Abstract
We identified three novel members of the R2R3‐MYB clade of anthocyanin regulators in the genome of the purple flowering Petunia inflata S6 wild accession, and we called them ANTHOCYANIN SYNTHESIS REGULATOR (ASR). Two of these genes, ASR1 and ASR2, are inactivated by two different single base mutations in their coding sequence. All three of these genes are absent in the white flowering species P. axillaris N and P. parodii, in the red flowering P. exserta, and in several Petunia hybrida lines, including R27 and W115. P. violacea and other P. hybrida lines (M1, V30, and W59) instead harbor functional copies of the ASR genes. Comparative, functional and phylogenic analysis of anthocyanin R2R3‐MYB genes strongly suggest that the ASR genes cluster is a duplication of the genomic fragment containing the other three R2R3‐MYB genes with roles in pigmentation that were previously defined, the ANTHOCYANIN4‐DEEP PURPLE‐PURPLE HAZE (AN4‐DPL‐PHZ) cluster. An investigation of the genomic fragments containing anthocyanin MYBs in different Petunia accessions reveals that massive rearrangements have taken place, resulting in large differences in the regions surrounding these genes, even in closely related species. Yeast two‐hybrid assays showed that the ASR proteins can participate in the WMBW (WRKY, MYB, B‐HLH, and WDR) anthocyanin regulatory complex by interacting with the transcription factors AN1 and AN11. All three ASRs can induce anthocyanin synthesis when ectopically expressed in P. hybrida lines, but ASR1 appeared to be the most effective. The expression patterns of ASR1 and ASR2 cover several different petunia tissues with higher expression at early stages of bud development. In contrast, ASR3 is only weakly expressed in the stigma, ovary, and anther filaments. The characterization of these novel ASR MYB genes completes the picture of the MYB members of the petunia anthocyanin regulatory MBW complex and suggests possible mechanisms of the diversification of pigmentation patterns during plant evolution.
Keywords: anthocyanin synthesis, evolution, Petunia, R2R3‐MYBs, transcription factor
1. INTRODUCTION
Pigmentation is a suitable model to study how new patterns are generated during the evolution of species. The study of regulatory genes involved in the formation of spots on Drosophila wings revealed how mutations contributed to the diversification of sexual preference and thus to genetic separation and the appearance of new species (Gompel, Prud'homme, Wittkopp, Kassner, & Carroll, 2005). The diversification of the coat color in mammals has been shown to be related to fitness in different environments (Hoekstra, 2006), and the understanding of the mechanism behind such variation is a key to the unraveling of adaptation mechanisms. In plants, the formation of different pigmentation patterns is often related to reproduction since many species display color to attract animals for the dispersal of pollen and seeds (Galliot, Stuurman, & Kuhlemeier, 2006) but also contribute to the adaptation to different growth conditions (Albert et al., 2014; Anderson, Willis, & Mitchell‐Olds, 2011; Steyn, Wand, Holcroft, & Jacobs, 2002). The most widely diffused plant pigments are anthocyanins. Their biosynthesis is one of the best‐studied metabolic pathways, making it very attractive to use these pigments as a model to understand how patterns are generated during the evolution of a species.
Anthocyanins are flavonoid pigments providing blue/violet pigmentation to foliage, fruit, and flowers and they fulfill a variety of physiological functions (Tanaka, Sasaki, & Ohmiya, 2008; Winkel‐Shirley, 2001). The synthesis of anthocyanins is regulated by a network of transcription factors determining tissue specificity and response to the stimuli of the pigment accumulation. In all species examined to date, these transcription factors are R2R3‐MYB, bHLH, and WD40 proteins forming an MBW protein complex, which activates the promoters of the anthocyanins synthesis structural genes (Spelt, Quattrocchio, Mol, & Koes, 2002; Koes, Verweij, & Quattrocchio, 2005; Ramsay & Glover, 2005; Gonzalez, Zhao, Leavitt, & Lloyd, 2008; ;Albert et al., 2014). The complex is boosted by the participation of a WRKY transcription factor, which also confers specificity for other sets of target genes involved in, for example, vacuolar hyperacidification (Verweij et al., 2016). The WDR (WD40) regulators are highly conserved, even among animals and plants (de Vetten, Quattrocchio, Mol, & Koes, 1997). To date, a single gene in all species, Transparent Testa Glabra1 (TTG1) in Arabidopsis, PAC in maize and AN11 in petunia, is known to encode the WD40 member of the MBW complex (Carey, Strahle, Selinger, & Chandler, 2004; de Vetten et al., 1997; Walker et al., 1999).
The bHLH anthocyanin regulators can instead be grouped into at least two phylogenetic clades, and most species have members belonging to each clade. One group includes maize B, Lc, and R (Purugganan & Wessler, 1996), petunia JAF13 (Quattrocchio, Wing, van der Woude, Mol, & Koes, 1998), and Arabidopsis GL3 and EGL3 (Bernhardt et al., 2003). The Arabidopsis TT8 protein groups in a distinct clade (Consonni, Geuna, Gavazzi, & Tonelli, 1993; Hernandez, Feller, Morohashi, Frame, & Grotewold, 2007) together with the petunia AN1 (Spelt, Quattrocchio, Mol, & Koes, 2000) and the maize IN factor (Burr et al., 1996). The WRKY factor is encoded by a single gene in all studied species (Amato et al., 2017; Johnson, Kolevski, & Smyth, 2002; Verweij et al., 2016). All components of the WMBW complex are essential to efficiently activate anthocyanin synthesis, as shown by the loss (or reduction) of pigmentation in mutants for each of these factors.
R2R3‐MYB components of the WMBW complex determine the set of target genes that the complex will activate. The gene families are classified into several subgroups with different functions in plant‐specific processes, such as development, signal transduction, resistance to pathogens, and metabolism (including anthocyanin synthesis) (Dubos et al., 2010). The members of the group represented by the petunia AN2 is characterized by the R2R3‐MYB Sub‐Group 6 (called SG6), sharing a short amino acid signature for anthocyanin regulating MYBs (Stracke, Werber, & Weisshaar, 2001; Zimmermann, Heim, Weisshaar, & Uhrig, 2004). SG6 MYBs are encoded in each species by a small family of genes with different expression patterns, contributing to the color of different plant parts (Albert et al., 2011; Gonzalez et al., 2008; Quattrocchio et al., 1998; Schwinn et al., 2006). The spotted pattern of the petals of Lilium hybrids has been shown to be associated with LhMYB6 or LhMYB12 (Yamagishi, Shimoyamada, Nakatsuka, & Masuda, 2010), while in Clarkia, the expression domain of an SG6 MYB, CgMYB1, determines the position of a spot in the flower (Martins, Berg, Blinka, Rausher, & Baum, 2013; Martins, Jiang, & Rausher, 2017). These examples show that duplication and diversification of R2R3‐MYB genes controlling anthocyanin synthesis result in different expression patterns of these regulators, leading to considerable possibilities of variation in temporal and spatial anthocyanin accumulation in plants (Albert et al., 2011; Bombarely et al., 2016; Schwinn et al., 2006).
Petunia hybrida has a long history as a genetic model system (Vandenbussche, Chambrier, Rodrigues Bento, & Morel, 2016), particularly in the genetics of pigmentation. The MYB member of the WMBW complex regulating anthocyanin accumulation in petunia was thought to be one of four MYBs: AN2, AN4, DEEP PURPLE (DPL), and PURPLE HAZE (PHZ). All of these genes encode similar proteins but differ in expression patterns controlling pigmentation in distinct tissues or under different conditions. AN2 is expressed in the petal limb and tube, whereas AN4 is expressed in the anthers at early developmental stages (Supplemental note in Bombarely et al., 2016). PHZ is a light‐regulated MYB gene, whereas DPL regulates venation patterning in petunia flowers (Albert et al., 2011, 2014). These R2R3‐MYBs determine the timing, localization, and degree of anthocyanin accumulation.
