Abstract
Eukaryotic cells organize their intracellular components into organelles that can be membrane-bound or membraneless. A large number of membraneless organelles, including nucleoli, Cajal bodies, P-bodies, and stress granules, exist as liquid droplets within the cell and arise from the condensation of cellular material in a process termed liquid–liquid phase separation (LLPS). Beyond a mere organizational tool, concentrating cellular components into membraneless organelles tunes biochemical reactions and improves cellular fitness during stress. In this review, we provide an overview of the molecular underpinnings of the formation and regulation of these membraneless organelles. This molecular understanding explains emergent properties of these membraneless organelles and shines new light on neurodegenerative diseases, which may originate from disturbances in LLPS and membraneless organelles.
Keywords: subcellular organelle, organelle, amyotrophic lateral sclerosis (ALS) (Lou Gehrig disease), RNA binding protein, chaperone, Cajal body, disaggregase, liquid–liquid phase separation, nucleolus, stress granule
Introduction
In The Origin of Life, Soviet biochemist Alexander Oparin (1) proposed that life originated as coacervate drops of organic materials. The theory was grounded in the simple observation that droplets of organic molecules coalesce spontaneously from an otherwise dilute solution. Oparin's coacervate idea eventually lost support because it failed to account for the membrane barriers that all cells use to separate inside from out and that eukaryotic cells use to further compartmentalize their cellular biochemistry inside membrane-bound organelles (1). However, cells also organize components into nonmembrane-bound organelles, suggesting that Oparin's coacervate idea deserves a second look (2–4). In fact, many cellular organelles are condensates of protein, nucleic acid, or both. In the nucleus, these include nucleoli, Cajal bodies, nuclear speckles, paraspeckles, histone–locus bodies, nuclear gems, and promyelocytic leukemia (PML)2 bodies (5–7). The cytoplasm also contains several membraneless organelles, including P-bodies, stress granules, and germ granules (6, 8). In this review, we highlight advances in our understanding of the molecular language of these membraneless organelles with respect to how they form, what functions they serve, what rules regulate them, and how their dysregulation may contribute to human disease.
Membraneless organelles are liquids that organize the cell
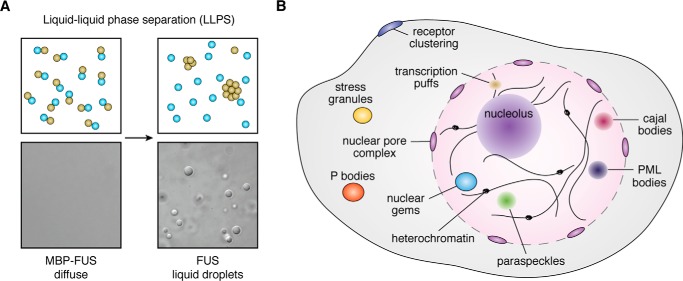
Early evidence that membraneless organelles may behave as liquids came from study of the Caenorhabditis elegans germ granule, or P granule. P granules are collections of RNA and RNA-binding proteins (RBPs) that accumulate on the posterior side of the C. elegans zygote before the cell divides into a posterior and anterior cell (9, 10). By fluorescently labeling a constitutive P-granule protein, Hyman and co-workers (9) discovered that P granules display liquid-like properties: the granules are spherical, fuse with one another, deform under shear stress, have fast internal rearrangement as assessed by recovery after photobleaching, and drip off the surface of the nucleus like a liquid. These observations led to the conclusion that P granules are liquid droplets inside the cell that form via a process called liquid–liquid phase separation (LLPS) (Fig. 1A). Burgeoning evidence now suggests that a wide range of membraneless structures–from ribonucleoprotein (RNP) granules like the nucleolus to centrosomes and clusters of signaling molecules on membranes (Fig. 1B)–exhibit liquid-like properties and coalesce through an LLPS mechanism (11–18).
Figure 1.
LLPS phase separation in vitro and in vivo. A, in a mixture of two types of molecules, LLPS leads to the formation of two phases akin to droplets of oil appearing from a mixture of oil and water. Proteins can undergo a similar phase separation. In this case, the RBP FUS (olive circles) undergoes LLPS upon cleavage of the maltose-binding protein (MBP) tag (cyan circles) and forms liquid droplets that are enriched in FUS compared with the surrounding medium. B, LLPS underpins the biogenesis of a wide array of membraneless organelles within cells. Depicted here is a nonexhaustive list of these organelles.
Phase separation and transition: liquids, gels, and crystals
The example of a salad dressing illustrates a simplified version of LLPS (16). Even after a vigorous shake, the oil and water in the salad dressing separate into a demixed two-phase system that has a lower free energy than the fully mixed state. This type of demixing is often called LLPS or a phase transition. Two types of interactions contribute to the process: the homotypic interactions between two molecules of oil or two molecules of water and the heterotypic interactions between a water and oil molecule. Entropy-driven mixing is disfavored due to the higher strength of the homotypic interactions over the heterotypic interactions, which leads to a phase-separated two-state system of lower free energy (16, 19). This simple example of phase separation extends more generally to solutions of polymers, for which the physics of LLPS has been well described (20–23). As polymers, proteins and nucleic acids are subject to the same underlying physics of LLPS (Fig. 1A) (19).
The concept that proteins undergo phase transitions is not novel, especially not to protein crystallographers whose work relies on coaxing proteins into crystals and who often observe gels, aggregations, and phase separation of proteins as side products of the crystallization process. As an example, lysozyme undergoes LLPS, gelation, and crystallization depending on certain conditions of temperature, salt concentration, and protein concentration (24, 25). Wang et al. (26) have more recently demonstrated that oligomeric peptides undergo LLPS in vitro, with LLPS stimulated by low temperature, crowding agents such as polyethylene glycol (PEG), and pH close to the pI. Together, the observations that both peptides and well-folded globular proteins undergo liquid demixing in vitro indicate that many if not all proteins can undergo LLPS in conducive environmental conditions. It is not our intention to trivialize the finding that proteins undergo LLPS but rather to point out that this property extends to all proteins and polymers in general. Indeed, RNAs can also phase separate in vitro (27, 28). The critical question then is to understand what makes phase separation biologically consequential and achievable within the cellular environment, which we address in the next two sections.
Biological consequences of phase separation
Life has harnessed the ability of proteins and other biopolymers to phase separate into liquids and, in some cases, further transition to gels and solids. We highlight some of the emergent biological properties of phase-separated compartments below.
Organization
First, LLPS serves as a dynamic organizing principle that enables cells to spatiotemporally compartmentalize specific biochemistry, provide specific infrastructure, or both (29). LLPS enables compartmentalization within a boundary while still allowing for both internal rearrangement and diffusion of biomolecules into and out of the compartment (29). For example, neurons have postsynaptic densities (PSDs), which are protein-rich compartments on the intracellular side of the postsynaptic plasma membrane that undergo remodeling in protein composition in response to long-term potentiation, i.e. the persistent strengthening of synapses due to recent patterns of activity, which underlies learning and memory (30, 31). Zeng et al. (30) propose a phase-separation model for the formation and remodeling of PSDs with supporting evidence that two major protein components of the PSD, SynGAP and PSD-95, can form liquid droplets in vitro. Because neurons have large surface areas, and consequently a large space for protein diffusion, spatially confining molecules involved in the same biochemical pathways poses a challenge. The formation of a PSD through a phase-separation mechanism allows neurons to locally concentrate protein without having to globally up-regulate protein synthesis.
In a similar vein, neuronal mRNP-granule assembly mediated by LLPS of low-complexity domains of ataxin 2 is critical for long-term memory formation in Drosophila (32, 33). This mRNP-granule–driven mechanism of long-term plasticity differs from how another RBP, CPEB/Orb2, underlies long-term potentiation. CPEB/Orb2 forms self-templating amyloid or prion conformers that directly stimulate synaptic mRNA translation (34–36). Thus, different RBPs may function via distinct assembly mechanisms and different material phases to encode long-term memories.
Neurons also utilize LLPS for functional purposes in the tight but dynamic clustering of neurotransmitter-laden synaptic vesicles (SVs) at synapses (37). These clusters serve as a replenishable pool of SVs, which can be rapidly mobilized for exocytosis during periods of heightened synaptic activity. It had remained unclear how SVs could remain motile while being confined in these clusters. It is now suggested that the physiological mediator of SV clustering, synapsin, forms a liquid phase that connects and recruits SVs in these clusters (37). This phase can be rapidly dispersed via synapsin phosphorylation by calcium/calmodulin-dependent protein kinase II (CaMKII) (37), which would emancipate SVs en masse for rapid bursts of exocytosis upon synaptic stimulation.
Phase separation is also implicated in transcription, both at the level of transcriptional activation and repression. In Drosophila polytene cells, for instance, the application of stresses like heat shock induces formation of transcription puffs at the sites of heat-shock protein (Hsp) genes where active transcription occurs (38–40). Studies on the recruitment of proteins, such as RNA polymerase II and topoisomerase, to these sites led Zobeck et al. (40) to originally propose a model in which a porous transcription compartment forms at the Hsp gene loci. In retrospect, these data support a phase-separation model for transcriptional control (41). A phase-separation model has also been proposed for the clustering of enhancer elements in DNA together with coactivator proteins to form super enhancers (42–45). Moreover, prion-like domains (PrLDs) of transcription factors can cluster into dynamic hubs that stabilize DNA binding and recruit chromatin-remodeling factors and RNA polymerase II (46–49). These hubs can manifest as phase-separated structures at elevated transcription-factor expression levels (46–49). LLPS also functions in transcriptional repression. For example, heterochromatin-mediated gene silencing is driven via compartmentalization of condensed chromatin into phase-separated liquid droplets formed by heterochromatin protein 1α (50, 51). The involvement of phase separation in regulating genome architecture and transcriptional output provides an exciting new avenue of research.
