Abstract
The genetic characteristics and diversity of 21 experimental chicken lines registered with the National BioResource Project of Japan were examined using mitochondrial D-loop sequences and 54 microsatellite DNA markers. A total of 12 haplotypes were detected in the 500-bp mitochondrial DNA sequences of the hypervariable segment I for 349 individuals of 21 lines. The 12 haplotypes belonged to three (A, D, and E) haplogroups, out of the eight (A‒H) common haplogroups in domestic chickens and red junglefowls. The haplogroups A and D were widely represented in indigenous chickens in the Asian and Pacific regions, and the haplogroup E was the most prevalent in domestic chickens. Genetic clustering by discriminant analysis of principal components with microsatellite markers divided 681 individuals of 21 lines into three groups that consisted of Fayoumi-, European-, and Asian- derived lines. In each of the cladograms constructed with Nei’s genetic distances based on allele frequencies and the membership coefficients provided by STRUCTURE and with the genetic distance based on the proportion of shared alleles, the genetic relationships coincided well with the breeding histories of the lines. Microsatellite markers showed remarkably lower genetic heterozygosities (less than 0.1 observed heterozygosity) for eight lines (GSP, GSN/1, YL, PNP, BM-C, WL-G, BL-E, and #413), which have been maintained as closed colonies for more than 40 years (except for #413), indicating their usefulness as experimental chicken lines in laboratory animal science research.
Keywords: chicken bioresource, genetic diversity, microsatellite DNA marker, mitochondrial DNA
Introduction
Thirty-eight species of birds and mammals are currently considered as livestock animals for food and agriculture in the world [14]. Long-term selective breeding for high productivity, resistance to diseases and environmental stress, high quality of stockbreeding products, and easy rearing have produced livestock animals with a wide variety of genetic characteristics. However, the recent selection bias has caused the centralization and/or specialization of certain breeds, leading to the decrease in genetic variations that otherwise ensure genetic diversity, flexibility, and adaptability. A total of 8,774 breeds have been recorded as animal genetic resources in the Global Databank of Food and Agriculture Organization of the United Nations (FAO); however, 17% of them are currently at risk of extinction within decades [14].
Chickens, one of the most ubiquitous domesticated animals, were bred for egg and meat production. The present-day chicken was domesticated from a wild progenitor called the red junglefowl (Gallus gallus) over 8,000 years ago [66]. At present, FAO’s Global Databank reports the presence of 1,729 chicken breeds, which account for 20% of the total number of livestock breeds. A great variety of chicken breeds were raised as a domestic animal for amusement, based primarily on certain unique phenotypic characters, such as a variety of morphologies (figure, body size, comb, wattle, tail feather, plumage color, etc.), crowing sound, and aggressive rooster behavior during fights. However, according to an assessment by the FAO, about 21% of chicken breeds are at a risk of extinction; these include breeds that are classified as critical, critical-maintained, endangered, and endangered-maintained breeds [14]. The Avian Genetic Resources Task Force (AGRTF) also reported that in just four years, from 1995 to 1998, 50 chicken stocks in research institutions across the USA and Canada were lost [48]. Commercial chickens are also commonly used for biological research owing to their easy availability; however, these chickens have a high degree of genetic heterogeneity between individuals, which could cause experimental errors. To minimize the errors and to ensure the accuracy and reproducibility of experimental results, the use of genetically homogenized chicken lines with a known genetic background is indispensable. Furthermore, it is essential to clarify the origins and genetic characteristics of chicken resources (breeds or lines) for selecting lines that are suitable for experimental purposes and designs. The Avian Bioscience Research Center (ABRC) at Nagoya University contributes to the advancement of avian science research by serving as a core facility of chicken resources that collects, maintains, and preserves resources with various origins and genetic backgrounds, subsequently making them available to the science community. It is supported by the National BioResource Project (NBRP) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and Japan Agency for Medical Research and Development (AMED), Japan (https://www.agr.nagoya-u.ac.jp/~nbrp/en/index.html). Actually, NBRP chicken resources have been used in a wide variety of research fields, such as brain science and behavioral biology [55, 56], infectious disease medicine [67], developmental biology [39, 68], genetics [22, 27, 28, 57], evolutionary developmental biology [21, 53], etc. Therefore, characterization by genetic monitoring using DNA markers is important to evaluate the utility of chicken experimental lines and to assure their quality, which lead to the preservation of valuable chicken resources and a stable supply of them to the science community. In this study, we examined the genetic characteristics of 21 chicken lines maintained at the ABRC, including their genetic diversity and relationships based on mitochondrial D-loop sequences and 54 microsatellite DNA markers. This study is the first research model for the large scale genetic monitoring of chicken lines to evaluate their usefulness as experimental animals.
Materials and Methods
Ethics statement
Animal care and all experimental procedures were conducted according to the guidelines for the care and use of experimental animals of Nagoya University. The animal protocols were approved by the Animal Experiment Committee, Graduate School of Bioagricultural Sciences, Nagoya University.
Rearing condition
The chickens were housed in stainless steel bird cages at a constant temperature (25°C) with 16 h light/8 h dark cycle and were given ad libitum access to food and water under conventional conditions. We made vaccination of 9 infectious diseases to all lines in each generation, and microbial testing was carried out regularly for 14 items using a microbiological testing service (see Appendix).
DNA extraction
The 21 chicken lines used in this study were as follows: a line of wild ancestral species (red junglefowl, RJF); lines derived from Fayoumi (GSP, GSN/1, YL, and PNP), Black Minorca (BM-C), White Leghorn (WL-G and WL-M/O), Brown Leghorn (BL-E), Rhode Island Red (RIR), Game Bantam (GB), Dandarawi (DD), White Silkie (SIL), Chahn (CHN), Ehime-jidori (EJ), Japanese Bantam (JB), and Cochin Bantam (CB) breeds; and the following mutant lines: an albino line (CAL) derived from the White Wyandotte, a dwarf mutant line Petit-cocco (PC) derived from the Rhode Island Red, a mutant line of muscular dystrophy (#413) derived from the Rhode Island Red, and the Obese line (OS) derived from the White Leghorn. Their origins and breeding histories are summarized in Appendix. Blood was collected from the wing vein using heparinized syringes. Genomic DNA was extracted from 20 µl of whole blood using 300 µl of DNAzol BD reagent (Molecular Research Center, Tokyo, Japan). We examined the mitochondrial D-loop sequences and the microsatellite DNA markers of 349 (Table 1) and 681 (Table 2) individuals, respectively.
Table 1. List of D-loop haplotypes found for 21 chicken lines.
| Line | n | Accession number | Haplotype name | Identical sequence on Genbank (% identity) |
Haplogroupa | Haplotypeb |
|---|---|---|---|---|---|---|
| GSP | 16 | LC330917 | NU-Gg_DL01 | EF586879 (100) | E | ‒ |
| GSN/1 | 16 | LC330918 | NU-Gg_DL01 | EF586879 (100) | E | ‒ |
| YL | 8 | LC330919 | NU-Gg_DL01 | EF586879 (100) | E | ‒ |
| PNP | 16 | LC330920 | NU-Gg_DL01 | EF586879 (100) | E | ‒ |
| BM-C | 8 | LC330921 | NU-Gg_DL02 | GU447978 (100) | E | E07 |
| WL-G | 8 | LC330922 | NU-Gg_DL03 | AB007723 (100) | E | E04 |
| WL-M/O | 15 | LC330923 | NU-Gg_DL04 | GU447782 (100) | A | A02 |
| BL-E | 16 | LC330924 | NU-Gg_DL05 | GU448836 (100) | E | E08 |
| RIR | 15 | LC330925 | NU-Gg_DL06 | GU449084 (100) | E | E11 |
| GB | 16 | LC330926 | NU-Gg_DL07 | GU448297 (100) | E | E01 |
| CAL | 16 | LC330927 | NU-Gg_DL08 | GU449063 (100) | A | A01 |
| DD | 16 | LC330928 | NU-Gg_DL07 | GU448297 (100) | E | E01 |
| SIL | 10 | LC330929 | NU-Gg_DL09 | GU261683 (100) | D | D13 |
| CHN | 16 | LC330930 | NU-Gg_DL09 | GU261683 (100) | D | D13 |
| LC330931 | NU-Gg_DL10 | AB007741 (100) | D | D04 | ||
| EJ | 16 | LC330932 | NU-Gg_DL08 | GU449063 (100) | A | A01 |
| LC330933 | NU-Gg_DL09 | GU261683 (100) | D | D13 | ||
| PC | 13 | LC330934 | NU-Gg_DL07 | GU448297 (100) | E | E01 |
| JB | 30 | LC330935 | NU-Gg_DL09 | GU448767 (100) | D | D13 |
| CB | 15 | LC330936 | NU-Gg_DL11 | AB268508 (99) | E | ‒ |
| #413 | 18 | LC330937 | NU-Gg_DL07 | GU448297 (100) | E | E01 |
| OS | 12 | LC330938 | NU-Gg_DL12 | GU449088 (100) | E | E03 |
| RJF | 53 | LC330939 | NU-Gg_DL07 | GU448297 (100) | E | E01 |
Table 2. Genetic diversity of 21 chicken lines estimated using 54 microsatellite DNA markers.
