Abstract
Background:
Akkermansia muciniphila (AM) is a Gram-negative, mucin-degrading bacteria inhabiting the gastrointestinal tract associated with host phenotypes and disease states.
Objective:
Explore characteristics of overweight and obese female early-stage (0-II) breast cancer (BC) patients with low AM relative abundance (LAM) vs. high (HAM) enrolled in a presurgical weight-loss trial.
Design:
Secondary analysis of pooled participants in a randomized controlled trial (NCT02224807).
Participants/setting:
In 2014–2017, 32 female BC patients were randomized to weight-loss or attention-control arms from time of diagnosis-to-lumpectomy (average 30±9 days).
Intervention:
All were instructed to correct nutrient deficiencies via food sources and on upperbody exercises. The weight-loss group received additional guidance to promote 0.5–1 kg/week weight-loss via energy restriction and aerobic exercise.
Main outcome measures:
At baseline and follow-up, sera, fecal samples, two-24 hour dietary recalls and dual x-ray absorptiometry were obtained. Bacterial DNA was isolated from feces and PCR (16S) amplified; inflammatory cytokines were measured in sera.
Statistical analyses performed:
Differences between LAM and HAM were analyzed using t-tests and non-parametric tests. Spearman correlations explored relationships between continuous variables.
Results:
Participants were 61±9 (mean±SD) years old with body mass index 34.8±6 kg/m2. Mean AM relative abundance was 0.02% (0.007–0.06%) and 1.59% (0.59–13.57%) for LAM and HAM, respectively. At baseline, women with HAM vs. LAM had lower fat mass (38.9±11.2kg vs. 46.4±9.0kg, p=0.044). Alpha diversity (species richness) was higher in women with HAM (360.8±84.8 vs. 282.4±69.6, p=0.008) at baseline, but attenuated after weight-loss (p=0.058). At baseline, Interleukin-6 was associated with species richness (ρ=-0.471, p=0.008) and fat mass (ρ=0.529, p=0.002), but not AM. Change in total dietary fiber was positively associated with A. Muciniphila in LAM (ρ=0.626, p=0.002), but not HAM (ρ=0.436, p=0.180).
Conclusions:
Among women with early-stage BC, body composition is associated with AM, microbiota diversity, and IL-6; AM may mediate the effects of dietary fiber in improving microbiota composition.
Keywords: breast neoplasms, Akkermansia muciniphila, gastrointestinal microbiome, cytokines, weight loss, diet, obesity
Introduction
Breast cancer continues to be the most prevalent cancer in the United States, with 266,120 new cases expected in women in 2018.1 While many factors are involved in the development of the disease, the role of inflammation and hormones, such as estrogen and insulin, are at the forefront particularly in postmenopausal breast cancer risk and prognosis.2–4 Recent advances have highlighted the role that the gut microbiota may play in altering these factors to affect oncogenesis and progression. Several bacterial species exhibit beta-glucuronidase activity, which affects the metabolism of sex hormones, and as such may be involved in estrogen-dependent (ER+) breast cancer.5, 6 Similarly, other microbes like Proteobacteria are able to stimulate a systemic inflammatory response that has been associated with several cancer types.7–12 Conversely, emerging evidence suggests some bacteria may play a key role in protecting the gut barrier and reducing inflammation and other metabolic disorders.13–15
Akkermansia muciniphila
(AM) is a Gram-negative mucin-degrading microbe that inhabits the gastrointestinal tract of humans and other mammals. Though accounting for a relatively low proportion of the microbiome, the abundance of AM in the colon and in stool samples has been inversely associated with several health outcomes including obesity, diabetes and systemic inflammation.15–22 Though specific animal models have found detrimental effects of increased AM abundance,23, 24 AM is generally accepted as a symbiont with the gut epithelium. AM primarily metabolizes mucin into short-chain fatty acids, which stimulate goblet cells to produce more mucin.14, 20, 25, 26 Because the colonic epithelial cells form a single-layer barrier between the lumen and lamina propria, the mucus layer is integral in helping prevent permeability and potential translocation of a range of small molecules and even bacteria to the lymphatic system.23, 27
As AM is increasingly recognized for its beneficial properties, efforts have been made to supplement AM as a probiotic,18, 25, 28, 29 as well as determine which substrates (prebiotics) best increase the abundance of AM.22, 25 While many factors throughout life affect various microbe populations, from feeding in infancy30 to antibiotic use31 and habitual diet,32 richness and diversity of species remain the best metrics for assessing microbiome health.33 Hence, it is important to utilize observational studies when clinically relevant to diseases such as obesityassociated cancers. The objective of this study was to utilize longitudinal data from a randomized controlled weight loss trial in breast cancer survivors to explore the relationships of AM with body composition, diet and inflammatory markers.
Subjects and Methods
Participants in this study were women diagnosed with stage 0-II breast cancer enrolled in a presurgical weight-loss trial (clinicaltrials.gov ID: NCT02224807), which was approved by the Institutional Review Board of the University of Alabama at Birmingham (UAB) as previously described.34 Thirty-two women were recruited through the UAB Breast Health Center from 2014 to 2017. Eligibility criteria included a body mass index (BMI) ≥ 25 kg/m2, at least three weeks lag time until surgery, no neoadjuvant treatment or conditions that would affect weight status, and not currently participating in a weight-loss program. All women provided written informed consent; fecal samples, phlebotomy, dietary recalls, and anthropometrics were obtained at baseline visits (shortly after diagnosis) and follow-up (1–3 days prior to tumor excision).
