Abstract
Camellia is an economically, ecologically and phylogenetically valuable genus in the family Theaceae. The frequent interspecific hybridization and polyploidization makes this genus phylogenetically and taxonomically under controversial and require detailed investigation. Chloroplast (cp) genome sequences have been used for cpDNA marker development and genetic diversity evaluation. Our research newly sequenced the chloroplast genome of Camellia japonica using Illumina HiSeq X Ten platform, and retrieved five other chloroplast genomes of Camellia previously published for comparative analyses, thereby shedding lights on a deeper understanding of the applicability of chloroplast information. The chloroplast genome sizes ranged in length from 156,607 to 157,166 bp, and their gene structure resembled those of other higher plants. There were four categories of SSRs detected in six Camellia cpDNA sequences, with the lengths ranging from 10 to 17bp. The Camellia species exhibited different evolutionary routes that lhbA and orf188, followed by orf42 and psbZ, were readily lost during evolution. Obvious codon preferences were also shown in almost all protein-coding cpDNA and amino acid sequences. Selection pressure analysis revealed the influence of different environmental pressures on different Camellia chloroplast genomes during long-term evolution. All Camellia species, except C. crapnelliana, presented the identical rate of amplification in the IR region. The datasets obtained from the chloroplast genomes are highly supportive in inferring the phylogenetic relationships of the Camellia taxa, indicating that chloroplast genome can be used for classifying interspecific relationships in this genus.
Introduction
Camellia, containing about 280 species, is a genus with high economic, ecological and phylogenetic values in the family Theaceae [1, 2]. It is native to Southern, Eastern Asia and China, which possess more than 80% of the species and are the center of species diversity [3]. Besides the abundance in species diversity and phylogenetic significance, people pay more attention to this genus, for their commercial and ornamental values. For example, C. sinensis var. sinensis and C. sinensis var. assamica have the highest economic value in Camellia. Tea leaves have been proven to be beneficial for human health as they contain over 700 chemical constituents [1, 4]. Camellia is also known as ornamental trees for urban gardening. The cultivation history of Camellia has been at least 1300 years in China [3]. Today, a group of yellow flowers named golden Camellia, e.g. C. chrysantha, are grown for ornamental purposes, with thirteen to sixteen petals of a flower and blooming several times in a year. Many other Camellia species, e.g. C. japonica, also had local uses. The other most economically valuable species, C. oleifera and C. reticulata, are used for edible oil and cooking in China [5, 6]. At present, more than 3 million hectares are used for Camellia oil production, and the yield of the Camellia nearly 164,000 tons of edible oil [3, 7]. Although the Camellia is native to Asia, because of its variety use, the cultivated species are now found all over the world [1, 8–10]. However, the genus Camellia is phylogenetically and taxonomically under controversial that detailed investigation is required, as a result of frequent interspecific hybridization and polyploidization. Whereas classification of the genus Camellia is traditionally based on morphology [11–16], the result of this systematics is often unreliable and made lots of controversy as morphology is often affected by environmental factors [2]. As a result, it is urgent to seek other methods for rebuilding the classification of Camellia.
Being relatively stable and not easily affected by the environment, molecular methods can provide useful information for taxonomic classification and phylogenetic. Molecular methods, e.g. DNA and RNA sequences [10, 17–24], internal transcribed spacer [10, 18], simple sequence repeats (SSR) [25, 26], ribosomal DNA [27] and several DNA loci [28], have been involved to better understand the evolution of the Camellia. A number of studies focus on the taxonomy, species identification and phylogenetics of the Camellia, but still have not get a satisfied resolution. A recent study used complete chloroplast genomes in several Camellia species [2, 29–31] and got more information of this species. Camellia japonica has been present in Qingdao, Shandong province since the tertiary, it has evolved in dependently after that. A recent research shows that the early flower development sequence placed C. japonica (Naidong) in a most primitive branch of the phylogenetic tree compared to other species [32]. The taxonomy of C. japonica with other Camellia species is in dispute.
In plant cells, the chloroplast is not only the most important and universal organelle, but also one of the major genetic systems (the other two are nucleus and mitochondria). It is involved in photosynthesis and associated with metabolism, such as fatty acid and amino acid synthetic pathways [33–35]. As an independent organelle, the chloroplast has its own genome. It has a covalently closed circular DNA structure and exists in multi-copies in plant cell. It has a conserved circular DNA arrangement [36]. Since the chloroplast genome is self-replicating and has a relatively independent evolutionary process, it has been used for resolving the source populations during species evolution [8, 34, 37–45].
Here, we report newly sequenced complete chloroplast genomes of C. japonica using next generation sequencing technology and genomic comparative analysis with other five published chloroplast genome sequence download from the NCBI. This study aims to deeply analyze the chloroplast genomes of six Camellia species and to determine their (especially the C. japonica) phylogenetic positions.
Materials and methods
Ethics statement
College of Landscape Architecture and Forestry, Qingdao Agricultural University has had a permit from local forestry authorities (Qingdao forestry bureau http://ly.qingdao.gov.cn/) to collect the sample. This research was carried out in compliance with the laws of People’s Republic of China.
Plant materials and genomic DNA isolation
We collected fresh leaves from an adult C. japonica tree growing in the Daguan Island (Jimo District Qingdao city, Shandong, China) (N36°14′, E120°46′, Altitude 10m). The leaves were dried immediately with silicagel. Total genomic DNA was extracted according to Wiland-Szymańska [46].
Chloroplast genome sequencing, assembly and annotation
We used an ultrasonicator to randomly fragment the extracted genomic DNA into 400–600 bp. The NEBNext Ultra DNA Library Prep Kit was used to construct an Illumina paired-end cpDNA library. Paired-end sequencing (2 × 150 bp) was run on an Illumina HiSeq X Ten platform. After filtering the raw data, the cp genomes were assembled according to the following steps. Frist the clean reads were used to assembled into contigs using SOAPdenovo 2.017. Then, the contigs were aligned to the relative species (C. sinensis JQ975030) and get the relative location of the contig sequences and the structure diagram of cp genomes were obtained. The software Gap Closer 1.12 were used to fill the gaps. Finally, the complete cp genome sequence were obtained. The chloroplast genome sequences were annotated with CpGAVAS software and DOGMA software, and then manually corrected.
Molecular marker development
We conducted a sliding window analysis and used DnaSP (DNA Sequences Polymorphism version 5.10.01) software to calculate the nucleotide diversity (Pi) of the six complete Camellia chloroplast genomes [47]. We set the step size to 200 bp with a window length of 600bp.
For different lengths of SSRs, including mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides, minimum numbers (thresholds) were 10, 5, 4, 3, 3, and 3, respectively. We manually verified all the repeats found, and removed unwanted results.