Finally, it is important to note that not all membraneless organelles are fully liquids; many likely exist along a continuum from more liquid-like to more gel-like, depending on the interaction strength of the constituents (29). On the gel side of the spectrum is an additional example of an organizational role for phase separation: the nuclear-pore complex (NPC). The central channel of the NPC is a gel-like, phase-separated structure that organizes the cell by acting as a barrier to diffusion of molecules above 30–40 kDa into or out of the nucleus (52–54). Similar selective-permeability barriers also form at the base of primary cilia (55, 56). Thus, depending on the structural, functional, or organizational need, the cell employs phase separation that spans from more dynamic, liquid-like compartments to more static, gel-like compartments. For example, globular S-crystallin proteins of different sizes assemble into a gel of varying density, thereby establishing a refractive-index gradient that forms the parabolic lens of the squid eye (57). At the extreme end of the spectrum, stable solid phases composed of amyloid or prion conformers are utilized as with CPEB/Orb2 prions in long-term potentiation (35, 36), Xvelo amyloids in Balbiani bodies that specify germline identity (58), or transient Rim4 amyloids in meiotic control (59–61).
Tuning reactions
Membraneless organelles likely tune and accelerate biochemical reactions in vivo in a manner akin to how various synthetic chemical reactions can be accelerated in microdroplets in vitro (62). The specific microenvironment within the liquid phase may serve to tune reaction rates and biochemical activities inside membraneless organelles. Phase separation can increase the concentration of certain molecules within dense liquid condensates compared with the surrounding solution by as much as 2 orders of magnitude (13). Given the dependence of reaction rates on reactant concentrations, achieving a locally high concentration of molecules due to phase separation can be a biological mechanism for increasing reaction rates. This prediction has been demonstrated in vitro by using an aqueous two-phase system to concentrate RNA substrate into liquid droplets and measuring the rate of substrate cleavage by a ribozyme (63). Concentrating the RNA and ribozyme into dense liquid droplets increased the reaction rate, suggesting that coacervation inside a cell can have a similar effect (63). Nuclear RNP granules called Cajal bodies provide one such in vivo example. Cajal bodies are the sites of assembly of the U4/U6·U5 tri-snRNP complex, which forms 11 times more efficiently within Cajal bodies than in the surrounding nucleoplasm (64).
Beyond increasing reaction rates, LLPS may also tune a biochemical process by acting as a filter to regulate which molecules enter a liquid droplet and which molecules stay out. In the case of Cajal bodies, only the fully formed U4/U6·U5 tri-snRNP complex can leave the nuclear body, whereas the di-snRNP complex cannot, which enables selective accumulation of a reactant into a confined space (65). A model of membraneless organelles acting as a filter also applies to the partitioning of RNA, which can tune the type of RNA chemistry that occurs in the organelle. RNAs can influence the compositional specificity of intracellular phases, with Langdon et al. (66) showing that RNA structure and RNA–RNA interactions affect which RNAs partition into liquid droplets. There is other evidence that the length of RNA affects which RNAs become more concentrated in liquid droplets, with longer RNAs partitioning more effectively into the droplet phase (63). Meanwhile, Nott et al. (67) have discovered that the microenvironment within phase-separated liquid droplets favors melting of double-stranded nucleic acids, stabilization of single-strand RNA secondary structure, and partitioning of RNA into droplets based on the stability of folding rather than the length (68). The discrepancy in the length dependence of RNA may be because Nott et al. (67) used liquid droplets arising from RBPs for their study, whereas Strulson et al. (63) used an aqueous two-phase system. Regardless, these findings present important steps toward understanding the molecular determinants of phase separation, some of which will be discussed later. The length dependence of RNA partitioning into liquid droplets is particularly interesting in light of data that local protein concentration and RNA length alter the binding mode and RNA-remodeling activity of the RNA helicases LAF-1 and DDX3X, both of which partition into membraneless organelles in vivo (69). Examining how tuning protein and RNA partitioning into membraneless organelles can modulate organelle activity will be an important avenue of further research.
Cellular fitness
One of the emergent properties of LLPS is that it is environmentally tunable and can thus play a cytoprotective role by sensing and responding to stress (29). Protein folding within the crowded intracellular environment is a challenge that is accentuated by cellular stresses that may trigger protein misfolding (35). The formation of reversible, phase-separated structures enables cells to store their proteins and RNAs temporarily in a manner that allows for their rapid recovery after dissipation of the stress. In yeast, the prion protein, Sup35, acts as a pH sensor and forms liquid condensates that undergo a phase transition to gels in response to a stress-induced drop in cytoplasmic pH (70). Upon stress, the formation of these Sup35 gels are protective and allow the yeast to better recover from the stress (70). In a similar manner, yeast poly(A)-binding protein (Pab1) acts as a sensor for pH and thermal stresses (71). In response to stress, Pab1 releases its bound mRNAs (which enables translation of key stress-response transcripts) and forms reversible, phase-separated hydrogels (71). Complementary findings have been made with another yeast RBP, Pub1 (72). Indeed, in response to thermal stress, yeasts form assemblages of functional proteins held together by weak interactions (73). These assemblages dissolve when the stress subsides, allowing for recovery of cellular proteins without widespread misfolding or degradation (73). Depending on the type of stress, assemblage dissolution can be spontaneous or may require protein disaggregases, such as Hsp104 (70, 72, 74). This controlled and reversible phase separation of mature proteins likely represents an adaptive strategy to stress, and it contrasts with previous models where stress-induced aggregates were thought to be disordered accumulations of misfolded, denatured proteins (73). Results in yeast also extend to mammalian cells, where stress-induced stalling of translation leads to the condensation of protein and RNA into stress granules, which are dissolved after stress by Hsp110, Hsp70, and Hsp40 (8, 75). Importantly, stress granules protect against cellular senescence by sequestering PAI-1, an established promoter of senescence (76). Overall, the ability of cells to form assemblages of proteins and nucleic acids in response to stress appears to be a conserved mechanism for cells to weather deleterious conditions.
Molecular language of phase separation
Although many if not all proteins can undergo phase transitions in vitro, not all proteins do so under physiological conditions. One of the common features of proteins that undergo phase separation in a biologically meaningful manner is the presence of multivalent binding domains, which we discuss below.
Multivalency: the key principle
The overarching property of proteins that phase separate is multivalency in interacting partners. Li et al. demonstrated this important principle by creating model proteins composed of tandem repeats of either a ligand or its binding partner (13). Combining repeats of an SH3 domain and its proline-rich motif (PRM) binding partner readily initiated phase separation of the proteins into liquid droplets (13). Increasing the interaction strength of the proteins by increasing the number of repeats of the two domains led to gelation of the liquid droplets (13). In this system, it is specific multivalent protein–protein interactions that drive phase separation.
Intrinsically disordered domains
Multivalency can arise from protein–protein interactions between ordered domains (13). However, intrinsically-disordered domains represent another method for achieving multivalency and often contain multiple short-linear motifs (SLiMs) that mediate protein–protein interactions (13). Our understanding of the molecular determinants of phase transitions increased with the discovery that biotinylated isoxasole reversibly precipitates many components of RNP granules (77, 78). The presence of low complexity PrLDs and RNA-recognition motifs (RRMs) are common denominators for many of the proteins precipitated, and in the case of TIA-1, the presence of a PrLD was sufficient for precipitation (77). This observation highlighted the importance of disordered regions, especially PrLDs, as a determinant of phase separation.
PrLDs represent a subset of low-complexity domains that show similar amino acid composition to yeast prion domains (79–85). These domains are enriched in polar, uncharged amino acids, such as asparagine (Asn), glutamine (Gln), tyrosine (Tyr), and serine (Ser), as well as glycine (Gly) (79–85). Yeast prion domains enable certain yeast proteins such as Sup35, Ure2, and Rnq1 to form prions, infectious proteins that usually propagate via self-templating amyloid forms (34). Typically, amyloid fibrils are highly-stable cross–β-structures, which represent an extreme form of phase separation to solid phases that are difficult to reverse (35). Indeed, specialized protein disaggregases such as Hsp104 or Hsp110, Hsp70, and Hsp40 are typically required to reverse their assembly (86–89). The precise features of prion domains and PrLDs that enable them to form phase-separated liquids, gels, or prions are still being delineated (79, 90–95).
In humans, of the 240 genes that encode proteins with a PrLD, a remarkable 72 encoded RBPs (84). These include FUS, TDP-43, TAF15, EWSR1, hnRNPA1, hnRNPA2, and TIA-1, which are components of RNP granules that are heavily implicated in neurodegenerative disease (82) and are precipitated by the biotinylated isoxasole compound (77). At high protein concentrations, the PrLD mediates the phase transition of FUS and hnRNPA1 into hydrogels in vitro that bind the PrLD of other RNP granule components. This observation led Kato et al. (77) to posit that the ability of low-complexity domains to reversibly form labile amyloid-like states lies at the crux of RNP granule formation. Numerous studies have since corroborated the importance of intrinsically-disordered domains, especially of PrLDs, in the formation of phase-separated membraneless organelles (11, 96–103). In some cases, deletion of the PrLD of key RBPs (e.g. TIA-1 and FUS) completely abrogates the formation of RNP granules (102, 104, 105). The natural tendency of PrLDs to engage in promiscuous interactions and aggregation promotes phase separation.
Evidence has also emerged that PrLDs may interact with another type of intrinsically disordered domain, termed RGG domains, to drive phase separation (106–110). RGG domains are enriched for arginine and glycine residues (111), can bind RNA and (112, 113), and are often found in RBPs with PrLDs (85). Indeed, for FUS and related RBPs, LLPS is elicited effectively via multivalent interactions between PrLD tyrosines and RGG arginines (106–108). These contacts are, in turn, modulated by negatively charged residues (107). Glycines confer liquidity, whereas glutamines and serines elicit gelation (107). Thus, a precise molecular grammar for phase separation by FUS and related RBPs begins to materialize (107).
