Abstract
Objective
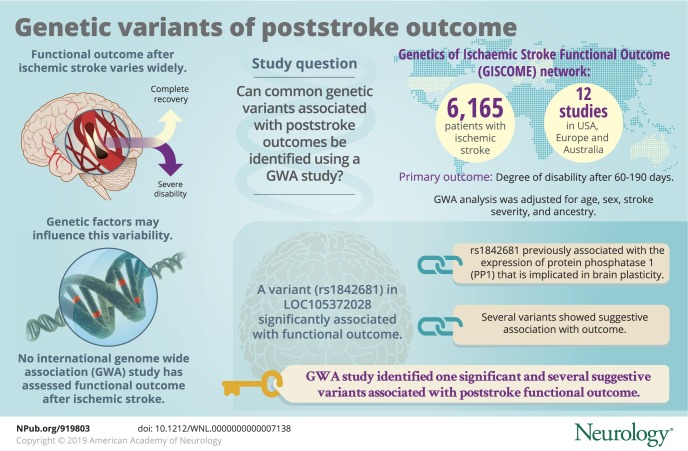
To discover common genetic variants associated with poststroke outcomes using a genome-wide association (GWA) study.
Methods
The study comprised 6,165 patients with ischemic stroke from 12 studies in Europe, the United States, and Australia included in the GISCOME (Genetics of Ischaemic Stroke Functional Outcome) network. The primary outcome was modified Rankin Scale score after 60 to 190 days, evaluated as 2 dichotomous variables (0–2 vs 3–6 and 0–1 vs 2–6) and subsequently as an ordinal variable. GWA analyses were performed in each study independently and results were meta-analyzed. Analyses were adjusted for age, sex, stroke severity (baseline NIH Stroke Scale score), and ancestry. The significance level was p < 5 × 10−8.
Results
We identified one genetic variant associated with functional outcome with genome-wide significance (modified Rankin Scale scores 0–2 vs 3–6, p = 5.3 × 10−9). This intronic variant (rs1842681) in the LOC105372028 gene is a previously reported trans-expression quantitative trait locus for PPP1R21, which encodes a regulatory subunit of protein phosphatase 1. This ubiquitous phosphatase is implicated in brain functions such as brain plasticity. Several variants detected in this study demonstrated suggestive association with outcome (p < 10−5), some of which are within or near genes with experimental evidence of influence on ischemic stroke volume and/or brain recovery (e.g., NTN4, TEK, and PTCH1).
Conclusions
In this large GWA study on functional outcome after ischemic stroke, we report one significant variant and several variants with suggestive association to outcome 3 months after stroke onset with plausible mechanistic links to poststroke recovery. Future replication studies and exploration of potential functional mechanisms for identified genetic variants are warranted.
Functional outcomes after ischemic stroke have a wide range of interindividual variability, from complete recovery to persistent severe disability.1 Although a large number of factors influence recovery after ischemic stroke, such as age and initial stroke severity, a discernible portion of this interindividual variation remains unexplained by clinical factors.1 The unexplored mechanisms behind this variation represent potential keys in the search for a personalized poststroke management approach that would include accurate prognostic prediction as well as patient-tailored treatment, secondary prevention, and rehabilitation.
Genetic factors may account for part of the variability in stroke outcomes. Studies in humans and animals support a genetic influence on recovery after brain injury.2–4 In addition, genetic variants identified through candidate gene studies have been reported to associate with functional outcome after stroke, e.g., within the brain-derived neurotrophic factor and cyclooxygenase-2 genes,5–7 although these candidate gene study results have been inconsistent and need replication.1 To date, no genome-wide association (GWA) study has been published on functional outcome after overall ischemic stroke. Such a hypothesis-free approach may identify variants indicating novel pathways for poststroke pathophysiologic processes or recovery, and thereby suggest new targets for interventions and drug development.
The Genetics of Ischaemic Stroke Functional Outcome (GISCOME) network aims to identify genetic variants that may influence functional outcome after ischemic stroke.8 Here, we present the results from a GWA study of functional outcome as assessed by the modified Rankin Scale (mRS) at 3 months after ischemic stroke.
Methods
Study population
The GISCOME study population and analysis plan have been previously described in detail.8 In brief, the GISCOME study population consists of patients with ischemic stroke of mainly European ancestry aged 18 years and older from 12 study locations in Europe, the United States, and Australia. After the publication of the GISCOME study protocol,8 2 additional sets of data from (1) the Sahlgrenska Academy Study on Ischemic Stroke, and (2) the Malmö Diet and Cancer Study have been added (data available from Dryad, supplemental Methods and tables e-1 and e-2, doi.org/10.5061/dryad.s38kf65). The GISCOME study includes centers with mRS and genotype data contributing to the International Stroke Genetics Consortium and the National Institute of Neurological Disorders and Stroke – Stroke Genetics Network (SiGN) study efforts studying genetics of stroke risk. We did not have any additional cohorts available at the time of this study for replication.
Standard protocol approvals, registrations, and patient consents
All participants provided written informed consent to participate. For participants who were unable to communicate, consent was obtained from their next of kin. Local ethics committees approved the individual studies.
Outcome
The mRS as close as possible to 90 days (60–190 days permitted) was selected to assess functional outcome, as described in the analysis plan.8 The majority of the included studies (≈80%) assessed mRS at 3 months ± 2 weeks.8 In most studies, this was done by face-to-face interviews. In 3 studies (Lund Stroke Register, Malmö Diet and Cancer Study, and parts of the Sahlgrenska Academy Study on Ischemic Stroke), data from the Swedish quality register for stroke (Riksstroke) were used to assess mRS by a validated translation algorithm.8,9 This approach prevented a differentiation between the mRS scores 0, 1, and 2.
Based on our a priori analysis plan, we analyzed mRS as 2 dichotomous variables, (1) mRS 0–2 vs 3–6 and (2) mRS 0–1 vs 2–6, and also as the full ordinal scale variable. The mRS 0–2 vs 3–6 and ordinal scale analyses included a larger number of participants. Analyses of mRS 0–1 vs 2–6 were performed to explore whether a second dichotomization could identify any strong associations that were not picked up by the other analyses. In addition, we made an explorative effort to investigate potential associations with infarcts in subcortical and cortical locations separately. Since information about infarct location was not available, we divided the cases into small vessel disease (referred to as lacunar stroke) and other subtypes (referred to as nonlacunar stroke), according to subtype classifications in TOAST (Trial of ORG 10172 in Acute Stroke Treatment).10 A total of 992 patients had lacunar stroke, 3,991 patients had nonlacunar stroke, and for 1,182 individuals, this information was missing.
