Abstract
The colorful heliconiine butterflies are distasteful to predators due to their content of defense compounds called cyanogenic glucosides (CNglcs), which they biosynthesize from aliphatic amino acids. Heliconiine larvae feed exclusively on Passiflora plants where ~30 kinds of CNglcs have been reported. Among them, some CNglcs derived from cyclopentenyl glycine can be sequestered by some Heliconius species. In order to understand the evolution of biosynthesis and sequestration of CNglcs in these butterflies and its consequences for their arms race with Passiflora plants, we analyzed the CNglc distribution in selected heliconiine and Passiflora species. Sequestration of cyclopentenyl CNglcs is not an exclusive trait of Heliconius, since these compounds were present in other heliconiines such as Philaethria, Dryas and Agraulis, and in more distantly related genera Cethosia and Euptoieta. Thus, it is likely that the ability to sequester cyclopentenyl CNglcs arose in an ancestor of the Heliconiinae subfamily. Biosynthesis of aliphatic CNglcs is widespread in these butterflies, although some species from the sara‐sapho group seem to have lost this ability. The CNglc distribution within Passiflora suggests that they might have diversified their cyanogenic profile to escape heliconiine herbivory. This systematic analysis improves our understanding on the evolution of cyanogenesis in the heliconiine–Passiflora system.
Keywords: coevolution, cyanide, Heliconius, Lepidoptera, Passiflora, specialized metabolites
1. INTRODUCTION
Land plants have been exposed to herbivores for over 430 million years. To cope with this, plants produce a remarkable diversity of specialized metabolites that act as chemical protections (Fürstenberg‐Hägg, Zagrobelny, & Bak, 2013). In turn, specialist herbivores have evolved under the selection pressure from the chemical defenses of their hosts and adapted to handle their toxicity and even to utilize these metabolites for their own benefit (Nishida, 2014).
The distasteful and colorful butterflies of the Heliconiini tribe selectively feed as larvae on plants from the Passiflora genus regardless of the plants’ chemical defenses which, effective against most other herbivores. Due to their larval‐feeding specialization, heliconiines are also called passion vine butterflies. The species diversity (more than 70 heliconiine and 600 passion vines) and multiplicity of feeding guilds found in the heliconiine–Passiflora system offer a unique comparative potential to address many intriguing questions on evolutionary and chemical ecology (Gilbert, 1991; Jiggins, 2017). The basal heliconiine genera Podotricha, Philaethria, Dryas, Dryadula, Dione, Agraulis, and Eueides (Kozak et al., 2015) are overall generalists, feeding on many Passiflora species. In contrast, different degrees of host specialization are observed within Heliconius, the most diverse heliconiine genus, with some close phylogenetic associations between infragenic groups of Heliconius and their Passiflora hosts (Arias et al., 2016; Engler‐Chaouat & Gilbert, 2007). He liconius species that perform pupal mating (erato and sara‐sapho groups), an unusual behavior where adult males search for female pupae for mating and either penetrate the pupa or mate the female as soon as it emerges, are characterized by being specialists on Passiflora plants of the subgenera Decaloba or Astrophea. Species of melpomene, silvaniforms, and primitive groups (aoede, doris, wallacei), which comprise the nonpupal mating clade, are overall specialists on Passiflora species of the subgenus Passiflora (Benson, Brown, & Gilbert, 1975).
The chemical defense of the genus Passiflora is comprised of different types of cyanogenic glucosides (CNglcs), and the great success of heliconiines feeding on Passiflora could be due to the prior ability of these butterflies to biosynthesize CNglcs (Nahrstedt & Davis, 1981). Subsequently, it has been hypothesized that the ability to handle the toxicity of CNglcs was one of the crucial traits that allowed the ancestor of heliconiines to feed on these plants, implying that the ability to detoxify CNglcs preceded their ability to sequester these compounds and perhaps even older than the ability to biosynthesize them (de Castro et al., 2018).
CNglcs are some of the most ancient and widespread defense compounds produced by plants: Whereas other defense compounds are typically restricted to a specific plant group, such as glucosinolates in Brassicaceae, CNglcs are broadly distributed in 2,500 species from ferns to flowering families (Gleadow & Møller, 2014). In insects, CNglc distribution is restricted to a few lineages within Coleoptera and Hemiptera, which seem to obtain these compounds from their diet, and to some lepidopterans (moths and butterflies) where both biosynthesis and/or sequestration is rather widespread (Zagrobelny, Castro, Møller, & Bak, 2018).
CNglcs are glycosylated cyanide‐containing compounds that are not intrinsically poisonous as glucosides. However, tissue damage caused by, for example, herbivore or predator attack leads to CNglcs coming in contact with hydrolytic enzymes (β‐glucosidases and α‐hydroxynitrile lyases), which convert these compounds into toxic hydrogen cyanide (HCN) and aglycones (Pentzold et al., 2017). Protein and nonprotein amino acids are precursors for the biosynthesis of CNglcs and can accordingly be classified as aliphatic, aromatic, or cyclopentenoid (Figure 1). The aromatic and aliphatic CNglcs are derived from protein amino acids like phenylalanine and valine and are broadly distributed in the Plant Kingdom (Zagrobelny et al., 2004). Contrarily, cyclopentenyl CNglcs are synthesized from the nonprotein amino acid cyclopentenyl glycine and have been so far found in five closely related plant families of the Order Malpighiales: Passifloraceae, Turneraceae, Achariaceae, Salicaceae, and Violaceae (Bjarnholt et al., 2008; Tober & Conn, 1985).
Figure 1.
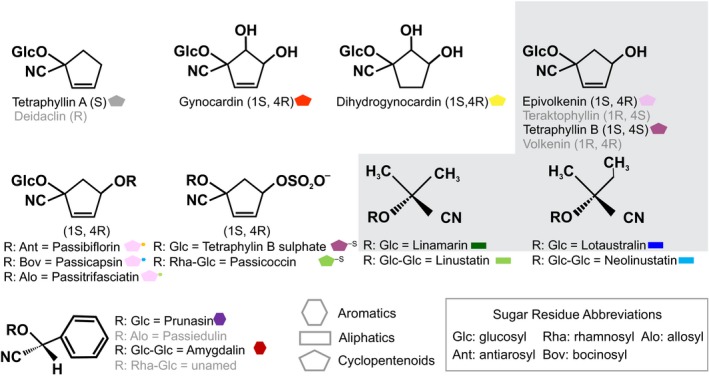
Cyanogenic glucoside (CNglcs) structures reported in Passiflora species. Compounds with a gray background were also reported in heliconiines butterflies (Nahrstedt & Davis, 1981; Engler, Spencer, & Gilbert, 2000). Structures with name in gray are enantiomers
The biosynthetic pathway of aromatic and aliphatic CNglcs has been characterized in several plant species. For example, in Lotus japonicus the biosynthetic pathway of the aliphatic CNglcs linamarin and lotaustralin consists of two cytochromes P450 (P450s) that convert valine and isoleucine into their respective α‐hydroxynitriles, and an UDP‐glycosyltransferase (UGT) that catalyzes the addition of a sugar residue to these molecules (Takos et al. 2011). In insects, this pathway has been characterized only in the burnet moth Zygaena filipendulae which also contain linamarin and lotaustralin. Here, the pathway is composed of the two P450s CYP405A2 and CYP332A3, and UGT33A1 (Jensen et al., 2011). Although the pathways in plants and insects share the same catalytic reactions and enzyme types, the genes encoding the enzymes are not orthologues, confirming that the ability to produce aliphatic CNglcs arose independently in these two Kingdoms. Heliconiine butterflies also synthesize the aliphatic CNglcs linamarin and lotaustralin through the same enzymatic steps as Zygaena (Davis & Nahrstedt, 1987), and genes homologous to ZfCYP405A2 and ZfCYP332A3 have been found in the H. melpomene genome (Chauhan, Jones, Wilkinson, Pauchet, & ffrench‐Constant, 2013) and other Heliconius species (Zagrobelny, Castro et al., 2018) although, they have not yet been functionally characterized. This indicates that the biosynthetic pathway of linamarin and lotaustralin has originated in a common ancestor of butterflies and moths (Zagrobelny, Castro et al., 2018).
Some lepidopterans are known to selectively sequester CNglcs from their larval host plant. For example, Z. filipendulae sequester linamarin and lotaustralin from Lotus corniculatus, possibly to reduce the energetic cost associated with the biosynthesis of these compounds (Fürstenberg‐Hägg et al., 2014). Furthermore, Apollo butterflies (Parnassius) are thought to sequester, as well as biosynthesize, sarmentosin, a bitter compound related to aliphatic CNglcs (Bjarnholt et al., 2012). In contrast to these lepidopterans, some Heliconius species, especially from the sara‐sapho group, have been reported to sequester the cyclopentenyl CNglc epivolkenin from Passiflora plants (Engler et al., 2000), a CNglc that differs from the aliphatic CNglc they can biosynthesize. Sequestration of cyclopentenyl CNglcs is also found in other species of the Heliconiinae subfamily, for example, in larvae of Euptoieta hegesia (tribe Argynnini) which were more cyanogenic when fed on cyanogenic Turnera ulmifolia plants (Turneraceae) (Schappert & Shore, 1999; Tober & Conn, 1985). Additionally, Acraea horta butterflies (tribe Acraeini) contained the cyclopentenyl CNglc gynocardin when fed on the plant Kigellaria africana (Achariaceae), which produces this CNglc (Raubenheimer, 1989). Indeed, most species of the Heliconiinae subfamily feed on Passifloraceae plants and closely related families containing cyclopentenyl CNglcs as larval hosts (Silva‐Brandão et al., 2008).
