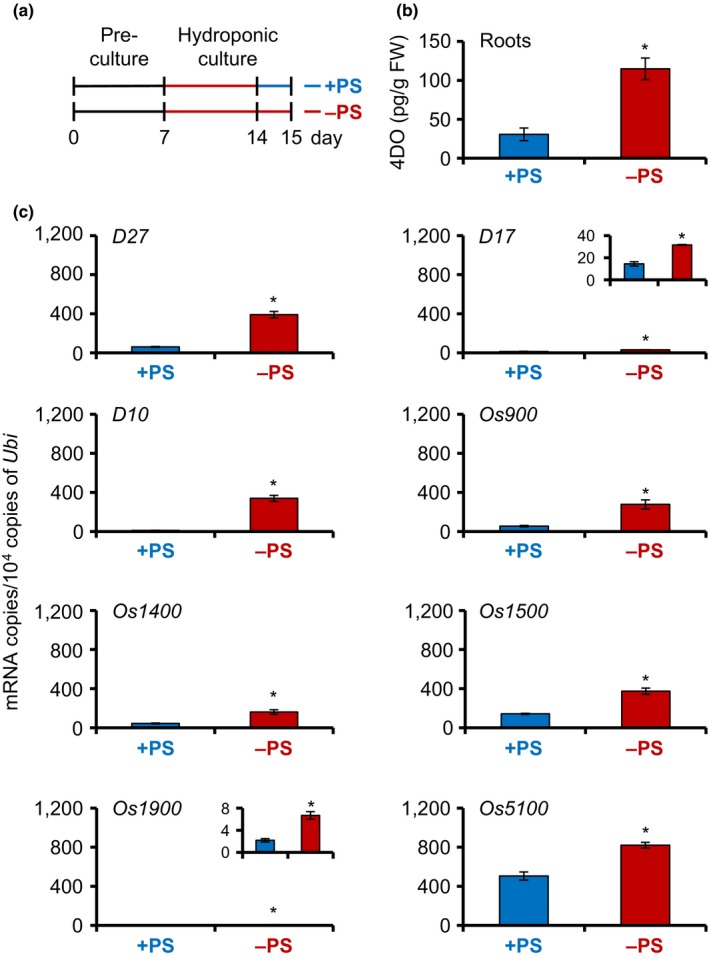
SL levels and expression of SL biosynthesis genes in WT rice seedlings under +PS or −PS conditions. (a) Schematic diagram showing the experimental conditions. Black and gray bars indicate +PS and −PS conditions, respectively. Seedlings were transferred to new +PS or −PS media on the day 14 of culture. 4DO levels and gene expression were analyzed on day 15. (b) 4DO levels in roots. Data are means ± SE (n = 4). (c) Transcript levels of SL biosynthesis genes in roots. Data are means ± SE (n = 3). *p < .05 (Student's t test)
3.4. Effect of S deficiency on tiller bud outgrowth and leaf senescence
Shoot branching is suppressed and chlorosis is accelerated under S deficiency (Dobermann & Fairhurst, 2001). To evaluate whether shoot branching inhibition and chlorosis are caused by SL produced under S deficiency, we grew WT, the SL biosynthesis mutant d10 and d27, and the SL signaling mutant d14 in hydroponic culture medium with or without S for 22 days after preculture (Figure S5A). In WT, the number of outgrowing tillers over 2 mm gradually increased from 14 to 29 days under +S, but no tiller outgrowth was observed under −S (Figures 6a and S5B). In d mutants, the number of outgrowing tillers also gradually increased from 14 to 29 days under +S and was significantly reduced under −S (Figures 6a and S5B). The tiller bud outgrowth of d27 was slower than those of other d mutants under +S, but achieved to similar levels at day 29 (Figure 6a). Under −S, the tiller number of d27 was higher than that of WT but lower than those of other d mutants (Figure 6a).
Figure 6.