| Line | n | ARa | MNAb | Nec | Hod |
|---|---|---|---|---|---|
| GSP | 39 | 1.19 | 1.04 | 1.03 | 0.01 |
| GSN/1 | 36 | 1.02 | 1.00 | 1.00 | 0.00 |
| YL | 34 | 1.47 | 1.22 | 1.11 | 0.04 |
| PNP | 34 | 1.18 | 1.06 | 1.02 | 0.01 |
| BM-C | 20 | 1.07 | 1.07 | 1.05 | 0.03 |
| WL-G | 45 | 1.32 | 1.24 | 1.11 | 0.08 |
| WL-M/O | 23 | 1.82 | 1.74 | 1.48 | 0.28 |
| BL-E | 26 | 1.17 | 1.17 | 1.12 | 0.07 |
| RIR | 41 | 1.60 | 1.46 | 1.28 | 0.18 |
| GB | 27 | 1.67 | 1.61 | 1.34 | 0.18 |
| CAL | 21 | 1.84 | 1.76 | 1.44 | 0.25 |
| DD | 38 | 1.56 | 1.54 | 1.21 | 0.12 |
| SIL | 33 | 1.72 | 1.59 | 1.32 | 0.18 |
| CHN | 24 | 2.19 | 1.98 | 1.55 | 0.27 |
| EJ | 19 | 3.69 | 3.56 | 2.45 | 0.51 |
| PC | 39 | 1.74 | 1.70 | 1.41 | 0.26 |
| JB | 30 | 2.85 | 2.80 | 2.02 | 0.44 |
| CB | 41 | 1.72 | 1.69 | 1.30 | 0.18 |
| #413 | 18 | 1.30 | 1.20 | 1.13 | 0.09 |
| OS | 40 | 1.64 | 1.46 | 1.25 | 0.14 |
| RJF | 53 | 2.88 | 2.83 | 1.90 | 0.37 |
aAR: Allelic richness. bMean number of alleles per locus. cMean number of effective alleles per locus, Ne=1 / (Sum pi^2), where Sum pi^2 is the sum of squared population allele frequencies. dObserved heterozygosity.
Analysis of mitochondrial D-loop sequences
The mitochondrial DNA sequences of the hypervariable segment I (HVS-I) fragments were amplified using primers L16750 (5’-AGGACTACGGCTTGAAAAGC-3’) [17] and H522 (5’-ATGTGCCTGACCGAGGAACCAG-3’) [16]. PCR amplification was performed in a total volume of 10 µl consisting of a reaction mixture containing 50 ng genomic DNA, 10 pmol of each primer, and 5 µl AmpliTaq Gold® 360 Master Mix (Life Technologies, Tokyo, Japan). The cycling conditions for PCR were as follows: initial denaturation at 95°C for 10 min, followed by 35 cycles at 95°C for 30 s, 60°C for 30 s, and 70°C for 20 s, and a final extension for 5 min at 72°C. Double-stranded PCR products were purified using a 13% polyethylene glycol/2.5 mol NaCl precipitation method. The purified products were reacted with a BigDye Terminator cycle sequencing kit (ver 3.1; Life Technologies) and sequenced using an ABI 3130 automated sequencer (Applied Biosystems, Foster, CA, USA).
DNA sequences were aligned using Clustal W [63] implemented in MEGA 7.0.14 [30] and edited using PROSEQ 2.9.1 [15]. The software MitotoolPy [47] was used for the D-loop sequences to determine the haplotypes matching those of domestic chickens around the world (haplotypes A-I, X, Y, and Z) [38]. Neighbor-joining and Bayesian phylogenetic trees were constructed using MEGA 7.0.14 [30] and BEAST 1.8.3 [9], respectively. For phylogenetic analyses, the TrN+I model [62] based on Akaike information criterion (AIC) and the HKY+I model [19] based on Bayesian information criterion (BIC) were selected as best-fitting substitution models using jModeltest 2.1.10 [7]. We only used TrN+I model for constructing the neighbor-joining tree because MEGA 7.0.14 did not implement the HKY+I model. Branch supports for the neighbor-joining tree were calculated using 1,000 bootstrap replications. The Bayesian phylogenetic analysis was performed using each of the TrN+I and HKY+I models. The analysis ran for 10 million Markov chain Monte Carlo (MCMC) generations, and one tree was saved for every 1,000 generations. The convergence of MCMC generations was verified using Tracer 1.6.0 [51]. After removing the first 10% of the saved 10,000 trees as a burn-in, Bayesian posterior probabilities were calculated from the remaining trees using Tree Annotator 1.8.3 (available in the BEAST package). The Bayesian phylogenetic tree with posterior probability was visualized using FigTree 1.4.2 (http://beast.bio.ed.ac.uk/FigTree).
Analysis of microsatellite DNA markers
Fifty-four microsatellite DNA markers were used for the genetic characterization of the 21 lines (Supplementary Table S1). Details of these markers are available from the ArkDB Database Website developed by the Roslin Bioinformatics Group (http://www.theark-db.org/). Twenty-eight of 54 markers used in this study were selected from 30 markers that are recommended by the FAO working group [13] for biodiversity studies on chickens.
PCR amplification of microsatellite DNA markers was performed in a total volume of 10 µl of reaction mix containing approximately 50 ng of genomic DNA, 10 pmol of each primer, and 5.0 µl of Taq Gold 360 Master Mix (Applied Biosystems). The cycling conditions for PCR were as follows: initial denaturation at 95°C for 10 min, followed by 42 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 25 s, and a final extension for 5 min at 72°C. PCR products were electrophoresed with a GeneScan 600 LIZ Size Standard (Applied Biosystems) in Hi-Di formamide (Applied Biosystems) using an ABI 3130 automated sequencer (Applied Biosystems). Allele sizes were determined using GENEMAPPER Software version 4.1 (Applied Biosystems).
Genetic diversity indices, including allelic richness (AR), the number of alleles per locus (Na), and F statistics (FIS, FST, and FIT) were calculated for each of the 54 microsatellite DNA markers using MICROSATELLITE ANALYSER 4.05 [8]. Then, genetic diversity indices were examined for each of the 21 lines as follows: AR was calculated using MICROSATELLITE ANALYSER 4.05 [8]; the mean number of alleles (MNA) and the mean number of effective alleles (Ne, the number of alleles weighted by the square of the allele frequency) were calculated using GENALEX 6.5 [46]; and the observed heterozygosity (Ho) was calculated using ARLEQUIN 3.5.1.2 [12]. MICRO-CHECKER 2.2.3 [65] was used to determine null allele markers. The chi-square test was used to test for Hardy-Weinberg equilibrium (HWE); and the calculation of a fixation index (F) for each line was carried out using GENALEX 6.5 [46]. A diagnostic marker, which shows a line-specific allele that is fixed at 100% frequency within a line, was determined using GENALEX6.5 [46]. The exploratory discriminant analysis of principal components (DAPC) [25] was performed using the R package ‘adegenet’ [24] under the condition that the numbers of principal components (PC) and clusters were set at 13 and 23, respectively, according to the optimal α-score and BIC values.
Bayesian clustering analysis was performed to infer the number of genetic clusters for 21 lines using STRUCTURE 2.3 [50]. The Log probability value (Ln P[D]) from K=2 to K=22 was estimated by running a sampling period of 500,000 MCMC generations after a burn-in period of 500,000 generations under the admixture model and the correlated allele frequency model [49]. Ten independent MCMC runs were performed for each K. Any MCMC run showing the variance of Ln likelihood more than double that of other MCMC runs within the same K was excluded from subsequent analyses. The results of the remaining runs were analyzed to extract one major clustering pattern in each K using CLUMPAK [29]. Then the optimal K value was determined using the Evanno method [11] implemented in STRUCTURE HARVESTER 0.6.94 [10].
Genetic relationships between lines were assessed using Nei’s angular genetic distance based on allele frequencies (Da) [42] and the genetic distance based on the proportion of shared alleles (Dps) [2], using 54 microsatellite DNA marker, which were calculated using MICROSATELLITE ANALYSER 4.05 [8]. We also constructed a cladogram using the Nei’s genetic distance between lines, which were calculated with the membership coefficients (here, for convenience, it is called Dmc) of 21 chicken lines from K=2 to K=21 by the following methods: 1) The membership coefficient matrix, which consists of rows for the 21 lines and columns for K clusters, was generated using the program CLUMPP [23]; 2) the matrix was expediently regarded as the allele frequency matrix, with rows for 21 lines and with columns for K alleles; 3) 20 pseudo-allele frequency matrices, which had rows for 21 lines and columns for each of the 2 to 21 alleles, were generated; 4) the matrices were converted into the input file of the software POPTREE2 [61]; next, we constructed a neighbor-joining tree.
Results
We determined nucleotide sequences of the 500-bp fragments in the mitochondrial DNA HVS-I region for 349 individuals of 21 chicken lines and detected a total of 12 haplotypes (NU-Gg_DL01‒NU-Gg_DL12, Accession No. LC330917‒LC330939) (Table 1) defined by 19 variable sites that consisted of 7 singletons and 12 parsimony informative sites. Each of 19 lines except for CHN and EJ had one haplotype. The four Fayoumi lines, GSP, GSN1, YL, and PNP, shared the same haplotype, NU-Gg_DL01, which was identical to a haplotype of Fayoumi chickens registered in GenBank (EF586879, Ramadan et al.; direct submission to GenBank, 2007). BM-C, WL-G, WL-M/O, BL-E, RIR, CHN, CB, and OS each had a line-specific haplotype, NU-Gg_DL02, NU-Gg_DL03, NU-Gg_DL04, NU-Gg_DL05, NU-Gg_DL06, NU-Gg_DL10, NU-Gg_DL11, and NU-Gg_DL12, respectively. NU-Gg_DL07 was assigned to five lines, GB, DD, PC, #413, and RJF. NU-Gg_DL08 was assigned to two lines, CAL and EJ. EJ also had another haplotype, NU-Gg_DL09, which it shared with SIL, CHN, EJ, and JB. Eleven of the 12 haplotypes (the exception was NU-Gg_DL11 that was found for CB) corresponded to D-loop sequences registered in GenBank (Table 1).