Remarkably, almost 30 types of CNglcs have been reported in the Passiflora genus, and it has been hypothesized that these plants have diversified the structures of CNglcs to specifically evade heliconiine herbivory (Spencer, 1988). However, a detailed comparison between the distribution of the different CNglcs within Passiflora and the heliconiine host‐plant preferences has not previously been carried out. Moreover, it is not yet known if basal genera of heliconiines can sequester cyclopentenyl CNglcs from Passiflora. The analyses of the cyanogenic potential of Heliconius and other heliconiine species have mostly been made with cyanide release measurements (Arias et al., 2016; Cardoso & Gilbert, 2013; Hay‐Roe & Nation, 2007), and this generic technique is unable to show which chemical types of CNglcs are present in each species.
In order to understand the evolution of biosynthesis and sequestration of CNglcs in these butterflies, information regarding the CNglc profiles of heliconiines and their Passiflora host are necessary. Here, we investigate the CNglcs profile of selected heliconiines species and combine it with phylogenetic comparisons. Additionally, we overlap the CNglc profile of 42 Passiflora species with the butterflies’ host‐plant preferences, in order to elucidate how cyanogenesis has influenced the arms race between heliconiines and their host plants.
2. METHODS
2.1. Butterfly and plant samples
Pupae from 19 species from the Heliconiinae subfamily were bought from the Costa Rica Entomological Supply (CRES) or Stratford‐Upon‐Avon Butterfly Farm and reared at the greenhouse facilities of the Department of Plant and Environmental Sciences, University of Copenhagen. Pupae were kept in separate cages and maintained under controlled conditions (24–28°C, 80% humidity, 14‐hr light). Cages were inspected every day and emerged butterflies collected in plastic bags, weighed, frozen in liquid nitrogen, and stored at −80°C. All butterflies were collected within 24 hr of eclosion to standardize age, mating history (virgins), and adult food consumption (unfed). Philaethria dido, Philaethria wernickei, and Euptoieta hegesia are not usually bred by butterfly farms, so these species were field‐captured in Brazil, in the Jiqui woods maintained by EMPARN (Parnamirin‐RN) (for more information, see Cardoso & De Lima, 2017) and in forested sites belonging to Miriri Food and Bioenergy S/A (Santa Rita municipality, state of Paraíba). These butterflies were captured with a net and collected in tubes containing 4 ml 80% methanol (v/v).
Passiflora samples from this study were from the Copenhagen Botanical Garden and the greenhouse facilities of the University of Copenhagen. Leaves of each species were collected in individual plastic bags, frozen in liquid nitrogen, and kept at −80°C for further analyses.
2.2. Methanol extraction
A cold extraction method was used for all butterfly samples as described previously (Zagrobelny, Bak, Olsen, & Møller, 2007). Samples were homogenized with ice‐cold mortars and pestles in 1 or 1.4 ml of a solution containing 55% (v/v) methanol and 0.1% (v/v) formic acid. Samples collected in the field were homogenized in the solution where they were soaked: 4 ml 80% (v/v) methanol. A boiling extraction method (Lai et al., 2015) was used for the plant samples, where leaf pieces of each species were added to microtubes containing 0.5 ml 85% (v/v) methanol and boiled for 5 min in a water bath. All samples were subsequently centrifuged at 10,000 g for 5 min and the supernatant filtered (Anapore 0.45 µm, Whatman) to remove insoluble components.
2.3. Liquid Chromatography‐Mass Spectrometry (LC‐MS/MS)
Analytical LC‐MS was carried out using an Agilent 1100 Series LC (Agilent Technologies, Germany) hyphenated to a Bruker HCT‐Ultra ion trap mass spectrometer (Bruker Daltonics, Bremen, Germany). Chromatographic separation was carried out using a Zorbax SB‐C18 column (Agilent; 1.8 μM, 2.1 × 50 mm) at a flow rate of 0.2 ml/min. Two solvents were used as mobile phases, A—containing 0.1% (v/v) formic acid with 50 μM NaCl and B—composed of acetonitrile with 0.1% (v/v) formic acid. The gradient program was: 0–0.5 min, isocratic 2% B; 0.5–7.5 min, linear gradient 2%–40% B; 7.5–8.5 min, linear gradient 40%–90% B; 8.5–11.5 min isocratic 90% B; and 11.6–17 min, isocratic 2% B. The flow rate was increased to 0.3 ml/min in the interval 11.2–13.5 min. The oven temperature was maintained at 35°C.
Mass spectral data were analyzed with the native data analysis software. Sodium adducts of tetraphyllin A (RT 5.5 min, [M+Na]+ at m/z 294), tetraphyllin B (RT 1.3 min, [M+Na]+ at m/z 310), epivolkenin (RT 2.3 min, [M+Na]+ at m/z 310), gynocardin (RT 1.4 min, [M+Na]+ at m/z 326), linamarin (RT 2.4 min, [M+Na]+ at m/z 270), lotaustralin (RT 5.5 min, [M+Na]+ at m/z 284), prunasin (RT 7 min, [M+Na]+ at m/z 317), and amygdalin (RT 6.6 min, [M+Na]+ at m/z 480) were detected and their RTs compared to authentic standards (Engler et al., 2000; Jaroszewski et al., 2002; Møller, Olsen, & Motawia, 2016). The total amount of each compound was estimated based on extracted ion chromatogram (EIC) peak areas and quantified based on calibration curves of linamarin, lotaustralin, and amygdalin. Linustatin (RT 3 min, [M+Na]+ at m/z 432), dihydrogynocardin (RT 1.4 min, [M+Na]+ at m/z 328), tetraphyllin B sulfate (RT 1.3 min, [M+Na]+ at m/z 390), passicapsin (6.5 min, [M+Na]+ at m/z 440), and passibiflorin (RT 5.8 min, [M+Na]+ at m/z 456) were identified by their m/z, fragmentation pattern (MS/MS), and comparison with data reported in the literature regarding these compounds. Quantification of CNglcs present in the butterfly samples was based on a regression equation calculated from a standard curve.
2.4. Comparative analyses
MANOVA and pairwise comparisons using Geomorph v.3.0.5 package (Adams, 2014) in R (R Core Team, 2017) were performed to analyze infraspecific (sexual dimorphism) and interspecific differences in CNglc concentrations. Data were not normally distributed; therefore, the analyses were performed with square root transformed data, as well as with the raw data for comparison. Preliminary analyses did not support sexual dimorphism in CNglc composition of the butterflies; thus, female and males were not distinguished in further investigations.
Phylogenetic comparative methods were performed using the tree‐hypothesis proposed by Kozak et al. (2015). Phytools 0.6‐20 package (Revell, 2012) in R (R Core Team, 2017) was used for the ancestral reconstruction of CNglc biosynthesis (aliphatic) and sequestration (cyclopentenoids) in heliconiines (Cethosia cyane was used as an outgroup). The phylogenetic signal of the butterflies’ CNglc profiles was measured utilizing the k mult approach (Adams, 2014), a generalization of the K statistic (Blomberg, Garland, & Ives, 2003). Values of K mult < 1 imply that taxa resemble each other phenotypically less than expected under Brownian motion, whereas values of K mult > 1 imply that close relatives are more similar to one another phenotypically than expected under Brownian motion.
Additionally, to build a phylomorphospace (Sidlauskas, 2008), we performed a principal component analyses (PCA) of the butterflies’ CNglc profiles and projected the first two components on the phylogeny. Both analyses were conducted using Geomorph 3.0.5 package (Adams, 2014) in R (R Core Team, 2017).
3. RESULTS
3.1. Cyanogenic glucoside distribution within heliconiines
Although all heliconiines were thought to be cyanogenic to some degree, the CNglc composition of only a few species has been reported to date. Therefore, we identified and quantified the CNglcs present in male and female adults of 22 species that belong to the subfamily Heliconiinae.
The aliphatic CNglcs linamarin and lotaustralin were found in almost all species analyzed, confirming the widespread occurrence of CNglc synthesis in the tribe Heliconiini (Figure 2). Interestingly, some species of the sara‐sapho clade (H. sapho, H. hewitsoni, and H. antiochus), specialized on plants of the Astrophea subgenus as larval hosts, lacked these compounds. Instead, these species contained epivolkenin, a compound derived from sequestration because insects are not known to biosynthesize its precursor cyclopentenyl glycine.
Figure 2.
