Diverse and highly variable systems involved in biological conflicts and self-versus-nonself discrimination are ubiquitous in bacteria but much less studied in archaea. We performed comprehensive comparative genomic analyses of the archaeal systems that share components with analogous bacterial systems and propose an approach to identify new systems that could be involved in these functions. We predict polymorphic toxin systems in 141 archaeal genomes and identify new, archaea-specific toxin and immunity protein families. These systems are widely represented in archaea and are predicted to play major roles in interactions between species and in intermicrobial conflicts. This work is expected to stimulate experimental research to advance the understanding of poorly characterized major aspects of archaeal biology.
KEYWORDS: archaea, archaeocins, polymorphic toxins, quorum sensing
ABSTRACT
Numerous, diverse, highly variable defense and offense genetic systems are encoded in most bacterial genomes and are involved in various forms of conflict among competing microbes or their eukaryotic hosts. Here we focus on the offense and self-versus-nonself discrimination systems encoded by archaeal genomes that so far have remained largely uncharacterized and unannotated. Specifically, we analyze archaeal genomic loci encoding polymorphic and related toxin systems and ribosomally synthesized antimicrobial peptides. Using sensitive methods for sequence comparison and the “guilt by association” approach, we identified such systems in 141 archaeal genomes. These toxins can be classified into four major groups based on the structure of the components involved in the toxin delivery. The toxin domains are often shared between and within each system. We revisit halocin families and substantially expand the halocin C8 family, which was identified in diverse archaeal genomes and also certain bacteria. Finally, we employ features of protein sequences and genomic locus organization characteristic of archaeocins and polymorphic toxins to identify candidates for analogous but not necessarily homologous systems among uncharacterized protein families. This work confidently predicts that more than 1,600 archaeal proteins, currently annotated as “hypothetical” in public databases, are components of conflict and self-versus-nonself discrimination systems.
INTRODUCTION
Prokaryotes (bacteria and archaea) inhabit complex environments, where they interact and compete with other organisms, both prokaryotic and eukaryotic. Multiple offense and defense systems emerged during evolution to combat resource competitors and parasites (1–3). These systems are involved in incessant arms races and therefore are typically among the fastest-evolving genes (4). They are relatively well studied in bacteria, but the data on such systems in archaea is scarce.
The peptide antibiotic compounds, which form a prominent arm of the microbial defense systems, can be classified into two major types: ribosomally and non-ribosomally synthesized peptides (RiPPs and NRPs, respectively) (5, 6). Both types of peptides are typically further processed (maturated) and chemically modified. The RiPPs are the only antimicrobial peptides that have been experimentally characterized in archaea. They are known as archaeocins (by analogy with bacteriocins) and so far have been identified only in a few Halobacteria and Sulfolobales species (7).
In addition to antibiotics, bacteria also deploy large, multidomain protein toxins in conflicts with other organisms. The polymorphic toxin systems (PTSs) that are typically deployed against closely related strains or species are large proteins with distinct trafficking mechanisms from which the toxin domain, often an enzyme, is cleaved off upon entry into the target cell (3, 8). The toxins deployed in PTSs are extremely diverse and attack a variety of cellular components, primarily RNA and DNA, and in some cases proteins and lipids (3). However, different types of toxin domains can be coupled in the same polypeptide to domains mediating one or more distinct mechanisms of trafficking/delivery (3, 9). Among these mechanisms, the delivery of a toxin through a phage tail apparatus is the most complex because it requires dozens of genes that encode phage tail components, toxins that often contain a Zn-dependent processing metallopeptidase (MPTase) and the toxin domain itself, as well as immunity proteins and regulatory components. This machinery is referred to as type VI secretion (9, 10) and PVC (Photorhabdus virulence cassettes) systems (3). Recently, the term “tailocins” was coined to denote type VI secretion and PVC systems, emphasizing the origin of both from phage tails (11).
Another type of toxin system consists of several large multidomain components that collectively make a pore in the membrane, attach to a target cell, and then deliver and cleave the toxin domain off once inside the target cell. These systems are typified by entomotoxins TcABC (toxin complex ABC) from Photorhabdus species that target eukaryotic cells via modification of Rho GTPases (3, 12). Some toxins are secreted outside the cell through dedicated secretion systems that either recognize specific signal sequences or use dedicated chaperones to target these toxins for export (9). Finally, many toxins are secreted through either the twin-arginine translocation (Tat) pathway translocating folded proteins (13) or through the Sec system as unfolded proteins (14).
Homologs of several bacterial PTSs have been identified in archaea (3), but the genes comprising these systems have never been accordingly annotated in any of archaeal genomes available in public databases or studied experimentally. The scarcity of information on intermicrobial conflict mechanisms in archaea prompted us to revisit the protein families that comprise predicted archaeal conflict systems. Here, we present an attempt to comprehensively characterize the representation of the PTS and related systems in archaea and to predict new varieties of such systems on the basis of observed sequence and contextual features of the respective genomic loci.
RESULTS AND DISCUSSION
Archaeal polymorphic toxin systems.
As a result of an iterative “guilt-by association” procedure (see Fig. S1 in the supplemental material for details) and extensive manual curation, we report here 1,909 genes, in 377 genomic islands from 141 archaeal genomes, that are predicted to encode protein components of PTS and related systems (see Tables S1 and S2 in the supplemental material).
Iterative “guilt-by-association” procedure to identify polymorphic toxin systems in archaea. Download FIG S1, PDF file, 0.1 MB (135.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Phyletic patterns for archaeal offense systems and key components of putative self-nonself recognition system. Download Table S1, XLSX file, 0.1 MB (81.3KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Genomic loci for archaeal offense systems and key components of putative self-nonself recognition systems. Download Table S2, XLSX file, 1.5 MB (1.5MB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
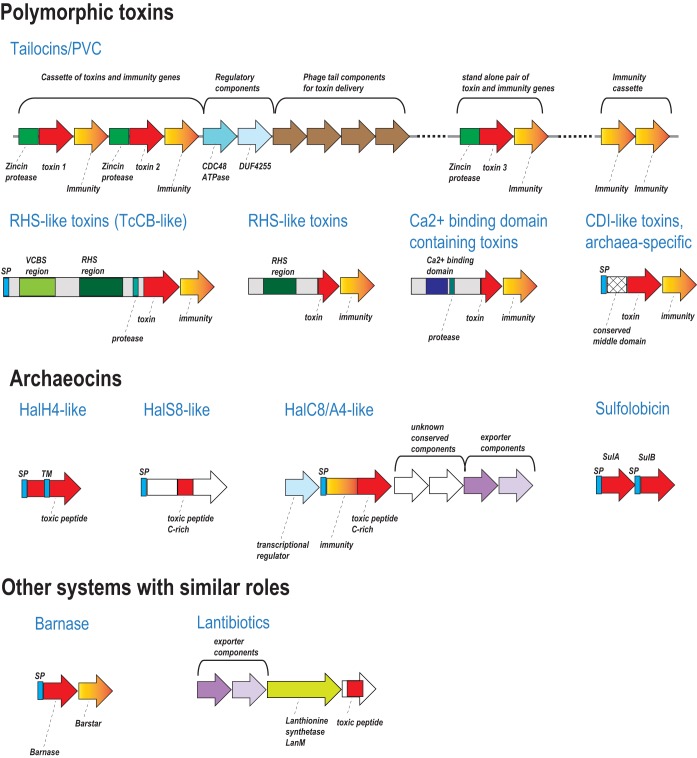
These systems could be classified into four distinct types based on their architectures or the trafficking-related domains with which the toxins are combined: (i) PVC-like systems or tailocins; (ii) RHS (rearrangement hot spot) systems, which often include a large, multidomain toxin protein, with a toxin domain located C terminal of the RHS repeats, coupled with entomotoxin TcB and TcC components; (iii) previously uncharacterized toxins with an N-terminal regions containing Ca2+-binding domains; and (iv) predicted archaea-specific PTSs that consist of multidomain proteins with signal peptides, indicating secretion via the general secretory pathway. To the N terminus of the toxin domain, these proteins contain a distinct domain, often with a prominent hydrophobic region, that might be involved in their maturation and delivery (see below) either via contact with rival cells or through some other protein-protein interaction (Fig. 1 and Table S1). Finally, there is the barnase-barstar system, which is a minimal, invariant version of the classic PTS.
FIG 1.
General organization of offense and self-versus-nonself discrimination systems identified in archaeal genomes. Genes are shown by block arrows. Distinct protein families are indicated. Identified domains are shown inside the arrows. Abbreviations: SP, signal peptide; TM, transmembrane helices.
PVC-like systems.
Complete PVC-like systems, with contractile phage tails implicated in toxin injection (Fig. 1 and Table S1), were identified in the genomes of 32 archaea, all mesophiles. Similarly to the previously described systems (3, 15), most of these genomes contain a large locus, with multiple genes encoding components of the phage tail, baseplate, tail assembly regulatory components, toxins, and putative immunity proteins (Fig. 1 and 2). Typically, toxin genes are followed by immunity genes, either within these large loci or in separate toxin-immunity cassettes. Several genomes also contain stand-alone poly-immunity loci that string together multiple immunity proteins that could counter the toxin from these PVC systems (3, 16) (Fig. 1 and 2; Table S2).
FIG 2.