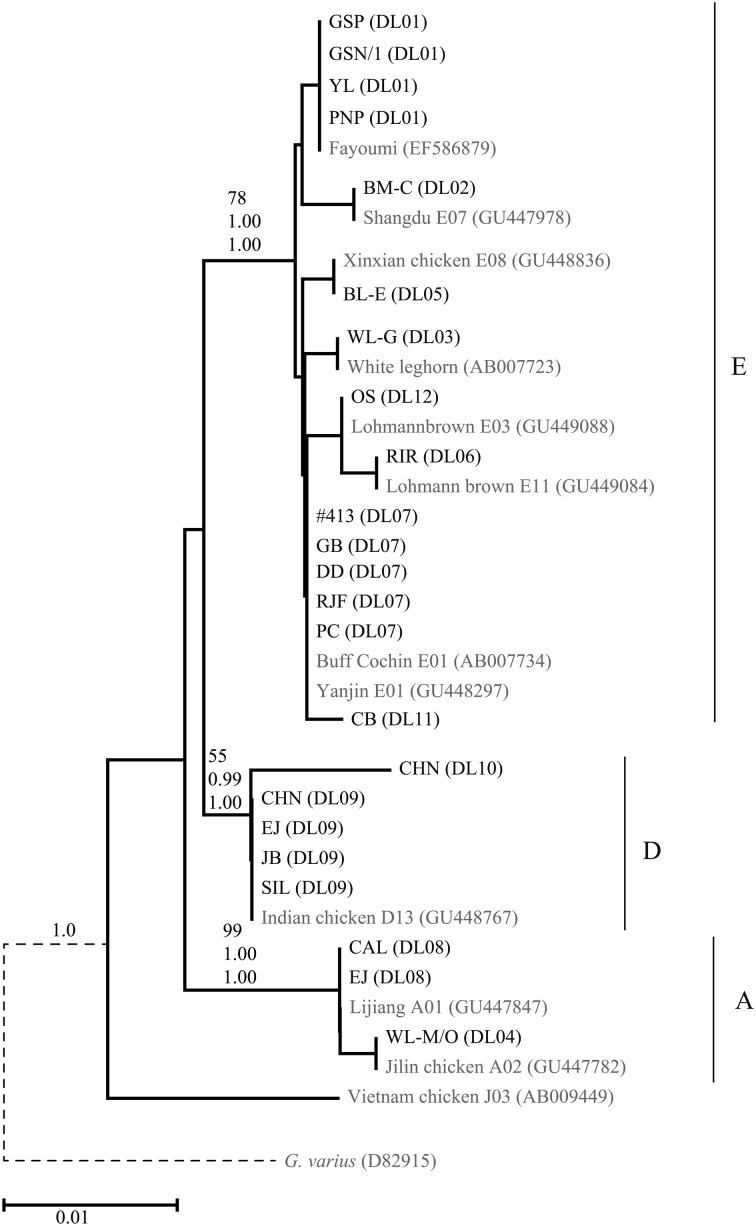
The 12 haplotypes belonged to three haplogroups, A, D, and E, which were previously defined by Liu et al. (2006) [33] (Table 1). The topologies of the neighbor-joining trees based on the TrN+I model and Bayesian trees based on the TrN+I and HKY+I models were fundamentally the same, and the neighbor-joining tree is shown in Fig. 1 as a representative tree. Eight haplotypes (NU-Gg_DL01‒ NU-Gg_DL03, NU-Gg_DL05‒ NU-Gg_DL07, NU-Gg_DL11, and NU-Gg_DL12) from 15 lines (GSP, GSN/1, YL, PNP, BM-C, WL-G, BL-E, RIR, GB, DD, PC, CB, #413, OS, and RJF) belonged to haplogroup E. NU-Gg_DL04 (WL-M/O) and NU-Gg_DL08 (CAL and EJ) were assigned to haplogroup A. Haplogroup D contained two haplotypes to which were assigned four lines representing Japanese indigenous chicken breeds (NU-Gg_DL09, SIL, CHN, JB and EJ; NU-Gg_DL10, CHN). The clades of each haplogroup of the neighbor-joining tree and the Bayesian trees were supported with high bootstrap and posterior probability values, respectively, although haplogroup D showed weak bootstrap support (55%) in the neighbor-joining tree (Fig. 1).
Fig. 1.
Neighbor-joining tree of mitochondrial DNA D-loop haplotypes of 21 chicken lines. Because the neighbor-joining tree and the Bayesian trees generated basically the same topologies, the neighbor-joining tree is shown as a representative tree. Sequences obtained from GenBank are indicated in gray. The branch support values on the primary branch of each haplogroup are shown in the following order from top to bottom: bootstrap values for the neighbor-joining tree based on the TrN+I model, posterior probabilities for the Bayesian tree based on the TrN+I model, and posterior probabilities for the Bayesian tree based on the HKY+I model. The branch of the outgroup species (Gallus varius, dashed line) is shortened because the branch lengths was too long to be shown in the figure.
The genetic diversity indices for 54 microsatellite DNA markers are shown in Supplementary Table S1. The AR for each marker ranged from 2.00 for MCW0098 to 13.12 for ADL0257 (6.36 on average). The Na for each locus across populations ranged from 2 for MCW0098 to 25 for LEI0228 (9.50 on average). The FIS varied from −0.15 for MCW0233 to 0.50 for MCW0331 (0.04 on average). The FST and FIT values fell within the range from 0.39 (MCW0222) to 0.92 (ADL273) and 0.42 (MCW0222) to 0.93 (ADL273 and MCW0331), respectively (FST=0.75 and FIT=0.76 on average). MCW0222 showed considerably lower genetic diversity than the other markers. All loci showed signs of null alleles within two lines, except for LEI0228 and LEI0075 where signs of null alleles were detected for four lines (Supplementary Table S2). Diagnostic loci, i.e., the loci with line-specific alleles, were detected for 13 of 21 lines, namely, GSP, YL, PNP, BM-C, WL-M/O, BL-E, RIR, DD, SIL, CHN, CB, OS, and RJF (Supplementary Table S3).
The genetic diversity indices of the 21 chicken lines are shown in Table 2. Four Fayoumi lines (GSP, GSN/1, YL, and PNP) and BM-C exhibited remarkably higher genetic homogeneity than the other 16 lines (AR, 1.02 for GSN/1 to 1.47 for YL; MNA, 1.00 for GSN/1 to 1.22 for YL; Ne, 1.00 for GSN/1 to 1.11 for YL; Ho, 0.00 for GSN/1 to 0.04 for YL). Among the remaining 16 lines, two Leghorn lines, WL-G and BL-E, also showed high homozygosity: AR, 1.32 (WL-G) and 1.17 (BL-E); MNA, 1.24 (WL-G) and 1.17 (BL-E); Ne, 1.11 (WL-G) and 1.12 (BL-E); and Ho, 0.08 (WL-G) and 0.07 (BL-E), whereas the EJ line showed the highest genetic heterogeneity among the 21 lines (AR=3.69; MNA=3.56; Ne=2.45; Ho=0.51).
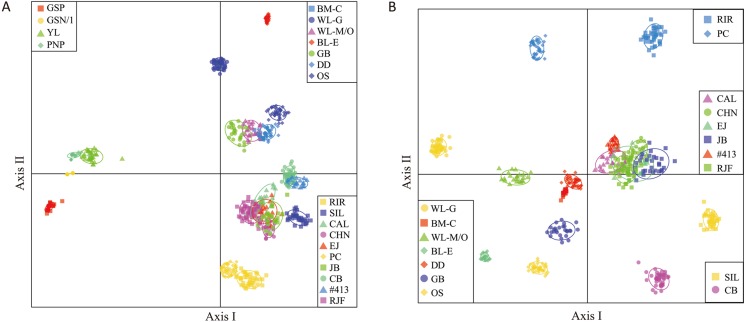
DAPC analysis suggests that the 21 lines may be clustered into three groups (Fig. 2A). The four Fayoumi lines are genetically differentiated clearly from the other 17 lines along Axis I. The remaining 17 lines were divided into the following two groups by Axis II: 1) a group at the right upper side of Axis II consisting of seven lines (BM-C, WL-G, WL-M/O, BL-E, GB, DD, and OS), which originated in European countries; and a group on the lower right side of Axis II consisting of 10 lines (RIR, SIL, CAL, CHN, EJ, PC, JB, CB, #413, and RJF), which originated in Asia or are related to Asian chickens. When the four Fayoumi lines were removed for further subdivision of the other lines, seven lines of the European group became distributed on the lower left side of the DAPC plot (Fig. 2B). Even though WL-G, WL-M/O, BL-E, and OS originated from Leghorn breeds, these four lines are not clustered together. Of the 10 lines of the Asian group, six (CAL, CHN, EJ, JB, #413, and RJF) are concentrated around the center on the right side. SIL and CB are located on the lower right, and PC and RIR are positioned on the upper side.
Fig. 2.
Discriminant analysis of principal components (DAPC) plots using 54 microsatellite DNA markers. (A) DAPC plot of 21 chicken lines. The populations are subdivided into three groups: four Fayoumi-derived lines, seven lines of European origin, and ten lines of Asian origin. (B) DAPC of 17 lines excluding four Fayoumi lines. Each population group is enclosed by a circle, which covers 80% of the distribution of individuals of each population.