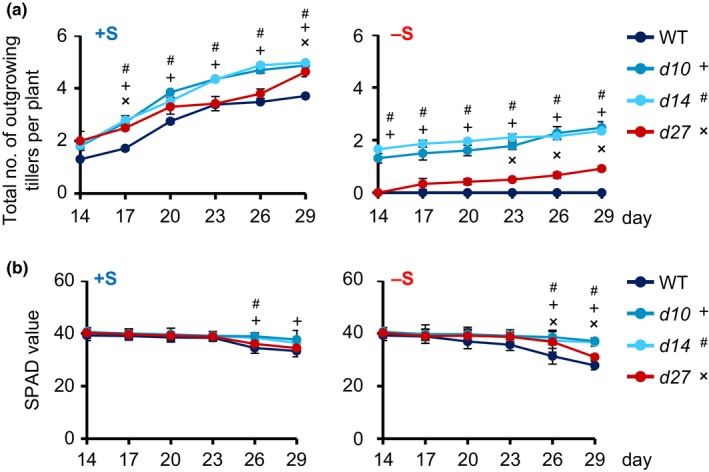
Phenotypical analysis of WT and SL mutants under +S and −S. (a) Total number of outgrowing tillers. Outgrowing tillers over 2 mm were counted in 21‐day‐old seedlings. Data are means ± SE (n = 3). (b) Comparison of the SPAD values of WT and the d27, d10, and d14 mutants. Data are means ± SE (n = 3). Significant differences between WT and d mutants are shown (ANOVA, p < .05)
Next, we pre‐cultured WT, d10, d14, and d27 for 1 week, then grew them in hydroponic culture medium under +S for 1 week and then under −S or +S for 15 days (Figure S6A) and measured SPAD value in the third leaves (the SPAD value indicates total chlorophyll content, and low SPAD values indicate leaf senescence) (Ata‐Ul‐Karim et al., 2016). The SPAD value tended to gradually decrease with time and was significantly lower under −S than under +S from day 20 to day 29 in WT, and on day 29 in d14 and d27, but did not differ significantly between +S and −S in d10 (Figures 6b and S6B). The SPAD values were mostly similar from day 14 to day 29 between WT and d mutants under +S, but the SPAD values of WT were lower than d mutants on days 26 and 29 (Figures 6b and S6B). Interestingly, the SPAD values of d27 were similar to WT and other d mutants under +S, but lower than that in other d mutants under −S (Figure 6b). To check a possibility that d27 may produce slight levels of 4DO, we confirmed 4DO levels in d27 using LC‐MS/MS. We could not detect 4DO in d27 under either −S, −P, or nutrient‐sufficient conditions (Figure S7). The reduction in the number of outgrowing tiller buds and SPAD value in d27 under −S was not observed under −P (Figure S8).
4. DISCUSSION
In this study, we found that 4DO levels increased under −S in addition to −P, and −N conditions in WT rice seedlings (Figure 2). Among SL biosynthesis genes, only D27 (not D10, D17, or OsMAX1s) was highly expressed and thus contributed to SL production under −S (Figure 3). Thus, SL production under S deficiency is due to a marked increase in the expression of D27. In this time, we cannot exclude a possibility that activity of SL metabolism is low under −S because we still do not know the SL metabolic pathway. On the other hand, gene expression of D3 and D14, which are the SL signaling genes, is rather upregulated under +S indicate that rice seedlings may increase the SL sensitivity because SL levels are low in S sufficient condition (Figures S3 and S4). When the d27 mutant was grown under −S, it had fewer outgrowing tiller buds than other d mutants, and its chlorophyll content was lower than those of other d mutants (Figure 6). These results suggest that D27 is involved in −S response and may maintain plant growth and development in −S conditions.