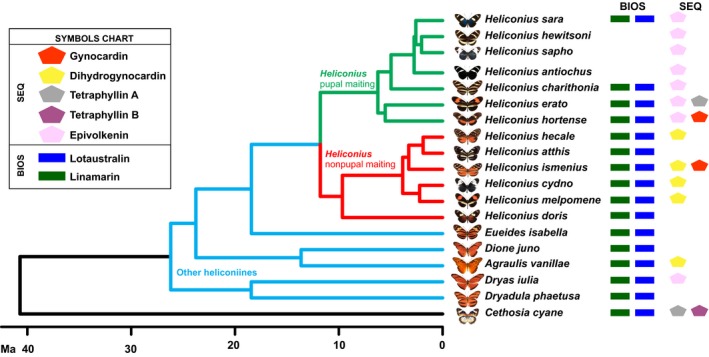
CNglc distribution in the Heliconiini tribe and in the outgroup Cethosia cyane. Phylogenetic dendrogram is according to Kozak et al. (2015)
Surprisingly, cyclopentenyl CNglcs were not found exclusively in Heliconius species as previously reported (Engler‐Chaouat & Gilbert, 2007). They were also present in A. vanillae, D. iulia, in the outgroup C. cyane (Figure 2), and in wild‐caught Philaethria dido and Euptoieta hegesia (data not shown). These findings indicate that sequestration of cyclopentenyl CNglcs is a broadly distributed trait within the subfamily Heliconiinae and is not a derived trait of Heliconius. Besides epivolkenin, the additional cyclopentenyl CNglcs dihydrogynocardin, gynocardin, tetraphyllin B, and tetraphyllin A were found in some species. We have not found earlier reports for the presence of tetraphyllin B, tetraphyllin A, and dihydrogynocardin in butterflies, or of gynocardin in Heliconius. Tetraphyllin B is a diastereomer of epivolkenin, whereas tetraphyllin A has a nonhydroxylated cyclopentenoid ring and gynocardin an extra hydroxylation in C5 (Figure 1). Dihydrogynocardin is the only cyclopentenylglycine‐derived CNglc containing a cyclopentanoid ring.
Across all Heliconius species, epivolkenin was present in the entire pupal mating clade although only traces of this compound were found in H. charithonia. In addition, H. hortense and H. erato also contained gynocardin and tetraphyllin A, respectively (Figure 2). Epivolkenin was conspicuously absent in the nonpupal mating clade, which almost exclusively sequestered dihydrogynocardin, although gyanocardin was found in H. ismenius and H. atthis, and H. doris did not contain any cyclopentenyl CNglcs.
3.2. Total CNglc concentration in heliconiines and host‐plant specialization
Our results suggest that the CNglc composition of heliconiine butterflies correlates with their larval‐diet specialization (Figure 3). The species with the highest total concentration of CNglcs were H. sapho (7.45 µg/mg) and H. antiochus (6.17 µg/mg), which are both Astrophea specialists and contained only epivolkenin (Figure 3). In the pairwise comparisons between species, the total CNglc concentration in H. sapho was significantly higher than most heliconiines, except Eueides isabela (4.17 µg/mg) and H. atthis (4.61 µg/mg) which contained only the biosynthesized CNglcs linamarin and lotaustralin, and H. antiochus (Figure 3 and Supporting Information Table S1). Heliconius species that were Passiflora and Decaloba specialists had similar CNglc concentrations, apart from H. atthis that had significantly higher concentrations than H. melpomene (2.56 µg/mg). Although E. isabela had more linamarin and lotaustralin and Agraulis vanillae contained additional cyclopentenyl CNglcs in its composition, there were no significant differences in the total concentration of CNglcs between other heliconiines which utilize many passion vine as larval host (Figure 3 and Supporting Information Table S1). The genus Heliconius tend to have more CNglcs than other heliconiine genera although differences were not statistically significant (F 1,133 = 4.12, p = 0.053).
Figure 3.
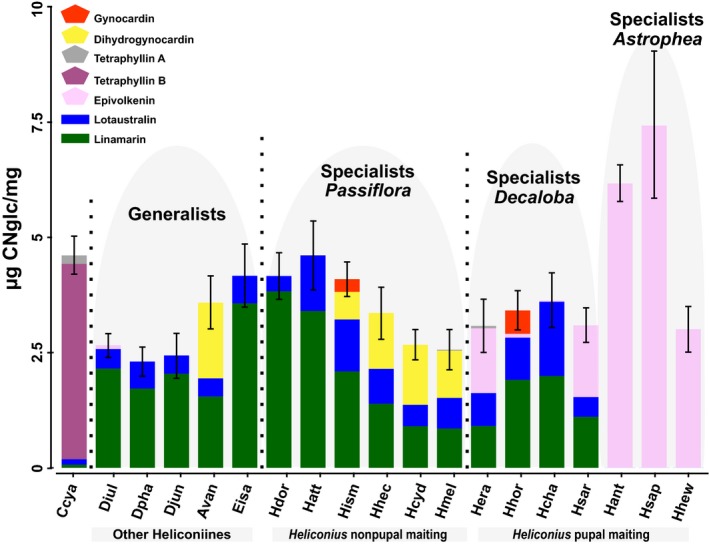
CNglc concentrations in heliconiine species and in the outgroup Cethosia cyane. CNglc composition of heliconiine butterflies is also categorized by their larval‐feeding strategy: generalists, Passiflora specialists, Decaloba specialists, and Astrophea specialists
Overall, there were no significant differences in CNglc concentration between male and female butterflies in this study (data not shown). Nevertheless, sequestration of epivolkenin in D. iulia and H. charithonia was observed only in female butterflies. Additionally, cyclopentenyl CNglcs were absent in most of the males of H. hecale (data not shown).
3.3. Biosynthesis of aliphatic versus sequestration of cyclopentenyl CNglcs
The heliconiine species analyzed in this study could be sorted into three groups based on their source of CNglcs: biosynthesis only, sequestration only, or both (Figure 2). The highest CNglc concentrations were found in species that either performed biosynthesis only (E. isabela, H. atthis, and H. doris) or sequestration only (H. sapho and H. antiochus) (Figure 3). In the Heliconius species that used both strategies, we observed a potential trade‐off between these two processes: species that sequestered cyclopentenyl CNglcs tended to have lower amounts of biosynthesized linamarin and lotaustralin (Figure 4). Preferences for biosynthesis or sequestration are apparently phylogenetic related (Figure 4) possibly due to a direct link with larval‐diet specialization (Figure 3).
Figure 4.
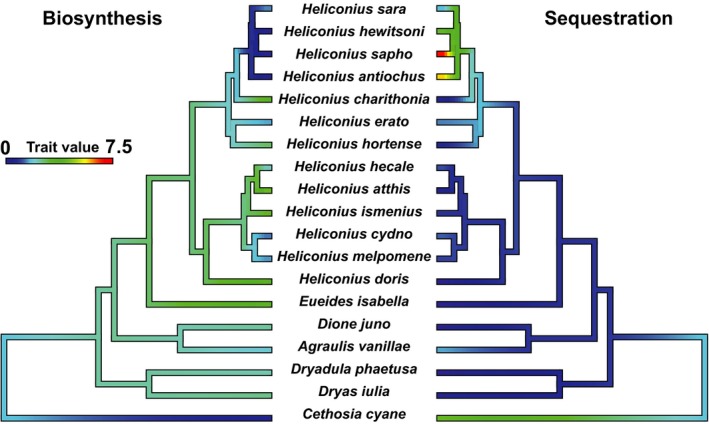
Ancestral reconstruction state of biosynthesis of aliphatic CNglcs and sequestration of cyclopentenyl CNglcs illustrating the balance between these two processes in heliconiines (+C. cyane). Trait value refers to the total mean CNglc biosynthesized or sequestered (μg/mg) by species.
Cethosia cyane and H. sapho, H. antiochus, and H. hewitsoni (Heliconius–Astrophea specialists) had significant differences in the concentration of biosynthesized CNglcs compared to the other species. Only traces of linamarin and lotaustralin were found in C. cyane, while in Heliconius–Astrophea specialists, these compounds were absent (Figures 3 and 4, Supporting Information Table S2). Contrarily, C. cyane and Heliconius–Astrophea specialists contained greater concentrations of sequestered CNglcs than all other species.
Within Decaloba specialists, H. charithonia had significantly higher concentrations of biosynthesized CNglcs than H. sara and H. erato, but similar to H. hortensis. In contrast, H. erato and H. sara had comparable concentrations of sequestered CNglcs and differed from all other analyzed species. (Figure 3 and Supporting Information Table S2).
The total concentration of CNglcs did not differ between the Passiflora specialists that performed both biosynthesis and sequestration (H. ismenius, H. hecale, H. cydno, and H. melpomene) (Figure 3 and Supporting Information Table S2). However, H. doris and H. atthis, which only biosynthesize linamarin and lotaustralin, have significantly higher concentration of these compounds than the remaining species in the group, except for H. ismenius.
There were no significant differences in the concentration of biosynthesized CNglcs within the heliconiine species with generalist‐feeding preferences. Additionally, Agraulis vanillae was the sole species showing significant differences regarding CNglc sequestration due to the presence of dihydrogynocardin (Figures 3).
3.4. Phylogenetic divergences in the CNglc profile of heliconiines
Among basal heliconiines, CNglcs were obtained mainly through biosynthesis, while sequestration was performed only by some species (Figure 2). The ability to sequester CNglc seems to gain more importance through the Heliconius genus until it becomes the sole source of CNglcs in the most specialized species in the sara‐sapho group (H. sapho, H. hewitsoni, H. antiochus) which are Astrophea specialists. CNglc sequestration is also the main strategy performed by the outgroup Cethosia cyane (Figure 4), an Asian species of the Heliconiinae subfamily, which does not belong to the Heliconiini tribe.