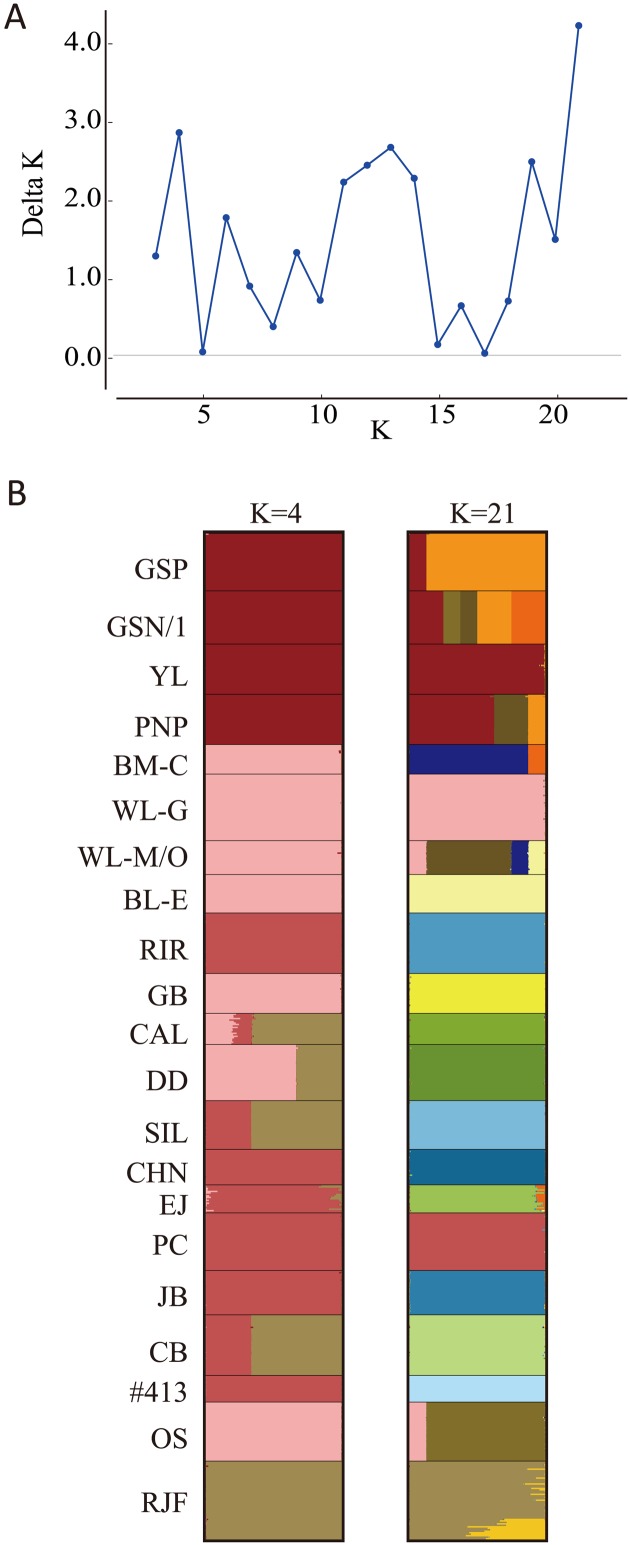
The analysis with STRUCTURE HARVESTER indicates that K=21 was optimal for the 21 lines (Delta K=4.33), and the Delta K value of 2.93 was the second highest at K=4 (Fig. 3A). Bayesian clustering divided the 21 lines into four groups at K=4: the Fayoumi group, European group, Asian group, and the RJF group (Fig. 3B). The European group was composed of BM-C, WL-G, WL-M/O, BL-E, GB, DD, and OS. The six Asian lines were RIR, CHN, EJ, PC, JB, and #413. CAL, SIL, and CB showed high membership coefficients to the RJF group. At K=21, GSP diverged from the other three Fayoumi lines as an independent cluster, and GSN/1 showed the state of mixing of the GSP cluster (painted in orange), the PNP and YL cluster (dark brown), and other clusters that were found in BM-C (dark orange) or WL-M/O (olive). In RJF, two different clusters were mixed; however, the genetic contribution of the cluster painted in ocher was much smaller than the other cluster (dark grass green). The other 16 lines almost independently formed their own clusters.
Fig. 3.
Bayesian clustering of 21 chicken lines. (A) Delta K values at K=3 to K=21 generated by STRUCTURE HARVESTER. (B) Group memberships of 21 chicken lines at K=4 and K=21 are shown by different colors, which were produced using CLUMPAK.
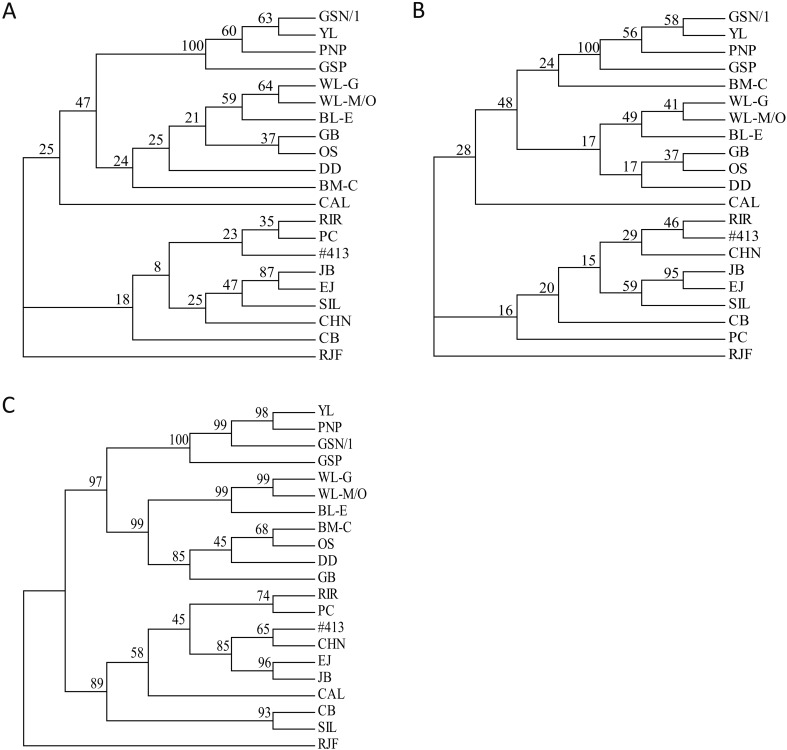
The three cladograms constructed from pairwise comparisons of Nei’s genetic distances based on allele frequencies (Da) (Fig. 4A) and the membership coefficients provided by STRUCTURE (Dmc) (Fig. 4C) and the genetic distance based on the proportion of shared alleles (Dps) (Fig. 4B) all showed similar genetic relationships between the 21 lines; the four Fayoumi lines formed a monophyletic clade; seven lines of European origin (BM-C, WL-G, WL-M/O, BL-E, GB, OS, and DD) were monophyletic with the Fayoumi lines. Eight lines of the Asian group (RIR, PC, #413, JB, EJ, SIL, CHN, and CB) were also monophyletic. EJ and JB showed a close genetic relationship with each other in all three cladograms (Fig. 4A). CAL was placed within the clade of the Asian group in the Dmc cladogram; however, it was assigned to an independent clade that was rather closely related to the European group in the Da and Dps cladograms.
Fig. 4.
Cladograms of 21 chicken lines. (A, B) Cladograms constructed with the Nei’s genetic distance based on allele frequencies (Da) [42] (A) and the genetic distance based on the proportion of shared alleles (Dps) [2] (B) using 54 microsatellite DNA markers, which were calculated using MICROSATELLITE ANALYZER. (C) Cladogram constructed by POPTREE2 with Nei’s genetic distances between lines based on the membership coefficients (Dmc) Dmc generated at K=2 to K=21 using CLUMPAK. Numbers on branches indicate bootstrap values of 1,000 replicates.
Discussion
Genetic diversity and relationships among chicken breeds have been studied actively in the past decade using mitochondrial DNA sequences [6, 31, 43, 45, 58] and microsatellite DNA markers [18, 35, 44, 59, 60, 69]. Even though high throughput genotyping methods have become available recently, mitochondrial and/or microsatellite (autosomal) DNA markers are still preferred because of their cost-effectiveness and ease of experimental manipulation. The application of both types of DNA markers is preferable to using either one of them alone for characterizing genetic structures of populations at a fine level because inheritance patterns are different between the mitochondrial genome (maternal inheritance) and the nuclear genome (biparental inheritance) [32, 40, 41]. In the present study, the genetic characterization of 21 experimental chicken lines of the NBRP was successfully performed using both mitochondrial DNA sequences and microsatellite DNA markers.
The 21 chicken lines belonged to one of the three D-loop haplogroups (A, D, and E) out of the eight common haplogroups (A–H) that had been previously defined using data from 347 commercial chickens, 4,385 indigenous chickens from around the world, and 206 red junglefowls [39]. Among the eight common haplogroups, haplogroup E was the most predominant, followed by haplogroup A. Haplogroup E is considered to have originated from the Indian subcontinent and is contemporary prevailed in the global chicken populations [6, 26, 33, 34, 36, 37, 45]. Haplogroup A may be of Chinese origin and includes indigenous chickens from China, Japan, and South and Southeast Asia [1, 26, 33]. Haplogroup D, which is distributed in populations from Africa, Madagascar, South to East Asia, and the Pacific islands [26, 38, 40], is thought to be associated with the propagation of the culture of cockfighting.
Rosenberg et al. (2001) [52] suggested that the probability of success in clustering analysis with STRUCTURE is more than 90% when at least 15 highly variable microsatellite DNA markers are genotyped for more than 15 individuals per population. In their study, they used 27 markers for 600 individuals from 20 breeds to show that the number of alleles range from 2 to 41 (12.07 on average) and FST values range from 0.189 to 0.501 (0.312 on average). Carlsson (2008) [3] also indicated that if more than 20 microsatellite DNA markers are used and the FST value is more than 0.10 for each marker, then the assignment accuracy of STRUCTURE analysis reaches nearly 100%, irrespective of the frequencies of null alleles. In this study, the number of alleles found for 54 markers ranged from 2 to 25 per marker (9.50 on average) in 21 lines, in which more than 18 individuals were examined for each line and their FST values ranged from 0.39 to 0.92 (0.75 on average). Thus, we consider our genetic clustering of 21 lines using 54 microsatellite DNA markers to be accurate. The results we provide are similar to those reported by Granevitze et al. (2009) [18], in which STRUCTURE analysis at K=3 subdivided 65 chicken populations into three groups as follows: 1) European group, 2) Asian group, and 3) a third group that contains broiler lines, Brown egg layers, the C line (inbred line), Green-legged Partridge, and Fayoumi.
