Abstract
Through serial promoter exchanges, we isolated several novel polyenes, the aspernidgulenes, from Aspergillus nidulans and uncovered their succinct biosynthetic pathway involving only four enzymes. An enoyl reductase (ER)-less highly reducing polyketide synthase (HR-PKS) putatively produces a 5,6-dihydro-α-pyrone polyene, which undergoes bisepoxidation, epoxide ring opening, cyclization, and hydrolytic cleavage by three tailoring enzymes to generate aspernidgulene A1 and A2. Our findings demonstrate the prowess of fungal-tailoring enzymes to transform a polyketide scaffold concisely and efficiently into complex structures. Moreover, comparison with citreoviridin and aurovertin biosynthesis suggests that methylation of the α-pyrone hydroxy group by methyltransferase (CtvB or AurB) is the branching point at which the biosynthesis of these two classes of compounds diverge. Therefore, scanning for the presence or absence of the gatekeeping α-pyrone methyltransferase gene in homologous clusters might be a potential way to classify the product bioinformatically as belonging to methylated α-pyrone polyenes or polyenes containing rings derived from the cyclization of the unmethylated 5,6-dihydro-α-pyrone, such as 2,3-dimethyl-γ-lactone and oxabicyclo[2.2.1]heptane.
Keywords: aspernidgulenes, biosynthesis, natural products, polyenes, polyketides
The rapid accumulation of publicly available genome data and recent advancements in molecular biology have opened up unprecedented opportunities to discover new chemical entities from natural sources and to investigate their biosynthesis.[1] In filamentous fungi, the vast majority of secondary metabolite gene clusters remain unelucidated owing to their cryptic nature.[2] For even a well-studied model fungus such as Aspergillus nidulans, less than half of its secondary metabolite gene clusters have been linked to downstream products despite intensive research,[3] and this suggests that the potential for secondary metabolites to be a source of antibiotics and therapeutics is vast and still largely unexploited.[4]
The genome of A. nidulans encodes 27 polyketide synthases (PKSs), 15 of which are nonreducing polyketide synthases (NR-PKSs) and 12 of which are highly reducing polyketide synthase (HR-PKSs).[3] The microarray transcription data of Aspergillus nidulans indicated that one enoyl reductase (ER)-less HR-PKS AN1784 is co-regulated with three other putative biosynthesis genes.[5] This four-gene cluster attracted our attention for two reasons. First, three of its four genes share sequence homology with the biosynthetic genes of citreoviridin (ctv)[6] and aurovertin (aur)[7] (Table 1). Citreoviridin and aurovertin belong to a class of potent α-pyrone polyene F1-ATPase inhibitors and have been tested as antitumor agents.[8–11] We thus speculated that the cluster could produce a novel variant given that A. nidulans was not known to produce any α-pyrone polyenes at the time of this study.[3] Second, the genes in the cluster are divergently transcribed. AN1784 and AN1785 are adjacent genes that have opposite orientations, and so are AN1786 and AN1787 (Figure 1). Divergent transcription would allow us to activate the entire cluster quickly in just two rounds of transformation with a previously described strategy.[12,13] Here, we report the discovery and isolation of the aspernidgulenes, which unexpectedly fall into an entirely different class of compounds than citreoviridin and aurovertin, from this cluster (sdg).
Table 1.
Aspernidgulene biosynthesis genes in A. nidulans, their homologs in other species, and gene function predictions.
| gene | putative function | Aspergillus terreus var. aureus | Calcarisporium arbuscula | Alternaria solani |
|---|---|---|---|---|
| AN1783 | dehydrogenase | --- | --- | Sol3 (25, 45)a |
| sdgA (AN1784) | highly reducing polyketide synthase | CtvA (44, 59)a | AurA (45, 63)a | Sol1 (28, 44)a |
| sdgD (AN1785) | hydrolase | CtvD (24, 40)a | AurD (23, 41)a | --- |
| sdgC (AN1786) | flavin-dependent monooxygenase (FMO) | CtvC (39, 56)a | AurC (37, 54)a | --- |
| sdgF (AN1787) | FAD linked oxidase | --- | --- | Sol5 (29, 44)a |
Numbers in parentheses represent the percentage of identity and similarity to Sdg proteins.
