Abstract
Expression of the mammalian DNA demethylase enzyme TET3 in plants can be used to induce hypomethylation of DNA. In tomato lines that express a TET3 transgene, we observed distinct phenotypes including an increase in the length and number of leaves of primary shoots. As these changes resemble phenotypes observed in plants with strong expression of SELF PRUNING (SP), a member of the PEBP/CETS family, we investigated in TET3 lines the expression levels of members of the PEBP/CETS gene family, which affect shoot architecture and growth of sympodial units in tomato. We did not detect any changes in SP expression in TET3 lines, but for CEN1.1, a putative family member that has not been functionally characterized, we identified changes in gene expression that corresponded to hypomethylation in the upstream region. In tomato wild type, CEN1.1 is expressed in roots, petals, and shoot apices but not in mature leaves. In contrast, in TET3 transformants, the CEN1.1 gene became hypomethylated and activated in leaves. Ectopic expression of CEN1.1 in tomato caused similar phenotypes to those seen in TET3 transformants. Vegetative growth was increased, resulting both in a delay in inflorescence development and in an instability of the inflorescences, which frequently reverted to a vegetative state. Ectopic expression of CEN1.1 in Arabidopsis thaliana also caused floral repression. Our data suggest that the phenotypes observed in TET3 lines are a consequence of ectopic activation of CEN1.1, which promotes vegetative growth, and that CEN1.1 expression is sensitive to DNA methylation changes.
Keywords: CETS gene, DNA demethylation, DNA methylation, floral repressor, PEBP protein, Solanum lycopersicum, Ten‐Eleven‐Translocation (TET) proteins
1. INTRODUCTION
Organization of shoot architecture in flowering plants is extremely important for the normal development of the plant, both under usual environmental conditions and when the plant is under stress. For crop plants such as tomato, shoot architecture also has great economic importance, with different patterns being preferred for different purposes. For example, for mechanically harvested processing tomatoes, tomato plants with determinate growth have a higher yield, while tomato varieties that grow indeterminately are better suited to produce tomatoes that are eaten fresh and require continuous market delivery (Jiang et al., 2013). Tomato is an example of a plant species with a sympodial growth pattern, composed of a series of determinate meristems. The primary shoot of tomato terminates with an inflorescence after 8–12 compound leaves (McGarry & Ayre, 2012), but growth continues from the uppermost axillary meristem (Lifschitz et al., 2006). After this point, the shoot is formed from repeating sympodial units consisting of three leaves and terminating with an inflorescence. Upward growth of the shoot is again continued from the most proximal axillary bud of the previous sympodial unit in an indeterminate fashion (Lifschitz et al., 2006).
The establishment of this pattern relies on the balance between the expression levels of genes in the tomato PEBP gene family (phosphatidylethanolamine‐binding protein), also called the CETS (CENTRORADIALIS/TERMINAL FLOWER 1/SELF PRUNING) gene family after its founding members (Shalit et al., 2009). This family is present in a large variety of species where it plays a role in mechanisms as diverse as bulb induction in onions and formation of needles in Norway spruce (Karlgren, Gyllenstrand, Clapham, & Lagercrantz, 2013; Lee, Baldwin, Kenel, McCallum, & Macknight, 2013; Wickland & Hanzawa, 2015). SFT (SINGLE FLOWER TRUSS), the tomato homolog of the Arabidopsis thaliana gene FT (FLOWERING LOCUS T; Lifschitz et al., 2006), and SP (SELF PRUNING), the tomato homolog of the Arabidopsis gene TFL1 (TERMINAL FLOWER 1; Pnueli et al., 1998), are the best described of the genes in this family in tomato. Mutations in SFT result in delayed flowering (Lifschitz et al., 2006) while sp tomato mutants initially flower after the normal number of leaves has been produced but afterward flowers switch to determinate growth (Shalit et al., 2009). Overexpression of the SFT gene causes early flowering, the opposite phenotype to overexpressing SP, which results in delayed termination of the primary shoot and increased numbers of leaves per sympodial unit (Lifschitz et al., 2006; McGarry & Ayre, 2012; Pnueli et al., 1998). Analysis of double mutants indicates that SP counteracts the florigenic effect of SFT in a dosage‐responsive manner (Molinero‐Rosales, Latorre, Jamilena, & Lozano, 2003; Shalit et al., 2009). In addition to SP and SFT, there are several other recognized members of the CETS gene family in tomato (Cao et al., 2016; Carmel‐Goren, Liu, Lifschitz, & Zamir, 2003). Three of these (SP5G, SP5G2, and SP5G3) have been shown to have a role in delaying flowering, with knockdown lines of these genes showing early flowering and overexpression in Arabidopsis causing delayed flowering (Cao et al., 2016; Chitwood et al., 2013). Expression of SP5G, SP5G2, and SP5G3 is affected by day length (Cao et al., 2016). Understanding the role of the genes in this family is an important tool to improve tomato crop yield or harvest index (yield per plant weight; Park et al., 2014; Soyk et al., 2017).
The likelihood of gene expression is frequently affected by epigenetic modifications to the gene, such as histone modifications and DNA methylation (Zilberman, Gehring, Tran, Ballinger, & Henikoff, 2007). DNA methylation occurs through the action of DNA methyltransferases and the presence of DNA methylation in the promoter of a gene is usually repressive, resulting in the silencing of that gene. Given the importance of tomato as a crop plant and the involvement of methylation in the ripening process of tomato (Liu et al., 2015; Zhong et al., 2013), a better understanding of the role of methylation in tomato is extremely important. Expression of the catalytic domain of the mammalian DNA demethylase TET3 (TET3c) in Arabidopsis has previously been shown to be capable of causing DNA demethylation (Hollwey, Watson, & Meyer, 2016).
Here, by transforming the TET3c construct into tomato, we observed specific phenotypes and demonstrated that expression of CEN1.1, a member of the CETS gene family, is affected by DNA methylation upstream of the start codon. We show that hypomethylation caused by TET3c results in the activation of this CETS family member. We demonstrate that ectopic expression of either TET3c or CEN1.1 causes common phenotypes in tomato plants, including an instability of the transition to an inflorescence, delayed growth, and an increase in the number of leaves between inflorescences. Ectopic expression of CEN1.1 in Arabidopsis thaliana also results in an increase in the number of rosette leaves and a delay in flowering.
