Abstract
N 6-Methyladenosine (m6A) is a dynamic mRNA modification which regulates protein expression in various posttranscriptional levels. Functional studies of m6A in nervous system have focused on its writers and erasers so far, whether and how m6A readers mediate m6A functions through recognizing and binding their target mRNA remains poorly understood. Here, we find that the expression of axon guidance receptor Robo3.1 which plays important roles in midline crossing of spinal commissural axons is regulated precisely at translational level. The m6A reader YTHDF1 binds to and positively regulates translation of m6A-modified Robo3.1 mRNA. Either mutation of m6A sites in Robo3.1 mRNA or YTHDF1 knockdown or knockout leads to dramatic reduction of Robo3.1 protein without affecting Robo3.1 mRNA level. Specific ablation of Ythdf1 in spinal commissural neurons results in pre-crossing axon guidance defects. Our findings identify a mechanism that YTHDF1-mediated translation of m6A-modified Robo3.1 mRNA controls pre-crossing axon guidance in spinal cord.
INTRODUCTION
The key step of neural circuit formation is the navigation of axons to their targets. The developing axons encounter intermediate targets before reaching their final targets. These intermediate targets, known as ‘choice points’, provide guidance cues for axon pathfinding. One of the most thoroughly characterized model system regarding axon guidance and intermediate targets is spinal commissural axons and floor plate (1). For the past decades, multiple guidance cues (attractive and repulsive) and their receptors have been discovered in this system, and their functions have been examined extensively in mediating initial axon attraction to the floor plate and following repulsion from it (2–4). The divergent member of the Roundabout (Robo) family of axon guidance receptors, Robo3 (5), plays a key role in precisely switching commissural axons from being attracted to being repulsed in vertebrates (6).
Alternative splicing of Robo3 generates two isoforms with different N terminus—Robo3A and Robo3B (7), and two isoforms with distinct C terminal domains—Robo3.1 and Robo3.2 (8). Robo3.1 is expressed in pre-crossing (before crossing the midline) and crossing commissural axons to facilitate crossing by suppressing Slit-mediated repulsion, while Robo3.2 is expressed in post-crossing commissural axons to promote repulsion from midline and block re-crossing (8). What are the mechanisms regulating the spatiotemporal expression of Robo3.1 and Robo3.2 isoforms? Because the ratio of the two isoform transcripts remains constant during commissural axon guidance (E10.5, E11.5 and E12.5) (8), the expression control of Robo3.1 and Robo3.2 isoforms is likely to take place after mRNA splicing (9). Alternative retention of intron 26 during Robo3 mRNA splicing results in a premature stop codon that is not located in the 3′-most exon (8), which makes Robo3.2 mRNA a predicted target of nonsense-mediated decay pathway (10). Our previous studies have shown that Robo3.2 is locally translated in post-crossing commissural axons, and NMD regulates Robo3.2 synthesis by inducing the degradation of Robo3.2 transcript in axons encountering the floor plate (10). However, the mechanisms regulating elimination of Robo3.1 isoform in post-crossing commissural axons remain to be explored (11).
N 6-Methyladenosine (m6A) is the most widely distributed internal modification in mRNA (12–14). m6A modification of mRNA is a dynamic and reversible process which occurs in nuclear speckles where the methyltransferases (‘writers’) such as METTL3/METTL14 complexes and demethylases (‘erasers’) such as FTO and ALKBH5 are concentrated (15). Our recent study also provided an example showing how non-nuclear pool of FTO regulates dynamic m6A modification and local translation of mRNA in axons (16). m6A modification mediates its effects on mRNAs primarily by recruiting proteins, known as ‘readers’ (17). One of the first identified and characterized reader protein family is the YTH domain-containing family protein (YTHDF), including YTHDF1, YTHDF2 and YTHDF3 which are all enriched in the cytoplasm (18–22). YTHDF2 was found to mediate m6A-associated mRNA instability (18), while YTHDF1 was reported to enhance translational efficiency of m6A-modifed mRNAs (19). Interestingly, YTHDF3 was shown to have dual roles which combine features of YTHDF2 and YTHDF1 upon binding to its target m6A-mRNAs (20,22). There are other m6A readers which have been shown to have diverse functions in m6A-modified mRNAs (23–25). However, the targets and functions of m6A readers in nervous system remain to be discovered. Recent studies have shown that m6A modification is detected in various brain regions (26) and can regulate neuronal development such as proliferation and differentiation (27–31), axon regeneration (32), and synapse function (33). However, whether and how m6A modification can regulate axon guidance is still unknown.
In this study, we found that the elimination of Robo3.1 in post-crossing commissural axons was controlled by floor plate through translational regulation. We provided evidence showing Robo3.1 mRNA was modified by m6A and bound by YTHDF1. YTHDF1 could promote Robo3.1 translation in an m6A-depdendent manner because Robo3.1 with m6A sites mutated lost its translational control by YTHDF1. We further showed that expression of YTHDF1 was controlled by floor plate. Using Ythdf1 conditional knockout (cKO) mice, we demonstrated that YTHDF1 was required for Robo3.1 expression and pre-crossing axon pathfinding. These findings reveal a novel mechanism for m6A modification and its reader YTHDF1 to regulate Robo3.1 translation in axon guidance.
MATERIALS AND METHODS
Animals and generation of Ythdf1 cKO mice
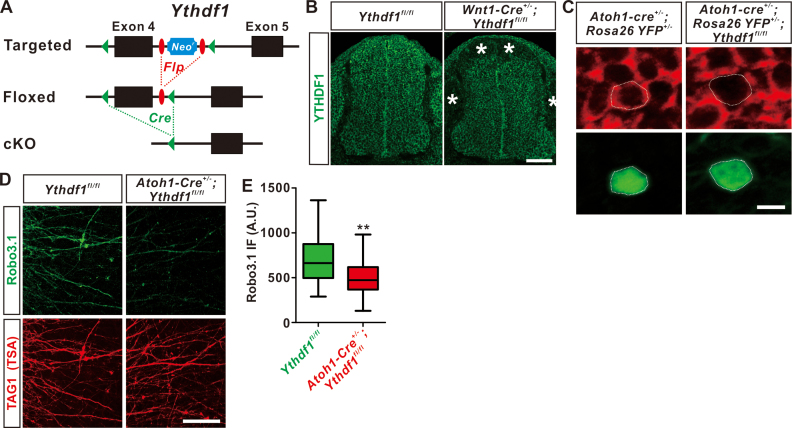
For generation of Ythdf1 conditional knockout (cKO) mice, exon 4 of mouse Ythdf1 gene was targeted with the consideration that exon 4 encodes the YTD domain. A loxP site and an FRT-flanked neomycin resistance gene (Neor) coupled with a loxP site were inserted in intron 3 and intron 4, respectively (Figure 5A). After electroporation, selection and screening for homologous recombination of ES cells, chimeric mice were generated and then crossed with ubiquitous Flp mice to remove Neor via FRT site recombination. The resultant Ythdf1fl/+mice and corresponding Cre mice lines were used to generate Ythdf1 cKO and littermate control embryos. Genotyping primers are as following: the first Ythdf1-loxP site, 5′-TAGTGCATTGTTAAGGCTGTCCTCGT-3′ and 5′-CTGCTGTCTCAAAGCACAAAGCCT-3′; the second Ythdf1-loxP site, 5′-CTTAGAAATCAGTGTTTGTGGCCCA-3′ and 5′-CCTGCCTCAACACACCATTCTCTTT-3′. Atoh1-Cre lines (34,35) and Gli2-/- line (36) were from Jackson Laboratory. Rosa26-mT/mG (37), Rosa26-YFP (38) and Wnt1-Cre (39) mice were from Nanjing Biomedical Research Institute of Nanjing University. For timed pregnancy, embryos were identified as E0.5 when a copulatory plug was observed. To induce Cre activity for Atoh1-CreERT2-derived Ythdf1 cKO in commissural neurons, 8 mg tamoxifen (Cayman Chemical) was given orally to E8.5 pregnant mice with an animal gauge feeding needle. All experiments using mice were carried out following animal protocols approved by the Laboratory Animal Welfare and Ethics Committee of Southern University of Science and Technology.
Figure 5.
