Abstract
Background
CD34+ stem/progenitor cells are involved in vascular homeostasis and in neovascularization of ischemic tissues. The number of circulating CD34+ stem cells is a predictive biomarker of adverse cardiovascular outcomes in diabetic patients. Here, we provide evidence that hyperglycemia can be “memorized” by the stem cells through epigenetic changes that contribute to onset and maintenance of their dysfunction in diabetes mellitus.
Methods and Results
Cord‐blood–derived CD34+ stem cells exposed to high glucose displayed increased reactive oxygen species production, overexpression of p66shc gene, and downregulation of antioxidant genes catalase and manganese superoxide dismutase when compared with normoglycemic cells. This altered oxidative state was associated with impaired migration ability toward stromal‐cell–derived factor 1 alpha and reduced protein and mRNA expression of the C‐X‐C chemokine receptor type 4 (CXCR4) receptor. The methylation analysis by bisulfite Sanger sequencing of the CXCR4 promoter revealed a significant increase in DNA methylation density in high‐glucose CD34+ stem cells that negatively correlated with mRNA expression (Pearson r=−0.76; P=0.004). Consistently, we found, by chromatin immunoprecipitation assay, a more transcriptionally inactive chromatin conformation and reduced RNA polymerase II engagement on the CXCR4 promoter. Notably, alteration of CXCR4 DNA methylation, as well as transcriptional and functional defects, persisted in high‐glucose CD34+ stem cells despite recovery in normoglycemic conditions. Importantly, such an epigenetic modification was thoroughly confirmed in bone marrow CD34+ stem cells isolated from sternal biopsies of diabetic patients undergoing coronary bypass surgery.
Conclusions
CD34+ stem cells “memorize” the hyperglycemic environment in the form of epigenetic modifications that collude to alter CXCR4 receptor expression and migration.
Keywords: cardiovascular complications, CD34 stem cells, CXCR4, diabetes mellitus, DNA methylation, histone modifications, metabolic memory
Subject Categories: Stem Cells, Vascular Biology, Basic Science Research, Pathophysiology, Coronary Artery Disease
Clinical Perspective
What Is New?
Human CD34+ stem cells “memorize” metabolic stress in the form of epigenetic changes that contribute to the self‐perpetuating alteration of gene expression.
Epigenetic mechanisms might contribute to C‐X‐C chemokine receptor type 4/stromal‐cell–derived factor 1 alpha axis impairment of CD34+ stem cells in diabetic patients, which has a critical function in revascularization processes.
What Are the Clinical Implications?
Epigenetic mechanisms contribute to the self‐perpetuating alterations of genes involved in progenitor cell function, with direct implications on tissue homeostasis and repair in diabetes mellitus.
Presence of molecular modifications potentially inherited by daughter cells dramatically reduce the translational potential of these cells in regenerative medicine.
Investigation of epigenetic mechanisms underlying metabolic memory may be relevant in understanding the barriers that prevent the pharmacological regression of diabetes mellitus cardiovascular complications despite optimal management of hyperglycemia.
Introduction
Diabetes mellitus (DM) is a chronic and complex disease characterized by hyperglycemia and associated with a broad spectrum of micro‐ and macrovascular complications that constitute a major health burden in Western countries.1 Despite the progresses of pharmacological therapy, the cellular and molecular mechanisms underlying diabetic complications are not entirely understood and the associated cardiovascular risk is still far from eliminated.2, 3
Endothelial dysfunction induced by hyperglycemia is recognized as the major causal factor in the development and progression of diabetic vascular complications. Since the early 2000s, numerous studies showed a linear correlation between the decline in number and function of circulating progenitor cells such as CD34+ cells, severity of diabetic disease, and presence of vascular complications.4, 5 CD34+ stem cells are bone marrow (BM)‐derived stem cells, both representative for hematopoietic stem cells and endothelial progenitor cells contributing to preservation of an intact endothelial layer and neorevascularization of ischemic tissues.6, 7 The dramatic numeric and functional decay of these cells in the diabetic metabolic environment8, 9 is linked to severe endothelial dysfunction and elevated risk of adverse cardiovascular events. Consistently, level of the circulating CD34+ stem cell population has recently proved to be an efficient clinical‐grade–independent biomarker of cardiovascular risk in diabetic patients, capable of predicting long‐term development or progression of microangiopathy and cardiovascular events in type 2 DM (T2DM).10
A consistent body of literature has shown that diabetic patients can develop diabetic vascular complications even after intensive glycemic control, a phenomenon known as “metabolic memory.”11, 12, 13, 14 Hyperglycemia appears to be remembered in organs, such as blood vessels, heart, kidneys, and eyes, and the underlying mechanisms seem to rely on epigenetics. Epigenetics plays a critical role in regulating tissue‐specific gene expression; hence, alterations in its mechanisms may induce long‐term changes in gene function and metabolism, which can persist after the return to normality. Mechanistically, recent studies clearly indicated that DM drives epigenetic changes, such as DNA methylation and histone modifications, that result in aberrant and long‐lasting expression of pathological genes in a plethora of cell types, such as vascular smooth muscle cells, endothelial cells, and leukocytes,15, 16, 17, 18, 19, 20 which may play a substantial role in the pathophysiology of vascular complications associated with DM. Persistence of functional defects in a variety of progenitor cell populations, described in both preclinical and human DM after glucose normalization, suggests the presence of this phenomenon also in stem cells.21 However, to date, no studies have established, at a single‐gene level, the implication of epigenetic mechanisms in hyperglycemia‐induced dysfunction of CD34+ stem cells. Here, we investigated, by an in vitro model, whether epigenetic mechanisms contribute to C‐X‐C chemokine receptor type 4 (CXCR4)/stromal‐cell–derived factor 1 alpha (SDF‐1α) axis impairment, which has a critical function in revascularization processes, in diabetic patients.
We show, for the first time, that umbilical cord blood (UCB)‐derived CD34+ stem cell alterations induced in vitro by hyperglycemia are not completely reversible and that epigenetic modifications resulting from high‐glucose (HG) challenge are involved in persistent downregulation of the CXCR4 gene and impaired migratory capacity despite recovery to normoglycemia (NG). Moreover, we have confirmed identical epigenetic findings in BM‐CD34+ stem cells isolated from sternal biopsies from diabetic patients undergoing coronary bypass surgery.
Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request. Additional methods can be found in Data S1.
Study Design
We aimed the present study at investigating the epigenetic mechanisms contributing to the dysfunctional phenotype of CD34+ stem cells in diabetic patients. To avoid readout misinterpretations derived from aging or other risk factors, we first evaluated the effects of HG on the epigenetic makeup of naïve UCB‐derived CD34+ stem cells (Figure 1). This allowed the discrimination between epigenetic modifications established as a direct consequence of hyperglycemia exposure, rather than those related to confounding variables such as age and cardiovascular risk factors. As depicted in Figure 1, we expanded CD34+ stem cells in HG conditions until CD34+ stem cells lost glucose tolerance. Afterward, in order to reproduce metabolic memory in vitro, cells were recovered in NG for 3 days, as previously described.22 We then performed molecular and epigenetic analyses of the gene(s) involved in CD34+ stem cell dysfunction upon HG and NG recovery. Results have been validated in BM‐derived CD34+ stem cells isolated from patients with coronary artery disease (CAD)±DM.
Figure 1.
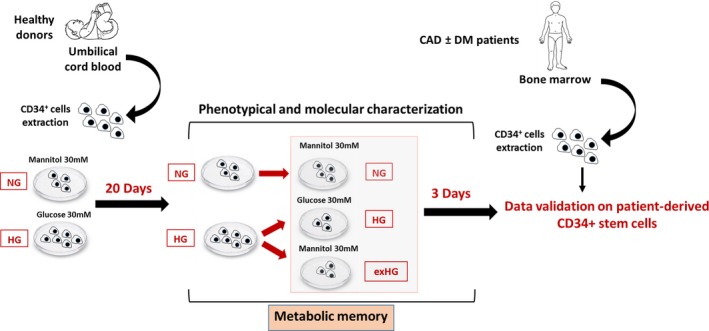
Experimental design. Schematic representation of the study. UCB‐derived CD34+ stem cells were expanded in HG conditions for up to 20 days. Afterward, part of the HG‐CD34+ stem cell population was returned to NG conditions for 3 days. At the end of the experiment, cells were used for functional and molecular characterization. CAD±DM indicates coronary artery disease with or without diabetes mellitus; exHG, ex‐high‐glucose conditions; HG, high‐glucose; NG, normal‐glucose; UCB, umbilical cord blood.
Study Participants
All experiments have been carried out upon approval of local ethic committees (No. 2015/ST/232 and R196/14—CCM 205). Informed written consent was obtained from all patients before BM harvesting. CAD (n=7) and CAD‐DM (n=7) subjects have been selected by stringent matching of: age, pharmacological treatments, and major risk factors. At admission, CAD‐DM patients were treated with sulfonylureas or metformin±insulin. Clinical characteristics of patients are shown in Table.