2. MATERIALS AND METHODS
2.1. Vector construction and plant transformation
The TET3c vector was constructed as described in Hollwey et al. (2016). The CEN1.1 vector was constructed by amplification of the CEN1.1 genomic region from tomato DNA using primers GGGAAGCTTGGCACGTTGATTGGTTTTTCG + GGGAATTCACAAGCAAATGAGTAGGACAAACA. It was then cloned into the HindIII/EcoRI site of pGreen II 0029. The vectors were transferred into Agrobacterium tumefaciens for leaf disk transformation (Rai et al., 2012) of a EZCBT1 tomato variety and floral dip transformation of Arabidopsis thaliana (Col‐0; Clough & Bent, 1998). Tomato transformation was carried out at the premises of ENZA ZADEN, Enkhuizen, The Netherlands.
2.2. Plant material
Plants were grown in a growth chamber under long day conditions (16 hr light, 8 hr dark, 23°C, 42% humidity). At the age of 5 weeks, tomato plants were transferred to a glasshouse. All samplings for nucleic acid extractions were done between 8 and 10 a.m. to avoid possible circadian variations in gene expression or DNA methylation.
2.3. Expression analyses
RNA for expression analysis was extracted as described in Stam et al. (2000). DNA was removed using the TURBO DNase kit (Ambion applied Biosystems) and converted to cDNA using M‐MLV reverse transcriptase and oligo‐dT primers (Invitrogen) according to the manufacturer's instructions. Semiquantitative PCR was carried out using MyTaq Red DNA Polymerase (Bioline) and qPCR was carried out using SsoFast Eva Green Supermix (Bio‐Rad) according to the manufacturer's instructions. cDNA levels were normalized using eukaryotic translation initiation factor 3 primers GAGCGATGGATGGTGAATCT + TTGTACGTGCGTCCAGAAAG.
CEN1.1 expression was analyzed using primers GACCCTGATGCTCCAAGTCC + TGGCTGCAGTTTCTCTCTGG.
2.4. DNA methylation analysis
Genomic DNA for bisulfite sequencing was extracted according to Vejlupkova and Fowler (2003) with some modifications. Tissue for the SAP methylation analysis was isolated using a dissection microscope from FFPE sections of tomato shoot apices made according to Vitha, Baluška, Jasik, Volkmann, and Barlow (2000). Bisulfite treatment was carried out using the EZ DNA Methylation‐Lightning kit (Zymo Research). Bisulfite‐treated DNA was amplified using primers AAYTTTTGGGGTGTGAGTTAGA + TCCACCCATTTCATTAACCACC and GTGAGGTGGGGTGTTAAAGAATGA + CACCRATRTAACACTCCACCT to amplify part of the region upstream of the CEN1.1 gene. Oxidative bisulfite sequencing was performed as described in (Booth et al., 2013) to quantify levels of 5‐methylcytosine and subtracted from bisulfite sequencing data (which contains 5‐methylcytosine and 5‐hydroxymethylcytosine) to calculate levels of 5‐hydroxymethylcytosine; 10–20 clones were sequenced per sample. Sequencing data were analyzed using the online CYMATE tool (Hetzl, Foerster, Raidl, & Scheid, 2007) and the program SequenceFileConverter (J. Royle).
3. RESULTS
3.1. TET3c tomato plants display abnormal growth phenotypes and ectopically express CETS family genes
The TET3c construct, which consists of the catalytic domain of the mammalian DNA demethylase TET3 under a constitutive 35S promoter, has previously been shown to induce DNA hypomethylation in Arabidopsis (Hollwey et al., 2016). TET3c was transformed into tomato plants in order to identify genes and processes affected by DNA methylation in tomato. Transgenic tomato plants, which strongly expressed TET3c, displayed a broad range of phenotypes, in particular an increase in primary shoot length and in the number of leaves in the primary shoot (Fig. S1a–d). These phenotypes have previously been observed in 35S::SP plants (Shalit et al., 2009), and we therefore analyzed cDNA from TET3c tomato for changes in expression of SP and a selection of other genes from the CETS/PEBP gene family (Fig. S2a). Gene expression changes are seen in three genes, only two of which, CEN1.1 and SP9D, showed a consistent increase in its expression in all TET3c lines in comparison with wild type. CEN1.1 (Solyc03 g026050.2.1) and SP9D (Solyc09 g009560.1.1) were not expressed in leaves from 5‐week‐old wild‐type tomato plants, but were expressed in leaves from 5‐week‐old TET3c tomato transformants (Figure 1a, Fig. S2a).
Figure 1.