Specific ablation of Ythdf1 from dorsal commissural neurons results in decrease of Robo3.1 protein level. (A) Schematic drawings are shown for the genetic deletion strategy for Ythdf1. Exon 4 which contains YTH domain-coding sequence is deleted after Cre-mediated recombination. (B) Depletion of YTHDF1 protein in the dorsal spinal cord of Wnt1-Cre+/-;Ythdf1fl/fl cKO mouse embryos. Anti YTHDF1 immunostaining of E11.5 spinal cord sections confirmed cKO of YTHDF1 protein from dorsal spinal cord and dorsal root ganglia (DRG), illustrated by asterisks. (C) Specific ablation of YTHDF1 protein from Atoh1-Cre+ commissural neurons. Anti YTHDF1 immunostaining of E11.5 spinal cord sections confirmed cKO of YTHDF1 protein in YFP+ commissural neurons in Atoh1-Cre+/-;Rosa26-YFP+/-;Ythdf1fl/fl cKO mouse embryos, while YTHDF1 expression was intact in Atoh1-Cre+/-;Rosa26-YFP+/- control embryos. (D) Ythdf1 cKO with Atoh1-Cre led to dramatic reduction of Robo3.1 protein from dorsal commissural axons. E10.5 pre-crossing DSC explants was dissected and cultured in vitro. Anti-Robo3.1 IF showed significant decline of Robo3.1 protein level in TAG1 (TSA)-positive commissural axons. Representative images are shown from eight Ythdf1fl/fl and nine Atoh1-Cre+/-;Ythdf1fl/fl embryos, respectively. (E) Quantification of Robo3.1 IF in commissural axons of cultured DSC explants from Ythdf1 cKO mouse embryos and their littermate controls. All data are mean ± S.E.M. and represented as box and whisker plots: Ythdf1fl/fl (n = 30 confocal fields) versus Atoh1-Cre+/-;Ythdf1fl/fl (n = 47 confocal fields), **P = 0.0014; by unpaired Student's t test. Scale bars, 100 μm (B and D) and 10 μm (C).
Explant and neuronal culture
All reagents used for neuronal and cell cultures were from Thermo Fisher Scientific (USA) unless otherwise specified. Explants and dissociated neurons of mouse embryonic dorsal spinal cord (DSC) were dissected and cultured following previously described methods (40,41). The culturing medium recipe is neurobasal medium supplemented with B27 (1×), Penicillin-Streptomycin (1×) and GlutaMAX-1 (1×). Netrin-1 (R&D Systems, 250 ng/ml) was added to stimulate outgrowth of commissural axons in DSC explants. Conditioned medium was prepared following previously described methods (42). After DSC explants were cultured for 48 h, the medium was replaced with floor plate-conditioned medium, control conditioned medium plus cycloheximide (CHX) (Sigma, 10 μM) or MG-132 (Selleck, 10 μM). Explants with axons were fixed and analyzed by immunofluorescence 8 h after treatments.
Immunofluorescence and immunostaining
For all immunofluorescence (IF) experiments using cultured DSC explants, dissociated commissural neurons or cell lines, samples were fixed with 4% paraformaldehyde (PFA)/phosphate-buffered saline (PBS). Permeabilization and blocking were done with PBS/0.1% Triton X-100/1% BSA for 20 min at room temperature (RT). Then samples were incubated with primary antibodies overnight at 4°C followed by incubation with secondary antibodies for 1 h at RT. TSA Plus Cyanine 3 System (PerkinElmer) was used to enhance TAG1 IF signals following the manufacturer's manual. For immunostaining of tissues sections, mouse embryonic spinal cords were dissected, fixed, sectioned and stained with antibodies as described previously (43). Detection of Robo3.1 immunoreactivity was performed using the well-described antibody (a gift from Marc Tessier-Lavigne) reported previously (8), and a custom antibody made by Everest Biotech (UK) using the same synthetic peptide (QSQSQRPGRNRREEPR) as immunogen, each of which generated identical immunostaining patterns for Robo3.1 in spinal cord (Supplementary Figure S1C). The dilutions and sources of antibodies are as following: Robo3.1 (1:500), GFP (1:500, Abcam), YTHDF1 (1:1000, Proteintech), Isl1/2 (1:500, DSHB), Lhx2 (1:500, Santa Cruz Biotechnology), Lhx2 (1:500, Abcam), Lhx9 (1:200, Santa Cruz Biotechnology), TAG1 (1:1000, R&D Systems), TAG1 (1:200, DSHB). Alexa Fluor-conjugated secondary antibodies (Thermo Fisher Scientific) were used at 1:1000 (555) or 1:500 (488). Fluorescent images were acquired using laser-scanning confocal microscopes Nikon A1R with NIS software for neurons and explants, and Leica SP8 with LASX software for cell lines. All images were collected with identical settings for each group in the same experiment. Quantification of immunofluorescence signals was performed using ImageJ software for dissociated neurons and cell lines, and using Imaris software for reconstructed 3D confocal images of DSC explants.
Plasmid construction and cell assays
The coding sequence (CDS) of wild-type (WT) Robo3.1 was amplified from E11.5 mouse embryonic cDNA by PCR with following primers: 5′-CGGAATTCATGCTGCGCTACCTGCTTAAAAC-3′ and 5′-TTGGCGCGCCAATGAAGGGTCATCTTGGTTCCTC-3′. The CDS of Robo3.1 with mutated m6A sites (MTm6A: A1505C, A2071T, A2149T, A2199C, A3797C) was synthesized by Genscript (China). pCAG-HA-Robo3.1, pCS2-HA-Robo3.1-WT, and pCS2-HA-Robo3.1-MTm6A were constructed with expression vectors reported previously (44). The CDS of Ythdf1 and Ythdf2 were amplified from E11.5 mouse embryonic cDNA by PCR with following primers: Ythdf1, 5′-GCTTGGCCGGCCAATGTCGGCCACCAGCGTG-3′ and 5′-TTGGCGCGCCAATGGCTTGTTCTTATTGTTTGTTTCG-3′; Ythdf2, 5′-GCTTGGCCGGCCAATGTCGGCCAGCAGCCTC-3′ and 5′-TTGGCGCGCCAATCTATTTCCCACGACCTTGACG-3′. pCAGGS-YTHDF1-IRES-eGFP and pCAGGS-YTHDF2-IRES-eGFP were constructed with an expression vector reported previously (16).
COS-7 or HEK293T cells were cultured in DMEM medium supplemented with 10% FBS, 1× Penicillin-Streptomycin and 1× GlutaMAX-1. Cells were transfected with expression constructs using Lipofectamine 3000 according to the manual. For Robo3.1 half-life assay, COS-7 cells were transfected with pCAG-HA-Robo3.1 and cultured for 48 h before adding CHX. Then cells were collected at different timepoints using RIPA lysis buffer with cOmplete Protease Inhibitor Cocktail (Roche) and protein levels were analyzed by Western Blotting (WB) using antibodies against HA (Abcam) and β-Actin (Abcam). For Robo3.1 and YTHDF1 co-expression assays, COS-7 (Figures 2H, 3A and B) or HEK293T (Figure 3C and D) cells were co-transfected with pCAGGS-YTHDF1-IRES-eGFP and pCS2-HA-Robo3.1-WT or pCS2-HA-Robo3.1-MTm6A. Then cells were harvested 48 h after transfection and analyzed by IF or WB.
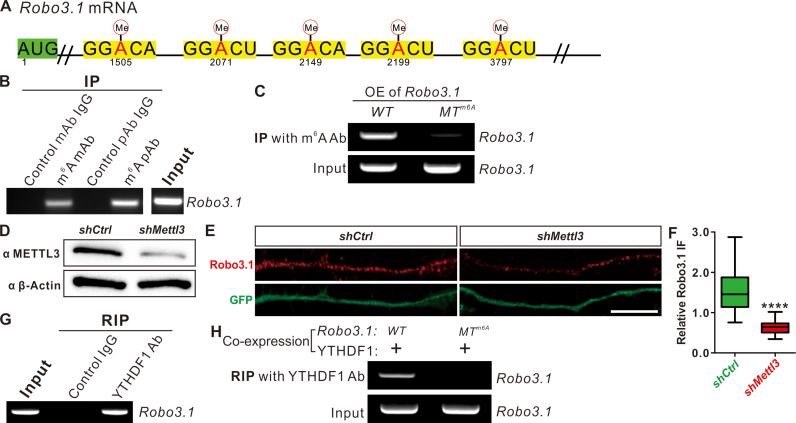
Figure 2.
Robo3.1 mRNA is modified by m6A and bound by the m6A reader YTHDF1. (A) Predicted m6A sites in Robo3.1 mRNA by SRAMP program. (B) Anti-m6A IP pulled down Robo3.1 mRNA from RNA of mouse embryonic spinal cord using two different m6A antibodies (one mAb and another pAb) with corresponding IgG as controls. RT-PCR was performed to detect Robo3.1 mRNA in elutes. (C) Verification of m6A sites in Robo3.1 mRNA. Anti-m6A IP failed to pull down Robo3.1 mRNA from COS-7 cells expressing Robo3.1 with m6A sites mutated (Robo3.1-MTm6A) compared with Robo3.1-WT. (D) Knockdown of METTL3 in commissural neurons. Dissociated commissural neurons from E10.5 mouse dorsal spinal cord was infected with lenti virus expressing shMettl3, marked by eGFP labeling. Knockdown by shMettl3 for 48 h resulted in dramatic decrease of METTL3 protein levels in neurons, compared with shCtrl. (E) Knockdown of METTL3 led to significant decreases of Robo3.1 protein levels in commissural axons compared with shCtrl. Scale bar, 10 μm. (F) Quantification of relative Robo3.1 IF to eGFP in (E). All data are mean ± S.E.M. and are represented as box and whisker plots: ****P = 1.18E–5 (n = 15 axons for shCtrl; n = 19 axons for shMettl3), by unpaired Student's t test. (G) RNA IP (RIP) pulled down Robo3.1 mRNA from mouse embryonic spinal cord lysate with YTHDF1 antibody, but not with control IgG. (H) Binding of YTHDF1 with Robo3.1 mRNA is m6A-dependent. RIP using YTHDF1 antibody failed to pull down Robo3.1 mRNA from COS-7 cells co-expressing YTHDF1 and Robo3.1 with m6A sites mutated (MTm6A) compared with WT Robo3.1.