GWA analysis and meta-analysis
The methods for genotyping, imputation, and quality control of genotype data are described in data available from Dryad (supplemental Methods and tables e-1 and e-2, doi.org/10.5061/dryad.s38kf65). Multivariable models were used for analyses of each outcome variable, under an additive genetic model. We aimed to explore genetic variants associated with functional outcome that were independent of stroke severity. In this primary model, results were adjusted for age, sex, ancestry (up to the first 5 principal components [data available from Dryad, table e-3]), and baseline stroke severity as assessed by the NIH Stroke Scale (NIHSS) at 0 to 10 days post stroke onset, with preference to as close to day 0–1 as possible.8 In addition, models without adjustment for baseline stroke severity were performed for each outcome for comparison; however, unless otherwise stated, all results in this report refer to the primary model as described above.
All dichotomized analyses were performed with logistic regression using PLINK software version 1.90b4.6.11 The full mRS was analyzed with ordinal logistic regression under a cumulative logit model using the MATLAB mnrfit algorithm (MATLAB Statistics and Machine Learning Toolbox Release 2016b; The MathWorks, Inc., Natick, MA). Under the proportional odds assumption, the effect of a predictor single-nucleotide polymorphism (SNP) is invariant of the choice of outcome categories. As a result, estimates from all study cohorts, including those that did not separate mRS 0–2, are comparable in ordinal regression. Furthermore, one unit change in the dosage of a variant with regression coefficient β suggests a change in the odds of an mRS score lower than or equal to x vs higher than x by a factor 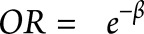 , equally for all scores x. We tested deviations from proportional odds for all reported variants from the ordinal models. No significant deviations were found.
, equally for all scores x. We tested deviations from proportional odds for all reported variants from the ordinal models. No significant deviations were found.
Inverse variance-weighted fixed-effects meta-analysis was performed using METAL software.12 Variants with minor allele frequency <0.01 were excluded. Variants that were missing in >50% of cohorts were also excluded. After filtering, approximately 8.5 million SNPs were included in each of the final meta-analyses. Quantile-quantile plots are shown in figures 1 and 2 and in data available from Dryad (figures e-1 to e-4, doi.org/10.5061/dryad.s38kf65), and genomic inflation factors (λ) are displayed in data available from Dryad (table e-4). There was no sign of population stratification based on these measures. Heterogeneity of SNP effects between studies was tested in the meta-analysis.
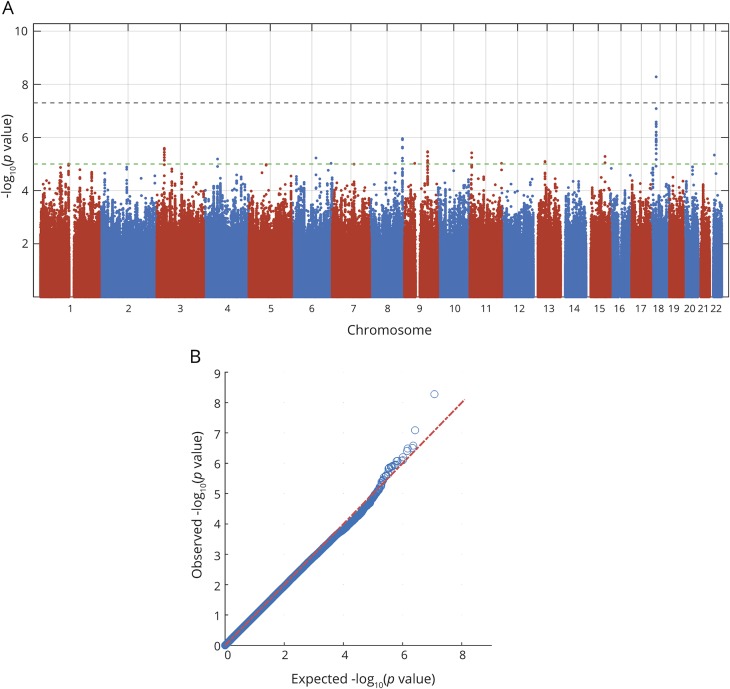
Figure 1. Manhattan and quantile-quantile plots of analysis for associations with dichotomized mRS at 3 months.
Outcome was measured as mRS 0–2 vs 3–6 at 3 months after ischemic stroke onset. Dotted lines show genome-wide significance (black, p <5 × 10−8) and suggestive association level (green, p < 10−5). Results are adjusted for age, sex, principal components, and baseline NIH Stroke Scale score. mRS = modified Rankin Scale.
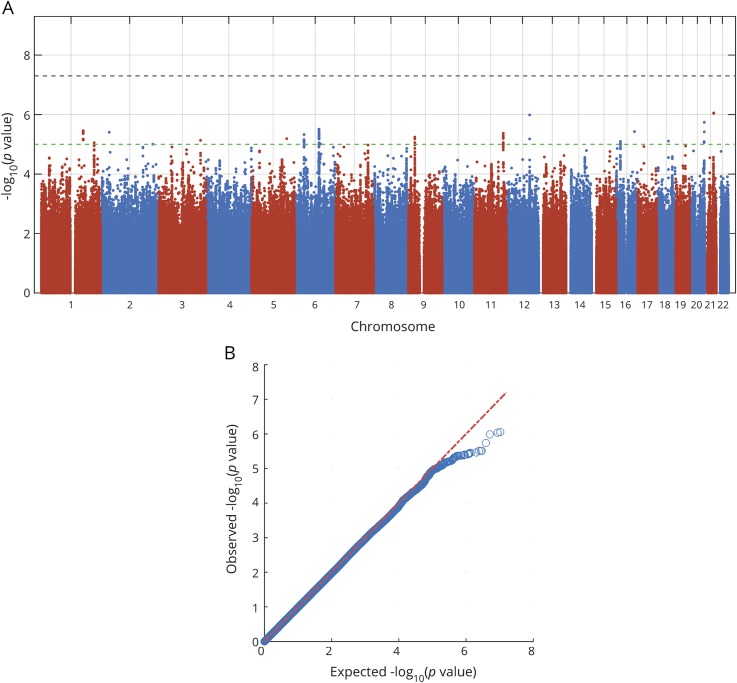
Figure 2. Manhattan and quantile-quantile plots of analysis for associations with ordinal mRS at 3 months.
Outcome was measured as ordinal mRS at 3 months after ischemic stroke onset. Dotted lines show genome-wide significance (black, p <5 × 10−8) and suggestive association level (green, p < 10−5). Results are adjusted for age, sex, principal components, and baseline NIH Stroke Scale score. mRS = modified Rankin Scale.
Markers with p values <5 × 10−8 were considered significant for association with outcome, while markers with p values <1 × 10−5 were considered suggestive. To facilitate comparison of the results from the dichotomized and ordinal analyses, we present all effect sizes as odds ratios (ORs) per copy of the minor allele; an OR >1 indicates a higher mRS score (worse outcome) per copy of the minor allele and an OR <1 indicates a lower mRS score.