RNA- and DNA-binding domains
RBPs present a special class of proteins that have biologically relevant phase behaviors. Many membraneless organelles are RNP granules that perform various RNA-processing activities, consist of RBPs and RNA, and assemble via LLPS of RBPs and RNA (8, 114). The RBPs within these granules contain multiple multivalent domains, including RRMs and intrinsically-disordered regions, which work together synergistically to modulate phase behavior (106–108, 110, 113, 115). For many of these RBPs, the purified proteins alone undergo LLPS in vitro (11, 101, 116, 117), and the intrinsically-disordered regions of these proteins are sufficient for droplet formation (99, 118). However, phase separation by the intrinsically-disordered region alone can lack the additional levels of regulation that arise from the presence additional multivalent domains like RRMs, RGG domains, and oligomerization domains (106, 107, 110, 113, 115, 119). The ability to bind to multivalent scaffolds, such as DNA and RNA, through RRMs, zinc fingers, or other nucleic acid–binding domains presents another common characteristic of proteins that undergo LLPS (29). The role of RNA as a scaffold for phase separation is evident from studies on several RBPs, including FUS (103) and Whi3, a fungal RBP that regulates nuclear division and cell polarity (120). In vitro, RNAs that bind Whi3 promote Whi3 phase separation (120) and encipher RNP granule identity (66). Mutations in the Whi3 RRM that abrogate RNA binding also prevent RNA-stimulated phase transitions of Whi3, suggesting that the RRM enables multiple Whi3 proteins to bind to the same RNA (120).
Oligomerization domains
Protein valency increases with the presence of oligomerization domains. For example, TDP-43, a highly expressed nuclear RBP, contains an N-terminal domain that forms oligomers (117, 121–124). Recently, Wang et al. (117) established that polymerization of the N-terminal domain promotes LLPS of TDP-43 in vitro and that a single phosphomimetic mutant in the N-terminal domain can reduce the propensity of TDP-43 to phase separate. The ability of oligomerization domains to nucleate a locally high concentration of a protein to promote phase separation has been used by Shin et al. (125) to form optogenetically controlled liquid droplets in vivo. Here, the intrinsically-disordered regions of several RBPs are fused to Cry2, a protein that oligomerizes in response to blue light (125). Oligomerization of Cry2 elicited by blue light nucleates intracellular droplets of the fusion proteins (125). Thus, environmentally-responsive oligomerization domains can promote phase separation in response to specific environmental cues.
Weak interactions maintain membraneless organelles in phase
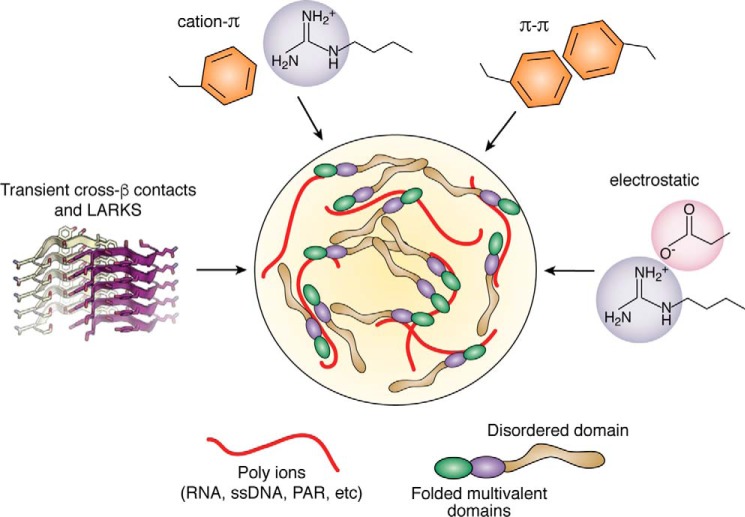
The essential physics of polymer phase separation are well-established and help inform biological phase separation (19). Concentrating molecules into a confined space can carry an energetic cost. Numerous weak interactions work together to counteract the entropic cost for phase separation as well the interfacial free energy cost to create a phase boundary. The molecular interactions found to be important in phase separation include π–π stacking, cation–π interactions, charge–charge interactions, and transient cross–β-contacts (Fig. 2).
Figure 2.
Critical interactions that drive LLPS. The interactions important in LLPS include cation–π, π–π, electrostatic, and transient cross–β-contacts. Proteins that undergo LLPS are enriched for low-complexity disordered regions and multivalent domains. Polymers of ions, such as RNA, may additionally act as scaffolds or molecular seeds for LLPS. The image for transient cross–β-contacts and LARKS comes from Hughes et al. (134).
π–π interactions
Aromatic residues tyrosine (Tyr), tryptophan (Trp), and phenylalanine (Phe) as well as residues arginine (Arg), glutamine (Gln), asparagine (Asn), aspartic acid (Asp), and glutamic acid (Gln) contain delocalized π electrons in their side chains that can engage in π–π stacking (126). Work to understand the sequence features of phase-separating proteins has uncovered π–π interactions as critical (126). Using a comprehensive mutagenesis approach, for example, Pak et al. (127) uncovered that phase separation of nephrin intracellular domain depends strongly on the presence of tyrosine residues, as missense mutations to those residues reduced the ability of the protein to form liquid droplets in cells. Similar observations have been made with FUS and hnRNPA2 PrLDs in vitro (128, 129). The gel-like state of the nuclear-pore complex results from π–π interactions between phenylalanine residues in the FG repeats of nucleoporins (52, 53). Remarkably, Vernon et al. (126) have established that long-range π–π contact propensity alone can identify the majority of known phase-separating proteins, highlighting the critical role for π–π interactions in LLPS.
Cation–π interactions
Cation–π interactions occur between the positively-charged amino acids lysine and arginine and the electron-rich aromatic groups. These interactions have also gained importance as drivers of LLPS (106–108, 110, 113). For the RNA-helicase, Ddx4, cation–π interactions between FG and RG regions of the protein are drivers of protein phase separation in vitro and in vivo (130). As a caveat, interaction between Phe and Arg could also include π–π interactions, which likely contributed to the phase separation of Ddx4 as well. Surprisingly, short-range cation–π interactions are strong enough to overcome long-range charge–charge repulsion and cause two positively charged polymers to coacervate in vitro (131). It is also interesting to note that an emergent property of multiple cation–π and π–π interactions that drive LLPS is the ability to melt nucleic acid duplexes by disrupting the π–π interactions that maintain them (67).
Charge–charge neutralization
Charge–charge interactions have also gained attention as important drivers of phase separation. Oppositely charged polymers when brought together can coacervate into liquid droplets through charge neutralization, as has been shown for mixtures of RNA and cationic peptides (4, 132). Although this may be a simplified artificial system, the phenomenon of long-range charge–charge interactions driving phase separation has also been observed in proteins in vitro and in vivo (127, 130). An emerging theme is that it is not the presence of charged residues per se, but rather the arrangement of charged resides into stretches that is important for phase separation (127, 130, 133). Working in this manner, clusters of charged residues act akin to a multivalent domain to promote phase separation.
LARKs and transient cross–β-contacts
Several RBPs that undergo LLPS contain PrLDs. In the RBP FUS, for example, a portion of the PrLD forms fibrils in which stretches of amino acids assemble into intermolecular cross–β-sheets as typically found in amyloid fibrils (35, 95). However, recent crystallographic studies of fibrils formed by short segments of the PrLDs of RBPs that undergo LLPS have uncovered a structural difference compared with classic amyloid fibrils (134–136). Although amyloid fibrils tend to have cross–β-sheets with interdigitated amino acids that form steric zippers, fibrils formed by short segments of PrLDs of RBPs that undergo LLPS have kinked cross–β-sheets termed low-complexity aromatic–rich kinked segments (LARKS) (134). These kinked β-sheets are less thermodynamically stable than the β-sheets of amyloid fibrils, and proteins with PrLDs enriched for LARKS are found in membraneless organelles that assemble via LLPS (134). Together, these findings suggest that weak, transient cross–β-contacts might contribute to LLPS, whereas more stable cross–β-contacts contribute to pathological amyloidogenesis.
Modulators of phase separation
One of the fundamental principles of a living organism is the ability to adapt to change. Cells must constantly tune their biochemistry in response to environmental cues, and the membraneless organelles within a cell must similarly be responsive to intra- and extracellular signals. To regulate phase separation, cells rely on several processes, including post-translational modifications and seeding mechanisms.
Post-translational modification (PTMs)
PTMs provide cells with a powerful means to facilitate or antagonize LLPS in response to environmental signals (137). Indeed, SLiMs often mediate protein–protein interactions that drive phase separation and are frequently the target of regulation by PTMs (13, 138). PTMs can promote LLPS, for example, by increasing the effective valency of a protein. In the nucleus, there are membraneless organelles called PML bodies for which the PML protein acts as a scaffold (Fig. 1B). SUMOylation of PML is necessary for proper formation of PML bodies because the small ubiquitin-like modifier acts as a binding ligand that recruits other proteins, such as Daxx, into the membraneless organelle (139–141). Similarly, tyrosine phosphorylation of the protein nephrin promotes phase separation of nephrin with the protein NCK because the phosphotyrosine acts as a docking site for NCK (13). Phosphorylation of serines in the FUS PrLD fluidizes FUS droplets (115, 142). In contrast, phosphorylation can promote the disruption of phases as with the dissolution of Rim4 assemblies by Ime2 (60) and the dissolution of various nuclear membraneless organelles during mitosis by Dyrk3 (143). Likewise, arginine methylation of the RBPs Ddx4, hnRNPA2, and FUS can antagonize phase separation (99, 106, 113, 130). This list of PTMs involved in regulation of phase transitions is by no means exhaustive, but rather represents a small subset of the numerous ways that PTMs can modulate LLPS.