In this study, we characterized three novel petunia R2R3‐MYB genes of the SG6 clade, ASR1 to ASR3, and examined their roles in regulating anthocyanin accumulation and their possible involvement in creating pigmentation patterns in petunia. ASRs, similar to other SG6 MYB members, are also involved in the WMBW regulatory complex, where they interact with AN1 and AN11 and contribute to anthocyanin synthesis by upregulating anthocyanin structural genes.
2. MATERIALS AND METHODS
2.1. Plant materials and growing conditions
The petunia species Petunia axillaris N S26, P. inflata S6, P. exserta S25, P. parodii S8, P. violacea S9, and all of the Petunia hybrida lines used in this work (Supporting Information Table S1) come from the collection of the University of Amsterdam. Plants were grown at standard greenhouse conditions. As the M1 and R27 lines are not easily transformable, they were crossed to generate M1×R27/F1 progeny, which performed well in the production of stable transformants.
2.2. Identification and phylogenetic analysis of ASR genes in Petunia
The ASR genes were identified by screening the genome of Petunia axillaris N and Petunia inflata S6 with known plant SG6 R2R3‐MYB sequences by BLAST search. The entire candidate sequences identified in this search were then used in a phylogenetic analysis, and ASR1, ASR2, and ASR3 were selected because they clustered in the clade of other well characterized SG6 MYBs. Sequence alignments were generated via multiple sequence alignment using DNAMAN software, and phylogenetic trees were constructed on‐line (http://www.phylogeny.fr/index.cgi) based on multiple alignments of DNA or predicted amino acid sequences. The sequences of Arabidopsis genes were obtained from TAIR and those of other plants from other databases as referred to by the accession numbers. The accession numbers of all genes or proteins appearing in this paper are shown in Supporting Information Table S3.
2.3. Synteny analysis of genomic fragment containing MYBs cluster
All of the genomic data of Solanaceae and Petunia were mined from the Sol Genomics Network (https://solgenomics.net/). We manually annotated some of the genes.
2.4. Generation of constructs
All constructs in this study were made using the Gateway cloning system. The cDNA or gDNA of ASR genes and other genes were amplified by PCR and the fragments were inserted into entry vectors (pDONR207, pDONR221) by BP reaction. The entry vectors were then used in recombination reactions with the destination vectors pK2GW2.0/rfa (OE), pKGWFS7.0/rfa (Promoter:GUS), pGBKT7/GW (Y2H), and pGADT7/GW (Y2H) by LR reaction. All constructs are described in Supporting Information Table S4.
2.5. Petunia transformation
Constructs were transformed into Agrobacterium tumefaciens strain Agl1. Stable transformants of Petunia hybrida M1×R27 or W115 were generated by Agrobacterium‐mediated leaf‐disc transformation. The regenerated plants were checked by PCR for the presence of the construct and by RT‐PCR for its expression.
2.6. pH measurement
pH measurements were performed as described previously (Verweij et al., 2008).
2.7. Determination of anthocyanin content by HPLC
HPLC analysis was carried out by Scistd Testing Co., LTD. (Qingdao, China). Anthocyanins were extracted using 50 mg of powdered petal (flower development stage 2–3) suspended in 5 ml of 0.5% (v/v) HCl‐methanol and incubated for 2 hr at 4°C in the dark. The extracts were then centrifuged for 15 min at 14,000 rpm, and the supernatants were collected and stored at −20°C. The supernatant was diluted with methanol to 5 ml and filtered through a cellulose acetate membrane (Sartorius, Göttingen, Germany). HPLC was performed with Agilent 1260 Infinity LC (Agilent Technologies, USA). Anthocyanidin profile analysis was quantified at the 530 nm wavelength by using a calibration curve from commercial standards of anthocyanidins (pelargonidin, cyanidin, delphinidin, petunidin, peonidin, and malvidin, Sigma, USA).
2.8. RT‐PCR and qRT‐PCR analysis
To determine the expression of the genes of interest, we analyzed their expression profiles by RT‐PCR or qRT‐PCR (Real‐time RT‐PCR) methods. Total RNA was extracted via the TRIzol method, and its quantity and quality was measured by Nanodrop (Thermo Fisher Scientific, USA). cDNA synthesis was performed with PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara, Japan) and RT‐PCR analysis was carried out as previously described (Zhang, Yin, & Xia, 2008). qRT‐PCR was performed using the Bio‐Rad CFX Connect (Bio‐Rad, USA) with a SYBR‐Green PCR Master Mix Kit (SYBR® Premix Ex Taq™, Japan). Transcript abundance was calculated relative to actin by using the 2−ΔΔCt method. The primers used for these PCR analyses are listed in Supporting Information Table S5.
2.9. Isolation of ASR genes promoters and GUS reporter studies
The promoters of the ASR1, ASR2, and ASR3 genes from P. inflata S6 were amplified from genomic DNA using primers 323 and 325 (pASR1), 404 and 321 (pASR2), and 416 and 317 (pASR3). The promoters of ASR1 and ASR2 were isolated from the petunia M1 line using a Genome walking kit (Takara biomed, Takara, Japan) with gene specific primers and the universal primer AP4. The promoters were amplified and cloned in front of the GUS reporter gene in the pKGWFS7.0/rfa vector using the Gateway system. M1×R27F1 hybrid petunia plants were used as hosts for the generation of stable transformants. Histochemical GUS staining was performed as described previously (Albert et al., 2014). Petunia tissues were pretreated with diethyl ether for 30 s, immersed for 1 min in 90% ice cold acetone and then rinsed twice in 50 mM phosphate buffer (pH 7). The samples were then incubated in X‐gluc staining solution (with or without 1 mM K3Fe(CN)6/K4Fe(CN)6) overnight at 37°C. Seedlings were then decolored by treatment with 70% ethanol.
2.10. Yeast two‐hybrid assay
The BD or AD constructs for the expression of the fusion proteins in yeast were first transformed into the yeast strain AH109 and the yeast transformants were then screened by growth on SC‐Leu (for AD) or SC‐Trp (for BD) plates (2–3 days at 30°C). The transformant colonies that grew under these conditions were then used for cotransformation with the second construct for the interaction assay and plated on SC‐Leu/Trp media plates. Yeast double transformants were diluted in ddH2O, plated again on selective SC‐Leu/Trp/Ade/His medium at different dilutions (10×, 500×, and 5,000×), and grown for 2–4 days. All transformants containing BD‐ASR constructs displayed self‐activation (detectable by growth on selective medium also in the presence of an empty AD‐ vector). Therefore, only AD‐ASR constructs were used to test interactions with other proteins expressed as BD‐fusions.
3. RESULTS
3.1. Three novel members of the anthocyanin R2R3‐MYB family in Petunia
R2R3‐MYB proteins, such as the petunia AN2, AN4, PHZ, and DPL (Albert et al., 2011; Quattrocchio et al., 1999), the Arabidopsis PAP1 and PAP2 (Maier et al., 2013; Teng, Keurentjes, Bentsink, Koornneef, & Smeekens, 2005), the grape VvMYBA1 and VvMYBA2 (Walker et al., 2007), and the apple MdMYB1 and 10 (Espley et al., 2007; Takos et al., 2006), have been demonstrated to play crucial roles in the regulation of anthocyanin synthesis. Searching the genomes of the petunia wild species P. axillaris and P. inflata for anthocyanin R2R3‐MYBs resulted in the identification of three new members of this clade in P. inflata S6, which are absent in P. axillaris. Because of their similarity to known R2R3‐MYB anthocyanin regulators, we named these three genes ASR (ANTHOCYANIN SYNTHESIS REGULATOR). In the genome of P. inflata, these genes are arranged in a cluster (present within a single scaffold) (Figure 1a). All three ASR genes have a structure identical to AN2, AN4, DPL, and PHZ containing two introns (Figure 1b). Comparison of the genomic regions containing these MYB clusters in tomato (Solanum lycopersicum), wild tomato (S. pennellii), and potato (S. tuberosum), with P. inflata and P. axillaris scaffolds containing AN2, AN4‐DPL‐PHZ, and ASR1‐ASR2‐ASR3 reveals some levels of shared synteny. The arrangement of the genes surrounding the MYB genes in all of these species supports their common origin suggested by the results of the phylogeny analysis and implicating a series of duplications that have generated the numerous members of this family in each Solanacea species. However, synteny analysis also suggests that many of these rearrangements took place in the short time separating these closely related species.
