Abstract
Tunneling nanotubes (TNTs) are membrane conduits that mediate long-distance intercellular cross-talk in several organisms and play vital roles during development, pathogenic transmission, and cancer metastasis. However, the molecular mechanisms of TNT formation and function remain poorly understood. The protein MSec (also known as TNFα-induced protein 2 (TNFAIP2) and B94) is essential for TNT formation in multiple cell types. Here, using affinity protein purification, mass spectrometric identification, and confocal immunofluorescence microscopy assays, we found that MSec interacts with the endoplasmic reticulum (ER) chaperone ERp29. siRNA-mediated ERp29 depletion in mammalian cells significantly reduces TNT formation, whereas its overexpression induces TNT formation, but in a strictly MSec-dependent manner. ERp29 stabilized MSec protein levels, but not its mRNA levels, and the chaperone activity of ERp29 was required for maintaining MSec protein stability. Subcellular ER fractionation and subsequent limited proteolytic treatment suggested that MSec is associated with the outer surface of the ER. The ERp29–MSec interaction appeared to require the presence of other bridging protein(s), perhaps triggered by post-translational modification of ERp29. Our study implicates MSec as a target of ERp29 and reveals an indispensable role for the ER in TNT formation, suggesting new modalities for regulating TNT numbers in cells and tissues.
Keywords: cell-cell interaction, endoplasmic reticulum (ER), chaperone, plasma membrane, protein stability, protein-disulfide isomerase, post-translational modification (PTM), chaperone, endoplasmic reticulum, intercellular communication, MSec, tunneling nanotubes, endoplasmic reticulum protein 29 (ERP29), TNFα-induced protein 2 (TNAIP2), interactome
Introduction
Eukaryotic cells communicate by several means, depending on organism and cell type. Intercellular communication plays an important role in the physiological processes of multicellular organisms. Diverse signaling pathways have been documented for the exchange of molecular information between cells, such as direct interaction (1, 2), gap junctions (3, 4), extracellular vesicles and exosomes (5, 6), as well as plasmodesmata (7). A novel mechanism for intercellular communication named tunneling nanotubes (TNTs)4 was recently discovered in several organisms and cell types (8, 9). TNTs constitute a diverse variety of long, membrane-enclosed, tubular cytoplasmic conduits that connect eukaryotic cells several hundred micrometers apart (8, 10, 11) and mediate a wide spectrum of critical cellular functions. These include ion signaling (12), antigen presentation (13), intercellular organelle transfer (14), morphogen transfer during development (15, 16), astrocyte–neuron communication (17), metastatic cell homing (18), intercellular viral transmission (19–21), prion protein transfer (22), and bacterial surfing between cells (10). TNTs are also induced in response to cellular stress (23, 24). TNTs are usually open at both ends with a propensity to “hang” between the connected cells above the substratum in culture and use distinct actin regulators for their formation (25–27), which are some of the properties that distinguish them from other similar cellular extensions like filopodia and cytonemes (in Drosophila), even though they are morphologically similar. TNTs thus represent a novel mode of intercellular communication across various in vitro and in vivo systems that are rapidly gaining widespread importance in biology (15, 16, 28–35).
Despite their demonstrated importance in health and disease as exemplified above, there is little mechanistic understanding of the formation, growth, maintenance, and function of TNTs. Several questions of importance remain unanswered. Which proteins regulate TNT formation? What are the reliable biochemical markers for TNTs? What is the order, strength, and nature of interactions between these proteins? What is the mechanism of membrane addition in elongating TNTs? Are these molecules and mechanisms conserved across evolution? Recently, the protein MSec (also called TNFAIP2/B94) was reported to be essential for TNT formation (36, 37). MSec interacts with a few key proteins that help mediate TNT formation, including the exocyst complex, the small GTPase RalA, and LST1 in macrophages (36, 38). The GTPase activity of RalA is required for TNT formation through the engagement of the exocyst complex, which is a known effector of RalA (36). RalA–exocyst is known to induce membrane addition in other contexts, such as epithelial membrane trafficking and anchorage-independent growth signaling (39, 40). It is therefore likely that other protein(s), such as MSec, may recruit the RalA–exocyst complex specifically at sites of TNT formation on the plasma membrane. Given the wide spectrum of functions performed by TNTs in a variety of cell lines and organisms (41), it is likely that a larger set of cellular proteins interacts with MSec to assist it in TNT formation and possibly to enable TNTs to function as transport conduits. In an effort to understand the molecular mechanisms of TNT formation, we determined the cellular interactome of MSec from a human osteosarcoma cell line (U2OS) by mass spectrometric analysis. Here, we characterize the function and mechanism of one of the novel interactors of MSec, the endoplasmic reticulum (ER)-based chaperone ERp29 and demonstrate that it is required for the induction of TNT formation. ERp29 is a 29-kDa ER-resident protein and is well-studied as an unconventional chaperone of the protein-disulfide isomerase (PDI) family that binds with, stabilizes, and escorts various protein substrates (42–45). In this study, we show that ERp29 regulates TNT formation through stabilization of MSec and thus reveal a new role for the ER in TNT induction. Our study suggests that the underlying mechanisms of TNT formation span a wider molecular spectrum than so far anticipated.
Results
Stably expressing TAP-tagged MSec induces TNT formation in U2OS cells
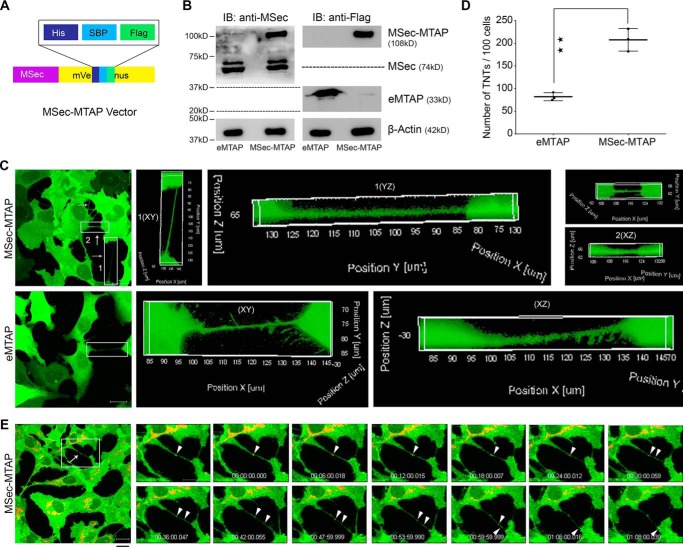
Earlier studies have demonstrated that MSec is required for TNT formation (41). Depletion of MSec from cells leads to a reduction in TNT numbers, whereas overexpression leads to increased TNT numbers (36). Given the importance of MSec-induced TNTs across a diverse functional spectrum, we hypothesized that MSec interacts with a larger set of cellular proteins to mediate TNT formation than is currently known. To identify new interaction partners of MSec, we generated a mammalian U2OS cell line (human osteosarcoma origin) stably expressing MSec fused with a multifunctional tag containing a C-terminal yellow fluorescent protein tag with an embedded tandem affinity purification tag consisting of octa-His, FLAG, and streptavidin-binding peptide (SBP) biochemical tags (mVenus-MTAP (46)) that has been used successfully for interactomic as well as imaging studies in human cells. Mouse MSec (residues 42–691; kind gift from H. Ohno (36)) complementary DNA (cDNA) was cloned into this vector to generate MSec-MTAP (Fig. 1A and Table S1). This fragment of mouse MSec can complement the loss of human MSec and can efficiently induce TNT formation (36). We determined the expression levels of both endogenous MSec and transgenic MSec-MTAP by Western blotting with an antibody that recognizes MSec and chose a clonally selected cell line (clone 1) that stably expressed the transgene at levels equivalent to endogenous MSec (Fig. 1B (left and right top panels) and Fig. S1). We also generated a U2OS cell line stably expressing the empty MTAP (eMTAP) vector as a negative control (Fig. 1B, right bottom panel). The tagged MSec-MTAP protein induced TNTs significantly, as reported earlier for MSec with other tags (36, 37) compared with the tag (empty vector) alone (Fig. 1, C and D), demonstrating that the transgenic MSec-MTAP construct was functional with respect to its ability to induce TNT formation.
Figure 1.
Stably expressed affinity-tagged MSec induces TNT formation in U2OS cells. A, MSec was cloned into a mammalian expression vector that imparts a multiaffinity tag (His-SBP-FLAG) embedded inside a fluorescent (mVenus) tag. B, expression levels of stably expressing empty vector (eMTAP) and exogenous mouse MSec (MSec-MTAP) equivalent to the endogenous human MSec assessed by immunoblotting (IB). C, live confocal microscope images taken from stably expressing MSec-MTAP and empty MTAP cells (both green). TNTs are indicated by arrows. The analyzed TNTs were not adhered to the substratum. Shown is an x-z/y-z visualization of confocal z-stacks of TNTs that were counted in both cell lines (MSec-MTAP and eMTAP). Scale bar, 20 μm. D, quantification of the number of TNTs per 100 cells from the confocal images. Data represent mean ± S.D. (error bars) based on three independent experiments, 100 cells counted per experiment (paired t test, two-tailed; **, p < 0.01). E, time lapse confocal images showing the transport of mitochondria from one cell to another. Arrowheads, position of the mitochondria.
We performed a series of standard assays to validate that the tubes under study were indeed TNTs. First, we established through x-z/y-z visualization of confocal z-stacks that the tubes counted in both cell lines (MSec-MTAP and eMTAP) did not contact the substratum, but rather “hung” between the two connected cells (Fig. 1C and Fig. S2), a well-documented characteristic of bona fide TNTs (26, 36). To ascertain the function of these tubes, we performed live-cell, time-lapse confocal imaging to visualize the transport of mitochondria (labeled with Mitotracker red), a well-known cargo transported through TNTs (47, 48). We could clearly observe the transport of mitochondria from one cell to another in both stable cell lines (Fig. 1E and Movies S1 and S2). Another well-documented cargo of TNTs is the HIV-1 virus (21). We infected U2OS cells with HIV-1 (JRFL envelope pseudotyped virus) and immunostained for the virus using an established antibody (49). We could clearly observe HIV-1 immunofluorescence in TNTs emanating from infected cells, but not in TNTs arising from uninfected cells (Fig. S3A). y-z projection clearly showed the TNTs “hanging” between the two cell bodies with the HIV-1 signal appearing to transfer from the infected toward the uninfected cell through the TNT (Fig. S3A, box). In addition, TNT formation in U2OS cells was reduced significantly among cells treated with a validated TNT-inhibitor (Fig. S3, B and C) (21). Together, the above experiments confirmed the identity of these tubes in our study as TNTs.
ERp29 is a novel interactor of MSec
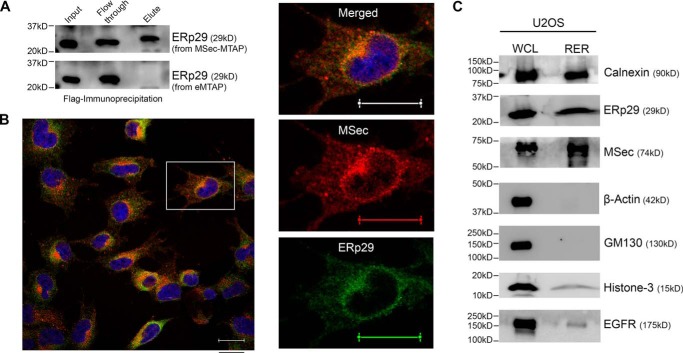
We used MSec-MTAP as the bait to identify its cellular interactome using methods adapted from earlier reports that have successfully used such multifunctional tags for interactomic analyses (46, 50–54). We lysed the U2OS cells stably expressing either MSec-MTAP or the MTAP tag alone under cryogenic conditions to help preserve the integrity of protein–protein interactions and subjected cell lysates to SBP affinity purification on a Streptactin column. We analyzed trypsinized eluates by electrospray ionization (ESI)-LC-MS (Fig. S4A) and evaluated the MS data with optimal search parameters (see “Materials and methods”) for the identification of the MSec interactome. The interactome obtained using the control tag (eMTAP) was subtracted from the MSec-MTAP interactome (Fig. S4B). Analysis of the unique interactome of MSec appearing in at least three experiments (Table S2 and Fig. S4C) with the help of the STRING (http://www.string-db.org)5 (105) and PANTHER (http://www.pantherdb.org)5 databases revealed that the interactors grouped into various functional classes (Fig. S4D and Tables S3 and S4), suggesting that MSec has a broad cross-talk with various functional pathways in the cell. One of the interacting proteins of MSec that appeared consistently and specifically only in the MSec-MTAP experiments was the endoplasmic reticulum chaperone ERp29, belonging to the class of unconventional protein chaperones belonging to the PDI-like family (42, 55). We confirmed the specificity of the interaction of ERp29 with MSec by immunoprecipitation, which validated that ERp29 interacted specifically with MSec-MTAP, but not with the empty vector (Fig. 2A). ERp29 is well-known as a marker for the ER and is hence highly enriched in this compartment at the perinuclear region (42, 56). We observed strong ER enrichment of MSec in U2OS cells as well as prominent co-localization with ERp29 at the perinuclear boundary through confocal microscopy (Fig. 2B), suggesting that MSec is associated with the ER. These data are supported by distinct ER-like localization of MSec-MTAP as well (Fig. S3B, bottom). To biochemically ascertain whether MSec localized with the ER, we performed subcellular fractionation of cell lysates and isolated the rough ER (as explained under “Materials and methods”). Western blot analysis confirmed that MSec was present in the rough ER fraction in U2OS cells, MSec-MTAP-U2OS cells, HeLa cells, and MDAMB231 cells (Fig. 2C and Fig. S5). These observations suggested that a substantial fraction of cellular MSec is associated strongly with the ER in multiple human cell types.