Table 1.
Clinical Characteristics of Patients
| CAD (n=7) | CAD‐DM (n=7) | P Value | |
|---|---|---|---|
| Age, y | 64±4 | 68±3 | 0.44 |
| BMI, kg/m2 | 26.7±1.1 | 25.6±0.7 | 0.41 |
| Glycemia, mg/dL | 110±3 | 162±13* | 0.0021 |
| LDL, mg/dL | 112±18 | 91±15 | 0.39 |
| HDL, mg/dL | 42±4 | 50±5 | 0.27 |
| Total cholesterol, mg/dL | 176±21 | 162±15 | 0.6 |
| Cardiovascular comorbidities | |||
| Hypertension (n) | 5 | 5 | |
| Dyslipidemia (n) | 4 | 5 | |
| Glucose‐lowering drugs | |||
| Insulin (n) | 0 | 2 | |
| Oral antidiabetic drug (n) | 0 | 5 | |
| Other therapies | |||
| Antihypertensive drugs (n) | 3 | 4 | |
| Lipid‐lowering drugs (n) | 4 | 5 | |
CAD±DM, coronary artery disease with or without diabetes mellitus; BMI indicates body mass index; HDL, high‐density lipoprotein; LDL, low‐density lipoprotein.
*P<0.01 vs CAD.
UCB CD34+ Stem Cell Isolation and Expansion
UCB was collected from the umbilical cord of healthy full‐term deliveries. CD34+ stem cells were isolated and expanded as previously described, with some slight modification.23 Briefly, mononuclear cell fraction was isolated from UCB by density gradient centrifugation using Ficoll‐Paque (Lymphoprep; Sentinel Diagnostics, SpA, Milan, Italy). CD34+ stem cells were then magnetically sorted from the mononuclear cell fraction using the MidiMACS system (CD34 Microbead Kit; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). Isolated cells were expanded up to 20 days in SFEM medium (Voden Medical Instruments, SpA, Meda, Italy) containing 20 ng/mL of interleukin‐6, 20 ng/mL of interleukin‐3, 50 ng/mL of fms‐like tyrosine kinase 3, and 50 ng/mL of stem cell factor (PeproTech EC Ltd., London, UK), in hyperglycemic (30 mmol/L of glucose; HG) and normoglycemic (30 mmol/L of mannitol; NG) conditions. To reproduce the metabolic memory phenomenon in vitro, HG‐CD34+ stem cells were returned to physiological glucose conditions for 3 days (exHG‐CD34+), as previously described.22
Sternal BM Biopsy and CD34+ Stem Cell Isolation
Sternal blood from the BM of 14 patients with CAD±DM was obtained by needle aspiration. Aspirates were suspended in saline buffer. Mononuclear cell fraction as well as CD34+ stem cells were isolated as aforementioned.
CD34+ Stem Cell Growth Curves
CD34+ stem cells were seeded at an initial density of 2.0×105 cells/well and cultured for up to 30 days in HG and NG conditions. Cells were counted on days 10, 20, and 30.
Migration Assays
Cell migration was determined by the use of Transwell culture inserts (5‐μm pore membrane; Corning Incorporated, Corning, NY), according to the manufacturer's instructions. In brief, 1×105 cells/well CD34+ stem cells were seeded onto the upper chamber and allowed to migrate toward the lower chamber containing, or not, SDF‐1α (50 ng/mL; PeproTech EC Ltd.). Transwells were incubated at 37°C, 5% CO2, for 4 hours. Migrated cells were counted and results are expressed as a migration index.
Flow Cytometric Assays
CD34+ stem cells were incubated for 30 minutes with allophycocyanin‐conjugated monoclonal antihuman CXCR4 antibody (BD Biosciences, San Jose, CA), Annexin V (BD Biosciences), and a CellROX green fluorescent assay kit (Life Technologies Italia, Monza, italy) for detection of the CXCR4 receptor, early apoptosis, and reactive oxygen species (ROS), respectively. The Becton‐Dickinson FACS Calibur platform (Becton‐Dickinson, Franklin Lakes, NJ) was used to analyze samples by use of appropriate physical gating. At least 104 events in the indicated gates were acquired.
Western Blot Analyses
Protein kinase B (AKT) phosphorylation was evaluated by stimulating for 15 minutes CD34+ stem cells (1.2×106/sample) with 50 ng/mL of SDF‐1α. Cells were then lysed in Laemmli buffer and 30 μL of proteins resolved on 10% SDS‐PAGE. Whole‐cell lysate (80 μg) was used to evaluate manganese superoxide dismutase, catalase, p66shc, DNA methyltransferase 1 (DNMT1), and DNA methyltransferase 3B (DNMT3B) expression. Proteins were transferred to PVDF membranes (Millipore, Merck SpA, Milan, Italy) and then incubated with the relevant primary antibody (Table S1). After washing, membranes were incubated with secondary antibody, which was linked to horseradish peroxidase (Pierce Antibody; Pierce Biotechnology, Rockford, IL) and revealed by ECL detection (Pierce). Results were quantified using the Alliance 9.7 Western Blot Imaging System 8 (UVItec Ltd., Cambridge, UK).
DNA Extraction and Bisulfite Modification
Genomic DNA was isolated by the PureLink Genomic DNA kit (Invitrogen, Carlsbad, CA), following the manufacturer's protocols. Nucleic acid samples were quantified by NanoDrop, and integrity was analyzed by 1% agarose gel electrophoresis. Bisulfite conversion and subsequent purification was performed with the MethylCode Bisulfite Conversion Kit (Invitrogen), starting from 300 ng of total DNA.
RNA Extraction, cDNA Preparation, and Quantitative Polymerase Chain Reaction Reactions
Total RNA from CD34+ stem cells was isolated by use of the Direct‐zol RNA Kit (Zymo Research, Irvine, CA), following the manufacturer's protocols. Total RNA (500 ng) was converted to cDNA with the Superscript III kit (Life Technologies, Carlsbad, CA), according to the manufacturer's protocol. Retrotrascribed RNA was used to quantify gene expression. Data, expressed as fold‐change (2−ΔΔCT) over NG after normalization to each housekeeping gene, were log2‐trasformed before analysis. Primers are reported in Table S2. All reactions were performed with SYBR Green Supermix 2X (Bio‐Rad Laboratories, Hercules, CA) on CFX96 Real–Time System PCR (Bio‐Rad).
Quantitative Polymerase Chain Reaction Analysis of CXCR4 Promoter Methylation
Methylation of the CXCR4 promoter was evaluated on bisulfite‐treated DNA by “2‐step SYBR Green–based polymerase chain reaction (PCR),” a new technique devised by our laboratory and described in detail by Bianchessi et al.24
CXCR4 Promoter Sequencing
The CXCR4 promoter was amplified from bisulfite‐treated DNA as previously described.24 Converted DNA was cloned into the pCR4‐TOPO‐TA cloning vector (Invitrogen) and transformed in Escherichia coli strain DH5α. Ten colonies for each sample were randomly picked and directly used for PCR amplification to verify vector insertion by T3 and T7 primers. PCR products were Sanger sequenced with the T3 primer with the help of an external service (GATC Biotech, Konstanz, Germany). Alignment (multiple sequence ClustalW alignment) and analysis of sequences was performed with BioEdit software and data analyzed and represented by QUMA software (http://quma.cdb.riken.jp/) for CpG methylation and Methtools 2.05 (http://genome.imbjena.de/methtools/) for non‐CpG methylation.
Evaluation of Bisulfite Conversion Efficiency and Normalization of Non‐CpG Methylation
Genomic DNA extracted from primary UCB‐derived CD34+ stem cells was whole‐genome amplified by using the REPLI‐g Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer's protocol. The whole‐genome amplified product was purified by the QIAquick PCR Purification Kit (Qiagen) and quantified with NanoDrop. The whole‐genome amplified product was used as fully unmethylated DNA.25 After bisulfite conversion, the CXCR4 promoter was amplified, cloned, and Sanger sequenced as before.24 Results were analyzed by Methtools 2.05 software and used for normalization of non‐CpG methylation data with the formula:
Chromatin Immunoprecipitation Assays
Treated cells were washed and cross‐linked using 1% formaldehyde for 20 minutes. After stopping cross‐linking by addition of 0.1 M of glycine, cell lysates were sonicated with Episonic 2000 (Epigentek Group Inc., Farmingdale, NY) and centrifuged for 10 minutes at 10 000g. Supernatants were immunoprecipitated using the EpiTect ChIP OneDay Kit (Qiagen), following the manufacturer's protocols. The list of antibodies used is reported in Table S1. Recovered DNA fragments were amplified for the CXCR4 gene by qPCR with 3 couples of primers spanning 3 different regions of the gene. qPCR values were normalized to input DNA and to the values obtained with immunoglobulin G isotype. The data are expressed as fold‐change over NG.