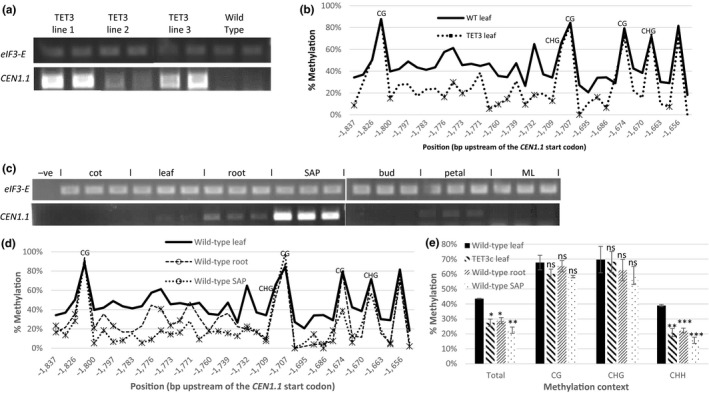
CEN1.1 expression correlates with reduced methylation at CHH sites in TET3c and wild‐type tomato. (a) Ectopic expression of CEN1.1 can be seen in the leaves of different lines of TET3c tomato plants (5 weeks old) using semiquantitative RT‐PCR on cDNA pools (n > 5). cDNA levels were normalized using the constitutively expressed eukaryotic translation initiation factor 3 subunit E. Two replicates are shown for each pool. (b) Reduced DNA methylation levels upstream of the CEN1.1 gene were seen in TET3c leaves in comparison with wild‐type leaves. A 206‐bp region was analyzed by bisulfite sequencing. Three biological replicates were averaged for each genotype. CG and CHG sites are indicated; all unlabeled sites are CHH sites. Points where methylation is significantly different are marked with a star (p < .05, calculated using Student's two‐tailed t test). (c) CEN1.1 is expressed in the shoot apex of wild‐type tomato, as well as roots and petals. Semiquantitative RT‐PCR was used to analyze expression of CEN1.1 in a collection of cDNA pools (n > 3) from different wild‐type tomato tissues. cDNA levels were normalized using the constitutively expressed eukaryotic translation initiation factor 3 subunit E. The negative control (−ve) corresponds to a lane where DNA was not added. Cot = cotyledon, leaf = true leaves of 5‐week‐old tomato, SAP = shoot apices, ML = leaves of a mature (20 weeks old) tomato. (d) Reduced methylation levels were seen in root and SAP DNA where CEN1.1 is expressed using the same 206‐bp region previously analyzed in TET3c tomato. Bisulfite sequencing was used to analyze methylation levels in 5‐week‐old leaf, root, and shoot apex from wild‐type tomato. Three biological replicates were averaged for each tissue. CG and CHG sites are indicated; all unlabeled sites are CHH sites. Points where methylation is significantly different are marked with a star (p < .05, calculated using Student's two‐tailed t test). (e) Methylation levels were reduced in the CHH context in TET3c leaves, wild‐type roots and wild‐type SAP. Graphs show averages with error bars representing standard error. *p < .05, **p < .005, ***p < .0005, ns = not significant, calculated using Student's two‐tailed t test
3.2. TET3c causes demethylation upstream of the CEN1.1 gene
We screened the tomato database (Zhong et al., 2013) for methylation patterns of the known CETS gene family members and found that several members of the CETS gene family are methylated in the first 3 kb upstream of the transcriptional start site. With 37% total methylation, CEN1.1 shows the highest levels of DNA methylation of all CETS genes analyzed. This includes 31% methylation of cytosines in a CHH (H=C, T or A) context, the context which shows the lowest methylation levels in plants. In comparison, SP9D had low levels of methylation upstream of the transcriptional start site, and therefore, further analysis was focused on CEN1.1. To investigate methylation levels upstream of CEN1.1, bisulfite sequencing was performed on DNA from leaf tissue of wild‐type and TET3c plants. A 200‐bp region with dense methylation in the tomato methylation database and homology to the tomato RK01 TRIM retrotransposon was chosen for analysis. DNA from wild‐type leaf tissue where CEN1.1 was not expressed showed methylation levels of at least 40% for most CHH sites in this region. In TET3c plants with ectopic expression of CEN1.1, methylation levels were reduced by at least 50% for most CHH sites (Figure 1b, Fig. S2b). To confirm that the reduction in 5mC levels was caused by TET3c, we screened the region for 5‐hydroxymethylcytosine, a derivative of 5‐methylcytosine produced by TET3 oxidation, which serves as a marker for TET3c‐mediated demethylation (Ito et al., 2010). Oxidative bisulfite sequencing showed that a significant increase in levels of 5‐hydroxymethylcytosine occurred in TET3c tissue compared to wild‐type tissue (Fig. S2c).
3.3. Tissue‐specific expression of CEN1.1 correlates with DNA methylation
CEN1.1 is characterized as a TFL1‐like member of the CETS/PEBP family based on its DNA sequence (Cao et al., 2016; Chardon & Damerval, 2005). Overexpression of other TFL1‐like genes, including SP in tomato and RCN1/2 in rice, causes a delay in flowering, as does TFL1 itself (Nakagawa, Shimamoto, & Kyozuka, 2002; Pnueli et al., 1998; Ratcliffe et al., 1998), suggesting that CEN1.1 may be the cause of the phenotype observed in the 35S::TET3c plants.
We used semiquantitative RT‐PCR to analyze the expression patterns of CEN1.1 in wild‐type tomato. CEN1.1 was not expressed in plant leaves in both juvenile (5 weeks old) and mature (20 weeks old) tomato plants, but was expressed strongly in the shoot apex and also weakly in roots and petals (Figure 1c). Bisulfite sequencing was used to analyze whether expression of CEN1.1 correlated with hypomethylation in wild‐type tissues as it does in TET3c plants. Methylation levels were reduced in root and shoot apex (SAP) tissue where CEN1.1 is expressed, in comparison with leaf tissue where CEN1.1 is silenced (Figure 1d, Fig. S2d). In TET3c, root and SAP tissues, hypomethylation was observed at CHH sites, while overall CG and CHG methylation did not change significantly (Figure 1e).
3.4. Ectopic expression of CEN1.1 causes increased vegetative meristematic identity
As discussed earlier, TET3c tomato plants ectopically expressing CEN1.1 displayed increased primary stem length and increased stem thickness. To determine if these phenotypes were being caused by CEN1.1 expression, the CEN1.1 gene was cloned behind the constitutive 35S promoter in a plant expression vector and transformed into tomato. Transformants were selected that expressed the CEN1.1 transgene, and progeny plants were analyzed 18 weeks after germination.
35S::CEN1.1 tomato plants displayed an increased propensity for vegetative growth in comparison with control plants, as had the TET3c plants. Increases in primary shoot length, the number of leaves between inflorescences, and stem circumference were again observed (Figure 2a‐c). Despite this increase in the number of leaves between inflorescences, 35S::CEN1.1 plants were smaller overall than control plants, due to a significant reduction in the number of inflorescences present in the plant at 18 weeks (Figure 2d), a phenotype which had not been observed in TET3c plants. The increased level of vegetative growth could also be seen elsewhere. Vegetative meristems grew from the rachis of complex leaves in 73% of 35S::CEN1.1 plants (n = 33), but not in the tomato control plants (n = 25; Figure 3a). Stems of 35S::CEN1.1 plants were frequently fasciated (Figure 3b), a possible cause of their increased circumference (Figure 2c). In the inflorescence, we observed unusual vegetative growth. Leafy inflorescences on the 35S::CEN1.1 plants produced fruits and then switched back to a vegetative state for a time, before returning to an inflorescent state (Figure 3c). This pattern was reiterated on multiple branches that emerged in the inflorescences, suggesting that neither floral nor vegetative identity could be stably maintained. The reversion to vegetative meristematic growth even continued in a small number of tomato fruit, with a vegetative meristem growing from the top of the fruit in 0.8% of fruit (n = 352; Figure 3d).
