Abstract
Pantothenate kinase generates 4′‐phosphopantothenate in the first and rate‐determining step of coenzyme A (CoA) biosynthesis. The human genome encodes three well‐characterized and nearly identical pantothenate kinases (PANK1‐3) plus a putative bifunctional protein (PANK4) with a predicted amino‐terminal pantothenate kinase domain fused to a carboxy‐terminal phosphatase domain. Structural and phylogenetic analyses show that all active, characterized PANKs contain the key catalytic residues Glu138 and Arg207 (HsPANK3 numbering). However, all amniote PANK4s, including human PANK4, encode Glu138Val and Arg207Trp substitutions which are predicted to inactivate kinase activity. Biochemical analysis corroborates bioinformatic predictions—human PANK4 lacks pantothenate kinase activity. Introducing Glu138Val and Arg207Trp substitutions to the human PANK3 and plant PANK4 abolished their robust pantothenate kinase activity. Introducing both catalytic residues back into human PANK4 restored kinase activity, but only to a low level. This result suggests that epistatic changes to the rest of the protein already reduced the kinase activity prior to mutation of the catalytic residues in the course of evolution. The PANK4 from frog, an anamniote living relative encoding the catalytically active residues, had only a low level of kinase activity, supporting the view that HsPANK4 had reduced kinase activity prior to the catalytic residue substitutions in amniotes. Together, our data show that human PANK4 is a pseudo‐pantothenate kinase—a catalytically deficient variant of the catalytically active PANK4 found in plants and fungi. The Glu138Val and Arg207Trp substitutions in amniotes (HsPANK3 numbering) completely deactivated the pantothenate kinase activity that had already been reduced by prior epistatic mutations.
Keywords: pantothenate kinase, PANK4, coenzyme A, kinase, evolution, pseudoenzyme
Abbreviations
- Clade 1
monofunctional human PANK3‐like pantothenate kinases
- Clade 2
bifunctional kinase/phosphatases and monofunctional human PANK4‐like pantothenate kinases
Introduction
Pantothenate kinase (PANK) catalyzes the phosphorylation of pantothenate to generate 4′‐phosphopantothenate in the first and rate‐determining step of coenzyme A (CoA) synthesis.1 CoA is an essential cofactor for a variety of metabolic processes including the tricarboxylic acid cycle, β‐oxidation, and lipid metabolism. Three distinct structural families of PANK have been characterized.2, 3, 4 Type I and type III PANKs are found only in bacteria. Type II PANKs are found in both bacteria and eukaryotes, although the bacterial type II PANK is only distantly related to the eukaryotic PANKs.3, 5 There are three human PANK genes (PANK1‐3) that encode four characterized Type II PANK isoforms.1, 6 These isoforms have highly related catalytic cores fused to ancillary sequence motifs that direct the proteins to different subcellular compartments.7 A fundamental characteristic of mammalian PANKs is their stringent feedback regulation by acetyl‐CoA and other CoA thioesters.8, 9 High‐resolution crystal structures of all the intermediates in the catalytic cycle and the inhibited acyl‐CoA complexes provide a detailed picture of the mechanisms of substrate binding, catalysis, and allosteric regulation of HsPANK3.10, 11 Genetic and chemical manipulations of PANK activities in mice illustrate the importance of the PANKs and CoA in supporting oxidative metabolism.12, 13, 14 Mutations in the human PANK2 gene cause Pantothenate Kinase‐Associated Neurodegeneration.15, 16
There is a fourth human gene called PANK4, which encodes a putative bifunctional protein with a predicted amino‐terminal Type II PANK domain and a carboxy‐terminal phosphatase domain (DUF89).17 PANK4 homologs are found in animals, fungi, and plants (PANK4 is called PanK2 in Arabidopsis thaliana).18 A recent publication characterized the phosphatase specificity of the isolated carboxy‐terminal domains of human and plant PANK4 proteins,17 and proposed that PANK4 has regulatory activity that contributes to the control of cellular CoA levels. This proposal was based on the assumption that the N‐terminal domain is an active PANK. The evidence as to whether the human PANK4 is a functional PANK is mixed. Genetic experiments reported that HsPANK4 complemented an Escherichia coli mutant strain with a temperature‐sensitive coaA(Ts) allele19 and extended the lifespan of a Drosophila PANK hypomorphic mutant.20 However, those reports did not provide data showing that the PANK4 was indeed expressed in the heterologous systems, nor did the authors assay for PANK activity. There are no positive biochemical data in the literature demonstrating that indeed PANK4 is active as a kinase. HsPANK4 has also been positively implicated in a variety of cellular pathways from screening assays, and the assumption that HsPANK4 is a functional PANK has led to speculations about the role of PANK activity and CoA biosynthesis in those pathways. An earlier biochemical analysis of the HsPANK4 expressed in HEK 293T cells concluded that this protein lacked PANK activity.13 Thus, the function of PANK4 is an open question.
In this article, we show definitely that HsPANK4 is a catalytically inactive pseudo‐PANK, and PANK4's implicated role in various pathways is not due to its kinase activity. Phylogenetic analysis indicates that the HsPANK4 is more closely related to the PANK4s found in plants and fungi, and only distantly related to the active HsPANK1‐3. Structural and biochemical analyses show that all active PANKs encode two conserved active site residues, numbered Glu138 and Arg207 in HsPANK3. Most PANK4 kinases, including AtPANK4, include these active site residues and are robust PANKs that are feedback regulated by CoA thioesters. However, we found that all amniote PANK4s harbor Glu138Val and Arg207Trp substitutions and are predicted to be enzymatically inactive. We found that the Glu138Val mutation in HsPANK3 and AtPANK4 abrogates the kinase function in those enzymes, while restoring the catalytic residues results in a low level of PANK activity in HsPANK4. The frog PANK4 is a close relative to the amniote and human PANK4s, but retains the catalytic residues and has a low level of PANK activity. These results show that the HsPANK4 is a pseudoenzyme, that is, a catalytically deficient variant of the active PANK4 found in plants and fungi. The complete loss of PANK activity in HsPANK4 is attributed to the Glu138Val and Arg207Trp substitutions in amniotes, although some reduction in activity due to additional substitutions in other portions of the kinase domain during evolution likely preceded the replacement of these key catalytic residues. This work shows that the various speculations about the biological role(s) of human and amniote PANK4s are not due to PANK activity as previously believed. HsPANK4 may have an alternative function(s) and should be considered a pseudoenzyme.21, 22, 23, 24
Results
HsPANK4 kinase domain has catalytically inactivating amino acid substitutions
The amino‐terminal domains of the bifunctional HsPANK4 and plant AtPANK4 are classified as PANKs based on their high degree of similarity to known Type II PANKs in the NCBI Conserved Domain Database (Fig. 1). The conserved regions with established roles in catalysis or substrate binding are highlighted in the sequence alignment (Fig. 1) and are based on the detailed structural and biochemical analysis of HsPANK3.10, 11 The DIGGS motif (α1) is responsible for interacting with the phosphate groups of the substrate ATP. The GSGVS region (α2) interacts with the phosphate groups of ATP while the terminal serine interacts with the carbonyl of the pantothenate amide. The conserved GGGT region (α3) interacts with the ribose of adenosine triphosphate (ATP). The FEE (α4) sequence in HsPANK3 is responsible for binding to the nucleotide ring of the ATP, and is similar (FDE) in HsPANK4. All of these essential primary sequences and structural motifs are found in HsPANK4 and AtPANK4, explaining why the bifunctional PANK4 kinase domains are classified as Type II PANKs.
Figure 1.

Multiple sequence alignment of the human PANK3, human PANK4, Arabidopsis thaliana PANK4, and Aspergillus nidulans PANK. Key sequences required for the function of human HsPANK3 are conserved in eukaryotic pantothenate kinase domains. The catalytic kinase domain (KD) of Clade 2 HsPANK3, A. nidulans (fungal) Clade 1 monofunctional kinase, A. thaliana Clade 1 HsPANK4‐related kinase/phosphatase (AtPANK4, called plant Pank2), and Clade 1 HsPANK4 were aligned to compare HsPANK4 to the sequences of three eukaryotic proteins known to possess pantothenate kinase activity. Regions of sequence conservation are observed over the entire catalytic domain. There are four highly conserved regions with established roles in the interaction with ATP.10 They are the DIGGS motif (α1), the VNΦGSGVSΦ region (α2), the ΦGGGTΦ region (α3), and the short FE(D)E (α4) sequence. Two key active site residues in HsPANK3, A. nidulans monofunctional pantothenate kinase, and AtPANK4 that are not conserved in HsPANK4 are highlighted. Glu138 (S1) is essential for phosphate transfer in the kinase reaction, and Arg207 (S2) is responsible for interacting with the carboxyl and the amide carbonyl of pantothenate.
Functional PANK proteins encode two key catalytic residues—Glu138 and Arg207 (Fig. 1) (HsPANK3 numbering). Glu138 functions as the catalytic base essential for phosphate transfer in the kinase reaction, and it also contributes to the binding of magnesium.11 Arg207 interacts with the carboxyl and the amide carbonyl of the substrate pantothenate, and the pathway feedback inhibitor, acetyl‐CoA.11 A close‐up of the HsPANK3 active site shows the structural orientation of these two residues in the active site and their interactions with substrates [Fig. 2(a)]. Alignment of the AtPANK4 and HsPANK4 with HsPANK3 shows that Glu138 and Arg207 are conserved in AtPANK4, but are Val and Trp in the HsPANK4, respectively (Fig. 1). The Glu138Val and Arg207Trp mutations are introduced to the HsPANK3 structure to illustrate the predicted effects on substrate and inhibitor binding [Fig. 2(b,d)]. Previous work shows that mutating Glu138 to alanine abolishes HsPANK3 activity,11 and a similar outcome is predicted with the valine substitution. The tryptophan that replaces arginine lacks the hydrogen bond interactions with the pantothenate carboxyl and amide carbonyl [Fig. 2(b)]. Glu138 does not play a role in the binding of the allosteric inhibitor, acetyl‐CoA, but the Arg207 side chain interacts with the two carbonyl groups of acetyl‐CoA [Fig. 2(c)].11 These hydrogen bond interactions are predicted to be absent if Arg207 is replaced with a tryptophan [Fig. 2(d)]. The consequences of the Arg207Trp substitution are predicted to be less catastrophic and result in decreased binding affinity for both the substrate pantothenate and the inhibitor, acetyl‐CoA. Based on this alignment, AtPANK4 is predicted to be a functional PANK that is feedback‐regulated by acetyl‐CoA. HsPANK4 is predicted to lack PANK activity due to the Glu138Val substitution.