Statistical Analysis
Results are given as mean±SEM. All experiments were performed at least in triplicate, unless stated otherwise. All data expressed as fold‐change were log2‐transformed before analysis. The data were tested for the normality by using the Shapiro–Wilk normality test. Differences between data were evaluated by paired or unpaired Student t test (2‐group comparisons), 1‐way, 2‐way repeated‐measures ANOVA followed by the post‐hoc Newman–Keuls multiple comparison test, as appropriate. Correlations were calculated using Pearson's coefficient. A value of P≤0.05 was considered significant. All statistical analysis was performed using GraphPad Prism software (GraphPad Software Inc., La Jolla, CA).
Results
HG Affects Proliferation and Oxidative State of CD34+ Stem Cells
In order to establish standard conditions under which HG effects may be reliably reproduced in vitro, cells were cultured in 30 mmol/L of glucose and counted after 10, 20, and 30 days. The analysis of cellular growth curves revealed significant differences between HG‐CD34+ and NG‐CD34+ stem cells. Twenty days after seeding, HG‐CD34+ stem cells showed a significant decrease in proliferation rates when compared with their osmotic controls (30 mmol/L of mannitol; Figure 2A). This loss of glucose tolerance was associated with a significant increase in mitochondrial ROS production (Figure 2B and 2C). Consistently, we found a significant reduction of the antioxidant enzymes, catalase and manganese superoxide dismutase, expression (Figure 3A, 3B, 3D, and 3E), whereas p66shc, a gene involved in ROS generation and linked to hyperglycaemic memory, was upregulated26 (Figure 3C and 3F).
Figure 2.
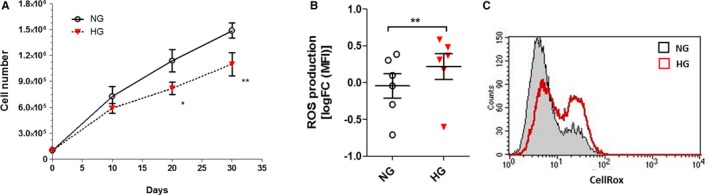
Effect of HG on proliferation and oxidative state of CD34+ stem cells. A, Proliferation curves of CD34+ stem cells exposed to HG concentrations (30 mmol/L; n=6; *P<0.05; **P<0.01; 2‐way rmANOVA). B, Flow cytometric quantification of ROS production in HG‐treated cells. Data are reported as log2 fold‐change (FC) of MFI over control (NG; n=6; **P<0.01 vs NG; paired t test). C, Representative flow cytometry histograms of ROS quantification. HG indicates high glucose; MFI, mean fluorescence intensity; NG, normal glucose; ROS, radical oxygen species.
Figure 3.
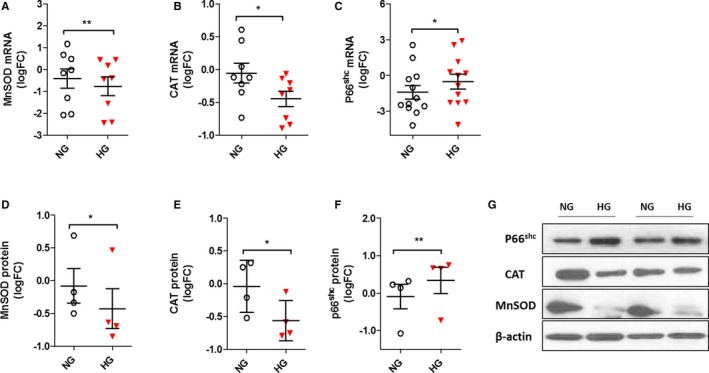
Effect of HG on antioxidant gene expression in CD34+ stem cells. A through C, Expression of MnSOD, CAT, and p66shc genes evaluated by qPCR. Data are from at least 6 independent experiments (*P<0.05; **P<0.01 vs NG; paired t test) and are expressed as log2 fold‐change (FC). D through F, Evaluation of MnSOD, CAT, and p66shc protein expression by western blot. Analysis of at least 3 independent experiments. Data after loading normalization (β‐actin) are expressed as log2 FC over control (NG; *P<0.05; **P<0.01 vs NG; paired t test). G, Representative immunoblot images are shown. CAT indicates catalase; HG, high glucose; MnSOD, manganese superoxide dismutase; NG, normal glucose; p66shc, Src homolog and collagen homologs (shc) adaptor protein.
HG Induces Persistent Impairment of CXCR4/SDF‐1α Axis in CD34+ Stem Cells
The CXCR4/SDF‐1α axis is primarily involved in CD34+ stem cell mobilization and migration from the BM to sites of ischemia and endothelial injury.27 A large body of evidence shows that expression and downstream pathways of CXCR4 receptor are impaired in diabetic patients.28, 29, 30 In our hands, after 20 days, HG‐CD34+ stem cells exhibited a significant reduced migration toward human recombinant SDF‐1α (50 ng/mL; Figure 4A). The analysis of CXCR4 downstream signaling by western blot revealed a significant decrease of AKT phosphorylation in HG‐CD34+ stem cells after SDF‐1α stimulation (Figure 4B and 4C). In addition, expression of CXCR4, both at mRNA and protein levels, was also found to be significantly downregulated, as assessed by qPCR and flow cytometry, respectively (Figure 4D through 4F). Taken together, these findings indicated that an HG challenge induces a profound SDF‐1α/CXCR4/phosphoinositide 3‐kinase/AKT signaling pathway impairment in naïve UCB‐derived CD34+ stem cells. Notably, upon recovery for 3 days in NG conditions, CD34+ stem cells (exHG‐CD34+ stem cells) still displayed functional and molecular alterations (Figure 4A through 4F), which was associated with enduring high ROS production (Figure S1). These data represent the first evidence of in vitro–induced metabolic memory in naïve CD34+ stem cells.
Figure 4.
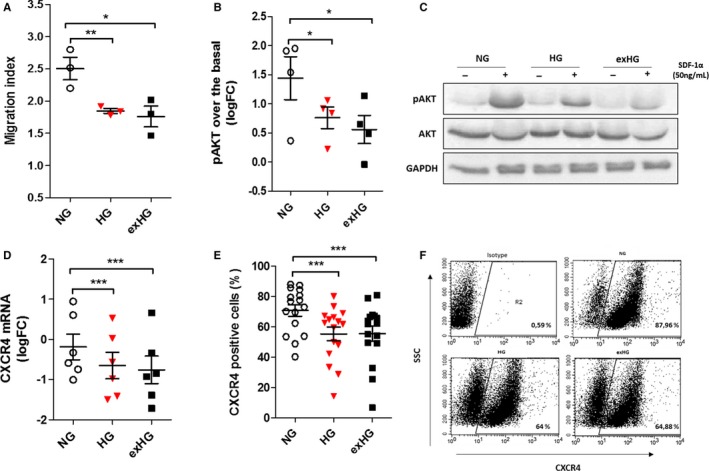
Evaluation of CXCR4/SDF‐1α axis in HG and exHG‐CD34+ stem cells. A, Migration ability of NG, HG, and exHG‐CD34+ stem cells toward SDF‐1α chemokine (n=3; *P<0.05; **P<0.01 vs NG). B, Evaluation of CXCR4 downstream AKT signalling pathway activation after SDF‐1α stimulation in NG, HG, and exHG‐CD34+ stem cells. Analysis of at least 3 independent experiments are shown (*P<0.05 vs NG). Quantification of western blot was normalized on total AKT expression, and data are expressed as pAKT log2 fold‐change (FC) over basal (unstimulated). C, Representative western blot image. D, qPCR analysis of CXCR4 mRNA expression in NG, HG, and exHG‐CD34+ stem cells (n=6; ***P<0.001 vs NG). Data are reported as log2 FC. E, CXCR4 membrane expression level in NG, HG, and exHG‐CD34+ stem cells by flow cytometric analysis (n=16; ***P<0.001 vs NG). Data are expressed as percentage of positive cells. F, Representative flow cytometry dot‐plots of CXCR4 quantification. Significant differences were evaluated by 1‐way ANOVA followed by Newman–Keuls post‐hoc analysis. AKT indicates protein kinase B; CXCR4, C‐X‐C chemokine receptor type 4; exHG, ex‐high‐glucose; GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; HG, high‐glucose; NG, normal‐glucose; pAKT, phospho‐AKT; SDF‐1α, stromal‐cell–derived factor 1 alpha.