Figure 2.
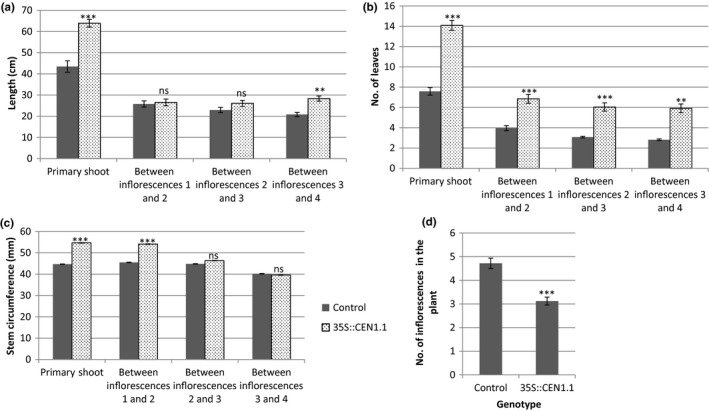
35S::CEN1.1 plants have similar increased vegetative growth features as observed in 35S::TET3c plants and also show a reduction in the number of sympodial units. (a, b) 35S::CEN1.1 plants (n = 34) have more leaves between each inflorescence than the control (n = 25), and also a greater primary shoot length. The number of leaves and distance between each inflorescence is shown separately, with the distance to the first inflorescence labeled as “Primary Shoot.” (c) Stem circumference is increased in 35S:CEN1.1 plants in comparison with tomato control plants. (d) 35S::CEN1.1 plants have a reduced number of inflorescences. Graphs show averages with error bars representing standard error. **p < .005, ***p < .0005, ns = not significant, calculated by Student's two‐tailed t test
Figure 3.
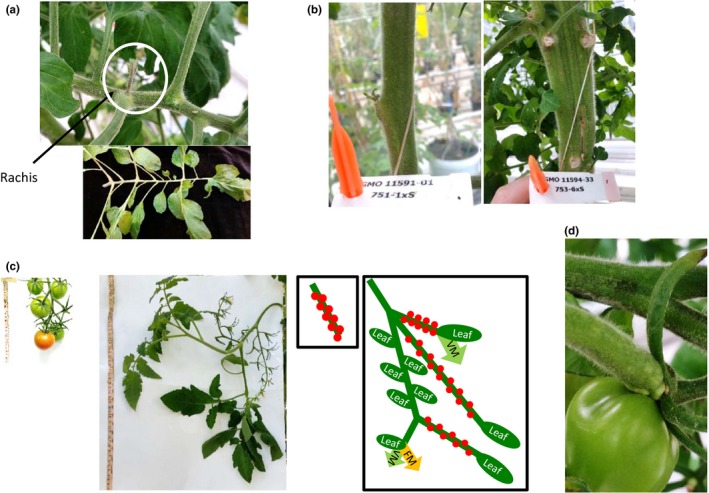
Increased vegetative growth in 35S::CEN1.1 plants results in abnormal phenotypes in a number of tissues. (a) Meristems grow directly out of the rachis of 35S::CEN1.1 tomato leaves (above), indicated by the white circle. (b) 35S::CEN1.1 plants (right) frequently show stem fasciation in older stems, not present in the control (left), resulting in increased stem circumference. (c) Images and diagrams comparing normal inflorescence growth and the leafy inflorescences of 35S::CEN1.1 plants. Normal growth of a tomato inflorescence is shown on the left. A leafy inflorescence from a plant ectopically expressing CEN1.1 is shown on the right. Both inflorescences are the second inflorescence on the plant. The measuring tape is included as a size marker. VM = growth of a vegetative meristem, FM = growth of an inflorescence meristem. (d) Ectopic vegetative meristems emerging from the fruit of a 35S::CEN1.1 tomato
3.5. CEN1.1 leafy inflorescences switch between the inflorescence and vegetative state resulting in an increased number of flowers
Large numbers of inflorescences of 35S::CEN1.1 plants (76%, n = 76) were leafy in comparison with control plants (0%, n = 17), a phenotype which had also been seen in 18% of TET3c inflorescences (n = 51; Fig. S1d). Inflorescences were classified as leafy when they contained multiple leaves and at least one vegetative meristem. Inflorescences containing leaves have also been described for lines that overexpress SP, although the reported effects are less severe than the ones we observed (Pnueli et al., 1998), and in sft, macrocalyx, or jointless mutants (Quinet, 2006; Vrebalov et al., 2002). Expression of these genes remains unchanged in the 35S::CEN1.1 tomato, which argues against CEN1.1 overexpression altering their expression (Fig. S3). While these abnormal, leafy inflorescences contained large quantities of vegetative material, they also produced a larger number of flowers due to the large branched nature of the inflorescence. Therefore, 35S::CEN1.1 inflorescences also produce more flowers on average than wild‐type inflorescences (Figure 4a), although the number of flowers on an inflorescence varied significantly, ranging from 11 to 60. This phenotype becomes more obvious when vegetative material is removed during the development of the inflorescence (Figure 4b). While it required more time for 35S::CEN1.1 lines to produce fully ripe flowers (Figure 4c), there was no significant difference in fruit size or weight (Figure 4d). Mutants of COMPOUND INFLORESCENCE (S) or ANANTHA (AN) can also produce branched inflorescences with an increased number of flowers (Lippman et al., 2008), but expression of these genes was unchanged in the CEN1.1 tomato (Fig. S3). Among the plants that we had selected on the basis that they contained the 35S::CEN1.1 construct, we identified three plants that no longer expressed the transgene. Silencing of transgenes after successful transformation can subsequently become silenced in plants for many reasons (Meyer & Heidmann, 1994). All three plants resembled the wild‐type phenotype (Figure 4e, Fig. S4), providing further support that the observed phenotypes result from ectopic CEN1.1 expression.