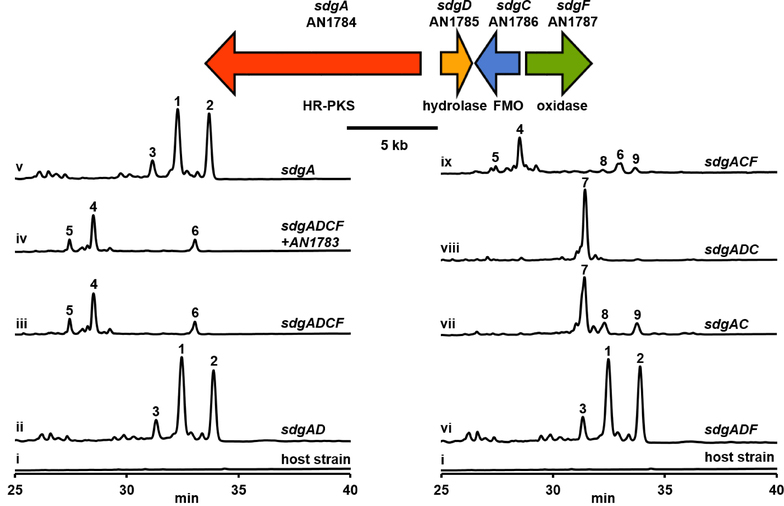
Figure 1.
Organization of the aspernidgulene biosynthesis gene cluster and total scan UV-Vis HPLC profiles of culture media of A. nidulans strains expressing aspernidgulene biosynthesis genes under the control of alcA(p).
Similar to the ctv cluster, the sdg cluster encodes an ER-less HR-PKS and two enzymes putatively responsible for cyclization of the polyketide product (Table 1). The HR-PKS AN1784 is homologous to ctvA, and the encoded enzymes share the same domain architecture of KS-AT-DH-CMet-KR-ACP (KS, ketosynthase; AT, acyltransferase; DH, dehydratase; CMet, C-methyltransferase; KR, ketoreductase; ACP, acyl carrier protein).[6,14] We thus named AN1784 sdgA. AN1785 encodes a putative membrane-associated hydrolase and exhibits homology with ctvD, so it was named sdgD. AN1786 encodes a flavin-dependent monooxygenase and is homologous to ctvC; thus, it was named sdgC. Previous work demonstrated that CtvC and CtvD and their homologs AurC and AurD are responsible for the assembly of the tetrahydrofurane ring in citreoviridin and the 2,6-dioxabicyclo[3.2.1]-octane ring in aurovertin, respectively (Table 1).[6,7] Unlike the ctv cluster, however, the sdg cluster does not contain methyltransferase (ctvB) or resistance (ctvE) genes. Instead, AN1787, which we named sdgF, encodes a putative FAD-linked oxidase and shows homology with sol5 of the solanapyrone biosynthesis cluster (Table 1).[15]
We first replaced the promoters of both sdgA and sdgD with the inducible alcA promoter [alcA(p)] in a single transformation[12,13] to create an alcA(p)-sdgAD strain (see Table S2 in the Supporting Information for the designation and genotype of strains created in this study). Under inducing conditions, three new peaks representing compounds 1 – 3 not present in the host strain were detected in its media (Figure 1, i and ii). Next, we replaced the promoters of the other divergently transcribed pair, sdgC and sdgF, to generate an alcA(p)-sdgADCF strain. In this strain, three new peaks representing compounds 4 – 6 emerged (Figure 1, iii). In addition, we also replaced the promoter of AN1783, which encodes a putative dehydrogenase and shows homology with sol3 from the solanopyrone pathway (Table 1).[15] However, we did not detect any new peaks in the alcA(p)-sdgADCF + AN1783 strain under the inducing conditions (Figure 1, iv); this suggests that AN1783 is not part of the sdg cluster as the transcription data predicted.[5] Therefore, 4 – 6 are likely the final products.