HG Increases DNA Methylation of the CXCR4 Promoter in CD34+ Stem Cells
Numerous studies in tumor biology have demonstrated that DNA methylation, an epigenetic modification associated with gene silencing, regulates CXCR4 expression in cancer cells.31, 32, 33 We therefore postulated that such a repressive epigenetic modification was also involved in the downregulation of CXCR4 mRNA in HG‐CD34+ stem cells. Thus, we evaluated the CpG methylation status (5mCpG) of a region of the CXCR4 gene promoter encompassing −1349 to −738 nucleotides relative to the +1 transcription start site (Figure 5A). Bisulfite‐treated DNA from HG and NG‐CD34+ stem cells was first analyzed for CXCR4 methylation density by a 2‐step qPCR methylation method. This assay, designed in our laboratory for the quantitative evaluation of DNA methylation status of genes,24 showed a significant increase of CXCR4 promoter methylation in HG‐CD34+ stem cells (Figure 5B). Results were then validated by bisulfite Sanger sequencing, the “gold‐standard” technology for DNA methylation studies. In accord with our method, bisulfite sequencing gave similar readouts (Figure 5C and 5D). Remarkably, after normalization against bisulfite conversion efficiency, sequencing data analysis by Methtools 2.05 (http://genome.imbjena.de/methtools/) revealed a 1.8‐fold increase of non‐CpG methylation (5mCpN) in HG‐CD34+ stem cells when compared with NG‐CD34+ stem cells (Figure 5E and 5F).
Figure 5.
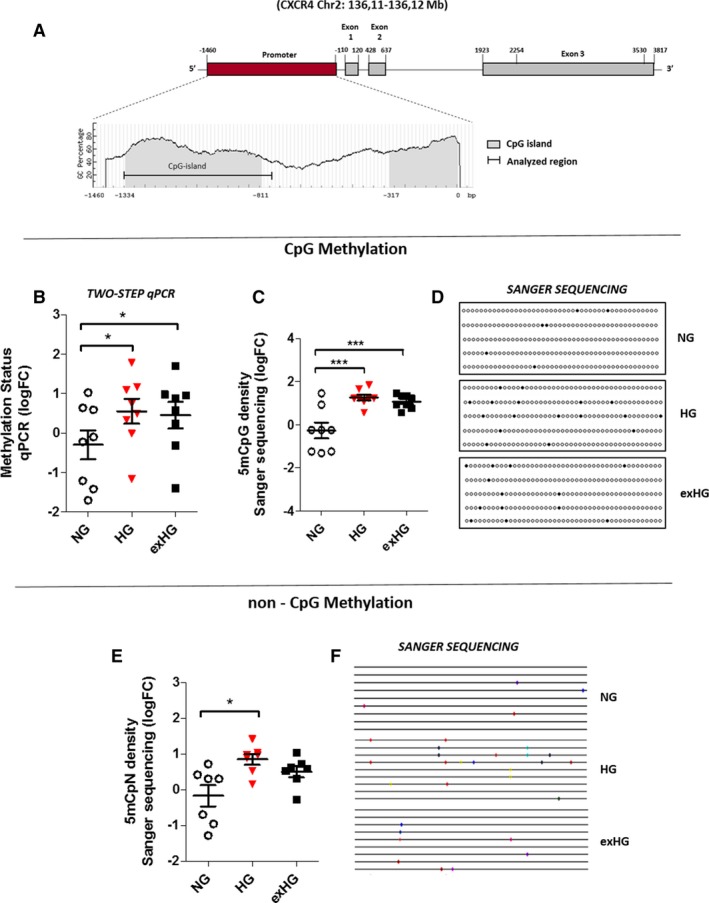
HG Increases CXCR4 promoter methylation in CD34+ stem cells. A, Schematic representation of the human CXCR4 gene. The CXCR4 gene contains a promoter region (1460 nt) and 3 exons separated by intronic sequences. The analysis of the CXCR4 promoter with MethPrimer software shows the main CpG island (from −1334 to −811 bp) within the promoter and part of the second CpG island (represented in gray). The black line in the CpG island indicates the analyzed region (from −1349 to −738 bp). B, Quantification of CpG methylation density of the CXCR4 promoter by a 2‐step qPCR method in NG, HG, and exHG‐CD34+ stem cells (n=8; *P<0.05 vs NG). Data are expressed as log2 fold‐change (FC) over NG. C, Quantification of CXCR4 promoter methylation levels by bisulfite Sanger sequencing in NG, HG, and exHG‐CD34+ stem cells (n=8; ***P<0.001 vs NG). The data, expressed as log2 FC over NG, are the result of 8 independent experiments where 10 colonies for each single sample were sequenced. D, Representative visualization of 5‐bisulfite sequencing results as analyzed by QUMA software (http://quma.cdb.riken.jp/). E, Non‐CpG methylation density of the CXCR4 promoter in NG, HG, and exHG‐CD34+ stem cells (n=7; *P<0.05 vs NG). The data, expressed as log2 FC over NG, are the results of Methtools 2.05 software analysis after normalization on bisulfite conversion efficiency. As reported for panel C, 10 clones for each cell treatment of 8 independent experiments were sequenced. F, Representative visualization of 9‐bisulfite sequencing results. Significant differences were evaluated by 1‐way ANOVA followed by Newman–Keuls post‐hoc analysis. CXCR4 indicates C‐X‐C chemokine receptor type 4; 5mCpG, methylation at the carbon 5 position of a cytosine ring in CpG (5′‐cytosine‐phosphate‐guanine‐3′); 5mCpN, methylation at the carbon 5 position of a cytosine ring in 5′‐C‐phosphate‐nucleotide‐3′; exHG, ex‐high‐glucose; HG, high‐glucose; NG, normal‐glucose; qPCR, quantitative polymerase chain reaction.
Notably, both CpG and non‐CpG methylation changes, although the latter did not reach statistical significance, were still present after 3‐days in NG concentrations (exHG‐CD34+ stem cells; Figure 5B through 5F).
Increased DNA Methylation Associates With a More‐Inactive Chromatin Conformation and Reduced Engagement of RNA Polymerase II to the CXCR4 Promoter
DNA methylation and histone modification systems are highly inter‐related and mechanistically rely on each other for chromatin function. Hence, we evaluated whether increased methylation of the CXCR4 promoter was associated with alterations in histone modification pattern. To this end, we performed a chromatin immunoprecipitation assay to assess changes on repressive histone 3 lysine 9 trimethylation and histone 3 lysine 27 trimethylation (often associated with DNA methylation) and activating acetyl‐H4 histone modifications. We found that whereas histone 3 lysine 9 trimethylation was not affected by HG exposure (Figure S2), there was a significant increase in repressive histone 3 lysine 27 trimethylation modification and reduced pan‐acetyl‐H4 level in HG‐CD34+ stem cells (Figure 6B and 6C). Taken together, these results provide evidence that DNA methylation at the CXCR4 promoter correlates with a more‐inactive chromatin conformation that better explains reduced gene expression. Indeed, when we plotted the qPCR data of CXCR4 expression against their respective DNA promoter methylation percentage and performed correlation analysis, we found that CXCR4 mRNA content negatively correlated with promoter methylation in CD34+ stem cells (Pearson r=−0.76; P=0.004; Figure 7A). Moreover, to determinate whether overall epigenetic modifications reduced RNA polimerase II recruitment to the CXCR4 promoter in HG‐CD34+ stem cells, we performed chromatin immunoprecipitation assay with an antibody specific for RNA polymerase II. Quantitative real‐time PCR analysis of the CXCR4 gene after chromatin immunoprecipitation assay, with 3 couples of primers landing up‐ and downstream of the transcription start site, displayed a significant reduction of RNA polymerase II binding to the CXCR4 gene in HG‐CD34+ stem cells compared with NG‐CD34+ stem cells (Figure 7B).
Figure 6.
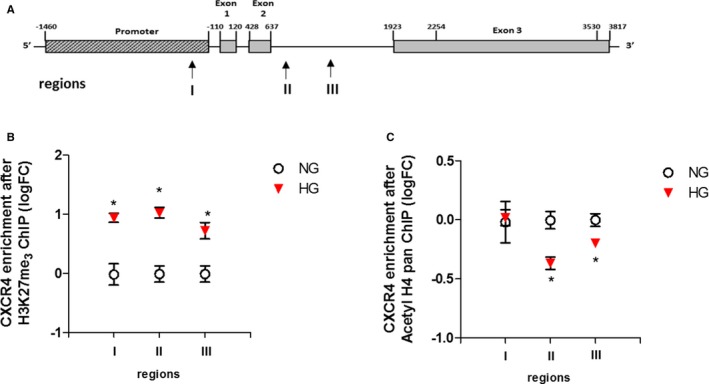
CXCR4 promoter methylation associates with a closer chromatin conformation. A, Schematic representation of the CXCR4 gene with mapped sequences (arrows, region I, II, and III). B and C, ChIP of the CXCR4 promoter by H3K27me3 and pan‐acetyl H4 after HG exposure of CD34+ stem cells. The data after input normalization are expressed as log2 fold‐change (FC) over the NG (n=3; *P<0.05 vs NG; paired t test). ChIP indicates chromatin immunoprecipitation; CXCR4, C‐X‐C chemokine receptor type 4; H3K27me3, trimethylation on lysine 27 of histone H3; pan‐acetyl H4, acetyl‐histone H4; HG, high‐glucose; NG, normal glucose.
