Significance
This paper demonstrates that aminoglycoside antibiotics induce the production of the Ap4A in bacteria. Increased intracellular Ap4A, in turn, promotes bacterial cell killing by this class of antibiotics, which correlated well with elevated damage to the bacterial membrane upon aminoglycoside treatment. These findings reveal a striking connection between aminoglycoside killing and the Ap4A production particularly under conditions of oxidative stress. Importantly, the results of this study suggest that targeting Ap4A degradation or inducing its hypersynthesis during therapy with aminoglycosides might help solve the well-known toxicity issue associated with this class of antibiotics by reducing the level of drug needed for effective treatment.
Keywords: antibiotic action, aminoglycoside, kanamycin, diadenosine tetraphosphate Ap4A, alarmone
Abstract
Second messenger molecules play important roles in the responses to various stimuli that can determine a cell's fate under stress conditions. Here, we report that lethal concentrations of aminoglycoside antibiotics result in the production of a dinucleotide alarmone metabolite–diadenosine tetraphosphate (Ap4A), which promotes bacterial cell killing by this class of antibiotics. We show that the treatment of Escherichia coli with lethal concentrations of kanamycin (Kan) dramatically increases the production of Ap4A. This elevation of Ap4A is dependent on the production of a hydroxyl radical and involves the induction of the Ap4A synthetase lysyl-tRNA synthetase (LysU). Ectopic alteration of intracellular Ap4A concentration via the elimination of the Ap4A phosphatase diadenosine tetraphosphatase (ApaH) and the overexpression of LysU causes over a 5,000-fold increase in bacterial killing by aminoglycosides. This increased susceptibility to aminoglycosides correlates with bacterial membrane disruption. Our findings provide a role for the alarmone Ap4A and suggest that blocking Ap4A degradation or increasing its synthesis might constitute an approach to enhance aminoglycoside killing potency by broadening their therapeutic index and thereby allowing lower nontoxic dosages of these antibiotics to be used in the treatment of multidrug-resistant infections.
Antimicrobial resistance has become a global crisis due to the rapid emergence and spread of drug-resistant bacteria as well as the reduced efforts by the pharmaceutical industry to provide a robust new pipeline of antibiotic candidates for clinical evaluation. The situation is expected to get worse as indicated by a recent governmental report showing that, if no effective prevention and controlling methods are taken, about 10 million people will die per year from the infections by antimicrobial-resistant organisms by the year 2050 (1). This crisis calls out for increased efforts to find new antibiotics or ways to use older antibiotics more safely and effectively.
Aminoglycosides, such as Kan and streptomycin (Str), constitute one of the oldest classes of antibiotics being used in current clinical practice, especially on multidrug-resistant gram-negative pathogenic bacteria (2). Unlike other antibiotics that simply block protein synthesis and are bacteriostatic, aminoglycosides cause tRNA mismatching which drives mistakes during translation, production of aberrant proteins, and bactericidal outcomes for the cell (3, 4). Mistranslated proteins caused by aminoglycosides can ultimately trigger the formation of the reactive oxygen species (ROS), especially hydroxyl radicals that can further oxidize proteins (5–7). Mistranslated or oxidized proteins tend to be misfolded and expose hydrophobic regions that interact with membranes or bind to other cellular components forming potentially toxic aggregates that can be detrimental to cells (8, 9). The protein aggregates can be transient or are sequestered into structures called sequestrosomes by bacteria (10, 11). However, it remains unclear how overwhelming levels of misfolded proteins produced by the lethal concentration of bactericidal aminoglycosides actually cause cell death but clearly multiple pathways are involved.
One positive characteristic of aminoglycosides is that they retain good activity against many multidrug-resistant bacteria, such as Acinetobacter spp. and Pseudomonas aeruginosa (2). However, the clinical usage of aminoglycosides has been limited due to their nephrotoxicity and ototoxicity at higher dosages (12, 13), which can induce extended cortical necrosis and overt renal dysfunction (14) as well as permanent hearing loss or balance disorders (2). Lowering the administration dose of aminoglycosides can reduce their toxicities, however, the effectiveness of these drugs would likely be compromised, and resistance could also be promoted (15). One possible solution is to find a potentiator that can enhance aminoglycosides’ potency. The combination of such a potentiator with an aminoglycoside might reduce the amount of drug needed for therapeutic efficacy, allowing adverse drug effects to be avoided and thus broadening clinical usage of these powerful antibiotics.
Ap4A, a dinucleotide metabolite that consists of two adenosines joined in 5′-5′ linkage by four phosphates (SI Appendix, Fig. S1A), was discovered 50 y ago and is considered to be an alarmone due to its induction under oxidative stress (16, 17). Ap4A is synthesized by aminoacyl-tRNA synthetases, among which LysU is the major one producing Ap4A upon oxidative stress in E. coli (18). Intracellular Ap4A in E. coli is degraded by ApaH, and null mutations in apaH cause the accumulation of Ap4A inside the bacterial cells (19). In eukaryotic cells, Ap4A disassociates histidine triad nucleotide-binding protein 1 from microphthalmia-associated transcription factor, the latter of which then activates the transcription of its target genes (20–22). However, the function of Ap4A in bacteria has yet to be fully addressed biologically. On the other hand, dicyclic and oligo ribonucleotides are increasingly being recognized as the key second messengers involved in triggering cell responses to various cellular stresses and may result in cell death under ill-defined environmental conditions (23–26).
In this paper, we investigated the bacterial response to aminoglycosides by a metabolomic profiling and identified Ap4A as being a major metabolite that is elevated upon Kan treatment. We further show that Ap4A production requires the generation of hydroxyl radicals. Importantly, the synthetic alterations of intracellular Ap4A concentration via the elimination of the Ap4A phosphatase ApaH and the overexpression of the Ap4A synthetase LysU result in over a 5,000-fold increase in sensitivity to aminoglycosides. The increased antibiotic susceptibility in the Ap4A overproduction strain correlated well with the enhanced damage to the bacterial membrane during the Kan killing. Our findings that Ap4A promoting bacterial killing by this class of antibiotic provides a new path toward potentiator discovery that might reduce the toxicity of aminoglycosides through increasing their killing potency.
Results
Metabolomic Profiling Identifies Ap4A as Being Elevated upon Kan Exposure.