Investigation of expression quantitative trait loci
We explored associations of the markers with p values <1 × 10−5 and proxy SNPs (r2 > 0.8 in 1000 Genomes Phase 1 European population) with expression quantitative trait loci (eQTLs) in publicly available datasets encompassing numerous tissues: Genotype-Tissue Expression (GTEx) V6,13 GRASP2,14,15 Human Genetic Variation Database (HGVD),16 and BIOS.17 For eQTLs, p < 10−4 was considered significant.
Gene-based analysis
Gene-based tests were performed for each meta-analysis using VEGAS2 with linkage disequilibrium structure based on the European population.18 All SNPs within ±10 kbp from the untranslated regions 3′ and 5′ of each gene were included, to account for potential regulatory variants.18 The number of genes included was approximately 23,000, which corresponds to a Bonferroni-corrected significance threshold of p < 2.2 × 10−6.
Data availability
The datasets generated and analyzed during the current study are available on reasonable request.
Results
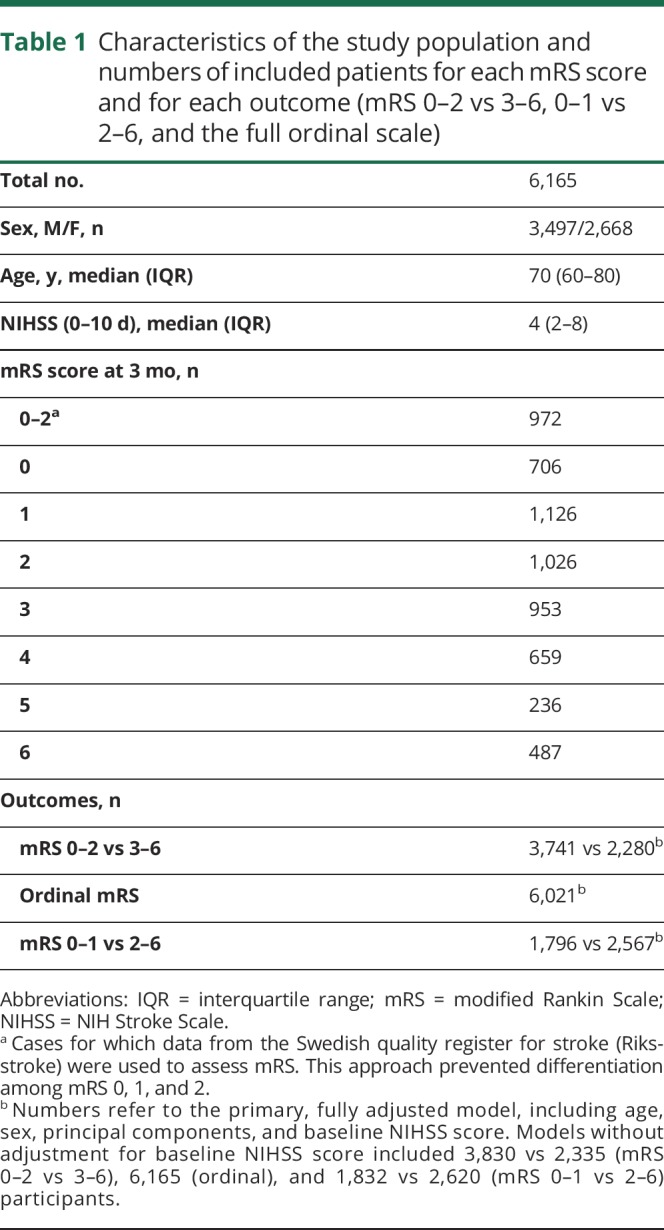
Characteristics such as age, sex, and stroke severity, as well as the numbers of included cases for each mRS score and in each outcome analysis, are displayed in table 1. The analyses of mRS 0–2 vs 3–6 and the ordinal analyses included 6,021 stroke cases, whereas analyses of mRS 0–1 vs 2–6 included 4,363 cases.
Table 1.
Characteristics of the study population and numbers of included patients for each mRS score and for each outcome (mRS 0–2 vs 3–6, 0–1 vs 2–6, and the full ordinal scale)
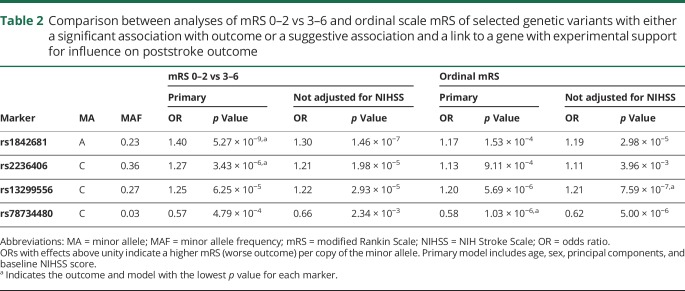
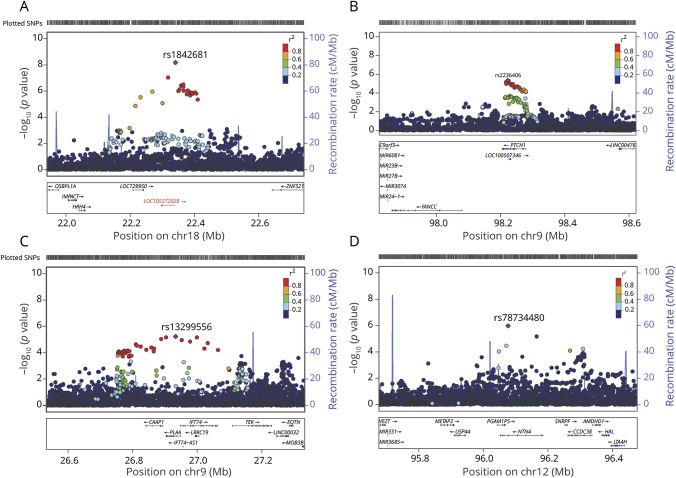
One common variant on chromosome 18q11.2 (rs1842681, minor allele frequency: 0.23) was associated at genome-wide significance with outcome defined as mRS 0–2 vs 3–6 (OR for minor allele [A]: 1.40, p = 5.3 × 10−9) (table 2, figures 1, 3, and 4). The effect was similar with and without adjustment for stroke severity (table 2), and the association was observed in the same direction, but with a somewhat lower effect size, for ordinal mRS (OR: 1.17, p = 1.5 × 10−4) and mRS 0–1 vs 2–6 (OR: 1.12, p = 7.4 × 10−2). In line with this, the distribution of the minor allele count for rs1842681 over mRS categories shows a threshold between mRS 2 and 3 (data available from Dryad, figure e-5, doi.org/10.5061/dryad.s38kf65). The variant is located in an intron of the gene LOC105372028 (long noncoding RNA synonymous with RP11-449D8.5 [Genome Reference Consortium Human Build 38/hg38]) and is in a putative binding site of several regulatory proteins (HaploReg, March 21, 2018). This variant has no eQTL reported in GTEx (June 1, 2018). However, in HGVD, it has a trans-eQTL for KLRAQ1 (also known as PPP1R21, pHGVD = 1.67 ×10−7). PPP1R21 is expressed in the brain (GTEx, June 1, 2018).