The phylomorphospace in Figure 5 allows a two‐dimensional visualization of the phylogeny in the morphospace (PCA axes). In this case, we correlated the concentration of CNgcls in heliconiines and the outgroup Cethosia cyane with their phylogenetic distances. Sequestration was the main strategy in most pupal mating Heliconius species and C. cyane (Figure 4), although the compounds obtained by them have a different chemical structure. Most of the pupal mating Heliconius species (except H. hortense and H. charithonia) are on the negative PC1 axis, which correspond to the presence of the sequestered CNglc epivolkenin (Figure 5 and inset). Nonpupal mating Heliconius and other heliconiines are in the positive extreme, because they perform biosynthesis of linamarin and lotaustralin and sequestration of other cyclopentenyl CNglcs besides epivolkenin (gynocardin, dihydrogynocardin, tetraphyllin A). Cethosia cyane is totally separated from the heliconiines in the positive extreme of PC2 axis, due to its sequestration of tetraphyllin B and its phylogenetic distance. Closely related species are chemically more similar among them than with more distant species (K mult = 0.49, p = 0.001), supporting the segregation pattern found in the phylomorphospace (Figure 5). Low phylogenetic signal is probably a consequence of chemical resemblances between some Heliconius species and basal heliconiines (e.g., H. charithonia and H. hortense which are separated in the multidimensional space of the pupal mating group and overlapping with other heliconiines).
Figure 5.
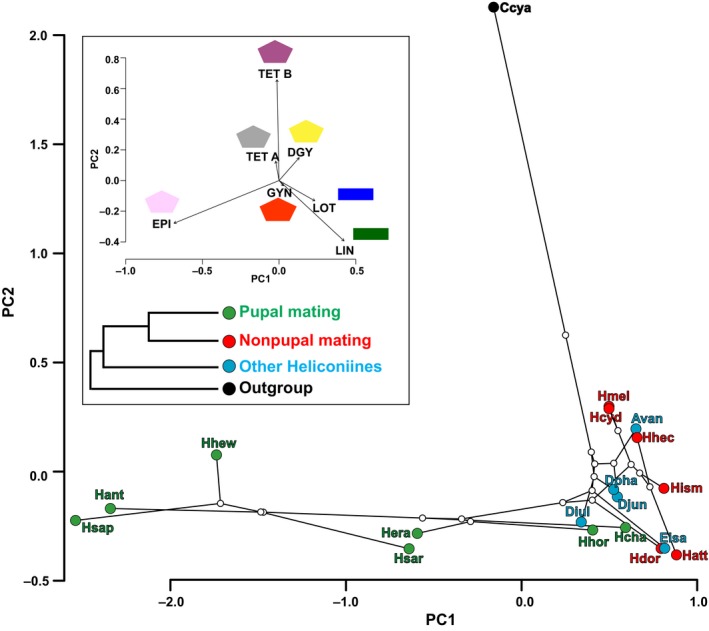
Phylomorphospace correlating the concentration of each CNglcs in heliconiines + Cethosia cyane (outgroup) with their phylogenetic distances. Each colored point represents a concentration value of CNglc by species and white points the hypothesized ancestral phenotype. Lines connect related species through hypothetical ancestors. Different colors represent the phylogenetic groups in box. The phylogeny (Kozak et al., 2015) has been pruned to include only species used in our study. Hsap = Heliconius sapho; Hant = H. antiochus; Hhew = H. hewitsoni; Hsar = H. sara; Hcha = H. charithonia; Hera = H. erato; Hhor = H. hortense; Hmel = H. melpomene; Hcyd = H. cydno; Hhec = H. hecale; Hatt = H. atthis; Hism = H. ismenius; Hdor = H. doris; Eisa = Eueides isabella; Avan = Agraulis vanillae; Djun = Dione juno; Diul = Dryas iulia; Dpha = Dryadula phaethusa; Ccya = Cethosia cyane
3.5. Distribution of CNglcs within Passiflora
The CNglc profiles of 22 Passiflora species were analyzed in order to document which compounds would be available to heliconiine larvae from their host plants. These results are shown in Table 1, together with information compiled from the literature regarding the CNglc composition of 20 other Passiflora species.
Table 1.
CNglcs distribution among Passiflora species
| Subgenus | Section | Serie | Species | CNglcs | References |
|---|---|---|---|---|---|
| Deidamioides | Discophora | P. discophora | TEB | Jaroszewski et al. (2002) | |
| Astrophea | P. amoena | LIN, LOT, LNT | This work | ||
| Passiflora | Stipulata | Grannadillastrum | P. foetida | TEA, TEB | This work |
| P. caerulea | TEB, TEB(S) | This work Jaroszewski and Fog (1989) | |||
| P. violacea | TEB(S) | Jaroszewski et al. (2002) | |||
| P. subpeltata | LIN | Olafsdottir et al. (1989) | |||
| P. menispermifolia | EPI, TEA | This work | |||
| Passiflora | Calopathanthus | P. racemosa | TEB(S) | Jaroszewski and Fog (1989) | |
| P. mathewsii | nd. | This work | |||
| Passiflora | P. incarnata | GYN | Spencer and Seigler (1984) | ||
| P. edulis | PRU, AMY | This work | |||
| P. cincinnata | PCP | This work | |||
| Coccinea | P. coccinea | PCO(S) | Kevin Spencer and Seigler (1985) | ||
| P. vitifolia | TEB(S) | This work | |||
| Laurifolia | Tiliifoliaa | P. serratodigitata | EPI, PBF | This work | |
| P. platyloba | PRU, AMY | This work | |||
| P. maliformis | PRU, AMY | This work | |||
| Quadrangulares | P. quadrangularis | EPI, TEA, TEB(S) | Jaroszewski and Fog (1989) | ||
| Laurifoliae | P. ligularis | nd. | This work | ||
| P. laurifolia | nd. | This work | |||
| P. riparia | nd. | This work | |||
| Tetrapathea | P. tetrandra | TEA, TEB | Olafsdottir et al. (1989) | ||
| Decaloba | Hahmopathanthus | P. guatemalensis | EPI, GYN, DGY | This study, Jaroszewski et al. (2002) | |
| Disemma | P. herbertiniana | EPI, TEA | Jaroszewski et al. (2002) | ||
| Multiflora | P. holosericea | EPI, TEA | This study | ||
| Auriculata | P. auriculata | EPI, GYN | This study | ||
| P. jatunsachenis | EPI | This study | |||
| Cieca | P. coriacea | EPI, TEB | This study | ||
| P. suberosa | EPI, GYN | This study | |||
| Bryonioides | P. morifolia | LIN, LOT | Olafsdottir et al. (1989) | ||
| P. pendens | LIN, LOT | Spencer et al. (1986) | |||
| P. adenopoda | LIN, LOT | Spencer et al. (1986) | |||
| Decaloba | Xerogama | P. capsularis | EPI, PCP | Olafsdottir et al. (1989) | |
| P. citrina | PSC | Jaroszewski et al. (2002) | |||
| Decaloba | Decaloba | P. lutea | PBF, LIN, LOT | Spencer and Seigler (1985) | |
| P. indecora | PBF | Jaroszewski et al. (2002) | |||
| P. kalbreyeri | PBF | Jaroszewski et al. (2002) | |||
| P. apetala | PBF | Jaroszewski et al. (2002) | |||
| P. trifasciata | PTF | Olafsdottir, Jaroszewski, & Seigler, (1991) | |||
| P. biflora | PBF, PCP | This work, Olafsdottir et al. (1989) | |||
| P. colivauxii | PBF, PCP | This work | |||
| P. cuneata | PBF | Jaroszewski et al. (2002) | |||
| P. talamascensis | PBF | Spencer and Seigler (1985) | |||
| P. murucuja | PBF | Jaroszewski et al. (2002) | |||
| P. perfoliata | PBF | Jaroszewski et al. (2002) |
Aliphatic CNglc: LIN = linamarin; LOT = lotaustralin; LNT = linustatin; Aromatic CNglcs: AMY = amygdalin; PRU = prunasin; CNglcs bisglycosylated with unusual sugars: PBF = passibiflorin, PCP = passicapsin, PTF = passitrifasciatin; nd = CNglcs not detected; Sulphated: PCO(S) = passicoccin; TEB(S) = tetraphyllin B sulfate.
For chemical structure of these compounds see Figure 1.
Linamarin, lotaustralin, and linustatin were found in P. amoena, and epivolkenin reported in P. pittieri, both from the Astrophea subgenus. In addition, tetraphyllin B and tetraphyllin A have been reported in P. discophora, which belong to the Deidaminoides subgenus (Jaroszewski et al., 2002).
Cyclopentenyl CNglcs were found in the Passiflora subgenus, but in most species, these compounds are sulfated. For example, P. caerulea and P. racemosa have tetraphyllin B sulfate (Jaroszewski & Fog, 1989), and passicoccin, a diglycoside and sulfated version of epivolkenin, was discovered in P. coccinea (Spencer & Seigler, 1985). Aliphatic CNglcs seem to be very rare in this subgenus, being reported only in two species. In addition, the aromatic CNglcs prunasin and amygdalin were found in P. platyloba, P. maliformis, and P. edulis. Although all Passiflora are thought to be cyanogenic, cyanogenic glucosides were not detected in our analyses in three species of the Passiflora subgenus, specifically P. mathewsii, P. laurifolia, and P. riparia (Table 1).