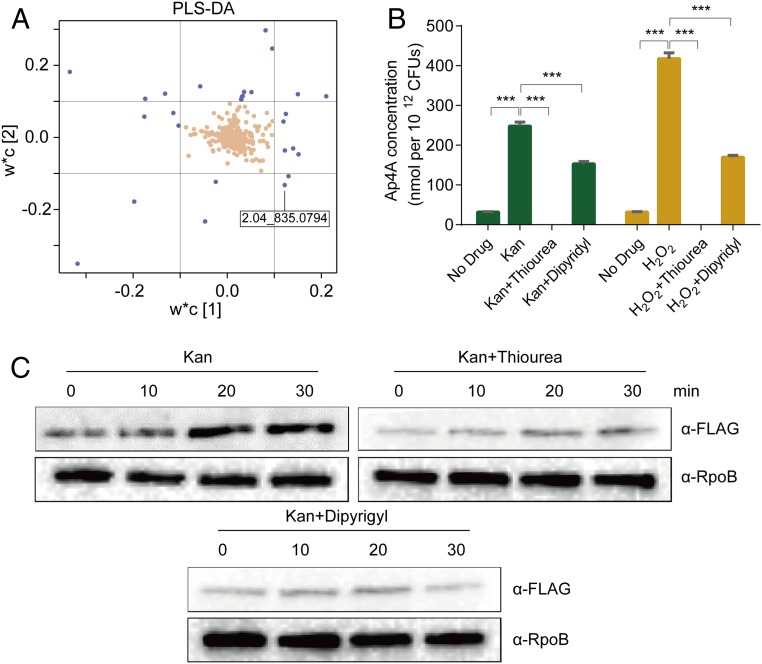
To investigate the bacterial response to aminoglycoside exposure, we treated midlog phase wild-type (WT) E. coli MG1655 with a lethal concentration of Kan (100 μg/mL), under which the majority (about 99.9%) of bacteria in the culture were killed after 3 h. Metabolomic profiling of cells upon 30-min treatment was performed with ultraperformance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry (UPLC/ESI-Q-TOF/MS) (SI Appendix, Fig. S1B). A partial least-squares discriminant analysis (PLS-DA) was performed, and the corresponding loading plot demonstrated 26 ions with the most prominent increase or decrease in abundance upon treatment with w*c [1] > |0.1| and w*c [2] > |0.1|, respectively (Fig. 1A and SI Appendix, Table S1). We focused on one ion (M13: m/z = 835.0794 at 2.04 min), which matched the Ap4A mass. The tandem mass spectrometry (MS/MS) fragmentation of the corresponding ion showed a fragmentation pattern that was the same as authentic Ap4A (SI Appendix, Fig. S1C) and consistent with a previous report on its chemical characterization (27). Based on a standard curve, built using authentic Ap4A (SI Appendix, Fig. S1D), the detected Ap4A increased about 20-fold in cells after 30 min of Kan treatment (Fig. 1B).
Fig. 1.
Ap4A is induced in E. coli upon Kan killing. (A) Loading plot of the PLS-DA showing the 26 ions (blue) with the most prominent increase or decrease in abundance upon 30-min Kan treatment with w*c [1] > |0.1| and w*c [2] > |0.1|. Each point represents a metabolic feature detected in the sample using UPLC/ESI-Q-TOF/MS. (B) Quantification of Ap4A in cells at 30-min post-treatment with 100-μg/mL Kan or 20-mM H2O2 as a control. Thiourea (300 mM) or 2,2′-dipyridyl (3 mM) was used to treat the cells. Each bar represents the mean ± SD from one representative experiment performed in triplicate. At least three biological replicates were performed. Significance was determined by one-way ANOVA. ***P < 0.001. (C) LysU is induced by Kan. lysU::flag was treated with 100-μg/mL Kan (Top Left); 100-μg/mL Kan plus 300-mM thiourea (Top Right); 100-μg/mL Kan plus 3-mM 2,2′-dipyridyl (Bottom). Cells were treated for 10, 20, and 30 min, and proteins were extracted for Western blot analysis. RNA polymerase (RpoB) was used as a control to show equal loading levels.
Elevation of Ap4A Involves Induction of Hydroxyl Radicals.
Kan targets the ribosome and corrupts protein synthesis, leading to the production of mistranslated aberrant proteins and the formation of hydroxyl radicals through a complex regulatory cascade that alters cellular metabolism (3, 5, 7). We examined whether Ap4A induction was related to intracellular hydroxyl radicals produced as a result of Kan treatment. Thiourea, a hydroxyl radical scavenger (28), and 2,2′-dipyridyl, an ion chelator (29), were used to quench hydroxyl radicals and to inhibit hydroxyl radical formation via Fenton reactions, respectively. The induction of Ap4A upon Kan treatment was abolished or significantly reduced in the presence of thiourea and 2,2′-dipyridyl, respectively (Fig. 1B).
Ap4A is known to be synthesized by several aminoacyl-tRNA synthetases in E. coli, among which LysU is the most active one (18). To further prove that thiourea and 2′-dipyridyl specifically reduced the Ap4A level in bacteria treated with Kan (Fig. 1B), we examined the expression level of LysU, the most active synthetase for Ap4A in E. coli (18), by Western blot analysis with the strains containing chromosomally FLAG tagged LysU (lysU::flag). We found that LysU production was induced upon Kan exposure and this was blocked by the presence of thiourea and 2,2′-dipyridyl (Fig. 1C and SI Appendix, Fig. S2), suggesting that elevated hydroxyl radicals likely play a role in the transcriptional activation of the lysU gene. These data are consistent with conclusions that aminoglycosides induce oxidative stress in E. coli (5) but also identify Ap4A as a new metabolic signal associated with exposure to this class of antibiotics.
Ap4A Promotes Death of E. coli in the Context of Aminoglycoside Exposure.
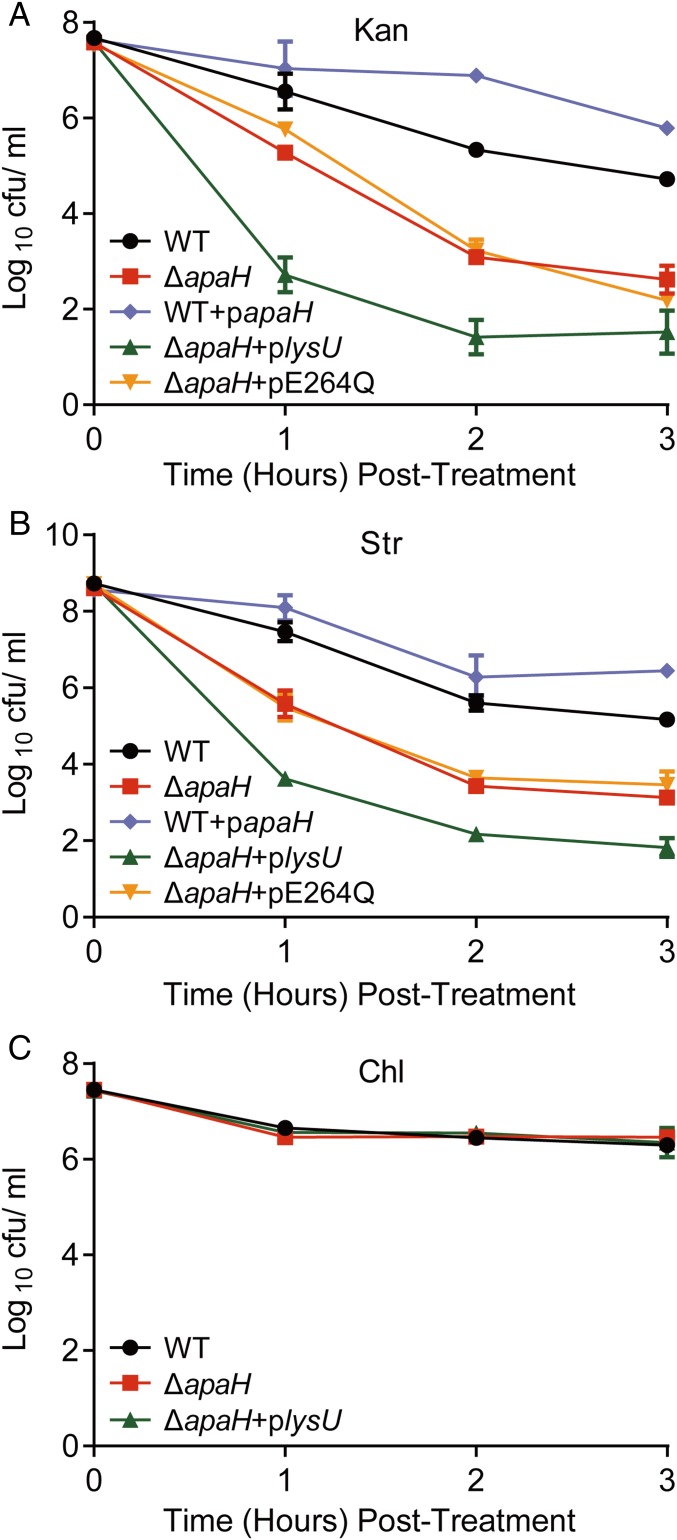