Figure 2.
ERp29 interacts with MSec. A, FLAG immunoprecipitates of the MSec-MTAP and empty-MTAP U2OS cell lysates probed for ERp29 by immunoblotting. B, confocal images showing co-localization of MSec (red) with ERp29 (green) in the ER region. Pearson's correlation coefficient (Rcoloc) of the whole frame is 0.6766, and that of the selected cell is 0.6866. Scale bar, 20 μm. C, Western blotting of the whole-cell lysate (WCL) and rough endoplasmic reticulum (RER) fractions from U2OS cells probed for the respective proteins as indicated. Shown are ER-positive markers (calnexin and ERp29), cytosolic marker (β-actin), Golgi complex marker (GM130), nuclear marker (histone-3), and cell membrane marker (EGFR).
ERp29 is required for TNT formation
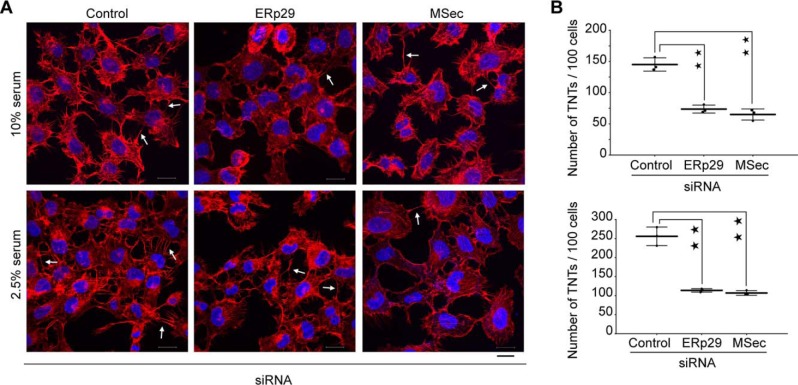
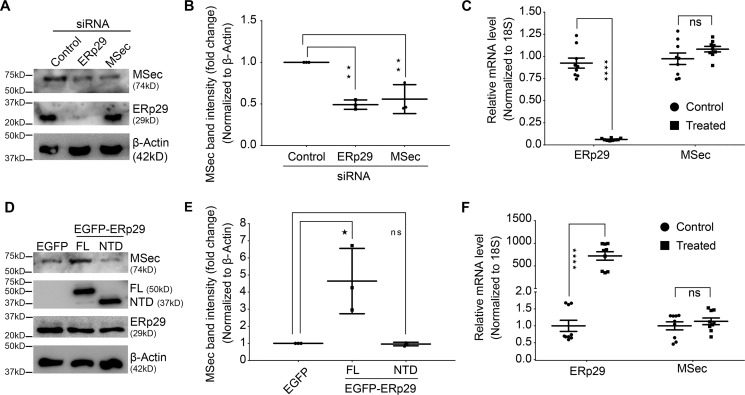
To probe whether ERp29 plays a functional role in TNT formation, we depleted it from U2OS cells using sequence-specific siRNAs. We confirmed depletion of protein levels using Western blotting (Fig. S6). Confocal microscopic analysis revealed that ERp29 depletion led to a significant loss in the number of TNTs formed by these cells (Fig. 3, A and B). Conditions such as serum deprivation, oxidative stress, pH changes, and nutrient shortage are known to induce the formation of TNTs (23, 24, 57–59), although the detailed cellular mechanisms have not been elucidated. We assessed whether ERp29 was required for induction of TNTs when cells were subjected to stress due to serum starvation. Control siRNA-treated cells cultured under stress conditions (2.5% serum, low-serum) induced more TNTs as compared with the optimal serum levels (10% serum). However, whereas serum starvation induced a marked increase in TNT numbers in control siRNA-treated cells, it failed to elicit this response upon either ERp29 depletion or MSec depletion (Fig. 3, A and B). Notably, we observed similar reduction in TNT numbers upon ERp29 depletion as seen for MSec depletion. Similarly, we repeated this experiment in HeLa cells, which also showed a significant reduction in their ability to form TNTs upon the depletion of ERp29 (Fig. S7).
Figure 3.
ERp29 is required for TNT formation. A, confocal microscopy images of cells co-stained with Alexa FluorTM 594–conjugated phalloidin (red, to visualize F-actin) and 4′,6-diamidino-2-phenylindole (blue, to visualize the nucleus) after 48 h of the indicated siRNA transfection. Arrows, TNTs connecting neighboring cells. Scale bar, 20 μm. B, quantification of the number of TNTs per 100 cells from the confocal images in both normal (10% serum) and stress (2.5% serum) conditions, respectively. Data represent mean ± S.D. (error bars) based on three independent experiments, 100 cells counted per experiment (paired t test, two-tailed; **, p < 0.01).
ERp29 influences TNT formation through its action on MSec
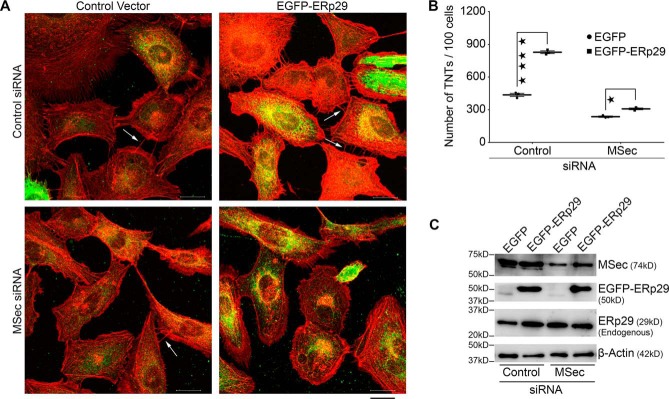
The above data clearly demonstrated that ERp29 was required for TNT formation. As a corollary, we tested whether ERp29 overexpression could induce the formation of a higher number of TNTs. We therefore transfected an exogenous ERp29 construct (EGFP-ERp29 (43)) into U2OS cells and compared the ability of the cells to induce TNT formation compared with cells transfected with a vector expressing EGFP alone. Confocal imaging of the cells followed by quantification showed that exogenous expression of ERp29 in U2OS cells led to a significant (almost 2-fold) increase in the number of TNTs formed per cell compared with cells treated with the empty vector alone (Fig. 4, A (top) and B). This result confirmed that the ability of the cells to form TNTs was directly proportional to the cellular expression levels of ERp29. We also asked whether exogenous expression of ERp29 could induce TNT formation even in the absence of MSec. To test this, we depleted MSec in U2OS cells using siRNA treatment and simultaneously overexpressed exogenous ERp29 (Fig. 4C). Despite robust expression of the exogenous ERp29 (Fig. 4C), induction of TNTs was dramatically reduced upon depletion of MSec (Fig. 4, A (bottom), B, and C). This observation suggested that ERp29 could induce TNTs only in the presence of MSec in the cell. We performed live cell, time-lapse confocal microscopy in eMTAP cells and observed that most of the TNTs formed were through the dislodgement of adjacent cells in culture, a mode of TNT formation already documented (27). ERp29-depleted cells appeared to move apart to shorter distances than the control cells, leading to the formation of shorter TNTs on average (Fig. S8A). This effect could be due to the direct effect of ERp29 on MSec, which has been shown to have a role in cell migration (60). We observed that ERp29 depletion led to a reduction in the overall lifetime of TNTs (Fig. S8B), suggesting that ERp29 may modulate TNT stability. We also measured the rate of elongation of growing TNTs, which did not show any measurable differences between the control and ERp29 depleted cells (Fig. S8C). These results together suggested that ERp29 may not be required for the elongation of growing TNTs, but may instead be needed for stabilizing already formed TNTs, in addition to its effect on TNT initiation. Further detailed investigation would be needed to build upon these observations on the precise role of ERp29 in regulating TNT stability.
Figure 4.
ERp29 overexpression induces TNTs in an MSec-dependent manner. A, U2OS cells transiently expressing either EGFP-ERp29 or luminal EGFP (control vector) were treated with siRNAs against MSec or luciferase (control) and co-stained with anti-GFP antibody (green) and phalloidin conjugated with Alexa FluorTM 594 (red). Cells were imaged using confocal microscopy. Arrows indicate TNTs connecting neighboring cells. Scale bar, 20 μm. B, quantification of the number of TNTs per 100 cells from the confocal images. Data represent mean ± S.D. (error bars) based on three independent experiments, 100 cells counted per experiment (two-way ANOVA, Sidak's multiple-comparison test; *, p < 0.05; ****, p < 0.0001). C, efficiency of overexpression of EGFP-ERp29 and depletion of MSec analyzed by immunoblotting with their corresponding antibodies.
ERp29 stabilizes MSec protein but not its mRNA
ERp29 is reported as a predominantly ER-resident protein and is an atypical chaperone that stabilizes and escorts various protein substrates. These include integral plasma membrane proteins like connexin 43, E-cadherin, occludin, cystic fibrosis transmembrane receptor, and epithelial sodium channel and secreted proteins like thyroglobulin (43, 44, 61, 62). A third major class of ERp29 substrates are localized at the cortex in the cytosol (e.g. β-catenin, PAR3, scribble, and zona occludens 1 (ZO1)) (61). ERp29 shares structural similarity with the PDI family of chaperones. However, it lacks the cysteine-binding residues common to PDIs and also does not have the ability to hydrolyze ATP, a common feature of several cellular chaperones, due to which it is named an unconventional PDI-like chaperone (63, 64). The mode of action of ERp29 appears to be to bind, stabilize, and escort its substrates to the correct destinations in the cell. Given that the effect of ERp29 on TNT formation depended on the presence of MSec (Fig. 4), we checked whether the levels of MSec were reduced upon ERp29 depletion in U2OS cells. Western blot analysis following siRNA treatment revealed that the levels of MSec protein in cell lysates reduced considerably upon ERp29 depletion, almost mimicking siRNA-mediated depletion of MSec itself (Fig. 5, A and B). However, the converse was not true; depletion of MSec did not change the protein levels of ERp29 (Fig. 5A), indicating that the loss of ERp29 could be destabilizing MSec but not vice versa.
Figure 5.
ERp29 stabilizes MSec protein levels but not its mRNA. A, siRNA-mediated depletion of ERp29 or MSec in U2OS cells using sequence-specific siRNAs. Control was luciferase siRNA. Efficiency of knockdown was analyzed by immunoblotting with anti-ERp29, anti-MSec, and anti-β-actin (loading control) antibodies, respectively. B, -fold change in protein expression of MSec quantified from the immunoblots using densitometry. C, relative mRNA expression levels of ERp29 and MSec (for the samples in A) quantified by RT-qPCR after respective siRNA treatment. D, immunoblots depicting exogenous expression of EGFP-ERp29 or EGFP-ERp29-NTD, endogenous ERp29 levels, and endogenous MSec levels in U2OS cells. Loading control was β-actin. E, -fold change in protein expression of MSec quantified from the immunoblots using densitometry. F, relative mRNA expression levels of ERp29 and MSec (for the samples in D) quantified by RT-qPCR after respective siRNA treatment. Data in B and E were analyzed by ordinary one-way ANOVA and Dunnett's multiple-comparison test and represent mean ± S.D. (error bars) based on three independent experiments. Data in C and F were analyzed by two-way ANOVA and Sidak's multiple-comparison test and represent mean ± S.E. (error bars) based on three independent experiments with three technical replicates each (*, p < 0.05; **, p < 0.01; ****, p < 0.0001; ns, not significant).