Within the Decaloba subgenus, cyclopentenyl CNglcs were present in all species examined, except in the Bryonioides section reported to have exclusively aliphatic CNglcs. However, in the Decaloba section, the most diverse section of the Decaloba subgenus, the cyclopentenyl CNglcs were bisglycosylated (sugar added in two different positions of the aglycone) with unusual sugars, and they seem to be derived from epivolkenin. Passibiflorin is present in several species, including P. biflora, and it is glycosylated with an antiarosyl sugar residue. Passicapsin, found in P. capsularis and P. citrina, has a boivinosyl residue, while passitrifasciatin, which is reported only in P. trifasciata, has an allosyl. Simple cyclopentenyl CNglcs (monoglycosides), such as epivolkenin, are present mainly in the basal species of the Decaloba subgenus and also in the sister subgenus Tetrapathea in P. tetrandra.
4. DISCUSSION
4.1. Cyanogenesis in the Heliconiinae: The chicken and egg paradox
Our results show that sequestration of cyclopentenyl CNglcs is not an exclusive ability of a few Heliconius species as previously hypothesized. It is, in fact, widespread, not only among Heliconius butterflies, but also across related genera, such as Dryas, Philaethria, and Agraulis. Cyclopentenyl CNglcs were found even in species outside the Heliconiini tribe such as Euptoieta hegesia (tribe Argynnini) and Cethosia cyane. Accordingly, sequestration of cyclopentenyl CNglcs probably arose in a common ancestor of the Heliconiinae subfamily or even earlier. In fact, most species of the Heliconiinae subfamily use plants from families where cyclopentenyl CNglcs have been reported as larval host plants. The only exceptions are the American genera of the tribe Acraeini, Actinote, and Altinote, hypothesized to have shifted preference to Asteraceae plants to avoid competition with heliconiines in America (Brown & Francini, 1990).
Since most butterflies of the Heliconiinae subfamily can biosynthesize aliphatic CNglcs and also sequester cyclopentenyl CNglcs from their larval host, the obvious question is—Which came first in the evolutionary process, biosynthesis or sequestration?
Whereas sequestration of cyclopentenyl CNglcs has been reported only in butterflies of the Heliconiinae subfamily, biosynthesis of linamarin and lotaustralin has been demonstrated in several species of butterflies and moths belonging to many taxonomically distinct families, such as Zygaenidae, Limacodidae, Heterogynidae, Nymphalidae, and Lycaenidae (Davis & Nahrstedt, 1982; Nahrstedt, 1988; Zagrobelny, Castro et al., 2018). The biosynthetic pathway in the burnet moth Z. filipendulae is encoded by the genes ZfCYP405A2, ZfCYP332A3, and ZfUGT33A1 (Jensen et al., 2011) and putative homologues of these P450s have been found in the genome of the postman butterfly H. melpomene, HmCYP405A4, HmCYP405A5, HmCYP405A6, and HmCYP332A1 (Chauhan et al., 2013) and in other Heliconius species (Zagrobelny, Jensen, Vogel, Feyereisen, & Bak, 2018). Therefore, it is likely that the ability to biosynthesize aliphatic CNglcs appeared during the early radiations of the Lepidoptera in a common ancestor of butterflies and moths over 150 MYA and was later lost in many lepidopteran species. Assuming that the biosynthesis of aliphatic CNglcs arose in the early radiation of Lepidoptera and sequestration of cyclopentenyl CNglcs in an ancestor of the Heliconiinae subfamily, biosynthesis would be the ancestral trait.
Nevertheless, to confirm this hypothesis it is necessary to identify which adaptations are involved in the sequestration of cyclopentenyl CNglcs by heliconiines and the sequestration of different CNglcs present in other insects. Yu, Fang, Zhang, and Jiggins (2016) demonstrated that larvae of H. melpomene expressed different sets of transporters when reared on Passiflora species with different CNglc profiles, and characterization of these transporters would improve our understanding of the sequestration of cyclopentenyl CNglcs.
A further consideration is the ability to detoxify cyanide to avoid intoxication, which enables both sequestration and biosynthesis of CNglcs in Lepidopterans and most likely predates these two processes. Butterflies, moths, and mites detoxify cyanide using β‐cyanoalanine synthase (CAS) which converts cysteine and HCN into β‐cyanoalanine and H2S (Zagrobelny, Jensen et al., 2018). The CAS gene is shown to have been horizontally transferred from bacteria to a common ancestor of Lepidoptera, allowing them to colonize cyanogenic plants (Wybouw et al., 2014). Three putative CAS genes have been found in the H. melpomene genome (Wybouw et al., 2014) emphasizing the importance of cyanide metabolism for these butterflies. It is still not known if the CAS genes are also present in other heliconiines and Heliconius species.
4.2. The balance between biosynthesis and sequestration of CNglcs
Our results suggest that there is a potential trade‐off between biosynthesis of aliphatic CNglc and sequestration of cyclopentenyl CNglcs in heliconiines (Figure 4). This trade‐off is probably due to the fact that biosynthesizing CNglcs is costlier than sequestering them from the larval host plants. This would explain why some highly specialized species of the sara‐sapho group seem to have lost CNglc biosynthesis in favor of epivolkenin sequestration (Figure 2).
This balance between biosynthesis and sequestration of cyanogenic glucosides has also been found in larvae of Zygaena filipendulae, which produce linamarin and lotaustralin, as well as sequester them from their host Lotus corniculatus. Although larvae of Z. filipendulae had similar concentrations of linamarin and lotaustralin when reared on high‐cyanogenic, low‐cyanogenic, and acyanogenic L. corniculatus, their growth and body mass at pupation was greatly reduced when these compounds were low or absent in their diet (Zagrobelny, Bak, Ekstrøm, Olsen, & Møller, 2007). This indicates that Z. filipendulae larvae increase CNglc biosynthesis when these compounds are not available in their host to be sequestered, and that biosynthesis is costly for their development. Similarly, the wings of Heliconius erato vary in size according to their larval host plant (Jorge, Cordeiro‐Estrela, Klaczko, Moreira, & Freitas, 2011) which could be a consequence of the energy expended on CNglc production due to the amount of these compounds available in their diet.
Moreover, Fürstenberg‐Hägg et al. (2014) demonstrated that Z. filipendulae larvae upregulate gene expression of its CNglc biosynthetic pathway (CYP405A2, CYP332A3, and UGT33A1) when these compounds are not available in their host plant. CYP405A2, the first enzyme of the pathway, is controlled at both transcriptional and enzyme steady state level by the concentration of linamarin and lotaustralin in the diet of Z. filipendulae and regulates the intensity of biosynthesis. Contrary to Z filipendulae, heliconiines sequester different kinds of CNglcs from their larval host than they can biosynthesize, so further investigations are needed to understand how the interplay of these two processes is regulated in these butterflies.
4.3. Evolution of cyanogenesis in heliconiines is associated with their larval host specialization
Across all species analyzed, H. sapho had the highest total concentration of CNglcs. This is in line with a previous report by Engler‐Chaouat and Gilbert (2007), who also found that H. sapho, and closely related species, were the most cyanogenic species within the Heliconius genus. Similar to H. sapho, all species of the sara‐sapho group feeding on Astrophea plants contain only epivolkenin, and it is possible that they may have lost the ability to synthesize aliphatic CNglcs to become more specialized in sequestration as suggested by Engler‐Chaouat and Gilbert (2007). Yet, apparently functional P450 genes associated with the biosynthesis of linamarin and lotaustralin are present in the genome of H. sapho (Zagrobelny, Castro et al., 2018) suggesting that the lack of biosynthesis could be due to transcriptional and/or translational mechanisms. On the other hand, large amounts of linamarin and lotaustralin were found in H. sara and H. charithonia which, although belonging to the same group as H. sapho, are not Astrophea specialists but Decaloba specialists. Thus, specialization for sequestration of cyclopentenyl CNglcs and feeding on woody Astrophea species seem to be related and could have happened in the recent radiation of the sara‐sapho group.
The fact that cyclopentenyl CNglcs were not found in H. atthis, H. doris, E. isabella, D. phatethusa, D. juno, and P. wernickei does not necessarily mean that these species lack the ability to sequester these compounds from Passiflora. They could simply have been feeding on Passiflora species devoid of cyclopentenyl CNglcs or on tissues where these compounds are not abundant to allow sequestration. Indeed, two of these species that lacked cyclopentenyl CNglcs, H. doris and H. atthis, are reported to use as larval host P. laurifolia, P. riparia, and P. subpeltata which do not produce these compounds (Benson et al., 1975) (Figure 6, Table 1). Since the butterflies used in this study were collected in the field or raised from pupae provided by commercial butterfly farms, their larval host plant is unfortunately unknown.
Figure 6.