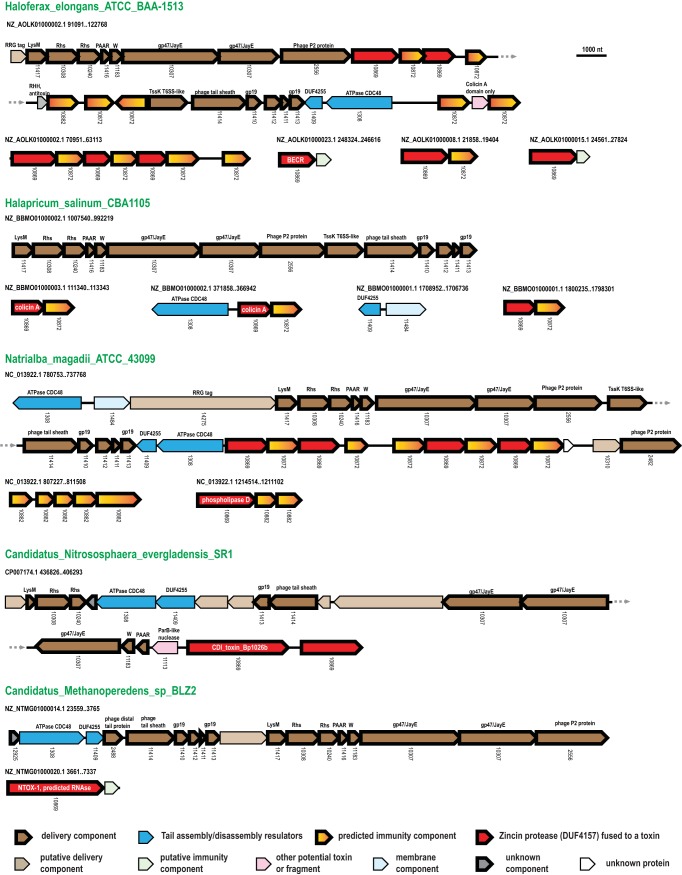
Selected gene neighborhoods of the PVC/tailocin polymorphic toxin system. For each gene neighborhood, the organism name, genome partition, and coordinates of the locus are indicated. Genes are shown by block arrows, with the length roughly proportional to the size of the corresponding gene. Genes are colored according to the general function (see color code at the bottom of the figure). The arCOG numbers are indicated beneath the respective genes (when assigned). Brief annotations of the selected genes are shown above the arrows. Identified toxin families are indicated within respective arrows. Abbreviations: SP, signal peptide; TM, transmembrane helices.
The phage tail in both PVC and type VI secretion systems consists of the tube-sheath complex and baseplate components, but the tail-related components and the disassembly ATPase in these systems appear to have originated from different phage families (3, 17). The tube is assembled from the tail tube proteins (arCOG11410 and arCOG11413) and is covered by the sheath protein (arCOG11414). The baseplate wedge consists of phage baseplate assembly protein (arCOG11183), gp47/JayE protein (arCOG10307), and phage P2-like protein (arCOG02556), which are homologous to phage Mu baseplate assembly proteins p46, p47, and p48, respectively (15). Baseplate central hub components, namely, an LysM domain-containing protein (arCOG11417) and RHS repeat-containing protein (arCOG10308), correspond to the phage Mu proteins p43 and p44, respectively (15). Another conserved component of the baseplate is a spike protein (arCOG10240), the counterpart to the phage Mu protein p45 (15). Some of these systems also contain the PAAR domain, typified by an amino acid motif that typically contains proline-alanine-alanine-arginine. The PAAR domains are typical of the type 6 secretion system (T6SS), in which they mediate the recruitment of stand-alone toxins (3, 18). The presence of these domains in the archaeal PVC systems suggests that they could be similarly used to diversify the toxin repertoire beyond those toxins that are fused with the PVC-metallopeptidase (MPTase) within the same multidomain protein. Several other proteins in the predicted archaeal PVC-like systems are not similar to any known phage proteins, so that their functions remain unclear, but because these proteins are encoded within the tail assembly module, they can be tentatively assigned to the toxin delivery apparatus (Fig. 2; Table S2). One of such proteins is specific to Halobacteria (arCOG14275). These proteins are variable in length, but all contain an arginine-rich C-terminal region with the conserved “RRG” motif (see Fig. S2 in the supplemental material). This motif has not been described previously but, like many other terminal motifs, might represent a protein targeting signal (19, 20). Additionally, these loci include genes for two essential regulators of the tail assembly, the CDC48-like AAA+ ATPase (arCOG01308) and the tail terminator protein (arCOG11409, DUF4255 family) (15).
Sequence logo for the “RRG” C-terminal motif specific for Halobacteria. Download FIG S2, PDF file, 1.0 MB (1,016.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The larger loci typically encode a toxin. All complete toxins contain an N-terminal metallopeptidase (PVC-MPTase) domain that is diagnostic of the PVC systems (3) and various C-terminal domains that represent known or putative toxins. Altogether, we identified 170 proteins containing the PVC-MPTase domain, but several of these are fragments rather than complete, functional proteins. The typical size of the PVC-MPTase-toxin fusions is about 350 amino acids (aa), but they could be as large as 1,106 aa (NTE_00486 in Nitrososphaera evergladensis SR1) when fused with additional domains. In the majority of these proteins, the C-terminal domains (i.e., the putative toxins) are not similar to any known toxins. We could identify sequence similarity with known toxins only for 56 of these proteins (Table S2). The most common identified toxins in the PVC systems are colicin A, a membrane-perforating toxin, and BECR RNases (barnase, EndoU, colicin D, RelE fold metal-independent RNases with a core 4-stranded sheet) of the colicin D family. We also identified variants of some of the PVC-like system protein families containing the HNH fold nucleases (in particular, the XHH family: for example, WP_013440589 from Halogeometricum borinquense) that are typical of bacterial PTSs but have not been previously observed in archaea (3). Seven of the 8 identified new toxin families containing 3 or more proteins are associated with the PVC systems (see Fig. S3 in the supplemental material). At least some of these new toxin families, such as arTOX1, arTOX2, arTOX4, arTOX5, and arTOX7, are likely to represent highly derived variants of colicin A because they are predicted to adopt a secondary structure compatible with the colicin A fold and contain one or two glycine-rich alpha-helices that are typical of the structurally characterized pore-forming toxins (3, 21) (Fig. S3). The arTOX6 family also, most likely, includes pore-forming toxins (Fig. S3) given the limited but significant similarity it shows to the hemolysin B component of enterotoxin from Bacillus cereus (e.g., PDB no. 2NRJ) (Fig. S3). All PVC toxin domains are expected to be cleaved off by the preceding PVC-MPTase before they are delivered into the target cell.
Supporting alignments for key components of archaeal offense systems or putative self-nonself recognition systems. Download FIG S3, DOCX file, 0.2 MB (242.3KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Each toxin is expected to be complemented by a respective immunity protein that protects host cells from autotoxicity or from toxin produced by other cells of the same clone (3, 9, 16). However, within the PVC-related loci, we identified only a few known immunity genes, namely, coding for four Imm60 family immunity proteins and one colicin D inhibitor (Table S2). This lack of homologs of known bacterial immunity proteins implies that archaea possess distinct, so far completely uncharacterized, immunity proteins. Indeed, we detected multiple, homologous, uncharacterized proteins (primarily, arCOG10872 and arCOG10882) encoded in most of the PVC loci in Halobacteria (Table S2). These genes are typically located next to those encoding toxins of the DUF4157 family but, in several cases, form a separate gene cassette located outside the PVC loci. It appears highly likely that these genes encode immunity proteins. Given that these homologous, putative immunity proteins are encoded next to unrelated toxins, they can be predicted to target a common mechanism of toxin delivery rather than a specific family of toxins.
Another class of likely immunity components encoded in the PVC loci in Halobacteria are ankyrin repeat-containing proteins (arCOG04004), which have been previously associated with bacterial PTS and implicated in immunity (3). Moreover, in several archaeal genomes, ankyrin genes are next to known immunity genes, such as those coding for the SUKH 6 immunity protein (WP_008454742 in Natrinema gari) or an SUFU family immunity protein (WP_048112428 in “Candidatus Methanoplasma termitum” MpT1) (16). Combining several immunity genes in the so-called poly-immunity loci is typical of bacterial polymorphic toxin and immunity systems (3, 16), which further strengthens the hypothesis that ankyrins are involved in immunity. The identities of immunity proteins in some other mesophilic archaea with genes encoding the PVC systems, such as Methanomicrobia and Thaumarchaeota, remain unclear. Typically, there are some uncharacterized proteins encoded next to the predicted PVC-toxin proteins in these genomes, but unlike the case of Halobacteria, they do not belong to any characterized large protein families. This implies that either these organisms possess completely different immunity mechanisms, such as the specific immunity proteins against each toxin family that have been identified in bacteria (3, 16), or these toxins are deployed against distantly related organisms.
Additionally, in several Halobacteria, putative immunity proteins of arCOG10872 are encoded adjacent to a membrane protein (arCOG07767) homologous to TrbL, which is present in the trb locus of Agrobacterium conjugative plasmid Ti (22). TrbL is related to TraG and VirB6 proteins, pore-forming proteins that are essential for conjugative DNA transfer (23). Moreover, in several archaeal genomes, a conjugative transfer ATPase gene, virB4 (23), is contained in the vicinity of trbL/arCOG10872 genes (e.g., WP_014030626 in Haloarcula sp.). These observations suggest that, in Halobacteria, genes for immunity proteins are propagated via conjugative plasmids independently from the toxins and delivery genes.
RHS-like systems.