Figure 4.
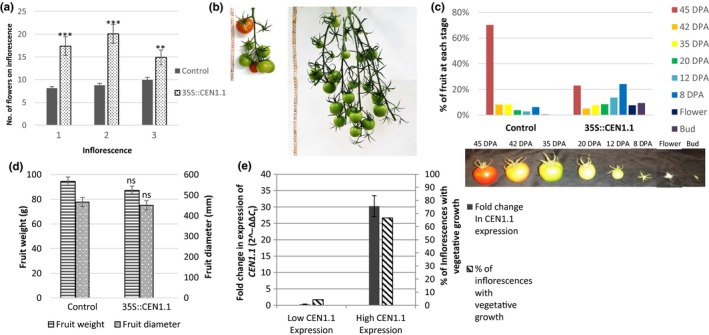
Inflorescences of 35S::CEN1.1 plants produced an increased number of flowers but were delayed in development. (a) Inflorescences of CEN1.1 (n = 34) produced an increased number of flowers compared to the control (n = 28). (b) An inflorescence from a 35S::CEN1.1 plant from which all leaves and vegetative meristems were removed as the inflorescence developed and a control inflorescence with a normal number of fruit. Both are the first inflorescence on the plant. (c) Fruits on 35S::CEN1.1 plants (n = 208) were less developed than fruits on the equivalent inflorescence of a control plant (n = 352). Fruits on the first inflorescence were categorized according to ripeness, or recorded as flowers. DPA—days postanthesis. (d) Ripe fruits were similar in size and weight on control and 35S::CEN1.1 plants. The widest diameter of each tomato fruit was used as a measurement; 11 plants were measured each for the control and CEN1.1. (e) Ectopic expression of CEN1.1 correlates with the leafy inflorescence phenotype. Plants categorized as “High CEN1.1 Expression” (greater than fivefold increase in CEN1.1 expression in comparison with the control, n = 18) were compared to plants categorized as “Low CEN1.1 Expression” (CEN1.1 expression did not increase in comparison with the control, n = 3). Graphs show averages with error bars representing standard error. *p < .05, **p < .005, ***p < .0005, ns = not significant, calculated by Student's two‐tailed t test
3.6. CEN1.1 expression in Arabidopsis thaliana delays or prevents flowering
To further verify the action of CEN1.1 as a floral repressor, the 35S::CEN1.1 construct was transferred into Arabidopsis thaliana. Eight independent transformant lines were grown under long day conditions. Four plants failed to flower completely, dying after 12 weeks without flowering. The other four plants did flower at late stages. While Col‐0 wild type flowered on average 39 days after germination when seven rosette leaves had been produced, 35S::CEN1.1 transformants flowered on average 65 days after germination when 53 rosette leaves had been produced (Figure 5).
Figure 5.
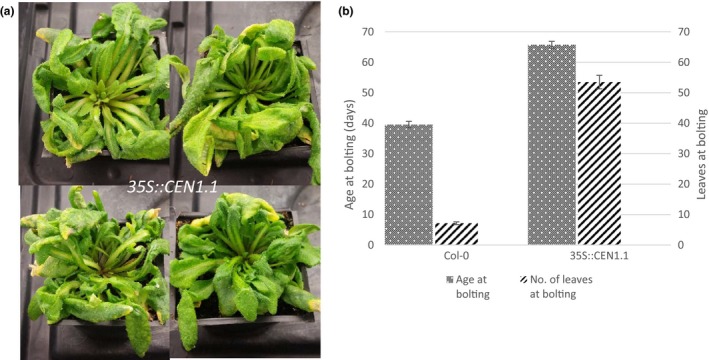
Arabidopsis thaliana transformed with 35S::CEN1.1 showed an increase in vegetative growth and flowering was delayed or absent. (a) Arabidopsis plants containing 35S::CEN1.1 had not flowered after 9 weeks but continued to produce rosette leaves. (b) 50% of 35S::CEN1.1 Arabidopsis (n = 8) were able to flower. Those that did flower, flowered late and produced a higher number of rosette leaves prior to bolting (n = 4) in comparison with Col‐0 (n = 12). Graphs show averages with error bars representing standard error
4. DISCUSSION
4.1. CEN1.1 is the first tomato CETS gene with a demonstrated link to DNA methylation
Thirteen members of the CETS/PEBP gene family are characterized in tomato, and five of these genes have been shown to act in the control of shoot architecture and timing of the floral transition (Cao et al., 2016; Shalit et al., 2009). Our analysis of CEN1.1 demonstrates that it also plays a role in this process and that it is the first of these genes to be shown to be affected by DNA methylation. The normal biological function of CEN1.1 is unknown, but its expression pattern (strong expression only in the shoot apex) suggests that it may act in the regulation of shoot architecture. Intensity of activation of CEN1.1 expression correlates with increasing hypomethylation in its promoter, suggesting that the expression of CEN1.1 is connected to DNA methylation of CHH sites in the promoter. The CHH methylation levels upstream of the CEN1.1 gene are unusually high, compared to an overall level of 8.6% in the tomato genome (Zhong et al., 2013). High levels of methylation in all three contexts is known as dense methylation, which has been shown to be dependent on the MET1 gene in some genes in Arabidopsis (Watson, Hawkes, & Meyer, 2014), but it is unknown if this is the case in tomato.
4.2. CEN1.1 acts as a floral repressor in tomato and Arabidopsis
Unsurprisingly, given its similarity and close phylogenetic relationship to SP, ectopic expression of CEN1.1 has similar effects to ectopic expression of SP. Expression of both genes causes delayed termination of the primary shoot, with an increase in the number of leaves in the primary shoot and in subsequent sympodial units. In Arabidopsis, expression of CEN1.1 either delayed or prevented flowering. Similarly, strong effects of CETS genes have been reported in other species; for example, the expression of the Antirrhinum floral repressor gene CEN in tobacco resulted in significant delays in flowering, with some plants being delayed for over 10 months, and one never flowering at all (Amaya, Ratcliffe, & Bradley, 1999). The delay in flowering caused by CEN1.1 in Arabidopsis was more severe than was observed when the Arabidopsis CEN1.1 homologues, TFL1 and BFT, were expressed under the 35S promoter (Mimida et al., 2001; Yoo et al., 2010). As would be expected, ectopic expression of the tomato SFT gene in Arabidopsis has the opposite effect to CEN1.1, causing early flowering after the production of four rosette leaves (Cao et al., 2016).