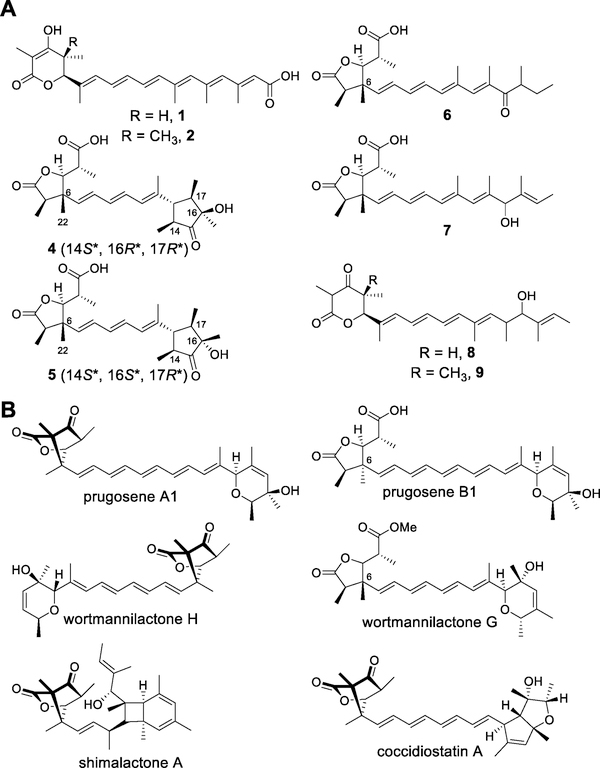
We purified compounds 4 – 6 from large-scale fermentation and elucidated their structures on the basis of their spectroscopic data. To our surprise, 4 – 6 bear little structural resemblance to either citreoviridin or aurovertin. Instead, all three compounds contain a 2,3-dimethyl-γ-lactone substituted with a 1-carboxyethyl group (Scheme 1A), a moiety found in several other fungal natural products (Scheme 1B).[16–19] Compound 4 had a molecular formula of C24H34O6. Its 1H, 13C, DEPT, and HSQC NMR spectra exhibit signals for seven methyl groups, three olefins, and three carbonyl groups. The conjugated triene moiety could be assigned unambiguously from the 1H-1H COSY and 1H-13C HMBC correlations, whereas the trimethylcyclopentanone and 2,3-dimethyl-γ-lactone moieties were constructed mainly from the 1H-13C HMBC correlations. From the NOESY correlations, we assigned relative configurations of (2R*,5S*,6R*) and (14S*,16R*,17R*), as shown in Scheme 1A (see the Supporting Information for detailed structural elucidation of all compounds). Compound 5 has the same molecular formula as 4 and the NMR spectral data of the former are very similar to that of the latter. From these data and the NOESY correlations, we established 5 to be a 16-epimer of 4. We named compounds 4 and 5 aspernidgulene A1 and A2, respectively. Compound 6 had a molecular formula of C24H34O5. Its 1H, 13C, and 2D NMR spectral data indicate that 6 also contains the same 2,3-dimethyl-γ-lactone as that found in 4 and 5 but lacks the trimethylcyclopentanone moiety and is a tetraene instead of a triene. We named it aspernidgulene B1.
Scheme 1.
(A) New compounds isolated from the study. (B) Representative compounds containing the oxabicyclo[2.2.1]heptane unit or their derivatives.
