Abstract
The post-replicative mismatch repair (MMR) system has anti-recombination activity that limits interactions between diverged sequences by recognizing mismatches in strand-exchange intermediates. In contrast to their equivalent roles during replication-error repair, mismatch recognition is more important for anti-recombination than subsequent mismatch processing. To obtain insight into this difference, ectopic substrates with 2% sequence divergence were used to examine mitotic recombination outcome (crossover or noncrossover; CO and NCO, respectively) and to infer molecular intermediates formed during double-strand break repair in Saccharomyces cerevisiae. Experiments were performed in an MMR-proficient strain, a strain with compromised mismatch-recognition activity (msh6Δ) and a strain that retained mismatch-recognition activity but was unable to process mismatches (mlh1Δ). While the loss of either mismatch binding or processing elevated the NCO frequency to a similar extent, CO events increased only when mismatch binding was compromised. The molecular features of NCOs, however, were altered in fundamentally different ways depending on whether mismatch binding or processing was eliminated. These data suggest a model in which mismatch recognition reverses strand-exchange intermediates prior to the initiation of end extension, while subsequent mismatch processing that is linked to end extension specifically destroys NCO intermediates that contain conflicting strand-discrimination signals for mismatch removal.
INTRODUCTION
Double-strand breaks (DSBs) are among the most lethal types of mitotic DNA damage in eukaryotes and are repaired either by homologous recombination (HR) or nonhomologous end joining (NHEJ). Because NHEJ can cause sequence loss at DSB sites or join ends created by different DSBs, it is generally considered to be a relatively error-prone repair pathway. By contrast, HR copies information from a homologous duplex DNA and is often of no genetic consequence. Although a sister chromatid is the preferred repair template when available (1,2), HR also can involve a homologous chromosome and lead to loss of heterozygosity. Finally, because recombination is a homology-driven process, non-allelic or ectopic interactions can occur between dispersed repeats. Such interactions can either create new hybrid genes or lead to various types of clinically relevant genome rearrangements that include inversions, duplications, deletions and translocations (3). It thus is of critical importance that an appropriate template is used and ectopic interactions are limited when HR occurs.
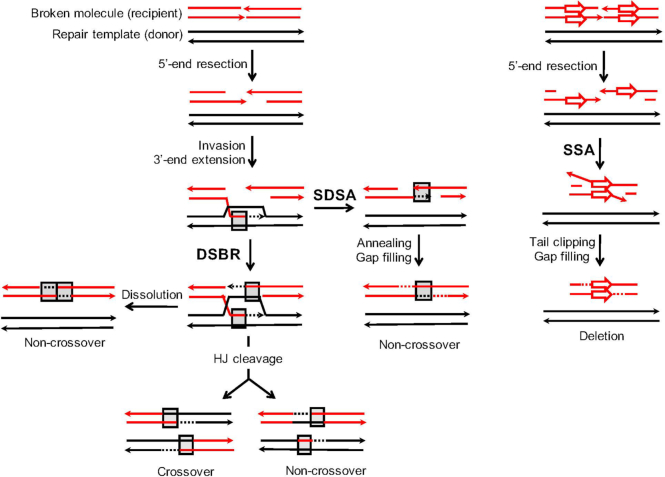
Current recombination models are based primarily on studies done in Saccharomyces cerevisiae (4), and yeast is used here to examine the regulation of ectopic interactions. Commitment to HR begins with resection of the DSB ends to generate long 3′ tails (5). A key step during HR is the invasion of duplex DNA by a 3′ end, which pairs with the complementary strand and displaces the homologous strand as a displacement (D)-loop. In the canonical DSB repair (DSBR; left side of Figure 1) model, the invading end is extended by a DNA polymerase, which enlarges the D-loop until annealing to the complementary 3′ tail on the other side of the initiating break occurs. Alternatively, both ends can independently invade the donor. Regions of pairing between single strands derived from different duplexes are referred to as heteroduplex DNA (hetDNA) and are indicated by gray boxes in Figure 1. Following the filling of gaps and ligation of ends, two Holliday junctions are formed that can be resolved by cleavage into either crossover or noncrossover products (COs and NCOs, respectively). In the DSBR model, there is a region of hetDNA on each side of the initiating break, one of which is present in each cleavage product. As an alternative to Holliday junction cleavage, the junctions can be dissolved by migrating towards each other. In this case, hetDNA is expected on each side of the break in the repaired duplex and the donor remains unaltered.
Figure 1.
Mechanisms of DSB repair. Shown on the left are models that require invasion of a donor repair template (black lines) by a resection-generated 3′ tail of the broken molecule (red lines; arrowheads represent 3′ ends). This creates a D-loop that is extended by DNA synthesis (dotted lines that are the same color as the template strand). The canonical DSB (DSBR) repair model requires that both broken ends engage the donor (gray boxes indicate hetDNA created by pairing between black and red strands) and a double Holliday junction (HJ) is generated. Following HJ cleavage, each product contains a single, break-adjacent hetDNA tract. Although HJ cleavage is assumed to create equal numbers of CO and NCO products, cleavage-generated NCOs are rare in the current system. HJ dissolution gives rise to only NCOs and hetDNA is confined to the recipient; a similar pattern is produced by a double SDSA event. If the D-loop collapses prior to interaction with the second end of the DSB, the extended end pairs with the 3′ tail on the other side of break. The resulting synthesis-dependent strand annealing (SDSA) product contains a single hetDNA tract on the annealing side of break, which corresponds to DNA synthesized prior to D-loop collapse. Single-strand annealing (SSA) is illustrated on the right side and occurs between direct repeats. Resection of the broken ends uncovers complementary strands of the repeats that then anneal to each other. Clipping of the 3′ tails removes the region between the repeats, resulting in its deletion and retention of a single copy of the repeat unit.
In addition to annealing to the second end of the break, a D-loop can be dismantled by helicases (6–8) or dissolved by Sgs1-Top3-Rmi1 (9), which releases the extended end. This end can then pair with the 3′ tail on the other side of the DSB in a process referred to as synthesis-dependent strand annealing (SDSA). SDSA results only in NCOs that are characterized by a single hetDNA tract on the annealing side of the break; the donor molecule is not altered. A third pathway that initiates with a strand invasion is break-induced replication (not shown). In this pathway, only a single end engages the donor and DNA synthesis continues to the end of the repair template via a migrating D-loop, resulting in a half-CO or non-reciprocal translocation product (10). In addition to the HR pathways that initiate with invasion of duplex DNA, there is an invasion-independent pathway that deletes the region between direct repeats and is referred to as single-strand annealing (SSA; right side of Figure 1). During SSA, the resection of broken ends uncovers complementary strands of the repeats, which then anneal to each other. The annealed region is flanked by 3′ tails that must be removed before gap filling and ligation complete the process (11).
There are two features of repeated sequences that limit mitotic interactions in yeast: the total length of homology (12–14) and the degree of sequence identity (14,15). Seminal studies in bacteria demonstrated that the identity barrier derives from anti-recombination activity of the post-replicative mismatch repair (MMR) system (16), which recognizes the mismatches created when strands from nonidentical duplexes pair. Similar anti-recombination activity of MMR has been documented in yeast (17–20), flies (21), plants (22–24) and mammals (25,26). Although the current study focuses on mitotic recombination, the MMR system is also a potent barrier to meiotic recombination between the diverged chromosomes of different yeast species (27).
There are two core components of the MMR system in all organisms: a MutS complex that recognizes and binds to mismatches and a MutL complex that is required for subsequent mismatch processing (28). In yeast, the major MutS complex (MutSα) is a heterodimer of Msh2 and Msh6 while the major MutL complex (MutLα) is a heterodimer of Pms1 and Mlh1. Whereas the effect of MutSα and MutLα loss is generally indistinguishable in mutation assays that measure base substitutions (29), the effect of MutLα loss on anti-recombination is variable and system specific. MutLα, for example, has anti-recombination activity equivalent to that of MutSα in a plasmid-chromosome assay (30) but exhibits less activity than MutSα in an inverted-repeat assay (19). In the specific case of SSA, MutLα has little, if any, effect on recombination fidelity (20). Although it has no known role during the repair of replication errors, the Sgs1 helicase forms a complex with MutSα (31) and is important during anti-recombination in yeast (20,32,33). While there is general agreement that mismatch-containing hetDNA can be dismantled by Sgs1 helicase activity (hetDNA rejection), the role of MutLα is less clear. In those events that escape anti-recombination, MMR can subsequently correct mismatches within hetDNA to generate gene conversion events. The choice between anti-recombination and mismatch correction, however, is not well understood (31). For mutation avoidance during replication, it is critical that the nascent DNA strand that contains the mistake is targeted for removal and re-synthesis. Strand discrimination is largely provided by PCNA, which tethers DNA polymerase to a growing 3′ end and has an inherent asymmetry that directs strand cleavage by MutLα (34,35). During recombination, analogous strand discrimination removes information from the broken strand rather than from the repair template, resulting in gene conversion (31,36).
In the current study, ectopic recombination substrates with 2% sequence divergence were used to examine the anti-recombination effects of MutSα and MutLα (msh6Δ and mlh1Δ mutants, respectively) during the repair of a defined chromosomal DSB. An analysis of NCO and CO frequencies coupled with characterization of donor sequence transferred to NCO products suggests distinct anti-recombination activities of MutSα and MutLα. While the anti-recombination activity of MutSα primarily affected repair events that require donor engagement by both ends of the DSB, that of MutLα affected only SDSA events. We suggest a two-stage model in which mismatch binding by MutSα leads to the dismantling of hetDNA intermediates prior to 3′-end extension, with subsequent processing by MutLα specifically destroying SDSA intermediates after DNA synthesis initiates.