Genetic characteristics of the Fayoumi group
The GSP, GSN/1, YL, and PNP lines are derived from the Fayoumi chicken that is an ancient chicken breed originating in Egypt around 3,000 years ago. To establish these lines, we repeatedly mated two or three pairs for more than 10 generations and have maintained the products as closed colonies for more than or close to 40 years (see Appendix for details). It is for this reason that these Fayoumi lines have high levels of genetic homogeneity. The remarkable genetic homogeneities of these Fayoumi lines, especially GSP and GSN/1, make them ideal as standard experimental chicken lines, since genetic homogeneity ensures the accuracy and reproducibility of experiments. The 100% similarity of the D-loop haplotype (NU-Gg_DL01) with that of Fayoumi chickens in Egypt (EF586879, Ramadan et al.; direct submission to GenBank, 2007) verifies the Egyptian origin of these lines. The exclusive genetic clustering of the four Fayoumi lines, placing them far from the other lines in DAPC and STRUCTURE plots, agrees well with the results reported by Granevitze et al. (2009) [18], in which a Fayoumi population was assigned into a cluster that was different from both the European and Asian chicken populations. These results indicate that the genetic difference of our Fayoumi lines from the other lines is not artifactual owing to the selective breeding for increasing their genetic homozygosity, but rather our long-term close breeding may have enhanced the unique genetic characteristics of these lines.
Genetic characteristics of the European group
The BM-C line, derived from the Minorca chicken, is thought to be a local chicken breed originating from the Island of Minorca in the Mediterranean Sea. The E07 haplotype of BM-C exists only in indigenous chickens of China, South Korea, and Japan [38]. Thus, the Minorca chicken has probably received genetic contributions from chickens that originated in the Far East region during the process of establishment, although the Mediterranean (European) origin of BM-C is evident in the DAPC and STRUCTURE plots. The BM-C line shows a level of genetic homogeneity as high as that of the four Fayoumi lines, indicating that it would also be ideal as a standard experimental line among our chicken resources.
WL-G, WL-M/O, BL-E, and OS are lines derived from the Leghorn chicken that was developed in the Mediterranean area. WL-G was established from a White Leghorn breed at Nagoya University and has been maintained as a closed colony since 1969. WL-M/O, which has been maintained at the Nippon Institute for Biological Science since 1975, was introduced into Nagoya University in 2006. BL-E has been maintained as a closed colony since it was introduced from the Edinburgh Poultry Research Center in 1960. OS (the Obese line) originated from a mutant with autoimmune thyroiditis that occurred in the Cornell line (C-line) of White Leghorn at Cornell University in the 1950s [5]. These Leghorn-derived chicken lines all showed the following different haplotypes: NU-Gg_DL03 of WL-G is identical to haplotype E04 reported in Miao et al. (2013) [38]; NU-Gg_DL04 of WL-M/O is identical to A02; NU-Gg_DL05 of BL-E is identical to E08; and NU-Gg_DL12 of OS is identical to E03. In addition to the difference between their D-loop haplotypes, the widely scattered distributions of the four Leghorn-derived lines in DAPC plots suggest that different source populations of Leghorn chickens have independently contributed to the establishment of these four lines.
The GB line was derived from a bantam race of the Modern Game chicken that originated in England. The D-loop region of the Modern Game Bantam has been examined for only four individuals in Australia [31]; three of them were of the haplotype KC347727 that is identical to haplotype NU-Gg_DL07 (E01 in Miao et al., 2013 [38]) of GB. The remaining individual had the haplotype KC347725 that is identical to NU-Gg_DL08 (A01 in Miao et al., 2013 [38]) of CAL and EJ. Since GB also groups with European chickens in STRUCTURE and DAPC plots, it represents a line that has conserved the genetic characteristics of the Modern Game Bantam.
The DD line originated from a pair of Dandarawi chickens, an indigenous Egyptian chicken breed. The E01 haplotype has been found in all individuals of Dandarawi originating from Egypt (1 individual), Kenya (45 individuals), and Nigeria (1 individual) (Mwacharo et al.; direct submission to GenBank, 2008; Miao et al. 2013 [38]). In a study by Osman et al. (2016) [45], eight haplotypes belonging to the haplogroup E were detected in Dandarawi chickens; one of the two major haplotypes was E01. These D-loop sequence data indicate that DD is an ideal laboratory line representing the Dandarawi chicken.
Genetic characteristics of the Asian group
We have maintained a Rhode Island Red (RIR) line and two RIR-related lines (PC and #413). The RIR line has been maintained as a closed colony since 1980. Petit-Cocco (PC) is a recessive dwarf mutant line derived from the Rhode Island Red breed. The #413 line is a mutant line with hereditary muscular dystrophy that occurred in the New Hampshire breed that was developed from Rhode Island Red to obtain the traits of early maturity and quick feathering [20]. Even though the D-loop haplotypes of these three lines differ from each other, their close genetic relationships shown in the Da cladogram confirm the involvement of Rhode Island Red in the establishment of PC and the New Hampshire breed. In addition, our analysis using microsatellite DNA markers reveals significant genetic affinities of these three lines with the lines of Asian origin, such as EJ, JB, and CHN. Granevitze et al. (2009) [18] demonstrated that Rhode Island Red belongs to the Asian group at K=3 of a STRUCTURE plot. The results of previous studies and our present study confirm its breeding history; i.e., Road Island Red was developed by crossing chickens of Asian origin, such as the Shanghai (Cochin), Malay, and Java breeds, with Brown Leghorn chickens [20].
CAL is an albino line developed by crossbreeding albino chickens derived from the White Wyandotte that was transferred from the Avian Disease and Oncology Laboratory (USDA-Agricultural Research Service, USA) [64], with a local Japanese chicken breed, Nagoya, at the ABRC. Therefore, the NU-Gg_DL08 haplotype of CAL is identical to that found for Nagoya (AB263971, Wada et al., direct submission to GenBank, 2008; HQ022887, [4]; AB007748, Miyake, direct submission to GenBank, 2000). In addition, a chicken breed of Chinese origin, Cochin, is known to have contributed to the establishment of White Wyandotte. This breeding history of CAL is supported by its close genetic relationship with the other lines of Asian origin, such as CHN, EJ, and JB, in the DAPC plot and the Dmc cladogram. This is a good example of the use of both mitochondrial and nuclear DNA markers for genetic monitoring to clarify the origins and histories of chicken breeds.
The SIL, CHN, EJ, and JB lines are derived from indigenous Japanese chicken breeds, the Japanese Silkie, Chahn (Utaichahn), Ehime-jidori, and Japanese Bantam, respectively. They share the NU-Gg_DL09 D-loop haplotype that is identical to the D13 haplotype in Miao et al. (2013) [38]. The D13 haplotype has been found only in indigenous chicken breeds in Japan, China, India and Southeast Asian countries [38], suggesting that any of these areas is the origin of each of the four lines. Their close genetic relationships, except for SIL, were also shown in STRUCTURE and DPAC plots constructed using microsatellite DNA markers. Less is known regarding the origin of the Ehime-jidori, except that it is a local breed of Ehime prefecture, Japan. The Chahn and Japanese Bantam are thought to have been introduced into Japan from the Southeast Asian region [54], while the Japanese Silkie is known to have been introduced from China. Therefore, we speculate that the genetic difference between SIL and the other three lines (EJ, JB, and CHN) shown by microsatellite DNA markers is due to the differences in their geographic origin, namely, Chinese or Southeast Asian origins. In fact, SIL showed a close genetic relationship with the CB line, which is a bantam type of the Cochin breed that originated in China. The RJF line originated in a population of red junglefowls, which was established from one cock and two hens from the Sumatra Island in Indonesia and had been maintained at Kagoshima University. The small number of female founders might explain why only a single D-loop haplotype (NU-Gg_DL07) is associated with the line. However, as described in Appendix, the RJF line had been crossed with red junglefowls from other regions in Asia. This would be a reason why a genetic subdivision within the RJF line occurred at K=21 in the STRUCTURE plot.
The genetic characteristics of 21 chicken lines and their breeding histories were well supported by three cladograms (Da, Dps, and Dmc). Among these, the genetic relationships shown in the Da cladogram seems to have the most support, and the only one in which RIR, PC, and #413 form a monophyletic clade. In the Dps cladogram, there were some inconsistencies with the other two cladograms; for example, BM-C was included in the same clade as the four Fayoumi lines; and PC formed a lineage that was distant from RIR. On the contrary, the Dmc cladogram supported certain breeding histories that were not shown in either Da or Dps cladograms; for example, CAL was categorized into the Asian group only in the Dmc cladogram; and this cladogram describes a close genetic relationship between SIL and CB, both of which are known to be of Chinese origin.
In this study, we revealed the genetic characteristics and diversity of 21 chicken lines that are registered as chicken resources with the National BioResource Project of Japan, by a large scale genetic monitoring effort using mitochondrial D-loop sequences and 54 microsatellite DNA markers. All the lines have been genetically controlled well, and their genetic characteristics confirm their origins and breeding histories. Four Fayoumi lines (GSP, GSN/1, YL, and PNP), BM-C, and two Leghorn-derived lines (WL-G and BL-E) exhibited high genetic homogeneities, which makes them ideal candidates as standard experimental chicken lines for avian biological research.