Figure 1.
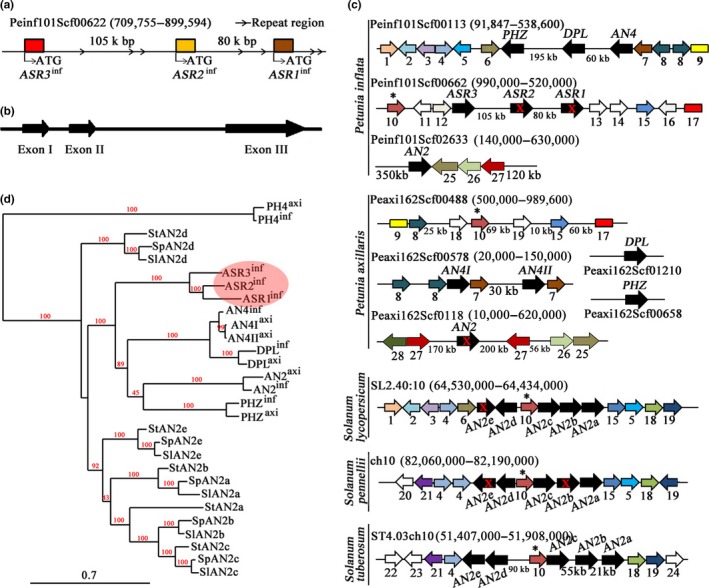
ASR genes cluster in Petunia. (a) The genomic fragment containing the ASR cluster in P. inflata. (b) Architecture of ASR genes in P. inflata. Exons are shown as arrow boxes. (c) Synteny analysis of the genomic regions containing SG6 MYB genes (black arrows) in Petunia and in three Solanum species. Arrows indicate genes and lines gene‐poor regions. Drawings are not to scale. Conserved genes are indicated by the same color. Red crosses indicate mutations in coding sequences that result in inactive proteins. The genes or sequences appearing here were summarized in Supporting Information Table S2. Among these genes, a much conserved gene (asterisk) for a heavy metal associated protein (10) is located in the genomic fragment in which SG6 genes are present in both Petunia and all Solanum species included in this analysis. (d) Phylogenetic analysis of the anthocyanin MYB genes in three Solanum and two Petunia species (genomic DNA sequences including introns). All of the Solanum genomic sequences were obtained from the SOL Genomic Network (https://solgenomics.net/). More information about the sequences used here is given in Supporting Information Table S3
The two copies of AN4 (and of some surrounding genes) in P. axillaris could be the reminiscence of the duplication that generated the ASR1, ASR2, ASR3 cluster, which in this lineage was followed by further rearrangements leading to partial loss of the cluster. Even though both P. axillaris and P. inflata contain a copy of the AN2 gene, these genes are contained in genomic fragments that were rearranged, resulting in a different distribution of surrounding genes in the two species. For example, on the right of AN2, the serine protease family protein (25), ARMADILLO/BETA‐CATENIN repeat family protein (26), and the serine/threonine protein kinase (27), are conserved, but their positions were rearranged. In addition, the P. axillaris AN2 allele contains a mutation that destroys the coding sequence (Quattrocchio et al., 1999), whereas the P. inflata allele encodes a wild type protein (Figure 1c).
The phylogenetic relations among these R2R3‐MYBs in petunia and different species of the Solanum genus show that all anthocyanin MYB genes of petunia are originated by duplication events that happened in the Petunia genus itself (Figure 1d). This finding further supports the previously proposed scenario that describes the evolution of specific sets of anthocyanin R2R3‐MYBs by recent duplications occurred in each species independently, thus generating species specific sets of SG6 genes and, consequently, unique pigmentation patterns (Supplemental note in Bombarely et al., 2016).
3.2. ASR1‐ASR2‐ASR3 cluster of anthocyanin regulators is different in distinct petunia accessions
Analysis of different petunia wild species revealed that all three ASR genes are present in the P. violacea accession and all of the three (the complete cluster) are missing in P. parodii and P. exserta. Therefore, it is not surprising that some Petunia hybrida lines, generated by repeated crosses among different wild species, have the genomic fragments containing these three genes (M1, V30, W59) and others do not (W115 and R27) (Figure 2a, Supporting Information Table S1). In P. inflata, a one base mutation in ASR1 inf at position 150 bp from the ATG and a one base insertion at position 148 bp in ASR2 inf (Figure 2b), interrupt the reading frame in these two genes. However, these mutations are absent in the ASR1 and ASR2 alleles of the P. hybrida lines M1, V30, and W59, which encode full size proteins. This finding brings up the previously proposed possibility that other wild petunia species, in addition to P. inflata and P. axillaris, contributed to the genomes of the P. hybrida lines (Bombarely et al., 2016). We indeed found that the purple flowering wild accession P. violacea has two sets of ASRs: one copy of each gene is similar to the P. inflata allele, and the other is similar to that of the P. hybrida line M1. Because the P. violacea material that we used is highly inbred, it is improbable that the two sequences we obtained belong to different alleles of the same locus, and we therefore concluded that these genes are present in two copies. This finding suggests that a genome from the P. violacea lineage contributed to the crosses from which (some of) the Petunia hybrida lines were generated.
Figure 2.
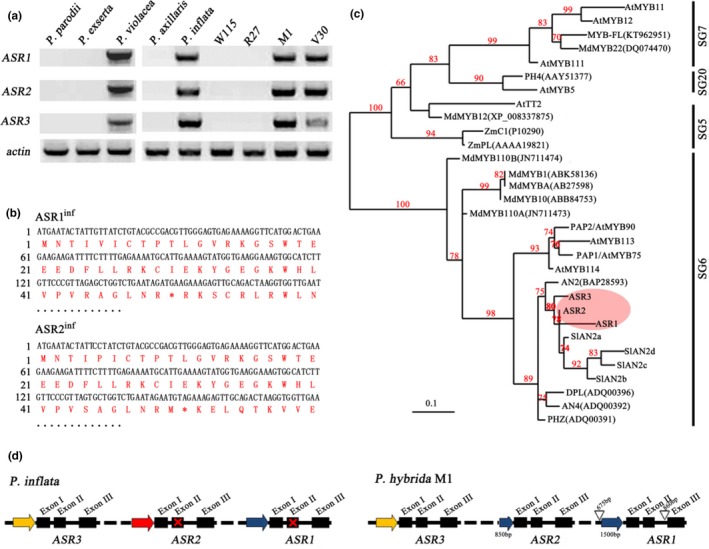
ASRs encode SG6 R2R3‐MYB proteins. (a) The ASR gene cluster is present in P. inflata, P. violacea, and in the P. hybrida lines M1 and V30 but is absent in P. axallaris, P. parodii, P. exserta, and in the P. hybrida lines W115, R27. A PCR method was used to amplify the specific ASR genes with their specific primers from DNA. (b) ASR1 and ASR2 encode truncated proteins in P. inflata. The mutations are indicated by an asterisk in the sequences. (c) Phylogenetic analysis of SG6 proteins in different species. The tree was constructed with maximum likelihood based on multiple alignments of the amino acid sequence of the R2R3‐motif only. (d) ASR genes from P. inflata and the P. hybrida line M1. Exons in ASR genes are indicated as solid boxes; similar promoter regions are shown as solid arrows of the same color, open triangles indicate insertions. Drawings are not to scale. Red crosses indicate mutation interrupting coding sequences
The phylogenic tree of the deduced amino acid sequences of ASRs and other R2R3‐MYBs in petunia further confirms that they encode SG6 MYBs and indicates that the different members of this group originate from duplications of a common ancestor and subsequent independent rearrangements in the different petunia species (Figure 2c). The comparison of the sequences of the ASR1‐ASR2‐ASR3 cluster from P. inflata and the P. hybrida line M1 shows several rearrangements. As shown in Figure 2d, a comparison between the ASR genes in P. inflata and in the P. hybrida line M1 reveals a 675 bp insertion present in the ASR1 M1 promoter region, approximately 1,500 bp upstream of the start codon. The ASR2 M1 promoter contains instead an 850 bp fragment, similar to a fragment of the ASR1 promoter. An 860 bp insertion is also found in the second intron of ASR1 M1.