MATERIALS AND METHODS
Yeast strain constructions
A complete list of yeast strains is provided in Supplementary Table S1. All haploid strains were derived from SJR3659 and SJR3782 [strains MC42-2d and HKL1-42-1c, respectively, in (37)], both of which are RAD5 CAN1 derivatives of W303 [rad5-535 leu2-3,112 his3-11,15 ura3-1 ade2-1 trp1-1 can1-100; (38)]. The recombination system in experimental strains contained three components: an I-SceI-cleavable lys2 recipient allele (hisG-lys2-6Amut::ISceI allele) at the endogenous LYS2 locus on chromosome II; a galactose-inducible I-SceI gene (his3Δ::hph-pGAL-ISceI) at the HIS3 locus on chromosome XV; and a diverged or identical lys2Δ3′-donor allele at the CAN1 locus on chromosome V. The diverged donor allele contained a mutated (non-cleavable) I-SceI site and shared 98% sequence identity with the recipient allele (can1::lys2Δ3′-98%,ISceInc-URA3-hisG). Strains with the 98%-identical donor were used to determine recombination frequencies, CO/NCO proportions, and hetDNA profiles. Strains containing a 100%-identical donor allele (can1::lys2Δ3′-100%,ISceInc-URA3-hisG) were used to determine recombination frequencies and CO/NCO proportions.
The hisG-lys2-6Amut::I-SceI recipient allele (abbreviated hisG/recipient) was a derivative of the recipient allele used in an earlier study [strain SJR 3714; (39)]. First, a 6A run near the I-SceI cleavage site was mutated (Supplementary Figure S1) to reduce the occurrence of polymerase slippage events in mismatch repair (MMR)-defective backgrounds. To introduce this modification, a CORE-UK cassette (40) was inserted close to the I-SceI cut site of SJR3714, yielding SJR4015. Replacement of the CORE-UK cassette converted the 6A run to AATCAA (SJR4194). Next, a copy of the bacterial hisG gene was inserted immediately upstream of the recipient allele to allow phenotypic distinction between CO and NCO products. To insert the hisG allele, SJR4194 was transformed with a PCR-amplified hisG-URA3-hisG fragment from pNKY51 (41). Following selection of Ura+ transformants, segregants that lost the URA3 marker during subsequent non-selective growth were identified on 5-FOA medium (SJR4195).
The sequence of the donor component of can1::lys2Δ3′-98%,I-SceInc-URA3-hisG donor allele (hereafter referred to as 98% donor/hisG) was the same as that used in a previous study (39). The allele was modified, however, by inserting a URA3-hisG cassette amplified from pNKY51 (41) immediately downstream of the donor allele. The resulting strain (SJR4031) was crossed with SJR4195 and a haploid segregant with both the recipient and 98% donor alleles was obtained (SJR4196). Finally, a his3Δ::hph-pGAL-ISceI (abbreviated as I-SceI) fragment amplified from pGSHU (40) was transformed into SJR4196, thereby creating a strain with all three components of the 98% recombination system (SJR4258). To preserve the hetDNA footprint in Lys+ colonies MLH1 (SJR4535), MSH6 (SJR4691) or both MLH1 and MSH6 (SJR4982) were deleted from SJR4258. MLH1 was replaced with an mlh1Δ:loxP-TRP1-loxP fragment amplified from pSR954, a loxP-TRP1-loxP plasmid constructed by replacing the kanamycin-resistance marker of pUG6 (42) with a TRP1 marker from pFA6-TRP1 (43). The MSH6 gene was replaced with an msh6Δ::loxP-kanMX-loxP fragment amplified from pUG6.
The first step in construction of the can1::lys2Δ3′-100%,I-SceInc-URA3-hisG (100% donor/hisG) allele was the PCR-mediated assembly of a fragment containing a mutated 6A run and a noncleavable I-SceI cut site using high-fidelity Phusion polymerase (New England BioLabs) and the recipient allele in SJR3714 as a template. The mutated 6A run and noncleavable I-SceI site were introduced using primers 5′-GGGCGCTCCCATTCGCCATTCAGGCTGCGCTTTGGAAAAGGCAAAACTATCGAAGG and 5′ ACAACTGAGGGGTCCTTTCCAGATCTAGGGATAAATAACAGGGTAATTGATCTTGGCAACTTTGAATTGATTCAGCTAGC, generating an 2.3 kb fragment containing the first half of the eventual donor allele. A 2.0 kb amplicon containing the downstream portion of the incipient donor allele was generated using primers 5′-CCCTAGATCTGGAAAGGACCCCTCAGTTGT and 5′-CTGGAAAGCGGGCAGTGAAAGGAAGGCCCATGAGGCCCAGATCTTTTCTTTAAGAAAGTC. The amplicons shared 30 bp of sequence overlap and were assembled into a single fragment by PCR. The resultant amplicon contained a 100% lys2Δ3′ donor allele that was then inserted into the MscI-digested pSR797 (39), which contains nucleotides 20-1141 of the CAN1 sequence. The resulting plasmid (pSR1154) was digested with PshAI-XcmI and used to transform SJR4195. Selection for canavanine resistance yielded SJR5000, which contained both the recipient and 100%-identical donor alleles. A URA3-hisG cassette amplified from pNKY51 was then inserted downstream of the donor allele, generating SJR5003. SJR5003 was crossed with SJR4535 to obtain SJR5007, which contained the hisG/recipient and 100% donor/hisG alleles in an mlh1Δ background. SJR5007 was then crossed with SJR4691 to produce haploid segregants that contained the three recombination components (hisG/recipient, 100% donor/hisG, and galactose regulated I-SceI) in an MMR-proficient (SJR5036), mlh1Δ (SJR5037), msh6Δ (SJR5038), or mlh1Δ msh6Δ (SJR5039) background.
Media and growth conditions
Strains stored at −80°C were streaked onto YEPD (1% Bacto-yeast extract, 2% peptone, 2% dextrose; 1.5% agar for plates) supplemented with 500 μg/ml adenine hemisulfate. All growth was at 30°C. Cells were pre-grown in YEPR liquid medium (2% dextrose replaced with 2% raffinose) prior to addition of galactose. Following galactose induction, cells were plated nonselectively on YEPD medium to determine the total number of viable cells. Lys+ recombinants were selected on synthetic complete (SC) medium lacking lysine and containing 2% dextrose as a carbon source (SC-lys). 5-FOA medium (SC-uracil supplemented with 2% dextrose, 0.5 g uracil and 1 g 5-fluoroorotic acid per liter) was used to select Ura− segregants.
I-SceI induction and determination of recombination frequencies
Individual YEPR liquid cultures were inoculated with yeast colonies grown for two days on YEPD plates. Following overnight expansion, cells were diluted to an optical density (OD) of 0.2 in fresh YEPR medium. To ensure experiments were performed using exponentially growing cells, diluted cultures were re-grown to an OD of 0.8–1 before galactose addition (final concentration of 0.1%). As previously determined, induction with 0.1% galactose for 45 minutes generally resulted in cleavage of only one sister chromatid (39). Following I-SceI induction, appropriate dilutions were plated on YEPD and SC-lys plates. After 2 days of growth, recombination frequencies were calculated by dividing the total number of Lys+ colonies by the total number of viable cells. For each genetic background, recombination frequency was determined using data obtained using multiple biological replicates from each of at least three separate days. The average Lys+ frequency and 95% confidence interval were calculated for each strain and then normalized to the Lys+ frequency obtained with the 98% substrates in the WT background.
Determination of CO-NCO proportions and frequencies
The CO-NCO distribution among Lys+ colonies was determined using two procedures: stability of the URA3 marker downstream of the donor allele and PCR using primer pairs diagnostic of CO and NCO products. Lys+ colonies were directly inoculated (without colony purification) into SC-lys medium in 96-well plates. After overnight expansion, 3 μl were spotted onto YEPD medium and plates were incubated overnight. Cells were then replica plated onto SC-lys, SC-ura and 5-FOA media. COs were scored based on frequent loss (more than five colonies on 5-FOA medium) of the URA3 marker flanked by hisG direct repeats in the V:II translocation product. Genomic DNA was extracted from the remaining cells and aliquots were used for CO-NCO partitioning by PCR; the remainder was used for hetDNA analysis (see below). NCO-specific primers (5′-ATGGTTGGGAAGTCATGGAA and 5′-TTGGGAGTTGGGAATTGAAG) detected the full-length LYS2 allele on chromosome II; CO-specific primers (5′-ATGGTTGGGAAGTCATGGAA and 5′-TCACTTTTGCCCTGGAACTT) amplified the truncated allele of the V:II translocation product.
As expected, most replica-plated patches of cells were Lys+ Ura+ and produced either no or many colonies on 5-FOA plates. A small number of patches, however, had confluent growth on 5-FOA and were Ura− on SD-ura plates (Supplementary Figure S2A). This suggested complete loss of the lys2Δ3′ allele and URA3 marker on the V:II translocation product by a second recombination event between the flanking hisG repeats, which leaves behind a single copy of hisG. In addition to this anomaly, PCR analysis revealed in a small number of colonies the presence of the V:II translocation product, which carries the lys2Δ3′ allele, as well as a copy of chromosome II. Subsequent Southern analysis (Supplementary Figure S2B) revealed that these colonies were of two types: (1) those containing two normal plus two translocation chromosomes and (2) those containing only a normal chromosome II and the V:II translocation. These can be explained by cleavage of both chromosome II sister chromatids by I-SceI, with one break repaired as an NCO and the other as a CO (Supplementary Figure S3). The breakage of both sisters could reflect either replication of a chromosome cleaved by I-SceI in G1 (44), or the cleavage of both chromatids following replication. Random segregation of the resulting chromatids has two predicted outcomes. If the reciprocal translocation products segregate into the same daughter cell, then half of the Lys+ colony is expected to contain CO chromosomes and the other half to contain NCO chromosomes. If the translocation products segregate into different cells, however, one cell will contain chromosomes V and II:V and the other will contain chromosomes II and V:II. The former will be inviable due to the loss of essential information from chromosome II, but the latter will survive because the CAN1-distal end of chromosome V is not required for viability (45). Because the CO-specific V:II translocation contains the lys2 truncation allele, a NCO-generated LYS2 allele residing on chromosome II is required to confer the Lys+ phenotype.