Table 2.
Comparison between analyses of mRS 0–2 vs 3–6 and ordinal scale mRS of selected genetic variants with either a significant association with outcome or a suggestive association and a link to a gene with experimental support for influence on poststroke outcome
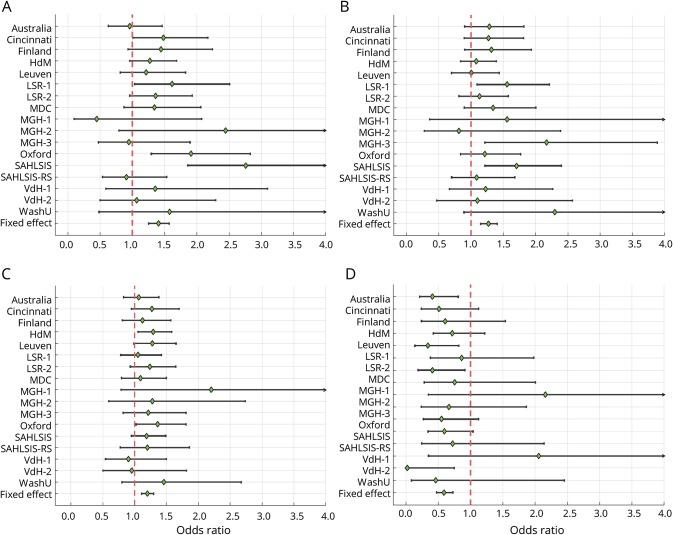
Figure 3. Forest plots for functional outcome at 3 months by study cohort.
The plots show ORs and 95% confidence intervals for minor allele of (A) rs1842681 (mRS 0–2 vs 3–6), (B) rs2236406 (mRS 0–2 vs 3–6), (C) rs13299556 (ordinal mRS), and (D) rs78734480 (ordinal mRS). ORs with effects above unity indicate a higher mRS score (worse outcome) per copy of the minor allele. Results are adjusted for age, sex, principal components, and baseline NIH Stroke Scale score. For cohort abbreviations, see data available from Dryad (table e-1, doi.org/10.5061/dryad.s38kf65). mRS = modified Rankin Scale; OR = odds ratio.
Figure 4. Regional association plots for outcome at 3 months after ischemic stroke onset.
(A) Significant locus (rs1842681) showing association with mRS 0–2 vs 3–6, (B) rs2236406 (intron variant in the PTCH1 gene) showing suggestive association with mRS 0–2 vs 3–6, (C) rs13299556 (intron variant in the PLAA gene with eQTL for TEK, pGTEx = 1.6 × 10−6) showing suggestive association with ordinal mRS, and (D) rs78734480 (intron variant in the NTN4 gene) showing suggestive association with ordinal mRS. Results are adjusted for age, sex, principal components, and baseline NIH Stroke Scale score. LOC105372028 (indicated in red, panel A) has been overlaid from the Genome Reference Consortium Human Build (GRCh) 38/hg38, as it was missing from GRCh37/hg19. The rs1842681 variant is intronic of the LOC105372028 gene (chromosome 18: 24725781–24766645). Position for rs1842681 in GRCh38/hg38, 18:24761199. eQTL = expression quantitative trait locus; GTEx = Genotype-Tissue Expression; mRS = modified Rankin Scale; SNP = single-nucleotide polymorphism.
No other SNP was significantly associated with mRS 0–2 vs 3–6, or with ordinal scale mRS (figures 1 and 2). The results for the lead SNPs of the top 10 independent loci for each outcome are displayed in data available from Dryad (tables e-5 and e-6, doi.org/10.5061/dryad.s38kf65). The results from analyses without severity adjustment are displayed in data available from Dryad (tables e-7 and e-8 and figures e-1, e-2, e-6, and e-7). There were no signs of heterogeneity for the effects of the reported SNPs. All I2 values were <0.35 and all p for heterogeneity >0.08, except for one SNP (rs58448576) in the analysis of mRS 0–1 vs 2–6, which had an I2 of 0.48 and a p value of 0.03 (without adjustment for multiple testing). In the gene-based analyses, no gene reached the predefined threshold for significance (data available from Dryad, table e-9).
The analysis of mRS 0–1 vs 2–6 yielded no significant associations. These results are presented in data available from Dryad (tables e-12 and e-13 and figures e-3, e-4, and e-8, doi.org/10.5061/dryad.s38kf65).
Suggestive associations with outcome
Thirty-three SNPs in 12 different loci (with at least 1 Mbp distance) were suggestively (p < 10−5) associated with mRS 0–2 vs 3–6 (excluding the SNPs in the significant locus on chromosome 18q11.2), and 75 SNPs in 17 different loci with ordinal mRS (figures 1 and 2). Of these 29 independent loci, the top SNPs of 16 loci have either significant eQTLs for and/or are located within or near (<100 kbp) genes that are expressed in the brain (GTEx, June 1, 2018). Five genes that were not reported to be expressed in brain tissue were expressed in arteries or lymphocytes (GTEx, June 1, 2018).
Among the suggestive associations, 3 are linked to genes with experimental evidence of influence on outcome from animal models of stroke and are discussed below.19–21 First, rs2236406, an intron variant in the PTCH1 gene, was identified in the mRS 0–2 vs 3–6 analysis (table 2, figures 3) and 4). The gene-based analysis also showed a suggestive association for PTCH1 with mRS 0–2 vs 3–6 (p = 6.8 × 10−5). Rs2236406 is an eQTL for long noncoding RNA RP11-435O5.5 (pGTEx = 5.7 × 10−7), which overlaps the PTCH1 gene. Second, in the ordinal analysis, a suggestive association with mRS was found for rs13299556 (table 2, figures 3) and 4). Associations at low p values were also found for this variant with dichotomous mRS, although not reaching the level for suggestive association (table 2). rs13299556 is an intron variant in the PLAA gene. There was no strong association for PLAA in the gene-based analysis (ordinal mRS; p = 0.072). However, this variant, and variants in high linkage disequilibrium with it (figure 4), are reported as eQTLs for the nearby genes TEK and LRRC19 (pGTEx = 1.6 × 10−6 for both). The p values from the gene-based analysis for those genes were 0.0019 and 0.084, respectively. The variant is also predicted to alter a putative regulatory motif sequence (HaploReg, March 21, 2018). Third, a suggestive association with ordinal mRS was found for an intron variant in NTN4 (rs78734480; table 2, figures 3 and 4). This variant was also associated with dichotomous mRS at a low p value, although not below the cutoff for suggestive association (table 2).