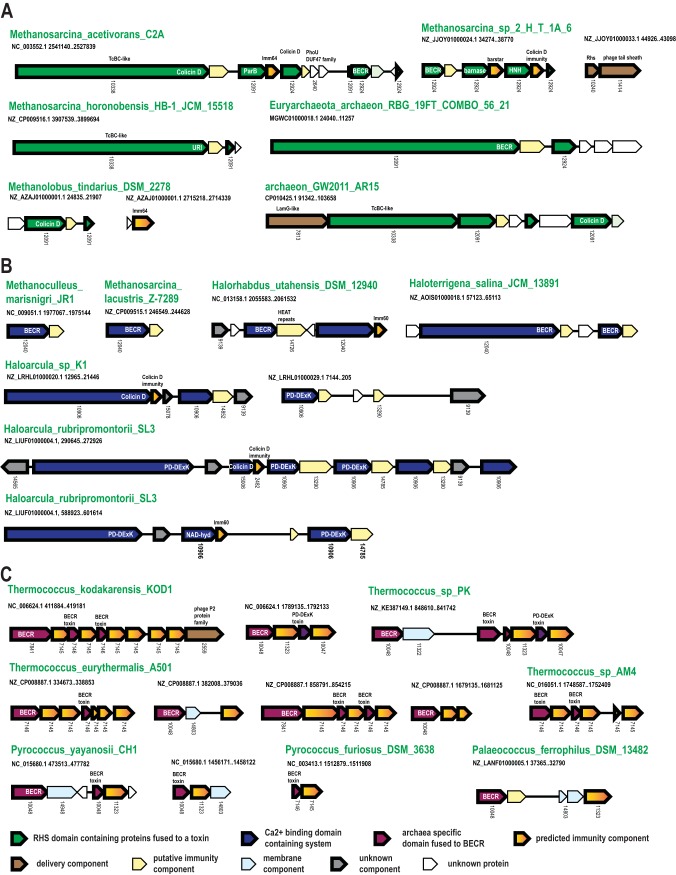
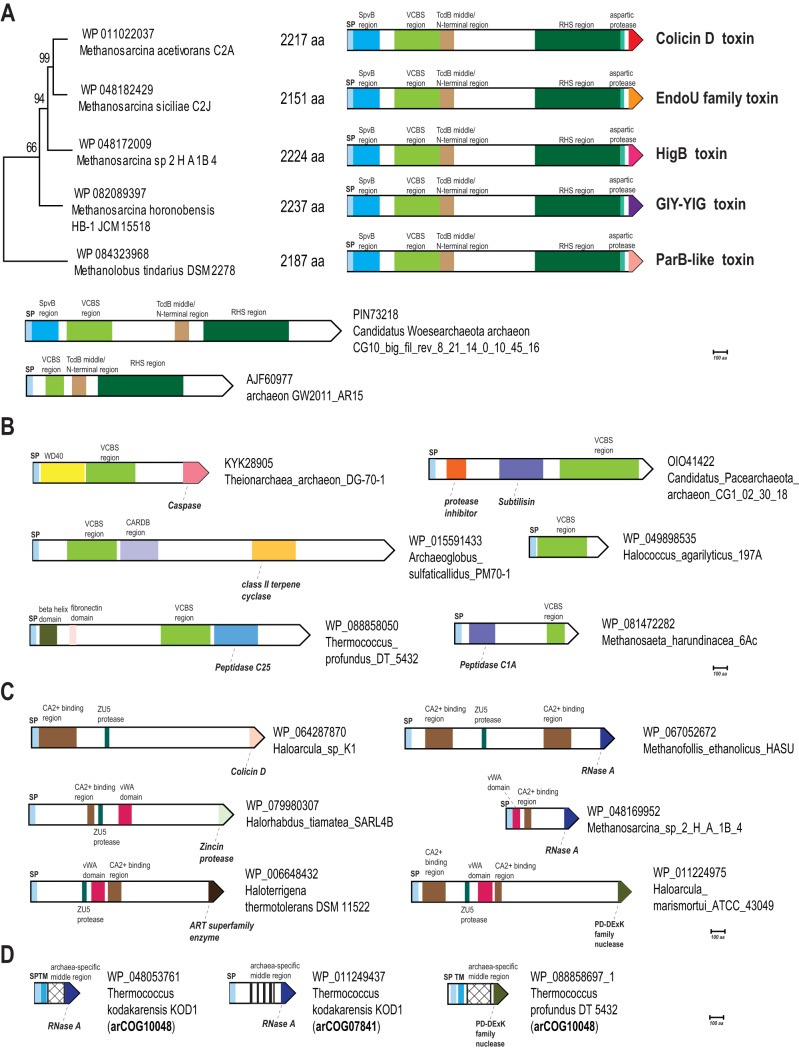
The RHS (YD) repeats are 28-aa sequences that typically contain the signature motif GxxxRYxYDxxGRL[I/T], where x is any amino acid, and were named after the rearrangement hot spot proteins of Escherichia coli (24). The RHS is the most common N-terminal element found in PTSs in general (3). Tandem RHS repeats form a large, hollow structure encapsulating the toxin for delivery (25). Additional domains fused to the RHS element vary substantially, but the C-terminal domain is usually a variable toxin (3, 8). In the PTS, these toxins are paired with a dedicated immunity protein, which is usually encoded by the gene next to the RHS element-encoding gene. These features of RHS toxins are shared by both bacterial and archaeal versions (Fig. 1 and 3A) (3). We identified RHS systems in 24 archaeal genomes—mostly Methanomicrobiales and several uncultured archaea (Table S1). Many RHS element-containing proteins also contain a Tat signal peptide, suggesting that they are secreted through the Tat system in the folded state. In most genomes, we identified at least one large RHS repeat-containing protein that also contained additional domains predicted to form the proximal part of the hollow toxin delivery structure (Fig. 3A). The N-terminal regions of the RHS proteins in four Methanosarcina species and in Methanolobus tindarius have the same domain organization as the B component of the entomotoxin from Yersinia entomophaga, for which the structure has been solved (25). Specifically, this protein contains the Salmonella virulence plasmid 65-kDa protein B similarity region (pfam03534), VCBS (a repetitive domain in Vibrio, Colwellia, Bradyrhizobium, and Shewanella [pfam13517]), and the middle/N-terminal region of the TcdB toxin (pfam12256). The 5 highly similar archaeal proteins have the same domain organization, but their C-terminal toxin domains are all different, indicating that, similarly to their bacterial counterparts, toxin domain polymorphism via recombination with stand-alone toxin-immunity cassettes also occurs in archaeal RHS systems (Fig. 4A). Typically, the large RHS protein (primary locus) is encoded adjacent to the respective immunity genes and several RHS cassettes (Fig. 3A; Table S2). These cassettes (mostly from arCOG12091 and arCOG12924) lack an N-terminal signal peptide but have a C-terminal toxin domain and, like in bacteria, likely recombine into the primary RHS-encoding genes, to generate variants that differ in their C-terminal toxin domains.
FIG 3.
Selected gene neighborhoods of the RHS, Ca_vWA, and CDI-like systems. (A) Selected gene neighborhoods of RHS systems. (B) Selected gene neighborhoods of Ca_vWA systems. (C) Selected gene neighborhoods of CDI-like systems. Designations are the same as in Fig. 2.
FIG 4.
Domain organization of proteins of each identified PTS. (A) RHS domain-containing proteins. The tree shows the relationships between close species of Methanomicrobia. All RHS-containing proteins have similar domain architectures, with the exception of the C-terminal toxin domains, which are different in all 5 proteins. (B) VCBS domain-containing proteins. These proteins are abundant in archaea, but their involvement in microbial conflict systems is unclear. (C) Ca2+-binding module-containing proteins. (D) Proteins associated with a CDI-like system specific for thermococci. Proteins are shown by block arrows with the length proportional to the size of the corresponding protein. For each protein, the GenBank nucleotide contig or genome partition accession number and the organism are indicated. The identified domains are shown inside the arrows approximately according to their location and are briefly annotated. Homologous domains are shown by the same color or pattern. Abbreviations: SP, signal peptide; TM, transmembrane helices.
Altogether, we identified 69 putative archaeal RHS proteins, of which 29 contain C-terminal toxin domains. These toxins belong to previously characterized endonuclease families of the HNH, BECR, and ParB folds or metallopeptidases (zincin or Tox-MPTase) (Table S2). Additionally, we defined a previously unrecognized family of predicted endo-RNases with the BECR fold (e.g., in Methanosarcina sp. strain 2.H.T.1A.15; WP_048139393.1), which is also present in several bacterial PTSs (Fig. S3). The majority of the toxin-containing RHS proteins also contain an aspartyl autopeptidase domain located downstream of the RHS repeat region (Fig. 4A). This is an autopeptidase of the same family as the autopeptidase in the TcC component of the Yersinia entomophaga entomotoxin (25) (Fig. S3). Additionally, several of the archaeal RHS toxins contain a previously unreported predicted pretoxin domain that is shared with several bacterial PTSs (e.g., WP_048139393.1). This domain is characterized by two hydrophobic helices with 3 highly conserved negatively charged residues and might be involved in processing of the respective toxins or their delivery to the target cells (Fig. S3). As expected, in several cases, a known toxin gene is followed by a recognizable immunity protein against this particular toxin: e.g., in Methanosarcina sp., barnase RNase is followed by the barstar inhibitor (26) (Fig. 3A; Table S2). Thus, numerous uncharacterized genes contained immediately downstream of the RHS element genes are likely to encode immunity proteins against either known or yet unknown toxins encoded by these RHS genes (Fig. 3A).
Whereas the RHS and the TcdB toxin that comprise the N-terminal and middle regions of the RHS proteins, respectively, appear to be specific to these systems, the VCBS repeat domain is found in many archaeal proteins that are typically encoded by genes outside the recognizable contexts associated with polymorphic toxins. These genes encode proteins of various sizes and domain organizations that are predicted to be secreted and often contain an enzymatic domain, such as different proteases and terpene cyclases (Fig. 4B; Table S1). Some of these enzymatic domains have been occasionally identified in PTSs, especially, in association with type II secretion systems (3). These proteins potentially represent distinct, uncharacterized archaeal offense systems, with their enzymatic domains being toxins. However, the absence of potential immunity protein genes in the vicinity of the respective genes suggests that they are either deployed against distantly related species or perform a different function, such as processing cell surface biomolecules.
Calcium-binding domain-containing toxins.