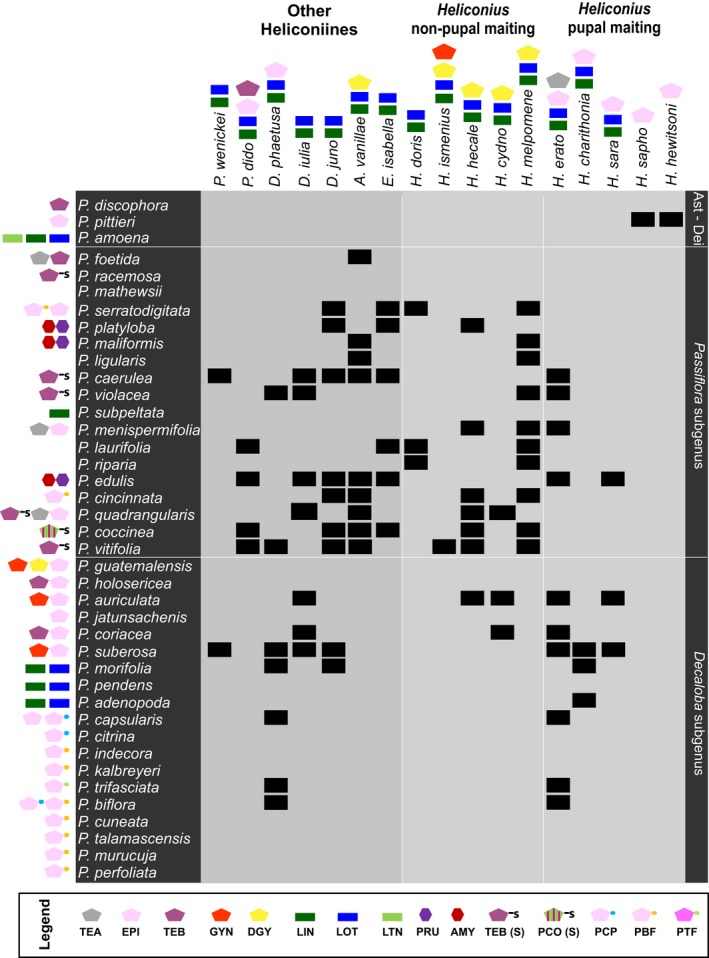
Hit‐map overlaying the host‐plant utilization by different heliconiines species described by Benson et al. (1975) and the chemical composition of the Passiflora species revised in this study
All Heliconius species which are Decaloba specialists sequestered epivolkenin. Actually, epivolkenin was found to be very common in the Decaloba subgenus, present in all the species analyzed, a part of the sections Bryonioides and Decaloba. Interestingly, there was no epivolkenin in the nonpupal mating Heliconius species, which generally contained dihydrogynocardin. Dihydrogynocardin was not common in Passiflora in our analyses, only present in P. guatemalensis which is not reported as a host of heliconiines. However, since the Passiflora genus has over 500 species and only 40 have had their chemical profile analyzed here, these butterflies could have fed on a species that produces dihydrogynocardin which was not present in our study. Another explanation is that nonpupal mating Heliconius convert epivolkenin into dihydrogynocardin, through a reduction and a hydroxylation reactions. Indeed, it has been shown that after host shifting, H. melpomene larvae change the expression of several P450s (Yu et al., 2016), and one of them could be involved in catalyzing this conversion. Further analyses are necessary to confirm the origin of dihydrogynocardin in these butterflies.
Heliconius butterflies are thought to be more toxic than basal heliconiines because they are generally more distasteful to avian predators (Chai, 1990). Cardoso and Gilbert (2013) observed that freshly emerged butterflies of H. charithonia have a higher cyanide emission after tissue disruption than A. vanillae and D. iulia. In our analyses, Heliconius species tended to have higher CNglc concentrations than other heliconiines; however, the differences were not statistically significant. Also, sequestration of cyclopentenyl CNglcs seems to be more common in Heliconius than in other genera. Spencer (1988) suggested that the aglycones resulting from the degradation of cyclopentenyl CNglcs were more toxic than the aglycones derived from aliphatic CNglcs, and this could contribute to the higher unpalatability of Heliconius.
Resource partitioning could be the reason why sequestration of CNglcs is less common in the basal heliconiines. Most of them feed on mature leaves of Passiflora plants (Benson et al., 1975), which as many cyanogenic plants—for example, Sorghum bicolor (Busk & Lindberg Møller, 2002) and Lotus japonicus (Forslund et al. 2004)—probably have lower concentrations of defense compounds and are nutritionally poorer than the new leaves and meristems preferred by Heliconius (Gilbert, 1991). Avoidance of competition with Heliconius larvae seems to drive larvae of D. iulia to leaves of lower quality (Millan, Borges, Rodrigues, & Moreira, 2013).
4.4. Structural diversification of CNglcs in Passiflora—a tool to evade heliconiine herbivory?
From the ~60 structures of CNglcs reported in plants, 27 are found in Passifloraceae and many of them are exclusive of the Passiflora genus. This led to the questions: What forces drove this structural diversification? Could it be associated with their coevolution with heliconiines? All CNglcs (α‐hydroxynitrile glucosides) are able to release HCN upon degradation—if cyanogenesis is the sole bioactivity of these compounds, then what is the purpose of biosynthesizing CNglcs with many different structures?
Most plants used as larval hosts by heliconiines belong to the Decaloba and Passiflora subgenera, and curiously, 80% of the CNglc structures reported in the Passiflora genus are in species of these two subgenera. The addition of a sulfate moiety to cyclopentenyl CNglcs and the biosynthesis of aromatic CNglcs are examples of structural diversifications of CNglc in the Passiflora subgenus. Additionally, some species of the Passiflora subgenus have become acyanogenic, presumably as a way to avoid sequestering specialists. Schappert and Shore (1999) found that many populations of Turnera ulmifolia are acyanogenic, most likely because the presence of CNglcs does not deter oviposition and herbivory by E. hegesia, its principal herbivore. Indeed, CNglc biosynthesis has high energetic costs to T. ulmifolia, impacting flower production and consequently reproduction, which in the absence of herbivores, will result in selection against cyanogenesis (Schappert & Shore, 2000). This could also be the case for other species in the Passiflora subgenus.
The diversification of CNglc structures in the Decaloba subgenus followed a different path than in the Passiflora subgenus. Aliphatic and simple cyclopentenyl CNglcs occur in the basal sections of the subgenus, whereas in the most advanced, the cyclopentenyl CNglcs are bisglycosylated with unusual sugars. Aliphatic CNglcs are present mainly in the Bryonioides section, which are also called the hooked trichome group and are only used as host by few heliconiine species.
Aliphatic and simple cyclopentenyl CNglcs were found in the basal subgenus Astrophea and Deidamioides, which seems to be the ancestral cyanogenic traits of the Passiflora genus. Even though Astrophea is the most basal subgenus of Passiflora, molecular clock analyses suggest that it was the last subgenus of Passiflora to diversify (Muschner, Zamberlan, Bonatto, & Freitas, 2012). Remarkably, specialization for feeding on Astrophea plants also happened in the advanced radiation of Heliconius, in the sara‐sapho group (Kozak et al., 2015). Perhaps, the modified CNglc composition of the Decaloba and Passiflora subgenus could have forced these Heliconius to change preferences to Astrophea species that produce simple cyclopentenyl CNglcs. It is not yet known if the modified cyclopentenyl CNglcs (sulfated and bisglycosylated) and aromatic CNglcs can be sequestered by heliconiinae larvae. Indeed, none of these compounds were found in adults of the heliconiine species analyzed.
We constructed a hit‐map with the larval host preferences of the heliconiines butterflies used in this study (Benson et al., 1975), overlaying it with the CNglc composition of the Passiflora plants (Figure 6) to identify if heliconiines have preference or avoidance for specific cyanogenic glucoside structures. Most Decaloba plants that have only bisglycosylated cyclopentenyl CNglcs seem to have escaped heliconiine herbivory, although P. biflora and P. trisfasciata which have these compounds exclusively are common hosts for many of these butterflies. Passiflora species with aromatic and sulfated cyclopentenyl CNglcs are also popular hosts for nonpupal mating Heliconius and basal heliconiines.
Despite that heliconiines are adapted to host plants with high amounts of CNglcs, modification of the structure of their aglycones and addition of unusual sugars might not deter the feeding of these butterflies. However, it could obstruct the sequestration of these compounds and thus might discourage some heliconiines to evolve preferences for the plants with modified compounds. Unfortunately, there is no obvious overlap between the host preferences of heliconiines and the CNglc profiles of Passiflora species. Nevertheless, there is a pattern in the distribution of modified CNglcs within the genus Passiflora which could have evolved in the plants to evade herbivory.
In conclusion, we demonstrated that the sequestration of cyclopentenyl CNglcs probably arose in a common ancestor of the Heliconiinae subfamily, whereas the biosynthesis of aliphatic CNglcs seems to have appeared before the divergence between butterflies and moths. In heliconiines, the profile and amount of CNglcs is related to their feeding strategy on Passiflora plants. In the future, it will be important to establish which CNglcs can be sequestered by heliconiine larvae in order to understand the role of cyanogenesis in the arms race between these butterflies and their Passiflora hosts. In addition, since many heliconiines vary their host preference according to their locality, and plants vary their metabolite profile in different environments, population studies regarding the chemical ecology of heliconiines and their Passiflora hosts might reveal different patterns which we could not observe in this study.
AUTHOR CONTRIBUTIONS
The research project that resulted in this manuscript was conceptualized by EC, MZ, MC, RF, and SB. EC conducted the sample collection assisted by MC and the chemical analyses under supervision of MZ. JZ developed the phylogenetic and statistical analyses. All authors contributed to the discussion of results, hypothesis development, and writing process of this manuscript.