Figure 2.
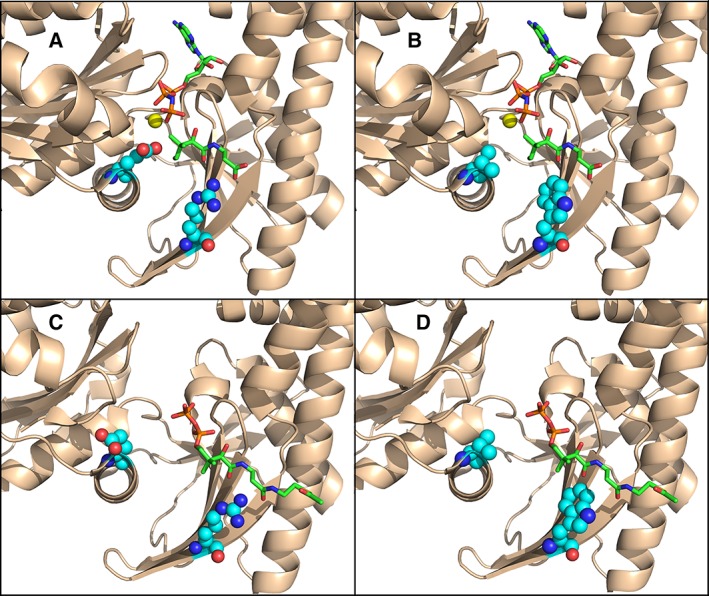
Predicted impact of Glu138Val and Arg207Trp mutations to HsPANK3. The carbons of residues 138 and 207 are represented as cyan balls, the carbons of the ATP and pantothenate ligands are green sticks, and Mg2+ is a gold ball. Oxygen atoms are colored red, nitrogen atoms blue, and phosphorus orange. (a) the HsPANK3•AMPPNP•Mg2+•pantothenate active site (PDBID: 5KPR). (b) the HsPANK3•AMPPNP•Mg2+•pantothenate active site with the E138V and R207W substitutions modeled onto the structure using PyMOL. (c) The active site in the HsPANK3•acetyl‐CoA complex (PDBID: 2I7N) with Glu138 and Arg207 depicted as cyan balls, and the carbons of the acetyl‐CoA ligand residues are colored as green sticks. (d) The active site in the HsPANK3•acetyl‐CoA complex with the E138V and R207W substitutions modeled onto the structure and represented as cyan balls.
Evolutionary history of PANK
A phylogenetic tree was constructed using eukaryotic type II PANK protein sequences to understand the relationship between HsPANK1‐3 and HsPANK4 as well as when the Glu138 and Arg207 substitutions arose in evolution (Fig. 3). Predicted type II PANK sequences were collected from the NCBI Reference Sequence Database and the Ensembl Genomic Database to contain representative sequences from all branches of eukaryotes. Multiple sequence alignment was generated using the DECIPHER package and the maximum likelihood phylogenetic tree was constructed using the phangorn package in R.25, 27 The modelTest function in phangorn found that the LG + Γ (4) + I model26 was best fitting by the Bayesian information criteria out of the WAG, JTT, LG, and Dayhoff models with and without gamma distributed rate variation among sites (Γ) and invariant sites (I). The bootstrap method (1000 replicate trees) was used to determine the confidence of the tree topology. The resulting tree was rooted at the midpoint because we are only trying to determine the relationship between the monofunctional and bifunctional eukaryotic PANK sequences. The overall maximum likelihood, protein phylogenetic tree containing all the eukaryotic PANK sequences are shown in Figure 3 and Figure S1. Figures 4 and 5 contain subtrees of the overall phylogenetic tree that focus on the phylogeny of the plant, fungi, and metazoan monofunctional and bifunctional PANKs, respectively.
Figure 3.
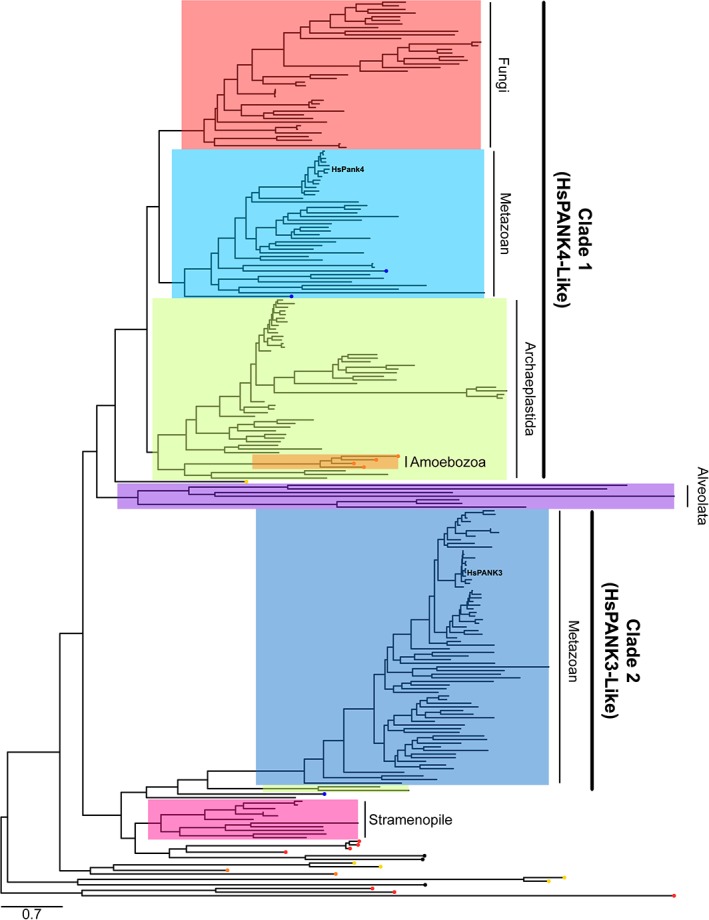
Phylogenetic tree of eukaryotic, Type II pantothenate kinase protein sequences. The phylogenetic tree was inferred through building a maximum likelihood phylogenetic tree using the phangorn package in R.25 Evolutionary distances were modeled using the LG + Γ (4) + I amino acid replacement matrix.26 Human PANK1‐3 belongs to Clade 2 while human PANK4 belongs to Clade 1, and are evolutionarily divergent. The Clade 1 PANK sequences from metazoan are highlighted in light blue. The Clade 1 PANK sequences from Archaeplastida are highlighted in green. The Clade 1 PANK sequences from fungi are highlighted in red. The Clade 2 PANK sequences from metazoan are highlighted in dark blue. PANK sequences from Stramenopile is highlighted in pink, while the PANK sequences from Alveolata is highlighted in purple. Many predicted PANK sequences from other supergroups are divergent from Clade 1 and Clade 2 PANKs as well as each other. PANK sequences from Excavata end with a yellow circular node. PANK sequences from Amoebozoa terminate with an orange circular node, and PANK sequences from Haptophyceae are indicated by a black circular node at intersecting lineages. Choanoflagellida and Filasterea are the closest relatives to metazoans, and their PANK sequences end with a blue circular node. Figure S1 contains more details with accession numbers and parent organism for each PANK sequence as well as the bootstrap values for each branch node.
Figure 4.
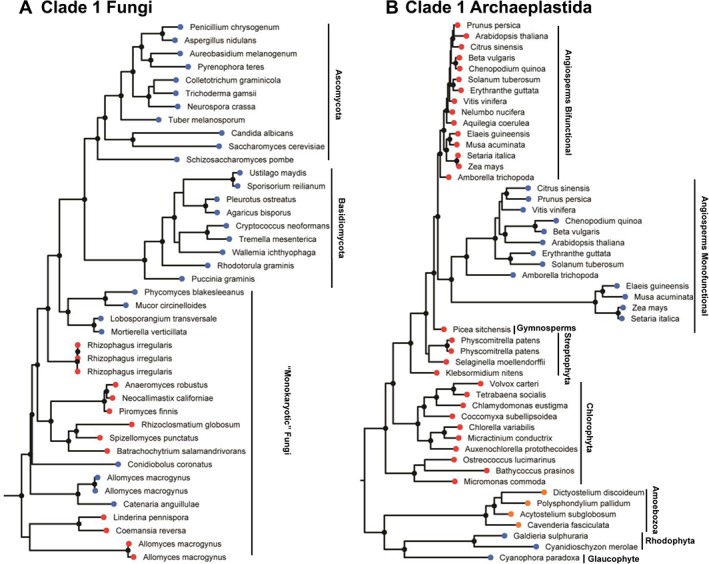
Phylogenetic tree of fungi and Archaeplastida Clade 1 pantothenate kinase protein sequence. The sequences terminating with red circular nodes encode a Clade 1 bifunctional kinase/phosphatase and sequences terminating with a blue circular node represent Clade 1 monofunctional kinases. Branches with bootstrap values greater than 70% have a black circular node. (a) The fungi Clade 1 PANK subtree. (b) The Archaeplastida Clade 1 PANK subtree. Amoebozoa are not Archaeplastida but some Amoebozoan monofunctional Clade 1 pantothenate kinases are phylogenetically related to the Archaeplastida Clade 1 PANK and terminate in orange circular nodes.
Figure 5.