To test whether the reduction of MSec protein levels was due to transcriptional down-regulation or due to regulation at the level of translation, we quantified the mRNA levels of both genes upon depletion of the other using RT-qPCR. Both MSec and ERp29 mRNA levels remained essentially unchanged upon siRNA-mediated depletion of the other mRNA using sequence-specific siRNAs (Fig. 5C), indicating that loss of ERp29 protein led only to reduced MSec protein levels. We tested whether loss of ERp29 could also impact the stabilization of other MSec-associated proteins required for TNT formation, namely subunits of the exocyst complex and the small GTPase RalA (36). We depleted ERp29 in U2OS cells and checked the protein levels of RalA and exocyst complex components Exoc2 (Sec5), Exoc3 (Sec6), and Exoc4 (Sec8). Western blot analysis revealed that depletion of ERp29 did not affect the protein stability of RalA and exocyst complex (Fig. S8). Conversely, we also tested whether increased expression of ERp29 would stabilize MSec and increase its cellular mRNA or protein levels. Indeed, exogenous expression of EGFP-ERp29 elevated the protein levels of endogenous MSec significantly while not affecting the endogenous ERp29 levels (Fig. 5, D and E). RT-qPCR analysis revealed that overexpression of EGFP-ERp29 did not enhance the mRNA expression of MSec (Fig. 5F), demonstrating that the higher levels of MSec observed were not due to transcriptional up-regulation. ERp29 has been reported to bind to protein substrates through its C-terminal domain (43, 55, 65). We therefore tested whether an ERp29 construct with a truncated C terminus (EGFP-ERP29-NTD, residues 33–153 (43)) could also stabilize MSec in a manner similar to full-length ERp29. We observed that overexpression of EGFP-ERp29-NTD failed to lead to an increase in MSec protein levels (Fig. 5D). Taken together, the above results suggest that ERp29 stabilized the MSec protein while not impacting its transcription and that the C-terminal domain of ERp29 was important for this function.
The chaperone function of ERp29 is required for TNT formation
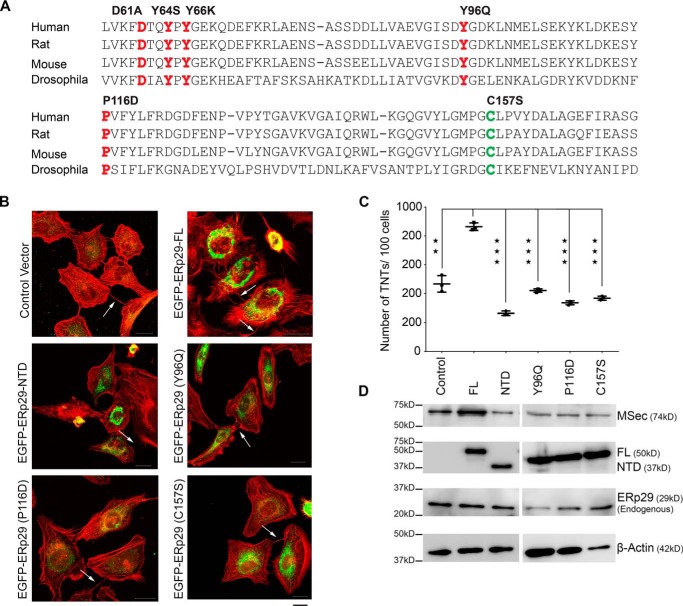
ERp29 has been well-studied as an unconventional chaperone that binds with and stabilizes various protein substrates (42–45). We therefore predicted that the chaperone function of ERp29 may be required for it to stabilize MSec. Whereas the C-terminal domain of ERp29 is required for stabilizing the MSec protein, several amino acid residues within the N-terminal domain have been implicated in its chaperone-like activity or in stabilization of the ERp29 protein itself (43, 65, 66). Mutations at these residues are known to abrogate or reduce the chaperone activity and/or stability of ERp29 (45, 62, 65). We generated individual site-directed mutants of ERp29 at these various conserved residues (chaperone mutants D61A, Y64S, Y66K, Y96Q, and P116D and a destabilizing mutant, C157S; Fig. 6A) in a mammalian expression vector with an N-terminal EGFP tag. The mutated fusion constructs when transfected individually in U2OS cells expressed and localized to the perinuclear region like the native WT construct (ERp29 FL), indicative of normal ER localization (Fig. 6B). We performed an analysis of the number of TNTs formed upon expression of each of these constructs. Expression of the WT EGFP-ERp29 led to a marked increase in the ability of cells to induce TNTs (Figs. 4 and 6 (B and C)). In comparison, none of the mutants was able to induce any appreciable increase in the number of TNTs over control EGFP (vector only)-transfected cells (Fig. 6 (B and C)) despite the proteins being expressed (Fig. 6, B and D) for most of the mutants. Closer analysis revealed that the protein levels of the D61A, Y64S, and Y66K mutants of ERp29 appeared to be lower than the WT and the other mutants (data not shown). These mutants of ERp29 have been shown earlier to co-degrade with their substrates through the proteasome (45). To test this possibility, we treated cells with the proteasome inhibitor MG132 and observed that the expression levels of these three mutants were restored to levels similar to the WT ERp29 and its other mutants (data not shown). We tested whether the chaperone mutants were capable of stabilizing endogenous MSec protein levels the way full-length ERp29 could (Fig. 5), by assessing the levels of MSec expression by Western blotting. None of the chaperone mutants was able to stabilize MSec protein levels over control (luminal EGFP vector alone) levels, suggesting that their chaperone activity was abolished (Fig. 6D). These results together confirmed that the chaperone activity of ERp29 was required for induction of TNT formation through the stabilization of the MSec protein.
Figure 6.
The chaperone activity of ERp29 is required for TNT formation. A, sequence alignment of ERp29 from multiple species showing conservation of chaperone (highlighted in red) and stabilization (highlighted in green) residues. B, confocal microscopy images of cells expressing either luminal EGFP (control) or the various chaperone point mutants of EGFP-ERp29 and co-stained with Alexa FluorTM 594–conjugated phalloidin (red, to visualize F-actin) and anti-GFP antibody (green, to visualize the EGFP/EGFP-ERp29 proteins). Arrows, TNTs connecting neighboring cells. Scale bar, 20 μm. C, quantification of the number of TNTs per 100 cells from the confocal images. Data were analyzed by the paired two-tailed t test and represent mean ± S.D. (error bars) based on three independent experiments, 100 cells counted per experiment. **, p < 0.01; ***, p < 0.001. D, immunoblots showing expression levels of endogenous MSec, EGFP-ERp29 constructs, and endogenous ERp29 probed with the respective antibodies as indicated upon expression of the EGFP-ERp29 WT and various mutant constructs. Loading control was β-actin.
Characterization of the ERp29–MSec interaction
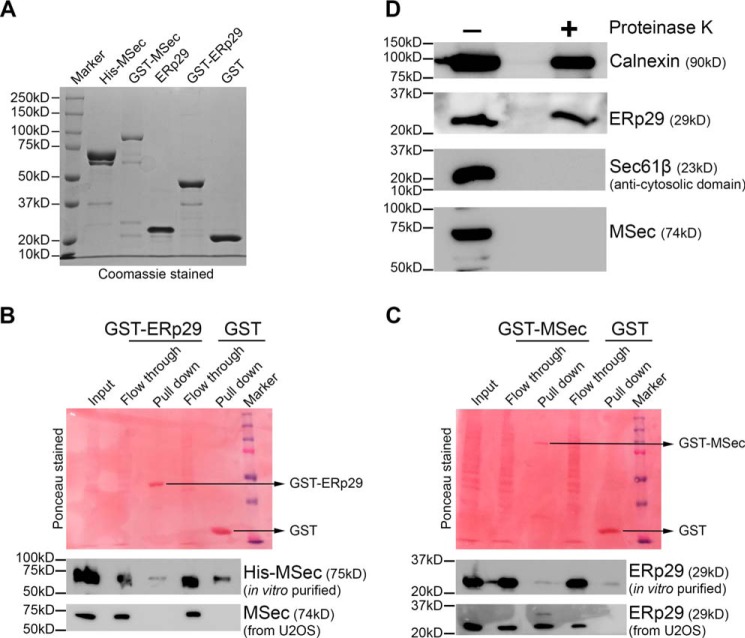
The strong binding of ERp29 to MSec (Fig. 2A), its ability to stabilize MSec levels (Fig. 5), and its requirement for TNT formation (Fig. 6) prompted us to characterize its physical interaction with MSec. To examine whether ERp29 interacts with MSec directly, we purified hexahistidine-tagged MSec (His-MSec in the pET28a vector), GSH-S-transferase–tagged MSec (GST-MSec), and GST-tagged ERp29 (GST-ERp29) as recombinant proteins as well as GST alone as control (Fig. 7A). We first ascertained whether the bacterially purified recombinant proteins would directly bind to each other. Surprisingly, purified MSec did not bind to GST-ERp29 with any higher efficiency than to the GST tag alone (Fig. 7B), precluding a direct interaction between the two proteins. We reasoned that the apparent absence of any detectable interaction could be due to the need for post-translational modification(s) (PTM(s)) on either or both proteins, which is absent during bacterial expression. To test whether PTM(s) on MSec could be important for this interaction, we passed U2OS cell lysates on beads that had GST-ERp29 immobilized. We again found that the MSec from cell lysates did not bind to GST-ERp29 (Fig. 7B). We therefore tested the opposite possibility, namely whether purified GST-MSec immobilized on beads could capture ERp29 from U2OS cell lysates. Indeed, we found that ERp29 from cell lysates could bind to bacterially purified GST-MSec, whereas the GST tag alone could not (Fig. 7C). This result suggested that PTM(s) of ERp29 is likely to be required for its interaction with MSec. It also left open the possibility of one or more additional proteins assisting the MSec–ERp29 interaction, perhaps triggered by the initial PTM(s) on ERp29. However, the exact identities of the PTM(s) and of any other potential protein(s) remain to be determined.
Figure 7.
Characterization of the ERp29–MSec interaction. A, SDS-PAGE profiles (Coomassie-stained) of the purified reombinant MSec and ERp29 proteins as indicated. B, purified ERp29 shows no interaction with either purified MSec or MSec from cell lysate. The bottom strips show immunoblots with the respective antibodies as indicated. C, purified MSec does not interact with purified ERp29 but interacts with ERp29 from cell lysate. The bottom strips show immunoblots with the respective antibodies as indicated. D, protease protection assay with proteinase K reveals that MSec is associated with the ER surface toward its cytosolic side. Markers used in immunoblots were calnexin (ER lumen) and Sec61β-cytosolic side (ER surface).
MSec is strongly associated with the ER fraction (Fig. 2 and Figs. S3B (bottom) and S5). However, MSec is reported as a predominantly cytosolic protein in the literature (21, 36, 37, 67), whereas ERp29 is believed to be mainly localized to the ER lumen (42, 56, 68). We therefore asked whether the ERp29–MSec interaction occurs in the ER lumen or in the cytosol, or whether it could occur across the ER membrane. To test this, we isolated ER microsomes by subcellular fractionation from U2OS cell lysates and subjected them to a protease protection assay. Briefly, ER microsomes were subjected to limited proteolysis with proteinase-K to digest all proteins/protein domains associated with the outer ER membrane facing the cytosol (69, 70). As controls, we confirmed through Western blot analysis that the cytosolic domain of the Sec61β subunit of the translocon was completely digested, whereas the luminal ER protein calnexin remained intact, as it was inaccessible to the protease (Fig. 7D). Similar to the Sec61β cytosolic domain, MSec was also almost completely digested by the protease, both in plain U2OS cells (Fig. 7D) and in U2OS cells stably expressing MSec-MTAP or HA-MSec (Fig. S10). These results confirmed that MSec is associated with the ER toward its cytosolic site and is unlikely to enter the ER lumen. Overall, these results suggest that ERp29 (which largely resides in the ER lumen) interacts with MSec (which is largely cytosolic) indirectly through a third (ER transmembrane) protein, which may require certain PTM(s) on ERp29.
Discussion
TNTs mediate the long-distance communication between animal cells and serve to facilitate the exchange of intracellular material between cells in normal physiological conditions as well as in disease conditions like cancer (13, 71–76). These structures are also implicated in the intercellular transmission of a variety of pathogenic organisms (10, 19–21, 77–81) and could serve as transport channels used by pathogens to evade extracellular defense responses of the body in combating pathogenic infection. It is therefore of prime importance to elucidate the (presently poorly understood) molecular mechanisms that underlie the formation and function of these enigmatic structures. Such knowledge would help to gain a fundamental understanding of this new form of intercellular communication as well as to devise strategies to modulate their numbers as a way of controlling the spread of several diseases.