Another type of predicted polymorphic toxin systems (here Ca_vWA) was identified in 49 archaea, mostly mesophiles. The key feature of this system is the presence of Ca2+-binding module that is related to type 3 thrombospondin repeats (27) (Fig. 3B; Table S1). These repeats contain a characteristic DxDxDGxxDxx[DE] motif, lack secondary structure, and are organized around a core of calcium ions (27). In addition to this Ca2+-binding module, these proteins often contain a von Willebrand factor type A (vWA) domain (Fig. 4C). The vWA domain is a peptide-binding domain that functions in several adhesion-related processes (28, 29). Among known polymorphic toxin delivery components, several include unrelated calcium-binding domains, such as repeats-in-toxin (RTX) domains (30) and the EF-hand calcium-binding domain of the protective antigen component of anthrax toxin (31, 32). As with the RHS systems, the Ca_vWA loci encompass several putative toxin-containing genes (mostly, from arCOG10906 and arCOG12040), each of which is typically followed by a known or putative immunity gene (Fig. 3B). Often, one of the toxin-containing proteins is much larger than the others. Most of these large proteins contain signal peptides, suggesting that these proteins are secreted. About 74% (48 of 65) of these predicted toxin proteins that are larger than 500 aa contain a ZU5 autopeptidase domain (e.g., WP_079891644.1 [366 to 479 aa]) that has been previously implicated in the maturation of bacterial PTSs (3). The smaller proteins usually lack a signal peptide and might form a part of the toxin delivery machinery.
We identified homologs of known toxin domains in the C-terminal regions of 92 of the 122 putative toxins in the Ca_vWA systems (Fig. 3B and 4C; Table S2). Most of the predicted toxin domains belong to one of the 6 families: 3 are the RNase domain of the BECR fold, one is a distant homolog of RNase A (NTox41) that has been previously identified in the contact-dependent growth inhibition toxin (CdiA) of Yersinia kristensenii (33), and the other two are the previously characterized Tox-ColD1 and Ntox49 (Table S2), whose active site configurations are similar to that of colicin D RNases compared to the other families of BECR RNases (3). Another toxin in these systems is an NAD(P)+-degrading enzyme or ADP-ribosyltransferase of the ART superfamily (Ntox48), which is also found in many bacterial effectors deployed against animals and plants (34, 35). Additionally, two predicted toxin families, namely a PD-DExK superfamily (also known as REase [restriction endonuclease] fold) enzyme and a zincin metallopeptidase (Tox-MPTase), have not yet been experimentally characterized. However, representatives of these families have been identified in analogous bacterial systems in the previous comparative genomics study (3) (Fig. 4C).
Surprisingly, only a few known immunity proteins were identified in the Ca_vWA systems. These include 6 colicin D immunity proteins, which are encoded next to the BECR RNase toxin-containing proteins (Table S2). Three additional families have been predicted as immunity proteins previously (3), namely, Imm49 family proteins, which are encoded next to the PD-DExK (REase) toxins, Imm60, often encoded next to ART family toxins, and HEAT repeat-containing proteins, encoded next to RNase A-like BECR toxins (Table S2). Many other putative immunity proteins appear to be specific for archaea, belong to arCOG13290, and can be predicted to be involved in immunity against the PD-DExK (REase) toxins (Table S2).
Thermococcus-specific PTS.
We identified a putative archaea-specific PTS in 18 species of Thermococcales (Fig. 1; Table S1). Typically, at least one of these loci in each genome contains a gene coding for a full-size toxin (usually ∼350 to 470 aa), which apparently contains all the domains required for the toxin delivery and cleavage. Other predicted toxins encoded in these loci seem to be diverse C-terminal fragments homologous to the full-size toxins (Fig. 3D). The full-size toxins belong to two distinct families, arCOG07841 and arCOG10048, whereas most of the partial ones belong to arCOG07146 (Table S2). All full-size predicted toxins contain a signal peptide, an uncharacterized middle region, and a C-terminal toxin domain (Fig. 4D; Fig. S3). The middle regions of arCOG07841 and arCOG10048 proteins are either unrelated or extremely diverged. The C-terminal domains of some of these proteins belong to the BECR fold of RNases with the same catalytic residues (histidine, arginine, and tyrosine) as in the RNase A-related toxin from the CDI (contact-dependent growth inhibition) polymorphic toxin system characterized in Yersinia kristensenii (33) or the PD-DExK (REase) toxins of bacterial PTSs (Fig. S3). Altogether, we have identified a BECR RNase domain (sometimes partial or degraded) in 38 proteins and PD-DExK superfamily toxin domains (sometimes partial) in 8 proteins of the 46 predicted toxins of this type. The organizations of these loci are similar to those of the RHS and Ca_vWA systems, suggesting that the smaller toxin domain-containing genes recombine with the principal, full-length toxin gene, resulting in diversification of the toxin domain (Fig. 3D). Proteins of arCOG07146 are typically encoded next to the BECR fold RNase toxins and can be predicted to confer immunity to these toxins. Indeed, an HHpred search identified limited sequence similarity between members of arCOG07146 and the CdiI immunity proteins that are known to neutralize related RNase toxins in CDI systems (36) (Fig. S3). Because RNase toxin domains and immunity components of these systems share similarity with CDI systems, we refer to these systems as “CDI-like.” The PD-DExK (REase) toxins are relatively rare in these systems but are likely neutralized by dedicated immunity proteins of arCOG10047 and arCOG11323 that are typically encoded next to the corresponding toxins (Fig. 3D).
Examination of the domain architecture of the complete toxins of this class yields hints regarding their trafficking and delivery. The presence of an N-terminal signal peptide indicates that these toxins are secreted from the producing cells via the general secretory pathway. The middle region of these proteins shows a tripartite structure, which includes a linker region located directly upstream of the toxin. The linker is preceded by a domain containing a GXG or GXD signature flanked by two predicted hydrophobic alpha-helices (Fig. S3). A similar element is present in several bacterial and methanococcal PTSs, where it is typically associated with the N-terminal RHS and is believed to help localize the toxin to the target cell membrane for delivery (3). Therefore, it seems likely that it plays an analogous role in the distinct thermococcal PTS described here. In the bacterial and methanococcal homologs, this domain is preceded by a predicted aspartyl autopeptidase domain containing an [ND]PxxxxDP motif, which mediates the autopeptidase activity (25). Although this autopeptidase motif is absent in the thermococcal PTS, the sequence conservation of the region upstream of the above domain and immediately C-terminal to the signal peptide featuring a distinct G[ED] motif suggests that this portion of the protein might facilitate similar cleavage, either directly or in conjunction with a general secretory peptidase (Fig. S3).
The barnase-barstar system.
This secreted RNase toxin barnase and the corresponding immunity protein barstar comprise one of the simplest versions of the class of conflict systems that is extensively elaborated in various PTSs. The barnase-barstar system was first identified in bacteria (37, 38) and has since been detected in a wide range of prokaryotes (3). Although barnase-barstar homologs are present, respectively, as the toxin tip and the cognate immunity protein in the RHS systems described above (Fig. 1), unlike the typical polymorphic systems, the solo barnase-barstar system contains no additional, recombining toxin cassettes. The barnase endo-RNase domain is one of the founding members of the BECR fold, which also includes other metal-independent toxin endo-RNases, such as RelE, colicin E5, EndoU, colicin D, and others, that are represented both in the PTS and in classical type II toxin-antitoxin systems (3). The solo barnase-barstar gene dyad was identified in 9 archaeal genomes, all from the Methanobacteria lineage (Table S1). These genes show no significant association with other genes in the respective genomic neighborhoods, in accord with the notion that barnase and barstar jointly comprise a stand-alone conflict system (Table S2).
Archaeocins.
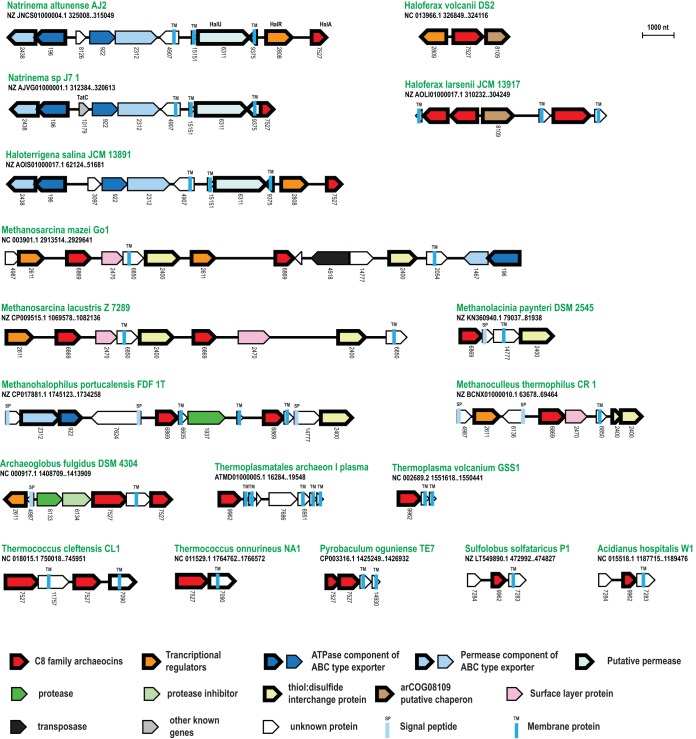
Archaeal bacteriocin-like proteins so far have been identified only in Halobacteria and Sulfolobales (7). To date, 8 distinct halocins have been isolated (39), but only five of these have been mapped to proteins encoded in the respective genomes. These halocins belong to three distinct families: C8/A4, S8/R1, and H4. Two more genes, sulA and sulB, comprise the only known crenarchaeal archaeocin genes, coding for sulfolobicin, in Sulfolobales species (Fig. 1; Table S1) (40). Typically, these proteins are secreted and expressed as precursors that are further processed by unknown peptidases, to release an active toxic peptide (41) and, in the case of C8, also the immunity protein HalI (42). Additional genes encoding transcriptional regulators and peptide exporters are sometimes present in the same loci and are assumed to be involved in immunity, regulation, and export of the halocins (7) (Fig. 1).