The mVISTA[48] program was used to compare the complete chloroplast genome of C. japonica to other five published chloroplast genomes of the genus Camellia, i.e., C. huana (KY_626042), C. crapnelliana (KF_753632), C. azalea (KY_856741), (KY_626042), C. liberofilamenta (KY_626041) with the shuffle-LAGAN mode [49], so that inter-and intra-specific variations were shown. We used Mega 6.0 software [50] to determine the variable and parsimony-informative base sites across the complete chloroplast genomes, and LSC, SSC and IR regions of the six chloroplast genomes.
DnaSP software was used for manual detection of insertions/deletions. To estimate selection pressures, the 78 protein coding genes in the chloroplast genomes were combined. We used PAML with the yn00 program to calculate the Ka and Ks rates of the combined sequences [51].
Selection pressure analysis
We used KaKs_Calculator 2.0 [52] to determine the Ka and Ks values of genes containing SNP variations, so that we can analyze how different Camellia have evolved under different environmental pressures. We also analyzed the codon preference, and mapped them by R software.
Phylogenies were constructed using the 19 cp genome of the Camellia species sequences from the NCBI Organelle Genome and Nucleotide Resources database: C. crapnelliana (KF_753632), C. azalea (KY_856741), C. luteoflora (KY_626042), C. huana (KY_626042), C. liberofilamenta (KY_626041), C. oleifera (JQ_975031), C. taliensis (KF_156836), C. yunnanensis (KF_156838), C. cuspidate (KF_156833), C. reticulate (KJ_806278), C. pitardii (KF_156837), C. danzaiensis (KF_156837), C. petelotii (KJ_806276), C. leptophylla (KJ_806275), C. impressinervis (KF_156835), C. grandibracteata (KJ_806274), C. sinensis (JQ_975030), C. pubicosta (KJ_806277). We implemented maximum likelihood (ML) analyses on the CIPRES cluster1[53]. GTR+I+R was selected as the nucleotide substitution model. This model was determined from jModel Test v2.1.4 [54]. This model is used to obtain the dataset from the chloroplast genome. For protein-coding regions, it is also used as a partitioned model.
We also used PAUP v4b10 to analyze maximum parsimony (MP). We treated gaps as missing, and character states as unordered. We selected MULPARS option when performing Heuristic search. Further steps include tree bisection-reconnection branch swapping, and random stepwise addition with 1,000 replications.
Results
Basic characteristics of the Camellia chloroplast
A total of with 10.44 Gb clean data were generated by Illumina HiSeq X Ten platform. After data filtering with mean Q20 higher than 94.70% and the mean length was 150 bp. The chloroplast genome sizes of the six Camellia species ranged from 156,607 bp (C. japonica) to 157,166 bp (C. luteoflora). The structure of all chloroplast genomes is quadripartite, which is typical of angiosperm cpDNA. Each chloroplast genome consists of a large single copy region (86,258–86,719bp) and a small single copy region (18,203–18,406bp), separated by two inverted repeat regions (25,967–26,077bp) (Table 1). The GC content of three Camellia species (C. japonica, C. huana and C. liberofilamenta) was 37.32% and the others were 37.30%. The average of GC content is almost similar among the six Camellia species. The well-conserved genomic structure resembled those of other higher plants, including gene number and gene order (Fig 1 and Table 2). The complete C. japonica cp genome sequence has been submitted to GenBank with the accession number PRJNA510919.
Table 1. Statistics on the basic features of the chloroplast genomes of six Camellia species.
| C. japonica | C. crapnelliana | C. azalea | C. luteoflora | C. huana | C. liberofilamenta | |
|---|---|---|---|---|---|---|
| Length (bp) | 156607 | 156997 | 157039 | 157166 | 156903 | 156865 |
| GC content (%) | 37.32 | 37.30 | 37.30 | 37.30 | 37.32 | 37.32 |
| AT content (%) | 62.68 | 62.70 | 62.70 | 62.70 | 62.68 | 62.68 |
| LSC length (bp) | 86258 | 86655 | 86674 | 86719 | 86568 | 86579 |
| SSC length (bp) | 18415 | 18406 | 18281 | 18293 | 18203 | 18236 |
| IR length (bp) | 25967 | 25968 | 26042 | 26077 | 26066 | 26025 |
| Gene number | 134 | 136 | 135 | 133 | 133 | 133 |
| Gene number in IR regions | 36 | 35 | 36 | 35 | 35 | 35 |
| Pseudogene number | 1 | 0 | 3 | 1 | 1 | 1 |
| Pseudogene (%) | 0.75 | 0 | 2.22 | 0.75 | 0.75 | 0.75 |
| Protein-coding gene number | 89 | 89 | 87 | 87 | 87 | 87 |
| Protein-coding gene (%) | 66.42 | 65.44 | 64.44 | 65.41 | 65.41 | 65.41 |
| rRNA gene number | 8 | 8 | 8 | 8 | 8 | 8 |
| rRNA (%) | 5.97 | 5.88 | 5.93 | 6.02 | 6.02 | 6.02 |
| tRNA gene number | 36 | 39 | 37 | 37 | 37 | 37 |
| tRNA (%) | 26.87 | 28.68 | 27.41 | 27.82 | 27.82 | 27.82 |
Fig 1. Gene map of Camellia japonica.
The genes inside and outside of the circle are transcribed in the clockwise and counterclockwise directions, respectively. Genes belonging to different functional groups are shown in different colors. The thick lines indicate the extent of the inverted repeats (IRa and IRb) that separate the genomes into small single-copy (SSC) and large single-copy (LSC) regions.
Table 2. Genes identified in the chloroplast genome of Camellia species.