3.3. ASR MYBs ectopic expression induces anthocyanin synthesis in different petunia organs
To test the role of ASR genes in anthocyanin synthesis, we generated ASR1, 2, and 3 overexpression (OE) lines containing constructs in which the coding sequences (from the Petunia hybrida line M1, ASRs M1 or from Petunia inflata, ASRs inf) are expressed from the CaMV35S promoter. As a host, we used the F1 hybrid between the two P. hybrida lines M1 and R27. Over 50 independent lines were generated for each construct. As shown in Figure 3a, even the calli of ASR1 M1‐OE, ASR2 M1‐OE, ASR3 M1‐OE, and ASR3 inf‐OE were pigmented, which was not observed for ASR1 inf‐OE and ASR2 inf‐OE as predicted because ASR1 inf and ASR2 inf encode truncated proteins. In the ASRs M1‐OE lines, flower buds showed pigmentation in very early stages of floral development when buds of the untransformed control are completely acyanic. However, after the buds opened, pigmentation did not differ significantly from that of the untransformed control lines. AN2 reaches high expression in petals shortly before bud opening (Quattrocchio et al., 1998), explaining why ectopic expression of the ASR genes induces early pigmentation of the young flower bud (when no AN2 activity is present) but does not contribute to increasing it further in the open flowers. Although overexpression of each of the three ASR genes affected pigmentation, ASR1 M1 ‐OE plants showed the most intense pigmentation, which included the stamens, stamen filaments, and gynoecium of the flower (Supporting Information Figure S2a). We could not measure obvious differences in the pH of the crude petal extract for flowers of any of the ASR M1‐OE lines compared to untransformed controls, suggesting that ASRs do not regulate the structural genes involved in vacuolar hyperacidification (Faraco et al., 2014).
Figure 3.
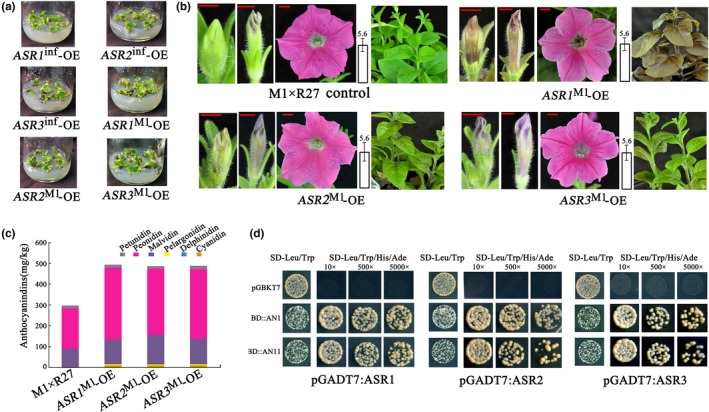
ASR1 M1, ASR2 M1, and ASR3 M1 all induce anthocyanin accumulation in different plant parts, and when ectopically expressed, are functionally similar. (a) Calli transformed with 35S:ASR1 M1 are strongly pigmented, whereas those transformed with 35S:ASR1 inf (inactive allele) show no pigmentation. (b) Pigmentation induced by the expression of ASRs in seedlings of transgenic lines in the hybrid M1×R27 between two Petunia hybrida lines. Petal phenotype at development stage 2, 4, and 7. Red size bars equal 5 mm. On the right of each panel is the pH value of the crude petal extract at development stage 7 (after bud opening). (c) Anthocyanin accumulation in ASRs M1‐OE petals at development stage 2–3, when no obvious color is yet visible in buds of untransformed controls. Anthocyanin levels are indicated as the mean of three biological replicates. Variation in biological replicates is less than 5%; therefore no error bar is shown here. (d) Interaction of ASRsM1 with AN1 and AN11 tested by a yeast two‐hybrid assay. The dilution series (10×, 500×, and 5,000×) were grown for 3 days before being photographed [Correction added on 31 January 2019, after first online publication: in the original version of Fig. 3a the same image was inadvertently used for ASR1M1‐OE and ASR2M1‐OE. This has now been amended.]
To further evaluate the effect of ASR expression on pigmentation, we measured the anthocyanidin content of flower petals at bud stage 2–3 in the overexpression lines. At this stage, no obvious coloration is detectable in buds of untransformed M1×R27 plants, while in transgenic plants, young buds are already colored. Petals of transgenic lines accumulate mainly peonidin and malvidin derivatives, similar to open buds of untransformed controls, which confer a magenta color to the tissue. ASRs M1‐OE young petals have higher anthocyanidin content than petals of the same age in controls, as is already clear by visual investigation (Figure 3c). In conclusion, expression of ASR genes induces anthocyanin accumulation.
Several studies previously showed that R2R3‐MYBs of the SG6 clade interact with bHLH factors and, in this way, participate to form WMBW regulatory complexes. Alignment of the deduced protein sequences for ASR1, ASR2, and ASR3 (for the ASR1 and ASR2 alleles of P. inflata, we have manually corrected the coding sequence to restore the reading frame destroyed by the mutation) showed that all three of them, as well as AN2, AN4, DPL, and PHZ, contain the conserved [DLx2Rx3Lx6Lx3] motif required for interactions with bHLH proteins (Supporting Information Figure S3) (Zimmermann et al., 2004). We used yeast two‐hybrid (Y2H) assays to examine the interaction of ASR1, ASR2, and ASR3 (from the Petunia hybrida line M1) with AN1 and AN11. This assay showed that the ASRs interact with both AN1 and AN11 (Figure 3d), implying that the ASR proteins can potentially participate in the WMBW complex that regulates anthocyanin biosynthesis.
3.4. ASR MYBs upregulate the expression of anthocyanin structural genes
We compared the expression profiles of anthocyanin structural genes in young bud petals of untransformed M1×R27 hybrids and ASRs M1‐OE lines by qRT‐PCR (Figure 4). Transcript levels of PALa, CHSaI, CHSj, F3H, F3′5′H, DFR, ANS, RT, MT, 5GT, and AT are increased in the overexpression lines for ASRs M1 compared to untransformed controls. Among these genes, most of the so‐called Late Biosynthetic Genes (LBGs), operating in the pathway starting from the step controlled by DFR, are strongly activated by the ASR regulators. However, the expression of the EBGs (Early Biosynthetic Genes), C4Ha, CHIa, 4CLa, and F3′H, were not affected. The Methylation at Five (MF) gene (Provenzano et al., 2014) responds differently to the different ASR MYBs. The tannin‐related genes ANR and LAR were not affected by ASRs M1 overexpression, indicating that these MYBs are specific for the anthocyanin pathway. Similarly, no expression of the flavonol synthase gene (FLS) was induced in ASRs‐OE lines, and no induction was observed for the expression of PH1 and PH5, the two P‐ATPases required for vacuolar hyperacidification in the epidermis of petals (Faraco et al., 2014, 2017; Quattrocchio et al., 2006; Verweij et al., 2008), consistent with the pH values measured in petals.
Figure 4.