Analysis of lacunar and nonlacunar stroke
To evaluate whether any strong associations could be identified specifically in patients with lacunar stroke (small vessel disease strokes, n = 992), or in nonlacunar stroke (other etiologic subtypes including cortical infarcts, n = 3,991), we analyzed those groups separately. No genome-wide significant association was detected (all p values >6 × 10−7). The top 5 findings differed between these subgroups and are shown in data available from Dryad (tables e-10 and e-11, doi.org/10.5061/dryad.s38kf65).
Discussion
This GWA study on functional outcome after ischemic stroke, including more than 6,000 patients, identified one significant locus and several suggestive variants related to genes with a potential mechanism for influencing stroke recovery and outcomes. Replication of these findings in independent datasets as they become available is the essential next step. The effect sizes of these genetic variants were generally small, and further studies should include even larger samples to identify additional variants associated with stroke outcome and to enable subgroup analyses.
The genome-wide significant SNP (rs1842681) for mRS 0–2 vs 3–6 is an intronic variant in the gene LOC105372028 (long noncoding RNA), the function of which remains to be established. However, expression analyses show that the gene expression of LOC105372028 is highest in brain tissue (National Center for Biotechnology Information, ncbi.nlm.nih.gov/gene/?term=LOC105372028) and rs1842681 is located in a putative regulatory element binding site (HaploReg, March 21, 2018). Furthermore, rs1842681 is a trans-eQTL for KLRAQ1, also known as PPP1R21, which encodes a regulatory subunit of protein phosphatase 1 (PP1). PP1 is a ubiquitous phosphatase implicated in many brain functions including learning and memory formation.22,23 PP1 is also a key regulator of Ca2+/calmodulin (CaM)-dependent protein kinase II (CaMKII) signaling, which is crucial for Ca2+-mediated neuronal plasticity in the brain.24 Thus, although speculative, rs1842681 may modulate expression of PPP1R21, which in turn could affect brain plasticity and thereby outcome post stroke, a hypothesis that requires validation through functional experiments. The association for rs1842681 with outcome was observed after adjusting for baseline NIHSS score, suggesting a mechanism that is independent of initial stroke severity. However, additional data from further studies are clearly needed to corroborate this finding and clarify the potential role of this locus for functional outcome after stroke.
Twenty-nine independent loci were associated with ischemic stroke outcome at the predefined suggestive association level. Three of these loci are linked to genes with experimental evidence of influence on outcome from animal models of stroke.19–21 First, the intronic variant in PLAA (rs13299556) was both one of the top findings in the ordinal analysis and associated with the dichotomized outcome at a low p value. It is located in a putative transcription factor binding site and is reported to be an eQTL for the expression of both LRRC19 and TEK. LRRC19 encodes a protein with a potential role in regulating neurite outgrowth,25 and might thus influence stroke recovery. TEK encodes a tyrosine kinase predominantly expressed in endothelial cells (ECs). Focal upregulation of TEK has been demonstrated in capillaries at the border of infarcted myocardium, and expression is induced by hypoxia and proinflammatory cytokines in human ECs in vitro.26 TEK is activated by angiopoietin-1 and this promotes migration, sprouting, and survival of ECs.27,28 Of note, results from a rodent ischemic stroke model suggest that upregulation of angiopoietin-1 and TEK improves stroke outcome,19 and one clinical study reported an association between high plasma levels of angiopoietin-1 and poor outcome as assessed by mRS at 3 months after ischemic stroke.29 Angiopoietin-2 is the antagonist of angiopoietin-1.30 Angiopoietin-2 gain-of-function mice have enhanced blood-brain barrier permeability and increased brain infarct sizes upon permanent middle cerebral occlusion compared to wild-type mice, and both phenotypes were rescued by activation of TEK signaling.31 The same study reported increased circulating serum concentrations of angiopoietin-2 in patients with acute ischemic stroke compared to controls.31 Finally, in an ischemic stroke model, mice with type 2 diabetes mellitus showed increased angiopoietin-2 but decreased angiopoietin-1/TEK protein expression compared to wild-type mice, suggesting that the TEK signaling may be involved in diabetes-induced vascular damage post stroke.32
Second, an intronic variant in the PTCH1 gene showed suggestive association with mRS 0–2 vs 3–6. PTCH1 is involved in sonic hedgehog signaling, a pathway that for instance reduces oxidative stress on neurons and regulates ischemia-induced neuronal progenitor proliferation.33,34 In rats with experimentally induced middle cerebral artery occlusion, inhibition of the sonic hedgehog signaling caused increased infarct size, and PTCH1 was downregulated when sonic hedgehog signaling was inhibited.20,35 The association between PTCH1 and stroke outcome observed in the present study may thus potentially be explained by an increased infarct size. However, the effect of this variant was somewhat stronger when adjusting for baseline NIHSS score, which may suggest a potential influence also on recovery processes.
Third, a variant in NTN4 showed suggestive association with ordinal mRS and was also associated with dichotomous mRS at a low p value. NTN4 encodes a member of the netrin family of proteins expressed in brain tissue (ncbi.nlm.nih.gov/gene/59277) and with functions in processes with biologically plausible roles in stroke recovery such as angiogenesis, neurite growth, and migration.36,37 In a mouse model, NTN4 was upregulated in blood vessels and astrocytes in the ischemic core after stroke, and intracerebroventricular administration of NTN4 led to improvements both in angiogenesis and in poststroke outcome, possibly through increased collateral blood flow leading to improved survival of neurons partially affected by ischemia.21 In another study, NTN4 contributed to thalamocortical axon branching in rodents and the expression was altered by neuronal activity in the cortex, implying that NTN4 might act as a positive regulator for thalamocortical axon branching through activity-dependent expression.38 Although highly speculative, this gene could thus have a role in the plastic process of axonal outgrowth and restoration of synaptic architecture post stroke.