All medications were recorded at baseline and follow-up appointments.35, 36 Two-24 hour dietary recalls (one weekend and one week day with the most recent recall corresponding to the day before fecal sample collection) were obtained at baseline and at follow-up by a registered dietitian using a multiple pass method and entered into the Nutrition Data System for Research (NDSR 2014, Minneapolis, Minnesota, USA).37 NDSR reports macro- and micronutrients derived primarily from United States Department of Agriculture National Nutrient Database for Standard Reference.38 Calories, nutrients, and food groups from the two dietary recalls at each time point were averaged for analysis.
Anthropometrics were obtained using standard procedures,39 and Dual X-ray Absorptiometry (DXA) (Lunar iDXA, GE Healthcare, Waukeha, Wisconsin, USA) was performed with patients wearing light clothing or gown as appropriate. Blood samples were collected after an 8-hour fast. Serum was separated and stored at -80°C until analysis. Interleukin-6 (IL-6) and Tumor Necrosis Factor-alpha (TNFα) were measured using a SECTOR imager 2400 (Meso Scale Diagnostics, Rockville, MD, USA).
Participants used sterile wipes to collect fecal samples, which were stored in their home freezer in plastic bags until the time of their appointment; samples were then stored at -80°C until processed. Microbiota analysis methods were previously reported by Demark-Wahnefried et al.34, 40 Microbe DNA was extracted using a ZR Fecal DNA Miniprep, and the V4 region of the 16S rRNA gene was PCR-amplified and sequenced with 500 cycle v2 kits with 250 bp paired-end read length using the Illumina Miseq, as previously described.41, 42 Quality was assessed via fastQC; reads where 80% of bases with a Phred score of less than 20 were removed. Analyses were performed with the Quantitative Insight into Microbial Ecology (QIIME) suite, version 1.9,43 and the QWRAP wrapper pipeline.41 Raw reads were clustered into operational taxonomic units (OTUs) using USEARCH at 97% similarity. The most common sequence in the cluster was the sequence assigned to that OTU which was given a taxonomic identification via QIIME’s RDP classifier. These identifications attempt to reach species level specificity for all OTUs however some can only be resolved to the genus, family, etc.44 In this dataset, 55 species were identified, including A. muciniphila. Taxonomic assignments to OTUs were performed with the Ribosomal Database Project classifier,45 Greengenes 16S database (version gg_13_8).46 Singletons were removed and OTUs with a total observation (sequence) count less than 0.005% were removed. The sequences were rarefied to the level of the sample with the lowest read count; all but one sample were above 10,000 so the rarefaction level was set at 50,000 (Supplementary Figure 1). The final filtered OTU table was used to calculate alpha and beta diversity. Alpha diversity measures were calculated using Chao1, observed species, Shannon Index, and whole tree Phylogenetic Diversity.33, 47 Beta diversity was calculated using BrayCurtis and weighted and unweighted UniFrac clustering of donors based on their microbial composition.48 Principal coordinates analysis (PCoA) plots were generated by QIIME to visualize the beta diversity among the samples.
After completing baseline appointments, participants were randomized to either weight loss or attention control arms and completed follow-up visits 1–3 days prior to surgery.34 All participants received resistance bands and were provided guidance on performing prehabilitation exercise for the biceps, triceps, and deltoids 2–3 times per week. All participants were advised on American Cancer Society Guidelines on nutrition and physical activity.49 Additionally, baseline dietary recall data from NDSR were used to determine any potential nutrient deficiencies, and participants were provided with guidance on correcting these deficiencies using food sources. Women randomized to the weight loss arm were prescribed a healthful, nutritionally adequate, energy-restricted diet, and encouraged to exercise at least 30 minutes/day to promote weight loss of 1 kilogram per week. Diet and exercise guidance was provided to the intervention group through twice-weekly in-person and telephone contact with a registered dietitian and an exercise physiologist.
Statistics
Exploratory analyses determined there were no differences between study arms in AM at baseline (p=0.924) or over the course of the study (p=0.585); therefore, participants from both arms were combined and then dichotomized per the median relative abundance of AM at baseline. All continuous variables were tested for normality and AM relative abundance was the only non-normally distributed variable. Descriptive statistics for the two groups, high AM (HAM) and low (LAM), were obtained at baseline and follow-up and compared using t-tests for normally distributed continuous variables (Mann-Whitney U-tests for all other continuous variables). Chi-square tests tested differences for categorical variables at baseline and follow-up. Changes in diet, anthropometric and alpha diversity data were compared between HAM and LAM using ANOVA. Spearman correlations were used to explore longitudinal relationships between AM and nutrients in the whole sample as well as within groups. P-values of 0.05 or less were considered statistically significant. Bonferroni correction was used for multiple comparisons in each set of analyses.
Operational Taxonomic Units (OTUs) were tested for significant differences on all taxonomic levels individually in frequency between LAM and HAM (Kruskal-Wallis p<0.05 with false discovery rate [FDR] correction).
Results
Participant characteristics
Thirty-three women were enrolled in the study, however, there was one postrandomization exclusion due to advanced disease within three days of randomization. Thirty-two women completed the trial and were included in this analysis. Study participants were 61±9 (mean ± standard deviation) years old with BMI 34.8±6 and evenly distributed between LAM (n=16) and HAM (n=16) (Table 1). Roughly half of the women were non-Hispanic white and were generally healthy, with few having a history of cardiovascular disease (n=5) or gastrointestinal disease (gastroesophageal reflux, n=3; ulcerative colitis, n=1; irritable bowel syndrome, n=1). None of the women took antibiotics over the course of the study. Roughly onethird of participants had type 2 diabetes; with two women taking metformin in the LAM and three in the HAM groups. No statistically significant differences were found in microbiota alpha diversity (observed species) between women with and without gastrointestinal disease (p=0.863), nor were there differences between women with and without diabetes (p=0.447), or those taking or not taking metformin (p=0.482). Nine women reporting taking proton pump inhibitors (PPI), six of which were in the LAM group. There were no statistically significant differences in AM (p=0.281) or observed species (p=0.175) between women taking PPI vs. not taking PPI. Twentynine of the women were postmenopausal, and the vast majority of participants had estrogen receptor positive and progesterone receptor positive disease. There were no differences between LAM and HAM for any of these variables. At baseline, HAM had lower fat mass (38.9±11.2kg vs. 46.4±9.0kg, p=0.044) than LAM.