D27 was originally characterized in rice as a gene encoding an iron‐containing protein that localizes in chloroplasts (Lin et al., 2009). d27 mutants fail to produce SL, have increased tiller number and reduced plant height, and their phenotypes are rescued by exogenously applied SL, indicating that D27 is required for SL biosynthesis (Lin et al., 2009). In vitro experiments demonstrated that the D27 protein is an isomerase that converts all‐trans‐β‐carotene to 9‐cis‐β‐carotene in the first step of SL biosynthesis (Alder et al., 2012). In rice roots, high expression of most SL biosynthesis genes (D27, D17, D10, and OsMAX1s) increases SL levels under −P (Umehara et al., 2010). The expression levels of D10 and D17 increase under −N, but that of D27 is not affected (Sun et al., 2014). In contrast, only D27 is highly expressed under −S, and its level is 10 times that under −P (Umehara et al., 2010) (Figure 3). Under −PS, expression levels of D27 decreased but that of the other SL biosynthetic genes increased compared with −S, resulting in increasing the flow of SL biosynthesis (Figures 3 and 5). Thus, 4DO content of PS deficiencies might be higher than that of S deficiency. D27 in Arabidopsis, AtD27, is located in plastids and acts upstream of MAX1 in the SL biosynthetic pathway (Waters, Brewer, Bussell, Smith, & Beveridge, 2012). Microarray data demonstrate that the expression level of AtD27 in Arabidopsis roots increases under low S condition, but the expression of MAX1 and MAX3 do not (Iyer‐Pascuzzi et al., 2011; Maruyama‐Nakashita, Nakamura, Tohge, Saito, & Takahashi, 2006). These data support our results that D27 expression is responsive to −S.
Sulfur is a macro‐element required for plant growth because it is a component of biologically important compounds such as the amino acids Cys and Met, antioxidant tripeptide glutathione, S‐adenosyl methionine, sulfolipids, and many secondary metabolites (Amtmann & Armengaud, 2009). In rice, S deficiency reduces chlorophyll content in young leaves, the number of tillers, and plant height (Dobermann & Fairhurst, 2001). These symptoms are similar to the phenotypes of SL biosynthesis‐ or signaling‐deficient mutants. In WT, tiller bud outgrowth was completely suppressed under −S, whereas tiller buds of d mutants still grew (Figure 6a). The SPAD values of d10 and d14 were higher than that of WT under −S (Figure 6b). Thus, SLs regulate shoot branching and leaf senescence in response to S deficiency. However, number of tiller buds and SPAD value of d mutants were smaller under −S than under +S, indicating that SLs only partially mediate the regulation of shoot branching and leaf senescence, and effect of nutrient deficiency are involved directly.
In SL biosynthesis, the transcription factors NSP1 and NSP2 are indispensable in Medicago and rice (Liu et al., 2011). D27 expression is upregulated by NSP1 and NSP2 under −P and in rhizobium symbiotic signaling (Liu et al., 2011; van Zeijl et al., 2015). However, the expression levels of NSP1 and NSP2 did not increase in the absence of N or P (Liu et al., 2011). NSP1 and NSP2 were originally found as the GRAS family transcription factors regulating nodule formation (Kalo et al., 2005; Smit et al., 2005). Later, they were also found to be components of the Myc signaling pathway of AM fungi, in addition to the Nod signaling pathway (Delaux, Becard, & Combier, 2013; Lauressergues et al., 2012). In rice, only D27 transcript increase in AM colonized roots compared with non‐AM colonized roots among SL biosynthetic genes (Kobae et al., 2018). Secreted SLs activate hyphal branching of AM fungi and trigger their symbiotic interactions with the host plants (Akiyama et al., 2005). AM fungi can supply S as well as N and P to the host (Allen & Shachar‐Hill, 2009; Govindarajulu et al., 2005; Smith & Read, 2008). Therefore, expression of rice D27 is involved in −S response, and D27 might play an important role in effective S absorption via AM fungi. To understand the interactions between SL production under −S condition and symbiosis with AM fungi, further research would be required in the future.
CONFLICT OF INTERESTS
None declared.
AUTHOR CONTRIBUTIONS
M.S. and M.U. performed the experiments and analyzed the data. K.S. and S.Y. contributed to the experimental design. M.U. directed the research and designed the experiments. M.S. and M.U. wrote the manuscript.