Supplementary Material
Acknowledgments
We would like to thank Makoto Mizutani, Takeo Uemura, and Kumiko Okada, and other Avian Bioscience Research Center, Nagoya University for their technical assistance. This study was carried out with support of the National BioResource Project (NBRP) “Chicken/Quail” from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and Japan Agency for Medical Research and Development (AMED), Japan.
Appendix
Summary of breeding histories of 21 chicken lines and their remarkable characteristics
GSP (Fayoumi)
GSP is a highly inbred line derived from the Fayoumi breed, which is native to Egypt and was introduced in 2004 by the Nippon Institute of Biological Science (NIBS) of Japan. This line originated from one cock and two hens that were introduced to NIBS from the Okazaki National Livestock Breeding Station in Japan in 1971. This line was maintained by pair mating of 2 ‒ 5 pairs per generation for 12 generations from 1972 to 1980, and it has been maintained as a closed colony consisting of 4 ‒ 5 cocks and 4 ‒ 6 hens for nearly 40 years. Skin grafts between individuals within the line and between this line and the GSN/1 line are acceptable.
GSN/1 (Fayoumi)
This is a highly inbred line derived from the Fayoumi breed, which was introduced by NIBS in 2004. This line originated from three cocks and two hens that were introduced from the Okazaki National Livestock Breeding Station to NIBS in 1971. This line was maintained by pair mating of 2 ‒ 5 pairs per generation for 12 generations from 1972 to 1980, and it has been maintained as a closed colony consisting of 5 ‒ 8 cocks and 5 ‒ 10 hens for nearly 40 years. Skin grafts between individuals within the line and between this line and the GSP line are acceptable.
PNP/DO (Fayoumi)
This is a highly inbred line derived from the Fayoumi breed, which was introduced by NIBS in 2004. This line originated from three cocks and two hens that were introduced to NIBS from the Okazaki National Livestock Breeding Station in 1971. This line was maintained by pair mating with 1 ‒ 4 pairs per generation for 10 generations from 1971 to 1979, and it has been maintained as a closed colony consisting of 2 ‒ 5 cocks and 5 ‒ 10 hens. Oviducts of females of PNP/DO are developed in both sides but the developmental grade of the right oviduct is different between individuals. Skin grafts between individuals is acceptable.
YL (Fayoumi)
YL originated from the Fayoumi breed that was introduced to NIBS by the Okazaki National Livestock Breeding Station in 1971. A mutant chicken with corneal degeneration was found in 1976, and YL was established from one pair of founder mutants. YL develops corneal degeneration in chickens older than 10 months. The line was maintained by pair mating of 1 ‒ 4 pairs per generation for 4 generations from 1976 to 1980, and it has been maintained as a closed colony consisting of 1 ‒ 5 cocks and 5 ‒ 10 hens. This line was introduced to Nagoya University by NIBS in 2004.
BM-C (Black Minorca)
The Black Minorca originated from the Island of Minorca, which is part of the Balearic Islands in the Mediterranean Sea. This line was initially introduced to Nagoya University from Connecticut University, USA in 1959. A pedigree breeding started with one cock and two hens in 1971, and this line was maintained with pair mating of 1 ‒ 4 pairs per generation for 5 generations from 1971 to 1975, and it has been maintained as a closed colony consisting of 3 ‒ 5 cocks and 5 ‒ 10 hens. Skin graft between individuals is acceptable
WL-G (White Leghorn)
The White Leghorn is the most common color type of the Leghorn breeds. This line is one of the oldest lines maintained at ABRC, whose breeding started as a closed colony at Nagoya University in 1969. Its MHC haplotype is fixed to B12, and skin graft is acceptable between individuals.
WL-M/O (White Leghorn)
WL-M/O was established from the White Leghorn, and it has been maintained as a closed colony at Nippon Institute for Biological Science since 1975. This line was introduced to Nagoya University in 2006. WL-M/O is susceptible to all subgroup viruses (A–E) of the avian leukosis virus (ALV).
BL-E (Brown Leghorn)
The Brown Leghorn is a color variant of the Leghorn layer chicken. This line was introduced to Nagoya University from Edinburgh Poultry Research Center, UK in 1960. This line is the oldest line maintained as a closed colony at Nagoya University.
RIR (Rhode Island Red)
The Rhode Island red is a dual-purpose breed for egg and meat production. RIR-Y8/NU was introduced to Nagoya University by the Okazaki National Livestock Breeding Station in 1980, and it has been maintained as a closed colony. Even though the MHC haplotype seems to be fixed to B12, some chickens showed rejection against skin graft from other individuals.
GB (Game Bantam)
The Game Bantam chicken, which originated from the modern Game Bantam breed, was introduced from a chicken fancier in the Miyagi Prefecture of Japan in 2008, and it has been maintained as a closed colony. This line has a small body size and is applicable for research on dwarf genes or genes that control the upright posture, which is a characteristic trait of game type chickens.
CAL (derived from White Wyandot)
This line was an albino mutant derived from the White Wyandot breed, which was introduced to Keio University, Japan from the Avian Disease and Oncology Laboratory (USDA-Agricultural Research Service, USA) in 1993, and it was then introduced to Nagoya University in 2006. After mating with the Nagoya breed (a local chicken breed in Japan), the c pigment locus of the line was fixed in a homozygous condition for an albino allele (ca/ca). This line has a normal auditory sense and is sensitive to anesthetics.
DD (Dandarawi)
Dandarawi is an Egyptian indigenous chicken breed. DD originated from one pair of Dandarawi chickens, which were introduced to Nagoya University from a chicken fancier from the Ehime Prefecture in 2008.
SIL (White Silkie)
The Silkie is a Japanese indigenous chicken breed that originated in China, which is famous for its various unique phenotypic traits, such as black skin, polydactyly, hairy crest, and silky feathers. SIL was introduced from a chicken fancier from the Aichi Prefecture in 1994. White plumage of SIL is controlled by a recessive gene (c) on the c locus, which is dominant against an albino allele (ca) on the same locus.
CHN (Chahn)
Chahn or the Okinawa crower is an indigenous chicken breed in Okinawa, Japan. This line was developed from one cock that was introduced from a chicken fancier from the Ehime Prefecture in 2007 and from three hens that were introduced from the Society for the Preservation of Chahn from Uruma city in the Okinawa Prefecture in 2009. This line exhibits several unique traits, such as a variety of plumage colors, a long-lasting crowing pattern, and a beard.
EJ (Ehime-jidori)
Ehime-jidori is a Japanese indigenous chicken breed from the Ehime Prefecture. This line was established from one cock and 13 hens introduced from a chicken fancier from the Ehime Prefecture in 2007, and hatching eggs of Ehime-jidori were introduced from a chicken fancier from the Fukui prefecture 2013. A variety of mutant genes related to plumage coloration have been maintained as a polymorphic status in this line.
PC (derived from Rhode Island Red)
PC (Petit-Cocco) is a dwarf mutant line derived from the Rhode Island red breed. This line was introduced by the Kochi Prefectural Livestock Experiment Station in Japan through the Faculty of Agriculture, Shinshu University, Japan in 2006.
JB (Japanese Bantam)
Japanese Bantam (Chabo) is a Japanese indigenous chicken breed. Japanese Bantam is considered to be a true Bantam chicken because this breed has much shorter legs (the semi-lethal creeper trait) than other bantam chicken races, such as the game bantam and the Polish bantam. A variety of plumage coloration is also characteristic of the Japanese bantam. Founder chickens of the JB line were introduced to Nagoya University from a chicken fancier in 2013, and this line has been maintained as a closed colony.
CB (Cochin Bantam)
Cochin Bantam or Pekin Bantam is a Bantam race of the Cochin breed that originated in China and was exported to England and the USA as a fancy chicken. The CB line originated from one pair of Cochin bantam chickens with a buff plumage color, which was introduced from a chicken fancier in 2014. CB has many mutant traits such as heavily feathered legs, ptilopody (feathered shank), and a vulture hock.
413 (derived from New Hampshire)
The #413 line was introduced by NIBS in 2008. This line exhibits muscular dystrophy disorder, which is not associated with the dystrophin that causes human Duchenne muscular dystrophy. Much research has been conducted to explore the causative genes, and the ubiquitin ligase gene (WWP1) is currently the most likely candidate. Excessive accumulation of caveolin-3 in the muscles is the likely cause for abnormal muscles in this mutant.
OS (derived from White Leghorn)
The OS (obese) line was introduced from Innsbruck University, Austria in 2007, which was originally developed at Cornell University, USA in the 1950s. This line exhibits autoimmune thyroiditis and is expected to be useful as a model animal for human Hashimoto’s disease, in which the thyroid gland is gradually destroyed by its own immune system. Many pathological, genetic, immunological, and endocrinology studies have been reported for OS chickens, but its definitive etiology is still unknown.
RJF (Red junglefowl)
One pair of red junglefowls were imported to the Kagoshima University, Japan from the Sumatra Island (Indonesia) and were mated with red junglefowls from Vietnam or Thailand. This line has been maintained as a closed colony for more than 20 years. The RJF line is derived from the descendant of the fowls described above, which were introduced to Nagoya University from a chicken fancier from the Yamanashi Prefecture in 2004.
Vaccination
The vaccination of the following 11 items for nine infectious diseases was made to all lines in each generation: Newcastle disease virus, infectious bursal disease virus, egg drop syndrome virus, Avibacterium (Haemophilus) paragallinarum A, Avibacterium (Haemophilus) paragallinarum C, Mycoplasma gallisepticum (Kyoto Biken Laboratories, Inc,. Kyoto, Japan), avian encephalomyelitis virus, infectious bronchitis virus, infectious bursal disease virus, and fowlpox virus (Nisseiken Co., Ltd, Tokyo, Japan), Marek’s disease virus (Kyoritsu Seiyaku Co., Ltd., Tokyo, Japan).