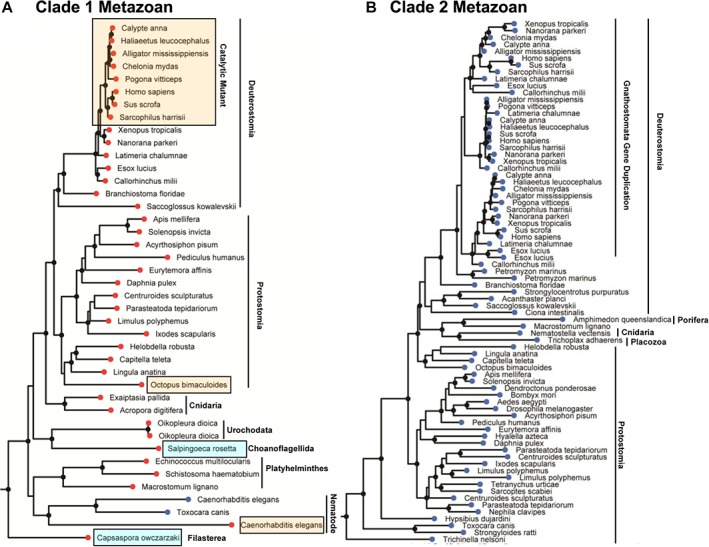
Phylogenetic tree of metazoan Clade 1 and Clade 2 pantothenate kinase protein sequences. Branches with bootstrap values greater than 70% have a black circular node. (a) Clade 1 metazoan PANK tree. Sequences terminating with red circular nodes encode for a Clade 1 bifunctional PANK/phosphatase, and the two sequences ending with blue circular nodes encode for Clade 1 monofunctional PANK. PANK sequences harboring substitutions to the key catalytic residues are highlighted by the yellow boxes. Choanoflagellida and Filasterea are single cell eukaryotes that are the closest living relatives to metazoans (blue boxes). (b) Monofunctional metazoan Clade 2 PANK subtree. Organisms belonging to the Gnathostomata lineage encode for 2–3 Clade 2 PANK genes with a recent common ancestor, suggesting a duplication of the Clade 2 PANK gene in the common ancestor of Gnathostomata.
The resulting PANK tree shows that HsPANK1‐3 and HsPANK4 belong to two separate clades (Fig. 3 and Fig. S1). HsPANK4 is a part of Clade 1 that contains other bifunctional metazoan PANK4s (light blue highlight) as well as the monofunctional and bifunctional PANKs from Archaeplastida (plants and algae) (green highlight) and fungi (red highlight). HsPANK1‐3 belongs to Clade 2, which only contains monofunctional metazoan PANKs (blue highlight). Clade 1 and Clade 2 PANK sequences are evolutionarily divergent with a deeply rooted common ancestor in the phylogenetic tree (Fig. 3 and Fig. S1). Many metazoans encode genes belonging to both clades, suggesting that the common metazoan ancestor possessed separate genes encoding Clade 1 HsPANK4‐like and Clade 2 HsPANK3‐like genes.
There are more than two clades of PANKs in our tree. The predicted PANKs from Alveolata form a separate clade (Fig. 3; purple highlight). The predicted PANK sequences from Stramenopile also form a distinct clade (Fig. 3; pink highlight). Many Amoebozoan PANKs cluster with the Archaeplastida clade, while other Amoebozoan PANKs are not a part of a clade. Many PANK sequences such as from Excavata and Haptophyceae as well as some known fungi outliers are also not a part of a clade. Although these sequences are predicted to be Type II PANKs based on the conservation of key structural features such as the DIGGS motif and the catalytic Glu138 and Arg207 residues, there is no experimental verification for these predictions. These sequences may have acquired another function and may not be PANKs, explaining their divergent evolution and outlier placement. They may also have a different evolutionary origin or result from lateral gene transfer.28 The focus of this article is to understand the evolution of the human PANK proteins, so the various questions surrounding the identity of the enzymes encoded by these sequences were not explored further. More detailed illustrations of the species distributions among Clades 1 and 2 are depicted in Figures 4 and 5.
Fungi and Archaeplastida only encode Clade 1 kinases
Fungi encode a Clade 1 HsPANK4‐like kinase, and do not contain Clade 2 monofunctional HsPANK3‐like kinases (Fig. 3). The Clade 1 PANK domains are either attached to a carboxy‐terminal phosphatase domain or exist as a standalone kinase. The Clade 1 sequences from the fungi cluster together and share a common ancestor [Fig. 4(a)] suggesting that the standalone PANKs (blue node) were derived from the loss of the phosphatase domain. The topology of the fungi Clade 1 PANK subtree [Fig. 4(a)] is consistent with the established fungi tree of life.29 Clade 1 sequences from the “Monokaryotic” fungi taxa diverge at the root of the tree. The monokaryotic fungi contain taxa that encode either a Clade 1 kinase (Mucoromycotina, Mortierellomycotina), a Clade 1 kinase/phosphatase (Chytridiomycota and Glomeromycotina), or both (Allomyces macrogynus of Blastocladiomycota). The two distinct groups of the higher fungi (Basidiomycota and Ascomycota) only encode a Clade 1 kinase lacking a phosphatase domain. The fungal Clade 1 PANKs are predicted to be active PANKs because they possess the active site Glu138 and Arg207 residues and they are the only PANKs in the essential CoA biosynthetic pathway found in some fungi. The monofunctional Clade 1 PANK in Aspergillus nidulans (of Ascomycota fungi) was previously demonstrated to be a robust PANK,30 thus supporting the prediction.
Archaeplastida (plants and algae) also encode only HsPANK4‐related Clade1 kinases, which cluster together and share a common ancestor [Fig. 4(b)]. The overall topology of the Archaeplastida Clade 1 PANK subtree is consistent with the established Archaeplastida tree of life, with the Archaeplastida common ancestor splitting into two branches: Glaucophyta/Rhodophyta and green algae/plants.31 Glaucophyta and Rhodophyta encode only a monofunctional Clade 1 kinase, while green algae/plants encode a Clade 1 kinase/phosphatase. Angiosperms are the exception because they encode both a Clade 1 kinase/phosphatase and a Clade 1 monofunctional kinase. The two different forms share a recent common ancestor, suggesting that the monofunctional angiosperm Clade 1 PANK gene arose from gene duplication and subsequent loss of the phosphatase domain from the Clade 1 kinase/ phosphatase. Genetic evidence suggests that both angiosperm Clade 1 proteins are PANKs,18 and the bifunctional PANK4 from Arabidopsis thaliana is experimentally verified as a robust pantothenate later in this report.
Duplication and loss of Clade 1 and Clade 2 PANKs in metazoans
Metazoans (animals) contain both Clade 1 and Clade 2 genes (Fig. 3). Some metazoan groups encode only a Clade 2 kinase, other groups encode only a Clade 1 kinase/phosphatase, and others encode both clades (Fig. 5). The loss of one of these clades occurs throughout the Metazoan kingdom. For example, the Insecta class contain members that encode only a Clade 2 kinase (Dendroctonus ponderosae, Aedes aegypti, Drosophila melanogaster, and Bombyx mori) as well as members that encode both a Clade 1 kinase/phosphatase and Clade 2 kinase (Acyrthosiphon pisum, Apis mellifera, Solenopsis invicta, and Pediculus humanus). The metazoan lineages closest to the root of the tree express Clade 1 or Clade 2 genes or both. Porifera (1 out of 1 available sequence) and Placozoa (1 out of 1 available sequence) encode only a Clade 2 monofunctional kinase. Cnidaria contains taxa that encode only a Clade 2 kinase (1 out of 3 available sequences) or only a Clade 1 kinase/phosphatase (2 out of 3 available sequences). Choanoflagellida are the closest living relatives to the metazoans and encode both clades. These observations suggest that the common metazoan ancestor most likely encoded both a Clade 1 HsPANK4‐like kinase/phosphatase and a Clade 2 HsPANK3‐like kinase. Because many metazoan taxa lost one or the other of the clades, the PANK activities of the two clades were possibly redundant in the metazoan common ancestor.
The metazoan Clade 1 PANK4 subtree is shown in Figure 5(a). Choanoflagellida and Filasterea are single cell eukaryotes that are the closest living relatives to metazoans (highlighted by blue boxes).32 Choanoflagellida encode both a Clade 1 kinase/phosphatase and a Clade 2 kinase, while Filasterea only encode a Clade 1 kinase/phosphatase. Some nematode genes are predicted to encode a monofunctional Clade 1 kinase (blue circle) that aligns closest to PANK domains of other metazoan Clade 1 PANK4 kinase/phosphatases, suggesting the loss of the phosphatase domain in these cases. Substitutions to the catalytic residue(s) in the Clade 1 kinase/phosphatase are observed in Caenorhabditis elegans (E138A), Octopus bimaculoides (R207W), and the entire amniote lineage (E138V and R207W) [Fig. 5(a), yellow highlight].
The metazoan Clade 2 monofunctional kinase subtree is shown in Figure 5(b). Most metazoan groups encode a single copy of a Clade 2 gene. However, Gnathostomata (jawed vertebrates, including cartilaginous fish, bony fish, amphibians, reptiles, birds, and mammals) encode for two or three Clade 2 genes that share a recent common origin, suggesting that the Clade 2 sequences in humans arose from gene duplication events in the common ancestor to the Gnathostomata. The three different Clade 2 genes in humans have differential cellular localizations and tissue expression levels, allowing for fine control of the kinase activity and cellular CoA levels.7
Amniotes encode clade 1 pseudo‐PANKs
Multiple sequence alignment of the 247 representative Clade 1 and Clade 2 sequences (Fig. 5) showed that they all encode the catalytic glutamate and arginine active site residues, except for all Clade 1 genes in the amniote lineage (see below) and in two outliers. Caenorhabditis elegans is one metazoan outlier that encodes two Clade 1 genes. One gene encodes a Glu138Ala substitution and has lost a portion of the amino‐terminal protein sequence, including the conserved DIGGS motif required for ATP binding.10 These mutations suggest a complete loss of ATP binding, and thus loss of kinase function. The second C. elegans Clade 1 sequence lost the phosphatase domain and became a monofunctional Clade 1 PANK. Octopus bimaculoides encodes both a Clade 2 kinase and a Clade 1 kinase/phosphatase. The bifunctional kinase encodes an Arg207Trp substitution. The effect of the Arg207Trp mutation is biochemically characterized in the human PANK3 later in this article, and leads to the decreased pantothenate and CoA binding affinity. The genomic sequence data are sparse in these two evolutionary branches making it difficult to determine the scope of Clade 1 sequence divergence.
The representative sequences from birds, reptiles, and mammals (amniotes) used to construct the metazoan phylogenetic tree (Fig. 5) all contained the Glu138Val and Arg207Trp substitutions in the Clade 1 HsPANK4‐like kinase/phosphatase. There are no examples of Clade 1 standalone kinases in amniotes. A detailed sequence analysis looking for the Glu138Val and Arg207Trp substitutions was conducted against all high‐quality amniote genome sequences in the NCBI Reference Sequence Database. In Sauropsidan (birds and reptiles), 92 of 92 genomes encoded both substitutions in the Clade 1 PANK4 genes. All 130 of 130 mammalian genomes encoded both substitutions in the Clade 1 PANK4s as well. All of the closest relatives to amniotes in the RefSeq database encode Clade 1 PANK genes containing the Glu and Arg active site residues (7/7 of amphibians, 1/1 of actinistia, and 125/125 of actinopterygii). There is evidence of other changes to the PANK4 protein sequence. For example, the Clade 1 PANK4 of Tursiops truncatus (dolphin) has lost the amino‐terminal PANK domain entirely and encodes a standalone phosphatase. These changes were not further examined due to the lack of high‐quality sequences in related organisms.