Having identified the products of the sdg cluster, we proceeded to investigate the function of each individual gene. First, we created two promoter replacement strains: alcA(p)-sdgA and alcA(p)-sdgADF. We found that the metabolite profiles of both strains were identical to that of the alcA(p)-sdgAD strain (Figure 1, ii and v, vi). These data indicate that SdgD and SdgF do not act immediately downstream of SdgA and that the HR-PKS SdgA alone is sufficient for the production of the peaks representing compounds 1 – 3. Purification of compounds 1 – 3 was difficult, possibly because of the instability of polyenes.[20] However, we were able to deduce the chemical structures of 1 and 2 from the spectra we obtained (see the Supporting Information for details). They were 5,6-dihydro-α-pyrone-containing hexaenoic acids, which we named preaspernidgulene A1 (1) and A2 (2) (Scheme 1A). As SdgA does not contain an oxygenase domain, it is likely that the terminal carboxylic acid functional groups on 1 and 2 were installed by unknown endogenous enzymes.[21] Despite repeated attempts, we were unable to obtain compound 3 for NMR spectral analysis owing to its instability.
After concluding that SdgD and SdgF do not act immediately downstream of SdgA, we turned our attention to SdgC. We created an alcA(p)-sdgAC strain, which produced compounds 7 – 9 (Figure 1, vii). Compound 7 has the same molecular formula as 6. Both molecules have similar 1H and 13C NMR spectral data, except for signals from the polyene portion. We elucidated the structure of 7 on the basis of 1H-13C HMBC correlations, as shown in Scheme 1A, and we named 7 aspernidgulene B2 as a result of its structural similarity to aspernidgulene B1 (6). The structures of 8 and 9 were revealed by NMR characterization (Scheme 1A, see the Supporting Information for details). As there compounds resemble 1 and 2, we named 8 and 9 preaspernidgulene A3 and A4, respectively. That all three compounds contain a secondary alcohol on their side chain suggests that SdgC acts on the polyene tail.
Lastly, to establish the roles of the predicted hydrolase SdgD and the oxidase SdgF, we generated two strains: alcA(p)- sdgADC and alcA(p)-sdgACF. The alcA(p)-sdgADC strain produced 7 without detectable amounts of 8 and 9 (Figure 1, viii), unlike the alcA(p)-sdgAC strain (Figure 1, vii). These data suggest that SdgD works in concert with SdgC to facilitate the complete conversion of the polyketide precursor into 7 and that 8 and 9 are presumably shunt products. In the alcA(p)-sdgACF strain, we observed the reemergence of 4 – 6 (Figure 1, ix); this indicates that SdgF is involved in trimethylcyclopentanone synthesis. Interestingly, the alcA(p)-sdgACF strain also produced minor products such as 8 and 9 as well as 10, 13, and 16 – 19 that were not found in the alcA(p)-sdgADCF strain (Figure S1). Compounds 10 and 13 are likely epimers of 4 and 5 on the basis of their UV-Vis and mass spectra (Figure S2 and S3), thus suggesting that SdgD could contribute to the stereoselectivity.