| Category for genes | Group of gene | Name of gene |
|---|---|---|
| Genes for photosynthesis | ATP synthase | atpA,atpF(pseudogene),atpH,atpI,atpE,atpB |
| NADH-dehydrogenase | ndhJ,ndhK,ndhC,ndhB,ndhF,ndhD,ndhE,ndhG,ndhI,ndhA,ndhH,ndhB | |
| cytochrome b/f complex | petN,petA,petL,petG,petB,petD | |
| photosystem I | psaB,psaA,psaI,psaJ,psaC | |
| photosystem II | psbA,psbK,psbI,psbM,psbD,psbC,psbZ,psbJ,psbL,psbF,psbE,psbB,psbT,psbN,psbH | |
| Rubisco | rbcL | |
| Transcription and translation related genes | transcription | rpoC2,rpoC1,rpoB,rpoA |
| ribosomal proteins | rps12, rps16, rps2, rps14, rps4, rps18, rps12, rps11, rps8, rps3, rps19, rps7,rps15,rps7,rpl33,rpl20,rpl36,rpl14,rpl16,rpl22,rpl2,rpl23,rpl32,rpl23,rpl2 | |
| RNA genes | ribosomal RNA | rrn16S,rrn23S,rrn4.5S,rrn5S,rrn5S,rrn4.5S,rrn23S,rrn16S |
| transfer RNA | trnH-GUG, trnK-UUU,trnQ-UUG,trnS-GCU,trnR-UCU,trnC-GCA,trnD-GUC,trnY-GUA, trnE-UUC,trnT-GGU,trnS-UGA,trnG-UCC,trnM-CAU,trnS-GGA,trnT-UGU,trnL-UAA, trnF-GAA, trnV-UAC,trnM-CAU,trnW-CCA,trnP-UGG,trnI-CAU,trnL-CAA,trnV-GAC, trnI-GAU, trnA-UGC,trnR-ACG,trnN-GUU,trnL-UAG,trnN-GUU,trnR-ACG,trnA-UGC, trnI-GAU,trnV-GAC,trnL-CAA,trnI-CAU | |
| Other genes | RNA processing | matK |
| carbon metabolism | cemA | |
| fatty acid synthesis | accD | |
| proteolysis | clpP | |
| Genes of unkown function | Conserved open reading frames | ycf3,ycf4,ycf2,ycf15,ycf15,ycf1,ycf1,ycf15,ycf15,ycf2 |
Comparative analysis of the Camellia chloroplast genomes
Chloroplast simple sequence repeats (cpSSRs) play a crucial role in studying phylogeny and population genetics[55]. We analyzed cpSSRs in the chloroplast genomes (S1 and S2 Tables). The number of cpSSRs ranged from 67 (C.azalea) to 74 (C.huana) among the six camellia taxa. The number of nucleotide repeats had no significant difference among the six camellia taxa (Fig 2).
Fig 2. Comparison of simple sequence repeats among six chloroplast genomes.
a. Numbers of SSRs detected in ten Camellia chloroplast genomes; b. Frequencies of identified SSRs in LSC, IR and SSC regions; c. Numbers of SSR types detected in ten Camellia chloroplast genomes.
The majority of the 420 SSR loci reside in LSC regions (286 loci, 68.10%). Only a minor portion are located in the SSC regions (72 loci, 17.14%) and IR regions (62 loci, 14.76%). Same as previously reported the SSR loci exhibited a significantly variable distribution among all regions in each of the six Camellia chloroplast genomes [34, 56]. The lowest value (17) was between C. huana and C. luteoflora, while the highest value (58) of nucleotide substitutions was observed between C. huana and C. liberofilamenta, showing a wider range of variability according to the sequence alignment of the six chloroplast genomes (Table 3). The values of Ka/Ks ranged from 0.2342 to 0.5971. The lowest value was between C. crapnelliana and C. luteoflora. The highest value was between C. huana and C. liberofilamenta (Table 3). The result that the Ka/Ks ratio is below 1 indicated negative selection as the selection model for the related gene regions.
Table 3. Pairwise substitution rate between the Camellia chloroplast gemomes based on the 78 protein-coding gene sequences.
| C. japonica | C. crapnelliana | C. azalea | C. luteoflora | C. huana | C. liberofilamenta | |
|---|---|---|---|---|---|---|
| C. japonica | 35 | 33 | 37 | 34 | 33 | |
| C. crapnelliana | 0.4200 | 26 | 34 | 58 | 54 | |
| C. azalea | 0.4791 | 0.3613 | 20 | 44 | 40 | |
| C. luteoflora | 0.3872 | 0.2342 | 0.3663 | 17 | 18 | |
| C. huana | 0.4870 | 0.3610 | 0.5096 | 0.4993 | 23 | |
| C. liberofilamenta | 0.4301 | 0.3470 | 0.4450 | 0.4605 | 0.5971 |
Chloroplast gene gain-loss events
Despite high conservation of chloroplast genome sequences, structural variations, gene loss, and metastasis occur in some species as a result of evolution. This study compared nineteen Camellia species (Table 4). We found that lhbA and orf188, followed by orf42 and psbZ, were readily lost during evolution. The results also showed that psaJ, psbF, psbH and psbZ were lost in C.danzaiensis, compared with the other eighteen species. Moreover, C.japonica had lost trnfM-CAU gene compared the other species. Gene loss events also occur in other plants, e.g. the loss of infA in the Fagales chloroplast genome [55], and the loss of rpl32 in the Paeonia obovata chloroplast genome [57]. Among the rpsl6, ndh, infA, and ycf2 genes, some have disappeared in some angiosperms, and in some legumes, gene loss events have occurred to all of them [58].
Table 4. Genes from the chloroplast genomes of Camellia.
| Name of species | lhbA | orf188 | orf42 | psaJ | psbF | psbH | psbZ | trnfM-CAU |
|---|---|---|---|---|---|---|---|---|
| C.japonica | - | - | - | + | + | + | + | - |
| C.azalea | - | - | + | + | + | + | + | + |
| C.luteoflora | - | - | - | + | + | + | + | + |
| C.liberofilamenta | - | - | - | + | + | + | + | + |
| C.huana | - | - | - | + | + | + | + | + |
| C.reticulate | - | - | + | + | + | + | + | + |
| C.pubicosta | - | - | + | + | + | + | + | + |
| C.petelotii | - | - | + | + | + | + | + | + |
| C.leptophylla | - | - | + | + | + | + | + | + |
| C.grandibracteata | - | - | + | + | + | + | + | + |
| C.crapnelliana | - | + | + | + | + | + | + | + |
| C.yunnanensis | + | + | + | + | + | + | - | + |
| C.pitardii | + | + | + | + | + | + | - | + |
| C.taliensis | + | + | + | + | + | + | - | + |
| C.impressinervis | + | + | + | + | + | + | - | + |
| C.danzaiensis | + | + | + | - | - | - | - | + |
| C.cuspidate | + | + | + | + | + | + | - | + |
| C.oleifera | - | - | + | + | + | + | + | + |
| C.sinensis | - | - | + | + | + | + | + | + |
| Total number of missing gene | 13 | 12 | 4 | 1 | 1 | 1 | 6 | 1 |
Analysis of codon preference
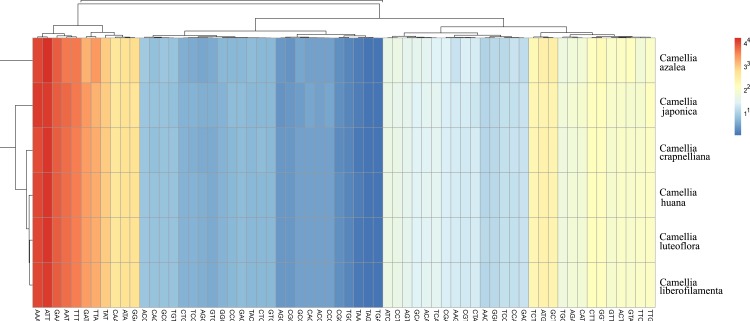
69.59% of the Camellia chloroplast genome sequence was gene coding, of which the vast majority was protein coding. The analytic varieties provided by statistical analyses of all protein-coding cpDNA and amino acid sequences showed obvious codon preferences. It also showed the similarity of protein codons in the six Camellia species, of which AAA, ATT, GAA, AAT, and TTT had the highest frequencies, and the TGA, TAG, TAA, TGC, CGC had the lowest frequencies (Fig 3). The third codon showed a high A/T preference, which is a common phenomenon in higher plant chloroplast genomes [59–61].