Supporting information
ACKNOWLEDGMENTS
This work was in part supported by grants from Toyo University (the Inoue Enryo Memorial Foundation for Promoting Sciences to M.S.) and from the Japan Society for the Promotion of Science (KAKENHI Grant Nos. 26450144 and 17K07650 to M.U.). We thank Dr. Junko Kyozuka (Tohoku University) for providing rice seeds, Dr. Kohki Akiyama (Osaka Prefecture University) for providing d 1‐labeled 4DO, Mr. Atsushi Hanada (Sciex) for his technical supports of LC‐MS/MS, and Drs. Shosaku Kashiwada, Yuichi Iwasaki, and Uma Maheswari Rajagopalan (Toyo University) for constructive comments. This study was in part supported by the Research Center for Life and Environmental Sciences, Toyo University.
Shindo M, Shimomura K, Yamaguchi S, Umehara M. Upregulation of DWARF27 is associated with increased strigolactone levels under sulfur deficiency in rice. Plant Direct. 2018;2:1–9. 10.1002/pld3.50
Funding information
This work was in part supported by grants from Toyo University (the Inoue Enryo Memorial Foundation for Promoting Sciences) to M.S. and from the Japan Society for the Promotion of Science (KAKENHI Grant Nos. 26450144 and 17K07650 to M.U.).
REFERENCES
- Abe, S. , Sado, A. , Tanaka, K. , Kisugi, T. , Asami, K. , Ota, S. , … Nomura, T. (2014). Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proceedings of the National Academy of Sciences of the United States of America, 111, 18084–18089. 10.1073/pnas.1410801111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agusti, J. , Herold, S. , Schwarz, M. , Sanchez, P. , Ljung, K. , Dun, E. A. , … Greb, T. (2011). Strigolactone signaling is required for auxin‐dependent stimulation of secondary growth in plants. Proceedings of the National Academy of Sciences of the United States of America, 108, 20242–20247. 10.1073/pnas.1111902108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama, K. , Matsuzaki, K. , & Hayashi, H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature, 435, 824–827. 10.1038/nature03608 [DOI] [PubMed] [Google Scholar]
- Alder, A. , Jamil, M. , Marzorati, M. , Bruno, M. , Vermathen, M. , Bigler, P. , … Al‐Babili, S. (2012). The path from β‐carotene to carlactone, a strigolactone‐like plant hormone. Science, 335, 1348–1351. 10.1126/science.1218094 [DOI] [PubMed] [Google Scholar]
- Allen, J. W. , & Shachar‐Hill, Y. (2009). Sulfur transfer through an Arbuscular mycorrhiza . Plant Physiology, 149, 549–560. 10.1104/pp.108.129866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann, A. , & Armengaud, P. (2009). Effects of N, P, K and S on metabolism: New knowledge gained from multi‐level analysis. Current Opinion in Plant Biology, 12, 275–283. 10.1016/j.pbi.2009.04.014 [DOI] [PubMed] [Google Scholar]
- Arite, T. , Iwata, H. , Ohshima, K. , Maekawa, M. , Nakajima, M. , Kojima, M. , … Kyozuka, J. (2007). DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. The Plant Journal, 51, 1019–1029. 10.1111/j.1365-313X.2007.03210.x [DOI] [PubMed] [Google Scholar]
- Arite, T. , Umehara, M. , Ishikawa, S. , Hanada, A. , Maekawa, M. , Yamaguchi, S. , & Kyozuka, J. (2009). d14, a strigolactone‐insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant and Cell Physiology, 50, 1416–1424. 10.1093/pcp/pcp091 [DOI] [PubMed] [Google Scholar]