Seeding mechanisms
Phase transitions are concentration-dependent, switch-like phenomena that occur above a certain local critical concentration (29). Cells can promote phase separation through a nucleator that seeds a locally higher concentration of certain biomolecules to reach the necessary critical concentration. We highlight three nucleators below.
RNA
Although RBPs receive a lot of attention in phase separation, RNAs also play key roles in the formation of various membraneless organelles (28, 66, 105). The function of several membraneless organelles is intimately centered around RNA, such as mRNA decay in P-bodies, mRNA storage in stress granules, mRNA splicing in nuclear speckles, and rRNA synthesis in nucleoli (29). Indeed, RNA acts as a potent, biologically important nucleator of intracellular phase separation. As examples, RNA induces the phase separation of MEG-3, a key scaffold protein in C. elegans P granules (144), stalling of translation during a stress response exposes free mRNAs that act as nucleators for stress granules (28, 145), and Men ϵ/β noncoding (nc) RNAs seed the formation of nuclear paraspeckles (146, 147). It is noteworthy that there are many ncRNAs for which biological functions are not well known (148). It is possible that these RNAs regulate phase transition events in cells like the Men ϵ/β ncRNA.
Poly(ADP-ribose)
Besides RNA, the cell also utilizes RNA-like molecules to seed LLPS. One such molecule is poly(ADP-ribose) or PAR, a polymer of ADP-ribose monomers that is involved in the formation of several stress-triggered membraneless organelles (149). For example, PAR recruits transcription factors to the heat-shock protein locus in Drosophila polytene cells in response to heat shock and recruits DNA-damage repair factors to sites of DNA damage (40, 101, 150, 151). PAR is both a PTM and an RNA-like scaffold that assembles proteins, including FUS and TDP-43, into membraneless organelles via LLPS (101, 150, 152). PAR is also found in stress granules (153). Thus, PAR has a wide-reaching role in stress-triggered assembly of membraneless organelles.
Polyphosphates
In light of the importance of charge–charge interactions and polyanion seeds like RNA in the molecular language of phase separation, it seems plausible that other polyanions, like polyphosphates for instance, may also act as seeds for phase separation. Cremers et al. (154) have previously elucidated a role for polyphosphates in nucleating amyloids, whereas Racki et al. (155) have uncovered that polyphosphate granules assemble and coalesce during starvation-induced stress response in bacteria. The uncanny similarity between RNP-granule biogenesis via LLPS and polyphosphate granule assembly merits further exploration.
Proline cis-trans isomerization
A key feature of several phase-separating RBPs is the presence of a PrLD (84), which can often contain sporadic proline residues. Given that proline introduces kinks as a result of its constrained side-chain geometry and given that PrLDs can aggregate into amyloid fibrils, prolines may serve as a natural fluidizer in PrLDs and other low-complexity domains to prevent aberrant aggregation. Proline isomerization may then serve a possible role in regulating phase transitions mediated by low-complexity domains. Indeed, peptidyl-prolyl cis-trans isomerases colocalize with stress granules, bind hydrogels formed from the PrLDs of RBPs, and increase the solvent accessibility of certain residues in hnRNPA2 as it assembles into fibrils (129). Importantly, peptidyl-prolyl cis-trans isomerases can also function as protein disaggregases with activity against amyloid fibrils (87, 156). Thus, proline cis-trans isomerization may be another mechanism by which cells modulate the phase behavior of proteins.
Aberrant phase transitions in neurodegenerative disease
A hallmark of several neurodegenerative diseases is aberrant protein aggregation: α-synuclein aggregates in Parkinson's disease, β-amyloid and tau in Alzheimer's disease, and TDP-43 and the FET family of proteins in amyotrophic later sclerosis (ALS) and frontotemporal dementia (FTD) (35). For RBPs implicated in ALS and FTD, LLPS provides a mechanistic link between normal cellular function and disease phenotypes.
FUS, TDP-43, hnRNPA1, and TIA-1 are among the RBPs that are associated with ALS and FTD, which coalesce into membraneless organelles called stress granules (83). Observations of purified FUS, hnRNPA1, and TIA-1 uncovered that these proteins form dynamic liquid droplets in vitro that age over time to become more static, fibrillar aggregates (11, 101, 157). The conversion from a liquid state to a more aggregated state has been termed an aberrant phase transition (101). The final aggregated form of the protein bears resemblance to the protein aggregates found in patients with ALS and FTD. Fibrillization can occur within the condensed liquid state, suggesting that concentrating these RBPs in membraneless organelles via LLPS as part of normal cellular biology may have the inadvertent effect of triggering protein aggregation over time (11, 83, 101). Indeed, data indicating that aggregates of these RBPs are immunoreactive for other components of stress granules have provided further evidence that stress granules may be the sites of disease biogenesis (83, 158). However, RBPs with PrLDs that are connected to neurodegenerative disease like FUS, TDP-43, TAF15, EWSR1, and hnRNPA1 are intrinsically aggregation-prone (81, 109, 159–161). Thus, pathological aggregation could also be nucleated outside of stress granules. Pathological aggregates could then subsequently sequester specific stress-granule proteins.
Additional evidence connecting aberrant phase transitions to disease comes from analysis of mutations in these RBPs that are associated with hereditary forms of neurodegenerative disease. Disease-associated mutations often exacerbate protein aggregation and alter the phase behavior of the protein (11, 101). For example, ALS and multisystem proteinopathy-associated mutations in the PrLD of hnRNPA1 and hnRNPA2 increase the amyloidogenicity of these proteins and accelerate fibrillization (81). Additionally, ALS-linked mutations in TDP-43 also promote aggregation and alter TDP-43 phase behavior (118, 152, 159). The PrLDs of these proteins normally form weak, transient interactions with each other in the liquid droplets. Some disease-associated mutations strengthen the otherwise transient interactions in the PrLD, leading to less dynamic droplets and RNP granules (11, 101, 157, 162). Likewise, the arginine-rich, dipeptide-repeat proteins, poly-PR and poly-GR, produced by repeat-associated non-ATG translation of the ALS/FTD-causing G4C2 repeat expansion of C9orf72 also accelerate aberrant phase transitions of RBPs with PrLDs and perturb the phases of several membraneless organelles (132, 163–165). The protein aggregates seen in disease likely represent an end-stage phenotype after aberrant phase separation has overwhelmed the cellular machinery that ordinarily reverses these altered phases.
Counteracting neurodegenerative diseases with knowledge of phase separation
Neurodegenerative disease like ALS and FTD lack effective therapies. Recent advances in our understanding of how altered phase transitions contribute to these disorders reveal several potential avenues for therapeutics. These include: 1) enhancing the machinery already present inside cells to maintain RNP-granule dynamics; and 2) targeting the factors that recruit RBPs to RNP granules.
The cell has various molecular chaperones that remodel misfolded proteins and contribute to proper maintenance of RNP-granule dynamics (166). Nuclear-import receptors also act as chaperones and dissolvases that reverse LLPS and aberrant phase separation of their RBP cargo (106, 110, 113, 119). Small-molecule enhancers of these chaperones or de novo–designed chaperone proteins with enhanced disaggregase activity thus present promising approaches for targeting neurodegenerative diseases (86, 87, 167–170).
Targeting the specific factors that recruit neurodegenerative disease-associated RBPs to RNP granules may also therapeutically tune the accumulation of these RBPs inside stress granules. For example, knockdown of Ataxin 2 reduces accumulation of TDP-43 in stress granules and is therapeutic in reducing TDP-43 toxicity in several ALS models (171, 172). Additionally, molecular seeds like PAR that nucleate RNP granules can also be potential targets for therapies using antisense oligonucleotides or small-molecule inhibitors of specific PAR polymerases (101, 152) or methods to up-regulate specific PAR glycohydrolases (173). Finally, RNA acts both as a molecular seed in the cell as well as a safeguard against aberrant phase separation in the nucleus where RNA concentration is higher (174). Thus, expression or delivery of certain RNAs that are particularly effective at reducing aberrant protein phase separation may also be therapeutic. Overall, we anticipate that advances in our understanding the molecular language of phase separation will ultimately enhance efforts to combat neurodegenerative diseases
Acknowledgments
We thank Lin Guo, Bede Portz, and Zach March for feedback on the manuscript, and Lin Guo for the images in Fig. 1A.
This article is part of the thematic series, Phase separation of RNA-binding proteins in physiology and disease. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PML
- promyelocytic leukemia
- LLPS
- liquid–liquid phase separation
- SV
- synaptic vesicle
- RBP
- RNA-binding protein
- ALS
- amyotrophic later sclerosis
- FUS
- fused in sarcoma
- FTD
- frontotemporal dementia
- RRM
- RNA-recognition motif
- PrLD
- prion-like domain
- PSD
- postsynaptic density
- SLiM
- short-linear motif
- PAR
- poly(ADP-ribose)
- PTM
- post-translational modification
- RNP
- ribonucleoprotein
- snRNP
- small nuclear ribonucleoprotein
- SV
- synaptic vesicle
- NPC
- nuclear-pore complex
- LARKS
- low-complexity aromatic–rich kinked segment
- nc
- noncoding.