We used PSI-BLAST and the guilt-by-association approach to identify archaeocin homologs and explore their gene neighborhoods. Halocin H4 is rare, being present in only 7 Halobacteria and two species from other archaeal lineages, Methanosarcina soligelidi and Thermococcus piezophilus. This gene does not appear to be linked to any other genes in the respective genomic neighborhoods (Table S2). The C8/A4 and S8 halocins could be unified into a single, large superfamily. Specifically, a PSI-BLAST search initiated with WP_084813291 from Haloterrigena thermotolerans, a representative of the halocin C8 family, recovers halocin S8 from Halobacterium salinarum (ALF62560.2) in iteration 4, with an E value of 10−3. This finding was confirmed by a hidden Markov model (HMM) search using HMMER3 starting with an alignment of bacterial and archaeal halocin C8, which recovered H. salinarum halocin S8, with an E value of 10−4. We identified homologs of these halobacterial proteins in 87 archaeal genomes, including representatives of Desulfurococcales, Thermoproteales, Methanomicrobia, Sulfolobales, Theionarchaea, Archaeoglobi, Thermococcales, and Thermoplasmata (Table S1). Furthermore, we detected members of the C8/A4/S8 halocin superfamily in bacteria, mostly in firmicutes, as well as some members of other phyla, such as actinobacteria, chloroflexi, and proteobacteria (Table S1). To our knowledge, C8 family halocins have not been previously identified outside the halobacterial lineage, and most of these newly identified proteins are annotated as “hypothetical.” These toxins typically contain a signal peptide, followed by a poorly conserved, strongly charged central region, followed by a C-terminal conserved globular domain containing 6 to 8 cysteines (Fig. S3). Notably, in some chloroflexi, for example, Nitrolancea hollandica (e.g., WP_008479876.1), this C-terminal globular domain is coupled with RHS repeats at the toxin tip of an RHS-type PTS. Thus, the toxic activity of the C8/A4 toxins is likely mediated by this C-terminal domain, consistent with the fact that the previously characterized mature versions from Halobacteria are derived from this region (41).
The S8 halocins were detected only in 9 Halobacteria and are typically encoded by a stand-alone gene (Tables S1 and S2). The more prevalent C8/A4 family genes are found in variable loci that often include additional genes that are likely to be involved in halocin expression regulation and transport (Fig. 5). Among the most common proteins encoded in these neighborhoods are multidrug transporter ABC-ATPase (arCOG00196), a DsbD-like membrane-associated disulfide-bond isomerase of the thioredoxin fold (arCOG02400), transcriptional regulators of arCOGs 2611, 2808, and 2809, membrane proteins of the YIP1 family, presumably mediating trafficking (arCOG02054), and others (Table S2). HalR, the transcriptional regulator from the originally characterized C8 locus (42), belongs to arCOG02808. Among the uncharacterized genes in these loci, two are most common. The first one encodes a Halobacteria-specific, uncharacterized membrane protein (arCOG06311, or HalU) with 14 transmembrane (TM) helices, possibly, a permease. The second gene also encodes a Halobacteria-specific protein (arCOG08109) that contains two predicted TM helices with a conserved CxxC motif at the N terminus and a C-terminal domain with 5 cysteines that might adopt the Zn ribbon fold (Fig. S3). Genes of arCOG08109 are present in the majority of halobacterial genomes and do not follow the distribution patterns of C8 halocins (Table S1), suggesting that this protein is likely to perform some important function in the respective organisms. Given the conservation of the configuration of the cysteines in these proteins, they could facilitate the maturation of the halocin in conjunction with the DsbD-like disulfide bond isomerase by ensuring the proper formation of disulfide bonds.
FIG 5.
Selected gene neighborhoods of predicted C8 halocin homologs. Designations are the same as in Fig. 2. Gene products HalA, -R, and -U are indicated for the Natrinema altunense AJ2 locus as the closest to the originally described locus in Haloarcula hispanica (42). Signal peptides and TM helices are indicated only for uncharacterized genes present in the loci.
Finally, we identified SulA and SulB homologs only in 3 species in our data set, all in the Sulfolobales lineage (Table S1). Notably, in Acidianus hospitalis, the SulA component was not detected, but there are two separately encoded, stand-alone sulB genes. Also, as noted previously (40), in Sulfolobus tokodaii, the sulB gene is tandemly duplicated.
Although different known archaeocins are typically encoded in different loci, even when present in the same genome, many halocin C8 loci contain genes coding for other secreted proteins. It appears likely that at least some of these proteins are unidentified archaeocins. Indirectly, this can be inferred from the fact that, in several genomes, there are additional, dissimilar halocin C8 precursor genes contained in the same loci (e.g., in Archaeoglobus fulgidus, Thermococcus cleftensis, Methanosarcina lacustris, Methanohalophilus portucalensi, Methanosarcina soligelidi, Haloferax larsenii, Picrophilus torridus, Theionarchaea archaeon, and others (Fig. 5; Table S2), suggesting that C8 halocin family genes are prone to duplication and diversification, similarly to some polymorphic toxin families described above.
Archaeal lantibiotic systems.
Another system that has been characterized in bacteria and identified in 19 halobacterial genomes produces lantibiotics (“lanthionine-containing antibiotic”), ribosomally synthesized and posttranslationally modified toxin peptides containing lanthionine (an atypical amino acid) and dehydrated derivatives of threonine and serine (5). The minimal system consists of two genes encoding the lantibiotic cyclase LanM and a small protein, the lantocin precursor. LanM, the signature protein for type 2 lantibiotics, is an enzyme that catalyzes both dehydration and cyclization of the precursor peptide (5) (Fig. 1). We identified 10 lantocin precursors in 19 genomes, 8 of which belong to the same protein family (Table S2). All these lantibiotic precursors contain several cysteines and threonines in the C-terminal region that are involved in lanthionine synthesis and cyclization (Fig. S3). Some of these proteins were missed by the gene prediction pipelines, and the rest are currently annotated as hypothetical. The loci encoding LanM often also contain genes for multidrug transporters and, less frequently, genes for a hydrolase of the TIKI superfamily that includes metallopeptidases and erythromycin esterases. These could either process the lantocin precursor or confer immunity cleaving lantocin peptides (Table S2).
Candidates for novel archaeocins, archaeal toxins, and other systems involved in interspecies conflicts or self-versus-nonself recognition.
Among the 524 analyzed archaeal genomes, 168 encompass at least one of the (predicted) offense systems described above. It appears highly unlikely that the remaining archaea entirely lack offense or self-nonself recognition systems. This being the case, such systems should be archaea specific, without readily detectable bacterial counterparts. In our previous comparative analyses of archaeal genomes, we noticed several archaea-specific systems that possess some features suggestive of their involvement in interspecies interactions, self-nonself recognition, or quorum sensing (43). For example, one of such predicted systems in Thermococcales includes multiple paralogous members of the family of secreted proteins (encoded by the TK2175, TK2176, and TK2177 genes in Thermococcus kodakarensis) and a zincin metallopeptidase fused to an immunoglobulin-like domain, TK2178, which might cleave these putative uncharacterized archaeocins, by analogy with some bacteriocin systems (43).
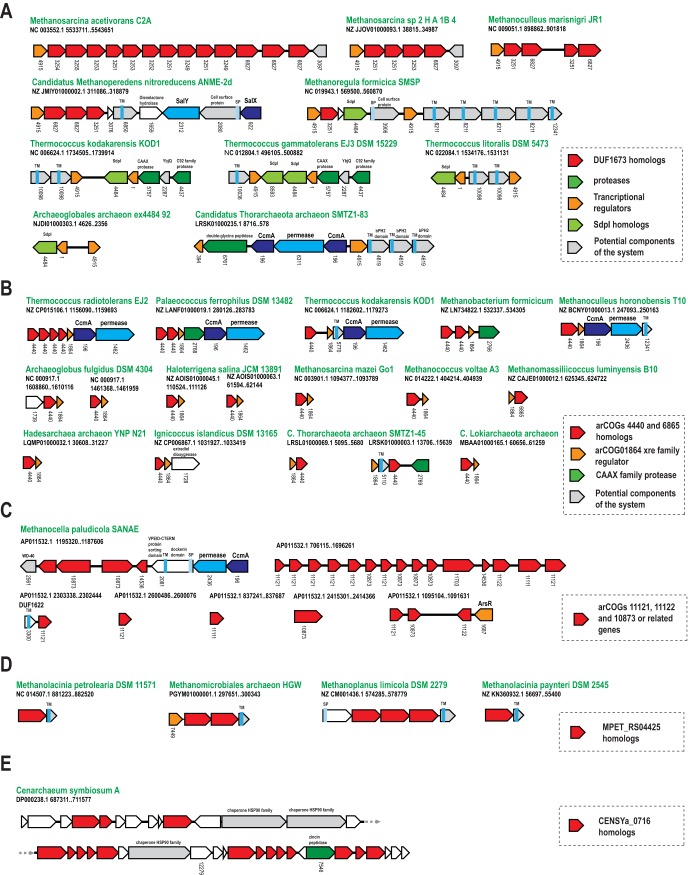
Taking into account sequence features of the known archaeocins, bacteriocins, and polymorphic toxins and the organization of the respective genomic loci, we sought to identify uncharacterized genes in archaeal genomes that could be involved in similar functions. In particular, we searched for uncharacterized protein families with a combination of several of the following features: (i) are prone to tandem duplications, (ii) are present in several archaeal lineages, (iii) have a patchy distribution across the genomes from those lineages, (iv) contain cysteine-rich regions, (v) contain either predicted signal peptides and/or TM helices, or (vi) are encoded next to (predicted) peptide exporters, peptidases, and/or transcriptional regulators (see Table S3 in the supplemental material). Examples from the list of candidates that were obtained by searching archaeal genomes for combinations of these features are shown in Fig. 6.
FIG 6.