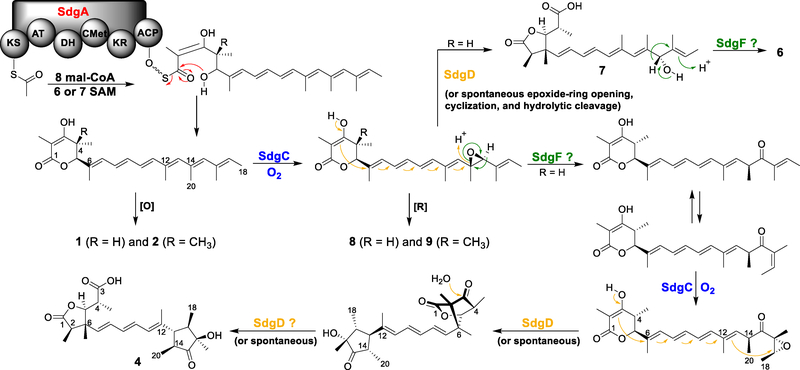
On the basis of our data, we propose a biosynthetic pathway of aspernidgulene A1 (4) (Scheme 2). The carbon backbone is synthesized by the HR-PKS SdgA, which accepts acetyl-CoA as the starter unit and performs malonyl-CoA extensions as well as regioselective methylation and reduction. The resulting nonaketide offloads the HR-PKS by intramolecular lactonization to yield a 5,6-dihydro-α-pyrone polyene. SdgC then installs the first epoxide on the penultimate double bond. Subsequently, a ketone intermediate is generated through Meinwald rearrangement involving a hydride shift. As 4 and 5 are only detected in sdgF promoter replacement strains, SdgF is presumably involved in this rearrangement. Next, SdgC introduces another epoxide on the last olefin of the ketone intermediate after E/Z isomerization, reminiscent of the actions of its homologs CtvC and AurC in iterative polyene epoxidation.[6,7] SdgD then catalyzes stereospecific cyclization of the 5,6-dihydro-α-pyrone and opening of the epoxide ring to form an oxygenated trimethylcyclopentanone and an oxabicyclo[2.2.1]heptane unit. Finally, the bicyclic unit undergoes hydrolytic cleavage, either spontaneously or catalyzed by SdgD, to assemble the dimethyl-γ-lactone moiety in compound 4. The presence of 4 and its epimers in strains not expressing sdgD (Figure 1, ix and Figure S1) likely results from non-stereoselective, spontaneous cyclization of the epoxidated ketone intermediate and subsequent spontaneous hydrolytic cleavage of the oxabicyclo[2.2.1]heptane. This hypothesis is supported by the results of Sofiyev et al., who obtained oxabicyclo[2.2.1]heptanes by treating the β-keto lactone substrate with camphorsulfonic acid.[22] Moreover, treatment of prugosene A1 with diluted NaOH led to the production of prugosene B1 (Scheme 1B).[16] Thus, bicyclic unit formation and hydrolysis can happen non-enzymatically under acidic and basic conditions, respectively.
Scheme 2.
Proposed biosynthetic pathway for asperniduglene A1 (4).
Interestingly, SdgD is implicated in several stereocontrol steps. The relative configurations of the trimethylcyclopentanone moieties in compounds 4 (14S*,16R*,17R*) and 5 (14S*,16S*,17R*) are determined by the configurations of the last two double bonds and the two epoxides installed on them. Given that each double bond could be either E or Z and could be epoxidized in two different orientations, 16 combinations are possible (Figure S4 – S7). Of them, four would result in the configurations of 4 and another four would result in the configurations of 5. The conformation depicted in Scheme 2 represents only one of the possible ways by which 4 could be formed. How SdgD works in concert with SdgC in isomerization and epoxidation to achieve the stereoselectivity remains to be investigated biochemically. Additionally, the NOESY data indicated that the methyl groups attached to C-6 in compounds 4 and 5 (CH3-22) (Table S8 and Figure S9) and wortmannilactone G are all on the β face, whereas the methyl group on C-6 in prugosene B1 is on the α face. The stereochemistry at C-6 is determined by how the β-keto lactone intercepts the positive charge on C-6 after opening of the epoxide ring. Because no C-6 epimers were isolated in any of the three studies,[16,17] this reaction is also presumably enzymatically controlled by SdgD and its homologs in wortmannilactone and prugosene biosynthesis.
Perhaps the most interesting step in our proposed biosynthesis is the Meinwald rearrangement following the first SdgC epoxidation. Until the recent discovery of PenF by Zhou et al., no enzyme had been known to perform Meinwald rearrangement.[23] Although our data indicate that SdgF could play a role in this rearrangement, further biochemical verification is needed because SdgF is predicted to be a FAD-containing dehydrogenase whereas PenF belongs to the CrtC (carotenoid 1,2-hydratase) superfamily. Furthermore, compounds 6 – 9 were off-pathway products that were likely generated before Meinwald rearrangement. Reduction of the epoxide intermediate, presumably by endogenous enzymes, would lead to the formation of 8 and 9. Spontaneous cyclization and hydrolytic cleavage of the epoxide intermediate would produce 7, after which hydride shift, possibly catalyzed by SdgF, would yield 6.