CEN1.1 may bind the same targets as SP, resulting in activation of the same pathway or may act through a different route. Like the rest of the CETS gene family, CEN1.1 possesses a PEBP domain, but the role of this domain in the function of the gene family has not yet been clarified. SP has been shown to interact with several proteins in tomato including a kinase, 14‐3‐3 proteins and a putative bZIP transcription factor (Pnueli et al., 2001; as does FT in Arabidopsis (Abe et al., 2005)), but the full pathway has not been elucidated and it is not known how the other known floral repressors in the tomato CETS gene family exert their influence (Cao et al., 2016). The development of leaves on inflorescences induced by ectopic expression of CEN1.1 had also been observed in sft, macrocalyx, and jointless mutants (Quinet, 2006; Vrebalov et al., 2002). There was no indication that any of these genes altered their expression in CEN1.1 transformants, which argues against their involvement in the phenotype observed in the transformants.
4.3. Increased vegetative growth caused by CEN1.1 appears differently in various tissues and paradoxically increases total fruit yield
Ectopic expression of CEN1.1 also stimulates vegetative growth elsewhere in tomato plants, which seems to be more severe than similar phenotypes described in 35S::SP tomato plants (Quinet, 2006). This presents differently in different plant tissues. In leaves, ectopic expression results in the presence of vegetative meristems emerging from the leaf. In stems, ectopic expression results in fasciation of the stem, and thus thicker stems. Inflorescences with ectopic expression of 35S::CEN1.1 are unable to finally commit to the inflorescent state, but switch repeatedly between a vegetative and an inflorescent state. This results in a greater quantity of fruit from a single tomato inflorescence, which could be advantageous. The average 35S::CEN1.1 inflorescence produces 22 flowers, in comparison with the 10 produced by a control inflorescence. Despite the reduced number of inflorescences per plant (3.1 c.f. 4.7), this still results in an increased yield of fruit from a single tomato plant, with an average of 68 fruits per 35S::CEN1.1 plant and only 47 fruits per control plant. Fruits from 35S::CEN1.1 plants, once ripe, have the same size as fruits from control plants, but more time is required for the fruits to become fully ripe. The median fruit on the first inflorescence of an 18‐week‐old control plant is ready to be removed (45 days postanthesis (D.P.A), while the median fruit on the first inflorescence of a 35S::CEN1.1 plant is still small and green (12 D.P.A.) on an 18‐week‐old plant. 35S::CEN1.1 plants would therefore require an extra 5 weeks for fruit to fully ripen, or 26% of the total growth time.
35S::CEN1.1 plants produce 45% more fruit than the control, although for them to ripen takes 26% longer. The reduced number of inflorescences per plant also means that 35S::CEN1.1 tomato plants are smaller despite the increase in the number of leaves between inflorescences, and therefore, more tomatoes can be produced in a smaller glasshouse space. However, pruning will be required to prevent effects on the harvest index (total yield per plant weight) due to the vegetative growth on the inflorescence.
4.4. CEN1.1 was identified using TET3c, which could be of use in identifying other methylation‐linked genes in tomato and other species
Expression of the mammalian demethylase TET3c in tomato facilitated the identification of the CEN1.1 gene. Phenotypes seen in 35S::CEN1.1 plants had already been observed in TET3c plants, although often at a lower frequency or intensity. This is to be expected, given that CEN1.1 expression due to TET3c‐mediated demethylation is likely to be less intense than the strong, constitutive expression under the 35S promoter.
Identification of the CEN1.1 gene illustrates that TET3c expression is a useful tool to discover previously unknown plant genes that are affected by DNA methylation changes. These may be otherwise difficult to detect, especially in species which are particularly susceptible to changes in DNA methylation. Arabidopsis mutants of the main methyltransferases are still viable. This allows high‐throughput analysis of changes in DNA methylation and gene expression, which can identify genes controlled by methylation. In contrast, species such as tomato and rice appear to be more sensitive to DNA methylation changes as they show more adverse effects when the enzymes involved in DNA methylation are lost (Liu et al., 2015; Ono et al., 2012). In tomato, null mutations of SlNRPE1, a component of the RdDM pathway, are lethal (Gouil & Baulcombe, 2016), and MET1 RNAi lines are not viable (Watson, 2013), making the identification of genes and processes affected by DNA methylation changes more challenging. TET3c expression may therefore offer an alternative in these species to gain a better understanding of which processes are controlled by DNA methylation.
AUTHOR CONTRIBUTIONS
P.M. and I.H. supervised the overall study, which was conducted largely by E.H with input from M.R.W. Tomato transformation and plant cultivation were carried out by M.R.W. and S.O. E.H. wrote the article with input from P.M. and I.H. All authors read and approved the final manuscript.
Supporting information
Hollwey E, Out S, Watson MR, Heidmann I, Meyer P. TET3‐mediated demethylation in tomato activates expression of a CETS gene that stimulates vegetative growth. Plant Direct. 2017;00:1–10. 10.1002/pld3.22
Funding information
E.H. was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) and Doctoral Training Programme (DTP) CASE studentship (#1360869) in connection with Enza Zaden.
Significance Statement:This study demonstrates the value of the mammalian demethylase TET3 as a tool to induce genes that are sensitive to DNA hypomethylation, while also providing new information on the process of shoot architecture determination in tomato.