- Ata‐Ul‐Karim, S. T. , Cao, Q. , Zhu, Y. , Tang, L. , Rehmani, M. I. , & Cao, W. (2016). Non‐destructive assessment of plant nitrogen parameters using leaf chlorophyll measurements in rice. Frontiers in Plant Science, 7, 1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer, F.‐D. , Germain, Ad S , Pillot, J.‐P. , Pouvreau, J.‐B. , Chen, V. X. , Ramos, S. , … Rameau, C. (2012). Structure‐activity relationship studies of strigolactone‐related molecules for branching inhibition in garden pea: Molecule design for shoot branching. Plant Physiology, 159, 1524–1544. 10.1104/pp.112.195826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaux, P. M. , Becard, G. , & Combier, J. P. (2013). NSP1 is a component of the Myc signaling pathway. New Phytologist, 199, 59–65. 10.1111/nph.12340 [DOI] [PubMed] [Google Scholar]
- Dobermann, A. , & Fairhurst, T. (2001). Rice: Nutrient disorders & nutrient management. Singapore: PPI/PPIC, IRRI, Philippines. [Google Scholar]
- Gomez‐Roldan, V. , Fermas, S. , Brewer, P. B. , Puech‐Pages, V. , Dun, E. A. , Pillot, J. P. , … Rochange, S. F. (2008). Strigolactone inhibition of shoot branching. Nature, 455, 189–194. 10.1038/nature07271 [DOI] [PubMed] [Google Scholar]
- Govindarajulu, M. , Pfeffer, P. E. , Jin, H. R. , Abubaker, J. , Douds, D. D. , Allen, J. W. , … Shachar‐Hill, Y. (2005). Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature, 435, 819–823. 10.1038/nature03610 [DOI] [PubMed] [Google Scholar]
- Ishikawa, S. , Maekawa, M. , Arite, T. , Onishi, K. , Takamure, I. , & Kyozuka, J. (2005). Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant and Cell Physiology, 46, 79–86. 10.1093/pcp/pci022 [DOI] [PubMed] [Google Scholar]
- Iyer‐Pascuzzi, A. S. , Jackson, T. , Cui, H. , Petricka, J. J. , Busch, W. , Tsukagoshi, H. , & Benfey, P. N. (2011). Cell identity regulators link development and stress responses in the Arabidopsis root. Developmental Cell, 21, 770–782. 10.1016/j.devcel.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalo, P. , Gleason, C. , Edwards, A. , Marsh, J. , Mitra, R. M. , Hirsch, S. , … Oldroyd, G. E. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science, 308, 1786–1789. 10.1126/science.1110951 [DOI] [PubMed] [Google Scholar]
- Kamachi, K. , Yamaya, T. , Mae, T. , & Ojima, K. (1991). A role for glutamine‐synthetase in the remobilization of leaf nitrogen during natural senescence in rice leaves. Plant Physiology, 96, 411–417. 10.1104/pp.96.2.411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka, H. , Dun, E. A. , Lopez‐Obando, M. , Brewer, P. B. , de Saint, Germain A. , Rameau, C. , … Kyozuka, J. (2016). Phloem transport of the receptor, DWARF14 protein, is required for full function of strigolactones. Plant Physiology, 172, 1844–1852. 10.1104/pp.16.01212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik, Y. , Delaux, P. M. , Resnick, N. , Mayzlish‐Gati, E. , Wininger, S. , Bhattacharya, C. , … Koltai, H. (2011). Strigolactones affect lateral root formation and root‐hair elongation in Arabidopsis . Planta, 233, 209–216. 10.1007/s00425-010-1310-y [DOI] [PubMed] [Google Scholar]
- Kobae, Y. , Kameoka, H. , Sugimura, Y. , Saito, K. , Ohtomo, R. , Fujiwara, T. , & Kyozuka, J. (2018). Strigolactone biosynthesis genes of rice is required for the punctual entry of arbuscular mycorrhizal fungi into the roots. Plant and Cell Physiology, 59, 544–553. 10.1093/pcp/pcy001 [DOI] [PubMed] [Google Scholar]
- Kohlen, W. , Charnikhova, T. , Liu, Q. , Bours, R. , Domagalska, M. A. , Beguerie, S. , … Ruyter‐Spira, C. (2011). Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis . Plant Physiology, 155, 974–987. 10.1104/pp.110.164640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues, D. , Delaux, P. M. , Formey, D. , Lelandais‐Briere, C. , Fort, S. , Cottaz, S. , … Combier, J. P. (2012). The microRNA miR171 h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. The Plant Journal, 72, 512–522. 10.1111/j.1365-313X.2012.05099.x [DOI] [PubMed] [Google Scholar]