Figure 7.
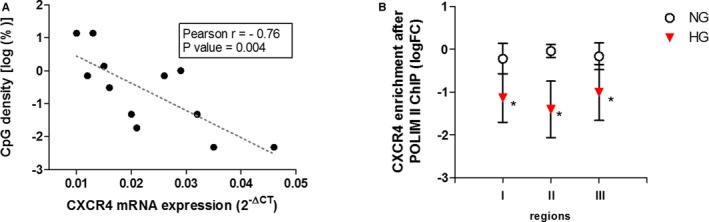
CXCR4 promoter methylation negatively correlates with CXCR4 mRNA expression and reduces engagement of RNA POL II to the gene. A, Correlation analysis between normalized quantification of CXCR4 mRNA (2−ΔCt) and log2 percentage of methylated CXCR4 promoter (Sanger sequencing). Pearson r and P value are indicated on the graph. B, ChIP of CXCR4 promoter by RNA POL II after HG exposure of CD34+ stem cells. The data, after input normalization are expressed as log2 fold‐change (FC) over the NG (n=6; *P<0.05 vs NG; paired t test). CpG indicates methylation in 5′‐cytosine‐phosphate‐guanine‐3′; ChIP, chromatin immunoprecipitation; CXCR4, C‐X‐C chemokine receptor type 4; HG, high‐glucose; NG, normal glucose; RNA POL II, RNA polymerase II.
HG Affects Gene Expression Pattern of the “DNA Methylation Machinery” in CD34+ Stem Cells
In mammals, the DNA methylation pattern is a fine‐tuned process in which DNA methyltransferases (DNMTs), namely DNMT1, DNMT3A, and DNMT3B, and demethylation enzymes, the ten eleven translocation proteins (TETs), take part. DNMT1 associates with S‐phase replication foci and acts primarily as a maintenance methyltransferase, whereas DNMT3A and DNMT3B are essential for de novo methylation.34, 35 Based on our observations, we assessed the variation of DNMT enzymes, in HG‐CD34+ stem cells. Surprisingly, both qPCR and western blot analyses revealed a significant downregulation of DNMT1 (Figure 8A, 8B, and 8C) that was counteracted by an increased expression of the DNMT3B enzyme (Figure 8D through 8F). No differences were found for DNMT3A expression (Figure S3). On the other hand, both mRNA expression levels of the TET2 and TET3 enzymes were significantly reduced (Figure 8G and 8H). Nevertheless, although these results implied an HG‐induced dysregulation in the “DNA methylation machinery,” we did not find any changes in global DNA methylation level and DNMT activity in HG‐CD34+ stem cells (Figure S4A and S4B), suggesting a gene‐specific epigenetic modification pattern induced by hyperglycemia.
Figure 8.
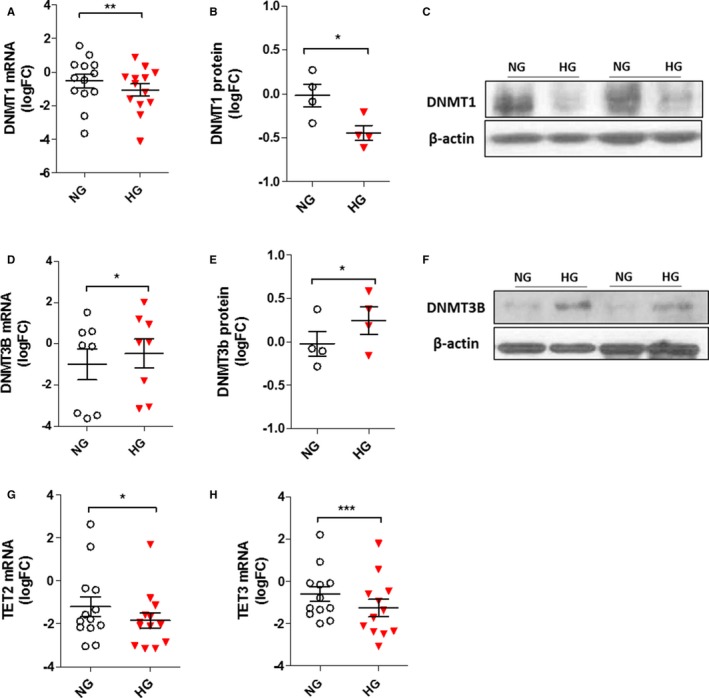
Effect of HG on gene‐expression pattern of DNA methylation machinery in CD34+ stem cells. A, D, G, and H, Analysis of DNMT1, DNMT3B, TET2, and TET3 expression in CD34+ stem cells by qPCR. The data from at least 6 independent experiments (*P<0.05; **P<0.01; ***P<0.001 vs NG; paired t test) are expressed as log2 fold‐change (FC). B and E, Evaluation of DNMT1 and DNMT3B protein expression by western blot. Analysis of at least 3 independent experiments. The data after loading normalization (β‐actin) are expressed as log2 FC over the control (NG; *P<0.05 vs NG). C and F, Representative immunoblot images are shown. DNMT1 indicates DNA methyltransferase 1; DNMT3B, DNA methyltransferase 3B; HG, high glucose; NG, normal glucose; qPCR, quantitative polymerase chain reaction; TET2, tet methylcytosine dioxygenase 2; TET3, tet methylcytosine dioxygenase 3.
Translation of Epigenetic Findings on CXCR4 Promoter of CD34+ Stem Cells From Diabetic Patients
Next, to validate our in vitro observations in humans, we investigated whether methylation of the CXCR4 promoter and gene expression were altered in BM‐derived CD34+ stem cells from diabetic patients. A cohort of NG‐tolerant and T2DM male CAD patients (CAD±DM) undergoing bypass surgery was enrolled in the study. Importantly, the 2 groups of patients were age‐matched in order to exclude any possible confounding effect of age, given that aging is associated with methylation events.36 Patients’ profiles and laboratory parameters are shown inTable. CD34+ stem cells isolated from sternal BM biopsies were analyzed by flow cytometry for expression of the CXCR4 receptor. As shown in Figure 9A, CD34+ stem cells from CAD‐DM patients showed a lower expression of the CXCR4 receptor when compared with the normoglycemic CAD group. We then analyzed the DNA methylation density of the CXCR4 promoter by 2‐step qPCR and bisulfite Sanger sequencing techniques. Consistent with our in vitro results, both methods corresponded in unveiling a significant increase of CpG methylation density at the level of the CXCR4 promoter in BM‐derived CD34+ stem cells from CAD‐DM patients (Figure 9B through 9D). Moreover, similarly to what we found in our in vitro model, we detected, although not significantly, higher non‐CpG methylation levels in the CXCR4 promoter in CAD‐DM patients’ CD34+ stem cells compared with the normoglycemic CAD group (Figure 9E and 9F). Again, when CXCR4 expression data were plotted against corresponding promoter methylation density data, a significant negative correlation was evident (Pearson r=−0.57; P=0.03; Figure 9G). Overall, these results describe, for the first time, a direct impact of hyperglycemia on methylation density of the CXCR4 promoter in human‐derived CD34+ stem cells.
Figure 9.