Intracellular Ap4A in E. coli is degraded by ApaH (19). To explore the role of Ap4A in aminoglycoside-mediated bactericidal activity, we compared the survival of WT and ∆apaH mutant cells when treated with lethal concentrations of these antibiotics. Although these two strains had comparable growth rates and sensitivities to Kan when determined in a standard minimal inhibitory concentration assay (MIC) (SI Appendix, Fig. S3 A and B), the ∆apaH mutant displayed an ∼100-fold reduction in survival compared with WT when exposed to either Kan or Str treatment (Fig. 2 A and B). The survival defect in ΔapaH was complemented with a plasmid containing WT apaH (SI Appendix, Fig. S3C). Furthermore, when the level of intracellular Ap4A was further elevated by overexpressing LysU in the context of an apaH mutant (ΔapaH + plysU), bacteria were 5,000–100,000 more susceptible to aminoglycoside toxicity compared with WT. The enhanced susceptibility of the ΔapaH + plysU mutant was likely a result of increased levels of intracellular Ap4A because the overexpression of the LysU derivative E264Q, a mutant defective in Ap4A synthesis (30) failed to increase the susceptibility of the ΔapaH mutant to either Kan or Str (Fig. 2 A and B). Also, reduction of intracellular Ap4A by overexpression of ApaH phosphatase (WT + papaH) significantly enhanced bacterial tolerance to Kan or Str killing (Fig. 2 A and B). Furthermore, the contribution of Ap4A to bacterial death was limited only to the mistranslation-inducing aminoglycosides but not to Chl, a bacteriostatic ribosome inhibitor, because the survival rate of either ΔapaH or ΔapaH + plysU was similar to WT upon treatment with this drug (Fig. 2C). Since bacteria become sensitive to aminoglycosides in strains with elevated levels of Ap4A and tolerant in strains with decreased levels of Ap4A, we conclude that the accumulation of Ap4A observed during treatment with aminoglycosides contributes directly to drug-dependent bacterial cell death rather than simply being the consequence of cell death (Figs. 1 A and B and 2). Together our results suggest that the baseline level of killing of E. coli cells by an aminoglycoside is determined, in part, by the level of Ap4A in the cells with higher levels of this alarmone driving increased sensitivity to this class of antibiotics.
Fig. 2.
Ap4A contributes to aminoglycoside killing on E. coli. Survival of E. coli WT and its derivatives after treatment with antibiotics. Bacterial cultures were treated with 100-μg/mL Kan (A), 100-μg/mL Str (B), or 30-μg/mL chloramphenicol (Chl) (C). Aliquots were taken from each sample at 1–3-h postantibiotic treatment for colony forming unit (CFU) enumeration. Both WT and ΔapaH contain a pBAD24 empty plasmid. Each point represents the mean ± SD from one representative experiment performed with triplicate samples. At least three biological replicates were performed.
High Intracellular Ap4A Correlates with Increased Bacterial Membrane Damage.
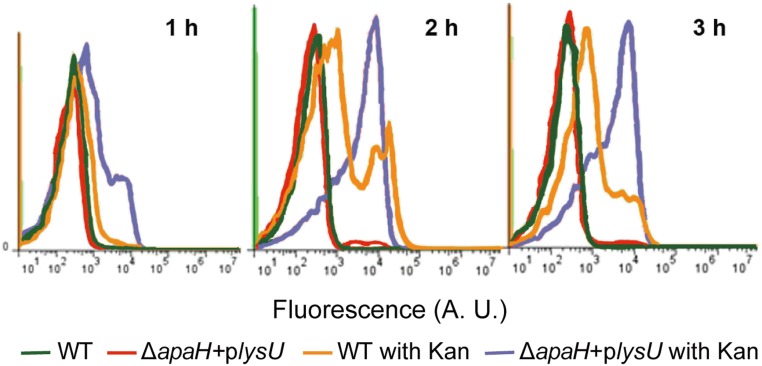
One of the hallmark phenotypes of aminoglycoside-mediated cell death is the incorporation of mistranslated proteins into the bacterial membrane, which leads to membrane damage and decreased membrane potential (3, 4). Such membrane damage can be measured through an increase in the fluorescent staining of bacterial cells with the dye DiBAC4(3) (31). We investigated whether an increase in intracellular Ap4A in the ΔapaH + plysU mutant affects the bacterial membrane potential during Kan treatment by flow cytometry. We found that, compared with WT cells, the ΔapaH + plysU mutant showed much higher DiBAC4(3) staining after Kan treatment (Fig. 3); this result correlated with the reduced survival of this mutant compared with WT after Kan treatment (Figs. 2A and 3). Therefore, our results suggest that production of Ap4A is a critical factor that drives cell death and this outcome is correlated with membrane damage. Because the ΔapaH + plysU mutant grows normally, we conclude that the alarmone Ap4A is not inherently toxic but rather enhances lethal outcomes that are associated with translational misreading following aminoglycoside exposure.
Fig. 3.
Flow cytometry profiles showing the membrane potential of WT and ΔapaH + plysU treated with or without 100-μg/mL Kan. Sample aliquots were harvested at 1–3-h post-treatment and preincubated with DiBAC4 (3).
Deletion of apaH in Other Bacterial Species and Enhancement of Aminoglycoside Bactericidal Activity.