MSec, also known as TNFAIP2 and B94, is a protein with well-established functions in vasculogenesis, inflammation, wound healing, focal adhesion formation, cell migration, invasion, and metastasis during cancer (60, 82–92). Thus, MSec plays key roles in vital physiological processes. Interestingly, MSec was also established as a key component in the machinery required for TNT formation in multiple cell lines (21, 36–38, 67), implicating MSec as one of the most important proteins required for TNT formation. Tubulogenesis from the plasma membrane is a complex cellular process that requires the concerted action of various cellular mechanisms (93–96). We hypothesized that the small list of known cellular interactors of MSec (38, 67) would not be sufficient to explain its role in TNT formation, especially in the context of the upstream regulation of MSec itself. We therefore performed whole-proteome interactomic analyses for MSec, which unveiled the role of the ER chaperone ERp29 in the formation of TNTs (Fig. 2).
ERp29 appeared as a reproducible novel interactor of MSec in our interactomic experiments (Figs. 2 and S4 and Tables S2–S4) and is well-known to localize in the ER (42, 56). We observed the co-localization of MSec with ERp29 at the perinuclear region, strongly reminiscent of the ER (Fig. 2B and Fig. S3B (bottom)). The subcellular rough ER fraction also showed an enrichment of both endogenous MSec and MSec-MTAP in our biochemical assays in multiple cell lines (Fig. 2C and Fig. S5). These results together with the observation of a cellular interaction between the two proteins suggests that MSec interacts with ERp29 as a prerequisite for it to eventually localize at the plasma membrane and initiate TNT formation. It is worth noting that ERp29 is unable to exert any measurable influence on TNT formation in the absence of MSec (Fig. 4). This observation again confirmed that the interaction of MSec with ERp29 is required prior to the initiation of TNT formation. It was instructive that we observed similar levels of disruption of TNT formation upon ERp29 depletion as seen for MSec depletion (Fig. 3). Thus, interaction of MSec with ERp29 appears to be a requirement for TNT formation.
The pronounced effect of ERp29 on the protein levels of MSec but not on its mRNA levels suggested that the action of ERp29 on MSec was as a protein chaperone (Figs. 5 and 6), an activity of ERp29 previously demonstrated and characterized on several other substrates (42–45). The chaperone activity was consistently observed upon either depletion of ERp29 (leading to reduction in MSec protein levels; Fig. 5) or, conversely, upon exogenous expression of ERp29 (leading to increased MSec protein levels; Figs. 5 and 6). C-terminally truncated ERp29 (ERp29-NTD) could not stabilize MSec levels the way full-length ERp29 could (Figs. 5D and 6D), with cell death observed upon exogenous expression of this construct (empirical observations; not shown), suggesting a dominant negative effect (43). We generated point mutants of ERp29 known to abolish its chaperone activity. Expression of each of these chaperone-defective mutants of full-length ERp29 led to lower cellular levels of MSec protein as well as to significantly lower TNT numbers (Fig. 6). Interestingly, the point mutants of ERp29 at sites D61A, Y64S, and Y66K themselves showed lower protein levels, as also seen for the substrate MSec, suggesting that these ERp29 mutants might co-degrade along with MSec (data not shown). This phenomenon has been reported for another ERp29 substrate as well (45). Indeed, upon MG132 treatment to block proteasome activity and hence prevent protein degradation, the levels of these mutants and of MSec went back to normal (data not shown). These experiments also revealed that in the absence of ERp29, MSec is degraded by the proteasome, as has been shown for other ERp29 substrates like Pipe (45), presumably because MSec does not get properly folded.
The ERp29–MSec interaction observed from cell lysates (Fig. 2A) was not recapitulated with the purified proteins or when MSec from cell lysates was incubated with purified ERp29 (Fig. 7B). Interestingly, purified MSec could only interact with ERp29 from cellular lysates (Fig. 7C). These results suggest that the MSec–ERp29 interaction could require post-translational modification(s) on ERp29. The exact identity and site(s) of this PTM(s) would be an interesting and valuable future pursuit to delineate the mechanism of MSec stabilization by ERp29. These results could also be interpreted to conclude that there is possibly a third protein from the cell lysate that is required for this interaction and/or stabilization, perhaps dependent on the initial PTM(s). Our imaging and biochemical experiments suggested that MSec is strongly present either inside the lumen or on the surface of the ER or possibly both (Fig. 2 (B and C) and Figs. S3B and S5). However, limited proteolytic treatment of ER microsomes to “shave” off ER-associated surface proteins facing the cytosol revealed that MSec is present only on the surface of the ER and not in the lumen (Fig. 7D and Fig. S10). These results suggest two possible models for the action of ERp29 on MSec. The largely ER lumen–resident ERp29 could interact with ER surface–localized MSec through a bridging transmembrane–ER protein(s) that remains to be identified. An alternative, although less likely, possibility is that a small cytosolic fraction of ERp29 (97, 98) stabilizes MSec. Further detailed studies will be needed to reveal the exact mechanism of MSec stabilization by ERp29. However, our study possibly suggests a generic mechanism that could be employed by ERp29 to stabilize a cohort of substrates that are known to localize at the cortex in the cytoplasm (e.g. β-catenin, ZO1, scribble, PAR3).
The stabilization of MSec by ERp29 is reminiscent of the well-documented interactions of ERp29 with other important proteins that reach the vicinity of the plasma membrane like connexin 43, thyroglobulin, PAR3, scribble, zona occludens 1 (ZO-1), occludin, and β-catenin, which are all substrates of ERp29 (43, 61, 65). It has been suggested that several of these substrates of ERp29 may follow broadly similar routes to traffic through the cell from the ER to the plasma membrane following stabilization through their interactions with ERp29 (61). However, at or near the plasma membrane, these proteins reach one among the following distinct destinations: 1) integral to the plasma membrane (connexin 43, cystic fibrosis transmembrane receptor, occludin (43, 44, 61)); 2) secreted out of the cell (thyroglobulin (42, 65)), or 3) localized to the subcortical region of the plasma membrane (MSec (this study); β-catenin, scribble, PAR3, ZO1 (61)). It is intuitive to rationalize that the first two of these classes of proteins would be accessed by ERp29 in the lumen of the ER (43, 44, 61, 65). Following stabilization, these proteins follow the classical secretory pathway and reach the plasma membrane (43, 44) or the exterior of the cell (65). However, if cortical substrates belonging to the third class first enter the ER lumen for interaction with ERp29, it is unclear how they would exit the endomembrane system to reach the cytosol following stabilization by ERp29 (61). Most of these proteins are also not reported to show ER surface localization as observed for MSec (Fig. 2B and Fig. S3B). The possibility that a small fraction of ERp29 may be present in the cytosol, probably coming through the endoplasmic reticulum–associated protein degradation pathway, to stabilize MSec as well as other substrates cannot be ruled out (97, 98). However, it is not clear how and where these proteins access ERp29 for their stabilization, and this is a subject worthy of detailed independent investigation.
Among the proteins known so far to be important for TNT formation (exocyst complex, RalA, Cdc42, LST1, filamin, connexin 43 (17, 36, 67, 99)), only connexin 43 is a known substrate of ERp29 (43). Our study adds MSec as a new member in this category, making it attractive to hypothesize that ERp29, and perhaps the endoplasmic reticulum itself, could be a vital cog in the cellular machinery required for TNT formation through its action on multiple protein substrates. However, it remained possible that other known cortical inducers of TNT formation like RalA and the exocyst complex are also chaperone substrates of ERp29. However, our results unequivocally show that protein levels of neither RalA nor multiple exocyst complex subunits are affected upon ERp29 depletion (Fig. S9), suggesting that MSec is one of the central targets of ERp29 in the regulation of TNT formation in addition to connexin 43.
Overall, this study reveals a novel role for the endoplasmic reticulum in directly controlling TNT formation through one of its chaperones ERp29. We show that MSec interacts with ERp29 and is stabilized by the resulting interaction, which is a prerequisite for MSec to be properly localized at the plasma membrane. In the absence of this stabilizing interaction, MSec is incapable of inducing TNT formation. We establish a hitherto unknown but essential role for the chaperone activity of ERp29 in inducing TNTs, by using an array of chaperone-defective ERp29 point mutants. We also show that in the absence of MSec, ERp29 has virtually no role in TNT formation. Thus, our study delineates an essential upstream molecular mechanism that regulates TNT formation through stabilization of MSec. The work highlights that the molecular mechanisms of TNT formation and function may be more complex than so far anticipated. Further fundamental mechanistic studies would reveal the contributions of other important proteins and processes in TNT biology. This knowledge offers an opportunity for the design and development of precisely targeted methods of modulating TNT numbers both for basic studies and for therapeutic purposes in TNT-mediated diseases.
Materials and methods
Cell lines
The U2OS cell line (kind gift from Stephen J. Doxsey, University of Massachusetts Medical School), HeLa cells, and MDAMB231 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1× antibiotic solution (100×, 10,000 units of penicillin and 10 mg of streptomycin per ml (HiMedia)). Cells were maintained at 37 °C with 5% CO2 and 95% humidity.
Generation of stably expressing MSec cell lines
Mouse MSec (NM_009396.2; nucleotides 124–2076) was cloned into the mammalian expression vector pcDNA4-TO-Hygromycin-mVenus-MAP (Addgene), which we then named the MSec-MTAP construct (Table S1). Sanger sequencing was performed to rule out any inadvertent mutations. The recombinant construct was transfected into U2OS cells using the X-tremeGENE HP DNA transfection reagent (Roche Applied Science/Sigma). Cells stably expressing MSec-MTAP were selected after culturing the transfected cells under selection medium containing antibiotic (hygromycin B, 300 μg/ml, TOKU-E) over a month. Surviving cells were sorted by serial dilution (one cell per well in a 24-well plate) into individual clones expressing different levels of the transgene MSec-MTAP and confirmed by Western blotting (Fig. S1). From these clones, we selected a clone (clone 1) expressing the transgene MSec-MTAP at levels comparable with endogenous MSec, which was used for proteomic and functional studies. A U2OS cell line stably expressing the empty vector (eMTAP) was generated in parallel to use for control experiments.
Transfection of HIV-1 (JRFL) into U2OS
For making the pseudotyped virus, we co-transfected U2OS cells with two plasmids, one that expresses HIV-1 JRFL envelope (gp160) and one (pSG3deltaenv) that expresses all of the genes of HIV-1 with a premature stop codon at the env gene as described previously (100–102). Transfection was performed in a 6-well cell culture plate using a FuGENE6 transfection kit (catalogue no. E2691, Promega Inc.). After 48 h of transfection, the supernatant was removed, and cells were washed twice with 1× PBS and immmunostained and imaged as explained under “Immunofluorescence and confocal microscopy.”
Experimental design and statistical rationale for proteomics
Affinity purification
The stably expressing (MSec-MTAP) U2OS cells were cultured in large scale in selection medium for affinity purification. Cultured cells were harvested in resuspension buffer containing 50 mm Tris-HCl, pH 7.5, 125 mm NaCl, 1 mm EDTA, 5% glycerol, Igepal CA-630 (0.025–0.2%) and a protease inhibitor mixture (Roche Applied Science). Harvested cells were lysed cryogenically by grinding them (using a mortar and pestle under liquid nitrogen) to preserve the integrity of protein complexes. The total cell lysate was centrifuged at 14,000 × g for 30 min at 4 °C to separate insoluble material. The supernatant was subjected to SBP-based affinity purification using a StrepTrap HP 1-ml column (GE Healthcare). The affinity column was equilibrated with resuspension buffer at a flow rate of 0.5 ml/min, and the supernatant was applied to it at a flow rate of 0.3 ml/min. The supernatant was incubated for 30 min and drained, and the column was subsequently washed with wash buffer containing 50 mm Tris-HCl, pH 7.5, 250 mm NaCl, Igepal CA-630 (0.05%), and protease inhibitor mixture. Bound protein was eluted in elution buffer containing 25 mm Tris-HCl, pH 7.5, 125 mm NaCl, 2.5 mm desthiobiotin, and protease inhibitor mixture. Four biological experiments were performed at varying Igepal CA-630 concentrations from 0.025 to 0.2% to identify protein interactions consistently observed over this range of detergent concentrations.
Mass spectrometry
After affinity elution, the protein solution (eluate) was concentrated using Amicon-Ultra centrifugal filters (Millipore). Concentrated protein was estimated by using the bicinchoninic acid kit (BCA; Thermo Scientific) and subsequently processed by the filter-assisted sample preparation (FASP protocol (103)) followed by digestion with trypsin using Trypsin Gold (Promega) at 37 °C for 14–16 h. Trypsinized peptides were desalted using ZipTips (Millipore) and analyzed by ESI-LC-MS by injecting a total of 1 μg of peptides on to a nano-reverse phase HPLC (RP-HPLC) C18 column (Thermo Scientific) equilibrated with buffer A (2% acetonitrile + 0.1% formic acid in water). Peptides were eluted using a gradient of buffer B (98% acetonitrile + 0.1% formic acid in water) applied at a flow rate of 200 nl/min over 80 min.