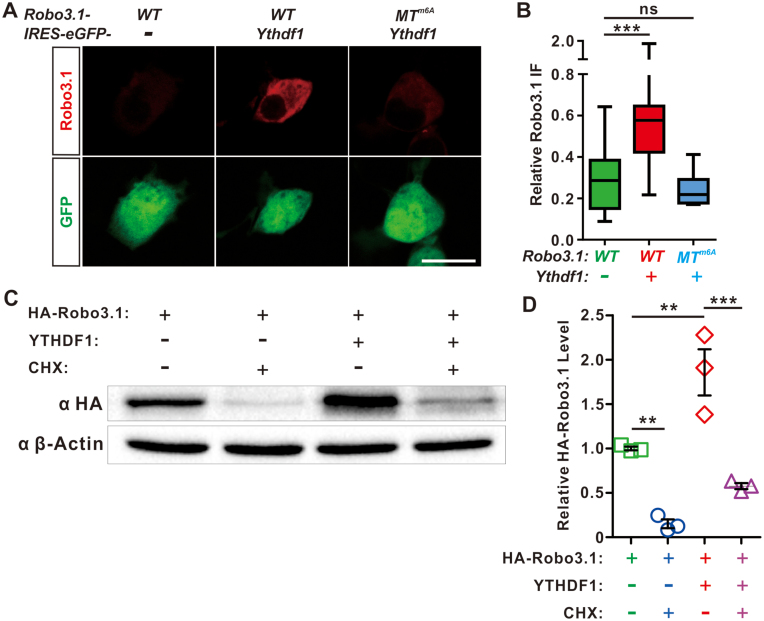
Figure 3.
The m6A reader YTHDF1 controls translation of Robo3.1 mRNA in an m6A-dependent manner. (A) YTHDF1 could enhance translation of Robo3.1. Co-expression of WT Robo3.1 with YTHDF1 in COS-7 cells resulted in a dramatic increase of Robo3.1 protein level by IF, compared with an empty vector expressing eGFP only. However, YTHDF1 failed to increase translation of Robo3.1 with m6A sites mutated (Robo3.1-MTm6A). Scale bar, 25 μm. (B) Quantification of relative Robo3.1 IF to eGFP in (A). (C) Western blotting analysis showing regulation of Robo3.1 protein levels by YTHDF1 is through translational control. Protein synthesis inhibitor CHX blocked translation of Robo3.1 in HEK293T cells expressing HA-Robo3.1. Similar effects were found with cells co-expressing HA-Robo3.1 and YTHDF1. (D) Quantification of relative HA-Robo3.1 levels to β-actin in (C). All data are mean ± S.E.M. Data of IF quantification (B) are represented as box and whisker plots: ***P = 5.11E–04, ‘Robo3.1-WT + IRES-eGFP’ (n = 16 cells) versus ‘Robo3.1-WT + Ythdf1-IRES-eGFP’ (n = 17 cells); ns, not significant (P = 0.41), ‘Robo3.1-WT + IRES-eGFP’ (n = 15 cells) vs ‘Robo3.1-MTm6A + Ythdf1-IRES-eGFP’ (n = 18 cells); by one-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test. WB quantification data (D, n = 3 replicates) are represented as dot plots: **P = 0.002 (‘HA-Robo3.1’ versus ‘HA-Robo3.1 + CHX’); **P = 0.002 (‘HA-Robo3.1’ versus ‘HA-Robo3.1 + YTHDF1’); ***P = 1.41E-04 (‘HA-Robo3.1 + YTHDF1’ versus ‘HA-Robo3.1 + YTHDF1 + CHX’); by one-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test.
RT-qPCR
Total RNA was extracted from tissues or cells with TRIzol Reagent (Thermo Fisher Scientific). 1 ug of total RNA was used for reverse transcription with PrimeScript™ RT Master Mix (Takara). Synthesized cDNA was then subjected to real-time quantitative PCR with 2 × ChamQ™ Universal SYBR qPCR Master Mix (Vazyme) using StepOnePlus™ Real-Time PCR System (ABI). Primers used in qPCR are as following: mouse Robo3.1 used in assays with tissues, 5′-GCTCTACCGCTGGTAGCAAT-3′ and 5′- TGCACAAAACAAGCAGGGAC-3′; mouse GAPDH used in assays with tissues, 5′- CAAGGAGTAAGAAACCCTGGAC-3′ and 5′-GGATGGAAATTGTGAGGGAG-3′; mouse Robo3.1 used in assays with cell lines (COS-7 and HEK293T), 5′-TGGCCCCGTACTCTCCTATC-3′ and 5′-TGGGGGAGTCATCTCTCCAG-3′; African green monkey GADPH for COS-7 assays, 5′-ACAACAGCCTCAAGATCGTCAGC-3′ and 5′-GTGGCAGTGATGGCGTGGAC-3′; human GAPDH for HEK293T assays, 5′-GGAAGGTGAAGGTCGGAGTC-3′ and 5′-TGGAATTTGCCATGGGTGGA-3′.
Anti-m6A immunoprecipitation and RNA immunoprecipitation (RIP)
For anti-m6A immunoprecipitation, total RNA was extracted from mouse embryonic spinal cords and COS-7 cells transfected with pCS2-HA-Robo3.1-WT or pCS2-HA-Robo3.1-MTm6A, respectively. Immunoprecipitation of m6A-modified RNA was carried out using two specific m6A antibodies (pAb from Synaptic Systems #202003; mAb from Abcam #ab190886) with corresponding control IgG following a published protocol (16).
RNA immunoprecipitation (RIP) was performed using EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore) following the manufacturer's protocol with some modifications. Briefly, mouse embryonic DSC tissues or COS-7 cells co-transfected with pCAGGS-YTHDF1-IRES-GFP and pCS2-HA-Robo3.1-WT or pCS2-HA-Robo3.1-MTm6A were homogenized in lysis buffer supplemented with Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific) and RNase inhibitor (Promega). Beads were incubated with YTHDF1 antibody (Proteintech) or normal IgG at room temperature for 0.5 h, and then incubated with lysate supernatant in IP Buffer supplemented with EDTA overnight at 4°C. After extensive washing with IP Buffer, the beads were treated with proteinase K for 0.5 h at 55°C with occasional shaking. RNA was purified from the supernatant using TRIzol Reagent following the manufacturer's instructions. Robo3.1 mRNA levels in the elutes were measured by RT-PCR with primers (mouse Robo3.1, 5′-AGCCTGTTCAAACCCAGGAC-3′ and 5′-TCGATTGAGGTGGAATCGGC-3′) and by RT-qPCR.
Knockdown or overexpression using lentiviral system
The lentiviral knockdown vector pLKO.1-eGFP and the lentiviral overexpression vector pLVX-IRES-eGFP for YTHDF1 were constructed and virus were prepared as described previously (16). Neurons were analyzed by immunofluorescence 48 h after virus infection. The target sequences of shRNA are as following: shYthdf1-2: 5′-GGACATTGGTACTTGGGATAA-3′; shYthdf1-3: 5′-GCACACAACCTCTATCTTTGA-3′; shMettl3: 5′-CGTCAGTATCTTGGGCAAATT-3′; shYthdf3: 5′-GCACCTAAACCAACTTCTTGG-3′; shCtrl: 5′-GCATAAACCCGCCACTCATCT-3′.
DiI tracing of commissural axons
Ythdf1 cKO and littermate control embryos were collected at E11.5. Open-book preparations of embryonic spinal cord in forelimb and thoracic levels were dissected and washed in cold PBS, followed by fixation with cold 4% PFA for 30 min at 4°C. After DiI injection, open-books of spinal cords were left in PBS for at least three days at 4°C, and then mounted and examined using Nikon A1R confocal microscope.