Microbial testing
Microbial testing is carried out regularly for the following 14 items (Shokukanken Inc., Gunma, Japan): Newcastle disease virus, avian encephalomyelitis virus, infectious laryngotracheitis virus, infectious bronchitis virus, infectious bursal disease virus, egg drop syndrome virus, turkey rhinotracheitis virus, Avibacterium (Haemophilus) paragallinarum, Mycoplasma gallisepticum, Mycoplasma synoviae, Salmonella pullorum, Salmonella enteritidis, avian influenza virus, and leucocytozoonosis.
References
- 1.Berthouly-Salazar C., Rognon X., Van T., Gély M., Chi C.V., Tixier-Boichard M., Bed’Hom B., Bruneau N., Verrier E., Maillard J.C., Michaux J.R.2010. Vietnamese chickens: a gate towards Asian genetic diversity. BMC Genet. 11: 53. doi: 10.1186/1471-2156-11-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowcock A.M., Ruíz-Linares A., Tomfohrde J., Minch E., Kidd J.R., Cavalli-Sforza L.L.1994. High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368: 455–457. doi: 10.1038/368455a0 [DOI] [PubMed] [Google Scholar]
- 3.Carlsson J.2008. Effects of microsatellite null alleles on assignment testing. J. Hered. 99: 616–623. doi: 10.1093/jhered/esn048 [DOI] [PubMed] [Google Scholar]
- 4.Chang C.S., Chen C.F., Berthouly-Salazar C., Chazara O., Lee Y.P., Chang C.M., Chang K.H., Bed’Hom B., Tixier-Boichard M.2012. A global analysis of molecular markers and phenotypic traits in local chicken breeds in Taiwan. Anim. Genet. 43: 172–182. doi: 10.1111/j.1365-2052.2011.02226.x [DOI] [PubMed] [Google Scholar]
- 5.Cole R.K.1966. Hereditary hypothyroidism in the domestic fowl. Genetics 53: 1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dancause K.N., Vilar M.G., Steffy R., Lum J.K.2011. Characterizing genetic diversity of contemporary pacific chickens using mitochondrial DNA analyses. PLoS One 6: e16843. doi: 10.1371/journal.pone.0016843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darriba D., Taboada G.L., Doallo R., Posada D.2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9: 772. doi: 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dieringer D., Schlötterer C.2003. Microsatellite analyzer (MSA): a platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes 3: 167–169. doi: 10.1046/j.1471-8286.2003.00351.x [DOI] [Google Scholar]
- 9.Drummond A.J., Suchard M.A., Xie D., Rambaut A.2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29: 1969–1973. doi: 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earl D.A., von Holdt B.M.2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4: 359–361. doi: 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- 11.Evanno G., Regnaut S., Goudet J.2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14: 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- 12.Excoffier L., Lischer H.E.L.2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10: 564–567. doi: 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- 13.Food and Agriculture Organization2011. Molecular genetic characterization of animal genetic resources. In: FAO Animal Production and Health Guidelines No. 9. Rome. [Google Scholar]
- 14.Food and Agriculture Organization2015. The Second Report on the State of the World’s Animal Genetic Resources for Food and Agriculture (Scherf, B.D. and Pilling, D. eds.), FAO Commission on Genetic Resources for Food and Agriculture Assessments, Rome. [Google Scholar]
- 15.Filatov D.A.2002. PROSEQ: A software for preparation and evolutionary analysis of DNA sequence data sets. Mol. Ecol. Notes 2: 621–624. doi: 10.1046/j.1471-8286.2002.00313.x [DOI] [Google Scholar]
- 16.Fu Y., Niu D., Luo J., Ruan H., He G.Q., Zhang Y.P.2001. [Studies of the origin of Chinese domestic fowls]. Yi Chuan Xue Bao 28: 411–417(in Chinese). [PubMed] [Google Scholar]
- 17.Fumihito A., Miyake T., Sumi S., Takada M., Ohno S., Kondo N.1994. One subspecies of the red junglefowl (Gallus gallus gallus) suffices as the matriarchic ancestor of all domestic breeds. Proc. Natl. Acad. Sci. USA 91: 12505–12509. doi: 10.1073/pnas.91.26.12505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granevitze Z., Hillel J., Feldman M., Six A., Eding H., Weigend S.2009. Genetic structure of a wide-spectrum chicken gene pool. Anim. Genet. 40: 686–693. doi: 10.1111/j.1365-2052.2009.01902.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasegawa M., Kishino H., Yano T.1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22: 160–174. doi: 10.1007/BF02101694 [DOI] [PubMed] [Google Scholar]
- 20.Hawksworth D.1982. British Poultry Standards, 4th ed., Butterworth Scientific, London. [Google Scholar]
- 21.Ishishita S., Kinoshita K., Nakano M., Matsuda Y.2016. Embryonic development and inviability phenotype of chicken-Japanese quail F1 hybrids. Sci. Rep. 6: 26369. doi: 10.1038/srep26369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishishita S., Takahashi M., Yamaguchi K., Kinoshita K., Nakano M., Nunome M., Kitahara S., Tatsumoto S., Go Y., Shigenobu S., Matsuda Y.2018. Nonsense mutation in PMEL is associated with yellowish plumage colour phenotype in Japanese quail. Sci. Rep. 8: 16732. doi: 10.1038/s41598-018-34827-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakobsson M., Rosenberg N.A.2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23: 1801–1806. doi: 10.1093/bioinformatics/btm233 [DOI] [PubMed] [Google Scholar]
- 24.Jombart T.2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405. doi: 10.1093/bioinformatics/btn129 [DOI] [PubMed] [Google Scholar]
- 25.Jombart T., Devillard S., Balloux F.2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11: 94. doi: 10.1186/1471-2156-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawabe K., Worawut R., Taura S., Shimogiri T., Nishida T., Okamoto S.2014. Genetic diversity of mtDNA D-loop polymorphism in Laotian native fowl populations. Asian-Australas. J. Anim. Sci. 27: 19–23. doi: 10.5713/ajas.2013.13443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinoshita K., Akiyama T., Mizutani M., Shinomiya A., Ishikawa A., Younis H.H., Tsudzuki M., Namikawa T., Matsuda Y.2014. Endothelin receptor B2 (EDNRB2) is responsible for the tyrosinase-independent recessive white (mo(w) ) and mottled (mo) plumage phenotypes in the chicken. PLoS One 9: e86361. doi: 10.1371/journal.pone.0086361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinoshita K., Shimogiri T., Ibrahim H.R., Tsudzuki M., Maeda Y., Matsuda Y.2016. Identification of TENP as the gene encoding chicken egg white ovoglobulin G2 and demonstration of its high genetic variability in chickens. PLoS One 11: e0159571. doi: 10.1371/journal.pone.0159571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopelman N.M., Mayzel J., Jakobsson M., Rosenberg N.A., Mayrose I.2015. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 15: 1179–1191. doi: 10.1111/1755-0998.12387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S., Stecher G., Tamura K.2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33: 1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langford S S.M., Kraitsek S., Baskerville B., Ho S.Y.W., Gongora J.2013. Australian and Pacific contributions to the genetic diversity of Norfolk Island feral chickens. BMC Genet. 14: 91. doi: 10.1186/1471-2156-14-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao Y., Mo G., Sun J., Wei F., Liao D.J.2016. Genetic diversity of Guangxi chicken breeds assessed with microsatellites and the mitochondrial DNA D-loop region. Mol. Biol. Rep. 43: 415–425. doi: 10.1007/s11033-016-3976-0 [DOI] [PubMed] [Google Scholar]
- 33.Liu Y.P., Wu G.S., Yao Y.G., Miao Y.W., Luikart G., Baig M., Beja-Pereira A., Ding Z.L., Palanichamy M.G., Zhang Y.P.2006. Multiple maternal origins of chickens: out of the Asian jungles. Mol. Phylogenet. Evol. 38: 12–19. doi: 10.1016/j.ympev.2005.09.014 [DOI] [PubMed] [Google Scholar]
- 34.Luzuriaga-Neira A., Villacís-Rivas G., Cueva-Castillo F., Escudero-Sánchez G., Ulloa-Nuñez A., Rubilar-Quezada M., Monteiro R., Miller M.R., Beja-Pereira A.2017. On the origins and genetic diversity of South American chickens: one step closer. Anim. Genet. 48: 353–357. doi: 10.1111/age.12537 [DOI] [PubMed] [Google Scholar]
- 35.Lyimo C.M., Weigend A., Msoffe P.L., Eding H., Simianer H., Weigend S.2014. Global diversity and genetic contributions of chicken populations from African, Asian and European regions. Anim. Genet. 45: 836–848. doi: 10.1111/age.12230 [DOI] [PubMed] [Google Scholar]
- 36.Lyimo C.M., Weigend A., Msoffe P.L., Hocking P.M., Simianer H., Weigend S.2015. Maternal genealogical patterns of chicken breeds sampled in Europe. Anim. Genet. 46: 447–451. doi: 10.1111/age.12304 [DOI] [PubMed] [Google Scholar]