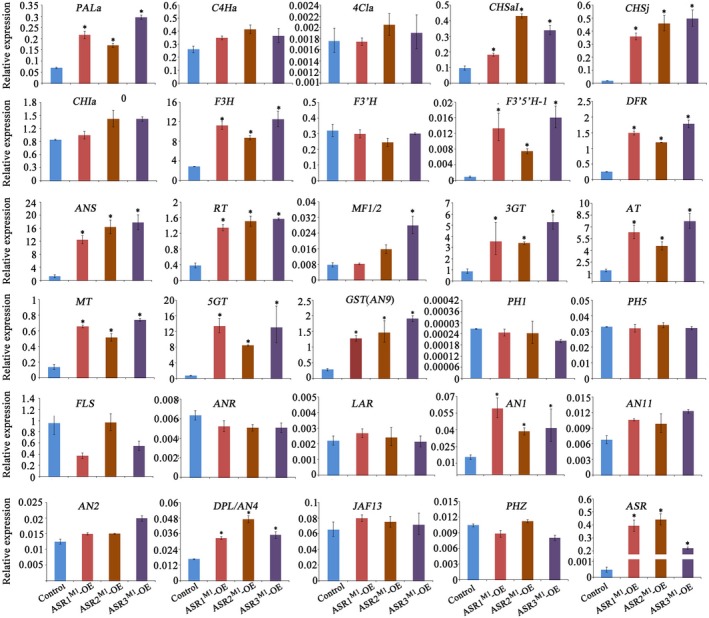
Expression of ASRs M1 induced pigmentation genes. Real‐time RT‐PCR analysis of the expression of anthocyanin structural and regulatory genes as well as vacuolar hyperacidification genes in petals of buds at developmental stage 2–3 from the untransformed control (M1×R27), ASR1 M1‐OE,ASR2 M1‐OE, and ASR3 M1‐OE. Relative expression is indicated as the mean ± SD of three biological replicates. Statistical significance was determined by one‐way ANOVA and significant differences between means is indicated by an asterisk (p‐value < 0.05)
To test the effect of ASR MYBs in a background with reduced activity of other anthocyanin R2R3‐MYBs, we also overexpressed ASR1 M1, ASR2 M1, and ASR3 M1 in the W115 Petunia hybrida line. This mutant is a double an2 an4 mutant that lacks the ASR cluster (similar to P. axillaris). Both petals and anthers of the flowers are acyanic in this line; therefore, the contribution of ASR genes to the pigmentation of flower organs can be more easily assessed in this genetic background. All lines ectopically expressing ASRs M1 in W115 had dusky bronze vegetative organs; ASR1 M1‐OE plants were the most intensely pigmented and ASR2 M1‐OE plants the least (Figure 5a). We further observed that OE of ASRs M1 increased pigmentation in young flower buds, especially at the early stages of floral development. Upon opening, however, all flowers were white, possibly because of the presence of the dominant allele of the FADING locus in W115 (Passeri, Koes, & Quattrocchio, 2016; de Vlaming & van Eekeres, 1982). The anthers of ASR M1 ‐OE plants were also (partially) pigmented, whereas the filaments and pistils did not differ in color from those of the untransformed controls. In addition, the seed pods in the early stages of development are pigmented in ASRs M1 overexpression lines in this background, while in the M1×R27 background, this finding was not observed, probably due to the weak hf1 allele (Supporting Information Figure S2). Analysis of anthocyanidin content in the young leaves of transgenic plants detected only highly substituted malvidin and petunidin derivatives, while no cyanidin‐related products were detected in these ASR‐OE plants, which was consistent with significant induction of expression of the F3′5′H gene (Figure 5b,c). We can conclude from this analysis that ASR1, ASR2, and ASR3 are indeed anthocyanin regulators sharing similar functions with other MYBs of the SG6 clade. However, although all of these genes are all effective in inducing anthocyanin accumulation, differences are detected in their effect on specific target genes.
Figure 5.
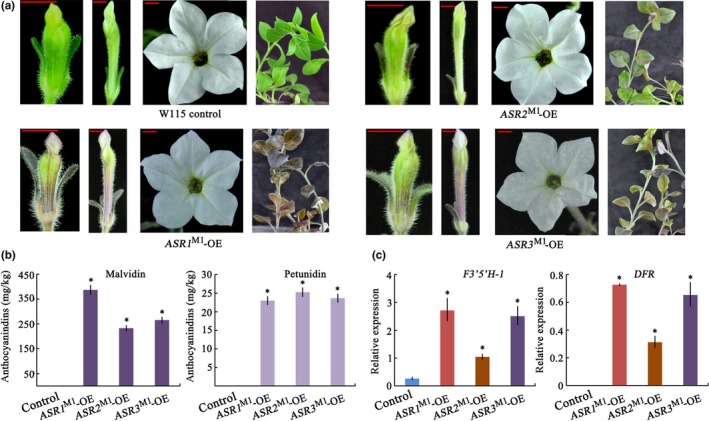
Overexpression of ASRs M1 enhance anthocyanin levels in the an2 an4 double mutant (Petunia hybrida line W115). (a) Flowers at development stages of 2, 4, and 7, and vegetative plant parts. Red size bars equal 5 mm. (b) Anthocyanin content in young leaves of untransformed control W115 and ASRs M1‐OE plants indicated as the mean ± SD of three biological replicates. (c) Analysis of the expression of F3′5′H and DFR genes in young leaves of W115 untransformed control and ASRs M1‐OE plant. Relative expression is indicated as the mean ± SD of three biological replicates. Statistical significance was determined by one‐way ANOVA and significant differences between means is indicated by an asterisk (p‐value < 0.05)
3.5. ASR genes are driven by highly rearranged promoters that confer different expression patterns
We analyzed the expression pattern of ASR genes using GUS reporter genes driven by their promoter regions. We generated stable transformants harboring these reporters in the M1×R27 hybrid background. The plants positive for GUS expression by PCR analysis were further analyzed for GUS activity. pASR1inf and pASR2inf drive GUS activity in several different tissues of young buds: petals, stigma, ovary, and anther filaments (Figure 6a,b). Instead, pASR3inf showed only weak expression in stigma, ovary, and anther filaments (Figure 6c). The expression pattern of pASR1M1 is similar to that of pASR1inf and pASR2inf, although the P. inflata alleles encode inactive proteins (Figure 6d). Mature leaves, stems, and sepals did not display GUS staining in any of the transgenic lines. T1 seedlings of pASR1inf:GUS, pASR2inf:GUS, and pASR1M1:GUS plants germinated on MS medium supplied with antibiotics for 15 d showed strong staining in several organs, while in pASR3inf:GUS seedlings grown under the same conditions, no GUS activity was detected (Figure 6e).
Figure 6.
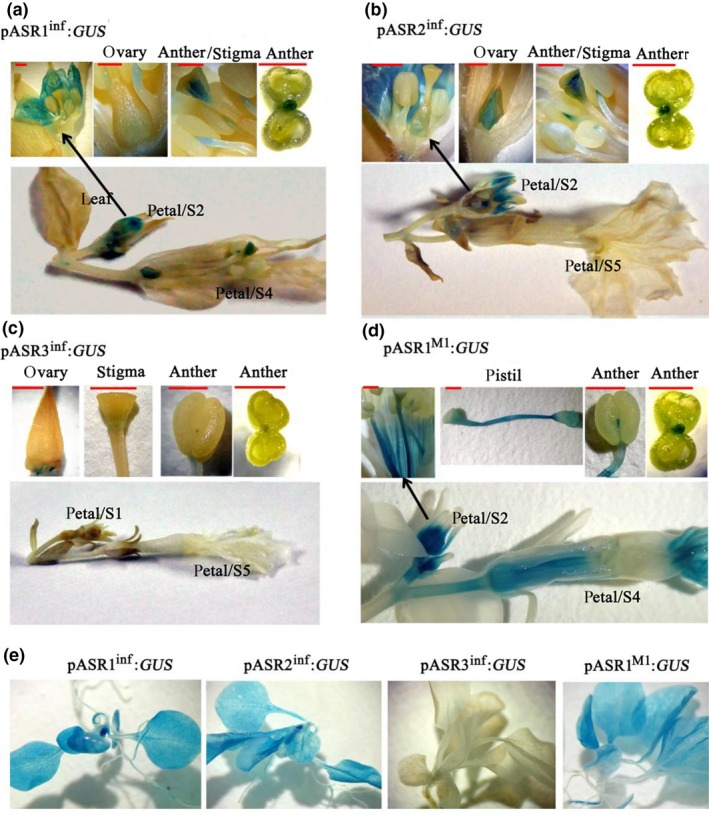
Promoter activity of ASRs in different petunia organs. Histochemical localization of GUS activity in flower organs from pASR1inf:GUS (a), pASR2inf:GUS (b), pASR3inf:GUS (c), and pASR1M1:GUS transgenic plants (d). Histochemical localization of GUS activity in T1 seedlings for the different Promoter:GUS fusions (e). Red size bars equal 1 mm
We confirmed the expression of each ASR gene in different tissues by RT‐PCR in P. inflata, and in the hybrids M1×R27 and M1×V30. The results were consistent with the promoter activity analysis. ASR1 and ASR2 were expressed throughout the reproductive organs with higher expression at early stages of floral development. In contrast, ASR3 was only expressed in stamens and pistils. Furthermore, we found that ASR1 and ASR2 were also expressed in leaves and stems (Supporting Information Figure S4a). Interestingly, after the exposure of the plants to intense light conditions, the total expression levels of ASRs was induced in vegetative tissues (Supporting Information Figure S4b), which is similar to what had been previously reported for DPL and PHZ (Albert et al., 2011, 2014). These results further support a possible common origin of ASR MYBs and DPL and PHZ from relatively recent duplication events.