Statistical analysis
Data are mean ± S.E.M. All experiments were conducted at a minimum of three independent biological replicates in the lab. Graphs and statistical analysis were performed using software GraphPad Prism 6.0 and SPSS. The settings for all box and whisker plots are: 25th–75th percentiles (boxes), minimum and maximum (whiskers), and medians (horizontal lines). One-way analysis of variance (ANOVA) with Tukey's post test was employed to the comparison of three or more groups after the homogeneity of variance was tested. Unpaired Student's t test was performed for comparison of changes between two groups except in the experiment of Figure 4H where paired Student's t test was used. P values less than 0.05 were considered as statistically significant: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
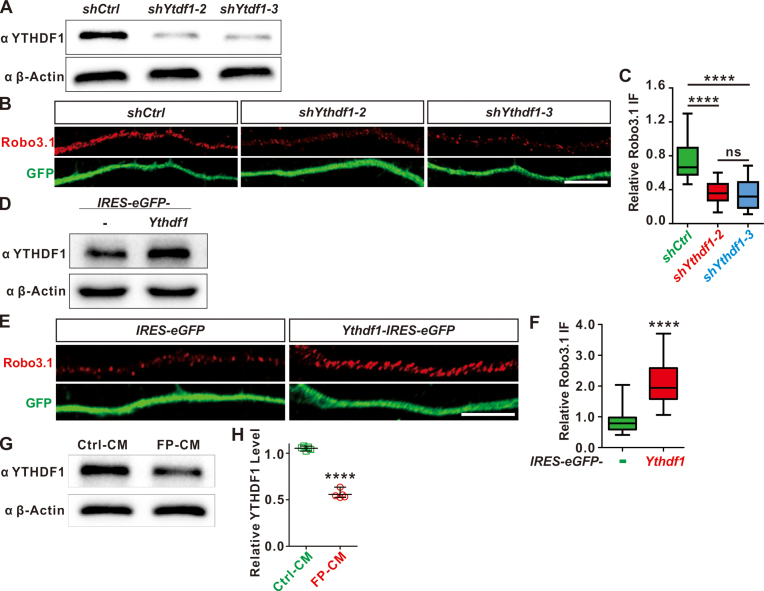
Figure 4.
YTHDF1 regulates translation of endogenous Robo3.1 in commissural neurons and is controlled by floor plate. (A) Knockdown of YTHDF1 in commissural neurons. Dissociated commissural neurons from E10.5 mouse spinal cord was infected with lenti virus expressing shYthdf1-2 and shYthdf1-3, respectively, and marked by eGFP labeling. Knockdown by shYthdf1 for 48 h resulted in dramatic decrease of YTHDF1 protein levels in commissural axons, compared with shCtrl. (B) Knockdown of YTHDF1 led to significant decreases of Robo3.1 protein levels in commissural axons compared with shCtrl. (C) Quantification of relative Robo3.1 IF to eGFP in (B). n = 16 axons for shCtrl; n = 17 axons for shYthdf1-2; n = 16 axons for shYthdf1-3. (D) Overexpression of YTHDF1 in commissural neurons. Dissociated commissural neurons from E10.5 mouse spinal cord was infected with lenti virus expressing YTHDF1, marked by eGFP labeling. Overexpression of YTHDF1 resulted in dramatic increase of YTHDF1 protein levels in commissural axons, compared with control. (E) Overexpression of YTHDF1 led to significant increases of Robo3.1 protein levels in commissural axons compared with eGFP control. (F) Quantification of relative Robo3.1 IF to eGFP in (E). n = 14 axons for IRES-eGFP; n = 16 axons for Ythdf1-IRES-eGFP. (G) Regulation of YTHDF1 expression by floor plate. DSC explants from E10.5 mouse embryonic spinal cords were cultured with FP-CM or Ctrl-CM. WB analysis was carried out to measure YTHDF1 protein levels. (H) Quantification of WB signals in (G). All data are mean ± S.E.M. Data of IF quantification (C, F) are represented as box and whisker plots: For C, ****P = 8.05E-7 (shYthdf1-2 versus shCtrl), ****P = 3.83E-7 (shYthdf1-3 versus shCtrl), ns, not significant (P = 0.76, shYthdf1-2 versus shYthdf1-3); for F, ****P = 3.35E–6; by unpaired Student's t test. WB quantification data (H, n = 5 replicates) are represented as dot plots: ****P = 1.68E–8; by paired Student's t test. Scale bars, 10 μm (B and E).
RESULTS
Elimination of Robo3.1 in post-crossing commissural axons is floor plate-dependent and through translational regulation
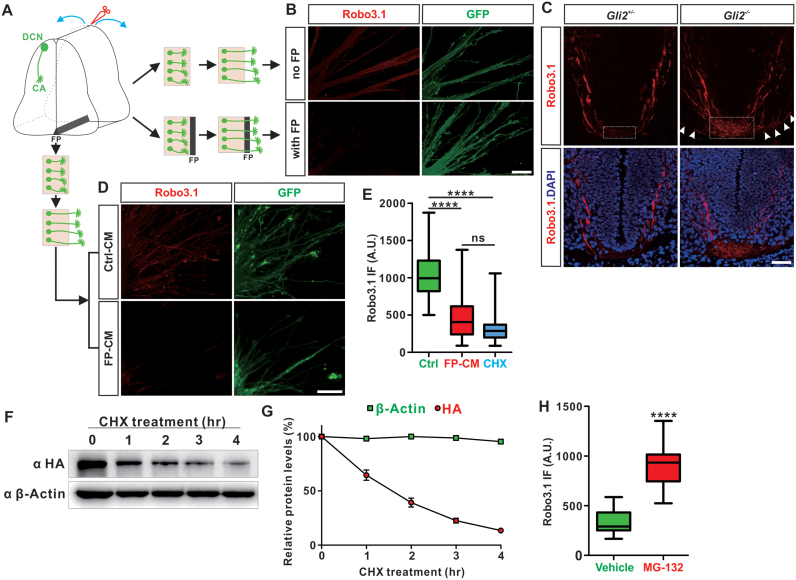
To explore the mechanisms regulating elimination of Robo3.1 from post-crossing spinal commissural axons, we first developed a strategy to label commissural axons (CA). Using a dorsal commissural neuron (DCN)-specific Cre line–Atoh1-Cre and a GFP reporter–Rosa26-mT/mG, we labelled commissural axons with GFP (Supplementary Figure S1A). Co-immunostaining with a Robo3.1-specific antibody (8) showed that Robo3.1 was expressed only in pre-crossing and midline-crossing commissural axons, but not post-crossing axons (Supplementary Figure S1B and C). The correlation of temporal Robo3.1 expression with commissural axon midline crossing stages suggested that the vanishing of Robo3.1 in post-crossing axons might be floor plate-dependent. To test this, we developed an in vitro assay in which the dorsal spinal cord (DSC) explants were dissected from pre-crossing E10.5 spinal cord and cultured with or without floor plate attached (Figure 1A). After commissural axons grew out, we monitored Robo3.1 protein levels in axons. As shown in Figure 1B, Robo3.1 protein was lost in commissural axons growing through floor plate, compared with axons growing out of the explants without contacting floor plate. To further confirm this in vivo, we used a floor plate-deficient spinal cord model by Gli2 knockout (KO) (36). As shown in Figure 1C, Robo3.1 protein level was elevated in ‘crossing’ axons and maintained in ‘post-crossing’ axons in floor plate-deficient spinal cord of Gli2-/- embryos, compared with Gli2+/- control littermates. These results suggest that floor plate is required for elimination of Robo3.1 protein from post-crossing commissural axons. To further test whether floor plate is sufficient to induce elimination of Robo3.1 expression, we carried out another in vitro assay using DSC explants without floor plate attached (Figure 1A). We prepared conditioned medium from floor plate (FP-CM) (42), and then added FP-CM to DSC explants (Figure 1D). Compared with control conditioned medium (Ctrl-CM), FP-CM application resulted in a loss of Robo3.1 protein in axons (Figure 1D). Taken together, these data show that floor plate is necessary as well as sufficient for Robo3.1 elimination in post-crossing commissural axons.
Figure 1.
Elimination of Robo3.1 in post-crossing commissural axons is floor plate-dependent and through translational regulation. (A) Schematic drawings showing in vitro explant culture assays. Dorsal spinal cord (DSC) explants containing dorsal commissural neuron (DCN) from E10.5 spinal cord were dissected and cultured with or without floor plate (FP) till commissural axons grew out. (B) DSC explants of E10.5 Atoh1-Cre+/-;Rosa26-mT/mG+/- embryonic spinal cords were dissected and cultured as shown in (A). Robo3.1 protein was largely lost in commissural axons growing through floor plate while its expression was maintained in explants without floor plate. (C) Robo3.1 expression was examined in the floor plate-deficient Gli2-/- embryos and their heterozygous control littermates at E11.5. As shown, Robo3.1 expression was elevated in crossing commissural axons in Gli2-/- spinal cord (white dotted box) compared with literate control (white dotted box), and maintained in post-crossing commissural axons (white arrowheads). (D) DSC explants of E10.5 Atoh1-Cre+/-;Rosa26-mT/mG+/- embryonic spinal cords without floor plate (as shown in A) were cultured with conditioned medium. Treatment of floor plate-conditioned medium (FP-CM) resulted in dramatic decrease of Robo3.1 in commissural axons compared with Ctrl-CM. (E) Quantification of Robo3.1 immunofluorescence (IF) in commissural axons of cultured DSC explants showing that treatments by FP-CM and protein synthesis inhibitor cycloheximide (CHX) had similar effects in eliminating Robo3.1 protein (n = 24 confocal fields for Ctrl, n = 29 confocal fields for FP-CM, n = 27 confocal fields for CHX). (F) Robo3.1 protein levels were measured by anti-HA WB after HA-Robo3.1 was expressed in COS-7 cells which were collected at different timepoints after CHX treatment. (G) Quantification of results in (F) and calculation of half-life for Robo3.1 protein (n = 3 replicates). (H) Quantification of Robo3.1 IF in commissural axons of cultured DSC explants showing that treatment by proteasome inhibitor MG-132 led to dramatic accumulation of Robo3.1 protein (n = 10 confocal fields for Vehicle, n = 24 confocal fields for MG-132). All data are mean ± S.E.M. Data of IF quantification (E, H) are represented as box and whisker plots. For E: Ctrl versus FP-CM, ****P = 9.06E–09; Ctrl versus CHX, ****P = 5.11E–09; FP-CM versus CHX, ns, not significant (P = 0.21); by one-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test. For H: Vehicle versus MG-132, ****P = 4.46E-11; by unpaired Student's t test. Scale bars, 50 μm (B–D).