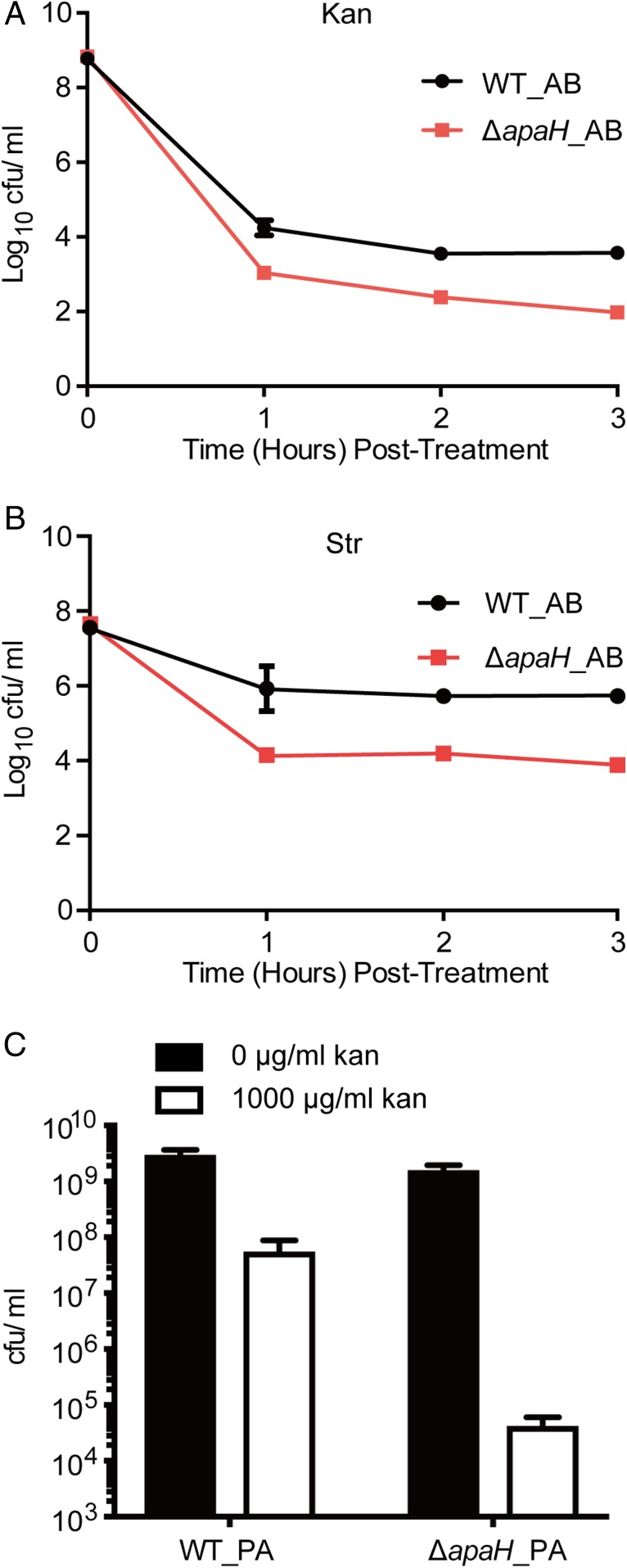
We then asked whether elevated intracellular Ap4A could increase susceptibility to aminoglycoside killing in other bacterial species. Acinetobacter baumannii is one of the most challenging nosocomial pathogens and is highly resistant to commonly used antibiotics (32). It is listed by WHO as a priority 1 (critical) pathogen for which there is an urgent need for more effective antibiotic treatment regimens (33). To examine whether Ap4A could also increase bacterial susceptibility to aminoglycoside killing in A. baumannii, we constructed a ΔapaH in ATCC17978, and compared the bacterial susceptibility of the WT and ΔapaH mutant upon Kan treatment. The A. baumannii ΔapaH mutant was more susceptible to Kan and Str killing and showed over a 50-fold greater reduction in bacterial survival than WT after 3 h of antibiotic treatment (Fig. 4 A and B). We did not attempt to further increase intracellular Ap4A by overexpressing LysU or other aminoacyl-tRNA synthetase as it is unknown which enzyme actively synthesizes Ap4A in A. baumannii. In addition, we also observed that deletion of apaH in P. aeruginosa caused approximately a 1,000-fold increase in sensitivity to Kan (Fig. 4C), suggesting that hypersensitivity to aminoglycosides caused by elevated Ap4A levels might be a widely conserved phenotype among gram-negative bacterial species.
Fig. 4.
Ap4A enhances aminoglycoside killing in A. baumannii and P. aeruginosa. (A and B) The survival of A. baumannii WT (WT_AB) and apaH mutant ΔapaH_AB upon treatment with 100-µg/mL Kan (A) or 200-µg/mL Str (B). (C) The survival of P. aeruginosa PAO1 WT (WT_PA) and ΔapaH (ΔapaH_PA) upon 3.5-h treatment with 1,000-μg/mL Kan. Both PAO1 WT and ΔapaH contain a copy of an empty pPSV37 plasmid. Cells were grown to midlog before treatment.
Discussion
Antibiotic resistance has emerged as a global crisis that demands new solutions. Aminoglycosides, one of the oldest classes of antibiotics, remain a valuable asset in this battle simply because they retain activity against many multidrug-resistant bacteria. However, the toxicities associated with aminoglycosides treatment in human patients limit their widespread clinical use. In this paper, we show that production of the alarmone Ap4A is dramatically elevated by aminoglycoside treatment of E. coli and that elevated levels of this metabolite promote cell death during aminoglycoside intoxication (Fig. 5). Elevation of Ap4A levels by either inactivation of the ApaH phosphatase or overexpression of the Ap4A synthetase LysU both caused 100–5,000 fold increases in Kan sensitivity. Because alteration of Ap4A levels using these synthetic approaches did not profoundly affect the MIC of Kan for E. coli, Ap4A may not affect adaption to low inhibitory but not bactericidal levels of this drug (SI Appendix, Fig. S3B). Nonetheless, our finding suggests that targeting Ap4A degradation or inducing its elevated synthesis might be an efficient way to enhance the killing potency of aminoglycosides for bacteria.
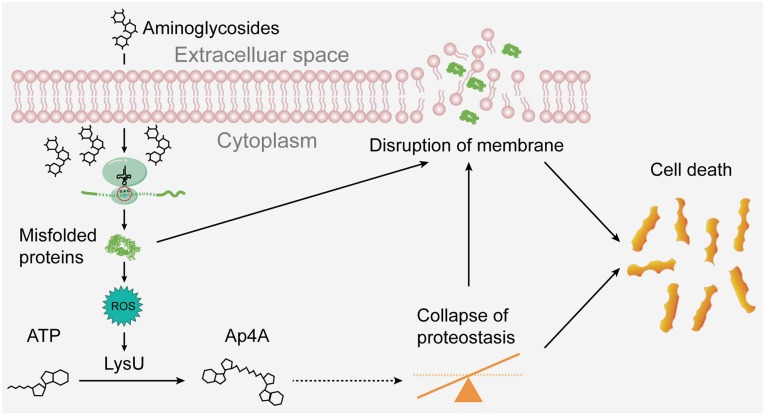
Fig. 5.
A model of Ap4A in promoting aminoglycoside killing. Treatment of bacteria with aminoglycosides leads to the production of misfolded proteins, which cause the disruption of the bacterial membrane and the generation of ROS. ROS induces LysU expression and its production of Ap4A. Ap4A might, in turn, inhibit intracellular proteostasis and aggravate the disruption of the membrane thereby promoting bacterial cell death. The dotted line arrow thus denotes only a potential pathway that Ap4A might contribute to cell death during aminoglycide exposure and was not directly demonstrated in this paper.