REFERENCES
- Abe, M. , Kobayashi, Y. , Yamamoto, S. , Daimon, Y. , Yamaguchi, A. , Ikeda, Y. , … Araki, T. (2005). FD, a bZIP Protein mediating signals from the floral pathway integrator FT at the shoot apex. Science, 309, 1052–1056. 10.1126/science.1115983 [DOI] [PubMed] [Google Scholar]
- Amaya, I. , Ratcliffe, O. J. , & Bradley, D. J. (1999). Expression of CENTRORADIALIS (CEN) and CEN‐like genes in tobacco reveals a conserved mechanism controlling phase change in diverse species. Plant Cell, 11, 1405–1417. 10.1105/tpc.11.8.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth, M. J. , Ost, T. W. B. , Beraldi, D. , Bell, N. M. , Branco, M. R. , Reik, W. , & Balasubramanian, S. (2013). Oxidative bisulfite sequencing of 5‐methylcytosine and 5‐hydroxymethylcytosine. Nature Protocols, 8, 1841–1851. 10.1038/nprot.2013.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, K. , Cui, L. , Zhou, X. , Ye, L. , Zou, Z. , & Deng, S. (2016). Four tomato FLOWERING LOCUS T‐like proteins act antagonistically to regulate floral initiation. Frontiers in Plant Science, 6, 1213 10.3389/fpls.2015.01213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel‐Goren, L. , Liu, Y. S. , Lifschitz, E. , & Zamir, D. (2003). The SELF‐PRUNING gene family in tomato. Plant Molecular Biology, 52, 1215–1222. 10.1023/B:PLAN.0000004333.96451.11 [DOI] [PubMed] [Google Scholar]
- Chardon, F. , & Damerval, C. (2005). Phylogenomic analysis of the PEBP gene family in cereals. Journal of Molecular Evolution, 61, 579–590. 10.1007/s00239-004-0179-4 [DOI] [PubMed] [Google Scholar]
- Chitwood, D. H. , Kumar, R. , Headland, L. R. , Ranjan, A. , Covington, M. F. , Ichihashi, Y. , … Sinha, N. R. (2013). A quantitative genetic basis for leaf morphology in a set of precisely defined tomato introgression lines. Plant Cell, 25, 2465–2481. 10.1105/tpc.113.112391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S. J. , & Bent, A. F. (1998). Floral dip: A simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . The Plant Journal, 16, 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Gouil, Q. , & Baulcombe, D. C. (2016). DNA methylation signatures of the plant chromomethyltransferases. PLoS Genetics, 12, e1006526 10.1371/journal.pgen.1006526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzl, J. , Foerster, A. M. , Raidl, G. , & Scheid, O. M. (2007). CyMATE: A new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. The Plant Journal, 51, 526–536. 10.1111/j.1365-313X.2007.03152.x [DOI] [PubMed] [Google Scholar]
- Hollwey, E. , Watson, M. , & Meyer, P. (2016). Expression of the C‐terminal domain of mammalian TET3 DNA dioxygenase in Arabidopsis thaliana induces heritable methylation changes at rDNA loci. Advances in Bioscience and Biotechnology, 07, 243–250. 10.4236/abb.2016.75023 [DOI] [Google Scholar]
- Ito, S. , D'Alessio, A. C. , Taranova, O. V. , Hong, K. , Sowers, L. C. , & Zhang, Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES‐cell self‐renewal and inner cell mass specification. Nature, 466, 1129–1133. 10.1038/nature09303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, K. , Liberatore, K. L. , Park, S. J. , Alvarez, J. P. , & Lippman, Z. B. (2013). Tomato yield heterosis is triggered by a dosage sensitivity of the florigen pathway that fine‐tunes shoot architecture. PLOS Genetics, 9, e1004043 10.1371/journal.pgen.1004043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlgren, A. , Gyllenstrand, N. , Clapham, D. , & Lagercrantz, U. (2013). FLOWERING LOCUS T/TERMINAL FLOWER1‐like genes affect growth rhythm and bud set in Norway spruce. Plant Physiology, 163, 792–803. 10.1104/pp.113.224139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R. , Baldwin, S. , Kenel, F. , McCallum, J. , & Macknight, R. (2013). FLOWERING LOCUS T genes control onion bulb formation and flowering. Nature Communications, 4, 2884 10.1038/ncomms3884 [DOI] [PubMed] [Google Scholar]
- Lifschitz, E. , Eviatar, T. , Rozman, A. , Shalit, A. , Goldshmidt, A. , Amsellem, Z. , … Eshed, Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proceedings of the National Academy of Sciences of the USA, 103, 6398–6403. 10.1073/pnas.0601620103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman, Z. B. , Cohen, O. , Alvarez, J. P. , Abu‐Abied, M. , Pekker, I. , Paran, I. , … Zamir, D. (2008). The making of a compound inflorescence in tomato and related nightshades. PLoS Biology, 6, e288 10.1371/journal.pbio.0060288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, R. , How‐Kit, A. , Stammitti, L. , Teyssier, E. , Rolin, D. , Mortain‐Bertrand, A. , … Gallusci, P. (2015). A DEMETER‐like DNA demethylase governs tomato fruit ripening. Proceedings of the National Academy of Sciences of the USA, 112, 10804–10809. 10.1073/pnas.1503362112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry, R. C. , & Ayre, B. G. (2012). Manipulating plant architecture with members of the CETS gene family. Plant Science, 188–189, 71–81. 10.1016/j.plantsci.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Meyer, P. , & Heidmann, I. (1994). Epigenetic variants of a transgenic petunia line show hypermethylation in transgene DNA: An indication for specific recognition of foreign DNA in transgenic plants. Molecular & General Genetics: MGG, 243, 390–399. 10.1007/BF00280469 [DOI] [PubMed] [Google Scholar]
- Mimida, N. , Goto, K. , Kobayashi, Y. , Araki, T. , Ahn, J. H. , Weigel, D. , … Sakamoto, W. (2001). Functional divergence of the TFL1‐like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes to Cells, 6, 327–336. 10.1046/j.1365-2443.2001.00425.x [DOI] [PubMed] [Google Scholar]