Fig 3. Codon distribution of all merged protein-coding genes.
Color key: Red indicates a higher frequency and blue indicates a lower frequency.
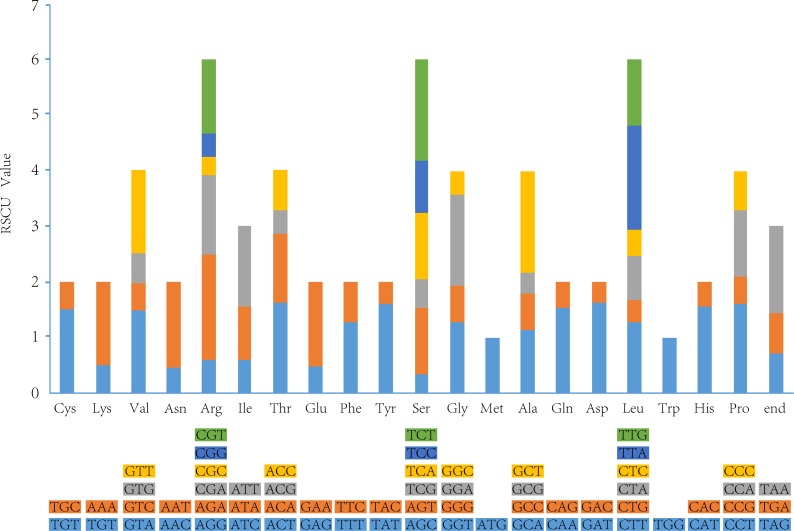
The pattern of the codon preference has important significance in studying species evolution. We used the relative synonymous codon usage (RSCU) as a relative intuitionistic to measure the extent of codon bias [62]. The synonymous codon preference is partitioned into four models: high preference (RSCU>1.3), moderate preference (1.2≤RSCU≤1.3), low preference (1.0<RSCU<1.2) and no preference (RSCU≤1.0).
Among the protein-coding chloroplast genes in the six Camellia species, the 20 amino acids were encoded by 64 codons, in which most of the amino acids had codon preferences except tryptophan (Fig 4). As a total, 30 codon preferences were identified, with 18 amino acids and one stop codon involved. Among the preferred codons, 70.00% exhibited high preferences. This result further revealed the relative conservation of Camellia chloroplast genomes, as high codon preference is also a common phenomenon in higher plants.
Fig 4. Codon content of 20 amino acid and stop codons in all protein-coding genes.
IR contraction analysis
In the chloroplast genome, the IR region is considered as the most conserved region. However, genome size variations often occur in its expansion/contraction regions among various plant lineages, which can be used to study the phylogenetic classification of plants [63]. We compared the IR-SSC and IR-LSC boundaries information of six Camellia were compared (Fig 5). The LSC/IRa boundaries was located within the coding region of rps 19 and created a pseudogene of 279bp at LSC/IRa border. The ycf1 gene spanned the IRb/SSC region and the length of ycf1 was from 936bp to 1069bp. Furthermore, the TrnH-GUG gene (75bp) was located in the LSC. However, the gene trn-GUU and ndhf was not observed in Camellia except C. crapnelliana, that means they contribute little to the overall size variations in the chloroplast genomes of Camellia plants.
Fig 5. Inverted repeat region contraction analysis of various plant species.
Genome divergence between the Camellia species
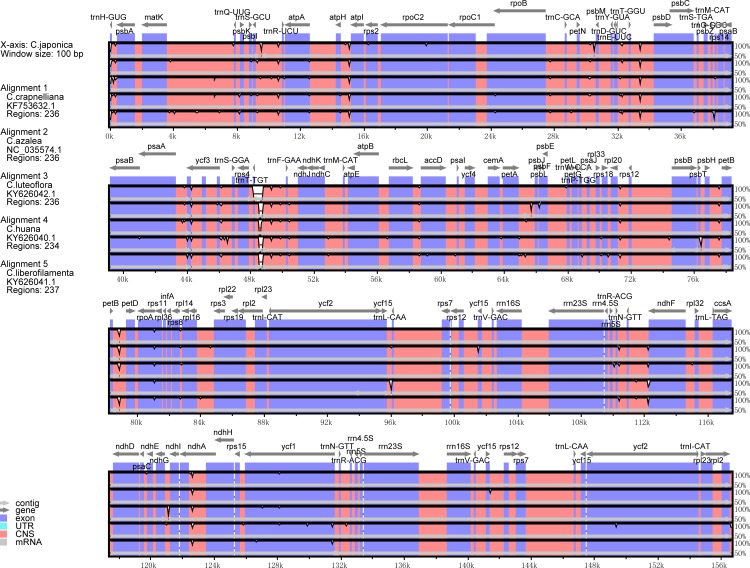
A sequence identity analysis based on mVISTA was performed between six Camellia species, and the reference was the C. japonica chloroplast genome (Fig 6). The aligned sequences that exhibit high similarity showed higher conservation than the remaining sequences across the whole chloroplast genome. Lower divergence levels were exhibited in IR and coding regions than in SC and non-coding regions, respectively.
Fig 6. Identity plot comparing the chloroplast genomes of six Camellia taxa.
The vertical scale indicates the percentage of identity, ranging from 50% to 100%. The horizontal axis indicates the coordinates within the chloroplast genome. Genome regions are color coded as protein-coding, rRNA, tRNA, intron, and conserved non-coding sequences.