Ap4A has been long known to be induced by oxidative stress (17) and most recently antibiotics, such as aminoglycosides, have also been identified as elicitors of oxidative stress (5–7). In theory, the production of the Ap4A alarmone and oxidative stress induced by aminoglycoside exposure could be a coincidence and not functionally linked to the biology of how cell death occurs after exposure to this class of antibiotics. However, a key obfuscating factor is that aminoglycosides cause translational misreading and thus these antibiotics trigger production of aberrant proteins. Mistranslated proteins are inherently toxic, in part, because they cause membrane damage that triggers production of reactive oxygen metabolites (e.g., hydroxide anion) that further exacerbate cellular injury (5). Our results add another layer of complexity in understanding how aminoglycosides kill bacterial cells. Production of Ap4A after aminoglycoside exposure is dependent on the production of hydroxyl radicals in E. coli. Thus, the reactive oxygen metabolites produced after aminoglycoside exposure not only damage macromolecules (such as protein or DNA) of the bacterial cells (5), but also trigger the production of the alarmone Ap4A (Figs. 1 A and B and 5).
Second messengers are used by cells to trigger suitable responses to environmental or cellular signals that lead to improved survival, growth, or other outcomes. In the bacterial stringent response, for example, amino acid starvation provokes the production of (p)ppGpp, and accumulated intracellular (p)ppGpp then interacts with RNA polymerase, and drives large-scale transcriptional alterations that repress the expression of genes required for bacterial rapid growth and concomitantly activate the genes involved in amino acid biosynthesis, nutrient acquisition, and stress survival (34). The dinucleotide cyclic-di-GMP is known to regulate bacterial adaptation to environmental changes, and cyclic-GMP-AMP regulates the mammalian innate immune response (26, 35, 36). Other cyclic dinucleotides have been implicated in triggering cell death by activating toxic effector proteins (24, 25). Whereas the target(s) and detailed mechanism of action of Ap4A upon induction by aminoglycosides are not yet fully understood, our results suggest that the elevation of Ap4A may not be a strategy to rescue individual bacterial cells undergoing aminoglycoside-induced stress. Rather, our paper suggests that accumulation of intracellular Ap4A concentration promoted the lethal effects of aminoglycosides (Fig. 2). Bacterial strains lacking a functional ApaH are incapable of Ap4A degradation but have also been shown to display enhanced sensitivity to heat, starvation, antibiotics (37, 38), reduced formation of antibiotic persisters (39), and a weakened stress-induced mutagenesis response (40). The correct level of this alarmone might help cells cope with some cellular stresses, but high levels of Ap4A levels may actually drive bacterial death when the severity of stress reaches a certain threshold. Thus, Ap4A might be the signal for “the point of no return” for irreversably damaged cells experiencing overwhelming stress conditions. Alternatively, Ap4A could function simply as a toxic “damage metabolite” (41, 42): The oxidative stress caused by aminoglycosides induces the production and side reaction of aminogacyl tRNA synthetase (e.g., LysU) to generate Ap4A, which likely, in turn, inhibits the functions of some proteins that are essential for bacterial survival under stress, resulting in the enhanced bacterial susceptibility to aminoglycosides. Nevertheless, it is unclear why bacteria would evolve an active mechanism to enhance aminoglycoside killing through elevated synthesis of Ap4A. It is possible that selection through evolution developed the mechanism of action of aminoglycosides to take advantage of the Ap4A responses specifically to enhance the toxicity of mistranslated proteins in target bacterial species.
Aminoglycoside antibiotics bind directly to rRNA of the 30S subunit of the ribosome and corrupt the translation of messenger RNA (43). Mistranslated and misfolded proteins tend to aggregate or insert in cell membranes, both of which are detrimental to cells. Protein aggregation is significantly elevated in E. coli by aminoglycoside action and can be prevented by hydrogen peroxide scavengers (11). On the other hand, protein quality control (PQC) systems refold or degrade misfolded proteins as well as spatially sequestrate them into aggregates to maintain intracellular proteostasis (44, 45). For example, large protein aggregates are formed upon ampicillin treatment and have been found to be associated with drug tolerance and dormancy (46). Clearly there is still much we need to understand when it comes to proteostasis and the effects that antibiotics have on this process in bacteria.
Although the mechanism underlying how Ap4A promotes aminoglycoside killing needs to be further investigated, it is possible that Ap4A interrupts the function of PQC systems. Indeed, several key components of PQC, such as DnaK, ClpB, and GroEL, have been proposed to interact with Ap4A (37, 47). If so, the inhibition of the PQC system could collapse intracellular proteostasis and result in enhanced accumulation of misfolded proteins caused by aminoglycoside on the bacterial membrane and decrease its membrane potential. On the other hand, the intracellular misfolded and aberrant toxic proteins accumulated due to the collapse of proteostasis could also interrupt other essential cellular functions, promoting bacterial death. Thus, it is likely that bacterial membrane disruption and collapse of intracellular proteostasis altogether contribute to the elevated bacterial susceptibility to aminoglycosides (Fig. 5). Further investigation will be needed to define whether Ap4A alters the function of the PQC system during aminoglycoside exposure. In addition, Ap4A interacts with a large number of other proteins, such as SurA and NADPH:flavin oxidoreductase, as well as some transcriptional regulators (48, 49), suggesting that Ap4A likely has other broad effects on bacterial physiology that might also enhance sensitivity to aberrant mistranslated proteins.
Despite the urgent need for new drugs to treat resistant bacteria, the clinical usage of aminoglycosides has been limited due to their nephrotoxicity and ototoxicity at higher dosages (12, 13). Genetic screens for targets that might potentiate aminoglycoside activity have been performed with some promising success (50, 51). Our findings suggest a drug that alters Ap4A degradation or causes its overproduction might provide a new approach to enhance aminoglycoside killing potency, allowing lower nontoxic dosages of these antibiotics to be used in the treatment of multidrug-resistant infections. However, the establishment of a screening method for such a drug would require a better understanding of genes/proteins that are controlled or targeted by Ap4A, a topic that warrants further studies.
Materials and Methods
Bacterial Strains and Culture Conditions.
The bacterial strains and plasmids used in this study are listed in SI Appendix, Table S2 and S3. LB broth was used for all of the experiments except for bacterial survival measurements in which M9 medium (8.55-mM NaCl, 18.7-mM NH4Cl, 22-mM KH2PO4, 47.8-mM Na2HPO4, 10-mM CH3COONa, 1-mM MgSO4, 0.1-mM CaCl2, and 0.000005% thiamine) or M63 medium (28.7-mM K2HPO4, 22-mM KH2PO4, 15.13-mM (NH4)2SO4, 1.79-mM FESO4 heptahydrate, 1-mM MgSO4, 0.2% glycerol, and 0.5% casamino acids) were used for E. coli and P. aeruginosa, respectively. The antibiotics Kan (BP906-5; Fisher Scientific), ampicillin (BP1760; Fisher Scientific), gentamicin (G1264; Sigma-Aldrich), Chl (BP904; Fisher Scientific), and Str (BP910; Fisher Scientific) were used for the experiments described here.