In conclusion, using serial promoter replacement, we discovered eight polyketides, the aspernidgulenes, that are new to science and elucidated their succinct biosynthesis. Conversion of the 5,6-dihydro-α-pyrone polyene into 4 and 5 required only three enzymes; this demonstrated the conciseness of the Sdg biosynthetic pathway. As similar enzymes are likely employed in the biosynthesis of related natural products such as the prugosenes,[16] the wortmannilactones,[17,24] shimalactone A,[18] and coccidiostatin A,[19] this study paves the way for future genome mining endeavors. Moreover, the characterization of SdgA contributes to the growing database of fungal ER-less HR-PKS gene-product connections (e.g., the first characterization of a mushroom ER-less HR-PKS was recently described[25]) which will lay the foundation for predicting polyene structure from phylogenetic analysis.
Notably, although the sdg cluster exhibits significant homology with the ctv and aur clusters, it lacks an O-methyltransferase gene. In compounds related to citreoviridin and aurovertin, O-methylation of the α-pyrone is universally observed.[26] In fact, downstream tailoring enzymes have been shown to be selective for methylated as opposed to unmethylated α-pyrones.[6,7] On the other hand, our results here demonstrated that unmethylated 5,6-dihydro-α-pyrone could undergo cyclization to form another class of compounds. Thus, α-pyrone methylation is a biosynthetic branching point at which remarkable chemical diversity is generated by homologues of ctvC/sdgC and ctvD/sdgD as well as of sdgF. We propose that for clusters homologous to ctv and sdg (also containing homologs of ctvC/sdgC and ctvD/sdgD), the O-methyltransferase gene is a genetic indicator of whether the product belongs to methylated α-pyrone polyenes or polyenes that undergo further 5,6-dihydro-α-pyrone cyclization to generate moieties including dimethyl-γ-lactone and oxabicyclo[2.2.1]heptane. If proven to be applicable, such accurate and rapid structural prediction from genetic information would bypass laborious genetic and chemical interrogation, and would be essential for the fully and efficient untapping of the potential of secondary metabolite as a source of novel chemicals.[27]
Supplementary Material
Acknowledgements
We thank Alireza Delfarah and Dr. Nicholas Graham for helping us obtain high-resolution mass spectrometry data. We thank Allan Kershaw for helping us with the Varian 600 MHz NMR spectrometer. We thank Stephanie F. Loekman for helping with experiments. The Project described was supported in part by the National Institute of General Medical Sciences (PO1GM084077) to C.C.C.W.
References
- [1].Cacho RA, Tang Y, Chooi Y-H, Front Microbiol 2014, 5, 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chiang Y-M, Lee K-H, Sanchez JF, Keller NP, Wang CCC, Nat Prod Commun 2009, 4, 1505–1510. [PMC free article] [PubMed] [Google Scholar]
- [3].Yaegashi J, Oakley BR, Wang CCC, J. Ind. Microbiol. Biotechnol 2014, 41, 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nielsen JC, Grijseels S, Prigent S, Ji B, Dainat J, Nielsen K, Frisvad J, Workman M, Nielsen J, Nature Microbiology 2017, 2, 17044. [DOI] [PubMed] [Google Scholar]