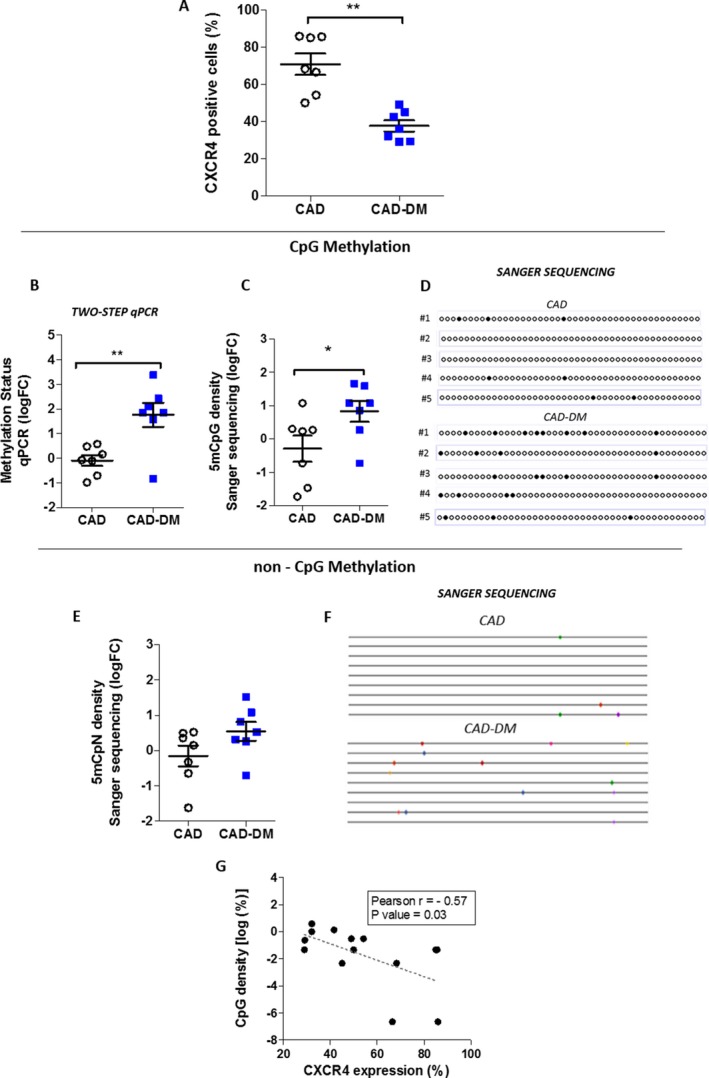
CXCR4 gene methylation and expression in CAD‐DM patients. A, CXCR4 membrane expression level in CD34+ stem cells derived from CAD and CAD‐DM patients by flow cytometric analysis (n=14). Data are expressed as a percentage of positive cells. B, Quantification of CpG methylation density of the CXCR4 promoter by a 2‐step qPCR method in CD34+ stem cells of CAD and CAD‐DM patients. The data are expressed as log2 fold‐change (FC) over CAD. C, Bisulfite Sanger sequencing quantification of CXCR4 promoter methylation in CAD and CAD‐DM patients. The data are expressed as log2 FC over CAD. Ten colonies for each patient's sample were sequenced. D, Representative visualization of 5‐bisulfite sequencing results as analyzed by QUMA software (http://quma.cdb.riken.jp/). E, Non‐CpG methylation density of the CXCR4 promoter in CAD and CAD‐DM patients. The data, expressed as log2 FC over the CAD, are the results of Methtools 2.05 software analysis after normalization on bisulfite conversion efficiency. As reported for panel C, 10 clones from each patient's sample were sequenced. F, Representative visualization of 9‐bisulfite sequencing results. Significant differences between sample patients were evaluated by unpaired t test (CAD, n=7; CAD‐DM, n=7; **P<0.01; *P<0.05 vs CAD). G, Correlation analysis between percentage of CXCR4 expression and log2 percentage of CXCR4 promoter methylation (Sanger sequencing). Pearson r and P value are indicated on the graph. CXCR4 indicates C‐X‐C chemokine receptor type 4; 5mCpG, methylation at the carbon 5 position of a cytosine ring in CpG (5′‐cytosine‐phosphate‐guanine‐3′); 5mCpN, methylation at the carbon 5 position of a cytosine ring in 5′‐C‐phosphate‐nucleotide‐3′; CAD±DM, coronary artery disease with or without diabetes mellitus; qPCR, quantitative polymerase chain reaction.
Discussion
Overall, our work shows that epigenetic alterations in the CXCR4 promoter of human CD34+ stem cells induced by hyperglycemia have detrimental effects on their functional properties, which endure after normoglycemia recovery. This evidence has been obtained both in UCB‐derived CD34+ stem cells exposed to hyperglycemia versus normoglycemia recovery as well as in BM‐derived CD34+ stem cells harvested from CAD‐DM patients undergoing state‐of‐the‐art antidiabetic therapy compared with age‐matched normoglycemic controls. Such findings show, for the first time, that human CD34+ stem cells “memorize” the hyperglycemic environment in the form of epigenetic changes that, by contributing to the self‐perpetuating alteration of gene expression, may be potentially responsible for progression of microangiopathy and cardiovascular events in T2DM subjects despite glycamia correction.10 In particular, through our in vitro cell model, we provided the first experimental evidence of an epigenetic contribution to CXCR4/SDF‐1α axis impairment.
Previous landmark articles demonstrated in vitro and animal models mimicking metabolic memory increases in the expression of nuclear factor kappa light chain enhancer of activated B cells p65, oxidant stress, and inflammatory genes in vascular cells.18, 37, 38, 39 These changes, which persisted after return of NG conditions, were dependent on epigenetic histone modifications such as H3 lysine methylation. Moreover, numerous studies have shown different DNA methylation patterns between individuals with DM and matched controls in multiple target tissues that may play a substantial role in the pathophysiology of DM and its associated vascular complications.20, 40, 41 However, although the interest on DM‐mediated epigenetic changes and their role in metabolic memory is growing, no data are available with regard to CD34+ stem cells.21 It is worth noting that the levels of circulating CD34+ stem cells have been recently proved to be an efficient clinical‐grade independent biomarker of cardiovascular risk in diabetic patients,10 able to predict on the long‐term the development or the progression of microangiopathy and cardiovascular events in T2DM.10 Our study adds information on epigenetic mechanisms underlying the exhaustion of reparative vascular homeostasis in DM.
Stem and progenitor cells are known to intrinsically express high levels of antioxidant enzymes that make them more resistant than mature cells to oxidative stress.42, 43 The antioxidant defense capacity explains how progenitor cells can execute a regenerative program in an unfavourable oxidative environment, such as that generated by ischemia reperfusion, and inflammation and why, in our hands, CD34+ stem cells exhibited signs of metabolic exhaustion after 20 days of chronic glucose challenge. Indeed, only after this long period of metabolic overload, cells started showing increased ROS production along with reduced expression of antioxidant enzyme genes catalase and manganese superoxide dismutase. Interestingly, HG exposure also induced expression of p66shc gene in naïve CD34+ stem cells. P66shc is a Src homolog and collagen homologs (shc) adaptor protein that plays a crucial role in development of diabetic vascular complications and mainly contributes to phenomenon of metabolic memory.26 The pathways described for its pro‐oxidant action operate on both mitochondrial and nuclear levels. In mitochondria, p66shc promotes ROS generation,44 whereas in the nucleus, in agreement with our findings, p66shc inhibits expression of the ROS‐scavenging enzymes, catalase and manganese superoxide dismutase.45 In our hands, such dampening of intrinsic antioxidant mechanisms and increased ROS production resulted in CD34+ stem cell dysfunction. Specifically, HG‐CD34+ stem cells displayed impaired chemotaxis toward SDF‐1α, and this defect was associated with decreased expression of CXCR4 receptor. Additionally, the downstream pathway, known for mediating the effect of SDF‐1α on its respective receptor, CXCR4, was also impaired. All these defects, that persisted despite the return in normoglycemic conditions, were associated with an endured high ROS production. This finding, in line with the increased expression of p66shc in HG‐CD34+ stem cells, strengthened the involvement of this gene in the mechanisms of metabolic memory in our cells.
Expression of chemokine receptor CXCR4 is transcriptionally regulated by DNA methylation.32 Alteration of this fine‐tuned modulation in diseases, such as cancer and primary myelofibrosis, is responsible for the pathological phenotype of tumor cells.33, 46, 47
Our findings provide evidence that HG exposure influences per se DNA methylation of CXCR4 promoter in HG‐CD34+ stem cells and negatively affects migration ability toward SDF‐1α. Importantly, both the epigenetic modification and functional defects persisted after a 3‐day recovery in normoglycemic conditions (exHG‐CD34+). Notably, the CXCR4 promoter also displayed a significant non‐CpG methylation increase. DNA methylation in mammals is predominantly reported on the cytosine of the dinucleotide sequence, CpG. However, non‐CpG methylation was described in embryonic cells,48 and a recent study by Barres et al provided evidence that non‐CpG methylation was present in skeletal muscle of T2DM subjects.49 Interestingly, we found that the transcription pattern of enzymes involved in DNA methylation was affected by HG exposure. In particular, DNMT3B was upregulated, thus suggesting, in agreement with Barre et al's findings,49 the involvement of this enzyme in increased methylation of the CXCR4 promoter. However, further investigations will be necessary to understand the biological role of non‐CpG methylation and DNMT3B on gene transcription regulation in the diabetic context.
DNA methylation and histone modification systems are known to be highly inter‐related and mechanistically rely on each other for chromatin function.50 We found, by chromatin immunoprecipitation assay, that DNA methylation of the CXCR4 promoter was associated with a significant increase of repressive histone 3 lysine 27 trimethylation modification and reduced level of activating pan‐acetyl‐H4 in HG‐CD34+ stem cells. Taken together, these findings provide evidence that HG exposure promotes a more‐inactive chromatin conformation, which decreases DNA accessibility, contributing to reduction of CXCR4 expression.