There were four types of Lys+ colonies based on phenotypic and physical criteria: (i) colonies that reflected an NCO event, (ii) colonies containing only CO chromosomes, (iii) colonies containing both CO and NCO chromosomes and (iv) a CO event followed by additional recombination between the CO-generated hisG direct repeats. When crossing over occurs between nonhomologous chromosomes in a haploid background, random chromosome segregation predicts that only half of the events will be recoverable as Lys+ colonies (46). We thus doubled the sum of the CO-only and Ura− colonies to obtain a partially corrected number of CO events. Those colonies that contained both CO and NCO products and hence had two independent events, were then added to both the NCO and CO-only classes (all COs were recovered in this case) to obtain the final numbers of NCO and CO events. The raw and corrected numbers as well as the corrected proportions of NCO and CO events are in Supplementary Table S2. In the main text, the corrected proportions are given. NCO and CO frequencies were calculated by multiplying the corrected proportions and the corresponding total Lys+ frequency.
Southern analysis
Representative colonies corresponding to CO and/or NCO outcomes based on phenotype and PCR analysis were analyzed by Southern blots. Cells was grown in 5 ml YEPD medium overnight prior to genomic DNA extraction. Approximately 2 μg of genomic DNA were digested with XhoI and fragments were separated on a 1% agarose gel run at 40 volts at 4°C for 15–17 h. Following electrophoresis, fragments were denatured and transferred to a positively charged nylon membrane (Roche) via capillary action. The membrane was then hybridized to ∼600 bp LYS2 probes labeled with digoxigenin (DIG)-dUTP using primers 5′-TGAAGCCTTCCCAGAGAGAA and 5′-GCCAAGGAAAAATGTCTACCA (Roche DIG DNA Labeling and Detection Kit). Due to the presence of SNPs between the donor and recipient alleles, two probes were synthesized using pSR1045 and pSR1072 [lys2::ISceI recipient and lys2::I-SceInc,98% donor alleles, respectively; (39)] as templates and were mixed in equal proportions for the hybridization. Hybridization of the DIG probe to the membrane was detected by chemiluminescence using an anti-DIG antibody conjugated to alkaline phosphatase. XhoI-digested chromosomes II, V, II:V and V:II generate 10 kb, 14 kb, 9 kb, and 15 kb fragments, respectively.
hetDNA analysis
The ∼4 kb hetDNA profiles for NCOs recovered from MMR-deficient strains as well as the gene conversion profile for the MMR-proficient strain were obtained using single-molecule real-time (SMRT) sequencing as previously described (39,47). NCO recipient alleles were amplified from genomic DNA using Phusion polymerase (New England Biolabs). Forward (5′-ATGGTTGGGAAGTCATGGAAGTCG) and reverse (5′-GCTTGGGAGTTGGGAATTGAAGTT) primers were conjugated to 16-nt barcodes so that each NCO was amplified using a unique primer pair. The full list of unique barcodes used is available at https://github.com/PacificBiosciences/Bioinformatics-Training/blob/master/barcoding/pacbio_384_barcodes.fasta. ImageJ (https://imagej.nih.gov/ij/) was utilized to determine amplicon concentrations and equivalent amounts of individual amplicons were pooled and purified using the GeneJet PCR Purification Kit (Thermo Scientific). SMRT library construction and sequencing (PacBio RSII system) was done at the Duke Center for Genomic and Computational Biology.
PacBio-generated circular consensus sequence (CCS) reads in FASTA format were sorted by unique barcode pairs using a previously described pipeline (48). The donor and recipient reference sequences used for sequence alignment and hetDNA calling are in Supplementary Figure S1. hetDNA was defined as regions that contained both the recipient and donor SNPs, with the minor species constituting at least 10% of the total informative reads; only barcode pairs for which there were at least 20 CCS reads were further considered. In addition, those colonies that contained 3–4 distinct species of CCS reads were not included in the analyses, as these presumably represented two independent NCO events. For the MMR-defective strains, Lys+ colonies that lacked hetDNA were excluded. For the WT strain, Lys+ colonies with a gene conversion tract (39/268 colonies) that did not extend beyond the most break-proximal SNP [which most often reflects activity of the Pol δ exonuclease activity; (39)] were not considered when assigning tract directionality. Tract lengths were calculated as previously described (47).
Statistical analyses
Three types of statistical tests were used during data analysis. First, the frequency data in Figure 3A were analyzed using one-way ANOVA (P < 0.001) prior to performing post-hoc unpaired Student's t-tests between pairs of strains. Second, hetDNA length distributions were tested using the Kruskal–Wallis test (P < 0.01) prior to performing post-hoc Mann–Whitney U tests. Obtaining a significant value in an omnibus test (ANOVA or the Kruskal–Wallis test) before proceeding to pairwise comparisons eliminated the need to correct for multiple comparisons. Finally, the distributions of CO-NCO and unidirectional-bidirectional hetDNA tracts in NCOs were compared using contingency Chi-square tests and the significance level for p values was adjusted using the Bonferroni correction. In the case of CO-NCO distribution, five comparisons were done and this shifted the significance level from P = 0.05 to P = 0.01. For the unidirectional-bidirectional hetDNA distributions in NCOs, three comparisons were done and the P-value for significance was reduced from 0.05 to 0.017.
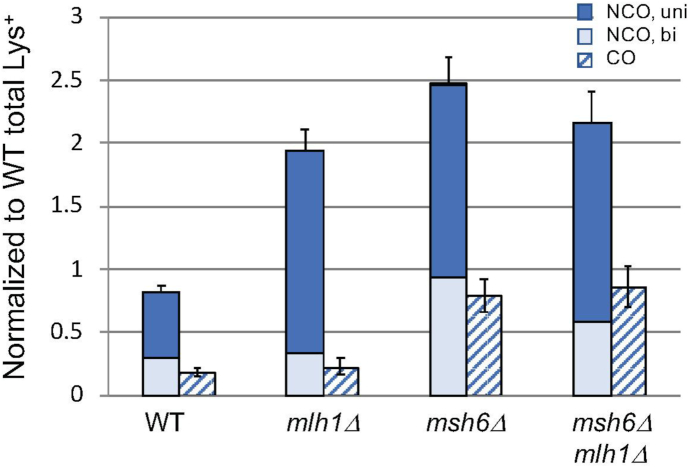
Figure 3.
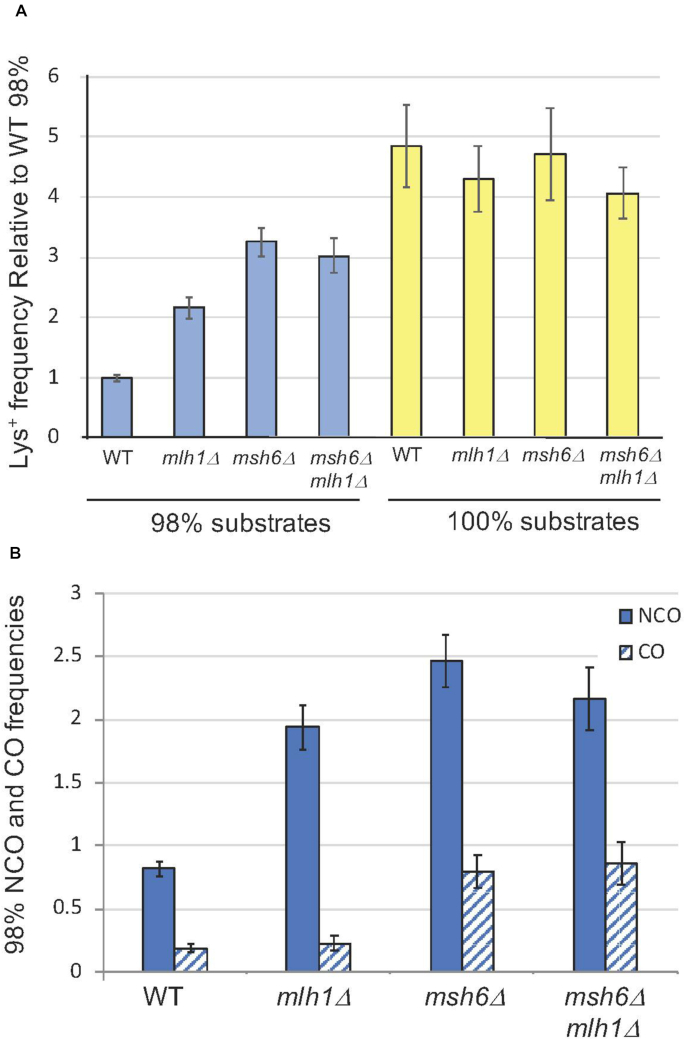
DSB repair frequencies and outcomes. (A) Mean Lys+ frequencies between diverged and identical substrates (blue and yellow bars, respectively). All frequencies were normalized to that obtained with diverged substrates in the WT background. For the 98% substrates, the number of independent inductions (N) for the WT was 100, N = 68 for mlh1Δ, N = 100 for msh6Δ and N = 39 for msh6Δ mlh1Δ. For the 100% substrates, N = 25 for WT, N = 22 for mlh1Δ, N = 22 for msh6Δ and N = 22 in msh6Δ mlh1Δ. (B) Mean NCO and CO frequencies (solid and cross-hatched bars) between the diverged substrates, which were calculated by multiplying the corrected proportion of the event type by the normalized Lys+ frequency. Error bars correspond to 95% confidence intervals. NCO and CO data are in Supplementary Table S2. Unusual CO events are described in Materials and Methods and Supplementary Figures S2 and S3. A complete strain list is provided in Supplementary Table S1.
When combining Lys+ frequencies and CO/NCO distributions to calculate CO/NCO frequencies, the 95% confidence interval (CI) of each derived frequency was calculated by taking into account the 95% CI of the component Lys+ frequency and of the CO/NCO proportion (vassarstats.net). The errors of both measurements were combined to obtain the upper and lower bounds for the derived 95% CI through the right triangle rule (49). The CIs thus calculated were used in the relevant figures.