Supporting information
ACKNOWLEDGMENTS
EC, MZ, and SB thank the Independent Research Fund Denmark | Natural Sciences for the financial support (DFF – 1323‐00088). MC and EC acknowledge the Brazilian National Council for Scientific Development (CNPq) for the funding provided (proc. 306985/2013‐6). EC and JZ acknowledge, respectively, Science Without Borders and CAPES for their PhD scholarships. All authors thank the Botanical Garden of Copenhagen University, in especial the gardener Sue Dix for providing the plant samples utilized in this study. We also thank EMPARN (Jiqui Woods) and Miriri Food and Bioenergy S/A for allowing us to collect butterflies in their forest reserve. We acknowledge Dr. Mohammed Saddik Motawie, Dr. Jerzy W Jaroszewski (in memoriam), and Dr. Kevin Spencer for synthesizing most standards utilized in this study and Professor Larry E Gilbert and Dr. Helene Engler‐Chaouat for donating to us some of these standards. We are also very grateful for all the contributions made by the Heliconius Research Community to this project and for the suggestions made by the three anonymous reviewers of this manuscript.
Pinheiro de Castro ÉC, Zagrobelny M, Zurano JP, Zikan Cardoso M, Feyereisen R, Bak S. Sequestration and biosynthesis of cyanogenic glucosides in passion vine butterflies and consequences for the diversification of their host plants. Ecol Evol. 2019;9:5079–5093. 10.1002/ece3.5062
DATA ACCESSIBILITY
The raw chemical data are archived at the Department of Plant and Environmental Science of Copenhagen University and is also publically available on Dryad (https://doi.org/10.5061/dryad.2r23j1q).
REFERENCES
- Adams, D. C. (2014). A generalized K statistic for estimating phylogenetic signal from shape and other high‐dimensional multivariate data. Systematic Biology, 63, 685–697. 10.1093/sysbio/syu030 [DOI] [PubMed] [Google Scholar]
- Arias, M. , Meichanetzoglou, A. , Elias, M. , Rosser, N. , de‐Silva, D. L., Nay, B., & Llaurens, V. (2016). Variation in cyanogenic compounds concentration within a Heliconius butterfly community: Does mimicry explain everything? BMC Evolutionary Biology, 16, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, W. W. , Brown, K. S. J. , & Gilbert, L. E. (1975). Coevolution of plants and herbivores: Passion flower butterflies. Evolution, 29, 659–680. 10.1111/j.1558-5646.1975.tb00861.x [DOI] [PubMed] [Google Scholar]
- Bjarnholt, N. , Nakonieczny, M. , Kedziorski, A. , Debinski, D. M. , Matter, S. F. , Olsen, C. E. , & Zagrobelny, M. (2012). Occurrence of sarmentosin and other hydroxynitrile glucosides in Parnassius (Papilionidae) butterflies and their food plants. Journal of Chemical Ecology, 38, 525–537. 10.1007/s10886-012-0114-x [DOI] [PubMed] [Google Scholar]
- Bjarnholt, N. , Rook, F. , Motawia, M. S. , Cornett, C. , Jørgensen, C. , Olsen, C. E. , … Møller, B. L. (2008). Diversification of an ancient theme: Hydroxynitrile glucosides. Phytochemistry, 69, 1507–1516. 10.1016/j.phytochem.2008.01.022 [DOI] [PubMed] [Google Scholar]
- Blomberg, S. P. , Garland, T. Jr , & Ives, A. R. (2003). Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution, 57, 717–745. [DOI] [PubMed] [Google Scholar]
- Brown, K. S. , & Francini, R. B. (1990). Evolutionary strategies of chemical defense in aposematic butterflies: Cyanogenesis in Asteraceae‐feeding American Acraeinae. Chemoecology, 1, 52–56. 10.1007/BF01325228 [DOI] [Google Scholar]
- Busk, P. K. , & Lindberg Møller, B. (2002). Dhurrin Synthesis in Sorghum Is Regulated at the Transcriptional Level and Induced by Nitrogen Fertilization in Older Plants. Plant physiology, 10.1104/pp.000687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso, M. Z. , & De Lima, L. L. F. (2017). Dissecting the nature of subtle phenotypic variation in wing colour elements of Müllerian co‐mimics. Journal of Tropical Ecology, 33, 188–196. 10.1017/S0266467417000074 [DOI] [Google Scholar]
- Cardoso, M. Z. , & Gilbert, L. E. (2013). Pollen feeding, resource allocation and the evolution of chemical defence in passion vine butterflies. Journal of Evolutionary Biology, 26, 1254–1260. 10.1111/jeb.12119 [DOI] [PubMed] [Google Scholar]
- Chai, P . (1990). Relationships between visual characteristics of rainforest butterflies and responses of a specialised insectivorous bird In Wicksten M. (Ed.), Adaptive coloration in invertebrates. Proc. Symp. Spon. Am. SOC. Zool. (pp. 31–60). Galveston, Texas: Texas A&M University. [Google Scholar]
- Chauhan, R. , Jones, R. , Wilkinson, P. , Pauchet, Y. , & ffrench‐Constant, R. H. (2013). Cytochrome P450‐encoding genes from the Heliconius genome as candidates for cyanogenesis. Insect Molecular Biology, 22, 532–540. [DOI] [PubMed] [Google Scholar]
- Davis, R. H. , & Nahrstedt, A. (1982). Occurrence and variation of the cyanogenic glucosides linamarin and lotaustralin in species of the Zygaenidae (Insecta, Lepidoptera). Comparative Biochemistry and Physiology, 71, 329–332.6121656 [Google Scholar]
- Davis, R. H. , & Nahrstedt, A. (1987). Biosynthesis of cyanogenic glucosides. Insect Biochem., 17, 689–693. [Google Scholar]
- de Castro, É. C. P. , Zagrobelny, M. , Cardoso, M. Z. , & Bak, S. (2018). The arms race between heliconiine butterflies and Passiflora plants ‐ new insights on an ancient subject. Biological Reviews, 93, 555–573. [DOI] [PubMed] [Google Scholar]
- Engler, H. S. , Spencer, K. C. , & Gilbert, L. E. (2000). Preventing cyanide release from leaves. Nature, 406, 144–145. 10.1038/35018159 [DOI] [PubMed] [Google Scholar]
- Engler‐Chaouat, H. S. , & Gilbert, L. E. (2007). De novo synthesis vs. sequestration: Negatively correlated metabolic traits and the evolution of host plant specialization in cyanogenic butterflies. Journal of Chemical Ecology, 33, 25–42. [DOI] [PubMed] [Google Scholar]
- Forslund, K. , Morant, M. , Jørgensen, B. , Olsen, C. E. , Asamizu, E. , Sato, S. , … Bak, S. (2004). Biosynthesis of the Nitrile Glucosides Rhodiocyanoside A and D and the Cyanogenic Glucosides Lotaustralin and Linamarin in Lotus japonicus. Plant physiology, 10.1104/pp.103.038059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürstenberg‐Hägg, J. , Zagrobelny, M. , & Bak, S. (2013). Plant defense against insect herbivores. International Journal of Molecular Sciences, 14, 10242–10297. 10.3390/ijms140510242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürstenberg‐Hägg, J. , Zagrobelny, M. , Erik, C. , Jørgensen, K. , Lindberg, B. , & Bak, S. (2014). Transcriptional regulation of de novo biosynthesis of cyanogenic glucosides throughout the life‐cycle of the burnet moth Zygaena filipendulae (Lepidoptera). Insect Biochemistry and Molecular Biology, 49, 80–89. 10.1016/j.ibmb.2014.04.001 [DOI] [PubMed] [Google Scholar]
- Gilbert, L. E. (1991). Biodiversity of a Central American Heliconius community: Pattern, process, and problems In Price P., Lewinsohn T., Fernandes T., & Benson W. (Eds.), Plant‐animal interactions: Evolutionary ecology in tropical and temperate regions (pp. 403–427). New York, NY: John Wiley & Sons. [Google Scholar]
- Gleadow, R. M. , & Møller, B. L. (2014). Cyanogenic glucosides: Synthesis, physiology, and phenotypic plasticity. Annual Review of Plant Biology, 65, 155–185. [DOI] [PubMed] [Google Scholar]
- Hay‐Roe, M. M. , & Nation, J. (2007). Spectrum of cyanide toxicity and allocation in Heliconius erato and Passiflora host plants. Journal of Chemical Ecology, 33, 319–329. 10.1007/s10886-006-9234-5 [DOI] [PubMed] [Google Scholar]
- Jaroszewski, J. W. , & Fog, E. (1989). Sulphate esters of cyclopentenoid cyanohydrin glucosides. Phytochemistry, 28, 1527–1528. [Google Scholar]
- Jaroszewski, J. W. , Olafsdottir, E. S. , Wellendorph, P. , Christensen, J. , Franzyk, H. , Somanadhan, B. , & B. a Budnik, L. B. Jørgensen, and V. Clausen., (2002). Cyanohydrin glucosides of Passiflora: Distribution pattern, a saturated cyclopentane derivative from P. guatemalensis, and formation of pseudocyanogenic alpha‐hydroxyamides as isolation artefacts. Phytochemistry, 59, 501–511. [DOI] [PubMed] [Google Scholar]