- Lin, H. , Wang, R. X. , Qian, Q. , Yan, M. X. , Meng, X. B. , Fu, Z. M. , … Wang, Y. H. (2009). DWARF27, an iron‐containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. The Plant Cell, 21, 1512–1525. 10.1105/tpc.109.065987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Kohlen, W. , Lillo, A. , Op den Camp, R. , Ivanov, S. , Hartog, M. , … Geurts, R. (2011). Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS‐Type transcription factors NSP1 and NSP2. The Plant Cell, 23, 3853–3865. 10.1105/tpc.111.089771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Raez, J. A. , Charnikhova, T. , Gomez‐Roldan, V. , Matusova, R. , Kohlen, W. , De Vos, R. , … Bouwmeester, H. (2008). Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytologist, 178, 863–874. 10.1111/j.1469-8137.2008.02406.x [DOI] [PubMed] [Google Scholar]
- Maruyama‐Nakashita, A. , Nakamura, Y. , Tohge, T. , Saito, K. , & Takahashi, H. (2006). Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell, 18, 3235–3251. 10.1105/tpc.106.046458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D. R. , Schuler, M. A. , Paquette, S. M. , Werck‐Reichhart, D. , & Bak, S. (2004). Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiology, 135, 756–772. 10.1104/pp.104.039826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proust, H. , Hoffmann, B. , Xie, X. , Yoneyama, K. , Schaefer, D. G. , Yoneyama, K. , … Rameau, C. (2011). Strigolactones regulate protonema branching and act as a quorum sensing‐like signal in the moss Physcomitrella patens . Development, 138, 1531–1539. 10.1242/dev.058495 [DOI] [PubMed] [Google Scholar]
- Rasmussen, A. , Mason, M. , De Cuyper, C. , Brewer, P. B. , Herold, S. , Agusti, J. , … Beveridge, C. A. (2012). Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiology, 158, 1976–1987. 10.1104/pp.111.187104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter‐Spira, C. , Kohlen, W. , Charnikhova, T. , van Zeijl, A. , van Bezouwen, L. , de Ruijter, N. , … Bouwmeester, H. (2011). Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiology, 155, 721–734. 10.1104/pp.110.166645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto, Y. , Sado, A. , Asami, K. , Hanada, A. , Umehara, M. , Akiyama, K. , & Yamaguchi, S. (2014). Carlactone is an endogenous biosynthetic precursor for strigolactones. Proceedings of the National Academy of Sciences of the United States of America, 111, 1640–1645. 10.1073/pnas.1314805111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, P. , Raedts, J. , Portyanko, V. , Debelle, F. , Gough, C. , Bisseling, T. , & Geurts, R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor‐induced transcription. Science, 308, 1789–1791. 10.1126/science.1111025 [DOI] [PubMed] [Google Scholar]
- Smith, S. E. , & Read, D. J. (2008). Mycorrhizal symbiosis, 3rd ed. Cambridge, MA: Academic Press. [Google Scholar]
- Sugimoto, Y. , & Ueyama, T. (2008). Production of (+)‐5‐deoxystrigol by Lotus japonicus root culture. Phytochemistry, 69, 212–217. 10.1016/j.phytochem.2007.06.011 [DOI] [PubMed] [Google Scholar]
- Sun, H. , Tao, J. , Liu, S. , Huang, S. , Chen, S. , Xie, X. , … Xu, G. (2014). Strigolactones are involved in phosphate‐ and nitrate‐deficiency‐induced root development and auxin transport in rice. Journal of Experimental Botany, 65, 6735–6746. 10.1093/jxb/eru029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, H. , & Kusaba, M. (2015). Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis . Plant Physiology, 169, 138–147. 10.1104/pp.15.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara, M. (2011). Strigolactone, a key regulator of nutrient allocation in plants. Plant Biotechnology, 28, 429–437. 10.5511/plantbiotechnology.11.1109a [DOI] [Google Scholar]