RESULTS
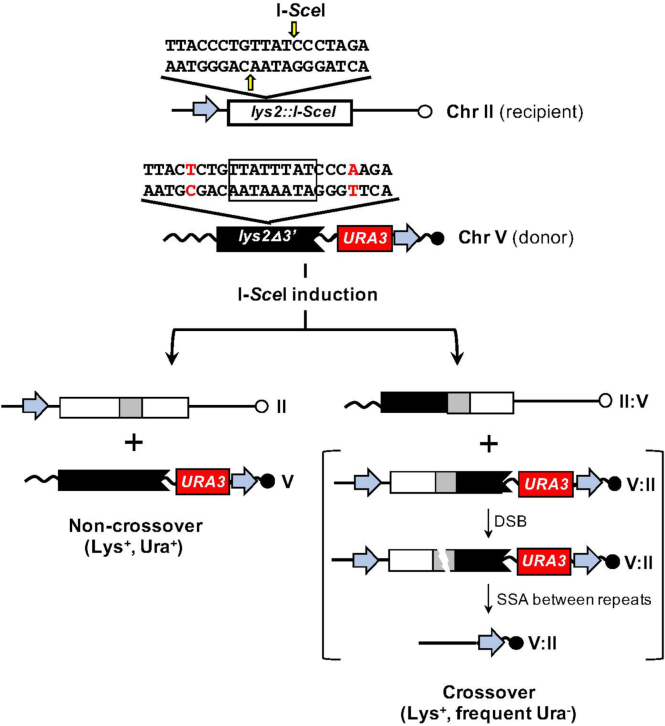
The role of the MMR machinery in regulating mitotic interactions between diverged sequences was examined using a LYS2-based ectopic system described previously (Figure 2; (39). In this system, an I-SceI cleavage site was inserted into the endogenous LYS2 locus on chromosome II, which coincidentally created a –1 frameshift mutation. As a donor template for repair, a 4.2 kb fragment containing a 3′-truncated lys2 allele (lys2Δ3′) was inserted into the CAN1 locus on chromosome V. The donor allele contained an I-SceI site inactivated by duplicating the 4 bp flanked by I-SceI-generated nicks. Copying the additional 4 bp of the donor allele into the broken, recipient allele during repair reverts the frameshift mutation and generates a selectable Lys+ phenotype. Finally, silent single-nucleotide polymorphisms (SNPs) were engineered into the donor allele at ∼50 bp intervals (2% sequence divergence; Supplementary Figure S1) in order to track the transfer of strands between the donor and recipient alleles. Although unrepaired mismatches within heteroduplex DNA segregate at the next cell division to form sectored colonies, the component SNPs can be detected by sequencing PCR products amplified from entire Lys+ colonies.
Figure 2.
Ectopic recombination assay. The sequence inserted into the LYS2 locus contains an I-SceI cleavage site, with the vertical yellow arrows indicating positions of nicks made by the enzyme. The insertion creates a –1 frameshift allele (lys2::I-SceI). The truncated donor allele (lys2Δ3′) on chromosome V is refractory to I-SceI cleavage because the region flanked by I-SceI nicks is duplicated (boxed); the break proximal SNPs on each side are in red. The addition of 4 bp during DSB repair reverts the frameshift allele and results in a Lys+ phenotype. Filled gray areas in recombination products indicate hetDNA. The blue arrows correspond to hisG and the red box to URA3, which were used to distinguish CO and NCO outcomes. Following a CO, the lys2Δ3′ allele is flanked by hisG direct repeats, which destabilizes the intervening URA3 marker and results in frequent Ura− segregants. By contrast, the URA3 marker remains stable following a NCO event.
An individual repair event can be resolved as either an NCO or a CO and one goal was to examine if/how sequence divergence affects recombination outcome. COs generally comprise only 10–15% of recombinants in ectopic assays (50) and their identification usually is through product analysis by either CO-specific PCR or Southern blots. For the current study, we modified the system to also allow COs and NCOs to be distinguished phenotypically. This was accomplished by inserting one copy of the bacterial hisG gene upstream of the recipient allele and a second copy downstream of the donor allele (blue arrows in Figure 2). The region between the donor allele and the hisG sequence additionally contained a URA3 marker. Following a CO event, the 3′-truncated product and the URA3 marker are flanked by hisG direct repeats, which provide the homology for subsequent single-strand annealing and deletion of the intervening segment (41). As a result, CO events frequently give rise to Ura− segregants that form colonies on medium containing 5-fluoroorotic acid (5-FOA), while NCOs have a stable Ura+ phenotype (Figure 2 and Supplementary Figure S2A). Because a CO generates a reciprocal translocation between chromosomes II and V, the presence of both products is expected to be required for subsequent cell viability.
Because the current study was designed to examine how sequence divergence affects DSB repair in the presence/absence of MMR components, we constructed an analogous system in which the donor sequence was devoid of SNPs and thus identical to the recipient allele except for the 4-bp difference at the position of the DSB (100% substrates). The effect of sequence divergence on DSB repair efficiency (Lys+ frequency) and CO-NCO outcome was determined using the 98%- and 100%-identical substrates in wild-type (WT) cells, in msh6Δ cells compromised for mismatch detection, and in mlh1Δ cells that retain mismatch recognition but are unable to initiate subsequent mismatch removal. In addition, the 98% substrates were used to determine how MMR activities affect the extent of strand exchange during NCO formation.
Effects of sequence divergence on DSB repair efficiency
In the MMR-proficient WT background containing the 98%-identical substrates, the Lys+ frequency following DSB induction (0.1% galactose for 45 minutes) was 1.3 × 10−2. In Figure 3A, Lys+ frequencies in the other strains examined were normalized to this frequency. In the msh6Δ background compromised for mismatch binding activity, the Lys+ frequency was elevated 3.3-fold (P < 0.001 relative to WT by Student's t-test). In the mlh1Δ background that retains mismatch binding, however, the Lys+ frequency was elevated only 2.2-fold (P < 0.001 when compared to WT or msh6Δ). These results are consistent with earlier analyses demonstrating that mismatch binding alone (mlh1Δ background) is sufficient for some anti-recombination activity (19). Recombination between the diverged substrates was elevated upon deletion of MSH6, but the Lys+ frequency was less than that observed between the 100%-identical substrates. Although it is generally assumed that MutSα initiates the repair of all base-base mismatches, the yeast MutSβ (Msh2-Msh3) complex has activity against some of the base-base mismatches that are expected to be present in the 98%-identical substrates (51). Disruption of MSH6 rather than MSH2 was used in the current analyses in order to preserve the additional role of MutSβ in processing recombination intermediates (11). As expected, MSH6 was epistatic to MLH1, with the repair frequency in the msh6Δ mlh1Δ double mutant being the same as in the msh6Δ single mutant (P = 0.25). In contrast to the 98% substrates, recombination between the identical substrates in the WT, msh6Δ (P = 0.77 relative to WT by Student t-test), mlh1Δ (P = 0.21), and msh6Δ mlh1Δ (P = 0.06) backgrounds showed no significant difference, indicating that the MMR system does not generally affect the recombination process and that anti-recombination effects with the 98% substrates are due to mismatch-containing heteroduplex intermediates.
Effect of sequence divergence and MMR status on CO-NCO frequencies
At least 200 Lys+ colonies from each of the 100% and 98% strains were analyzed (Supplementary Table S2) and each colony was assigned to one of four recombinant classes based on phenotype and PCR analysis: (i) an NCO event, (ii) a CO event, (iii) occurrence of a CO and NCO event in the same cell or (iv) a CO with additional deletion of the region between the hisG direct repeats (Ura− colony). CO and NCO proportions were determined using corrections to the raw data that considered (a) the failure to detect CO events in which the translocation chromosomes segregated into different daughter cells and (b) the co-occurrence of CO and NCO products in the same Lys+ colony (Supplementary Table S2; see Materials and Methods for details). For the 100% substrates, the corrected proportion of CO-type Lys+ colonies in the WT background was 0.24. The CO-NCO product distribution between the identical substrates was not affected by deletion of MLH1 or MSH6 (P = 0.35 and P = 0.25, respectively, by contingency chi-square test). Relative to the 100% substrates, the proportion of COs obtained with the 98% substrates in a WT background was reduced to 0.18 (P = 0.06). Whereas deletion of MLH1 or MSH6 had no effect on the CO-NCO distribution with the 100% substrates, the loss of either had a significant effect on the distribution obtained with the diverged substrates. In the mlh1Δ strain containing the 98% substrates, the CO proportion decreased to 0.10 (P < 0.001 relative to WT); in the msh6Δ background the CO proportion increased to 0.24 (P = 0.02 relative to WT; not significant with the Bonferroni correction).
Because sequence divergence affected recombination frequencies as well as repair outcomes, the data were combined to obtain NCO and CO frequencies (Figure 3B). Relative to WT, loss of mismatch recognition (msh6Δ mutant) elevated the NCO and CO frequencies 3.0- and 4.3-fold, respectively. Although the increases were similar, the slightly greater increase in CO frequency is suggestive of more MMR-associated anti-recombination activity with respect to COs. In the mlh1Δ background where mismatch recognition was retained, however, only the frequency of NCO events was elevated (2.4-fold relative to WT). Altogether, these data indicate that mismatch recognition alone is sufficient to suppress CO events and that mismatch processing primarily affects NCOs.
The presence of CO and NCO products in a single Lys+ colony, which represents cleavage of both sister chromatids, was relatively rare. This, along with the predicted loss of 50% of CO-only events, was considered when calculating the ‘corrected’ proportions of CO and NCO events in the above analyses. Because making these corrections could potentially skew the results and alter interpretations, we also calculated CO and NCO frequencies without incorporating these corrections (Supplementary Figure S4). The trends were the same as those in Figure 3B and the conclusions were unaltered.