We next analyzed BM‐derived CD34+ stem cells of control CAD and CAD‐DM patients receiving antidiabetic drugs. Remarkably, we were able to confirm the epigenetic changes observed in HG‐CD34+ stem cells. In CAD‐DM subjects receiving pharmacological glycemia control versus age‐matched nondiabetic subjects, a significant increase of CpG methylation at the level of the CXCR4 promoter was evident and which negatively correlated with CXCR4 protein expression. Intriguingly, in line with previous data, non‐CpG methylation was also increased in CAD‐DM patients, although not significantly.
We acknowledge that this study, despite introducing unprecedented information, has some limitations. First, the sample size of CAD‐DM patients and controls is relatively small. However, in an attempt to partially overcome this shortcoming, enrolled subjects have been selected by accurate matching for age and major risk factors. We are aware that this study does not provide a full mechanistic explanation of the link between CXCR4 methylation, mRNA expression, and cell migration impairment. We partly overcame this issue by showing that increased DNA methylation is associated with a closer chromatin structure that reduces RNA polymerase II recruitment at the CXCR4 promoter level. Further mechanistic insights will be the object of future studies.
In summary, the observations reported here show, for the first time, that CD34+ stem cells are reminiscent of diabetic milieu and that epigenetic mechanisms induced by hyperglycemia might concur to the pathogenesis of CD34+ stem cell dysfunction having direct implications on tissue homeostasis and repair in DM. We believe this study provides a proof of concept opening new perspectives in investigation on epigenetic mechanisms involved in diabetic progenitor cell dysfunction. This line of investigation may be relevant in understanding the barriers that prevent the pharmacological regression of DM cardiovascular complications despite optimal management of hyperglycemia.
Sources of Funding
This work was supported by Ricerca Finalizzata, Ministero della Salute (PE‐2011‐02348537); Bianchessi and Vigorelli's fellowships were supported by Ricerca Finalizzata, Ministero della Salute (PE‐2011‐02348537).
Disclosures
None.
Supporting information
Data S1. Supplemental Methods.
Table S1. Antibody List
Table S2. Primer List
Figure S1. Flow cytometric quantification of ROS production in HG and exHG‐CD34+ stem cells.
Figure S2. A, Schematic representation of the CXCR4 gene with mapped sequences (arrows, region I, II, and III). B, ChIP analysis of the CXCR4 promoter for H3K9me3 modification after hyperglycemia exposure of CD34+ stem cells.
Figure S3. Analysis of DNMT3A, expression in CD34+ stem cells by qPCR.
Figure S4. A, Effect of HG exposure on global 5‐mC methylation level in CD34+ stem cells. The data are expressed as 5‐mC% (5‐mC/total DNA [A+G+C+T]). B, DNMT activity in HG‐CD34+ stem cells after HG exposure.
Acknowledgments
We are grateful to Dr Angela Raucci for helpful discussion of the data and to Dr Aoife Gowran, for language editing and critical comments. We also thank Dr Chiesa and the Unit of Biostatistics of Centro Cardiologico Monzino for their support in statistical data analysis.
(J Am Heart Assoc. 2019;8:e010012 DOI: 10.1161/JAHA.118.010012.)
References
- 1. Mangiapane H. Cardiovascular disease and diabetes. Adv Exp Med Biol. 2012;771:219–228. [DOI] [PubMed] [Google Scholar]
- 2. Bornfeldt KE, Tabas I. Insulin resistance, hyperglycemia, and atherosclerosis. Cell Metab. 2011;14:575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Paneni F, Beckman JA, Creager MA, Cosentino F. Diabetes and vascular disease: pathophysiology, clinical consequences, and medical therapy: part I. Eur Heart J. 2013;34:2436–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hayek SS, MacNamara J, Tahhan AS, Awad M, Yadalam A, Ko YA, Healy S, Hesaroieh I, Ahmed H, Gray B, Sher SS, Ghasemzadeh N, Patel R, Kim J, Waller EK, Quyyumi AA. Circulating progenitor cells identify peripheral arterial disease in patients with coronary artery disease. Circ Res. 2016;119:564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Werner N, Kosiol S, Schiegl T, Ahlers P, Walenta K, Link A, Bohm M, Nickenig G. Circulating endothelial progenitor cells and cardiovascular outcomes. N Engl J Med. 2005;353:999–1007. [DOI] [PubMed] [Google Scholar]
- 6. Takahashi T, Kalka C, Masuda H, Chen D, Silver M, Kearney M, Magner M, Isner JM, Asahara T. Ischemia‐ and cytokine‐induced mobilization of bone marrow‐derived endothelial progenitor cells for neovascularization. Nat Med. 1999;5:434–438. [DOI] [PubMed] [Google Scholar]
- 7. Losordo DW, Kibbe MR, Mendelsohn F, Marston W, Driver VR, Sharafuddin M, Teodorescu V, Wiechmann BN, Thompson C, Kraiss L, Carman T, Dohad S, Huang P, Junge CE, Story K, Weistroffer T, Thorne TM, Millay M, Runyon JP, Schainfeld R; Autologous CD34+ Cell Therapy for Critical Limb Ischemia Investigators . A randomized, controlled pilot study of autologous CD34+ cell therapy for critical limb ischemia. Circ Cardiovasc Interv. 2012;5:821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tepper OM, Galiano RD, Capla JM, Kalka C, Gagne PJ, Jacobowitz GR, Levine JP, Gurtner GC. Human endothelial progenitor cells from type ii diabetics exhibit impaired proliferation, adhesion, and incorporation into vascular structures. Circulation. 2002;106:2781–2786. [DOI] [PubMed] [Google Scholar]
- 9. Zhang L, Xu Q. Stem/progenitor cells in vascular regeneration. Arterioscler Thromb Vasc Biol. 2014;34:1114–1119. [DOI] [PubMed] [Google Scholar]
- 10. Fadini GP, Rigato M, Cappellari R, Bonora BM, Avogaro A. Long‐term prediction of cardiovascular outcomes by circulating CD34+ and CD34+CD133+ stem cells in patients with type 2 diabetes. Diabetes Care. 2017;40:125–131. [DOI] [PubMed] [Google Scholar]
- 11. Engerman RL, Kern TS. Progression of incipient diabetic retinopathy during good glycemic control. Diabetes. 1987;36:808–812. [DOI] [PubMed] [Google Scholar]
- 12. Action to Control Cardiovascular Risk in Diabetes Study Group , Gerstein HC, Miller ME, Byington RP, Goff DC Jr, Bigger JT, Buse JB, Cushman WC, Genuth S, Ismail‐Beigi F, Grimm RH Jr, Probstfield JL, Simons‐Morton DG, Friedewald WT. Effects of intensive glucose lowering in type 2 diabetes. N Engl J Med. 2008;358:2545–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nathan DM, Cleary PA, Backlund JY, Genuth SM, Lachin JM, Orchard TJ, Raskin P, Zinman B, Diabetes C; Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications (DCCT/EDIC) Study Research Group . Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes. N Engl J Med. 2005;353:2643–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Duckworth W, Abraira C, Moritz T, Reda D, Emanuele N, Reaven PD, Zieve FJ, Marks J, Davis SN, Hayward R, Warren SR, Goldman S, McCarren M, Vitek ME, Henderson WG, Huang GD; VADT Investigators . Glucose control and vascular complications in veterans with type 2 diabetes. N Engl J Med. 2009;360:129–139. [DOI] [PubMed] [Google Scholar]
- 15. Babu M, Durga Devi T, Makinen P, Kaikkonen M, Lesch HP, Junttila S, Laiho A, Ghimire B, Gyenesei A, Yla‐Herttuala S. Differential promoter methylation of macrophage genes is associated with impaired vascular growth in ischemic muscles of hyperlipidemic and type 2 diabetic mice: genome‐wide promoter methylation study. Circ Res. 2015;117:289–299. [DOI] [PubMed] [Google Scholar]
- 16. Paneni F, Costantino S, Battista R, Castello L, Capretti G, Chiandotto S, Scavone G, Villano A, Pitocco D, Lanza G, Volpe M, Luscher TF, Cosentino F. Adverse epigenetic signatures by histone methyltransferase set7 contribute to vascular dysfunction in patients with type 2 diabetes mellitus. Circ Cardiovasc Genet. 2015;8:150–158. [DOI] [PubMed] [Google Scholar]
- 17. Wegner M, Neddermann D, Piorunska‐Stolzmann M, Jagodzinski PP. Role of epigenetic mechanisms in the development of chronic complications of diabetes. Diabetes Res Clin Pract. 2014;105:164–175. [DOI] [PubMed] [Google Scholar]