Search parameters and acceptance criteria (for MS/MS data)
The results obtained from the MS experiments (peptide and fragmentation spectra) were analyzed through the peaklist-generating software, ProteinPilotTM (version 4.5) and Mascot search engine (version 2.3.02) with optimal parameters as follows. The sequence database used for protein search was SwissProt 57.15, which contained 515,203 sequences (181,334,896 residues) in the database. The taxonomy (species) of the database used to analyze the data was Homo sapiens (human). The protease used to generate peptides was Trypsin Gold (Promega, V528A). The number of missed and/or nonspecific cleavages permitted was one. Fixed and variable modifications considered were carbamidomethyl (C) and oxidation (M), respectively. Mass tolerance for precursor and fragment ions was ±0.5 and ±0.2 Da, respectively. The threshold score (confidence threshold) for accepting individual spectra was 5%. The estimated false discovery rates for peptide and protein identifications were 5% based on the significance threshold (p < 0.05). The positive hits present in the bait (MSec-MTAP) experiments with a significant score from each of the four experimental conditions were overlapped using Venny version 2.1 software (http://bioinfogp.cnb.csic.es/tools/venny/index.html)5 and selected for further classification into various functional categories by using the PANTHER database (http://www.pantherdb.org/)5 and to assess for protein-protein interactions through the STRING database (http://www.string-db.org)5 (105). Further details of the unique MSec interactome appearing in at least three experiments are listed in Table S2. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (104) partner repository with the data set identifier PXD012441.
Co-immunoprecipitation
MSec-MTAP cells were harvested and washed twice with 10 volumes of ice-cold 1× PBS. Cells were lysed in 1× lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Igepal 630, 5 mm 2-mercaptoethanol, 1 mm EDTA, 5% glycerol) containing protease inhibitor mixture tablet (Roche Applied Science) and phosphatase inhibitor cocktails 2 and 3 (Sigma). Cells were lysed by sonication on ice three times for 10-s pulses each with an amplitude of 50% and 0.5 cycles. Lysed (sonicated) samples were centrifuged for 20 min at 14,000 × g and 4 °C, and the supernatant was transferred to a prechilled microcentrifuge tube (kept on ice). In parallel, 50 μl of preconjugated agarose-anti-FLAG M2 beads (Sigma) were washed three times with 1× lysis buffer. The beads were incubated with supernatant at 4 °C overnight with gentle rotation (5 rpm). The lysate-bead-antibody conjugate mixture was washed three times with 1× lysis buffer to remove nonspecifically adhered proteins. 2× sample buffer was added to the beads and boiled at 95 °C for 5 min. Boiled beads were centrifuged at 14,000 × g for 1 min, and the supernatant was resolved by SDS-PAGE, transferred onto a polyvinylidene difluoride (PVDF) membrane, and analyzed by Western blotting using appropriate antibodies against the respective antigens.
RNAi and RT-qPCR
Cells growing at ∼40% confluence were transfected with 67 nm gene-specific (ERp29/MSec/control luciferase) siRNAs (Dharmacon) using the Lipofectamine RNAiMax (Invitrogen) transfection reagent. After 48 h of transfection, cells were harvested, and the efficiency of knockdown was confirmed by Western blotting. Alternatively, total RNA was extracted using a NucleoSpin RNA isolation kit (Macherey Nagel) after 48 h of siRNA transfection. 1 μg of total RNA was used to synthesize cDNA using the iScript cDNA synthesis kit (Bio-Rad). RT-qPCR was conducted using a 7500 fast real-time PCR system (Applied Biosystems/Life Technologies). 100 ng of cDNA/reaction was used for real-time amplification using SYBR® Premix Ex TaqTM II reagents (TaKaRa). PCR conditions used were as follows: initial denaturation at 95 °C for 2 min, followed by denaturation at the cycling stage at 95 °C for 5 s, and annealing and extension at 60 °C for 30 s for a total of 40 cycles. The CT value was normalized to that of 18S mRNA. Three biological experiments (each with three technical replicates) were performed. Primers used for qPCR amplification are listed in Table S5.
Western blotting
Cultured cells were harvested and lysed, and the total protein concentration was estimated using the CB-XTM protein assay kit (G-Biosciences), and an equal amount of total protein was loaded in each lane. Proteins were resolved by SDS-PAGE and transferred onto a PVDF membrane. The PVDF membranes were incubated in blocking buffer (5% skimmed milk and 0.1% Tween 20 in 1× PBS) at room temperature for 2–3 h and then probed with specific primary antibodies (at 4 °C overnight) and secondary antibodies (at room temperature for 1 h). The antibodies used were rabbit anti-TNFAIP2 (1:200; Santa Cruz Biotechnology, Inc., catalogue no. sc-30138), mouse anti-TNFAIP2-F6 (1:200; Santa Cruz Biotechnology, catalogue no. sc-28318), rabbit anti-ERp29 (1:2000; Abcam, catalogue no. ab11420), mouse anti-β-actin (1:2000; Sigma, catalogue no. A3853), rabbit anti-GFP (1:2000; Abcam, catalogue no. ab6556), rabbit anti-calnexin (1:4000; Abcam, catalogue no. ab22595), mouse anti-FLAGM2 (1:4000; Sigma, catalogue no. F1804), rabbit anti-GM130 (1:2000; Abcam, catalogue no. ab52649), rabbit anti-histone-3 (1:20,000; Abcam, catalogue no. ab1791), rabbit anti-EGFR (1:2000; Abcam, catalogue no. ab52894), mouse anti-RalA (1:4000; BD Transduction Laboratory, catalogue no. 610221), rabbit anti-Sec5 (1:1000; Proteintech, catalogue no. 12751-1-AP), mouse anti-Sec6 (1:200; Thermo Fisher Scientific, catalogue no. MA1-25480), mouse anti-Sec8 (1:1000; ENZO Life Sciences, catalogue no. ADI-VAM-SV016), mouse anti-Sec61β (1:100; Santa Cruz Biotechnology, catalogue no. sc-393633), anti-mouse m-IgGk BP-HRP (1:2000; Santa Cruz Biotechnology, catalogue no. sc-516102), donkey anti-mouse IgG-HRP (1:10,000; Jackson Immunoresearch, catalogue no. 715-035-150), and donkey anti-rabbit IgG-HRP (1:10,000; Jackson Immunoresearch, catalogue no. 711-035-152). Chemiluminescent signals were developed using HRP substrate (Merck Millipore, catalogue no. WBLUF0500), and the signals were analyzed using an ImageQuantTM LAS-4000 gel documentation system (GE Healthcare). Protein band intensity was quantified using the ImageJ software.
Immunofluorescence and confocal microscopy
Cells were cultured in 6-well plates on sterilized glass coverslips. After 48 h of incubation, cells were washed twice with 1× PBS and fixed with 3.7% paraformaldehyde for 4–5 min. Alternatively, MSec-MTAP cells were fixed with chilled methanol for 30 min at −20 °C. After two washes with 1× PBS, cells were permeabilized with 0.1% Triton X-100 for 10 min. After two more washes with 1× PBS, cells were blocked with 5% (w/v) BSA in 1× PBS for 45 min. Cells were incubated with primary antibodies (VRC01 (1:200; obtained from the National Institutes of Health AIDS reagent program (49), mouse anti-TNFAIP2-F6 (1:100; Santa Cruz Biotechnology, catalogue no. sc-28318), rabbit anti-ERp29 (1:200; Abcam, catalogue number ab11420), and rabbit anti-GFP antibody (1:1000; Abcam, catalogue number ab6556, for visualizing EGFP-ERp29 constructs)) for 60–90 min, followed by three washes with 1× PBS. Coverslips were incubated with the corresponding secondary antibodies (goat anti-human (1:250; Jackson Immunoresearch, catalogue no. 109-095-064), m-IgGk BP-CFL 555 (1:100; Santa Cruz Biotechnology, sc-516177)), donkey anti-rabbit DL 488 (1:800; Jackson Immunoresearch, catalogue no. 711-485-152) for 60 min, followed by three washes with 1× PBS. F-actin filaments were stained with Alexa FluorTM 594–conjugated phalloidin (1:20; Invitrogen, catalogue no. A12381) for 40–60 min. Cells were then incubated with 4′,6-diamidino-2-phenylindole to stain the nucleus for 2–3 min. After three final washes with 1× PBS, the coverslips were mounted onto glass slides using ProLongTM Gold or ProLongTM Diamond antifade mounting medium (Invitrogen). All steps were performed at ambient room temperature, and all antibody dilutions were made in 5% BSA in 1× PBS. Both fixed and live cells were imaged using a Leica TCS SP8 confocal microscope (Leica Microsystems). Mitochondria were labeled with Mitotracker red (Thermo Scientific, catalogue no. M22425) for 30 min, and live cell imaging was performed subsequently. The TNT inhibitor was used at 10 μm for 24 h as described previously (21). Image analysis was performed by the LASX offline analysis software (Leica), ImageJ, and the IMARIS software suite (Bitplane).
Isolation of ER microsomes and proteinase K treatment
Rough ER was isolated by the protocol outlined in the Sigma catalogue (product no. ER0100), with manual preparation of the recommended solutions required for isolation. Briefly, cells were harvested and washed with 10 volumes of ice-cold 1× PBS. The cell pellet was resuspended in three volumes of 1× hypotonic buffer and incubated at 4 °C for 20 min. Swollen cells were microcentrifuged at 600 × g for 5 min at 4 °C, and the cell pellet was resuspended in two volumes of 1× isotonic buffer. Cells were disrupted (lysed) using a micropestle, and the cell homogenate was microcentrifuged at 1000 × g for 10 min at 4 °C. The post-nuclear supernatant was microcentrifuged at 12,000 × g for 15 min at 4 °C to separate the PMF. The PMF was then titrated dropwise with 8 mm CaCl2 (7.5 times the volume of PMF) with gentle mixing with a suitable magnetic spin bar. After titration, the sample was microcentrifuged at 8000 × g for 10 min at 4 °C to pellet down the rough ER microsomes, which were then resuspended in 2× Laemmli sample buffer and boiled at 95 °C for 5 min. Alternatively (for the protease protection assay), the PMF was ultracentrifuged at 1,00,000 × g in a preparative ultracentrifuge (Hitachi, model no. CP100WX) at 4 °C for 60 min. The pellet containing the ER microsomes was resuspended in the 1× isotonic buffer and subjected to proteinase K treatment (final concentration 20 ng/μl; Sigma, catalogue no. P2308) for 10 min on ice. After incubation, 2× Laemmli sample buffer was added, and the sample was boiled at 95 °C for 5 min. The samples were then resolved by SDS-PAGE, transferred onto a PVDF membrane, and analyzed by Western blotting.
Generation of ERp29 mutant constructs by site-directed mutagenesis
The recombinant human ERp29 full-length construct (EGFP-ERp29FL) was a kind gift from Dr. Michael Koval (43). Using this construct as a template, various ERp29 single point mutant (D61A, Y64S, Y66K, Y96Q, P116D, and C157S) constructs were generated using a standard site-directed mutagenesis protocol with primers incorporating the desired mutations. Primers used are listed in Table S5.