4. DISCUSSION
The understanding of how gene expression patterns are generated and how they diverged during evolution is an important question in both developmental and evolution biology. Pigmentation genes provide an attractive model for such studies as their underlying genetic machineries are relatively simple compared to genetic networks regulating body shape and architecture and because changes in pigmentation are easy to score visually (Dembeck, Huang, Carbone, & Mackay, 2015; Gompel et al., 2005; Hoekstra, 2006). In plants, a good description of the pathway for the production of flavonoid pigments makes the use of this system even more attractive (Jaakola, 2013; Koes et al., 2005).
R2R3‐MYBs play in plants a central role in the regulation of the spatiotemporal pattern of expression of structural genes involved in the synthesis and accumulation of pigments, such as anthocyanins (Quattrocchio et al., 1999), condensed tannins (Baudry et al., 2004), betalains (Hatlestad et al., 2015), and the copigments flavonols (Sheehan et al., 2016), and are therefore main players in determining pigmentation patterns. A group of SG6 R2R3‐MYBs has been identified as regulators of the biosynthesis of anthocyanins in different plant parts in all plant species. In this report, we described a newly identified cluster of SG6 MYB genes in P. inflata, ASR1‐ASR2‐ASR3. These genes complete the picture of the SG6 phylogenic clade of anthocyanin regulators in this species, together with the already described AN4‐DPL‐PHZ cluster and the AN2 gene in petunia. The MYB proteins encoded by these genes are all very similar and all belong to the SG6 clade, which is clearly distinct from the clades containing MYBs involved in other aspects of pigmentation, such as vacuolar acidification (PhPH4) (Quattrocchio et al., 2006) and tannin accumulation (AtTT2) (Nesi, Jond, Debeaujon, Caboche, & Lepiniec, 2001) (Figure 2c).
The set of SG6 MYB genes in petunia consists of several members that probably originated by repeated events of duplication that were accompanied by heavy rearrangements of the genomic regions containing the different copies. Such events resulted in the expansion of this MYB group and in the diversification of their promoter regions. Apparently, the ASR genes, similar to other anthocyanin MYBs, reside in a very variable region of the genome that is subject to frequent rearrangements (Supplemental note in Bombarely et al., 2016). The amplification of the SG6 MYB family in petunia has occurred after the separation of the Solanum and Petunia genus, resulting in a petunia‐specific set of anthocyanin MYBs. The synteny of the genomic regions containing these SG6 genes in two wild petunia accessions, P. axillaris and P. inflata, revealed massive rearrangements that occurred in the time since the separation of these two evolutionary very closely related species, estimated at 0.9 Myr ago (Bombarely et al., 2016). It is conceivable that after the first duplications, and the consequent generation of repeats with highly similar sequences, duplication and rearrangement events happened relatively easily and frequently, due to (local) chromosome miss‐pairing. It is also likely that such events have been under strong positive selection when new flower color patterns generated new (successful) pollination syndromes in (for instance) Petunia.
In P. axillaris and P. inflata, the pattern of deposition of anthocyanins reflects, as expected, the function of the SG6 MYB regulators. In P. axillaris, AN2 is interrupted by a frame shift mutation, the whole ASR cluster is lost and AN4 is not expressed. As a consequence, P. axillaris can only synthesize anthocyanins in specific cells of the petal tube, but not in the petal limb and anthers (Supplemental note in Bombarely et al., 2016). In contrast, the P. inflata genome contains active copies of AN2, AN4, DPL, and PHZ, while ASR1 and ASR2 harbor inactivating mutations. The flowers of P. inflata have pigmented petals and colored anthers, while the flower pedicels do not show any anthocyanin accumulation. This finding is in agreement with the idea that the variation in expression pattern of SG6 MYB regulators drives the formation of anthocyanin pigmentation patterns.
The Petunia hybrida lines originated from manual crosses between different wild petunia species, which lead to the mixing and sorting of the different genomes. P. hybrida lines of the Amsterdam petunia collection analyzed in this work contain none, one or two ASR clusters. Rearrangement of the ASR2 promoter region and new insertions in the second intron of the M1 allele of this gene suggests that the dynamic nature of this genomic region born in wild petunias is conserved in the P. hybrida lines and possibly resulted in rearrangements even in the short time since people started breeding these species. However, it is also still possible that other (to us unknown) genomes participated in the breeding of Petunia hybrida and that the M1 allele originated from one of those. In fact, the wild petunia species P. violacea probably contains two copies of the ASR cluster, one of which contains ASR genes similar to the alleles found in the P. hybrida line M1 and the other genes containing the same mutations found in the P. inflata ASR cluster. This again supports the idea that patterns of coloration in petunia have been generated by repeated gain (and maybe loss) of new regulators occurring by duplications and differentiation of MYB genes, as well as by repeated loss of function for some of these copies. The differences in promoter regions, the expression patterns of single SG6 MYB genes in petunia, and the effect of mutation and overexpression reported here and in previous studies have shown that these genes differentiated substantially from each other. They can confer pigmentation to different tissues and under different conditions and result in the activation of a partially different set of target genes in the anthocyanin pathway.
The participation of MYBs in the WMBW complex regulating anthocyanin synthesis has been previously shown (Quattrocchio et al., 2006), and in this study, we assessed that this holds also for the three new petunia members of this group of regulators. The display of pigments by plant cells involves, next to the synthesis of anthocyanin molecules, other processes that determine the color of the tissue. One of these is the hyperacidification of the vacuolar lumen that is regulated by a WMBW complex containing a different R2R3‐MYB, encoded in petunia by the PH4 gene (Quattrocchio et al., 2006), and inducing transcription of the heterodimeric proton pump encoded by the PH1 and PH5 genes (Faraco et al., 2014). Similarly, the accumulation of other pigments and copigments that contribute to the coloration of plant tissues is regulated by yet other R2R3‐MYBs, for example, TT2 for tannins (Baudry et al., 2004), MYB‐FL for flavonols (Sheehan et al., 2016), and Y for betalaines (Hatlestad et al., 2015).
All of these MYBs are closely related to each other but differentiated in (a) their function, as they activate distinct sets of target genes, and (b) their expression patterns. The differences in sets of target genes for different MYB members of the WMBW complex are probably not merely due to the evolution of the promoters of the target genes. The acquired specificity is supported by the observation that MYBs of the SG6 clade cannot activate the PH4 targets PH1 and PH5. MYBs of these different clades (e.g., SG5 and SG20) can take part in the WMBW complex, conferring on it large flexibility in target gene specificity and giving rise to a never‐ending variation in pigmentation patterns via the activation of genes involved in the synthesis of different pigments and copigments and acidification of the vacuolar lumen where the pigments are accumulated (ref. PH4 and ref. TT2).
Thus, the characterization of the new R2R3‐MYB cluster containing the genes ASR1, ASR2, and ASR3 completes the picture of the petunia MYB genes involved in processes related to the differentiation of petal epidermal cells. These three genes can participate in the formation of WMBW complexes, probably playing a role in the acquisition of pigmentation in very young buds and in other plant parts when these are exposed to very high light. The presence in all plants of numerous genes encoding similar MYB proteins associated with strong rearrangement activity of the genomic region around them provides a genetic basis for the generation of new patterns of pigmentation and might have contributed to the differentiation of species within the Petunia genus by affecting their pollination syndromes.