References
- 1. Oparin A. I. (1938) The Origin of Life, McMillan, New York [Google Scholar]
- 2. Feric M., Vaidya N., Harmon T. S., Mitrea D. M., Zhu L., Richardson T. M., Kriwacki R. W., Pappu R. V., and Brangwynne C. P. (2016) Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brangwynne C. P., Mitchison T. J., and Hyman A. A. (2011) Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U.S.A. 108, 4334–4339 10.1073/pnas.1017150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aumiller W. M. Jr., and Keating C. D. (2016) Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat. Chem. 8, 129–137 10.1038/nchem.2414 [DOI] [PubMed] [Google Scholar]
- 5. Mao Y. S., Zhang B., and Spector D. L. (2011) Biogenesis and function of nuclear bodies. Trends Genet. 27, 295–306 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mitrea D. M., and Kriwacki R. W. (2016) Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 14, 1 10.1186/s12964-015-0125-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Handwerger K. E., and Gall J. G. (2006) Subnuclear organelles: new insights into form and function. Trends Cell Biol. 16, 19–26 10.1016/j.tcb.2005.11.005 [DOI] [PubMed] [Google Scholar]
- 8. Decker C. J., and Parker R. (2012) P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 4, a012286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., and Hyman A. A. (2009) Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- 10. Voronina E., Seydoux G., Sassone-Corsi P., and Nagamori I. (2011) RNA granules in germ cells. Cold Spring Harb. Perspect. Biol. 3, a002774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A. P., Kim H. J., Mittag T., and Taylor J. P. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li X. H., Chavali P. L., Pancsa R., Chavali S., and Babu M. M. (2018) Function and regulation of phase-separated biological condensates. Biochemistry 57, 2452–2461 10.1021/acs.biochem.7b01228 [DOI] [PubMed] [Google Scholar]
- 13. Li P., Banjade S., Cheng H. C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J. V., King D. S., Banani S. F., Russo P. S., Jiang Q. X., Nixon B. T., and Rosen M. K. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Toretsky J. A., and Wright P. E. (2014) Assemblages: functional units formed by cellular phase separation. J. Cell Biol. 206, 579–588 10.1083/jcb.201404124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Banjade S., and Rosen M. K. (2014) Phase transitions of multivalent proteins can promote clustering of membrane receptors. Elife 3, 10.7554/eLife.04123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hyman A. A., Weber C. A., and Jülicher F. (2014) Liquid–liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- 17. Berry J., Weber S. C., Vaidya N., Haataja M., and Brangwynne C. P. (2015) RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl. Acad. Sci. U.S.A. 112, E5237–E5245 10.1073/pnas.1509317112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Woodruff J. B., Ferreira Gomes B., Widlund P. O., Mahamid J., Honigmann A., and Hyman A. A. (2017) The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169, 1066–1077.e10 10.1016/j.cell.2017.05.028 [DOI] [PubMed] [Google Scholar]
- 19. Brangwynne C. P., Tompa P., and Pappu R. V. (2015) Polymer physics of intracellular phase transitions. Nat. Phys. 11, 899–904 10.1038/nphys3532 [DOI] [Google Scholar]
- 20. Nojima S., Shiroshita K., and Nose T. (1982) Phase separation process in polymer systems. II. Microscopic studies on a polystyrene and diisodecyl phthalate mixture. Polymer. J. 14, 289–294 10.1295/polymj.14.289 [DOI] [Google Scholar]
- 21. Bansil R. (1993) Phase separation in polymer solutions and gels. J. Phys. IV France 3, C1-225–C1-235 10.1051/jp4:1993119 [DOI] [Google Scholar]
- 22. Flory P. J., Orwoll R. A., and Vrij A. (1964) Statistical thermodynamics of chain molecule liquids. II. Liquid mixtures of normal paraffin hydrocarbons. JACS 86, 3507–3514 10.1021/ja01071a023 [DOI] [Google Scholar]
- 23. Flory P. J. (1953) Principles of Polymer Chemistry, Cornell University Press, Ithaca, NY [Google Scholar]
- 24. Muschol M., and Rosenberger F. (1997) Liquid–liquid phase separation in supersaturated lysozyme solutions and associated precipitate formation/crystallization. J. Chem. Phys. 107, 1953–1962 10.1063/1.474547 [DOI] [Google Scholar]
- 25. Dumetz A. C., Chockla A. M., Kaler E. W., and Lenhoff A. M. (2008) Protein phase behavior in aqueous solutions: crystallization, liquid–liquid phase separation, gels, and aggregates. Biophys. J. 94, 570–583 10.1529/biophysj.107.116152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y., Lomakin A., Kanai S., Alex R., and Benedek G. B. (2017) Liquid–liquid phase separation in oligomeric peptide solutions. Langmuir 33, 7715–7721 10.1021/acs.langmuir.7b01693 [DOI] [PubMed] [Google Scholar]
- 27. Jain A., and Vale R. D. (2017) RNA phase transitions in repeat expansion disorders. Nature 546, 243–247 10.1038/nature22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van Treeck B., Protter D. S. W., Matheny T., Khong A., Link C. D., and Parker R. (2018) RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. U.S.A. 115, 2734–2739 10.1073/pnas.1800038115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boeynaems S., Alberti S., Fawzi N. L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L., Tompa P., and Fuxreiter M. (2018) Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28, 420–435 10.1016/j.tcb.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zeng M., Shang Y., Araki Y., Guo T., Huganir R. L., and Zhang M. (2016) Phase transition in postsynaptic densities underlies formation of synaptic complexes and synaptic plasticity. Cell 166, 1163–1175.e12 10.1016/j.cell.2016.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sheng M., and Kim E. (2011) The postsynaptic organization of synapses. Cold Spring Harb. Perspect. Biol. 3, a005678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bakthavachalu B., Huelsmeier J., Sudhakaran I. P., Hillebrand J., Singh A., Petrauskas A., Thiagarajan D., Sankaranarayanan M., Mizoue L., Anderson E. N., Pandey U. B., Ross E., VijayRaghavan K., Parker R., and Ramaswami M. (2018) RNP-granule assembly via Ataxin-2 disordered domains is required for long-term memory and neurodegeneration. Neuron 98, 754–766.e4 10.1016/j.neuron.2018.04.032 [DOI] [PubMed] [Google Scholar]
- 33. Becker L. A., and Gitler A. D. (2018) Ataxin-2 is Droppin' some knowledge. Neuron 98, 673–675 10.1016/j.neuron.2018.05.002 [DOI] [PubMed] [Google Scholar]
- 34. Shorter J., and Lindquist S. (2005) Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 6, 435–450 10.1038/nrg1616 [DOI] [PubMed] [Google Scholar]
- 35. Chuang E., Hori A. M., Hesketh C. D., and Shorter J. (2018) Amyloid assembly and disassembly. J. Cell Sci. 131, jcs189928 10.1242/jcs.189928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Si K., and Kandel E. R. (2016) The role of functional prion-like proteins in the persistence of memory. Cold Spring Harb. Perspect. Biol. 8, a021774 10.1101/cshperspect.a021774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Milovanovic D., Wu Y., Bian X., and De Camilli P. (2018) A liquid phase of synapsin and lipid vesicles. Science 361, 604–607 10.1126/science.aat5671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ashburner M., and Bonner J. J. (1979) The induction of gene activity in Drosophila by heat shock. Cell 17, 241–254 10.1016/0092-8674(79)90150-8 [DOI] [PubMed] [Google Scholar]
- 39. Bonner J. J. (1981) Induction of Drosophila heat-shock puffs in isolated polytene nuclei. Dev. Biol. 86, 409–418 10.1016/0012-1606(81)90199-8 [DOI] [PubMed] [Google Scholar]
- 40. Zobeck K. L., Buckley M. S., Zipfel W. R., and Lis J. T. (2010) Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol. Cell 40, 965–975 10.1016/j.molcel.2010.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harlen K. M., and Churchman L. S. (2017) The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 18, 263–273 10.1038/nrm.2017.10 [DOI] [PubMed] [Google Scholar]
- 42. Pott S., and Lieb J. D. (2015) What are super-enhancers? Nat. Genet. 47, 8–12 10.1038/ng.3167 [DOI] [PubMed] [Google Scholar]
- 43. Hnisz D., Shrinivas K., Young R. A., Chakraborty A. K., and Sharp P. A. (2017) A phase separation model for transcriptional control. Cell 169, 13–23 10.1016/j.cell.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sabari B. R., Dall'Agnese A., Boija A., Klein I. A., Coffey E. L., Shrinivas K., Abraham B. J., Hannett N. M., Zamudio A. V., Manteiga J. C., Li C. H., Guo Y. E., Day D. S., Schuijers J., Vasile E., et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cho W. K., Spille J. H., Hecht M., Lee C., Li C., Grube V., and Cisse I. I. (2018) Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 10.1126/science.aar4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G. M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X., and Tjian R. (2018) Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, eaar2555 10.1126/science.aar2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boulay G., Sandoval G. J., Riggi N., Iyer S., Buisson R., Naigles B., Awad M. E., Rengarajan S., Volorio A., McBride M. J., Broye L. C., Zou L., Stamenkovic I., Kadoch C., and Rivera M. N. (2017) Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell 171, 163–178.e19 10.1016/j.cell.2017.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shorter J. (2017) Prion-like domains program Ewing's sarcoma. Cell 171, 30–31 10.1016/j.cell.2017.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kwon I., Kato M., Xiang S., Wu L., Theodoropoulos P., Mirzaei H., Han T., Xie S., Corden J. L., and McKnight S. L. (2013) Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 10.1016/j.cell.2013.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Larson A. G., Elnatan D., Keenen M. M., Trnka M. J., Johnston J. B., Burlingame A. L., Agard D. A., Redding S., and Narlikar G. J. (2017) Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Strom A. R., Emelyanov A. V., Mir M., Fyodorov D. V., Darzacq X., and Karpen G. H. (2017) Phase separation drives heterochromatin domain formation. Nature 547, 241–245 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Frey S., Richter R. P., and Görlich D. (2006) FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817 10.1126/science.1132516 [DOI] [PubMed] [Google Scholar]