The key result is that the Glu138Val and Arg207Trp substitutions occurred at the point where amniotes and anamniotes diverged. As previously noted, Gnathostomata (jawed vertebrates which includes amniotes and anamniotes) encode multiple Clade 2 monofunctional kinase sequences, suggesting that the PANK function of the Clade 1 kinase/ phosphatase potentially became unnecessary following the gene duplication of monofunctional Clade 2 PANKs in jawed vertebrates.
The full phylogenetic tree is illustrated in Figure S1.
Critical roles of Glu138 and Arg207 in HsPANK3
Although we would like to directly interrogate the biochemical function of the HsPANK4 protein, we were unable to successfully express and purify either the full‐length protein or the kinase domain of the HsPANK4 protein despite trying a variety of technologies. Therefore, we modeled the catalytic mutations on the HsPANK3 protein. HsPANK3 is the best characterized PANK with extensive biochemical and structural data.8, 11 The Glu138Val and Arg207Trp substitutions found in HsPANK4 were evaluated in HsPANK3, keeping in mind that HsPANK4 and HsPANK3 are sequentially distinct. The goal was to understand the impact of these catalytic mutations on activity. This approach did not account for the other residues that are different between the HsPANK3 and HsPANK4 enzymes that may also affect activity. The HsPANK3(E138V) and HsPANK3(R207W) mutant proteins were overexpressed, purified, and characterized. Like the catalytically inactive HsPANK3(E138A) protein,11 HsPANK3(E138V) was expressed and purified but was completely inactive as a PANK (not shown) and was not studied further. The purified HsPANK3(R207W) protein [Fig. 6(a)] was biochemically characterized and we found that the mutated protein had a lower apparent Km (Km*) for ATP of 16 ± 1 μM compared to 175 ± 13 μM for wild‐type HsPANK3 at 180 μM pantothenate [Fig. 6(b)]. HsPANK3(R207W) also exhibited a higher Km* for pantothenate, 833 ± 66 μM compared to 17 ± 1 μM for wild‐type HsPANK3 at 5 mM ATP [Fig. 6(c)]. The kcat of the HsPANK3(R207W) mutant was not significantly different from wild type (6.1 ± 0.1 seconds−1 vs. 5.5 ± 0.1 seconds−1 for HsPANK3). A second important feature of HsPANK3(R207W) was its resistance to acetyl‐CoA inhibition. HsPANK3 was potently inhibited by acetyl‐CoA, but HsPANK3(R207W) was refractory to acetyl‐CoA inhibition under the same assay conditions [Fig. 6(d)]. The binding of HsPANK3(R207W) to ATP and acetyl‐CoA was interrogated directly using protein thermal stabilization assays [Fig. 6(e,f)]. HsPANK3(R207W) exhibited a greater affinity for ATP (K 0.5 = 0.15 ± 0.01 mM) compared to HsPANK3 (0.5 ± 0.1 mM) [Fig. 6(e)], consistent with the kinetic analysis [Fig. 6(b)]. Thermal stabilization assays showed that HsPANK3(R207W) also had lower binding affinity for acetyl‐CoA (K 0.5 = 44 ± 3 μM vs. 9.4 ± 0.4 μM for PANK3) [Fig. 6(f)]. ATP and acetyl‐CoA compete for binding to HsPANK3.8, 11 Thus, the combination of increased ATP affinity and decreased acetyl‐CoA affinity explains the significant resistance of HsPANK3(R207W) to acetyl‐CoA inhibition. The biochemical characterization of HsPANK3(E138V) and HsPANK3(R207W) enzymes predicts that these substitutions compromise both substrate binding and catalysis.
Figure 6.
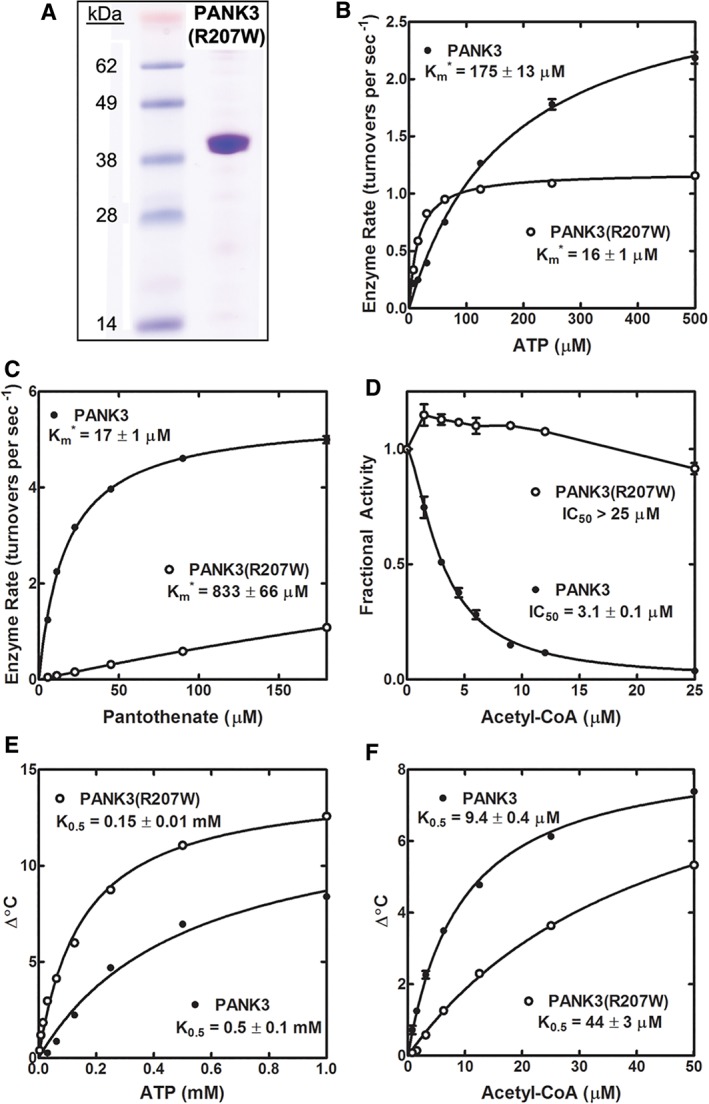
Purification and kinetics of HsPANK3(R207W). (a) HsPANK3(R207W) was purified by affinity chromatography to >95% purity based on sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gel electrophoresis. (b) Initial velocities were determined in duplicate as a function of ATP concentration at 180 μM of pantothenate for the wild‐type HsPANK3 and HsPANK3(R207W). The average and the range are plotted in GraphPad Prism. For some data points, no range bars appear because the range bar is shorter than the size of the symbol. The data points were fit to the Michaelis–Menten nonlinear regression equation using GraphPad Prism software, and the fitted line is shown on the graph. The apparent Km (Km*) values are shown in the figure panels. (c) Initial velocities were determined in duplicate as a function of pantothenate concentration at 5 mM ATP. (d) Initial velocities were determined in duplicate as a function of acetyl‐CoA concentration at 0.5 mM ATP and 45 μM pantothenate for HsPANK3 and HsPANK3(R207W). Inhibition curve was fit to the data using GraphPad Prism software to calculate the IC50. (e) Binding of ATP and acetyl‐CoA to HsPANK3 and HsPANK3(R207W) was assessed by protein thermal stabilization. Data shown are the mean of triplicate measurements and are fitted to the Michaelis–Menten equation represented by the lines in the figure panels. No range bars appear for some data because the range bar is shorter than the size of the symbol. Concentration dependent thermal stabilization of HsPANK3 and HsPANK3(R207W) by ATP. (f) Concentration dependent thermal stabilization by acetyl‐CoA.
AtPANK4, but not HsPANK4, is a robust PANK
Because we were not successful in the expression and purification of the full‐length HsPANK4 protein or the HsPANK4 kinase domain (HsPANK4‐KD) from bacteria or insect cells, we employed a mammalian cell expression system to compare the activities of the protein constructs (Figs. 7 and 8). PANK is the rate‐determining step in CoA synthesis, and its activity following expression in mammalian cells was determined by measuring the cellular CoA levels in HEK 293T cells 40 hours after transfection. The expression of the kinase isoform was verified by Western blot analysis of transfected cell lysates. The HsPANK3, HsPANK3(E138V), and HsPANK3(R207W) constructs were used to validate the sensitivity of this assay [Fig. 7(a)]. The cells expressing the empty vector (control) had a CoA level of 54 ± 2 pmol/mg protein. The cells expressing the wild‐type HsPANK3 had a CoA level of 1203 ± 72 pmol/mg protein, 20 times higher than control. The HEK 293T cells expressing the HsPANK3(E138V) mutant had a CoA level of 70 ± 3 pmol/mg protein, consistent with previous results showing that Glu138 has an essential catalytic role, and mutation to an aliphatic amino acid eliminates PANK activity.11 The cells expressing the HsPANK3(R207W) mutant had a CoA level of 3387 ± 167 pmol/mg protein, showing that the mutant had higher cellular activity than the wild‐type HsPANK3. Although HsPANK3(R207W) had reduced pantothenate binding [Fig. 6(c)], it is also refractory to feedback regulation by acetyl‐CoA [Fig. 6(d)]. Pantothenate is present in excess in cell culture media, and this result illustrates the power of acetyl‐CoA regulation of PANK3 in the control of cellular CoA levels. The cellular assay is a sensitive measure of the PANK activity of an expressed protein.
Figure 7.