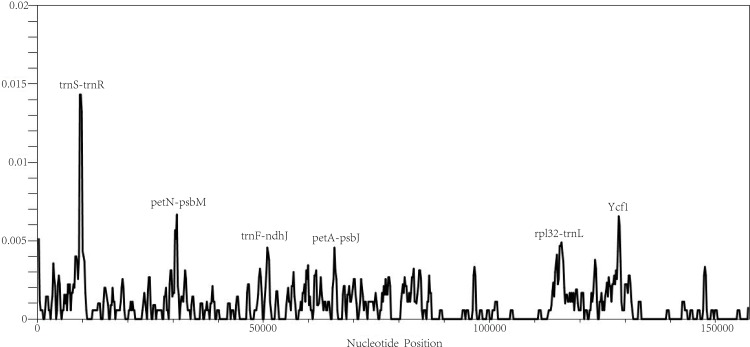
We conducted a sliding window analysis and DnaSP software to calculate the nucleotide diversity of the six complete Camellia chloroplast genomes (Fig 7) Among the six Camellia taxa, C. japonica had the most nucleotide substitutions and insertions/deletions, while C. huana had least nucleotide diversity, and the smallest numbers of nucleotide substitutions and insertions/deletions.
Fig 7. Sliding window analysis of the whole chloroplast genomes of six Camellia taxa (window length: 600 bp, step size: 200bp).
X-axis, position of the midpoint of a window; Y-axis, nucleotide diversity of each window.
According to the chloroplast genome sequence alignment of the six Camellia taxa, six hyper-variable regions, trnS-trnR, petN-psbM, trnF-ndhJ, petA-psbJ, rpl32-trnL, ycf1 were discovered (Fig 7). These six sequences could be used as DNA markers for classification and revealing the genetic divergence of the Camellia taxa, with a high discrimination success ranging from 60% to 100% (Table 5). The sequences of the petN-psbM and ycf1 are two most rapidly evolving regions were able to discriminate all the taxa investigated in this study. In those most rapidly evolving regions, 121 and 122 variable base sites were detected, respectively, of which, 61 and 62 informative base sites, made up 2.86–2.99% in each of the sequences. Comparatively, the commonly recommended DNA fragments (rbcL and matK) achieved only 40% and 80% of discrimination success respectively.
Table 5. Variability of six hyper-variable markers and universal chloroplast DNA barcodes (rbcL and matK) in Camellia.
| Maker | length | Variable base sites | Informative base sites | Mean distance | Discriminationsuccess(%) based on Distance method | ||
|---|---|---|---|---|---|---|---|
| Number | Percentage (%) | Number | Percentage (%) | ||||
| trnS-trnR | 1024 | 55 | 5.37 | 29 | 2.78 | 0.0172 | 60 |
| petN-psbM | 2038 | 121 | 5.94 | 61 | 2.99 | 0.0157 | 100 |
| trnF-ndhJ | 867 | 48 | 5.53 | 25 | 2.88 | 0.0167 | 80 |
| petA-psbJ | 1547 | 86 | 5.60 | 44 | 2.84 | 0.0165 | 80 |
| rpl32-trnL | 2017 | 108 | 5.35 | 55 | 2.72 | 0.0178 | 80 |
| ycf1 | 2168 | 122 | 5.63 | 62 | 2.86 | 0.0181 | 100 |
| rbcL | 1401 | 31 | 2.22 | 17 | 1.18 | 0.0063 | 40 |
| matK | 1535 | 49 | 3.19 | 26 | 1.66 | 0.0091 | 80 |
Phylogenetic analysis
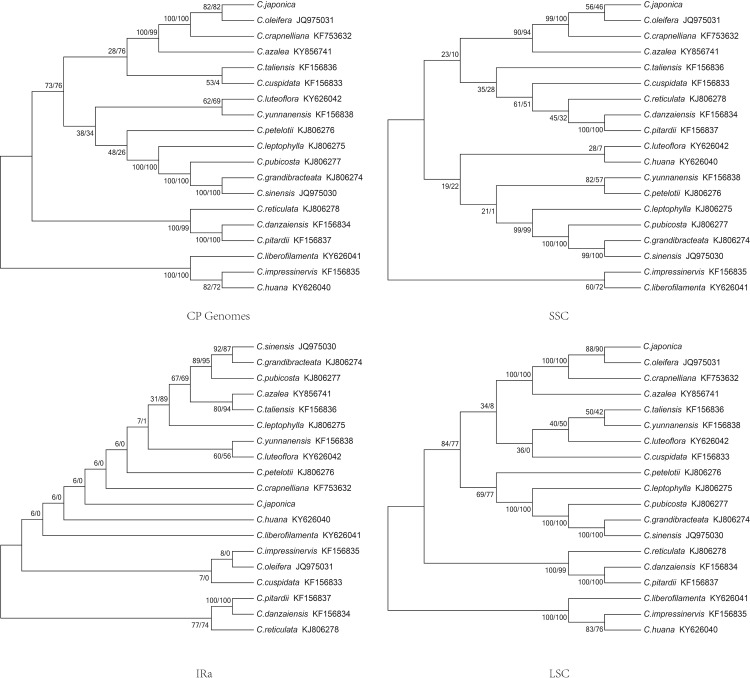
Previous studies have fairly well resolved the relationships between Camellia species, but have not well studied the position of Camellia [29, 30, 64]. Six data partitions including coding regions, large single-copy region, the small single copy region, IR region, the inverted repeat region, introns and spacers and the complete cp DNA sequences from the 19 Camellia were used for phylogenetic analyses. All six datasets produced similar phylogenetic trees with moderate to high support, whereas the IR dataset had poor support (Fig 8). The reconstructed phylogeny divided the species into clads based on maximum likelihood (ML) and Maximum parsimony methods (MP).
Fig 8. Phylogenetic relationships of the nineteen Camellia species constructed from the complete chloroplast genome sequences using maximum likelihood (ML) and maximum parsimony methods (MP).
The phylogenetic tree reveals that C. japonica is most related with C. oleifera. Furthermore, the phylogenetic result is consistent with the section-level classification by Raven [65]. The chloroplast resource will be helpful for the conservation, taxonomy, and breeding programs of the genus Camellia.
Chloroplast genome variation and evolution
In this study, Illumina next-generation technology was used to completely sequence the chloroplast genome of C. japonica and compared with the previously reported chloroplast genomes in Camellia. The chloroplast genomes of C. japonica displayed the typical quadripartite structure of flowering plants, were conservative in gene order and gene content, in comparison with the most lineages of angiosperms. The chloroplast genome sizes ranged from 156,607 to 157,166 bp in length. IR regions are considered as the most conserved region, which considered to be the primary mechanisms affecting length variation of angiosperm chloroplast genomes. Only minor variations were detected at the SC/IR boundaries of six Camellia. Occurrence of indels was the main factor effecting the variation of the length in Camellia chloroplast genomes. the Camellia chloroplast genomes contained more AT content than GC content, which is a common phenomenon in higher plant chloroplast genomes [59–61].
SSRs are widely used in phylogenetic analyses and population genetics and polymorphism investigations. A total of 420 SSR loci were identified and the number of SSRs ranged from 67 to74 in Camellia. the mono-nucleotide repeats are the most common SSRs in chloroplast genomes, which make more contributions to the genetic variation than the longer SSRs. Since the structure of chloroplast genomes are conservative, SSR primers are transferable across species and genera. Information involving SSRs in this study will provide useful sources for estimating the phylogenetic relationships among species and genera.