Selected gene neighborhoods of candidate genes for self-versus-nonself recognition systems. (A) Neighborhoods including DUF1673- or arCOG04915-related genes. The tree shows the relationships between close species of Methanomicrobia. All the proteins have similar domain architectures, with the exception of the C-terminal toxin domains, which are different in all 5 proteins. (B) Neighborhoods including arCOG04440- or arCOG01864-related genes. (C) Neighborhood including arCOG11121, -11122-, and -10873-related genes in Methanocella. (D) Neighborhood including MPET_RS04425 homologs in several Methanomicrobia species. (E) Neighborhood including CENSYa_737 homologs in Cenarchaeum symbiosum. Insets for each panel describe key components of the respective system; otherwise, designations are the same as in Fig. 5.
Sequence and genomic organization features for archaeal clusters. Download Table S3, XLSX file, 0.2 MB (243.4KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The most prominent case of tandem duplications was identified in Methanomicrobia genomes for the DUF1673 (pfam07895) family (arCOGs 3249, 3255, 3251, 3253, 3252, 3254, 6827, and 6845). The patchy distribution in Methanomicrobia suggests that these genes are not involved in an essential function. This protein family is characterized by an N-terminal globular domain of about 60 to 63 aa with a strictly conserved hGWCPN motif. This domain is often followed by a C-terminal variable domain that contains 4 TM helices and is predicted to anchor the protein in the membrane. The presence of the conserved cysteine is reminiscent of the cysteine in the thioester domain that functions as a thiol chemical harpoon to attach bacteria to target cells via a covalent linkage through the active cysteine (44). An analogous function appears possible for this the expanded protein family in Methanomicrobia. Alternatively, the conserved cysteine could serve as a site for a covalent lipid modification.
Most of the respective genomic neighborhoods include a gene for a predicted transcriptional regulator of the cI/cro helix-turn-helix (cHTH) family (arCOG04915), which is a common component of toxin-antitoxin systems (45, 46). In addition, regulators of this family have been identified in the context of antimicrobial genes (47) and quorum-sensing systems: e.g., RsaL from Pseudomonas aeruginosa (48), which was the best hit for arCOG04915 in the HHpred search against the PDB database. Members of arCOG04915 are also present in other archaeal genomes, where their genes are contained in the vicinity of genes for proteins with four TM helices that might be functional DUF1673 analogs or homologs that diverged beyond recognition (Fig. 6A). Among these proteins, there are members of arCOG04484, and analysis of these proteins provides a clue to the likely involvement of the entire system in interspecies conflicts or recognition. The membrane proteins from this arCOG are homologs of sporulation-delaying protein I (SdpI), which confers immunity to the cannibalism peptide SdpC in bacilli (49). This system has been studied in detail in Bacillus subtilis, where it causes a delay in spore formation by cannibalizing siblings of the respective bacterium under nutrient-limiting conditions (49, 50). It consists of 5 proteins, SdpABCIR, where the SdpA and -B components are necessary for toxin maturation, SdpI is an immunity protein, and SdpR is a transcriptional regulator. Components SdpB, -C, and -I are unrelated membrane proteins, and SdpI contains four TM helices, similar to DUF1673 (Table S2). Thus, it appears likely that the transcriptional regulators of arCOG04915 are functionally analogous to SdpR. The membrane proteins encoded in the respective loci could be either toxins or immunity proteins, or both if they contain both immunity and toxin domains, such as, for example, C8 family halocins (Fig. 1 and 6A). The key transcriptional regulator (arCOG04915) is present in 59 archaeal genomes from Methanomicrobia, Thermococcales, Theionarchaea, Archaeoglobales, and members of the Asgard group, in different genomic contexts (Fig. 6A; Tables S1 and S2). Further detailed investigation of the respective gene neighborhoods could lead to identification of other components of this system and its extension to other archaeal genomes. It has been noticed previously that SdpI homologs are present in several archaea (51) that, as we find now, lack arCOG04915. In our present searches, SdpI homologs from arCOGs 4484, 6110, and 8593 were identified in 152 additional archaeal genomes that encode neither DUF1673 nor the transcriptional regulator of arCOG04915, suggesting that different variants of this putative offense system are widespread in archaea (Table S1).
Another putative archaeal offense system seems to be a mimic of the preceding one, although without such pronounced tandem duplications, which occur mostly in Thermococcales and Methanomicrobia (Fig. 6B). As in the case of the DUF1673/arCOG04915 system, the two main components are a protein with four predicted TM helices (arCOGs 4440 and 6865) and a cHTH family transcriptional regulator (arCOG01864). This system also shows a patchy distribution but is widespread in archaea. It was identified in 161 genomes, including several members of the Asgard group, DPANN superphylum, all major euryarchaeal lineages, and a few crenarchaea (Tables S1 and S2). In most genomes, only these two genes are present in putative operons—sometimes, in two or more genomic loci (Fig. 6B; Table S2). Also, similarly to the DUF1673/arCOG04915 system (Fig. 6A), the transcriptional regulator is most closely related to the quorum-sensing regulator RsaL (Fig. S3), but unlike the arCOG04915 proteins, lacks a Zn ribbon. Furthermore, an ABC family multidrug exporter and a CAAX family protease are often encoded in these neighborhoods (Fig. 6B). The CAAX family proteases are involved in the processing of and immunity against bacteriocins (52), which further suggests that this system is a strong candidate for an either offense or, more generally, a self-versus-nonself discrimination function.
The next family is specific for three closely related Methanocella species and is not found in other archaea. Multiple paralogs are encoded in each of the three genomes, often as tandem duplications that mostly belong to arCOGs 11121, 11122, and 10873 (Fig. 6C; Table S2). These proteins are present in several loci in each genome, and in one of these loci, they are encoded next to an ABC-type multidrug exporter (Fig. 6C). The paralogous proteins are highly variable and contain a signal peptide and a glycine-rich C-terminal region that is weakly predicted as a TM helix, suggesting that this protein could be a pore-forming archaeocin (Fig. S3).
Secreted proteins that are enriched in cysteines similarly to C8 and S8 archaeocins and the entire class of bacterial cysteine-rich toxic peptides known as thiazole/oxazole-modified microcins (5) appear to be potential candidates for similar functions. One such family of proteins containing 3 to 6 cysteines is present in four Methanomicrobia genomes (Fig. S3), in two of which these genes are duplicated (Fig. 6D), and also, sporadically, in certain bacteria (e.g., “Candidatus Peregrinibacteria”). Typically, these proteins are encoded next to an uncharacterized membrane protein (e.g., MPET_RS04425) with four TM helices, likely a component of the same system. In Methanomicrobiales archaeon HGW, another gene (arCOG07449) is present in the same predicted operon (Fig. 6D). An HHpred search for the proteins from this arCOG shows significant sequence similarity to Escherichia coli antitoxin MqsA (Fig. S3). The arCOG07449 proteins share a Zn ribbon and a cHTH domain with MqsA but, additionally, contain another Zn ribbon at the C terminus. MqsA, together with the MqsR toxins, forms a motility quorum-sensing (MQS) type II toxin-antitoxin system (53, 54). Thus, the connection identified here might indicate involvement of this archaeal protein family in quorum sensing.
The final example we address here is a remarkable locus with multiple tandem paralogous genes that thus far had only been identified in Cenarchaeum symbiosum. The complete versions of the proteins encoded in this locus contain two N-terminal TM helices (e.g., ABK77370 protein) which are connected via a linker consisting of a variable number of short repeats to a C-terminal domain. The locus also contains at least 7 stand-alone genomes encoding the C-terminal domain alone, resembling the toxin cassettes of the PTS that recombine with the corresponding full-length genes (Fig. 6E). The C-terminal domain shows considerable variability, suggesting that it might possess an effector activity analogous to the PTS (Fig. S3). Furthermore, this locus encodes a predicted metallopeptidase, which might process these proteins. It would be of obvious interest to investigate if this locus plays a role in the interaction of C. symbiosum with its animal host.
Concluding remarks.
Like most other organisms, archaea are part of ubiquitous webs of interactions with cohabitating kin, as well as closely and distantly related species. As free-living microbes, archaea need to distinguish self from nonself, to form colonies or biofilms, to exchange genetic material, and to leverage conflicts among interacting organisms. Bacteria possess numerous systems that are involved in self-versus-nonself discrimination and offense against competing organisms (3, 5, 8, 55, 56). In contrast, these functionalities are poorly characterized in archaea, although they are likely to face ecological challenges comparable to those of bacteria and microbial eukaryotes. The current knowledge is limited to the identification of a few archaeocins (7) and several studies on quorum-sensing signaling molecules in Halobacteria (57). Although several PTSs have been identified in archaea by comparative genomics methods (3), their annotation has not propagated to the respective archaeal genomes, and they remain unexplored experimentally.
Here, we expand the previous work and present a comprehensive in silico analysis of the potential biological conflict systems encoded in 524 archaeal genomes. We identify PTSs in 141 archaeal genomes and classify them into four types based on the delivery components. The range of complexity among these PTSs varies from the simplest barnase-barstar systems all the way to elaborate systems with a multicomponent delivery apparatus. Three of these types—PVC/tailocins, RHS, and Ca_vWMA—encompass the same maturation/trafficking components as the corresponding bacterial systems and therefore are likely of bacterial origin. The fourth type of PTS identified in this work shares the toxin and immunity components with bacterial CDI PTS but might employ an archaea-specific maturation mechanism. Furthermore, we predicted several previously unknown families of toxins and immunity proteins that appear to be archaea specific. Similar to the bacterial counterparts, the predicted archaeal PTSs are highly variable and show evidence of diversification of the toxin domains and immunity proteins via recombination with stand-alone cassettes within each system.
We also unified the C8 and S8 halocins into a single superfamily and expanded their phyletic horizon beyond Halobacteria by identifying representatives of this family in 5 other major archaeal and several bacterial lineages. Additionally, in many archaeal genomes, we identified modified peptide toxins that are counterparts of the bacterial lantibiotics along with the corresponding enzyme LanM, which is responsible for their modification.