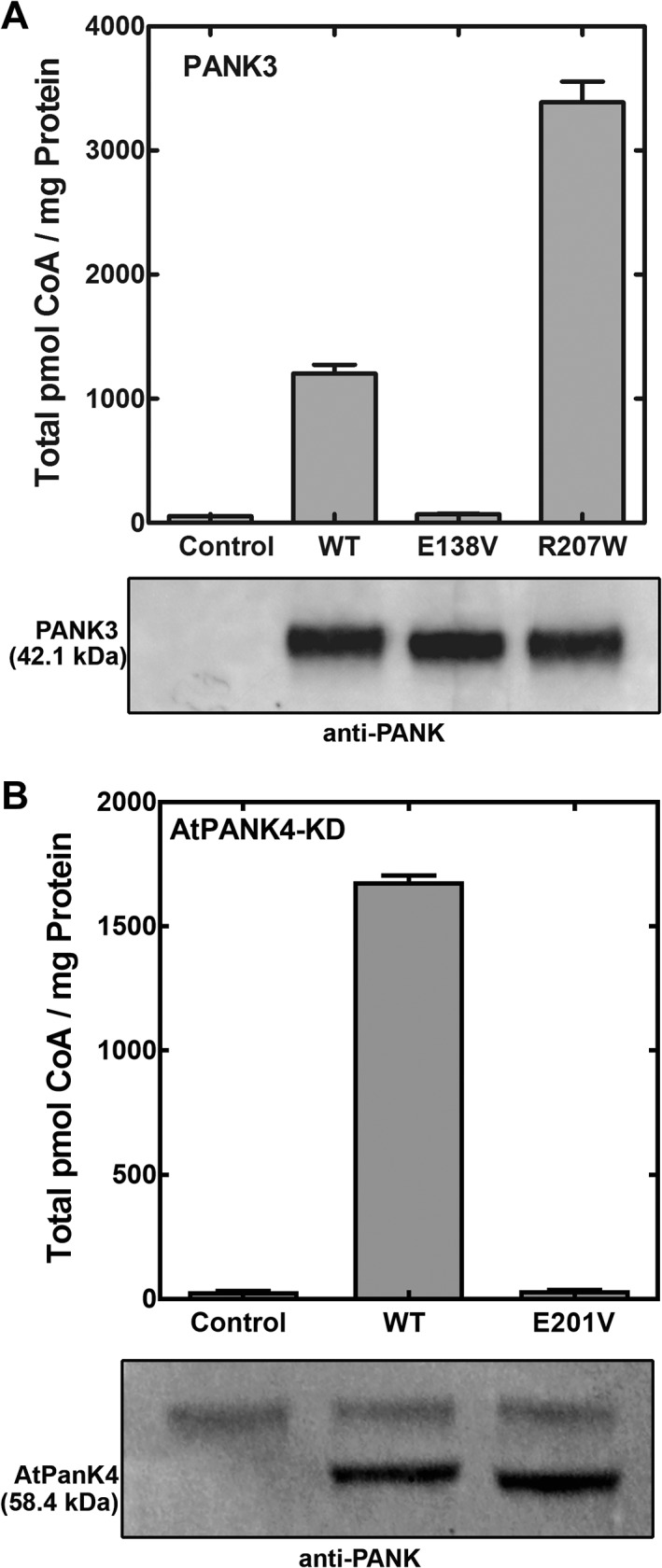
Cell‐based assay for pantothenate kinase activity. HEK 293T cells were transfected with plasmids directing the expression of various protein constructs, and the intracellular CoA levels were quantified 40 hours later. Controls were from cells transfected with the empty vector. Cell lysates were immunoblotted with anti‐PANK antibody to validate protein expression. (a) Cellular CoA levels in response to the expression of human HsPANK3, HsPANK3(E138V), and HsPANK3(R207W) proteins. (b) Cellular CoA levels in response to the expression of the kinase domains of plant PANK4 kinase domain (AtPANK4‐KD) and AtPANK4(E201V)‐KD mutant (equivalent to E138V in HsPANK3). Experiments were performed in triplicate and mean ± SEM are plotted.
Figure 8.
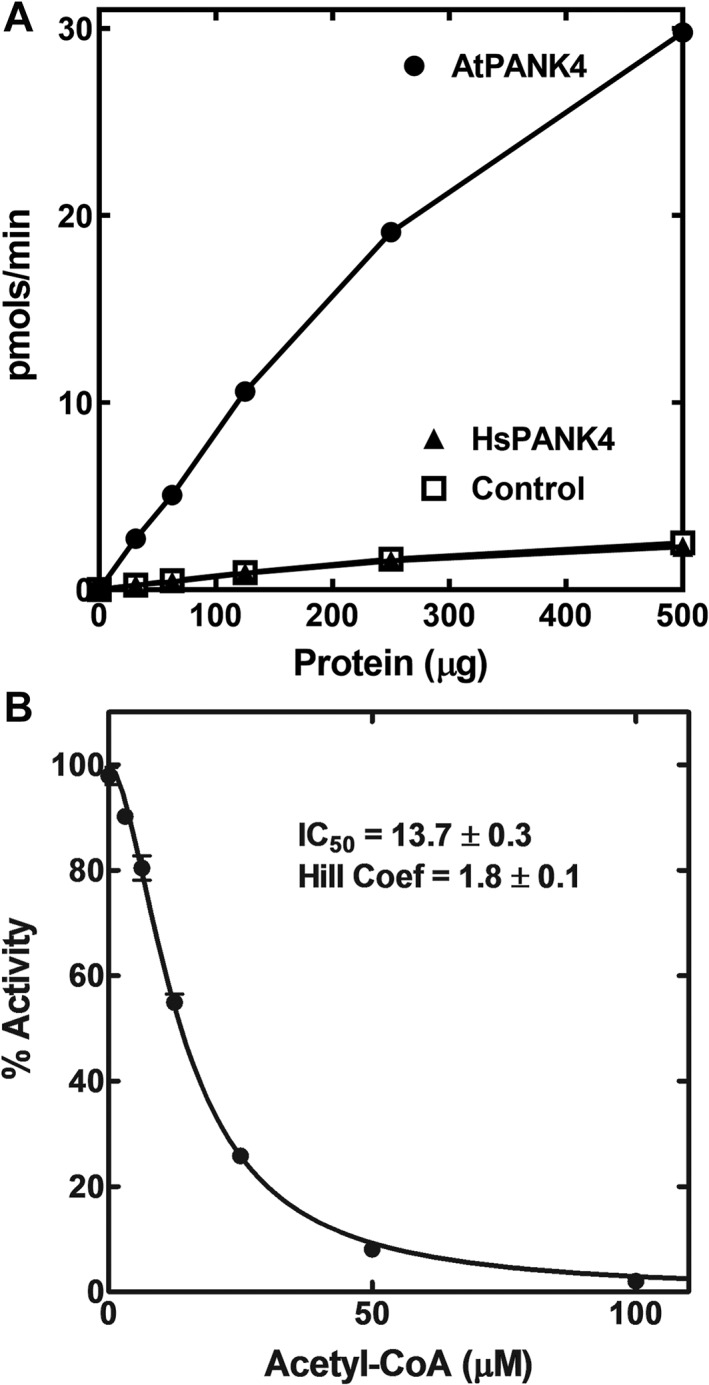
Plant AtPANK4‐KD kinase domain is feedback‐regulated by acetyl‐CoA. (a) Lysates were prepared from control (empty vector) and HEK 293T cells expressing AtPANK4‐KD or HsPANK4‐KD domains and the pantothenate kinase activities in the cell extracts were measured using a biochemical assay. (b) AtPANK4‐KD activity was inhibited by acetyl‐CoA (IC50 = 13.7 ± 0.3 μM; Hill number = 1.8 ± 0.1). Initial velocities were determined in duplicate and the error bars indicate the range of data. No range bars appear for some data because the range bar is shorter than the size of the symbol.
The activities of plant AtPANK4‐KD (kinase domain), HsPANK4 full‐length protein, and HsPANK4‐KD were measured using this assay. The AtPANK4‐KD exhibited robust PANK activity in cells, elevating CoA from 32 ± 1 to 1681 ± 23 pmol/mg protein [Fig. 7(b)]. These data show that the isolated kinase domain of AtPANK4 is a highly active PANK. The effect of the Glu138Val mutation was also tested on the AtPANK4. Mutating the catalytic glutamate into a valine (E201V in AtPANK4 numbering) abolished the PANK activity (36 ± 1 pmol/mg protein) [Fig. 7(b)]. Extracts of cells transfected with the AtPANK4‐KD construct had significantly elevated specific activity [Fig. 8(a)]. The strong inhibition of AtPANK4‐KD activity by acetyl‐CoA was highly cooperative [Fig. 8(b)], illustrating that AtPANK4 has the allosteric and feedback inhibition properties associated with previously characterized PANKs.33
Neither the full‐length HsPANK4 nor HsPANK4‐KD raised the cellular CoA levels (46 ± 1 pmol/mg protein for full‐length HsPANK4 and 48 ± 1 pmol/mg protein for HsPANK4 kinase domain vs. 48 ± 2 pmol/mg protein for control) indicating the absence of kinase activity [Fig. 9(a)]. These data were confirmed by enzyme assays that showed PANK activity was not elevated in the cell extracts expressing HsPANK4‐KD [Fig. 8(a)]. Phylogenetic and structural analysis predicted that the Glu138Val and Arg207Trp substitutions would inactivate the kinase function, so the catalytic Glu and Arg residues were restored in the HsPANK4‐KD [Fig. 9(a)]. The HsPANK4‐KD encoding either single catalytic residue mutation did not exhibit higher activity compared to control (44 ± 1 pmol/mg protein for HsPANK4(V155E) and 45 ± 1 pmol/mg protein HsPANK4(W219R)). However, the HsPANK4 encoding both substitutions exhibited a small, but significant increase in cellular CoA (113 ± 5 pmol/mg protein) indicating a low level of PANK activity [Fig. 9(a)]. HsPANK1‐3/HsPANK4 heterodimers are not expected to form because HsPANK4 is sequential divergent from HsPANK1‐3 and is not known to interact with HsPANK1‐3.34
Figure 9.