- 53. Schmidt H. B., and Görlich D. (2015) Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. Elife 4, 10.7554/eLife.04251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schmidt H. B., and Görlich D. (2016) Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem. Sci. 41, 46–61 10.1016/j.tibs.2015.11.001 [DOI] [PubMed] [Google Scholar]
- 55. Kee H. L., Dishinger J. F., Blasius T. L., Liu C. J., Margolis B., and Verhey K. J. (2012) A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat. Cell Biol. 14, 431–437 10.1038/ncb2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Endicott S. J., and Brueckner M. (2018) NUP98 sets the size-exclusion diffusion limit through the ciliary base. Curr. Biol. 28, 1643–1650.e3 10.1016/j.cub.2018.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cai J., Townsend J. P., Dodson T. C., Heiney P. A., and Sweeney A. M. (2017) Eye patches: protein assembly of index-gradient squid lenses. Science 357, 564–569 10.1126/science.aal2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Boke E., Ruer M., Wühr M., Coughlin M., Lemaitre R., Gygi S. P., Alberti S., Drechsel D., Hyman A. A., and Mitchison T. J. (2016) Amyloid-like self-assembly of a cellular compartment. Cell 166, 637–650 10.1016/j.cell.2016.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Berchowitz L. E., Kabachinski G., Walker M. R., Carlile T. M., Gilbert W. V., Schwartz T. U., and Amon A. (2015) Regulated formation of an amyloid-like translational repressor governs gametogenesis. Cell 163, 406–418 10.1016/j.cell.2015.08.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Carpenter K., Bell R. B., Yunus J., Amon A., and Berchowitz L. E. (2018) Phosphorylation-mediated clearance of amyloid-like assemblies in meiosis. Dev. Cell 45, 392–405.e6 10.1016/j.devcel.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ford A. F., and Shorter J. (2015) Fleeting amyloid-like forms of Rim4 ensure meiotic fidelity. Cell 163, 275–276 10.1016/j.cell.2015.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stroberg W., and Schnell S. (2018) Do cellular condensates accelerate biochemical reactions? Lessons from microdroplet chemistry. Biophys. J. 115, 3–8 10.1016/j.bpj.2018.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Strulson C. A., Molden R. C., Keating C. D., and Bevilacqua P. C. (2012) RNA catalysis through compartmentalization. Nat. Chem. 4, 941–946 10.1038/nchem.1466 [DOI] [PubMed] [Google Scholar]
- 64. Novotný I., Blažíková M., Staněk D., Herman P., and Malinsky J. (2011) In vivo kinetics of U4/U6.U5 tri-snRNP formation in Cajal bodies. Mol. Biol. Cell 22, 513–523 10.1091/mbc.e10-07-0560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schaffert N., Hossbach M., Heintzmann R., Achsel T., and Lührmann R. (2004) RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 23, 3000–3009 10.1038/sj.emboj.7600296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Langdon E. M., Qiu Y., Ghanbari Niaki A., McLaughlin G. A., Weidmann C. A., Gerbich T. M., Smith J. A., Crutchley J. M., Termini C. M., Weeks K. M., Myong S., and Gladfelter A. S. (2018) mRNA structure determines specificity of a polyQ-driven phase separation. Science 360, 922–927 10.1126/science.aar7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nott T. J., Craggs T. D., and Baldwin A. J. (2016) Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nat. Chem. 8, 569–575 10.1038/nchem.2519 [DOI] [PubMed] [Google Scholar]
- 68. Shorter J. (2016) Membraneless organelles: phasing in and out. Nat. Chem. 8, 528–530 10.1038/nchem.2534 [DOI] [PubMed] [Google Scholar]
- 69. Kim Y., and Myong S. (2016) RNA Remodeling activity of DEAD box proteins tuned by protein concentration, RNA length, and ATP. Mol. Cell 63, 865–876 10.1016/j.molcel.2016.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Franzmann T. M., Jahnel M., Pozniakovsky A., Mahamid J., Holehouse A. S., Nüske E., Richter D., Baumeister W., Grill S. W., Pappu R. V., Hyman A. A., and Alberti S. (2018) Phase separation of a yeast prion protein promotes cellular fitness. Science 359, eaao5654 10.1126/science.aao5654 [DOI] [PubMed] [Google Scholar]
- 71. Riback J. A., Katanski C. D., Kear-Scott J. L., Pilipenko E. V., Rojek A. E., Sosnick T. R., and Drummond D. A. (2017) Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168, 1028–1040.e19 10.1016/j.cell.2017.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kroschwald S., Munder M. C., Maharana S., Franzmann T. M., Richter D., Ruer M., Hyman A. A., and Alberti S. (2018) Different material states of Pub1 condensates define distinct modes of stress adaptation and recovery. Cell Rep. 23, 3327–3339 10.1016/j.celrep.2018.05.041 [DOI] [PubMed] [Google Scholar]
- 73. Wallace E. W., Kear-Scott J. L., Pilipenko E. V., Schwartz M. H., Laskowski P. R., Rojek A. E., Katanski C. D., Riback J. A., Dion M. F., Franks A. M., Airoldi E. M., Pan T., Budnik B. A., and Drummond D. A. (2015) Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162, 1286–1298 10.1016/j.cell.2015.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kroschwald S., Maharana S., Mateju D., Malinovska L., Nüske E., Poser I., Richter D., and Alberti S. (2015) Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife 4, e06807 10.7554/eLife.06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cherkasov V., Hofmann S., Druffel-Augustin S., Mogk A., Tyedmers J., Stoecklin G., and Bukau B. (2013) Coordination of translational control and protein homeostasis during severe heat stress. Curr. Biol. 23, 2452–2462 10.1016/j.cub.2013.09.058 [DOI] [PubMed] [Google Scholar]
- 76. Omer A., Patel D., Lian X. J., Sadek J., Di Marco S., Pause A., Gorospe M., and Gallouzi I. E. (2018) Stress granules counteract senescence by sequestration of PAI-1. EMBO Rep. 19, e44722 10.15252/embr.201744722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kato M., Han T. W., Xie S., Shi K., Du X., Wu L. C., Mirzaei H., Goldsmith E. J., Longgood J., Pei J., Grishin N. V., Frantz D. E., Schneider J. W., Chen S., Li L., et al. (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Han T. W., Kato M., Xie S., Wu L. C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., and McKnight S. L. (2012) Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- 79. Alberti S., Halfmann R., King O., Kapila A., and Lindquist S. (2009) A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137, 146–158 10.1016/j.cell.2009.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cushman M., Johnson B. S., King O. D., Gitler A. D., and Shorter J. (2010) Prion-like disorders: blurring the divide between transmissibility and infectivity. J. Cell Sci. 123, 1191–1201 10.1242/jcs.051672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kim H. J., Kim N. C., Wang Y. D., Scarborough E. A., Moore J., Diaz Z., MacLea K. S., Freibaum B., Li S., Molliex A., Kanagaraj A. P., Carter R., Boylan K. B., Wojtas A. M., Rademakers R., et al. (2013) Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 10.1038/nature11922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. King O. D., Gitler A. D., and Shorter J. (2012) The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 1462, 61–80 10.1016/j.brainres.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li Y. R., King O. D., Shorter J., and Gitler A. D. (2013) Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372 10.1083/jcb.201302044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. March Z. M., King O. D., and Shorter J. (2016) Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res. 1647, 9–18 10.1016/j.brainres.2016.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Harrison A. F., and Shorter J. (2017) RNA-binding proteins with prion-like domains in health and disease. Biochem. J. 474, 1417–1438 10.1042/BCJ20160499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Shorter J. (2016) Engineering therapeutic protein disaggregases. Mol. Biol. Cell 27, 1556–1560 10.1091/mbc.E15-10-0693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Shorter J. (2017) Designer protein disaggregases to counter neurodegenerative disease. Curr. Opin. Genet. Dev. 44, 1–8 10.1016/j.gde.2017.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Duennwald M. L., Echeverria A., and Shorter J. (2012) Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol. 10, e1001346 10.1371/journal.pbio.1001346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sweeny E. A., Jackrel M. E., Go M. S., Sochor M. A., Razzo B. M., DeSantis M. E., Gupta K., and Shorter J. (2015) The Hsp104 N-terminal domain enables disaggregase plasticity and potentiation. Mol. Cell 57, 836–849 10.1016/j.molcel.2014.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Afsar Minhas F. U. A., Ross E. D., and Ben-Hur A. (2017) Amino acid composition predicts prion activity. PLoS Comput. Biol. 13, e1005465 10.1371/journal.pcbi.1005465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cascarina S. M., Paul K. R., Machihara S., and Ross E. D. (2018) Sequence features governing aggregation or degradation of prion-like proteins. PLoS Genet. 14, e1007517 10.1371/journal.pgen.1007517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Toombs J. A., McCarty B. R., and Ross E. D. (2010) Compositional determinants of prion formation in yeast. Mol. Cell. Biol. 30, 319–332 10.1128/MCB.01140-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Toombs J. A., Petri M., Paul K. R., Kan G. Y., Ben-Hur A., and Ross E. D. (2012) De novo design of synthetic prion domains. Proc. Natl. Acad. Sci. U.S.A. 109, 6519–6524 10.1073/pnas.1119366109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Khan T., Kandola T. S., Wu J., Venkatesan S., Ketter E., Lange J. J., Rodríguez Gama A., Box A., Unruh J. R., Cook M., and Halfmann R. (2018) Quantifying nucleation in vivo reveals the physical basis of prion-like phase behavior. Mol. Cell 71, 155–168.e7 10.1016/j.molcel.2018.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Murray D. T., Kato M., Lin Y., Thurber K. R., Hung I., McKnight S. L., and Tycko R. (2017) Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell 171, 615–627.e16 10.1016/j.cell.2017.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lin Y., Protter D. S., Rosen M. K., and Parker R. (2015) Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Uversky V. N., Kuznetsova I. M., Turoverov K. K., and Zaslavsky B. (2015) Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett. 589, 15–22 10.1016/j.febslet.2014.11.028 [DOI] [PubMed] [Google Scholar]