Although we identified numerous archaeal counterparts of bacterial offense and self-versus-nonself discrimination systems, these are represented in only a minority of the available archaeal genomes. These systems are predominantly found in mesophilic archaea as opposed to hyperthermophiles. A similar trend has been previously noticed in bacteria (3). One possibility is that the high-temperature environments lower the levels of interorganismal conflict as only a select set of species can thrive under these conditions. Additionally, the high-energy environments might disfavor the biochemistry of certain toxins. However, given that ecological studies point to high densities of life even at high temperatures (58) and that such functionalities appear to be essential for free-living organisms, multiple, unknown, and most likely, archaea-specific conflict and kin discrimination systems could exist. We searched for such cases using the characteristic features of the components of the known systems and the organization of the respective genomic loci, including the tendency to form tandem duplications, the presence of predicted signal peptides and/or TM helices, and others. This search resulted in the identification of several candidate systems that might be implicated in either interspecies conflicts or quorum sensing. However, the analysis presented here cannot be considered exhaustive, and the computational strategies employed for the prediction of uncharacterized systems involved in intermicrobial conflicts and communication remain to be refined. Nevertheless, the present study prompts multiple experimental directions that can be expected to move forward this important area of microbial biology.
MATERIALS AND METHODS
Comparative genomics framework.
Genome sequences of 524 archaea with complete or nearly complete genomes were downloaded from the NCBI FTP site (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/). Sequences were assigned to the 2014 arCOGs using PSI-BLAST (59) with the arCOG alignments as the position-specific scoring matrix (PSSM) sources as previously described (60). Phyletic patterns (i.e., patterns of the presence or absence of protein families) were derived from the respective arCOG assignments.
General sequence analysis.
Iterative profile searches using PSI-BLAST (59), with a cutoff E value of 0.01 and composition-based statistics and low-complexity filtering turned off, were employed to search for distantly similar sequences in either the NR (nonredundant) database or the protein sequence database of 524 archaeal genomes. Hidden Markov model searches were run after constructing an HMM from a multiple-sequence alignment of the predicted toxin domain with an initial seed set of representatives. These were then run against NR or a database of 2,700 prokaryotic genomes using the HMMsearch program from the HMMER3 package. Alternatively, iterative HMM searches were run using the JACKHMMER program with an inclusion threshold for the next iteration of 0.001 (61). Additionally, other sensitive methods for distant sequence similarity detection were used, including a CDD search (62), with a cutoff E value of 0.01 and low-complexity filtering turned off, and HHpred search with default parameters against the PDB, Pfam, and CDD profile databases (63).
Transmembrane helices were predicted using TMHMM v.2.0c with default parameters (64). Signal peptides were predicted using SignalP v.4.1c; the union of the three predictions (Gram-negative, Gram-positive, and eukaryotic models) was used (65). Protein secondary structure was predicted using Jpred 4 (66). Approximate maximum likelihood phylogenetic trees were constructed using FastTree with default parameters (67).
Search for PTSs.
The iterative procedure used for the delineation of polymorphic loci is shown in Fig. S1. Briefly, 6 arCOGs (seeds) predicted to be involved in the polymorphic toxin delivery system by the PVC systems (3) were initially used to map the respective genes in archaeal genomes. For further gene neighborhood analysis, 20 genes located upstream and downstream of the seeds were extracted. Genomic islands around the mapped genes were trimmed manually based on several criteria: the flanking genes were discarded if arCOG assignments of the surrounding genes were incompatible with offense functions based on arCOG functional category assignments, all genes located in the same strand within a 100-nucleotide (nt) distance were retained, and “orphan” genes not assigned to arCOGs also were retained regardless of their direction. In the second iteration, the most frequent arCOGs from the trimmed genomic islands were used as new seeds. The new islands were analyzed as described above. Sequences of known and predicted toxin domains within these islands were searched against the arCOG database using PSI-BLAST (4 iterations or until convergence). All proteins with similarity to a toxin domain with an E value of <0.001 were used as new seeds.
Analysis of archaeocins.
Six previously detected archaeocin sequences were used as queries for PSI-BLAST searches, with 5 iterations (Table S2). For gene neighborhood analysis, 5 or 10 genes located upstream and downstream of the archaeocin homologs were extracted. Proteins found to be encoded in the same strand as the archaeocin homologs were searched against the arCOG database using PSI-BLAST. For all genes in the neighborhood, signal peptides and TM helices were predicted.
Prediction of previously unknown conflict systems.
All sequences in the database of 524 archaeal genomes were classified into superclusters, based on the sequence similarity as detected by reciprocal PSI-BLAST hits. To that end, a multiple alignment of the protein sequences from each cluster (arCOG) was used as the query in a search against a database of cluster (arCOG) consensus sequences. Superclusters were retained for further analysis if they satisfied the following criteria: (i) they had a narrow phylogenetic representation (present in at most 2 major lineages of archaea), (ii) they had a patchy phyletic distribution (present in at most half of the genomes in each of the respective lineages), and (iii) they were classified as a gene with unknown function, had general function prediction only, or were a defense gene. All instances of genes of the same supercluster (paralogs) and occurring in the same genome partition or contig within a distance of 5 genes from each other were recorded as tandem blocks. For each supercluster, the root mean square length of the block, the entropy of the block length distribution, and the total number of tandem blocks were calculated. The mean number of predicted TM helices and the fraction of proteins containing a signal peptide were calculated for each supercluster. For each cluster alignment within a supercluster, the number of cysteine residues conserved in least 50% of the sequences was calculated and the weighted mean was calculated for the supercluster. The genomic context (5 upstream and 5 downstream genes) of each gene within a supercluster was examined for the presence of known transporters, transporter regulators, and peptidases; the fraction of contexts positive for these gene categories was recorded. All data are available in Table S3.
ACKNOWLEDGMENT
The authors’ research is supported by the NIH Intramural Research Program at the National Library of Medicine, U.S. Department of Health and Human Services.
Footnotes
Citation Makarova KS, Wolf YI, Karamycheva S, Zhang D, Aravind L, Koonin EV. 2019. Antimicrobial peptides, polymorphic toxins, and self-nonself recognition systems in archaea: an untapped armory for intermicrobial conflicts. mBio 10:e00715-19. https://doi.org/10.1128/mBio.00715-19.
Contributor Information
Christa M. Schleper, University of Vienna.
Rotem Sorek, Weizmann Institute of Science.
R. Thane Papke, University of Connecticut at Storrs.
REFERENCES
- 1.Iyer LM, Burroughs AM, Anand S, de Souza RF, Aravind L. 2017. Polyvalent proteins, a pervasive theme in the intergenomic biological conflicts of bacteriophages and conjugative elements. J Bacteriol 199:e00245-17. doi: 10.1128/JB.00245-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind L, Anantharaman V, Zhang D, de Souza RF, Iyer LM. 2012. Gene flow and biological conflict systems in the origin and evolution of eukaryotes. Front Cell Infect Microbiol 2:89. doi: 10.3389/fcimb.2012.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. 2012. Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang D, Iyer LM, Burroughs AM, Aravind L. 2014. Resilience of biochemical activity in protein domains in the face of structural divergence. Curr Opin Struct Biol 26:92–103. doi: 10.1016/j.sbi.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, et al. 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh CT. 2016. Insights into the chemical logic and enzymatic machinery of NRPS assembly lines. Nat Prod Rep 33:127–135. doi: 10.1039/c5np00035a. [DOI] [PubMed] [Google Scholar]
- 7.Besse A, Peduzzi J, Rebuffat S, Carre-Mlouka A. 2015. Antimicrobial peptides and proteins in the face of extremes: lessons from archaeocins. Biochimie 118:344–355. doi: 10.1016/j.biochi.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Jamet A, Nassif X. 2015. New players in the toxin field: polymorphic toxin systems in bacteria. mBio 6:e00285. doi: 10.1128/mBio.00285-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes CS, Aoki SK, Low DA. 2010. Bacterial contact-dependent delivery systems. Annu Rev Genet 44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen VS, Douzi B, Durand E, Roussel A, Cascales E, Cambillau C. 2018. Towards a complete structural deciphering of type VI secretion system. Curr Opin Struct Biol 49:77–84. doi: 10.1016/j.sbi.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Ghequire MGK, De Mot R. 2015. The tailocin tale: peeling off phage tails. Trends Microbiol 23:587–590. doi: 10.1016/j.tim.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 12.Jank T, Lang AE, Aktories K. 2016. Rho-modifying bacterial protein toxins from Photorhabdus species. Toxicon 116:17–22. doi: 10.1016/j.toxicon.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Berks BC. 2015. The twin-arginine protein translocation pathway. Annu Rev Biochem 84:843–864. doi: 10.1146/annurev-biochem-060614-034251. [DOI] [PubMed] [Google Scholar]
- 14.Green ER, Mecsas J. 2016. Bacterial secretion systems: an overview. Microbiol Spectr 4. doi: 10.1128/microbiolspec.VMBF-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buttner CR, Wu Y, Maxwell KL, Davidson AR. 2016. Baseplate assembly of phage Mu: defining the conserved core components of contractile-tailed phages and related bacterial systems. Proc Natl Acad Sci U S A 113:10174–10179. doi: 10.1073/pnas.1607966113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D, Iyer LM, Aravind L. 2011. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res 39:4532–4552. doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherrak Y, Rapisarda C, Pellarin R, Bouvier G, Bardiaux B, Allain F, Malosse C, Rey M, Chamot-Rooke J, Cascales E, Fronzes R, Durand E. 2018. Biogenesis and structure of a type VI secretion baseplate. Nat Microbiol 3:1404–1416. doi: 10.1038/s41564-018-0260-1. [DOI] [PubMed] [Google Scholar]