ACCESSION NUMBERS
Sequence data from this paper have been deposited in GenBank (Supporting Information Table S3).
AUTHORS’ CONTRIBUTIONS
H.Z., R.K., and F.M.Q. designed the experiments and wrote the manuscript, H. S., L.W., Y.L (Yanbang Li), and J.G. completed gene identification and sequence analysis, X.D. and H. W. cared for the plants, J.Z. and V. P. performed DNA and RNA experiments, H. J. and Z.F. performed RT‐PCR analysis, Y.L (Yanmin Li) generated the transgenic petunia.
Supporting information
ACKNOWLEDGMENTS
We thank Professor Kai He in Lanzhou University for the vectors of pGBKT7/GW (Y2H) and pGADT7/GW (Y2H). We thank Wiley Editing Services Support for editing the English text of a draft of this manuscript.
Zhang H, Koes R, Shang H, et al. Identification and functional analysis of three new anthocyanin R2R3‐MYB genes in Petunia . Plant Direct. 2019;3:1–13. 10.1002/pld3.114
Contributor Information
Hechen Zhang, Email: zhc5128@126.com.
Francesca M. Quattrocchio, Email: f.quattrocchio@uva.nl
REFERENCES
- Albert, N. W. , Davies, K. M. , Lewis, D. H. , Zhang, H. , Montefiori, M. , Brendolise, C. , … Schwinn, K. E. (2014). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell, 26(3), 962–980. 10.1105/tpc.113.122069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert, N. W. , Lewis, D. H. , Zhang, H. , Schwinn, K. E. , Jameson, P. E. , & Davies, K. M. (2011). Members of an R2R3‐MYB transcription factor family in petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. The Plant Journal, 65(5), 771–784. 10.1111/j.1365-313X.2010.04465.x [DOI] [PubMed] [Google Scholar]
- Amato, A. , Cavallini, E. , Zenoni, S. , Finezzo, L. , Begheldo, M. , Ruperti, B. , & Tornielli, G. B. (2017). A grapevine TTG2‐Like WRKY transcription factor is involved in regulating vacuolar transport and flavonoid biosynthesis. Frontiers in Plant Science, 7, 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J. T. , Willis, J. H. , & Mitchell‐Olds, T. (2011). Evolutionary genetics of plant adaptation. Trends in Genetics, 27(7), 258–266. 10.1016/j.tig.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry, A. , Heim, M. A. , Dubreucq, B. , Caboche, M. , Weisshaar, B. , & Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana . The Plant Journal, 39(3), 366–380. 10.1111/j.1365-313X.2004.02138.x [DOI] [PubMed] [Google Scholar]
- Bernhardt, C. , Lee, M. M. , Gonzalez, A. , Zhang, F. , Lloyd, A. , & Schiefelbein, J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development, 130, 6431–6439. 10.1242/dev.00880 [DOI] [PubMed] [Google Scholar]
- Bombarely, A. , Moser, M. , Amrad, A. , Bao, M. , Bapaume, L. , Barry, C. S. , … Kuhlemeier, C. (2016). Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nature Plants, 2(6), 16074 10.1038/nplants.2016.74 [DOI] [PubMed] [Google Scholar]
- Burr, F. A. , Burr, B. , Scheffler, B. E. , Blewitt, M. , Wienand, U. , & Matz, E. C. (1996). The maize repressor‐like gene intensifier1 shares homology with the r1/b1 multigene family of transcription factors and exhibits missplicing. Plant Cell, 8, 1249–1259. 10.1105/tpc.8.8.1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey, C. C. , Strahle, J. T. , Selinger, D. A. , & Chandler, V. L. (2004). Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana . Plant Cell, 16, 450–464. 10.1105/tpc.018796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni, G. , Geuna, F. , Gavazzi, G. , & Tonelli, C. (1993). Molecular homology among members of the R gene family in maize. The Plant Journal, 3(2), 335–346. 10.1111/j.1365-313X.1993.tb00185.x [DOI] [PubMed] [Google Scholar]
- Dembeck, L. M. , Huang, W. , Carbone, M. A. , & Mackay, T. F. (2015). Genetic basis of natural variation in body pigmentation in Drosophila melanogaster. Fly (Austin), 9(2), 75–81. 10.1080/19336934.2015.1102807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vetten, N. , Quattrocchio, F. , Mol, J. , & Koes, R. (1997). The an11 locus controlling flower pigmentation in petunia encodes a novel WD‐repeat protein conserved in yeast, plants, and animals. Genes & Development, 1(11), 1422–1434. 10.1101/gad.11.11.1422 [DOI] [PubMed] [Google Scholar]
- de Vlaming, P. , & van Eekeres, J. E. M. (1982). A gene for flower color fading in Petunia hybrida. Theoretical and Applied Genetics, 61, 41–46. 10.1007/BF00261508 [DOI] [PubMed] [Google Scholar]
- Dubos, C. , Stracke, R. , Grotewold, E. , Weisshaar, B. , Martin, C. , & Lepiniec, L. (2010). MYB transcription factors in Arabidopsis . Trends in Plant Science, 15(10), 573–581. 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Espley, R. V. , Hellens, R. P. , Putterill, J. , Stevenson, D. E. , Kutty‐Amma, S. , & Allan, A. C. (2007). Red colorations in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. The Plant Journal, 49(3), 414–427. 10.1111/j.1365-313X.2006.02964.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraco, M. , Li, Y. , Li, S. , Spelt, C. , Di Sansebastiano, G. P. , Reale, L. , … Quattrocchio, F. M. (2017). A tonoplast P3B‐ATPase mediates fusion of two types of vacuoles in petal cells. Cell Reports, 19(12), 2413–2422. 10.1016/j.celrep.2017.05.076 [DOI] [PubMed] [Google Scholar]
- Faraco, M. , Spelt, C. , Bliek, M. , Verweij, W. , Hoshino, A. , Espen, L. , … Quattrocchio, F. M. (2014). Hyperacidification of vacuoles by the combined action of two different P‐ATPases in the tonoplast determines flower color. Cell Reports, 6(1), 32–43. 10.1016/j.celrep.2013.12.009 [DOI] [PubMed] [Google Scholar]
- Galliot, C. , Stuurman, J. , & Kuhlemeier, C. (2006). The genetic dissection of floral pollination syndromes. Current Opinion in Plant Biology, 9(1), 78–82. 10.1016/j.pbi.2005.11.003 [DOI] [PubMed] [Google Scholar]
- Gompel, N. , Prud'homme, B. , Wittkopp, P. J. , Kassner, V. A. , & Carroll, S. B. (2005). Chance caught on the wing: Cis‐regulatory evolution and the origin of pigment patterns in Drosophila . Nature, 433(7025), 481–487. 10.1038/nature03235 [DOI] [PubMed] [Google Scholar]
- Gonzalez, A. , Zhao, M. , Leavitt, J. M. , & Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal, 53, 814–827. 10.1111/j.1365-313X.2007.03373.x [DOI] [PubMed] [Google Scholar]
- Hatlestad, G. J. , Akhavan, N. A. , Sunnadeniya, R. M. , Elam, L. , Cargile, S. , Hembd, A. , … Lloyd, A. M. (2015). The beet Y locus encodes an anthocyanin MYB‐like protein that activates the betalain red pigment pathway. Nature Genetics, 47(1), 92–96. 10.1038/ng.3163 [DOI] [PubMed] [Google Scholar]
- Hernandez, J. M. , Feller, A. , Morohashi, K. , Frame, K. , & Grotewold, E. (2007). The basic helix loop helix domain of maize R links transcriptional regulation and histone medications by recruitment of an EMSY‐related factor. Proceedings of the National Academy Science of the United States of America, 104, 17222–17227. 10.1073/pnas.0705629104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra, H. E. (2006). Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity, 97(3), 222–234. 10.1038/sj.hdy.6800861 [DOI] [PubMed] [Google Scholar]
- Jaakola, L. (2013). New insights into the regulation of anthocyanin biosynthesis in fruit. Trends in Plant Science, 18(9), 477–483. 10.1016/j.tplants.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Johnson, C. S. , Kolevski, B. , & Smyth, D. R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell, 14, 1359–1375. 10.1105/tpc.001404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes, R. , Verweij, W. , & Quattrocchio, F. (2005). Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science, 10(5), 236–242. 10.1016/j.tplants.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Maier, A. , Schrader, A. , Kokkelink, L. , Falke, C. , Welter, B. , Iniesto, E. , … Hoecker, U. (2013). Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis . The Plant Journal, 74, 638–651. 10.1111/tpj.12153 [DOI] [PubMed] [Google Scholar]