Table 1.
Baseline characteristics of 32 women with breast cancer participating in a presurgical weight loss trial.
| Total (n=32) Mean (sd) | Low A. Muciniphila (n=16) Mean (sd) | High A. Muciniphila (n=16)] Mean (sd) | p | |
|---|---|---|---|---|
|
| ||||
| A. muciniphila relative abundancea | 3.38% (6.25%) | 0.04% (0.05%) | 6.72% (7.55%) | |
| Median | 0.14% | 0.02% | 1.59% | <0.0001 |
| (25th percentile, 75th percentile) | (0.002–6.25%) | (0.007–0.06%) | (0.59–13.57%) | |
| Microbiota Alpha Diversity | ||||
| Observed speciesb | 321.6 (86.1) | 282.4 (69.6) | 360.8 (84.8) | 0.008 |
| Chaolc | 380.0 (89.0) | 341.6 (71.2) | 418.4 (90.2) | 0.012 |
| Whole tree phylogenyd | 23.2 (5.7) | 20.7 (4.3) | 25.8 (5.8) | 0.010 |
| Shannon Indexe | 5.065 (0.722) | 4.735 (0.729) | 5.395 (0.559) | 0.007 |
| Randomized to weight loss group | 17 (53.1) | 9 (56.3) | 8 (50.0) | 1.000 |
| Age in years | 60.9 (9) | 58.1 (8.1) | 63.8 (9.9) | 0.090 |
| Body Mass Index | 34.8 (6) | 36.3 (5.5) | 33.3 (5.8) | 0.146 |
| Race | ||||
| Non-Hispanic Black [N (%)] | 13 (40.6%) | 7 (43.8%) | 6 (37.5%) | 0.934 |
| Non-Hispanic White [N (%)] | 17 (53.1%) | 8 (47.1%) | 9 (56.3%) | |
| More than one race [N (%)] | 2 (6.3%) | 1 (6.3%) | 1 (6.3%) | |
| Cardiovascular Disease [N (%)] | 5 (16%) | 2 (13%) | 3 (19%) | 1.000 |
| Gastrointestinal Disease [N (%)] | 5 (16%) | 4 (25%) | 1 (6%) | 0.166 |
| Type 2 Diabetes [N (%)] | 9 (28%) | 2 (13%) | 7 (44%) | 0.113 |
| Current smoker [N (%)] | 2 (6%) | 0 | 2 (13%) | 0.484 |
| Hormone receptor status per | ||||
| biopsy immunohistochemistry | 29 (91%) | 14 (88%) | 15 (94%) | 0.500 |
| ER+f | 23 (72%) | 11 (69%) | 12 (75%) | |
| PR+g | ||||
|
| ||||
A. muciniphila relative abundance: Akkermansia muciniphila bacterial species relative abundance in fecal samples provided by study participants
Observed species: number of unique species observed per sample
Chao1: estimate of species richness including rare species
Whole tree phylogeny: metric of phylogenetic diversity among species
Shannon Index: estimate of species richness
Estrogen Receptor Positive
Progesterone Receptor Positive
The median (interquartile range) relative abundance of AM was 0.148% (0.002–6.251%) at baseline; mean AM relative abundance was 0.019% (0.007–0.06%) and 1.594% (0.59113.570%) for LAM and HAM, respectively. All four alpha diversity metrics indicated greater microbial richness and diversity in HAM (p<0.05 for all), with HAM participants having approximately 25% more species present in stool samples at baseline (p=0.008). Comparing LAM and HAM, relative of abundance of AM did not change significantly over the study period (p=0.419), however, 2 LAM and 1 HAM would have been reclassified at follow-up.
Beta diversity analysis.
Kruskal-Wallis tests indicated statistically significant differences in microbiota composition between LAM and HAM. In addition to differences in AM (FDR p=0.000219), an additional 40 OTUs were different between LAM and HAM (FDR p<0.2 for all). Noteworthy among these OTUs were higher Prevotella and Lactobacillus and lower Clostridium, Campylobacter and Helicobacter genera in HAM vs. LAM. PCoA plots (Supplementary Figure 2) were generated to visualize differences between LAM in HAM at baseline. Though all betadiversity tests yielded significant differences between LAM and HAM, Bray-Curtis dissimilarity yielded the most visual and statistical (p=0.002) differences between the two groups.
Relationships among AM, anthropometrics and diet.
Over the course of the study, which lasted 30±9 days, average weight loss was 2.2±2.0 kg among all participants. Women in LAM and HAM lost significant body weight and fat mass (p<0.005 for all); HAM had a significant decrease in percent body fat (p=0.005), though not statistically greater than LAM (Table 2). Weight change was not associated with AM change (p=0.905).
Table 2.