Potential cpDNA barcodes
Camellia is the largest genus in its family, including more than 280 species all over the world. For effective exploration, conservation, and domestication, accurately identified wild species would provide a clear genetic background of this genus. However, the taxonomic inventory of genus Camellia is still under controversial, because of the vast amount of species with extensive global distribution and interspecific hybridization. DNA barcoding has been widely used in identify unknown species [66]. The rbcL and matK is considered as core universal DNA barcodes in many species. Therefore, genomic comparative researches of more complete chloroplast genome sequences have become necessary for developing variable DNA barcodes. These mutation “hotspot” regions can be used to develop novel DNA barcodes [67]. The six potential mutational hotspots (trnS-trnR, petN-psbM, trnF-ndhJ, petA-psbJ, rpl32-trnL, ycf1) identified in this study could be suitable barcodes for plant classification in Camillia. In previous reports, the gene ycf1 was recommended as core DNA barcode for plants because of the high divergence [68]. Ycf1 gene has been widely applied in plant phylogeny and DNA barcoding studies [69–70].
Recently, using the chloroplast genome as a super-barcode for plant species identification was discussed [71]. The analyses on chloroplast genome sequence divergence showed that it may indeed be useful as a super-barcode for species identification of Camellia. Further research is necessary to investigate whether these hyper-variable regions or complete chloroplast genome sequences could be used as reliable and effective DNA barcodes for species of Camellia. The results obtained in this study have significant value for future studies on global genetic diversity assessment, phylogeny, and population genetics of Camellia.
Perspectives of persimmon research in future
It is important to elucidate the genetic relationship of Camellia taxa for germplasm conservation, breeding strategies of Camellia. The accurate classification of sect. Thea have widely been acknowledged to be complex. For example, the taxonomy of C. pubicosta still has a dispute. Min et al considered that the C. pubicosta belongs to sect. Corallina [71], while Chang and Huang insisted it belongs to sect. Thea. [29, 72]. In our research the C. pubicosta was close to C. sinensis and C. grandibracteata supporting C. pubicosta might be classified into sect. Thea. Previous studies reported that species of sect. Thea can be divided into two groups, agreeing with the locule ovary number [73,74]. However, our results showed that the classification of this species was not entirely consistent with previous studies [74,75]. For instance, the C. taliensis and C. cuspidate, C. grandibracteata and C. sinensis were supported as monophyletic respectively. However, the C. taliensis and C. grandibracteata have 5 ovaries, while C. cuspidata and C. sinensis have 3 ovaries.
The C. japonica population in Qingdao, Shandong province is the only one in temperate areas in China. While this population has been present in this area since the tertiary, after the quaternary glacier most thermophyilic species extinction or migration to warmer regions. In contrast, C. japonica adapted to temperate climate. Since then, it has evolved independently and no gene exchanges with the distribution center species. Zhang et al considered that the C. japonica was the relative evolutionary species. The results of phylogenetic analysis support that C. japonica and C.oleifera as monophyletic, However the C. japonica have 2–3 ovaries and the C.oleifera have 3–5 ovaries. Our results indicated that the classification of Camellia species using locule ovary number may be reconsidered. The combination of traditional classification methods, molecular markers and sequencing of more complete cp genomes of Camellia are necessary to solve the problem of Camellia classification in the future research.
Conclusions
We reported the complete chloroplast genome sequences of C. japonica were reported based on the Illumina HiSeq X Ten platform. C. japonica chloroplast genomes exhibited a typical quadripartite and circular structure with 156607bp.We investigated the variation of repeat sequences, SSRs among the six complete Camellia cp genomes. Selection pressure analysis revealed the influence of different environmental pressures on different Camellia chloroplast genomes during long-term evolution. Obvious codon preferences were shown in almost all protein-coding cDNA and amino acid sequences. Lower divergence levels were exhibited in IR and Coding regions than in SC and Non-coding regions, respectively. The results of phylogenetic showed that C. japonica has the closest relationship with C. oleifera. Therefore, chloroplast genome resources will be helpful for taxonomic studies, conservation, and breeding programs of the genus Camellia.
Supporting information
(DOCX)
(DOCX)
Data Availability
The complete C. japonica cp genome sequence has been submitted to GenBank with the accession number PRJNA510919. All other relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the Forestry Science & Technology Innovation Project of Shangdong Province (LYCX01-2018-05) to KW; National Natural Science Foundation of China (No.31500264) to XG and the Collection and Protection of Featured, Rare and Endangered Forest Tree Germplasm Resources (2016LZGC038) to KW. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vijayan K, Zhang W, Tsou C. Molecular taxonomy of Camellia (Theaceae) inferred from nrITS sequences. Am J Bot. 2009;96(7):1348–1360. 10.3732/ajb.0800205 [DOI] [PubMed] [Google Scholar]
- 2.Yang JB, Yang SX, Li HT, Yang J, Li DZ. Comparative chloroplast genomes of Camellia species. PLoS One. 2013;8(8):e73053 10.1371/journal.pone.0073053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao JY, Parks CR and Du YQ. Collected species of the genus Camellia and illustrated outline. Zhejiang: Zhejiang Science and Technology Press; 2005. [Google Scholar]
- 4.Khan N, Mukhtar H. Tea polyphenols for health promotion. Life Sciences. 2007;81(7):519–533. 10.1016/j.lfs.2007.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang D, Yu J, Chen Y, Zhang R. Ornamental Tea oil Camellia Cultivars and Their Hypocotyl Graft Propagation. SNA Research Conference. 2007; 52: 257–260. [Google Scholar]
- 6.Flora of China Editorial Committee. Flora of China. Beijing: Science Press; 2004. [Google Scholar]
- 7.Ming T. A systematic synopsis of the genus Camellia[J]. Acta Botanica Yunnanica, 1999;21(2):149–159. [Google Scholar]
- 8.Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA. 2010;107(10):4623–4628. 10.1073/pnas.0907801107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wachira F, Tanaka J, Takeda Y. Genetic variation and differentiation in tea (Camellia sinensis) germplasm revealed by RAPD and AFLP variation. J Hortic Sci Biotech. 2001;76(5):557–563. [Google Scholar]
- 10.Tian M, Li JY, Ni H, Fan ZQ, Li XL. Phylogenetic Study on Section Camellia Based on ITS Sequences Data. Acta Horticulturae Sinica. 2008;35(11):1685–1688. [Google Scholar]
- 11.Sealy JR. A Revision of the Genus Camellia. The Royal Horticultural Society Press; 1958. [Google Scholar]
- 12.Pi E, Peng QF, Lu HF, Shen JB, Du YQ, Huang FL, et al. Leaf morphology and anatomy of Camellia section Camellia (Theaceae). Bot J Linn Soc. 2009;159(3):456–476. [Google Scholar]
- 13.Lu HF, Shen JB, Lin XY, Fu JL. Relevance of Fourier Transform Infrared Spectroscopy and Leaf Anatomy for Species Classification in Camellia (Theaceae). Taxon. 2008;57(4):1274–1288. [Google Scholar]