Next we wanted to explore how floor plate controlled Robo3.1 protein level. Because Robo3.1 transcript level is not decreased but instead, dramatically increased in crossing and post-crossing stages (E11.5 and E12.5) compared with pre-crossing stage (E10.5) (8), elimination of Robo3.1 protein was not likely due to decrease of transcription, splicing or stability of Robo3.1 mRNA. Indeed, treatment of DSC explants by floor plate-conditioned medium (FP-CM) did not change Robo3.1 mRNA levels (Supplementary Figure S1D), which supported this idea. We hypothesized that there are two possible posttranscriptional mechanisms that could be adopted by floor plate to eliminate Robo3.1 protein. Mechanism 1: Robo3.1 protein has a short half-life and continuous translation is a pre-requisite to maintain Robo3.1 protein level; repression of its translation could result in depletion of this protein rapidly. Mechanism 2: Robo3.1 protein has a long half-life and does not need continuous synthesis; activation of degradation pathway could lead to its elimination.
To distinguish these two models, we carried out the following experiments. Firstly, we treated DSC explants with protein synthesis inhibitor cycloheximide (CHX) and found that Robo3.1 protein in commissural axons was similarly eliminated as FP-CM treatment (Figure 1E). Also similarly as FP-CM, CHX treatment did not change Robo3.1 mRNA levels (Supplementary Figure S1D). These results implied that floor plate regulated Robo3.1 levels through translational regulation. Secondly, we measured the half-life of Robo3.1 protein. HA(hemagglutinin)-tagged Robo3.1 was expressed in COS-7 cells and cell lysate was collected at different timepoints after CHX treatment. Then Robo3.1 protein levels were measured by anti-HA Western Blotting. As shown, Robo3.1 protein was eliminated much more rapidly than β-Actin when protein synthesis was inhibited (Figure 1F and G). The calculated half-life for Robo3.1 protein is 87 ± 4 min, which classifies Robo3.1 as a short-lived protein according to established standards (45,46). Thirdly, we found that treatment of DSC explants with MG-132, a proteasome inhibitor, led to significant accumulation of Robo3.1 protein in commissural axons without affecting Robo3.1 mRNA levels (Figure 1H and Supplementary S1E), supporting that Robo3.1 is continuously synthesized. All together, these data support a model that Robo3.1 protein has a short half-life and its levels are tightly controlled through translational regulation: continuous translation maintains Robo3.1 protein levels in pre-crossing and crossing commissural axons; repression of its translation by floor plate results in a rapid elimination of Robo3.1 protein so commissural axons can exit floor plate, becoming post-crossing axons.
The m6A reader YTHDF1 binds to and controls translation of m6A-modified Robo3.1 mRNA
To further explore the mechanisms regulating Robo3.1 protein in translational level, we tested the involvement of an important posttranscriptional regulation–m6A modification. m6A modification and its readers including YTHDF1, YTHDF2 and YTHDF3 play key roles in regulating mRNA translation and stability (18–22). The tight translational control of Robo3.1 (Figure 1) prompted us to wonder whether this occurs through m6A modification mechanism. To test this, we first checked whether Robo3.1 mRNA is modified by m6A or not. Published m6A mapping data in brain (26,28,29,31,33) did not give us much information about m6A modification of Robo3.1, likely due to the low and restricted expression of Robo3.1. The mammalian m6A site predictor named SRAMP (sequence-based RNA adenosine methylation site predictor) (47) is a powerful tool and has successfully predicted m6A sites in mRNAs (16). Analysis of Robo3.1 mRNA with SRAMP program predicted five High Confidence m6A sites (Figure 2A and Supplementary S2A). We further confirmed this by experiments. Anti-m6A immunoprecipitation using two different m6A antibodies pulled down Robo3.1 mRNA from RNA of mouse embryonic spinal cord (Figure 2B). Mutation of the predicted m6A sites (Figure 2A) resulted in a near complete loss of m6A modification in Robo3.1 mRNA, shown by failure to pull down m6A-mutated Robo3.1 mRNA from COS-7 cells expressing Robo3.1-MTm6A compared with cells expressing Robo3.1-WT (Figure 2C). These results suggest that Robo3.1 mRNA is modified by m6A. To test whether m6A modification is involved in regulation of Robo3.1 translation, we monitored the effects of knocking down the m6A writer METTL3. Dissociated spinal commissural neurons were cultured and then infected with lenti virus expressing shMettl3 which led to dramatic decrease of METTL3 protein levels in commissural neurons compared with control shRNA (Figure 2D). Knockdown of METTL3 led to suppression of Robo3.1 translation which was indicated by significant decline of Robo3.1 protein level in commissural axons (Figure 2E and F), without changing Robo3.1 mRNA levels (Supplementary Figure S2B). These data indicate that m6A modification is required for Robo3.1 translation.
We continued to test whether m6A-modified Robo3.1 mRNA could be recognized and bound by m6A readers. RNA Immunoprecipitation (RIP) of mouse embryonic spinal cord lysate with YTHDF1 antibody detected Robo3.1 mRNA, but not with control IgG (Figure 2G). RIP experiments performed in COS-7 cells co-expressing YTHDF1 and WT Robo3.1 (Robo3.1-WT) detected Robo3.1 mRNA, but not with m6A-mutated Robo3.1 (Robo3.1-MTm6A) (Figure 2H; Supplementary Figure S2C), suggesting that binding of YTHDF1 with Robo3.1 mRNA is m6A-dependent.
YTHDF1 has been shown to increase translational efficiency of m6A-modifed mRNAs (19). Therefore, we wanted to know whether YTHDF1 could regulate translation of Robo3.1. Co-transfection of pCS2-HA-Robo3.1-WT with pCAGGS-Ythdf1-IRES-eGFP into COS-7 cells led to a dramatic increase of Robo3.1 protein levels compared with pCAGGS-IRES-eGFP control (Figure 3A and B), without affecting Robo3.1 mRNA levels (Supplementary Figure S3A), suggesting that YTHDF1 can enhance translation of Robo3.1. Similar experiments were done using pCAGGS-YTHDF2-IRES-eGFP which showed no upregulation of Robo3.1 translation by YTHDF2 (Supplementary Figure S3B and C), suggesting translational regulation of Robo3.1 is an YTHDF1-specific mechanism. Interestingly, this positive regulation of Robo3.1 translation by YTHDF1 was lost when the predicted m6A sites were mutated in Robo3.1 (pCS2-HA-Robo3.1-MTm6A) (Figure 3A and B), indicating this translational regulation is m6A-dependent. To further confirm that upregulation of Robo3.1 protein level by YTHDF1 is mediated by translational control, we used CHX to inhibit Robo3.1 synthesis, which resulted in a decrease of Robo3.1 protein level in HEK293T cells expressing HA-Robo3.1 (Figure 3C and D, the first two conditions with no YTHDF1). Similar inhibition of HA-Robo3.1 synthesis by CHX was found in cells co-expressing YTHDF1 (Figure 3C and D, the last two conditions with YTHDF1), suggesting that YTHDF1 indeed increases Robo3.1 protein levels through translational control, but not other mechanisms such as affecting Robo3.1 protein stability, or Robo3.1 mRNA stability (Supplementary Figure S3D).
Taken together, these data support that Robo3.1 mRNA is modified by m6A, and recognized and bound by YTHDF1 which can enhance its translation.