Longitudinal dual x-ray absorptiometry body composition measures of 32 women with breast cancer participating in a presurgical weight loss triala having low versus high Akkermansia muciniphila relative abundance at baseline.
| Low A. muciniphila (n=16) | High A. muciniphila (n=16) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline | Follow-up | Baseline | Follow-up | ||||||||
|
| |||||||||||
| Mean | SD | Mean | SD | pb | Mean | SD | Mean | SD | pb | pc | |
| Weight (kg) | 96.98 | 13.57 | 94.71 | 13.20 | 0.0003 d | 85.40 | 17.59 | 83.34 | 17.62 | 0.001 d | 0.766 |
| Lean body mass (kg) | 47.23 | 5.55 | 46.56 | 5.32 | 0.026 | 43.63 | 6.72 | 42.99 | 6.83 | 0.046 | 0.420 |
| Fat mass (kg) | 46.44 | 9.00 | 44.87 | 8.70 | 0.003 d | 38.91 | 11.17 | 37.02 | 11.42 | 0.003 d | 0.849 |
| Fat percentage | 49.3% | 3.6% | 48.8% | 3.5% | 0.097 | 46.5% | 4.4% | 45.5% | 4.6% | 0.005 d | 0.499 |
|
| |||||||||||
Mean time from baseline to follow-up was 30±9 days.
Within group differences from baseline to follow-up
Differences between groups from baseline to follow-up
Significant difference from baseline to follow-up within group: p<0.05/4 = 0.0125
At baseline, there were no significant differences between the nutrients and diet quality measures listed in Table 3. Both groups reduced total calories, carbohydrates and glycemic load from baseline to follow-up (p<0.003 for all), however HAM additionally reduced total and all major types of fats (p<0.0003 for all), though not statistically greater than LAM. Post-hoc analyses explored whole sample and within group correlations between AM and food groups. Unadjusted correlations between changes in AM and total fat, carbohydrate, vegetable protein and monounsaturated fatty acids were observed (p<0.05 for all), however polyunsaturated fatty acids (PUFA) (ρ=0.512, p=0.003) and total fiber (ρ=0.489, p=0.005) were statistically significant after correction for multiple comparisons. Further exploratory analyses revealed that among PUFAs, linolenic acid (ρ=0.5442, p=0.011) and linoleic acid ρ=0.509, p=0.003) were associated with change in AM, and among fiber types, change in AM and insoluble fiber (ρ=0.478, p=0.006) was significant, with pectins (ρ=0.409, p=0.020) trending as well. Change in total dietary fiber was positively associated with AM in LAM (ρ=0.626, p=0.002), but not HAM (ρ=0.436, p=0.180). Within group analyses indicate LAM had significant changes in AM associated with change in total and animal protein, as well as soluble fiber (p<0.003 for all). Conversely, no significant associations between diet and AM were observed in HAM.
Table 3.
Longitudinal dietary intake reported by women with breast cancer participating in a presurgical weight loss triala having low versus high Akkermansia muciniphila abundance.
| Low A. muciniphila (n=16) | High A. muciniphila (n=16) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline | Follow-up | Baseline | Follow-up | ||||||||
| Mean | SD | Mean | SD | pb | Mean | SD | Mean | SD | pb | Pc | |
|
| |||||||||||
| Energy (kcal)e | 1549.9 | 359.5 | 1088.1 | 545.6 | 0.002 d | 1335.9 | 412.8 | 889.2 | 238.7 | 0.000 d | 0.920 |
| Total Fat (g)f | 68.3 | 17.5 | 47.0 | 28.2 | 0.006 | 59.5 | 23.1 | 33.7 | 9.7 | 0.000 d | 0.590 |
| Total Carbohydrate (g)f | 166.6 | 53.3 | 112.6 | 58.5 | 0.002 d | 148.0 | 49.4 | 101.5 | 39.5 | 0.002 d | 0.692 |
| Total Protein (g)f | 64.4 | 19.9 | 55.1 | 21.7 | 0.078 | 56.0 | 19.9 | 49.4 | 18.6 | 0.253 | 0.706 |
| Animal Protein (g)f | 43.7 | 14.9 | 39.9 | 17.8 | 0.280 | 39.6 | 17.8 | 35.6 | 18.1 | 0.450 | 0.976 |
| Vegetable Protein (g)f | 20.7 | 7.1 | 15.1 | 8.1 | 0.036 | 16.4 | 7.3 | 13.8 | 4.9 | 0.210 | 0.344 |
| Total Saturated Fatty Acids (g)f | 21.5 | 8.5 | 14.6 | 9.6 | 0.007 | 17.3 | 6.3 | 11.1 | 4.3 | 0.001 d | 0.791 |
| Total Monounsaturated Fatty Acids (g)f | 23.4 | 6.6 | 17.4 | 11.7 | 0.024 | 21.1 | 8.9 | 12.6 | 4.0 | 0.002 d | 0.434 |
| Total Polyunsaturated Fatty Acids (g)f | 18.0 | 5.2 | 11.1 | 6.5 | 0.006 | 16.6 | 8.4 | 7.1 | 2.5 | 0.000 d | 0.378 |
| Total Dietary Fiber (g)f | 14.6 | 4.5 | 14.4 | 6.9 | 0.939 | 13.7 | 8.5 | 14.3 | 6.4 | 0.675 | 0.735 |
| Soluble Dietary Fiber (g)f | 5.3 | 1.9 | 4.7 | 2.5 | 0.230 | 4.0 | 1.5 | 4.7 | 2.5 | 0.106 | 0.048 |
| Insoluble Dietary Fiber (g)f | 9.3 | 3.4 | 9.8 | 5.0 | 0.702 | 9.7 | 7.5 | 9.5 | 4.7 | 0.877 | 0.701 |
| Pectins (g)f | 1.9 | 0.7 | 2.0 | 1.0 | 0.864 | 1.7 | 0.8 | 1.9 | 0.7 | 0.404 | 0.595 |
| Glycemic Indexg | 59.0 | 6.0 | 57.5 | 5.5 | 0.292 | 60.6 | 4.4 | 60.3 | 5.7 | 0.855 | 0.570 |
| Glycemic Loadg | 91.1 | 34.6 | 55.9 | 28.3 | 0.000 d | 82.0 | 30.8 | 52.5 | 24.7 | 0.001 d | 0.591 |
| Cholesterol (mg)h | 259.0 | 122.5 | 217.7 | 168.8 | 0.336 | 213.3 | 113.5 | 140.8 | 65.9 | 0.018 | 0.535 |
|
| |||||||||||
Mean time from baseline to follow-up was 30±9 days.