- 37.Meydan H., Jang C.P., Yıldız M.A., Weigend S.2016. Maternal origin of Turkish and Iranian native chickens inferred from mitochondrial DNA D-loop sequences. Asian-Australas. J. Anim. Sci. 29: 1547–1554. doi: 10.5713/ajas.15.1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miao Y.W., Peng M.S., Wu G.S., Ouyang Y.N., Yang Z.Y., Yu N., Liang J.P., Pianchou G., Beja-Pereira A., Mitra B., Palanichamy M.G., Baig M., Chaudhuri T.K., Shen Y.Y., Kong Q.P., Murphy R.W., Yao Y.G., Zhang Y.P.2013. Chicken domestication: an updated perspective based on mitochondrial genomes. Heredity 110: 277–282. doi: 10.1038/hdy.2012.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murai H., Tadokoro R., Sakai K., Takahashi Y.2015. In ovo gene manipulation of melanocytes and their adjacent keratinocytes during skin pigmentation of chicken embryos. Dev. Growth Differ. 57: 232–241. doi: 10.1111/dgd.12201 [DOI] [PubMed] [Google Scholar]
- 40.Mwacharo J.M., Bjørnstad G., Mobegi V., Nomura K., Hanada H., Amano T., Jianlin H., Hanotte O.2011. Mitochondrial DNA reveals multiple introductions of domestic chicken in East Africa. Mol. Phylogenet. Evol. 58: 374–382. doi: 10.1016/j.ympev.2010.11.027 [DOI] [PubMed] [Google Scholar]
- 41.Mwacharo J.M., Nomura K., Hanada H., Han J.L., Amano T., Hanotte O.2013. Reconstructing the origin and dispersal patterns of village chickens across East Africa: insights from autosomal markers. Mol. Ecol. 22: 2683–2697. doi: 10.1111/mec.12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nei M., Tajima F., Tateno Y.1983. Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J. Mol. Evol. 19: 153–170. doi: 10.1007/BF02300753 [DOI] [PubMed] [Google Scholar]
- 43.Oka T., Ino Y., Nomura K., Kawashima S., Kuwayama T., Hanada H., Amano T., Takada M., Takahata N., Hayashi Y., Akishinonomiya F.2007. Analysis of mtDNA sequences shows Japanese native chickens have multiple origins. Anim. Genet. 38: 287–293. doi: 10.1111/j.1365-2052.2007.01604.x [DOI] [PubMed] [Google Scholar]
- 44.Osman S.A.M., Sekino M., Kuwayama T., Kinoshita K., Nishibori M., Yamamoto Y., Tsudzuki M.2006. Genetic variability and relationships of native Japanese chickens based on microsatellite DNA polymorphisms -focusing on the natural monuments of Japan. J. Poult. Sci. 43: 12–22. doi: 10.2141/jpsa.43.12 [DOI] [Google Scholar]
- 45.Osman S.A.M., Yonezawa T., Nishibori M.2016. Origin and genetic diversity of Egyptian native chickens based on complete sequence of mitochondrial DNA D-loop region. Poult. Sci. 95: 1248–1256. doi: 10.3382/ps/pew029 [DOI] [PubMed] [Google Scholar]
- 46.Peakall R., Smouse P.E.2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research--an update. Bioinformatics 28: 2537–2539. doi: 10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng M.S., Fan L., Shi N.N., Ning T., Yao Y.G., Murphy R.W., Wang W.Z., Zhang Y.P.2015. DomeTree: a canonical toolkit for mitochondrial DNA analyses in domesticated animals. Mol. Ecol. Resour. 15: 1238–1242. doi: 10.1111/1755-0998.12386 [DOI] [PubMed] [Google Scholar]
- 48.Pisenti J.M., Delany M.E., Taylor R.L., Abbott U.K., Abplanalp H., Arthur J.A., Bakst M.R., Baxter-Jones C., Bitgoof J.J., Bradley F.A., Cheng K.M., Dietert R.R., Dodgson J.B., Donoghe A.M., Emsley A.B., Etches R.J., Frahm R.R., Gerritz R.J., Goetinck P.F., Grunder A.A., Harry D.E., Lamont S.J., Martin G.R., McGuire P.E., Moberg G.P., Pierro L.L., Qualset C.O., Qureshu M.A., Shultz F.T., Wilson B.W.1999. Avian genetic resources at risk: An assessment and proposal for conservation of genetic stocks in the USA and Canada. Report no. 20. Univ. California Div. Agric. Nat. Res., Genet. Resources Conserv. Program, Davis, CA.
- 49.Porras-Hurtado L., Ruiz Y., Santos C., Phillips C., Carracedo A., Lareu M.V.2013. An overview of STRUCTURE: applications, parameter settings, and supporting software. Front. Genet. 4: 98. doi: 10.3389/fgene.2013.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pritchard J.K., Stephens M., Donnelly P.2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rambaut A., Suchard M.A., Xie D., Drummond A.J.2014. Tracer v1.6, Available from http://beast.bio.ed.ac.uk/Tracer.
- 52.Rosenberg N.A., Burke T., Elo K., Feldman M.W., Freidlin P.J., Groenen M.A., Hillel J., Mäki-Tanila A., Tixier-Boichard M., Vignal A., Wimmers K., Weigend S.2001. Empirical evaluation of genetic clustering methods using multilocus genotypes from 20 chicken breeds. Genetics 159: 699–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seki R., Li C., Fang Q., Hayashi S., Egawa S., Hu J., Xu L., Pan H., Kondo M., Sato T., Matsubara H., Kamiyama N., Kitajima K., Saito D., Liu Y., Gilbert M.T., Zhou Q., Xu X., Shiroishi T., Irie N., Tamura K., Zhang G.2017. Functional roles of Aves class-specific cis-regulatory elements on macroevolution of bird-specific features. Nat. Commun. 8: 14229. doi: 10.1038/ncomms14229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shinjo A., Oikawa T., Sasanuma K.1985. The body measurements and morpho-genetic characteristics in Utaichahn (Okinawa native chicken). Science bulletin of the College of Agriculture. Univ. Ryukyus 32: 91–98(in Japanese with English abstract). [Google Scholar]
- 55.Shimmura T., Yoshimura T.2013. Circadian clock determines the timing of rooster crowing. Curr. Biol. 23: R231–R233. doi: 10.1016/j.cub.2013.02.015 [DOI] [PubMed] [Google Scholar]
- 56.Shimmura T., Ohashi S., Yoshimura T.2015. The highest-ranking rooster has priority to announce the break of dawn. Sci. Rep. 5: 11683. doi: 10.1038/srep11683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shinomiya A., Kayashima Y., Kinoshita K., Mizutani M., Namikawa T., Matsuda Y., Akiyama T.2012. Gene duplication of endothelin 3 is closely correlated with the hyperpigmentation of the internal organs (Fibromelanosis) in silky chickens. Genetics 190: 627–638. doi: 10.1534/genetics.111.136705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Storey A.A., Athens J.S., Bryant D., Carson M., Emery K., deFrance S., Higham C., Huynen L., Intoh M., Jones S., Kirch P.V., Ladefoged T., McCoy P., Morales-Muñiz A., Quiroz D., Reitz E., Robins J., Walter R., Matisoo-Smith E.2012. Investigating the global dispersal of chickens in prehistory using ancient mitochondrial DNA signatures. PLoS One 7: e39171. doi: 10.1371/journal.pone.0039171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suh S., Sharma A., Lee S., Cho C.Y., Kim J.H., Choi S.B., Kim H., Seong H.H., Yeon S.H., Kim D.H., Ko Y.G.2014. Genetic diversity and relationships of korean chicken breeds based on 30 microsatellite markers. Asian-Australas. J. Anim. Sci. 27: 1399–1405. doi: 10.5713/ajas.2014.14016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tadano R., Nishibori M., Nagasaka N., Tsudzuki M.2007. Assessing genetic diversity and population structure for commercial chicken lines based on forty microsatellite analyses. Poult. Sci. 86: 2301–2308. doi: 10.3382/ps.2007-00233 [DOI] [PubMed] [Google Scholar]
- 61.Takezaki N., Nei M., Tamura K.2010. POPTREE2: Software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Mol. Biol. Evol. 27: 747–752. doi: 10.1093/molbev/msp312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tamura K., Nei M.1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10: 512–526. [DOI] [PubMed] [Google Scholar]
- 63.Thompson J.D., Higgins D.G., Gibson T.J.1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. doi: 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tobita-Teramoto T., Jang G.Y., Kino K., Salter D.W., Brumbaugh J., Akiyama T.2000. Autosomal albino chicken mutation (ca/ca) deletes hexanucleotide (-deltaGACTGG817) at a copper-binding site of the tyrosinase gene. Poult. Sci. 79: 46–50. doi: 10.1093/ps/79.1.46 [DOI] [PubMed] [Google Scholar]
- 65.Van Oosterhout C., Hutchinson W.F., Wills D.P.M., Shipley P.2004. Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4: 535–538. doi: 10.1111/j.1471-8286.2004.00684.x [DOI] [Google Scholar]
- 66.West B., Zhou B.X.1989. Did chickens go north? New evidence for domestication. Worlds Poult. Sci. J. 45: 205–218. doi: 10.1079/WPS19890012 [DOI] [Google Scholar]
- 67.Yamada K., Wanchun J., Ohkura T., Murai A., Hayakawa R., Kinoshita K., Mizutani M., Okamoto A., Namikawa T., Ohta M.2013. Detection of methicillin-resistant Staphylococcus aureus using a specific anti-PBP2a chicken IgY antibody. Jpn. J. Infect. Dis. 66: 103–108. doi: 10.7883/yoken.66.103 [DOI] [PubMed] [Google Scholar]
- 68.Yoshimura K., Kinoshita K., Mizutani M., Matsuda Y., Saito N.2012. Inheritance and developmental pattern of cerebral hernia in the crested Polish chicken. J. Exp. Zoolog. B Mol. Dev. Evol. 318: 613–620. doi: 10.1002/jez.b.22464 [DOI] [PubMed] [Google Scholar]
- 69.Zanetti E., De Marchi M., Dalvit C., Cassandro M.2010. Genetic characterization of local Italian breeds of chickens undergoing in situ conservation. Poult. Sci. 89: 420–427. doi: 10.3382/ps.2009-00324 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.