- Jensen, N. B. , Zagrobelny, M. , Hjernø, K. , Olsen, C. E. , Houghton‐Larsen, J. , Borch, J. , … Bak, S. (2011). Convergent evolution in biosynthesis of cyanogenic defence compounds in plants and insects. Nature Communications, 2, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins, C. D. (2017). The ecology and evolution of Heliconius butterflies: A passion for diversity. Oxford, UK: Oxford University Press. [Google Scholar]
- Jorge, L. R. , Cordeiro‐Estrela, P. , Klaczko, L. B. , Moreira, G. R. P. , & Freitas, A. V. L. (2011). Host‐plant dependent wing phenotypic variation in the neotropical butterfly Heliconius erato . Biological Journal of the Linnean Society, 102, 765–774. 10.1111/j.1095-8312.2010.01610.x [DOI] [Google Scholar]
- Kozak, K. M. , Wahlberg, N. , Neild, A. F. E. , Dasmahapatra, K. K. , Mallet, J. , & Jiggins, C. D. (2015). Multilocus species trees show the recent adaptive radiation of the mimetic Heliconius butterflies. Systematic Biology, 64, 505–524. 10.1093/sysbio/syv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, D. , Pičmanová, M. , Abou Hachem, M. , Motawia, M. S. , Olsen, C. E. , Møller, B. L. , … Takos, A. M. (2015). Lotus japonicus flowers are defended by a cyanogenic β-glucosidase with highly restricted expression to essential reproductive organs. Plant Molecular Biology, 10.1007/s11103-015-0348-4. [DOI] [PubMed] [Google Scholar]
- Millan, C. , Borges, S. S. , Rodrigues, D. , & Moreira, G. R. P. (2013). Behavioral and life‐history evidence for interspecific competition in the larvae of two heliconian butterflies. Naturwissenschaften, 100, 901–911. 10.1007/s00114-013-1089-3 [DOI] [PubMed] [Google Scholar]
- Møller, B. L. , Olsen, C. E. , & Motawia, M. S. (2016). General and stereocontrolled approach to the chemical synthesis of naturally occurring cyanogenic glucosides. Journal of Natural Products, 79, 1198–1202. 10.1021/acs.jnatprod.5b01121 [DOI] [PubMed] [Google Scholar]
- Muschner, V. C. , Zamberlan, P. M. , Bonatto, S. L. , & Freitas, L. B. (2012). Phylogeny, biogeography and divergence times in Passiflora (Passifloraceae). Genet. Mol. Biol., 35, 1036–1043. 10.1590/S1415-47572012000600019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahrstedt, A. , & Davis, R. H. (1981). Cyanogenic glucosides in butterflies: Detection and synthesis of linamarin and lotaustralin in the Heliconiinae. Planta Medicine, 42, 124–125. Thieme. [DOI] [PubMed] [Google Scholar]
- Nahrstedt, A. (1988). Cyanogenesis and the role of cyanogenic compounds in insects. Ciba Foundation Symposium, 140, 131–150. [DOI] [PubMed] [Google Scholar]
- Nishida, R. (2014). Chemical ecology of insect–plant interactions: Ecological significance of plant secondary metabolites. Bioscience, Biotechnology, and Biochemistry, 78, 5079–13. 10.1080/09168451.2014.877836 [DOI] [PubMed] [Google Scholar]
- Olafsdottir, E. S. , Cornett, C. , Jaroszewski, J. W. , Songstad, J. , Lönnberg, H. , Colacio, E. , … Tekenbergs-Hjelte, L. (1989). Cyclopentenoid Cyanohydrin Glycosides with Unusual Sugar Residues. Acta Chemica Scandinavica, 43, 51–55. [DOI] [PubMed] [Google Scholar]
- Olafsdottir, E. S. , Jaroszewski, J. W. , & Seigler, D. S. (1991). Cyanohydrin glycosides with unusual sugar residues: Revised structure of passitrifasciatin. Phytochemistry, 30, 867–869. [DOI] [PubMed] [Google Scholar]
- Pentzold, S. , Jensen, M. K. , Matthes, A. , Olsen, C. E. , Petersen, B. L. , Clausen, H. , … Zagrobelny, M. (2017). Spatial separation of the cyanogenic β‐glucosidase ZfBGD2 and cyanogenic glucosides in the haemolymph of Zygaena larvae facilitates cyanide release. Royal Society Open Science, 4, 170262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell, L. J. (2012). phytools: An R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution, 3, 217–223. 10.1111/j.2041-210X.2011.00169.x [DOI] [Google Scholar]
- Raubenheimer, D. (1989). Cyanoglycoside gynocardin from Acraea horta (L.) (Lepidoptera: Acraeinae) - Possible implications for evolution of acraeine host choice. Journal of Chemical Ecology, 15, 2177–2189. [DOI] [PubMed] [Google Scholar]
- R Core Team (2017). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Schappert, P. J. , & Shore, J. S. (1999). Effects of cyanogenesis polymorphism in Turnera ulmifolia on Euptoieta hegesia and potential Anolis predators. Journal of Chemical Ecology, 25, 1455–1479. [Google Scholar]
- Schappert, P. J. , & Shore, J. S. (2000). Cyanogenesis in Turnera ulmifolia L. (Turneraceae): II. Developmental expression, heritability and cost of cyanogenesis. Evolutionary Ecology Research, 2, 337–352. [Google Scholar]
- Sidlauskas, B. (2008). Continuous and arrested morphological diversification in sister clades of characiform fishes: A phylomorphospace approach. Evolution, 62, 3135–3156. 10.1111/j.1558-5646.2008.00519.x [DOI] [PubMed] [Google Scholar]
- Silva-Brandão, K. L. , Wahlberg, N. , Francini, R. B. , Azeredo-Espin, A. M. L. , Brown, K. S. , Paluch, M. , … Freitas, A. V. L. (2008). Phylogenetic relationships of butterflies of the tribe Acraeini (Lepidoptera, Nymphalidae, Heliconiinae) and the evolution of host plant use. Molecular Phylogenetics and Evolution, 46, 515–531. [DOI] [PubMed] [Google Scholar]
- Spencer, K. C. (1988). Chemical mediation of coevolution in the Passiflora‐Heliconius interaction In Spencer K. C. (Ed.), Chemical Mediation of Coevolution (pp. 167–240). San Diego, CA: Academic Pres. [Google Scholar]
- Spencer, K. C. , & Seigler, D. S. (1984). Gynocardin from Passiflora. Planta Medica, 50, 356–357. [DOI] [PubMed] [Google Scholar]
- Spencer, K. , & Seigler, D. S. (1985). Passicoccin: A sulphated cyanogenic glucoside from Passiflora coccinea . Phytochemistry, 24, 2615–2617. [Google Scholar]
- Takos, A. M. , Knudsen, C. , Lai, D. , Kannangara, R. , Mikkelsen, L. , Motawia, M. S. , … Rook, F. (2011). Genomic clustering of cyanogenic glucoside biosynthetic genes aids their identification in Lotus japonicus and suggests the repeated evolution of this chemical defence pathway. The Plant Journal, 68, 273–286. [DOI] [PubMed] [Google Scholar]
- Tober, I. , & Conn, E. E. (1985). Cyclopentenylglycine, a precursor of deidaclin in Turnera ulmifolia . Phytochemistry, 24, 1215–1218. 10.1016/S0031-9422(00)81104-8 [DOI] [Google Scholar]
- Wybouw, N. , Dermauw, W. , Tirry, L. , Stevens, C. , Grbić, M. , Feyereisen, R. , & Van Leeuwen, T. (2014). A gene horizontally transferred from bacteria protects arthropods from host plant cyanide poisoning. Elife, 3, e02365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Q. Y. , Fang, S. M. , Zhang, Z. , & Jiggins, C. D. (2016). The transcriptome response of Heliconius melpomene larvae to a novel host plant. Molecular Ecology, 25, 4850–4865. [DOI] [PubMed] [Google Scholar]
- Zagrobelny, M. , Bak, S. , Ekstrøm, C. T. , Olsen, C. E. , & Møller, B. L. (2007). The cyanogenic glucoside composition of Zygaena filipendulae (Lepidoptera: Zygaenidae) as effected by feeding on wild‐type and transgenic lotus populations with variable cyanogenic glucoside profiles. Insect Biochemistry and Molecular Biology, 37, 10–18. 10.1016/j.ibmb.2006.09.008 [DOI] [PubMed] [Google Scholar]
- Zagrobelny, M. , Bak, S. , Olsen, C. E. , & Møller, B. L. (2007). Intimate roles for cyanogenic glucosides in the life cycle of Zygaena filipendulae (Lepidoptera, Zygaenidae). Insect Biochemistry Molecular Biology, 37, 1189–1197. [DOI] [PubMed] [Google Scholar]
- Zagrobelny, M. , Bak, S. , Rasmussen, A. V. , Jørgensen, B. , Naumann, C. M. , & Møller, B. L. (2004). Cyanogenic glucosides and plant‐insect interactions. Phytochemistry, 65, 293–306. 10.1016/j.phytochem.2003.10.016 [DOI] [PubMed] [Google Scholar]
- Zagrobelny, M. , de Castro, É. , Møller, B. , & Bak, S. (2018). Cyanogenesis in Arthropods: From chemical warfare to nuptial gifts. Insects, 9, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagrobelny, M. , Jensen, M. K. , Vogel, H. , Feyereisen, R. , & Bak, S. (2018). Evolution of the biosynthetic pathway for cyanogenic glucosides in lepidoptera. Journal of Molecular Evolution, 86, 379–394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw chemical data are archived at the Department of Plant and Environmental Science of Copenhagen University and is also publically available on Dryad (https://doi.org/10.5061/dryad.2r23j1q).