Construction of Bacterial Strains and Plasmids.
In-frame deletion mutants and the strain with chromosomal FLAG tagged LysU (lysU::flag) were generated by the SacB-based allelic exchange as previously described (52). Plasmid pDS132 (53) was used for mutant construction in E. coli, and pEXG2 (54) was used in A. baummanni and P. aeruginosa. For construction of ΔapaH in P. aeruginosa, 1145-bp fragments upstream and 800-bp downstream of the apaH gene were generated that contained a linker allowing Gibson assembly of the two fragments. EcoRI and HindIII linearized pEXG2, and both fragments were ligated using Gibson assembly cloning kit (New England BioLabs), followed by transformation into DH5α. Correct clones were identified by sequencing. The pEXG2-ΔapaH plasmid was transformed into SM10λpir and conjugated with P. aeruginosa PAO1 with subsequent sucrose selection for a double-crossover event. Correct deletion of the apaH sequence was confirmed by sequencing. A similar protocol was used to construct other mutants with an In-Fusion HD Cloning kit (Clontech Laboratories, Inc.). Plasmid pBAD24 (55) was used for complementation (SI Appendix, Table S3). Site-directed mutagenesis of lysU was achieved by standard overlap PCR approaches followed by ligation of PCR fragment into the appropriate plasmid. All of the primers used for strain and plasmid construction are provided in SI Appendix, Table S4.
Bacterial Survival Measurements.
The antibiotic concentration used for lethality assays was optimized to reduce the CFUs of cultures by 2 to 3 logs after 3 h of treatment at a concentration of ∼108 CFU/mL Under our laboratory conditions, this corresponded to 100-μg/mL Kan or Str. Briefly, bacteria were grown in 3-mL LB medium (224620; BD Difco) in 15-mL Falcon tubes (352059; Corning) at 37 °C, 220 rpm, on an orbital shaker (ZQTY-70S; Zhichu). The overnight bacterial culture was then diluted at 1:100 into 15-mL M9 medium in 50-mL tubes (430829; Corning) and was cultured to OD600 ∼ 0.2 at 37 °C, 220 rpm, and 0.02% arabinose (Sigma-Aldrich) was added to induce the expression of lysU. Bacteria were grown further to OD600 ∼ 0.5 and were then treated with antibiotics. A volume of 100-μL treated cells were taken out at the indicated time point, serially diluted, and plated on LB agar plates for enumeration of surviving bacterial cells. For P. aeruginosa, overnight cultures in LB medium were back-diluted to OD600 0.05 into 1-mL M63 and grown to midlog in deep-well plates at 800 rpm. Some 1,000 μg/mL Kan was added and incubated at 37 °C for 3.5 h. Cultures were serially diluted and plated on LB agar for enumation of viable CFU. The minimal inhibition concentrations (MIC50s) were determined with the twofold serial microtiter broth dilution method (56). Bacteria were cultured in LB medium, and the growth of bacteria was measured by Tecan Infinite M200 Pro microplate readers at OD600 after 14-h incubation at 37 °C. The MIC50 values were calculated using Graphpad Prism 6.
Nontargeted Metabolomic Profiling.
For metabolomic profiling, bacteria (OD600 ∼ 0.5) were treated with or without 100-μg/mL Kan for 30 min. A volume of 30-mL cultured bacteria was pelleted at 4,000 ×g for 5 min at 4 °C, resuspended in 250-μL extraction buffer (methanol: acetonitrile: water = 40:40:20 with 0.1-N formic acid), and kept at −20 °C for 30 min to allow for the lysis of bacterial cells. Bacterial lysates were centrifuged at 15,000 ×g for 10 min at 4 °C, and the supernatant was collected and stored at −20 °C as the primary extract; the corresponding cell debris pellets were resuspended in 125-μL extraction buffer, stored at −20 °C for 15 min, and centrifuged at 15,000 ×g for 5 min at 4 °C to yield a secondary extract supernatant which, after thawing at 4 °C, were combined with the primary extracts. The mixtures were centrifuged at 15,000 ×g for 5 min to remove any bacterial debris. A volume of a 100-μL sample of the supernatant was transferred into a fresh tube. Extracted metabolites were analyzed by a C18 column (ACQUITY BEH C18, 100 × 2.1 mm, 1.7 μm, Waters) with a Waters ultraperformance liquid chromatography (UPLC) system conjugated with electrospray ionization. Accurate mass time of flight (Xevo G2-XS) (UPLC/ESI-TOF/MS) with the following settings: negative mode; capillary voltage: 2.5 kV; sampling cone voltage: 20 V; source temperature: 120 °C; desolvation gas temperature, 350 °C; cone gas flow 50 L/h; desolvation gas flow: 400 L/h; data were collected over the mass range of 400–1000; The mobile solvent A consisted of 0.1% N,N-dimethylhexylamine in 1-mM ammonium formate prepared in LC-MS grade water. The pH of phase A was adjusted to 9.0 with formate acid. Solvent B was HPLC grade acetonitrile, and the injection volume was 10 μL. The following gradient was used for elution of metabolites: 0 min, 5% B; 0 to 1 min, 5% B; 1.0–1.5 min, 25% B; 1.5–5.5 min, 25–95% B; 5.5–6.5 min 95% B. The primary flow was 0.4 mL/min, and the temperature of the column was maintained at 40 ± 5 °C.
Data processing and multivariate data analysis were performed as described previously with minor modifications (57). Briefly, centroided and integrated chromatographic mass data from 400 to 1000 m/z were processed by MarkerLynx (Waters) to build a multivariate data matrix. Pareto-scaled MarkerLynx matrices including information on sample identity were analyzed by principal components analysis and PLS-DA using SIMCA-P+ 12 (Umetrics). For identification of the nontargeted metabolites, the raw data were first converted to mzML format using ProteoWizard 3.0. The mzML files were uploaded to the XCMS website to create individual datasets for peak alignment and data filtering. The datasets were then compared using pairwise analysis with the UPLC/ESI-Q-TOF/MS parameter. The results were filtered using the following parameters: P value threshold at 0.05; fold change threshold ≥1.5; and maximum intensity ≥500. The m/z ratio from the species that fit the parameters was entered into the E. coli biosource online website search tool of Metlin with the following parameters: [M − H]− and [M ± Cl]− (for negative run), mass tolerance of 0.02 Da.
Quantification of Ap4A Concentration.
E. coli cells were grown in LB medium at 37 °C to OD600 ∼ 0.5. Cultures were treated with antibiotics (or H2O2 as a control) as indicated in the text. Treated bacteria were divided into two: One part of the sample was used for quantification of Ap4A by UPLC/ESI-TOF/MS following the same method used for nontargeted metabolites profiling. The other part was used for total RNA extraction with a RNeasy Mini Kit (QIAGEN) according to the product manual. A qRT-PCR for each RNA sample was performed with SYBR Premix Ex TaqII(TAKARA) to examine the level of the rpoB gene in the total RNA extracted from the bacterial samples. Primers used are listed in SI Appendix, Table S4.