Mismatches in repair products are associated with an additional round of recombination
As described above, a CO event generates hisG direct repeats that flank the truncated product and a URA3 marker (Figure 2). Recombination between the flanking hisG repeats is expected to occur at a frequency of ∼10−4 (52), which results in rare Ura− papillae when Lys+ CO recombinants are plated on 5-FOA medium. Using URA3 instability for phenotypic scoring of CO versus NCO outcomes revealed an unanticipated class: Lys+ colonies that grew confluently on 5-FOA medium and failed to grow in the absence of uracil (Supplementary Figure S2A). Physical analysis of fully Ura− colonies by PCR and Southern blots confirmed loss of the URA3 marker together with the lys2Δ3′ allele on the V:II translocation chromosome and retention of a single copy of hisG (Supplementary Figure S2B). Repair of the I-SceI induced DSB as a CO was thus associated with a very early recombination event that removed the region between the hisG direct repeats in all progeny cells. The fully Ura− recombinants were observed at a low frequency following recombination between the diverged substrates (18/1742 Lys+ colonies screened from all backgrounds), but none were detected when identical substrates were used (0/1145 Lys+ colonies screened; P = 0.001 by contingency chi-square). This indicates a specific association of a secondary recombination event with mismatch-containing hetDNA. Furthermore, the Ura− colonies detected with the 98% substrates were more frequent in the WT than in the MMR-defective backgrounds (13/578 and 5/1234 Ura− among the Lys+ colonies screened, respectively; P < 0.001). We suggest that the recognition/processing of mismatches creates a second DSB that initiates deletional recombination between the hisG direct repeats that flank the V:II CO product.
hetDNA position in NCOs is regulated by the MMR machinery
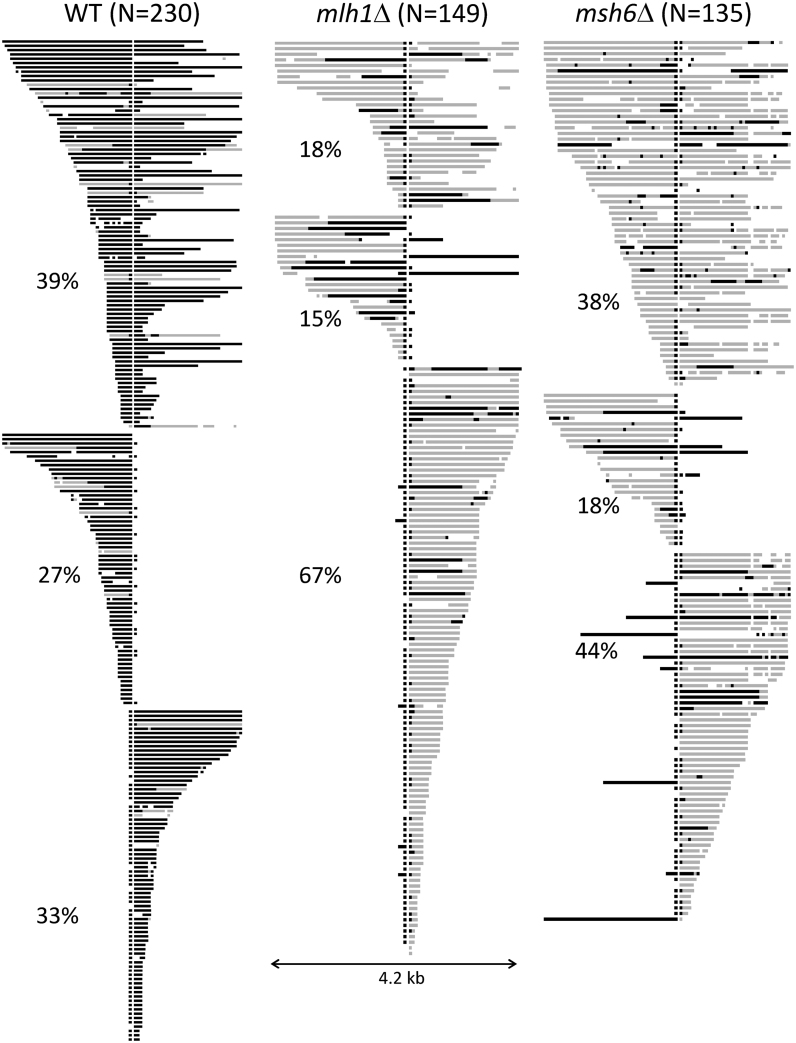
To obtain insight into the mechanism(s) of MMR-associated anti-recombination, we sequenced the recipient allele of NCO recombinants isolated in the WT, mlh1Δ and msh6Δ backgrounds. The sequence profiles of individual events are presented in Figure 4; black bars correspond to regions of gene conversion (GC) that had only donor SNPs and gray bars to hetDNA comprised of a mix of donor and recipient SNPs. hetDNA/GC tracts were divided into three groups based on their position relative to the initiating DSB. In the first group, there was a tract on each side of the break (bidirectional), a pattern consistent either with dissolution of a double Holliday junction or with a double SDSA event. The second and third groups had a hetDNA/GC tract on only one side of the DSB (unidirectional), the pattern expected of an SDSA event. It should be noted that frequent conversion of the DSB-proximal SNP was observed in all three genetic backgrounds but was not considered when assigning tract directionality. We previously reported that such single-SNP conversions in this system reflect the proofreading activity of DNA polymerase δ rather than MMR activity (39). In the MMR-defective backgrounds, we also made the assumption that uninterrupted GC tracts extending from the break site reflected gap expansion; these also were not considered when assigning tract directionality.
Figure 4.
hetDNA/GC tracts in NCO products. Each horizontal line represents an independent NCO event and the vertical white line marks the position of the DSB. Black bars correspond to donor sequence and gray bars to hetDNA. Tracts are grouped by whether they are present on both sides of the initiating DSB, are only upstream of the break, or are only present downstream of the break; the percentage of each type is given. For each NCO, conversion of the break proximal SNP on each side of the DSB is due to proofreading activity of Pol δ and was not considered when assigning tract directionality. In MMR-defective backgrounds, an uninterrupted GC tract extending from the DSB was assumed to reflect gap expansion. N, total number of NCOs.
In a WT background, GC events reflect NCO intermediates that escaped anti-recombination activity of the MMR system and were subsequently subjected to mismatch correction. Of NCOs sequenced, 230 (88%) contained regions of GC and/or hetDNA (Figure 4); the remainder contained neither and were considered uninformative. With regard to the distribution of tracts relative to the DSB, 39% (90/230) were bidirectional and 61% (140/230) were unidirectional. The positions of the unidirectional tracts were random: 63 were confined to the upstream (promoter proximal) side of the break and 77 to the downstream side (P = 0.27 by Chi-square goodness of fit). Among the 153 tracts that were upstream of the DSB (90 from bidirectional and 63 from unidirectional events, respectively), 110 had a continuous GC tract and 14 had a continuous hetDNA tract; the remaining 29 tracts were a discontinuous mix of GC, hetDNA and/or restoration (restoration is defined as recipient-only SNPs within a GC or hetDNA tract). Similar proportions were seen among the 167 downstream tracts: 130 continuous GC tracts, 16 continuous hetDNA tracts and 21 mixed tracts (P = 0.29 by contingency Chi-square). These data demonstrate that (1) 80–90% of hetDNA tracts that escaped anti-recombination were nevertheless repaired and (2) ∼20% of repaired tracts were discontinuous. That hetDNA tracts were converted to GC tracts is consistent with the prior conclusion that anti-recombination and mismatch correction are separable activities of the MMR machinery (20,53,54). Discontinuities within tracts could reflect patchy repair, template switching during 3′-end extension (47,55) or invasion of multiple donors by a single 3′ end (56).
In the absence of MSH6, the proportions of bidirectional (52/135) versus unidirectional (83/135) tracts were similar to those observed in WT (P = 0.92 by contingency Chi-square). The unidirectional tracts, however, were biased to the downstream side of the initiating break; there were 59 tracts downstream of the break but only 24 upstream (P < 0.001 relative to WT). The significance of this altered distribution is unclear. A feature of hetDNA tracts in the msh6Δ background was their frequent punctuation by a GC or restoration event involving only a single SNP. We suggest that binding of a mismatch by MutSα may shield it from recognition and processing by an alternative, short-patch repair pathway. The existence of such a pathway in budding yeast was previously suggested (57) and in both Schizosaccharomyces pombe and Drosophila, the nucleotide excision repair pathway mediates mismatch removal (58,59). At least in the assay used here, however, the repair of single mismatches persisted in a msh6Δ rad14Δ background in which nucleotide excision repair was additionally eliminated (data not shown).
In the mlh1Δ background, the distribution of unidirectional versus bidirectional hetDNA tracts was dramatically different from that observed in the WT and msh6Δ backgrounds (Figure 4). Only 18% (27/149) of tracts were bidirectional in the mlh1Δ background, whereas ∼40% were bidirectional in WT and msh6Δ strains (P < 0.001 when compared to either). As observed in the msh6Δ background, the positions of unidirectional tracts were not random in the mlh1Δ background and were strongly biased to the downstream side of the break (22 upstream and 100 downstream tracts, respectively; P < 0.001 relative to WT). Although the bias appeared stronger than in the msh6Δ strain, it was not significantly different (P = 0.057 by contingency Chi-square).
Because the overall DSB repair frequency and hetDNA positions among NCOs varied as a function of MMR status, we calculated the frequencies of unidirectional and bidirectional tracts. In Figure 5, these tract frequencies are presented together with the corresponding CO frequencies in order to highlight the similarity between the CO and bidirectional NCO-tract frequencies in each background. Relative to the WT background, loss of mismatch recognition (msh6Δ strain) was associated with an ∼3-fold increase in both unidirectional and bidirectional NCO frequencies. In the mlh1Δ background, however, only the frequency of the unidirectional NCOs was elevated and their frequency was similar to that observed in the msh6Δ background. The same correlations were seen when the frequencies of unidirectional and bidirectional NCOs were calculated using an uncorrected data set (Supplementary Figure S4). The lack of an increase in bidirectional hetDNA in the mlh1Δ background mirrors the lack of an increase in CO-type events in this background. Altogether, these data suggest that mismatch binding alone is sufficient to reduce COs as well as NCOs with bidirectional hetDNA, while mismatch processing specifically reduces NCOs with unidirectional hetDNA.
Figure 5.