- Martins, T. R. , Berg, J. J. , Blinka, S. , Rausher, M. D. , & Baum, D. A. (2013). Precise spatiotemporal regulation of the anthocyanin biosynthetic pathway leads to petal spot formation in Clarkia gracilis (Onagraceae). New Phytologist, 197, 958–969. 10.1111/nph.12062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins, T. R. , Jiang, P. , & Rausher, M. D. (2017). How petals change their spots: Cis‐regulatory re‐wiring in Clarkia (Onagracea). New Phytologist, 216(2), 510–518. 10.1111/nph.14163 [DOI] [PubMed] [Google Scholar]
- Nesi, N. , Jond, C. , Debeaujon, I. , Caboche, M. , & Lepiniec, L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell, 13, 2099–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passeri, V. , Koes, R. , & Quattrocchio, F. M. (2016). New challenges for the design of high value plant products: Stabilization of anthocyanins in plant vacuoles. Frontiers in Plant Science, 7, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano, S. , Spelt, C. , Hosokawa, S. , Nakamura, N. , Brugliera, F. , Demelis, L. , … Koes, R. (2014). Genetic control and evolution of anthocyanin methylation. Plant Physiology, 65(3), 962–977. 10.1104/pp.113.234526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan, M. D. , & Wessler, S. R. (1996). Molecular evolution of the plant R regulatory gene family. Genetics, 138, 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio, F. , Verweij, W. , Kroon, A. , Spelt, C. , Mol, J. , & Koes, R. (2006). PH4 of petunia is an R2R3‐MYB protein that activates vacuolar acidification through interaction with basic‐helix‐loop‐helix transcription factors of the anthocyanin pathway. Plant Cell, 18(5), 1274–1291. 10.1105/tpc.105.034041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio, F. , Wing, J. F. , van der Woude, K. , Mol, J. N. M. , & Koes, R. (1998). Analysis of bHLH and MYB‐domain proteins: Species‐specific regulatory differences are caused by divergent evolution of target anthocyanin genes. The Plant Journal, 13, 475–488. 10.1046/j.1365-313X.1998.00046.x [DOI] [PubMed] [Google Scholar]
- Quattrocchio, F. , Wing, J. , van der Woude, K. , Souer, E. , de Vetten, N. , Mol, J. , & Koes, R. (1999). Molecular analysis of the anthocyanin2 gene of Petunia and its role in the evolution of flower color. Plant Cell, 11, 1433–1444. 10.1105/tpc.11.8.1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay, N. A. , & Glover, B. J. (2005). MYB‐bHLH‐WD40 protein complex and the evolution of cellular diversity. Trends in Plant Science, 10(2), 63–70. 10.1016/j.tplants.2004.12.011 [DOI] [PubMed] [Google Scholar]
- Schwinn, K. , Venail, J. , Shang, Y. , Mackay, S. , Alm, V. , Butelli, E. , … Martin, C. (2006). A small family of MYB‐regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum . Plant Cell, 18(4), 831–851. 10.1105/tpc.105.039255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan, H. , Moser, M. , Klahre, U. , Esfeld, K. , Dell'Olivo, A. , Mandel, T. , … Kuhlemeier, C. (2016). MYB‐FL controls gain and loss of floral UV absorbance, a key trait affecting pollinator preference and reproductive isolation. Nature Genetics, 48(2), 159–166. 10.1038/ng.3462 [DOI] [PubMed] [Google Scholar]
- Spelt, C. , Quattrocchio, F. , Mol, J. N. , & Koes, R. (2000). Anthocyanin1 of petunia encodes a basic helix‐loop‐helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell, 12(9), 1619–1632. 10.1105/tpc.12.9.1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt, C. , Quattrocchio, F. , Mol, J. , & Koes, R. (2002). ANTHOCYANIN1 of petunia controls pigment, synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. Plant Cell, 14(9), 2121–2135. 10.1105/tpc.003772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn, W. J. , Wand, S. J. E. , Holcroft, D. M. , & Jacobs, G. (2002). Anthocyanins in vegetative tissues: A proposed unified function in photoprotection. New Phytologist, 155, 349–361. 10.1046/j.1469-8137.2002.00482.x [DOI] [PubMed] [Google Scholar]
- Stracke, R. , Werber, M. , & Weisshaar, B. (2001). The R2R3‐MYB gene family in Arabidopsis thaliana . Current Opinion in Plant Biology, 4(5), 447–456. 10.1016/S1369-5266(00)00199-0 [DOI] [PubMed] [Google Scholar]
- Takos, A. M. , Jaffé, F. W. , Jacob, S. R. , Bogs, J. , Robinson, S. P. , & Walker, A. R. (2006). Light‐induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiology, 142(3), 1216–1232. 10.1104/pp.106.088104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T. , Sasaki, N. , & Ohmiya, A. (2008). Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. The Plant Journal, 54, 733–749. 10.1111/j.1365-313X.2008.03447.x [DOI] [PubMed] [Google Scholar]
- Teng, S. , Keurentjes, J. , Bentsink, L. , Koornneef, M. , & Smeekens, S. (2005). Sucrose‐specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiology, 139, 1840–1852. 10.1104/pp.105.066688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche, M. , Chambrier, P. , Rodrigues Bento, S. , & Morel, P. (2016). Petunia, your next supermodel. Frontiers in Plant Science, 7, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij, W. , Spelt, C. E. , Bliek, M. , de Vries, M. , Wit, N. , Faraco, M. , … Quattrocchio, F. M. (2016). Functionally similar WRKY proteins regulate vacuolar acidification in Petunia and hair development in Arabidopsis . Plant Cell, 28(3), 786–803. 10.1105/tpc.15.00608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij, W. , Spelt, C. , Di Sansebastiano, G. P. , Vermeer, J. , Reale, L. , Ferranti, F. , … Quattrocchio, F. (2008). An H+ P‐ATPase on the tonoplast determines vacuolar pH and flower colour. Nature Cell Biology, 10(12), 1456–1462. 10.1038/ncb1805 [DOI] [PubMed] [Google Scholar]
- Walker, A. R. , Davison, P. A. , Bolognesi‐Winfield, A. C. , James, C. M. , Srinivasan, N. , Blundell, T. L. , … Gray, J. C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell, 11, 1337–1350. 10.1105/tpc.11.7.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, A. R. , Lee, E. , Bogs, J. , McDavid, D. A. , Thomas, M. R. , & Robinson, S. P. (2007). White grapes arose through the mutation of two similar and adjacent regulatory genes. The Plant Journal, 49(5), 772–785. 10.1111/j.1365-313X.2006.02997.x [DOI] [PubMed] [Google Scholar]
- Winkel‐Shirley, B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology, 126(2), 485–493. 10.1104/pp.126.2.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi, M. , Shimoyamada, Y. , Nakatsuka, T. , & Masuda, K. (2010). Two R2R3‐MYB genes, homologs of Petunia AN2, regulate anthocyanin biosyntheses in flower Tepals, tepal spots and leaves of asiatic hybrid lily. Plant Cell and Physiology, 51, 463–474. 10.1093/pcp/pcq011 [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Yin, W. , & Xia, X. (2008). Calcineurin B‐Like family in Populus: Comparative genome analysis and expression pattern under cold, drought and salt stress treatment. Plant Growth Regulation, 56, 129–140. 10.1007/s10725-008-9293-4 [DOI] [Google Scholar]
- Zimmermann, I. M. , Heim, M. A. , Weisshaar, B. , & Uhrig, J. F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B‐like BHLH proteins. The Plant Journal, 40, 22–34. 10.1111/j.1365-313X.2004.02183.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