Several additional variants with suggestive association to outcome in this study are linked to genes that may potentially be mechanistically involved in processes affecting stroke outcomes. For instance, RUNX1 encodes a runt-related proangiogenic transcription factor with high expression in arteries (GTEx, June 1, 2018) that is upregulated after stroke.39 In mice, expression of RUNX1 is induced in putative neural stem or progenitor cells after brain injury and suggested to promote neuronal differentiation.40 TNR encodes a protein that is involved in neuronal plasticity and is highly and exclusively expressed in the brain.41,42 Examples of genes with a possible stroke subtype-specific significance among suggestive findings are MTHFS, which is involved in folate and homocysteine metabolism, pathways with potential influence on cerebral small vessel disease,43 and SCML4, in which the variant rs74514008 was suggestively associated with ordinal mRS in the present study. SCML4 has shown association with coronary artery disease in a recent GWA study, and subsequent functional characterization suggested a role in atherosclerosis.44
To further explore the SNPs associated with outcomes in different subgroups, such as certain stroke subtypes, patients with diabetes, or those receiving recanalization therapies would be of great interest. Mechanisms of neuron injury and recovery probably have similarities regardless of the etiology, but mechanism of recovery after cortical and subcortical stroke may be different. Therefore, we analyzed mRS 0–2 vs 3–6 in patients with lacunar and nonlacunar stroke separately. The top 5 independent loci differed between the 2 groups, but no genome-wide significant association was detected. Given the small sample sizes, especially for lacunar stroke, these analyses are clearly exploratory. Thus, in further studies with a higher number of patients, an important aim will be to further explore the genetic associations in specific subgroups.
The strengths of the present study include a relatively large sample size, well defined endpoints, extensive genetic data, and that analyses were adjusted for NIHSS score to assess the influence on outcome independent of baseline stroke severity. We performed dichotomous outcome analyses to identify genetic variants associated with being dependent or independent in activities of daily living, which reflects a clinically important difference in functional outcome. We also performed ordinal analysis, which aims to identify variants that have a similar effect across different degrees of functional outcome. To detect such variants, the ordinal model provides greater power.8 Another strength of the ordinal approach is the ability to compensate for any differences in mRS assessment between studies.
There are also several limitations that should be considered. Although this study included more than 6,000 patients with ischemic stroke, only one significant locus was identified, and thus, the findings do not explain the assumed genetic variation for ischemic stroke outcomes. This implies that the effect sizes of individual SNPs on outcome of ischemic stroke are small and that our sample size might be insufficient even to detect common genetic variants. Another important limitation is the explorative nature of the results, which clearly require future replication in independent cohorts as they become available. The use of the mRS as the primary outcome metrics is a clear limitation in that it is a crude measurement of outcome, especially when dichotomized. However, the mRS represents a well-established, formalized, and easily available measure of poststroke functional outcome, and the dichotomizations represent differences of great importance clinically and for the patient. The mRS scores were assessed at different time points ranging from 60 to 190 days, potentially diluting any detectable associations, as functional outcome status may vary over time. However, the majority of included studies assessed mRS at 3 months ± 2 weeks after stroke onset. We did not have information on premorbid mRS and this would have provided a valuable means to calculate the change in functional ability. Moreover, stroke severity was scored at different time points. However, in a majority of cases, NIHSS was scored early and in only 160 individuals later than day 3 after admission. As previously described,8 we lack data on some important factors that influence outcomes such as acute therapies and rehabilitation, which is why these factors could not be accounted for in the analyses. However, data from the cohorts with available data on treatment with tissue plasminogen activator (about half of the cohorts) show that this treatment was given to 14% of the patients. We can also not exclude the possibility of selection bias since participation in the study required individual informed consent, and data entry also requested availability of functional status follow-up data. In line with this, our study sample mainly reflects milder strokes (median NIHSS score of 4, table 1), which may hamper the detection of factors influencing a greater dynamic range of recovery. Lastly, because our study population was of European ancestry, the results are not necessarily generalizable to other stroke populations. Further studies should ideally include a series of prespecified collection time points, specific clinical variables of importance for stroke outcome, and a battery of outcome metrics that assess several domains of recovery including motor impairment, aphasia, neglect, and cognitive impairment.
In this first large international GWA study on functional outcome after overall ischemic stroke, we report one significant variant and several variants with suggestive association to outcome at 3 months after stroke onset with plausible links to poststroke recovery. Future studies on common variants should include larger sample sizes, enabling subgroup analyses, as well as replication of the present results and elaboration of potential mechanisms.
Author Affiliation
From the Department of Clinical Sciences Malmö (M.S.) and Department of Clinical Sciences Lund, Neurology (B.N., A.L.), Lund University; Department of Neurology and Rehabilitation Medicine (M.S., B.N., A.L.), Neurology, Skåne University Hospital, Lund and Malmö; Institute of Biomedicine (A.P., T.M.S., M.O., C.J.) and Department of Clinical Neuroscience Institute of Neuroscience and Physiology (K.J., T.T.), Sahlgrenska Academy at the University of Gothenburg; Department of Clinical Genetics and Genomics (A.P., C.J.) and Department of Neurology (K.J.), Sahlgrenska University Hospital, Gothenburg; Bioinformatics Core Facility (E.L.), University of Gothenburg, Sweden; School of Life Sciences (S.B.), University of Lincoln, UK; Department of Neurology (J.C.), University of Maryland School of Medicine and Baltimore VAMC; Stroke Pharmacogenomics and Genetics (I.F.-C.), Institut d'investigació Biomedica de Sant Pau, Hospital de Sant Pau; Neurovascular Research Laboratory and Neurovascular Unit (I.F.-C.), Neurology and Medicine Departments, Universitat Autònoma de Barcelona, Vall d'Hebrón Hospital, Barcelona, Spain; Medical School (G.J.H.), The University of Western Australia, Perth; Department of Neurology (J.J.-C.), Institut Hospital del Mar d'Investigació Mèdica (IMIM), Barcelona; Universitat Autònoma de Barcelona (J.J.-C.), Barcelona, Spain; Department of Neurology (J.-M.L.), Washington University School of Medicine, St. Louis, MO; Department of Neurosciences (R.L.), Experimental Neurology, KU Leuven, University of Leuven; Center for Brain & Disease Research (R.L.), Laboratory of Neurobiology, VIB, Leuven; Department of Neurology (R.L.), University Hospitals Leuven, Belgium; School of Medicine and Public Health (C.L., R.S., J.S.), Hunter Medical Research Institute (C.L., R.S.), and Priority Research Centre for Stroke and Traumatic Brain Injury (C.L.), University of Newcastle, Australia; Department of Medicine (B.D.M.), University of Maryland, Baltimore; Geriatric Research, Education, and Clinical Center (B.D.M.), Veterans Affairs Medical Center, Baltimore, MD; Centre for Clinical Brain Sciences (K.R., C.S.), University of Edinburgh, UK; J. Philip Kistler Stroke Research Center, Department of Neurology (N.R., J.R.), Massachusetts General Hospital, Harvard Medical School, Boston; Program in Medical and Population Genetics (J.R.), Broad Institute of MIT and Harvard, Cambridge; Center for Genomic Medicine (J.R.), Massachusetts General Hospital, Boston; Stroke Prevention Research Unit (P.M.R.), Nuffield Department of Clinical Neurosciences, University of Oxford, UK; Department of Neurology (D.S., T.T.), Helsinki University Hospital, Finland; Department of Clinical Neurosciences (M.T.), University of Cambridge, UK; Stroke Division (V.T.), Florey Institute for Neuroscience and Mental Health, University of Melbourne, Heidelberg; Department of Neurology (V.T.), Austin Health, Heidelberg, Australia; Department of Neurology and Rehabilitation (D.W.), University of Cincinnati College of Medicine, OH; Departments of Neurology and Health Evaluation Sciences (B.B.W.), University of Virginia, Charlottesville; University of Technology Sydney (J.M.M.), Faculty of Health, Australia; and Hunter Medical Research Centre (J.M.M.), PRC Stroke and Brain Injury, Newcastle, Australia.