To normalize the measured Ap4A concentration in each bacterial sample, a stand curve was built between bacterial CFU and rpoB mRNA levels (by Ct value of qRT-PCR): E. coli WT were grown in LB broth at 37 °C to OD600 ∼ 0.5. Some 10-fold serial dilutions were conducted. The diluted bacterial samples were divided into two parts: A 1-mL sample was used for total RNA extraction; another 1 mL of the same sample was used for CFU counting (diluted accordingly if necessary). A qRT-PCR for each RNA sample was performed with SYBR Premix Ex TaqII to examine the level of rpoB gene in the total RNA extracted from the bacterial samples. The same primers mentioned above were used (SI Appendix, Table S4). The Ct value for each sample was used to build a stand curve with the CFU number of each samples. To estimate CFU, we compared the Ct of the rpoB transcript in the antibiotic treated sample to the standard. The Ap4A concentration from UPLC/ESI-TOF/M was normalized based on the estimated CFU. A schematic for the whole procedure is provided in SI Appendix, Fig. S4.
Expression Analysis.
For analysis of the lysU expression level, bacteria were treated with Kan or elevated temperature as a control. A concentration of 300-mM thiourea or 3-mM 2,2′-dipyridyl was added when necessary. After treatment, cultures were centrifuged at 15,000 ×g for 2 min. Pellets were resuspended in lysis buffer (12.5-mM Tris pH 6.8, 4% SDS) and lysed by boiling for 10 min at 95 °C. The concentration of extracted proteins was determined using the Bio-Rad Protein Assay. An equal amount of protein from different samples was analyzed by Western blot analysis with an anti-FLAG antibody (Sigma-Aldrich).
Membrane Potential Measurement by Flow Cytometry.
The bacterial membrane potential was measured as described previously (31). A fresh E. coli culture was grown in LB broth to OD600 of 0.2–0.3 in 15-mL Falcon tubes at 37 °C, 220 rpm. Arabinose at a final concentration of 0.2% was then provided to induce the expression of lysU. Bacteria were grown further to OD600 ∼ 0.5 and were then treated with 100-μg/mL Kan for 1–3 h. A volume of 1 mL of antibiotic treated bacteria cells were collected and washed with 1 × PBS, followed by staining with 5-μg/mL DiBAC4 (3) for 15 min. After being filtered through a 10-μm filter, the cells were analyzed by flow cytometry (BD Accuri C6 Cytometer, laser excitation: 488 nm; emission detection: FL1 533/30 nm). GFP-expressing cells were used to determine the appropriate gates for single cells and the setting for forward scatter, side scatter, and cell density. The adjusted setting and gates were used for all subsequent experiments. Data were processed using the FlowJo 7.6 software.
Supplementary Material
Acknowledgments
We thank the Metabolomics core, Faculty of Health Sciences, University of Macau, for the technical assistance in metabolic profiling. This work was supported by the Macau Science and Technology Development Fund (Grants FDCT/066/2015/A2 and FDCT/0058/2018/A2), and Research Committee of University of Macau (Grants MYRG2016-00073-FHS and MYRG2016-00199-FHS, to J. Zheng), as well as a grant from the National Institute of Allergy and Infectious Disease, USA (AI-26289, to J.J.M.). A.-S.S. was supported by the German Research Association (DFG) with the Project STO 1208/1-1.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1822026116/-/DCSupplemental.
References
- 1.O’Neill J. HM Government; London: 2014. Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations The Review on Antimicrobial Resistance. [Google Scholar]
- 2.Pagkalis S, Mantadakis E, Mavros MN, Ammari C, Falagas ME. Pharmacological considerations for the proper clinical use of aminoglycosides. Drugs. 2011;71:2277–2294. doi: 10.2165/11597020-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 3.Davis BD. Mechanism of bactericidal action of aminoglycosides. Microbiol Rev. 1987;51:341–350. doi: 10.1128/mr.51.3.341-350.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis BD, Chen LL, Tai PC. Misread protein creates membrane channels: An essential step in the bactericidal action of aminoglycosides. Proc Natl Acad Sci USA. 1986;83:6164–6168. doi: 10.1073/pnas.83.16.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 7.Dwyer DJ, et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci USA. 2014;111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dukan S, et al. Protein oxidation in response to increased transcriptional or translational errors. Proc Natl Acad Sci USA. 2000;97:5746–5749. doi: 10.1073/pnas.100422497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 10.Vaubourgeix J, et al. Stressed mycobacteria use the chaperone ClpB to sequester irreversibly oxidized proteins asymmetrically within and between cells. Cell Host Microbe. 2015;17:178–190. doi: 10.1016/j.chom.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling J, et al. Protein aggregation caused by aminoglycoside action is prevented by a hydrogen peroxide scavenger. Mol Cell. 2012;48:713–722. doi: 10.1016/j.molcel.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu WJ, Sha SH, Schacht J. Recent advances in understanding aminoglycoside ototoxicity and its prevention. Audiol Neurotol. 2002;7:171–174. doi: 10.1159/000058305. [DOI] [PubMed] [Google Scholar]
- 13.Mingeot-Leclercq MP, Tulkens PM. Aminoglycosides: Nephrotoxicity. Antimicrob Agents Chemother. 1999;43:1003–1012. doi: 10.1128/aac.43.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker RA, Bennett WH, Porter GA. Animal models in the study of aminoglycoside nephrotoxicity. In: Whelton A, Neu HC, editors. The Aminoglycosides: Microbiology, Clinical Use and Toxi- Cology. Marcel Dekker, Inc.; New York: 1982. pp. 235–267. [Google Scholar]
- 15.Mogre A, Sengupta T, Veetil RT, Ravi P, Seshasayee AS. Genomic analysis reveals distinct concentration-dependent evolutionary trajectories for antibiotic resistance in Escherichia coli. DNA Res. 2014;21:711–726. doi: 10.1093/dnares/dsu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Randerath K, Janeway CM, Stephenson ML, Zamecnik PC. Isolation and characterization of dinucleoside tetra- and tri-phosphates formed in the presence of lysyl-sRNA synthetase. Biochem Biophys Res Commun. 1966;24:98–105. doi: 10.1016/0006-291x(66)90416-5. [DOI] [PubMed] [Google Scholar]
- 17.Bochner BR, Lee PC, Wilson SW, Cutler CW, Ames BN. AppppA and related adenylylated nucleotides are synthesized as a consequence of oxidation stress. Cell. 1984;37:225–232. doi: 10.1016/0092-8674(84)90318-0. [DOI] [PubMed] [Google Scholar]
- 18.Charlier J, Sanchez R. Lysyl-tRNA synthetase from Escherichia coli K12. Chromatographic heterogeneity and the lysU-gene product. Biochem J. 1987;248:43–51. doi: 10.1042/bj2480043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farr SB, Arnosti DN, Chamberlin MJ, Ames BN. An apaH mutation causes AppppA to accumulate and affects motility and catabolite repression in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:5010–5014. doi: 10.1073/pnas.86.13.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee YN, Nechushtan H, Figov N, Razin E. The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FcepsilonRI-activated mast cells. Immunity. 2004;20:145–151. doi: 10.1016/s1074-7613(04)00020-2. [DOI] [PubMed] [Google Scholar]