YTHDF1 regulates translation of endogenous Robo3.1 in commissural neurons and YTHDF1 expression is controlled by floor plate
Next we tested whether YTHDF1 could regulate translation of endogenous Robo3.1 in commissural neurons. Dissociated spinal commissural neurons were cultured and then infected with lenti virus knocking down or overexpressing Ythdf1 (Figure 4A–F). Knocking down YTHDF1 with two shRNAs against Ythdf1 (shYtdhf1-2 and shYthdf1-3) resulted in dramatic decrease of YTHDF1 protein levels in commissural neurons compared with control shRNA (Figure 4A). Because Robo3.1 mRNA was not detected in commissural axons and not locally translated in axons (10), the regulation of Robo3.1 translation will take place in neuronal soma and Robo3.1 protein will be transported to axons to exert its functions. Then we monitored Robo3.1 protein levels in commissural axons. Knockdown of YTHDF1 led to suppression of Robo3.1 translation which was indicated by significant decline of Robo3.1 protein level in commissural axons (Figure 4B and C), with Robo3.1 mRNA levels not affected (Supplementary Figure S4A). Overexpression of YTHDF1 in commissural neurons (Figure 4D) led to significant increases of Robo3.1 protein levels (Figure 4E and F), without changing Robo3.1 mRNA levels (Supplementary Figure S4B). These results suggest that YTHDF1 can enhance translation of endogenous Robo3.1 in commissural neurons.
Floor plate could eliminate Robo3.1 protein from post-crossing commissural axons through translational regulation (Figure 1 and Supplementary S1). The m6A reader YTHDF1 could enhance translation of Robo3.1 in commissural neurons (Figure 4A-F and Supplementary Figure S4). These results prompted us to hypothesize that floor plate might downregulate YTHDF1 to negatively control Robo3.1 translation. To test this, we cultured dissociated DSC neurons and then treated them with floor plate-conditioned medium (FP-CM). As shown in Figure 4G and H, FP-CM treatment significantly reduced YTHDF1 protein levels compared with Ctrl-CM. Consistent with these results, endogenous YTHDF1 expression showed continuous drop from pre-crossing to post-crossing stages (Supplementary Figure S4C and D). These data support a mechanism that floor plate-derived signal(s) downregulate YTHDF1 expression to inhibit Robo3.1 translation in post-crossing commissural axons.
Robo3.1 protein is reduced and axon guidance is disturbed in Ythdf1-deficient commissural neurons
In order to physiologically confirm the mechanisms that YTHDF1 regulates Robo3.1 translation, we generated Ythdf1 cKO mice (Figure 5A). Ythdf1fl/fl mouse was validated using Wnt1-Cre mouse (48) by anti YTHDF1 immunostaining (Figure 5B). As shown in Figure 5B, YTHDF1 is widely expressed in whole spinal cord of Ythdf1fl/fl embryos and is knocked out efficiently in dorsal spinal cord of Wnt1-Cre+/-;Ythdf1fl/fl embryos. However, the ubiquitous expression of YTHDF1 in spinal cord and the wide-range expression of Wnt1-Cre in dorsal spinal cord raise the possibility that embryonic spinal cord development and neural patterning might be disturbed in Wnt1-Cre-derived Ythdf1 cKO embryos. To avoid this, we used Atoh1-Cre mouse to specifically ablate Ythdf1 from spinal commissural neurons (see Supplementary Figure S1A for specificity of Atoh1-Cre). Atoh1-Cre drives Cre-mediated recombination in postmitotic commissural neurons (49), which makes it possible to determine the functions of YTHDF1 in Robo3.1 translational regulation and commissural axon guidance without disturbing neuronal specification in spinal cord. Loss of YTHDF1 from Atoh1-Cre+ commissural neuronal soma which was indicated by a Rosa26-YFP reporter was validated (Figure 5C). Robo3.1 protein level was significantly reduced in commissural axons growing from Ythdf1-deficient DSC explants compared with littermate controls (Figure 5D and E). Robo3.1 mRNA level was not affected in spinal cord of Ythdf1 cKO embryos (Supplementary Figure S5A). These results demonstrate that Robo3.1 protein but not Robo3.1 mRNA is reduced in spinal cord of Ythdf1 cKO embryos, which are consistent with the in vitro data, suggesting that YTHDF1 physiologically regulates Robo3.1 translation.
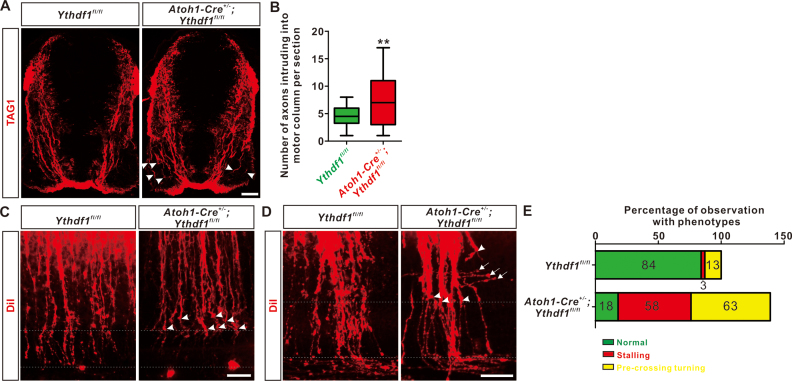
Loss-of-function of Robo3.1 led to defects in pre-crossing commissural axon guidance (8). The fact that Robo3.1 protein is reduced in commissural Ythdf1-deficient embryos prompted us to further test whether pre-crossing axon guidance was disturbed in these embryos. Immunostaining of TAG1, a pre-crossing commissural axon marker in spinal cord sections showed that there were significantly more commissural axons misprojecting to motor column in Ythdf1 cKO embryos compared with their littermate controls (Figure 6A and B). Further analysis using DiI labeling in open-book of spinal cord showed that ablation of Ythdf1 from commissural neurons caused pre-crossing axon guidance defects, which was indicated by many abnormal premature turning and stalling of pre-crossing axons in Ythdf1 cKO embryos compared with their littermate controls (Figure 6C–E). As controls, we confirmed that patterning of spinal cord and development of dI1 commissural neurons were not affected in Ythdf1 cKO embryos (Supplementary Figure S5B-I).
Figure 6.
Ythdf1 cKO embryos exhibit defects in pre-crossing commissural axon guidance. (A) Misprojection of pre-crossing commissural axons into motor columns in Ythdf1 cKO embryos. TAG1 marks commissural axons in E11.5 embryonic sections and there are significantly more misprojecting axons into motor columns (arrowheads) in Ythdf1 cKO embryos compared with their littermate controls. Representative images are shown from three Ythdf1fl/fl and three Atoh1-Cre+/-;Ythdf1fl/fl embryos, respectively. (B) Quantification of commissural axons intruding into motor columns. All data are mean ± S.E.M. and represented as box and whisker plots: Ythdf1fl/fl (n = 15 sections) versus Atoh1-Cre+/-;Ythdf1fl/fl (n = 12 sections), **P = 0.0096; by unpaired Student's t test. (C and D) DiI tracing of commissural axons with open-book preparations. Pre-crossing axon guidance defects including stalling (arrowheads) and pre-crossing turning (arrows) were observed in Ythdf1 cKO embryos. Representative images of E10.5–11 (C) and E11.5 (D) were shown. (E) Quantification of phenotypes in (C and D). Total 31 DiI injections with four Ythdf1fl/fl embryos and 40 DiI injections with three Atoh1-Cre+/-;Ythdf1fl/fl embryos were analyzed. Percentage of observations with phenotypes was calculated. Note that the summed percentage for Atoh1-Cre+/-;Ythdf1fl/fl is >100 because some of DiI injections were found with both stalling and pre-crossing turning phenotypes. Scale bars, 50 μm (A, C and D).
Taken together, these results suggest that YTHDF1 can physiologically regulate Robo3.1 translation and consequently control guidance of pre-crossing commissural axons in embryonic spinal cord.
DISCUSSION
Posttranscriptional regulation plays important roles in axon guidance, and diverse RNA-binding proteins have been shown to be involved in these mechanisms (50). For example, RNA-binding protein IMP2 is localized in spinal commissural axons and could regulate axon pathfinding by controlling local translation of axon guidance-related mRNAs in axons (51). RNA-binding protein Nova controls axon guidance by regulating alternative splicing of Dcc in spinal cord (52), and a set of axon guidance related genes in different brain regions (53). RNA-binding protein Msi1 has been shown to bind Robo3 mRNA and promote its translation (54). In the hindbrain of Msi1 knockout mice, midline crossing of precerebellar neurons and axons are severely impaired (54). Surprisingly, in Msi1-deficient spinal cord, Robo3 expression and midline crossing of commissural axons were not affected (54). Thus there might be other molecules and mechanisms exerting post-transcriptional regulation of Robo3 expression in spinal cord.
In the present study, we found that Robo3.1 protein has a short half-life and maintenance of its protein levels requires continuous translation of Robo3.1 mRNA. Robo3.1 protein is depleted from post-crossing commissural axons because its translation is inhibited in a floor plate-dependent manner. We further provided evidence showing that Robo3.1 mRNA is modified by m6A and bound by the m6A reader YTHDF1, an RNA-binding protein which positively regulates translation of Robo3.1 mRNA. Floor plate controls elimination of Robo3.1 protein from post-crossing commissural axons by downregulating YTHDF1 expression. We generated Ythdf1 cKO embryos in which Robo3.1 protein level is reduced in spinal commissural axons and consequently axon guidance is disturbed in pre-crossing commissural axons.