- 98. Wei M. T., Elbaum-Garfinkle S., Holehouse A. S., Chen C. C., Feric M., Arnold C. B., Priestley R. D., Pappu R. V., and Brangwynne C. P. (2017) Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem. 9, 1118–1125 10.1038/nchem.2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ryan V. H., Dignon G. L., Zerze G. H., Chabata C. V., Silva R., Conicella A. E., Amaya J., Burke K. A., Mittal J., and Fawzi N. L. (2018) Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol. Cell 69, 465–479.e7 10.1016/j.molcel.2017.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Protter D. S. W., Rao B. S., Van Treeck B., Lin Y., Mizoue L., Rosen M. K., and Parker R. (2018) Intrinsically disordered regions can contribute promiscuous interactions to RNP granule assembly. Cell Rep. 22, 1401–1412 10.1016/j.celrep.2018.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Patel A., Lee H. O., Jawerth L., Maharana S., Jahnel M., Hein M. Y., Stoynov S., Mahamid J., Saha S., Franzmann T. M., Pozniakovski A., Poser I., Maghelli N., Royer L. A., Weigert M., et al. (2015) A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- 102. Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L. M., and Anderson P. (2004) Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15, 5383–5398 10.1091/mbc.e04-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Burke K. A., Janke A. M., Rhine C. L., and Fawzi N. L. (2015) Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell 60, 231–241 10.1016/j.molcel.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Shelkovnikova T. A., Robinson H. K., Troakes C., Ninkina N., and Buchman V. L. (2014) Compromised paraspeckle formation as a pathogenic factor in FUSopathies. Hum. Mol. Genet. 23, 2298–2312 10.1093/hmg/ddt622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. West J. A., Mito M., Kurosaka S., Takumi T., Tanegashima C., Chujo T., Yanaka K., Kingston R. E., Hirose T., Bond C., Fox A., and Nakagawa S. (2016) Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J. Cell Biol. 214, 817–830 10.1083/jcb.201601071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Qamar S., Wang G., Randle S. J., Ruggeri F. S., Varela J. A., Lin J. Q., Phillips E. C., Miyashita A., Williams D., Ströhl F., Meadows W., Ferry R., Dardov V. J., Tartaglia G. G., Farrer L. A., et al. (2018) FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation–π interactions. Cell 173, 720–734.e15 10.1016/j.cell.2018.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wang J., Choi J. M., Holehouse A. S., Lee H. O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D., Poser I., Pappu R. V., Alberti S., and Hyman A. A. (2018) A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 10.1016/j.cell.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bogaert E., Boeynaems S., Kato M., Guo L., Caulfield T. R., Steyaert J., Scheveneels W., Wilmans N., Haeck W., Hersmus N., Schymkowitz J., Rousseau F., Shorter J., Callaerts P., Robberecht W., et al. (2018) Molecular dissection of FUS points at synergistic effect of low-complexity domains in toxicity. Cell Rep. 24, 529–537.e4 10.1016/j.celrep.2018.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sun Z., Diaz Z., Fang X., Hart M. P., Chesi A., Shorter J., and Gitler A. D. (2011) Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 9, e1000614 10.1371/journal.pbio.1000614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yoshizawa T., Ali R., Jiou J., Fung H. Y. J., Burke K. A., Kim S. J., Lin Y., Peeples W. B., Saltzberg D., Soniat M., Baumhardt J. M., Oldenbourg R., Sali A., Fawzi N. L., Rosen M. K., and Chook Y. M. (2018) Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell 173, 693–705.e22 10.1016/j.cell.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Thandapani P., O'Connor T. R., Bailey T. L., and Richard S. (2013) Defining the RGG/RG motif. Mol. Cell 50, 613–623 10.1016/j.molcel.2013.05.021 [DOI] [PubMed] [Google Scholar]
- 112. Chong P. A., Vernon R. M., and Forman-Kay J. D. (2018) RGG/RG motif regions in RNA binding and phase separation. J. Mol. Biol. 430, 4650–4665 10.1016/j.jmb.2018.06.014 [DOI] [PubMed] [Google Scholar]
- 113. Hofweber M., Hutten S., Bourgeois B., Spreitzer E., Niedner-Boblenz A., Schifferer M., Ruepp M. D., Simons M., Niessing D., Madl T., and Dormann D. (2018) Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719.e13 10.1016/j.cell.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 114. Uversky V. N. (2017) Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin Struct. Biol. 44, 18–30 10.1016/j.sbi.2016.10.015 [DOI] [PubMed] [Google Scholar]
- 115. Monahan Z., Ryan V. H., Janke A. M., Burke K. A., Rhoads S. N., Zerze G. H., O'Meally R., Dignon G. L., Conicella A. E., Zheng W., Best R. B., Cole R. N., Mittal J., Shewmaker F., and Fawzi N. L. (2017) Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36, 2951–2967 10.15252/embj.201696394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C. C., Eckmann C. R., Myong S., and Brangwynne C. P. (2015) The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 112, 7189–7194 10.1073/pnas.1504822112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Wang A., Conicella A. E., Schmidt H. B., Martin E. W., Rhoads S. N., Reeb A. N., Nourse A., Ramirez Montero D., Ryan V. H., Rohatgi R., Shewmaker F., Naik M. T., Mittag T., Ayala Y. M., and Fawzi N. L. (2018) A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 37, e97452 10.15252/embj.201797452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Conicella A. E., Zerze G. H., Mittal J., and Fawzi N. L. (2016) ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure 24, 1537–1549 10.1016/j.str.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Guo L., Kim H. J., Wang H., Monaghan J., Freyermuth F., Sung J. C., O'Donovan K., Fare C. M., Diaz Z., Singh N., Zhang Z. C., Coughlin M., Sweeny E. A., DeSantis M. E., Jackrel M. E., Rodell C. B., Burdick J. A., King O. D., Gitler A. D., Lagier-Tourenne C., Pandey U. B., Chook Y. M., Taylor J. P., and Shorter J. (2018) Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677–692.e20 10.1016/j.cell.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zhang H., Elbaum-Garfinkle S., Langdon E. M., Taylor N., Occhipinti P., Bridges A. A., Brangwynne C. P., and Gladfelter A. S. (2015) RNA controls polyQ protein phase transitions. Mol. Cell 60, 220–230 10.1016/j.molcel.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Chang C. K., Wu T. H., Wu C. Y., Chiang M. H., Toh E. K., Hsu Y. C., Lin K. F., Liao Y. H., Huang T. H., and Huang J. J. (2012) The N terminus of TDP-43 promotes its oligomerization and enhances DNA binding affinity. Biochem. Biophys. Res. Commun. 425, 219–224 10.1016/j.bbrc.2012.07.071 [DOI] [PubMed] [Google Scholar]
- 122. Tsoi P. S., Choi K. J., Leonard P. G., Sizovs A., Moosa M. M., MacKenzie K. R., Ferreon J. C., and Ferreon A. C. M. (2017) The N-terminal domain of ALS-linked TDP-43 assembles without misfolding. Angew. Chem. Int. Ed. Engl. 56, 12590–12593 10.1002/anie.201706769 [DOI] [PubMed] [Google Scholar]
- 123. Afroz T., Hock E. M., Ernst P., Foglieni C., Jambeau M., Gilhespy L. A. B., Laferriere F., Maniecka Z., Plückthun A., Mittl P., Paganetti P., Allain F. H. T., and Polymenidou M. (2017) Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat. Commun. 8, 45 10.1038/s41467-017-00062-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zhang Y. J., Caulfield T., Xu Y. F., Gendron T. F., Hubbard J., Stetler C., Sasaguri H., Whitelaw E. C., Cai S., Lee W. C., and Petrucelli L. (2013) The dual functions of the extreme N terminus of TDP-43 in regulating its biological activity and inclusion formation. Hum. Mol. Genet. 22, 3112–3122 10.1093/hmg/ddt166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Shin Y., Berry J., Pannucci N., Haataja M. P., Toettcher J. E., and Brangwynne C. P. (2017) Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168, 159–171.e14 10.1016/j.cell.2016.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Vernon R. M., Chong P. A., Tsang B., Kim T. H., Bah A., Farber P., Lin H., and Forman-Kay J. D. (2018) π–π contacts are an overlooked protein feature relevant to phase separation. Elife 7, e31486 10.7554/eLife.31486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Pak C. W., Kosno M., Holehouse A. S., Padrick S. B., Mittal A., Ali R., Yunus A. A., Liu D. R., Pappu R. V., and Rosen M. K. (2016) Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell 63, 72–85 10.1016/j.molcel.2016.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Lin Y., Currie S. L., and Rosen M. K. (2017) Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 292, 19110–19120 10.1074/jbc.M117.800466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Xiang S., Kato M., Wu L. C., Lin Y., Ding M., Zhang Y., Yu Y., and McKnight S. L. (2015) The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell 163, 829–839 10.1016/j.cell.2015.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Nott T. J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T. D., Bazett-Jones D. P., Pawson T., Forman-Kay J. D., and Baldwin A. J. (2015) Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kim S., Huang J., Lee Y., Dutta S., Yoo H. Y., Jung Y. M., Jho Y., Zeng H., and Hwang D. S. (2016) Complexation and coacervation of like-charged polyelectrolytes inspired by mussels. Proc. Natl. Acad. Sci. U.S.A. 113, E847–E853 10.1073/pnas.1521521113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Boeynaems S., Bogaert E., Kovacs D., Konijnenberg A., Timmerman E., Volkov A., Guharoy M., De Decker M., Jaspers T., Ryan V. H., Janke A. M., Baatsen P., Vercruysse T., Kolaitis R. M., Daelemans D., et al. (2017) Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e5 10.1016/j.molcel.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Brady J. P., Farber P. J., Sekhar A., Lin Y. H., Huang R., Bah A., Nott T. J., Chan H. S., Baldwin A. J., Forman-Kay J. D., and Kay L. E. (2017) Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc. Natl. Acad. Sci. U.S.A. 114, E8194–E8203 10.1073/pnas.1706197114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Hughes M. P., Sawaya M. R., Boyer D. R., Goldschmidt L., Rodriguez J. A., Cascio D., Chong L., Gonen T., and Eisenberg D. S. (2018) Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science 359, 698–701 10.1126/science.aan6398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Luo F., Gui X., Zhou H., Gu J., Li Y., Liu X., Zhao M., Li D., Li X., and Liu C. (2018) Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation. Nat. Struct. Mol. Biol. 25, 341–346 10.1038/s41594-018-0050-8 [DOI] [PubMed] [Google Scholar]