- 14.Luna I, Ochoterena H. Phylogenetic relationships of the genera of Theaceae based on morphology [Review]. Cladistics-the International Journal of the Willi Hennig Society. 2004;20(3):223–270. [DOI] [PubMed] [Google Scholar]
- 15.Jiang B, Peng QF, Shen ZG, Möller M, Pi EX, Lu HF. Taxonomic treatments of Camellia (Theaceae) species with secretory structures based on integrated leaf characters. Plant Sys & Evol. 2010;290(1–4):1–20. [Google Scholar]
- 16.Lu H, Wu J, Ghiassi M, Sean L, Mantri N. Classification of Camellia(Theaceae) Species Using Leaf Architecture Variations and Pattern Recognition Techniques. Plos One. 2012;7(1):e29704 10.1371/journal.pone.0029704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Li H, Yang S, Li D, Yang Y. The application of four DNA sequences to studying molecular phylogeny of Camellia (Theaceae). Acta Botanica Yunnanica. 2006; 28(2):108–114. [Google Scholar]
- 18.Tang S, Zhong Y. A Phylogenetic Analysis of nrDNA ITS Sequences from Ser. Chrysantha (Sect. Chrysantha, Camellia, Theaceae). J Genet Mol Bio. 2002;13(2):105–107. [Google Scholar]
- 19.Yao QY. EST SSR development and identification of candidate genes related to triacylglycerol and pigment biosynthesis and photoperiodic flowering in Camellia reticulata by RNA-seq. Thesis, The University of Yunnan. 2003.
- 20.Xiao TJ, Parks CR. Molecular analysis of the genus Camellia. 2003; (35):57–65. [Google Scholar]
- 21.Su MH, Hsieh CF, Tsou CH. The confirmation of Camellia formosensis (Theaceae) as an independent species based on DNA sequence analyses. Botanical Studies. 2011;50(4):477–485. [Google Scholar]
- 22.Prince LM, Parks CR. Phylogenetic Relationships of Theaceae Inferred from Chloroplast DNA Sequence Data. Am J Bot. 2001;88(12):2309–2320. [PubMed] [Google Scholar]
- 23.Liu Y, Yang SX, Ji PZ, Gao LZ. Phylogeography of Camellia taliensis (Theaceae) inferred from chloroplast and nuclear DNA: insights into evolutionary history and conservation. BMC Evol Biol. 2012;12(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei SJ, Lu YB, Ye QQ, Tang SQ. Population Genetic Structure and Phylogeography of Camellia flavida (Theaceae) Based on Chloroplast and Nuclear DNA Sequences. Front Plant Sci. 2017;8:718 10.3389/fpls.2017.00718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Ruan CJ, Ding GJ, Mopper S. Genetic relationships in a germplasm collection of Camellia japonica and Camellia oleifera using SSR analysis. Gene Mol Res. 2017;16(1):1–14. [DOI] [PubMed] [Google Scholar]
- 26.Zhao DW, Yang JB, Yang SX, Kato K, Luo JP. Genetic diversity and domestication origin of tea plant Camellia taliensis (Theaceae) as revealed by microsatellite markers. BMC Plant Biol. 2014;14(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu J, Xu Y, Yonezawa T, Li L, Hasegawa M, Lu F, et al. Polymorphism and evolution of ribosomal DNA in tea (Camellia sinensis, Theaceae). Molecular Phylogenetics & Evolution. 2015;89:63–72. [DOI] [PubMed] [Google Scholar]
- 28.Fang W, Yang JB, Yang SX, Li DZ. Phylogeny of Camellia sects. Longipedicellata, Chrysantha and Longissima (Theaceae) Based on Sequence Data of Four Chloroplast DNA Loci. Acta Botanica Yunnanica. 2010;32(1):1–13. [Google Scholar]
- 29.Huang H, Shi C, Liu Y, Mao SY, Gao LZ, et al. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol Biol. 2014, 14(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang G, Luo Y, Hou N, Deng LX. The complete chloroplast genomes of three rare and endangered camellias (Camellia huana, C. liberofilamenta and C. luteoflora) endemic to Southwest China. Conserv Genet Resour. 2017:1–3. [Google Scholar]
- 31.Liu Y, Han Y. The complete chloroplast genome sequence of endangered camellias (Camellia pubifurfuracea). Conserv Genet Resour. 2017(15):1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q, Hao Q, Guo X, Liu Q, Sun Y, Liu Q, et al. Anther and ovule development in Camellia japonica (Naidong) in relation to winter dormancy: Climatic evolution considerations. Flora. 2017;233:127–139. [Google Scholar]
- 33.Neuhaus HE, Emes MJ. Nonphotosynthetic metabolism in plastids. Annual Review of Plant Physiology & Plant Molecular Biology. 2000;51(51):111–140. [DOI] [PubMed] [Google Scholar]
- 34.Dong W, Xu C, Li D, Jin X, Li R, Lu Q, et al. Comparative analysis of the complete chloroplast genome sequences in psammophytic Haloxylon species (Amaranthaceae). Peerj. 2016;4(2):e2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gurusamy R, Seonjoo P. The Complete Chloroplast Genome Sequence of Ampelopsis: Gene Organization, Comparative Analysis, and Phylogenetic Relationships to Other Angiosperms. Front Plant Sci. 2016;7(32). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansen RK, Raubeson LA, Boore JL, Depamphilis CW, Chumley TW, Haberle RC, et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Method in Enzymol. 2005;395(6):348–384. [DOI] [PubMed] [Google Scholar]
- 37.Small RL, Cronn RC, Wendel JF. Use of nuclear genes for phylogeny reconstruction in plants. Aust Syst Bot. 2004;17(2):145–170. [Google Scholar]
- 38.Dong W, Xu C, Li C, Sun J, Zuo Y, Shi S, et al. ycf1, the most promising plastid DNA barcode of land plants. Sci Rep. 2015;5:8348 10.1038/srep08348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suo Z, Chen L, Dong P, Jin X, Zhang H. A new nuclear DNA marker from ubiquitin ligase gene region for genetic diversity detection of walnut germplasm resources. Biotechnol Rep. 2015;5:40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suo Z, Li WY, Jin XB, Zhang HJ. A New Nuclear DNA Marker Revealing Both Microsatellite Variations and Single Nucleotide Polymorphic Loci: A Case Study on Classification of Cultivars in Lagerstroemia indica L. Journal of Microbial & Biochemical Technology. 2016;8(4):266–271. [Google Scholar]
- 41.Song Y, Dong W, Liu B, Xu C, Yao X, Gao J, et al. Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus yunnanensis and Machilus balansae in the family Lauraceae. Front Plant Sci. 2015;6:662 10.3389/fpls.2015.00662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Downie SR, Jansen RK. A Comparative Analysis of Whole Plastid Genomes from the Apiales: Expansion and Contraction of the Inverted Repeat, Mitochondrial to Plastid Transfer of DNA, and Identification of Highly Divergent Noncoding Regions. Syst Bot. 2016;40(1):336–351. [Google Scholar]
- 43.Curci PL, De PD, Danzi D, Vendramin GG, Sonnante G. Complete chloroplast genome of the multifunctional crop globe artichoke and comparison with other Asteraceae. Plos One. 2015;10(3):e0120589 10.1371/journal.pone.0120589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebensmack J, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 2007;104(49):19369–19374. 10.1073/pnas.0709121104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parks M, Cronn R, Liston A. Parks M, Cronn R, Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009; 7: 84 10.1186/1741-7007-7-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Wang S, Jing Y, Ling W. A Modified CTAB Protocol for Plant DNA Extraction. Chinese Bulletin of Botany. 2013;48(1):72–78. [Google Scholar]
- 47.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 48.Thiel T, Michalek W, Varshney RK, Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet. 2003;106(3):411–422. 10.1007/s00122-002-1031-0 [DOI] [PubMed] [Google Scholar]
- 49.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32(suppl-2):273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- 52.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom, Proteom Bioinf. 2010;8(1):77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller MA, Pfeiffer W, Schwartz T. The CIPRES science gateway: a community resource for phylogenetic analyses. Teragrid Conference: Extreme Digital Discovery; 2011.