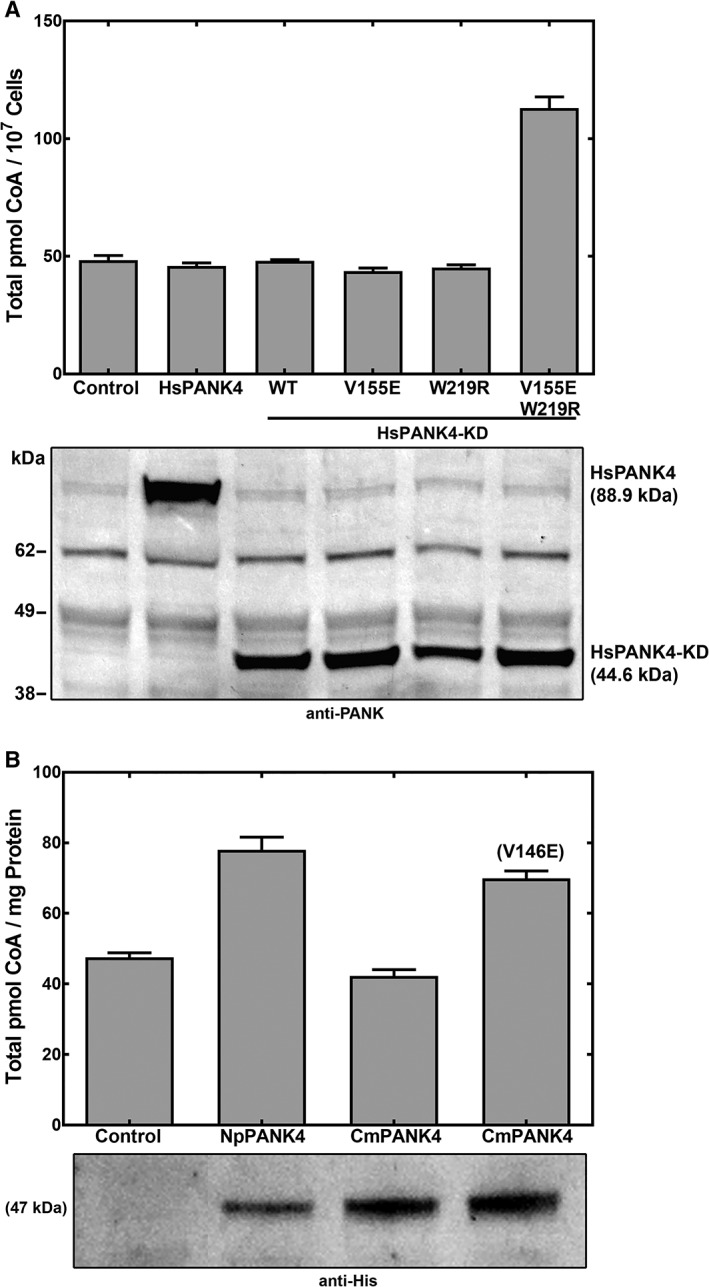
Pantothenate kinase activities of amniote PANK4s using the cell‐based assay. Mutations were made in the equivalent positions to Glu138 and Arg207 in HsPANK3 in human and turtle PANK4‐KD. (a) Cellular CoA levels in response to the expression of HsPANK4 full‐length protein, and four HsPANK4‐KD domains. Expressed proteins were Full‐length HsPANK4, HsPANK4 kinase domain (HsPANK4‐KD; WT), HsPANK4(V155E)‐KD, HsPANK4(W219R)‐KD, and HsPANK4(V155E, W219R)‐KD. Residues 155 and 219 of HsPANK4‐KD are equivalent to residues E138 and R207 in HsPANK3. (b) Cellular CoA levels in response to the expression of frog (NpPANK4) and turtle (CmPANK4) PANK4 kinase domains, and the CmPANK4(V146E) mutant (equivalent to E138V in HsPANK3). Protein expression levels were determined by immunoblotting with the anti‐His‐tag antibody. Experiments were performed in quadruplicate and the mean ± SEM are plotted.
Although mutation(s) of the catalytic residue(s) is the most common mechanism for the loss of enzymatic activity in pseudoenzymes, mutations to other parts of the protein that would alter the shape of the active site could also reduce the activity.35, 36 The low activity of the HsPANK4 with the restored catalytic residues suggests that other changes in amino acid sequence contribute to the absence of activity in the native protein. This raises the possibility that the Clade 1 PANK4s in anamniotes may have substantially lower PANK activity despite encoding the active Glu138 and Arg207 residues. The PANK activities of PANK4 from Nanorana parkeri (frog, Np) and Chelonia mydas (turtle, Cm), close living relatives on the two sides of the anamniote/amniote divergence, were determined to test this hypothesis [Fig. 9(b)]. The anamniote NpPANK4‐KD raised cellular CoA levels only slightly (78 ± 4 pmol/mg protein vs. 48 ± 1 pmol/mg protein control), showing that its activity was not robust [Fig. 9(b)]. Similarly, the kinase domain of CmPANK4‐KD did not increase cellular CoA (42 ± 2 pmol/mg protein), as expected for an amniote sequence, but the Val138Glu reactivating mutation was sufficient to restore a detectable increase in CoA (70 ± 2 pmol/mg protein) [Fig. 9(b)]. Together, these data indicated that the HsPANK4‐like proteins had substantially lower PANK activity before the introduction of the Glu138Val and Arg207Trp substitutions in amniotes.
It was reported that the HsPANK4 complemented the defect of strain ts9, an E. coli coaA(Ts) mutant,19 suggesting it was a functional PANK. However, we could not reproduce this result. Escherichia coli missense mutants often undergo reversion under selection pressure, and false positive complementation results are possible under these circumstances.37, 38 HsPANK4 was also reported to extend the lifespan of fumble (PANK1) mutants in Drosophila, but not as effectively as the active PANK3.20 Some function of HsPANK4, other than PANK activity, may be at play in the context of Drosophila development.
Discussion
Our results show that HsPANK4 (and amniote PANK4s) is a pseudoenzyme—a catalytically deficient variant of the active PANKs found in plants and fungi. Clade 1 HsPANK4‐related PANKs are widely distributed in metazoans, plants, and fungi (Fig. 3), and are proteins with either a predicted amino‐terminal PANK domain fused to a carboxy‐terminal phosphatase domain or a predicted monofunctional PANK. These Clade 1 kinase predictions are based on homology with the well‐characterized Clade 2 human PANKs. The Clade 1 PANK from fungi was also previously characterized as an active PANK,30 while the kinase activity of AtPANK4 was biochemical verified in this work. In contrast, the Clade 1 PANK4s in the amniote lineage, including HsPANK4, have Glu138Val and Arg207Trp substitutions at two key catalytic residues that are conserved in all active PANK proteins. Glu138 functions as the catalytic base essential for the kinase reaction, and Arg207 binds the pantothenate substrate.11 Introducing the Glu138Val mutation to the catalytically active HsPANK3 and AtPANK4 deactivates those enzymes, while restoring the Glu138 and Arg207 residues in HsPANK4 results in low level activity. Based on the absence of these conserved, essential residues and the lack of PANK activity in HsPANK4, all amniote Clade 1 PANK4 kinase/phosphatases are predicted to possess a pseudo‐PANK domain. Furthermore, the low activities of both the HsPANK4 catalytic revertant and the anamniote frog PANK4 with the catalytically active residues suggest that ancillary changes to other parts of the PANK4 protein significantly decreased the activity prior to the catalytic residue mutations in amniotes.
Pseudoenzymes often function as regulators in biological systems.35, 36, 39, 40, 41 Pseudoenzymes contain a protein domain that resembles a catalytically active counterpart, but lacks the predicted enzymatic activity.39, 40 At least 10% of mammalian enzyme domains are predicted to be catalytically inactive.35, 41 Well‐described classes of pseudoenzymes include pseudokinases and pseudophosphatases, which affect biology through their often poorly understood regulatory activities.22, 39 HsPANK4 has been implicated in several intracellular pathways. HsPANK4 overexpression in an insulin‐secreting cell line inhibits apoptosis by decreasing pro‐caspase‐9 transcription.42 PANK4 expression is upregulated by glucose in rat muscle and the M2‐type pyruvate kinase is one of its protein partners.43 PANK4 was identified as a host gene required for influenza virus replication in multiple influenza‐genome screens,44, 45, 46, 47 and the participation of PANK4 in flu proliferation was validated by siRNA‐mediated knockdown.47 Knockdown of PANK4 expression restores the plasma membrane localization of a mutant cystic fibrosis transmembrane conductance regulator.48 Our work demonstrates that these functions are not due to the PANK activity. HsPANK4 also has very low phosphatase activity against 4′‐phosphopantothenate and slightly higher activity toward damaged or oxidized forms of 4′‐phosphopantetheine.17 These data suggest that HsPANK4 also lacks specific 4′‐phosphopantothenate phosphatase activity and support the conclusion that HsPANK4 is not involved enzymatically in CoA biosynthesis.
A cellular function for HsPANK4 is apparent from two studies that identify congenital disease associated with amplification or reduction of PANK4 expression.49, 50 In particular, reduced expression due to intronic mutation was causative for congenital cataracts in both humans and mice.50 HsPANK4 has been reported to directly interact with LDHA, LDHB, HSP90AB1, MYBL2, TUBA1B, NUP133, and GCC1,51 and these protein–protein interactions are candidates for PANK4 cellular activity. Also, Pank4 −/− mice exhibited significant transcriptional variation among crystallin protein family members,50 suggesting a regulatory feedback onto gene expression. It is unknown if the catalytically active PANK4s (such as from plants and fungi) serve functions in addition to phosphorylating pantothenate. Many enzymes such as delta‐crystallin have distinct structural and enzymatic functions, known as gene‐sharing.52 It is a possibility that catalytically active PANK4s from plants, fungi, and other metazoans also have a regulatory or structural role.
The best‐described pseudoenzymes typically originate from a gene duplication of the parent enzyme, and subsequent evolution of one copy into a pseudoenzyme.35 The parent enzyme is usually an enzyme involved in signaling, and the pseudoenzyme is often thought to regulate the same pathway of the parent enzyme because it retains the same tissue and cellular expression pattern.35 In contrast, HsPANK4 is a kinase involved in metabolism rather than signaling, and the evolution into a pseudoenzyme occurred in the original, single copy of the gene. Our data show that the Clade 2 monofunctional PANK gene was duplicated in Gnathostomata (jawed vertebrates including amniote and anamniotes), raising the possibility that the duplication of the Clade 2 PANK may have made the PANK activity of PANK4 dispensable and enabled the evolution of PANK4 into a pseudoenzyme in amniote and anamniotes. It is interesting that the Keio collection of E. coli gene knockouts listed a coaA mutant that would have an inactivated PANK (http://cgsc2.biology.yale.edu/Mutation.php?ID=100356). However, the putative knockout strain is not available for distribution to investigators as it contains an intact coaA gene duplication that maintains PANK activity. This observation indicates that gene modifications such as duplication occur readily in response to selection pressures and may underly the positive genetic complementation in E. coli 19 that suggested HsPANK4 to be catalytically active. The Drosophila hypomorphic mutant called “fumble” (fbl) had global defects in cell division that became evident when maternally derived stores of mRNA and protein were depleted.20 We speculate that overexpression of HsPANK4 in the fumble mutant fly larvae perhaps stabilized a protein(s) involved in meiosis or mitosis, or extended the half‐life of the endogenous maternal PANK expression. The list of identified pseudoenzymes is expanding rapidly but the alternate functions, if any, are yet to be determined for most pseudoenzymes.