- Molinero‐Rosales, N. , Latorre, A. , Jamilena, M. , & Lozano, R. (2003). SINGLE FLOWER TRUSS regulates the transition and maintenance of flowering in tomato. Planta, 218, 427–434. 10.1007/s00425-003-1109-1 [DOI] [PubMed] [Google Scholar]
- Nakagawa, M. , Shimamoto, K. , & Kyozuka, J. (2002). Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. The Plant Journal, 29, 743–750. 10.1046/j.1365-313x.2002.01255.x [DOI] [PubMed] [Google Scholar]
- Ono, A. , Yamaguchi, K. , Fukada‐Tanaka, S. , Terada, R. , Mitsui, T. , & Iida, S. (2012). A null mutation of ROS1a for DNA demethylation in rice is not transmittable to progeny. The Plant Journal, 71, 564–574. 10.1111/j.1365-313X.2012.05009.x [DOI] [PubMed] [Google Scholar]
- Park, S. J. , Jiang, K. , Tal, L. , Yichie, Y. , Gar, O. , Zamir, D. , … Lippman, Z. B. (2014). Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nature Genetics, 46, 1337–1342. 10.1038/ng.3131 [DOI] [PubMed] [Google Scholar]
- Pnueli, L. , Carmel‐Goren, L. , Hareven, D. , Gutfinger, T. , Alvarez, J. , Ganal, M. , … Lifschitz, E. (1998). The SELF‐PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development, 125, 1979–1989. [DOI] [PubMed] [Google Scholar]
- Pnueli, L. , Gutfinger, T. , Hareven, D. , Ben‐Naim, O. , Ron, N. , Adir, N. , & Lifschitz, E. (2001). Tomato SP‐interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell, 13, 2687–2702. 10.1105/tpc.010293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinet, M. (2006). Characterization of tomato (Solanum lycopersicum L.) mutants affected in their flowering time and in the morphogenesis of their reproductive structure. Journal of Experimental Botany, 57, 1381–1390. 10.1093/jxb/erj117 [DOI] [PubMed] [Google Scholar]
- Rai, G. K. , Rai, N. P. , Kumar, S. , Yadav, A. , Rathaur, S. , & Singh, M. (2012). Effects of explant age, germination medium, pre‐culture parameters, inoculation medium, pH, washing medium, and selection regime on Agrobacterium‐mediated transformation of tomato. In Vitro Cellular & Developmental Biology‐Plant, 48, 565–578. 10.1007/s11627-012-9442-3 [DOI] [Google Scholar]
- Ratcliffe, O. J. , Amaya, I. , Vincent, C. A. , Rothstein, S. , Carpenter, R. , Coen, E. S. , & Bradley, D. J. (1998). A common mechanism controls the life cycle and architecture of plants. Development, 125, 1609–1615. [DOI] [PubMed] [Google Scholar]
- Shalit, A. , Rozman, A. , Goldshmidt, A. , Alvarez, J. P. , Bowman, J. L. , Eshed, Y. , & Lifschitz, E. (2009). The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proceedings of the National Academy of Sciences of the USA, 106, 8392–8397. 10.1073/pnas.0810810106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyk, S. , Müller, N. A. , Park, S. J. , Schmalenbach, I. , Jiang, K. , Hayama, R. , … Lippman, Z. B. (2017). Variation in the flowering gene SELF PRUNING 5G promotes day‐neutrality and early yield in tomato. Nature Genetics, 49, 162–168. 10.1038/ng.3733 [DOI] [PubMed] [Google Scholar]
- Stam, M. , De Bruin, R. , Van Blokland, R. , Van Der Hoorn, R. A. L. , Mol, J. N. M. , & Kooter, J. M. (2000). Distinct features of post‐transcriptional gene silencing by antisense transgenes in single copy and inverted T‐DNA repeat loci. The Plant Journal, 21, 27–42. 10.1046/j.1365-313x.2000.00650.x [DOI] [PubMed] [Google Scholar]
- Vejlupkova, Z. , & Fowler, J. (2003). Maize DNA preps for undergraduate students: A robust method for PCR genotyping. Maize Genetics Cooperation News Letter, 77, 24–25. [Google Scholar]
- Vitha, S. , Baluška, F. , Jasik, J. , Volkmann, D. , & Barlow, P. W. (2000). Steedman's wax for F‐actin visualization In Staiger C. J., Baluška F., Volkmann D. & Barlow P. W. (Eds.), Actin: A dynamic framework for multiple plant cell functions (pp. 619–636). Developments in plant and soil sciences Dordrecht: Springer; 10.1007/978-94-015-9460-8_35 [DOI] [Google Scholar]
- Vrebalov, J. , Ruezinsky, D. , Padmanabhan, V. , White, R. , Medrano, D. , Drake, R. , … Giovannoni, J. (2002). A MADS‐box gene necessary for fruit ripening at the tomato ripening‐inhibitor (rin) locus. Science, 296, 343–346. 10.1126/science.1068181 [DOI] [PubMed] [Google Scholar]
- Watson, M. R. (2013). Heritable epigenetic variation of DNA methylation targets in plants. PhD Thesis, University of Leeds, UK.
- Watson, M. , Hawkes, E. , & Meyer, P. (2014). Transmission of epi‐alleles with MET1‐dependent dense methylation in Arabidopsis thaliana . PLoS One, 9, e105338 10.1371/journal.pone.0105338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickland, D. P. , & Hanzawa, Y. (2015). The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: Functional evolution and molecular mechanisms. Molecular Plant, 8, 983–997. 10.1016/j.molp.2015.01.007 [DOI] [PubMed] [Google Scholar]
- Yoo, S. J. , Chung, K. S. , Jung, S. H. , Yoo, S. Y. , Lee, J. S. , & Ahn, J. H. (2010). BROTHER OF FT AND TFL1 (BFT) has TFL1‐like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. The Plant Journal, 63, 241–253. 10.1111/j.1365-313X.2010.04234.x [DOI] [PubMed] [Google Scholar]
- Zhong, S. , Fei, Z. , Chen, Y.‐R. , Zheng, Y. , Huang, M. , Vrebalov, J. , … Giovannoni, J. J. (2013). Single‐base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nature Biotechnology, 31, 154–159. 10.1038/nbt.2462 [DOI] [PubMed] [Google Scholar]
- Zilberman, D. , Gehring, M. , Tran, R. K. , Ballinger, T. , & Henikoff, S. (2007). Genome‐wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nature Genetics, 39, 61–69. 10.1038/ng1929 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