Frequencies of repair types. Frequencies of NCO events with unidirectional tracts (dark blue bars), NCO events with bidirectional tracts (light blue bars) and CO events (cross-hatched bars) are shown. Frequencies were calculated using only those NCO events with informative tracts; error bars are 95% confidence intervals for CO and NCO frequencies.
hetDNA/GC tracts are shortened by MMR
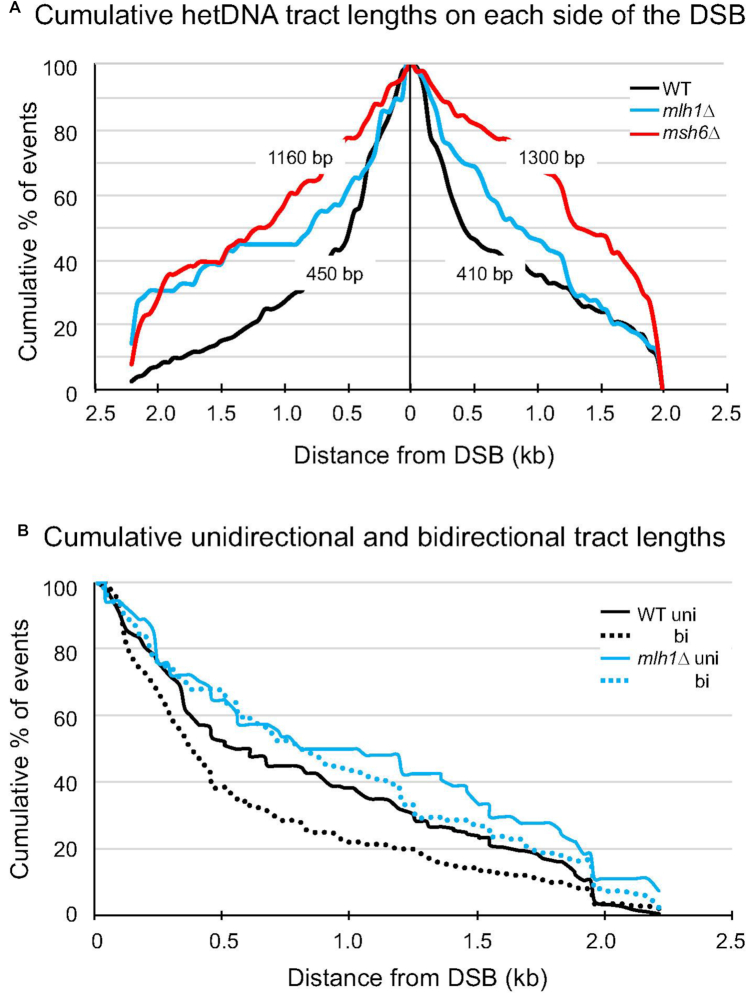
The length of hetDNA/GC on each side of the initiating break was determined for each NCO event in Figure 4. For patchy/mixed tracts, the most break distal position where donor sequence was acquired was used for the length determination. There were a total of 153 upstream and 167 downstream tracts for the WT strain and the cumulative distribution of tract lengths on each side of the break is shown in Figure 6A (black lines). Upstream of the DSB, the median tract length was 450 bp; downstream of the break, the median length was 410 bp. Among the 135 NCOs sequenced from the msh6Δ background, there were a total of 76 upstream and 111 downstream tracts that were used to construct the tract-length distributions (Figure 6A, red lines). Upstream of the DSB, the median hetDNA length was 1160 bp compared to only 450 bp in WT (P < 0.0001 Mann–Whitney U test); downstream of the break, the median hetDNA length was 1300 bp in the msh6Δ background compared to 410 bp in WT (P < 0.0001). Finally, the median hetDNA tract lengths in the mlh1Δ background were 760 bp upstream and 830 bp downstream of the DSB (Figure 6A, blue lines), which were intermediate between those observed in the WT and msh6Δ backgrounds. In contrast to mismatch binding, which is expected to affect all hetDNA tracts, frequency measurements indicated that mismatch processing only affected the unidirectional tracts diagnostic of SDSA (Figure 5). We thus examined the unidirectional and bidirectional tracts separately in the WT and mlh1Δ backgrounds. As shown in Figure 6B, the medians of the unidirectional and bidirectional tracts were similar in the absence of MLH1 (799 bp for unidirectional and 909 bp for bidirectional tracts; P = 0.44 by Mann–Whitney). By contrast, the unidirectional tracts in the WT background were significantly shorter than the bidirectional tracts (367 bp for unidirectional and 561 bp for bidirectional tracts; P = 0.012 by Mann–Whitney). These data are consistent with the selective destruction of SDSA intermediates by Mlh1-mediated mismatch processing.
Figure 6.
Cumulative GC/hetDNA tracts lengths. (A) The cumulative distributions of tract lengths on each side the initiating DSB (vertical line). The median tract lengths on each side of the break in the WT and msh6Δ strains are indicated. The black, blue and red lines are WT, mlh1Δ and msh6Δ distributions, respectively. (B) Cumulative distributions of unidirectional versus bidirectional (uni and bi, respectively) tracts relative to the initiating DSB (0 kb). The black and blue lines are WT and mlh1Δ distributions, respectively.
DISCUSSION
In the current study, we used 4.2 kb ectopic lys2 chromosomal substrates to examine the effects of 2% sequence divergence and MMR status on the repair of an I-SceI-induced DSB. Repair was assessed by measuring the frequency of Lys+ colonies, the proportions of Lys+ colonies that were COs versus NCOs, and the length/position of hetDNA/GC tracts in NCO products relative to the initiating DSB. The major findings were that (i) gene conversion tracts in WT are much shorter than hetDNA tracts in MMR-defective strains, (ii) mismatch binding reduces the frequency of COs as well as NCO events with unidirectional or bidirectional hetDNA and (iii) mismatch processing reduces the frequency only of NCOs with unidirectional hetDNA, with little effect on NCOs with bidirectional hetDNA or COs. These results provide novel insight into how sequence divergence affects mitotic DSB repair and distinguish the anti-recombination effects of mismatch binding from those of mismatch processing.
In an MMR-proficient WT background, 2% sequence divergence was associated with an ∼5-fold reduction in the Lys+ frequency (Figure 3). In terms of MMR-directed anti-recombination, early in vitro studies demonstrated that bacterial MutS blocks strand transfer between diverged DNA sequences (60) and more recent work has shown that MutS and MutL together can promote helicase (UvrD)-catalyzed unwinding of a D-loop intermediate formed between diverged sequences (61). Additionally, human MutSα efficiently recognizes mismatches in the context of a model D-loop structure (62). Beyond its anti-recombination role, the repair activity of the MMR system is required to convert mismatch-containing hetDNA to gene conversion (GC) tracts. The latter activity is similar to the strand-directed repair that occurs in the context of replication, which biases hetDNA repair to GC rather than restoration during recombination. The anti-recombination and replication-editing activities of MMR can be separated based on their genetic requirements. The 3′>5′ Sgs1 helicase, for example, is important for anti-recombination but has no known role in mismatch removal during recombination or replication (20,32,33). By contrast, PCNA is critical for coupling MMR to the replication fork and facilitates strand discrimination during replication-error repair (34,35), but a role during heteroduplex rejection has not been detected (31,36). The identification of pms1 alleles with defects only in anti-recombination further supports a mechanistic distinction between anti-recombination and mismatch correction (63). In the E. coli system, a mutS mutant with a similar separation-of function phenotype has been described (64).
In considering the results reported here, it is important to note that heteroduplex rejection can, in principle, be of two types that are predicted to be temporally distinct. The first occurs early during recombination and presumably involves the mismatch-triggered unwinding of hetDNA by Sgs1. The second occurs after the initiation of 3′-end extension when MMR switches into a repair mode that requires PCNA (65). Although repair usually converts rather than restores the information contributed by the broken molecule, it has the potential to be destructive if excision tracts are introduced into complementary strands. Genetic studies in bacterial cells are consistent with two distinct types of MMR-mediated anti-recombination that work early and late (66), as well a more prominent anti-recombination role for mismatch binding than for mismatch processing (16). Whereas mismatch repair in yeast depends on MutLα, anti-recombination has exhibited variable dependence.
As reported previously in other systems (17,19,20), we observed that overall recombination was elevated more in a msh6Δ background where mismatch recognition was compromised than in an mlh1Δ background where mismatch binding persisted (Figure 3). A key observation emerged from subsequent partitioning of events into NCOs and COs and from defining hetDNA tracts in NCOs as either unidirectional or bidirectional. Relative to WT, deletion of MSH6 had a similar stimulatory effect on each class, demonstrating that mismatch binding is required for all of the observed anti-recombination activity (compare WT and msh6Δ in Figure 5). By contrast, loss of mismatch processing elevated only the frequency of NCO products that contained unidirectional hetDNA (compare WT and mlh1Δ in Figure 5). Stated another way, mismatch binding alone was sufficient for the reduction in CO events and specifically those NCO events containing bidirectional hetDNA (compare msh6Δ and mlh1Δ in Figure 5). What these two classes have in common is that both ends of the broken molecule interact with a donor repair template. The lack of a processing requirement is consistent with the reversal/unwinding of an early hetDNA intermediate. By contrast, mismatch processing only affects the NCO events with unidirectional hetDNA, which presumably occur via SDSA and require that only one end of the broken molecule engage the donor.