Experimental Procedures
Bioinformatic analysis
The HsPANK4 protein sequence was aligned against protein sequences in the NCBI databases using BLAST.53 Multiple sequence alignments were generated using ClustalW with an identity weight matrix.53 Protein domain homology was analyzed via NCBI's Conserved Domain Database. Structural analysis was carried out using PyMOL. Analysis of active site mutations was conducted through using the mutagenesis function in PyMOL.
Phylogeny
Predicted Type II PANK sequences were collected from the NCBI Reference Sequence Database and the Ensembl Genomic Database to contain representative sequences from all branches of the eukaryotic tree of life. Maximum likelihood phylogenetic trees were constructed using the DECIPHER and phangorn packages in R.25, 27 The final tree as well as the R script and protein sequences used to generate the phylogenetic tree are available on request. Briefly, sequences were aligned using the AlignSeqs and StaggerAlignment function in DECIPHER. The modelTest function in phangorn was used to evaluate which amino acid replacement matrix (from WAG, JTT, LG, and Dayhoff models with and without gamma distributed rate variation among sites [Γ] and invariant sites [I]) is the best fit model. The LG + Γ (4) + I model26 was best fitting by the Bayesian information criteria. The phylogenetic tree was constructed following standard protocol using phangorn. The starting neighbor‐joining tree was constructed using the distance matrix. The maximum likelihood tree was generated from the starting tree using the LG + Γ (4) + I model with stochastic branch rearrangement. The bootstrap method (1000 replicate trees) was used to determine the confidence of the tree topology. The maximum likelihood tree was saved with the bootstrap percentage in the Newick format and visualized using Figtree 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) and ggtree.54
Preparation of kinase expression vectors
The HsPANK3 cDNA was described earlier.55 The PANK3 with the His tag was cut from pET28a‐PANK3 with BsrBI‐XhoI and ligated into EcoRV‐XhoI‐digested pcDNA3.1 to obtain pPJ569. The full‐length HsPANK4 (pKM29) construct was used as described previously.13 To obtain the HsPANK4 kinase domain, HsPANK4 residues 64–1117 bp codon optimized for E. coli and containing an amino‐terminal 6x‐histidine tag were ordered as a gene string and cloned into the NdeI‐EcoRI site of pcDNA3.1 to obtain pPJ576. The kinase domains were identified by their homology to HsPANK3 and began at the amino terminus of each protein and ended just before the low complexity region (GSSGL) that marks the transition between the kinase and phosphatase domains. A gene string for AtPANK4 was obtained and TOPO‐cloned into pCR2.1 blunted vector. Residues 1–1467 were PCR amplified and cloned into BamHI‐NotI sites of pcDNA3.1‐His to obtain pPJ591. NpPANK4 (1–1140 bp) and CmPANK4 (1–1149 bp) were obtained as gene strings and cloned into BamHI‐NotI sites of pcDNA3.1‐His to obtain pPJ525 and pPJ524, respectively. Gene strings were purchased from GeneArt and their sequences verified. The corresponding E to V and R to W mutations were generated by mutagenizing the parent plasmids. The plasmids used in this study are listed in Table 1.
Table 1.
Plasmid list
| Plasmid | Description | Source |
|---|---|---|
| pPJ569 | pCDNA3.1‐PANK3 | This study |
| pKM29 | pCDNA3.1‐His HsPANK4‐FL | 13 |
| pPJ576 | pCDNA3.1‐His HsPANK4‐KD | This study |
| pPJ591 | pCDNA3.1‐His AtPANK4‐KD | This study |
| pPJ524 | pCDNA3.1‐His CmPANK4‐KD | This study |
| pPJ525 | pCDNA3.1‐His NpPANK4‐KD | This study |
| pPJ570 | pCDNA3.1‐PANK3 E138V | This study |
| pPJ571 | pCDNA3.1‐PANK3 R207W | This study |
| pPJ577 | pCDNA3.1‐His HsPANK4‐KD V155E | This study |
| pPJ578 | pCDNA3.1‐His HsPANK4‐KD W219R | This study |
| pPJ579 | pCDNA3.1‐His HsPANK4‐KD V155EW219R | This study |
| pJW9 | pCDNA3.1‐His AtPANK4‐KD E201V | This study |
| pJW11 | pCDNA3.1‐His CmPANK4‐KD V146E | This study |
Human PANK3 kinetic analysis
The pET28a‐PANK3(R207W) construct was generated through mutagenesis of the previously described pET28a‐PANK3 parent plasmid.11 The construct was expressed in BL21(DE3) cells, with overnight induction (1 mM IPTG) at 16°C. The protein was purified using standard nickel chelation chromatography. The fractions containing protein were collected and dialyzed against 20 mM Tris, pH 8.0, 10 mM EDTA, and 150 mM NaCl at 4°C overnight. A pure protein (>95%) running at ∼41 kDa (theoretical average mass of 41.1 kDa with an amino‐terminal methionine) was observed on a NuPAGE 10% bis‐Tris gel (Life Technologies, Inc.). Final yield of purified protein was approximately 3 mg/L culture. HsPANK3 was purified as previously described.11 Enzyme kinetic and thermal stabilization analyses of the wild type and mutant PANK3 were performed as previously described.11
The lysates were tested for PANK activity as described earlier.56 Briefly the HEK 293T cells were transfected for 48 hours with the plasmids expressing the genes of interest. The frozen pellets were resuspended in lysis buffer; 20 mM Tris–HCl (pH 7.5), 2 mM EDTA, 2 mM DTT, 0.1% Triton X‐100, and protease inhibitor and lysed by sonication. Lysates were added to a reaction mixture containing 100 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 2.5 mM ATP, and 45 μM [14C]pantothenate. The reaction was incubated for 10 minutes and 37°C and spotted on DE81 disks, which were washed three times in 95% ethanol containing 1% acetic acid. The radioactive phosphopantothenate was quantified by scintillation counting. For testing acetyl‐CoA inhibition, the concentration of ATP was 0.5 mM and acetyl‐CoA was titrated from 0 to 200 μM. All assays were done in duplicate.
HEK 293T cell expression
The HsPANK3 full‐length gene, the HsPANK4 full‐length gene, the HsPANK4 amino‐terminal PANK domain, the N. parkeri (frog) amino‐terminal PANK domain, C. mydas (turtle) amino‐terminal PANK domain, and the A. thaliana (plant) PANK4 amino‐terminal PANK domain were cloned into pcDNA3.1(+) and used in the transfection experiments. The corresponding E138V and R207W mutations were generated by mutagenizing the parent plasmids. The HEK 293T cells at 40% confluence were transfected with the respective pcDNA expression constructs using the FuGENE 6 Transfection Reagent (Promega) following manufacturer instructions (12 μg plasmid per 100‐mm dish). Cells were grown for 40 hours and harvested. The total CoA levels were determined using monobromobimane derivatization and HPLC analysis as previously described.57
The expression of the constructs was checked via Western blot analysis using either a mouse monoclonal anti‐polyhistidine‐alkaline phosphatase antibody (Sigma, A5588) or a rabbit polyclonal, affinity purified anti‐PANK antibody58 coupled with goat anti‐rabbit IgG‐alkaline phosphatase from Sigma (A3687) and exposed using the ECF substrate (GE Healthcare). The anti‐PANK antibody recognized all human PANK isoforms and AtPANK4. The anti‐PANK antibody was validated by immunoblotting serial dilutions of the purified antibody preparations against purified recombinant mouse PanK1α, PanK1β, PanK2, and PanK3 proteins.58 The CmPANK4 and the NpPANK4 were blotted with anti‐His probe. All the experiments were done in triplicate.
Conflict of Interest
The authors declare that they have no conflict of interest with the contents of this article.
Supporting information
Figure S1.
Acknowledgments
We thank Pam Jackson, Jina Wang, and Matthew Frank for their expert technical assistance.