Within group differences from baseline to follow-up
Differences between groups from baseline to follow-up
Significant difference from baseline to follow-up within group: p<0.05/16 = 0.0031
kcal: kilocalories
g: grams
glucose reference
mg: milligrams.
Alpha diversity
To determine these nutrients’ effects on microbiota composition, correlations between changes in the nutrients and alpha diversity were undertaken. Change in observed species and whole tree phylogeny were significantly associated with change in polyunsaturated fats (p=0.001 for both), but not fiber. Because PUFAs were reduced significantly in the entire sample throughout the study, it is unclear whether increasing PUFA would increase AM or diversity. Over the course of the study, alpha diversity diverged between LAM and HAM, with LAM increasing and HAM decreasing nonsignificantly. Between group change ANOVA p-values for Chao1 and Shannon Index were 0.036 and 0.017, respectively (Supplementary Table 1).
Inflammatory cytokines
Inflammatory cytokines did not change significantly in the whole sample of 32 women over the study period. Mean IL-6 levels were 1.26±0.61 pg/mL and 1.50±1.09 at baseline and follow-up, respectively (p=0.165). Mean TNFα levels were 2.90±0.70 pg/mL and 2.92±0.66 at baseline and follow-up, respectively (p=0.808). No significant relationships were observed between TNFα and AM, microbiota diversity or body composition at baseline or over the course of the study. However, IL-6 was associated with species richness (ρ=-0.471, p=0.008) and fat mass (ρ=0.529, p=0.002), but not AM at baseline (Figure 1).
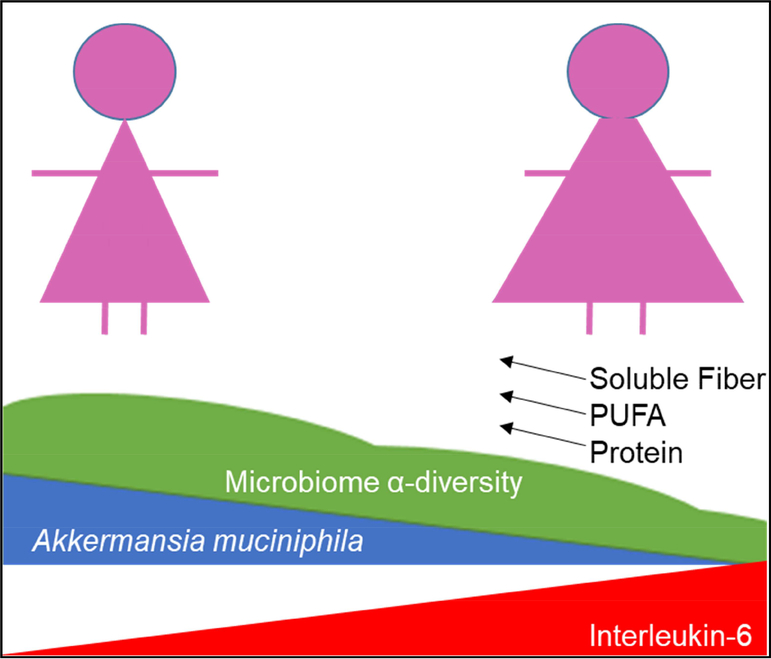
Figure 1.
In 32 overweight and obese women with early stage breast cancer participating in a presurgical weight loss trial, body fat is inversely associated with fecal Akkermansia muciniphila (AM) and microbiota diversity, and positively associated with Interleukin-6. In women with lower AM, change in dietary soluble fiber, polyunsaturated fatty acids (PUFA), and protein were positively associated with microbiota diversity and AM abundance.
Discussion
To date, this is the first longitudinal study investigating the relationship between diet, body composition and the fecal microbiota in women with breast cancer. Most notably, the relative abundance of Akkermansia muciniphila, had a bimodal distribution (Supplementary Figure 3), which correlated with relevant health outcome measures and was associated with favorable dietary changes. Relative abundance of AM in this sample was similar to those previously reported. Healthy adults have been reported to have 1–5% AM in several studies,50–52 though men and women with diabetes have been observed to have lower abundance without metformin, and 15–20% relative abundance while taking metformin.53 Low and high AM groups were observed to be ecologically different as measured through alpha- and beta diversity analyses, indicating AM may be indicative of microbe diversity.
In this sample, women with higher body fat had lower AM relative abundance. This phenotype has been observed in humans16 and preclinical models,15 and similar associations were observed with metabolic health and inflammation. The mechanisms by which AM may play a causal role have been explored in several mouse models, indicating that its relative abundance increases cecal antimicrobial peptides and improves mucus layer thickness in most animal models, two functions that serve to prevent bacterial translocation, endotoxemia, and subsequent systemic inflammation.14 Similarly, supplementing AM in chow-fed mice prevented weight gain and reduced inflammation via the same mechanisms.54 To date, these results have not been replicated in humans, though AM supplementation has been determined to be safe.55
Preclinical studies have also indicated mechanisms by which increased AM abundance may be deleterious to the mucus layer and increase inflammation. One study found that feeding gnotobiotic mice colonized with human flora a fiber-free diet led to degradation of the mucus layer and increased susceptibility to pathogens.23 Another study explored the role of microbes in heme-induced hyperproliferation of coloncytes, finding that antibiotics prevented cytotoxicity by eliminating sulfide- and mucin-degrading bacteria, including AM. Taken together, it is plausible in humans that increased AM may promote epithelial damage when dietary intake is suboptimal (i.e. lacking fiber and/or rich in heme). Future studies are needed to translate these studies in humans.