- 21.Yannay-Cohen N, et al. LysRS serves as a key signaling molecule in the immune response by regulating gene expression. Mol Cell. 2009;34:603–611. doi: 10.1016/j.molcel.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 22.Motzik A, et al. Post-translational modification of HINT1 mediates activation of MITF transcriptional activity in human melanoma cells. Oncogene. 2017;36:4732–4738. doi: 10.1038/onc.2017.81. [DOI] [PubMed] [Google Scholar]
- 23.Danilchanka O, Mekalanos JJ. Cyclic dinucleotides and the innate immune response. Cell. 2013;154:962–970. doi: 10.1016/j.cell.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Severin GB, et al. Direct activation of a phospholipase by cyclic GMP-AMP in El Tor Vibrio cholerae. Proc Natl Acad Sci USA. 2018;115:E6048–E6055. doi: 10.1073/pnas.1801233115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whiteley AT, et al. Bacterial cGAS-like enzymes synthesize diverse nucleotide second messengers. Nature. 2019;567:194–199. doi: 10.1038/s41586-019-0953-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou W, et al. Structure of the human cGAS-DNA complex reveals enhanced control of immune surveillance. Cell. 2018;174:300–311. doi: 10.1016/j.cell.2018.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulz A, Jankowski V, Zidek W, Jankowski J. Highly sensitive, selective and rapid LC-MS method for simultaneous quantification of diadenosine polyphosphates in human plasma. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;961:91–96. doi: 10.1016/j.jchromb.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 28.Repine JE, Fox RB, Berger EM. Hydrogen peroxide kills Staphylococcus aureus by reacting with staphylococcal iron to form hydroxyl radical. J Biol Chem. 1981;256:7094–7096. [PubMed] [Google Scholar]
- 29.Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, et al. Multiple catalytic activities of Escherichia coli lysyl-tRNA synthetase (LysU) are dissected by site-directed mutagenesis. FEBS J. 2013;280:102–114. doi: 10.1111/febs.12053. [DOI] [PubMed] [Google Scholar]
- 31.Goltermann L, Good L, Bentin T. Chaperonins fight aminoglycoside-induced protein misfolding and promote short-term tolerance in Escherichia coli. J Biol Chem. 2013;288:10483–10489. doi: 10.1074/jbc.M112.420380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie R, Zhang XD, Zhao Q, Peng B, Zheng J. Analysis of global prevalence of antibiotic resistance in Acinetobacter baumannii infections disclosed a faster increase in OECD countries. Emerg Microbes Infect. 2018;7:31. doi: 10.1038/s41426-018-0038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.World Health Organization 2017 WHO publishes list of bacteria for which new antibiotics are urgently needed. Available at www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/ Accessed April 6, 2019.
- 34.Gaca AO, Colomer-Winter C, Lemos JA. Many means to a common end: The intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol. 2015;197:1146–1156. doi: 10.1128/JB.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenal U, Reinders A, Lori C. Cyclic di-GMP: Second messenger extraordinaire. Nat Rev Microbiol. 2017;15:271–284. doi: 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 36.Du M, Chen ZJ. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361:704–709. doi: 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnstone DB, Farr SB. AppppA binds to several proteins in Escherichia coli, including the heat shock and oxidative stress proteins DnaK, GroEL, E89, C45 and C40. EMBO J. 1991;10:3897–3904. doi: 10.1002/j.1460-2075.1991.tb04959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou J, et al. Studies on aminoglycoside susceptibility identify a novel function of KsgA to secure translational fidelity during antibiotic stress. Antimicrob Agents Chemother. 2018;62:e00853-18. doi: 10.1128/AAC.00853-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen S, Lewis K, Vulić M. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob Agents Chemother. 2008;52:2718–2726. doi: 10.1128/AAC.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al Mamun AA, et al. Identity and function of a large gene network underlying mutagenic repair of DNA breaks. Science. 2012;338:1344–1348. doi: 10.1126/science.1226683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Linster CL, Van Schaftingen E, Hanson AD. Metabolite damage and its repair or pre-emption. Nat Chem Biol. 2013;9:72–80. doi: 10.1038/nchembio.1141. [DOI] [PubMed] [Google Scholar]
- 42.Despotović D, et al. Diadenosine tetraphosphate (Ap4A)–An E. coli alarmone or a damage metabolite? FEBS J. 2017;284:2194–2215. doi: 10.1111/febs.14113. [DOI] [PubMed] [Google Scholar]
- 43.Magnet S, Blanchard JS. Molecular insights into aminoglycoside action and resistance. Chem Rev. 2005;105:477–498. doi: 10.1021/cr0301088. [DOI] [PubMed] [Google Scholar]
- 44.Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- 45.Mogk A, Bukau B, Kampinga HH. Cellular handling of protein aggregates by disaggregation machines. Mol Cell. 2018;69:214–226. doi: 10.1016/j.molcel.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Pu Y, et al. ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol Cell. 2018;73:143–156. doi: 10.1016/j.molcel.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 47.Fuge EK, Farr SB. AppppA-binding protein E89 is the Escherichia coli heat shock protein ClpB. J Bacteriol. 1993;175:2321–2326. doi: 10.1128/jb.175.8.2321-2326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azhar MA, Wright M, Kamal A, Nagy J, Miller AD. Biotin-c10-AppCH2ppA is an effective new chemical proteomics probe for diadenosine polyphosphate binding proteins. Bioorg Med Chem Lett. 2014;24:2928–2933. doi: 10.1016/j.bmcl.2014.04.076. [DOI] [PubMed] [Google Scholar]
- 49.Guo W, et al. Isolation and identification of diadenosine 5′,5′''-P1,P4-tetraphosphate binding proteins using magnetic bio-panning. Bioorg Med Chem Lett. 2011;21:7175–7179. doi: 10.1016/j.bmcl.2011.09.070. [DOI] [PubMed] [Google Scholar]
- 50.Gallagher LA, Shendure J, Manoil C. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. MBio. 2011;2:e00315-10. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gallagher LA, Lee SA, Manoil C. Importance of core genome functions for an extreme antibiotic resistance trait. MBio. 2017;8:e01655-17. doi: 10.1128/mBio.01655-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng J, Tung SL, Leung KY. Regulation of a type III and a putative secretion system in Edwardsiella tarda by EsrC is under the control of a two-component system, EsrA-EsrB. Infect Immun. 2005;73:4127–4137. doi: 10.1128/IAI.73.7.4127-4137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid. 2004;51:246–255. doi: 10.1016/j.plasmid.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 54.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2005;102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jorgensen JH, Turnidge JD. Manual of Clinical Microbiology. 11th Ed. American Society of Microbiology; Washington DC: 2015. Susceptibility test methods: dilution and disk diffusion methods; pp. 1253–1273. [Google Scholar]
- 57.Cho JY, et al. Urinary metabolomics in Fxr-null mice reveals activated adaptive metabolic pathways upon bile acid challenge. J Lipid Res. 2010;51:1063–1074. doi: 10.1194/jlr.M002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.