References
- 1. Leonardi R, Zhang Y‐M, Rock CO, Jackowski S (2005) Coenzyme A: Back in action. Prog Lipid Res 44:125–153. [DOI] [PubMed] [Google Scholar]
- 2. Brand LA, Strauss E (2005) Characterization of a new pantothenate kinase isoform from Helicobacter pylori . J Biol Chem 280:20185–20188. [DOI] [PubMed] [Google Scholar]
- 3. Leonardi R, Chohnan S, Zhang Y‐M, Virga KG, Lee RE, Rock CO, Jackowski S (2005) A pantothenate kinase from Staphylococcus aureus refractory to feedback regulation by coenzyme A. J Biol Chem 280:3314–3322. [DOI] [PubMed] [Google Scholar]
- 4. Yun M, Park C‐G, Kim J‐Y, Rock CO, Jackowski S, Park H‐W (2000) Structural basis for the feedback regulation of Escherichia coli pantothenate kinase by coenzyme A. J Biol Chem 275:28093–28099. [DOI] [PubMed] [Google Scholar]
- 5. Hong BS, Yun MK, Zhang Y‐M, Chohnan S, Rock CO, White SW, Jackowski S, Park HW, Leonardi R (2006) Prokaryotic type II and type III pantothenate kinases: the same monomer fold creates dimers with distinct catalytic properties. Structure 14:1251–1261. [DOI] [PubMed] [Google Scholar]
- 6. Dansie LE, Reeves S, Miller K, Zano SP, Frank M, Pate C, Wang J, Jackowski S (2014) Physiological roles of the pantothenate kinases. Biochem Soc Trans 42:1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alfonso‐Pecchio A, Garcia M, Leonardi R, Jackowski S (2012) Compartmentalization of mammalian pantothenate kinases. PLoS One 7:e49509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rock CO, Calder RB, Karim MA, Jackowski S (2000) Pantothenate kinase regulation of the intracellular concentration of coenzyme A. J Biol Chem 275:1377–1383. [DOI] [PubMed] [Google Scholar]
- 9. Rock CO, Karim MA, Zhang Y‐M, Jackowski S (2002) The murine Pank1 gene encodes two differentially regulated pantothenate kinase isozymes. Gene 291:35–43. [DOI] [PubMed] [Google Scholar]
- 10. Hong BS, Senisterra G, Rabeh WM, Vedadi M, Leonardi R, Zhang YM, Rock CO, Jackowski S, Park HW (2007) Crystal structures of human pantothenate kinases. Insights into allosteric regulation and mutations linked to a neurodegeneration disorder. J Biol Chem 282:27984–27993. [DOI] [PubMed] [Google Scholar]
- 11. Subramanian C, Yun MK, Yao J, Sharma LK, Lee RE, White SW, Jackowski S, Rock CO (2016) Allosteric regulation of mammalian pantothenate kinase. J Biol Chem 291:22302–22314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leonardi R, Rehg JE, Rock CO, Jackowski S (2010) Pantothenate kinase 1 is required to support the metabolic transition from the fed to the fasted state. PLoS One 5:e11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang YM, Chohnan S, Virga KG, Stevens RD, Ilkayeva OR, Wenner BR, Bain JR, Newgard CB, Lee RE, Rock CO, Jackowski S (2007) Chemical knockout of pantothenate kinase reveals the metabolic and genetic program responsible for hepatic coenzyme A homeostasis. Chem Biol 14:291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garcia M, Leonardi R, Zhang YM, Rehg JE, Jackowski S (2012) Germline deletion of pantothenate kinases 1 and 2 reveals the key roles for CoA in postnatal metabolism. PLoS One 7:e40871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou B, Westaway SK, Levinson B, Johnson MA, Gitschier J, Hayflick SJ (2001) A novel pantothenate kinase gene (PANK2) is defective in Hallervorden‐Spatz syndrome. Nat Genet 28:345–349. [DOI] [PubMed] [Google Scholar]
- 16. Taylor TD, Litt M, Kramer P, Pandolfo M, Angelini L, Nardocci N, Davis S, Pineda M, Hattori H, Flett PJ, Cilio MR, Bertini E, Hayflick SJ (1996) Homozygosity mapping of Hallervorden‐Spatz syndrome to chromosome 20p12.3‐p13. Nat Genet 14:479–481. [DOI] [PubMed] [Google Scholar]
- 17. Huang L, Khusnutdinova A, Nocek B, Brown G, Xu X, Cui H, Petit P, Flick R, Zallot R, Balmant K, Ziemak MJ, Shanklin J, de Crecy‐Lagard V, Fiehn O, Gregory JF 3rd, Joachimiak A, Savchenko A, Yakunin AF, Hanson AD (2016) A family of metal‐dependent phosphatases implicated in metabolite damage‐control. Nat Chem Biol 12:621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tilton GB, Wedemeyer WJ, Browse J, Ohlrogge J (2006) Plant coenzyme A biosynthesis: characterization of two pantothenate kinases from Arabidopsis . Plant Mol Biol 61:629–642. [DOI] [PubMed] [Google Scholar]
- 19. Hörtnagel K, Prokisch H, Meitinger T (2003) An isoform of hPANK2, deficient in pantothenate kinase‐associated neurodegeneration, localizes to mitochondria. Hum Mol Genet 12:321–327. [DOI] [PubMed] [Google Scholar]
- 20. Wu Z, Li C, Lv S, Zhou B (2009) Pantothenate kinase‐associated neurodegeneration: insights from a Drosophila model. Hum Mol Genet 18:3659–3672. [DOI] [PubMed] [Google Scholar]
- 21. Byrne DP, Foulkes DM, Eyers PA (2017) Pseudokinases: update on their functions and evaluation as new drug targets. Future Med Chem 9:245–265. [DOI] [PubMed] [Google Scholar]
- 22. Reiterer V, Eyers PA, Farhan H (2014) Day of the dead: pseudokinases and pseudophosphatases in physiology and disease. Trends Cell Biol 24:489–505. [DOI] [PubMed] [Google Scholar]
- 23. Boudeau J, Miranda‐Saavedra D, Barton GJ, Alessi DR (2006) Emerging roles of pseudokinases. Trends Cell Biol 16:443–452. [DOI] [PubMed] [Google Scholar]
- 24. Kung JE, Jura N (2016) Structural basis for the non‐catalytic functions of protein kinases. Structure 24:7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schliep KP (2011) Phangorn: phylogenetic analysis in R. Bioinformatics 27:592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. [DOI] [PubMed] [Google Scholar]
- 27. Wright ES (2015) DECIPHER: harnessing local sequence context to improve protein multiple sequence alignment. BMC Bioinf 16:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sieber KB, Bromley RE, Dunning Hotopp JC (2017) Lateral gene transfer between prokaryotes and eukaryotes. Exp Cell Res 358:421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Choi J, Kim SH (2017) A genome tree of life for the fungi kingdom. Proc Natl Acad Sci U S A 114:9391–9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Calder RB, Williams RSB, Ramaswamy G, Rock CO, Campbell E, Unkles SE, Kinghorn JR, Jackowski S (1999) Cloning and characterization of a eukaryotic pantothenate kinase gene (panK) from Aspergillus nidulans . J Biol Chem 274:2014–2020. [DOI] [PubMed] [Google Scholar]
- 31. Palmer JD, Soltis DE, Chase MW (2004) The plant tree of life: an overview and some points of view. Am J Bot 91:1437–1445. [DOI] [PubMed] [Google Scholar]
- 32. Burki F (2014) The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb Perspect Biol 6:a016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y‐M, Rock CO, Jackowski S (2005) Feedback regulation of murine pantothenate kinase 3 by coenzyme A and coenzyme A thioesters. J Biol Chem 280:32594–32601. [DOI] [PubMed] [Google Scholar]
- 34. Warde‐Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38:W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adrain C, Freeman M (2012) New lives for old: evolution of pseudoenzyme function illustrated by iRhoms. Nat Rev Mol Cell Biol 13:489–498. [DOI] [PubMed] [Google Scholar]
- 36. Starr TN, Thornton JW (2016) Epistasis in protein evolution. Protein Sci 25:1204–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vallari DS, Rock CO (1987) Isolation and characterization of temperature‐sensitive pantothenate kinase (coaA) mutants of Escherichia coli . J Bacteriol 169:5795–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jackowski S, Rock CO (1981) Regulation of coenzyme A biosynthesis. J Bacteriol 148:926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eyers PA, Murphy JM (2016) The evolving world of pseudoenzymes: proteins, prejudice and zombies. BMC Biol 14:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Murphy JM, Farhan H, Eyers PA (2017) Bio‐Zombie: the rise of pseudoenzymes in biology. Biochem Soc Trans 45:537–544. [DOI] [PubMed] [Google Scholar]
- 41. Pils B, Schultz J (2004) Inactive enzyme‐homologues find new function in regulatory processes. J Mol Biol 340:399–404. [DOI] [PubMed] [Google Scholar]
- 42. Xiang RL, Yang YL, Zuo J, Xiao XH, Chang YS, De Fang F (2007) PanK4 inhibits pancreatic beta‐cell apoptosis by decreasing the transcriptional level of pro‐caspase‐9. Cell Res 17:966–968. [DOI] [PubMed] [Google Scholar]
- 43. Li Y, Chang Y, Zhang L, Feng Q, Liu Z, Zhang Y, Zuo J, Meng Y, Fang F (2005) High glucose upregulates pantothenate kinase 4 (PanK4) and thus affects M2‐type pyruvate kinase (Pkm2). Mol Cell Biochem 277:117–125. [DOI] [PubMed] [Google Scholar]
- 44. Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y (2008) Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia‐Sastre A, Young JA, Palese P, Shaw ML, Chanda SK (2010) Human host factors required for influenza virus replication. Nature 463:813–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shapira SD, Gat‐Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N (2009) A physical and regulatory map of host‐influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bakre A, Andersen LE, Meliopoulos V, Coleman K, Yan X, Brooks P, Crabtree J, Tompkins SM, Tripp RA (2013) Identification of host kinase genes required for influenza virus replication and the regulatory role of microRNAs. PLoS One 8:e66796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trzcinska‐Daneluti AM, Chen A, Nguyen L, Murchie R, Jiang C, Moffat J, Pelletier L, Rotin D (2015) RNA interference screen to identify kinases that suppress rescue of ΔF508‐CFTR. Mol Cell Proteomics 14:1569–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu F, Zhang YN, Cheng DH, Tan K, Zhong CG, Lu GX, Lin G, Tan YQ (2014) The first patient with a pure 1p36 microtriplication associated with severe clinical phenotypes. Mol Cytogenet 7:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun M, Chen C, Hou S, Li X, Wang H, Zhou J, Chen X, Liu P, Kijlstra A, Lin S, Ye J (2019) A novel mutation of PANK4 causes autosomal dominant congenital posterior cataract. Hum Mutat 40:380–391. [DOI] [PubMed] [Google Scholar]
- 51. Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, Hyman AA, Mann M (2015) A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163:712–723. [DOI] [PubMed] [Google Scholar]
- 52. Piatigorsky J, O'Brien WE, Norman BL, Kalumuck K, Wistow GJ, Borras T, Nickerson JM, Wawrousek EF (1988) Gene sharing by delta‐crystallin and argininosuccinate lyase. Proc Natl Acad Sci U S A 85:3479–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hung JH, Weng Z (2016) Sequence alignment and homology search. Cold Spring Harb Protoc 2016:937–940. [DOI] [PubMed] [Google Scholar]
- 54. Yu GC, Smith DK, Zhu HC, Guan Y, Lam TTY (2017) GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. [Google Scholar]
- 55. Leonardi R, Zhang Y‐M, Yun M‐K, Zhou R, Zeng F‐Y, Lin W, Cui J, Chen T, Rock CO, White SW, Jackowski S (2010) Modulation of pantothenate kinase 3 activity by small molecules that interact with the substrate/allosteric regulatory domain. Chem Biol Drug Des 17:892–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang Y‐M, Rock CO, Jackowski S (2006) Biochemical properties of human pantothenate kinase 2 isoforms and mutations linked to pantothenate kinase‐associated neurodegeneration. J Biol Chem 281:107–114. [DOI] [PubMed] [Google Scholar]
- 57. Zano SP, Pate C, Frank M, Rock CO, Jackowski S (2015) Correction of a genetic deficiency in pantothenate kinase 1 using phosphopantothenate replacement therapy. Mol Genet Metab 116:281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sharma LK, Subramainian C, Yun M, Frank MW, White SW, Rock CO, Lee RE, Jackowski S (2018) A therapeutic approach to pantothenate kinase associated neurodegeneration. Nat Commun 9:4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.