- 54.Posada D. jModelTest: Phylogenetic Model Averaging. Mol Biol Evol. 2008;25(7):1253–1256. 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- 55.Liu LX, Li R, Worth J, Li X, Li P, Cameron KM, et al. The Complete Chloroplast Genome of Chinese Bayberry (Morella rubra, Myricaceae): Implications for Understanding the Evolution of Fagales. Front Plant Sci. 2017;8:968 10.3389/fpls.2017.00968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Y, Zhou T, Duan D, Yang J, Feng L, Zhao G. Comparative Analysis of the Complete Chloroplast Genomes of Five QuercusSpecies. Front Plant Sci. 2016;7:959 10.3389/fpls.2016.00959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong W, Xu C, Cheng T, Lin K, Zhou S. Sequencing Angiosperm Plastid Genomes Made Easy: A Complete Set of Universal Primers and a Case Study on the Phylogeny of Saxifragales. Geno Bio Evol. 2013;5(5):989–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolfe KH, Mordent CW, Ems SC, Palmer JD. Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: loss or accelerated sequence evolution of tRNA and ribosomal protein genes. J Mol Evol. 1992;35(4):304–317. [DOI] [PubMed] [Google Scholar]
- 59.Nie XJ, Lv SZ, Zhang YX, Du XH, Wang L, Biradar SS, et al. Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora). Plos One. 2012;7(5):e36869 10.1371/journal.pone.0036869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yi DK, Kim KJ. Complete Chloroplast Genome Sequences of Important Oilseed Crop Sesamum indicum L. Plos One. 2012;7(5):e35872 10.1371/journal.pone.0035872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuo LH, Shang AQ, Zhang S, Yu XY, Ren YC, Yang MS, et al. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: Genome comparative and taxonomic position analysis. Plos One. 2017;12(2):e0171264 10.1371/journal.pone.0171264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharp PM, Li WH. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–1295. 10.1093/nar/15.3.1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang RJ, Cheng CL, Chang CC, Wu CL, Su TM, Chaw SM. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol. 2008;8(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang JB, Yang SX, Li HT, Yang J, Li DZ. Comparative Chloroplast Genomes of Camellia Species. Plos One. 2013;8(8):e73053 10.1371/journal.pone.0073053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu ZY, Raven PH, Hong DY. Flora of China Vol. 12: Hippocastanaceae through Theaceae. 2007. [Google Scholar]
- 66.Hebert PDN, Cywinska A, Ball SL, DeWaard JR. Biological identifications through DNA barcodes. Proc Royal Soc London Series B-Biological Sciences.2003;270:313–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dong WP, Liu J, Yu J, Wang L, Zhou SL. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One. 2012;7:e35071 10.1371/journal.pone.0035071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dong WP, Xu C, Li CH, Sun JH, Zuo YJ, Shi S, Cheng T, Guo JJ, Zhou SL. ycf1, the most promising plastid DNA barcode of land plants. Sci Rep. 2015;5:8348 10.1038/srep08348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang J, Vazquez L, Chen X, Li H, Zhang H, Liu Z, Zhao G. Development of chloroplast and nuclear DNA markers for Chinese oaks (Quercus subgenus Quercus) and assessment of their utility as DNA barcodes. Front Plant Sci.2017;8:816 10.3389/fpls.2017.00816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dastpak A, Osaloo SK, Maassoumi AA, Safar KN. Molecular phylogeny of Astragalus sect. Ammodendron (Fabaceae) inferred from chloroplast ycf1 gene. Ann Bot Fenn. 2018;55:75–82. [Google Scholar]
- 71.Hernandez-Leon S, Gernandt DS, Perez de la Rosa JA, Jardon-Barbolla L. Phylogenetic relationships and species delimitation in Pinus section Trifoliae inferred from plastid DNA. PLoS One. 2013;8:e70501 10.1371/journal.pone.0070501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Min TL: A revision of Camellia sect. Thea Acta Bot Yunnanica 1992, 14:115–132. [Google Scholar]
- 73.Chang HD, Ren SX: Flora of China. Science Press. Tomus 1998, 49(3):1–251. [Google Scholar]
- 74.Chen L, Yamaguchi S, Wang PS, Xu M, Song WX, Tong QQ: Genetic polymorphism and molecular phylogeny analysis of section Thea based on RAPD markers. J Tea Sci 2002, 22:19–24. [Google Scholar]
- 75.Li XH, Zhang CZ, Liu CL, Shi ZP, Luo JW, Chen X: RAPD analysis of the genetic diversity in Chinese tea germplasm. Acta Hort Sin 2007, 34:507–508. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
The complete C. japonica cp genome sequence has been submitted to GenBank with the accession number PRJNA510919. All other relevant data are within the manuscript and its Supporting Information files.