Translocation of Gram-negative bacteria (most notably, E. coli) in the colon induces endotoxemia,56, 57 which results in production of TNFα by macrophages.58 Because AM generally serves to reinforce the gut barrier, an inverse relationship between the two was anticipated. Similarly, it was hypothesized that IL-6 would be reflective of gut-derived inflammation.26, 59 Though IL-6 was not associated with AM relative abundance, its relationship to body composition and microbiota alpha diversity was a significant finding that warrants further investigation in a larger population.60
Previous reports have indicated metformin (Glucophage) may alter the composition of the gut microbiome.36 In this study, there were seven patients with type 2 Diabetes in HAM, only three of these were prescribed and taking metformin over the course of the study, compared to both patients with diabetes taking metformin in LAM. Recently, a longitudinal study observed microbial changes in patients undergoing metformin treatment which included increases in AM and other beneficial microbes.61 Transfer of these patients’ stools samples to germ-free mice increased glucose tolerance, indicating a potential benefit of AM to metabolic health. It is unknown whether these benefits are sustained over years, which may be why a microbial benefit from metformin was not present in this study.
Microbiota alpha diversity was widely distributed in this study sample with LAM and HAM diverging over the study period. These results are supported by positive associations with alpha diversity and PUFA,62 protein,63 and more commonly, dietary fiber64 that have been observed in other studies. Mixed results have been reported in regard to the effects of weight loss on microbiota diversity,65, 66 however, it seems most probable that a diet reducing microbial substrates would prevent expansion of the populations studied.23, 67 These data suggest that dietary intervention may increase the alpha diversity of AM in individuals with lower levels of AM; future studies are warranted.
Limitations
There are four main limitations to this secondary analysis. First, the sample size was small. Though the sample was racially diverse, study participants were women in the state of Alabama, and thus these findings may not be generalizable to the larger population. Second, this was not a controlled feeding study, so dietary recall data is subject to error and may have been influenced by recommendations to improve nutrient deficiencies. Additionally, only two days of recall data were collected at each time point; therefore, these may not have represented a sufficient sampling to represent habitual intake.68 Similarly, dietary habits at study initiation may have been affected by stress associated with a recent diagnosis of breast cancer. Third, this was a brief study, and some of the dietary changes required to maintain target weight loss may not have been sustainable in the long term; more specifically, it is unknown whether the changes observed in microbe composition were maintained after participants completed the study. Additionally, fecal samples were used to quantify AM, which is in greater abundance in the mucus layer; thus generalizations from fecal data to colonic contents cannot be assumed. Mucosal contents of feces were not quantified, and data on stool consistency were not collected, which may have been a significant factor in the microbe composition. Nonetheless, several findings herein support previous observations and warrant further investigation.
Conclusion
In this sample of overweight and obese women with early stage breast cancer, Akkermansia muciniphila was associated with body composition and microbiota alpha diversity. Alpha diversity, but not AM was inversely associated with the inflammatory cytokine IL-6, which suggests a lesser role for AM in mitigating systemic inflammation. Additionally, these data suggest that in women with negligible AM composition, increasing dietary fiber and protein during weight loss may result in increases in AM; a benefit not observed in those with higher AM at baseline. Overall, this secondary analysis conducted among women with breast cancer supports previous observations in the general population that AM is associated with microbiota diversity and body composition, and its relative abundance can change in conjunction with diet.
Supplementary Material
Research Question:
Is the relative abundance of Akkermansia muciniphila in fecal samples associated with body composition, diet and inflammatory markers in women with early stage breast cancer participating in a randomized controlled weight loss trial?
Key Findings:
A. muciniphila had a bimodal distribution; women with higher abundance of the bacterial species had an average 7.5kg less fat mass, significantly higher microbiota alpha diversity, and no differences in inflammatory markers at baseline. Change in total dietary fiber intake was positively associated with change in A. muciniphila relative abundance, but only for those with low abundance at baseline.
Acknowledgments
This study was supported by the National Cancer Institute R21 (CA178359), P30 (CA013148), and R25 (CA047888). The following are acknowledged for their support of the Microbiome Resource at the University of Alabama at Birmingham: School of Medicine, Comprehensive Cancer Center (P30AR050948), Center for AIDS Research (5P30AI027767), Center for Clinical Translational Science (UL1TR000165, UL1TR001417), and Heflin Center for Genomic Sciences.
Footnotes
The authors have no potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Andrew D. Frugé, Email: fruge@auburn.edu, Department of Nutrition, Dietetics and Hospitality Management, Auburn University, 260 Lem Morrison Drive, Auburn, AL 36849 334844-3271.
William Van der Pol, Email: liamvdp@uab.edu, Department of Computational Biology and Bioinformatics, University of Alabama at Birmingham (UAB), Birmingham, AL 205-934-3727.
Laura Q. Rogers, Email: rogersl@uab.edu, Department of Nutrition Sciences, UAB, Birmingham, AL 205-975-1667.
Casey D. Morrow, Email: caseym@uab.edu, Department of Cell, Developmental & Integrative Biology, UAB, Birmingham, AL 205-934-5705.
Yuko Tsuruta, Email: tsuru@uab.edu, Department of Nutrition Sciences, UAB, Birmingham, AL 205-975-2418.
Wendy Demark-Wahnefried, Email: demark@uab.edu, Department of Nutrition Sciences, UAB, Birmingham, AL 205-975-4022.