We did observe that the axon guidance defects in Ythdf1 cKO using Atoh1-Cre were less severe compared with Robo3 mutants which showed a marked reduction of commissure thickness (55). We think the following reasons may explain this phenotype difference. Robo3 mutant is a complete knockout of Robo3 itself and has all neurons expressing Robo3 affected. As for Ythdf1 cKO using Atoh1-Cre, Robo3 gene itself is intact and one of its splicing isoform—Robo3.1 is affected only in translational level (indeed, Robo3.1 protein level is reduced but not completely lost in Ythdf1 cKO embryos) and only in a small population of dorsal spinal neurons which express Atoh1-Cre (Supplementary Figure S1A). Nevertheless, we observed significant defects in pre-crossing axon guidance in Ythdf1 cKO embryos in which Robo3.1 translation was impaired.
Our previous studies demonstrated that Robo3.1 mRNA was not detected in commissural axons and was not locally translated in axons (10). Thus the regulation of Robo3.1 translation by YTHDF1 likely takes place in commissural neuronal soma. The current study and our previous findings (10) suggest that the spatiotemporal expression of Robo3.1 and Robo3.2 are under control of different mechanisms: Robo3.2 mRNA which is a target of non-sense mediated decay (NMD) is locally translated in post-crossing commissural axons while translation of Robo3.1 mRNA which is modified by m6A is controlled by m6A reader YTHDF1 in pre-crossing neuronal soma.
Developing axons encounter intermediate targets before reaching their final targets. In addition to providing guidance cues for axon navigation, these targets can also regulate neuronal development, neural circuit formation and regeneration by activating retrograde signals (56–58). Target tissue-derived signals identified so far include neurotrophins (including BDNF, NGF, NT-3), growth factors (including BMP, FGF, progranulin), and axon guidance cues (including semaphorins, Slits) (59–65). In the present study, our experiments and results support such a model that spinal cord-derived signal(s) downregulates YTHDF1 to finely control the timing of Robo3.1 elimination from post-crossing commissural axons. It would be interesting to test known signal molecules derived from spinal cord to see whether they are responsible for reduction of YTHDF1 expression.
YTHDF1 expression was only mildly down-regulated in post-crossing or floor plate-conditioned medium-treated DSC neurons (Supplementary Figure S4C and D; Figure 4G and H) while Robo3.1 protein is lost in post-crossing (8) or floor plate-conditioned medium-treated commissural axons (Figure 1D). In addition to YTHDF1 downregulation in post-crossing commissural neurons, are there additional mechanisms in m6A modification pathway which help facilitate efficient blocking of Robo3.1 translation? We further tested whether m6A modification of Robo3.1 mRNA is changed in commissural neurons during midline crossing. We performed anti m6A IP and found that m6A modification levels of Robo3.1 mRNA also decreased through E10.5 to E12.5 (Supplementary Figure S6A). These data support a coincident mechanism that decreases of both m6A modification of Robo3.1 mRNA and its reader YTHDF1 in post-crossing commissural neurons ensures an efficient inhibition of Robo3.1 translation in post-crossing commissural axons. We continued to check the possible involvement of other m6A readers. The current working model for YTHDF2 is that it causes instability of its target m6A-modified mRNAs (18), and eventually leads to down-regulation of translation. Thus YTHDF2 is not compatible with Robo3.1 translational control which is enhanced by m6A modification. As for YTHDF3, we found that knockdown of YTHDF3 did not change Robo3.1 protein or mRNA levels (Supplementary Figure S6B–E), suggesting that it is not involved in regulation of Robo3.1 translation.
Growing studies have shown that m6A modification plays important roles in neuronal development and regeneration. However, how m6A modification works through its readers and what are the neuronal target mRNAs for m6A readers remain to be investigated. Here, we provided evidence showing how the m6A reader YTHDF1 physiologically regulates expression of an important guidance molecule Robo3.1 and controls axon guidance. In addition to Robo3.1 mRNA, it would be interesting to identify m6A-modified neural mRNAs targeted by YTHDF1 (and other m6A readers as well) in the transcriptomic level. The following characterization of these mRNAs will help elucidate functions and mechanisms of m6A modification in nervous system.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Samie R. Jaffrey for instructions, discussions and supports when initiating this project; members of Ji lab for technical support, helpful discussions and comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Science Fund of Guangdong Province [2016A030313638 to S.-J.J.]; Basic Research Grant [JCYJ20160331115633182 to S.-J.J.] and Technology Innovation Grant of Peacock Plan [KQJSCX2017032815460815 to S.-J.J.] from Science and Technology Innovation Commission of Shenzhen Municipal Government. S.-J.J. is a scholar in China Thousand Talent Program for Young Outstanding Scientists. Running cost of Ji lab was covered by startup funds from Southern University of Science and Technology and Peacock Plan of Shenzhen Municipal Government. Funding for open access charge: Basic Research Grant from Science and Technology Innovation Commission of Shenzhen Municipal Government.
Conflict of interest statement. None declared.
REFERENCES
- 1. Colamarino S.A., Tessier-Lavigne M.. The role of the floor plate in axon guidance. Annu. Rev. Neurosci. 1995; 18:497–529. [DOI] [PubMed] [Google Scholar]
- 2. Chedotal A. Further tales of the midline. Curr. Opin. Neurobiol. 2011; 21:68–75. [DOI] [PubMed] [Google Scholar]
- 3. Nawabi H., Castellani V.. Axonal commissures in the central nervous system: how to cross the midline?. Cell Mol. Life Sci. 2011; 68:2539–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Ramon Francas G., Zuniga N.R., Stoeckli E.T.. The spinal cord shows the way - how axons navigate intermediate targets. Dev. Biol. 2017; 432:43–52. [DOI] [PubMed] [Google Scholar]
- 5. Sabatier C., Plump A.S., Le M., Brose K., Tamada A., Murakami F., Lee E.Y., Tessier-Lavigne M.. The divergent Robo family protein rig-1/Robo3 is a negative regulator of slit responsiveness required for midline crossing by commissural axons. Cell. 2004; 117:157–169. [DOI] [PubMed] [Google Scholar]
- 6. Beamish I.V., Kennedy T.E.. Robo3: the road taken. Dev. Cell. 2015; 32:3–4. [DOI] [PubMed] [Google Scholar]
- 7. Camurri L., Mambetisaeva E., Davies D., Parnavelas J., Sundaresan V., Andrews W.. Evidence for the existence of two Robo3 isoforms with divergent biochemical properties. Mol. Cell Neurosci. 2005; 30:485–493. [DOI] [PubMed] [Google Scholar]
- 8. Chen Z., Gore B.B., Long H., Ma L., Tessier-Lavigne M.. Alternative splicing of the Robo3 axon guidance receptor governs the midline switch from attraction to repulsion. Neuron. 2008; 58:325–332. [DOI] [PubMed] [Google Scholar]
- 9. Black D.L., Zipursky S.L.. To cross or not to cross: alternatively spliced forms of the Robo3 receptor regulate discrete steps in axonal midline crossing. Neuron. 2008; 58:297–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Colak D., Ji S.J., Porse B.T., Jaffrey S.R.. Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell. 2013; 153:1252–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Friocourt F., Chedotal A.. The Robo3 receptor, a key player in the development, evolution, and function of commissural systems. Dev. Neurobiol. 2017; 77:876–890. [DOI] [PubMed] [Google Scholar]
- 12. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al.. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 13. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roundtree I.A., Evans M.E., Pan T., He C.. Dynamic RNA modifications in gene expression regulation. Cell. 2017; 169:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee M., Kim B., Kim V.N.. Emerging roles of RNA modification: m(6)A and U-tail. Cell. 2014; 158:980–987. [DOI] [PubMed] [Google Scholar]
- 16. Yu J., Chen M., Huang H., Zhu J., Song H., Zhu J., Park J., Ji S.J.. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 2018; 46:1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patil D.P., Pickering B.F., Jaffrey S.R.. Reading m(6)A in the Transcriptome: m(6)A-Binding proteins. Trends Cell Biol. 2018; 28:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. et al.. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C.. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015; 161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li A., Chen Y.S., Ping X.L., Yang X., Xiao W., Yang Y., Sun H.Y., Zhu Q., Baidya P., Wang X. et al.. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017; 27:444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.B.. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015; 526:591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C.. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017; 27:315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.B., Jaffrey S.R.. 5′ UTR m(6)A promotes Cap-Independent translation. Cell. 2015; 163:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kennedy E.M., Bogerd H.P., Kornepati A.V., Kang D., Ghoshal D., Marshall J.B., Poling B.C., Tsai K., Gokhale N.S., Horner S.M. et al.. Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe. 2016; 19:675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xiao W., Adhikari S., Dahal U., Chen Y.S., Hao Y.J., Sun B.F., Sun H.Y., Li A., Ping X.L., Lai W.Y. et al.. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 2016; 61:507–519. [DOI] [PubMed] [Google Scholar]