- 18.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. 2013. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haft DH. 2015. Using comparative genomics to drive new discoveries in microbiology. Curr Opin Microbiol 23:189–196. doi: 10.1016/j.mib.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haft DH, Payne SH, Selengut JD. 2012. Archaeosortases and exosortases are widely distributed systems linking membrane transit with posttranslational modification. J Bacteriol 194:36–48. doi: 10.1128/JB.06026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iacovache I, van der Goot FG, Pernot L. 2008. Pore formation: an ancient yet complex form of attack. Biochim Biophys Acta 1778:1611–1623. doi: 10.1016/j.bbamem.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 22.Li PL, Hwang I, Miyagi H, True H, Farrand SK. 1999. Essential components of the Ti plasmid trb system, a type IV macromolecular transporter. J Bacteriol 181:5033–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christie PJ, Whitaker N, Gonzalez-Rivera C. 2014. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta 1843:1578–1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill CW, Sandt CH, Vlazny DA. 1994. Rhs elements of Escherichia coli: a family of genetic composites each encoding a large mosaic protein. Mol Microbiol 12:865–871. doi: 10.1111/j.1365-2958.1994.tb01074.x. [DOI] [PubMed] [Google Scholar]
- 25.Busby JN, Panjikar S, Landsberg MJ, Hurst MR, Lott JS. 2013. The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature 501:547–550. doi: 10.1038/nature12465. [DOI] [PubMed] [Google Scholar]
- 26.Hartley RW. 1989. Barnase and barstar: two small proteins to fold and fit together. Trends Biochem Sci 14:450–454. doi: 10.1016/0968-0004(89)90104-7. [DOI] [PubMed] [Google Scholar]
- 27.Kvansakul M, Adams JC, Hohenester E. 2004. Structure of a thrombospondin C-terminal fragment reveals a novel calcium core in the type 3 repeats. EMBO J 23:1223–1233. doi: 10.1038/sj.emboj.7600166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Springer TA. 2006. Complement and the multifaceted functions of VWA and integrin I domains. Structure 14:1611–1616. doi: 10.1016/j.str.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whittaker CA, Hynes RO. 2002. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell 13:3369–3387. doi: 10.1091/mbc.e02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linhartová I, Bumba L, Mašín J, Basler M, Osička R, Kamanová J, Procházková K, Adkins I, Hejnová-Holubová J, Sadílková L, Morová J, Sebo P. 2010. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev 34:1076–1112. doi: 10.1111/j.1574-6976.2010.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawasaki H, Kretsinger RH. 2017. Structural and functional diversity of EF-hand proteins: evolutionary perspectives. Protein Sci 26:1898–1920. doi: 10.1002/pro.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friebe S, van der Goot FG, Burgi J. 2016. The ins and outs of anthrax toxin. Toxins (Basel) 8:E69. doi: 10.3390/toxins8030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Batot G, Michalska K, Ekberg G, Irimpan EM, Joachimiak G, Jedrzejczak R, Babnigg G, Hayes CS, Joachimiak A, Goulding CW. 2017. The CDI toxin of Yersinia kristensenii is a novel bacterial member of the RNase A superfamily. Nucleic Acids Res 45:5013–5025. doi: 10.1093/nar/gkx230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aravind L, Zhang D, de Souza RF, Anand S, Iyer LM. 2015. The natural history of ADP-ribosyltransferases and the ADP-ribosylation system. Curr Top Microbiol Immunol 384:3–32. doi: 10.1007/82_2014_414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon NC, Aktories K, Barbieri JT. 2014. Novel bacterial ADP-ribosylating toxins: structure and function. Nat Rev Microbiol 12:599–611. doi: 10.1038/nrmicro3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morse RP, Willett JL, Johnson PM, Zheng J, Credali A, Iniguez A, Nowick JS, Hayes CS, Goulding CW. 2015. Diversification of beta-augmentation interactions between CDI toxin/immunity proteins. J Mol Biol 427:3766–3784. doi: 10.1016/j.jmb.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartley RW. 1988. Barnase and barstar. Expression of its cloned inhibitor permits expression of a cloned ribonuclease. J Mol Biol 202:913–915. doi: 10.1016/0022-2836(88)90568-2. [DOI] [PubMed] [Google Scholar]
- 38.Buckle AM, Schreiber G, Fersht AR. 1994. Protein-protein recognition: crystal structural analysis of a barnase-barstar complex at 2.0-A resolution. Biochemistry 33:8878–8889. doi: 10.1021/bi00196a004. [DOI] [PubMed] [Google Scholar]
- 39.Mazguene S, Rossi M, Gogliettino M, Palmieri G, Cocca E, Mirino S, Imadalou-Idres N, Benallaoua S. 2018. Isolation and characterization from solar salterns of north Algeria of a haloarchaeon producing a new halocin. Extremophiles 22:259–270. doi: 10.1007/s00792-017-0994-3. [DOI] [PubMed] [Google Scholar]
- 40.Ellen AF, Rohulya OV, Fusetti F, Wagner M, Albers SV, Driessen AJ. 2011. The sulfolobicin genes of Sulfolobus acidocaldarius encode novel antimicrobial proteins. J Bacteriol 193:4380–4387. doi: 10.1128/JB.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price LB, Shand RF. 2000. Halocin S8: a 36-amino-acid microhalocin from the haloarchaeal strain S8a. J Bacteriol 182:4951–4958. doi: 10.1128/JB.182.17.4951-4958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun C, Li Y, Mei S, Lu Q, Zhou L, Xiang H. 2005. A single gene directs both production and immunity of halocin C8 in a haloarchaeal strain AS7092. Mol Microbiol 57:537–549. doi: 10.1111/j.1365-2958.2005.04705.x. [DOI] [PubMed] [Google Scholar]
- 43.Makarova KS, Wolf YI, Forterre P, Prangishvili D, Krupovic M, Koonin EV. 2014. Dark matter in archaeal genomes: a rich source of novel mobile elements, defense systems and secretory complexes. Extremophiles 18:877–893. doi: 10.1007/s00792-014-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walden M, Edwards JM, Dziewulska AM, Bergmann R, Saalbach G, Kan SY, Miller OK, Weckener M, Jackson RJ, Shirran SL, Botting CH, Florence GJ, Rohde M, Banfield MJ, Schwarz-Linek U. 2015. An internal thioester in a pathogen surface protein mediates covalent host binding. eLife 4. doi: 10.7554/eLife.06638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Makarova KS, Wolf YI, Koonin EV. 2013. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res 41:4360–4377. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anantharaman V, Aravind L. 2003. New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol 4:R81. doi: 10.1186/gb-2003-4-12-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gebhard S. 2012. ABC transporters of antimicrobial peptides in Firmicutes bacteria—phylogeny, function and regulation. Mol Microbiol 86:1295–1317. doi: 10.1111/mmi.12078. [DOI] [PubMed] [Google Scholar]
- 48.Kang H, Gan J, Zhao J, Kong W, Zhang J, Zhu M, Li F, Song Y, Qin J, Liang H. 2017. Crystal structure of Pseudomonas aeruginosa RsaL bound to promoter DNA reaffirms its role as a global regulator involved in quorum-sensing. Nucleic Acids Res 45:699–710. doi: 10.1093/nar/gkw954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R. 2006. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell 124:549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- 50.Gonzalez-Pastor JE, Hobbs EC, Losick R. 2003. Cannibalism by sporulating bacteria. Science 301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- 51.Povolotsky TL, Orlova E, Tamang DG, Saier MH Jr.. 2010. Defense against cannibalism: the SdpI family of bacterial immunity/signal transduction proteins. J Membr Biol 235:145–162. doi: 10.1007/s00232-010-9260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kjos M, Snipen L, Salehian Z, Nes IF, Diep DB. 2010. The abi proteins and their involvement in bacteriocin self-immunity. J Bacteriol 192:2068–2076. doi: 10.1128/JB.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol 6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown BL, Wood TK, Peti W, Page R. 2011. Structure of the Escherichia coli antitoxin MqsA (YgiT/b3021) bound to its gene promoter reveals extensive domain rearrangements and the specificity of transcriptional regulation. J Biol Chem 286:2285–2296. doi: 10.1074/jbc.M110.172643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whiteley M, Diggle SP, Greenberg EP. 2017. Progress in and promise of bacterial quorum sensing research. Nature 551:313–320. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang D, Burroughs AM, Vidal ND, Iyer LM, Aravind L. 2016. Transposons to toxins: the provenance, architecture and diversification of a widespread class of eukaryotic effectors. Nucleic Acids Res 44:3513–3533. doi: 10.1093/nar/gkw221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Montgomery K, Charlesworth JC, LeBard R, Visscher PT, Burns BP. 2013. Quorum sensing in extreme environments. Life (Basel) 3:131–148. doi: 10.3390/life3010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robertson CE, Harris JK, Spear JR, Pace NR. 2005. Phylogenetic diversity and ecology of environmental Archaea. Curr Opin Microbiol 8:638–642. doi: 10.1016/j.mib.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 59.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Makarova KS, Wolf YI, Koonin EV. 2015. Archaeal Clusters of Orthologous Genes (arCOGs): an update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales. Life (Basel) 5:818–840. doi: 10.3390/life5010818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson LS, Eddy SR, Portugaly E. 2010. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res 37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 65.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 66.Drozdetskiy A, Cole C, Procter J, Barton GJ. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Iterative “guilt-by-association” procedure to identify polymorphic toxin systems in archaea. Download FIG S1, PDF file, 0.1 MB (135.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Phyletic patterns for archaeal offense systems and key components of putative self-nonself recognition system. Download Table S1, XLSX file, 0.1 MB (81.3KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Genomic loci for archaeal offense systems and key components of putative self-nonself recognition systems. Download Table S2, XLSX file, 1.5 MB (1.5MB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Sequence logo for the “RRG” C-terminal motif specific for Halobacteria. Download FIG S2, PDF file, 1.0 MB (1,016.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Supporting alignments for key components of archaeal offense systems or putative self-nonself recognition systems. Download FIG S3, DOCX file, 0.2 MB (242.3KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Sequence and genomic organization features for archaeal clusters. Download Table S3, XLSX file, 0.2 MB (243.4KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.