- Umehara, M. , Hanada, A. , Magome, H. , Takeda‐Kamiya, N. , & Yamaguchi, S. (2010). Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant and Cell Physiology, 51, 1118–1126. 10.1093/pcp/pcq084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara, M. , Hanada, A. , Yoshida, S. , Akiyama, K. , Arite, T. , Takeda‐Kamiya, N. , … Yamaguchi, S. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature, 455, 195–200. 10.1038/nature07272 [DOI] [PubMed] [Google Scholar]
- Umehara, M. , Mengmeng, C. , Akiyama, K. , Akatsu, T. , Seto, Y. , Hanada, A. , … Yamaguchi, S. (2015). Structural requirements of strigolactones for shoot branching inhibition in rice and Arabidopsis . Plant and Cell Physiology, 56, 1059–1072. 10.1093/pcp/pcv028 [DOI] [PubMed] [Google Scholar]
- van Zeijl, A. , Liu, W. , Xiao, T. T. , Kohlen, W. , Yang, W. C. , Bisseling, T. , & Geurts, R. (2015). The strigolactone biosynthesis gene DWARF27 is co‐opted in rhizobium symbiosis. BMC Plant Biology, 15, 260 10.1186/s12870-015-0651-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, M. T. , Brewer, P. B. , Bussell, J. D. , Smith, S. M. , & Beveridge, C. A. (2012). The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiology, 159, 1073–1085. 10.1104/pp.112.196253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X. N. , Yoneyama, K. , & Yoneyama, K. (2010). The strigolactone story. Annual Review of Phytopathology, 48, 93–117. 10.1146/annurev-phyto-073009-114453 [DOI] [PubMed] [Google Scholar]
- Yamada, Y. , Furusawa, S. , Nagasaka, S. , Shimomura, K. , Yamaguchi, S. , & Umehara, M. (2014). Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta, 240, 399–408. 10.1007/s00425-014-2096-0 [DOI] [PubMed] [Google Scholar]
- Yoneyama, K. , Awad, A. A. , Xie, X. N. , Yoneyama, K. , & Takeuchi, Y. (2010). Strigolactones as germination stimulants for root parasitic plants. Plant and Cell Physiology, 51, 1095–1103. 10.1093/pcp/pcq055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama, K. , Xie, X. , Kim, H. I. , Kisugi, T. , Nomura, T. , Sekimoto, H. , … Yoneyama, K. (2012). How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta, 235, 1197–1207. 10.1007/s00425-011-1568-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama, K. , Xie, X. , Kisugi, T. , Nomura, T. , & Yoneyama, K. (2013). Nitrogen and phosphorus fertilization negatively affects strigolactone production and exudation in sorghum. Planta, 238, 885–894. 10.1007/s00425-013-1943-8 [DOI] [PubMed] [Google Scholar]
- Yoneyama, K. , Xie, X. , Kusumoto, D. , Sekimoto, H. , Sugimoto, Y. , Takeuchi, Y. , & Yoneyama, K. (2007). Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5‐deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta, 227, 125–132. 10.1007/s00425-007-0600-5 [DOI] [PubMed] [Google Scholar]
- Yoneyama, K. , Yoneyama, K. , Takeuchi, Y. , & Sekimoto, H. (2007). Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta, 225, 1031–1038. 10.1007/s00425-006-0410-1 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , van Dijk, A. D. , Scaffidi, A. , Flematti, G. R. , Hofmann, M. , Charnikhova, T. , … Bouwmeester, H. J. (2014). Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nature Chemical Biology, 10, 1028–1033. 10.1038/nchembio.1660 [DOI] [PubMed] [Google Scholar]
- Zou, J. , Zhang, S. , Zhang, W. , Li, G. , Chen, Z. , Zhai, W. , … Zhu, L. (2006). The rice HIGH‐TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. The Plant Journal, 48, 687–698. 10.1111/j.1365-313X.2006.02916.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