Purification of recombinant (His-, GST-tagged) proteins and GST pulldown assay
ERp29 cDNA was cloned into a bacterially expressed pGEX6P1 vector, and MSec cDNA was cloned into both pGEX6P1 and pET28a vectors to impart the respective tags (GST or His6) for affinity purification and for subsequent interaction studies. After sequencing to rule out inadvertent mutations, these recombinant vectors were transformed and expressed in Rossetta cells. GST-tagged proteins (GST-ERp29, GST-MSec, and GST alone) were purified by using a GSTPrepTM FF 16/10 column (GE Healthcare). Briefly, the GSTPrep column was equilibrated with 10 column volumes of resuspension/binding buffer (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 5 mm 2-mercaptoethanol, protease inhibitors, PMSF, and pepstatin). The supernatant containing bacterially expressed recombinant proteins was applied by pumping it onto the column using the sample pump (of the FPLC, AKTA Explorer, GE Healthcare) at a flow rate of 0.3 ml/min. The column was washed with 10 column volumes of resuspension buffer at a flow rate of 3 ml/min to remove unbound protein. Bound protein was eluted with five column volumes of elution buffer (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 5 mm 2-mercaptoethanol, 10 mm reduced GSH) at a flow rate of 2 ml/min. Fractions containing GST-tagged protein were collected by monitoring UV absorption at 280 nm into a prechilled 50-ml tube. To cleave the GST tag from the protein of interest, PreScission protease (GE Healthcare) was added to GST-tagged protein (eluate) and subjected to dialysis against a buffer containing 50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 5 mm 2-mercaptoethanol, 1 mm EDTA, 0.02% Igepal CA-630 overnight at 4 °C, with buffer changes at regular intervals. After overnight dialysis, the sample was applied to an equilibrated GSTprep column to remove the GST tag from the protein of interest. The flow-through (containing the protein of interest) was collected by monitoring UV absorption at 280 nm into a prechilled 50-ml tube. The purity of the eluted protein was analyzed by SDS-PAGE. Fractions containing highly pure (>95%) protein of interest were pooled into a prechilled container. Protein was concentrated using Amicon-Ultra centrifugal filters (Millipore). His-tagged MSec was purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography. Briefly, Ni-NTA beads (Qiagen) were equilibrated with resuspension/binding buffer and incubated with the supernatant (as prepared above) for 2–3 h at 4 °C with gentle rotation. The lysate/Ni-NTA mixture was loaded into a filter column, and the flow-through was collected. Protein-bound Ni-NTA beads were washed with wash buffer (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 5 mm 2-mercaptoethanol, 20 mm imidazole), and bound protein was eluted in elution buffer (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 5 mm 2-mercaptoethanol, 500 mm imidazole) and dialyzed (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 5 mm 2-mercaptoethanol) to remove imidazole. Dialyzed His-MSec was further purified by anion-exchange chromatography (Mono-Q 10/100, GE Healthcare). Briefly, the anion-exchange column was equilibrated with 10 column volumes of buffer containing 50 mm Tris-HCl, pH 8.0, 5 mm 2-mercaptoethanol, 100 mm NaCl. The dialyzed sample was applied to the anion-exchange column at a flow rate of 1–2 ml/min. Unbound protein was washed out with five column volumes of the equilibration buffer. A linear gradient of 100–500 mm NaCl (in the equilibration buffer) over 20 column volumes was used to elute the protein. Eluted fractions containing the protein of interest were collected by monitoring UV absorption at 280 nm into fresh microcentrifuge tubes. The purity of the eluted protein was analyzed by SDS-PAGE. Fractions containing highly pure (>95%) protein of interest were pooled into a prechilled tube. Both His-tagged and GST-tagged proteins were further purified by size-exclusion chromatography (Superdex 200 16/60 GL (preparative) and Superdex 200 10/300 GL (analytical), GE Healthcare). Briefly, size-exclusion columns were equilibrated with one column volume of buffer C (50 mm Tris-HCl, pH 8.0, 5 mm 2-mercaptoethanol, 150 mm NaCl) at a flow rate of 0.5 ml/min. A concentrated protein sample of interest was injected onto the column and run at a flow rate of 0.5 ml/min. Eluted fractions containing the protein of interest were collected, analyzed by SDS-PAGE, pooled, and concentrated as above. Protein concentration was estimated using the bicinchoninic acid (BCA) assay kit (Thermo Scientific, catalogue no. 23227). 10% glycerol was added as cryoprotectant and stored in the −80 °C freezer after snap freezing in liquid nitrogen until further use.
Statistical analysis
All results of cell culture experiments were obtained from three independent biological repeats and expressed as the mean ± S.D. RT-qPCR experiments were carried out in triplicates and repeated at least three times and expressed as mean ± S.E. Paired test (two-tailed) and one-way or two-way ANOVA were used to analyze the significance of variance, and p < 0.05 was considered as significant. GraphPad Prism version 7 software was used for analysis and plotting the graphs.
Author contributions
P. R. and S. D. designed and conducted experiments and analyzed data. H. K. and R. K. conducted experiments. J. B. helped in the conceptualization and execution of HIV-1 experiments. S. V. S. M. conceptualized and supervised the entire study, designed experiments, and analyzed the data. P. R. and S. V. S. M. wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Hiroshi Ohno for the gift of the mouse MSec construct (residues 42–691), Vandana Thakur for help with cloning MTAP-MSec, and Dr. Michael Koval for sharing ERp29 constructs (EGFP-ERp29-FL and EGFP-ERp29-NTD). The HIV-1 SG3 ΔEnv noninfectious molecular clone was obtained through the National Institutes of Health AIDS Reagent Program, Division of AIDS, NIAID, NIH, from Drs. John C. Kappes and Xiaoyun Wu. We thank Dr. Nirpendra Singh, M. Madhava Rao, and G. Nagavaraprasad for advice and assistance with MS and Suraj Tewari for assistance with confocal microscopy. We are grateful to the Regional Centre for Biotechnology (RCB) for providing the requisite infrastructure and the members of the Laboratory of Cellular Dynamics, RCB, for critical comments and suggestions during the study.
This work was supported by institutional funding from the Regional Centre for Biotechnology (RCB) (to S. V. S. M.). The HIV experiments were supported by research funding from DBT National Bioscience Grant Award BT/HRD/NBA/34/01/2012 (to J. B.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S5, Figs. S1–S10, and Movies S1 and S2.
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (104) partner repository with the data set identifier PXD012441.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- TNT
- tunneling nanotube
- ERp29
- endoplasmic reticulum protein 29
- TNF
- tumor necrosis factor
- TNFAIP2
- TNFα-induced protein 2
- LST1
- leukocyte-specific transcript 1
- U2OS
- osteosarcoma
- MTAP
- multifunctional tandem affinity purification
- eMTAP
- empty MTAP
- SBP
- streptavidin-binding peptide
- ESI
- electrospray ionization
- RP-HPLC
- reverse-phase HPLC
- PANTHER
- protein analysis through evolutionary relationships
- PDI
- protein-disulfide isomerase
- EGFP
- enhanced green fluorescent protein
- EGFR
- epidermal growth factor receptor
- ZO1
- zona occludens 1
- RT-qPCR
- real-time quantitative PCR
- GST
- glutathione S-transferase
- PMF
- post-mitochondrial fraction
- PTM
- post-translational modification
- PVDF
- polyvinylidene difluoride
- BCA
- bicinchoninic acid
- ER
- endoplasmic reticulum
- cDNA
- complementary DNA
- HRP
- horseradish peroxidase
- Ni-NTA
- nickel-nitrilotriacetic acid
- ANOVA
- analysis of variance.
References
- 1. Kornberg T. B., and Roy S. (2014) Cytonemes as specialized signaling filopodia. Development 141, 729–736 10.1242/dev.086223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. González-Méndez L., Seijo-Barandiarán I., and Guerrero I. (2017) Cytoneme-mediated cell-cell contacts for Hedgehog reception. Elife 6, e24045 10.7554/eLife.24045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nielsen M. S., Axelsen L. N., Sorgen P. L., Verma V., Delmar M., and Holstein-Rathlou N. H. (2012) Gap junctions. Compr. Physiol. 2, 1981–2035 10.1002/cphy.c110051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huettner J. E., Lu A., Qu Y., Wu Y., Kim M., and McDonald J. W. (2006) Gap junctions and connexon hemichannels in human embryonic stem cells. Stem Cells 24, 1654–1667 10.1634/stemcells.2005-0003 [DOI] [PubMed] [Google Scholar]
- 5. Maas S. L. N., Breakefield X. O., and Weaver A. M. (2017) Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 27, 172–188 10.1016/j.tcb.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Colombo M., Raposo G., and Théry C. (2014) Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- 7. Brunkard J. O., and Zambryski P. C. (2017) Plasmodesmata enable multicellularity: new insights into their evolution, biogenesis, and functions in development and immunity. Curr. Opin. Plant Biol. 35, 76–83 10.1016/j.pbi.2016.11.007 [DOI] [PubMed] [Google Scholar]
- 8. Rustom A., Saffrich R., Markovic I., Walther P., and Gerdes H. H. (2004) Nanotubular highways for intercellular organelle transport. Science 303, 1007–1010 10.1126/science.1093133 [DOI] [PubMed] [Google Scholar]
- 9. Hurtig J., Chiu D. T., and Onfelt B. (2010) Intercellular nanotubes: insights from imaging studies and beyond. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2, 260–276 10.1002/wnan.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Onfelt B., Nedvetzki S., Benninger R. K. P., Purbhoo M. A., Sowinski S., Hume A. N., Seabra M. C., Neil M. A. A., French P. M. W., and Davis D. M. (2006) Structurally distinct membrane nanotubes between human macrophages support long-distance vesicular traffic or surfing of bacteria. J. Immunol. 177, 8476–8483 10.4049/jimmunol.177.12.8476 [DOI] [PubMed] [Google Scholar]
- 11. Austefjord M. W., Gerdes H. H., and Wang X. (2014) Tunneling nanotubes: diversity in morphology and structure. Commun. Integr Biol. 7, e27934 10.4161/cib.27934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smith I. F., Shuai J., and Parker I. (2011) Active generation and propagation of Ca2+ signals within tunneling membrane nanotubes. Biophys. J. 100, L37–L39 10.1016/j.bpj.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Watkins S. C., and Salter R. D. (2005) Functional connectivity between immune cells mediated by tunneling nanotubules. Immunity 23, 309–318 10.1016/j.immuni.2005.08.009 [DOI] [PubMed] [Google Scholar]
- 14. Plotnikov E. Y., Khryapenkova T. G., Galkina S. I., Sukhikh G. T., and Zorov D. B. (2010) Cytoplasm and organelle transfer between mesenchymal multipotent stromal cells and renal tubular cells in co-culture. Exp. Cell Res. 316, 2447–2455 10.1016/j.yexcr.2010.06.009 [DOI] [PubMed] [Google Scholar]
- 15. Sanders T. A., Llagostera E., and Barna M. (2013) Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nature 497, 628–632 10.1038/nature12157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pyrgaki C., Trainor P., Hadjantonakis A. K., and Niswander L. (2010) Dynamic imaging of mammalian neural tube closure. Dev. Biol. 344, 941–947 10.1016/j.ydbio.2010.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang X., Bukoreshtliev N. V., and Gerdes H. H. (2012) Developing neurons form transient nanotubes facilitating electrical coupling and calcium signaling with distant astrocytes. PLoS One 7, e47429 10.1371/journal.pone.0047429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Connor Y., Tekleab S., Nandakumar S., Walls C., Tekleab Y., Husain A., Gadish O., Sabbisetti V., Kaushik S., Sehrawat S., Kulkarni A., Dvorak H., Zetter B. E., Edelman E. R., Sengupta S. (2015) Physical nanoscale conduit-mediated communication between tumour cells and the endothelium modulates endothelial phenotype. Nat. Commun. 6, 8671 10.1038/ncomms9671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eugenin E. A., Gaskill P. J., and Berman J. W. (2009) Tunneling nanotubes (TNT) are induced by HIV-infection of macrophages: a potential mechanism for intercellular HIV trafficking. Cell. Immunol. 254, 142–148 10.1016/j.cellimm.2008.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martinez M. G., and Kielian M. (2016) Intercellular extensions are induced by the alphavirus structural proteins and mediate virus transmission. PLoS Pathog. 12, e1006061 10.1371/journal.ppat.1006061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hashimoto M., Bhuyan F., Hiyoshi M., Noyori O., Nasser H., Miyazaki M., Saito T., Kondoh Y., Osada H., Kimura S., Hase K., Ohno H., and Suzu S. (2016) Potential role of the formation of tunneling nanotubes in HIV-1 spread in macrophages. J. Immunol. 196, 1832–1841 10.4049/jimmunol.1500845 [DOI] [PubMed] [Google Scholar]