Acknowledgment
The authors thank the Swedish quality register for stroke, Riksstroke, collaboration for providing information about outcome measures for some of the included studies.
Glossary
- EC
endothelial cell
- eQTL
expression quantitative trait locus
- GISCOME
Genetics of Ischaemic Stroke Functional Outcome
- GTEx
Genotype-Tissue Expression
- GWA
genome-wide association
- HGVD
Human Genetic Variation Database
- mRS
modified Rankin Scale
- NIHSS
NIH Stroke Scale
- OR
odds ratio
- PP1
protein phosphatase 1
- SNP
single-nucleotide polymorphism
Appendix. Authors
Footnotes
Editorial, page 549
Study funding
The Stroke Genetics Network (SiGN) study was funded by a cooperative agreement grant from the US National Institute of Neurological Disorders and Stroke (NINDS), NIH (U01 NS069208 and R01 NS100178). SAHLSIS was supported by the Swedish Heart and Lung Foundation (HLF-20160316), the Swedish Research Council (K2014-64X-14605-12-5), the Swedish Stroke Association, the Swedish state (under the “Avtal om Läkarutbildning och Medicinsk Forskning, ALF”) (ALFGBG-720081). Australian Stroke Genetics Collaboration study was supported by the National Health and Medical Research Council, Australia. Stroke Pharmacogenomics and Genetics group was supported by Invictus plus network, Generation project, and Miguel Servet programme from Instituto de Salud Carlos III, GODs project and Epigenesis project from Marató de TV3 Foundation and Agaur from Generalitat de Catalunya Government. Arne Lindgren was supported by the Swedish Heart and Lung Foundation, Region Skåne, Skåne University Hospital, the Freemasons Lodge of Instruction EOS in Lund, Lund University, the Foundation of Färs & Frosta—one of Sparbanken Skåne's ownership Foundations, and the Swedish Stroke Association. Martin Söderholm was supported by grants from the Swedish Stroke Association, the Foundation of Färs & Frosta—one of Sparbanken Skåne's ownership Foundations, and the Swedish government (under the “Avtal om Läkarutbildning och Medicinsk Forskning, ALF”). Annie Pedersen was supported by grants from the Swedish government (under the “Avtal om Läkarutbildning och Medicinsk Forskning, ALF”) and the Gothenburg Foundation for Neurological Research. Natalia Rost was in part supported by NIH-NINDS (R01NS086905 and R01NS082285). Daniel Strbian was supported by the Finnish Subsidiary Governmental Fund (VTR). The authors thank NINDS for funding the genotyping of patients included in the SiGN study (U01 NS069208 and R01 NS100178) and Sólveig Grétarsdóttir for genotyping a subsample of the SAHLSIS cohort.
Disclosure
M. Söderholm, A. Pedersen, E. Lorentzen, T. Stanne, S. Bevan, M. Olsson, J. Cole, I. Fernandez-Cadenas, G. Hankey, J. Jimenez-Conde, K. Jood, J. Lee, R. Lemmens, C. Levi, B. Mitchell, B. Norrving, K. Rannikmäe, N. Rost, J. Rosand, P. Rothwell, R. Scott, D. Strbian, J. Sturm, C. Sudlow, M. Traylor, V. Thijs, T. Tatlisumak, D. Woo, B. Worrall, and J. Maguire report no disclosures relevant to the manuscript. A. Lindgren reports personal fees for advisory board, speech, seminar participation from Bayer, AstraZeneca, Boehringer Ingelheim, BMS Pfizer, ReNeuron. C. Jern reports no disclosures relevant to the manuscript. Go to Neurology.org/N for full disclosures.
References
- 1.Lindgren A, Maguire J. Stroke recovery genetics. Stroke 2016;47:2427–2434. [DOI] [PubMed] [Google Scholar]
- 2.McAllister TW. Genetic factors modulating outcome after neurotrauma. PM R 2010;2:S241–S252. [DOI] [PubMed] [Google Scholar]
- 3.Pearson-Fuhrhop KM, Burke E, Cramer SC. The influence of genetic factors on brain plasticity and recovery after neural injury. Curr Opin Neurol 2012;25:682–688. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg A, Curtis CL, Kleim JA. Linking genes to neurological clinical practice: the genomic basis for neurorehabilitation. J Neurol Phys Ther 2015;39:52–61. [DOI] [PubMed] [Google Scholar]
- 5.Zhao J, Wu H, Zheng L, Weng Y, Mo Y. Brain-derived neurotrophic factor G196A polymorphism predicts 90-day outcome of ischemic stroke in Chinese: a novel finding. Brain Res 2013;1537:312–318. [DOI] [PubMed] [Google Scholar]
- 6.Stanne TM, Tjärnlund-Wolf A, Olsson S, Jood K, Blomstrand C, Jern C. Genetic variation at the BDNF locus: evidence for association with long-term outcome after ischemic stroke. PLoS One 2014;9:e114156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maguire J, Thakkinstian A, Levi C, et al. Impact of COX-2 rs5275 and rs20417 and GPIIIa rs5918 polymorphisms on 90-day ischemic stroke functional outcome: a novel finding. J Stroke Cerebrovasc Dis 2011;20:134–144. [DOI] [PubMed] [Google Scholar]
- 8.Maguire J, Bevan S, Stanne TM, et al. GISCOME—Genetics of Ischaemic Stroke Functional Outcome network: a protocol for an international multicentre genetic association study. Eur Stroke J 2017;2:229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eriksson M, Appelros P, Norrving B, Terent A, Stegmayr B. Assessment of functional outcome in a national quality register for acute stroke: can simple self-reported items be transformed into the modified Rankin Scale? Stroke 2007;38:1384–1386. [DOI] [PubMed] [Google Scholar]
- 10.Adams HP Jr, Bendixen BH, Kappelle LJ, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke 1993;24:35–41. [DOI] [PubMed] [Google Scholar]
- 11.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eicher JD, Landowski C, Stackhouse B, et al. GRASP v2.0: an update on the Genome-Wide Repository of Associations between SNPs and phenotypes. Nucleic Acids Res 2015;43:D799–D804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leslie R, O'Donnell CJ, Johnson AD. GRASP: analysis of genotype-phenotype results from 1,390 genome-wide association studies and corresponding open access database. Bioinformatics 2014;30:i185–i194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higasa K, Miyake N, Yoshimura J, et al. Human genetic variation database, a reference database of genetic variations in the Japanese population. J Hum Genet 2016;61:547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonder MJ, Luijk R, Zhernakova DV, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet 2017;49:131–138. [DOI] [PubMed] [Google Scholar]