- 18. El‐Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, Cooper ME, Brownlee M. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med. 2008;205:2409–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cencioni C, Spallotta F, Greco S, Martelli F, Zeiher AM, Gaetano C. Epigenetic mechanisms of hyperglycemic memory. Int J Biochem Cell Biol. 2014;51:155–158. [DOI] [PubMed] [Google Scholar]
- 20. Simar D, Versteyhe S, Donkin I, Liu J, Hesson L, Nylander V, Fossum A, Barres R. DNA methylation is altered in B and NK lymphocytes in obese and type 2 diabetic human. Metabolism. 2014;63:1188–1197. [DOI] [PubMed] [Google Scholar]
- 21. Rodrigues M, Wong VW, Rennert RC, Davis CR, Longaker MT, Gurtner GC. Progenitor cell dysfunctions underlie some diabetic complications. Am J Pathol. 2015;185:2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paneni F, Mocharla P, Akhmedov A, Costantino S, Osto E, Volpe M, Luscher TF, Cosentino F. Gene silencing of the mitochondrial adaptor p66(Shc) suppresses vascular hyperglycemic memory in diabetes. Circ Res. 2012;111:278–289. [DOI] [PubMed] [Google Scholar]
- 23. Burba I, Colombo GI, Staszewsky LI, De Simone M, Devanna P, Nanni S, Avitabile D, Molla F, Cosentino S, Russo I, De Angelis N, Soldo A, Biondi A, Gambini E, Gaetano C, Farsetti A, Pompilio G, Latini R, Capogrossi MC, Pesce M. Histone deacetylase inhibition enhances self renewal and cardioprotection by human cord blood‐derived CD34 cells. PLoS One. 2011;6:e22158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bianchessi V, Lauri A, Vigorelli V, Toia M, Vinci MC. Evaluating the methylation status of CXCR4 promoter: a cost‐effective and sensitive two‐step PCR method. Anal Biochem. 2017;519:84–91. [DOI] [PubMed] [Google Scholar]
- 25. Bianchessi V, Vinci MC, Nigro P, Rizzi V, Farina F, Capogrossi MC, Pompilio G, Gualdi V, Lauri A. Methylation profiling by bisulfite sequencing analysis of the mtDNA non‐coding region in replicative and senescent endothelial cells. Mitochondrion. 2016;27:40–47. [DOI] [PubMed] [Google Scholar]
- 26. Paneni F, Volpe M, Luscher TF, Cosentino F. SIRT1, p66(Shc), and Set7/9 in vascular hyperglycemic memory: bringing all the strands together. Diabetes. 2013;62:1800–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cencioni C, Capogrossi MC, Napolitano M. The SDF‐1/CXCR4 axis in stem cell preconditioning. Cardiovasc Res. 2012;94:400–407. [DOI] [PubMed] [Google Scholar]
- 28. Egan CG, Lavery R, Caporali F, Fondelli C, Laghi‐Pasini F, Dotta F, Sorrentino V. Generalised reduction of putative endothelial progenitors and CXCR4‐positive peripheral blood cells in type 2 diabetes. Diabetologia. 2008;51:1296–1305. [DOI] [PubMed] [Google Scholar]
- 29. Ceradini DJ, Yao D, Grogan RH, Callaghan MJ, Edelstein D, Brownlee M, Gurtner GC. Decreasing intracellular superoxide corrects defective ischemia‐induced new vessel formation in diabetic mice. J Biol Chem. 2008;283:10930–10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hamed S, Brenner B, Abassi Z, Aharon A, Daoud D, Roguin A. Hyperglycemia and oxidized‐LDL exert a deleterious effect on endothelial progenitor cell migration in type 2 diabetes mellitus. Thromb Res. 2010;126:166–174. [DOI] [PubMed] [Google Scholar]
- 31. Sato N, Matsubayashi H, Fukushima N, Goggins M. The chemokine receptor CXCR4 is regulated by DNA methylation in pancreatic cancer. Cancer Biol Ther. 2005;4:70–76. [DOI] [PubMed] [Google Scholar]
- 32. Ramos EA, Grochoski M, Braun‐Prado K, Seniski GG, Cavalli IJ, Ribeiro EM, Camargo AA, Costa FF, Klassen G. Epigenetic changes of CXCR4 and its ligand CXCL12 as prognostic factors for sporadic breast cancer. PLoS One. 2011;6:e29461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bogani C, Ponziani V, Guglielmelli P, Desterke C, Rosti V, Bosi A, Le Bousse‐Kerdiles MC, Barosi G, Vannucchi AM; Myeloproliferative Disorders Research Consortium . Hypermethylation of CXCR4 promoter in CD34+ cells from patients with primary myelofibrosis. Stem Cells. 2008;26:1920–1930. [DOI] [PubMed] [Google Scholar]
- 34. Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Rep. 2011;12:647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30:733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekstrom TJ, Harris TB, Launer LJ, Eiriksdottir G, Leppert MF, Sapienza C, Gudnason V, Feinberg AP. Intra‐individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brasacchio D, Okabe J, Tikellis C, Balcerczyk A, George P, Baker EK, Calkin AC, Brownlee M, Cooper ME, El‐Osta A. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene‐activating epigenetic marks that coexist on the lysine tail. Diabetes. 2009;58:1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reddy MA, Villeneuve LM, Wang M, Lanting L, Natarajan R. Role of the lysine‐specific demethylase 1 in the proinflammatory phenotype of vascular smooth muscle cells of diabetic mice. Circ Res. 2008;103:615–623. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39. Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci USA. 2008;105:9047–9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou Z, Sun B, Li X, Zhu C. DNA methylation landscapes in the pathogenesis of type 2 diabetes mellitus. Nutr Metab. 2018;15:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Muka T, Nano J, Voortman T, Braun KVE, Ligthart S, Stranges S, Bramer WM, Troup J, Chowdhury R, Dehghan A, Franco OH. The role of global and regional DNA methylation and histone modifications in glycemic traits and type 2 diabetes: a systematic review. Nutr Metab Cardiovasc Dis. 2016;26:553–566. [DOI] [PubMed] [Google Scholar]
- 42. He T, Peterson TE, Holmuhamedov EL, Terzic A, Caplice NM, Oberley LW, Katusic ZS. Human endothelial progenitor cells tolerate oxidative stress due to intrinsically high expression of manganese superoxide dismutase. Arterioscler Thromb Vasc Biol. 2004;24:2021–2027. [DOI] [PubMed] [Google Scholar]
- 43. Rossig L, Urbich C, Dimmeler S. Endothelial progenitor cells at work—not mature yet, but already stress‐resistant. Arterioscler Thromb Vasc Biol. 2004;24:1977–1979. [DOI] [PubMed] [Google Scholar]
- 44. Bonfini L, Migliaccio E, Pelicci G, Lanfrancone L, Pelicci PG. Not all Shc's roads lead to RAS. Trends Biochem Sci. 1996;21:257–261. [PubMed] [Google Scholar]
- 45. Nemoto S, Finkel T. Redox regulation of forkhead proteins through a p66shc‐dependent signaling pathway. Science. 2002;295:2450–2452. [DOI] [PubMed] [Google Scholar]
- 46. Robledo MM, Bartolome RA, Longo N, Rodriguez‐Frade JM, Mellado M, Longo I, van Muijen GN, Sanchez‐Mateos P, Teixido J. Expression of functional chemokine receptors CXCR3 and CXCR4 on human melanoma cells. J Biol Chem. 2001;276:45098–45105. [DOI] [PubMed] [Google Scholar]
- 47. Scotton CJ, Wilson JL, Scott K, Stamp G, Wilbanks GD, Fricker S, Bridger G, Balkwill FR. Multiple actions of the chemokine CXCL12 on epithelial tumor cells in human ovarian cancer. Can Res. 2002;62:5930–5938. [PubMed] [Google Scholar]
- 48. Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non‐CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA. 2000;97:5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Barres R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, Krook A, Zierath JR. Non‐CpG methylation of the PGC‐1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–198. [DOI] [PubMed] [Google Scholar]
- 50. Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochem Biophys Acta. 2014;1839:1362–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supplemental Methods.
Table S1. Antibody List
Table S2. Primer List
Figure S1. Flow cytometric quantification of ROS production in HG and exHG‐CD34+ stem cells.
Figure S2. A, Schematic representation of the CXCR4 gene with mapped sequences (arrows, region I, II, and III). B, ChIP analysis of the CXCR4 promoter for H3K9me3 modification after hyperglycemia exposure of CD34+ stem cells.
Figure S3. Analysis of DNMT3A, expression in CD34+ stem cells by qPCR.
Figure S4. A, Effect of HG exposure on global 5‐mC methylation level in CD34+ stem cells. The data are expressed as 5‐mC% (5‐mC/total DNA [A+G+C+T]). B, DNMT activity in HG‐CD34+ stem cells after HG exposure.