- 136. Guenther E. L., Cao Q., Trinh H., Lu J., Sawaya M. R., Cascio D., Boyer D. R., Rodriguez J. A., Hughes M. P., and Eisenberg D. S. (2018) Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation. Nat. Struct. Mol. Biol. 25, 463–471 10.1038/s41594-018-0064-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Bah A., and Forman-Kay J. D. (2016) Modulation of intrinsically disordered protein function by post-translational modifications. J. Biol. Chem. 291, 6696–6705 10.1074/jbc.R115.695056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Bhowmick P., Guharoy M., and Tompa P. (2015) Bioinformatics approaches for predicting disordered protein motifs. Adv. Exp. Med. Biol. 870, 291–318 10.1007/978-3-319-20164-1_9 [DOI] [PubMed] [Google Scholar]
- 139. Zhong S., Müller S., Ronchetti S., Freemont P. S., Dejean A., and Pandolfi P. P. (2000) Role of SUMO-1-modified PML in nuclear body formation. Blood 95, 2748–2752 [PubMed] [Google Scholar]
- 140. Shen T. H., Lin H.-K., Scaglioni P. P., Yung T. M., and Pandolfi P. P. (2006) The mechanisms of PML-nuclear body formation. Mol. Cell 24, 331–339 10.1016/j.molcel.2006.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Banani S. F., Rice A. M., Peeples W. B., Lin Y., Jain S., Parker R., and Rosen M. K. (2016) Compositional control of phase-separated cellular bodies. Cell 166, 651–663 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Shorter J. (2017) Liquidizing FUS via prion-like domain phosphorylation. EMBO J. 36, 2925–2927 10.15252/embj.201798078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Rai A. K., Chen J. X., Selbach M., and Pelkmans L. (2018) Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559, 211–216 10.1038/s41586-018-0279-8 [DOI] [PubMed] [Google Scholar]
- 144. Smith J., Calidas D., Schmidt H., Lu T., Rasoloson D., and Seydoux G. (2016) Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. Elife 5, e21337 10.7554/eLife.21337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Bounedjah O., Desforges B., Wu T. D., Pioche-Durieu C., Marco S., Hamon L., Curmi P. A., Guerquin-Kern J. L., Piétrement O., and Pastré D. (2014) Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res. 42, 8678–8691 10.1093/nar/gku582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Mao Y. S., Sunwoo H., Zhang B., and Spector D. L. (2011) Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 13, 95–101 10.1038/ncb2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Shevtsov S. P., and Dundr M. (2011) Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 13, 167–173 10.1038/ncb2157 [DOI] [PubMed] [Google Scholar]
- 148. Palazzo A. F., and Lee E. S. (2015) Non-coding RNA: what is functional and what is junk? Front. Genet. 6, 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Schreiber V., Dantzer F., Ame J. C., and de Murcia G. (2006) Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7, 517–528 10.1038/nrm1963 [DOI] [PubMed] [Google Scholar]
- 150. Altmeyer M., Neelsen K. J., Teloni F., Pozdnyakova I., Pellegrino S., Grøfte M., Rask M. B., Streicher W., Jungmichel S., Nielsen M. L., and Lukas J. (2015) Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6, 8088 10.1038/ncomms9088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Liu C., Vyas A., Kassab M. A., Singh A. K., and Yu X. (2017) The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res. 45, 8129–8141 10.1093/nar/gkx565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. McGurk L., Gomes E., Guo L., Mojsilovic-Petrovic J., Tran V., Kalb R. G., Shorter J., and Bonini N. M. (2018) ADP-ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress-granule localization. Mol. Cell 71, 703–719.e9 10.1016/j.molcel.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Leung A. K., Vyas S., Rood J. E., Bhutkar A., Sharp P. A., and Chang P. (2011) Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 42, 489–499 10.1016/j.molcel.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Cremers C. M., Knoefler D., Gates S., Martin N., Dahl J. U., Lempart J., Xie L., Chapman M. R., Galvan V., Southworth D. R., and Jakob U. (2016) Polyphosphate: a conserved modifier of amyloidogenic processes. Mol. Cell 63, 768–780 10.1016/j.molcel.2016.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Racki L. R., Tocheva E. I., Dieterle M. G., Sullivan M. C., Jensen G. J., and Newman D. K. (2017) Polyphosphate granule biogenesis is temporally and functionally tied to cell cycle exit during starvation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 114, E2440–E2449 10.1073/pnas.1615575114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Baker J. D., Shelton L. B., Zheng D., Favretto F., Nordhues B. A., Darling A., Sullivan L. E., Sun Z., Solanki P. K., Martin M. D., Suntharalingam A., Sabbagh J. J., Becker S., Mandelkow E., Uversky V. N., et al. (2017) Human cyclophilin 40 unravels neurotoxic amyloids. PLoS Biol. 15, e2001336 10.1371/journal.pbio.2001336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Mackenzie I. R., Nicholson A. M., Sarkar M., Messing J., Purice M. D., Pottier C., Annu K., Baker M., Perkerson R. B., Kurti A., Matchett B. J., Mittag T., Temirov J., Hsiung G. R., Krieger C., Murray M. E., et al. (2017) TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron 95, 808–816.e9 10.1016/j.neuron.2017.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. McGurk L., Lee V. M., Trojanowksi J. Q., Van Deerlin V. M., Lee E. B., and Bonini N. M. (2014) Poly-A binding protein-1 localization to a subset of TDP-43 inclusions in amyotrophic lateral sclerosis occurs more frequently in patients harboring an expansion in C9orf72. J. Neuropathol. Exp. Neurol. 73, 837–845 10.1097/NEN.0000000000000102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Johnson B. S., Snead D., Lee J. J., McCaffery J. M., Shorter J., and Gitler A. D. (2009) TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 284, 20329–20339 10.1074/jbc.M109.010264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Couthouis J., Hart M. P., Erion R., King O. D., Diaz Z., Nakaya T., Ibrahim F., Kim H. J., Mojsilovic-Petrovic J., Panossian S., Kim C. E., Frackelton E. C., Solski J. A., Williams K. L., Clay-Falcone D., et al. (2012) Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 21, 2899–2911 10.1093/hmg/dds116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Couthouis J., Hart M. P., Shorter J., DeJesus-Hernandez M., Erion R., Oristano R., Liu A. X., Ramos D., Jethava N., Hosangadi D., Epstein J., Chiang A., Diaz Z., Nakaya T., Ibrahim F., et al. (2011) A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl. Acad. Sci. U.S.A. 108, 20881–20890 10.1073/pnas.1109434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Gopal P. P., Nirschl J. J., Klinman E., and Holzbaur E. L. (2017) Amyotrophic lateral sclerosis-linked mutations increase the viscosity of liquid-like TDP-43 RNP granules in neurons. Proc. Natl. Acad. Sci. U.S.A. 114, E2466–E2475 10.1073/pnas.1614462114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Lin Y., Mori E., Kato M., Xiang S., Wu L., Kwon I., and McKnight S. L. (2016) Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167, 789–802.e12 10.1016/j.cell.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Shi K. Y., Mori E., Nizami Z. F., Lin Y., Kato M., Xiang S., Wu L. C., Ding M., Yu Y., Gall J. G., and McKnight S. L. (2017) Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc. Natl. Acad. Sci. U.S.A. 114, E1111–E1117 10.1073/pnas.1620293114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Lee K. H., Zhang P., Kim H. J., Mitrea D. M., Sarkar M., Freibaum B. D., Cika J., Coughlin M., Messing J., Molliex A., Maxwell B. A., Kim N. C., Temirov J., Moore J., Kolaitis R. M., Shaw T. I., Bai B., Peng J., Kriwacki R. W., and Taylor J. P. (2016) C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788.e17 10.1016/j.cell.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Protter D. S. W., and Parker R. (2016) Principles and properties of stress granules. Trends Cell Biol. 26, 668–679 10.1016/j.tcb.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Vashist S., Cushman M., and Shorter J. (2010) Applying Hsp104 to protein-misfolding disorders. Biochem. Cell Biol. 88, 1–13 10.1139/O09-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Shorter J. (2011) The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE 6, e26319 10.1371/journal.pone.0026319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Jackrel M. E., DeSantis M. E., Martinez B. A., Castellano L. M., Stewart R. M., Caldwell K. A., Caldwell G. A., and Shorter J. (2014) Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell 156, 170–182 10.1016/j.cell.2013.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Mack K. L., and Shorter J. (2016) Engineering and evolution of molecular chaperones and protein disaggregases with enhanced activity. Front. Mol. Biosci. 3, 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Becker L. A., Huang B., Bieri G., Ma R., Knowles D. A., Jafar-Nejad P., Messing J., Kim H. J., Soriano A., Auburger G., Pulst S. M., Taylor J. P., Rigo F., and Gitler A. D. (2017) Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544, 367–371 10.1038/nature22038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Elden A. C., Kim H. J., Hart M. P., Chen-Plotkin A. S., Johnson B. S., Fang X., Armakola M., Geser F., Greene R., Lu M. M., Padmanabhan A., Clay-Falcone D., McCluskey L., Elman L., Juhr D., Gruber P. J., et al. (2010) Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075 10.1038/nature09320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Leung A. K. (2014) Poly(ADP-ribose): an organizer of cellular architecture. J. Cell Biol. 205, 613–619 10.1083/jcb.201402114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Maharana S., Wang J., Papadopoulos D. K., Richter D., Pozniakovsky A., Poser I., Bickle M., Rizk S., Guillén-Boixet J., Franzmann T. M., Jahnel M., Marrone L., Chang Y. T., Sterneckert J., Tomancak P., et al. (2018) RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 10.1126/science.aar7366 [DOI] [PMC free article] [PubMed] [Google Scholar]