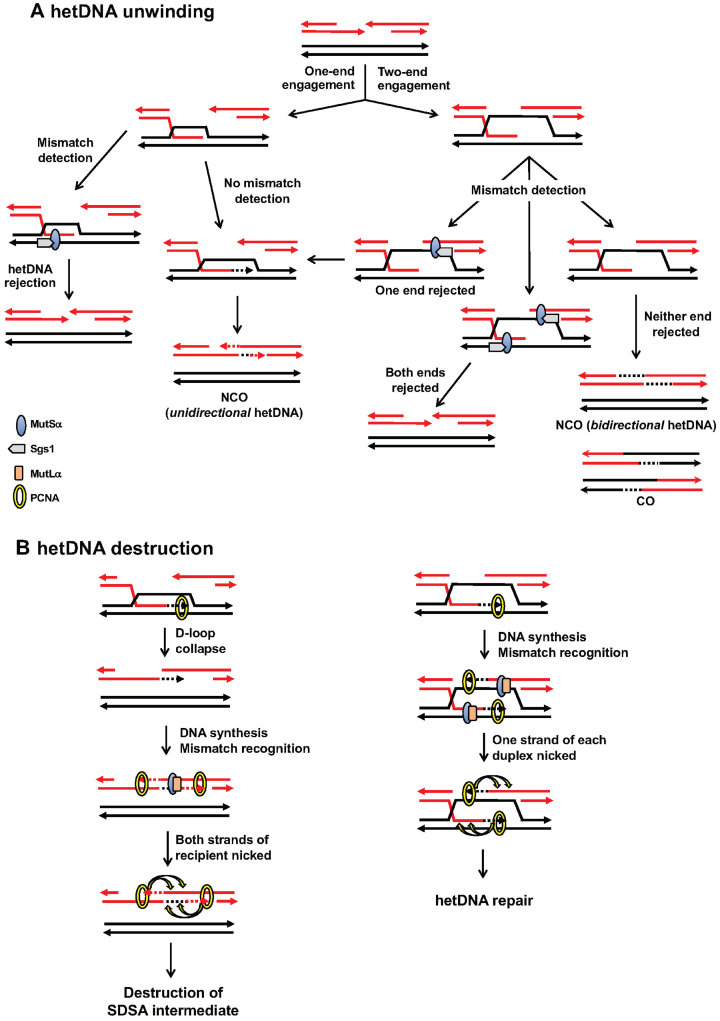
The above data suggest a model in which the anti-recombination effect observed in msh6Δ versus mlh1Δ background is related to whether one or both ends of the broken molecule engage the donor (Figure 7A). This model assumes that each end engages the donor independently and that each engagement has the same probability of escaping mismatch detection (X) by MutSα or of being detected (1 – X). If both ends are engaged, the probability that neither will be rejected/unwound is X2, and this results in a CO or a bidirectional NCO. In the current system the proportion of bidirectional events that escaped the initial hetDNA rejection (X2) is 0.327 (the ratio of the WT to msh6Δ frequency for COs plus bidirectional NCOs) and X = 0.57. The probability that both ends are rejected is (1 – X)2 or 0.18; the probability that only one end is rejected is 2X(1 – X) or 0.49. If only one end is rejected, the outcome is the equivalent of an one-ended SDSA intermediate that escapes hetDNA rejection. Although ∼50% of single-end invasion events are also expected to be eliminated by hetDNA rejection, they will be replenished by the two-end engagement events that had only a single end rejected. Whether the rejected events attempt one or more additional rounds of DSB repair is not known.
Figure 7.
Models for anti-recombination activity of the MMR machinery. (A) Mismatch detection by MutSα before DNA synthesis initiates from an invading end triggers unwinding of the D-loop by Sgs1. This primarily eliminates those events in which both of the broken ends engage the donor, which corresponds to CO and NCO intermediates with hetDNA on both sides of the DSB. (B) The conflicting strand discrimination signals conferred by PCNA during extension of both ends of an annealed SDSA intermediate results in destruction by MutLα endonuclease activity. See main text for details.
A second key observation concerns the processing-specific anti-recombination activity associated with MutLα. Deletion of MLH1 alone elevated only the frequency of unidirectional NCOs, with no discernible effect either on COs or on bidirectional-tract NCOs. This suggests a unique feature of an SDSA intermediate that triggers its destruction when MMR switches to a repair mode. As noted previously, repair likely occurs after DNA synthesis is initiated and there is assumed to be strand discrimination, which biases exonucleolytic removal of the nascent strand. During SDSA (Figure 7B; left side), the pairing of the extended end with the other side of the break creates a hetDNA tract that is flanked by gaps. Extension of the top strand during gap filling will bias mismatch removal from the top strand, while extension of the bottom strand will trigger a similar process on the bottom strand. We suggest that the net effect is destruction of the intermediate via overlapping excision tracts. In the case of a CO or NCO intermediate formed by two-end engagement (Figure 7B; right side), there will be only a single gap (rather than flanking gaps) adjacent to each hetDNA tract. An alternative explanation for the Mlh1-dependent loss of SDSA events is that the unwinding of annealed strands requires Mlh1 in addition to Msh6, while the unwinding of strand-invasion intermediates only requires Msh6. This explanation, however, is not consistent with the Mlh1 independence of anti-recombination during SSA, which involves only strand annealing. Further support of a repair-based destructive mechanism of anti-recombination is provided by the rare Lys+ COs that were completely Ura−. That these were observed only with the 98% substrates is consistent with the initiation of a second, mismatch-dependent recombination (SSA) event. We suggest that such secondary events are due to rare failures in normal strand discrimination that result in overlapping excision tracts. Similar secondary recombination events triggered by sequence divergence were previously observed in meiotic studies (67), and these similarly required MutLα (68). In addition, early studies in E. coli indicated that loss of strand discrimination during MMR results in recombination-initiating DSBs (69,70). Finally, the nuclease activity of MutLα is partially required for its mitotic anti-recombination activity in an inverted repeat assay (71).
A destruction-based model provides an explanation for why SDSA events in the current system are uniquely sensitive to Mlh1-dependent anti-recombination activity. Although one might expect SSA intermediates to be similarly affected, Mlh1 plays little, if any, anti-recombination role during SSA (20). We suggest that this reflects the fundamentally different structures of the annealed intermediates that arise during SDSA versus SSA. During SSA, each strand of the annealed segment contains a long 3′ tail/flap (Figure 1). These tails are critical for the anti-recombination activity of Msh6 and it has been suggested that they provide an entry point for Sgs1-mediated unwinding (31). Following the escape of an intermediate from anti-recombination activity, the 3′ tails must be removed by Rad1–Rad10 before filling of the adjacent gaps (11). If there is no coordination between the removal of these tails, then the filling of the gaps may be temporally separated. This, in turn, would reduce or preclude the simultaneous repair of mismatches directed by the two different extending ends. The presence of a nonhomologous 3′ tail at an invading end during break-induced replication also promotes hetDNA rejection between mismatched substrates (72). The system used here, however, was designed to preclude such tails and their potential effects on HR.
NCO events in a WT background had GC tracts that were 2–3 fold shorter than hetDNA tracts in the msh6Δ background (Figure 6). This suggests that shorter hetDNA tracts are more likely to escape anti-recombination and is consistent with the cumulative negative effect of mismatches on recombination rates in both yeast (15) and E. coli (73). In SDSA events, GC/hetDNA tracts reflect DNA synthesis from the invading end, and we have previously suggested that this, in turn, is proportional to the size of the D-loop formed during invasion of the donor (39). The size of the D-loop cannot exceed the length of homology between the 3′ tail and the repair template, and this may limit new DNA synthesis in an ectopic system. In this model, D-loops are unwound from their back end as they are extended by DNA synthesis at the front end to create a migrating D-loop. The reverse direction of hetDNA unwinding may be highly favored when there is a nonhomologous 3′ tail (as in SSA). It will be interesting to examine if and how the presence of a 3′ tail affects anti-recombination in the current system. It should be noted that for spontaneous events involving ectopic substrates, a tail would be present if the initiating DSB occurs outside the region of homology.
With ectopic substrates the length of homology between the recombining substrates is limited and an anti-recombination effect of the MMR system is evident. By contrast, there is little, if any, effect of MMR-associated anti-recombination on mitotic loss-of-heterozygosity (LOH) involving diverged, homologous chromosomes in diploid strains (74). This difference could reflect less sequence divergence in the LOH system and/or the presence of unlimited homology. Nevertheless, the results reported here have broad relevance to interactions between diverged, repetitive elements that are abundant in more complex eukaryotic genomes. Genome rearrangements with endpoints in such elements are associated with human genetic diseases and also may contribute to clinically relevant tumor evolution in cancers with underlying MMR defects.
DATA AVAILABILITY
PacBio sequencing was employed in this study. Unprocessed CCS reads and barcodes used in this work are available at https://data.mendeley.com/datasets/z38×9nvwmg/1.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tom Petes and members of the SJR lab for discussions throughout the course of this work and for comments on the manuscript.