References:
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: A Cancer Journal for Clinicians. 2018:n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 2.Madeddu C, Gramignano G, Floris C, Murenu G, Sollai G, Macciò A. Role of inflammation and oxidative stress in post-menopausal oestrogen-dependent breast cancer. Journal of Cellular and Molecular Medicine. 2014;18:2519–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agnoli C, Grioni S, Pala V, et al. Biomarkers of inflammation and breast cancer risk: a case-control study nested in the EPIC-Varese cohort. Scientific Reports. 2017;7:12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pierce BL, Ballard-Barbash R, Bernstein L, et al. Elevated Biomarkers of Inflammation Are Associated With Reduced Survival Among Breast Cancer Patients. Journal of Clinical Oncology. 2009;27:3437–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwa M, Plottel CS, Blaser MJ, Adams S. The Intestinal Microbiome and Estrogen Receptor–Positive Female Breast Cancer. JNCI: Journal of the National Cancer Institute. 2016;108:djw029–djw029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goedert JJ, Jones G, Hua X, et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: a population-based case-control pilot study. Journal of the National Cancer Institute. 2015;107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goedert JJ, Gong Y, Hua X, et al. Fecal Microbiota Characteristics of Patients with Colorectal Adenoma Detected by Screening: A Population-based Study. EBioMedicine. 2015;2:597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain S, Suklabaidya S, Das B, Raghav SK, Batra SK, Senapati S. TLR4 activation by lipopolysaccharide confers survival advantage to growth factor deprived prostate cancer cells. The Prostate. 2015;75:1020–1033. [DOI] [PubMed] [Google Scholar]
- 9.Bultman SJ. Emerging roles of the microbiome in cancer. Carcinogenesis. 2014;35:249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Candela M, Turroni S, Biagi E, et al. Inflammation and colorectal cancer, when microbiota-host mutualism breaks. World Journal of Gastroenterology : WJG. 2014;20:908–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hullar MAJ, Burnett-Hartman AN, Lampe JW. Gut Microbes, Diet, and Cancer. Cancer treatment and research. 2014;159:377–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nistal E, Fernández-Fernández N, VIVAS S, Olcoz JL. Factors determining colorectal cancer: the role of the intestinal microbiota. Frontiers in Oncology. 2015;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zitvogel L, Daillere R, Roberti MP, Routy B, Kroemer G. Anticancer effects of the microbiome and its products. Nat Rev Micro. 2017;advance online publication. [DOI] [PubMed] [Google Scholar]
- 14.Everard A, Belzer C, Geurts L, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9066–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneeberger M, Everard A, Gómez-Valadés AG, et al. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Scientific reports. 2015;5:16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dao MC, Everard A, Aron-Wisnewsky J, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2015. [DOI] [PubMed] [Google Scholar]
- 17.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55. [DOI] [PubMed] [Google Scholar]
- 18.Anhê FF, Pilon G, Roy D, Desjardins Y, Levy E, Marette A. Triggering Akkermansia with dietary polyphenols: A new weapon to combat the metabolic syndrome? Gut Microbes. 2016;7:146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cani PD, de Vos WM. Next-Generation Beneficial Microbes: The Case of Akkermansia muciniphila. Frontiers in Microbiology. 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derrien M, Belzer C, de Vos WM. Akkermansia muciniphila and its role in regulating host functions. Microbial Pathogenesis. 2017;106:171–181. [DOI] [PubMed] [Google Scholar]
- 21.Ottman N, Geerlings SY, Aalvink S, de Vos WM, Belzer C. Action and function of Akkermansia muciniphila in microbiome ecology, health and disease. Best Practice & Research Clinical Gastroenterology. 2017. [DOI] [PubMed] [Google Scholar]
- 22.Plovier H, Everard A, Druart C, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23:107–113. [DOI] [PubMed] [Google Scholar]
- 23.Desai Mahesh S, Seekatz Anna M, Koropatkin Nicole M, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167:1339–1353.e1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ijssennagger N, Belzer C, Hooiveld GJ, et al. Gut microbiota facilitates dietary hemeinduced epithelial hyperproliferation by opening the mucus barrier in colon. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:10038–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gómez-Gallego C, Pohl S, Salminen S, Vos WMD, Kneifel W. Akkermansia muciniphila: a novel functional microbe with probiotic properties. Beneficial Microbes. 2016;7:571–584. [DOI] [PubMed] [Google Scholar]
- 26.Reunanen J, Kainulainen V, Huuskonen L, et al. Akkermansia muciniphila Adheres to Enterocytes and Strengthens the Integrity of the Epithelial Cell Layer. Appl Environ Microbiol. 2015;81:3655–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carding S, Verbeke K, Vipond DT, Corfe BM, Owen L. Dysbiosis of the gut microbiota in disease. Microbial Ecology in Health and Disease. 2015;26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou K Strategies to promote abundance of Akkermansia muciniphila, an emerging probiotics in the gut, evidence from dietary intervention studies. Journal of Functional Foods. 2017;33:194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen W, Shen M, Zhao X, et al. Anti-obesity Effect of Capsaicin in Mice Fed with HighFat Diet Is Associated with an Increase in Population of the Gut Bacterium Akkermansia muciniphila. Frontiers in Microbiology. 2017;8:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bäckhed F, Roswall J, Peng Y, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:690–703. [DOI] [PubMed] [Google Scholar]