- 18.Mishra A, Macgregor S. VEGAS2: software for more flexible gene-based testing. Twin Res Hum Genet 2015;18:86–91. [DOI] [PubMed] [Google Scholar]
- 19.Zheng Q, Zhu D, Bai Y, Wu Y, Jia J, Hu Y. Exercise improves recovery after ischemic brain injury by inducing the expression of angiopoietin-1 and Tie-2 in rats. Tohoku J Exp Med 2011;224:221–228. [DOI] [PubMed] [Google Scholar]
- 20.Ji H, Miao J, Zhang X, et al. Inhibition of sonic hedgehog signaling aggravates brain damage associated with the down-regulation of Gli1, Ptch1 and SOD1 expression in acute ischemic stroke. Neurosci Lett 2012;506:1–6. [DOI] [PubMed] [Google Scholar]
- 21.Hoang S, Liauw J, Choi M, Choi M, Guzman RG, Steinberg GK. Netrin-4 enhances angiogenesis and neurologic outcome after cerebral ischemia. J Cereb Blood Flow Metab 2009;29:385–397. [DOI] [PubMed] [Google Scholar]
- 22.Giese KP, Mizuno K. The roles of protein kinases in learning and memory. Learn Mem 2013;20:540–552. [DOI] [PubMed] [Google Scholar]
- 23.Heroes E, Lesage B, Gornemann J, Beullens M, Van Meervelt L, Bollen M. The PP1 binding code: a molecular-lego strategy that governs specificity. FEBS J 2013;280:584–595. [DOI] [PubMed] [Google Scholar]
- 24.Shioda N, Fukunaga K. Physiological and pathological roles of CaMKII-PP1 signaling in the brain. Int J Mol Sci 2017;19:E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aruga J, Mikoshiba K. Identification and characterization of Slitrk, a novel neuronal transmembrane protein family controlling neurite outgrowth. Mol Cell Neurosci 2003;24:117–129. [DOI] [PubMed] [Google Scholar]
- 26.Willam C, Koehne P, Jurgensen JS, et al. Tie2 receptor expression is stimulated by hypoxia and proinflammatory cytokines in human endothelial cells. Circ Res 2000;87:370–377. [DOI] [PubMed] [Google Scholar]
- 27.Suri C, Jones PF, Patan S, et al. Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell 1996;87:1171–1180. [DOI] [PubMed] [Google Scholar]
- 28.Suri C, McClain J, Thurston G, et al. Increased vascularization in mice overexpressing angiopoietin-1. Science 1998;282:468–471. [DOI] [PubMed] [Google Scholar]
- 29.Golledge J, Clancy P, Maguire J, et al. Plasma angiopoietin-1 is lower after ischemic stroke and associated with major disability but not stroke incidence. Stroke 2014;45:1064–1068. [DOI] [PubMed] [Google Scholar]
- 30.Maisonpierre PC, Suri C, Jones PF, et al. Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science 1997;277:55–60. [DOI] [PubMed] [Google Scholar]
- 31.Gurnik S, Devraj K, Macas J, et al. Angiopoietin-2-induced blood-brain barrier compromise and increased stroke size are rescued by VE-PTP-dependent restoration of Tie2 signaling. Acta Neuropathol 2016;131:753–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui X, Chopp M, Zacharek A, Ye X, Roberts C, Chen J. Angiopoietin/Tie2 pathway mediates type 2 diabetes induced vascular damage after cerebral stroke. Neurobiol Dis 2011;43:285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai RL, Zhu SY, Xia YP, et al. Sonic hedgehog protects cortical neurons against oxidative stress. Neurochem Res 2011;36:67–75. [DOI] [PubMed] [Google Scholar]
- 34.Sims JR, Lee SW, Topalkara K, et al. Sonic hedgehog regulates ischemia/hypoxia-induced neural progenitor proliferation. Stroke 2009;40:3618–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji H, Zhang X, Du Y, Liu H, Li S, Li L. Polydatin modulates inflammation by decreasing NF-kappaB activation and oxidative stress by increasing Gli1, Ptch1, SOD1 expression and ameliorates blood-brain barrier permeability for its neuroprotective effect in pMCAO rat brain. Brain Res Bull 2012;87:50–59. [DOI] [PubMed] [Google Scholar]
- 36.Koch M, Murrell JR, Hunter DD, et al. A novel member of the netrin family, beta-netrin, shares homology with the beta chain of laminin: identification, expression, and functional characterization. J Cell Biol 2000;151:221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C, Meng F, Wang C, et al. Identification of a novel alternative splicing form of human netrin-4 and analyzing the expression patterns in adult rat brain. Brain Res Mol Brain Res 2004;130:68–80. [DOI] [PubMed] [Google Scholar]
- 38.Hayano Y, Sasaki K, Ohmura N, et al. Netrin-4 regulates thalamocortical axon branching in an activity-dependent fashion. Proc Natl Acad Sci USA 2014;111:15226–15231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buga AM, Margaritescu C, Scholz CJ, Radu E, Zelenak C, Popa-Wagner A. Transcriptomics of post-stroke angiogenesis in the aged brain. Front Aging Neurosci 2014;6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Logan TT, Rusnak M, Symes AJ. Runx1 promotes proliferation and neuronal differentiation in adult mouse neurosphere cultures. Stem Cell Res 2015;15:554–564. [DOI] [PubMed] [Google Scholar]
- 41.Hargus G, Cui Y, Schmid JS, et al. Tenascin-R promotes neuronal differentiation of embryonic stem cells and recruitment of host-derived neural precursor cells after excitotoxic lesion of the mouse striatum. Stem Cell 2008;26:1973–1984. [DOI] [PubMed] [Google Scholar]
- 42.Xu JC, Xiao MF, Jakovcevski I, et al. The extracellular matrix glycoprotein tenascin-R regulates neurogenesis during development and in the adult dentate gyrus of mice. J Cell Sci 2014;127:641–652. [DOI] [PubMed] [Google Scholar]
- 43.Rutten-Jacobs LC, Traylor M, Adib-Samii P, et al. Association of MTHFR C677T genotype with ischemic stroke is confined to cerebral small vessel disease subtype. Stroke 2016;47:646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Wang DW, Chen Y, et al. Genome-wide association and functional studies identify SCML4 and THSD7A as novel susceptibility genes for coronary artery disease. Arterioscler Thromb Vasc Biol 2018;38:964–975. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are available on reasonable request.