Notes
Present address: Yee Fang Hum, Department of Cancer Biology, Abramson Family Cancer Research Institute, Basser Center for BRCA, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health grant [R35GM118077] to S.J.R.; American Heart Association [15PRE25510022] to Y.F.H.. Funding for open access charge: National Institutes of Health grant [R35GM118077].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kadyk L.C., Hartwell L.H.. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992; 132:387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhao Y., Dominska M., Petrova A., Bagshaw H., Kokoska R.J., Petes T.D.. Properties of mitotic and meiotic recombination in the tandemly-repeated CUP1 gene cluster in the yeast Saccharomyces cerevisiae. Genetics. 2017; 206:785–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carvalho C.M., Lupski J.R.. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016; 17:224–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Symington L.S., Rothstein R., Lisby M.. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014; 198:795–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Symington L.S., Gautier J.. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011; 45:247–271. [DOI] [PubMed] [Google Scholar]
- 6. Ira G., Malkova A., Liberi G., Foiani M., Haber J.E.. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003; 115:401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prakash R., Satory D., Dray E., Papusha A., Scheller J., Kramer W., Krejci L., Klein H., Haber J.E., Sung P. et al.. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009; 23:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mitchel K., Lehner K., Jinks-Robertson S.. Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS Genet. 2013; 9:e1003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fasching C.L., Cejka P., Kowalczykowski S.C., Heyer W.D.. Top3-Rmi1 dissolve Rad51-mediated D loops by a topoisomerase-based mechanism. Mol. Cell. 2015; 57:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sakofsky C.J., Malkova A.. Break induced replication in eukaryotes: mechanisms, functions, and consequences. Crit. Rev. Biochem. Mol. Biol. 2017; 52:395–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lyndaker A.M., Alani E.. A tale of tails: insights into the coordination of 3′ end processing during homologous recombination. Bioessays. 2009; 31:315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jinks-Robertson S., Michelitch M., Ramcharan S.. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol. Cell Biol. 1993; 13:3937–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sugawara N., Ira G., Haber J.E.. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell Biol. 2000; 20:5300–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Inbar O., Kupiec M.. Homology search and choice of homologous partner during mitotic recombination. Mol. Cell Biol. 1999; 19:4134–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Datta A., Hendrix M., Lipsitch M., Jinks-Robertson S.. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:9757–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rayssiguier C., Thaler D.S., Radman M.. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989; 342:396–401. [DOI] [PubMed] [Google Scholar]
- 17. Selva E.M., New L., Crouse G.F., Lahue R.S.. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics. 1995; 139:1175–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bailis A.M., Rothstein R.. A defect in mismatch repair in Saccharomyces cerevisiae stimulates ectopic recombination between homeologous genes by an excision repair dependent process. Genetics. 1990; 126:535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Datta A., Adjiri A., New L., Crouse G.F., Jinks-Robertson S.. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell Biol. 1996; 16:1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sugawara N., Goldfarb T., Studamire B., Alani E., Haber J.E.. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:9315–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Do A.T., LaRocque J.R.. The role of Drosophila mismatch repair in suppressing recombination between diverged sequences. Sci. Rep. 2015; 5:17601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Emmanuel E., Yehuda E., Melamed-Bessudo C., Avivi-Ragolsky N., Levy A.A.. The role of AtMSH2 in homologous recombination in Arabidopsis thaliana. EMBO Rep. 2006; 7:100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dion E., Li L., Jean M., Belzile F.. An Arabidopsis MLH1 mutant exhibits reproductive defects and reveals a dual role for this gene in mitotic recombination. Plant J. 2007; 51:431–440. [DOI] [PubMed] [Google Scholar]
- 24. Trouiller B., Schaefer D.G., Charlot F., Nogue F.. MSH2 is essential for the preservation of genome integrity and prevents homeologous recombination in the moss Physcomitrella patens. Nucleic Acids Res. 2006; 34:232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Elliott B., Jasin M.. Repair of double-strand breaks by homologous recombination in mismatch repair-defective mammalian cells. Mol. Cell Biol. 2001; 21:2671–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Wind N., Dekker M., Berns A., Radman M., te Riele H.. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995; 82:321–330. [DOI] [PubMed] [Google Scholar]
- 27. Hunter N., Chambers S.R., Louis E.J., Borts R.H.. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 1996; 15:1726–1733. [PMC free article] [PubMed] [Google Scholar]
- 28. Modrich P., Lahue R.. Mismatch repair in replication fidelity, genetic recombination and cancer biology. Ann. Rev. Biochem. 1996; 65:101–133. [DOI] [PubMed] [Google Scholar]
- 29. Harfe B.D., Jinks-Robertson S.. DNA mismatch repair and genetic instability. Ann. Rev. Genet. 2000; 34:359–399. [DOI] [PubMed] [Google Scholar]
- 30. Welz-Voegele C., Jinks-Robertson S.. Sequence divergence impedes crossover more than noncrossover events during mitotic gap repair in yeast. Genetics. 2008; 179:1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chakraborty U., George C.M., Lyndaker A.M., Alani E.. A delicate balance between repair and replication factors regulates recombination between divergent DNA sequences in Saccharomyces cerevisiae. Genetics. 2016; 202:525–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spell R.M., Jinks-Robertson S.. Examination of the roles of the Sgs1 and Srs2 helicases in the enforcement of recombination fidelity in Saccharomyces cerevisiae. Genetics. 2004; 168:1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Myung K., Datta A., Chen C., Kolodner R.D.. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nature Genet. 2001; 27:1–4. [DOI] [PubMed] [Google Scholar]
- 34. Pluciennik A., Dzantiev L., Iyer R.R., Constantin N., Kadyrov F.A., Modrich P.. PCNA function in the activation and strand direction of MutLalpha endonuclease in mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:16066–16071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goellner E.M., Smith C.E., Campbell C.S., Hombauer H., Desai A., Putnam C.D., Kolodner R.D.. PCNA and Msh2-Msh6 activate an Mlh1-Pms1 endonuclease pathway required for Exo1-independent mismatch repair. Mol. Cell. 2014; 55:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stone J., Gealy R., Petes T., Jinks-Robertson S.. Role of PCNA interactions in the mismatch repair-dependent processing of mitotic and meiotic recombination intermediates in yeast. Genetics. 2008; 178:1221–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Craven R.J., Greenwell P.W., Dominska M., Petes T.D.. Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR genes. Genetics. 2002; 161:493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thomas B.J., Rothstein R.. Elevated recombination rates in transcriptionally active DNA. Cell. 1989; 56:619–630. [DOI] [PubMed] [Google Scholar]
- 39. Guo X., Hum Y.F., Lehner K., Jinks-Robertson S.. Regulation of hetDNA length during mitotic double-strand break repair in yeast. Mol. Cell. 2017; 67:539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Storici F., Durham C.L., Gordenin D.A., Resnick M.A.. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:14994–14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alani E., Cao L., Kleckner N.. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987; 116:541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guldener U., Heck S., Fielder T., Beinhauer J., Hegemann J.H.. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996; 24:2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R.. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998; 14:953–961. [DOI] [PubMed] [Google Scholar]
- 44. Lee P.S., Petes T.D.. Mitotic gene conversion events induced in G1-synchronized yeast cells by gamma rays are similar to spontaneous conversion events. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:7383–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen C., Kolodner R.D.. Gross chromosome rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 1999; 23:81–85. [DOI] [PubMed] [Google Scholar]
- 46. Chua P., Jinks-Robertson S.. Segregation of recombinant chromatids following mitotic crossing over in yeast. Genetics. 1991; 129:359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hum Y.F., Jinks-Robertson S.. DNA strand-exchange patterns associated with double-strand break-induced and spontaneous mitotic crossovers in Saccharomyces cerevisiae. PLoS Genet. 2018; 14:e1007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guo X., Lehner K., O’Connell K., Zhang J., Dave S.S., Jinks-Robertson S.. SMRT sequencing for parallel analysis of multiple targets and accurate SNP phasing. G3 (Bethesda). 2015; 5:2801–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moore A., Dominska M., Greenwell P., Aksenova A.Y., Mirkin S., Petes T.. Genetic control of genomic alterations induced in yeast by intersitital telomeric sequences. Genetics. 2018; 209:425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jinks-Robertson S., Petes T.D.. High-frequency meiotic gene conversion between repeated genes on nonhomologous chromosomes in yeast. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:3350–3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Harrington J.M., Kolodner R.D.. Saccharomyces cerevisiae Msh2-Msh3 acts in repair of base-base mispairs. Mol. Cell Biol. 2007; 27:6546–6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scherer S., Davis R.W.. Recombination of dispersed repeated DNA sequences in yeast. Science. 1980; 209:1380–1384. [DOI] [PubMed] [Google Scholar]
- 53. Mitchel K., Zhang H., Welz-Voegele C., Jinks-Robertson S.. Molecular structures of crossover and noncrossover intermediates during gap repair in yeast: implications for recombination. Mol. Cell. 2010; 38:211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li S., Wehrenberg B., Waldman B.C., Waldman A.S.. Mismatch tolerance during homologous recombination in mammalian cells. DNA Repair. 2018; 70:25–36. [DOI] [PubMed] [Google Scholar]
- 55. Tsaponina O., Haber J.E.. Frequent interchromosomal template switches during gene conversion in S. cerevisiae. Mol Cell. 2014; 55:615–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Piazza A., Wright W.D., Heyer W.D.. Multi-invasions are recombination byproducts that induce chromosomal rearrangements. Cell. 2017; 170:760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Coic E., Gluck L., Fabre F.. Evidence for short-patch mismatch repair in Saccharomyces cerevisiae. EMBO J. 2000; 19:3408–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fleck O., Lehmann E., Schar P., Kohli J.. Involvement of nucleotide-excision repair in msh2 pms1-independent mismatch repair. Nat. Genet. 1999; 21:314–317. [DOI] [PubMed] [Google Scholar]
- 59. Crown K.N., McMahan S., Sekelsky J.. Eliminating both canonical and short-patch mismatch repair in Drosophila melanogaster suggests a new meiotic recombination model. PLoS Genet. 2014; 10:e1004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Worth L.J., Clark S., Radman M., Modrich P.. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:3238–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tham K.C., Hermans N., Winterwerp H.H., Cox M.M., Wyman C., Kanaar R., Lebbink J.H.. Mismatch repair inhibits homeologous recombination via coordinated directional unwinding of trapped DNA structures. Mol. Cell. 2013; 51:326–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Honda M., Okuno Y., Hengel S.R., Martin-Lopez J.V., Cook C.P., Amunugama R., Soukup R.J., Subramanyam S., Fishel R., Spies M.. Mismatch repair protein hMSH2-hMSH6 recognizes mismatches and forms sliding clamps within a D-loop recombination intermediate. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Welz-Voegele C., Stone J.E., Tran P.T., Kearney H.M., Liskay R.M., Petes T.D., Jinks-Robertson S.. Alleles of the yeast PMS1 mismatch-repair gene that differentially affect recombination- and replication-related processes. Genetics. 2002; 162:1131–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Calmann M.A., Nowosielska A., Marinus M.G.. Separation of mutation avoidance and antirecombination functions in an Escherichia coli mutS mutant. Nucleic Acids Res. 2005; 33:1193–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li X., Stith C.M., Burgers P.M., Heyer W.D.. PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase δ. Mol. Cell. 2009; 36:704–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stambuk S., Radman M.. Mechanism and control of interspecies recombination in Escherichia coli. I. Mismatch repair, methylation, recombination and replication functions. Genetics. 1998; 150:533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Borts H.R., Haber J.E.. Meiotic recombination in yeast: alteration by multiple heterozygosities. Science. 1987; 237:1459–1465. [DOI] [PubMed] [Google Scholar]
- 68. Borts R.H., Leung W.-Y., Kramer W., Kramer B., Williamson M., Fogel S., Haber J.E.. Mismatch repair-induced meiotic recombination requires PMS1 gene product. Genetics. 1990; 124:573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Glickman B.W., Radman M.. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc. Natl. Acad. Sci. U.S.A. 1980; 77:1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McGraw B.R., Marinus M.G.. Isolation and characterization of Dam+ revertants and suppressor mutations that modify secondary phenotypes of dam-3 strains of Escherichia coli K-12. Mol. Gen. Genet. 1980; 178:309–315. [DOI] [PubMed] [Google Scholar]
- 71. Erdeniz N., Nguyen M., Deschenes S.M., Liskay R.M.. Mutations affecting a putative MutLα endonuclease motif impact multiple mismatch repair functions. DNA Repair. 2007; 6:1463–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Anand R., Beach A., Li K., Haber J.. Rad51-mediated double-strand break repair and mismatch correction of divergent substrates. Nature. 2017; 544:377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vulic M., Dionisio F., Taddei F., Radman M.. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:9763–9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yin Y., Dominska M., Yim E., Petes T.D.. High-resolution mapping of heteroduplex DNA formed during UV-induced and spontaneous mitotic recombination events in yeast. Elife. 2017; 6:e28069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PacBio sequencing was employed in this study. Unprocessed CCS reads and barcodes used in this work are available at https://data.mendeley.com/datasets/z38×9nvwmg/1.