- 26. Chang M., Lv H., Zhang W., Ma C., He X., Zhao S., Zhang Z.W., Zeng Y.X., Song S., Niu Y. et al.. Region-specific RNA m6A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 2017; 7:170166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma C., Chang M., Lv H., Zhang Z.W., Zhang W., He X., Wu G., Zhao S., Zhang Y., Wang D. et al.. RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018; 19:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang C.X., Cui G.S., Liu X., Xu K., Wang M., Zhang X.X., Jiang L.Y., Li A., Yang Y., Lai W.Y. et al.. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 2018; 16:e2004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y., Li Y., Yue M., Wang J., Kumar S., Wechsler-Reya R.J., Zhang Z., Ogawa Y., Kellis M., Duester G. et al.. N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 2018; 21:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li M., Zhao X., Wang W., Shi H., Pan Q., Lu Z., Perez S.P., Suganthan R., He C., Bjoras M. et al.. Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 2018; 19:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yoon K.J., Ringeling F.R., Vissers C., Jacob F., Pokrass M., Jimenez-Cyrus D., Su Y., Kim N.S., Zhu Y., Zheng L. et al.. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell. 2017; 171:877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weng Y.L., Wang X., An R., Cassin J., Vissers C., Liu Y., Liu Y., Xu T., Wang X., Wong S.Z.H. et al.. Epitranscriptomic m(6)A regulation of axon regeneration in the adult mammalian nervous system. Neuron. 2018; 97:313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Merkurjev D., Hong W.T., Iida K., Oomoto I., Goldie B.J., Yamaguti H., Ohara T., Kawaguchi S.Y., Hirano T., Martin K.C. et al.. Synaptic N(6)-methyladenosine (m(6)A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 2018; 21:1004–1014. [DOI] [PubMed] [Google Scholar]
- 34. Machold R., Fishell G.. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005; 48:17–24. [DOI] [PubMed] [Google Scholar]
- 35. Matei V., Pauley S., Kaing S., Rowitch D., Beisel K.W., Morris K., Feng F., Jones K., Lee J., Fritzsch B.. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev. Dyn. 2005; 234:633–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bai C.B., Joyner A.L.. Gli1 can rescue the in vivo function of Gli2. Development. 2001; 128:5161–5172. [DOI] [PubMed] [Google Scholar]
- 37. Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L.. A global double-fluorescent Cre reporter mouse. Genesis. 2007; 45:593–605. [DOI] [PubMed] [Google Scholar]
- 38. Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F.. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001; 1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Danielian P.S., Muccino D., Rowitch D.H., Michael S.K., McMahon A.P.. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 1998; 8:1323–1326. [DOI] [PubMed] [Google Scholar]
- 40. Langlois S.D., Morin S., Yam P.T., Charron F.. Dissection and culture of commissural neurons from embryonic spinal cord. J. Vis. Exp. 2010; e1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moore S.W., Kennedy T.E.. Dissection and culture of embryonic spinal commissural neurons. Curr.Protoc. Neurosci. 2008; doi:10.1002/0471142301.ns0320s45. [DOI] [PubMed] [Google Scholar]
- 42. Nawabi H., Briancon-Marjollet A., Clark C., Sanyas I., Takamatsu H., Okuno T., Kumanogoh A., Bozon M., Takeshima K., Yoshida Y. et al.. A midline switch of receptor processing regulates commissural axon guidance in vertebrates. Genes Dev. 2010; 24:396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ji S.J., Zhuang B., Falco C., Schneider A., Schuster-Gossler K., Gossler A., Sockanathan S.. Mesodermal and neuronal retinoids regulate the induction and maintenance of limb innervating spinal motor neurons. Dev Biol. 2006; 297:249–261. [DOI] [PubMed] [Google Scholar]
- 44. Ji S.J., Periz G., Sockanathan S.. Nolz1 is induced by retinoid signals and controls motoneuron subtype identity through distinct repressor activities. Development. 2009; 136:231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen W., Smeekens J.M., Wu R.. Systematic study of the dynamics and half-lives of newly synthesized proteins in human cells. Chem. Sci. 2016; 7:1393–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jiang X., Coffino P., Li X.. Development of a method for screening short-lived proteins using green fluorescent protein. Genome Biol. 2004; 5:R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou Y., Zeng P., Li Y.H., Zhang Z., Cui Q.. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016; 44:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Charron F., Stein E., Jeong J., McMahon A.P., Tessier-Lavigne M.. The morphogen sonic hedgehog is an axonal chemoattractant that collaborates with netrin-1 in midline axon guidance. Cell. 2003; 113:11–23. [DOI] [PubMed] [Google Scholar]
- 49. Yamauchi K., Phan K.D., Butler S.J.. BMP type I receptor complexes have distinct activities mediating cell fate and axon guidance decisions. Development. 2008; 135:1119–1128. [DOI] [PubMed] [Google Scholar]
- 50. Stoeckli E. Where does axon guidance lead us [version 1; referees: 2 approved]. F1000Res. 2017; 6:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Preitner N., Quan J., Li X., Nielsen F.C., Flanagan J.G.. IMP2 axonal localization, RNA interactome, and function in the development of axon trajectories. Development. 2016; 143:2753–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leggere J.C., Saito Y., Darnell R.B., Tessier-Lavigne M., Junge H.J., Chen Z.. NOVA regulates Dcc alternative splicing during neuronal migration and axon guidance in the spinal cord. Elife. 2016; 5:e14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Saito Y., Miranda-Rottmann S., Ruggiu M., Park C.Y., Fak J.J., Zhong R., Duncan J.S., Fabella B.A., Junge H.J., Chen Z. et al.. NOVA2-mediated RNA regulation is required for axonal pathfinding during development. Elife. 2016; 5:e14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kuwako K., Kakumoto K., Imai T., Igarashi M., Hamakubo T., Sakakibara S., Tessier-Lavigne M., Okano H.J., Okano H.. Neural RNA-binding protein Musashi1 controls midline crossing of precerebellar neurons through posttranscriptional regulation of Robo3/Rig-1 expression. Neuron. 2010; 67:407–421. [DOI] [PubMed] [Google Scholar]
- 55. Jaworski A., Long H., Tessier-Lavigne M.. Collaborative and specialized functions of Robo1 and Robo2 in spinal commissural axon guidance. J Neurosci. 2010; 30:9445–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. da Silva S., Wang F.. Retrograde neural circuit specification by target-derived neurotrophins and growth factors. Curr. Opin. Neurobiol. 2011; 21:61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rishal I., Fainzilber M.. Axon-soma communication in neuronal injury. Nat. Rev. Neurosci. 2014; 15:32–42. [DOI] [PubMed] [Google Scholar]
- 58. Tao H.W., Poo M.. Retrograde signaling at central synapses. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:11009–11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ji S.J., Jaffrey S.R.. Intra-axonal translation of SMAD1/5/8 mediates retrograde regulation of trigeminal ganglia subtype specification. Neuron. 2012; 74:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hodge L.K., Klassen M.P., Han B.X., Yiu G., Hurrell J., Howell A., Rousseau G., Lemaigre F., Tessier-Lavigne M., Wang F.. Retrograde BMP signaling regulates trigeminal sensory neuron identities and the formation of precise face maps. Neuron. 2007; 55:572–586. [DOI] [PubMed] [Google Scholar]
- 61. Sharma N., Deppmann C.D., Harrington A.W., St Hillaire C., Chen Z.Y., Lee F.S., Ginty D.D.. Long-distance control of synapse assembly by target-derived NGF. Neuron. 2010; 67:422–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Patel T.D., Kramer I., Kucera J., Niederkofler V., Jessell T.M., Arber S., Snider W.D.. Peripheral NT3 signaling is required for ETS protein expression and central patterning of proprioceptive sensory afferents. Neuron. 2003; 38:403–416. [DOI] [PubMed] [Google Scholar]
- 63. Terauchi A., Johnson-Venkatesh E.M., Bullock B., Lehtinen M., Umemori H.. Retrograde fibroblast growth factor 22 (FGF22) signaling regulates insulin-like growth factor 2 (IGF2) expression for activity-dependent synapse stabilization in the mammalian brain. Elife. 2016; 5:e12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Uesaka N., Uchigashima M., Mikuni T., Nakazawa T., Nakao H., Hirai H., Aiba A., Watanabe M., Kano M.. Retrograde semaphorin signaling regulates synapse elimination in the developing mouse brain. Science. 2014; 344:1020–1023. [DOI] [PubMed] [Google Scholar]
- 65. Wu H., Barik A., Lu Y., Shen C., Bowman A., Li L., Sathyamurthy A., Lin T.W., Xiong W.C., Mei L.. Slit2 as a beta-catenin/Ctnnb1-dependent retrograde signal for presynaptic differentiation. Elife. 2015; 4:e07266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.