- 22. Gousset K., Schiff E., Langevin C., Marijanovic Z., Caputo A., Browman D. T., Chenouard N., de Chaumont F., Martino A., Enninga J., Olivo-Marin J. C., Männel D., and Zurzolo C. (2009) Prions hijack tunnelling nanotubes for intercellular spread. Nat. Cell Biol. 11, 328–336 10.1038/ncb1841 [DOI] [PubMed] [Google Scholar]
- 23. Wang Y., Cui J., Sun X., and Zhang Y. (2011) Tunneling-nanotube development in astrocytes depends on p53 activation. Cell Death Differ. 18, 732–742 10.1038/cdd.2010.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thayanithy V., Babatunde V., Dickson E. L., Wong P., Oh S., Ke X., Barlas A., Fujisawa S., Romin Y., Moreira A. L., Downey R. J., Steer C. J., Subramanian S., Manova-Todorova K., Moore M. A., and Lou E. (2014) Tumor exosomes induce tunneling nanotubes in lipid raft-enriched regions of human mesothelioma cells. Exp. Cell Res. 323, 178–188 10.1016/j.yexcr.2014.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abounit S., Delage E., and Zurzolo C. (2015) Identification and characterization of tunneling nanotubes for intercellular trafficking. Curr. Protoc. Cell Biol. 67, 12.10.1–21 10.1002/0471143030.cb1210s67 [DOI] [PubMed] [Google Scholar]
- 26. Gousset K., Marzo L., Commere P. H., and Zurzolo C. (2013) Myo10 is a key regulator of TNT formation in neuronal cells. J. Cell Sci. 126, 4424–4435 10.1242/jcs.129239 [DOI] [PubMed] [Google Scholar]
- 27. Ariazi J., Benowitz A., De Biasi V., Den Boer M. L., Cherqui S., Cui H., Douillet N., Eugenin E. A., Favre D., Goodman S., Gousset K., Hanein D., Israel D. I., Kimura S., Kirkpatrick R. B., et al. (2017) Tunneling nanotubes and gap junctions—their role in long-range intercellular communication during development, health, and disease conditions. Front. Mol. Neurosci. 10, 333 10.3389/fnmol.2017.00333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Caneparo L., Pantazis P., Dempsey W., and Fraser S. E. (2011) Intercellular bridges in vertebrate gastrulation. PLoS One 6, e20230 10.1371/journal.pone.0020230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takahashi A., Kukita A., Li Y. J., Zhang J. Q., Nomiyama H., Yamaza T., Ayukawa Y., Koyano K., and Kukita T. (2013) Tunneling nanotube formation is essential for the regulation of osteoclastogenesis. J. Cell Biochem. 114, 1238–1247 10.1002/jcb.24433 [DOI] [PubMed] [Google Scholar]
- 30. Wang T., Xu Z., Jiang W., and Ma A. (2006) Cell-to-cell contact induces mesenchymal stem cell to differentiate into cardiomyocyte and smooth muscle cell. Int. J. Cardiol. 109, 74–81 10.1016/j.ijcard.2005.05.072 [DOI] [PubMed] [Google Scholar]
- 31. Chinnery H. R., Pearlman E., and McMenamin P. G. (2008) Cutting edge: membrane nanotubes in vivo: a feature of MHC class II+ cells in the mouse cornea. J. Immunol. 180, 5779–5783 10.4049/jimmunol.180.9.5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seyed-Razavi Y., Hickey M. J., Kuffová L., McMenamin P. G., and Chinnery H. R. (2013) Membrane nanotubes in myeloid cells in the adult mouse cornea represent a novel mode of immune cell interaction. Immunol. Cell Biol. 91, 89–95 10.1038/icb.2012.52 [DOI] [PubMed] [Google Scholar]
- 33. McKinney M. C., Stark D. A., Teddy J., and Kulesa P. M. (2011) Neural crest cell communication involves an exchange of cytoplasmic material through cellular bridges revealed by photoconversion of KikGR. Dev. Dyn. 240, 1391–1401 10.1002/dvdy.22612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Teddy J. M., and Kulesa P. M. (2004) In vivo evidence for short- and long-range cell communication in cranial neural crest cells. Development 131, 6141–6151 10.1242/dev.01534 [DOI] [PubMed] [Google Scholar]
- 35. Niedenberger B. A., Cook K., Baena V., Serra N. D., Velte E. K., Agno J. E., Litwa K. A., Terasaki M., Hermann B. P., Matzuk M. M., and Geyer C. B. (2018) Dynamic cytoplasmic projections connect mammalian spermatogonia in vivo. Development 145, dev161323 10.1242/dev.161323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hase K., Kimura S., Takatsu H., Ohmae M., Kawano S., Kitamura H., Ito M., Watarai H., Hazelett C. C., Yeaman C., and Ohno H. (2009) M-Sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nat. Cell Biol. 11, 1427–1432 10.1038/ncb1990 [DOI] [PubMed] [Google Scholar]
- 37. Kimura S., Yamashita M., Yamakami-Kimura M., Sato Y., Yamagata A., Kobashigawa Y., Inagaki F., Amada T., Hase K., Iwanaga T., Ohno H., and Fukai S. (2016) Distinct roles for the N- and C-terminal regions of M-Sec in plasma membrane deformation during tunneling nanotube formation. Sci. Rep. 6, 33548 10.1038/srep33548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kimura S., Hase K., and Ohno H. (2012) Tunneling nanotubes: emerging view of their molecular components and formation mechanisms. Exp. Cell Res. 318, 1699–1706 10.1016/j.yexcr.2012.05.013 [DOI] [PubMed] [Google Scholar]
- 39. Sugihara K., Asano S., Tanaka K., Iwamatsu A., Okawa K., and Ohta Y. (2002) The exocyst complex binds the small GTPase RalA to mediate filopodia formation. Nat. Cell Biol. 4, 73–78 10.1038/ncb720 [DOI] [PubMed] [Google Scholar]
- 40. Pawar A., Meier J. A., Dasgupta A., Diwanji N., Deshpande N., Saxena K., Buwa N., Inchanalkar S., Schwartz M. A., and Balasubramanian N. (2016) Ral-Arf6 crosstalk regulates Ral dependent exocyst trafficking and anchorage independent growth signalling. Cell. Signal. 28, 1225–1236 10.1016/j.cellsig.2016.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sisakhtnezhad S., and Khosravi L. (2015) Emerging physiological and pathological implications of tunneling nanotubes formation between cells. Eur. J. Cell Biol. 94, 429–443 10.1016/j.ejcb.2015.06.010 [DOI] [PubMed] [Google Scholar]
- 42. Sargsyan E., Baryshev M., Szekely L., Sharipo A., and Mkrtchian S. (2002) Identification of ERp29, an endoplasmic reticulum lumenal protein, as a new member of the thyroglobulin folding complex. J. Biol. Chem. 277, 17009–17015 10.1074/jbc.M200539200 [DOI] [PubMed] [Google Scholar]
- 43. Das S., Smith T. D., Sarma J. D., Ritzenthaler J. D., Maza J., Kaplan B. E., Cunningham L. A., Suaud L., Hubbard M. J., Rubenstein R. C., and Koval M. (2009) ERp29 restricts Connexin43 oligomerization in the endoplasmic reticulum. Mol. Biol. Cell 20, 2593–2604 10.1091/mbc.e08-07-0790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Suaud L., Miller K., Alvey L., Yan W., Robay A., Kebler C., Kreindler J. L., Guttentag S., Hubbard M. J., and Rubenstein R. C. (2011) ERp29 regulates DeltaF508 and wild-type cystic fibrosis transmembrane conductance regulator (CFTR) trafficking to the plasma membrane in cystic fibrosis (CF) and non-CF epithelial cells. J. Biol. Chem. 286, 21239–21253 10.1074/jbc.M111.240267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barnewitz K., Guo C., Sevvana M., Ma Q., Sheldrick G. M., Söling H. D., and Ferrari D. M. (2004) Mapping of a substrate binding site in the protein disulfide isomerase-related chaperone wind based on protein function and crystal structure. J. Biol. Chem. 279, 39829–39837 10.1074/jbc.M406839200 [DOI] [PubMed] [Google Scholar]
- 46. Ma H., McLean J. R., Chao L. F., Mana-Capelli S., Paramasivam M., Hagstrom K. A., Gould K. L., and McCollum D. (2012) A highly efficient multifunctional tandem affinity purification approach applicable to diverse organisms. Mol. Cell Proteomics 11, 501–511 10.1074/mcp.O111.016246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jackson M. V., Morrison T. J., Doherty D. F., McAuley D. F., Matthay M. A., Kissenpfennig A., O'Kane C. M., and Krasnodembskaya A. D. (2016) Mitochondrial transfer via tunneling nanotubes is an important mechanism by which mesenchymal stem cells enhance macrophage phagocytosis in the in vitro and in vivo models of ARDS. Stem Cells 34, 2210–2223 10.1002/stem.2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang X., and Gerdes H. H. (2015) Transfer of mitochondria via tunneling nanotubes rescues apoptotic PC12 cells. Cell Death Differ 22, 1181–1191 10.1038/cdd.2014.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu X., Yang Z. Y., Li Y., Hogerkorp C. M., Schief W. R., Seaman M. S., Zhou T., Schmidt S. D., Wu L., Xu L., Longo N. S., McKee K., O'Dell S., Louder M. K., Wycuff D. L., et al. (2010) Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 10.1126/science.1187659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cheeseman I. M., and Desai A. (2005) A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE 2005, pl1 [DOI] [PubMed] [Google Scholar]
- 51. Kobayashi T., Morone N., Kashiyama T., Oyamada H., Kurebayashi N., and Murayama T. (2008) Engineering a novel multifunctional green fluorescent protein tag for a wide variety of protein research. PLoS One 3, e3822 10.1371/journal.pone.0003822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Olinares P. D., Dunn A. D., Padovan J. C., Fernandez-Martinez J., Rout M. P., and Chait B. T. (2016) A robust workflow for native mass spectrometric analysis of affinity-isolated endogenous protein assemblies. Anal Chem. 88, 2799–2807 10.1021/acs.analchem.5b04477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. LaCava J., Molloy K. R., Taylor M. S., Domanski M., Chait B. T., and Rout M. P. (2015) Affinity proteomics to study endogenous protein complexes: pointers, pitfalls, preferences and perspectives. BioTechniques 58, 103–119 10.2144/000114262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cristea I. M., Williams R., Chait B. T., and Rout M. P. (2005) Fluorescent proteins as proteomic probes. Mol. Cell Proteomics 4, 1933–1941 10.1074/mcp.M500227-MCP200 [DOI] [PubMed] [Google Scholar]
- 55. Rainey-Barger E. K., Mkrtchian S., and Tsai B. (2009) The C-terminal domain of ERp29 mediates polyomavirus binding, unfolding, and infection. J. Virol. 83, 1483–1491 10.1128/JVI.02057-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Steven S. D., and Hubbard M. J. (2002) ERp29 is a ubiquitous resident of the endoplasmic reticulum with a distinct role in secretory protein production. J. Histochem. Cytochem. 50, 557–566 10.1177/002215540205000413 [DOI] [PubMed] [Google Scholar]
- 57. Rustom A. (2016) The missing link: does tunnelling nanotube-based supercellularity provide a new understanding of chronic and lifestyle diseases? Open Biol. 6, 160057 10.1098/rsob.160057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhu D., Tan K. S., Zhang X., Sun A. Y., Sun G. Y., and Lee J. C. (2005) Hydrogen peroxide alters membrane and cytoskeleton properties and increases intercellular connections in astrocytes. J. Cell Sci. 118, 3695–3703 10.1242/jcs.02507 [DOI] [PubMed] [Google Scholar]
- 59. Marlein C. R., Zaitseva L., Piddock R. E., Robinson S. D., Edwards D. R., Shafat M. S., Zhou Z., Lawes M., Bowles K. M., and Rushworth S. A. (2017) NADPH oxidase-2 derived superoxide drives mitochondrial transfer from bone marrow stromal cells to leukemic blasts. Blood 130, 1649–1660 [DOI] [PubMed] [Google Scholar]
- 60. Jia L., Zhou Z., Liang H., Wu J., Shi P., Li F., Wang Z., Wang C., Chen W., Zhang H., Wang Y., Liu R., Feng J., and Chen C. (2016) KLF5 promotes breast cancer proliferation, migration and invasion in part by upregulating the transcription of TNFAIP2. Oncogene 35, 2040–2051 10.1038/onc.2015.263 [DOI] [PubMed] [Google Scholar]
- 61. Bambang I. F., Lee Y. K., Richardson D. R., and Zhang D. (2013) Endoplasmic reticulum protein 29 regulates epithelial cell integrity during the mesenchymal-epithelial transition in breast cancer cells. Oncogene 32, 1240–1251 10.1038/onc.2012.149 [DOI] [PubMed] [Google Scholar]