- 31.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4554–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graf D, Di Cagno R, Fåk F, et al. Contribution of diet to the composition of the human gut microbiota. Microb Ecol Health Dis. 2015;26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuruta Y, Rogers LQ, Krontiras H, et al. Exploring effects of presurgical weight loss among women with stage 0–II breast cancer: protocol for a randomised controlled feasibility trial. BMJ Open. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogers MA, Aronoff DM. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect. 2016;22:178.e171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Keyzer W, Huybrechts I, De Vriendt V, et al. Repeated 24-hour recalls versus dietary records for estimating nutrient intakes in a national food consumption survey. Food Nutr Res. 2011;55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bodner-Montville J, Ahuja JKC, Ingwersen LA, Haggerty ES, Enns CW, Perloff BP. USDA Food and Nutrient Database for Dietary Studies: Released on the web. J Food Comp Anal. 2006;19, Supplement:S100–S107. [Google Scholar]
- 39.Lohman T, Roache A, Martorell R. Anthropometric Standardization Reference Manual. Medicine and science in sports and exercise. 1992;24:952. [Google Scholar]
- 40.Demark-Wahnefried W, Nix JW, Hunter GR, et al. Feasibility outcomes of a presurgical randomized controlled trial exploring the impact of caloric restriction and increased physical activity versus a wait-list control on tumor characteristics and circulating biomarkers in men electing prostatectomy for prostate cancer. BMC Cancer. 2016;16:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar R, Eipers P, Little RB, et al. Getting started with microbiome analysis: sample acquisition to bioinformatics. Current protocols in human genetics / editorial board, Jonathan L. Haines ... [et al.]. 2014;82:18.18.11–18.18.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daft JG, Ptacek T, Kumar R, Morrow C, Lorenz RG. Cross-fostering immediately after birth induces a permanent microbiota shift that is shaped by the nursing mother. Microbiome. 2015;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America. 2011;108 Suppl 1:4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. [DOI] [PubMed] [Google Scholar]
- 45.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faith DP, Baker AM. Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol Bioinform Online. 2006;2:121–128. [PMC free article] [PubMed] [Google Scholar]
- 48.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kushi LH, Doyle C, McCullough M, et al. American Cancer Society Guidelines on nutrition and physical activity for cancer prevention: reducing the risk of cancer with healthy food choices and physical activity. CA Cancer J Clin. 2012;62:30–67. [DOI] [PubMed] [Google Scholar]
- 50.Belzer C, de Vos WM. Microbes inside—from diversity to function: the case of Akkermansia. The ISME Journal. 2012;6:1449–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gobert AP, Sagrestani G, Delmas E, et al. The human intestinal microbiota of constipated-predominant irritable bowel syndrome patients exhibits anti-inflammatory properties. Scientific Reports. 2016;6:39399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. [DOI] [PubMed] [Google Scholar]
- 53.de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, et al. Metformin Is Associated With Higher Relative Abundance of Mucin-Degrading Akkermansia muciniphila and Several Short-Chain Fatty Acid–Producing Microbiota in the Gut. Diabetes Care. 2017;40:54–62. [DOI] [PubMed] [Google Scholar]
- 54.Zhao S, Liu W, Wang J, et al. Akkermansia muciniphila improves metabolic profiles by reducing inflammation in chow diet-fed mice. Journal of molecular endocrinology. 2017;58:1–14. [DOI] [PubMed] [Google Scholar]
- 55.Brodmann T, Endo A, Gueimonde M, et al. Safety of Novel Microbes for Human Consumption: Practical Examples of Assessment in the European Union. Front Microbiol. 2017;8:1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neal MD, Leaphart C, Levy R, et al. Enterocyte TLR4 Mediates Phagocytosis and Translocation of Bacteria Across the Intestinal Barrier. The Journal of Immunology. 2006;176:3070–3079. [DOI] [PubMed] [Google Scholar]
- 57.Cani PD, Amar J, Iglesias MA, et al. Metabolic Endotoxemia Initiates Obesity and Insulin Resistance. Diabetes. 2007;56:1761–1772. [DOI] [PubMed] [Google Scholar]
- 58.Patel PN, Shah RY, Ferguson JF, Reilly MP. Human Experimental Endotoxemia in Modeling the Pathophysiology, Genomics, and Therapeutics of Innate Immunity in Complex Cardiometabolic Diseases. Arteriosclerosis, Thrombosis, and Vascular Biology. 2015;35:525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nature Immunology. 2015;16:448. [DOI] [PubMed] [Google Scholar]
- 60.Sindhu S, Thomas R, Shihab P, Sriraman D, Behbehani K, Ahmad R. Obesity Is a Positive Modulator of IL-6R and IL-6 Expression in the Subcutaneous Adipose Tissue: Significance for Metabolic Inflammation. PLoS ONE. 2015;10:e0133494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu H, Esteve E, Tremaroli V, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;advance online publication. [DOI] [PubMed] [Google Scholar]
- 62.Menni C, Zierer J, Pallister T, et al. Omega-3 fatty acids correlate with gut microbiome diversity and production of N-carbamylglutamate in middle aged and elderly women. Scientific Reports. 2017;7:11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clarke SF, Murphy EF, O'Sullivan O, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63:1913–1920. [DOI] [PubMed] [Google Scholar]
- 64.Simpson HL, Campbell BJ. Review article: dietary fibre–microbiota interactions. Alimentary pharmacology & therapeutics. 2015;42:158–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Furet J-P, Kong L-C, Tap J, et al. Differential Adaptation of Human Gut Microbiota to Bariatric Surgery–Induced Weight Loss. Links With Metabolic and Low-Grade Inflammation Markers. 2010;59:3049–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seganfredo FB, Blume CA, Moehlecke M, et al. Weight-loss interventions and gut microbiota changes in overweight and obese patients: a systematic review. Obesity Reviews. 2017;18:832–851. [DOI] [PubMed] [Google Scholar]
- 67.Deehan EC, Walter J. The Fiber Gap and the Disappearing Gut Microbiome: Implications for Human Nutrition. Trends in Endocrinology & Metabolism. 2016;27:239–242. [DOI] [PubMed] [Google Scholar]
- 68.Ma Y, Olendzki BC, Pagoto SL, et al. Number of 24-Hour Diet Recalls Needed to Estimate Energy Intake. Annals of Epidemiology. 2009;19:553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.