- [5].Andersen MR, Nielsen JB, Klitgaard A, Petersen LM, Zachariasen M, Hansen TJ, Blicher LH, Gotfredsen CH, Larsen TO, Nielsen KF, et al. , Proc. Natl. Acad. Sci. U.S.A 2013, 110, E99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lin T-S, Chiang Y-M, Wang CCC, Org. Lett 2016, 18, 1366–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mao X-M, Zhan Z-J, Grayson MN, Tang M-C, Xu W, Li Y-Q, Yin W-B, Lin H-C, Chooi Y-H, Houk KN, et al. , J. Am. Chem. Soc 2015, 137, 11904–11907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wu Y-H, Hu C-W, Chien C-W, Chen Y-J, Huang H-C, Juan H-F, PLoS ONE 2013, 8, e70642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hong S, Pedersen PL, Microbiol Mol Biol Rev 2008, 72, 590–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Huang T-C, Chang H-Y, Hsu C-H, Kuo W-H, Chang K-J, Juan H-F, Proteome Res J. 2008, 7, 1433–1444. [DOI] [PubMed] [Google Scholar]
- [11].Chang H-Y, Huang T-C, Chen N-N, Huang H-C, Juan H-F, Cell Death & Disease 2014, 5, e1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ahuja M, Chiang Y-M, Chang S-L, Praseuth MB, Entwistle R, Sanchez JF, Lo H-C, Yeh H-H, Oakley BR, Wang CCC, J. Am. Chem. Soc 2012, 134, 8212–8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yeh H-H, Ahuja M, Chiang Y-M, Oakley CE, Moore S, Yoon O, Hajovsky H, Bok J-W, Keller NP, Wang CCC, et al. , ACS Chem. Biol 2016, 11, 2275–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hertweck C, Angew. Chem. Int. Ed. Engl 2009, 48, 4688–4716. [DOI] [PubMed] [Google Scholar]
- [15].Kasahara K, Miyamoto T, Fujimoto T, Oguri H, Tokiwano T, Oikawa H, Ebizuka Y, Fujii I, Chembiochem 2010, 11, 1245–1252. [DOI] [PubMed] [Google Scholar]
- [16].Lang G, Wiese J, Schmaljohann R, Imhoff JF, Tetrahedron 2007, 63, 11844–11849. [Google Scholar]
- [17].Yuesheng Dong, Jie Lin, Xinhua Lu, Zhihui Zheng, Xiao Ren, Hua Zhang, Jiangong He, Junshan Yang, Helvetica Chimica Acta 2009, 92, 567–574. [Google Scholar]
- [18].Wei H, Itoh T, Kinoshita M, Kotoku N, Aoki S, Kobayashi M, Tetrahedron 2005, 61, 8054–8058. [Google Scholar]
- [19].Jayasuriya H, Guan Z, Dombrowski AW, Bills GF, Polishook JD, Jenkins RG, Koch L, Crumley T, Tamas M. Dubois, et al. , J. Nat. Prod 2007, 70, 1364–1367. [DOI] [PubMed] [Google Scholar]
- [20].Ma SM, Li JW-H, Choi JW, Zhou H, Lee KKM, Moorthie VA, Xie X, Kealey JT, Da Silva NA, Vederas JC, et al. , Science 2009, 326, 589–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liu T, Sanchez JF, Chiang Y-M, Oakley BR, Wang CCC, Org. Lett 2014, 16, 1676–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sofiyev V, Navarro G, Trauner D, Org. Lett 2008, 10, 149–152. [DOI] [PubMed] [Google Scholar]
- [23].Zou Y, Garcia-Borràs M, Tang MC, Hirayama Y, Li DH, Li L, Watanabe K, Houk KN, Tang Y, Nature Chemical Biology 2017, 13, 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu WC, Wang YY, Liu JH, Ke AB, Zheng ZH, Lu XH, Luan YS, Xiu ZL, Dong YS, Bioorg Med Chem Lett 2016, 26, 5328–5333. [DOI] [PubMed] [Google Scholar]
- [25].Brandt P, García-Altares M, Nett M, Hertweck C, Hoffmeister D, Angew. Chem. Int. Ed. Engl 2017, 56, 5937–5941. [DOI] [PubMed] [Google Scholar]
- [26].Li W, Ma Z, Chen L, Yin W-B, Appl Microbiol Biotechnol 2018, 102, 6373–6381. [DOI] [PubMed] [Google Scholar]
- [27].Chooi Y-H, Tang Y, J. Org. Chem 2012, 77, 9933–9953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.