- 62. Grumbach Y., Bikard Y., Suaud L., Chanoux R. A., and Rubenstein R. C. (2014) ERp29 regulates epithelial sodium channel functional expression by promoting channel cleavage. Am. J. Physiol. Cell Physiol. 307, C701–C709 10.1152/ajpcell.00134.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Barak N. N., Neumann P., Sevvana M., Schutkowski M., Naumann K., Malesević M., Reichardt H., Fischer G., Stubbs M. T., and Ferrari D. M. (2009) Crystal structure and functional analysis of the protein disulfide isomerase-related protein ERp29. J. Mol. Biol. 385, 1630–1642 10.1016/j.jmb.2008.11.052 [DOI] [PubMed] [Google Scholar]
- 64. Mkrtchian S., and Sandallova T. (2006) ERp29, an unusual redox-inactive member of the thioredoxin family. Antioxid. Redox Signal. 8, 325–337 10.1089/ars.2006.8.325 [DOI] [PubMed] [Google Scholar]
- 65. Baryshev M., Sargsyan E., and Mkrtchian S. (2006) ERp29 is an essential endoplasmic reticulum factor regulating secretion of thyroglobulin. Biochem. Biophys. Res. Commun. 340, 617–624 10.1016/j.bbrc.2005.12.052 [DOI] [PubMed] [Google Scholar]
- 66. Rainey-Barger E. K., Mkrtchian S., and Tsai B. (2007) Dimerization of ERp29, a PDI-like protein, is essential for its diverse functions. Mol. Biol. Cell 18, 1253–1260 10.1091/mbc.e06-11-1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schiller C., Diakopoulos K. N., Rohwedder I., Kremmer E., von Toerne C., Ueffing M., Weidle U. H., Ohno H., and Weiss E. H. (2013) LST1 promotes the assembly of a molecular machinery responsible for tunneling nanotube formation. J. Cell Sci. 126, 767–777 10.1242/jcs.114033 [DOI] [PubMed] [Google Scholar]
- 68. MacLeod J. C., Sayer R. J., Lucocq J. M., and Hubbard M. J. (2004) ERp29, a general endoplasmic reticulum marker, is highly expressed throughout the brain. J. Comp. Neurol. 477, 29–42 10.1002/cne.20222 [DOI] [PubMed] [Google Scholar]
- 69. Vidugiriene J., Menon A. K. (1994) The GPI anchor of cell-surface proteins is synthesized on the cytoplasmic face of the endoplasmic reticulum. J. Cell Biol. 127, 333–341 10.1083/jcb.127.2.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vashist S., and Ng D. T. (2004) Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 165, 41–52 10.1083/jcb.200309132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Guescini M., Leo G., Genedani S., Carone C., Pederzoli F., Ciruela F., Guidolin D., Stocchi V., Mantuano M., Borroto-Escuela D. O., Fuxe K., and Agnati L. F. (2012) Microvesicle and tunneling nanotube mediated intercellular transfer of G-protein coupled receptors in cell cultures. Exp. Cell Res. 318, 603–613 10.1016/j.yexcr.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 72. Arkwright P. D., Luchetti F., Tour J., Roberts C., Ayub R., Morales A. P., Rodríguez J. J., Gilmore A., Canonico B., Papa S., and Esposti M. D. (2010) Fas stimulation of T lymphocytes promotes rapid intercellular exchange of death signals via membrane nanotubes. Cell Res. 20, 72–88 10.1038/cr.2009.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Thayanithy V., Dickson E. L., Steer C., Subramanian S., and Lou E. (2014) Tumor-stromal cross talk: direct cell-to-cell transfer of oncogenic microRNAs via tunneling nanotubes. Transl. Res. 164, 359–365 10.1016/j.trsl.2014.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lou E., Fujisawa S., Barlas A., Romin Y., Manova-Todorova K., Moore M. A., and Subramanian S. (2012) Tunneling nanotubes: a new paradigm for studying intercellular communication and therapeutics in cancer. Commun. Integr. Biol. 5, 399–403 10.4161/cib.20569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lou E., Fujisawa S., Morozov A., Barlas A., Romin Y., Dogan Y., Gholami S., Moreira A. L., Manova-Todorova K., and Moore M. A. (2012) Tunneling nanotubes provide a unique conduit for intercellular transfer of cellular contents in human malignant pleural mesothelioma. PLoS One 7, e33093 10.1371/journal.pone.0033093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Polak R., de Rooij B., Pieters R., and den Boer M. L. (2015) B-cell precursor acute lymphoblastic leukemia cells use tunneling nanotubes to orchestrate their microenvironment. Blood 126, 2404–2414 10.1182/blood-2015-03-634238 [DOI] [PubMed] [Google Scholar]
- 77. Sowinski S., Jolly C., Berninghausen O., Purbhoo M. A., Chauveau A., Köhler K., Oddos S., Eissmann P., Brodsky F. M., Hopkins C., Önfelt B., Sattentau Q., and Davis D. M. (2008) Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat. Cell Biol. 10, 211–219 10.1038/ncb1682 [DOI] [PubMed] [Google Scholar]
- 78. Sherer N. M., Lehmann M. J., Jimenez-Soto L. F., Horensavitz C., Pypaert M., and Mothes W. (2007) Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat. Cell Biol. 9, 310–315 10.1038/ncb1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Roberts K. L., Manicassamy B., and Lamb R. A. (2015) Influenza A virus uses intercellular connections to spread to neighboring cells. J. Virol. 89, 1537–1549 10.1128/JVI.03306-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mukerji J., Olivieri K. C., Misra V., Agopian K. A., and Gabuzda D. (2012) Proteomic analysis of HIV-1 Nef cellular binding partners reveals a role for exocyst complex proteins in mediating enhancement of intercellular nanotube formation. Retrovirology 9, 33 10.1186/1742-4690-9-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Peralta B., Gil-Carton D., Castaño-Diez D., Bertin A., Boulogne C., Oksanen H. M., Bamford D. H., and Abrescia N. G. (2013) Mechanism of membranous tunnelling nanotube formation in viral genome delivery. PLoS Biol. 11, e1001667 10.1371/journal.pbio.1001667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rusiniak M. E., Yu M., Ross D. T., Tolhurst E. C., and Slack J. L. (2000) Identification of B94 (TNFAIP2) as a potential retinoic acid target gene in acute promyelocytic leukemia. Cancer Res. 60, 1824–1829 [PubMed] [Google Scholar]
- 83. Einstein M. H., Cruz Y., El-Awady M. K., Popescu N. C., DiPaolo J. A., van Ranst M., Kadish A. S., Romney S., Runowicz C. D., and Burk R. D. (2002) Utilization of the human genome sequence localizes human papillomavirus type 16 DNA integrated into the TNFAIP2 gene in a fatal cervical cancer from a 39-year-old woman. Clin. Cancer Res. 8, 549–554 [PubMed] [Google Scholar]
- 84. Chen L. C., Chen C. C., Liang Y., Tsang N. M., Chang Y. S., and Hsueh C. (2011) A novel role for TNFAIP2: its correlation with invasion and metastasis in nasopharyngeal carcinoma. Mod. Pathol. 24, 175–184 10.1038/modpathol.2011.22 [DOI] [PubMed] [Google Scholar]
- 85. Kondratiev S., Duraisamy S., Unitt C. L., Green M. R., Pinkus G. S., Shipp M. A., Kutok J. L., Drapkin R. I., and Rodig S. J. (2011) Aberrant expression of the dendritic cell marker TNFAIP2 by the malignant cells of Hodgkin lymphoma and primary mediastinal large B-cell lymphoma distinguishes these tumor types from morphologically and phenotypically similar lymphomas. Am. J. Surg. Pathol. 35, 1531–1539 10.1097/PAS.0b013e31822bd476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liu Z., Wei S., Ma H., Zhao M., Myers J. N., Weber R. S., Sturgis E. M., and Wei Q. (2011) A functional variant at the miR-184 binding site in TNFAIP2 and risk of squamous cell carcinoma of the head and neck. Carcinogenesis 32, 1668–1674 10.1093/carcin/bgr209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xu Y., Ma H., Yu H., Liu Z., Wang L. E., Tan D., Muddasani R., Lu V., Ajani J. A., Wang Y., and Wei Q. (2013) The miR-184 binding-site rs8126 T>C polymorphism in TNFAIP2 is associated with risk of gastric cancer. PLoS One 8, e64973 10.1371/journal.pone.0064973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chen C. C., Liu H. P., Chao M., Liang Y., Tsang N. M., Huang H. Y., Wu C. C., and Chang Y. S. (2014) NF-κB-mediated transcriptional upregulation of TNFAIP2 by the Epstein-Barr virus oncoprotein, LMP1, promotes cell motility in nasopharyngeal carcinoma. Oncogene 33, 3648–3659 10.1038/onc.2013.345 [DOI] [PubMed] [Google Scholar]
- 89. Zhang J., Yu H., Zhang Y., Zhang X., Zheng G., Gao Y., Wang C., and Zhou L. (2014) A functional TNFAIP2 3′-UTR rs8126 genetic polymorphism contributes to risk of esophageal squamous cell carcinoma. PLoS One 9, e109318 10.1371/journal.pone.0109318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cheng Z., Wang H. Z., Li X., Wu Z., Han Y., Li Y., Chen G., Xie X., Huang Y., Du Z., and Zhou Y. (2015) MicroRNA-184 inhibits cell proliferation and invasion, and specifically targets TNFAIP2 in glioma. J. Exp. Clin. Cancer Res. 34, 27 10.1186/s13046-015-0142-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Barzilai S., Blecher-Gonen R., Barnett-Itzhaki Z., Zauberman A., Lebel-Haziv Y., Amit I., and Alon R. (2016) M-sec regulates polarized secretion of inflammatory endothelial chemokines and facilitates CCL2-mediated lymphocyte transendothelial migration. J. Leukoc. Biol. 99, 1045–1055 10.1189/jlb.3VMA0915-427R [DOI] [PubMed] [Google Scholar]
- 92. Xie Y., and Wang B. (2017) Downregulation of TNFAIP2 suppresses proliferation and metastasis in esophageal squamous cell carcinoma through activation of the Wnt/β-catenin signaling pathway. Oncol. Rep. 37, 2920–2928 10.3892/or.2017.5557 [DOI] [PubMed] [Google Scholar]
- 93. Armijo-Weingart L., and Gallo G. (2017) It takes a village to raise a branch: Cellular mechanisms of the initiation of axon collateral branches. Mol. Cell Neurosci. 84, 36–47 10.1016/j.mcn.2017.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mattila P. K., and Lappalainen P. (2008) Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9, 446–454 10.1038/nrm2406 [DOI] [PubMed] [Google Scholar]
- 95. Sainath R., and Gallo G. (2015) Cytoskeletal and signaling mechanisms of neurite formation. Cell Tissue Res. 359, 267–278 10.1007/s00441-014-1955-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Rytlewski J. A., Alejandra Aldon M., Lewis E. W., and Suggs L. J. (2015) Mechanisms of tubulogenesis and endothelial phenotype expression by MSCs. Microvasc. Res. 99, 26–35 10.1016/j.mvr.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hubbard M. J., McHugh N. J., and Carne D. L. (2000) Isolation of ERp29, a novel endoplasmic reticulum protein, from rat enamel Cells: evidence for a unique role in secretory-protein synthesis. Eur. J. Biochem. 267, 1945–1957 10.1046/j.1432-1327.2000.01193.x [DOI] [PubMed] [Google Scholar]
- 98. Guo W., Qu F., Xia L., Guo Q., Ying X., and Ding Z. (2007) Identification and characterization of ERp29 in rat spermatozoa during epididymal transit. Reproduction 133, 575–584 10.1530/REP-06-0301 [DOI] [PubMed] [Google Scholar]
- 99. Lock J. T., Parker I., and Smith I. F. (2016) Communication of Ca2+ signals via tunneling membrane nanotubes is mediated by transmission of inositol trisphosphate through gap junctions. Cell Calcium 60, 266–272 10.1016/j.ceca.2016.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Patil S., Kumar R., Deshpande S., Samal S., Shrivastava T., Boliar S., Bansal M., Chaudhary N. K., Srikrishnan A. K., Murugavel K. G., Solomon S., Simek M., Koff W. C., Goyal R., Chakrabarti B. K., and Bhattacharya J. (2016) Conformational epitope-specific broadly neutralizing plasma antibodies obtained from an HIV-1 clade C-infected elite neutralizer mediate autologous virus escape through mutations in the V1 loop. J. Virol. 90, 3446–3457 10.1128/JVI.03090-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wei X., Decker J. M., Liu H., Zhang Z., Arani R. B., Kilby J. M., Saag M. S., Wu X., Shaw G. M., and Kappes J. C. (2002) Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46, 1896–1905 10.1128/AAC.46.6.1896-1905.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wei X., Decker J. M., Wang S., Hui H., Kappes J. C., Wu X., Salazar-Gonzalez J. F., Salazar M. G., Kilby J. M., Saag M. S., Komarova N. L., Nowak M. A., Hahn B. H., Kwongk P. D., and Shaw G. M. (2003) Antibody neutralization and escape by HIV-1. Nature 422, 307–312 10.1038/nature01470 [DOI] [PubMed] [Google Scholar]
- 103. Potriquet J., Laohaviroj M., Bethony J. M., and Mulvenna J. (2017) A modified FASP protocol for high-throughput preparation of protein samples for mass spectrometry. PLoS One 12, e0175967 10.1371/journal.pone.0175967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 10.1093/nar/gkv1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Szklarczyk D., Morris J. H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N. T., Roth A., Bork P., Jensen L. J., and von Mering C. (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.